- 1Key Laboratory of Marine Environment and Ecology, Ministry of Education, Ocean University of China, Qingdao, China
- 2Laboratory for Marine Ecology and Environmental Science, Pilot National Laboratory for Marine Science and Technology (Qingdao), Qingdao, China
- 3College of Environmental Science and Engineering, Ocean University of China, Qingdao, China
- 4Frontiers Science Center for Deep Ocean Multispheres and Earth System, and Key Laboratory of Marine Chemistry Theory and Technology, Ministry of Education, Ocean University of China, Qingdao, China
- 5School of Resources & Environment and Safety Engineering, University of South China, Hengyang, China
Introduction: Denitrification and anammox play the crucial role for N removal processes in coastal ecosystems, but the ecological distribution of denitrifying and anammox microorganisms and their N removal rates in the Yangtze Estuary and its adjacent sea are required in-depth analysis.
Methods: Here, we utilized high-throughput sequencing, qPCR, and 15N isotope to reveal the community structure and function of denitrifying and anammox microorganisms in the surface sediments from Yangtze Estuary and adjacent sea.
Results: The results suggested that the gene abundances of nirS and nirK for denitrifiers were higher than AMX 16S rRNA for anammox bacteria. The genera composition of nirS- and nirK-encoding denitrifiers communities showed different distribution patterns. Furthermore, Candidatus Anammoximicrobium dominated the anammox community, implying the anammox oxidation capacity of the other genera should be noted in marine sediments.
Discussion: Compared to anammox, denitrification was the dominant contributor of nitrogen removal process and contributed 73.5% on average. The sediment Chla was the key factor to regulate denitrification and anammox rates, indicating the fresh organic matter was more labile and easier to be utilized by NOx− removal processes.
Introduction
Nitrogen is a necessary nutrient element for all kinds of life. However, due to the rapid expansion of various anthropogenic N input, numerous estuaries and adjacent seas have high nitrogen loading, causing serious environmental problems such as eutrophication globally (Howarth, 2008), increased harmful algal blooms (Gobler, 2020), biodiversity loss (Worm et al., 2006), and increasing nitrous oxide (N2O) emission (Murray et al., 2015; Lin and Lin, 2022). Excess anthropogenic N inputs have severely affected N-balance in coastal ecosystems (Zhao et al., 2020b), thus, N pollution has been widely concerned in global.
Both denitrification and anammox are the critical pathways in microbially-mediated N removal processes, which can remove excess nitrogen in estuarine and coastal ecosystems. Under hypoxic or anaerobic conditions, denitrification convert nitrate (NO3−) to N2O or N2 into the atmosphere, while anammox can oxide ammonium (NH4+) with nitrite (NO2−) to N2 (Thamdrup and Dalsgaard, 2002; Medhi et al., 2017). It has been reported that ~45% of reactive N can be removed by denitrification in estuarine sediments (Seitzinger et al., 2006), which was considered as the main N removal process in aquatic ecosystems before the discovery of anammox (Trimmer et al., 2003; Fernandes et al., 2012). In 1995, the anammox process was found in a wastewater treatment plant (Mulder et al., 1995). Additionally, it has also been detected in marine sediments, freshwater sediments, and anaerobic water column (Thamdrup and Dalsgaard, 2002; Trimmer and Nicholls, 2009; Lisa et al., 2014). Some studies showed that anammox was the main N removal process in the marine environments, contribution up to 67% of N2 production (Dalsgaard and Thamdrup, 2002; Kuypers et al., 2003). Hou et al. (2015) reported that 20.7% of inorganic nitrogen can be removed by anammox in coastal wetland ecosystems in China, reflecting a non-negligible role of anammox to N removal in marine ecosystems. However, denitrification and anammox have varied in their contribution to N removal in marine ecosystems, low anammox rates and even lower denitrification rates were found in most sites, while high denitrification rates were detected in a few isolated places (Babbin et al., 2014). C/N ratio was the crucial environmental factor to regulate the balance between denitrification and anammox, resulting in the different ratios of denitrification to anammox (Babbin et al., 2014). In addition, the two nitrogen removal processes have different effects on N2O emissions, as well on C mineralization (Ravishankara et al., 2009; Babbin et al., 2020). Therefore, further explorations about denitrification and anammox are critical for maintaining and conserving the health of coastal ecosystems.
Different molecular markers have been broadly utilized to understand and identify denitrifying and anammox microorganisms. Two nitrite reductases (cytochrome cd 1 and copper-containing nitrite reductases) catalyze NO2− to form a gaseous product NO, which are the key enzymes in the denitrification process (Beman, 2014). The genes (nirS or nirk) encoding nitrite reductase are widespread and efficient biomarker for precise and direct identification of denitrifying communities in numerous environments, such as the aquatic ecosystems and sediments (Abell et al., 2010; Mosier and Francis, 2010; Xie et al., 2020). The16S rRNA gene and the other functional genes (hzo and hzs) are both used to investigate and detect the community structure and abundances of anammox bacteria (Harhangi et al., 2012; Yang et al., 2020). Due to high specificity and good amplification efficiency, 16S rRNA gene was broadly employed to identify anammox bacteria in diverse environments and enrichment cultures (Li et al., 2010; Yang et al., 2020). In addition, the abundance, community composition and rates of denitrification and anammox can be regulated by multiple environmental parameters, for instance, salinity (Bernhard et al., 2005), pH (Nicol et al., 2008), nitrogen availability (Di et al., 2010), and sediment total organic carbon (TOC) (Zheng et al., 2019). Thus, it is essential to common research of the diversity, abundance, and activity of denitrification and anammox under different geographic environmental factors, to better understand the contribution and role of N removal via denitrification and anammox, and further reveal the niche of denitrifiers and anammox bacteria in coastal ecosystems.
The Yangtze Estuary and its adjacent sea, the East China Sea (ECS), was chosen as the research region to investigate the characteristics of denitrifiers and anammox bacteria. Yangtze River, the greatest river in Asian regions, can transport amounts of anthropogenic nitrogen via estuary region to adjacent ECS (Chen et al., 2016). Increasing nitrogen has resulted in significant eutrophication and the appearance of toxic algal blooms in coastal ecosystem (Smith and Schindler, 2009; Anderson et al., 2012). In addition, the Yangtze River also transports most terrestrial sediments through currents into the ECS (Lim et al., 2007). The benthic sediments serve as the significant sink for reactive nitrogen and play the vital role in the N biogeochemical cycle (Lin et al., 2017). Meanwhile, the complex benthic environment favors the growth and survival of microorganisms (Basha et al., 2018). Liu and Wang demonstrated that the diversity of nirK- gene-encoded encoding denitrifiers was lower than that of nirS-encoding denitrifiers by high-throughput sequencing within sediments from northern ECS (Liu et al., 2018; Wang et al., 2019). Lin et al. (2017) confirmed that denitrification rates were higher than anammox via 15N isotope tracer incubation experiment in surface sediment from Yangtze Estuary and its adjacent sea. Currently, molecular detection is used to understand the microbial community, and 15N isotope tracer is used to study the denitrification and anammox rates. However, few studies have focused on common research of the community structure, abundance, and activity of anammox and denitrifiers bacteria in coastal ecosystems, to better understand the role of denitrifiers and anammox bacteria in N removal processes.
Herein, we studied the characteristics of denitrifiers and anammox bacteria and environmental parameters in surface sediments (0–5 cm) from the Yangtze Estuary and its adjacent sea. The community structures and abundances were analyzed using the high-throughput sequencing and qPCR, as well as the rates of denitrification and anammox were tested by 15N isotope tracer technology. The main objective of this work is to reveal possible roles of denitrifiers and anammox bacteria in nitrogen removal processes as well as how these microorganisms respond to various environmental factors in coastal ecosystems.
Materials and methods
Sampling collection and environmental factor analysis
The bottom water and surface sediments have been gathered from 16 stations in the Yangtze Estuary and adjacent sea from 1 to 6 March 2022 (Figure 1). At each station, surface sediments (0–5 cm) was extracted from the box cores, mixed homogeneously, and placed into air-tight sterile plastics bags. The sediment samples were then subdivided into two parts for different analyses. One part immediately stored at 4°C for denitrification and anammox rates incubation. Another was stored on board (-20°C) and then transferred to laboratory storage (−80°C) for DNA extraction and molecular analysis. The bottom water was filtered through a filter (0.45 μm) and stored in polyethylene bottles (-20°C). The salinity was determined onsite by HQ40d Water Analyzer (Hach, Loveland, USA). The sediment water content was calculated using wet-dry weight method (Klute, 1986). The NH4+, NO3− and PO43− in bottom water were analyzed using QuAAtro nutrient autoanalyzer (Seal Analytical, Germany). 2 M KCl was used to extract the exchangeable NH4+, NO2− and NO3− from the sediments and then determined by QuAAtro nutrient autoanalyzer (Seal Analytical, Germany). Sediment TN and TOC were detected by the elemental analyzer (Vario EL cube, Germany). A laser particle sizer analyzer (LS 13320, Beckman, USA) was used to measure sediment grain size (MФ). The sediment Fe(II) and Fe(III) contents were measured by phenanthroline spectrophotometric method (Lovley and Phillips, 1987). Under the dark condition of 4°C for 24 hours, Sediment Chla can be extracted with 80% (v/v) acetone. A UV-5200 visible spectrophotometer was used to measure the absorbance of Chla after centrifuging these extracts at 2200 g for 10 min.
