- 1Key Laboratory of Tropical Translational Medicine of Ministry of Education, School of Pharmacy, Hainan Medical University, Haikou, China
- 2Shenzhen Key Lab of Marine Genomics, Guangdong Provincial Key Lab of Molecular Breeding in Marine Economic Animals, BGI Academy of Marine Sciences, Shenzhen Huahong Marine Biomedicine Co. Ltd., BGI Marine, BGI, Shenzhen, China
- 3Institute for Molecular Bioscience, University of Queensland, St. Lucia, QLD, Australia
Sea anemone venom is a marine drug resource library with pharmacological and biotechnology value, and it contains complex and diverse functional peptide neurotoxins. However, the venom components of only a limited number of sea anemone species have been globally evaluated by transcriptomics and proteomics. In this study, 533 putative protein as well as peptide toxin sequences were found on a large scale from dissimilar developmental stages of sea anemone Exaiptasia diaphana, which can be divided into 75 known superfamilies according to the predicted functions. Among them, the proportion of protein is 72.98%, and its main families are metalloproteases, chymotrypsinogen like, collagen, pancreatic lipase-associated protein like, and G-protein coupled receptor, while the proportion of peptides is 27.02%, and main families are ShK domain, thrombin, Kunitz-type, defensin, as well as insulin-like peptide. Finally, typical anemone peptide neurotoxins were screened, and the 3D structure and pharmacological activity of these anemone peptide neurotoxins were predicted by homology modeling. We elucidate on a valuable high-throughput approach for obtaining sea anemone proteins and peptides. Our findings form the basis for targeted studies on the diversity as well as pharmacological effects of sea anemone peptide neurotoxins.
Introduction
Sea anemones, which are phylum Cnidaria members, are among the oldest extant lineage of venomous animals, with fossil and molecular data placing their origins before the Ediacaran period ~750 million years ago (Jouiaei et al., 2015; Columbus-Shenkar et al., 2018). The venom produced by sea anemone is mainly used for defense, depredation, and intra- and interspecific competition (Frazao et al., 2012; Fu et al., 2021). Different from other poisonous animals, the venom of sea anemone is generally concentrated in structures called nematocysts (a type of cnidae), which is distributed in different areas of polyps, such as actinopharynx, tentacles, column, acrorhagi, mesenterial filaments, but with a high abundance of tentacles (Fautin, 2009; Madio et al., 2018; Prentis et al., 2018). However, Nv1, the Type I neurotoxin of the sea anemone Nematostella vectensis, appears in ectodermal gland cells rather than nematocysts, which reveals another alternative mechanism of venom delivery in sea anemones (Moran et al., 2012). Among sea anemone peptides, there are neurotoxins that block either voltage-gated sodium channels (VGSCs), acid-sensing ion channels (ASICs), voltage-gated potassium channels (VGPCs), transient receptor potential vanilloid 1 (TRPV1), and phospholipases A2 (PLA2) (Diochot and Lazdunski, 2009; Martins et al., 2009; Moran et al., 2009; Rodriguez et al., 2014; Monastyrnaya et al., 2016; Cristofori-Armstrong and Rash, 2017). The ShK toxin from sun anemone (Stichodactyla helianthus) has been widely investigated (Castaneda et al., 1995). This peptide can block Kv1.3 channels of T lymphocytes, which suppresses their activations thereby acting as treatment options for autoimmune disorders (Beeton et al., 2011; Chandy and Norton, 2017; Wang et al., 2019). This peptides’ analogue (ShK-186) is a first-inclass clinical candidate dalazatide and phase I clinical trials completed for psoriasis treatment (Chi et al., 2012; Tarcha et al., 2012). Other types of toxins have exhibited activities on pain-associated channels, including the first sea anemone (Anthopleura elegantissima) toxin specific for the HERG channel (APETx1), which suppresses ASIC3, an acid pain sensor, and is involved in inflammatory pathways (Zhang et al., 2007; Peigneur et al., 2012). π-AnmTX Hcr 1b-2, the first peptide that is able to inhibit both homomeric ASIC1a and ASIC3, was isolated from hydrophobic 20% ethanol fraction of Heteractis crispa, which showed anti-hyperalgesic effect and significantly reduced the pain threshold of experimental animals in the acid-induced muscle pain model (Kalina et al., 2018). Analgesic peptides 1-3 (APHC1-3) from Heteractis crispa, a sea anemone, suppresses TRPV1, which has various roles such as epilepsy, peripheral neuropathic and cancer pain (Philyppov et al., 2012; Andreev et al., 2013).
Aiptasia pallida (recently renamed Exaiptasia diaphana) was used as a model system for studies of cnidarian-dinoflagellate symbiosis because of its comparatively fast growth rate, abilities for sexual and asexual reproduction, which will help us better understand how environmental pressure affects the survival and growth of corals (Weis et al., 2008; Sunagawa et al., 2009; Lehnert et al., 2012; Grajales and Rodriguez, 2016). In addition, Exaiptasia diaphana can also provide a basis for the future study of regeneration ability, fission, immunity, especially the ability of biological adhesion (Davey et al., 2019). We evaluated the venom of Exaiptasia diaphana through transcriptomic approach. We reveal a set of transcript data that show previously unreported peptide diversity in Exaiptasia diaphana, including neurotoxins that may act on VGSCs or VGPCs, cytotoxic components, protease inhibitors, PLA2, proteases, lectins, the Cysteine-rich secretory proteins, Antigen 5, as well as Pathogenesis-associated 1 proteins (CAP) superfamily and various hydrolases. Moreover, we reveal the complexity of peptides in Exaiptasia diaphana venom as well as the applications of these compounds in the pharmaceutical industry. The identified novel peptides and proteins will be subjects for further structural, functional as well as venom evolution studies.
Results
Transcriptome Sequence Assembly
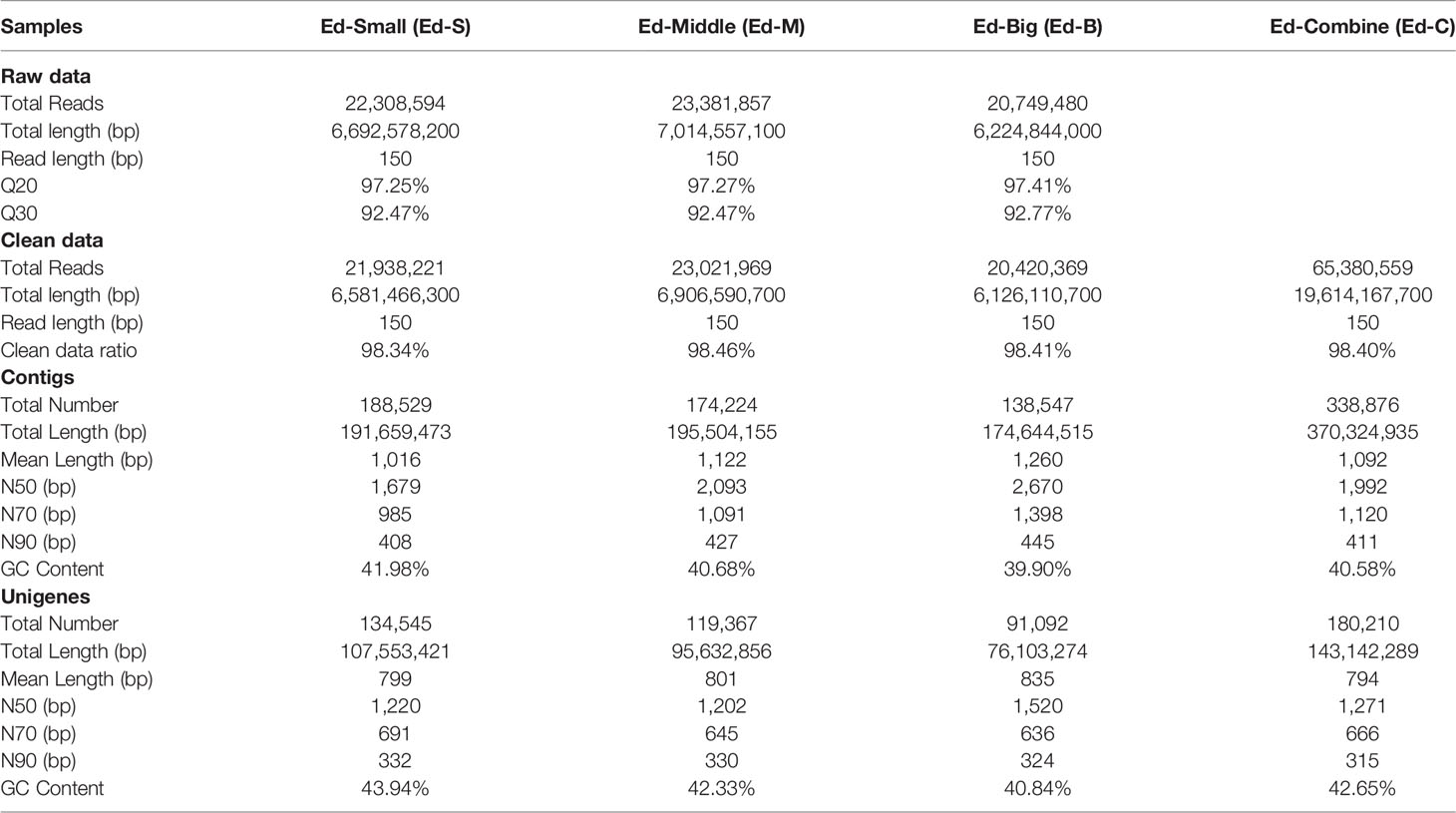
Sequencing as well as assembly of the transcriptome of Exaiptasia diaphana was developed and deposited at the National Center for Biotechnology Information (NCBI) database (BioProject: PRJNA694232, and SRA accession: SRP303227). High-throughput sequencing revealed about 22.31, 23.38 and 20.75 million paired-end short reads with a length of 150 bp and Q30 were 92.47%, 92.47% and 92.77% for small, middle and big sea anemones respectively (Table 1). After filtration of low-quality reads as well as adapters, 21.94, 23.02 and 20.42 gigabases (Gb) of clean data were obtained for every sample. Clean data ratio was projected to be 98.34%, 98.46% and 98.41% respectively (Table 1). For the whole transcriptome, De novo assembly of short reads produced 180210 unigenes, and N50 was determined to be 1271 bp with mean lengths of 794 bp. Average respective GC levels of contigs and unigenes for the transcriptome were 40.58% and 42.65% (Table 1). To assess the functions of unigenes, annotations were done based on 4 databases. The 180210 unigenes were allocated into the databases, Uniprot (48716 unigenes), Nr (48631 unigenes), KOG (25973 unigenes), and KEGG (21364 unigenes), while 124166 more unigenes were not annotated in these databases (Supplementary Figure 1).
About 27769 genes were enriched in 34 KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways, and were allocated to 5 primary categories: environmental information processing (3844), cellular processes (3892), genetic information processing (3804), organismal systems (6936) and metabolism (9293) (Supplementary Figure 2). Most of the unigenes were allocated to metabolism, with the global as well as overview maps exhibiting the highest counts of annotated unigenes (3414). Another GO analysis illustrated that 139662 unigenes were annotated into 3 categories: cellular components (37080), biological processes (58038), and molecular functions (44504) with binding to be the most enriched one (18132), respectively (Supplementary Figure 3). We also annotated the functions of the predicted proteins using Cluster of KOG database. A total of 23202 unigenes were allotted KOG annotations and were classified into 25 molecular families (Supplementary Figure 4).
Venom Components Identified in the Transcriptome
In this study, 533 identified transcripts were assigned into 75 groups based on predicted functions, which were projected based on sequence homologies at amino acid levels with peptides as well as proteins from UniprotKB databases, among which 72.98% were proteins and 27.02% were peptides (Figure 1 and Supplementary Table 1). Majority of protein constituents corresponded to metalloproteases, chymotrypsinogen like, pancreatic lipase-related protein like (PLRP like), G-protein coupled receptor, and collagen, while peptide components corresponded to the ShK domain, thrombin, Kunitz-type, and insulin-like peptide and defensin (Figure 1 and Supplementary Tables 2, 3). In general, a similar finding has been reported in transcriptomes of other sea anemones (Madio et al., 2017; Ramirez-Carreto et al., 2019).
