
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 31 January 2022
Sec. Microbial Symbioses
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.801344
This article is part of the Research Topic Interaction between Marine Invertebrates and Symbiotic Microbes in a Changing Environment: Community Structure and Ecological Functions View all 10 articles
There is a lot of evidence indicating pioneer microbes in early life having various effects on later host biology. Because of the influential phylogenetic position of sea cucumber, which is a deep branching clade in Deuterostomia, the attention on the microbiome in sea cucumber has been increasing. Although microbes in sea cucumber have been reported in several studies, there is a lack of knowledge regarding the pioneer microbiota in the early life stages of sea cucumber. In this study, microbiota changes during the larval development of sea cucumber were assessed using a laboratory rearing system. Microbial community structure was likely to be related to the developmental stage and significant alterations were detected in the late auricularia stage. The relative abundances of Oceanospirillales, Alteromonadales, and Rhodobacterales significantly varied after gut formation. A total of 257 strains were isolated from larval developmental stages of sea cucumber and affiliated to 124 ASVs in the metagenomic analysis. This data demonstrates for the first-time dynamic changes of sea cucumber microbiota in the developmental stages in early life.
Since the introduction of holobiont and hologenome concept, comprehensive studies on how holobiont assembly starts, in particular, which kinds of microorganisms first colonize when establishing the holobiont have been the subject of discussion. These first colonizers are called pioneer microbes, and have been widely studied in Deuterostomia animals with a view to investigating their composition and function in holobionts (Wopereis et al., 2014; Arrieta et al., 2015; Schokker et al., 2015; Gensollen et al., 2016). In humans, Therodor Escherich (1857–1911) was the pioneer in the progress of understanding human gut microflora, demonstrating the relationship between intestinal bacteria and physiology of digestion in infants (Escherich, 1886; Bettelheim, 1986; Shulman et al., 2007). Escherich’s observations have been strengthened by the evidence that early life microbiota plays an important role in shaping the immune system (Gomez de Agüero et al., 2016; Korpela and de Vos, 2018), and in decreasing risks in developing allergic diseases (Koenig et al., 2011; Arrieta et al., 2014; Gensollen et al., 2016), with the development of modern sequencing technology and metagenomics today. A recent study in infant gut microbiome indicated there are dynamic changes in early life microbiome during the first year after birth, and the newborns’ gut microbiome gradually develops and resembles adult-like microbiomes with Firmicutes and Bacteroidetes as the most abundant taxa, followed by Actinobacteria and Proteobacteria (Bäckhed et al., 2015).
As many studies already indicate that pioneer microbiome in early life is crucial in Deuterostomia, an increasing number of studies focus on the influence of pioneer microbes on host biology which could then be used to benefit agriculture and aquaculture. Pioneer microbes’ colonization in bird chicks showed an impact on the day of hatching through alteration of intestinal proteome which affects gastrointestinal tract immunity and cellular development (Wilson et al., 2019). The pioneer gut microbiota of tilapia larvae varied between individuals in different rearing environments (Giatsis et al., 2014). Although many previous studies have suggested the importance of early life microbiota in host biology, the structure and function of pioneer microbiota in marine Deuterostomia invertebrate has not been fully elucidated yet.
Sea cucumber (Echinodermata, Holothuroidea) is a marine invertebrate within Deuterostomia linage, and is a worldwide popular fishery and aquaculture resource, especially in Asian regions, due to its nutritive and medicinal value. In Asian region, sea cucumber has high commercial value and 200,000 tons a year in China and 6,500 tons a year in Japan are produced, which show that Apostichopus japonicus Selenka is one of the most consumed species (Ministry of Agriculture and Rural Affairs Fishery Administration, 2003-2012; MAFF, 2020). However, the fisheries of wild A. japonicus have declined worldwide and the animal has been placed on the red list of endangered species. Wild A. japonicus in China had already been reported as being overfished in the 1950s (Zhang and Liu, 1984; Hamel and Mercier, 2013). The huge demands for sea cucumber in recent decades has resulted in overfishing and over-exploitation in more than 70% of regions across the world (González-Wangüemert et al., 2018). Many studies have recommended regulation and management of sea cucumber fisheries and to develop restocking programs of overfished species in various regions including Brazil, Mexico, the Philippines, the Mediterranean, and Northeast Atlantic (González-Wangüemert et al., 2018; Jontila et al., 2018; Rogers et al., 2018; Gamboa-Álvarez et al., 2020). Due to the growing commercial demand for sea cucumber and decreasing wild stocks, aquaculture of sea cucumber has emerged and developed. A. japonicus is one of the most popular species in sea cucumber aquaculture, and artificial breeding was first attempted in 1937 and total output reached 170,830 tons in China, 2012 and 6,611 tons in Japan, 2019 (Inaba, 1937; Ministry of Agriculture and Rural Affairs Fishery Administration, 2003-2012).
In order to develop sea cucumber aquaculture, larval development of sea cucumber have been intensively studied (Chen and Chian, 1990; Sewell and McEuen, 2002; Ramofafia et al., 2003; Hu et al., 2010; Soliman et al., 2013). There are six major developmental stages in the early life of A. japonicus after the egg is fertilized; blastula, gastrula, auricularia, doliolaria, pentactula, and juvenile stages (Soliman et al., 2013; Supplementary Figure 1). Fundamental organs (buccal cavity, esophagus, intestine, cloaca, ciliary bands) appear in the auricularia stage and mature in the juvenile stage within 17 days on average after fertilization under optimal conditions. Primary tentacles and primary podium are used for attaching to habitat in the pentactula stage and then fully develop to tentacles and foot tube in the juvenile stage. Although some studies indicate food, culture conditions and bacterial control may relate to growth and survival rates of sea cucumber larvae, no certain links have been confirmed (Ramofafia et al., 2003; Hu et al., 2010).
Sea cucumber microbiome has been vigorously studied in recent decades in the aspects of diseases and host physiology. Rotting-edges syndrome caused by Vibrio lentus (Zhang et al., 2010) and stomach ulcer disease caused by Vibrio splendidus (Wang et al., 2006) occur in the auricularia to doliolaria stage. Off-plate syndrome is a serious disease caused by several bacteria that rapidly spreads and has a high mortality rate during the pentactula stage (Zhang et al., 2009). Probiotics have also been studied as potentially improving growth and stimulating the immune system in juvenile sea cucumbers (Ma et al., 2019). Intestinal microbial community composition altered by dietary supplementation benefits the growth of sea cucumber (Yang et al., 2015b). Rhodobacterales retaining PHB metabolism genes may play a key role in cultured sea cucumber growth (Yamazaki et al., 2016). Molecular ecological network analysis also indicates the stability of the intestinal community ecosystem is improved by probiotics and florfenicol has a positive impact on sea cucumber growth (Yang et al., 2017). All these studies suggest that microbiome in sea cucumber larvae development might be a crucial factor in developing seed production for sustainable aquaculture, however, there is a lack of knowledge of the structure and function of pioneer microbes of sea cucumber larvae and the dynamic changes during the larval development.
In this study, we set up a laboratory rearing system and performed Meta16S analysis in order to assess the changes to the microbiome during the larval development of sea cucumber and to detect the pioneer microbiota in the early developmental stages. Our results demonstrate that detection of pioneer microbiome in A. japonicus and stepwise changed microbiota that significantly related to organogenesis and larval development.
Fertilized eggs of the sea cucumber A. japonicus were prepared at 18.7°C at a farm in Hokkaido Aquaculture Promotion Corporation Kumaishi Branch, Japan (42.12574, 139.99966) on 11:00 am 5 August 2019. These eggs were transferred to the laboratory of Microbiology, Faculty of Fisheries Sciences, Hokkaido University whilst being kept at 18°C for 2 h, and then used immediately for experiments. Density of the fertilized eggs was set at 7,500 eggs/L in an 8 L volume aquarium after manual counting of these eggs in 0.1 mL seawater using a microscope (AxioImager Z2, Zeiss, Oberkochen, Germany) and reared at 18°C. The aquarium was prepared using a sterilized 8 L glass bottle (Ishizuka glass Co. Ltd., Aichi, Japan) set in an incubator (MLR-352-PJ, PHC Corp., Tokyo, Japan). Each bottle was filled with 7.5 L natural filtrated seawater using a 50 μm mesh cartridge filter (SWP50P10, AS One, Osaka, Tokyo) used in the Kumaishi farm. Each 80 mL (7,500 eggs) of fertilized egg suspensions, corresponding to the final egg density of around 1 egg/mL, was added to each bottle, and rearing was started with aeration (SPP-25GA, Techno Takatsuki Co., Ltd., Osaka, Japan). Feeding started at 48 h after fertilization (on 7 August 2020), when early auricularia morphogenesis was observed in over 80% of individuals. A commercially available diatom, Chaetoceros gracilis (Hakodate Fisheries Research, Japan), was fed to the sea cucumber larvae daily.
Sea cucumber larvae at five major developmental stages, gastrula, early and late auricularia, doliolaria, and pentactula after being confirmed by a microscopic observation (Supplementary Figure 1) were used for characterization of microbiomes and microbial isolations. Five liters of rearing seawater including sea cucumber larvae were filtered using a sterilized 40 μm nylon mesh (Falcon Cell Strainer, Durham, NC, United States), and washed once using 0.22 μm filter-sterilized seawater. Sterivex filter (SterivexTM-GV Sterile Vented Filter Unit 0.22 μm, EMD Millipore, Billerica, MA, United States) was used to prepare this sterilized seawater. The larvae on nylon filters were immediately frozen at −80°C until DNA extraction.
To prepare microbial fractions in rearing water, five liters of rearing water after passing through nylon mesh was filtered through a 0.22 μm Sterivex filter by positive pressure using filtered (0.22 μm) N2 gas. These Sterivex filters were preserved at −80°C until DNA extraction.
Microbial DNA extraction from sea cucumber was performed using the NucleoSpin Soil Kit (MACHEREY-NAGEL, Düren, Germany), according to the manufacturer’s protocol. Microbial DNA extraction from seawater was performed using the NucleoSpin Tissue kit (MACHEREY-NAGEL), according to the modified manufacturer’s protocol. In brief, seawater samples were heated at 55°C for 1 h to add an active cell lysis process in TE buffer (10 mM Tris–HCl, 1 mM EDTA) containing 20% SDS and proteinase K (20 mg mL–1) instead of buffer T1. In the third step Lyse Sample, 1 mL buffer B3 was used instead of 200 μL amount of the buffer.
The hypervariable V1-V2 region of the 16S rRNA gene was amplified by PCR with barcoded 27Fmod and 338R primers with Illumina adaptor sequences (Yamazaki et al., 2019). PCR amplicons were purified using AMPure XP magnetic purification beads (Beckman Coulter, Brea, CA, United States), and quantified using the Quant-iT PicoGreen dsDNA Assay Kit (Life Technologies Japan). Equal amount of each PCR amplicon was mixed and then sequenced using MiSeq Reagent Kit v3 (600-cycles) with the MiSeq Illumina platform. Based on sample specific barcodes, obtained reads were assigned to each sample.
The paired-end sequence data with quality scores (i.e., Fastq files) was analyzed using Quantitative Insights Into Microbial Ecology 2 (QIIME 2, version 2018.11) (Bolyen et al., 2019). Quality controls (e.g., trimming primers and denoising sequences, removing chimeric sequences) and merging paired-end sequences were performed using DADA2 (Callahan et al., 2016). Reads with 100% similarity constituted an amplicon sequence variance (ASV). Unlike the method to cluster sequences into operational taxonomic units (OTUs) with fixed threshold (usually 97%), this quality control method using DADA2 allows us to detect even a single nucleotide difference. ASVs were assigned to taxonomy using the Naive Bayes classifier and Greengenes database. Using subsampled reads, unweighted UniFrac distances as beta-diversity were calculated and visualized in prinicipal coordinate analysis (PCoA) plots (Lozupone et al., 2011). Significant differences of unweighted UniFrac distance were tested by permutational multivariate analysis of variance (PERMANOVA) (FDR-corrected p < 0.05). The phylogenic tree was generated by FastTree (Price et al., 2009). Z-score was calculated by genefilter package in R (Gentleman et al., 2021). R studio software (Version 1.2.5019) was used for heatmaps construction, Z-score calculation by genefilter package (Gentleman et al., 2021) and statistical significance of t-test to pick up the key ASVs.
