- 1College of Chemistry and Environmental Engineering, Shenzhen University, Shenzhen, China
- 2Key Laboratory of Geographic Information Science (Ministry of Education), School of Geographical Sciences, East China Normal University, Shanghai, China
- 3Institute of Eco-Chongming, East China Normal University, Shanghai, China
- 4State Key Laboratory of Estuarine and Coastal Research, East China Normal University, Shanghai, China
Most studies of whether and how nanoparticles (NPs) affect antibiotic resistance genes (ARGs) focus on testing single NPs type. In this study, we determined the combined effect of Cu- and ZnO- NPs in the water samples collected from the Yangtze River Estuary and found the effect differs greatly from that produced by individual NPs. The results showed that the Cu- and ZnO- NPs co-exposure resulted in an enrichment of ARGs, whereas individual Cu- and ZnO- NPs exposure decreased the abundance of ARGs. Furthermore, the co-exposure of Cu- and ZnO- NPs induced obvious changes in the microbial communities compared to the control communities. Redundancy analysis suggested that the microbial community contributed the most (43.5%) to the ARG profiles, followed by dissolved metal ions (25.7%), MRGs, (19.4%), and MGEs (4.4%). Network analysis found several potential hosts (such as Mycobacterium and Escherichia coli) and implied the extent of the risk of ARG transmission into various environmental niches by these common microbes.
Introduction
The very large production and utilization of engineered nanoparticles (NPs) in a variety of fields may directly or indirectly lead to an increase in residual NP concentrations in the environment, which may pose potential effects on various organisms in aquatic or terrestrial ecosystems and attracts more attention on their ecological risks (Bundschuh et al., 2018). For example, due to their unique structural and optical properties (Collins et al., 2012), copper (Cu) NPs are widely added to catalysts and electronics. One of the first commercial NPs, zinc oxide (ZnO) NPs are widely used in textile, cosmetic, pigment, food additive, and medical industries (Piccinno et al., 2012). It has been reported that metal-based NPs (M-NPs) are commonly found as colloids in aquatic environments, and dissolved metal ions or small inorganic complexes produced by M-NPs may be toxic to tested organisms (Liu et al., 2016; Turan et al., 2019). The effect of exposure to single M-NPs, such as Cu or ZnO NPs, has been investigated, and in contrast to studies that suggesting that the dissolved metal species of Cu and ZnO NPs were more toxic than particle forms (Barjhoux et al., 2012; Adam et al., 2014; Hua et al., 2014), it has been reported that the cytotoxic effects were most likely due to the particulate forms of Cu and ZnO NPs (Fernaíndez-Cruz et al., 2013; Song et al., 2014). Notably, the exposure of organisms to multiple contaminants is likely to take place in the natural environment (Wang et al., 2016; Guo et al., 2018a). Nevertheless, it is a phenomenon that Cu and ZnO NPs naturally occurred in the environment; for instance, in freshwater, soil, air, and landfills (Keller and Lazareva, 2014; Pu et al., 2016), but the knowledge of the joint effects of Cu and ZnO NPs on organisms is still lacking.
Antibiotic resistance contamination has become a worldwide challenge since nearly all classes of antibiotics and a wide range of antibiotic-resistant bacteria (ARB) and antibiotic resistance genes (ARGs) have been observed in various environmental matrices (Berglund, 2015; Amarasiri et al., 2019; Mills and Lee, 2019), among which surface water is an important reservoir of ARGs and ARB (Stoll et al., 2012; Zhang et al., 2016). In surface water, ARGs and ARB could be easily transmitted via microorganisms between humans and other animal species, and ARGs could also be spread by microbial community succession and mobile gene elements (MGEs); i.e., integrons, plasmids, and transposons (MacLean and San Millan, 2019).
There has been emerging evidence that single M-NPs such as Al2O3, TiO2, Fe2O3, CuO, and ZnO could promote antibiotic resistance spread via horizontal gene transfer (HGT) in pure culture (Qiu et al., 2012; Qiu et al., 2015; Wang et al., 2018; Zhang et al., 2019), suggesting the potential risk of ARG dissemination in the presence of M-NPs. Moreover, the metal ions released from M-NPs (e.g., Ag, CuO, ZnO) likely contribute to the promotion of conjugation frequencies, and the conjugation of ARGs facilitated by M-NPs has occurred through mechanisms including intracellular reactive oxygen species production, the SOS response, and cell membrane permeability (Wang et al., 2018; Zhang et al., 2019; Lu et al., 2020). Additionally, the natural environment is a complex heterogeneous matrix comprised of heterogenous chemical and bacterial compositions: determining the effects of M-NPs on the propagation of ARGs in a natural environment is a topic of importance. Recent studies, although limited, had reported that microbial communities and ARG profiles could be altered in various environmental matrices (e.g., wastewater, estuarine water, sludge) when exposed to specific M-NPs (e.g., Au, Ag, CuO, ZnO) (Ma et al., 2016; Metch et al., 2018; Chen et al., 2019; Zhang et al., 2020). Moreover, it is likely that the mixture of various pollutants may have a greater impact on the test organism than that of individual chemicals (Huang et al., 2018; Malandrakis et al., 2020).
Estuary and nearby coastal areas have great ecological and economic significance and often receive considerable pollutant inputs (e.g., ARGs, ARB, NPs, heavy metals, organic pollutants) from urban runoff, river runoff, and sewage outfalls (Czekalski et al., 2014; Guo et al., 2018b; Tou et al., 2021). ARG pollution in estuaries worldwide has been investigated, among which the Yangtze River Estuary is the most frequently examined in China, due to its significant economic and ecological value (Lin et al., 2015; Guo et al., 2018b). However, it is currently unclear how the presence of multiple M-NPs co-exposure influences the microbial communities and ARGs in the estuarine environment. It is crucial to better understand the ARG dissemination risk in estuarine waters under the stress of M-NP co-exposure.
In the current study, Cu- and ZnO- NPs were selected as representative M-NPs to investigate the changes in microbial community and ARG abundance influenced by the coexistence of those two NPs under environmentally relevant concentrations in the Yangtze River Estuary. Those two M-NPs were chosen on the basis that (1) Cu and Zn are heavy metals of great environmental concern that show significant correlations with the concentrations of ARGs in various environment matrices (e.g., water, sediment, and biofilm) (Ji et al., 2012; Guo et al., 2018a; Chen et al., 2020), and (2) Cu and ZnO are the most commonly used nanomaterials in the coatings, chemical sensors, catalysis, and cosmetics industries (Collins et al., 2012; Piccinno et al., 2012). In particular, ZnO NPs have been widely detected in the water and sediment of the Yangtze River Estuary (Chen et al., 2020; Tou et al., 2021). To this end, the associations among the concentrations of the released metal ions, MGEs, and ARGs as well as the microbial community were investigated. The objectives of this study were (1) to evaluate the effect of the coexistence of Cu and ZnO NPs on variations in the microbial community and ARG profiles in estuarine waters and (2) to explore the underlying mechanisms of the selection of ARGs during Cu and ZnO NPs co-exposure.
Materials and methods
Estuarine water samples
Water samples were collected from the Yangtze River Estuary in March 2018. The sampling site (longitude: 121°25′59.8” East, latitude: 31°28′52.1” North) (Figure S1) is located near a sewage outfall of a wastewater treatment plant along the Yangtze River. The specific information of water properties is presented in the Table S1.
Nanoparticle preparation
Commercial Cu NPs (50 nm, powder) and ZnO NPs (60 nm, powder) were purchased from the Chaowei Nanotechnology Co., Ltd. (Shanghai, China). The size and morphology of NPs were characterized by a transmission electron microscope (JEM-2100F, JEOL, Japan) according to our previous study (Niu et al., 2018) and the detailed information are listed in Figure S2. The 1,000 mg/L stock solutions were prepared and ultrasonically dispersed (100 W, 40 kHz) in filtration-sterilized ultrapure water for 30 min (temperature controlled by crushed ice) before use.
