- 1Institute of Marine Science and Technology, Shandong University, Qingdao, China
- 2Joint Laboratory for Ocean Research and Education at Dalhousie University, Shandong University and Xiamen University, Qingdao, China
- 3Southern Marine Science and Engineering Guangdong Laboratory, Zhuhai, China
- 4Key Laboratory of Tropical Marine Bio-resources and Ecology, South China Sea Institute of Oceanology, Chinese Academy of Sciences, Guangzhou, China
Cold surges result in a rapid drop in air temperature and freezing of seawater, which was likely to impact bacterial communities. We examined the differences in bacteria abundance and bacterial community composition in the sea ice and seawater during a cold surge along Aoshan Bay, southern Yellow Sea in January 2021. Results showed that the differences in bacteria abundance between sea ice and seawater likely resulted from the physical impact of ice formation. The parent water played a key role in bacterial community composition in the early phase of ice formation, in which bacterial community compositions at class level were similar, but the relative abundances were different between sea ice and seawater. The Gammaproteobacteria dominated in sea ice, and the relative abundances of Verrucomicrobiae were also significantly higher, possibly due to the high concentration of algal-derived DOM in coastal areas. The predicted functional profiles suggested the lower abundance of functional genes related to ATP-binding cassette transporters in sea ice than in seawater, which might be due to the bacteria not requiring varieties of functional genes of ATP-binding cassette transporters in restricted sea ice brine.
Introduction
Human activities have caused a dramatic increase in atmospheric carbon dioxide (CO2) concentration. Rising levels of atmospheric CO2, in turn, have led to global temperature rise and climate change (Alexander et al., 2006; Hansen et al., 2006). Previous studies have indicated that the frequent cold surges in Eurasia are closely associated with the decreases in autumn-winter Arctic Sea ice resulting from rising temperature (Takaya and Nakamura, 2005; Petoukhov and Semenov, 2010; Park et al., 2011; Kug et al., 2015; Johnson et al., 2018). The outbreak of cold surges will be accompanied by cooling, strong winds, rain, snow, and other extreme weather phenomena, causing frost, rime, and other disasters (Liu et al., 2012). Temperature is one of the most important factors affecting microbial microorganism growth, and the suitable temperature is within a certain range for bacteria (Ewert and Deming, 2014). The occurrence of cold surges is coupled with a sharp drop in temperature, which may have an inevitable impact on microorganisms, due to the inability to adapt quickly to sustain their regular metabolic functions (Shivaji and Prakash, 2010; Subramanian et al., 2011). Low temperature also creates favorable conditions for the formation of sea ice. In high-latitude regions, it was found that both bacteria and algae experienced a strong metabolic inhibition during the ice formation (Grossmann and Gleitz, 1993), and the ice formation and growth reshaped bacterial community structure in drift ice (Eronen-Rasimus et al., 2015). However, few studies were conducted in mid-latitude regions (Xu et al., 2012). During the formation and growth of sea ice, organic and inorganic components dissolved in seawater are concentrated into the brine (Duprat et al., 2020). Internal channels with highly saline brine establish and create distinct habitats for microbial communities, encompassing members such as algae, bacteria, and viruses from seawater (Lund-Hansen et al., 2020). The activity of microbes in sea ice is greatly affected by environmental variables such as temperature, brine salinity, nutrients, and organic matter, which are different from seawater (Torstensson et al., 2018; Piontek et al., 2020). As the most abundant cellular lives in the ocean, bacteria play an essential role in the marine microbial loop (Azam et al., 1983). The transformation of various forms of carbon by bacterial activities is an important regulator of global carbon fluxes in marine environments and is of profound importance for marine ecosystems (Azam et al., 1983; Jiao et al., 2010, 2014; Zhao et al., 2019). Bacteria are carried from seawater into the formed sea ice matrix, and succession and development of bacterial communities were found along with ice-type changes (Eronen-Rasimus et al., 2015). Compared to the bacteria in high-latitude regions which may have adapted to the low temperature and the formation of sea ice, the bacterial community in mid-latitude regions where sea ice barely happened may have different strategies to the formation of sea ice, especially when the ice formation happened in a very short period.
In the context of global climate change, how bacterial communities vary from seawater to sea ice during the abrupt temperature drop and icing caused by cold surges in mid-latitude regions, where sea ice formation was seldom observed in the past, is poorly known. In this study, we examined the differences in bacterial community between the sea ice and seawater directly after the cold surge in January 2021 to understand the impact of ice formation on bacteria abundance (BA) and community composition.
Materials and Methods
Site Description and Sample Collection
Aoshan Bay is a semi-enclosed coastal inlet located in southern Yellow Sea, China. Given its geographical location and climatic conditions, the minimum air temperature in winter around the bay is typically above −5°C (Guo et al., 2014), and the sea fluidity and the sunlight radiation during the day merely meet the conditions for ice formation. However, a cold surge occurred in East Asia on January 6th, 2021 and lasted for 3 days, during which the minimum air temperature in this bay reached −14°C and massive sea ice appeared on the shore and lasted for 7 days. On January 11th, the ice and seawater samples around Aoshan Bay were collected to investigate the change of bacterial community. Samples were taken from three sites, among which two sites (S1 and S2) were located in the inner bay and one (S3) in the outer bay (Figure 1). Ice thickness ranged from 30 to 50 mm, and the in situ temperature of ice was measured by inserting a needle temperature probe (TP101) into ice core, with a precision of ± 0.1°C. The sea ice was collected using a stainless-steel saw to cut chunks from the thin ice and placed into polypropylene bags. Air was gently removed from the bag using a vacuum pump (H1, Reelanx). In addition, under-ice seawater samples were directly collected using 1-L HDPE bottles.
