- 1Ninghai Institute of Mariculture Breeding and Seed Industry, Zhejiang Wanli University, Ninghai, China
- 2Key Laboratory of Exploration and Utilization of Aquatic Genetic Resources, Ministry of Education, Shanghai Ocean University, Shanghai, China
- 3Key Laboratory of Aquatic Germplasm Resource of Zhejiang, College of Biological and Environmental Sciences, Zhejiang Wanli University, Ningbo, China
Accumulation of excessive ammonia is a big threat to aquatic animals, which causes adverse effects on the health, production reduction, and even high mortality. The razor clam Sinonovacula constricta, a bivalve living in intertidal mudflat with a deep-burrowing lifestyle, often faces a high concentration of ambient ammonia. However, there is less available information concerning the toxic effects of ammonia on razor clam and its molecular mechanisms of adaptation to ammonia stress. The aim of this study was to investigate the effects of ammonia exposure on the gill and hepatopancreas of razor clam by transcriptome sequencing. The results showed that the median lethal concentration of ammonia was 244.55 mg/L for 96 h. A total of 1,415 and 306 differentially expressed genes (DEGs) were identified in the gill and hepatopancreas, respectively. The functional annotation showed that DEGs of the gill were mainly involved in the regulation of nitrogen compound metabolic process, nitrogen compound transport, and amide transport. The DEGs of the hepatopancreas were mostly enriched in oxidation-reduction process, response to stress, and amine metabolic process. The expression levels of NH3/NH4+ transporting channels and H+ excreting-related genes, including Rhesus glycoproteins (Rh), Na+/K+-ATPase (NKA), Na+/H+ exchanger, V-ATPase (VHA), and carbonic anhydrase (CA), were upregulated significantly in the gill (p < 0.05). In addition, the expression levels of glutamine and urea synthesis-related genes that played vital roles in ammonia detoxification, such as glutamine synthetase (GS), arginase (ARG), and argininosuccinate synthetase (ASS), were also increased obviously in the hepatopancreas (p < 0.05). Taken together, our results indicate that the synergistic action of ammonia excretion in the gill and ammonia metabolism in the hepatopancreas might be the mechanism through which the clams tolerate to environmental ammonia. This study provides a molecular basis for the better evaluation of the responding mechanism of ammonia tolerance.
Introduction
With the development of intensive aquaculture and high feed inputs, ammonia has become a particularly harmful stress factor to aquatic animals. Generally, there are mainly two chemical forms of ammonia in water, namely, ionized (NH4+) and unionized (NH3) form. The latter is more toxic as the NH3 diffuse more easily across the cell membrane for its lipid solubility and non-polarity (Cong et al., 2018). Meantime, other environmental factors also affect the ratio of two forms of ammonia. Higher pH and temperature (Miron et al., 2008; Kir et al., 2015) can lead to higher ratio of NH3, as well as lower salinity (Kir et al., 2019). Previous studies have shown that excessive ammonia causes physiological dysfunctions, such as decreased growth rate (Harris et al., 1998; Hu et al., 2018), changed histological structure in gill (Cong et al., 2017), disturbed energy and amino acid metabolism (Miranda-Filho et al., 2009), and suppressed immunity response (Cui et al., 2017). It also resulted in massive mortality (Chen et al., 2019). Therefore, clarifying the strategies of ammonia detoxification would provide reliable data for practical guidance of reducing ammonia toxicological effects and protecting aquaculture environment.
The detoxification mechanisms have been explored more thoroughly in fishes and crustaceans than in molluscs. Researchers pointed out that ammonotelism was one of the primary strategies for detoxification in most teleosts, including Nile tilapia (Oreochromis niloticus) (Zhu et al., 2019), large-scale loach (Paramisgurnus dabryanus) (Shang et al., 2021), and mudskipper (Boleophthalmus pectinirostris) (Chew and Ip, 2014). You et al. (2018) found that NH4+/NH3 transporting channels and H+ excreting-related genes were involved in ammonia excretion in the gill of mudskippers. Meanwhile, non/less-toxic glutamine synthesis by glutamine synthetase (GS) has been noted as another traditional strategy to cope with high level of ammonia in teleosts (Ip et al., 2004). In addition, an unnegligible pathway of detoxifying ammonia into urea, which had been commonly found in the liver of ureogenic animals, maintained low concentration of ammonia. It occurs principally through ornithine-urea cycle involving catalysis of crucial enzymes, including arginase (ARG), argininosuccinate synthetase (ASS), and GS (Mommsen and Walsh, 1989; You et al., 2018; Sun et al., 2021). Although the study on molecular mechanism underlying regulation expose to ammonia in molluscs is still at its infancy, preliminary studies have been reported. The ammonia detoxification metabolism in triangle sail mussel (Hyriopsis cumingii) may transfer ammonia into glutamine, alanine, and aspartate (Zhao et al., 2021). In razor clam Sinonovacula constricta, the potential protective strategies of glutamine formation, which are mediated by glutamine dehydrogenase (GDH) and GS, convert ammonia into non/less-toxic nitrogenous compounds under ammonia stress (Zhang H. et al., 2020). Taken together, more attention should be paid to the comprehensive study on molecular response mechanism of ammonia stress in mollusks, especially in terms of ammonia excretion.
RNA sequencing is a well-established comprehensive subject to explore molecular mechanisms in responding to abiotic stresses, especially effective for blank areas of studies (Gao et al., 2018; Stark et al., 2019). Recently, more and more studies have investigated the complex interplay among cellular pathways when exposed to ammonia (Yu et al., 2019; Xue et al., 2021). However, only limited works have been reported in bivalves. In Manila clam Ruditapes philippinarum, short- and long-tern exposure to ammonia displayed the different responding patterns in the gill via digital gene expression analysis. It induced pathways of notch signaling and protein processing in endoplasmic reticulum in short-term exposure and pathways of gap junction, immunity, and key substance metabolism in long-term exposure (Cong et al., 2018). Digestive gland transcriptome analysis of response mechanism to ammonia in Asian clam Corbicula fluminea revealed that the immunity and apoptotic signaling pathways play important roles during stress (Zhang et al., 2019). Although abovementioned studies revealed a mass of important genes responding to ammonia stress in a few species, the potential molecular mechanisms were not fully elucidated for most economic molluscs.
The razor clam S. constricta is one of the most economically important marine bivalves, which is widely distributed in the intertidal zones and estuarine areas of China, Japan, Korea, and Vietnam along the western Pacific Ocean with a deep-burrowing lifestyle (Dong et al., 2020). According to the statistical data from U.S. Environmental Protection Agency (USEPA), shellfish are more sensitive to ammonia nitrogen. Of the top 10 sensitive genera, eight genera belong to molluscs (USEPA, 2013). However, the clam-fish/shrimps polyculture makes razor clam to face to high concentration of ambient ammonia (Sun et al., 2021). Therefore, S. constricta is a suitable model species to explore the molecular mechanism of higher tolerance to ammonia in molluscs. To date, the molecular mechanism of ammonia toxicity has not been understood comprehensively in razor clam. In this study, the influence of high concentration ammonia stress was systematically evaluated by profiling the gill and hepatopancreas transcriptomes of S. constricta, which were subjected to acute challenge of 180 mg/L ammonia for 72 h. This study provides insights into the molecular regulatory mechanism of ammonia tolerance in bivalve.
Materials and Methods
Clam Culture
The healthy razor clams (wet weight 9.43 ± 1.60 g, shell length 63.42 ± 1.57 mm) were collected from aquafarm in Xiangshan (Ningbo, China). Before the experiment, the clams were acclimated for 1 week in 1,000 L tanks containing filtered seawater (salinity 22) with aeration at the temperature (14–16°C). During the acclimatization and ammonia exposure periods, the clams were fed twice per day at 8:00 and 18:00 on a diet of microalgae Chlorella vulgaris and Chaeroeeros moelleri. All the seawater was renewed two times daily. The whole experiment was maintained under a photoperiod of 12 h light and 12 h dark.
Toxicity and Exposure Experiments of Ammonia
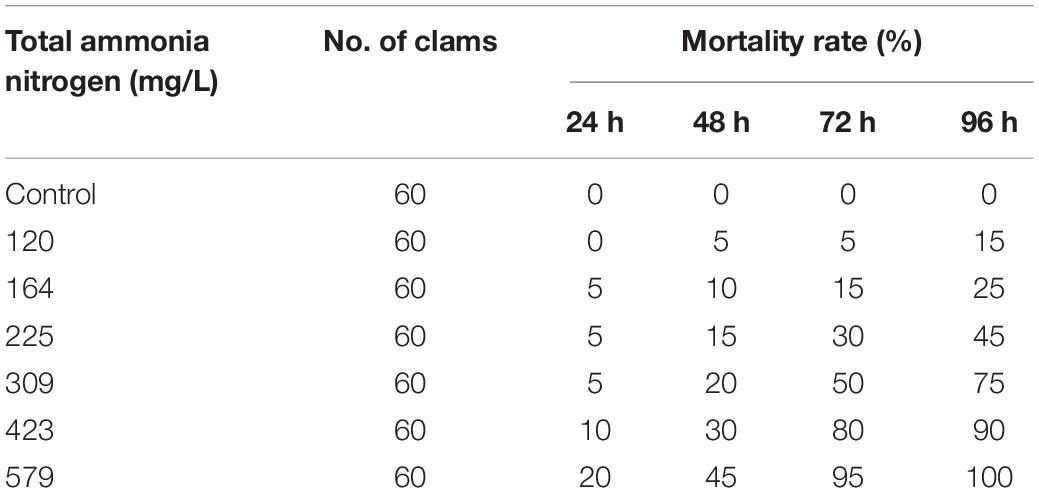
The total ammonia nitrogen (TAN) concentrations were designed by equal space between logarithm with the gradients of 0, 120, 164, 225, 309, 423, and 579 mg/L. The stock solution of 1,000 mg/L TAN (stock solution was disposed of ammonium chloride NH4Cl; CNW, Shanghai, China, ACS reagent grade >99%) was used for producing the desired final concentrations of TAN. A total of 420 clams were randomly divided into seven groups with three replicates in each different ammonia concentration and cultured in plastic tanks with 30 L of filtered seawater (20 individuals in each replicate). Water quality parameters were tested daily throughout the experiment. The pH and dissolved oxygen were measured using HACH HQ30d pH meter (Hach Company, CO, United States). The salinity and temperature were tested by AZ8371 Salinometer (Hengxing, Taiwan, China) and fish tank thermometer, respectively. During 96 h duration of ammonia exposure, observation was performed every 12 h. To calculate the median lethal concentrations (LC50), the mortality rates were measured every 24 h until it reached 100%.
