- 1College of Marine Science and Environment Engineering, Dalian Ocean University, Dalian, China
- 2Key Laboratory of Environment Controlled Aquaculture (Dalian Ocean University), Ministry of Education, Dalian, China
- 3College of Fisheries and Life Science, Dalian Ocean University, Dalian, China
Successful stock enhancement refers to the maintenance of economic profit, whilst minimizing negative genetic and ecological effects. As hatchery environments differ from natural habitats, post-release identification of hatchery-reared juveniles, and estimation of their adaptability are essential steps in conducting responsible stock enhancement. The black rockfish is an important fishery species in Japan, South Korea and China. Overfishing has dramatically decreased natural resources and a stock enhancement program was first initiated in the Liaodong peninsula in 2019. We traced this program and used nine microsatellite markers to identify 279 individuals as being hatchery-reared, out of 710 captured fish, with a resource contribution rate of 39.30%. These results indicated that stock enhancement of this species is likely to maintain a positive economic performance. To understand the potential genetic impacts on the local wild population, the genetic patterns of S. schlegelii stocks (wild controls from 2017, wild controls from 2018, female broodstock from 2019, recaptured hatchery-reared fish from 2019 and recaptured non-hatchery-reared fish from 2019 stocks) from the same habitat were evaluated before and after artificial stocking. We found that the S. schlegelii population was able to maintain high genetic diversity and showed weak genetic differentiation, but potential genetic introgression derived from stock enhancement should be concerned. The hatchery-reared individuals showed good adaptability, as shown by feeding condition and growth status. There was no obvious morphology difference between hatchery-reared fish and their wild counterparts, but hatchery-reared fish seemed to be bigger in size (significantly higher in MDB and BW), had a larger head (significantly larger in PRL/HL and ID/HL) and smaller paired fins (significantly smaller PCFL/BL), when compared with wild individuals across all investigation months. Our results provided insights into management of black rockfish enhancement in the future.
Introduction
The development of aquaculture meets the requirement of providing seed stocks for restoring natural resources and deals with the declining natural productivity of capture fisheries by stock restoration, sea ranching and stock enhancement (Grant et al., 2017). Amongst these processes, stock enhancement is a practice that uses the release of hatchery-reared juveniles to support natural stocks, for the resolution of overexploitation and habitat degradation (Salayo et al., 2020). It is estimated that the sea is restocked each year with more than 26 billion hatchery-reared seed stocks, covering 180 marine species from over 20 countries and regions (Kitada, 2018). Although stock enhancement has become a popular short-term method to increase catch production in the wild (Kitada, 2020), it also raises several concerns. The main concerns of marine fishery researchers include (1) the economic effects of releasing hatchery-reared seed stocks into the wild, (2) the adaptation of hatchery-reared seed stocks after release and their ecological interaction with their wild counterparts, (3) the genetic impacts of released individuals on the local wild populations. Consideration of each of these factors is essential to build a responsible resource restoration process. To date, the release of hatchery-reared juveniles includes finfish, crustaceans, mollusks and echinoderms (Molony et al., 2005; Stoner, 2019; Salayo et al., 2020; Lee et al., 2021). A sharp increase in stock enhancement can be observed in China, where over 94 billion hatchery-reared juveniles were released from 2004 to 2013, and a large proportion of these stock enhancement activities were on a massive scale (Luo and Zhang, 2014). However, most of the stock enhancement activities lack investigation and monitoring, with no economic, genetic or ecological data available for evaluation.
Although actually the recapture rate was widely applied in the determination of economic efficiency of stock enhancement programs, estimation of recapture contribution rate (RCR) during fishery investigations is also beneficial for estimation of economic return rate per release, in case of absence of recapture rate (Fang et al., 2021). Identifying the differentiation between hatchery-reared fish and their wild counterparts is the first step in calculating the RCR. In a limited number of species, i.e., the hatchery-reared Japanese flounder (Paralichthys olivaceus), an excess of pigmentation is observed on the blind side, in contrast to their wild counterparts (Zhu et al., 2004). The hatchery-reared red sea bream (Pagrus major) seeds exhibit deformity of the internostril epidermis (Kitada et al., 2019). Both morphological variations can be used in the identification of hatchery-reared individuals in recapture investigations. However, most other enhanced species lack such stable morphological variations. Hence, DNA profiling can be used to identify hatchery-reared seeds to assess economic efficiency. A set of successful identifications have been reported in finfish species, such as red drum (Sciaenops ocellatus) (Karlsson et al., 2008) and silver carp (Hypophthalmichthys molitrix) (Fang et al., 2021), and in crustaceans, such as Chinese shrimp (Fenneropenaeus chinensis) (Wang et al., 2014) and swimming crab (Portunus trituberculatus) (Liu et al., 2018).
Genetic and ecological effects of aquaculture stock releases are also a concern (Pinter et al., 2019). Hatchery-reared steelhead (Oncorhynchus mykiss) juveniles show DNA differences from wild fish and adaptation to captivity, rather than the natural environment. This can occur after just one generation in a hatchery environment (Christie et al., 2012). Moreover, due to a limited number of broodstock, hatchery-reared seed stocks may have a low level of genetic diversity and a high level of homozygosity (Araki and Schmid, 2010). Captive rearing can induce different epigenetic modifications between hatchery coho salmon (Oncorhynchus kisutch) and their wild counterparts (Le Luyer et al., 2017). Different DNA methylation patterns have been observed in the sperm of captive-reared Atlantic salmon (Salmo salar) when compared with wild-born ones (Barreto et al., 2019). The above DNA and DNA methylation changes may result in phenotypic changes (e.g., body shapes: see Fragkoulis et al., 2017; Sánchez-González and Nicieza, 2017), as well as adaptive changes (e.g., survival: see Stoner and Davis, 1994; the returned number for spawning: see Jonsson et al., 2019; reproductive success: see Janowitz-Koch et al., 2019; empty stomach rate: see Fischer et al., 2019; food items: see Davis et al., 2018) in hatchery-reared seed stocks, in comparison to their wild counterparts (Kitada, 2020). Consequently, continuous hatchery stocking allows introgression with the wild gene pool or even the replacement of wild genes, which causes a decline in adaptation and a decrease in the production of post-mixed wild populations when stock enhancement programs are undertaken (Jonsson et al., 2019). In contrast, several investigations have revealed that long-term hatchery-reared stock enhancements do not significantly affect wild populations, e.g., on genetic impacts (Lehnert et al., 2020), which indicates the variable impacts of stock enhancement. Hence, fishery investigations and evaluations are an emergent requirement during stock enhancement programs to reveal the conditions of post-released hatchery-reared individuals and the impacts on wild populations.
The black rockfish (Sebastes schlegelii), which belongs to the Sebastidae family of the Scorpaeniformes order, is one of the economically important viviparous marine teleost species. It is regarded as delicious seafood, with a high market value and additional recreational value. This fish is found along the shallow coasts of Japan, South Korea and northern China (Onodera et al., 2001), where it inhabits the rocky reef areas. Individuals show strong site fidelity and limited capacity for movement (Zhang et al., 2015). However, the larval dispersal pattern of S. schlegelii seems to be more widespread that is influenced by floating seaweed (Gao et al., 2016). S. schlegelii is internal fertilization and has long-term sperm storage characteristics in the female ovary. In the wild, the male and female can mate in November and December, while fertilization occurs in April and May next year, around 6 months after copulation (Mori et al., 2003; He et al., 2019). Hence, only adult female ones were collected every year for artificial breeding or even enhancement. Although the natural resources have dramatically decreased, the hatchery industry for this species is growing rapidly due to the advances in aquaculture technology. This has provided sufficient seed stock for stock enhancement to begin in China (enhancement programs were gradually launched in Shandong, Hebei and Jiangsu provinces from 2016 to 2019), where the enhancement area is mostly restricted to the Shandong peninsula waters, belonging to the Bohai Sea and South Yellow Sea. From 2015 to 2019, an average of over 30 million S. schlegelii (account for 97.9% of total released number in China) was released each year in Shandong peninsula waters. However, little research had evaluated the impacts on wild S. schlegelii populations. We only found that Wang et al. (2020) reported that no marked decrease in the hatchery-released stock, low but significant differentiation between the hatchery-released and wild stocks, as well as the wild stock before enhancement and the mixed stock after enhancement in Lidao bay, Shandong peninsula. Aside from the above enhancement events, there has been no release of S. schlegelii in the Liaodong peninsula before 2019. Hence, it is a suitable place for assessment of the different impacts arising from stock enhancement. In 2019, a hatchery-reared S. schlegelii enhancement program was initiated in the Liaodong peninsula. We traced the process before and after enhancement, sampled all the female broodstock and carried out fishery investigations. We also distinguished the hatchery-reared fish from wild-born individuals by DNA profiling and analyzed the genetic background, growth, morphology and feeding ecology in hatchery-reared individuals and their wild counterparts. This study aimed to evaluate (1) whether hatchery-reared S. schlegelii contributes economically via the enhancement programs, (2)whether hatchery-reared fish can maintain natural levels of genetic diversity to avoid a potentially negative genetic effect on the wild population, although comprehensive detection of such genetic effects may cast over generations and (3) whether hatchery-reared fish exhibit different growth, morphological characteristics and feeding ecology when compared with the wild population. Our results represent the first assessment of the multiple impacts of S. schlegelii stock enhancement and will be valuable for responsible stock enhancement programs.
Materials and Methods
Ethics Statement
This research was approved by the Care and Use of Laboratory Animals in Fishery Resources Enhancement Laboratory of Dalian Ocean University, Dalian. All animal treatment protocols were performed in accordance with Chinese laws, regulations and ethics.
Fish Samples
From the end of April to the beginning of May 2019, approximately 800 mature female S. schlegelii (age: approximate 4–6 age), with an obvious increase in abdomen size, were collected from the natural habitat of Dalian by fishing trap. On May the 7th, each of 30 females were placed in 50 m3 tanks and allowed to spawn in a shady environment. Finally, four hundred and eighty-nine fish spawned. The female fish were then removed and the hatchery-reared larvae were reared until their total length reached five centimeters. On July the 30th, 2.05 million first-generation offspring (seed cost: 0.29 million US dollars) were released into the coastal water of Dalian, with 1.05 million of these juveniles (seed cost: 0.15 million US dollars) were released covering our investigation area in the North Yellow Sea area. Fishery investigations were conducted from August the 28th to December the 25th and 710 S. schlegelii were caught by fishing trap (mesh size: 3.5 cm, 10 m long, 25 cm height and 30 cm width) in the North Yellow Sea (Figure 1 and Supplementary Table 1). Before stock enhancement was first initiated, we had collected 86 and 46 mature S. schlegelii (age: approximate 4–6 years old, total length: 38–49 cm) by fishing trap from the same Dalian waters of the Liaodong peninsula, in May, 2017 and May, 2018, respectively, to use as wild controls. Consequently, a total of 1,331 S. schlegelii were used in this study. These included the wild controls from 2017 (86 individuals, WC2017), the wild controls from 2018 (46 individuals, WC2018), female broodstock from 2019 (489 individuals, FB2019) and recaptured individuals from 2019 (710 individuals, RI2019).