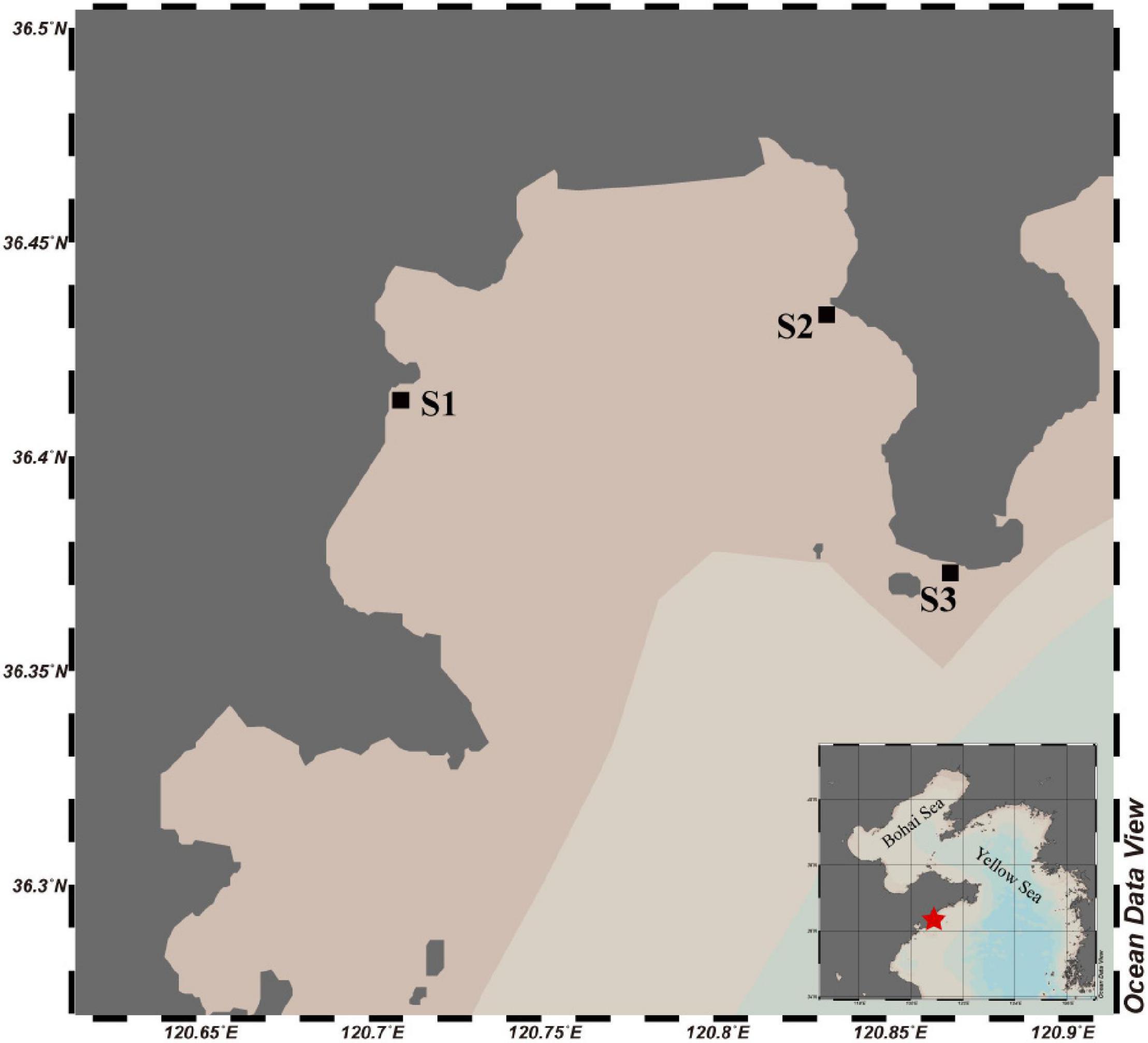
Figure 1. Map of Aoshan Bay with the squares showing the sampling stations. The water color indicates the relative depth of the water column.
Sample Processing and Analysis
After returning to laboratory, the sea ice was melted in the dark at room temperature and processed immediately after melting completely. The salinity of the melted sea ice and seawater was measured with a salinity meter (Orion star A212, Thermo Scientific, United States). Brine salinity (Sbrine) was calculated from ice temperature using the equation: Sbrine = 1,000/[1-(54.11/T)] (Cox and Weeks, 1983). After this, the melted ice and seawater samples were separately filtered through 0.2-μm-pore-size polycarbonate membrane filters (25 mm diameter; Millipore), and the filters were then stored in cryotubes at −80°C for later DNA extraction. At the same time, 30 mL of melted sea ice and seawater were separately filtered through 0.45-μm-pore-size membrane filters (Millipore), and the filtrates were stored at −20°C for nutrients analysis. Nutrient concentrations including nitrite, nitrate, ammonium, silicate and phosphate, were measured using a segmented flow analyzer (SEAL Analytical Ltd., AA3 HR Autoanalyzer) according to the classical colorimetric methods (Grasshoff et al., 1999). The detection limits for all channels were 0.1 μmol kg–1. 500 mL of melted sea ice or seawater was filtered on a pre-combusted glass fiber filter (25-mm, Whatman GF/F), and the filter and aliquots of 30 mL of filtration were stored at −20°C for chlorophyll a (Chl-a) and dissolved organic carbon (DOC) analysis, respectively. Chl-a was extracted overnight by immersing the GF/F filter into 90% acetone solution and was measured with a Cary Eclipse spectrofluorometer (Agilent Technologies, Santa Clara, CA) (Pinhassi et al., 2004). DOC was measured using the high-temperature combustion method with a TOC-L analyzer (Shimadzu, Japan) (Liu et al., 2020). Samples of seawater and melted sea ice were pre-filtrated through a 20-μm nylon mesh, then fixed with glutaraldehyde at 1% final concentration and stored at −80°C for BA analysis. After staining with SYBR Green I Nucleic Acid Gel Stain (Invitrogen) for 10 min in the dark, the bacteria cell counting was performed with a flow cytometer (Accuri C6, Becton-Dickinson, United States), according to side scattering light (SSC) and green fluorescence (FL1) (Li et al., 2018). Nutrients, Chl-a, DOC, and BA data of sea ice were normalized to brine concentration to correct for dilution during melting, and normalized salinity (Cbrine) were calculated following the equation of Cbrine = Cbulk (Sbrine/Sbulk), where Cbulk was the measured concentration in bulk sea ice; Sbrine was the brine salinity and Sbulk was the measured salinity of the melted ice (Cox and Weeks, 1983). The sample at site S3 for nitrate determination in sea ice was contaminated, so the data was discarded.
DNA Extraction Amplification and Sequencing
DNA was extracted from 0.2-μm-pore-size membrane filters (Millipore) using DNeasy PowerSoil Kit (Qiagen, Germany) (Zhao et al., 2021). The V3–V4 region of 16S rRNA genes was amplified using primer pairs 341F (5′-CCTAYGGGRBGCASCAG-3′) and 806R (5′-GGACTACNNGGGTATCTAAT-3′) (Liu et al., 2019), and the amplicons were sequenced using the Illumina MiSeq platform.
Bioinformatic and Statistical Analysis
Demultiplexing and quality filtering of raw sequences were conducted in QIIME2 (Bolyen et al., 2019), where amplicon sequencing variants (ASVs) were generated with DADA2 and classified against the SILVA 138 database (Robeson et al., 2020). Sequences assigned to archaea, chloroplast and mitochondrion were removed from the dataset. Sequencing data from each sample were normalized based on the smallest sample size to avoid potential bias caused by sequencing depth. All sequence data were rarefied to 66,141 sequences per sample for bacterial diversity analyses. To determine whether alpha diversity differs across samples, a variety of alpha diversity indices were calculated, including Shannon (diversity) and Pielou (evenness) index (Shannon, 1948). To estimate similarity among samples, hierarchical cluster analysis was also conducted based on a matrix of different ASVs and their abundance in each sample using Bray-Curtis dissimilarity calculated using vegan (R package) and a dendrogram inferred with the unweighted pair-group average algorithm (UPGMA) (Ortega-Retuerta et al., 2013). The BAs of sea ice and seawater were compared using a two-group White’s non-parametric t-test in STAMP (Parks et al., 2014). To compare the differences in the bacterial community composition between sea ice and seawater, Bray-Curtis distance-based principal coordinate analysis (PCoA) was conducted in the R package “vegan” (Oksanen et al., 2019). The correlation matrix among different environment parameters was obtained using the function “cor,” and the plot was obtained using the function “corrplot” of the “corrplot” package on R4.1.1. The predicted functional analysis was performed by Tax4Fun2 v. 1.1.5 software to explore the functional gene content in the bacterial community based on the 16S rRNA sequencing data (Wemheuer et al., 2020). The differences of functional genes or pathways between sea ice and seawater were compared using t-test in STAMP (Parks et al., 2014).
