
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 04 May 2022
Sec. Marine Fisheries, Aquaculture and Living Resources
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.856126
This article is part of the Research TopicComposition, Functions and Modulation of Gut Microbiota in Maricultural AnimalsView all 8 articles
 Dongwei Hou1
Dongwei Hou1 Renjun Zhou1
Renjun Zhou1 Dongdong Wei2
Dongdong Wei2 Shenzheng Zeng1
Shenzheng Zeng1 Shaoping Weng2
Shaoping Weng2 Qingyun Yan3
Qingyun Yan3 Jianguo He1,2*
Jianguo He1,2* Zhijian Huang1*
Zhijian Huang1*Unraveling the assembly mechanism is a core research topic of microbial ecology. Abundant and rare microbial communities are crucial for diversity, function and host health in a given ecosystem, but few studies focused on their assembly strategies. Here, we explored the microbial diversity of abundant and rare communities of water, shrimp intestine and sediment habitats in the shrimp cultural ponds. Our results found that the numbers of rare operational taxonomic units (OTUs) (6,003, 4,566 and 8,237 OTUs of water, intestine and sediment) was dozens of times more than abundant ones (only 199, 157 and 122 OTUs of water, intestine and sediment). The community diversity of abundant and rare microbial taxa was markedly different, as well as their taxonomic composition. Despite different diversity, similar abundance-occupancy relationship and biogeographic patterns between the abundant and rare microbial communities were observed, with much stronger obvious distance-decay relationships for rare community than abundant community. Furthermore, stochastic processes dominated the community assemblies of both abundant and rare microbial taxa, and deterministic process contributed more microbial community variation to rare taxa than abundant taxa. All the findings advance our understanding on the community assembly strategies of abundant and rare microbial taxa and prompt the contributions of abundant and rare microbial community to the aquatic ecosystems, which will improve aquaculture management strategy.
Microorganisms in ecosystems usually present a skewed abundance distribution, because a mass of rare species coexisting alongside relatively few abundant ones (Sogin et al., 2006). Generally, abundant and rare taxa exhibit disparate distribution patterns and functional traits (Pedrós-Alió, 2012). Abundant taxa are considered to be the most important for ecosystem functions (Cottrell and Kirchman, 2003), but rare taxa are also indispensable that serve as limitless repositories of phylogenetic diversity and play a disproportionate role in functions (Jousset et al., 2017; Jia et al., 2018). Thus, distinguishing community assemblies between abundant and rare taxa is helpful for understanding microbial ecosystem processes and functions.
Different ecological processes regulate the microbial assemblies of abundant and rare communities. One study showed a more deterministic assembly of abundant bacterial community but a more stochastic assembly of rare community in water from subtropical bays (Mo et al., 2018). By contrast, stochastic processes play a key role in shaping the microeukaryotic variation of both abundant and rare communities of water in river (Chen et al., 2019), as well as of water in lake, reservoir and bay (Liu et al., 2015; Zhao et al., 2017; Zhang et al., 2019). These studies have deepened our understanding of different patterns and assemblies of abundant and rare communities in aquatic ecosystems, but they have focused primarily on a single habitat. In fact, the aquatic ecosystem is a complex system of multiple habitats, abundant and rare communities of multiple habitats constitute a metacommunity, and that contribute to the overall microbial diversity and functions (Escalas et al., 2013; Bolnick et al., 2014; Escalas et al., 2015). Therefore, it is necessary to clarify the microbial assembly mechanisms of abundant and rare communities among multiple habitats within a metacommunity framework in a given ecosystem.
Aquaculture is a typical artificial production model, and has become the third largest animal protein source globally, but frequent occurrence of diseases has threatened its development (FAO, 2018). Microorganisms of multiple habitats (e.g., water, sediment and animal intestines) in aquaculture ecosystems modulate many ecosystem services, including the water quality, animal health and disease control (Moriarty, 1997; Zhou et al., 2009; Assefa and Abunna, 2018). Many studies show that the microbial assembly and relationships of animal intestines and environments in aquaculture ecosystems is closely related to the host health (Xiong et al., 2017; Xiong et al., 2018; Huang et al., 2020; Huang et al., 2021), and ultimately affects the aquaculture output. Thus, revealing assembly mechanisms and relationship of animal intestine and environment communities could facilitate microbiota management for promoting the productivity in aquaculture. Importantly, microorganisms of animal intestines and environments in aquaculture ecosystems also present a skewed abundance distribution. Microbial species tend to be rare taxa in one habitat but abundant in another, and vice versa; and many species (especially opportunistic pathogens) that are rare taxa in environments tend to be abundant taxa in animal intestines, which can cause disease outbreaks (Schryver and Vadstein, 2014; Del’Duca et al., 2015; Zhang et al., 2020). Therefore, it is necessary to distinguish the distribution characteristics and assembly mechanisms of abundant and rare microbial communities in aquaculture ecosystems.
In this study, we took the Litopenaeus vannamei (the most widely marine cultured shrimp species in the world) cultural pond ecosystems as the research object and aimed to investigate the microbial patterns at abundant and rare levels of water, shrimp intestine and sediment habitats and expound the key ecological processes that driving abundant and rare community assembly of three habitats. We mainly found that (i) the rare taxa are served as huge repositories of microbial diversity in the cultural ponds, and the community diversity, taxonomic composition and sources of abundant and rare taxa were markedly different; (ii) similar abundance-occupancy relationship and biogeographic patterns between the abundant and rare microbial communities were observed, with much stronger obvious distance-decay relationships for rare community than that of abundant community; (iii) the stochastic processes dominated the community assemblies of both abundant and rare taxa, and the deterministic process contributed more community variation to rare taxa than that to abundant taxa. Our results could be helpful understanding the microbial diversity for abundant and rare communities in aquatic ecosystems, and contribute to the microbiota management in aquaculture.
Forty L. vannamei cultural ponds at five regional sites in China (19.20°-24.06° N, 108.56°-117.86° E) were chosen, which with the similar pond area, water depth and stocking density (Table S1). Three parallel water/shrimp intestine/sediment samples were collected form each pond, so a total of 360 samples were collected. Sample collections (Hou et al., 2018a and Hou et al., 2018b) and environmental factor analyses of water and sediment samples (Hou et al., 2017 and Hou et al., 2021) were detailed in our previous study. Physicochemical factor results were described in the Tables S2, S3, respectively.
Water and intestine/sediment DNA was extracted using the Water DNA Isolation Kit (Omega Bio-tek, Doraville, GA, USA) and PowerFecal/Soil DNA Isolation Kit (Mobio, Carlsbad, CA, USA), respectively. The V3-V4 regions of 16S rRNA gene were amplified by the primer set of 338F (5’-ACTCCTACGGGAGGCA GCAG-3’) and 806R (5’-GGACTACHVGGGTWTCTAAT-3’), and then the PCR products were combined equally and sequenced by the Illumina MiSeq platform (Illumina, San Diego, USA). To completely cover the microbes, we mixed the sequencing data of three parallel water/intestine/sediment samples in each pond for further analysis. The FLASH was used to merge the paired-end sequences, and then the QIIME (version 1.9.0) was employed to process the merged sequences (Caporaso et al., 2010; Magoč and Salzberg, 2011). Then, the chimeric sequences were removed by the UCHIME (Edgar et al., 2011). A distance-based identity of sequences with ≥ 97% were grouped into operational taxonomic units (OTUs) using the UCLUST. Finally, we used the RDP Classifier algorithm (Silva SSU database 132) to select the most abundant sequence from OTU as a representative and classified into a closed reference genome. Sequence data were stored in the NCBI under the accession number PRJNA545396.
We classified all OTUs of each habitat into six categories: always abundant taxa (AAT, with a relative abundance ≥ 1% in all samples of each habitat); conditionally abundant taxa (CAT, ≥ 0.01% in all samples and ≥ 1% in some samples); always rare taxa (ART, < 0.01% in all samples); conditionally rare taxa (CRT, < 0.01% in some samples but never ≥ 1% in any sample); moderate taxa (MT, from 0.01% to 1% in all samples); conditionally rare and abundant taxa [CRAT, ranging from rare (< 0.01%) to abundant (≥ 1%)] (Logares et al., 2014; Liu et al., 2015; Chen et al., 2017). Thus, abundant OTUs included the AAT, CAT and CRAT, and rare OTUs covered the ART and CRT.
