
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 01 March 2022
Sec. Marine Fisheries, Aquaculture and Living Resources
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.847585
This article is part of the Research TopicMollusk Breeding and Genetic ImprovementView all 25 articles
Marine heatwaves (MHWs) can severely affect bivalves and ecosystems they support. Heat shock proteins (HSPs) are a group of molecular chaperones playing a critical role in the cellular protection and thermo tolerance and thereby constraining physiological responses of marine bivalves to MHWs. Here, we cloned the full-length of HSP70 cDNA from the Pinctada maximal (PmHSP70) and evaluated the expression of PmHSP70 in pearl oysters under acute and repeatedly occurring MHWs conditions. The full-length of PmHSP70 is 2,474 bp, containing an ORF of 1,956 bp encoding 655 amino acids with a predicted molecular weight of 71.23 kDa and 5.26 theoretical isoelectric point. Under the scenario of acute MHWs, the expression of PmHSP70 was significantly highly expressed at 32 and 36°C, and reached the highest at 12 and 72 h, respectively, indicating that pearl oysters rapidly up-regulated the expression of HSP70 in response to MHWs. In the repeatedly occurring MHWs scenario, the thermal response of pearl oysters was alleviated, as best exemplified by significantly lowered expression levels of PmHSP70. Therefore, we speculate that long-term and repeated MHWs can alleviate the thermal stress of pearl oysters. This finding is encouraging and will provide us with meaningful insights into the acclimation of marine bivalves to extreme environments in the future.
Marine heatwaves (MHWs) are prolonged discrete anomalously climatic events that can last for days or longer (Hobday et al., 2016). Since 2011, MHWs have been retrospectively and contemporaneously observed in the world’s oceans and now are recognized to become more frequent, intense and longer-lasting over a wide range of spatio-temporal scales (Holbrook et al., 2020). Although MHWs can be short-term and discrete events, they can have devastating impacts on marine ecosystems (Smale et al., 2019). Accumulating evidence indicates devastating impacts of MHWs on marine organisms, especially sessile species such as corals (Fordyce et al., 2019; Rendina et al., 2019) and mollusks (Caputi et al., 2016; Chandrapavan et al., 2019; Zhao et al., 2019; Amorim et al., 2020; Scanes et al., 2020) because they must endure anomalously high seawater temperatures during MHWs events. Stress responses caused by seawater temperature sudden changes due to MHWs can affect the innate immune system of marine bivalves (Nie et al., 2017; Rahman and Rahman, 2021). Sessile marine bivalves are vulnerable to heatwaves because they cannot physically remove themselves and seek refuge (Pörtner, 2001; Somero, 2005), and several studies have demonstrated that MHWs causes the mass mortality events in mytilid mussels such as Mytilus californianus (Harley, 2008), M. galloprovincialis (Petes et al., 2007), and M. edulis (Seuront et al., 2019). In response to the challenges associated with temperature changes caused by MHWs, some aquatic organisms employ biochemical, physiological, and molecular mechanisms for self-protection at extreme temperatures (Dong et al., 2020), such as increasing the activity of enzymes and up-regulating the expression of HSPs genes (He et al., 2021; Xu et al., 2021, 2022).
Heat shock protein (HSP) expression is a key cellular mechanism of acclimation to environmental temperature fluctuations (Bedulina et al., 2013). Under thermal stress, HSPs perform critical protein-stabilizing functions (Tomanek and Somero, 2002) and thus play a crucial role in the development of thermotolerance (Clegg et al., 1998). HSP is a type of conservative family protein (Hu et al., 2012), playing an important role in protein folding and biosynthesis as a molecular chaperone (Zhang and Xie, 2019). Among HSPs, the proteins of molecular mass 70 kDa, termed heat shock protein 70 (HSP70), are the most abundant and highly conserved type of protein (Hassan et al., 2019). HSP70 is an important biomolecule for biological resistance, and is most sensitive to environmental degradation. When the cells of the organism are stressed by the external environment, it interacts with many key regulators of signal transduction pathways to control cell homeostasis, proliferation, differentiation, and cell death (Mayer and Bukau, 2005). In particular, HSP70 proteins play an important role in the adaptation of poikilothermic organisms to adverse environmental (Evgen’ev et al., 2007).
Pinctada maxima is an important economic species and is widely distributed in coastal areas such as Australia and the South China Sea (FAO, 2014; Xu et al., 2021). The “South China Sea” pearls cultivated from P. maxima are widely popular in the world because of the large diameter, thick nacre and beautiful (Jones et al., 2013). The Beibu Gulf is a hot spot for MHWs in the South China Sea (Zhang et al., 2020). However, in the context of global climate change and intensified human activities, the duration, frequency and intensity of MHWs have increased sharply in recent decades (Li et al., 2019; Yao et al., 2020; Liu et al., 2021). The coastal waters where pearl oysters live have undergone unprecedented changes, especially MHWs posing a great threat to the survival of P. maxima. Although studies have been conducted to evaluate the effects of temperature on the expression level of HSP genes in some marine invertebrates (Park et al., 2015; Chen et al., 2018; Liu et al., 2018; Zhao et al., 2020), little is known about thermal resistant gene expression responses of the P. maxima to MHWs. Therefore, it is particularly important to explore the molecular mechanism of pearl oysters’ adaptability to MHWs.
In this study, we cloned the full-length of HSP70 by RACE technology, and then evaluated the tissue expression specificity of HSP70 and expression when exposed to acute and repeatedly occurring MHWs. We also attempted to discover the role of HSP70 in thermal resistance ability and plasticity during the process of MHWs acclimation in P. maxima. To clarify the potential contributions of HSP70 in P. maxima to against environmental stress. The results of this study provide useful information on the response of P. maxima to MHWs, and this information can be used to improve the ability of pearl oysters to withstand such adverse environments as MHWs in an aquaculture environment.
The pearl oysters P. maxima (1-year-old; 34.69 ± 5.25 mm shell length, 3.33 ± 0.99 g wet weight) were collected from the Liusha Bay, Beibu Gulf, South China Sea. After arriving in the laboratory, they were carefully cleaned to remove epiphytes on the shell surface and then temporarily cultivate in the laboratory to acclimate to laboratory conditions. The gills (Gi), hepatopancreas (He), mantle (M), and adductor muscle (A) were selected from five healthy individuals for gene tissue distribution, and the tissues were quickly frozen in liquid nitrogen and then stored in a refrigerator at −80°C until RNA extraction.
In order to explore the impact of MHWs on HSPs, we adopted the definition of MHWs by Hobday et al. (2016), who defined the MHWs event is considered a long-term discrete abnormal warm water event, which can be described in terms of intensity, duration, spatial scope, and evolution speed. The specific experimental design is shown in Xu et al. (2022). Specifically, the temperature of 24°C represents contemporaneously annual average sea surface temperature in the Beibu Gulf as control group, and temperature regimes of 28, 32, 36°C correspond to instrumental data recorded during periods of MHWs (Yao et al., 2020; Zhang et al., 2020). In the MHWs experiment, the pearl oysters experienced two MHWs stimulation and two recovery scenarios (Figure 1). In the acute MHWs experiment, 5 gills in each group were carefully dissected at 6, 12, 24, and 72 h after the temperature stabilized. During repeatedly occurring MHWs experiments, the gills of 5 individuals were sampled at 3, 7, 10, and 14 days in repeatedly occurring MHWs experiments, and immediately placed in liquid nitrogen and stored at −80°C pending analysis.