Sea cucumber larvae at each life stage were also used for microbial isolations. Larvae were filtered with 40 μm nylon mesh (Falcon Cell Strainer, Durham, NC, United States), washed once with sterilized seawater, and then homogenized in 1 mL filter-sterilized natural seawater for 60 s manually. A 10-fold serial dilution of these homogenates was prepared using a filter-sterilized natural seawater, and then the dilutions were cultured on 1/5 strength ZoBell 2216E agar plate (0.1% polypeptone, 0.02% yeast extract, 1.5% agar, 75% natural seawater collected at Kumaishi farm) at 18°C. After calculating viable bacterial counts, ca. 30 bacterial colonies per plate were purified using the same agar plate.
The16S rRNA gene sequences of each isolate was amplified by colony PCR using GoTaq Green Mater Mix (Promega, Madison, WI, United States), 27F primer (20 pmol), 1492R primer (20 pmol) with the following thermal profile; initial denaturation at 96°C for 3 min, followed by 30 cycles of denaturation at 95°C for 1 min, annealing at 50°C for 1 min and extension at 72°C for 2 min, and final extension at 72°C for 7 min. PCR products of ∼1500 bp were visualized by electrophoresis with 1% agarose gels. PCR products were purified using a Wizard SV Gel and PCR clean-up system (Promega). PCR amplicons were directly sequenced by Hokkaido System Science (Sapporo, Japan). Sequences were assembled using ChromasPro (Version 2.1.8, Technelysium Pty. Ltd., Australia). Almost complete sequences of 16S rRNA gene of 257 isolates were obtained in this study. In order to determine the phylogenetic position of isolates, the sequences were aligned using Clustal X (Version 2.1) (Larkin et al., 2007) with the top five most similar sequences indicated by megaBLAST and sequences of type strains retrieved from RDP release 11 as reference sequences (Cole et al., 2014). The phylogenetic tree was reconstructed using Maximum-Likelihood algorism with MEGA X (Version 10.1.7) after performing the best model selection, and was further edited by an online tool Interactive Tree of Life (Letunic and Bork, 2019).
Integration of Meta16S sequences and 16S rRNA gene sequences of isolates were performed using QIIME2. Identification and taxonomic analysis of representative were also performed during the comparison. Dominate ASVs were identified and isolated and after combination with identification of isolated strains sequenced by 16S rRNA sequencing, taxonomic analysis reach to both gene and species level.
A total of 974,753 Meta16S sequence reads were obtained from sea cucumber and seawater samples collected from fertilized eggs (FE), gastrula (GL), early auricularia (EA), late auricularia (LA), pentactula (PT), and juveniles (JN) (Supplementary Table 1). Reads passed quality control and removed eukaryotic reads (e.g., mitochondria and chloroplast) were used for microbial diversity analyses and taxonomic assignments; 106,713 and 134,940 qualified reads were generated from sea cucumber samples and seawater samples, respectively (Supplementary Table 1).
Unweighted UniFrac distance analysis revealed the dynamic changes of microbiotas associated with host development. The 2D PCoA plot based on unweighted UniFrac analysis obtained sea cucumber larvae and rearing seawater samples at each developmental stage indicates that (1) microbiotas between seawater and sea cucumber were not grouped, (2) microbiota between FE, GL, and EA juveniles were grouped but FE, GL, and EA seawater microbiotas were not, and (3) microbiota between PT and JN sea cucumber larvae were grouped and LA, PT, and JN seawater microbiota were grouped (Figure 1). Microbial community of sea cucumber samples according to the PCoA plot reveals a large variation between early and late auricularia stages, the time point before and after gut developed. The UniFrac distance matrix also showed similar tendency in microbiota grouping between sea cucumber and seawater (Figures 2B,C). A decreased alpha diversity based on Shannon index and increased beta diversity was observed after gut development (Figures 2A,D), indicating a less complex but more dissimilar microbial community after the digestive system had developed.
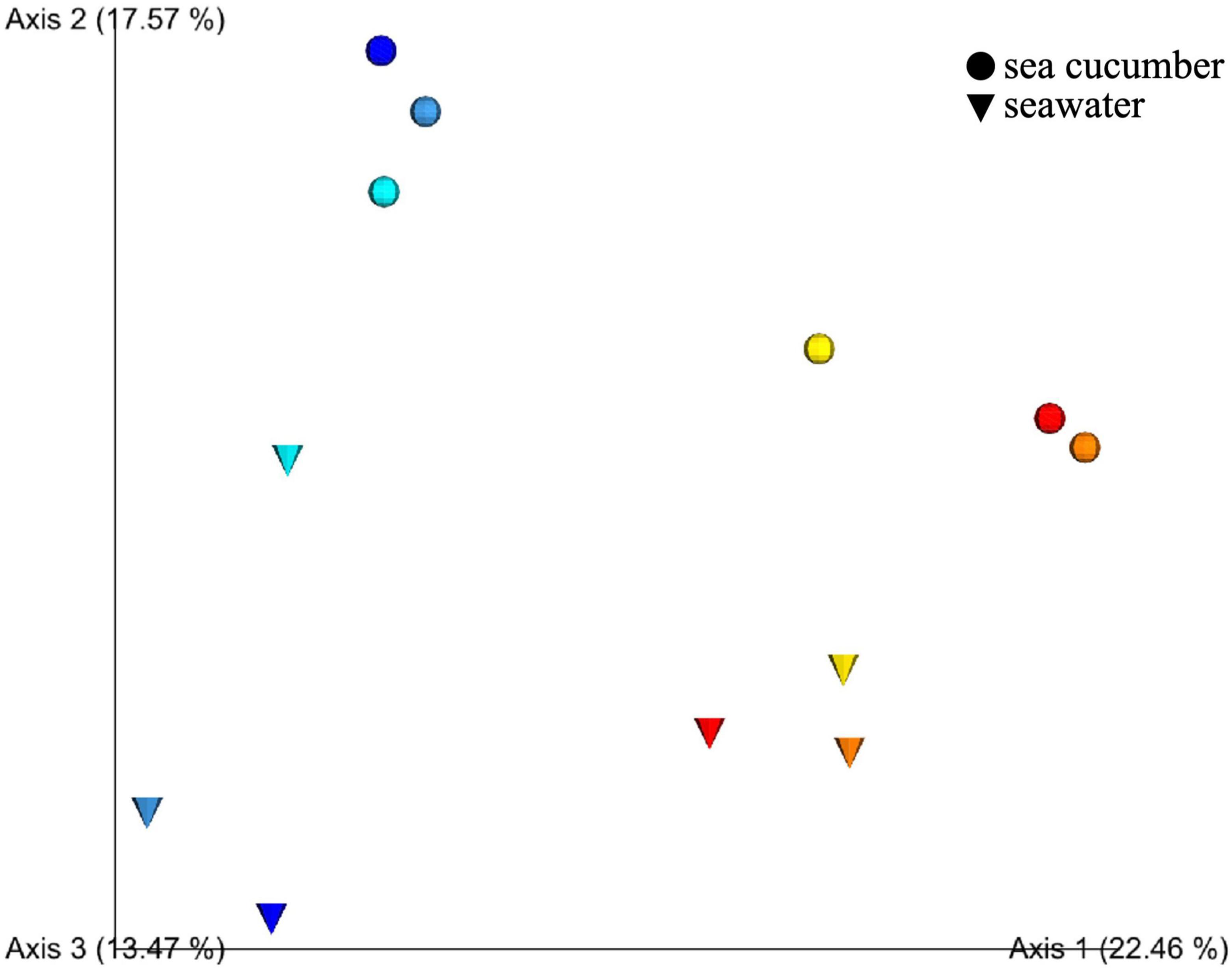
Figure 1. Prinicipal coordinate analysis (PCoA) plot based on unweighted UniFrac distances obtained by comparison of microbiotas at each developmental stage of sea cucumber larva. Blue, fertilized egg; light blue, gastrula; cyan, early auricularia; yellow, late auricularia group; orange, pentactula group; red, juvenile. Circle and cone indicated microbiota from sea cucumber and rearing seawater, respectively.
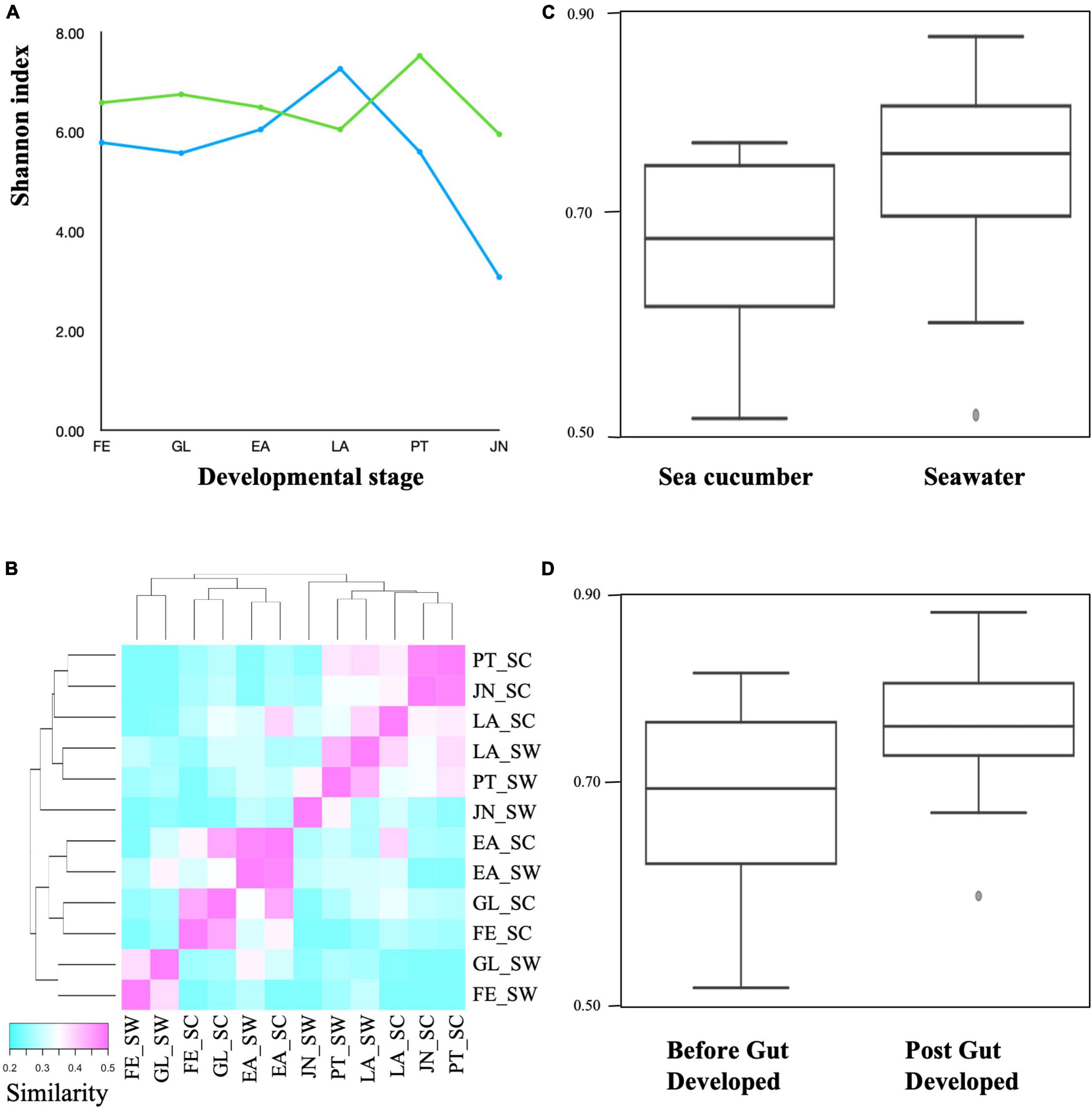
Figure 2. Alpha and beta diversity among sea cucumber and seawater samples. (A) Shannon index. Green, sea cucumber samples; blue, seawater samples. (B) Heatmap based on unweighted UniFrac distance (p < 0.05, q < 0.05). Scale represent similarity within samples. FESC, Fertilized egg; GLSC, Gastrula; EASC, Early auricularia; LASC, Late auricularia; PTSC, Pentactula; JNSC, Juvenile; FESW, Fertilized egg rearing seawater; GLSW, Gastrula rearing seawater; EASW, Early auricularia rearing seawater; LASW, Late auricularia rearing seawater; PTSW, Pentactula rearing seawater; JNSW, Juvenile rearing seawater. (C) Unweighted Unifrac distance plot between sea cucumber and seawater (p < 0.05, q < 0.05). (D) Unweighted Unifrac distance plot between samples before and post-gut developed (p < 0.05, q < 0.05).
