- 1School of Ecology and Nature Conservation, Beijing Forestry University, Beijing, China
- 2 Department of Civil and Environmental Engineering, Faculty of Science and Technology, University of Macau, Macao, Macao SAR, China
- 3State Key Laboratory of Quality Research in Chinese Medicine and institute of Chinese Medical Sciences, University of Macau, Macao, Macao SAR, China
Mangroves are extreme inter-tidal environments containing rich microbial communities. Actinobacteria from mangroves have an unprecedented ability to biosynthesize secondary metabolites. Therefore, the secondary metabolite production potential of actinobacteria sourced from mangroves warrants further exploration to determine whether they offer new sources of natural products. For this purpose, we selected a strain of Mycobacterium saopaulense (Actinobacteria phylum) from the mangroves in Macau for whole genome sequencing and tandem mass spectrometry analysis. The results showed that the 5,376,881-bp genome from this strain contains 5,391 protein-coding genes and a coding density of 90.53%. The main participating KEGG pathway was “Metabolism”. Altogether, the 81 gene clusters identified in its genome were associated with the production of 23 secondary metabolites. Sixteen of them were classified as antibiotics and three of them as bioactive compounds. Furthermore, two of the predicted secondary metabolites from the fermentation process were clavulanic acid and streptomycin. Both of these antibiotics were initially found to be produced by the M. saopaulense strain. This study shows that mangrove-derived actinobacteria have a large number of biosynthetic pathways with the potential to produce a range of biologically active secondary metabolites.
Introduction
The natural products produced by microorganisms are important sources of many drugs in wide-spread use today for treating various diseases and infectious agents, such as cancer, immunosuppression, bacteria, and viruses (Gullo et al., 2006; Newman and Cragg, 2016). Among these drugs, 75% are natural products derived from actinobacteria, a Gram-positive bacteria with a high genomic G+C content (Abdel-Razek et al., 2020). Actinobacteria produce structurally diverse secondary metabolites such as polyketides, terpenes, macrolides, and alkaloids, which among others have remarkable biological activities (Rateb et al., 2018). However, recent genomics studies have found that the number of biosynthetic gene clusters in actinobacteria is far greater than the number of compounds that have been obtained from them (Hu et al., 2018; Hu et al., 2019; Hu et al., 2020). Therefore, identifying all of the gene clusters and exploring the potentials of secondary metabolic biosynthesis in actinobacteria will not only help to enrich the resource pool of natural products, but also to provide more lead compounds for drug development.
Our previous work and some other studies found that mangrove-derived actinobacteria contain a higher number of gene clusters and have greater potential to produce more bioactive compounds than those from terrestrial environments (Huang et al., 2016; Pei et al., 2021). In general, actinobacteria isolated from mangroves have a higher number of clusters for secondary metabolites than those obtained from terrestrial habitats (Bentley et al., 2002; Liu et al., 2012; Komatsu et al., 2013; Hu et al., 2018; Hu et al., 2019; Ser et al., 2019; Hu et al., 2020; Ser et al., 2020). Mangroves are unique plant communities that grow in the intertidal zones of tropical and subtropical coasts and are submerged by periodic tides (Collins et al., 2017). Actinobacteria are generally best suited to semi-arid environments (Ghorbani-Nasrabadi et al., 2013); hence, it appears that the actinobacteria that live in mangroves are likely to have unique molecular adaptations because these environments have been characterized by humid, highly acidic and high-salt conditions for long periods of time. As a consequence, mangrove-derived actinobacteria have broad biosynthetic potential and merit further exploration.
Many well-known antibiotics were initially sourced from actinobacterial strains, among which many are from Streptomyces, the largest genus in the Actinobacteria phylum (Raja and Prabakarana, 2011). Streptomyces species can produce most of the known aminoglycosides, many beta-lactams, non-ribosomal peptides, and a great number of macrolides (Nagarajan et al., 1971; Challis and Ravel, 2000). Streptomyces-derived macrolides tend to be nystatin, natamycin and amphotericin B (Linke et al., 1974; Brautaset et al., 2000; Farid et al., 2000). Aminoglycosides that are produced by the Streptomyces group include streptomycin, kanamycin, and neomycin (Shier et al., 1969; Distler et al., 1987). However, the secondary bioactive metabolite production capacities of non-Streptomyces or rare genera may be completely underestimated. Without doubt, the search for antibiotic production potential in rare actinobacterial species for pharmaceutical development is of great interest (Demain, 2014; Lee et al., 2015; Indupalli et al., 2018; Ser et al., 2018). In common with the findings from our previous study, it was first reported that mangrove-derived non-Streptomyces Mycobacterium species can produce asukamycin and apramycin antibiotics (Hu et al., 2019), and that Micromonospora aurantiaca can produce the kanamycin antibiotic, the yield of which may be greater than that of the original actinobacteria that produced it (Hu et al., 2020). Thus, we propose that other mangrove-derived non-Streptomyces species have unknown secondary metabolite production potential worth exploring.
Therefore, the aim of this study was to explore the potential of secondary metabolite production of mangrove plant-derived actinobacteria. We selected one actinobacteria species, Mycobacterium saopaulense, which was isolated from mangroves, with which to conduct whole genome sequencing to identify its biosynthetic gene clusters and corresponding compounds. The fermentation products of this M. saopaulense strain were analyzed by mass spectrometry to determine which predicted compounds were actually produced. Our results augment current understanding about the secondary metabolite production ability of actinobacteria, and provide possible bacterial-derived resources for the development of natural products.
Materials and Methods
Isolation of the M. saopaulense Strain
The M. saopaulense strain was isolated from a mangrove plant (Kandelia candel) in Macau. It shares 99.90% 16S rRNA sequence similarity with M. saopaulense EPM 10906T (Hu et al., 2020). The isolate was stored in 20% glycerol at -80°C in the State Key Laboratory of Quality Research in Chinese Medicine, University of Macau.
