- 1School of Medicine, Shanghai University, Shanghai, China
- 2Department of Pharmacy, Shanghai Pudong New Area People’s Hospital, Shanghai, China
- 3Department of Critical Care Medicine, Shanghai Tenth People’s Hospital, School of Medicine, Tongji University, Shanghai, China
The emergence of antibiotic-resistant bacteria has become a major challenge in current clinical treatment. As essential natural molecules involved in innate immunity, antimicrobial peptides (AMPs) have the potential to break the limits of antibiotic resistance. While AMPs are widely presented in various organisms on this planet, the marine environment is unique in generating a large number of AMPs that exhibit characteristic structures and functions. This review summarizes the structures and mechanisms of action of AMPs derived from invertebrates, fish, amphibians, reptiles, and mammals in the ocean. The comparison of AMPs from organisms in different habitats demonstrates the preference of Arg (arginine) and Leu (leucine) in marine AMPs, which might be associated with the adaptation to the unique features of bacterial membranes in marine environments. In addition, the potential and applications of marine AMPs for the development of novel antibiotics are also described. These summaries are expected to provide a reference for the development and utilization of marine resources in drug development or aquiculture.
1. Introduction
In 2017, the World Health Organization (WHO) released a list of super-resistant bacteria that urgently need new antibiotics. Antibiotic resistance has been identified by WHO as one of the great threats to health, food safety and social development. Common antibiotics are mainly derived from metabolites or related derivatives, and kill bacteria by inhibiting the synthesis of bacterial cell walls, proteins, and nucleic acids, or by affecting bacterial metabolism. The bacteria can change the target gene expression, creating resistance. Penicillin-binding proteins (PBPs) are the targets of penicillin G. After bacteria such as Staphylococcus aureus acquire the gene “mecA”, the structure of PBPs changes, and the affinity with Penicillin G decreases, resulting in antibiotic resistance (Liu et al., 1985). Differing from traditional antibiotics, antimicrobial peptides (AMPs) selectively act on the acidic head group of phospholipids. This approach makes it difficult for bacteria to evolve resistance. AMP is thus a promising alternative to antibiotics.
AMPs is an amphiphilic cationic polypeptide with 3-50 amino acids (Javia et al., 2018). By electrostatic interaction, positively charged peptides are absorbed by negatively charged bacterial cell membranes. That interaction causes the permeability of the membrane to change so that intracellular ions and metabolites leak out. In final, the cell membrane crack (Falanga et al., 2016). AMPs can have biological activity not only on bacteria, but also on viruses, fungi, and others. In 1981, Steiner, (1981) first isolated and described a AMP from mammalian macrophages, named cecropins. Subsequently, Nakamura et al. (1988) first discovered tachyplesin, a AMP from marine organisms. Since then, the process of marine AMP research has been carried out.
The ocean, the cradle of human life, occupies 71 percent of the earth’s surface area and is rich in biological resources. Marine species diversity accounts for about 2/3 of life on earth (Ziko et al., 2022). Marine animals can be divided into vertebrates and invertebrates based on the presence or absence of the dorsal spine. Invertebrates include cephalochordates, cauda chordates, mollusca annelids, arthropods, echinoderms, and cnidarians. Of these, cephalochordates, caudal chordates, echinoderms, and most cnidarians are endemic to the oceans. Vertebrates include fish, reptiles, and mammals. They need to produce bioactive molecules with unique structures to adapt to extreme marine environments such as high salt, high pressure, and low temperature. Marine bioactive molecules play a crucial role in biomedical research and drug development. These molecules can be used exclusively as leads for drug or inspired chemical drug synthesis (Molinski et al., 2009; Nalini et al., 2018). Therefore, the ocean is also known as the “medicine bank of the new millennium”. According to the AMP database, 96% of AMPs are currently derived from terrestrial organisms. Therefore, rich marine AMP resources need to be exploited.
In this review, we discuss AMPs from different organisms in the ocean, their structures, mechanisms of action, development strategies, and potential applications. We hope to provide a reference for the exploitation and utilization of marine resources.
2. AMPs from marine organisms
Currently, marine AMPs have been found in fish, invertebrates, amphibious reptiles, and mammals. The discovered polypeptides and their structural simulations are shown in Figures 1A, 2. The AMP families of marine and terrestrial animals are nearly distinct. They share only the family of defensin and cathelicidins. The majority of marine AMPs are rich in cysteines (Ding and Zhang, 2013). In addition, AMPs from marine animals are predominantly expressed constitutively.
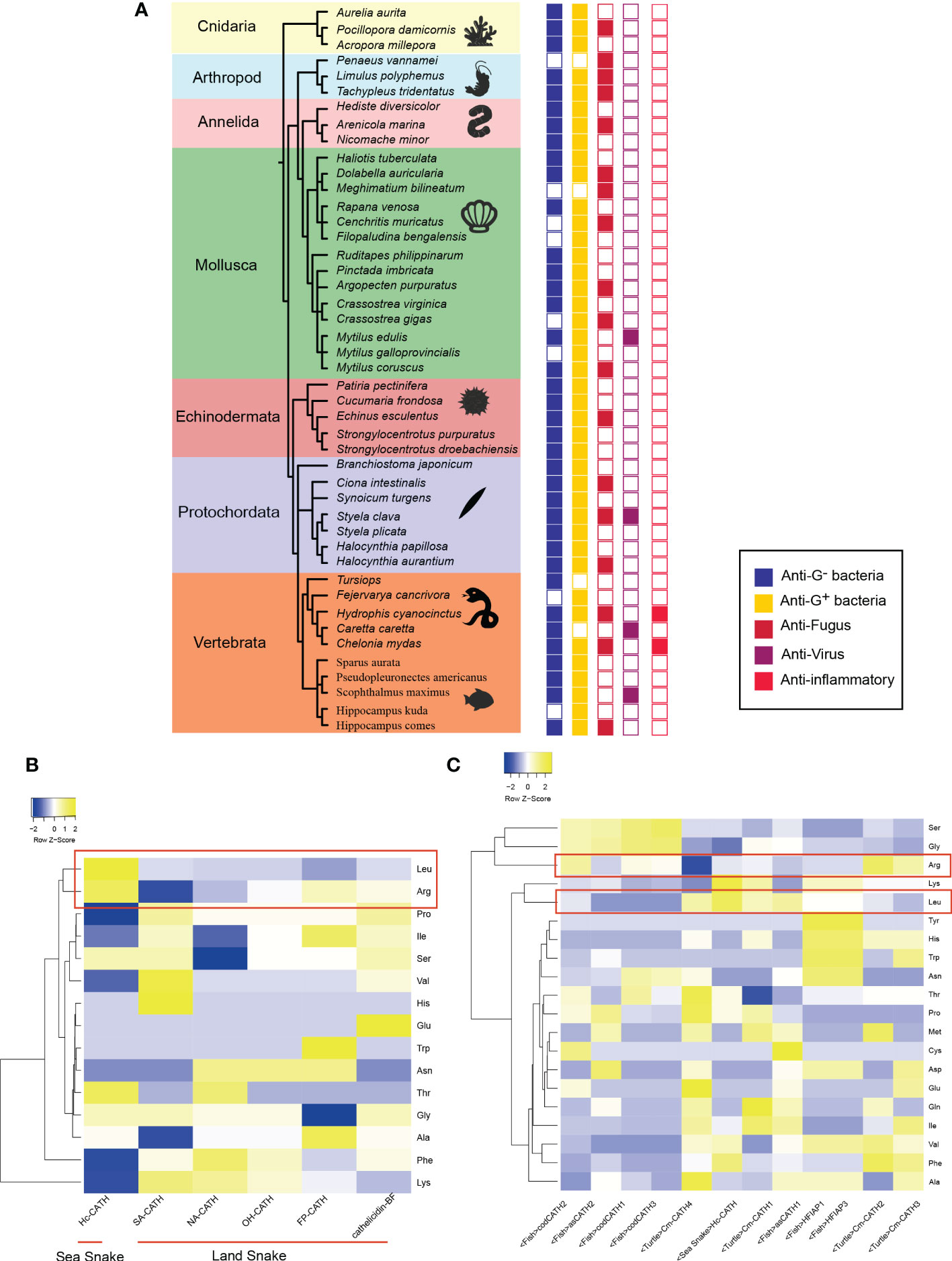
Figure 1 Activity and characteristic of marine AMPs (A) shows the activity of marine AMPs. The color of boxes from left to right correlates with activity for Gram-negative, Gram-positive, fungus, virus, and inflammatory. Empty squares indicate no activity or activity has yet been found. (B) is the results of comparison between sea snake AMPs and land snake AMPs. Color represents contents about different amino acids in AMP. (C) exhibits the comparison of marine AMPs. Hc, Hydrophis cyanocinctus; SA, Trimerodytes annularis; NA, Naja atra; OH, Ophiophagus hannah; FP, Deinagkistrodon acutus; BF, Bungarus fasciatus;Cm, Chelonia mydas; as, Salmo salar; cod, Gadus morhua; HFI, Myxine glutinosa.
