
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 17 March 2022
Sec. Aquatic Physiology
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.854140
This article is part of the Research Topic Homeostasis and Physiological Regulation in the Aquatic Animal during Osmotic Stress View all 7 articles
Fish can maintain a stable intracellular concentration of ions in environments of variable salinities through osmoregulation. In this study, we focused on obscure puffer Takifugu obscurus (T. obscurus), an anadromous fish with high commercial value and rapid depression of wide populations, to investigate changes at molecular and physiological levels underlying salinity tolerance through multifaceted analyses integrating transcriptomics and micrography. We found that with the increase of salinity, the structure of the three main osmoregulation organs, i.e., gill, intestine, and kidney had remarkable changes. The results of transcriptome demonstrated that the ATP-binding box transporter and cyclic adenosine monophosphate (cAMP) signaling pathway in the gill and kidney were significantly reduced. Growth hormone, prolactin, and cortisol regulated more transporters of body composition and stimulated chloride cell proliferation and differentiation, which change the capacity for membrane transport between ion and water molecules. Adenosine-activating protein kinase and thyroid hormone signaling pathway were also significantly upregulated. These transcriptional levels changes of T. obscurus combined with ultrastructure in response to salinity increase indicated that osmoregulation is a complex process involving multiple organs and signaling pathways. Overall, this study can deepen the understanding of osmotic regulation during fish migration.
Salinity has great influence on the growth and development of fish; some species have specific optimal salinity ranges that others could easily transition. When the osmotic pressure of water is close to that of fish body fluid, the energy consumed in regulating osmotic pressure of fish body to adapt to external environment is the least, which is most conducive to the survival and growth of fish (Giffard-Mena et al., 2020). As an anadromous species, obscure puffer [Takifugu obscurus (T. obscurus)] confronts a variety of different ambient salinities during its life cycle, which makes it a typical euryhaline fish species (Wang et al., 2016, 2018). T. obscurus can maintain normal feeding, absorption, transformation, and growth efficiency in a wide salinity range (Gao et al., 2017). It has a strong adaptability to the environment with high or low osmotic pressure and has a better osmoregulation mechanism than stenohaline fish (Kato et al., 2005). The Takifugu species have an advantage as animal model, i.e., they have a short genome (∼400 Mb) compared to those of other vertebrates (Venkatesh et al., 2000). Thus, T. obscurus is an enormously useful tool for studying molecular mechanism of osmoregulation. As T. obscurus has very high commercial and economic value and the wide populations are experiencing rapid depression due to overfishing pressure, it has become an important and precious breed species in aquaculture (Yang and Chen, 2005, 2006; Kim et al., 2008). A comprehensive research on osmoregulation mechanism of T. obscurus could help to facilitate the understanding of the physiological change of euryhaline fish during migration.
The osmotic pressure in the euryhaline fish is regulated by the main osmoregulation organs (such as gill, kidney, and intestine) to maintain the balance of internal and external osmotic pressure, thus ensuring the normal operation of all the biochemical and physiological processes in organism (Pérez-Robles et al., 2019). The gill and kidney are the main organs for osmoregulation in fish. Intestinal tract is used to maintain the balance of ion concentration in fish through water reabsorption and electrolyte ingestion in food and it is also an important organ of osmoregulation in fish (Barany et al., 2020). The osmoregulation mechanism of fish mainly includes two processes: (1) rapid adaptation to changes in osmotic pressure environment depends on the osmoregulation functions of the organism, including epithelial cells and ion transporters and (2) regulating the neuroendocrine system allows epithelial cells to differentiate and synthesis more ion transporters, leading to changes in the structure and function of osmotic pressure-regulated organs (McCormick, 2001). Osmotic adaptation mechanism of aquatic organisms is based on complex gene interaction network (Autenrieth et al., 2018). Many enzymes and transporters play important roles in maintaining osmotic pressure and ion homeostasis in the process of salinity adaptation and regulation (Hwang and Lee, 2007; Nguyen et al., 2016). Carrier proteins related to ion transport, such as Na+/K+-ATPase (NKA), Na+/K+/2Cl– cotransporter (NKCC), cystic fibrosis transmembrane conductance regulator (CFTR), Na+/Cl– cotransporter (NCC), and Na+/H+ exchange protein (NHE), play an important role in maintaining osmotic pressure balance in teleosts (Marshall, 2011; Regish et al., 2018; Shaughnessy and McCormick, 2020).
The functions of osmotic organs, such as gill, kidney, and intestine, are closely related to the mediation of neuroendocrine system (Moorman et al., 2014). Fish secrete hormones, including growth hormone (GH), prolactin (PRL), insulin-like growth factor-1 (IGF-1), cortisol, etc., to synthesize more transporters and stimulate the proliferation and differentiation of chloride cells, thus changing the transmembrane transport capacity of ions and water molecules (Deane and Woo, 2007). GH can regulate the number and volume of chloride cells in gill and promote the activity of NKA and NKCC plasma transporters. The GH level increases significantly in Oncorhynchus mykiss from freshwater to hypertonic environment (Sakamoto and McCormick, 2006). When exposed to exogenous GH and cortisol, largemouth bass exhibits stronger osmoregulation and significantly increases the tolerance of salinity (Yi et al., 2021). PRL plays an important regulatory role in cell apoptosis and proliferation of osmotic regulator organs such as gill and intestinal tract as well as in the proliferation and differentiation of skin and mucosal cells and is an important regulatory hormone for fish adapting to hypoosmotic environment (Breves et al., 2020). Cortisol is mainly related to biological stress response and glycometabolism. At the same time, cortisol can regulate the excretion of excess ions by regulating the number of chloride cells in gill epithelium and the proliferation of intestinal cells (Sadoul et al., 2021). It can be seen that the osmotic pressure changes in the environment can be adapted by the synergistic action of hormone and osmoregulation in the organism of euryhaline teleosts.
Salinity adaptation mechanism is the most important link in the evolution of the euryhaline teleosts and it is of great significance to reveal their migration habits. This mechanism not only involves ion and water transport and regulation of neuroendocrine system, but also depends on energy metabolism to maintain the auxiliary role of various osmoregulation substances such as the activity of transporter proteins and free amino acids, which is a complex regulatory network (Lam et al., 2014; Drenner et al., 2018). Although many studies analyzed the transcriptome of osmoregulation in aquatic organisms, few of them are combined with ultrastructural changes in tissues (Lv et al., 2013; Hui et al., 2014). In this study, we performed transcriptome analysis and ultrastructural observation on the gill, kidney, and intestine of T. obscurus under different salinities to further reveal the osmoregulation mechanism. In addition, considering the ecological and economic importance of this species, histological and molecular knowledge about the environmental tolerance in the tissues of obscure puffer will be helpful for improving, managing, and culturing this species.
All the experiments were approved by and carried out in accordance with the guidelines of the Institutional Animal Care and Use Committee (IACUC) of Nanjing Normal University, Nanjing, China (Research permit number: SYXK2015-0028).
Adult T. obscurus was collected from a stock farm in Yangzhong city, Jiangsu Province, China. They were transferred to the laboratory and maintained at 23 ± 1°C in an aquarium with continuous aeration. After 7 days, adults were randomly selected to assess the effect of salinity on their osmoregulation. In the experiment, the control group (0 ppt) and the treatment group (30 ppt) were setup and each group replicated in triplicate. Three adults in each replicate were placed in an aquarium (100 cm × 50 cm × 60 cm) filled with 200 L of test solution. The treatment group increased 5 ppt every 12 h; when reached the target salinity, the adults continued to adapt 7 days. During exposure, normal commercial pelletized feeds with a 44.0% protein content (Tongwei Corporation Ltd., Nantong, China) were given in both the groups twice a day at a dose of 4% of fish body weight. Water quality parameters (salinity, temperature, pH, and the concentrations of nitrite and dissolved oxygen) were measured using a multifunctional water quality analyzer (YSI 650MDS, Yellow Springs, United States) and maintained at constant levels (temperature: 23 ± 1°C; pH: 7.5 ± 0.4; light:dark = 12:12 h; dissolved oxygen >5 mg l–1; and nitrite concentration: < 0.01 mg l–1) by replacing half of each test solution everyday. Seven days later, one healthy adult in each aquarium was selected and all the six fishes (three fishes from 0 ppt and three fishes from 30 ppt) were euthanized with a lethal dose of NaOH-neutralized MS-222. Then, gill, kidney, and intestine were removed: one part was used for microscopic observation and the other part was rinsed with cold physiological saline (0.6% NaCl), dried on filter paper, frozen in liquid nitrogen, and stored at −80°C, for subsequent total RNA extraction.
