- 1School of Ocean, Yantai University, Yantai, China
- 2Fishery College, Zhejiang Ocean University, Zhoushan, China
Intertidal organisms are more sensitive to temperature stresses (whether high or low temperatures). As an intertidal crustacean, the optimal survival temperature ranges of Charybdis japonica are from 20 to 27°C. In this study, C. japonica was selected as the research species to better explore the molecular regulatory mechanisms of intertidal crustaceans to temperature stresses. The transcriptomes of C. japonica exposed to three temperature gradients (12, 20, and 28°C) were sequenced. A total of 69.22 Gb clean transcriptome reads were obtained from nine libraries and then de novo assembled to 52,972 unigenes with a mean length of 1080.23 bp and an N50 length of 1,775 bp. A total of 20,121 unigenes were successfully matched with at least one protein database. The transcriptome structure was predicted, and 12,125 coding sequences and 12,854 simple sequence repeats (SSRs) were obtained. The gene expression level of C. japonica at 20°C was used as control, and 548 and 90 unigenes were significantly differentially expressed at 28 and 12°C, respectively. A total of 720 unigenes were significantly differentially expressed at 28°C compared with 12°C. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) annotation showed that genes related to cell structure, metabolism, and protein folding and hormone synthesis might be involved in the regulation of temperature stress in C. japonica. Our results reveal for the first time the response of C. japonicas to low- and high- temperature stresses at the transcriptome level. The results provide fundamental information for revealing the temperature regulation mechanisms of C. japonica and other intertidal crustaceans. Furthermore, the present study enhances our understanding of how temperature fluctuations will affect the survival of marine crustaceans.
Introduction
Although oceans have a higher specific heat capacity than terrenes, the temperature fluctuations (temperature rises or falls) caused by tides, circadian rhythms, human activities, and other factors are still observable (Edwards and Richardson, 2004; Perry et al., 2005). Changes in sea temperature have also been shown to negatively affect the behavior, survival, distribution, and demographics of many marine organisms (Saucedo et al., 2004; Scott and Johnston, 2012; Molinos et al., 2016; Poloczanska et al., 2016; Boyd et al., 2018). In fact, marine organisms can quickly copy with temperature fluctuations by fleeing, but there is no denying that it can expose them to new biotic or abiotic stresses (Holt, 1990; Parmesan, 2006; Whitehead et al., 2011). The genetic regulatory mechanisms developed during the long evolutionary process of marine organisms seem to be more conducive to improving the tolerance of marine organisms to temperature fluctuations (Kleinjan and Van, 2005; Wray, 2007; Romero et al., 2012). Therefore, the effects of temperature fluctuations on the genetic regulatory mechanisms of marine organisms are necessary to investigate.
Crustacean represents one of the most species-rich taxa in the ocean and has been particularly successful in colonizing a variety of habitats, including ocean trenches, hydrothermal vents, intertidal mud flats, and terrenes (Greenaway, 2003; Schram, 2013; Stein and Harzsch, 2021). The high invasiveness of crustaceans seems to indicate that these species can tolerate large temperature fluctuations. However, temperature changes can directly affect the physiological processes and natural behaviors of many crustaceans, because these species are poikilotherms and have no organs for temperature regulation (Figure 1; Soofi et al., 2014). Previous studies further support the idea that temperature regulates the basal metabolism and osmotic pressure of crustaceans by altering the oxygen solubility and salinity of seawater (Whiteley and Taylor, 2015; Whiteley et al., 2018). Although crustaceans having a wide range of behavioral, physiological, and other adaptation responses to temperature changes (Stein and Harzsch, 2021), the regulatory mechanisms are not yet clear. Notably, intertidal temperatures exhibit substantial diurnal and seasonal variations (Pörtner, 2010; Somero, 2012), and intertidal crustaceans may be closer to their survival temperature limits (cold and warm) than subtidal crustaceans, and thus intertidal crustaceans are more sensitive to additional water temperature changes (Stillman and Somero, 2000; Somero, 2010; Legrand et al., 2018). With this in mind, assessing how low- or high- temperature stresses affect intertidal crustaceans is particularly imperative to our understanding of how temperature fluctuations impact marine crustaceans (Somero, 2012).
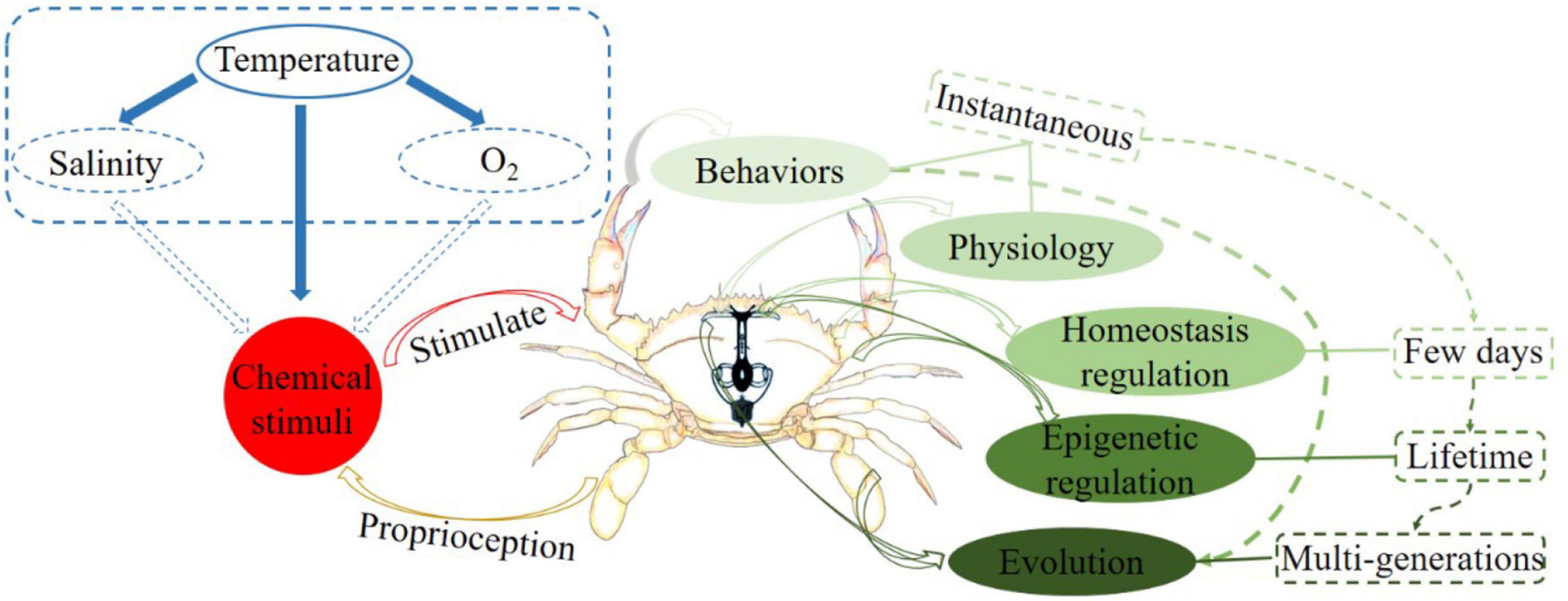
Figure 1. Diagram of the interaction between temperature fluctuations and crustacean regulatory mechanisms.
Charybdis japonica (Milne Edwards, 1861) belongs to the genus Charybdis of the family Portunidae (Decapoda, Crustacea). It is a eurythermic and euryhaline marine crab and predominantly distributed in China, Japan, the Korean Peninsula, and Southeast Asian countries (Yu et al., 2004). As a delicious aquatic product, C. japonica is rich in amino acids and unsaturated fatty acids and therefore has high economic and medicinal values (Yu et al., 2004). This fishery resource has been developed in recent years (Zheng, 2015). It can survive at temperatures between 5 and 30°C, and its optimum survival temperature ranges from 20 to 27°C. Notably, C. japonica is considered sensitive to temperature changes because it is always distributed in the intertidal zone (Zheng et al., 2013). Therefore, C. japonica can be used as an ideal biological model for investigating the response of intertidal crustaceans to temperature changes. The emergence and widespread use of RNA sequencing (RNA-seq) provide the opportunity to explore the gene-level homeostasis regulation of C. japonica subjected to temperature pressures (Humphreys et al., 2005; Wang et al., 2009), but relevant research has not been carried out so far. At present, RNA-seq has successfully identified the gene expression patterns of many crustaceans responses to temperature changes, such as Oratosquilla oratoria (Lou et al., 2019) and Shinkaia crosnieri (Cheng et al., 2019). Therefore, RNA-seq may help us explain how C. japonica responds to temperature stress through gene expression regulation.
Here, RNA-seq was applied to capture the gene expression levels of C. japonica exposed to acute low- and high- temperatures. Furthermore, differentially expressed genes (DEG) and related functions in C. japonica at different temperatures were obtained by comparative transcriptome analysis. We believe that the present results would provide fundamental information for revealing the crucial temperature regulation mechanisms of C. japonica. The present study aimed to enhance the prediction of how temperature fluctuations will contribute to C. japonica and other intertidal crustaceans.
Materials and Methods
Ethics Approval and Consent to Participate
C. japonica is an invertebrate and is not endangered or protected. During the sampling process, all C. japonica individuals were frost anaesthetized to minimize the suffering of animals.
Charybdis japonica Collection
Thirty healthy female C. japonica individuals (mean carapace length: 50.43 mm; mean carapace width: 72.33 mm; mean weight: 72.85 g) were collected from the coastal water (Temperature: 19.89°C; Salinity: 33.13‰) of Zhoushan, China, in Nov 2018. All C. japonica individuals were equally distributed into three plastic aquariums (length × width × height: 100 cm × 70 cm × 40 cm) and acclimated over 48 h. Considering the optimum temperature (20–27°C) and salinity (15–27‰) of C. japonica, the temperature and salinity of domesticated seawater were set at 20°C and 20‰, respectively. To reduce the influence of other factors on RNA-seq, all C. japonica individuals were not fed and oxygenated continuously throughout the breeding process. It is worth noting that the entire farming process of C. japonica is carried out under natural light, and we keep the water clean by replacing it with seawater with the same salinity and temperature.
