
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 15 March 2022
Sec. Microbial Symbioses
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.808696
The ability of symbionts to recolonize their hosts after a period of dysbiosis is essential to maintain a resilient partnership. Many cnidarians rely on photosynthate provided from a large algal symbiont population. Under periods of thermal stress, symbiont densities in host cnidarians decline, and the recovery of hosts is dependent on the re-establishment of symbiosis. The cellular mechanisms that govern this process of colonization are not well-defined and require further exploration. To study this process in the symbiotic sea anemone model Exaiptasia diaphana, commonly called Aiptasia, we developed a non-invasive, efficient method of imaging that uses autofluorescence to measure the abundance of symbiont cells, which were spatially distributed into distinct cell clusters within the gastrodermis of host tentacles. We estimated cell cluster sizes to measure the occurrence of singlets, doublets, and so on up to much larger cell clusters, and characterized colonization patterns by native and non-native symbionts. Native symbiont Breviolum minutum rapidly recolonized hosts and rapidly exited under elevated temperature, with increased bleaching susceptibility for larger symbiont clusters. In contrast, populations of non-native symbionts Symbiodinium microadriaticum and Durusdinium trenchii persisted at low levels under elevated temperature. To identify mechanisms driving colonization patterns, we simulated symbiont population changes through time and determined that migration was necessary to create observed patterns (i.e., egression of symbionts from larger clusters to establish new clusters). Our results support a mechanism where symbionts repopulate hosts in a predictable cluster pattern, and provide novel evidence that colonization requires both localized proliferation and continuous migration.
The ecological success of many cnidarian species relies on their nutritional symbiosis with endosymbiotic dinoflagellate algae (family Symbiodiniaceae; LaJeunesse et al., 2018) to provide nutrient-rich photosynthate in nutrient-poor environments (Muscatine and Porter, 1977). These photosynthetic algae reside in vesicles called symbiosomes inside gastrodermal cells of the host cnidarian (Fitt and Trench, 1983; Wakefield et al., 2000). In the majority of symbiotic cnidarians, symbionts are newly acquired with each host generation (Hartmann et al., 2017). Once ingested into the cnidarian gastrovascular cavity, algal symbionts colonize the cnidarian gastrodermis via host cell phagocytic pathways (Colley and Trench, 1983; Davy et al., 2012). Symbiont populations then increase to populate the entire host gastrodermis. This intracellular relationship is vulnerable to environmental stress throughout the life history of the host. Altered temperature or nutrient conditions can result in dysbiosis, known as cnidarian bleaching, wherein algal populations decrease within host tissues to the point where the host cnidarian can die from a lack of nutrition (Oakley and Davy, 2018). As environmental changes occur more rapidly worldwide, the mechanisms governing these host-symbiont dynamics and the ability to recover from dysbiosis has become consequential for the fate of coral reef ecosystems across the planet.
The establishment of algal populations within cnidarians can either occur through initial colonization or colonization after symbiont population depletion. During initial colonization, algae are acquired by host larvae or juvenile polyps (Schwarz et al., 1999). After episodes of dysbiosis, algae can recolonize adult hosts with differential success in a process which can result in “shuffled” symbiont population proportions (Cunning et al., 2018). These colonization events are fundamental to the fitness and survival of symbiotic cnidarians; the ability for the host to be rescued by thermotolerant symbiont colonization represents one of several solutions for coral species resilience and survival during the climate crisis (Berkelmans and van Oppen, 2006; Hoegh-Guldberg et al., 2018). It is therefore critical to study and model how cnidarian-dinoflagellate symbiosis operates with non-native thermotolerant symbiont populations under ambient and elevated temperature regimes.
Many experimental studies have measured the dynamics of symbiont populations on an organismal level (e.g., symbiont cells per larva) or after tissue homogenization (e.g., symbiont cells per mg host protein). In general, symbiont species are more rapid colonizers when derived from the same host cnidarian species (i.e., native symbionts). This differential success has been measured in larvae and juvenile coral polyps, by inoculations of native species resulting in robust colonization and inoculation of non-native species resulting in more varied colonization outcomes (Weis et al., 2001; Little et al., 2004; Harii et al., 2009; Wolfowicz et al., 2016). Host-symbiont specificity is hypothesized to be more restrictive in the colonization of adult aposymbiotic hosts (Coffroth et al., 2001; Baker, 2003; Abrego et al., 2009). This restriction has implications for colonization of adults during recovery from bleaching. In examining successful symbiont colonization events in aposymbiotic adults, native algal symbiont populations colonize at faster rates resulting in larger symbiont populations (Schoenberg and Trench, 1980; Davy et al., 1997; Belda-Baillie et al., 2002; Gabay et al., 2018; Tortorelli et al., 2020). Non-native algal colonization often comes with a slower initial colonization rate, and success depends on symbiont species. In some studies, differences in colonization between native and non-native symbionts have been correlated with significant differences in oxidative stress response and apoptotic pathway activation (Dunn and Weis, 2009; Matthews et al., 2017).
Although the overall symbiont population dynamics during bleaching are well-documented, mechanisms for how algal populations decrease and subsequently recover in hosts are not well-understood (Gates et al., 1992; Weis, 2008; Bieri et al., 2016). For example, it is still not clear whether symbiont populations originate externally from the water column or surrounding sediment, or if population recovery is solely from algae remaining within the depleted host. In some coral hosts, thermotolerant species such as Durusdinium trenchii have been found to occur at low proportions prior to bleaching events, persist during periods of bleaching, and become the predominant symbiont after bleaching events, presumably through lack of competition from other thermally sensitive symbiont species (Silverstein et al., 2015; Bay et al., 2016; Manzello et al., 2019). However, these thermotolerant species appear to be replaced over longer periods of time by species that were dominant at pre-bleaching timepoints (Thornhill et al., 2006). Accurate predictions of responses to rapidly changing ocean environments by threatened cnidarian hosts and their algal symbionts requires further examination of these cellular mechanisms of symbiont population depletion and re-establishment (Davy et al., 2012).
Despite the considerable number of colonization studies in cnidarians, the spatial population dynamics of symbionts within their cnidarian hosts remains relatively uncharacterized. Gabay et al. (2018) examined these spatial patterning in the symbiotic sea anemone Exaiptasia diaphana (commonly called Aiptasia) across the entire host polyp in combination with inoculation of several symbiont species. Patterns of symbiont occurrence across host body sections were uniformly “patchy” for all observed symbiont species, appearing first in tentacles and oral disks, and then spreading to column and pedal disk. The authors suggest this rapid increase in colonized patches of host tissue is a result of symbiont migration after rapid mitotic cell division, whereby either algae move into the gastrovascular cavity and are reacquired by other gastrodermal cells, or they move directly through gastrodermal tissue. This hypothesis suggests that the size of a colonized patch of host tissue represents a history of localized population dynamics, one in which smaller patches originate from recent colonization/migration events and larger patches originate from older colonization/migration events. Spatial population dynamics of symbionts may therefore aid in understanding the different constraints governing host-symbiont interactions under environmental change.
In anthozoans, which include scleractinian corals, it is presumed that symbiont proliferation occurs within cnidarian gastrodermal host cells, and that levels of host cell-specific symbiont density vary from species to species (Muscatine et al., 1998). Due to the small size of anthozoan host cells, however, cell-specific symbiont population studies are extraordinarily difficult to perform (Davy et al., 2012). Therefore, most of our understanding of host cell-specific symbiont dynamics comes from the larger gastrodermal cells of hydroids Hydra and Myrionema (McAuley and Cook, 1994; Fitt, 2000; Fitt and Cook, 2001). Despite the relatively uncharacterized cellular relationship between symbiont and host cell proliferation in anthozoans, there are a number of hypotheses for host cell modulation of symbiont colonization, including augmenting symbiont populations via host cell cycle dynamics (Camaya, 2020; Tivey et al., 2020), constraining symbiont populations by restricting nitrogen availability (Krueger et al., 2020; Xiang et al., 2020), and physical expulsion of dividing symbionts in fully populated hosts (Baghdasarian and Muscatine, 2000). Though some of these physiological mechanisms are well-studied on the organismal scale, many questions about the underlying cellular mechanisms governing spatial population dynamics on the tissue level and cellular level remain unanswered.
In this study, we characterized and modeled the spatial patterns of native and non-native symbiont population dynamics in Aiptasia. Aiptasia provides a tractable model to study these fundamental cellular mechanisms in depth (Weis et al., 2008; Hambleton et al., 2014; Goldstein and King, 2016). The thin, translucent epidermal and gastrodermal tissue layers of Aiptasia polyps enable flexible imaging techniques to characterize the location and proliferation of autofluorescent symbionts (Tortorelli et al., 2020). In addition, Aiptasia is readily culturable in a laboratory setting and can be maintained indefinitely in an aposymbiotic state after rapid bleaching from either thermal stress or menthol treatment (Belda-Baillie et al., 2002; Matthews et al., 2016). Aposymbiotic adult Aiptasia polyps can, to varying degrees, be colonized with both native and non-native symbionts, which allows for symbiont species specificity comparisons of spatial patterns within host tissue.
In this study, we quantified spatiotemporal proliferation dynamics of algal symbionts in individual host anemones during both periods of colonization and heat-stress-induced bleaching. We developed and compared two non-invasive imaging methods for tracking symbiont population patterns over time throughout the tentacles of Aiptasia. We further used this methodology to compare the patterning and population dynamics of native symbiont species (Breviolum minutum) and non-native species (Symbiodinium microadriaticum and D. trenchii) under ambient and elevated temperatures. Finally, we compared our data with models of symbiont population growth to better understand the relationship between local symbiont proliferation and migration within gastrodermal tissue. We find that migratory events after initial inoculation play a critical role throughout symbiont colonization, and that thermal stress impacts symbiont density on a localized scale in a species-specific manner.
Algal cultures of Symbiodiniaceae were maintained at 26°C under 12:12 L:D cycle and grown in f/2 media at a light intensity of 50 μmol quanta ∙ m–2 ∙ s–1. Cultures included one native symbiont of Aiptasia, Breviolum minutum (culture IDs: FLAp2 Mf1.05b), and two competent but non-native species, Symbiodinium microadriaticum (CCMP 2467) and Durusdinium trenchii (Ap2). All cultures were genotyped prior to experiments to verify the identification of symbionts (data not shown). Experimental Aiptasia polyps were generated from animal stocks of the clonal H2 strain containing their native symbiont species B. minutum, originally isolated from a single individual collected from Coconut Island, Kaneohe, Hawaii (Xiang et al., 2013). Aposymbiotic sea anemones were generated using a menthol bleaching protocol (Matthews et al., 2016). Sea anemones were maintained in filtered sea water (FSW) in an aposymbiotic state in the dark for longer than 2 months prior to use in experiments and fed Artemia nauplii three times per week until 1 week prior to experiments. Anemones were plated into individual wells of multi-well plates 1 week in advance of experiments, moved to 26°C incubators under 12:12 L:D cycle and checked for symbionts to ensure that the polyps remained symbiont-free using an inverted epifluorescence Zeiss AxioObserver A1 microscope with an Axiovert ICm1 camera (Carl Zeiss AG, Jena, Germany).
