
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 01 March 2022
Sec. Marine Biology
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.805209
Neuropeptides, as neurotransmitters and neuromodulators, have a variety of physiological functions in the mollusk. Here, a PRQFVamide-related peptide gene was cloned from cuttlefish Sepiella japonica (designated as SjPRQFVRP, GenBank Accession No: OK999997). The full length of SjPRQFVRP is 1748 bp, including an open reading frame (ORF) of 738 bp encoding 245 amino acids. The putative precursor protein comprises one signal peptide and four different mature pentapeptides: fourteen copies of PMEFLamide, three copies of RMEFLamide, one copy of AMEFLamide and GMEFLamide. Multiple alignments showed SjPRQFVRP shared 71% identity with that of Octopus vulgaris and supported the phylogenetic analysis. The spatio-temporal expression pattern showed that SjPRQFVRP mRNA was widely expressed among the 13 tissues and primarily abundantly expressed in the brain and optic lobe during the whole development stage. In situ hybridization data indicated that SjPRQFVRP was detected in the vertical lobe, subvertical lobe, anterior basal lobe, anterior pedal lobe, and optic lobes of the brain. Subcellular localization analysis revealed that the SjPRQFVRP protein was localized in the cytoplasm of HEK293 cells. Collectively, the results will provide a foundation for further exploring the mechanism of SjPRQFVRP function in cephalopods.
Neuropeptides are signaling molecular produced and released primarily by neurons and engaged in diverse physiological processes (Wang et al., 2015). A large number of bioactive neuropeptides have been found in vertebrates with multiple functions in the regulation of learning and memory, reproduction, energy homeostasis, cardiovascular activity, and stress response (Corbière et al., 2019). Since the first neuropeptide eledoisin was sequenced from octopus Eledone moschata (Erspamer and Anastasi, 1962), neuropeptides have been extensively studied in multiple invertebrates, including mollusks (Price and Greenberg, 1977; Pulst et al., 1988), nematodes (Davenport et al., 1988; Keating et al., 1995), and arthropods (Marder et al., 1986; Nambu et al., 1988). These neuropeptides have also been linked to various physiological and behavioral processes such as learning and memory (Beets et al., 2012), metabolism (Kaufmann and Brown, 2008; Wang et al., 2013), longevity (Waterson et al., 2014), and reproduction (Garrison et al., 2012).
Diverse neuropeptides have been identified in cephalopods. It was reported that they play a vital role in regulating feeding (Zhang and Tublitz, 2013), reproduction (Di Cristo et al., 2005), muscle contraction (Go et al., 2011), memory (Bardou et al., 2010), chromatophore activity (Zhang et al., 2012), and heart activities (Springer et al., 2004). FMRFamide (Phe-Met-Arg-Phe-NH2) and FMRFamide-related peptides (FaRPs) were identified in Loligo pealei (Burbach et al., 2013), Sepia officinalis (Chrachri, 2020), and Sepia pharaonis (Zhu et al., 2020); GnRH (Gonadotropin-releasing hormone) was reported in S. officinalis (Di Cristo et al., 2009), Sepia lycidas (Murata et al., 2021) and S. pharaonis (Song et al., 2021); APGWamide (Ala-Pro-Gly-Trp-NH2) was reported in Octopus vulgaris (Di Cristo et al., 2005), and Idiosepius pygmaeus (Sirinupong et al., 2011); Small cardioactive peptide (sCAP) was identified in Octopus minor (Iwakoshi et al., 2000), O. vulgaris (Kanda and Minakata, 2006), and Sepiella japonica (Li et al., 2019).
The -FVamide neuropeptides are purified from Aplysia with -FVamide in their C-terminus. These neuropeptides contain AMRPs (Fujisawa et al., 1999), enterins (Furukawa et al., 2001), and PRQFVamide (Furukawa et al., 2003). PRQFVamide was firstly identified from the central nervous system (CNS) and gut of Aplysia californica (Furukawa et al., 2003). Through immunohistochemistry, the PRQFVamide immunopositive signals were observed in the gut, especially where related to the vasculature (Furukawa et al., 2003). Moreover, synthetic PRQFVamide not only could inhibit contractions of the gut and vasculature but also could reduce the excitability of B4/5 and B31/32 neurons of the buccal feeding circuit (Furukawa et al., 2003). Later, the same research group discovered that activation of PRQFVamide-containing neurons could lead to the organized rhythmic output of the feeding central pattern generator (CPG) (Dembrow et al., 2003). It is poorly studied when we search for the PRQFVamide gene or protein from the National Center for Biotechnology Information (NCBI). Only one or two sequences from Deroceras reticulatum, A. californica, O. vulgaris, Mizuhopecten yessoensis, and Lingula anatina can be found. And another sequence from Charonia tritonis was only found in Uniprot. PRQFVamide was only reported in two cephalopods, S. officinalis and O. vulgaris. In S. officinalis, the incomplete PRQFVamide preprohormones showed very large precursors (Zatylny Gaudin et al., 2016). Moreover, researchers obtained five mature products from the hemolymph of the cuttlefish, and one of them was the pentapeptide PMEFLamide (Zatylny Gaudin et al., 2016). A study on O. vulgaris indicated that the PRQFVamide was expressed in the gastric ganglion. The gene expression level was suppressed with the higher levels of the octopuses with “high” Aggregata parasite load, while those with “low” parasite load showed an increased expression (Baldascino et al., 2017). However, the underlined function and regulatory mechanisms of the PRQFVamide in cephalopod are poorly understood.
The common Chinese cuttlefish S. japonica used to be one of the four traditional major marine fishery species in the East China Sea (Wu et al., 2010b). Because of great economic importance and ecological value, S. japonica is a promising aquaculture species. However, due to overexploitation and the deterioration of environmental conditions, the resources of S. japonica have been severely damaged since the mid-1970s. Great efforts have been poured on it to recover its ecological resources. Although artificial breeding and some achievements have been made in China, precocious puberty and disease arose in cuttlefish during artificial breeding (Wu et al., 2010a; Lü et al., 2019). Thus, exploring the potential physiological functions of PRQFVamide-related peptide in S. japonica (termed as SjPRQFVRP) is urgently needed and our work has important scientific and practical significance.
In this study, the main objectives aim to (1) clone the full-length cDNA of SjPRQFVRP; (2) detect the spatio-temporal expression during the whole developmental stages with quantitative Real-time PCR (qRT-PCR); (3) investigate the tissues distribution profiles by in situ hybridization (ISH); (4) observe the subcellular localization in HEK293 cells. The results will inform us the follow up studies based on different approaches that the gene might be involved in various physiological functions in S. japonica.
Healthy S. japonica was collected from a local aquaculture base (29°53′ N, 122°18′ E, Xixuan Island, Zhejiang Province, China). Cuttlefishes were cultured in aerated seawater at approximately 25°C on a 14/10 h light/dark cycle and fed with shrimp twice daily. On the basis of the growth time and gonad appearance, cuttlefishes were divided into six different developmental stages (stage I–VI) as previously reported (Jiang et al., 2007; Luo et al., 2014). Three cuttlefishes were used for the tissue distribution analysis at each developmental stage: stage I-II (17.3 ± 4.7 g), stage III (52.7 ± 5.1 g), stage IV (60.8 ± 4.3 g), stage V (70.0 ± 9.4 g), stage VI (99.1 ± 6.4 g). After anesthetized on ice for 1 min, experimental cuttlefishes were dissected. The brain, optic lobe, heart, liver, intestine, stomach, pancreas, gill, muscle, skin, ovary, nidamental gland, and accessory nidamental gland were removed separately and put in RNAstore (CoWin Biosciences, Jiangsu, China) at −80°C. The brain of stage I-II was fixed in 4% paraformaldehyde (PFA) overnight at 4°C. All experiments were conducted according to the protocols and guidelines approved by the Ethics Committee of Zhejiang Ocean University and the Academy of Experimental Animal Center of Zhejiang Ocean University.
To isolate total RNA from tissues, Trizol reagent (Takara, Kyodo, Japan) was used as described in previous studies (Li et al., 2019). Then the quality, purity, and integrity of RNAs were assessed by UV spectroscopy (A260/A280) on Nanodrop 2000 (Thermo Fisher Scientific, Waltham, MA, United states) and agarose gel electrophoresis. The mRNA was reverse transcribed into first-strand cDNA using PrimeScript™ RT reagent Kit with gDNA Eraser (Perfect Real Time) (Takara Bio Inc., Kyodo, Japan). The first-strand cDNA was used as the template.
To amplify the SjPRQFVRP sequence, PRQFVRP-F/R primers (Table 1) were designed in accordance with the unigene annotated in S. japonica transcription (Lü et al., 2016). The PCR amplification reaction was conducted with 2 × Easy Taq Mix (Sangon Biotech, Shanghai, China). The target band was gel-purified and ligated to pMD19-T Vector (Takara Bio Inc., Kyodo, Japan). The insert was sent to the company for sequence determination (Sangon Biotech, Shanghai, China).
