- 1Jiangsu Key Laboratory of Marine Bioresources and Environment, Jiangsu Ocean University, Lianyungang, China
- 2Co-Innovation Center of Jiangsu Marine Bio-industry Technology, Jiangsu Ocean University, Lianyungang, China
- 3Jiangsu Key Laboratory of Marine Biotechnology, Jiangsu Ocean University, Lianyungang, China
This study aims to determine the genetic structure of rainbow clam Moerella iridescens in different sea areas of China. Seventeen pairs of microsatellite primers (SSR) were used to amplify the SSRs of rainbow clam in Lianyungang of Haizhou Bay, Chongming of Shanghai, Ningde of Fujian, Daishan of Zhoushan, and Cixi and Wenzhou of Zhejiang. A total of 1,146 alleles were detected in 310 individuals from the 17 SSR loci. The average observed heterozygosity of six populations was 0.4381−0.6139, the average expected heterozygosity was 0.5897−0.7325, and the average Shannon diversity index was 1.2655−1.7998. The clams exhibited rich genetic diversity, and the FST of the genetic differentiation index of the six populations was 0.0470, indicating low genetic differentiation among the populations. The results indicated that rainbow clams along the coasts of China exhibited high diversity and low population differentiation.
Highlights
- This study aims to determine the genetic structure and diversity of rainbow clam Moerella iridescens in different sea areas of China.
- The clams exhibited rich genetic diversity and the FST of the genetic differentiation index of the six populations was 0.0470, indicating low genetic differentiation amongst the populations.
- The results indicated that rainbow clam along China coasts exhibited high diversity and low population differentiation.
Introduction
The rainbow clam Moerella iridescens is an economically important small-sized marine clam due to its delicious taste and high nutritional value. The clam is mainly distributed on the West Pacific coast and northern Australia. In recent years, the rainbow clam has been increasingly used as high-valued seafood and is preferred by consumers of China, and the output of the rainbow clams mainly comes from marine fishing. However, in the past 2 years, the development of the shrimp and crab aquaculture industry and the overfishing of wild rainbow clams have exacerbated the destruction of the habitat of rainbow clams. Moreover, the status of genetic diversity of the germplasm resources of rainbow clams remains unclear. Thus, the protection of rainbow clam resources is of great importance. Several authors have explored the biological characteristics and morphological differences of rainbow clams (Ji et al., 2007; Lv et al., 2012). To date, however, reports on the genetic diversity and relationships among clam populations are limited (Xu et al., 2016).
Microsatellites, also known as simple repetitive sequences (SSRs), present the advantages of co-dominance, single locus, high polymorphism, and easy operation and can be screened from different populations to complete genetic diversity studies (Cui et al., 2011). Microsatellite marker technology has been widely used in the structural analysis of the population genetics of aquatic animals, e.g., the rainbow trout (Zhao et al., 2010), the scallop Patinopecten yessoensis (Chang et al., 2007), and the triangle pearl Hyriopsis cummingii (Bai et al., 2015). So far, few reports are available on the genetic diversity of the population of rainbow clams. In the present study, the genetic diversity of six populations of rainbow clams in Lianyungang, Chongming Island, Ningde, Zhoushan, Cixi, and Wenzhou was analyzed by using microsatellite markers. The results obtained can provide a scientific basis for assessing germplasm resources and genetic diversity protection of rainbow clams.
Materials and Methods
Materials
Rainbow clams were collected from the six coastal mudflats, namely, Lianyungang Haizhou Bay (LYG), Chongming Island Dongtan (CM), Ningde Fu’an (ND), Zhoushan Daishan (ZS), Hangzhou Bay Cixi (CX), and Wenzhou Yueqing Bay (WZ). Fifty samples were randomly obtained from each population. Information on the samples collected is presented in Table 1 and Figure 1. The sampled rainbow clams were stored at −70°C until analysis.