Temperature Stress Schemes
In the present study, 20°C was set as the control temperature. The water temperature of one plastic aquarium was increased from 20 to 28°C, whereas the water temperature of another plastic aquarium was decreased from 20 to 12°C. The heating and cooling processes were completed within 1 h. The water temperature of the remaining plastic aquarium was maintained at 20°C. The temperature changing and maintenance processes were carried out under the constant temperature device (Sunsun, China). The temperature maintenance time was set to 12 h to reduce the change of C. japonica regulation mechanisms caused by circadian rhythm. No remarkable differences in other factors except the water temperatures of all plastic aquariums were introduced. Additionally, dying or dead C. japonica individuals were removed immediately throughout the breeding process. After the experiment, three C. japonica individuals were randomly selected from each plastic aquarium, and their gills were dissected immediately after frost anesthesia. Considering that temperature changes can alter the dissolved oxygen and salinity, the gill tissue, which is critical organ for respiration and osmotic regulation, was obtained and then separately snap-frozen in liquid nitrogen and stored at −80°C prior to the subsequent experiments.
Total RNA Extraction and Illumina Sequencing
The total RNAs of nine C. japonica individuals were extracted using the standard Trizol Reagent Kit (Huayueyang Biotech Co., Ltd., Beijing, China) and following the manufacturer’s protocol. The Agilent 2100 bioanalyzer (Agilent Technologies, Santa Clara, CA, United States) was applied to quantify the extracted total RNAs. MRNAs were further purified using RNA Purification Beads (Illumina, San Diego, CA, United States) to deplete rRNA from each total RNA and then cleaned three times with Beads Binding Buffer (Illumina, San Diego, CA, United States). After the mRNAs were incubated for 8 min at 94°C, fragmented mRNAs were combined with 8 μL of First Strand Synthesis Act D Mix (Illumina, San Diego, CA, United States) and SuperScript II Reverse Transcriptase (Invitrogen, Carlsbad, CA, United States). The conjugated product was incubated at 25°C for 50 min to synthesize first-strand cDNA. Furthermore, 5 μL of End Repair Control (Illumina, San Diego, CA, United States) and 20 μL of Second Strand Marking Master Mix (Illumina, San Diego, CA, United States) were mixed and incubated at 16°C for 60 min to complete the second-strand cDNA synthesis. The reaction was then purified using 90 μL of AMPure XP beads (Beckman Coulter, Beverly, United States). A-Tailing Control and Ligation Control were used to add A-tailing and adapter ligation for each double-stranded cDNA. Subsequently, we enriched the cDNA fragment. The enriched reaction system comprised 5 μL of PCR Primer Cocktail (Beckman Coulter, Beverly, United States) and 25 μL of PCR Master Mix (Beckman Coulter, Beverly, United States). The PCR program was as follows: initial denaturing at 98°C for 30 s, 15 cycles of denaturing at 98°C for 10 s, annealing at 60°C for 30 s, extension at 72°C for 30 s and final extension at 72°C for 5 min. All RNA-seq libraries were diluted to 10 pM and quantified using an Agilent 2100 bioanalyzer (Agilent Technologies, Santa Clara, CA, United States). Finally, nine RNA-seq libraries were sequenced on an Illumina HiSeq 2000 platform across one lane with 150 bp paired-end.
Transcriptome de novo Assembly, Functional Annotation, and Transcriptome Structure Analyses
Low-quality sequencing reads, including reads with sequencing adaptors, unknown nucleotide ratio > 10% and quality score ≤ 20, were removed from all raw reads to obtain high-quality clean reads. High-quality clean data from all samples were combined into a multifasta file, and the transcripts were de novo assembled using Trinity software (version 2.4.0; Grabherr et al., 2011) based on the K-mer algorithm using min_kmer_cov 3. According to the number of reads mapped to the transcripts, Corset software (version 1.05; Nadia and Alicia, 2014) was applied to hierarchically cluster the transcripts to obtain unigenes for subsequent analyses, and default parameters were used for software. Furthermore, Basic Local Alignment Search Tool software (Altschul, 1990) was used to compare unigene sequences with those in National Center for Biotechnology Information (NCBI) Non-redundant (NR), Swiss-Prot, Gene Ontology (GO), Eukaryotic Othologous Groups (KOG), eggNOG and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases (E < 1e–5) and finally obtain the gene function information of C. japonica. Additionally, TransDecoder software was used to predict the coding sequences (CDSs) of unigenes, and the obtained CDSs were translated into amino acid sequences from 5′ to 3′. The simple sequence repeats (SSRs) of unigenes were predicted using MISA (version 1.0), and the repetition numbers of mononucleotide, dinucleotide, trinucleotide, tetranucleotide, pentanucleotide, and hexanucleotide were 10, 6, 5, 5, 5, and 5, respectively.
Gene Regulatory Mechanisms Related to Temperature Stress
The gene expression levels of C. japonica at 20°C were treated as the control, and differences in the gene expression levels of C. japonica at 12 and 28°C were analyzed to explore the potential functional genes of C. japonica exposed to temperature stresses. DEGs are thought to be potentially involved in the homeostasis regulation of C. japonica under temperature stresses. Specifically, BWA-mem (Li and Durbin, 2009) was used to map all unigenes into the multifasta file. Then, the expression level of each unigenes was normalized based on DESeq2 (Leng et al., 2013) with default parameters, and the standardized expression level was expressed using fragments per kilobase of exon model per million mapped fragments. Furthermore, false discovery rate ≤ 0.01 and | log2 fold change (FC)| ≥ 2 (which corresponds to FC = 4) were used as the filtering criteria of DEGs. A Venn diagram was constructed to visualize the number of DEGs. Additionally, the characteristics, products and biochemical metabolic pathways of the DEGs were predicted using the Blast2GO software (E < 1e–5; Conesa et al., 2005).
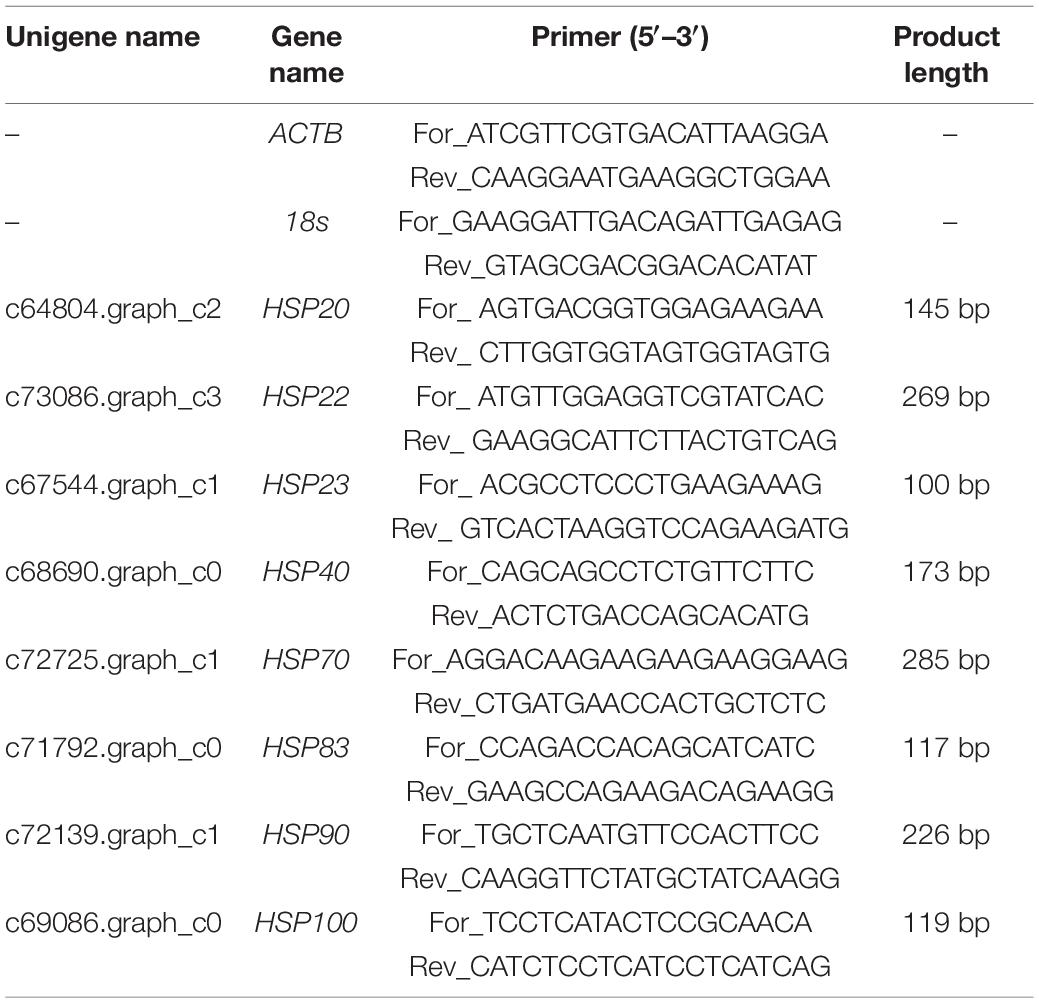
Quantitative Reverse Transcription PCR Validation of RNA-Seq Data
In the present study, the accuracy of RNA-seq data was verified by Quantitative Reverse Transcription PCR (qRT-PCR). Eight heat shock protein (HSP) genes (HSP20, HSP22, HSP23, HSP40, HSP70, HSP83, HSP90, and HSP100) associated with temperature stresses were selected for qRT-PCR. Primer Premier 6.0 software was applied to design primers specific to each gene (Table 1). Meanwhile, β-actin (ACTB) and 18s were chosen as the reference genes for internal standardization, and the primer of each gene is shown in Table 1. Nine cDNA samples were diluted by 20 folds using nuclease-free water according to the standard curves and further used for the subsequent qRT-PCR experiments. QRT-PCR was performed in an ABI PRISM 7300 Real-time PCR System following the manufacturer’s instructions for SYBR® Premix Ex Taq™ (Tli RNaseH Plus) RR420A. The 25 μL reaction system comprised 2.0 μL of diluted cDNA template, 12.5 μL of SYBR Premix Ex Taq (2×), 0.5 μL of each of forward and reverse primers and 9.5 μL of nuclease-free water. The amplification process consisted of a holding stage of 30 s at 95°C, followed by 40 cycles of 5 s at 95°C and 20 s at 56°C. Three biological replicates for each cDNA template were performed to improve the accuracy of the qRT-PCR results. Finally, the relative expression level of each HSP gene we calculated based on 2–ΔΔCT (ΔCT = CTtarget gene – CTreference gene, ΔΔCT = ΔCTtreatment – ΔCTcontrol) analysis.