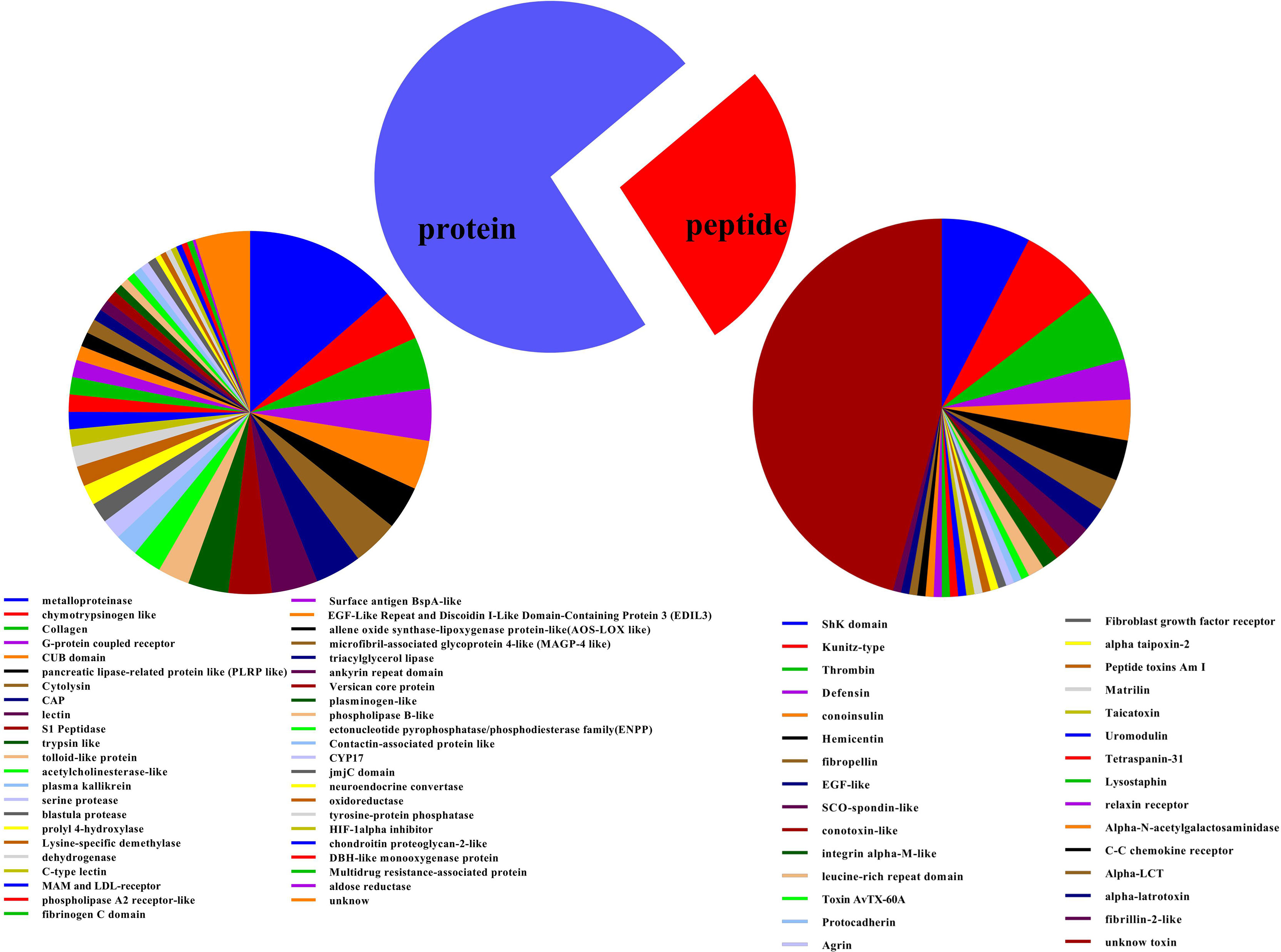
Figure 1 Putative protein as well as peptide toxin families in transcriptome of Exaiptasia diaphana. The 533 protein sequences with substantial BLAST hits to manually curated lists of animal toxins in UniProt (www.uniprot.org/program/Toxins) were allocated into various toxin families based on their amino acid sequences and cysteine scaffolds.
Protein and Peptide Toxins From Various Developmental Stages
A total of 166, 181, 148 putative proteins and peptide toxin precursors were respectively identified from Small, Middle, and Big datasets from three body-sized Exaiptasia diaphana. Comparative distributions of protein as well as peptide toxins is shown in Figure 2A. Of these putative proteins as well as toxin precursors, 31 were detected in all three datasets (Ed-S, Ed-M and Ed-B), 15 were common to both Ed-S and Ed-M, 16 were shared by Ed-S and Ed-B, and 24 were common to both Ed-M and Ed-B. The 31 common toxins precursors were classified into 18 superfamilies, such as CAP, ShK domain, metalloproteinase, triacylglycerol lipase, and defensin.
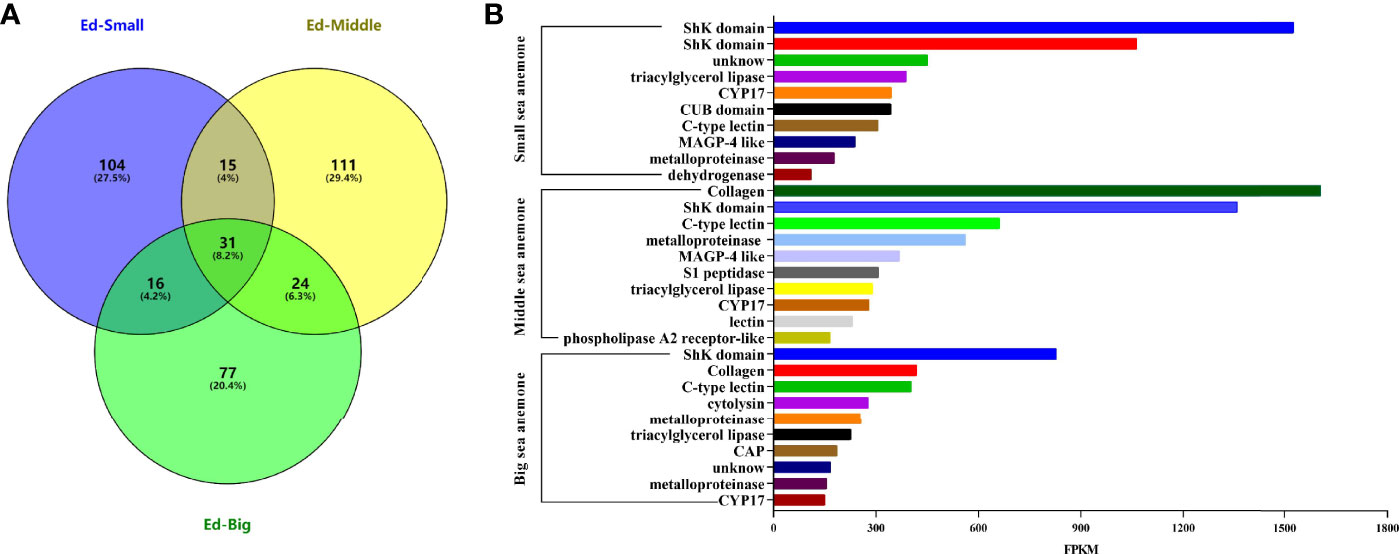
Figure 2 Comparison of protein as well as peptide toxins transcripts from various Exaiptasia diaphana developmental stages. (A) Relationship of the putative protein and peptide toxins from Ed-Small, Ed-Middle and Ed-Big datasets of Exaiptasia diaphana. (B) Ten most highly transcribed protein and peptide transcripts from various Exaiptasia diaphana developmental stages.
Fragments per kilobase of exon per million mapped fragment (FPKM) values were evaluated to denote transcription levels for every protein and peptide toxin. The top 10 protein as well as peptide toxins (with highest FPKM values) were chosen from every dataset (Figure 2B), and ShK domain, triacylglycerol lipase, CYP17, metalloproteinase, as well as C-type lectin were transcribed in 3 various developmental stages of Exaiptasia diaphana. The ShK domain dominantly expressed. Moreover, in the middle development stage of Exaiptasia diaphana, collagen levels were the highest, and were comparatively high in the mature stage.
Typical Sea Anemone Peptide Toxins
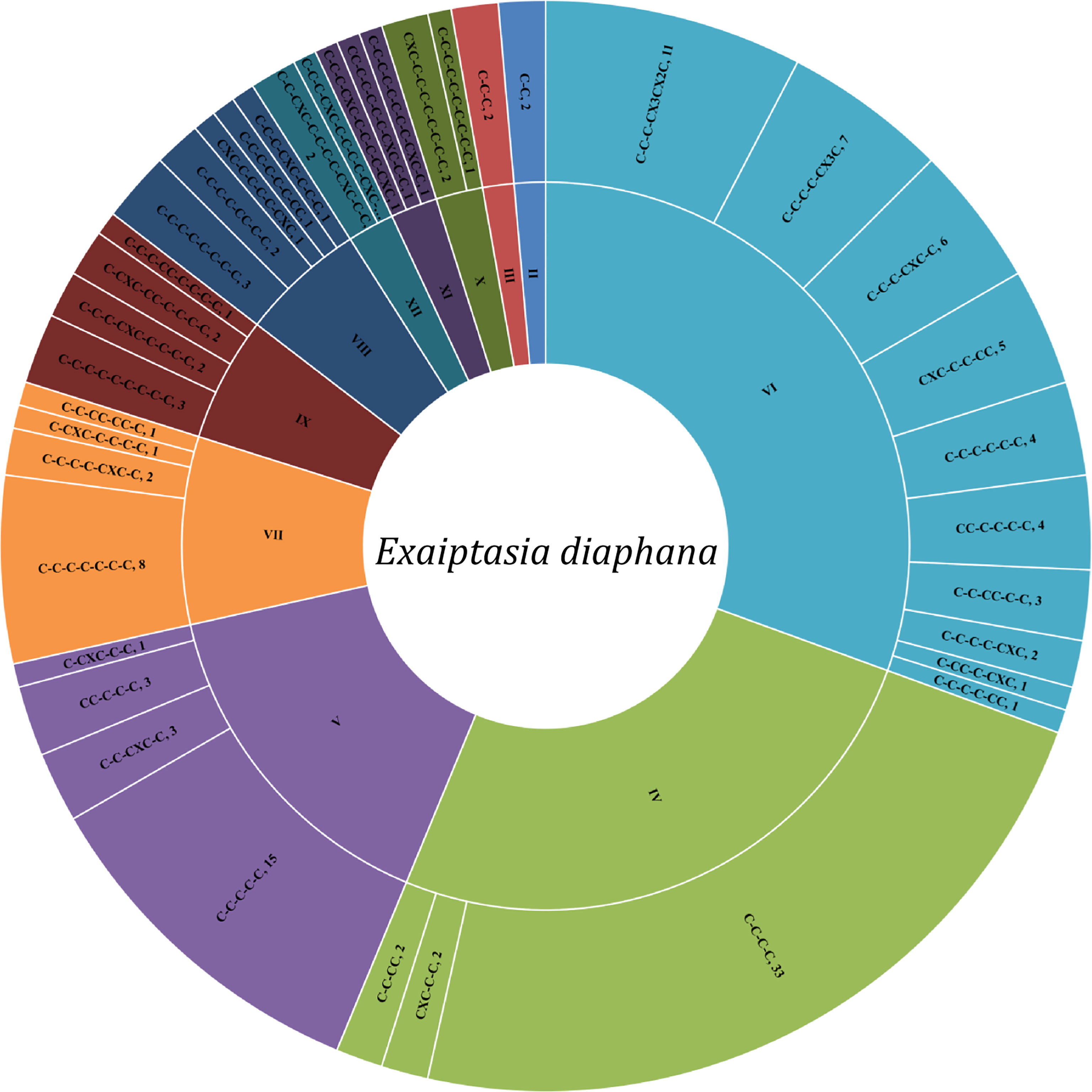
Sea anemones are sessile animals, so peptide neurotoxins are very important in immobilization of prey as well as defenses against predators. Peptide neurotoxins are among the best characterized constituents of sea anemone venoms in terms of their action mechanisms (Madio et al., 2019). They associate with various ion channels, such as VGSCs (Bosmans and Tytgat, 2007), VGPCs (Castaneda and Harvey, 2009), ASICs (Cristofori-Armstrong and Rash, 2017), and TRP channels (Monastyrnaya et al., 2016; Logashina et al., 2017). Here, 144 peptide toxins were obtained from the Small, Middle, Big, and Combine datasets of Exaiptasia diaphana, and named as Ed-001 to 144 in order (Supplementary Table 3). Cysteine-rich peptides are present in almost all sea anemone neurotoxins, and diversities of structural scaffolds among them is notable. The cysteine patterns of the 144 peptides were divided into 11 major categories, each with different subclasses (Figure 3). The peptides containing 4 cysteines forming 2 disulfide bond patterns (IV) and 6 cysteines forming 3 disulfide bond patterns (VI) are the most abundant, and their numbers are 37 and 44, respectively. In addition to peptides with even number of cysteines, there are also a large number of peptides with odd number of cysteines.
So far, the 3D structure and/or cysteine-pattern of nine typical anemone peptide toxins have been identified, which are ShK, Kunitz-domain, β-defensin-like, Anemonia sulcata toxin III (ATX-III), boundless β-hairpin (BBH), inhibitor cystine-knot (ICK), epidermal growth factor-like (EGF-like), proline-hinged asymmetric β-hairpin (PHAB), and small cysteine-rich peptides (SCRiPs) respectively (Madio et al., 2019). The putative peptide toxins with typical and unique homologues were ShK domain (11 homologues), Kunitz-type peptides (10 homologues), β-defensin-like (5 homologues), and EGF-like (3 homologues).
The ShK domain was named after the ShK toxin, which had been identified from the venom of Stichodactyla helianthus (Castaneda et al., 1995). This toxin is a member of Kv type 1 family, which blocks Kv1.1, Kv1.2, Kv1.3, Kv1.6, Kv3.2, and Kca3.1 channels (Kalman et al., 1998; Rauer et al., 1999; Yan et al., 2005). ShK blocks Kv1.3, which has important functions in T and B lymphocyte subsets involved in autoimmune conditions. Therefore, ShK is potential immune modulator for treatment of autoimmune diseases (Pennington et al., 1995). ShK analogs have been developed to be Kv1.3-specific (Beeton et al., 2011). The ShK-186 analogue is being developed as a treatment for autoimmune diseases (Chi et al., 2012). BgK, an ShK domain peptide from Bunodosoma granulifera, a sea anemone, blocks Kv1.1, Kv1.2, as well as Kv1.3 potassium channels (Cotton et al., 1997). Here, eleven homologous sequences with ShK and BgK were identified, and their cysteine pattern was C-C-C-CX3CX2C, including the connection mode of disulfide bond as C1-C6, C2-C4, C3-C5 (Figure 4A). Among these sequences, by using blast alignment in NCBI, Ed-079, Ed-080 and Ed-081 are the same as previously reported sequences (GenBank No. KXJ15362.1, XP_020909625.1 and KXJ06239.1) from Exaiptasia diaphana in the database, respectively. The homology comparison results show that the sequence identity of Ed-079 and ShK is 43.75%, and the sequence identity of Ed-081 and BgK is 33.33%. The homology modeling prediction results show that Ed-079 and Ed-81 have similar 3D structures, indicating that they may also act on potassium channels (Figure 4B). As analogues of ShK and BgK, they are worthy of in-depth study in the future.