A total of 1,440 ASVs were obtained and further affiliated to 75 bacterial orders using a similarity threshold of 99% sequence identity; 144, 219, 157, 120, 327, and 254 ASVs, and 29, 30, 19, 28, 39, and 35 bacterial orders, were affiliated in FE, GL, EA, LA, PT, and JN stage, respectively (Supplementary Table 2). Oceanospirillales and Alteromonadales were the most abundant in FE, GL, and EA stages with relative abundance ranging from 24.1 to 29.9% and 32.6 to 46.2%, respectively (Figure 3 and Supplementary Table 2). However, abundances of Oceanospirillales dramatically decreased to 3.2% in the LA stage and gradually increased in the PT (9.8%) and JN (49.1%) stages. Such a fluctuation in abundance was also observed in Alteromonadales, 32.5–46.2% in the FE to EA stages, but those decreased after EA (14.8, 10.6, and 6.3% in LA, PT, and JN stage, respectively) (Figure 3 and Supplementary Table 2). Vibrionales, Pseudomonadales, and Rhodobacterales were the third most abundant taxa in FE, GL, and EA stages before gut organogenesis. Vibrionales was detected in FE (7.2%), GL (13.6%), and EA (12.1%) stages than LA (4.7%), PT (1.0%), and JN (0.3%). Likewise, Pseudomonadales was at FE (7.7%), but relative lower amounts in GL (4.6%), EA (2.4%), LA (5.0%), PT (1.0%), and JN (0.3%). In contrast, Rhodobacterales was observed in LA (26.0%), PT (37.0%), and JN (22.8%) with higher fractions than in the early three stages in FE (4.9%), GL (7.0%), and EA (9.8%). Relative abundance of Flavobacteriales was much lower in early stages of FE (3.6%), GL (2.1%), and EA (1.5%) but increased in LA (25.5%), PT (14.2%), and JN (3.4%).
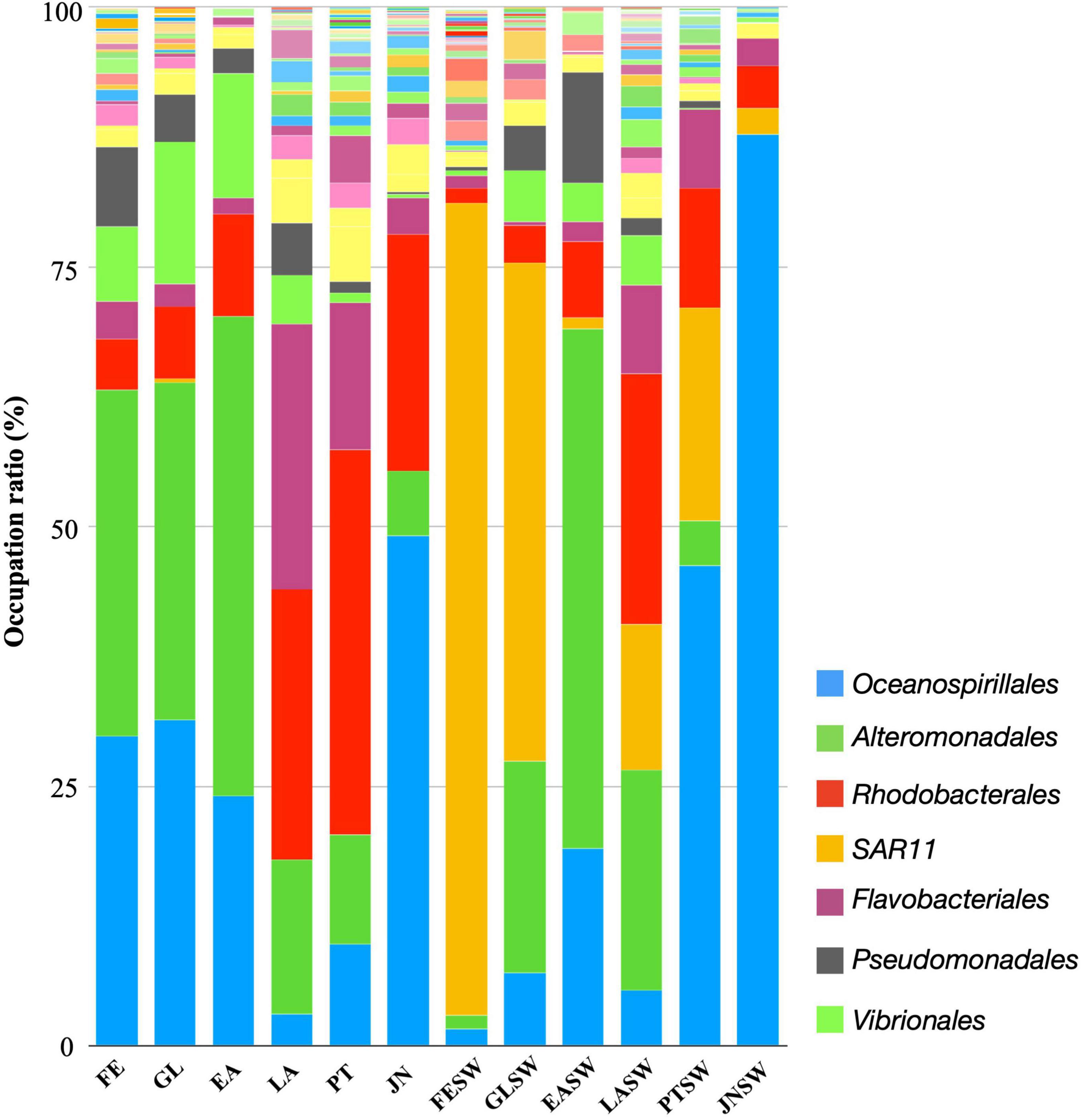
Figure 3. Order-level taxonomic distribution among sea cucumber and seawater samples. Bars represent the relative percentage of each bacterial orders. FE, Fertilized egg; GL, Gastrula; EA, Early auricularia; LA, Late auricularia; PT, Pentactula; JN, Juvenile; FESW, Fertilized egg rearing seawater; GLSW, Gastrula rearing seawater; EASW, Early auricularia rearing seawater; LASW, Late auricularia rearing seawater; PTSW, Pentactula rearing seawater; JNSW, Juvenile rearing seawater.
On the other hand, the microbial community in seawater is significantly different from those associated with sea cucumber hosts (Figure 2B). A total of 232, 84, 139, 277, 163, and 69 features were detected in fertilized egg rearing seawater (FESW), gastrula seawater (GLSW), early auricularia seawater (EASW), late auricularia seawater (LASW), pentactula seawater (PTSW), and juvenile seawater (JNSW), respectively (Figures 3, 4). Alteromonadales and Oceanospirillales, which were dominant at early three stages of sea cucumber samples, were rarely detected at FESW (1.3%, 1.7%), but significantly increased to become the second and third most abundant at GLSW (20.3%, 7.1%), followed by rising to the first and second most abundant at EASW (50.1%, 19.0%). Similar to sea cucumber samples, the abundance of Oceanospirillales also dropped in LASW (5.3%) but increased in PTSW (46.2%) and appeared to be present in JNSW (87.7%). The abundance of Alteromonadales in LASW (21.3%) decreased below a detectable limit (0.1%) in JNSW. Vibrionales and Pseudomonadales were also abundant in GLSW (5.0%, 4.3%) and EASW (3.7%, 10.7%), but relatively low in later stage PTSW (0.2%, 0.7%) and undetectable in JNSW. Rhodobacterales increased in later stages LASW and PTSW but limited at FE, GL and EA stage, during the host development progress. Different from the host bacterial community, Flavobacteriales was rarely detected in seawater samples, ranging from 0.3 to 8.5%. SAR11 clade, belonging to class Alphaproteobacteria, was observed in all samples of seawater, in particular, rich in FE and GL seawaters with 78.2 and 48.0% frequency, but low relative abundance in sea cucumbers (0.3% in GL) (Figure 3 and Supplementary Table 2).
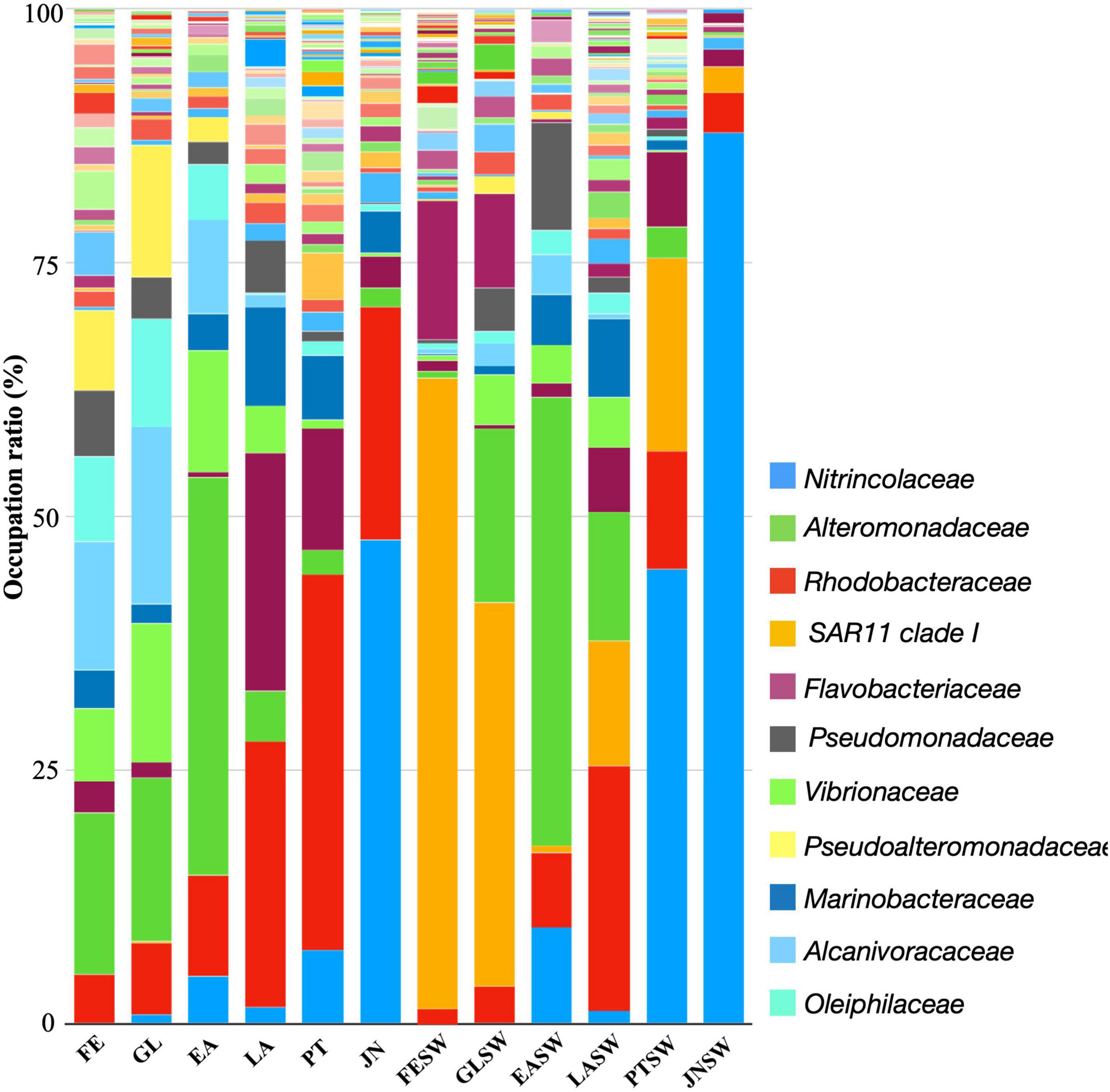
Figure 4. Family-level taxonomic distribution among sea cucumber and seawater samples. Bars represent the relative percentage of each bacterial orders. FE, Fertilized Egg; GL, Gastrula; EA, Early Auricularia; LA, Late Auricularia; PT, Pentactula; JN, Juvenile; FESW, Kuma-shi seawater; GLSW, Gastrula rearing seawater; EASW, Early Auricularia rearing seawater; LASW, Late Auricularia rearing seawater; PTSW, Pentactula rearing seawater; JNSW, Juvenile rearing seawater.
