
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 16 December 2022
Sec. Marine Biogeochemistry
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.1100289
This article is part of the Research Topic Advances in Estuarine and Coastal Nitrogen Cycle View all 17 articles
 Yanjing Song1
Yanjing Song1 Lan Ma1
Lan Ma1 Haiyang Zhang1
Haiyang Zhang1 Rao Fu1
Rao Fu1 Xiaoyan Liang1
Xiaoyan Liang1 Junlin Li1
Junlin Li1 Jiajia Li1
Jiajia Li1 Meng Li1
Meng Li1 Yan Shan1
Yan Shan1 Jieshan Cheng2,3
Jieshan Cheng2,3 Xiangyu Wang1*
Xiangyu Wang1* Hongxia Zhang1,2,3*
Hongxia Zhang1,2,3*The diversity and community structure of rhizospheric microbes are largely affected by soil physicochemical properties and plant species. In this work, high throughput sequencing and quantitative real-time PCR targeting nifH gene were used to assess the abundance and diversity of diazotrophic community in the coastal saline soils of Yellow River Delta (YRD). We demonstrated that the copy number of nifH gene encoding the Fe protein subunit of the nitrogenase in the nitrogen fixation process was significantly affected by soil physiochemical factors, and the abundance of diazotrophs in the rhizospheric soil samples collected from different locations was positively related with soil physicochemical properties. Soil salinity (P=0.003) and moisture (P=0.003) were significantly co-varied with the OTU-based community composition of diazotrophs. Taxonomic analysis showed that most diazotrophs belonged to the Alphaproteobacteria, Gammaproteobacteria and Deltaproteobacteria. Linear discriminant analysis (LDA) effect size (LEfSe) and canonical correspondence analysis (CCA) showed that diazotrophic community structure significantly varied with soil salinity, moisture, pH and total nitrogen, carbon, sulphur and nitrite (NO2–N) content. Our findings provide direct evidence toward the understanding of different effects of soil physicochemical properties and host plant traits such as halophytes types, life span and cotyledon type, on the community composition of diazotrophic populations in the rhizosphere of plants grown in coastal saline soils.
Nitrogen (N) is one of the essential primary macronutrients for plant growth and production (Li et al., 2018). Through the reduction of atmospheric N2 to biologically available ammonium, about 70% of total N is fixed (Galloway et al., 2004; Lin et al., 2016). An efficient way to introduce N2 into the biosphere is N fixation, since plants cannot directly utilize gaseous N2. In the rhizosphere of plants, a wide variety of diazotrophs, including free living bacteria, sulfate reducing bacteria and symbiotic diazotrophs, usually dominated by Proteobacteria, Cyanobacteria and Firmicutes, inhabited (Wang et al., 2016; Zhang et al., 2021). These diazotrophs shared the same operon in which the nifH gene encoding the Fe protein subunit of the nitrogenase in the nitrogen fixation process has been widely used for phylogenetic analyses (Ininbergs et al., 2011; Ospina-Betancourth et al., 2020; Dong et al., 2022). Plant diversity could affect the composition and function of microbial community, which in return, increased soil N supply to plants (Zak et al., 2003). Due to the high genetic diversity, rhizospheric diazotrophs could play multiple roles in plant growth and development (Vejan et al., 2016; Oliveira et al., 2017). Generally, diazotrophs provided readily available N sources to host plants, and in return, host plants supplied carbon sources to diazotrophs through photosynthesis (Gupta et al., 2006; Liu et al., 2019).
The diversity and composition of microbial communities in the rhizosphere of plants were affected by soil properties (Garbeva et al., 2008; Tkacz et al., 2015; Jiang et al., 2017; Li et al., 2021). In some studies, a greater impact of host plant species on microbial assemblage composition, than of abiotic parameters, has been observed, since the selection of microbial communities in rhizospheric soils was dependent on the root morphology, root exudation and nutrient competition of plants (Rodrigo et al., 2006; Ladygina and Hedlund, 2010). In some studies, diazotrophic abundance and community composition were found to be mainly affected by soil characteristics, such pH, inorganic N and carbon/nitrogen (C/N) ratio, especially salinity and water content (Moisander et al., 2007; Collavino et al., 2014; Levy-Booth et al., 2014; Wang et al., 2017; Che et al., 2018; Lin et al., 2018). The effects of plants on microbial community in plant rhizosphere were species specific, and a synergistic relationship between plant species and environmental conditions existed (Morina et al., 2018). In addition, diazotroph assemblages were also strongly influenced by seasons, abiotic environmental parameters and host plants (Debra et al., 2011; Lin et al., 2017).
One of the major causes for soil salinization in the coastal region was the intrusion of seawater into coastal aquifers (Omuto et al., 2020). With the increase of distance from the vertical coastline, soil salinity and moisture of coastal wetlands changed significantly (Xian et al., 2019). In coastal sediments, higher rate of sediment N-fixation, which could be a significant N source, has been detected (Mortazavi et al., 2012; Bhavya et al., 2016). In coastal seagrass beds, microbial N-fixing provided 50% of the N requirements (Whiting et al., 1986). The biological N fixation (BNF) rates in coastal region can be affected by many biotic and abiotic factors, such as diazotroph diversity, soil C:N ratio and soil P level (Dang et al., 2013; Huang et al., 2016). Microbially-mediated N-dynamics were particular important to the biogeochemical functions in coastal marine wetlands (Affourtit et al., 2001; Moisander et al., 2007; Moseman-Valtierra et al., 2009). Nitrogen-fixing bacteria constituted a key functional group of microorganisms relevant to the functions of wetland ecosystems (Moseman et al., 2009). Meanwhile, human activity also generated a great amount of nitrogen, causing numerous ecological and environmental problems, such as eutrophication, in coastal region (Newell et al., 2016; Huang et al., 2021; Lin and Lin, 2022). Salinization could affect the transformation and uptake of N, and ultimately restrain the growth and decrease the production of plants (Midgley, 2012). To adapt to the salt stress condition, halophyte plants could shape specific rhizospheric microbiomes and increase the microbial diversities (Wang et al., 2010; Anburaj et al., 2012; Qiu et al., 2022). The growth of halophyte plants is usually limited by the availability of N, which is supplemented by the introduction of ‘new’ N in the ecosystems through plant associated diazotrophs (Debra et al., 2011; Rejmánková et al., 2018; Zhang et al., 2020; Qiu et al., 2022). The rate of microbial N fixation is correlated to plant photosynthetic activity, and the structure of diazotroph assemblage is significantly influenced by the variety of carbon sources associated with plant species. According to the mechanisms of salt tolerance, halophytes are divided into secretohalophyte, pseudohalophyte and euhalophyte. Dicotyledonous plants have more developed root system than monocotyledonous plants, and annual halophyte plants have developed a set of mechanisms safer than those of perennial halophyte plants, including seed dormancy when subjected to high salt stress, fast germination after rehydration, polymorphism in morphology and germination, and persistent seed bank and plastic resource allocation (Cao et al., 2021).
Since the 1980s, the coastal zone of Laizhou Bay-Yellow River Estuary, which cover an area of 2,870 km2, has become one of the most serious seawater intruded regions in China (Guo and Gong, 2014). In this area, coastal saline soils show a gradient of salinity and water content from land to sea, accompanied with an apparent species succession of native plants. Although some studies on the characterization and evolution of vegetation in this area have been carried out for saline soil bioremediation, limited work has been done on the microbial community structure in the rhizosphere of plants grown in this area (Zhang et al., 2006; Ravit et al., 2007; Cao et al., 2015; Jing et al., 2018). In this study, we investigated the physicochemical factors, as well as the diversity and composition of rhizospheric N-fixing microbes in the coastal wetland. We demonstrate that the key driving factors affecting diazotrophic community structure in the coastal saline soil are physicochemical factors but not host plant traits.
The research area was located at the coastal zone of Laizhou Bay - Yellow River Estuary in China, with an average annual rainfall of 415.9 -842 mm and a mean annual temperature of 12.3°C-12.9°C (Figure 1A). Four sampling sites, Changyi (CY) and Shouguang (SG), which possess silt and low or non-saline soils, and Dongying (DY) and Binzhou (BZ), which possess rigid and high salinity soils, were chosen in this study (Figure 1B). For sample collection, surface soil was removed from each individual plant in a given plot, and plant roots together with rhizospheric soil were carefully excavated. Rhizospheric soil was obtained by shaking plants vigorously to separate the soil that not tightly adhered to the roots. Soils remaining attached to the roots after this were considered as rhizospheric soils (Jing et al., 2018). Finally, a total number of 18 samples corresponding to 8 different plant species, including Chenopodium L., Phragmites australis, Tamarix austromongolica Nakai, Tamarix austromongolica Nakai, Suaeda salsa, Limonium sinens, Imperata sp., and Typha sp., were collected (Figure 1C; Table 1). Host plants were morphologically identified and classified based their salt tolerance (6 halophytic and 2 non-halophytic), life style (3 annual and 5 perennial) and cotyledon number (5 dicotyledon and 3 monocotyledon), according to the Subject Database of Chinese Plant (http://www.plant.csdb.cn/) and Plant Collection Database (http://www.plantpic.csdb.cn/), as described previously (Zhang et al., 2006).