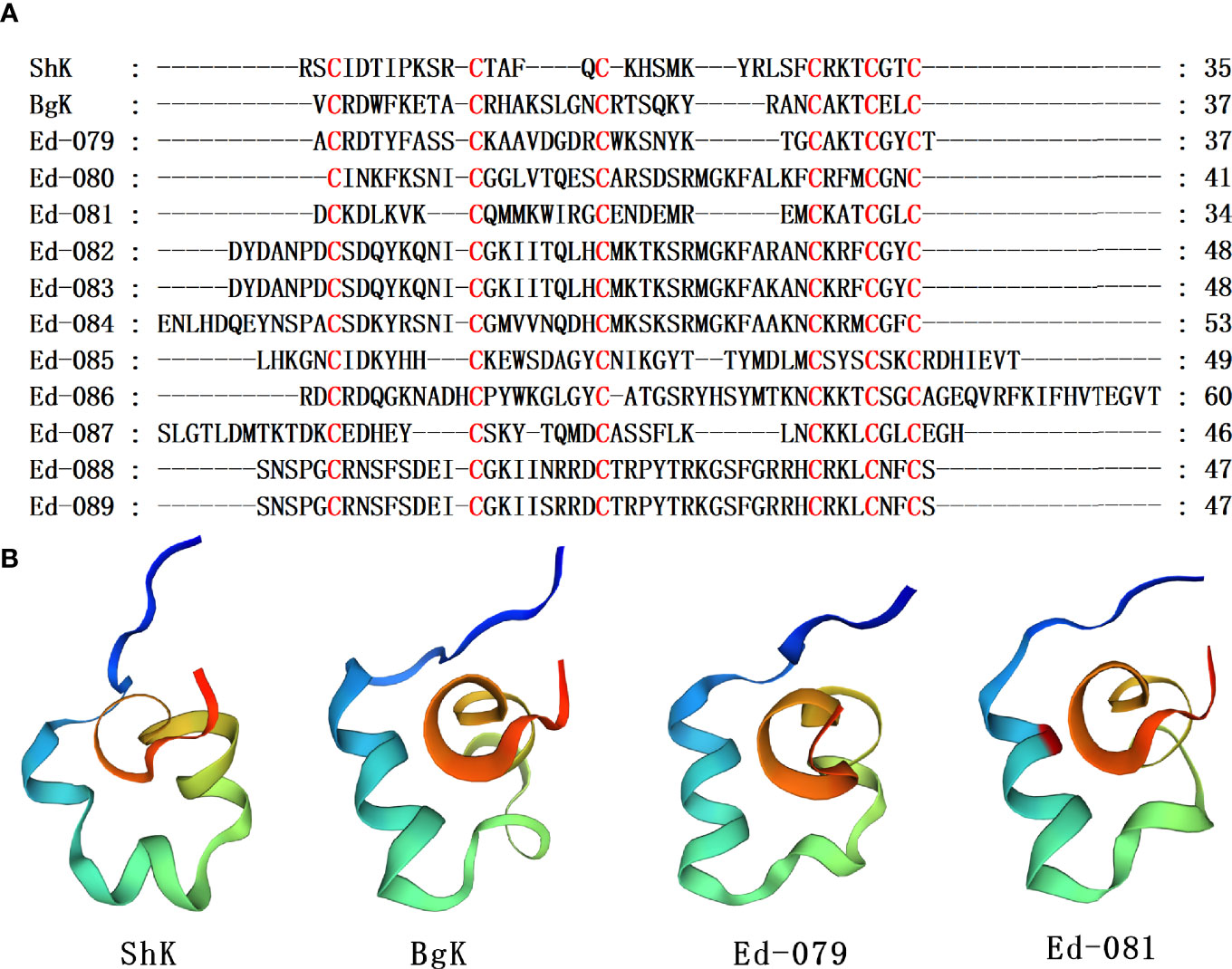
Figure 4 Representative sea anemone mature peptide sequences with ShK domain. (A) Alignments of sea anemone mature peptides with ShK domain. Cysteines are highlighted in red. (B) Homology modeling prediction of the representative sea anemone mature peptide Ed-079 and Ed-081 with ShK (κ-stichotoxin-She3a, PDB 4LFQ) and BgK (κ-actitoxin-Bgr1a, PDB 1BGK).
Epidermal growth factor (EGF) family-associated proteins that have been identified in invertebrates. Among them, L-EGF is a growth factor secreted from Lymnaea stagnalis, a gastropod mollusk. Gigantoxin I (ω-stichotoxin-Sgt1a), a peptide toxin from Stichodactyla giganteus, a sea anemone, can paralyze crabs (Hermann et al., 2004). With regards to sequence homology, this toxin has EGF activities as shown by rounding of human epidermoid carcinoma A431 cells (Shiomi et al., 2003). Moreover, gigantoxin I modulates the activities of TRPV1 channels (Cuypers et al., 2011), however, these activities result from involvement of the EGF receptor/PLA2/arachidonic acid/lipoxygenase pathway in indirect TRPV1 activation. Gigantoxin I was the first toxin that initiated this effect. Therefore, these toxins may elucidate on TRPV1 channel regulation (Cuypers et al., 2011). Three homologous sequences with Gigantoxin I were identified, and their cysteine pattern was C-C-C-CXC-C (Figure 5A). The homology comparison results show that the sequence identity of Gigantoxin I, Ed-94, Ed-95 and Human EGF are 35.71%, 36.36% and 34.09%, respectively. Taking Human EGF as the template, the homology modeling prediction results show that Gigantoxin I, Ed-93,Ed-94, and Ed-95 have similar three-dimensional structures, respectively (Figure 5B). Given that mammalian EGF as well as its family members are regularly associated with neoplastic diseases, the invertebrate EGF family members may inform the designs of erbB receptor antagonists.
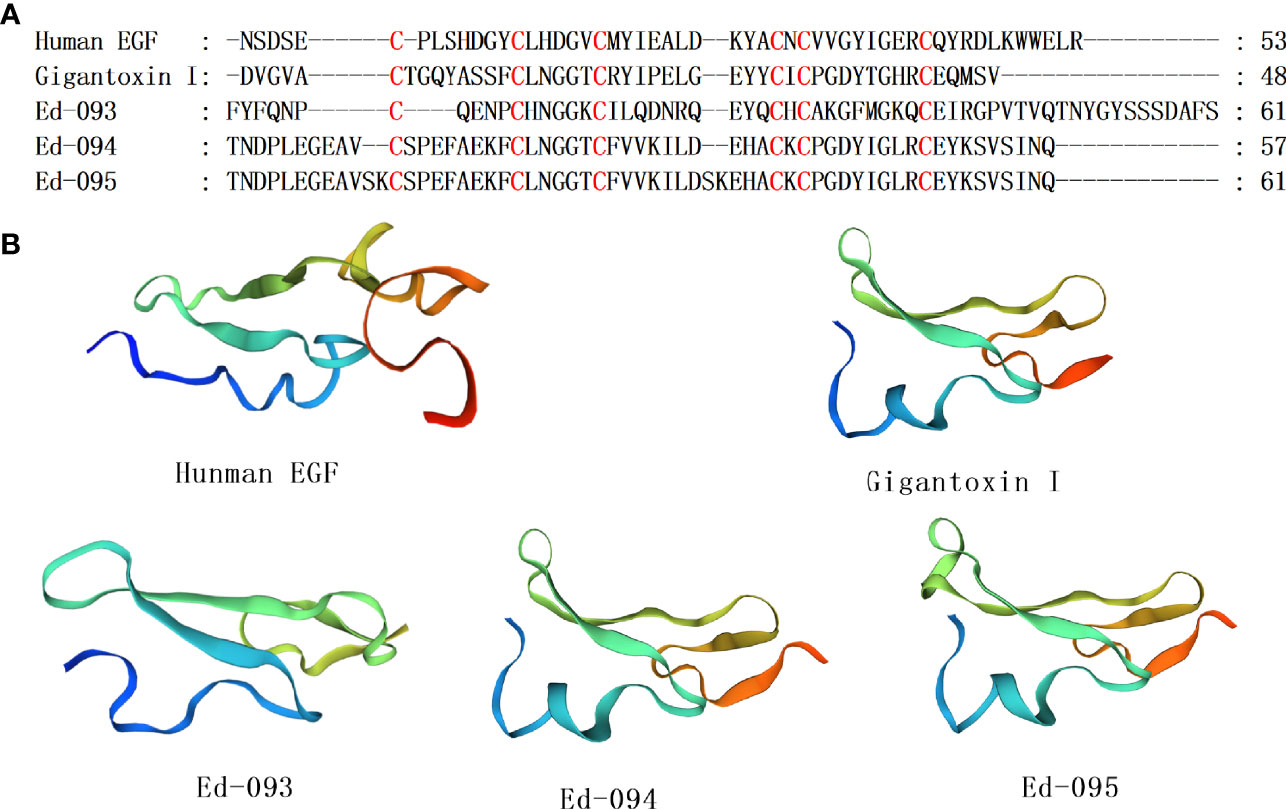
Figure 5 Representative sea anemone mature peptide sequences with EGF. (A) Alignment of sea anemone mature peptides with EGF. Cysteines are highlighted in red; (B) Homology modeling prediction of the representative sea anemone mature peptide Gigantoxin I, Ed-093, Ed-094 and Ed-095 with Human EGF.
Kunitz-type peptides have been discovered from venoms as well as glands of various venomous animals, where they are involved in protease inhibition and ion channel modulation (Mishra, 2020). In sea anemones, Kunitz-type peptides act on TRPV and type II Kv channels (Monastyrnaya et al., 2016; Gladkikh et al., 2020). Various sea anemone type II Kv channel toxins have dual activities, where they acts as blockers of potassium channels as well as serine protease inhibitors, which allows them to serve as defense molecules as well as neurotoxins for prey immobilization (Honma et al., 2008; Mishra, 2020). The AsKC1 and ShPI-I are some of the toxins that suppress proteases and Kv channels. The AsKC1 and ShPI-I belong to Kunitz-type peptide, which are derived from Anemonia sulcata and Stichodactyla haddoni, respectively (Schweitz et al., 1995; Garcia-Fernandez et al., 2016). Seven homologous sequences with ShPI-I and AsKC-1 were identified, and their cysteine pattern was CX8CX15CX7CX12CX3C, including 3 disulfide bridges with C1-C6, C3-C5, C2-C4, connectivities (Figure 6A). Ed-100 was 98.28% similar to the thyroglobulin (GenBank XP_020905524.1) from Exaiptasia diaphana in the NCBI database. The homology comparison results show that the sequence identity of Ed-100, Ed-097 and SHPI-1 is both 51.85%. Taking ShPI-I as the template, the homology modeling prediction results show that AsKC-1, Ed-100, and Ed-097 are described by an α/β/α motif with a compact hydrophobic core (Figure 6B). To date, several Kunitz-type peptides have been retrieved from sea anemone’s, tentacles, secrete mucus, whole bodies and aggressive organs, including acrorhagi in various species of the Actiniidae family (Minagawa et al., 2008).
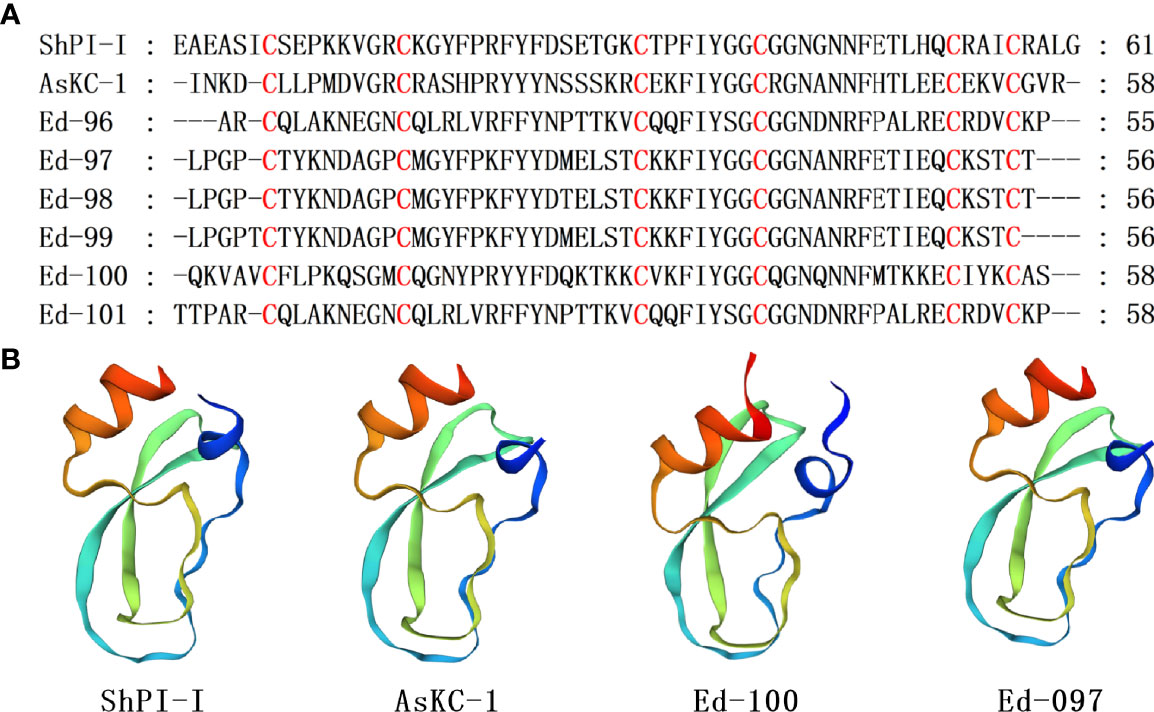
Figure 6 Representative sea anemone mature peptide sequences with Kunitz-type peptides. (A) Alignment of sea anemone mature peptides with Kunitz-type peptides. Cysteines are highlighted in red; (B) Homology modeling prediction of the representative sea anemone mature peptide AsKC-1,Ed-100 and Ed-097 with ShPI-I (π-stichotoxin-She2a; PDB 3OFW).