As part of a pilot experiment, tiny (1 mm oral disc diameter) aposymbiotic anemones were selected and plated into 24-well plates. Anemones were visually inspected under bright light and under laser excitation using the Cy3 red filter to capture symbiont autofluorescence and verified to contain no algal symbionts. Animals were then inoculated with 1 × 104 cells ∙ mL–1 of B. minutum (Culture FLAp2). After 24 h of exposure, anemones were rinsed with FSW, moved to a 96-well plate, and imaged daily for up to 7 days after inoculation (n = 10). Animals were kept at 26°C for the duration of the experiment. Prior to imaging, animals were placed in a relaxing solution of 1:1 FSW mixed with 0.37 M MgCl2 in FSW. After 5 min, the animal was plated onto a depression slide for confocal imaging at 10x magnification on a Zeiss LSM 780 NLO Confocal Microscope System (Carl Zeiss AG, Jena, Germany) in the Center for Genome Research and Biocomputing at Oregon State University. Z-stack images (850 × 850 μm) were taken of the tentacles and columns of the anemone under bright light only. Quantification of the number of singlet, doublet, triplet, and quadruplet algal cells in the tentacles and in the body column/oral disc was performed for each Z-stack image using Fiji (ImageJ2) (Schindelin et al., 2012; Rueden et al., 2017).
To broaden our understanding of symbiont population dynamics in Aiptasia tentacles over a longer duration in larger areas of tentacle tissue, we changed our imaging approach to epifluorescence microscopy and developed a more rapid imaging method, which captured symbiont populations using z-stack videos of symbiont autofluorescence without a need for animal anaesthetization. For this method, medium-sized (0.25 cm oral disc diameter) aposymbiotic sea anemones were selected and plated into 24-well plates. This animal size ensured that the full-length of any chosen tentacle could be imaged under a 10x objective. To determine if there were residual symbiont populations, animals were visually inspected under bright light and under laser excitation using the Cy3 red filter to capture symbiont autofluorescence. In this experiment, very low algal densities consisting of singlet algal cells (approximately 5–15 cells per tentacle) were found to have persisted in Aiptasia tentacles after 6 months of darkness post-menthol bleaching (Supplementary Figure 1).
To examine patterns of symbiont colonization, we inoculated these Aiptasia with their native symbiont B. minutum and examined proliferation patterns under two temperature regimes. All animals were inoculated with 5 × 104 cells ∙ mL–1 of B. minutum culture in 2 mL of FSW in 24-well plates. After 24 h, polyps were rinsed with FSW (d = 0) and moved into fresh FSW-filled 24-well plates to restrict the timing of inoculation. Animals were imaged and maintained at 26°C for 1 week before dividing them into an ambient treatment (26°C) and an elevated heat treatment (32°C) (n = 24 anemones per treatment). Animals were maintained at either 26°C or 32°C for an additional 2 weeks. Since heat and ambient treatment of anemones required incubation in two separate incubators, each 24-well plate was housed in identical chambers with LED lighting strips (LEDENET 24V cold white 6500K-7000K LED strip, Enet Light Technology Co., Limited, China) covered with nylon mesh for a final light intensity of 10 μmol quanta ∙ m–2 ∙ s–1 across the entire plate. Periodically, over 3 weeks, an inverted epifluorescence Zeiss AxioObserver A1 microscope with an Axiovert ICm1 camera (Carl Zeiss AG, Jena, Germany) was used to take videos of three tentacles per anemone at 35 ms exposure under bright light and under laser excitation (Cy3 red filter) to capture algal cell chlorophyll autofluorescence (Figure 1A). For each tentacle, videos were taken by scrolling through the focus from the tip to the base of a tentacle using a 10x objective, simulating a confocal z-stack (Figure 1B). Brightfield video was taken first to determine the depth of the tentacle, and a video was taken immediately after using darkfield laser excitation.
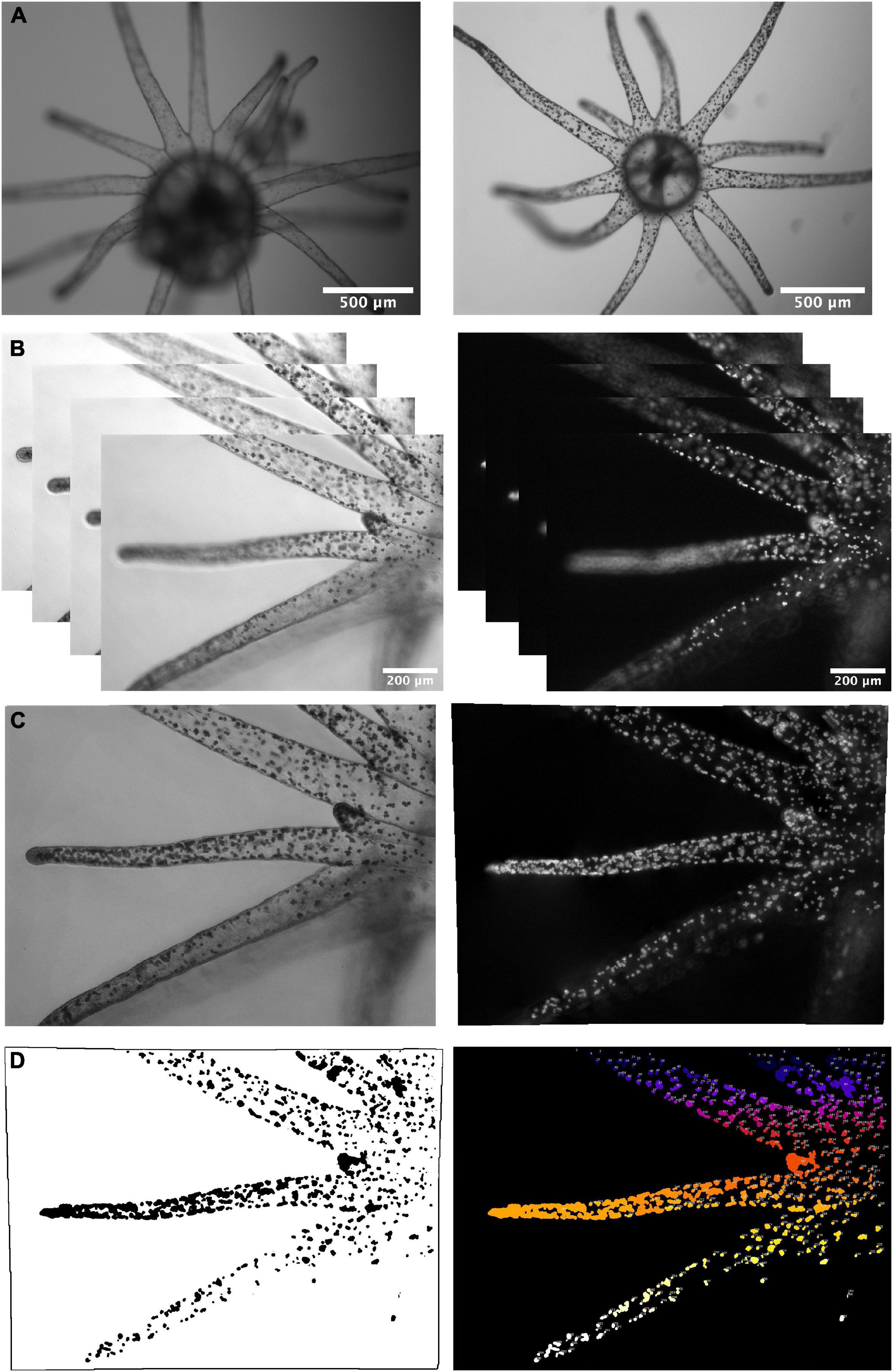
Figure 1. Imaging colonization dynamics in Aiptasia polyps using epifluorescence microscopy. (A) Bleached Aiptasia polyps (left: day 0) were colonized with algal symbionts over a period of 4 weeks (right: day 10) and imaged under brightfield and epifluorescence to track symbiont autofluorescence. (B) Videos were taken as representative Z-stack images under brightfield (left) and laser excitation (right) to provide focus for each z-plane of the tentacles. (C) Videos were merged into 2D images, and (D) the epifluorescence images were processed in Fiji (ImageJ2) to create a 2D mask of symbiont presence (left), which was used to identify size and location of symbiont cluster objects (right). The number of cells within each cluster was determined based on object size cutoffs.
To compare patterns of host colonization by different symbionts species, we inoculated medium-sized (0.25 cm oral disc diameter) Aiptasia with one of three species of Symbiodiniaceae: native B. minutum, and non-native S. microadriaticum or D. trenchii. Prior to inoculations, animals were visually inspected under bright light and under laser excitation using the Cy3 red filter to capture symbiont autofluorescence and verified to contain no algal symbionts. Inoculations were performed using 5 × 105 cells ∙ mL–1 for all species over 2 days, tenfold higher than the symbiont density of the previous experiment, to increase the success of non-native inoculation. After 48 h of inoculation, anemones were rinsed and moved to new 24-well plates. All sea anemones were maintained at 26°C for 2 weeks before separating into an ambient treatment (26°C) and an elevated heat treatment (32°C) (n = 12 each). Animals were maintained at either 26 or 32°C for an additional 3 weeks. Environmental conditions and epifluorescence imaging were identical to the previous experiment.
For experiments performed on the epifluorescence microscope (i.e., Experiment 2 and 3), videos were saved and exported into jpeg stacks using the batch export function in the ZEN software package (Black Edition v. 3.0). Photo stacks contained either all autofluorescent images or all brightfield images. Photo stacks with more than 50 images were flagged and truncated to delete images that were out of focus at the top and bottom of the image stack. Image stacks were then imported into Adobe Photoshop CC 2019 (v. 20.0.3) to undergo autoblend in order to merge the entire image stack into one 2D image of a tentacle (Figure 1C). Once all of the stacks were merged into single images, brightfield and fluorescent merged images were separated into two folders. Fluorescent merged images were analyzed in FIJI using intensity thresholds (Figure 1D) and the 3D object counter plugin (Bolte and Cordelieres, 2006). Each sample image resulted in a list of object sizes corresponding to clusters of autofluorescent symbionts. These tables containing cluster sizes for each sample were imported into R, binned as count data according to cluster object area size, and combined into a dataframe with sample metadata for further analysis. To bin clusters into singlets, doublets, and larger clusters, a range of size cutoffs was used. To ensure accuracy, manual counting was performed for 39 samples across experimental timepoints (Supplementary Table 1). The manually counted numbers of singlets, doublets, triplets, quadruplets, quintuplets, total clusters, and total cells were correlated into the best fitting cutoff range for size-based segregation. After assigning size cutoffs and binning clusters into cell numbers, a subset of clusters (n = 177) was used to directly compare the identification of each cluster between hand-counting and automatic binning methods (Supplementary Figure 2). Overall, automatic binning connected neighboring clusters more frequently, resulted in 19% fewer clusters overall compared to hand-counting, however, there was a tight correlation on total clusters for each sample (R = 0.94, p < 2.2 × 10–16, Supplementary Figure 2). Cell clusters containing more than two cells were binned into groups of three-to-four cells, five-to-ten cells, and greater than ten cells to signify increased difficulty in discriminating between larger cluster sizes with increased cell-cell obscuration.
To assess differences between count data in Experiment 2, linear mixed models were applied for each cluster size to test the interaction between days and temperature treatment, with plate treated as a random effect. To assess differences between count data in Experiment 3, linear models were used to test the interaction between days and temperature treatment for each cluster size of each symbiont species. For both experiments, each model required a square root or log transformation to normalize count data. To assess pairwise relationships, Tukey’s tests were constructed using the general linear hypothesis test (glht) function, which was corrected for multiple comparisons by the single-step method. All statistical analyses were performed in R and are made available in Supplementary Tables 2, 3.
