- UGC-CAS Department of Biosciences, Saurashtra University, Rajkot, India
Deserts in general, and Indian deserts in particular, are less attended for microbial diversity. The Little Rann of Kutch (LRK), a coastal saline desert, is characterized by a unique combination of both dry and wet features. This study represents the first report on the extensive isolation, spatial distribution, 16S rRNA gene-based phylogeny, and identification of novel taxa. A total of 87 isolates were obtained from three different study sites in LRK. Based on the full 16S rRNA gene sequences, the isolates were grouped into 44 different phylotypes of four phyla: Firmicutes, Proteobacteria, Actinobacteria, and Euryarchaeota. These in turn were represented by 19 different genera. Halomonas, Gracilibacillus, Thalassobacillus, Piscibacillus, Salimicrobium, Alkalibacillus, Bhargavaea, Proteus, Marinobacter, Pseudomonas, Kocuria, Corynebacterium, Planococcus, Micrococcus and Natronococcus identified in this study had never before been reported from this habitat. A majority of the isolates displayed broad salt and pH tolerance. The bacterial diversity of Venasar and Jogad closely resembled with each other. While Bacillus, Virgibacillus, Gracillibacillus, and Bhargavaea were common genera in all sites, six putative novel taxa of different phylogenetic groups were identified. Available nitrogen, pH, Organic carbon, TDS, and EC were the main environmental variables affecting the microbial diversity. Analysis of the geographical distribution revealed that a majority of the phylotypes had cosmopolitan distribution, followed by the saline and marine distribution, while ∼13% were affiliated with only LRK. The phylotypes associated with marine distribution decreased with increasing distance from the Gulf of Kutch, suggesting their endemism to marine environments. The study established the taxonomic novelty and prospects for the discovery of unique products and metabolites.
Introduction
Arid regions cover large areas of the terrestrial surface and yet are the most understudied biome (Bhatt and Singh, 2016; Bhatt et al., 2018). Deserts face a variety of extreme conditions such as limited water availability, nutrients deficiency, temperature fluctuation, and high exposure to UV irradiation from the sun (An et al., 2013). Deserts are the most understudied biome as compared with other biomes. Greater attention should be given to the microbial communities in these unusual and yet-to-be explored environments to investigate their diversity, ecological significance, and potential applications (Chaudhary et al., 2019; Bhatt and Singh, 2020; Bodor et al., 2020).
The Indian deserts are relatively less explored compared to other deserts of the world. The Little Rann of Kutch (LRK) is a saline desert of varied demography covering a large area of 4953.7 km2 (Ishnava et al., 2011; Gupta and Ansari, 2014). The north head of the Gulf of Kutch adjoins LRK with the regular flow of saline water during tides or through the water drifting from the south-west winds, making it a saline coastal desert (Gupta and Ansari, 2014; Bhatt and Singh, 2016; Bhatt et al., 2018). Unlike other deserts, major portions of the LRK consists of 60% clay (Gupta and Ansari, 2014). The LRK has been nominated as a “Biosphere Reserve” characterized as terrestrial and coastal ecosystems (UNESCO’s Man and Biosphere (MAB) program, http://whc.unesco.org/en/tentativelists/2105/). Despite its unique enigmatic terrain of ecological significance, not much is known about its microbial diversity and ecology.
Many of the studies so far have focused on the diversity of the microbial community of the biological soil crust (Abed et al., 2010; Li et al., 2013), endolithic communities of translucent stones and gypsum deposits (Dong et al., 2007), and shrubs (Saul-Tcherkas et al., 2013). The unvegetated soil of the deserts worldwide have been investigated in a limited sense only (Bhatt et al., 2018). Therefore, in the current study, we attempted to study microbial diversity of unvegetated soil of saline desert.
Some studies on the microbial diversity of saline habitats are based on culture-independent molecular techniques (Purohit and Singh, 2009; Siddhapura et al., 2010; Caton and Schneegurt, 2012; Vavourakis et al., 2016; Bachran et al., 2018; Binayke et al., 2018; Raiyani and Singh, 2020). While metagenomics reveals a great extent of the taxonomic and genetic diversity of the microbial world of a given habitat, it needs to be complemented and validated with cultivable approaches linked with biotechnological, ecological, and taxonomic significance. High throughput cultivation methods have been developed using synthetic media of low concentrations of nutrients, mimicking oligotrophic conditions (Bruns et al., 2002; Connon and Giovannoni, 2002; Cho and Giovannoni, 2004) with prolonged growth time (Stevenson et al., 2004; Davis et al., 2005). The improved and modified cultivation strategies help to cultivate novel organisms at a relatively low cost.
Haloalkaliphilic bacteria have been isolated from varied saline environments (Bhatt et al., 2018). The interest in haloalkaliphilic microorganisms is not only due to the understanding of the mechanisms of adaptation to multiple stresses and detecting their diversity, but also due to their possible applications in biotechnology. Enzymes from haloalkaliphilic microorganisms have gained considerable interest in recent years due to their excellent stability and activity in high salt, pH, and temperature (Bhatt and Singh, 2020). Recent studies on the extracellular enzymes of the microorganisms dwelling in extreme habitats of high salinity and alkaline pH have established their ecological and biotechnological significance (Purohit and Singh, 2011; Sinha and Khare, 2013; Raval et al., 2015; Baweja et al., 2016; Raval et al., 2018; Si et al., 2018; Thakrar and Singh, 2019; Bhatt and Singh, 2020; Dwivedi et al., 2021; Rathore and Singh, 2021).
In light of the above facts, the current study focused on the isolation strategies, phylogenetic analysis, and identification of novel lineages of the bacterial community of the unvegetated yet unexplored saline coastal desert of the Little Rann of Kutch. The abiotic factors associated with the dominance and distribution of bacterial genera among different sites has also been investigated. In addition, global geographical distribution of the phylotypes has been analyzed and interpreted. To the best of our knowledge, this is the first report on extensive isolation, spatial diversity, and phylogenetic analysis of bacteria from the saline desert of Little Rann of Kutch, Gujarat, India.
Materials And Methods
Study Sites and Sample Collection
Little Rann of Kutch is roughly triangular in shape, located between 22° 55’’ to 24° 35’’ North latitudes and 70° 30’’ to 71° 45’’ East longitudes near the Great Rann of Kutch, Gujarat, with an average annual rainfall of below 400 mm (Ishnava et al., 2011). In summer, temperatures reach highs of 48°C and lows of 10°C during the winter, while the average temperature is 30-35°C (Datta et al., 2020). Geologically, this area was a part of oceanic floor and has emerged in the recent past. The Rann of Kutch was a gulf of the sea with surrounding coastal towns (Frere, 1870). A fairly low rain fall coupled with evaporation led to the receding of water, subsequently leaving behind a crust of halite and gypsum crystals that converted into the clay and sands.
Soil samples were collected from three different study sites of the LRK: Surajbari (N 23° 11’ 16.373, E 070° 43’ 03.929”), Venasar (N 23° 09’ 57.327”, E 070° 55’ 14.575”), and Jogad (N 23° 10’ 38.992”, E 071° 15’ 04.999”) (Figure 1). A total of three subsamples were collected from each site at distances of 100 - 200 m. The collected samples were designated as SB, VS, and JO for Surajbari, Venasar, and Jogad, respectively. One hundred grams of three subsamples were collected from each site as far as possible from plants to avoid rhizosphere effects and at a depth of 10 to 15 cm in order to obtain a sample minimally impacted by aeolian dispersion. Samples were collected into sterile polythene bags, transported to the laboratory, and stored at 4°C until further analysis.
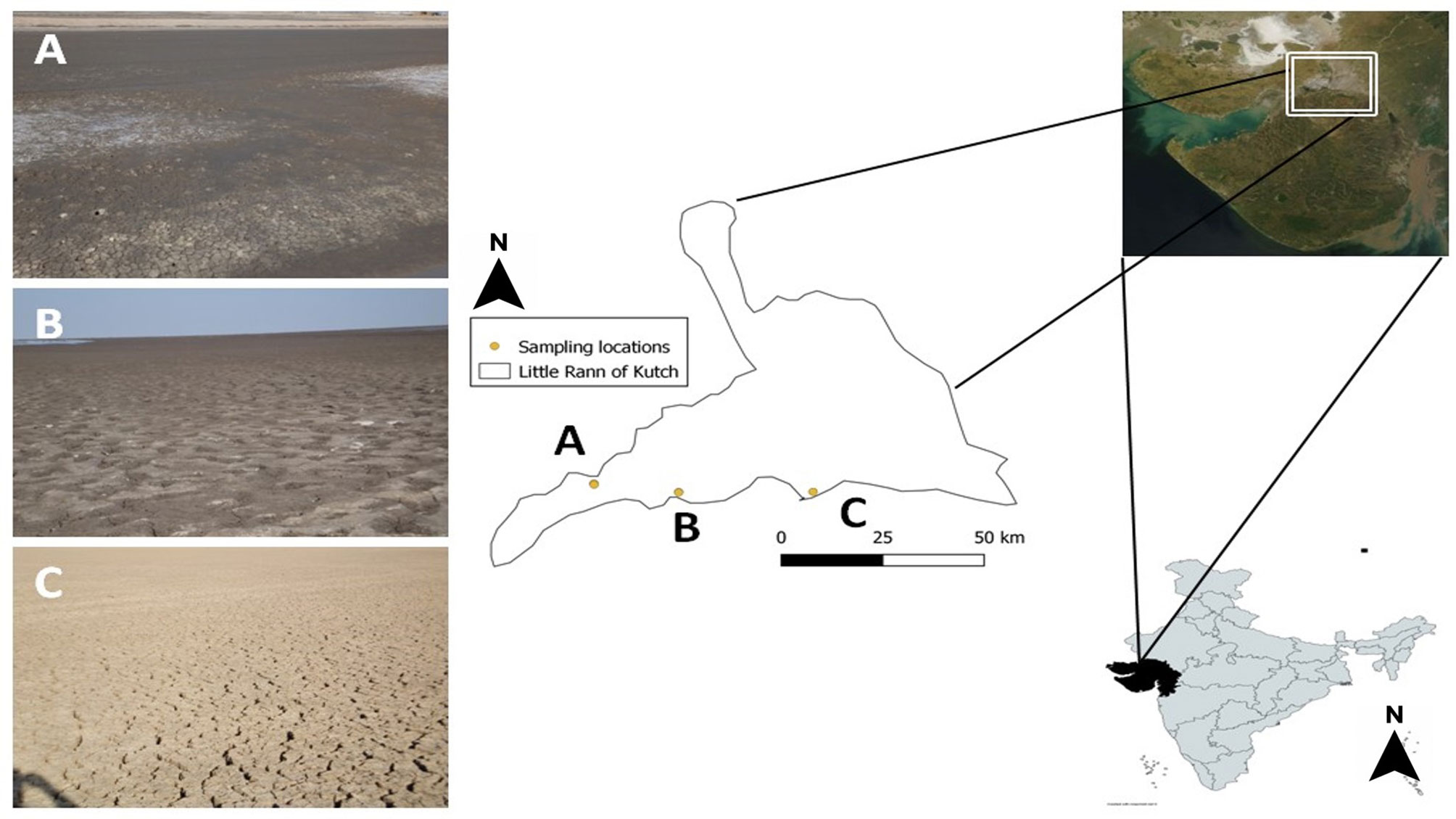
Figure 1 Location map of sampling sites in Little Rann of Kutch, Gujarat, India. Dots and alphabets indicate the sampling sites: (A) Surajbari (N 23° 11’ 16.373, E 070° 43’ 03.929”) (B) Venasar (N 23° 09’ 57.327”, E 070° 55’ 14.575”) (C) Jogad (N 23° 10’ 38.992”, E 071° 15’ 04.999”).
Soil Analysis
The physicochemical properties of the soil samples were analyzed. The soil pH and salinity were measured in a 1:5 (wt/wt) aqueous solution, while the conductivity was measured in dS/m by digital conductivity meter (CON700, Eutech instruments, Singapore). Estimation of the oxidizable organic carbon was based on the Walkely and Black Method (Walkley and Black, 1934), while phosphorous (P) was estimated by the extraction with sodium bicarbonate (Olsen et al., 1954). Available nitrogen (N) was measured by alkaline permanganate method (Asija and Subbiah, 1956), while potassium and sodium were measured by the flame photometric method and sulphur by turbidity method (Chesnin and Yien, 1951; Jackson, 1974). Exchangeable calcium and magnesium were estimated using versanate EDTA Method. Total dissolved solid (TDS) of soil was measured as described earlier (Jackson, 1974). Chlorine was estimated by titration method (Jackson, 1974). Cu, Fe, Mn, Zn, and B were estimated using MP-AES (Microwave Plasma Atomic Emission Spectrometry) method (Hettipathirana, 2011).
Isolation of Aerobic Heterotrophic Bacteria
Two approaches, direct plating and Liquid enrichment technique, were used to isolate bacteria from the desert soil of the LRK. In direct plating, five different media (Modified SP medium consisting, (g/l): (NaCl, 49; KCl, 1; MgSO4.7H2O, 0.5; CaCl2.2H2O, 0.18; NaHCO3, 0.03; NaBr, 0.115; FeCl3.6H2O, 0.5; Bacto trypton, 5; Yeast extract, 10; Glucose, 1; Agar, 30) (Caton et al., 2004), Reasoner’s 2A (R2A) agar (HiMedia Laboratories, India) consisting, (g/l): (Casein acid hydrolysate, 0.5; yeast extract, 0.5; protease peptone, 0.5; dextrose, 0.5; starch, 0.5; KH2PO4, 0.3; MgSO4, 0.024; Sodium pyruvate, 0.30; NaCl, 50; Agar, 30) (Reasoner and Geldreich, 1985), Soil Extract medium (SE) consisting, (g/l): (Glucose, 1; K2HPO4, 0.5; Soil extract, 17.75; NaCl, 50; Agar, 15) (Yadav et al., 2015), Complex medium (CM) agar consisting, (g/l): (Glucose, 10; peptone, 5; yeast extract, 5; KH2PO4, 5; NaCl, 150; Agar, 30) (Bhatt et al., 2018) and Halophilic medium (HM) agar consisting, (g/l): [NaCl, 220; MgSO4.7H2O, 10; KCl, 5; Sodium citrate, 3; KNO3, 1; CaCl2.2H2O, 0.20, Trace minerals, 0.5; Bacto tryptone, 5; Yeast extract, 1; Agar, 30) (Post, 1977)] were used to capture and isolate the diverse microbiota of the desert soil. Moreover, serial dilution technique was used with two different diluents i.e., 1) Distilled water 2) Winogradsky’s salt solution consisting, (g/l): K2HPO4, 0.25; MgSO4, 0.125; NaCl, 0.125; Fe2(SO4)3, 0.0025; MnSO4, 0.0025). For bacterial isolation, a composite sample was prepared out of the three subsamples collected from each site. The composite soil sample (1g) was thoroughly mixed and suspended in 9 ml of sterile distilled water and Winogradsky’s salt solution using a Vortex mixer. The samples were then diluted from 10-1 to 10-7, and 0.1 ml aliquots from each dilution were spread on the five different media as described above and incubated at 37°C for 7 days. After a week of incubation, the bacterial colonies macroscopically differing in morphology, size, and pigmentation were sub-cultured, isolated, and purified using the respective culture medium. Only the plates containing 30-300 colonies were used to calculate total bacterial counts. The results were expressed as mean colony forming units (cfu) per gram weight of soil. In addition, enrichment technique was used to enrich soil samples using three different media: SP medium, CMB medium, and HM medium. In enrichment technique, after inoculation, enrichment media was incubated on the orbitek shaker at 180 rpm and 37°C and the growth was periodically observed. After 72h of incubation, the cultures were serially diluted up to 10-7 and 0.1 ml of an appropriately diluted culture was spread over the respective agar plate and incubated at 37°C. After 7 days of the incubation, distinct isolated colonies were selected, and pure cultures obtained.
pH and Salt Tolerance
In order to evaluate the salt tolerance of the bacteria, the isolates were spot-inoculated onto the surface of the respective culture media with varying salt concentrations in the range of 0-25% at pH 8. The pH tolerance was examined at 37°C in respective growth medium used for the isolation and the pH was adjusted to pH 7.0–11.0 with an interval of 1 pH units by using separately autoclaved Na2CO3 (20%, w/v) for pH 6–8, 1 M glycine/NaOH buffer for pH 9.0–10.0, and 1M NaOH or 1M HCl for pH 11. The growth was monitored at 24 hr. intervals and considered positive on appearance after 4 days of incubation for fast-growing bacteria and 7 days of incubation for slow-growing archaea at 37°C.