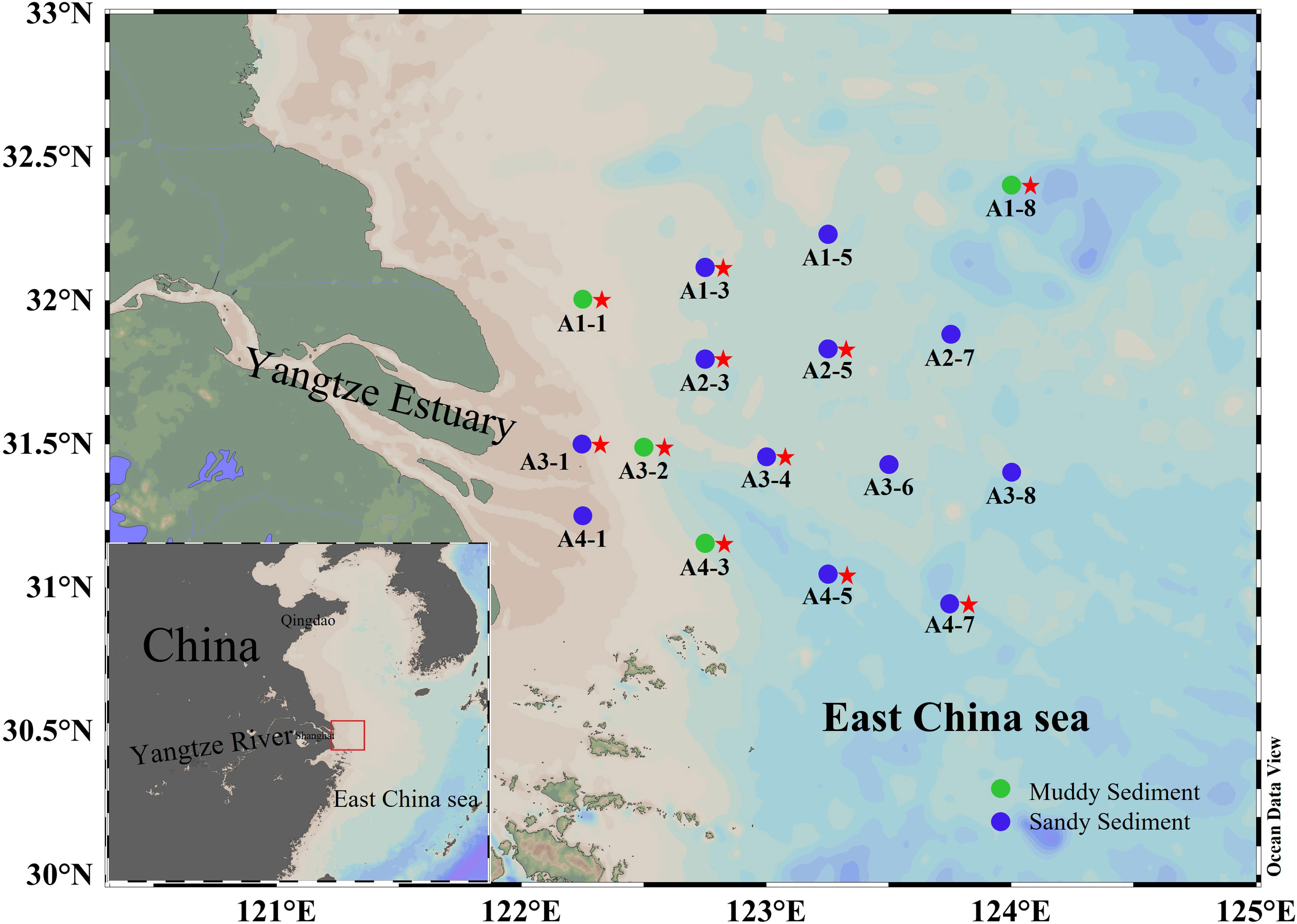
Figure 1 The map of sediment sampling stations. The dots represented sampling sites (green dots denoted muddy sediments and blue dots denoted sandy sediments). The red stars represent that the key functional genes were analyzed by high-throughput sequencing in these sampling sites.
Measurement of denitrification and anammox rates
The potential denitrification and anammox rates were measured by 15N isotope-tracing technique (Lin et al., 2017). Briefly, the laboratory slurry incubation experiments were made with the in situ benthic sterilized seawater and the sediments at a seawater/sediment mass ratio of 7:1, and the homogeneous mixture was fed with helium gas for 30 min before transferring into 12-mL vials (Labco Exetainers, UK). Subsequently, these 12-mL vials were in dark preincubated for 24 h to eliminate residual dissolved oxygen, nitrite, and nitrate. After the preincubation, three treatments were given to these vials as follows: (1) 15 NH4+ (100 μmol N L−1), (2) 15 NO3− (100 μmol N L−1), and (3) 15 NH4+ + 14 NO3− (100 μmol N L−1). After 0 h and 8 h incubation, these vials were spiked with ZnCl2 solution (200-μL 50%) to stop microbial activity. Produced 29N2 and 30N2 concentrations during incubation were determined by the membrane inlet mass spectrometry (MIMS, Hiden Analytical Ltd, UK), and denitrification and anammox rates were calculated according to previous methods (Thamdrup and Dalsgaard, 2002; Song et al., 2016).
DNA extraction and quantification for denitrifiers and anammox bacteria
Total DNA extraction was performed from 0.5 g of homogenized sediment samples by PowerSoil® DNA Isolation Kit (MoBio, USA). The obtained DNA was used as the template for real-time qPCR analysis with the primer pairs cad3Af/R3cd for the nirS gene, and FlaCu/R3Cu for nirK gene, and Amx-808-F/Amx-1040-R for Anammox 16S rRNA gene (Supplementary Table 1). Reactions were carried out using ABI PRISM®7500 Sequence Detection System (Applied Biosystems, Foster City, CA, USA). The 20-μL qPCR mixture reaction system contained FastStart Universal SYBR Green Master (ROX) (10 μL, Roche, Germany), ddH2O (6.6 μL), each primer (0.6 μL, 10 μM), bovine serum albumin (0.2 μL, 20 mg·mL−1), and template (2.0 μL). The qPCR reactions of nirS, nirK, and Anammox 16S rRNA gene were performed as the following: 50°C for 2 minutes, 95°C for 10 minutes, 45 cycles of 95°C for 30 seconds, and 58°C for 40 seconds, 72°C for 60 seconds (Throbäck et al., 2004; Hamersley et al., 2007). The significant linear relationships (R2 > 0.996) were produced with a 10-fold dilution series (102–109 copies) of standard plasmids containing the key functional genes, and the qPCR efficiency was 84%, 110%, and 92%, respectively. The abundance of the key functional genes can be calculated by the standard curves. The melting curves contained a single peak, and non-specific amplification was not found by agarose gel electrophoresis, demonstrating amplification specificity.
High-throughput sequencing
11 sampling stations have been picked from 16 stations according to the class of muddy and sandy sediments (the value of median grain size (> 64 μm) was classified to sandy sediment). The PCR amplicons of key functional genes were from the same primer pairs with qPCR (cd3aF/R3cd and FlaCu/R3Cu for denitrifiers, and Amx-808-F/Amx-1040-R for anammox bacteria). The PCR conditions were as follows: initial denaturation was performed at 98°C for 2 min, followed by control at 98°C for 15 s, 55°C for 30 s and 72°C for 30 s (25-30 cycles), and finally extended at 72°C for 5 min. The Vazyme VAHTSTM DNA Clean Beads (Vazyme, Nanjing, China) and Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen, Carlsbad, CA, USA) were used for the purification and quantification of PCR amplicons, respectively. The Illumina novaSeq PE250 platform (Shanghai, China) was used for the paired-ended sequenced of high-throughput sequencing of nirS/nirK and AMX 16S rRNA genes. The QIIME2 (2019.4) was used to incorporate, quality-filter, and dereplicate the sequences. High quality sequences with 97% similarity level were clustered into OTUs based on Vsearch (v2.13.4_linux_x86_64), and then representative OTUs were output. The raw sequences obtained in this study for nirS, nirK and AMX 16S rRNA was kept in NCBI under the accession numbers of PRJNA891470, PRJNA891480, and PRJNA891491.
Statistical analyses
The map of the sampling stations and the distribution of environmental parameters were drawn using Ocean Data View (ODV 5.2.1). The differences of nirS, nirK and AMX 16S rRNA gene abundances in the same station were studied using one-way analysis of variance (ANOVA). Correlation between the three microbial communities and environmental factors were identified with CCA/RDA analyses with Monte Carlo tests by Canoco 5 software. The Pearson and Spearman correlation analysis of denitrifiers and anammox bacteria with environmental parameters was created by IBM SPSS statistic 26 and R (version 4.2.1, R package corrplot) (Wei and Simko. 2021). MEGA11, TBtools and Origin 2022b software were used for graphics. Finally, the phylogenetic tree was visualized and beautified by Evolview online tool (http://www.evolgenius.info/evolview-v2/).
Results
Station characteristics
The physicochemical parameters of bottom water and surface sediments were given in Figure 2. The salinity ranged from 27.3 to 37.1 (Figure 2A). Bottom water NH4+ (0.1–3.15 μM), NO3− (3.55–23.93 μM), PO43− (0.21–0.71 μM) values varied across different sampling sites (Figures 2B–D). The average concentrations of NH4+ and NO3− in the sediments varied from 0.32–1.20 μg N g–1, 0.04–0.86 μg N g–1, respectively (Figure 2E, F). The sediment Fe(II)/Fe(III) ratios were in a range of 0.29–6.19 (Figure 2G). The contents of sediment TN and TOC ranged from 0.23 to 1.19 mg N g−1 with an average of 0.51 ± 0.25 mg N g−1 and from 1.05 to 6.97 mg C g−1 with an average of 2.62 ± 1.65 mg C g−1, respectively (Figures 2H, I). The sediment median grain size showed a greater change. The sampling stations of A1-1, A1-8, A3-2, and A4-3 were muddy sediments (8.83–44.27 μm), while other stations were sandy sediments (93.32–333.41 μm) (Figure 2L). Meanwhile, the C/N ratios (TOC/TN) were higher in muddy sediments (A1-1, A1-8, A3-2, and A4-3) than in sandy sediments (Figure 2J). The average content of Chla in sediment was in a range of 0.12–1.75 μg g−1, with obvious spatial pattern (Figure 2K).
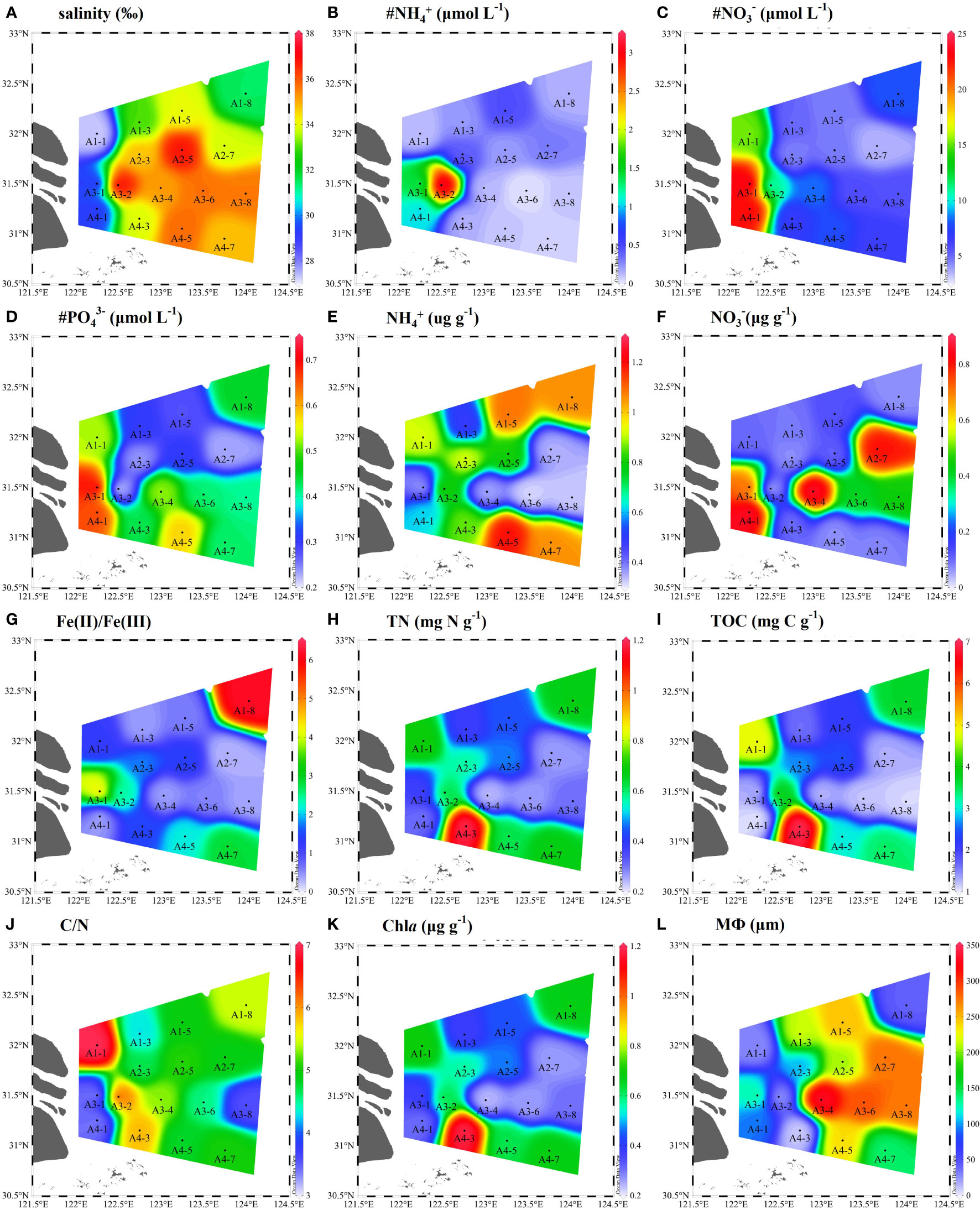
Figure 2 Spatial distributions of physicochemical parameters in bottom water and surface sediments of the Yangtze Estuary and East China Sea (ECS) (A-L). # represents the bottom water properties (# NH4+, # NO3−, # PO43−) and MФ denote median grain size.
