
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 14 March 2022
Sec. Marine Fisheries, Aquaculture and Living Resources
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.851098
This article is part of the Research TopicMollusk Breeding and Genetic ImprovementView all 25 articles
Hypoxia is considered to be one of the key factors affecting the survival of ocean organisms, it is necessary to parse the molecular processes involved in response to hypoxia. As a potential breeding species, the hybrid of Crassostrea sikamea (♀) × Crassostrea gigas (♂) shows valuable heterosis in survival and growth traits. Thus, RNA de novo was deployed in this study to analyze the molecular processes in the hybrids under hypoxia stress. The hybrids were cultured in occluded water, then the dissolved oxygen was gradually consumed by oysters, and the gill tissue of hybrids was sampled at the very beginning and the lowest respiration point in the experiment. In the current study, 901 significant differentially expressed genes (DEGs) were identified under hypoxia compared to normoxia, among which 432 DEGs were downregulated, and the other 469 DEGs were upregulated. A total of 27 GO terms were significantly enriched, such as an integral component of membrane, extracellular region, immune response, tumor necrosis factor receptor binding, and neurotransmitter: sodium symporter activity. Besides, 19 KEGG pathways were significantly enriched, such as apoptosis, Th1 and Th2 cell differentiation, complement, and coagulation cascades, antigen processing and presentation, notch signaling pathway, and cytokine–cytokine receptor interaction. The current results showed that the TRAIL genes were downregulated, but the HSP70 and LIGHT genes were upregulated, which indicated the inhibition of Apoptosis, and the activity of innate immunity in oysters under hypoxia. This study provides preliminary insight into the molecular response to hypoxia in the gill of hybrids.
Mollusks, including oysters, serve as indeed important aquaculture resources providing considerable economic value for the coastal areas. Hybridization could be a considerable way to enhance yield or survival due to better performance than the half-siblings, which is known as heterosis (Hedgecock and Davis, 2007). Recently, the studies on oyster interspecific crossbreeding have been well documented (Xu et al., 2011, 2019; Huo et al., 2014; Zhang et al., 2017), and some of these hybrids have shown adaptabilities or better growth traits. Previous studies only focused on growth performance, while identifying the changes in physiological mechanisms that respond to environmental stress is critical to help determine the growth and survival of individuals and the yield potential in aquaculture (Furr et al., 2021).
In recent years, with the development of the oyster industry summer mortality has been one of the most severe questions in farming practice. Previous studies indicated that the two major reasons for bivalve mass mortality were temperature increasing and dissolved oxygen decreasing (Parthasarathy et al., 1992; Joos et al., 2003; Soon and Zheng, 2019; Zhao et al., 2019; He et al., 2021), and hypoxia is considered to be one of the key factors affecting the survival of ocean organisms (Gu et al., 2019; Andreyeva et al., 2021). Benthic intertidal communities have developed appropriate survival mechanisms due to daily exposure into the air during low tide (Larade and Storey, 2002; Zhao et al., 2020), but over-farming, sediment covering, or algae blooming (Wu, 2002) could also cause water hypoxia. Recent studies showed that the fluctuations of dissolved oxygen frequently influence physiological, metabolic, and cellular processes in organisms, and in this situation, oxidative stress becomes one of the reasons that cause cellular damage (Nogueira et al., 2017; Andreyeva et al., 2021).
The heterosis of survival and maternal comparable growth traits have been reported in Crassostrea gigas and Crassostrea sikamea hybrids (Xu et al., 2019). According to our previous study, the hybrids have lower mortality than both parents when they were cultured in southern China, particularly the sire-siblings, where the annual water temperature is high (Zhang et al., 2021). Therefore, hybrids could be a potential breeding species. Gills of bivalves are in direct contact with seawater and therefore are the first issue facing the damaging effect of hypoxia, besides, the gill is an important organ involved in the innate immunity of shellfish among a variety of organs (Seo et al., 2013; Guo et al., 2015; Wang et al., 2018). The antioxidant complex in the gills of bivalves has been fully characterized in several studies (Bo-Mi et al., 2018; Box et al., 2020), and the expressions of stress response-related adapting genes could also be triggered by environmental threats in oyster gills (Guo et al., 2015).
The purpose of this experiment was to explore the molecular mechanism of hypoxia tolerance in gills of C. sikamea × C. gigas hybrids. RNA de novo was used to analyze the transcriptome in gills under hypoxia stress. This study provides preliminary insight into the molecular response to hypoxia in the gill of hybrids.
The hybrids of C. sikamea (♀) × C. gigas (♂) that were 12 months old were used in the experiment with a shell length of 43.19 ± 6.71 mm and weight of 15.17 ± 3.71 g (mean ± SD). The hybrids were cultured in Beihai, China, in 2018, as described by Zhang et al. (2021). A total of 60 healthy non-injured hybrids were selected and reared in filtered, aerated water with the temperature of 26.00 ± 0.11°C, dissolved oxygen of 6.12 ± 0.11 mg/L, the salinity of 30.56 ± 0.02 ppt, and pH 8.18 ± 0.02 for 2 weeks of acclimatization, and fed with a mixture of Isochrysis galbana and Chaetoceros muelleri.
The experiment was performed in June 2019 and the oysters were placed in three 30 L tanks with filtered seawater. Three tanks were used for each control group and test group. About 20 oysters were placed in each tank in separate small cases. Control groups were 3 sealed tanks without oysters to test the changes of dissolved oxygen in the water itself. All groups were being aerated till the system was in a steady state, then stop aerating. To avoid the exchange of air, floating plastic membranes were placed on the surface of the water. Hypoxia condition occurred in the 3 test tanks due to dissolved oxygen consumed by oysters. Each tank was connected to a dissolved oxygen detector to monitor the water condition. The experiment lasted for 60 h and recorded the dissolved oxygen every 2 h. As dissolved oxygen decreases, the oxygen consumption rate of the oyster changes. It was observed that the dissolved oxygen in the occluded water was lower than 3.0 mg/L after 36 h, and decreased to less than 2.5 mg/L after 48 h, and then the oxygen consumption level of oysters was extremely low (Figure 1). This occluded water was chosen to mimic the hypoxia water conditions. According to the national standard of water quality, the ammonia–nitrogen in water was below 0.02 mg/L (minimum detection limit) before and after the experiment determined by gas-phase molecular absorption spectrometry (Standard No: HJ/T 195-2005).
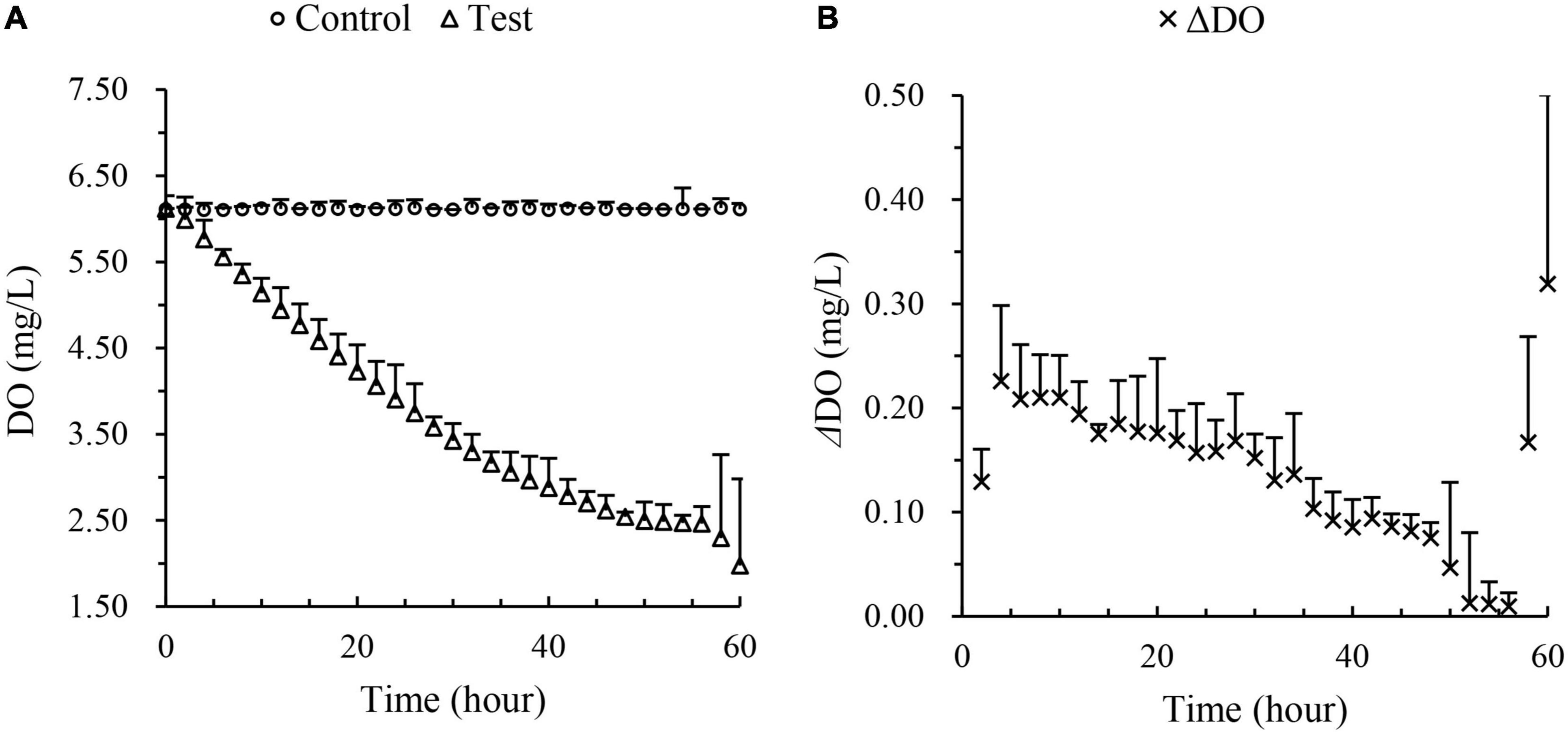
Figure 1. Dissolved oxygen in occluded water with time: (A) The average DO of control groups and test groups; (B) the average Δ DO in the test groups between two successive recording points.