A further family level taxonomic distribution and heatmap based on relative abundance of top 50 abundant bacterial group revealed that (1) bacterial groups such as Alteromonadaceae (Alteromonadales), Alcanivoracaceae (Oceanospirillales), Pseudoalteromonadaceae (Alteromonadales), Oleiphilaceae (Oceanospirillales) first appeared and were abundant at FE stage, (2) Rhodobacteriaceae (Rhodobacterales), Flavobacteriaceae (Flavobacteriales), and Marinobacteraceae (Alteromonadales) increased from LA stage, and (3) Colwelliaceae (Alteromonadales) and Halieaceae (Cellvibrionales) increased at PT stage (Figures 4, 5). Moreover, JN represents a unique microbiota compared to other life stages; Nitrincolaceae (Oceanospirilales), the population showed low levels in other life stages (0–7.3%), was significantly dominant in PT with 47.8% relative abundance. Rhodobacteriaceae (Rhodobacterales) was still the second most abundant family in JN, which appeared to be the most abundant family from the LA stage (22.8–37.0%) (Figures 4, 5).
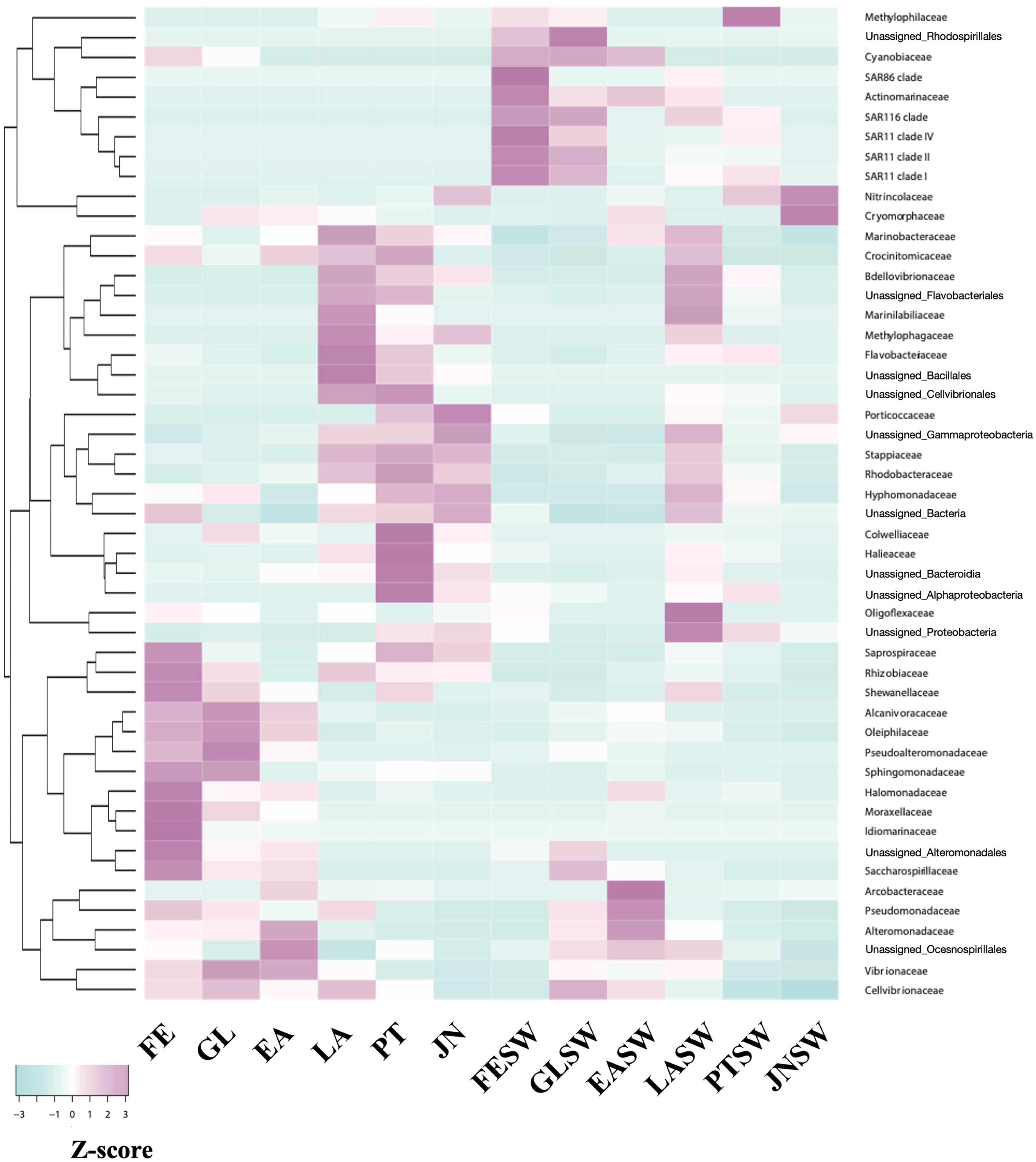
Figure 5. Dynamics of signature families at each stage. Scale represents relative abundance after Z-score calculation. FE, Fertilized egg; GL, Gastrula; EA, Early auricularia; LA, Late auricularia; PT, Pentactula; JN, Juvenile; FESW, Fertilized egg rearing seawater; GLSW, Gastrula rearing seawater; EASW, Early auricularia rearing seawater; LASW, Late auricularia rearing seawater; PTSW, Pentactula rearing seawater; JNSW, Juvenile rearing seawater.
Dynamics profiling based on ASVs containing more than 500 reads revealed that 14 key ASVs significantly changed at different developmental stages in sea cucumber larvae (Figures 6, 7 and Supplementary Figure 2); these were assigned into 5 bacterial families, 4, 2, 4, 2, and 2 ASVs were assigned to Nitrincolaceae (Oceanospirillales), Alcanivoracaceae (Oceanospirillales), Rhodobacteriaceae (Rhodobacterales), Flavobacteriaceae (Flavobacteriales), and Alteromonadaceae (Alteromonadales). The ASVs assigned to Nitrincolaceae and Rhodobacteraceae showed different variation trends with corresponding seawater, but the relative abundance of them changed at specific developmental stages in sea cucumber larvae (Figure 8). In addition, 2 ASVs of ASV0013 and ASV0051 belonging to Alteromonadaceae and Cellvibrionaceae, respectively, were defined as core microbiome that present in sea cucumber’s microbiome during whole developmental process, which also detected in rearing seawater (Supplementary Figure 3).
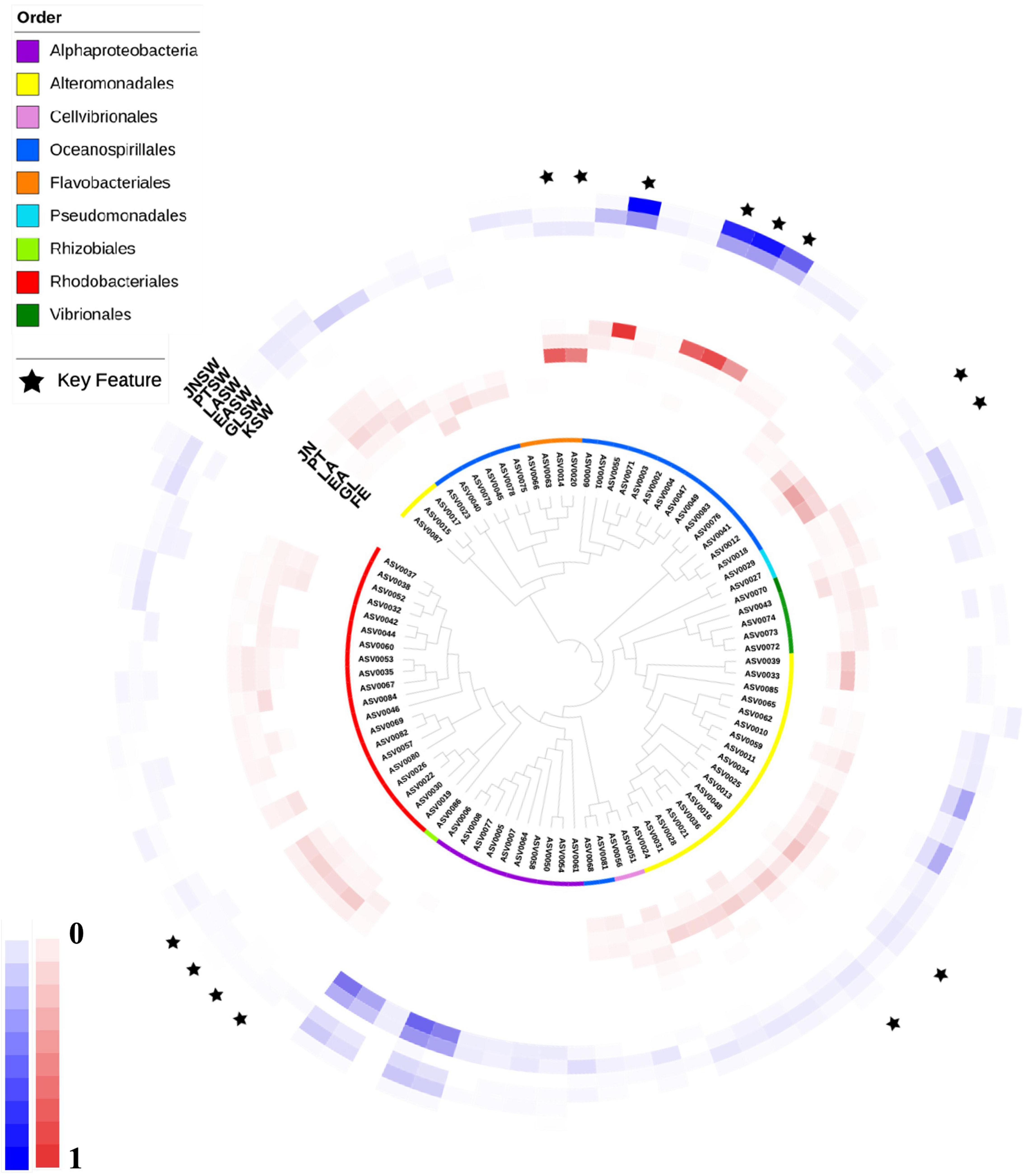
Figure 6. A phylogenetic tree based on features having >500 reads with relative abundance. Inner circles represent taxonomic analysis at order lever. Red colored heatmap represent relative abundance of features in sea cucumber samples. Blue colored heatmap showed relative abundance of features in seawater samples. Key features were labeled with black stars. FE, Fertilized egg; GL, Gastrula; EA, Early auricularia; LA, Late auricularia; PT, Pentactula; JN, Juvenile; FESW, Fertilized egg rearing seawater; GLSW, Gastrula rearing seawater; EASW, Early auricularia rearing seawater; LASW, Late auricularia rearing seawater; PTSW, Pentactula rearing seawater; JNSW, Juvenile rearing seawater.