16S rRNA Gene Sequencing and Identification
For bacteria, 16S rRNA genes were amplified using forward and reverse primer pairs 27F (5’-AGAGTTTGATCMTGGCTCAG-3’) and 1492R (5’- TACGGYTACCTTGTTACGACTT-3’), respectively, followed by the sequencing of each amplified product after purification. Whereas for archaea, forward and reverse primer pairs 21F (5-TTCCGGTTGATCCYGCCGGA-3) and 1492R (5’-GGTTACCTTGTTACGACTT-3’) were used for the amplification. The amplified 16S rRNA genes were sequenced on the Applied Biosystems Automatic Sequencer (ABI3730XL). Identification of the phylogenetic neighbors and calculation of pairwise 16S rRNA gene sequence similarity was achieved using the EzTaxon-e server (Kim et al., 2012). The CLUSTAL W algorithm of MEGA 6 software was used for sequence alignments and MEGA 6 software for phylogenetic analysis of the individual sequences (Tamura et al., 2013). Distances were calculated using the Kimura correction in a pairwise deletion manner. Maximum Likelihood (ML) method was used to construct phylogenetic tree. Percentage support values were obtained using a bootstrap procedure based on 1000 replications. The 16S rRNA gene sequences of 87 isolates are deposited in the NCBI GenBank (Accession No. MF321815- MF321853, MK785115 - MK785132, MK779774 - MK779775, MK779747, MK779714, MK779742, MK779796, MK779859 - MK779864, KT008286, and KT008288 - KT008300).
Multivariate Statistical Analysis
The influence of soil chemical properties on the microbial diversity was determined by the CCA analysis using PAST v3.02 software (Hammer et al., 2001). Before the multivariate analyses, to use the same units for all the environmental variables, the values of all variables were normalized by subtracting the mean of the raw data and dividing by the standard deviation of the raw data to better conform to normality (Pagaling et al., 2009). Ordination triplots were used to represent the effect of environmental variables on the bacterial community structure. The environmental factors were represented as lines. The CCA plots were generated using the PAST v3.02 software (Hammer et al., 2001). Cluster analysis based on genera distribution among sampling sites was carried out using Bray-curtis similarity measure and UPGMA algorithm.
Diversity Measures
The Good’s non-parametric coverage estimator was calculated according to the equation C = 1 - (n1/N), where n1 is the number of phylotypes for which only one isolate was recovered, and N is the total number of isolates (Good, 1953). The Shannon biodiversity index and Margalef index were also calculated (Magurran, 1988).
Geographic Distribution of the Phylotypes Recovered
In order to assess the geographic distribution of the phylotypes, 16S rRNA gene sequences were compared with the sequences of the cultured strains as well as environmental sequences (from metagenomics and high-throughput sequencing) available in public databases (EMBL and NCBI) using BLAST (Blast Local Alignment Search Tool) (Altschul et al., 1990). In BLAST analysis, only high scoring entries with ≥ 98.7% sequence similarity were considered to obtain geographical data as per the species delineation criteria (Stackebrandt and Ebers, 2006). Data on geographic location of high scoring hits (≥ 98.7%) was obtained, such as isolation source and environment from where strains were isolated, in order to assess any concordance in biome or habitat type. Based on the geographical location data of the high scoring entries (≥ 98.7%), the phylotypes were labeled as: 1) LRK (If no high scoring sequences from non-LRK origin), 2) Marine (Only high scoring sequences from any marine environment – For instance, Sea water/Sea sediments), 3) Saline (At least one high scoring sequence from inland saline habitat/athalassohaline habitat - For instance, saline lake or saline desert), or 4) Cosmopolitan (Phylotypes having similarity with at least one high scoring sequence from non-LRK/non-marine/non-saline environment - For instance, garden soil/fresh water/rhizosphere etc.).
Results
Physicochemical Properties of the Soil Samples
Soil samples from all the three study sites have shown considerable variation with respect to their physicochemical properties. However, many properties of Surajbari and Venasar were similar. The detailed physicochemical properties for the three soil samples are shown in Table 1.
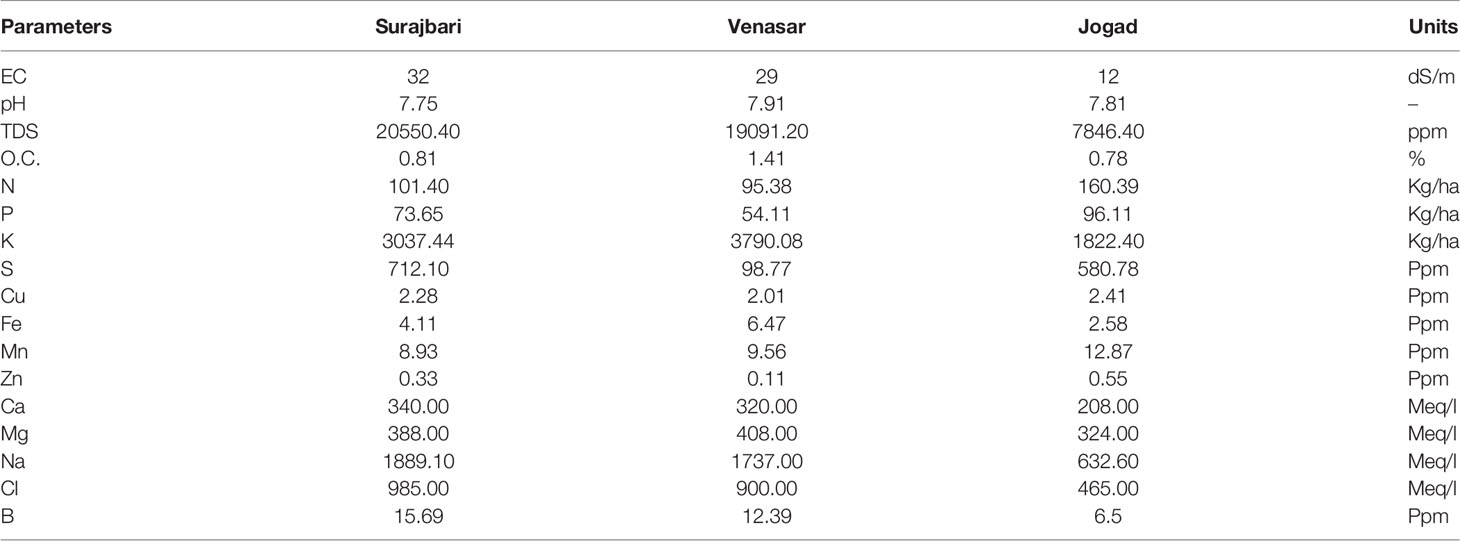
Table 1 Physico-chemical parameters observed in the soil samples collected from Little Rann of Kutch.
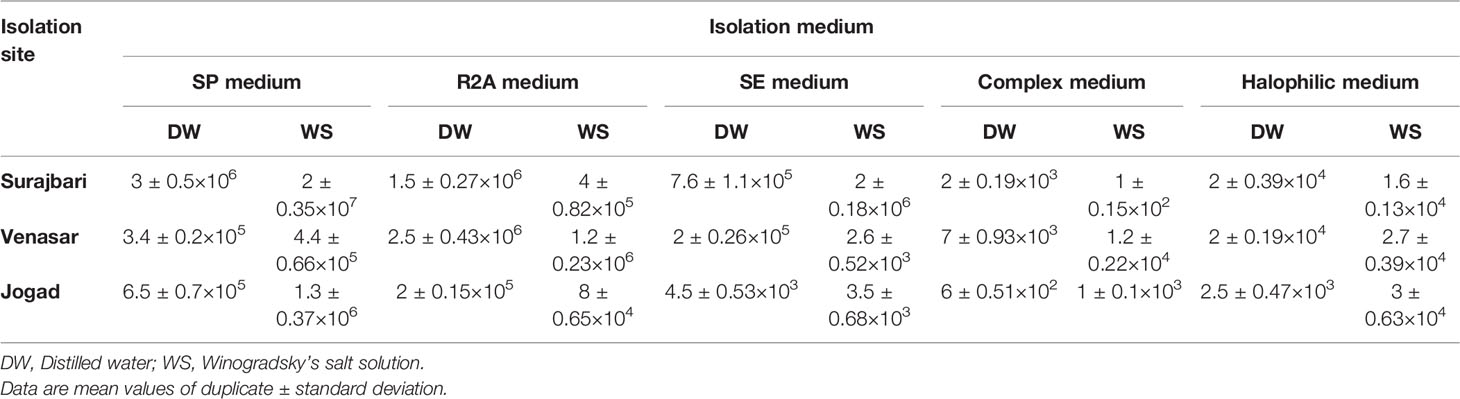
Viable Bacterial Counts
The viable counts in the Surajbari site varied from 1 ± 0.15×102 cfu/g of soil in CMB medium to 2 ± 0.35×107 cfu/g of soil in SP medium. In the Venasar site, viable counts varied from 2.6 ± 0.52×103 cfu/g of soil in SE medium to 2.5 ± 0.43×106 cfu/g of soil in R2A medium. Whereas, in the Jogad site, viable counts varied from 6 ± 0.51×102 cfu/g of soil in CMB medium to 1.3 ± 0.37×106 cfu/g of soil in SP medium. Overall, the cfu values were high in SP and R2A medium compared to other media used in this study. Moreover, cfu count was higher when Winogradsky’s salt solution was used compared to distilled water as diluents (Table 2).
Isolation of Heterotrophic Bacteria
Cultivation strategies yielded 87 isolates from three sampling sites at the Little Rann of Kutch (Table 3). Out of all isolates, 82.75% (n=72) were obtained by direct plating technique, while the remaining 17.24% (n=15) by enrichment technique. In direct plating approach, maximum isolates were obtained on SP medium (43.05%) followed by R2A medium (30.55%), SE medium (11.11%), Complex medium (8.33%), and Halophilic medium (6.94%). In addition, 65.27% of the isolates were obtained when distilled water was used as diluent, while 34.72% isolates were obtained with Winogradsky’s salt solution as diluent. In enrichment technique, out of 15 isolates, nearly one third were obtained on SP medium, Halophilic medium, and Complex medium.
Salt Profile of the Isolates
The bacterial isolates can be diversified based on their growth behavior in different salt concentrations. Overall, the isolates displayed a broad range of salt tolerance. Among the isolates, a majority (∼98%) could grow at 7.5% NaCl, ∼75% at 12% NaCl, and ∼36% at 25% NaCl. In the Surajbari site, all the isolates grew in 3-7.5% NaCl, whereas in Venasar 7.5% NaCl supported growth of all the isolates (Figure 2). While∼ 93% of the Jogad isolates grew in the range of 3-7.5% NaCl. Further, ∼ 73% of the Venasar isolates displayed a broader salt tolerance of 0-25% NaCl (Figure 2). Isolates growing without salt were maximally represented in Jogad site followed by Surajbari and Venasar. Moreover, the number of isolates growing at 15% NaCl were maximum in Venasar (∼73%) followed by Surajbari (∼50%) and Jogad (∼26%). Isolates SB-7, SB-27, SB-28, and SB-29 of Surajbari, VS-7, VS-27, VS-30, VS-34, and VS-36 of Venasar, and JO-30, JO-39, and JO-44 of Jogad could not grow without salt, indicating their true halophilic nature.
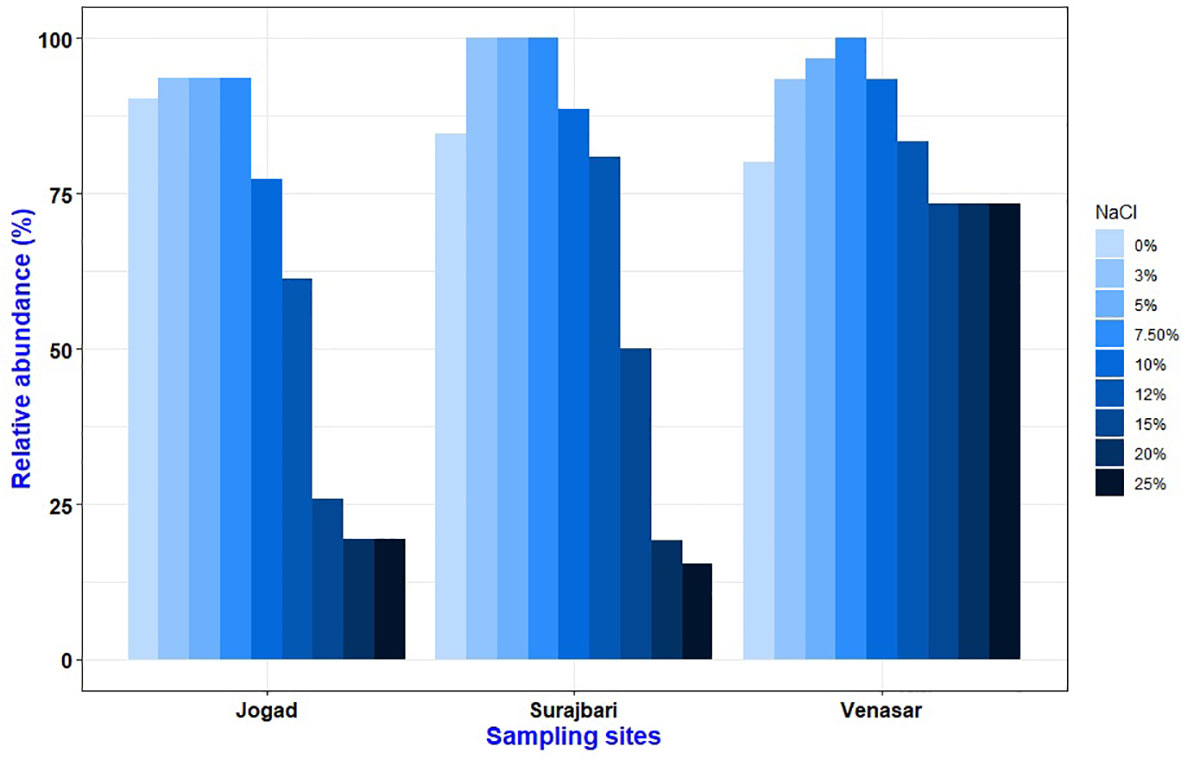
Figure 2 Comparative profile showing effect of NaCl concentration (%, w/v) on growth of isolates from Surajbari, Venasar, and Jogad study sites.
pH Profile of the Isolates
In the present study, a majority of the bacterial isolates grew over a wide range of pH 7-10. Out of 87 isolates, a majority (>90%) could grow at pH 8-9 followed by pH 7, 10, and 11. All the isolates of the Surajbari site grew at pH 8-9, while the isolates of Venasar displayed growth at pH 7 and 8 (Figure 3). On the other hand, the majority of the Jogad isolates were able to grow in the pH range of 7-10. Interestingly, ∼93% of the Venasar isolates displayed a broader salt range for growth at pH 7-10 (Figure 3). The isolates growing at neutral pH were largely from Venasar study site followed by Surajbari and Jogad. While the maximum number of the isolates growing at pH 11 belonged to Venasar (∼73%) followed by Jogad (∼45%) and Surajbari (∼42%) (Figure 3). Isolates SB-2 of Surajbari and JO-23, JO-35, and JO-39 of Jogad did not grow at pH 7, indicating their true alkaliphilic nature.