Experimental setup of nanoparticle exposure
The exposure experiments were conducted in conical flasks that were sequentially cleaned with tap water, ultrapure water, and acetone. Due to our previous results found that Zn2+ and Cu2+ were significantly correlated to ARGs (Guo et al., 2018b) and ZnO NPs (0.2 and 1 mg/L) could induce the selection of ARGs in this study area (Chen et al., 2019), the ZnO NPs was selected and its individual final working concentration was set to 0.5 mg/L. For comparison with ZnO NPs, the elemental Cu NPs was chosen and its concentration was also set to 0.5 mg/L. The combination group contained 0.25 mg/L of ZnO and Cu NPs, respectively and the blank control group without any NPs was set at the same time. The collected water samples were mixed first and then separated into sub-samples before use. After the preparation, the flasks with 800 mL estuarine water were placed in a shaker (150 rpm) at the temperature of 25°C in a 12 h dark/12 h light cycle. In our previous publications, the results showed an obvious time-dependent variation in ARG profiles and the microbial community over a 24 h period when exposed to ZnO NPs (Chen et al., 2019), especially within 3 h. Therefore, samples were collected at 0 h, 1 h, 3 h, 6 h, 12 h, and 24 h. The collected samples were immediately vacuum filtered through a filter membrane (0.22 μm), and then the membranes were stored at –20°C until DNA extraction. All these culture assays were conducted in triplicates. In order to eliminate artificial disturbances and to better mimic the environmental conditions, any nutrients were not added to the samples during the experiments.
Metal ion control groups
In this study, the effects of metal ions released from NPs on variations in microbial community and ARGs were investigated. When the water samples in the NP exposure experiments were collected, 5 mL subsamples were withdrawn from each treatment at a certain exposure time (0 h, 1 h, 3 h, 6 h, 12 h, and 24 h) as well. Once collected, the samples were centrifuged at 12,000 rpm for 30 min, then the supernatants were subjected to analyze the concentration of metal ions dissolved from M-NPs with an inductively coupled plasma mass spectrometry (Perkin-Elmer, NexION 350D, USA). In accordance with the detected concentrations, CuSO4·5H2O and ZnSO4·7H2O (AR, Sinopharm Chemical Reagent Co., Ltd., Shanghai, China) were prepared and used as the source of Cu2+ and Zn2+ in the metal ion controls to mimic nanoparticle dissolution. Likewise, samples on the filter membranes were taken for microbial community and ARG analysis. All the procedures were similar to those of the NP exposure experiment described in Section “Experimental Setup of Nanoparticle Exposure”, and the culture assays were conducted in triplicates.
DNA extraction and quantitative polymerase chain reaction analysis
The DNA of all the samples was isolated from the filter membranes by a soil DNA kit (Mag-Bind, Omega Bio-Tek, USA) according to the manufacturer’s instructions. Following extraction, high-quality DNA samples (A260/A280: 1.8–2.0) were stored at –20°C for further analysis of the ARGs and the microbial community. Given our previous studies in the Yangtze River Estuary, six most prevalent ARGs which consisted of two sulfonamide resistance genes (sul1 and sul2), two tetracycline resistance genes (tetA and tetW), one macrolide resistance gene (ermB) and one aminoglycoside resistance gene (aac(6’)-Ib) together with class 1 integron (intI1), conjugative transposon (Tn916/1545), zinc and copper resistance genes (zntA, zntB and copA), and 16S rRNA were chosen as target genes for quantitative PCR analysis (Guo et al., 2018a; Guo et al., 2018b). The assays were conducted in a 25 μL volume reaction that consisted of 12.5 μL of 2 × SGExcel FastSYBR mixture (with ROX) (Sangon, China), 1 μL of standard plasmid or DNA template, 0.5 μL of each primer (10 μM), and 10.5 μL of ddH2O. The primer sequences, amplification protocols, and standard curves for all target genes are listed in Tables S2, S3.
Microbial community analysis
Illumina MiSeq sequencing (Verogen, USA) was used to assess the microbial community composition of the water samples during Cu and ZnO NP exposure, and the 16S rRNA gene (V3–V4 region) was amplified with details shown in the supplementary material. Then, the amplicons were analysed on an Illumina MiSeq platform by Personal Biotechnology (Shanghai, China). The sequence data were analyzed by QIIME software (v1.8.0). Then, the resulting sequences were clustered into operational taxonomic units (OTUs) by USEARCH method with a similarity level of 97% (Edgar, 2010). The original paired Illumina MiSeq reads were submitted to the NCBI sequence Read Archive (SRA) database (accession number SRP409221).
Statistical analysis
Comparisons of individual gene abundance were done by one-way analysis of variance using SPSS 19.0 software (IBM, USA) and the Spearman correlation analysis was conducted to analyze the correlations between ARGs, MRGs, MGEs, and dissolved ions. P < 0.05 indicates significant correlation or significant difference. The heatmap analysis, the principal component analysis (PCA), and the redundancy discriminant analysis (RDA) were conducted using the R base packages. Network analysis based on a Spearman’s correlation analysis between ARGs, MRGs, MGEs and genera were performed using Gephi.
Results
Response of target ARGs to stresses of individual and combined Cu and ZnO NPs
The relative abundances of the six target ARGs in all the treatment groups ranged from –5.4 to –1.4 log10 (copies/16S rRNA) (Figure 1A). Notably, sul1 and sul2 with the relative abundance ranging from –3.1 to –1.4 log10 (copies/16S rRNA) were consistently found to be the dominant ARGs compared to the other four ARGs and accounted for 64%–90% of the six ARGs. By contrast, there was no apparent predominance concerning the relative abundances of tetA, tetW, aac(6’)-Ib, and ermB, which were in the range of –5.4 to –2.1 log10 (copies/16S rRNA) (Figure 1A).
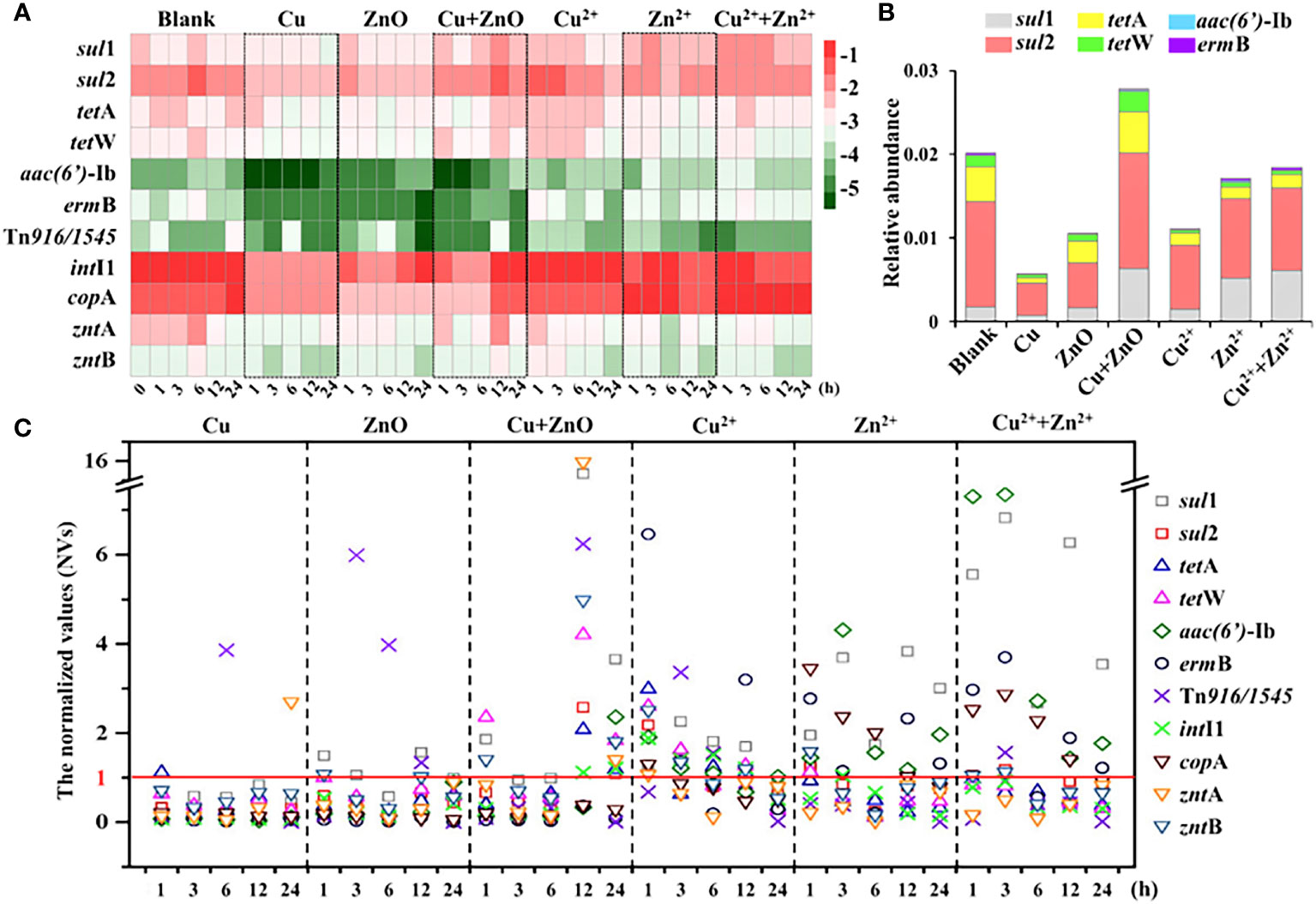
Figure 1 The abundance of target genes. (A) the relative abundance (log10 (copies/16S rRNA)) of target genes in all the samples at different exposure time; (B) the relative abundance (ARGs copies/16S rRNA copies) of ARGs at 24h; (C) The normalized values (NVs) of ARGs during exposure experiment in all the treatments. The NV >1 indicated the abundances of ARGs were enhanced when exposed to NPs or metal ions, at certain time; the NV <1 indicated the abundances of ARGs were reduced when exposed to NPs or metal ions, at certain time. The labels of Cu, ZnO and Cu+ZnO represent the groups with Cu NPs, ZnO NPs and Cu+ZnO NPs, respectively.