Microsatellite Primer
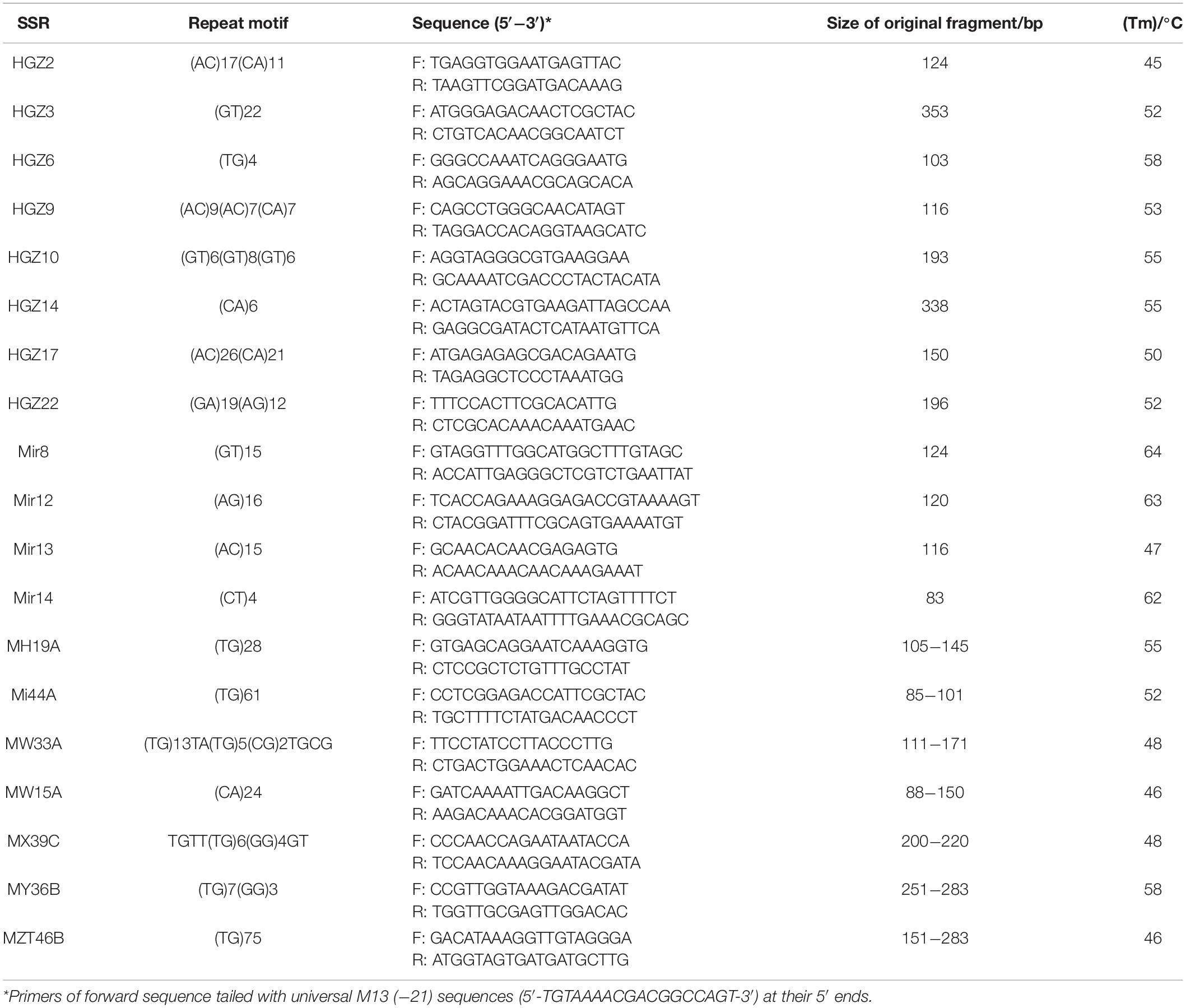
The M13 (−21) universal primer sequence of 5′-TGTAAAACGACGGCCAGT-3′ reported by Schuelke (2000) was adopted in this study to elongate short primers for economic consideration in genotyping. We used two fluorescent labels, FAM and HEX, for forwarding primes, which were abbreviated as FAM-M13 and HEX-M13. Nineteen SSRs were used for PCR, and the information of these primers is shown in Table 2.