Oysters were sampled under the steady state (normoxia) and DO less than 2.5 mg/L (hypoxia) in occluded water (around 54 h culturing), respectively. The gills of six oysters were sampled, pooled, and stored at −80°C until total RNA was extracted.
Total RNA was extracted from the normoxia and hypoxia gills using TRIzol (Invitrogen, Carlsbad, CA, United States). RNA samples were qualified and quantified using a NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE, United States) and the Agilent Technologies 2100 Bioanalyzer (Thermo Fisher Scientific, Waltham, MA, United States).
The mRNA was purified by removing potential DNA pollution with DNase I, removing possible rRNA pollution with RNase H, and finally recovered with Oligo(dT)-attached magnetic beads (Illumina, San Diego, CA, United States). Purified mRNA was fragmented with a fragment buffer at an appropriate temperature. Then, first-/second-strand cDNA was generated by polymerase chain reaction. The A-Tailing Mix and RNA index adapters were added and incubated to carry out end-repair. The cDNA fragments with adapters were amplified, and the products were purified with Ampure XP beads. The library was validated on the Agilent Technologies 2100 Bioanalyzer for quality control. The final library was amplified with phi29 to create DNA nanoballs (DNB), containing more than 300 copies of the original template. The DNBs were loaded into the patterned nanoarray, and pair-end 100 base reads were generated on a BGISEQ500 platform (BGI, Shenzhen, China).
The filtering software SOAPnuke v1.4.0 developed by BGI was used for statistical analysis and filtering. Raw reads with joint contamination and high numbers of unknown N bases were removed before the data analysis to ensure the reliability of the results. Then, the Q20/Q30 and GC-content of the clean data were measured. Clean reads were assembled with Trinity v2.0.6 software, and the transcripts were clustered with Tgicl v 2.1 to generate unigenes. The quality of the assembled transcripts was assessed using Benchmarking Universal Single-Copy Orthologs (BUSCO), a single-copy lineal homology database, and the integrity of the transcriptome assembly was demonstrated by comparison with conserved genes.
The fragments per kilobase per million reads (FPKM) method was used to calculate the expression levels. Bowtie2 v2.2.5 software was used to compare clean reads with the reference gene sequence, and then RSEM was used to calculate the expression levels of the unigenes and transcripts. The prediction and annotation data of all unigenes were compared with protein databases, including NCBI non-redundant protein sequences and nucleotide sequences (Nr&Nt),1 Gene Ontology (GO),2 Kyoto Encyclopedia of Genes and Genomes (KEGG),3 Clusters of Orthologous Groups of proteins/eukaryotic Orthologous Groups (COG/KOG),4 Swiss-Prot,5 and Protein family (Pfam).6 Hmmscan v3.0, Blast v2.2.23, and Blast2GO software were used to obtain the annotation information for the unigenes. When the annotation of different databases conflicted, the priority order of alignments from the Nr, Nt, KEGG, Swiss-Prot, GO, COG, and Pfam databases were followed.
In this study, the FPKM method was used to calculate the expression of unigenes so that to compare gene expression levels between samples. The FDR (false discovery rate) was applied to identify the threshold of the p-value in multiple tests (Mortazavi et al., 2008). The threshold of significantly expressed genes was set as FPKM ≥ 1, |log2(foldchange)| ≥ 2, FDR < 0.05. Then, GO and KEGG enrichment was performed to analyze the main biochemical and signal transduction pathways by using p < 0.01 as the threshold of significantly enriched differentially expressed genes (DEGs).
Total RNA was reverse-transcribed into cDNA with the PrimeScript RT Reagent Kit (Takara, Beijing, China). Eight typical DEGs, tumor necrosis factor (TNF) superfamily member 10 (TNFSF10), meprin B (MEP1B), histone deacetylase 11 (HDAC11), Notch 1 (NOTCH1), caspase 7 (CASP7), mucin-2 (MUC2), plasminogen (PLG), interferon regulatory factor 2 (IRF2), and a reference gene (tubulin α, TUBG) were selected for verification by qRT-PCR.
Gene-specific primers were designed with Primer Premier 5.0 software. All real-time PCR experiments were carried out in 96-well PCR plates. Each 20 μL amplification reaction was conducted using the StepOne Plus system (ABI, Foster City, CA, United States) in triplicate. The PCR process included 95°C for 3 min; 45 cycles of 95°C for 5 s, and 60°C for 30 s; followed by a melting curve. The differentiation expression was calculated with the method by Livak and Schmittgen (2001). The expression fold change (2–ΔΔCT) of the qRT-PCR data and Log2(foldchange) of the RNA-seq data were used, and correlation was calculated in Spearman analysis.
The statistical information of sequencing data for the normoxia and hypoxia samples is given in Table 1. The average number of total raw reads generated from each library was 66.84 million, and the average clean reads were 64.03 million. The total mapping rate of the library was 81.11–83.21%, and the average Q30 value was 89.49. In total, 84,359 unigenes were assembled into six libraries with an average unigene number of 54,010, with a mean length of 932 bp. The average GC percentage of unigene was 39.67%, and the N50 was 1,640 bp.
According to the evaluation of transcript assembly, more than 85% of unigenes matched or partially matched into the BUSCO database, and only around 2.31% were unmatched (Figure 2). The results showed that transcription data could be used for annotation analysis. All raw reads were deposited in the NCBI Short Read Archive database with an Accession number of PRJNA587775.
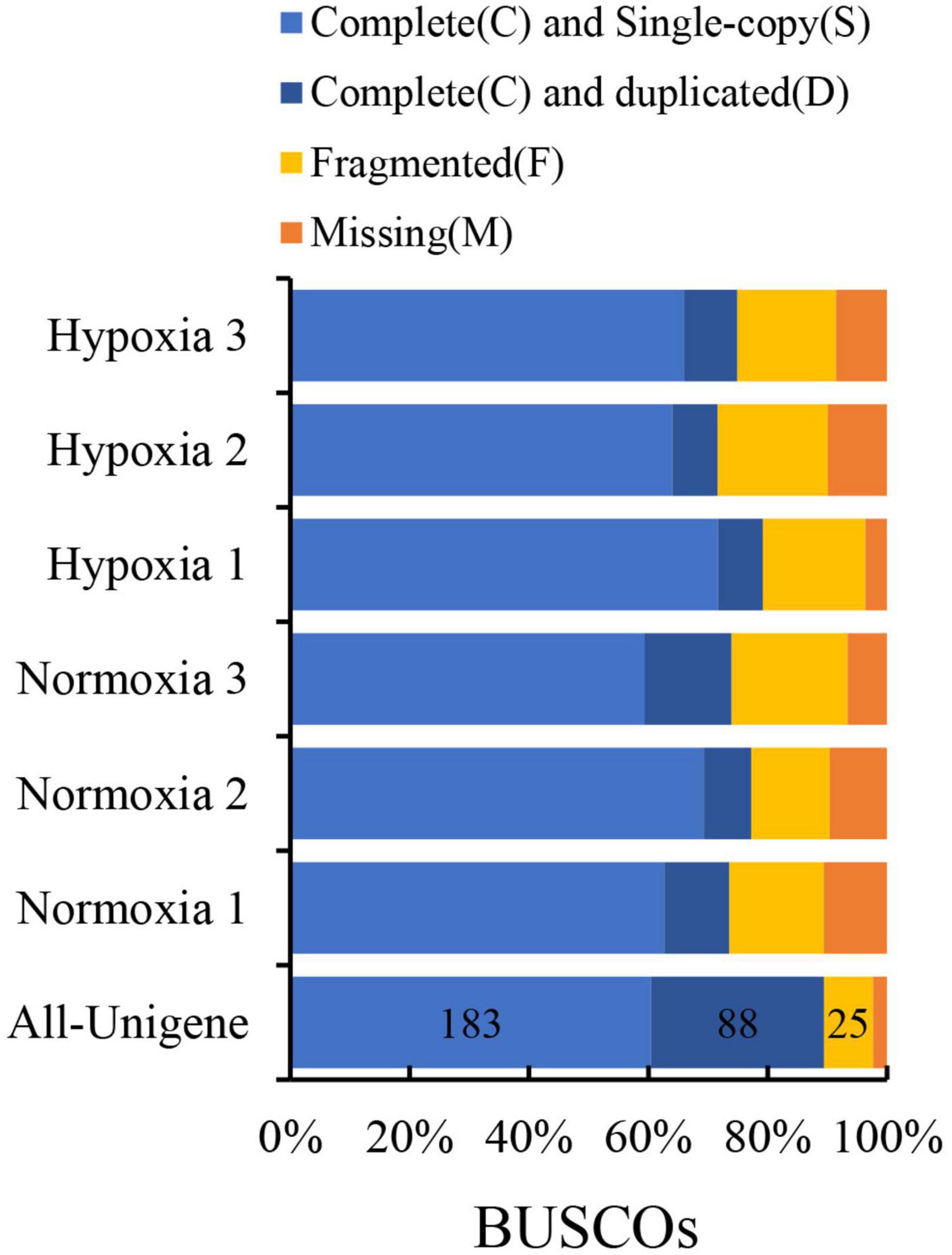
Figure 2. Evaluation of transcript assembly by Benchmarking Universal Single-Copy Orthologs (BUSCO).
To identify the categories and functions of transcriptional sequences, all unigenes were blasted into seven functional databases. Among which 56,071 unigenes in the NR protein sequence database (66.47%), 67,797 unigenes in the NT database (80.37%), 32,377 unigenes in the Swiss-Prot (38.38%), 39,082 unigenes in the KEGG (46.33%), 29,614 unigenes in the KOG (35.10%), 38,026 unigenes in the Pfam (45.08%), and 29,415 unigenes in the GO (34.87%) were annotated. Overall, 72,724 (86.21%) unigenes matched to at least one database, and 15,016 (17.80%) unigenes were annotated into all seven databases (Table 2). There were five species in the NR annotations, including C. gigas, Crassostrea virginica, Mizuhopecten yessoensis, Lottia gigantea, and Apostichopus japonicus, with the annotation rates of 89.88% (50,396), 6.39% (3,581), 0.65% (365), 0.14% (81), and 0.11% (64), respectively (Supplementary Table 1).