Abundances of denitrifiers and anammox bacteria and their influencing factors
The nirS/nirK gene and AMX 16S rRNA gene were utilized to quantify denitrifiers and anammox bacteria abundances in surface sediments, respectively. The abundances of nirS gene were ranging between 9.87 × 105 to 2.71 × 107 copies g−1 with the mean value of (1.28 ± 0.93) × 107 copies g−1 (Figure 3A, B). The nirS gene abundance had negative correlation with NO3− concentration in the bottom water (r = -0.576, p< 0.05) (Figure 4). The nirK gene abundances ranged from 4.64 × 105 to 1.26 × 107 copies g−1 with the mean value of (4.52 ± 3.34) × 106 copies g−1 (Figure 3A, B). Compared to nirS/nirK gene, the abundances of AMX 16S rRNA gene were obtained between 1.03× 105 to 2.97 × 106 copies g−1with the mean value of (1.40 ± 0.84) × 106 copies g−1 (Figure 3A, B). In all surface sediment samples, nirS abundances were the highest, followed by nirK abundances and then AMX 16S rRNA gene.
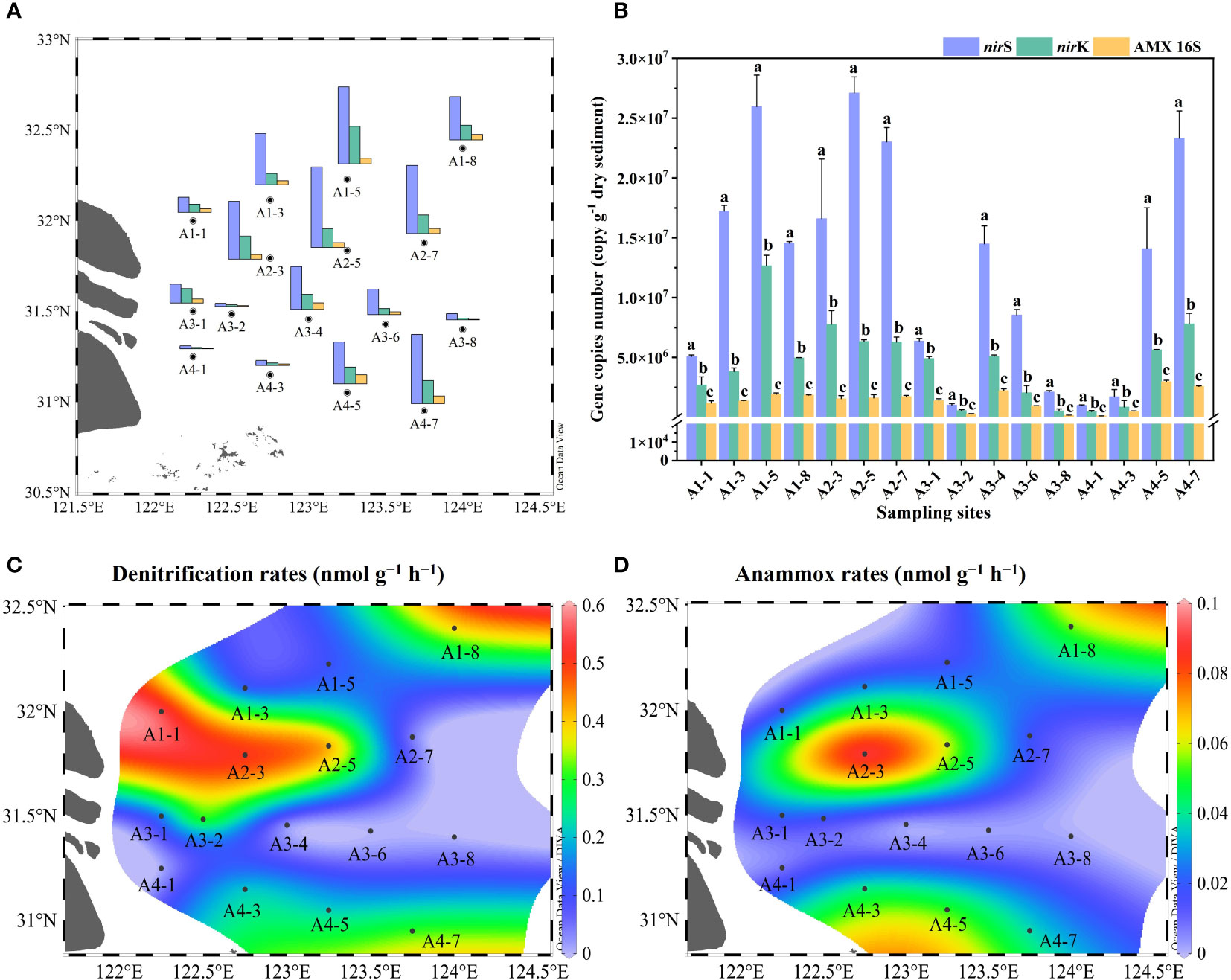
Figure 3 (A) Spatial distributions of gene abundances in the Yangtze Estuary and the adjacent sea; (B) Gene copies numbers of nirS, nirK and 16S rRNA. Error bars in the column represented the standard deviations of the triplicate qPCR reactions, and different letters indicated a significant difference between gene abundances in the same sampling sites (p< 0.05); (C, D) Denitrification and anammox rates.
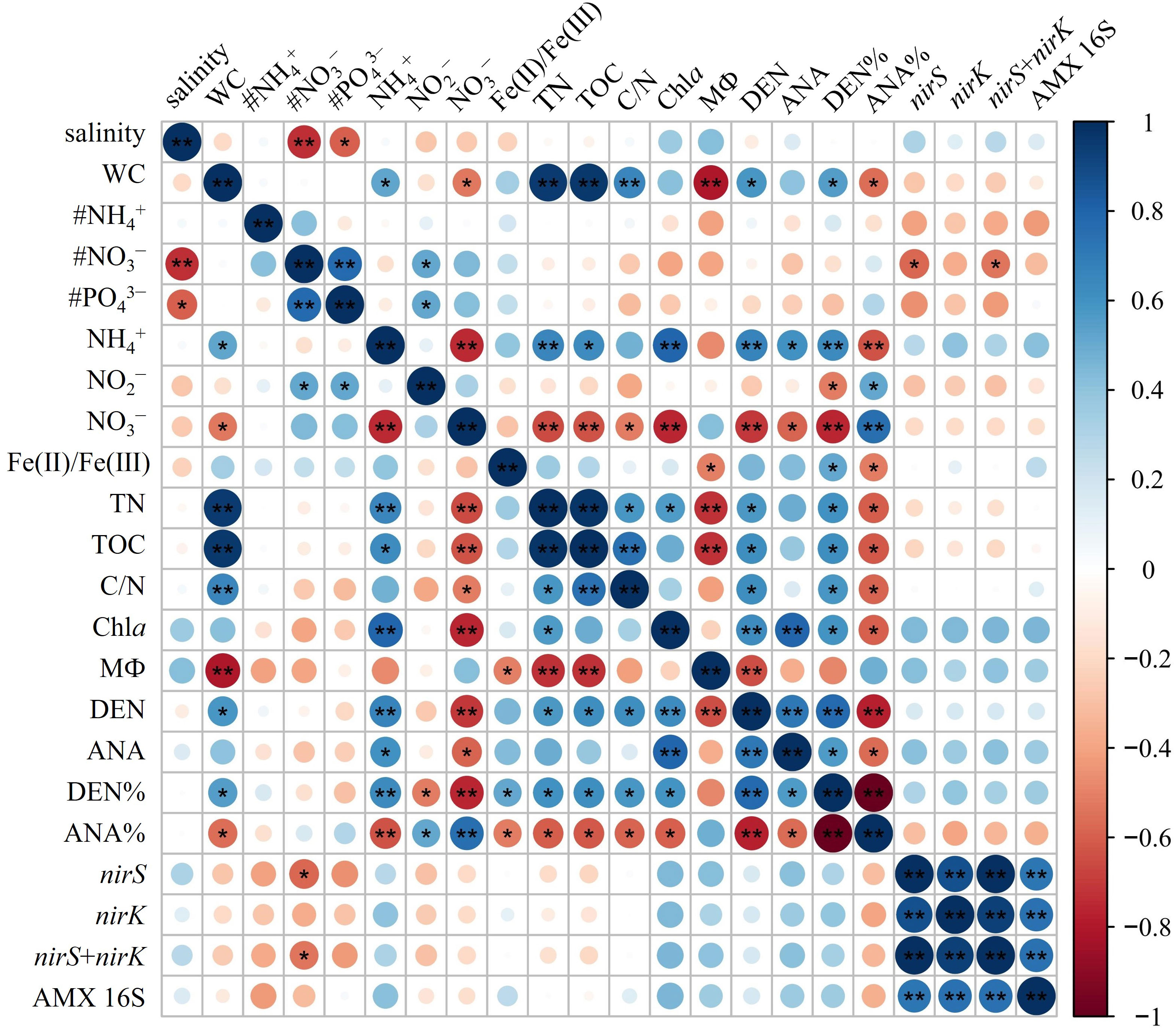
Figure 4 Pearson’s correlations of environmental factors with the denitrification/anammox and related gene abundances. * significant at p< 0.05, ** significant at p< 0.01. # represents bottom water parameter (# NH4−, # PO3−, # PO43−). WC, water content; Chla, Chlorophyll a; DEN, denitrification rates; ANA, anammox rates; DEN%, the contribution of denitrification to N removal; ANA%, the contribution of anammox to N removal.
Potential rates of denitrification and anammox and their influencing factors
The potential rates of denitrification and anammox were analyzed in all sampling site of the surface sediments (n = 16), which varied considerably, ranging from 0–0.56 nmol N g−1 h−1 and 0–0.091 nmol N g−1 h−1, respectively (Figure 3C, D). The highest denitrification rate was detected at A1-1, whereas the lowest denitrification rate was observed at A3-8. The potential denitrification rates had positive association with sediment water content, NH4+, TN, TOC, C/N, and Chla, while negatively regulated by sediment NO3− and sediment grain size (Figure 4, Supplementary Table 5). Compared to denitrification rates, the highest anammox rate was detected at A2-3, whereas the lowest anammox rate was observed at A3-4. Anammox was positively correlated with NH4+ and Chla, and negatively correlated with NO3− (Figure 4, Supplementary Table 5). Moreover, Pearson’s correlation analyses showed that anammox was significantly related to denitrification (r = 0.710, p< 0.01) (Figure 4, Figure 5A; Supplementary Table 5). The contribution of denitrification to N removal (DEN%) was higher than anammox in most sampling sites except A3-4, A3-6, A3-8, and A4-1 (Figure 5B), with the contribution of 73.5% on average. In addition, DEN% was positively related to water content, NH4+, Fe(II)/Fe(III), TN, TOC, C/N, and Chla, and negatively regulated by NO2−.
