- 1Key Laboratory for Sustainable Utilization of Marine Fisheries Resources, Ministry of Agriculture and Rural Affairs, Yellow Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Qingdao, China
- 2Function Laboratory for Marine Fisheries Science and Food Production Processes, Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
- 3Institute of Biomedical Engineering, College of Life Sciences, Qingdao University, Qingdao, China
Saline-alkali water is distributed all over the world and affects the development of fisheries. The ridge tail white prawn Exopalaemon carinicauda is an economically important shrimp in China, which has excellent environmental tolerance. However, due to its complex genetic structure, there have been few studies on its alkalinity-adaptation mechanisms. In order to explore the molecular mechanisms of E. carinicauda in adapting to the alkaline water, mRNA and miRNA transcriptomes from the gills of E. carinicauda were determined. The results showed that after alkalinity stress, the structures of the gill and hepatopancreas were disorganized; however, E. carinicauda could still maintain vital signs. Transcriptome results showed that ATP binding protein and carbonic anhydrase played an important role in alkalinity-adaptation. At the same time, a large number of immune-related genes were up-regulated, which protect E. carinicauda from bacterial infection. MiRNAs also played an important role in alkalinity-adaptation. A total of 24 miRNAs were identified as differentially expressed after alkalinity stress, and up-regulated miRNAs might active the GnRH signaling pathway and accelerate the synthesis and secretion of aldosterone, which might maintain the balance of osmotic pressure in E. carinicauda to adapt to alkaline environment. These results provide a better understanding of the alkalinity-adaptation mechanism of economic aquatic animals and provide theoretical basis for breeding in the future.
Introduction
Saline-alkali water is distributed all over the world with high alkalinity, high salinity, high pH, and complex ion composition. There were 45.87 million hm2 of saline-alkali water in China, which is distributed across 19 provinces and autonomous regions. Generally, most aquatic animals cannot grow and reproduce in saline-alkali water, which makes saline-alkali waters a global low-yielding water resource, which greatly hinders the utilization of saline-alkali water resources. In order to make the best use of saline-alkali water resources, an increasing amount of saline-alkali water has been used for fisheries. To date, underground brackish water, brackish water, and surface brackish water have been used for aquaculture of prawns, freshwater fish, sea cucumbers, and flatfish in China’s Yellow River Delta, Liao River Delta, Hebei, Tianjin, Jilin, Inner Mongolia, and other inland saline-alkali water areas. To effectively use saline-alkali water resources, it is urgent to breed more saline-alkali-resistant aquaculture species and study the saline-alkali stress mechanism of aquatic organisms.
Exopalaemon carinicauda, an important economic shrimp in China, has great adaptive capacity to extreme environments, especially for pH and carbonate alkalinity (Xu et al., 2010; Liu et al., 2018; Ge et al., 2019), and has been recognized as an excellent model for both physiological and evolutionary studies of saline-alkali adaptation in crustaceans. At present, E. carinicauda has been successfully cultivated in coastal saline-alkali waters in Cangzhou, Hebei Province (approximate carbonate alkalinity 3.5–13.0 mmol/L, salinity 5-15, pH 8.3–9.2) and Dongying, Shandong province (approximate carbonate alkalinity 1.4–8.0 mmol/L, salinity 5–8, pH 8.5–10.0). However, the mechanism of alkalinity tolerance in E. carinicauda remained unknown. To date, comparative transcriptome studies on E. carinicauda adapted to thermal stress and viral infection have been performed, and significant differential gene expression that encoded modulators of stress adaptation and tolerance, including heat shock proteins (e.g., heath shock protein 70, heat shock protein 90, heat shock protein 40, and heat shock protein 10), pattern recognition proteins (e.g., lectins, lipopolysaccharide β-1,3-glucan binding protein, and β-1,3-glucan binding protein), antioxidant enzymes (e.g., glutathione-S-transferase, glutamine synthetase, glutathione peroxidase, and copper/zinc superoxide dismutase) (Ge et al., 2017). These results revealed that there are substantial gene expression changes that allow E. carinicauda to adapt to extreme environments. However, the mechanism of alkalinity tolerance has remained unclear in recent years.
MicroRNAs (miRNAs) were well-studied small single-stranded RNAs that regulated gene expression by base-pairing with target mRNAs, which leaded to translational repression and exonucleolytic mRNA decay (Kim et al., 2009; Berezikov, 2011; Urbarova et al., 2018; Liu et al., 2019). MiRNAs were initially transcribed as RNA polymerase II transcripts and then processed by RNases Drosha and Dicer into precursor miRNAs and mature miRNAs, respectively (Gregory et al., 2005; Bartel, 2009). Crustacean miRNAs appeared to contain plant-like features in post-transcriptional gene silencing, and expression profiling indicated that miRNAs were involved in environmental stress and gonadal development (Moran et al., 2014; Meng et al., 2018; Liu et al., 2019). However, few studies were currently available concerning miRNAs during acute carbonate alkalinity stress in E. carinicauda.
High-throughput RNA sequencing was widely used to explore stress resistance mechanisms in aquatic animals (Collins et al., 2015; Hu et al., 2015; Liu et al., 2018; Chen et al., 2019; Shi et al., 2019, 2020; Zhang et al., 2019; Shang et al., 2021). Identification and characterization of the miRNAs and differentially expressed genes (DEGs) in E. carinicauda was an important step for a thorough understanding of the adaptation mechanism to acute carbonate alkalinity stress. In this study, we investigated the mRNA and miRNA transcriptomes from the gills of E. carinicauda using Illumina deep sequencing technology and found 96 and 272 DEGs after 24 h and 48 h alkalinity stress respectively. At the same time, a total of 24 miRNAs were identified as differentially expressed after alkalinity stress. This study provided a better understanding of the function of mRNA and miRNAs after acute carbonate alkalinity stress in E. carinicauda.
Materials and Methods
Sampling and Histology
Two hundreds E. carinicauda, weighting 0.51 ± 0.18 g, were obtained from Haichen Aquaculture Co., Ltd., in Rizhao, China. Before the experiment, the prawns were acclimated at a salinity of 30 and temperature of 23 ± 0.5°C aerated seawater for one week, and 50% of the water was renewed once daily. The prawns were checked every day to confirm that no prawns died before the experiment.
During the experimental process, the carbonate alkalinity of water was maintained at 8.26 mmol/L according to previous study (Qin et al., 2021), adjusted by NaHCO3, and determined by acid titration (Millero et al., 1993). The experiment involved three tanks with 30 prawns per tank (Figure 1). Samples were taken at 0, 24, and 48 h under stress after the carbonate alkalinity of water was adjusted to 8.26 mmol/L. At each time point, four prawns were taken from each tank, and the gill tissues were dissected and mixed into one sample. Then, all samples were rapidly frozen in liquid nitrogen and stored at −80°C until RNA extraction. Thus, nine samples were prepared for the RNA-seq experiments. At the same time, one prawn was taken from each tank at 0, 24, and 48 h, and the gill and hepatopancreas tissues were dissected and placed in 4% paraformaldehyde in PBS for gill and hepatopancreas histology for 24 h. The samples were dehydrated with ethanol (70, 90, 95, and 100%), embedded in paraffin wax serially, obtained a solid paraffin block after cooling, and sectioned at 5 μm. Then, the slide sections were stained with hematoxylin and eosin and observed using a Nikon ECLIPSE microscope (Nikon, Tokyo, Japan).
