- 1Fisheries College of Jimei University, Xiamen, China
- 2Qingdao Key Laboratory of Mariculture Epidemiology and Biosecurity, Key Laboratory of Maricultural Organism Disease Control, Ministry of Agriculture, Function Laboratory for Marine Fisheries Science and Food Production Processes, Yellow Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
- 3Yangtze River Fisheries Research Institute, Chinese Academy of Fishery Sciences, Wuhan, China
- 4School of Science and Medicine, Lake Superior State University, Sault Ste. Marie, MI, United States
Covert mortality nodavirus (CMNV) recently became more prevalent, severely affecting farmed shrimps, and also other invertebrates and teleost fish, in the coastal areas of China. For better understanding of its prevalence and susceptibility of large yellow croaker Larimichthys crocea to CMNV, an epidemiological survey was carried out in the main producing areas (Ningbo, Zhejiang, China, and Ningde, Fujian, China) and its offshore feeding grounds in the Southern Yellow Sea. The collected cultured L. crocea individuals showed clinical symptoms such as anorexia and abnormal swimming and diagnosed for CMNV infection by using RT-nPCR assay. The positive rates for CMNV in the collected L. crocea samples from Ningbo, Ningde, and the Yellow Sea were 14.29% (4/28), 20.00% (7/35), and 16.67% (1/6), respectively. Furthermore, CMNV diagnosis using TaqMan probe-based reverse transcription quantitative PCR (TaqMan RT-qPCR) yielded positive rates of 25.00% (7/28), 22.86% (8/35), and 16.67% (1/6) respectively in the above-mentioned samples, and with a range of 12.73–3,108.33 of CMNV copy numbers/μg total RNA. Phylogenetic tree analysis based on the RNA-dependent RNA polymerase (RdRp) and capsid protein genes showed that CMNV isolates from L. crocea samples were clustered tightly with the original isolate of CMNV from Penaeus vannamei. The histopathological studies indicated that severe cellular necrosis occurred in the heart, liver, spleen, eye, and gill of naturally infected fish. Stronger positive signals for CMNV-specific probes appeared in the necrotic cells and tissues in the in situ hybridization (ISH) analysis. Transmission electron microscopy (TEM) revealed the presence of numerous 30 nm diameter CMNV-like viral particles. The purified CMNV suspension could cause a significant cytopathic effect (CPE) in epithelioma papilloma cyprini (EPC) cells and resulted in the formation of typical inclusion bodies, and also visible CMNV particles around the lysed EPC cells under the TEM. Taken together, all results of this study demonstrated that L. crocea is susceptible to CMNV. The prevalence of CMNV and its potential impacts on the wild population of L. crocea see the need for further investigations.
Introduction
Covert mortality nodavirus (CMNV), an emerging alphanodavirus, severely impacted the shrimp aquaculture industries in China and Southeast Asian countries in the past decade (Zhang et al., 2014; Flegel, 2015; Thitamadee et al., 2016). Epizootic investigations recorded CMNV prevalence in farmed shrimp at the annual rates of 45.93%, 27.91%, 20.85%, 26.8%, and 16.3% respectively during the period of 2013–2017 in the major shrimp producing provinces in China (Zhang et al., 2017). Except for wide geographical prevalence, CMNV has also shown much broader host ranges. Previous studies had confirmed that CMNV could infect the main cultured crustaceans, including Penaeus vannamei, Penaeus chinensis, Marsupenaeus japonicus, Penaeus monodon, and Macrobrachium rosenbergii, and result in multiple tissue or organ damages (Zhang et al., 2014, 2017). Recent studies demonstrated that CMNV could cross species barriers to infect a variety of cultured and wild fish, such as cultured Japanese flounder Paralichthys olivaceus, wild gobiid fish Mugilogobius abei, and wild crucian carp Carassius auratus, which were collected from shrimp ponds or drainage channel affected by CMNV (Zhang et al., 2018; Wang et al., 2019a,b). More recently, varying prevalence (from 7 to 18%) of CMNV infection was reported in wild small yellow croaker Larimichthys polyactis sampled from coastal water of China in 2018–2019 (Xu et al., 2021). Furthermore, zebrafish (Danio rerio) also could be artificially infected with CMNV and the infected fish would exhibit abnormal spiraling swimming activity, with a cumulative mortality rate of 53.33% within 14 days of postchallenge injection (Wang et al., 2021). The above studies revealed that CMNV has been widely distributed in the farmed aquatic animals of coastal areas, and also the wild hosts from the offshore waters. The appearance of certain negative effects on teleost fishes has raised concerns about the prevalence of CMNV in both cultured and wild fish from the coastal areas.
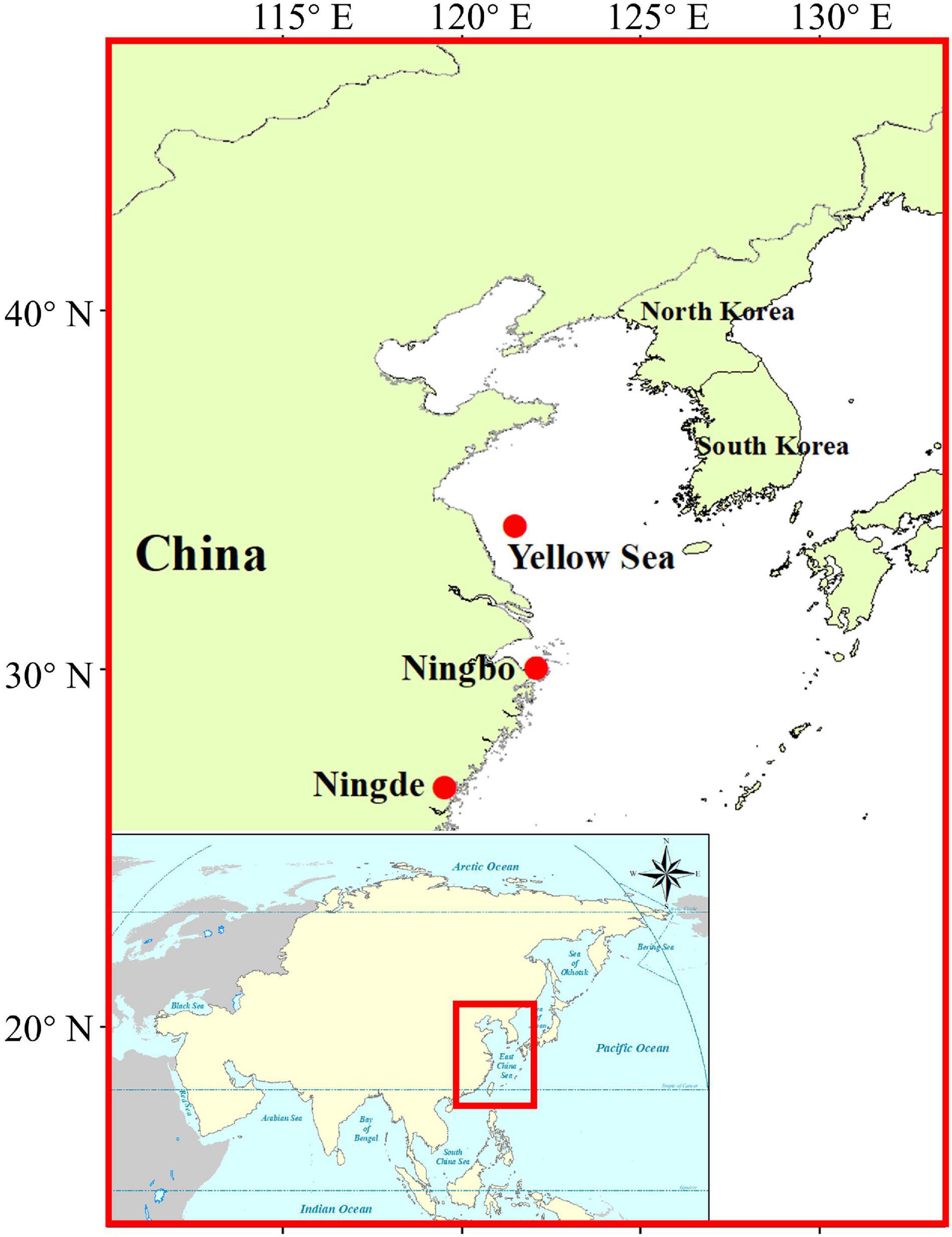
The large yellow croakers (Larimichthys crocea) mainly distribute in the southeast coast of China, including the Southern Yellow Sea, the East China Sea, and the Northern South China Sea (Li and Yue, 2019; Wang et al., 2019). Due to overfishing, the natural resources of L. crocea have been seriously damaged and even on the verge of exhaustion (Chen et al., 2020). In recent years, with the success of artificial breeding, the L. crocea aquaculture industry has rapidly developed and L. crocea has become one of the most economically important marine fish species, especially in the provinces of Fujian (mainly in Ningde) and Zhejiang (mainly in Ningbo), the top two largest L. crocea producing provinces in China (Ministry of Agriculture and Rural Affairs of the People’s Republic of China, 2021). However, more and more new emerging and reemerging infectious diseases, caused by bacteria, parasites, and viruses, have become the most important restricting factor for the healthy and sustainable development of L. crocea mariculture in China (Chen et al., 2003; You and Li, 2003; Shen et al., 2004; Wang et al., 2010; An, 2013; Xu et al., 2015). With big concern on the recent extremely high prevalence of CMNV in coastal aquaculture animals in China, in this study, we investigated the potential CMNV prevalence, and its histopathological characteristics in farmed and wild L. crocea both in the main L. crocea farming areas (Ningbo, Zhejiang, China and Ningde, Fujian, China) and on offshore feeding grounds in the Southern Yellow Sea.