DNA Extraction and Whole Genome Sequencing
DNA extraction, library construction and whole genome sequencing were performed in accordance with the methods of a previous study (Hu et al., 2018). Genomic DNA from the M. saopaulense strain was extracted using the IANamp Bacteria DNA Kit (TIANGEN Biotech Co. LTD). The library was constructed using the NEBNext Ultra II DNA library Prep Kit. Sequencing was conducted on the Illumina NovaSeq HiSeq 4000 instrument. IBDA was used to assemble and annotate the reads obtained after sequencing. MetaGeneMark (http://topaz.gatech.edu/GeneMark/), tRNAscan-SE (http://trna.ucsc.edu/software/) and rnammer (http://www.cbs.dtu.dk/services/RNAmmer/) were used for gene prediction, tRNA prediction and rRNA prediction, respectively. Gene functions were determined by comparison with the Kyoto Encyclopedia of Genes and Genomes (KEGG, https://www.genome.jp/kegg/) database. Mega software was used to construct a phylogenetic tree. The 16S rRNA sequences of the M. saopaulense strain were extracted from the annotation results. The top 20 closely related type strains’ 16S rRNA sequences were downloaded from the NCBI database. These species were M. saopaulense strain EPM10906 (GeneBank accession number: KM973037.1, similarity: 99.90%), M. abscessus strain WH572 (GenBank accession number: KR995262.1, similarity: 99.89%), M. abscessus strain WH834 (GenBank accession number: KR995258.1, similarity: 99.89%), M. chelonae strain CH 3/4 (GenBank accession number: MN577459.1, similarity: 99.78%), M. saopaulense strain FMS-15 (GenBank accession number: MK396591.1, similarity: 99.78%), M. chelonae strain JNBP 03213 (GenBank accession number: LC082312.1, similarity: 99.78%), Mycobacterium sp. strain MA45 (GenBank accession number: MH699146.1, similarity: 99.78%), Mycobacterium sp. strain TM-A149 (GenBank accession number: MH698675.1, similarity: 99.78%), Mycobacterium sp. strain TM-A163 (GenBank accession number: MH698674.1, similarity: 99.78%), Mycobacterium sp. strain TM-A160 (GenBank accession number: MH698673.1, similarity: 99.78%), M. chelonae subsp. bovis strain QIA-37 (GenBank accession number: NR158073.1, similarity: 99.67%), M. abscessus strain JCM 6390 (GenBank accession number: LC149865.1, similarity: 99.67%), Mycobacterium sp. G1 (GenBank accession number: LC637768.1, similarity: 99.67%), Mycobacterium sp. G2 (GenBank accession number: LC637767.1, similarity: 99.67%), M. abscessus strain AF7 (GenBank accession number: MW332182.1, similarity: 99.67%), M. abscessus strain AF6 (GenBank accession number: MW332181.1, similarity: 99.67%), M. abscessus strain WH750 (GenBank accession number: KR995261.1, similarity: 99.67%), M. abscessus strain WH999 (GenBank accession number: KR995259.1, similarity: 99.67%), Mycobacterium sp. strain TM-B145 (GenBank accession number: MH698713.1, similarity: 99.57%), M. chelonae strain JNBP 03238 (GenBank accession number: LC082323.1, similarity: 99.57%) and Escherichia coli strain U 5/41 (GeneBank accession number: NR 024570.1). Online anti-SMASH (https://antismash.secondarymetabolites.org) software was used to profile the biosynthetic gene clusters and secondary metabolites.
Fermentation Culture and Tandem Mass Spectrometry (MS/MS) Analysis
The fermentation culture and chemical analysis were conducted in accordance with Hu et al. (2018). ISP2 medium, which was selected as the culture medium, contained 4.0 g/L yeast, 10.0 g/L malt extract, 4.0 g/L glucose and 20.0 g/L agar, and pH 7.2. The stored isolate was streaked out onto ISP2 solid medium, and cultured at 28°C for four days. A single colony was selected and inoculated into 20 mL of ISP2 liquid medium and shaken (28°C, 7 days, 250 rpm). Ethyl acetate (60mL) was added to the fermentation products, the culture was centrifuged, and the supernatant was collected and dried at 60°C. The extracts were dissolved in 3mL of methanol for chemical analysis. The 4000 Q Trap MS/MS instrument was used to identify the fermentation products. The full scan mode was used first for detecting compounds falling into the 100–1500 m/z range under the positive ion mode. The conditions were set as follows: capillary voltage, 3.0 kV; cone voltage, 20V; 0.6S per spectrum. A full secondary scan, which was conducted to detect the fragments of ions, was compared with the Human Metabolome Database (HMDB, https://hmdb.ca/) to finally determine the compound.
Results and Discussion
Phylogenetic Analysis
A phylogenetic analysis of the isolated strain to illustrate its evolutionary relationship with the top 20 closely related type strains (Figure 1). The 16S sequences based phylogenetic tree showed that the M. saopaulense strain and M. abscessus strain WH572, M. abscessus strain WH999, M. abscessus strain WH750, M. abscessus strain AF6, M. abscessus strain AF7, Mycobacterium sp. G2, Mycobacterium sp. G1, M. abscessus strain JCM 6390, M. chelonae subsp. bovis strain QIA-37, Mycobacterium sp. strain TM-A160, Mycobacterium sp. strain TM-A163, Mycobacterium sp. strain TM-A149, Mycobacterium sp. strain MA45, M. chelonae strain JNBP 03213, M. saopaulense strain FMS-15, M. abscessus strain WH834, M. saopaulense strain EPM10906 and M. chelonae strain CH 3/4 were located on the same branch in the tree, suggesting that they should have a close relationship, but had a relatively long distance from Mycobacterium sp. strain TM-B145 and M. chelonae strain JNBP 03238. It is reasonable that the M. saopaulense strain and 2 type strains of Mycobacterium sp. strain TM-B145 and M. chelonae strain JNBP 03238 with low 16S sequence similarity (99.57%) were located on the different branch in the tree. But the relationship between the M. saopaulense strain and the other type strains did not show the closest phylogenetic distance in combination with the highest 16S sequence similarity, which might resulted from that the sequences used to construct the tree were obviously short (1300bp-1500bp).

Figure 1 Comparison of 16S rRNA sequences from the M. saopaulense strain with 20 other orthologous sequences. Escherichia coli was used as the root of the phylogenetic tree. The method of neighbor-joinging was used to construct the tree. The number of bootstrap replications was set to 1000.
General Genome Features and Function Annotations
The de novo assembly showed that the full length genome from the M. saopaulense strain was 5,376,881-bp long with an average read length of 21,085 bp, and a G+C content of 63.91%. A total of 6,881,851 reads and 255 scaffolds were generated. The genome contains 5,391 protein-coding genes, 72 tRNA genes, 3 rRNA genes, and has a coding density of 90.53%. The average coding sequence length was 903 bp. In the KEGG database, 80.44% of the genes were annotated as “Metabolism”, and their main metabolic pathways involved “global and overview maps”, “carbohydrate metabolism”, and “amino acid metabolism” (Figure 2).