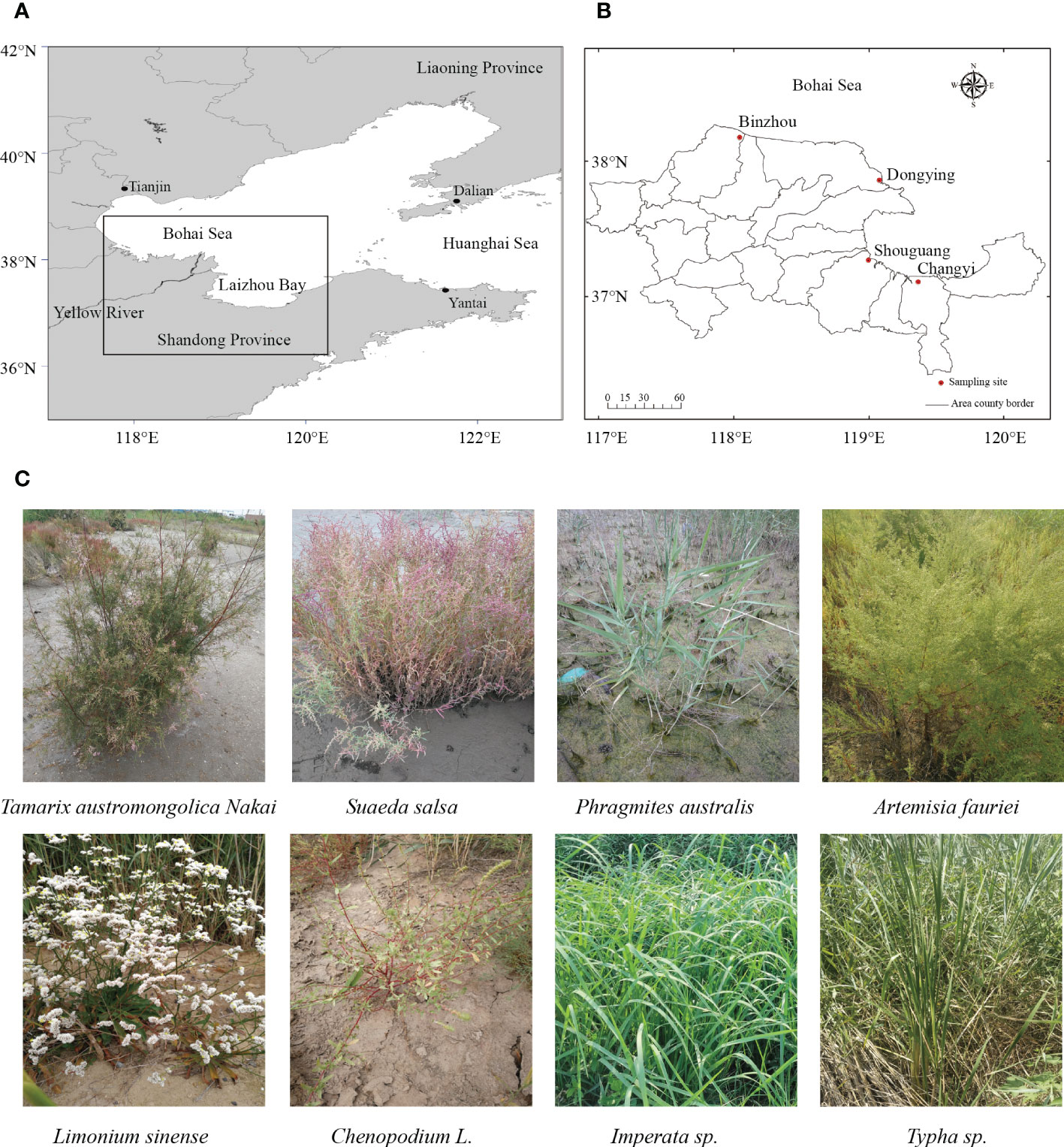
Figure 1 Images showing the locations and plants of sampling sites. (A, B) Geographic maps showing the sampling positions at the south coastal plain of the Laizhou Bay and the mouth of the Yellow River in China. (C) Phenotypes of representative plants for ryhizospheric soil samplings.
Soil samples with different soil salinity and moisture were collected in April, 2021. In each sampling area, five replication blocks (10×10 m) were established, and the rhizospheric soils of the most abundant plant species (4–5) available in the blocks were sampled. Fresh soil pH and salinity were determined with a soil to CaCl2 (0.01mol L-1) ratio of 1:5 (w:v). Soil moisture was measured gravimetrically as described previously (Gardner, 1986). Total carbon (TC), total sulphur (TS) and total N (TN) were determined using the Thermo Scientific™ FlashSmart™ Elemental Analyzers. Concentrations of nitrate (NO3--N), nitrite (NO2--N) and ammonium (NH4+-N) in the soils were extracted with KCl solution (2 mol L-1) and determined with the auto-analyzer (Seal, Germany) (Mulvaney, 1996). Soil types was classified as non-saline (Sal<1.2‰), slight saline (1.2‰–2.4‰), moderate saline (2.4‰–4.8‰), strong saline (4.8‰–9.6‰) and severe saline (Sal>9.6‰) as described previously (Richards, 1954). Soil moisture was divided into low moisture (<8%), medium moisture (8%–12%) and high moisture (>12%).
Total soil DNA was isolated using the FastDNA® SPIN Kit for Soil (MP Biomedicals, Solon, OH, USA) following the manufacturer’s protocol. DNA concentrations were determined with spectrophotometer Nanodrop 2000c (Thermo-Fisher, USA). To quantify the abundance of nifH gene in the rhizospheric soils, quantitative real-time PCR (qPCR) was performed with the forward (5’-TGCGAYCCSAARGCBGACTC-3’) and reverse (5’-ATSGCCATCATYTCRCCGG A-3’) primers (Poly et al., 2001). For each reaction, a total volume of 20 μL TB Green™ Premix Ex Taq™ II (TaKaRa, Japan) supplemented with 0.8 μL of each primer and 1μL of soil DNA template was generated. Thermal cycling conditions was as follows: pre-incubation at 50°C for 2 min, pre-denaturation at 95°C for 10min, then reaction for 40 cycles consisting of denaturation at 94°C for 30 s, annealing at 60°C for 30 s and extension at 72°C for 60 s, followed by melting curve analysis at 65-95°C (0.5°C per reading). The reactions were carried out in an BioRAD CFX96 fast real-time PCR system (Applied Biosystems, USA). Standard curves for the genes were obtained using serial dilutions of linearized plasmids (pTZ57R/T, Fermentas, USA) containing the target gene amplified from environmental clones (R2 = 0.99 for all standard curves).
For high-throughput sequencing analysis, the forward and reverse primers (5’-TGYGAYCCNAARGCNGA-3’ and 5’-ADNGCCATCATYTCNCC-3’) were used to amplify the nifH gene with the barcode. All PCR reactions were carried out in 30μL reactions with 15μL of Phusion®High-Fidelity PCR Master Mix (New England Biolabs), 0.2μM of forward and reverse primers, and about 10 ng template DNA. Thermal cycling consisting of initial denaturation at 98°C for 1 min, followed by 30 cycles of denaturation at 98°C for 10 s, annealing at 50°C for 30s, and elongation at 72°C for 60s, was performed as described previously (Gaby and Buckley, 2012). PCR products was purified with GeneJET Gel Extraction Kit (Thermo Scientific) and sequenced on the Illumina MiSeq PE250 platform at Biozeron Company (Shanghai, China).
Sequence analysis were performed with UPARSE software package using the UPARSE-OTU and UPARSE-OTUref algorithms. In-house Perl scripts were used to analyze alpha (within samples) and beta (among samples) diversity. Sequences with ≥97% similarity were assigned to the same operational taxonomic units (OTUs). A representative sequence for each OTU was picked and classified using the Ribosomal Database Project (RDP) classifier. Alpha diversity indexes (Chao 1, Shannon, Simpson) were calculated using the R software. Beta diversity was calculated based on Bray-Curtis dissimilarities and visualized using non-metric multidimensional scaling (NMDS) in PRIMER v.6 (Primer-E, UK). To confirm the difference in the abundance of individual taxonomy between the two groups, Metastats 2.0 was used. LEfSe was taken for the quantitative analysis of biomarkers in different groups. This method was designed to analyze data in which the number of species is much higher than the number of samples, and to provide biological class explanation to establish statistical significance, biological consistency and effect-size estimation of predicted biomarkers. To identify the differences of microbial communities between the two groups, ANOSIM were performed based on the Bray-Curtis dissimilarity distance matrices (Laurent et al., 2013).
The normality of all variables was tested using Shapiro-Wilk analysis. One-way analysis of variance (ANOVA) (for normally distributed variables) and non-parametric Kruskal-Wallis test (for the variables showing non-normal distribution) were performed to identify the differences between environmental factors, alpha diversity estimators, and soil salinity and moisture levels. Spearman’s correlations between diazotrophs abundance, alpha diversity and environmental factors were performed using SPSS v.11.5 (SPSS, Chicago, IL, United States). Following detrended correspondence analysis determining the length of the environmental gradient, canonical correspondence analysis (CCA) was performed to establish which environmental parameters played an important role in the variation of diazotrophic compositions using Canoco 5.0.
To assess the diversity and community composition of diazotrophic microbes in the rhizospheres of plants grown in the coastal saline area, rhizospheric soil samples from four sites, Changyi (CY), Shouguang (SG), Dongying (DY) and Binzhou (BZ), located at the coastal zone of Laizhou Bay-Yellow River Estuary in China, were collected (Figures 1A, B). In this sampling area, plant community is dominated by halophyte plants such as Suaeda salsa, Phragmites australis, Tamarix austromongolica Nakai, Chenopodium L., Artemisia fauriei, Limonium sinense, and some non-halophyte plants such as Imperata sp. and Typha sp. (Figure 1C; Table 1). Accompanied with the gradient of salinity and water moisture from land to sea, the host plant community showed obvious species succession. Since halophytes can survive in saline soils with low nutrient loads and water holding capacity, we first compared the property of the soil samples collected from different saline sites. Generally, all rhizospheric soil samples were slightly alkaline, with pH values ranged from 7.18 to 7.77, and their salinity and water content varied greatly, ranged from 0.5‰ to 15.14‰ and 1.10‰ to 13.38%, respectively (Table 1). Soils from DY and BZ sampling sites showed strong and severe salinity (5.38‰~15.14‰), and medium and high moisture (8.96%~13.38%), whereas soils from CY and SG sampling sites showed low saline (0.50‰~0.99‰) and moisture (1.1%~4.13%). The fertility of each sampling site was poor and the content of inorganic N was low, ranged from 2.78 to 14.05 mg·kg-1, with NO3–N as the dominant inorganic N ranged from 2.25 to 9.18 mg·kg-1 (Table 1). The highest salinity and moisture, with a percentage of 15.14% and 13.38%, was detected in the soil samples collected from DY-01 and DY-02, respectively. Whereas the lowest salinity and moisture, with a percentage of 0.50% and 1.10%, was detected in the soil samples collected from SG-03 and SG-01, respectively. The highest TN and TC, with a percentage of 0.069% and 2.007%, respectively, was detected in the soil samples collected from BZ-03. Whereas the lowest TN (0.016%) and TC (0.635%) was detected in the soil samples collected from SG03 and CY02, respectively (Table 1).
To assess the abundance of diazotrophic microbes in the rhizosphere of plants grown in the coastal saline area, we examined the copy numbers of nifH gene in the collected soil samples. The copy numbers of nifH gene varied widely, ranged from 1.99×106 to 3.80×108 copies g-1 dry soil across all the collected samples, with the highest nifH gene copy number detected in the rhizospheric soil samples of Tamarix austromongolica Nakai collected in Binzhou area (Figure 2). Further ANOVA and Kruskal-Wallis analyses revealed that the copy numbers of nifH gene were significantly affected by soil salinity and moisture, not plant traits (Table 2). A higher nifH gene copy number was observed in soil samples with high salinity and moisture than in soil samples with non-saline and low moisture. Spearman’s correlation analysis exhibited that the copy numbers of nifH gene were significantly related to pH value, soil salinity, moisture, and the levels of TN, TC, TS (total sulphur), NH4+-N and NO3–N (Table 3).