Materials and Methods
Sample Collection
A total of 28 and 35 cultured L. crocea (20–30 cm in length) were collected from marine cages in Ningbo, Zhejiang, and Ningde, Fujian, respectively, during the period from 2019 to 2021 (Figure 1). Six wild L. crocea (10–14 cm in length) were collected from the southern Yellow Sea (34°N, 121.5°E) of China by bottom trawl in 2019 (Figure 1). The collected cultured L. crocea individuals showed clinical symptoms such as anorexia and abnormal swimming. The heart, liver, kidney, spleen, eye, and gill tissues were dissected from each individual, part of which was stored in 4% paraformaldehyde (4% PFA) solution (Sinopharm, Beijing, China) for in situ RNA hybridization and histopathological analysis, or in the RNAstore solution (Tiangen, Beijing, China) for molecular biological detection and 2.5% glutaraldehyde solution (Sinopharm, Beijing, China) for transmission electron microscopy (TEM) analysis, respectively. The remaining part was then stored in dry ice for virus purification. The collected wild L. crocea samples were stored in liquid nitrogen and brought back to the laboratory for further analysis.
Virus Isolation
The frozen tissues of L. crocea were first homogenized thoroughly in TN buffer (20 mM Tris/HCl, 400 mM NaCl, pH 7.4) and then subjected to the purification of viral particles according to a previously reported protocol (Xu et al., 2020). The purified virus particles were filtered through a 0.22-μm membrane prior to further use.
Cell Inoculation
The fish cell line [epithelioma papilloma cyprini (EPC)] was used for virus proliferation (Xiao et al., 2012). EPC cells were cultured in Eagle’s minimal essential medium (EMEM, Sigma-Aldrich, United States) in a 25-cm2 flask (Corning, United States) with 10% FBS at 25°C. One hundred milliliter of purified virus suspension was inoculated into a confluent monolayer of EPC cells. The same volume of EMEM was used as a mock infection in EPC cells as control. After adsorption for 1 h at 25°C, 5 mL of medium supplemented with 2% FBS was added into the flasks, and the infected cells were incubated at 25°C in the incubator (SANYO, Japan). The infected cells were examined daily under an inverted phase-contrast microscope (Nikon, Japan). When 70–80% of cultured cells showed cytopathic effect (CPE), the cultures were collected in 2.5% glutaraldehyde for TEM analysis or in an RNAstore solution for RNA extraction.
RNA Extraction
Approximately 40 mg tissues or cell cultures fixed in RNAstore solution and liquid nitrogen were used for total RNA extraction by TIANamp Marine Animal RNAprep pure Tissue Kit (TIANGEN Biotech, Beijing, China) according to the manufacturer’s instructions. The concentration and purity of the extracted RNA was determined by Nanodrop 2000c (Thermo Scientific, Waltham, MA, United States).
Detection of Covert Mortality Nodavirus by RT-nPCR
By using the total RNA as template, the first-step PCR of the RT-nPCR was conducted by using the PrimeScript One Step RT-PCR Kit (Version 2.0) (TaKaRa, Dalian, China) in total of 25 μL reaction mixture, including 12.5 μL first-step buffer, 1 μL PrimeScript first-step enzyme mix, 1 μL forward primer (Noda-F1: 5′-AAATACGGCGATGACG-3′, 10 μM), 1 μL reverse primer (Noda-R1: 5′-ACGAAGTGCCCACAGAC-3′, 10 μM), 1 μL RNA template, and 8.5 μL ddH2O. The programmed reaction was then performed for the reverse transcription at 50°C for 30 min, predenaturation at 94°C for 3 min followed by 25 cycles of denaturation at 94°C for 30 s, annealing at 52°C for 30 s, extension at 72°C for 50 s, and the final extension at 72°C for 5 min. The second-step PCR of the RT-nPCR was carried out by using the TaKaRa Premix Taq™ (Ex Taq™ Version 2.0) in a 25 μL reaction mixture, which included 12.5 μL Premix Taq, 1 μL forward primer (CMNV-D-F1: 5′-TCGCGTATTCGTGGAT-3′, 10 μM), 1 μL reverse primer (CMNV-D-R1: 5′-TAGGGTCAAAAGGTGTAGT-3′, 10 μM), 1 μL product of the first-step RT-PCR, and 9.5 μL ddH2O. The second-step PCR was performed in a similar way as the first-step PCR except for the incubation for the reverse transcription.
Two target CMNV-specific RNA-dependent RNA polymerase (RdRp) gene fragments of 619 and 413 bp (GenBank accession number KM112247) were amplified by the first- and second-step PCRs, respectively. The PCR products were sequenced by using the Sanger sequencing method.
Detection of Covert Mortality Nodavirus by TaqMan Probe-Based Reverse Transcription Quantitative PCR
All the samples for TaqMan probe-based reverse transcription quantitative PCR (TaqMan RT-qPCR) were analyzed and quantified in three duplicates for targeting the same gene as previously described (Li et al., 2018).
Histopathological Assay
The samples that were positive for CMNV from above TaqMan RT-qPCR assay were subjected to histopathological analysis with hematoxylin and eosin-phloxine (H&E) staining by using regular histological methods as previously described (Lightner, 1996).
In situ RNA Hybridization
The samples positive for CMNV in TaqMan RT-qPCR assay were subjected to in situ hybridization (ISH) analysis according to the previous procedures (Piette et al., 2008; Chen et al., 2014; Zhang et al., 2017). Nuclear Fast Red solution (Sigma-Aldrich, St. Louis, MO, United States) was added dropwise on the surface of the sections and dyed the nucleus for 2 min for counterstain postcolor development of BCIP/NBT in the ISH. The sections were visualized and photographed by using the Nikon Eclipse E80i microscope (Nikon Co., Tokyo, Japan).
Transmission Electron Microscopy
The samples preserved in 2.5% glutaraldehyde with the corresponding number were taken out and treated with 1% osmic acid for 2 h, and then dehydrated and embedded in Spurr’s resin and stained with uranyl acetate and lead citrate following the protocols of Panphut et al. (2011). All specimens were visualized under JEOL JEM-1200 transmission electron microscopy at 80 kV.
