- 1College of Biological and Food Engineering, Guangdong University of Petrochemical Technology, Maoming, China
- 2Key Laboratory for Water Quality and Conservation of the Pearl River Delta, Ministry of Education, Institute of Environmental Research at Greater Bay Area, Guangzhou University, Guangzhou, China
The oxidation of nitrite to nitrate as the second step of nitrification is vital for the global nitrogen cycle, and the genus Nitrospira is the most widespread nitrite-oxidizing bacteria (NOB) in diverse natural ecosystems. However, our understanding of Nitrospira distribution and their response to dynamic environmental parameters is still limited in the subterranean estuary (STE), a special environment of the mixed zone between land and sea. In this study, Nitrospira communities were collected within 5–10 cm depth intervals of sediments with the physiochemical gradients along the transections of the Daya Bay (DYB) from the landward member to the seaward member. The abundance, community structure of Nitrospira, and their potentially influencing environmental factors were investigated using Q-PCR, the high throughput sequencing targeting Nitrospira nxrB gene, and biostatistics analyses. The abundance of Nitrospira showed uniform distribution along the transections, but significantly decreasing variations were observed from the surface (<20 cm) to the bottom (>20 cm) of sediments. The community diversity and structure of Nitrospira also displayed a remarkable vertical distribution in the DYB (STE), although no significant differences were observed along the transections of STE. Nitrospira marina (N. marina) predominated in the sediments, Candidatus Nitrospira bockiana (Ca. N. bockiana), and Candidatus Nitrospira defluvii (Ca. N. defluvii) were the main Nitrospira species, and Candidatus Nitrospira lenta (Ca. N. lenta) and Nitrospira japonica (N. japonica) also existed with relatively low abundance in the DYB (STE). These findings revealed that Nitrospira species (Ca. N. defluvii, Ca. N. lenta, and N. japonica) derived from activated sludge was also widespread in natural habitats and deduced that the STE may be affected by the pollution derived from terrigenous human activities. The statistical analysis combined with the STE dynamic variation indicated that dissolved organic carbon (DOC), salinity, and ammonium along the sediment depths attributed to the vertical community distribution of the Nitrospira species. In summary, the vertical distribution of Nitrospira and their response to the dynamic physicochemical parameters imply their important role in the nitrite oxidation of the STE and provide insights into the niche differentiation and diversely physiologic metabolism of NOB.
Introduction
The subterranean estuary (STE), an unconfined aquifer mixed with land fresh water and seawater, is the transition zone between land underground runoff and the ocean in the surface estuary (Moore, 1999). Due to the influence of tides, waves, and other geochemical factors, the STE forms a dynamic redox state, which leads to the occurrence of complex biogeochemical reactions and makes the STE become a very active and strong area of biogeochemical reactions (Santos et al., 2009). The inorganic nitrogen in the STE comes from both the input of freshwater groundwater and the mineralization products of organic matter input through seawater circulation (Santos et al., 2009). Therefore, the STE is a key “Filter” of nitrogen compounds derived from terrestrial and marine ecosystems. In recent years, some studies have confirmed that groundwater discharge, especially the submarine groundwater discharge (SGD) of the STE, contributed a large part of the nutrients (especially the nitrogen budget) input to coastal water and has an important impact on the coastal ecosystem (Moore, 1996; Hwang et al., 2005; Lee and Kim, 2007; Smith and Swarzenski, 2012; Sáenz et al., 2012).
A large amount of nitrogen transformation occurred in the STE based on the analysis of geochemical processes in multiple sites (Santos et al., 2009). Nitrification is a process in which microorganisms oxidize ammonia to nitrite and further oxidize nitrite to nitrate. It is an important part of the global nitrogen cycle and plays an important role in maintaining the operation of natural ecosystems (Zehr and Kudela, 2011; Kuypers et al., 2018). Nitrification is generally considered to be catalyzed by two kinds of functional microorganisms: ammonia-oxidizing microorganisms (AOM) and nitrite-oxidizing bacteria (NOB). Compared with the research on AOM, little cognition about NOB has seriously restricted our understanding of the nitrification process and related functional groups.
Nitrite oxidizing bacteria includes seven effectively recognized genera (Daims et al., 2016). Among these NOB genera, Nitrospira belongs to phylum Nitrospirae (Ehrich et al., 1995), which forms an independent branch in phylogenetic analysis, and is considered to be the most diverse and ubiquitous in natural and engineered ecosystems (Fujitani et al., 2013; Park et al., 2017), such as soils, marine sponge, geothermal spring, acid mine drainage, estuary, and subtropical forest (Bartosch et al., 2002; Off et al., 2010; Lebedeva et al., 2011; Ramanathan et al., 2017; Hou et al., 2018; Han et al., 2020). In addition, Nitrospira may be involved in soil nitrification more actively than Nitrobacter by DNA-stable isotope probing (SIP) investigation (Xia et al., 2011). These results suggest Nitrospira plays a predominant role in the nitrite oxidation process and ecological balance in natural ecosystems (Han et al., 2021). However, the community structure and diversity distribution characteristics of Nitrospira are still limited in the STE.
A series of primers targeting the 16S rRNA gene was used to analyze Nitrospira in the environments, but the genetic diversity of Nitrospira makes most primers unable to amplify all subgroups or may amplify non-specific sequences (Pester et al., 2014). Functional genes nxrA, nxrB, and nxrC of NOB, respectively, encode the expression of subunits α, β, and γ of nitrite oxidoreductase (NXB). The multicopies of the nxrB gene in Nitrospira may benefit the synthesis of NXB and the transmission of the electronic respiratory chain and accelerate the metabolism and consumption of nitrite in the environments (Hong et al., 2020). In addition, when the nitrite increased in the environment after the starvation treatment, the nxrB expression of Candidatus Nitrospira defluvii (Ca. N. defluvii) increased significantly, while the 16S rRNA expression did not change significantly (Lücker et al., 2010). This result further indicates that the functional gene nxrB is more suitable than 16S rRNA for the biological analysis of Nitrospira.
In this study, sediment samples were collected from the DYB (STE) and the community characteristics of Nitrospira were investigated using the nxrB gene of Nitrospira by high throughput sequencing. Q-PCR assays were employed to analyze the abundance of Nitrospira. The potential key environmental factors influencing the Nitrospira distribution and abundance in the STE sediments were identified by multivariate statistical and correlation analyses. The understanding of the community structure and characteristics of Nitrospira in the special habitat of the STE will expand the current cognition of the NOB distribution characteristics.
Materials and Methods
Sample Collection
The STE mentioned in this study belongs to the shallow type. Sedimentary columns were obtained from the STE in Renshan District, DYB (E 114.80° and N 22.82°) in September 2016. The DYB is a semi open bay in the north of the South China Sea, and its northeast Bay is also a small semi open bay with a maximum width of 2 km (Jiao et al., 2018). The intertidal zone selected in this study is located in the lower reaches of farmland and the population accumulation area of Renshan. Due to the confluence of two small seasonal rivers near the DYB, the overlying water of the intertidal zone is actually saltwater with a mix of terrestrial fresh water and seawater.
After obtaining the sediment column by vibracoring technique, the sediment columns were pushed out, placed on the plastic paper, and marked, respectively, as D1, D2, D3, and D4 from the landward member to the seaward member. The sediments in the sampling profile were heterogeneous. Some areas were mainly sand and gravel, while some areas were mainly silt. Therefore, the depths of different sediment columns were limited by the sediment characters in some areas under the same conditions. The depths of four sedimentary columns D1–D4 were 110, 90, 90, and 80 cm, respectively. The surface and the bottom sediments were, respectively, defined as depth <20 cm and >20 cm. The sampling location and sampling map are shown in Figure 1 as our previous report (Jiao et al., 2018). The name of the sample was marked by the sediment column and depth, as shown in Figure 1. For core D1, every 5 cm sediment section from 5 to 90 cm was mixed as one sample named from D1_5 to D1_90, while every 10 cm sediment section from 90 to 110 cm was named D1_100 and sample D1_110, respectively. For core D2, core D3, and core D4, every 10 cm sediment section was mixed as one sample with the same naming policy. The above sediment column of 5 or 10 cm length was taken as a sample, collected, and sealed in a plastic bag, stored in a refrigerator at −20°C, and transported to the laboratory. After the samples were transported to the laboratory, they were separated into two parts for use. One part of the samples were centrifuged to obtain pore water, which was used to determine the physical and chemical parameters of sediments. The other samples were stored at −80°C for DNA extraction and molecular biological analysis.