β-Defensins are essential antimicrobial peptides secreted as innate immune system constituents in various organisms (Suarez-Carmona et al., 2015; Shafee et al., 2016). But, in the sea anemone venom, β-defensin-like peptides are potential neurotoxins that can block ligand- and voltage-gated ion channels including NaV types 1, 2 and 4, KV type 3, and ASIC (Cariello et al., 1989; Ishida et al., 1997; Diochot et al., 1998; Diochot et al., 2004). APETx1 and CgNa belong to β-Defensin-like peptids. APETx1 is a 42-amino-acid peptide toxin of the sea anemone Anthopleura elegantissima, which suppresses voltage-dependent activation of hERG (Matsumura et al., 2021). CgNa was purified from Condylactis gigantea, sea anemone. It inhibits Nav types I and II (Salceda et al., 2007). Five homologous sequences with sea anemone toxin APETx1 and CgNa were identified, and their cysteine pattern was CXC-C-C-CC (Figure 7A). These cysteines are paired in a C1–C5, C2–C4, and C3–C6 fashion to form disulfide bonds that are importance for maintenance of the compact core β-defensins configuration. The homology comparison results show that the sequence identity of Ed-103 and APETx1, Ed-105 and CgNa are 37.84% and 36.59%, respectively. Taking APETx1 and CgNa as the template, the homology modeling prediction results show that Ed-103 and APETx1, Ed-105 and CgNa have similar 3D structures, respectively (Figure 7B).
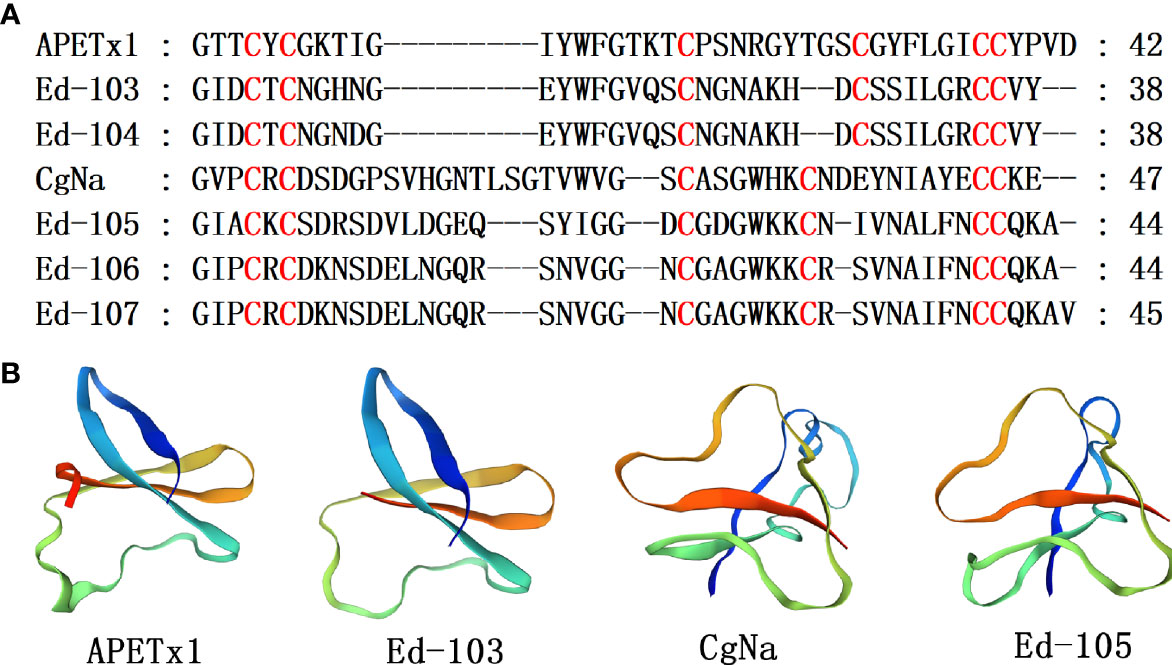
Figure 7 Representative sea anemone mature peptide sequences with β-defensin-like peptides. (A) Alignment of sea anemone mature peptides with β-defensin-like peptides. Cysteines are highlighted in red; (B) Homology modeling prediction of the representative sea anemone mature peptide Ed-103 and Ed-105 with APETx1 (κ-actitoxin-Ael2a; PDB7BWI) and CgNa (△-actitoxin-Cgg1a; PDB 2H9X).
Discovery of Insulin-Like Peptide
The insulin-like peptides (ILPs) have been extensively characterized in invertebrates and vertebrates, with their genes constituting a bug multi-gene family, including genes that encode peptides in fishes, cone snails, amphibians, and mammals (Conlon et al., 1998; Caruso and Sheridan, 2011; Viscarra et al., 2013; Ahorukomeye et al., 2019). In vertebrates, the physiological role and structure of insulin are highly conserved (Conlon, 2001). In contrast, invertebrate insulins are highly variable with several functions, such as haemolymph glucose level regulation, neuronal signaling, reproduction, memory, and growth (Smit et al., 1998; Safavi-Hemami et al., 2016). In general, a majority of these ILP genes encode a single pre-propeptide with a signal peptide and adjacent B, C, and A peptides. In the process of peptide processing, signal peptides are first removed to form the main active proinsulin. Next, the proinsulin is transformed into mature peptide insulin by forming disulfide bonds between cysteine in A peptide and B peptide, and the C peptide was then removed by a proteolytic enzyme (Thompson and Di Gregorio, 2015).
Here, five ILPs (ILP-Ap01~05) were obtained from a transcriptome data of Exaiptasia diaphana, and the alignment sequence is shown in Figure 8 and Supplementary Table 1. Multiple sequence alignment of insulin family members, involving cone snails, fishes, amphibians and mammals, showed that primary sequences of insulin family members were poorly conserved, particularly among invertebrates, apart from the stringently conserved cysteines (Lu et al., 2020). All ILPs from sea anemones exhibited classic cysteine characteristics shared by the entire protein family, while sequence identities for other amino acids with ILPs of other species were low. Two more cysteines were found in B chain-like domain as a phylum-specific character for ILPs in platyhelminths, which markedly differed from findings in other phyla. In addition, compared with human insulin, a short additional propeptide between B chain and signal peptides were predicted in all sea anemone ILPs (Figure 8). ILP-Ap01, ILP-Ap02 and ILP-Ap03 are similar to human insulin and have contiguous B, C and A peptides. However, A-chain was inserted between pro-peptide and B-chain in ILP-Ap04 and ILP-Ap05, resulting in contiguous A, B and C peptides, even the IPL-Ap05 had no C peptide. Therefore, these novel findings indicate that the C chain is not necessary for the formation of insulin.
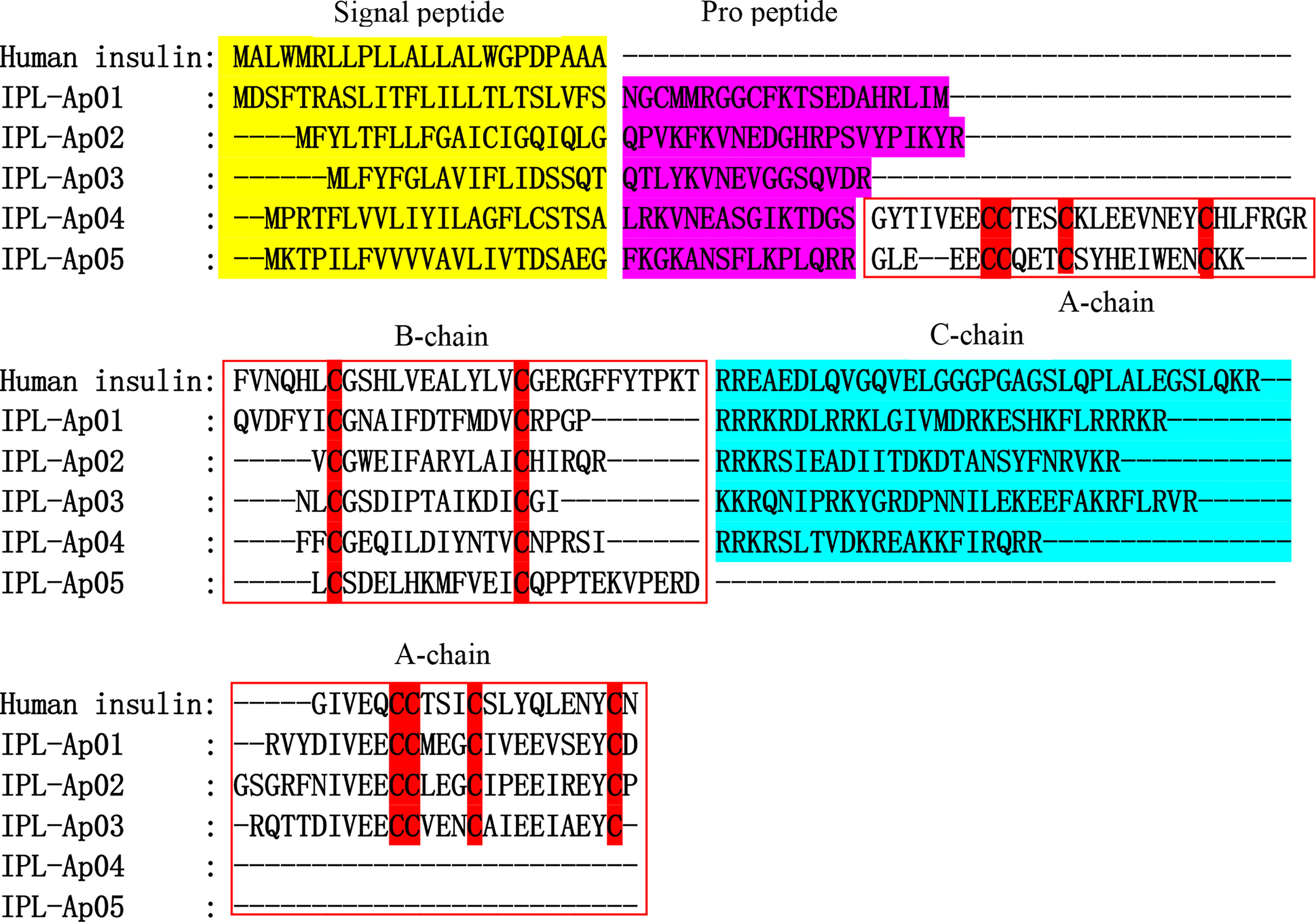
Figure 8 Alignments of the human insulin and five insulin-like peptides (ILPs) from Exaiptasia diaphana. The conserved cysteines are highlighted in red shade. Signal peptide, propeptide and C chain are respectively shown in yellow, purple, and blue. The A chain and B chain boundaries are shown in the red box.
The homologous map of six ILPs shows a basic insulin structure, including residues that maintain an insulin-type hydrophobic core and consisting of A and B chains covalently linked by one intrachain disulfide bond and two interchain disulfide bonds (Mao et al., 2019). At the same time, the conserved cysteines with the characteristic amino acid sequence CCX3CX8C was also shown, which was the result of the retention distance in A peptide (Mayer et al., 2007). The projected tertiary structures for ILPs from sea anemone are shown in Figure 9. The models established by the SWISS-MODLE (Waterhouse et al., 2018) and I-TASSER server (Zheng et al., 2019) for every ILP are presented only with the model exhibiting the highest C-core. The predicted tertiary ILP structures exhibited great similarities to those of proinsulin from human. The tertiary ILP structure was assigned into 3 main α-helices, and a C-domain loop separating helices 1 and 2. The typical hydrophobic core, which had been formed by the 3 α-helices was present in all ILP tertiary structures modeled by SWISS-MODEL and I-TASSER. However, all of the projected models exhibited same conserved helices as well as functional hydrophobic core-features that were present in all insulin family members.