Cloning and Analysis of Covert Mortality Nodavirus Genes
Five and two pairs of primers (Table 1) were designed based on the original sequences of CMNV RdRp and capsid protein genes, respectively (GenBank accession numbers MT270124 and MT270123). cDNAs were synthesized by using the extracted total RNA of L. crocea samples which were positive for CMNV with SMART® MLV-Reverse Transcriptase (TaKaRa) according to the procedure described by Xu et al. (2020). The reaction mixture and procedures for CMNV genes cloning PCR were referred to the previous report (Xu et al., 2020) with modifications of annealing temperatures and extension time (Table 1). The PCR product was subjected to electrophoresis on a 2% agarose gel and sequenced by the Sanger method (Sangon Biotech, Shanghai, China). Sequence analysis was conducted by using BioEdit (Ver. 7.1.3) and BLAST.1 The phylogenetic tree based on the RdRp and capsid protein genes was generated using the neighbor-joining method of MEGA 7.0 (Tamura et al., 2011).
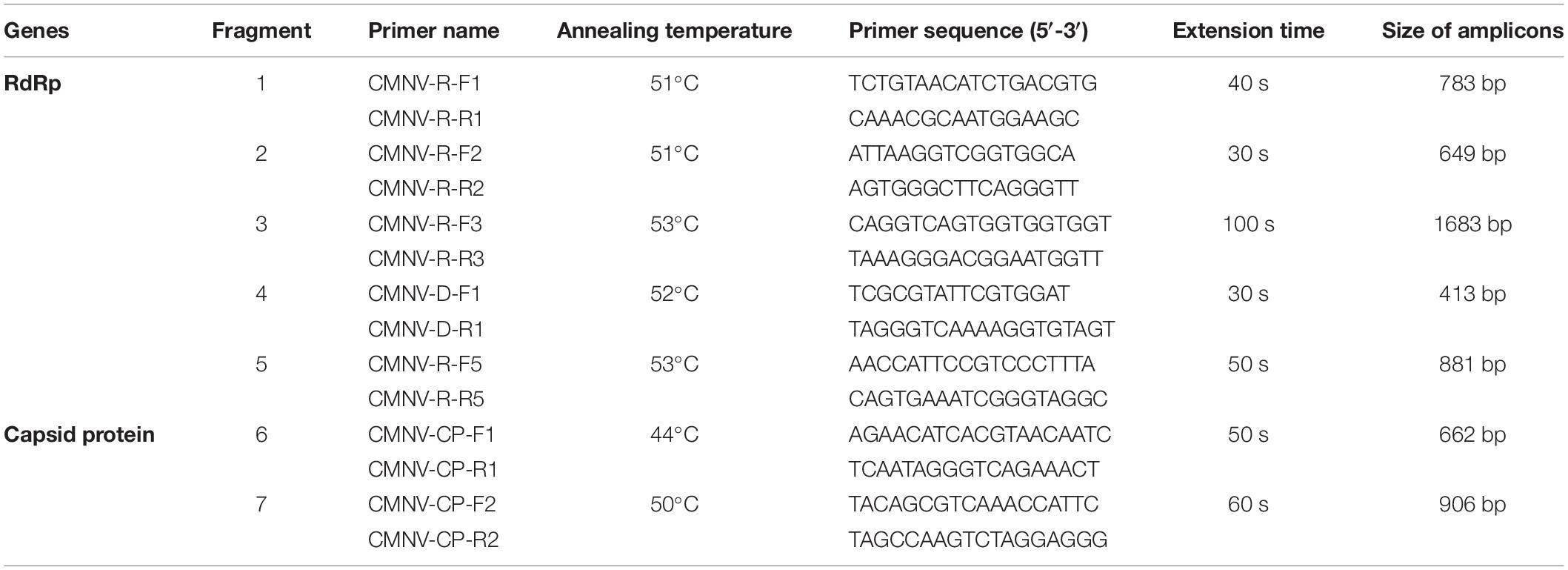
Table 1. PCR primer sequences, annealing temperature, and extension duration used to amplify the RdRp and capsid protein gene sequences of CMNV.
Results
Detection of Covert Mortality Nodavirus by RT-nPCR and TaqMan Probe-Based Reverse Transcription Quantitative PCR
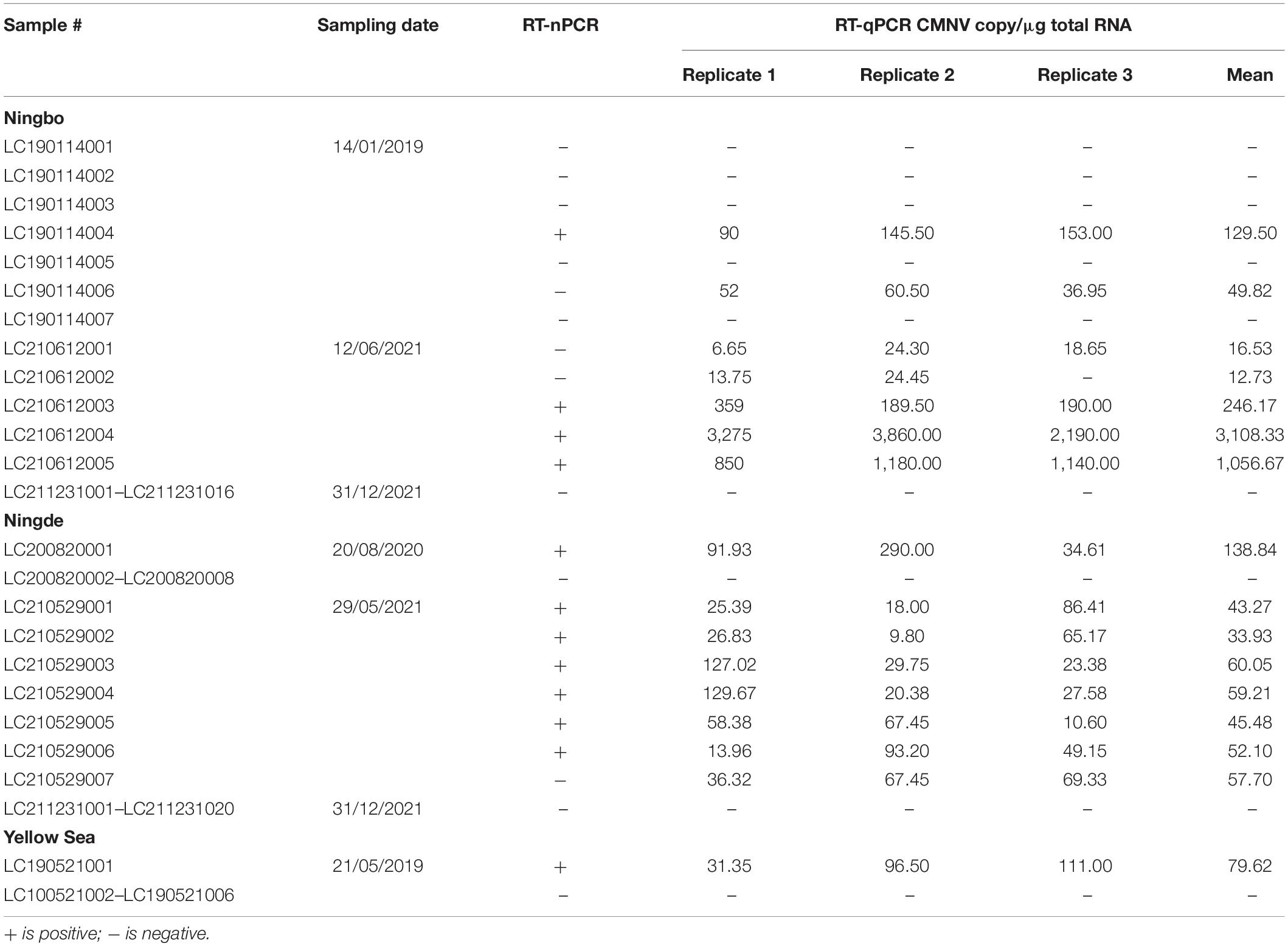
The results of RT-nPCR assay indicated that CMNV-positive rates of the collected L. crocea samples from Ningbo, Ningde, and the Yellow Sea were 14.29% (4/28), 20.00% (7/35), and 16.67% (1/6), respectively. The RT-qPCR test yielded 25.00% (7/28), 22.86% (8/35), and 16.67% (1/6) CMNV-positive rates in L. crocea collected from Ningbo, Ningde, and the Yellow Sea, respectively, with a range of 12.73–3,108.33 copy numbers/μg of total RNA (Table 2). The standard curve showed that the RT-qPCR had a high correlation coefficient (R2 = 0.999) within the range of 2.21 × 109 – 2.21 × 100 RNA copies/reaction (Supplementary Figure 1).