Results
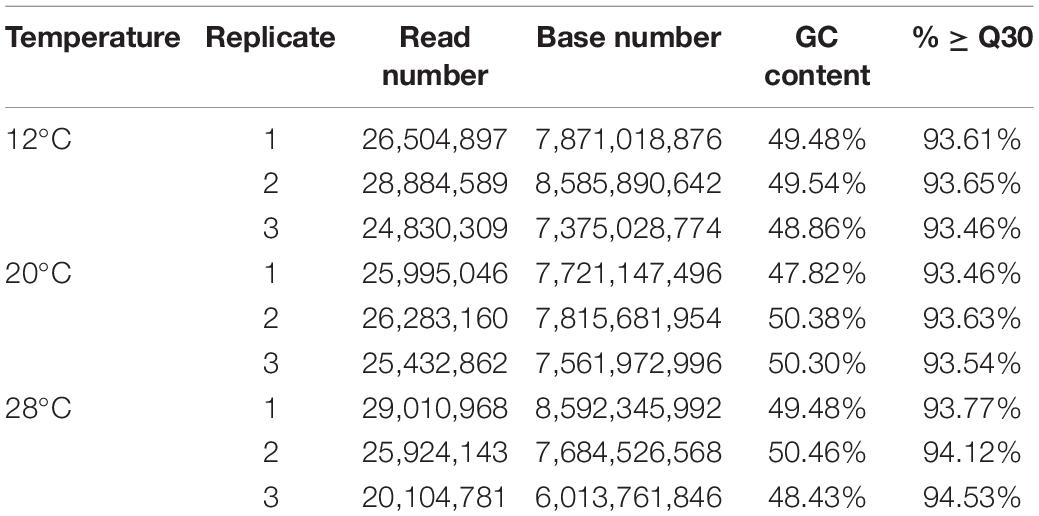
RNA-Seq Reads
All raw RNA-seq reads of nine C. japonica individuals were deposited in NCBI Sequence Read Archive under the accession number of SRR1742011, SRR1742010, SRR17422004, SRR17422003, SRR17422002, SRR17422001, SRR17422000, SRR17421999, and SRR17421998 under BioProject PRJNA793994. After low-quality raw reads were filtered out, 69.22 Gb clean reads were obtained, and the statistical results of the clean reads are shown in Table 2.
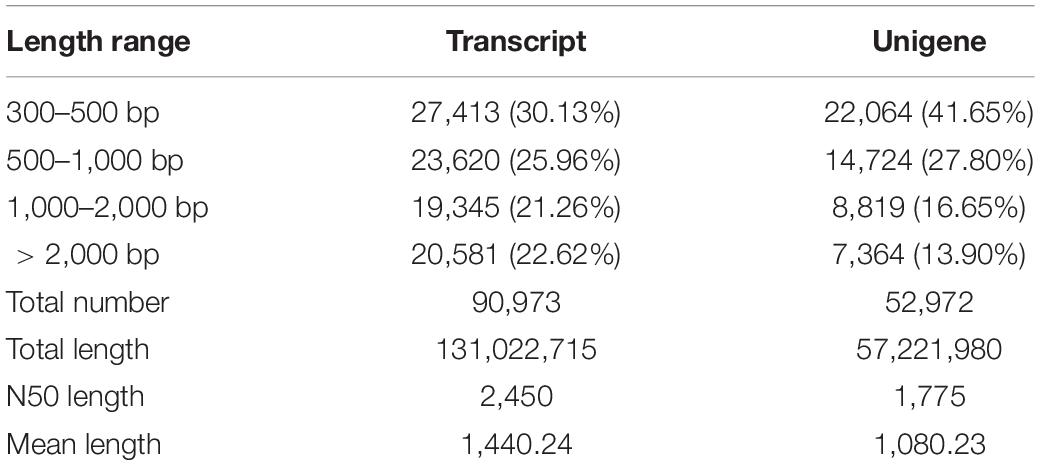
De novo Assembly, Functional Annotation and Transcriptome Structure
The clean reads of nine C. japonica individuals were de novo assembled using Trinity software, and 90,973 transcripts with a mean length of 1,440.24 bp and an N50 length of 2,450 bp were obtained. Furthermore, 52,972 unigenes were clustered by removing all redundant transcripts. The mean and N50 lengths of the unigenes was 1,080.23 and 1,775 bp, respectively. The statistical results of the transcripts and unigenes are shown in Table 3. All unigenes were then compared with the sequences in protein databases to predict the gene function of C. japonica. A total of 20,121 unigenes were annotated in protein databases. Moreover, 8,503, 10,436, 13,494, 15,676, 11,742, 17,795, and 18,466 unigenes had remarkable matches with sequences in the GO, KEGG, KOG, Pfam, Swiss-Prot, eggNOG, and NR databases, respectively. Additionally, 12,125 CDSs and 12,854 SSRs were predicted in the unigenes. The number of mononucleotides, dinucleotides, trinucleotides, tetranucleotides, pentanucleotides, and hexanucleotides were 4,373, 4,468, 2,960, 179, 8, and 2, respectively.
Quantification of the Number of Differentially Expressed Genes in Charybdis japonica at Different Temperatures
The gene expression levels of C. japonica at 20°C were used as the control, and the DEGs of C. japonica at 12 and 28°C were extracted (Figure 2). Compared with 20°C, 548 (up-regulated of 73.18%; such as HSP20, HSP40, HSP70) and 90 (up-regulated of 36.67%; such as HSP83, HSP90, HSP100) unigenes were significantly differentially expressed at 28 and 12°C, respectively. A total of 720 (524 up- and 196 down-regulated) unigenes were significantly differentially expressed at 28°C compared with 12°C.
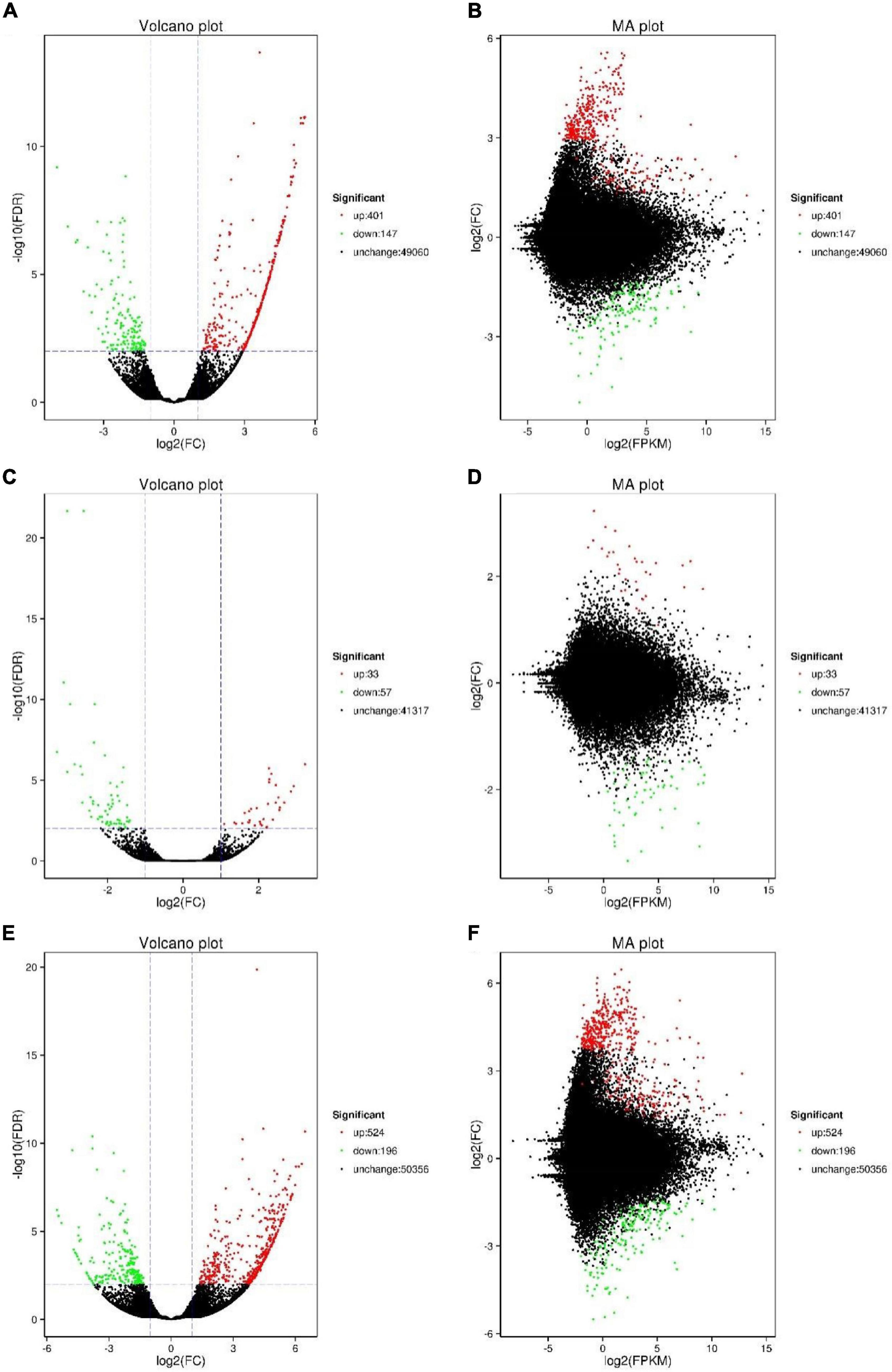
Figure 2. Number of DEGs of C. japonica in three temperature gradients. (A,B) Represent the DEGs between 20 and 28°C; (C,D) represent the DEGs between 20 and 12°C; (E,F) represent the DEGs between 12 and 28°C.
