
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 04 May 2022
Sec. Marine Fisheries, Aquaculture and Living Resources
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.856674
Identification of the two sympatric species, Alloteuthis media and Alloteuthis subulata, has long relied on a set of identifying morphometric parameters and descriptive guidelines. To resolve taxonomic status of Alloteuthis in the Eastern Adriatic, we used morphological and molecular approach on a dataset collected during MEDITS expeditions sampling the entire Eastern Adriatic over consecutive summers. Phylogenetic analyses inferred from mitochondrial DNA cytochrome oxidase I (COI) gene sequences confirmed presence of both species in the Eastern Adriatic, with A. subulata occurring only in its central and southern parts. Analyses of genetic diversity showed that A. subulata samples in the Eastern Adriatic shared a single haplotype while A. media showed high haplotype diversity. Comparison of Eastern Adriatic A. media samples and populations from other regions showed statistically significant genetic differentiation between the Atlantic haplotypes and each of the Adriatic, Aegean, and Ionian populations. Conversely, A. subulata had low genetic diversity with only two haplotypes present across samples collected globally. There was no single morphometric character with strong enough power to discriminate between species, however, when morphological traits were looked as a composite metric rather than in isolation, the majority of individuals were correctly classified into one of three groups (A. media males or females and A. subulata).
Alloteuthis (Loliginidae) is a genus of slender loliginid squids occurring in the East Atlantic Ocean and Mediterranean Sea. The genus is comprised of three species, Alloteuthis media, Alloteuthis subulata and Alloteuthis africana. These squids are of little fishery importance due to their small body size and are usually caught as bycatch in trawl fisheries (Roper et al., 1984; Jereb and Roper, 2010). Genus Alloteuthis is morphologically easily distinguished from other loliginids due to the characteristic long, sharply pointed “tail” that extends from the mantle (Nesis, 1987) and the characteristic heart-shaped fins (Naef, 1921; Roper et al., 1984). However, morphometric distinction within the genus is less apparent and largely debated (Laptikhovsky et al., 2002; Anderson et al., 2008; Gebhardt and Knebelsberger, 2015).
Alloteuthis africana has almost no geographic overlap with the other two Alloteuthis species, as its range is restricted to eastern Atlantic Ocean, extending from southern Angola to southern Morocco (Roper et al., 1984). In addition, A. africana is characterized by a larger head width compared to A. media and A. subulata (Anderson et al., 2008), which allows easier species identification based on sampling location and morphology. However, A. media and A. subulata have a highly overlapping distribution range in the eastern Atlantic and Mediterranean (Jereb and Roper, 2010). The distribution range of A. subulata extends from northwest Africa to the southern coast of Norway and is considered the most prevalent squid species in the North Sea (Zuev and Nesis, 2003). It is more commonly found in deeper waters and is less common in the Mediterranean and the southern part of its range (Zuev and Nesis, 2003). In the Atlantic, A. media is considered to occur predominantly between Morocco and Scotland, while only occasionally occurring as far north as the Irish Sea and the southern part of the North Sea, and is considered to be a relatively warm-water species (Zuev and Nesis, 2003). Historically, species identification has relied primarily on two indices: the ratio between the fin length (FL) and dorsal mantle length (DML; but see Anderson et al., 2008), and the ratio between the diameter of the largest tentacular club sucker (CCS) and head width (HW; Grimpe, 1925; Roper et al., 1984; Nesis, 1987). Firstly, the length of slender, elongated tail (i.e. the posterior section of the mantle and fins) exceeds 50% of the total DML in A. subulata and A. africana, whereas it is shorter and accounts for less than 50% of the DML in A. media (Nesis, 1987). Secondly, A. subulata and A. africana have smaller tentacular club suckers relative to head width (<8%) while the diameter of the largest club sucker in A. media is between 9-14% of head width (Naef, 1921; Nesis, 1987). Adults exhibit sexual dimorphism in which females of A. media are larger than males, whereas the opposite is true for A. subulata and A. africana, with males having longer mantle and tail length than females. In addition, mature A. subulata and A. africana males (DML up to 20 cm) are larger than A. media males (DML<14 cm; Fischer, 1987).
Recent efforts to resolve Alloteuthis taxonomy and systematics using molecular approaches show that species identification based on established morphological parameters is not accurate and appears to be of low taxonomic value (Anderson et al., 2008; Lefkaditou et al., 2012; Gebhardt and Knebelsberger, 2015). Using the tail length index (TLI; calculated as FL vs DML ratio) as an identifying parameter, Anderson et al. (2008) categorized most of the Atlantic samples as A. subulata and most of the Mediterranean samples as A. media. However, molecular analyses of mtDNA in the same study showed that A. subulata occurred only in the southern Adriatic while all Atlantic samples were actually A. media. The authors emphasized that TLI is not a reliable parameter for species identification and suggested the diameter of the largest club sucker as the more appropriate morpho-character for A. media and A. subulata discrimination, albeit the latter is more difficult to measure in the field, especially in smaller individuals. Conversely, Lefkaditou et al. (2012) identified arm length as a variable best suited for successful discrimination between A. media and A. subulata. Thus, the identification of Alloteuthis species remains confusing and the exact species distribution unknown, as the available occurrence data are predominantly based on morphological identification of collected specimens.
To date, molecular studies of the genus Alloteuthis have confirmed the occurrence of A. media in the Atlantic (Anderson et al., 2008; Olmos-Pérez et al., 2018), English Channel (Anderson et al., 2008), West Mediterranean (Anderson et al., 2008), Ionian Sea (Lefkaditou et al., 2012), Aegean Sea (Lefkaditou et al., 2012) and the Adriatic (Anderson et al., 2008). Alloteuthis subulata specimens were recorded only in the southern Adriatic (Anderson et al., 2008), Ionian Sea (Lefkaditou et al., 2012) and the central eastern Atlantic (Olmos-Pérez et al., 2018). Data from previous Mediterranean International Trawl Surveys (MEDITS) indicate the presence of both A. media and A. subulata in the Eastern Adriatic, with A. media present throughout Eastern Adriatic while A. subulata is restricted to the southern part (Krstulovic Sifner et al., 2005). Furthermore, Petrić et al. (2014) reported finding 36 A. media individuals and no A. subulata in a fisheries restricted area (Jabuka Pit) in the central Adriatic. Traditionally, these occurrence data were based on species identification by morphology, which can be misleading. The aim of this study was therefore to use both molecular and morphological data to (i) investigate the co-occurrence and distribution of both Mediterranean Alloteuthis species in the Eastern Adriatic, (ii) to assess the level of genetic structure and connectivity of local dems and Alloteuthis populations from other geographic regions, and (iii) to evaluate the systematic contribution of morphological characters, as a useful tool for the identification of A. media and A. subulata.
Squid samples were collected during the MEDITS trawl survey conducted between May and August for two consecutive years, 2014 (n=32) and 2015 (n=21), covering the whole Eastern Adriatic coast (Figure 1). Specimens were caught using the bottom trawl net GOC 73 with 20 mm mesh size at predefined stations. The duration of hauls was set at 30 min at depths less than 200 m and 1 hour at depths greater than 200 m (Bertrand et al., 2002). Samples were frozen on board and transported to the laboratory.