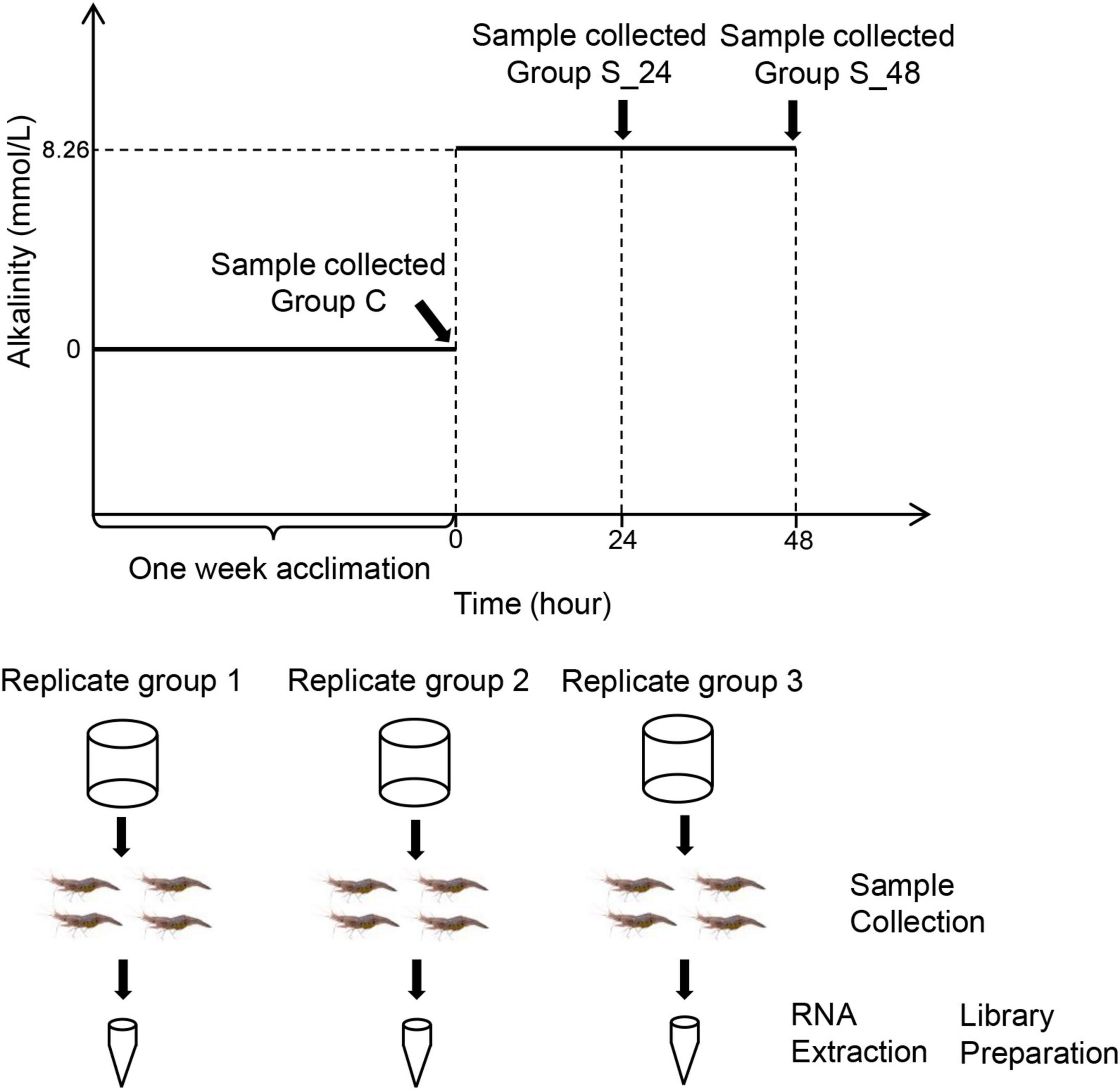
Figure 1. Schematic representation of the acute saline-alkali stress experiment and sample collection, the saline-alkali stress concentration as 10 mmol/L. Samples were collected before the experiment (control group), at 24 h (24 h of saline-alkali stress), and at 48 h (48 h of saline-alkali stress).
RNA Isolation, Sequencing, and Library Preparation
Total RNA was extracted from the gills using TRIzol reagent according to the manufacturer’s instructions (Invitrogen, United States). A 1% agarose gel was used to detect RNA degradation and contamination. RNA purity was determined using a NanoPhotometer® spectrophotometer (Implen, CA, United States). RNA concentration was measured using a Qubit® 2.0 fluorometer (Life Technologies, CA, United States) using the Qubit® RNA detection kit. RNA Nano 6000 detection kit from Agilent Bioanalyzer 2100 System (Agilent Technologies, CA, United States) was used to assess RNA integrity.
The sequencing library was generated using the NEB Next® Ultra™ RNA Library Preparation Kit from Illumina® (NEB, Boston, MA, United States), as recommended by the manufacturer. The mRNA and sRNA-seq libraries were sequenced on an Illumina Novae 6000 and were constructed according to the manuals provided by Illumina by Neogene Company (Beijing, China). Library quality was assessed on an Agilent Bioanalyzer 2100 (Agilent Technologies, CA, United States) using DNA High Sensitivity Chips. All original transcriptome data were deposited in the National Center for Biotechnology Information (NCBI), with the accession number PRJNA782370.
Bioinformatic Analysis
The initial raw reads for Illumina sequencing were filtered to remove adaptor sequences and low-quality reads, and Q20, Q30, and GC content were calculated. All downstream analyses were based on high-quality, clean reads. The clean reads of each RNA-seq library were aligned to the reference transcriptome in a previous study using STAR (version 2.5.0b) to obtain a perfect match and only one mismatch read for further annotation and analysis. The false discovery rate (FDR)-corrected P value was corrected using the EBseq package version 1.24.0, and corrected P value < 0.05, and | log2(fold change)| ≥ 1 was set as the threshold for significant differential expression.
Gene ontology (GO) function and KEGG pathway enrichment analyses were performed on all DEGs in each comparison using the R packages of GOseq and KOBAS software version 3.3.0. Only the GO terms and KEGG pathways were identified as highly expressed in DEGs with the corrected P value < 0.05.
All clean reads were searched against the Rfam and GenBank databases to exclude rRNAs, tRNAs, snRNAs, and snoRNAs. The distribution of small RNA read lengths was determined and reads within the range of 18--35 nt were selected for the following analysis. Then, the conserved miRNAs were identified using the miRBase database (version 21.01). Additionally, the Mirdeep2 software tool was used to analyze potential novel miRNAs (Friedlnder et al., 2012).
The sequences over the length of 18 bp were submitted to Mirdeep2 to get the mapping sequences for further analysis. To obtain the differentially expressed miRNAs between samples, the expression levels of miRNAs were normalized as previously described (Liu et al., 2019). Audic-Claverie was used to analyze differentially expressed miRNAs (DEMs) among libraries (Tiňo, 2009). The criteria for significant expression were defined as P < 0.05, and transcripts per million > 2.
ORF Finder2 was used to identify the 5′ UTR, 3′ UTR, and protein-coding regions. Potential target genes of the DEMs were analyzed using Miranda software (S ≥ 150, ΔG ≤ −30 kcal/mol) using 3′UTR data extracted from the transcriptomes of E. carinicauda (John et al., 2004). In addition, GO enrichment analysis (corrected P value < 0.05) and KEGG pathway analysis (P < 0.05) were conducted to categorize the function and to analyze the pathway of putative target genes of DEMs.
Quantitative Real-Time-PCR Analysis
To further validate our DEG library, qRT-PCR analysis was performed on RNA samples originally used for transcriptome sequencing. Six genes with large expression differences were selected, including acetaldehyde dehydrogenase (ALDH), ATP citrate lyase (ACL), actin (ACT), sarcoplasmic reticulum Ca2+-ATP enzyme (SER2a), blood coagulating protein (BC), DEAD-box (DEA), and Premier v5 (Premier Biosoft International, Palo Alto, CA, United States) to design specific primer pairs. The first strand of cDNA was synthesized from 1 mg of RNA using the Prime-Script™ RT Reagent Kit with gDNA Eraser (Takara, Japan). qPCR was performed using SYBR Premix Ex Taq (TaKaRa, Japan) on the ABI PRISM 7500 Sequence Detection system (Applied Biosystems, United States) according to the manufacturer’s instructions, and, 18s rRNA (GenBank accession number: GQ369794) was used to detect the expression level of genes, and all experiments were repeated three times (Ge et al., 2017). The PCR program was set at 95°C for 30 s, then 40 cycles of 95°C for 5 s and 60°C for 34 s, followed by 1 cycle of 95°C for 15 s, 60°C for 1 min, and 95°C for 15 s. The fold change in target gene expression level was calculated using a relative quantitative method. Data were statistically analyzed using SigmaStat software (Jandel Scientific, San Rafael, CA, United States). One-way analyses of variance were assessed between-group comparisons, followed by Student-Newman-Keuls tests. Values with P < 0.05 were considered statistically significant.