Detection of Covert Mortality Nodavirus in Larimichthys crocea Samples by in situ Hybridization
For further confirmation of the CMNV infection in the CMNV-positive L. crocea samples, ISH was conducted by using a CMNV-specific RNA probe. The results showed that positive purple hybridization signals of CMNV probe could be observed in the cardiac muscle (Figures 2A,B), hepatocytes (Figures 2E,F), spleen (Figures 2I,J), bipolar cell of the retina (Figures 2M,N), and chloride cells of gill filament (Figures 2Q,R). No positive hybridization signals appeared on the sections from the same samples without CMNV-specific RNA probes, or from the CMNV negative samples determined by RT-nPCR (Supplementary Figures 2, 3).
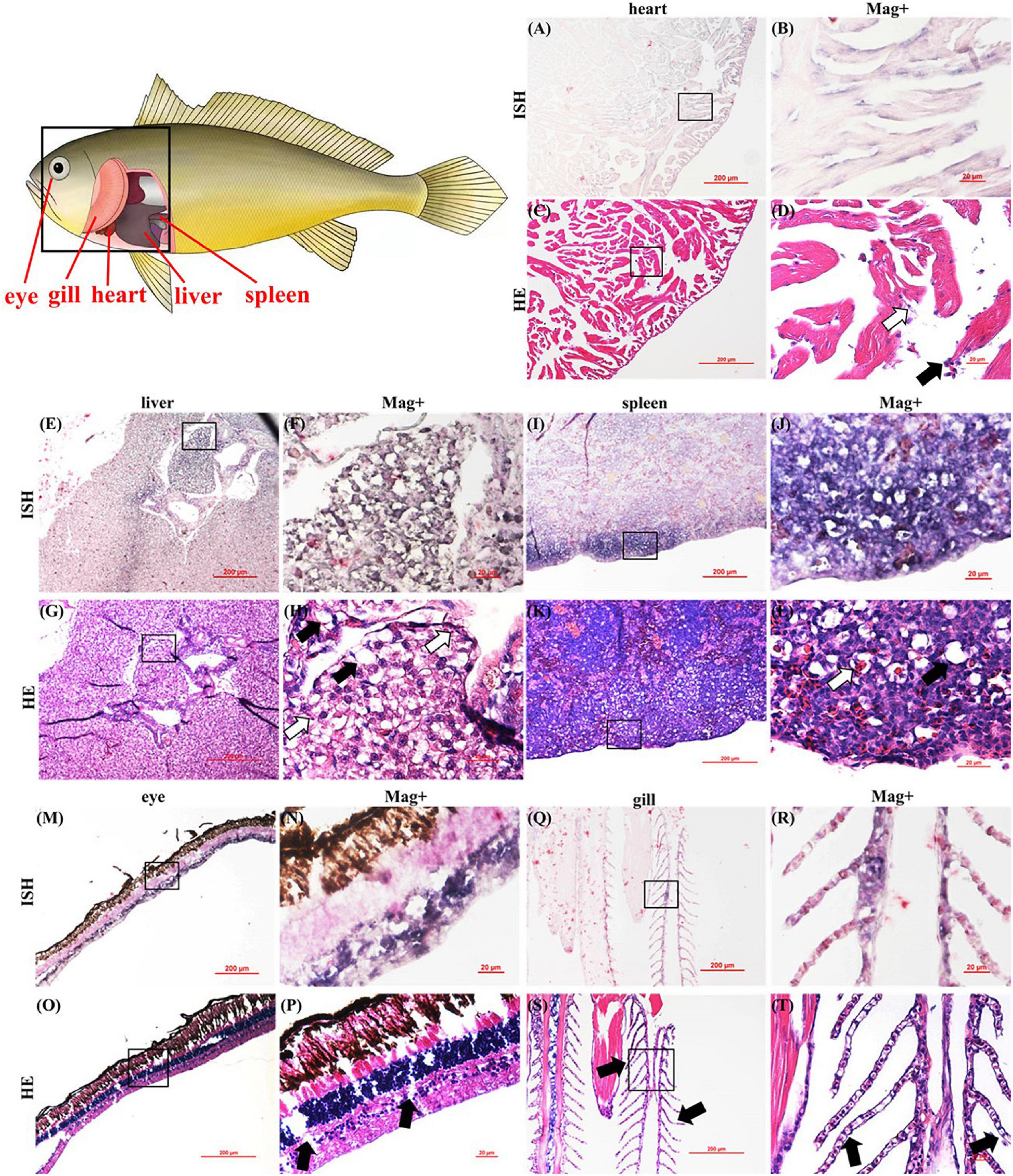
Figure 2. Pattern diagram of Larimichthys crocea and micrographs of in situ hybridization (ISH) and HE staining for heart, liver, spleen, eye, and gill of the L. crocea naturally infected with CMNV. (A) Micrograph of ISH for the heart. (B) Magnified micrograph of the cardiac muscle in the black frame in (A). Note the purple signals of CMNV probe in the necrotic cardiac muscle. (C) Micrographs of HE staining for the heart. (D) Magnified micrograph of the cardiac muscle in the black frame in (C). Note the dissolved necrosis (pointed by white arrows) and mild hemocyte infiltration in the fibromuscular stroma (pointed by black arrows) of the cardiac muscle. (E) Micrograph of ISH for the liver of the L. crocea. (F) Magnified micrograph of the zone in the black frame in (E). (G) Micrographs of HE staining for the liver. (H) Magnified micrograph of the zone in the black frame in (G). Note the serious vacuolation (pointed by black arrows) and karyopyknosis (pointed by white arrows) of the hepatocytes. An intense CMNV probe hybridization signal could be observed at necrotic hepatocytes. (I) Micrographs of ISH for spleen. (J) Magnified micrograph of the zone in the black frame in (I). (K) Micrographs of HE staining for spleen. (L) Magnified micrograph of the zone in the black frame in (K). Note the moderate vacuolation (pointed by black arrows) and degenerated erythrocytes (pointed by white arrows) of the spleen. Intensive purple hybridization signals of CMNV probe could be observed at the spleen. (M) Micrographs of ISH for the eye. (N) Magnified micrograph of the zone in the black frame in (M). (O) Micrographs of HE staining for the eye. (P) Magnified micrograph of the zone in the black frame in (O). Note the serious vacuolation (pointed by black arrows) at the bipolar cells layer. Intensive purple hybridization signals of the CMNV probe could be observed at the bipolar cells layer. (Q) Micrographs of ISH for gill. (R) Magnified micrograph of the zone in the black frame in (Q). (S) Micrographs of HE staining for gill. (T) Magnified micrograph of the zone in the black frame in (S). Note the disorganized secondary lamella (pointed by black arrows). Intensive purple hybridization signals of the CMNV probe could be observed at the secondary lamella. Scale bars (A,C,E,G,I,K,M,O,Q,S), 200 μm and (B,D,F,H,J,L,N,P,R,T), 20 μm.