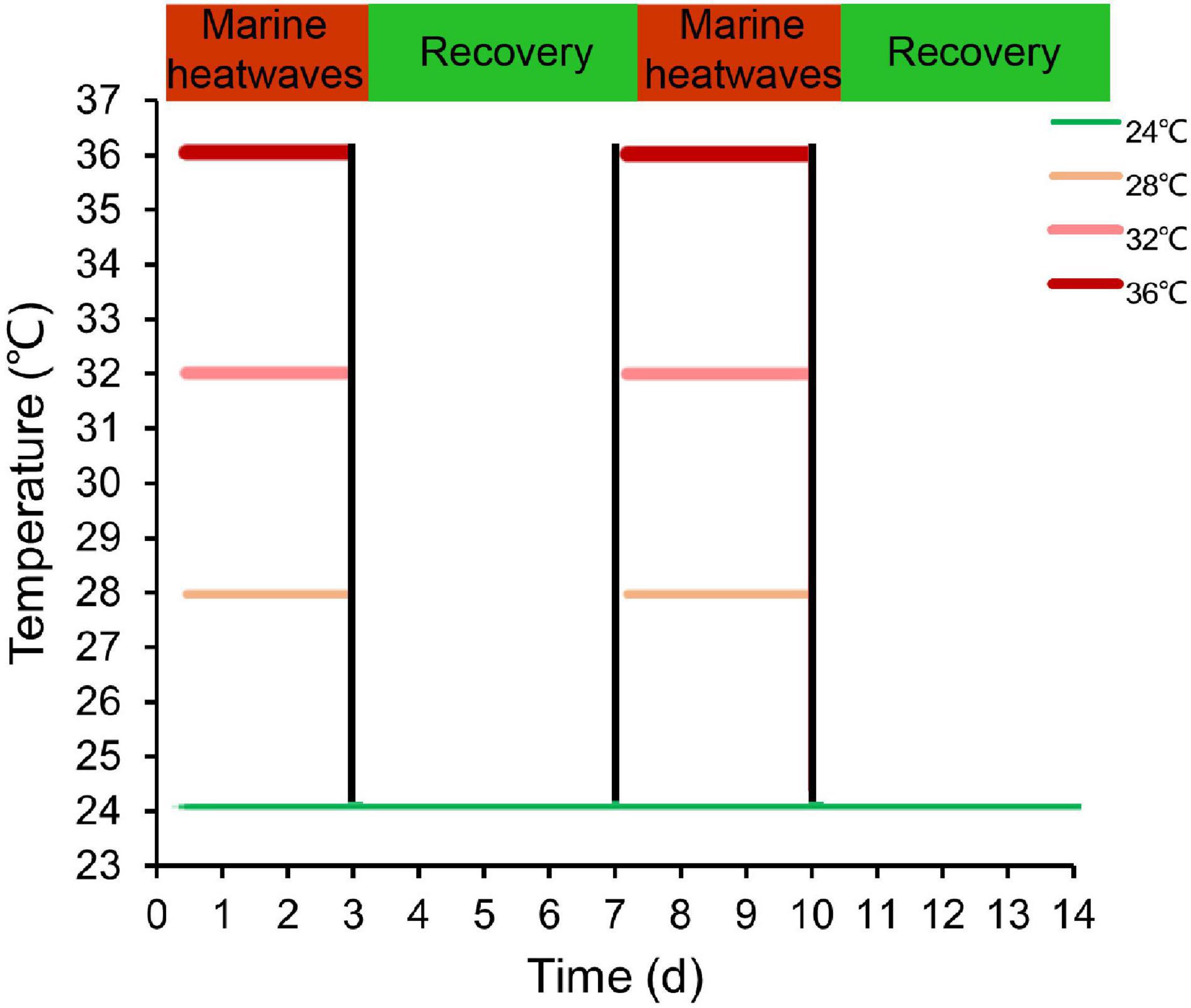
Figure 1. Overview of the experimental design illustrating MHWs scenarios varying in frequency and intensity. Green line indicates the temperature regime designated in the control and color intensity of red lines represents the intensity of temperature regimes designated under MHWs conditions.
The total RNA was extracted from the tissues using the Trizol Reagent (Invitrogen, United States) according to the manufacturer’s instructions. The RNA quantity, purity and integrity were verified by 1% agarose gel electrophoresis and Nano Drop ND1000 ultraviolet spectrophotometer. Preparing 5′RACE and 3′RACE templates according to the instructions of SMARTer™ RACE cDNA Amplification kit follow the instructions of Reverse Transcriptase M-MLV (RNaseH) to synthesize cDNA templates for RT-qPCR.
The partial sequence of HSP70 was obtained from the P. maxima transcriptome database (data of our research group), and we used Primer Premier 5 software to design specific primers (Table 1). Using RACE technology to obtain 3′- and 5′-ends sequences, polymerase chain reaction amplification was performed for intermediate fragment verification. The amplified products were detected by 1% agarose gel electrophoresis. The target gene fragments separated and purified by the purification and recovery kit were connected to the PMD19-T vector and transformed into DH5α competent cells. The positive cloned bacteria were selected by LA (containing ampicillin Apm +) and sequenced by Sangon (Sangon Biotech, Shanghai, China). Then the 3′- and 5′-RACE PCR fragment sequences were aligned to assemble the full nucleotide sequence of the pearl oysters putative HSP70 cDNA. The resulting sequences were verified by the amplification of the whole full length and further subjected to cluster analysis.
The full-length sequence of PmHSP70 was spliced with DNAMAN. The ORF Finder1 in the NCBI database was used to predict the open reading frame and amino acid sequence of the PmHSP70. The physical and chemical properties were determined by Expasy-ProtParam online tool2 and ExPASy-ProtScale3 for amino acid hydrophilicity analysis, use PSITE-Search for Prosite patterns with statistics4 for predicting amino acid sub-basic functional sites. SignalP 4.15 predict amino acid sequence signal peptide and domains were identified with SMART6 with the three-dimensional structure built with the SWISS-MODEL7. The similarity analysis of nucleotide and protein sequence was carried out using NCBI online BLAST,8 and then Clustal W was used to compare multiple sequences of PmHSP70 protein. A phylogenetic tree was constructed using MEGA 6.0 software.
Total RNA was extracted using Trizol reagent (Invitrogen, United States) in accordance with the manufacturer’s instructions. cDNA was synthesized using PrimeScript RT Reagent Kit (TaKaRa, Dalian, China). RT-qPCR was conducted using a Roche LightCycler 480 RT-PCR system with SYBR(R) Premix Ex Taq™ (TOYOBO, Japan) following the manufacturer’s protocol. The reactions were carried out in a total volume of 10 μL containing 5 μL of SYBR mixture, 0.4 μL cDNA, 3.8 μL of ddH2O, 0.4 μL of forwarding primer, and 0.4 μL of the reverse primer (Wang et al., 2019). The comparative Ct method (2–ΔCt) was used to analyze the relative expression of these genes by taking β-actin as an endogenous control.
All data were analyzed using the software SPSS 22.0. Shapiro-Wilk’s test and Levene’s F-test were, respectively, performed to examine the normal distribution and homogeneity of experimental data. Two-way analysis of variance (ANOVA) was applied to test whether and how various MHWs scenarios stimulated in duration, frequency and intensity significantly affect the expression levels of genes. Statistical significance was set at P < 0.05.
The cDNA of PmHSP70 gene was cloned by RACE technology with a full length of 2,474 bp, containing an open reading frame (ORF) of 1,956 bp, encoding 655 amino acids, a 138 bp 5′-terminal untranslated region (UTR) and a 363 bp 3′-UTR. PmHSP70 had a predicted molecular weight of 71.23 kDa, a 5.26 theoretical isoelectric point. ExPASy-ProtParam analysis showed that the aliphatic index of PmHSP70 protein is 80.48, and the instability index is 34.69, indicating the stability of HSP70. Amino acid sub-basic functional sites predicted found PmHSP70 has an ATP/GTP-binding site (AEAYLGKT), three HSP70 family signature (IDLGTTYS, IFDLGGGTFDVSIL, IVLVGGSTRIPKIQK) and the cytoplasmic characteristic motif EEVD. The average coefficient of hydrophilicity (GRAVY) is −0.490, which is a hydrophilic protein. SMART analysis revealed that PmHSP70 has an HSP70 domain at positions 6–612 (Figure 2).