When exposed to Cu NPs alone, the abundances of the six ARGs all showed a decreasing trend relative to the control (Figure 1A). Notably, the normalized values (NVs) (the abundance value of the ARGs at each exposure time normalized to the corresponding blank control) were introduced to reduce the influence caused by self-variations in the microbes over time (Chen et al., 2019). During the exposure of Cu NPs, the NVs of six ARGs were almost lower than 1, and the NVs of most ARGs showed a conspicuous trend of first decreasing (within 6 h), then increasing, and then decreasing with the exposure time, except for aac(6’)-Ib, which showed a trend of an initial decrease (within 12 h) followed by an increase during the exposure (Figure 1C). A similar decreasing trend of ARGs was also found in single-ZnO NP exposure (Figure 1A). The NVs of six ARGs ranged from 0.01 to 1.56, and the NVs of most ARGs also decreased first (within 6 h), then increased and then decreased with the exposure time (Figure 1C). Notably, only the NVs (at exposure times of 1, 3, and 12 h) of sul1 were greater than 1, and the abundance of sul1 did not show significant variations compared to the control at the end of exposure (24 h) (P > 0.05) (Figure 1C). In contrast, when exposed to Cu and ZnO NPs simultaneously, the abundance of most ARGs showed a trend of first decreasing and then increasing compared to the blank (Figure 1A). With respect to the variations in ARGs present in single Cu NPs and ZnO NP exposure, most ARGs (except ermB) were enriched with the NVs in the range of 1.09 to 3.66 at the end of the exposure (24 h) under the coexistence of Cu and ZnO NPs (Figure 1B). Moreover, the relative abundance (ARGs copies/16S rRNA copies) of total ARGs at 24 h in the group of Cu and ZnO NPs was significantly higher than those with single Cu NPs or ZnO NP exposure (Figure 1B). Relative to the single NP exposure, a significant decrease (P < 0.05) in biomass (16S rRNA gene copies) was observed in the Cu and ZnO NP co-exposure group (Figure S3).
Effects of released Cu2+ and Zn2+ from Cu and ZnO NPs on ARGs variation
As shown in Figure S4, the concentrations of released Cu2+ and Zn2+ from Cu and ZnO NPs ranged from 6 to 12 μg/L and 25 to 130 μg/L respectively. According to the exposure experiments with single Cu2+ and Zn2+ at the concentration of 12 μg/L and 130 μg/L, respectively, and with Cu2+ and Zn2+ co-exposure at the concentration of 6 μg/L and 65 μg/L, respectively, the shifts of ARG abundances with the presence of Cu2+, Zn2+, and both ions are shown in Figure 1A. With respect to Cu2+ exposure alone, the abundances of most ARGs showed a trend of first increasing (within 1 h) and then decreasing compared to the blank group. In the Zn2+ exposure group, the abundances of sul2, tetA, and tetW showed a decreasing trend compared to the blank group, while the other three ARGs (sul1, aac(6’)-Ib, and ermB) showed an increasing trend. Besides, in the Cu2+ and Zn2+ co-exposure group, the trend of ARG variations was similar to that of the Zn2+. Among the three ion exposure groups, the variations in the NVs of individual ARGs fluctuated over the 24 h exposure period in all the samples, of which the NVs of three ARGs (sul1, aac(6’)-Ib, and ermB) at different exposure times were almost greater than 1 (Figure 1C). When it comes to the end of the exposure (24 h), Cu2+ was observed to cause attenuations (NVs <1) of more ARGs (sul1, sul2, tetA, tetW, and ermB) than Zn 2+ (sul2, tetA, and tetW) and Cu2+ and Zn2+ co-exposure (sul2, tetA, and tetW) did (Figures 1B, C).
Response of target MRGs and MGEs to single and combined stress of Cu and ZnO NPs
Compared with the blank group, the abundance of these three MRGs showed a downward trend in most treatment groups (Figure 1A) with NVs of less than 1 at the end of the exposure (Figure 1C). It is worth noting that under the combined exposure of Cu and ZnO NPs, the NVs of zntA and zntB were 1.4 and 1.81 at 24 h, respectively (Figure 1C) and they were comparable to the variations in most ARGs (except for ermB), of which the NVs were in the range of 1.09 to 3.66. Moreover, the correlation analysis showed that most ARGs showed significant positive correlations with three MRGs (P < 0.01) (Table S4). In contrast, Cu2+ and Zn2+ concentrations released from Cu and ZnO NPs in the estuarine water samples showed a negative correlation with some ARGs and MRGs, including sul2, aac(6’)-Ib, ermB, and copA.
In terms of the variations in two MGEs, compared with the blank group, the abundances of Tn916/1545 and intI1 also showed a decreasing trend (Figure 1C), and most of the NVs were less than 1 (Figure 1C). In particular, under the co-exposure to Cu and ZnO NPs, the intI1was slightly enriched at the end of exposure, with an NV of 1.21 (Figure 1C). Moreover, the correlation coefficients between ARGs and MGEs showed that MGEs were significantly positively correlated with most ARGs (P < 0.05) (Table S4).
Changes in microbial community under exposure of individual and combined Cu and ZnO NPs
In the current study, variations in the microbial community in estuarine waters were analyzed through Illumina MiSeq sequencing, and the results suggested that the presence of Cu NPs and ZnO NPs individually or together affected the diversity and richness of microbial communities (Table S5). Additionally, the microbial communities in these samples were scattered into two groups in the axes of PC1 (Figure S5) from the PCA analysis, denoted as Group A and Group B. In Group A there were three samples, including single Cu NPs, ZnO NPs, and Cu2+ exposure that were not separated from the control group, whereas samples in Group B were closely clustered and separated from the control group, including Cu and ZnO NP co-exposure, Cu2+and Zn2+ co-exposure, and Zn2+ exposure.
Overall, Proteobacteria, Cyanobacteria, Actinobacteria, and Bacteroidetes were the most abundant phyla, roughly accounting for 81.5% to 94.3% of the total 24 identified phyla (Figure S6). In order to investigate the variation characteries of microbes during stimuli exposure in this study, we introduced the absolute abundance metric as discussed in our previous study (Chen et al., 2019). The relative abundances of Proteobacteria increased over time (Figure S6), but the NV values decreased over time in all the treatments except for the Cu NP sole exposure (Table S6). The presence of NPs likely caused an increase in the absolute abundance of Bacteroidetes (Table S6). For Cyanobacteria, the relative abundances decreased over time (Figure S6), but Cyanobacteria was more abundant in the samples in Group A than in those of the corresponding blanks (NVs >1) (Table S6). The relative abundance of Actinobacteria in the samples in Group B showed an increasing trend over time, and the NVs were 1.19- to 4.75-fold higher than those in the corresponding blank groups (Table S6).
In order to further analyze the response of bacterial communities to M-NPs, the relative abundances of the top ten abundant genera in each treatment were selected and summarized in the heatmap. As shown in Figure 2, 27 genera were analyzed, and they were clearly divided into the above-mentioned Groups A and B. Compared with the control group, the abundance of the 12 genera (marked in red) in Group B showed an increasing trend, and most of those 12 genera belonged to the Proteobacteria and Actinobacteria phyla (Table S7). In this study, the relative abundance of Nocardioides in Group B increased significantly relative to those in the corresponding controls. In addition, the genera Panacagrimonas and Nevskia, belonging to the Proteobacteria phylum, showed an increasing trend in relative abundance that was similar to that of Nocardioides (Figure 2). In terms of the absolute abundances of Nevskia, Panacagrimonas, and Nocardioides in the samples of Group B, they also showed an increasing trend (Figure S7) with NVs ranging from 1.13 to 1.28 at the end of exposure (Table S8).