Histopathology of Larimichthys crocea Infected by Covert Mortality Nodavirus
The impact of CMNV infection on the heart, eye, liver, spleen, and gill tissues of L. crocea was investigated by histopathology of H&E-stained sections as shown in Figure 2. Moderate dissolution-like necrotic muscle fibers and hemocyte infiltration in the fibromuscular stroma were observed in cardiac muscle (Figures 2C,D). Severe vacuolation and karyopyknosis were found in hepatocytes (Figures 2G,H) and bipolar cells layer of the retina (Figures 2O,P). Moderate vacuolation and degenerated erythrocytes were observed in the spleen (Figures 2K,L). Disorders of the secondary lamella were also viewed in the gills (Figures 2S,T).
Detection of Covert Mortality Nodavirus in Larimichthys crocea Samples by Transmission Electron Microscopy
Under the TEM, massive CMNV-like particles with approximate 28–32 nm in diameter and spherical to icosahedral shapes were observed in the blood cells in the kidney (Figures 3A–C) and cardiac muscle cells of the heart of the L. crocea (Figures 3D–F).
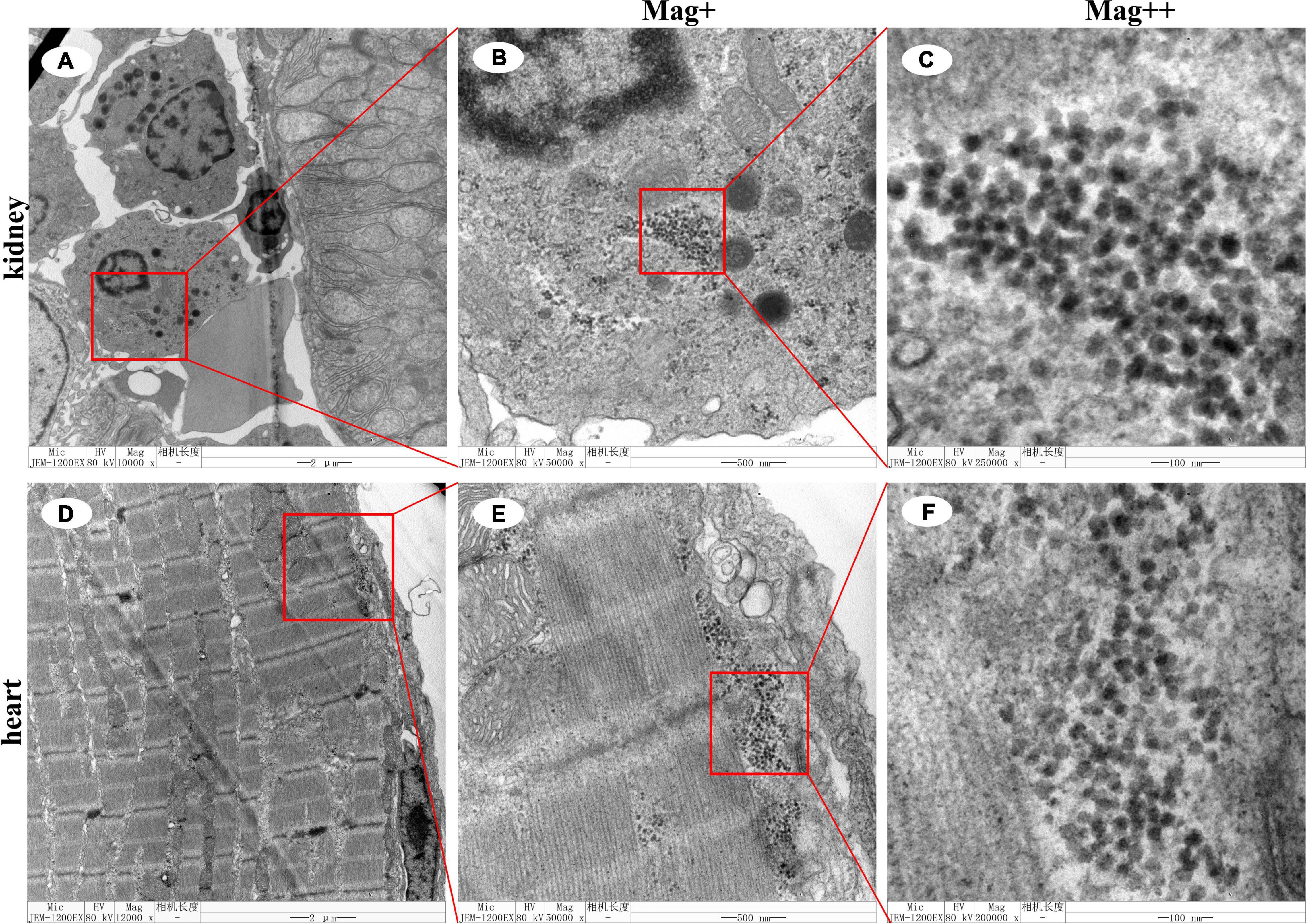
Figure 3. Transmission electron microscopic micrographs of ultrathin section of kidney and heart of the Larimichthys crocea. (A) The blood cells of the kidney of the L. crocea. (B) Magnified micrograph of the blood cells in the red frame in (A). Note mass CMNV-like particles distributed in the blood cells. (C) Magnified micrograph of the CMNV-like particles in the red frame in (B). (D) The cardiac muscle of the heart of the L. crocea. (E) Magnified micrograph of the cardiac muscle in the red frame in (D). Note mass CMNV-like particles distributed in the cardiac muscle. (F) Magnified micrograph of the CMNV-like particles in the red frame in (E). Scale bars, (A,D), 2 μm, (B,E), 500 nm, and (C,F), 100 nm.
Nucleotide Sequence Analysis of RNA-Dependent RNA Polymerase and Capsid Protein Genes of Covert Mortality Nodavirus Isolated From Larimichthys crocea
Five amplicons of the nucleotide sequence of RdRp, with the sizes of 783, 649, 1,683, 413, and 881 bp were obtained by PCR (Figure 4A, lanes 1–5). Two amplicons of the nucleotide sequence of capsid protein gene, with the sizes of 662 and 906 bp, were generated by PCR (Figure 4A, lanes 6 and 7). Then, by sequencing and assembling the overlapping amplicons, the nucleotide sequences of RdRp and capsid protein genes of CMNV isolated from L. crocea were determined, which were 3,132 and 1,314 bp in length, respectively.