Table 2. The annotation number and proportion of unigenes in NR, NT, Swiss-prot, KEGG, KOG, Pfam, and GO databases.
The classification of 25 KOG gene homologs was identified, and the top three annotated terms were signal transduction mechanisms (5,451), general function prediction only (5,066), and post-translational modifications, protein turnover, and chaperones (3,088) (Figure 3).
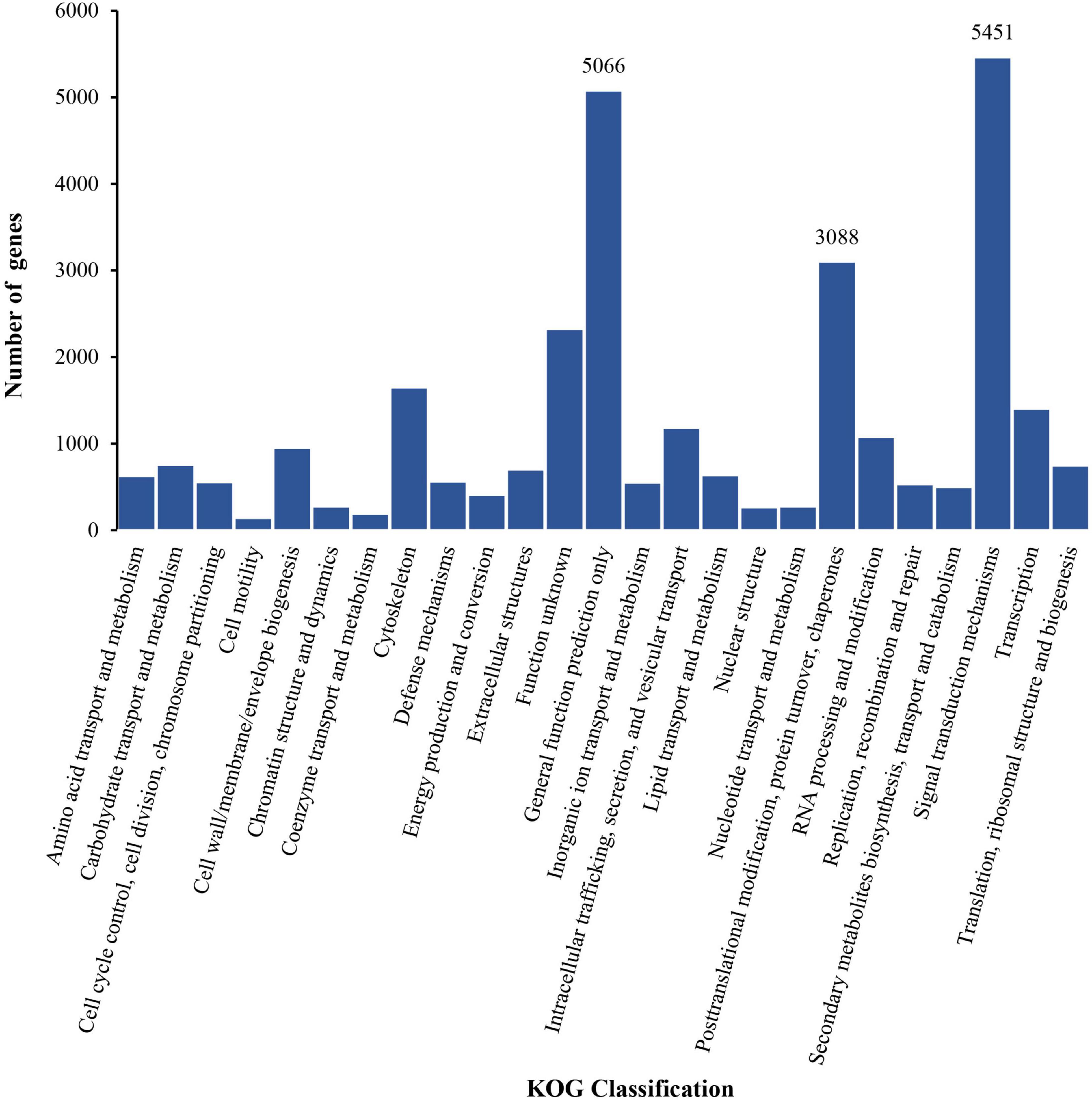
Figure 3. The annotated classification information of unigene sequence on the gene homologs from the Eukaryotic Orthologous Groups (KOG) database.
In total, 901 DEGs were identified in the analysis between the normoxia and hypoxia groups. Among these genes, 432 DEGs were downregulated and 469 were upregulated (Figure 4). The GO annotation assigns all DEGs into three major functional categories: molecular functions, cellular components, and biological processes. Overall, 325 genes were grouped into molecular functions, that is, catalytic activity (116 genes) and binding sectors (154 genes); 531 genes were grouped into cellular components, that is, membrane part (163 genes) and membrane (172 genes); 292 genes were grouped into biological processes, that is, metabolic process (56 genes) and cellular process (73 genes), respectively (Figure 5).
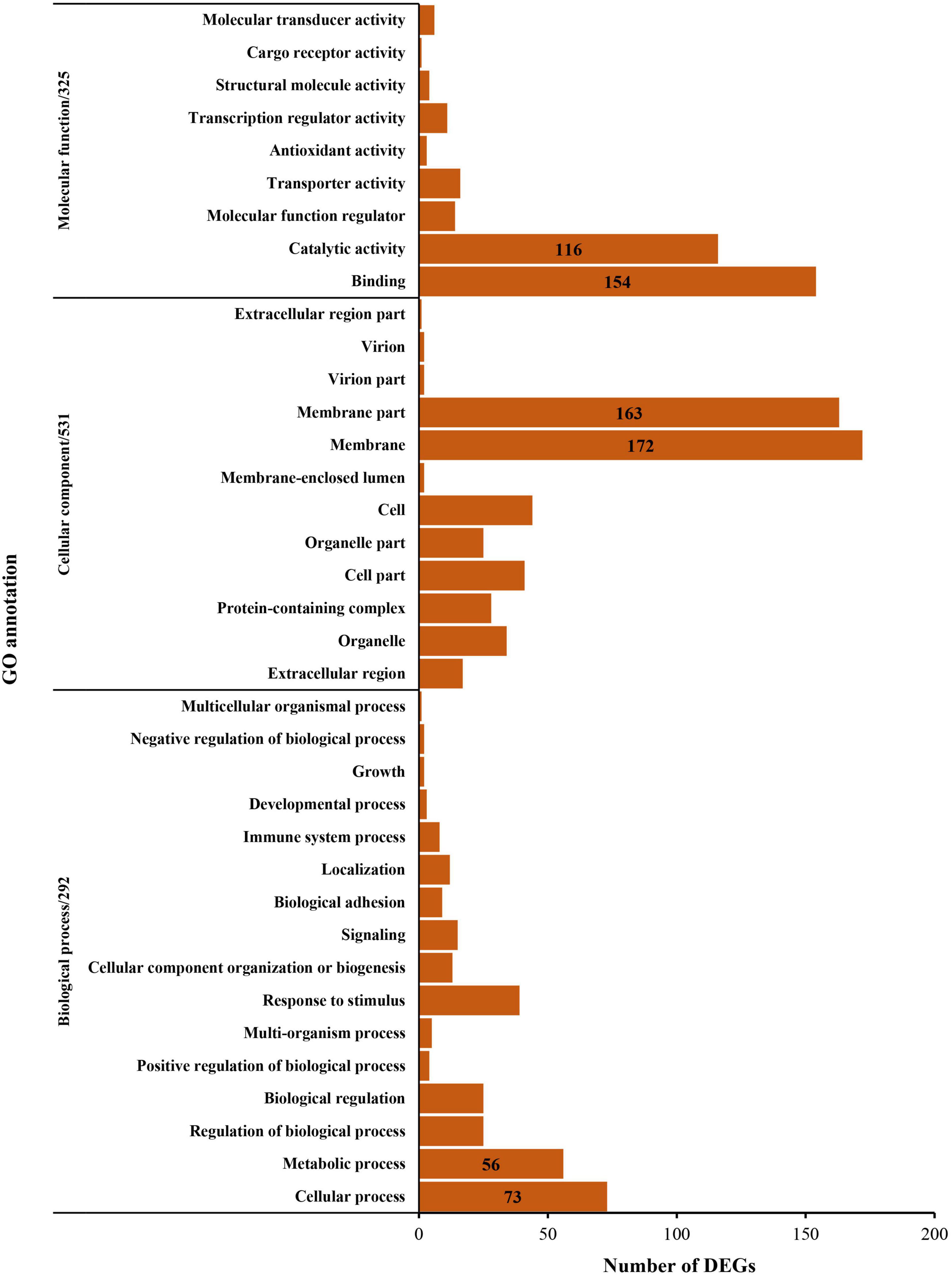
Figure 5. Gene Ontology analysis of the differentially expressed genes between the normoxia and hypoxia groups.
The KEGG annotation assigns all DEGs into six major functional categories: organismal systems, metabolism, human diseases, genetic information processing, environmental information processing, and cellular processes. Overall, 317 genes were grouped into organismal systems, that is, endocrine system (72 genes) and immune system (108 genes); 136 genes were grouped into a metabolism, that is, global overview maps (51 genes); 444 genes were grouped into human diseases, that is, cancers: specific types (71 genes), cancers: overview (77 genes), and infectious diseases: viral (77 genes); 70 genes were grouped into genetic information processing, that is, folding, sorting, and degradation (31 genes); 147 genes were grouped into environmental information processing, that is, signal transduction (109 genes); 139 genes were grouped into cellular processes, that is, transport catabolism (46 genes), respectively (Figure 6).