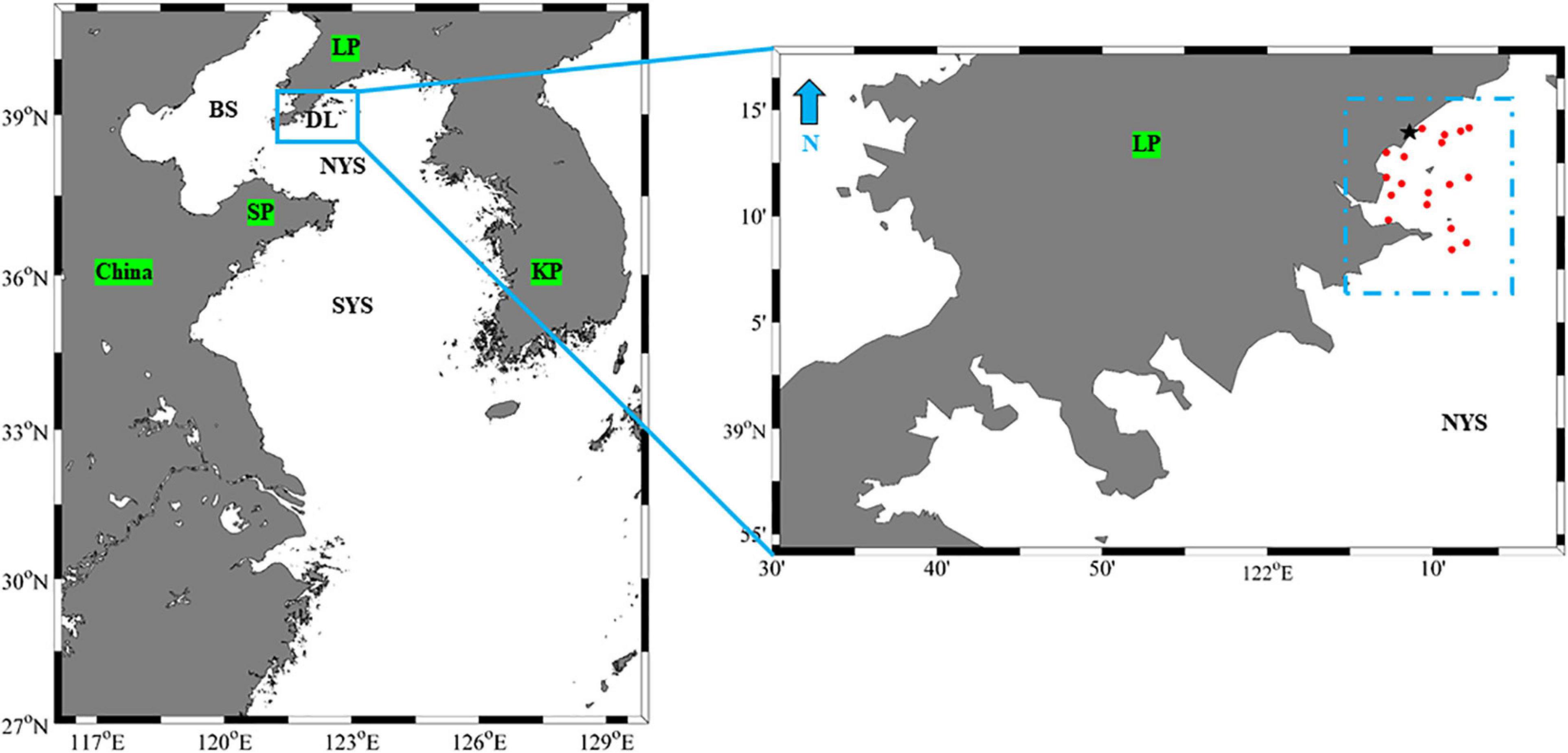
Figure 1. The recapture investigations of S. schlegelii in the North Yellow Sea in 2019. Dotted lines indicate the fishery recapture area. Red dots indicate the locations of fishing trap for collecting RA2019 and NA2019 samples. WC2017, WC2018 and FB2019 were collected in the blue dotted lines area. Black star indicates the North Yellow Sea area of releasing S. schlegelii in 2019. DL, Dalian; NYS, North Yellow Sea; SYS, South Yellow Sea; BS, Bohai Sea; LP, Liaodong Peninsula; SP, Shandong Peninsula; KP, South Korea Peninsula.
Genomic DNA Extraction and Genotyping
Muscle tissue samples were dissected, total genomic DNA was extracted using the TIANamp Marine Animals DNA Kit (Tiangen, China). The concentration and purity of DNA were measured using 1.0% agarose gel electrophoresis and a spectrophotometer. A total of nine high polymorphic microsatellite loci were selected, based on reported genotypes (Supplementary Table 2). All loci were amplified in each individual. The PCR procedure was carried out as previously reported (Liu et al., 2018). In brief, the 15 μL PCR reactions contained 20 ng of genomic DNA, 1.2 μL of MgCl2 (2.5 mM), 1.6 μL of 10 × PCR buffer, 0.12 μL of Taq polymerase, 1.2 μL of dNTPs (2.5 mM), 1.2 μL of forward primer labeled with either FAM, HEX, or TAMRA (5 μM) and 1.2 μL of unlabeled reverse primer (5 μM). Amplification conditions consisted of an initial denaturation step at 94°C for 3 min, followed by 35 cycles of denaturation at 94°C for 45 s, annealing at an optimal temperature (specified in Supplementary Table 2) for 45 s, extension at 72°C for 45 s and a final extension at 72°C for 10 min. The protocol ended with an infinite hold at 12°C. The PCR products were subsequently diluted and pooled into one of three groups (A, B and C) for multiple sequencing (Supplementary Table 2). The genotyping was scored using an ABI Prism 3730XL system (Applied Biosystems, Foster City, CA, United States). The ROX-500 size standard was used.
Parentage Assignment
Genotyping errors and null alleles were first assessed using Microchecker 2.2.3 (Van Oosterhout et al., 2004), followed by a manual check. A second reaction was performed on an individual if the genotype was not clear. Parentage assignment was estimated between the 489 female broodstock (FB) and the 710 recaptured S. schlegelii samples using Cervus 3.0.7 (Kalinowski et al., 2007) based on likelihood approach. Simulation of parentage analysis and maternity analysis was performed. In order to maintain the accuracy of assignment, parentage was assigned using a minimum of eight loci, 489 candidate mothers, 100,000 offspring, 1% genotyping error, 1% error rate in likelihood calculations were simulated under an 80% confidence interval. Non-exclusion probability (NE) and the power of exclusion (PE) were obtained using Cervus 3.07 and the combined power of exclusion (CPE) of all the loci was then derived using the following formula:
where PEk is the power of exclusion (PE) of the kth microsatellite allele.
The recaptured 2019 individuals (RI2019) can be distinguished as recaptured hatchery-reared S. schlegelii (RA2019) and non-hatchery-reared S. schlegelii (NA2019). The RCR was defined as the proportion of recaptured hatchery-reared S. schlegelii (RA2019) to total recaptured individuals of 2019. The economic return rate was defined as the estimated market landed value (sales) divided by the total seed costs (Nakagawa, 2008). Consequently, our collected samples can be classified as different stock units (WC2017, WC2018, FB2019, RA2019, and NA2019), covering before and after release in Liaodong peninsula.
Potential Genetic Effect Estimation
Genetic Diversity
To compare the genetic diversity among the five stocks, deviations from Hardy-Weinberg equilibrium (HWE) were evaluated for each locus and stock using Arlequin (Excoffier et al., 2005). The standard genetic diversity parameters, such as the number of alleles (Na), effective number of alleles (Ne-allele), observed heterozygosity (Ho), expected heterozygosity (He), unbiased expected heterozygosity (uHe) and allele frequency pattern were evaluated using GenAlEx 6.503 (Peakall and Smouse, 2012). Allelic richness (Ar) was calculated by HP Rare 1.0, with consideration of different sample sizes (Kalinowski, 2005). The inbreeding coefficient (Fis) was estimated at each locus, for each stock, using the Genepop 4.7 software package (Rousset, 2008). The polymorphism information content (PIC) and null allele frequency (Null) were estimated by Cervus 3.0.7 (Kalinowski et al., 2007). Estimations of effective population size (Ne) in S. schlegelii stocks were performed with NeEstimator v.2.01, based on the linkage disequilibrium methods (Waples and Do, 2008). The confidence intervals were determined using the jackknife method and 95% confidence interval. The relatedness of each stock was calculated using ML-Relate (Kalinowski and Taper, 2006). The 95% confidence intervals were added by SPSS 24.0 (IBM, Armonk, NY, United States). Significance was adjusted using the sequential Bonferroni correction in multiple tests (Gaetano, 2018). To compare the genetic parameter variations between each stock, pairwise comparisons of Na, Ne-allele, uHe, Ar and Fis, following the sequential Bonferroni correction, were made via the paired t-test or Wilcoxon signed-rank test, to account for the normal distribution of the genetic diversity data. All tests were conducted in SPSS 24.0 and statistical significance was assigned using P < 0.05.
Genetic Structure
To analyze the genetic structure before and after S. schlegelii stock enhancement, several approaches were used. Molecular variation analysis of variance (AMOVA), pairwise fixation index (Fst) and gene flow (Nm) tests were performed using Arlequin 3.5 (Excoffier et al., 2005). Structure v2.3.4 software was used to divide the stocks into subgroups (Falush et al., 2003). Initially, each run included a burn-in of 1,00,000 Markov Chain Monte Carlo (MCMC) generations, following an initial burn-in of 1,000,000 steps. After five repeats, the most probable value of K was determined by structure harvester, based on the Delta K (ΔK) method (Earl, 2012). We performed a (Principal Coordinates Analysis) PCoA using a genetic distance matrix generated from allele frequencies, using GenAlEx 6.503 (Peakall and Smouse, 2012). We retained the first two PCoA axes to generate a plot. For evaluation of a recent bottleneck effect, the Bottleneck 1.2.02 program was used (Piry et al., 1999). The sign test, the standardized differences test, Wilcoxon’s signed-rank test and a qualitative test of the model shift were used for the bottleneck analysis.
Morphology and Growth Status
For comparison of morphological changes between hatchery-reared individuals and their wild counterparts, 18 morphological characteristics were examined. These included number of dorsal fins, number of pectoral fins, number of pelvic fins, number of anal fins, preorbital length (PRL, cm), eye diameter (ED, cm), postocular length (PL, cm), interorbital distance (ID, cm), head length (HL, cm), body length (BL, cm), total length (TL, cm), maximum depth of body (MDB, cm), length of pectoral fin base (PLFB, cm), pectoral fin length (PLFL, cm), pelvic fin length (PCFL, cm), length of pelvic fin base (PCFB, cm), pre-anal fin length (PRFL, cm) and body weight (BW, g). The liver of each fish was dissected out and the wet weight was determined (Wl, g). The hepatosomatic index (HSI) was calculated as previously reported (Sulistyo et al., 2000), where HSI = 100% (Wl/BW). The weight of the intestine (Wi, g) was measured and the intestinosomatic index (ISI) was estimated as ISI = 100% (Wi/BW). The condition factor (CF) was calculated as: CF = 100%(BW/BL3).
Feeding Adaptation of Captured Hatchery-Reared Individuals, When Compared With Their Wild Counterparts
The stomach of each fish was dissected out and the wet weight was determined (Ws, g). The stomach content index (SCI) was calculated as SCI = 100% (Ws/BW). Fullness of the stomach was used to assign feeding intensity to one of five levels; 0: empty; 1: 1/4 full; 2: 1/2 full; 3: 3/4 full; and 4: full with stomach wall extended.
Differences in the morphology, growth status and feeding adaptation, between the hatchery-reared individuals and their wild counterparts, were analyzed before the normality test. The independent samples t-test in the parametric test or non-parametric independent samples t-test, followed by the Mann-Whitney U-test, were selected to test the significance.
Results
Identification of Hatchery-Reared Individuals
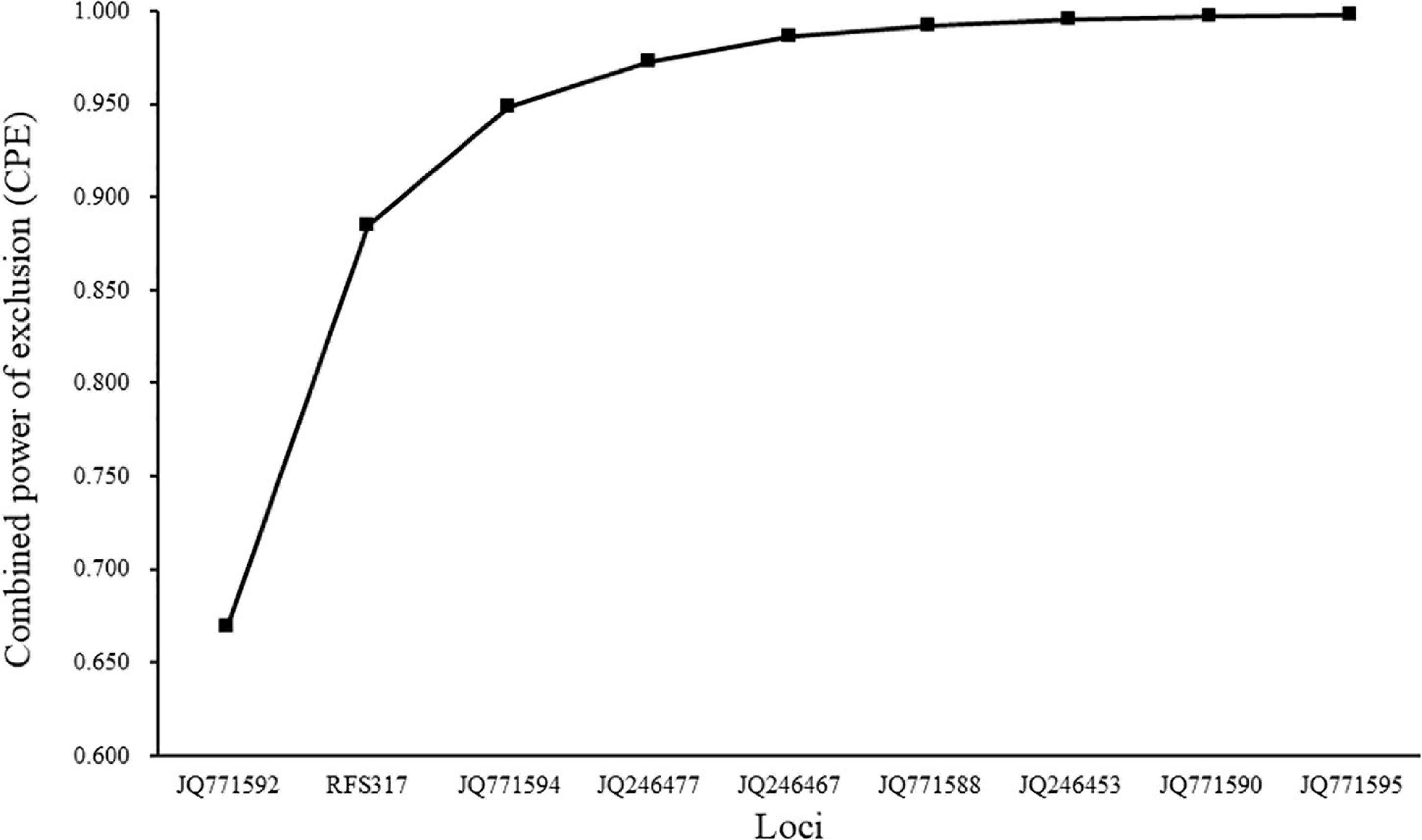
A total of 1,331 individuals were genotyped using nine polymorphic microsatellite markers. No evidence was found for genotyping errors caused by stuttering or large allele dropout. Cervus was used to identify 279 individuals, out of the 710 recaptured individuals, as hatchery-reared (RA2019). The LOD (logarithm of the odds) of parentage assignment ranged from 3.021 to 13.017 (Supplementary Table 3). The remaining 431 recaptured S. schlegelii could not be assigned maternity, so were considered to be wild stock. The PE of each locus ranged from 0.334 to 0.669 (mean of 0.489) and the CPE value in our parentage analysis, for nine loci, reached 99.8% (Figure 2 and Supplementary Table 4). These results indicated that our identification of hatchery-reared individuals was effective and reliable. Overall, the 1,331 fish could be classified into five stocks; WC2017 (86), WC2018 (46), FB2019 (489), RA2019 (279), and NA2019 (431), which were used in the following analyses.
