- 1National Oceanic and Atmospheric Administration, National Marine Fisheries Service (NOAA/NMFS) National Systematics Lab., National Museum of Natural History, Washington, DC, United States
- 2Retired, Durham, NC, United States
- 3Department of Invertebrate Zoology, Smithsonian Museum of Natural History, Washington, DC, United States
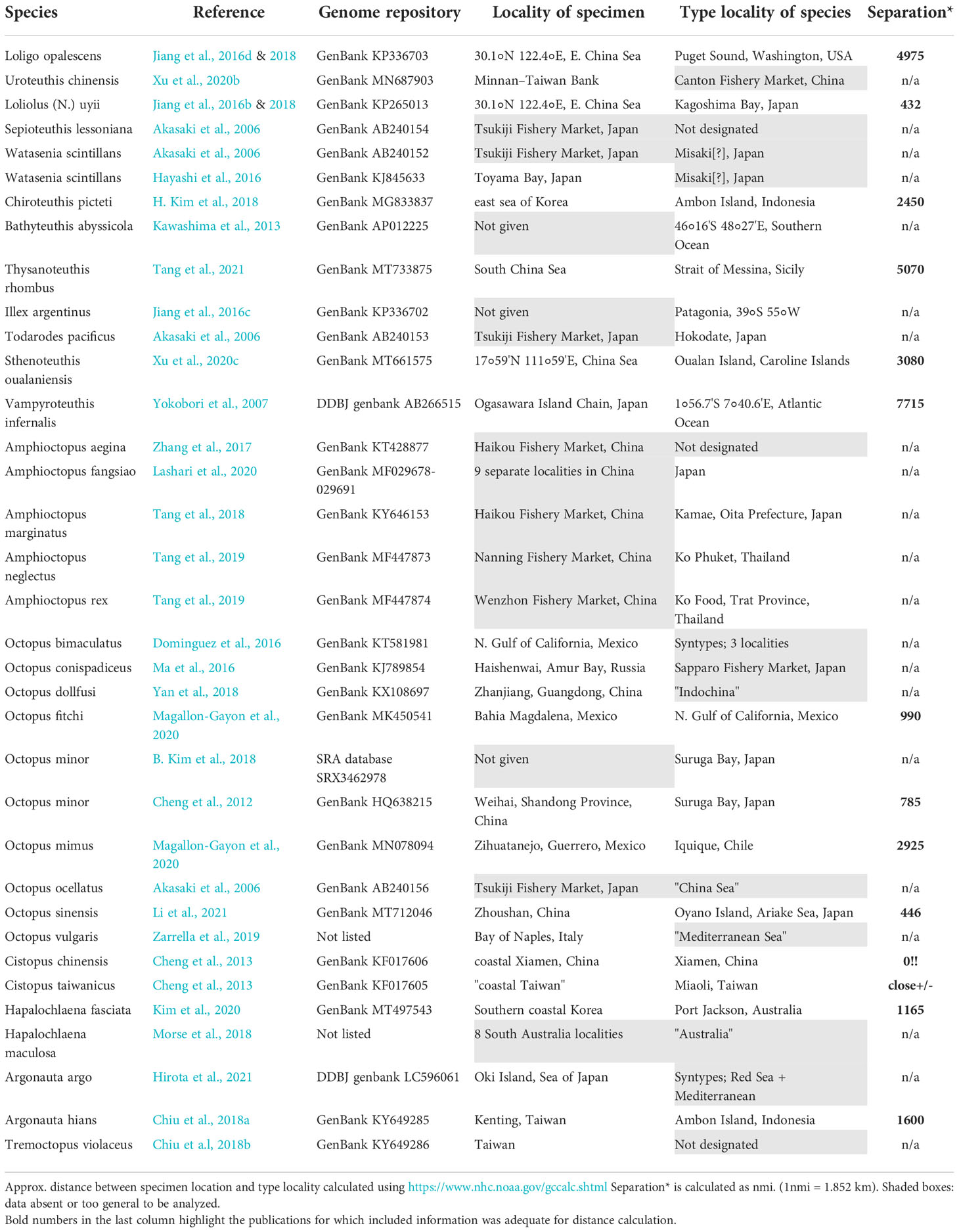
Publications describing genomes of various cephalopod species have recently proliferated. Some papers have involved large geographic distances between the collection locality of sequenced specimens and the type locality of the presumed species. However, cryptic species have been demonstrated in many cephalopods. Therefore, even if the sequenced specimen is very similar morphologically to the species in question, the likelihood that it is a member of the species in question decreases with increasing distance from the type locality. An associated problem is that many publications do not provide information adequate to determine the source locality for the genomic sequence. We reviewed a decade of literature on mitochondrial genomes of cephalopods and found a total of 43 publications containing 48 species within 23 genera. Of the 48 species, only 17 could be evaluated for our geographic question. Distances between sampling locality and type locality of the named species ranged from 0 nautical miles (sampled at type locality) to half-way around the world. Where data were present for distance calculation, the average for the 17 species was 3785 km (2044 nmi).