Method
Total DNA was extracted with the sodium dodecyl sulphate (SDS) phenol-chloroform method and according to the detailed protocol for the DNA extraction as previously described by Li et al. (2020). The PCR system and program in this study was done as previously described by Zhang et al. (2019). Specifically, the used PCR system consisted of 1 μl of template DNA (about 30 ng), 2 μl of primer mix, 1 μl of universal fluorescent primers, 10 μl of 2 × Taq PCR Master Mix (TaKaRa, Dalian, China), and 6 μl of ddH2O to form a total volume of 20 μl. The PCR program was as follows: pre-denaturation at 90°C for 5 min, 30 cycles, denaturation at 94°C for 3 min, annealing at 53°C for 1 min and at 72°C for 30 s, and a final extension at 72°C for 10 min at the end of the cycle. The PCR products were subjected to capillary electrophoresis, and the electrophoresis patterns were genotyped by GeneMapper 3.7 and Peak Scanner software.
Data Processing
Pop32 software was utilized to calculate the number of effective alleles (Ne), expected heterozygosity (He), observed heterozygosity (Ho), and gene flow (Nm = 1− FST/4 FST, where the FST represent genetic differentiation index), genetic distance (Ds), and Shannon diversity index (Raymond and Rousset, 1995). Population clustering was analyzed using the unweighted pair-population method with arithmetic means (UPGMA) of the MEGA 3.0 software. Analysis of molecular variance (AMOVA) was performed using Arlequin 3.11software, and the genetic diversity and genetic differentiation index (FST) were computationally analyzed.
Results
Genetic Diversity of the Rainbow Clam
There were 1,146 alleles that were successfully detected from the genomes of rainbow clam population via 17 SSR loci scanning in the six populations. Despite this, the HGZ22 in the Wenzhou population failed to be amplified by PCR. Except for the alleles detected at the two loci of HGZ2 and HGZ14, the remaining alleles were highly polymorphic loci, which indicated that the 17 microsatellite loci could be used to study the population genetics of rainbow clam (Table 3).
The average number of alleles among the 17 loci in the six populations was 5.5000−22.5000 (17 loci average, 11.9039), the Ne was 1.2538−13.3411 (average, 5.1287), the Ho was 0.0100−0.9933 (average, 0.5171), the He was 0.1820−0.9176 (average, 0.6847), and the Shannon diversity index was 0.3847−2.6472 (average, 1.6914).
The average number of alleles of the six populations was 8.3125−12.8235, the Ne was 3.8135−5.7762, and the Ho was 0.4381−0.6139 (average, 0.5121). Among the populations studied, the Ho of CX was the highest, whereas the Ho of CM was the lowest. The average He was 0.6266−0.7325 (average, 0.6847). The Zhoushan population was the highest, whereas the Wenzhou population was the lowest. The average Shannon diversity indices were 1.7998 (ZS), 1.7451 (CX), 1.6603 (ND), 1.6941 (LYG), 1.6355 (CM), and 1.2655 (WZ). These results showed that, although the genetic diversities of the six wild populations of rainbow clam were different, the overall genetic diversity was high (Table 3).
Phylogenetic Relationships of the Six Populations of Rainbow Clams
Analysis of Molecular Variance
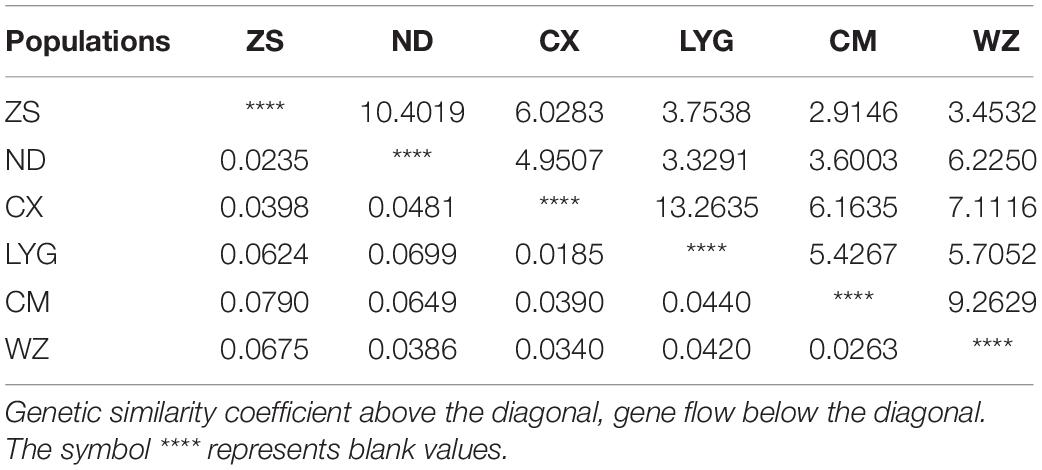
Analysis of molecular variance (AMOVA) of the six different geographical populations of rainbow clams showed that 4.70% of their genetic variation could be derived from the population, while 95.30% of their variation was from within the population. The FST of the populations was 0.0470, indicating that the degree of genetic differentiation among the populations was low (Table 4).