For light microscopy, tissues were fixed by immersion in Bouin’s fluid for 24 h, embedded in paraffin, and serially sectioned at 5 μm thickness. Sections were dried for 24 h and stained with H&E and periodic acid-Schiff (PAS). Observation and photographing were performed using digital slice scanner (Pannoramic MIDI, 3DHistech, Budapest, Hungary). For ultrastructural studies, tissues were cut into small pieces (1 mm × 1 mm × 1 mm) and fixed at 4°C for 2–4 h and then rinsed three times with 0.1 mol/l phosphate buffer (pH 7.4) for 15 min each time. After dehydration, the tissues were soaked with acetone for 2–4 h and percolated overnight. The pure 812 embedding agent was poured into the embedding plate for 5–8 h. The samples were inserted into the embedding plate and then overnight in 37°C. After 48 h of polymerization in 60°C oven, the ultra-thin slices of 60–80 nm were made by Leica UC7 (Leica, Wetzlar, Germany). Finally, after double staining of uranium and lead (2% uranium acetate, saturated alcohol solution, and lead citrate, each stained for 15 min), slices were dried overnight at room temperature. The stained sections were examined in a transmission electron microscopy (TEM) (HT7700, Hitachi, Tokoyo, Japan).
The total RNA was extracted from gill, kidney, and intestine using TRIzol Reagent (Takara, Shiga, Japan) according to the manufacturer’s instructions. The total RNA quality and quantity of the two groups of samples were evaluated by electrophoresis on 1.2% agarose gels and the absorbance at 260/280 nm was determined using a spectrophotometer (Nanodrop, ThermoFisher, Waltham, United States). Only samples with OD260/OD280 = 1.8–2.0, OD260/OD230 ≥ 2.0, and RIN ≥ 8.0 were used for sequencing. Further details of RNA sampling and complementary DNA (cDNA) generation were provided in the Supporting Information. The cDNA library was sequenced on the Illumina HiSeq 2500 Sequencing Platform that generated approximately 100 bp paired-end (PE) raw reads by LC Sciences (Houston, United States). The full data sets have been submitted to the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under accession SUB6140175, Bioproject: PRJNA562326.
To analyze the differentially expressed genes (DEGs) in T. obscurus under freshwater and seawater, each transcript was measured as the number of clean reads mapped to its sequence. The mapped clean read number was normalized to reads per kilobase per million (RPKM) by using RSEM version 1.2.3. We used DESeq to determine the false discovery rate (FDR) threshold. FDR < 0.05 and fold change > 2 were considered to indicate significant expression abundance. DEGs were further annotated by the Gene Ontology (GO) functional enrichment and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis, as demonstrated in the Supporting Information.
To validate the RNA Sequencing (RNA-seq) results, four upregulated and four downregulated genes from gill were randomly selected to confirm the expression profiles of T. obscurus in response to salinity by quantitative PCR (qPCR). The T. obscurus was treated as described above. qPCR was performed with the SYBR Green Master Kit (Takara, Japan) according to the manufacturer’s protocol. The experiments were carried out in triplicate with a total volume of 20 μl [10 μl of 2 × SYBR Premix Ex Taq, 1 μl cDNA mix, 0.5 μl of each primer (10 μM), and 8 μl of sterile distilled H2O]. The PCR was programmed at 95°C for 30 s, followed by 40 cycles of 95°C for 5 s and 60°C for 30 s. qPCR method with β-actin as an internal control was used to explore messenger RNA (mRNA) expression levels. The expression level was calculated by 2–ΔΔCt method and subjected to statistical analysis.
Microscopic comparisons were performed on the differences in the structure of tissues at different salinities. The results showed that with the increase of salinity, the number of chloride-secreting cells on gill filaments and gill patches of T. obscurus increased significantly, the cells also enlarged, and the staining of H&E deepened (Figure 1). The chloride-secreting cells tended to diffuse to the middle of adjacent gill patches and there were more chloride-secreting cells in the base of gill patches. According to TEM observation, in freshwater, tubular network is underdeveloped and chloride-secreting cells are small (Figure 2). The volume and nucleus of chloride-secreting cells increased significantly in saline solution and the number of mitochondria in chloride-secreting cells increased significantly. The reticulum in the cytoplasm was well-developed and the diameter of the tube was larger than that in freshwater.
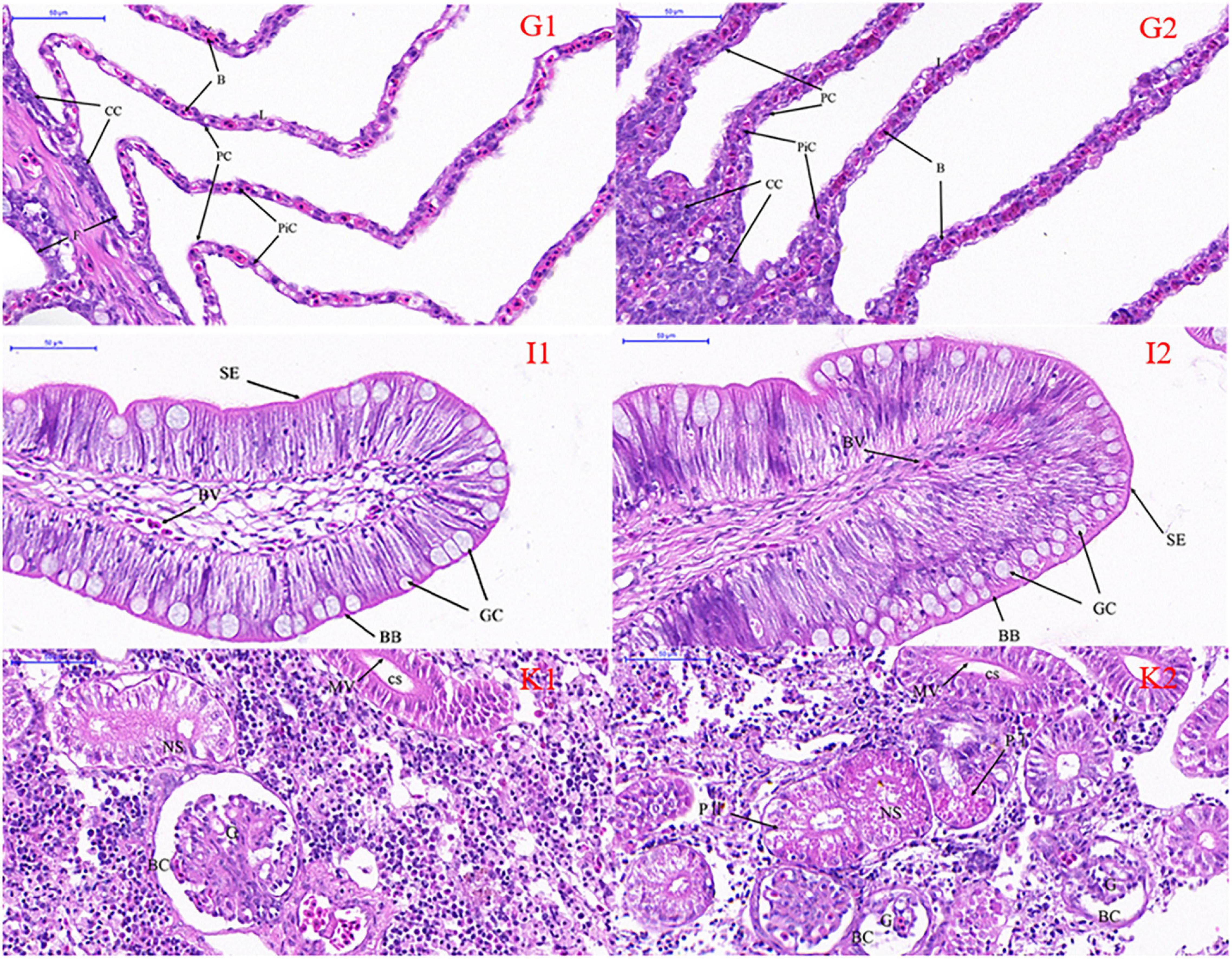
Figure 1. Light micrography of gill (G), intestine (I), and kidney (K) in Takifugu obscurus adapted to freshwater (1) and saline water (2). B, blood channel; CC, chloride cells; F, filament; L, lamellae; PC, pavement cells; PiC, pillar cells; BB, brush border; BV, blood vessel; GC, goblet cell; SE, serosa; BC, bowman capsule; CS, collecting segment; G, glomeruli; NS, neck segment; MV, microvillus; P I, the first proximal segment; P II, the second proximal segment.