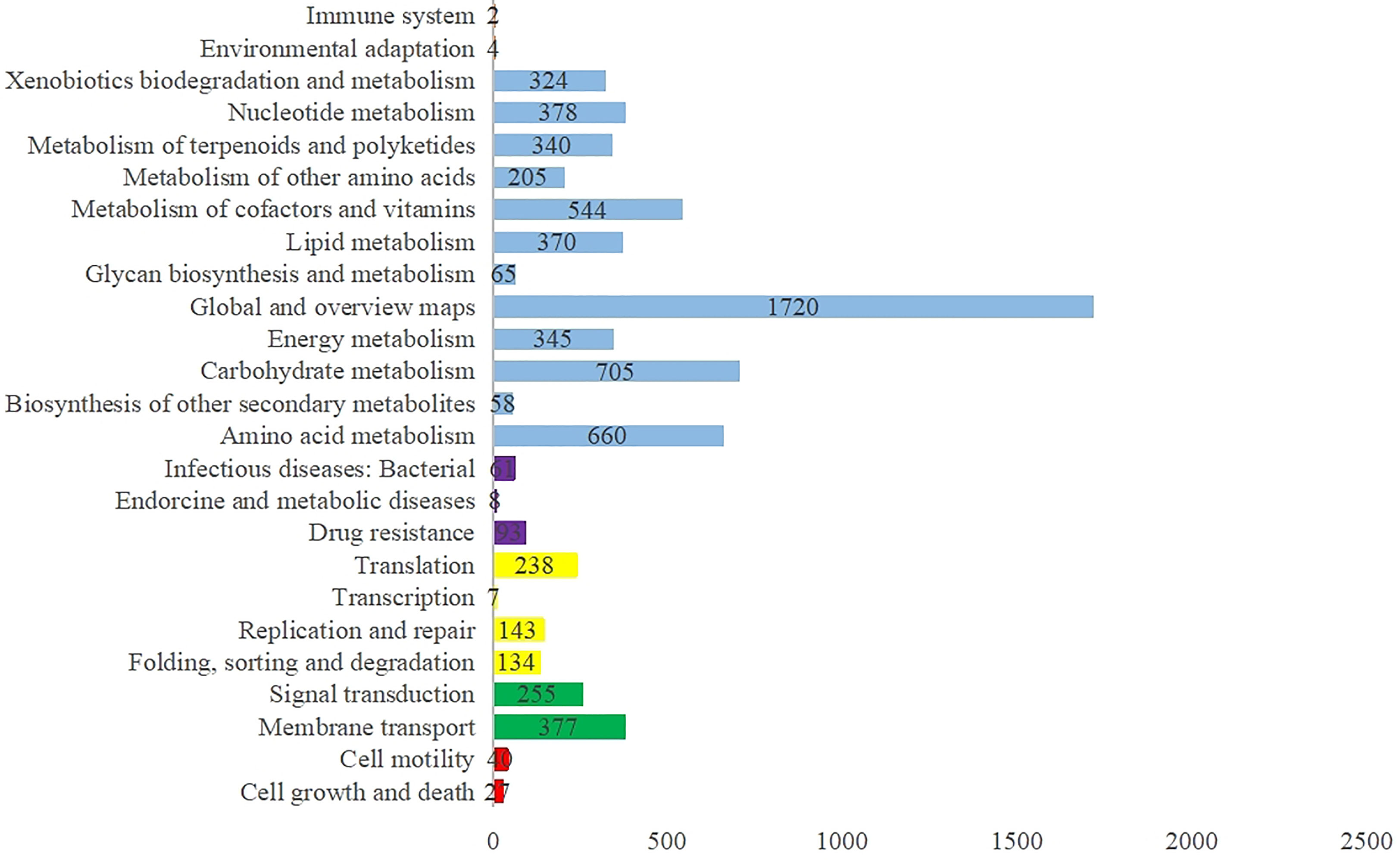
Figure 2 Distribution of KEGG pathways in the M. saopaulense strain’s genome, showing the top five KEGG Orthology (KO) categories in the assigned sequences, which include organismal systems (orange), metabolism (blue), human diseases (purple), genetic information processing (yellow), environmental information processing (green), and cellular process (red).
Identification of Gene Clusters and Secondary Metabolites
Altogether, 81 biosynthetic gene clusters were identified in the genome from the M. saopaulense strain; these were linked to bacteriocin, ectoine, fatty acid, indole, nonribosomal peptides (NRPS), polyketide synthase (PKS), saccharide and others. Worth noting is the possibility that the 54 gene clusters in the genome that fall within the putative-type category encode unique natural products (Dahal et al., 2021). These gene clusters putatively produce 23 known secondary metabolites (Table 1), from which 16 are antibiotics, three are bioactive compounds, leaving four others. Until now, no known antibiotics have been found to be produced by M. saopaulense sp., but the genome results suggest that the mangrove-derived M. saopaulense strain’s genome has huge potential to biosynthesize antibiotics. With the exception of mycobactin, which is produced by actinobacteria in the Mycobacterium genus, the other predicted antibiotics are derived from non-Mycobacterium genera, such as Actinomadura, Amycolatopsis, Saccharothrix, Salinispora, Streptomyces, and Micromonospora (Table 1). This indicates that the extreme environment of mangroves acts not only to enrich the gene clusters of the actinobacteria that live there, but has also enabled them to evolve different biosynthetic pathways.