Figure 2. The nucleotide sequence analysis of Pm-HSP70. 5′ UTR and 3′ UTR are indicated with small letters; The deduced amino acid sequences are indicated with capital letters; The start codon in the 5′-UTR was in a box and the stop codons (TAA) was indicated with an asterisk (*). Three signatures were green background; a putative ATP-GTP binding site was yellow background; the cytoplasmic characteristic motif was gray background and the underlined indicates the HSP70 domain.
The species information of multiple alignment and phylogenetic tree are shown in Table 2. The multiple sequence alignment based on the amino acid sequence alignment of PmHSP70 and other bivalves HSP70 shows that HSP70 has high homology among bivalves, and Pinctada fucada martensii HSP70 has the highest homology with PmHSP70, which is 95.70% (Figure 3). The phylogenetic tree shows that PmHSP70 clusters with bivalves and is most closely related to P. f. martensii and Pteria penguin, and is farther from mammals and fish (Figure 4).
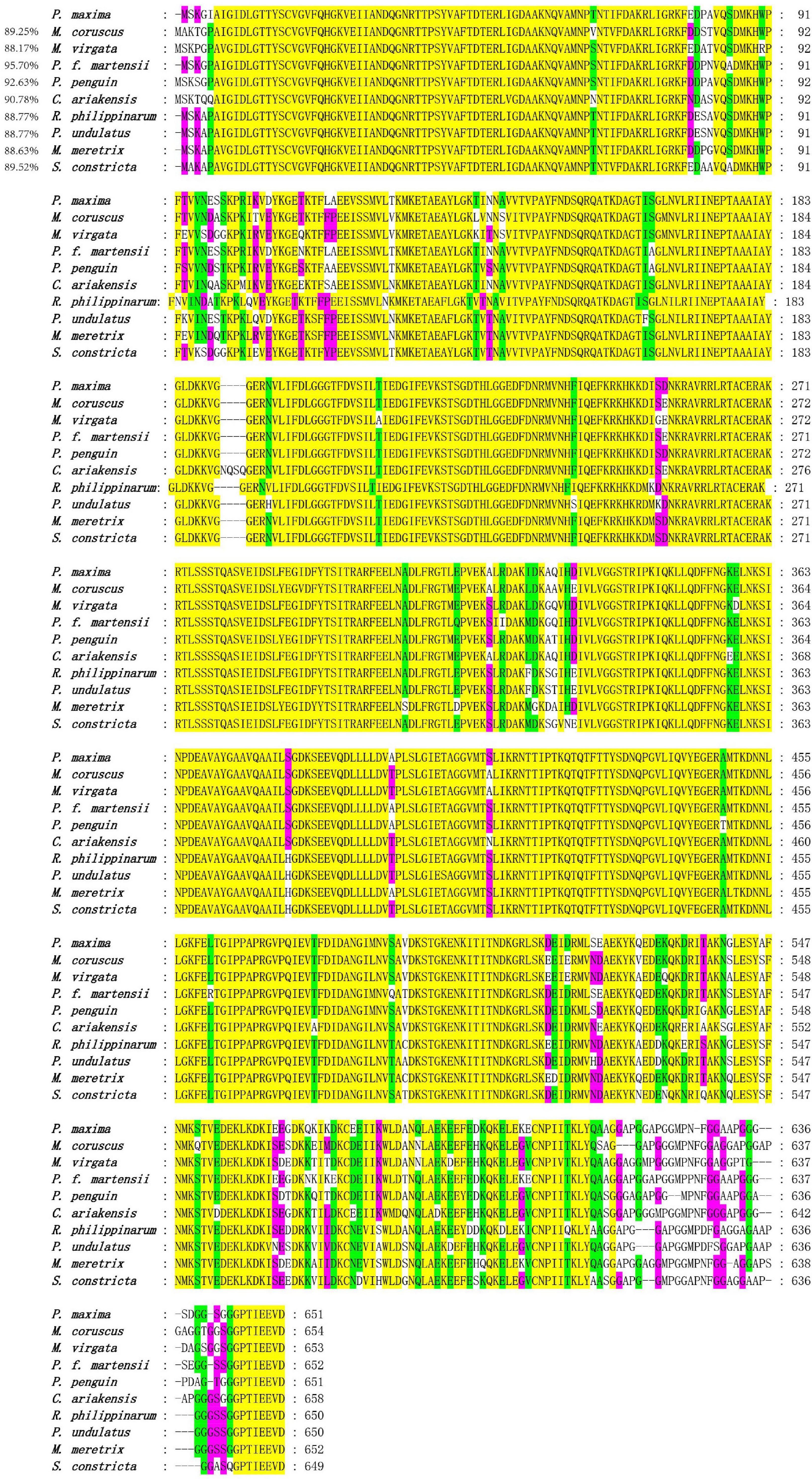
Figure 3. Multiple alignment of the deduced amino acid sequences of HSP70 from P. maxima and other organisms. Yellow means 100% homology, green means homology greater than 90%, purple means homology greater than 80%.
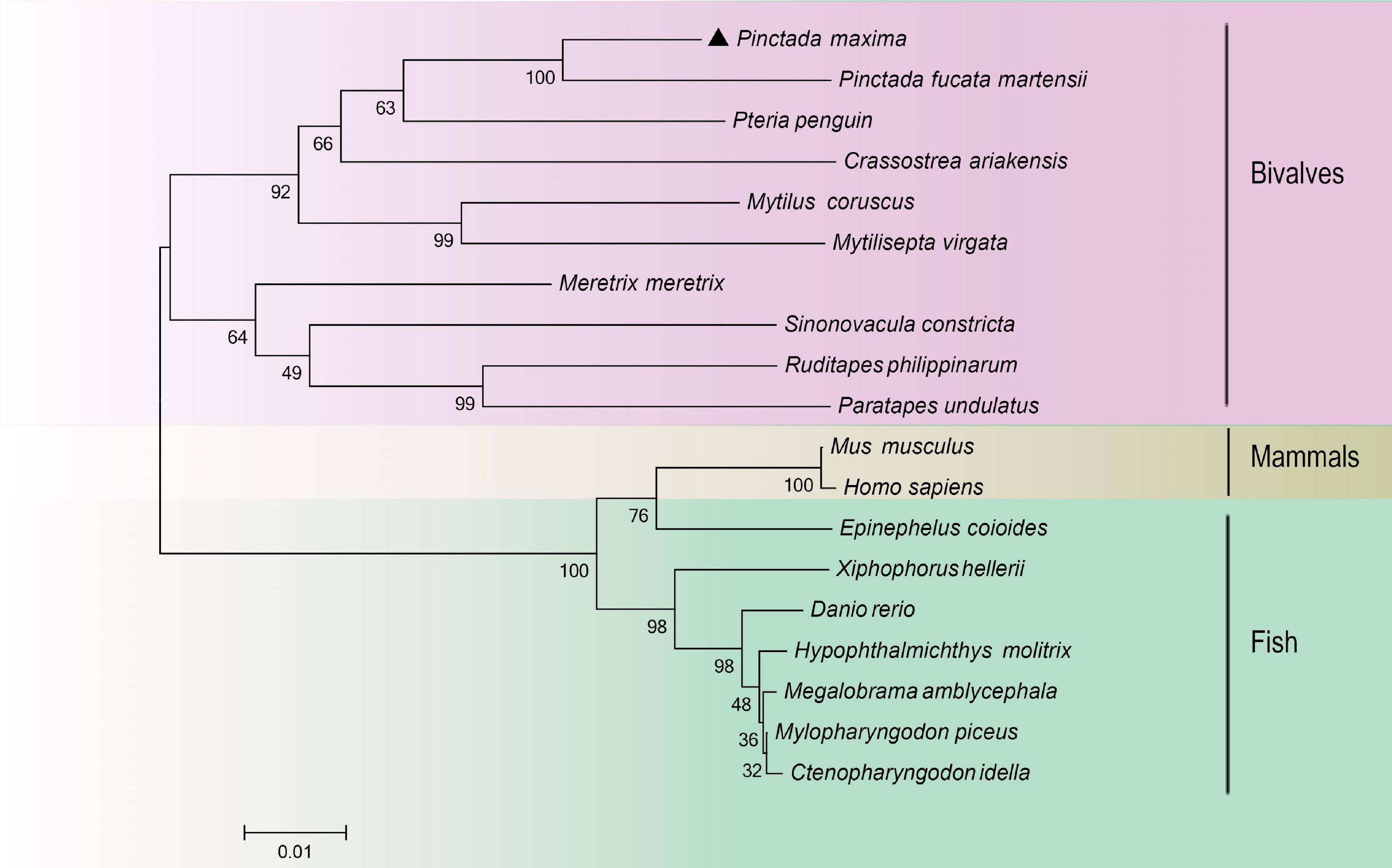
Figure 4. A phylogenetic tree constructed using the neighbor-joining method based on amino acids sequence deduced from HSP70 of Pinctada maxima and other species. Pinctada maxima was labeled with “▲”. The GenBank accession numbers of all the HSP70 were shown in Table 2.