To test the effect of migration among symbiont clusters during colonization, simulated symbiont populations were created from inoculation data from Experiment 3. Three random subsamples of symbiont cluster populations from each anemone host were generated from total cell clusters present at day four. Each subsample contained the mean number of cell clusters present per host anemone at day four. A regression model was then constructed by measuring the observed summed symbiont cell population in each sample image as a function of days post-inoculation: and fitting a self-starting nls logistic model. To account for variation between the symbiont populations of sample anemone hosts, a unique self-starting nls logistic model was fit to each anemone host (Supplementary Table 4). The logistic growth models were then applied equally to every cell present in the newly generated samples at day four. To create a zero-migration model, only these clusters present at day four were allowed to grow in cluster size at this linear growth rate over time. To create a simulated migration model, each cell was given the same daily probability of migration, independent of cluster size, and subjected to a Bernoulli trial where the cell would either migrate from a cluster or remain in the cluster. For each positive migration outcome, the cell would be subtracted from the cluster and added as a newly formed cluster.
After the migration process was completed for every cell in a sample, a daily growth rate was applied equally to all cells. An end day cell population was determined by the fixed population number from the corresponding logistic growth model (Supplementary Table 4). The difference in cell number between the starting day and end day cell population was then allocated to each cell proportionally. To further test the influence of symbiont cluster size-dependent growth in the model, the distribution of this allocation was root-transformed according to the cell cluster size, which added proportionally more cells to smaller cell clusters and fewer cells to larger clusters. An array of root transformations (un-transformed, √2, √4, √8, √16, log2) and migration probabilities (0, 0.5, 1, 1.5, 2, 3, 4, 7, 10%) were used to simulate symbiont populations. Each simulated dataset was compared against the observed dataset using a chi-square goodness-of-fit test, which additively measured Chi-square values between simulated and observed cluster frequencies for each biological replicate at each timepoint (df = 127). Chi-square values were measured for total clusters to determine the best fit simulation (Supplementary Table 5). Chi-square values were also measured for clustered bins (one, two, three to four, five to ten, over ten) to determine modeling accuracy for differing cluster sizes (Supplementary Table 6). After choosing the best fit simulations, the cluster size abundances of best fit simulated populations over time were compared to observed experimental data to infer the effects of migration and growth rate application among clusters.
Using brightfield confocal microscopy, we took repeated measurements and tracked symbiont cell cluster populations of native symbiont Breviolum minutum during 1 week of colonization in Aiptasia polyps (oral disk < 0.5 mm; Figures 2A,B). After a relatively low symbiont inoculation concentration (1 × 104 cells ∙ mL–1), tentacles were populated with low levels of singlet symbionts primarily in the tentacles (Figure 2B). Over the course of 1 week of colonization, singlet cells remained in tentacles at a constant rate [F(6, 63) = 0.93, p = 0.48], whereas the occurrence of doublets grew exponentially (Figure 2B) [F(6, 63) = 1.85, p = 0.10]. Though the appearance of triplets was recorded as early as 2 days post-inoculation, multiple triplets and quadruplets only began to appear at day seven of the experiment [F(6, 63) = 4.18, p = 0.001; F(6, 63) = 2.7, p = 0.02]. The area including the oral disk and column of inoculated Aiptasia had much lower symbiont densities and remained at low density through the duration of the experiment (Supplementary Figure 3).
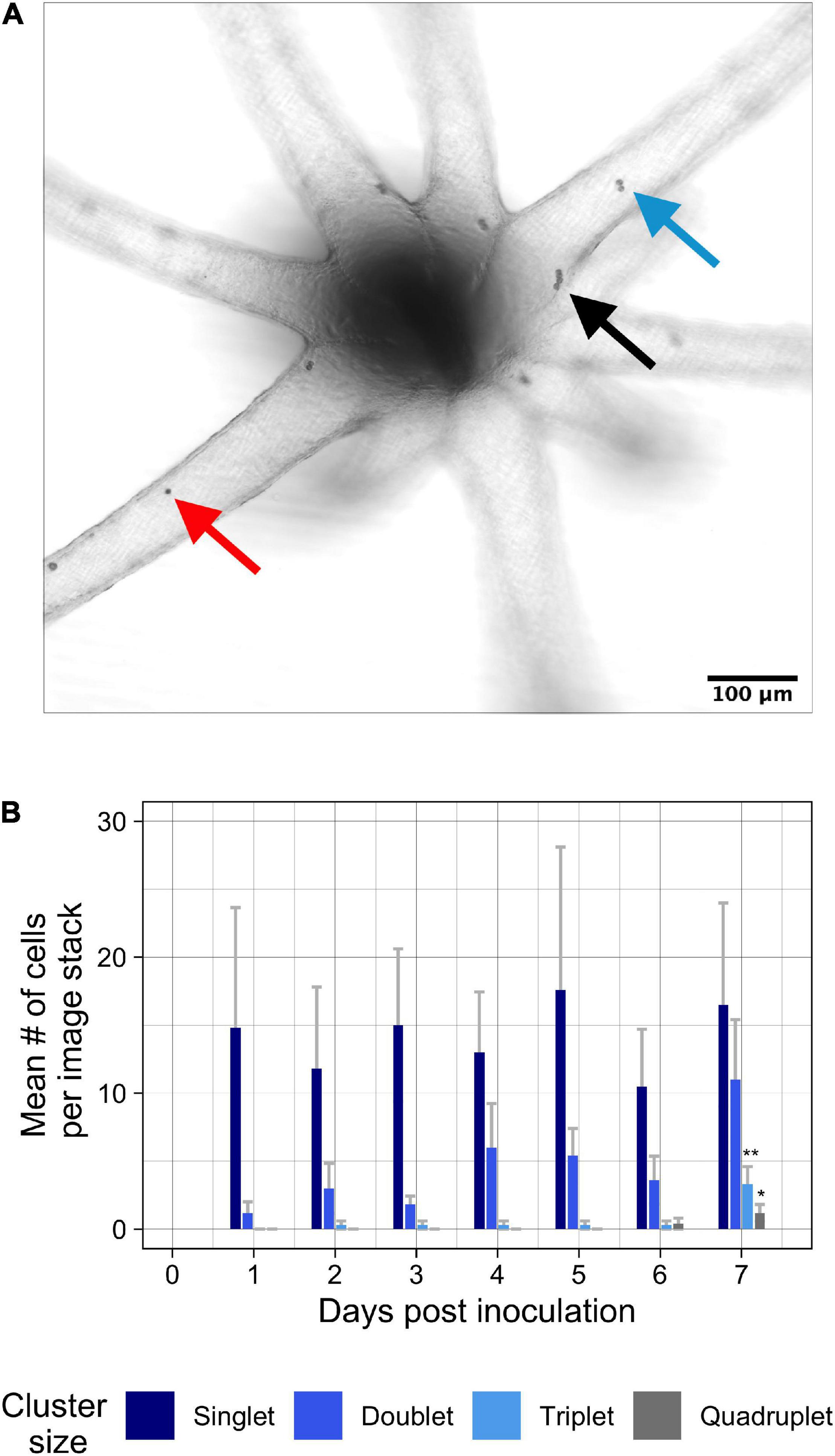
Figure 2. Colonization by B. minutum over 1 week at 26°C. (A) Colonizing symbionts were tracked within the tentacles of tiny Aiptasia polyps with the genetic host strain background of H2 (n = 7 anemones). For each day, one brightfield confocal z-stack per anemone was used to capture all information between the focal planes of the closest and furthest tissue. Clusters of symbiont cells were identified over a period of 7 days. (B) The cluster sizes of singlets (red arrow), doublets (blue arrow), triplets (not shown) and quadruplets (black arrow) were summarized within tentacles to find mean abundances of cells per cluster size over time. Aiptasia were inoculated with a concentration of 1 × 104 cells ∙ mL–1. Error bars signify standard error. Asterisks represent statistically significant differences after ANOVA and Tukey post hoc test: ** p < 0.005 for triplets at 7 days compared to all other days; *p < 0.05 for quadruplets at 7 days compared to days 1–5.
Epifluorescence microscopy and automated imaging processes (Figure 1) allowed us to collect and analyze symbiont patterns within host Aiptasia more efficiently and with less damage to the host than the confocal imaging used in Experiment 1. Based on the increased colonization of tentacles compared to the oral disk/column area in Experiment 1 (Supplementary Figure 3), we chose to focus on imaging only tentacles. With these methods, we were able to capture symbiont proliferation dynamics in Aiptasia tentacles over an extended period of colonization. After scaling our symbiont inoculation concentration to accommodate for a larger animal size, compared to the anemones used in Experiment 1, initial symbiont uptake of B. minutum occurred throughout the length of the tentacle with no discernible differences in density along the proximal-distal axis (data not shown). During the colonization process at 26°C, the cell populations within our sample images experienced consistent growth throughout the experiment (Figure 3A). In comparison, the total number of clusters became constant at day 9 (Figure 3B). Similarly to the total cells and clusters, each measured cluster size at 26°C increased in abundance up to day 9, but abundance trends differed between cluster sizes after day 9 (Figure 3C). The frequency of singlet populations increased significantly from day two to day six of colonization (d2C-d6C, p < 0.001, Tukey post hoc test). Doublets and triplets/quadruplets were the most numerous cluster sizes present in host tissue within the first week. As expected, larger clusters of five-ten and over ten cells increased with each day post-inoculation except for day 16 (p < 0.001, Tukey post hoc test), when both the total number of cells and clusters slightly decreased (Figures 3A,B).