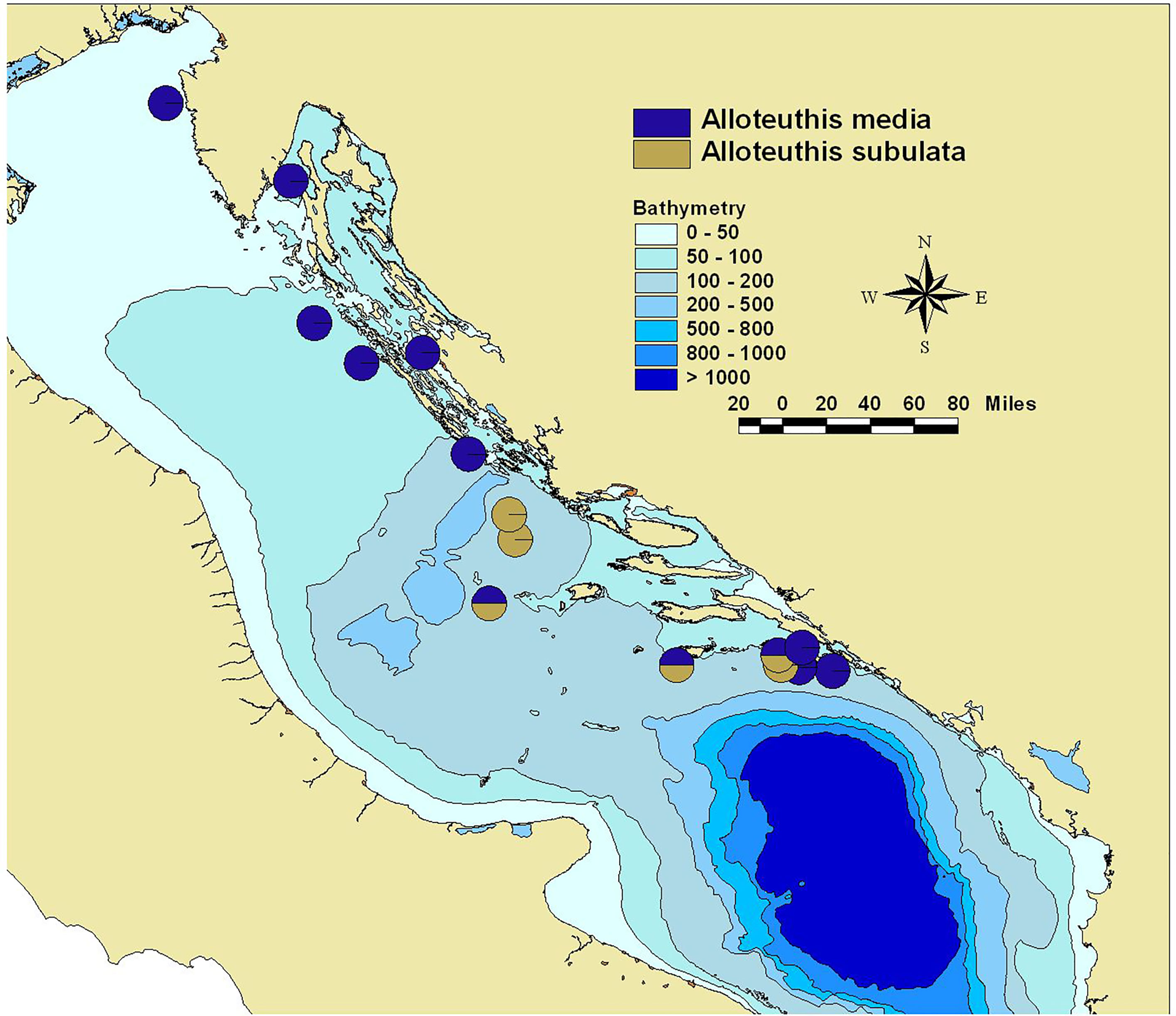
Figure 1 Collection sites in the Eastern Adriatic during MEDITS surveys in 2014 and 2015 for A. media (blue) and A. subulata (yellow).
DNA was extracted from mantle tissue samples previously preserved in absolute ethanol and stored at -20°C, following the modified extraction protocol of Turtinen and Juran (1998). A small amount of each tissue sample was first digested in 180 μL cell lysis buffer (0.2% SDS, 0.01 M TrisBase, 0.01 M EDTA, 0.15 M NaCl) and 4 μL proteinase K (10 mg/mL) and incubated overnight at 55°C on a heat block (BioSan Thermo-Shaker TS-100C, Riga, Latvia). The purity and quantity of DNA preparations were examined using the Genova Nano spectrophotometer (Jenway, Staffordshire, UK). Approximately 650bp long region of cytochrome oxidase subunit I (COI) mitochondrial gene was amplified using universal primers (LCO1490: 5’GGTCAACAAATCATAAAGATATTGG3’ and HC02198: 5’TAAACTTCAGGGTGACCAAAAAATCA3’; Folmer et al., 1994). The PCR reaction consisted of 1 U of Taq polymerase (Sigma Aldrich, D1806), 1 X PCR incubation buffer, 0.2 mM dNTP, 3 mM MgCl2, 0.4 µM of each primer, 50 ng of DNA template and distilled water to the final volume (25 µL). Amplification was performed using the MyCycler Thermal Cycler (Bio-Rad, CA, USA) and PCR conditions were as follows: 3 min at 94°C; 5 cycles of 94°C for 30 s, 45°C for 90 s, 72°C for 60 s; 30 cycles of 94°C for 30 s, 50°C for 90 s, 72°C for 30 s; and a final extension of 7 min at 72°C. The success of amplification was visually examined on a 1% agarose gel. PCR products were sent to the sequencing laboratory Macrogen Europe, where forward sequencing was performed on the ABI 3100 automated sequencer.
Sequence quality was assessed using the Geneious prime software v2020.2.4 (Kearse et al., 2012). Sequences were aligned with the Geneious aligner (Kearse et al., 2012), using default parameters, and uploaded to GenBank with accession numbers MW349463-89 for A. media and MW349517-37 for A. subulata. All additional COI mtDNA sequences from A. media and A. subulata that were available on GenBank and for which the sampling location was indicated, were downloaded, as listed in Table S1. Analyses were initially performed using only the samples from this study (hereafter, “present dataset”). Subsequently, the A. media sequences were combined with those downloaded from GenBank and classified into six broader categories based on sampling location: Adriatic (n=37 of which 30 were sampled in this study and 7 were downloaded from GenBank), Aegean (n=6), Atlantic (n=32), Ionian Sea (n=4), North Sea (n=30) and West Mediterranean (n=2). Similarly, A. subulata sequences were grouped into three categories: Adriatic (n=24 of which 21 were sampled in this study and 3 were downloaded from GenBank), Atlantic (n=3) and Ionian Sea (n=1).
The number of haplotypes (NHap), haplotype diversity (Hd), number of polymorphic sites (S), nucleotide diversity (π) and average number of nucleotide differences (k) were calculated using DnaSp 6.12.3.0 (Nei, 1987; Librado and Rozas, 2009) for each species. Haplotype networks were constructed using the median-joining distance method in PopART 1.7 software (Leigh and Bryant, 2015) while population differentiation index (FST) was calculated using ARLEQUIN 3.5.2.2 (Excoffier and Lischer, 2010). Statistical significance of FST indices was adjusted using Bonferroni correction to account for multiple comparisons between populations. Demographic history changes were also examined in ARLEQUIN 3.5.2.2 where Tajima’s D (Tajima, 1989) and Fu’s FS (Fu, 1997) indices were calculated, together with simulation of mismatch distribution of DNA pairwise difference using the sudden demographic expansion model (Rogers and Harpending, 1992). Phylogenetic analyses were performed using MrBayes 2.2.4 (Huelsenbeck and Ronquist, 2001). For Bayesian analysis, the best-fit model of sequence evolution was selected with jModelTest 2.1.10 (Posada, 2008) and the phylogenetic tree of the present dataset samples was constructed using the Hasegawa-Kishino-Yano model (Hasegawa et al., 1985) with a proportion of invariable sites. Afrololigo mercatolis (EU668101) was used as the outgroup for phylogenetic analyses.