Results
Gill and Hepatopancreas Morphology
The results of hematoxylin-eosin staining of gill and hepatopancreas sections from E. carinicauda are shown in Figure 2. Normal gill tissue of the E. carinicauda was phyllodes, the gill filaments were arranged in order with clear structure, the cuticle was smooth and flat, and the epithelial cells were regular, the gap at the end of branchial filament was flat or round, and there were blood cells in the microvasculature (Figure 2A). After 24 h of alkalinity stress, the gill filaments were enlarged, the cuticle was deformed, and the space under the cuticle was expanded. At the same time, the epithelial cells were damaged and the number of blood cells were decreased (Figure 2B). After 48 h of alkaline stress, the gill filaments were arranged disorderly, the cuticle and epithelial cells were necrotic and exfoliated, the space under the stratum corneum was seriously deformed, and the structure of the capillary network was ruptured (Figure 2C). The hepatopancreatic tissues of E. carinicauda showed that normal hepatopancreatic cells were closely packed and structurally intact with small vacuoles (Figure 2D). After 24 h of alkaline stress, the hepatopancreas was loosely arranged, and the vacuoles were enlarged (Figure 2E). After 48 h of alkaline stress, the vacuoles in the hepatopancreas was enlarged and the hepatopancreas cells lost its basic structure (Figure 2F).
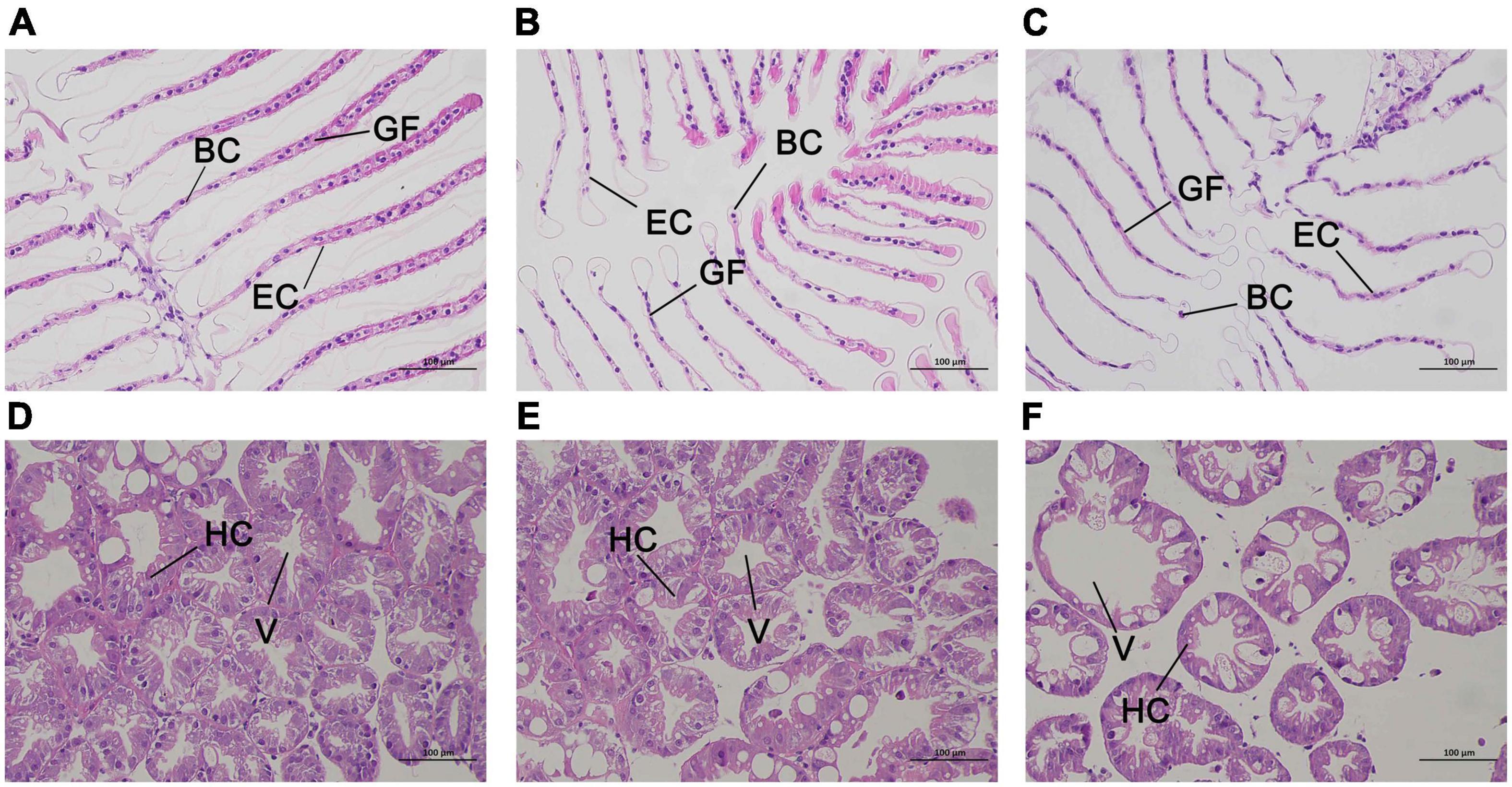
Figure 2. Representative HE staining and TEM photomicrographs of Exopalaemon carinicauda gill (A–C) and hepatopancreas (C–E) after saline-alkali stress. Among them, panels (A,D) were control group, panels (B,E) were saline- alkali stress for 24 h, panels (C,F) were saline-alkali stress for 48 h. BC, blood cell; GF, gill filament; EC, epithelial cell; HC, hepatopancreatic cells; V, vacuole.
Raw Sequencing Data and Quality Statistics
To identify the differences in gene expression of E. carinicauda after exposure to alkalinity stress, nine cDNA libraries were constructed using the Illumina HiSeq 4000 sequencing platform. A total of 543,504,644 clean reads filtered from 54,547,064 paired-end raw reads were identified, with Q30 (%) above 93.74% (Supplementary Table 1). After mapping these clean reads to reference sequences of E. carinicauda, an average of 55,460,344 (63.01%), 62,453,121 (63.14%), and 63, 254, and 750 (68.37%) were found to be unique matches for groups C, S24, and S48, respectively (Supplementary Table 2). A total of 14,112 unigenes with a total length of 41,067,851 bp, ranging from 92 to 10,458 bp (with an average of 2,910 bp) were obtained (Figure 3A).