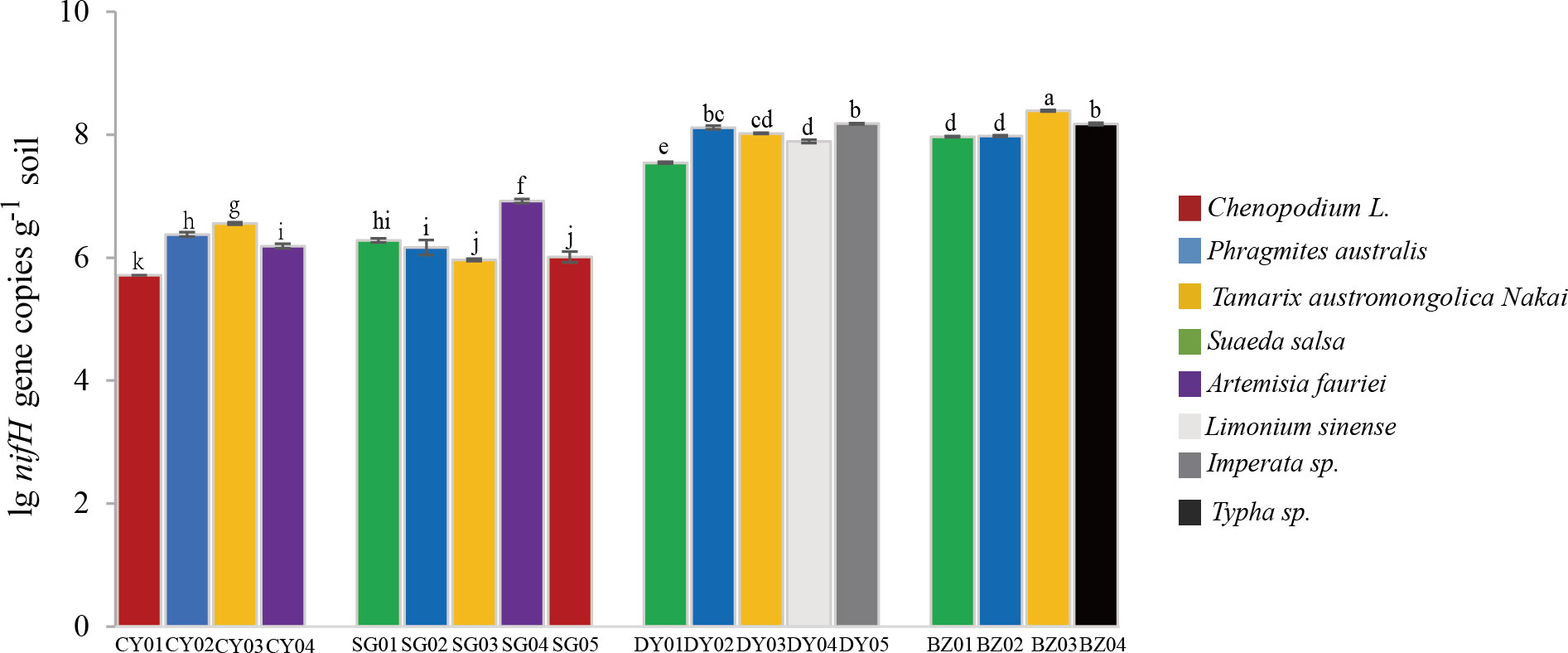
Figure 2 Quantification of nifH gene in the rhizospheric soil samples collected at different locations. Four sampling locations, Changyi (CY) and Shouguang (SG) with silt and low or non-saline soils, and Dongying (DY) and Binzhou (BZ) with rigid and high salinity soils, were selected. CY01, CY02, CY03, CHY04, SG01, SG02, SG03, SG04, SG05, DY01, DY02, DY03, DY04, DY05, BZ01, BZ02, BZ03 and BZ04 represent the soil samples respectively collected from the four locations. Values were means and standard deviations of three biological replicates (n=3). Lowercase letters denote significant differences of nifH gene copy numbers (Student’s t-test, P<0.05).
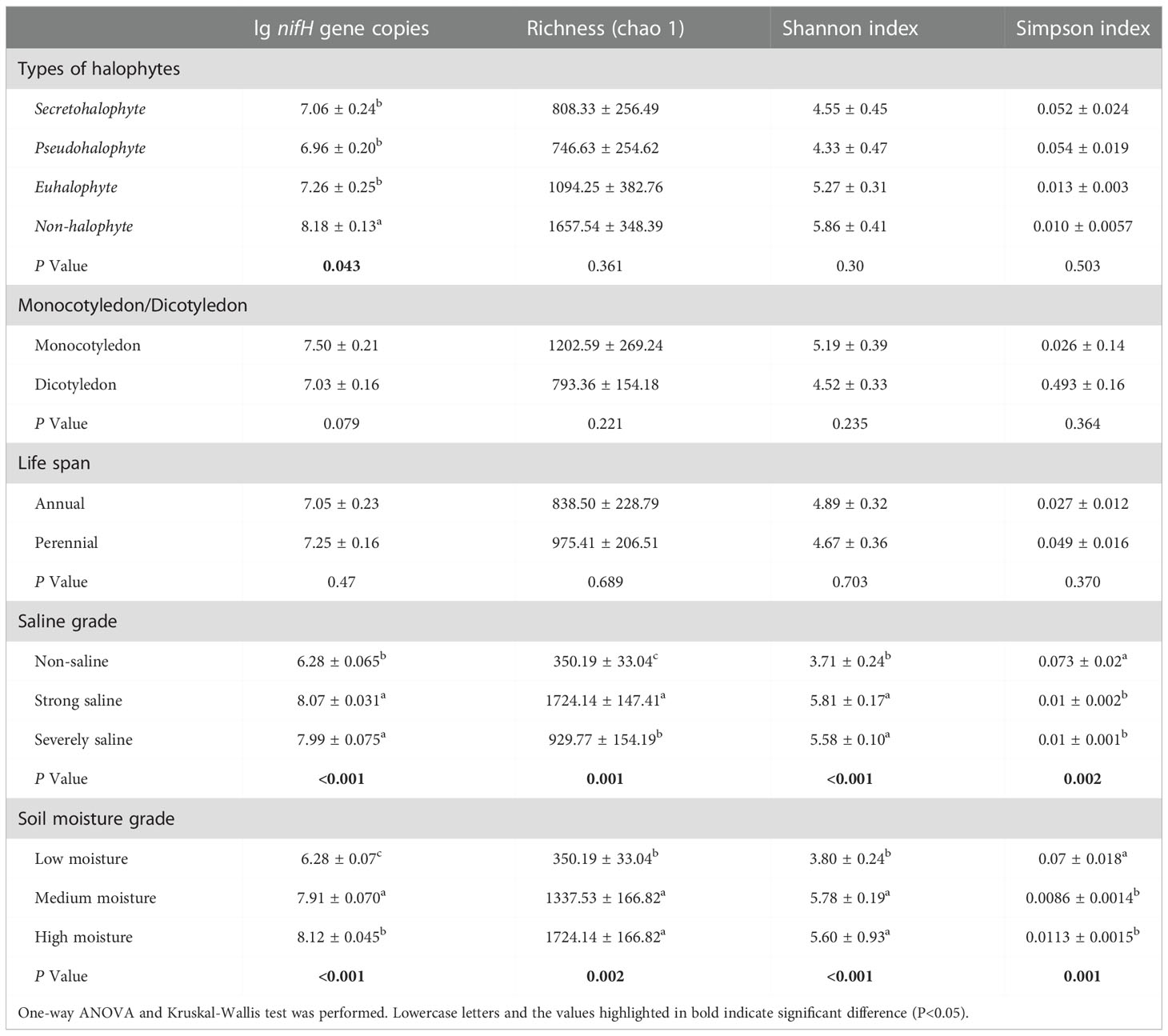
Table 2 Comparison of the abundance and diversity of nitrogen-fixing bacteria from the rhizosphere of different plant species.
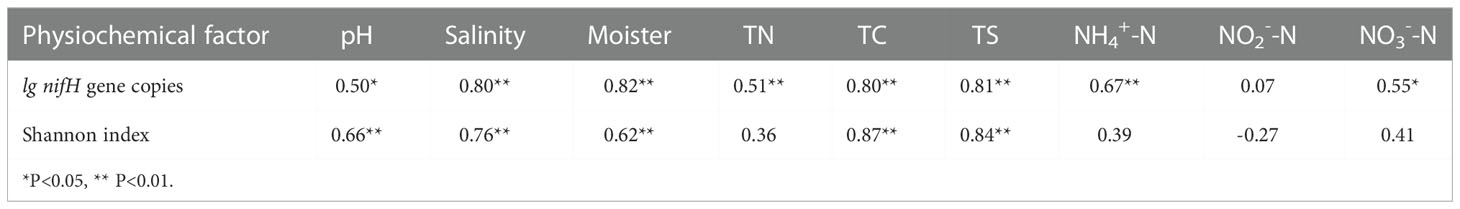
Table 3 Spearman’s correlation between the physiochemical factors and community structure of nitrogen-fixing bacteria.
To further understand the richness and diversity of diazotrophic communities in the rhizosphere of plants grown in the selected coastal saline area, we carried out high-throughput sequencing analysis. A total number of 812554 raw reads were obtained from 18 samples. At a cut-off of 97% sequence similarity, 5007 OTUs representing 10 phyla were obtained. We analyzed the diversity (Shannon and Simpson) and richness (Chao1) indices with two-way ANOVA and Kruskal-Wallis tests, and found that Shannon, Simpson and Chao1 indices were remarkably influenced by soil salinity and moisture, whereas alpha-diversities were not significantly affected by plant traits such as halophyte type, life span and cotyledon type (Table 2). Overall, the most abundant phylum was Proteobacteria (mean ± SE, 85.0% ± 13.1%; n = 18), followed by Cyanobacteria (6.71% ± 11.3%), unclassified phylum (5.86% ± 3.61%) and Firmicutes (1.21% ± 1.96%). Two phyla, Chloroflexi and Fibrobacteres, appeared to be the minor components (<1%) across all the soil samples (Figure S1). At genus level, Bradyrhizobium was found to be the most abundant genus (16.1%), followed by Halorhodospira (8.36%), Desulfovibrio (6.81%) and Azospirillum (5.98%), in the soil samples (Figure 3A).