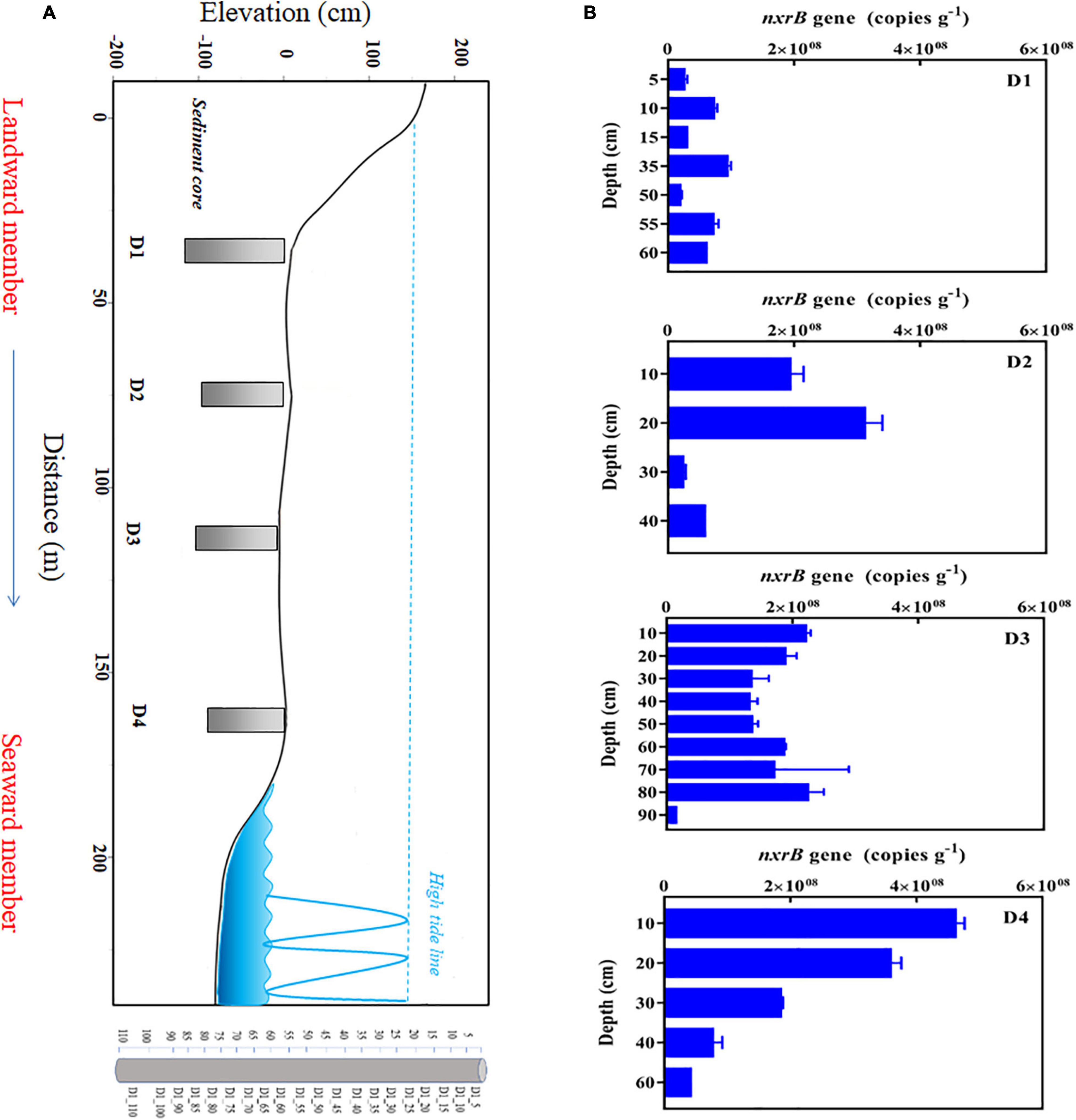
Figure 1. Sampling sites (A) and nxrB gene abundance of Nitrospira the DYB (STE) (B). D1, D2, D3, and D4 (from landward member to seaward member) stands for sediment cores with a depth of 110, 90, 90, and 80 cm, respectively. As shown in core D1, eveiy 5 cm sediment section was mixed as one sample named from D1_5 to D1_90, while eveiy 10 cm sediment section from 90 to 110 cm respectively named D1_100 and sample D1_110. For core D2, Core D3 and core D4, eveiy 10 cm sediment section was mixed as one sample with the same naming policy.
Determination of Environmental Parameters
Oxidation reduction potential (ORP) of sediment samples was measured by SG2 pH meter (SevenGo™, Mettler Toledo, Switzerland). The nitrogen nutrients were measured after the samples were transported back to the laboratory. The concentrations of NO3–, NO2–, and NH4+ were determined by the continuous and rapid determination method with small systems established by our laboratory (Wu et al., 2016; Guan et al., 2017).
DNA Extraction
About 0.3 g (wet weight) sediment from each sample was used for DNA extraction using the PowerSoil DNA Isolation Kit (MO BIO Laboratories, Inc., Switzerland). The total genomic DNA extracted was dissolved in 50 μl nuclease-free water, and the concentration and quality of DNA were determined by Nanodrop™ Lite spectrophotometer (Thermo Scientific, Wilmington, ME, United States), and then stored in a refrigerator at −20°C for further study.
Quantitative PCR
The quantitative PCR (Q-PCR) for functional gene nxrB of Nitrospira was carried out by Roche LightCycler 480 Real-Time PCR using SYBR Green as a fluorescence dye (Promega, Switzerland) with primer set Nxr f27 (5′-GAGGAGCGGCATCCGAAGT-3′), Nxr r617 (5′-GGCABCGRTTCTGGTCG-3′) as optimized in the previous study, which has proven showing good coverage of Nitrospira in environmental samples (Hong et al., 2020). The correlation coefficient R2 of the Q-PCR standard curve was in the range of 0.989–0.995, and the amplification efficiency was 94.9–101%.
PCR Amplification and High Throughput Sequencing
Functional gene nxrB of Nitrospira was amplified using primers Nxr f27/Nxr r617 which can effectively explore the diversity of Nitrospira (Hong et al., 2020). Barcode and adaptor sequences were designed and ligated to the 5′-end of the sequencing forward primers for distinguishing samples in the pyrosequencing runs. The PCR reaction mixtures contained the following components: 12.5 μl GoTaq Green Master Mix (Promega, United States), 0.5 μl primers, 1 μl DNA, and 10.5 μl H2O. The PCR protocol was as follows: initial denaturation at 95°C for 2 min, 32 cycles at 95°C for 45 s, 62°C for 45 s, 72°C for 45 s, and a final extension at 72°C for 5 min. The PCR mixture products were purified using the Agarose Gel DNA Extraction Kit (TaKaRa MiniBEST) and were then prepared for the Illumina MiSeq sequencing.
The quality of purified PCR products was controlled by a 2200 tape station system (Agilent Technologies, Santa Clara, CA, United States). The sequencing process was performed by the Ion Torrent PGM sequencer (life technologies, Grand Island, NY, United States) and the sample library with a barcode on 316 chips. The sequences were assigned to different samples according to barcodes after a series of high-quality controls.
Processing and Analysis of High Throughput Sequencing Data
Nitrospira functional gene nxrB sequences were analyzed by standard operating procedures using Mothur (V.1.35.1)1 (Schloss et al., 2009). In short, the effective original data can be obtained by de-barcoding, de-noising, and pruning the sample sequences. At first, the sequences less than 380 bp in length can be removed, the quality of the sequence was controlled by utilizing the command of the screen.seqs and unique.seqs, and the search and removal of chimera were conducted by the command combinations of pre.cluster, chimera.uchime, and remove.seqs. The effective sequences were compared with the reference database established before, and the final sequences obtained from preprocessing were divided into different operational taxonomic units (OTUs) by the cut-off value of 91% similarity established before (Hong et al., 2020). After removing the rare OTU (sequence number <0.1 ‰ total sequence number), the Shannon–Wiener curve and α diversity-related parameters, such as Chao1 index, Shannon index, and Simpson index, can be generated from the files generated by Mothur. The Shannon index curve was drawn by OriginPro 2017 to verify the sampling depth.
Phylogenetic Analysis
Operational taxonomic units with representative sequences of more than 0.1% of total sequences were defined as the dominant OTUs. The dominant OTUs and seed database were integrated into a FASTA file and imported into MEGA7. The sequences were aligned by running ClustalW. The phylogenetic analysis of the sequences was carried out by using the neighbor-joining method and the phylogenetic tree was constructed. The bootstrap test was set to repeat 1,000 times, and the bootstrap value was not less than 50%. Then, phylogenetic analysis of the representative sequences from dominant OTUs was customized by Evolview (He et al., 2016). Combining the abundance distribution of OTU in each sample with the clustering results, the heat map was drawn to identify similarities and differences in species distribution among samples by the software GraphPad Prism 7.
Biostatistical Analysis
The physical and geochemical parameters of each sample and the clustering and abundance distribution information of dominant OTUs in each sample were integrated into two sheets of the same excel file and imported into Canoco (version: 5.0). Principal component analysis (PCA) was used to analyze the distribution characteristics of the Nitrospira community structure. Redundancy analysis (RDA) was used to analyze the main physical and geochemical parameters affecting the distribution of the Nitrospira community structure. Pearson correlation analysis was used to analyze statistically significant correlations between the abundance distribution of dominant OTUs, the abundance of nxrB gene, and physical and geochemical parameters by R language. The differentiation of the Nitrospira abundance and the diversities between the surface and the bottom sediments was tested by the one-way ANOVA using SPSS (version 22.0). A two-tailed t-test was used to detect the significant effects of environmental parameters on the diversity, abundance, and distribution of Nitrospira in different samples with P-values of 0.05 or 0.01 by using SPSS (version 22.0). The differences among community structures of the nxrB gene were tested by multiresponse permutation procedure (MRPP; Zimmerman et al., 1985) analysis using Vegan in R package with permutation 999.
Nucleotide Sequence Accession Numbers
The accession number CAR005397 of sequences generated in this study has been reported to GSA DATA ADMIN.