Introduction
Determination of genetic sequences has revolutionized understanding of evolutionary relationships. Increasingly sophisticated methods have allowed this revolution to progress greatly throughout the last few decades to include inferences about entire genomes. Accordingly, the literature describing cephalopod genomes, especially those of the mitochondria, has increased greatly over the past 10 years (O'Brien, 2018). The primary goal of most of these publications has been to resolve phylogenetic relationships within extant Cephalopoda.
Another result of widespread use of genetic sequencing, including “barcodes” and other sequences shorter than entire genomes, has been an increasing recognition that species with distributions once considered to be very broad or even global were actually complexes of morphologically similar species with geographic ranges resembling a patchwork within the broad range of the species complex. Some examples include taxa within the families Sepiolidae (Fernandez-Alvarez et al., 2021), Loliginidae (Sales et al., 2017), Chtenopterygidae (Escanez et al. (2018), Ommastrephidae (Fernandez-Alvarez et al., 2020; Xu et al., 2020a), Spirulidae (Hoffmann et al., 2021), and Octopodidae (Avendano et al., 2020; Amor and Hart, 2021). Because of these species complexes, both currently recognized and possibly to be discovered in the future, a substantial potential exists for misidentification of specimens collected for genomic sequencing (e.g., Lima et al., 2017; Salvi et al., 2021). This misidentification potential is especially true if the genomic specimen is not collected within the normal range of nominal sequenced species (i.e., named based on morphological identification). We are concerned that authors, using specimens from the nearest convenient area to sample a presumed species or from sources where the actual collection locality cannot be verified (e.g., fish markets, aquarium dealers), could be using a different species than what they report and, as a result, sequences in genomic databases may be misidentified.
Materials and methods
We surveyed the past decade of genomic literature on Cephalopoda for comparison of collection locality with designated type localities of the nominal species to determine the extent of this potential problem. Only publications describing complete mitochondrial genomes were analyzed. Each publication was examined to determine the collection locality of the specimen used for genomic analysis. Type localities for the nominal species are available online. We converted both sample locality and type locality to latitude/longitude and then calculated distance between them using NOAA Latitude/Longitude Distance Calculator (https://www.nhc.noaa.gov/gccalc.shtml). The repository and accession numbers for published genome sequences were also recorded (Table 1).
Results
An online search of the previous ten years of Cephalopoda genomic literature found a total of 58 genomic descriptions within 43 publications containing 48 different species in 23 genera (Table 1). For many species sequenced (70%), either collection locality or type locality (from the original description) was missing or was too general (e.g., Australia). In addition, if either locality was indeterminate (e.g., Tsukiji fishery market); or there were multiple type localities (ex. syntypes); or the genome was derived from combined specimens from multiple localities, the sequence was not included in our distance analysis.
Of the 48 species sequenced, only 17 could be evaluated for our geographic question (Table 1). Distances calculated ranged from 0 km (sampled at type locality) to half-way around the world in a different ocean basin. The average distance between sampling locality and type locality for the 17 species for which data were adequate for distance calculation, was 3785 km (2044 nm).