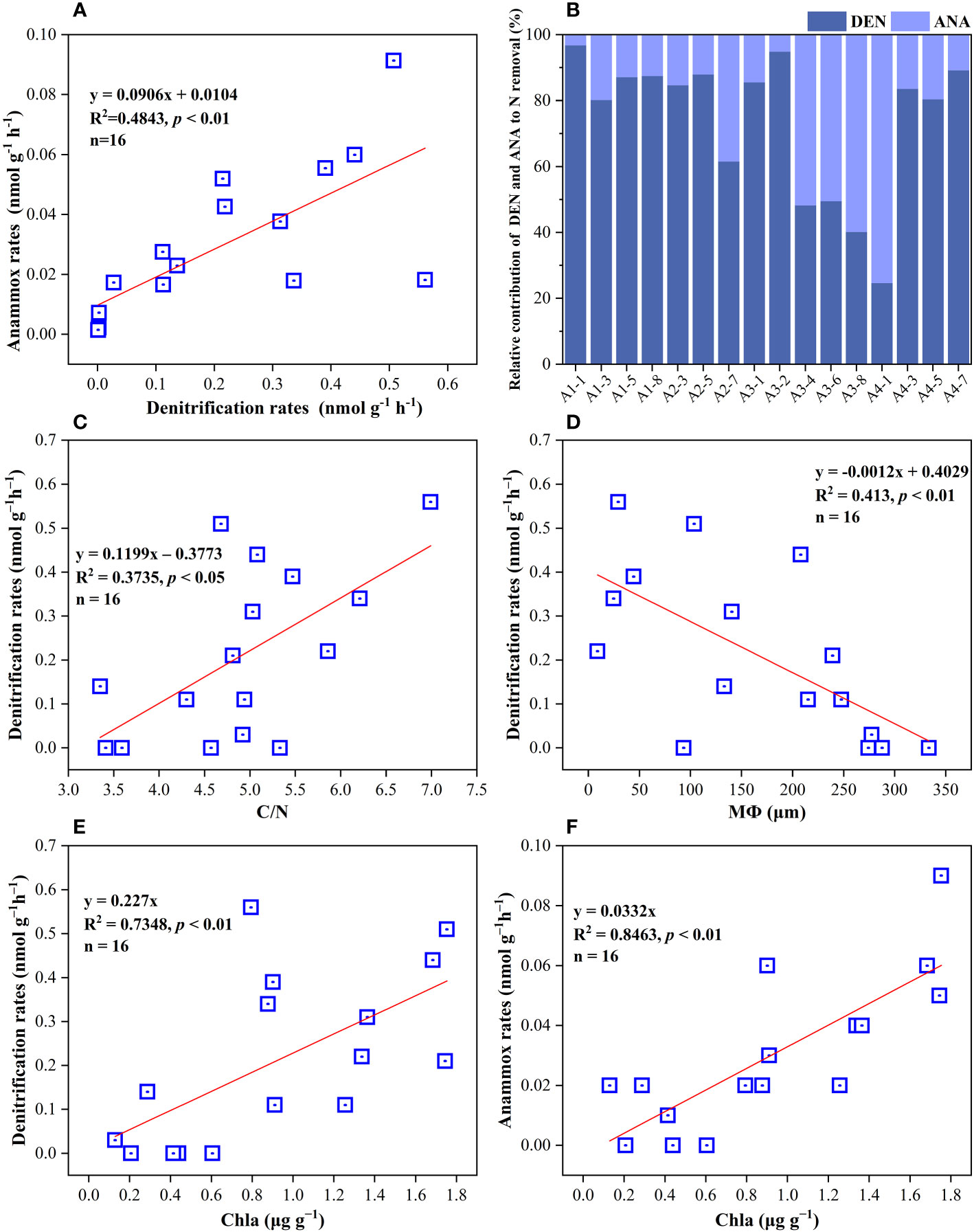
Figure 5 (A) Correlations analysis of anammox rates with denitrification rates in surface sediments. (B) The percentage of denitrification and anammox to NOx− removal in all sampling sites. (C–F) their regulating factors in sediments of the Yangtze Estuary and adjacent sea.
Community diversity and composition of nirS- and nirK-encoding denitrifiers
Totally, 1,219,102 filtered, high-quality nirS sequences were acquired from 11 surface sediments samples, ranging from 73,783–172,218 for each sample (Supplementary Table 2). The effective sequences were clustered into 7,536 OTUs based on the 97% similarity level. Additionally, Good’s coverage (99.73–99.91%) indicated that most of the nirS-encoding denitrifiers had been obtained. The highest OTU numbers (1,186) and Chao1 richness estimators (1267.16) were observed in the samples A4-7. Higher Shannon diversity index and Simpson indices were found in samples A2-3, A4-5 and A4-7 (Supplementary Table 2). Moreover, the diversity of nirS gene decreased with the increasing the NH4+ concentration in the bottom water (Shannon: r = -0.732, p< 0.05; Simpson: r = -0.897, p< 0.01) (Supplementary Table 6). Compared to the nirS gene, 763,468 high-quality reads of nirK gene sequences were acquired and 3,151 OTUs were identified (Supplementary Table 3). The diversity and richness of nirK-encoding were lower than nirS-encoding denitrifiers in all samples. In additionally, the richness of nirK gene declined with the increasing NO3− concentrations in the bottom water (Chao1: r = -0.606, p< 0.05) and increased with the sediment total nitrogen concentration (Chao1: r = 0.603, p< 0.05) (Supplementary Table 6). The rarefaction curves showed that the majority of the nirS- and nirK-encoding denitrifiers was captured in the current study (Supplementary Figures 1A, B).
A total of 50 dominant OTUs of the nirS gene were phylogenetically classified into 10 different clusters (Figures 6A, B; Supplementary Figures 2, 3). Within these 10 clusters, only C9 belonged to α-proteobacteria and C5, C6, and C7 affiliated to β-Proteobacteria, whereas the other 6 clusters were belonged to γ-Proteobacteria. C3 only contained OTU01, which was existed exclusively in the A3-2 station, closely affiliating to Pseudomonas (Figures 6A, B, Supplementary Figure 3). C8 (belonging to Marinobacter), C9 (belonging to Paracoccus) and C10 (belonging to Chromohalobacter) were observed almost in muddy sediments (Figures 6A, B). According to a redundancy analysis (RDA) with the dominant OTUs, the first two axes (RDA1 and RDA2) explained 79.97% of the cumulative variance in the nirS-encoding denitrifiers communities (Figure 6C). The bottom water NH4+ (pseudo-F = 3.1 and p< 0.01), NO3− (pseudo-F = 2.7 and p< 0.01), and C/N (pseudo-F = 2.6 and p< 0.05) were the major factors influencing the nirS gene community. At genus levels, Pseudomonas was dominant genera only in A3-2, while Marinobacter was dominant genera and observed in all sampling stations (Supplementary Figure 4B). Spearman’s correlation analysis showed that Pseudomonas was positively related with in the bottom water (p< 0.01) (Supplementary Table 7), and Marinobacter was positively correlated with sediment NH4+ (p< 0.05).
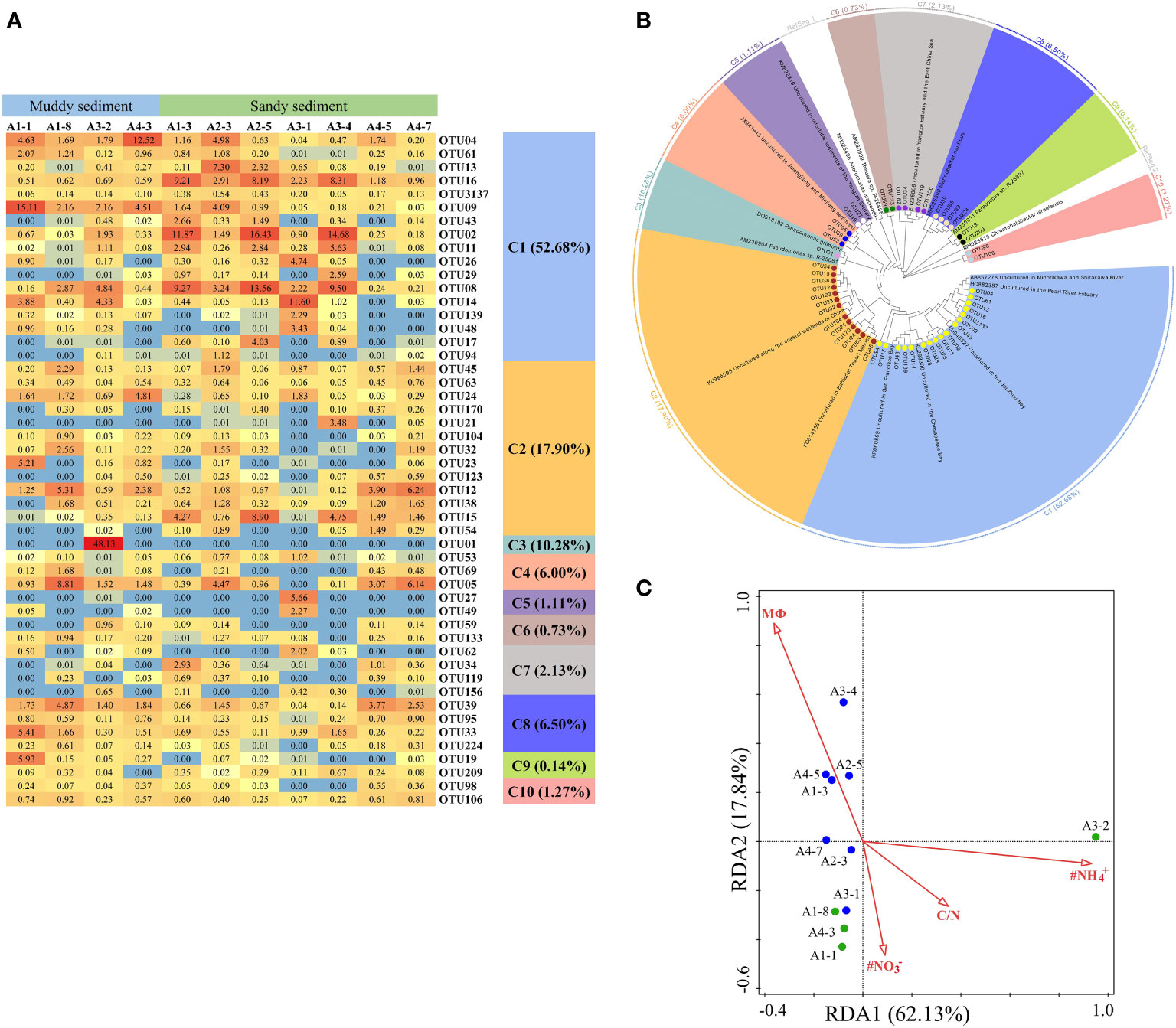
Figure 6 (A) The heat map of the dominant denitrification OTUs for the nirS gene (top 50 OTUs, 97% cutoff). (B) Reconstructed phylogenetic tree with neighbor-joining method of partial nirS-gene sequences (bootstrap 1000). (C) The redundancy analysis (RDA) for environmental factors and the nirS gene-encoded denitrifying communities.