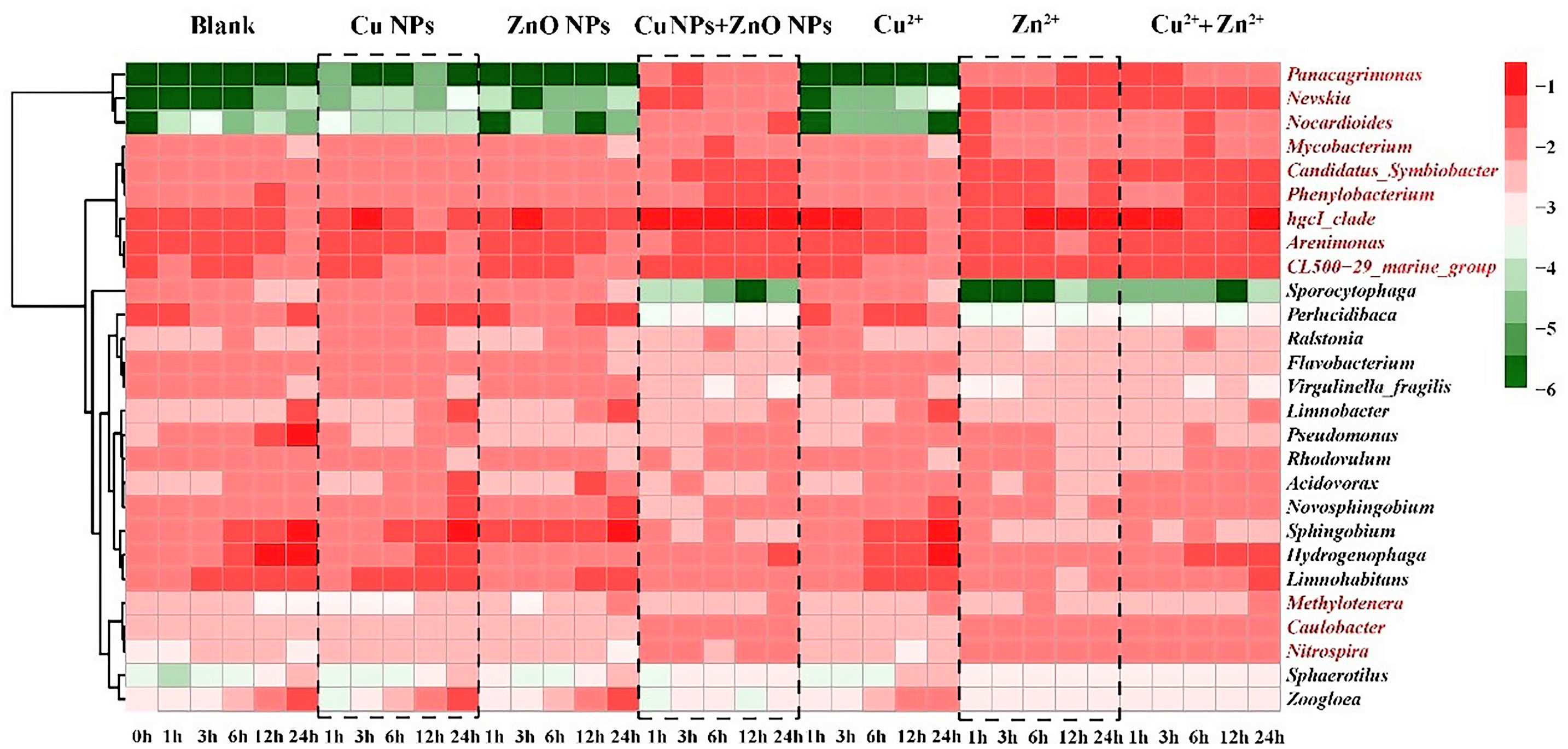
Figure 2 Heatmap of relative abundance of top 10 genera (values were log10- transformed), presenting the evolution of the microbial community in all the samples at different exposure time. The redder, the higher the values, and the bluer, the lower the values. The genera marked in red are the 12 genera with higher abundances comparing to the corresponding controls in Group B.
Factors influencing the variations in ARGs in the presence of individual and combined Cu and ZnO NPs
The RDA analysis showed that the selected variables including metal ions, MRGs, MGEs, and the microbial community (relative abundance >1%) accounted for 74% of the total ARG variables (Figure 3). To determine the key explanatory factor and to separate the contributions of selected variables (e.g., the microbial community, MGEs, MRGs, and metal ions), a partial RDA was done. In general, the microbial community contributed the most (43.5%) to the ARG profiles, followed by dissolved metal ions (25.7%), MRGs, (19.4%), and MGEs (4.4%). Considering the microbial community, among the 11 phyla, Actinobacteria (Pr, 0.039) and Acidobacteria (Pr, 0.009) were significantly related to sul1, ermB, and aac(6’)-Ib. As shown in Figure 3, intI1 (Pr, 0.001), zntA (Pr, 0.002), and zntB (Pr, 0.001) are significantly correlated with ARGs (sul2, tetA, and tetW). Regarding the network analysis, 90 potential host bacteria were identified for ARGs (Figure 4). The sul1 had the highest diversity of host bacteria, with 80 potential hosts, of which 57 bacterial genera belonged to Proteobacteria. Among the 80 potential hosts, 9 belonged to the top 10 bacterial genera summarized in Figure 2. For example, Panacagrimonas and Mycobacterium were identified as potential hosts of sul1. Another SAs-ARG, sul2, has three potential hosts. In particular, Arenimonas, one of the dominant genera in all the samples, was identified as a potential host for sul2. In addition, Chlorella sp. CC-Bw-9 was identified as a potential host for tetW.
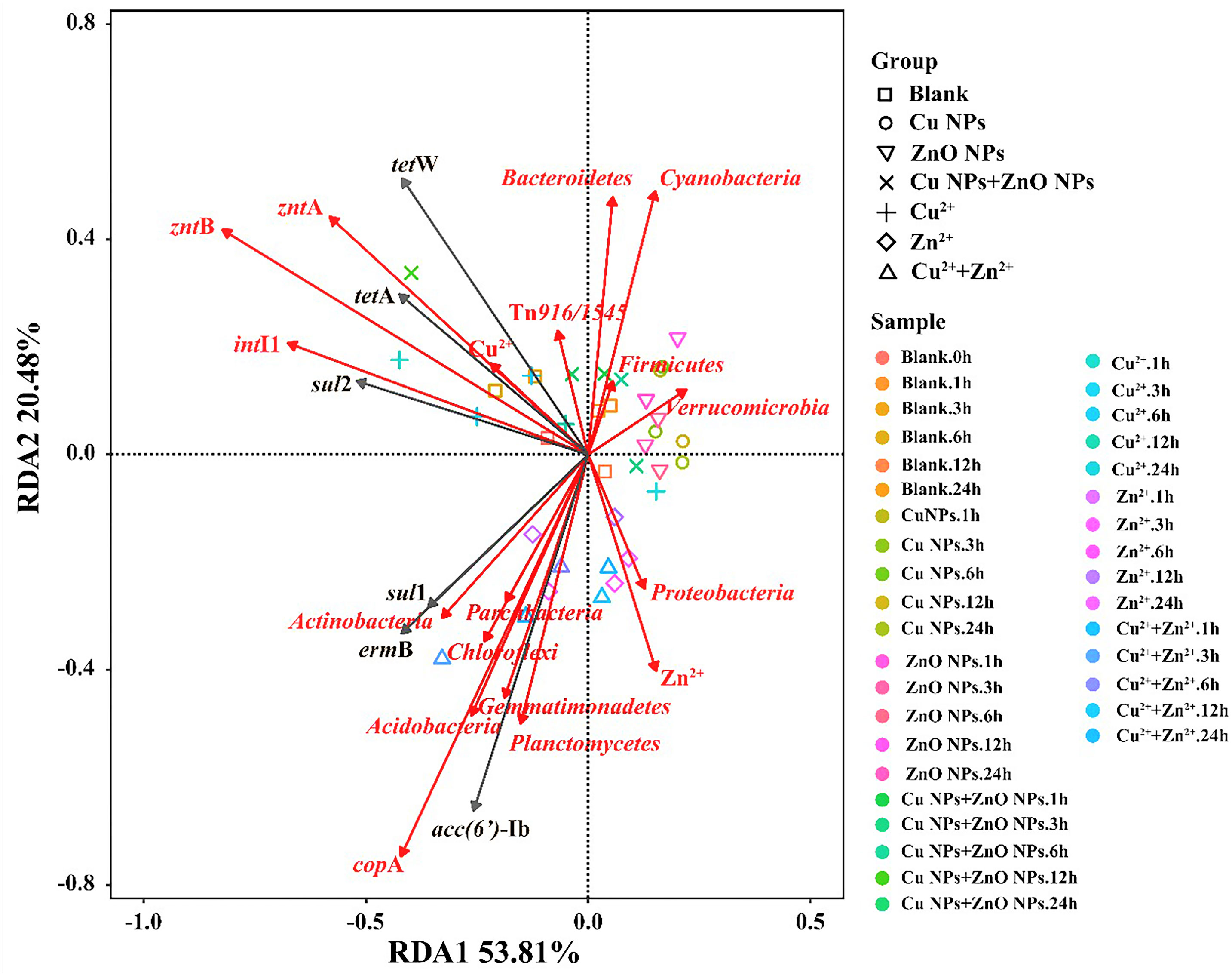
Figure 3 RDA analysis of microbial community (phylum), MGEs, MRGs, Cu2+, Zn2+ and ARG abundances in all the samples.