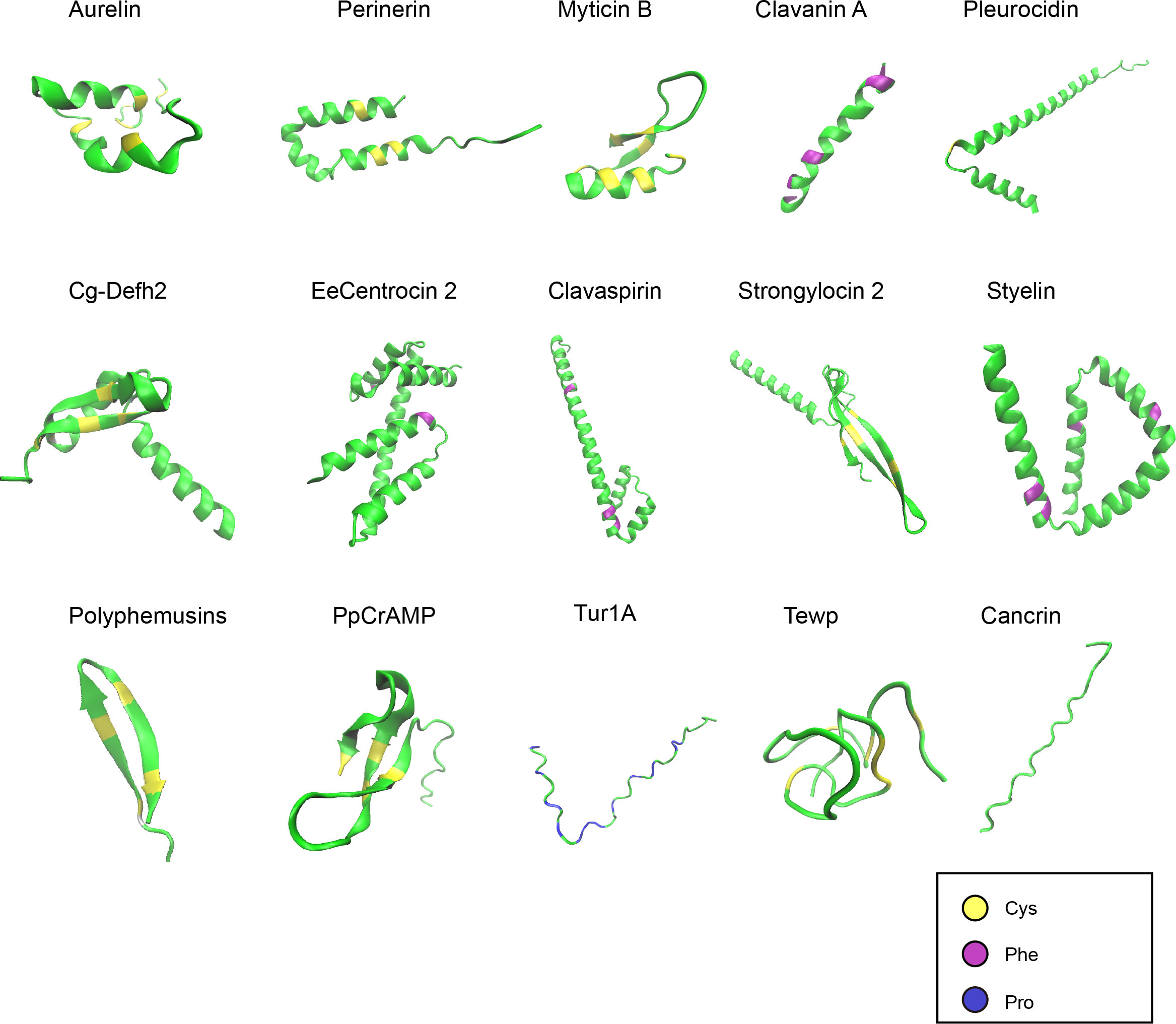
Figure 2 Structures of representative marine AMPs. The AMPs demonstrated are from different taxa of marine animals. The peptide structures were predicted by AlphaFold and shown in green. Cysteines, phenylalanines, and prolines are highlighted in yellow, purple, and blue, respectively.
2.1. AMPs from fish
Fish are the lowest vertebrates in the ocean, and the body’s defense depends primarily on the innate immune system. The fish’s innate immunity as the first line of defense is the mucus layer, which contains lysozyme, AMPs, and other molecules. It can quickly kill pathogenic microorganisms. The mucus layer is also the primary source of AMPs in fish. According to the structural characteristics, AMPs can be divided into five types: Piscidins, Cathelicidins, β-defensins, Hepcidins, and Histone-derived (Ding and Zhang, 2013; Masso-Silva and Diamond, 2014; Chen et al., 2020).
Precursors of Cathelicidins have a conserved “cathelin” domain and there is almost no sequence homology between mature peptides, but they have cationic, amphiphilic, and broad-spectrum antibacterial activity fundamentally. It is worth noting that this species appears to have some relation to the activity of AMPs. Cod AMPs have high activity against Gram-negative bacteria and fungi (Candida albicans), but have little activity against Gram-positive bacteria, while blind eel AMPs have activity against Gram-positive bacteria and negative bacteria, but have no effect on Candida. The Piscidins family of AMPs is a unique type of fish, which is homologous to the Cathelicidins family. Pleurocidin (Cole et al., 1997) is the first peptide identified in the piscidins family, derived from the skin and mucosa of the soleus (Pleuronectes americanus). It has strong antibacterial activity against Gram-negative bacteria (such as Pseudomonas, and Vibrio) and Gram-positive bacteria (such as Streptococcus, and Bacillus).
Defensin is a general term for cationic AMPs rich in disulfide bonds (one to six pairs). It is found in plants, animals, fungi, and other species. According to the position and amounts of disulfide bonds, it can be divided into α-defensins, β-defensins, and θ-defensins. β-defensins were discovered in fish databases (Zou et al., 2007). Its precursors include signal peptides, propeptides, and mature peptides. The mature peptide is located downstream of the precursor peptide (Nam et al., 2010). In addition to Gram-negative and Gram-positive bacteria, defensins are also active against fish-specific viruses (Jin et al., 2010).
Fish hepcidins is a cysteine-rich polypeptides, generally composed of a β-sheet hairpin with six to eight cysteine, and have an uncommon adjacent bridge at the hairpin corner (Hunter et al., 2002). It has two types, HAMP1 and HAMP2, which are constitutively expressed and inducible (Liang et al., 2013). Some studies have shown that hepcidins are active against fish pathogens and cancer cells. It can inhibit the proliferation and migration of tumor cells such as Hela, HepG2, and HT1080 in a dose-dependent manner (Chang et al., 2011; Cai et al., 2012).
Histone-derived peptides are similar to N- and C-terminal fragments of histones and have inhibitory activity against pathogens in humans and fish. It is widely distributed in tissues such as the skin, gills, and intestines, which are involved in the defense of the fish’s body.
2.2. AMPs from invertebrate
Marine invertebrates have existed for more than 450 million years, indicating that they have a strong and effective immune system (Tincu and Taylor, 2004). Invertebrates lack acquired immunity based on T lymphocyte subsets and rely entirely on innate immune mechanisms. Hemolymph is an important component of innate immunity in marine invertebrates. Circulating hemolymph contains a variety of active components such as complement, lectin, coagulation factor, and AMPs, which defend against invading microorganisms. Notably, AMPs from marine invertebrates have been found to account for 67% of marine AMPs (statistics as of December 2022).