We employed the mean nearest taxon distance measure to determine which ecological processes govern the microbial assembly. The weighted beta nearest taxon index (β-NTI) was calculated (Webb et al., 2002; Kembel, 2009) and OTUs was separated into two pairwise communities. Then, the β-NTI combined with Bray-Curtis-based Raup-Crick (RCBray) was employed to quantify the influence of five major processes governing the communities (Stegen et al., 2013). When β-NTI > 2/< 2, the community turnover determined by the heterogeneous selection/homogeneous selection (Dini-Andreote et al., 2015). When |β-NTI | < 2, RCBray > 0.95/< -0.95 represented the influences of dispersal limitation/homogenizing dispersal, while | RCBray| < 0.95, indicated the ecological drift (Stegen et al., 2015). The Sloan neutral community model (SNCM) (Sloan et al., 2006) was also used to predict the relationship between OTUs detection frequency and their abundance. We separately used the datasets of abundant and rare OTUs from each habitat, and the OTUs from each dataset were subsequently separated into three partitions with whether they occurred more/less/within frequently (above/below/neutral partition) than the 95% confidence interval of the SNCM predictions. All computations were performed in the R (version 3.2.3) (R Core Team, 2017).
The non-metric multidimensional scaling (NMDS) and analysis of similarity (ANOSIM) were explored the differences in abundant/rare community of each habitat. The relationships among microbial communities of three habitats were analyzed by the Venn and Chord Diagram analyses. Then, the contributions of different sources to community of each habitat were estimated using the fast expectation-maximization microbial source tracking (FEAST) (Shenhav et al., 2019), with abundant/rare microbial OTUs of one habitat as the sink, and the whole community of other two habitats as the sources. The Spearman’s rank correlation was analyzed the relationship between dissimilarity of abundant/rare community and geographic distance of each habitat, as well as abundance-occupancy relationship. The Mantel test was used to examine the correlations between the environmental factors and water/sediment community structure (999 permutations) (Oksanen et al., 2015).
In total, 199 (the relative abundance with 75.14% of total sequences), 157 (83.39%), and 122 (30.71%) OTUs were considered to be abundant taxa of water, intestine and sediment habitats, while 6,003 (24.60%), 4,566 (16.61%), and 6,948 (68.52%) OTUs were classified as rare taxa (Table 1). For abundant community, only 5 (5.66%), 2 (3.52%) and 5 (2.57%) OTUs were CAT, while 194 (69.94%), 155 (79.88%) and 122 (26.29%) OTUs were CRAT, and for rare community, 3,719 (only 0.69%), 2,514 (only 0.56%) and 1,289 (only 0.66%) OTUs were ART, while 2,284 (23.91%), 2,052 (16.04%) and 6,948 (67.86%) OTUs were CRT (Table 1). Accordingly, the numbers of rare OTUs was dozens of times more than abundant ones, but their relative abundance was only about one-fifth that of abundant ones in water and shrimp intestine; and differently, the numbers of rare OTUs was also dozens of times more than abundant ones with their relative abundance was more than twice as abundant ones in sediment. Further, the great mass of OTUs of three habitats were assigned to Proteobacteria, Bacteroidetes, Cyanobacteria, Actinobacteria and Chloroflexi, and the relative abundance of Proteobacteria was highest in intestine (Table S4). For abundant or rare taxa, the most diverse OTUs of three habitats were assigned to different phyla (Figure S1). At the genus level, the most abundant OTUs were assigned to Cyanobium, Vibrio, Photobacterium, Candidatus Bacilloplasma and Shewanella, and Cyanobium was the highest in water, whereas other four genera that of intestine (Table S5). Thus, the rare taxa are served as huge repositories of microbial diversity in the shrimp cultural ponds, and the community diversity and taxonomic composition of abundant and rare taxa of each habitat was markedly different.
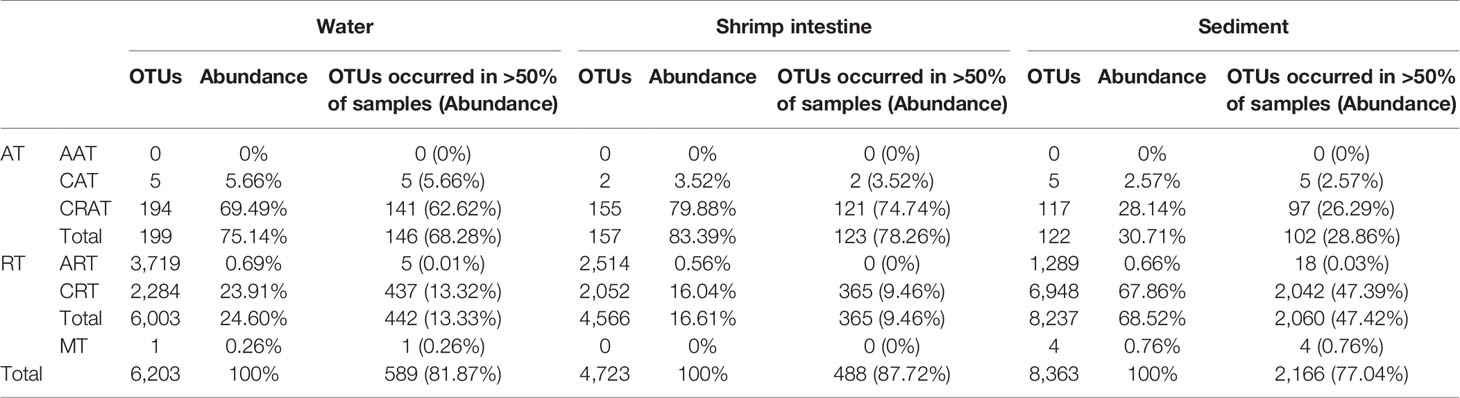
Table 1 General description of abundant and rare microbial OTUs datasets in water, shrimp intestine and sediment habitats.
Venn analysis results found that lots of OTUs shared in three habitats: total 3,876 OTUs were commonly present in three habitats, and the number of OTUs was found to be in any two habitats: 3,950 (water, 83.63%) or 4,604 (sediment, 97.48%) out of 4,723 intestine OTUs, 3,950 (intestine, 63.68%) or 6,074 (sediment, 97.92%) out of 6,203 water OTUs, and 4,604 (intestine, 55.05%) or 6,074 (water, 72.63%) out of 8,363 sediment OTUs (Figure S2). Then, the Chord Diagram results indicated that, for the microbial OTUs that shared in any two habitats, most of rare OTUs in one habitat were also belonged to rare OTUs in another but some of them were belonged to abundant, while OTUs that were abundant in one habitat were almost always rare in another (Figure 1A). Of interest, the Vibrio OTUs were as abundant taxa in shrimp intestine but as rare taxa in water and sediment (Table S6). Such close relationships were observed among abundant and/or rare communities of three habitats. The FEAST results further showed that, for abundant or rare community of water, the most dominant source was sediment (an average of 64.09% or 31.92%), followed by shrimp intestine (24.62% or 19.94%) (Figure 1B). For abundant community of intestine, the water and sediment source accounted for nearly a quarter; but for rare community, the sediment source was more than half (Figure 1C). While for abundant community of sediment, the most dominant source was intestine (51.26%), and the source from water and intestine attributed 25.02% and 24.12% for rare community of sediment (Figure 1D). These results suggest that the sources of abundant and rare communities of each habitat were markedly different, and sediment community was the most important source for abundant community of water and rare community of shrimp intestine.