The RT-qPCR was used to investigate the tissue distribution and expression level of PmHSP70 under normal conditions. The results showed that the PmHSP70 was expressed in the mantle, gills, hepatopancreas, and adductor muscle of P. maxima, with the highest expression level in the hepatopancreas and gills, and the lowest expression level in the adductor muscle (Figure 5).
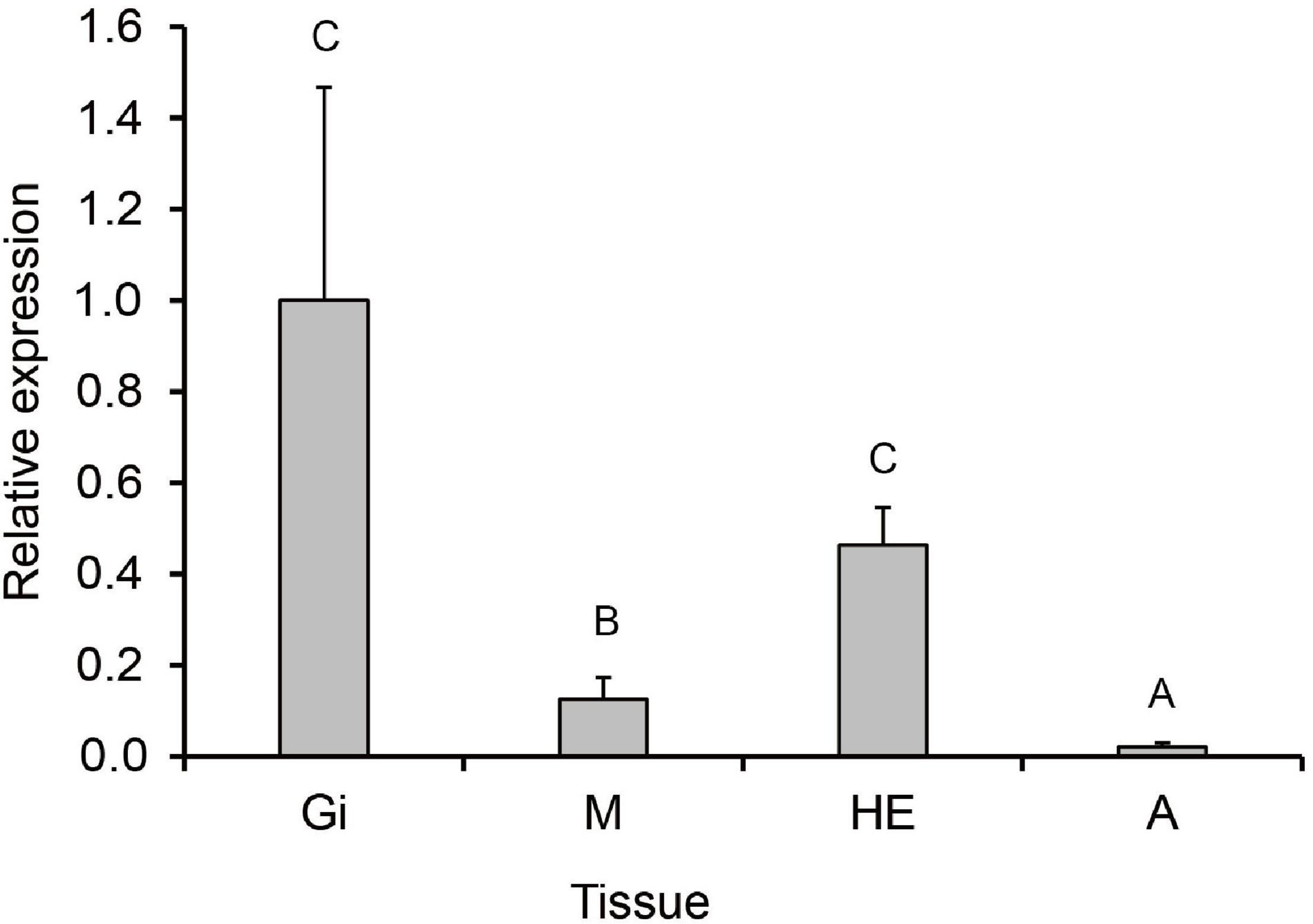
Figure 5. Tissue distribution of PmHSP70 mRNA expression. Different capital letters indicate significant differences (P < 0.05). The four tissues include gills (Gi), hepatopancreas (He), mantle (M), and adductor muscle (A).
During the acute exposure, in gills, compared with the control groups, the expression level was significantly up-regulated in the experimental groups especially in the 36°C groups. After being stimulated by the MHWs, the expression level of PmHSP70 in the 28°C groups reached the highest level at 72 h, while the expression level of the 32°C groups reached the highest level at 12 h (Figure 6). Analysis of variance shows that both temperature of MHWs and stress time made significant impact on the gene expression level (P < 0.05), and their interaction was significant (P < 0.05) (Table 3).