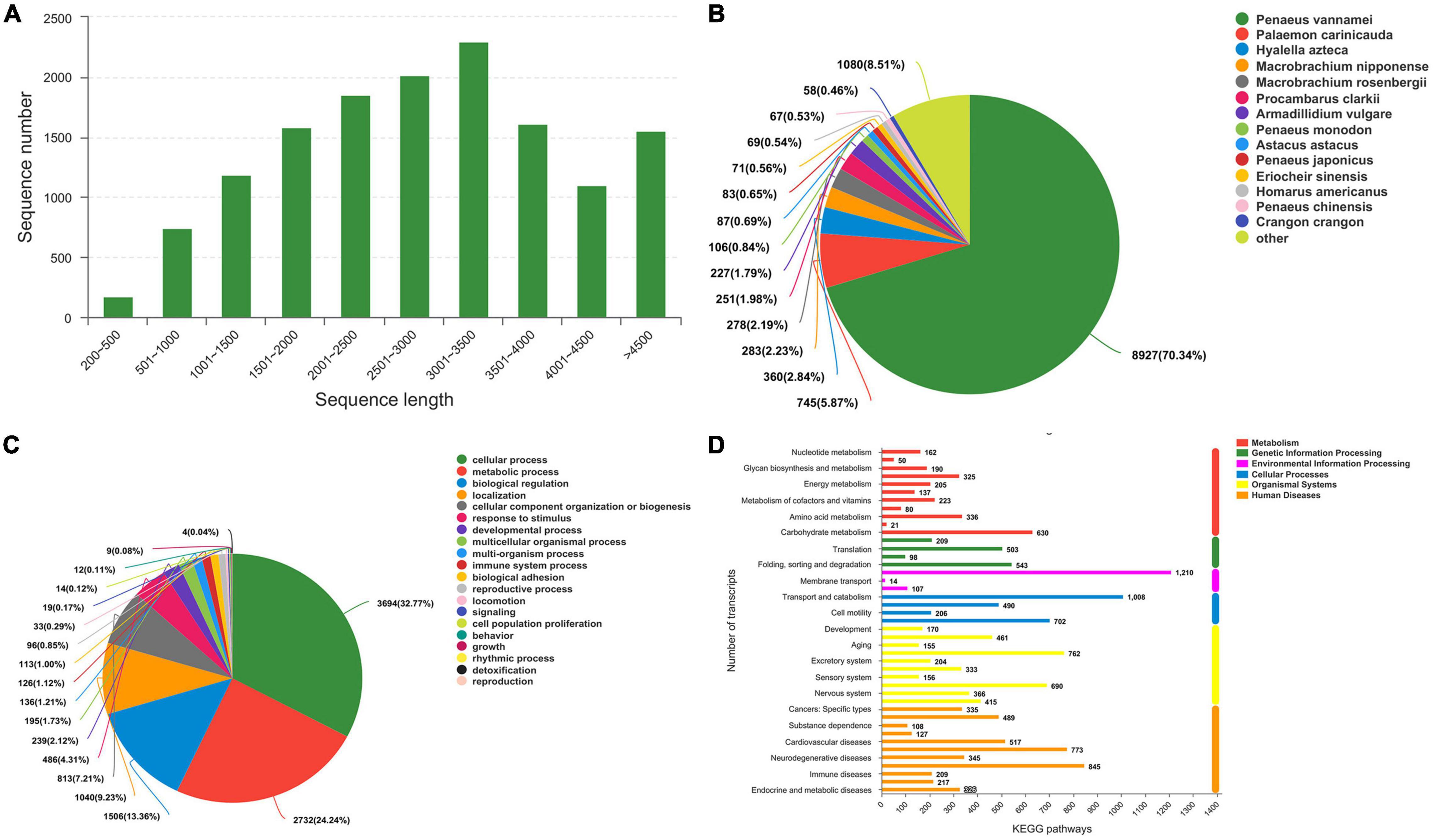
Figure 3. Transcriptome sequence length distribution and annotation. (A) Sequence length distribution. (B) Species Distribution. (C) GO Classification_BP. (D) Histogram of KEGG.
The potential functions of transcripts were annotated using BLAST against the nr databases with an E-value cutoff of 1 e–5. Of the 13, 704 transcripts, 12, 383 (90.36%) matched nr proteins, and the rest had no match (Supplementary Table 3). The highest matched species was Penaeus vannamei with 70.34% of the sequences, while only 5.87% were similar to the available E. carinicauda (Figure 3B). This result could be explained by the fact that the P. vannamei genome was completely sequenced (ASM378908v1).
Analysis of Differentially Expressed Genes
A total of 10,431 annotated transcripts were classified into the GO categories (Supplementary Table 3). The top four categories of molecular function were cellular process (32.77%), metabolic process, biological regulation, and localization (Figure 3C). A total of 7,788 annotated transcripts were classified into the KEGG categories (Figure 3D and Supplementary Table 3). Candidate genes involved in alkalinity stress responses were identified from DEGs that significantly enriched GO terms and KEGG pathways.
A total of 368 genes were identified as differentially expressed in pairwise comparisons (C vs. S24, and C vs. S48, log2[fold change] > 1, corrected P value < 0.05). Among all DEGs, 96 DEGs were identified after 24 h of alkaline stress; among them, 59 genes were up-regulated and 40 genes were down-regulated; 272 DEGs were identified after 48 h of alkalinity stress, among which 147 genes were up-regulated and 125 genes were down regulated (Figure 4A). A Venn diagram of DEGs at each time point is shown in Figure 4B. There were 25 genes were significantly expressed at both 24 and 48 h alkalinity stress.
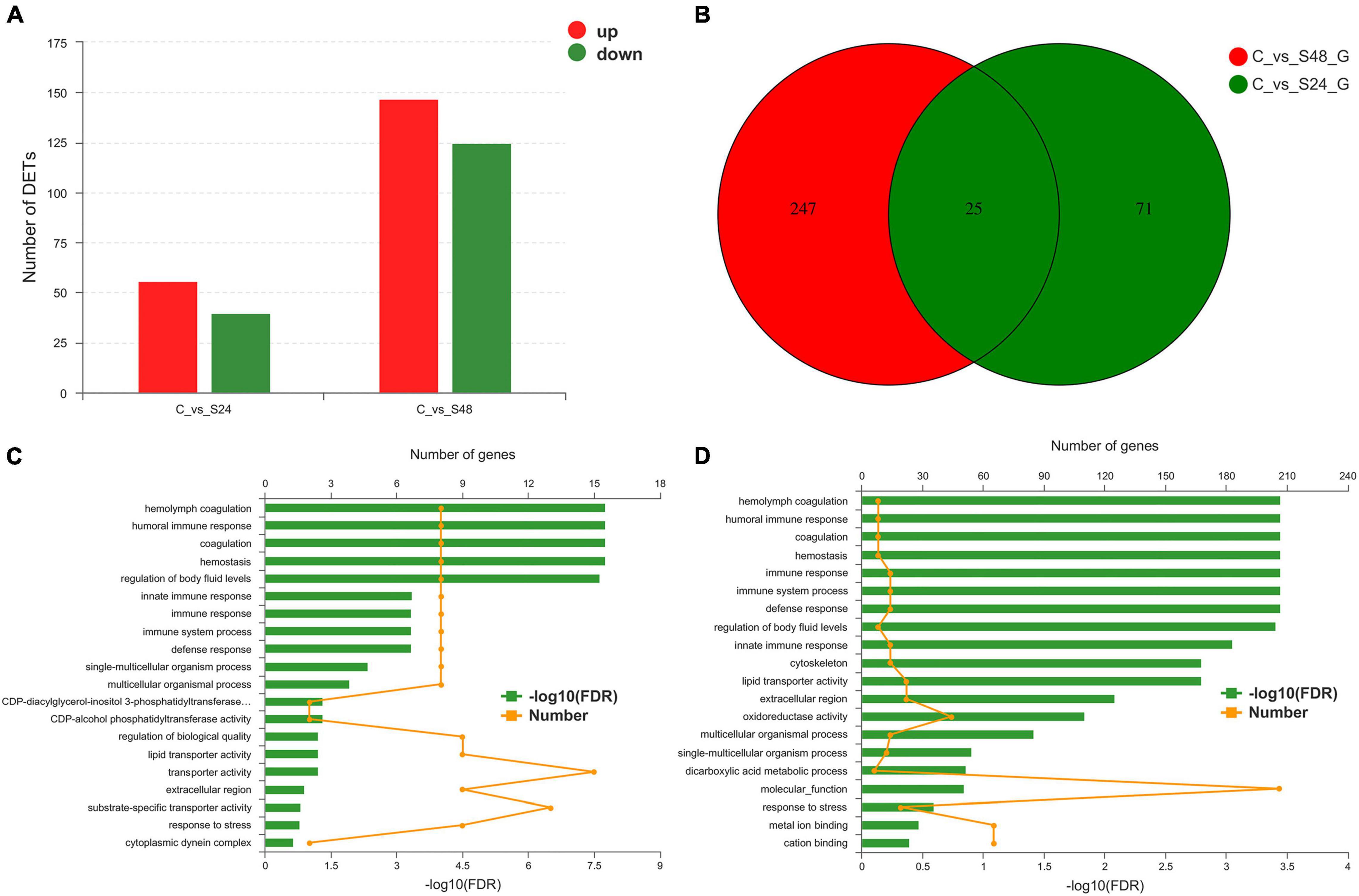
Figure 4. Statistical analysis of differentially expressed genes. (A) Differentially expressed genes statistics in group C vs. S24 and C vs. S48 (C, control group; S24, acute saline-alkali stress for 24 h; S48 acute saline-alkali stress for 48 h). (B) DEGs number and Venn diagram of overlap of the different group. (C,D) The most enriched Gene Ontology (GO) term analysis and function classifications of differentially expressed genes after 24 h (C) and 48 h (D) acute heat stress.