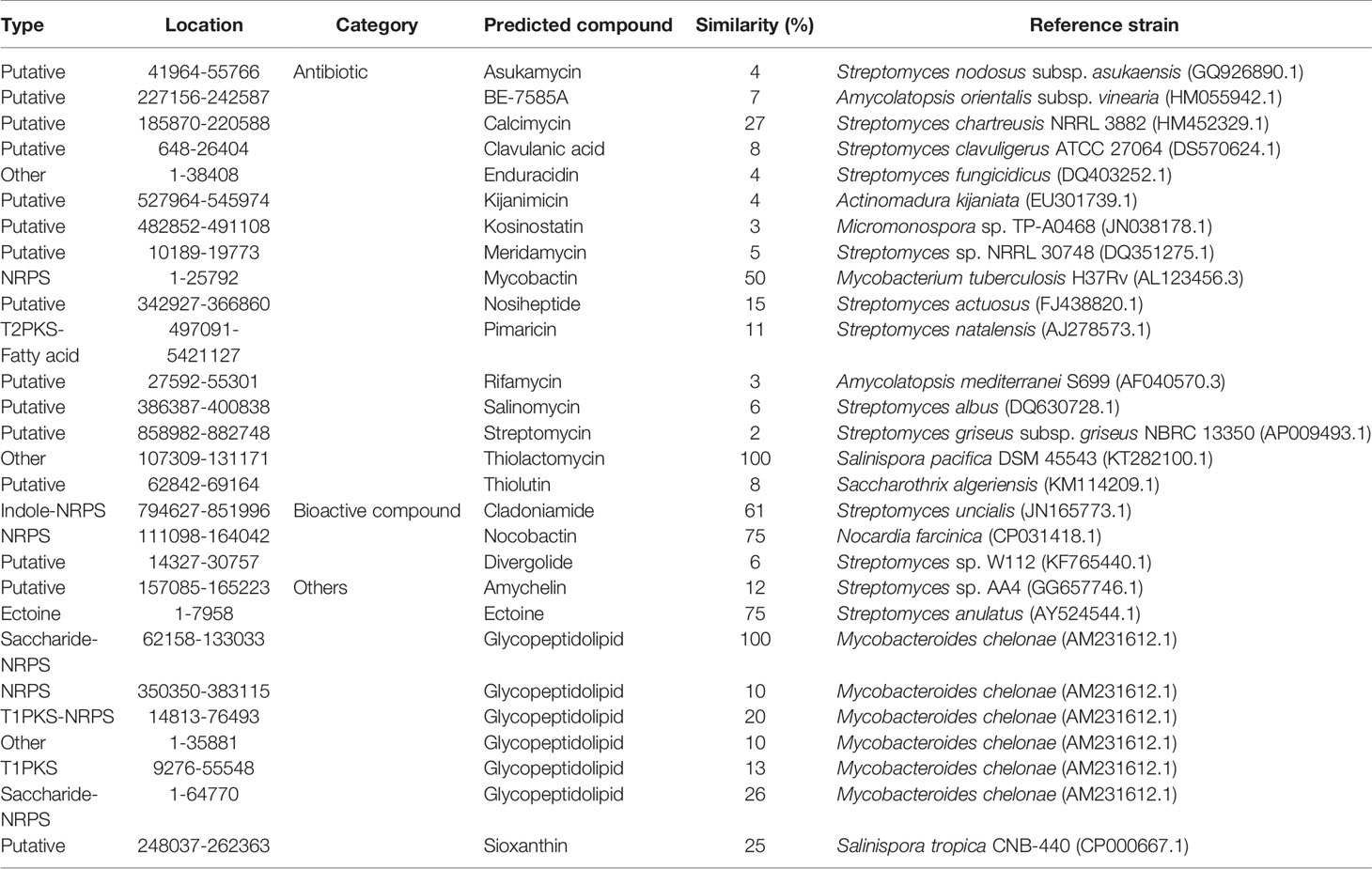
Table 1 Predicted gene clusters and their corresponding secondary metabolites in the M. saopaulense strain.
Detecting Fermentation Products in the M. saopaulense Strain.
To move beyond genome prediction, fermentation and mass spectrometry experiments were performed to determine which of the secondary metabolites predicted by genomic analysis were actually produced by the M. saopaulense strain. The secondary metabolites were detected using the same previously reported methods (Hu et al., 2018; Hu et al., 2019; Hu et al., 2020).
We first calculated the theoretical m/z values of the 23 predicted secondary metabolites displayed in Table 2. An MS/MS system was used to separate the secondary metabolites produced by the M. saopaulense strain for analysis. By comparing the theoretical m/z values with the mass spectrometry’s full scan atlas, the compounds with molecular ion peaks close to or consistent with the theoretical values were selected. Eight predicted secondary metabolites were detected in the full-scan mode (Table 2). We then conducted a secondary mass spectrometry scan of the eight secondary metabolites, and the fragments’ information we obtained was compared with that in the HMDB to further determine the identities of the tested compounds. Table 2 shows that a compound with a molecular ion at 200.3 m/z was identified in the spectrum from the first scan, indicating that it could be clavulanic acid. Its in-source fragments at m/z 110.4, 136.1, 154.2, 156.3, 158.1, 170.1 and 182.3 identified by the secondary scanning mode (Figure 3) matched the MS/MS spectrum (20V, positive) of clavulanic acid from the HMDB. Table 2 also shows that the compound with a molecular ion at m/z 582.5, which was identified in the spectrum from the first scan, could be streptomycin. Its in-source fragments at m/z 202.1, 261.3, 371.4 and 522.4, which were identified in the secondary scan mode (Figure 4), matched the reference MS/MS spectrum (20V, positive) in the HMDB. However, the fragment information of kijanimicin, mycobactin, pimaricin, thiolactomycin, thiolutin and ectoine was not identified in the secondary scan mode. Hence, the tentative production of clavulanic acid and streptomycin is suggested by the MS spectra but needs to be confirmed by isolation and characterization of the pure compound (LC-MS), bioassays and quantification of the compounds.

Table 2 Theoretical and detected m/z values for potential secondary metabolites from the M. saopaulense strain.