2.2.1. AMPs from arthropod
Arthropods are one of the most successful animals in evolution (Wu et al., 2021). AMPs should play a part role in the process of adapting to the marine environment. AMPs from Crustaceans (shrimp and crab) and Chelicerata (Horseshoe crab) have already been fully researched. Penaeidins, Tachyplesin I, and Polyphemusins were discovered.
Shigenaga et al. (1993) isolated Tachyplesin I and II from hemocytes of Limulus amebocyte lysate. They belong to the cations-rich family that C-terminal amidated, which has an amphiphilic anti-parallel β-sheet structure (positive charge and hydrophobic amino acids are distributed on both sides of the β-sheet). The molecule is localized to small granules in hemocytes and released at sites of microbial infection, which can remove Mycobacterium tuberculosis from infected macrophages (Nakamura et al., 1988; Kushibiki et al., 2014; Priya et al., 2022). Polyphemusins I and II were found in the hemocytes of the Limulus polyphemus. Polyphemusins have two disulfide bonds and C-terminal amidation like Tachyplesin, but Polyphemusins contain an additional arginine residue at the N-terminal. It can inhibit the growth and reproduction of Candida albicans (Miyata et al., 1989). In addition, AMPs from Limulus mostly have the characteristics of binding chitin and stimulating chitin formation. It may inhibit cell growth by binding chitin in the cell wall of invading microorganisms, and may also be beneficial to wound healing.
Litopenaeus vannamei is an important aquaculture species in China. It grows rapidly and is rich in nutrition. And, it is also highly resistant to disease. Penaeidins are cationic polypeptides isolated and purified from the hemolymph of L. vannamei, with a molecular weight of 5.5-6.6 kDa, rich in proline at the N-terminus and stable at the C-terminus by cross-linking of three intramolecular disulfide bonds (Destoumieux et al., 2000). It is resistant to filamentous fungi, Fusarium oxysporum, and Vibrio, but has no activity against C.albicans and Saccharomyces cerevisiae (Destoumieux et al., 1999).
2.2.2. AMPs from annulata
Marine annulata mainly consists mainly of Worms and Polychaetes, such as sand silkworms. Safronova, (2022) found Capitellacin in marine Worms. The mature Capitellacin is highly homologous to Tachyplesins and Polyphemusins that belong to the β-hairpin AMPs family of the H. crab. It exists in an aqueous solution in the form of a monomeric right-handed β-hairpin. Compared with Tachyplesin-1, it has higher safety and selectivity.
Perinerin was isolated and purified from the homogenate of Perinereis aibuhitensis, which has four cysteines and a unique helix-turn-helix structure. It has strong antibacterial activity against Gram-positive and Gram-negative bacteria and fungi (Pan et al., 2004). Hedistin, isolated from the coelomocytes of Nereis diversicolor, also has helix-turn-helix. The helix fragment can bind to the lipid head region and enter the lipid bilayer (Xu et al., 2009), which may play a major role in its action.
2.2.3. AMPs from mollusca
Mollusca is the largest and most widely distributed group of animals in the oceans. And it is one of the most studied taxa in marine AMPs (Williams, 2017). AMPs can be divided into Mytilin, Myticin, and Mytimycin based on their primary structure and disulfide bond types. Despite the lack of similarity in the primary sequence, except Mytimycin, they contain eight cysteine residues, forming four disulfide bonds, and are cationic-rich amphiphilic peptides (Greco et al., 2020).
Each family has different subsets with different activities. Mytilin includes 5 types: A, B, C, D, and G.Mytilin A and B have significant inhibitory activity against Gram-positive strains including Bacillus megaterium and Micrococcus flavus (Charlet et al., 1996). Mytilin C, D, and G are characterized by a complementary range of action (Mitta et al., 2000) and antiviral activity (Roch et al., 2008). Myticin is divided into three subtypes: A, B, and C. Both Myticin A and Myticin B have inhibitory activity against Gram-positive bacteria, but Myticin B also has Gram-negative bacteria activity and antifungal activity (Anderson and Beaven, 2001). Myticin C has a high degree of molecular polymorphism and is expressed in the larval stage of mussels to carry immune protection (Balseiro et al., 2011).
Unlike the other three AMPs, Mytimycin contains six conserved cysteines that inhibit the growth of Neurospora crassa (Charlet et al., 1996; Sonthi et al., 2012).
2.2.4. AMPs from cnidaria
Cnidaria is a marine zooplankton with an open body cavity. The contact area between organisms and the sea is close, so has to face a variety of pathogenic bacteria. It relies on innate immune mechanisms for defense, which must be associated with its body containing a large number of AMPs.
Aurelin was purified from the mucilage of Aurelia asiaticus (Ovchinnikova et al., 2006), composed of six cysteines to form three disulfide bonds. The structure is similar to K+-channel blocking toxin but does not have the function of channel blocking (Shenkarev et al., 2012). Aurelin is active against both Gram-positive and Gram-negative bacteria. Damicornin, similar to Aurelin, contains three disulfide bonds formed by six cysteines. However, it is active against Gram-positive bacteria and fungi (Vidal-Dupiol et al., 2011). As technology evolves, Hernández-Elizárraga, (2022) has excavated the antibacterial fragment of “Fire Coral” Millepora complanata transcriptome data and found that the purified mitochondrial gene has inhibitory activity against Salmonella enteritidis, Bacillus subtilis, and Pseudomonas aeruginosa.
2.2.5. AMPs from protochordata
As representatives of protochordates, Ascidians are similarly connected to oval sacs and have high AMPs content. Clavanin from the blood cells of Styela clava is a histidine-rich polypeptide with four α-helixes. It has activity against methicillin-resistant S. aureus (Ding and Zhang, 2013). Two phenylalanine-rich AMPs: sphingosine A (StyelinA) and sphingosine B (StyelinB), were also found in S. clava, which have inhibitory activity against common Gram-negative bacteria and Gram-positive bacteria.
2.2.6. AMPs from echinoderms
Strongylocins and centrocins have been identified in the coelomocytes of sea urchins. Strongylocin is rich in cysteine, contains three disulfide bonds, and has a molecular weight of 5.6 - 5.8 kDa. Centrocins are a family of heteronuclear AMPs with masses between 4.4 kDa and 4.5 kDa, consisting of a heavy chain and a light chain. The heavy chain is a cationic peptide, which not only has inhibitory activity against bacteria and fungi but also has anti-inflammatory effects (Solstad et al., 2016). Kim et al. (2018) discovered the first Patiria pectinifera AMPs, named PpCrAMP, which has 38 amino acid residues and is rich in cysteine. Apart from other echinoderms, there is no significant homology with known AMPs, suggesting that it may be a unique AMPs family in echinoderms.
2.3. AMPs from amphibious and reptile
2.3.1. AMPs from marine amphibians
Marine amphibians mainly include sea frogs (crab-eating frogs), living in shoals and mangroves that are similar to most bacterial habitats. To avoid bacterial invasion, amphibian skin glands secrete a large number of active molecules. Lu et al. (2008) fractionated and purified the skin secretions of the crab-eating frog and found a 19-peptide Cancrin with antibacterial activity. The precursor of Cancrin includes a signal peptide, an N-terminal spacer containing aspartic acid and glutamic acid residues, and a mature AMP at the C-terminus, which is very similar to the structure of AMPs precursors of other amphibians. But it is worth noting that the mature peptides are almost completely different. Gram-positive bacteria, especially S. aureus, are extremely sensitive to Cancrin. After co-incubation, even if the AMPs are removed, the strain cannot resume growth on the plate. When the concentration is as high as 100μg/ml, it has almost no hemolytic activity against mammalian blood cells. The discovery of Cancrin provides references for the design of new anti-infective drugs.
2.3.2. AMPs from reptile
Reptiles are among the ancient species, which include mainly sea snakes and turtles. Over millennia of evolution, they have been highly adapted to complex and changeable environments and have a strong immune system (van Hoek, 2014).