Results
Physical and Geochemical Environmental Parameters of the Study Profile
The physical and geochemical parameters of pore water in the sedimentary column were shown in Supplementary Table 1 as our previous report (Jiao et al., 2018). The salinity of the pore water in this study profile ranged from 19 to 30 psu. The salinity of bottom sediments was higher than that of upper sediments, and that of landward member (D1) was higher than that of seaward member (D4). The ORP presented an increasing trend along with the depth and ranged between −385 mv and −76 mv. The concentrations of NO3– varied from 0.00 to 44.07 μmol L–1, and the average concentration of the study profile was 4.90 μmol L–1. The concentrations of NO2– varied from 0.13 to 27.29 μmol L–1, and the average concentration of the study profile was 2.60 μmol L–1. The nitrogen concentrations were high (i.e., NO3– concentration 44.07 μmol L–1 and NO2– – concentration 27.29 μmol L–1) in the surface sediment (0–10 cm) of D3_10, which may be due to the sampling randomness. In the whole study profile, the NH4+ concentrations varied from 0.78 to 19.18 μmol L–1 with an average concentration of 5.15 μmol L–1.
Abundance Distribution of Nitrospira nxrB Gene in the Daya Bay Subterranean Estuary
The abundance distributions of the Nitrospira nxrB gene in the DYB (STE) are shown in Figure 1. Overall, the abundances of the nxrB gene in the sediments of the DYB (STE) ranged from 1.47 × 107 to 4.61 × 108 copies g–1. Among the four sediment columns, the abundance of nxrB gene in sediment column D1 was relatively low compared with the other sediment columns and ranged from 1.90 × 107 copies g–1 to 9.36 × 107 copies g–1 along the sediment depths. The nxrB gene cope number in sediment column D2 ranged from 2.57 × 107 copies g–1 to 3.14 × 108 copies g–1, and showed a reducing trend from the surface to bottom sediments. The abundance of the nxrB gene in sediment column D3 was relatively high and fluctuated in the range of 108 orders of magnitude except for the lowest abundance at 80–90 cm profile. The abundance of the nxrB gene fluctuated from 4.61 × 108 copies g–1 in the surface layer (D4_10) to 4.07 × 107 copies g–1 in the deep layer (D4_60) in sediment column D4 and showed the decreasing trend with depth. There was insignificant variation (F = 2.88, P > 0.05) in the abundance variation of the nxrB gene among the four sediment columns, which indicated that Nitrospira bacteria were uniformly distributed in the DYB (STE) from the landward member to the seaward member. However, the nxrB gene abundance varied significantly along the sediment depth in the DYB (STE) (F = 5.96, P < 0.05) with higher abundance in the surface sediments than the bottom sediments.
Diversity and Richness Analysis of Nitrospira in DYB (Subterranean Estuary) Based on High-Throughput Sequencing of nxrB Gene
Every sediment sample screened 2,500 sequences with a total of 25 sediment samples for diversity analysis. Some samples with too few sequences (sequence number <1000) were removed to avoid affecting the subsequent diversity analysis. A total of 68,594 high-quality sequences were obtained through the pretreatment of the Mothur platform. The sequences with similarities greater than 91% were classified as the same OTU (Hong et al., 2020) and 689 OTUs were obtained by cluster analysis. After removing the rare OTU (sequence number <6), Nitrospria in the sediment samples of the DYB (STE) could be divided into 62 OTUs according to the nxrB gene, accounting for 98.82% of the total sequence number. The top 20 OTUs in sediments of the DYB (STE) (the sequence number of OTUs more than 0.1%) were defined as the dominant OTUs, which accounted for 97.63% of the total sequence number. The calculation results of sample richness and diversity index (Supplementary Table 2) showed that the coverage of Nitrospira in all samples was higher than 99.55%, which reflected the reliability of diversity index results. Moreover, the Shannon curve tended to be flat with the increase of sequencing depth as shown in Supplementary Figure 1, which indicated that the amount of sequencing data in the sediments of the DYB (STE) was large enough to reflect the vast majority of microbial information in the samples and can be used for the subsequent diversity and community structure analysis of Nitrospira.
The OTU number of Nitrospira ranged from 16 to 40 among the sediment samples. Although the OTU number did not show significant variations from the landward member to the seaward member (F = 1.95, P = 0.16), it generally decreased from the surface to the bottom sediments (F = 5.49, P < 0.05) with the exception of sediment column D1_5 with the lowest value 16. The highest OTU number was detected in sediment column D2_10. In general, the Shannon index also decreased from the surface to the bottom sediment (F = 15.18, P < 0.05) except for sediment column D1_5 with the lowest Shannon index of 1.08. The higher Shannon indexes were, respectively, observed in sediment columns D3 (D3_10) and D4 (D4_10) with values of 2.08 and 2.02. The Chao1 and Simpson indexes showed a similar changing variation as the OTU number and Shannon index among samples from the surface to the bottom sediment (F = 5.49, P < 0.05; F = 8.01, P < 0.05). Overall, the Nitrospira diversities of the surface sediments were generally higher than those of the bottom sediments in the DYB (STE).
Phylogenetic and Community Distribution Analysis of Nitrospira in DYB (Subterranean Estuary)
The representative sequences of dominant OTUs and reference sequences were extracted and the phylogenetic tree was constructed, as shown in Figure 2. The obtained nxrB genes of Nitrospira shared 83.58 to 94.17% identity with each other and 84.92 to 95.84% similarity with the sequences matched with the known Nitrospira genus in the Genbank. All retrieved sequences could be divided into seven branches that could be affiliated to the known genus Nitrospira. OTU04 was affiliated to Nitrospira marina (N. marina), with 95.84% similarity. OTU01, OTU02, and OTU12 also showed a close relationship with N. marina with up 90.42% similarity. OTU06 was closely related to Candidatus Nitrospira bockiana (Ca. N. bockiana). OTU07 was affiliated with Candidatus Nitrospira defluvii (Ca. N. defluvii) with 93.33% similarity. OTU11 was clustered with Candidatus Nitrospira nitrifcans (Ca. N. nitrifcans) with 89.38% similarity. OTU9 and OTU15 were closely related to Nitrospira japonica (N. japonica) with 90.83 and 94.38% similarity, respectively. OTU16 was affiliated with Nitrospira calida (N. calida) with 91.25% similarity. OTU18 was closely affiliated to Candidatus Nitrospira lenta (Ca. N. lenta) with 90.00% similarity. The remaining OTUs (OTU03, 05, 08, 10, 13, 14, 17, 19, and 20) shared low similarity and did not group closely with the known Nitrospira genus, whereas they still clustered distinctly with Nitrospira. A heat map based on the abundance distribution and clustering results of dominant OTUs (Figure 3) showed that OTU01, OTU02, OTU03, OTU04, OTU05, and OTU08 were widespread in all samples, while OTU06 and OTU07 mainly existed in the bottom and surface sediments, respectively, which suggested that the Nitrospira community should be distinct along the sediment depth.
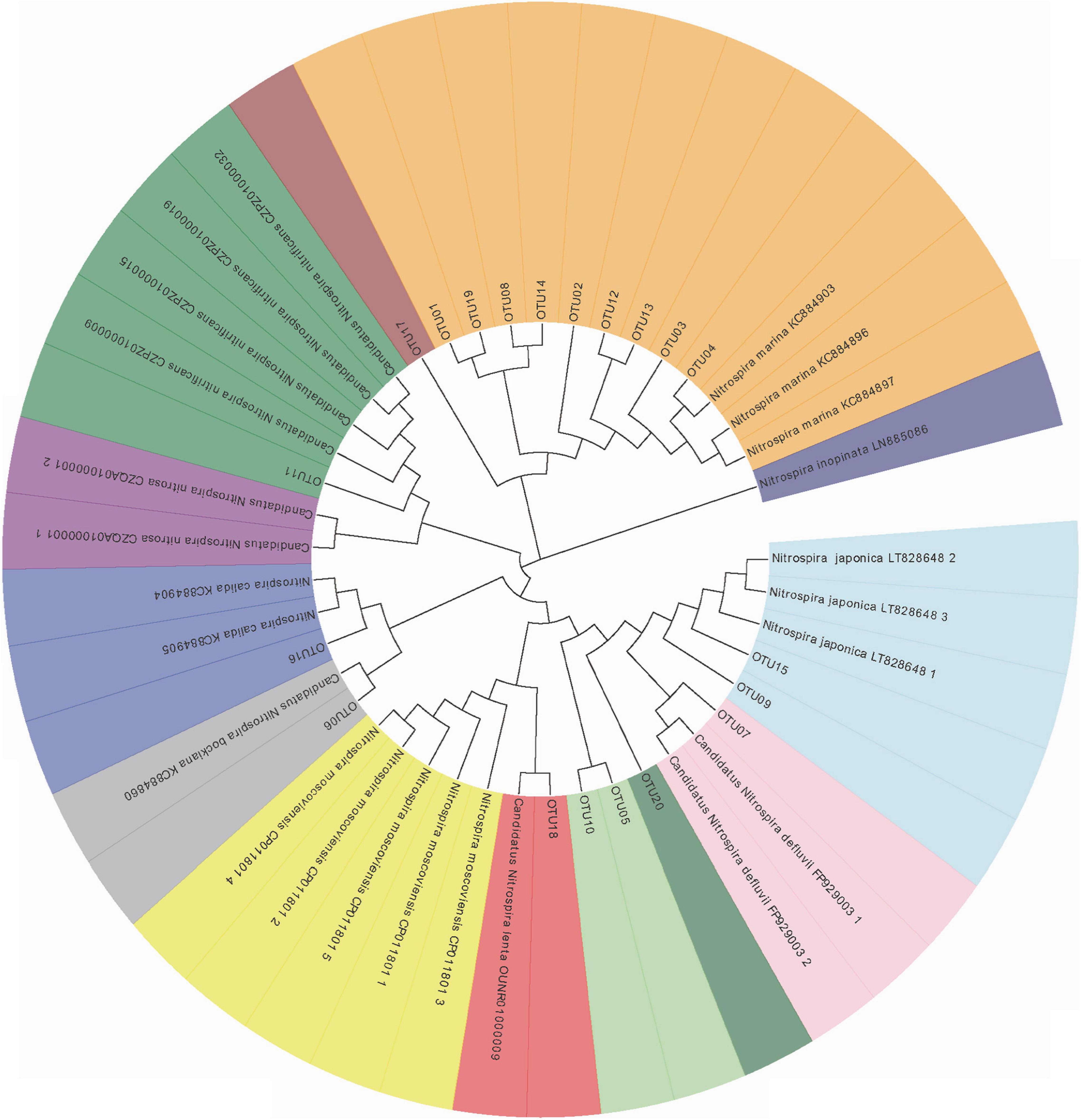
Figure 2. Neighbor-joining phylogenetic tree exhibiting the clustering relationship between the known Nitrosopira species and representative sequences of dominant OTUs obtained from the DYB (STE). Sequences with bootstrap value (1000 replicates) higher than 50% were grouped together and are shape with the same color.