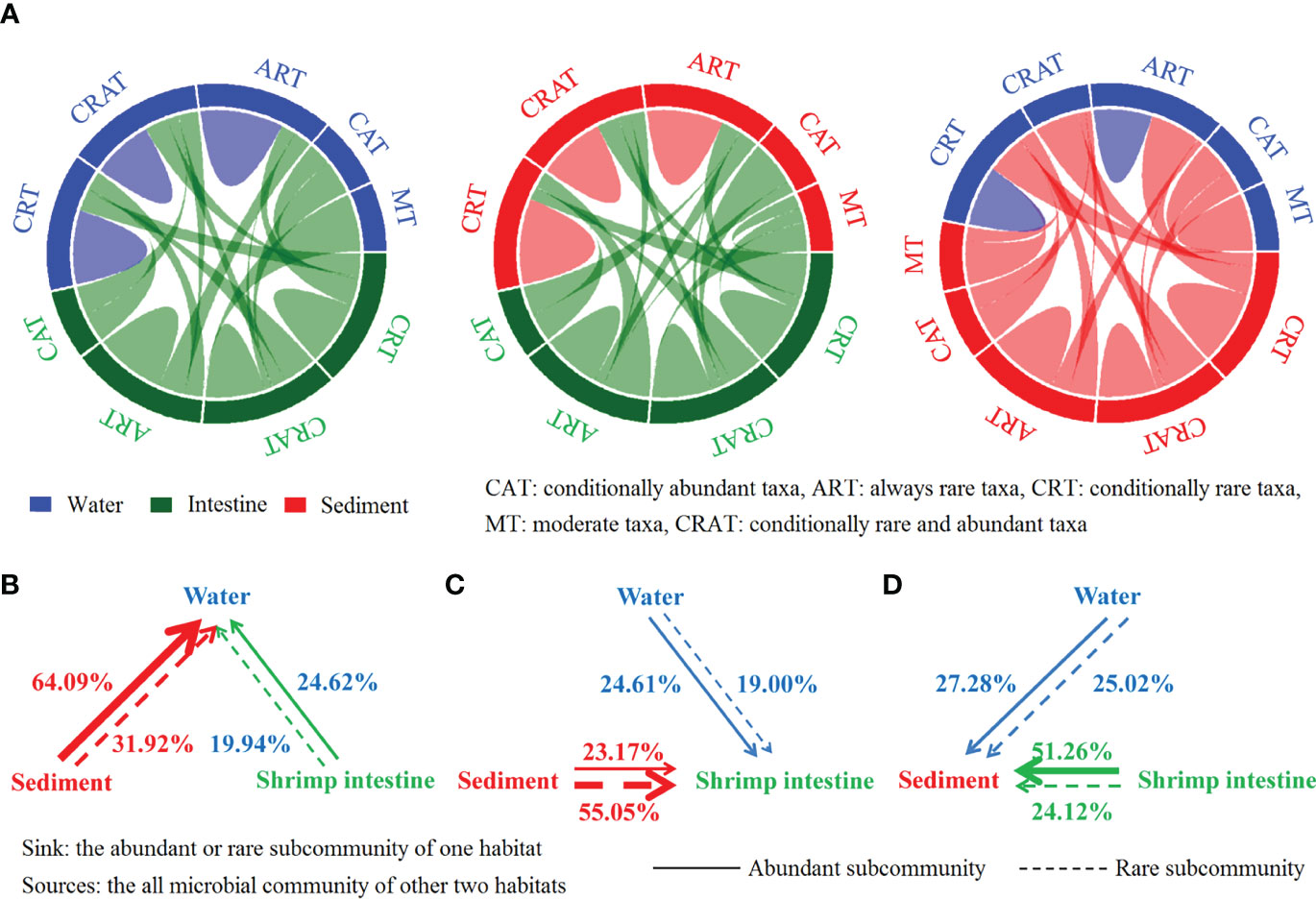
Figure 1 Relationship among communities of three habitats. (A) The Chord Diagram analysis showed that the close relationships among abundant and/or rare communities among water, shrimp intestine and sediment habitats. (B–D) The FEAST analysis results of the contributions of whole communities in water, intestine and sediment to each other’s abundant community or rare community.
Then, we investigated the abundance-occupancy relationship of each habitat, and found that the relative abundance of abundant (r2 = 0.663, 0.549 and 0.767 of water, shrimp intestine and sediment, respectively) or rare (r2 = 0.830, 0.850 and 0.784, respectively) OTUs and sites occupied were significantly (P < 0.001 in all cases) positively correlated (Figure 2). To be specific, the most abundant OTUs occupied >50% of water (146 out of 199 OTUs with relative abundance accounted for 68.28%), intestine (123 out of 157 with 78.26%) and sediment (102 out of 122 with only 28.86%) samples (Table 1). By contrast, only few rare OTUs were identified in >50% of water or intestine samples (only 442 or 365 out of 6,003 or 4,556 OTUs with 13.33% or 9.46%), but 2,060 OTUs (out of 8,237 OTUs with 47.42%) occupied >50% of sediment samples (Table 1).
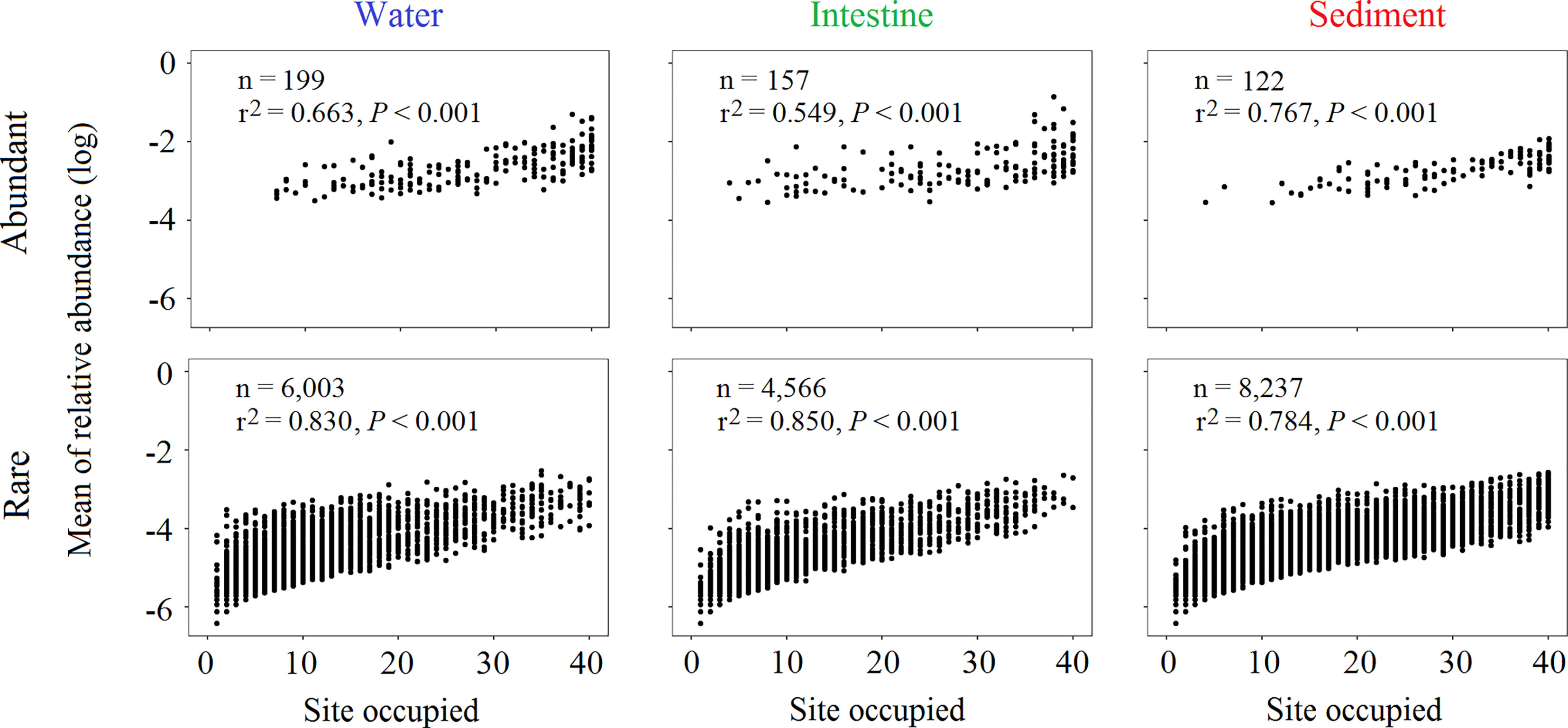
Figure 2 Abundance-occupancy relationship of abundant and rare OTUs. Spearman’s rank correlation between the mean relative abundance of abundant and rare OTUs and number of samples occupied (n is the number of OTUs).