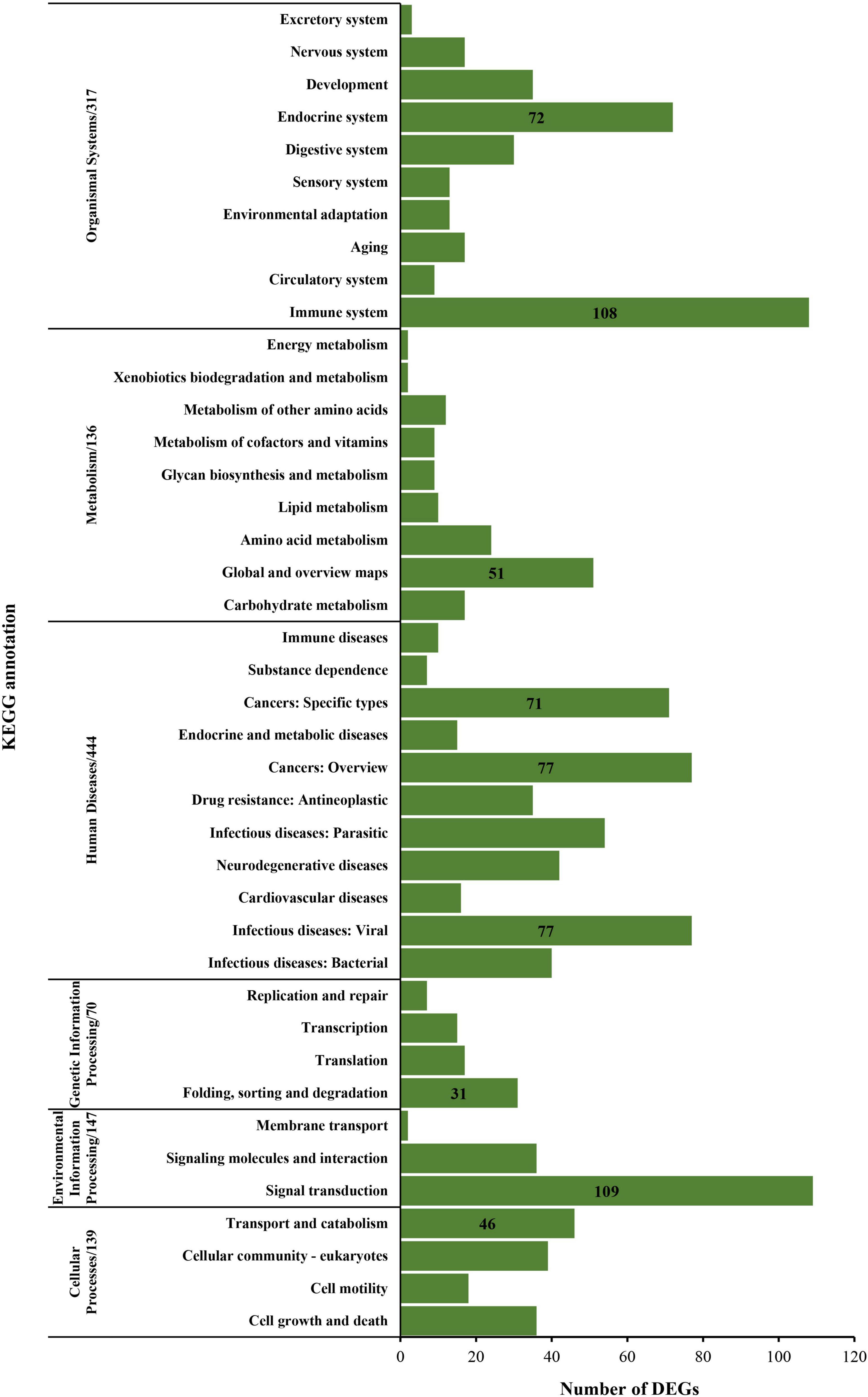
Figure 6. KEGG annotation of the differentially expressed genes between the normoxia and hypoxia groups.
The GO analysis was deployed to clarify the changes of the DEGs in the hybrids under hypoxia stress. Overall, 27 GO terms were significantly enriched, and the order of the GO term enriching number in the different categories was BP (13 terms) > MF (12 terms) > CC (2 terms). The primary enriched GO terms that included the GO:0016021_Integral component of membrane (e.g., GLYT, NCOR2, and SR140), GO:0005576_Extracellular region (e.g., TIMP3, MUC2, and FAT4), GO:0006955_Immune response (e.g., DPCD and TRAIL), GO:0005164_Tumor necrosis factor receptor binding (e.g., TNF and LIGHT), GO:0005328_Neurotransmitter: sodium symporter activity (e.g., PROT and SLC6A5_9), and the enriched number of DEGs into the above GO terms counting for 157, 16, 7, 7, and 5 DEGs, respectively (Figure 7). The lists of these representative DEGs and their functional results are shown in Supplementary Tables 3, 4.
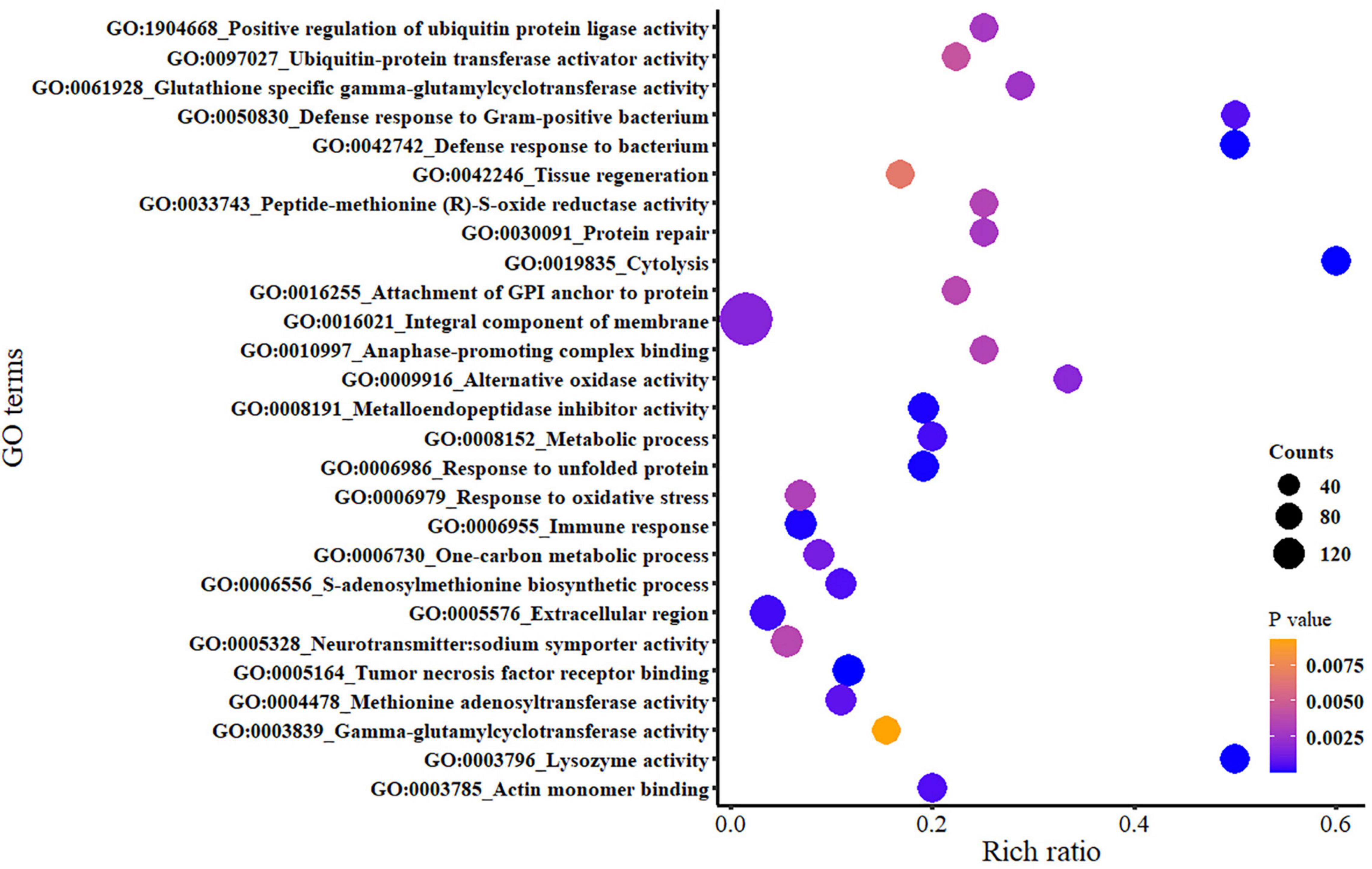
Figure 7. The Gene Ontology enrichment of the differentially expressed genes (DEGs) between the normoxia group and hypoxia group. The color of the marker represents the significance; the square area represents the number of DEGs enriched in each pathway; the x coordinate represents the rich ratio (the ratio of the number of genes annotated to an item in the selected gene set to the number of genes annotated to the item by the species), similarly hereinafter.
The KEGG analysis was deployed to investigate the predicted functional information of DEGs in the gills under hypoxia stress. The KEGG analysis mainly focused on cellular processes, environmental information processing, genetic information processing, human diseases, metabolism, and organismal systems in KEGG classification level 1. The analysis of the KEGG enrichment analysis provides an overview of the pathway regulation.