Tewp was found in turtle albumen, consisting of three disulfide bonds at 4-30, 8-29, and 12-24 (Chattopadhyay et al., 2006). Moreover, Tewp contains two anti-parallel β-sheets. Although the three-dimensional structure is similar to human β-defensin-3 (hbd-3), the disulfide bond is quite different from the defensin family. Tewp has strong antimicrobial activity against Gram-negative bacteria, including E. coli and S.typhimurium. Furthermore, it also has activity against the human pathogen Chandipura virus.
Hydrophis cyanocinctus have been recorded as medicine in China. Wei et al., (2015) obtained a polypeptide of the Cathelicidins family, Hc-CATH, from Hydrophis cyanocinctus. The Hc-CATH consists of 30 amino acids and has an α-helix structure in the membrane simulated environment. Studies suggest it may have an effect by destroying cell membranes and dissolving bacterial cells. It has broad-spectrum and rapid bactericidal activity, highly stable structural activity, and is non-toxic to mammalian cells. Derivatives of Hc-CATH show high inhibitory activity against drug-resistant Acinetobacter baumannii in vitro and in vivo. In addition, Hc-CATH is involved in immune regulation. It can induce the production of chemokines and recruit immune cells (Gao et al., 2020).
2.4. AMPs from mammal
Marine mammals mainly include cetaceans, fin-foot, sea otters, and other animal groups. They all originated from terrestrial ancestors and experienced major changes in adapting to water life. Unfortunately, due to the scarcity of food sources, human activity, and other reasons, gray whales, Chinese white dolphins, and North Sea dogs have been listed as critical protected wildlife. At present, there are few AMPs studies on this kind of animal. Tur1A was found in Tursiops truncatus (bottlenose dolphin) genomic data from the 2011 mammalian genome project. Tur1A belongs to the proline-rich AMPs (PrAMP) family, which has low toxicity. Tur1A is able to inhibit bacterial protein synthesis by internalizing membrane proteins into bacteria. It has a higher membrane permeability than Bac7 from the same family as Bovine.
3. Comparison of AMPs from different organisms
The Cathelicidin family has good potential for development of antimicrobial drugs. Several Cathelicidins from human and snakes are under clinical tests, among which the phase II clinical trial of Indolicidin from bow has been finished (Andrès, 2012). Therefore, taking Cathelicidin family as an example, we compared the amino acids of AMPs from organisms living in different environments (Figures 1B,C). Surprisingly, we found that AMPs from marine organisms may have a clear preference for Arg (arginine) and Leu (leucine) in relation to terrestrial organisms. A comparison of sea snake AMPs and terrestrial snake AMPs shows that sea snake AMPs tend to harbor higher levels of Arg and Leu than terrestrial snake AMPs. Nevertheless, the antimicrobial spectrum between AMPs from terrestrial snakes and sea snakes do not differ significantly. Since the hydrophilic residue Arg and the hydrophobic residue Leu contribute to the amphiphilic property of AMPs, which influences the action on bacterial cell membranes implicated in AMPs’ antibacterial mechanisms, this natural bias of amino acid usage may be related to adaptation to different characteristics of the bacterial membranes from distinct environments. The structures of Arg and Leu may be more useful for sea snake AMPs to act with marine bacterial membranes.
Furthermore, we compared the AMPs from snakes, turtles, and fish in the sea. Except for sea turtle Cm-CATH4, the Arg ratios in other sea life AMPs are even higher than that in the sea snake AMP, suggesting that marine AMPs do have a significant usage bias for Arg. The pI (5.51), sequence composition and length of Cm-CATH4 are significantly different with those of other turtle AMPs. Cm-CATH4 only has anti-microbial bioactivity against Gram-positive bacteria, which is however, weaker than other Cm-CATH peptides. It was speculated that Cm-CATH4 and other Cm-CATH peptides work in concert in sea turtles. Noteworthily, the level of Leu in marine AMPs varies by organisms or environments. Cod AMPs have lower levels of Leu than sea snakes and hagfish. Turtle and Atlantic salmon AMPs have both high and low levels of Leu. The differences are likely to be related with varied ecological environments. Sea turtles need to go to shore to lay eggs, and Atlantic salmon are migratory, so this environmental change may affect amino acid composition of AMPs. Cod generally live 20 meters below the surface of sea, and the temperature is extremely low. Additionally, hagfish AMPs contain a high percentage of tyrosine, histidine, tryptophan, and asparagine. This ratio may be related to its saprophytic lifestyle and the unique distribution of the peptides (gut).
4. Comparison of the higher-order structures and post-translational modifications of AMPs
Like AMPs from other sources, marine AMPs can also contain the canonical secondary structures of α-helix, β-sheet, β-turn, and random coil (Figure 2). The tertiary and higher-order structures of marine AMPs are folded upon disulfide bond formation or at the presence of specific amino acid resides such as prolines. The structures of marine AMPs seem to be more conserved within families than within species. This may be to some extent related to the classification criteria of different AMP families.
According to the reported data, the post-translational modifications and processing of AMPs in various marine animals were summarized (Table 1). AMP precursor proteins contain the regions of propeptides and mature peptides. After the removal of the propeptide and chemical modification, the mature antibacterial peptide can be obtained. It can be found that the modification of amino acid residues tends to be more complex in AMPs from lower organisms. Dihydroxylysine, hydroxylysine, and methionine sulfoxide appear in AMPs of protozoans. The only reported residue modification of AMPs in Ethinodermata is bromotryptophan. Moreover, arthropod-derived Penaeidins carry pyrrolidone carboxylic acid at the N-terminal. These differences in post-translational modification may be related to the distinct degrees of functional importance of AMPs in different species.
5. Action mechanisms of AMPs
As a part of innate immunity, AMPs are evolutionarily conserved. According to the mode of action, AMPs can be divided into two categories: targeting cell membrane peptides and non-targeting cell membrane peptides (Mookherjee et al., 2020). Research showed that multiple and complementary actions can be applied in the process that AMPs causes bacterial death. Targeting cell membrane peptides mainly include cationic peptides, which kill cells by destroying membrane permeability. While non-targeting cell membrane peptides do not destroy membrane integrity and play a role within cells after transmembrane transport (Hancock and Patrzykat, 2002). The outer membrane of Eukaryotic cells consists of zwitterionic phosphatidylcholine and sphingomyelin. However, the outer membrane of Prokaryotic cells possesses negatively charged because it is rich in components such as lipopolysaccharide or teichoic acid. Therefore, cationic AMPs can selectively interact with negatively charged microbial membranes (Sani and Separovic, 2016) without affecting Eukaryotic cells (da Costa et al., 2015).
5.1. The mechanism of AMPs that target cell membrane
At present, there are three models to explain the mechanism: the carpet model, barrel plate model, and ring pore model (Boparai and Sharma, 2020). In the “Carpet model”, as shown in Figure 3A, AMPs accumulate on the cell membrane surface by electrostatic interaction in a direction parallel to the membrane (Pouny et al., 1992). This is the case for Ovispirin (Yamaguchi et al., 2001). Ovispirin is electrostatically attracted by the anionic phospholipid head group first and covers the membrane in a “carpet” manner to form a “peptide carpet”. When the peptide concentration reaches the threshold, a ring-shaped instantaneous pore is formed on the membrane, and other peptides can enter the cell membrane, eventually destroying the bimolecular layer to form micelles and cracking the cell membrane (Oren and Shai, 1998; Bechinger, 1999).