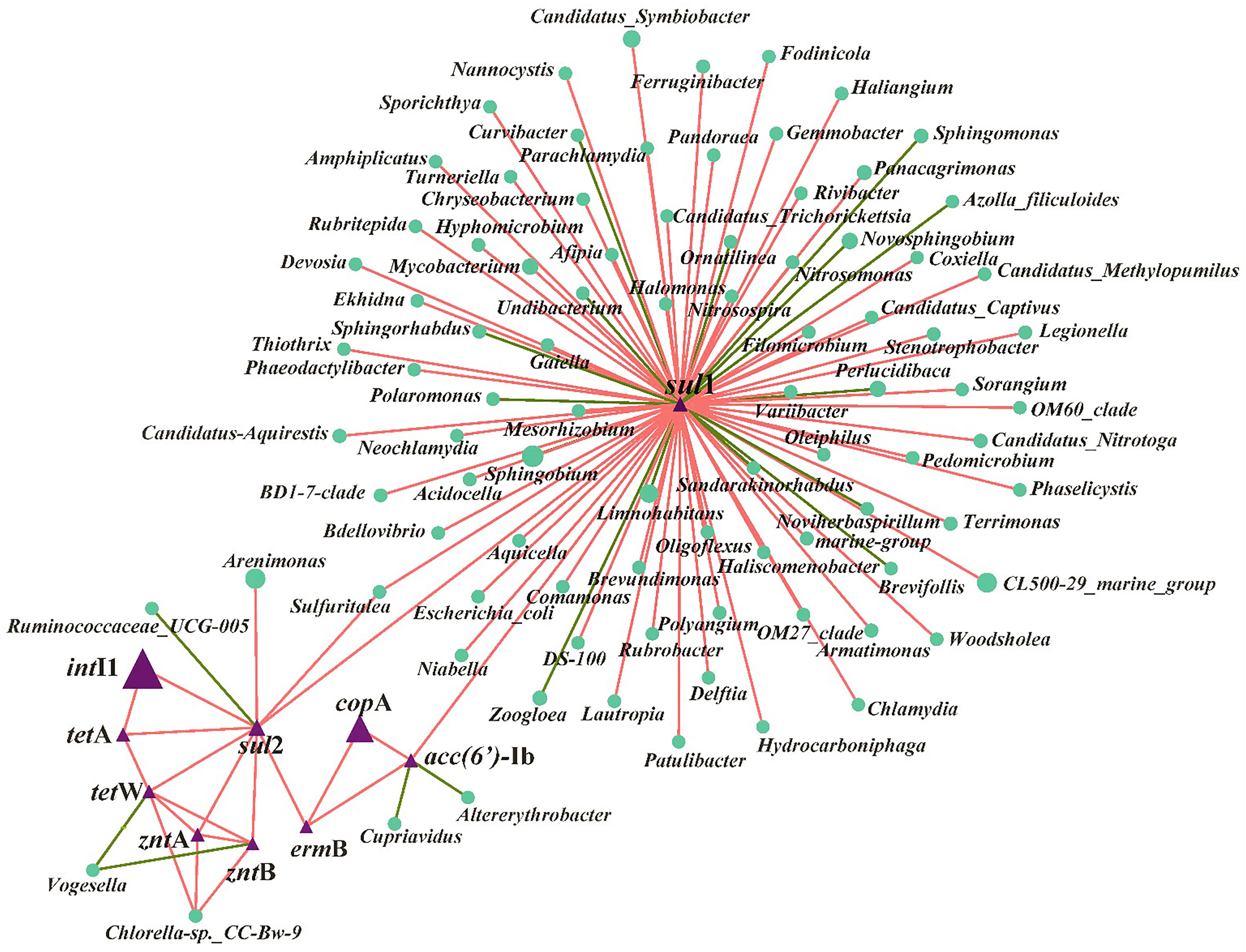
Figure 4 Network analysis based on Spearman correlation analysis among ARGs, MGEs, MRGs and their potential host bacteria in all the samples. The connection between two nodes represents a significant positive correlation (P < 0.05), the red color means positive correlation and green color means negative correlation.
Discussion
Previous studies posited that the M-NPs were involved in the shaping of ARG profiles (Chen et al., 2019; Su et al., 2019; Shi et al., 2019), but effect of M-NPs co-exposure on ARG profiles in natural environment is still lacking. In this study, effect of Cu and ZnO NPs co-exposure on ARG profiles and microbial community in the Yangtze River Estuary was investigated. Considering our previous study, the 22 ARGs corresponding to five antibiotic classes were commonly found in the waters collected from the Yangtze River Estuary (Guo et al., 2018b). Therefore, those 22 ARGs were analyzed, and the results showed that only six ARGs (sul1, sul2, tetA, tetW, ermB, and aac(6’)-Ib) were abundant in the samples; consequently, they were selected for our further study.
In the present study, the presence of Cu NPs reduced the ARGs. Previous study also reported that Cu NPs can inhibit ARG (sul1 and aadA1) dissemination in leachate (Su et al., 2019). In addition, our results suggested that the existence of ZnO NPs (at a concentration of 0.5 mg/L) caused a decrease in the ARG abundance. In previous publications, ZnO NPs with tested concentrations in a wide magnitude ranging from 0.2 mg/L (an environmentally relevant concentration) to 500 mg/L (a relatively high concentration) were observed to have an increased ARG abundance in natural waters, sludge, and landfill leachate (Chen et al., 2019; Huang et al., 2019; Shi et al., 2019), and ZnO NPs could facilitate dissemination of ARGs via enhancing the conjugative frequencies of plasmid RP4 (Wang et al., 2018). This difference may have been caused by the physicochemical differences among these environmental matrices that led to a diverse microbial community which is the major host and transmitter of ARGs and the different fates of NPs (aggregation, bioavailability, or toxicity), which would influence their effects toward microbes. Additionally, the density of donors and recipients directly influenced the formation of new transconjugants (Sorensen et al., 2005). The bacteria density in the present study was lower (by at least two orders of magnitude) than those in the previous studies, which may be resulted in the lower HGT frequencies of ARGs during the NP exposure. Nevertheless, a similar observation was found by Su et al. (2019), which implied that the ZnO NP (5, 50, and 100 mg/L) exposure prompted the rate of ARG attenuation and reduced the abundance of leachate microbiota. Considering these facts, there were multiple possible mechanisms involved in the ZnO NPs affecting ARG dynamics in various environments. Therefore, more studies are required to better understand the ARG dissemination risk in various environments under the stress of M-NP exposure.
Notably, the mixture of Cu and ZnO NPs was likely to induce the enrichment of ARGs at the exact concentration of 0.5 mg/L of NP exposure. Also, it has been reported that M-NPs, such as CuO NPs, ZnO NPs, nano-Al2O3, and nano-TiO2 exposures can enhance the conjugative transfer of ARGs, which is mainly contributed to the oxidative stresses caused by NPs or the released ions, with cell membrane permeability increasing, SOS response activation, and genes involved in conjugative transfer up-regulation (Qiu et al., 2012; Qiu et al., 2015; Wang et al., 2018). The biomass (16S rRNA gene copies) in the Cu and ZnO NP co-exposure group was significantly lower than single NP exposure (Figure S3), which could be attributed to the enhanced toxicity in the present study. Therefore, the enhanced toxicity of the Cu and ZnO NP co-exposure may have induced more oxidative stress on the microbes and stimulated the ARG transfer in the microbial community, which resulted in the ARG enrichment under the co-exposure of Cu and ZnO NPs. Furthermore, the natural aquatic environment is complex, involving multiple concentration ratios of Cu and ZnO NPs and various environmental matrices and bacterial compositions. Therefore, to determine the combined effects of multiple NPs on ARGs and the underlying mechanism in the natural environment, more studies concerning controlled single and multiple environmental factors are required.