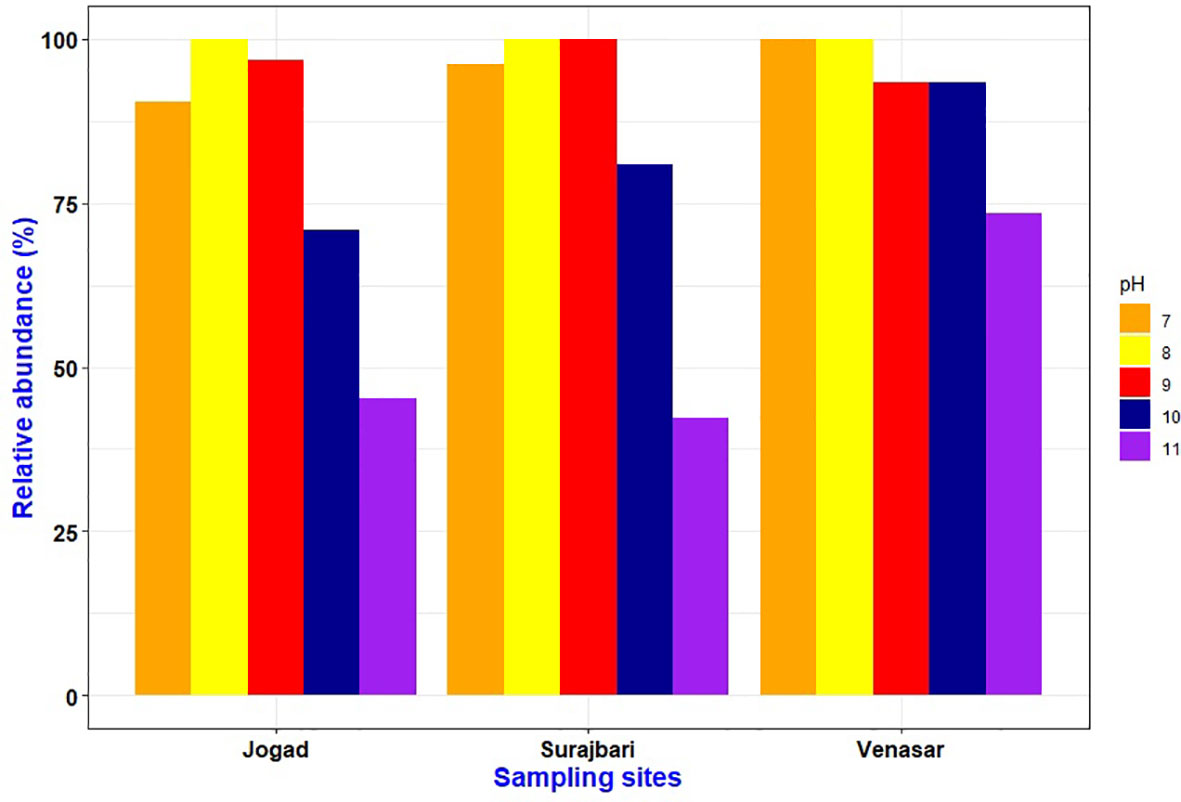
Figure 3 Comparative profile showing effect of pH on growth of isolates from Surajbari, Venasar, and Jogad study sites.
Identification and Phylogenetic Analyses
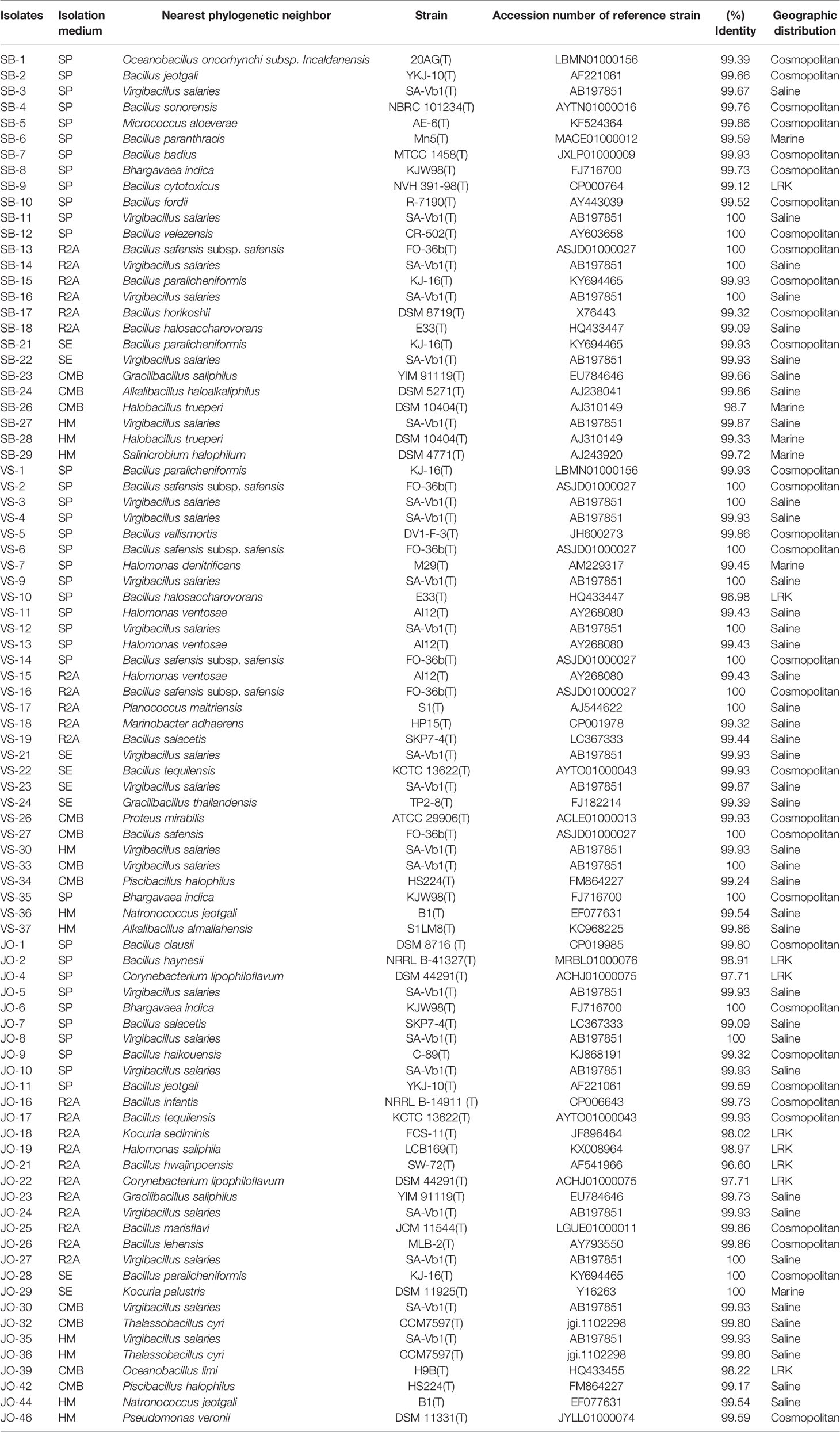
The isolates identified and analyzed on the basis of 16S rRNA gene sequences were classified in the kingdom bacteria and archaea. The EZ taxon server analysis suggested homology of the isolates in the range of 96.6-100% with the reference strains of the NCBI database. The isolates were distributed in four phyla, namely Firmicutes, Proteobacteria, Actinobacteria, and Euryarchaeota, belonging to 19 different genera: Bacillus, Virgibacillus, Halomonas, Gracilibacillus, Thalassobacillus, Piscibacillus, Halobacillus, Oceanobacillus, Salimicrobium, Alkalibacillus, Bhargavaea, Proteus, Marinobacter, Kocuria, Corynebacterium, Planococcus, Pseudomonas, Micrococcus, and Natronococcus. Overall, 90.81% isolates were Gram positive while the rest displayed a Gram-negative character. The relatively abundant genera were Bacillus (33 isolates: 37.9%), Virgibacillus (21 isolates: 24.1%), Halomonas (5 isolate: 5.74%), Bhargavaea (3 isolate: 3.44%), Gracillibacillus (3 isolate: 3.44%), Kocuria (2 isolate: 2.29%), Corynebacterium (2 isolate: 2.29%), Piscibacillus (2 isolate: 2.29%), Oceanobacillus (2 isolate: 2.29%), Thalassobacillus (2 isolate: 2.29%), Halobacillus (2 isolate: 2.29%), Alkalibacillus (2 isolate: 2.29%), Natronococcus (2 isolate: 2.29%), Micrococcus (1 isolate: 1.14%), Salimicrobium (1 isolate: 1.14%), Pseudomonas (1 isolate: 1.14%), Marinobacter (1 isolate: 1.14%), Proteus (1 isolate: 1.14%), and Planococcus (1 isolate: 1.14%).
Based on the phylogenetic analysis, a majority of the isolates belong to the phylum Firmicutes, which included Low G+C Gram-positive bacteria of different families. Nine genera (Bacillus, Virgibacillus, Gracillibacillus, Thalassobacillus, Piscibacillus, Alkalibacillus, Halobacillus, Oceanobacillus, and Salimicrobium) belonged to the family Bacillaceae, while Planococcus and Bhargavaea were associated with Planococcaceae (Figure 4). The second most abundant phylum was Proteobacteria, which included gram negative bacteria of various genera; Proteus, Marinobacter, Halomonas, and Pseudomonas belonged to the families Enterobacteriaceae, Alteromonadaceae, Halomonadaceae, and Pseudomonadaceae, respectively (Figure 5).
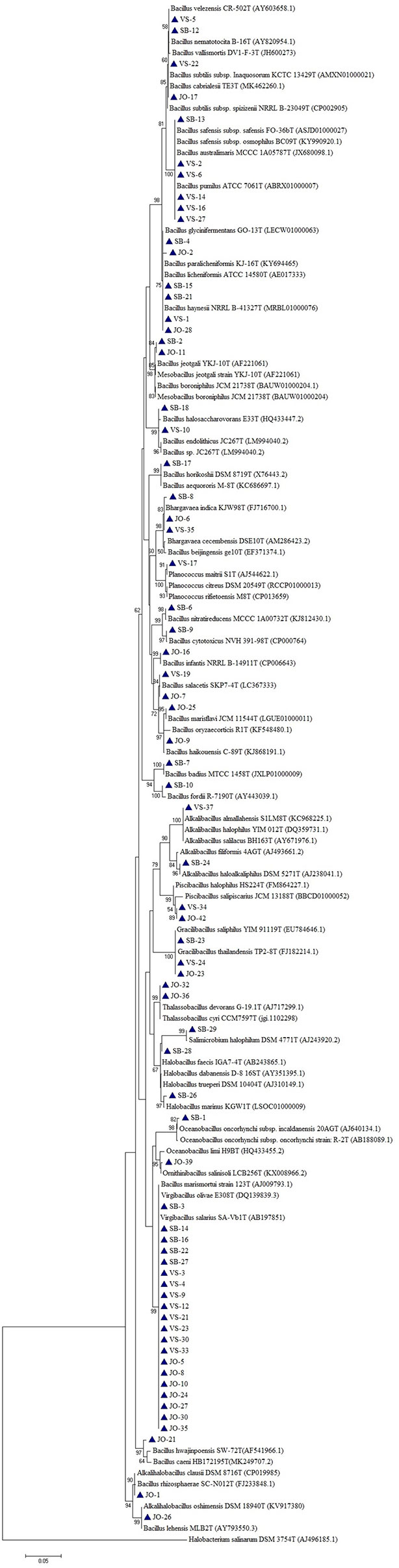
Figure 4 Phylogenetic tree constructed using 16S rRNA gene sequences of LRK isolates belonging to Firmicutes phylum and their closest phylogenetic relatives. The tree was reconstructed by the Maximum Likelihood method using MEGA 6 software. Halobacterium salinarum DSM 3754T was used as an outgroup. The numbers on the tree indicate the percentages of bootstrap sampling derived from 1,000 replications. Bar, 5 nt substitution per 100 nt. Blue triangles indicates the strains isolated in this study.
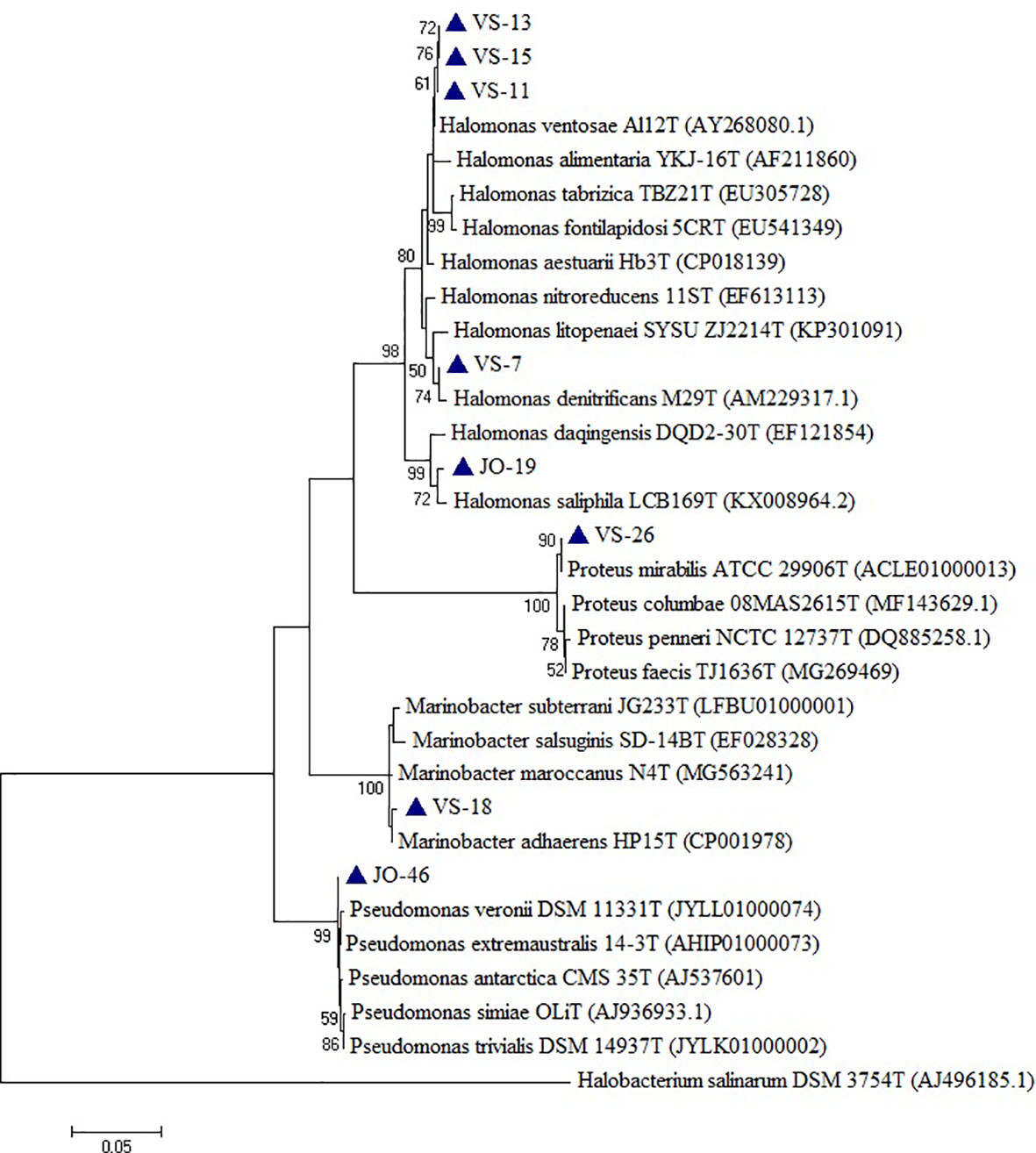
Figure 5 Phylogenetic tree constructed using 16S rRNA gene sequences of LRK isolates belonging to Proteobacteria phylum and their closest phylogenetic relatives. The tree was reconstructed by the Maximum Likelihood method using MEGA 6 software. Halobacterium salinarum DSM 3754T was used as an outgroup. The numbers on the tree indicate the percentages of bootstrap sampling derived from 1,000 replications. Bar, 5 nt substitution per 100 nt. Blue triangles indicates the strains isolated in this study.