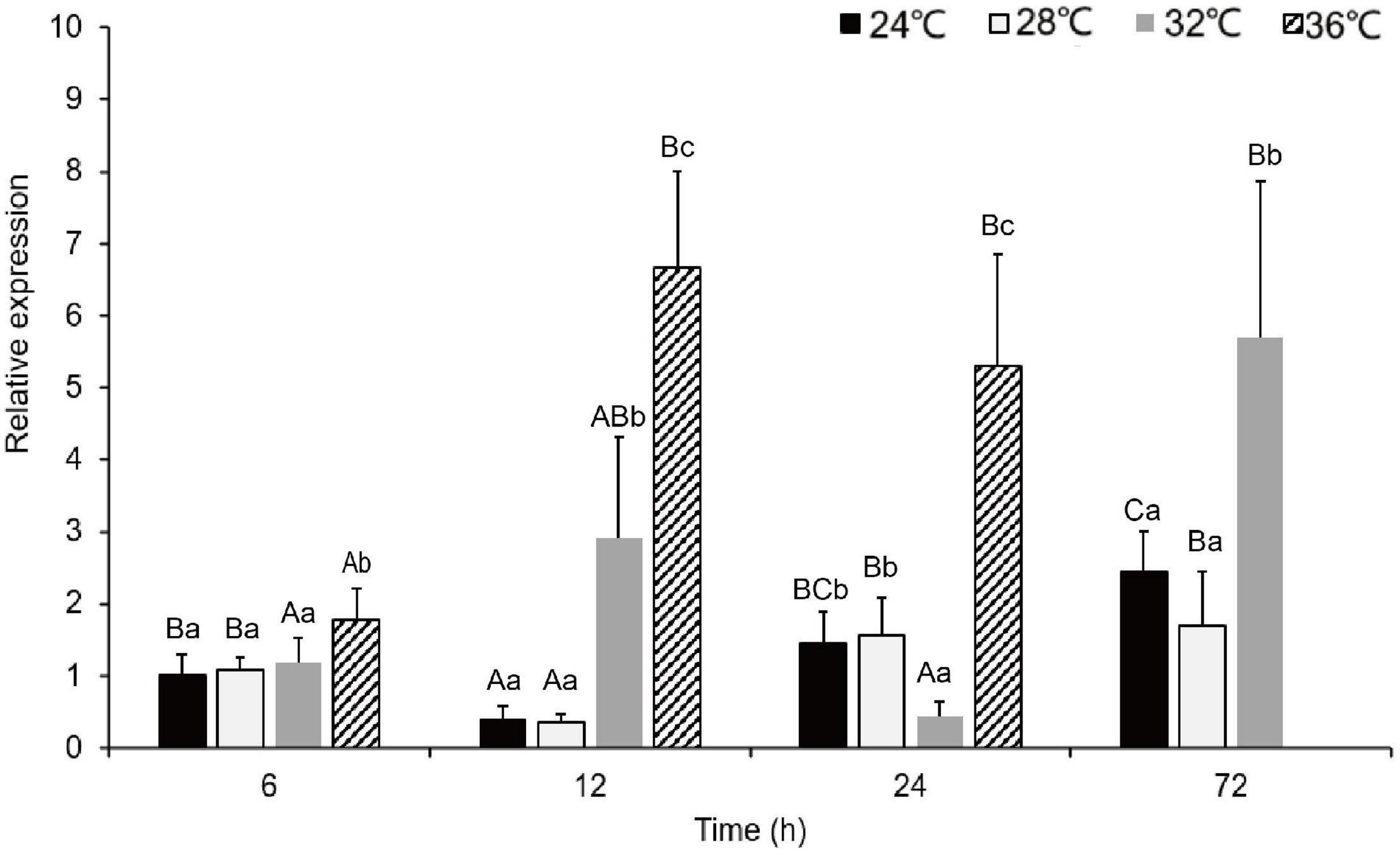
Figure 6. The expression levels of PmHSP70 mRNA in gill from P. maxima after acute MHWs stress. Small letters indicate significant differences between four temperature levels within each time, and capital letters indicate significant difference between four time within each temperature treatment (P < 0.05). As pearl oysters exposed to the temperature anomaly of 36°C suffered 100% mortality at 24 h, the experiment was terminated accordingly.
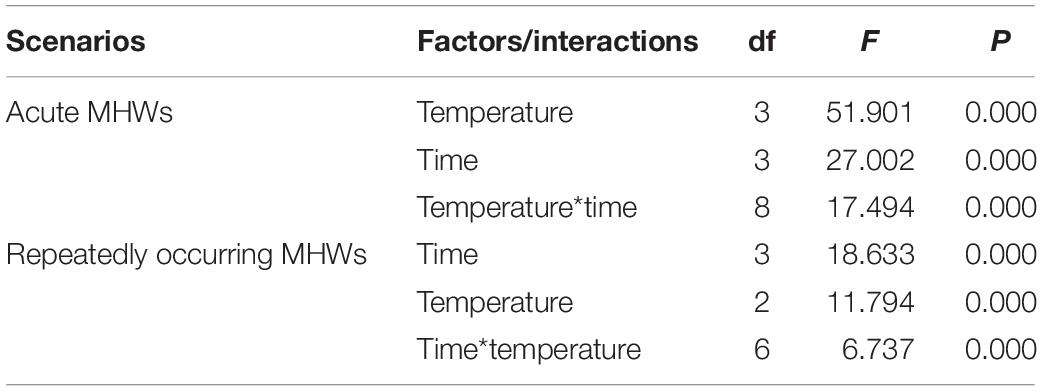
Table 3. Two-way ANOVA results for relative expression of HSP70 in pearl oysters exposed to acute and repeatedly occurring MHWs scenarios accounting for temperature and sampling time.
Following repeat exposure to MHWs, we found the expression level of HSP70 in the second MHWs, the fold changes between the experimental groups and the control groups were lower than those in the first MHWs (Figure 7). Analysis of variance shows that both temperature of MHWs and stress time made significant impact on the gene expression level (P < 0.05), and their interaction was significant (P < 0.05) (Table 3).
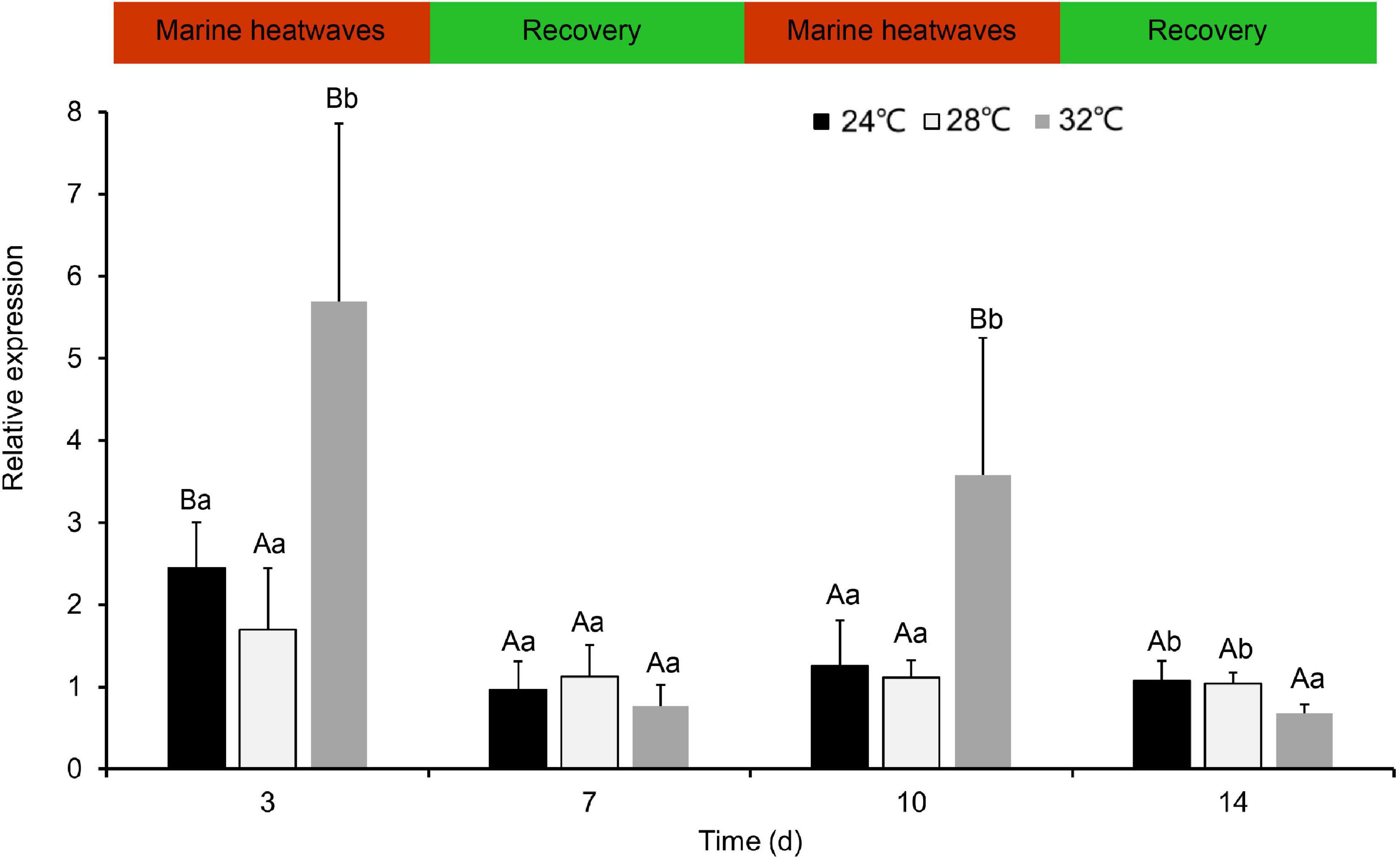
Figure 7. The expression levels of PmHSP70 mRNA in gill from P. maxima after exposed to repeatedly occurring MHWs scenarios. Small letters indicate significant differences between four temperature levels within each time, and capital letters indicate significant difference between different time within each temperature treatment (P < 0.05).