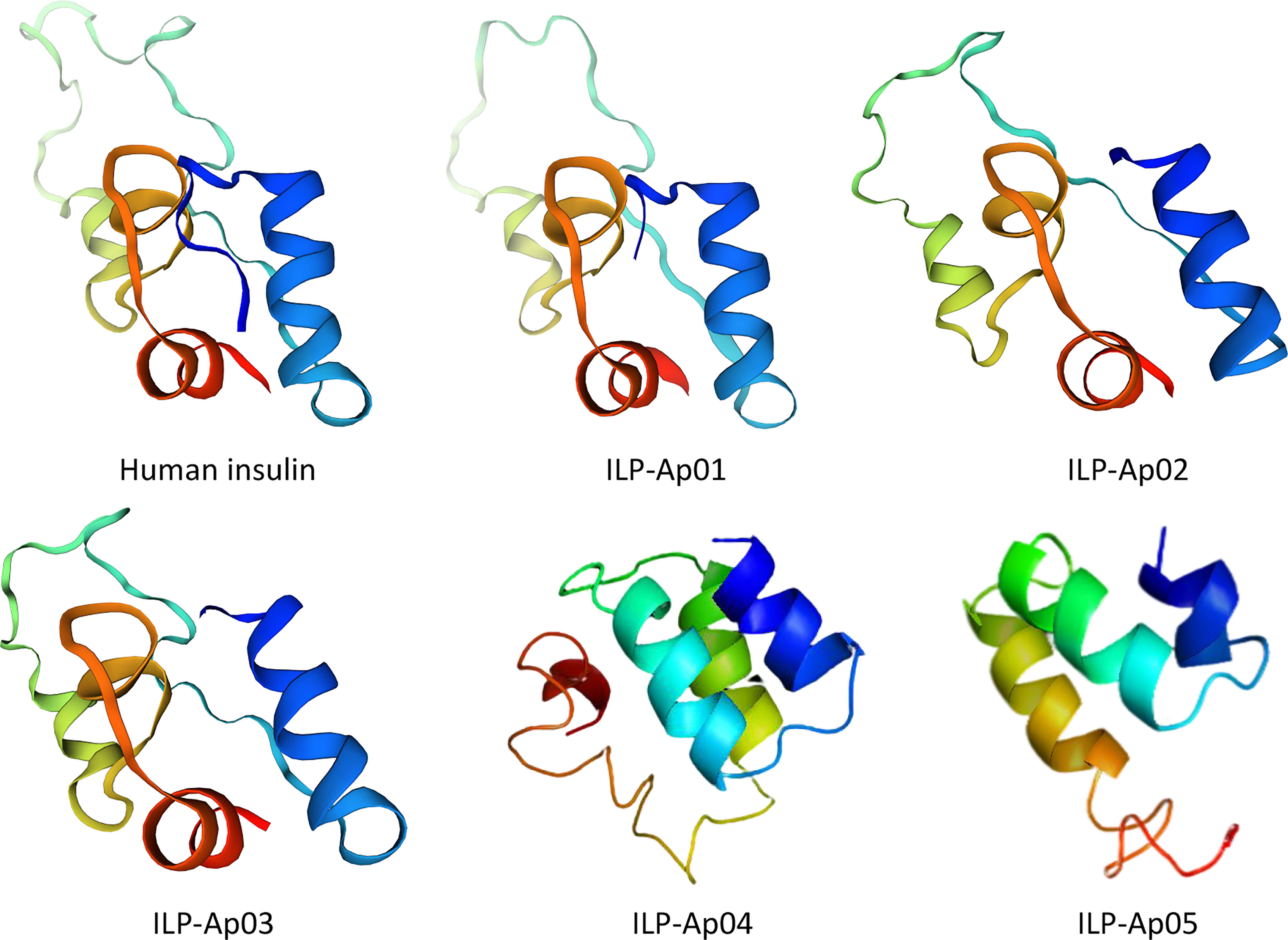
Figure 9 Comparisons of tertiary structures for human insulin with predicted 3D models for ILPs from Exaiptasia diaphana. 3D structure for human pro-insulin was downloaded from Protein Data Bank (PDB ID: 2kqp). The 3 functional helices as well as hydrophobic cores are shown. Colors denote the A chain (blue), the C chain (yellow-cyan) and the B chain (red) for all peptides. All ILP models were predicted without signal peptides or the propeptide before the B chain.
Phylogenetic analyses of ILPs from different species are shown in Figure 10. All ILPs from different species (sea anemone, fish, Conus, mammals, and amphibians) were clearly divided into five groups, and sea anemone ILPs had close evolutionary associations with Conus ILPs, relative to vertebrates ILPs.
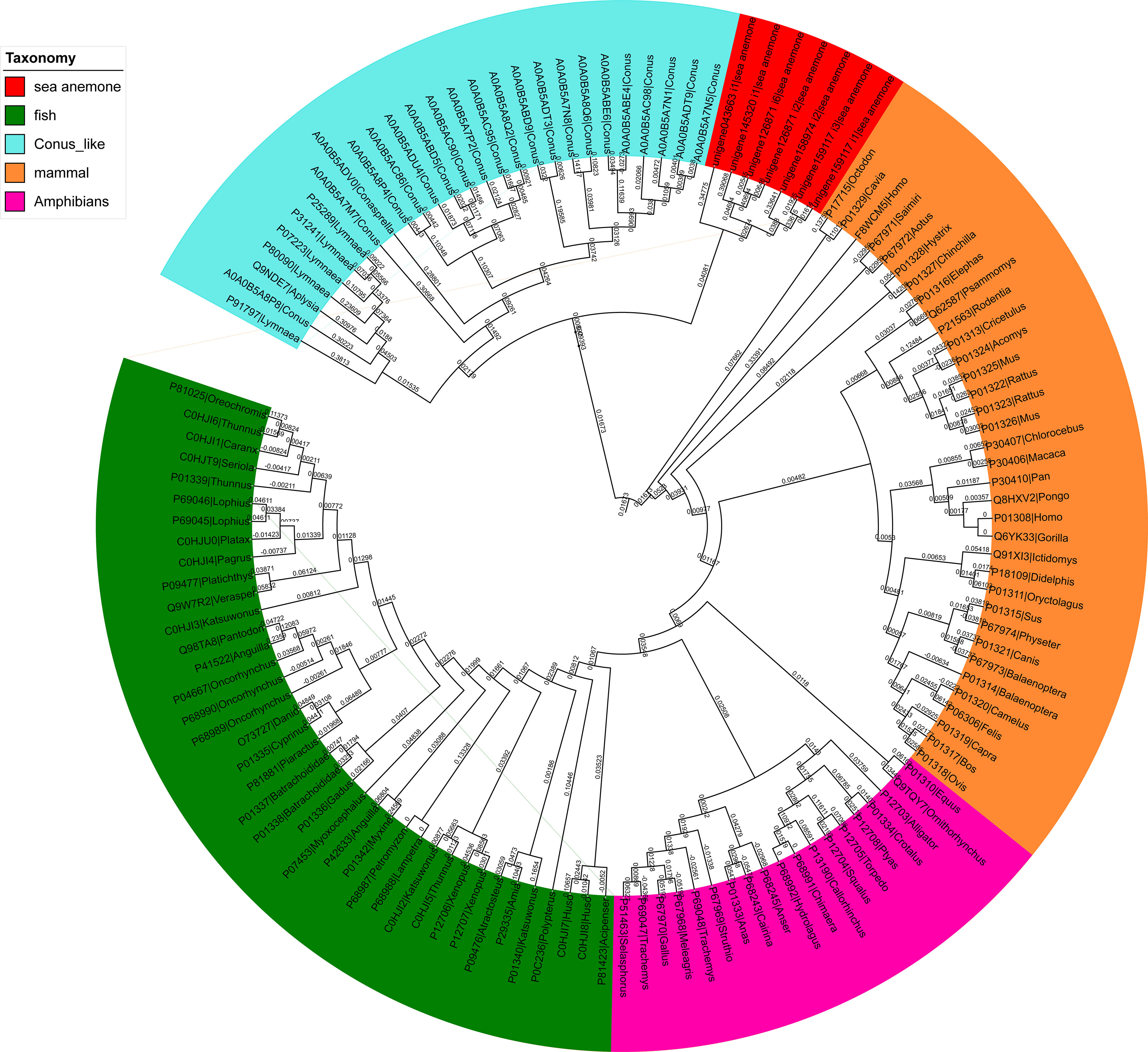
Figure 10 A phylogenetic analysis of the insulin-like peptides (ILPs) from different species. The tree was built via the NJ method. The reliability of every branch was evaluated using 1000 bootstrap replications.
Discussion
Transcriptome technology have recently been applied to explore protein and neuropeptide from several sea anemone species, such as Stichodactyla haddoni, Anemonia sulcata, Exaiptasia pallida, Anthopleura elegantissima, Anthopleura dowii, Heteractis crispa, Cnidopus japonicus, Oulactis sp., Nematostella vectensis, Megalactis griffithsi, and so on (Elran et al., 2014; Macrander et al., 2015; Macrander et al., 2016; Ayala-Sumuano et al., 2017; Madio et al., 2017; Grafskaia et al., 2018; Davey et al., 2019; Mitchell et al., 2020). Exaiptasia diaphana has been commonly used as coral symbiosis study models (Poole et al., 2016; Maire et al., 2021), but responses of its neuropeptide toxins for predation and defense have been barely explored. (Lehnert et al., 2012) assembled a transcriptome of clonal adult population of the aposymbiotic (dinoflagellate-free) Aiptasia pallida from ~208 million reads, yielding 58,018 contigs. They have the resource to identify single-nucleotide variants (SNVs) in the clonal anemone population, approximate Aiptasia genome sizes to be ~421 M, and detect potential neuropeptide-encoding genes. The focus of this transcriptome will form the basis for future studies to investigate changes in gene expressions accompanying the relationship with dinoflagellate endosymbionts, regulate how symbiotic partners respond to various stress factors, assess the suitability of this model system to corals, and complete Aiptasia genome assembly as well as annotation.
With regards to the variety of sea anemone toxins in pale anemones (Exaiptasia diaphana) at different developmental stages, 533 transcripts were detected and grouped into 75 groups based on their possible functions, and peptide toxins with 72.98% being proteins and 27.02% being peptides, this result is similar to the published transcriptome analysis of Stichodactyla haddoni (Madio et al., 2017). Among them, 166, 181, and 148 putative protein and toxins transcripts are derived from the three datasets for Small, Middle, and Big body-sized Exaiptasia diaphana, respectively. The three datasets suggest that sea anemone Exaiptasia diaphana at produced the most proteins and toxins in the middle of development, followed by the early developmental stage, and the least in the late stage of development. By comparing each two of the three transcriptomes, of these putative protein and toxins, 31 were identified in all the three datasets, which were classified into 18 superfamilies. Majority of the protein constituents corresponded to metalloproteases, chymotrypsinogen like, PLRP like, collagen, as well as G-protein coupled receptor, while peptide constituents corresponded to Kunitz-type, EGF-like, defensin, ShK domain, and insulin-like. Among these proteins and peptides, the ShK domain was dominantly expressed, and its roles involved capturing prey, defense as well as in intraspecific competition. Moreover, collagen levels were highest in the middle developmental stage of Exaiptasia diaphana, and relatively elevated in the mature developmental stage. Elevated collagen levels may be associated with adhesive abilities of sea anemones. (Davey et al., 2019) found that adhesions of Exaiptasia pallida pedal disc may be achieved via secretions of extracellular matrix-like proteinaceous matrices made up of collagen, proteoglycans, glycoproteins, and lectins. Venom is already present in the early life stages of the sea anemone, and its composition can change dramatically throughout its life cycle (Sachkova et al., 2019). Columbus-Shenkar and his team study the changes of different toxin expression levels in different developmental stages of Nematostella vectensis, and suggested that venom composition changes across development and that the changes were related to the differences of interspecific interactions in its life stage (Columbus-Shenkar et al., 2018). Recent studies have found that the functional toxin profiles of different anatomical regions in sea anemones are related to their ecological function (Ashwood et al., 2022). Because sea anemones do not have a centralized venom system, but have nematocysts distributed in the entire body, it is possible to realize the functional specialization of venom in a specific tissue based on the function of the tissue (Mccabe and Mackessy, 2015; Surm et al., 2019).
All sea anemone neurotoxins are virtually made up of cysteine-rich peptides, and structural scaffold diversity among these is significant. To data, there is no literature report on the systematic classification and nomenclature of cysteine patterns of sea anemone neurotoxin peptides. In this study, a total of 144 cysteine-rich peptides were found from the transcriptomic data of sea anemone Exaiptasia diaphana. We tried to simply sort and classify the cysteine patterns of these peptides. According to the amount of cysteine, the cysteine patterns of these peptides can be divided into 11 major categories, using II-XII respectively, because there is no single cysteine. Further, cysteine pattern is divided into different subclasses according to the amino acids between cysteine. The peptides containing 4 cysteines forming 2 disulfide bond patterns (IV-type) and 6 cysteines forming 3 disulfide bond patterns (VI-type) are the most abundant. Usually, even numbers of cysteine rich peptides are obtained from venom to form intramolecular disulfide bonds, while a large number of odd cysteines are found in sea anemone neuropeptides, which may exist in the form of polymers to form intermolecular disulfide bonds.