We further observed that the abundant or rare community of each habitat significantly (P < 0.001 in all case) differed between any two of compared sites (Figure S3). We further calculated distance decay curves and found that the community similarity and geographic distance showed significantly (P < 0.05 in all cases) negative correlations for both the abundant (r2 = 0.072, 0.008 and 0.063 of water, intestine and sediment, respectively) and rare communities (r2 = 0.199, 0.025 and 0.107, respectively) (Figure 3). Thus, abundant and rare communities of each habitat exhibit the significant distance-decay relationship (DDRs), and the DDRs of rare community were much stronger than that of abundant of each habitat.
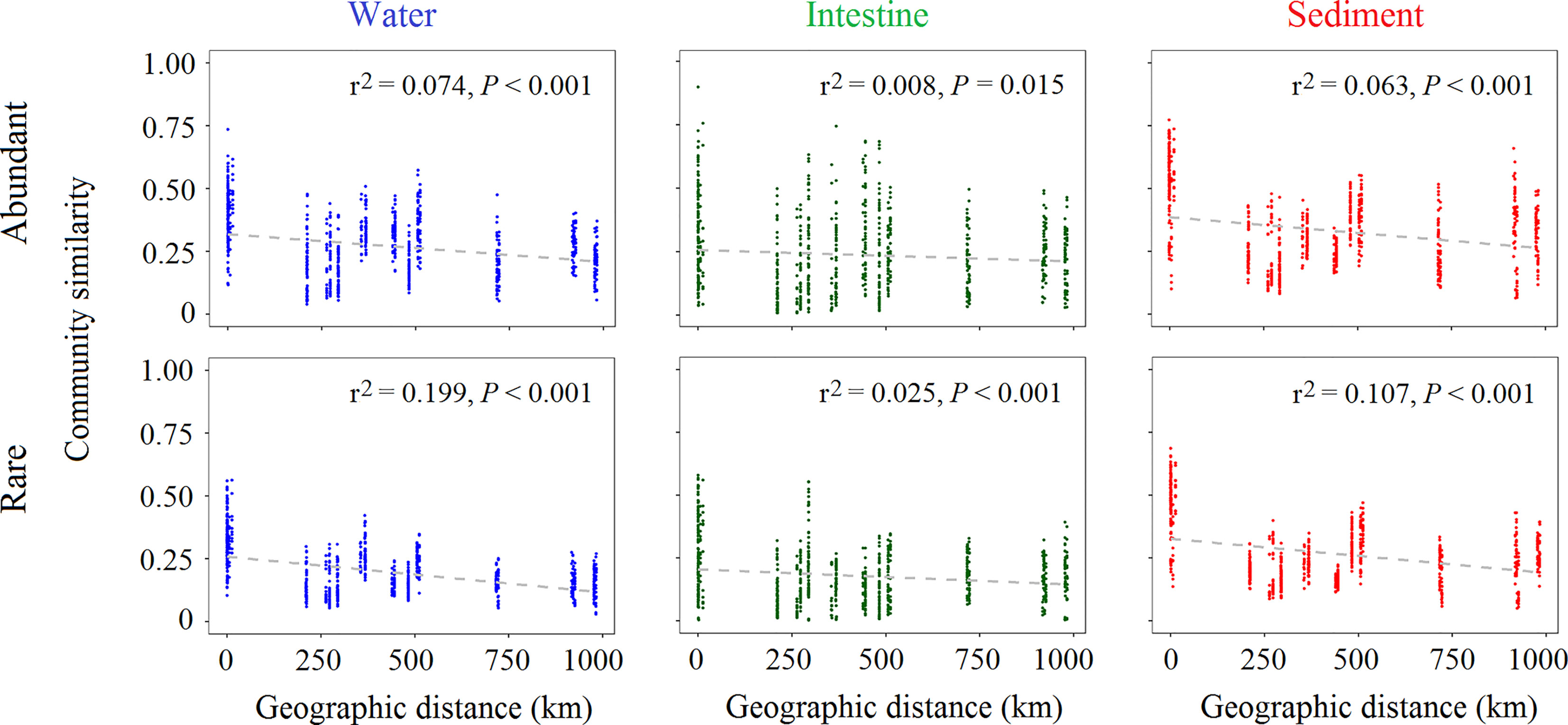
Figure 3 The Spearman’s rank correlations between the community similarity of abundant or rare microbial taxa and geographic distance.
Finally, we found that ~70% of microbial variations at abundant and rare taxa levels of all three habitats were controlled by the dispersal (dispersal limitation contributed over 60%) and drift, followed by the selection; and interestingly, a litter higher microbial variations controlled by the dispersal limitation for abundant community than rare (Figure 4). Thus, stochastic processes dominated the community assembly of abundant and rare taxa of each habitat, which were also confirmed by the SNCM results (Figure S4). Additionally, much higher microbial variations were controlled by the deterministic process for rare community than that for abundant (Figure 4). The Mantel test results further revealed that spatial and physicochemical factors had significantly (P < 0.05 in all cases) influence on the community structure (Table 2). Especially, salinity (r = 0.301 and 0.543) and temperature (r = 0.313 and 0.432) were the best predictors for community structure in both abundant and rare taxa of water, while TN (r = 0.340 and 0.403) for that of sediment (Table 2).
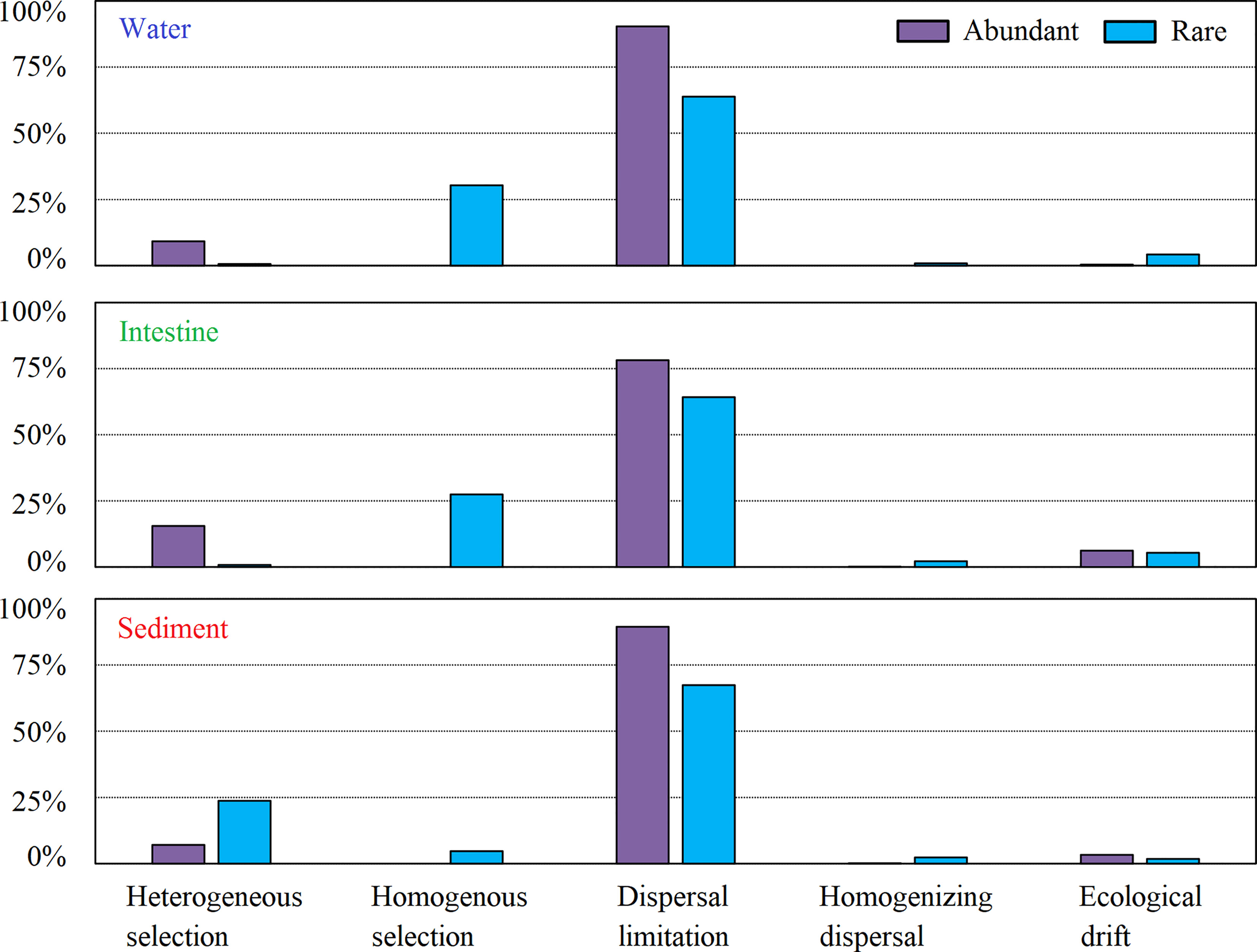
Figure 4 Microbial assembly mechanisms of abundant and rare community of three habitats in the shrimp cultural ponds.