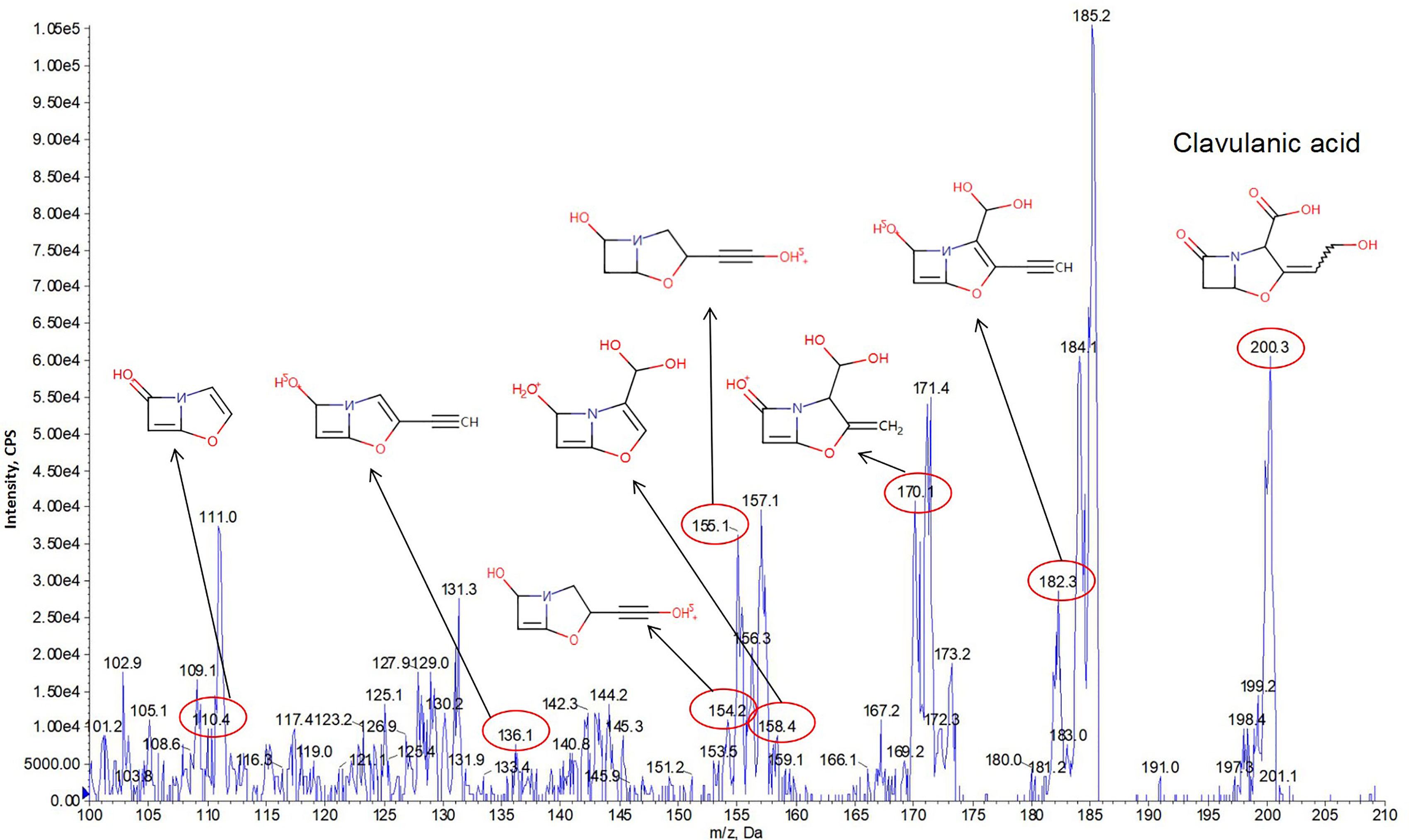
Figure 3 The in-source fragments from clavulanic acid (m/z 200.3) at m/z 110.4, 136.1, 154.2, 156.3, 158.1, 170.1 and 182.3 were identified in the secondary scan mode by MS/MS analysis.
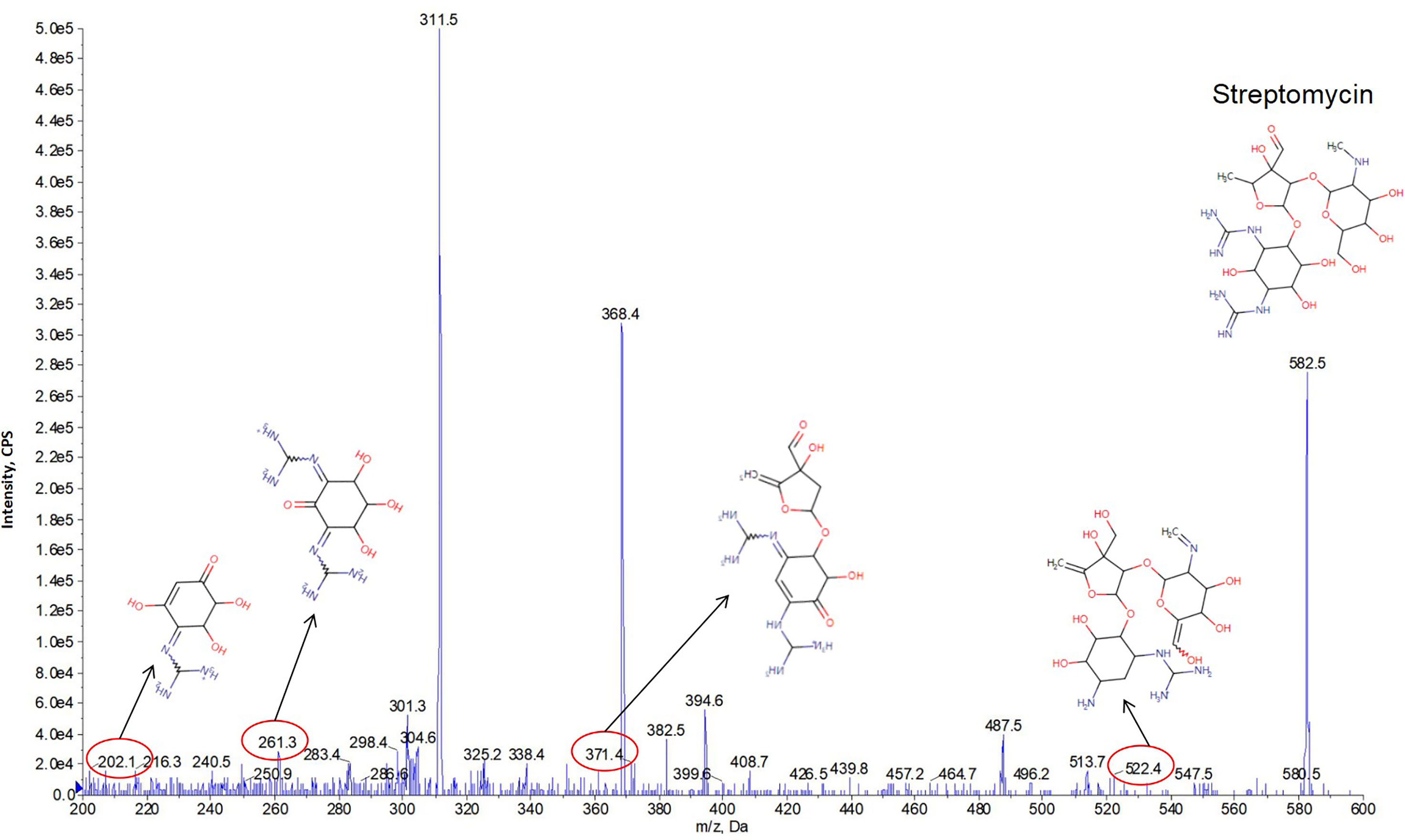
Figure 4 The in-source fragments from streptomycin (m/z 582.5) at m/z 202.1, 261.3, 371.4 and 522.4 were identified in the secondary scan mode by MS/MS analysis.