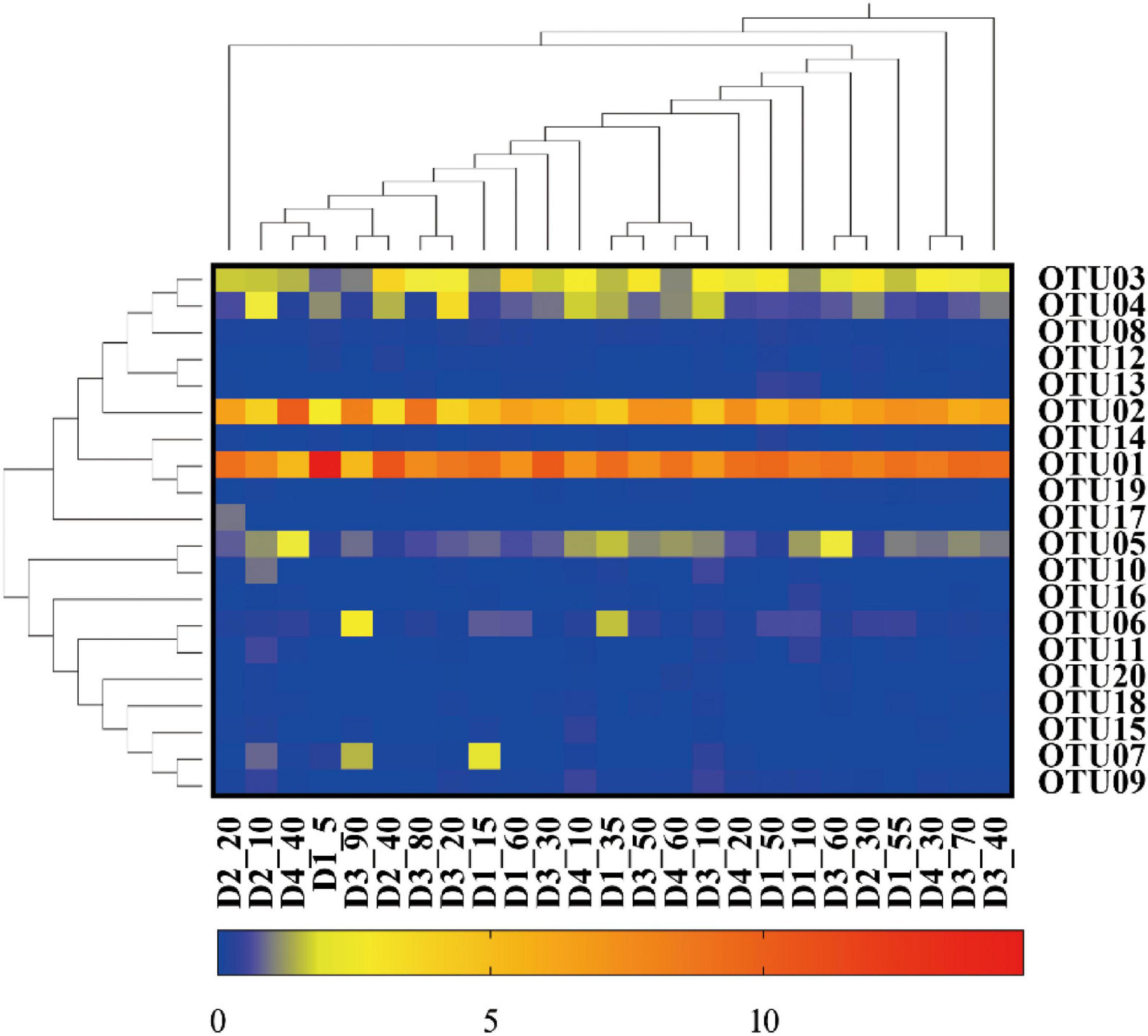
Figure 3. Heat map of Nitrospira in sediment of the DYB (STE). Dl, D2, D3, and D4 stands for sediment cores with a depth of 110, 90, 90, and 80 cm, respectively. As shown in core Dl, every 5 cm sediment section was mixed as one sample named from D15 to Dl_90, while sample Dl_100 and sample Dl_110 represent sediment sections from 90 to 100 cm and from 100 to 110 cm, respectively. For core D2, Core D3 and core D4, every 10 cm sediment section was mixed as one sample with the same naming policy. The 20 dominant OTUs in sediments of the DYB (STE) (OTUs of sequence number >0.1%) were clustered, which accounted for 97.63% of the total sequence number.
The overall community composition and relative abundance of Nitrospira species based on phylogenetic clustering analysis were exhibited in Supplementary Figure 2. Among the seven known classified Nitrospira genus, N. marina, Ca. N. bockiana and Ca. N. defluvii were the main Nitrospira species, with the relative abundance proportion 78.41, 2.21, and 1.06%, respectively. N. marina is commonly distributed and predominated in all sediment samples. Although the distribution of Ca. N. bockiana was also ubiquitous in all sediment samples, its heterogeneous distribution among the surface and bottom sediments was exhibited with relatively high abundance in the bottom sediments of D1_35, D1_60, D2_30, D3_90, and D4_40. However, Ca. N. defluvii did not ubiquitously distribute in all samples and exhibited the opposite changing trend compared with Ca. N. bockiana. The relative abundance of Ca. N. defluvii was higher in the surface sediments of D1_15, D2_10, D3_10, and D4_10 with the exception of D3_90, and Ca. N. defluvii was not detected in nine samples from the bottom sediments. Furthermore, a significant difference (P < 0.05, Supplementary Table 3) was discovered in the community composition of Nitrospira between the surface sediments and bottom sediments, although no significant difference (D1/D2, D1/D3, D1/D4, D2/D3, D2/D4, D3/D4, all of P > 0.05) existed in the community composition of Nitrospira from the landward member D1 to the seaward member D4.
The difference of community structure among samples was performed by PCA as shown in Figure 4. The contribution rates of the first and second components in PCA were 31.92 and 19.72%, respectively. The surface sediment columns of D1, D2, D3, and D4 (D1_10, D2_10, D2_20, D3_10, and D4_10) except D1_5 and D1_15 were gathered together, while most of the bottom sedimentary samples except D3_90 were grouped together. The analysis of MRPP indicated that the Nitrospira community structure from the DYB (STE) was not significantly different on the total level (all, P > 0.05, Supplementary Table 3), whereas there was a significant inconsistent community structure of Nitrospira between the surface and the bottom sediments (Surface/Bottom, P = 0.044).