Samples were defrosted at room temperature and wet body mass (BW) was measured using analytical balance down to ±0.1 mg. Following Roper and Voss (1983), nine morphological traits were measured for each specimen: dorsal mantle length (DML), mantle width (MW), fin length (FL), fin width (FW), head length (HL), head width (HW), tentacle length (TL), tentacular club length (TCL) and length of longest arm (AL). In addition, the tail length index (TLI) was calculated as FL/DML. The TCL/TL ratio was also calculated as two types of tentacular clubs were observed in the samples: longer, wider clubs with larger suckers, typical for A. media, and short and narrow clubs with extremely small suckers, typical for A. subulata. The diameter of the largest club sucker was not measured as freezing of samples affected the quality of the soft and delicate club sucker tissue. Finally, specimens were sexed and categorized into adults or sub-adults based on the ontogenetic stage, following a modified protocol of Jereb and Ragonese (1995).
To investigate the discriminatory potential of morphological characteristics between A. subulata and A. media, we combined principal component analysis (PCA) and quadratic discriminant analysis (QDA). Firstly, we used PCA analyses to examine which morphological variables explained most of the variation in the data. The PCA was performed using the R package ‘FactoMineR’ (Lê et al., 2008). Visualization of PCA outputs was done using the ‘factoextra’ R package (Kassambara and Mundt, 2016). Principal components were retained according to the Kaiser-Guttman criterion (eigenvalues >1; Jackson, 1993). Variables with loadings > 0.4 were considered to contribute to a principal component (Tabachnick et al., 2007). Secondly, quadratic discriminant analysis was used as a constrained approach that discriminates multivariate data among a priori groups. QDA was run using the R package ‘MASS’ (Ripley et al., 2013) with the 80/20 split for training and testing data. In addition to multivariate analyses, a two-sample t-test and Wilcoxon rank-sum test was used to specifically examine species differences in TLI and TCL/TL ratio, respectively, as these indices have previously been shown to be important for species discrimination based on morphology. Data were analyzed in R Studio (RStudio Team, 2020) for R (3.5.0).
Sequences for the cytochrome oxidase I (COI) gene were obtained for 53 individuals, two of which were excluded from further analyses due to poor sequence quality. According to the BLAST search (Altschul et al., 1990) and using ≥97% cut-off for sequence identity, 30 individuals were identified as A. media and 21 as A. subulata. Phylogenetic reconstruction was later applied to support the species identification results (Figure 2). The A. media alignment performed on the present dataset included 30 sequences and was 529 bp long. A total of 14 haplotypes were identified in the present A. media dataset, indicating high haplotype diversity (Hd=0.89) of populations from the Eastern Adriatic, with nine haplotypes represented by a single individual. Nine individuals shared the most common haplotype while two haplotypes comprised of four individuals each, and two haplotypes of 2 individuals each. Genetic diversity estimates are shown in Table 1. The A. subulata alignment comprised of 21 sequences and was 522 bp long. However, all of A. subulata samples in the present dataset shared the same haplotype with no polymorphic sites present, thus the observed genetic diversity was zero (Table 1).
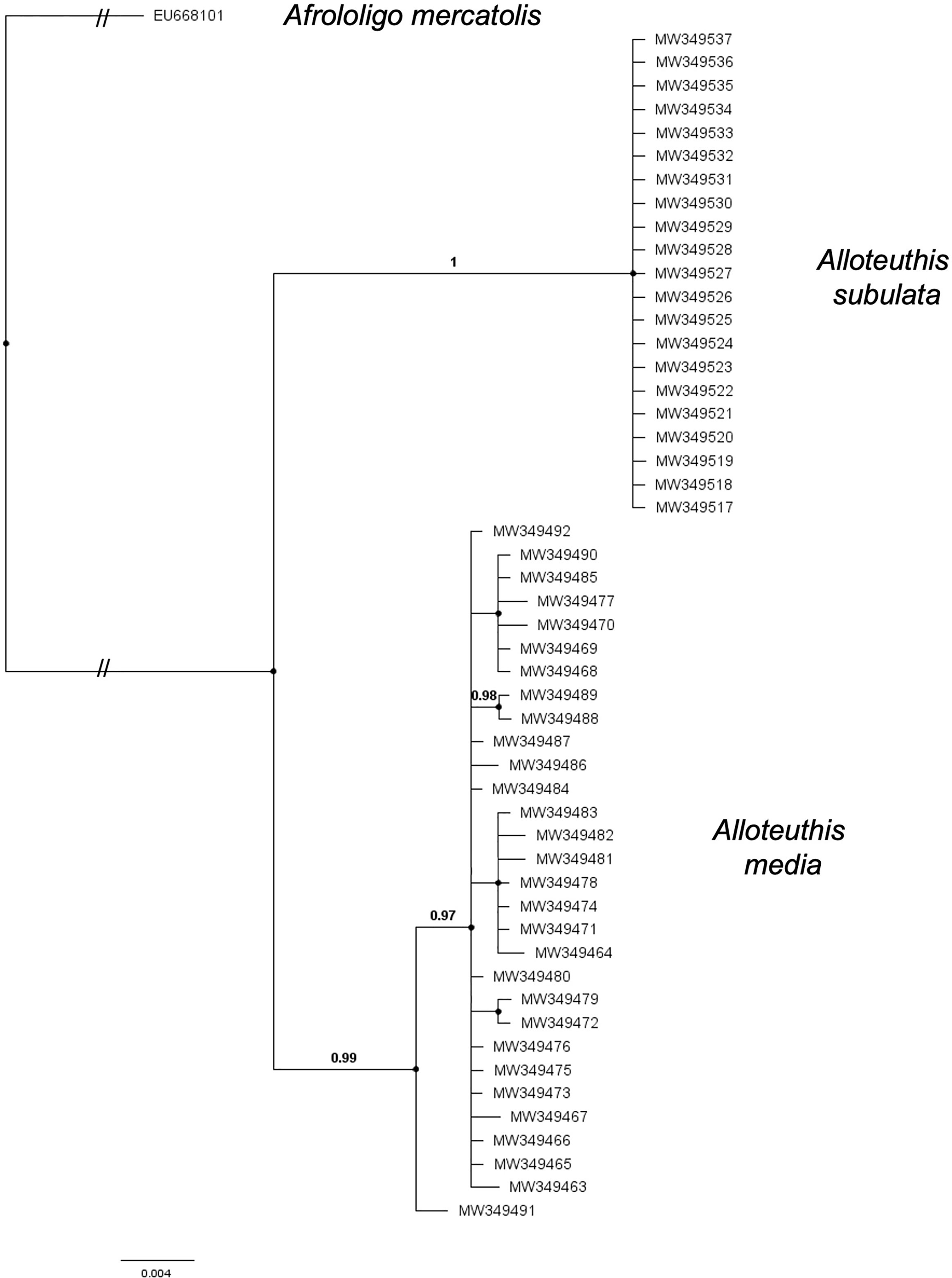
Figure 2 Bayesian tree topology based on COI haplotypes of A. media, A. subulata sampled in the Eastern Adriatic in 2014 and 2015 and Afrololigo mercatolis as outgroup. Bayesian posterior probabilities are included at the nodes.
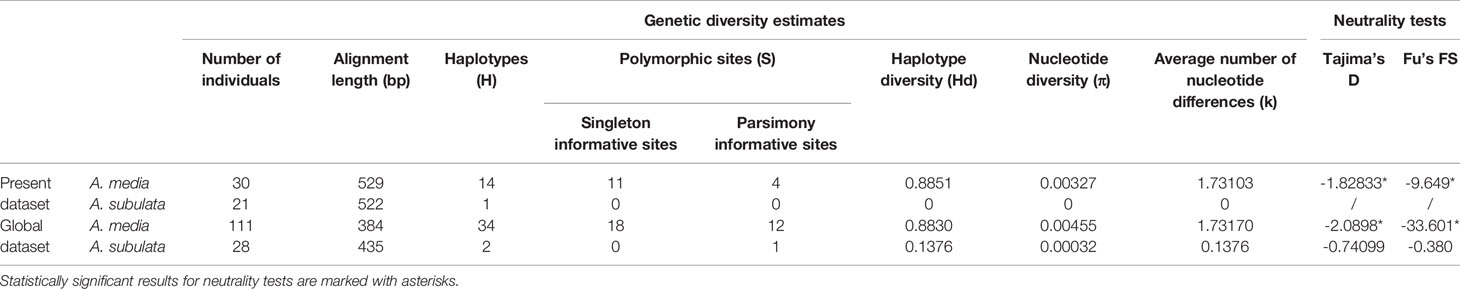
Table 1 Genetic diversity and neutrality tests for the mitochondrial DNA COI sequences of Alloteuthis media and A. subulata in the Eastern Adriatic (“Present dataset”) and across all sequences available on GenBank (“Global dataset”).
