
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 14 February 2022
Sec. Microbial Symbioses
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.808132
 Ya-Fan Chan1
Ya-Fan Chan1 Chia-Yu Chen2
Chia-Yu Chen2 Chih-Ying Lu2,3,4
Chih-Ying Lu2,3,4 Yung-Chi Tu2
Yung-Chi Tu2 Kshitij Tandon2
Kshitij Tandon2 Shinya Shikina5,6
Shinya Shikina5,6 Sen-Lin Tang2*
Sen-Lin Tang2*
Endozoicomonas, a core bacterial group in corals, may also be a coral symbiont. Endozoicomonas communities often decrease rapidly in corals under heat stress. However, how the bacteria respond to changes in temperature and coral host during heat stress is unknown. Here, we employed the cultivable, dominant species E. montiporae as a working organism to explore how Endozoicomonas responds to heat stress. We designed two experiments to clarify the extent to which E. montiporae is influenced by temperature and coral host. We detected differentially expressed protein (DEP) profiles in this bacterium at 31 and 33°C compared to 25°C by tandem mass tags-based quantitative proteome analysis. Fifty DEPs, including many heat shock proteins, were detected when the temperature changed. The expression of antioxidant defense proteins and key pyruvate synthase proteins decreased, suggesting that E. montiporae were in a physiology of stress at 33°C. Furthermore, some proteins were differentially expressed because of the heat-stress-treated coral lysate specifically, suggesting that not only heat but also heat-induced host factors can affect the protein expression of the bacterium. This study provides an in-depth analysis of how the molecular mechanisms of Endozoicomonas are affected by heat stress and coral host.
The coral holobiont is composed of coral and various associated microorganisms, including algae, bacteria, archaea, fungi, protists, and viruses. Temperature is a crucial factor governing the composition of this holobiont (Hughes et al., 2017). When the seawater temperature increases, heat stress impacts not only the coral host but also all of the coral microbial partners in the coral holobiont (Bourne et al., 2016). The most typical example is that coral-algae symbiosis can be dissociated by high temperature (Kemp et al., 2014; Hughes et al., 2018). Along with symbiotic algae, the impact of heat stress on the coral bacteria has recently received increasing attention due to these bacteria’s indispensable ecological contribution to coral hosts (Bourne et al., 2008; Mouchka et al., 2010; McDevitt-Irwin et al., 2017).
Bacteria in the coral holobiont are highly sensitive to heat stress. For instance, Ziegler et al. (2017) demonstrated that coral bacteria were significantly affected by short-term heat stress (35°C for 1 h) within 20 h of incubation. Similarly, Lee et al. (2015) showed rapid changes in the composition of coral bacteria between coral mucus and tissue under heat stress. For example, the relative abundance of α-Proteobacteria and Verrucomicrobiaceae became dominant in the mucus from 29 to 31°C; in contrast, the relative abundance of γ-Proteobacteria dropped in both mucus and tissue. Moreover, Shiu et al. (2017) even showed that the coral bacterial community quickly changed from normal to pathogenic- or stress-associated microbiota within 12 h of a short-term heat stress experiment (30°C), whereas the density and photosynthetic efficiency (Fv/Fm) of Symbiodiniaceae remained unchanged.
Although heat stress usually elicits a clear shift in bacterial community, we still do not know how coral bacteria respond to heat stress, or their underlying biochemical or molecular mechanisms. This is because it is difficult to detect the response of a specific bacterial group in corals, as many studies have shown that coral-associated bacterial communities are diverse and dynamic in various corals with environmental variations. Hence there is a need to establish a bacterial manipulable model to explain in-depth physiological and biochemical characteristics that change when bacteria are exposed to specific biotic or abiotic factors. A similar idea was also proposed recently by van Oppen and Blackall (2019): using pure bacterial cultures for molecular approaches could provide insights into the in-depth, molecular mechanisms of coral bacteria and their interactive relationships with their coral host under a specific environmental condition, such as heat stress. Gao et al. (2021) used a cultivable coral pathogenic bacterium, Vibrio coralliilyticus, to investigate transcriptional responses when bacteria interact with coral mucus, and furthermore to elucidate a potential behavior and infection mode of the V. coralliilyticus that helps explain the pathogenesis of V. coralliilyticus and coral disease. However, there is no working model to assess the health associated bacteria in coral for such an advanced study.
To establish the working model, the first need is to find an appropriate coral bacterium strain that may represent specific functionally important or dominant bacteria in the coral microbiota. Of the vast number of coral bacteria, the genus Endozoicomonas (Gammaproteobacteria, Oceanospirillales, Hahellaceae or Endozoicomonaceae) is the best studied and is recognized to be a functionally important, dominant bacterium in the core microbiome of various Scleractinian corals, such as common species of Acropora, Stylophora, Pocillopora, and Porites (Bayer et al., 2013; Mendoza et al., 2013; Meyer et al., 2014; Tandon et al., 2020). Endozoicomonas has long been suggested to be a potential bacterial symbiont (Bourne et al., 2016; Neave et al., 2016). Hence, it is a suitable bacterium for establishing such a working model. To date, there are only three cultivable Endozoicomonas species isolated from Scleractinian corals: E. montiporae, E. acroporae Acr-14T, and E. coralli (Yang et al., 2010; Sheu et al., 2017; Chen et al., 2019). Of them, only E. montiporae (E. montiporae CL-33T) has had its complete genome sequenced. The genomic features also indicate that E. montiporae is a facultative symbiont (Ding et al., 2016; Neave et al., 2017).
Furthermore, in Lee et al. (2015), E. montiporae prevailed in Acropora sp., and the former’s relative abundance decreased rapidly in the latter – from 80% to less than 20% – when the temperature was increased to 31 and 33°C. Besides, a shift in the associated bacterial community (the E. montiporae was the second most dominant in the community) happened in the coral mucus when the sugar composition of coral mucus was altered by heat stress (Lee et al., 2016). Based on these tight relationships between coral host and this bacterium, we believed that E. montiporae is the most suitable model microorganism for developing a working model to investigate the in-depth molecular physiological response of coral bacteria under heat stress (Shiu et al., 2017).
In this study, we used E. montiporae as a model organism and detected differentially expressed protein profiles of this bacterium in vitro at two high temperatures, 31 and 33°C, based on the previous study (Lee et al., 2015). Furthermore, we assumed that some of the heat stress-induced responses in the bacterium can be affected not only directly by temperature but also indirectly by the host factors, also induced by heat stress. Thus, the differentially expressed protein profiles of the bacteria were also detected by co-incubating with coral lysates after high temperature treatments. We found that temperature was the main factor explaining the changes in most proteins of E. montiporae. However, some proteins were affected by factors of temperature-induced coral lysates. Further discussion is provided on the molecular response of the specific coral bacterium under heat stress and influence from heat-induced coral hosts.
Endozoicomonas montiporae CL-33T was cultivated in an MMBv4 medium at 25°C on a shaker with 200 rpm as previously described (Ding et al., 2016).
The initial E. montiporae CL-33T culture was grown at 25°C to an optical density at 600 nm (OD) of 0.6, and then diluted 1/200 in total of nine Falcon centrifuge tubes with 20 mL fresh MMBv4 medium (Ding et al., 2016). Nine bottles were incubated at 25, 31, and 33°C (n = 3 for each temperature) (Figure 1A). Bacterial growth was monitored by a spectrophotometer, measuring the OD value after 16, 24 and 48 h from each tube. A 1 mL subsample from each cultural bottle was centrifuged at 9,000 g at 4°C. After discarding the supernatant, the pellet was washed with 1 mL 1× phosphate-buffered saline (PBS) three times. A total of 27 sample tubes were flash-frozen using liquid nitrogen immediately and stored at −80°C until total protein extraction.
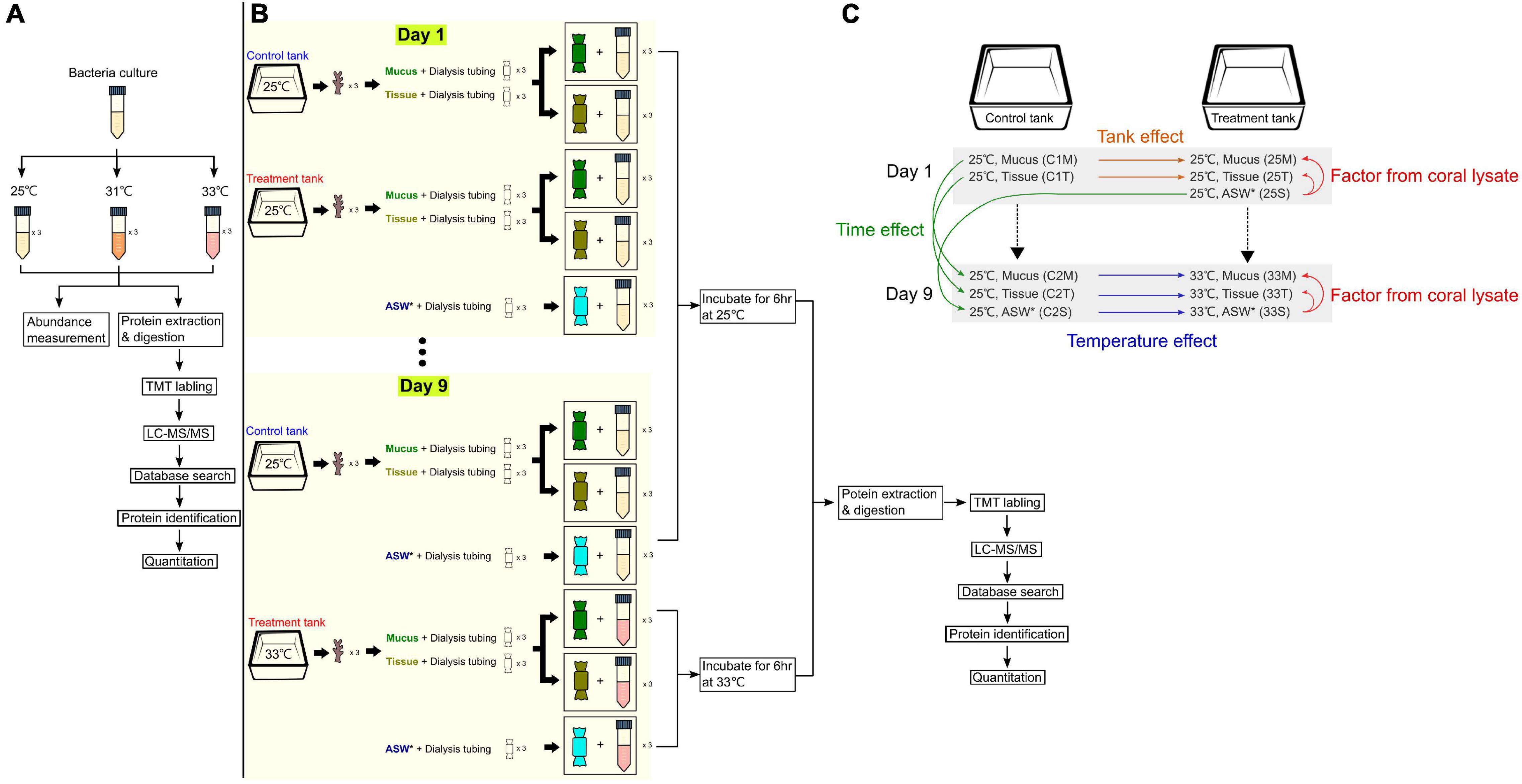
Figure 1. Experimental designs and setup. (A) Experiment to identify differentially expressed proteins when the bacterial culture (Endozoicomonas montiporae CL-33T) was exposed to different heat stresses. (B) Setup for coral lysate experiment, where coral mucus and tissue were incubated with cultures of E. montiporae CL-33T for 9 days, with temperature increased by 1°C per day (25 to 33°C). (C) Overview of the different factors analyzed throughout the coral lysate experiment. * denotes artificial sea water.