The total RCR was 39.30% and ranged from 35.18 to 43.50% per month (Supplementary Table 3). The market price of naturally captured S. schlegelii at the Dalian Fish Market is higher for size larger fish. In December 2019, our captured S. schlegelii were still too small to be sold and could only return 4.632 US dollar/kg. Using the total release scale of 1.05 million in the North Yellow Sea, seed cost 0.15 million US dollars and RCR of 42.03% in December, a primary economic return rate of 0.30 was estimated.
Genetic Diversity
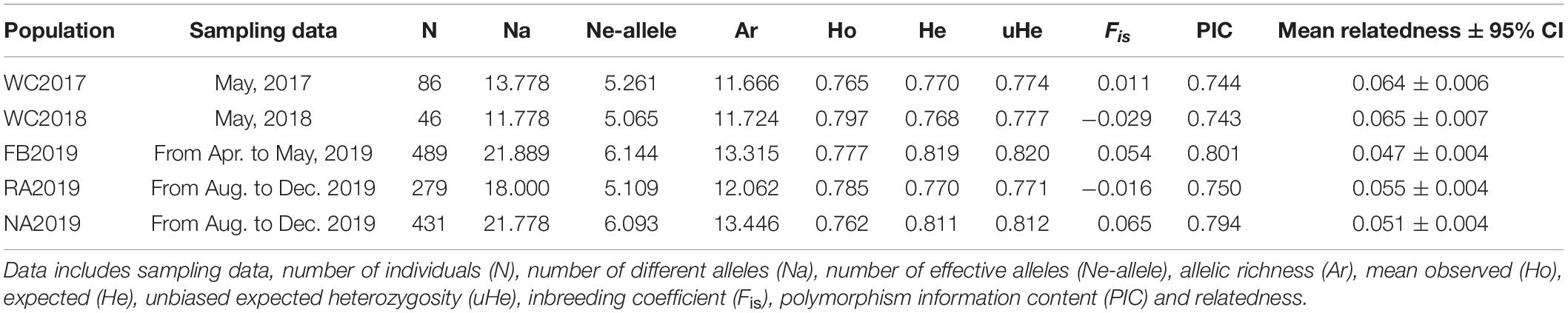
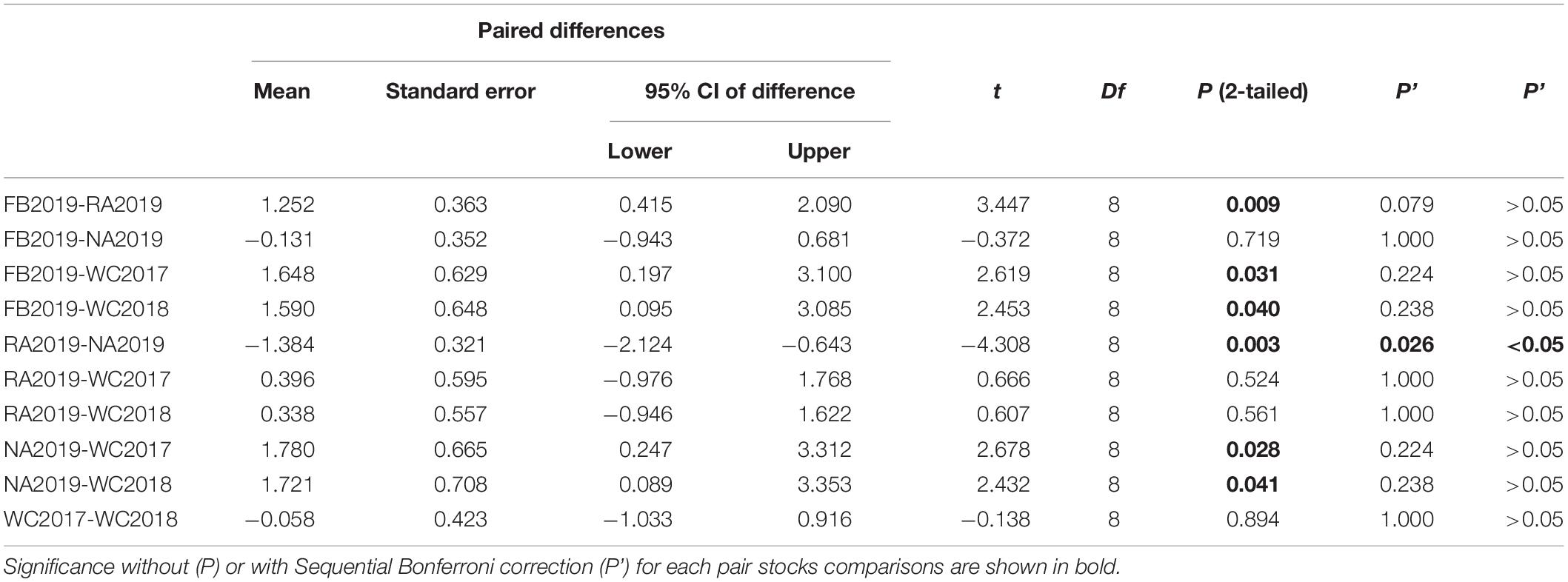
The allele frequency plot of the five stocks is shown in Supplementary Figure 1. The genetic diversity for each stock was evaluated using the allele distribution, which is shown in Table 1 and Supplementary Tables 5–9. No loci exhibited a null allele frequency (Null) of greater than 0.2 in any of the five stocks. Consequently, we used all the loci data in the following analyses. The average number of alleles (Na) and effective alleles (Ne-allele) per stock ranged from 11.778 (WC2018) to 21.889 (FB2019) and 5.065 (WC2018) to 6.144 (FB2019), respectively. The average PIC values ranged from 0.743 (WC2018) to 0.801 (FB2019) (Table 1). Our results showed that the loci were highly polymorphic. The allelic richness for the five stocks ranged from 11.666 (WC2017) to 13.446 (NA2019). We observed HWE disequilibrium, using the exact test (following Sequential Bonferroni correction), at six loci (RFS317, JQ771594, JQ246477, JQ246467, JQ246453, and JQ771595) in FB2019, at one locus (JQ246453) in RA2019 and five loci (JQ771592, JQ246477, JQ246467, JQ246453, and JQ771595) in NA2019 (P < 0.05). However, there was no departure from HWE in the wild control groups of WC2017 and WC2018. Homozygous excess was observed in WC2017, FB2019 and NA2019 when all loci were used to calculate Fis. The maximum value of Fis was found in NA2019 (0.065, Table 1). There was no significant difference in Na, Ne-allele and Ho between each pair of stocks, following sequential Bonferroni correction (P > 0.05) (Tables 2–4). However, RA2019 exhibited significantly smaller values for Ar than NA2019 (Table 5). The S. schlegelii stocks were characterized by a relatedness range of 0.047–0.065 (Table 1). Hence, the local population exhibited very low genetic similarity both before and after stock enhancement. Evaluation of Ne showed large values (> 1,727) for all local stocks distributed in the North Yellow Sea of the Liaodong peninsula and the confidence intervals reached infinity when using a Pcrit of 0.02 or 0.05, based on the linkage disequilibrium method (Table 6). Overall, the local S. schlegelii population maintained high levels of genetic diversity before and after the first stock enhancement in Liaodong peninsula but the significantly decreased Ar value of RA2019, when compared with their wild neighbor, NA2019, could be of concern.
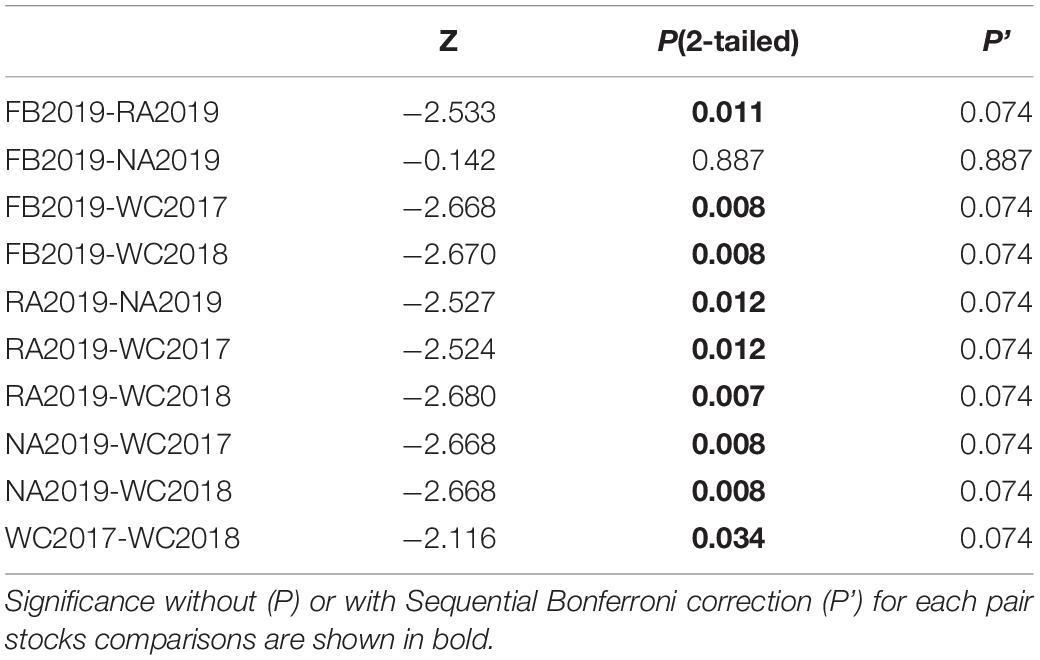
Table 2. Wilcoxon signed-rank test to compare the number of alleles (Na) between each pair of S. schlegelii stocks.
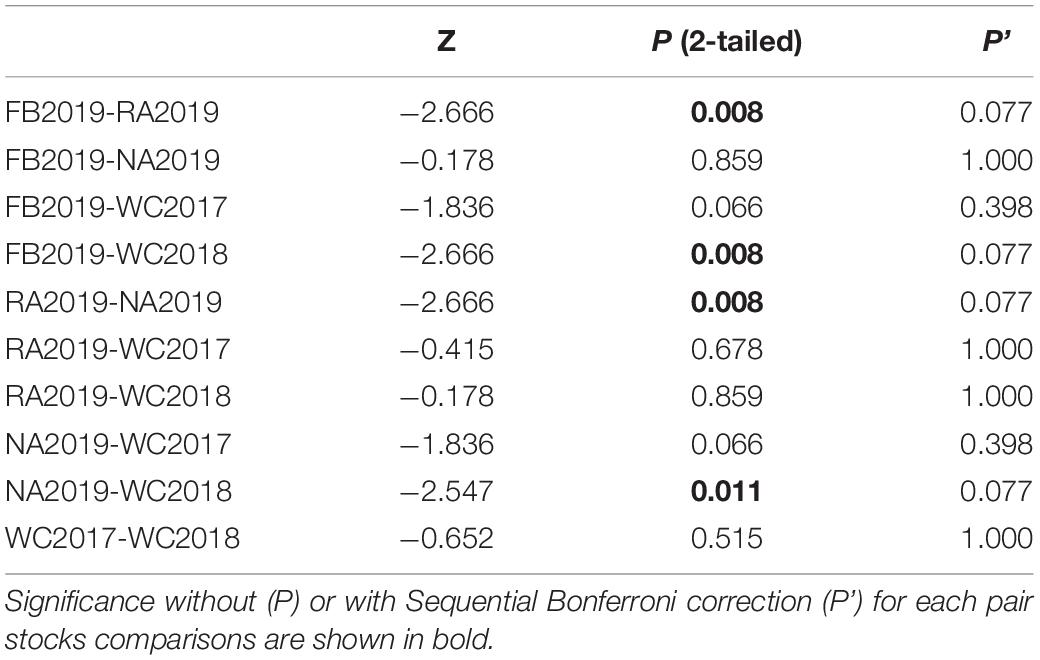
Table 3. Wilcoxon signed-rank test to compare the number of effective alleles (Ne) between each pair of S. schlegelii stocks.