Global A. media dataset (i.e., pooled dataset comprising of present dataset plus Genbank sequences) consisted of 111 sequences 384 bp long (Table 1). A total of 34 haplotypes were identified, indicating high haplotype diversity (Hd=0.88) with 22 haplotypes represented by a single individual. The majority of individuals shared one of the three most common haplotypes, of which one haplotype represented predominantly samples from East Atlantic and North Sea, second one represented samples from the Adriatic, Ionian and Aegean Sea, while the third haplotype included samples from most regions (Figure 3). Moderate and significant global FST (0.064) was found among the samples of different basins. Pairwise FST values indicated significant heterogeneity when comparing samples from Atlantic and North Sea with any basin within Mediterranean (p<0.001; Table 2). The mismatch distribution showed unimodal distribution of pairwise DNA differences, following Rogers and Harpending’s model (Rogers and Harpending, 1992) of demographic expansion (SSD = 0.011, p > 0.05). Demographic expansion is also evident from the haplotype network star-like shape, with dominant ancestral haplogroups that connect to a network of new haplogroups separated by one or two mutations. Significant negative Tajima’s D (-2.09; p<0.001) and Fu’s FS (-27.70; p<0.001) values indicate an excess of rare haplotypes and presence of recent population expansion (Tajima, 1989; Fu, 1997).
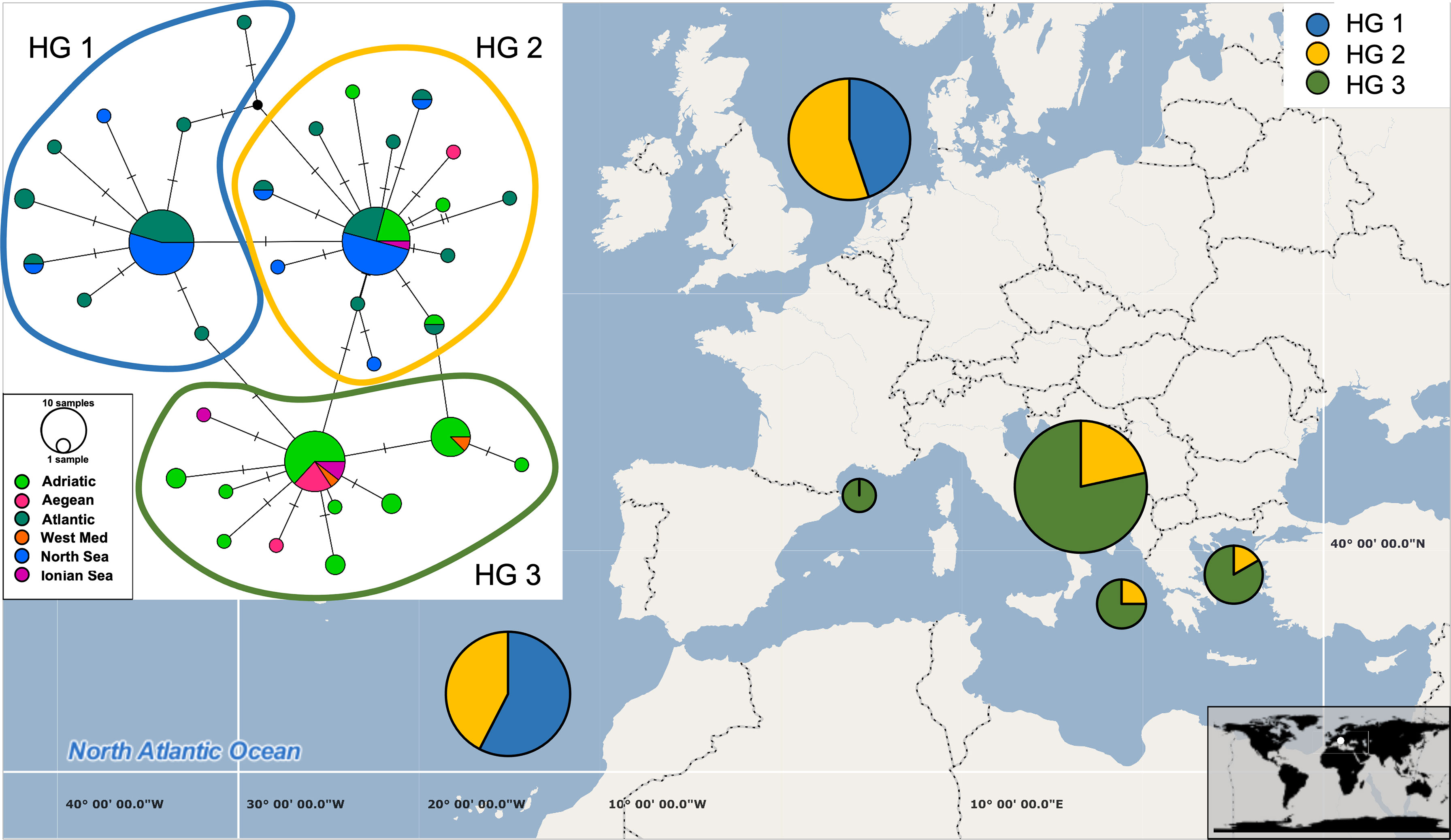
Figure 3 Median-joining network based on COI haplotypes (inset) and distribution of haplotypes across six geographical categories (Adriatic Sea, Aegean Sea, Ionian Sea, West Mediterranean, North Sea and Atlantic Ocean). The size of each circle corresponds to the number of sequences belonging to each haplotype and short lines correspond to the number of base changes between two haplotypes. The pie charts on the map display the haplotype group frequencies found in each of the six regions.
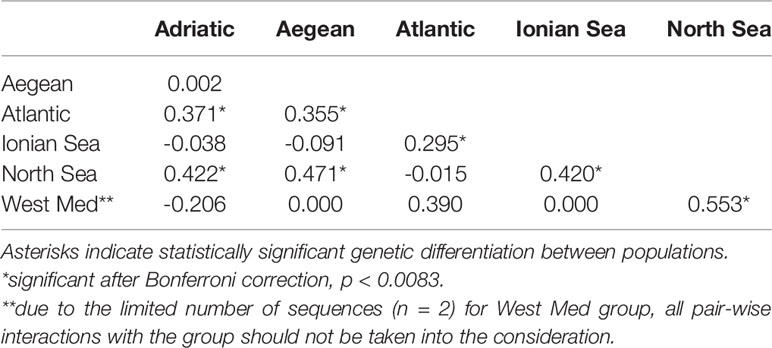
Table 2 Pairwise FST values showing genetic differentiation between populations of Alloteuthis media.
The length of the global A. subulata alignment included 28 sequences and was 435 bp long. Genetic diversity analysis revealed the presence of a single polymorphic site (Table 1). Therefore, only two haplotypes were present in the total dataset; 26 samples from the Adriatic, Ionian and Atlantic Seas had the same haplotype while two Atlantic samples shared a different haplotype where a T/C base change occurred. Nucleotide diversity for A. subulata across all sampled locations was thus very low (Hd=0.0003) with the average number of nucleotide differences of 0.1376.