Incidentally, as we reviewed this literature for geographic information, we also noticed that very few of the publications included any indication that voucher specimens or unprocessed tissue were preserved in established archival collections for future research. For example, of the 17 species mentioned above, only 5 (29.4%) had vouchered specimens. Thus, 10.4% of species accounts included both adequate geographic information and archived specimens.
Discussion
Our point here is not that any of these publications is wrong. Rather, we want to highlight the potential for taxonomic errors in publications where the sampling area is very distant from the species’ type locality. As pointed out by one of the reviewers, for coastal cephalopod species in complex habitats, such errors are possible even at very small distances. Any taxonomic error introduced by this geographic mismatch may be compounded when the sequence is archived in a genomic database and the database is used for other investigations.
We therefore recommend selection of specimens for genomic sequencing collected from as close to the type locality of the species as possible. Although we recognize that it may not always be possible to sample the type locality, we recommend that the genomic sample be from the same biogeographic province (e.g., GOODS, 2009 or subsequent modifications by various authors) or “Large Marine Ecosystem” (LME – Sherman and Duda, 2011) as the type locality. The collecting locality should always be included in any publication resulting from DNA sequencing. Furthermore, specimens should not be selected for sequencing from a source where the actual collecting locality cannot be determined confidently (e.g., not from fishery landings, etc.). Also, although our primary purpose here is to highlight the need for sequenced specimens to come from as close to the type locality as possible, we also recommend that specimens sequenced and any unprocessed tissue be vouchered in an established archival collection. Relevant information about archived material (e.g., museum catalogue number) should be included in resulting publications.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Ethics statement
Ethical review and approval was not required for the animal study because this is a literature review. No animals (live or preserved) were used.
Author contributions
MS conceived the idea. MV and MS analyzed the data. PR accumulated the references. MV wrote the first draft. All authors contributed to the final manuscript. All authors contributed to the article and approved the submitted version.
Funding
NOAA, NMFS which employs MV. US Gov’t is committed to open-access publication.
Acknowledgments
The manuscript was improved by the comments from three reviewers and the Associate Editor.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Akasaki T., Nikaido M., Tsuchiya K., Segawa S., Hasegawa M., Okada N. (2006). Extensive mitochondrial gene arrangements in coleoid Cephalopoda and their phylogenetic implications. Molec. Phylogen. Evol. 38, 648–658. doi: 10.1016/j.ympev.2005.10.018
Amor M. D., Hart A. M. (2021). Octopus djinda (Cephalopoda: Octopodidae): a new member of the Octopus vulgaris group from southwest Australia. Zootaxa 5061, 145–156. doi: 10.11646/zootaxa.5061.1.7
Avendano O., Roura A., Cedillo-Robles C. E., Gonzalez A. F., Rodriguez-Canul R., Velazquez-Abunader I., et al. (2020). Octopus americanus: a cryptic species of the O. vulgaris species complex redescribed from the Caribbean. Aq. Ecol. 54, 909–925. doi: 10.1007/s10452-020-09778-6