Results
Biochemical Environmental Parameters
The brine salinity of sea ice varied between 35.64 and 55.84, and the salinity in seawater ranged from 32.23 to 40.37 (Table 1). The temperature in seawater was around −1.1°C, while the temperature in sea ice varied from −3.2 to −2.0°C. Salinity between the seawater and sea ice (brine salinity) were similar at site S1, while the brine salinity of sea ice was higher than that of seawater at site S2 and S3. Nitrate concentrations in seawater were different among the three sampling sites, whereas the nitrite, ammonium, silicate, and phosphorus were more homogenous. The nutrient concentration of sea ice was higher, particularly at site S2. The DOC concentrations of seawater and sea ice ranged from 168.1 to 219.0 and from 211.7 to 552.2 μmol kg–1, respectively. Higher DOC concentrations in seawater and ice were found at sites S1 and S2, individually. Seawater at sites S2 and S3 showed a high concentration of Chl-a. The Chl-a concentration in sea ice was lower than that in seawater, and that at site S1 was the lowest among the three sites. The BA in seawater varied from 1.4 × 106 to 1.6 × 106 cells mL–1, and the maximum abundance was observed at site S2 (Figure 2). Brine-scaled sea-ice BA distribution was similar to bacteria in seawater and the abundance of bacteria at site S2 was the highest, and BA in seawater was lower than that observed in sea ice.
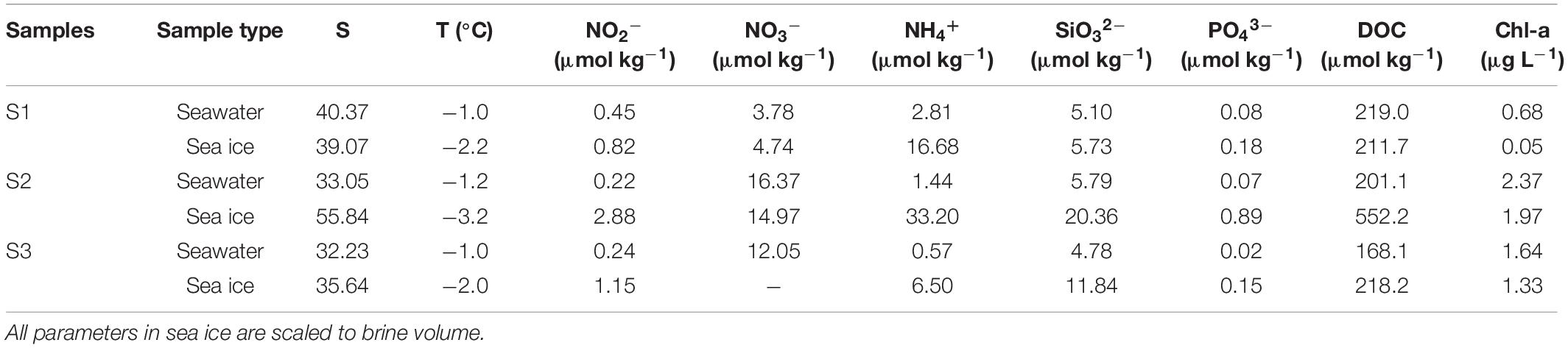
Table 1. Salinity (S), temperature (T), nutrients (μmol kg–1), dissolved organic carbon (DOC), and Chl-a concentrations in seawater and sea ice.
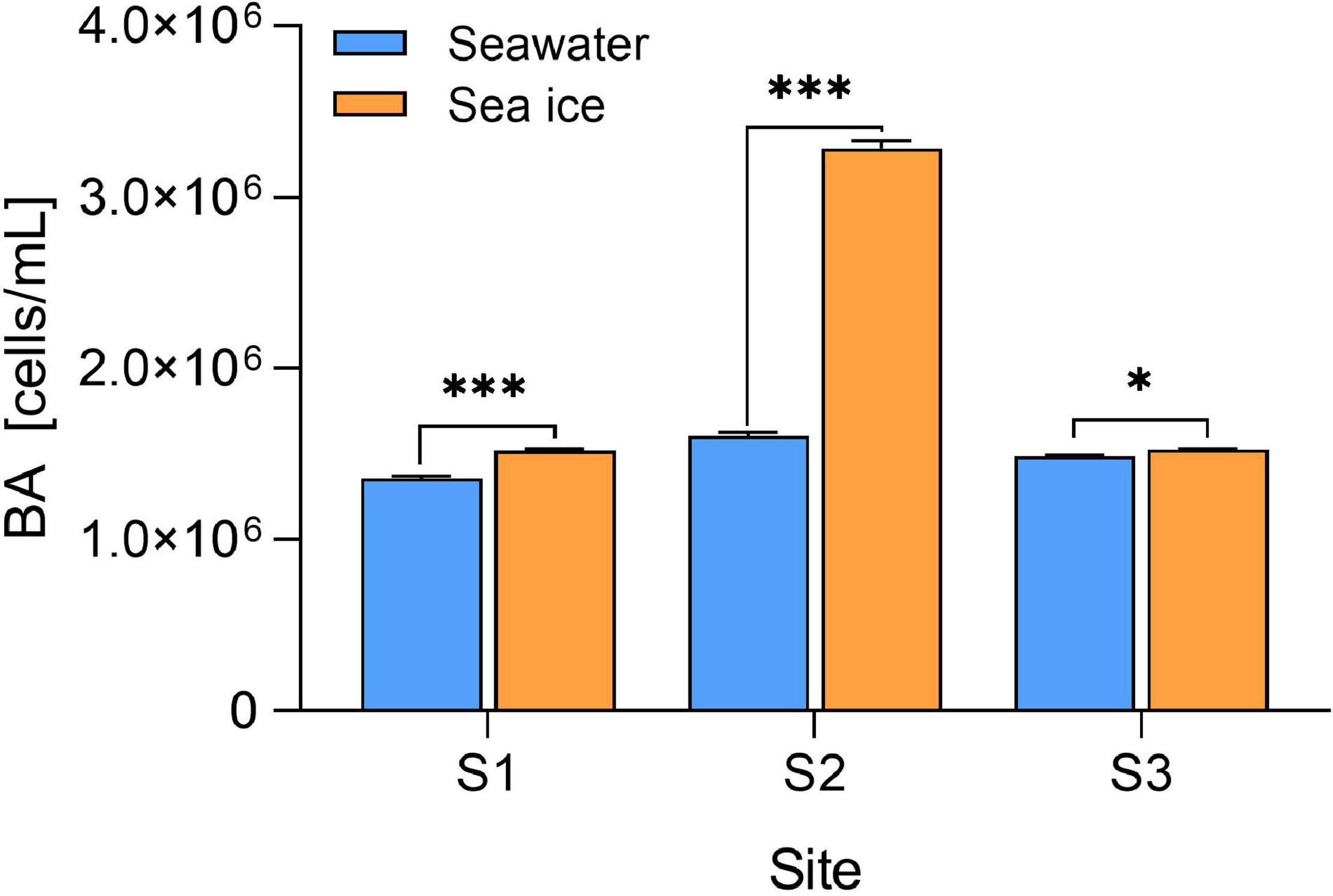
Figure 2. Bacteria abundance (BA) in seawater and sea ice. The BA in sea ice is scaled to brine volume. Error bars are SDs (n = 3). Level of significance: *p < 0.05; ***p < 0.001.
