
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 12 January 2023
Sec. Marine Fisheries, Aquaculture and Living Resources
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.1088176
This article is part of the Research Topic Aquatic Microorganism and Their Response to Environment: Virulence and Antimicrobial Resistance View all 10 articles
The increasing abundance of antibiotic resistance genes (ARGs), which are regarded as new pollutants, has raised public health concern. The use of antibiotics in aquaculture has promoted the evolution and spread of ARGs. The occurrence and abundance of ARGs in aquaculture has attracted extensive attention. However, the distribution and transmission of ARGs in aquaculture require further study. This study analyzed water and sediment from intensive culture of hybrid snakehead fish farm in Zhongshan, South China. Twenty-two types of ARGs were detected in all environmental samples. The relative abundance of sulfonamide resistance genes (sul1 and sul2) was the highest, ranging from 3.37×10-2 to 8.53×100 copies/16S rRNA gene. High occurrence of quinolones, phenicols, tetracycline resistance genes, and class 1 integrase gene (intI1) was also observed in pond water samples. This implies that pond water is one of the main reservoirs and origins of ARGs in the aquatic environment. Proteobacteria was the most abundant phylum in all the environmental samples, and its relative abundance ranged from 24.05% to 41.84%. Network and canonical correspondence analyses showed that a high abundance of ARGs (int1, sul1, sul2, qacEΔ1, aac6, and oqxA) was positively correlated with Proteobacteria, Cyanobacteria, and Bacteroidetes, and the abundance of Proteobacteria, Actinobacteria, and Patescibacteria was positively correlated with environmental factors (sulfide, nitrite nitrogen, pH, free chlorine, and ammonia nitrogen). These findings demonstrate the prevalence and persistence of ARGs in intensive fish farming in southern China. This suggests that ARG levels and microbiological community composition in aquaculture should be conventionally determined to assess potential risks to public health.
The hybrid snakehead fish (Channa maculate ♀ × C. argus ♂) is an important commercial fish with high market value in Chinese aquaculture. The production of hybrid snakehead fish has increasingly developed towards intensive and high-density aquaculture in China with the continual improvement of aquaculture technology (Liu et al., 2017). In the past ten years, rough culture has changed to a modern refined aquaculture mode by increasing the stocking density and feeding compounds or providing complete feed to sustain the rapid growth of this industry. However, negative effects related to nutrient discharge may cause eutrophication, deterioration of water quality, and other potential environmental risks (Liu et al., 2014). High-stock density aquaculture involves the rearing of fish in congested and stressful environments. These fishes are more susceptible to bacterial infections, resulting in significant production losses. Antibiotics have been widely used to reduce disease burden in China since the 1980s because of their low cost, convenience of use, and remarkable curative effects. Antibiotics are extensively used as therapeutic drugs for humans, livestock, aquaculture, and as feed additives in farming (Su et al., 2018a). Several studies have reported that improper use of antibiotics in aquaculture exerts selection pressure on the natural bacterial population and enhances their ability to produce antibiotic-resistant bacteria or antibiotic resistance genes (ARGs) in aquatic environments (Hossain et al., 2022).
ARGs are defined as a new type of environmental pollutants because of their persistence and easy diffusivity. This has become the biggest challenge to human health in the 21st century (Berendonk et al., 2015). Antibiotic resistance genes are key factors in the development of bacterial drug resistance. They are enriched in medical wastewater, riverbed sludge, pig farm soil, and drinking water treatment systems. They can be horizontally transferred through mobile genetic elements (MGEs) (Gao et al., 2012; Li et al., 2015; Zhou et al., 2017; Hu et al., 2018; Su et al., 2018b). Moreover, foodborne pathogens that carry ARGs pose a potential threat to human health when they enter the food chain (Zhao et al., 2018). The aquatic ecosystem is an enrichment pool for many types of ARGs, and the pollution of the system by exogenous antibiotics is an important factor that results in the enrichment and diffusion of ARGs (Allen et al., 2010). Several studies have also reported the high presence of antibiotics in the environment, including sulfonamides, tetracyclines, quinolones, and the most frequently detected antibiotic resistance genes along with their respective antibiotic classes are sulfonamides (sul1 and sul2), tetracyclines (tetA, tetB, tetK, and tetM), and quinolones (qnrA, qnrB, and qnrS) (Zhou et al., 2017; Hu et al., 2018).
Aquaculture is considered to be a “genetic hotspot” for the transmission of resistance genes. Several ARGs have been detected in aquaculture environments and farming organisms in China, India, Finland, Nigeria, and Chile (Su et al., 2017). Antibiotic-resistant genes are widely distributed in aquaculture environments and in aquatic animals (aquatic products). It has been speculated that the use of antibiotics in aquaculture may promote the enrichment and migration of ARGs in the environment and animals. Nutrients have been suggested to directly or indirectly promote ARGs dissemination (Ning et al., 2022). Environmental factors such as total phosphorus and total nitrogen affect the abundance and diversity of ARGs in shrimp and fishponds (Su et al., 2017). Furthermore, bacterial communities in different aquaculture modes affect the distribution and abundance of ARGs (Huang et al., 2017; Wang et al., 2022). These findings indicated that antibiotic use and environmental factors affect the occurrence and distribution of ARGs.
Hybrid snakehead fish farming is a typical example of intensive aquaculture, which may result in a series of negative effects including the production of large amounts of antibiotic-resistant bacteria and ARGs pollution. This study evaluated the risk of ARGs in intensive aquaculture by quantitatively analyzing 22 typical ARGs in the breeding environment of hybrid snakehead fish using real-time quantitative PCR (RT-qPCR). In addition, the correlation between bacterial communities at different culture stages, ARG abundance, and environmental factors was elucidated, and co-occurrence patterns between ARGs and bacterial taxa were identified. This study provides information on antibiotic resistance in intensive aquaculture and advances the development of a healthy, green, and sustainable freshwater aquaculture industry.
The hybrid snakehead fish farm (22.602° N, 113.503° E) selected for this study is located in Zhongshan, Guangdong Province, China. This farm has over 70 mud ponds with a total feeding area of 50 ha. Each pond has an area of approximately 0.4–0.6 ha with a depth of approximately 1.8–2.5 m. Three ponds at the sampling sites (ponds A, B, and C) were selected for this study. The seasons in Guangdong Province allowed three different fish harvests. The first crop of hybrid fish involved stocking fingerlings before May and harvesting in November of the same year with a rearing period of no more than six months. The stocking density of the fingerlings was 10 thousand per ha (1,500 fish/kg), and the harvest size of the adult fish was usually 500–750 g. Different antibiotics, including florfenicol, doxycycline, sulfonamides, and enrofloxacin, have been used as therapeutic and prophylactic agents against bacterial infections because of their high stock density. Oral administration is usually mixed with feed; the usage limits for sulfonamides, florfenicol, doxycycline, and enrofloxacin are 100-200 mg/kg BW, 10-15 mg/kg BW, 20-50 mg/kg BW, and 2500 mg/kg feed, respectively (Liu et al., 2017).
Sediment and water samples were collected in June (fingerlings, X), September (rearing, Y), and November (harvesting, Z) 2020. The rearing and harvesting periods corresponded to rapid growth and upcoming sales, respectively. Five hundred milliliters of water samples (500 mL) were collected from the four corners of each pond (10–15 cm below the surface) using a stainless-steel sampler. Samples from each pond were then completely mixed and transferred to sterile polyethylene bottles to avoid differences in heterogeneity caused by single sampling. Sediment samples (500 g with four replicates of each sample) were collected from 0-10 cm depth using a grab sampler and then mixed thoroughly in sterile polyethylene bottles. All samples were stored in a portable refrigerator and transported to the laboratory for pre-treatment within 24 h.
Each sample was subsequently divided into two equal parts, which were used for the quantitative PCR analysis and high-throughput sequencing of the bacterial community. Water samples (0.5–1 L) were filtered through a 0.22-μm micro-pore membrane (Pall, Michigan, USA) using vacuum filtration for DNA extraction. The membranes were aseptically stored at -80°C for DNA extraction using the PowerSoil DNA isolation kit (Qiagen Biotech, Germany) according to the manufacturer’s instructions. The sediment samples were lyophilized, ground, and sieved through an 80-mesh screen. Approximately 0.25 g of each sediment sample was used for DNA extraction using the PowerSoil DNA isolation kit (Qiagen Biotech, Germany) according to the manufacturer’s protocol. The DNA concentration and purity were monitored using 1% agarose gels and an ultramicro ultraviolet spectrophotometer (Gene Company Limited, China).