Referencing with genomic information, the gene cluster of clavulanic acid was composed of core biosynthetic genes, additional biosynthetic genes, transport-related genes, regulatory genes and some other genes. The core biosynthetic genes with the length of 2,520 nt were responsible for the functions of beta-lactam synthetase and clavaminate synthase. The additional biosynthetic genes, transport-related genes, regulatory genes and some other genes were related to the functions of hypothetical protein, carboxyethylarginine synthetase, proclavaminate amidinohydrolase, glutamate N-acetyltransferase 2, oligopeptide-binding lipoprotein, transcription activator, clavaldehyde dehydrogenase, cytochrome P450-SU2, ferredoxin, beta-lactamase, efflux protein, acetyltransferase, oligopeptide-binding lipoprotein, conserved hypothetical protein, glycylclavaminate synthase, penicillin-binding protein PBP, penicillin-binding protein, cytochrome P450 hydroxylase, RNA polymerase sigma factor SigL, histidine kinase, two-component system response regulator and transcriptional regulator. Besides, the core biosynthetic genes, additional biosynthetic genes, transport-related genes and some other genes were observed in the gene cluster of streptomycin. The core biosynthetic genes with the length of 2,505 nt were associated with the functions of putative dTDP-dihydrostreptose–streptidine-6-phosphate dihydrostreptosyltransferase and putative streptomycin-6-phosphate phosphatase. The functions of rest genes were related to putative NAD(P)-dependent oxidoreductase, putative ABC-type multidrug transport system ATPase and permease component, putative ABC-type multidrug transport system ATPase and permease component, putative L-glutaminyl-2-N-methyltransferase, StsF protein, putative ATP:scyllo-inosamine phosphotransferase, putative aminosugar-converting enzyme, putative L-glutamine:scyllo-inosose aminotransferase, putative NADH:N-amidino-scyllo-inosamine oxidoreductase, putative L-alanine:N-amidino-3-keto-scyllo-inosamine aminotransferase, putative glucose-1-phosphate thymidylyltransferase, putative N-amidionostreptamine 6-phosphotransferase, putative scyllo-inosamine-4-phosphate amidinotransferase, putative dTDP-4-dehydrorhamnose 3,5-epimerase, putative dTDP-4-dehydrorhamnose reductase, putative dTDP-glucose 4,6-dehydratase, putative glucose-1-phosphate thymidylyltransferase, streptomycin biosynthesis operon regulator, streptomycin 6-phosphotransferase, scyllo-inosamine-4-phosphate amidinotransferase, StrF protein, StrG protein, putative NAD:myo-inositol oxidoreductase, putative N-methyl-L-glucosamine biosynthetic aminotransferase and putative oxidoreductase.
As reported, clavulanic acid, which was first found to be produced by S. clavuligerus, is an antibiotic with a weak antibacterial effect and has been used on several livestock species (Putecova et al., 2021). It has the important ability of inhibiting β-lactamase, and is therefore an appropriate supplement for use with other β-lactam antibiotics, such as amoxicillin and ticarcillin, and it can enhance their antibacterial effects (Putecova et al., 2021; Shin et al., 2021). The commercial drugs, Augmentin™ and Timentin™, which are mixtures of clavulanic acid and β-lactam antibiotics, have been prescribed in many countries (Elander, 2003). Many studies have attempted to identify ways of increasing clavulanic acid production. For example, adding glycerol to bacterial cultures can enhance clavulanic acid production by approximately two-fold (Chen et al., 2003). Furthermore, stimulating the expression of additional copies of the glycerol utilization operon was found to increase the yield of clavulanic acid by approximately 4.5-fold. Further supplementation with glycerol was found to enhance clavulanic acid production by about 7.5-fold (Banos et al., 2009). Hence, the M. saopaulense strain is another promising clavulanic acid-producing with the potential to be utilized as an alternative strain to produce clavulanic acid identified herein. Our M. saopaulense strain was also found to produce streptomycin, an antibiotic first isolated from S. griseus in 1943 and subsequently submitted for clinical trial in 1994. Streptomycin is notable for its effective control of tuberculosis in humans, and with its broad spectrum antimicrobial effect it has been widely used in clinical settings (Waksman, 1953; Pawar et al., 2019). This study is the first to report that a strain of M. saopaulense can produce streptomycin. The fact that actinobacteria isolated from mangrove swamps have unknown antibiotic producing abilities, which makes them worthy of immediate examination.
Our results confirm that M. saopaulense derived from the mangrove environment has a huge gene pool related to the biosynthesis of secondary metabolites. To our best knowledge, this is the first study predicted the production of clavulanic acid and streptomycin by M. saopaulense. Thus, it appears that the mangrove environment hosts Actinobacteria capable of producing bioactive secondary metabolites that can be used in biotechnology applications such as antibiotic production.