Gene Ontology Analysis of Significant Differentially Expressed Genes
Differentially expressed genes were enriched in 20 GO terms after 24 and 48 h of alkalinity stress (corrected P < 0.05). Of these GO terms, hemolymph coagulation, humoral immune response, coagulation, hemostasis, and regulation of body fluid levels were significantly enriched after 24 h of alkaline stress (Figure 4C). Hemolymph coagulation, humoral immune response, coagulation, hemostasis, immune response, immune system process, defense response, and regulation of body fluid levels were significantly enriched after 48 h of alkaline stress (Figure 4D).
Computational Identification of MicroRNA From sRNA Libraries
The length distribution of the identified miRNAs showed a similar pattern, and the predominant length of miRNAs was 22 nt (Supplementary Figure 1). To obtain the details of the matching sRNA, all the sRNAs were compared with the sequence in the specified range of miRBase, and a total of 22 mature miRNAs and 23 precursor miRNAs were obtained (Supplementary Table 4). All sRNAs were compared and annotated; among them, the proportion of known sRNAs in all sRNAs ranged from 16 to 50% (Supplementary Figure 2). Then, the distribution preferences of the bases in different locations were analyzed. Among them, most of the miRNA bases were initiated by U and ended by G (Supplementary Figure 3). Subsequently, the signature hairpin structure of the miRNA precursor was used to predict new miRNAs, and a total of 69 mature RNAs and 70 precursor RNAs were predicted (Supplementary Table 5). The expression levels of miRNAs were analyzed using miRDeep2. The top 10 expressed miRNAs are shown in Figure 5A, and dre-miR-100-5p had the highest expression level. Principal component analysis (PCA) showed that alkalinity stress was an effective factor for miRNA expression (Figure 5B).
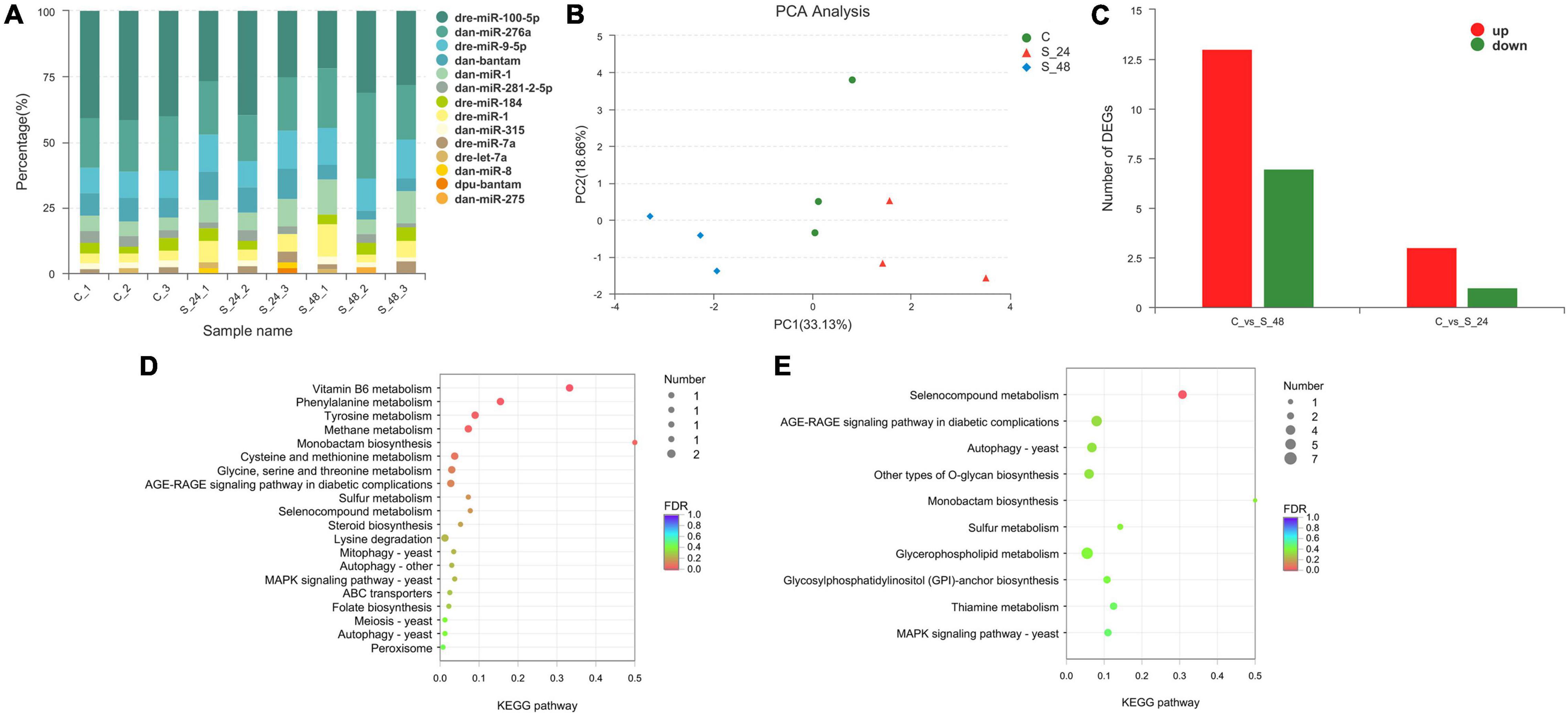
Figure 5. Analysis of the miRNAs in Exopalaemon carinicauda. (A) Top10 expressed miRNAs in different experiment group. (B) Principal component analysis plot of the expression quality of different experiment group. The green circle showed the control group, and the red triangle showed the group after 24 h saline-alkali stress, while the blue rhombus showed the group after 48 h saline-alkali stress. (C) Differentially expressed miRNAs statistics in group C vs. S24 and C vs. S48 (C, control group; S24, acute saline-alkali stress for 24 h; S48 acute saline-alkali stress for 48 h). (D) KEGG analysis of the target genes for differentially expressed miRNAs after 24 h saline-alkali stress. (E) KEGG analysis of the target genes for differentially expressed miRNAs after 48 h saline-alkali stress.
Identification of Differentially Expressed MicroRNAs
A total of 24 miRNAs were identified as differentially expressed in pairwise comparisons (C vs. S24, and C vs. S48, log2[fold change] > 1, corrected P value < 0.05). Among all differentially expressed miRNAs, only 4 miRNAs were identified after 24 h of alkalinity stress; among them, three miRNAs were up-regulated and one miRNA was down-regulated; 20 DEGs were identified after 48 h of alkalinity stress, among which 13 miRNAs were up-regulated and 7 miRNAs were down regulated (Figure 5C). The details of the differentially expressed miRNAs are shown in Table 1. Among them, 11 differentially expressed miRNAs were identified. The target genes for differentially expressed miRNAs were analyzed to understand the molecular functions of miRNAs after alkaline stress. KEGG analysis results are shown in Figures 5D,E, where the predicted target genes of 24 differentially expressed miRNAs were involved in various pathways, among which Vitamin B6 metabolism, phenylalanine metabolism, tyrosine metabolism, methane metabolism, and monobactam biosynthesis were the top five pathways of predicted target genes after 24 h of alkalinity stress, and selenocompound metabolism was the top pathway of predicted target genes after 48 h of alkalinity stress.
Quantitative Real-Time-PCR Analysis
According to the importance of potential gene function, six DEGs (ALDH, acetaldehyde dehydrogenase; ACL, ATP citrate lyase; ACT, actin; SER2a, sarcoplasmic reticulum Ca2+-ATPase; BC, blood coagulating protein; DEA, DEAD-box) were selected for further validation using qRT-PCR. The results of quantitative real-time PCR showed that all up-regulated and down-regulated genes showed the same trend as the high-throughput sequencing data (Figure 6).
