- 1Department of Aquaculture, National Taiwan Ocean University, Keelung, Taiwan
- 2Doctoral Degree Program in Marine Biotechnology, National Taiwan Ocean University, Keelung, Taiwan
- 3Doctoral Degree Program in Marine Biotechnology, Academia Sinica, Taipei, Taiwan
- 4Marine Research Station, Institute of Cellular and Organismic Biology, Academia Sinica, Taipei, Taiwan
- 5Center of Excellence for the Oceans, National Taiwan Ocean University, Keelung, Taiwan
Some cephalopods carry microorganisms in two specialized organs, the light organ and the accessory nidamental gland (ANG). For the light organ, comprehensive mechanisms have been described for winnowing (bacterial selection) and maintenance of the symbiotic luminescent bacterium Vibrio fischeri (V. fischeri). However, the mechanisms controlling bacterial selection and maintenance during bacterial colonization of the ANG are open biological issues with physiological significance. Our recent study on bigfin reef squid (Sepioteuthis lessoniana) already showed that the ANG bacterial community shifts gradually and exhibits decreased diversity throughout maturation. This study further describes a potential role of an innate immunity-involved molecule, peptidoglycan recognition proteins (PGRPs), in the ANG of bigfin reef squid during bacterial transmission and colonization. First, we found that four homologs of the PGRP family are expressed in the ANG of bigfin reef squid (slPGRP2-5), but only slPgrp2 transcript levels are highly correlated with ANG development and bacterial colonization. Besides, slPgrp2 transcripts are mainly expressed in the epithelial cells of certain secondary tubules of ANG, and the expression levels are varied in the epithelial cells of other secondary tubules. This data reveals that slPgrp2 transcripts may associate with the composition of bacterial consortium and its secretary factors. Moreover, recombinant slPGRP2 had a negative effect of Escherichia coli (E.coli) which inhibited bacterial growth in culture. Therefore, our data suggest slPgrp2 expression in the epithelial cells of secondary tubules in the ANG may have an essential role in the winnowing and maintenance of holobiont homeostasis in bigfin reef squid.
Introduction
Animals often harbor large numbers of beneficial microorganisms on tissue surfaces, in digestive organs or in specialized organs. Some cephalopods have two specialized organs that house microbiota, including the light organ and accessory nidamental gland (ANG) (reviewed in Belcaid et al., 2019). Unlike the light organ, which is found in both sexes of bobtail squid (Sepiolidae), the ANG is found only in the females of most squid and cuttlefish (Decapodiformes) species; it is absent in some squid (Oegopsidae), all octopuses (Octopodiformes) and all nautiluses (Nautiloidea) (Lindgren et al., 2012). The ANG is thought to play an important role in the delivery of bacteria from the parent to the egg capsule by preventing fungal fouling (Kerwin et al., 2019). In contrast with the single bacterial population (Vibrio fischeri) hosted in the light organ of the bobtail squid (Ruby and Asato, 1993; Nyholm et al., 2002), a complex microbial consortium is hosted in the ANG of most squid and cuttlefish (Grigioni et al., 2000; Pichon et al., 2005; Collins et al., 2012; Kerwin and Nyholm, 2017; Yang et al., 2021). This bacterial consortium is populated by horizontal transmission from the environmental bacterial community (Kaufman et al., 1998; Li et al., 2019). Furthermore, the ANG bacterial community shifts gradually and decreases in diversity during maturation of the bigfin reef squid (Yang et al., 2021) and Hawaiian bobtail squid (Kerwin et al., 2021). Therefore, the establishment and maintenance of the complex consortium in the ANG appear to comprise an intricate process governed by host selection during the initial bacterial transmission and later bacterial colonization. However, a comprehensive understanding of bacterial selection mechanisms during colonization of the ANG is still lacking.
The animal immune system not only defends against microbial infection but also accommodates colonization by symbiotic microorganisms to maintain microbiota-host homeostasis (Cerf-Bensussan and Gaboriau-Routhiau, 2010; Royet et al., 2011). In invertebrates, the innate immune system includes pattern recognition receptors that recognize motifs uniquely present in microorganisms and absent in the host. One family of such pattern recognition receptors is the peptidoglycan recognition proteins (PGRPs), which recognize structural components of bacterial cell walls such as peptidoglycan (most bacteria), lipoteichonic acid (Gram-positive bacteria), and lipopolysaccharide (Gram-negative bacteria) (Dziarski, 2004). PGRPs are conserved in vertebrates and many invertebrates, but no member of the PGRP family has been found in nematodes (Dziarski and Gupta, 2006; Montaño et al., 2011) or crustaceans (Liu et al., 2011; Bai et al., 2020). Unlike mammals (Pglyrp1-4) and fish (pglyrp2, 5, and 6), which have a few conserved PGRP members with a narrow range of functions, insects usually have many Pgrp genes and mainly involved in bacterial recognition and control, such as the 13 Pgrp genes in fruit fly (Dorsophila melanogaster) and 14 Pgrp genes in silkworms (Bombyx mori) (Dziarski and Gupta, 2006; Montaño et al., 2011). The diverse function of PGRPs is advantageous in invertebrates, due to the absence of adaptive immunity, in contrast to the moderate repertoire in vertebrates (Montaño et al., 2011). Vertebrates have evolved a large number of other pattern recognition receptors, such as C-type lectin receptors, nucleotide-binding oligomerization domain (NOD)-like receptors, RIG (retinoic acid-inducible gene)-I-like receptors, Toll-like receptors, scavenger receptors, and the mannose receptors (Royet et al., 2011). Moreover, vertebrates possess many secreted recognition molecules, such as complement, collectins, pentraxins and C-reactive protein (Royet et al., 2011).
Based on the structural characteristics, most PGRPs contain a C-terminal PGRP domain, which is homologous to bacteriophage type 2 amidase (Dziarski, 2004; Royet et al., 2011). Furthermore, PGRPs have been shown into three forms: secreted proteins (with a signal peptide), transmembrane receptors, and intracellular proteins (Royet et al., 2011). The diverse functions of PGRPs within a host’s immune system can include both direct bactericidal effects and indirect effects, such as the induction of antimicrobial compound production and the modulation of inflammation and immune responses (Royet et al., 2011). Furthermore, PGRPs are known to be involved in the maintenance of beneficial and normal gut microbiota in mammals (Saha et al., 2010) and insects (Bischoff et al., 2006; Zaidman-Rémy et al., 2006). In the vent mussel (Bathymodiolus septemdierum), PGRPs have been suggested to regulate the homeostasis of gill symbionts (Ikuta et al., 2019). Moreover, PGRPs are thought to participate in the winnowing and maintenance of luminescent bacteria (Vibrio fischeri) in the light organ of the Hawaiian bobtail squid (Euprymna scolopes) (Nyholm and Graf, 2012; McAnulty and Nyholm, 2017). Therefore, PGRPs could serve essential biological roles in the control of commensal bacteria and in defense against pathogens.
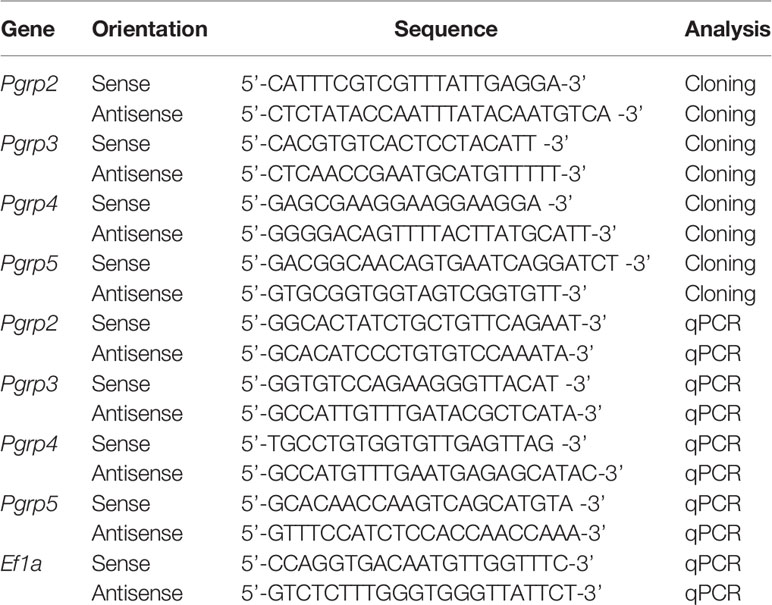
The bigfin reef squid (also called oval squid, Sepioteuthis lessoniana) is closely associated with coral reef habitats, and lays jelly-like egg strings in strip-like structures such as seagrasses, sea fans, coral rubble, and reefs (Norman, 2003). The development of the ANGs has been studied in female bigfin reef squid (Li et al., 2019) and other coleoid cephapods (Lum-Kong, 1992; Kaufman et al., 1998; Kerwin et al., 2021). During the ANG development, the ANG gradually transfers bacteria from the outer epithelial cell layer, invaginating bacteria from the marine environments into the ANG and forming tubule structures (contains primary tubules in the outer layer and secondary tubules in the inner layer of the ANG) for bacterial flora (Lum-Kong, 1992; Kaufman et al., 1998; Li et al., 2019; Kerwin et al., 2021). During the female maturation, four developmental stages of ANGs were categorized on the basis of morphological and histological characteristics in bigfin reef squid (Figure 1): juvenile squid with a colorless ANG without visible bacterial flora (stage 1), immature squid with a colorless ANG with bacterial flora (stage 2), maturing squid with a white/light-orange ANG with large numbers of bacterial flora (stage 3), and mature squid with a pigmented ANG with large numbers of bacterial flora (stage 4) (Li et al., 2019). Furthermore, our previously study suggest that robust transferrin expression in the outer layer of the ANG plays an important role in microbe selection by the squid during bacterial transmission (Li et al., 2019). However, the regulatory mechanism of squid’s immune system in the ANG to maintain holobiont homeostasis during the female maturation in bigfin reef squid remains unknown.