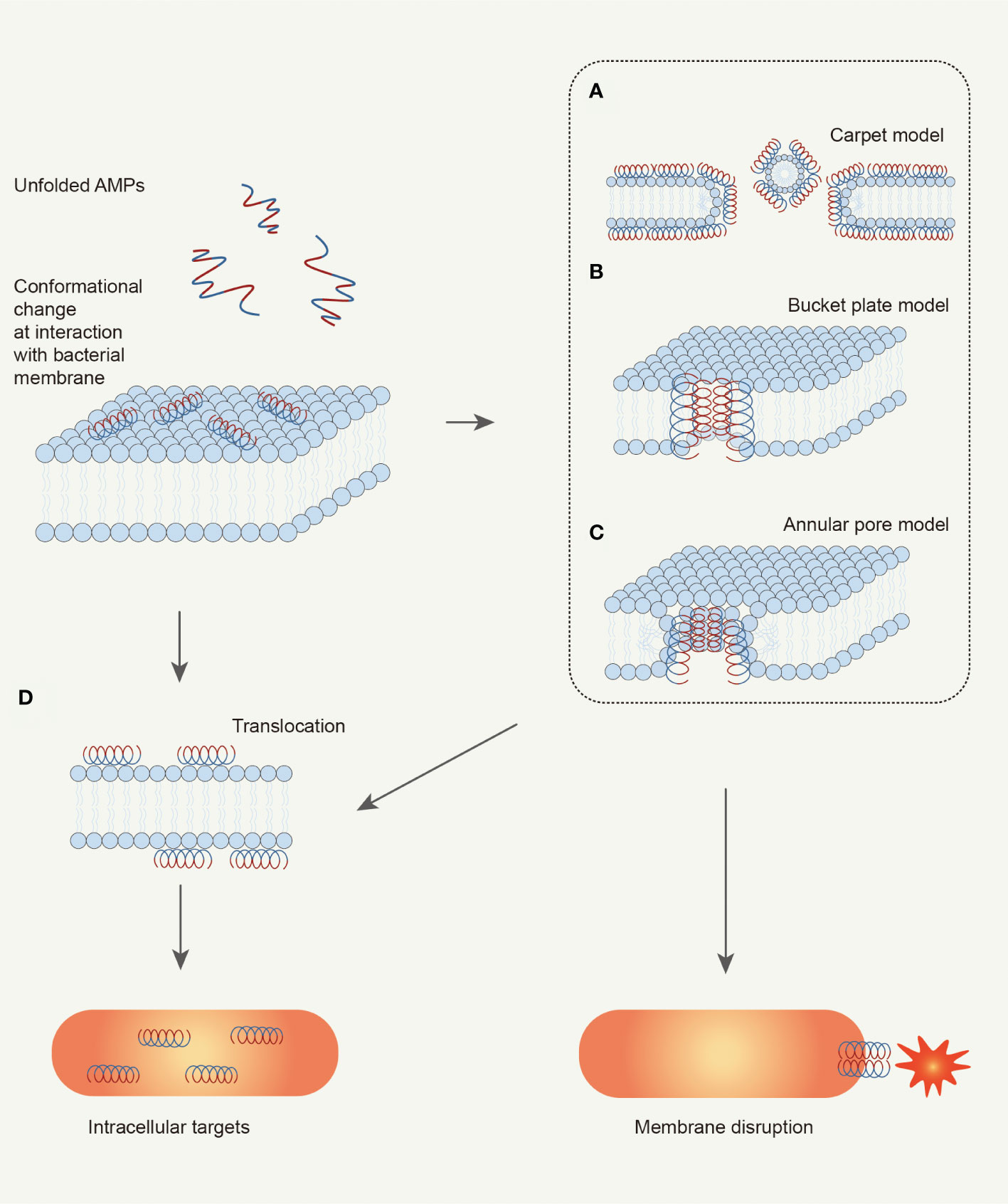
Figure 3 Schematic illustration of bacteria-killing mechanisms of AMPs. AMPs kill bacteria depending on the interaction with microbial membrane. AMPs would change their conformation from random coil to helix when interacting with bacterial membrane. The helix structure with amphipathy can promote the adsorption of AMPs by the membrane. Then, AMPs change membrane permeabilization by disturbing membrane structure, which can be explained by three models: (A) Carpet model; (B) Bucket plate model; (C) Annular pore model. Finally, the bacterial cell is disrupted because of ion leakage. Besides, the alteration of membrane permeabilization can also take some AMPs into the cell: (D) Translocation. These AMPs can target key cellular processes to kill the bacterial cell (Brogden, 2005; Mahlapuu et al., 2016).
In the “Barrel stave model” (Figure 3B), helical peptides gather around like a wooden plate, and the hydrophobic region binds to the lipid region of the lipid bilayer. The hydrophilic region constitutes the inner surface of the transmembrane pore, forming a barrel-shaped transmembrane pore with a central cavity on the membrane (Ehrenstein and Lecar, 1977; Yang et al., 2001). The mechanism of alamethicin can be explained by the “Barrel stave model”. Clarithromycin is a curved helical linear peptide that induces the formation of a transmembrane pore with an inner diameter of about 1.8 nm and an outer diameter of 4.0 nm (He et al., 1995; Spaar et al., 2004). The thickness of the transmembrane pore is about 1.1 nm, approximately the same as the diameter of the helical peptide itself (Fox and Richards, 1982; He et al., 1996; Yang et al., 2001).
In the “Toroidal pore model” (Figure 3C), the surface-attached AMPs are vertically inserted into the membrane and induce the bending of the lipid monolayer molecule. The inserted helical peptide and the phospholipid head group together to form a transmembrane pore. Melittin (Matsuzaki et al., 1996; Yang et al., 2001; Hallock et al., 2003) can induce the formation of this type of transmembrane pore. During the formation of circular pores, the hydrophobic surface of the helical peptide is always associated with the polar head group of the lipid layer (Yamaguchi et al., 2002). The lipid monolayer then bends continuously from the top lobule to the bottom lobule, forming a circular pore together with the helical peptide. Notably, different from the “Barrel stave model”, when the polypeptide in the ring-pore model is vertically inserted into the lipid bilayer (Yang et al., 2001), the phospholipid head group of the polypeptide and the lipid molecular layer also always act together.
5.2. The mechanism of AMPs that do not target cell membrane
Non-targeting cell membrane peptides are also known as intracellularly active AMPs (Boparai and Sharma, 2020). Intracellular active peptides (Figure 3D) can pass through the lipid bilayer and target intracellular components, including binding to intracellular targets such as DNA, RNA, and proteins, to inhibit protein and nucleic acid synthesis and disrupt enzyme and protein activity (Dennison et al., 2015). In other words, it exerts antibacterial effects through intracellular inhibitory mechanisms.
5.3. The mechanism of AMPs that regulate immunity
AMPs can not only kill bacteria directly, but also recruit and activate immune cells to reduce inflammation or kill bacteria. SpCrus8 has no antibacterial activity, but can significantly reduce bacterial load by promoting blood cell phagocytosis (Jiang et al., 2022). Some AMPs can induce multiple immune responses: activation, attraction, and differentiation of leukocytes; stimulate angiogenesis; reduce inflammation by reducing the expression of pro-inflammatory chemokines; control the production of chemokines and reactive oxygen species (de Barros et al., 2019). For example, human-derived LL-37 can not only maintain skin immune balance by regulating chemokines and vascular endothelial growth factor levels but also recruit immune cells such as white blood cells and mast cells (Sigurgrimsdottir et al., 2021).
6. Research and development potential of AMPs
Due to the many advantages of AMPs such as broad antimicrobial spectrum, high efficiency, low probability of inducing bacterial resistance, and low toxicity, their development as drugs are underway. To date, a total of 3,324 AMPs and 721 Chinese related patents can be retrieved in the database worldwide. There are more than 70 AMPs available as drug candidates worldwide, with 42 in clinical trials. The indications include Gram-positive bacterial infection, fungal infection, diabetic foot ulcer, periodontal disease, HIV and burn infection, and so on.
At present, the research progress on marine AMPs is slow, but there are still some exciting reports. Wei et al., (2015) found Hc-CATH from a sea snake, which has biological activity against 48 kinds of microorganisms including Gram-negative bacteria, positive bacteria, and fungi. Its rate of sterilization is better than the current first-line bacteriostatic drug, meropenem. Taking it as the lead compound (Gao et al., 2020), the derivate HC1 has activity against drug-resistant bacteria. Meantime, drug-resistance tendency of bacteria was not induced. This suggests that development potential of marine AMPs is evident.
However, it cannot be ignored that the search for marine active molecules often suffers from problems such as few materials, low content, similar structures, and difficult separation. Omics technology may be a solution to effectively produce new peptides with unique pharmacological potential (Giordano et al., 2018). Recent advances in genomics, transcriptomics, proteomics, and other omics technologies have provided a new platform for AMPs discovery. Zhang was applying for a patent for a peptide from transcriptomes and proteomes of sea snakes, which was capable to inhibit LPS infections from different sources. In addition, several studies have found AMPs sequences that may have antibacterial properties through Hippocampus erectus genome and transcriptome data (Chen et al., 2019), and the next activity study is underway. Li et al. (2021) have completed the genome sequencing of H. cyanocinctus, which provides conditions for the direct mining of AMPs.