Bacterial Diversity and Community Composition
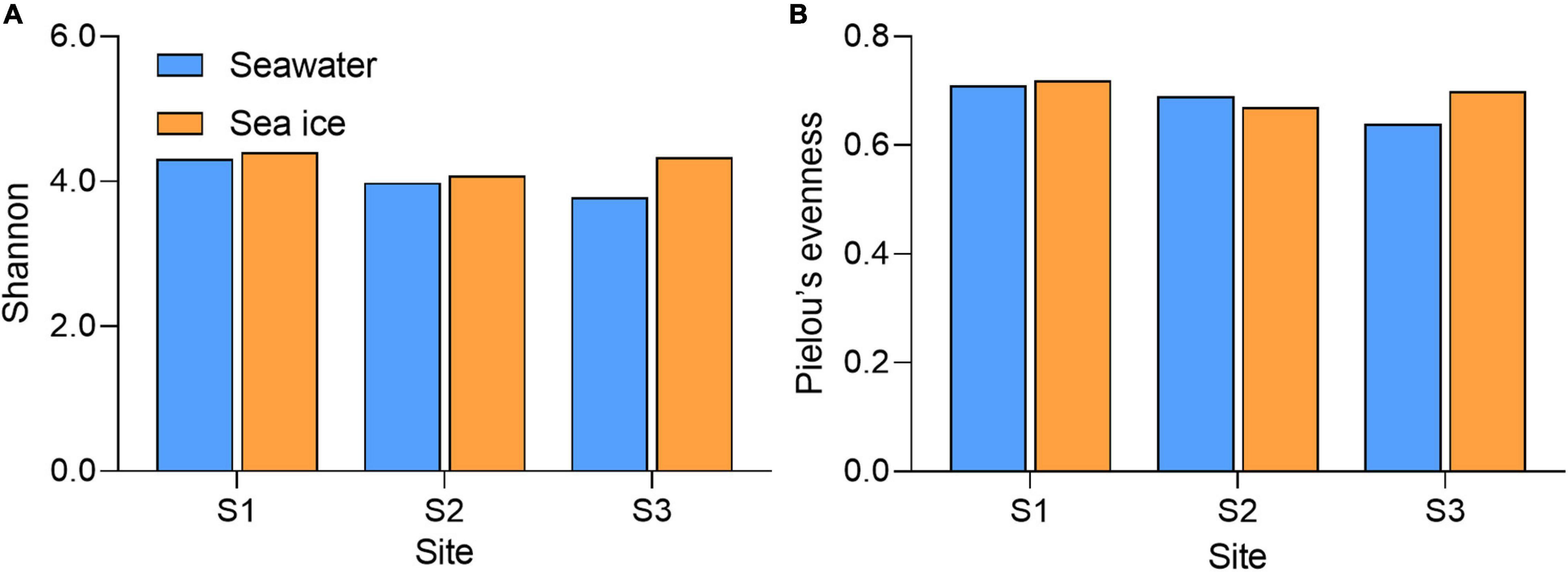
Community diversity based on Shannon diversity index and evenness of the community reflected by the Pielou’s index showed that diversity and evenness of the sea ice bacterial assemblages were similar to those of corresponding seawater assemblages (Figure 3). A total of nine bacterial phyla were identified in all samples. Proteobacteria were the dominant phylum existed in both sea ice and seawater samples, accounting for more than 70% of the whole bacterial community (Figure 4A). Phyla of Bacteroidota, Actinobacteriota, and Verrucomicrobiota were also detected in each sample, but their relative abundances were much lower. Actinobacteriota were more abundant in seawater, while Verrucomicrobiota were more abundant in sea ice. Alphaproteobacteria (31.7–60.0%), Gammaproteobacteria (20.6–48.9%), Bacteroidia (7.2–10.6%), Verrucomicrobiae (1.8–7.0%), Acidimicrobiia (1.1–7.3%), and Actinobacteria (0.7–2.4%) were determined as six major classes of bacteria among six samples (Figure 4B). The relative abundance of Alphaproteobacteria in seawater (45.0–60%) was higher than that in sea ice (31.7–38.9%), as well as Acidimicrobiia. While Gammaproteobacteria were more abundant in sea ice. Verrucomicrobiae followed the same pattern as Gammaproteobacteria. Bacteroidia and Actinobacteria showed variable abundance patterns in the samples. PCoA was used to evaluate the overall differences between sea ice and seawater bacterial communities based on Bray-Curtis distance (Figure 5A). PCo1 and PCo2 explained 52 and 33%, respectively, of the variance among the 6 samples, giving a total of 85% of the variance (Figure 5A). PCo1 grouped the samples into two major components: the samples of sea ice with negative PCo1 values and the samples of seawater (S2sw and S3sw) with positive PCo1 values, except for the sample of S1sw with a negative PCo1 value. On the PCo2 axis, S1ice and S1sw (with high positive PCo2 values) and S2ice and S3ice (with high negative PCo2 values) samples were mainly responsible for the variance. Seawater samples were clustered together but separated from ice samples, indicating high similarity among the bacterial communities in sea ice or seawater. In addition, UPGMA clustering dendrogram supported the result of the PCoA analysis by revealing the distinctiveness of bacterial communities in sea ice and seawater, except for the samples at site S1, where the seawater sample was clustered with sea ice (Figure 5B). The most significant differences (p < 0.05) between seawater and sea ice at class level were related to Alphaproteobacteria and Gammaproteobacteria. Alphaproteobacteria were significantly more abundant in seawater, whereas Gammaproteobacteria, Verrucomicrobiae, Bacilli, and Desulfobulbia were significantly more abundant in sea ice (Figure 5C).