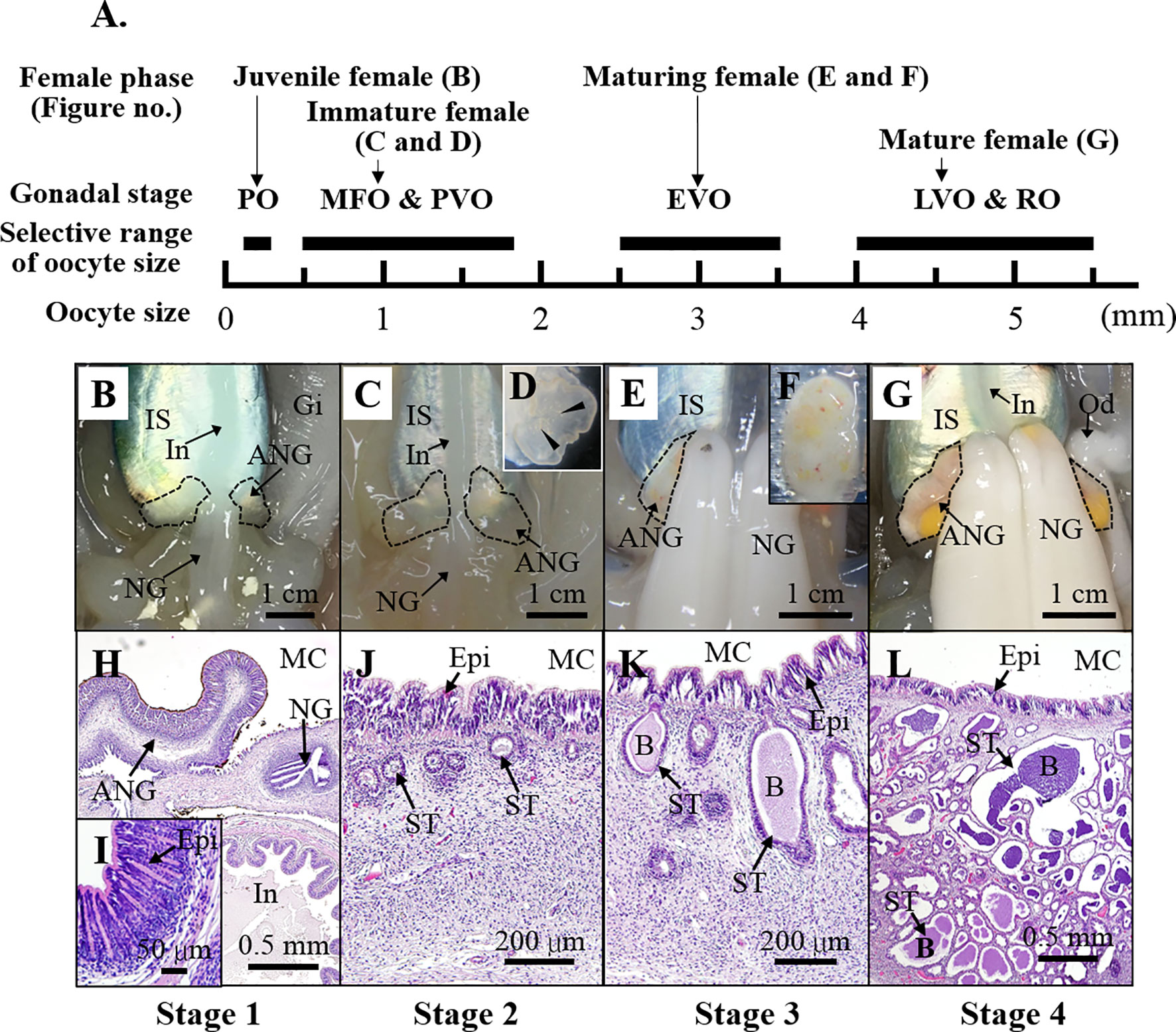
Figure 1 The characteristics of ANG during female squid maturation. (A) The profiles of female growth, including the juvenile female (primary oocyte stage), immature female (multiple follicular oocyte stage and previtellogenic oocyte stage), maturing female (early vitellogenic oocyte stage), and mature female (late vitellogenic oocyte stage and ripe oocytes). (B–G) Morphological changes during ANG growth. (B) juvenile squid with a colorless ANG (stage 1); (C, D) immature squid with a colorless ANG several white spots (stage 2); (E, F) maturing squid with a white/light-orange ANG (stage 3); (G) mature squid with a pigmented ANG (stage 4). (H–L) Histological changes during ANG growth. (H, I) juvenile squid with a colorless ANG without visible bacterial flora (stage 1); (J) immature squid with a colorless ANG with bacterial flora (stage 2); (K) maturing squid with a white/light-orange ANG with large numbers of bacterial flora (stage 3); (L) mature squid with a pigmented ANG with large numbers of bacterial flora (stage 4). PO, primary oocyte; MFO, multiple follicular oocyte; PVO, previtellogenic oocyte; EVO, early vitellogenic oocyte; LVO, late vitellogenic oocyte stage; RO, ripe oocyte; IS, ink sac; In, intestine; Gi, gill; NG, nidamental gland; ANG, accessory nidamental gland; Od, oviduct; MC, mantle cavity; Epi, epithelial cell; ST, secondary tubule; B, bacteria.
In Hawaiian bobtail squid, five homologs of the PGRP family (Pgrp1-5) were reported to be expressed in the light organ (Goodson et al., 2005) and hemocytes (Collins et al., 2012). PGRP family genes have also been identified in other cephalopods as well (reviewed in Castillo et al., 2015). Therefore, this study especially aims to determine possible physiological significances of bigfin reef squid’s PGRPs in the ANG. We further characterize the potential involvement of bacterial transmission and colonization in the ANG to maintain holobiont homeostasis during female maturation in bigfin reef squid.
Materials and Methods
Squid Collection
Bigfin reef squids were purchased from a fisherman on Heping Island, Keelung city, Taiwan. The squids were collected by hand jigging from a boat off the northeast coast of Taiwan. The squids were maintained in a seawater system on the boat, then transferred to a seawater tank until tissue sampling (less than 2 hr). The squids were anesthetized in seawater containing 5% ethanol at room temperature and then decapitated after tissue sampling. All procedures and investigations were approved by the National Taiwan Ocean University Institutional Animal Care and Use Committee and were performed in accordance with standard guidelines.
Tissue Histology and Gram Staining
The reproductive phase of female was determined based on ovarian stage by oocyte size and morphological characteristic (mature eggs in oviduct), as described previously (Chen et al., 2018). The oocyte size was examined by hematoxylin and eosin (H&E) staining and stereomicroscope for small ovary (oocyte size smaller than 1.5 mm) and others (oocyte size lager than 1.5 mm), respectively. The oocyte stage was determined based on the oocyte size, in accordance with our previous study (Chen et al., 2018). The oocyte stage and relative oocyte diameter as follow: primary oocyte stage (35-300 µm), multiple follicular oocyte stage (0.3-1.2 mm), previtellogenic oocyte stage (1.2-1.8 mm), early vitellogenic oocyte stage (1.8-3 mm), late vitellogenic oocyte stage (3-5 mm), and ripe oocyte stage (5-5.5 mm). The developmental status of most ANG (36 of 38 ANGs) was examined by H&E staining, as described previously (Chen et al., 2018). The ANGs were fixed with 4% paraformaldehyde in PBS at 4°C for 16 hr, then dehydrated in methanol and stored at -20°C. Dehydrated ANGs were transferred to xylene and then embedded in paraffin. Sections (5 μm in thickness) were rehydrated with PBS, treated with HistoVT One (Nacalai Tesque), and stained with hematoxylin and eosin. The ANG stage was determined based on morphological characteristics and confirmed by histology, in accordance with our previous study (Li et al., 2019). Four developmental stages of ANGs were used for RNA analysis in this study (Figure 1): stage 1 (n = 4), stage 2 (n = 14), stage 3 (n = 12), and stage 4 (n = 9). The characteristics of sampled squids (n = 39) was shown in Supplemental Table 1 (Table S1).
The bacterial distribution was examined with ASK® Rapid Gram Stain Kit (Tonyar Biotech. Inc., Taoyuan, Taiwan). Gram staining was performed according to the manufacturer’s protocol. Rehydrated sections (5 μm in thickness) were stained with crystal violet and Lugol’s solution, then stained with safranin and counterstained with tartrazine solution (HT3028, Sigma). The bacterial species were divided into two large groups according to color, violet Gram-positive bacteria and pink Gram-negative bacteria.
Total RNA Extraction and cDNA Synthesis
Tissues were homogenized in Trizol reagent (Invitrogen). Extraction of nucleic acids and protein was performed according to the manufacturer’s protocol. The nucleoprotein complexes were mixed with chloroform to respectively separate the DNA, RNA, and protein into a lower phenol-chloroform phase, an upper aqueous phase, and an interphase. Isopropanol was used to precipitate the total RNA from the aqueous phase. The quality and quantity of isolated total RNA were determined by gel electrophoresis and NanoDrop™ 1000 spectrophotometer (Thermo Fisher Scientific), respectively. First-strand cDNA was synthesized from 1 μg total RNA with oligo(dT)12-18 primers (Promega) using Superscript III reverse transcriptase (Invitrogen). The resultant cDNA was used for gene cloning and RNA analysis.
Cloning of the Bigfin Reef Squid Pgrp Genes
The ANG (stage 2) from an immature female squid was used to isolate total RNA. The cDNA synthesis, cDNA library construction, and illumine sequencing were done by Welgene, Inc. Paired-end sequencing (150bp) was performed on a HiSeq 2000 sequencer (Illumina). A de novo transcriptome assembly was performed using CLC Genome Workbench 8.0 and used in our previous study (Li et al., 2019). This transcriptomic database was used to identify a fragment of the target gene in this study. A local blast on CLC Genome workbench was used to detect homologs of Pgrp in bigfin reef squid. The sequences of Pgrp genes were confirmed by cloning. The cloning primers were listed in Table 1. Full-length (containing open reading frame) cDNA sequences of slPgrp2 (GenBank accession no. MZ781965), slPgrp3 (GenBank accession no. MZ781966), slPgrp4 (GenBank accession no. MZ781967), and slPgrp5 (GenBank accession no. MZ781968) were obtained in this study.
Sequence Alignment and Phylogenetic Analysis
For the phylogenetic analysis, a subset of PGRP sequences from various taxa was retrieved from GenBank and compared with the PGRPs identified from the bigfin reef squid. The consensus PGRP domain of PGRPs was determined by SMART sequence analysis (http://smart.embl-heidelberg.de/) and used for analysis. Alignments were performed using MUSCLE in MEGA11. The phylogenetic tree was constructed using the neighbor-joining method in MEGA11. The number at each node represents the bootstrap probability (% from 1000 replicates). The accession numbers of the sequences used for analysis are listed in Table S2.
RNA Analysis
Quantitative real-time PCR (qPCR) analysis was modified from our previous study (Chen et al., 2018). Elongation factor 1 alpha (Ef1a; GenBank accession no. MG924746) was used as an internal control to normalize the gene expression levels. Specific qPCR primers for target genes and amplicon sizes are listed in Table 1. Gene expression quantification for standards, samples, and controls was conducted simultaneously by qPCR (CFX Connect™ Real-Time PCR Detection System; Bio-Rad Laboratories) with the SYBR Green Master Mix (Bio-Rad Laboratories). The thermal cycling conditions were as follows: one cycle of 95°C for 10 min, then 40 cycles of 95°C for 15 sec, and 60°C for 1 min. The PCR specificity was confirmed by a single melting curve in unknown samples and template-contained controls. No signal was detected by qPCR in non-template controls. The reaction efficiency of different genes was evaluated by serially diluted standards. The data were analyzed according to the 2-ΔΔCt method. The relative expression values of target genes in all samples were normalized to Ef1a, and the highest value of the target genes was defined as 100%.