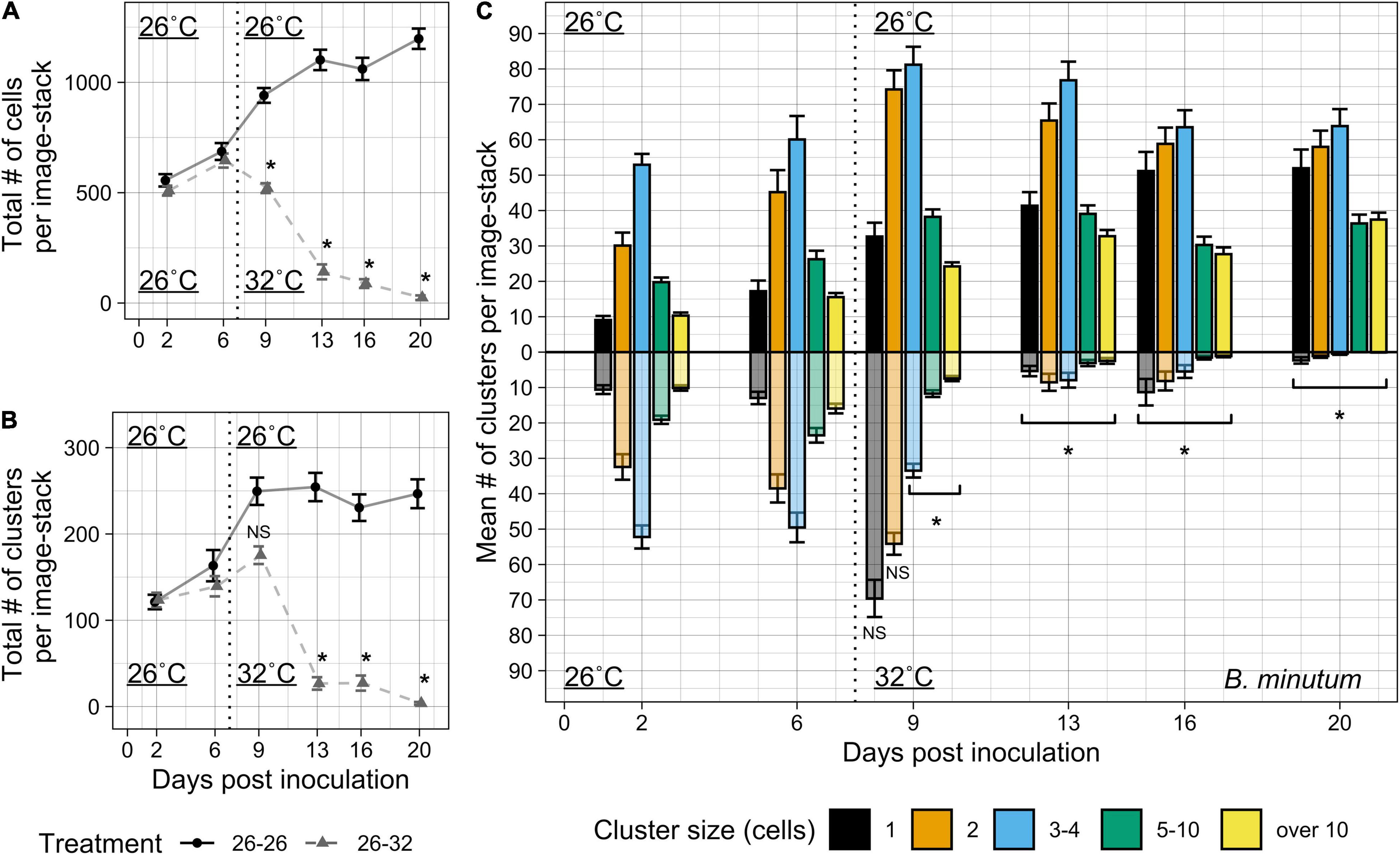
Figure 3. Colonization by B. minutum over 3 weeks at 26 and 32°C. Algal symbiont populations were monitored over 3 weeks in the tentacles of Aiptasia polyps under ambient and elevated temperature regimes. (A) The total numbers of cells per sample image-stack were measured over a 3-week period (3 sample image-stacks per anemone, 24 anemones per treatment). During the first week both treatments received the same 26°C temperature treatment to promote colonization. After 7 days, one treatment remained at 26°C and the other treatment was moved to 32°C to compare colonization and bleaching phenotypes. (B) The total numbers of clusters per sample image-stack were measured over 3 weeks under ambient and bleaching conditions. (C) The mean number of cell clusters per image-stack were binned into discrete cell cluster sizes measured over 3 weeks. 26°C treatment for 3 weeks is shown on the positive y-axis, while 26–32°C treatment is shown on the negative y-axis, reflected across the x-axis for comparison. The beginning of the difference in treatment conditions is indicated by a vertical dotted line at 7 days post-inoculation. Aiptasia were inoculated with a concentration of 5 × 104 cells ∙ mL–1. Error bars signify standard error for the total abundance of clusters or cells. Asterisks indicate significant differences between temperature treatments at each timepoint by Tukey post hoc tests (p < 0.01 in all cases).
After 1 week of colonization at 26°C (26d7), a heat treatment of 32°C was applied to half of the sample animals in Experiment 2 (26d7 + 32d0). Two days of 32°C heat treatment was enough to significantly reduce the total number of symbiont cells (26d7 + 32d2) compared to samples in the 26°C treatment (26d9) (p < 0.01, post hoc Tukey test; Figure 3A and Supplementary Table 2). Though the total number of cells immediately decreased compared to counts just prior to elevated temperature onset, there was a temporary increase of cell clusters during the bleaching response (d7C + d0H, Figures 3A,B). The increase in total cluster abundance was explained by the dramatic increase of singlets and retention of doublet clusters during this immediate bleaching response (Figure 3C). In contrast to smaller cluster sizes, all clusters containing four or more cells immediately decreased compared to their previous measurements (d7C + d0H) and compared to samples from the 26°C (d9C-d20C) temperature treatment (Figure 3C, p < 0.01 for all pairwise post hoc Tukey tests). Singlets, doublets, and triplets/quadruplets were the last type of clusters to remain after 1 week of elevated temperature. All cell cluster sizes declined after 2 weeks of heat treatment, and only the occasional singlet or doublet symbiont cluster was found in tentacles. There was no locational difference in colonization or bleaching along the length of the tentacle (data not shown).
To explore how colonization patterns of hosts by symbionts differ across symbiont species, we compared colonization of Aiptasia using three species of Symbiodiniaceae: native B. minutum, and non-native Symbiodinium microadriaticum, and Durusdinium trenchii (Figure 4). Aposymbiotic Aiptasia were verified to contain no residual native symbiont populations. In comparison to Experiment 2, symbiont inoculation concentration was increased to ensure successful symbiont uptake of non-native species. Despite this increase in inoculation concentration, the initial cell populations of B. minutum during the first week of colonization were threefold lower in Experiment 3 (26d7, Figure 4A) compared to Experiment 2 (26d6, Figure 3A). As a result, we were able to capture growing populations of symbionts across a longer timeframe than the previous experiment. The initial uptake of native symbiont B. minutum populations resulted in symbiont populations twice as large as non-native symbiont populations S. microadriaticum and D. trenchii (26d4, Figure 4A). As in the previous experiment, polyps colonized with B. minutum showed consistent linear growth of large cell clusters (10 cells or more) (Figure 4B). Triplet/quadruplet populations were again the most abundant at each timepoint during this experiment. Though overall trends of B. minutum colonization were consistent across experiments, cluster patterns exhibited higher variation in Experiment 3 compared to Experiment 2, most likely as a result of lower inoculation numbers and halving the sample size.
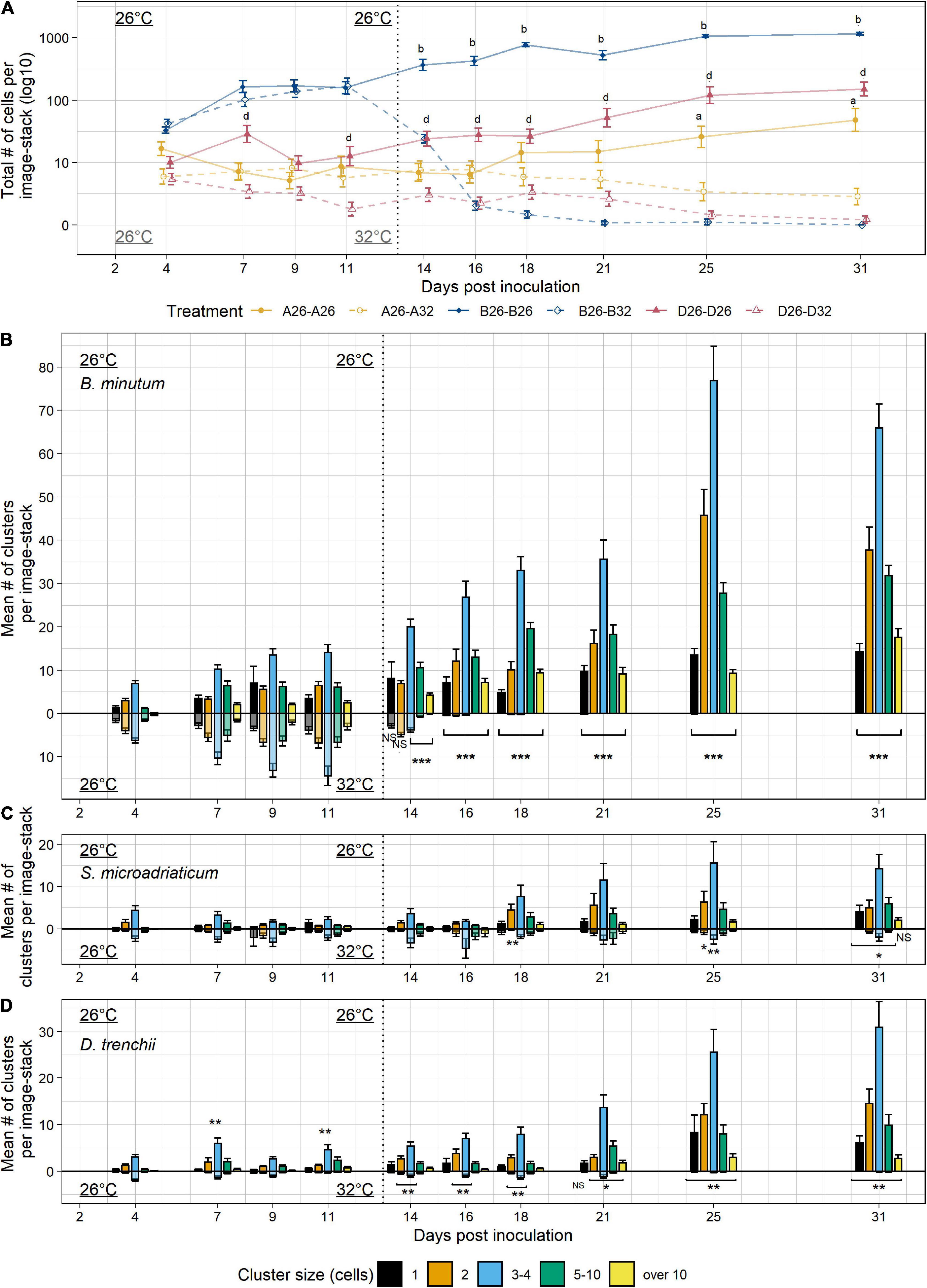
Figure 4. Colonization by B. minutum, S. microadriaticum, and D. trenchii over 4 weeks at 26 and 32°C. (A) Bleached Aiptasia polyps were colonized with one of three species of algal symbionts: B. minutum, S. microadriaticum, and D. trenchii and subjected to a temperature treatment of a constant 26°C or a temperature shift from 26 to 32°C to compare colonization and bleaching phenotypes. The total numbers of cells per sample image-stack were measured over a 4 week period for each symbiont (3 sample image-stacks per anemone host, 12 anemones per treatment). During the first week both treatments received the same 26°C temperature treatment to promote colonization. After 7 days, one temperature treatment remained at 26°C and the other temperature treatment was moved to 32°C. Aiptasia were inoculated with a concentration of 5 × 105 cells ∙ mL–1. Y-axis is log10 transformed for clarity. (B–D) The mean number of cell clusters per image-stack were binned into discrete cell cluster sizes measured over 3 weeks for each symbiont species, (B) B. minutum, (C) S. microadriaticum, and (D) D. trenchii. 26°C treatment for 3 weeks is shown on the positive y-axis, while 26–32°C treatment is shown on the negative y-axis, reflected across the x-axis for comparison. The beginning of the difference in treatment conditions is indicated by a vertical dotted line at 13 days post-inoculation. Error bars signify standard error for the total abundance of clusters or cells. Letters (A) and asterisks (B–D) indicate significant differences between temperature treatments within symbiont species at each timepoint by Tukey post hoc tests (*p < 0.05, **p < 0.01, ***p < 0.001).
Both non-native symbionts colonized animals at much lower levels and slower rates compared to native symbionts (Figure 4A). In polyps colonized by S. microadriaticum, low levels of initial symbiont uptake by animal hosts during the inoculation period was predictive of overall slow colonization (Figures 4C). The total number of cells and clusters did not significantly change within the first 2 weeks of colonization. Substantial colonization correlated with increases in singlet, doublet, and triplet populations during the third week of colonization. Mean number of cells and clusters doubled between day 16 and day 18 of colonization (26d16-26d18, Figures 4A,C), with stronger colonization patterns appearing from day 25 (26d25) onward.
Colonization by D. trenchii also resulted in reduced levels of symbiont uptake and took longer to establish in Aiptasia polyps (Figure 4D). After remaining low, total cells tripled and total clusters doubled from day 18 to day 21, beginning a pattern of consecutive increases whereby effective colonization occurred (Figure 4A). Sudden population growth was marked by increases in triplet and quadruplets, followed by increases in singlets and doublets. After growth of these smaller clusters, larger clusters of five-to and over ten cells began to form relatively quickly (26d21).