Based on the median lethal concentrations (LC50-96 h) of ammonia established in the toxicity tests, the clams were subjected to a 96 h ammonia exposure at three concentrations of 100, 140, and 180 mg/L comparing with 0 mg/L with three replicate tanks for each treatment (100 clams per tank). Daily acclimation was performed referred from Zhang H. et al. (2020). Six individuals were randomly sampled from each treatment at 0, 1, 3, 6, 9, 12, 24, 48, 72, and 96 h time points, respectively. For each sampled clam, hemolymph was immediately collected from adductor using a 2 mL disposable sterilized syringe for pH value and ammonia content analysis. The pH value was measured using HACH HQ30d Portable Dissolved Oxygen Tester Conductivity Meter pH meter (Hach Company). The ammonia content was measured with the commercial kits (no. A086-1-1) of Nanjing Jiancheng Bioengineering Co. (Nanjing, China) according to the instructions of the manufacturer. Meanwhile, the gill and hepatopancreas were dissected separately and frozen in liquid nitrogen immediately for RNA extraction. Finally, the tissues obtained at 72-h exposure to 180 mg/L group were considered for RNA-seq analysis.
RNA Extraction and Transcriptome Sequencing
Total RNA was extracted from the gill and hepatopancreas at each time point using TRIzol reagent (Omega, Norcross, GA, United States) according to the instructions of the manufacturer. Then RNA concentration and integrity was assessed using NanoDrop 2000 (Thermo Scientific, Waltham, MA, United States). The cDNA libraries were constructed by a NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (NEB, Ipswich, MA, United States) and then assessed on the Agilent Bioanalyzer 2100 system. After quality assessment, the sequencing was performed on Illumina Hiseq 2000 platform.
Transcriptome Assembly and Gene Expression Quantification
Clean data were filtered out as reads containing adapter, poly-N, and low-quality reads. The sequence quality was assessed by Q20, Q30, and GC content. Trinity was used for clean reads assembly (Grabherr et al., 2013). Subsequently, the clean reads were aligned to the S. constricta reference genome (Dong et al., 2020) using Tophat (Trapnell et al., 2009).
The gene expression levels were estimated using RSEM (Dewey and Li, 2011). Then, gene expression was normalized using the reads per kilobase per million mapped reads (RPKM). Genes with low read counts (<10 reads) were filtered out before gene expression analysis by using featureCounts software (Liao et al., 2014). Differential expression analysis between ammonia-N group (TG) and control group (CG) was performed using the DESeq2. Differentially expressed genes (DEGs) were defined with a fold change >2 and a cutoff false discovery rate (FDR) of 0.05.
Functional Annotation and Enrichment of Differentially Expressed Genes
Gene function was annotated based on the following databases including NR (NCBI non-redundant protein sequences)1, Pfam (Protein family),2 KOG/COG (Clusters of Orthologous Groups of proteins),3 Swiss-Prot4, KEGG (Kyoto Encyclopedia of Genes and Genomes),5 and GO (Gene Ontology).6
To identify GO terms and KEGG pathways among the DEGs under ammonia stress, enrichment analyses were implemented by the software GOseq (Young et al., 2010) and KOBAS (Chen et al., 2011), respectively, with a cutoff q-value of 0.05. The significantly enriched DEGs were subject to further scrutiny to identify genes potentially involved in ammonia stress response.
Validation of RNA-Seq Data by Quantitative Real-Time PCR
To confirm the gene expression profiles from RNA-seq analysis, six DEGs were selected to perform quantitative real-time PCR (qRT-PCR) for tissues of gill and hepatopancreas, respectively. Specific primers for candidate genes and the housekeeping gene (Rs9) were designed based on the unigene sequences using Primer Premier 5.0 software (Table 1).
Total RNA was extracted from the gill and hepatopancreas tissues using the Omega RNA-Solv Reagent and was visualized with agarose gel electrophoresis. The qRT-PCR was performed using the SYBR® PremixEXTaq™II (Takara, Dalian, China) kit on the Roche LightCycler® 480 System (Roche Diagnostics, Basel, Switzerland). The PCR reaction was carried out in a total volume of 20 μl, containing 10.0 μl of 2 × SYBR Green Master Mix (Applied Biosystems), 2.0 μl 500 ng of first-strand cDNA, 1.0 μl of each primer (10 μmol/L), and 6.0 μl of PCR-grade DEPC water. Then, the amplification parameters were set as follows: initial denaturation at 95°C for 10 min, followed by 40 cycles of 95°C for 10 s, 60°C for 60 s, and 40°C for 5 s in 96-well optical reaction plates (Roche, Basel, Switzerland). All reactions were performed in quadruplicate. The gene expression levels were quantitatively analyzed and converted to fold differences by the comparative CT method (2–ΔΔCT method).
Statistical Analyses
The median lethal concentrations (LC50-96 h) of ammonia were calculated by the probit method (Denham, 2016). The relationship between concentration of ammonia and mortality was subjected to linear regression analysis. All data were expressed as means ± SD. The gene expression levels were subjected to one-way ANOVA followed by least significant difference (LSD) analysis. Statistical significance was defined at p ≤ 0.05. All statistical analyses were performed using the SPSS version 13.0.
Results
Median Lethal Concentration (LC50) of Total Ammonia–Nitrogen and NH3–N
To calculate LC50 of TAN, we recorded the mortality per 24 h for a total of 96 h ammonia exposure till all clams died. As expected, the mortality rate was increased with the increased concentration of TAN and exposure time (Table 2). No death occurred in the CGs as well as the groups treated with 120 mg/L of TAN for 24 h. All clams died after 96 h of exposure in 579 mg/L of TAN. According to the statistical method, the corresponding LC50 of unionized ammonia (NH3–N) was calculated (Table 3). The LC50 value of TAN determined at 96 h was 244.55 mg/L and the corresponding LC50 value of NH3–N was 9.69 mg/L.
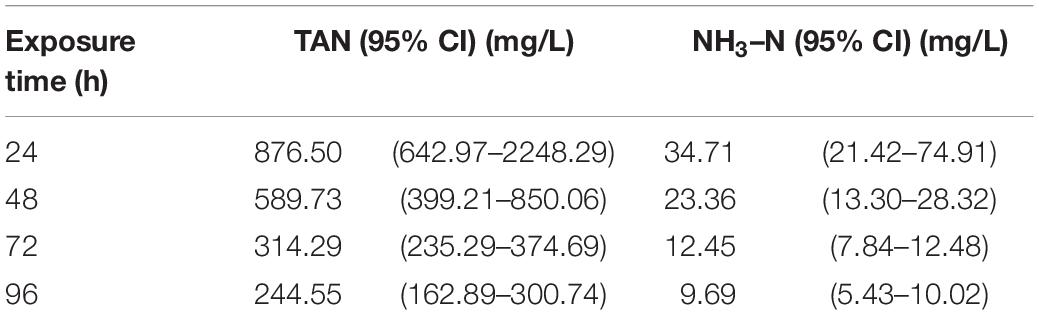
Table 3. Median lethal concentration (LC50) of total ammonia-N (TAN) and unionized ammonia-N (NH3–N) exposed to S. constricta.
Effects of Ammonia Content and pH in Hemolymph
The ammonia content in the hemolymph of S. constricta increased dramatically under ammonia exposure. After 1 h of exposure, the ammonia content increased sharply and then the level reached a plateau occurred after 24 h exposure in all treatment groups. Finally, the highest value was up to 3,331.2 μmol/L at 96 h in 100 mg/L group. It is worthy to note that ammonia contents showed an opposite decrease trend with increased ammonia concentration during 12–96 h (Figure 1A). Moreover, the pH of hemolymph showed significant decreases in all treatment groups (Figure 1B).