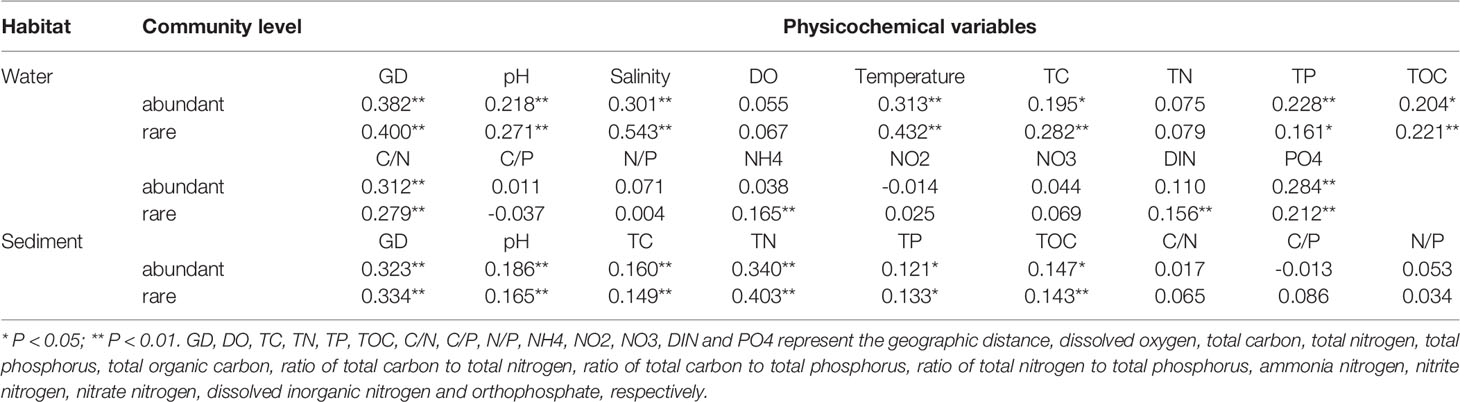
Table 2 The Mantel test results of physicochemical variables against the microbial community structure of abundant and rare taxa of water or sediment habitat. .
The knowledge on abundant and rare taxa has accumulated rapidly in recent years, but little is known about their community diversity and assembly mechanism of multiple habitats in a given ecosystem. In this study, we emphasized the importance of rare taxa to the microbial diversity in aquaculture cultural ponds, and provided the evidences for exploring differences in microbial diversity and assembly mechanism between abundant and rare communities of multiple habitats in aquatic ecosystems.
Typically, the distribution of microbial abundance in environments was skewed. For instance, Mo et al. (2018) reported that only 0.44% of bacterial OTUs (over 50% of average relative abundance) were considered to be abundant taxa, but ~80% OTUs (only 6%) were classified as rare taxa of water in subtropical bays. These results indicated that the rare taxa are served as huge repositories of microbial diversity in ecosystems, and abundant taxa accounted the majority of all species’ abundance, which was consistent with our results for water and shrimp intestine. However, this phenomenon was not universal, as we observed in sediment: the numbers of rare OTUs was also dozens of times more than abundant ones, with their relative abundance was more than twice as abundant ones. Therefore, the microbial diversity of abundant and rare taxa was markedly different.
Our study also revealed that the sources of abundant and rare communities of each habitat were markedly different in cultural ponds; and especially, the most important source for shrimp intestine rare community was sediment. Theoretically, the intestine microbiota of aquatic animals is directly influenced by surrounding microorganisms (Yan et al., 2016; Xiong et al., 2019; Xiong et al., 2020). Thus, our results can be largely due to sediment features and host lifestyles. On one hand, the sediment serves as the largest microbial repositories in the shrimp cultural ponds and has the potential to be an important microbial source for host intestines. On the other hand, the activities of L. vannamei (as a planktobenthos, mainly lives at the bottom of water) are more related to sediment, and it sometimes feed from sediment that ingestion of particulate matter into the host intestines. We also found that Vibrio OTUs were abundant taxa in intestine but rare taxa in water and sediment. Vibrio species is known as common opportunistic pathogens and which in aquatic animal intestines could be from surroundings (Vadstein et al., 2004); and when these species in surroundings was killed by disinfection, those can also spread into surroundings as “seeds” through excretion of hosts (Gustafson and Bowen, 1997), making the challenging to control the opportunistic pathogens (Pérez-Sánchez et al., 2018). Thus, we should establish more ecological management strategies to restrain the proliferation of opportunistic pathogens in aquaculture.
Despite with different diversity, similar biogeographic patterns (obvious DDRs for abundant and rare communities, and stronger DDRs of rare than of abundant) between abundant and rare communities of each habitat were observed in the shrimp cultural ponds. Extensive studies also found that the similar biogeographic patterns between abundant and rare communities of water and soil (Logares et al., 2013; Liu et al., 2015; Mo et al., 2018; Chen et al., 2019; Jiao and Lu, 2019a). In 2006, Pedrós-Alió hypothesized that, due to infinite potential for dispersal and low loss rate, rare taxa have a worldwide distribution (Pedrós-Alió, 2006). However, biogeographic patterns for “rare biosphere” clearly refuted the applicability of “everything is everywhere” (Baas-Becking, 1934) in most cases. Thus, so barriers for dispersal exist in rare taxa, and conversely, abundant taxa tend toward more cosmopolitan distributions. Indeed, some previous (Liu et al., 2015; Zhang et al., 2018) and our studies found that a higher proportion of abundant OTUs were distributed in more samples, resulting that the abundant taxa might have a widespread or ubiquitous distribution. One possible reason is that abundant taxa grow on a wider spectrum of resources compared with the rare taxa. Thus, the abundant taxa can reach the high population levels in a wider range of environments (Hambright et al., 2015).
Furthermore, we found that stochastic processes were dominant in both abundant and rare communities for all three habitats in the cultural ponds. One reason can explain our phenomenon is that the regular management of artificial systems allows a dynamic community turnover that lowers the homogeneity of the system over time (Li et al., 2020). The importance of stochastic process on the microbial assembly was also reflected in significantly strong distance-decay patterns. Accordingly, due to dispersal limitation, community similarity was predicted to decrease along spatial gradients (Hubbell, 2001). Our results supported one study reporting that the important role of stochastic factors in shaping both the abundant and rare microeukaryotic community assemblies of water in a lotic river (Chen et al., 2019). By contrast, stochastic and deterministic processes dominated fungal abundant and rare communities of soil in fields, respectively (Jiao and Lu, 2019b), but the trend is the opposite in water of bay (Mo et al., 2018). Such differences were likely due to the discrepancies of habitats and geography (Shi et al., 2018). We also found that the contribution of dispersal limitation to abundant was slightly higher than that to rare community of all habitats, but that of selection to rare community was much higher. Some studies also found that the rare communities in a variety of environments were dominated by the deterministic processes (Liu et al., 2015; Wu et al., 2017; Liang et al., 2020), suggesting that rare taxa may be subjected to stronger environmental selection and therefore less widely distributed than abundant taxa.