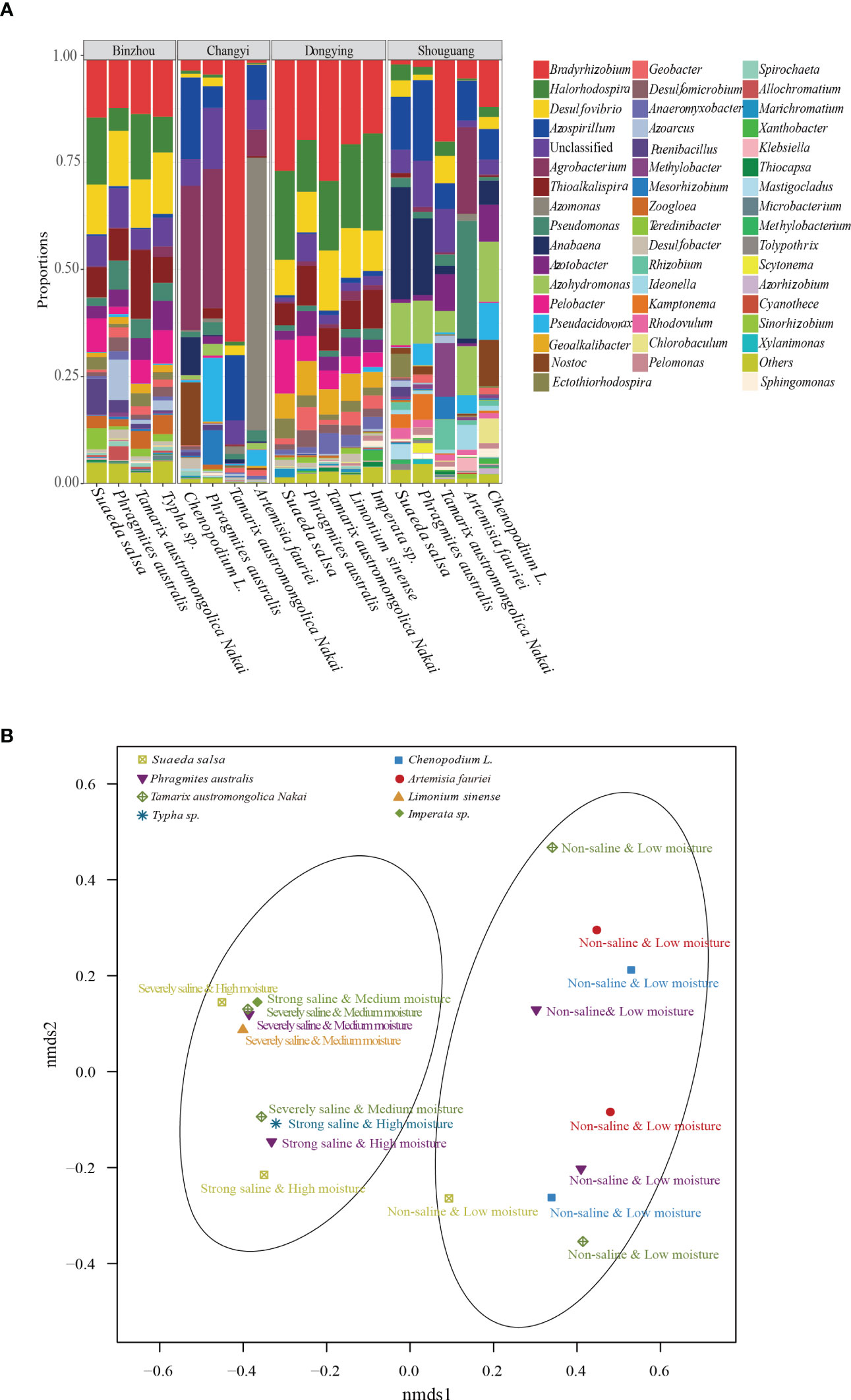
Figure 3 Diazotrophic community structure assays. (A) Diazotrophic genera in different soil samples collected from the four locations. Dominant diazotrophic genera with high relative abundance (>1%) were shown. (B) Non-metric multidimensional scaling analysis (nNMDS). The diazotrophic community dissimilarities based on OTUs from the nifH gene sequences were shown. All the samples were clearly divided into two groups based on soil salinity and moisture levels.
Based on the OTUs from the 16S rRNA gene sequences, we further generated non-metric multidimensional scaling (NMDS) ordination plot of diazotrophic community dissimilarities. We observed that diazotrophic communities in all the soil samples were clearly divided into two groups based on their soil salinity and moisture levels (Figure 3B). Further ANOSIM test also demonstrated that the diversity of diazotrophic community was significantly affected by soil salinity and soil moisture, but not by plant traits (Table 4). A higher diversity of diazotrophic community was observed in the soil samples with strong saline and high moisture, whereas no significant difference was observed in the overall diazotrophic composition among the soil samples collected from the rhizosphere of plants with different traits (Table 4).
We also performed Venn diagram analysis and a total number of 495 shared OTUs among the soil samples with different salinity and moisture levels were identified (Figures 4A, B). Samples collected at high salinity site showed the highest numbers of unique OTUs. Among the samples collected from the rhizospheric soils of different plant species, the highest number of unique OTUs (732) was observed in the rhizospheric soil samples of Secretohalophyte (Figure 4C). Higher numbers of unique OTUs were also observed in the rhizospheric soil samples of perennial and dicotyledonous plants than in those of annual and monocotyledonous plants, with a number of 4359 versus 2895, and 3970 versus 3555, respectively (Figures 4D, E). We further compared the unique OTUs in the soil samples collected from the rhizosphere of the same plant species grown at different sites. Higher numbers of unique OTUs were observed in the rhizospheric soil samples of Suaeda salsa, Phragmites australis and Tamarix austromongolica Nakai at high salinity and moisture sites in Binhzou and Dongying (Figures 4F-H).
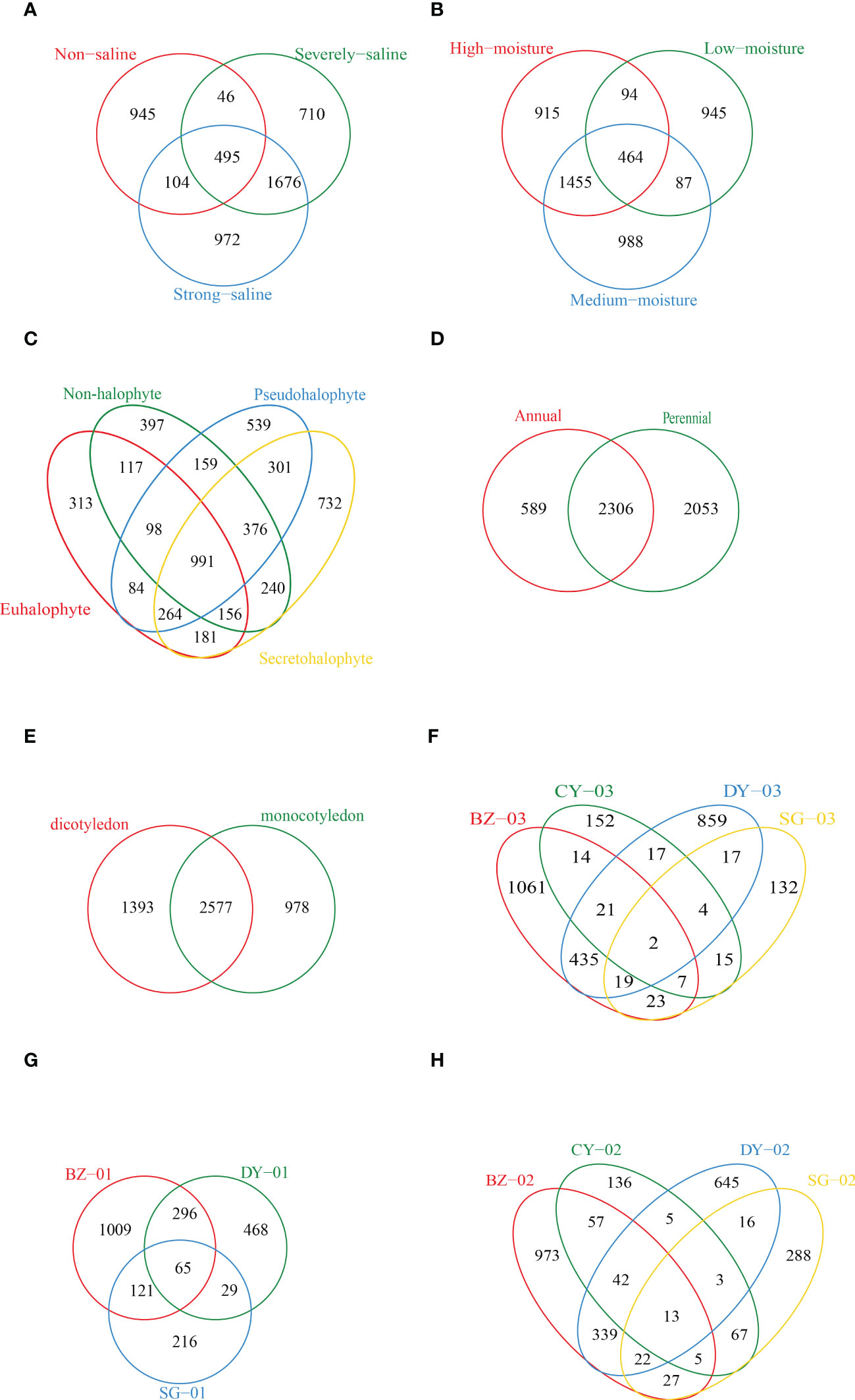
Figure 4 Venn diagrams showing the numbers of shared and unique diazotrophic OTUs among the soil samples. (A) Different salinity levels. (B) Different soil moistures. (C) Different plant types based on salt resistance. (D) Different plant types based on life span. (E) Different plant types based on cotyledon number. (F-H) Soil samples respectively collected from the rhizosphere of Tamarix austromongolica Nakai, Suaeda salsa and Phragmites australis grown at different sampling locations. CY02, CY03, SG01, SG02, SG03, DY01, DY02, DY03, BZ01, BZ02 and BZ03 represent the soil samples respectively collected from Changyi (CY), Shouguang (SG), Dongying (DY) and Binzhou (BZ).
To verify the potential discriminating species in the relative abundance between different groups, we carried out Linear discriminant analysis (LDA) effect size (LEfSe) analysis. A total number of 92 and 87 potential diazotrophic biomarkers distinguishing soil salinity and moisture levels were identified (Figures 5A, B). Among them, Cyanobacteria, Spirochaetes and Euryarchaeota was respectively found to be the main biomarker for the diazotrophic community in the samples collected at non-saline and low soil moisture, strong saline and medium soil moisture, and severe saline and high soil moisture sites (Figures 5A, B).