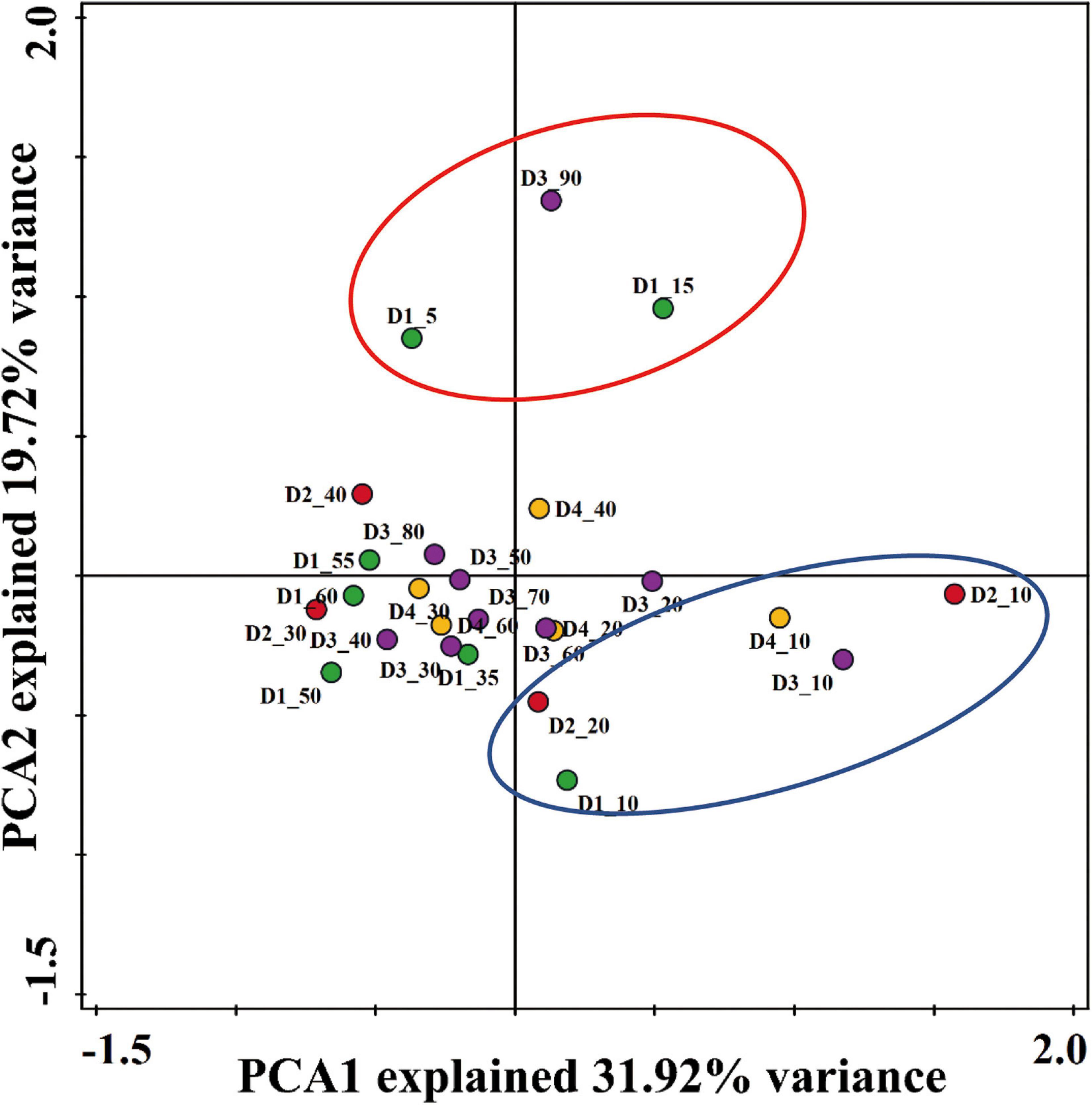
Figure 4. Principal component analysis (PCA) of Nitrospira nxrB gene sequences retrieved from sediments of the DYB (STE). Horizontal and vertical axes represent the first two dimensions, axis 1 and axis 2, respectively.
Relationship Between Environmental Factors and Nitrospira Community Structure in the DYB (Subterranean Estuary)
The correlation between the community structure of Nitrospira based on main OTUs and environmental factors was analyzed by RDA, which showed that the analysis model was invalid based on the Monte Carlo test (F = 1.2, p > 0.05). That is to say, the environmental factors determined in this study could not adequately explain the composition of Nitrospira community structure in the sediments of Daya Bay (STE), and other environmental factors which were not determined might affect the distributions of Nitrospira community structure. The correlations between environmental factors and diversity indexes of Daya Bay STE were plotted and shown in Figure 5. Shannon index was positively correlated with nitrite (r = 0.41, p < 0.05). The number of OTU was negatively correlated with salinity (r = −0.57, p < 0.01), and significantly decreased with depth (r = −0.40, p < 0.05). The abundance of nxrB gene was positively correlated with nitrate (r = 0.407, p < 0.05) and significantly negatively correlated with ammonium (r = −0.42, p < 0.05) and salinity (r = −0.48, p < 0.05), but did not significantly correlate with nitrite.
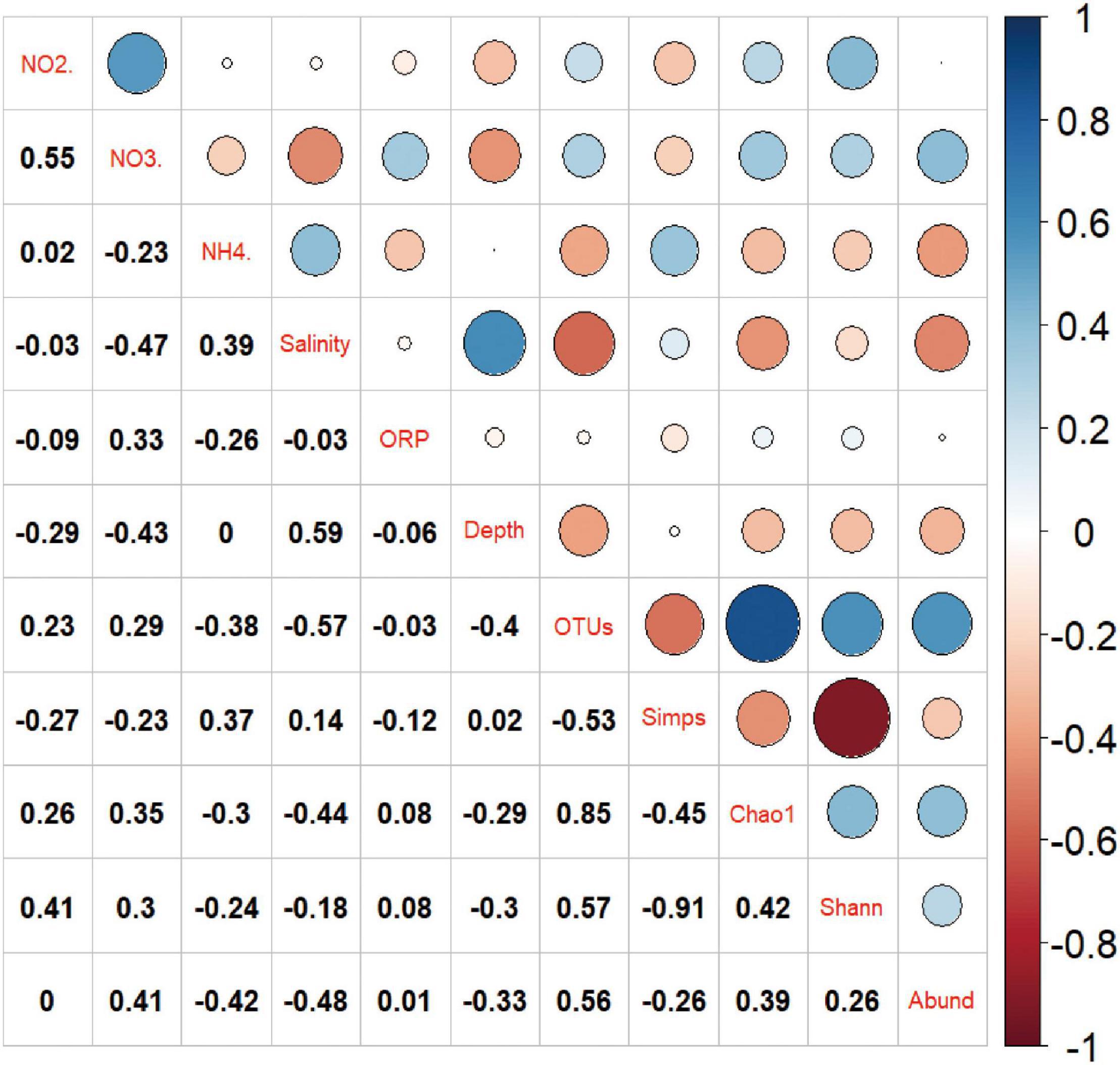
Figure 5. Pearson correlation analysis of environmental parameters and diversity. Chaol stands for Chaol estimator, Shann stands for Shannon index, Simps stands for Simpson index, N02 stands for nitrite, N03 stands for nitrate and NH4 represents ammonia, OTUs represents the number of OTU and Abund stands for the abundance of nxrB gene. Blue and red denote positive and negative correlations, respectively. The r-values of pearson correlation analysis were shown in the grid.
The correlations between environmental factors and dominant OTUs were calculated by the Pearson correlation analysis. The data with statistically significant correlation was shown in Supplementary Table 4 in the Supplementary Information. OTU02, OTU08, and OTU14 were positively correlated with depth, ammonium, and salinity, respectively, while OTU04 was negatively correlated with depth. OTU06 was positively correlated with salinity and increased with depth. OTU09 was positively correlated with nitrate and nitrite and decreased with depth. OTU18 was positively correlated with nitrite concentration.