In order to assess whether the main cause of ARG variation was the dissolved metals or the NPs, the concentrations of Cu2+ and Zn2+ released from Cu and ZnO NPs were determined in estuarine waters, and the effects of these two metal ions on ARGs abundances were investigated. The results showed that dissolved Cu2+ caused most ARG attenuations followed by Zn2+ and Cu2+ and Zn2+ co-exposure (Figures 1B, C). Previous study also observed that under the exposure of the same doses (5, 50, and 100 mg/L) of Cu or ZnO NPs, the corresponding dissolved Cu2+ reduced the ARG abundances to a greater extent than Zn2+ did (Su et al., 2019). Taken together, due to the different trends between the groups with the exposure of metal NPs and their corresponding metal ions, the dissolved metal ions from M-NPs may not be the dominant factor for ARG profiles in this study.
It has been reported that genes involved in heavy metal and antibiotic resistance are likely to be located on the same MGEs, which could promote the HGT of ARGs between microbial communities (Zhang et al., 2018a; Ding et al., 2019). Correspondingly, changes in MRGs and MGEs may indicate a variation in the ARGs. Therefore, we investigated three MRGs corresponding to Cu and Zn (Guo et al., 2018b; Adekanmbi and Adeleke, 2020) and two MGEs which were generally occurred in the estuarine environment (Chen et al., 2019). The co-selection of ARGs and MRGs may have occurred and MGEs may participate in the dissemination of these ARGs in the estuarine water in the presence of Cu and ZnO NPs individually or together in the present study. Similarly, the co-selection of heavy metal and ARGs has been commonly reported in various environments such as soil (Zhao et al., 2019), water (Zhang et al., 2018a) and gut microbiota (Ding et al., 2019).
Changes in the composition of the microbial community are closely related to the variations in the abundance of ARGs, MRGs, and MGEs (Guo et al., 2018b; Chen et al., 2019; Shi et al., 2019). The variations in the microbial community under the co-exposure of Cu and ZnO NPs were mainly attributed to the metal ion dissolution and that the Zn2+ contributed by the ZnO NPs had more significant effects than Cu2+ did. Proteobacteria, one of the dominant bacterial phyla, showed a decreasing trend in most treatment (Table S6). Likewise, previous studies have reported that abundance of Proteobacteria decreased under stress from the M-NPs (e.g., ZnO and Fe0 NP) in OCO reactor and landfill leachate (Liu et al., 2018; Shi et al., 2019). The trend of Actinobacteria changes was similar to ARGs changes, such as sul1, aac(6’)-Ib, and ermB, which implies that Actinobacteria may have been responsible for the increasing trend of ARG in the samples. These findings were also consistent with those of our previous study, which indicated that Actinobacteria are important hosts for carrying and disseminating ARGs in the Yangtze River Estuary (Chen et al., 2019). As for the response of top ten abundant genera in each treatment, most genera were found to belong to the Proteobacteria and Actinobacteria phyla (Table S7). These two bacterial phyla were considered to be important hosts of ARGs and participate in the spread of ARGs (Zhang et al., 2018b). For example, the Nocardioides genus, which belongs to the Actinobacteria phylum, has been identified as an important host of ARGs (e.g., sul1, tetA, tetW, ermB) and MGEs (intI1) in our previous studies (Guo et al., 2018b; Chen et al., 2019). Nevskia belonging to the Proteobacteria phylum was resistant to trimethoprim (Low et al., 2016). In this study, the abundance of Nocardioides, Nevskia, and Panacagrimonas (belonging to Proteobacteria phylum) showed an increasing trend in the samples of Group B, which suggests that these genera may be involved in the enrichment of ARGs in samples in Group B.
According to the RDA (Figure 3) and partial RDA analysis, the microbial community was found to be the key contributor to the variance in ARG profiles, and changes in the abundance of sul1, ermB, and aac(6’)-Ib are likely to be related to the microbes classified into Actinobacteria and Acidobacteria. Xu et al. (2020) reported that most chosen ARG numbers (e.g., sul1, sul2, tetA, ermB) were correlated to Acidobacteria in a drinking water system. The dissemination of ARGs through HGT and co-selection commonly exists in the environment (Bengtsson-Palme et al., 2018). In this study, sul2, tetA, and tetW are significantly correlated with intI1, zntA, and zntB, which indicates that the shifts of these three ARGs may be related to intI1, zntA, and zntB, which were involved in the HGT of ARGs and co-selected by metals. Accordingly, whether Cu NPs or ZnO NPs were exposed alone or together, the variance in the ARG profiles was mainly affected by the abundance of potential host bacteria and the HGT of ARG among microbial communities in estuarine water (Chen et al., 2019; Shi et al., 2019). In terms of the potential host bacteria for ARGs (Figure 4), most bacterial genera belong to Proteobacteria were identified for sul1, which indicates that the changes in the abundance of Proteobacteria may be responsible for sul1. The result was consistent with the conclusion of a previous study, which reported that Proteobacteria was significantly correlated to SAs-ARGs in a large aquaculture pond (Shen et al., 2020). As other hosts for sul1, the abundance of Panacagrimonas and Mycobacterium in the samples in Group B showed an increasing trend, which were likely corresponded to the increasing of sul1 in Group B. The results indicate that Panacagrimonas and Mycobacterium in the estuary water may be involved in the regulation of the sul1 variation. In addition, Mycobacterium was found to be significantly related to a variety of ARGs (sul1, sul2, ermB) in a pig farm (He et al., 2019). Moreover, Haliangium, one kind of moderately halotolerant bacterium able to survive extreme conditions, was found to be related to sul1, resulting in a high risk of ARG dissemination. It is well known that Escherichia coli is the most common enteric pathogen in our daily life, and antibiotic resistance in E. coli is of particular concern (Adekanmbi and Adeleke, 2020). One previous study has reported that multi-resistant E. coli is widespread in tropical estuaries and India, and more than 95% of the isolated strains are resistant to more than three antibiotics (Chandran et al., 2008). In this study, it was found that E. coli was significantly related to sul1. Therefore, the presence of such pathogenic bacteria in natural waters will enhance the direct or indirect risks of human infection. The abundance of Arenimonas in the samples in Group A showed a decreasing trend over time, whereas the abundance of Arenimonas showed a trend of first decline and then arise, in the samples in Group B, which implies that Arenimonas may be related to the variations in sul2 during the NP exposure. Cyanobacteria was demonstrated to act as a significant reservoir for diverse ARGs (e.g., tetA, tetW) dissemination in aquatic environments (Wang et al., 2020). Consistently, Chlorella sp. CC-Bw-9 belonging to Cyanobacteria was identified as a potential host for tetW.
Conclusions
It is unclear the concentrations of Cu and ZnO NPs that are present in the estuarine environment, however, the production of NPs is exponentially increasing, making possible higher inputs into the aquatic environments in the future. In this study, the effects of Cu and ZnO NPs exposure individually or combined under environmentally relevant concentrations on the changes in ARG abundance and microbial composition in estuarine waters were investigated. It was found that under the exact doses (0.5 mg/L) of NP exposure, Cu NPs or ZnO NPs exposed alone mainly resulted in the attenuation of target ARGs in the estuarine water, while a mixture of Cu NPs and ZnO NPs was likely to induce the enrichment of ARGs. Further analysis indicated that changes in the microbial community were the dominant factors driving ARG propagation, followed by dissolved metal ions, MRGs, and MGEs. Network analysis showed that most potential hosts belonged to the Proteobacteria and Actinobacteria phyla, such as Mycobacterium and E. coli., which are likely to be pathogenic to humans and animals. Therefore, in contrast to individual Cu- and ZnO-NPs, combined Cu and ZnO NPs seemed to induce the increase of ARGs, which would increase the risk of ARG transmission into different environmental niches including normal human intestinal flora.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI Sequence Read Archive, SRP409221.
Author contributions
Y-RC: Methodology, validation, formal analysis, investigation, data curation, writing - original draft, writing - review & editing, visualization. R-RS: Investigation. X-LS: Investigation. X-PG: Formal analysis, investigation, data curation, writing - review & editing, supervision, project administration, funding acquisition. YY: Writing - review & editing, supervision. All authors contributed to the article and approved the submitted version.