Colonization with either S. microadriaticum or D. trenchii resulted in higher variation in symbiont colonization between individual anemones compared to colonization with B. minutum (Supplementary Figures 4–6). For 26°C treatment, all polyps with B. minutum ended up being substantially colonized (Supplementary Figure 4), whereas only three of twelve polyps were substantially colonized with S. microadriaticum (Supplementary Figure 5) and eight of twelve polyps were substantially colonized with D. trenchii (Supplementary Figure 6). Substantial colonization was determined by the presence of continuous symbiont population growth with average symbiont abundances consistently above 50 clusters (Supplementary Figures 4–6). In polyps inoculated with S. microadriaticum, higher levels of early inoculation resulted in polyps with substantial symbiont populations (26d4-26d7, Supplementary Figure 5). In contrast, there were no obvious differences in inoculation levels between rapid and slower colonizations of D. trenchii at early time points immediately following inoculation (26d4-26d7, Supplementary Figure 6).
After 2 weeks of colonization (26d13), a heat stress temperature of 32°C was applied to half of the anemones undergoing colonization. In animals recolonized by native B. minutum, algal populations again declined in overall cell and cluster number after the temperature was increased (26d13 + 32d1, Figures 4A,B). There was a decline in all clusters containing three or more cells at 1 day post heat stress (26d13 + 32d1) compared to day 11 (26d11 + 32d0) and compared to the 26°C treatment (26d14) (p < 0.001 for all pairwise Tukey post hoc tests). Similar to Experiment 2, singlet and doublet populations did not immediately decline under heat stress. After 1 week post-bleaching exposure (26d14 + 32d7), however, most tentacles had no clusters of symbionts present in their tentacles.
In contrast, in animals recolonized by S. microadriaticum, algal population numbers remained relatively constant under thermal stress conditions (Figure 4B and Supplementary Figure 5). Symbiont clusters did not significantly change throughout the heat treatment, with symbiont populations averaging between 50 and 100 symbionts per sample (Figures 4A,C and Supplementary Table 3). Cell number and cluster populations only significantly declined at day 31 after 2 weeks of heat stress (26d13 + 32d18, p < 0.05, Tukey post-hoc test; Figure 4C). Polyps colonized by S. microadriaticum remained symbiotic at the end of the experiment.
In animals recolonized by D. trenchii, symbiont populations also initially persisted under thermal stress conditions (Figure 4D and Supplementary Figure 6). D. trenchii inoculations resulted in the lowest levels of algal colonization of hosts prior to bleaching, with fewer than ten symbionts on average per sample image. These small populations persisted under thermal stress for 1 week in host polyps before showing population reductions in all clusters at day 25 (26d13 + 32d12, p < 0.001, Tukey post hoc test; Figure 4D). By day 33 (26d13 + 32d20, Figure 4D), all symbionts had been lost from host polyps.
To better understand the mechanisms governing population growth of symbiont clusters, we created population simulations to test the effect of migration rate, i.e., egress of algae from existing clusters to form new clusters, on symbiont colonization patterns. Self-starting nls logistic models were fit to Experiment 3 colonization by native symbiont B. minutum under 26°C conditions by using summed cell populations per sample image from between day 4 and day 31 (Figure 5A). Logistic growth models generated three numeric parameters for each biological replicate (n = 12), representing the asymptote, the inflection point of the logistic curve, and y-axis scaling parameter in the equation y = asymptote/[1 + exp((inflection-x)/scale)] (Supplementary Table 4). A linear model described the data within this period but overestimated population growth in the days following, whereas a logistic model provided a better estimate of the slowing population growth in the tentacles (Supplementary Table 4).
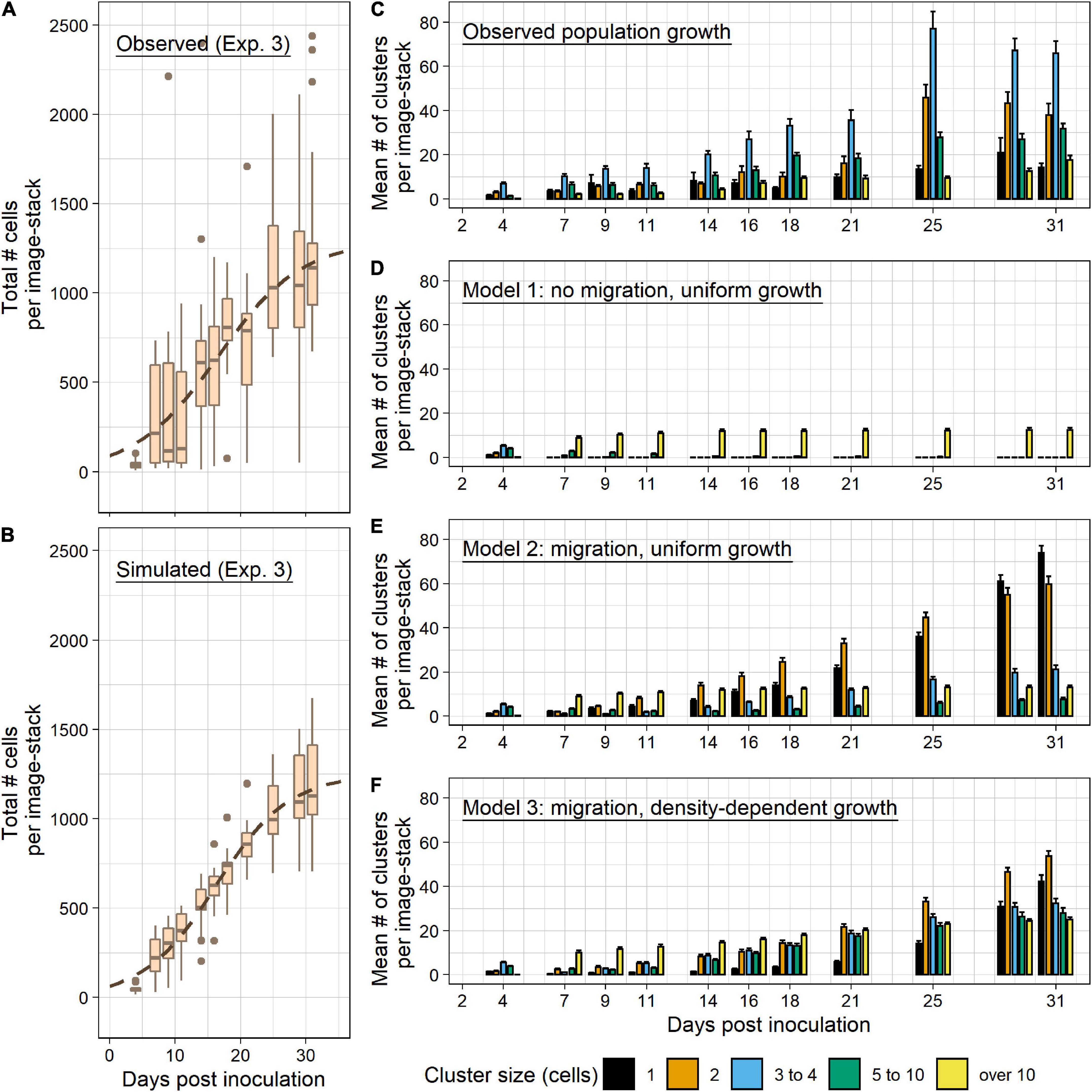
Figure 5. Population growth and migration models. (A) Data from Experiment 3 (B. minutum populations at 26°C) were used to create simulated population growth models. Total symbiont populations for each sample over time are represented by boxplots, with points showing outliers at each timepoint. The dashed line depicts a representative summary logistic model for all samples [y = 1249.146/(1 + e((15.916–x)/5.815)]. (B) Simulated data was created using logistic model parameters generated from observed populations for each biological replicate. Boxplots indicate total symbiont populations for each simulated sample over time. The same representative summary logistic model as above is indicated by the dashed line. The modeled data shown was generated using a 1% migration rate and a cluster density-dependent growth rate. (C–F) The mean number of clusters per sample image for each cluster size are displayed for observed and simulated data. (C) Observed data replicated from Figure 4B for comparison. (D) Simulated data with no migration allowed and a logistic growth rate unique to each host anemone applied uniformly applied across sample populations. (E) Simulated data with cells allowed to egress from clusters to create new singlet clusters. Each cell was given a daily 1% probability of migration and had a uniformly applied growth rate. (F) Simulated data with a 1% probability of migration and a cell density-dependent transformation, in which daily population growth is allotted to cells using a square-root transformation to slow the growth of large clusters and instead apply population growth to smaller sized clusters. Error bars signify standard error.
These parameters were then used to simulate new symbiont populations for each biological replicate. Three random samples of clusters per biological replicate were taken from observed data at day four, with each sample containing the mean number of clusters per replicate. Using parameters specific to each biological replicate, a growth rate was applied to these cell populations each day up to day 33 (Figure 5B). Daily total population growth was distributed proportionally to each cell so that each cell grew at the same rate. Comparison of total observed population growth (Figure 5A) and a simulation of total population growth (Figure 5B) follow the same summary logistic model, but with substantially more variation in observed data.
To visualize the effect of allowing population growth but no migration, observed data (Figure 5C) was compared with simulated data showing growth of only the original clusters from day 4 (Figure 5D). This type of growth was not representative of the cluster sizes found in our observed population (χ2 = 23419.03, df = 127, p < 1 ⋅ 10–5; Supplementary Table 5). To more closely replicate observed data, a daily cell migration probability was set for each simulated cell population to model cells egressing from clusters to establish new clusters. Briefly, a set migration probability was applied to each cell in a sample population for each day. If the resulting outcome was migration, then the cell was subtracted from its cluster and added back into the sample population as a new singlet cluster. Migration probabilities between 0.5 and 10% were tested. A chi-square goodness of fit test was applied between observed data set and each migration simulation to compare total cluster number across timepoints (Supplementary Table 5). The closest migration rate matching observed data was 1% per day (χ2 = 5257.04, df = 127, p < 1 ⋅ 10–5; Figure 5E and Supplementary Table 5). However, the migration model consistently overestimated singlet clusters and underestimated cluster sizes between three and ten (Figure 5E and Supplementary Table 6).
To model a more rapid transition of singlet clusters into larger-sized clusters, the daily total population growth was instead distributed among individual clusters using root transformations of cluster size, which resulted in a faster growth rate for smaller clusters and a slower growth rate for large clusters. A series of root transformations were used in conjunction with migration rates between 0.5 and 10% to generate population simulations. All simulated datasets including non-transformed data (n = 63) were compared against observed data using chi-square goodness of fit tests (Supplementary Tables 5, 6). Among all simulations tested, the simulation that most consistently estimated observed cluster growth was a square-root transformation with a migration rate of 1% (Figure 5F). The dataset was still significantly different from observed data (χ2 = 5039.69, df = 127, p < 1 ⋅ 10–5; Supplementary Table 5). Singlets and clusters with over 10 cells remained elevated compared to observed data, but the overall distribution of cells among cluster sizes was more representative of measured populations (Figure 5F and Supplementary Table 6).
This study is a first step in describing symbiont populations within the context of their dynamic spatial landscape of colonized cnidarian tissue. Our imaging methods are a simple and affordable method for capturing symbiont proliferation patterns under colonizing and bleaching conditions. Using this dataset, we were able to model and infer the role of symbiont migration (i.e., egress and subsequent inoculation) within cnidarian tissue. We found that the colonization process is not a discrete set of ordered steps but rather an iterative process that involves continual inoculation of aposymbiotic tissue, most likely sustained by symbiont egress and migration, coupled with localized symbiont growth and proliferation.