In general, the application of AMPs in the treatment of infection is successful, and with the iterative update of biotechnology, the continuously excavated marine AMPs will also enter the vision of major pharmaceutical companies. Actually, a few AMPs have now entered the global market, such as Omiganan. Omiganan has been used in the treatment of condyloma acuminatum and acne vulgaris. Undoubtedly, the need for new approaches to infection control, together with the importance of AMPs in innate immunity, will continue to bring a large number of peptide antibiotics into clinical research.
Discussion
The unique marine environment provides the conditions for the production of bioactive molecules, which is a natural treasure house for drug discovery. Some AMPs have high levels of Arg and Leu, which may be helpful to kill bacteria in seawater and may offer a reference for aquaculture. However, the study of AMPs in marine organisms represents only 3% of the total AMPs because experimental material is not readily available. At present, with the development of biotechnology, many research teams use transcriptome and proteome analysis to mine active molecules (Li et al., 2021), which can solve the bottleneck problems in marine biological research. Shortly, more AMPs from marine organisms will inevitably appear. It is the current trend of drug discovery to select natural products with new structures and exact pharmacological effects or known synthetic drugs as lead compounds, and then modify and simplify their chemical structures through drug design. While most AMPs that have entered clinical studies are derived from humans, cattle, and pigs, the molecular skeletons of marine AMPs are unique and valuable. The development of anti-infective drugs such as HC1 against drug-resistant bacteria is of great significance and potential. All in all, marine-derived AMPs promise to be a new generation of antibiotics that could play a huge role in the anti-infective field.
Author contributions
YL and YQ conceived and designed the research; SW and LF contributed to the literature search and wrote the paper; HP and YYL prepared figures. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (No. 42076094, No.82273844), the Shanghai Science and Technology Innovation Action Plan (Grant No.21S11902400), Guilin Technological Innovation Guidance plan (No.2020010907), and Funded by Outstanding Leaders Training Program of Pudong Health Bureau of Shanghai (Grant No. PWRl2020-08).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer LW declared a shared affiliation with the author YL to the handling editor at the time of review.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Anderson R. S., Beaven A. E. (2001). Antibacterial activities of oyster (Crassostrea virginica) and mussel (Mytilus edulis and Geukensia demissa) plasma. Aquat. Living Resour. 14 (6), 343–349. doi: 10.1016/S0990-7440(01)01143-3
Andrès E. (2012). Cationic antimicrobial peptides in clinical development, with special focus on thanatin and heliomicin. Eur. J. Clin. Microbiol. Infect. Dis. 31 (6), 881–888. doi: 10.1007/s10096-011-1430-8
Balseiro P., Falcó A., Romero A., Dios S., Martínez-López A., Figueras A., et al. (2011). Mytilus galloprovincialis myticin c: a chemotactic molecule with antiviral activity and immunoregulatory properties. PloS One 6 (8), e23140. doi: 10.1371/journal.pone.0023140
Bechinger B. (1999). The structure, dynamics and orientation of antimicrobial peptides in membranes by multidimensional solid-state NMR spectroscopy. Biochim. Biophys. Acta 1462 (1-2), 157–183. doi: 10.1016/s0005-2736(99)00205-9
Boparai J. K., Sharma P. K. (2020). Mini review on antimicrobial peptides, sources, mechanism and recent applications. Protein Pept. Lett. 27 (1), 4–16. doi: 10.2174/0929866526666190822165812
Brogden K. A. (2005). Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 3 (3), 238–250. doi: 10.1038/nrmicro1098
Cai L., Cai J. J., Liu H. P., Fan D. Q., Peng H., Wang K. J. (2012). Recombinant medaka (Oryzias melastigmus) pro-hepcidin: Multifunctional characterization. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 161 (2), 140–147. doi: 10.1016/j.cbpb.2011.10.006
Chang W. T., Pan C. Y., Rajanbabu V., Cheng C. W., Chen J. Y. (2011). Tilapia (Oreochromis mossambicus) antimicrobial peptide, hepcidin 1-5, shows antitumor activity in cancer cells. Peptides 32 (2), 342–352. doi: 10.1016/j.peptides.2010.11.003
Charlet M., Chernysh S., Philippe H., Hetru C., Hoffmann J. A., Bulet P. (1996). Innate immunity. isolation of several cysteine-rich antimicrobial peptides from the blood of a mollusc. Mytilus edulis. J. Biol. Chem. 271 (36), 21808–21813. doi: 10.1074/jbc.271.36.21808
Chattopadhyay S., Sinha N. K., Banerjee S., Roy D., Chattopadhyay D., Roy S. (2006). Small cationic protein from a marine turtle has beta-defensin-like fold and antibacterial and antiviral activity. Proteins 64 (2), 524–531. doi: 10.1002/prot.20963
Chen X., Yi Y., Bian C., You X., Shi Q. (2020). Putative antimicrobial peptides in fish: Using zebrafish as a representative. Protein Pept. Lett. 27 (11), 1059–1067. doi: 10.2174/0929866527666200517104610
Chen X., Yi Y., You X., Liu J., Shi Q. (2019). High-throughput identification of putative antimicrobial peptides from multi-omics data of the lined seahorse (Hippocampus erectus). Mar. Drugs 18 (1). doi: 10.3390/md18010030
Cole A. M., Weis P., Diamond G. (1997). Isolation and characterization of pleurocidin, an antimicrobial peptide in the skin secretions of winter flounder. J. Biol. Chem. 272 (18), 12008–12013. doi: 10.1074/jbc.272.18.12008
da Costa J. P., Cova M., Ferreira R., Vitorino R. (2015). Antimicrobial peptides: an alternative for innovative medicines? Appl. Microbiol. Biotechnol. 99 (5), 2023–2040. doi: 10.1007/s00253-015-6375-x
de Barros E., Gonçalves R. M., Cardoso M. H., Santos N. C., Franco O. L., Cândido E. S. (2019). Snake venom cathelicidins as natural antimicrobial peptides. Front. Pharmacol. 10 1415. doi: 10.3389/fphar.2019.01415
Dennison S. R., Mura M., Harris F., Morton L. H., Zvelindovsky A., Phoenix D. A. (2015). The role of c-terminal amidation in the membrane interactions of the anionic antimicrobial peptide, maximin H5. Biochim. Biophys. Acta 1848 (5), 1111–1118. doi: 10.1016/j.bbamem.2015.01.014
Destoumieux D., Bulet P., Strub J. M., Van Dorsselaer A., Bachère E. (1999). Recombinant expression and range of activity of penaeidins, antimicrobial peptides from penaeid shrimp. Eur. J. Biochem. 266 (2), 335–346. doi: 10.1046/j.1432-1327.1999.00855.x
Destoumieux D., Munoz M., Bulet P., Bachère E. (2000). Penaeidins, a family of antimicrobial peptides from penaeid shrimp (Crustacea, decapoda). Cell Mol. Life Sci. 57 (8-9), 1260–1271. doi: 10.1007/pl00000764