In this study, the DEGs were assigned to 19 KEGG pathways, among which 43, 40, 28, 26, 24, 16, 11, and 8 genes were grouped into signal transduction, immune system, endocrine system, development, cell growth and death, folding, sorting and degradation, aging, and signaling molecules and interaction, respectively, in KEGG classification level 2. The main enriched pathways included the ko04210_Apoptosis (e.g., IAP/XIAP, CASP7, and PARP), ko04658_Th1 and Th2 cell differentiation (e.g., NOTCH1/2/3), ko04610_Complement and coagulation cascades (e.g., A2M, PLG, and C1QG), ko04612_Antigen processing and presentation (e.g., HSPA1s, and CTSL), ko04330_Notch signaling pathway (e.g., NUMBL, NOTCH, and SMRT), ko04060_Cytokine–cytokine receptor interaction (e.g., TNFSF10/14, XDEAR, and SF19). Among the listed pathways above, the ko04330_Notch signaling pathway enriched the most number of genes as 25 (Figure 8). The lists of these representative DEGs and their functional results are shown in Supplementary Tables 5, 6.
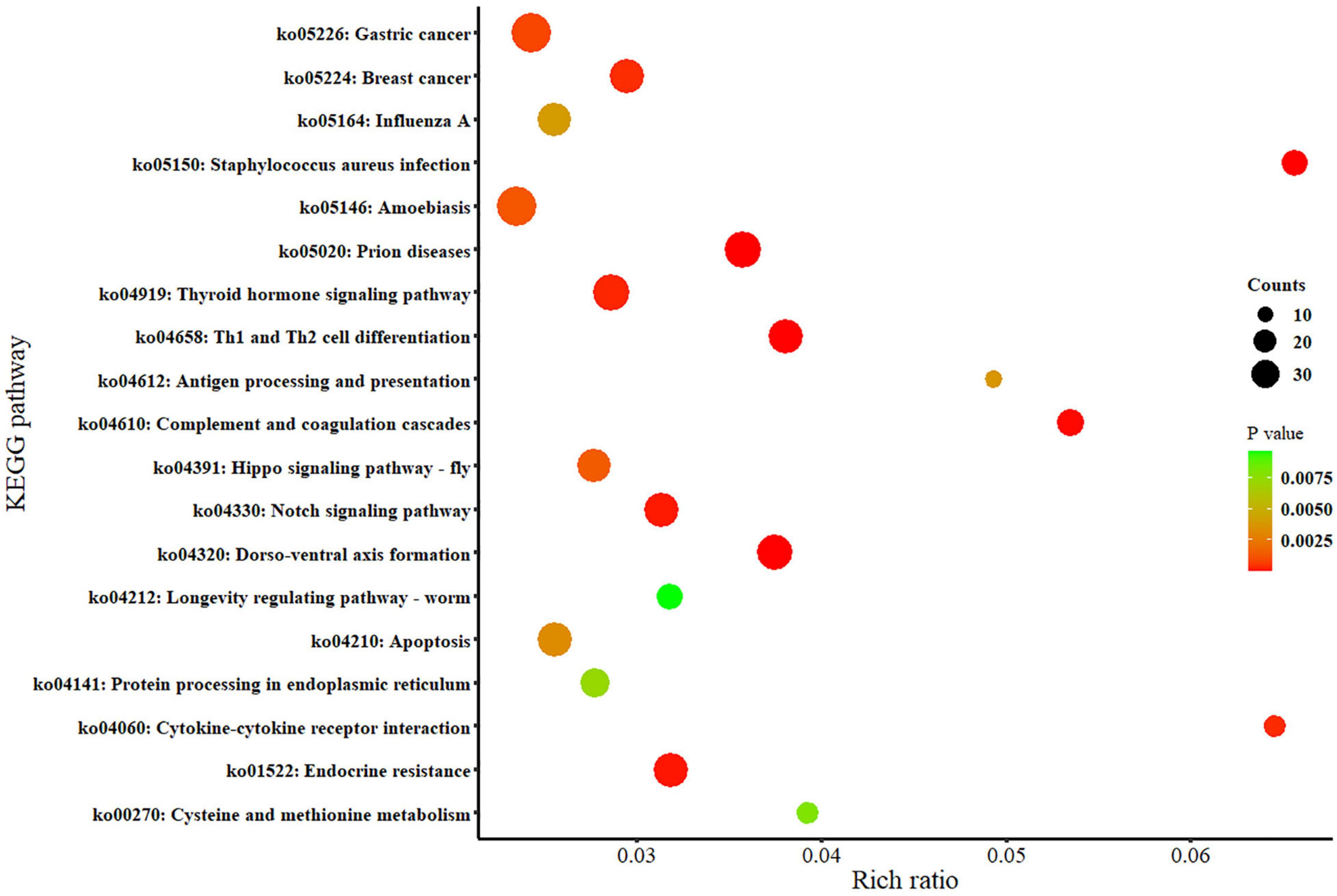
Figure 8. Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment of the DEGs between the normoxia and hypoxia groups.
The comparison of the qRT-PCR gene and RNAseq expression is shown in Figure 9. The qRT-PCR expression pattern was consistent with RNA-seq (r = 0.8095, p < 0.05; Figure 9 and Supplementary Table 7) that validated the reliability and accuracy of the transcriptome data.
Summer mortality has been of great concern in aquaculture all over the world. It has been observed in many countries since the 1970s, recently hypoxic stress was proved that played an important role in summer mortality events (Parache, 1989; Sussarellu et al., 2010). In our previous study, hybrids of C. sikamea (♀) × C. gigas (♂) had lower mortality in summer than the half-siblings (Zhang et al., 2021). Thus, we deployed an occluded water experiment to assess hybrids’ transcriptome responses to hypoxia stress. In this study, 901 DEGs were found involved in the resistance to environmental hypoxia in hybrid oysters, in which there were 432 downregulated DEGs and 469 upregulated DEGs. According to GO annotation, the cellular component term grouped the most numbered DEGs, and the KEGG annotated 108 DEGs in the immune system and 109 DEGs in signal transduction, respectively. A previous study identified 616 DEGs in C. gigas gills, mantle, and digestive gland after 7–10 and 24 days of hypoxia stress with 30% O2-saturation that was associated with 12 major cellular physiological functions, including protein synthesis and degradation, transcription, cell cycle regulation, and metabolism of nucleic acid components, cellular matrix, and cytoskeleton, immune system, membrane receptors, and so on (David et al., 2005). Our study partially matches their result, the cell cycle regulation, and metabolism of nucleic acid components, cellular matrix, and cytoskeleton-related processes have not been identified in this study which may be related to the duration of hypoxia.
The GO contains a set of terms to describe the activity and actions of gene products with three sectors: cellular component, molecular function, and biological process (Roncaglia et al., 2013; Ding et al., 2018). In our study, the integral component of membrane (GO:0016021) and extracellular region (GO:0005576) in the cellular component category enriched the most numbered DEGs, which indicated that the gill tissue is actively involved in the response to hypoxic stress. In a previous study, extracellular region term was also significantly enriched in oyster gills under aerial exposure (Zhang et al., 2015), which further supports our finding. In addition, another study has also shown that extracellular region term was involved in the osmotic pressure regulation of C. virginica (Eierman and Hare, 2014). Moreover, the integral component of the membrane term is more discussed in oyster larval development (Xu and Zhang, 2020; Durland et al., 2021), which needs a further comparative study between larvae and adult oysters under hypoxia stress. The immune response (GO:0006955) is defined as an immune system process that functions in the calibrated response of an organism to a potential internal or invasive threat (Dunwoodie et al., 2018), which is the only significantly enriched GO term under the biological process category in hypoxia stress. In previous studies, it has been shown to also participate in Saccostrea glomerata threatened by environmental stressors (Ertl et al., 2016) and in Ruditapes philippinarum under lipopolysaccharide challenge (Zuo et al., 2020). Under the molecular function category, neurotransmitter:sodium symporter activity (GO:0005328) and tumor necrosis factor receptor binding (GO:0005614) were both highly enriched that involved in membrane transport and receptor recognition binding, separately. The GO:0005328 term related to the transport function of sodium ions in the neural structure indicated that the information transmission function in the gill tissue of the hybrid oyster was more excited and activated under hypoxia stress, and the function of information collection and transmission was strengthened in response to stress (Adhikary et al., 2017), and this term was also enriched in the study of mechanisms of heat and hypoxia defense in hard clam (Hu et al., 2022). The GO:0005614 was also reported as highly enriched in the transcriptome analysis of Aequipecten opercularis exposure to Pseudo-nitzschia (Ventoso et al., 2019) that referred to a group of multifunctional inflammatory cytokines which indicates that hypoxia stress might cause various pathological and immune processes (Zheng et al., 2020).
The innate immune is a vital defensive system to protect oysters from the environment (Guo et al., 2015). The regulation of innate immunity relies on the pathways of the ko04610_complement and coagulation cascades (Bajic et al., 2015), ko04658_Th1 and Th2 cell differentiation (Dong and Flavell, 2000), and ko04612_antigen processing and presentation (Gaudino and Kumar, 2019). The complement system is a non-specific defense mechanism against pathogens, and the effector molecules of complement could be generated by enzymatic activity. In the present study, the immune system-related pathways, such as ko4610, ko4658, and ko4612, were highly enriched with DEGs, indicating that hybrid oysters rely heavily on innate immunity in coping with hypoxic stress. The fact that hypoxia stress could be a factor in activating the complement and coagulation cascades pathway has also been reported in Pacific abalone, which supports our findings (Shen et al., 2019). Organism immune to different microorganisms is regulated by separate lineages of effector T helper cells, that differentiate from primal CD4 + precursor cells in response to cues provided by antigen-presenting cells and include T helper type 1 and type 2 (Amsen et al., 2009; Knosp and Johnston, 2012), and also the transporter associated with antigen processing is essential for the adaptive immune system (Abele and Tampe, 2004). In the study of solid tumors, hypoxia was generally taken as a common biological determinant of immune exclusion (Doedens et al., 2013; Pietrobon and Marincola, 2021). Hypoxia enhances the immune response and leads to cellular stabilization of hypoxia-inducible factor 1α, resulting in a synergistic effect with the key inflammatory transcription factor nuclear factor of activated B cells (Saikumar et al., 1998; Kiers et al., 2016). Therefore, we hypothesized that cell damage caused by hypoxia could also be recognized in some forms by the antigen processing and presentation pathway in hybrid oyster gills involved in the immune response. Notch signaling is an evolutionarily conserved pathway, and it is a key intercellular signaling mechanism for embryonic development (Hansson et al., 2004). The notch signaling pathway also participates in environmental information processing. Jiao et al. (2019) reported that the notch signaling pathway participates in the recovery of pearl oysters after allograft and xenograft transplantations, and it was also actively involved in the regulation of Crassostrea gigas and Crassostrea hongkongensis under low osmotic stress (Zhao et al., 2014). In the present study, 25 DEGs were enriched into the notch signaling pathway, thus the environmental information processing function of the notch signaling pathway may be highly activated by hypoxia (Hiyama et al., 2011; Danza et al., 2012). Cytokines are soluble extracellular proteins or glycoproteins that are key modulators of inflammation, participating in cells engaged in innate immunity via a complex and sometimes seemingly contradictory network of interactions (Turner et al., 2014). The latest study on Onchidium reevesii revealed that cytokine–cytokine receptor interaction pathway involved in resisting the low-frequency noise stress (Tu et al., 2021), and Zhao et al. (2016) analyzed the gene expression profiles of oyster gills that have found that cytokine–cytokine receptor interaction pathway was also involved in salinity tolerance. The lack of available oxygen contributes to a bioenergetic crisis within cells, which in turn can lead to cellular necrosis-promoting inflammatory processes in a non-specific manner through the release of intracellular contents into the surrounding space (Nizet and Johnson, 2009; Taylor and Colgan, 2017). In this experiment, it was found that the hypoxia stress might induce adaptive inflammation in the gill tissue of hybrid oyster, thus activating the cytokine–cytokine receptor, which was consistent with the above conclusions.