In the family classification of 144 peptides, the most abundant and pharmacologically valuable sea anemone peptide family is ShK domain, Kunitz-type, EGF-like, and defensin. The ShK domain peptides blocked Kv channels, especially potential Kv1.3, which has an important role in T and B lymphocyte subsets involved in autoimmune diseases (Chi et al., 2012). Therefore, ShK domain peptides are probable immune modulators for autoimmune disease treatment. Shafee et al. studied the evolution of the biophysical properties of sequences within the ShK domain (Shafee et al., 2019). This chemical spatial analysis can be used as a guide for selecting sequences for functional studies, and can help to understand the evolution of these highly divergent sequences with ancient conserved folds. EGF peptides modulate TRPV1 channel activities, however, these activities result from involvement of the EGF receptor/PLA2/arachidonic acid/lipoxygenase pathway in indirect TRPV1 activation (Cuypers et al., 2011). Therefore, this toxin group might be important in elucidation of regulation of TRPV1 channels. Kunitz-type peptides act on TRPV and type II Kv channels (Andreev et al., 2008; Peigneur et al., 2011). Some type II Kv channel toxins exhibit dual activities, as they act as suppressors of serine proteases and potassium channel blockers, which enables them to serve as defense molecules as well as neurotoxins for prey immobilizations (Honma et al., 2008; Mishra, 2020). β-defensin-like peptides are neurotoxins for blocking voltage- and ligand-gated ion channels including Kv type 4, Nav type 1, 2 and 4, and ASIC (Cariello et al., 1989; Ishida et al., 1997; Diochot et al., 1998; Diochot et al., 2004), and also inhibit the voltage-dependent activation of hERG (Matsumura et al., 2021).
Interestingly, five novel ILPs were identified from the transcriptomic data of sea anemone Exaiptasia diaphana. ILP-Ap01~03 are similar to human insulin and have contiguous B, C and A peptides. However, A-chain was inserted between propeptide and B-chain in ILP-Ap04 and ILP-Ap05, resulting in contiguous A, B and C peptides, even the IPL-Ap05 had no C peptide. Therefore, these findings indicate that the C chain is not necessary for the formation of insulin. With similar research, several ILPs were identified in the tentacle transcriptomes of Oulactis sp. (Mitchell et al., 2020). The representative ILP-IlO1-i1did not bind to the insulin orinsulin-like growth factor receptors, but showed weak activity against KV1.2, 1.3, 3.1, and 11.1 (hERG) channels, as well as NaV1.4 channels (Mitchell et al., 2021). In addition, two fish-hunting cone snails, Conus tulipa and Conus geographus, have special insulins that are major constituents of their venoms (Safavi-Hemami et al., 2016). These insulins have great similarity to fish insulins compared to molluscan hormones and are unique because of the presence of post-translational modifications that are characteristic of conotoxins. When venom insulin is injected into fish, it induces hypoglycemic shock, which is associated with suppressed blood glucose. Therefore, for various fish-hunting cone snails, insulin is a weapon for prey capture. The cluster tree showed that the relationship between sea anemone insulin and conoinsulin was very close, indicating that sea anemone ILPs was also used for predation. Studies should elucidate on structural elements of insulin that are important for subtle regulations of these hormones.
Materials And Methods
Sample Collection and RNA Extraction
Exaiptasia diaphana were collected in offshore areas of Lingshui City (18°40 E, 109°95 N), Hainan province, China, on August 17, 2020. A total of 3 Exaiptasia diaphana were collected at each developmental stage (including early stage, middle stage and mature stage). The mixture of 3 anemone whole body was used for total RNA extraction, respectively. RNA integrity number (RIN) values of extracted RNA were assessed by an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). Then, 3 Illumina cDNA libraries were separately build using qualified RNA from various developmental stages. The libraries were sequenced on an Illumina HiSeq4000 platform (Illumina, San Diego, CA, USA) at BGI-Tech (BGI, Shenzhen, Guangdong, China).
Sequence Analyses and Assembly
SOAPnuke software (Li et al., 2008) was employed to filter reads with sequencing adaptors, as well as reads with > 10% of non-sequenced bases or > 50% of low-quality bases (≤10 was the base quality score). Trinity v2.5.1 (Haas et al., 2013) was used for assembling the remaining clean reads into contigs, which were then clustered based on sequence similarities and thereafter assembled into consensus unigenes via TGICL v2.1 (Pertea et al., 2003). We used the a Bowtie 2 read aligner to align clean reads with the de novo assembled assembly to evaluate values of gene transcriptions in assembled transcriptomes (Langmead and Salzberg, 2012). The established alignments were analyzed by RNA-Seq by Expectation Maximization (RSEM) v1.2.31 for transcript abundance estimation in FPKM terms (Li and Dewey, 2011).
Functional Annotations of Transcripts
This objective was achieved by searching various public databases (with E-value ≤ 10−5 as the threshold) including InterPro (Finn et al., 2017), Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa and Goto, 2000), NCBI Nr and Nt, UniProtKB/Swiss-Prot (Boeckmann et al., 2003), Clusters of Orthologous Groups (COG) (Tatusov et al., 2000) and Gene Ontology (GO). Blast2GO v4.1 (Conesa et al., 2005) was used to conduct GO annotations of Nr blast findings.
Identification of Protein and Peptide Toxins
Homolog searches as well as an ab initio prediction approach was used for predicting sea anemone protein and peptide toxins from 3 transcriptome datasets. With regards to homologous predictions, sea anemone protein and peptide toxins from the BLAST database were used to build a local reference dataset. Then, BLASTX (with an E-value of 1e-5) was used to run the assembled sequences against a local dataset. Unigenes with best hits in BLASTX data were transformed into aa sequences. The three datasets of sea anemone protein and peptide toxins were allocated into various classes based on superfamily as well as family classification in BLAST database.
Classification of Protein and Peptide Toxins Superfamilies
Manual inspection of predicted sea anemone protein and peptide transcripts was done by the BLAST database. Transcripts with duplicate or truncated mature region sequences were eliminated. Gene superfamilies, signal peptides, as well as cysteine frameworks of the peptide and protein toxins were checked for verification. Based on 75% identity of highly conserved signal peptide sequences, protein and peptide toxins were allocated into known superfamilies in the BLAST database. Those protein and peptide toxins with low similarity were classified as unknown groups.
Alignment, Homology Modelling as Well as Phylogenetic Analyses
Concatenated amino-acid alignments of the signal domains of all superfamilies were performed using MUSCLE v.3.8.3 (Edgar, 2004). Before phylogenetic analyses, divergent and indistinctly aligned blocks were eliminated by the Gblocks software. Then, JModelTest and Prottest3.2 (Darriba et al., 2011) were used for selection of the best-fitted model. Phylogenetic analyses were performed by the maximum likelihood approach with RAxML8.1, and the trees was visualized and annotated using the tree viewer of MEGA4 (Stamatakis, 2006). The 1,000 bootstrap pseudoreplicates were used to assess statistical supports.
The 3D structure model of putative peptides were developed from primary amino acid sequences via homology modelling on a SWISS-Model server in combination with threading-based LOMETS on an I-TASSER server (Waterhouse et al., 2018; Zheng et al., 2019). In the SWISS-Model server, the homologous sequences with sequence identify than 50% were selected as the template, and the QMEAN score was used to evaluate the model quality.
Sequence alignments as well as phylogenetic analyses of members of insulin/relaxin family from sea anemone, fish, Conus, mammals, and amphibians were acquired from UniProt (Supplementary Table 3; www.uniprot.org/);. Boundary identifications for A and B chains of the peptides were conducted according to sequence annotations in this database. Sequence alignments were done by CLUSTALW, after which they were manually checked using the BIOEDI software. Phylogenetic analyses of platyhelminth ILPs were conducted using NJ and maximum likelihood approaches implemented in MEGA. The settings were: Poisson substitution models and bootstrap test approached with 1000 replications.
Conclusions
In this study, 533 protein and neuropeptide toxin sequences were obtained, which were divided into 75 known families. Majority of the proteins were metalloproteases, PLRP like, collagen, chymotrypsinogen like, and G-protein coupled receptor, while the peptides corresponded to ShK domain, defensin, thrombin, Kunitz-type, and insulin-like peptides. Collagen and ShK domain were the dominant expression, and collagen was associated with adhesive abilities of sea anemones, and neuropeptide toxins are for predation, defense and intraspecific competition. Our findings form a basis for further studies on genetic resources, diversity and genetic evolution of the functional proteins and neuropeptide toxins of Exaiptasia diaphana at various developmental stages.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI (accession: PRJNA694232).
Author Contributions
BG conceived and designed the project. CP, YH, and JF analyzed the data. JF and BG wrote the paper. BG, QS, YL, CP, AJ, and TT revised the manuscript. All authors approved submission of the manuscript.
Funding
This work is supported by National Natural Science Foundation of China (no. 82060686) and Hainan Provincial Natural Science Foundation of China (no. 820RC636), Special scientific research project of Hainan academician innovation platform (no. YSPTZX202132), and Shenzhen Dapeng Special Program for Industrial Development (no. KJYF202001-17).
Conflict of Interest
Authors YH, CP, and QS were employed by Shenzhen Huahong Marine Biomedicine Co. LTD.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.856501/full#supplementary-material
Supplementary Figure 1 | Comparison of the number of Exaiptasia diaphana gene annotations between different databases.
Supplementary Figure 2 | KEGG pathway of the sea anemone Exaiptasia diaphana.
Supplementary Figure 3 | GO analysis of the sea anemone Exaiptasia diaphana.
Supplementary Figure 4 | KEG analysis of Exaiptasia diaphana.
Supplementary Table 1–3 | Protein as well as peptide sequences annotation from various developmental stages of Exaiptasia diaphana.
References
Ahorukomeye P., Disotuar M. M., Gajewiak J., Karanth S., Watkins M., Robinson S. D., et al. (2019). Fish-Hunting Cone Snail Venoms Are a Rich Source of Minimized Ligands of the Vertebrate Insulin Receptor. Elife 8, e41574. doi: 10.7554/eLife.41574
Andreev Y. A., Kozlov S. A., Korolkova Y. V., Dyachenko I. A., Bondarenko D. A., Skobtsov D. I., et al. (2013). Polypeptide Modulators of TRPV1 Produce Analgesia Without Hyperthermia. Mar. Drugs 11 (12), 5100–5115. doi: 10.3390/md11125100
Andreev Y. A., Kozlov S. A., Koshelev S. G., Ivanova E. A., Monastyrnaya M. M., Kozlovskaya E. P., et al. (2008). Analgesic Compound From Sea Anemone Heteractis Crispa Is the First Polypeptide Inhibitor of Vanilloid Receptor 1 (TRPV1). J. Biol. Chem. 283 (35), 23914–23921. doi: 10.1074/jbc.M800776200
Ashwood L. M., Undheim E., Madio B., Hamilton B. R., Daly M., Hurwood D. A., et al. (2022). Venoms for All Occasions: The Functional Toxin Profiles of Different Anatomical Regions in Sea Anemones Are Related to Their Ecological Function. Mol. Ecol. 31 (3), 866–883. doi: 10.1111/mec.16286
Ayala-Sumuano J. T., Licea-Navarro A., Rudino-Pinera E., Rodriguez E., Rodriguez-Almazan C. (2017). Sequencing and De Novo Transcriptome Assembly of Anthopleura Dowii Verril), From Mexico. Genom. Data 11, 92–94. doi: 10.1016/j.gdata.2016.11.022
Beeton C., Pennington M. W., Norton R. S. (2011). Analogs of the Sea Anemone Potassium Channel Blocker ShK for the Treatment of Autoimmune Diseases. Inflamm. Allergy Drug Targets 10 (5), 313–321. doi: 10.2174/187152811797200641
Boeckmann B., Bairoch A., Apweiler R., Blatter M. C., Estreicher A., Gasteiger E., et al. (2003). The SWISS-PROT Protein Knowledgebase and its Supplement TrEMBL in 2003. Nucleic Acids Res. 31 (1), 365–370. doi: 10.1093/nar/gkg095
Bosmans F., Tytgat J. (2007). Sea Anemone Venom as a Source of Insecticidal Peptides Acting on Voltage-Gated Na+ Channels. Toxicon 49 (4), 550–560. doi: 10.1016/j.toxicon.2006.11.029
Cariello L., de Santis A., Fiore F., Piccoli R., Spagnuolo A., Zanetti L., et al. (1989). Calitoxin, a Neurotoxic Peptide From the Sea Anemone Calliactis Parasitica: Amino Acid Sequence and Electrophysiological Properties. Biochemistry 28 (6), 2484–2489. doi: 10.1021/bi00432a020
Caruso M. A., Sheridan M. A. (2011). New Insights Into the Signaling System and Function of Insulin in Fish. Gen. Comp. Endocrinol. 173 (2), 227–247. doi: 10.1016/j.ygcen.2011.06.014
Castaneda O., Harvey A. L. (2009). Discovery and Characterization of Cnidarian Peptide Toxins That Affect Neuronal Potassium Ion Channels. Toxicon 54 (8), 1119–1124. doi: 10.1016/j.toxicon.2009.02.032
Castaneda O., Sotolongo V., Amor A. M., Stocklin R., Anderson A. J., Harvey A. L., et al. (1995). Characterization of a Potassium Channel Toxin From the Caribbean Sea Anemone Stichodactyla Helianthus. Toxicon 33 (5), 603–613. doi: 10.1016/0041-0101(95)00013-c
Chandy K. G., Norton R. S. (2017). Peptide Blockers of Kv1.3 Channels in T Cells as Therapeutics for Autoimmune Disease. Curr. Opin. Chem. Biol. 38, 97–107. doi: 10.1016/j.cbpa.2017.02.015
Chi V., Pennington M. W., Norton R. S., Tarcha E. J., Londono L. M., Sims-Fahey B., et al. (2012). Development of a Sea Anemone Toxin as an Immunomodulator for Therapy of Autoimmune Diseases. Toxicon 59 (4), 529–546. doi: 10.1016/j.toxicon.2011.07.016
Columbus-Shenkar Y. Y., Sachkova M. Y., Macrander J., Fridrich A., Modepalli V., Reitzel A. M., et al. (2018). Dynamics of Venom Composition Across a Complex Life Cycle. Elife 7, e35014. doi: 10.7554/eLife.35014
Conesa A., Gotz S., Garcia-Gomez J. M., Terol J., Talon M., Robles M. (2005). Blast2GO: A Universal Tool for Annotation, Visualization and Analysis in Functional Genomics Research. Bioinformatics 21 (18), 3674–3676. doi: 10.1093/bioinformatics/bti610
Conlon J. M. (2001). Evolution of the Insulin Molecule: Insights Into Structure-Activity and Phylogenetic Relationships. Peptides 22 (7), 1183–1193. doi: 10.1016/s0196-9781(01)00423-5
Conlon J. M., Abdel-Wahab Y. H., O'Harte F. P., Nielsen P. F., Whittaker J. (1998). Purification and Characterization of Insulin, Glucagon, and Two Glucagon-Like Peptides With Insulin-Releasing Activity From the Pancreas of the Toad, Bufo Marinus. Endocrinology 139 (8), 3442–3448. doi: 10.1210/endo.139.8.6139
Cotton J., Crest M., Bouet F., Alessandri N., Gola M., Forest E., et al. (1997). A Potassium-Channel Toxin From the Sea Anemone Bunodosoma Granulifera, an Inhibitor for Kv1 Channels. Revision of the Amino Acid Sequence, Disulfide-Bridge Assignment, Chemical Synthesis, and Biological Activity. Eur. J. Biochem. 244 (1), 192–202. doi: 10.1111/j.1432-1033.1997.00192.x
Cristofori-Armstrong B., Rash L. D. (2017). Acid-Sensing Ion Channel (ASIC) Structure and Function: Insights From Spider, Snake and Sea Anemone Venoms. Neuropharmacology 127, 173–184. doi: 10.1016/j.neuropharm.2017.04.042
Cuypers E., Peigneur S., Debaveye S., Shiomi K., Tytgat J. (2011). TRPV1 Channel as New Target for Marine Toxins: Example of Gigantoxin I, a Sea Anemone Toxin Acting Via Modulation of the PLA2 Pathway. Acta Chim. Slov 58 (4), 735–741.