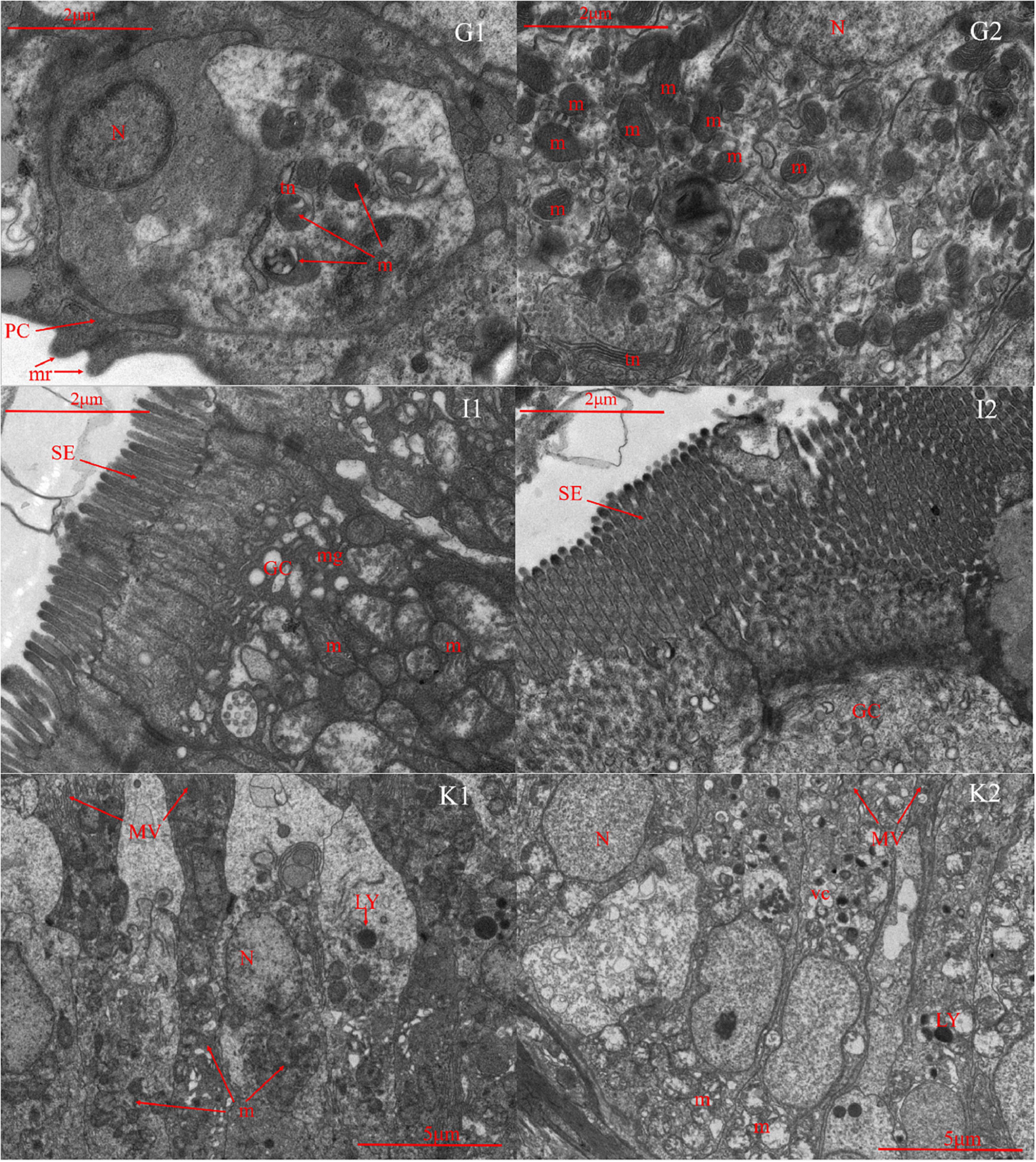
Figure 2. Transmission electron microscope of gill (G), intestine (I), and kidney (K) in Takifugu obscurus adapted to freshwater (1) and saline water (2). N, nucleus; m, mitochondria; mr, microridges; PC, pavement cells; tn, tubular network; GC, goblet cell; m, mitochondria; mg, mucous granule; SE, serosa; LY, lysosome; m, mitochondria; MV, microvillus; N, nucleus; vc, vacuole.
Light microscopy observation showed that with the increase of salinity, there was a marked increase in goblet cells in the intestine and the blood vessels also became dense (Figure 1). There was an increase of the mitochondria and mucous granules in goblet cells increased and the density and length of serosa increased at high salinity (Figure 2).
In freshwater, most of the glomeruli are full, but in the salinity group, the capillaries in the glomerulus are underfilled and the volume of the whole glomeruli is smaller than that in the freshwater group. Additionally, all the parts of the glomeruli in salinity group had varying degrees of atrophy (Figure 1). There are a lot of mitochondria in the cytoplasm of freshwater and a few lysosomes in the mitochondria, while the number of mitochondria in saline water decreased, arranged irregularly, and the number of lysosomes increased significantly (Figure 2).
The cDNA libraries of gills, kidneys, and intestines were sequenced under 0 and 30 ppt treatments and the raw reads were filtered to get clean reads. More than 40 million clean reads were obtained in each sample. The Q30 (%) of the cDNA libraries of gill, kidney, and intestine in freshwater and seawater was higher than 85%, which means that the sequencing quality was high (Supplementary Table 1). After the clean reads of each sample were treated and quality controlled, the clean reads were assembled and stitched. The transcript sequence obtained by Trinity stitching was used as the reference gene sequence for subsequent analysis. At the same time, the longest transcript of each gene is unigene. A total of 193,511 unigenes were obtained and 75,920 unigenes were annotated after aligning with the reference genome (mainly the genome of the Takifugu rubripes). The transcripts and the length of unigene were counted and the results are shown in Supplementary Figure 1. After assembly, all the unigenes were used for BLAST analysis in the NCBI nucleotide sequences (NT), the NCBI non-redundant protein sequences (NR), Pfam protein domain (Pfam), clusters of the orthologous groups for eukaryotic complete genomes (KOG), Swiss-Prot, the KEGG, and the GO databases, respectively. The results showed that at least 39.23% of all the unigenes can be annotated in one database (Supplementary Table 2).
The unigenes annotated successfully in the GO database were classified and a total of 44,990 GO functional notes of unigene were obtained (Supplementary Table 2). The GO databases are divided into three classes: biological process (BP), cellular component (CC), and molecular function (MF). Genes annotated under the each GO category are classified into different secondary categories. The results showed that the most abundant genes in BP are involved in cell process and metabolic process, the most abundant genes in CC are involved in cell and cell part, and the most abundant genes in MF are involved in binding and catalytic activity (Supplementary Figure 2).
A total of 25,284 unigenes were successfully annotated and divided into 26 subcategories in KOG database (Supplementary Table 2). The results showed that the largest subgroup of unigenes annotations is T: signal transduction mechanism, accounting for about 18%; followed by R: general function prediction only; and O: posttranslational modification, protein turnover, and chaperones (Supplementary Figure 3).