Five strains, namely JO-4, JO-18, JO-22, JO-29, and SB-5, were affiliated to phylum Actinobacteria, comprising high G+C Gram-positive bacteria of two different families, Corynebacteriaceae and Micrococcaceae, and three different genera, Corynebacterium, Kocuria, and Micrococcus (Figure 6). Strains VS-36 and JO-44 were identified as archaea affiliated to phylum Euryarchaeota, family Halobacteriaceae, and genus Natronococcus.
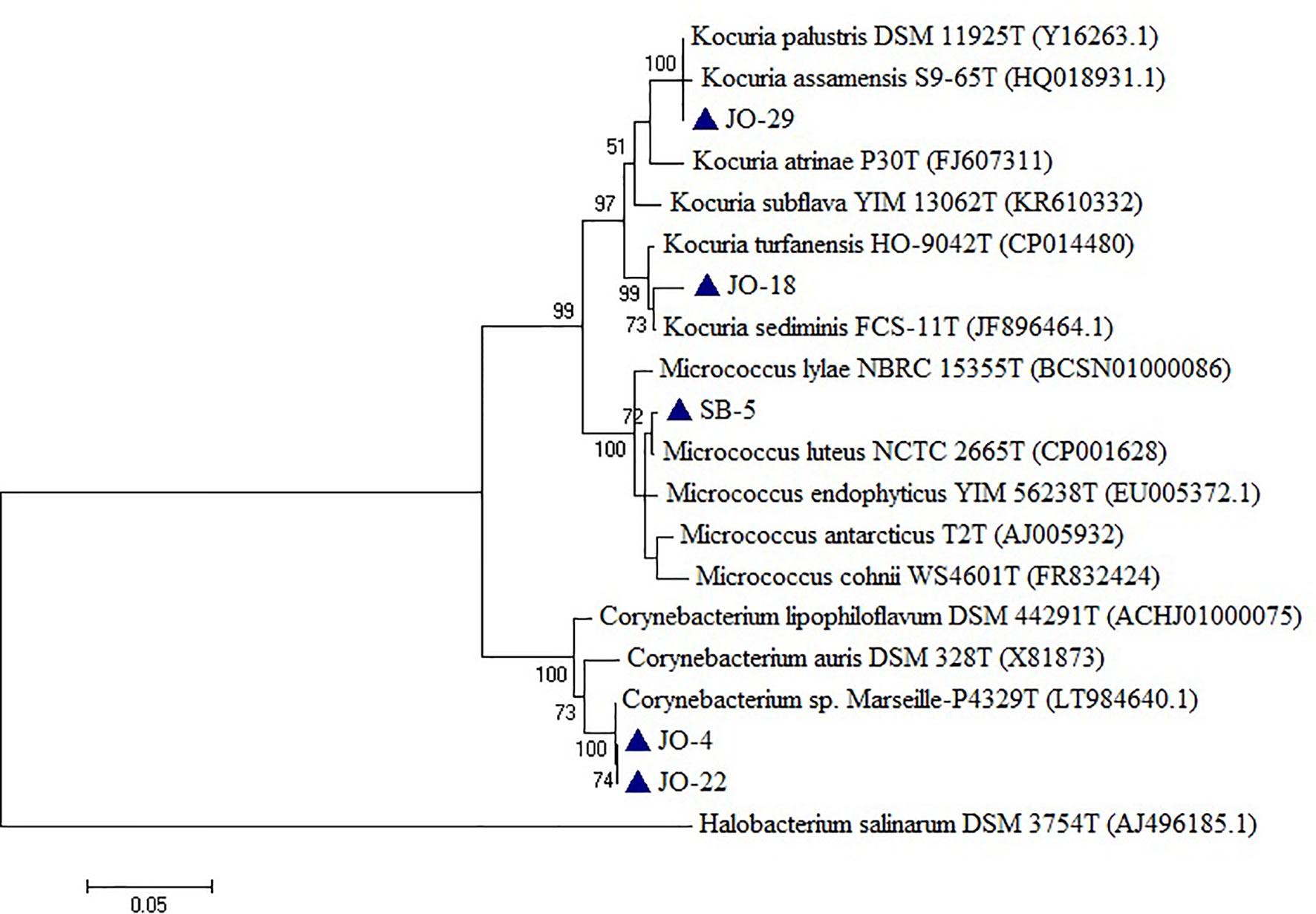
Figure 6 Phylogenetic tree constructed using 16S rRNA gene sequences of LRK isolates belonging to Actinobacteria phylum and closest phylogenetic relatives. The tree was reconstructed by the Maximum Likelihood method using MEGA 6 software. Halobacterium salinarum DSM 3754T was used as an outgroup. The numbers on the tree indicate the percentages of bootstrap sampling derived from 1,000 replications. Bar, 5 nt substitution per 100 nt. Blue triangles indicates the strains isolated in this study.
Spatial Distribution in Surajbari, Venasar, and Jogadstudy Sites
The isolates of the Surajbari were classified in the kingdom bacteria and phylum Firmicutes and Actinobacteria, which included Gram-positive bacteria related to the families of Bacillaceae, Planococcaceae, and Micrococcaceae. The isolates were distributed among nine different genera with 20 species. The relative abundance of different genera in three study sites is as depicted in Figure 7.
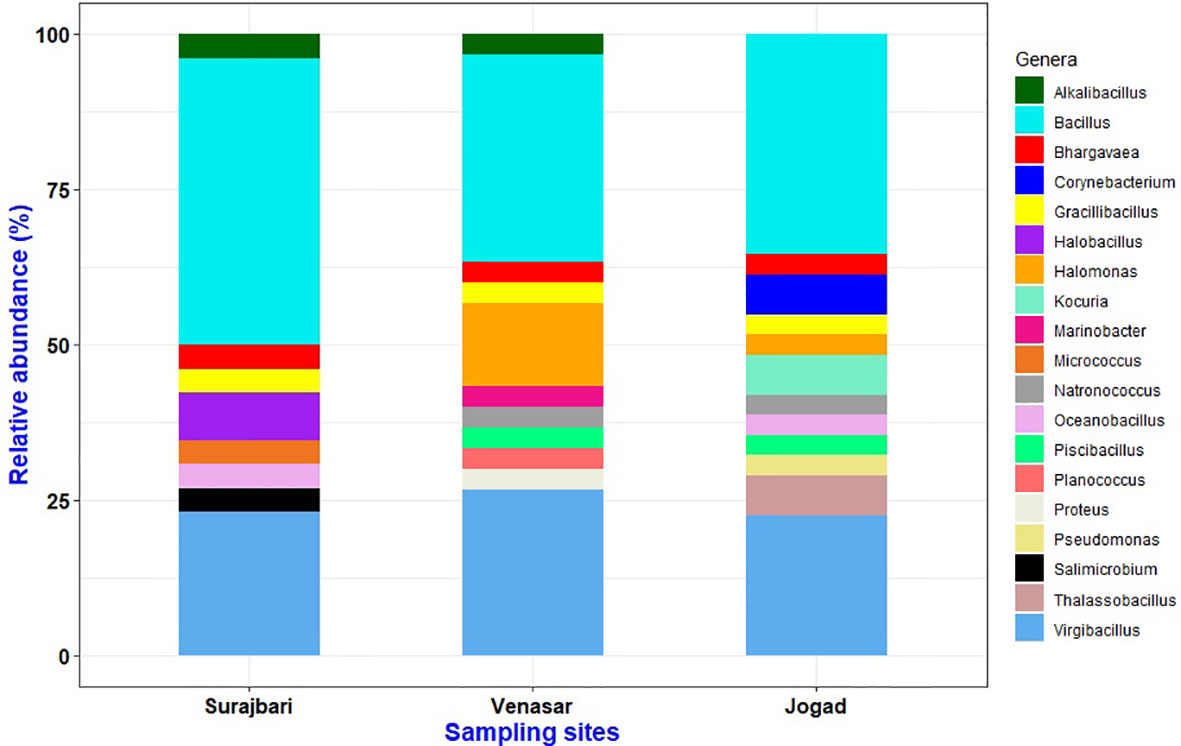
Figure 7 Relative abundance profile of genera obtained from three sampling sites Surajbari, Venasar, and Jogad.
The isolates of the Venasar study site were classified in the two kingdoms, bacteria and archaea, with phyla from Firmicutes, Proteobacteria, and Euryarchaeota, which overall included Gram-positive bacteria belonging to the families of Bacillaceae and Planococcaceae and Gram-negative bacteria related to families Halomonadaceae, Enterobacteriaceae, and Alteromonadaceae as well as members belonging to the family of Halobacteriaceae. The isolates were distributed among 11 different genera, displaying 17 different species (Figure 7).
Similarly, the isolates of the Jogad study site were classified in two kingdoms, bacteria and archaea, with phylums from Firmicutes, Proteobacteria, Actinobacteria, and Euryarchaeota which overall included Gram-positive bacteria related to the families of Bacillaceae, Planococcaceae, Micrococcaceae, and Corynebacteriaceae and Gram-negative bacteria related to families Halomonadaceae, Pseudomonadaceae, and members of the family Halobacteriaceae. The isolates were represented by 11 different genera associated with 23 different species (Figure 7).
Out of 19 genera isolated from all the three study sites, 5.26% (1 genus: Alkalibacillus) was common among Surajbari and Venasar, while 15.78% (3 genera: Halomonas, Piscibacillus, and Natronococcus) were common among Venasar and Jogad. Similarly, 5.26% were represented by 1 genus, Oceanobacillus, which was common between Surajbari and Jogad. While 21% (4 genera: Bacillus, Virgibacillus, Gracilibacillus, and Bhargavaea) were common in all study sites (Figure 8).
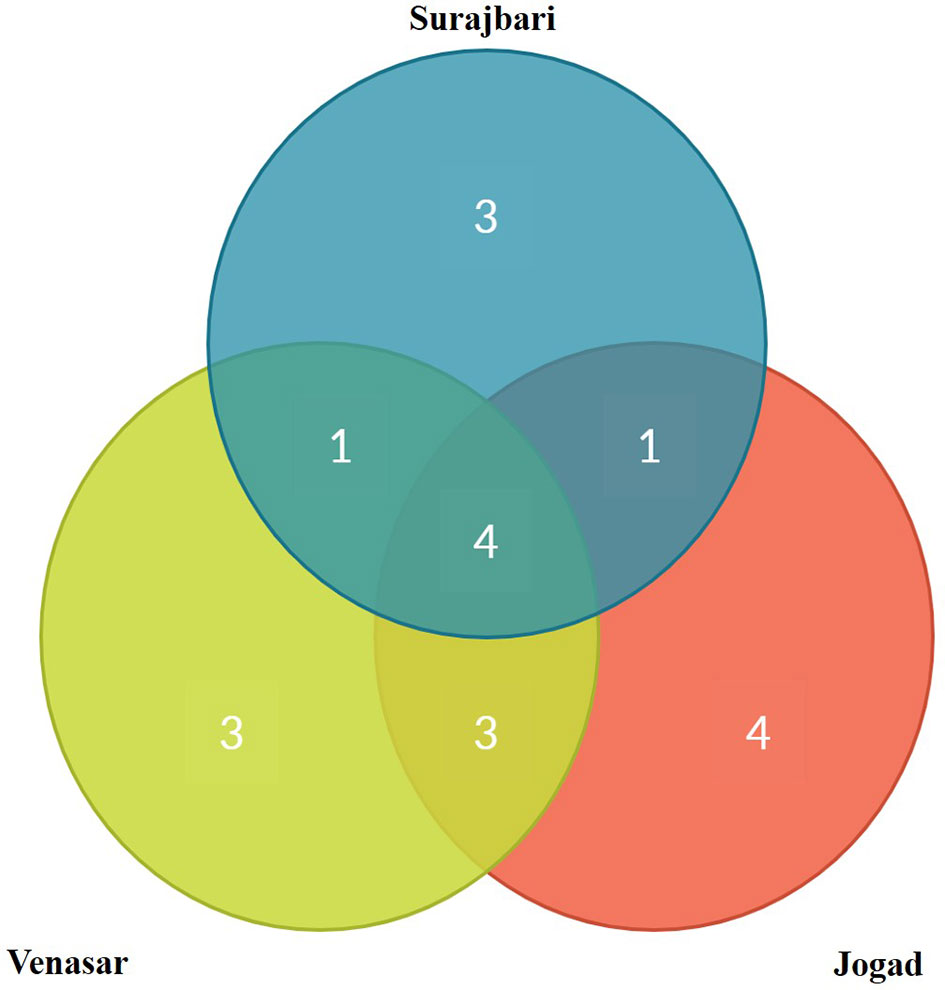
Figure 8 Venn diagram representation of shared and unique genera across three different study sites of LRK.
Further, some genera were exclusively confined to a specific study site. Halobacillus, Salimicrobium, and Micrococcus were exclusively identified in Surajbari, while Proteus, Planococcus, and Marinobacter were present only in Venasar. On the other hand, Corynebacterium, Kocuria, Thalassobacillus, and Pseudomonas were exclusively present in Jogad (Figure 8).
Hierarchical clustering using UPGMA method and Bray-Curtis similarity index indicated more than 68% similarity between the three study sites based on the spatial distribution of genera (Supplementary Figure S1). Venasar and Jogad sites displayed less variation in genera distribution across the spatial profile compared to the Surajbari study site.
Detection of Putative Novel Taxa
Six putative novel taxa were identified in the phylum Firmicutes, Actinobacteria, and Proteobacteria (Figure 9). They had 16S rRNA gene sequence identities below conservative threshold (98.7% identity) with their closest relatives for species level identification (Stackebrandt and Ebers, 2006). Four novel taxa (JO-4 and JO-22, JO-18, JO-21, and JO-39) from Jogad, one from Venasar (VS-10), and one from Surajbari (SB-26) each were obtained.