Cheng R., Zheng X., Lin X., Yang J., Li Q. (2012). Determination of the complete mitochondrial DNA sequence of Octopus minor. Molec. Biol. Rep. 39, 3461–3470. doi: 10.1007/s11033-011-1118-2
Cheng R., Zheng X., Ma Y., Li Q. (2013). The complete mitochondrial genomes of two octopods Cistopus chinensis and Cistopus taiwanicus: Revealing the phylogenetic position of the genus Cistopus within the order octopoda. PLoS One 8 (12), e84216. doi: 10.1371/journal.pone.0084216
Chiu Y. W., Chang C. W., Lin H. D., Shen K. N. (2018a). The complete mitogenome of the winged argonaut Argonauta hians and its phylogenetic relationships in octopoda. Conserv. Gen. Res. 10, 359–362. doi: 10.1007/s12686-017-0824-z
Chiu Y. W., Chang C. W., Shen K. N., Ju Y. M., Lin H. D. (2018b). Complete mitochondrial genome and the phylogenetic position of the pelagic octopus Tremoctopus violaceus (Mollusca: Tremoctopodidae). Mitochon. DNA Part B 3, 1248–1249. doi: 10.1080/23802359.2018.1532347
Dominguez-Contreras J. F., Munguia-Vega A., Ceballos-Vazquez B. P., Garcia-Rodriguez F. J., Arellano-Martinez M. (2016). The complete mitochondrial genome of Octopus bimaculatus verrill 1883 from the gulf of California. Mitochon. DNA Part A 27, 4584–4585. doi: 10.3109/19401736.2015.1101575
Escanez A., Roura A., Riera R., Gonzalez A. F., Guerra A. (2018). New data on the systematics of comb-fin squids Chtenopteryx spp. (Cephalopoda: Chtenopterygidae) from the canary islands. Zool. Stud. 57. doi: 10.6620/ZS.2018.57-40
Fernandez-Alvarez F. A., Braid H. E., Nigmatullin C. M., Bolstad K. S., Haimovici M., Sanchez S., et al. (2020). Global biodiversity of the genus Ommastrephes (Ommastrephidae: Cephalopoda): an allopatric cryptic species complex. Zool. J. Linn. Soc 190, 460–482. doi: 10.1093/zoolinnean/zlaa014
Fernandez-Alvarez F. A., Sanchez P., Villanueva R. (2021). Morphological and molecular assessments of bobtail squids (Cephalopoda: Sepiolidae) reveal a hidden history of biodiversity. Front. Mar. Sci. 7. doi: 10.3389/fmars.2020.632261
GOODS (2009). “Global open oceans and deep seabed biogeographic classification,” in Intergovernmental oceanographic commission of Paris, UNESCO-IOC, vol. 64. .
Guo B., Chen Y., Zhang C., Lv Z., Xu K., Ping H., et al. (2018). Characterization of complete mitochondrial genome and phylogeny of Sepia lycidas (Sepioidea, sepiidae). Pak. J. Zool. 50, 1497–1508. doi: 10.17582/journal.pjz/2018.50.4.1497.1508
Guo B., Wang W., Qi P., Wu C., Chen Y., Lv Z. (2016). Complete mitochondrial genome of the needle cuttlefish Sepia aculeata (Sepioidea, sepiidae). Mitochon. DNA Part A 27, 67–68. doi: 10.3109/19401736.2013.873903
Hayashi K., Kawai Y. L., Yura K., Yoshida M. A., Ogura A., Hata K., et al. (2016). Complete genome sequence of the mitochondrial DNA of the sparkling enope squid, Watasenia scintillans. Mitoch. DNA Part A 27, 1842–1843. doi: 10.3109/19401736.2014.971251
Hirota K., Yoshida M., Itoh T., Toyoda A., Setiamarga D. H. E. (2021). The full mitochondrial genome sequence of the greater argonaut Argonauta argo (Cephalopoda, argonautoidea) and its phylogenetic position in octopodiformes. Mitochon. DNA Part B 6, 1451–1453. doi: 10.1080/23802359.2021.1911710
Hoffmann R., Weinkauf M. F. G., Fuchs D., Lukeneder A. (2021). Is there more than one species in the genus Spirula (Cephalopoda: Decabrachia): evidence for an Atlantic–pacific divide. J. Moll. Stud. 87, eyab001. doi: 10.1093/mollus/eyab001