Across our experiments with native B. minutum colonizing Aiptasia polyps, we found predictive symbiont patterns that were broadly consistent across three distinct colonization timelines. Colonization resulted from localized symbiont proliferation centered around locations of symbiont inoculation events, as well as from what appeared to be new migration events (e.g., new singlet clusters) occurring throughout the colonization timeline. In each experiment, increased frequencies of both singlet and doublet populations were short-term predictors of overall colonization establishment; these recurring migration events during colonization preceded immediate increases in overall population growth. Larger symbiont cluster sizes were also useful predictors; clusters containing more than 10 cells grew in number at a consistent rate and were accurate long-term indicators of increased symbiont density and colonization establishment.
Although patterning of colonization was consistent across experiments, the rate of symbiont population growth depended strongly on the initial number of symbionts and clusters. In low-density symbiont inoculation of tiny anemones (Figure 2), symbiont populations only grew if there were at least two symbiont cells per tentacle. In larger polyps, colonization success could be measured by consistent increases in both symbiont numbers and clusters (Figure 3C). In measuring this success, an important timepoint in the progression of native B. minutum colonization was an increased level of larger cluster sizes (greater than five cells) and a symbiont density of between 100 and 200 cells and 25 cell clusters per sample image, as measured on day 2 of Experiment 2 (Figure 3A) and day 7 of Experiment 3 (Figure 4A and Supplementary Figure 4). Between these two experiments, these strong differences in the timing of overall symbiont population growth correlated with significant differences in levels of native B. minutum uptake within the first 4 days of initial inoculation. Differences in this uptake could have arisen from very small residual symbiont populations in aposymbiotic Aiptasia used in Experiment 2. Alternatively, there could have been differences in growth rate and density of B. minutum cultures, or differences in menthol bleaching history of aposymbiotic Aiptasia. Despite these differing colonization timelines, native B. minutum experienced similar early proliferation cluster patterns. These first 2 weeks of colonization are a critical time of symbiosis establishment, prior to log-phase growth of symbionts within hosts. Early proliferation has been a common indicator of colonization success in many studies, and is a characteristic of colonization by native symbionts (Schoenberg and Trench, 1980; Davy et al., 1997; Belda-Baillie et al., 2002; Gabay et al., 2018). Furthermore, past studies have shown that the initial symbiont density at 2 weeks appeared predictive of overall length and intensity of colonization (Belda-Baillie et al., 2002).
Non-native symbiont species S. microadriaticum and D. trenchii recolonized aposymbiotic Aiptasia adults more slowly and with significantly higher variation across animals than did B. minutum (Figure 4). Despite the stochastic failure to robustly colonize with non-native species, in general the same symbiont cluster patterning and growth occurs irrespective of symbiont species. For each symbiont species, doublet and triplet clusters each averaged greater than 10 clusters in polyps with rapid symbiosis establishment (i.e., continuous population growth over time), and averaged fewer than 10 clusters in polyps with slow symbiosis establishment (Supplementary Figures 4–6). Therefore, in our experiments the most important stage of symbiont incompatibility occurred during initial inoculation (especially with S. microadriaticum). Similar to native species, an important moment in colonization was the consistent measurement of larger clusters (greater than five cells) coupled with a symbiont density of between 100 and 200 cells and 25 cell clusters per sample image, as found on day 25 for S. microadriaticum (Figures 4A,C), and day 21 for D. trenchii (Figures 4A,D). This benchmark acted as a threshold for non-native species; before reaching this level of colonization, non-native species increased relatively slowly over the first few weeks, whereas afterward symbiont growth was much more robust. In our experiments, only a handful of inoculations of S. microadriaticum proliferated with a slower but similar pattern to B. minutum, resulting in the lowest colonization averages overall (Figure 4A and Supplementary Figure 5). A large proportion of D. trenchii inoculations began substantial proliferation of symbiont clusters after 3 weeks (Supplementary Figure 6). Even in replicates with the most robust initial colonization, non-native symbiont populations more closely matched the slowest growing native populations (Supplementary Figures 4–6).
Lower amounts of symbiont uptake and slower rates of host colonization by these non-native species were consistent with previous Aiptasia studies (Colley and Trench, 1983; Belda-Baillie et al., 2002; Gabay et al., 2018). Gabay et al. (2018) specifically examined the colonization patterns of symbionts within adult aposymbiotic Aiptasia and found that unsuccessful colonization with non-competent symbiont Effrenium voratum failed from the beginning, and that those with competent but non-native symbionts, S. microadriaticum and D. trenchii, start slowly and take twice as long to recolonize hosts than do B. minutum. These slower rates of colonization of a cnidarian host by non-native symbiont species have been observed in the colonization of the scyphozoan aposymbiotic Cassiopeia xamachana, a scyphozoan, and sea anemone Cereus pedunculatus, a sea anemone (Colley and Trench, 1983; Davy et al., 1997).
Polyps recolonized by native symbiont B. minutum experienced consistent symbiont cluster patterns during thermal stress. Larger cluster sizes decreased, and smaller cluster sizes temporarily remained stable or increased before subsequently decreasing through algal loss (Figures 3B, 4B). The initial winnowing of larger symbiont clusters may be a function of local tissue stress, which would result from local effects of reactive oxygen species production from both high algal densities and the host cells that contain these multiple algal cells (Lesser, 1996; Perez and Weis, 2006; Hawkins and Davy, 2012). This stress could also result directly in the temporary stability/increase of singlet clusters in B. minutum populations immediately post-bleaching, as increased exit of larger clusters may leave neighboring host cells containing only singlets. The subsequent rapid disappearance of B. minutum singlet populations indicated the relatively poor tolerance of the B. minutum—Aiptasia symbiosis to heat stress. Additional video observations were gathered during the time-point immediately following thermal stress treatment (Movie 1). In many samples, circulation of large symbiont clusters was observed within the gastrovascular cavity of the column and tentacles as evidenced by autofluorescence of the clusters. As a consequence of environmental stress, there are a variety of cellular mechanisms that may be acting to decrease symbiont density, including symbiont expulsion, host cell detachment, and apoptosis; however, the prevalence and co-occurrence of these mechanisms are not clear (Gates et al., 1992; Dunn et al., 2007; Bieri et al., 2016; Oakley and Davy, 2018). In our video observations, the gastrovascular circling of symbiont clusters between 1 and 5 cells in size provides evidence for the continued presence of a host membrane surrounding these circulating algal cells; whether this membrane is an intact host cell or a symbiosome membrane remains unclear.
The reduction of non-native symbiont populations in Aiptasia polyps as a result of increased temperature was tempered in hosts harboring non-native symbiont species. In contrast to the poor tolerance of native B. minutum populations to heat stress, S. microadriaticum and D. trenchii populations did not initially change in abundance as a result of heat stress, but instead remained at low population densities in cluster sizes ranging from 1 to 10 cells (Figure 4). Unlike 26°C treatments, populations of singlets and doublets did not significantly increase over time, therefore resulting in relatively stable symbiont densities.
The continued presence of these non-native species under elevated temperatures is consistent with their physiology in culture. These symbiont species differ significantly in their production of nitric oxide (NO) and tolerance to heat stress; B. minutum produces increased NO and experiences high photoinhibition under heat stress, whereas S. microadriaticum is a much more thermally tolerant species (Hawkins and Davy, 2012). In addition, coral monitoring during elevated temperatures has shown that thermally tolerant symbiont species such as D. trenchii are able to opportunistically recolonize coral hosts during and after bleaching events (Berkelmans and van Oppen, 2006; LaJeunesse et al., 2009; Boulotte et al., 2016; Cunning et al., 2018).
In Aiptasia, it appears that during ambient conditions, these opportunistic thermally tolerant symbionts are either unproductive or selfish, requiring hosts to upregulate their own catabolism of lipids and carbohydrates (Matthews et al., 2017, 2018). Host transcriptional responses also included upregulated innate immunity and stress pathways. The lack of an immediate bleaching response to elevated thermal conditions supports their hypothesized role as thermally tolerant, opportunistic, and less-productive symbionts (Stat et al., 2008). Despite these thermally tolerant phenotypes, S. microadriaticum and D. trenchii populations were not able to expand under thermal stress in our study. Our results are similar to previous work showing limited colonization success when attempting inoculation during periods of thermal stress (Gabay et al., 2019). Further exploration is needed to better understand how thermal stress limits proliferation of non-native species but not their persistence within hosts.
The simulations of native B. minutum symbiont population growth support the inclusion of both proliferation and migration in establishing symbiont colonization patterns (Figure 5). Migration was necessary in the models to create new clusters over time, otherwise populations developed a handful of large clusters that looked nothing like observed colonization patterns. Likewise, modeled cluster populations fit better when adjusting for the potential of density-dependent negative feedback on symbiont growth (Supplementary Tables 5, 6). Therefore, by using three simple inputs: The initial inoculum, a density-dependent growth rate, and a migration probability, we were able to more accurately model the colonization patterning observed in anemones. Despite limitations, our models did find better fit when taking into account a density-dependent effect on growth rate within symbiont clusters. This fit corresponds with previous literature where the biomass of individual cells is strongly negatively correlated with density and access to nitrogen. Previous studies have shown that algal growth and division within host cnidarians is strongly tied to nitrogen availability (Falkowski et al., 1993; Jones and Yellowlees, 1997), and that there is density-dependent nutrient competition among neighboring symbiont cells when approaching high population densities (Krueger et al., 2020). Metabolites from dense symbiont populations resemble metabolites from N-depleted algal cultures, and transcriptional data from dense symbiont populations show increased expression of genes involved in nitrogen acquisition (Xiang et al., 2020). Further studies could use our method to track proliferation patterns and experimentally manipulate the predicted metabolic balance of nitrogen acquisition and glucose production during the establishment of symbiont colonization (Cui et al., 2019).
The spatial organization of algal colonial population dynamics within cnidarians remains largely unstudied. In sea anemones such as Aiptasia, only a few studies have directly examined spatial patterning of algae within hosts during colonization and bleaching (Gabay et al., 2018; Tortorelli et al., 2020), and none have examined spatial dynamics in hospite at a cellular level. Another handful of studies have used techniques such as tissue maceration to isolate symbiotic cells and estimate densities of symbionts within each gastrodermal host cell (Muscatine et al., 1998; Ferrier-Pagès et al., 2001; Lecointe et al., 2013). Symbiodiniaceae population dynamics and movement have also been studied in organisms where spatial organization in the host plays a more clear role in colonization and/or bleaching. For example, in octocorals, proliferation dynamics of algal symbionts and their migration from polyps into stolons helps to mitigate the symbiont loss from thermal bleaching (Parrin et al., 2016). In symbioses with molluscs, the colonization of algal symbionts not only influences host development (Banaszak et al., 2013; Gula and Adams, 2018), but also creates a structured environment optimized for photosynthesis within the sunlight-facing mantle of giant clams by spatially orienting algae into micropillars (Holt et al., 2014).