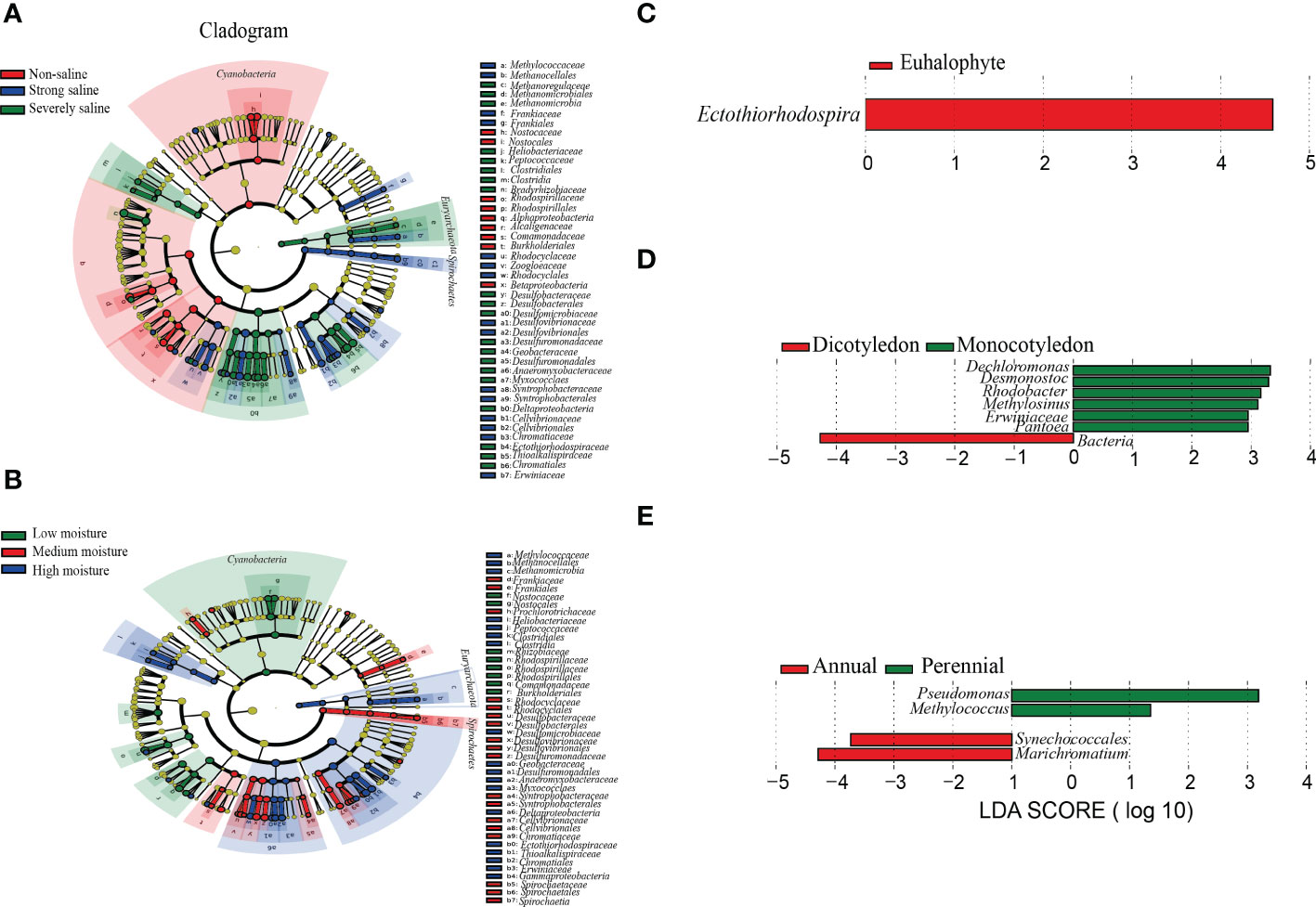
Figure 5 Phylogenetic dendrograms showing the biomarkers of diazotrophic microbes in the rhizospheric soil samples collected from Changyi (CY), Shouguang (SG), Dongying (DY) and Binzhou (BZ). (A) Different salinity levels. (B) Different soil moistures. (C) Different plant types based on salt resistance. (D) Different plant types based on cotyledon number. (E) Different plant types based on life span. The diazotrophic taxonomic levels from phylum to genus were exhibited with the circles from inside to outside. Diazotrophic abundance with no significant difference among individual soil samples was marked with yellow dots. The biomarkers were classified with different colors and shown on the right.
In the samples collected from the rhizospheric soils of euhalophyte plants, the abundance of Ectothiorhodospira was found to be significantly different (Figure 5C). Dechloromonas, Desmonostoc, Rhodobacter, Methylosinus, Erwiniaceae and Pantoea were specifically identified in the samples collected from the rhizospheric soils of monocotyledonous plants (Figure 5D). In addition, four potential diazotrophic biomarkers distinguishing the rhizospheric soil samples of annual and perennial plants were identified (Figure 5E). In the samples collected from the rhizospheric soils of annual plants, the biomarkers were mostly clustered in Cyanobacteria and Gammaproteobacteria, including Synechococcale and Marichromatium, whereas in those of perennial plants, the biomarkers were mostly clustered in Gammaproteobacteria, including Pseudomonas and Methylococcus (Figure 5E).
To dissect the relationships between diazotrophic communities and environmental parameters, we performed canonical correlation analysis (CCA). A remarkable correlation between diazotrophic communities and environmental parameters was observed. As shown in the CCA results, a percentage of 28.5% and 24.2% variation in the diazotrophic community was respectively described in the two CCA canonical axes, suggesting a remarkable correlation between diazotrophic communities and environmental parameters (Figure 6). The major environmental parameters closely correlated with diazotrophic community were soil salinity (P=0.002), soil moisture (P=0.002), TC (P=0.002), TN (P=0.008), TS (P=0.002) and pH value (P=0.002).
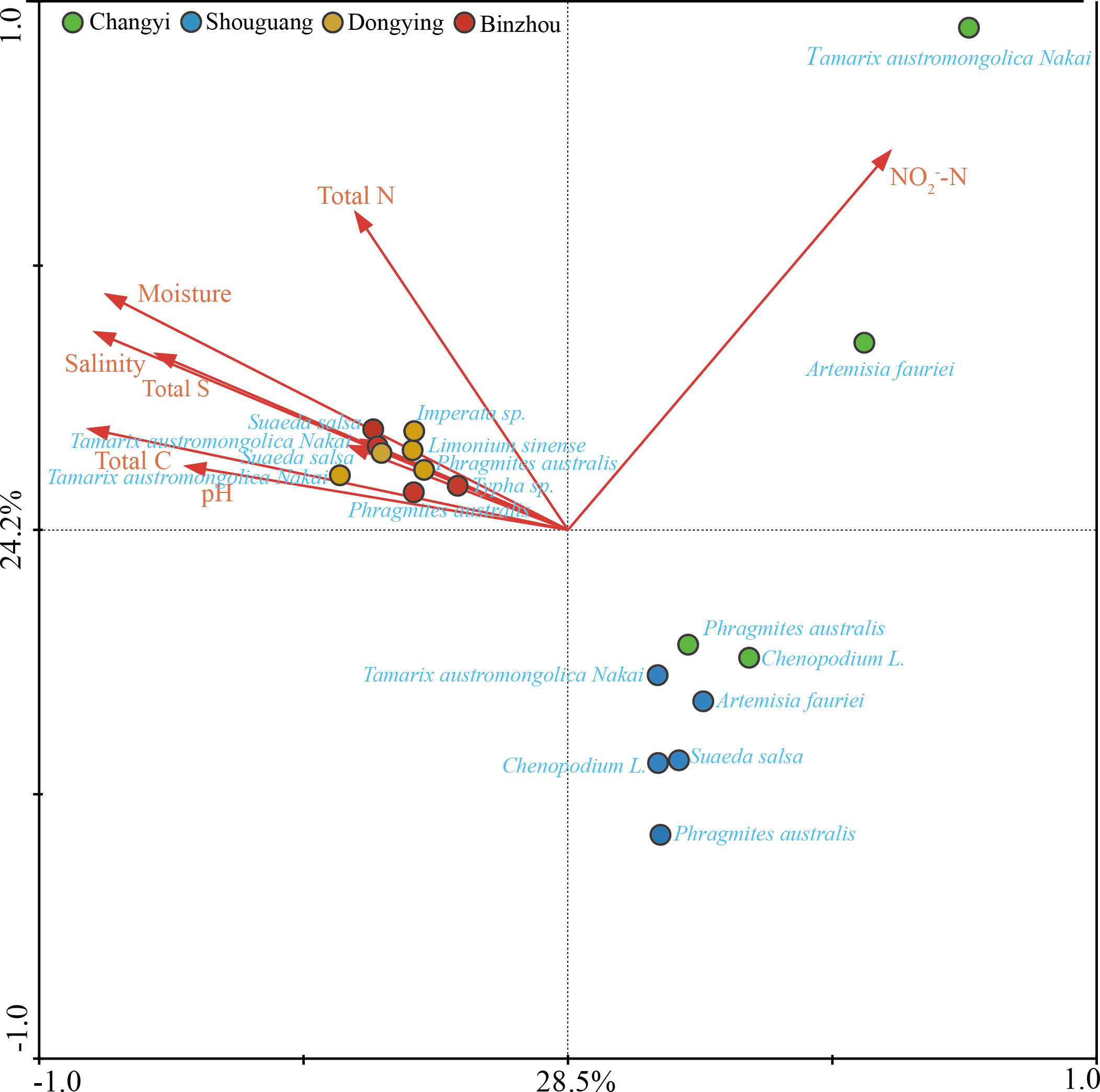
Figure 6 Canonical correspondence analysis (CCA) based on diazotrophic microbes and environmental parameters in different plant host habitats at Changyi (CY), Shouguang (SG), Dongying (DY) and Binzhou (BZ).
The growth and health of plants are largely affected by soil physiochemical properties and microbial community structure (Marasco et al., 2018; Jing et al., 2018; Wang et al., 2020). And plant invasion and N enrichment can considerably affect BNF (Xu et al., 2012; Huang et al., 2016). To date, a number of studies based on the diversity and abundance of diazotrophic community in the soils of different habitats, such as paddy field, forest and desert, have been carried out (Wang et al., 2016; Meng et al., 2019; Wang et al., 2021).However, the knowledge about the diazotrophic community structures in the rhizosphere of plants grown in coastal saline soil is limited. In this work, we assess the diversity and community composition of diazotrophic microbes in the rhizosphere of plants grown at the coastal zone of Laizhou Bay-Yellow River estuary in China (Figures 1A, B). Similar to that of other coastal lands in the north part of China, the host plant community is mainly composed of various halophytes in this area, and the soil physiochemical properties are obviously varied at different sampling locations (Figure 1C; Table 1).
Previously it was found that the abundance of nifH gene was also significantly related to the BNF rates of in the temperate coastal region of the northwestern North Pacific (Shiozaki et al., 2015). Consistent with the soil physiochemical properties, we found that nifH gene copy numbers in the collected soil samples varied widely, with the highest nifH gene copy number observed in the rhizospheric soils of plants grown in high salinity and medium moister area (Figure 2; Table 2). To further understand the correlation between the nifH gene copy numbers and soil physiochemical properties, we performed Spearman’s correlation analysis. The results demonstrated that, in addition to soil salinity and moister, the copy numbers of nifH gene were also closely related to the value of soil pH, and the levels of TN, TC, TS, NH4+-N and NO3--N (Table 3).