Jiang L., Ge C., Liu W., Wu C., Zhu A. (2016a). Complete mitochondrial genome of the squid Loligo duvaucelii. Mitochon. DNA Part A 27, 2723–2724. doi: 10.3109/19401736.2015.1046164
Jiang L., Kang L., Wu C. (2017a). Complete mitochondrial genome of the squid Loligo japonica. Mitochond. DNA Part B 2, 493–494. doi: 10.1080/23802359.2017.1361349
Jiang L., Kang L., Wu C., Chen M., Lu Z. (2018). A comprehensive description and evolutionary analysis of 9 loliginidae mitochondrial genomes. Hydrobiol. 808, 115–124. doi: 10.1007/s10750-017-3377-y
Jiang L., Liu W., Zhang J., Wu C., Zhu A. (2016b). Complete mitochondrial genome of the squid Loliolus (Nipponololigo) uyii. Mitochond. DNA Part A 27, 3122–3123. doi: 10.3109/19401736.2015.1007294
Jiang L., Liu W., Zhang J., Zhu A., Wu C. (2016c). Complete mitochondrial genome of the Argentine shortfin squid (Illex argentinus). Mitochond. DNA Part A 27, 3335–3336. doi: 10.3109/19401736.2015.1018210
Jiang L., Liu W., Zhu A., Zhang J., Wu C. (2016d). Complete mitochondrial genome of the squid Loligo opalescence. Mitochond. DNA Part A 27, 3337–3338. doi: 10.3109/19401736.2015.1018211
Jiang L., Wu. C., Liu W., Chen M. (2016e). Complete mitochondrial genome of the squid Loligo beka. Mitochond. DNA Part A 27, 4278–4279. doi: 10.3109/19401736.2015.1082093
Kawashima Y., Nishihara H., Akasaki T., Nikaido M., Tsuchiya K., Segawa S., et al. (2013). The complete mitochondrial genomes of deep-sea squid (Bathyteuthis abyssicola), bob-tail squid (Semirossia patagonica) and four giant cuttlefish (Sepia apama, S. latimanus, S. lycidas and S. pharaonis), and their application to the phylogenetic analysis of decapodiformes. Molec. Phylogen. Evol. 69, 980–993. doi: 10.1016/j.ympev.2013.06.007
Kim B. M., Kang S., Ahn D. H., Jung. S. H., Rhee H., Yoo J. S., et al. (2018). The genome of common long-arm octopus Octopus minor. GigaScience 7, 1–7. doi: 10.1093/gigascience/giy119
Kim H. S., Kim K. Y., Kim I. H., Yang D., Kim H. (2020). The complete mitochondrial genome of blue-lined octopus Hapalochlaena fasciata (Hoyle 1886) (Octopodiformes; octopoda; octopodidae). Mitochond. DNA Part B 5, 3340–3341. doi: 10.1080/23802359.2020.1778557
Kim H., Yu C., Kim H. J., Kang D. W., Jung Y. H. (2018). The complete mitochondrial genome of the oceanic squid: Chiroteuthis picteti (Oegopsida, chiroteuthidae). mitochond. DNA Part B 3, 229–230. doi: 10.1080/23802359.2018.1437831
Lashari P., Wei C., Gong L., Liu L., Jiang L., Liu B., et al. (2020). A mitogenomic phylogeny and genetic history of Amphioctopus fangsiao (d’Orbigny 1839-1841) from China. Surv. Fish. Sci. 6, 1–16. doi: 10.18331/SFS2020.6.2.1
Lee H. T., Liao C. H., Huang C. W., Ma C. H., Hsu T. H. (2021). The complete mitochondrial genome of Metasepia tullbergi (Cephalopoda: Sepiidae). Mitochond. DNA Part B 6, 1192–1193. doi: 10.1080/23802359.2021.1902873
Li F., Liu Y., Qin B., Bian L., Ge J., Chang Q., et al. (2021). Sequence and phylogenetic analysis of the mitochondrial genome for the East Asian common octopus, Octopus sinensis (Octopodidae: Octopoda). Mitochond. DNA Part B 6, 2120–2122. doi: 10.1080/23802359.2021.1944360
Lima F. D., Berbel-Filho W. M., Leite T. S., Rosas C., Lima S. M. (2017). Occurrence of Octopus insularis leite and haimovici 2008 in the tropical northwestern Atlantic and implications of species misidentification to octopus fisheries management. Mar. Biodi. 47, 723–734. doi: 10.1007/s12526-017-0638-y