Genetic Distance
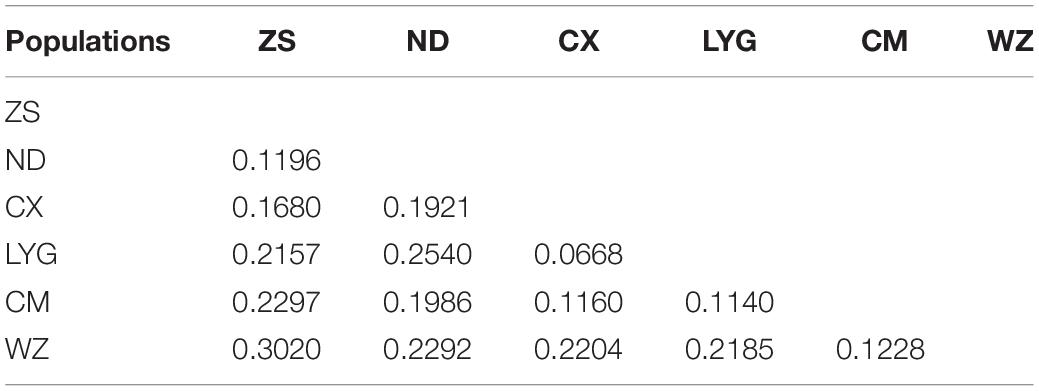
The Ds among the six populations of rainbow clam was 0.0668−0.3020. The Ds between the Wenzhou and Zhoushan populations was the largest (0.3020), and their genetic relationship was far. In contrast, the Ds between the Lianyungang and Cixi populations was the smallest (0.0668), and their genetic relationship was relatively close (Table 5).
Genetic Differentiation Index and Gene Flow
Among those of the different populations, the FST between the Chongming and Zhoushan populations was the highest (0.0790), whereas that between the Cixi and Lianyungang populations was the lowest (0.0185). The Nm values between the Lianyungang and Cixi populations and those between the Ningde and Zhoushan populations were 13.2635 and 10.4019, respectively. The Nm between the Chongming and Zhoushan populations was 2.9146. The total FST between populations was 0.04702. FST < 0.05 indicates a low degree of genetic differentiation, which, in turn, reveals rich genetic diversity. The overall differentiation degree among populations was low (Table 6).
Cluster Analysis
Based on the genetic distances among the populations, a clustering map was constructed using the UPGMA method of the MEGA3.0 software. The six populations of rainbow clam could be divided into the following three branches: LYG and CX in the first branch, CM and WZ in the second branch, and ZS and ND in the third branch (Figure 2).