Due to its strong persistence across various soil environments, Bradyrhizobium has been shown to be the dominant diazotroph in soil system (Pereira et al., 2013; Piromyou et al., 2015). Bradyrhizobium was generally detected in the rhizosphere of plants forming root nodules with legumes for N fixation or plants participating in N fixation under non-symbiotic condition (Davis et al., 2011; Meng et al., 2019; Wang et al., 2020; Wang et al., 2021). Halorhodospira was also dominantly identified in the rhizospheric soils of coastal salt tolerant plants due to its strong salinity resistance (Imhoff and Truper, 1997). Azospirillum has been found to be associated with a variety of crops, and performed an important role in the non-symbiotic N fixation (Steenhoudt and Vanderleyden, 2000; Di Salvo et al., 2018). Our results expanded the distribution range of Bradyrhizobium and Azospirillum to the rhizosphere of coastal halophyte plants. In addition, Alpha- and Gammaproteobacteria have been identified as the dominant N fixers in coastal saline soil ecosystem (Yousuf et al., 2014). Similarly, a total number of 10 phyla, with Bradyrhizobium, Desulfovibrio, Halorhodospira and Azospirillum were the dominant diazotrophic groups at genus level, were identified in the collected rhizospheric soil samples (Figures 3A, B).
The growth and photosynthetic activity of plants driven by the energy from the sulfide oxidation in root tissues was stimulated by rhizodeposition (Mendelssohn and Morris, 2000; Rolando et al., 2022). In our study, some sulfate reducing bacteria, such as Desulfovibrio, Desulfomicrobium and Desulfobacter, were observed in the diazotrophic community (Figure 3A). This is consistent with previous reports that plant rhizodeposition stimulated N fixation by root-associated sulfate reducers in marsh and hypersaline soda lake environments, and Desulfovibrio, Desulfobulbus and Desulfatitalea coupled sulfate or sulfur respiration to N fixation (Gandy and Yoch, 1988; Tourova et al., 2014; Thajudeen et al., 2017; Rolando et al., 2022). A percentage of 17% energy produced in sulfate reduction was provided to the nitrogen fixation process (Nielsen et al., 2001). Thioalkalispira, the typical haloalkaliphilic sulfur-oxidizing bacteria (SOB), could remove the toxic sulphides from the root zone of plants, and couple S oxidation with C and N fixation (Barbieri et al., 2010; Petersen et al., 2016; Thajudeen et al., 2017). In all the collected rhizospheric soil samples, Thioalkalispira, were found to be the core diazotrophs (Figure 3A). Similar results were also observed in the coastal sediments of Bohai Bay, China (Wang et al., 2015). Furthermore, in the rhizosphere of each coastal host species grown under different salinity and moisture conditions, a high proportion of unclassified taxa (5.86%) was observed, suggesting that a substantially high and novel diversity of diazotrophic taxa existed in the coastal ecosystem (Figures 3A, B). This is possibly attributed to the special soil physicochemical and plant community property of the coastal ecosystem. Indeed, a total number of 495 shared OTUs among the soil samples with different salinity levels were identified, as indicated by the Venn diagram analysis (Figures 4A-H).
Under stress condition, specific root related microbial community could be formed in the rhizosphere of a certain plant. And in return, this microbiota may help to improve plant resistance to the corresponding stress (Bai et al., 2022). Ectothiorhodospira species were found in all kinds of aquatic environments, especially in the stagnant littoral, estuarine and highly saline area (Trüper & Imhoff, 1981). They were also found in the rhizospheric soils of plants grown on Ebinur Lake Wetlands, but not in the paddy fields (Davis et al., 2011; Wang et al., 2020; Zhang et al., 2021). In coastal alkaline soil, Ectothiorhodospira attached to anoxygenic purple sulfur bacteria and played a role in the process of N fixation (Tourova et al., 2014; Zhou et al., 2021). We observed that the community structure of diazotrophic microbes in the tested rhizospheric soil samples were significantly affected by the types of halophytes, and as indicated by the high throughput sequencing analysis, Ectothiorhodospira was significantly different in the diazotrophic microbial community of halophyte rhizosphere (Table 2; Figures 5A-C). No significant difference was found in the diazotrophic abundance and diversity between monocotyledon and dicotyledon, or perennial and annual plant rhizosphere (Table 2; Figures 5D, E).
When either NO3--N or NH4+-N as N source was insufficient, N fixation occurred (Zhivotchenko et al., 1995). In the rhizosphere of annual halophytes, Synechococcales and Marichromatium might play an important role in N fixation, especiall in coastal saline soils (Rigonato et al., 2013). We observed that, for the community structure of diazotrophic microbes, more diazotrophic genus, such as Dechloromonas, Desmonostoc, Rhodobacter, Methylosinus, Erwiniaceae and Pantoea, were found in the rhizospheric soils of monocotyledonous plants, and the biomarkers were mostly clustered in Synechococcale and Marichromatium in the rhizospheric soils of annual plants (Figures 5D, E). These results may be largely explained by the adventitious root systems of monocotyledonous plants, the main organs for the absorption of nutrient and water, and for the recruiting of specific N-fixing microbes in the rhizosphere.
Previous study has showed that low salinity promoted, whereas high salinity inhibited, the growth of soil N-fixing bacteria (Hou et al., 2018). In addition to the salinity, soil electrical conductivity (EC) also functioned as the main factor driving the variation of microbial community composition (Wang et al., 2020). In our study, an influence of soil salinity and moisture on the abundance and community composition of diazotrophic microbes was observed (Table 2; Figure 3B). In coastal saline soils, the influence of soil salinity and moisture on diazotrophic community could be explained by several reasons (Herbert et al., 2015; Rolando et al., 2022). First, salinization increased NH4+ release through ion substitution, which subsequently inhibited N fixation, leading to decreased nifH copies under severely saline environment. Second, seawater infiltration increased soil SO42- content, which affected the coupling of chemolithotrophic S oxidation with N fixation, a sulfate reduction process by diazotrophic microorganisms. Third, salinity changed the stability of microbial extracellular enzymes, and affected the mineralization and decomposition of macromolecular organic matter. And fourth, the increase of ionic strength reduced the adsorption of organic matters, and affected the availability of organic substrates for the growth of heterotrophs including diazotrophs.
In conclusion, using high throughput sequencing and quantitative real-time PCR targeting nifH genes, we investigated the diazotrophic abundance and community composition in the coastal saline soils of Laizhou Bay-Yellow River estuary. A strong correlation between diazotrophic abundance and soil physicochemical factors was observed. The physicochemical factors were significantly co-varied with the OTU based community composition of diazotrophs. Our findings demonstrate that in coastal saline soils, the community structure of diazotrophic microbes in the rhizosphere of plants grown in coastal saline soil is mainly affected by soil physicochemical factors but not by the host plant traits.
The datasets presented in this study can be found at NCBI Sequence Read Archive, with accession number SRP411732.
YS, LM, HYZ, RF, XL, YS, ML and JuL conducted the experiments. YS, RF and JL analyzed data. YS, JC and HXZ wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work has been jointly supported by the following grants: the National Key Research & Development Program of China (2021YFD1900903; 2019YFD1002703); the Natural Science Foundation of Shandong Province, China (ZR2020QC062; ZR2021QD157); the Science and Technology Demonstration Project of “Bohai Granary” of Shandong Province (2019BHLC002); the Science and Technology Innovation Project of Shandong Academy of Agricultural Sciences (CXGC2022F06; CXGC2022E19); the National Natural Science Foundation of China (41807083; 31870576, 32071733).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.1100289/full#supplementary-material
Affourtit J., Zehr J. P., Paerl H. W. (2001). Distribution of nitrogen-fixing microorganisms along the neuse river estuary, north Carolina. Microb. Ecol. 41, 114–123. doi: 10.1007/s002480000090
Anburaj R., Nabeel M. A., Sivakumar T., Kathiresan K. (2012). The role of rhizobacteria in salinity effects on biochemical constituents of the halophyte sesuvium portulacastrum. Russ. J. Plant Physl. 59 (1), 115–119. doi: 10.1134/s1021443712010025
Bai B., Liu W. D., Qiu X. Y., Zhang J., Zhang J. Y., Bai Y. (2022). The root microbiome: Community assembly and its contributions to plant fitness. J. Integr. Plant Biol. 64, 230–243. doi: 10.1111/jipb.13226
Barbieri E., Ceccaroli P., Saltarelli R., Guidi C., Potenza L., Basaglia M., et al. (2010). New evidence for nitrogen fixation within the Italian white truffle tuber magnatum. Fungal Biol. 114 (11-12), 936–942. doi: 10.1016/j.funbio.2010.09.001
Bhavya P. S., Kumar S., Gupta G. V. M., Sudheesh V., Sudharma K. V., Varrier D. S., et al. (2016). Nitrogen uptake dynamics in a tropical eutrophic estuary (Cochin, India) and adjacent coastal waters. Estuaries Coasts 39, 54–67. doi: 10.1007/s12237-015-9982-y
Cao J., Lv X. Y., Chen L., Xing J. J., Lan H. Y. (2015). Effects of salinity on the growth, physiology and relevant gene expression of an annual halophyte grown from heteromorphic seeds. AoB Plants. 7, plv112. doi: 10.1093/aobpla/plv112
Cao J. J., Wang H. R., Holden N. M., Adamowski J. F., Biswas A., Zhang X. F., et al. (2021). Soil properties and microbiome of annual and perennial cultivated grasslands on the qinghai–Tibetan plateau. Land Degrad. Dev. 32 (18), 5306–5321. doi: 10.1002/ldr.4110
Che R. X., Deng Y. C., Wang F., Wang W. J., Xu Z. H., Hao Y. B. (2018). Autotrophic and symbiotic diazotrophs dominate nitrogen-fixing communities in Tibetan grassland soils. Sci. Total Environ. 639, 997–1006. doi: 10.1016/j.scitotenv.2018.05.238
Collavino M. M., Tripp H. J., Frank I. E., Vidoz M. L., Calderoli P. A., Donato M., et al. (2014). nifH pyrosequencing reveals the potential for location-specific soil chemistry to influence N2-fixing community dynamics. Environ. Microbiol. 16 (10), 3211–3223. doi: 10.1111/1462-2920.12423
Dang H., Yang J., Li J., Luan X., Zhang Y., Gu G., et al. (2013). Environment-dependent distribution of the sediment nifH-harboring microbiota in the northern south China Sea. Appl. Environ. Microbio. 79 (1), 121–132. doi: 10.1128/AEM.01889-12
Davis D. A., Gamble M. D., Bagwell C. E., Bergholz P. W., Lovell C. R. (2011). Responses of salt marsh plant rhizosphere diazotroph assemblages to changes in marsh elevation, edaphic conditions and plant host species. Microb. Ecol. 61 (2), 386–398. doi: 10.2307/41489060
Debra A. D., Megan D. G., Christopher E. B., Peter W. B., Charles R. L. (2011). Responses of salt marsh plant rhizosphere diazotroph assemblages to changes in marsh elevation, edaphic conditions and plant host species. Microb. Ecol. 61 (2), 386–398. doi: 10.1007/s00248-010-9757-8
Di Salvo L. P., Ferrando L., Fernández-Scavino A., García de S., Inés E. (2018). Microorganisms reveal what plants do not: Wheat growth and rhizosphere microbial communities after Azospirillum brasilense inoculation and nitrogen fertilization under field conditions. Plant Soil. 424, 405–417. doi: 10.1007/s11104-017-3548-7
Dong J. Y., Zhang J. Q., Liu Y. H., Jing H. C. (2022). How climate and soil properties affect the abundances of nitrogen-cycling genes in nitrogen-treated ecosystems: a meta-analysis. Plant Soil 477(1), 389–404. doi: 10.1007/s11104-022-05420-6
Gaby J. C., Buckley D. H. (2012). A comprehensive evaluation of PCR primers to amplify the nifH gene of nitrogenase. PLoS One 7, e42149. doi: 10.1371/journal.pone.0042149
Galloway J. N., Dentener F. J., Capone D. G., Boyer E. W., Howarth R. W., Seitzinger S. P., et al. (2004). Nitrogen cycles: Past, present, and future. Biogeochemistry 70 (2), 153–226. doi: 10.2307/4151466
Gandy E. L., Yoch D. C. (1988). Relationship between nitrogen-fixing sulfate reducers and fermenters in salt marsh sediments and roots of spartina alternifora. Appl. Environ. Microb. 54, 2031–2036. doi: 10.1002/bit.260320421
Garbeva P., van Elsas J. D., van Veen J. A. (2008). Rhizosphere microbial community and its response to plant species and soil history. Plant Soil 302, 19–32. doi: 10.2307/42951751
Gardner W. H. (1986). “Water content,” in Methods of soil analysis. part i. physical and mineralogical methods. Ed. Klute A. (Madison: American Society of Agronomy).