To test the protein profiles of E. montiporae co-incubating with coral lysates, we prepared and placed coral lysates into dialysis tubing (D9777, 14 kDa for molecular weight cutoff; Sigma, Aldrich, MN, United States) and co-incubated the dialysis tubing in the bacterial culture at a specific temperature. The details of the experiment are described in the following paragraphs.
For the dialysis tubing preparation, the tubing was cut into 25-cm long pieces, boiled in the washing solution (1 mM EDTA, 0.2% w/v sodium bicarbonate) for 10 min and washed three times with sterilized water. The treated tubing was then moved into another glass bottle with boiled-EDTA solution (1 mM, pH = 8.0) for 10 min. The tubing was stored at 4°C until use.
To prepare the coral lysate, nubbins of the coral Acropora spp. (about 3 cm long for each) were collected from one colony from the northeast coast of Taiwan (25°02′N, 121°98′E). Coral nubbins (n = 12) were acclimated in a 30-liter aquarium filled with filtered nature seawater for 2 weeks at 25°C. The temperature of the water baths was maintained by Hailea 150A aquarium chillers. Light intensity was about 150-μ mol photons m–2 s–1 under 12/12 h light and dark cycles.
The temperature in one treatment tank (with six nubbins) was raised by 1°C per day for 9 days from 25 to 33°C. One control tank (with 6 nubbins) was maintained at 25°C throughout the experiment (Figure 1B). HOBO temperature loggers were placed in the tanks to monitor the water temperature diving- pulse amplitude modulated (PAM) (Walz, Germany, MI: 11, SI: 8, saturation width: 0.8 s, Gain: 10∼12, Damp: 2) was used to monitor the change in coral photosystem health. Dark-adapted yield values (Fv/Fm) were estimated from six randomly chosen coral nubbins from each tank. Coral mucus and tissue were collected from three randomly chosen coral nubbins on Days 1 and 9 from the two tanks. The detailed method for coral mucus and tissue collection is described below.
First, the nubbins were gently rinsed with artificial seawater (ASW). When the nubbins were upside-down, the mucus was dropped from the nubbins and added to a 5 mL tube. Later, we sprayed the tissues (with some mucus) from the same nubbin with ASW. In the laminar hood, we transferred mucus (1 mL) and tissue (1 mL) into two separate dialysis tubings. These dialysis tubings (one for mucus and the other for tissue) were placed into two 50 mL Falcon tubes containing 15 mL bacteria cultures, separately. A total of 12 samples from the treatment (mucus: n = 3, tissue: n = 3) and control (mucus: n = 3, tissue: n = 3) tanks were incubated at 25°C at 200 rpm on Day 1 (Figure 1B). Another dialysis bag, which contained ASW (1 mL), was put into another 15 mL bacteria culture with three replicates, which acted as controls (25°C and 200 rpm). After 6 h of incubation, all the bacteria culture (n = 15) from each tube were collected into several 2 mL Eppendorf tubes and centrifuged at 9,000 × g for 10 min. The supernatant was discarded, and the pellet was stored at −80°C until total protein extraction. Bacteria growth (1 mL) was monitored by measuring the OD values at the same time.
On the ninth day, coral mucus and tissue were collected using the same approach described above – except that the sample tubes, which had the coral extract (mucus or tissue) from the treatment tank, were now at 33°C (Figure 1B). Three of the ASW sample tubes were incubated at 33°C as well. For the control tank, those sample tubes (containing mucus, tissue or ASW) were incubated at 25°C. Thus, there were a total of 18 samples on the ninth day. After 6 h, all of the 15 mL bacteria cultures were collected from the Falcon tubes and the OD value was measured.
Total protein was extracted using ExtractPRO Protein Extraction Reagent (VisualProtein, Energenesis Biomedical, Taiwan) following the manufacturer’s instructions. The protein pellet was firstly suspended in 8 M Urea in 50 mM Tris–HCl (pH 8.0) for the extraction.
Protease digestion and labeling before LC-MS analysis were performed as described in Lan et al. (2011). Protein concentrations were measured using a Pierce 660 nm protein Assay kit (Thermo Fisher Scientific, Rockford, IL, United States). Proteins (100 μg) from each sample were reduced in 10 mM dithiothreitol for 1 h at 37°C and alkylated in 50 mM iodoacetamide at room temperature for 30 min in the dark. The protein solution was then diluted with 4 M urea with 50 mM Tris–HCl, pH 8.5 and digested with 250 units/ml of Benzonase (Sigma-Aldrich, St. Louis, MO, United States) at room temperature for 2 h, followed by lysyl endopeptidase (Wako, Japan) digestion [1:200 (w/w)] at room temperature for 4 h. The protein solution was further diluted down to less than 2 M urea with 50 mM Tris–HCl (pH 8) and incubated with 2 μg of modified trypsin (w/w, 1:50) (Promega, Madison, WI, United States) at 37°C overnight. This protease digested solution was acidified with 10% trifluoroacetic acid, desalted using an Oasis HLB cartridge (Waters) and dried with a SpeedVac (Thermo Fisher Scientific, Rockford, IL, United States).
After protein digestion, the peptide mixture was labeled with TMT 10-plex and 11-plex isobaric tandem mass tags (catalog nos. 90110 and A34808, Thermo Fisher Scientific, Rockford, IL, United States) following the manufacturer’s instruction. The labeled peptide samples were then pooled and lyophilized in a vacuum concentrator. The final dried pellet was re-dissolved in 10 of 3% ACN/0.1% formic acid and its protein profile analyzed by LC-MS/MS (Orbitrap Fusion™ Tribrid™ Mass Spectrometer).
Peptide and protein identification were performed using the Proteome Discoverer software (v2.2, Thermo Fisher Scientific) with SEQUEST search engine (MacCoss et al., 2002; Muthusamy et al., 2017). Protein identification used 10 ppm and a fragment ion mass tolerance of 0.02 Da. The q-value threshold of 0.01 was used to filter the peptides. In addition, the minimal required peptide length was set to two peptides per protein. Only the proteins that were exhibited in all three replicate samples were kept.
The identified proteins were subjected to Gene Ontology (GO) annotation using the analytical system of Generic Gene Ontology Term Finder1. The biological pathways of the proteins were acquired from the KEGG Pathway database2 coupled with UniProtKB annotation (UniProtKB database3). Protein profiles were analyzed for statistical significance analysis with the Proteome Discoverer software. Proteins with a log2 fold change (log2FC) < 1.5 (or > 1.5) and p < 0.05 were considered significantly different in the protein quantification (one-way ANOVA). Furthermore, significantly expressed proteins were clustered into different function categories using the Cluster of Orthologous Groups of proteins (COG) database4. Hierarchical clustering analysis and nMDS analysis were used to visualize the pairwise dissimilarity in protein composition between different samples after transformation [Log (x + 1)] by Primer 6 (PRIMER-E, Lutton, Ivybridge, United Kingdom) (Clarke and Gorley, 2005).
In the coral lysate experiment, comparisons among or within the treatment and control experiments revealed the proteins that were differentially produced across the bacteria that were affected by four different factors (coral lysate, tank, temperature, and time) (Figure 1C). (1) The differences in the proteins that the bacteria produced between the artificial seawater and mucus (or tissue) represented the effect of coral lysate in 25 or 33°C treatment experiments (25°C mucus/25°C artificial seawater: 25M/25S, 25°C tissue/25°C artificial seawater: 25T/25S, 33°C mucus/33°C artificial seawater: 33M/33S, and 33°C tissue/33°C artificial seawater: 33T/33S). (2) Comparisons between control and treatment experiments on the first day (control 1) were represented as tank effects (25°C mucus/control 1 25°C mucus: 25M/C1M and 25°C tissue/control 1 25°C tissue: 25T/C1T). (3) The differences in the proteins that the bacteria produced between the control and treatment experiments on the last day (control 2) represented the effects of temperature (33°C mucus/control 2 33°C mucus: 33M/C2M, 33°C Tissue/control 2 33°C tissue: 33T/C2T, and 33°C artificial seawater/control 2 33°C artificial seawater: 33S/C2S). (4) Time also appeared to have an effect, with the first and last day of the control experiments yielding different proteins (control 2 25°C mucus/control 1 25°C mucus: C2M/C1M, control 2 25°C tissue/control 1 25°C tissue: C2T/C1T, and control 2 25°C artificial seawater/control 1 25°C artificial seawater: C2S/25S).