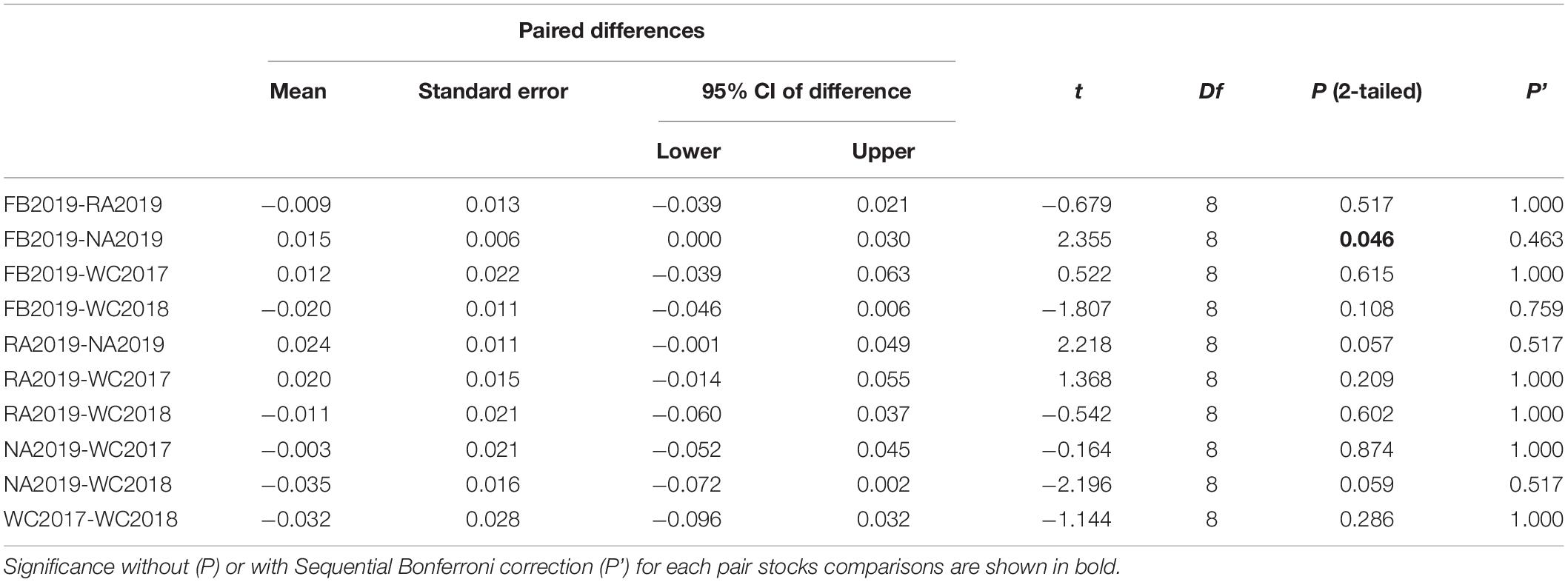
Table 4. Paired t-test of the observed heterozygosity (Ho) between each pair of S. schlegelii stocks.
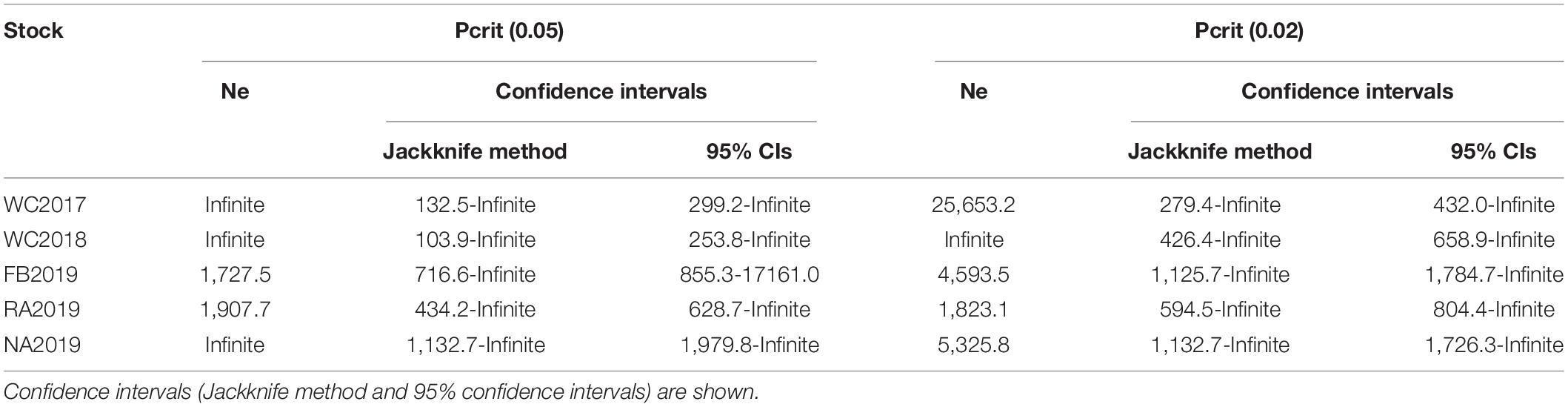
Table 6. Effective population size (Ne) based on linkage disequilibrium method in each stock of S. schlegelii.
Genetic Divergence
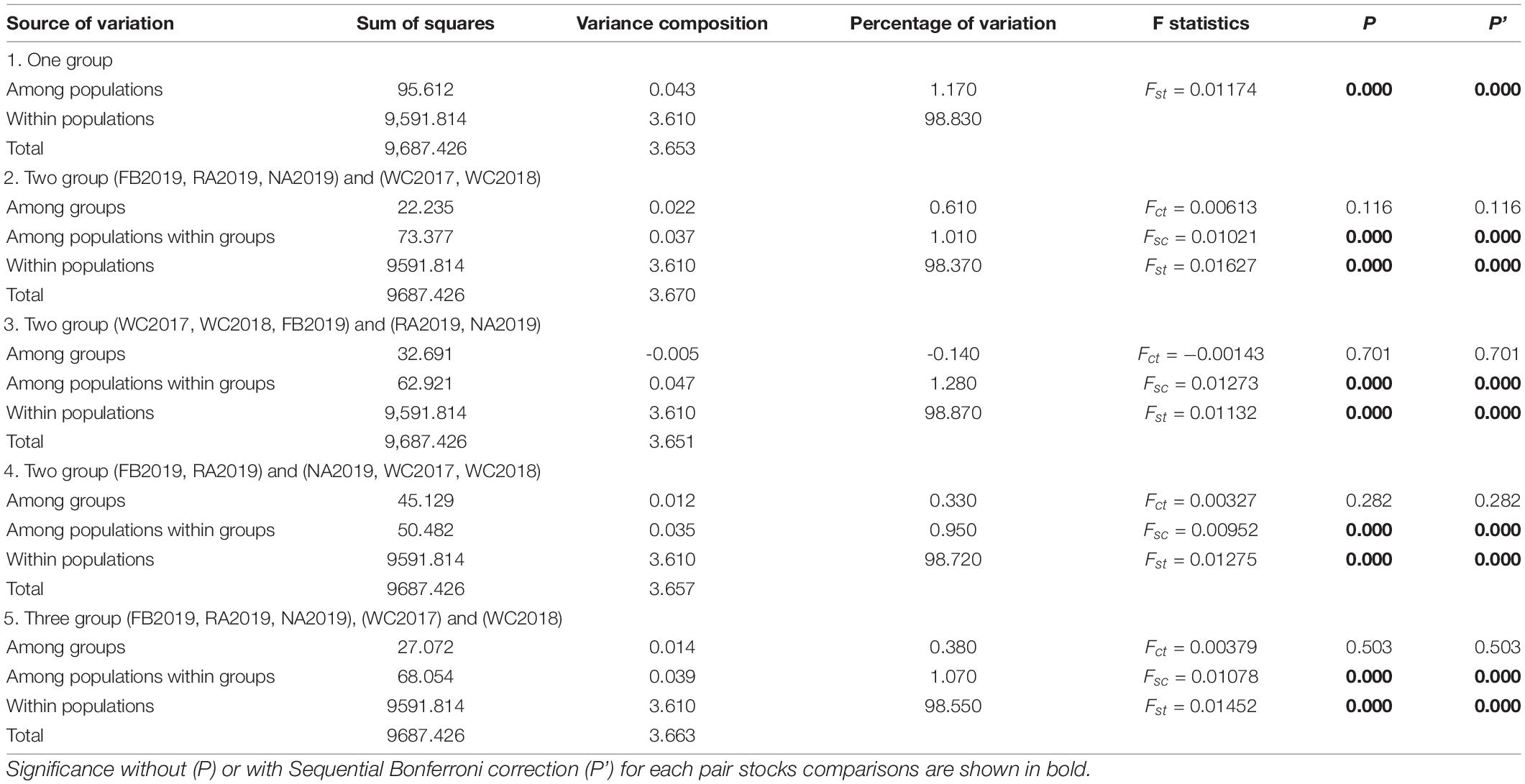
The bottleneck test was applied via infinite allele model (IAM) and two-phased model of mutation (TPM) models. The results are presented in Supplementary Table 10. None of the stock showed significant heterozygosity excess in these two models and no significant bottleneck effect was observed in the recent past, before or after stock enhancement. Furthermore, a normal L-shaped distribution is shown in Supplementary Figure 2. The genetic differentiation between stocks was initially evaluated by pairwise Fst. The values ranged from 0.003 to 0.027 (P < 0.05), which revealed very weak genetic differentiation between the local populations, before and after stock enhancement. The low but significant differentiation was found, except for NA2019 vs. WC2018 and WC2017 vs. WC2018 (Figure 3). This phenomenon may be due to the strong gene flow (Nm range was 9.122–75.623) that occurred in the S. schlegelii populations. The genetic structure was also investigated by the Structure program. The results revealed that the highest delta K-value obtained was K = 2 (Supplementary Figure 3). The large percentage of individuals from WC2017, WC2018, and NA2019 consisted of the first cluster, whereas the individuals from FB2019 and RA2019 were assigned to the first cluster and the second cluster (Figure 4). The analysis of molecular variance (AMOVA) test was also consistent with the above results as most of the variance (> 98%) was explained by the differences among individuals, despite the S. schlegelii stocks being assigned to different groups (Table 7). Moreover, the PCoA did not separate all individuals into different clusters and also revealed that there was no obvious genetic differentiation (Figure 5). In general, we observed very limited genetic differentiation in the S. schlegelii population before and after stock enhancement.