After KOG annotation of unigene, unigenes were annotated and classified in the KEGG pathway database. The KEGG metabolic pathways involved by unigene were divided into five branches: A: cellular processes, B: environmental information processing, C: genetic information processing, D: metabolism, and E: organismal systems. Similar to the results of KOG annotation, the most abundant pathway was signal transduction pathway in B class and 3,954 unigenes were enriched in this pathway. Next were endocrine system in E class and translation path in C class (Supplementary Figure 4).
In gills of T. obscurus in seawater (30 ppt), 60 genes were significantly upregulated and 433 genes were significantly downregulated compared to T. obscurus in freshwater (0 ppt) (Figure 3). The most 30 upregulated and downregulated significant DEGs in gill are given in Supplementary Tables 3, 4. The significantly upregulated genes included: (1) glycosyl and other substance transporters GLUT1 (c73272_g5), CFTR protein (c77540_g2), and other transmembrane substance transporters; (2) aspartate protease gene Asp (c70542_g1) and many other cell membrane-related genes; (3) sucrose nonfermenting 2 (SNF2) family genes (c78072_g3 and c78072_g2), E-twenty six-related transcription factors (ETS-related transcription factors); and (4) G protein-coupled receptor family genes (c77520_g1, c74771_g2, and c69879_g2). In addition, claudin gene, a calcium-activated neutral protease, was significantly upregulated, which plays an important role in the tight junction structure and extracellular secretion of gill filament cells. The downregulated genes were mainly membrane related (Supplementary Table 4) such as: (1) calpain family genes (c73409_g1, c67931_g2, and c40381_g1); (2) neuronal cell adhesion factor 1 [L1 cell adhesion molecule (L1CAM)] gene (c77667_g2), excitatory amino acid transporter (EAAT) gene (c31080_g2), etc.; and (3) KCC gene (c6363_g1 and c58965_g1).
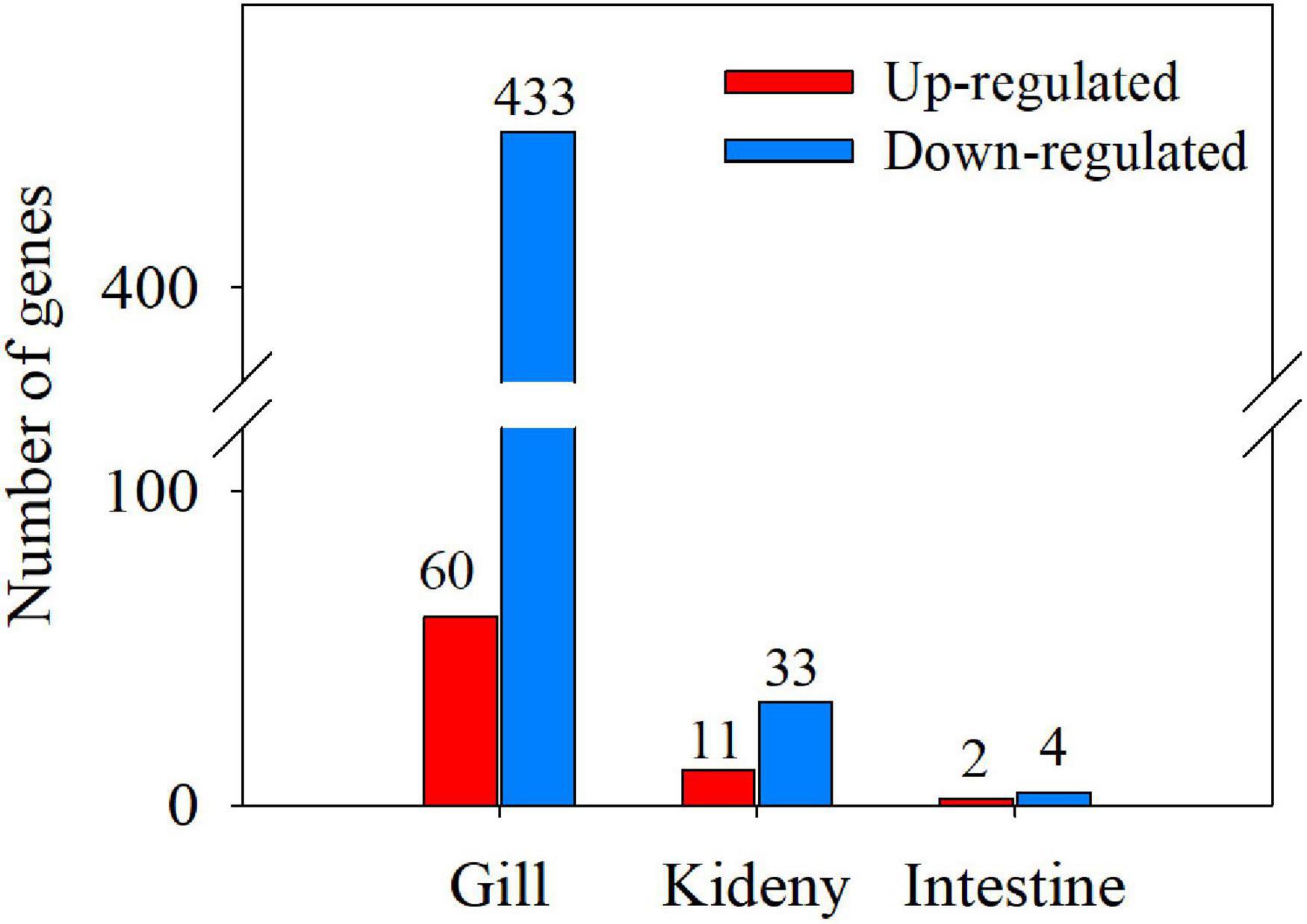
Figure 3. Numbers of differentially expressed genes (DEGs) in gill, kidney, and intestine (T vs. C).
Compared with the kidneys of T. obscurus under the seawater (30 ppt) and freshwater (0 ppt), 11 genes were significantly upregulated and 33 genes were significantly downregulated (Supplementary Table 5). The most upregulated gene is the domain of anaphylatoxin (c72623_g1). ABC transmembrane transporter protein (c75748_g1) related to transmembrane transport of substances was significantly upregulated. Deoxyuridine triphosphate kinase (dUTPase) (c66890_g1) and ligand-gated ion channel (c66391_g4) related to glutamate receptor 2 were significantly upregulated. Tumor necrosis factor (TNF) (c77455_g2) and glutathione S-transferase (GST) (c57788_g1) genes related to disease resistance were significantly upregulated. Among the downregulated genes, protein tyrosine phosphatase (c61724_g4) was the gene with the largest percentage of downregulation (Supplementary Table 6). Inward rectifier potassium channel/ion transport protein (c74159_g1) was significantly downregulated. In addition, corticotropin-releasing factor ryanodine receptor1 (RYR1) (c76257_g1) was also significantly downregulated.
There were few genes with significant differences in intestines, i.e., only two genes were significantly upregulated and four genes were significantly downregulated (Supplementary Table 7). The upregulated genes were type I phosphodiesterase (c69796_g2) and glycoside hydrolases family (c70353_g1), which were related to substance metabolism. The downregulated genes were calcium-activated potassium channel (c71878_g1) and pyridoxal-dependent decarboxylase (c72686_g1).
Because there are few DEGs in kidney and intestine, only DEGs in gill can be analyzed by the GO annotation (Figure 4). Among the GO classifications of DEGs, the primary metabolic process (GO:0044238) was the most abundant in the second class, indicating that the changes of osmotic pressure environment had a significant impact on the basic life processes such as protein and sugar metabolism of T. obscurus. The most abundant genes were intracellular organelles (GO:0043229), organelles (GO:0043231), and polymer complexes (GO:0032991). There are fewer genes enriched in the major categories of MF and the most abundant group is structural molecular activity (GO:0005198).