Lu Z., Cui W., Liu L., Pang Z., Zhang Y. (2019). The complete mitochondrial genome of Sepia latimanus (Sepiidae, sepioidea) and its phylogenetic implications. Mitochond. DNA Part B 4, 1002–1003. doi: 10.1080/23802359.2019.1583543
Magallon-Gayon E., del Río-Portilla M. A., de los Angeles Barriga-Sosa I. (2020). The complete mitochondrial genomes of two octopods of the eastern pacific ocean: Octopus mimus and ‘Octopus’ fitchi (Cephalopoda: Octopodidae) and their phylogenetic position within octopoda. Molec. Biol. Rep. 47, 943–952. doi: 10.1007/s11033-019-05186-8
Ma Y., Zheng X., Cheng R., Li Q. (2016). The complete mitochondrial genome of Octopus conispadiceus (Sasaki 1917) (Cephalopoda: Octopodidae). Mitochond. DNA Part A 27, 1058–1059. doi: 10.3109/19401736.2014.928866
Morse P., Kjeldsen S. R., Meekan M. G., McCormick M. I., Finn J. K., Huffard C. L., et al. (2018). Genome-wide comparisons reveal a clinal species pattern within a holobenthic octopod - the Australian southern blue-ringed octopus, Hapalochlaena maculosa (Cephalopoda: Octopodidae). Ecol. Evol. 8, 2253–2267. doi: 10.1002/ece3.3845
O’Brien C. E., Roumbedakis K., Winkelmann I. E. (2018). The current state of cephalopod science and perspectives on the most critical challenges ahead from three early-career researchers. Front. Physiol. 700. doi: 10.3389/fphys.2018.00700
Sales J. B. D. L., Rodrigues-Filho L. F. D. S., Ferreira Y. D. S., Carneiro J., Asp N. E., Shaw P. W., et al. (2017). Divergence of cryptic species of Doryteuthis plei blainville 1823 (Loliginidae, Cephalopoda) in the Western Atlantic ocean is associated with the formation of the Caribbean Sea. Molec. Phylog. Evol. 106, 44–54. doi: 10.1016/j.ympev.2016.09.014
Salvi D., Berrilli E., Garzia M., Mariottini P. (2021). Yet another mitochondrial genome of the pacific cupped oyster: the published mitogenome of Alectryonella plicatula (Ostreinae) is based on a misidentified Magallana gigas (Crassostreinae). Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.741455
Sherman K., Duda A. M. (2011). Large Marine ecosystems: An emerging paradigm for fishery sustainability. Fisheries 24, 15–26. doi: 10.1577/1548-8446(1999)024<0015:LME>2.0.CO;2
Song W., Li R., Zhao Y., Migaud H., Wang C., Bekaert M. (2021). Pharaoh cuttlefish, Sepia pharaonis, genome reveals unique reflection camouflage gene set. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.639670
Strugnell J. M., Hall N. E., Vecchione M., Fuchs D., Allcock A. L. (2017). Whole mitochondrial genome of the ram’s horn squid shines light on the phylogenetic position of the monotypic order spirulida (Haeckel 1896). Molec. Phylog. Evol. 109, 296–301. doi: 10.1016/j.ympev.2017.01.011
Takemoto K., Yamashita M. (2012). Complete nucleotide sequences of mitochondrial DNA of the long-finned squid Loligo edulis. Fish. Sci. 78, 1031–1039. doi: 10.1007/s12562-012-0541-1
Tang Y., Zhang X., Ma Y., Zheng X. (2021). Descriptive study of the mitogenome of the diamondback squid (Thysanoteuthis rhombus troschel 1857) and the evolution of mitogenome arrangement in oceanic squids. J. Zool. Syst. Evol. Res. 59, 981–991. doi: 10.1111/jzs.12478
Tang Y., Zheng X., Ma Y., Cheng R., Li Q. (2018). The complete mitochondrial genome of Amphioctopus marginatus (Cephalopoda: Octopodidae) and the exploration for the optimal DNA barcoding in octopodidae. Conserv. Gen. Res. 10, 115–118. doi: 10.1007/s12686-017-0777-2