The Notch family has four transmembrane receptors (Notch1–Notch4) and five ligands (Kadesch, 2004). In the previous studies, fibropellin-1, related to the Notch protein, drastically increased when post chronic hypoxia in the clam Ruditapes philippinarum (Nie et al., 2020) and the NOTCHs were upregulated in Hermodice carunculata under hypoxia (Grimes et al., 2021). Notch signaling was originally identified as a pleiotropic mediator of cell fate in invertebrates, which has recently emerged as an important regulator of immune cells (Radtke et al., 2013; Vanderbeck and Maillard, 2021). In the current study, several upregulated DEGs were annotated into the notch signaling pathway, which participates in the response to hypoxia stress by regulating the innate immunity in oyster gills.
Heat shock proteins (HSPs) are a group of molecular chaperones that can be induced by hypoxia (Wang et al., 2019), temperature (Zhang et al., 2021), and a variety of other stresses (Murphy, 2013; Cheng et al., 2016). HSPA1s contain the heat shock 70 kDa protein 1, 2, 6, and 8. Hsp70 proteins are central components of molecular chaperones and folding catalysts, and they assist a large variety of protein folding processes in the cell (Mayer and Bukau, 2005). The HSP70 gene was significantly upregulated in the study. Hypoxia could induce protein misfolding due to the lack of oxygen required for the formation of disulfide linkages, which leads to endoplasmic reticulum stress and the activation of unfolded protein response (Bartoszewska and Collawn, 2020; Díaz-Bulnes et al., 2020). As a chaperone protein, the hsp70 proteins facilitate the proper folding of proteins and DNA repair (Duan et al., 2014). In a previous study, HSP70 genes were also increased in Anadara broughtonii when exposed to sulfide and hypoxia (Wang et al., 2019), which further supports our finding. All findings suggest that the HSP70 genes might play key roles in bivalves dealing with different stress factors.
Apoptosis is another evolution-conserved process in organisms, which is also an important process in the molluskan immune system (Zhang et al., 2021). The TNF family has been verified as a key inducer in apoptosis, both LIGHT and TRAIL are important members of it. LIGHT is also named TNFSF14 (tumor necrosis factor superfamily member 14) which has two spliced transcript variants encoding distinct isoforms (Locksley et al., 2001). TRAIL is a cytokine that is produced and secreted by most normal tissue cells, which induces apoptosis by binding the DR4 and DR5 receptors that result in trimerization of the receptor and clustering of the receptor’s intracellular DD, leading to the formation of the death-inducing signaling (Wang and El-Deiry, 2003; Szliszka et al., 2009). In this study, both TRAIL and CASP7 were significantly downregulated, which indicated that the normal apoptosis might be inhibited due to hypoxia stress and the hypoxia has not caused cell damage that required the apoptotic process in this experiment. Previous studies have also verified that TRAIL/LIGHT could rapidly induce apoptosis in diverse origin cell lines, and LIGHT could be ligands for herpesvirus entry mediators (Wiley et al., 1995; Mauri et al., 1998; McGrath et al., 2011).
Gray et al. (2002) demonstrated that marine bivalves have the least sensitivity in the classified marine organisms according to their tolerance to hypoxia. Sussarellu et al. (2010) infer that the hypoxic effects appeared late in such a low oxygen-tolerant species, and their study indicated that all of the complexes of the electron transport chain were involved. In the current study, the typical genes of the respiratory chain, like NADH-ubiquinone oxidoreductase chains, succinate dehydrogenase, cytochromes b and c, and cytochrome c oxidase subunits weren’t overexpressed, which echoes his point of view at some layer. The LIGHTs and TRAILs play a part in apoptosis, and the downregulation of TRAILs and CASP7 indicates the inhibiting of the apoptosis process. Therefore, in this experiment, the LIGHTs may only be involved in the adaptive inflammatory defenses through the cytokine–cytokine receptor interaction process, which was upregulated. HSP70 interacts with NOTCHs and contributes to the activity of Notch signaling, and the interaction represents a mechanism regulating Notch signaling (Juryńczyk et al., 2015). In our study, the typical immune genes, such as NOTCH1, HSP70s, and LIGHTs were all upregulated, were found to be involved in the immune response of oyster gills to hypoxia, by which the innate immune process is mediated. However, the TRAILs, which also come from the TNF family with LIGHTs, were downregulated in this study, suggesting that the apoptosis was inhibited under hypoxia, and the downregulated CASP7 also confirmed this conclusion.
This study is the first transcriptome report of C. sikamea ♀ × C. gigas ♂ hybrids under hypoxia stress. In total, 902 DEGs were identified in gill tissue of the hybrids under hypoxia stress, among which 432 downregulated and 469 upregulated DEGs were identified, respectively. In addition, several immune-related pathways were significantly enriched, such as Th1 and Th2 cell differentiation; complement and coagulation cascades, and antigen processing and presentation, which indicated that the innate immunity of oysters plays an important role in coping with hypoxic stress. Our findings demonstrated a gradually complicated immune pathway network in the gill tissue of hybrids under hypoxia stress. In conclusion, these results provide a basis for genetic breeding and transcriptome study of the hybrid oyster.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI (accession: PRJNA587775).
XuZ, CF, and ZW gathered, analyzed, and interpreted data, discussed the results, and co-wrote the manuscript. JL, XiZ, and QL contributed constructively ideas for the work. All authors contributed to the article and approved the submitted version.
This study was supported by the National Natural Science Foundation of China (Grant No. 31172403), and the Science and Technology Major Project of Guangxi (Grant/Award No. GuiAA17204094-3).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.851098/full#supplementary-material
Supplementary Table 1 | The proportion of different species that are annotated onto the transcription.
Supplementary Table 2 | Total differentially expressed genes.
Supplementary Table 3 | Gene Ontology categorization of the unigenes in a cellular component, biological process, and molecular function from the transcriptome.
Supplementary Table 4 | DEGs that were assigned to the significantly enriched GO terms.
Supplementary Table 5 | Kyoto Encyclopedia of Genes and Genomes (KEGG) assignment of unigenes in transcriptome under hypoxia stress.
Supplementary Table 6 | DEGs that were assigned to the significantly enriched KEGG pathways.
Supplementary Table 7 | Primers designed and used in qRT-PCR validation of hypoxia-responsive genes identified by RNA-seq analysis.
Abele, R., and Tampe, R. (2004). The ABCs of immunology: structure and function of TAP, the transporter associated with antigen processing. Physiology. 19, 216–224. doi: 10.1152/physiol.00002.2004
Adhikary, S., Deredge, D. J., Nagarajan, A., Forrest, L. R., Wintrode, P. L., and Singh, S. K. (2017). Conformational dynamics of a neurotransmitter: sodium symporter in a lipid bilayer. Proc. Natl. Acad. Sci. U.S.A. 114, E1786–E1795. doi: 10.1073/pnas.1613293114
Amsen, D., Spilianakis, C. G., and Flavell, R. A. (2009). How are TH1 and TH2 effector cells made? Curr. Opin. Immunol. 21, 153–160.
Andreyeva, A. Y., Gostyukhina, O., Kladchenko, E., Afonnikov, D., Rasskazov, D., Lantushenko, A., et al. (2021). Hypoxia exerts oxidative stress and changes in expression of antioxidant enzyme genes in gills of Mytilus galloprovincialis (Lamarck, 1819). Mar. Biol. Res. 17, 369–379.