A total of 17 pathways were significantly upregulated in the KEGG enrichment of DEGs in gills (Supplementary Figure 5). The two most abundant pathways were bile secretion (ko04976) and thyroid hormone synthesis (ko04918), followed by steroid hormone biosynthesis (ko00140). ATP-binding cassette transporter pathway was significantly upregulated, which provides energy released from ATP hydrolysis to various molecules for transmembrane transport. The significantly upregulated CFTR gene in ATP pathway is a chloride ion channel regulated by cyclic adenylate signaling pathway [cyclic adenosine monophosphate (cAMP)], which mainly participates in the transport of electrolytes and liquids and the cAMP pathway is also significantly enriched. In addition, the neuroactive ligand-receptor interaction was significantly upregulated (ko04080). The most significant downregulation pathway enriched by the KEGG is the ribosomal pathway (ko03010), in which 47 genes are significantly downregulated (Supplementary Figure 6), suggesting that protein biosynthesis in T. obscurus is affected by hypertonic environment. Phosphatidylinositol 3-kinase-AKT signaling pathway (Ko4151) has 30 genes significantly downregulated. Phosphatidylinositol 3-kinase (PI3Ks) protein family is involved in the regulation of cell proliferation, differentiation, apoptosis, and glucose transport. Nineteen genes in the tight junction (ko04530) pathway were significantly downregulated. The downregulation of ko04530 pathway plays an important role in the regulation of ionic secretion and the permeability of gill epithelial tissue is stronger. In addition, significant decreases were found in the tricarboxylic acid cycle (TCA) (ko00020) and glycolysis/gluconeogenesis (ko00010).
In kidney, 29 pathways were significantly upregulated in the KEGG enrichment (Supplementary Figure 7). Drug metabolism-cytochrome P450 (ko00982), cytochrome P450 (ko0080), glutathione metabolic pathway (ko00480), and others were significantly upregulated. The significant downregulated pathway was mainly metabolic pathway (Supplementary Figure 8). Five environmental signaling pathways were significantly downregulated, including PI3K-Akt, janus tyrosine kinase-janus tyrosine kinase (JAK-STAT), and calcium signaling pathways. RYR1, a downregulated gene of calcium pathway, is a calcium release pathway in calcium signaling pathway.
In intestine tissues, there were a few genes with significant differences and the KEGG-enriched pathways were all the material metabolism pathways (Supplementary Table 8). Sphingolipid metabolism (ko00600) and aminoglycoside and nucleoside metabolism (ko00520) were significantly upregulated, while glutathione metabolism (ko00480) and arginine and proline metabolism (ko00330) pathways were significantly downregulated. Among them, sphingolipid phosphodiesterase (EC: 3.1.4.12) is a gene significantly upregulated in sphingolipid metabolic pathway, which directly leads to the upregulation of ceramide synthesis.
In order to verify the results of the RNA-seq analysis, four upregulated or downregulated genes were selected from gill and kidney, respectively, for quantitative reverse transcription-PCR (qRT-PCR). All the tested genes showed significantly differential expression (Figure 5). Even though most qRT-PCR results indicated smaller differences compared with the RNA-seq analysis, there was a consistent expression tendency between the two results, which verified the accuracy of the RNA-seq.
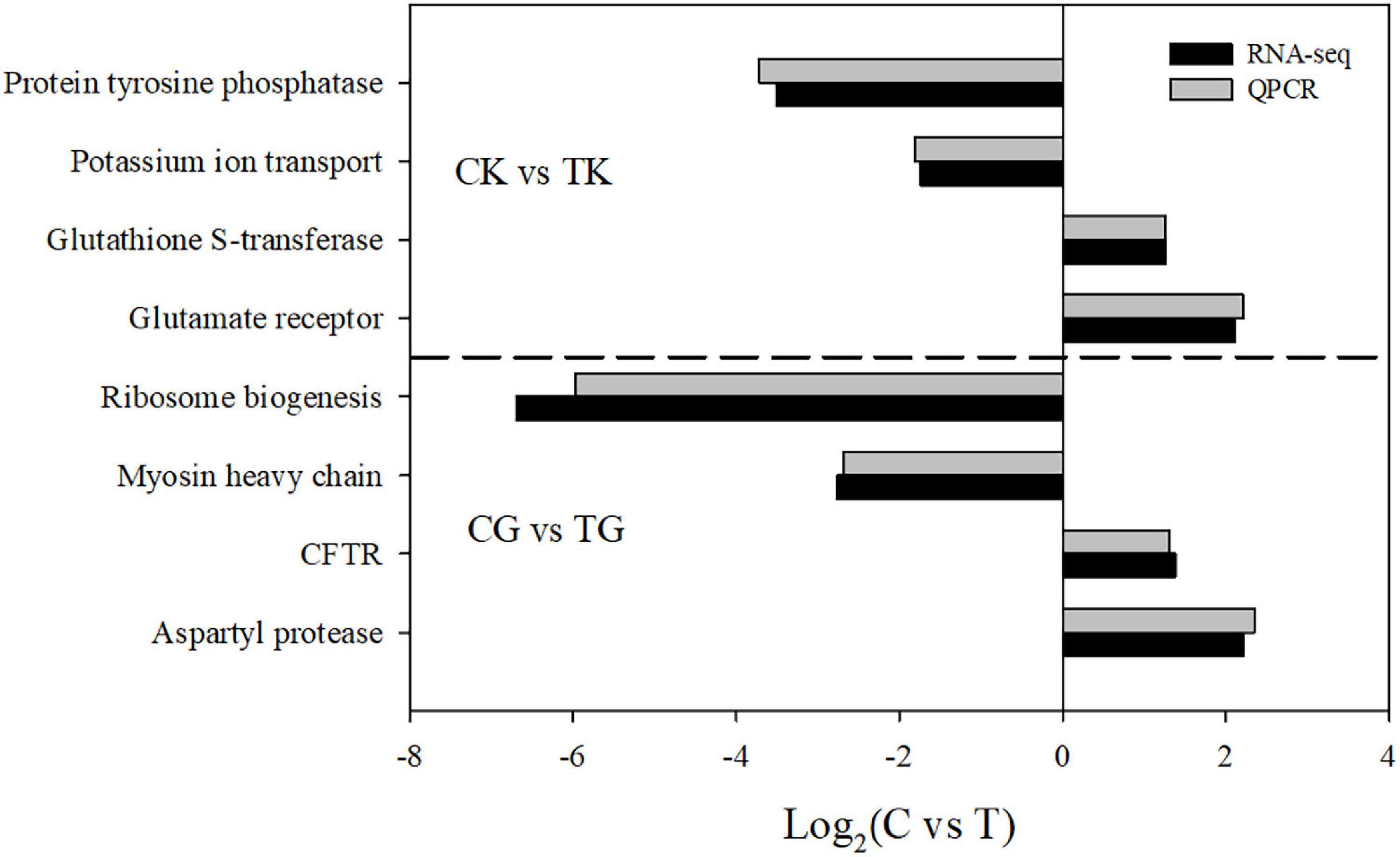
Figure 5. A comparison of the expression profiles in gill and kidney between the control and treatment groups determined by RNA-sequencing and quantitative reverse transcription-PCR (qRT-PCR).
The osmoregulation is needed to maintain the relative stability of osmotic pressure in euryhaline teleosts when they are migrated from freshwater to seawater, thus they can adapt to the changes of osmotic pressure in external water. This study illustrated: (1) substantial changes of the structure of gill, intestine, and kidney; (2) significant expression of signaling pathways [i.e., cAMP, AMPK, and thyroid hormone (TH) signaling pathway] and protein pathways in different tissues; and (3) different modes of neuroendocrine hormone regulation in two saline conditions. This study proved that T. obscurus has a sensitive and changeable osmoregulation function, which is of great significance to understand the mechanism of salinity adaption in teleost fishes and the migratory biology of T. obscurus.
The gill of fish is not only the respiratory organ of fish, but also plays an important role in regulating osmotic pressure balance. In saline water, chloride cells increased and concentrated in the base of gill lamellae and the number and volume of chloride-secreting cells increased significantly. At the same time, the number of mitochondria in the cells was more abundant than that in the freshwater treatment group and the tubular network system was much more developed than that in the freshwater treatment group. Mitochondria are closely related to the tubular network (Fridman, 2020). NKA on the tubular network system indirectly provides energy for excreting excessive Cl– from the body (de Fonseka et al., 2022).