The 21 most common target ARGs in the water and sediment samples were quantified using quantitative PCR. These included three sulfonamide resistance genes (sul1, sul2, and sul3), four tetracycline resistance genes (tetA, tetB, tetM, and tetW), six quinolone resistance genes (qnrA, qnrB, qnrC, qnrS, oqxA, and aac(6)-Ib-cr), three β-lactams (blaCTX-M, blaSHV, and blaTEM), three phenicol resistance genes (fexA, floR, and cmlA), two macrolide resistance genes (mefA and ermB), together with a class 1 integron integrase gene (intI1), a quaternary ammonium compound resistance gene (qacEΔ1), and 16S rRNA gene. Table S1 lists the annealing temperatures, specific primers, and expected amplicon sizes of target genes. Calibration standard curves for positive controls and quantification were constructed using plasmid DNA (cloned ARGs) extracted from Escherichia coli DH5α, as previously described (Li et al., 2015). All qPCR assays were performed on a QuantStudio 6 Flex Real-Time Fluorescence PCR instrument (Applied Biosystems, USA) equipped with real-time PCR-based gene expression profiling. Quantitative PCR was performed using TB Green® Premix Ex Taq™ (Takara), according to the manufacturer’s instructions. Each reaction was performed in triplicate. The standard curve for each qPCR assay consisted of a 10-fold dilution series from 0.5×109 copies·mL-1 to 0.5×103 copies·mL-1. The amplification efficiencies ranged from 78.918% to 104.98%, and the R2 values were between 0.982 and 1.000 for all the standard curves. The abundance of ARGs was determined as the copy number per milliliter of the water sample or the copy number per gram of the sediment sample (dry weight). Relative copy number of the resistance gene (relative abundance gene) = copy number of resistance gene/copy number of the 16S rRNA gene.
Dissolved oxygen (DO), water temperature (WT), pH, ammonia nitrogen (NH3-N), nitrite nitrogen (NO2-N), sulfide (SO4), phosphate (PO4), free chlorine (Cl), copper (Cu), chromium (Cr), total iron (Fe), total alkalinity (TOA), and total hardness (TOH) of the water samples were determined using a water quality detector (Octadem W-II, USA).
High-throughput sequencing of the bacterial community was performed using an Illumina HiseqTM 2500/4000 by Gene Denovo Biotechnology Co., Ltd. (Guangzhou, China) to further analyze the microbial community composition of the environmental samples. Specific primers (341F, CCTACGGGNGGCWGCAG; 806R, GGACTACHVGGGTATCTAAT) were used to amplify the V3 - V4 regions of 16S rRNA gene. PCR products (466 bp) were recorded to construct a library of amplicons, and amplicon sequencing was performed on an OmicSmart platform (Illumina, USA). The original sequencing data contained a large amount of low-quality data or data without biological significance (such as chimeras) owing to PCR and sequencing errors. Therefore, strict quality control was performed in multiple data processing processes, including read utilization, tag splicing, and operational statistical unit (OTU) clustering, to ensure the statistical reliability and biological effectiveness of subsequent analyses. The sequences date reported in this study was archived in Sequence Read Archive (SRA) with the accession number PRJNA907527.
The copy numbers calculated by qPCR were defined as absolute gene abundance. A heatmap showing the natural logarithm-transformed values of normalized ARG copies was constructed using GraphPad Prism 7 (San Diego, CA, USA). Bacterial community data, including taxonomic composition, community comparison, and diversity indices (Shannon, Simpson, Chao1, and ACE), were obtained using Omicsmart (http://www.omicsmart.com). Principal coordinate analysis (PCoA) was used to compare and visualize the differences in the abundance of ARGs and microbial communities based on a Jaccard distance matrix. Visualizing the network topology in Gephi (0.9.2) using the Frucherman-Reingold algorithm. Canonical correspondence analysis (CCA) was performed using the R language vegan package to compare the bacterial communities and environmental variables. Statistical analyses were performed using SPSS (version 25.0; IBM Corp., Armonk, NY, USA). The evaluations were based on a significance level of 5%. Statistical significance was set at P < 0.05.
The abundance of twenty-two resistance genes, intI1 and 16S rRNA gene, was investigated to analyze the distribution of ARGs in hybrid snakehead fish farms. A significantly higher abundance of ARGs (sulfonamides, quinolones, and phenicols) was detected in water samples than in sediment samples (Figure 1). The absolute abundances of sulfonamides, quinolones, and phenolics in the water samples were 1.61×102–2.34×109, 2.90×103–4.64×107 and 1.48×102–2.97×107 copies·mL-1, respectively. The absolute abundances of sulfonamides, quinolones, and phenols in the sediment samples were 2.26×102–7.77×108, 1.20×103–1.38×107 and 2.93×102–1.71×107 copies·g-1, respectively. The most abundant antibiotic resistance genes were sulfonamide resistance genes (sul1 and sul2) in the snakehead fish farm, with a total abundance of 7.36×106–2.34×109 copies·mL-1 (copies·g-1) in all samples. Quinolones, tetracycline, and phenicol resistance genes were also abundant on the aquatic farm, with a total abundance of 1.20×103–4.64×107 copies·mL-1 (copies·g-1), 5.83×100–5.00×106 copies·mL-1 (copies·g-1), and 1.48×102–2.97×107 copies·mL-1 (copies·g-1), respectively. β-lactam resistance genes (blaCTX-M and blaSHV) showed the lowest absolute abundance compared with the other types of ARGs. The total absolute abundance of β-lactam resistance genes was 1.78×10-2 to 1.76×105 copies·mL-1 (copies·g-1) (Table S2). The absolute abundance of qacEΔ1 ranged from 2.51×104 to 6.60 ×105 copies·mL-1 (copies·g-1). The abundance of ARGs did not differ significantly during the fingerling, rearing, and harvesting stages (Figure 1). Pearson’s test showed that there was a prominent correlation between intI1 and eight ARGs: sul1, sul2, aac6, qnrA, tetM, ermB, blaCTX-M, and qacEΔ1 (r > 0.5, P < 0.05).
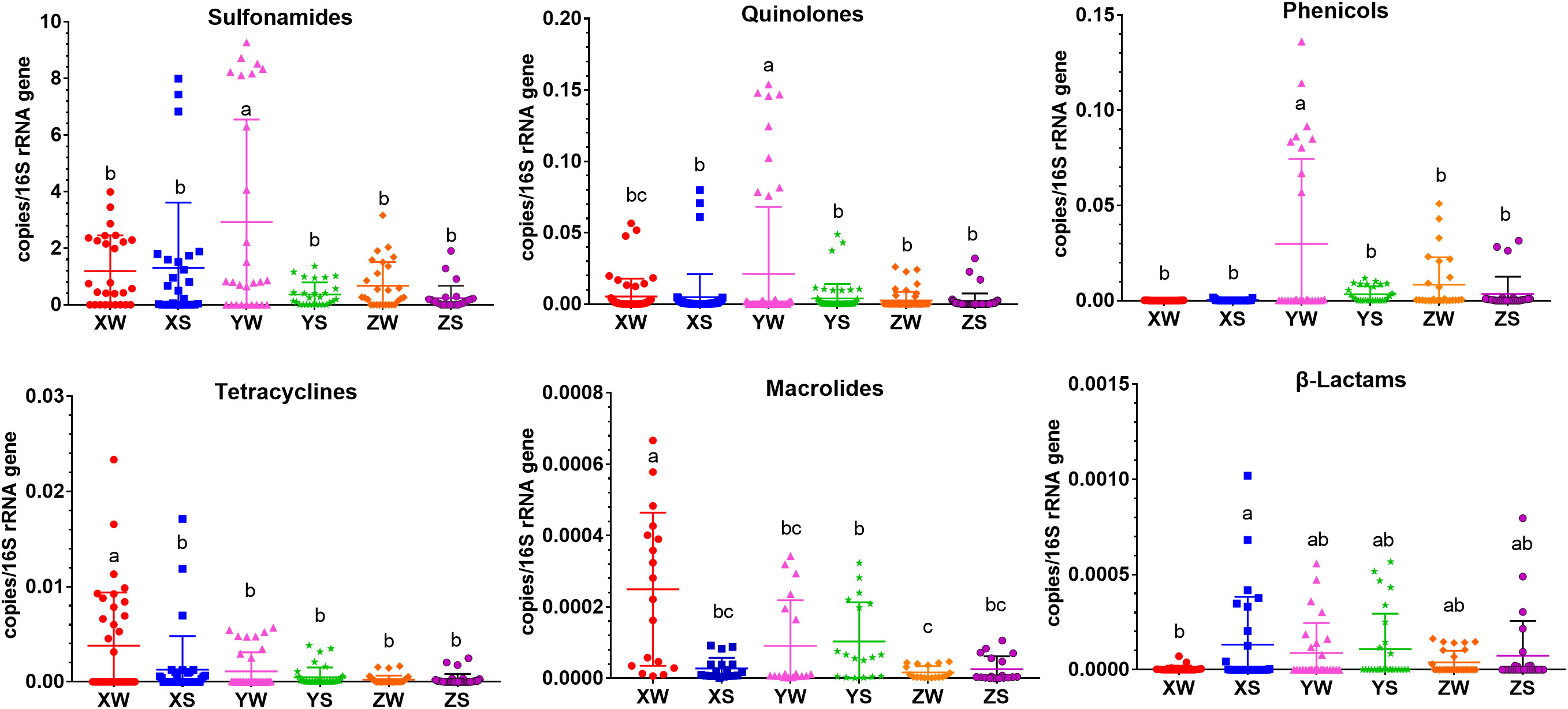
Figure 1 Distribution of different classes of ARGs in the water and sediment samples. X, Y, and Z indicate the fingerling, rearing, and harvesting stages, respectively, and S and W indicate the pond water and sediment samples, respectively. a,b and c indicate different significance at P < 0.05.