To monitor the growth conditions of E. montiporae under thermal stress and determine the sampling time for the second experiment (the coral lysate experiment), we estimated the OD values of E. montiporae during different growth phases in the three temperatures treatments. The maximum OD value of E. montiporae was 1.28 at 25°C and 1.2 at 31°C after 36 h. However, the growth curve at 33°C was lower than those of the other two temperatures, especially after 24 h of incubation (Figure 2A); therefore, E. montiporae grew slower at 33°C than at the other two temperatures.
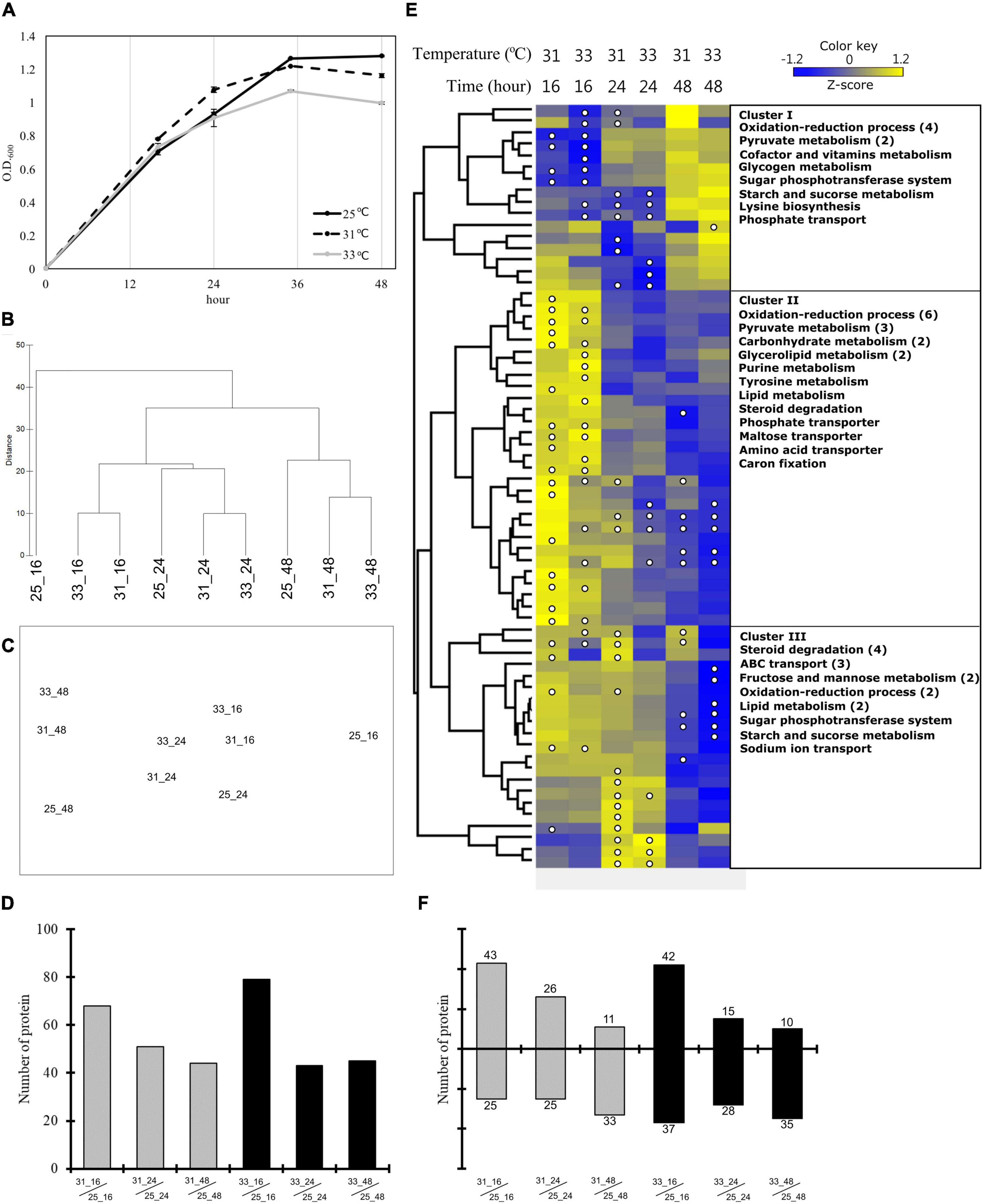
Figure 2. Growth pattern and quantitative proteomics during heat stress. (A) Growth curves of Endozoicomonas montiporae CL-33T at 25, 31, and 33°C in heat stress experiments. The quantitative proteomics analysis of heat stress experiments (B–F). (B) Hierarchical clustering and (C) nMDS ordination represent the similarity between the protein profiles from different samples. (D) The number of significantly changed proteins at each incubation time point at 31 and 33°C. (E) Hierarchical clustering based heatmap visualization of the metabolism-related (from COG functional categories) proteins that showed significantly changes in any one of the samples. The heatmap displays the relative protein abundance (z-score transformed) from low (blue) to high (yellow) per row. The columns represent three time points (16, 24, and 48 h) at two incubation temperatures (31 and 33°C). The significantly changed proteins were marked by a white circle. The right side of the image show the protein functions by category. Numbers in brackets indicate the number of proteins when there are multiple proteins in one category. (F) The number of proteins up-expression and down-expression in each incubation time at 31 and 33°C.
A total of 1,477,327 unique spectra and 33,841 unique peptides were detected by the LC-MS/MS, and 2,017 unique proteins were identified across the 24 samples. Approximately 41% of the 4,910 predicted open reading frames in the genome of E. montiporae CL-33T were identified from all 24 samples. Most proteins are functionally related to the categories in the cellular metabolic process, nitrogen compound metabolic process, and primary metabolic process according to GO (Supplementary Figure 1). Noteworthily, 59 proteins were annotated in GO as responding to stress – e.g., chaperone proteins and catalase (Supplementary Figure 1).
Results of hierarchical clustering and nMDS analyses showed that the protein profiles of the E. montiporae grown for 48 h were clearly distinct from the profiles of those grown for 16 and 24 h (Figures 2B,C and Supplementary Figure 2). Comparing within the same incubation time, the protein profiles from 25°C were different from those at 31 and 33°C (Figure 2B). Bacteria grown for 16 h at 25°C were especially separate from other samples.
The number of differentially expressed proteins (DEPs) decreased with increasing incubation time (Figure 2D and Data Set 1). At 16 h, the number of DEPs in the culture at 33°C (79 proteins) was more than the culture at 31°C (68 proteins). After 24 h, both E. montiporae cultured at 31 and 33°C showed about 43 to 51 DEPs. In the time course, up-expression proteins were detected less and less in E. montiporae cultured at 31 and 33°C (Figure 2F). Furthermore, the up-expression proteins in E. montiporae cultured at 33°C were less than that at 31°C at every time point. In contrast, E. montiporae cultured at 33°C had more down-expression proteins than E. montiporae cultured at 31°C. Sixty-six of the total 154 DEPs were annotated by COGs (Supplementary Table 1). Except for proteins with unknown function, most of the COG annotated-proteins were in the metabolism category (43.52%).
A heatmap (z-score transformed) was made to detect expression patterns of the DEPs in the metabolism category (Figure 2E and Supplementary Figure 3). Cluster I shows down-expression proteins after 16 h at 31 and 33°C, respectively. Those down-expression proteins are mainly involved in the oxidation-reduction process and pyruvate metabolism. Cluster II shows the up-expression proteins after 16 h at 31 and 33°C. These up-expression proteins were involved in the oxidation-reduction process, pyruvate metabolism, glycerolipid metabolism, carbohydrate metabolism, transport, and others. Furthermore, Cluster III shows down-expression proteins after 48 h, especially the culture at 33°C. These proteins are mainly involved in steroid degradation and ABC transport.
We categorized the proteins related to heat shock-related responses into two groups: (1) heat shock protein and co-chaperone proteins, and (2) antioxidant defense proteins (Supplementary Table 2). The first group of proteins were up-expressed after 16 h at 31 and 33°C. In particular, the small heat shock protein IbpA (log2FC 2.81), chaperone protein ClpB (log2FC 1.76), lon protease (log2FC 1.72), and periplasmic serine endoprotease DegP (log2FC 1.93) were significantly up-expressed after 16 h at 33°C when tricorn protease was significantly down-expressed after 48 h. In the category of antioxidant defense proteins, catalase proteins were down-expressed after 24 and 48 h in the E. montiporae culture at 33°C (log2FC: −2.08 and −2.1).
The average PAM values decreased from 0.7 to 0.6 following an increase in the water temperature in the experimental tank (Supplementary Figure 4) (coral color card E3). However, the average PAM value in the control tank was consistent at 0.67 throughout the experiment. In addition, the color of the coral nubbins remained dark brown in the control tank (Supplementary Figure 4) (coral color card E5).
The average OD values of bacterial cultures changed from 0.5 to 0.9 during incubation (6 h in total), indicating that the bacterial growth stayed in the exponential phase throughout the experimental time (Supplementary Figure 5).
A total of 2,950,378 spectra and 33,582 unique peptides were detected. Among them, 1,901 unique proteins were identified across all samples, accounting for 38.7% of the 4,910 predicted open reading frames in the E. montiporae CL-33T genome. Most proteins were involved in organic substance metabolic processes, cellular metabolic processes, primary metabolic processes and nitrogen compound metabolic processes (Supplementary Figure 6).
In the nMDS analysis, one replicate (25M_1) from the mucus treatment at 25°C was not clustered with the other two; it was deemed an outlier (Supplementary Figure 7A) and removed from subsequent analyses (Supplementary Figure 7B). Clustering samples were clearly separated by temperature (33 and 25°C) (Figure 3 and Supplementary Figure 7C). Furthermore, the samples from the first day in the control tank (C1M and C1T) and treatment tank (25M, 25T, and 25S) were clustered together (Figure 3A).