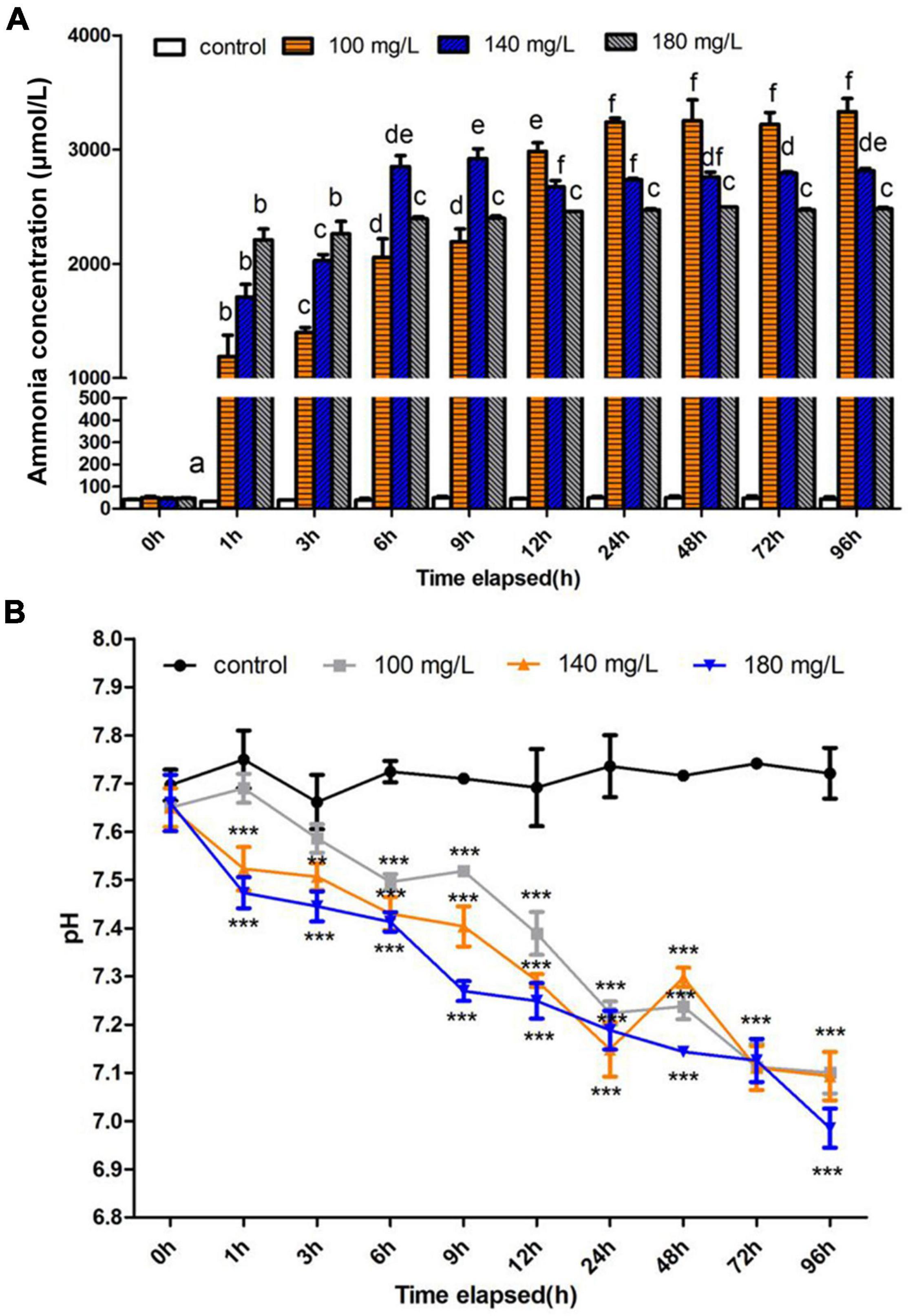
Figure 1. Effects of ammonia concentration and pH in Sinonovacula constricta hemolymph under ammonia stress. (A) Ammonia concentration is mean ± SD (n = 3). Data with different letters are significantly different in the same concentration group (p < 0.05). (B) pH value is mean ± SD (n = 3). Asterisk (*) indicates a significant difference between the exposed group and its respective control (*p < 0.05; **p < 0.01; ***p < 0.001).
Characterization of Transcriptome Data
The details on the transcriptome data for each sample are summarized in Supplementary Table 1 and the raw sequence data were deposited at the NCBI Sequence Read Archive (SRA) with the accession numbers of SRR9943679–SRR9943690. After data filtering, approximately 28.44 and 28.51 million high-quality clean reads were obtained from the gill and hepatopancreas, respectively. In total of 12 libraries, the Q20 and Q30 were >97.07% and >92.83%. And the GC content ranged from 40.10 to 43.13%. Among the samples from the gill, 66.99–71.71% of the clean reads could be mapped onto the reference genome, and the mapping rates to the reference genome for hepatopancreas reads were 67.10–69.90%.
Analysis of Differentially Expressed Genes in Gill and Hepatopancreas
With the criteria of a FDR ≤ 0.05, and |fold change of expression level > 2, we identified 1,415 DEGs in the gill after 72 h ammonia exposure, which included 1,029 upregulated and 386 downregulated genes. Meanwhile, there were 306 DEGs in the hepatopancreas, which included 248 upregulated and 58 downregulated genes (Figure 2A). In addition, overlapping analysis of data sets from both two tissues revealed 85 common DEGs in response to ammonia-N stress, of which 79 genes were upregulated and four genes were downregulated in both the gill and the hepatopancreas. The remaining two genes were downregulated in the gill but were upregulated in the hepatopancreas (Figure 2B and Supplementary Table 2).
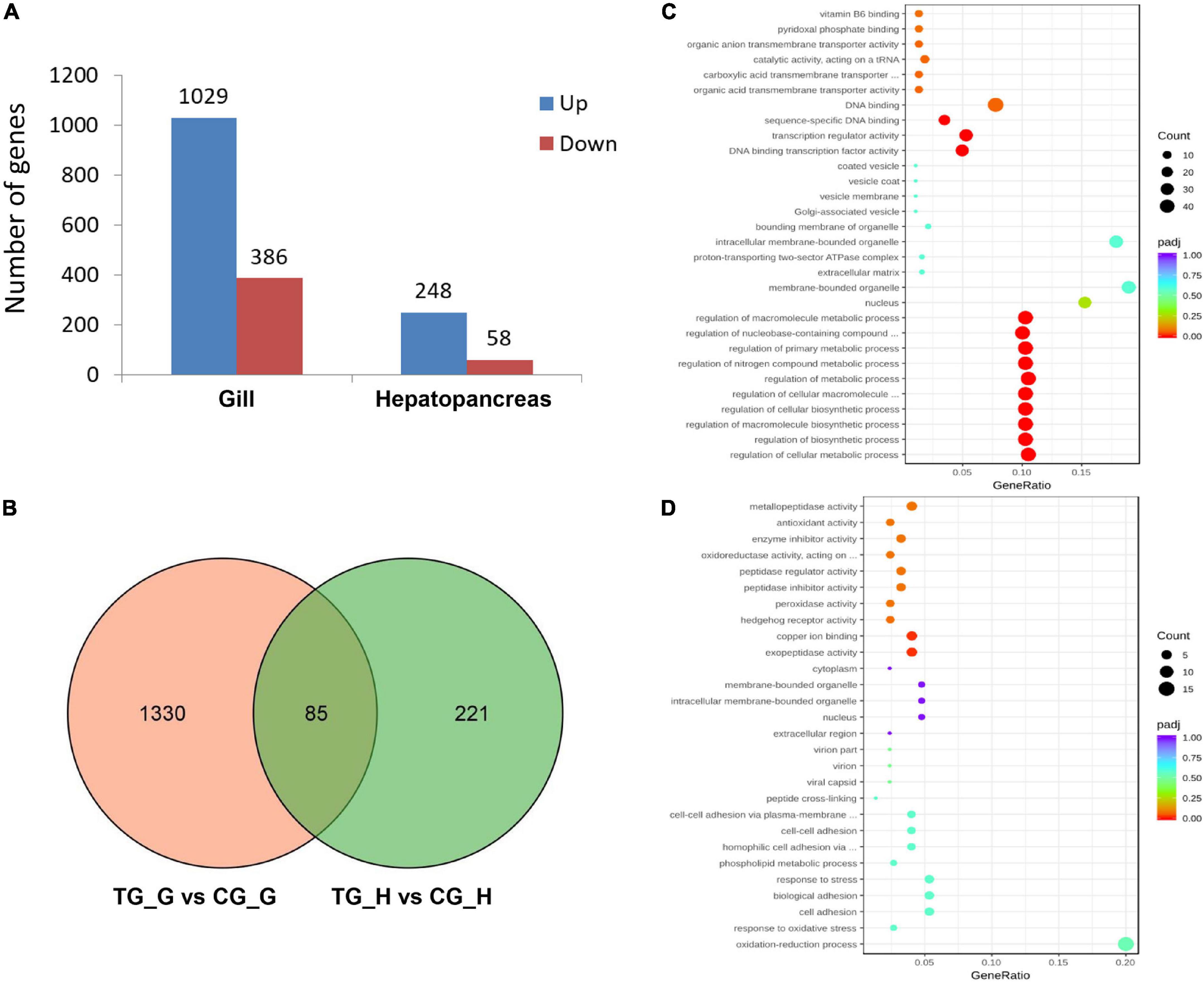
Figure 2. A summary of differentially expressed genes (DEGs) of the gill and hepatopancreas in S. constricta exposure to acute ammonia. (A) The numbers of DEGs in the gill and hepatopancreas. (B) Venn diagram of DEGs shared between gill and hepatopancreas. G represents gill and H represents hepatopancreas. (C) Enriched gene ontology (GOs) in the gill. (D) Enriched GOs in the hepatopancreas.
Gene Ontology and Kyoto Encyclopedia of Genes and Genomes Pathway Enrichment of Differentially Expressed Genes
Gene ontology analysis was conducted to profile the functional annotation of DEGs in the gill and hepatopancreas. The DEGs were assigned into three GO terms including biological process (BP), molecular function (MF), and cellular component (CC), and the significantly enriched items were illustrated (Figure 2C and Supplementary Figures 1, 2). In the gill, DEGs primarily enriched in BP, including regulation of cellular metabolic process (GO:0031323), regulation of biosynthetic process (GO:0009889), regulation of nitrogen compound metabolic process (GO:0051171), organic substance transport (GO:0071702), nitrogen compound transport (GO:0071705), organic anion transport (GO:0015711), and amide transport (GO:0042886) (Figure 2C). In the hepatopancreas, DEGs were mainly enriched in oxidation-reduction process (GO:0055114), response to oxidative stress (GO:0006979), response to stress (GO:0006950), amine metabolic process (GO:0009308), copper ion binding (GO:0005507), enzyme inhibitor activity (GO:0004857), and antioxidant activity (GO:0016209) (Figure 2D).