Functional Annotation of Differentially Expressed Genes in Charybdis japonica at Different Temperatures
GO analyses can be used to determine the functional properties of DEGs and their gene products. Based on the GO annotation information (Figure 3), the gene functions of the DEGs were divided into biological processes, cellular components and molecular functions. Compared with 20°C, the up- and down-regulated unigenes of C. japonica at 28°C were remarkably enriched in cell (GO: 0005623) and cell part (GO: 0044464) under cellular component; in binding (GO: 0005488) and catalytic activity (GO: 0003824) under molecular function and in metabolic process (GO: 0009987) and cellular process (GO: 0008152) under biological processes. Under cellular component, the up-regulated unigenes of C. japonica at 12°C were remarkably enriched in the cell (GO: 0005623) and cell part (GO: 0044464), and the down-regulated unigenes were remarkably enriched in the membrane (GO: 0016020) and membrane part (GO: 0044425). Moreover, the up- and down-regulated unigenes of C. japonica at 12°C were remarkably enriched in binding (GO: 0005488) and catalytic activity (GO: 0003824) under molecular function and in cellular process (GO: 0008152) and single-organism process (GO: 0044699) under biological process. Compared with 12°C, the up-regulated unigenes of C. japonica at 28°C were remarkably enriched in the cell (GO: 0005623) and cell part (GO: 0044464) under cellular component, and the down-regulated unigenes were significantly enriched in the cell (GO: 0005623) and membrane (GO: 0016020). Furthermore, the up- and down-regulated unigenes of C. japonica at 28°C were remarkably enriched in binding (GO: 0005488) and catalytic activity (GO: 0003824) under molecular function and in metabolic process (GO: 0009987) and cellular process (GO: 0008152) under biological process.
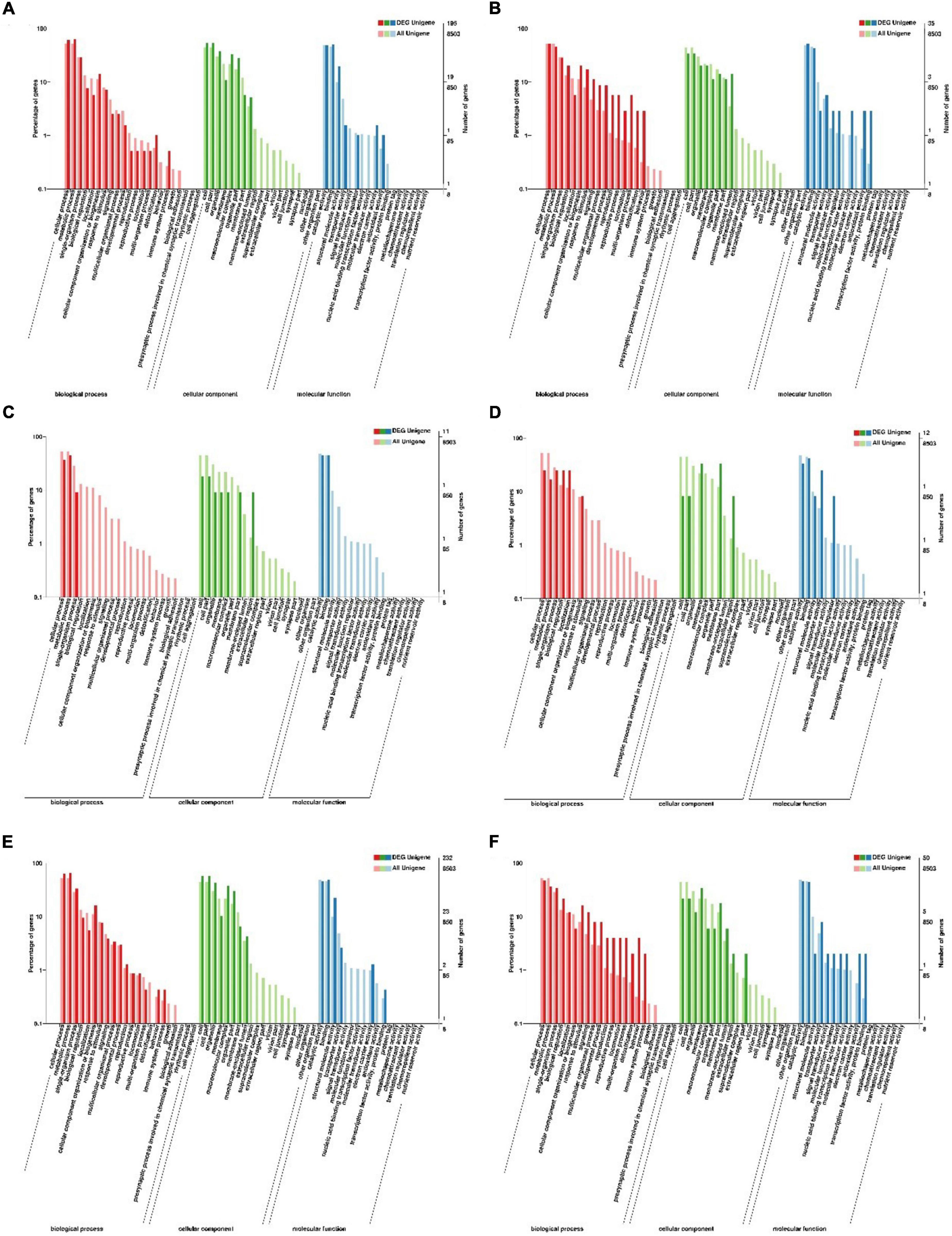
Figure 3. GO enrichment analysis of the DEGs of C. japonica exposed to three temperature gradients. (A,B) Represent the GO terms of DEGs between 20 and 28°C; (C,D) represent the GO terms of DEGs between 20 and 12°C; (E,F) represents the GO terms of DEGs between 12 and 28°C.
Kyoto Encyclopedia of Genes and Genomes Pathway Annotation of Differentially Expressed Genes in Charybdis japonica at Different Temperatures
The functions of gene products in cells and their potential metabolic pathways were analyzed by KEGG annotation (Figure 4). Compared with 20°C, the up-regulated unigenes of C. japonica at 28°C were significantly enriched in protein processing in the endoplasmic reticulum pathway (ko04141; P = 0.00185), and the down-regulated unigenes were significantly enriched in the ribosome biogenesis in the eukaryote pathway (ko03008; P = 0.009317). The down-regulated unigenes of C. japonica at 12°C were significantly enriched in the insect hormone biosynthesis pathway (ko00981; P = 0.000461), whereas the up-regulated unigenes were not significantly enriched in any pathway. Compared with 12°C, the up-regulated unigenes of C. japonica at 28°C were significantly enriched in protein processing in the endoplasmic reticulum pathway (ko04141; P = 0.001476) and ribosome pathway (ko03010; P = 3.16e-7), whereas the down-regulated unigenes were significantly enriched in the glycerophospholipid metabolism pathway (ko00564; P = 0.001643) and ether lipid metabolism (ko00565; P = 0.019503).
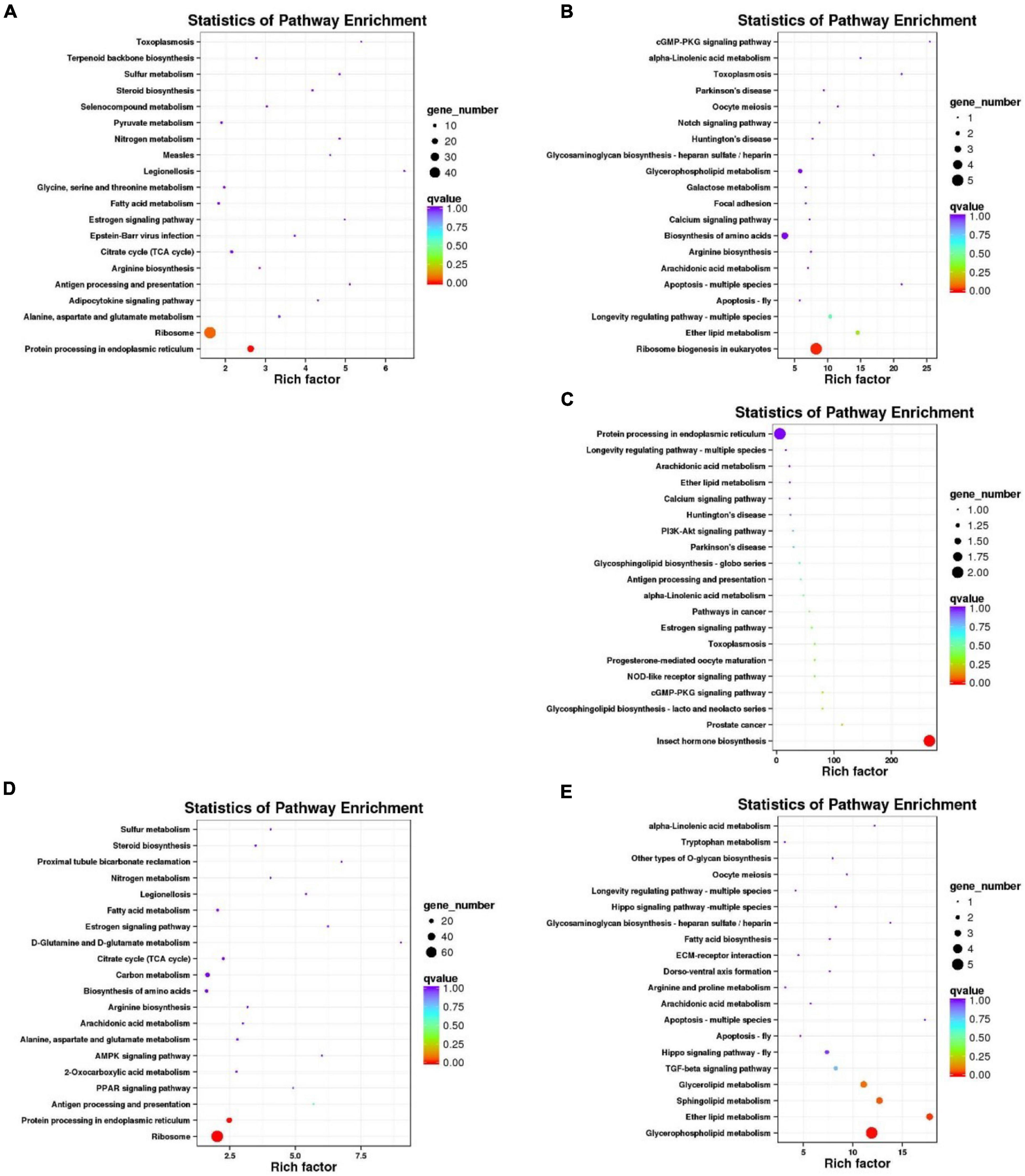
Figure 4. KEGG pathway enrichment analysis of DEGs of C. japonica exposed to three temperature gradients. KEGG pathways of (A) C. japonica unigenes up-regulated and (B) down-regulated at 28°C compared with 20°C; (C) C. japonica unigenes down-regulated at 12°C compared with 20°C and (D) C. japonica unigenes up-regulated and (E) down-regulated at 28°C compared with 12°C.