HSP70 genes in the pearl oyster P. maxima was characterized for the first time in present study. The PmHSP70 has a typical domain HSP70 and three characteristic sequences (IDLGTTYS, IFDLGGGTFDVSIL, IVLVGGSTRIPKIQK) of the HSP70 family (Gupta and Singh, 1994), which are well conserved in the eukaryotic HSP70 family (Gupta and Singh, 1994). And we observed an ATP/GTP-binding site (AEAYLGKT) in PmHSP70, which demonstrates that the activity regulation of PmHSP70 requires the participation of ATP (Sousa, 2012). The highly conserved intracellular specific motif EEVD ends in PmHSP70, indicating that PmHSP70 belongs to cytoplasmic HSP70 family (Boutet et al., 2003; Liu et al., 2015). The results of multiple sequence alignments show that HSP70 in various bivalve species is highly conserved. The similarity of these sequence and structure suggest that PmHSP70 is functionally conserved. The construction of the phylogenetic tree found that PmHSP70 and HSP70 of bivalves such as P. f. martensii clustered together.
The distribution patterns of PmHSP70 gene expression in different tissues indicate that PmHSP70 was ubiquitously expressed in all tested tissues, including gills, hepatopancreas, adductor muscle and mantle, where the highest expression in gills. These results are consistent with previous findings of Ruditapes philippinarum (Nie et al., 2017) and Chlamys nobilis (Cheng et al., 2019). Studies have found that heat stress has the potential to destroy cell and tissue integrity, and impair the physiological functions of gills in marine bivalves (Rahman and Rahman, 2021). This is because bivalves are filter-feeding organisms, and the gills are both their respiratory and feeding organs which can directly sense environmental changes more than other tissues (Cheng et al., 2019), so in the face of MHWs, the expression of HSP70 in the gills was significantly higher than that in other tissues.
In the present study, the expression of PmHSP70 mRNA in gills of MHWs was measured at, 6, 12, 24, and 72 h after acute MHWs. As expected, PmHSP70 is significantly up-regulated and correlated with temperature, and was highly expressed at 32 and 36°C groups, and the relative expression reached the highest at 72 and 12 h, respectively. These results are consistent with previous findings in R. philippinarum, C. nobilis and Laternula elliptica (Clark et al., 2008; Nie et al., 2017; Cheng et al., 2019). It is well known that when organisms are exposed to high temperatures, they produce amounts of reactive oxygen species (ROS) (Wang et al., 2018), which severely impairs normal cell functions. HSP70 is a universal molecular chaperone that participates in repairing or degrading damaged proteins (Feder and Hofmann, 1999), thereby protecting cells from environmental stress. The previous study showed that the HSPs of oysters is up-regulated under adverse environmental pressures and may be central to the oyster defense against all stresses (Zhang et al., 2012). We speculated that under the conditions of MHWs, the protein synthesis of pearl oysters was wrong, and too many harmful polymers were produced. The upregulation of PmHSP70 expression in the pearl oysters corrects the misfolded peptide chain, promotes the degradation of harmful polymers in the cell, resists harsh environments and responds to changes in environmental temperature.
It is worth noting that the relative expression of PmHSP70 in experimental groups was significantly lower than that at first MHWs, when pearl oysters are exposed to repeated MHWs. The expression level of HSP70 has no significant difference between the second MHWs and the recovery period. These findings indicate that long-term and repeated occurring MHWs stimulate relieve the thermal stress of pearl oysters caused by MHWs. The previous study showed that in the Crassostrea gigas a moderate elevation of temperature promotes the induction of members of the HSP gene family, which makes cells more resistant to any further challenge with heat stress (Li and Werb, 1982; Hamdoun et al., 2003). Similar phenomena was found in our previous studies, where the activity of various antioxidant enzymes was significantly lower than that of the first MHWs during repeated MHWs, and the expression of some genes related to metabolism was also alleviated during the recurring MHWs (He et al., 2021; Xu et al., 2021, 2022).
In recent years, due to the unprecedented speed of MHWs, whether marine bivalves can keep up and adapt to rapid environmental changes has attracted widespread concern. Results in present study indicated PmHSP70 expression in pearl oysters was inducible under MHWs stress. Plasticity of the heat-shock response is apparently a common feature in sessile intertidal invertebrates (Tomanek and Somero, 1999). In the laboratory, other bivalve species such as Mytilus trossulus and C. gigas exhibit significant plasticity in various features of the heat shock response (Buckley et al., 2001; Hamdoun et al., 2003). HSP family plays a critical role in rapid adaptation to novel thermal environments by preventing the degradation of intracellular proteins, and the high correlation between the induction of this gene family and the strong thermotolerance mechanism contributes to the environmental adaptation of aquatic animals (Kim et al., 2017). This adaptive plasticity will help marine bivalves adapt to changing marine environments and buffer the extinction of intertidal populations.
This study was the first report of full-length cloning, characterization and inducible expression of HSP70 of P. maxima. The results of the phylogenetic tree and multiple sequence alignment showed that the sequence of the PmHSP70 was relatively conserved among species and had high homology with the HSP70 of other species. The tissue expression characteristics showed that the PmHSP70 was expressed the highest in the gills. The expression level of HSP70 in the acute MHWs scenario indicates that pearl oyster can respond to the adverse effects of MHWs by rapidly up-regulating the expression of PmHSP70 under MHWs stimulation. It is worth noting that when faced with recurring MHWs, compared with the first MHWs period the expression of PmHSP70 was significantly down-regulated and had no significant difference between the recovery period. We speculated that the pearl oysters have adaptation potential under the stress of long-term and repeatedly occurring. Our findings will provide reference materials for the adaptation of marine organisms to the extreme marine environment. But we have only done one generation of impact. Further investigations are needed to elucidate transmission of carryover effects of MHWs within and across generations.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
YX: methodology, investigation, and writing—original draft preparation. JL, GH, and FM: methodology. XL: methodology, formal analysis. KY: resources and formal analysis. YD: conceptualization. LZ: conceptualization, writing—original draft preparation, and project administration. All authors contributed to the article and approved the submitted version.