The results of KEGG enrichment analyses are summarized in Table 4. Specifically, the upregulated DEGs in the gill were related to protein processing in endoplasmic reticulum and endocytosis (spu04141), autophagy/mitophagy (spu04140 and spu04137), and aminoacyl-tRNA biosynthesis (spu00970). The downregulated DEGs in the gill were related to nucleotide/base excision repair (spu03420 and spu03410) and lysine degradation (spu00310). At the same time, DEGs in the hepatopancreas were assigned to Wnt signaling pathway (spu04310) and amino acid metabolism, including arginine, proline, tyrosine, and phenylalanine biosynthesis and metabolism (i.e., spu00220, spu00330, spu00350, and spu00360). Despite no KEGG pathways were significantly enriched, the DEGs were also analyzed according to their KEGG pathways to describe the toxic effect and response mechanism of ammonia in S. constricta, which are summarized in Table 5. Ammonia exposure obviously induced significant expressed changes of genes that were related to nitrogen compounds-related metabolism and excretion, pH regulation, immune, and apoptosis in both tissues. In aforementioned pathways, DEGs involved in glutamine-arginine-urea cycle such as ARG, ASS, and GS were mostly upregulated in both the hepatopancreas and the gill, while glutathione S-transferase (GST) related to antioxidant activity was significantly downregulated. In addition, the RNA-seq data showed that ammonium transporter (AMT) responsibility for ammonia excretion was significantly downregulated after 72 h ammonia exposure in gill, which is the primary organ for ammonia excretion. In contrast, carbonic anhydrase (CA) related to nitrogen metabolism showed upregulation in the gill. The abundant expression of immune response- and apoptosis-related genes suggested that their corresponding pathways were activated during ammonia stress.
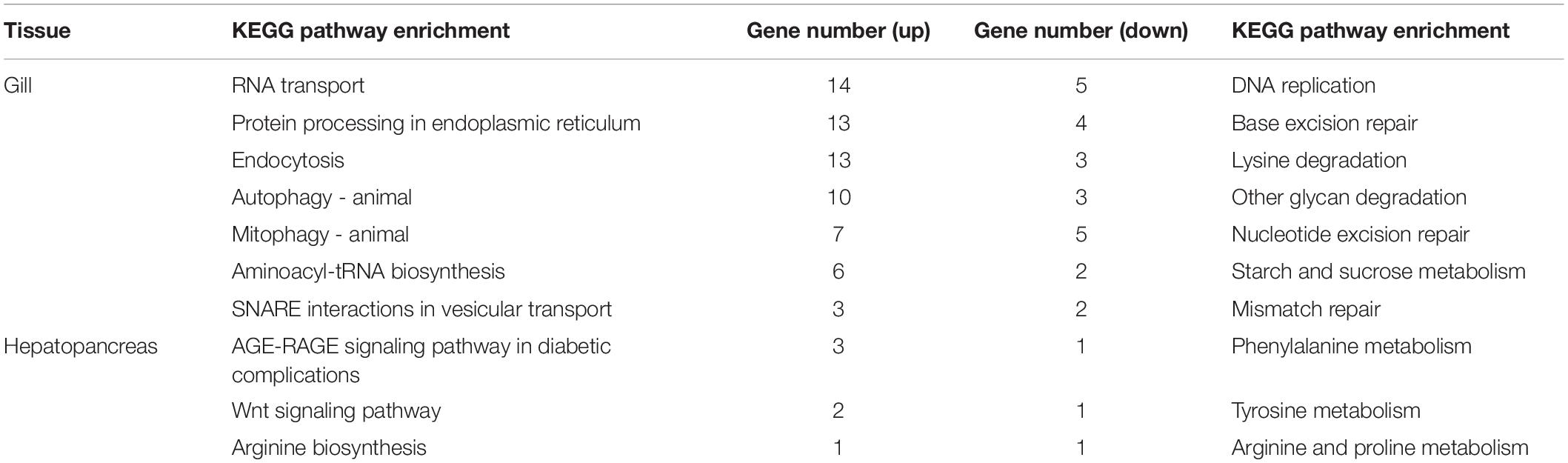
Table 4. Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment of differentially expressed genes (DEGs) in the gill and hepatopancreas of S. constricta under ammonia exposure.
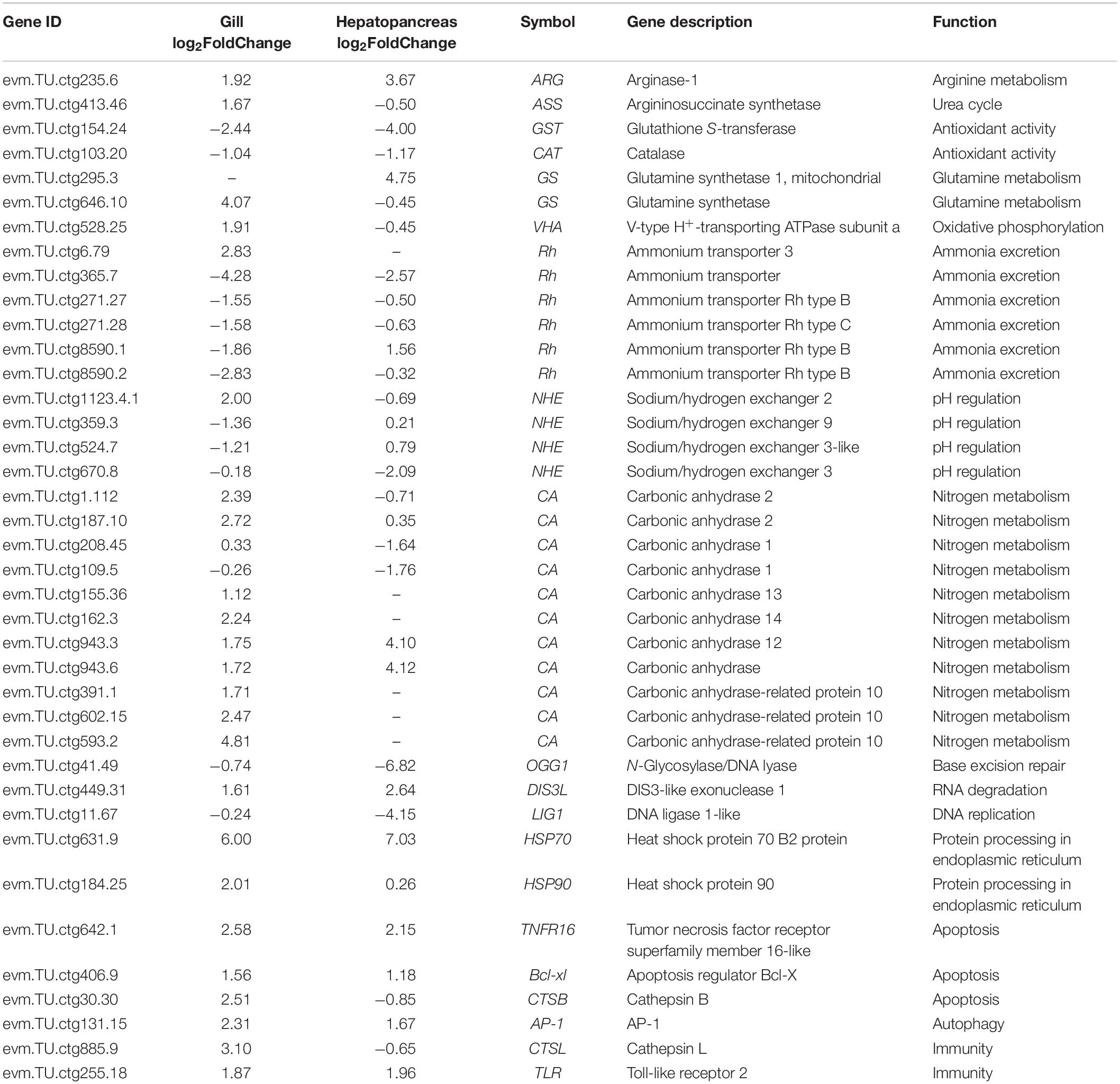
Table 5. List of DEGs of the gill and hepatopancreas involved in responding to ammonia stress in S. constricta.
Validation of Differentially Expressed Genes by Quantitative Real-Time PCR
Four genes related to ammonia excretion (i.e., Rh, NKA, V-ATP, and NHE) in the gills and two genes related to ammonia metabolism (i.e., ARG and ASS) in the hepatopancreas were selected to verify the accuracy of RNA-seq data by qRT-PCR. Rs9 were chosen as reference genes. As shown in Figure 3, four genes in the gill (i.e., Rh, NKA, V-ATP, and NHE), as well as ARG in the hepatopancreas, all presented the same trend with earlier increase and later decrease when exposed to 180 mg/L ammonia. Unlike the abovementioned genes, the expression of ASS was increased along whole time points and reached the maximum at 96 h. It should be noted that the expression of Rh in the treatment group was significantly decreased at 96 h when compared with CG.