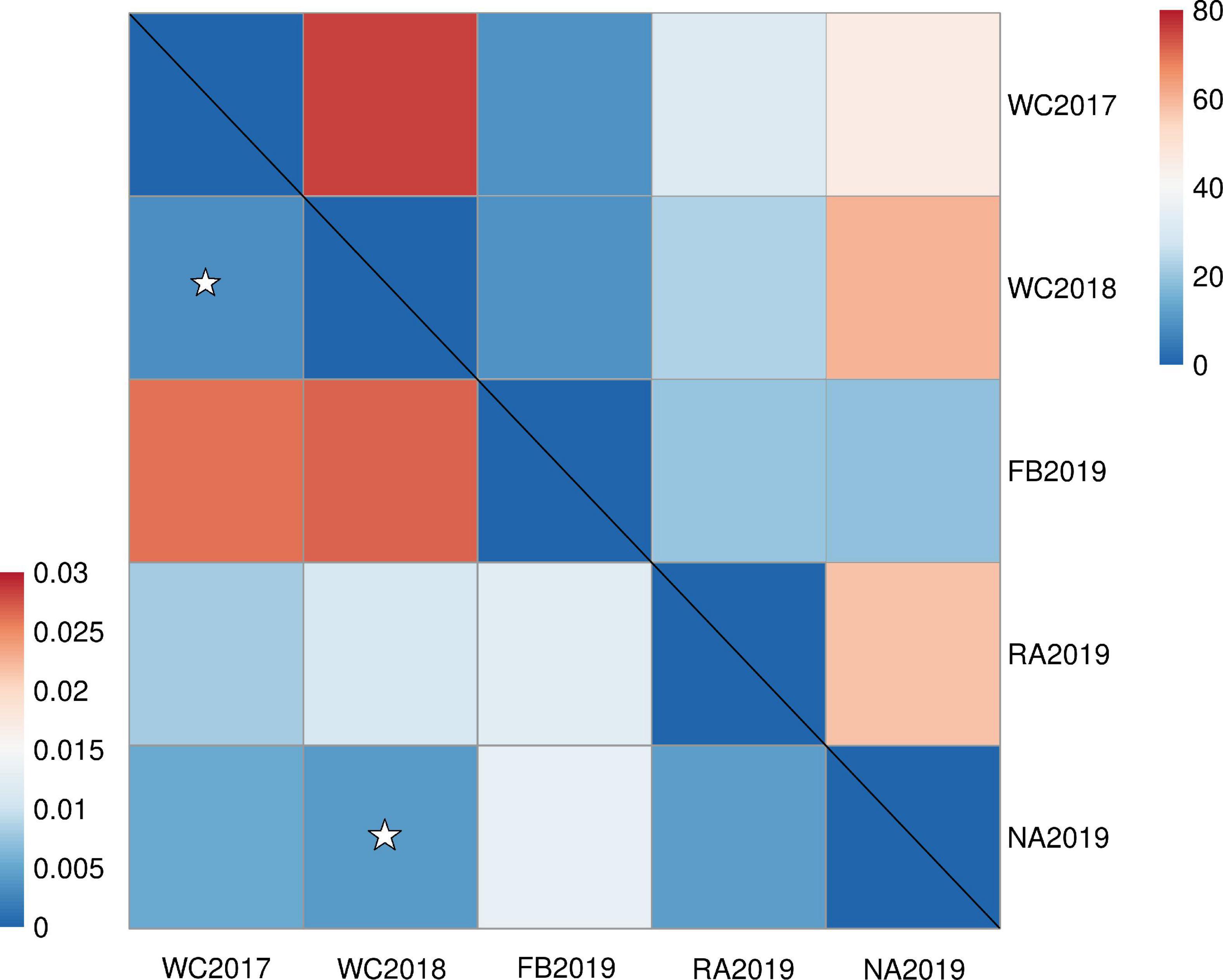
Figure 3. The heat map of pairwise genetic divergence (pairwise Fst, lower triangle) and gene flow (Nm, upper triangle). All the pairwise Fst-values between each pair of stock (apart from NA2019 vs. WC2018 and WC2018 vs. WC2019) were significant under sequential Bonferroni correction.
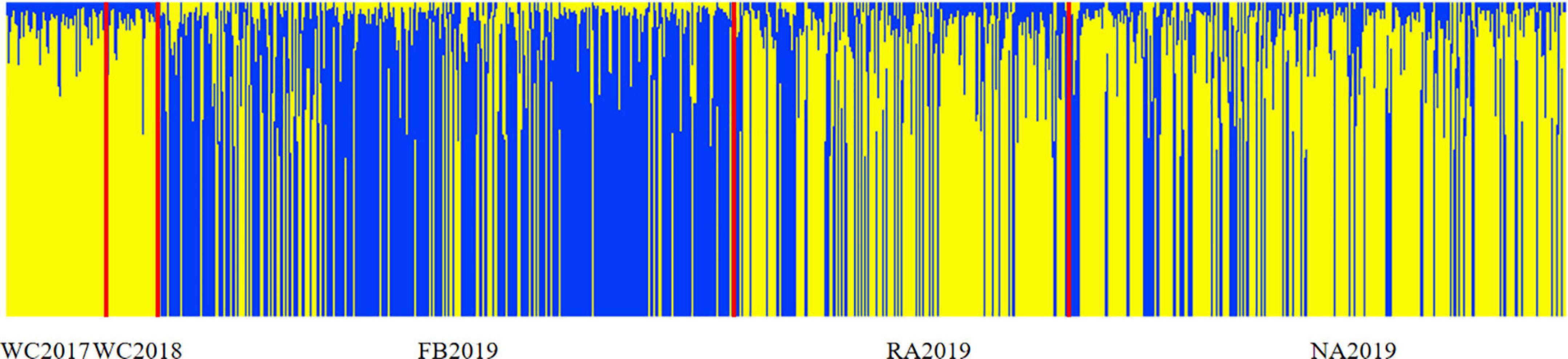
Figure 4. Results of Structure analysis for S. schlegelii stocks before and after stock enhancement. Structure plots showed individual assignments to K = 2, which were determined as the optimal cluster of K, estimated by Delta K.
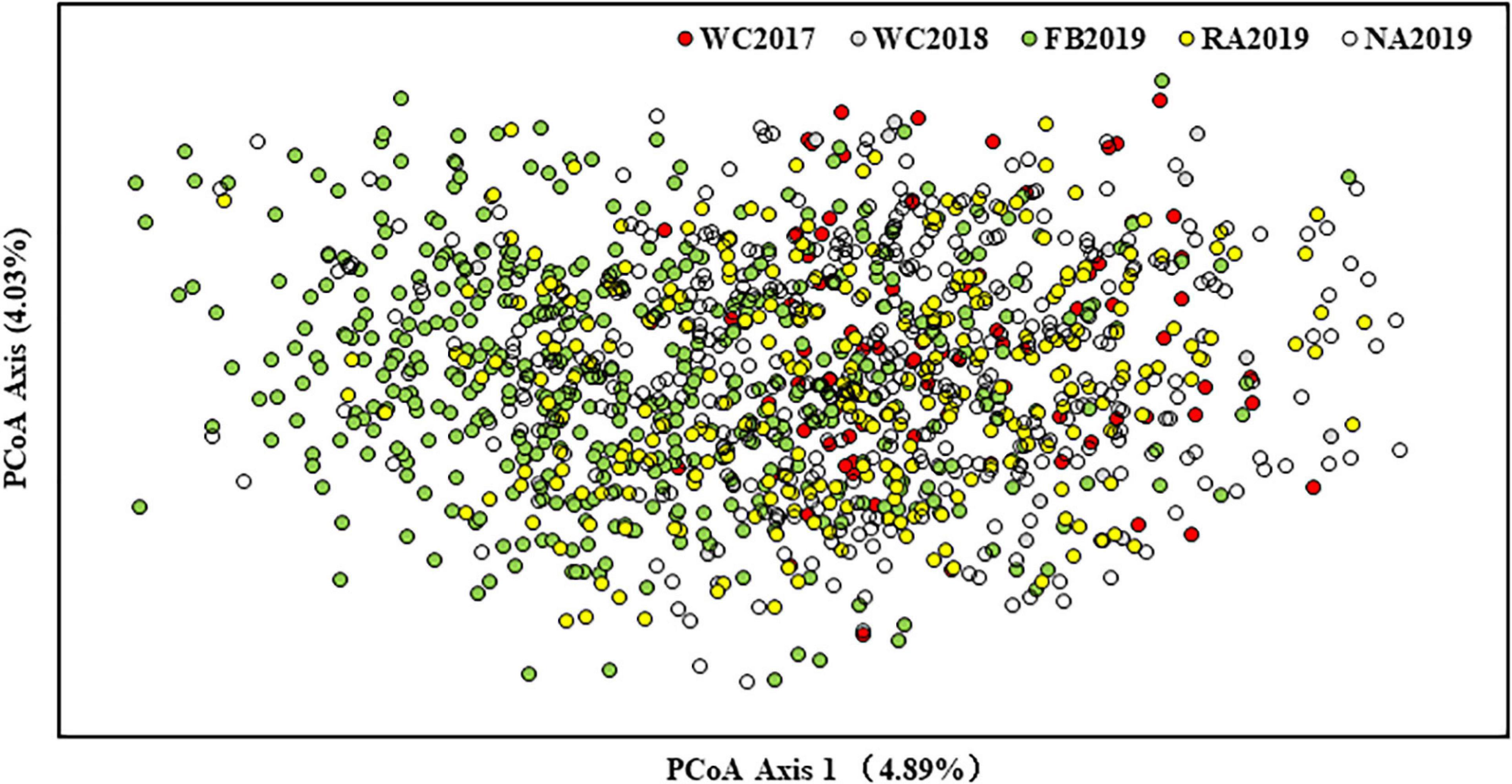
Figure 5. Genetic relationships among S. schlegelii stocks based on PCoA. Each data point (individual) is colored in accordance with the stock.
Growth Status and Morphology Comparison Between Hatchery-Reared Individuals and Their Wild Counterparts
The monthly growth difference comparison between RA2019 and NA2019 is shown in Supplementary Table 11. In December, the RA2019 stock exhibited significantly higher MDB, BW and CF than NA2019 (P < 0.05) and a significant difference was also detected in MDB and BW during the whole investigation period, from August to December. However, there was no difference in the other growth parameters. Our results seemed to indicate that the hatchery-reared individuals may adapt to the wild environment following release. They showed a similar growth status, when compared with wild individuals, and appeared to have grown even bigger than their wild counterparts 5 months after release. The difference in morphology between hatchery-reared and wild S. schlegelii is shown in Supplementary Tables 12, 13. In general, the results of the two independent sample tests indicated that there are five cases (BW/BL in December, PRL/HL in September, PLFB/BL in October, PCFL/BL in November and PCFB/BL in October), out of 60 morphology comparisons, that showed significant differences. However, across all investigation months (August to December), significant differences were only found in PRL/HL, ID/HL and PCFL/BL (P < 0.05). Moreover, there was no difference in number of fin rays between RA2019 and NA2019. Overall, there was no obvious morphology difference between hatchery-reared fish and their wild counterparts but the hatchery-reared fish were seemed to be larger, had a larger head and smaller paired fins.
Feeding Ecology Between Hatchery-Reared Individuals and Their Wild Counterparts
The differences in feeding ecology between the released and wild S. schlegelii in the natural environment are shown in Table 8. In each month, there was no significant difference in SCI values between the two groups. Moreover, for all individuals, level 0 of stomach fullness was not found, regardless of whether the individual was hatchery-born or wild-born. Our feeding ecology results also seemed to reveal that hatchery-reared individuals can adapt to the natural environment and showed similar feeding conditions to their wild counterparts.
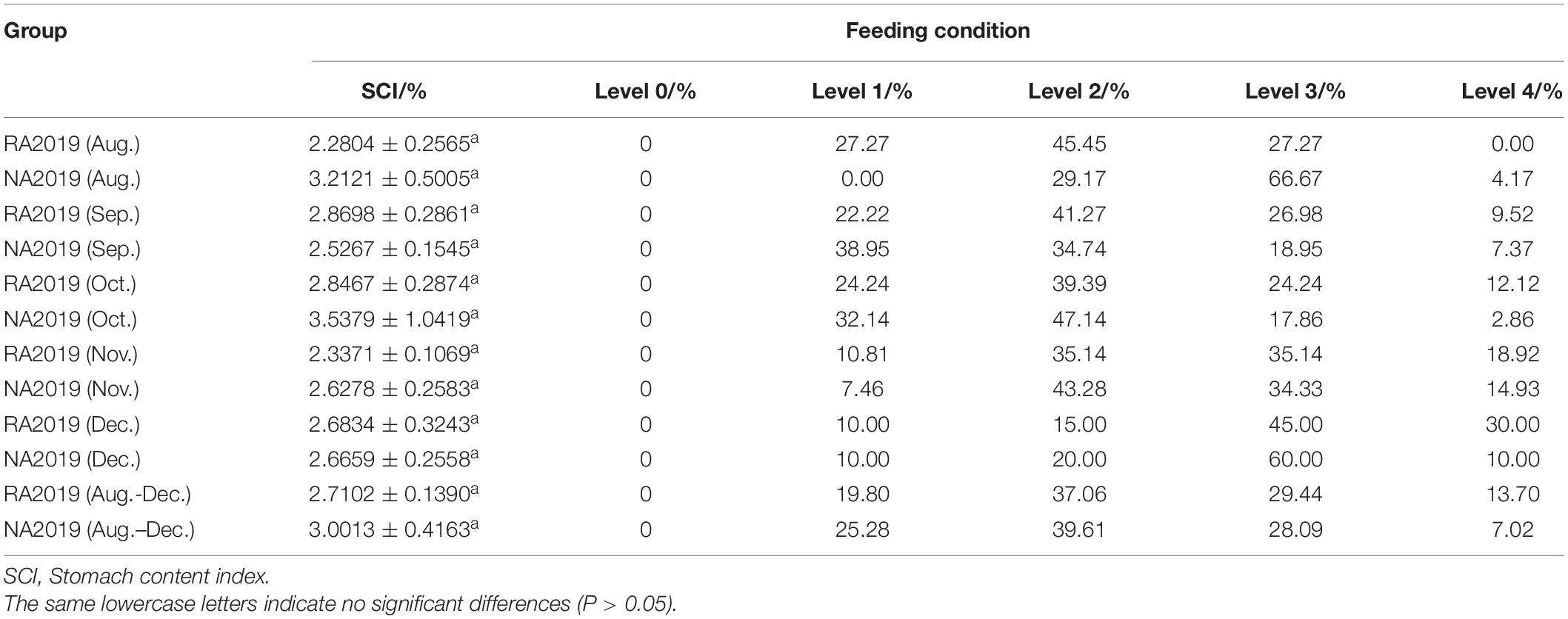
Table 8. Comparison of feeding conditions, each month, between RA2019 and NA2019 S. schlegelii stocks (mean ± S.E.).