Guo X. H., Gong J. (2014). Differential effects of abiotic factors and host plant traits on diversity and community composition of root-colonizing arbuscular mycorrhizal fungi in a salt-stressed ecosystem. Mycorrhiza 24, 79–94. doi: 10.1007/s00572-013-0516-9
Gupta V. V. S. R., Roper M. M., Roget D. K. (2006). Potential for non-symbiotic N2-fixation in different agroecological zones of southern Australia. Aust. J. Soil Res. 44 (4), 343–354. doi: 10.1071/SR05122
Herbert E. R., Boon P., Burgin A. J., Neubauer S. C., Franklin R. B., Ardon. M., et al. (2015). A global perspective on wetland salinization: Ecological consequences of a growing threat to freshwater wetlands. Ecosphere 6 (10), art206. doi: 10.1890/ES14-00534.1
Hou L. J., Wang R., Yin G. Y., Liu M., Zheng Y. L. (2018). Nitrogen fixation in the intertidal sediments of the Yangtze estuary: Occurrence and environmental implications. J. Geophys. Res-Biogeo. 123 (3), 936–944. doi: 10.1002/2018JG004418
Huang F., Lin X., Hu W., Zeng F., He L., Yin K. (2021). Nitrogen cycling processes in sediments of the pearl river estuary: Spatial variations, controlling factors, and environmental implications. Catena 206, 105545. doi: 10.1016/j.catena.2021.105545
Huang J., Xu X., Wang M., Nie M., Qiu S. Y., Quan Z. X., et al. (2016). Responses of soil nitrogen fixation to Spartina alterniflora invasion and nitrogen addition in a Chinese salt marsh. Sci. Rep. 6, 20384. doi: 10.1038/srep20384
Imhoff J. F., Truper H. G. (1997). Ectothiorhodospira halochloris sp. nov: a new extremely halophilic phototrophic bacterium containing bacteriochlorophyll. Arch. Microbiol. 114, 115–121. doi: 10.1007/bf00410772
Ininbergs K., Bay G., Rasmussen U., Wardle D. A., Marie-Charlotte W. (2011). Composition and diversity of nifH genes of nitrogen-fixing Cyanobacteria associated with Boreal forest feather mosses. New Phytol. 192, 507–517. doi: 10.1111/j.1469-8137.2011.03809.x
Jiang Y. J., Li S. Z., Li R. P., Zhang J., Liu Y. H., Lv L. F., et al. (2017). Plant cultivars imprint the rhizosphere bacterial community composition and association networks. Soil Biol. Biochem. 109, 145–155. doi: 10.1016/j.soilbio.2017.02.010
Jing C. L., Xu Z. X., Zou P., Tang Q., Li Y. Q., You X. W., et al. (2018). Coastal halophytes alter properties and microbial community structure of the saline soils in the yellow river delta, China. Appl. Soil Ecol. 134, 1–7. doi: 10.1016/j.apsoil.2018.10.009
Ladygina N., Hedlund K. (2010). Plant species influence microbial diversity and carbon allocation in the rhizosphere. Soil Biol. Biochem. 42 (2), 162–168. doi: 10.1016/j.soilbio.2009.10.009
Laurent P., Aymé S., Catherine H., David B., Florian B., Jones C. M., et al. (2013). Loss in microbial diversity affects nitrogen cycling in soil. ISME J. 7 (8), 1609–1619. doi: 10.1038/ismej.2013.34
Levy-Booth D. J., Prescott C. E., Grayston S. J. (2014). Microbial functional genes involved in nitrogen fixation, nitrification and denitrification in forest ecosystems. Soil Biol. Biochem. 75, 11–25. doi: 10.1016/j.soilbio.2014.03.021
Li J. Y., Chen Q. F., Li Q., Zhao C. S., Feng Y. (2021). Influence of plants and environmental variables on the diversity of soil microbial communities in the yellow river delta wetland, China. Chemosphere 274, 129967. doi: 10.1016/j.chemosphere.2021.129967
Li Y. Y., Chen X. H., Xie Z. X., Li D. X., Wu P. F., Kong L. F., et al. (2018). Bacterial diversity and nitrogen utilization strategies in the upper layer of the northwestern pacific ocean. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.00797
Lin X. B., Hou L. J., Liu M., Li X. F., Zheng Y. L., Yin G. Y., et al. (2016). Nitrogen mineralization and immobilization in sediments of the East China Sea: Spatiotemporal variations and environmental implications. J. Geophys. Res-Biogeo. 121, 2842–2855. doi: 10.1002/2016JG00349
Lin X. B., Li X. F., Gao D. Z., Liu M., Lv C. (2017). Ammonium production and removal in the sediments of shanghai river networks. J. Geophys. Res-Biogeo 122, 2461–2478. doi: 10.1002/2017JG003769
Lin G. M., Lin X. B. (2022). Bait input altered microbial community structure and increased greenhouse gases production in coastal wetland sediment. Water Res. 30, 218. doi: 10.1016/j.watres.2022.118520
Lin Y. X., Ye G. X., Liu D. Y., Ledgard S., Luo J. F., Fan J. B., et al. (2018). Long-term application of lime or pig manure rather than plant residues suppressed diazotroph abundance and diversity and altered community structure in an acidic ultisol. Soil Biol. Biochem. 123, 218–228. doi: 10.1016/j.soilbio.2018.05.018
Liu X. Y., Liu C., Gao W. H., Xue C., Guo Z. H., Jiang L., et al. (2019). Impact of biochar amendment on the abundance and structure of diazotrophic community in an alkaline soil. Sci. Total Environ. 688, 944–951. doi: 10.1016/j.scitotenv.2019.06.293
Marasco R., Mosqueira M. J., Fusi M., Ramond J. B., Merlino G., Booth J. M., et al. (2018). Rhizosheath microbial community assembly of sympatric desert speargrasses is independent of the plant host. Microbiome 6 (1), 215. doi: 10.1186/s40168-018-0597-y
Mendelssohn I. A., Morris J. T. (2000). “Eco-physiological controls on the productivity of spartina alternifora loisel,” in Concepts and controversies in tidal marsh ecology. Eds. Weinstein M. P., Kreeger D. A. (Kluwer Academic Publishers: Boston), 59–80.
Meng H., Zhou Z. C., Wu R. N., Wang Y. F., Gu J. D. (2019). Diazotrophic microbial community and abundance in acidic subtropical natural and re-vegetated forest soils revealed by high-throughput sequencing of nifH gene. Appl. Microbiol. Biot. 103, 995–1005. doi: 10.1007/s00253-018-9466-7
Midgley G. F. (2012). Biodiversity and ecosystem function. Science 335 (6065), 174. doi: 10.1126/science.1217245
Moisander P. H., Morrison A. E., Ward B. B., Jenkins B. D., Zehr J. P. (2007). Spatial-temporal variability in diazotroph assemblages in Chesapeake bay using an oligonucleotide nifH microarray. Environ. Microbial. 9 (7), 1823–1835. doi: 10.1111/j.1462-2920.2007.01304.x
Morina J. C., Morrissey E. M., Franklin R. B. (2018). Vegetation effects on bacteria and denitrifier abundance in the soils of two tidal freshwater wetlands in Virginia. Appl. Environ. Soil Sci. 2018, 1–10. doi: 10.1155/2018/4727258
Mortazavi B., Riggs A. A., Caffrey J. M., Genet H., Phipps S. W. (2012). The contribution of benthic nutrient regeneration to primary production in a shallow eutrophic estuary, weeks bay, Alabama. Estuaries Coasts 35 (3), 862–877. doi: 10.1007/s12237-012-9478-y
Moseman-Valtierra S. M., Johnson R., Zhang R., Qian P. Y. (2009). Differences in cordgrass structure between a mature and developing marsh reflect distinct N2-fixing communities. Wetlands 29, 919–930. doi: 10.1672/08-222.1
Moseman S. M., Zhang R., Qian P. Y., Levin L. A. (2009). Diversity and functional responses of nitrogen-fixing microbes to three wetland invasions. Biol. Invasions. 11, 225–239. doi: 10.1007/s10530-008-9227-0
Mulvaney R. L. (1996). “Nitrogen: Inorganic forms,” in Methods of soil analysis. Ed. Sparks D. L. (Madison, WI: SSSA Book Ser. 5. SSSA), 1123–1184.
Newell S. E., McCarthy M. J., Gardner W. S., Fulweiler R. W. (2016). Sediment nitrogen fixation: a call for re-evaluating coastal N budgets. Estuaries Coasts 39, 1626–1638. doi: 10.1007/s12237-016-0116-y
Nielsen L. B., Finster K., Welsh D. T., Donelly A., Herbert R. A., Wit R., et al. (2001). Sulphate reduction and nitrogen fixation rates associated with roots, rhizomes and sediments from Zostera noltii and Spartina maritima meadows. Environ. Microbiol. 3 (1), 63–71. doi: 10.1046/j.1462-2920.2001.00160.x
Oliveira J. T. C., Figueredo E. F., Diniz W. P., da S., Oliveira L. F. P., de, Andrade P. A. M., et al. (2017). Diazotrophic bacterial community of degraded pastures. Appl. Environ. Soil Sci. 1, 1–10. doi: 10.1155/2017/2561428
Omuto C. T., Vargas R. R., El Mobarak A. M., Mohamed N., Viatkin K., Yigini Y. (2020). Mapping of salt-affected soils: Lesson 3-spatial modelling of soil indicators (Properties) related salt problems (Rome: FAO).