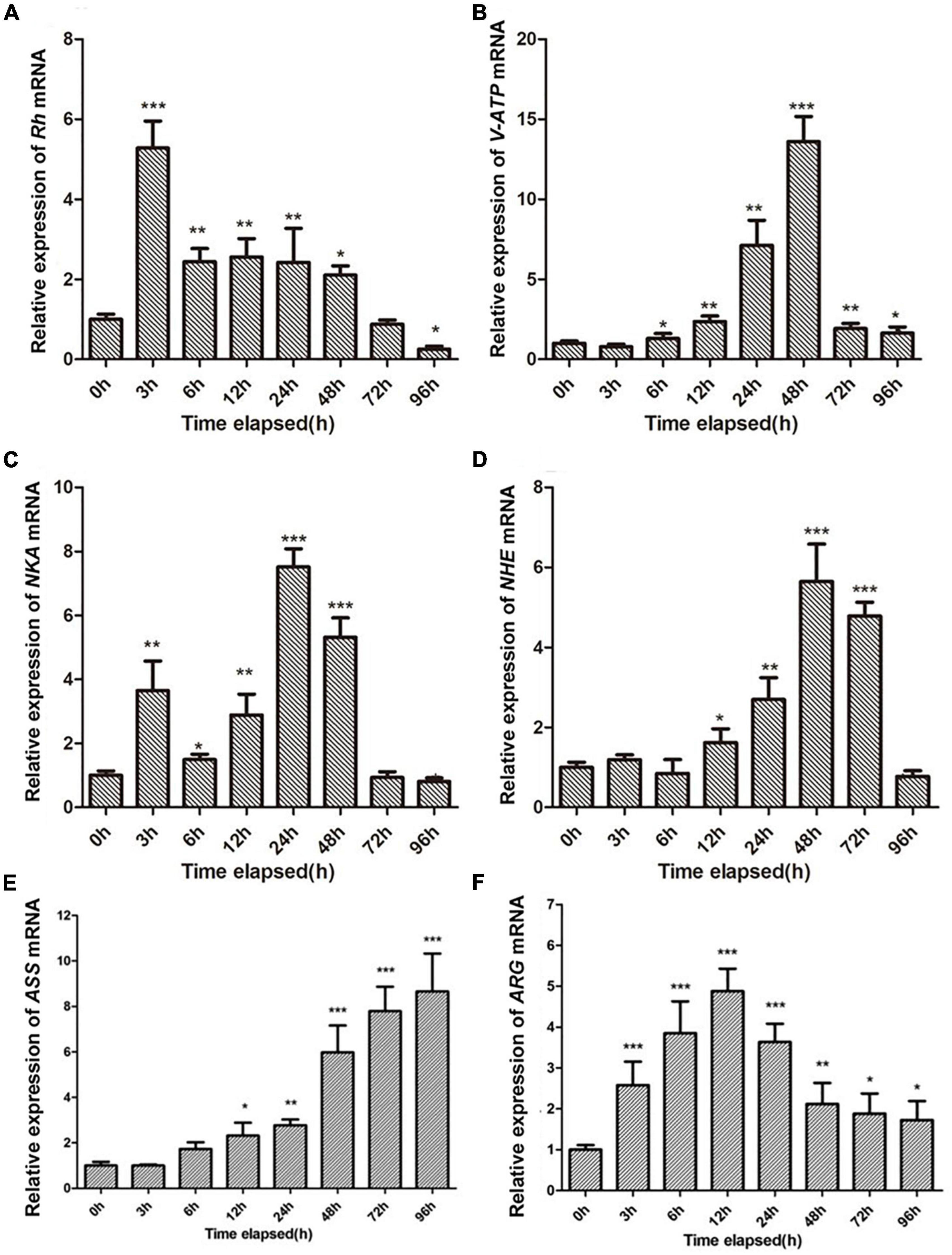
Figure 3. Validation of RNA-seq profiles by qRT-PCR. (A–D) Represent the expression patterns of Rh, NKA, V-ATP, and NHE genes in the gill of S. constricta after exposure to 180 mg/L ammonia for 96 h, respectively. (E,F) Panels represent the expression patterns of the ARG and ASS genes in the hepatopancreas after exposure to 180 mg/L ammonia for 96 h, respectively. Vertical bars represent the mean ± SD (n = 4). Asterisk (*) indicates a significant difference between the exposed shellfish and its respective control (*p < 0.05; **p < 0.01; ***p < 0.001).
Discussion
Tolerate Capacity to Ammonia Adapt to the Deep-Burrowing Habit
Ammonia is one of major contaminants in aquaculture environment. Its excessive accumulation would limit animal health and production in aquaculture (Shang et al., 2021; Shen et al., 2021). Therefore, it is necessary to know the tolerance and lethal concentrations of ammonia nitrogen for aquatic animals and controlling the ammonia nitrogen to safe concentration in water environment.
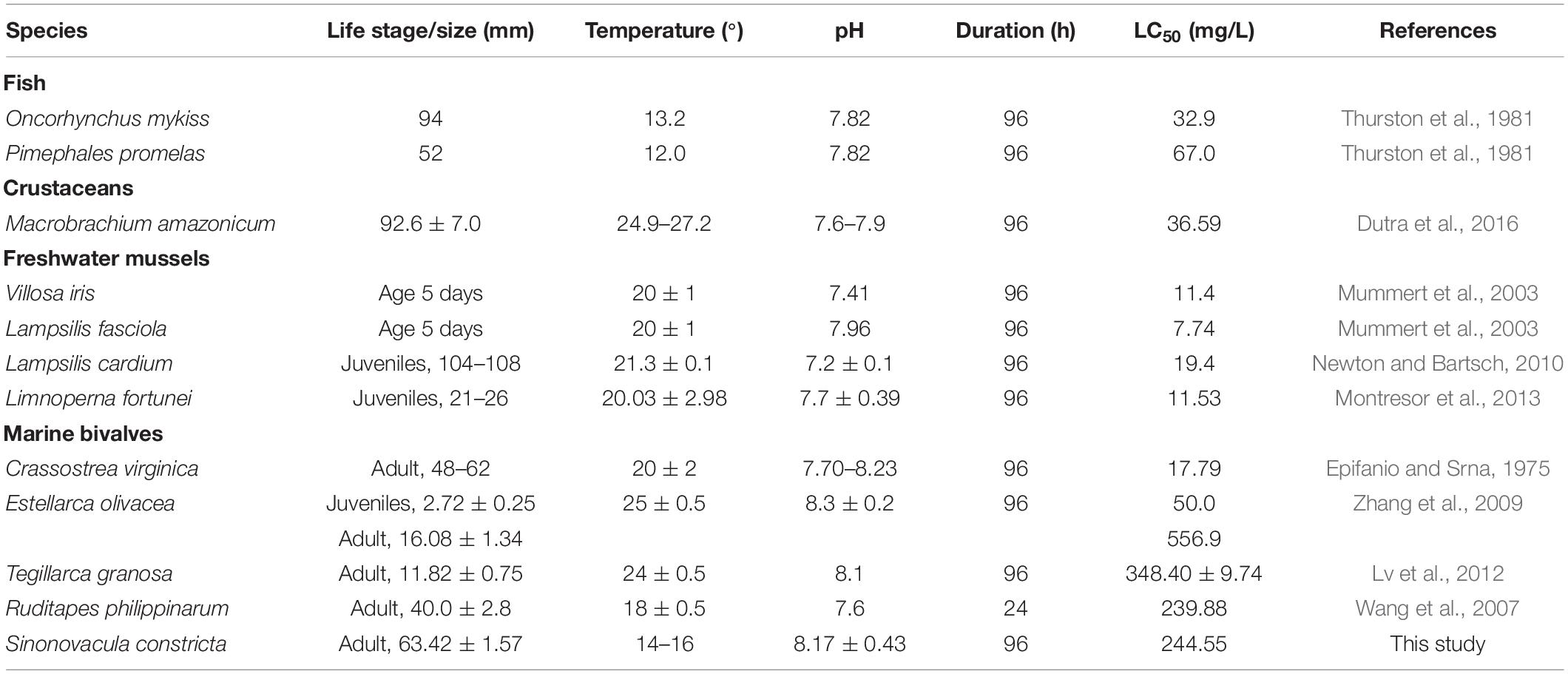
In fact, the USEPA has accomplished the comparison of available toxicity data for 69 genera and concluded that shellfish may be more sensitive to ammonia than other aquatic animals (USEPA, 2013; Zhang et al., 2019). However, only freshwater molluscs, but no saltwater ones, were collected in their report. Recently, many tests about the sublethal effects of ammonia have been performed on saltwater animals, including the shellfish. The available data are presented in Table 6. The ammonia tolerance displayed a distinct difference among species. Particularly, excellent ammonia tolerance had been found in marine bivalves such as clam Estellarca olivacea (Zhang et al., 2009), blood clam Tegillarca granosa (Lv et al., 2012), R. philippinarum (Wang et al., 2007), and S. constricta (this study, 96-h LC50: 244.55 mg/L). Notably, these four species are all benthic bivalves. Weihrauch et al. (2004) had confirmed that benthic animals (crabs and bivalves) faced to higher ambient ammonia compared with those living free in the water. The razor clam is one of typical deep-burrowing bivalves living in intertidal zones, where the ammonia concentration is high in water (Newton and Bartsch, 2010). Therefore, we speculated that the deep-burrowing habitat possibly promoted the acquisition of greater capacity to tolerate higher ambient ammonia in razor clam. Hence, it is considered to be a suitable model to explore the response mechanism of ammonia toxicity. In this study, the molecular mechanism on response to high ammonia tolerance was investigated through transcriptomic analysis.
Response to Ammonia Toxicity in Sinonovacula constricta
Previous studies have shown that ammonia stress caused a variety of adverse effects, such as energy metabolism disturbance, induced oxidative stress and immune defense, increased sensitivity of the animal to pathogens, and even death (Cheng et al., 2015; Shang et al., 2021; Shen et al., 2021). In this study, our results also demonstrated that ammonia exposure induced the change of metabolic process, oxidative stress, immune response, and apoptosis-related signaling pathways in S. constricta.
Oxidative stress, one of the toxicity mechanisms of environmental stress on aquatic organisms, is usually coupled with DNA damage (Wang et al., 2016). In this study, the genes encoding antioxidant enzymes, i.e., GST and catalase (CAT), were significantly downregulated, suggesting a weakness in compensatory mechanism due to excessive reactive oxygen species (ROS). The excessive ROS might destruct cell membranes with the formation of lipid peroxides and oxidized proteins (Liang et al., 2016). In contrast, the accumulation of free radicals resulted in the ischemia and hypoxia in cells, which further aggravated the metabolic disorders and tissue damage (Zhang et al., 2016). In addition, the gene encoding N-glycosylase/DNA lyase, which was involved in base excision repair (Wyatt, 2009), was also significantly downregulated after ammonia exposure (log2FoldChange = −6.82). The altered expressions of DIS3-like exonuclease and DNA ligase 1 involved in RNA degradation and DNA replication indicated the disorder of genetic information processing (Staals et al., 2010), which might produce “protein-oaf” causing metabolic impairments (Hasenbein et al., 2014). Besides, previous study has been demonstrated that ammonia has severe impacts on the immune system (Lu et al., 2016), and functioned as predisposing factors, which accompanied with low levels of pathogen infections, could lead to high mortalities (Hasenbein et al., 2014). The DEGs related to immune defense, including heat shock protein, toll-like receptor, and cathepsin L, presented significant upregulation exposure to ammonia in S. constricta. The similar findings have been reported in some invertebrates, such as abalone Haliotis discus hannai (Shen et al., 2015), Pacific white shrimp Litopenaeus vannamei (Liu et al., 2020), and freshwater shrimp Macrobrachium nipponense (Yu et al., 2019).