To gain an advanced understanding of microbial ecology, it is essential to reveal the deterministic factors shaping the community assembly (Zhou and Ning, 2017). Previous studies have shown that physicochemical factors were important on influencing the water and sediment microbiota in aquaculture ponds (Li et al., 2017; Huang et al., 2018; Wei et al., 2020; Hou et al., 2021), but little is known about their influences on the abundant and rare communities. We found that the physicochemical factors significantly affected abundant and rare communities, and especially, salinity and temperature/TN were the best predictors for abundant and rare communities of water/sediment. Our previous (Hou et al., 2017 and Hou et al., 2021) and other studies (Huang et al., 2018) also found that salinity, temperature and TN were important factors shaping the water and sediment community structures in shrimp cultural ponds. Indeed, salinity and temperature are key factors that influence the microbiota (Székely et al., 2013; Sunagawa et al., 2015). Salinity can affect production of microorganisms by enhancing nutrient availability (Pinckney et al., 1995). While based on metabolic theory of ecology, temperature can affect species diversity, metabolic activity and population growth rates (Zhou et al., 2016). The shrimp culture ponds in our study are in an outdoor environment, where the salinity will change due to rainfall and evaporation, and temperature will be strongly affected by climate factors. These may be the reason why the salinity and temperature are important factors affecting the microbial community structure of shrimp culture ponds. Moreover, TN was very important for sediment microbiota, which might be related to the continuous accumulation of nitrogen caused by continuous high protein feed residues and fecal deposition in shrimp cultural ponds. Our results suggested that it may be possible to regulate physicochemical properties to manipulate the microbiota in aquaculture.
Our study proposed a paradigm portraying the microbial diversity and assembly mechanism of multiple habitats within a metacommunity framework in shrimp cultural ponds at the abundant and rare community levels (Figure 5). The rare taxa are served as huge repositories of microbial diversity, and abundant and rare microbial taxa had distinct community diversity, taxonomic composition and sources. Stochastic process dominated the community assemblies of both abundant and rare taxa in three habitats, and deterministic process contributed to the assembly of rare community was much higher than that of abundant community. Salinity and temperature were the best physicochemical predictors for abundant and rare community composition of water, while TN for that of sediment. These findings could promote our understanding of microbial diversity from the perspective of abundant and rare taxa, and facilitate the microbiota management for healthy aquaculture.
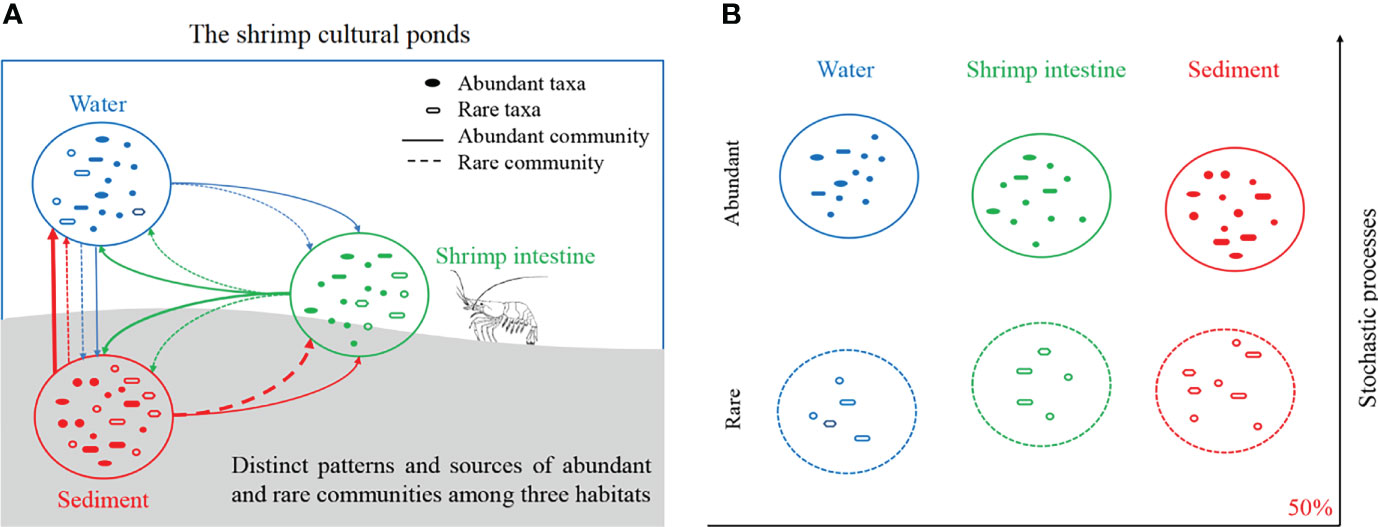
Figure 5 Illustration of community diversity and assembly. (A) Patterns and sources of abundant and rare communities of water/shrimp intestine/sediment habitats in the cultural ponds. (B) Similarities and differences of microbial assembly mechanisms between the abundant and rare communities of each habitat.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA545396.
The original concept was conceived by ZH, JH, and DH; experimental strategies and sampling design were developed by DH, SW, ZH, and JH; sample collections, DNA extraction, DNA sequencing and data analyses were performed by DH, ZH, RZ, SZ, DW, ZH, and JH; the manuscript was written by DH, QY, ZH, and JH. All authors contributed to the article and approved the submitted version.
This work was supported by the Key-Area Research and Development Program of Guangdong Province (2021B0202040001 and 2020B0202010009); National Natural Science Foundation of China (31902392); Southern Marine Science and Engineering Guangdong Laboratory (Zhuhai) (SML2021SP203); and China Agriculture Research System of MOF and MARA.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.856126/full#supplementary-material
Supplementary Figure 1 | Taxonomic compositions of abundant and rare taxa were markedly different of water/shrimp intestine/sediment habitat in the cultural ponds.
Supplementary Figure 2 | The Venn analysis of microbial community compositions of water, shrimp intestine and sediment habitats in the cultural ponds.
Supplementary Figure 3 | The NMDS and ANOSIM evaluated the overall differences in microbial structure for water/shrimp intestine/sediment habitat in the cultural ponds based on the Bray-Curtis distance. The abundant or rare community of each habitat significantly differed between any two of the compared regional sites.
Supplementary Figure 4 | The SNCM estimated the relationship between the occurrence frequency of abundant or rare OTUs and their variations of water/shrimp intestine/sediment habitat in the cultural ponds. Parameter r2 represents the overall fit to this neutral model. Calculation of 95% confidence intervals around all fitting statistics was done by the bootstrapping with 1,000 bootstrap replicates.
Assefa A., Abunna F. (2018). Maintenance of Fish Health in Aquaculture: Review of Epidemiological Approaches for Prevention and Control of Infectious Disease of Fish. Vet. Med. Int. 26, 5432497. doi: 10.1155/2018/5432497
Baas-Becking L. G. M. (1934). Geobiologie of Inleiding Tot De Milieukunde (The Hague: The Netherlands: WP Van Stockum & Zoon).
Bolnick D. I., Snowberg L. K., Hirsch P. E., Lauber C. L., Knight R., Caporaso J. G., et al. (2014). Individuals’ Diet Diversity Influences Gut Microbial Diversity in Two Freshwater Fish (Threespine Stickleback and Eurasian Perch). Ecol. Lett. 17, 979–987. doi: 10.1111/ele.12301
Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., et al. (2010). QIIME Allows Analysis of High-Throughput Community Sequencing Data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Chen W., Pan Y., Yu L., Yang J., Zhang W. (2017). Patterns and Processes in Marine Microeukaryotic Community Biogeography From Xiamen Coastal Waters and Intertidal Sediments, Southeast China. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.01912
Chen W., Ren K., Isabwe A., Chen H., Liu M., Yang J. (2019). Stochastic Processes Shape Microeukaryotic Community Assembly in a Subtropical River Across Wet and Dry Seasons. Microbiome 7, 138. doi: 10.1186/s40168-019-0749-8
Cottrell M. T., Kirchman D. L. (2003). Contribution of Major Bacterial Groups to Bacterial Biomass Production (Thymidine and Leucine Incorporation) in the Delaware Estuary. Limnol. Oceanogr. 48, 168–178. doi: 10.4319/lo.2003.48.1.0168
Del’Duca A., Cesar D. E., Abreu P. C. (2015). Bacterial Community of Pond’s Water, Sediment and in the Guts of Tilapia (Oreochromis Niloticus) Juveniles Characterized by Fluorescent In Situ hybridization Technique. Aquac. Res. 46, 707–715. doi: 10.1111/are.12218
Dini-Andreote F., Stegen J. C., van Elsas J. D., Salles J. F. (2015). Disentangling Mechanisms That Mediate the Balance Between Stochastic and Deterministic Processes in Microbial Succession. Proc. Natl. Acad. Sci. U. S. A. 112, E1326–E1332. doi: 10.1073/pnas.1414261112
Edgar R. C., Haas B. J., Clemente J. C., Quince C., Knight R. (2011). UCHIME Improves Sensitivity and Speed of Chimera Detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Escalas A., Bouvier T., Mouchet M. A., Leprieur F., Bouvier C., Troussellier M., et al. (2013). A Unifying Quantitative Framework for Exploring the Multiple Facets of Microbial Biodiversity Across Diverse Scales. Environ. Microbiol. 15, 2642–2657. doi: 10.1111/1462-2920.12156
Escalas A., Troussellier M., Yuan T., Bouvier T., Bouvier C., Mouchet M. A., et al. (2015). Functional Diversity and Redundancy Across Fish Gut, Sediment and Water Bacterial Communities. Environ. Microbiol. 19, 3268–3282. doi: 10.1111/1462-2920.13822
FAO (2018). Fishery and Aquaculture Statistics (Rome: Food and Agriculture Organization of the United Nations).