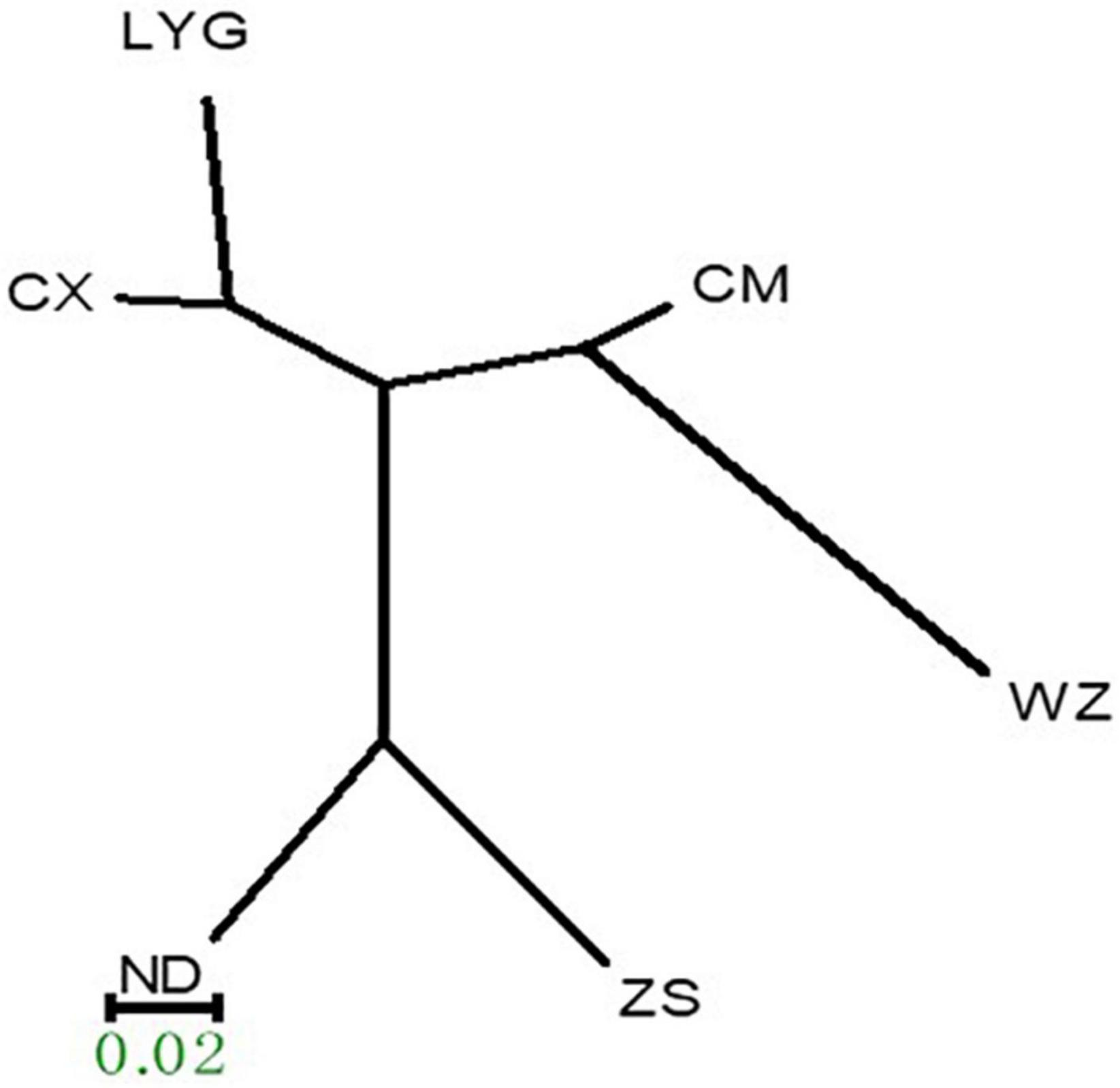
Figure 2. Rainbow clam six populations of unweighted pair-population method with arithmetic means (UPGMA) trees.