The copy numbers of ARGs were normalized to the corresponding copy numbers of 16S rRNA gene to calculate the relative abundance of ARGs (Figure 2). The relative abundance of sulfonamide resistance gene sul2 from the water sample was the highest (8.53 ×100 ± 2.73×10-1 copies/16S rRNA genes), followed by sul1 from the water sample (2.38×100 ± 2.27×100 copies/16S rRNA genes). The following ARGs had lower relative copies/16S rRNA genes: tetW (6.07×10-8–1.40×10-4), tetB (5.65×10-6–3.12×10-4), ermB (3.80×10-7–2.01×10-4), blaTEM (5.67×10-6–1.08×10-4), blaCTX-M (8.10×10-8–1.98×10-6), and blaSHV (8.99×10-11–1.32×10-7). The lowest concentration of ARGs was for blaCTX-M (copies/16S rRNA genes). The relative abundance of each resistance gene is shown in Table S3.
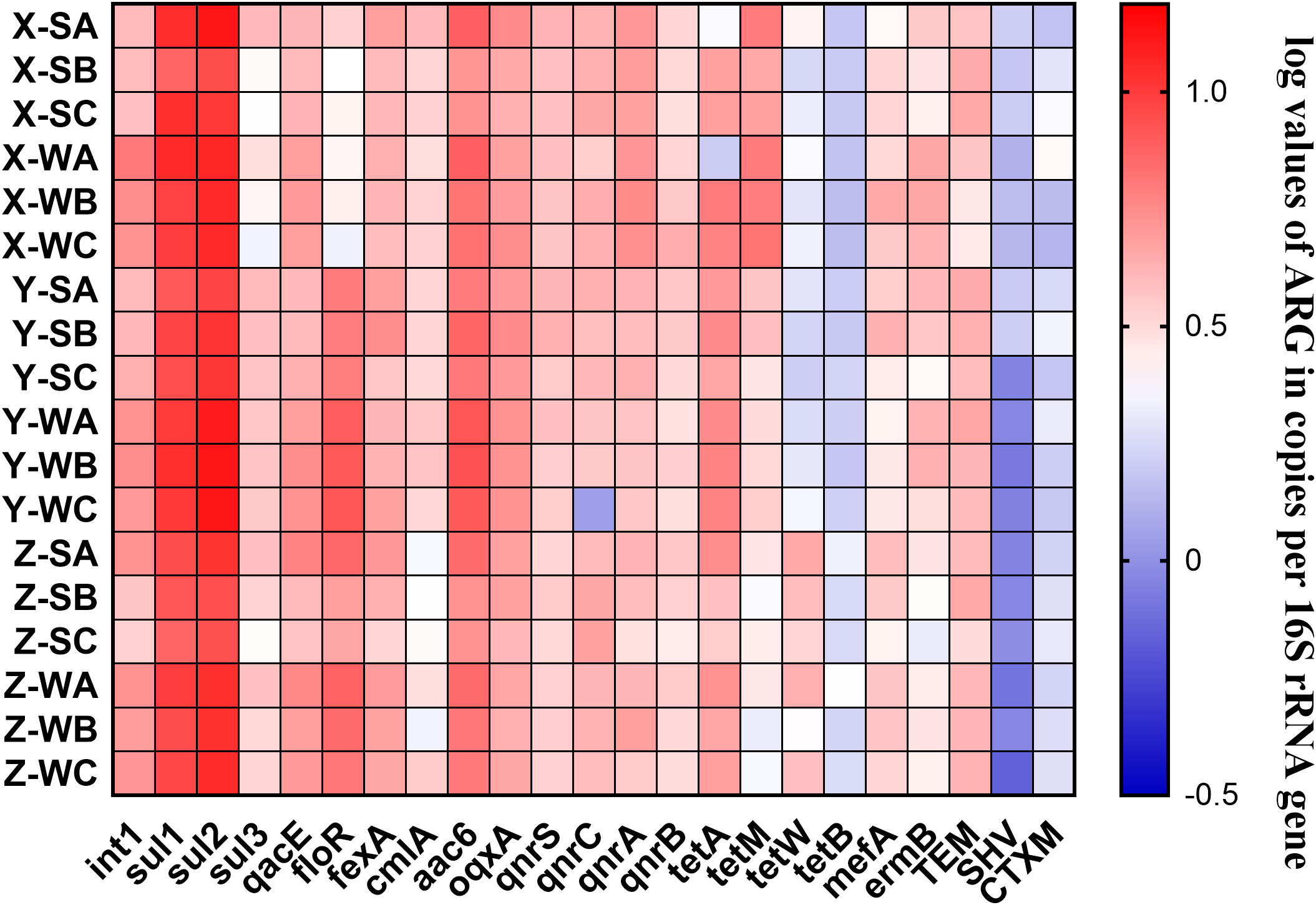
Figure 2 Heat map of the relative abundance of ARGs in each sample. X, Y, and Z indicate the fingerling, rearing, and harvesting stages, respectively; S and W indicate pond water and sediment samples, respectively; and A, B, and C indicate three different ponds.
Overall, the pH value and concentrations of ammonia nitrogen, phosphate, and total alkalinity were significantly different among the three culture stages, whereas the concentrations of dissolved oxygen, nitrite nitrogen, sulfide, free chlorine, copper, chromium, total iron, total hardness, and water temperature were not significantly different (Figure 3). Most of these parameters were within the acceptable ranges except for ammonia nitrogen, which ranged from 0.036 to 5.65 mg·L-1 (acceptable ranges ≤4.0 mg·L-1) (Liu Q et al., 2014). The pH was maintained between 6.27 and 7.51, which was slightly lower during the harvesting stage. The temperature was maintained between 28°C and 31°C. The concentration of nitrite nitrogen ranged from 0.176 to 0.55 mg·L-1. High concentration of dissolved oxygen was determined from the surface water (10.49–17.83 mg·L-1), while low concentrations of sulfide, phosphate, free chlorine, copper and total iron were detected during fingerlings to the harvesting stage. The details of the environmental factors are listed in Table S4.