Validation of Gene Expression Level by Quantitative Reverse Transcription PCR
QRT-PCR was performed to determine the expression levels of eight HSP genes. The expression trends of eight candidate HSP genes based on qRT-PCR and transcriptome were consistent, although they did not match exactly (Figure 5). This result means that the present transcriptome results are reliable. The expression levels of HSP20, HSP40 and HSP70 increased at 28°C (relative expression level > 1) but decreased at 12°C (relative expression level < 1). Furthermore, although the expression levels of HSP22 and HSP23 decreased (relative expression level < 1) with temperature change (12 or 28°C), the expression levels of HSP83 and HSP90 increased (relative expression level > 1). Additionally, the expression levels of HSP100 increased at 12°C (relative expression level > 1) but decreased at 28°C (relative expression level < 1).
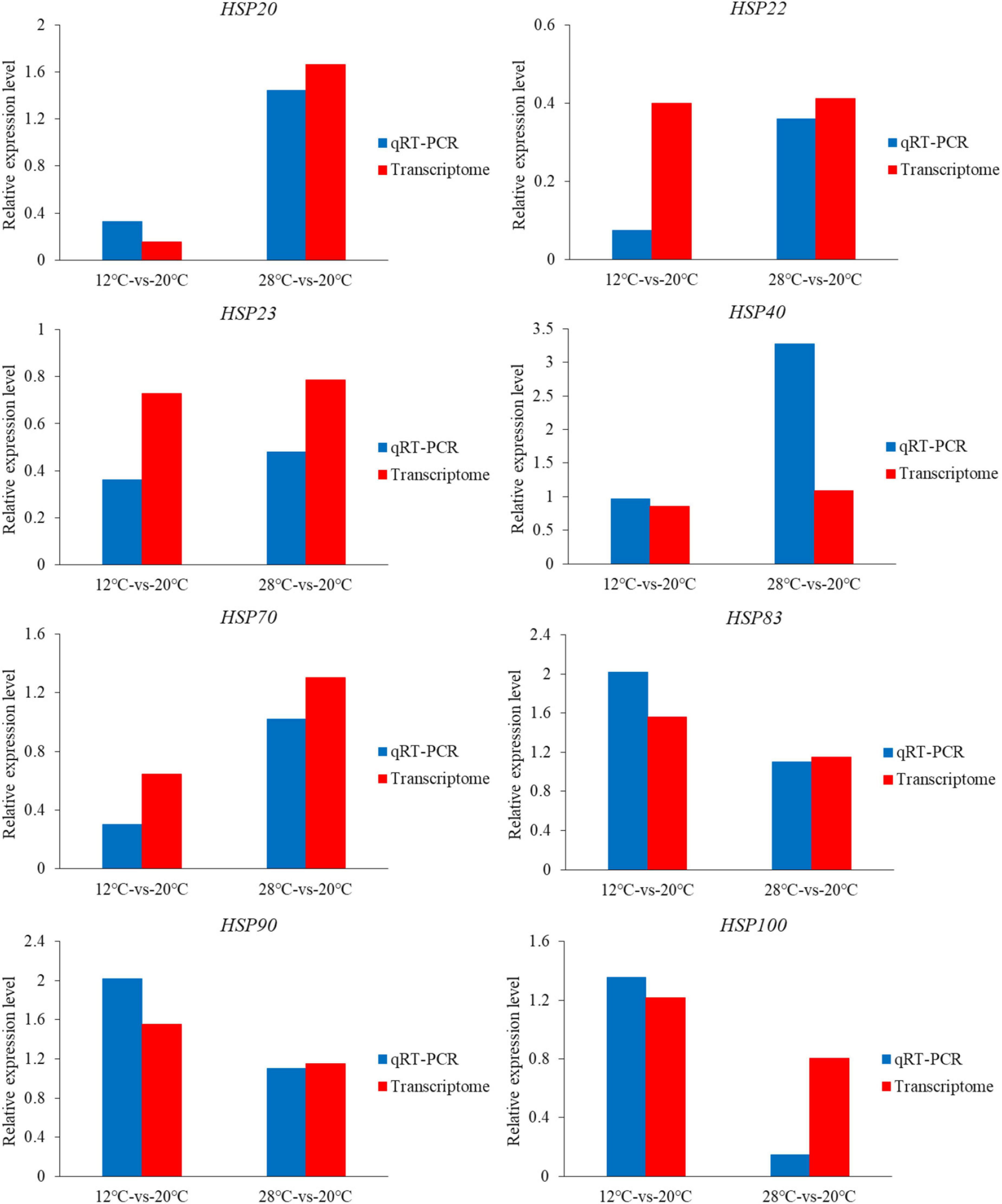
Figure 5. Relative expression levels of eight HSP genes based on transcriptome data and qRT-PCR data.
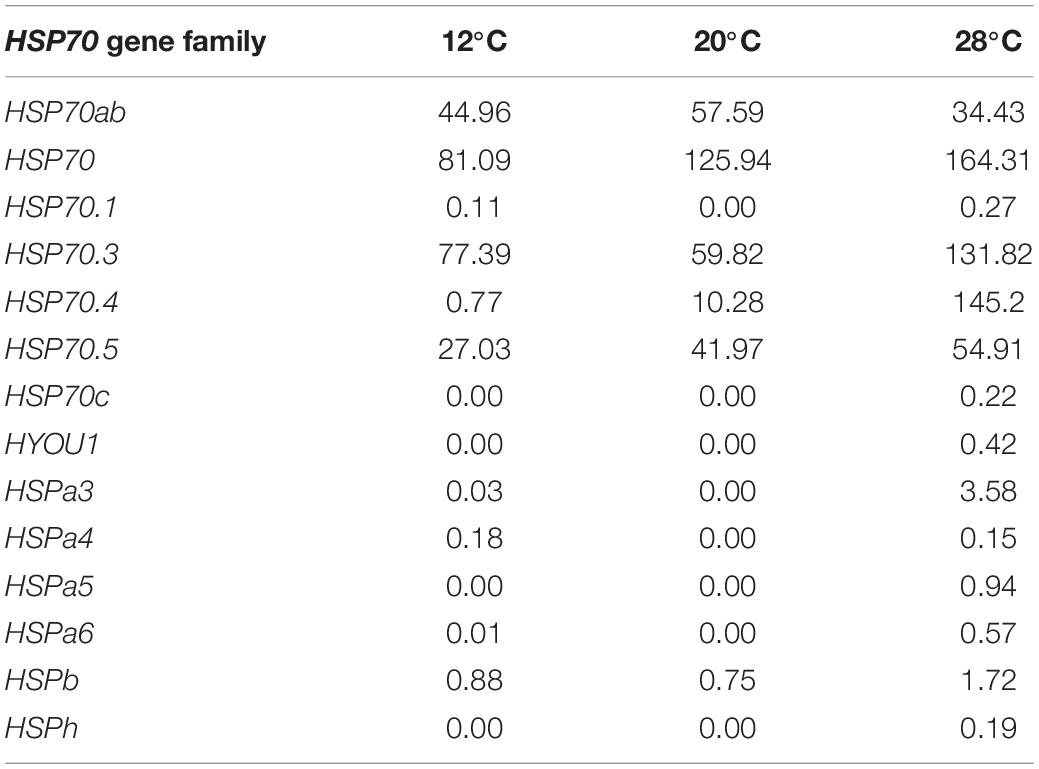
Expression Levels of HSP70 Gene Family in the Charybdis japonicas at Different Temperatures
The expression levels of the HSP70 gene family in the C. japonicas at three temperature gradients were calculated in the present study (Figure 6 and Table 4). Results showed that the expression levels of the HSP 70.1, HSP70.3, HSPa3, HSPa4, HSPa6, and HSPb increased at either 12 or 28°C, and the expression levels of the HSP70, HSP70.4, and HSP70.5 decreased at 12°C but increased at 28°C. Meanwhile, the expression levels of the HSP70c, HYOU1, HSPa5, and HSPh did not change at 12°C, but increased at 28°C. Additionally, only the expression levels of the HSP70ab gene decreased at 12 and 28°C.