Discussion
Genetic differentiation index (FST) is an important parameter for measuring the degree of genetic differentiation in a population. A large FST value indicates a high degree of differentiation between populations. No differentiation exists when FST is 0−0.05, moderate differentiation is observed when FST is 0.05−0.15, high differentiation is obtained when FST is 0.15−0.25, and great differentiation is found when FST > 0.25 (Hartl and Clark, 1997). In this study, the FST of the six populations under study was 0.047 (<0.05). Overall, the differentiation between the populations was minimal at best. However, the FST values of the ZS−LYG, ZS−CM, ZS−WZ, LYG−ND, and CM−ND populations were in the range of 0.0624−0.0790 (0.05), which indicates that moderate genetic differentiation existed between the Zhoushan and each of the Lianyungang, Chongming, and Wenzhou populations and between the Ningde and each of the Lianyungang and Chongming populations of rainbow clams. Nm can indicate the degree of genetic differentiation in a population. If Nm < 1, genetic differentiation occurs among populations. If Nm > 1, genetic differentiation is relatively low. If Nm > 4, genetic differentiation is very low (Ratnaningrum et al., 2017). The results of AMOVA showed that the Nm values among the six populations of rainbow clams lay between 2.9146 and 13.2635, and the Nm values among the ZS−LYG, ZS−CM, ZS–WZ, LYG−ND, and CM−ND populations were greater than 1 and less than 4. These results indicate low genetic differentiation among these populations. In the present study, the sampling area of rainbow clams was distributed in the Yellow Sea and East China Seas. LYG and CM are in the Yellow Sea with a geographical distance (about 1,000 km coastline), and no differentiation between these populations was observed (FST = 0.0040 < 0.05). Although the geographical distance between the Chongming and Zhoushan populations is close, the Yangtze estuary is situated between them. The inflow of the Yangtze River lowers the salinity of the seawater and, hence, obstructs the passage of larvae and hinders gene communication to some extent (the Nm for CM−ZS is 2.9146). Therefore, moderate genetic differentiation occurs between the CM and ZS populations (FST = 0.0790). Xu et al. (2016) analyzed the population morphology and genetic diversity of rainbow clams from Zhejiang and revealed that the Zhoushan population has great variation from the Yueqing, Taizhou, and Wenling populations (GST = 0.2479). The present study also indicated that genetic differentiation occurred in the Zhoushan population (FST = 0.2479), which was consistent with Xu et al. (2016), who showed that the Zhoushan population of rainbow clams presents moderate genetic differentiation.
This study also found that the degree of genetic differentiation between rainbow clam populations is not related to the geographical distance. Although the geographical distance between LYG and CX was relatively far away, the FST value between the LYG and CX populations of the rainbow was shallow, only 0.0180 (<0.05), which indicated a lack of genetic differentiation between the two populations. The gene exchange between the two populations occurred frequently (Nm = 13.2635 > 4). The inconsistency of the relationship between the degree of genetic differentiation and geographical distance has previously been observed in different populations of Coelomactra antiquata. Meng et al. (2013) showed that the population genetic differentiation between the C. antiquata from the Southeastern Sea and that from the Yellow Sea in Lianyungang and Rizhao is not apparent based on the results of ITS2 and 16S rRNA for six populations of C. antiquata in the coasts of China. By contrast, the difference between the Guangxi and Fujian populations, which have a relatively close geographical distance, reached the interspecies level. The real reason may be from human activity, such as artificial farming and introduction, but more detailed reasons remain unknown so far.
Genetic diversity, including the degree of genetic variation and the genetic structure of the population, is an important basis for evaluating the status of genetic resources. A higher genetic diversity of a population results in a stronger adaptability to the living environment and a greater potential for evolution (Tian et al., 2013). The genetic diversity of a population is mainly manifested in two aspects: heterozygosity and the number of alleles (Yu et al., 2012). Many reports have explored the genetic diversity of aquatic animals by using SSR markers. Li et al. (2011) studied the population genetics of Portunus trituberculatus by microsatellite markers and found an average Ho of 0.2222−1.0000 and an average He of 0.4367−0.9099. Li et al. (2009) used SSR technology to measure the average Ho (0.32−0.49) of wild and cultured populations of scallops and found an average He of 0.37−0.55. Chang et al. (2007) analyzed the genetic diversity of five populations of scallops P. yessoensis in China and abroad and found an average Ho of 0.2708−0.3292 and an average He of 0.3620−0.4595 for the five populations. Tian et al. (2013) determine the Ho and He of four populations of Scapharca broughtonii by using the 20 microsatellite markers, and the results showed that the ranges of Ho and He was 0.667–0.9667 and 0.6198–0.9318, respectively. An et al. (2012) studied the genetic structure of five wild-type populations of Ruditapes philippinarum in two sea areas of Korea by using seven SSR markers and noted high genetic diversity among the clams (total He = 0.813). The results of the present study revealed that the average Ho of the six populations of rainbow clam was between 0.4381 and 0.6139 and that the He was 0.6266−0.7325. Specifically, the value of He was as follows: ZS (0.7325), CX (0.7162), ND (0.6839), LYG (0.6786), CM (0.6701), and WZ (0.6266). The average number of alleles of the 17 loci in the six populations ranged from 5.5000 to 22.500, and the Ne was between 1.2538 and 13.3411. The genetic diversity of the rainbow clams was higher than those of bay scallop Argopecten irradians and scallop P. yessoensis. This particular genetic diversity is similar to the He of four populations of clams T. quinquefasciatus and is lower than that of five populations of clam R. philippinarum. Generally, the He (0.7325) of the present study and Shannon diversity index (1.7998) were the highest in the Zhoushan population, which indicated that the Zhoushan population has the richest genetic diversity among the clam populations studied.