In situ Hybridization
Digoxigenin-11-UTP (DIG)-labeled antisense and sense probes were synthesized using cDNA fragments of Pgrp2 (nucleotides -28-654, 682bp). The DIG-labeled antisense RNA probe was used to detect the localization of Pgrp2. A DIG-labeled sense RNA probe was used as a control for non-specific signals. Whole mount in situ hybridization (ISH) was performed as described in our previous study (Guo et al., 2021). Fixed ANGs were dissected into small pieces by vibratome sectioning (0.2 mm in thickness). Then, the sections were rehydrated, and processed for ISH. The ANG samples were incubated with DIG-labeled RNA probes at 60°C overnight. mRNA expression was detected with ANG-pre-adsorbed alkaline phosphatase-conjugated sheep anti-DIG antibody (11093274910, Roche) and visualized using the NBT/BCIP Detection System. The samples were then fixed with 4% paraformaldehyde and dehydrated in ethanol. The dehydrated samples were transferred from ethanol to infiltration buffer, and then embedded in plastic (Technovit 3040). The sections (5 μm in thickness) were rehydrated in water and counterstained with Fast red to label the cell nucleus.
Turbidity Assay
The ORF of slPgrp2 was cloned into the isopropyl-β-D-thiogalactopyranoside (IPTG)-inducible expression plasmid, pET-28a(+) DNA (69684-3CN, Novagen). The plasmid was transformed into ECOS™21 BL21 (DE3) competent cells (Yestern Biotechnology). The E. coli were grown at 37°C, and the recombinant slPGRP2 (rslPGRP2) was induced by the addition of 1 mM of IPTG, followed by culture at 37°C for 5 hr in LB medium. The growth rate of E. coli was evaluated by spectrophotometer (absorbance at 600 nm). E.coli carrying an IPTG-inducible orange-spotted grouper (Epinephelus coioides) dmrt1 (ecdmrt1) expression cassette were used as reference control.
Data Analysis
Data were shown as mean ± standard deviation (SD). The Shapiro-Wilk test was used to test the normal distribution (variance > 0.5 indicates normality). The Games-Howell test was used to check the homogeneity of different groups. Student’s t-test was used to check the significance of difference between two groups. With One-way ANOVA followed by a Games-Howell test (homogeneity of variance < 0.05) was used to check the significance of differences among three or more groups. Statistical Package for the Social Sciences (SPSS) software was used. In all cases, P < 0.05 indicated significant difference.
Results
Molecular Identification and Phylogenetic Analysis of Bigfin Reef Squid slPGRPs
Both vertebrates and invertebrates, PGRPs have a highly conserved C-terminal region of the PGRP domain. Therefore, the deduced amino sequences of the PGRP domain sequence from Pgrp genes (Pgrp1-5) in bobtail squid were used as a reference to search for Pgrp genes in bigfin reef squid. Four PGRP domain-containing transcripts were found in a draft transcriptome database for the ANG of bigfin reef squid. The sequences of these four genes were determined by cDNA cloning. The deduced amino acid sequences of the genes showed conservation of the PGRP domain (Amidase 2). A phylogenetic analysis of the PGRP domains revealed that bigfin reef squid PGRP domain-contained genes cluster with PGRPs in cephalopods (Figure 2A). One deduced amino sequence of gene (named slPgrp2) was located in the same group as PGRP1 and PGRP2 of Hawaiian bobtail squid, but it was not grouped specifically with either branch (Figure 2A). The predicted protein structure of this gene includes a signal peptide that is expected to cause its secretion (Figure 2B). This characteristic (secretion) was observed for PGRP2 but not PGRP1 of the Hawaiian bobtail squid (Goodson et al., 2005). Three deduced amino sequence of genes (named slPgrp3, slPgrp4, and slPgrp5) belong to the same branch as PGRP3, PGRP4, and PGRP5 in Hawaiian bobtail squid (Figure 2A). Similar to the genes in Hawaiian bobtail squid (Collins et al., 2012), the predicted protein structures of these genes included a glycophosphatidylinositol (GPI)-anchoring site in slPGRP3, a transmembrane receptor domain in slPGRP4, and a secretion signal in slPGRP5 (Figure 2B).
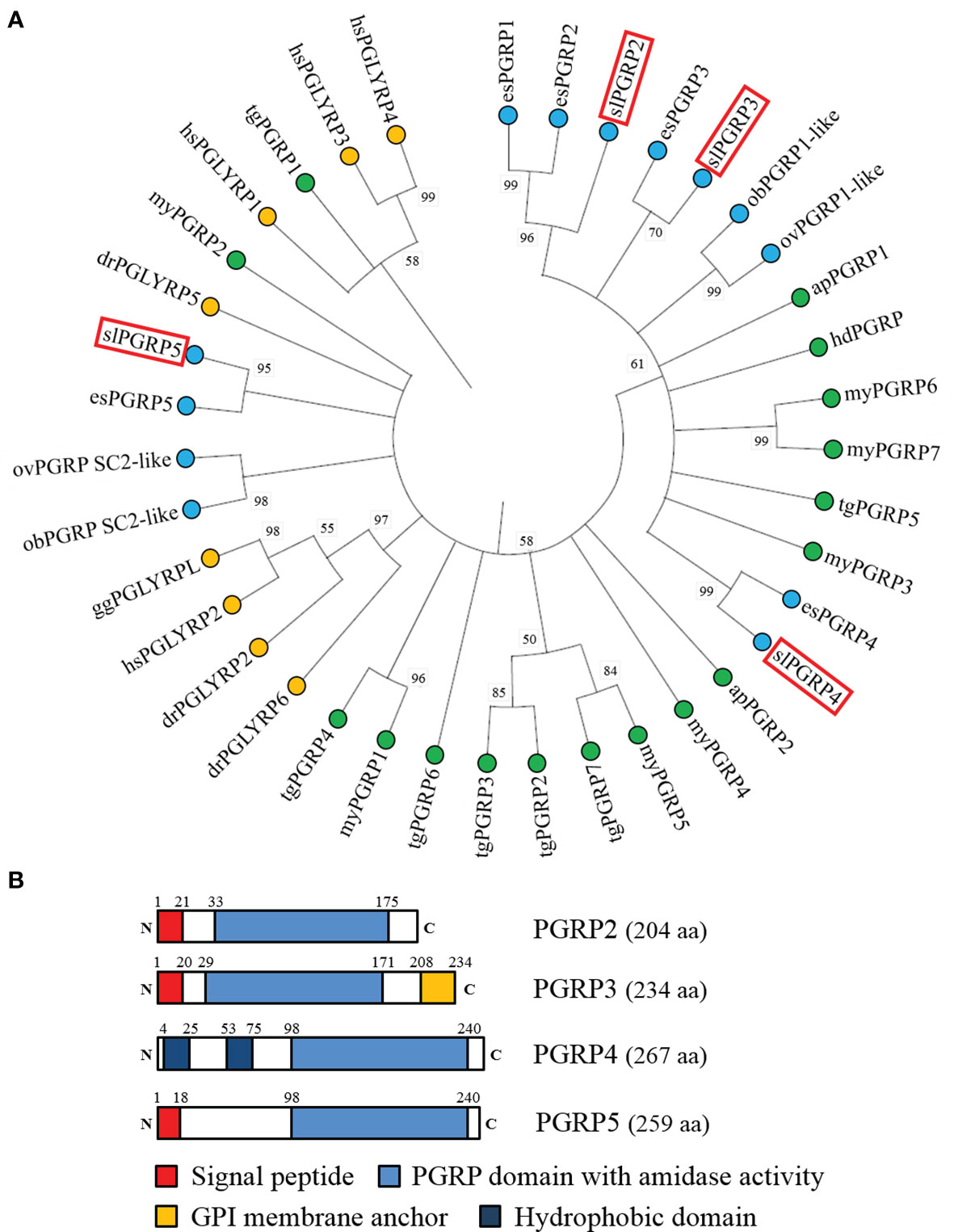
Figure 2 Phylogenetic analysis of PGRP family proteins from various taxa and domain structure of PGRPs in the bigfin reef squid. (A) The phylogenetic analysis of PGRPs in various species. The PGRP domain (with amidase) was used for analysis. The number at each node represents the bootstrap probability (% of 1000 replicates). Branches correspond to values of 50% and higher. The PGRPs of bigfin reef squid (slPGRP2, slPGRP3, slPGRP4, and slPGRP5) are indicated by red boxes. The PGRPs from vertebrates, bivalves and gastropoda, and cephalopods are highlighted in yellow, green and blue circles, respectively. The gene names and accession numbers are listed in Table S2. (B) The predicted structure of slPGRPs in the bigfin reef squid. Red denotes the signal peptide. Blue denotes PGRP domain with amidase activity. Yellow denotes glycosylphosphatidylinositol (GPI) membrane anchor. Indigo denotes hydrophobic domain (predicted transmembrane domain).
Expression Profiles of slPgrp Genes During Female Growth
The slPgrp family (slPgrp2, slPgrp3, slPrgp4, and slPgrp5) transcript levels were then analyzed in various tissues of mature females. According to the qPCR results, slPgrp2 was predominantly expressed in the hepatopancreas, but it was also detected in the ANG and ovary (Figure S1). Only slight or no expression of slPgrp2 was found in other tissues (Figure S1). Compared to the gene expression level of slPgrp2 (highest transcription level in hepatopancreas, 3.4E-02), slPgrp4 (highest transcription level in hemolymph-isolated hemocytes, 0.127), and slPgrp5 (highest transcription level in hemocytes, 4.11) in various tissues, slPgrp3 showed weak or no expression in various tissues (highest transcription level in gills, 1.19E-06) (Figure S1). slPgrp4 and slPgrp5 were predominantly expressed in hemocytes and exhibited slight expression in other tissues (Figure S1). Based on the weak levels of gene expression in ANG, we did not analyze the slPgrp3 expression.
We investigated whether the expression of slPgrp family (slPgrp2, slPgrp4, and slPgrp5) is correlated to the growth of female bigfin reef squid. Based on histological characteristics of the ovary, gene transcript levels were analyzed at the primary oocyte stage of immature females (stage 2 of ANG) and vitellogenic oocyte stage of mature females (stage 4 of ANG). Results from qPCR showed that ovarian slPgrp2 expression levels were high in the immature females, and the expression level was significantly decreased in the mature females (Figure 3A). The expression level of slPgrp2 was low in the ANG of immature females, but the expression level was significantly increased in the mature females (Figure 3A). No differences in slPgrp2 expression were found between age groups in the hepatopancreas, gills, mantle, and NG (Figure 3A). qPCR results also showed that slPgrp4 expression level in gills, NG, and ANG were high in immature females and that the expression levels were significantly decreased in the mature females (Figure 3B). No differences in slPgrp4 expression were found between age groups in the hepatopancreas, mantle, and ovary (Figure 3B). Furthermore, slPgrp5 expression level in hepatopancreas was low in the immature females and significantly increased in the mature females (Figure 3C). The slPgrp5 expression level in ANG was high in the immature females and significantly decreased in mature females (Figure 3C). No differences in slPgrp5 expression were identified between age groups in the gills, mantle, ovary, and NG (Figure 3C). Thus, among slPgrp genes expressed in female-specific tissues (ovary, NG, and ANG), only the expression of slPgrp2 in the ANG was positively correlated with growth of female bigfin reef squid.