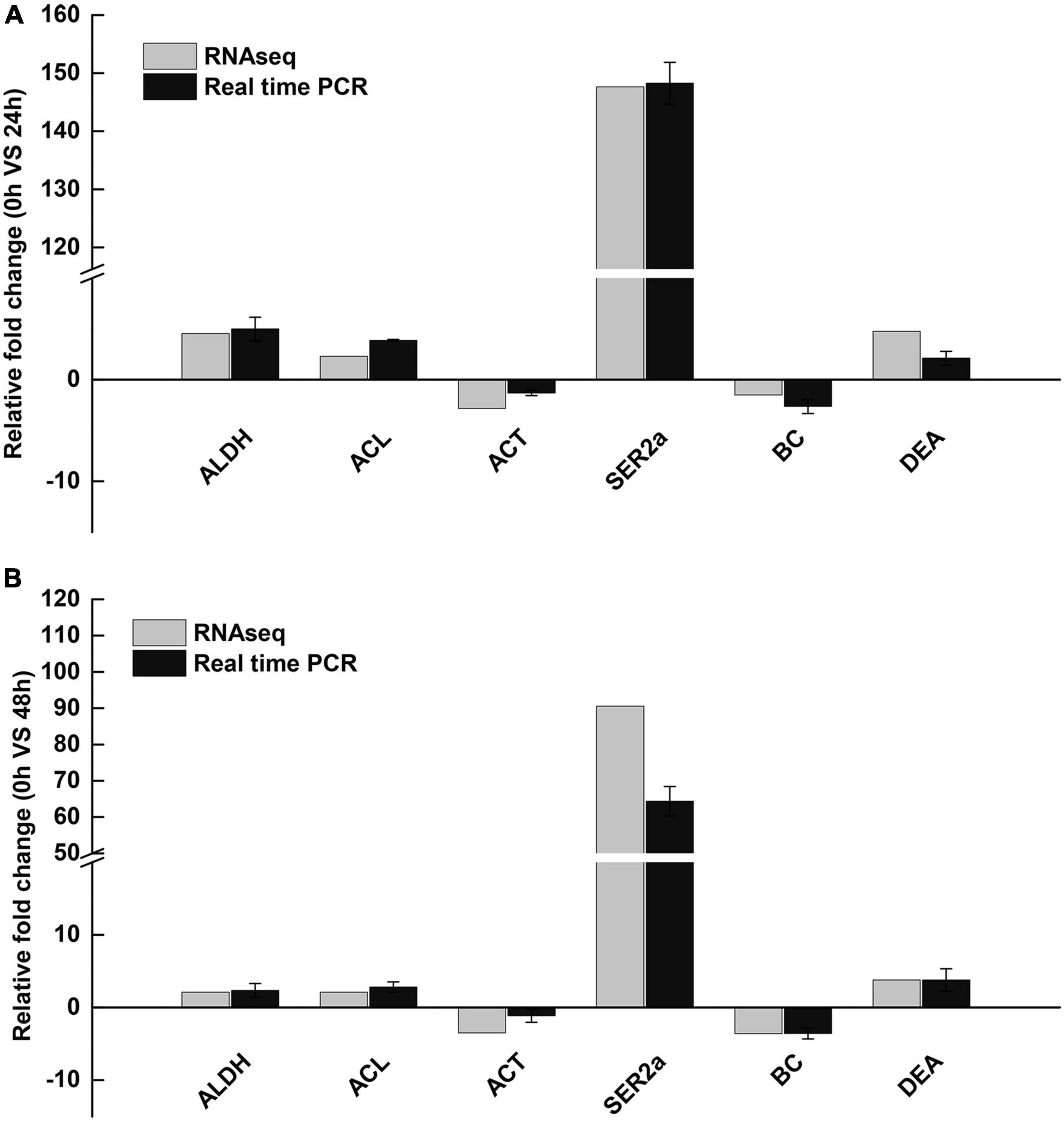
Figure 6. Validation of RNA-Seq results using qRT-PCR. (A) 24 h saline-alkali stress, (B) 48 h saline-alkali stress. ALDH, acetaldehyde dehydrogenase; ACL, ATP citrate lyase; ACT, actin; SER2a, sarcoplasmic reticulum Ca2+-ATPase; BC, blood coagulating protein; DEA, DEAD-box.
Discussion
The gill and hepatopancreas are important metabolic organs of E. carinicauda. The main function of gill tissue is to exchange gas and maintain ionic equilibrium (Henry et al., 2012), which is vulnerable to toxic substances (Bhavan and Geraldine, 2000). The results of this study showed that the gill tissue of E. carinicauda was damaged under alkaline stress, the gill filaments arranged disorderly, the stratum corneum epithelial cells were necrotic and shedding, and the capillary network ruptured with increasing alkalinity stress, which seriously affected the osmotic pressure regulation function of the gill tissue and eventually resulted in the death of E. carinicauda. The histological lesions in gills may result of the stress within short period of time. The similar results were found that when Chalcalburnus tarichi migrate from alkaline water to fresh water, which indicate that these damage in gill tissue may due to the changing alkaline (Oguz, 2015). At the same time, the hepatopancreas was also damaged under alkaline stress, which influences the function of detoxification, excretion, and metabolism of E. carinicauda (Caceci et al., 1988). Previous study also showed the result of epithelium disorganization in the hepatopancreas tubules, and hepatopancreas cells suffer from oxidative damage with high pH stress, which indicated the stress of carbonate alkalinity might have the same mechanism with pH stress (Wang et al., 2018).
There was limited study concerning the adaptation mechanism of crustacean alkalinity in previous studies. In this study, the genes including ATP binding protein kinase and carbonic anhydrase were significantly up-regulated in E. carinicauda, and these genes played a key role in the response to alkalinity stress in E. carinicauda. Carbonic anhydrase, a Zn-containing metal enzyme, played an important role in the regulation of osmotic pressure and ion transport in aquatic animals (Shang et al., 2021). The main role of carbonic anhydrase was to provide H+ for the neutral exchange of Na+/H+ and participates in Cl–/HCO3– transportation (Jillette et al., 2010). Ali et al. (2015) found through transcriptome sequencing that carbonic anhydrase played an important role in red crayfish after pH stress. Ge et al. (2019) found that the carbonic anhydrase gene was significantly up-regulated after pH stress in E. carinicauda, which proved that carbonic anhydrase played an important role in the regulation of osmotic pressure in E. carinicauda. At the same time, carbonic anhydrase played an important role in salinity and pH stress in previous studies on L. vannamei and Portunus trituberculatus (Liu et al., 2015; Pan et al., 2016). In this study, the up-regulated carbonic anhydrase and ATP binding protein kinase might accelerate ion excretion of E. carinicauda, which help E. carinicauda to adapt to the alkalinity environment as soon as possible.
Alkalinity stress may also affect the immune system of aquatic animals (Zhao et al., 2020). In this study, some immune-related genes, like T-cell immunomodulatory protein, HSP90, antimicrobial peptide type 2, zinc finger protein were up-regulated after alkalinity exposure. Transcriptome analysis also revealed that the NOD-receptor signaling pathway and IL-17 signaling pathway were significant enrichment after alkalinity exposure. One explanation was that the alkalinity exposure of E. carinicauda triggered the immune response of the prawn, and up-regulated immune-related genes to protect the prawn from infection. In addition, alkalinity exposure may affect the diet of organism and trigger the intestinal inflammation, then resulting in the change of immune network (Shang et al., 2021).
Gene ontology enrichment analysis of the DEGs of E. carinicauda after alkalinity stress showed that after 24 h of alkalinity stress, hemolymph coagulation, humoral immune response, hemostasis, and humoral regulation were significantly enriched, after 48 h of alkalinity stress, immune response, and stress response were significantly enriched. Many immune-related genes of E. carinicauda were up-regulated after alkalinity stress, including chitinase-related protein genes and antibacterial peptides. Chitinase not only participates in molting and digestion, but also has immune functions (Gayathri et al., 2018). Antimicrobial peptides are natural antibiotics with antibacterial potential and are also used for immune regulation by activating mast cells (Amparyup et al., 2020). Zhao et al. (2020) analyzed the changes in the transcriptome of Nile tilapia under alkaline stress and found that the immune-related genes cytosolic 5-nucleotidase, L-threonine dehydrogenase, and transmembrane superfamily members were significantly up-regulated. Acute carbonate alkalinity stress breaks the balance between the individual and the environment of E. carinicauda, which leads to a disorder in its physiological function. Up-regulation of immune-related genes could be interpreted as an attempt to improve the immune system to adapt to highly alkaline environments, which was consistent with the results of this study (Gao et al., 2021). The results suggested that in order to maintain the stability of the internal environment and normal life activities, the over-expression of immune-related genes was a protective mechanism.