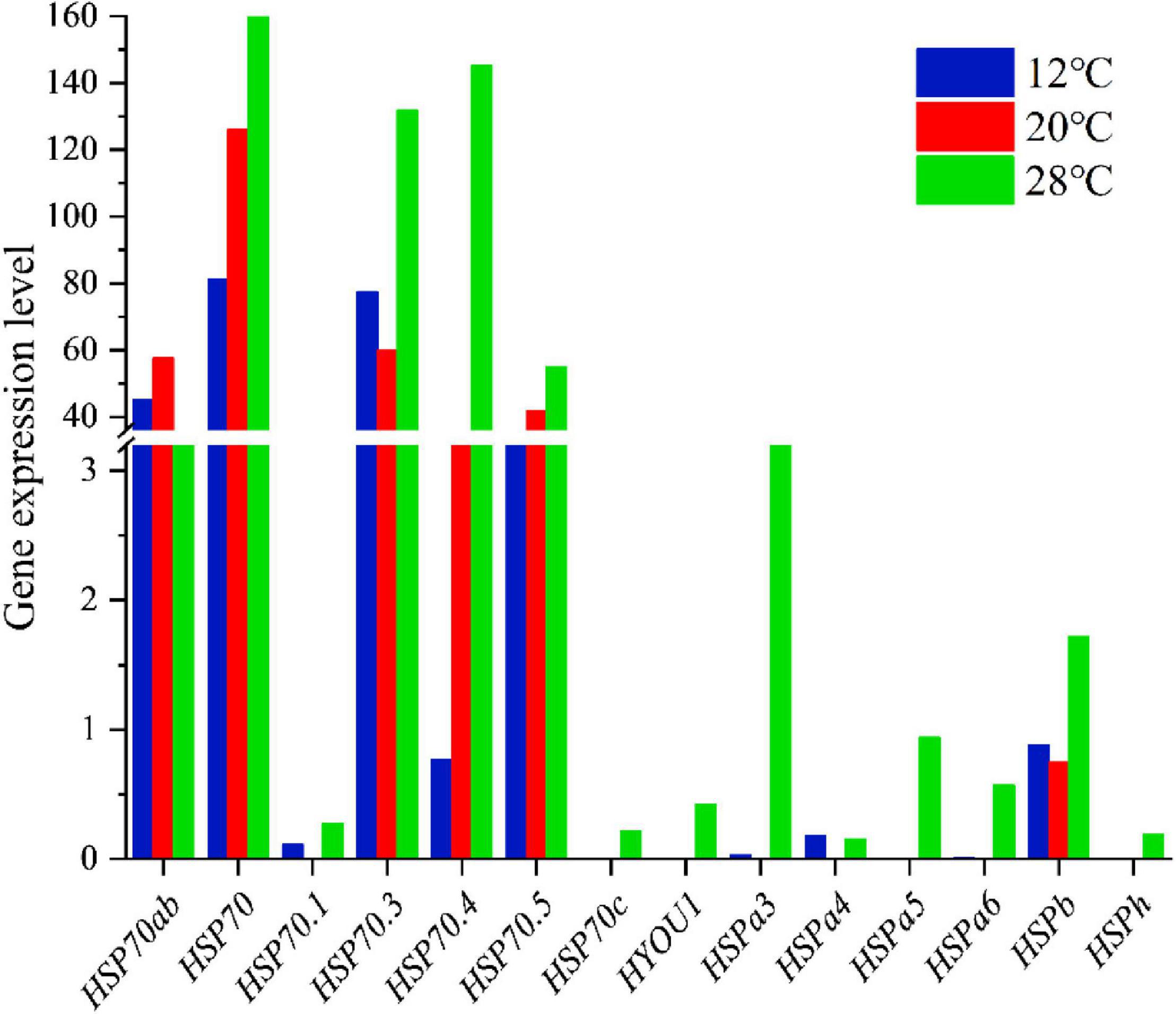
Figure 6. The histogram visualizes the expression level of the HSP70 gene family in the C. japonicas at three temperature gradients.
Discussion
The occurrence time of low tide and sunshine radiation act on temperature fluctuations in the intertidal zone. Specifically, the intertidal zone heats up dramatically with the combination of low tide time and intense solar radiation, but the intertidal zone cools dramatically with the low tide occurring at night (Helmuth and Hofmann, 2001; Helmuth, 2002). Therefore, the ambient temperature in the intertidal zone has very complex temporal and spatial variation, which further shapes the specific temperature adaptation ability of the intertidal organisms (Huey et al., 2012). In fact, intertidal organisms that undergo special temperature domestication also have the ability to adapt to the surrounding temperature fluctuations, thus contributing to their adaptability to temperature fluctuations (Stillman and Tagmount, 2009; Marshall et al., 2011; Ng et al., 2017). The present study chose C. japonica as the research object to explore how intertidal crustaceans respond to temperature fluctuations.
Considering that C. japonica can adjust its physiological state by changing gene expression level in time and further realize homeostasis under temperature stresses, the transcriptomes of C. japonica at different temperatures are meaningful to investigate. A comparative transcriptomic analysis of C. japonica subjected to 12, 20, and 28°C was performed to obtain sufficient evidence to explain the potential effects of temperature stresses on the gene regulation of C. japonica. With the help of improved sequencing techniques and assembly methods, this study obtained transcriptome data with higher assembly efficiency and accuracy and provided new perspectives for identifying the temperature stresses-related gene expression levels of C. japonica. However, a low proportion (37.98%) of unigenes were successfully mapped in the protein databases, probably because the genome resources of crustaceans are relatively scarce.
The transcriptome data of C. japonica individuals at three temperature gradients provide valuable resources for exploring the critical genes and related regulatory mechanisms of C. japonica under temperature stresses. As expected, the expression levels of numerous unigenes varied remarkably with temperature changes (from 20 to 12 or 28°C). This result may mean that C. japonica subjected to temperature stresses at 28 and 12°C have a promoted co-expression of multiple functional genes to produce an effective temperature regulation mechanism (Liu et al., 2013; Jeffries et al., 2014). The number of DEGs in C. japonica between 20 and 28°C was much higher than that between 20 and 12°C. Moreover, the C. japonica individuals exposed to 12 and 28°C also had more DEGs. Similar gene expression trends were found in Larimichthys crocea exposed to high- and low- temperature stresses (Qian and Xue, 2016). Furthermore, most unigenes in C. japonica at 28°C were up-regulated relative to those at 20 and 12°C. This seems to mean that C. japonica is more susceptible to high temperatures in their survival temperature range (5–30°C), and some function genes were thus activated. In fact, previous studies have confirmed that the intertidal environment is more likely to bring organisms closer to the thermal maximum than the subtidal environment; thus, intertidal organisms may be less able to increase their upper thermal tolerance limits (Stillman and Somero, 2000; Hofmann and Todgham, 2010). Compared with 20°C, a small number of DEGs and up-regulated unigenes were found in C. japonica at 12°C. That’s probably because the physiological functions (such as food intake, metabolic capacity of digestive system, enzyme activity, and binding capacity of iron ions) of organisms under low temperature are reduced, which may indirectly and passively inhibit gene expression (Liu et al., 2013; Jeffries et al., 2014). In conclusion, we suspected that 12°C was thought to inhibit the physiological functions of C. japonica and indirectly affected the active expression of some functional genes, and the expression at 28°C showed completely opposite trends. Considering the survival temperature (5–30°C) of C. japonica, it is not easy to conclude that high temperature poses a more significant threat to C. japonica than low temperature, although higher temperature has been confirmed to affect biological survival directly or indirectly by altering dissolved oxygen (Wulff et al., 2007) and salinity (Durack et al., 2012). Therefore, setting lower temperatures is necessary for future research.
The annotation information indicates that DEGs were enriched in GO terms such as cell, cell part, membrane, membrane part, binding, catalytic activity, metabolic process and cellular process, because the cellular structure and function of C. japonica exposed to temperature stresses (whether high- or low- temperatures) will change. At this time, some functional enzymes and structural proteins will be activated to help C. japonica resist external stresses (Robinson et al., 2010). Moreover, the increased metabolism of C. japonica under temperature stresses (whether high- or low- temperature) also helps them to be more efficient in coping with the increased energy expenditure (Adeghate et al., 2011). Compared with 12 or 20°C, the DEGs of C. japonica at 28°C were remarkably enriched in protein processing in the endoplasmic reticulum pathway. Similar results were found in our previous study of O. oratoria (Lou et al., 2019). Proteins in C. japonica cells may have misfolded or unfolded because of high-temperature stress. Endoplasmic reticulum (ER) stress is activated as a protective stress response in cells, which reduces the concentration of unfolded proteins, prevents the accumulation of misfolded proteins, and ultimately restores the normal function of cells (Ron and Walter, 2007; Imrie and Sadler, 2012). Compared with 20°C, the DEGs of C. japonica at 12°C were remarkably enriched in the insect hormone biosynthesis pathway, probably because C. japonica activates certain critical hormonal pathways to produce hormones that protect them from low-temperature damage, such as adrenocorticotrophic hormone (Stein and Harzsch, 2021). Compared with 28°C, the C. japonica unigenes up-regulated at 12°C were enriched in the lipid metabolism-related pathways, which may provide energy for C. japonica under low-temperature stress which may be because C. japonicas under 12°C need to consume a large amount of energy to ensure the rapid response of regulatory mechanism, and lipid metabolism provides guarantee for energy supply.
As molecular chaperones, the HSPs produced by the HSP gene families are usually induced by various biotic and abiotic stressors (Song et al., 2016). Previous studies have confirmed that HSPs possess three principal biochemical activities, namely, chaperonin activity, cellular redox state regulation, and protein turnover (Georgopoulos and Welch, 1993; Parsell and Lindquist, 1993; Otterbein and Choi, 2000). In the present study, eight HSP genes were selected, and their expression levels were analyzed at the transcriptome and qRT-PCR levels, respectively. Several HSP genes (HSP20, HSP40, and HSP70) were up-regulated at 28°C, and HSP83 and HSP90 were up-regulated at 12 and 28°C. This result could mean that temperature stress-induced an increase in the expression levels of these HSP genes, and the produced HSPs may play essential roles in cellular protection, stress response, bacterial infection, parasitization, and inflammation (Molina et al., 2000; Qin et al., 2013). Moreover, the expression levels of HSP20, HSP40, and HSP70 at 12°C may be reduced by a decrease in the basal metabolic rate of C. japonica. The HSP100 gene was only up-regulated at 12°C. Notably, up-regulation of the HSP100 gene improves heat and cold resistance in plants (Xu et al., 2008). Therefore, the up-regulation of the HSP100 gene may help improve C. japonica’s tolerance at 12°C. However, this finding does not mean that the HSP100 gene is not effective against the thermal stress of C. japonica. Additionally, the remaining HSP genes (HSP22 and HSP23) were down-regulated at 12 and 28°C. Two reasons were suggested: (i) the HSPs produced by these HSP genes may act as housekeeping proteins and be constitutively expressed in unstressed cells; therefore, they are down-regulated in response to temperature stress (Song et al., 2016); (ii) these HSP genes have missed or not reached their peak expression. In conclusion, our results confirmed for the first time that HSP genes might provide an effective cellular stress-buffering system and protect C. japonica against temperature stresses. However, we do not deny that the expression of gene members of the HSP genes is functionally and temporally specific, and further confirmation is needed in future research.