Dominant OTUs (top 50 OTUs) of nirK gene were classified into 9 clusters, all belonging to the Proteobacteria (Figures 7A, B, Supplementary Figures 5, 6). Among the 9 clusters, C2, C5, and C7 belonged to the β-Proteobacteria, whereas the other 6 clusters were intimately related to the α-Proteobacteria (Figure 7B, Supplementary Figure 6). Moreover, C7 (with the lowest abundances of top 50 OTUs) only contained OTU32 which was detected in the A3-2 and A3-4 sample, showing some similarities with the sediment denitrifying microorganisms in the Yangtze lakes (Jiang et al., 2017). There were no obvious differences in the relative abundance of OTUs between muddy sediments and sandy sediments (Figure 7A). From the CCA (Figure 7C), the first two axes explained 36.10% of the cumulative variance, and NH4+ in the bottom water (pseudo-F = 2.0 and p< 0.01), PO43− (pseudo-F = 2.1 and p< 0.05), sediment NO3− (pseudo-F = 2.2 and p< 0.05), and MФ (pseudo-F = 2.0 and p< 0.05) had significant influences on the nirK-encoding denitrifiers community. At genus level, Mesorhizobium was the dominant genera in A2-5 and A3-1 sample with no other genera observed, however, Bradyrhizobium was detected in most sampling stations except A2-5 and A3-1 (Supplementary Figure 7B). Spearman’s correlation analysis showed that Mesorhizobium was positively related with sediment Fe(Ⅱ)/Fe(III) (p< 0.05) (Supplementary Table 7).
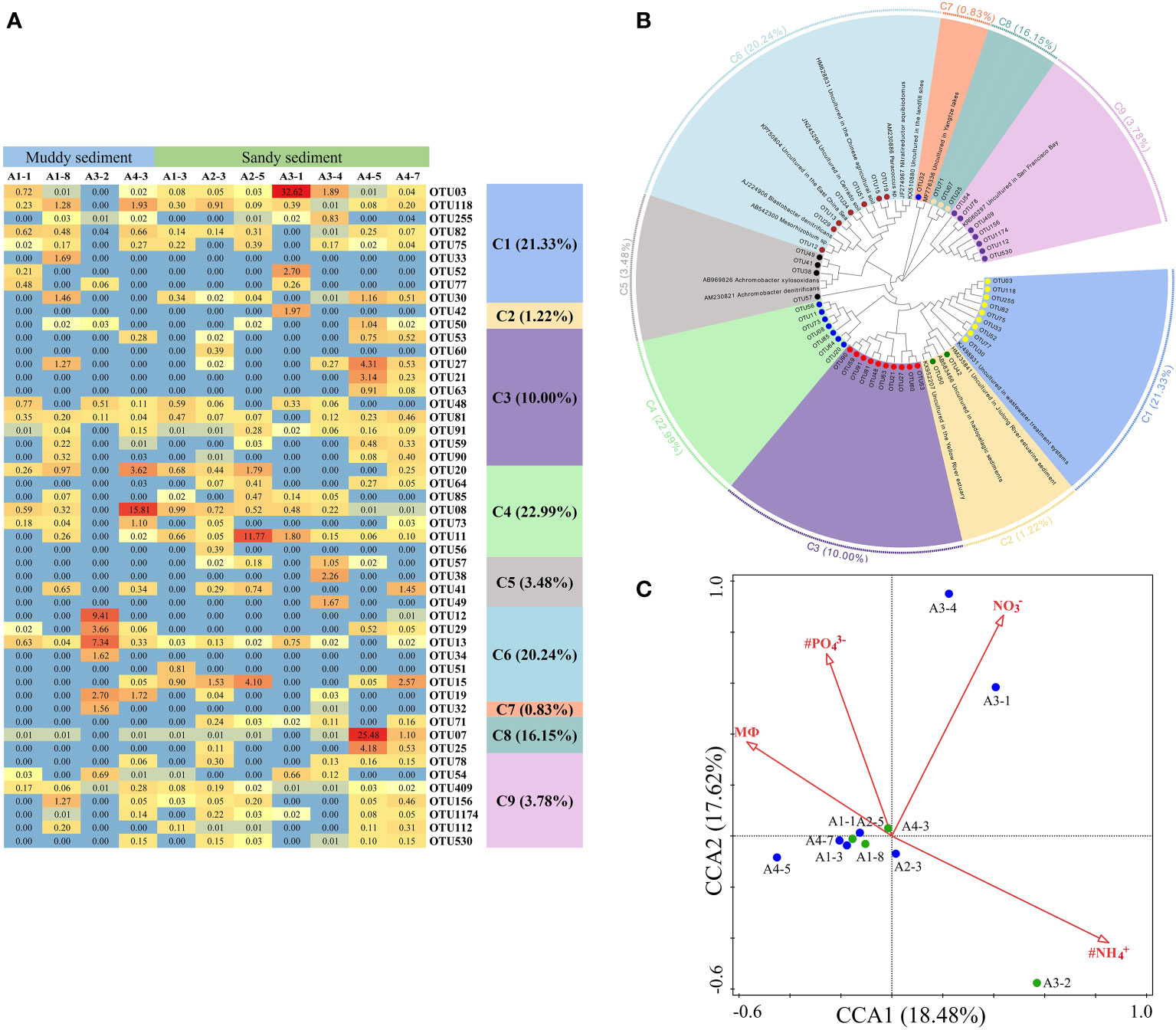
Figure 7 (A) The heat map of the dominant denitrification OTUs for the nirK gene (top 50 OTUs, 97% cutoff). (B) Reconstructed phylogenetic tree with neighbor-joining method of nirK-gene sequences (bootstrap 1000). (C) The canonical correspondence analysis (CCA) for environmental factors and the nirK gene-encoded denitrifying communities.
Community diversity and composition of anammox bacteria
For the anammox 16S rRNA gene (AMX 16S), a total of 479,390 high-quality reads were gained, which ranged from 40,134–47,445 for each sample (Supplementary Table 4). Based on the 0.03 genetic distance (97% as cutoff), 23,950 OTUs (2177 ± 440 OTUs per samples, n=11) were detected. The highest OTU numbers, Chao1 and Shannon index were found in station A4-7, showing the higher diversity and richness of the anammox 16S rRNA gene in surface sediment of station A4-7 than the other stations. In addition, Pielou’s evenness (from 0.76 to 0.84) and Good’s coverage values (from 98% to 99%) can be observed from each sample, demonstrating the dependability of the sequencing accuracy. The community diversity of the AMX 16S rRNA gene decreased with the increasing concentration in the bottom water (Chao1: r = -0.681, p< 0.05; Observed species: r = -0.710, p< 0.05), decreasing sediment NH4+ (Chao1: r = 0.677, p< 0.05; Observed species: r = 0.678, p< 0.05), and decreasing Chla concentration (Chao1: r = 0.730, p< 0.05; Observed species: r =0.722, p< 0.05) (Supplementary Table 6). The rarefaction curves indicated that the majority of the anammox bacteria was captured in the current study (Supplementary Figure 1C).
The dominant OTUs (top 10 OTUs) of AMX 16S rRNA gene and their related sequences from the GenBank were phylogenetically analyzed to 4 clusters, including Ca. Anammoximicrobium moscowii and the other uncultured clusters (Figures 8A, B; Supplementary Figures 8, 9). The OTUs (OTU173, OTU159, and OTU65) were closely related to Ca. Anammoximicrobium moscowii, and the OTU abundances were higher in muddy sediment than those in sandy sediment. The other clusters were related to uncultured anammox bacteria, showing a very wide range and matched with marine habitats and activated sludge (Figure 8B). According to a redundancy analysis (RDA), the link between AMX 16S communities and environmental factors may be described by the first two axes (65.75% of the total variance) (Figure 8C). The sediment MФ (pseudo-F = 6.1 and p< 0.01), Chla (pseudo-F = 2.3 and p< 0.05), and bottom water NO3− (pseudo-F = 3.4 and p< 0.05), had huge impact on the anammox community. At genus level, Candidatus Anammoximicrobium was the dominant genera in all sampling stations except A3-2, while the dominant genera in A3-2 was Rhodoplanes (Supplementary Figure 10B). Spearman’s correlation analysis revealed that Candidatus Anammoximicrobium was negatively related with sediment water content (p< 0.05), TOC (p< 0.05), and positively correlated with sediment grain size (Supplementary Table 7).
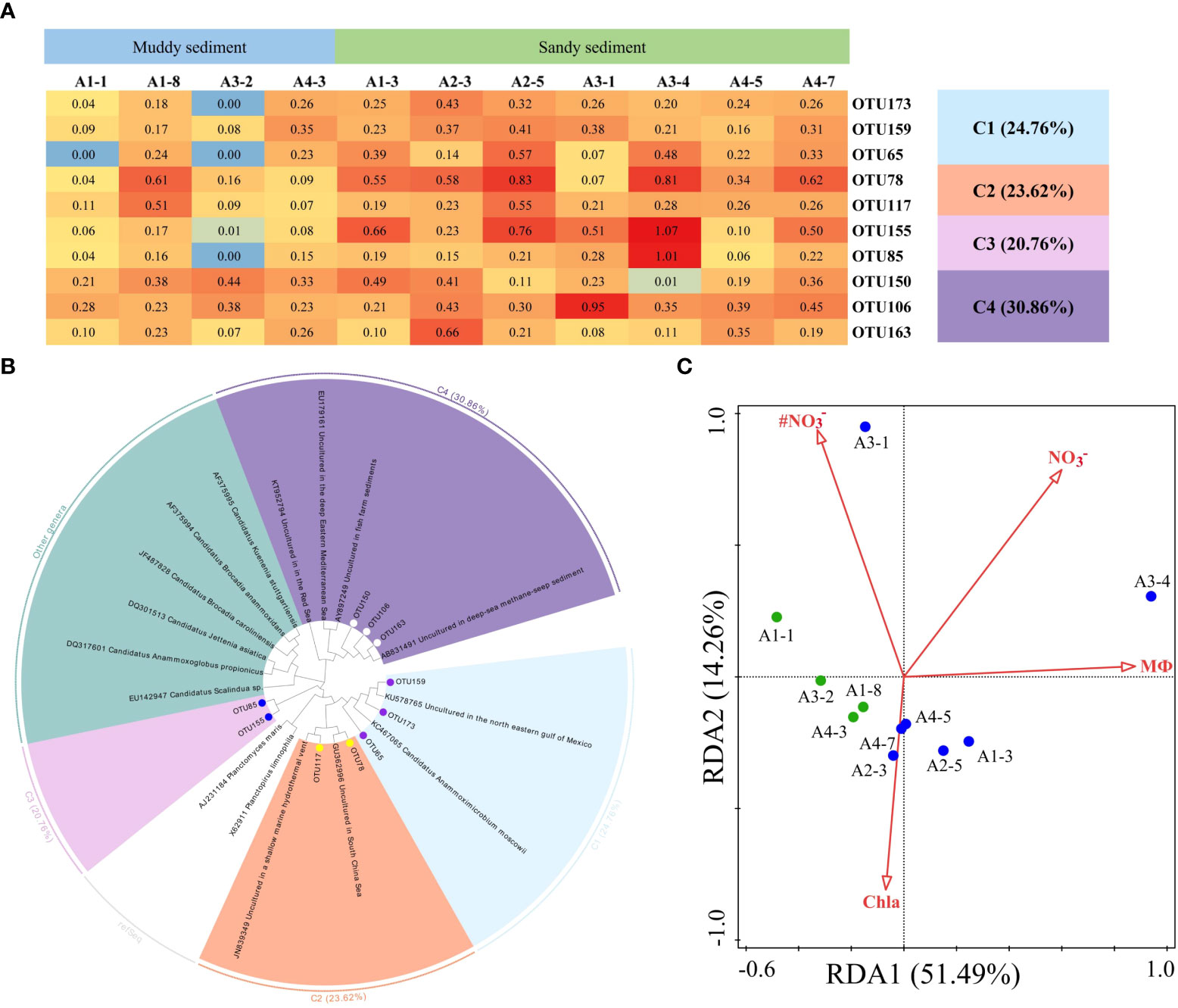
Figure 8 (A) The heat map of the dominant anammox OTUs for the 16S rRNA gene (top 10 OTUs, 97% cutoff). (B) Reconstructed phylogenetic tree with neighbor-joining method of 16S rRNA gene sequences (bootstrap 1000). (C) The redundancy analysis for environmental factors and the AMX 16S gene-encoded anammox bacterial communities.