Funding
This study was funded by the National Natural Science Foundation of China (42107384, 42125102). Additional funding for this work was provided by the China Postdoctoral Science Foundation (2019M661426) and the Shanghai Post-doctoral Excellence Program (2019066).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.1086606/full#supplementary-material
References
Adam N., Leroux F., Knapen D., Bals S., Blust R. (2014). The uptake of ZnO and CuO nanoparticles in the water-flea Daphnia magna under acute exposure scenarios. Environ. pollut. 194, 130–137. doi: 10.1016/j.envpol.2014.06.037
Adekanmbi A. O., Adeleke O. J. (2020). Occurrence of metal and antibiotic resistant Escherichia coli harbouring zntA and copA genes in selected surface water in ibadan, south-west Nigeria. Int. J. Environ. Stud. 77, 876–885. doi: 10.1080/00207233.2020.1719804
Amarasiri M., Sano D., Suzuki S. (2019). Understanding human health risks caused by antibiotic resistant bacteria (ARB) and antibiotic resistance genes (ARG) in water environments: Current knowledge and questions to be answered. Crit. Rev. Environ. Sci. Technol. 50, 2016–2059. doi: 10.1080/10643389.2019.1692611
Barjhoux I., Baudrimont M., Morin B., Landi L., Gonzalez P., Cachot J. (2012). Effects of copper and cadmium spiked-sediments on embryonic development of Japanese medaka (Oryzias latipes). Ecotoxicol. Environ. Saf. 79, 272–282. doi: 10.1016/j.ecoenv.2012.01.011
Bengtsson-Palme J., Kristiansson E., Larsson D. G. J. (2018). Environmental factors influencing the development and spread of antibiotic resistance. FEMS Microbiol. Rev. 42, 68–80. doi: 10.1093/femsre/fux053
Berglund B. (2015). Environmental dissemination of antibiotic resistance genes and correlation to anthropogenic contamination with antibiotics. Infect. Ecol. Epidemiol. 5, 28564. doi: 10.3402/iee.v5.28564
Bundschuh M., Filser J., Luderwald S., McKee M. S., Metreveli G., Schaumann G. E., et al. (2018). Nanoparticles in the environment: where do we come from, where do we go to? Environ. Sci. Eur. 30, 6. doi: 10.1186/s12302-018-0132-6
Chandran A., Hatha A. A. M., Varghese S., Sheeja K. M. (2008). Prevalence of multiple drug resistant Escherichia coli serotypes in a tropical estuary, India. Microbes Environ. 23, 153–158. doi: 10.1264/jsme2.23.153
Chen Y. R., Guo X. P., Feng J. N., Lu D. P., Niu Z. S., Tou F. Y., et al. (2019). Impact of ZnO nanoparticles on the antibiotic resistance genes (ARGs) in estuarine water: ARG variations and their association with the microbial community. Environ. Sci. Nano 6, 2405–2419. doi: 10.1039/C9EN00338J
Chen Y. R., Guo X. P., Niu Z. S., Lu D. P., Sun X. L., Zhao S., et al. (2020). Antibiotic resistance genes (ARGs) and their associated environmental factors in the Yangtze estuary, China: From inlet to outlet. Mar. pollut. Bull. 158, 111360. doi: 10.1016/j.marpolbul.2020.111360
Collins D., Luxton T., Kumar N., Shah S., Walker V. K., Shah V. (2012). Assessing the impact of copper and zinc oxide nanoparticles on soil: a field study. PloS One 7, 42663. doi: 10.1371/journal.pone.0042663
Czekalski N., Gascon D. E., Burgmann H. (2014). Wastewater as a point source of antibiotic-resistance genes in the sediment of a freshwater lake. ISME J. 8, 1381–1390. doi: 10.1038/ismej.2014.8
Ding J., An X. L., Lassen S. B., Wang H. T., Zhu D., Ke X. (2019). Heavy metal-induced co-selection of antibiotic resistance genes in the gut microbiota of collembolans. Sci. Total Environ. 683, 210–215. doi: 10.1016/j.scitotenv.2019.05.302
Edgar R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461.
Fernaíndez-Cruz M. L., Lammel T., Connolly M., Conde E., Barrado A. I., Derick S., et al. (2013). Comparative cytotoxicity induced by bulk and nanoparticulated ZnO in the fish and human hepatoma cell lines PLHC-1 and hep G2. Nanotoxicology 7, 935–952. doi: 10.3109/17435390.2012.676098
Guo H. H., Gu J., Wang X. J., Tuo X. X., Yu J., Zhang R. R. (2018b). Key role of cyromazine in the distribution of antibiotic resistance genes and bacterial community variation in aerobic composting. Bioresour. Technol. 274, 418–424. doi: 10.1016/j.biortech.2018.12.005
Guo X. P., Yang Y., Lu D. P., Niu Z. S., Feng J. N., Chen Y. R., et al. (2018a). Biofilms as a sink for antibiotic resistance genes (ARGs) in the Yangtze estuary. Water Res. 129, 277–286. doi: 10.1016/j.watres.2017.11.029
He L. Y., He L. K., Liu Y. S., Zhang M., Zhao J. L., Zhang Q. Q., et al. (2019). Microbial diversity and antibiotic resistome in swine farm environments. Sci. Total Environ. 685, 197–207. doi: 10.1016/j.scitotenv.2019.05.369
Huang H., Chen Y., Yang S., Zheng X. (2019). CuO and ZnO nanoparticles drive the propagation of antibiotic resistance genes during sludge anaerobic digestion: possible role of stimulated signal transduction. Environ. Sci. Nano 6, 528–539. doi: 10.1039/C8EN00370J
Huang W., Wang C., Duan H., Bi Y., Wu D., Du J., et al. (2018). Synergistic antifungal effect of biosynthesized silver nanoparticles combinedwith fungicides. Int. J. Agric. Biol. 20, 1225–1229. doi: 10.17957/IJAB/15.0595
Hua J., Vijver M. G., Ahmad F., Richardson M. K., Peijnenburg W. J. (2014). Toxicity of different-sized copper nano- and submicron particles and their shed copper ions to zebrafish embryos. Environ. Toxicol. Chem. 33, 1774–1782. doi: 10.1002/etc.2615
Ji X., Shen Q., Liu F., Ma J., Xu G., Wang Y., et al. (2012). Antibiotic resistance gene abundances associated with antibiotics and heavy metals in animal manures and agricultural soils adjacent to feedlots in shanghai; China. J. Hazard. Mater. 235-236, 178–185. doi: 10.1016/j.jhazmat.2012.07.040
Keller A. A., Lazareva A. (2014). Predicted releases of engineered nanomaterials: from global to regional to local. Environ. Sci. Technol. Lett. 1, 65–70. doi: 10.1021/ez400106t
Lin L., Yuan K., Liang X. M., Chen X., Zhao Z. S., Yang Y., et al. (2015). Occurrences and distribution of sulfonamide and tetracycline resistance genes in the Yangtze river estuary and nearby coastal area. Mar. pollut. Bull. 100, 304. doi: 10.1016/j.marpolbul.2015.08.036
Liu Y., Baas J., Peijnenburg W. J., Vijver M. G. (2016). Evaluating the combined toxicity of Cu and ZnO nanoparticles: utility of the concept of additivity and a nested experimental design. Environ. Sci. Technol. 50, 5328–5337. doi: 10.1021/acs.est.6b00614
Liu Z., Zhou H., Liu J., Huang M., Yin X., Liu Z., et al. (2018). Evaluation of performance and microbial community successional patterns in an integrated OCO reactor under ZnO nanoparticle stress. RSC Adv. 8, 26928–26933. doi: 10.1039/C8RA05057K
Low A., Ng C., He J. Z. (2016). Identification of antibiotic resistant bacteria community and a GeoChip based study of resistome in urban watersheds. Water Res. 106, 330–338. doi: 10.1016/j.watres.2016.09.032
Lu J., Wang Y., Jin M., Yuan Z., Bond P., Guo J. (2020). Both silver ions and silver nanoparticles facilitate the horizontal transfer of plasmid-mediated antibiotic resistance genes. Water Res. 169, 115229. doi: 10.1016/j.watres.2019.115229
MacLean R. C., San Millan A. (2019). The evolution of antibiotic resistance. Science 365, 1082–1083. doi: 10.1126/science.aax3879