Conclusions
In this study, a strain of M. saopaulense sp. isolated from the mangrove environment was successfully investigated using a combination of whole genome sequencing and mass spectrometric analysis. The outstanding biosynthetic secondary metabolite-producing potential of mangrove-derived actinobacteria was underestimated before M. saopaulense sp. was first found to produce clavulanic acid and streptomycin antibiotics. Thus, the results of this study further augment research on the biosynthetic properties of actinobacteria from extreme mangrove environments, which are undoubtedly promising sampling sites for obtaining bacteria to produce secondary metabolites for medicinal use.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author Contributions
DH and SML designed the study. DH and MKM conducted the experiment and analyzed the data. DH, KL and MKM wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by The International S&T Cooperation (2016YFE0122000), the Macau Science and Technology Development Fund (FDCT) and the Ministry of Science and Technology of China (MOST) joint funding scheme (No. FDCT 017/2015/AMJ), Research Committee, University of Macau (MYRG2016-00056-FST, MYRG2015-00182-ICMS-QRCM, MYRG139(Y1-L4)-ICMS12-LMY, and MYRG2016-00129-ICMS-QRCM) and China Postdoctoral Science Foundation (No. 2020TQ0047).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdel-Razek A. S., EI-Naggar M. E., Allam A., Morsy O. M., Othman S. I. (2020). Microbial Natural Products in Drug Discovery. Processes 8, 470. doi: 10.3390/pr8040470
Banos S., Perez-Redondo R., Koekman B., Liras P. (2009). Glycerol Utilization Gene Cluster in Streptomyces Clavuligerus. Appl. Environ. Microb. 75 (9), 2991–2995. doi: 10.1128/AEM.00181-09
Bentley S. D., Chater K. F., Cerdeño-Tárraga A. M., Challis G. L., Thomson N., James K. D., et al. (2002). Complete Genome Sequence of the Model Actinomycete Streptomyces coelicolor A3 (2). Nature 417 (6885), 141. doi: 10.1038/417141a
Brautaset T., Sekurova O. N., Sletta H., Ellingsen T. E., Strøm A. R., Valla S., et al. (2000). Biosynthesis of the Polyene Antifungal Antibiotic Nystatin in Streptomyces noursei ATCC 11455: Analysis of the Gene Cluster and Deduction of the Biosynthetic Pathway. Chem. Biol. 7 (6), 395–403. doi: 10.1016/S1074-5521(00)00120-4
Challis G. L., Ravel J. (2000). Coelichelin, a New Peptide Siderophore Encoded by the Streptomyces Coelicolor Genome: Structure Prediction From the Sequence of its Non-Ribosomal Peptide Synthetase. FEMS Microbiol. Lett. 187 (2), 111–114. doi: 10.1111/j.1574-6968.2000.tb09145.x
Chen K. C., Lin Y. H., Wu J. Y., Hwang S. C. J. (2003). Enhancement of Clavulanic Acid Production in Streptomyces clavuligerus With Ornithine Feeding. Enzyme Microb. Tech. 32 (1), 152–156. doi: 10.1016/S0141-0229(02)00280-6
Collins D. S., Avdis A., Allison P. A., Johnson H. D., Hill J., Piggott M. D., et al. (2017). Tidal Dynamics and Mangrove Carbon Sequestration During Oligo-Miocene in the South China Sea. Nat. Commun. 8, 15698. doi: 10.1038/ncomms15698
Dahal R. H., Chaudhary D. K., Kim J. (2021). Genome Insight and Description of Antibiotic Producing Massilia antibiotica sp. nov., Isolated From Oil-Contaminated Soil. Sci. Rep. UK 11 (1), 6695. doi: 10.1038/s41598-021-86232-z
Demain A. L. (2014). Importance of Microbial Natural Products and the Need to Revitalize Their Discovery. J. Ind. Microbiol. Biot. 41 (2), 185–201. doi: 10.1007/s10295-013-1325-z
Distler J., Ebert A., Mansouri K., Pissowotzkri K., Stockmann M., Piepersberg W. (1987). Gene Cluster for Streptomycin Biosynthesis in Streptomyces griseus: Nucleotide Sequence of Three Genes and Analysis of Transcriptional Activity. Nucleic. Acids Res. 15 (19), 8041–8056. doi: 10.1093/nar/15.19.8041
Elander R. P. (2003). Industrial Production of β-Lactam Antibiotics. Appl. Microbiol. Biot. 61, 385–392. doi: 10.1007/s00253-003-1274-y
Farid M. A., El-Enshasy H. A., El-Diwany A. I., El-Sayed E. S. A. (2000). Optimization of the Cultivation Medium for Natamycin Production by Streptomyces Natalensis. J. Basic. Microb. 40 (3), 157–166. doi: 10.1002/1521-4028(200007)40:3<157::AID-JOBM157>3.0.CO;2-1
Ghorbani-Nasrabadi R., Greiner R., Alikhnai H. A., Hamedi J., Yakhchali B. (2013). Distribution of Actinomycetes in Different Soil Ecosystems and Effect of Media Composition on Extracellular Phosphatase Activity. J. Soil. Sci. Plant Nutt. 13 (1), 223–236. doi: 10.4067/S0718-95162013005000020
Gullo V. P., McAlpine J., Lam K. S., Baker D., Petersen F. (2006). Drug Discovery From Natural Products. J. Ind. Microbiol. Biot. 33 (7), 523–531. doi: 10.1007/s10295-006-0107-2
Huang C., Leung R. K., Guo M., Tuo L., Guo L., Yew W. W., et al. (2016). Genome-Guided Investigation of Antibiotic Substances Produced by Allosalinactinospora lopnorensis CA15-2T From Lop Nor Region, China. Sci. Rep. 6, 20667. doi: 10.1038/srep20667
Hu D., Chen Y., Sun C., Jin T., Fan G., Liao Q., et al. (2018). Genome Guided Investigation of Antibiotics Producing Actinomycetales Strain Isolated From a Macau Mangrove Ecosystem. Sci. Rep. UK 8 (1), 1–12. doi: 10.1038/s41598-018-32076-z
Hu D., Gao C., Sun C., Jin T., Fan G., Mok K. M., et al. (2019). Genome-Guided and Mass Spectrometry Investigation of Natural Products Produced by a Potential New Actinobacterial Strain Isolated From a Mangrove Ecosystem in Futian, Shenzhen, China. Sci. Rep. UK 9, 1–12. doi: 10.1038/s41598-018-37475-w