Discussion
Community characteristics of nirS/nirK-encoding denitrifiers
In coastal aquatic ecosystems, denitrification in sediments can reduce bioavailable nitrogen from anthropogenic and natural inputs, thus, sediments are recognized as hotspot regions of N loss (Seitzinger et al., 2006). Lin et al. (2017) reported that the contribution of denitrification to N loss (2.2 × 106 t N yr –1) was higher than anammox (4.6 × 105 t N yr–1) in the surface sediments from the East China Sea. The biogeochemical cycle of nitrogen is driven by microorganisms, and the microbial communities are constantly changing to adapt to the complex coastal environments (Dang and Lovell, 2016; Hutchins and Fu, 2017). Thus, a further understanding of the microbial processes of N loss is still highly significant. Here, potential rates of denitrification and anammox, and their related gene abundances and community composition, along with influencing environmental factors were explored in the surface sediments from the Yangtze Estuary and the adjacent sea.
In the present study, the average abundance of nirS gene (1.28 × 107 copies g−1) in the surface sediment was greater than that of nirK gene (4.52× 106 copies g−1), which were similar to the observations in the San Francisco Bay (1.50 × 107 copies g−1 for nirS and 3.20× 106 copies g−1 for nirK) (Mosier and Francis, 2010). Some previous studies showed that the mRNA abundance of nirS was also higher compared with nirK, suggesting a main role in N removal processes of nirS-encoding denitrifiers (Graham et al., 2010; Liu et al., 2021). The differential abundance between nirS and nirK may be due to the competition for the substrates required by different enzyme synthesis (Huang et al., 2011). Moreover, compared to nirK-encoding denitrifiers, the variety of nirS-encoding denitrifiers was also greater, which is consistent with previous studies (Shi et al., 2019b; Ming et al., 2021). In general, nirS- and nirK-encoding denitrifiers had different respond to environmental parameters, and nirS denitrifiers had better adaptation to the coastal environments (Jones and Hallin, 2010; Likhitrattanapisal et al., 2021).
In consistent to our study (Figure 3), the abundance of nirS and nirK have the same change trends across the sampling sites (Zhang et al., 2019), which was affected by both the microbial interactions between nirS and nirK- denitrifiers, as well as the different preferences in structure components of corresponding nitrite reductases (Cutruzzolà et al., 2001). Nitrite reduction is the key step in denitrification, and both the nirS and nirK- encoded nitrite reductase are functionally identical while being evolutionarily distinct (Shrewsbury et al., 2016). The previous studies reported that both enzymes are often absent in the same bacterium, and the estimated number of denitrifiers in the soil sites was determined using the summation of nirS and nirK copy counts (Philippot et al., 2011; Zhang et al., 2019). However, Pseudomonas was found in nirS- and nirK-encoding denitrifiers at genus level in our study (Supplementary Figures 4B, 7B). Furthermore, there other different genera were detected in nirS- and nirK-encoding denitrifiers, which have capabilities of denitrification (Kim et al., 2008; Borrero‐De Acuña et al., 2017; Gao et al., 2021). Thus, marker genes of nirS and nirK should be used to detect denitrifiers together. In addition, there are another four different genes involved in the denitrification pathway, nitric oxide reduction (norB and qnorB), and nitrous oxide reduction (nosZI and nosZII). A single gene does not capture the entire denitrifying microbial community, due to several maker genes involved in denitrification pathway (Shrewsbury et al., 2016). Furthermore, Zhang et al. (2019) found that the contribution of denitrification to N removal was positively related to the relative abundance of nosZ/nirS, implying that marker genes from different denitrification steps should be considered simultaneously.
According to phylogenetic analysis, the dominant OTUs of nirS showed some similarities to multiple estuarine and marine sediments (Dang et al., 2008; Hong et al., 2014; Zheng et al., 2015; Gao et al., 2016). In contrast to nirS, the nirK-encoding denitrifiers were found to originate from several habitats, such as marine sediments, fresh sediments, agricultural soil, and wastewater treatment systems (Figure 7B, Supplementary Figure 6). Numerous studies have revealed that nirS and nirK- denitrifiers occupy different niches or have distinct habitat preferences in the environment (Smith and Ogram, 2008; Hallin et al., 2009; Knapp et al., 2009). The communities of nirS and nirK gene were observed to clearly separate in saline environments, and nirS-encoding denitrifiers predominated in marine environment (Jones and Hallin, 2010), in consistent with our study. Furthermore, nirK denitrifiers might exhibit more habitat selection, showing positive correlations with organic matter content (Smith and Ogram, 2008). In the present study, RDA/CCA demonstrated that C/N ratio and MФ was the key factor affecting the community of nirS and nirK, respectively (Figure 6C, Figure 7C). High level of C/N ratio may promote the growth of denitrification bacteria, as previous studies showed that the denitrifiers OTUs and Shannon index decreased with decreasing organic matter and N contents (Chen et al., 2018; Wu et al., 2021). Meanwhile, the dominant OTUs of nirS- and nirK-encoding denitrifiers were affiliated with Proteobacteria (Figures 6B, 7B, Supplementary Figures 3, 6), which was consistent with previous reports (Kim et al., 2016; Xie et al., 2020). Furthermore, γ- Proteobacteria and α-Proteobacteria was the dominant class of nirS- and nirK-encoding denitrifiers, respectively. Some studies showed that denitrification can be performed by phylogenetically different microbes, and the most of denitrifiers early reported are belonging to subphyla of the Proteobacteria (Peralta et al., 2010; Wu et al., 2021). This phylum of Proteobacteria are widely distributed in aquatic environments because of high phylogenetic and phenotypic diversities (Liu et al., 2015; He and Zhang, 2016; Shi et al., 2019a). With the Proteobacteria community, Pseudomonas and Mesorhizobium were the dominant genera of nirS- and nirK-encoding denitrifiers, respectively. Pseudomonas stutzeri strain XL-2 and strain XL-2 were identified as the main denitrifiers which can remove 97.9% of nitrate (Dang and Lovell, 2016). In nirS bacterial community, Pseudomonas was mainly affected by NH4+ in the bottom water; while in nirK bacterial community, Mesorhizobium was significantly influenced by with sediment Fe(II)/Fe(III). In addition, it has been reported that the higher abundance of Mesorhizobium was observed in sandy soil (Li et al., 2019), in consistent to our study that Mesorhizobium was the dominant genera in A2-5 and A3-1 with high median grain size.
Community characteristics of anammox bacteria
In the present study, the abundance of AMX 16S rRNA gene was ranging from 1.03× 105 to 2.97 × 106 copies g−1 (Figures 3A, B), which was lower than that of nirS and nirK gene in all sampling sites. The gene abundances of AMX 16S rRNA gene were like to the values in the Pearl River Estuary (Wu et al., 2020), but 1-2 orders magnitude lower than those in paddy soil (Zhu et al., 2011) and the coastal wetland sediments of China (Hou et al., 2015). Furthermore, anammox bacterial abundance was no obvious spatial heterogeneity at the sampling sites (Figure 3A).
The high-throughput sequencing of the 16S rRNA gene of anammox bacteria could provide adequate data, as compared to data from cloning libraries, to identify community changes and even recognize unusual bacteria with low relative abundance (Zhao et al., 2013; Shen et al., 2016). Based on the OTUs and the Shannon index, the AMX 16S genes in the surface sediment represented a very varied community of anammox bacteria (Supplementary Table 4). A wide variety of anammox bacteria was found in the surface sediment of Yangtze Estuary and the adjacent sea, in similarity to other estuarine sediments (Dale et al., 2009; Hong et al., 2014; Lisa et al., 2014). Salinity is a significant environmental factor that affects the diversity of anammox bacteria, according to earlier research (Schmid et al., 2007; Hamersley et al., 2009; Hu et al., 2012). However, in this present study, the Chao1 had significantly correlated with NH4+ concentration in bottom water and sediment (Supplementary Table 6). These results implied that the substrates contents might be the main limiting factors for anammox bacteria.
In this study, the dominant OTUs were closely related to Ca. Anammoximicrobium moscowii and Candidatus Anammoximicrobium was the dominant genera at genus level (Supplementary Figures 9, 10B). In contrast to previous studies (Schmid et al., 2007; Fernandes et al., 2016; Fu et al., 2019; Wu et al., 2021), Ca. Scalindua are the dominant anammox bacteria in marine environments. Some studies reported that Ca. Scalindua has a relatively high tolerance for salinity and it can adapt to high hypersaline environment (Oshiki et al., 2017; Speth et al., 2017). Recently, the other genera have been found and Ca. Scalindua was not the only genus in the marine environments (Wu et al., 2019). Ca. Anammoximicrobium moscowii also can carry out anaerobic ammonium oxidation, which was firstly isolated in the Moscow region (Khramenkov et al., 2013), and the key role of this genus in N loss process cannot be ignored. Some studies reported that the different anammox bacteria lineages can become the dominant populations based on the slight change in the environment (Wang et al., 2021). In addition, the community composition study of anammox bacteria can be directly impacted by primer sets and sequencing depth. In the present study, considering the primer efficiency, Amx808F and Amx1040R was the appropriate choice for capturing anammox bacteria with good coverage, however, the specificity was not strong for Ca. Scalindua detection. From the RDA analysis, the MФ and bottom water NO3− significantly affected anammox bacteria community. In addition, according to Spearman’s correlation analysis, Candidatus Anammoximicrobium was negatively related by TOC.