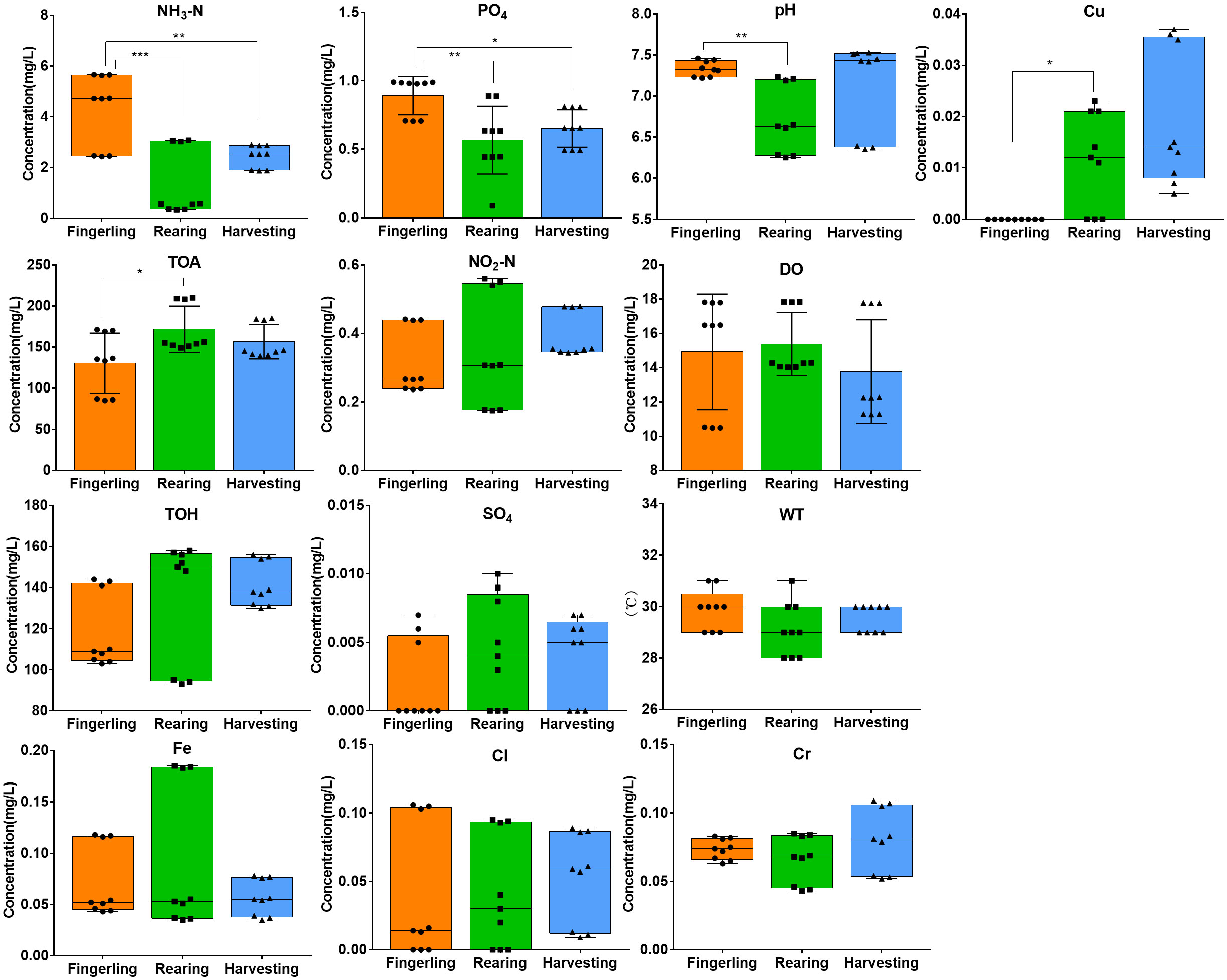
Figure 3 Concentrations of NH3-N, PO4, Cu, TOA, NO2-N, DO, TOH, SO4, Fe, Cl, and Cr and values of water temperature and pH in the water samples. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Eighteen libraries of bacterial 16S rRNA genes from pond environmental samples were sequenced using the Illumina high-throughput sequencing platform to explore the shift in bacterial communities during snakehead fish culture. After assembly and filtration, the samples had an average of 116,921.93 ± 132 and 116,907.93 ± 137 clean tags in the sediment and water samples, respectively. The α-diversity indices of the microbial communities in the sediment and water of the fish culture ponds are shown in Figure 4 and Table S5. The Shannon, Simpson, Chao1, and ACE indices considered both the species richness and relative abundance distribution. Greater population richness and more diverse bacterial communities were observed in the sediment samples than in the water samples from the snakehead fish farm (Figure 4). Population richness was abundant in the sediment samples during the fingerling and harvesting stages, but was slightly lower during the rearing stage.
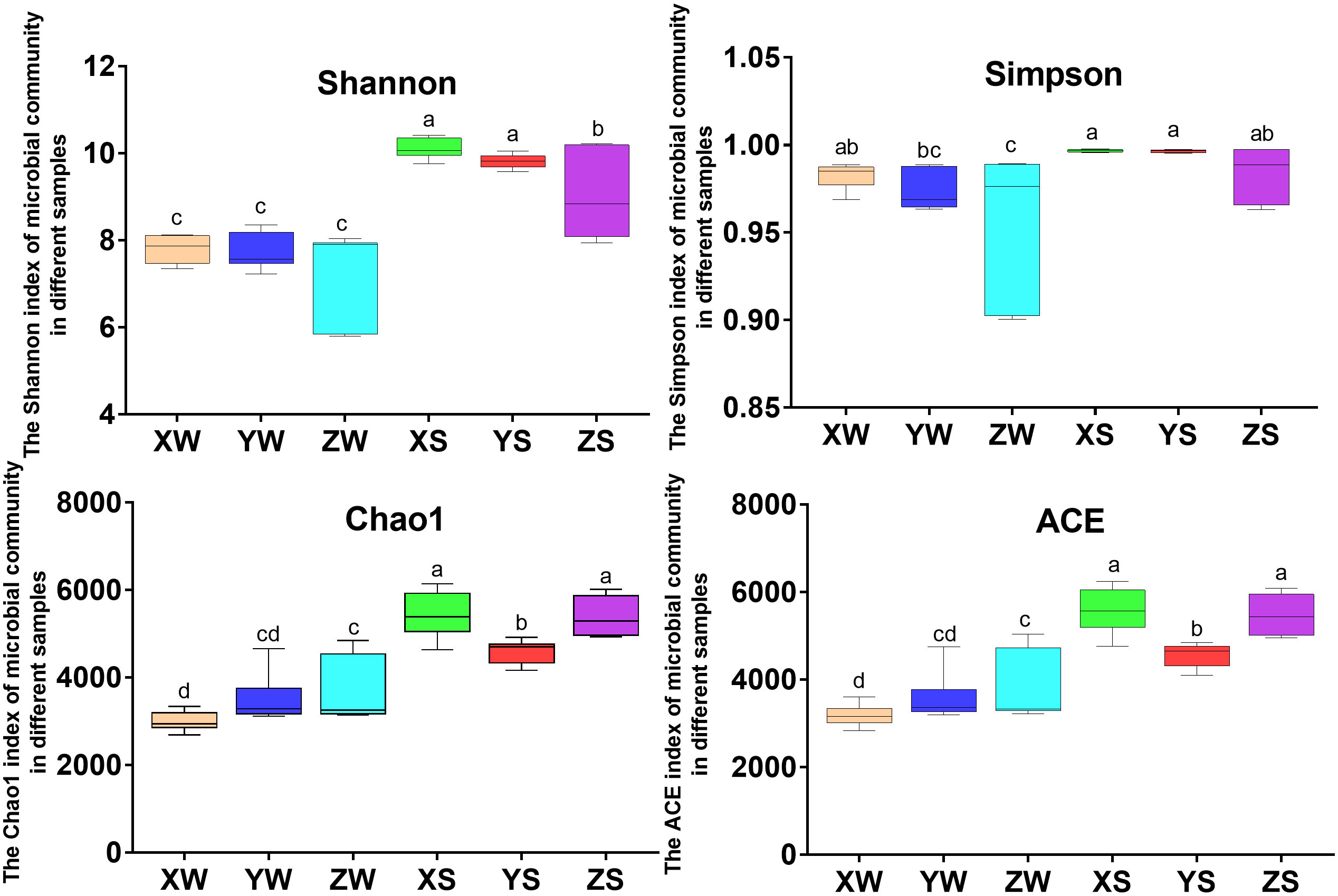
Figure 4 Differences and temporal trends of α-diversity indices between water and sediment samples in the hybrid snakehead fish culture system. X, Y, and Z indicate the fingerling, rearing, and harvesting stages, respectively, and S and W indicate the pond water and sediment samples, respectively. a,b and c indicate different significance at P < 0.05.
Proteobacteria and Cyanobacteria were the most abundant phyla in water sample and sediment samples, with relative abundances ranging from 38.77% to 44.92% (Figure 5). The second most abundant phylum in water samples was Cyanobacteria, followed by Planctomycetes, Bacteroidetes, Chloroflexi, Actinobacteria, and Verrucomicrobia. The second-most dominant phylum in the sediment samples was Chloroflexi, followed by Actinobacteria, Planctomycetes, Acidobacteria, Verrucomicrobia, Bacteroidetes, and Gemmatimonadetes. Although the bacterial community structures were different between the environmental samples in the three ponds, the most abundant phyla (Cyanobacteria, Proteobacteria, Bacteroidetes, Actinobacteria, and Chloroflexi) were similar in the environmental samples, with total relative abundances of 49.22% and 85.12% (Table S6).