Gustafson R. H., Bowen R. E. (1997). Antibiotic Use in Animal Agriculture. J. Appl. Microbiol. 83, 531–541. doi: 10.1046/j.1365-2672.1997.00280.x
Hambright K. D., Beyer J. E., Easton J. D., Zamor R. M., Easton A. C., Hallidayschult T. C. (2015). The Niche of an Invasive Marine Microbe in a Subtropical Freshwater Impoundment. ISME J. 9, 256–264. doi: 10.1038/ismej.2014.103
Hou D., Huang Z., Zeng S., Liu J., Wei D., Deng X., et al. (2017). Environmental Factors Shape Water Microbial Community Structure and Function in Shrimp Cultural Enclosure Ecosystems. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.02359
Hou D., Huang Z., Zeng S., Liu J., Wei D., Deng X., et al. (2018a). Intestinal Bacterial Signatures of White Feces Syndrome in Shrimp. Appl. Microbiol. Biotechnol. 102, 3701–3709. doi: 10.1007/s00253-018-8855-2
Hou D., Huang Z., Zeng S., Liu J., Weng S., He J. (2018b). Comparative Analysis of the Bacterial Community Compositions of the Shrimp Intestine, Surrounding Water and Sediment. J. Appl. Microbiol. 125, 792–799. doi: 10.1111/jam.13919
Hou D., Zhou R., Zeng S., Wei D., Deng X., Xing C., et al. (2021). Stochastic Processes Shape the Bacterial Community Assembly in Shrimp Cultural Pond Sediments. Appl. Microbiol. Biotechnol. 105, 5013–5022. doi: 10.1007/s00253-021-11378-9
Huang Z., Hou D., Zhou R., Zeng S., Xing C., Wei D., et al. (2021). Environmental Water and Sediment Microbial Communities Shape Intestine Microbiota for Host Health: The Central Dogma in an Anthropogenic Aquaculture Ecosystem. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.772149
Huang F., Pan L., Song M., Tian C., Gao S. (2018). Microbiota Assemblages of Water, Sediment, and Intestine and Their Associations With Environmental Factors and Shrimp Physiological Health. Appl. Microbiol. Biotechnol. 102, 8585–8598. doi: 10.1007/s00253-018-9229-5
Huang Z., Zeng S., Xiong J., Hou D., Zhou R., Xing C., et al. (2020). Microecological Koch’s Postulates Reveal That Intestinal Microbiota Dysbiosis Contributes to Shrimp White Feces Syndrome. Microbiome 8, 32. doi: 10.1186/s40168-020-00802-3
Hubbell S. P. A. (2001). The Unified Neutral Theory of Biodiversity and Biogeography (Princeton: Princeton University Press).
Jia X., Dini-Andreote F., Salles J. F. (2018). Community Assembly Processes of the Microbial Rare Biosphere. Trends. Microbiol. 26, 738–747. doi: 10.1016/j.tim.2018.02.011
Jiao S., Lu Y. (2019a). Soil pH and Temperature Regulate Assembly Processes of Abundant and Rare Bacterial Communities in Agricultural Ecosystems. Environ. Microbiol. 22, 1052–1065. doi: 10.1111/1462-2920.14815
Jiao S., Lu Y. (2019b). Abundant Fungi Adapt to Broader Environmental Gradients Than Rare Fungi in Agricultural Fields. Glob. Change. Biol. 26, 4506–4520. doi: 10.1111/gcb.15130
Jousset A., Bienhold C., Chatzinotas A., Gallien L., Gobet A., Kurm V., et al. (2017). Where Less May Be More: How the Rare Biosphere Pulls Ecosystems Strings. ISME J. 11, 853–862. doi: 10.1038/ismej.2016.174
Kembel S. W. (2009). Disentangling Niche and Neutral Influences on Community Assembly: Assessing the Performance of Community Phylogenetic Structure Tests. Ecol. Let. 12, 949–960. doi: 10.1111/j.1461-0248.2009.01354.x
Liang Y., Xiao X., Nuccio E. E., Yuan M., Zhang N., Xue K., et al. (2020). Differentiation Strategies of Soil Rare and Abundant Microbial Taxa in Response to Changing Climatic Regimes. Environ. Microbiol. 22, 1327–1340. doi: 10.1111/1462-2920.14945
Li T., Li H., Gatesoupe F. J., She R., Lin Q., Yan X., et al. (2017). Bacterial Signatures of “Red-Operculum” Disease in the Gut of Crucian Carp (Carassius Auratus). Microb. Ecol. 74, 510–521. doi: 10.1007/s00248-017-0967-1
Liu L., Yang J., Yu Z., Wilkinson D. M. (2015). The Biogeography of Abundant and Rare Bacterioplankton in the Lakes and Reservoirs of China. ISME J. 9, 2068–2077. doi: 10.1038/ismej.2015.29
Li X., Zhu H., Geisen S., Bellard C., Hu F., Li H., et al. (2020). Agriculture Erases Climate Constraints on Soil Nematode Communities Across Large Spatial Scales. Glob. Change. Biol. 26, 919–930. doi: 10.1111/gcb.14821
Logares R., Audic S., Bass D., Bittner L., Boutte C., Christen R., et al. (2014). Patterns of Rare and Abundant Marine Microbial Eukaryotes. Curr. Biol. 24, 813–821. doi: 10.1016/j.cub.2014.02.050
Logares R., Lindström E. S., Langenheder S., Logue J. B., Paterson H., Laybourn-Parry J., et al. (2013). Biogeography of Bacterial Communities Exposed to Progressive Long-Term Environmental Change. ISME J. 7, 937–948. doi: 10.1038/ismej.2012.168
Magoč T., Salzberg S. L. (2011). FLASH: Fast Length Adjustment of Short Reads to Improve Genome Assemblies. Bioinformatics 27, 2957–2296. doi: 10.1093/bioinformatics/btr507
Moriarty D. J. W. (1997). The Role of Microorganisms in Aquaculture Ponds. Aquaculture 151, 333–349. doi: 10.1016/S0044-8486(96)01487-1
Mo Y., Zhang W., Yang J., Lin Y., Yu Z., Lin S. (2018). Biogeographic Patterns of Abundant and Rare Bacterioplankton in Three Subtropical Bays Resulting From Selective and Neutral Processes. ISME J. 12, 2198–2210. doi: 10.1038/s41396-018-0153-6
Oksanen J., Blanchet F. G., Kindt R., Legendre P., Minchin P. R., O’Hara R. B., et al. (2015). Vegan: Community Ecology Package (Oulu: R package version 2.0-10).