According to the SjPRQFVRP sequence, specific primers (Table 1) for 5′- and 3′-RACE were designed. RACE reactions were conducted following the instructions of SMARTer™ RACE cDNA Amplification Kit (Clontech, Fitchburg, MA, United states) as a previous study (Zheng et al., 2016). UPM (10 × Universal Primer A Mix, Table 1) and 5′-PRQFVRP-RACE (Table 1) were used to perform the 5′-RACE of the SjPRQFVRP. The final reaction volume of 50 μl contained 41.5 μl Master Mix, 2.5 μl cDNA of the brain, 1.0 μl 5′-PRQFVRP-RACE, and 5 μl UPM. The PCR reaction program was 25 cycles of 94°C for 30 s, 68°C for 30 s, and 72°C for 2 min. The next steps were the same as conventional PCR. The annealing temperature was adjusted according to the primer temperature. The 3′-RACE of the SjPRQFVRP used 3′-PRQFVRP-RACE (Table 1) and UPM. The PCR reaction was similar to that mentioned above for 5′-RACE. All the products were purified using DNA gel extraction kit (Sangon Biotech, Shanghai, China) and ligated into the pMD19-T vector for sequence determination. All primers used in the present study are listed in Table 1.
To get the target sequence, three conserved nucleotide sequences (XM 029790420.1, NM 001204600.1, KY659273.1) (Supplementary Table 1) were acquired from the NCBI database1 and were aligned with the Blast (Blastx and Blastn). Once gaining the full length of SjPRQFVRP, the open reading frame (ORF) of the SjPRQFVRP gene was predicted in NCBI. Online software NetPhos 3.1 Server2 was used to predict the phosphorylation sites. The polyadenylation signal, cleavage sites, amidation site, and mature peptides were marked as mentioned in previous studies (Veenstra, 2000; Proudfoot, 2011; Zatylny Gaudin et al., 2016). Online software Expasy-ProtParam3 was used to translate it into amino acid sequence and predict the molecular weight (MW) and theoretical isoelectric point (pI) of the protein. The signal peptide was predicted by SignalP 5.0.4 Multiple alignments were performed through software DNAMAN (Lynnon biosoft, San Ramon, CA, United states). The phylogenic tree was established by the maximum likelihood method (MEGA-X) and the bootstrap test was assessed from 1,000 replications to assure reliability.
qRT-PCR was performed with primers PRQFVRP-RT-F/R (Table 1) as previously described (Li et al., 2018) on a CFX Connect Real-time PCR amplifier (Bio-Rad, Richmond, VA, United states) with TB Green® Premix Ex Taq™ II (Tli RNaseH Plus) (Takara, Kyodo, Japan). GAPDH (Huo et al., 2018) and β-actin (JN564496.1) were taken as double internal controls. The reactions were performed with three individual repeats and three independent experiments in each tissue. PCR specificity was assessed in the terms of a melting curve.
The SjPRQFVRP mRNA levels were analyzed using the threshold and Ct (threshold cycle) values. The mRNA expression level detected in the muscle tissue was used as the reference value according to the 2–ΔΔCt method (Livak and Schmittgen, 2001). All data were normalized with the mRNA expression level in the muscle and presented as mean ± standard deviation (SD) (n = 3). The Least Significant Difference (LSD) multiple comparison test was performed by SPSS 21. Differences were considered significant at p < 0.05.
The specific primers (PRQFVRP-ISH-F/R, Table 1) were designed for ISH. The PCR was carried out with brain cDNA as a template. The gel-purified PCR products were ligated into the pGEM-T Easy vector (Promega, Madison, WI, United states) and sequenced. The sense and anti-sense probes were synthesized by in vitro transcription with the DIG RNA Labeling kit (Roche Diagnostics, Mannheim, Germany) as previously described (Li et al., 2019).
The fixed brain at stage I-II was washed with 1 × PBST (0.01 M PBS, 1% Tween-20) and dehydrated with gradient methanol (25, 50, 75, and 100%). The dehydrated tissue was embedded into the paraffin and sectioned with an ultra-microtome at a thickness of 7 μm.
In situ hybridization procedure was described as a previous study (Li et al., 2019). Briefly, on the first day, sections were dewaxed in xylene and rehydrated in gradient alcohol. Then, the slices were digested with Proteinase K (1 μg/ml) for 8 min at 37°C. Later, the slices were transferred to Tris/Glycine buffer to stop the reaction and fixed in 4% PFA. Pre-hybridization was conducted to reduce non-specific hybridization for 2 h at 42°C. Then slices were incubated at 55°C for 14–16 h with sense or anti-sense RNA probes (3 μg/ml). On the second day, the slices were washed at 37°C with gradient saline sodium citrate (SSC). Following the wash, sections were blocked for 2 h at room temperature (RT) with blocking buffer (2% goat serum, 2 mg/ml BSA, 0.01 M PBS). Next, slides were incubated in anti-DIG-AP Fab fragments (1:1000, Roche Diagnostics, Mannheim, Germany) overnight at 4°C. On the third day, sections were stained with NBT/BCIP (Roche Diagnostics, Mannheim, Germany). Signals were checked every 30 min. Then stained slides were mounted in glycerol and observed with a microscope (Nikon, Tokyo, Japan).
To construct recombinant plasmids SjPRQFVRP-EGFP, PCR was used to amplify the ORF of SjPRQFVRP with specific primers (Table 1) containing restriction sites XhoI and HindIII. Then, the PCR products were ligated into pEGFP-N1 and sequenced.
Human embryonic kidney 293 (HEK293) cells were cultured at 37°C with 5% CO2 for 24 h before transfection. The medium was discarded and 2 ml Opti-MEM (Gibco, Waltham, MA, United states) was added to the plate. The recombinant plasmids SjPRQFVRP-EGFP and Lipo6000™ Transfection Reagent (Beyotime Biotechnology, Shanghai, China) were diluted with Opti-MEM for 5 min at RT, respectively. The mixture of two diluents was transfected into HEK293 cells and the cells were incubated for 6 h at 37°C. Then, the medium was removed and the cells were cultured with a fresh medium for 24 h. After fixation with 4% PFA overnight, the cytomembrane and nucleus were stained with DiI (7 μM) (Beyotime Biotechnology, Shanghai, China) and DAPI (5 μg/ml) (Beyotime Biotechnology, Shanghai, China), respectively. The stained cells were visualized under TCS SP5II laser scanning confocal microscope (Leica, Wetzlar, Germany).
The full-length cDNA of SjPRQFVRP is 1748 bp, including 428 bp untranslated region (UTR) in the 5′-terminus, 582 bp of UTR in the 3′-terminus, and a 738 bp ORF encoding 245 amino acids (aa) (Figure 1). The putative precursor protein had a signal peptide of 19 aa and four different pentapeptides: 14 copies of PMEFLamide, three copies of RMEFLamide, one copy of AMEFLamide and GMEFLamide. The sequence MEFL was followed by a glycine which is an amidation site. The predicted MW was 28.6 kDa and the theoretical pI was 9.47.

Figure 1. Nucleotide sequence and deduced amino acid sequence of SjPRQFVRP. The initiation codon (ATG), stop codon (TAA), and polyadenylation signal (AATAAA) are boxed in black. The deduced signal peptide is underlined in black. The putative mature peptides are shaded in gray. The cleavage sites are identified in green color, and glycine used for C-terminus amidation is marked in red. The predicted phosphorylation sites are shown in colorful frames (Tyr site in red, Thr site in blue, and Ser site in green).
The amino acid sequences of PRQFVamide downloaded from the NCBI database from the other four representative species and the SjPRQFVRP were used for alignment analysis. The results indicated that SjPRQFVRP shared 71% identity to the PRQFVamide-like of O. vulgaris (XP 029646280.1), and 47, 46, 45% identity to those of M. yessoensis (OWF49118.1), A. californica (NP 001191529.1), and D. reticulatum (ARS01374.1), respectively. The multiple alignments results showed that SjPRQFVRP had some conserved amino acids with other PRQFVamide, such as cleavage sites (KR/R) and partial mature peptide amino acids (FL/FV) (Figure 2).