Bajic, G., Degn, S. E., Thiel, S., and Andersen, G. R. (2015). Complement activation, regulation, and molecular basis for complement-related diseases. EMBO J. 34, 2735–2757. doi: 10.15252/embj.201591881
Bartoszewska, S., and Collawn, J. F. (2020). Unfolded protein response (UPR) integrated signaling networks determine cell fate during hypoxia. Cell. Mol. Biol. Lett. 25, 1–20. doi: 10.1186/s11658-020-00212-1
Bo-Mi, K., Haque, M. D. N., Do-Hee, L., Sang-Eun, N., and Jae-Sung, R. (2018). Comparative toxicokinetics and antioxidant response in the Microcystin-LR-Exposed gill of two marine bivalves, Crassostrea gigas and Mytilus edulis. J. Shellfish Res. 37, 497–506. doi: 10.2983/035.037.0305
Box, A., Capó, X., Tejada, S., Catanese, G., Grau, A., Deudero, S., et al. (2020). Reduced antioxidant response of the fan mussel Pinna nobilis related to the presence of Haplosporidium pinnae. Pathogens 9:932. doi: 10.3390/pathogens9110932
Cheng, J., Xun, X., Kong, Y., Wang, S., Yang, Z., Li, Y., et al. (2016). Hsp70 gene expansions in the scallop Patinopecten yessoensis and their expression regulation after exposure to the toxic dinoflagellate Alexandrium catenella. Fish Shellfish Immunol. 58, 266–273. doi: 10.1016/j.fsi.2016.09.009
Danza, G., Di Serio, C., Rosati, F., Lonetto, G., Sturli, N., Kacer, D., et al. (2012). Notch signaling modulates hypoxia-induced neuroendocrine differentiation of human prostate cancer cells. Mol. Cancer Res. 10, 230–238. doi: 10.1158/1541-7786.MCR-11-0296
David, E., Tanguy, A., Pichavant, K., and Moraga, D. (2005). Response of the Pacific oyster Crassostrea gigas to hypoxia exposure under experimental conditions. FEBS J. 272, 5635–5652. doi: 10.1111/j.1742-4658.2005.04960.x
Díaz-Bulnes, P., Saiz, M. L., López-Larrea, C., and Rodríguez, R. M. (2020). Crosstalk between hypoxia and ER stress response: a key regulator of macrophage polarization. Front. Immunol. 10:2951. doi: 10.3389/fimmu.2019.02951
Ding, R., Qu, Y., Wu, C. H., and Vijay-Shanker, K. (2018). Automatic gene annotation using GO terms from cellular component domain. BMC Med. Inform. Decision Making 18:119. doi: 10.1186/s12911-018-0694-7
Doedens, A. L., Phan, A. T., Stradner, M. H., Fujimoto, J. K., Nguyen, J. V., Yang, E., et al. (2013). Hypoxia-inducible factors enhance the effector responses of CD8+ T cells to persistent antigen. Nat. Immunol. 14, 1173–1182. doi: 10.1038/ni.2714
Dong, C., and Flavell, R. A. (2000). Cell fate decision: t-helper 1 and 2 subsets in immune responses. Arthritis Res. Ther. 2:179. doi: 10.1186/ar85
Duan, Y., Huang, S., Yang, J., Niu, P., Gong, Z., Liu, X., et al. (2014). HspA1A facilitates DNA repair in human bronchial epithelial cells exposed to Benzo[a]pyrene and interacts with casein kinase 2. Cell Stress Chaperones 19, 271–279. doi: 10.1007/s12192-013-0454-7
Dunwoodie, L. J., Poehlman, W. L., Ficklin, S. P., and Feltus, F. A. (2018). Discovery and validation of a glioblastoma co-expressed gene module. Oncotarget 9:10995. doi: 10.18632/oncotarget.24228
Durland, E., De Wit, P., Meyer, E., and Langdon, C. (2021). Larval development in the Pacific oyster and the impacts of ocean acidification: differential genetic effects in wild and domesticated stocks. Evolution. Appl. 14, 2258–2272. doi: 10.1111/eva.13289
Eierman, L. E., and Hare, M. P. (2014). Transcriptomic analysis of candidate osmoregulatory genes in the eastern oyster Crassostrea virginica. BMC Genomics 15:503. doi: 10.1186/1471-2164-15-503
Ertl, N. G., O’Connor, W. A., Papanicolaou, A., Wiegand, A. N., and Elizur, A. (2016). Transcriptome analysis of the sydney rock oyster, Saccostrea glomerata: insights into molluscan immunity. PLoS One 11:e0156649. doi: 10.1371/journal.pone.0156649
Furr, D., Ketchum, R. N., Phippen, B. L., Reitzel, A. M., and Ivanina, A. V. J. (2021). Physiological variation in response to vibrio and hypoxia by aquacultured eastern oysters in the Southeastern United States. Integrat. Comparat. Biol. 61, 1715–1729. doi: 10.1093/icb/icab176
Gaudino, S. J., and Kumar, P. (2019). Cross-talk between antigen presenting cells and T cells impacts intestinal homeostasis, bacterial infections, and tumorigenesis. Front. Immunol. 10:360. doi: 10.3389/fimmu.2019.00360
Gray, J. S., Wu, R. S.-S., and Or, Y. Y. (2002). Effects of hypoxia and organic enrichment on the coastal marine environment. Mar. Ecol. Progr. Series 238, 249–279.
Grimes, C. J., Petersen, L. H., and Schulze, A. (2021). Differential gene expression indicates modulated responses to chronic and intermittent hypoxia in corallivorous fireworms (Hermodice carunculata). Sci. Rep. 11:11110. doi: 10.1038/s41598-021-90540-9
Gu, H., Shang, Y., Clements, J., Dupont, S., Wang, T., Wei, S., et al. (2019). Hypoxia aggravates the effects of ocean acidification on the physiological energetics of the blue mussel Mytilus edulis. Mar. Pollut. Bull. 149:110538. doi: 10.1016/j.marpolbul.2019.110538
Guo, X., He, Y., Zhang, L., Lelong, C., and Jouaux, A. (2015). Immune and stress responses in oysters with insights on adaptation. Fish Shellfish Immunol. 46, 107–119. doi: 10.1016/j.fsi.2015.05.018
Hansson, E. M., Lendahl, U., and Chapman, G. (2004). Notch signaling in development and disease. Semin. Cancer Biol. 14, 320–328.
He, G., Liu, X., Xu, Y., Liang, J., Deng, Y., Zhang, Y., et al. (2021). Repeated exposure to simulated marine heatwaves enhances the thermal tolerance in pearl oysters. Aquatic Toxicol. 239:105959. doi: 10.1016/j.aquatox.2021.105959
Hedgecock, D., and Davis, J. P. (2007). Heterosis for yield and crossbreeding of the Pacific oyster Crassostrea gigas. Aquaculture 272, S17–S29.
Hiyama, A., Skubutyte, R., Markova, D., Anderson, D. G., Yadla, S., Sakai, D., et al. (2011). Hypoxia activates the notch signaling pathway in cells of the intervertebral disc: implications in degenerative disc disease. Arthritis Rheumatism 63, 1355–1364. doi: 10.1002/art.30246
Hu, Z., Feng, J., Song, H., Zhou, C., Yu, Z.-L., Yang, M.-J., et al. (2022). Mechanisms of heat and hypoxia defense in hard clam: insights from transcriptome analysis. Aquaculture 549:737792. doi: 10.1016/j.aquaculture.2021.737792
Huo, Z., Wang, Z., Yan, X., and Yu, R. (2014). Hybridization between Crassostrea hongkongensis and Crassostrea ariakensis at different salinities. J. World Aquacult. Soc. 45, 226–232.
Jiao, Y., Yang, S., Cao, Y., Zheng, Z., Deng, Y., Wang, Q., et al. (2019). Genome and transcriptome analyses providing insight into the immune response of pearl oysters after allograft and xenograft transplantations. Fish Shellfish Immunol. 90, 109–117. doi: 10.1016/j.fsi.2019.04.061
Joos, F., Plattner, G. K., Stocker, T. F., Körtzinger, A., and Wallace, D. W. (2003). Trends in marine dissolved oxygen: implications for ocean circulation changes and the carbon budget. Eos Trans. Am. Geophys. Union 84, 197–201.
Juryńczyk, M., Lewkowicz, P., Domowicz, M., Mycko, M. P., and Selmaj, K. W. (2015). Heat shock protein 70 (Hsp70) interacts with the Notch1 intracellular domain and contributes to the activity of Notch signaling in myelin-reactive CD4 T cells. J. Neuroimmunol. 287, 19–26. doi: 10.1016/j.jneuroim.2015.08.007
Kadesch, T. (2004). Notch signaling: the demise of elegant simplicity. Curr. Opin. Genet. Dev. 14, 506–512. doi: 10.1016/j.gde.2004.07.007
Kiers, H. D., Scheffer, G.-J., van der Hoeven, J. G., Eltzschig, H. K., Pickkers, P., and Kox, M. (2016). Immunologic consequences of hypoxia during critical illness. Anesthesiology 125, 237–249. doi: 10.1097/ALN.0000000000001163
Knosp, C. A., and Johnston, J. A. (2012). Regulation of CD4+ T-cell polarization by suppressor of cytokine signalling proteins. Immunology 135, 101–111. doi: 10.1111/j.1365-2567.2011.03520.x
Larade, K., and Storey, K. (2002). Chapter 3—a profile of the metabolic responses to anoxia in marine invertebrates. Cell. Mol. Response Stress 3, 27–46. doi: 10.1016/s1568-1254(02)80005-5
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408.
Locksley, R. M., Killeen, N., and Lenardo, M. (2001). The TNF and TNF receptor superfamilies: integrating mammalian biology. Cell 104, 487–501. doi: 10.1016/s0092-8674(01)00237-9
Mauri, D. N., Ebner, R., Montgomery, R. I., Kochel, K. D., Cheung, T. C., Yu, G.-L., et al. (1998). LIGHT, a new member of the TNF superfamily, and lymphotoxin α are ligands for herpesvirus entry mediator. Immunity 8, 21–30. doi: 10.1016/s1074-7613(00)80455-0
Mayer, M. P., and Bukau, B. (2005). Hsp70 chaperones: cellular functions and molecular mechanism. Cell. Mol. Life Sci. 62:670. doi: 10.1007/s00018-004-4464-6
McGrath, E. E., Marriott, H. M., Lawrie, A., Francis, S. E., Sabroe, I., Renshaw, S. A., et al. (2011). TNF-related apoptosis-inducing ligand (TRAIL) regulates inflammatory neutrophil apoptosis and enhances resolution of inflammation. J. Leukocyte Biol. 90, 855–865. doi: 10.1189/jlb.0211062
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L., and Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628. doi: 10.1038/nmeth.1226
Nie, H., Wang, H., Jiang, K., and Yan, X. (2020). Transcriptome analysis reveals differential immune related genes expression in Ruditapes philippinarum under hypoxia stress: potential HIF and NF-κB crosstalk in immune responses in clam. BMC Genomics 21:318. doi: 10.1186/s12864-020-6734-6
Nizet, V., and Johnson, R. S. (2009). Interdependence of hypoxic and innate immune responses. Nat. Rev. Immunol. 9, 609–617. doi: 10.1038/nri2607
Nogueira, L., Mello, D. F., Trevisan, R., Garcia, D., da Silva Acosta, D., Dafre, A. L., et al. (2017). Hypoxia effects on oxidative stress and immunocompetence biomarkers in the mussel Perna perna (Mytilidae, Bivalvia). Mar. Environ. Res. 126, 109–115. doi: 10.1016/j.marenvres.2017.02.009
Parache, A. (1989). Growth performance of oyster Crassostrea angulata and Crassostrea gigas reared in Arcachon Bay between 1950 and 1986: first results. Haliotis 19, 227–236.