Imaging small Aiptasia using confocal microscopy provided highly accurate data within the first week of inoculation. By analyzing brightfield confocal Z-stacks, we captured symbiont cell proliferation in hosts with high resolution. Despite this fidelity, imaging symbiont autofluorescence was slow and unreliable using this method. In general, confocal imaging took much longer than epifluorescence imaging. In addition, repeated anesthetic MgCl2 treatment for confocal imaging was detrimental to Aiptasia polyps. At the end of the experiment, MgCl2-relaxed Aiptasia tentacles appeared unhealthy, as evidenced by permanently contracted tentacles columns (data not shown). Finally, manual counting of symbionts within brightfield image stacks was needed to quantify symbiont cluster number and size. Though manual counts provided accurate symbiont cluster data, after 1 week of host colonization by symbionts, for groups of successfully colonized anemones, the increased abundance of symbionts made counting infeasible without automation.
Conversely, imaging colonization of Aiptasia by symbionts using epifluorescence microscopy provided consistent, repeatable measurements across all samples. Animals were minimally disrupted during imaging; the brightfield and epifluorescence imaging process was rapid and samples experienced no photobleaching as a result of fluorescent excitation. One important limitation was data storage; videos and their representative photo stacks required considerable storage space. All image analysis steps were able to be run in batch format and assessed afterward. Time-intensive steps occurred primarily during image analysis, specifically during image stitching into a single merged photo, which took approximately 2 min per sample. Image analysis steps could optionally take advantage of manual thresholding and tentacle gating if one is interested in symbiont density and tentacle location.
In general, the collapse of three dimensions into two dimensions sacrificed information on tentacle depth and likely resulted in overlap between symbiont clusters at higher symbiont densities during later weeks of colonization. Growth of patches, especially in the tentacle tips, is difficult to accurately assess based only on 2D area, which was one reason why cluster sizes greater than five were binned into cell size ranges. As a result of this limitation, it remains difficult to draw any conclusions about factors that limit the densities of symbiont populations within cnidarian hosts. The methods used here are therefore most useful during the initial weeks of colonization before cluster analysis of dense symbiont populations requires 3D imaging.
With our study, we provide a unique, in-depth view into the spatial organization and dynamics of algal symbionts within cnidarian host tissue. Here we show consistent growth in abundance and size of cell clusters pattern in the host during the initial weeks of colonization, while elevated temperature affects larger clusters and native symbionts more rapidly. Further studies in these areas of spatial dynamics of partnerships will help us better understand the responses of Symbiodiniaceae within their hosts to environmental challenges on the horizon. We hope the imaging method developed here enables others to use similar rapid and inexpensive assays to test how different winnowing mechanisms affect initial colonization, including altering host-symbiont nutrition or host-symbiont recognition pathways such as glycan-lectin signaling. In conclusion, there is valuable information to gain between cellular and organismal scales, and understanding these dynamic patterns inside symbiotic host tissue can help better integrate these two spatial levels of understanding.
The datasets and code presented in this study can be accessed at the online repository GitHub (https://github.com/trtivey/aiptasia-symbiont-colonization-patterns).
TT, TC, and VW conceived and designed the experiments. TT and TC performed the experiments and analyzed the data. TT wrote the manuscript and prepared the figures. TT and VW made revisions to the manuscript. All authors contributed to the article and approved the submitted version.
This work was funded by award IOB1529059 from the National Science Foundation awarded to VW. Support for the confocal microscope was from award 1337774 from the National Science Foundation, MI: Acquisition of Confocal and Two-Photon Excitation Microscope. We acknowledge the Confocal Microscopy Facility of the Center for Quantitative Life Sciences at Oregon State University.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer RC declared a past co-authorship with one of the authors VW to the handling editor.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Milan Sengthep and Debbie B. Kim for their work in experimental data collection and animal care. We thank John Parkinson and Shumpei Maruyama for assistance with undergraduate training and mentoring. This work would not be possible without the support of Oregon State University undergraduate research initiatives including URSA Engage and the SURE Science Program.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.808696/full#supplementary-material
Abrego, D., Van Oppen, M. J. H., and Willis, B. L. (2009). Highly infectious symbiont dominates initial uptake in coral juveniles. Mole. Ecol. 18, 3518–3531. doi: 10.1111/j.1365-294X.2009.04275.x
Baghdasarian, G., and Muscatine, L. (2000). Preferential expulsion of dividing algal cells as a mechanism for regulating algal-cnidarian symbiosis. Biol. Bull. 199, 278–286. doi: 10.2307/1543184
Baker, A. C. (2003). Flexibility and specificity in coral-algal symbiosis: Diversity, ecology, and biogeography of Symbiodinium. Annu. Rev. Ecol. Evolut. System. 34, 661–689. doi: 10.1146/annurev.ecolsys.34.011802.132417
Banaszak, A. T., García Ramos, M., and Goulet, T. L. (2013). The symbiosis between the gastropod Strombus gigas and the dinoflagellate Symbiodinium: An ontogenic journey from mutualism to parasitism. J. Exp. Mar. Biol. Ecol. 449, 358–365.
Bay, L. K., Doyle, J., Logan, M., and Berkelmans, R. (2016). Recovery from bleaching is mediated by threshold densities of background thermo-tolerant symbiont types in a reef-building coral. Roy. Soc. Open Sci. 3:160322. doi: 10.1098/rsos.160322
Belda-Baillie, C. A., Baillie, B. K., and Maruyama, T. (2002). Specificity of a Model Cnidarian-Dinoflagellate Symbiosis. Biol. Bull. 202, 74–85.
Berkelmans, R., and van Oppen, M. J. H. (2006). The role of zooxanthellae in the thermal tolerance of corals: a ‘nugget of hope’ for coral reefs in an era of climate change. Proc. Roy. Soc. B 273, 2305–2312. doi: 10.1098/rspb.2006.3567
Bieri, T., Onishi, M., Xiang, T., Grossman, A. R., and Pringle, J. R. (2016). Relative Contributions of Various Cellular Mechanisms to Loss of Algae during Cnidarian Bleaching. PLoS One 11:e0152693. doi: 10.1371/journal.pone.0152693
Bolte, S., and Cordelieres, F. P. (2006). A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224, 213–232. doi: 10.1111/j.1365-2818.2006.01706.x
Boulotte, N. M., Dalton, S. J., Carroll, A. G., Harrison, P. L., Putnam, H. M., Peplow, L. M., et al. (2016). Exploring the Symbiodinium rare biosphere provides evidence for symbiont switching in reef-building corals. ISME J. 10, 2693–2701. doi: 10.1038/ismej.2016.54
Camaya, A. P. (2020). Stages of the symbiotic zooxanthellae–host cell division and the dynamic role of coral nucleus in the partitioning process: a novel observation elucidated by electron microscopy. Coral Reefs 39, 929–938. doi: 10.1007/s00338-020-01912-y
Coffroth, M. A., Santos, S. R., and Goulet, T. L. (2001). Early ontogenetic expression of specificity in a cnidarian-algal symbiosis. Mar. Ecol. Progr. Ser. 222, 85–96. doi: 10.3354/meps222085
Colley, N. J., and Trench, R. K. (1983). Selectivity in Phagocytosis and Persistence of Symbiotic Algae by the Scyphistoma Stage of the Jellyfish Cassiopeia xamachana. Proc. Roy. Soc. Lon. Ser. B Biol. Sci. 219, 61–82. doi: 10.1098/rspb.1983.0059
Cui, G., Liew, Y. J., Li, Y., Kharbatia, N., Zahran, N. I., Emwas, A.-H., et al. (2019). Host-dependent nitrogen recycling as a mechanism of symbiont control in Aiptasia. PLoS Genet. 15:e1008189. doi: 10.1371/journal.pgen.1008189
Cunning, R., Silverstein, R. N., and Baker, A. C. (2018). Symbiont shuffling linked to differential photochemical dynamics of Symbiodinium in three Caribbean reef corals. Coral Reefs 37, 145–152. doi: 10.1007/s00338-017-1640-3
Davy, S. K., Allemand, D., and Weis, V. M. (2012). Cell Biology of Cnidarian-Dinoflagellate Symbiosis. Microbiol. Mole. Biol. Rev. 76, 229–261. doi: 10.1128/MMBR.05014-11
Davy, S. K., Lucas, I. A. N., and Turner, J. R. (1997). Uptake and Persistence of Homologous and Heterologous Zooxanthellae in the Temperate Sea Anemone Cereus pedunculatus (Pennant). Biol. Bull. 192, 208–216. doi: 10.2307/1542715
Dunn, S. R., Schnitzler, C. E., and Weis, V. M. (2007). Apoptosis and autophagy as mechanisms of dinoflagellate symbiont release during cnidarian bleaching: every which way you lose. Proc. Roy. Soc. B 274, 3079–3085. doi: 10.1098/rspb.2007.0711
Dunn, S. R., and Weis, V. M. (2009). Apoptosis as a post-phagocytic winnowing mechanism in a coral-dinoflagellate mutualism. Environ. Microbiol. 11, 268–276. doi: 10.1111/j.1462-2920.2008.01774.x
Falkowski, P. G., Dubinsky, Z., Muscatine, L., and McCloskey, L. (1993). Population control in symbiotic corals: Ammonium ions and organic materials maintain the density of zooxanthellae. BioScience 43, 606–611. doi: 10.2307/1312147
Ferrier-Pagès, C., Schoelzke, V., Jaubert, J., Muscatine, L., and Hoegh-Guldberg, O. (2001). Response of a scleractinian coral, Stylophora pistillata, to iron and nitrate enrichment. J. Exp. Mar. Biol. Ecol. 259, 249–261. doi: 10.1016/S0022-0981(01)00241-6
Fitt, W. K. (2000). Cellular Growth of Host and Symbiont in a Cnidarian-Zooxanthellar Symbiosis. Biol. Bull. 198, 110–120. doi: 10.2307/1542809
Fitt, W. K., and Cook, C. B. (2001). The effects of feeding or addition of dissolved inorganic nutrients in maintaining the symbiosis between dinoflagellates and a tropical marine cnidarian. Mar. Biol. 139, 507–517. doi: 10.1007/s002270100598
Fitt, W. K., and Trench, R. K. (1983). Endocytosis of the symbiotic dinoflagellate symbiodinium microadriaticum Freudenthal by endodermal cells of the scyphistomae of Cassiopeia xamachana and resistance of the algae to host digestion. J. Cell Sci. 64, 195–212. doi: 10.1111/j.1469-8137.1983.tb03456.x