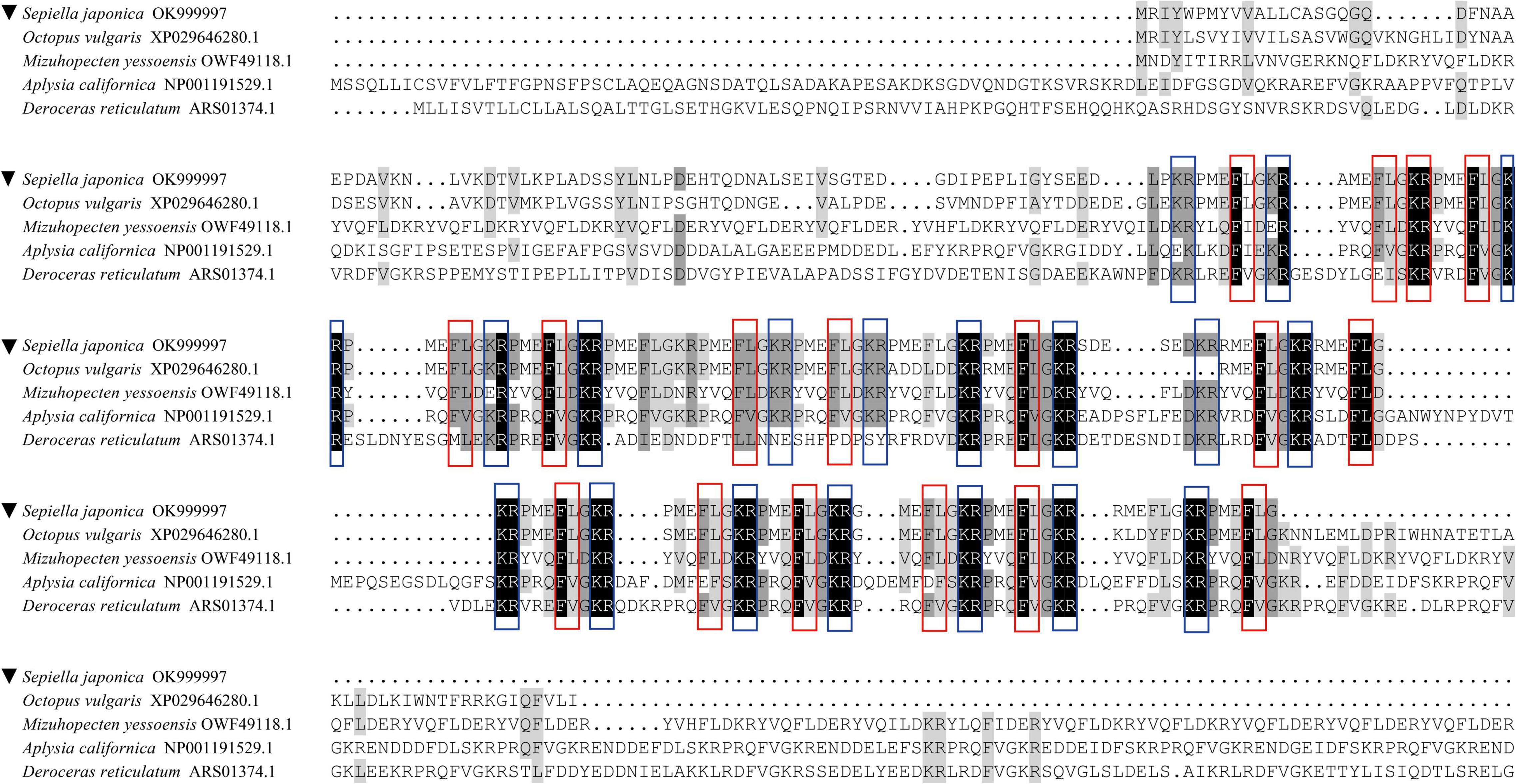
Figure 2. Multiple alignments of the SjPRQFVRP with PRQFVamide of other species. The SjPRQFVRP is noted by ▼. The GenBank accession numbers of the sequences are as follows: Sepiella japonica (OK999997), Octopus vulgaris (XP 029646280.1), Mizuhopecten yessoensis (OWF49118.1), Aplysia californica (NP 001191529.1), and Deroceras reticulatum (ARS01374.1). Identical residues in all sequences are showed with black boxes. Conservative substitutions are showed with gray boxes. The red boxes are used to indicate the conservative position in the predicted mature peptide (FX). The blue boxes are used to indicate the conservative cleavage sites (KR/R).
To determine the evolutionary status of the SjPRQFVRP, a phylogenetic tree was generated from SjPRQFVRP and the other five PRQFVamide sequences by the maximum likelihood method (MEGA-X) (Figure 3). Phylogenetics reveals that the PRQFVamide from five mollusks were clustered. Moreover, a close relationship between SjPRQFVRP and PRQFVamide-like of O. vulgaris was observed at a high confidence value (99%), suggesting they were close relatives. The PRQFVamide of brachiopod L. anatina (XP 013402951.1), as an outgroup, was distantly related to those of the mollusks.
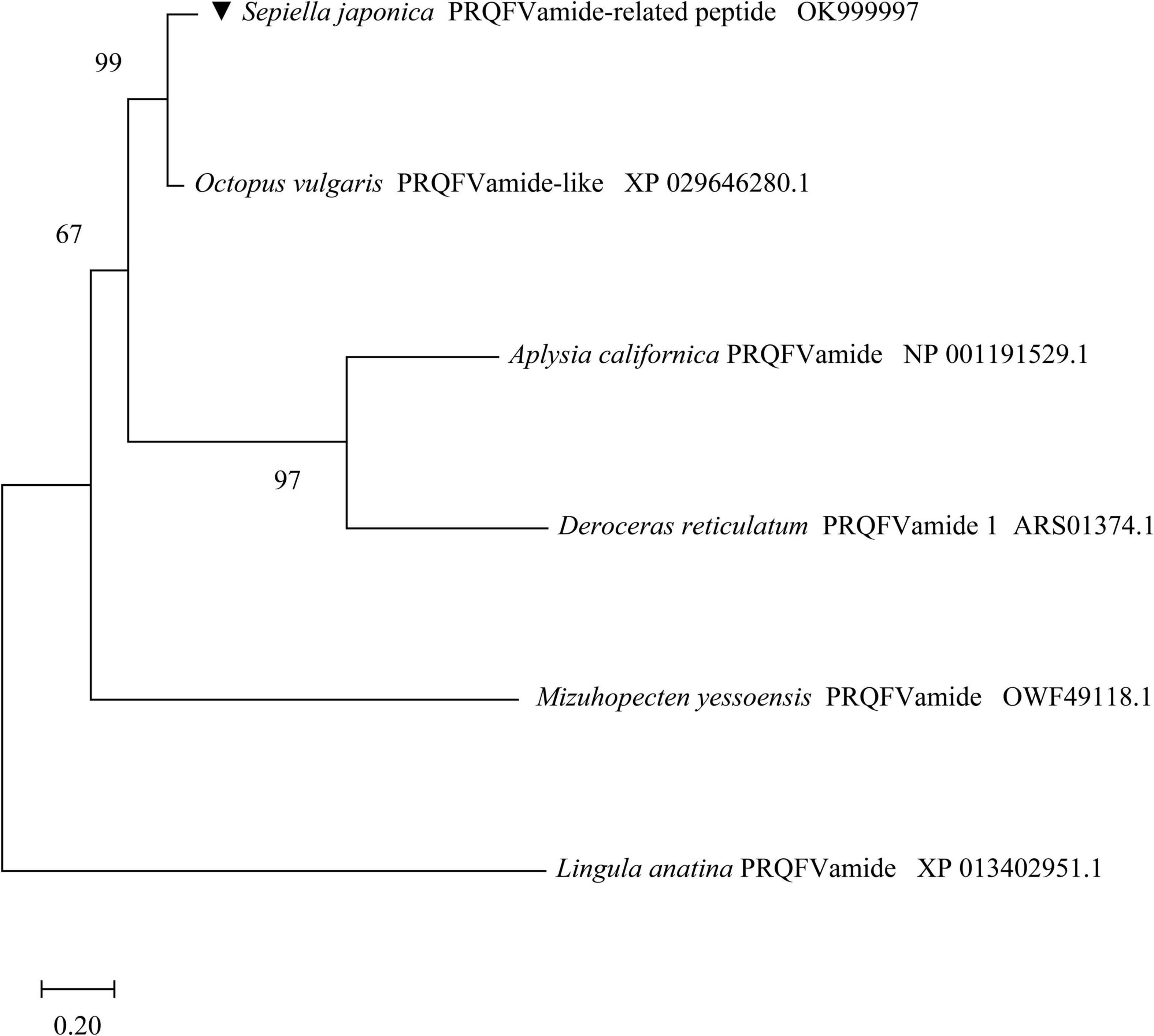
Figure 3. Phylogenetic tree of SjPRQFVRP with five other PRQFVamide using the maximum likelihood method. The SjPRQFVRP is noted by ▼. All protein sequences are obtained from GenBank of the NCBI, and the GenBank accession numbers are as follows: Sepiella japonica (OK999997), Octopus vulgaris (XP 029646280.1), Mizuhopecten yessoensis (OWF49118.1), Aplysia californica (NP 001191529.1), Deroceras reticulatum (ARS01374.1), and Lingula anatina (XP 013402951.1). The PRQFVamide of L. anatina serves as the outgroup. The topological stability of the tree is achieved by running 1,000 bootstrap replications. Bootstrap values (%) are indicated by numbers at the nodes.
As shown in Figure 4, the mRNA expression level of SjPRQFVRP at different development stages from females was detected. The transcripts of the SjPRQFVRP in the muscle were taken as the reference.