With the increase of salinity, the number of goblet cells in the mucosa increased significantly. Although goblet cells are isolated cells, they have significant secretory function. They can actively absorb glucose and amino acids and store the glycoproteins in the goblet (Zheng et al., 2015). It is presumed that in salinity environment, T. obscurus needs more energy to maintain osmotic balance, even to resist the stress response caused by salinity. Therefore, as the salinity increases, the goblet cells in intestine also increase. Hajirezaee et al. (2017) found that Persian sturgeon fingerlings rely more on oxidation of glucose to meet energetic requirements of osmoregulation than oxidation of lipids. TEM showed that serosa was longer and more abundant in the salinity group than that in the freshwater group because serosa could increase the absorption area, thus increasing the absorption of water and nutrients to maintain stable osmotic pressure (Ando et al., 2000).
The kidney of fish is another important organ of osmoregulation. In this study, we found that the structure of kidney in freshwater is substantial, the glomerulus is developed, and the renal tubule is strong. Under salinity treatment, the kidney tissue was degenerated and the glomerulus was atrophied. The number of mitochondria in the epithelial cells of first proximal convoluted tubule decreased with the increase of salinity. It is believed that the high osmotic pressure in seawater environment leads to the loss of water in fish and the decrease of the excretion function of kidney (Hallow et al., 2017). The reabsorption function of proximal convoluted tubule is degraded, which is also an adaptation of fish to different water environments.
When T. obscurus migrates from freshwater to seawater, it inevitably faces a hypertonic environment. It was found that ATP-binding cassette transport pathway and cAMP signaling pathway were significantly downregulated in gill and kidney. Extracellular signals (usually hormones) bind to receptors, regulate the level of cAMP, a common second messenger in cells, then induce cell reactions, and finally affect cell metabolism and behavior, which is an important process for cells to respond to extracellular signals quickly (Stutts et al., 1995; Jia and Liu, 2018). ATP-binding cassette family is intramembrane proteins superfamily. The energy released by ATP hydrolysis is supplied to various molecules for transmembrane transport, most of which were molecular channels regulated by cAMP. In hyperosmotic environment, teleost fish transports ions across the membrane through gill epithelial cells, enhancing chloride secretion efficiency of chloride cells, and reducing excessive loss of ions into cells. Maintaining water-salt balance relies on water reabsorption and excretion of hypertonic urine by kidney. Therefore, in hypertonic environment, the gills and kidneys of T. obscurus mainly rely on ATP-binding cassette transporter pathway for transmembrane transport of substances. CFTR gene in ATP-binding cassette family pathway was significantly upregulated. CFTR gene is a chloride channel regulated by cAMP and plays an important role in the transport of electrolytes and liquids (Hiroi and McCormick, 2012; Narayanan et al., 2020).
Under hypertonic conditions, the number of DEGs is highest in the gill. As T. obscurus was at a high-quality food environment during the experiment, the influence of diet on ionoregulatory processes might take a place. Liew et al. (2020) demonstrated common carp, Cyprinus carpio, prefers branchial ionoregulation at high feeding rates and kidney ionoregulation when food supply is limited. Functional annotation of DEGs in gills, kidneys, and intestines revealed that the number and expression genes of aquaporin family genes in three tissues significantly increased under hypertonic conditions. Among them, c75932_g1 was the highest expression and the greatest difference gene in gills and the predicted protein structure showed that the gene was aquaporin-9; c70360_g1 was the greatest differentiated gene in intestine, i.e., aquaporin-11. The number of expression aquaporin genes in kidney increased under hypertonic condition, but no significantly upregulated genes were found. The expression level of CFTR was different in three tissues. In gill and kidney, CFTR was significantly upregulated by hyperosmotic environment, while in intestine, the expression level of CFTR was significantly downregulated (mainly c70380_g1 and c72674_g1). In addition, the expression of other genes of solute carrier family, such as NKCC and NCC, was also analyzed. It was found that the expression of NKCC was the highest in intestine under hypertonic condition, mainly in NKCC1 gene: c75258_g1 and c75258_g2. The highest expression of NCC was also found in the intestine under hypertonic stress, which was NCC2 gene: c75258_g1 and c75258_g2. The previous study had shown that the high expression of NKCC and NCC in the foregut and hindgut of eel under hypertonic conditions plays an important role in the process of absorbing water by reducing the absorption of solutes in food and swallowing seawater (Watanabe et al., 2011). NKCC, NCC, and NHE use NKA to form Na+ concentration gradient difference and transport Na+ and Cl– inside and outside the cell (Shaughnessy and McCormick, 2020).
In teleost fish, hormones regulate osmotic organs to acquire the ability to adapt to environmental osmotic pressure. A large number of DEGs and pathways in the neuroendocrine system were found by enrichment and differential analysis of the transcriptional library of T. obscurus. In gill and kidney, the neuroactive ligand-receptor interaction pathway was significantly upregulated in hypertonic environment. In this study, we found that genes encoding hormones such as GH, PRL, IGF-1, and cortisol were significantly increased with the increase of salinity. These hormones regulated the synthesis of more transporter proteins and stimulate the proliferation and differentiation of chloride cells, thus changing the transmembrane transport capacity of ions and water molecules (Deane and Woo, 2007). Neuroendocrine pathways such as sphingolipid metabolism, steroid hormone synthesis, insulin secretion, and thyroid hormone signaling in gills were significantly upregulated. In this study, the expression of growth hormone-related hormone (YGHL1) and prolactin-related gene (PRLR1) in freshwater and seawater were both at high level, whereas the prolactin receptor gene (c67245_g1) was significantly downregulated in gill and upregulated in kidney and intestine under hypertonic stress, but the difference was not significant. PRL is an important regulatory hormone for euryhaline teleosts to adapt hypoosmotic environment. It can reduce the permeability of various organs and prevent excessive loss of ions in the body. Therefore, it has a higher expression level in freshwater than in seawater (Fuentes et al., 2010). IGF-1 receptor (IGF-1R) (c60000_g1) was significantly upregulated in gill, but not in other tissues. In addition, the upregulated Notch signaling pathway was also enriched in gill under hypertonic conditions. Notch gene encodes a highly conserved cell surface receptor, which affects many processes of normal cell morphogenesis, including differentiation of pluripotent progenitor cells, apoptosis, cell proliferation, and cell boundary formation. It may play a role in the differentiation of chloride cells in gill epithelium (Wong et al., 2017).
Takifugu obscurus can adapt to different salinity environments by its osmoregulation, which was relied on the active transport of solutes and water molecules (Shabala and Cuin, 2006). The transmembrane transport of substances is mostly accompanied by ATP output (Xu et al., 2017). It was found in this study that AMPK signaling pathway and TH signaling pathway were significantly upregulated in gill under hypertonic environment. AMPK plays an important role in coordinating metabolism and energy balance in cells and its activity is mainly regulated by the ratio of AMP to ATP (Hardi et al., 2012). The important role of AMPK in resisting hypertonic environment had been identified in Caenorhabditis elegans and yeast (Possik et al., 2015; Schutt and Moseley, 2017). TH signaling pathway is one of the indispensable signaling pathways to regulate the growth, development, and energy metabolism of organisms (Hönes et al., 2017). TH can promote growth and development, affect metabolism, increase oxygen consumption rate of most tissues, and increase thermogenic effect. This thermogenic effect may be due to the increased synthesis and activity of Na+-K+ pumps on cell membranes by THs (Rasheed et al., 2018). In addition, TCA pathway, the most important pathway of energy metabolism, is significantly upregulated by hypertonic environment in the kidney. This means that even with strong osmoregulation, euryhaline fish needs to consume a lot of capacity to adapt with hypertonic environment.
The changes in ultrastructure of gill also showed that the number of chloride-secreting cells increased significantly in saline solution. The increase of mitochondrial number in chloride-secreting cells may be related to the upregulation of CFTR gene, which regulates the metabolism of Cl– and transports Cl– to the outside of the organism in hypertonic environment. In kidney, the significant decrease in the biosynthesis of various amino acids in salinity may be associated with the atrophy of glomeruli and tubules.