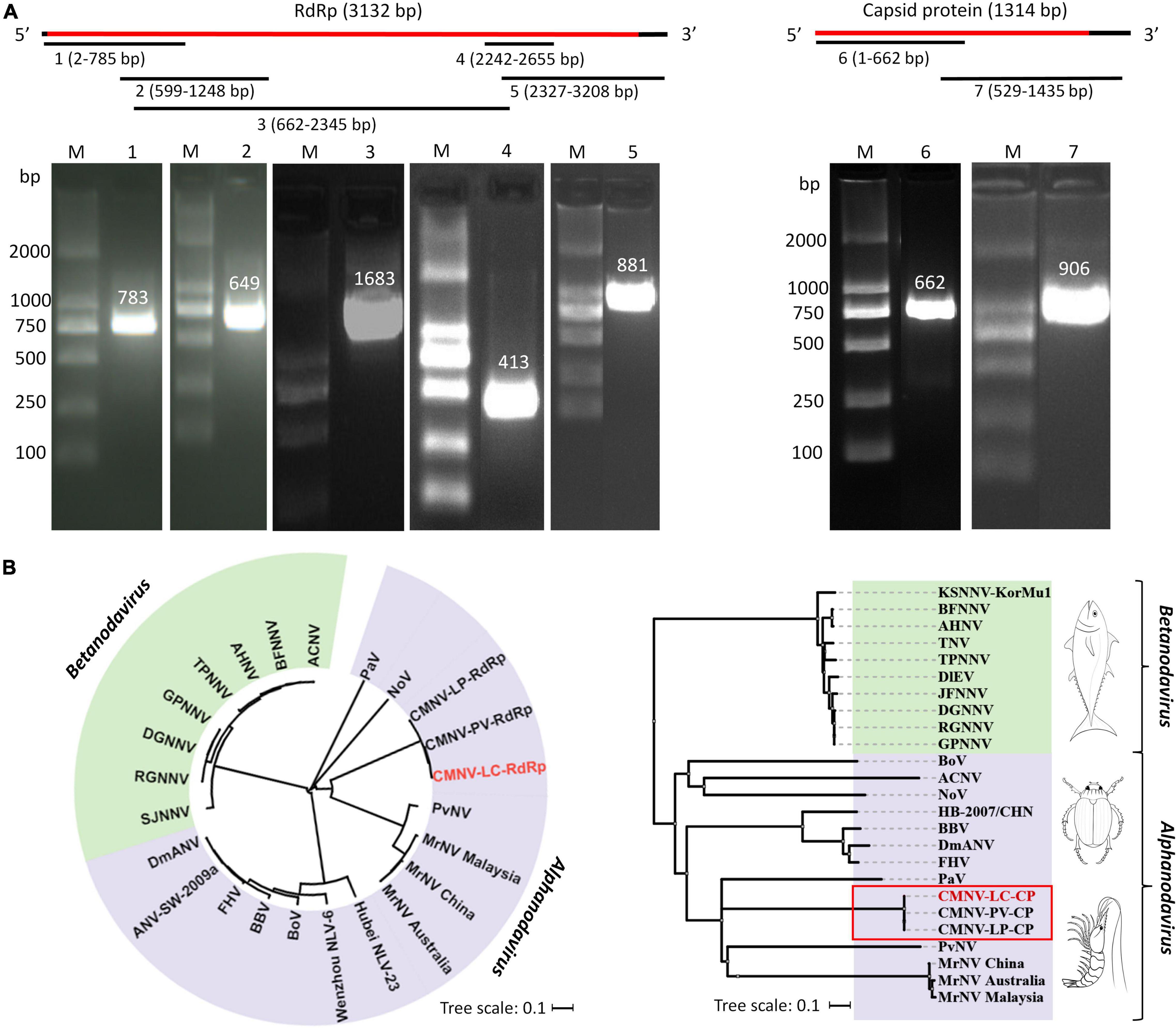
Figure 4. The cloning and sequences analysis of RdRp gene and capsid protein gene of CMNV isolated from Larimichthys crocea. (A) Assembly strategy and agarose gel electrophoresis of PCR amplicons in cloning RdRp and capsid protein genes from L. crocea. The numbers 1–7 represent the seven PCR amplicons produced in cloning the CMNV genome. The images are cropped from the original image (Supplementary Figure 4). Lane M: 2000 bp marker; lanes 1–5: amplicons of the sequence of RdRp; lanes 6 and 7: amplicons of the sequence of capsid protein. (B) Phylogenetic diversity of RNA-dependent RNA polymerase (RdRp) and capsid protein genes from different CMNV isolates and other nodaviruses (for virus abbreviations, see Supplementary Table 1). CMNV-LC-RdRp and CMNV-LC-CP represented the RdRp and capsid protein gene of CMNV isolated from L. crocea, respectively. The phylogenetic tree was generated by the neighbor-joining method using the MEGA 7.0 program, and the scale bar was 0.1.
Searching results of BLASTx showed that the obtained sequences of RdRp and capsid protein genes of CMNV isolated from L. crocea had 99% similarities to the corresponding gene sequences of CMNV isolates from P. vannamei (MT270124 and MT270123) and L. polyactis (MW625911 and MW625912). The phylogenetic analysis indicated that both the RdRp and capsid protein gene sequences of CMNV isolated from L. crocea were clustered tightly with the corresponding sequences of other CMNV isolates. However, it showed a high variation rate with the other members of Alpha Nodavirus, including Drosophila melanogaster American nodavirus (DmANV), flock house virus (FHV), black beetle virus (BBC), P. vannamei nodavirus (PvNV), and M. rosenbergii nodavirus (MrNV), etc. (Figure 4B).
Cytopathic Effect in Epithelioma Papilloma Cyprini Cells Infected With Covert Mortality Nodavirus
The CPE was first observed by light microscopy at 4 days postinoculation (dpi) of tissue filtrates. The cells shrank, their boundaries became invisible, and a degree of cell fusion appeared. The cell monolayer was destroyed on day four (Figure 5A). After three consecutive passages, the CPE became more consistent. Cell cultures were positive for CMNV by PCR assay (Figure 5B). The expected fragment with 619 and 413 bp in size of the CMNV partial RdRp gene was cloned and sequenced from the inoculated cells. The phylogenetic analysis showed that CMNV isolated from EPC cell cultures was clustered into the same branch with the known CMNV isolates (Figure 5C). Typical CMNV-like virus inclusion bodies could be observed around the lysed cells under the TEM (Figure 5D).
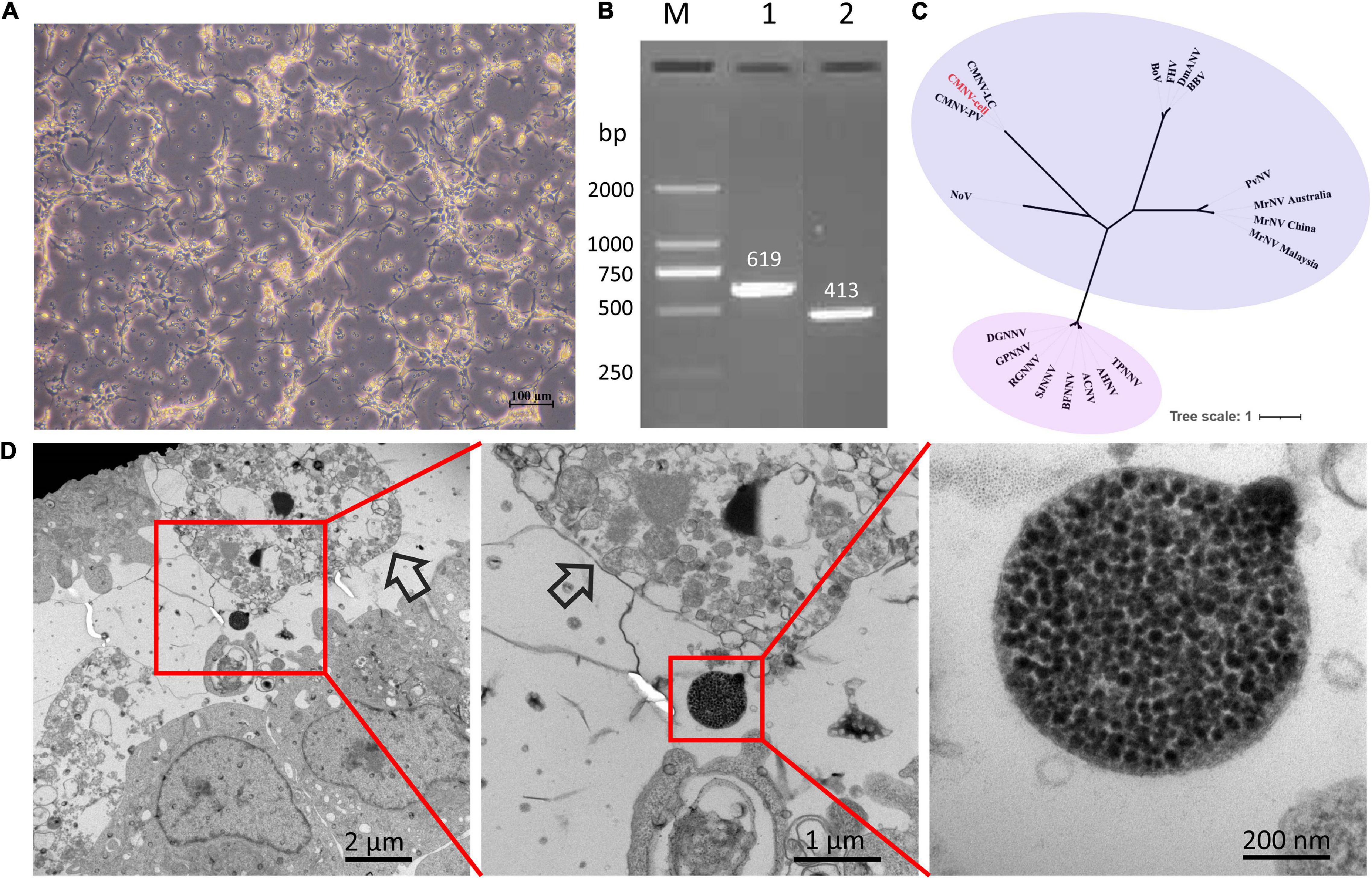
Figure 5. The analysis results of CMNV infection in the EPC cells. (A) Cytopathic effect in EPC cells infected with CMNV; (B) Analysis of agarose gel electrophoresis of CMNV RT-nPCR amplicons; (C) phylogenetic tree analysis of the partial sequences of RNA-dependent RNA polymerase (RdRp) gene from different CMNV isolates and other nodaviruses (for virus abbreviations see Supplementary Table 1). The phylogenetic tree based on the partial sequences of RdRp gene was generated by the neighbor-joining method using the MEGA 7.0 program, and the scale bar was 1. (D) Transmission electron microscopic micrographs of ultrathin sections of EPC cells infected with CMNV. Scale bars, 2 μm, 1 μm, and 200 nm.