Gabay, Y., Parkinson, J. E., Wilkinson, S. P., Weis, V. M., and Davy, S. K. (2019). Inter-partner specificity limits the acquisition of thermotolerant symbionts in a model cnidarian-dinoflagellate symbiosis. ISME J. 13, 2489–2499. doi: 10.1038/s41396-019-0429-5
Gabay, Y., Weis, V. M., and Davy, S. K. (2018). Symbiont Identity Influences Patterns of Symbiosis Establishment, Host Growth, and Asexual Reproduction in a Model Cnidarian-Dinoflagellate Symbiosis. Biol. Bull. 234, 1–10. doi: 10.1086/696365
Gates, R. D., Baghdasarian, G., and Muscatine, L. (1992). Temperature stress causes host cell detachment in symbiotic cnidarians: implications for coral bleaching. Biol. Bull. 182, 324–332. doi: 10.1007/BF00255468
Goldstein, B., and King, N. (2016). The Future of Cell Biology: Emerging Model Organisms. Trends Cell Biol. 26, 818–824. doi: 10.1016/j.tcb.2016.08.005
Gula, R. L., and Adams, D. K. (2018). Effects of Symbiodinium Colonization on Growth and Cell Proliferation in the Giant Clam Hippopus hippopus. Biol. Bull. 234, 130–138. doi: 10.1086/698265
Hambleton, E. A., Guse, A., and Pringle, J. R. (2014). Similar specificities of symbiont uptake by adults and larvae in an anemone model system for coral biology. J. Exp. Biol. 217, 1613–1619. doi: 10.1242/jeb.095679
Harii, S., Yasuda, N., Rodriguez-Lanetty, M., Irie, T., and Hidaka, M. (2009). Onset of symbiosis and distribution patterns of symbiotic dinoflagellates in the larvae of scleractinian corals. Mar. Biol. 156, 1203–1212. doi: 10.1007/s00227-009-1162-9
Hartmann, A. C., Baird, A. H., Knowlton, N., and Huang, D. (2017). The Paradox of Environmental Symbiont Acquisition in Obligate Mutualisms. Curr. Biol 27, 3711–3716.e3. doi: 10.1016/j.cub.2017.10.036
Hawkins, T. D., and Davy, S. K. (2012). Nitric Oxide Production and Tolerance Differ Among Symbiodinium Types Exposed to Heat Stress. Plant Cell Physiol. 53, 1889–1898. doi: 10.1093/pcp/pcs127
Hoegh-Guldberg, O., Kennedy, E. V., Beyer, H. L., McClennen, C., and Possingham, H. P. (2018). Securing a Long-term Future for Coral Reefs. Trends Ecol. Evolut. 33, 936–944. doi: 10.1016/j.tree.2018.09.006
Holt, A. L., Vahidinia, S., Gagnon, Y. L., Morse, D. E., and Sweeney, A. M. (2014). Photosymbiotic giant clams are transformers of solar flux. J. R. Soc. Interf. 11:20140678. doi: 10.1098/rsif.2014.0678
Jones, R. J., and Yellowlees, D. (1997). Regulation and control of intracellular algae (= zooxanthellae) in hard corals. Philosoph. Transact Roy. Soc. B 352, 457–468. doi: 10.1098/rstb.1997.0033
Krueger, T., Horwitz, N., Bodin, J., Giovani, M.-E., Escrig, S., Fine, M., et al. (2020). Intracellular competition for nitrogen controls dinoflagellate population density in corals. Proc. Roy. Soc. B 287:20200049. doi: 10.1098/rspb.2020.0049
LaJeunesse, T. C., Parkinson, J. E., Gabrielson, P. W., Jeong, H. J., Reimer, J. D., Voolstra, C. R., et al. (2018). Systematic Revision of Symbiodiniaceae Highlights the Antiquity and Diversity of Coral Endosymbionts. Curr. Biol. 28, 2570–2580.e6. doi: 10.1016/j.cub.2018.07.008
LaJeunesse, T. C., Smith, R. T., Finney, J., and Oxenford, H. (2009). Outbreak and persistence of opportunistic symbiotic dinoflagellates during the 2005 Caribbean mass coral ‘bleaching’ event. Proc. Roy. Soc. B 276, 4139–4148. doi: 10.1098/rspb.2009.1405
Lecointe, A., Cohen, S., Gèze, M., Djediat, C., Meibom, A., and Domart-Coulon, I. (2013). Scleractinian coral cell proliferation is reduced in primary culture of suspended multicellular aggregates compared to polyps. Cytotechnology 65, 705–724. doi: 10.1007/s10616-013-9562-6
Lesser, M. P. (1996). Elevated temperatures and ultraviolet radiation cause oxidative stress and inhibit photosynthesis in symbiotic dinoflagellates. Limnol. Oceanogr. 41, 271–283. doi: 10.4319/lo.1996.41.2.0271
Little, A. F., Oppen, M. J. H. V., and Willis, B. L. (2004). Flexibility in Algal Endosymbioses Shapes Growth in Reef Corals. Science 304, 1492–1494. doi: 10.1126/science.1095733
Manzello, D. P., Matz, M. V., Enochs, I. C., Valentino, L., Carlton, R. D., Kolodziej, G., et al. (2019). Role of host genetics and heat-tolerant algal symbionts in sustaining populations of the endangered coral Orbicella faveolata in the Florida Keys with ocean warming. Global Change Biol. 25, 1016–1031. doi: 10.1111/gcb.14545
Matthews, J. L., Crowder, C. M., Oakley, C. A., Lutz, A., Roessner, U., Meyer, E., et al. (2017). Optimal nutrient exchange and immune responses operate in partner specificity in the cnidarian-dinoflagellate symbiosis. Proc. Natl. Acad. Sci. U.S.A. 114, 13194–13199. doi: 10.1073/pnas.1710733114
Matthews, J. L., Oakley, C. A., Lutz, A., Hillyer, K. E., Roessner, U., Grossman, A. R., et al. (2018). Partner switching and metabolic flux in a model cnidarian–dinoflagellate symbiosis. Proc. Roy. Soc. B 285:20182336. doi: 10.1098/rspb.2018.2336
Matthews, J. L., Sproles, A. E., Oakley, C. A., Grossman, A. R., Weis, V. M., and Davy, S. K. (2016). Menthol-induced bleaching rapidly and effectively provides experimental aposymbiotic sea anemones (Aiptasia sp.) for symbiosis investigations. J. Exp. Biol. 219, 306–310. doi: 10.1242/jeb.128934
McAuley, P. J., and Cook, C. B. (1994). Effects of host feeding and dissolved ammonium on cell division and nitrogen status of zooxanthellae in the hydroid Myrionema amboinense. Mar. Biol. 121, 343–348. doi: 10.1007/BF00346743
Muscatine, L., Ferrier-Pagès, C., Blackburn, A., Gates, R. D., Baghdasarian, G., and Allemand, D. (1998). Cell specific density of symbiotic dinoflagellates in tropical anthozoans. Coral Reefs 17, 329–337. doi: 10.1007/S003380050133
Muscatine, L., and Porter, J. W. (1977). Reef Corals: Mutualistic Symbioses Adapted to Nutrient-Poor Environments. BioScience 27, 454–460. doi: 10.2307/1297526
Oakley, C. A., and Davy, S. K. (2018). “Cell Biology of Coral Bleaching,” in Coral Bleaching. Ecological Studies (Analysis and Synthesis), eds M. J. H. van Oppen and J. M. Lough (Cham: Springer), 189–211. doi: 10.1007/978-3-319-75393-5_8
Parrin, A. P., Goulet, T. L., Yaeger, M. A., Bross, L. S., McFadden, C. S., and Blackstone, N. W. (2016). Symbiodinium migration mitigates bleaching in three octocoral species. J. Exp. Mar. Biol. Ecol. 474, 73–80. doi: 10.1016/j.jembe.2015.09.019
Perez, S., and Weis, V. M. (2006). Nitric oxide and cnidarian bleaching: an eviction notice mediates breakdown of a symbiosis. J. Exp. Biol. 209, 2804–2810. doi: 10.1242/jeb.02309
Rueden, C. T., Schindelin, J., Hiner, M. C., DeZonia, B. E., Walter, A. E., Arena, E. T., et al. (2017). ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinform. 18:529. doi: 10.1186/s12859-017-1934-z
Schindelin, J., Arganda-carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. doi: 10.1038/nmeth.2019
Schoenberg, D. A., and Trench, R. K. (1980). Genetic Variation in Symbiodinium (=Gymnodinium) microadriaticum Freudenthal, and Specificity in its Symbiosis with Marine Invertebrates. III. Specificity and Infectivity of Symbiodinium microadriaticum. Proc. Roy. Soc. Lon. Ser. B 207, 445–460. doi: 10.1098/rspb.1980.0031
Schwarz, J. A., Krupp, D. A., and Weis, V. M. (1999). Late larval development and onset of symbiosis in the scleractinian coral Fungia scutaria. Biol. Bull. 196, 70–79. doi: 10.2307/1543169
Silverstein, R. N., Cunning, R., and Baker, A. C. (2015). Change in algal symbiont communities after bleaching, not prior heat exposure, increases heat tolerance of reef corals. Global Change Biol. 21, 236–249. doi: 10.1111/gcb.12706
Stat, M., Morris, E., and Gates, R. D. (2008). Functional diversity in coral-dinoflagellate symbiosis. Proc. Natl. Acad. Sci. U.S.A. 105, 9256–9261. doi: 10.1073/pnas.0801328105
Thornhill, D. J., LaJeunesse, T. C., Kemp, D. W., Fitt, W. K., and Schmidt, G. W. (2006). Multi-year, seasonal genotypic surveys of coral-algal symbioses reveal prevalent stability or post-bleaching reversion. Mar. Biol. 148, 711–722. doi: 10.1007/s00227-005-0114-2
Tivey, T. R., Parkinson, J. E., and Weis, V. M. (2020). Host and Symbiont Cell Cycle Coordination Is Mediated by Symbiotic State, Nutrition, and Partner Identity in a Model Cnidarian-Dinoflagellate Symbiosis. mBio 11, e2626–e2619. doi: 10.1128/mBio.02626-19
Tortorelli, G., Belderok, R., Davy, S. K., McFadden, G. I., and van Oppen, M. J. H. (2020). Host Genotypic Effect on Algal Symbiosis Establishment in the Coral Model, the Anemone Exaiptasia diaphana, From the Great Barrier Reef. Front. Mar. Sci. 6:833. doi: 10.3389/fmars.2019.00833
Wakefield, T. S., Farmer, M. A., and Kempf, S. C. (2000). Revised description of the fine structure of in situ “Zooxanthellae” genus Symbiodinium. Biol. Bull. 199, 76–84. doi: 10.2307/1542709
Weis, V. M. (2008). Cellular mechanisms of Cnidarian bleaching: stress causes the collapse of symbiosis. J. Exp. Biol. 211, 3059–3066. doi: 10.1242/jeb.009597
Weis, V. M., Davy, S. K., Hoegh-Guldberg, O., Rodriguez-Lanetty, M., and Pringle, J. R. (2008). Cell biology in model systems as the key to understanding corals. Trends Ecol. Evolut. 23, 369–376. doi: 10.1016/j.tree.2008.03.004
Weis, V. M., Reynolds, W. S., DeBoer, M. D., and Krupp, D. A. (2001). Host-symbiont specificity during onset of symbiosis between the dinoflagellates Symbiodinium spp. and planula larvae of the scleractinian coral Fungia scutaria. Coral Reefs 20, 301–308. doi: 10.1007/s003380100179
Wolfowicz, I., Baumgarten, S., Voss, P. A., Hambleton, E. A., Voolstra, C. R., Hatta, M., et al. (2016). Aiptasia sp. larvae as a model to reveal mechanisms of symbiont selection in cnidarians. Scientific Rep. 6:32366. doi: 10.1038/srep32366
Xiang, T., Hambleton, E. A., DeNofrio, J. C., Pringle, J. R., and Grossman, A. R. (2013). Isolation of clonal axenic strains of the symbiotic dinoflagellate Symbiodinium and their growth and host specificity 1. J. Phycol. 49, 447–458. doi: 10.1111/jpy.12055
Keywords: Symbiodiniaceae, cnidarian, coral reefs, dinoflagellate, microscopy, symbiosis
Citation: Tivey TR, Coleman TJ and Weis VM (2022) Spatial and Temporal Patterns of Symbiont Colonization and Loss During Bleaching in the Model Sea Anemone Aiptasia. Front. Mar. Sci. 9:808696. doi: 10.3389/fmars.2022.808696
Received: 03 November 2021; Accepted: 15 February 2022;
Published: 15 March 2022.
Edited by:
Russell T. Hill, University of Maryland, Baltimore County, United StatesReviewed by:
Fan Zhang, Baylor College of Medicine, United StatesCopyright © 2022 Tivey, Coleman and Weis. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Trevor R. Tivey, dHJ0NDNAY29ybmVsbC5lZHU=
†Present address: Trevor R. Tivey, Boyce Thompson Institute, Ithaca, NY, United States
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.