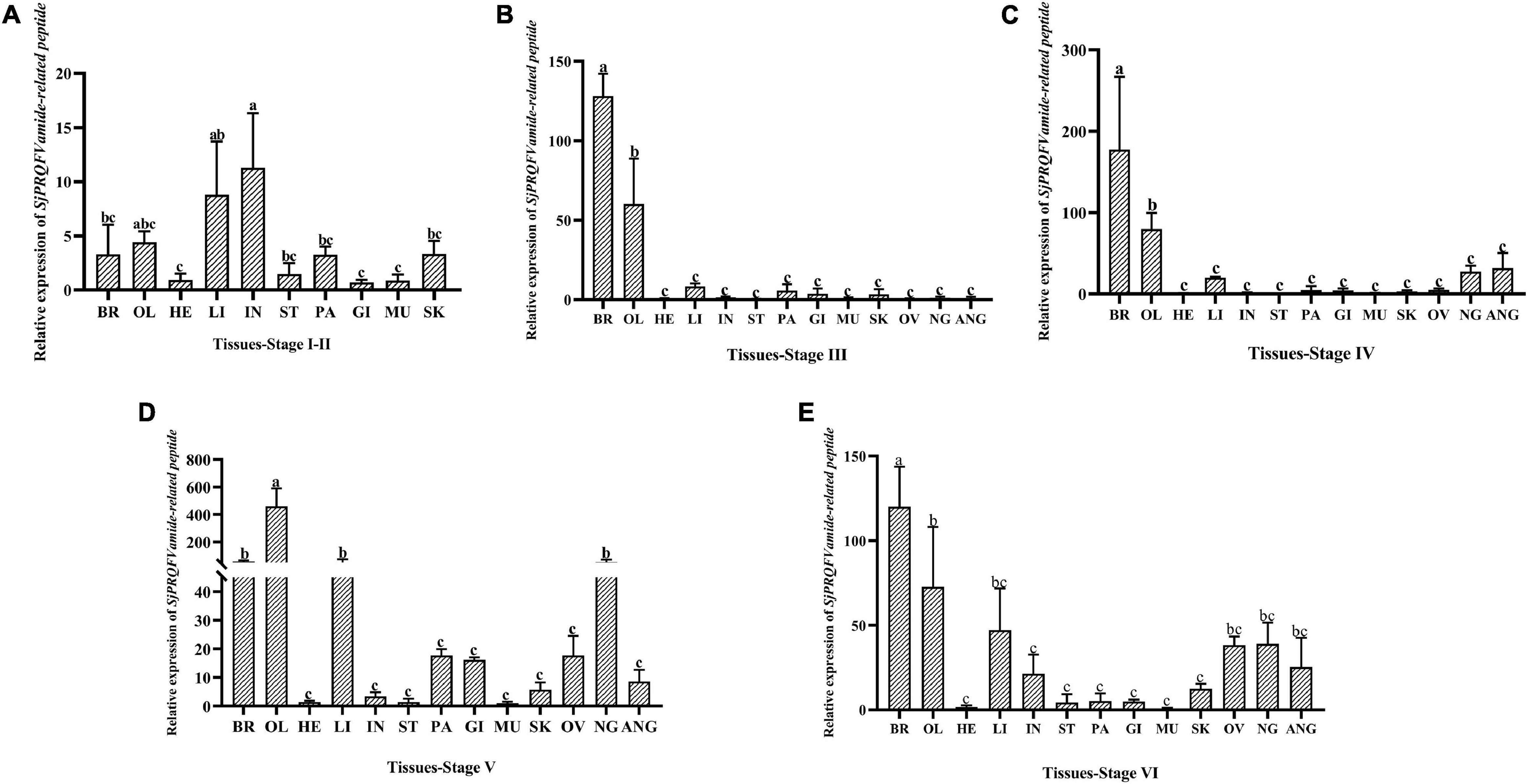
Figure 4. Expression analysis of SjPRQFVRP from diverse tissues at different development stages of female cuttlefish. (A) Stage I-II; (B) stage III; (C) stage IV; (D) stage V; (E) stage VI. In each case, the transcripts of the muscle were served as the reference value. Vertical bars represent the mean ± SD (n = 3). The analyses were conducted in tissues including the brain (BR), optic lobe (OL), heart (HE), liver (LI), intestine (IN), stomach (ST), pancreas (PA), gill (GI), muscle (MU), skin (SK), ovary (OV), nidamental gland (NG), and accessory nidamental gland (ANG). GAPDH and β-actin served as reference genes. Different letters on the bars indicate statistically significant differences (p < 0.05).
At stage I-II (Figure 4A), the expression of SjPRQFVRP in the intestine was approximately 10 times that of the muscle, followed in the liver, which was about 8 times higher (p < 0.05). At stage III (Figure 4B), the expression level of SjPRQFVRP in the brain and optic lobe was significantly higher than that of other tissues, with the levels were approximately 125 times and 60 times that of the muscle, respectively (p < 0.05). At stage IV (Figure 4C), the expression pattern is very similar to that of stage III, with the brain and optic lobe showing very high levels (p < 0.05). The difference with a slightly increasing level in the nidamental gland and accessory nidamental gland has been detected. At stage V (Figure 4D), the optic lobe had higher expression level than that of other tissues and was around 450 times that of the muscle (p < 0.05), followed by the brain, liver, and nidamental gland. At stage VI (Figure 4E), the relative expression in the brain was significantly higher than other tissues and was around 120 times that of the muscle (p < 0.05).
To better elucidate our data, the stage-point expression levels of SjPRQFVRP in one single tissue were re-quantified in Figure 5. As shown in Figure 5A, for the brain tissue, the expression level of SjPRQFVRP showed a steady increase from stage I-II to IV and reached the peak at stage IV (p < 0.05), then the expression level decreased at stage V and recovered to the level of stage III at stage VI. In the optic lobe (Figure 5B) and pancreas (Figure 5E), the transcripts of SjPRQFVRP at stage V were significantly higher than that of other stages (p < 0.05). In the liver (Figure 5C), it showed an increasing trend, and the peak was at stage V and VI (p < 0.05). In the ovary (Figure 5F), from stage III to VI, the expression showed a continuous increase. In the nidamental gland (Figure 5G), from stage III to stage V, the expression increased until stage V reached the peak (p < 0.05). In the accessory nidamental gland (Figure 5H), the highest expression of SjPRQFVRP appeared at stage IV (p < 0.05).
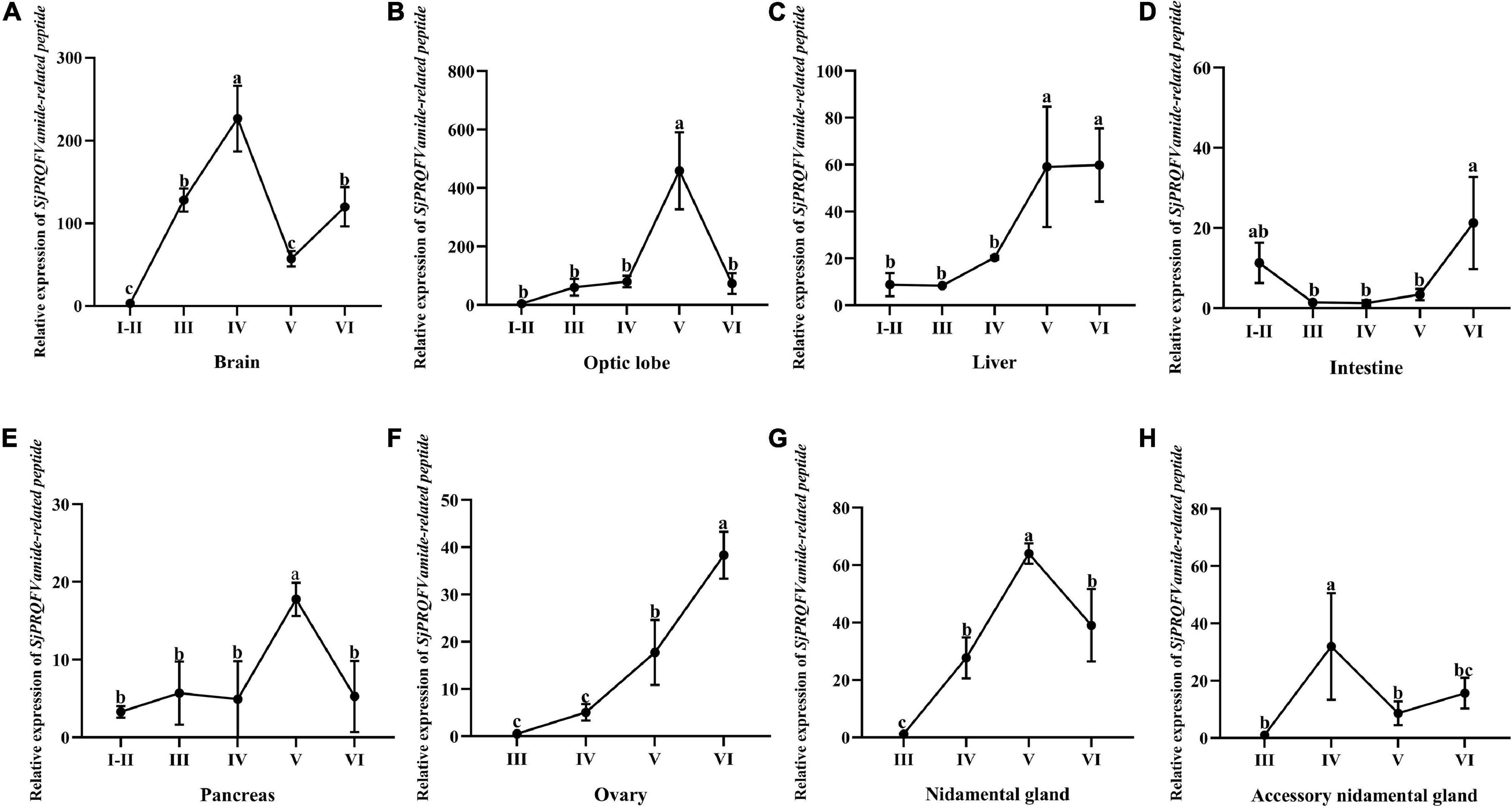
Figure 5. The expression level of SjPRQFVRP of females in different tissues at different stages. Vertical bars represent the mean ± SD (n = 3). GAPDH and β-actin served as reference genes. Different letters indicate statistically significant differences (p < 0.05). (A) Expression trend of SjPRQFVRP in the brain. (B) Expression trend of SjPRQFVRP in the optic lobe. (C) Expression trend of SjPRQFVRP in the liver. (D) Expression trend of SjPRQFVRP in the intestine. (E) Expression trend of SjPRQFVRP in the pancreas. (F) Expression trend of SjPRQFVRP in the ovary. (G) Expression trend of SjPRQFVRP in the nidamental gland. (H) Expression trend of SjPRQFVRP in the accessory nidamental gland.