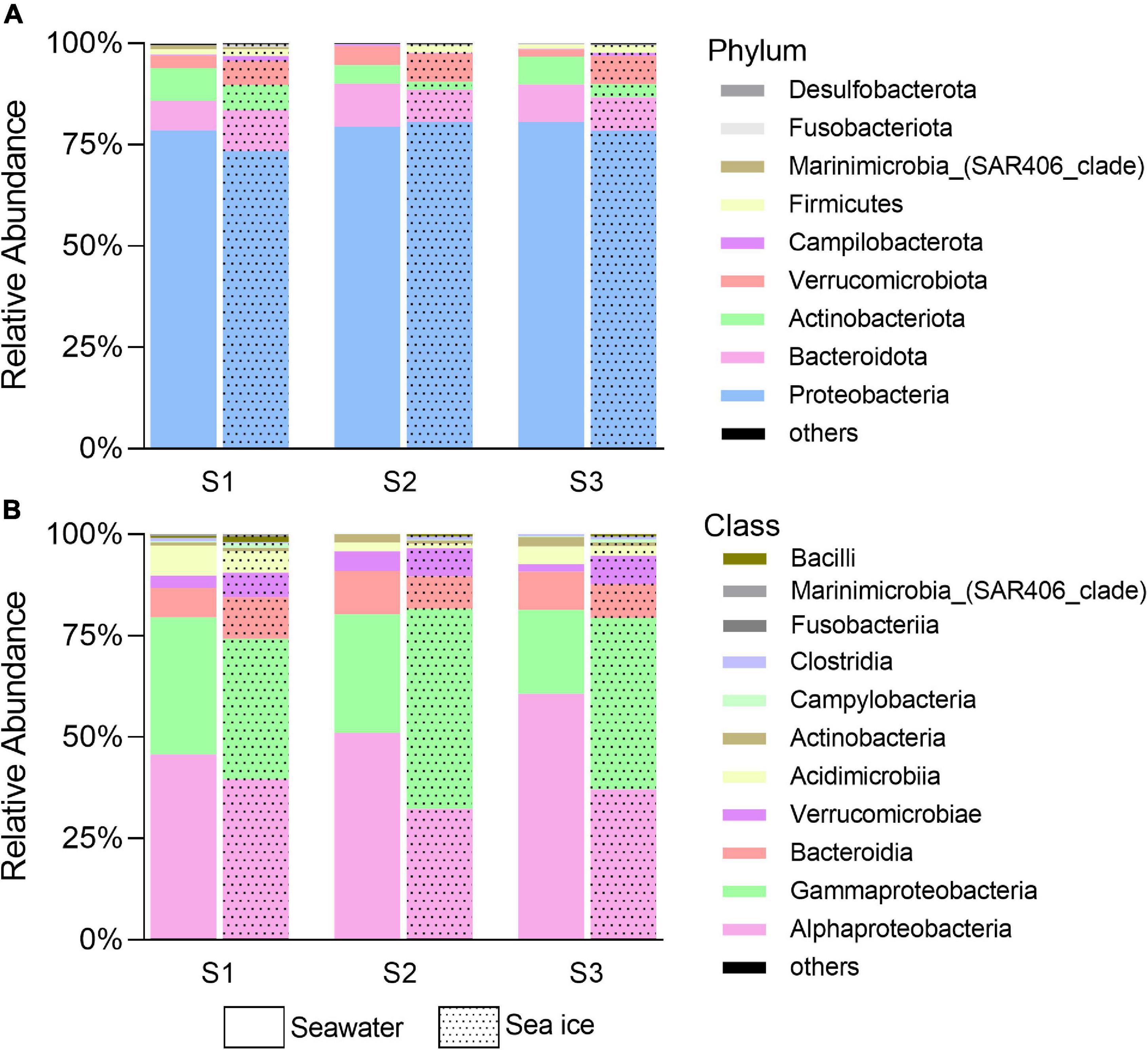
Figure 4. Bacterial community in phylum (A) and class (B) level in seawater and sea ice. Others denote the sum of rare bacteria arbitrarily defined as having a frequency of < 0.5% of total sequences.
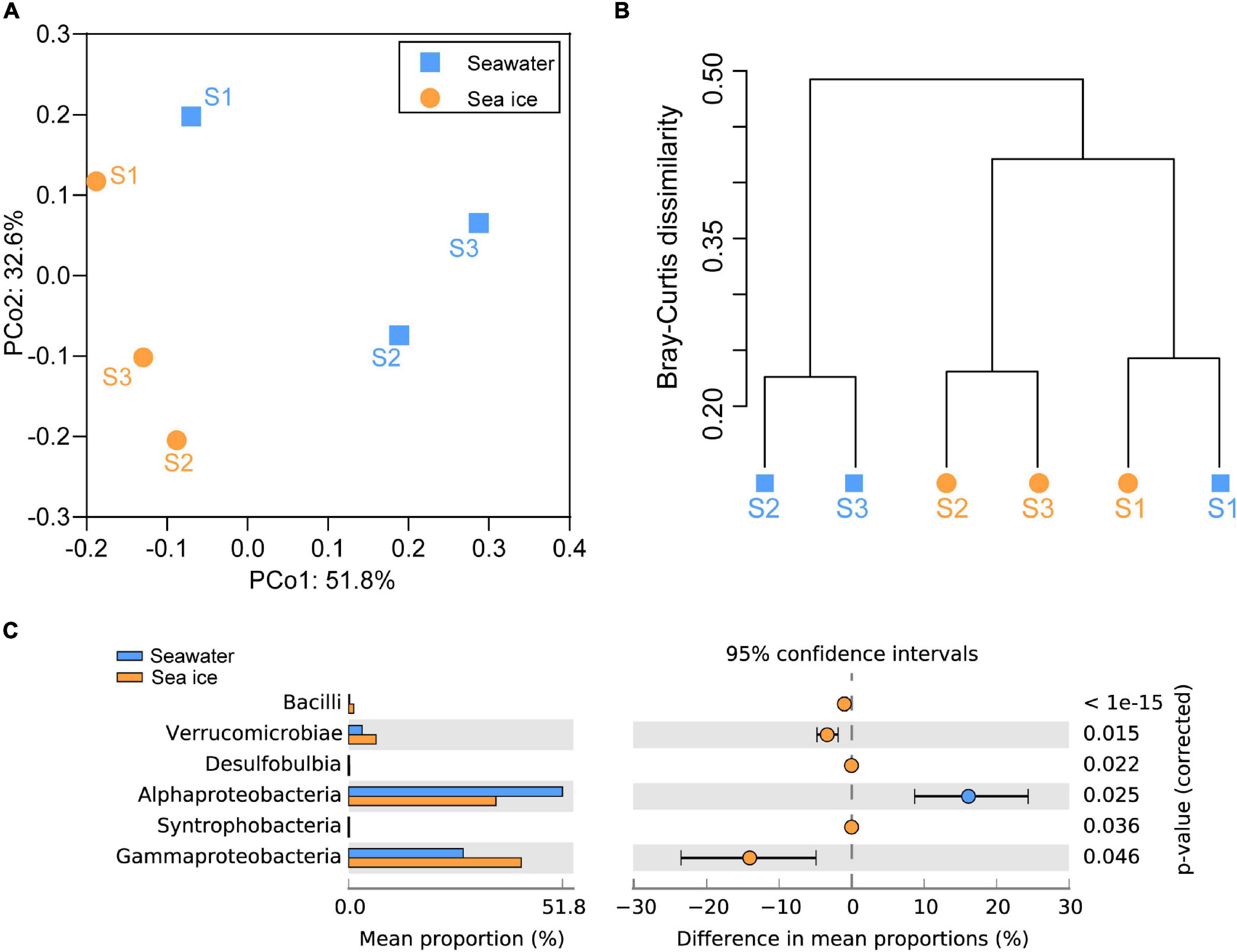
Figure 5. Relationships between the bacterial communities in seawater and sea ice. (A) Principal coordinates analysis (PCoA) plot of Bray–Curtis distances for bacterial communities in seawater and sea ice. (B) Dendrogram from the cluster analysis of the relationship between bacterial communities in seawater and sea ice samples. (C) Comparison (White’s non-parametric t-test in STAMP) of the bacterial classes between seawater and sea ice. Plots compare the bacteria in seawater (blue) to that in sea ice (orange). Only differences with a p-value less than 0.05 are shown.
The predicted functional analyses of 6 samples were carried out by Tax4Fun2 tools. A total of 46 KEGG subsystems presented at level 2 were found in samples, of which a total of 359 pathways were included in the subsystems. The pathways of the top 20 relative abundance were shown in Supplementary Figure 1, and the pathways with relatively high abundance obtained in different samples include: metabolic pathways, biosynthesis of secondary metabolites, biosynthesis of antibiotics, membrane transport, and ATP-binding cassette (ABC) transporters. Stamp analysis and t-test revealed the pathway of ABC transporters was significantly different between seawater and sea ice. Further analysis of the functional genes involved in ABC transporters indicated that genes encoding for oligopeptide transport system ATP-binding protein (oppF), oligopeptide transport system ATP-binding protein (malK), dipeptide transport system permease protein (dppB), raffinose/stachyose/melibiose transport system permease protein (msmF), and putative aldouronate transport system permease protein (lplB) were more abundant in seawater (Figure 6).