Microbial processes for nitrogen loss and their environmental implications
For microbial mediated N transformation, the surface sediment are usually the most vibrant with greater potential N loss rates, particularly the denitrification rates; however, the potential rates of denitrification and anammox were lower than previous studies (Hou et al., 2015; Lin et al., 2017; Wu et al., 2021), denitrification and anammox can exhibit strongly spatiotemporal variabilities, and the rates might be affected by biotic and abiotic factors in sampling sites. The bulk of the samples belong to sandy sediments, with the low organic matter content and low N removal rates. In our study, denitrification was the main contributor of N loss, dominating the N loss process in surface sediment in most sampling sites (Figure 5B). These results were in accordance with those in the previous studies in estuarine and coastal habitats (Fernandes et al., 2012; Cao et al., 2017; Lin et al., 2017; Huang et al., 2021). Moreover, Huang et al. (2021) found that denitrification rates were substantially correlated with organics and median grain size (MФ), implying that both MФ and organic matter were the crucial factors regulating denitrification activities. High surface-to-volume ratios in fine-grained sediments typically result in greater organic matter absorption (Keil et al., 1994). Meanwhile, organic matter (as an electron donor) can directly or indirectly boost N removal processes by providing energy via the oxidation of organics (Devol, 2015). Additionally, we verified that denitrification rates were substantially correlated with MФ and the C/N ratio (Figures 5C, D). Higher specific surface area in sediment with little grains was more suited to microbial colonization, and the required energy for the growth of heterotrophic denitrifiers were provided by organic matter Moreover, the contribution of anammox was higher than denitrification in few sampling stations with lower C/N ratio and higher median grain size, further implying that sediment texture can control the N loss rates via affecting the residence time of organic matter (Huang et al., 2021).
Anammox rates and denitrification rates have a positive correlation in this study (Figure 5A), suggesting that denitrification was probably the main source of NO2− for anammox. Coherent to many previous researches in the aquatic environment (Brin et al., 2017; Zhao et al., 2020a), anammox metabolism requires the substrates of NO2−, which are main supplied by denitrification rather than nitrification in these anaerobic environments. This coupling between anammox and denitrification may be mediated by an imbalance in NO3− and organic carbon needed for denitrification. Furthermore, in the present study, NO2− was negatively related to DEN%, but positively correlated to ANA%, implying the coupling of denitrification and anammox. However, there also studies showing that anammox coupled with nitrification rather than denitrification in estuarine sediments in winter (Hou et al., 2015; Lin et al., 2017). The uncertainties in the relationships between anammox and denitrification might be attributed to organic acids, and anammox bacteria can convert NO3− to NO2− in the presence of organic acids (Brin et al., 2014). Thus, coupled anammox and nitrification or denitrification play a vital role in N removal processes from the surface of marine sediments.
Previous studies have revealed that NH4+ and NO3− play the crucial and central role in N removal (Lin et al., 2017; Huang et al., 2021), which were as substrates in denitrification and anammox processes. In consistent to our study, NH4+ and NO3− were the key factors to influence denitrification and anammox rates. In the present study, Chla was the key environmental regulator for denitrification rates (p< 0.01) (Figure 4, Figure 5E, Supplementary Table 5). The sediment Chla is an indicator for the availability of labile organic matter which is obtained from phytoplankton deposited from upper waters, along as phytobenthos and benthic algal debris in the shallow marginal system (Franco et al., 2007; Kitidis et al., 2017). The sediment organic matter correlates positively with Chla in sediment and water column (Li et al., 2013), in similarity to our study (Figure 4, Supplementary Table 5). The growth of phytoplankton may be aided by light intensity and water temperature in March (Shi et al., 2019b), indicating that many phytoplankton cells might be destroyed to release labile DOC (Zhang et al., 2018), which could enhance denitrification by sinking down to anaerobic sediment layers. Meanwhile, anammox rates also positively correlated with Chla (p< 0.01) (Figure 4, Figure 5F, Supplementary Table 5). The main reason for that was the substrates (NH4+ and NO2−) of anammox originated from the mineralization of organic matters (Zhang et al., 2018). Thus, both the quality and amount of organic matter in sediment are essential to manage estuarine N-cycling because fresh organics has a higher lability and simpler to be utilized by NOx− removal processes (Westrich and Berner, 1984; Henrichs and Doyle, 1986).
There were no significant correlations between N removal rates and related gene abundances in this study. The gene abundance obtained from the DNA levels has limitations. On the one hand, since DNA extracts include a wide variety of cells (e.g., active cells, inactive cells and dead cells) and its degraded or lysed extracellular DNA, gene abundance does not always indicate activity (Gaidos et al., 2011). On the other hand, the number of microbials just indicates potential microbial metabolism and has no direct impact on the activities of N transformation (Dai et al., 2008). It has been reported that the related functional gene transcript abundance is reflective of N transformation process potential, further revealing N transformation potential had a close relationship with active microbials (Li et al., 2022). Thus, to gain an insight into changes in denitrification and anammox potential, the functional transcript abundance combining with N removal rates are needed.
Conclusion
Taken as a whole, the current work described the distribution and diversity of denitrifiers and anammox bacteria in the Yangtze Estuary and the adjacent sea, and the potential rates of denitrification and anammox were measured together. We found the abundance and diversity of nirS-denitrifiers were higher than those of nirK-denitrifiers, and nirK-denitrifiers were more sensitive to the environments. For anammox bacteria, Ca. Anammoximicrobium moscowii has been found in surface sediment, and the other genera of anammox bacteria cannot be neglected in marine ecosystems. Compared to anammox, denitrification was the dominant contributor of N removal process and contributed 73.5% on average. Denitrification and anammox rates were regulated by multiple environmental factors, and the median grain size (MФ), C/N ratio, and Chla were the key factors.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
ZT: conceptualization,investigation, methodology, data curation, formal analysis, and writing-original draft. YZ: conceptualization, writing-review and editing, supervision, and funding acquisition. TM: writing-review and supervision. ZY: conceptualization, writing-review and editing, and supervision. TC: writing-review and editing. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (No. 42130410, 41620104001, 41976133).
Acknowledgments
We thank Han Zhang and crew members on the R/V Blue Sea 201 for the assistance provided in the collection of samples during the cruise.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.1078653/full#supplementary-material
References
Abell G. C. J., Revill A. T., Smith C., Bissett A. P., Volkman J. K., Robert S. S. (2010). Archaeal ammonia oxidizers and nirS-type denitrifiers dominate sediment nitrifying and denitrifying populations in a subtropical macrotidal estuary. ISME J. 4 (2), 286–300. doi: 10.1038/ismej.2009.105
Anderson D. M., Cembella A. D., Hallegraeff G. M. (2012). Progress in understanding harmful algal blooms: paradigm shifts and new technologies for research, monitoring, and management. Annu. Rev. Mar. Sci. 4, 143–176. doi: 10.1146/annurev-marine-120308-081121
Babbin A. R., Buchwald C., Morel F. M. M., Wankel S. D., Ward B. B. (2020). Nitrite oxidation exceeds reduction and fixed nitrogen loss in anoxic pacific waters. Mar. Chem. 224, 103814. doi: 10.1016/j.marchem.2020.103814
Babbin A. R., Keil R. G., Devol A. H., Ward B. B. (2014). Organic matter stoichiometry, flux, and oxygen control nitrogen loss in the ocean. Science 344 (6182), 406–408. doi: 10.1126/science.1248364
Basha S., Lingamgunta L. K., Kannali J., Gajula S. K., Bandikari R., Dasari S., et al. (2018). Subsurface endospore-forming bacteria possess bio-sealant properties. Sci. Rep. 8 (1), 1–13. doi: 10.1038/s41598-018-24730-3
Beman J.M. (2014). Activity, abundance, and diversity of nitrifying archaea and denitrifying bacteria in sediments of a subtropical estuary: Bahía del tóbari, Mexico. Estuar. Coasts 37 (6), 1343–1352. doi: 10.1007/s12237-013-9716-y
Bernhard A. E., Donn T., Giblin A. E., Stahl D. A. (2005). Loss of diversity of ammonia-oxidizing bacteria correlates with increasing salinity in an estuary system. Environ. Microbiol. 7 (9), 1289–1297. doi: 10.1111/j.1462-2920.2005.00808.x
Borrero-De Acuña J. M., Timmis K. N., Jahn M., Jahn D. (2017). Protein complex formation during denitrification by pseudomonas aeruginosa. Microbial. Biotechnol. 10 (6), 1523–1534. doi: 10.1111/1751-7915.12851
Brin L. D., Giblin A. E., Rich J. J. (2014). Environmental controls of anammox and denitrification in southern new England estuarine and shelf sediments. Limnol. Oceanogr. 59 (3), 851–860. doi: 10.4319/lo.2014.59.3.0851
Brin L. D., Giblin A. E., Rich J. J. (2017). Similar temperature responses suggest future climate warming will not alter partitioning between denitrification and anammox in temperate marine sediments. Global Change Biol. 23 (1), 331–340. doi: 10.1111/gcb.13370
Cao W., Guan Q., Li Y., Wang M., Liu B. (2017). The contribution of denitrification and anaerobic ammonium oxidation to N2 production in mangrove sediments in southeast China. J. Soils Sediments 17 (6), 1767–1776. doi: 10.1007/s11368-017-1653-0
Chen F., Hou L., Liu M., Zheng Y., Yin G., Lin X., et al. (2016). Net anthropogenic nitrogen inputs (NANI) into the Yangtze river basin and the relationship with riverine nitrogen export. J. Geophys. Res.: Biogeosci. 121 (2), 451–465. doi: 10.1002/2015JG003186
Chen S., Wang F., Zhang Y., Qin S., Wei S., Wang S., et al. (2018). Organic carbon availability limiting microbial denitrification in the deep vadose zone. Environ. Microbiol. 20 (3), 980–992. doi: 10.1111/1462-2920.14027