Darriba D., Taboada G. L., Doallo R., Posada D. (2011). ProtTest 3: Fast Selection of Best-Fit Models of Protein Evolution. Bioinformatics 27 (8), 1164–1165. doi: 10.1093/bioinformatics/btr088
Davey P. A., Rodrigues M., Clarke J. L., Aldred N. (2019). Transcriptional Characterisation of the Exaiptasia Pallida Pedal Disc. BMC Genomics 20 (1), 581. doi: 10.1186/s12864-019-5917-5
Diochot S., Baron A., Rash L. D., Deval E., Escoubas P., Scarzello S., et al. (2004). A New Sea Anemone Peptide, APETx2, Inhibits ASIC3, a Major Acid-Sensitive Channel in Sensory Neurons. EMBO J. 23 (7), 1516–1525. doi: 10.1038/sj.emboj.7600177
Diochot S., Lazdunski M. (2009). Sea Anemone Toxins Affecting Potassium Channels. Prog. Mol. Subcell Biol. 46, 99–122. doi: 10.1007/978-3-540-87895-7_4
Diochot S., Schweitz H., Beress L., Lazdunski M. (1998). Sea Anemone Peptides With a Specific Blocking Activity Against the Fast Inactivating Potassium Channel Kv3.4. J. Biol. Chem. 273 (12), 6744–6749. doi: 10.1074/jbc.273.12.6744
Edgar R. C. (2004). MUSCLE: Multiple Sequence Alignment With High Accuracy and High Throughput. Nucleic Acids Res. 32 (5), 1792–1797. doi: 10.1093/nar/gkh340
Elran R., Raam M., Kraus R., Brekhman V., Sher N., Plaschkes I., et al. (2014). Early and Late Response of Nematostella Vectensis Transcriptome to Heavy Metals. Mol. Ecol. 23 (19), 4722–4736. doi: 10.1111/mec.12891
Fautin D. G. (2009). Structural Diversity, Systematics, and Evolution of Cnidae. Toxicon 54 (8), 1054–1064. doi: 10.1016/j.toxicon.2009.02.024
Finn R. D., Attwood T. K., Babbitt P. C., Bateman A., Bork P., Bridge A. J., et al. (2017). InterPro in 2017-Beyond Protein Family and Domain Annotations. Nucleic Acids Res. 45 (D1), D190–D199. doi: 10.1093/nar/gkw1107
Frazao B., Vasconcelos V., Antunes A. (2012). Sea Anemone (Cnidaria, Anthozoa, Actiniaria) Toxins: An Overview. Mar Drugs 10 (8), 1812–1851. doi: 10.3390/md10081812
Fu J., Liao Y., Jin A. H., Gao B. (2021). Discovery of Novel Peptide Neurotoxins From Sea Anemone Species. Front. Biosci. (Landmark Ed.) 26 (11), 1256–1273. doi: 10.52586/5022
Garcia-Fernandez R., Peigneur S., Pons T., Alvarez C., Gonzalez L., Chavez M. A., et al. (2016). The Kunitz-Type Protein ShPI-1 Inhibits Serine Proteases and Voltage-Gated Potassium Channels. Toxins (Basel) 8 (4), 110. doi: 10.3390/toxins8040110
Gladkikh I., Peigneur S., Sintsova O., Lopes Pinheiro-Junior E., Klimovich A., Menshov A., et al. (2020). Kunitz-Type Peptides From the Sea Anemone Heteractis Crispa Demonstrate Potassium Channel Blocking and Anti-Inflammatory Activities. Biomedicines 8 (11), 473. doi: 10.3390/biomedicines8110473
Grafskaia E. N., Polina N. F., Babenko V. V., Kharlampieva D. D., Bobrovsky P. A., Manuvera V. A., et al. (2018). Discovery of Novel Antimicrobial Peptides: A Transcriptomic Study of the Sea Anemone Cnidopus Japonicus. J. Bioinform. Comput. Biol. 16 (2), 1840006. doi: 10.1142/S0219720018400061
Grajales A., Rodriguez E. (2016). Elucidating the Evolutionary Relationships of the Aiptasiidae, a Widespread Cnidarian-Dinoflagellate Model System (Cnidaria: Anthozoa: Actiniaria: Metridioidea). Mol. Phylogenet. Evol. 94 (Pt A), 252–263. doi: 10.1016/j.ympev.2015.09.004
Haas B. J., Papanicolaou A., Yassour M., Grabherr M., Blood P. D., Bowden J., et al. (2013). De Novo Transcript Sequence Reconstruction From RNA-Seq Using the Trinity Platform for Reference Generation and Analysis. Nat. Protoc. 8 (8), 1494–1512. doi: 10.1038/nprot.2013.084
Hermann P. M., Schein C. H., Nagle G. T., Wildering W. C. (2004). Lymnaea EGF and Gigantoxin I, Novel Invertebrate Members of the Epidermal Growth Factor Family. Curr. Pharm. Des. 10 (31), 3885–3892. doi: 10.2174/1381612043382503
Honma T., Kawahata S., Ishida M., Nagai H., Nagashima Y., Shiomi K. (2008). Novel Peptide Toxins From the Sea Anemone Stichodactyla Haddoni. Peptides 29 (4), 536–544. doi: 10.1016/j.peptides.2007.12.010
Ishida M., Yokoyama A., Shimakura K., Nagashima Y., Shiomi K. (1997). Halcurin, a Polypeptide Toxin From the Sea Anemone Halcurias Sp., With a Structural Resemblance to Type 1 and 2 Toxins. Toxicon 35 (4), 537–544. doi: 10.1016/s0041-0101(96)00143-2
Jouiaei M., Yanagihara A. A., Madio B., Nevalainen T. J., Alewood P. F., Fry B. G. (2015). Ancient Venom Systems: A Review on Cnidaria Toxins. Toxins (Basel) 7 (6), 2251–2271. doi: 10.3390/toxins7062251
Kalina R., Gladkikh I., Dmitrenok P., Chernikov O., Koshelev S., Kvetkina A., et al. (2018). New APETx-Like Peptides From Sea Anemone Heteractis Crispa Modulate ASIC1a Channels. Peptides 104, 41–49. doi: 10.1016/j.peptides.2018.04.013
Kalman K., Pennington M. W., Lanigan M. D., Nguyen A., Rauer H., Mahnir V., et al. (1998). ShK-Dap22, a Potent Kv1.3-Specific Immunosuppressive Polypeptide. J. Biol. Chem. 273 (49), 32697–32707. doi: 10.1074/jbc.273.49.32697
Kanehisa M., Goto S. (2000). KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28 (1), 27–30. doi: 10.1093/nar/28.1.27
Langmead B., Salzberg S. L. (2012). Fast Gapped-Read Alignment With Bowtie 2. Nat. Methods 9 (4), 357–359. doi: 10.1038/nmeth.1923
Lehnert E. M., Burriesci M. S., Pringle J. R. (2012). Developing the Anemone Aiptasia as a Tractable Model for Cnidarian-Dinoflagellate Symbiosis: The Transcriptome of Aposymbiotic A. Pallida. BMC Genomics 13, 271. doi: 10.1186/1471-2164-13-271
Li B., Dewey C. N. (2011). RSEM: Accurate Transcript Quantification From RNA-Seq Data With or Without a Reference Genome. BMC Bioinf. 12, 323. doi: 10.1186/1471-2105-12-323
Li R., Li Y., Kristiansen K., Wang J. (2008). SOAP: Short Oligonucleotide Alignment Program. Bioinformatics 24 (5), 713–714. doi: 10.1093/bioinformatics/btn025
Logashina Y. A., Mosharova I. V., Korolkova Y. V., Shelukhina I. V., Dyachenko I. A., Palikov V. A., et al. (2017). Peptide From Sea Anemone Metridium Senile Affects Transient Receptor Potential Ankyrin-Repeat 1 (TRPA1) Function and Produces Analgesic Effect. J. Biol. Chem. 292 (7), 2992–3004. doi: 10.1074/jbc.M116.757369
Lu A., Watkins M., Li Q., Robinson S. D., Concepcion G. P., Yandell M., et al. (2020). Transcriptomic Profiling Reveals Extraordinary Diversity of Venom Peptides in Unexplored Predatory Gastropods of the Genus Clavus. Genome Biol. Evol. 12 (5), 684–700. doi: 10.1093/gbe/evaa083
Macrander J., Broe M., Daly M. (2016). Tissue-Specific Venom Composition and Differential Gene Expression in Sea Anemones. Genome Biol. Evol. 8 (8), 2358–2375. doi: 10.1093/gbe/evw155
Macrander J., Brugler M. R., Daly M. (2015). A RNA-Seq Approach to Identify Putative Toxins From Acrorhagi in Aggressive and Non-Aggressive Anthopleura Elegantissima Polyps. BMC Genom. 16, 221. doi: 10.1186/s12864-015-1417-4
Madio B., King G. F., Undheim E. A. B. (2019). Sea Anemone Toxins: A Structural Overview. Mar. Drugs 17 (6), 325. doi: 10.3390/md17060325
Madio B., Peigneur S., Chin Y. K. Y., Hamilton B. R., Henriques S. T., Smith J. J., et al. (2018). PHAB Toxins: A Unique Family of Predatory Sea Anemone Toxins Evolving via Intra-Gene Concerted Evolution Defines a New Peptide Fold. Cell Mol. Life Sci. 75 (24), 4511–4524. doi: 10.1007/s00018-018-2897-6
Madio B., Undheim E. A. B., King G. F. (2017). Revisiting Venom of the Sea Anemone Stichodactyla Haddoni: Omics Techniques Reveal the Complete Toxin Arsenal of a Well-Studied Sea Anemone Genus. J. Proteomics 166, 83–92. doi: 10.1016/j.jprot.2017.07.007
Maire J., Blackall L. L., van Oppen M. J. H. (2021). Microbiome Characterization of Defensive Tissues in the Model Anemone Exaiptasia Diaphana. BMC Microbiol. 21 (1), 152. doi: 10.1186/s12866-021-02211-4
Mao R., Chen Y., Chi Z., Wang Y. (2019). Insulin and its Single-Chain Analogue. Appl. Microbiol. Biotechnol. 103 (21-22), 8737–8751. doi: 10.1007/s00253-019-10170-0
Martins R. D., Alves R. S., Martins A. M., Barbosa P. S., Evangelista J. S., Evangelista J. J., et al. (2009). Purification and Characterization of the Biological Effects of Phospholipase A(2) From Sea Anemone Bunodosoma Caissarum. Toxicon 54 (4), 413–420. doi: 10.1016/j.toxicon.2009.05.005
Matsumura K., Shimomura T., Kubo Y., Oka T., Kobayashi N., Imai S., et al. (2021). Mechanism of hERG Inhibition by Gating-Modifier Toxin, APETx1, Deduced by Functional Characterization. BMC Mol. Cell Biol. 22 (1), 3. doi: 10.1186/s12860-020-00337-3
Mayer J. P., Zhang F., DiMarchi R. D. (2007). Insulin Structure and Function. Biopolymers 88 (5), 687–713. doi: 10.1002/bip.20734
Mccabe T. M., Mackessy S. P. (2015). “Functional and Genetic Diversity of Toxins in Sea Anemones,” in Evolution of Venomous Animals and Their Toxins. Ed. Daly M. (Dordrecht: Springer), 87–101.