Discussion
Animal populations, such as large mammals, birds, reptiles and fish living in freshwater or the ocean, have been threatened with serious losses over the past 50 years (Ripple et al., 2017). Supplementation of hatchery-reared individuals is being introduced in order to help support many species and restore resources (Lehnert et al., 2020). The Code of Conduct for Responsible Fisheries was adopted by FAO members worldwide in 1995 (FAO, 1995) and the responsible use of fisheries and aquaculture resources has gradually been recognized and accepted. Responsible approaches to marine stock enhancement have also been proposed, to cover the key steps of the enhancement process (Lorenzen et al., 2010; Grant et al., 2017). However, it is difficult to coordinate and practice all the principles of stock enhancement for several reasons (mainly due to cost, take time and effort). Nonetheless, a consensus has been drawn on the most important steps to consider when determining whether enhancement programs have been a success (Kitada, 2020). In this study, our research traced the first-time stock enhancement of S. schlegelii in the Liaodong peninsula. A total of 1,331 individuals were analyzed and will provide data for the discussion of the potential economic, genetic and ecological effects of enhancement programs.
Assessment of Potential Economic Impact Based on Recapture Contribution Rate
Microsatellite markers provide a non-invasive molecular marker for accurate identification of hatchery-reared individuals following release (Fang et al., 2021). Effective parentage determination, based on microsatellite markers, has been reported in finfish, such as brown trout (Salmo trutta) (Kalayci et al., 2020), crustaceans, such as the swimming crab (Portunus trituberculatus) (Liu et al., 2018), and mollusk, such as Pacific abalone (Haliotis discus) (Sekino et al., 2019). Accuracy of parentage determination in S. trutta, using the eight most informative microsatellite markers, obtained an identification accuracy of 96.08%, whilst CPE was estimated to be 99.9% using 15 microsatellite loci (Kalayci et al., 2020). In order to save costs, only female, wild-caught S. schlegelii were chosen as broodstock for the stock enhancement in China. There was no male genetic background available for our research. However, previous research has revealed that this difficulty can be eliminated using polymorphic and stable microsatellite loci. The CPE can reach ≥ 99% when using four microsatellite loci in single-parent parentage identification of released Chinese shrimp (Fenneropenaeus chinensis) and RCR was around 2% (Wang et al., 2014). Liu et al. (2018) also performed single-parent parentage identification in the enhancement of P. trituberculatus using six microsatellite loci. This yielded a CPE of > 99.97% and the RCR was 7.2%. In this study, the parentage assignment provided a high CPE value (99.8%) and 279 individuals were identified as RA2019 (Supplementary Table 3). This indicated that the nine loci used in our maternity analysis of S. schlegelii were suitable and an RCR of 39.3% was estimated. This high value may be due to the unique characteristics of this species, such as location fidelity, limited movement capability after larvae and good temperature tolerance (Chen et al., 2021). The high recapture rate of released S. schlegelii was also reported in Miyako Bay, Japan (13.8–22.9%), from 1999 to 2001 (Nakagawa, 2008). Empirically, large-size juveniles show excellent adaptation and have a lower chance of predation when released. The high RCR in our study suggests that S. schlegelii fish with a total length of 5 cm are suitable for release. Similar enhancement results were also found in P. olivaceus and F. chinensis (Yamashita, 1999; Hou et al., 2015). However, due to the relatively short interval time and the small size of our captured S. schlegelii mean that the economic return rate can only be estimated as 0.30, based on the data from December. But if the release activities maintain a high RCR of half the level of December (21.015%), the average body weight would be approximately 225 g in May of the next year. The fish could sell for 7.715 US dollars in 2020 and the primary economic return rate would be estimated to be 2.504. Similarly, it has been reported that the economic return rate of S. schlegelii was 1.32, when fish with a total length of 4.5 cm were released in Miyako Bay in 2007 (Chin et al., 2013). An economic return rate of 1.18 was reported, based on the S. schlegelii market return rate of 22.9% in 2001 (Nakagawa, 2008). It is notable that our economic return rate is likely to be an underestimation as this value was calculated using the local market sales, rather than the high values of recreational fishing or other indirect commercial values, such as coastal tourism (Miret-Pastor et al., 2020). Overall, this enhancement program in the Liaodong peninsula led to population restoration, despite recapture investigations favoring much longer periods over which to evaluate long-term economic return rate. Moreover, in consideration of absence of male parent information in S. schlegelii stock enhancement activities, the hatchery-reared offspring before releasing would also be analyzed in order to well apply to the parentage assignment in S. schlegelii evaluation further.
Assessment of the Potential Genetic Impact
To evaluate whether hatchery-reared S. schlegelii can maintain high genetic diversity or whether large-scale releases have a potential negative genetic impact on the local population at present, we compared the genetic divergence in different stocks. We observed that the S. schlegelii population showed relatively high genetic diversities before and after stock enhancement. Similar results have been reported in the Shandong peninsula, China (Wang et al., 2020). There was no significant difference in Na, Ne-allele and Ho between different stocks but the hatchery-reared stock, RA2019, exhibited a significantly lower Ar value than NA2019, after sequential Bonferroni correction (P < 0.05). This phenomenon may be due to different stock sizes but our analysis process compensated for uneven stock size by using the rarefaction method (Kalinowski, 2005). The loss of genetic diversity should be of concern when considering continuous large-scale enhancement of S. schlegelii in the future. Hence, long-term genetic evaluation is necessary. Previous research has revealed that the Ne is the key parameter of populations to maintain genetic variation and to keep the power in evolution and adaptation (Hoban et al., 2021). The Ne of hatchery-reared stock is often much smaller than that of their wild neighbors. Thus, long-term stock enhancement, using hatchery stocks, may cause a depletion of genetic diversity in local populations through direct replacement or genetic drift (Christie et al., 2012). Finally, restoration of wild populations with stock enhancement can reduce the Ne of the entire mixed population, based on the Ryman-Laikre effect (Hagen et al., 2021). But in this study, the S. schlegelii population had large Ne values before and after release and the confidence intervals were infinite (Pcrit = 0.05 or 0.02). This may be due to the large number of female S. schlegelii (489) that were used as broodstock. Previous research reported that multiple paternity occurs in the wild in this species (Yoshida et al., 2001; Gao et al., 2018) and other species in the Sebastes genus (Hyde et al., 2008). This is considered to be an adaptive mechanism to relieve the restriction of limited individual exchange caused by short-distance migration. Luo et al. (2021) found that one female can have multiple mates, with an average of 3.56 males, and can store their sperm in the zona pellucida of the ovary until the ovary is mature. Hence, the actual estimated Ne in the present study may be very large (>1823.1), which suggests that hatchery-reared fish can maintain high genetic diversities (Table 6). Previous research has also indicated that in populations with a large Ne, the effects of gene flow may dilute the artificial selection, which means that hatchery-reared fish have the chance to respond to environmental change after release (Charlesworth, 2009). This explains why low levels of genetic relatedness (mean relatedness < 0.07), low levels of inbreeding (Fis = −0.029∼0.065) and no genetic bottleneck effect were observed in the current S. schlegelii population.
The AMOVA, pairwise Fst and PCoA tests indicated that the S. schlegelii population in our study showed very weak genetic divergence, regardless of stock enhancement. The AMOVA result suggested that the major portion of genetic variance is found within stocks (>98%), rather than between stocks. These results were supported by the low pairwise Fst-values (0.003–0.027), strong gene flow (all Nm values > 9) and lack of separation of individuals by PCoA. Very weak but significant levels of genetic differentiation were found in each pairwise comparison of stocks (apart from WC2017 vs. WC2018 and WC2018 vs. NA2019), based on Fst estimation. While the controversial point was that Structure analysis indicated that the stock of WC 2018, WC2019 seemed to show a different genetic structure in relation to FB2019, though such divergence was weak as mentioned above. Previous research indicated that marine fish have different life stages, from egg, larvae, juvenile to adults (Miller and Kendall, 2009). Unlike juveniles and adults, which showed limited movement ability, the dispersal of S. schlegelii larvae could be extended along with the complex oceanographic current systems in the northwestern Pacific due to their aggregation around drifting seaweed during early development (Gao et al., 2016), Similar phenomenon was reported in their relatives, marbled rockfish (Sebasticus marmoratus) (Liu et al., 2019). The huge-scale stock enhancement of S. schlegelii in Shandong peninsula may possible introgress in Liaoning peninsula and alter the genetic composition to some degree, although the populations of S. schlegelii are considered to share the same gene pool and show no significant genetic differentiation in different locations of northwestern Pacific waters (Gao et al., 2016; Zhang et al., 2016). Furthermore, our collected WC2017 and WC2018 individuals are the mature samples with 4–6 years old and could be considered as the wild individuals of Liaodong peninsula that do not experience the genetic introgression derived from stock enhancement. This can explain the weak genetic structure difference between WC2017, WC2018, and FB2019. Previous reports indicated that long-term stock enhancements may lead to substantial genetic structure changes in local wild populations, as seen in coho salmon (O. kisutch) (Eldridge and Naish, 2007), chum salmon (O. keta) (Sato et al., 2014) and Atlantic salmon (S. salar) (Ozerov et al., 2016). Our research inferred that the local population of S. schlegelii can maintain high genetic diversities before and after stock enhancement in Liaodong peninsula and there was no obvious genetic differentiation in the local S. schlegelii population in the present study. However, potential genetic introgression should be concerned possibly as a result of stock enhancement and long-term genetic evaluations are required for comprehensively understanding the genetic impacts on local populations.
Assessment of Adaptability
Previous research has indicated that growth status, morphology and feeding ecology are good indicators, for the discussion of post-release adaptability and the potential ecological interactions with wild individuals (Gil et al., 2014). In our study, significant growth differences were detected between hatchery-reared and wild fish in MDB and BW, over the whole investigation period. This suggested that hatchery-reared individuals are likely to be larger in size and heavier than wild fish. We did not find any fish with empty stomachs, regardless of whether they were from the hatchery or wild-born, and the well-fed condition is likely to lead us to conclude that released fish use unutilized resources after dispersal from the release site. Both hatchery and wild stocks demonstrated a similar stomach content index (SCI%), which showed that hatchery-reared juveniles adapted well to capture their prey although previous results also showed that it had to take < 16 days before hatchery-reared fish could capture prey in the wild (Furuta et al., 1997; Taylor et al., 2006). Hence, it seemed like the release of hatchery stock in the present study did not lead to environmental capacity being reached and the competition for food between hatchery and wild stock was very limited. Further investigations are also necessary to investigate the interactions between hatchery-reared and wild individuals and to determine whether such releases reduce the body size of wild individuals in the long term. This is required as there is evidence that the release of Japanese Spanish mackerel (Scomberomorus niphonius), of larger size than wild individuals, led to a reduction in the growth rate of wild individuals, which were replaced by the hatchery individuals as the population exceeded environmental capacity (Nakajima et al., 2013). On the other hand, large-scale releases could simultaneously reduce the growth of both hatchery and wild fish, in addition to other competing species, if the population goes beyond the carrying capacity (Kitada, 2018). Moreover, fish populations exhibit extensive morphological plasticity (Sommer, 2020) and the variation in the phenotype of a population is influenced by both genetic and environmental factors (Fusco and Minelli, 2010). For example, when compared with hatchery-reared fish, some wild fish have been found to have more streamlined body shapes because of moving around a larger area, in fast-flowing waters (Pakkasmaa and Piironen, 2001). Several studies have revealed that hatchery-reared juvenile salmonids show lower survival rates than wild individuals (Jokikokko et al., 2006). Morphometric differences observed in hatchery fish may be one of the major causes for the changes in adaptability in the natural habitat (Vehanen and Huusko, 2011) and can have a negative effect (Von Cramon-Taubadel et al., 2005). In this study, we did not observe significant morphometric differences throughout the whole investigation period but significantly smaller head shapes (PRL/HL and ID/HL) and larger paired fins (PCFL/BL) were exhibited in wild S. schlegelii, when compared with hatchery-reared fish. Sebastes schlegelii is not good at long-distance swimming and the fish often hide in the reef area to ambush their prey. Hence, smaller head shapes and larger paired fins may help to accelerate swimming over short distances, for predation, and control the body position. As a large number of adult females from the local habitat were utilized as broodstock for this release, the genetic factors that affect morphology variations were extremely weak and can be ignored. Hence, divergence in the environment (natural or artificial hatchery) is the most likely cause of the different morphometrics observed during the S. schlegelii stock enhancement. Similar results were also reported in brown trout (Salmo trutta) (Sánchez-González and Nicieza, 2017) and gilthead sea bream (Sparus aurata) (Rogdakis et al., 2011).