The present study has been made possible by the research grants from National Natural Science Foundation of China (42076121, M-0163), the Department of Education of Guangdong Province (2020KTSCX050) and the Starting Research Fund from Guangdong Ocean University (R20083) to LZ, and the Earmarked Fund for Modern Agro-industry Technology Research System (CARS-49) to YD.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Amorim, V., Gonçalves, O., Capela, R., Fernández-Boo, S., Oliveira, M., Dolbeth, M., et al. (2020). Immunological and oxidative stress responses of the bivalve Scrobicularia plana to distinct patterns of heatwaves. Fish Shellfish Immunol. 106, 1067–1077. doi: 10.1016/j.fsi.2020.09.024
Bedulina, D., Evgen’Ev, M., Timofeyev, M., Protopopova, M., Garbuz, D., Pavlichenko, V., et al. (2013). Expression patterns and organization of the hsp70 genes correlate with thermotolerance in two congener endemic amphipod species (Eulimnogammarus cyaneus and E. verrucosus) from Lake Baikal. Mol. Ecol. 22, 1416–1430. doi: 10.1111/mec.12136
Boutet, I., Tanguy, A., Rousseau, S., Auffret, M., and Moraga, D. (2003). Molecular identification and expression of heat shock cognate 70 (hsc70) and heat shock protein 70 (hsp70) genes in the Pacific oyster Crassostrea gigas. Cell Stress Chaperones 8:76. doi: 10.1379/1466-1268(2003)8<76:miaeoh>2.0.co;2
Buckley, B. A., Owen, M.-E., and Hofmann, G. E. (2001). Adjusting the thermostat: the threshold induction temperature for the heat-shock response in intertidal mussels (genus Mytilus) changes as a function of thermal history. J. Exp. Biol. 204, 3571–3579. doi: 10.1242/jeb.204.20.3571
Caputi, N., Kangas, M., Denham, A., Feng, M., Pearce, A., Hetzel, Y., et al. (2016). Management adaptation of invertebrate fisheries to an extreme marine heat wave event at a global warming hot spot. Ecol. Evol. 6, 3583–3593. doi: 10.1002/ece3.2137
Chandrapavan, A., Caputi, N., and Kangas, M. I. (2019). The decline and recovery of a crab population from an extreme marine heatwave and a changing climate. Front. Mar. Sci. 6:510. doi: 10.3389/fmars.2019.00510
Chen, T., Lin, T., Li, H., Lu, T., Li, J., Huang, W., et al. (2018). Heat shock protein 40 (HSP40) in pacific white shrimp (Litopenaeus vannamei): molecular cloning, tissue distribution and ontogeny, response to temperature, acidity/alkalinity and salinity stresses, and potential role in ovarian development. Front. Physiol. 9:1784. doi: 10.3389/fphys.2018.01784
Cheng, D., Liu, H., Zhang, H., Soon, T. K., Ye, T., Li, S., et al. (2019). Differential expressions of HSP70 gene between golden and brown noble scallops Chlamys nobilis under heat stress and bacterial challenge. Fish Shellfish Immunol. 94, 924–933. doi: 10.1016/j.fsi.2019.10.018
Clark, M. S., Fraser, K. P., and Peck, L. S. (2008). Antarctic marine molluscs do have an HSP70 heat shock response. Cell Stress Chaperones 13, 39–49. doi: 10.1007/s12192-008-0014-8
Clegg, J., Uhlinger, K., Jackson, S., Cherr, G., Rifkin, E., and Friedman, C. (1998). Induced thermotolerance and the heat shock protein-70 family in the Pacific oyster Crassostrea gigas. Mol. Mar. Biol. Biotechnol. 7, 21–30.
Dong, S., Nie, H., Ye, J., Li, D., Huo, Z., and Yan, X. (2020). Physiological and gene expression analysis of the Manila clam Ruditapes philippinarum in response to cold acclimation. Sci. Total Environ. 742:140427. doi: 10.1016/j.scitotenv.2020.140427
Evgen’ev, M., Garbuz, D., Shilova, V., and Zatsepina, O. (2007). Molecular mechanisms underlying thermal adaptation of xeric animals. J. Biosci. 32, 489–499. doi: 10.1007/s12038-007-0048-6
Feder, M. E., and Hofmann, G. E. (1999). Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Ann. Rev. Physiol. 61, 243–282.
Fordyce, A. J., Ainsworth, T. D., Heron, S. F., and Leggat, W. (2019). Marine heatwave hotspots in coral reef environments: physical drivers, ecophysiological outcomes, and impact upon structural complexity. Front. Mar. Sci. 6:498. doi: 10.3389/fmars.2019.00498
Gupta, R. S., and Singh, B. (1994). Phylogenetic analysis of 70 kD heat shock protein sequences suggests a chimeric origin for the eukaryotic cell nucleus. Curr. Biol. 4, 1104–1114. doi: 10.1016/s0960-9822(00)00249-9
Hamdoun, A. M., Cheney, D. P., and Cherr, G. N. (2003). Phenotypic plasticity of HSP70 and HSP70 gene expression in the Pacific oyster (Crassostrea gigas): implications for thermal limits and induction of thermal tolerance. Biol. Bull. 205, 160–169. doi: 10.2307/1543236
Harley, C. D. G. (2008). Tidal dynamics, topographic orientation, and temperature-mediated mass mortalities on rocky shores. Mar. Ecol. Prog. Ser. 371, 37–46.
Hassan, F. U., Nawaz, A., Srehman, M., Aali, M., Mrdilshad, S., and Yang, C. J. (2019). Prospects of HSP70 as a genetic marker for thermo-tolerance and immuno-modulation in animals under climate change scenario. Anim. Nutr. 5, 340–350. doi: 10.1016/j.aninu.2019.06.005
He, G., Liu, X., Xu, Y., Liang, J., Deng, Y., Zhang, Y., et al. (2021). Repeated exposure to simulated marine heatwaves enhances the thermal tolerance in pearl oysters. Aquatic Toxicol. 239:105959. doi: 10.1016/j.aquatox.2021.105959
Hobday, A. J., Alexander, L. V., Perkins, S. E., Smale, D. A., Straub, S. C., Oliver, E. C., et al. (2016). A hierarchical approach to defining marine heatwaves. Prog. Oceanography 141, 227–238. doi: 10.1016/j.pocean.2015.12.014
Holbrook, N. J., Gupta, A. S., Oliver, E. C., Hobday, A. J., Benthuysen, J. A., Scannell, H. A., et al. (2020). Keeping pace with marine heatwaves. Nat. Rev. Earth Environ. 1, 482–493.
Hu, M., Qiu, Z., Zhou, P., Xu, L., and Zhang, J. (2012). Proteomic analysis of ‘Zaosu’pear (Pyrus bretschneideri Rehd.) and its red skin bud mutation. Proteome Sci. 10, 1–15. doi: 10.1186/1477-5956-10-51
Jones, D. B., Jerry, D. R., Khatkar, M. S., Raadsma, H. W., and Zenger, K. R. (2013). A high-density SNP genetic linkage map for the silver-lipped pearl oyster, Pinctada maxima: a valuable resource for gene localisation and marker-assisted selection. BMC Genom. 14:810. doi: 10.1186/1471-2164-14-810
Kim, B.-M., Kim, K., Choi, I.-Y., and Rhee, J.-S. (2017). Transcriptome response of the Pacific oyster, Crassostrea gigas susceptible to thermal stress: a comparison with the response of tolerant oyster. Mol. Cell. Toxicol. 13, 105–113.
Li, G. C., and Werb, Z. (1982). Correlation between synthesis of heat shock proteins and development of thermotolerance in Chinese hamster fibroblasts. Proc. Natl. Acad. Sci. U S A. 79, 3218–3222. doi: 10.1073/pnas.79.10.3218
Li, Y., Ren, G., Wang, Q., and You, Q. (2019). More extreme marine heatwaves in the China Seas during the global warming hiatus. Environ. Res. Lett. 14:104010.