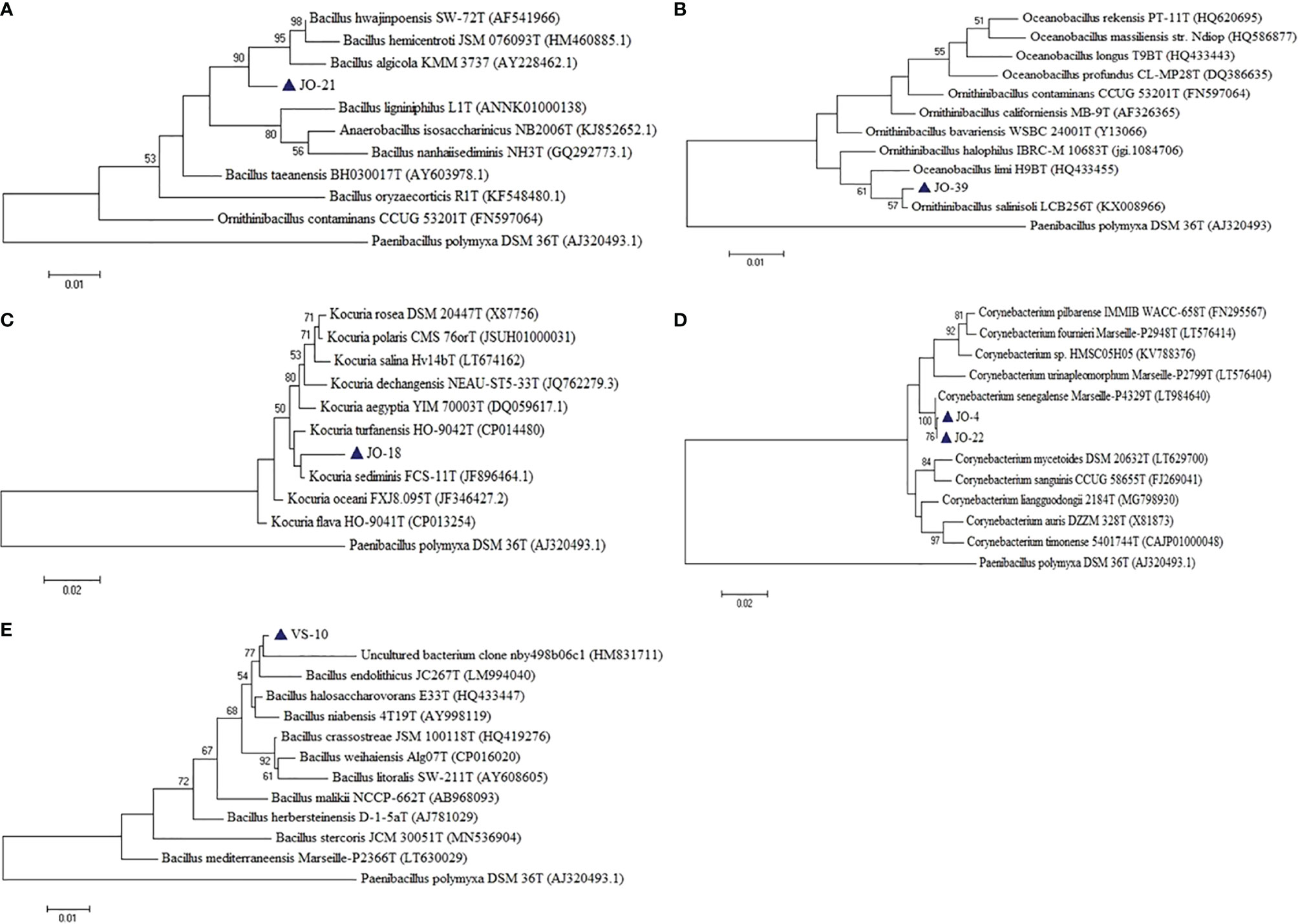
Figure 9 Phylogenetic tree constructed using 16S rRNA gene sequences of putative novel taxa (A) JO-21, (B) JO-39, (C) JO-18 (D) JO-4, and JO-22, (E) VS-10, and some of their closest phylogenetic relatives. The tree was reconstructed by the Maximum Likelihood method using MEGA 6 software. Paenibacillus polymyxa DSM 36T was used as an outgroup. The numbers on the tree indicate the percentages of bootstrap sampling derived from 1,000 replications. Blue triangles indicate the strains isolated in this study.
In the phylum Firmicutes, two putative novel genera and two novel species were identified, related to the family Bacillaceae. JO-21 had a low 16S rRNA gene sequence similarity of 96.60% with their closest phylogenetic relative Bacillus hwajinpoensis SW-72T followed by Bacillus algicola KMM 3737T (95.92%) and Bacillus hemicentroti JSM 076093T (95.77%) (Figure 9A). Moreover, the strain has a sequence similarity of 94.12% with the type species of genus Bacillus, Bacillus subtilis subsp. subtilis DSM10T. Strain JO-21 had equal low sequence similarity with the species of neighboring genera Ornithibacillus contaminans CCUG 53201T (95.53%), Anaerobacillus isosaccharinicus NB2006T (95.38%), Anaerobacillus alkaliphilus B16-10T (94.98%), Anaerobacillus alkalidiazotrophicus MS6T (94.97%), Pradoshia eiseniae EAG3T (94.84%), Fermentibacillus polygoni IEB3T (94.83%), and Desertibacillus haloalkaliphilus KJ1-10-99T (94.77%). As the sequence identities with the most closely related genera were equally low, it may be a new genus and thus further studies are required. Similarly, strain JO-39 had the highest 16S rRNA gene sequence similarity and clustered together with the species of two different genera, Oceanobacillus limi H9BT (98.22%) and Ornithinibacillus salinisoli LCB256T (98.11%), and thus could not be assigned to an existing genus (Figure 9B). They either belong to one of these genera or alternatively may represent new genera.
Strain VS-10 showed low 16S rRNA sequence similarity with their closest phylogenetic relatives, Bacillus halosaccharovorans E33T (96.98%), Bacillus endolithicus JC267T (96.92%), and Bacillus crassostreae JSM 100118T (96.53%) and thus represents novel species (Figure 9E). Strain SB-26 shared equally low 16S rRNA gene sequence similarity with two different species, Halobacillus trueperi (98.7%) and Halobacillus dabanensis (98.7%), thus representing novel species.
In the phylum Actinobacteria, two putative novel species were identified related to genera Corynebacterium and Kocuria. Strains JO-4 and JO-22 have shown 97.7% sequence similarity with their closest phylogenetic relative Corynebacterium auris DSM 328T (97.4%). However, in the phylogenetic tree, the two novel strains were not only placed far apart from their phylogenetic relatives but also formed a separate branch (Figure 9D). Owing to a low 16S rRNA gene sequence similarity and the tree topology, these two strains represent a single novel species. 16S rRNA gene sequence similarity among these two strains was 99.93%, meaning these two strains belong to a single species. Similarly, strain JO-18 showed low sequence similarity of 98.3% with Kocuria sediminis FCS-11T and 97.93% with Kocuria turfanensis HO-9042T and therefore represents a potential new species (Figure 9C). All these isolates are therefore indicative of new genera/species and should be studied further to establish their classification and taxonomic positions.
Effect of Soil Properties on the Bacterial Community
Multivariate analyses are used to elucidate the associations between the abundance of certain genera and environmental parameters. In this study, the Canonical Correspondence Analysis (CCA) (Ter Braak and Verdonschot, 1995) was used to analyze the relationships between the microbial communities of LRK as a function of the physicochemical variables of the habitat (Figure 10). Among the physicochemical parameters, pH, Electrical Conductivity (EC), TDS, organic carbon (OC), nitrogen (N), phosphorous (P), potassium (K), sulphur (S), copper (Cu), ferrous (Fe), Mg, Ca, Na, Cl, B, Zn, and Mn were considered.
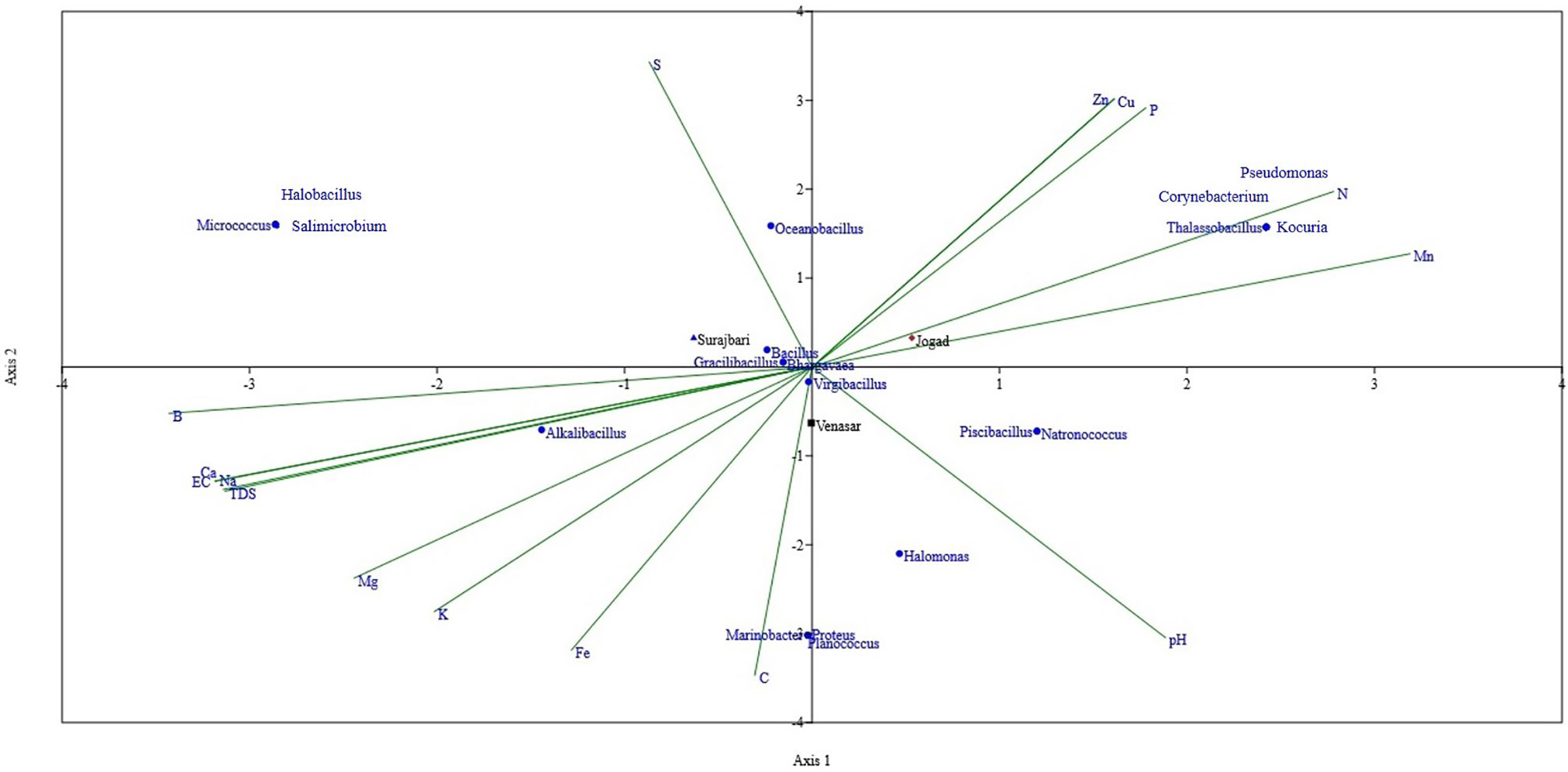
Figure 10 Canonical Correspondence Analysis (CCA) ordination diagram (Triplot) of bacterial structure composition in relation to soil properties of Surajbari, Venasar, and Jogad sites of LRK.
Overall, organic carbon and nitrogen content were found to be the main environmental variables affecting the microbial diversity (Figure 10). Chu et al. (2016) found that soil organic carbon is the best predictor of microbial community distribution. In this study, organic carbon was reported highest in the Venasar site followed by Surajbari and Jogad. Interestingly, a negative correlation between soil microbial diversity and organic carbon was observed. The results are in line with some previous reports (Yao et al., 2020; Cui et al., 2021). On the contrary, a positive correlation was observed between nitrogen content and microbial diversity indices. Nitrogen content was highest in jogad site followed by surajbari and venasar. The results clearly match with the trend of Shannon-Weaver Index (H’) among three sites.
Several genera, such as Thalassobacillus, Corynebacterium, Kocuria, and Pseudomonas, were most positively correlated with nitrogen followed by manganese and phosphorous, while Pseudomonas and Thalassobacillus showed a negative correlation with the conductivity, sodium, chlorine, calcium, and magnesium. A positive correlation was apparent between the carbon content and the occurrence of the Proteobacteria, i.e., Marinobacter, Proteus, and Halomonas. Similarly, Halomonas, Piscibacillus, and Natronococcus have shown a positive correlation with soil pH. A negative correlation existed with sulphur.
Alkalibacillus shows a positive correlation with the electrical conductivity (EC), TDS, calcium, sodium, and chlorine, while it was negatively correlated with nitrogen and phosphorous. Sulphur content had positive correlation with Oceanobacillus, while this genus was negatively correlated with carbon and pH. None of the physicochemical parameters were significantly correlated with the occurrence of Micrococcus, Halobacillus, or Salimicrobium. However, a marginal positive correlation with EC and negative correlation with pH was evident. The other bacterial genera, such as Bacillus, Virgibacillus, Gracillibacillus, and Bhargavaea, were closely aligned to the center of triplot, reflecting an insignificant correlation between their abundance and soil properties.
Sample Coverage and Diversity Measures
On the basis of the Good’s Estimator, 77%, 73.4%, and 77.5% of the cultivable diversity at the genus level was retrieved from the Surajbari, Venasar, and Jogad study sites, respectively. Overall, taking the entire three sites together, 93% of the cultivable diversity was retrieved. Shannon-Weaver Index (H’) was highest in Jogad (2.86) followed by Surajbari (2.79) and Venasar (2.46), while Margalef Index was found to be highest in Jogad (6.115) followed by Surajbari (5.832) and Venasar (4.704).