Ammonia Excretion in Gill of Sinonovacula constricta
Most teleost fishes excrete ammonia across branchial and/or epithelial surfaces to maintain appropriate internal ammonia concentration, which is an effective way to protect against ammonia toxicity under ammonia stress (You et al., 2018). The bulk of ammonia excretion mainly occurred in the gill is thought to depend on the cooperation of various transport proteins, including Rh glycoproteins, Na+/K+-ATPase, and so on (Sinha et al., 2015). As is well known, NH3 diffusion across the gills of ammoniotelic fish is facilitated by Rh glycoproteins (Wood et al., 2013), such as Pacific hagfish Eptatretus stoutii (Clifford et al., 2015), P. dabryanus (Shang et al., 2021), and gulf toadfish Opsanus beta (Rodela et al., 2011). In addition, ammonia has also been excreted as NH4+ (Ip et al., 2001). NH4+ possesses similar ionic characteristics, hydration shell sizes, and water mobility rates compared with K+, which allows it to compete against K+ to be transported through the K+-associated channels like NKA (Sinha et al., 2015). We found that mRNA expressions of Rh glycoproteins and NKA were increased significantly in the gill during ammonia stress, suggesting that ammonia excretion in vivo was activated together by NH3/NH4+ transport proteins in S. constricta. However, Rh was dramatically decreased after 96-h ammonia excretion. It is reported that the expression of Rh and NKA, together with activities of related enzymes in the gill tissue, decreased significantly with increased ammonia concentration (Yang et al., 2010; Ren et al., 2015). The reason leading to a decrease in the transcription and activity of related enzymes may be the gradual changes occurring in the gill morphology culminated in drastic lesions when long-term exposure to high concentrations of ammonia (Schram et al., 2010). Although the morphological change of gill was not investigated in this study, a large number of DEGs were enriched into the apoptosis-related signaling pathways, suggesting the occurrence of gill injury after ammonia exposure. The microstructure damage of gill tissue has been observed in the gill of R. philippinarum after subacute exposure of ammonia, which reinforces our assumption (Cong et al., 2017).
Ammonia excretion is augmented by acidic conditions in the water because any NH3 excreted into the water is rapidly converted to and trapped as NH4+ (Ip et al., 2001). There are two potential mechanisms for this acidification, namely, the hydration of CO2 (also crossing the membrane as CO2 gas) to form H+ and HCO3– and the active transport of protons via H+ transport like V-type-H+-ATPase (VHA) and Na+/H+ exchanges (NHE) (Clifford et al., 2015; Tresguerres et al., 2020). CA catalyzes the HCO3– and H+ reaction on the gill surface, which contributes to the secretion of acid and traps the excretion of NH4+ actively (Tresguerres et al., 2020). The significant upregulation of CA genes was found in both the tissues of TGs (Table 5). The expressions of VHA and NHE were significantly upregulated in the gill. These results confirmed that S. constricta could augment ammonia excretion by excreting acid through H+ pump, also like mudskipper Periophthalmus magnuspinnatus (You et al., 2018). In summary, the upregulated mRNA expression of Rh together with upregulated NKA, NHE, and V-ATP suggested that the coordinated protective response of ammonia excretion by NH3/NH4+ transport proteins and acid-trapping of ammonia by H+ transport contributed to maintain a relatively low ammonia concentration in the body fluids during ambient ammonia exposure in razor clam.
Detoxification of Endogenous Ammonia via Glutamine Synthesis and Urea Formation in Hepatopancreas
It has been reported that some fish convert excess ammonia into low toxic compound glutamine to avoid ammonia toxicity (Ip et al., 2001). The key enzymes in the pathway of glutamine synthesis are GS and GDH. Glutamine is produced from glutamate and NH4+, which was catalyzed by GS. The GDH catalyzes α-ketoglutarate and NH4+ to synthesize glutamate (Ip et al., 2001). Hence, one mole of glutamine synthesis is needed to consume two moles of NH4+, which is a highly efficient choice for the detoxification of ammonia. Previous study has shown that exposure to sublethal concentration of NH3 could induce high activities of GS and GDH enzymes in mudskipper Periophthalmodon schlosseri and Boleophthalmus boddaerti (Peng et al., 1998). Exposure to ammonia also results in significant increases in enzyme activity, protein content, and mRNA levels of GS in liver, intestine, and muscle to detoxify ammonia in the Chinese black sleeper Bostrichthys sinensis (Anderson et al., 2002). In accordance with previous results, the significant increased expressions of GS in both the hepatopancreas and the gill in our results suggested that the clam could also reduce the toxicity of ammonia by converting ammonia into glutamine, although the mRNA level of GDH showed no significant change in treatment group compared with CG. However, a recent study showed a dramatic increase in glutamate concentration in the hemocytes of S. constricta exposure to ammonia coupled with an increment of relative mRNA and protein expression of both GS and GDH (Zhang H. et al., 2020). Combined these results together, we believe that it is an important protective strategy for razor clam to convert ammonia into non- or less-toxic nitrogenous compounds such as glutamate formation when faced high ambient ammonia concentration (Zhang H. et al., 2020).
Accumulated ammonia could also be converted into urea to reduce ammonia toxicity, while only a few species could synthesize urea via the ornithine-urea cycle (Peng et al., 1998). The urea production from ornithine cycle has usually been suppressed in all freshwater teleosts, except for some airbreathers, such as the tilapia fish Oreochromis alcalicus graham, which is the only fish that can live in the high alkaline soda lake with pH 9.6–10 (Randall et al., 1989). Besides, urea biosynthesis increases in the estivating African lungfish Protopterus aethiopicus living in shallow waters and burrowing beneath the substratum to survive in estivation, as well as in Xenopus laevis under water-shortage condition (Janssens and Cohen, 1968). Interestingly, there was a significant increase in the urea content in the liver of P. schlosseri after 48 h of aerial exposure, but no changes were observed in B. boddaerti (Lim et al., 2001). Hence, these different results from different fishes indicate that ammonia detoxification through urea cycle is not a universal mechanism, but a unique adaptation to some special environmental circumstances (Ip et al., 2001). In this study, the significantly higher expression of ARG and ASS in the hepatopancreas indicated that the pathway of urea synthesis might have been activated in S. constricta when exposed to high environmental ammonia. As we known, the razor clam lives in intertidal mudflat amount of high ammonia, which place to undergo overlying seawater changing gradually to overlying air during low tide, particularly in summer with increased temperature (Zhang W. Y. et al., 2020). Therefore, it is supported that the retention of urea synthesis pathway contributes to the survival of S. constricta under high ammonia stress during low tide with the condition of extreme dry or heat. Since urea content has not detected in this study, further investigation of this issue is warranted.
Conclusion
This study carried out a preliminary analysis of the responding mechanisms under ammonia stress in S. constricta. The higher capacity of tolerance to ammonia was observed and the 96 h sublethal concentration of ammonia was 244.55 mg/L. A total of 1,415 and 306 DEGs were identified in the gill and hepatopancreas subjected to 180 mg/L TAN for up to 72 h. The DEGs in the gill were majorly enriched in metabolic processes, including regulation of nitrogen compound metabolic process, nitrogen compound transport, and amide transport. Most of DEGs in the hepatopancreas were enriched in oxidation-reduction process, response to stress, and amine metabolic process. In addition, the expressions of ammonia transport-related genes (e.g., Rh, VHA, NHE, and NKA) in the gill were significantly changed under ammonia stress (p < 0.05), indicating that the ammonia excretion by NH3/NH4+ transport and H+ transport proteins synergistically contributed to maintain ammonia at a relative low level during ammonia exposure. Besides, ammonia metabolism-related genes such as GS, ARG, and ASS were upregulated in the hepatopancreas. These results provided preliminary evidence for detoxification of endogenous ammonia via glutamine synthesis and urea formation in S. constricta. Furthermore, the pathways and genes identified in this study would facilitate further studies on the more detailed molecular mechanisms of the ammonia detoxification in molluscs and promote molecular selective breeding for ammonia-tolerance varieties.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: NCBI (accession: SRR9943679-SRR9943690).
Ethics Statement
The animal study was reviewed and approved by Institutional Animal Care and Use Committee (IACUC) of Zhejiang Wanli University.
Author Contributions
JR and ZL designed the experiments. LL and HZ performed the experiments. LL, JR, and HZ analyzed the data. LL, JR, YD, and CS wrote and revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Key Research and Development Program of China (2018YFD0901405), Zhejiang Major Program of Science and Technology (2021C02069-7), Ningbo Major Project of Science and Technology (2019B10005 and 2019C10046), and the Project of “3315” Innovative Team of Ningbo City.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.832494/full#supplementary-material
Footnotes
- ^ http://www.ncbi.nlm.nih.gov/
- ^ http://pfam.xfam.org/
- ^ http://www.ncbi.nlm.nih.gov/COG/
- ^ http://www.ebi.ac.uk/uniprot/
- ^ http://www.genome.jp/kegg/
- ^ http://www.geneontology.org/
References
Anderson, P. M., Broderius, M. A., Fong, K. C., Tsui, K. N. T., Chew, S. F., and Ip, Y. K. (2002). Glutamine synthetase expression in liver, muscle, stomach and intestine of Bostrichthys sinensis in response to exposure to a high exogenous ammonia concentration. J. Exp. Biol. 205, 2053–2065.