The SjPRQFVRP mRNA distribution in the cuttlefish brain at stage I-II was determined by ISH. As shown in Figure 6, no signal was detected with the sense probe (Figures 6A,C). In contrast, specific staining was detected in various regions of the cuttlefish brain with the anti-sense probe (Figures 6B,D). In the supraesophageal mass (Figure 6B), positive signals were intensively appeared in cells of the anterior basal lobes (abl) and followed in the vertical lobe (vl) and subvertical lobe (svl). In the suboesophageal mass, the positive signals were primarily observed in the anterior pedal lobe (apl) (Figure 6B). In other words, the positive signals were mainly distributed in the peripheral cells of the functional lobes of supraesophageal and suboesophageal masses. In the optic lobe (ol) (Figure 6D), the staining in the central medulla (med) was the most extensive and intense. The edge of the medulla cells was stained as weakly as the inner granulosa cells (inn. gr. cel) and outer granulosa cells (out. gr. cel). Moreover, the esophagus (Figure 6B) was also observed some weak signals at stage I-II.
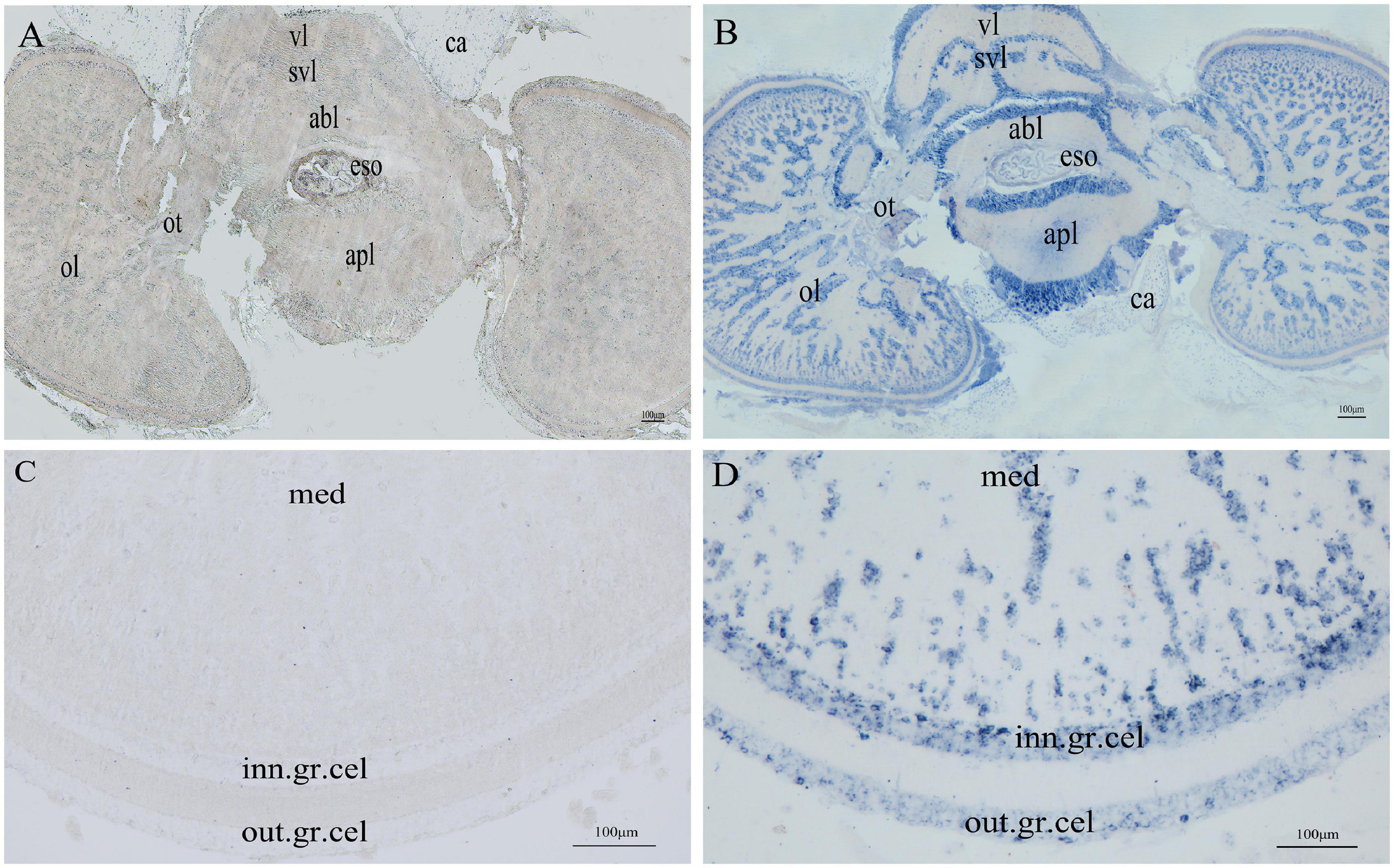
Figure 6. Distribution of SjPRQFVRP mRNA in the brain. (A) The brain of stage I-II was stained with sense SjPRQFVRP probe; (B) The brain of stage I-II was stained with anti-sense SjPRQFVRP probe; (C) The optic lobe was stained with the sense SjPRQFVRP probe; (D) The optic lobe was stained with the anti-sense SjPRQFVRP probe. Vl, vertical lobe; svl, subvertical lobe; abl, anterior basal lobe; eso, esophagus; apl, anterior pedal lobe; ca, cartilage; ot, optic tract; ol, optic lobe; med, medulla; out. gr. cel, outer granulosa cell; inn. gr. cel, inner granulosa cell.
Figure 7 showed the localization of SjPRQFVRP protein in the HEK293 cells. Strong and clear green signals were detected in the cytoplasm of the transfected cells (Figure 7A) but not in the membrane and nucleus (Figures 7B,C), indicating that SjPRQFVRP was localized in the cytoplasm of the cells (Figure 7D).