Ospina-Betancourth C., Acharya K., Allen B. D., Entwistle J., Head I. M., Sanabria J., et al. (2020). Enrichment of nitrogen fixing bacteria in a nitrogen deficient wastewater treatment system. Environ. Sci. Technol. 54, 3539–3548. doi: 10.1021/acs.est.9b05322
Pereira E., Silva M. C., Schloter-Hai B., Schloter M., van Elsas J. D., Salles J. F. (2013). Temporal dynamics of abundance and composition of nitrogen fixing communities across agricultural soils. PloS One 8, e74500. doi: 10.1371/journal.pone.0074500
Petersen J. M., Kemper A., Gruber-Vodicka H., Cardini U., van der Geest M., Kleiner M., et al. (2016). Chemosynthetic symbionts of marine invertebrate animals are capable of nitrogen fixation. Nat. Microbiol. 2, 16195. doi: 10.1038/nmicrobiol.2016.195
Piromyou P., Greetatorn T., Teamtisong K., Okubo T., Shinoda R., Nuntakij A., et al. (2015). Preferential association of endophytic Bradyrhizobia with different rice cultivars and its implications for rice endophyte evolution. Appl. Environ. Microbiol. 81 (9), 3049–3061. doi: 10.1128/AEM.04253-14
Poly F., Monrozier L. J., Bally R. (2001). Improvement in the RFLP procedure for studying the diversity of nifH genes in communities of nitrogen fixers in soil. Res. Microbiol. 152, 95–103. doi: 10.1016/S0923-2508(00)01172-4
Qiu L. P., Kong W. B., Zhu H. S., Zhang Q., Banerjee S., Ishii S., et al. (2022). Halophytes increase rhizosphere microbial diversity, network complexity and function in inland saline ecosystem. Sci. Total Environ. 831, 154944. doi: 10.1016/j.scitotenv.2022.154944
Ravit B., Ehrenfeld J. G., Häggblom M. M., Bartels M. (2007). The effects of drainage and nitrogen enrichment on Phragmites australis, Spartina alterniflora, and their root-associated microbial communities. Wetlands 27, 915–927. doi: 10.1672/0277-5212(2007)27[915:TEODAN]2.0.CO;2
Rejmánková E., Sirová D., Castle S. T., Bárta J., Carpenter H., Bradley R. (2018). Heterotrophic N2-fixation contributes to nitrogen economy of a common wetland sedge, schoenoplectus californicus. PloS One 13 (4), e0195570. doi: 10.1371/journal.pone.0195570
Richards L. A. (1954). Diagnosis and improvement of saline and alkali soil, U.S (Washington D. C: Salinity Lab. Staff, USDA HandBook 60).
Rigonato J., Kent A. D., Alvarenga D. O., Andreote F. D., Fiore M. F. (2013). Drivers of cyanobacterial diversity and community composition in mangrove soils in south-east Brazil. Environ. Microbiol. 15 (4), 1103–1114. doi: 10.1111/j.1462-2920.2012.02830.x
Rodrigo C., Monika G., Nicole M., Jana L., Gabriele B., Kornelia S. (2006). Effects of site and plant species on rhizosphere community structure as revealed by molecular analysis of MicrobialGuilds. FEMS Microbiol. Ecol. 56 (2), 236–249. doi: 10.1111/j.1574–6941.2005.00026.x
Rolando J. L., Kolton M., Song T., Kostka J. E. (2022). The core root microbiome of spartina alterniflora is predominated by sulfur-oxidizing and sulfate-reducing bacteria in Georgia salt marshes, USA. Microbiome 10, 37. doi: 10.1101/2021.07.06.451362
Shiozaki T., Nagata T., Ijichi M., Furuya K. (2015). Nitrogen fixation and the diazotroph community in the temperate coastal region of the northwestern north pacific. Biogeosciences 12 (15), 4751–4764. doi: 10.5194/bg-12-4751-2015
Steenhoudt O., Vanderleyden J. (2000). Azospirillum, a free-living nitrogen-fixing bacterium closely associated with grasses: Genetic, biochemical and ecological aspects. FEMS Microbiol. Rev. 24 (4), 487–506. doi: 10.1016/s0168-6445(00)00036-x
Thajudeen ,. J., Yousuf J., Veetil V. P., Varghese S., Singh A., Abdulla M. H. (2017). Nitrogen fixing bacterial diversity in a tropical estuarine sediments. World J. Microb. Biot. 33 (2), 41. doi: 10.1007/s11274-017-2205-x
Tkacz A., Cheema J., Go C., Grant A., Poole P. S. (2015). Stability and succession of the rhizosphere microbiota depends upon plant type and soil composition. ISME J. 9(11), 2349–2359. doi: 10.1038/ismej.2015.41
Tourova T. P., Slobodova N. V., Bumazhkin B. K., Sukhacheva M. V., Sorokin D. (2014). Diversity of diazotrophs in the sediments of saline and soda lakes analyzed with the use of the nifH gene as a molecular marker. Microbiology 83 (5), 634–647. doi: 10.1134/S002626171404016X
Trüper H. G., Imhoff J. F. (1981). “The genus ectothiorhodospira,” in The prokaryotes. Eds. Starr M. P., Stolp H., Trüper H. G., Balows A., Schlegel H. G. (Berlin, Heidelberg: Springer). doi: 10.1007/978-3-662-13187-915
Vejan P., Abdullah R., Khadiran T., Ismail S., Nasrulhaq Boyce A. (2016). Role of plant growth promoting rhizobacteria in agricultural sustainability–a review. Molecules 21 (5), 573–590. doi: 10.3390/molecules21050573
Wang J., Bao J. T., Li X. R., Liu Y. B. (2016). Molecular ecology of nifH genes and transcripts along a chronosequence in revegetated areas of the tengger desert. Microb. Ecol. 71 (1), 150–163. doi: 10.1007/s00248-015-0657-9
Wang H. H., Li X., Li X. Y., Li F. L., Su Z. C., Zhang H. W. (2021). Community composition and Co-occurrence patterns of diazotrophs along a soil profile in paddy fields of three soil types in China. Microb. Ecol. 82, 961–970. doi: 10.1007/s00248-021-01716-9
Wang X. G., Sun R. B., Tian Y. P., Guo K., Sun H. Y., Liu X. J., et al. (2020). Long-term phytoremediation of coastal saline soil reveals plant species-specific patterns of microbial community recruitment. mSystems 5 (2), e00741–19. doi: 10.1128/msystems.00741-19
Wang Z. Y., Xin Y. Z., Gao D. M., Li F. M., Morgan J., Xing B. S. (2010). Microbial community characteristics in a degraded wetland of the yellow river delta. Pedosphere 20 (4), 466–478. doi: 10.1016/s1002-0160(10)60036-7
Wang L. P., Zheng B. H., Lei K. (2015). Diversity and distribution of bacterial community in the coastal sediments of bohai bay, China. Acta Oceanol. Sin. 34 (10), 122–131. doi: 10.1007/s13131-015-0719-3
Wang C., Zheng M., Song W. F., Wen S. L., Wang B., Zhu C. Q., et al. (2017). Impact of 25 years of inorganic fertilization on diazotrophic abundance and community structure in an acidic soil in southern China. Soil Biol. Biochem. 113, 240–249. doi: 10.1016/j.soilbio.2017.06.019
Whiting G. J., Gandy E. L., Yoch D. C. (1986). Tight coupling of root-associated nitrogen fixation and plant photosynthesis in the salt marsh grass spartina alterniflora and carbon dioxide enhancement of nitrogenase activity. Appl. Environ. Microbiol. 52 (1), 108–113. doi: 10.1021/ie0002226
Xian X. X., Pang M. Y., Zhang J. L., Zhu M. K., Kong F. L., Xi M. (2019). Assessing the effect of potential water and salt intrusion on coastal wetland soil quality: Simulation study. J. Soils Sediments. 19, 2251–2264. doi: 10.1016/j.apsoil.2011.10.021
Xu C. W., Yang M. Z., Chen Y. J., Chen L. M., Zhang D. Z., Liang M., et al. (2012). Changes in non-symbiotic nitrogen-fixing bacteria inhabiting rhizosphere soils of an invasive plant ageratina adenophora. Appl. Soil Ecol. 54, 32–38. doi:10.1016/j.apsoil.2011.10.021
Yousuf B., Kumar R., Mishra A., Jha B. (2014). Differential distribution and abundance of diazotrophic bacterial communities across different soil niches using a gene-targeted clone library approach. FEMS Microbiol. Lett. 360 (2), 117–125. doi: 10.1111/1574-6968.12593
Zak D. R., Holmes W. E., White D. C., Peacock A. D., Tilman D. (2003). Plant diversity, soil microbial communities, and ecosystem function: Are there any links? Ecology 84 (8), 2042–2050. doi: 10.2307/3450029
Zhang M. J., Chai L. W., Huang M., Jia W. Q., Guo J. B., Huang Y. (2020). Deciphering the archaeal communities in tree rhizosphere of the qinghai-Tibetan plateau. BMC Microbiol. 20 (1), 235–248. doi: 10.1186/s12866-020-01913-5
Zhang X. L., Gu D. Q., Feng A. P., Xia D. X. (2006). Comparative research on characters and evolvement of vegetation of coastal wetlands of yellow river delta and southern laizhou bay. Bull. Soil Water Conserv. 26, 127–140. doi: 10.1016/S1872-2032(06)60050-4
Zhang X., Hu W., Jin X. T., Chen T., Niu Y. H. (2021). Diversity of soil nitrogen-fixing bacteria in the rhizosphere and non-rhizophere soils of ebinur lake wetland. Arch. Microbiol. 22–30), 1–14. doi: 10.1007/s00203-021-02363-x
Zhivotchenko A. G., Nikonova E. S., Jørgensen M. H. (1995). Effect of fermentation conditions on N2 fixation by methylococcus capsulatus. Microb. Ecol. 14 (1), 9–15. doi: 10.1007/bf00369847
Keywords: diazotroph, nifH gene, community structure, rhizosphere, halophyte
Citation: Song Y, Ma L, Zhang H, Fu R, Liang X, Li J, Li J, Li M, Shan Y, Cheng J, Wang X and Zhang H (2022) The diversity and structure of diazotrophic communities in the rhizosphere of coastal saline plants is mainly affected by soil physicochemical factors but not host plant species. Front. Mar. Sci. 9:1100289. doi: 10.3389/fmars.2022.1100289
Received: 16 November 2022; Accepted: 05 December 2022;
Published: 16 December 2022.
Edited by:
Xianbiao Lin, Ocean University of China, ChinaReviewed by:
Minjie Hu, Fujian Normal University, ChinaCopyright © 2022 Song, Ma, Zhang, Fu, Liang, Li, Li, Li, Shan, Cheng, Wang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiangyu Wang, Y3lzd2FuZ3hpYW5neXVAc2hhbmRvbmcuY24=; Hongxia Zhang, aHh6aGFuZ0BzaWJzLmFjLmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.