Discussion
Up to now, except for members in the genus of Betanodavirus, CMNV was the only virus in the family of Nodaviridae that is able to infect fish naturally (Zhang et al., 2018; Wang et al., 2019a,b, 2021; Xu et al., 2021). In this study, the possibility and prevalence of CMNV infection in farmed and wild L. crocea were evaluated.
The total RNA samples extracted from the tissues of naturally diseased fish were positive for CMNV based on the RT-nPCR and TaqMan RT-qPCR assays. Meanwhile, the CMNV isolates from L. crocea showed highly homologous with that of CMNV isolates from cultured P. vannamei in the phylogenetic tree analysis. It suggests that CMNV-L. crocea isolates might originate from the aquacultured shrimp. In addition, CMNV-like virus particles appeared in the cytoplasm of cardiac muscle and kidney under TEM observations, roughly spherical in shape, without an envelope, and about 30 nm in diameter. The morphological characteristics and size of viruses were generally consistent with the previous findings (Zhang et al., 2014; Wang et al., 2019a,b).
Recent studies have found that CMNV could infect the crucian carp C. auratus, the gobiid fish M. abei, the Japanese flounder P. olivaceus, and the small yellow croaker L. polyactis under natural conditions (Zhang et al., 2018; Wang et al., 2019a,b; Xu et al., 2021). In this study, CMNV infection in L. crocea could cause histopathological alteration and lesions, including extensive cardiac muscle necrosis and severe vacuolation in the liver, eye, and spleen, which were similar to the clinical signs found in other fishes with CMNV infections. In addition to the above-mentioned tissue damages, severe tissue necrosis also occurred in the gills of CMNV-positive L. crocea. Meanwhile, the intense purple positive hybridization signals of CMNV-specific RNA probes were present in the necrotic tissues in the ISH analysis. Moreover, the purified CMNV suspension could cause significant CPE in EPC cells, and typical CMNV-like inclusion bodies could be observed around the lysed EPC cells under the TEM. Those results provided reliable evidence to support that CMNV was a novel pathogen of L. crocea.
Larimichthys crocea is the fish of highest yield in mariculture in China (Ministry of Agriculture and Rural Affairs of the People’s Republic of China, 2021). However, with the rapid expansion of the culture, L. crocea has been suffering from several serious diseases, in which large yellow croaker iridovirus (LYCIV) is a viral disease that can cause high mortality up to 75% of cultured L. crocea, which results in severe economic losses (Chen et al., 2003). CMNV can naturally infect large yellow croakers and cause multiple histopathological damages, which undoubtedly further increases the risk of disease outbreaking and also the difficulty of breeding healthy large yellow croakers, and might lead to more threat to large yellow croakers farming. Hence, the disease caused by CMNV infection in large yellow croakers deserves further epidemiological investigation and research.
Stock enhancement has become a positive approach to restore the natural resource of L. crocea (Lin, 2006; Xu, 2009; Zhang et al., 2010; Ma, 2017). Although stock enhancement could amplify the abundance of target species (Bartley and Bell, 2008; Jiang et al., 2014; Lorenzen, 2015), it may also pose some risks for the recipient aquatic ecosystem (Wang et al., 2006). It was reported that the release of cultured Atlantic salmon in Norwegian offshore waters caused infections of local wild populations of Atlantic salmon with Gyrodactylus salaris and Aeromonas salmonicida, respectively (Heggberget et al., 1993). The present finding that CMNV naturally infected L. crocea and induced severe tissue necrosis in infected individuals suggests that CMNV should be regarded as a pathogen of serious concern to be screened before releasing L. crocea into the natural waters to minimize the adverse impact on stock enhancement.
Conclusion
In summary, the evidence of pathogen detection, histopathology, ISH, TEM, and cell inoculation consistently confirmed that L. crocea is a new susceptible host for CMNV, which highlights that threats due to CMNV infection should be seriously considered in the development of coastal aquaculture industry of L. crocea and the conservation of coastal ecosystems.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee of Yellow Sea Fisheries Research Institute.
Author Contributions
QZ and CY designed the experiments. TX, YF, TJ, CW, and WW executed the experiments. TX and QZ analyzed the data. TX and TJ contributed to sampling. TX wrote the manuscript draft. QZ, JL, and CY revised the manuscript. All authors interpreted the data, critically revised the manuscript for important intellectual contents, and approved the final version.
Funding
This work was supported by the Marine S&T Fund of Shandong Province for Pilot National Laboratory for Marine Science and Technology (Qingdao) (2018SDKJ0303-1), National Natural Science Foundation of China (32073016), Central Public-Interest Scientific Institution Basal Research Fund, CAFS (No. 2020TD39), Project of Species Conservation from the MARA-Marine Fisheries Resources Collection and Preservation, and Central Public-Interest Scientific Institution Basal Research Fund, YSFRI, CAFS (No. 20603022020005).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank Dr. Guoshi Xie and also the colleagues from the Division of Fishery Resources and Ecosystem, Yellow Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences for sampling.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.789128/full#supplementary-material
Footnotes
References
An, S. W. (2013). Study on the pathogen and histopathology of visceral white spot disease of large yellow croaker (Larimichthys crocea). Ningbo: Ningbo University.
Bartley, D. M., and Bell, J. D. (2008). Restocking, stock enhancement and sea ranching: arenas of progress. Rev. Fish. Sci. 16, 357–365. doi: 10.1080/10641260701678058
Chen, S., Zhang, G., Shao, C., Huang, Q., Liu, G., Zhang, P., et al. (2014). Whole genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nat. Genet. 46:253. doi: 10.1038/ng.2890
Chen, X. H., Lin, K. B., and Wang, X. W. (2003). Outbreaks of an iridovirus disease in maricultured large yellow croaker, Larimichthys crocea (Richardson), in China. J. Fish. Dis. 26, 615–619. doi: 10.1046/j.1365-2761.2003.00494.x
Chen, Y. L., Huang, W. Q., Shan, X. J., Chen, J., Weng, H. S., and Yang, T. (2020). Growth characteristics of cage-cultured large yellow croaker Larimichthys crocea. Aquac. Rep. 16:100242. doi: 10.1016/j.aqrep.2019.100242
Heggberget, T. G., Johnsen, B. O., Hindar, K., Jonsson, B., and Jensen, A. J. (1993). Interactions between wild and cultured Atlantic salmon: a review of the Norwegian experience. Fish. Res. 18, 123–146. doi: 10.1016/0165-7836(93)90044-8
Jiang, Y., Lin, N., Yang, L., and Cheng, J. H. (2014). The ecological risk of stock enhancement and the measures for prevention and control. J. Fish. Sci. China 21, 413–422.