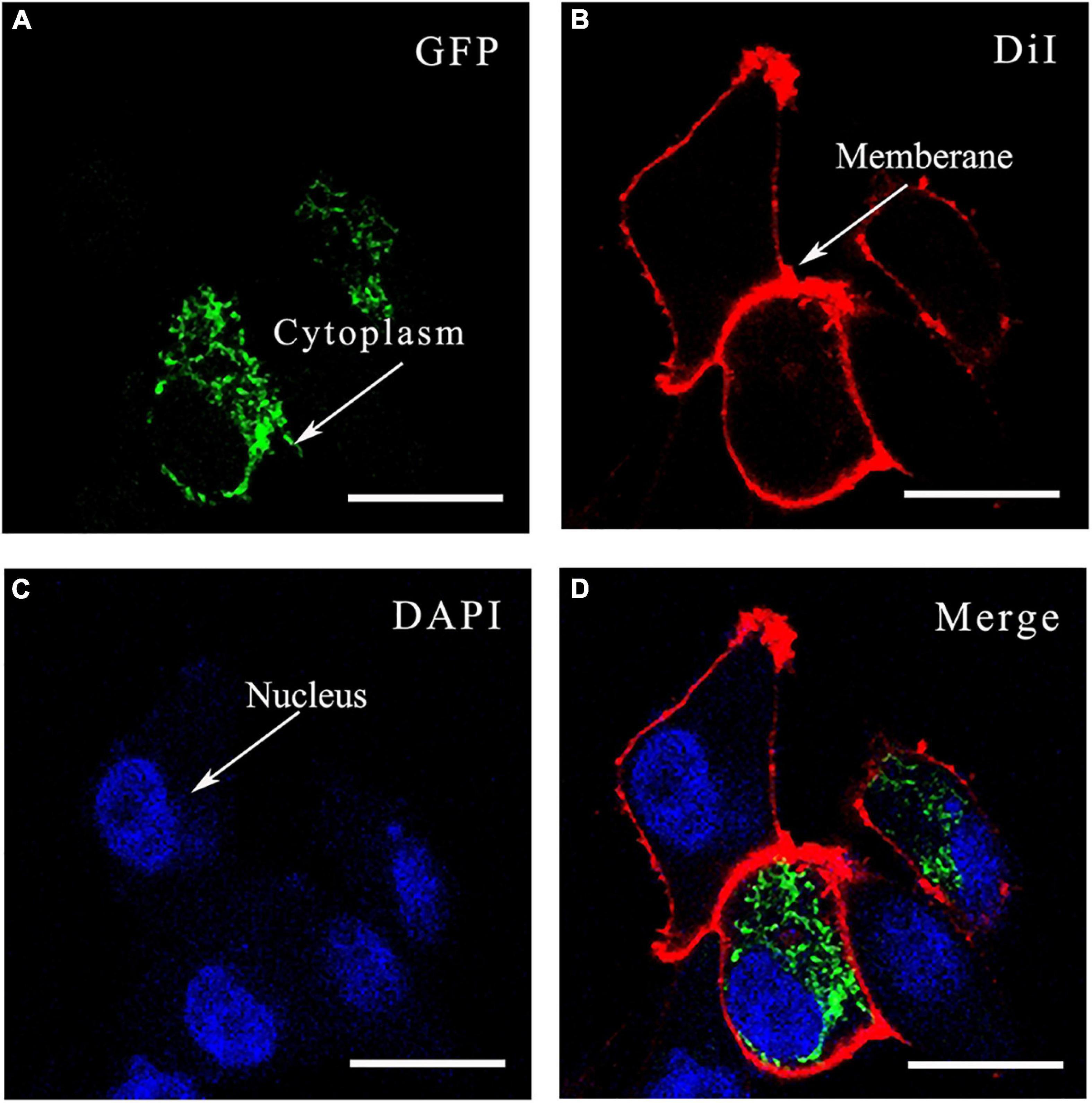
Figure 7. The localization of SjPRQFVRP-EGFP fusion protein in HEK293 cells. (A) GFP: SjPRQFVRP-EGFP fusion protein was stained green; (B) DiI: cytomembranes of cells were stained red with DiI; (C) DAPI: nucleus of cells was stained blue with DAPI; (D) Merge: SjPRQFVRP protein was mapped in the cytoplasm of HEK293 cells. Scale bars = 20 μm.
In this study, SjPRQFVRP contained four pentapeptides: 14 copies of PMEFLamide, three copies of RMEFLamide, one copy of AMEFLamide and GMEFLamide. In A. californica, PRQFVamide precursor protein contained PRQFVamide and other four kinds of pentapeptides: AREFVamide, VRDFVamide, VREFVamide, and IREFVamide (Furukawa et al., 2003). These five predicted pentapeptides were amidated and shared three immobile amino acids: the second amino acid is an arginine, the fourth is a phenylalanine, and the fifth is a valine (Furukawa et al., 2003). Compared with the PRQFVamide of A. californica, the predicted mature peptides of SjPRQFVRP did not contain PRQFVamide or the other four peptides. But similar to the PRQFVamide of A. californica, all four pentapeptides of SjPRQFVRP were amidated and shared four invariant amino acids (-MEFLamide). In addition, they had a similar structure feature at C-terminus: FXamide (X = hydrophobic amino acid). In S. officinalis, there were two sequences of incomplete PRQFVamide precursor with two different signal peptides and two termination codons at the C-terminus (Zatylny Gaudin et al., 2016). Moreover, five mature products were recovered from hemolymph in S. officinalis, including PMEFLamide (Zatylny Gaudin et al., 2016). However, in the predicted peptides of SjPRQFVRP, we did not find the other four mature peptides like the ones recovered in the hemolymph of S. officinalis. One possible explanation is that there are other transcripts of PRQFVamide in the cuttlefish, and this probably result from alternative splicing (Zatylny Gaudin et al., 2016).
Some interesting discoveries have been made by this study through the qRT-PCR data. 1) The SjPRQFVRP expression level was increased in the ovary from stage III to VI. 2) The expression in the nidamental gland was also increased from stage III to V but reduced at stage VI. 3) The expression in the liver increased and reached the peak at stage V and stage VI. The synthesis and release of neuropeptides were found to be a general feature of most cells of the nervous system (Kiss, 2011). However, researchers found that neuropeptides are also produced and released by non-neuronal cells such as glands or endocrine cells (Pirger et al., 2008; Kiss, 2011). Therefore, the expression of SjPRQFVRP in liver and nidamental gland might be related to its potential function. Nidamental gland is a typical secretory gland. It is mainly used to form a third layer of egg membrane to protect the fertilized eggs (Wang et al., 2021). Liver is an important organ for metabolism and innervated by a large number of afferent and efferent nerves (sympathetic and parasympathetic) in vertebrate (Yi et al., 2010). Otherwise, liver is also one of the synthesis organs of vitellogenin, which is transported to the ovary to be accumulated and stored in the form of vitellin (Tian et al., 2014). However, the function of the liver in cephalopods is still unclear. Given these interesting discoveries, more efforts are needed to further study in S. japonica.
A previous study reported that the cephalopod brain was divided into the supraesophageal mass, suboesophageal mass, and optic lobes (Yu et al., 2011). Various nerve lobes have been found in cephalopod brain with different functions. The vertical lobe stores memory information and accurately identifies the opposite position (Young, 1960). The subvertical lobe receives nerve impulses from the vertical lobe and transmits them to the optic lobe (Hochner et al., 2006). The reproductive nerve lobes are mainly the subpeduncle lobe and optic gland (Messenger, 1967; Froesch, 1974). The anterior and posterior basal lobes control the direction of steering movement and tentacle movement and color change by chromatophores (Shigeno and Yamamoto, 2002). The anterior and posterior pedal lobes also regulate tentacle movements, while the brachial lobe coordinates the movements of the arms (Shigeno and Yamamoto, 2002). It is reported that various neuropeptides distribute in different parts of the cephalopod brain, where is likely to be related to their function. In S. japonica, SjLFRFamide mRNA was highly expressed in the brain of both male and female cuttlefish. It was detected in several different functional lobes, mainly in the subpeduncle lobe, subvertical lobe, and all regions of suboesophageal mass. These findings suggested that SjLFRFamide might function in regulating reproduction and feeding (Cao et al., 2016). The distribution pattern of SjFMRFamide mRNA was similar to that of the SjLFRFamide in the S. japonica brain (Li et al., 2018). In S. officinalis, the SOFaRP2 gene was detected in the posterior chromatophore, anterior chromatophore, lateral basal, and optic lobes (Zhang et al., 2012; Zhang and Tublitz, 2013). These observations supported the idea that SOFaRP2 might be related to regulate chromatophore activity, feeding behavior, learning and memory. FMRFamide transcripts were also detected in the brain and expressed at the early life of S. pharaonis (Zhu et al., 2020). Additionally, strongly positive signals of GnRH were observed in the different lobes of the supraesophageal, suboesophageal mass, and the medulla of the optic lobe in S. pharaonis, suggesting that GnRH might have a role in regulating feeding and reproduction (Song et al., 2021). In this manuscript, the brain at stage I-II was selected to preliminarily explore its potential functionally relevant localization in cuttlefish at early developmental stages. The SjPRQFVRP was distributed in several functional lobes of the brain at stage I-II, including the vertical lobe, subvertical lobe, anterior basal lobe, anterior pedal lobe, and optic lobes. The distribution of SjPRQFVRP in the brain is similar to the distribution of neuropeptides mentioned above. However, to further explore the role of SjPRQFVRP during the growth and development of cuttlefish, more experiments are needed.
Moreover, in Aplysia, researchers described the distribution of PRQFVamide on the buccal and cerebral ganglion and found that the gene might have effect on some systems as a modulator, especially the feeding system (Dembrow et al., 2003; Furukawa et al., 2003). Whether SjPRQFVRP plays a part in the feeding system like that of Aplysia still needs to be confirmed by further experiments.
The subcellular localization suggests that SjPRQFVRP was in the cytoplasm of HEK293 cells. At present, the PRQFVamide receptor has not been found yet. In a future experiment, we can start with the PRQFVamide receptor and conduct a functional exploration of SjPRQFVRP in S. japonica.
In conclusion, our study described the full-length sequence of SjPRQFVRP cDNA in S. japonica. qRT-PCR analysis and ISH of the brain indicated the dominant expression of SjPRQFVRP throughout different developmental stages in S. japonica. In brief, the results informed us the follow up studies based on different approaches that the gene might be involved in various physiological functions in S. japonica. This study could also be of the theoretical basis for the restoration of cuttlefish germplasm resources.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, OK999997.
The animal study was reviewed and approved by the Ethics Committee of Zhejiang Ocean University and the Academy of Experimental Animal Center of Zhejiang Ocean University.