Global Geographic Distribution of the Phylotypes
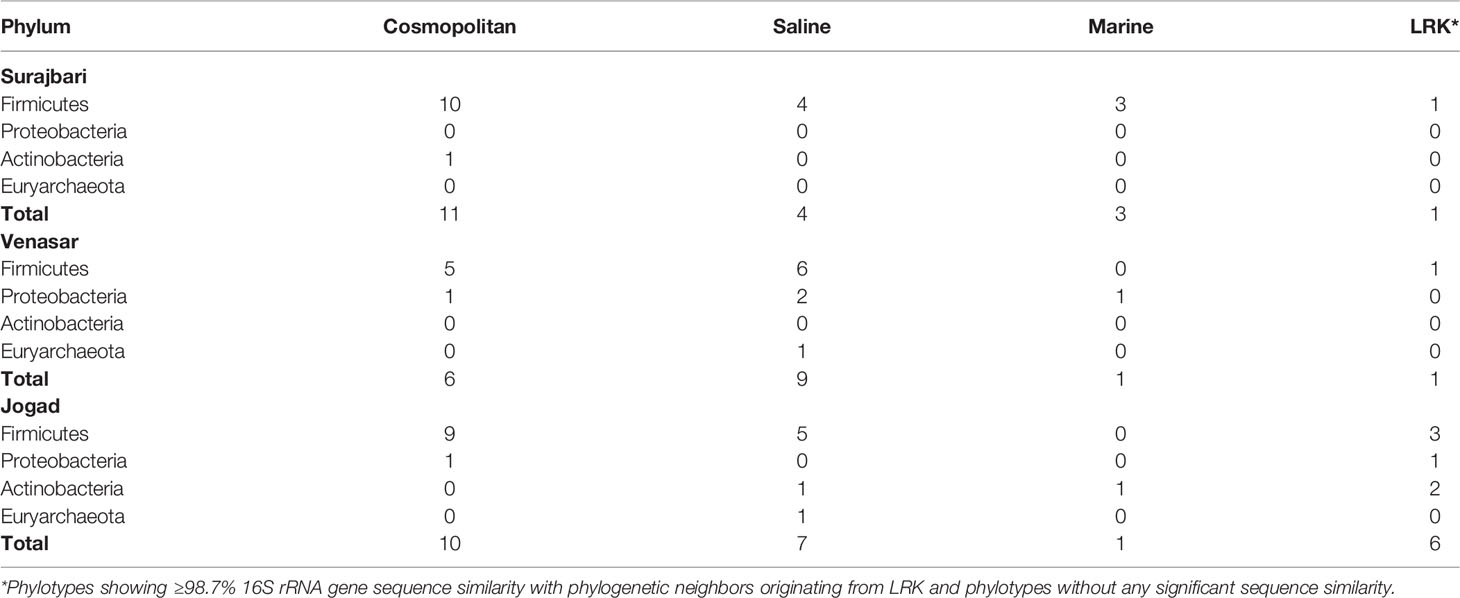
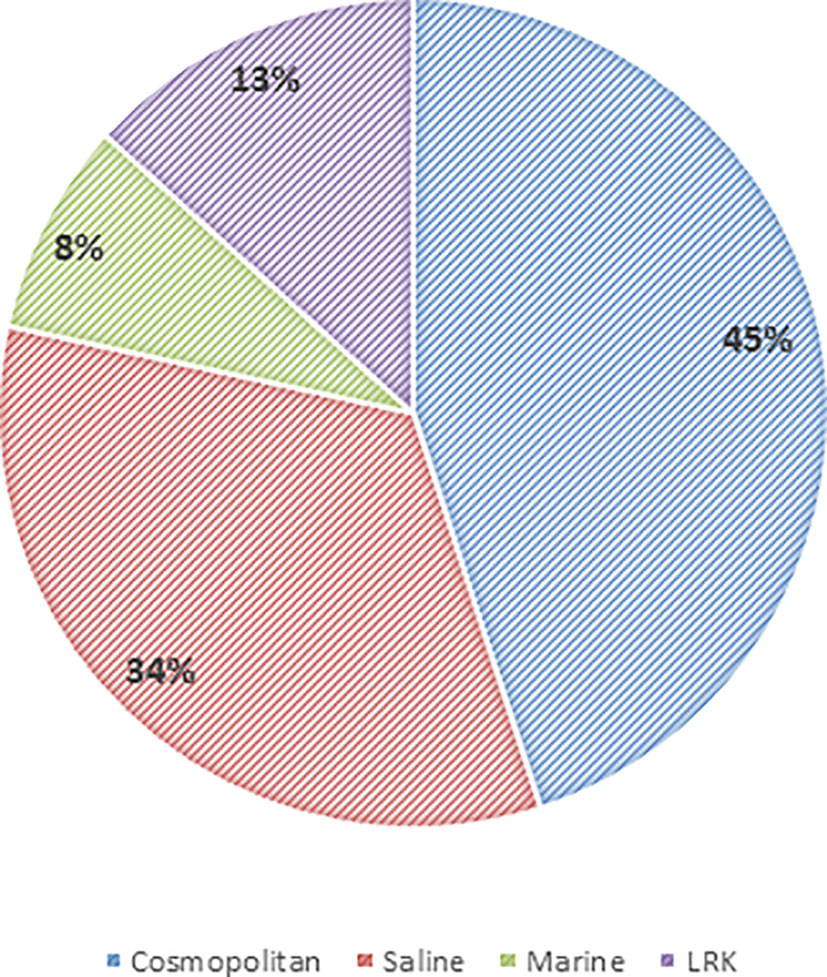
The phylotypes were clustered into four categories, namely LRK, Marine, Saline, and Cosmopolitan, as per the origin and habitat type (Tables 3, 4). Overall, among the three study sites, maximum phylotypes (45%) were mostly of a cosmopolitan nature in their distribution followed by Saline (33.3%), LRK (13.3%), and Marine (8.3%) (Figure 11).
Nearly 58% of the Surajbari phylotypes were associated with the cosmopolitan distribution followed by Saline (21%), Marine (16%), and LRK (5%). Similarly, a majority of the Jogad phylotypes (41.6%) had cosmopolitan distribution followed by Saline (29.1%), LRK (25%), and Marine (3.8%). However, among the Venasar phylotypes, 52.9% were of saline type followed by cosmopolitan (35.2%), Marine (5.8%), and LRK (5.8%).
In phylum Firmicutes, a majority of the phylotypes belonging to the genus Bacillus had cosmopolitan distribution (Table 3). However, Bacillus salecetis (JO-7 and VS-19) and Bacillus halosachharovorans (SB-18) had saline distribution while Bacillus paranthracis (SB-6) had marine distribution. Other genera, such as Oceanobacillus and Bhargavaea, were of the cosmopolitan type in their distribution. Further, the phylotypes belonging to the genera Virgibacillus, Planococcus, Gracilibacillus, Piscibacillus, Alkalibacillus, and Thalassobacillus had saline distribution, while Halobacillus and Salimicrobium had marine type distribution. On the other hand, the phylotypes VS-10, JO-21, JO-39, and SB-26 had no significant similarity with any sequences in the public databases and thus are at present known from LRK only.
Within the phylum Actinobacteria, Corynebacterium strains (JO-4 and JO-22) and Kocuria strain (JO-18) showed no significant similarity with any sequences in the public databases and thus are at present known from LRK only. The Phylotypes Micrococcus aloeverae (SB-5) and Kocuria palustris (JO-29) had cosmopolitan and marine distribution patterns, respectively (Table 3). Among the five Proteobacteria phylotypes, Halomonas ventosae (VS-11 and VS-13) and Marinobacter adhaerens (VS-18), Halomonas denitrificans (VS-7), Proteus mirabilis (VS-26), and Pseudomonas veronii (JO-46) had saline, Marine, and cosmopolitan distribution, respectively. Phylotype JO-19 belonging to Halomonas genus showed no significant similarity with any sequences in the public databases and, therefore, is currently associated with the LRK type only (Table 3). In phylum Euryarchaeota, the phylotype Natronococcus jeotgali (VS-36 and JO-44) was associated with the saline distribution.
Discussion
The Little Rann of Kutch, a saline coastal desert harboring unique flora and fauna, has a different demography from other deserts. It is an admixture of the saline, marshy, and coastal deserts (Gupta and Ansari, 2014; Bhatt et al., 2018). Despite being an enigmatic terrain, LRK has not yet been extensively explored for its microbial diversity. In this study, we investigated the culture dependent microbial diversity employing varied and extensive isolation approaches and 16S rRNA gene based phylogenetic analysis.
Although there were some earlier studies on the microbial diversity of different deserts worldwide, only limited reports are available in the context of Indian deserts (Subrahmanyam et al., 2014; Pandit et al., 2015; Tiwari et al., 2015; Rao et al., 2016). While some aspects of the non-cultivable microbial communities of the LRK have been studied using metagenomics (Patel et al., 2015), there is only one report on the cultivable microbial diversity using enrichment technique (Bhatt et al., 2018). Therefore, we investigated the saline desert of the LRK for the bacterial diversity using various isolation strategies to trap maximum bacteria and novel taxa.
As per the ‘Intermediate disturbance’ hypothesis, highly stable or highly unstable environments harbor only limited microbial diversity in comparison to the moderately unstable habitats (Connell, 1978; Petraitis et al., 1989; Kim et al., 2013). In LRK, during monsoon, marine water of the Gulf of Kutch flows towards the flat surface of the Rann, while it remains dry the rest of the time. Therefore, this desert reflects an intermediate level of disturbance and hence is expected to harbor high microbial diversity (Bhatt et al., 2018).
In order to access maximum diversity, different isolation strategies were employed. Both direct plating and enrichment techniques eventually resulted in capturing highly diverse bacterial flora. Further, Winogradsky’s salt solution along with distilled water was used for serial dilution. Serial dilution of soil using salt solution plays an important role in maintaining osmotic pressure (de Almeida Ribeiro et al., 2015; Sengupta et al., 2015). In direct plating approach, high CFU counts were obtained on different media, such as SP, SE, CM, and HM with the Winogradsky’s salt solution as diluent (Dabek-Szreniawska and Hattori, 1981). However, in R2A medium, the CFU counts were higher in distilled water compared to Winogradsky’s salt solution. Moreover, CFU as well as diversity in phylotypes were reduced in medium with high salt concentrations, such as Complex medium and Halophilic medium.
A majority of the isolates were able to grow in a broad range of pH and salt, reflecting wide opportunity for their applications as well as their ecological significance. However, the isolates growing at very high salt concentrations could grow in a rather narrow range of pH around neutrality. For instance, SB-27, SB-28, SB-29, JO-32, JO-35, JO-36, and JO-42 could grow only in range of 7-9 pH. This finding was previously supported and explained (Bowers and Wiegel, 2011). The haloalkaliphilic bacteria growing at the dual extremity of alkaline pH and high salt encounter more energetic problems as compared to alkaliphilic bacteria (Bowers and Wiegel, 2011). This complication may explain the more prevalent occurrence of halophilic microorganisms growing optimally in environments that are pH neutral or near neutral (Bowers and Wiegel, 2011). Moreover, the isolates with the ability to grow in high salt and alkaline pH were greater in Venasar as compared to the other two sites.
Phylogenetics
The phylogenetic analysis revealed four phyla, Firmicutes, Proteobacteria, Actinobacteria, and Euryarchaeota, with 19 different genera. Archaea dominates the microbial community of the hypersaline soil over bacteria (Hacěne et al., 2004). However, in this study, bacteria were dominant over archaea in LRK, a trend also reflected in a previous metagenomic study (Patel et al., 2015). Saline and hypersaline environments are known to possess the dominance of the gram-negative bacteria (Baati et al., 2010; Makhalanyane et al., 2015; Ronca et al., 2015). However, in the present investigation, the gram-positive bacteria were dominant.
Out of 19 genera reported in this study, Bacillus, Halobacillus, Oceanobacillus, and Virgibacillus were earlier reported (Bhatt et al., 2018), while the rest are being reported for the first time from the LRK. The Firmicutes were most dominant followed by the Proteobacteria, Actinobacteria, and Euryarchaeota, a trend also reflected in some earlier reports of the deserts (El Hidri et al., 2013; Li et al., 2017). However, Proteobacteria were reported as dominant in metagenomic studies of the LRK (Patel et al., 2015), the Great Rann of Kutch (Pandit et al., 2015), and Thar desert of India (Sivakala et al., 2018).
In Surajbari, the Firmicutes dominated followed by Actinobacteria, while Proteobacteria and Euryarchaeota were absent. The Firmicutes were also dominant in Venasar followed by Proteobacteria and Euryarchaeota with the absence of Actinobacteria. In Jogad, Firmicutes still dominated followed by Actinobacteria, Proteobacteria, and Euryarchaeota. The Firmicutes and Actinobacteria were reported as dominant phyla in two geographically distinct saline pan sediments of the Kalahari Desert of Southern Africa (Genderjahn et al., 2018). It is well established that spore forming bacteria can withstand harsh environmental conditions. Once the environmental conditions become favorable and water availability enhances, the cells begin to divide (Jones and Lennon, 2010; Crits-Christoph et al., 2013).
Phylogenetic analysis revealed that the identification of certain isolates was not straightforward, primarily due to the equally high sequence similarities with two different species. For instance, strains SB-28, SB-6, VS-5, and JO-46 display equally high sequence similarities with two different species, Halobacillus trueperi (98.3%) and Halobacillus dabanensis (98.3%), Bacillus paranthracis (99.5%) and Bacillus nitratireducens (99.5%), Bacillus vallismortis (99.8%) and Bacillus subtilis subsp. spizizenii (99.8%), and Pseudomonas veronii (99.4%) and Pseudomonas simiae (99.4%), in that order. Similarly, other isolates such as SB-2, JO-11, SB-23, VS-24, JO-17, VS-22, JO-32, and JO-36 shared equally high sequence similarities with two different species. Therefore, either they belong to one of the species or may represent a new species. However, in order to probe further and ascertain their taxonomic status, the polyphasic characterization is required. The bacterial diversity was highest in Jogad followed by Surajbari and Venasar. This was a fact also revealed by the Shannon-Weaver Index (H’) and Margalef Index.
Effect of Soil Properties on the Spatial Diversity Pattern
As revealed by the CCA analysis, many physicochemical parameters were positively correlated with the bacterial community. Organic carbon, pH, nitrogen, TDS, EC, and Mn were found to be the main environmental factors significantly affecting the microbial diversity. Organic carbon and nitrogen content were significantly high in all three sites.
In saline soils, the SOC content is influenced by two opposing factors: reduced plant inputs which may decrease SOC, and reduced rates of decomposition (and associated mineralization of organic C to CO2) (Datta et al., 2020). Despite the absence of any plant C input to soil of the salt desert, a considerably high amount of organic carbon might be due to the deposition of early Holocene sediments under estuarine deltaic environment (Datta et al., 2020). Furthermore, higher salt concentration improved the soil structure with good aggregation, which can retain organic carbon for a longer time in absence of decomposition due to lower microbial activity (Datta et al., 2020).
On the other hand, Fe, Zn, P, Cu, and B were not associated with the bacterial diversity (Figure 10). Bacillus, Virgibacillus, Gracilibacillus, and Bhargavaea were commonly present in all three study sites. However, Alkalibacillus was exclusively present in Surajbari and Venasar. CCA analysis suggests that Na+, Cl-, Ca2+, Mg2+, and TDS displayed positive correlation with Alkalibacillus. High concentrations of these ions in Surajbari and Venasar compared to Jogad further supports this correlation. Similarly, a positive correlation of Alkalibacillus with Na+ content in saline and alkaline soil of Cuatro Cienegas, Sayula, and San Marcos lakes was identified (Delgado-García et al., 2018).
Halomonas, Natronococcus, and Piscibacillus were exclusively shared between Venasar and Jogad. A positive correlation with pH might be one of the reasons for the exclusive presence of these two genera in these sites. pH in Venasar and Jogad was more alkaline compared to Surajbari. A negative correlation with EC would have played a role in the absence of Halomonas in Surajbari. Earlier, a positive correlation of pH with Halomonas was reported (Oueriaghli et al., 2014). The genus Oceanobacillus was exclusively present in Surajbari and Jogad due to a positive correlation between this genus and sulphur content (Figure 10).
Surajbari, having close proximity with the Gulf of Kutch, has a regular flow of sea water, subsequently causing higher salinity as compared to other sites located at farther distances. Halobacillus, Salimicrobium, and Micrococcus have a marginal positive correlation with the electrical conductivity (EC) and thus were exclusively identified in Surajbari. Proteus, Planococcus, and Marinobacter were exclusively present in Venasar. A positive correlation between the occurrences of these genera with the organic carbon explains their exclusive presence in Venasar. Earlier, a significant correlation between the organic carbon and Gammaproteobacteria were reported from the freshwater Poyang Lake in China (Ding et al., 2015). Further, the exclusive presence of Corynebacterium, Kocuria, Thalassobacillus, and Pseudomonas in Jogad is explained by a positive correlation between these genera, nitrogen (N), and manganese (Mn) contents of the habitat. Previous reports suggests that proteobacteria have a vital role in degrading sedimentary organic nitrogen (Zhou et al., 2013; Zhang et al., 2015). Similarly, Garcia-Lopez et al. (2019) reported a correlation between Pseudomonas and high levels of nitrogen content. Similarly, previous reports suggest the important role of manganese in sporulation of Bacillus sp. (Charney et al., 1951; Weinberg, 1964; Sinnelä et al., 2019). Therefore, spore forming Thalassobacillus might have shown a positive correlation with manganese.
The hierarchical clustering based on the genera distribution revealed considerable variation in the microbial diversity and abundance in different study sites. However, Venasar and Jogad clustered together while Surajbari was placed apart (Supplementary Figure 1). Since Surajbari is in close proximity with the Gulf of Kutch with a regular flow of sea water, its microbial diversity appears different than the other two sites. EC, TDS, Na+, Cl-, and Ca2+ decreased with the increasing distances from the sea, affecting the microbial community in the three study sites.