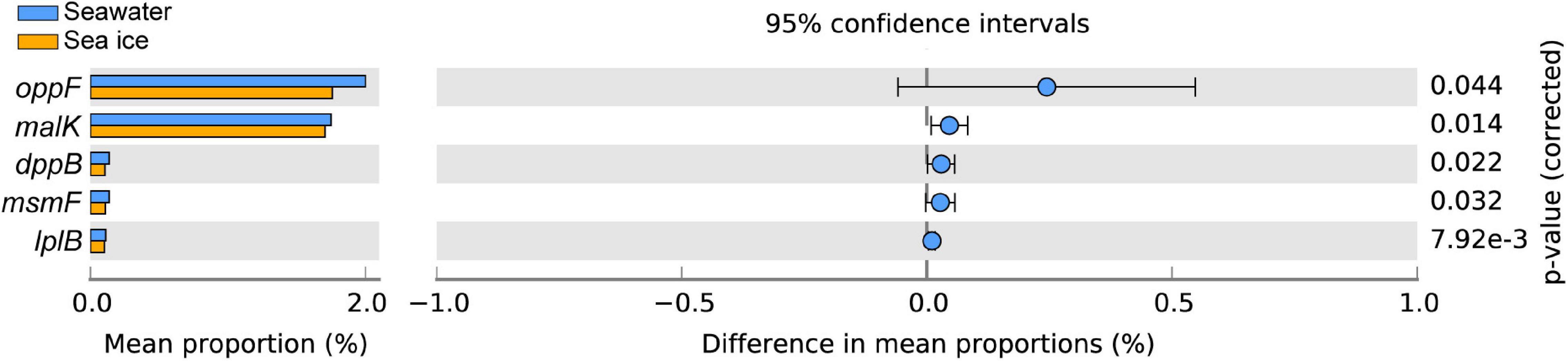
Figure 6. Statistical analysis of functional genes in pathway of ATP-binding cassette transporters in seawater and sea ice. Differences between samples are shown at 95% confidence intervals.
Discussion
Seawater and sea ice exhibited differences in terms of chemical properties and bacterial community. The decline of temperature reduced the fraction of the liquid remaining in sea ice with the result that the salinity and nutrients became increasingly concentrated in the brine, as well as dissolved organic components (Table 1). The high salinity of seawater at S1 might be due to the shallow water depth, where salt expulsion during sea ice formation caused an increase in salinity in the under-ice seawater column. The concentration of DOC in sea ice at site S2 was higher than that at the other two sites, which might be explained by the high concentration of Chl-a in sea ice (Eronen-Rasimus et al., 2014). Cold surges caused nearshore seawater to change from liquid to ice, where changes in physical conditions drove bacterial communities to adapt to the lifestyle in sea ice, and the community variations were consistent with previous studies in the Arctic (Grossmann and Gleitz, 1993; Eronen-Rasimus et al., 2015; Hatam et al., 2016). The BA in the sea ice was higher (p < 0.05) than that in the seawater at each site, which might be due to that the bacteria were concentrated in the brine inclusions during the ice formation (Eronen-Rasimus et al., 2014). In addition, the high BA of S2 may be related to the high DOC concentration in sea ice, suggesting that bacterial metabolism might be regulated by the DOC availability (Lu et al., 2015). Differences in the bacterial community composition were observed both at phylum and class levels between seawater and sea ice. The result suggested that bacterial community similarity within sea ice or seawater was higher than geographic distance. Pressure caused by ice formation was a stronger factor to shape the community compared to geographical distance (Eronen-Rasimus et al., 2015). However, the difference in community composition was not apparent as a result of the short time of ice formation. The same bacterial classes were found in both seawater and sea ice, while the difference between seawater and sea ice was mainly reflected in the relative abundance of different classes, indicating that the early stage of sea-ice bacterial communities was determined by seawater under the ice (Eronen-Rasimus et al., 2015). Due to the versatile metabolism, Proteobacteria were dominant in both sea ice and seawater in all locations, and formation of sea ice did not alter the proportion of Proteobacteria in entire bacterial community. However, due to the change of the condition, it is clear that the relative abundance at class level within Proteobacteria changed during the formation of sea ice. The classes Alphaproteobacteria and Gammaproteobacteria dominated the bacterial communities, consistent with the previous study of the seawater bacterial communities in the Yellow Sea (Guo et al., 2011; Liu et al., 2013). Gammaproteobacteria were the most abundant class in the sea ice, and Gammaproteobacteria dominance in the initial phases of ice formation was also found in an experimental study where the cultivation was enriched with algal-derived DOM (Eronen-Rasimus et al., 2014). Mid-latitude regions with high abundance of algae might stimulate the growth of Gammaproteobacteria. In addition to the dominant bacterial classes, a high relative abundance of Verrucomicrobiae was found in the sea ice compared with that in seawater (p < 0.05) (Figure 5C), and it could be found in sea ice in the polar regions (Bowman et al., 2012; Freitas et al., 2012; Hatam et al., 2014). Previous study indicated that class Verrucomicrobiae preferred a particle-attached life style (Chiang et al., 2018), and the phytoplankton concentrated in the brine during the formation of sea ice might stimulate bacterial growth. The sequences affiliated with the class Bacilli were in very low levels in both environments but more abundant in sea ice, probably due to the ability to survive in sea ice, which was also found in Arctic Sea ice (Han et al., 2014). The class Acidimicrobiia and Actinobacteria, of the phylum Actinobacteriota, were the other two major bacterial groups inhabiting sea ice and seawater, and the relative abundances in seawater were higher than in sea ice. Previous reports indicated that Actinobacteriota was not only commonly seen in seawater (Lu et al., 2015; Yu et al., 2018), but also present in sea ice (Brinkmeyer et al., 2004; Eronen-Rasimus et al., 2015). It was found that Actinobacteria favored a low salinity habitat, and the abundance of Actinobacteriota decreased in ice with high brine salinity (Eronen-Rasimus et al., 2015). The phenomenon of higher abundance of ABC transporters in seawater might be explained by that diverse substrates could be utilized in seawater and bacteria may therefore require various ABC systems (Garmory and Titball, 2004), while the substrates are more specific in the restricted young sea ice brine.
Conclusion
This study investigated the bacterial community in seawater and sea ice after a cold surge in the coastal area of the mid-latitude. The differences in bacteria abundance (BA) between sea ice and seawater might be associated with the physical impact of ice formation, and BA in sea ice was higher than in seawater, which was consistent with the findings in Arctic. The bacterial community composition in sea ice was similar to that in seawater, while the relative abundances were significantly different. The sea-ice bacterial community was dominated by Gammaproteobacteria, which could be capable of opportunistic growth in sea ice with high concentration of algal-derived DOM in coastal areas, and the high relative abundance of Verrucomicrobiae in sea ice might also be that. The relatively lower abundance of functional genes of ATP-binding cassette transporters in sea ice might result from the restricted environment in sea ice brine. More works need be done to investigate the impact of cold surges on microbial communities in mid-latitudes along with global climate change.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI (accession: PRJNA797534).
Author Contributions
YH designed the research. HR analyzed the data and drafted the manuscript. YH, JL, GL, and HR discussed the interpretation of the results. All authors agreed to authorship and approved the manuscript submission.