Challenges in Sebastes schlegelii Resource Restoration
Recovery of depleted populations cannot rely on yearly releases and the stock enhancement activities are too complex for all aspects to be considered. Kitada (2020) reviewed the stock enhancement of red sea bream (Pagrus major) in Japan and found that recovery of nursery habitats and reduction in fishing efforts were more effective than restocking for wild populations. In China, releases of artificial reef in coastal areas, to rebuild seaweed communities, and restriction of trawling have been initiated. The summer fishing moratoria was extended from May to August in the Bohai Sea and the Yellow Sea. For stock enhancement of S. schlegelii, a large number of spawning broodstock is required to maintain a high Ne. Previous simulations have inferred that 1% genetic diversity will be lost after ten generations, even when Ne = 500 (Frankham et al., 2014). In this study, 486 adult females were used as broodstock to contribute to the next generation and benefit stock enhancement. It is also recommended that wild female broodstock are caught from local areas and are replaced each year. Most importantly, reducing the number of releases should be discussed in the future. In 2019, over two million S. schlegelii juveniles have been released into our research area of Liaodong peninsula, without a combination of a much larger scale of enhancement in Shandong peninsula. This seems too large a number to handle, for example, easy to lead genetic introgression, regardless of whether the population has reached environmental capacity. Less than 10% of the whole local population is the recommended proportion of hatchery-reared juveniles (Waples et al., 2012), to avoid replacement of wild genes and fitness degradation (Segovia-Viadero et al., 2016; Kitada, 2020).
Conclusion
This study indicates that the first-time stock enhancement of S. schlegelii in Liaodong peninsula is likely to has economic effects, brought about by a high RCR. This release of hatchery-reared individuals showed high genetic diversity. There was no obvious genetic differentiation was observed before and after stock enhancement, but genetic introgression should be concerned. The captured hatchery individuals showed good adaptability, as indicated by feeding condition and growth status, but seemed to have larger heads and smaller paired fins than wild S. schlegelii. However, such mass stocking of S. schlegelii must be evaluated to establish whether they pose long-term threats to wild populations, to ensure responsible stock enhancement of this highly valued species.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The animal study was reviewed and approved by the Care and Use of Laboratory Animals in Fishery Resources Enhancement Laboratory of Dalian Ocean University.
Author Contributions
QL and HY designed the research. ZW, WL, QZ, BL, TT, and ML performed the research. QL and BL analyzed the data. QL wrote the manuscript. YL and HY revised the manuscript. All authors read and approved the final version of the manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (42030408), the Scientific, Technological and Innovation Program of Dalian (2018J12SN069), the R&D Program of Liaoning Province (2019JH2/10200015), and the Liaoning Education Department Project (LJKZ0712).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.800607/full#supplementary-material
References
Araki, H., and Schmid, C. (2010). Is hatchery stocking a help or harm?: Evidence, limitations and future directions in ecological and genetic surveys. Aquaculture 308, S2–S11. doi: 10.1016/j.aquaculture.2010.05.036
Barreto, D. R., Leaniz, C. G., Verspoor, E., Sobolewska, H., Coulson, M., and Consuegra, S. (2019). DNA methylation changes in the sperm of captive-reared fish: a route to epigenetic introgression in wild populations. Mol. Biol. Evol. 36, 2205–2211. doi: 10.1093/molbev/msz135
Charlesworth, B. (2009). Effective population size and patterns of molecular evolution and variation. Nat. Rev. Genet. 10, 195–205. doi: 10.1038/nrg2526
Chen, Y., Shan, X., Ovando, D., Yang, T., Dai, F., and Jin, X. (2021). Predicting current and future global distribution of black rockfish (Sebastes schlegelii) under changing climate. Ecol. Indic. 128:107799. doi: 10.1016/j.ecolind.2021.107799
Chin, B. S., Nakagawa, M., Noda, T., Wada, T., and Yamashita, Y. (2013). Determining optimal release habitat for black rockfish, Sebastes schlegelii: examining growth rate, feeding condition, and return rate. Rev. Fish. Sci. 21, 286–298. doi: 10.1080/10641262.2013.837364
Christie, M. R., Marine, M. L., French, R. A., and Blouin, M. S. (2012). Genetic adaptation to captivity can occur in a single generation. P. Natl. Acad. Sci. USA. 109, 238–242. doi: 10.1073/pnas.1111073109
Chu, G., Jiang, L., Yu, H., Wang, Y., Jiang, H., and Zhang, Q. (2013). Family assessment with microsatellite markers in the ovoviviparous black rock fish Sebastes schlegelii. J. Oceanol. Limnol. 44, 670–676.
Davis, M. J., Woo, I., Ellings, C. S., Hodgson, S., Beauchamp, D. A., and Nakai, G. (2018). Integrated diet analyses reveal contrasting trophic niches for wild and hatchery juvenile Chinook Salmon in a large river delta. T. Am. Fish. Soc. 147, 818–841. doi: 10.1002/tafs.10088
Earl, D. A. (2012). STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 4, 359–361. doi: 10.1007/s12686-011-9548-7
Eldridge, W. H., and Naish, K. A. (2007). Long-term effects of translocation and release numbers on fine-scale population structure among coho salmon (Oncorhynchus kisutch). Mol. Ecol. 16, 2407–2421. doi: 10.1111/j.1365-294X.2007.03271.x
Excoffier, L., Laval, G., and Schneider, S. (2005). Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol. Bioinform. 1:117693430500100003. doi: 10.1177/117693430500100003
Falush, D., Stephens, M., and Pritchard, J. K. (2003). Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164, 1567–1587. doi: 10.1093/genetics/155.2.945
Fang, D., Luo, Y., Xu, D., Yang, X., and Wang, X. (2021). Relationship between genetic risk and stock enhancement of the silver carp (Hypophthalmichthys molitrix) in the Yangtze River. Fish. Res. 235:105829. doi: 10.1016/j.fishres.2020.105829
Fischer, J. R., Kwak, T. J., Flowers, H. J., Cope, W. G., Rash, J. M., and Besler, D. A. (2019). Growth, condition, and trophic relations of stocked trout in Southern Appalachian Mountain streams. T. Am. Fish. Soc. 148, 771–784. doi: 10.1002/tafs.10170
Fragkoulis, S., Christou, M., Karo, R., Ritas, C., Tzokas, C., Batargias, C., et al. (2017). Scaling of body-shape quality in reared gilthead seabream Sparus aurata L. Consumer preference assessment, wild standard and variability in reared phenotype. Aquac. Res. 48, 2402–2410. doi: 10.1111/are.13076
Frankham, R., Bradshaw, C. J., and Brook, B. W. (2014). Genetics in conservation management: revised recommendations for the 50/500 rules, Red List criteria and population viability analyses. Biol. Conserv. 170, 56–63. doi: 10.1016/j.biocon.2013.12.036
Furuta, S., Watanabe, T., Yamada, H., and Miyanaga, T. (1997). Changes in feeding condition of released hatchery-reared Japanese flounder, Paralichthys olivaceus, and prey mysid density in the coastal area of Tottori Prefecture. Nippon. Suisan. Gakk. 63, 886–891. doi: 10.2331/suisan.63.886
Fusco, G., and Minelli, A. (2010). Phenotypic plasticity in development and evolution: facts and concepts. Biol. Sci. 2010, 547–556. doi: 10.1098/rstb.2009.0267
Gaetano, J. (2018). Holm-Bonferroni sequential correction: an excel calculator. Availble online at: https://bit.ly/2VQOqcn (accessed October 5, 2020).
Gao, T., Ding, K., Song, N., Zhang, X., and Han, Z. (2018). Comparative analysis of multiple paternity in different populations of viviparous black rockfish, Sebastes schlegelii, a fish with long-term female sperm storage. Mar. Biodivers. 48, 2017–2024. doi: 10.1007/s12526-017-0713-4
Gao, T., Han, Z., Zhang, X., Luo, J., Yanagimoto, T., and Zhang, H. (2016). Population genetic differentiation of the black rockfish Sebastes schlegelii, revealed by microsatellites. Biochem. Syst. Ecol. 68, 170–177. doi: 10.1016/j.bse.2016.07.013
Gil, M. D. M., Palmer, M., Grau, A., Deudero, S., Alconchel, J. I., and Catalán, I. A. (2014). Adapting to the wild: the case of aquaculture-produced and released meagres Argyrosomus regius. J. Fish. Biol. 84, 10–30. doi: 10.1111/jfb.12241
Grant, W. S., Jasper, J., Bekkevold, D., and Adkison, M. (2017). Responsible genetic approach to stock restoration, sea ranching and stock enhancement of marine fishes and invertebrates. Rev. Fish. Biol. Fisher. 27, 615–649. doi: 10.1007/s11160-017-9489-7
Hagen, I. J., Ugedal, O., Jensen, A. J., Lo, H., Holthe, E., Bjøru, B., et al. (2021). Evaluation of genetic effects on wild salmon populations from stock enhancement. ICES. J. Mar. Sci. 78, 900–909. doi: 10.1093/icesjms/fsaa235
He, Y., Chang, Y., Bao, L., Yu, M., Li, R., Niu, J., et al. (2019). A chromosome-level genome of black rockfish, Sebastes schlegelii, provides insights into the evolution of live birth. Mol. Ecol. Resour. 19, 1309–1321. doi: 10.1111/1755-0998.13034
Hoban, S., Paz-Vinas, I., Aitken, S., Bertola, L., Breed, M. F., and Bruford, M. (2021). Effective population size remains a suitable, pragmatic indicator of genetic diversity for all species, including forest trees. Biol. Conserv. 253:108906. doi: 10.1016/j.biocon.2020.108906
Hou, Z., Gu, Q., Li, Z., Pan, G., Xie, Z., and Qiu, S. (2015). Comparative effect of Chinese shrimp Fenneropenaeus chinensis with different size released in Wuleidao Bay. Aquat. Sci. 34, 282–287.
Hyde, J. R., Kimbrell, C., Robertson, L., Clifford, K., Lyn, E., and Vetter, R. (2008). Multiple paternity and maintenance of genetic diversity in the live-bearing rockfishes Sebastes spp. Mar. Ecol. Prog. Ser. 357, 245–253. doi: 10.3354/meps07296
Janowitz-Koch, I., Rabe, C., Kinzer, R., Nelson, D., Hess, M. A., and Narum, S. R. (2019). Long-term evaluation of fitness and demographic effects of a Chinook Salmon supplementation program. Evol. Appl. 12, 456–469. doi: 10.1111/eva.12725
Jokikokko, E., Kallio-Nyberg, I., Saloniemi, I., and Jutila, E. (2006). The survival of semi-wild, wild and hatchery-reared Atlantic salmon smolts of the Simojoki River in the Baltic Sea. J. Fish. Biol. 68, 430–442. doi: 10.1111/j.0022-1112.2006.00892.x
Jonsson, B., Jonsson, N., and Jonsson, M. (2019). Supportive breeders of Atlantic salmon Salmo salar have reduced fitness in nature. Conserv. Sci. Pract. 1:e85. doi: 10.1111/csp2.85
Kalayci, G., Ozturk, R. C., and Altinok, I. (2020). Parentage assignment in Salmo trutta strains and their crossbreeds with known mating. Environ. Biol. Fish. 103, 1391–1399. doi: 10.1007/s10641-020-01030-2
Kalinowski, S. T. (2005). Hp-rare 1.0: a computer program for performing rarefaction on measures of allelic richness. Mol. Ecol. 5, 187–189. doi: 10.1111/j.1471-8286.2004.00845.x
Kalinowski, S. T., and Taper, M. L. (2006). Maximum likelihood estimation of the frequency of null alleles at microsatellite loci. Conserv. Genet. 7, 991–995. doi: 10.1007/s10592-006-9134-9
Kalinowski, S. T., Taper, M. L., and Marshall, T. C. (2007). Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 16, 1099–1106. doi: 10.1111/j.1365-294X.2007.03089.x
Karlsson, S., Saillant, E., Bumguardner, B. W., Vega, R. R., and Gold, G. R. (2008). Genetic identification of hatchery-released red drum in Texas bays and estuaries. N. Am. J. Fish. Manage. 28, 1294–1304. doi: 10.1577/M07-181.1
Kitada, S. (2018). Economic, ecological and genetic impacts of marine stock enhancement and sea ranching: A systematic review. Fish. Fish. 19, 511–532. doi: 10.1111/faf.12271
Kitada, S. (2020). Lessons from Japan marine stock enhancement and sea ranching programmes over 100 years. Rev. Aquacult. 12, 1944–1969. doi: 10.1111/raq.12418
Kitada, S., Nakajima, K., Hamasaki, K., Shishidou, H., Waples, R. S., and Kishino, H. (2019). Rigorous monitoring of a large-scale marine stock enhancement program demonstrates the need for comprehensive management of fisheries and nursery habitat. Sci. Rep-UK. 9:5290. doi: 10.1038/s41598-019-39050-3
Le Luyer, J., Laporte, M., Beacham, T. D., Kaukinen, K. H., Withler, R. E., and Leong, J. S. (2017). Parallel epigenetic modifications induced by hatchery rearing in a Pacific salmon. P. Natl. Acad. Sci. 114, 12964–12969. doi: 10.1073/pnas.1711229114
Lee, Y. C., Chang, P. H., Shih, C. H., Shiao, J. C., Tzeng, T. D., and Chang, W. C. (2021). The impact of religious release fish on conservation. Glob. Ecol. Conserv. 27:e01556. doi: 10.1016/j.gecco.2021.e01556
Lehnert, S. J., Baillie, S. M., MacMillan, J., Paterson, I. G., Buhariwalla, C. F., Bradbury, I. R., et al. (2020). Multiple decades of stocking has resulted in limited hatchery introgression in wild brook trout (Salvelinus fontinalis) populations of Nova Scotia. Evol. Appl. 13, 1069–1089. doi: 10.1111/eva.12923
Liu, Q., Cui, F., Hu, P., Yi, G., Ge, Y., Liu, W., et al. (2018). Using of microsatellite DNA profiling to identify hatchery-reared seed and assess potential genetic risks associated with large-scale release of swimming crab Portunus trituberculatus in Panjin, China. Fish. Res. 207, 187–196. doi: 10.1016/j.fishres.2018.05.003
Liu, Q., Liu, L., Song, N., Wang, X., and Gao, T. (2019). Genetic structure in the marbled rockfish (Sebastiscus marmoratus) across most of the distribution in the northwestern Pacific. J. Appl. Ichthyol. 35, 1249–1259. doi: 10.1111/jai.13972
Lorenzen, K., Leber, K. M., and Blankenship, H. L. (2010). Responsible approach to marine stock enhancement: an update. Rev. Fish. Sci. 18, 189–210. doi: 10.1080/10641262.2010.491564
Luo, G., and Zhang, Z. D. (2014). Current situation of the development in stock enhancement of China. China Fish 12, 37–39. doi: 10.1186/s40249-018-0454-z
Luo, Z., Dong, J., Zhang, Z., Xu, X., and Zhang, X. (2021). Microsatellite-based parentage analysis of offspring conducted in different regions of the black rockfish (Sebastes schlegelii) ovary. J. Fish. Sci. Chin. 28, 391–402.