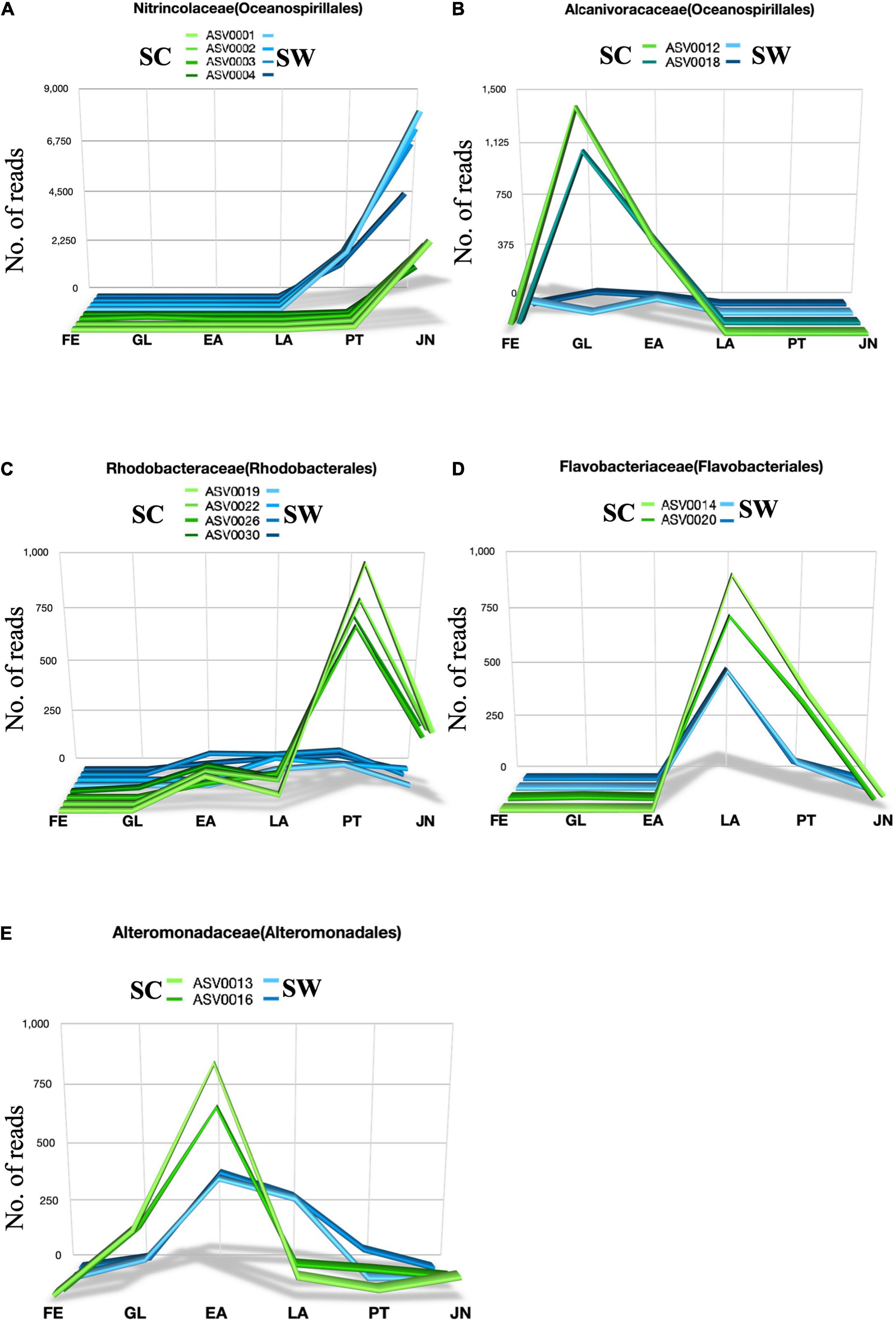
Figure 7. Dynamics of relative abundance of key ASVs based on heatmap. 14 key ASVs were shown in panels (A–E), respectively. X axis is developmental stage, and Y axis is number of reads. FE, Fertilized egg group; GL, Gastrula group; EA, Early auricularia group; LA, Late auricularia group; PT, Pentactula group; JN, Juvenile group.
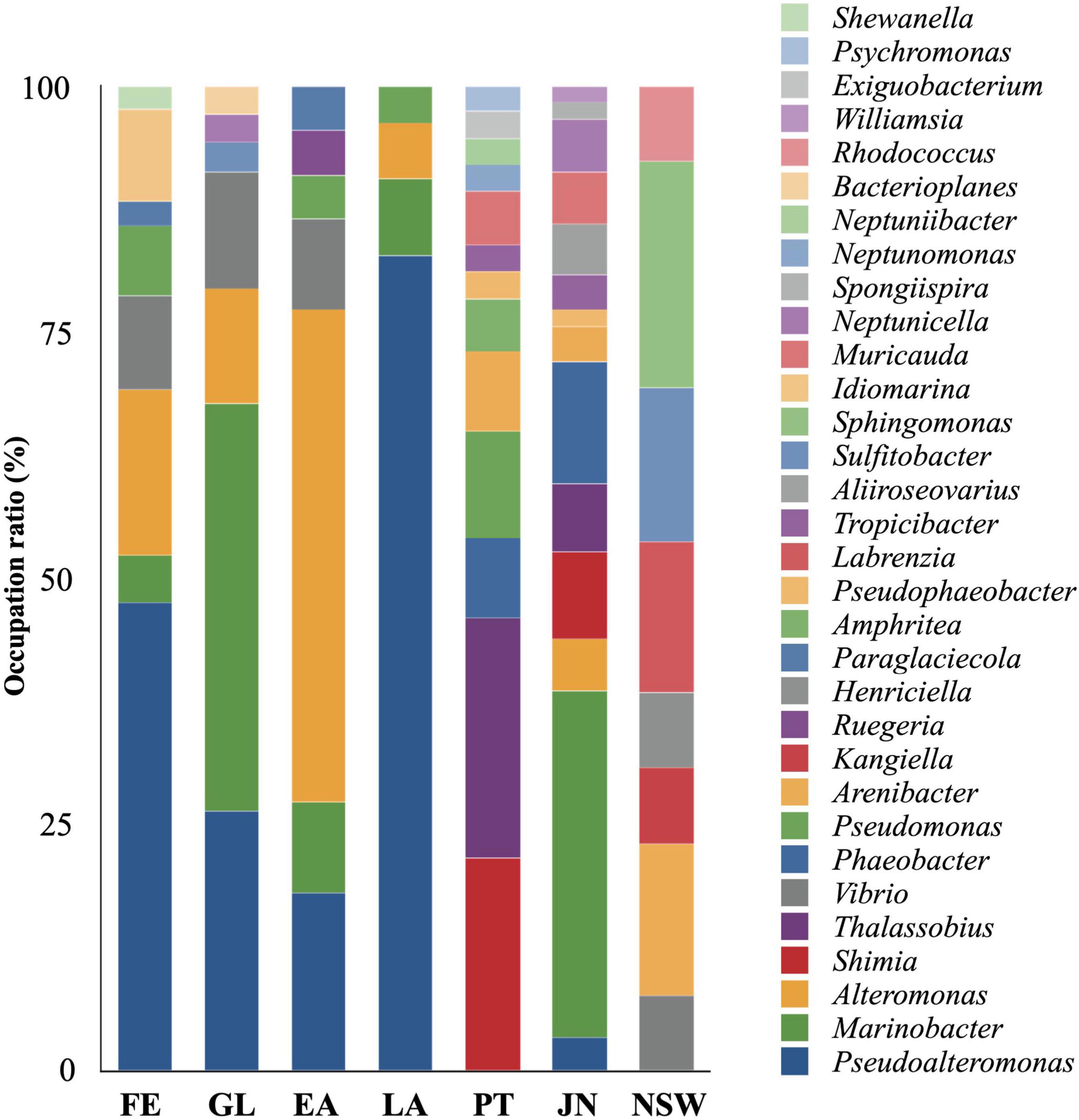
Figure 8. Genus-level taxonomic distribution of isolates from sea cucumber and seawater. Bars represent proportion of isolates. FE, Fertilized egg; GL, Gastrula; EA, Early auricularia; LA, Late auricularia; PT, Pentactula; JN, Juvenile; NSW, Kumaishi nature seawater.
A total of 257 strains were isolated from sea cucumber specimens and assigned to 45 species belonging to 32 genera, in which 89.1% of isolates were assigned to known species and 100% of isolates were assigned to known genera (Figures 8, 9 and Supplementary Table 4). A total of 42, 34, 22, 53, 37, 57, and 13 isolates were isolated from FE, GL, EA, LA, PT, JN, and fertilized egg rearing seawater (NSW), respectively, being counted up 102 to 104 CFU/g sample (Supplementary Table 4). A total of 20 of 42 isolates collected from FE specimen were identified as Pseudoalteromonas belonging to Alteromonadales, followed by Alteromonas (n = 7), Vibrio (n = 4), Idiomarina (n = 4), Pseudomonas (n = 3), Marinobacter (n = 2), Paraglaciecola (n = 1), and Shewanella (n = 1). Marinobacter (n = 14) also belonged to Alteromonadales was the most abundant genera in isolates collected from GL specimen, followed by Pseudoalteromonas (n = 9), Alteromonas (n = 4), Vibrio (n = 4), Neptunicella (n = 1), Sulfitobacter (n = 1), and Bacterioplanes (n = 1). The most abundant genera in EA isolates were Alteromonas (n = 11) and Pseudoalteromonas (n = 4), both of them belonging to Alteromonadales, subsequently Marinebacter (n = 2), Vibrio (n = 2), and Pseudomonas (n = 1) were also obtained in EA. In LA isolates, 43 of 53 isolates were assigned to Pseudoalteromonas, which is the significant dominant genera compared to other genus Marinobacter (n = 4), Alteromonas (n = 3), and Pseudomonas (n = 2).