Examination of the gonads revealed the presence of 20 female, seven male and three sub-adult A. media. Similarly, females formed most of the A. subulata sample with 16 individuals, in addition to four males and one sub-adult. Adult individuals were divided into four categories based on sex and species. To reduce the impact of ontogenetic allometry on study results, sub-adults were excluded from further morphometric analyses (Laptikhovsky et al., 2005). Morphometric variables were tested for correlations using ‘car’ function in ‘caret’ R package (Kuhn, 2008), to avoid multicollinearity. Highly correlated morphometric variables (>70%) were excluded and the final reduced dataset comprised of five variables: mantle width (MW), head width (HW), tentacular club length (TCL), TCL/TL index and TLI index. Principal component analysis of the morphological parameters showed that the first two principal components (PC1 and PC2) were significant and explained 83.7% of the total variance. PC1 showed significant positive loadings (0.62-0.88) for all retained variables, except the TCL/TL index, indicating that individuals with larger TLI also had larger MW, HW and TCL (Table 3). Mainly, the first axis separated larger A. media females from all other individuals, with A. media males and A. subulata females being characterized by intermediate sizes, and A. subulata males at the lower end of the size range (Figure 4). The second principal component mostly correlated with the TCL/TL ratio (strong positive loadings of 0.97) and TCL (positive loadings of 0.74). This axis predominantly explained the variation within A. media females as this was the only group that showed a substantial level of variation in TCL/TL ratio and TCL compared to other traits (Figure 4).
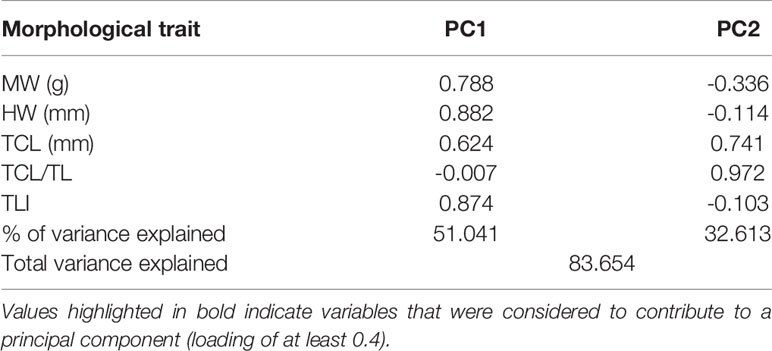
Table 3 Component loadings of morphological traits observed on two retained orthogonally rotated principal components (PC1 and PC2) from the PCA analysis combining Alloteuthis media and A. subulata samples from Eastern Adriatic.

Figure 4 Graphic representation of the PCA analysis combining Eastern Adriatic Alloteuthis media and A. subulata samples showing the individual placement along the first two principal components based on five retained morphological variables (MW, mantle width; HW, head width; TCL, tentacular club length; TLI, tail length index; TCL/TL, tentacle club length to tentacle length ratio). Group centroids are shown with dashed circles.
Due to the small number of males sampled for A. subulata (n=4), individuals were grouped into three categories for QDA analysis instead of four: A. media females, A. media males and A. subulata (pooled males and females). This is consistent with previous approaches in the literature (Lefkaditou et al., 2012) and was justified by the output of the PCA analysis (Figure 4) that showed substantial variation in morphometric characteristics in A. media males and females, but generally much less in A. subulata. Quadratic discriminant analysis of morphological traits correctly classified 87.5% of individuals to either A. media (males or females) or A. subulata species, suggesting that the measured morphological traits have relatively high power for accurate species delineation (Figure S1). In addition, we found significant variation in the TLI among species (t(37.2) = 4.13, p = 0.0002), the trait historically used for species classification based on morphology. However, there was no significant difference in the TCL/TL ratio between species (Z = -0.35, p = 0.74; Figure 5).
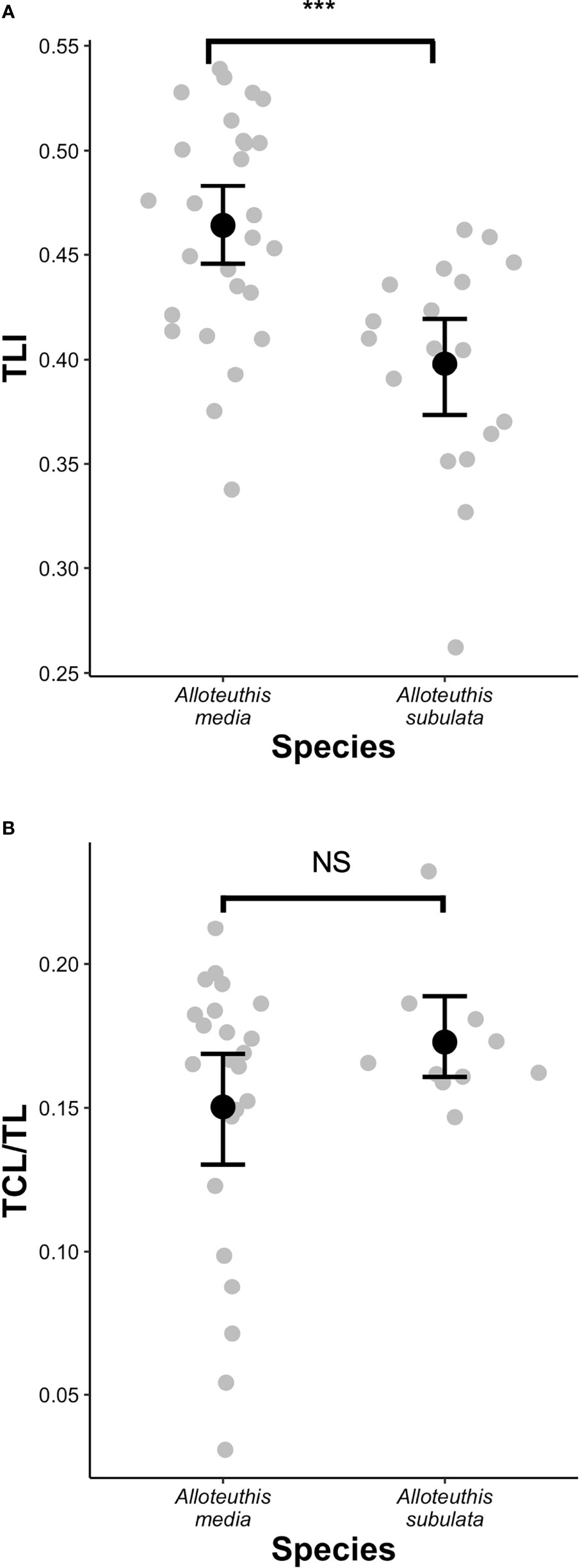
Figure 5 Species differences in (A) tail length index (TLI) and (B) tentacular club length to tentacle length ratio (TCL/TL). Black dots in the boxplots depict the mean and bars represent standard errors. ***p < 0.001, NS, not signifficant.
Sequencing data of the cytochrome oxidase I (COI) unit confirmed the presence of both Alloteuthis media and A. subulata in the Eastern Adriatic. While this study was limited to the use of a single marker (COI), its efficiency in separating closely related cephalopod species was previously demonstrated, outperforming alternative markers such as 18S rDNA (Gebhardt and Knebelsberger, 2015), 16S and 12S rRNA (Braid et al., 2014). In the north-eastern Adriatic, the genus Alloteuthis was exclusively represented by A. media (n=19; Figure 1). However, both species were found in the central and southern parts of the Eastern Adriatic, suggesting a potential structure of Alloteuthis species distribution patterns in the basin. These findings are consistent with previous reports of A. subulata in the Adriatic Sea, where three individuals were sampled off the southern Italian coast (Mola di Bari; Anderson et al., 2008) and with the results from MEDITS surveys where this species was reported only in the southern Adriatic (Krstulovic Sifner et al., 2005), albeit latter identification was based only on morphological parameters. Additionally, previous MEDITS surveys reported the occurrence of A. media throughout the Adriatic Sea (Krstulovic Sifner et al., 2005; Anderson et al., 2008), further supporting the present findings. The observed difference in the distribution patterns of A. media and A. subulata in the Eastern Adriatic is not surprising, as A. subulata is considered to occur predominantly in deeper waters (Zuev and Nesis, 2003; but see González and Sánchez, 2002), while shallow waters of the northern Adriatic, the most extensive continental shelf of the Mediterranean Sea with an average bottom depth of about 35 m (Trincardi et al., 1994), may not be suitable for this species. However, as the only samples previously identified in the Adriatic using molecular markers were seven A. media and three A. subulata individuals, all collected from a single location in Mola di Bari, Italy (Anderson et al., 2008), it remains to be ascertained whether the distribution of A. subulata is restricted to central and southern parts of the Adriatic basin.