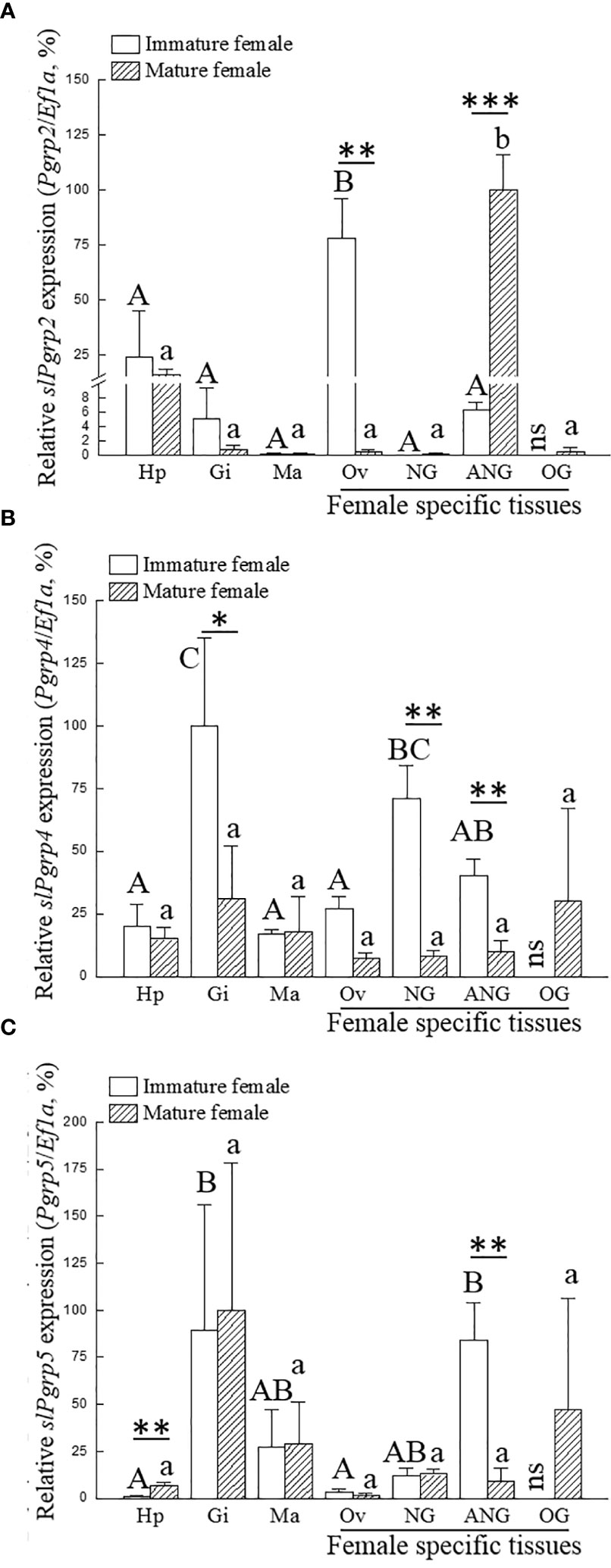
Figure 3 Expression of slPgrp transcripts in various tissues. The gonadal stage of female squid was distinguished by histology. Immature (n = 3; primary oocyte stage) and mature (n = 3; vitellogenic oocyte stage) females were used to analyze gene expression in different tissues. Three slPgrp genes were selected for analysis, including slPgrp2 (A), slPgrp4 (B), and slPgrp5 (C). Different capital letters indicate significant differences between tissues in immature female (P < 0.05). Different lower-case letters indicate significant differences between tissues in mature female (P < 0.05). Asterisks indicate significant difference between the immature and mature females (*P < 0.05, **P < 0.01, and ***P < 0.001, respectively). Hp, hepatopancreas, Gi, gill; Ma, mantle (muscle); Ov, ovary; NG, nidamental gland; ANG, accessory nidamental gland; OG, oviductal gland; ns, no sample.
Expression Profiles of slPgrp2 During ANG Growth
We next sought to clarify the correlation between slPgrp (slPgrp2, slPgrp4, and slPgrp5) transcript levels and distributions in the ANG. The relative portion of outer and inner layer of ANG was identified as Figure 4A. The outer layer of ANG was composed by outer epithelial cell layer and intermediate filament layer (Figures 4B, C). The inner layer of ANG was composed by inner epithelial layer (Figure 4D). From the mature female squid, the relative portion of outer and inner layers of ANGs (stage 4) were isolated under a stereomicroscope and analyzed separately, as in our previous study (Li et al., 2019). Histological result showed that the outer layer of the ANG was completely removed from the ANG in outer layer-isolated ANG in our previous study (Li et al., 2019). Thus, the relative portion of outer layer of the ANG contained outer epithelial cell layer with a small portion of inner layer of the ANG for RNA analysis. qPCR results showed that the outer layer had lower slPgrp2 expression than the inner layer of ANG in mature females (Figure 4E). In contrast, the outer layer had higher slPgrp4 and slPgrp5 expression than the inner layer (Figure 4E). Next, gene transcript levels were analyzed at different ANG stages. The histological features and bacterial distributions of different ANG stages in female squids are shown in Figure 1. According to qPCR, slPGRP2 expression levels were low at stage 1, stage 2, and stage 3 of ANG development, but the expression levels were increased significantly at stage 4 of ANG development (Figure 5). Thus, the data showed that slPgrp2 expression in the inner layer of ANG is correlated with bacterial colonization and pigmentation during ANG growth.
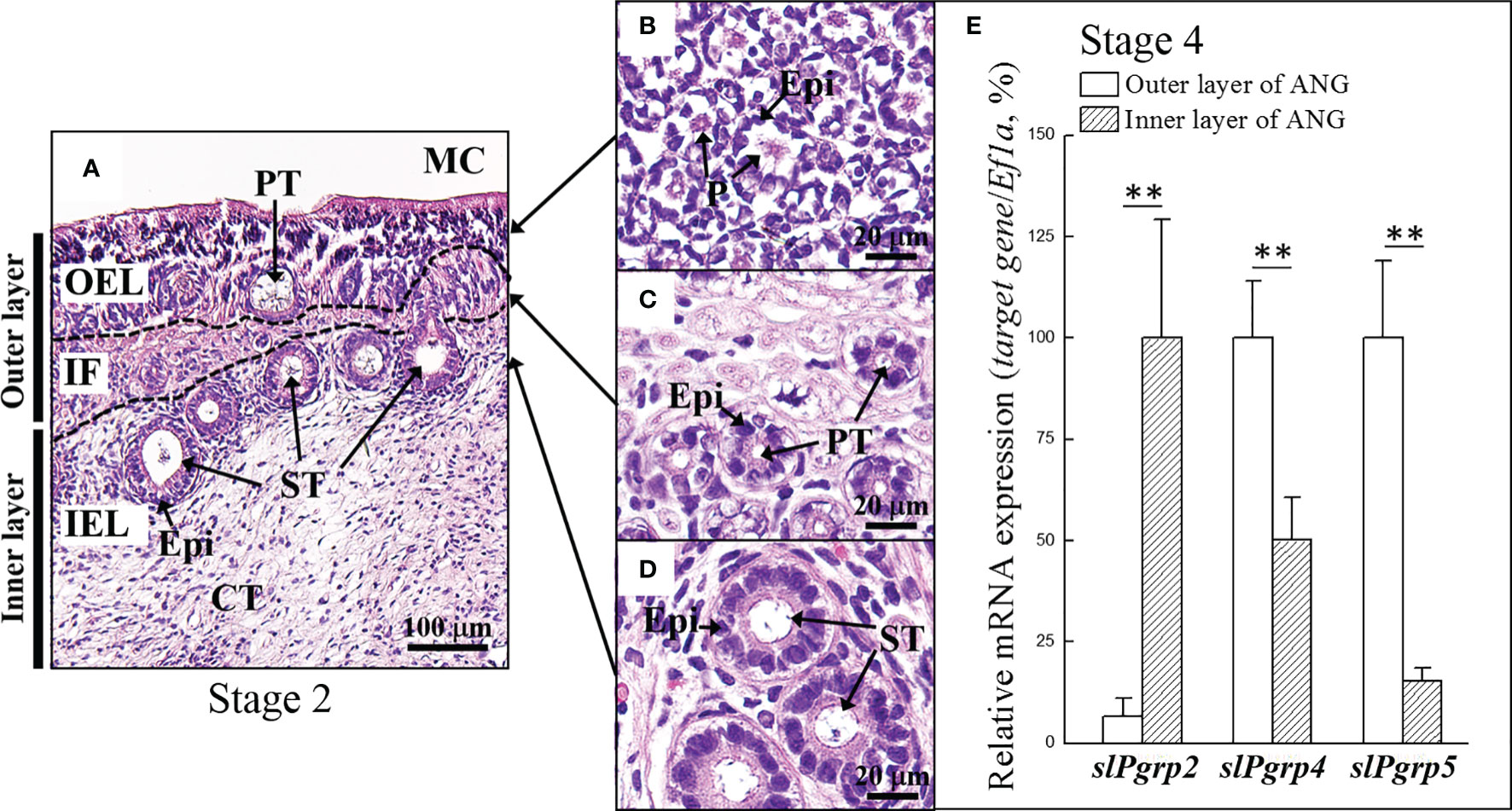
Figure 4 Expression of slPgrp transcripts in different layers of ANG. (A) The definition of outer and inner layer of ANG. (B, C) The outer layer of ANG was composed by outer epithelial cell layer and intermediate fiber layer, respectively. (D) The inner layer of ANG was composed by inner epithelial cell layer. The relative portion of outer and inner layer of ANGs from mature female squid (stage 4, n = 4) were isolated under a stereomicroscope and analyzed separately. (E) Expression levels of slPgrp2, slPgrp4, and slPgrp5 were analyzed by qPCR. **P < 0.01 comparing the outer and inner layer of ANGs. OEL, outer epithelial cell layer; IF, intermediate fiber layer; IEL, inner epithelial cell layer; MC, mantle cavity; CT, connective tissue; Epi, epithelial cell; PT, primary tubule; ST, secondary tubule; P, pore.