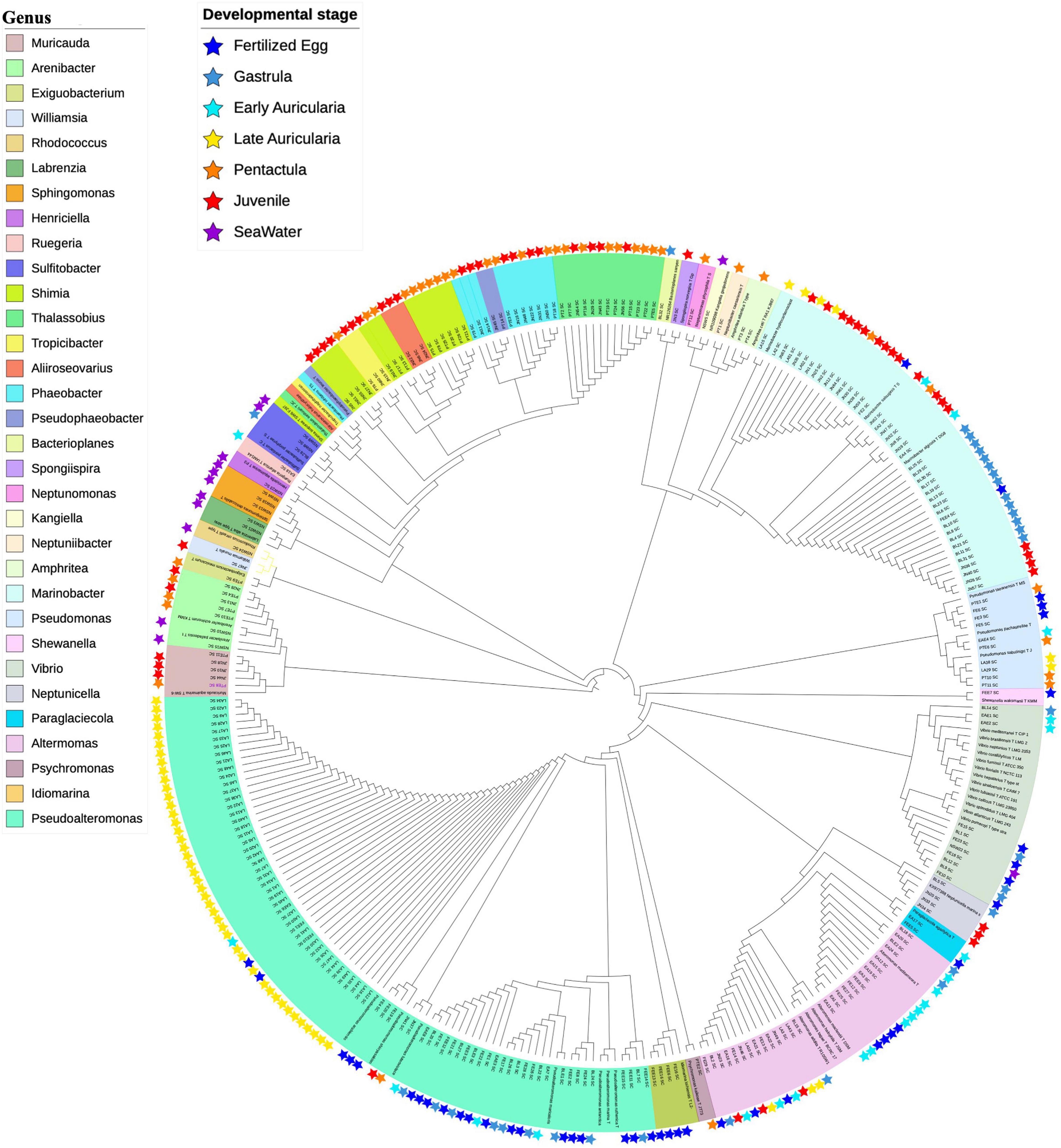
Figure 9. Maximum-Likelihood phylogenetic tree based on partial 16S rRNA gene sequences of isolates. Isolates were shown with stars in specific color. Different genera were labeled with different color. 257 isolates were assigned to 32 genera and 45 species.
The composition of isolates showed a variation between LA and PT stages, which Pseudoalteromonas, Marinobacter, and Alteromonas belonging to the order Alteromonadales were the most abundant genera in FE to LA but absent in PT (Figure 9). In addition, taxonomic diversity of isolates was richer in later stages such as PT and JN compared to earlier life stages. Likewise, Thalassobius (n = 9) and Shimia (n = 8) belonging to the order Rhodobacterales were the most abundant genera in PT, followed by Pseudomonas (n = 3), Arenibacter (n = 3), Phaeobacter (n = 3), Muricauda (n = 2), Amphritea (n = 2), and other 6 isolates assigned to 6 different genera. On the other hand, Rhodobacterales and Alteromonadales showed a higher possibility to be isolated in JN samples; a total of 20 of 57 isolates in JN were assigned to Marinobacter belonging to Alteromonadales; 7, 5, and 4 isolates were assigned to Phaeobacter, Shimia, and Thalassobius belonging to Rhodobacterales, respectively. The other isolates in JN were assigned to genera Muricauda (n = 3), Alteromonas (n = 3), Neptunicella (n = 3), Aliiroseovarius (n = 3), Arenibacter (n = 2), Tropicibacter (n = 2), Pseudoalteromonas (n = 2), Pseudophaeobacter (n = 1), Williamsia (n = 1), and Spongiispira (n = 1). Isolates from NSW were relatively unique compared to the sea cucumber samples. Genera Sphingomonas, Sulfitobacter, Labrenzia, Rhodococcus, Kangiella, and Henticiella were only isolated from NSW samples, which demonstrates a different microbial community structure.
Comparison of isolates with ASVs in Meta16S revealed a total of 124 ASVs were assigned to those sequences of isolates with 100% identity, including 51 dominant ASVs (>0.1% abundance) (Supplementary Table 3). In addition, 6 out of 14 key ASVs found in the Meta16S sequences were successfully isolated. ASV0013 and ASV0016 belonging to Alteromonadaceae, the relative abundance of these two key ASVs were 0.0090 and 0.0082, respectively. They showed a significant increase at the EA stage with the relative abundance of 0.046 and 0.037, assigning to 10 isolated strains from GL, EA, LA, and JN samples and identified as genus Alteromonas. One of them ASV0016 was absent at FE and then slightly increased in the gastrula stage. ASV0019, ASV0022, ASV0026, and ASV0030 belonging to Rhodobacteriaceae, the relative abundance of them were 0.0072, 0.0070, 0.0063, and 0.0060, respectively. They showed a significant increase at the PT stage with relative abundance of 0.033–0.040%, assigning to 13 isolated strains collected from JN and PT, which appeared from the early auricularia stage (Supplementary Table 3). Interestingly, these five key ASVs were not detected in the fertilized egg rearing seawater. In addition, although 75.8% sequences in Meta16S analysis were unassigned to known species or genera, the taxonomic level of 81 ASVs were improved to species or genus level after taxonomic analysis with isolated strains, in which one unassigned ASV1407 was affiliated to strains NSW5 identified as Kangiella geojedonens and 10 uncultured ASVs were assigned to known species (Supplementary Table 3).
Microbiotas of sea cucumber A. japonicus has been intensively studied over recent decades aiming to discover beneficial bacteria that could contribute to sea cucumber aquaculture, disease control, and bioremediation (Chi et al., 2014; Yang et al., 2015b; Zhao et al., 2016; Chen et al., 2018; Ma et al., 2019). After the recent findings of potential physiological roles in gut microbiomes of the sea cucumber A. japonicus (Yamazaki et al., 2016), dynamics of the microbiome, in particular How sea cucumbers shape their microbiomes? has been a central question (Yamazaki et al., 2020). Among a series of studies on the sea cucumber microbiome, first colonizers on the animal guts called Pioneer Microbes are worth studying in sea cucumbers as they potentially contribute to improving aquaculture technology and accumulate biological knowledge on gut microbiome evolution compared to those in humans (Wopereis et al., 2014). Using a laboratory rearing system, we succeeded in conducting Meta16S sequencing to characterize microbiotas of fertilized eggs at major developmental stages up to juvenile and key bacteria were successfully isolated. To our knowledge, this is the first study to investigate the dynamics of microbiota during the larval development of sea cucumber and the possible relationship between early life microbiota and host biology in marine invertebrates.
The pioneer microbiome in early life has been assessed mainly on human infant gut microbiome, and environmental factors, diets, developmental stage and genetics affecting microbiota shaping have been investigated (Nayak, 2010; Sullam et al., 2012; Mercier et al., 2015). Bifidobacterium and Lactobacillus, which are lactic acid utilizers, were prevalent in later infant microbiome (4–12 months) during breast feeding but decreased in adult-like microbiota when solid food was introduced (Bäckhed et al., 2015; Hill et al., 2017). Similar to human infants, sea cucumber larvae’s gut microbiota altered after feeding, which is probably linked to the changing of gut environment and its functional shifts. Many studies have also worked on pioneer microbiome using fish eggs and larvae to improve intensive aquaculture of larvae rearing (Vadstein et al., 2018), relationships to hatchability, and contribution to the formation of adult fish microbiota (Hansen and Olafsen, 1989, 1999; Califano et al., 2017). Whole-body microbiota of aquatic animals using fertilized eggs and post-hatch larvae of sea bream suggest that microbiotas of rearing water and diet affect the shaping of early life microbiota (Nikouli et al., 2019). The Meta16S analyses of sea cucumber during early developmental stages reveals several new findings on microbiotas changes related to host development and organogenesis; (1) significant changes of microbiotas in the late auricularia stage, (2) possible first colonizers in gut assigned to Rhodobacterales and Flavobacterales appeared from auricularia stage, (3) significantly different microbiota in juveniles, and (4) significantly different microbiotas between host associated and environmental water.
Such microbiotas changes are likely to be related to organogenesis of the digestive system. Blastopore is formed in blastula and observed until the early auricularia stage. As the primitive gut-like structure in gastrula disappears, the buccal cavity, esophagus, intestine, cloaca and ciliary bands related to digestion, feeding and locomotion can be observed from the early auricularia stage and are well developed in the late auriculaira. Stomach, axohydrocoel and hyaline sphere are also clearly observed in the late auriculaira. After the pentacutula stage, sea cucumbers start a benthic lifestyle with more organogenesis of tentacles and ossicales formation, which are necessary for feeding and habitat settlement in adult individuals. Early life microbiome could be dynamically changed during the organogenesis, which allow microbes to colonize first on the tissue of host sea cucumber as they shape to a matured form.
Oceanospirillales, Rhodobacteriales, Alteromonadales, and Flavobacterales colonize sea cucumber’s microbiota at different developmental stages and show significant changes during larval development. Many researchers have analyzed microbial communities and the impact of these individual bacteria on the host microbiome in sea cucumbers (Yang et al., 2015a; Yamazaki et al., 2016; Zhao et al., 2016; Kim et al., 2017; Li et al., 2018; Pagán-Jiménez et al., 2019; Ren et al., 2019; Yamazaki et al., 2019), but pioneer colonizers in fertilized eggs and larvae of sea cucumber have hardly been reported at all. Rhodobacterales, Flavobacterales, and Alteromonadales were also abundant in adult sea cucumber A. japonicus without similar microbial diversity (Yamazaki et al., 2016). Of particular interest in this study, Rhodobacterales was detected as the dominant bacterial order and key bacteria in microbiota in early life, which colonize in the early auricularia stage and significantly increase in the pentactula stage (Figures 3, 4). Rhodobacterales is known as one of the primary surface colonizers and key players of biogeochemical cycling in marine environments, consisting of more than 350 species (Dang et al., 2008; Simon et al., 2017). Complicated taxonomic subgroups and worldwide distribution of Rhodobacterales helped increase research into the understanding the community structures and molecular roles in Rhodobacterales (Fu et al., 2013; Rasmussen et al., 2018; Tavares et al., 2018). There are studies indicating Rhodobacterales could produce tropodithietic acid (TDA) against the pathogen Vibrio (Grotkjaer et al., 2016; Dittmann et al., 2020). In sea cucumber, Yamazaki et al. (2016) compared microbiota between larger and smaller individuals, in which Rhodobacterales was more abundant in larger sea cucumber, and it contains polyhydroxybutyrate (PHB) metabolism genes which could promote host growth (Yamazaki et al., 2016). Dietary β-glucan supplementation and dietary astragalus polysaccharide (APS) in sea cucumber A. japonicus have a positive impact on immune responses and enriched relative abundance of Rhodobacterales in microbiota through the NF-κB signaling pathway (Yang et al., 2015b; Song et al., 2019). Currently, 51 isolates are assigned to Rhodobacterales and 13 of the 51 isolates are assigned to key ASVs detected in Meta16S analysis. The success of Rhodobacterales isolation opens up a way for us to perform bioassay, meta-transcriptomic analysis, molecular studies on host-microbiome association in the future.
Culture environments and supplied diets are always considered as major factors resulting in alteration of microbiota, many studies reveal the relationship of microbiotas between animal gut and environments. In some studies, microbiota of eggs shows highest similarity of 77% to that of rearing water (Nikouli et al., 2019). However, a study on sea bream discusses the potential effects of rearing water and diet on microbiota of different days post-hatch larvae, and they indicated that both rearing water and diet differ from host microbiota but somehow contribute to microbial community (Nikouli et al., 2019). According to Hansen and Olafsen (1989), there is a variation between microbiota in sea cucumber and rearing sea water. The microbiota of fertilized egg and gastrula in sea cucumber were significantly different from rearing water in our results. Only 6.3 and 15.5% ASVs in fertilized egg and gastrula were shared with rearing seawater and five key ASVs significantly changed along with developmental stages were not detected in early life rearing seawater. Nonetheless, with the development of sea cucumber larvae, the sharing ASVs increase to 42.5% at late auricularia stage but decreased to 8.3% at juvenile stage, indicating the majority of larvae microbiome were not affected by rearing seawater.