Our study focused on the HSP70 gene family, and the expression levels of 14 gene family members in C. japonica with different temperature gradients were analyzed. HSP70 gene family is the most conserved and important non-specific cell-protective protein with many important functions. The established biological functions of the HSP70 gene family mainly include: (i) it can act as a molecular chaperone to bind to stable proteins to form unstable conformations that promote polypeptide chain folding; (ii) participates in transmembrane transport of organelle proteins (Kang et al., 1990; Nelson et al., 1992). In the present study, a large proportion of HSP70 gene family members (HSP70, HSP70.1, HSP70.3, HSP70.4, HSP70.5, HSP70c, HYOU1, HSPa3, HSPa4, HSPa5, HSPa6, HSPb, HSPh) were up-regulated at 12 or 28°C, especially at 28°C. We speculated that the up-regulation of these HSP70 genes in C. japonica at 28°C could enhance the resistance of cells to high-temperature damage, thus accelerating the degradation of abnormal proteins and enhancing the stability of intracellular structures (Franklin et al., 2005). Meanwhile, the HSP70 gene family has also been shown to be involved in multiple apoptotic signaling pathways, which may inhibit the apoptosis of C. japonica (Parcellier et al., 2003). Additionally, the HSP70 gene family also participates in immune responses and inhibits inflammatory responses. Up-regulation of the HSP70 gene family on the surface of C. japonica cells damaged by temperature stress will help the immune system recognize and eliminate toxic cells (Hu and Anselmo, 2000; Parent et al., 2009). Furthermore, the HYOU1 gene, which encodes a hypoxia up-regulated protein, was also found to be up-regulated at 28°C, confirming that high temperatures may lead to low oxidation of seawater. Therefore, up-regulated of the HYOU1 gene at 28°C may promote the adaptation of C. japonicas to hypoxia-induced stress caused by 28°C. In fact, hypoxia up-regulated protein is also a protein associated with the ER, and its expression can be regulated by ER stress (Ozawa et al., 2001; Stojadinovic et al., 2007). In conclusion, we hypothesized that the up-regulation of the HSP70 gene family plays a critical role in maintaining the normal biological activity and survival rate of C. japonica cells under temperature stress.
Conclusion
The behavior and physiology of intertidal crustaceans are very susceptible to temperature stresses. Gene regulation may contribute to the rapid homeostasis restoration of C. japonica under temperature stresses. The present study analyzed the genetic regulation differences of C. japonica at different temperatures according to the comparative transcriptome. These results can provide basic information for exploring the temperature regulation mechanism of intertidal crustaceans. Furthermore, our research is expected to provide valuable information for further understanding how temperature changes affect the survival of marine crustaceans.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, PRJNA793994.
Author Contributions
FL and ZH conceptualized the study and conducted the analyses. FL, YW, ZH, and BS wrote and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Zhejiang Provincial Natural Science Foundation of China (LR21D060003) and the National Natural Science Foundation of China (32070513).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adeghate, E., Adem, A., Hasan, M. Y., Tekes, K., and Kalasz, H. (2011). Medicinal chemistry and actions of dual and pan PPAR modulators. Open Med. Chem. J. 5, 93–98. doi: 10.2174/1874104501105010093
Altschul, S. F. (1990). Basic local alignment search tool (BLAST). J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Boyd, P. W., Collins, S., Dupont, S., Fabricius, K., Gattuso, J. P., Havenhand, J., et al. (2018). Experimental strategies to assess the biological ramifications of multiple drivers of global ocean change-A review. Glob. Chang Biol. 24, 2239–2261. doi: 10.1111/gcb.14102
Cheng, J., Hui, M., and Sha, Z. L. (2019). Transcriptomic analysis reveals insights into deep-sea adaptations of the dominant species, Shinkaia crosnieri (Crustacea: decapoda: anomura), in habiting both hydrothermal vents and cold seeps. BMC Genomics 20:388. doi: 10.1186/s12864-019-5753-7
Conesa, A., Götz, S., García-Gómez, J. M., Terol, J., Talón, M., and Robles, M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. doi: 10.1093/bioinformatics/bti610
Durack, P. J., Wijffels, S. E., and Matear, R. J. (2012). Ocean salinities reveal strong global water cycle intensification during 1950 to 2000. Science 336, 455–458. doi: 10.1126/science.1212222
Edwards, M., and Richardson, A. J. (2004). Impact of climate change on marine pelagic phenology and trophic mismatch. Nature 430, 881–884. doi: 10.1038/nature02808
Franklin, T. B., Krueger-Naug, A. M., Clarke, D. B., Arrigo, A. P., and Currie, R. W. (2005). The role of heat shock proteins Hsp70 and Hsp27 in cellular protection of the central nervous system. Int. J. Hyperthermia 21, 379–392. doi: 10.1080/02656730500069955
Georgopoulos, C., and Welch, W. J. (1993). Role of the major heat shock proteins as molecular chaperones. Annu. Rev. Cell Biol. 9, 601–634. doi: 10.1146/annurev.cb.09.110193.003125
Grabherr, M., Haas, B., Yassour, M., Levin, J., Thompson, D., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Greenaway, P. (2003). Terrestrial adaptations in the anomura (Crustacea: decapoda). Mem. Museum Victor. 60, 13–26. doi: 10.24199/j.mmv.2003.60.3
Helmuth, B. (2002). How do we measure the environment? Linking intertidal thermal physiology and ecology through biophysics. Integr. Comp. Biol. 42, 837–845. doi: 10.1093/icb/42.4.837
Helmuth, B. S. T., and Hofmann, G. E. (2001). Microhabitats, thermal heterogeneity, and patterns of physiological stress in the rocky intertidal zone. Biol. Bull. 201, 374–384. doi: 10.2307/1543615
Hofmann, G. E., and Todgham, A. E. (2010). Living in the now: physiological mechanisms to tolerate a rapidly changing environment. Annu. Rev. Physiol. 72, 127–145. doi: 10.1146/annurev-physiol-021909-135900
Holt, R. (1990). The microevolutionary consequences of climate change. Trends Ecol. Evol. 5, 311–315. doi: 10.1016/0169-5347(90)90088-U
Hu, J., and Anselmo, D. (2000). In vitro reconstitution of a functional duck hepatitis B virus reverse transcriptase: posttranslational activation by Hsp90. J. Virol. 74, 11447–11455. doi: 10.1128/JVI.74.24.11447-11455.2000
Huey, R. B., Kearney, M. R., Krockenberger, A., Holtum, J., and Williams, S. E. (2012). Predicting organismal vulnerability to climate warming: roles of behaviour, physiology and adaptation. Phil. Trans. R. Soc. B. 367, 1665–1679. doi: 10.1098/rstb.2012.0005
Humphreys, D. T., Westman, B. J., Martin, D. I. K., and Preiss, T. (2005). MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc. Nati. Acad. Sci. U. S. A. 102, 16961–16966. doi: 10.1073/pnas.0506482102
Imrie, D., and Sadler, K. C. (2012). Stress management: how the unfolded protein response impacts fatty liver disease. J. Hepatol. 57, 1147–1151. doi: 10.1016/j.jhep.2012.06.018
Jeffries, K. M., Hinch, S. G., Sierocinski, T., Pavlidis, P., and Miller, K. M. (2014). Transcriptomic responses to high water temperature in two species of Pacific salmon. Evol. Appl. 7, 286–300. doi: 10.1111/eva.12119
Kang, P. J., Osterrnann, J., Shilling, J., Neupert, W., Craig, E. A., and Pfanner, N. (1990). Requirement for hsp70 in the mitochondrial matrix for tran slocation and folding of precursor proteins. Nature 348, 137–143. doi: 10.1038/348137a0
Kleinjan, D., and Van, H. (2005). Long-range control of gene expression: emerging mechanisms and disruption in disease. Am. J. Hum. Genet. 76, 8–32. doi: 10.1086/426833
Legrand, E., Riera, P., Pouliquen, L., Bohner, O., Cariou, T., and Martin, S. (2018). Ecological characterization of intertidal rockpools: seasonal and diurnal monitoring of physicochemical parameters. Reg. Stud. Mar. Sci. 17, 1–10. doi: 10.1016/j.rsma.2017.11.003
Leng, N., Dawson, J. A., Thomson, J. A., Ruotti, V., Rissman, A. I., Smits, B. M. G., et al. (2013). EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics 29, 1035–1043. doi: 10.1093/bioinformatics/btt087
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Liu, S. K., Wang, X. L., Sun, F. Y., Zhang, J. R., Feng, J. B., Liu, H., et al. (2013). RNA-Seq reveals expression signatures of genes involved in oxygen transport, protein synthesis, folding, and degradation in response to heat stress in catfish. Physiol. Genomics 45, 462–476. doi: 10.1152/physiolgenomics.00026.2013
Lou, F. R., Gao, T. X., and Han, Z. Q. (2019). Transcriptome analyses reveal alterations in muscle metabolism, immune responses and reproductive behavior of Japanese mantis shrimp (Oratosquilla oratoria) at different cold temperature. Comp. Biochem. Phys. D 32:100615. doi: 10.1016/j.cbd.2019.100615
Marshall, D. J., Dong, Y. W., Mcquaid, C. D., and Williams, G. A. (2011). Thermal adaptation in the intertidal snail Echinolittorina malaccana contradicts current theory by revealing the crucial roles of resting metabolism. J. Exp. Biol. 214, 3649–3657. doi: 10.1242/jeb.059899