Malandrakis A. A., Kavroulakis N., Chrysikopoulos C. V. (2020). Synergy between Cu-NPs and fungicides against botrytis cinerea. Sci. Total. Environ. 703, 135557. doi: 10.1016/j.scitotenv.2019.135557
Ma Y., Metch J. W., Yang Y., Pruden A., Zhang T. (2016). Shift in antibiotic resistance gene profiles associated with nanosilver during wastewater treatment. FEMS Microbiol. Ecol. 92, 8. doi: 10.1093/femsec/fiw022
Metch J. W., Burrows N. D., Murphy C. J., Pruden A., Vikesland P. J. (2018). Metagenomic analysis of microbial communities yields insight into impacts of nanoparticle design. Nat. Nanotechnol. 13, 253–259. doi: 10.1038/s41565-017-0029-3
Mills M. C., Lee J. (2019). The threat of carbapenem-resistant bacteria in the environment: Evidence of widespread contamination of reservoirs at a global scale. Environ. pollut. 255, 113143. doi: 10.1016/j.envpol.2019.113143
Niu Z. S., Pan H., Guo X. P., Lu D. P., Feng J. N., Chen Y. R., et al. (2018). Sulphate-reducing bacteria (SRB) in the Yangtze estuary sediments: abundance, distribution and implications for the bioavailibility of metals. Sci. Total Environ. 634, 296–304. doi: 10.1016/j.scitotenv.2018.03.345
Piccinno F., Gottschalk F., Seeger S., Nowack B. (2012). Industrial production quantities and uses of ten engineered nanomaterials in Europe and the world. J. Nanopart. Res. 14, 1–11. doi: 10.1007/s11051-012-1109-9
Pu Y., Tang F., Adam P. M., Laratte B., Ionescu R. E. (2016). Fate and characterization factors of nanoparticles in seventeen subcontinental freshwaters: A case study on copper nanoparticles. Environ. Sci. Technol. 50, 9370–9379. doi: 10.1021/acs.est.5b06300
Qiu Z., Shen Z., Qian D., Jin M., Yang D., Wang J., et al. (2015). Effects of nano-TiO2 on antibiotic resistance transfer mediated by RP4 plasmid. Nanotoxicology 9, 895–904. doi: 10.3109/17435390.2014.991429
Qiu Z., Yu Y., Chen Z., Jin M., Yang D., Zhao Z., et al. (2012). Nanoalumina promotes the horizontal transfer of multiresistance genes mediated by plasmids across genera. Proc. Natl. Acad. Sci. U. S. A. 109, 4944–4949. doi: 10.1073/pnas.1107254109
Shen X. X., Jin G. Q., Zhao Y. J., Shao X. H. (2020). Prevalence and distribution analysis of antibiotic resistance genes in a large-scale aquaculture environment. Sci. Total Environ. 711, 134626. doi: 10.1016/j.scitotenv.2019.134626
Shi J. H., Su Y. L., Zhang Z. J., Wei H. W., Xie B. (2019). How do zinc oxide and zero valent iron nanoparticles impact the occurrence of antibiotic resistance genes in landfill leachate? Environ. Sci. Nano 6, 2141–2151. doi: 10.1039/C9EN00068B
Song L., Connolly M., Fernaíndez-Cruz M. L., Vijver M. G., Fernaíndez M., Conde E., et al. (2014). Species-specific toxicity of copper nanoparticles among mammalian and piscine cell lines. Nanotoxicology 8, 383–393. doi: 10.3109/17435390.2013.790997
Sorensen S. J., Bailey M., Hansen L. H., Kroer N., Wuertz S. (2005). Studying plasmid horizontal transfer in situ: a critical review. Nat. Rev. Microbiol. 3, 700–710. doi: 10.1038/nrmicro1232
Stoll C., Sidhu J. P., Tiehm A., Toze S. (2012). Prevalence of clinically relevant antibiotic resistance genes in surface water samples collected from Germany and Australia. Environ. Sci. Technol. 46, 9716–9726. doi: 10.1021/es302020s
Su Y. L., Wu D., Xia H. P., Zhang C. Y., Shi J. H., Wilkinson K. J., et al. (2019). Metallic nanoparticles induced antibiotic resistance genes attenuation of leachate culturable microbiota: The combined roles of growth inhibition, ion dissolution and oxidative stress. Environ. Int. 128, 407–416. doi: 10.1016/j.envint.2019.05.007
Tou F., Wu J., Fu J., Niu Z., Liu M., Yang Y. (2021). Titanium and zinc-containing nanoparticles in estuarine sediments: Occurrence and their environmental implications. Sci. Total. Environ. 754, 142388. doi: 10.1016/j.scitotenv.2020.142388
Turan N. B., Erkan H. S., Engin G. O., Bilgili M. S. (2019). Nanoparticles in the aquatic environment: usage, properties, transformation and toxicity-a review. Process Saf. Environ. Prot. 130, 238–249. doi: 10.1016/j.psep.2019.08.014
Wang Z., Chen Q., Zhang J., Guan T., Chen Y., Shi W. (2020). Critical roles of cyanobacteria as reservoir and source for antibiotic resistance genes. Environ. Int. 144, 106034. doi: 10.1016/j.envint.2020.106034
Wang Z., Wang S., Peijnenburg W. (2016). Prediction of joint algal toxicity of nano-CeO2/nano-TiO2 and florfenicol: Independent action surpasses concentration addition. Chemosphere 156, 8–13. doi: 10.1016/j.chemosphere.2016.04.072
Wang X. L., Yang F. X., Zhao J., Xu Y., Mao D. Q., Zhu X., et al. (2018). Bacterial exposure to ZnO nanoparticles facilitates horizontal transfer of antibiotic resistance genes. NanoImpact 10, 61–67. doi: 10.1016/j.impact.2017.11.006
Xu L., Campos L. C., Canales M., Ciric L. (2020). Drinking water biofiltration: Behaviour of antibiotic resistance genes and the association with bacterial community. Water Res. 182, 115954. doi: 10.1016/j.watres.2020.115954
Zhang X., Chen Z., Ma Y., Zhang N., Wei D., Zhang H., et al. (2020). Response of partial nitrification sludge to the single and combined stress of CuO nanoparticles and sulfamethoxazole antibiotic on microbial activity, community and resistance genes. Sci. Total. Environ. 712, 135759. doi: 10.1016/j.scitotenv.2019.135759
Zhang Y., Gu A. Z., Cen T., Li X., He M., Li D., et al. (2018a). Sub-Inhibitory concentrations of heavy metals facilitate the horizontal transfer of plasmid-mediated antibiotic resistance genes in water environment. Environ. pollut. 237, 74–82. doi: 10.1016/j.envpol.2018.01.032
Zhang L., Gu J., Wang X. J., Zhang R. R., Tuo X. X., Guo A. Y., et al. (2018b). Fate of antibiotic resistance genes and mobile genetic elements during anaerobic co-digestion of Chinese medicinal herbal residues and swine manure. Bioresour. Technol. 250, 799–805. doi: 10.1016/j.biortech.2017.10.100
Zhang S., Wang Y., Song H., Lu J., Yuan Z., Guo J. (2019). Copper nanoparticles and copper ions promote horizontal transfer of plasmid-mediated multi-antibiotic resistance genes across bacterial genera. Environ. Int. 129, 478–487. doi: 10.1016/j.envint.2019.05.054
Zhang X. H., Xu Y. B., He X. L., Huang L., Ling J. Y., Zheng L., et al. (2016). Occurrence of antibiotic resistance genes in landfill leachate treatment plant and its effluent-receiving soil and surface water. Environ. pollut. 218, 1255–1261. doi: 10.1016/j.envpol.2016.08.081
Keywords: Cu nanoparticles, ZnO nanoparticles, co-exposure, antibiotic resistance genes, microbial community
Citation: Chen Y-r, Sha R-r, Sun X-l, Guo X-p and Yang Y (2023) Combined effect of Cu- and ZnO- NPs on antibiotic resistance genes in an estuarine water. Front. Mar. Sci. 9:1086606. doi: 10.3389/fmars.2022.1086606
Received: 01 November 2022; Accepted: 29 December 2022;
Published: 16 January 2023.
Edited by:
Sujin Kim, Baylor University, United StatesReviewed by:
Sivalingam Periyasamy, National Research Council (CNR), ItalyIlunga Kamika, University of South Africa, South Africa
Copyright © 2023 Chen, Sha, Sun, Guo and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xing-pan Guo, eHBndW9AZ2VvLmVjbnUuZWR1LmNu
 Yu-ru Chen
Yu-ru Chen Rong-rong Sha2
Rong-rong Sha2 Xing-pan Guo
Xing-pan Guo