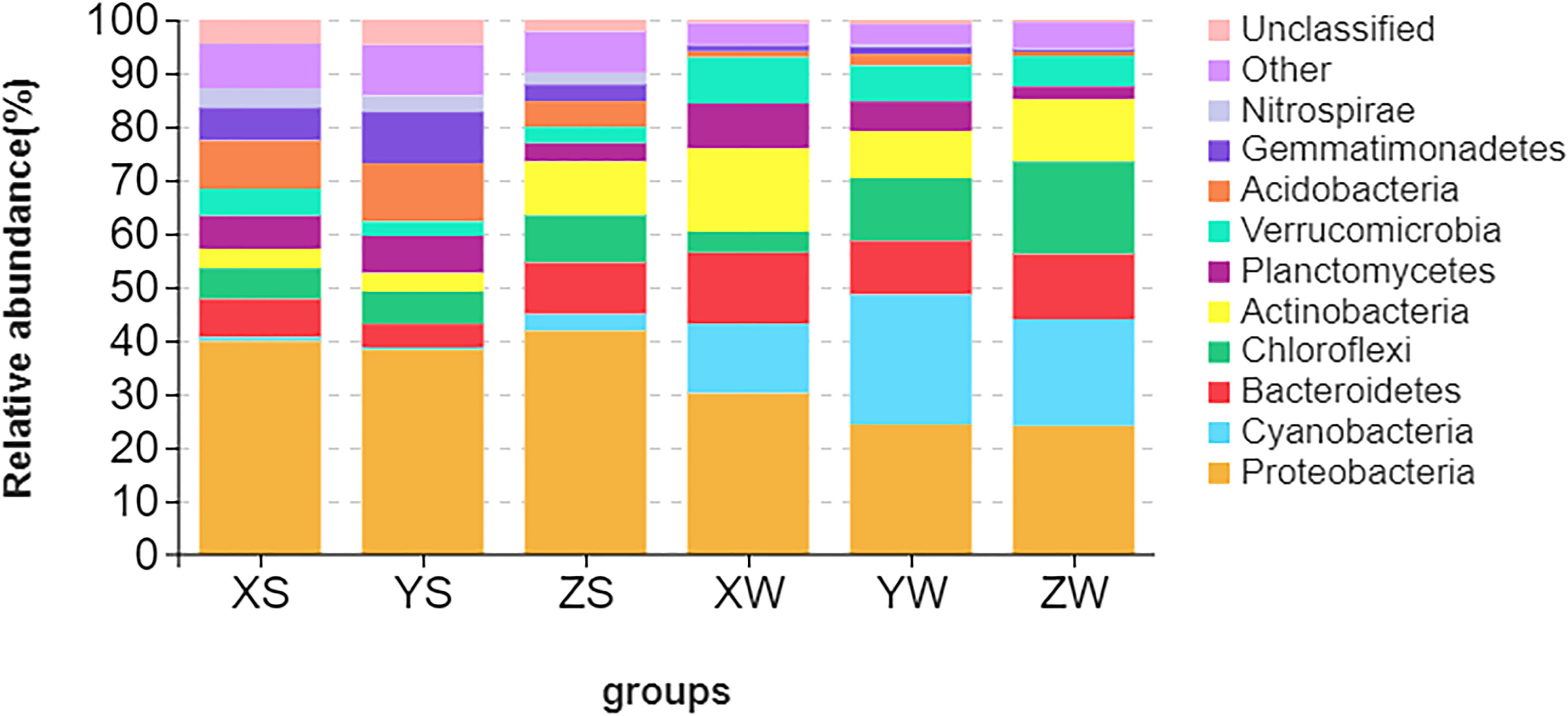
Figure 5 Relative abundance of the top 10 phyla in the water and sediment samples. X, Y, and Z indicate the fingerling, rearing, and harvesting stages, respectively, and S and W indicate the pond water and sediment samples, respectively.
Bacteria from water or sediment samples of fingerling, rearing, and harvesting stages clustered together in the principal coordinate analysis plot based on the Jaccard similarity index (OTUs presence and absence). However, the presence and abundance of bacterial communities differed between the water and sediment samples at the family level (Figure 6 and Table S7). In addition, the different colored nodes in Figure 6 represent samples from different breeding stages, implying that the microbial community structure greatly changed at different breeding stages.
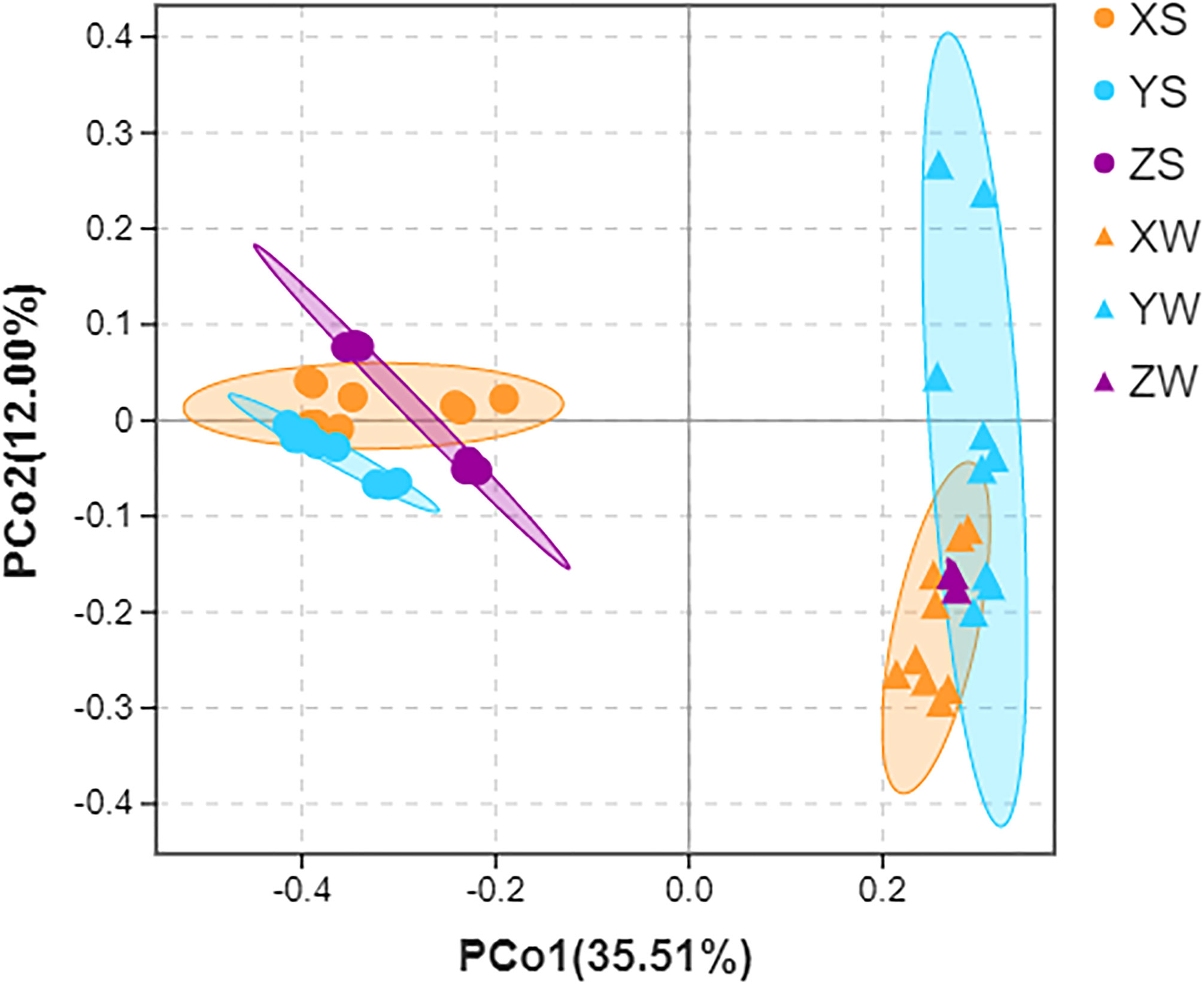
Figure 6 Principal coordinate analysis (PCoA) of Jaccard dissimilarity in water and sediment samples. X, Y, and Z indicate the fingerling, rearing, and harvesting stages, respectively, and S and W indicate the pond water and sediment samples, respectively.
Network analysis and CCA were performed to further explore the relationship between the bacterial community in the sediment and water samples, environmental factors, and ARG abundance in hybrid snakehead fish farms (Figures 7A, B). Network analysis showed that the co-occurrence patterns between the bacterial community and ARGs consisted of 41 nodes and 96 edges (Figure 7A). The modularity index was 0.448, suggesting that the network had a modular structure. Proteobacteria, Cyanobacteria, Bacteroidetes, Actinobacteria, and Verrucomicrobia were strongly correlated with ten ARG subtypes (intI1, sul2, sul1, aac6, cmlA, ermB, tetA, qacEΔ1, qnrA, and qnrS) (Figure 7A). This suggested that the spread of ARGs in aquaculture is significantly affected by the proliferation of environmental microorganisms.