Pedrós-Alió C. (2006). Marine Microbial Diversity: Can It Be Determined? Trends. Microbiol. 14, 257–263. doi: 10.1016/j.tim.2006.04.007
Pedrós-Alió C. (2012). The Rare Bacterial Biosphere. Annu. Rev. Mar. Sci. 4, 449–466. doi: 10.1146/annurev-marine-120710-100948
Pérez-Sánchez T., Mora-Sánchez B., Balcázar J. L. (2018). Biological Approaches for Disease Control in Aquaculture: advantages, limitations and challenges. Trends Microbiol 26, 896–903. doi: 10.1016/j.tim.2018.05.002
Pinckney J., Paerl H. W., Bebout B. M. (1995). Salinity Control of Benthic Microbial Mat Community Production in a Bahamian Hypersaline Lagoon. J. Exp. Mar. Bio. Ecol. 187, 223–237. doi: 10.1016/0022-0981(94)00185-G
R Core Team (2017). R: A Language and Environment for Statistical Computing (Vienna: R Foundation for Statistical Computing). Available at: http://www.R-project.org.
Schryver P. D., Vadstein O. (2014). Ecological Theory as a Foundation to Control Pathogenic Invasion in Aquaculture. ISME J. 8, 2360–2368. doi: 10.1038/ismej.2014.84
Shenhav L., Thompson M., Joseph T. A., Briscoe L., Furman O., Bogumil D., et al. (2019). FEAST: Fast Expectation-Maximization for Microbial Source Tracking. Nat. Methods 16, 627–632. doi: 10.1038/s41592-019-0431-x
Shi Y., Li Y., Xiang X., Sun R., Yang T., He D., et al. (2018). Spatial Scale Affects the Relative Role of Stochasticity Versus Determinism in Soil Bacterial Communities in Wheat Fields Across the North China Plain. Microbiome 6, 27. doi: 10.1186/s40168-018-0409-4
Sloan W. T., Lunn M., Woodcock S., Head I. M., Nee S., Curtis T. P. (2006). Quantifying the Roles of Immigration and Chance in Shaping Prokaryote Community Structure. Environ. Microbiol. 8, 732–740. doi: 10.1111/j.1462-2920.2005.00956.x
Sogin M. L., Morrison H. G., Huber J. A., Welch D. M., Huse S. M., Neal P. R., et al. (2006). Microbial Diversity in the Deep Sea and the Underexplored ‘‘Rare Biosphere’’. Proc. Natl. Acad. Sci. U. S. A. 103, 12115–12120. doi: 10.1073/pnas.0605127103
Stegen J. C., Lin X., Fredrickson J. K., Chen X., Kennedy D. W., Murray C. J., et al. (2013). Quantifying Community Assembly Processes and Identifying Features That Impose Them. ISME J. 7, 2069–2079. doi: 10.1038/ismej.2013.93
Stegen J. C., Lin X., Fredrickson J. K., Konopka A. E. (2015). Estimating and Mapping Ecological Processes Influencing Microbial Community Assembly. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.00370
Sunagawa S., Coelho L. P., Chaffron S., Kultima J. R., Labadie K., Salazar G., et al. (2015). Structure and Function of the Global Ocean Microbiome. Science 348, 1261359. doi: 10.1126/science.1261359
Székely A. J., Berga M., Langenheder S. (2013). Mechanisms Determining the Fate of Dispersed Bacterial Communities in New Environments. ISME J. 7, 61–71. doi: 10.1038/ismej.2012.80
Vadstein O., Mo T. A., Bergh Ø. (2004). “Microbial Interactions, Prophylaxis and Diseases,” in Culture of Cold-Water Marine Fishes. Eds. Moksness E., Kjørsvik E., Olsen Y. (Bath, UK: Blackwell Publishing Ltd), 28–72.
Webb C. O., Ackerly D. D., McPeek M. A., Donoghue M. J. (2002). Phylogenies and Community Ecology. Annu. Rev. Ecol. Syst. 33, 475–505. doi: 10.1146/annurev.ecolsys.33.010802.150448
Wei D., Zeng S., Hou D., Zhou R., Xing C., Deng X., et al. (2020). Community Diversity and Abundance of Ammonia-Oxidizing Archaea and Bacteria in Shrimp Pond Sediment at Different Culture Stages. J. Appl. Microbiol. 130, 1442–1455. doi: 10.1111/jam.14846
Wu W., Logares R., Huang B., Hsieh C. H. (2017). Abundant and Rare Picoeukaryotic Sub-Communities Present Contrasting Patterns in the Epipelagic Waters of Marginal Seas in the Northwestern Pacific Ocean. Environ. Microbiol. 19, 287–300. doi: 10.1111/1462-2920.13606
Xiong J., Dai W., Qiu Q., Zhu J., Yang W., Li C. (2018). Response of Host-Bacterial Colonization in Shrimp to Developmental Stage, Environment and Disease. Mol. Ecol. 27, 3686–3699. doi: 10.1111/mec.14822
Xiong J., Xuan L., Yu W., Zhu J., Qiu Q., Chen J. (2019). Spatiotemporal Successions of Shrimp Gut Microbial Colonization: High Consistency Despite Distinct Species Pool. Environ. Microbiol. 4, 1383–1394. doi: 10.1111/1462-2920.14578
Xiong J., Zhu J., Dai W., Dong C., Qiu Q., Li C. (2017). Integrating Gut Microbiota Immaturity and Disease-Discriminatory Taxa to Diagnose the Initiation and Severity of Shrimp Disease. Environ. Microbiol. 19, 1490–1501. doi: 10.1111/1462-2920.13701
Xiong C., Zhu Y., Wang J., Singh B., Han L., Shen J., et al. (2020). Host Selection Shapes Crop Microbiome Assembly and Network Complexity. N. Phytol. 229, 1091–1104. doi: 10.1111/nph.16890
Yan Q., Li J., Yu Y., Wang J., He Z., van Nostrand J. D., et al. (2016). Environmental Filtering Decreases With Fish Development for the Assembly of Gut Microbiota. Environ. Microbiol. 18, 4739–4754. doi: 10.1111/1462-2920.13365
Zhang H., Hou F., Xie W., Wang K., Zhou X., Zhang D., et al. (2019). Interaction and Assembly Processes of Abundant and Rare Microbial Communities During a Diatom Bloom Process. Environ. Microbiol. 22, 1707–1719. doi: 10.1111/1462-2920.14820
Zhang X., Li X., Lu J., Qiu Q., Chen J., Xiong J. (2020). Quantifying the Importance of External and Internal Sources to the Gut Microbiota in Juvenile and Adult Shrimp. Aquaculture 531, 735910. doi: 10.1016/j.aquaculture.2020.735910
Zhang W., Pan Y., Yang J., Chen H., Holohan B., Vaudrey J., et al. (2018). The Diversity and Biogeography of Abundant and Rare Intertidal Marine Microeukaryotes Explained by Environment and Dispersal Limitation. Environ. Microbiol. 20, 462–476. doi: 10.1111/1462-2920.13916
Zhao D., Xu H., Zeng J., Cao X., Huang R., Shen F., et al. (2017). Community Composition and Assembly Processes of the Freeliving and Particle-Attached Bacteria in Taihu Lake. FEMS Microbiol. Ecol. 93, fix062. doi: 10.1093/femsec/fix062
Zhou J., Deng Y., Shen L., Wen C., Yan Q., Ning D., et al. (2016). Temperature Mediates Continental-Scale Diversity of Microbes in Forest Soils. Nat. Commun. 7, 12083. doi: 10.1038/ncomms12083
Zhou Q., Li K., Bo L. (2009). Role and Functions of Beneficial Microorganisms in Sustainable Aquaculture. Bioresour. Technol. 100, 3780–3789. doi: 10.1016/j.biortech.2008.12.037
Keywords: microbial community, rare taxa, abundant taxa, multiple habitats, aquaculture pond ecosystem, shrimp
Citation: Hou D, Zhou R, Wei D, Zeng S, Weng S, Yan Q, He J and Huang Z (2022) Abundant and Rare Microbial Communities Respectively Contribute to an Aquaculture Pond Ecosystem. Front. Mar. Sci. 9:856126. doi: 10.3389/fmars.2022.856126
Received: 16 January 2022; Accepted: 11 April 2022;
Published: 04 May 2022.
Edited by:
Yanjiao Zhang, Ocean University of China, ChinaReviewed by:
Xiaoli Zhang, Chinese Academy of Sciences (CAS), ChinaCopyright © 2022 Hou, Zhou, Wei, Zeng, Weng, Yan, He and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianguo He, bHNzaGpnQG1haWwuc3lzdS5lZHUuY24=; Zhijian Huang, bHNzaHpoakBtYWlsLnN5c3UuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.