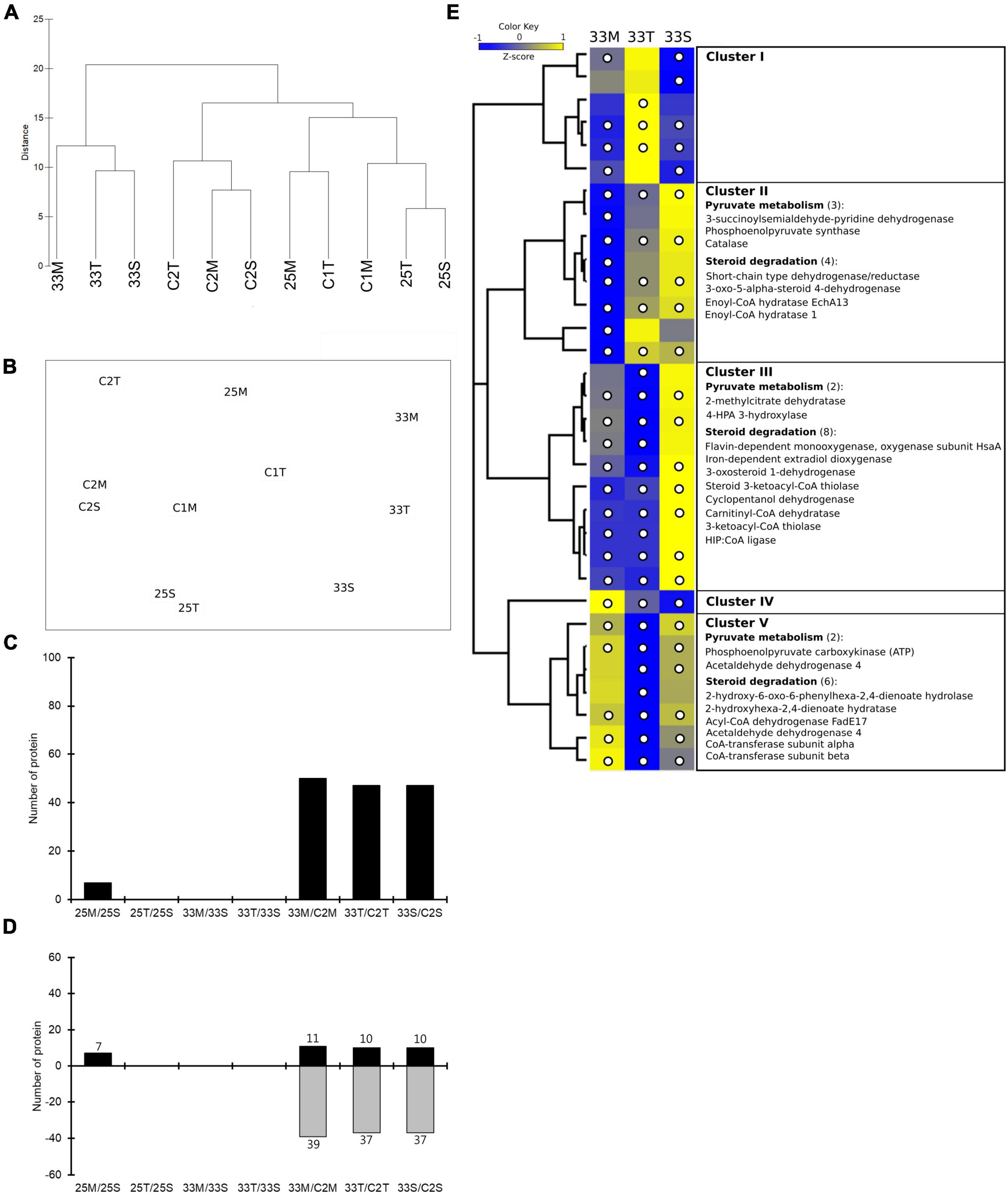
Figure 3. Quantitative proteomics analysis of coral lysate incubated with bacterial cultures at different temperatures. (A) Hierarchical clustering and (B) nMDS ordination of different samples similarities. (C) The number of significantly changed proteins that were affected by coral lysate (25M/25S, 25T/25S, 33M/33S, and 33T/33S) and temperature (33M/C2M, 33T/C2T, and 33S/C2S). (D) The number of proteins with increased and decreased expression in each comparison. (E) Hierarchical clustering heatmap visualization of the metabolism-related (from COG functional categories) proteins that have significantly changed in any of the three treatments (33M/C2M, 33T/C2T, and 33S/C2S). The columns names (33M, 33T, and 33S) represent three treatments. Relative protein abundance (z-score transformed) is graded from low (blue) to high (yellow) in each row. The bracketed numbers represent the number of proteins if there are multiple proteins in a category. The significantly changed proteins are marked by a white circle.
Comparing the effects of coral lysate and temperature, a high number of DEPs (approximately 50) were detected based on changes in temperature, and only a few (seven) were detected to have been caused by the coral lysate (Figure 3C and Data Set 2). Furthermore, most of the DEPs in temperature effect were down-expressed (Figure 3D). Five proteins, involved in two categories of bacterial heat stress response, were highly up- or down-expressed during the 6 h of incubation (Supplementary Table 3): the small heat shock protein IbpA, lon protease, catalase, aldehyde dehydrogenase and 4-hydroxyphenylacetate 3-monooxygenase oxygenase component.
Cluster of orthologous groups analysis was performed for all of the DEPs in the metabolism process category (Supplementary Table 4). In those proteins, the z-score heatmap displayed 18 down-expressed proteins related to steroid degradation in mucus and tissue treatments (Figure 3E and Supplementary Figure 8). Moreover, there were also eight down-expressed proteins involved in pyruvate metabolism (Figure 3E).
To gain more insights into the potentially important cellular mechanisms of bacteria-coral host interactions, proteins that underwent significant changes but had a log2FC < 1.5 were also discussed. Hence, if we consider all of the differential expression proteins (p < 0.05, regardless of their log2FC), 10 to 70 proteins were influenced by the coral lysate effect (Figure 4A and Supplementary Table 5). Furthermore, many differential expression proteins were down-expressed in the tissue treatment at 25°C (25T/25S), but up-expressed in the tissue treatment at 33°C (33T/33S) (Figure 4B). In addition, most of the log2FC of differential expression proteins were < 1 in the tissue treatments at 25 and 33°C (Figure 4C). Noteworthily, 14 differential expression proteins were detected in the mucus treatment at 25°C (25M/25S) and had a log2FC > 1; these 14 proteins were involved in the signal recognition particle pathway and toxin-antitoxin system (Table 1).
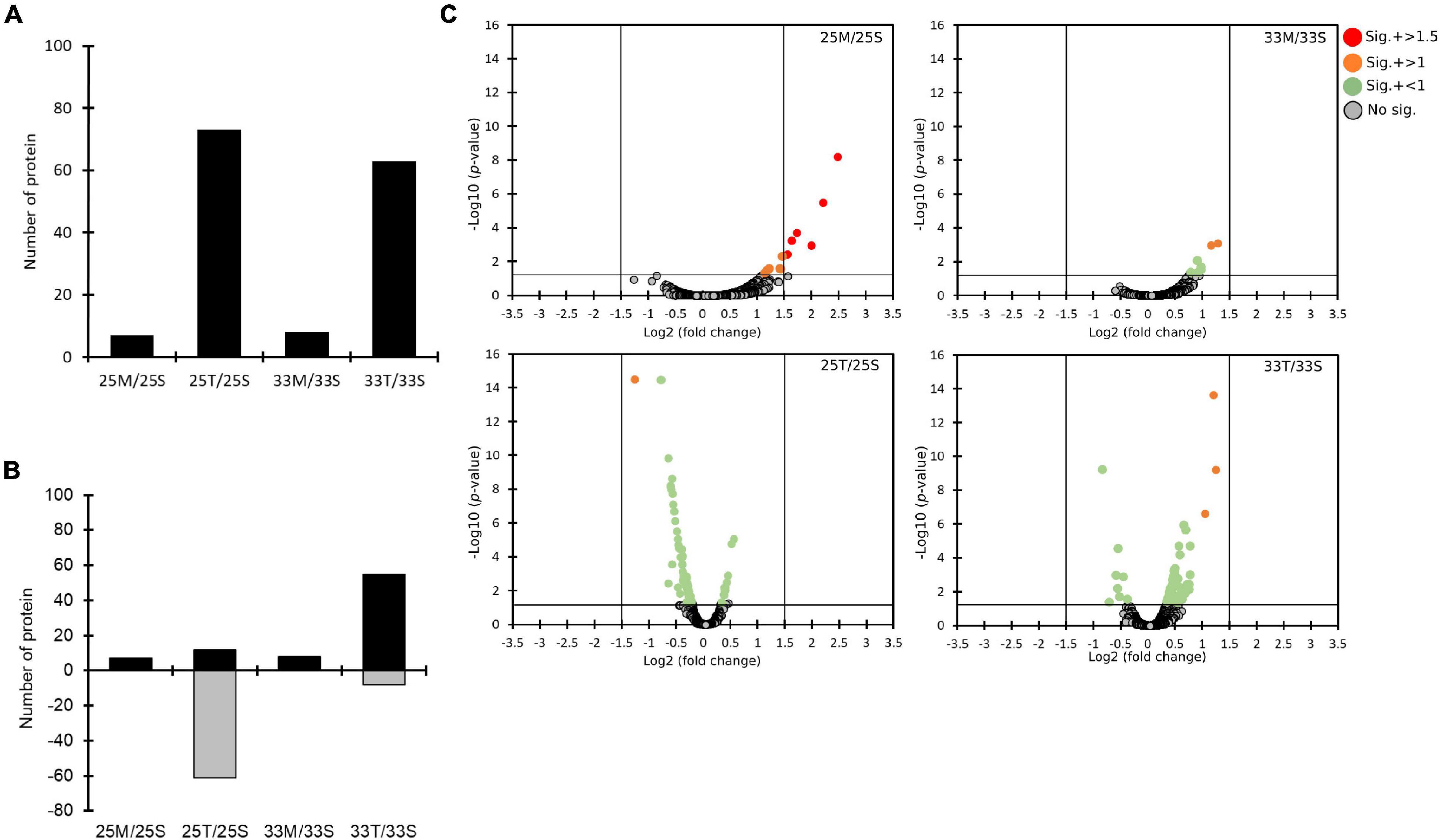
Figure 4. The quantitative proteomics analysis comparing the effects of different coral lysates (25M/25S, 25T/25S, 33M/33S, and 33T/33S). (A) Bar plots on counts of significantly changed proteins. (B) The numbers of proteins with increased (black) and decreased (gray) expression. (C) The distributions of statistical significance [-log10 (p-values)] and magnitude of change (Log2 fold change) for all the proteins in four different comparisons represented by volcano plots. The proteins for which there was no significant difference are marked in gray circles. Significantly changed proteins are marked in different colors based on different levels of log2 fold change. (red: log2 fold change > 1.5, orange: log2 fold change > 1, and green: log2 fold change < 1).