Li, X. J., and Yue, D. D. (2019). The export trade structure and international competitiveness of my country’s large yellow croaker. Ocean Dev. Manage. 36, 50–54.
Li, X. P., Wan, X. Y., Xu, T. T., Huang, J., and Zhang, Q. L. (2018). Development and validation of a TaqMan RT-qPCR for the detection of covert mortality nodavirus (CMNV). J. Virol. Methods 262, 65–71. doi: 10.1016/j.jviromet.2018.10.001
Lightner, D. V. (1996). A Handbook of Shrimp Pathology and Diagnostic Procedures for Diseases of Cultured Penaeid Shrimp. Baton Rouge: World Aquaculture Society.
Lin, Y. M. (2006). Review and summary of the reproduction and release of large yellow croaker (Larimichthys crocea) in Xiangshan Port, Zhejiang. Sci. Fish Farm 6:4.
Lorenzen, K. (2015). Understanding and managing enhancements: why fisheries scientists should care. J. Fish. Biol. 85, 1807–1829. doi: 10.1111/jfb.12573
Ma, X. L. (2017). Proliferation and Release of Large Yellow Croaker (Larimichthys crocea) in Dongtou Waters and its Preliminary Evaluation. Zhejiang: Zhejiang Ocean University.
Ministry of Agriculture and Rural Affairs of the People’s Republic of China (2021). Chinese National Fishery Statistics Yearbook. Beijing: China Agriculture Press.
Panphut, W., Senapin, S., Sriurairatana, S., Withyachumnarnkul, B., and Flegel, T. W. (2011). A novel integrase-containing element may interact with Laem-Singh virus (LSNV) to cause slow growth in giant tiger shrimp. BMC Vet. Res. 7:18. doi: 10.1186/1746-6148-7-18
Piette, D., Hendrickx, M., Willems, E., Kemp, C. R., and Leyns, L. (2008). An optimized procedure for whole-mount in situ hybridization on mouse embryos and embryoid bodies. Nat. Protoc. 3, 1194–1201. doi: 10.1038/nprot.2008.103
Shen, J. Y., Yu, X. P., Yin, W. L., Cao, Z., Pan, X. Y., and Zhu, J. L. (2004). Study on the pathogen of low temperature vibrio in cultured large yellow croaker (Larimichthys crocea). J. Fish. China 28, 130–134.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Thitamadee, S., Prachumwat, A., Srisala, J., Jaroenlak, P., Salachan, P. V., Sritunyalucksana, K., et al. (2016). Review of current disease threats for cultivated penaeid shrimp in Asia. Aquaculture 452, 69–87. doi: 10.1016/j.aquaculture.2015.10.028
Wang, C., Liu, S., Li, X. P., Hao, J. W., and Zhang, Q. L. (2019a). Infection of covert mortality nodavirus in Japanese flounder reveals host jump of the emerging alphanodavirus. J. Gen. Virol. 100, 166–175. doi: 10.1099/jgv.0.001177
Wang, C., Wang, X. H., Liu, S., Sang, S. W., and Zhang, Q. L. (2019b). Preliminary study on the natural infection of Carassius auratus with covert mortality nodavirus (CMNV). J. Fish. Sci. China 40, 25–32.
Wang, C., Liu, S., Xu, T. T., Li, X. P., Li, J., and Zhang, Q. L. (2021). Pathogenicity study of covert mortality nodavirus (CMNV) infection in zebrafish model. Aquaculture 546:737378. doi: 10.1016/j.aquaculture.2021.737378
Wang, F., Liao, B. C., Sun, M. Q., Lin, G. Q., Chen, H. Q., Huang, G. L., et al. (2019). Analysis of the development situation of Fujian large yellow croaker (Larimichthys crocea) industry. Fish. Sci. 3, 45–49.
Wang, G., Yuan, S., and Jin, S. (2010). Nocardiosis in large yellow croaker, Larimichthys crocea (Richardson). J. Fish. Dis. 28, 339–345. doi: 10.1111/j.1365-2761.2005.00637.x
Wang, Q. Y., Zhuang, Z. M., Deng, J. Y., and Ye, Y. M. (2006). Stock enhancement and translocation of the shrimp Penaeus chinensis in China. Fish. Res. 80, 67–79. doi: 10.1016/j.fishres.2006.03.015
Xiao, Y., Zheng, L., Xu, J., and Zhou, Y. (2012). Establishment and characterization of a cell line derived from fin of Cryprinus carpiod. Chin. J. Cell Biol. 34, 767–774.
Xu, T. T., Li, Y. X., Shan, X. J., Hao, J. W., Teng, G. L., Tang, K. F. J., et al. (2021). Natural infection of covert mortality Nodavirus in Small Yellow Croaker in coastal water. Front. Mar. Sci. 8:670831. doi: 10.3389/fmars.2021.670831
Xu, T. T., Liu, S., Li, X. P., and Zhang, Q. L. (2020). Genomic characterization of covert mortality nodavirus from farming shrimp: evidence for a new species within the family Nodaviridae. Virus Res. 286:198092. doi: 10.1016/j.virusres.2020.198092
Xu, W. J., Qin, Z. D., Shi, H., Jiang, N., Zhou, Y., Liu, X. L., et al. (2015). Mass mortality associated with a viral-induced anemia in cage-reared large yellow croaker, Larimichthys crocea (Richardson). J. Fish. Dis. 38, 499–502. doi: 10.1111/jfd.12270
Xu, Y. M. (2009). Fishery enhancement and release in Fujian Province has achieved good results. Fish. Inf. Strategy 7:32.
You, K. R., and Li, X. X. (2003). Common diseases and prevention of large yellow croaker cage culture. Spec. Econ. Anim. Plants 6, 000042–43. doi: 10.1111/jfd.13206
Zhang, Q. L., Liu, Q., Liu, S., Yang, H. L., Liu, S., Zhu, L. L., et al. (2014). A new nodavirus is associated with covert mortality disease of shrimp. J. Gen. Virol. 95, 2700–2709. doi: 10.1099/vir.0.070078-0
Zhang, Q. L., Shuang, L., Jun, L., Xu, T. T., Wang, X. H., Fu, G. M., et al. (2018). Evidence for Cross-Species Transmission of Covert Mortality Nodavirus to New Host of Mugilogobius abei. Front. Microbiol. 9:1447. doi: 10.3389/fmicb.2018.01447
Zhang, Q. L., Xu, T. T., Wan, X. Y., Liu, S., and Huang, J. (2017). Prevalence and distribution of covert mortality nodavirus (CMNV) in cultured crustacean. Virus Res. 233, 113–119. doi: 10.1016/j.virusres.2017.03.013
Keywords: covert mortality nodavirus (CMNV), large yellow croaker (Larimichthys crocea), cellular necrosis, cytopathic effect, susceptibility
Citation: Xu TT, Fan YD, Jia TC, Wang C, Wang W, Li J, Zhang QL and Yao CL (2022) Investigation on Natural Infection of Covert Mortality Nodavirus in Large Yellow Croaker, Larimichthys crocea. Front. Mar. Sci. 9:789128. doi: 10.3389/fmars.2022.789128
Received: 05 October 2021; Accepted: 09 February 2022;
Published: 11 March 2022.
Edited by:
Youji Wang, Shanghai Ocean University, ChinaReviewed by:
Najiah Musa, University of Malaysia Terengganu, MalaysiaChao Li, Qingdao Agricultural University, China
Copyright © 2022 Xu, Fan, Jia, Wang, Wang, Li, Zhang and Yao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingli Zhang, emhhbmdxbEB5c2ZyaS5hYy5jbg==; Cuiluan Yao, Y2x5YW9Aam11LmVkdS5jbg==
 Tingting Xu1,2
Tingting Xu1,2 Yuding Fan
Yuding Fan Chong Wang
Chong Wang Jun Li
Jun Li Qingli Zhang
Qingli Zhang