Chen, S., Yu, Y., Gao, Y., Yin, P., and Tian, L. (2019). Exposure to acute ammonia stress influences survival, immune response and antioxidant status of pacific white shrimp (Litopenaeus vannamei) pretreated with diverse levels of inositol. Fish Shellfish Immunol. 89, 248–256. doi: 10.1016/j.fsi.2019.03.072
Chen, X., Mao, X., Huang, J., Yang, D., Wu, J., Dong, S., et al. (2011). KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 39, 316–322. doi: 10.1093/nar/gkr483
Cheng, C. H., Yang, F. F., Ling, R. Z., Liao, S. A., Miao, Y. T., Ye, C. X., et al. (2015). Effects of ammonia exposure on apoptosis, oxidative stress and immune response in pufferfish (Takifugu obscurus). Aquatic Toxicology. 164, 61–71. doi: 10.1016/j.aquatox.2015.04.004
Chew, S. F., and Ip, Y. K. (2014). Excretory nitrogen metabolism and defence against ammonia toxicity in air-breathing fishes. J. Fish Biol. 84, 603–638. doi: 10.1111/jfb.12279
Clifford, A. M., Goss, G. G., and Wilkie, M. P. (2015). Adaptations of a deep sea scavenger: high ammonia tolerance and active NH4? excretion by the Pacific hagfish (Eptatretus stoutii). Comp. Biochem. Physiol. A Mol. Integr. Physiol. 182, 64–74. doi: 10.1016/j.cbpa.2014.12.010
Cong, M., Wu, H., Cao, T., Lv, J., Wang, Q., Ji, C., et al. (2018). Digital gene expression analysis in the gills of Ruditapes philippinarum exposed to short- and long-term exposures of ammonia nitrogen. Aquat. Toxicol. 194, 121–131. doi: 10.1016/j.aquatox.2017.11.012
Cong, M., Wu, H., Yang, H., Zhao, J., and Lv, J. (2017). Gill damage and neurotoxicity of ammonia nitrogen on the clam Ruditapes philippinarum. Ecotoxicology 26, 1–11. doi: 10.1007/s10646-017-1777-4
Cui, Y., Ren, X., Li, J., Zhai, Q., Feng, Y., Xu, Y., et al. (2017). Effects of ammonia-N stress on metabolic and immune function via the neuroendocrine system in Litopenaeus vannamei. Fish Shellfish Immunol. 64, 270. doi: 10.1016/j.fsi.2017.03.028
Denham, B. E. (2016). Categorical Statistics for Communication Research. Hoboken: Blackwell Publishing Ltd, 255–258. doi: 10.1002/9781119407201.ch12
Dewey, C. N., and Li, B. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. doi: 10.1186/1471-2105-12-323
Dong, Y., Zeng, Q., Ren, J., Yao, H., Lv, L., He, L., et al. (2020). The chromosome-level genome assembly and comprehensive transcriptomes of the razor clam (Sinonovacula constricta). Front. Genet. 11:664. doi: 10.3389/fgene.2020.00664
Dutra, F. M., Freire, C. A., Santos, A., Forneck, S. C., Brazão, C. C., and Ballester, E. L. C. (2016). Acute toxicity of ammonia to various life stages of the Amazon river prawn, Macrobrachium amazonicum, Heller, 1862. Aquaculture 453, 104–109. doi: 10.1007/s00128-016-1932-2
Epifanio, C. E., and Srna, R. F. (1975). Toxicity of ammonia, nitriteion, nitrateion, and orthophosphate to Mercenaria mercenaria and Crassostrea virginica. Mar. Biol. 33, 241–246. doi: 10.1007/BF00390928
Gao, M., Lv, M., Liu, Y., and Song, Z. (2018). Transcriptome analysis of the effects of Cd and nanomaterial-loaded Cd on the liver in zebrafish. Ecotoxicol. Environ. Saf. 164, 530–539. doi: 10.1016/j.ecoenv.2018.08.068
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., and Amit, I. (2013). Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Harris, J. O., Maguire, G. B., Edwards, S., and Hindrum, S. M. (1998). Effect of ammonia on the growth rate and oxygen consumption of juvenile greenlip abalone, Haliotis laevigata Donovan. Aquaculture 160, 259–272. doi: 10.1016/S0044-8486(97)00249-4
Hasenbein, M., Werner, I., Deanovic, L. A., Geist, J., Fritsch, E. B., Javidmehr, A., et al. (2014). Transcriptomic profiling permits the identification of pollutant sources and effects in ambient water samples. Sci. Total Environ. 468-469, 688–698. doi: 10.1016/j.scitotenv.2013.08.081
Hu, W., Zhao, B., Li, C., Han, S., and Zhang, S. (2018). Effects of chronic ammonia nitrogen stress on the feeding and digestive enzyme activities of sea cucumber (Apostichopus japonicas Selenka). J. Fish. Sci. China 25, 137–146. doi: 10.3724/SP.J.1118.2018.17117
Ip, Y. K., Chew, S. F., and Randall, D. J. (2001). Ammonia toxicity, tolerance, and excretion. Fish Physiol. 20, 109–148. doi: 10.1016/S1546-5098(01)20005-3
Ip, Y. K., Chew, S. F., Wilson, J. M., and Randall, D. J. (2004). Defences against ammonia toxicity in tropical air-breathing fishes exposed to high concentrations of environmental ammonia: a review. J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 174, 565–575. doi: 10.1007/s00360-004-0445-1
Janssens, P. A., and Cohen, P. P. (1968). Biosynthesis of urea in the estivating African lungfish and in Xenopus laevis under conditions of water-shortage. Comp. Biochem. Physiol. 24, 887–898. doi: 10.1016/0010-406x(68)90800-1
Kir, M., Sunar, M. C., and Gök, M. (2019). Acute ammonia toxicity and the interactive effects of ammonia and salinity on the standard metabolism of European sea bass (Dicentrarchus labrax). Aquaculture 511:734273. doi: 10.1016/j.aquaculture.2019.734273
Kir, M., Topuz, M., Sunar, M. C., and Topuz, H. (2015). Acute toxicity of ammonia in Meagre (Argyrosomus regius Asso, 1801) at different temperatures. Aquac. Res. 47, 3593–3598. doi: 10.1111/are.12811
Liang, Z., Liu, R., Zhao, D., Wang, L., and Sun, M. (2016). Ammonia exposure induces oxidative stress, endoplasmic reticulum stress and apoptosis in hepatopancreas of pacific white shrimp (Litopenaeus vannamei). Fish Shellfish Immunol. 54, 523–528. doi: 10.1016/j.fsi.2016.05.009
Liao, Y., Smyth, G. K., and Shi, W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. doi: 10.1093/bioinformatics/btt656
Lim, C. B., Chew, S. F., Anderson, P. M., and Ip, Y. K. (2001). Reduction in the rates of protein and amino acid catabolism to slow down the accumulation of endogenous ammonia: a strategy potentially adopted by mudskippers (Periophthalmodon schlosseri and Boleophthalmus boddaerti) during aerial exposure in constant darkness. J. Exp. Biol. 204, 1605–1614.
Liu, F., Li, S., Yu, Y., Sun, M., and Li, F. (2020). Effects of ammonia stress on the hemocytes of the Pacific white shrimp Litopenaeus vannamei. Chemosphere 239:124759. doi: 10.1016/j.chemosphere.2019.124759
Lu, X., Kong, J., Luan, S., Dai, P., Meng, X., Cao, B., et al. (2016). Transcriptome analysis of the hepatopancreas in the Pacific white shrimp (Litopenaeus vannamei) under acute ammonia stress. PLoS One 11:0164396. doi: 10.1371/journal.pone.0164396
Lv, Y. L., Zhang, Y. P., and Shan, S. S. (2012). Acute toxic effects of ammonia-N to Tegillarca granosa at different temperatures. J. Zhejiang Ocean Univ. 31, 54–58.