Discussion
Distributions of Nitrospira Community Structure in the DYB (Subterranean Estuary)
There are at least six lineages of Nitrospira according to the phylogenetic analysis of 16S rRNA gene sequences (Daims and Wagner, 2018). N. marina belonging to the so-called Nitrospira lineage IV (Daims et al., 2001) has been detected previously in saltwater aquaria (Hovanec et al., 1998), marine biofilters (Tal et al., 2003), and marine recirculation aquaculture system (Foesel et al., 2008). In addition, N. marina was also the major group in the sediments of Nanliu (NL) River Estuary (Zhao et al., 2021) and in the water column of the Pearl River in winter (Hong et al., 2021). N. marina was predominant and ubiquitous in the DYB (STE), which predicted the potentially important role of N. marina in nitrite oxidation during nitrification in the STE. The genome and proteome of the first marine Nitrospira isolate indicate that N. marina is highly metabolically versatile, which might enable it to adapt to life in the oxic marine environments and even survive under conditions unfavorable for aerobic nitrite oxidation with fluctuating levels of electron acceptors and donors (Bayer et al., 2021).
Candidatus Nitrospira bockiana and N. calida belonging to the Nitrospira lineages V and VI, respectively, are considered to be strict chemolithoautotrophic species and organotrophic growth of these two species is not observed (Lebedeva et al., 2008, 2011). Their relatively low abundances in the DYB (STE) may infer that rich organic matter exists in the land–sea interface, which might adversely affect the development and growth of these two species. Ca. N. defluvii belonging to Nitrospira lineages I was most important for sewage treatment (Lücker et al., 2010) and was also found in the downstream river of wastewater discharge area (Cébron and Garnier, 2005). Ca. N. defluvii was first detected in the waters of PRE which likely resulted from land source pollution (Hong et al., 2021). Ca. N. lenta and N. japonica belonged to Nitrospira lineages II mainly inhabited in activated sludge from wastewater treatment plants (Ushiki et al., 2013; Boris et al., 2015), while Ca. N. lenta also dominated in the waters of PRE (Hong et al., 2021). Ca. N. defluvii, Ca. N. lenta, and N. japonica were all present with relatively low abundance in Daya Bay (STE). These findings revealed that members of Nitrospira lineage I was also widespread in natural habitats, and confirmed the versatile distribution of representatives of Nitrospira lineage II (Boris et al., 2015). On the other hand, it can be deduced that STE may be affected by the pollution derived from terrigenous human activities. The discharge of domestic wastewater from the population and the agricultural gathering area may lead to organic pollution in the STE. The emergence of anammox Ca. Kuenenia in this study profile (Jiao et al., 2018) also proved this view.
The discovery of comammox (with the capability for the complete oxidation of ammonia to nitrate in a single organism) has fundamentally overthrown the conventional understanding of the nitrification process with two-step (Daims et al., 2015). All comammox Nitrospira found in natural ecosystems was so far identified in representatives of Nitrospira lineage II (Zhao et al., 2021). However, comammox Nitrospira generally dominates the ammonia oxidation in acidic soils (Hu et al., 2021), and appears to be absent in marine systems (Daims et al., 2015; Bayer et al., 2021). The general existence of comammox Nitrospira in a wide range of coastal wetlands and Chongming eastern intertidal sediments indicates the potential role of comammox Nitrospira in the estuarine coastal system (Sun et al., 2020, 2021). OTU11 affiliated to Ca. N. nitrifcans belonged to comammox with a low proportion in the DYB (STE). This may be due to the amplification limit of primers, and amoA gene of comammox might broaden our cognition of comammox in the STE. Groundwater flow and chemical transport in STE are potentially important implications for chemical fluxes from aquifers to coastal waters (Robinson et al., 2007). How different types of nitrifiers respond to the dynamic changes may provide new insights into the ecological niche specificity of nitrifying microorganisms in STE. Therefore, the detection of comammox Nitrospira in Daya Bay (STE) deserves further study.
Environmental Factors Affecting the Abundance and Distribution of Nitrospira in Subterranean Estuary
A distinctly vertical pattern of Nitrospira distribution was revealed considering the surface to the bottom zones of the STE sediments. The abundance of the nxrB gene was maintained in the range of 107–108 copies g–1 in the DYB (STE) sediments, which was 1–2 orders of magnitude lower than that of NOB in surface sediments (0–5 cm) in the Nanliu River estuary (108–1010) (Zhao et al., 2021). Moreover, apparent depth-related differentiation of abundance was detected in the DYB (STE), which resembles previous studies with the lowest microbial abundance in the anaerobic zone (depth of 80–100 cm) in sediments of STE located at Gloucester Point (GP-STE), the beach of Virginia, United States (Hong et al., 2019). In addition, the Nitrospira diversity indexes (Shannon, Chao1, and Simpson) were all obviously higher on the surface than the bottom sediments. Although the predominant of N. marina may lead to the little differentiation in Nitrospira community among the sediments from the landward member to the seaward member of the STE, the distribution of Ca. N. defluvii and Ca. N. bockiana showed the opposite changing trend along with depths. Moreover, the overall community structure exhibited significant differences along the sediment depths based on MRPP and PCA analysis. The vertical distribution of bacteria at GP-STE suggested the predictable changes in permeable STE sediment from surface to bottom layers (Hong et al., 2019). Therefore, the vertical distribution of Nitrospira seems to be shaped with the gradient variation of physiochemical parameters along with the sediments’ depths, such as salinity, organic matter, and nitrogen species.
Salinity is one of the most important factors which impacts the community composition and distribution of nitrifying microorganisms in ecosystems (Zhao et al., 2021). The lower diversity of Nitrospira is exhibited in bottom sediments with relatively high salinity at the DYB (STE). A less diverse microbiology community appeared to be associated with salinity in most cases (Hong et al., 2019). N. marina affiliated to lineage IV has widely adapted to variable salinity (Zhao et al., 2021), which may make it widely present in salinity ecosystems, including deep sea, estuary area, and saline soils (Pester et al., 2014; Hong et al., 2021; Li et al., 2021; Zhao et al., 2021). Ca. N. bockiana affiliated with LineageVmight also adapt to salinity, due to its positive relationship with salinity in DYB (STE) (Supplementary Table 3). Lineage II (N. japonica and Ca. N. lenta) were also the main groups with the adaption of variable salinity, while Lineage I (Ca. N. defluvii) appeared to be less adaptable salt than Lineage II and IV (Zhao et al., 2021). This may be also one of the reasons for the higher abundance of Ca. N. defluvii in the surface sediments of the STE with relatively low salinity. However, salinity could interact with the other environmental parameters. Salinity showed positive relationships with the contents of ammonium in this study (Figure 5). Salinity could control ammonium concentrations by influencing its absorption onto sediment and then play an important role in shaping the community distribution of nitrifying microorganisms (Li et al., 2015). In addition, different cations are also affected by salinity, which deserves attention (Zhao et al., 2021). Nitrospira community variation in agriculture soils was found to be significantly affected by potassium levels (Luo et al., 2020).
The study profile of the DYB (STE) was the typical submarine groundwater discharge (SGD) area. SGD is an important component of the global water balance and the rates of SGD play important role in biogeochemical cycling with coastal ecosystems (Smith et al., 2008; Anderson and Emanuel, 2010). Even though up to 96% of total SGD is marine water recirculating through the nearshore by the processes of wave setup and tidal oscillations (Anderson and Emanuel, 2010), fresh SGD is an important pathway of nutrients as a result of biogeochemical additions across the mixing zone of STE (Santos et al., 2009). About 25% of dissolved organic carbon (DOC) and 50% of total nitrogen (TN) inputs were transported by this pathway, and the remainder was associated with a one-dimensional vertical seawater recirculation along a seepage face (Santos et al., 2009). Considering that the sampling sites were located downstream of the population and agricultural accumulation areas, the abundant DOC and nitrogen species in the STE originated from fresh SGD transportation may be important factors affecting the growth, abundance, and distribution of Nitrospira. Han et al. (2018) found that manure could increase the activity and abundance of Nitrospira in the red soil by providing organic matter. N. marina benefit from additions of undefined organic carbon substrates, has adaptations to resist oxidative, osmotic, and is thus chemoorganotroph but not an obligate chemolithoautotroph (Bayer et al., 2021). In addition, the predominance of N. marina might be a general characteristic of ammonium-limited systems (Foesel et al., 2008). Ammonium concentration was relatively low throughout the transect in the DYB (STE), which may be due to the metabolization of ammonium along the flow path in the intertidal subsurface with permeability and long residue time (Jiao et al., 2018). The abundance of the nxrB gene and distribution of OTU02 were both significantly correlated with ammonium in the DYB (STE). Ammonium was also reported to be the most important factor influencing Nitrospira distribution in the water column of the PRE (Hong et al., 2021). Therefore, DOC and ammonium concentrations regulated by the dynamics of the STE may be the main factors driving the community composition and distribution of Nitrospira in the DYB (STE).
The pH, which was not determined in this study, may also be a factor affecting the growth of Nitrospira and was a potential factor affecting the distribution of Nitrospira in the DYB (STE). Low soil pH could accelerate the ionization of ammonium ions, which will affect the ammonia oxidation activity by reducing the available substrate for ammonia oxidizing microorganisms (Han et al., 2018). Due to the spatial dependence of ammonia oxidation and nitrite oxidation, low pH may affect soil NOB activity (Han et al., 2018). In addition, dissolved oxygen (DO) also plays important role in the niche differentiation of functional microorganisms (Hong et al., 2019). DO was responsible for the Nitrospira distribution in the PRE estuary (Hong et al., 2021). Ca. N. defluvii may be inhibited by the elevated DO, which is likely related to the evolutionary history, evolved from anaerobic ancestors (Lücker et al., 2010; Hong et al., 2021). Due to the permeability of the STE sediments (Santos et al., 2009), the gradient DO along the sediment depth may affect the Nitrospira distribution in the DYB (STE).