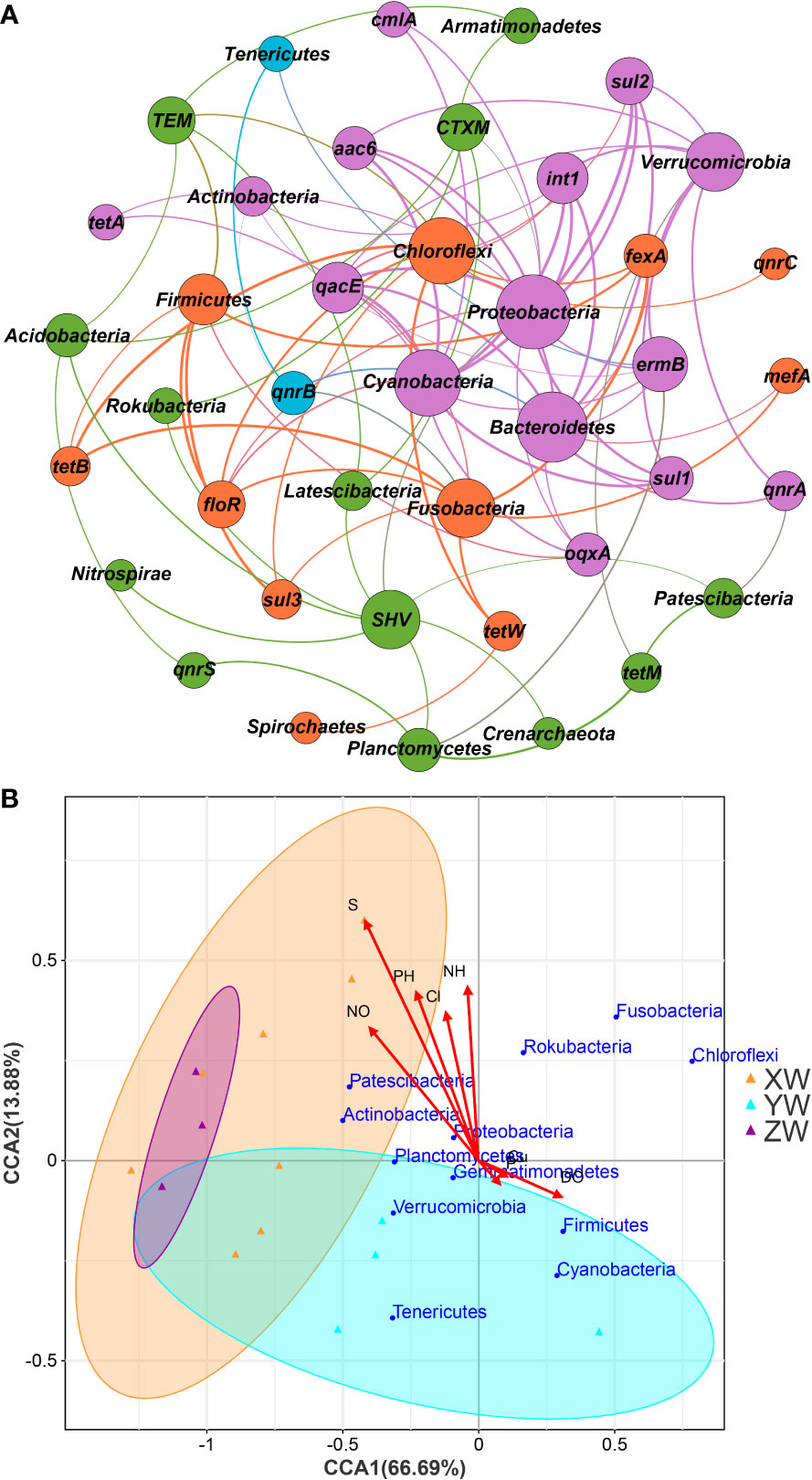
Figure 7 Correlations among the bacterial community, ARGs, and environmental factors. (A) Network analysis of ARGs and bacterial communities at the phylum level in the pond water. The linkage between the nodes indicates a correlation between the two factors, and the size of the node is proportional to the number of connections. At the same time, the thickness of the line was proportional to the correlation between the factors. (B) Canonical correspondence analysis of the environmental factors and bacterial communities at the phylum level.
The correlation between the target environmental factors and bacteria at the phylum level in fishpond water samples was determined using CCA (Figure 7B). At the phylum level, CCA1 and CCA2 explained 66.69% and 13.18% of total variance, respectively. Proteobacteria, Actinobacteria, Patescibacteria, Fusobacteria, Rokubacteria, Chloroflexi, and Planctomycetes were positively correlated with sulfide, nitrite nitrogen, pH, free chlorine, and ammonia nitrogen. In addition, copper and dissolved oxygen levels were positively correlated with Firmicutes, Cyanobacteria, Tenericutes, Verrucomicrobia, Fusobacteria, and Chloroflexi.
Twenty-one target ARGs and two integron-related genes (intI1 and qacEΔ1) were detected in environmental samples from the hybrid snakehead fish culture system at the three breeding stages. Consistent with previous studies, these results indicated the persistence of ARGs in aquaculture environments. (Shen et al., 2020; Suyamud et al., 2021). The high abundance of sulfonamide, tetracycline, quinolone, and phenicol resistance genes suggests that these bacteria may confer resistance to the most commonly used antibiotics in hybrid snakehead fish culture systems. Sulfonamide resistance genes (sul1 and sul2) were detected in high absolute abundance, followed by floR, aac6, and tetA. Coincidentally, sulfonamide and tetracycline resistance genes frequently occur in river environments and are accompanied by the presence of integrons (Kumar et al., 2020; Dong et al., 2022). Similarly, the ARG subtypes mentioned above have the highest abundance in sediment and water samples from shrimp and fishponds (Huang et al., 2017; Cheng et al., 2021). For instance, the abundance of the tetracycline resistance gene tetA(106–109 copies·mL-1) was the highest in sediment and water samples from freshwater fish farms (Huang et al., 2017). Concentrations of the sulfonamide resistance gene sul1 and tetracycline resistance gene tetW were the highest in 15 sediment samples collected from five Chinese provinces (Cheng et al., 2021). In addition, the quinolone resistance gene qnrB is abundant in six freshwater ponds (Wang et al., 2022). Integrons play an important role in the emergence and horizontal transfer of ARGs among bacteria and are the main transmissible vectors commonly found in different environments (Wang et al., 2019). The relatively high abundance of intI1 in the hybrid snakehead fish farm suggests potentially high mobility of ARGs. We also found a significant correlation between intI1 and eight resistance genes (r > 0.5, P < 0.05). The abundances of ARGs and intI1 measured in this study were comparable to those reported in other natural water bodies and aquaculture environments (Wang et al., 2019).
Several studies have confirmed the prevalence of ARGs in hospitals, pharmaceutical manufacturers, livestock farms, wastewater treatment plants, and rivers (Gao et al., 2012; Li et al., 2015; Zhou et al., 2017; Hu et al., 2018; Su et al., 2018a). Overuse of various antibacterial agents in humans and veterinarians, agriculture, and aquaculture has led to the emergence of antibiotic-resistant pathogens (Pham et al., 2018; Li et al., 2022). Water from nearby rivers brings polluting compounds from agricultural and domestic wastewater into aquaculture systems. In contrast, untreated aquaculture wastewater is pumped into rivers. High-density aquaculture involves rearing hybrid snakehead fishes in congested and stressful environments. The pond water is hardly exchanged, which allows antibiotics to impose selective pressure on bacteria. Our study demonstrated high ARGs and intI1 copy numbers in pond water samples. This implies that one of the main reservoirs and sources of ARG in the surrounding aquatic environment is the pond water.
The abundance of ARGs varies temporally during different seasons and breeding stages in aquaculture (Zhang et al., 2021; Zhou et al., 2022). A higher mean abundance of ARGs in aquaculture sediments was observed in autumn and winter, whereas the mean abundance of ARGs decreased significantly in spring and summer (Xu et al., 2022). A comparison of the different breeding stages of grass carp ponds showed that the relative abundance of sul2 in the harvesting and rearing culture stages was significantly higher than that in the larval culture stage, whereas the abundances of cfr and tetQ were significantly higher in the larval culture stage (Zhou et al., 2022). This study found that the abundance of ARGs did not differ significantly during the fingerling, rearing, or harvesting stages. Notably, the differential abundance of different ARG subtypes belonging to the same class of resistance genes may be irregularly expressed in aquaculture according to different culture systems, geographical locations, or drug usage.
The relationship between ARGs and microbial communities was comprehensively explored by analyzing 22 resistance genes, intI1, and identifying bacteria at the phylum level, based on previous studies (Su et al., 2018b; Han et al., 2022). Despite the presence of various ARGs in aquaculture environments, microorganisms are considered the main vectors and determinants of ARG distribution and transmission (Wang et al., 2019). Antibiotic resistance genes mainly exist in bacterial genomes and may be transmitted through mobile genetic elements, including plasmids, integrons, transposons, and insertion sequences (Suyamud et al., 2021). Proteobacteria is the predominant phylum in Litopenaeus vannamei pond water, followed by Bacteroides (Zheng et al., 2016). Proteobacteria is also the most abundant phylum in water and sediment samples from different natural environments, followed by Actinobacteria, Bacteroidetes, Chloroflexi, and Cyanobacteria (Fang et al., 2019). This study found that Proteobacteria and Cyanobacteria were the two predominant phyla in water samples from snakehead fish aquaculture, which is similar to the dominant bacterial phyla found in previous studies on fish ponds (Su et al., 2017; Klase et al., 2019). The present results indicate that the microbiota structure at the family or genus level significantly changed at different breeding stages despite the similar abundance of the predominant bacterial phyla. There are differences in breeding patterns and seasonal changes in the bacterial composition of different ponds (Xu et al., 2020). We also found that the presence and abundance of bacterial communities differed between water and sediment samples at the family level. These results may be attributed to differences in aquaculture patterns and regional differences.