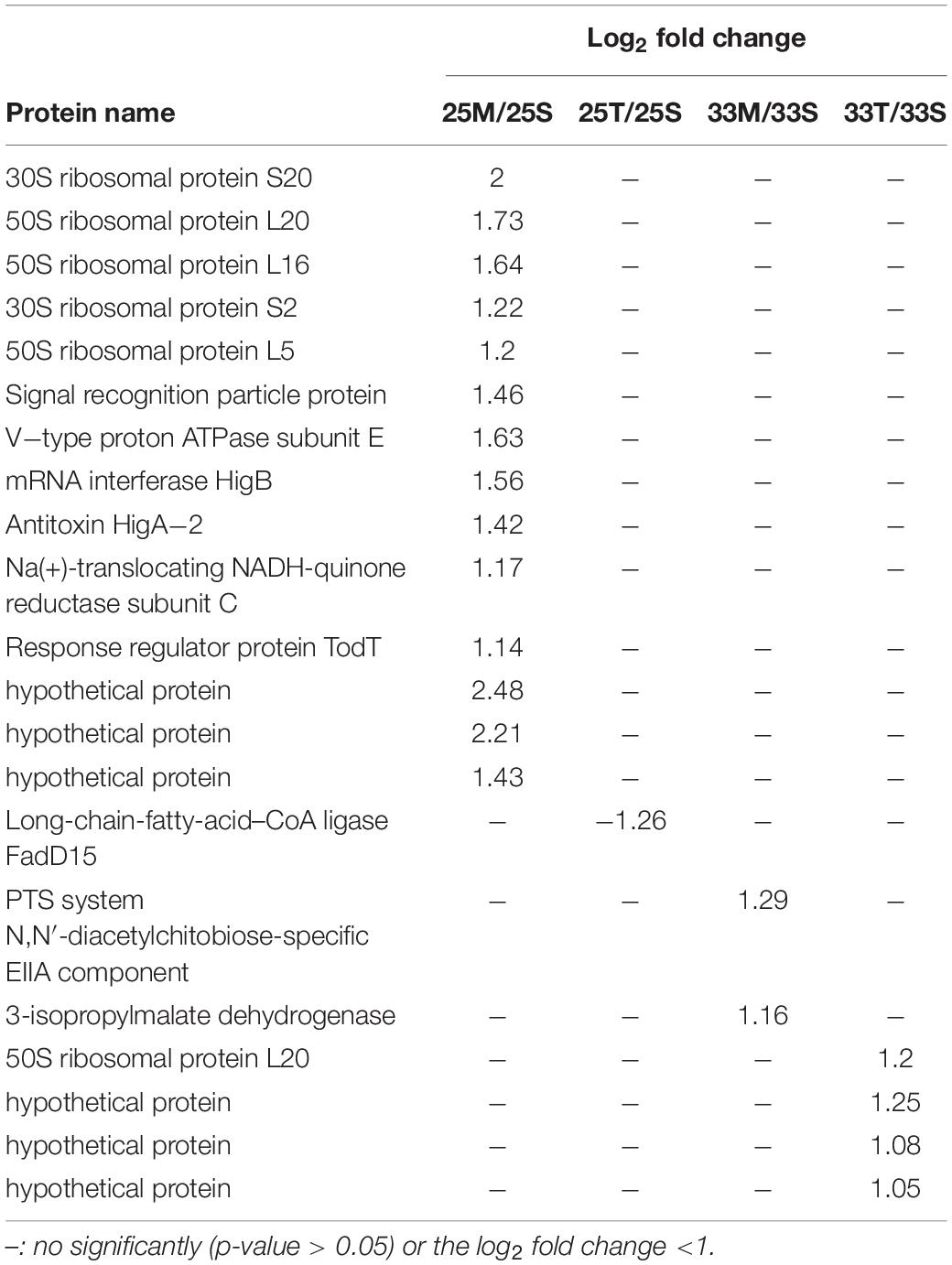
Table 1. List of significantly changed proteins (p-value < 0.05) which have log2 fold change >1 from the coral lysate comparisons.
The tank and time effects were very low because most proteins maintained a similar protein abundance (log2FC < 1.5; Supplementary Figures 9, 10).
Endozoicomonas is a core bacterial group in corals that plays a role in coral health, and often decreases rapidly in corals under heat stress (Bourne et al., 2008; McDevitt-Irwin et al., 2017; Shiu et al., 2017; Savary et al., 2021). However, how these bacteria respond to changes in temperature and coral hosts during heat stress were unknown until now. This study is the first to provide insights into the physiological changes of E. montiporae to heat stress and host factors using a proteomic approach. We designed two experiments to clarify how much temperature and the coral host influence the bacterium, and how bacterial metabolism is affected by both factors. Our study shows that temperature is the main factor affecting protein abundance in the bacterium, though the coral host also has some influence. More details are given in the following paragraphs.
Temperature was the main factor in this study affecting changes in the protein profiles of E. montiporae. Previous studies have shown that Endozoicomonas are highly sensitive to temperature changes in various corals (Bourne et al., 2008; Lee et al., 2015; Neave et al., 2016; Shiu et al., 2017; Maher et al., 2020). Most recent studies also show the same phenomenon – e.g., at 32°C, Endozoicomonas populations’ relative abundances rapidly halved in the coral Pocillopora damicornis within 48 h (Li et al., 2021). In the coral Stylophora pistillata, the resident Endozoicomonas population quickly decreased at 32 and 34.5°C over 6 h (Savary et al., 2021).
However, what exactly caused these reductions in Endozoicomonas is still mostly unknown. There are some possible reasons for the phenomenon. One is that heat stress made it difficult for the bacteria to live and grow in their host corals, and the bacteria consequently left for other habitats. Or other bacteria outcompeted the Endozoicomonas. Our molecular study clearly shows that a stressful physiological status is one of the major factors that causes Endozoicomonas populations to decline in corals. At 33°C, those crucial metabolisms such as the proteins for central metabolisms were mostly down-expressed in this study. In other words, Endozoicomonas physiology – including growth – was clearly influenced by heat stress. Protein expression changed starkly for a number of proteins, such as heat-shock proteins, stress-associated reactive oxygen species, and other important metabolism-related proteins. We elaborate on each of these below.
The heat shock-related response is one of the fundamental responses that most bacteria have to heat stress (Guo and Gross, 2014). How heat shock proteins (HSPs) help bacteria adapt and survive in thermal stress environments had been widely explored by previous studies (Sowell et al., 2008; Lüders et al., 2009; Ryno et al., 2014; Mosier et al., 2015; Lee et al., 2018). In this study, a number of HSPs and co-chaperones were significantly increased in E. montiporae in the two heat stress experiments; for example, small heat shock protein IbpA, chaperone protein ClpB, lon protease, and periplasmic serine endoprotease DegP were all up-expressed after 16 h at 33°C (log2FC values were about 2). Previous studies suggested that those proteins contribute to the resolution of protein aggregates to confer superior heat tolerance (Squires et al., 1991; Rosen and Ron, 2002; Kim and Sauer, 2014). In particular, we found that IbpA and lon protease were quickly expressed during the 6 h of incubation at 33°C in the coral lysate experiment (meaning log2FC of IbpA was 2.36 and lon protease was 1.16). The major consequences of increasing temperature for cells are protein unfolding and aggregation, with concomitant loss of function (Rosen and Ron, 2002). Those unfolding or aggregated proteins could be degraded and cleaned by both IbpA and Lon proteases under heat stress (Haslbeck and Vierling, 2015). Thus, E. montiporae responded quickly to the expression of HSPs to prevent the irreversible aggregation or misfolding of proteins in the early stages of the heat stress treatment, especially at 33°C.
Reactive oxygen species (ROS) are crucial to the coral holobiont, and they are tightly associated with coral physiology under heat stress and bleaching conditions (Downs et al., 2002; Cziesielski et al., 2019). Removing ROS is critical for corals to maintain their homeostasis during stress. In this study, we also detected a rapid change in the proteins that are responsible for ROS-related metabolisms. For example, catalases are mainly responsible for removing ROS in many bacterial cells. Similarly, E. montiporae expressed more catalases (Log2FC: 1.91) and other antioxidant defense proteins (i.e., hydroperoxy fatty acid reductase) at 31°C that would facilitate the bacterium to remove extra ROS from cells and prevent further damages by ROS. However, the expression of those proteins was decreased at 33°C, suggesting that the bacterium’s physiology was not functioning properly. The growth curve of the bacterial culture at 33°C also plateaued earlier than the other two temperature treatments, also indicating that the bacteria grew incompetently at 33°C. Additionally, we also noticed that energy-related metabolism under heat stress in the coral lysate experiment was significantly decreased – such as the key proteins, phosphoenolpyruvate synthase and phosphoenolpyruvate carboxykinase – also suggesting that the bacteria had poor cellular physiology at 33°C.
The temperature effect also greatly reduced ecotin levels in E. montiporae (meaning of log2FC was −2.51). Ecotin, which is usually located in periplasmic space of many bacteria, is both a serine protease inhibitor and functionally related to bacterial defenses against bacteriophages and other bacteria (Chung et al., 1983; Eggers et al., 2004; Clark et al., 2011; Ireland et al., 2014). A recent study found that ecotin plays a role in the defense of Escherichia coli against attacks from Vibrio cholera (Myint et al., 2021). The decrease in ecotin levels in E. montiporae at high temperature might weaken the bacterium’s defenses against other bacteria. Intriguingly, Lee et al. (2015) found that the E. montiporae population decreased at high temperatures while Vibrio spp. increased rapidly under the same conditions. Perhaps the phenomenon of these opposing changes in E. montiporae and Vibrio spp. was partially the result of E. montiporae becoming more defenseless at the high temperature, allowing Vibrio spp. to colonize the coral tissues. Myint et al. (2021) also showed that an E. coli ecotin mutant became more susceptible to V. cholera attack than did the wild type strain. However, the relationship between ecotin and the defense mechanism in E. montiporae requires mutation experiments to prove.