Although Nitrospira based on functional gene level could represent the diversity and community distribution of NOB in natural habitats, it is necessary to measure the rate of nitrite oxidation in situ and measure the flux during the nitrite oxidation process. On this basis, the contribution of Nitrospira to the process of nitrite oxidation and the internal relationship between Nitrospira and nitrite oxidation could be explored. At present, there are some controversies about the determination methods of nitrite oxidation rate (Daims et al., 2001) and the determination of nitrite oxidation rate needs to be further optimized in the future.
Conclusion
In this study, the abundance, diversity, and community structure of Nitrospira along sediment depths at the DYB (STE) were detected by using high-throughput sequencing of the Nitrospira nxrB gene. From the surface to the bottom sediments, the abundance and diversity of Nitrospira showed a vertical decreasing trend. Although the Nitrospira community showed little differentiation from landward member to seaward member of the DYB (STE), a prominent spatial shift in composition was detected between the surface and bottom sediments. Seven known Nitrospira species were detected in the DYB (STE), but N. marina were predominant, and Ca. N. bockiana and Ca. N. defluvii were the main groups. Such observation not only reflected the wide distribution of various Nitrospira species along the depths but might also be associated with the physiologies of Nitrospira with the adaption and response to the dynamic environmental parameters in the STE. Salinity, DOC, and ammonium were predicted to be the important factors affecting the Nitrospira distribution combined with statistical analysis and dynamic characteristics of SGD transportation in STE. These findings may broaden our new insights into the niche differentiation of Nitrospira under the dynamic and complex changes and provide a guide for future studies on the functional activity and ecological status of Nitrospira in STE environments.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: National Genomics Data Center (NGDC), part of the China National Center for Bioinformation (CNCB) [accession: CRA005397].
Author Contributions
YH was responsible for the conceptualization. LJ was responsible for the experimental operation. WS and LJ prepared and wrote the original draft. YH, WS, and MW wrote, reviewed, and edited the manuscript. JW and LJ are responsible for field sampling. YH and WS funded the acquisition. All authors read and approved the final manuscript.
Funding
This work was funded by the National Natural Science Foundation of China (Nos. 42006137 and 31870100), Basic and Applied Basic Research Foundation of Guangdong Province (Nos. 2019B1515120066 and 2020A1515110597), Guangdong Key Area R&D Program (2018B020205002), Scientific Research Fund of talent introduction of Guangdong University of Petrochemical Technology (2019rc108), and Demonstration of harmless treatment of shrimp tail water in high-place pond [Letter of Agricultural Bureau in Maoming, Guangdong (2019) NO. 440].
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors would like to thank the reviewers for their valued criticism and suggestions.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.822939/full#supplementary-material
Footnotes
References
Anderson, W. P., and Emanuel, R. E. (2010). Effect of interannual climate oscillations on rates of submarine groundwater discharge. Water Resour. Res. 46:W05503. doi: 10.1029/2009WR008212
Bartosch, S., Hartwig, C., Spieck, E., and Bock, E. (2002). Immunological detection of Nitrospira-like bacteria in various soils. Microb. Ecol. 43, 26–33. doi: 10.2307/4287571
Bayer, B., Saito, M. A., Mcilvin, M. R., Luecker, S., and Santoro, A. E. (2021). Metabolic versatility of the nitrite-oxidizing bacterium Nitrospira marina and its proteomic response to oxygen-limited conditions. ISME J. 15, 1025–1039. doi: 10.1038/s41396-020-00828-3
Boris, N., Sandra, O., Holger, D., and Eva, S. (2015). Improved isolation strategies allowed the phenotypic differentiation of two Nitrospira strains from widespread phylogenetic lineages. FEMS Microbiol. Ecol. 91, 1–11. doi: 10.1093/femsec/fiu031
Cébron, A., and Garnier, J. (2005). Nitrobacter and Nitrospira genera as representatives of nitrite-oxidizing bacteria: detection, quantification and growth along the lower seine river (France). Water Res. 39, 4979–4992. doi: 10.1016/j.watres.2005.10.006
Daims, H., Lebedeva, E. V., Pjevac, P., Han, P., Herbold, C., Albertsen, M., et al. (2015). Complete nitrification by Nitrospira bacteria. Nature 528, 504–509. doi: 10.1038/nature16461
Daims, H., Lücker, S., and Wagner, M. (2016). A new perspective on microbes formerly known as nitrite-oxidizing bacteria. Trends Microbiol. 24, 699–712. doi: 10.1016/j.tim.2016.05.004
Daims, H., Nielsen, J. L., Nielsen, P. H., Schleifer, K. H., and Wagner, M. (2001). In situ characterization of Nitrospira-like nitrite oxidizing bacteria active in wastewater treatment plants. Appl. Environ. Microbiol. 67, 5273–5284. doi: 10.1128/AEM.67.11.5273-5284.2001
Ehrich, S., Behrens, D., Lebedeva, E., Ludwig, W., and Bock, E. (1995). A new obligately chemolithoautotrophic, nitrite-oxidizing bacterium, Nitrospira moscoviensis sp. nov. and its phylogenetic relationship. Arch. Microbiol. 164, 16–23. doi: 10.1007/BF02568729
Foesel, B. U., Gieseke, A., Schwermer, C., Stief, P., Koch, L., Cytryn, E., et al. (2008). Nitrosomonas Nm143-like ammonia oxidizers and Nitrospira marina-like nitrite oxidizers dominate the nitrifier community in a marine aquaculture biofilm. FEMS Microbiol. Ecol. 63, 192–204. doi: 10.1111/j.1574-6941.2007.00418.x
Fujitani, H., Aoi, Y., and Tsuneda, S. (2013). Selective enrichment of two different types of Nitrospira-like nitrite-oxidizing bacteria from a wastewater treatment plant. Microbes. Environ. 28, 236–243. doi: 10.1264/jsme2.ME12209
Guan, F. J., Hong, Y. G., Wu, J. P., Wang, Y., Bin, L. Y., Tang, B., et al. (2017). A fast sodium hypobromite oxidation method for the sequential determination of ammonia nitrogen in small volume. Ecol. Sci. 36, 42–48.
Han, S. H., Zeng, L. Y., Luo, X. S., Xiong, X., Wen, S. H., Wang, B. R., et al. (2018). Shifts in Nitrobacter- and Nitrospira-like nitrite-oxidizing bacterial communities under long-term fertilization practice. Soil Biol. Biochem. 124, 118–125. doi: 10.1016/j.soilbio.2018.05.033
Han, S., Huang, Q., and Chen, W. (2021). Partitioning nitrospira community structure and co-occurrence patterns in a long-term inorganic and organic fertilization soil. J. Soil Sediment 21, 1–10. doi: 10.1007/s11368-020-02813-x
Han, S., Tan, S., Wang, A. C., Chen, W. L., and Huang, Q. Y. (2020). Deciphering belowground nitrifier assemblages with elevational soil sampling in a subtropical forest ecosystem (Mount Lu. China). FEMS Microbiol. Ecol. 96, 1–11. doi: 10.1093/femsec/fiz197
He, Z., Zhang, H., Gao, S., Lercher, M. J., Chen, W. H., and Hu, S. (2016). Evolview v2: an online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 44, W236–W241. doi: 10.1093/nar/gkw370
Hong, Y. G., Jiao, L. J., and Wu, J. P. (2020). New primers, taxonomic database and cut-off value for processing nxrB gene high-throughput sequencing data by MOTHUR. J. Microbiol. Meth. 173:105939. doi: 10.1016/j.mimet.2020.105939
Hong, Y. G., Wu, J. P., Jiao, L. J., Hu, Y. H., Ye, F., Wang, Y., et al. (2021). Shifts in the abundance and community composition of particle-associated and free-living Nitrospira across physicochemical gradients in the Pearl River Estuary. Estuar. Coast 44, 1931–1945. doi: 10.1007/s12237-021-00909-2
Hong, Y. G., Wu, J. P., Wilson, S., and Song, B. (2019). Vertical stratification of sediment microbial communities along geochemical gradients of a subterranean estuary located at the gloucester beach of virginia, united states. Front. Microbiol. 9:3343. doi: 10.3389/fmicb.2018.03343
Hou, L., Xie, X., Wan, X., Kao, S. J., Jiao, N., and Zhang, Y. (2018). Niche differentiation of ammonia and nitrite oxidizers along a salinity gradient from the Pearl River estuary to the South China Sea. Biogeosci. Discuss. 15, 5169–5187. doi: 10.5194/bg-2018-189