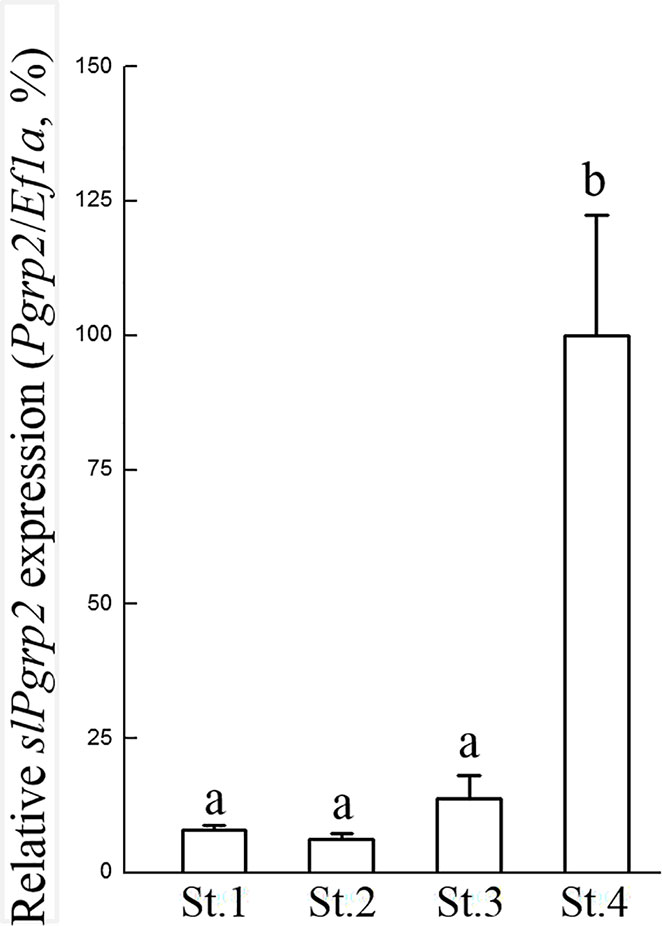
Figure 5 Gene expression profile of slPgrp2 during ANG growth. According to histology, four different stages of ANGs were used for gene expression analysis, including stage 1 (n = 4), stage 2 (n = 14), stage 3 (n = 12), and stage 4 (n = 9). The expression profile of slPgrp2 during ANG development was analyzed by qPCR. Different lower-case letters indicate significant differences between stages of ANGs (P < 0.05). St, stage.
Localization of slPgrp2 Expression
To analyze the distribution of slPgrp2 in the maturing ANG, ISH for slPgrp2 was performed on ANGs of different stages. In the stage 2 ANG, slPgrp2 transcripts were detected in the epithelial cells of primary tubules, but no signal was observed in the epithelial cells of secondary tubules (Figures 6A, B). No signal was observed from the control sense probe in stage 2 ANGs (Figure 6C). At stage 4, slPgrp2 transcripts were mainly expressed in the epithelial cells of certain secondary tubules of the ANG, but slight or no signal was observed in primary tubules and other secondary tubules at stage 4 of ANG development (Figures 6D, E). No signal was observed from the control sense probe for slPgrp2 at stage 4 of ANG development (Figure 6F). Thus, slPgrp2 transcripts showed differential expression levels in the epithelial cells of different secondary tubules of ANG.
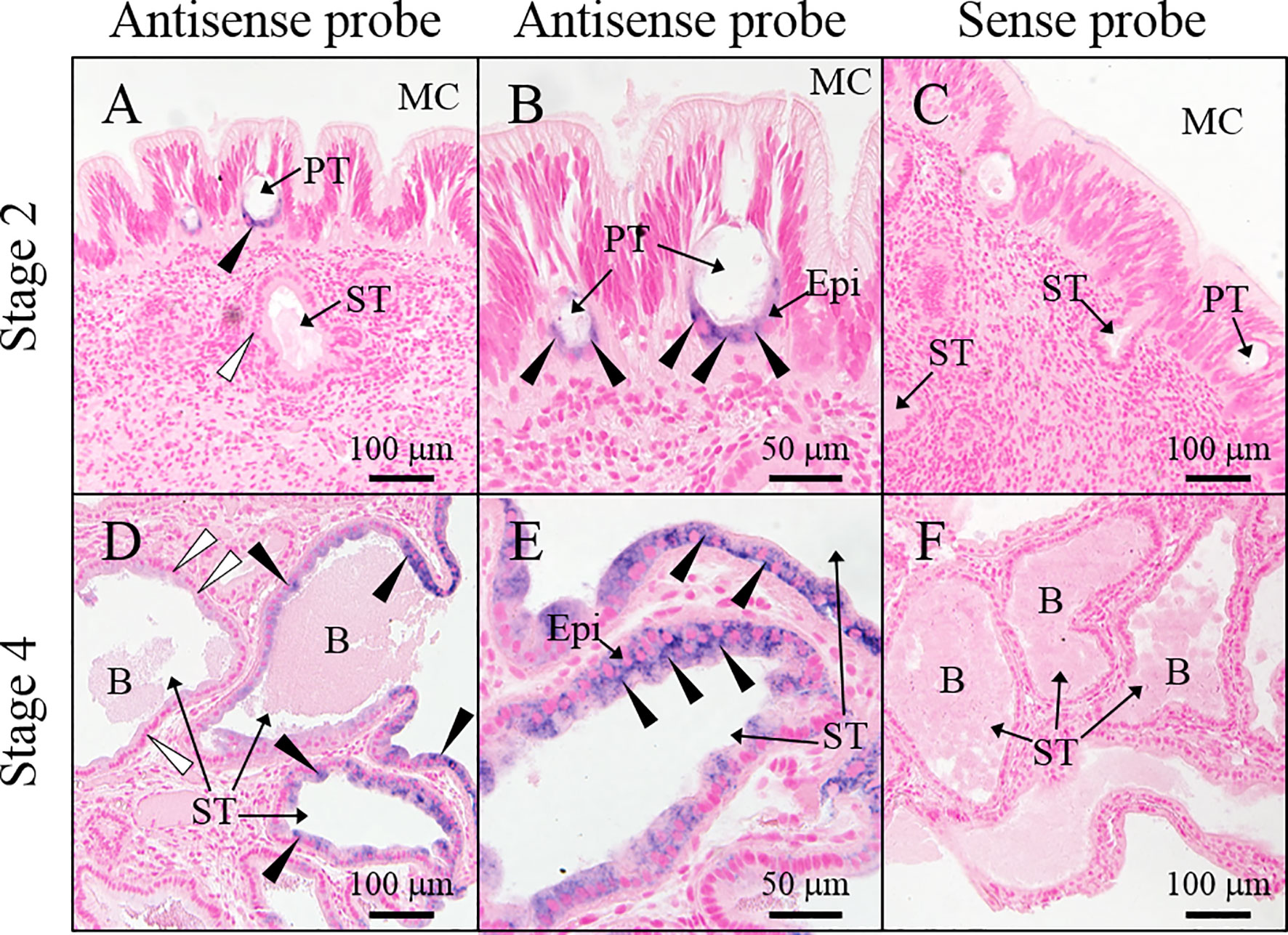
Figure 6 Location of slPgrp2 transcripts in ANG. (A, B) slPgrp2 transcripts were detected in epithelial cells of primary tubules in the ANG using an antisense probe in immature female squid (stage 2). (C) Background staining was evaluated using a sense probe for slPgrp2 in immature female squid (stage 2). (D, E) slPgrp2 transcripts were detected with an antisense probe in epithelial cells of secondary tubules of ANG in mature female squid (stage 4). (F) Background staining was evaluated using a sense probe for slPgrp2 in mature female squid (stage 4). Black arrowhead denotes slPgrp2 signal, and white arrowhead denotes areas without slPgrp2 signal in epithelial cells. B, bacteria; Epi, epithelial cells; MC, mantle cavity; PT, primary tubule; ST, secondary tubule.
slPgrp2 Expression in Different Areas of ANG
Next, we examined transcript levels and distributions of slPgrp2 in different secondary tubules at stage 4 of ANG development. slPgrp2 transcripts were differentially distributed in different regions of the ANG (Figures 7A–C). No signal was observed from the control sense probe for slPgrp2 in serial sections (Figure 7D). Thus, slPgrp2 expression was varied in the epithelial cells of different secondary tubules in the ANG. According to the HE staining (Figure 7E) and Gram staining (Figures 7F–H) results, different area of ANG showed the difference on colors and bacterial density. To analyze slPgrp2 expression in relation to different region of secondary tubules in the ANG, the different regions were identified by color, isolated under stereomicroscopy and analyzed separately. Eight regions were isolated from two ANGs (stage 4), including three white regions, one red region, two orange regions, and two yellow regions (Figure 7I). qPCR analysis showed differential slPgrp2 transcripts associated with different regions (Figure 7J). Furthermore, three mature female squid (stage 4) were used to examine the correlation between region colors and slPgrp2 transcript levels. Unexpectedly, the expression levels of slPgrp2 showed no apparent correlations with the different region colors (Figure 7K). These data confirmed our conclusion made from ISH results that slPgrp2 transcripts are differentially expressed in different secondary tubules of ANG. Importantly, these data also do not rule out the possibility that slPgrp2 expression in the epithelial cells of secondary tubules in the ANG might correspond to microenvironment characteristics, such as region colors and it bacterial community.