In addition to temperature, we found that host factors changed a number of proteins in the co-incubation experiment with coral lysates, suggesting that bacterial physiological responses to heat stress are constituted by both temperature and host factors. Under stressful conditions, coral host factors also affected or influenced the bacterium. Some intriguing proteins are discussed as follows.
First, N, N′-diacetylchitobiose-specific EIIA component (ChbA) of E. montiporae was up-expressed with a high value of log2FC (i.e., 1.29) in the thermal stressed-coral mucus samples. ChbA protein participates in transporting cellobiose and belongs to the phosphoenolpyruvate-dependent sugar phosphotransferase system (PTS system), a major carbohydrate active transport system in bacteria (Kotrba et al., 2001). Cellobiose is a sugar component of cellulose and the major composition of algal cells (Brady et al., 2015; Wahlström et al., 2020). A recent report shows that Symbiodiniaceae increased in coral mucus after incubating at 31°C (Wright et al., 2019), suggesting that the cellobiose concentration increases in the mucus at higher temperatures. In our study, the increase in ChbA levels in E. montiporae could be induced to transport increasing amounts of cellobiose. Moreover, high ChbA expression might also help in the removal of unwanted algal components in coral host by the PTS system, as suggested by Neave et al. (2017), who discovered many genes of the PTS system encoded in Endozoicomonas genomes.
Second, the proteins involved in steroid degradation in E. montiporae had more down-expression in the mucus and tissue treatments than in the seawater treatment under heat stress (Figure 3E). For example, the log2FCs of 3-oxosteroid 1-dehydrogenase were −2.01 and −2.3 in mucus and tissue treatments, but −1.69 in the seawater treatment. This implies that the thermal-stressed coral lysate had additional effects on the expression of proteins during the heat stress. Why did the temperature effect cause the down-expression of proteins that were involved in steroid degradation in E. montiporae under heat stress? The steroid uptake mechanisms in bacteria have remained unclear because of high physiological and structural differences among bacterial groups (Olivera and Luengo, 2019). A previous study, however, demonstrated that down-expression of steroid metabolism processes in the coral Acropora spp. and Symbiodiniaceae during the thermal stress (Hillyer et al., 2017). Hence, we speculate that the phenomenon that the proteins involved in steroid degradation in E. montiporae decreased could be caused by the lower concentration of steroids in the coral lysate after the heat stress treatment. This hypothesis, however, requires more experimentation to prove.
It is worth addressing that fewer proteins underwent significant changes in the coral lysate effect than the temperature effect in the coral lysate experiment. This could be partially due to a limitation in the experiment – that the dialysis tubing with a pore size of 14 kDa used to load coral lysate only allowed small molecules to pass into the bacterial culture. Molecules such as glucose, vitamin B12, inorganic salt, and cytochrome C can pass through the tubing, but larger molecules – such as liposome and starch – could not. Thus, the effect of host factors on E. montiporae are likely to be underestimated. Furthermore, this experimental treatment was performed in one tank instead of three independent tank systems, which might have led to some bias or potential errors in the results.
Coral mucus composition is dynamic of changes in host physiology and environmental factors. For example, Lee et al. (2015) demonstrated that there was a shift in the proportion of different sugar components in coral mucus after heat stress. Interestingly, we found that E. montiporae showed significant changes in protein expressions, and that log2FC values were higher than 1 when the bacteria were co-incubated with mucus at 25°C compared with the seawater samples (Table 1). Most of the proteins were involved in three types of protein groups: signal recognition particle pathway, type II toxin-antitoxin system and V-type ATPase. These significantly changed proteins were presented only with unheated coral mucus rather than heat stress induced-coral mucus. This indicates that specific factors in coral mucus must change during heat stress and affect the expression of those proteins in E. montiporae. In other words, those highly expressed protein groups are likely to be functionally related to coral mucus under regular conditions.
The first group comprised six significantly changed proteins involved in the signal recognition particle pathway (SRP). SRP is essential for delivering the nascent protein to properly localize the inner membrane and mediate secretory proteins (Akopian et al., 2013; von Loeffelholz et al., 2015). SRP depletion may cause the accumulation of mis-localized proteins in bacteria (Zhang et al., 2012). Many reports showed various evidence that the SPR pathway plays important roles in membrane integrity, cell physiology, and viability (Ulbrandt et al., 1997; Miyazaki et al., 2016; Fu et al., 2017; Sauerbrei et al., 2020). In this study, high expression of the SRP is specifically associated with coral mucus, suggesting that the SRP might be functionally crucial for E. montiporae living in or passing coral mucus during colonization.
The second group is two highly expressed proteins, HigB and HigA, also called Type II toxin-antitoxin (TA) system, which is present in many bacteria (Leplae et al., 2011; Yamaguchi and Inouye, 2011). They are a pair of proteins composed of a stable toxin and its cognate antitoxin protein. Under normal growth conditions, they form a tight and non-toxic complex. When the antitoxin is degraded by proteases under environmental stress, cell fundamental functions such as growth, gene regulation and biofilm formation are impaired (Ren et al., 2004; Wang et al., 2011). Hence, the high expression of HigB and HigA also indicates that E. montiporae was in a non-stressful condition. Moreover, the TA system is also functionally related to defense in bacteria. For example, Sberro et al. (2013) demonstrated that the TA system in E. coli provides resistance against the T7 phage. Our proteomic results suggest that the up-expression of the TA system would be helpful to E. montiporae in defense.
The third group is V-type ATPase. The log2FC of V-type ATPase subunit E in E. montiporae increased to 1.63.V-type ATPase is a membrane-embedded protein complex (Vasanthakumar and Rubinstein, 2020). V-type ATPase was found not only in eukaryotes but also archaea and bacteria, including Thermus thermophilus, Enterococcus hirae, Lactobacillus plantarum, and Clostridium fervidus (Lolkema et al., 2003; Yokoyama and Imamura, 2005; Saijo et al., 2011; Zhao et al., 2017; Vasanthakumar et al., 2019). V-type ATPase in bacteria have multiple functions, including proton pump, ion pump and ATP synthesis (Speelmans et al., 1994; Murata et al., 2000; Toei et al., 2007; Zhou and Sazanov, 2019). Moreover, V-type ATPase also plays a role in maintaining physiological homeostasis for bacteria (Bove et al., 2013; van den Nieuwboer et al., 2016; Roh and Kim, 2021). Although the function of V-type ATPase is still unclear in E. montiporae, highly expressed V-type ATPase in E. montiporae serves as potential evidence of a specific response that is functionally related to coral mucus in coral-E. montoporae interactions.
To establish a working model for the in-depth investigation of bacteria-coral interactions, this study used the cultivable, dominant coral-associated bacterium E. montiporae as a model organism to dissect changes in proteomic profiles of the bacteria, responding to heat stress and heat stressed-coral lysate. We successfully detected a number of pivotal proteins related to the heat-stress physiology of E. montiporae that provides new research directions in the molecular microbiology and microbial ecology of E. montiporae in the coral holobiont. This is one of few primary works exploring the molecular mechanisms underlying physiological changes in coral symbiotic bacteria, and we anticipate that this study will inspire more molecular studies on coral microbes that will better elucidate the detailed coral-bacteria interactions.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: ProteomeXchange (accession: PXD029816).
S-LT conceived and supervised the research, reviewed, and edited the manuscript. S-LT and Y-FC designed the experiments. Y-FC, C-YL, C-YC, and SS performed the experiments. Y-FC, C-YC, and Y-CT analyzed the data and the results. Y-FC and S-LT wrote the original draft of the manuscript. S-LT and KT modified the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the Ministry of Science and Technology (MOST) in Taiwan (Grant Nos. MOST107-2611-M-001-002, MOST108-2611-M-001-004, and MOST109-2611-M-001-002) and three Postdoctoral Research Fellows projects (Grant Nos. MOST107-2811-M-001-043, MOST108-2811-M-001-603, and MOST109-2811-M-001-586). Y-FC is also supported by the Ministry of Science and Technology, Taiwan (MOST 110-2611-M-031-001) and Research Grants for New Teachers of College of Science, Soochow University, Taiwan.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Yin-Chu Cheng and Tzu-Chieh Lin (Institute of Marine Environment and Ecology, National Taiwan Ocean University, Taiwan) for their help in collecting the corals.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.808132/full#supplementary-material
Akopian, D., Shen, K., Zhang, X., and Shan, S.-O. (2013). Signal recognition particle: an essential protein-targeting machine. Annu. Rev. Biochem. 82, 693–721. doi: 10.1146/annurev-biochem-072711-164732
Bayer, T., Arif, C., Ferrier-Pages, C., Zoccola, D., Aranda, M., and Voolstra, C. R. (2013). Bacteria of the genus Endozoicomonas dominate the microbiome of the Mediterranean gorgonian coral Eunicella cavolini. Mar. Ecol. Prog. Ser. 479, 75–84. doi: 10.3354/meps10197
Bourne, D. G., Morrow, K. M., and Webster, N. S. (2016). Insights into the coral microbiome: underpinning the health and resilience of reef ecosystems. Annu. Rev. Microbiol. 70, 317–340. doi: 10.1146/annurev-micro-102215-095440
Bourne, D., Iida, Y., Uthicke, S., and Smith-Keune, C. (2008). Changes in coral-associated microbial communities during a bleaching event. ISME J. 2, 350–363. doi: 10.1038/ismej.2007.112
Bove, P., Russo, P., Capozzi, V., Gallone, A., Spano, G., and Fiocco, D. (2013). Lactobacillus plantarum passage through an oro-gastro-intestinal tract simulator: carrier matrix effect and transcriptional analysis of genes associated to stress and probiosis. Microbiol. Res. 168, 351–359. doi: 10.1016/j.micres.2013.01.004
Brady, S. K., Sreelatha, S., Feng, Y., Chundawat, S. P., and Lang, M. J. (2015). Cellobiohydrolase 1 from Trichoderma reesei degrades cellulose in single cellobiose steps. Nat. Commun. 6:10149. doi: 10.1038/ncomms10149
Chen, W.-M., Lin, K.-R., and Sheu, S.-Y. (2019). Endozoicomonas coralli sp. nov., isolated from the coral Acropora sp. Arch. Microbiol. 201, 531–538. doi: 10.1007/s00203-018-1591-2
Chung, C., Ives, H., Almeda, S., and Goldberg, A. (1983). Purification from Escherichia coli of a periplasmic protein that is a potent inhibitor of pancreatic proteases. J. Biol. Chem. 258, 11032–11038.