Tang Y., Zheng X., Zhong H., Li Q. (2019). Phylogenetics and comparative analysis of the mitochondrial genomes of three violet-ringed octopuses. Zool. Scrip. 48, 482–493. doi: 10.1111/zsc.12359
Wang W., Guo B., Li J., Qi P., Wu C. (2014). Complete mitochondrial genome of the common cuttlefish Sepia pharaonis (Sepioidea, sepiidae). Mitochond. DNA Part A 25, 198–199. doi: 10.3109/19401736.2013.796462
Wang W., Guo B., Li J., Wang H., Qi P., Lv Z., et al. (2015). Complete mitochondrial genome of the spineless cuttlefish Sepiella inermis (Sepioidea, sepiidae). Mitochond. DNA Part A 26, 151–152. doi: 10.3109/19401736.2013.819498
Wang Y., Wang X., Li J., Tong X., Bi G., Han Y. (2018). The complete mitochondrial genome of Nautilus pompilius (Nautiloids: Nautilidae). Conserv. Genet. Res. 10, 437–440. doi: 10.1007/s12686-017-0843-9
Xu L., Liu P., Wang X., Van Damme K., Du F. (2020a). Phylogenetic relationships and cryptic species in the genus Sthenoteuthis (Cephalopoda: Ommastrephidae) in the south China Sea. Molec. Phylogen. Evol. 149, 106846. doi: 10.1016/j.ympev.2020.106846
Xu L., Wang X., Du F. (2020b). The complete mitochondrial genome of the loliginid squid (Uroteuthis chinensis) from minnan–Taiwan bank fishing ground. Mitochond. DNA Part B Resour. 5, 428–429. doi: 10.1080/23802359.2019.1703599
Xu L., Wang X., Huang D., Li Y., Wang L., Ning J., et al. (2020c). The complete mitochondrial genome of the middle-sized form of Sthenoteuthis oualaniensis (Cephalopoda: Ommastrephidae) from the south China Sea. Mitochond. DNA Part B 5, 3048–3050. doi: 10.1080/23802359.2020.1797562
Yan Y., Lu Z., Wang T., Chen Y., Yang J., Guo B., et al. (2018). Determination and analysis of the complete mitochondrial DNA sequence of Octopus dollfusi (Mollusca: Cephalopoda: Octopodidae) from China. Pak. J. Zool. 50, 463–472. doi: 10.17582/journal.pjz/2018.50.2.463.472
Yokobori S., Lindsay D. J., Yoshida M., Tsuchiya K., Yamagishi A., Maruyama T., et al. (2007). Mitochondrial genome structure and evolution in the living fossil vampire squid, Vampyroteuthis infernalis, and extant cephalopods. Molec. Phylogen. Evol. 44, 898–910. doi: 10.1016/j.ympev.2007.05.009
Zarrella I., Herten K., Maes G. E., Tai S., Yang M., Seuntjens E., et al. (2019). The survey and reference assisted assembly of the Octopus vulgaris genome. Sci. Data 6, 1–8. doi: 10.1038/s41597-019-0017-6
Zhang X., Zheng X., Ma Y., Li Q. (2017). Complete mitochondrial genome and phylogenetic relationship analyses of Amphioctopus aegina (Gray 1849) (Cephalopoda: Octopodidae). Mitochon. DNA Part A 28, 17–18. doi: 10.3109/19401736.2015.1106522
Keywords: biogeography, genomics, species complex, type locality, sampling
Citation: Vecchione M, Sweeney MJ and Rothman PL (2022) The geographic problem in cephalopod genomics. Front. Mar. Sci. 9:1090034. doi: 10.3389/fmars.2022.1090034
Received: 04 November 2022; Accepted: 21 November 2022;
Published: 08 December 2022.
Edited by:
Michael J. Kuba, Okinawa International University, JapanReviewed by:
Gianluca Polese, University of Naples Federico II, ItalyAnna Di Cosmo, University of Naples Federico II, Italy
Ryuta Nakajima, University of Minnesota Duluth, United States
Copyright © 2022 Vecchione, Sweeney and Rothman. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael Vecchione, dmVjY2hpb21Ac2kuZWR1
 Michael Vecchione
Michael Vecchione Michael J. Sweeney2
Michael J. Sweeney2