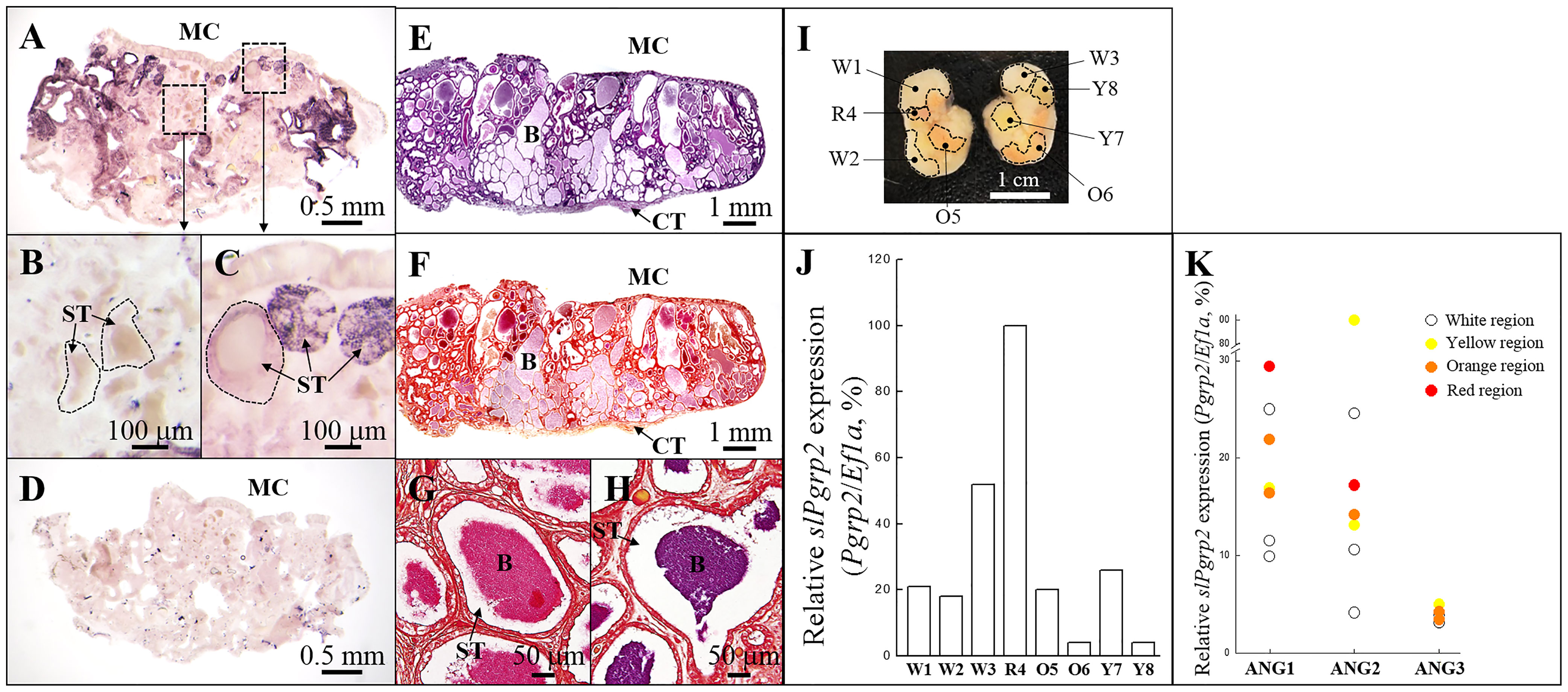
Figure 7 Expression of slPgrp2 transcripts in different areas of ANG in the mature female squid. (A–C) slPgrp2 mRNA expression of ANG was detected by an antisense probe in mature female squid (stage 4). (D) Background staining was evaluated using a sense probe for slPgrp2 in serial sections. (E, F) Histological characteristics and Gram staining of ANG in mature female squid, respectively. (G, H) Gram staining in different secondary tubules of ANG. (I) Different regions in ANG were distinguished by color and isolated under a stereomicroscope. Each color region was analyzed separately. (J) Expression of slPgrp2 mRNA was analyzed by qPCR in different colors regions, as shown in (I). (K) Four different colors of bacterial regions were detected and used for correlation with gene expression pattern: white, yellow, orange, and red. slPgrp2 mRNA expression showed high variability in areas with different regions, even between regions with the same color. Dash line in (B, C) denotes the border of secondary tubule. B, bacteria; MC, mantle cavity; ST, secondary tubule; W, white; R, red; Y, yellow; O, orange.
The Effect of Recombinant slPGRP2 on E. coli Growth
To analyze the potential bactericidal activity of slPGRP2, we evaluated the effect of rslPGRP2 expression on E.coli growth in culture. For the bacterial culture study, E.coli carrying an IPTG-inducible slPgrp2 expression cassette were treated with or without IPTG. According to turbidity measurements, cell density was increased in cultures of the control (without IPTG treatment) during the experimental period. In contrast, cell density was significantly lower in cultures with the expression of rslPGRP2 (with IPTG treatment) compared with the control (without IPTG treatment) (Figure 8). E.coli carrying an IPTG-inducible ecdmrt1 expression cassette were used as a reference control. Turbidity assay results showed that cell density was significantly lower in cultures with the expression of recDmrt1 (with TPTG treatment; OD600 = 1.71 ± 0.01) compared to the control (without IPTG treatment; OD600 = 1.13 ± 0.01) after 3hrs of IPTG treatment. Although the negative effect of E.coli growth was observed in cultures of both IPTG-inducible ecdmrt1 and slPgrp2 expression cassette. Our data also showed that cell density was significantly higher in cultures with the expression of recDmrt1 (OD600 = 1.13 ± 0.01) compared with the expression of rslPGRP2 (OD600 = 0.7 ± 0.02) after 3hrs of IPTG treatment. These data suggest that slPGRP2 has negatively effect in bacterial growth.
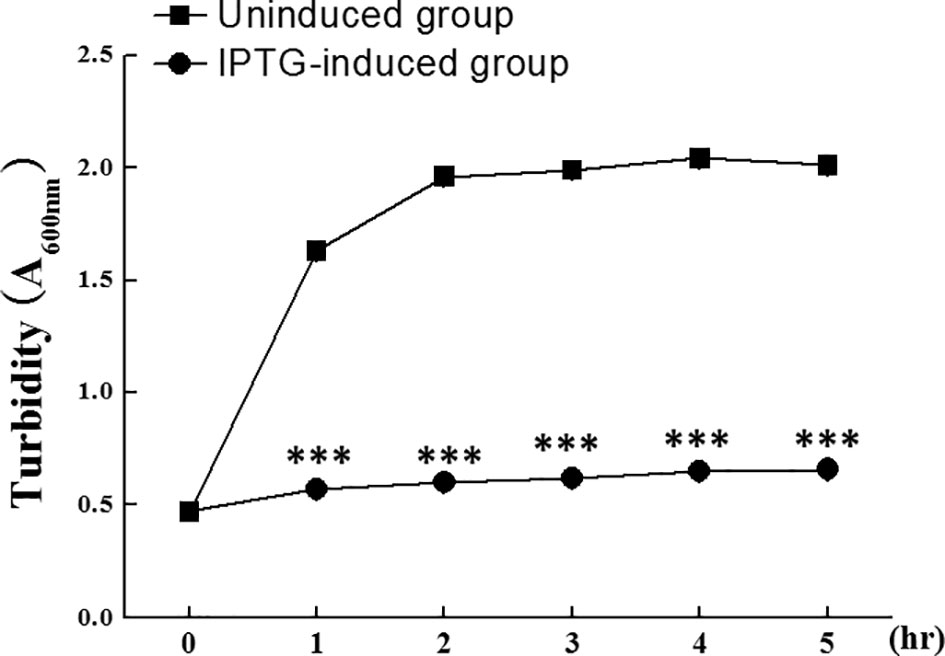
Figure 8 The effect of rslPGRP2 in bacterial growth. Recombinant slPGRP2 (rslPGRP2) was induced by the addition of IPTG. The potential effect of rslPGRP2 in bacterial growth was analyzed by growth rate of E. coli using turbidity assay (spectrophotometer absorbance at 600 nm). E.coli carrying an IPTG-inducible slVasa expression cassette were used as reference control to show that the vector did not negatively affect E. coli growth. Samples were collected at 1-hr intervals after IPTG addition in IPTG-induced group. E.coli without IPTG treatment (uninduced group) was used as a reference. The three asterisks indicate Student’s t-test (P < 0.001).
Discussion
According to this systematic study, we found four slPgrps in ANG transcriptome database. We analyzed the mRNA expression profile of these slPgrps in various tissues and different female phases, only the slPgrp2 expression profile was correlated to the ANG development. We further characterized the spatial and temporal expression profiles of slPgrp2 in ANG during the female maturation. We provide evidence that slPgrp2: 1) gene expression profile is correlated with ANG growth in the female squid; 2) localizes on epithelial cells of the ANG, including the outer epithelial cell layer (primary tubules) and the inner epithelial cell layer (secondary tubules); 3) has high variability in expression levels in the different inner epithelial cell layer (secondary tubule). However, the color of different regions (pigmentation), which represent bacteria-synthesized carotenoids, did not correlated with the slPgrp2 expression levels. These data suggest that the epithelial cell-synthesized slPGRP2 may be involved in bacterial selection during bacterial colonization in the female bigfin reef squid.
Both the slPgrp2 and slPgrp5 showed high similarity in predicted protein structures. The apparent roles for slPgrp2 are striking in contrast to slPgrp5. Unlike slPgrp5, which was mainly expressed in the outer epithelial cell layer, slPgrp2 was dominantly expressed in the inner epithelial cell layer. Thus, this study is beginning to reveal the role of different slPgrps in ANG growth.
Bacterial Community of ANG in Cephalopods
The ANG houses a bacterial consortium that is horizontally transmitted from the environmental bacterial community (Kaufman et al., 1998; Li et al., 2019). In the present (Figure 1) and our previous studies (Li et al., 2019), our data show that the amount of bacterial flora in ANG varied during ANG growth. Furthermore, our recent study showed that the ANG bacterial community shifts gradually and decreases in diversity during maturation of the bigfin reef squid (Yang et al., 2021). In the Hawaiian bobtail squid, the microbiota of ANG undergoes a reduction of complexity during squid maturation and development of ANG (Kerwin et al., 2021). Such changes in bacterial diversity in the ANG during maturation stages could be due to host selection.
The core bacterial taxa found in cephalopod ANGs are Gram-negative Alphaproteobacteria, but the compositions of bacterial communities vary among cephalopod species (Grigioni et al., 2000; Pichon et al., 2005; Collins et al., 2012; Kerwin and Nyholm, 2017; Kerwin et al., 2021; Yang et al., 2021). Notably, the ANG bacterial community in Hawaiian bobtail squids collected from the field was conserved across hosts and was not affected by maintenance in artificial conditions (Kerwin et al., 2017). We suspect that the bacteria consortium in ANG is strongly selected by the host and varies according to species.
The Importance of Symbiotic Bacteria in Squid ANG for Offspring
Marine environments contain many microorganisms, some of which are lethal. Animals that lay eggs without subsequent parental care must have some mechanisms to protect developing embryos from infection and biofouling. In some crustaceans, parents deposit bacteria on eggs, and these bacteria secrete antimicrobial compounds to inhibit biofouling (Gil-Turnes et al., 1989; Gil-Turnes and Fenical, 1992). Similarly, bacteria from the ANG are transmitted to the egg jelly coat in most squid and cuttlefish (Kerwin and Nyholm, 2017; Huang et al., 2018). In the Hawaiian bobtail squid, the bacteria composition of the jelly coat shows a high similarity to that in the ANG (Kerwin and Nyholm, 2017). Furthermore, eggs treated with antibiotics (suppress the bacteria deposited on the eggs) were susceptible to heavy biofouling and very low hatching rates compared to untreated eggs in Hawaiian bobtail squid (Kerwin et al., 2019). The growth of some fungi and yeast was inhibited by extracts from the symbiotic bacteria isolated from ANG and jelly coat (Kerwin et al., 2019; Suria et al., 2020). Therefore, delivery of ANG bacteria to the eggs play an important role in promoting embryo survival by preventing fungal fouling of the egg capsule for most squid and cuttlefish.