Clark, E. A., Walker, N., Ford, D. C., Cooper, I. A., Oyston, P. C., and Acharya, K. R. (2011). Molecular recognition of chymotrypsin by the serine protease inhibitor ecotin from Yersinia pestis. J. Biol. Chem. 286, 24015–24022. doi: 10.1074/jbc.M111.225730
Cziesielski, M. J., Schmidt-Roach, S., and Aranda, M. (2019). The past, present, and future of coral heat stress studies. Ecol. Evol. 9, 10055–10066. doi: 10.1002/ece3.5576
Ding, J.-Y., Shiu, J.-H., Chen, W.-M., Chiang, Y.-R., and Tang, S.-L. (2016). Genomic insight into the host–endosymbiont relationship of Endozoicomonas montiporae CL-33T with its coral host. Front. Microbiol. 7:251. doi: 10.3389/fmicb.2016.00251
Downs, C., Fauth, J. E., Halas, J. C., Dustan, P., Bemiss, J., and Woodley, C. M. (2002). Oxidative stress and seasonal coral bleaching. Free Radic. Biol. Med. 33, 533–543. doi: 10.1016/s0891-5849(02)00907-3
Eggers, C. T., Murray, I. A., Delmar, V. A., Day, A. G., and Craik, C. S. (2004). The periplasmic serine protease inhibitor ecotin protects bacteria against neutrophil elastase. Biochem. J. 379, 107–118. doi: 10.1042/BJ20031790
Fu, Y.-H. H., Huang, W. Y., Shen, K., Groves, J. T., Miller, T., and Shan, S.-O. (2017). Two-step membrane binding by the bacterial SRP receptor enable efficient and accurate Co-translational protein targeting. Elife 6:e25885. doi: 10.7554/eLife.25885
Gao, C., Garren, M., Penn, K., Fernandez, V. I., Seymour, J. R., Thompson, J. R., et al. (2021). Coral mucus rapidly induces chemokinesis and genome-wide transcriptional shifts toward early pathogenesis in a bacterial coral pathogen. ISME J. 15, 3668–3682. doi: 10.1038/s41396-021-01024-7
Guo, M. S., and Gross, C. A. (2014). Stress-induced remodeling of the bacterial proteome. Curr. Biol. 24, R424–R434. doi: 10.1016/j.cub.2014.03.023
Haslbeck, M., and Vierling, E. (2015). A first line of stress defense: small heat shock proteins and their function in protein homeostasis. J. Mol. Biol. 427, 1537–1548. doi: 10.1016/j.jmb.2015.02.002
Hillyer, K. E., Dias, D. A., Lutz, A., Wilkinson, S. P., Roessner, U., and Davy, S. K. (2017). Metabolite profiling of symbiont and host during thermal stress and bleaching in the coral Acropora aspera. Coral Reefs 36, 105–118. doi: 10.1007/s00338-016-1508-y
Hughes, T. P., Kerry, J. T., Álvarez-Noriega, M., Álvarez-Romero, J. G., Anderson, K. D., Baird, A. H., et al. (2017). Global warming and recurrent mass bleaching of corals. Nature 543, 373–377. doi: 10.1038/nature21707
Hughes, T. P., Kerry, J. T., Baird, A. H., Connolly, S. R., Dietzel, A., Eakin, C. M., et al. (2018). Global warming transforms coral reef assemblages. Nature 556, 492–496. doi: 10.1038/s41586-018-0041-2
Ireland, P. M., Marshall, L., Norville, I., and Sarkar-Tyson, M. (2014). The serine protease inhibitor Ecotin is required for full virulence of Burkholderia pseudomallei. Microb. Pathog. 67, 55–58. doi: 10.1016/j.micpath.2014.01.001
Kemp, D. W., Hernandez-Pech, X., Iglesias-Prieto, R., Fitt, W. K., and Schmidt, G. W. (2014). Community dynamics and physiology of Symbiodinium spp. before, during, and after a coral bleaching event. Limnol. Oceanogr. 59, 788–797. doi: 10.4319/lo.2014.59.3.0788
Kim, S., and Sauer, R. T. (2014). Distinct regulatory mechanisms balance DegP proteolysis to maintain cellular fitness during heat stress. Genes Dev. 28, 902–911. doi: 10.1101/gad.238394.114
Kotrba, P., Inui, M., and Yukawa, H. (2001). Bacterial phosphotransferase system (PTS) in carbohydrate uptake and control of carbon metabolism. J. Biosci. Bioeng. 92, 502–517. doi: 10.1263/jbb.92.502
Lan, P., Li, W., Wen, T.-N., Shiau, J.-Y., Wu, Y.-C., Lin, W., et al. (2011). iTRAQ protein profile analysis of Arabidopsis roots reveals new aspects critical for iron homeostasis. Plant Physiol. 155, 821–834. doi: 10.1104/pp.110.169508
Lee, K.-J., Jung, Y.-C., Park, S.-J., and Lee, K.-H. (2018). Role of heat shock proteases in quorum-sensing-mediated regulation of biofilm formation by Vibrio species. Mbio 9:e02086-17. doi: 10.1128/mBio.02086-17
Lee, S. T., Davy, S. K., Tang, S.-L., and Kench, P. S. (2016). Mucus sugar content shapes the bacterial community structure in thermally stressed Acropora muricata. Front. Microbiol. 7:371. doi: 10.3389/fmicb.2016.00371
Lee, S. T., Davy, S. K., Tang, S.-L., Fan, T.-Y., and Kench, P. S. (2015). Successive shifts in the microbial community of the surface mucus layer and tissues of the coral Acropora muricata under thermal stress. FEMS Microbiol. Ecol. 91:fiv142. doi: 10.1093/femsec/fiv142
Leplae, R., Geeraerts, D., Hallez, R., Guglielmini, J., Dreze, P., and Van Melderen, L. (2011). Diversity of bacterial type II toxin–antitoxin systems: a comprehensive search and functional analysis of novel families. Nucleic Acids Res. 39, 5513–5525. doi: 10.1093/nar/gkr131
Li, J., Long, L., Zou, Y., and Zhang, S. (2021). Microbial community and transcriptional responses to increased temperatures in coral Pocillopora damicornis holobiont. Environ. Microbiol. 23, 826–843. doi: 10.1111/1462-2920.15168
Lolkema, J. S., Chaban, Y., and Boekema, E. J. (2003). Subunit composition, structure, and distribution of bacterial V-type ATPases. J. Bioenerg. Biomembr. 35, 323–335. doi: 10.1023/a:1025776831494
Lüders, S., Fallet, C., and Franco-Lara, E. (2009). Proteome analysis of the Escherichia coli heat shock response under steady-state conditions. Proteome Sci. 7, 1–15. doi: 10.1186/1477-5956-7-36
MacCoss, M. J., Wu, C. C., and Yates, J. R. (2002). Probability-based validation of protein identifications using a modified SEQUEST algorithm. Anal. Chem. 74, 5593–5599. doi: 10.1021/ac025826t
Maher, R. L., Schmeltzer, E. R., Meiling, S., Mcminds, R., Ezzat, L., Shantz, A. A., et al. (2020). Coral microbiomes demonstrate flexibility and resilience through a reduction in community diversity following a thermal stress event. Front. Ecol. Evol. 8:356. doi: 10.3389/fevo.2020.555698
McDevitt-Irwin, J. M., Baum, J. K., Garren, M., and Vega Thurber, R. L. (2017). Responses of coral-associated bacterial communities to local and global stressors. Front. Mar. Sci. 4:262. doi: 10.3389/fmars.2017.00262
Mendoza, M., Guiza, L., Martinez, X., Caraballo, X., Rojas, J., and Aranguren, L. F. (2013). A novel agent (Endozoicomonas elysicola) responsible for epitheliocystis in cobia Rachycentrum canadum larvae. Dis. Aquat. Organ. 106, 31–37. doi: 10.3354/dao02636
Meyer, J. L., Paul, V. J., and Teplitski, M. (2014). Community shifts in the surface microbiomes of the coral Porites astreoides with unusual lesions. PLoS One 9:e100316. doi: 10.1371/journal.pone.0100316
Miyazaki, R., Yura, T., Suzuki, T., Dohmae, N., Mori, H., and Akiyama, Y. (2016). A novel SRP recognition sequence in the homeostatic control region of heat shock transcription factor σ 32. Sci. Rep. 6:24147. doi: 10.1038/srep24147
Mosier, A. C., Li, Z., Thomas, B. C., Hettich, R. L., Pan, C., and Banfield, J. F. (2015). Elevated temperature alters proteomic responses of individual organisms within a biofilm community. ISME J. 9, 180–194. doi: 10.1038/ismej.2014.113
Mouchka, M. E., Hewson, I., and Harvell, C. D. (2010). Coral-associated bacterial assemblages: current knowledge and the potential for climate-driven impacts. Integr. Comp. Biol. 50, 662–674. doi: 10.1093/icb/icq061
Murata, T., Igarashi, K., Kakinuma, Y., and Yamato, I. (2000). Na+ Binding of V-type Na+-ATPase inEnterococcus hirae. J. Biol. Chem. 275, 13415–13419. doi: 10.1074/jbc.275.18.13415
Muthusamy, S., Lundin, D., Mamede Branca, R. M., Baltar, F., González, J. M., Lehtiö, J., et al. (2017). Comparative proteomics reveals signature metabolisms of exponentially growing and stationary phase marine bacteria. Environ. Microbiol. 19, 2301–2319. doi: 10.1111/1462-2920.13725
Myint, S. L., Zlatkov, N., Aung, K. M., Toh, E., Sjöström, A., Nadeem, A., et al. (2021). Ecotin and LamB in Escherichia coli influence the susceptibility to Type VI secretion-mediated interbacterial competition and killing by Vibrio cholerae. Biochim. Biophys. Acta BBA Gen. Subj. 1865:129912. doi: 10.1016/j.bbagen.2021.129912
Neave, M. J., Apprill, A., Ferrier-Pagès, C., and Voolstra, C. R. (2016). Diversity and function of prevalent symbiotic marine bacteria in the genus Endozoicomonas. Appl. Microbiol. Biotechnol. 100, 8315–8324. doi: 10.1007/s00253-016-7777-0
Neave, M. J., Michell, C. T., Apprill, A., and Voolstra, C. R. (2017). Endozoicomonas genomes reveal functional adaptation and plasticity in bacterial strains symbiotically associated with diverse marine hosts. Sci. Rep. 7:40579. doi: 10.1038/srep40579
Olivera, E. R., and Luengo, J. M. (2019). Steroids as environmental compounds recalcitrant to degradation: genetic mechanisms of bacterial biodegradation pathways. Genes 10:512. doi: 10.3390/genes10070512
Ren, D., Bedzyk, L., Thomas, S., Ye, R., and Wood, T. K. (2004). Gene expression in Escherichia coli biofilms. Appl. Microbiol. Biotechnol. 64, 515–524.