Pioneer microbial colonization is related to birth process, gestational age, digestive system development and genetic factors (Benson et al., 2010; Karlsson et al., 2011; Gomez de Agüero et al., 2016; Borghi et al., 2019). In particular, genetic background of newborn individual influence gut microbial colonization, corresponding to functional development of digestive systems, were emerged in cod and chicken (Bakke et al., 2015; Schokker et al., 2015). Our results also show five key ASVs belonging to Rhodobacterales and Flavobacteriales are recognized as first colonizers in gut of sea cucumber’s microbiota at different life stages. These bacteria were not detected in rearing seawater at corresponding life stages and before the feeding process. Based on these findings, it is assumed that the process of organ formation along with larval developments impacts first colonizers in gut as an influential factor. Interestingly, another variation of early life microbiota was also observed in juvenile individuals, which possess important organ tentacles. Therefore, we assume that organogenesis occurring in sea cucumber larvae is one of the likely factors resulting in the alteration of microbiota, but more insights into the role of individual pioneer colonizer are necessary for deeper understanding.
The auricularia stage is the longest period that primary organs of the digestive system are formed during larval development of sea cucumber, and is sustained for 8–12 days as reported in another study (Soliman et al., 2013). We also observed that the larvae in the auricularia stage have been continuously detected for more than 17 days, to a maximum 19 days after fertilization. We started feeding after the early auricularia stage, and buccal cavity, mouth, esophagus, stomach, intestine, and cloaca, these digestive organs in sea cucumber were clearly observed in late auricularia individuals. The late auricularia microbiota was different from the those of individuals before and after feeding, which indicates that organogenesis accompanied with feeding are major factors impacting on early life gut microbiota. As there are many studies on sea cucumber that indicate the supplied diet could regulate intestinal microbiota and immunity (Yang et al., 2015b; Chen et al., 2018; Song et al., 2019), dietary microalgae could be considered as one of the major factors causing alteration of microbiota observed in the auricularia stage, in fact microbiotas of seawater after diet microalgae spikes were different of those before the spikes. However, even in microbiotas changes of seawater between before and after diets spikes, we still observed differences between host-associated and environmental microbiotas, which means there is still a presence of host factors selecting colonizers and shaping microbiotas.
The combination of culture-independent and culture-dependent methods can improve the accuracy of taxonomic assignment in metagenomics. Also, the acquisition of reference genome sequences of isolates provides the possibility of meta-transcriptomes to understand the functions of host-associated bacteria. In this study, we were able to isolate 13 strains belonging to Rhodobacterales and they were also assigned 4 key ASVs detected by Meta16S analysis, emphasizing the importance of culture dependent method. Rhodobacterales has been reported as possible bacteria for promoting A. japonicus growth (Yamazaki et al., 2016). Our study opens the way to investigating the effects of individual bacteria on host physiology and emphasizes the importance of the culture dependent method.
In this study, we analyzed dynamic changes of microbiome for the first time alongside sea cucumber developmental stages and investigated potential factors impacting on the alteration of the microbiome during host development. Our results indicate early life microbiota in sea cucumber significant changes at the auricularia stage and is distinct from environmental seawater. We also isolated crucial bacteria candidates including pioneer microbes on fertilized egg and first colonizers in gut such as Rhodobacterales and Alteromonadales showing significant difference among larval developmental stages. Further study of individual host-associated bacteria could contribute to a deeper understanding of the interaction between bacteria and host biology.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ddbj.nig.ac.jp/, DRR319863–DRR319874.
TS and JY designed the study. JY, TS, and YS performed experiments. JY and SM obtained and analyzed data. JY drafted the manuscript. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.801344/full#supplementary-material
Arrieta, M. C., Stiemsma, L. T., Amenyogbe, N., Brown, E. M., and Finlay, B. (2014). The intestinal microbiome in early life: health and disease. Front. Immunol 5:427. doi: 10.3389/fimmu.2014.00427
Arrieta, M. C., Stiemsma, L. T., Dimitriu, P. A., Thorson, L., Russell, S., Yurist-Doutsch, S., et al. (2015). Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Transl. Med. 7:307ra152. doi: 10.1126/scitranslmed.aab2271
Bäckhed, F., Roswall, J., Peng, Y., Feng, Q., Jia, H., Kovatcheva-Datchary, P., et al. (2015). Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 17, 690–703. doi: 10.1016/j.chom.2015.04.004
Bakke, I., Coward, E., Andersen, T., and Vadstein, O. (2015). Selection in the host structures the microbiota associated with developing cod larvae (Gadus morhua). Environ. Microbiol. 17, 3914–3924. doi: 10.1111/1462-2920.12888
Benson, A. K., Kelly, S. A., Legge, R., Ma, F., Low, S. J., Kim, J., et al. (2010). Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. PNAS 107, 18933–18938. doi: 10.1073/pnas.1007028107
Bettelheim, K. A. (1986). Commemoration of the publication 100 years ago of the papers by Dr. Th. Escherich in which are described for the first time the organisms that bear his name. Zbl. Bakt. Hyg. A. 261, 255–265. doi: 10.1016/s0176-6724(86)80043-8
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Borghi, E., Massa, V., Severgnini, M., Fazio, G., Avagliano, L., Menegola, E., et al. (2019). Antenatal microbial colonization of mammalian gut. Reprod. Sci. 26, 1045–1053. doi: 10.1177/1933719118804411
Califano, G., Castanho, S., Soares, F., Ribeiro, L., Cox, C. J., Mata, L., et al. (2017). Molecular taxonomic profiling of bacterial Communities in a gilthead seabream (Sparus aurata) hatchery. Front. Microbiol. 8:204. doi: 10.3389/fmicb.2017.00204
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Chen, C. P., and Chian, C. S. (1990). Larval development of the sea cucumber. Actinopyga echinites (Echinodermata: Holothuroidea). Bull. Inst. Zool. Acad. Sin. 29, 127–133.
Chen, J., Ren, Y., Li, Y., and Xia, B. (2018). Regulation of growth, intestinal microbiota, non-specific immune response and disease resistance of sea cucumber Apostichopus japonicus (Selenka) in biofloc systems. Fish Shellfish Immunol. 77, 175–186. doi: 10.1016/j.fsi.2018.03.053
Chi, C., Liu, J., Fei, S., Zhang, C., Chang, Y., Liu, X., et al. (2014). Effect of intestinal autochthonous probiotics isolated from the gut of sea cucumber (Apostichopus japonicus) on immune response and growth of A. japonicus. Fish Shellfish Immunol. 38, 367–373. doi: 10.1016/j.fsi.2014.04.001
Cole, J. R., Wang, Q., Fish, J. A., Chai, B., McGarrell, D. M., Sun, Y., et al. (2014). Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 42, D633–D642. doi: 10.1093/nar/gkt1244
Dang, H., Li, T., Chen, M., and Huang, G. (2008). Cross-ocean distribution of Rhodobacterales bacteria as primary surface colonizers in temperate coastal marine waters. Appl. Environ. Microbiol. 74, 52–60. doi: 10.1128/aem.01400-07
Dittmann, K. K., Rasmussen, B. B., Melchiorsen, J., Sonnenschein, E. C., Gram, L., and Bentzon-Tilia, M. (2020). Changes in the microbiome of mariculture feed organisms after treatment with a potentially probiotic strain of Phaeobacter inhibens. Appl. Environ. Microbiol. AEM.499–AEM.520. doi: 10.1128/aem.00499-20
Escherich, T. (1886). Die Darmbakterien des Säuglings und ihre Beziehungen Zur Physiologie der Verdauung. Stuttgart: Ferndinand Enke.
Fu, Y., Keats, K. F., Rivkin, R. B., and Lang, A. S. (2013). Water mass and depth determine the distribution and diversity of Rhodobacterales in an Arctic marine system. FEMS Microbiol. Ecol. 84, 564–576. doi: 10.1111/1574-6941.12085
Gamboa-Álvarez, M. Á, López-Rocha, J. A., Poot-López, G. R., Aguilar-Perera, A., and Villegas-Hernández, H. (2020). Rise and decline of the sea cucumber fishery in Campeche Bank, Mexico. Ocean Coast. Manag. 184:105011. doi: 10.1016/j.ocecoaman.2019.105011
Gensollen, T., Iyer, S. S., Kasper, D. L., and Blumberg, R. S. (2016). How colonization by microbiota in early life shapes the immune system. Science 352, 539–544. doi: 10.1126/science.aad9378
Gentleman, R., Carey, V., and Huber, W. (2021). genefilter: Genefilter: Methods for Filtering Genes from High-Throughput Experiments.” R Package Version 1.74.1.
Giatsis, C., Sipkema, D., Smidt, H., Verreth, J., and Verdegem, M. (2014). The colonization dynamics of the gut microbiota in tilapia larvae. PLoS One 9:e103641. doi: 10.1371/journal.pone.0103641
Gomez de Agüero, M., Ganal-Vonarburg, S. C., Fuhrer, T., Rupp, S., Uchimura, Y., Li, H., et al. (2016). The maternal microbiota drives early postnatal innate immune development. Science 351, 1296–1302. doi: 10.1126/science.aad2571
González-Wangüemert, M., Domínguez-Godino, J. A., and Cánovas, F. (2018). The fast development of sea cucumber fisheries in the Mediterranean and NE Atlantic waters: from a new marine resource to its over-exploitation. Ocean Coast. Manag. 151, 165–177. doi: 10.1016/j.ocecoaman.2017.10.002
Grotkjaer, T., Bentzon-Tilia, M., D’Alvise, P., Dourala, N., Nielsen, K. F., and Gram, L. (2016). Isolation of TDA-producing Phaeobacter strains from sea bass larval rearing units and their probiotic effect against pathogenic Vibrio spp. in Artemia cultures. Syst. Appl. Microbiol. 39, 180–188. doi: 10.1016/j.syapm.2016.01.005
Hamel, J. F., and Mercier, A. (2013). Apostichopus japonicus. The IUCN Red List of Threatened Species 2013: e.T180424A1629389. Gland: IUCN. doi: 10.2305/IUCN.UK.2013-1.RLTS.T180424A1629389.en
Hansen, G. H., and Olafsen, J. A. (1989). Bacterial colonization of cod (Gadus morhua L.) and halibut (Hippoglossus hippoglossus) eggs in marine aquaculture. Appl. Environ. Microbiol. 55, 1435–1446. doi: 10.1128/aem.55.6.1435-1446.1989
Hansen, G. H., and Olafsen, J. A. (1999). Bacterial interactions in early life stages of marine cold-water fish. Microb. Ecol. 38, 1–26. doi: 10.1007/s002489900158
Hill, C. J., Lynch, D. B., Murphy, K., Ulaszewska, M., Jeffery, I. B., O’Shea, C. A., et al. (2017). Evolution of gut microbiota composition from birth to 24 weeks in the infant met Cohort. Microbiome 5:4. doi: 10.1186/s40168-016-0213-y
Hu, C., Xu, Y., Wen, J., Zhang, L., Fan, S., and Su, T. (2010). Larval development and juvenile growth of the sea cucumber Stichopus sp. (Curry fish). Aquaculture 300, 73–79. doi: 10.1016/j.aquaculture.2009.09.033
Jontila, J. B. S., Monteclaro, H. M., Quinitio, G. F., Santander-de Leon, S. M., and Altamirano, J. P. (2018). Status of sea cucumber fishery and populations across sites with different levels of management in Palawan, Philippines. Ocean Coast. Manag. 165, 225–234. doi: 10.1016/j.ocecoaman.2018.08.025
Karlsson, C. L. J., Molin, G., Cilio, C. M., and Ahrné, S. (2011). The pioneer gut microbiota in human neonates vaginally born at term—a pilot study. Pediatr. Res. 70, 282–286. doi: 10.1203/PDR.0b013e318225f765
Kim, T. Y., Lee, J. J., Kim, B. S., and Choi, S. H. (2017). Whole-body microbiota of sea cucumber (Apostichopus japonicus) from south Korea for improved seafood management. J Microbiol. Biotechnol. 27, 1753–1762. doi: 10.4014/jmb.1707.07067
Koenig, J. E., Spor, A., Scalfone, N., Fricker, A. D., Stombaugh, J., Knight, R., et al. (2011). Succession of microbial consortia in the developing infant gut microbiome. PNAS 108(Suppl. 1), 4578–4585. doi: 10.1073/pnas.1000081107
Korpela, K., and de Vos, W. M. (2018). Early life colonization of the human gut: microbes matter everywhere. Curr. Opin. Microbiol. 44, 70–78. doi: 10.1016/j.mib.2018.06.003
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Letunic, I., and Bork, P. (2019). Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259. doi: 10.1093/nar/gkz239
Li, C., Ren, Y., Jiang, S., Zhou, S., Zhao, J., Wang, R., et al. (2018). Effects of dietary supplementation of four strains of lactic acid bacteria on growth, immune-related response and genes expression of the juvenile sea cucumber Apostichopus japonicus Selenka. Fish Shellfish Immunol. 74, 69–75. doi: 10.1016/j.fsi.2017.12.037
Lozupone, C., Lladser, M. E., Knights, D., Stombaugh, J., and Knight, R. (2011). UniFrac: an effective distance metric for microbial community comparison. ISME J. 5, 169–172. doi: 10.1038/ismej.2010.133
Ma, Y., Li, L., Li, M., Chen, W., Bao, P., Yu, Z., et al. (2019). Effects of dietary probiotic yeast on growth parameters in juvenile sea cucumber, Apostichopus japonicus. Aquaculture 499, 203–211. doi: 10.1016/j.aquaculture.2018.09.043
MAFF (2020). Japan statistics of Agriculture, Forestry and fisheries [Online]. Available online at: https://www.maff.go.jp/j/tokei/kouhyou/kaimen_gyosei/index.html [Accessed October 22, 2021].