The population differentiation of the existing rainbow clams is not obvious, and the genetic diversity of germplasm resources is relatively high, indicating that the effective population of rainbow clams in nature is large enough. At present, it is not very urgent to establish a germplasm resource reserve and its ex situ culture technology. In the future, the monitoring and evaluation of the germplasm resources of rainbow clams should be strengthened.
Conclusion
The six populations of rainbow clams all presented high genetic diversity, which reflect the good protection and development prospects of rainbow clam resources in China. These results would be helpful to genetic breeding practice and resources management.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Ethics Statement
All applicable International, National, and/or Institutional Guidelines for the Care and Use of Animals (invertebrates) were followed.
Author Contributions
XL conceived and designed the experiments. SG performed the experiments. MZ analyzed the data. XL and ZD wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported in part by grants from the Modern Agricultural Industry Technology System of China (No. CAR49), National Natural Science Foundation of China (No. 31101900), and Priority Academic Program Development of Jiangsu Higher Education Institutions.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We greatly acknowledge the following colleagues for their help and support in the process of investigation and sampling for rainbow clam: Feng Zhao from East China Sea Fisheries Institute of CAFS for the specimen in East Beach of Chongming Island, Binlun Yan from Huaihai Institute of Technology for the specimen in Sanyang harbor of Haizhou bay, Zehui Hu from Marine Fisheries Research Institute of Zhejiang of China for the specimen in Datian Bay at Daishan of Zhoushan, Chen from Xiabaishi at Sansha bay in Fu’an for the specimen in the site, Meizheng Wang from Institute of Cixi Fisheries for the specimen in Cixi Sanbei shoal at Hangzhou Bay, and Weicheng Lui from Zhejiang Mariculture Research Institute for the specimen in Simon Island at Yueqing Bay.
References
An, H. S., Kwang, K. J., Cho, K. C., Han, H. S., and Myeong, J. I. (2012). Genetic structure of Korean populations of the clam Ruditapes philippinarum inferred from microsatellite marker analysis. Biochem. Syst. Ecol. 44, 186–195.
Bai, Z., Han, X., Luo, M., Lin, J., Wang, J., and Li, J. (2015). Constructing a microsatellite-based linkage map and identifying QTL for pearl quality traits in triangle pearl mussel (Hyriopsis cumingii). Aquaculture 437, 102–110.
Chang, Y. Q., Chen, X. X., and Ding, J. (2007). Genetic diversity in five scallop populations of the Japanese scallop (Patinopecten yessoensis). Ecol. J. 27, 1145–1152.
Cui, H. Y., Ma, H. L., and Ma, C. Y. (2011). Genetic diversity among different families of mud crab Scylla paramamosain by microsatellite markers. Mar. Fish. 33, 274–281.
Hartl, D. L., and Clark, A. G. (1997). Principles of population genetics, 3rd Edn. Sunderland, MA: Sinauer Associates Inc.
Ji, Y. B., Li, T. W., and Su, X. R. (2007). Preliminary study on biological characteristics of rainbow clam in Zhejiang coast. Fish. Sci. 26, 494–496.