Minagawa S., Sugiyama M., Ishida M., Nagashima Y., Shiomi K. (2008). Kunitz-Type Protease Inhibitors From Acrorhagi of Three Species of Sea Anemones. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 150 (2), 240–245. doi: 10.1016/j.cbpb.2008.03.010
Mishra M. (2020). Evolutionary Aspects of the Structural Convergence and Functional Diversification of Kunitz-Domain Inhibitors. J. Mol. Evol. 88 (7), 537–548. doi: 10.1007/s00239-020-09959-9
Mitchell M. L., Hossain M. A., Lin F., Pinheiro-Junior E. L., Peigneur S., Wai D., et al. (2021). Identification, Synthesis, Conformation and Activity of an Insulin-Like Peptide From a Sea Anemone. Biomolecules 11 (12), 1785. doi: 10.3390/biom11121785
Mitchell M. L., Tonkin-Hill G. Q., Morales R. A. V., Purcell A. W., Papenfuss A. T., Norton R. S. (2020). Tentacle Transcriptomes of the Speckled Anemone (Actiniaria: Actiniidae: Oulactis Sp.): Venom-Related Components and Their Domain Structure. Mar Biotechnol. (N. Y.) 22 (2), 207–219. doi: 10.1007/s10126-020-09945-8
Monastyrnaya M., Peigneur S., Zelepuga E., Sintsova O., Gladkikh I., Leychenko E., et al. (2016). Kunitz-Type Peptide HCRG21 From the Sea Anemone Heteractis Crispa Is a Full Antagonist of the TRPV1 Receptor. Mar Drugs 14 (12), 229. doi: 10.3390/md14120229
Moran Y., Genikhovich G., Gordon D., Wienkoop S., Zenkert C., Ozbek S., et al. (2012). Neurotoxin Localization to Ectodermal Gland Cells Uncovers an Alternative Mechanism of Venom Delivery in Sea Anemones. Proc. Biol. Sci. 279 (1732), 1351–1358. doi: 10.1098/rspb.2011.1731
Moran Y., Gordon D., Gurevitz M. (2009). Sea Anemone Toxins Affecting Voltage-Gated Sodium Channels–Molecular and Evolutionary Features. Toxicon 54 (8), 1089–1101. doi: 10.1016/j.toxicon.2009.02.028
Peigneur S., Beress L., Moller C., Mari F., Forssmann W. G., Tytgat J. (2012). A Natural Point Mutation Changes Both Target Selectivity and Mechanism of Action of Sea Anemone Toxins. FASEB J. 26 (12), 5141–5151. doi: 10.1096/fj.12-218479
Peigneur S., Billen B., Derua R., Waelkens E., Debaveye S., Beress L., et al. (2011). A Bifunctional Sea Anemone Peptide With Kunitz Type Protease and Potassium Channel Inhibiting Properties. Biochem. Pharmacol. 82 (1), 81–90. doi: 10.1016/j.bcp.2011.03.023
Pennington M. W., Byrnes M. E., Zaydenberg I., Khaytin I., de Chastonay J., Krafte D. S., et al. (1995). Chemical Synthesis and Characterization of ShK Toxin: A Potent Potassium Channel Inhibitor From a Sea Anemone. Int. J. Pept. Protein Res. 46 (5), 354–358. doi: 10.1111/j.1399-3011.1995.tb01068.x
Pertea G., Huang X., Liang F., Antonescu V., Sultana R., Karamycheva S., et al. (2003). TIGR Gene Indices Clustering Tools (TGICL): A Software System for Fast Clustering of Large EST Datasets. Bioinformatics 19 (5), 651–652. doi: 10.1093/bioinformatics/btg034
Philyppov I. B., Paduraru O. N., Andreev Y. A., Grishin E. V., Shuba Y. M. (2012). Modulation of TRPV1-Dependent Contractility of Normal and Diabetic Bladder Smooth Muscle by Analgesic Toxins From Sea Anemone Heteractis Crispa. Life Sci. 91 (19-20), 912–920. doi: 10.1016/j.lfs.2012.09.001
Poole A. Z., Kitchen S. A., Weis V. M. (2016). The Role of Complement in Cnidarian-Dinoflagellate Symbiosis and Immune Challenge in the Sea Anemone Aiptasia Pallida. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.00519
Prentis P. J., Pavasovic A., Norton R. S. (2018). Sea Anemones: Quiet Achievers in the Field of Peptide Toxins. Toxins (Basel) 10 (1), 36. doi: 10.3390/toxins10010036
Ramirez-Carreto S., Vera-Estrella R., Portillo-Bobadilla T., Licea-Navarro A., Bernaldez-Sarabia J., Rudino-Pinera E., et al. (2019). Transcriptomic and Proteomic Analysis of the Tentacles and Mucus of Anthopleura Dowii Verril. Mar. Drugs 17 (8), 436. doi: 10.3390/md17080436
Rauer H., Pennington M., Cahalan M., Chandy K. G. (1999). Structural Conservation of the Pores of Calcium-Activated and Voltage-Gated Potassium Channels Determined by a Sea Anemone Toxin. J. Biol. Chem. 274 (31), 21885–21892. doi: 10.1074/jbc.274.31.21885
Rodriguez A. A., Salceda E., Garateix A. G., Zaharenko A. J., Peigneur S., Lopez O., et al. (2014). A Novel Sea Anemone Peptide That Inhibits Acid-Sensing Ion Channels. Peptides 53, 3–12. doi: 10.1016/j.peptides.2013.06.003
Sachkova M. Y., Singer S. A., Macrander J., Reitzel A. M., Peigneur S., Tytgat J., et al. (2019). The Birth and Death of Toxins With Distinct Functions: A Case Study in the Sea Anemone Nematostella. Mol. Biol. Evol. 36 (9), 2001–2012. doi: 10.1093/molbev/msz132
Safavi-Hemami H., Lu A., Li Q., Fedosov A. E., Biggs J., Showers Corneli P., et al. (2016). Venom Insulins of Cone Snails Diversify Rapidly and Track Prey Taxa. Mol. Biol. Evol. 33 (11), 2924–2934. doi: 10.1093/molbev/msw174
Salceda E., Perez-Castells J., Lopez-Mendez B., Garateix A., Salazar H., Lopez O., et al. (2007). CgNa, a Type I Toxin From the Giant Caribbean Sea Anemone Condylactis Gigantea Shows Structural Similarities to Both Type I and II Toxins, as Well as Distinctive Structural and Functional Properties(1). Biochem. J. 406 (1), 67–76. doi: 10.1042/BJ20070130
Schweitz H., Bruhn T., Guillemare E., Moinier D., Lancelin J. M., Beress L., et al. (1995). Kalicludines and Kaliseptine. Two Different Classes of Sea Anemone Toxins for Voltage Sensitive K+ Channels. J. Biol. Chem. 270 (42), 25121–25126. doi: 10.1074/jbc.270.42.25121
Shafee T. M., Lay F. T., Hulett M. D., Anderson M. A. (2016). The Defensins Consist of Two Independent, Convergent Protein Superfamilies. Mol. Biol. Evol. 33 (9), 2345–2356. doi: 10.1093/molbev/msw106
Shafee T., Mitchell M. L., Norton R. S. (2019). Mapping the Chemical and Sequence Space of the ShKT Superfamily. Toxicon Off. J. Int. Soc. Toxinol 165, 95–102. doi: 10.1016/j.toxicon.2019.04.008
Shiomi K., Honma T., Ide M., Nagashima Y., Ishida M., Chino M. (2003). An Epidermal Growth Factor-Like Toxin and Two Sodium Channel Toxins From the Sea Anemone Stichodactyla Gigantea. Toxicon 41 (2), 229–236. doi: 10.1016/s0041-0101(02)00281-7
Smit A. B., van Kesteren R. E., Li K. W., Van Minnen J., Spijker S., Van Heerikhuizen H., et al. (1998). Towards Understanding the Role of Insulin in the Brain: Lessons From Insulin-Related Signaling Systems in the Invertebrate Brain. Prog. Neurobiol. 54 (1), 35–54. doi: 10.1016/s0301-0082(97)00063-4
Stamatakis A. (2006). RAxML-VI-HPC: Maximum Likelihood-Based Phylogenetic Analyses With Thousands of Taxa and Mixed Models. Bioinformatics 22 (21), 2688–2690. doi: 10.1093/bioinformatics/btl446
Suarez-Carmona M., Hubert P., Delvenne P., Herfs M. (2015). Defensins: "Simple" Antimicrobial Peptides or Broad-Spectrum Molecules? Cytokine Growth Factor Rev. 26 (3), 361–370. doi: 10.1016/j.cytogfr.2014.12.005
Sunagawa S., Wilson E. C., Thaler M., Smith M. L., Caruso C., Pringle J. R., et al. (2009). Generation and Analysis of Transcriptomic Resources for a Model System on the Rise: The Sea Anemone Aiptasia Pallida and its Dinoflagellate Endosymbiont. BMC Genomics 10, 258. doi: 10.1186/1471-2164-10-258
Surm J. M., Smith H. L., Madio B., Undheim E., King G. F., Hamilton B. R., et al. (2019). A Process of Convergent Amplification and Tissue-Specific Expression Dominates the Evolution of Toxin and Toxin-Like Genes in Sea Anemones. Mol. Ecol. 28 (9), 2272–2289. doi: 10.1111/mec.15084
Tarcha E. J., Chi V., Munoz-Elias E. J., Bailey D., Londono L. M., Upadhyay S. K., et al. (2012). Durable Pharmacological Responses From the Peptide ShK-186, a Specific Kv1.3 Channel Inhibitor That Suppresses T Cell Mediators of Autoimmune Disease. J. Pharmacol. Exp. Ther. 342 (3), 642–653. doi: 10.1124/jpet.112.191890
Tatusov R. L., Galperin M. Y., Natale D. A., Koonin E. V. (2000). The COG Database: A Tool for Genome-Scale Analysis of Protein Functions and Evolution. Nucleic Acids Res. 28 (1), 33–36. doi: 10.1093/nar/28.1.33
Thompson J. M., Di Gregorio A. (2015). Insulin-Like Genes in Ascidians: Findings in Ciona and Hypotheses on the Evolutionary Origins of the Pancreas. Genesis 53 (1), 82–104. doi: 10.1002/dvg.22832
Viscarra J. A., Rodriguez R., Vazquez-Medina J. P., Lee A., Tift M. S., Tavoni S. K., et al. (2013). Insulin and GLP-1 Infusions Demonstrate the Onset of Adipose-Specific Insulin Resistance in a Large Fasting Mammal: Potential Glucogenic Role for GLP-1. Physiol. Rep. 1 (2), e00023. doi: 10.1002/phy2.23
Wang X., Li G., Guo J., Zhang Z., Zhang S., Zhu Y., et al. (2019). Kv1.3 Channel as a Key Therapeutic Target for Neuroinflammatory Diseases: State of the Art and Beyond. Front. Neurosci. 13. doi: 10.3389/fnins.2019.01393
Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., et al. (2018). SWISS-MODEL: Homology Modelling of Protein Structures and Complexes. Nucleic Acids Res. 46 (W1), W296–W303. doi: 10.1093/nar/gky427
Weis V. M., Davy S. K., Hoegh-Guldberg O., Rodriguez-Lanetty M., Pringle J. R. (2008). Cell Biology in Model Systems as the Key to Understanding Corals. Trends Ecol. Evol. 23 (7), 369–376. doi: 10.1016/j.tree.2008.03.004
Yan L., Herrington J., Goldberg E., Dulski P. M., Bugianesi R. M., Slaughter R. S., et al. (2005). Stichodactyla Helianthus Peptide, a Pharmacological Tool for Studying Kv3.2 Channels. Mol. Pharmacol. 67 (5), 1513–1521. doi: 10.1124/mol.105.011064
Zhang M., Liu X. S., Diochot S., Lazdunski M., Tseng G. N. (2007). APETx1 From Sea Anemone Anthopleura Elegantissima is a Gating Modifier Peptide Toxin of the Human Ether-a-Go-Go- Related Potassium Channel. Mol. Pharmacol. 72 (2), 259–268. doi: 10.1124/mol.107.035840
Keywords: sea anemone, Exaiptasia diaphana, transcriptomics, cnidarian, toxins, insulin-like peptide
Citation: Fu J, He Y, Peng C, Tang T, Jin A, Liao Y, Shi Q and Gao B (2022) Transcriptome Sequencing of the Pale Anemones (Exaiptasia diaphana) Revealed Functional Peptide Gene Resources of Sea Anemone. Front. Mar. Sci. 9:856501. doi: 10.3389/fmars.2022.856501
Received: 17 January 2022; Accepted: 01 March 2022;
Published: 21 March 2022.
Edited by:
Joseph Selvin, Pondicherry University, IndiaReviewed by:
Peter J. Prentis, Queensland University of Technology, AustraliaChoo Hock Tan, University of Malaya, Malaysia
Copyright © 2022 Fu, He, Peng, Tang, Jin, Liao, Shi and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiong Shi, c2hpcWlvbmdAZ2Vub21pY3MuY24=; Bingmiao Gao, Z2FvYmluZ21pYW9AaGFpbm1jLmVkdS5jbg==
 Jinxing Fu
Jinxing Fu Yanbin He
Yanbin He Chao Peng
Chao Peng Tianle Tang
Tianle Tang Aihua Jin
Aihua Jin Yanling Liao
Yanling Liao Qiong Shi
Qiong Shi Bingmiao Gao
Bingmiao Gao