Hu D., Sun C., Jin T., Fan G., Mok K. M., Li K., et al. (2020). Exploring the Potential of Antibiotic Production From Rare Actinobacteria by Whole-Genome Sequencing and Guided MS/MS Analysis. Front. Microbiol. 11, 1540. doi: 10.3389/fmicb.2020.01540
Indupalli M., Muvva V., Mangamuri U., Munaganti R. K., Naragani K. (2018). Bioactive Compounds From Mangrove Derived Rare Actinobacterium Saccharomonospora oceani VJDS-3. 3 Biotech. 8 (2), 1–9. doi: 10.1007/s13205-018-1093-6
Komatsu M., Komatsu K., Koiwai H., Yamada Y., Kozone I., Izumikawa M., et al. (2013). Engineered Streptomyces avermitilis Host for Heterologous Expression of Biosynthetic Gene Cluster for Secondary Metabolites. ACS Synth Biol. 2 (7), 384–396. doi: 10.1021/sb3001003
Lee L. H., Azman A. S., Zainal N., Yin W. F., Ab Mutalib N. S., Chan K. G. (2015). Sinomonas humi sp. Nov., an Amylolytic Actinobacterium Isolated From Mangrove Forest Soil. Int. J. Syst. Evol. Micr. 65, 996–1002. doi: 10.1099/ijs.0.000053
Linke H. A., Mechlinski W., Schaffner C. P. (1974). Production of Amphotericin B-14C by Streptomyces nodosus Fermentation, and Preparation of the Amphotericin B-14C-Methyl-Ester. J. Antibiot. 27 (3), 155–160. doi: 10.7164/antibiotics.27.155
Liu C. X., Zhang J., Wang X. J., Qian P. T., Wang J. D., Gao Y. M., et al. (2012). Antifungal Activity of Borrelidin Produced by a Streptomyces Strain Isolated From Soybean. J. Agr. Food. Chem. 60 (5), 1251–1257. doi: 10.1021/jf2044982
Nagarajan R., Boeck L., Gorman M., Hamill R. L., Higgens C. E., Hoehn M. M., et al. (1971). Beta.-Lactam Antibiotics From Streptomyces. J. Am. Chem. Soc. 93 (9), 2308. doi: 10.1021/ja00738a035
Newman D. J., Cragg G. M. (2016). Natural Products as Sources of New Drugs From 1981 to 2014. J. Nat. Prod. 79 (3), 629–661. doi: 10.1021/acs.jnatprod.5b01055
Pawar H. V., Tetteh J., Debrah P., Boateng J. S. (2019). Comparison of In Vitro Antibacterial Activity Streptomycin-Diclofenac Loaded Composite Biomaterial Dressings With Commercial Silver Based Antimicrobial Wound Dressings. Int. J. Biol. Macromol. 121, 191–199. doi: 10.1016/j.ijbiomac.2018.10.023
Pei S., Niu S., Xie F., Zhang S., Wang W., Zhang G. (2021). Complete Genome Sequence of Streptomyces Sp. HSG2 From Rhizosphere Soil of Mangrove in Qingmei Gang, Sanya. Arch. Microbiol. 203 (6), 3519–3524. doi: 10.1007/s00203-021-02339-x
Putecova K., Nedbalcova K., Bartejsova I., Zouharova M., Matiaskova K., Stastny K. (2021). Mass Spectrometric Identification and Quantification of the Antibiotic Clavulanic Acid in Broiler Chicken Plasma and Meat as a Necessary Analytical Tool in Finding Ways to Increase the Effectiveness of Currently Used Antibiotics in the Treatment of Broiler Chickens. Anal. Bioanal. Chem. 413 (13), 3561–3571. doi: 10.1007/s00216-021-03307-6
Raja A., Prabakarana P. (2011). Actinomycetes and Drug-an Overview. Amer. J. Dru. Dis. Dev. 1 (2), 75–84. doi: 10.3923/ajdd.2011.75.84
Rateb M. E., Ebel R., Jaspars M. (2018). Natural Product Diversity of Actinobacteria in the Atacama Desert. Anton. Leeuw. Int. J. G. 111, 1467–1477. doi: 10.1007/s10482-018-1030-z
Ser H. L., Law J. W. F., Tan W. S., Yin W. F., Chan K. G. (2020). Whole Genome Sequence of Streptomyces Colonosanans Strain MUSC 93JT Isolated From Mangrove Forest in Malaysia. Pro In Micro Mol. Bio. 3 (1), a0000061. doi: 10.36877/pmmb.a0000061
Ser H. L., Law J. W. F., Tan W. S., Yin W. F., Chan K. G., Lee L. H. (2019). Genome Sequence of Bioactive Streptomycete Isolated From Mangrove Forest in East Malaysia, Streptomyces monashensis MUSC 1jt. Prog. In Drug Discovery Bio Sci. 2 (1), a0000045. doi: 10.36877/pddbs.a0000045
Ser H. L., Tan W. S., Cheng H. J., Yin W. F., Chan K. G., Ab Mutalib N. S., et al. (2018). Draft Genome of Starch-Degrading Actinobacterium, Microbacterium mangrovi MUSC 115T Isolated From Intertidal Sediments. Prog. In Drug Discovery Bio Sci. 1 (1), a0000001. doi: 10.36877/pddbs.a0000005
Shier W. T., Rinehart K. L., Gottlieb D. (1969). Preparation of Four New Antibiotics From a Mutant of Streptomyces Fradiae. PNAS 63 (1), 198–204. doi: 10.1073/pnas.63.1.198
Shin C.-H., Cho H. S., Won H.-J., Kwon H. J., Kim C.-W., Yoon Y. J. (2021). Enhanced Production of Clavulanic Acid by Improving Glycerol Utilization Using Reporter-Guided Mutagenesis of an Industrial Streptomyces Clavuligerus Strain. J. Ind. Microbiol. Biot. 48(3–4):kuab004 doi: 10.1093/jimb/kuab004
Keywords: Mycobacterium, mangrove, whole genome, gene cluster, secondary metabolite, mass spectrometry
Citation: Hu D, Lee SM-Y, Li K and Mok KM (2022) Exploration of Secondary Metabolite Production Potential in Actinobacteria Isolated From Kandelia candel Mangrove Plant. Front. Mar. Sci. 9:700685. doi: 10.3389/fmars.2022.700685
Received: 28 April 2021; Accepted: 23 March 2022;
Published: 12 April 2022.
Edited by:
Johannes F. Imhoff, Helmholtz Association of German Research Centres (HZ), GermanyReviewed by:
Jodi Woan-Fei Law, Monash University Malaysia, MalaysiaRam Hari Dahal, Kyungpook National University, South Korea
Copyright © 2022 Hu, Lee, Li and Mok. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kai Li, bGlrYWlfc2lub0BzaW5hLmNvbQ==; Kai Meng Mok, a21tb2tAdW0uZWR1Lm1v
 Dini Hu
Dini Hu Simon Ming-Yuen Lee
Simon Ming-Yuen Lee Kai Li1*
Kai Li1*