Hovanec, T. A., Taylor, L. T., Blakis, A., and DeLong, E. F. (1998). Nitrospira like bacteria associated with nitrite oxidation in freshwater aquaria. Appl. Environ. Microbiol. 64, 258–264. doi: 10.1002/abio.370180412
Hu, J. H., Zhao, Y. X., Yao, X. W., Wang, J. P., Zheng, P., Xi, C. W., et al. (2021). Dominance of comammox nitrospira in soil nitrification - sciencedirect. Sci. Total Environ. 780:146558. doi: 10.1016/j.scitotenv.2021.146558
Hwang, D. W., Kim, G., Lee, Y. W., and Yang, H. S. (2005). Estimating submarine inputs of groundwater and nutrients to a coastal bay using radium isotopes. Mar. Chem. 96, 61–71. doi: 10.1016/j.marchem.2004.11.002
Jiao, L. J., Wu, J. P., He, X., Wen, X. M., Li, Y. B., and Hong, Y. G. (2018). Significant microbial nitrogen loss from denitrification and anammox in the land-sea interface of low permeable sediments. Int. Biodeter. Biodegr. 135, 80–89. doi: 10.1016/j.ibiod.2018.10.002
Kuypers, M., Marchant, H. K., and Kartal, B. (2018). The microbial nitrogen-cycling network. Nat. Rev. Microbiol. 16, 263–276. doi: 10.1038/nrmicro.2018.9
Lebedeva, E. V., Alawi, M., Maixner, F., Jozsa, P. G., Daims, H., and Spieck, E. (2008). Physiological and phylogenetic characterization of a novel lithoautotrophic nitrite-oxidizing bacterium, ‘Candidatus Nitrospira Bockiana’. Int. J. Syst. Evol. Micr. 58, 242–250. doi: 10.1099/ijs.0.65379-0
Lebedeva, E. V., Off, S., Zumbrägel, S., Kruse, M., Shagzhina, A., and Lücker, S. (2011). Isolation and characterization of a moderately thermophilic nitrite oxidizing bacterium from a geothermal spring. FEMS Microbiol. Ecol. 75, 195–204. doi: 10.1111/j.1574-6941.2010.01006.x
Lee, Y. W., and Kim, G. (2007). Linking groundwater-borne nutrients and dinoflagellate red-tide outbreaks in the southern sea of Korea using a Ra tracer. Estuar. Coast. Shelf S. 71, 309–317. doi: 10.1016/j.ecss.2006.08.004
Li, J., Nedwell, D. B., Beddow, J., Dumbrell, A. J., Mckew, B. A., Thorpe, E. L., et al. (2015). AmoA gene abundances and nitrification potential rates suggest that benthic ammonia-oxidizing bacteria and not archaea dominate N cycling in the Colne Estuary, United Kingdom. Appl. Environ. Microbiol. 81, 159–165. doi: 10.1128/AEM.02654-14
Li, X., Wan, W., Zheng, L., Wang, A., Luo, X., Huang, Q., et al. (2021). Community assembly mechanisms and co-occurrence patterns of nitrite-oxidizing bacteria communities in saline soils. Sci. Total Environ. 772:145472. doi: 10.1016/j.scitotenv.2021.145472
Lücker, S., Wagner, M., Maixner, F., Pelletier, E., Koch, H., Vacherie, B., et al. (2010). A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proc Natl. Acad. Sci. U.S.A. 107, 13479–13484. doi: 10.1073/pnas.1003860107
Luo, X., Wang, A., Hou, C., Han, S., Huang, Q., and Chen, W. (2020). The limited effects of carbonaceous material amendments on nitrite-oxidizing bacteria in an Alfisol. Sci. Total Environ. 734:139398. doi: 10.1016/j.scitotenv.2020.139398
Moore, W. S. (1996). Large groundwater inputs to coastal waters revealed by 226Ra enrichments. Nature 380, 612–614. doi: 10.1038/380612a0
Moore, W. S. (1999). The subterranean estuary: a reaction zone of ground water and sea water. Mar. Chem. 65, 111–125. doi: 10.1016/S0304-4203(99)00014-6
Off, S., Alawi, M., and Spieck, E. (2010). Enrichment and physiological characterization of a novel Nitrospira-like bacterium obtained from a marine sponge. Appl. Environ. Microbiol. 76, 4640–4646. doi: 10.1128/aem.00320-10
Park, M. R., Park, H., and Chandran, K. (2017). Molecular and kinetic characterization of planktonic Nitrospira spp. selectively enriched from activated sludge. Environ. Sci. Technol. 51, 2720–2728. doi: 10.1021/acs.est.6b05184
Pester, M., Maixner, F., Berry, D., Rattei, T., Koch, H., Lücker, S., et al. (2014). NxrB encoding the beta subunit of nitrite oxidoreductase as functional and phylogenetic marker for nitrite-oxidizing Nitrospira. Environ. Microbiol. 16, 3055–3071. doi: 10.1111/1462-2920.12300
Ramanathan, B., Boddicker, A. M., Roane, T. M., and Mosier, A. C. (2017). Nitrifier gene abundance and diversity in sediments impacted by acid mine drainage. Front. Microbiol. 8:2136. doi: 10.3389/fmicb.2017.02136
Robinson, C., Li, L., and Barry, A. D. (2007). Effect of tidal forcing on a subterranean estuary. Ade. Water Resour. 30, 851–865. doi: 10.1016/j.advwatres.2006.07.006
Sáenz, J. P., Hopmans, E. C., Dan, R., Henderson, P. B., Charette, M. A., Schouten, S., et al. (2012). Distribution of anaerobic ammonia-oxidizing bacteria in a subterranean estuary. Mar. Chem. 13, 7–13. doi: 10.1016/j.marchem.2012.04.004
Santos, I. R., Burnett, W. C., Dittmar, T., Suryaputra, I., and Chanton, J. (2009). Tidal pumping drives nutrient and dissolved organic matter dynamics in a gulf of mexico subterranean estuary. Geochim. Cosmochim. AC. 73, 1325–1339. doi: 10.1016/j.gca.2008.11.029
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microb. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Smith, C. G., and Swarzenski, P. W. (2012). An investigation of submarine groundwater-borne nutrient fluxes to the west florida shelf and recurrent harmful algal blooms. Limnol. Oceanogr. 57, 471–485. doi: 10.4319/lo.2012.57.3.0896
Smith, C. G., Cable, J. E., and Martin, J. B. (2008). Episodic high intensity mixing events in a subterranean estuary: effects of tropical cyclones. Limnol. Oceanogr. 53, 666–674. doi: 10.2307/40006450
Sun, D., Liu, M., Hou, L., Zhao, M., Tang, X., Zhao, Q., et al. (2021). Community structure and abundance of comammox nitrospira in chongming eastern intertidal sediments. J. Soil Sediment 21, 3213–3224. doi: 10.1007/s11368-021-02940-z
Sun, D., Tang, X., Zhao, M., Zhang, Z., Hou, L., Liu, M., et al. (2020). Distribution and diversity of comammox Nitrospira in coastal wetlands of China. Front. Microbiol. 11:589268. doi: 10.3389/fmicb.2020.589268
Tal, Y., Watts, J. E. M., Schreier, S. B., Sowers, K. R., and Schreier, H. J. (2003). Characterization of the microbial community and nitrogen transformation processes associated with moving bed bioreactors in a closed recirculated mariculture system. Aquaculture 215, 187–202. doi: 10.1016/S0044-8486(02)00372-1
Ushiki, N., Fujitani, H., Aoi, Y., and Tsuneda, S. (2013). Isolation of nitrospira belonging to sublineage ii from a wastewater treatment plant. Microbes. Environ. 28, 346–353. doi: 10.1264/jsme2.me13042
Wu, J. P., Hong, Y. G., Guan, F., Wang, Y., Tan, Y., Yue, W., et al. (2016). A rapid and high-throughput microplate spectrophotometric method for field measurement of nitrate in seawater and freshwater. Sci. Rep. 6:20165. doi: 10.1038/srep20165
Xia, W., Zhang, C., Zeng, X., Feng, Y., Weng, J., Lin, X., et al. (2011). Autotrophic growth of nitrifying community in an agricultural soil. ISME J. 5, 1226–1236. doi: 10.1038/ismej.2011.5
Zehr, J. P., and Kudela, R. M. (2011). Nitrogen cycle of the open ocean: from genes to ecosystems. Annu. Rev. Mar. Sci. 3, 197–225. doi: 10.1146/annurev-marine-120709-142819
Zhao, M. Y., Tang, X. F., Sun, D. Y., Hou, L. J., Liu, M., Zhao, Q., et al. (2021). Salinity gradients shape the nitrifier community composition in Nanliu River estuary sediments and the ecophysiology of comammox Nitrospira inopinata. Sci. Total Environ. 795:148768. doi: 10.1016/j.scitotenv.2021.148768
Keywords: nitrite oxidization, Nitrospira, diversity, subterranean estuary, Daya Bay
Citation: Sun W, Jiao L, Wu J, Wei M and Hong Y (2022) Distribution, Diversity, and Abundance of Nitrite Oxidizing Bacteria in the Subterranean Estuary of the Daya Bay. Front. Mar. Sci. 9:822939. doi: 10.3389/fmars.2022.822939
Received: 26 November 2021; Accepted: 18 January 2022;
Published: 17 February 2022.
Edited by:
Meilin Wu, South China Sea Institute of Oceanology, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Hui Wang, Shantou University, ChinaYuchun Yang, Sun Yat-sen University, China
Yonggang Yang, Guangdong Institute of Microbiology, Guangdong Academy of Science, China
Jing Wang, Tianjin Normal University, China
Copyright © 2022 Sun, Jiao, Wu, Wei and Hong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yiguo Hong, eWdob25nQGd6aHUuZWR1LmNu
 Wei Sun
Wei Sun Lijing Jiao2
Lijing Jiao2 Mingken Wei
Mingken Wei Yiguo Hong
Yiguo Hong