Control of Commensal Microbiota in Squid by PGRPs
PGRPs are conserved from insects to mammals and have diverse functions in antimicrobial defense as well as recognition and maintenance of symbiotic organisms (Royet et al., 2011). Unlike insects, which have many Pgrp genes, cephalopods and vertebrates have fewer Pgrp genes (Goodson et al., 2005; Collins et al., 2012). Only four Pgrp genes were found in the ANG of bigfin reef squid, including the secreted slPgrp2 and slPgrp5, GPI-anchored slPgrp3, and the slPgrp4 surface receptor (Figure 2). In bigfin reef squid, slPgrp2 mRNA are predominantly expressed in the ovary and ANG, while slPgrp4 and slPgrp5 mRNA are predominantly expressed in hemocytes (Figure 2 and Supplemental Figure 1). Furthermore, only the slPgrp2 gene expression levels of epithelial cells are correlated to the ANG growth (Figures 4–6). In Hawaiian bobtail squid, PGRPs participate in the winnowing and maintenance of the symbiotic luminescent bacterium V. fischeri within the light organ (Nyholm and Graf, 2012; McAnulty and Nyholm, 2017; Nyholm and McFall-Ngai, 2021). Both PGRP1 and PGRP2 play integral roles in the recognition of symbiotic bacteria during V. fischeri colonization of the Hawaiian bobtail squid light organ (Troll et al., 2009; Troll et al., 2010). After V. fischeri enters the light organ duct and then colonizes the crypts, the PGRP1 level decreases dramatically and ciliated epithelial cells undergo apoptosis to promote the loss of appendages during the morphogenesis of light organ (Troll et al., 2009). Furthermore, epithelial cell-synthesized PGRP2 is secreted into the lumen of the crypts, where it degrades peptidoglycan fragments secreted by V. fischeri (Troll et al., 2010). Pgrp5 mRNA is dominantly expressed in hemocytes of Hawaiian bobtail squid (Collins et al., 2012). Notably, the expression of Pgrp5 is decreased in hemocytes of Hawaiian bobtail squids upon removal of symbiotic bacteria (Collins et al., 2012). Thus, the colonization of the light organ influence Pgrp gene expression in the Hawaiian bobtail squid (Collins et al., 2012; Nyholm and McFall-Ngai, 2021). Taken together, these observations and ours suggest that slPgrp2 may participate in bacterial colonization during the ANG growth in female bigfin reef squid.
PGRP2 in both bigfin reef squid and Hawaiian bobtail squid share 67% identity and 88% similarity at the amino acid level. PGRP2 showed bactericidal activity in Hawaiian bobtail squid (Troll et al., 2010). PGRP2 also prevents the inflammatory effects of hydrolyzed peptidoglycan fragments released from symbiotic bacteria in the light organ (Troll et al., 2010). Bactericidal PGRPs are also found in other species, such as some bivalves (Kong et al., 2018; Huang et al., 2019; Yang et al., 2019), teleosts (Li et al., 2007; Kim et al., 2010; Li et al., 2020) and mammals (Lu et al., 2006). In zebrafish, knockdown of pglyrp5 and pglyrp6 expression during embryogenesis results in increased sensitivity to infections with pathogenic bacteria and poor survival (Chang and Nie, 2008; Li et al., 2007). Moreover, Pgrp-deficient mice (Pglyrp1-/-, Pglyrp2-/-, Pglyrp3-/-, and Pglyrp4-/-) are more sensitive to colitis than wild-type mice due to aberrations in the interferon-γ regulating pathway (Saha et al., 2010). Thus, mammalian PGRPs are important regulators of commensal microbiota and intestinal homeostasis, protecting the host from tissue damage and colitis. Our results speculate that inner epithelial cell layer-expressed slPgrp2 may participate in bacterial selection during the bacterial colonization of ANG. We suspect that slPGRPs may have a similar essential role in the defense against bacteria and further participate in the winnowing (bacterial selection) and maintenance of symbiotic bacteria in the bigfin reef squid ANG in comparison to Hawaiian bobtail squid ANG or light organ. In bigfin reef squid, slPgrp2 was highly expressed in ovary. In fish, pglyrp2 transcripts is expressed in the developing oocytes (Li et al., 2007). Moreover, high expression level of Pglyrp2 protein is found in mature eggs and developing embryos (Li et al., 2007). Fish Pglyrp2 proteins showed bactericidal activity (Li et al., 2007). Therefore, slPgrp2 may have essential role in the defense against bacteria during the early stage of embryonic development in bigfin reef squid. Furthermore, the potential roles of other homologs, slPgrp4 and slPgrp5 in hemocytes, should be examined with regard to the integral bacterial colonization of the ANG.
Conclusion and Future Directions
Although the mechanisms of colonization in the ANG association are not yet as well understood as those of the light organ, this study in ANG are able to provide a new insight of mechanisms of colonization and maintenance in complex consortia. In this study, we examined the temporal and spatial profiles of slPgrp2 transcripts. Unlike PGRP2, which is broadly distributed in the surface area (epithelial cell layer) of different organs in Hawaiian bobtail squid (Troll et al., 2010), we found epithelial cell-expressed slPgrp2 shows differentially expression levels in different regions of ANG. This data reveals that slPgrp2 transcripts may associate with the composition of bacterial consortium and it secretary factors. Further studies are needed to determine which bacteria species can trigger slPgrp2 synthesis and cause the secretion of slPGRP2 into secondary tubules of the ANG. A specific antibody for slPGRP2 is required to better explore the regulatory machinery underlying the interactions of microbiomes with host immunity. The eukaryotic cell-expressed rslPGRP2 is required to examine the bactericidal activity (against E. coli or other bacterial isolates from ANG of bigfin reef squid).
Recently, a study of the whole genome of Hawaiian bobtail squid provides new insight on organ-specific regulatory mechanism in light organ and ANG (Belcaid et al., 2019). The transcripts of light organ have high similarity with the eye and the ANG have high number of ANG-specific transcripts. These analyses show that the light organ and ANG are likely to have evolved via different mechanisms. Future studies are need to perform the ANG transcriptome analysis of bigfin reef squid that will make a better understanding of the regulatory mechanism of complex bacterial consortium.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by National Taiwan Ocean University Institutional Animal Care and Use Committee.
Author Contributions
H-WL and G-CW collected the samples. H-WL conducted the molecular cloning, RNA analysis, turbidity assay and WB experiments. W-LK conducted the ISH study. CC conducted the phylogenetic analysis. Y-CT gave comments. C-FC gave the comments and participated in compiling the main manuscript. G-CW designed and conducted experiments, analyzed the data, and participated in compiling the main manuscript. All authors reviewed and approved the manuscript.
Funding
This work was partly supported by grants from the Ministry of Science and Technology [MOST 104-2923-B-019-001-MY4] and by the Center of Excellence for the Oceans, National Taiwan Ocean University, from The Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) of Taiwan.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Dr. Marcus Calkins at Academia Sinica for reading and correcting of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.825267/full#supplementary-material
Supplementary Figure 1 | Tissue distributions of slPgrp gene expression in mature female squid. Mature female squid was used for gene expression analysis in various tissues. Four Pgrp genes were identified from the ANG transcriptomic database and selected for gene expression analysis, including slPgrp2 (A), slPgrp3 (C), slPgrp4 (B), and slPgrp5 (C). The relative gene expression level of each Pgrp is also shown in the tissue with highest expression. Ma, mantle (muscle); Te, tentacle; Ol, optic lobe; Br, brain; St, stomach; Hp, hepatopancreas; Gi, gill; He, heart; NG, nidamental gland; ANG, accessory nidamental gland; Od, oviduct; Og, oviductal gland; Hm, hemocytes.
Supplementary Table 1 | The characteristics of sampled female squids.
Supplementary Table 2 | List of gene names and accession numbers for phylogenetic analysis.
References
Bai L., Zhou K., Li H., Qin Y., Wang Q., Li W. (2020). Bacteria-Induced IMD-Relish-AMPs Pathway Activation in Chinese Mitten Crab. Fish Shellfish Immunol. 106, 866–875. doi: 10.1016/j.fsi.2020.08.046
Belcaid M., Casaburi G., McAnulty S. J., Schmidbaur H., Suria A. M., Moriano-Gutierrez S., et al. (2019). Symbiotic Organs Shaped by Distinct Modes of Genome Evolution in Cephalopods. Proc. Natl. Acad. Sci. U. S. A. 116, 3030–3035. doi: 10.1073/pnas.1817322116
Bischoff V., Vignal C., Duvic B., Boneca I. G., Hoffmann J. A., Royet J. (2006). Downregulation of the Drosophila Immune Response by Peptidoglycan-Recognition Proteins SC1 and SC2. PloS Pathog. 2, e14. doi: 10.1371/journal.ppat.0020014
Castillo M. G., Salazar K. A., Joffe N. R. (2015). The Immune Response of Cephalopods From Head to Foot. Fish Shellfish Immunol. 46, 145–160. doi: 10.1016/j.fsi.2015.05.029
Cerf-Bensussan N., Gaboriau-Routhiau V. (2010). The Immune System and the Gut Microbiota: Friends or Foes? Nat. Rev. Immunol. 10, 735–744. doi: 10.1038/nri2850
Chang M. X., Nie P. (2008). RNAi Suppression of Zebrafish Peptidoglycan Recognition Protein 6 (Zfpgrp6) Mediated Differentially Expressed Genes Involved in Toll-Like Receptor Signaling Pathway and Caused Increased Susceptibility to Flavobacterium Columnare. Vet. Immunol. Immunopathol. 124, 295–301. doi: 10.1016/j.vetimm.2008.04.003
Chen C., Li H. W., Ku W. L., Lin C. J., Chang C. F., Wu G. C. (2018). Two Distinct Vitellogenin Genes Are Similar in Function and Expression in the Bigfin Reef Squid Sepioteuthis Lessoniana. Biol. Reprod. 99, 1034–1044. doi: 10.1093/biolre/ioy131