Liu, K., Xu, K., Zhu, C., and Liu, B. (2021). Diversity of marine heatwaves in the South China Sea regulated by the ENSO phase. J. Climate 35, 877–893. doi: 10.1175/jcli-d-21-0309.1
Liu, T., Pan, L., Ca, I. Y., and Miao, J. (2015). Molecular cloning and sequence analysis of heat shock proteins 70 (HSP70) and 90 (HSP90) and their expression analysis when exposed to benzo(a)pyrene in the clam Ruditapes philippinarum. Gene 555, 108–118. doi: 10.1016/j.gene.2014.10.051
Liu, Z. M., Zhu, X. L., Lu, J., Cai, W. J., Ye, Y. P., and Lv, Y. P. (2018). Effect of high temperature stress on heat shock protein expression and antioxidant enzyme activity of two morphs of the mud crab Scylla paramamosain. Comp. Biochem. Physiol. Part A Mol. Integrative Physiol. 223, 10–17. doi: 10.1016/j.cbpa.2018.04.016
Mayer, M., and Bukau, B. (2005). Hsp70 chaperones: cellular functions and molecular mechanism. Cell. Mol. Life Sci. 62, 670–684. doi: 10.1007/s00018-004-4464-6
Nie, H., Liu, L., Huo, Z., Chen, P., Ding, J., Yang, F., et al. (2017). The HSP70 gene expression responses to thermal and salinity stress in wild and cultivated Manila clam Ruditapes philippinarum. Aquaculture 470, 149–156.
Park, K., Lee, J. S., Kang, J. C., Kim, J. W., and Kwak, I. S. (2015). Cascading effects from survival to physiological activities, and gene expression of heat shock protein 90 on the abalone Haliotis discus hannai responding to continuous thermal stress. Fish Shellfish Immunol. 42, 233–240. doi: 10.1016/j.fsi.2014.10.036
Petes, L. E., Menge, B. A., and Murphy, G. D. (2007). Environmental stress decreases survival, growth, and reproduction in New Zealand mussels. J. Exp. Mar. Biol. Ecol. 351, 83–91.
Pörtner, H. (2001). Climate change and temperature-dependent biogeography: oxygen limitation of thermal tolerance in animals. Naturwissenschaften 88, 137–146. doi: 10.1007/s001140100216
Rahman, M. S., and Rahman, M. S. (2021). Effects of elevated temperature on prooxidant-antioxidant homeostasis and redox status in the American oyster: signaling pathways of cellular apoptosis during heat stress. Environ. Res. 196:110428. doi: 10.1016/j.envres.2020.110428
Rendina, F., Bouchet, P. J., Appolloni, L., Russo, G. F., Sandulli, R., Kolzenburg, R., et al. (2019). Physiological response of the coralline alga Corallina officinalis L. to both predicted long-term increases in temperature and short-term heatwave events. Mar. Environ. Res. 150:104764. doi: 10.1016/j.marenvres.2019.104764
Scanes, E., Parker, L. M., O’Connor, W. A., Dove, M. C., and Ross, P. M. (2020). Heatwaves alter survival of the Sydney rock oyster, Saccostrea glomerata. Mar. Pollution Bull. 158:111389. doi: 10.1016/j.marpolbul.2020.111389
Seuront, L., Nicastro, K. R., Zardi, G. I., and Goberville, E. (2019). Decreased thermal tolerance under recurrent heat stress conditions explains summer mass mortality of the blue mussel Mytilus edulis. Sci. Rep. 9:17498. doi: 10.1038/s41598-019-53580-w
Smale, D. A., Wernberg, T., Oliver, E. C., Thomsen, M., Harvey, B. P., Straub, S. C., et al. (2019). Marine heatwaves threaten global biodiversity and the provision of ecosystem services. Nat. Climate Change 9, 306–312. doi: 10.1038/s41558-019-0412-1
Somero, G. N. (2005). Linking biogeography to physiology: evolutionary and acclimatory adjustments of thermal limits. Front. Zool. 2:1. doi: 10.1186/1742-9994-2-1
Sousa, R. (2012). A dancer caught midstep: the structure of ATP-bound Hsp70. Mol. Cell 48, 821–823. doi: 10.1016/j.molcel.2012.12.008
Tomanek, L., and Somero, G. N. (1999). Evolutionary and acclimation-induced variation in the heat-shock responses of congeneric marine snails (genus Tegula) from different thermal habitats: implications for limits of thermotolerance and biogeography. J. Exp. Biol. 202, 2925–2936. doi: 10.1242/jeb.202.21.2925
Tomanek, L., and Somero, G. N. (2002). Interspecific-and acclimation-induced variation in levels of heat-shock proteins 70 (hsp70) and 90 (hsp90) and heat-shock transcription factor-1 (HSF1) in congeneric marine snails (genus Tegula): implications for regulation of hsp gene expression. J. Exp. Biol. 205, 677–685. doi: 10.1242/jeb.205.5.677
Wang, J., Dong, B., Yu, Z.-X., and Yao, C.-L. (2018). The impact of acute thermal stress on green mussel Perna viridis: oxidative damage and responses. Comp. Biochem. Physiol. Part A: Mol. Integrat. Physiol. 222, 7–15. doi: 10.1016/j.cbpa.2018.04.001
Wang, Z., Liang, F., Huang, R., Deng, Y., and Li, J. (2019). Identification of the differentially expressed genes of Pinctada maxima individuals with different sizes through transcriptome analysis. Regional Stud. Mar. Sci. 26:100512. doi: 10.1016/j.rsma.2019.100512
Xu, Y., Wang, Z., Zhang, Y., Liang, J., He, G., Liu, X., et al. (2022). Transcriptome analysis reveals acclimation responses of pearl oysters to marine heatwaves. Sci. Total Environ. 810:151189. doi: 10.1016/j.scitotenv.2021.151189
Xu, Y., Zhang, Y., Liang, J., He, G., Liu, X., Zheng, Z., et al. (2021). Impacts of marine heatwaves on pearl oysters are alleviated following repeated exposure. Mar. Pollution Bull. 173:112932. doi: 10.1016/j.marpolbul.2021.112932
Yao, Y., Wang, J., Yin, J., and Zou, X. (2020). Marine heatwaves in china’s marginal seas and adjacent offshore waters: Past, Present, and Future. J. Geophys. Res.: Oceans 125:e2019JC015801.
Zhang, G., Fang, X., Guo, X., Li, L., Luo, R., Xu, F., et al. (2012). The oyster genome reveals stress adaptation and complexity of shell formation. Nature 490, 49–54. doi: 10.1038/nature11413
Zhang, W., Zheng, Z., Zhang, T., and Chen, T. (2020). Strengthened marine heatwaves over the Beibu Gulf coral reef regions from 1960 to 2017. Haiyang Xuebao 42, 41–49.
Zhang, X., and Xie, J. (2019). Analysis of proteins associated with quality deterioration of grouper fillets based on TMT quantitative proteomics during refrigerated storage. Molecules 24:2641. doi: 10.3390/molecules24142641
Zhao, L., Liu, L., Liu, B., Liang, J., Lu, Y., and Yang, F. (2019). Antioxidant responses to seawater acidification in an invasive fouling mussel are alleviated by transgenerational acclimation. Aquatic Toxicol. 217:10533. doi: 10.1016/j.aquatox.2019.105331
Keywords: Pinctada maxima, PmHSP70, extreme weather events, acclimation, climate change
Citation: Xu Y, Liang J, He G, Liu X, Yang K, Masanja F, Deng Y and Zhao L (2022) Responses of Pearl Oysters to Marine Heatwaves as Indicated by HSP70. Front. Mar. Sci. 9:847585. doi: 10.3389/fmars.2022.847585
Received: 03 January 2022; Accepted: 25 January 2022;
Published: 01 March 2022.
Edited by:
Hui Zhang, Institute of Oceanology (CAS), ChinaCopyright © 2022 Xu, Liang, He, Liu, Yang, Masanja, Deng and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liqiang Zhao, bHpoYW9AZ2RvdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.