Network analysis was performed to further analyze the co-occurrence of ARGs and dominant bacteria at the phylum level in the pond water and sediment samples. Proteobacteria, Cyanobacteria, Bacteroidetes, Actinobacteria, and Verrucomicrobia were strongly correlated with ten ARG subtypes (sul1, sul2, aac6, cmlA, ermB, tetA, qacE, qnrA, and qnrS) and intI1. This implies that these predominant bacteria may be responsible for the increased abundance of ARGs in the snakehead fish farms. Some Firmicutes bacterial taxa are major potential hosts of ARGs, which strongly co-occur with sul1, qnrS, tetM, and intl1 genes (Cheng et al., 2021). Meanwhile, the sulfonamide resistance gene sul1 and tetracycline resistance genes (tetW and tetG) are mainly associated with Actinomyces, Proteobacteria, and Actinomycetes. Among these, Proteobacteria have a high potential for hosting the sulfonamide resistance gene (Shen et al., 2020). These results indicate that changes in bacterial composition may lead to significant and strong correlation differences between ARG and predicted carrier bacteria in different environments.
The distribution of ARG and their potential carrier microorganisms is greatly affected by environmental factors such as nutrients and pollutants in water (Zhao et al., 2021; Ning et al., 2022). Changes in environmental factors, such as organic matter level, total nitrogen, temperature, and heavy metals, promote the transfer of bacterial communities and reproduction of ARG in aquatic environments (Xu et al., 2022). The present study found a significant correlation between the environmental factors of water and the bacterial communities in ponds. Proteobacteria, Actinobacteria, and Patescibacteria were positively correlated with sulfide, nitrite nitrogen, pH, free chlorine, and ammonia nitrogen concentrations. Ammonia nitrogen, nitrite nitrogen, and pH are important parameters for fish culture. Most fish species do well within a pH range of 6.5–9.5. Chronic pH levels below 6.5 may reduce fish reproduction and are associated with fish die-offs. The nitrate levels in pond water are also a major concern in fish culture. Exceedingly high nitrate-nitrogen and ammonia concentrations are health hazards. Most of these parameters, except for ammonia nitrogen, were within the acceptable range for snakehead fish cultures. This may be attributed to the high-density culture of fish and excessive feed in the pond. Long rearing times induce the accumulation of feeding residues that affect the levels of nitrogenous compounds (Chen et al., 2018), and prolonged exposure to nitrogenous compounds may result in immunological responses within fishponds. At the same time, heavy metal pollution causes the deterioration of water quality, which affects the ecological stability of flora in the water environment (Ai et al., 2022). Other pollutants with significant antibacterial activity can select antibiotic resistance genes, such as heavy metals (He et al., 2017), pesticides (Anjum and Krakat, 2016), and disinfectants (Chaturvedi et al., 2021). In addition, antibiotic resistance can be triggered and catalyzed by metal-driven co-selection, an indirect selection process with physiological and genetic consequences (Ai et al., 2022). In contrast, this study only detected low metal concentrations in pond water and their levels were not significantly correlated with the bacterial community. However, multiple factors in snakehead fish cultures with high stock densities pose a high potential risk of increasing antibiotic resistance in aquaculture. Controlling bacterial diseases in fish cultures has become more difficult because of the emergence of ARGs, especially those caused by multidrug-resistant bacteria.
This study showed an increased occurrence of sulfonamides, tetracyclines, quinolones, and phenicol resistance genes in pond water and sediments of the hybrid snakehead fish farm. Some bacterial taxa from the phyla Proteobacteria, Cyanobacteria, Bacteroidetes, Actinobacteria, and Verrucomicrobia may be major potential hosts of ARGs in this aquatic farm. In addition, the congested and stressful environment of snakehead fish cultures may cause pollution problems with negative effects on the propagation of resistant bacteria that drive the occurrence of ARGs. More attention should be paid to the better feeding and management of fish cultures. Moreover, the elimination of ARGs in wastewater from aquaculture environments should be considered to reduce the pollutants discharged into the natural environment. The levels of antibiotic resistance genes and microbial community composition in aquaculture should be tested regularly to assess potential risks to public health.
The raw sequencing date generated from this study have been deposited in NCBI SRA (http://www.ncbi.nlm.nih.gov/sra) under the accession number PRJNA907527.
XL and YD played an important role in this study. XL and AT completed a part of the experiment. FZ and WL performed the data analysis. XL wrote the first draft of the manuscript. AT has edited the final version of the manuscript. YD and ZH reviewed and edited the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by Guangdong Natural Science Foundation (No.2020A1515011584) and Central Public-interest Scientific Institution Basal Research Fund, CAFS (2022GH04, 2021SJ-XT3, 2020TD45).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.1088176/full#supplementary-material
Ai L., Ma B., Shao S., Zhang L., Zhang L. (2022). Heavy metals in Chinese freshwater fish: Levels, regional distribution, sources and health risk assessment. Sci. Total Environ. 853, 158455. doi: 10.1016/j.scitotenv.2022.158455
Allen H. K., Donato J., Wang H. H., Cloud-Hansen K. A., Davies J., Handelsman J. (2010). Call of the wild: Antibiotic resistance genes in natural environments. Nat. Rev. Microbio. 8 (4), 251–259. doi: 10.1038/nrmicro2312
Anjum R., Krakat N. (2016). Detection of multiple substances, biofilm formation, and conjugative transfer of b. cereus from contaminated soils. Curr. Microbio. 72 (3), 321–328. doi: 10.1007/s00284-015-0952-1
Berendonk T. U., Manaia C. M., Merlin C., Fatta-Kassinos D., Cytryn E., Bürgmann H., et al. (2015). Tackling antibiotic resistance: The environmental framework. Nat. Rev. Microbiol. 13 (5), 310–317. doi: 10.1038/nrmicro3439
Chaturvedi P., Shukla P., Giri B. S., Chowdhary P., Chandra R., Gupta P., et al. (2021). Prevalence and hazardous impact of pharmaceutical and personal care products and antibiotics in environment: A review on emerging contaminants. Environ. Res. 194, 110664. doi: 10.1016/j.envres.2020.110664
Cheng X., Lu Y., Song Y., Zhang R., ShangGuan X., Xu H. Z., et al. (2021). Analysis of antibiotic resistance genes, environmental factors, and microbial community from aquaculture farms in five provinces, china. Front. Microbio. 12. doi: 10.3389/fmicb.2021.679805
Chen B., Lin L., Fang L., Yang Y., Chen E., Yuan K., et al. (2018). Complex pollution of antibiotic resistance genes due to beta-lactam and aminoglycoside use in aquaculture farming. Water Res. 134, 200–208. doi: 10.1016/j.watres.2018.02.003
Dong J., Yan D., Mo K., Chen Q., Zhang J., Chen Y. C., et al. (2022). Antibiotics along an alpine river and in the receiving lake with a catchment dominated by grazing husbandry. J. Environ. Sci. 115, 374–382. doi: 10.1016/j.jes.2021.08.007
Fang H., Huang K., Yu J., Ding C., Wang Z., Zhao C., et al. (2019). Metagenomic analysis of bacterial communities and antibiotic resistance genes in the eriocheir sinensis freshwater aquaculture environment. Chemosphere. 224, 202–211. doi: 10.1016/j.chemosphere.2019.02.068
Gao P., Munir M., Xagoraraki I. (2012). Correlation of tetracycline and sulfonamide antibiotics with corresponding resistance genes and resistant bacteria in a conventional municipal wastewater treatment plant. Sci. Total Environ. 421-422, 173–183. doi: 10.1016/j.scitotenv.2012.01.061