Contrasting intraspecific genetic diversity pattern was observed in these two sympatric species. Based on the available data, A. subulata is constituted by a single panmictic unit within the Mediterranean. Namely, lack of haplotype diversity attributed to A. subulata in the Eastern Adriatic is consistent with patterns observed on a larger scale, with A. subulata occurring only in the eastern part of the Mediterranean, specifically in the southern Adriatic and Ionian Sea (Anderson et al., 2008; Lefkaditou et al., 2012) with whom eastern Adriatic samples share the same haplotype. Molecularly confirmed A. subulata samples from the eastern Atlantic (Olmos-Pérez et al., 2018) had a haplotype that differed only in a single base pair from those observed in the eastern Mediterranean. In addition to the general assumption that environmental stability of the deep sea supports low genetic diversity due to niche refinement (Bretsky and Lorenz, 1970), the low evolutionary rate for the coding region, and the loss of diversity following demographic collapses during the Messinian Salinity Crisis (MSC) or selective events, could explain the low haplotype diversity of A. subulata found in the present work. Similar level of diversity for mtDNA region was reported for Octopus vulgaris in the central Mediterranean (Fadhlaoui-Zid et al., 2012). It is still unclear whether A. subulata occurs in the rest of the Mediterranean, as research based on molecular markers is sparse resulting in low number of published sequences (n=3, nsamples=23; Laptikhovsky et al., 2002; Anderson et al., 2008; Lefkaditou et al., 2012), which could also partially explain the low observed genetic diversity in this species. Therefore, any conclusions regarding genetic variation and connectivity along the species distribution range would need to be validated by further research.
In contrast, 34 haplotypes across the A. media distribution range have been recorded in the present study. High haplotype diversity and low nucleotide diversity due to changes in a single nucleotide among haplotypes likely indicate sudden demographic expansion from a small initial population throughout recent demographic history. Three major haplotype clusters separated A. media individuals: those sampled west of Gibraltar (i.e., Atlantic and North Sea populations) from those sampled east of Gibraltar (i.e., Mediterranean samples; Figure 3). An intermediate haplotype cluster was formed with individuals from most sampling sites, but with dominance of the Eastern Atlantic collection (Figure 3). Glacial periods throughout history caused geographic isolation between Mediterranean and Atlantic populations, with subsequent genetic divergence observed in many marine invertebrates (Quesada et al., 1995; Pérez-Losada et al., 1999; Ríos et al., 2002). At present, Gibraltar Strait (GS) and the Almería‐Oran Front (AOF) represent oceanographic discontinuities caused by the difference in salinity between the colder less saline Atlantic waters that enter through the GS into the Mediterranean (warmer higher density waters). The presence of three haplotype clusters and the significant genetic differentiation between Atlantic and Adriatic, Aegean and Ionian populations found in this study suggest that the GS and/or the AOF might represent a geographic barrier that reduces gene flow between Atlantic and Mediterranean populations of A. media. Furthermore, despite the pelagic larval and adult stage, gene flow could be restricted by strong marine currents that occur at the narrow and shallow passage of the GS and along the Almería-Oran line (Pascual et al., 2017).
Although it has been commonly used as a species specific parameter, several recent studies have found TLI as potentially unreliable morphometric trait for the identification of Alloteuthis species (Laptikhovsky et al., 2002; Pilsits, 2007; Anderson et al., 2008; Lefkaditou et al., 2012). While our results indicate that the TLI index differs between A. media and A. subulata with the TLI being significantly greater in A. media, this also contradicts the guidelines for morphological species identification which indicate the TLI<0.5 in A. media (TL is less than 50% of DML) and >0.5 in A. subulata (Nesis, 1987). With the mean TLI of 0.46 in A. media and 0.40 in A. subulata, our results suggest that the TLI threshold of 0.5 is not appropriate for discriminating between these two species. In addition, TLI values of adults of each species included in this study vary considerably (Figure 5), indicating that the ratio of FL to DML should not be used for species differentiation. In addition, research conducted from 1990 until 2002 across the Aegean, Mediterranean and East Atlantic have shown that relative fin length increases with body length (Laptikhovsky et al., 2005), thus the smallest Alloteuthis specimens are likely to be identified as A. media while the largest get attributed to A. subulata, following the traditionally accepted species assignment based on the 0.5 TLI cut-off (Nesis, 1987). Similarly, in this study larger individuals had a higher TLI, regardless of species. However, A. media individuals (especially females) were larger than A. subulata (Table 4 and Figure 4).
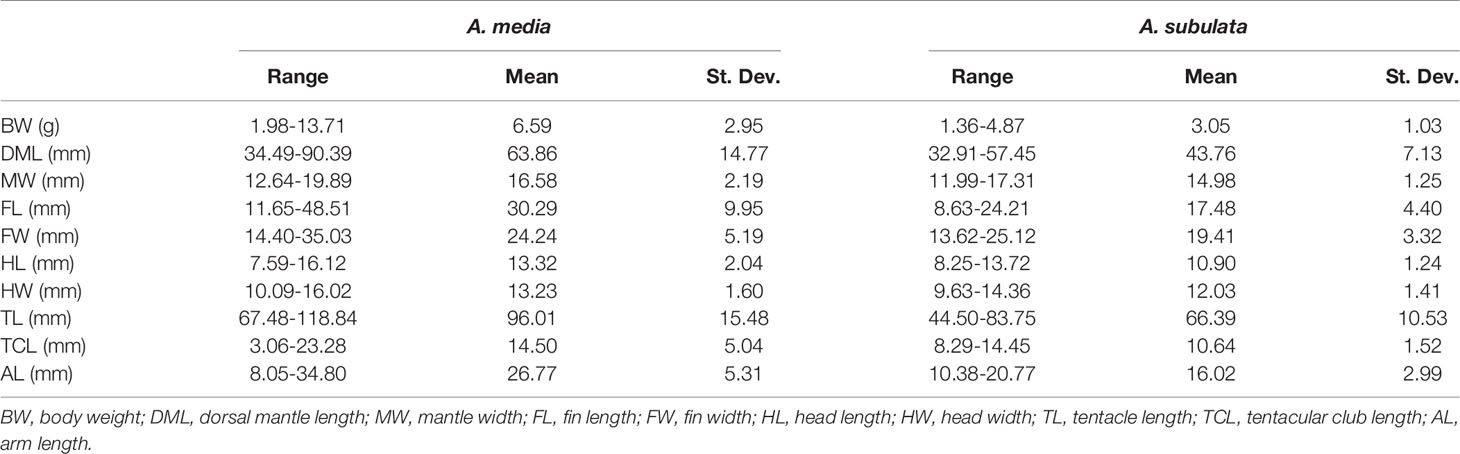
Table 4 Range, mean and standard deviation for morphometric variables measured in adult A. media and A. subulata in the Eastern Adriatic.