Milne Edwards, A. (1861). Études zoologiques sur les Crustacés récents de la famille des Portuniens. Archives du Muséum d’Histoire naturelle 10, 309–428. doi: 10.5962/bhl.title.10629
Molina, A., Biemar, F., Müller, F., Iyengar, A., Prunet, P., Maclean, N., et al. (2000). Cloning and expression analysis of an inducible HSP70 gene from tilapia fish. FEBS Lett. 474, 5–10. doi: 10.1016/S0014-5793(00)01538-6
Molinos, J. G., Halpern, B. S., Schoeman, D. S., Brown, C. J., Kiessling, W., Moore, P. J., et al. (2016). Climate velocity and the future global redistribution of marine biodiversity. Nat. Clim. Change 6:83. doi: 10.1038/nclimate2769
Nadia, M. D., and Alicia, O. (2014). Corset: enabling differential gene expression analysis for de novo assembled transcriptomes. Genome Biol. 15:410. doi: 10.1186/s13059-014-0410-6
Nelson, R. J., Ziegelhoffer, T., Nicolet, C., Werner-Washburne, M., and Craig, E. A. (1992). The translation machinery and 70 kd heat shock protein cooperate in protein synthesis. Cell 71, 97–105. doi: 10.1016/0092-8674(92)90269-I
Ng, T. P. T., Lau, S. L. Y., Seuront, L., Davies, M. S., Stafford, R., Marshall, D. J., et al. (2017). Linking behaviour and climate change in intertidal ectotherms: insights from littorinid snails. J. Exp. Mar. Biol. Ecol. 492, 121–131. doi: 10.1016/j.jembe.2017.01.023
Otterbein, L. E., and Choi, A. M. (2000). Heme oxygenase: colors of defense against cellular stress. Am. J. Physiol. Lung Cell Mol. Physiol. 279, L1029–L1037. doi: 10.1152/ajplung.2000.279.6.L1029
Ozawa, K., Kondo, T., Hori, O., Kitao, Y., Stern, D. M., Eisenmenger, W., et al. (2001). Expression of the oxygen-regulated protein ORP150 accelerates wound healing by modulating intracellular VEGF transport. J. Clin. Invest. 108, 41–50. doi: 10.1172/JCI11772
Parcellier, A., Gurbuxani, S., Schmitt, E., Solary, E., and Garrido, C. (2003). Heat shock proteins, cellular chaperones that modulate mitochondrial cell death pathways. Biochem. Biophys. Res. Commun. 304, 505–512. doi: 10.1016/S0006-291X(03)00623-5
Parent, R., Qu, X., Petit, M. A., and Beretta, L. (2009). The heat shock cognate protein 70 is associated with hepatitis C virus particles and modulates virus infectivity. Hepatology 49, 1798–1809. doi: 10.1002/hep.22852
Parmesan, C. (2006). Ecological and evolutionary responses to recent climate change. Annu. Rev. Ecol. Evol. Syst. 37, 637–669. doi: 10.1146/annurev.ecolsys.37.091305.110100
Parsell, D. A., and Lindquist, S. (1993). The function of heat-shock proteins in stress tolerance: degradation and reactivation of damaged proteins. Annu. Rev. Genet. 27, 437–496. doi: 10.1146/annurev.ge.27.120193.002253
Perry, A. L., Low, P. J., Ellis, J. R., and Reynolds, J. D. (2005). Climate change and distribution shifts in marine fishes. Science 308, 1912–1915. doi: 10.1126/science.1111322
Poloczanska, E. S., Burrows, M. T., Brown, C. J., Molinos, J. G., Halpern, B. S., Hoegh-Guldberg, O., et al. (2016). Responses of marine organisms to climate change across oceans. Front. Mar. Sci. 3:62. doi: 10.3389/fmars.2016.00062
Pörtner, H. O. (2010). Oxygen- and capacity-limitation of thermal tolerance: a matrix for integrating climate-related stressor effects in marine ecosystems. J. Exp. Biol. 213, 881–893. doi: 10.1242/jeb.037523
Qian, B., and Xue, L. (2016). Liver transcriptome sequencing and de novo annotation of the large yellow croaker (Larimichthy crocea) under heat and cold stress. Mar. Genomics 25, 95–102. doi: 10.1016/j.margen.2015.12.001
Qin, C., Zhao, D., Gong, Q., Qi, Z., Zou, Y., Yue, X., et al. (2013). Effects of pathogenic bacterial challenge after acute sublethal ammonia-N exposure on heat shock protein 70 expression in Botia reevesae. Fish Shellfish Immunol. 35, 1044–1047. doi: 10.1016/j.fsi.2013.07.007
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Romero, I., Ruvinsky, I., and Gilad, Y. (2012). Comparative studies of gene expression and the evolution of gene regulation. Nat. Rev. Genet. 13, 505–516. doi: 10.1038/nrg3229
Ron, D., and Walter, P. (2007). Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Bio. 7, 519–529. doi: 10.1038/nrm2199
Saucedo, E., Ocampo, L., Monteforte, M., and Bervera, H. (2004). Effect of temperature on oxygen consumption and ammonia excretion in the Calafia mother-of-pearl oyster, Pinctada mazatlanica (Hanley, 1856). Aquaculture 229, 377–387. doi: 10.1016/S0044-8486(03)00327-2
Schram, F. R. (2013). Comments on crustacean biodiversity and disparity of body plans. Nat Hist. Crustacea 1, 1–33. doi: 10.1093/acprof:osobl/9780195398038.003.0001
Scott, G. R., and Johnston, I. A. (2012). Temperature during embryonic development has persistent effects on thermal acclimation capacity in zebrafish. Proc. Natl. Acad. Sci. U. S. A. 109, 14247–14252. doi: 10.1073/pnas.1205012109
Somero, G. N. (2010). The physiology of climate change: how potentials for acclimatization and genetic adaptation will determine’ winners’ and’ losers’. J. Exp. Biol. 213, 912–920. doi: 10.1242/jeb.037473
Somero, G. N. (2012). The physiology of global change: linking patterns to mechanisms. Ann. Rev. Mar. Sci. 4, 39–61. doi: 10.1146/annurev-marine-120710-100935
Song, L., Li, C., Xie, Y. J., Liu, S. K., Zhang, J. R., Yao, J., et al. (2016). Genome-wide identification of HSP70 genes in channel catfish and their regulated expression after bacterial infection. Fish Shellfish Immunol. 49, 154–162. doi: 10.1016/j.fsi.2015.12.009
Soofi, W., Goeritz, M. L., Kispersky, T. J., Prinz, A. A., Marder, E., and Stein, W. (2014). Phase maintenance in a rhythmic motor pattern during temperature changes in vivo. J. Neurophysiol. 111, 2603–2613. doi: 10.1152/jn.00906.2013
Stein, W., and Harzsch, S. (2021). The Neurobiology of Ocean Change-insights from decapod crustaceans. Zoology 144:125887. doi: 10.1016/j.zool.2020.125887
Stillman, J. H., and Somero, G. N. (2000). A comparative analysis of the upper thermal tolerance limits of eastern Pacific porcelain crabs, genus Petrolisthes: influences of latitude, vertical zonation, acclimation, and phylogeny. Physiol. Biochem. Zool. 73, 200–208. doi: 10.1086/316738
Stillman, J. H., and Tagmount, A. (2009). Seasonal and latitudinal acclimatization of cardiac transcriptome responses to thermal stress in porcelain crabs, Petrolisthes cinctipes. Mol. Ecol. 18, 4206–4226. doi: 10.1111/j.1365-294x.2009.04354.x
Stojadinovic, A., Hooke, J. A., Shriver, C. D., Nissan, A., Kovatich, A. J., Kao, T. C., et al. (2007). HYOU1/Orp150 expression in breast cancer. Med. Sci. Monit. 13, BR231–BR239. doi: 10.1051/medsci/200723111063
Wang, Z., Gerstein, M. M., and Snyder, M. (2009). RNA-seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63. doi: 10.1038/nrg2484
Whitehead, A., Galvez, F., Zhang, S., Williams, L., and Oleksiak, M. (2011). Functional genomics of physiological plasticity and local adaptation in killifish. J. Hered. 2011, 499–511. doi: 10.1093/jhered/esq077
Whiteley, N. M., Suckling, C. C., Ciotti, B. J., Brown, J., Mccarthy, I. D., Gimenez, L., et al. (2018). Sensitivity to near-future CO2 conditions in marine crabs depends on their compensatory capacities for salinity change. Sci. Rep. 8:15639. doi: 10.1038/s41598-018-34089-0
Whiteley, N. M., and Taylor, E. (2015). “Responses to Environmental Stresses: oxygen, Temperature and pH”in Physiology. The Natural History of the Crustacea. Eds E. S. Chang and M. Thiel (New York: Oxford University Press). 320–358.
Wray, G. (2007). The evolutionary significance of cis-regulatory mutations. Nat. Rev. Genet. 8, 206–216. doi: 10.1038/nrg2063
Wulff, F., Savchuk, O. P., Sokolov, A., Humborg, C., and Morth, C. M. (2007). Management options and effects on a marine ecosystem: assessing the future of the Baltic. Ambio 36, 243–249. doi: 10.1579/0044-7447(2007)36[243:moaeoa]2.0.co;2
Xu, S. T., Sun, W. X., Tian, J. P., and Wang, C. Y. (2008). Plant heat shock protein HSP100/ClaB and its applications in improvement of heat and cold resistances in plants. Plant Physiol. Agric. Appl. 44, 804–810.
Yu, C. G., Song, H. T., and Yao, G. Z. (2004). Assessment of the crab stock biomass in the continental shelf waters of the East China Sea. J. Fish. China 28, 41–46.
Zheng, W. (2015). Molecular Phylogeography Study of Charybdis Japonica and Charybdis Bimaculata in East China Sea and Yellow Sea. China: Zhejiang Ocean University.
Keywords: Charybdis japonica, comparative transcriptome, temperature stress, gene regulation mechanism, adaption
Citation: Lou F, Wang Y, Han Z and Shui B (2022) Comparative Transcriptome Reveals the Molecular Regulation Mechanism of Charybdis japonica to High- and Low-Temperature Stresses. Front. Mar. Sci. 9:849485. doi: 10.3389/fmars.2022.849485
Received: 06 January 2022; Accepted: 31 January 2022;
Published: 17 February 2022.
Edited by:
Eduardo Almansa, Spanish Institute of Oceanography (IEO), SpainReviewed by:
Jose Maria Landeira, Instituto de Oceanografía y Cambio Global (IOCAG), SpainKhor Waiho, University of Malaysia Terengganu, Malaysia
Copyright © 2022 Lou, Wang, Han and Shui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhiqiang Han, ZDYzMzkxMjRAMTYzLmNvbQ==; Bonian Shui, c2h1aWJvbmlhbkAxNjMuY29t
 Fangrui Lou
Fangrui Lou Yuju Wang2
Yuju Wang2 Zhiqiang Han
Zhiqiang Han