In this study, we obtained 22 known and 69 novel miRNAs using high-throughput sequencing. In these miRNAs, we focused on DEMs to determine the adaptation mechanism of E. carinicauda. We found that dme-mir-9a-5p and dme-mir-1-3p were up-regulated after 24 and 48 h of alkaline stress, indicating that they may be closely related to the alkalinity adaptation of E. carinicauda. To gain insight into the potential function of DEMs, the predicted target genes of these miRNAs were subjected to KEGG pathway analyses. KEGG enrichment analysis of the target genes of the differentially expressed miRNAs of E. carinicauda after alkalinity stress showed that the main enriched pathways were the cholinergic synaptic pathway, the GnRH signaling pathway, and the synthesis and secretion of aldosterone. Among them, the expression of most miRNAs was up-regulated in the synthesis and secretion pathways of aldosterone, indicating that it might be related to the alkali-tolerance mechanism of E. carinicauda. The main function of aldosterone is to regulate the excretion and secretion of Na+ and K+ and maintain the balance of osmotic pressure in the organism, which is of positive significance for E. carinicauda in dealing with a highly alkaline environment (Rash and Lillywhite, 2019). In addition, the GnRH signaling pathway also has a great function in the adaptation of the alkalinity environment in crustaceans. Previous studies have shown that the GnRH signaling pathway played an important role in the reproduction of Macrobrachium rosenbergii (Suwansa-Ard et al., 2016), Penaeus japonicus (Amano et al., 2009), Penaeus monodon (Ngernsoungnern et al., 2008), Eriocheir sinensis (Yuan et al., 2019) and Procambarus clarkii (Guan et al., 2014). Based on this, we speculate that the miRNA enriched in the aldosterone synthesis and secretion pathway in this pathway may play a key role in the alkali resistance mechanism of E. carinicauda, and the specific functions need to be verified later.
Conclusion
Under alkaline stress, the E. carinicauda gill transcriptome results showed that 96 DEGs were identified after 24 h of alkaline stress and 272 DEGs were identified after 48 h of alkalinity stress. ATP binding protein, carbonic anhydrase and a large number of immune-related genes were up-regulated, which played an important role in alkalinity-adaptation of E. carinicauda. A total of 24 miRNAs were identified as differentially expressed after alkalinity stress, among which 11 differentially expressed miRNAs were newly discovered. Up-regulated miRNAs might active the GnRH signaling pathway and accelerate the synthesis and secretion of aldosterone, which might maintain the balance of osmotic pressure in E. carinicauda to adapt to alkaline environment. These results provide a better understanding of the alkalinity-adaptation mechanism of E. carinicauda.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI SRA BioProject, accession no: PRJNA782370.
Ethics Statement
The animal study was reviewed and approved by Institutional Animal Care and Use Committee of Yellow Sea Fisheries Research Institute. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author Contributions
JnL, PL, and JtL designed the research. KS and ML conducted the experiment, preformed the statistical and bioinformatics analyses, and wrote the manuscript. KS, ML, ZQ, and JW obtained the data. All authors reviewed the manuscript.
Funding
This research was supported by the National Key R&D Program of China (2018YFD0901302), National Natural Science Foundation of China (32072974), China Agriculture Research System of MOF and MARA (CARS-48), and the Central Public-interest Scientific Institution Basal Research Fund, CAFS (2020TD46).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.816932/full#supplementary-material
Footnotes
References
Ali, M. Y., Pavasovic, A., Mather, P. B., and Prentis, P. J. (2015). Transcriptome analysis and characterisation of gill-expressed carbonic anhydrase and other key osmoregulatory genes in freshwater crayfish Cherax quadricarinatus. Data Brief. 5, 187–193. doi: 10.1016/j.dib.2015.08.018
Amano, M., Okumura, T., Okubo, K., Amiya, N., and Oka, Y. (2009). Biochemical analysis and immunohistochemical examination of a GnRH-like immunoreactive peptide in the central nervous system of a decapod crustacean, the kuruma prawn (Marsupenaeus japonicus). Zoolog. Sci. 26, 840–845. doi: 10.2108/zsj.26.840
Amparyup, P., Charoensapsri, W., Samaluka, N., Chumtong, P., Yocawibun, P., and Imjongjirak, C. (2020). Transcriptome analysis identifies immune-related genes and antimicrobial peptides in Siamese fighting fish (Betta splendens). Fish Shellfish Immun. 99, 403–413. doi: 10.1016/j.fsi.2020.02.030
Berezikov, E. (2011). Evolution of microRNA diversity and regulation in animals. Nat. Rev. Genet. 12, 846–860. doi: 10.1038/nrg3079
Bhavan, P. S., and Geraldine, P. (2000). Histopathology of the hepatopancreas and gills of the prawn Macrobrachium malcolmsonii exposed to endosulfan. Aquat. Toxicol. 50, 331–339. doi: 10.1016/s0166-445x(00)00096-5
Caceci, T., Neck, K. F., Lewis, D. D. H., and Sis, R. F. (1988). Ultrastructure of the hepatopancreas of the Pacific white shrimp, Penaeus vannamei. J. Mar. Biol. Assoc. U. K. 68, 323–337. doi: 10.1017/s002531540005222x
Chen, B. H., Xu, J., Cui, J., Pu, F., Peng, W. Z., Chen, L., et al. (2019). Transcriptional differences provide insight into environmental acclimatization in wild amur ide (Leuciscus waleckii) during spawning migration from alkalized lake to freshwater river. Genomics 111, 267–276. doi: 10.1016/j.ygeno.2018.11.007
Collins, J. E., Wali, N., Sealy, I. M., Morris, J. A., and Busch-Nentwich, E. M. (2015). High-throughput and quantitative genome-wide messenger RNA sequencing for molecular phenotyping. BMC Genomics 16:578. doi: 10.1186/s12864-015-1788-6
Friedlnder, M. R., Mackowiak, S. D., Li, N., Chen, W., and Nikolaus, R. (2012). miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids. Res. 40, 37–52. doi: 10.1093/nar/gkr688
Gao, H., Ma, H. K., Sun, J. Q., Xu, W. Y., Gao, W., Lai, X. F., et al. (2021). Expression and function analysis of crustacyanin gene family involved in resistance to heavy metal stress and body color formation in Exopalaemon carinicauda. J. Exp. Zool. Parta. 336, 352–363. doi: 10.1002/jez.b.23025
Gayathri, R., Venkatesh, K., Arun, M., Arunkumar, D., and Aziz, A. (2018). Bactericidal and fungistatic activity of peptide derived from GH18 domain of prawn chitinase 3 and its immunological functions during biological stress. Int. J. Biol. Macromol. 106, 1014–1022. doi: 10.1016/j.ijbiomac.2017.08.098
Ge, Q. Q., Li, J., Wang, J. J., Li, J. T., Ge, H. X., and Zhai, Q. Q. (2017). Transcriptome analysis of the hepatopancreas in Exopalaemon carinicauda infected with an AHPND-causing strain of Vibrio parahaemolyticus. Fish Shellfish Immunol. 67, 620–633. doi: 10.1016/j.fsi.2017.06.047
Ge, Q. Q., Li, J., Wang, J. J., Li, Z., and Li, J. (2019). Characterization, functional analysis, and expression levels of three carbonic anhydrases in response to pH and saline-alkaline stresses in the ridgetail white prawn Exopalaemon carinicauda. Cell Stress Chaperones 24, 503–515. doi: 10.1007/s12192-019-00987-z
Gregory, R. I., Chendrimada, T. P., Cooch, N., and Shiekhattar, R. (2005). Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 123, 631–640. doi: 10.1016/j.cell.2005.10.022