Funding
This work was financially supported by the Key Research and Development Program of Shandong Province (2020ZLYS04), the National Natural Science Foundation of China (41806094), the National Key Research and Development Program of China (2020YFA0608300), the Joint Funds of the National Natural Science Foundation of China (U1906216), and the Young Scholars Program of Shandong University (2018WLJH43).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to especially thank Xianzhe Gong for the internal review of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.856110/full#supplementary-material
References
Alexander, L. V., Zhang, X., Peterson, T. C., Caesar, J., Gleason, B., Tank, A. M. G. K., et al. (2006). Global observed changes in daily climate extremes of temperature and precipitation. J. Geophys. Res. Atmos. 111:D05109. doi: 10.1029/2005jd006290
Azam, F., Fenchel, T., Field, J. G., Gray, J. S., Meyerreil, L. A., and Thingstad, F. (1983). The ecological role of water-column microbes in the sea. Mar. Ecol. Prog. Ser. 10, 257–263. doi: 10.3354/meps010257
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Bowman, J. S., Rasmussen, S., Blom, N., Deming, J. W., Rysgaard, S., and Sicheritz-Ponten, T. (2012). Microbial community structure of Arctic multiyear sea ice and surface seawater by 454 sequencing of the 16S RNA gene. ISME J. 6, 11–20. doi: 10.1038/ismej.2011.76
Brinkmeyer, R., Glockner, F. O., Helmke, E., and Amann, R. (2004). Predominance of beta-proteobacteria in summer melt pools on Arctic pack ice. Limnol. Oceanogr. 49, 1013–1021. doi: 10.4319/lo.2004.49.4.1013
Chiang, E., Schmidt, M. L., Berry, M. A., Biddanda, B. A., Burtner, A., Johengen, T. H., et al. (2018). Verrucomicrobia are prevalent in north-temperate freshwater lakes and display class-level preferences between lake habitats. PLoS One 13:e0195112. doi: 10.1371/journal.pone.0195112
Cox, G. F. N., and Weeks, W. F. (1983). Equations for determining the gas and brine volumes in sea-ice samples. J. Glaciol. 29, 306–316. doi: 10.3189/S0022143000008364
Duprat, L., Corkill, M., Genovese, C., Townsend, A. T., Moreau, S., Meiners, K. M., et al. (2020). Nutrient distribution in East Antarctic summer sea ice: a potential iron contribution from glacial basal melt. J. Geophys. Res. Oceans 125:e16130. doi: 10.1029/2020JC016130
Eronen-Rasimus, E., Kaartokallio, H., Lyra, C., Autio, R., Kuosa, H., Dieckmann, G. S., et al. (2014). Bacterial community dynamics and activity in relation to dissolved organic matter availability during sea-ice formation in a mesocosm experiment. Microbiologyopen 3, 139–156. doi: 10.1002/mbo3.157
Eronen-Rasimus, E., Lyra, C., Rintala, J. M., Jurgens, K., Ikonen, V., and Kaartokallio, H. (2015). Ice formation and growth shape bacterial community structure in Baltic Sea drift ice. FEMS Microbiol. Ecol. 91, 1–13. doi: 10.1093/femsec/fiu022
Ewert, M., and Deming, J. W. (2014). Bacterial responses to fluctuations and extremes in temperature and brine salinity at the surface of Arctic winter sea ice. FEMS Microbiol. Ecol. 89, 476–489. doi: 10.1111/1574-6941.12363
Freitas, S., Hatosy, S., Fuhrman, J. A., Huse, S. M., Welch, D. B., Sogin, M. L., et al. (2012). Global distribution and diversity of marine Verrucomicrobia. ISME J. 6, 1499–1505. doi: 10.1038/ismej.2012.3
Garmory, H. S., and Titball, R. W. (2004). ATP-binding cassette transporters are targets for the development of antibacterial vaccines and therapies. Infect. Immun. 72, 6757–6763. doi: 10.1128/IAI.72.12.6757-6763.2004
Grasshoff, K., Kremling, K., and Ehrhardt, M. (1999). Methods of Seawater Analysis. Hoboken, NJ: John Wiley & Sons.
Grossmann, S., and Gleitz, M. (1993). Microbial responses to experimental sea-ice formation: implications for the establishment of Antarctic sea-ice communities. J. Exp. Mar. Biol. Ecol. 173, 273–289. doi: 10.1016/0022-0981(93)90058-v
Guo, C., Li, F. C., Jiang, P., Liu, Z. P., and Qin, S. (2011). Bacterial diversity in surface water of the Yellow Sea during and after a green alga tide in 2008. Chin. J. Oceanol. Limnol. 29, 1147–1154. doi: 10.1007/s00343-011-0264-7
Guo, Y. L., Xiong, X. J., Chen, C., Yu, L., and Guo, Y. Q. (2014). Hydro-meteorological interpretation and cause analysis on Qingdao’s warmer winter. Coast. Eng. 33, 1–11.
Han, D., Kang, I., Ha, H. K., Kim, H. C., Kim, O. S., Lee, B. Y., et al. (2014). Bacterial communities of surface mixed layer in the Pacific sector of the western Arctic Ocean during sea-ice melting. PLoS One 9:e86887. doi: 10.1371/journal.pone.0086887
Hansen, J., Sato, M., Ruedy, R., Lo, K., Lea, D. W., and Medina-Elizade, M. (2006). Global temperature change. Proc. Natl. Acad. Sci. U.S.A. 103, 14288–14293. doi: 10.1073/pnas.0606291103
Hatam, I., Charchuk, R., Lange, B., Beckers, J., Haas, C., and Lanoil, B. (2014). Distinct bacterial assemblages reside at different depths in Arctic multiyear sea ice. FEMS Microbiol. Ecol. 90, 115–125. doi: 10.1111/1574-6941.12377
Hatam, I., Lange, B., Beckers, J., Haas, C., and Lanoil, B. (2016). Bacterial communities from Arctic seasonal sea ice are more compositionally variable than those from multi-year sea ice. ISME J. 10, 2543–2552. doi: 10.1038/ismej.2016.4
Jiao, N., Herndl, G. J., Hansell, D. A., Benner, R., Kattner, G., Wilhelm, S. W., et al. (2010). Microbial production of recalcitrant dissolved organic matter: long-term carbon storage in the global ocean. Nat. Rev. Microbiol. 8, 593–599. doi: 10.1038/nrmicro2386
Jiao, N., Robinson, C., Azam, F., Thomas, H., Baltar, F., Dang, H., et al. (2014). Mechanisms of microbial carbon sequestration in the ocean – future research directions. Biogeosciences 11, 5285–5306. doi: 10.5194/bg-11-5285-2014
Johnson, N. C., Xie, S. P., Kosaka, Y., and Li, X. (2018). Increasing occurrence of cold and warm extremes during the recent global warming slowdown. Nat. Commun. 9:1724. doi: 10.1038/s41467-018-04040-y
Kug, J. S., Jeong, J. H., Jang, Y. S., Kim, B. M., Folland, C. K., Min, S. K., et al. (2015). Two distinct influences of Arctic warming on cold winters over North America and East Asia. Nat. Geosci. 8, 759–762. doi: 10.1038/Ngeo2517
Li, G., Liu, J., Diao, Z., Jiang, X., Li, J., Ke, Z., et al. (2018). Subsurface low dissolved oxygen occurred at fresh- and saline-water intersection of the Pearl River estuary during the summer period. Mar. Pollut. Bull. 126, 585–591. doi: 10.1016/j.marpolbul.2017.09.061
Liu, J., Curry, J. A., Wang, H., Song, M., and Horton, R. M. (2012). Impact of declining Arctic sea ice on winter snowfall. Proc. Natl. Acad. Sci. U.S.A. 109, 4074–4079. doi: 10.1073/pnas.1114910109
Liu, M., Liu, L., Chen, H., Yu, Z., Yang, J. R., Xue, Y., et al. (2019). Community dynamics of free-living and particle-attached bacteria following a reservoir Microcystis bloom. Sci. Total Environ. 660, 501–511. doi: 10.1016/j.scitotenv.2018.12.414
Liu, M., Xiao, T., Sun, J., Wei, H., Wu, Y., Zhao, Y., et al. (2013). Bacterial community structures associated with a natural spring phytoplankton bloom in the Yellow Sea, China. Deep Sea Res. II Top. Stud. Oceanogr. 97, 85–92. doi: 10.1016/j.dsr2.2013.05.016
Liu, S., Baetge, N., Comstock, J., Opalk, K., Parsons, R., Halewood, E., et al. (2020). Stable isotope probing identifies bacterioplankton lineages capable of utilizing dissolved organic matter across a range of bioavailability. Front. Microbiol. 11:580397. doi: 10.3389/fmicb.2020.580397
Lu, X., Sun, S., Zhang, Y. Q., Hollibaugh, J. T., and Mou, X. (2015). Temporal and vertical distributions of bacterioplankton at the Gray’s reef national marine sanctuary. Appl. Environ. Microbiol. 81, 910–917. doi: 10.1128/AEM.02802-14
Lund-Hansen, L. C., Sgaard, D. H., Sorrell, B. K., Gradinger, R., and Meiners, K. M. (2020). Arctic Sea Ice Ecology: Seasonal Dynamics in Algal and Bacterial Productivity. Berlin: Springer.
Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P. R., O’Hara, R. B., et al. (2019). Vegan: community ecology package.
Ortega-Retuerta, E., Joux, F., Jeffrey, W. H., and Ghiglione, J. F. (2013). Spatial variability of particle-attached and free-living bacterial diversity in surface waters from the Mackenzie River to the Beaufort Sea (Canadian Arctic). Biogeosciences 10, 2747–2759. doi: 10.5194/bg-10-2747-2013
Park, T. W., Ho, C. H., Jeong, S. J., Choi, Y. S., Park, S. K., and Song, C. K. (2011). Different characteristics of cold day and cold surge frequency over East Asia in a global warming situation. J. Geophys. Res. Atmos. 116:D12118. doi: 10.1029/2010jd015369
Parks, D. H., Tyson, G. W., Hugenholtz, P., and Beiko, R. G. (2014). STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 30, 3123–3124. doi: 10.1093/bioinformatics/btu494
Petoukhov, V., and Semenov, V. A. (2010). A link between reduced Barents-Kara sea ice and cold winter extremes over northern continents. J. Geophys. Res. Atmos. 115:D21111. doi: 10.1029/2009jd013568
Pinhassi, J., Sala, M. M., Havskum, H., Peters, F., Guadayol, O., Malits, A., et al. (2004). Changes in bacterioplankton composition under different phytoplankton regimens. Appl. Environ. Microbiol. 70, 6753–6766. doi: 10.1128/AEM.70.11.6753-6766.2004
Piontek, J., Galgani, L., Nöthig, E. M., Peeken, I., and Engel, A. (2020). Organic matter composition and heterotrophic bacterial activity at declining summer sea ice in the central Arctic Ocean. Limnol. Oceanogr. 66, S343–S362. doi: 10.1002/lno.11639
Robeson, M. S., O’Rourke, D. R., Kaehler, B. D., Ziemski, M., Dillon, M. R., Foster, J. T., et al. (2020). RESCRIPt: reproducible sequence taxonomy reference database management for the masses. bioRxiv [Preprint] doi: 10.1101/2020.10.05.326504
Shannon, C. E. (1948). A mathematical theory of communication. Bell Syst. Tech. J. 27, 379–423. doi: 10.1002/j.1538-7305.1948.tb01338.x
Shivaji, S., and Prakash, J. S. (2010). How do bacteria sense and respond to low temperature? Arch. Microbiol. 192, 85–95. doi: 10.1007/s00203-009-0539-y
Subramanian, P., Joe, M. M., Yim, W.-J., Hong, B.-H., Tipayno, S. C., Saravanan, V. S., et al. (2011). Psychrotolerance mechanisms in cold-adapted bacteria and their perspectives as plant growth-promoting bacteria in temperate agriculture. Korean J. Soil Sci. Fert. 44, 625–636. doi: 10.7745/kjssf.2011.44.4.625
Takaya, K., and Nakamura, H. (2005). Mechanisms of intraseasonal amplification of the cold Siberian high. J. Atmos. Sci. 62, 4423–4440. doi: 10.1175/Jas3629.1
Torstensson, A., Fransson, A., Currie, K., Wulff, A., and Chierici, M. (2018). Microalgal photophysiology and macronutrient distribution in summer sea ice in the Amundsen and Ross Seas, Antarctica. PLoS One 13:e0195587. doi: 10.1371/journal.pone.0195587
Wemheuer, F., Taylor, J. A., Daniel, R., Johnston, E., Meinicke, P., Thomas, T., et al. (2020). Tax4Fun2: prediction of habitat-specific functional profiles and functional redundancy based on 16S rRNA gene sequences. Environ. Microbiome 15:11. doi: 10.1186/s40793-020-00358-7
Xu, Z. Y., Yang, Y. Z., Wang, G. F., Cao, W. X., Li, Z. J., and Sun, Z. H. (2012). Optical properties of sea ice in Liaodong Bay, China. J. Geophys. Res. Oceans 117:C03007. doi: 10.1029/2010jc006756
Yu, S. X., Pang, Y. L., Wang, Y. C., Li, J. L., and Qin, S. (2018). Distribution of bacterial communities along the spatial and environmental gradients from Bohai Sea to northern Yellow Sea. PeerJ 6:e4272. doi: 10.7717/peerj.4272
Zhao, D., Gao, P., Xu, L., Qu, L., Han, Y., Zheng, L., et al. (2021). Disproportionate responses between free-living and particle-attached bacteria during the transition to oxygen-deficient zones in the Bohai Seawater. Sci. Total Environ. 791:148097. doi: 10.1016/j.scitotenv.2021.148097
Keywords: cold surge, bacterial community, bacteria abundance, mid-latitude, sea ice
Citation: Ren H, Liu J, Li G and Hu Y (2022) Bacterial Community in Cold Surge-Caused Sea Ice Differs From Seawater in Mid-Latitude Region: A Case Study in Aoshan Bay, Southern Yellow Sea. Front. Mar. Sci. 9:856110. doi: 10.3389/fmars.2022.856110
Received: 16 January 2022; Accepted: 28 February 2022;
Published: 04 April 2022.
Edited by:
Valeria Chávez, National Autonomous University of Mexico, MexicoReviewed by:
Dolors Vaque, Spanish National Research Council (CSIC), SpainYuqiu Wei, Chinese Academy of Fishery Sciences (CAFS), China
Copyright © 2022 Ren, Liu, Li and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jihua Liu, bGl1amlodWExOTgyQHNkdS5lZHUuY24=; Yubin Hu, eXViaW5odUBzZHUuZWR1LmNu
 Hongwei Ren
Hongwei Ren Jihua Liu
Jihua Liu Gang Li
Gang Li Yubin Hu
Yubin Hu