Novel Taxa Recovered
Based on an earlier metagenomic study, some novel taxa were reported in the LRK (Patel et al., 2015). Two novel taxa, Desertibacillus haloalkaliphilus KJ1-10-99T (Bhatt et al., 2017) and Chryseobacterium salipaludis JC490T (Divyasree et al., 2018), were reported from the LRK. A 97.0% of the 16S rRNA gene sequence similarity is generally accepted as the threshold to regard bacterial species as different (Stackebrandt and Goebel, 1994). A sequence similarity in the range of 98.7–99.0% could be regarded as a threshold range above which DNA–DNA hybridization is required for species identification (Stackebrandt and Ebers, 2006). In the present study, nearly 15% of the phylotypes have shown <98.7% 16S rRNA gene sequence similarity with their phylogenetic neighbors. Based on 16S rRNA gene based phylogenetic analysis with the corresponding phylogenetic neighbors, six strains were identified as the putative novel species (Figure 9). Among these, a majority were from Jogad followed by Venasar and Surajbari These phylotypes are affiliated with the Firmicutes for Surajbari and Venasar while with the Firmicutes and Actinobacteria for Jogad.
Geographical Distribution of the Phylotypes
The 16S rRNA gene sequences of the isolates were compared with the public databases to assess the geographic distribution patterns of different phylotypes. A majority of the phylotypes had cosmopolitan distribution, followed by the saline habitats. Meanwhile, ∼13% of the phylotypes had no significant sequence similarity with their phylogenetic neighbors and thus were considered as the exclusive native of the LRK (Figure 11 and Table 4). Further, ∼8% phylotypes had their distribution confined to marine environment, despite being obtained from the desert, possibly due to the close proximity of the LRK with the Gulf of Kutch (Gupta and Ansari, 2014). In a similar study from Antarctica, 72% of the phylotypes were associated with the cosmopolitan distribution with only 23% being exclusive to Antarctica (Peeters et al., 2011).
A majority of the Firmicutes and Proteobacteria reflected a cosmopolitan and saline distribution (Tables 3 and 4), a trend which might be due to their spore forming ability enabling them to adapt in saline and non-saline environments. The phylotypes within the phylum Actinobacteria had equal distribution to four categories.
The marine distribution of phylotypes decreased with the increasing distance from the Gulf of Kutch, suggesting their endemic nature to the marine environment. It is important to mention that the distribution pattern highlighted in this study is based on the current knowledge of the bacterial diversity and ecology bound by limitations. Therefore, the taxa which are exclusively identified from LRK may turn out as cosmopolitan with the increasing data of the microbial diversity from other habitats.
In the era of metagenomics, there is still a need to improve the conventional cultivation methods to recover the yet unexplored majority of the microorganisms from unusual and unexplored habitats. Metagenomics will not be enough to understand the roles and structures of the microbial communities (Borsodi et al., 2005; Joshi et al., 2008; González-Rocha et al., 2017). In this regard, besides the discovery of potentially new taxa and genera, the availability of the cultures will facilitate detailed investigations including biotechnological avenues. The study also represents the cultivability and geographical distribution of the phylotypes of a saline desert.
Conclusion
This study represents the first extensive analysis of the cultivable bacterial diversity of the unexplored saline desert, the Little Rann of Kutch, Gujarat, India. The study revealed high bacterial diversity with novel taxa in the LRK. The strategies employed to trap maximum diversity and novel taxa yielded a significant outcome. Fifteen different genera are being reported in this study for the first time from LRK. Although the three study sites represented high spatial variation, the bacterial diversity in Jogad and Venasar as compared to that of the Surajbari were similar, arguably due to the variations in soil properties and geographical locations. The CCA analysis predicted the influence of nitrogen, organic carbon, TDS, EC, Sulphur, and aqueous concentrations of various ions on the spatial variation. A majority of the phylotypes were associated with the cosmopolitan distribution followed by those of the saline and marine habitats, while 13% are currently known from the LRK only. The number of phylotypes of marine distribution pattern decreased with the increasing distance from the Gulf of Kutch, suggesting their endemic nature to marine environments. The rich microbial diversity and occurrence of novel species/genera provides a strong base to expand the investigations on the ecological and biotechnological aspects.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
Both HB and SS contributed to the study conception and design. HB performed the experiments, analyzed data and wrote the manuscript. SS supervised the work, helped in analyzing the data and edited the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
HB is thankful to the Council of Scientific and Industrial Research (CSIR), New Delhi, India for the award of Direct Senior Research Fellowship (CSIR–SRF). HB also acknowledges the University Grants Commission (UGC) for the award of UGC BSR Meritorious Fellowship. Authors further acknowledge the infrastructural and financial support under the UGC-Centre of Advanced Study Program [No.F.5-4/2012 (SAP-II)] and DST-FIST program (N0.SR/FST/LSI-281/2006). DBT Multi-Institutional Project, MoES (Government of India) Net-Working Project, and UGC-BSR Faculty Fellowship-Project awarded to SPS are duly acknowledged. We also acknowledge the financial and infrastructural support from Saurashtra University. SS further acknowledges International Travel Supports from SERB-DST, UGC, CSIR, and DBT to present his research in Hamburg (Germany), Brisbane (Australia), Cape Town (South Africa), and Kyoto (Japan).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.769043/full#supplementary-material
Supplementary Figure 1 | Clustering of sampling sites based on genera distribution using Bray-curtis similarity measure and UPGMA algorithm.
References
Abed R. M., Al Kharusi S., Schramm A., Robinson M. D. (2010). Bacterial Diversity, Pigments and Nitrogen Fixation of Biological Desert Crusts From the Sultanate of Oman. FEMS Microbiol. Ecol. 72, 418–428. doi: 10.1111/j.1574-6941.2010.00854.x
Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic Local Alignment Search Tool. J. Mol. Biol. 215 (3), 403–410. doi: 10.1016/S0022-2836(05)80360-2
An S., Couteau C., Luo F., Neveu J., DuBow M. S. (2013). Bacterial Diversity of Surface Sand Samples From the Gobi and Taklamaken Deserts. Microb. Ecol. 66, 850–860. doi: 10.1007/s00248-013-0276-2
Asija G. L., Subbiah B. V. (1956). A Rapid Procedure for the Estimation of Available Nitrogen in Soils. Curr. Sci. 25, 259–260.
Baati H., Amdouni R., Gharsallah N., Sghir A., Ammar E. (2010). Isolation and Characterization of Moderately Halophilic Bacteria From Tunisian Solar Saltern. Curr. Microbiol. 60, 157–161. doi: 10.1007/s00284-009-9516-6
Bachran M., Kluge S., Lopez-Fernandez M., Cherkouk A. (2018). Microbial Diversity in an Arid, Naturally Saline Environment. Microb. Ecol. 78 (2), 494–505. doi: 10.1007/s00248-018-1301-2
Baweja M., Tiwari R., Singh P. K., Nain L., Shukla P. (2016). An Alkaline Protease From Bacillus Pumilus MP 27: Functional Analysis of Its Binding Model Toward its Applications as Detergent Additive. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.01195
Bhatt H. B., Begum M. A., Chintalapati S., Chintalapati V. R., Singh S. P. (2017). Desertibacillus Haloalkaliphilus Gen. Nov., Sp. Nov., Isolated From a Saline Desert. Int. J. Syst. Evol. Microbiol. 67, 4435–4442. doi: 10.1099/ijsem.0.002310
Bhatt H. B., Gohel S. D., Singh S. P. (2018). Phylogeny, Novel Bacterial Lineage and Enzymatic Potential of Haloalkaliphilic Bacteria From the Saline Coastal Desert of Little Rann of Kutch, Gujarat, India. 3 Biotech. 8, 53. doi: 10.1007/s13205-017-1075-0
Bhatt H. B., Singh S. P. (2016). “Phylogenetic and Phenogram Based Diversity of Haloalkaliphilic Bacteria From the Saline Desert,” in Microbial Biotechnology, Eds. Bhima Bhukya and Anjana Devi Tangutur (Apple Academic Press), 373–386. doi: 10.1201/b19978-24
Bhatt H. B., Singh S. P. (2020). Cloning, Expression, and Structural Elucidation of a Biotechnologically Potential Alkaline Serine Protease From a Newly Isolated Haloalkaliphilic Bacillus Lehensis JO-26. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.00941
Binayke A., Ghorbel S., Hmidet N., Raut A., Gunjal A., Uzgare A., et al. (2018). Analysis of Diversity of Actinomycetes From Arid and Saline Soils at Rajasthan, India. Environ. Sustain. 1, 61–70. doi: 10.1007/s42398-018-0003-5
Bodor A., Bounedjoum N., Vincze G. E., Kis Á.E., Laczi K., Bende G., et al. (2020). Challenges of Unculturable Bacteria: Environmental Perspectives. Rev. Environ. Sci. Biotechnol. 19 (1), 1–22. doi: 10.1007/s11157-020-09522-4
Borsodi A. K., Micsinai A., Rusznyak A., Vladar P., Kovacs G., Toth E. M., et al. (2005). Diversity of Alkaliphilic and Alkalitolerant Bacteria Cultivated From Decomposing Reed Rhizomes in a Hungarian Soda Lake. Microb. Ecol. 50, 9–18. doi: 10.1007/s00248-004-0063-1
Bowers K. J., Wiegel JA.. (2011). Temperature and pH Optima of Extremely Halophilic Archaea: A Mini-Review. Extremophiles 15 (2), 119–128. doi: 10.1007/s00792-010-0347-y
Bruns A., Cypionka H., Overmann J. (2002). Cyclic AMP and Acyl Homoserine Lactones Increase the Cultivation Efficiency of Heterotrophic Bacteria From the Central Baltic Sea. Appl. Environ. Microbiol. 68, 3978–3987. doi: 10.1128/AEM.68.8.3978-3987.2002
Caton T. M., Witte L.R., Ngyuen H.D., Buchheim J.A., Buchheim M.A., Schneegurt M.A. (2004). Halotolerant Aerobic Heterotrophic Bacteria From the Great Salt Plains of Oklahoma. Microb. Ecol. 48 (4), 449–462. doi: 10.1007/s00248-004-0211-7
Caton I. R., Schneegurt M. A. (2012). Culture-Independent Analysis of the Soil Bacterial Assemblage at the Great Salt Plains of Oklahoma. J. Basic Microbiol. 52, 16–26. doi: 10.1002/jobm.201100175
Charney J., Fisher W. P., Hegarty C. P. (1951). Manganese as an Essential Element for Sporulation in the Genus Bacillus. J. Bacteriol. 62, 145–148. doi: 10.1128/jb.62.2.145-148.1951
Chaudhary D. K., Khulan A., Kim J. (2019). Development of a Novel Cultivation Technique for Uncultured Soil Bacteria. Sci. Rep. 9 (1), 1–1. doi: 10.1038/s41598-019-43182-x
Chesnin L., Yien C. H. (1951). Turbidimetric Determination of Available Sulfates. Soil Sci. Soc. Am. J. 15 (C), 149–151. doi: 10.2136/sssaj1951.036159950015000C0032x
Cho J. C., Giovannoni S. J. (2004). Cultivation and Growth Characteristics of a Diverse Group of Oligotrophic Marine Gammaproteobacteria. Appl. Environ. Microbiol. 70, 432–440. doi: 10.1128/AEM.70.1.432-440.2004
Chu H., Sun H., Tripathi B.M., Adams J.M., Huang R., Zhang Y., et al (2016). Bacterial Community Dissimilarity Between the Surface and Subsurface Soils Equals Horizontal Differences Over Several Kilometers in the Western Tibetan Plateau. Environ. Microbiol. 18 (5), 1523–1533. doi: 10.1111/1462-2920.13236
Connell J. H. (1978). Diversity in Tropical Rain Forests and Coral Reefs. Science 199, 1302–1310. doi: 10.1126/science.199.4335.1302
Connon S. A., Giovannoni S. J. (2002). High-Throughput Methods for Culturing Microorganisms in Very-Low-Nutrient Media Yield Diverse New Marine Isolates. Appl. Environ. Microbiol. 68, 3878–3885. doi: 10.1128/AEM.68.8.3878-3885.2002
Crits-Christoph A., Robinson C. K., Barnum T., Fricke W. F., Davila A. F., Jedynak B., et al. (2013). Colonization Patterns of Soil Microbial Communities in the Atacama Desert. Microbiome 1, 28. doi: 10.1186/2049-2618-1-28
Cui Y., Dong Y., Lui H., Sun Z. (2021). Short-Term Grazing Exclusions Reduced Soil Organic Carbon But Not Bacterial Diversity in the Sagebrush Desert, Northwest Chinal. Glob. Ecol. Conserv. 31, e01872. doi: 10.1016/j.gecco.2021.e01872
Dabek-Szreniawska M., Hattori T. (1981). Winogradsky's Salts Solution as a Diluting Medium for Plate Count of Oligotrophic Bacteria in Soil. J. Gen. Appl. Microbiol. 27, 517–518. doi: 10.2323/jgam.27.517
Datta A., Mahato A. K. R., Jat H. S., Sharma P. C. (2020). Soil Organic Carbon Pools and Microbial Population in Extremely Saline Soils: A Case Study in Salt Desert of Rann of Kachchh, India. Eur. J. Environ. Earth Sci. 1 (3). doi: 10.24018/ejgeo.2020.1.3.24
Davis K. E., Joseph S. J., Janssen P. H. (2005). Effects of Growth Medium, Inoculum Size, and Incubation Time on Culturability and Isolation of Soil Bacteria. Appl. Environ. Microbiol. 71, 826–834. doi: 10.1128/AEM.71.2.826-834.2005
de Almeida Ribeiro P. R., dos Santos J. V., da Costa E. M., Lebbe L., Assis E. S., Louzada M. O., et al. (2015). Symbiotic Efficiency and Genetic Diversity of Soybean Bradyrhizobia in Brazilian Soils. Agric. Ecosyst. Environ. 212, 85–93. doi: 10.1016/j.agee.2015.06.017
Delgado-García M., Contreras-Ramos S. M., Rodríguez J. A., Mateos-Díaz J. C., Aguilar C. N., Camacho-Ruíz R. M. (2018). Isolation of Halophilic Bacteria Associated With Saline and Alkaline-Sodic Soils by Culture Dependent Approach. Heliyon 4, e00954. doi: 10.1016/j.heliyon.2018.e00954
Ding X., Peng X. J., Jin B. S., Xiao M., Chen J. K., Li B., et al. (2015). Spatial Distribution of Bacterial Communities Driven by Multiple Environmental Factors in a Beach Wetland of the Largest Freshwater Lake in China. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.00129
Divyasree B., Suresh G., Sasikala C., Ramana C. V. (2018). Chryseobacterium Salipaludis Sp. Nov., Isolated at a Wild Ass Sanctuary. Int. J. Syst. Evol. Microbiol. 68, 542–546. doi: 10.1099/ijsem.0.002536
Dong H., Rech J. A., Jiang H., Sun H., Buck B. J. (2007). Endolithic Cyanobacteria in Soil Gypsum: Occurrences in Atacama (Chile), Mojave (United States), and Al-Jafr Basin (Jordan) Deserts. J. Geophys. Res. Biogeosci 112 (G2). doi: 10.1029/2006JG000385
Dwivedi P., Sharma A. K., Singh S. P. (2021). Biochemical Properties and Repression Studies of an Alkaline Serine Protease From a Haloalkaliphilic Actinomycete, Nocardiopsis dassonvillei subsp. albirubida OK-14. Biocatal. Agric. Biotechnol. 35, 102059.