Parthasarathy, A., Srinivasan, S., Appleby, A. J., and Martin, C. R. (1992). Temperature dependence of the electrode kinetics of oxygen reduction at the platinum/Nafion ® interface—a microelectrode investigation. J. Electrochem. Soc. 139:2530.
Pietrobon, V., and Marincola, F. M. (2021). Hypoxia and the phenomenon of immune exclusion. J. Transl. Med. 19:9. doi: 10.1186/s12967-020-02667-4
Radtke, F., MacDonald, H. R., and Tacchini-Cottier, F. (2013). Regulation of innate and adaptive immunity by Notch. Nat. Rev. Immunol. 13, 427–437. doi: 10.1038/nri3445
Roncaglia, P., Martone, M. E., Hill, D. P., Berardini, T. Z., Foulger, R. E., Imam, F. T., et al. (2013). The Gene Ontology (GO) cellular component ontology: integration with SAO (Subcellular Anatomy Ontology) and other recent developments. J. Biomed. Semantics 4, 20–20. doi: 10.1186/2041-1480-4-20
Saikumar, P., Dong, Z., Weinberg, J. M., and Venkatachalam, M. (1998). Mechanisms of cell death in hypoxia/reoxygenation injury. Oncogene 17, 3341–3349. doi: 10.1038/sj.onc.1202579
Seo, J.-K., Lee, M. J., Go, H.-J., Do Kim, G., Do Jeong, H., Nam, B.-H., et al. (2013). Purification and antimicrobial function of ubiquitin isolated from the gill of Pacific oyster, Crassostrea gigas. Mol. Immunol. 53, 88–98. doi: 10.1016/j.molimm.2012.07.003
Shen, Y., Huang, Z., Liu, G., Ke, C., and You, W. (2019). Hemolymph and transcriptome analysis to understand innate immune responses to hypoxia in Pacific abalone. Comparat. Biochem. Physiol. Part D: Genomics Proteomics 30, 102–112. doi: 10.1016/j.cbd.2019.02.001
Soon, T. K., and Zheng, H. (2019). Climate change and bivalve mass mortality in temperate regions. Rev. Environ. Contamination Toxicol. 251, 109–129. doi: 10.1007/398_2019_31
Sussarellu, R., Fabioux, C., Le Moullac, G., Fleury, E., and Moraga, D. (2010). Transcriptomic response of the Pacific oyster Crassostrea gigas to hypoxia. Mar. Genomics 3, 133–143.
Szliszka, E., Mazur, B., Zydowicz, G., Czuba, Z. P., and Król, W. (2009). TRAIL-induced apoptosis and expression of death receptor TRAIL-R1 and TRAIL-R2 in bladder cancer cells. Folia Histochem. Cytobiol. 47, 579–585. doi: 10.2478/v10042-009-0111-2
Taylor, C. T., and Colgan, S. P. (2017). Regulation of immunity and inflammation by hypoxia in immunological niches. Nat. Rev. Immunol. 17, 774–785. doi: 10.1038/nri.2017.103
Tu, Z., Tang, L., Zhang, X., Jia, J., and Shen, H. (2021). Transcriptome analysis of the central nervous system of sea slug (Onchidium reevesii) exposed to low-frequency noise. Front. Mar. Sci. 8:807489. doi: 10.3389/fmars.2021.807489
Turner, M. D., Nedjai, B., Hurst, T., and Pennington, D. J. (2014). Cytokines and chemokines: at the crossroads of cell signalling and inflammatory disease. Biochim. Biophys. Acta (BBA) - Mol. Cell Res. 1843, 2563–2582. doi: 10.1016/j.bbamcr.2014.05.014
Vanderbeck, A., and Maillard, I. (2021). Notch signaling at the crossroads of innate and adaptive immunity. J. Leukocyte Biol. 109, 535–548. doi: 10.1002/JLB.1RI0520-138R
Ventoso, P., Pazos, A. J., Pérez-Parallé, M. L., Blanco, J., Triviño, J. C., and Sánchez, J. L. (2019). RNA-Seq transcriptome profiling of the queen scallop (Aequipecten opercularis) digestive gland after exposure to domoic acid-producing Pseudo-nitzschia. Toxins. 11, 1–26. doi: 10.3390/toxins11020097
Wang, L., Song, X., and Song, L. (2018). The oyster immunity. Dev. Comparat. Immunol. 80, 99–118. doi: 10.1016/j.dci.2017.05.025
Wang, S., and El-Deiry, W. S. (2003). TRAIL and apoptosis induction by TNF-family death receptors. Oncogene 22, 8628–8633. doi: 10.1038/sj.onc.1207232
Wang, Y., Zhou, S., Liu, T., Chen, M., Li, W., and Zhang, X. (2019). The transcriptomic responses of the ark shell, Anadara broughtonii, to sulfide and hypoxia exposure. Mol. Biol. Rep. 46, 4245–4257. doi: 10.1007/s11033-019-04879-4
Wiley, S. R., Schooley, K., Smolak, P. J., Din, W. S., Huang, C.-P., Nicholl, J. K., et al. (1995). Identification and characterization of a new member of the TNF family that induces apoptosis. Immunity 3, 673–682. doi: 10.1016/1074-7613(95)90057-8
Wu, R. S. (2002). Hypoxia: from molecular responses to ecosystem responses. Mar. Pollut. Bull. 45, 35–45.
Xu, F., Guo, X., Li, L., and Zhang, G. (2011). Effects of salinity on larvae of the oysters Crassostrea ariakensis, C. sikamea and the hybrid cross. Mar. Biol. Res. 7, 796–803.
Xu, F., and Zhang, G. (2020). Transcriptomic and proteomic dynamics during metamorphosis of Pacific oyster Crassostrea gigas. bioRxiv [preprint] 2020.2003.2025.004614. doi: 10.1101/2020.03.25.004614
Xu, H., Li, Q., Kong, L., Yu, H., and Liu, S. (2019). Fertilization, survival and growth of hybrids between Crassostrea gigas and Crassostrea sikamea. Fisheries Sci. 85, 821–828.
Zhang, X., Fan, C., Zhang, X., Li, Q., Li, Y., Ma, P., et al. (2021). Transcriptome analysis of Crassostrea sikamea (♀) × Crassostrea gigas (♂) hybrids under and after thermal stress. J. Ocean Univ. China 21, 213–224.
Zhang, Y., Sun, J., Mu, H., Li, J., Zhang, Y., Xu, F., et al. (2015). Proteomic basis of stress responses in the gills of the pacific oyster Crassostrea gigas. J. Proteome Res. 14, 304–317.
Zhang, Y., Zhang, Y., Li, J., Wang, Z., Yan, X., and Yu, Z. (2017). Incomplete sterility of hybrids produced by Crassostrea hongkongensis female× Crassostrea gigas male crosses. Aquaculture Res. 48, 1351–1358.
Zhao, L., Liu, L., Liu, B., Liang, J., Lu, Y., and Yang, F. (2019). Antioxidant responses to seawater acidification in an invasive fouling mussel are alleviated by transgenerational acclimation. Aquatic Toxicol. 217:105331.
Zhao, L., Shirai, K., Tanaka, K., Milano, S., Higuchi, T., Murakami-Sugihara, N., et al. (2020). A review of transgenerational effects of ocean acidification on marine bivalves and their implications for sclerochronology. Estuarine Coastal Shelf Sci. 235:106620.
Zhao, X., Yu, H., Kong, L., and Li, Q. (2016). Gene co-expression network analysis reveals the correlation patterns among genes in euryhaline adaptation of Crassostrea gigas. Mar. Biotechnol. 18, 535–544.
Zhao, X., Yu, H., Kong, L., Liu, S., and Li, Q. (2014). Comparative transcriptome analysis of two oysters, Crassostrea gigas and Crassostrea hongkongensis provides insights into adaptation to hypo-osmotic conditions. PLoS One 9:e111915. doi: 10.1371/journal.pone.0111915
Zheng, Y., Liu, Z., Wang, L., Li, M., Zhang, Y., Zong, Y., et al. (2020). A novel tumor necrosis factor in the Pacific oyster Crassostrea gigas mediates the antibacterial response by triggering the synthesis of lysozyme and nitric oxide. Fish Shellfish Immunol. 98, 334–341.
Keywords: oyster hybrids, gill, hypoxia, transcriptome, de novo
Citation: Zhang X, Fan C, Li J, Zhang X, Li Q and Wang Z (2022) Transcriptome Analysis of Crassostrea sikamea (♀) × Crassostrea gigas (♂) Hybrids Under Hypoxia in Occluded Water. Front. Mar. Sci. 9:851098. doi: 10.3389/fmars.2022.851098
Received: 09 January 2022; Accepted: 31 January 2022;
Published: 14 March 2022.
Edited by:
Yuehuan Zhang, South China Sea Institute of Oceanology (CAS), ChinaReviewed by:
Xizhi Huang, Johannes Gutenberg University Mainz, GermanyCopyright © 2022 Zhang, Fan, Li, Zhang, Li and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhaoping Wan, enB3YW5nQG91Yy5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.