Han Y., Hu Y., Wen H., Wu J., Liu Y., Zhang Y. C., et al. (2022). Occurrence and driving mechanism of antibiotic resistance genes in marine recreational water around qinhuangdao, China. Fron Mar. Sci. 9. doi: 10.3389/fmars.2022.976438
He X., Xu Y., Chen J., Ling J., Li Y., Huang L., et al. (2017). Evolution of corresponding resistance genes in the water of fish tanks with multiple stresses of antibiotics and heavy metals. Water Res. 124, 39–48. doi: 10.1016/j.watres.2017.07.048
Hossain A., Habibullah-Al-Mamun M., Nagano I., Masunaga S., Kitazawa D. (2022). Antibiotics, antibiotic-resistant bacteria, and resistance genes in aquaculture: Risks, current concern, and future thinking. Environ. Sci. pollut. Res. Int. 299 (8), 11054–11075. doi: 10.1007/s11356-021-17825-4
Huang L., Xu Y., Xu J., Ling J., Chen J., Zhou. J. L., et al. (2017). Antibiotic resistance genes (ARGs) in duck and fish production ponds with integrated or non-integrated mode. Chemosphere. 168, 1107–1114. doi: 10.1016/j.chemosphere.2016.10.096
Hu J., Zhao F., Zhang X., Li K., Li C., Ye L., et al. (2018). Metagenomic profiling of ARGs in airborne particulate matters during a severe smog event. Sci. Total Environ. 615, 1332–1340. doi: 10.1016/j.scitotenv.2017.09.222
Klase G., Lee S., Liang S., Kim J., Zo Y. G., Lee J. (2019). The microbiome and antibiotic resistance in integrated fish farm water: Implications of environmental public health. Sci. Total Environ. 649, 1491–1501. doi: 10.1016/j.scitotenv.2018.08.288
Kumar M., Sulfikar, Chaminda T., Patel A. K., Sewwandi H., Mazumder P. (2020). Prevalence of antibiotic resistance in the tropical rivers of Sri Lanka and India. Environ. Res. 188, 109765. doi: 10.1016/j.envres.2020.109765
Li J., Cheng W., Xu L., Strong P. J., Chen H. (2015). Antibiotic-resistant genes and antibiotic-resistant bacteria in the effluent of urban residential areas, hospitals, and a municipal wastewater treatment plant system. environ. Sci. pollut. Res. 22 (6), 4587–4596. doi: 10.1007/s11356-014-3665-2
Li S., Ondon B. S., Ho S. H., Jiang J., Li F. (2022). Antibiotic resistant bacteria and genes in wastewater treatment plants: From occurrence to treatment strategies. Sci. Total Environ. 838 (Pt 4), 156544. doi: 10.1016/j.scitotenv.2022.156544
Liu Q., Lai Z., Yang W., Gao Y., Wang C. (2014). Assessment of water quality of intensive ponds in the pearl river delta region. South China Fisheries Sci. 10, 36–43. doi: 10.3969/j.issn.2095–0780.2014.06.005
Liu X., Steele J. C., Meng X. (2017). Usage, residue, and human health risk of antibiotics in Chinese aquaculture: A review. Environ. Pollution. 223, 161–169. doi: 10.1016/j.envpol.2017.01.003
Ning K., Ji L., Zhang L., Zhu X., Wei H., Wang Z. (2022). Is rice-crayfish co-culture a better aquaculture model: From the perspective of antibiotic resistome profiles. Environ. pollut. 2929 (Pt B), 118450. doi: 10.1016/j.envpol.2021.118450
Pham T., Rossi P., Dinh H., Pham N., Tran P. A., Ho T., et al. (2018). Analysis of antibiotic multi-resistant bacteria and resistance genes in the effluent of an intensive shrimp farm (Long an, Vietnam). J. Environ. Manage. 214, 149–156. doi: 10.1016/j.jenvman.2018.02.089
Shen X. X., Jin G. Q., Zhao Y. Z., Shao X. H. (2020). Prevalence and distribution analysis of antibiotic resistance genes in a large-scale aquaculture environment. Sci. Total Environ. 711, 134626. doi: 10.1016/j.scitotenv.2019.134626
Su H., Hu X., Xu Y., Xu W., Huang X., Wen G., et al. (2018a). Persistence and spatial variation of antibiotic resistance genes and bacterial populations change in reared shrimp in south China. Environ. Int. 119, 327–333. doi: 10.1016/j.envint.2018.07.007
Su H., Liu S., Hu X., Xu X., Xu W., Xu Y., et al. (2017). Occurrence and temporal variation of antibiotic resistance genes (ARGs) in shrimp aquaculture: ARGs dissemination from farming source to reared organisms. Sci. Total Environ. 607-608, 357–366. doi: 10.1016/j.scitotenv.2017.07.040
Su H., Liu Y., Pan C., Chen J., He L., Ying G. G., et al. (2018b). Persistence of antibiotic resistance genes and bacterial community changes in drinking water treatment system: from drinking water source to tap water. Sci. Total Environ. 616-617, 453–461. doi: 10.1016/j.scitotenv.2017.10.318
Suyamud B., Lohwacharin J., Yang Y., Sharma V. K. (2021). Antibiotic resistant bacteria and genes in shrimp aquaculture water: identification and removal by ferrate(VI). J. Hazard Mater. 420, 126572. doi: 10.1016/j.jhazmat.2021.126572
Wang J. H., Lu J., Wu J., Zhang Y., Zhang C. (2019). Proliferation of antibiotic resistance genes in coastal recirculating mariculture system. Environ. pollut. 248, 462–470. doi: 10.1016/j.envpol.2019.02.062
Wang Q., Mao C., Lei L., Yan B., Yuan J., Guo Y. Y., et al. (2022). Antibiotic resistance genes and their links with bacteria and environmental factors in three predominant freshwater aquaculture modes. Ecotoxicol Environ. Saf. 241, 113832. doi: 10.1016/j.ecoenv.2022.113832
Xu M., Huang X. H., Shen X. X., Chen H. Q., Li C., Jin G. Q., et al. (2022). Metagenomic insights into the spatiotemporal responses of antibiotic resistance genes and microbial communities in aquaculture sediments. Chemosphere. 307 (Pt 1), 135596. doi: 10.1016/j.chemosphere.2022.135596
Xu C., Lv Z., Shen Y., Liu D., Fu Y., Zhou L., et al. (2020). Metagenomic insights into differences in environmental resistome profiles between integrated and monoculture aquaculture farms in China. Environ. Int. 144, 106005. doi: 10.1016/j.envint.2020.106005
Zhang R., Kang Y., Zhang R., Han M., Zeng W., Wang Y. H., et al. (2021). Occurrence, source, and the fate of antibiotics in mariculture ponds near the maowei Sea, south China: storm caused the increase of antibiotics usage. Sci. Total Environ. 752, 141882. doi: 10.1016/j.scitotenv.2020.141882
Zhao X., Su H., Xu W., Hu X., Xu Y., Wen G. L., et al. (2021). Removal of antibiotic resistance genes and inactivation of antibiotic-resistant bacteria by oxidative treatments. Sci. Total Environ. 778, 146348. doi: 10.1016/j.scitotenv.2021.146348
Zhao W., Wang B., Yu G. (2018). Antibiotic resistance genes in China: occurrence, risk, and correlation among different parameters. Environ. Sci. pollut. Res. 25 (22), 21467–21482. doi: 10.1007/s11356-018-2507-z
Zheng Y. F., Min Y., Yan L., Ying S., Tao X., Yu M. C., et al. (2016). Comparison of cultivable bacterial communities associated with pacific white shrimp ( litopenaeus vannamei ) larvae at different health statuses and growth stages. Aquaculture 451, 163169. doi: 10.1016/j.aquaculture.2015.09020
Zhou Y., Niu L., Zhu S., Lu H., Liu W. (2017). Occurrence, abundance, and distribution of sulfonamide and tetracycline resistance genes in agricultural soils across China. Sci. Total Environ. 599-600, 1977–1983. doi: 10.1016/j.scitotenv.2017.05.152
Zhou A., Xie S., Tang H., Zhang L., Zhang Y., Zuo Z. H., et al. (2022). The dynamic of the potential pathogenic bacteria, antibiotic-resistant bacteria, and antibiotic resistance genes in the water at different growth stages of grass carp pond. Environ. Sci. pollut. Res. Int. 29 (16), 23806–23822. doi: 10.1007/s11356-021-17578-0
Keywords: antibiotic resistance genes (ARGs), bacterial community, snakehead fish, public health, intensive aquaculture
Citation: Lin X, Tan A, Deng Y, Liu W, Zhao F and Huang Z (2023) High occurrence of antibiotic resistance genes in intensive aquaculture of hybrid snakehead fish. Front. Mar. Sci. 9:1088176. doi: 10.3389/fmars.2022.1088176
Received: 03 November 2022; Accepted: 15 December 2022;
Published: 12 January 2023.
Edited by:
Yiqin Deng, South China Sea Fisheries Research Institute (CAFS), ChinaReviewed by:
Shaowu Li, Heilongjiang River Fisheries Research Institute (CAFS), ChinaCopyright © 2023 Lin, Tan, Deng, Liu, Zhao and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuting Deng, ZGVuZ3l0QHByZnJpLmFjLmNu
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.