Miller, B., and Kendall, A. W. (2009). Early life history of marine fishes. Oakland, CA: University of California Press.
Miret-Pastor, L., Molina-García, A., García-Aranda, C., and Herrera-Racionero, P. (2020). The connection between recreational fishing and the traditional fishing sector in the emerging area of marine tourism: challenges and opportunities for diversification with the European Fisheries Fund (EFF). ICES. J. Mar. Sci. 77, 2369–2374. doi: 10.1093/icesjms/fsz150
Molony, B. W., Lenanton, R., Jackson, G., and Norriss, J. (2005). Stock enhancement as a fisheries management tool. Rev. Fish. Biol. Fisher. 13, 409–432. doi: 10.1007/s11160-004-1886-z
Mori, H., Nakagawa, M., Soyano, K., and Koya, Y. (2003). Annual reproductive cycle of black rockfish Sebastes schlegeli in captivity. Fish. Sci. 69, 910–923. doi: 10.1046/j.1444-2906.2003.00707.x
Nakagawa, M. (2008). Studies on the stock enhancement technology of the black rockfish, Sebastes schlegeli. Bull. Fish. Res. Agency 25, 223–287.
Nakajima, K., Kitada, S., Yamazaki, H., Takemori, H., Obata, Y., Iwamoto, A., et al. (2013). Ecological interactions between hatchery and wild fish: a case study based on the highly piscivorous Japanese Spanish mackerel. Aquacult. Env. Interac. 3, 231–243. doi: 10.3354/aei00065
Onodera, T., Kikuchi, Y., Mizuma, T., Sato, K., and Fujita, N. (2001). Estimation of the total number of black rockfish caught by commercial and recreational fishing boat and surf fishing. Bull. Tohok. Natl. Fish. Res. Inst. 2001, 53–60.
Ozerov, M. Y., Gross, R., Bruneaux, M., Vähä, J. P., Burimski, O., Pukk, L., et al. (2016). Genomewide introgressive hybridization patterns in wild Atlantic salmon influenced by inadvertent gene flow from hatchery releases. Mol. Ecol. 25, 1275–1293. doi: 10.1111/mec.13570
Pakkasmaa, S., and Piironen, J. (2001). Water velocity shapes juvenile salmonids. Evol. Ecol. 14, 721–730. doi: 10.1023/A:1011691810801
Peakall, R., and Smouse, P. E. (2012). GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28, 2537–2539. doi: 10.1111/j.1471-8286.2005.01155.x
Pinter, K., Epifanio, J., and Unfer, G. (2019). Release of hatchery-reared brown trout (Salmo trutta) as a threat to wild populations? A case study from Austria. Fish. Res. 219:105296. doi: 10.1016/j.fishres.2019.05.013
Piry, S., Luikart, G., and Cornuet, J. M. (1999). Computer note. BOTTLENECK: a computer program for detecting recent reductions in the effective size using allele frequency data. J. Hered. 90, 502–503.
Ripple, W. J., Wolf, C., Newsome, T. M., Hoffmann, M., Wirsing, A. J., and McCauley, D. J. (2017). Extinction risk is most acute for the world’s largest and smallest vertebrates. P. Natl. Acad. Sc. 114, 10678–10683. doi: 10.1073/pnas.1702078114
Rogdakis, Y., Koukou, K., Ramfos, A., Dimitriou, E., and Katselis, G. (2011). Comparative morphology of wild, farmed and hatchery-released gilthead sea bream (Sparus aurata) in western Greece. Int. J. Fish Aquacult. 3, 1–9.
Rousset, F. (2008). Genepop’007: a complete reimplementation of the Genepop software for Windows and Linux. Mol. Ecol. Resour. 8, 103–106. doi: 10.1111/j.1471-8286.2007.01931.x
Salayo, N. D., Azuma, T., Castel, R. J. G., Barrido, R. T., Tormon-West, D. H. M., and Shibuno, T. (2020). Stock enhancement of abalone, Haliotis asinina, in multi-use buffer zone of Sagay. Marine Reserve in the Philippines. Aquaculture. 523:735138. doi: 10.1016/j.aquaculture.2020.735138
Sánchez-González, J. R., and Nicieza, A. G. (2017). Phenotypic convergence of artificially reared and wild trout is mediated by shape plasticity. Ecol. Evol. 7, 5922–5929. doi: 10.1002/ece3.3156
Sato, S., Templin, W. D., Seeb, L. W., Seeb, J. E., and Urawa, S. (2014). Genetic structure and diversity of Japanese chum salmon populations inferred from single-nucleotide polymorphism markers. T. Am. Fish. Soc. 143, 1231–1246. doi: 10.1080/00028487.2014.901251
Segovia-Viadero, M. Serrão, E. A. Canteras-Jordana, J. C. Gonzalez-Wangueemert, M. (2016). Do hatchery-reared sea urchins pose a threat to genetic diversity in wild populations? Heredity 116, 378–383. doi: 10.1038/hdy.2015.109
Sekino, M., Nakamichi, R., Kurokawa, T., and Hoshikawa, H. (2019). Reproductive success of released hatchery stocks in the Pacific abalone Haliotis discus hannai. Aquaculture 504, 291–299. doi: 10.1016/j.aquaculture.2019.01.027
Sommer, R. J. (2020). Phenotypic plasticity: from theory and genetics to current and future challenges. Genetics 215, 1–13. doi: 10.1534/genetics.120.303163
Stoner, A. W. (2019). Forty years of field and laboratory experiments with hatchery-reared Queen Conch: The case for conserving natural stocks. Rev. Fish. Sci. Aquac. 27, 490–516. doi: 10.1080/23308249.2019.1628705
Stoner, A. W., and Davis, M.(1994). Experimental outplanting ofjuvenile queen conch, Strombus gigas: comparison of wildand hatchery-reared stocks. Fish Bull. 92, 390–411.
Sulistyo, I., Fontaine, P., Rincarh, J., Gardeur, J. N., Migaud, H., Capdeville, B., et al. (2000). Reproductive cycle and plasma level of steroid in male eurasian perch (Perca fluviatilis). Aquat. Living Resour. 13, 99–106. doi: 10.1016/S0990-7440(00)00146-7
Taylor, M. D., Fielder, D. S., and Suthers, I. M. (2006). Spatial and ontogenetic variation in the diet of wild and stocked mulloway (Argyrosomus japonicus, Sciaenidae) in Australian estuaries. Estuar. Coast. 29, 785–793. doi: 10.1007/BF02786529
Van Oosterhout, C., Hutchinson, W. F., Wills, D. P., and Shipley, P. (2004). MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. 4, 535–538. doi: 10.1111/j.1471-8286.2004.00684.x
Vehanen, T., and Huusko, A. (2011). Brown trout Salmo trutta express different morphometrics due to divergence in the rearing environment. J. Fish. Biol. 79, 1167–1181. doi: 10.1111/j.1095-8649.2011.03093.x
Von Cramon-Taubadel, N., Ling, E. N., Cotter, D., and Wilkins, N. P. (2005). Determination of body shape variation in Irish hatchery-reared and wild Atlantic salmon. J. Fish. Biol. 66, 1471–1482. doi: 10.1111/j.0022-1112.2005.00698.x
Wang, L., Wu, Z., Wang, Y., Liu, M., Song, A., Liu, H., et al. (2020). Genetic assessment of a black rockfish, Sebastes schlegelii, stock enhancement program in Lidao Bay, China based on mitochondrial and nuclear DNA analysis. Front. Mar. Sci. 7:94. doi: 10.3389/fmars.2020.00094
Wang, W., Zhang, K., Luo, K., Xiao, G., Song, W., Kong, J., et al. (2014). Assessment of recapture rates after hatchery release of Chinese shrimp Fenneropenaeus chinensis in Jiaozhou Bay and Bohai Bay in 2012 using pedigree tracing based on SSR markers. Fisheries. Sci. 80, 749–755. doi: 10.1007/s12562-014-0743-9
Waples, R. S., and Do, C. H. I. (2008). LDNE: a program for estimating effective population size from data on linkage disequilibrium. Mol. Ecol. Resour. 8, 753–756. doi: 10.1111/j.1755-0998.2007.02061.x
Waples, R. S., Hindar, K., and Hard, J. J. (2012). Genetic risks associated with marine aquaculture. Washington, D.C: NOAA Technical Memorandum NMFS-NWFSC-119.
Yamashita, Y. (1999). “Release strategy for Japanese flounder fry in stock enhancement programs,” in Stock Enhancement and Sea Ranching, eds B. R. Howell, E. Moksness, and T. Svasand (London: Blackwell Science Ltd), 191–204.
Yasuike, M., Noda, T., Fujinami, Y., and Sekino, M. (2013). Tri-, tetra-and pentanucleotide-repeat microsatellite markers for the Schlegel’s black rockfish Sebastes schlegelii: the potential for reconstructing parentages. Conserv. Genet. Resour. 5, 577–581. doi: 10.1007/s12686-012-9856-6
Yoshida, K., Nakagawa, M., and Wada, S. (2001). Pedigree tracing of a hatchery-reared stock used for aquaculture and stock enhancement based on DNA markers. Fish Genet. Breed. Sci. 30, 27–35.
Zhang, H., Yanagimoto, T., Zhang, X. M., Song, N., and Gao, T. X. (2016). Lack of population genetic differentiation of a marine ovoviviparous fish Sebastes schlegelii in Northwestern Pacific. Mitochondrial. DNA. A. 27, 1748–1754. doi: 10.3109/19401736.2014.963797
Zhang, Y., Xu, Q., Alós, J., Liu, H., Xu, Q., and Yang, H. (2015). Short-term fidelity, habitat use and vertical movement behavior of the black rockfish Sebastes schlegelii as determined by acoustic telemetry. PLoS One 10:e0134381. doi: 10.1371/journal.pone.0134381
Keywords: adaptability, ecological effect, genetic effect, black rockfish, stock enhancement
Citation: Liu Q, Wang Z, Li W, Zhang Q, Liu B, Li M, Liu Y, Tian T and Yan H (2022) Genetic Stock Identification and Adaptability of Hatchery-Reared Black Rockfish, Sebastes schlegelii, Released Into the North Yellow Sea waters. Front. Mar. Sci. 9:800607. doi: 10.3389/fmars.2022.800607
Received: 23 October 2021; Accepted: 07 February 2022;
Published: 15 March 2022.
Edited by:
Liping Liu, Shanghai Ocean University, ChinaReviewed by:
Te-Hua Hsu, National Taiwan Ocean University, TaiwanZhihao Wu, Key Laboratory of Experimental Marine Biology, Institute of Oceanology (CAS), China
Lijuan Wang, Institute of Oceanology (CAS), China
Copyright © 2022 Liu, Wang, Li, Zhang, Liu, Li, Liu, Tian and Yan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongwei Yan, eWFuaG9uZ3dlaV8xOTg1QGhvdG1haWwuY29t
 Qi Liu
Qi Liu Ziwei Wang1,2
Ziwei Wang1,2 Binwei Liu
Binwei Liu Ying Liu
Ying Liu Hongwei Yan
Hongwei Yan