For T. obscurus, when adapting to hypertonic environment, the structure of gill, intestine, and kidney changes substantially, which is the structure basis of osmoregulation and osmotic balance maintenance. Some signaling pathways (i.e., cAMP, AMPK, and TH signaling pathway), protein pathways, and various neuroendocrine hormones changed significantly in different tissues of T. obscurus, especially in gill and kidney, indicating that adapting to hypertonic environment is a complex process involving multiple organs and genes. This study provided essential information for enriching the basic theory of osmoregulation in euryhaline fish.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI (accession: PRJNA562326).
The animal study was reviewed and approved by the Institutional Animal Care and Use Committees (IACUC) of the Nanjing Normal University, Nanjing, China (Research permit number: SYXK2015-0028).
ZY and YC were responsible for the design of the experiments. JW and ZY wrote the manuscript. JW, XZ, YS, and LG performed the experiments. JW, YW, YC, and ZY conducted the data analysis and prepared figures and tables. All authors read and approved the final version of the manuscript.
This study was financially supported by the National Natural Science Foundation of China (52009082 and 31370429), the National Key Research and Development Project (2019YFC0408901), and the Priority Academic Program Development of Jiangsu Higher Education Institutions.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.854140/full#supplementary-material
Ando, M., Kim, H. T., Takase, I., and Kawahara, A. (2000). Imidazoline receptor contributes to ion and water transport across the intestine of the eel acclimated to sea water. Zool. Sci. 17, 307–312. doi: 10.2108/jsz.17.307
Autenrieth, M., Hartmann, S., Lah, L., Roos, A., Dennis, A. B., and Tiedemann, R. (2018). High quality whole genome sequence of an abundant Holarctic odontocete, the harbor porpoise (Phocoena phocoena). Mol. Ecol. Resour. 18, 1469–1481. doi: 10.1111/1755-0998.12932
Barany, A., Shaughnessy, C. A., Fuentes, J., Mancera, J. M., and McCormick, S. D. (2020). Osmoregulatory role of the intestine in the sea lamprey (Petromyzon marinus). Am. J. Physiol. Regul. Integr. Comp. Physiol. 318, 410–417. doi: 10.1152/ajpregu.00033.2019
Breves, J. P., Popp, E. E., Rothenberg, E. F., Rosenstein, C. W., Maffett, K. M., and Guertin, R. R. (2020). Osmoregulatory actions of prolactin in the gastrointestinal tract of fishes. Gen. Comp. Endocr. 298:113589. doi: 10.1016/j.ygcen.2020.113589
de Fonseka, R., Fjelldal, P. G., Sambraus, F., Nilsen, T. O., Remø, S. C., Stien, L. H., et al. (2022). Triploidy leads to a mismatch of smoltification biomarkers in the gill and differences in the optimal salinity for post-smolt growth in Atlantic salmon. Aquaculture 546:737350. doi: 10.1016/j.aquaculture.2021.737350
Deane, E. E., and Woo, N. Y. (2007). Impact of nitrite exposure on endocrine, osmoregulatory and cytoprotective functions in the marine teleost Sparus sarba. Aquat. Toxicol. 82, 85–93. doi: 10.1016/j.aquatox.2007.02.004
Drenner, S. M., Hinch, S. G., Furey, N. B., Clark, T. D., Li, S. R., Ming, T. B., et al. (2018). Transcriptome patterns and blood physiology associated with homing success of sockeye salmon during their final stage of marine migration. Can. J. Fish. Aquat. Sci. 75, 1511–1524. doi: 10.1139/cjfas-2017-0391
Fridman, S. (2020). Ontogeny of the Osmoregulatory Capacity of Teleosts and the Role of Ionocytes. Front. Mar. Sci. 7:709. doi: 10.3389/fmars.2020.00709
Fuentes, J., Brinca, L., Guerreiro, P. M., and Power, D. M. (2010). PRL and GH synthesis and release from the sea bream (Sparus auratus L.) pituitary gland in vitro in response to osmotic challenge. Gen. Comp. Endocr. 168, 95–102. doi: 10.1016/j.ygcen.2010.04.005
Gao, Q. X., Min, M. H., Zhang, C. J., Peng, S. M., and Shi, Z. H. (2017). Influence of salinity on juvenile obscure puffer Takifugu obscurus. J. Appl. Anim. Res. 45, 292–297. doi: 10.1080/09712119.2016.1174123
Giffard-Mena, I., Hernandez-Montiel, A. H., Perez-Robles, J., and David-True, C. (2020). Effects of salinity on survival and plasma osmolarity of Totoaba macdonaldi eggs, larvae, and juveniles. J. Exp. Mar. Biol. Ecol. 526:151339. doi: 10.1016/j.jembe.2020.151339
Hajirezaee, S., Mirvaghefi, A. R., Farahmand, H., and Agh, N. (2017). A metabolic approach to understanding adaptation to sea water by endangered Persian sturgeon, Acipenser persicus fingerlings. Aquac. Res. 49, 341–351. doi: 10.1111/are.13464
Hallow, K. M., Gebremichael, Y., Helmlinger, G., and Vallon, V. (2017). Primary proximal tubule hyperreabsorption and impaired tubular transport counterregulation determine glomerular hyperfiltration in diabetes: a modeling analysis. Am. J. Physiol. Renal. Physiol. 312, 819–835. doi: 10.1152/ajprenal.00497.2016
Hardi, D. G., Ross, F. A., and Hawley, S. A. (2012). AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat. Rev. Mol. Cell Biol. 13, 251–262. doi: 10.1038/nrm3311
Hiroi, J., and McCormick, S. D. (2012). New insights into gill ionocyte and ion transporter function in euryhaline and diadromous fish. Respir. Physiol. Neurobiol. 184, 257–268. doi: 10.1016/j.resp.2012.07.019
Hönes, G. S., Rakov, H., Logan, J., Liao, X. H., Werbenko, E., Pollard, A. S., et al. (2017). Noncanonical thyroid hormone signaling mediates cardiometabolic effects in vivo. Proc. Natl. Acad. Sci. U. S. A. 114, 11323–11332. doi: 10.1073/pnas.1706801115
Hui, M., Liu, Y., Song, C. W., Li, Y. D., Shi, G. H., and Cui, Z. X. (2014). Transcriptome changes in Eriocheir sinensis megalopae after desalination provide insights into osmoregulation and stress adaption in larvae. PLoS One 9:e114187. doi: 10.1371/journal.pone.0114187
Hwang, P. P., and Lee, T. H. (2007). New insights into fish ion regulation and mitochondrion-rich cells. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 148, 479–497. doi: 10.1016/j.cbpa.2007.06.416
Jia, Y. L., and Liu, X. (2018). Expression of Na+/K+-ATPase was affected by salinity change in pacific abalone Haliotis discus hannai. Front. Physiol. 9:1244. doi: 10.3389/fphys.2018.01244
Kato, A., Doi, H., Nakada, T., Sakai, H., and Hirose, S. (2005). Takifugu obscurus is a euryhaline fugu species very close to Takifugu rubripes and suitable for studying osmoregulation. BMC Physiol. 5:18. doi: 10.1186/1472-6793-5-18
Kim, J. H., Kim, S. J., Min, G. S., and Han, K. N. (2008). Nutritional condition determined using RNA/DNA ratios of the river pufferfish Takifugu obscurus under different salinities. Mar. Ecol. Prog. Ser. 372, 243–252. doi: 10.3354/meps07704
Lam, S. H., Lui, E. Y., Li, Z. J., Cai, S. J., Sung, W. K., Mathavan, S., et al. (2014). Differential transcriptomic analyses revealed genes and signaling pathways involved in iono-osmoregulation and cellular remodeling in the gills of euryhaline Mozambique tilapia, Oreochromis mossambicus. BMC Genomics 15:921. doi: 10.1186/1471-2164-15-921
Liew, H. J., Pelle, A., Chiarella, D., Faggio, C., Tang, C.-H., Blust, R., et al. (2020). Common carp, Cyprinus carpio, prefer branchial ionoregulation at high feeding rates and kidney ionoregulation when food supply is limited: additional effects of cortisol and exercise. Fish Physiol. Biochem. 46, 451–469. doi: 10.1007/s10695-019-00736-0
Lv, J. J., Liu, P., Wang, Y., Gao, B. Q., Chen, P., and Li, J. (2013). Transcriptome analysis of Portunus trituberculatus in response to salinity stress provides insights into the molecular basis of osmoregulation. PLoS One 8:e82155. doi: 10.1371/journal.pone.0082155
Marshall, W. S. (2011). Mechanosensitive signalling in fish gill and other ion transporting epithelia. Acta Physiol. 202, 487–499. doi: 10.1111/j.1748-1716.2010.02189.x
McCormick, S. D. (2001). Endocrine control of osmoregulation in teleost fish. Am. Zool. 41, 781–794.