Li, H. J., Liu, X., Du, X. D., Song, R., Zhang, G. F., Hu, J. J., et al. (2009). Development and isolation of microsatellite markers in bay scallop. Mar. Sci. 33, 3–8.
Li, X., Liu, P., and Song, X. (2011). Construction on enriched microsatellite library and characterization of microsatellite markers from swimming crab. J. Fish. Sci. China 18, 194–201.
Li, X. Y., Zhang, M., Lu, G. Z., Mao, S., Wu, R. X., Ge, H. X., et al. (2020). Population genetic analysis of Rainbow Clam Moerella iridescens by Using rDNA [J]. Isr. J. Aquacult-Bamid. 72:1120903.
Lv, G. T., Zhang, X. M., and Zhao, J. (2012). The Effect of Phenotypic and Morphometric Traits on Body Weight of Moerella iridescens. J. Zhejiang Ocean Univ. Nat. Sci. Edit. 31, 487–491.
Meng, X. P., Shen, X., Zhao, N. N., Tian, M., Zeng, Y., Chen, J. A., et al. (2013). Microsatellite analysis of genetic diversity in four geographic populations of Sca parc brughtonii. Prog. Fish. Sci. 34, 59–67.
Ratnaningrum, Y. W. N., Indrioko, S., Faridah, E., and Syahbudin, A. (2017). Gene flow and selection evidence of sandalwood (Santalum album) under various population structures in gunung sewu (Java, Indonesia), and its effects on genetic differentiation. Biodiversitas 18, 1493–1505. doi: 10.13057/biodiv/d180427
Raymond, M., and Rousset, F. (1995). GENEPOP (version3.3) population genetics software for exact tests and ecumenicism. J. Hered. 86, 248–249. doi: 10.1093/oxfordjournals.jhered.a111573
Schuelke, M. (2000). An economic method for the fluorescent labeling of PCR fragments. J. Nat. Biotechnol. 18, 233–234. doi: 10.1038/72708
Tian, J. T., Liu, Z. H., Yang, A. G., Wu, B., and Zhou, L. Q. (2013). Microsatellite analysis of genetic diversity of 4 geographic populations of Scapharca broughtonii. Prog. Fish. Sci. 6, 59–67.
Xu, H., Hu, Y. L., Xu, Y. P., Jin, K., and Chen, P. (2016). The analysis of genetic diversity and morphology of Moerella iridescens in Zhejiang Province. J. Shanghai Ocean Univ. 4, 508–514.
Yu, Z. F., Yan, X. C., Zhang, Y. H., Yang, F., Yang, F., and Zhang, G. F. (2012). EST-SSR diversity of Philippines clam in different age groups in Dalian. Acta Ecologica. Sinica. 1, 4673–4681. doi: 10.5846/stxb201105150625
Zhang, M., Wei, M., Dong, Z., Duan, H., Mao, S., Feng, S., et al. (2019). Fecal DNA isolation and degradation in clam Cyclina sinensis: noninvasive DNA isolation for conservation and genetic assessment. BMC. Biotechnol. 19:99. doi: 10.1186/s12896-019-0595-6
Keywords: Moerella iridescens, SSR, genetic diversity, China coasts, conservation management
Citation: Li X, Gao S, Zhao M and Dong Z (2022) SSR Marker-Based Genetic Resource Assessment of the Rainbow Clam Moerella iridescens Along the Coasts of China: Implications for Strategy of Conservation Management. Front. Mar. Sci. 9:843312. doi: 10.3389/fmars.2022.843312
Received: 25 December 2021; Accepted: 24 January 2022;
Published: 23 February 2022.
Edited by:
Zhongming Huo, Dalian Ocean University, ChinaReviewed by:
Zulin Zhang, The James Hutton Institute, United KingdomZhiyi Bai, Shanghai Ocean University, China
Copyright © 2022 Li, Gao, Zhao and Dong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhiguo Dong, ZHpnNzcxMkAxNjMuY29t
 Xiaoying Li1,2,3
Xiaoying Li1,2,3 Zhiguo Dong
Zhiguo Dong