C-FC and L-BZ conceived and designed the study, reviewed, revised, and edited the manuscript, and contributed the reagents, materials, and analytical tools. J-YQ performed the investigation, analyzed the data, and led the writing-original draft. All authors contributed critically to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (NSFC; No. 31872547) and Natural Science Foundation of Zhejiang Province, China (LY20C190007).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank Shuang Li for her help with the revision of the article. We thank Huilai Shi and Hongling Ping of the Zhejiang Marine Fisheries Research Institute for the supply and husbandry of S. japonica. We are grateful to Jingwen Yang for the assistance in HEK293 cells and Bin Wang for the assistance in cell culture.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.805209/full#supplementary-material
Baldascino, E., Di Cristina, G., Tedesco, P., Hobbs, C., Shaw, T. J., Ponte, G., et al. (2017). The gastric ganglion of Octopus vulgaris: preliminary characterization of gene and putative neurochemical-complexity, and the effect of Aggregata octopiana digestive tract infection on gene expression. Front. Physiol. 8:1001. doi: 10.3389/fphys.2017.01001
Bardou, I., Leprince, J., Chichery, R., Vaudry, H., and Agin, V. (2010). Vasopressin/oxytocin-related peptides influence long-term memory of a passive avoidance task in the cuttlefish, Sepia officinalis. Neurobiol. Learn. Mem. 93, 240–247. doi: 10.1016/j.nlm.2009.10.004
Beets, I., Janssen, T., Meelkop, E., Temmerman, L., Suetens, N., Rademakers, S., et al. (2012). Vasopressin/oxytocin-related signaling regulates gustatory associative learning in C. elegans. Science 338, 543–545. doi: 10.1126/science.1226860
Burbach, J. P. H., Grant, P., Hellemons, A. J., DeGiorgis, J. A., Li, K. W., and Pant, H. C. (2013). Differential expression of the FMRF gene in adult and hatchling stellate ganglia of the squid Loligo pealei. Biol. Open 3, 50–58. doi: 10.1242/bio.20136890
Cao, Z., Sun, L., Chi, C., Liu, H., Zhou, L., Lv, Z., et al. (2016). Molecular cloning, expression analysis and cellular localization of an LFRFamide gene in the cuttlefish Sepiella japonica. Peptides 80, 40–47. doi: 10.1016/j.peptides.2015.10.005
Chrachri, A. (2020). Effect of FMRFamide on voltage-dependent currents in identified centrifugal neurons of the optic lobe of the cuttlefish, Sepia officinalis. bioRxiv [Preprint]. doi: 10.1101/2020.09.29.318691
Corbière, A., Vaudry, H., Chan, P., Walet Balieu, M. L., Lecroq, T., Lefebvre, A., et al. (2019). Strategies for the identification of bioactive neuropeptides in vertebrates. Front. Neurosci. 13:948. doi: 10.3389/fnins.2019.00948
Davenport, T. R. B., Lee, D. L., and Isaac, R. E. (1988). Immunocytochemical demonstration of a neuropeptide in Ascaris suum (Nematoda) using an antiserum to FMRFamide. Parasitology 97, 81–88. doi: 10.1017/S0031182000066762
Dembrow, N. C., Jing, J., Proekt, A., Romero, A., Vilim, F. S., Cropper, E. C., et al. (2003). A newly identified buccal interneuron initiates and modulates feeding motor programs in Aplysia. J. Neurophysiol. 90, 2190–2204. doi: 10.1152/jn.00173.2003
Di Cristo, C., De Lisa, E., and Di Cosmo, A. (2009). GnRH in the brain and ovary of Sepia officinalis. Peptides 30, 531–537. doi: 10.1016/j.peptides.2008.07.008
Di Cristo, C., Van Minnen, J., and Di Cosmo, A. (2005). The presence of APGWamide in Octopus vulgaris: a possible role in the reproductive behavior. Peptides 26, 53–62. doi: 10.1016/j.peptides.2004.07.019
Erspamer, V., and Anastasi, A. (1962). Structure and pharmacological actions of eledoisin, the active endecapeptide of the posterior salivary glands of Eledone. Experientia 18, 58–59. doi: 10.1007/BF02138250
Froesch, D. (1974). The subpedunculate lobe of the octopus brain: evidence for dual function. Brain Res. 75, 277–285. doi: 10.1016/0006-8993(74)90747-1
Fujisawa, Y., Furukawa, Y., Ohta, S., Ellis, T. A., Dembrow, N. C., Li, L., et al. (1999). The Aplysia Mytilus inhibitory peptide-related peptides: identification, cloning, processing, distribution, and action. J. Neurosci. 19, 9618–9634. doi: 10.1523/JNEUROSCI.19-21-09618.1999
Furukawa, Y., Nakamaru, K., Sasaki, K., Fujisawa, Y., Minakata, H., Ohta, S., et al. (2003). PRQFVamide, a novel pentapeptide identified from the CNS and gut of Aplysia. J. Neurophysiol. 89, 3114–3127. doi: 10.1152/jn.00014.2003
Furukawa, Y., Nakamaru, K., Wakayama, H., Fujisawa, Y., Minakata, H., Ohta, S., et al. (2001). The enterins: a novel family of neuropeptides isolated from the enteric nervous system and CNS of Aplysia. J. Neurosci. 21, 8247–8261. doi: 10.1523/JNEUROSCI.21-20-08247.2001
Garrison, J. L., Macosko, E. Z., Bernstein, S., Pokala, N., Albrecht, D. R., and Bargmann, C. I. (2012). Oxytocin/vasopressin-related peptides have an ancient role in reproductive behavior. Science 338, 540–543. doi: 10.1126/science.1226201
Go, H. J., Jo, E. H., Seo, J. K., Hong, Y. K., Lee, H. H., Do Kim, G., et al. (2011). Isolation and characterization of a novel myoactive tetradecapeptide-related peptide isolated from the brain of the squid, Todarodes pacificus. Peptides 32, 447–453. doi: 10.1016/j.peptides.2010.11.016
Hochner, B., Shomrat, T., and Fiorito, G. (2006). The octopus: a model for a comparative analysis of the evolution of learning and memory mechanisms. Biol. Bull. 210, 308–317. doi: 10.2307/4134567
Huo, L., Bao, M., Lv, Z., Chi, C., Wang, T., and Liu, H. (2018). Identification, functional characterization and expression pattern of myeloid differentiation factor 88 (MyD88) in Sepiella japonica. Fish Shellfish Immunol. 79, 112–119. doi: 10.1016/j.fsi.2018.04.065
Iwakoshi, E., Hisada, M., and Minakata, H. (2000). Cardioactive peptides isolated from the brain of a Japanese octopus, Octopus minor. Peptides 21, 623–630. doi: 10.1016/S0196-9781(00)00201-1
Jiang, X., Fu, F., Li, Z., and Feng, X. (2007). Study on the oogenesis and ovarial development of Sepiella maindroni. J. Fish. China 31, 607–617. doi: 10.3321/j.issn:1000-0615.2007.05.007
Kanda, A., and Minakata, H. (2006). Isolation and characterization of a novel small cardioactive peptide-related peptide from the brain of Octopus vulgaris. Peptides 27, 1755–1761. doi: 10.1016/j.peptides.2005.12.006
Kaufmann, C., and Brown, M. R. (2008). Regulation of carbohydrate metabolism and flight performance by a hypertrehalosaemic hormone in the mosquito Anopheles gambiae. J. Insect Physiol. 54, 367–377. doi: 10.1016/j.jinsphys.2007.10.007
Keating, C. D., Holden-Dye, L., Thorndyke, M. C., Williams, R. G., Mallett, A., and Walker, R. J. (1995). The FMRFamide-like neuropeptide AF2 is present in the parasitic nematode Haemonchus contortus. Parasitology 111, 515–521. doi: 10.1017/S0031182000066026
Kiss, T. (2011). Diversity and abundance: the basic properties of neuropeptide action in molluscs. Gen. Comp. Endocrinol. 172, 10–14. doi: 10.1016/j.ygcen.2011.02.016
Li, Y., Cao, Z., Li, H., Liu, H., Lu, Z., and Chi, C. (2018). Identification, Characterization, and Expression Analysis of a FMRFamide-Like Peptide Gene in the Common Chinese Cuttlefish (Sepiella japonica). Molecules 23:742. doi: 10.3390/molecules23040742
Li, Y., Zhou, Y., Shan, Y., Huang, W., Liu, H., Ping, H., et al. (2019). Cloning, expression analysis and localization of neuropeptide sCAP in Sepiella japonica. Acta Hydrobiol. Sin. 43, 778–785. doi: 10.7541/2019.092