Moorman, B. P., Inokuchi, M., Yamaguchi, Y., Lerner, D. T., Grau, E. G., and Seale, A. P. (2014). The osmoregulatory effects of rearing Mozambique tilapia in a tidally changing salinity. Gen. Comp. Endocr. 207, 94–102. doi: 10.1016/j.ygcen.2014.03.013
Narayanan, V., Schappell, L. E., Mayer, C. R., Duke, A. A., Armiger, T. J., Arsenovic, P. T., et al. (2020). Osmotic gradients in epithelial acini increase mechanical tension across E-cadherin, drive morphogenesis, and maintain homeostasis. Curr. Biol. 30, 624–633. doi: 10.1016/j.cub.2019.12.025
Nguyen, T. V., Jung, H., Nguyen, T. M., Hurwood, D., and Mather, P. (2016). Evaluation of potential candidate genes involved in salinity tolerance in striped catfish (Pangasianodon hypophthalmus) using an RNA-Seq approach. Mar. Genom. 25, 75–88. doi: 10.1016/j.margen.2015.11.010
Pérez-Robles, J., Re-Araujo, A. D., Cortez-Garcia, M., Diaz, F., Ibarra-Castro, L., and Giffard-Mena, I. (2019). Fluctuating salinity effect on Sphoeroides annulatus (Jenyns 1842) physiological responses. Aquac. Res. 50, 198–208. doi: 10.1111/are.13884
Possik, E., Ajisebutu, A., Manteghi, S., Gingras, M. C., Vijayaraghavan, T., Flamand, M., et al. (2015). FLCN and AMPK confer resistance to hyperosmotic stress via remodeling of glycogen stores. PLoS Genet. 11:e1005520. doi: 10.1371/journal.pgen.1005520
Rasheed, E., Seheult, J., Gibney, J., and Boran, G. (2018). Does thyrotoxic periodic paralysis have a genetic predisposition? A case report. Ann. Clin. Biochem. 55, 713–716. doi: 10.1177/0004563218785395
Regish, A. M., Kelly, J. T., O’Dea, M. F., and McCormik, S. D. (2018). Sensitivity of Na+/K+-ATPase isoforms to acid and aluminum explains differential effects on Atlantic salmon osmoregulation in fresh water and seawater. Can. J. Fish. Aquat. Sci. 75, 1319–1328. doi: 10.1139/cjfas-2017-0198
Sadoul, B., Alfonso, S., Cousin, X., Prunet, P., Begout, M. L., and Leguen, I. (2021). Global assessment of the response to chronic stress in European sea bass. Aquaculture 544:737072. doi: 10.1016/j.aquaculture.2021.737072
Sakamoto, T., and McCormick, S. D. (2006). Prolactin and growth hormone in fish osmoregulation. Gen. Comp. Endocr. 147, 24–30. doi: 10.1016/j.ygcen.2005.10.008
Schutt, K. L., and Moseley, J. B. (2017). Transient activation of fission yeast AMPK is required for cell proliferation during osmotic stress. Mol. Biol. Cell 28, 1804–1814. doi: 10.1091/mbc.E17-04-0235
Shabala, S. N., and Cuin, T. A. (2006). “Osmoregulation vs osmoprotection: re-evaluating the role of compatible solutes,” in Floriculture, Ornamental and Plant Biotechnology – Advances and Topical Issues, ed. J. Teixeira da Silva (Tokyo: Global Science Books), 405–416.
Shaughnessy, C. A., and McCormick, S. D. (2020). Functional characterization and osmoregulatory role of the Na+-K+-2Cl– cotransporter in the gill of sea lamprey (Petromyzon marinus), a basal vertebrate. Am. J. Physiol. Regul. Integr. Comp. Physiol. 318, 17–29. doi: 10.1152/ajpregu.00125.2019
Stutts, M. J., Canessa, C. M., Olsen, J. C., Hamrick, M., Cohn, J. A., Rossier, B. C., et al. (1995). CFTR as a cAMP-dependent regulator of sodium channels. Science 269, 847–850. doi: 10.1126/science.7543698
Venkatesh, B., Gilligan, P., and Brenner, S. (2000). Fugu: a compact vertebrate reference genome. FEBS Lett. 476, 3–7. doi: 10.1016/S0014-5793(00)01659-8
Wang, J., Tang, H. X., Zhang, X. X., Xue, X. F., Zhu, X. X., Chen, Y. F., et al. (2018). Mitigation of nitrite toxicity by increased salinity is associated with multiple physiological responses: a case study using an economically important model species, the juvenile obscure puffer (Takifugu obscurus). Environ. Pollut. 232, 137–145. doi: 10.1016/j.envpol.2017.09.026
Wang, J., Zhu, X. X., Huang, X., Gu, L., Chen, Y. F., and Yang, Z. (2016). Combined effects of cadmium and salinity on juvenile Takifugu obscurus: cadmium moderates salinity tolerance; salinity decreases the toxicity of cadmium. Sci. Rep. 6:30968. doi: 10.1038/srep30968
Watanabe, S., Mekuchi, M., Ideuchi, H., Kim, Y. K., and Kaneko, T. (2011). Electroneutral cation-Cl–, cotransporters NKCC2β and NCCβ expressed in the intestinal tract of Japanese eel Anguilla japonica. Comp. Biochem. Phys. A Mol. Integr. Physiol. 159, 427–435. doi: 10.1016/j.cbpa.2011.04.009
Wong, M. K. S., Tsukada, T., Ogawa, N., Pipil, S., Ozaki, H., Suzuki, Y., et al. (2017). A sodium binding system alleviates acute salt stress during seawater acclimation in eels. Zool. Lett. 3:22. doi: 10.1186/s40851-017-0081-8
Xu, Y. Y., Seelig, A., and Berneche, S. (2017). Unidirectional Transport Mechanism in an ATP Dependent Exporter. ACS Central Sci. 3, 250–258. doi: 10.1021/acscentsci.7b00068
Yang, Z., and Chen, Y. F. (2005). Effect of temperature on incubation period and hatching success of obscure puffer Takifugu obscurus (Abe) eggs. Aquaculture 246, 173–179. doi: 10.1016/j.aquaculture.2004.12.030
Yang, Z., and Chen, Y. F. (2006). Salinity tolerance of embryos of obscure puffer Takifugu obscurus. Aquaculture 253, 393–397. doi: 10.1016/j.aquaculture.2005.08.014
Yi, H. D., Chen, X. L., Liu, S., Han, L. Q., Liang, J. H., Su, Y. Q., et al. (2021). Growth, osmoregulatory and hypothalamic–pituitary–somatotropic (HPS) axis response of the juvenile largemouth bass (Micropterus salmoides), reared under different salinities. Aquacult. Rep. 20:100727. doi: 10.1016/j.aqrep.2021.100727
Keywords: osmoregulation, salinity, Takifugu obscurus, transcriptome, ultrastructure
Citation: Wang J, Zhu XX, Sun YF, Gu L, Wu YP, Chen YF and Yang Z (2022) Changes in Transcriptome and Ultrastructure Reveal Salinity Tolerance of Obscure Puffer Takifugu obscurus. Front. Mar. Sci. 9:854140. doi: 10.3389/fmars.2022.854140
Received: 13 January 2022; Accepted: 10 February 2022;
Published: 17 March 2022.
Edited by:
Xiaodan Wang, East China Normal University, ChinaReviewed by:
Hon Jung Liew, Universiti Malaysia Terengganu, MalaysiaCopyright © 2022 Wang, Zhu, Sun, Gu, Wu, Chen and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yafen Chen, Y2hlbnlmQG5pZ2xhcy5hYy5jbg==; Zhou Yang, eWFuZ3pob3VAbmpudS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.