Collins A. J., Schleicher T. R., Rader B. A., Nyholm S. V. (2012). Understanding the Role of Host Hemocytes in a Squid/Vibrio Symbiosis Using Transcriptomics and Proteomics. Front. Immunol. 3. doi: 10.3389/fimmu.2012.00091
Dziarski R. (2004). Peptidoglycan Recognition Proteins (PGRPs). Mol. Immunol. 40, 877–886. doi: 10.1016/j.molimm.2003.10.011
Dziarski R., Gupta D. (2006). The Peptidoglycan Recognition Proteins (PGRPs). Genome Biol. 7, 232. doi: 10.1186/gb-2006-7-8-232
Gil-Turnes M. S., Fenical W. (1992). Embryos of Homarus Americanus Are Protected by Epibiotic Bacteria. Biol. Bull. 182, 105–108. doi: 10.2307/1542184
Gil-Turnes M. S., Hay M. E., Fenical W. (1989). Symbiotic Marine Bacteria Chemically Defend Crustacean Embryos From a Pathogenic Fungus. Science 246, 116–118. doi: 10.1126/science.2781297
Goodson M. S., Kojadinovic M., Troll J. V., Scheetz T. E., Casavant T. L., Bento Soares M., et al. (2005). Identifying Components of the NF-kappaB Pathway in the Beneficial Euprymna Scolopes-Vibrio Fischeri Light Organ Symbiosis. Appl. Environ. Microbiol. 71, 6934–6946. doi: 10.1128/AEM.71.11.6934-6946.2005
Grigioni S., Boucher-Rodoni R., Demarta A., Tonolla M., Peduzzi R. (2000). Phylogenetic Characterisation of Bacterial Symbionts in the Accessory Nidamental Glands of the Sepioid Sepia Officinalis (Cephalopoda: Decapoda). Marine Biol. 136, 217–222. doi: 10.1007/s002270050679
Guo C. Y., Tseng P. W., Hwang J. S., Wu G. C., Chang C. F. (2021). The Characteristics and Expression Profile of Gonadal Soma-Derived Factor (Gsdf/Gsdf) During Androgen-Induced Reversible Sex Change in the Protogynous Orange-Spotted Grouper, Epinephelus Coioides. Aquaculture 542, 736832. doi: 10.1016/j.aquaculture.2021.736832
Huang J. D., Lee S. Y., Chiang T. Y., Lu C. C., Lee M. F. (2018). Morphology of Reproductive Accessory Glands in Female Sepia Pharaonis (Cephalopoda: Sepiidae) Sheds Light on Egg Encapsulation. J. Morphol. 279, 1120–1131. doi: 10.1002/jmor.20835
Huang Y., Pan J., Li X., Ren Q., Zhao Z. (2019). Molecular Cloning and Functional Characterization of a Short Peptidoglycan Recognition Protein From Triangle-Shell Pearl Mussel (Hyriopsis Cumingii). Fish Shellfish Immunol. 86, 571–580. doi: 10.1016/j.fsi.2018.12.007
Ikuta T., Tame A., Saito M., Aoki Y., Nagai Y., Sugimura M., et al. (2019). Identification of Cells Expressing Two Peptidoglycan Recognition Proteins in the Gill of the Vent Mussel, Bathymodiolus Septemdierum. Fish Shellfish Immunol. 93, 815–822. doi: 10.1016/j.fsi.2019.08.022
Kaufman M. R., Ikeda Y., Patton C., van Dykhuizen G., Epel D. (1998). Bacterial Symbionts Colonize the Accessory Nidamental Gland of the Squid Loligo Opalescens via Horizontal Transmission. Biol. Bull. 194, 36–43. doi: 10.2307/1542511
Kerwin A. H., Gromek S. M., Suria A. M., Samples R. M., Deoss D. J., O’Donnell K., et al. (2019). Shielding the Next Generation: Symbiotic Bacteria From a Reproductive Organ Protect Bobtail Squid Eggs From Fungal Fouling. mBio 10, 02376–02319, e02376-19. doi: 10.1128/mBio
Kerwin A. H., McAnulty S. J., Nyholm S. V. (2021). Development of the Accessory Nidamental Gland and Associated Bacterial Community in the Hawaiian Bobtail Squid, Euprymna Scolopes. Biol. Bull 240, 205–218. doi: 10.1086/713965
Kerwin A. H., Nyholm S. V. (2017). Symbiotic Bacteria Associated With a Bobtail Squid Reproductive System are Detectable in the Environment, and Stable in the Host and Developing Eggs. Environ. Microbiol. 19, 1463–1475. doi: 10.1111/1462-2920.13665
Kim M. Y., Jang J. H., Lee J. W., Cho J. H. (2010). Molecular Cloning and Characterization of Peptidoglycan Recognition Proteins From the Rockfish, Sebastes Schlegeli. Fish Shellfish Immunol. 28, 632–639. doi: 10.1016/j.fsi.2009.12.023
Kong X., Liu H., Li Y., Zhang H. (2018). Two Novel Short Peptidoglycan Recognition Proteins (PGRPs) From the Deep Sea Vesicomyidae Clam Archivesica Packardana: Identification, Recombinant Expression and Bioactivity. Front. Physiol. 9. doi: 10.3389/fphys.2018.01476
Li H. W., Chen C., Kuo W. L., Lin C. J., Chang C. F., Wu G. C. (2019). The Characteristics and Expression Profile of Transferrin in the Accessory Nidamental Gland of the Bigfin Reef Squid During Bacteria Transmission. Sci. Rep. 9, 20163. doi: 10.1038/s41598-019-56584-8
Lindgren A. R., Pankey M. S., Hochberg F. G., Oakley T. H. (2012). A Multi-Gene Phylogeny of Cephalopoda Supports Convergent Morphological Evolution in Association With Multiple Habitat Shifts in the Marine Environment. BMC Evol. Biol. 12, 129. doi: 10.1186/1471-2148-12-129
Liu H., Wu C., Matsuda Y., Kawabata S. I., Lee B. L., Söderhäll K., et al. (2011). Peptidoglycan Activation of the proPO-System Without a Peptidoglycan Receptor Protein (PGRP)? Dev. Comp. Immunol. 35, 51–61. doi: 10.1016/j.dci.2010.08.005
Li X., Wang S., Qi J., Echtenkamp S. F., Chatterjee R., Wang M., et al. (2007). Zebrafish Peptidoglycan Recognition Proteins are Bactericidal Amidases Essential for Defense Against Bacterial Infections. Immunity 27, 518–529. doi: 10.1016/j.immuni.2007.07.020
Li X., Yuan S., Sun Z., Lei L., Wan S., Wang J., et al. (2020). Gene Identification and Functional Analysis of Peptidoglycan Recognition Protein From the Spotted Sea Bass (Lateolabrax Maculatus). Fish Shellfish Immunol. 106, 1014–1024. doi: 10.1016/j.fsi.2020.08.041
Lum-Kong A. (1992). A Histological Study of the Accessory Reproductive Organs of Female Loligo Forbesi (Cepahlopoda: Loliginidae). J. Zool. 226, 469–490. doi: 10.1111/j.1469-7998.1992.tb07493.x
Lu X., Wang M., Qi J., Wang H., Li X., Gupta D., et al. (2006). Peptidoglycan Recognition Proteins are a New Class of Human Bactericidal Proteins. J. Biol. Chem. 281, 5895–5907. doi: 10.1074/jbc.M511631200
McAnulty S. J., Nyholm S. V. (2017). The Role of Hemocytes in the Hawaiian Bobtail Squid, Euprymna Scolopes: A Model Organism for Studying Beneficial Host-Microbe Interactions. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.02013
Montaño A. M., Tsujino F., Takahata N., Satta Y. (2011). Evolutionary Origin of Peptidoglycan Recognition Proteins in Vertebrate Innate Immune System. BMC Evol. Biol. 11, 79. doi: 10.1186/1471-2148-11-79
Nyholm S. V., Deplancke B., Gaskins H. R., Apicella M. A., McFall-Ngai M. J. (2002). Roles of Vibrio Fischeri and Nonsymbiotic Bacteria in the Dynamics of Mucus Secretion During Symbiont Colonization of the Euprymna Scolopes Light Organ. Appl. Environ. Microbiol. 68, 5113–5122. doi: 10.1128/AEM.68.10.5113-5122.2002
Nyholm S. V., Graf J. (2012). Knowing Your Friends: Invertebrate Innate Immunity Fosters Beneficial Bacterial Symbioses. Nat. Rev. Microbiol. 10, 815–827. doi: 10.1038/nrmicro2894
Nyholm S. V., McFall-Ngai M. J. (2021). A Lasting Symbiosis: How the Hawaiian Bobtail Squid Finds and Keeps Its Bioluminescent Bacterial Partner. Nat. Rev. Microbiol. 19, 666–679. doi: 10.1038/s41579-021-00567-y
Pichon D., Gaia V., Norman M. D., Boucher-Rodoni R. (2005). Phylogenetic Diversity of Epibiotic Bacteria in the Accessory Nidamental Glands of Squids (Cephalopoda: Loliginidae and Idiosepiidae). Marine Biol. 147, 1323–1332. doi: 10.1007/s00227-005-0014-5
Royet J., Gupta D., Dziarski R. (2011). Peptidoglycan Recognition Proteins: Modulators of the Microbiome and Inflammation. Nat. Rev. Immunol. 11, 837–851. doi: 10.1038/nri3089
Ruby E. G., Asato L. M. (1993). Growth and Flagellation of Vibrio Fischeri During Initiation of the Sepiolid Squid Light Organ Symbiosis. Arch. Microbiol. 159, 160–167. doi: 10.1007/BF00250277
Saha S., Jing X., Park S. Y., Wang S., Li X., Gupta D., et al. (2010). Peptidoglycan Recognition Proteins Protect Mice From Experimental Colitis by Promoting Normal Gut Flora and Preventing Induction of Interferon-Gamma. Cell Host Microbe 8, 147–162. doi: 10.1016/j.chom.2010.07.005
Suria A. M., Tan K. C., Kerwin A. H., Gitzel L., Abini-Agbomson L., Bertenshaw J. M., et al. (2020). Hawaiian Bobtail Squid Symbionts Inhibit Marine Bacteria via Production of Specialized Metabolites, Including New Bromoalterochromides BAC-D/D'. mSphere 5, e00166-20. doi: 10.1128/mSphere.00166-20
Troll J. V., Adin D. M., Wier A. M., Paquette N., Silverman N., Goldman W. E., et al. (2009). Peptidoglycan Induces Loss of a Nuclear Peptidoglycan Recognition Protein During Host Tissue Development in a Beneficial Animal-Bacterial Symbiosis. Cell Microbiol. 11, 1114–1127. doi: 10.1111/j.1462-5822.2009.01315.x
Troll J. V., Bent E. H., Pacquette N., Wier A. M., Goldman W. E., Silverman N., et al. (2010). Taming the Symbiont for Coexistence: A Host PGRP Neutralizes a Bacterial Symbiont Toxin. Environ. Microbiol. 12, 2190–2203. doi: 10.1111/j.1462-2920.2009.02121.x
Yang S. H., Chen C., Hsieh Y. E., Yang S. Y., Li H. W., Ching T. Y., et al. (2021). Bacterial Dynamics in the Accessory Nidamental Gland of Sepioteuthis Lessoniana Throughout Maturation. Microbes Environ. 36, ME21030. doi: 10.1264/jsme2.ME21030
Yang D., Han Y., Liu Y., Cao R., Wang Q., Dong Z., et al. (2019). A Peptidoglycan Recognition Protein Involved in Immune Recognition and Immune Defenses in Ruditapes Philippinarum. Fish Shellfish Immunol. 88, 441–448. doi: 10.1016/j.fsi.2019.03.017
Keywords: PGRP, host immunity, symbiotic bacteria, ANG, cephalopod
Citation: Li H-W, Kuo W-L, Chen C, Tseng Y-C, Chang C-F and Wu G-C (2022) The Characteristics and Expression Profile of Peptidoglycan Recognition Protein 2 in the Accessory Nidamental Gland of the Bigfin Reef Squid During Bacterial Colonization. Front. Mar. Sci. 9:825267. doi: 10.3389/fmars.2022.825267
Received: 30 November 2021; Accepted: 02 May 2022;
Published: 30 May 2022.
Edited by:
Xiaotong Wang, Ludong University, ChinaReviewed by:
Tara Essock-Burns, University of Hawaii, United StatesGuoxing Nie, Henan Normal University, China
Copyright © 2022 Li, Kuo, Chen, Tseng, Chang and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guan-Chung Wu, gcwu@mail.ntou.edu.tw
 Hau-Wen Li1
Hau-Wen Li1