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quanti tative PCR and the 2 (-MC (T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lü, Z., Liu, W., Liu, L., Shi, H., Ping, H., Wang, T., et al. (2016). De novo assembly and comparison of the ovarian transcriptomes of the common Chinese cuttlefish (Sepiella japonica) with different gonadal development. Genomics Data 7, 155–158. doi: 10.1016/j.gdata.2015.12.011
Lü, Z., Zhu, K., Pang, Z., Liu, L., Jiang, L., Liu, B., et al. (2019). Identification, characterization and mRNA transcript abundance profiles of estrogen related receptor (ERR) in Sepiella japonica imply its possible involvement in female reproduction. Anim. Reprod. Sci. 211:106231. doi: 10.1016/j.anireprosci.2019.106231
Luo, J., Jiang, X. M., Liu, M. H., Tang, F., and Peng, R. B. (2014). Oogenesis and ovarian development in Sepia lycidas. Acta Hydrobiol. Sin. 38, 1107–1116. doi: 10.7541/2014.162
Marder, E., Hooper, S. L., and Siwicki, K. K. (1986). Modulatory action and distribution of the neuropeptide proctolin in the crustacean stomatogastric nervous system. J. Comp. Neurol. 243, 454–467. doi: 10.1002/cne.902430403
Messenger, J. B. (1967). The peduncle lobe: a visuo-motor centre in Octopus. Proc. R. Soc. Lond. B Biol. Sci. 167, 225–251. doi: 10.1098/rspb.1967.0025
Murata, R., Mushirobira, Y., Tanaka, Y., and Soyano, K. (2021). Expression profile of GnRH-like peptide during gonadal sex differentiation in the cephalopod kisslip cuttlefish, Sepia lycidas. Gen. Comp. Endocrinol. 304:113718. doi: 10.1016/j.ygcen.2021.113718
Nambu, J. R., Murphy-Erdosh, C., Andrews, P. C., Feistner, G. J., and Scheller, R. H. (1988). Isolation and characterization of a Drosophila neuropeptide gene. Neuron 1, 55–61. doi: 10.1016/0896-6273(88)90209-7
Pirger, Z., Nemeth, J., Hiripi, L., Toth, G., Kiss, P., Lubics, A., et al. (2008). PACAP has anti-apoptotic effect in the salivary gland of an invertebrate species, Helix pomatia. J. Mol. Neurosci. 36, 105–114. doi: 10.1007/s12031-008-9070-x
Price, D. A., and Greenberg, M. J. (1977). Structure of a molluscan cardioexcitatory neuropeptide. Science 197, 670–671. doi: 10.1126/science.877582
Proudfoot, N. J. (2011). Ending the message: poly (A) signals then and now. Genes Dev. 25, 1770–1782. doi: 10.1101/gad.17268411
Pulst, S. M., Gusman, D., and Maykri, E. (1988). Immunostaining for peptides of the egg-laying hormone/bag cell peptide precursor protein in the head ganglia of Aplysia. Neuroscience 27, 363–371. doi: 10.1016/0306-4522(88)90244-8
Shigeno, S., and Yamamoto, M. (2002). Organization of the nervous system in the pygmy cuttlefish, Idiosepius paradoxus Ortmann (Idiosepiidae, Cephalopoda). J. Morphol. 254, 65–80. doi: 10.1002/jmor.10020
Sirinupong, P., Suwanjarat, J., and van Minnen, J. (2011). Distribution of APGWamide-immunoreactivity in the brain and reproductive organs of adult Pygmy squid, Idiosepius pygmaeus. Invertebr. Neurosci. 11, 97–102. doi: 10.1007/s10158-011-0122-5
Song, C., Sun, L., Zheng, L., and Chi, C. (2021). Gonadotropin-releasing hormone-like gene in the cephalopod, Sepia pharaonis: characterization, expression analysis, and localization in the brain. Invertebr. Reprod. Dev. 65, 226–234. doi: 10.1080/07924259.2021.1935335
Springer, J., Ruth, P., Beuerlein, K., Westermann, B., and Schipp, R. (2004). Immunohistochemical localization of cardio-active neuropeptides in the heart of a living fossil, Nautilus pompilius L.(Cephalopoda, Tetrabranchiata). J. Mol. Histol. 35, 21–28. doi: 10.1023/B:HIJO.0000020934.70110.0f
Tian, H., Meng, Y., and Xiao, H. (2014). Advances in vitellogenin research of aquatic animals. South China Fish. Sci. 10, 91–96. doi: 10.3969/j.issn.2095-0780.2014.04.015
Veenstra, J. A. (2000). Mono−and dibasic proteolytic cleavage sites in insect neuroendocrine peptide precursors. Arch. Insect Biochem. Physiol. 43, 49–63. doi: 10.1002/(SICI)1520-6327(200002)43:2<49::AID-ARCH1<3.0.CO;2-M
Wang, H., Girskis, K., Janssen, T., Chan, J. P., Dasgupta, K., Knowles, J. A., et al. (2013). Neuropeptide secreted from a pacemaker activates neurons to control a rhythmic behavior. Curr. Biol. 23, 746–754. doi: 10.1016/j.cub.2013.03.049
Wang, Y., Wang, M., Yin, S., Jang, R., Wang, J., Xue, Z., et al. (2015). NeuroPep: a comprehensive resource of neuropeptides. Database 2015:bav038. doi: 10.1093/database/bav038
Wang, Z., Liu, C., Zhai, J., Lin, L., Zhang, S., Chen, S., et al. (2021). Ultrastructure of the accessory nidamental gland of adult Sepioteuthis lessoniana. J. Shanghai Ocean Univ. 56, 231–238. doi: 10.12024/jsou.20191202875
Waterson, M. J., Chung, B. Y., Harvanek, Z. M., Ostojic, I., Alcedo, J., and Pletcher, S. D. (2014). Water sensor ppk28 modulates Drosophila lifespan and physiology through AKH signaling. Proc. Natl. Acad. Sci. U. S. A. 111, 8137–8142. doi: 10.1073/pnas.1315461111
Wu, C., Dong, Z., Chi, C., and Ding, F. (2010b). Reproductive and spawning habits of Sepiella maindroni off Zhejiang, China. Oceanol. Limnol. Sin. 41, 39–46. doi: 10.3724/SP.J.1011.2010.01138
Wu, C., Chi, C., He, G., Lü, Z., and Xu, M. (2010a). Isolation via enrichment and characterization of ten polymorphic microsatellite loci in the cuttlefish, Sepiella maindroni de Rochebruns. Acta Oceanol. Sin. 29, 121–124. doi: 10.1007/s13131-010-0083-2
Yi, C.-X., La Fleur, S. E., Fliers, E., and Kalsbeek, A. (2010). The role of the autonomic nervous liver innervation in the control of energy metabolism. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 1802, 416–431. doi: 10.1016/j.bbadis.2010.01.006
Young, J. Z. (1960). The failures of discrimination learning following the removal of the vertical lobes in Octopus. Proc. R. Soc. Lond. B Biol. Sci. 153, 18–46. doi: 10.1098/rspb.1960.0085
Yu, X., Wu, C., and Chi, C. (2011). Brain microstructure and optic gland ultrastructure in Sepiella maindroni. Oceanol. Limnol. Sin. 42, 300–304. doi: 10.11693/hyhz201102022022
Zatylny Gaudin, C. L., Cornet, V. R., Leduc, A., Zanuttini, B., Corre, E., Le Corguillé, G., et al. (2016). Neuropeptidome of the cephalopod Sepia officinalis: identification, tissue mapping, and expression pattern of neuropeptides and neurohormones during egg laying. J. Proteome Res. 15, 48–67. doi: 10.1021/acs.jproteome.5b00463
Zhang, Z., and Tublitz, N. J. (2013). Expression of the SOFaRP2 gene in the central nervous system of the adult cuttlefish Sepia officinalis. Neuropeptides 47, 149–155. doi: 10.1016/j.npep.2013.01.003
Zhang, Z. B., Goodwin, E., Loi, P. K., and Tublitz, N. J. (2012). Molecular analysis of a novel FMRFamide-related peptide gene (SOFaRP2) and its expression pattern in the brain of the European cuttlefish Sepia officinalis. Peptides 34, 114–119. doi: 10.1016/j.peptides.2011.07.011
Zheng, L., Liu, Z., Wu, B., Dong, Y., Zhou, L., Tian, J., et al. (2016). Ferritin has an important immune function in the ark shell Scapharca broughtonii. Dev. Comp. Immunol. 59, 15–24. doi: 10.1016/j.dci.2015.12.010
Keywords: neuropeptide, Sepiella japonica, cuttlefish, cephalopod, PRQFVamide-related peptide
Citation: Qiu J-y, Zheng L-b and Chi C-f (2022) Identification, Characterization, and Expression of a PRQFVamide-Related Peptide in Cephalopod Sepiella japonica. Front. Mar. Sci. 9:805209. doi: 10.3389/fmars.2022.805209
Received: 30 October 2021; Accepted: 07 February 2022;
Published: 01 March 2022.
Edited by:
Xiaodong Zheng, Ocean University of China, ChinaReviewed by:
Qiong Shi, Beijing Genomics Institute (BGI), ChinaCopyright © 2022 Qiu, Zheng and Chi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li-bing Zheng, emhlbmdsYkB6am91LmVkdS5jbg==; Chang-feng Chi, Y2hpY2hhbmdmZW5nQGhvdG1haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.