Ding Y., Zhang S. (2013). Antimicrobial peptides from marine invertebrates and fish. Chin. J. Mar. Drugs 06, 10. doi: 10.13400/j.cnki.cjmd.2013.06.003
Ehrenstein G., Lecar H. (1977). Electrically gated ionic channels in lipid bilayers. Q Rev. Biophys. 10 (1), 1–34. doi: 10.1017/s0033583500000123
Falanga A., Lombardi L., Franci G., Vitiello M., Iovene M. R., Morelli G., et al. (2016). Marine antimicrobial peptides: Nature provides templates for the design of novel compounds against pathogenic bacteria. Int. J. Mol. Sci. 17 (5). doi: 10.3390/ijms17050785
Fox R. O. Jr., Richards F. M. (1982). A voltage-gated ion channel model inferred from the crystal structure of alamethicin at 1.5-a resolution. Nature 300 (5890), 325–330. doi: 10.1038/300325a0
Gao J., Zhang M., Zhang F., Wang Y., Ouyang J., Luo X., et al. (2020). Design of a Sea snake antimicrobial peptide derivative with therapeutic potential against drug-resistant bacterial infection. ACS Infect. Dis. 6 (9), 2451–2467. doi: 10.1021/acsinfecdis.0c00255
Giordano D., Costantini M., Coppola D., Lauritano C., Núñez Pons L., Ruocco N., et al. (2018). Biotechnological applications of bioactive peptides from marine sources. Adv. Microb. Physiol. 73, 171–220. doi: 10.1016/bs.ampbs.2018.05.002
Greco S., Gerdol M., Edomi P., Pallavicini A. (2020). Molecular diversity of mytilin-like defense peptides in mytilidae (Mollusca, bivalvia). Antibiotics (Basel) 9 (1). doi: 10.3390/antibiotics9010037
Hallock K. J., Lee D. K., Ramamoorthy A. (2003). MSI-78, an analogue of the magainin antimicrobial peptides, disrupts lipid bilayer structure via positive curvature strain. Biophys. J. 84 (5), 3052–3060. doi: 10.1016/s0006-3495(03)70031-9
Hancock R. E., Patrzykat A. (2002). Clinical development of cationic antimicrobial peptides: from natural to novel antibiotics. Curr. Drug Targets Infect. Disord. 2 (1), 79–83. doi: 10.2174/1568005024605855
He K., Ludtke S. J., Huang H. W., Worcester D. L. (1995). Antimicrobial peptide pores in membranes detected by neutron in-plane scattering. Biochemistry 34 (48), 15614–15618. doi: 10.1021/bi00048a002
He K., Ludtke S. J., Worcester D. L., Huang H. W. (1996). Neutron scattering in the plane of membranes: structure of alamethicin pores. Biophys. J. 70 (6), 2659–2666. doi: 10.1016/s0006-3495(96)79835-1
Hernández-Elizárraga V. H., Ocharán-Mercado A., Olguín-López N., Hernández-Matehuala R., Caballero-Pérez J., Ibarra-Alvarado C., et al. (2022). New insights into the toxin diversity and antimicrobial activity of the "Fire coral" Millepora complanata. Toxins (Basel) 14 (3). doi: 10.3390/toxins14030206
Hunter H. N., Fulton D. B., Ganz T., Vogel H. J. (2002). The solution structure of human hepcidin, a peptide hormone with antimicrobial activity that is involved in iron uptake and hereditary hemochromatosis. J. Biol. Chem. 277 (40), 37597–37603. doi: 10.1074/jbc.M205305200
Javia A., Amrutiya J., Lalani R., Patel V., Bhatt P., Misra A. (2018). Antimicrobial peptide delivery: an emerging therapeutic for the treatment of burn and wounds. Ther. Delivery 9 (5), 375–386. doi: 10.4155/tde-2017-0061
Jiang M., Chen R., Chen F., Zhu X., Wang K. J. (2022). A new crustin gene homolog SpCrus8 identified in Scylla paramamosain exerting In vivo protection through opsonization and immunomodulation. Front. Immunol. 13. doi: 10.3389/fimmu.2022.946227
Jin J. Y., Zhou L., Wang Y., Li Z., Zhao J. G., Zhang Q. Y., et al. (2010). Antibacterial and antiviral roles of a fish β-defensin expressed both in pituitary and testis. PloS One 5 (12), e12883. doi: 10.1371/journal.pone.0012883
Kim C. H., Go H. J., Oh H. Y., Park J. B., Lee T. K., Seo J. K., et al. (2018). Identification of a novel antimicrobial peptide from the sea star Patiria pectinifera. Dev. Comp. Immunol. 86, 203–213. doi: 10.1016/j.dci.2018.05.002
Kushibiki T., Kamiya M., Aizawa T., Kumaki Y., Kikukawa T., Mizuguchi M., et al. (2014). Interaction between tachyplesin I, an antimicrobial peptide derived from horseshoe crab, and lipopolysaccharide. Biochim. Biophys. Acta 1844 (3), 527–534. doi: 10.1016/j.bbapap.2013.12.017
Liang T., Ji W., Zhang G. R., Wei K. J., Feng K., Wang W. M., et al. (2013). Molecular cloning and expression analysis of liver-expressed antimicrobial peptide 1 (LEAP-1) and LEAP-2 genes in the blunt snout bream (Megalobrama amblycephala). Fish Shellfish Immunol. 35 (2), 553–563. doi: 10.1016/j.fsi.2013.05.021
Liu C., Hu X., Feng Y. (1985). Bacterial resistance : biochemical mechanisms and coping strategies. Biotechnol. Bull. 38 (09), 4–16. doi: 10.13560/j.cnki.biotech.bull.1985.2022-0759
Li A., Wang J., Sun K., Wang S., Zhao X., Wang T., et al. (2021). Two reference-quality Sea snake genomes reveal their divergent evolution of adaptive traits and venom systems. Mol. Biol. Evol. 38 (11), 4867–4883. doi: 10.1093/molbev/msab212
Lu Y., Ma Y., Wang X., Liang J., Zhang C., Zhang K., et al. (2008). The first antimicrobial peptide from sea amphibian. Mol. Immunol. 45 (3), 678–681. doi: 10.1016/j.molimm.2007.07.004
Mahlapuu M., Håkansson J., Ringstad L., Björn C. (2016). Antimicrobial peptides: An emerging category of therapeutic agents. Front. Cell Infect. Microbiol. 6. doi: 10.3389/fcimb.2016.00194
Masso-Silva J. A., Diamond G. (2014). Antimicrobial peptides from fish. Pharm. (Basel) 7 (3), 265–310. doi: 10.3390/ph7030265
Matsuzaki K., Murase O., Fujii N., Miyajima K. (1996). An antimicrobial peptide, magainin 2, induced rapid flip-flop of phospholipids coupled with pore formation and peptide translocation. Biochemistry 35 (35), 11361–11368. doi: 10.1021/bi960016v
Mitta G., Vandenbulcke F., Hubert F., Salzet M., Roch P. (2000). Involvement of mytilins in mussel antimicrobial defense. J. Biol. Chem. 275 (17), 12954–12962. doi: 10.1074/jbc.275.17.12954
Miyata T., Tokunaga F., Yoneya T., Yoshikawa K., Iwanaga S., Niwa M., et al. (1989). Antimicrobial peptides, isolated from horseshoe crab hemocytes, tachyplesin II, and polyphemusins I and II: chemical structures and biological activity. J. Biochem. 106 (4), 663–668. doi: 10.1093/oxfordjournals.jbchem.a122913
Molinski T. F., Dalisay D. S., Lievens S. L., Saludes J. P. (2009). Drug development from marine natural products. Nat. Rev. Drug Discovery 8 (1), 69–85. doi: 10.1038/nrd2487
Mookherjee N., Anderson M. A., Haagsman H. P., Davidson D. J. (2020). Antimicrobial host defence peptides: functions and clinical potential. Nat. Rev. Drug Discovery 19 (5), 311–332. doi: 10.1038/s41573-019-0058-8
Nakamura T., Furunaka H., Miyata T., Tokunaga F., Muta T., Iwanaga S., et al. (1988). Tachyplesin, a class of antimicrobial peptide from the hemocytes of the horseshoe crab (Tachypleus tridentatus). isolation and chemical structure. J. Biol. Chem. 263 (32), 16709–16713.