Guan, Z. B., Shui, Y., Liao, X. R., Xu, Z. H., and Zhou, X. (2014). Primary structure of a novel gonadotropin-releasing hormone in the ovary of red swamp crayfish Procambarus clarkii. Aquaculture 418, 67–71. doi: 10.1016/j.aquaculture.2013.10.010
Henry, R. P., Lucu, C., Onken, H., and Weihrauch, D. (2012). Multiple functions of the crustacean gill: osmotic/ionic regulation, acid-base balance, ammonia excretion, and bioaccumulation of toxic metals. Front. Physiol. 3:431. doi: 10.3389/fphys.2012.00431
Hu, D. X., Pan, L. Q., Zhao, Q., and Ren, Q. (2015). Transcriptomic response to low salinity stress in gills of the Pacific white shrimp, Litopenaeus vannamei. Mar. Genom. 24, 297–304. doi: 10.1016/j.margen.2015.07.003
Jillette, N., Cammack, L., Lowenstein, M., and Henry, R. P. (2010). Down-regulation of activity and expression of three transport-related proteins in the gills of the euryhaline green crab, Carcinus maenas, in response to high salinity acclimation. Comp. Pestic. Biochem. Phys. 158, 189–193. doi: 10.1016/j.cbpa.2010.10.024
John, B., Enright, A. J., Aravin, A., Tuschl, T., Sander, C., and Marks, D. S. (2004). Human MicroRNA Targets. PLoS Biol. 2:e363. doi: 10.1371/journal.pbio.0020363
Kim, V. N., Han, J., and Siomi, M. C. (2009). Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell. Biol. 10, 126–139. doi: 10.1038/nrm2632
Liu, M., Liu, S., Hu, Y., and Pan, L. (2015). Cloning and expression analysis of two carbonic anhydrase genes in white shrimp Litopenaeus vannamei, induced by pH and salinity stresses. Aquaculture 448, 391–400. doi: 10.1016/j.aquaculture.2015.04.038
Liu, X., Luo, B. Y., Feng, J. B., Zhou, L. X., Ma, K. Y., and Qiu, G. F. (2019). Identification and profiling of microRNAs during gonadal development in the giant freshwater prawn Macrobrachium rosenbergii. Sci. Rep U. K. 9, 2406. doi: 10.1038/s41598-019-38648-x
Liu, Y., Xin, Z. Z., Song, J., Zhu, X. Y., Liu, Q. N., Zhang, D. Z., et al. (2018). Transcriptome analysis reveals potential antioxidant defense mechanisms in Antheraea pernyi in response to zinc stress. J. Agr. Food. Chem. 66, 8132–8141. doi: 10.1021/acs.jafc.8b01645
Meng, X. L., Zhang, X. H., Li, J., and Liu, P. (2018). Identification and comparative profiling of ovarian and testicular microRNAs in the swimming crab Portunus trituberculatus. Gene 640, 6–13. doi: 10.1016/j.gene.2017.10.026
Millero, F. J., Zhang, J. Z., Lee, K., and Campbell, D. M. (1993). Titration alkalinity of seawater. Mar. Chem. 44, 153–165. doi: 10.1016/0304-4203(93)90200-8
Moran, Y., Fredman, D., Praher, D., Li, X. Z., Wee, L. M., Rentzsch, F., et al. (2014). Cnidarian microRNAs frequently regulate targets by cleavage. Genome Res. 24, 651–663. doi: 10.1101/gr.162503.113
Ngernsoungnern, P., Ngernsoungnern, A., Kavanaugh, S., Sobhon, P., Sower, S. A., and Sretarugsa, P. (2008). The presence and distribution of gonadotropin-releasing hormone-liked factor in the central nervous system of the black tiger shrimp, Penaeus monodon. Gen. Comp. Endocr. 155, 613–622. doi: 10.1016/j.ygcen.2007.08.012
Oguz, A. R. (2015). Histological changes in the gill epithelium of endemic Lake Van Fish (Chalcalburnus tarichi) during migration from alkaline water to freshwater. North West J. Zool. 11, 51–57.
Pan, L., Hu, D., Liu, M., and Hu, Y. (2016). Molecular cloning and sequence analysis of two carbonic anhydrase in the swimming crab Portunus trituberculatus and its expression in response to salinity and pH stress. Gene 576, 347–357. doi: 10.1016/j.gene.2015.10.049
Qin, Z., Ge, Q. Q., Wang, J. J., Li, M. D., Liu, P., Li, J., et al. (2021). Comparative transcriptomic and proteomic analysis of Exopalaemon carinicauda in response to alkalinity stress. Front. Mar. Sci. 8:759923. doi: 10.3389/fmars.2021.759923
Rash, R., and Lillywhite, H. B. (2019). Drinking behaviors and water balance in marine vertebrates. Mar. Biol. 166:122.
Shang, X. C., Geng, L. W., Yang, J., Zhang, Y. T., and Xu, W. (2021). Transcriptome analysis reveals the mechanism of alkalinity exposure on spleen oxidative stress, inflammation and immune function of Luciobarbus capito. Ecotoxicol. Environ. Saf. 225:112748. doi: 10.1016/j.ecoenv.2021.112748
Shi, K. P., Dong, S. L., Zhou, Y. G., Li, Y., Gao, Q. F., and Sun, D. J. (2019). RNA-seq reveals temporal differences in the transcriptome response to acute heat stress in the Atlantic salmon (Salmo salar). Comp. Biophys. D 30, 169–178. doi: 10.1016/j.cbd.2018.12.011
Shi, K. P., Li, J. T., Lv, J. J., Liu, P., Li, J., and Li, S. D. (2020). Full-length transcriptome sequences of ridgetail white prawn Exopalaemon carinicauda provide insight into gene expression dynamics during thermal stress. Sci. Total. Environ. 747:141238. doi: 10.1016/j.scitotenv.2020.141238
Suwansa-Ard, S., Zhao, M., Thongbuakaew, T., Chansela, P., Ventura, T., Cummins, S. F., et al. (2016). Gonadotropin-releasing hormone and adipokinetic hormone/corazonin-related peptide in the female prawn. Gen. Comp. Endocr. 236, 70–82. doi: 10.1016/j.ygcen.2016.07.008
Tiňo, P. (2009). Basic properties and information theory of Audic-Claverie statistic for analyzing cDNA arrays. BMC Bioinformatics 10:310. doi: 10.1186/1471-2105-10-310
Urbarova, I., Patel, H., Foret, S., Karlsen, B. O., Jorgensen, T. E., Hall-Spencer, J. M., et al. (2018). Elucidating the small regulatory RNA repertoire of the sea anemone Anemonia viridis based on whole genome and small RNA sequencing. Genome Biol. Evol. 10, 410–426. doi: 10.1093/gbe/evy003
Wang, Z., Cai, C. F., Cao, X. M., Zhu, J. M., He, J., Wu, P., et al. (2018). Supplementation of dietary astaxanthin alleviated oxidative damage induced by chronic high pH stress, and enhanced carapace astaxanthin concentration of Chinese mitten crab Eriocheir sinensis. Aquaculture 483, 230–237. doi: 10.1016/j.aquaculture.2017.10.006
Xu, W. J., Xie, J. J., Shi, H., and Li, C. W. (2010). Hematodinium infections in cultured ridgetail white prawns, Exopalaemon carinicauda, in eastern China. Aquaculture 300, 25–31.
Yuan, L. J., Peng, C., Liu, B. H., Feng, J. B., and Qiu, G. F. (2019). Identification and characterization of a luteinizing hormone receptor (LHR) homolog from the Chinese Mitten Crab Eriocheir sinensis. Int. J. Mol. Sci. 20:1736.
Zhang, Y., Li, Z. Y., Kholodkevich, S., Sharov, A., Feng, Y. J., Ren, N. Q., et al. (2019). Cadmium-induced oxidative stress, histopathology, and transcriptome changes in the hepatopancreas of freshwater crayfish (Procambarus clarkii). Sci. Total Environ. 666, 944–955. doi: 10.1016/j.scitotenv.2019.02.159
Keywords: alkalinity stress, Exopalaemon carinicauda, microRNA, adaptation mechanism, aquaculture
Citation: Shi K, Li M, Qin Z, Wang J, Liu P, Li J and Li J (2022) The Mechanism of Carbonate Alkalinity Exposure on Juvenile Exopalaemon carinicauda With the Transcriptome and MicroRNA Analysis. Front. Mar. Sci. 9:816932. doi: 10.3389/fmars.2022.816932
Received: 17 November 2021; Accepted: 03 January 2022;
Published: 31 January 2022.
Edited by:
Xiangli Tian, Ocean University of China, ChinaCopyright © 2022 Shi, Li, Qin, Wang, Liu, Li and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jitao Li, bGlqdEB5c2ZyaS5hYy5jbg==
†These authors share first authorship
 Kunpeng Shi1,2,3†
Kunpeng Shi1,2,3† Ping Liu
Ping Liu Jitao Li
Jitao Li