El Hidri D., Guesmi A., Najjari A., Cherif H., Ettoumi B., Hamdi C., et al. (2013). Cultivation-Dependant Assessment, Diversity, and Ecology of Haloalkaliphilic Bacteria in Arid Saline Systems of Southern Tunisia. BioMed. Res. Int. 2013. doi: 10.1155/2013/648141
Frere H. B. E. (1870). Notes on the Runn of Cutch and Neighbouring Region. J. R. Geogr. Soc Lond. 40, 181–207. doi: 10.2307/1798642
Garcia-Lopez E., Rodriguez-Lorente I., Alcazar P., Cid C. (2019). Microbial Communities in Coastal Glaciers and Tidewater Tongues of Svalbard archipelago, Norway. Front. Mar. Sci. 5, 512. doi: 10.3389/fmars.2018.00512
Genderjahn S., Alawi M., Mangelsdorf K., Horn F., Wagner D. (2018). Desiccation-And Saline-Tolerant Bacteria and Archaea in Kalahari Pan Sediments. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.02082
González-Rocha G., Muñoz-Cartes G., Canales-Aguirre C. B., Lima C. A., Domínguez-Yévenes M., Bello-Toledo H., et al. (2017). Diversity Structure of Culturable Bacteria Isolated From the Fildes Peninsula (King George Island, Antarctica): A Phylogenetic Analysis Perspective. PloS One 12, e0179390. doi: 10.1371/journal.pone.0179390
Good I. J. (1953). The Population Frequencies of Species and the Estimation of Population Parameters. Biometrika 40, 237–264. doi: 10.1093/biomet/40.3-4.237
Gupta V., Ansari A. A. (2014). Geomorphic Portrait of the Little Rann of Kutch. Arab. J. Geosci. 7, 527–536. doi: 10.1007/s12517-012-0743-y
Hacěne H., Rafa F., Chebhouni N., Boutaiba S., Bhatnagar T., Baratti J. C., et al. (2004). Biodiversity of Prokaryotic Microflora in El Golea Salt Lake, Algerian Sahara. J. Arid. Environ. 58, 273–284. doi: 10.1016/j.jaridenv.2003.08.006
Hammer Ø., Harper D. A., Ryan P. D. (2001). PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol. Electron. 4, 9.
Hettipathirana T. (2011). Determination of Metals in Soils Using the 4100 MP-AES (Melbourne, Australia, Agilent Technologies Melbourne, Australia), 667–683.
Ishnava K., Ramarao V., Mohan J. S. S., Kothari I. L. (2011). Ecologically Important and Life Supporting Plants of Little Rann of Kachchh, Gujarat. J. Ecol. Nat. Environ. 3, 33–38. doi: 10.5897/JENE.9000044
Jones S. E., Lennon J. T. (2010). Dormancy Contributes to the Maintenance of Microbial Diversity. PNAS 107, 5881–5886. doi: 10.1073/pnas.0912765107
Joshi A. A., Kanekar P. P., Kelkar A. S., Shouche Y. S., Vani A. A., Borgave S. B., et al. (2008). Cultivable Bacterial Diversity of Alkaline Lonar Lake, India. Microb. Ecol. 55, 163–172. doi: 10.1007/s00248-007-9264-8
Kim O. S., Cho Y. J., Lee K., Yoon S. H., Kim M., Na H., et al. (2012). Introducing EzTaxon-E: A Prokaryotic 16S rRNA Gene Sequence Database With Phylotypes That Represent Uncultured Species. Int. J. Syst. Evol. Microbiol. 62, 716–721. doi: 10.1099/ijs.0.038075-0
Kim M., Heo E., Kang H., Adams J. (2013). Changes in Soil Bacterial Community Structure With Increasing Disturbance Frequency. Microb. Ecol. 66, 171–181. doi: 10.1007/s00248-013-0237-9
Li Y., Chen J., Yang Z., Wang Y., Zhang Y., Zhou Y. (2017). Changes in Desert Steppe Soil Culturable Bacteria From Northwestern China and Correlation With Physicochemical Parameters. Toxicol. Environ. Chem. 99, 809–823. doi: 10.1080/02772248.2017.1323461
Li K., Liu R., Zhang H., Yun J. (2013). The Diversity and Abundance of Bacteria and Oxygenic Phototrophs in Saline Biological Desert Crusts in Xinjiang, Northwest China. Microb. Ecol. 66, 40–48. doi: 10.1007/s00248-012-0164-1
Magurran A. E. (1988). Ecological Diversity and its Measurements (New Jersey: Princeton University Press), 192.
Makhalanyane T. P., Valverde A., Gunnigle E., Frossard A., Ramond J. B., Cowan D. A. (2015). Microbial Ecology of Hot Desert Edaphic Systems. FEMS Microbiol. 39, 203–221. doi: 10.1093/femsre/fuu011
Olsen S. R., Cole C. V., Watanabe F. S., Dean L. A. (1954). Estimation of Available Phosphorus Insoils by Extraction With Sodium Bicarbonate (Washington: Circular 939, US Dept. of Agric.), 1–19.
Oueriaghli N., González-Domenech C. M., Martínez-Checa F., Muyzer G., Ventosa A., Quesada E., et al. (2014). Diversity and Distribution of Halomonas in Rambla Salada, a Hypersaline Environment in the Southeast of Spain. FEMS Microbiol. Ecol. 87, 460–474. doi: 10.1111/1574-6941.12237
Pagaling E., Wang H., Venables M., Wallace A., Grant W. D., Cowan D. A., et al. (2009). Microbial Biogeography of Six Salt Lakes in Inner Mongolia, China, and a Salt Lake in Argentina. Appl. Environ. Microbiol. 75, 5750–5760. doi: 10.1128/AEM.00040-09
Pandit A. S., Joshi M. N., Bhargava P., Shaikh I., Ayachit G. N., Raj S. R., et al. (2015). A Snapshot of Microbial Communities From the Kutch: One of the Largest Salt Deserts in the World. Extremophiles 19, 973–987. doi: 10.1007/s00792-015-0772-z
Patel R., Mevada V., Prajapati D., Dudhagara P., Koringa P., Joshi C. G. (2015). Metagenomic Sequence of Saline Desert Microbiota From Wild Ass Sanctuary, Little Rann of Kutch, Gujarat, India. Genom 3, 137–139. doi: 10.1016/j.gdata.2015.01.003
Peeters K., Hodgson D. A., Convey P., Willems A. (2011). Culturable Diversity of Heterotrophic Bacteria in Forlidas Pond (Pensacola Mountains) and Lundström Lake (Shackleton Range), Antarctica. Microb. Ecol. 62, 399. doi: 10.1007/s00248-011-9842-7
Petraitis P. S., Latham R. E., Niesenbaum R. A. (1989). The Maintenance of Species Diversity by Disturbance. Q. Rev. Biol. 64, 393–418. doi: 10.1086/416457
Post F. J. (1977). The Microbial Ecology of the Great Salt Lake. Microb. Ecol. 3, 143–165. doi: 10.1007/BF02010403
Purohit M. K., Singh S. P. (2009). Assessment of Various Methods for Extraction of Metagenomic DNA From Saline Habitats of Coastal Gujarat (India) to Explore Molecular Diversity. Lett. Appl. Microbiol. 49 (3), 338–344. doi: 10.1111/j.1472-765X.2009.02663.x
Purohit M. K., Singh S. P. (2011). Comparative Analysis of Enzymatic Stability and Amino Acid Sequences of Thermostable Alkaline Proteases From Two Haloalkaliphilic Bacteria Isolated From Coastal Region of Gujarat, India. Int. J. Biol. Macromol. 49, 103–112. doi: 10.1016/j.ijbiomac.2011.04.001
Raiyani N. M., Singh S. P. (2020). Taxonomic and Functional Profiling of the Microbial Communities of Arabian Sea: A Metagenomics Approach. Genomics 112 (6), 4361–4369. doi: 10.1016/j.ygeno.2020.07.024
Rao S., Chan Y., Bugler-Lacap D. C., Bhatnagar A., Bhatnagar M., Pointing S. B. (2016). Microbial Diversity in Soil, Sand Dune and Rock Substrates of the Thar Monsoon Desert, India. Indian J. Microbiol. 56, 35–45. doi: 10.1007/s12088-015-0549-1
Rathore D. S., Singh S. P. (2021). Kinetics of Growth and Co-Production of Amylase and Protease in Novel Marine Actinomycete, Streptomyces Lopnurensis Kam5. Folia Microbiol. 66 (3), 303–316. doi: 10.1007/s12223-020-00843-z
Raval V. H., Bhatt H. B., Singh S. P. (2018). “Adaptation Strategies in Halophilic Bacteria,” in Extremophiles. Eds. Durvasula R. V., Subba Rao D. V. (Boca Raton, FL: CRC Press), 137–164.
Raval V. H., Rawal C. M., Pandey S., Bhatt H. B., Dahima B. R., Singh S. P. (2015). Cloning, Heterologous Expression and Structural Characterization of an Alkaline Serine Protease From Sea Water Haloalkaliphilic Bacterium. Ann. Microbiol. 65 (1), 371–381. doi: 10.1007/s13213-014-0869-0
Reasoner D. J., Geldreich E. (1985). A New Medium for the Enumeration and Subculture of Bacteria From Potable Water. Appl. Environ. Microbiol. 49, 1–7. doi: 10.1128/aem.49.1.1-7.1985
Ronca S., Ramond J. B., Jones B. E., Seely M., Cowan D. A. (2015). Namib Desert Dune/Interdune Transects Exhibit Habitat-Specific Edaphic Bacterial Communities. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.00845
Saul-Tcherkas V., Unc A., Steinberger Y. (2013). Soil Microbial Diversity in the Vicinity of Desert Shrubs. Microb. Ecol. 65, 689–699. doi: 10.1007/s00248-012-0141-8
Sengupta S., Pramanik A., Ghosh A., Bhattacharyya M. (2015). Antimicrobial Activities of Actinomycetes Isolated From Unexplored Regions of Sundarbans Mangrove Ecosystem. BMC Microbiol. 15, 170. doi: 10.1186/s12866-015-0495-4
Siddhapura P. K., Vanparia S., Purohit M. K., Singh S. P. (2010). Comparative Studies on the Extraction of Metagenomic DNA From the Saline Habitats of Coastal Gujarat and Sambhar Lake, Rajasthan (India) in Prospect of Molecular Diversity and Search for Novel Biocatalysts. Int. J. Biol. Macromol. 47 (3), 375–379. doi: 10.1016/j.ijbiomac.2010.06.004
Si J. B., Jang E. J., Charalampopoulos D., Wee Y. J. (2018). Purification and Characterization of Microbial Protease Produced Extracellularly From Bacillus Subtilis FBL-1. Biotechnol. Bioprocess. Eng. 23, 176–182. doi: 10.1007/s12257-017-0495-3
Sinha R., Khare S. K. (2013). Characterization of Detergent Compatible Protease of a Halophilic Bacillus Sp. EMB9: Differential Role of Metal Ions in Stability and Activity. Bioresour. Technol. 145, 357–361. doi: 10.1016/j.biortech.2012.11.024
Sinnelä M. T., Park Y. K., Lee J. H., Jeong K. C., Kim Y. W., Hwang H. J., et al. (2019). Effects of Calcium and Manganese on Sporulation of Bacillus Species Involved in Food Poisoning and Spoilage. Foods 8, 119. doi: 10.3390/foods8040119
Sivakala K. K., Jose P. A., Anandham R., Thinesh T., Jebakumar S. R. D., Samaddar S., et al. (2018). Spatial Physicochemical and Metagenomic Analysis of Desert Environment. J. Microbiol. Biotechn. 28, 1517–1526. doi: 10.4014/jmb.1804.04005
Stackebrandt E., Ebers J. (2006). Taxonomic Parameters Revisited: Tarnished Gold Standards. Microbiol. Today 33, 152–155.
Stackebrandt E., Goebel B. M. (1994). Taxonomic Note: A Place for DNA-DNA Reassociation and 16S rRNA Sequence Analysis in the Present Species Definition in Bacteriology. Int. J. Syst. Evol. Microbiol. 44, 846–849. doi: 10.1099/00207713-44-4-846
Stevenson B. S., Eichorst S. A., Wertz J. T., Schmidt T. M., Breznak J. A. (2004). New Strategies for Cultivation and Detection of Previously Uncultured Microbes. Appl. Environ. Microbiol. 70, 4748–4755. doi: 10.1128/AEM.70.8.4748-4755.2004
Subrahmanyam G., Khonde N., Maurya D. M., Chamyal L. S., Archana G. (2014). Microbial Activity and Culturable Bacterial Diversity in Sediments of the Great Rann of Kachchh, Western India. Pedosphere 24, 45–55. doi: 10.1016/S1002-0160(13)60079-X
Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Ter Braak C. J., Verdonschot P. F. (1995). Canonical Correspondence Analysis and Related Multivariate Methods in Aquatic Ecology. Aquat. Sci. 57 (3), 255–289. doi: 10.1007/BF00877430
Thakrar F. J., Singh S. P. (2019). Catalytic, Thermodynamic and Structural Properties of an Immobilized and Highly Thermostable Alkaline Protease From a Haloalkaliphilic Actinobacteria, Nocardiopsis Alba TATA-5. Bioresour. Technol. 278, 150–158. doi: 10.1016/j.biortech.2019.01.058
Tiwari K., Upadhyay D. J., Mösker E., Süssmuth R., Gupta R. K. (2015). Culturable Bioactive Actinomycetes From the Great Indian Thar Desert. Ann. Microbiol. 65, 1901–1914. doi: 10.1007/s13213-014-1028-3
UNESCO. (2006). World Heritage Centre-Nomination Entry. Available at: http://whc.unesco.org/en/tentativelists/2105/ (Accessed 12 July 2019).
Vavourakis C. D., Ghai R., Rodriguez-Valera F., Sorokin D. Y., Tringe S. G., Hugenholtz P., et al. (2016). Metagenomic Insights Into the Uncultured Diversity and Physiology of Microbes in Four Hypersaline Soda Lake Brines. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.00211
Walkley A., Black I. A. (1934). An Examination of the Degtjareff Method for Determining Soil Organic Matter, and a Proposed Modification of the Chromic Acid Titration Method. Soil. Sci. 37, 29–38. doi: 10.1097/00010694-193401000-00003
Weinberg E. D. (1964). Manganese Requirement for Sporulation and Other Secondary Biosynthetic Processes of Bacillus. Appl. Microbiol. 12, 436–441. doi: 10.1128/am.12.5.436-441.1964
Yadav A. N., Verma P., Kumar M., Pal K. K., Dey R., Gupta A., et al. (2015). Diversity and Phylogenetic Profiling of Niche-Specific Bacilli From Extreme Environments of India. Ann. Microbiol. 65, 611–629. doi: 10.1007/s13213-014-0897-9
Yao X., Yu K., Deng Y., Liu J. (2020). Spatial Variability of Soil Organic Carbon and Total Nitrogen in the Hilly Red Soil Region of Southern China. J. For. Res. 6, 2385–2394. doi: 10.1007/s11676-019-01014-8
Zhang J., Yang Y., Zhao L., Li Y., Xie S., Liu Y. (2015). Distribution of Sediment Bacterial and Archaeal Communities in Plateau Freshwater Lakes. Appl. Microbiol. Biotechnol. 99 (7), 3291–3302. doi: 10.1007/s00253-014-6262-x
Keywords: spatial distribution analysis, bacterial diversity, coastal saline desert, Little Rann of Kutch, phylogenetic analysis, novel taxa, geographical distribution, haloalkaliphilic bacteria
Citation: Bhatt HB and Singh SP (2022) Diversity of Cultivable Bacteria in A Saline Desert of Little Rann of Kutch, India: A Phylogenetic Perspective. Front. Mar. Sci. 9:769043. doi: 10.3389/fmars.2022.769043
Received: 01 September 2021; Accepted: 15 March 2022;
Published: 19 April 2022.
Edited by:
Martin W. Hahn, University of Innsbruck, AustriaReviewed by:
Vojtech Kasalicky, Academy of Sciences of the Czech Republic (ASCR), CzechiaPrachi Singh, Sardar Patel University, India
Ashim Datta, Central Soil Salinity Research Institute (ICAR), India
Klára Řeháková, Czech Academy of Sciences, Czechia
Copyright © 2022 Bhatt and Singh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Satya P. Singh, c2F0eWFwc2luZ2hAeWFob28uY29t
 Hitarth B. Bhatt
Hitarth B. Bhatt Satya P. Singh
Satya P. Singh