Mercier, A., Yang, H., and Hamel, J. F. (2015). The Sea Cucumber Apostichopus japonicus : History, Biology and Aquaculture. Cambridge, MA: Academic Press.
Ministry of Agriculture and Rural Affairs Fishery Administration (2003-2012). China Fishery Statistical Yearbook. Beijing. CN: China Agriculture Press.
Nayak, S. K. (2010). Role of gastrointestinal microbiota in fish. Aquac. Res. 41, 1553–1573. doi: 10.1111/j.1365-2109.2010.02546.x
Nikouli, E., Meziti, A., Antonopoulou, E., Mente, E., and Kormas, K. A. (2019). Host-associated bacterial succession during the rarly embryonic stages and first feeding in farmed gilthead sea bream (Sparus aurata). Genes 10:483. doi: 10.3390/genes10070483
Pagán-Jiménez, M., Ruiz-Calderón, J. F., Dominguez-Bello, M. G., and García-Arrarás, J. E. (2019). Characterization of the intestinal microbiota of the sea cucumber Holothuria glaberrima. PLoS One 14:e0208011. doi: 10.1371/journal.pone.0208011
Price, M. N., Dehal, P. S., and Arkin, A. P. (2009). FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650. doi: 10.1093/molbev/msp077
Ramofafia, C., Byrne, M., and Battaglene, S. (2003). Development of three commercial sea cucumber Holothuria scabra. H. fuscogilva and Actinopyga mauritiana: larval structure and growth. Mar. Freshw. Res. 54, 657–667. doi: 10.1071/MF02145
Rasmussen, B. B., Erner, K. E., Bentzon-Tilia, M., and Gram, L. (2018). Effect of TDA-producing Phaeobacter inhibens on the fish pathogen Vibrio anguillarum in non-axenic algae and copepod systems. Microb. Biotechnol. 11, 1070–1079. doi: 10.1111/1751-7915.13275
Ren, H., Li, Z., Xu, Y., Wang, L., and Li, X. (2019). Protective effectiveness of feeding phage cocktails in controlling Vibrio parahaemolyticus infection of sea cucumber Apostichopus japonicus. Aquaculture 503, 322–329. doi: 10.1016/j.aquaculture.2019.01.006
Rogers, A., Hamel, J. F., Baker, S. M., and Mercier, A. (2018). The 2009–2016 Belize sea cucumber fishery: resource use patterns, management strategies and socioeconomic impacts. Reg. Stud. Mar. Sci. 22, 9–20. doi: 10.1016/j.rsma.2018.05.003
Schokker, D., Veninga, G., Vastenhouw, S. A., Bossers, A., de Bree, F. M., Kaal-Lansbergen, L. M. T. E., et al. (2015). Early life microbial colonization of the gut and intestinal development differ between genetically divergent broiler lines. BMC Genomics 16:418. doi: 10.1186/s12864-015-1646-6
Sewell, M. A., and McEuen, F. S. (2002). “Phylum Echinodermata: Holothuroidea,” in Atlas of Marine Invertebrate Larvae, ed. C. M. Young (San diego, CA: Academic Press), 513–530.
Shulman, S. T., Friedmann, H. C., and Sims, R. H. (2007). Theodor Escherich: the first pediatric infectious diseases physician? Clin. Infect. Dis. 45, 1025–1029. doi: 10.1086/521946
Simon, M., Scheuner, C., Meier-Kolthoff, J. P., Brinkhoff, T., Wagner-Döbler, I., Ulbrich, M., et al. (2017). Phylogenomics of Rhodobacteraceae reveals evolutionary adaptation to marine and non-marine habitats. ISME J. 11, 1483–1499. doi: 10.1038/ismej.2016.198
Soliman, T., Yamazaki, Y., Niiyama, H., and Tsunoda, K. (2013). Spontaneous captive breeding and larval development in the green and red variants of the Japanese sea cucumber Apostichopus japonicus (Selenka 1867). Aquac. Res. 44, 738–746. doi: 10.1111/j.1365-2109.2011.03078.x
Song, X., Feng, Z., Zhang, Y., and Zhu, W. (2019). Regulation of dietary astragalus polysaccharide (APS) supplementation on the non-specific immune response and intestinal microbiota of sea cucumber Apostichopus japonicus. Fish Shellfish Immunol. 94, 517–524. doi: 10.1016/j.fsi.2019.09.049
Sullam, K. E., Essinger, S. D., Lozupone, C. A., O’Connor, M. P., Rosen, G. L., Knight, R., et al. (2012). Environmental and ecological factors that shape the gut bacterial communities of fish: a meta-analysis. Mol. Ecol. 21, 3363–3378. doi: 10.1111/j.1365-294X.2012.05552.x
Tavares, N. K., VanDrisse, C. M., and Escalante-Semerena, J. C. (2018). Rhodobacterales use a unique L-threonine kinase for the assembly of the nucleotide loop of coenzyme B(12). Mol. Microbiol. 110, 239–261. doi: 10.1111/mmi.14100
Vadstein, O., Attramadal, K. J. K., Bakke, I., Forberg, T., Olsen, Y., Verdegem, M., et al. (2018). Managing the microbial community of marine fish larvae: a holistic perspective for larviculture. Front. Microbiol. 9:1820. doi: 10.3389/fmicb.2018.01820
Wang, Y., Sun, S., and Rong, X. (2006). Stomach ulcer disease in auricularia of sea cucumber (Apostichopus japonicus) and its etiological identification. Fish. Sci. China 13, 908–916.
Wilson, K. M., Rodrigues, D. R., Briggs, W. N., Duff, A. F., Chasser, K. M., and Bielke, L. R. (2019). Evaluation of the impact of in ovo administered bacteria on microbiome of chicks through 10 days of age. Poult. Sci. J. 98, 5949–5960. doi: 10.3382/ps/pez388
Wopereis, H., Oozeer, R., Knipping, K., Belzer, C., and Knol, J. (2014). The first thousand days – intestinal microbiology of early life: establishing a symbiosis. Pediatr. Allergy Immunol. 25, 428–438. doi: 10.1111/pai.12232
Yamazaki, Y., Meirelles, P. M., Mino, S., Suda, W., Oshima, K., Hattori, M., et al. (2016). Individual Apostichopus japonicus fecal microbiome reveals a link with polyhydroxybutyrate producers in host growth gaps. Sci. Rep. 6:21631. doi: 10.1038/srep21631
Yamazaki, Y., Sakai, Y., Mino, S., Suda, W., Hattori, M., Meirelles, P. M., et al. (2019). Repeated selective enrichment process of sediment microbiota occurred in sea cucumber guts. Environ. Microbiol. Rep. 11, 797–807. doi: 10.1111/1758-2229.12791
Yamazaki, Y., Sakai, Y., Yu, J., Mino, S., and Sawabe, T. (2020). Tracking the dynamics of individual gut microbiome of sea cucumber Apostichopus japonicus during gut regeneration. Peer J. 8:e10260. doi: 10.7717/peerj.10260.
Yang, G., Peng, M., Tian, X., and Dong, S. (2017). Molecular ecological network analysis reveals the effects of probiotics and florfenicol on intestinal microbiota homeostasis: an example of sea cucumber. Sci. Rep. 7:4778. doi: 10.1038/s41598-017-05312-1
Yang, G., Xu, Z., Tian, X., Dong, S., and Peng, M. (2015b). Intestinal microbiota and immune related genes in sea cucumber (Apostichopus japonicus) response to dietary β-glucan supplementation. Biochem. Biophys. Res. Commun. 458, 98–103. doi: 10.1016/j.bbrc.2015.01.074
Yang, G., Tian, X., Dong, S., Peng, M., and Wang, D. (2015a). Effects of dietary Bacillus cereus G19, B. cereus BC-01, and Paracoccus marcusii DB11 supplementation on the growth, immune response, and expression of immune-related genes in coelomocytes and intestine of the sea cucumber (Apostichopus japonicus Selenka). Fish Shellfish Immunol. 45, 800–807. doi: 10.1016/j.fsi.2015.05.032
Zhang, C. Y., Chen, G., Xu, Z., Yan, P. S., and Wang, Y. G. (2010). Etiology of rotting edges syndrome in cultured larval Apostichopus japonicus at auricularia stage and analysis of reservoir of pathogens. Acta. Microbiol. Sin. 50, 687–693.
Zhang, C., Chen, G., Xu, Z., Yan, P., Wang, G., and Wang, Y. (2009). Etiology of off-plate syndrome in cultured larval Apostichopus japonicus at attachment stage and analysis of reservoir of pathogens. Wei Sheng Wu Xue Bao 49, 631–637.
Zhang, Y., and Liu, Y. (1984). The review of research progress and exploration of resources proliferation of Stichopus japonicus. Mar. Fish. 2, 57–60.
Keywords: pioneer microbiome, gut microbiome, holobiont, sea cucumber, Apostichopus japonicus
Citation: Yu J, Sakai Y, Mino S and Sawabe T (2022) Characterization of Microbiomes Associated With the Early Life Stages of Sea Cucumber Apostichopus japonicus Selenka. Front. Mar. Sci. 9:801344. doi: 10.3389/fmars.2022.801344
Received: 25 October 2021; Accepted: 10 January 2022;
Published: 31 January 2022.
Edited by:
Fabiano Thompson, Federal University of Rio de Janeiro, BrazilReviewed by:
Francisco Vargas-Albores, Centro de Investigación en Alimentación y Desarrollo, Consejo Nacional de Ciencia y Tecnología (CONACYT), MexicoCopyright © 2022 Yu, Sakai, Mino and Sawabe. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tomoo Sawabe, c2F3YWJlQGZpc2guaG9rdWRhaS5hYy5qcA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.