Nalini S., Sandy Richard D., Mohammed Riyaz S. U., Kavitha G., Inbakandan D. (2018). Antibacterial macro molecules from marine organisms. Int. J. Biol. Macromol 115, 696–710. doi: 10.1016/j.ijbiomac.2018.04.110
Nam B. H., Moon J. Y., Kim Y. O., Kong H. J., Kim W. J., Lee S. J., et al. (2010). Multiple beta-defensin isoforms identified in early developmental stages of the teleost Paralichthys olivaceus. Fish Shellfish Immunol. 28 (2), 267–274. doi: 10.1016/j.fsi.2009.11.004
Oren Z., Shai Y. (1998). Mode of action of linear amphipathic alpha-helical antimicrobial peptides. Biopolymers 47 (6), 451–463. doi: 10.1002/(sici)1097-0282(1998)47:6<451::Aid-bip4>3.0.Co;2-f
Ovchinnikova T. V., Balandin S. V., Aleshina G. M., Tagaev A. A., Leonova Y. F., Krasnodembsky E. D., et al. (2006). Aurelin, a novel antimicrobial peptide from jellyfish Aurelia aurita with structural features of defensins and channel-blocking toxins. Biochem. Biophys. Res. Commun. 348 (2), 514–523. doi: 10.1016/j.bbrc.2006.07.078
Pan W., Liu X., Ge F., Han J., Zheng T. (2004). Perinerin, a novel antimicrobial peptide purified from the clamworm Perinereis aibuhitensis grube and its partial characterization. J. Biochem. 135 (3), 297–304. doi: 10.1093/jb/mvh036
Pouny Y., Rapaport D., Mor A., Nicolas P., Shai Y. (1992). Interaction of antimicrobial dermaseptin and its fluorescently labeled analogues with phospholipid membranes. Biochemistry 31 (49), 12416–12423. doi: 10.1021/bi00164a017
Priya A., Aditya A., Budagavi D. P., Chugh A. (2022). Tachyplesin and CyLoP-1 as efficient anti-mycobacterial peptides: A novel finding. Biochim. Biophys. Acta Biomembr 1864 (7), 183895. doi: 10.1016/j.bbamem.2022.183895
Roch P., Yang Y., Toubiana M., Aumelas A. (2008). NMR structure of mussel mytilin, and antiviral-antibacterial activities of derived synthetic peptides. Dev. Comp. Immunol. 32 (3), 227–238. doi: 10.1016/j.dci.2007.05.006
Safronova V. N., Panteleev P. V., Sukhanov S. V., Toropygin I. Y., Bolosov I. A., Ovchinnikova T. V. (2022). Mechanism of action and therapeutic potential of the β-hairpin antimicrobial peptide capitellacin from the marine polychaeta Capitella teleta. Mar. Drugs 20 (3). doi: 10.3390/md20030167
Sani M. A., Separovic F. (2016). How membrane-active peptides get into lipid membranes. Acc Chem. Res. 49 (6), 1130–1138. doi: 10.1021/acs.accounts.6b00074
Shenkarev Z. O., Panteleev P. V., Balandin S. V., Gizatullina A. K., Altukhov D. A., Finkina E. I., et al. (2012). Recombinant expression and solution structure of antimicrobial peptide aurelin from jellyfish Aurelia aurita. Biochem. Biophys. Res. Commun. 429 (1-2), 63–69. doi: 10.1016/j.bbrc.2012.10.092
Shigenaga T., Takayenoki Y., Kawasaki S., Seki N., Muta T., Toh Y., et al. (1993). Separation of large and small granules from horseshoe crab (Tachypleus tridentatus) hemocytes and characterization of their components. J. Biochem. 114 (3), 307–316. doi: 10.1093/oxfordjournals.jbchem.a124173
Sigurgrimsdottir H., Bjornsdottir E. O., Eysteinsdottir J. H., Olafsson J. H., Sigurgeirsson B., Agnarsson B. A., et al. (2021). Keratinocytes secrete multiple inflammatory and immune biomarkers, which are regulated by LL-37, in a psoriasis mimicking microenvironment. Scand. J. Immunol. 94 (6), e13096. doi: 10.1111/sji.13096
Solstad R. G., Li C., Isaksson J., Johansen J., Svenson J., Stensvåg K., et al. (2016). Novel antimicrobial peptides EeCentrocins 1, 2 and EeStrongylocin 2 from the edible Sea urchin Echinus esculentus have 6-Br-Trp post-translational modifications. PloS One 11 (3), e0151820. doi: 10.1371/journal.pone.0151820
Sonthi M., Cantet F., Toubiana M., Trapani M. R., Parisi M. G., Cammarata M., et al. (2012). Gene expression specificity of the mussel antifungal mytimycin (MytM). Fish Shellfish Immunol. 32 (1), 45–50. doi: 10.1016/j.fsi.2011.10.017
Spaar A., Münster C., Salditt T. (2004). Conformation of peptides in lipid membranes studied by x-ray grazing incidence scattering. Biophys. J. 87 (1), 396–407. doi: 10.1529/biophysj.104.040667
Steiner H., Hultmark D., Engström A., Bennich H., Boman H. G. (1981). Sequence and specificity of two antibacterial proteins involved in insect immunity. Nature 292 (5820), 246–248. doi: 10.1038/292246a0
Tincu J. A., Taylor S. W. (2004). Antimicrobial peptides from marine invertebrates. Antimicrob. Agents Chemother. 48 (10), 3645–3654. doi: 10.1128/aac.48.10.3645-3654.2004
van Hoek M. L. (2014). Antimicrobial peptides in reptiles. Pharm. (Basel) 7 (6), 723–753. doi: 10.3390/ph7060723
Vidal-Dupiol J., Ladrière O., Destoumieux-Garzón D., Sautière P. E., Meistertzheim A. L., Tambutté E., et al. (2011). Innate immune responses of a scleractinian coral to vibriosis. J. Biol. Chem. 286 (25), 22688–22698. doi: 10.1074/jbc.M110.216358
Wei L., Gao J., Zhang S., Wu S., Xie Z., Ling G., et al. (2015). Identification and characterization of the first cathelicidin from Sea snakes with potent antimicrobial and anti-inflammatory activity and special mechanism. J. Biol. Chem. 290 (27), 16633–16652. doi: 10.1074/jbc.M115.642645
Williams S. T. (2017). Molluscan shell colour. Biol. Rev. Camb Philos. Soc. 92 (2), 1039–1058. doi: 10.1111/brv.12268
Wu R., Patocka J., Nepovimova E., Oleksak P., Valis M., Wu W., et al. (2021). Marine invertebrate peptides: Antimicrobial peptides. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.785085
Xu G., Wu M., Wang L., Zhang X., Cao S., Liu M., et al. (2009). Conformational and dynamics simulation study of antimicrobial peptide hedistin-heterogeneity of its helix-turn-helix motif. Biochim. Biophys. Acta 1788 (12), 2497–2508. doi: 10.1016/j.bbamem.2009.10.001
Yamaguchi S., Hong T., Waring A., Lehrer R. I., Hong M. (2002). Solid-state NMR investigations of peptide-lipid interaction and orientation of a beta-sheet antimicrobial peptide, protegrin. Biochemistry 41 (31), 9852–9862. doi: 10.1021/bi0257991
Yamaguchi S., Huster D., Waring A., Lehrer R. I., Kearney W., Tack B. F., et al. (2001). Orientation and dynamics of an antimicrobial peptide in the lipid bilayer by solid-state NMR spectroscopy. Biophys. J. 81 (4), 2203–2214. doi: 10.1016/S0006-3495(01)75868-7
Yang L., Harroun T. A., Weiss T. M., Ding L., Huang H. W. (2001). Barrel-stave model or toroidal model? a case study on melittin pores. Biophys. J. 81 (3), 1475–1485. doi: 10.1016/S0006-3495(01)75802-X
Ziko L., AbdelRaheem O., Nabil M., Aziz R. K., Siam R. (2022). Bioprospecting the microbiome of red Sea Atlantis II brine pool for peptidases and biosynthetic genes with promising antibacterial activity. Microb. Cell Fact 21 (1), 109. doi: 10.1186/s12934-022-01835-z
Keywords: antimicrobial peptides, structures, marine organisms, drug resistance, drug discovery
Citation: Wang S, Fan L, Pan H, Li Y, Qiu Y and Lu Y (2023) Antimicrobial peptides from marine animals: Sources, structures, mechanisms and the potential for drug development. Front. Mar. Sci. 9:1112595. doi: 10.3389/fmars.2022.1112595
Received: 30 November 2022; Accepted: 30 December 2022;
Published: 19 January 2023.
Edited by:
Zhongshan Zhang, Huzhou University, ChinaReviewed by:
Hou-Wen Lin, Shanghai Jiao Tong University, ChinaXu-Wen Li, Shanghai Institute of Materia Medica (CAS), China
Lianghua Wang, Second Military Medical University, China
Copyright © 2023 Wang, Fan, Pan, Li, Qiu and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan Qiu, UWl1eWFuMjE4OUAxNjMuY29t; Yiming Lu, Ymx1ZXNsdXlpQHNpbmEuY29t
†These authors have contributed equally to this work
 Shuocun Wang1†
Shuocun Wang1† Yan Qiu
Yan Qiu Yiming Lu
Yiming Lu