Miranda-Filho, K. C., Pinho, G. L. L., Wasielesky, W., and Bianchini, A. (2009). Long-term ammonia toxicity to the pink-shrimp Farfantepenaeus paulensis. Comp. Biochem. Physiol. Toxicol. Pharmacol. 150, 377–382. doi: 10.1016/j.cbpc.2009.06.001
Miron, D., Moraes, B., Becker, A. G., Crestani, M., Spanevello, R., Loro, V. L., et al. (2008). Ammonia and pH effects on some metabolic parameters and gill histology of silver catfish, Rhamdia quelen (Heptapteridae). Aquaculture 277, 192–196. doi: 10.1016/j.aquaculture.2008.02.023
Mommsen, T., and Walsh, P. (1989). Evolution of urea synthesis in vertebrates: the piscine connection. Science 243, 72–75. doi: 10.1126/science.2563172
Montresor, L. C., Miranda-Filho, K. C., Paglia, A., Luz, D., Araújo, J., Silva, M., et al. (2013). Short-term toxicity of ammonia, sodium Hydroxide and a commercial biocide to golden mussel Limnoperna fortunei (Dunker, 1857). Ecotoxicol. Environ. Saf. 92, 150–154. doi: 10.1016/j.ecoenv.2013.03.016
Mummert, A. K., Neves, R. J., Newcomb, T. J., and Cherry, D. S. (2003). Sensitivity of juvenile freshwater mussels (Lampsilis fasciola, Villosa iris) to total and un-ionized ammonia. Environ. Toxicol. Chem. 22, 2545–2553. doi: 10.1897/02-341
Newton, T. J., and Bartsch, M. R. (2010). Lethal and sublethal effects of ammonia to juvenile Lampsilis mussels (Unionidae) in sediment and water-only exposures. Environ. Toxicol. Chem. 26, 2057–2065. doi: 10.1897/06-245R.1
Peng, K. W., Chew, S. F., Lim, C. B., Kuah, S., Kok, W. K., and Ip, Y. K. (1998). The mudskippers Periophthalmodon schlosseri and Boleophthalmus boddaerti can tolerate environmental NH3 concentrations of 446 and 36μM, respectively. Fish Physiol. Biochem. 19, 59–69. doi: 10.1023/A:1007745003948
Randall, D. J., Wood, C. M., Perry, S. F., Bergman, H. L., and Wright, P. A. (1989). Urea excretion as a strategy for survival in a fish living in a very alkaline environment. Nature 337, 165–166. doi: 10.1038/337165a0
Ren, Q., Pan, L., Zhao, Q., and Si, L. (2015). Ammonia and urea excretion in the swimming crab Portunus trituberculatus exposed to elevated ambient ammonia-N. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 187, 48–54. doi: 10.1016/j.cbpa.2015.04.013
Rodela, T. M., Esbaugh, A. J., McDonald, M. D., Gilmour, K. M., and Walsh, P. J. (2011). Evidence for transcriptional regulation of the urea transporter in the gill of the Gulf toadfish, Opsanus beta. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 160, 72–80. doi: 10.1016/j.cbpb.2011.06.004
Schram, E., Roques, J., Abbink, W., Spanings, T., de Vries, P., Bierman, S., et al. (2010). The impact of elevated water ammonia concentration on physiology, growth and feed intake of African catfish (Clarias gariepinus). Aquaculture 306, 108–115. doi: 10.1016/j.aquaculture.2010.06.005
Shang, Z. H., Huang, M., Wu, M. X., Mi, D., You, K., and Zhang, Y. L. (2021). Transcriptomic analyses of the acute aerial and ammonia stress response in the gill and liver of large-scale loach (Paramisgurnus dabryanus). Comp. Biochem. Physiol. C Toxicol. Pharmacol. 250:109185. doi: 10.1016/j.cbpc.2021.109185
Shen, C., Tang, D., Bai, Y., Luo, Y., and Wang, Z. (2021). Comparative transcriptome analysis of the gills of Procambarus clarkii provide novel insights into the response mechanism of ammonia stress tolerance. Mol. Biol. Rep. 48, 2611–2618. doi: 10.1007/s11033-021-06315-y
Shen, J. D., Cai, Q. F., Yan, L. J., Du, C. H., Liu, G. M., Su, W. J., et al. (2015). Cathepsin L is an immune-related protein in Pacific abalone (Haliotis discus hannai)–Purification and characterization. Fish Shellfish Immunol. 47, 986–995. doi: 10.1016/j.fsi.2015.11.004
Sinha, A. K., Zinta, G., AbdElgawad, H., Asard, H., Blust, R., and De Boeck, G. (2015). High environmental ammonia elicits differential oxidative stress and antioxidant responses in five different organs of a model estuarine teleost (Dicentrarchus labrax). Comp. Biochem. Physiol. C Toxicol. Pharmacol. 174-175, 21–31. doi: 10.1016/j.cbpc.2015.06.002
Staals, R. H. J., Bronkhorst, A. W., Schilders, G., Slomovic, S., and Pruijn, G. J. M. (2010). Dis3-like 1: a novel exoribonuclease associated with the human exosome. EMBO J. 29, 2358–2367. doi: 10.1038/emboj.2010.122
Stark, R., Grzelak, M., and Hadfield, J. (2019). RNA sequencing: the teenage years. Nat. Rev. Genet. 20, 631–656. doi: 10.1038/s41576-019-0150-2
Sun, G., Sun, C., He, J., Yao, H., Dai, W., Lin, Z., et al. (2021). Characterizing the role of glutamine synthetase gene on ammonia nitrogen detoxification metabolism of the razor clam Sinonovacula constricta. Front. Mar. Sci. 8:793118. doi: 10.3389/fmars.2021.793118
Thurston, R. V., Russo, R. C., and Vinogradov, G. A. (1981). Ammonia toxicity to fishes. Effect of pH on the toxicity of the unionized ammonia species. Environ. Sci. Technol. 15, 837–840. doi: 10.1021/es00089a012
Trapnell, C., Pachter, L., and Salzberg, S. L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. doi: 10.1093/bioinformatics/btp120
Tresguerres, M., Clifford, A. M., Harter, T. S., Roa, J. N., and Brauner, C. J. (2020). Evolutionary links between intra- and extracellular acid-base regulation in fish and other aquatic animals. J. Exp. Zool. A Ecol. Integr. Physiol. 333, 1–17. doi: 10.1002/jez.2367
USEPA (2013). Final Aquatic Life Ambient Water Quality Criteria for Ammonia-Freshwater. EPA-822-R-13-001. Washington: United States Environmental Protection Agency.
Wang, W. N., Zhou, J., Wang, P., Tian, T. T., Zheng, Y., Liu, Y., et al. (2016). Oxidative stress, DNA damage and antioxidant enzyme gene expression in the Pacific white shrimp, Litopenaeus vannamei when exposed to acute pH stress. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 150, 428–435. doi: 10.1016/j.cbpc.2009.06.010
Wang, W. Q., Jiang, L. X., Yang, N., Jian, L. I., and Wang, R. J. (2007). The effect of ammonia-N on immune activity of Ruditapes philippinarum. Mar. Sci. 31, 23–27.
Weihrauch, D., Morris, S., and Towle, D. W. (2004). Ammonia excretion in aquatic and terrestrial crabs. J. Exp. Biol. 207, 4491–4504. doi: 10.1242/jeb.01308
Wood, C. M., Nawata, C. M., Wilson, J. M., Laurent, P., Chevalier, C., Bergman, H. L., et al. (2013). Rh proteins and NH4(+)-activated Na+-ATPase in the Magadi tilapia (Alcolapia grahami), a 100% ureotelic teleost fish. J. Exp. Biol. 216, 2998–3007. doi: 10.1242/jeb.078634
Wyatt, M. (2009). Differential effects of reactive nitrogen species on DNA base excision repair initiated by the alkyladenine DNA glycosylase. Carcinogenesis 30, 2123–2129. doi: 10.1093/carcin/bgp256
Xue, S., Lin, J., Zhou, Q., Wang, H., and Han, Y. (2021). Effect of ammonia stress on transcriptome and endoplasmic reticulum stress pathway for common carp (Cyprinus carpio) hepatopancreas. Aquac. Rep. 20:100694. doi: 10.1016/j.aqrep.2021.100694
Yang, W., Xiang, F., Sun, H., Chen, Y., Minter, E., and Yang, Z. (2010). Changes in the selected hematological parameters and gill Na(+)/K(+) ATPase activity of juvenile crucian carp Carassius auratus during elevated ammonia exposure and the post-exposure recovery. Biochem. Syst. Ecol. 38, 557–562. doi: 10.1016/j.bse.2010.06.005
You, X., Chen, J., Bian, C., Yi, Y., Ruan, Z., Li, J., et al. (2018). Transcriptomic evidence of adaptive tolerance to high environmental ammonia in mudskippers. Genomics 110, 404–413. doi: 10.1016/j.ygeno.2018.09.001
Young, M. D., Wakefield, M. J., Smyth, G. K., and Oshlack, A. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11:R14. doi: 10.1186/gb-2010-11-2-r14
Yu, J., Sun, J., Zhao, S., Wang, H., and Zeng, Q. (2019). Transcriptome analysis of oriental river prawn (Macrobrachium nipponense) hepatopancreas in response to ammonia exposure. Fish Shellfish Immunol. 93, 223–231. doi: 10.1016/j.fsi.2019.07.036
Zhang, H., Sun, G., Lin, Z., Yao, H., and Dong, Y. (2020). The razor clam Sinonovacula constricta uses the strategy of conversion of toxic ammonia to glutamine in response to high environmental ammonia exposure. Mol. Biol. Rep. 47, 1–15. doi: 10.1007/s11033-020-06018-w
Zhang, W. Y., Storey, K., and Dong, Y.-W. (2020). Adaptations to the mudflat: insights from physiological and transcriptional responses to thermal stress in a burrowing bivalve Sinonovacula constricta. Sci. Total Environ. 710:136280. doi: 10.1016/j.scitotenv.2019.136280
Zhang, L., Zhao, Z., and Fan, Q. (2016). Effects of ammonia on growth, digestion and antioxidant capacity in juvenile yellow catfish Pelteobagrus fulvidraco (Richardson, 1846). J. Appl. Ichthyol. 32, 1205–1212. doi: 10.1111/jai.13203
Zhang, T., Yan, Z., Zheng, X., Fan, J., and Guo, S. (2019). Transcriptome analysis of response mechanism to ammonia stress in Asian clam (Corbicula fluminea). Aquat. Toxicol. 214:105235. doi: 10.1016/j.aquatox.2019.105235
Zhang, Y. P., Xiao, G. Q., Lin, L. Z., Zhang, J. M., and Chai, X. L. (2009). Effects of pH and ammonia-N on tolerance of Estellarca olivacea. Sichuan J. Zool. 28, 73–76. doi: 10.1360/972009-1142
Zhao, Q., Feng, K., Zhang, L., Bai, Y., and Yao, W. (2021). Effects of acute ammonia stress on antioxidant responses, histopathology and ammonia detoxification metabolism in triangle sail mussels (Hyriopsis cumingii). Water 13, 1–16. doi: 10.3390/w13040425
Keywords: ammonia tolerance, transcriptomic analysis, NH3/NH4+ transporter, ammonia excretion and metabolism, gene expression, Sinonovacula constricta
Citation: Lv L, Ren J, Zhang H, Sun C, Dong Y and Lin Z (2022) Transcriptomic Analysis of Gill and Hepatopancreas in Razor Clam (Sinonovacula constricta) Exposed to Acute Ammonia. Front. Mar. Sci. 9:832494. doi: 10.3389/fmars.2022.832494
Received: 09 December 2021; Accepted: 07 January 2022;
Published: 10 February 2022.
Edited by:
Liqiang Zhao, Guangdong Ocean University, ChinaReviewed by:
Jun Li, Key Laboratory of Marginal Sea Geology, South China Sea Institute of Oceanology, Chinese Academy of Sciences (CAS), ChinaXing Zheng, Hainan University, China
Copyright © 2022 Lv, Ren, Zhang, Sun, Dong and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianfeng Ren, amZyZW5Ac2hvdS5lZHUuY24=; Zhihua Lin, emhpaHVhOTk4OEAxMjYuY29t
 Liyuan Lv
Liyuan Lv Jianfeng Ren
Jianfeng Ren Huan Zhang3
Huan Zhang3 Changsen Sun
Changsen Sun Yinghui Dong
Yinghui Dong