While Anderson et al. (2008) indicated that the ratio of central club sucker to head width could be a good discriminatory parameter of the two species, Pilsits (2007) showed that the CCS/HW index is not reliable for species identification and reported lack of consensus between molecular and morphological assignment based on this index. In the present study, the CCS/HW index was not examined due to the delicate nature of club suckers severely affected by defrosting process, especially in individuals with smaller TCL. However, a simple visual inspection of these squids revealed that two types of tentacular clubs were observed in our sample (Figure S1), differing substantially in length and width of the tentacular club. Analyses of the TCL/TL index revealed that all A. subulata samples had long tentacular clubs (15-23% of TL) while a wide range of sizes was observed in A. media (3-21% of TL). Some A. media had extremely short and narrow tentacular clubs (Figure 5), which is usually considered a distinguishing feature of A. subulata. In addition to the higher observed variation in size and shape of tentacular clubs in A. media and the lack of significant differences in the TCL/TL index between the two species, the use of TCL or TCL/TL index alone as identifying parameter is rendered difficult since these structures can be easily damaged or often missing in frozen samples.
Given the lack of reliable indices for species identification, this study focused on examining a full set of morphological variables to try to determine if species can be classified into separate categories based on a combination of morphological parameters. Among the morphological parameters examined, mantle width, head width, TCL, TCL/TL index and TLI were selected as variables that best described interspecies variation. Overall, individuals with larger TLI also had larger MW, HW and TCL. Females of A. media were generally larger than male conspecifics, and A. subulata males and females, that were at the smaller end of the size spectrum. However, a wide variation in morphometric parameters was observed in females of A. media, mainly reflected by TCL/TL index and TCL. Nevertheless, when morphological traits were considered as a composite metric rather than in isolation, most individuals were correctly classified into one of the three groups (A. media males or females, and A. subulata). These results suggest that individual morphometric parameters are not accurate enough for species identification, but each of the groups studied is characterized by a specific combination of values of the morphological traits. To date, it is not clear what drives the observed variation in morphology between species, as well as within A. media females, where observed variation in morphology largely exceeds observed variation in A. subulata and A. media males. This morphological variability may explain why the search for a morphological key to species identification has not yielded conclusive results in the case of A. subulata and A. media.
While early research suggested that A. subulata is widespread in the Atlantic, and A. media is more common in the Mediterranean, molecular analyses conducted in the last decade largely contradict these conclusions. Judging from the frequent errors in morphological species assignment noted in studies using molecular data (Anderson et al., 2008; Lefkaditou et al., 2012), it is possible that the geographic distribution of these species is widely misinterpreted and it is unclear whether A. subulata is common in the Atlantic as originally assumed. Furthermore, assumptions about habitat preferences are largely based on occurrence data obtained from morphological identification of species, which raises the question of whether A. media is a warm-water species and A. subulata prefers deeper, colder waters, although this has been widely accepted in the literature (Zuev and Nesis, 2003). Therefore, to understand the global distribution patterns and ecology of A. media and A. subulate, a detailed review is needed that prioritizes molecular data to delineate the species and attempt to provide context for the observed morphological variability.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethical review and approval was not required for the animal study because squid samples were collected during the MEDITS trawl survey (fishing landings).
KA and MP designed the study. II and KA collected the data. ŽT provided protocols and reagents for DNA isolation. KA and TŠ-B performed statistical analyses. KA, MP, ŽT and TŠ-B participated in data discussion and interpretation. KA wrote the first draft of the manuscript. All authors contributed to manuscript editing and revision.
This study was supported throughout the research project “Data Collection Framework - DCF” funded by the Ministry of Agriculture of the Republic of Croatia.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors acknowledge the use of infrastructure and equipment provided by the University of Split and the Institute of Oceanography and Fisheries, Split, Croatia.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.856674/full#supplementary-material
Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic Local Alignment Search Tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Anderson F. E., Pilsits A., Clutts S., Laptikhovsky V., Bello G., Balguerías E., et al. (2008). Systematics of Alloteuthis (Cephalopoda: Loliginidae) Based on Molecular and Morphometric Data. J. Exp. Mar. Bio. Ecol. 364, 99–109. doi: 10.1016/j.jembe.2008.07.026
Bertrand J. A., de Sola L. G., Papaconstantinou C., Relini G., Souplet A. (2002). The General Specifications of the MEDITS Surveys. Sci. Mar. 66, 9–17. doi: 10.3989/scimar.2002.66s29
Braid H. E., McBride P. D., Bolstad K. S. R. (2014). Molecular Phylogenetic Analysis of the Squid Family Mastigoteuthidae (Mollusca, Cephalopoda) Based on Three Mitochondrial Genes. Hydrobiologia 725, 145–164. doi: 10.1007/s10750-013-1775-3
Bretsky P. W., Lorenz D. M. (1970). An Essay on Genetic-Adaptive Strategies and Mass Extinctions. Geol. Soc Am. Bull. 81, 2449–2456. doi: 10.1130/0016-7606(1970)81[2449:AEOGSA]2.0.CO;2
Excoffier L., Lischer H. E. L. (2010). Arlequin Suite Ver 3.5: A New Series of Programs to Perform Population Genetics Analyses Under Linux and Windows. Mol. Ecol. Resour. 10, 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
Fadhlaoui-Zid K., Knittweis L., Aurelle D., Nafkha C., Ezzeddine S., Fiorentino F., et al. (2012). Genetic Structure of Octopus Vulgaris (Cephalopoda, Octopodidae) in the Central Mediterranean Sea Inferred From the Mitochondrial COIII Gene. C. R. Biol. 335, 625–636. doi: 10.1016/j.crvi.2012.10.004
Fischer W. (1987). Fiches FAO D’identification Des Especes Pour Les Besoins De La Peche.(Rev 1). Mediterranee Et Mer Noire. Zone De Peche 37. Vertébrés 2, 761–1530.
Folmer O., Black M., Hoeh W., Lutz R., Vrijenhoek R. (1994). DNA Primers for Amplification of Mitochondrial Cytochrome C Oxidase Subunit I From Diverse Metazoan Invertebrates. Mol. Mar. Biol. Biotechnol. 3, 294–299. doi: 10.1071/ZO9660275
Fu Y.-X. (1997). Statistical Tests of Neutrality of Mutations Against Population Growth, Hitchhiking and Background Selection. Genetics 147, 915–925. doi: 10.1093/genetics/147.2.915
Gebhardt K., Knebelsberger T. (2015). Identification of Cephalopod Species From the North and Baltic Seas Using Morphology, COI and 18S rDNA Sequences. Helgol. Mar. Res. 69, 259–271. doi: 10.1007/s10152-015-0434-7
González M., Sánchez P. (2002). Cephalopod Assemblages Caught by Trawling Along the Iberian Peninsula Mediterranean Coast. Sci. Mar. 66, 199–208. doi: 10.3989/scimar.2002.66s2199
Grimpe G. (1925). Zur Kenntnis Der Cephalopodenfauna Der Nordsee. Wissenschaftliche Meeresuntersuchungen. Neue Folge. Abteilung Helgoland, Vol. 16, Part 1, Number 3. Ad. Littmann. Oldenbg. 34.
Hasegawa M., Kishino H., Yano T. (1985). Dating of the Human-Ape Splitting by a Molecular Clock of Mitochondrial DNA. J. Mol. Evol. 22, 160–174. doi: 10.1007/BF02101694
Huelsenbeck J. P., Ronquist F. (2001). MRBAYES: Bayesian Inference of Phylogenetic Trees. Bioinformatics 17, 754–755. doi: 10.1093/bioinformatics/17.8.754
Jackson D. A. (1993). Stopping Rules in Principal Components Analysis: A Comparison of Heuristical and Statistical Approaches. Ecology 74, 2204–2214. doi: 10.2307/1939574
Jereb P., Ragonese S. (1995). An Outline of the Biology of the Squid Illex Coindetii in the Sicilian Channel (Central Mediterranean). J. Mar. Biol. Assoc. United Kingdom 75, 373–390. doi: 10.1017/S0025315400018245
Jereb P., Roper C. F. E. (2010). “Cephalopods of the World-an Annotated and Illustrated Catalogue of Cephalopod Species Known to Date,” in Myopsid and Oegopsid Squids, vol. 2. (Washington, D.C: FAO). doi: 10.7312/nei-92038
Kassambara A., Mundt F. (2016). Factoextra: Extract and Visualize the Results of Multivariate Data Analyses. R Packag. version 1, 76. https://cran.r-project.org/web/packages/factoextra/index.html.
Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., et al. (2012). Geneious Basic: An Integrated and Extendable Desktop Software Platform for the Organization and Analysis of Sequence Data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Krstulovic Sifner S., Lefkaditou E., Ungaro N., Ceriola L., Osmani K., Kavadas S., et al. (2005). Composition and Distribution of the Cephalopod Fauna in the Eastern Adriatic and Eastern Ionian Sea. Isr. J. Zool. 51, 315–330. doi: 10.1560/4LT4-K01W-C9GF-7YK3
Kuhn M. (2008). Building Predictive Models in R Using the Caret Package. J. Stat. Softw 28, 1–26. doi: 10.18637/jss.v028.i05
Laptikhovsky V., Salman A., MoustahfId H. (2005). Morphological Changes at Maturation and Systematics in the Squid Genus Alloteuthis. Phuket Mar. Biol. Cent. Res. Bull. 66, 187–193.
Laptikhovsky V., Salman A., Önsoy B., Katagan T. (2002). Systematic Position and Reproduction of Squid of the Genus Alloteuthis (Cephalopoda: Loliginidae) in the Eastern Mediterranean. Mar. Biol. Assoc. United Kingdom. J. Mar. Biol. Assoc. United Kingdom 82, 983. doi: 10.1017/S0025315402006483
Lefkaditou E., Tsigenopoulos C. S., Alidromiti C., Haralabous J. (2012). On the Occurrence of Alloteuthis Subulata in the Eastern Ionian Sea and its Distinction From the Sympatric Alloteuthis Media. J. Biol. Res. 17, 169.
Leigh J. W., Bryant D. (2015). Popart: Full-Feature Software for Haplotype Network Construction. Methods Ecol. Evol. 6, 1110–1116. doi: 10.1111/2041-210X.12410
Lê S., Josse J., Husson F. (2008). FactoMineR: An R Package for Multivariate Analysis. J. Stat. Software 25, 1–18. doi: 10.18637/jss.v025.i01
Librado P., Rozas J. (2009). DnaSP V5: A Software for Comprehensive Analysis of DNA Polymorphism Data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Naef A. (1921). Fauna E Flora Del Golfo Di Napoli, Monograph No. 35, Cephalopoda. (Berlin: R. Friedländer & Sohn)
Nesis K. N. (1987). Cephalopods of the World: Squids, Cuttlefishes, Octopuses, and Allies. (Neptune, NJ: TFH Publications, Inc).
Olmos-Pérez L., Pierce G. J., Roura Á., González Á.F. (2018). Barcoding and Morphometry to Identify and Assess Genetic Population Differentiation and Size Variability in Loliginid Squid Paralarvae From NE Atlantic (Spain). Mar. Biol. 165, 1–18. doi: 10.1007/s00227-018-3387-y
Pascual M., Rives B., Schunter C., Macpherson E. (2017). Impact of Life History Traits on Gene Flow: A Multispecies Systematic Review Across Oceanographic Barriers in the Mediterranean Sea. PloS One 12, e0176419. doi: 10.1371/journal.pone.0176419
Pérez-Losada M., Guerra Á., Sanjuan A. (1999). Allozyme Differentiation in the Cuttlefish Sepia Officinalis (Mollusca: Cephalopoda) From the NE Atlantic and Mediterranean. Heredity (Edinb). 83, 280–289. doi: 10.1038/sj.hdy.6885520
Petrić M., Šifner S. K., Ferri J., Škeljo F., Brčić J., Krželj M. (2014). Cephalopods in the Trawl Catches in the Jabuka Pit Wider Area. Proceedings of the 49th Croatian & 9th International Symposium on Agriculture. Marić M., Lončarić Z. (eds) (Faculty of Agriculture, University of Josip Juraj Strossmayer in Osijek), 504–508, in Croatian.
Pilsits A. (2007). Phylogeny and Population Genetics of Allotheuthis (Loliginidae) and Discovery of a Cryptic Species. Honor. Theses 327.
Posada D. (2008). Jmodeltest: Phylogenetic Model Averaging. Mol. Biol. Evol. 25, 1253–1256. doi: 10.1093/molbev/msn083
Quesada H., Beynon C. M., Skibinski D. O. (1995). A Mitochondrial DNA Discontinuity in the Mussel Mytilus Galloprovincialis Lmk: Pleistocene Vicariance Biogeography and Secondary Intergradation. Mol. Biol. Evol. 12, 521–524. doi: 10.1093/oxfordjournals.molbev.a040227
Ríos C., Sanz S., Saavedra C., Pena J. B. (2002). Allozyme Variation in Populations of Scallops, Pecten Jacobaeus (L.) and P. Maximus (L.)(Bivalvia: Pectinidae), Across the Almeria–Oran Front. J. Exp. Mar. Bio. Ecol. 267, 223–244. doi: 10.1016/S0022-0981(01)00371-9
Ripley B., Venables B., Bates D. M., Hornik K., Gebhardt A., Firth D., et al. (2013). Package ‘Mass.’. Cran r 538, 113–120.
Rogers A. R., Harpending H. (1992). Population Growth Makes Waves in the Distribution of Pairwise Genetic Differences. Mol. Biol. Evol. 9, 552–569. doi: 10.1093/oxfordjournals.molbev.a040727
Roper C. F. E., Sweeney M. J., Nauen C. (1984). Cephalopods of the World. An Annotated and Illustrated Catalogue of Species of Interest to Fisheries. (Washington, D.C: National Museum of Natural History Smithsonian Institution)
Roper C. F. E., Voss G. L. (1983). Guidelines for Taxonomic Descriptions of Cephalopod Species. Biol. Resour. Potential Cephalopods. Mem. Natl. Museum Victoria 44, 48–63. doi: 10.24199/j.mmv.1983.44.03
RStudio Team (2020). RStudio: Integrated Development for R. RStudio. Boston, MA: PBC. Availabale at: http://www.rstudio.com/.
Tabachnick B. G., Fidell L. S., Ullman J. B. (2007). Using Multivariate Statistics (Pearson Boston, MA: Pearson Education, Inc).
Tajima F. (1989). Statistical Method for Testing the Neutral Mutation Hypothesis by DNA Polymorphism. Genetics 123, 585–595. doi: 10.1093/genetics/123.3.585
Trincardi F., Correggiari A., Roveri M. (1994). Late Quaternary Transgressive Erosion and Deposition in a Modern Epicontinental Shelf: The Adriatic Semienclosed Basin. Geo-mar Lett. 14, 41–51. doi: 10.1007/BF01204470
Turtinen L. W., Juran B. D. (1998). Protein Salting-Out Method Applied to Genomic DNA Isolation From Fish Whole Blood. Biotechniques 24, 238–239. doi: 10.2144/98242bm14
Keywords: Alloteuthis media, Alloteuthis subulata, Adriatic Sea, morphometry, DNA barcoding
Citation: Alujević K, Šegvić-Bubić T, Isajlović I, Trumbić Ž and Petrić M (2022) Distribution and Differentiation Patterns of Sympatric Squids Alloteuthis media and Alloteuthis subulata (Cephalopoda: Loliginidae) Using Morphological and Molecular Approaches. Front. Mar. Sci. 9:856674. doi: 10.3389/fmars.2022.856674
Received: 17 January 2022; Accepted: 08 April 2022;
Published: 04 May 2022.
Edited by:
Pedro M. Costa, New University of Lisbon, PortugalReviewed by:
Fernando Ángel Fernández-Álvarez, Institute of Marine Sciences (CSIC), SpainCopyright © 2022 Alujević, Šegvić-Bubić, Isajlović, Trumbić and Petrić. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Karla Alujević, YWx1amV2aWNrQGdtYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.