Roh, H., and Kim, D.-H. (2021). Genotypic and phenotypic characterization of highly alkaline-resistant Carnobacterium maltaromaticum V-Type ATPase from the dairy product based on comparative genomics. Microorganisms 9:1233. doi: 10.3390/microorganisms9061233
Rosen, R., and Ron, E. Z. (2002). Proteome analysis in the study of the bacterial heat-shock response. Mass Spectrom. Rev. 21, 244–265. doi: 10.1002/mas.10031
Ryno, L. M., Genereux, J. C., Naito, T., Morimoto, R. I., Powers, E. T., Shoulders, M. D., et al. (2014). Characterizing the altered cellular proteome induced by the stress-independent activation of heat shock factor 1. ACS Chem. Biol. 9, 1273–1283. doi: 10.1021/cb500062n
Saijo, S., Arai, S., Hossain, K. M., Yamato, I., Suzuki, K., Kakinuma, Y., et al. (2011). Crystal structure of the central axis DF complex of the prokaryotic V-ATPase. Proc. Natl. Acad. Sci. U.S.A. 108, 19955–19960. doi: 10.1073/pnas.1108810108
Sauerbrei, B., Arends, J., Schünemann, D., and Narberhaus, F. (2020). Lon protease removes excess signal recognition particle protein in Escherichia coli. J. Bacteriol. 202:e00161-20. doi: 10.1128/JB.00161-20
Savary, R., Barshis, D. J., Voolstra, C. R., Cárdenas, A., Evensen, N. R., Banc-Prandi, G., et al. (2021). Fast and pervasive transcriptomic resilience and acclimation of extremely heat-tolerant coral holobionts from the northern Red Sea. Proc. Natl. Acad. Sci. U.S.A. 118:e2023298118. doi: 10.1073/pnas.2023298118
Sberro, H., Leavitt, A., Kiro, R., Koh, E., Peleg, Y., Qimron, U., et al. (2013). Discovery of functional toxin/antitoxin systems in bacteria by shotgun cloning. Mol. Cell 50, 136–148. doi: 10.1016/j.molcel.2013.02.002
Sheu, S.-Y., Lin, K.-R., Hsu, M.-Y., Sheu, D.-S., Tang, S.-L., and Chen, W.-M. (2017). Endozoicomonas acroporae sp. nov., isolated from Acropora coral. Int. J. Syst. Evol. Microbiol. 67, 3791–3797. doi: 10.1099/ijsem.0.002194
Shiu, J.-H., Keshavmurthy, S., Chiang, P.-W., Chen, H.-J., Lou, S.-P., Tseng, C.-H., et al. (2017). Dynamics of coral-associated bacterial communities acclimated to temperature stress based on recent thermal history. Sci. Rep. 7:14933. doi: 10.1038/s41598-017-14927-3
Sowell, S. M., Norbeck, A. D., Lipton, M. S., Nicora, C. D., Callister, S. J., Smith, R. D., et al. (2008). Proteomic analysis of stationary phase in the marine bacterium “Candidatus Pelagibacter ubique”. Appl. Environ. Microbiol. 74, 4091–4100. doi: 10.1128/AEM.00599-08
Speelmans, G., Poolman, B., Abee, T., and Konings, W. N. (1994). The F-or V-type Na (+)-ATPase of the thermophilic bacterium Clostridium fervidus. J. Bacteriol. 176, 5160–5162. doi: 10.1128/jb.176.16.5160-5162.1994
Squires, C., Pedersen, S., Ross, B., and Squires, C. (1991). ClpB is the Escherichia coli heat shock protein F84. 1. J. Bacteriol. 173, 4254–4262. doi: 10.1128/jb.173.14.4254-4262.1991
Tandon, K., Lu, C.-Y., Chiang, P.-W., Wada, N., Yang, S.-H., Chan, Y.-F., et al. (2020). Comparative genomics: dominant coral-bacterium Endozoicomonas acroporae metabolizes dimethylsulfoniopropionate (DMSP). ISME J. 14, 1290–1303. doi: 10.1038/s41396-020-0610-x
Toei, M., Gerle, C., Nakano, M., Tani, K., Gyobu, N., Tamakoshi, M., et al. (2007). Dodecamer rotor ring defines H+/ATP ratio for ATP synthesis of prokaryotic V-ATPase from Thermus thermophilus. Proc. Natl. Acad. Sci. U.S.A. 104, 20256–20261. doi: 10.1073/pnas.0706914105
Ulbrandt, N. D., Newitt, J. A., and Bernstein, H. D. (1997). The E. coli signal recognition particle is required for the insertion of a subset of inner membrane proteins. Cell 88, 187–196. doi: 10.1016/s0092-8674(00)81839-5
van den Nieuwboer, M., Van Hemert, S., Claassen, E., and De Vos, W. M. (2016). Lactobacillus plantarum WCFS 1 and its host interaction: a dozen years after the genome. Microb. Biotechnol. 9, 452–465. doi: 10.1111/1751-7915.12368
van Oppen, M. J., and Blackall, L. L. (2019). Coral microbiome dynamics, functions and design in a changing world. Nat. Rev. Microbiol. 17, 557–567. doi: 10.1038/s41579-019-0223-4
Vasanthakumar, T., and Rubinstein, J. L. (2020). Structure and roles of V-type ATPases. Trends Biochem. Sci. 45, 295–307. doi: 10.1016/j.tibs.2019.12.007
Vasanthakumar, T., Bueler, S. A., Wu, D., Beilsten-Edmands, V., Robinson, C. V., and Rubinstein, J. L. (2019). Structural comparison of the vacuolar and Golgi V-ATPases from Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 116, 7272–7277. doi: 10.1073/pnas.1814818116
von Loeffelholz, O., Jiang, Q., Ariosa, A., Karuppasamy, M., Huard, K., Berger, I., et al. (2015). Ribosome–SRP–FtsY cotranslational targeting complex in the closed state. Proc. Natl. Acad. Sci. U.S.A. 112, 3943–3948. doi: 10.1073/pnas.1424453112
Wahlström, N., Edlund, U., Pavia, H., Toth, G., Jaworski, A., Pell, A. J., et al. (2020). Cellulose from the green macroalgae Ulva lactuca: isolation, characterization, optotracing, and production of cellulose nanofibrils. Cellulose 27, 3707–3725. doi: 10.1007/s10570-020-03029-5
Wang, X., Kim, Y., Hong, S. H., Ma, Q., Brown, B. L., Pu, M., et al. (2011). Antitoxin MqsA helps mediate the bacterial general stress response. Nat. Chem. Biol. 7, 359–366. doi: 10.1038/nchembio.560
Wright, R. M., Strader, M. E., Genuise, H. M., and Matz, M. (2019). Effects of thermal stress on amount, composition, and antibacterial properties of coral mucus. PeerJ 7:e6849. doi: 10.7717/peerj.6849
Yamaguchi, Y., and Inouye, M. (2011). Regulation of growth and death in Escherichia coli by toxin–antitoxin systems. Nat. Rev. Microbiol. 9, 779–790. doi: 10.1038/nrmicro2651
Yang, C.-S., Chen, M.-H., Arun, A., Chen, C. A., Wang, J.-T., and Chen, W.-M. (2010). Endozoicomonas montiporae sp. nov., isolated from the encrusting pore coral Montipora aequituberculata. Int. J. Syst. Evol. Microbiol. 60, 1158–1162. doi: 10.1099/ijs.0.014357-0
Yokoyama, K., and Imamura, H. (2005). Rotation, structure, and classification of prokaryotic V-ATPase. J. Bioenerg. Biomembr. 37, 405–410. doi: 10.1007/s10863-005-9480-1
Zhang, D., Sweredoski, M. J., Graham, R. L., Hess, S., and Shan, S. O. (2012). Novel proteomic tools reveal essential roles of SRP and importance of proper membrane protein biogenesis. Mol. Cell. Proteomics 11:M111.011585. doi: 10.1074/mcp.M111.011585
Zhao, J., Beyrakhova, K., Liu, Y., Alvarez, C. P., Bueler, S. A., Xu, L., et al. (2017). Molecular basis for the binding and modulation of V-ATPase by a bacterial effector protein. PLoS Pathog. 13:e1006394. doi: 10.1371/journal.ppat.1006394
Zhou, L., and Sazanov, L. A. (2019). Structure and conformational plasticity of the intact Thermus thermophilus V/A-type ATPase. Science 365:eaaw9144. doi: 10.1126/science.aaw9144
Keywords: Endozoicomonas montiporae, differentially expressed proteins, heat stress, coral lysate, coral symbiont
Citation: Chan Y-F, Chen C-Y, Lu C-Y, Tu Y-C, Tandon K, Shikina S and Tang S-L (2022) A First Insight Into the Heat-Induced Changes in Proteomic Profiles of the Coral Symbiotic Bacterium Endozoicomonas montiporae. Front. Mar. Sci. 9:808132. doi: 10.3389/fmars.2022.808132
Received: 10 November 2021; Accepted: 07 January 2022;
Published: 14 February 2022.
Edited by:
Zhiyong Li, Shanghai Jiao Tong University, ChinaReviewed by:
Christine Ferrier-Pagès, Centre Scientifique de Monaco, MonacoCopyright © 2022 Chan, Chen, Lu, Tu, Tandon, Shikina and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sen-Lin Tang, c2x0YW5nQGdhdGUuc2luaWNhLmVkdS50dw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.