Cutruzzolà F., Brown K., Wilson E. K., Bellelli A., Arese M., Tegoni M., et al. (2001). The nitrite reductase from pseudomonas aeruginosa: essential role of two active-site histidines in the catalytic and structural properties. Proc. Natl. Acad. Sci. 98 (5), 2232–2237. doi: 10.1073/pnas.041365298
Dai M., Wang L., Guo X., Zhai W., Li Q., He B., et al. (2008). Nitrification and inorganic nitrogen distribution in a large perturbed river/estuarine system: the pearl river estuary, China. Biogeosciences 5 (5), 1227–1244. doi: 10.5194/bg-5-1227-2008
Dale O. R., Tobias C. R., Song B. (2009). Biogeographical distribution of diverse anaerobic ammonium oxidizing (anammox) bacteria in cape fear river estuary. Environ. Microbiol. 11 (5), 1194–1207. doi: 10.1111/j.1462-2920.2008.01850.x
Dalsgaard T., Thamdrup B. (2002). Factors controlling anaerobic ammonium oxidation with nitrite in marine sediments. Appl. Environ. Microbiol. 68 (8), 3802–3808. doi: 10.1128/AEM.68.8.3802-3808.2002
Dang H., Lovell C. R. (2016). Microbial surface colonization and biofilm development in marine environments. Microbiol. Mol. Biol. Rev. 80 (1), 91–138. doi: 10.1128/MMBR.00037-15
Dang H., Zhang X., Sun J., Li T., Zhang Z., Yang G. (2008). Diversity and spatial distribution of sediment ammonia-oxidizing crenarchaeota in response to estuarine and environmental gradients in the changjiang estuary and East China Sea. Microbiology 154 (7), 2084–2095. doi: 10.1099/mic.0.2007/013581-0
Devol A. H. (2015). Denitrification, anammox, and N2 production in marine sediments. Annu. Rev. Mar. Sci. 7, 403–423. doi: 10.1146/annurev-marine-010213-135040
Di H. J., Cameron K. C., Shen J.-P., Winefield C. S., O’callaghan M., Bowatte S., et al. (2010). Ammonia-oxidizing bacteria and archaea grow under contrasting soil nitrogen conditions. FEMS Microbiol. Ecol. 72 (3), 386–394. doi: 10.1111/j.1574-6941.2010.00861.x
Fernandes S. O., Javanaud C., Michotey V. D., Guasco S., Anschutz P., Bonin P. (2016). Coupling of bacterial nitrification with denitrification and anammox supports n removal in intertidal sediments (Arcachon bay, France). Estuar. Coast. Shelf Sci. 179, 39–50. doi: 10.1016/j.ecss.2015.10.009
Fernandes S. O., Michotey V. D., Guasco S., Bonin P. C., Bharathi P. A. L. (2012). Denitrification prevails over anammox in tropical mangrove sediments (Goa, India). Mar. Environ. Res. 74, 9–19. doi: 10.1016/j.marenvres.2011.11.008
Franco M. A., De Mesel I., Diallo M.D., van der Gucht K., Van Gansbeke D., Van Rijswijk P., et al. (2007). Effect of phytoplankton bloom deposition on benthic bacterial communities in two contrasting sediments in the southern north Sea. Aquat. Microbial. Ecol. 48 (3), 241–254. doi: 10.3354/ame048241
Fu L., Chen Y., Li S., He H., Mi T., Zhen Y., et al. (2019). Shifts in the anammox bacterial community structure and abundance in sediments from the changjiang estuary and its adjacent area. Syst. Appl. Microbiol. 42 (3), 383–396. doi: 10.1016/j.syapm.2018.12.008
Gaidos E., Rusch A., Ilardo M. (2011). Ribosomal tag pyrosequencing of DNA and RNA from benthic coral reef microbiota: community spatial structure, rare members and nitrogen-cycling guilds. Environ. Microbiol. 13 (5), 1138–1152. doi: 10.1111/j.1462-2920.2010.02392.x
Gao J., Hou L., Zheng Y., Liu M., Yin G., Li X., et al. (2016). nirS-encoding denitrifier community composition, distribution, and abundance along the coastal wetlands of China. Appl. Microbiol. Biotechnol. 100 (19), 8573–8582. doi: 10.1007/s00253-016-7659-5
Gao Y., Mania D., Mousavi S. A., Lycus P., Arntzen M. Ø., Woliy K., et al. (2021). Competition for electrons favours N2O reduction in denitrifying bradyrhizobium isolates. Environ. Microbiol. 23 (4), 2244–2259. doi: 10.1111/1462-2920.15404
Gobler C. J. (2020). Climate change and harmful algal blooms: insights and perspective. Harmful Algae 91, 101731. doi: 10.1016/j.hal.2019.101731
Graham D. W., Trippett C., Dodds W. K., O’brien J. M., Banner E. B. K., Head I. M., et al. (2010). Correlations between in situ denitrification activity and nir-gene abundances in pristine and impacted prairie streams. Environ. pollut. 158 (10), 3225–3229. doi: 10.1016/j.envpol.2010.07.010
Hallin S., Jones C. M., Schloter M., Philippot L. (2009). Relationship between n-cycling communities and ecosystem functioning in a 50-year-old fertilization experiment. ISME J. 3 (5), 597–605. doi: 10.1038/ismej.2008.128
Hamersley M.R., Lavik G., Woebken D., Rattray J. E., Lam P., Hopmans E. C., et al. (2007). Anaerobic ammonium oxidation in the Peruvian oxygen minimum zone. Limnol. Oceanogr. 52 (3), 923–933. doi: 10.4319/lo.2007.52.3.0923
Hamersley M.R., Woebken D., Boehrer B., Schultze M., Lavik G., Kuypers M. M. M. (2009). Water column anammox and denitrification in a temperate permanently stratified lake (Lake rassnitzer, Germany). Syst. Appl. Microbiol. 32 (8), 571–582. doi: 10.1016/j.syapm.2009.07.009
Harhangi H. R., Le Roy M., Van Alen T., Hu B.-L., Groen J., Kartal B., et al. (2012). Hydrazine synthase, a unique phylomarker with which to study the presence and biodiversity of anammox bacteria. Appl. Environ. Microbiol. 78 (3), 752–758. doi: 10.1128/AEM.07113-11
Henrichs S. M., Doyle A. P. (1986). Decomposition of 14C-labeled organic substances in marine sediments 1. Limnol. Oceanogr. 31 (4), 765–778. doi: 10.4319/lo.1986.31.4.0765
He T., Zhang X. (2016). Characterization of bacterial communities in deep-sea hydrothermal vents from three oceanic regions. Mar. Biotechnol. 18 (2), 232–241. doi: 10.1007/s10126-015-9683-3
Hong Y., Xu X., Kan J., Chen F. (2014). Linking seasonal inorganic nitrogen shift to the dynamics of microbial communities in the Chesapeake bay. Appl. Microbiol. Biotechnol. 98 (7), 3219–3229. doi: 10.1007/s00253-013-5337-4
Hou L., Zheng Y., Liu M., Li X., Lin X., Yin G., et al. (2015). Anaerobic ammonium oxidation and its contribution to nitrogen removal in china’s coastal wetlands. Sci. Rep. 5 (1), 1–11. doi: 10.1038/srep15621
Howarth R. W. (2008). Coastal nitrogen pollution: a review of sources and trends globally and regionally. Harmful Algae 8 (1), 14–20. doi: 10.1016/j.hal.2008.08.015
Huang S., Chen C., Wu Y., Wu Q., Zhang R. (2011). Characterization of depth-related bacterial communities and their relationships with the environmental factors in the river sediments. World J. Microbiol. Biotechnol. 27 (11), 2655–2664. doi: 10.1007/s11274-011-0739-x
Huang F., Lin X., Hu W., Zeng F., He L., Yin K. (2021). Nitrogen cycling processes in sediments of the pearl river estuary: Spatial variations, controlling factors, and environmental implications. Catena 206, 105545. doi: 10.1016/j.catena.2021.105545
Hu B.-L., Shen L.-D., Zheng P., Hu A.-H., Chen T.-T., Cai C., et al. (2012). Distribution and diversity of anaerobic ammonium-oxidizing bacteria in the sediments of the qiantang river. Environ. Microbiol. Rep. 4 (5), 540–547. doi: 10.1111/j.1758-2229.2012.00360.x
Hutchins D. A., Fu F. (2017). Microorganisms and ocean global change. Nat. Microbiol. 2 (6), 1–11. doi: 10.1038/nmicrobiol.2017.58
Jiang X., Yao L., Guo L., Liu G., Liu W. (2017). Multi-scale factors affecting composition, diversity, and abundance of sediment denitrifying microorganisms in Yangtze lakes. Appl. Microbiol. Bbiotechnol. 101 (21), 8015–8027. doi: 10.1007/s00253-017-8537-5
Jones C. M., Hallin S. (2010). Ecological and evolutionary factors underlying global and local assembly of denitrifier communities. ISME J. 4 (5), 633–641. doi: 10.1038/ismej.2009.152
Keil R. G., Montluçon D. B., Prahl F. G., Hedges J. I. (1994). Sorptive preservation of labile organic matter in marine sediments. Nature 370 (6490), 549–552. doi: 10.1038/370549a0
Khramenkov S. V., Kozlov M. N., Kevbrina M. V., Dorofeev A. G., Kazakova E. A., Grachev V. A., et al. (2013). A novel bacterium carrying out anaerobic ammonium oxidation in a reactor for biological treatment of the filtrate of wastewater fermented sludge. Microbiology 82 (5), 628–636. doi: 10.1134/S002626171305007X
Kim H., Bae H.-S., Reddy K.R., Ogram A. (2016). Distributions, abundances and activities of microbes associated with the nitrogen cycle in riparian and stream sediments of a river tributary. Water Res. 106, 51–61. doi: 10.1016/j.watres.2016.09.048
Kim M., Jeong S.-Y., Yoon S. J., Cho S. J., Kim Y. H., Kim M. J., et al. (2008). Aerobic denitrification of pseudomonas putida AD-21 at different C/N ratios. J. Biosci. Bioeng. 106 (5), 498–502. doi: 10.1263/jbb.106.498
Kitidis V., Tait K., Nunes J., Brown I., Woodward E. M. S., Harris C., et al. (2017). Seasonal benthic nitrogen cycling in a temperate shelf sea: the celtic Sea. Biogeochemistry 135 (1), 103–119. doi: 10.1007/s10533-017-0311-3
Klute A. (1986). Water retention: laboratory methods. Methods Soil anal.: Part 1 Phys. Mineral. Methods 5, 635–662. doi: 10.2136/sssabookser5.1.2ed.c26
Knapp C. W., Dodds W. K., Wilson K. C., O’brien J. M., Graham D. W. (2009). Spatial heterogeneity of denitrification genes in a highly homogenous urban stream. Environ. Sci. Technol. 43 (12), 4273–4279. doi: 10.1021/es9001407
Kuypers M. M. M., Sliekers A.O., Lavik G., Schmid M., Jørgensen B. B., Kuenen J.G., et al. (2003). Anaerobic ammonium oxidation by anammox bacteria in the black Sea. Nature 422 (6932), 608–611. doi: 10.1038/nature01472
Li M., He H., Mi T., Zhen Y. (2022). Spatiotemporal dynamics of ammonia-oxidizing archaea and bacteria contributing to nitrification in sediments from bohai Sea and south yellow Sea, China. Sci. Total Environ. 825, 153972. doi: 10.1016/j.scitotenv.2022.153972
Li M., Hong Y., Cao H., Gu J. (2013). Community structures and distribution of anaerobic ammonium oxidizing and nirS-encoding nitrite-reducing bacteria in surface sediments of the south China Sea. Microbial. Ecol. 66 (2), 281–296. doi: 10.1007/s00248-012-0175-y
Li M., Hong Y., Klotz M. G., Gu J.-D. (2010). A comparison of primer sets for detecting 16S rRNA and hydrazine oxidoreductase genes of anaerobic ammonium-oxidizing bacteria in marine sediments. Appl. Microbiol. Biotechnol. 86 (2), 781–790. doi: 10.1007/s00253-009-2361-5
Likhitrattanapisal S., Siriarchawatana P., Seesang M., Chunhametha S., Boonsin W., Phithakrotchanakoon C., et al. (2021). Uncovering multi-faceted taxonomic and functional diversity of soil bacteriomes in tropical southeast Asian countries. Sci. Rep. 11 (1), 1–13. doi: 10.1038/s41598-020-79786-x
Lim D. I., Choi J. Y., Jung H. S., Rho K. C., Ahn K. S. (2007). Recent sediment accumulation and origin of shelf mud deposits in the yellow and East China seas. Prog. Oceanogr. 73 (2), 145–159. doi: 10.1016/j.pocean.2007.02.004
Lin G., Lin X. (2022). Bait input altered microbial community structure and increased greenhouse gases production in coastal wetland sediment. Water Res. 218, 118520. doi: 10.1016/j.watres.2022.118520
Lin X., Liu M., Hou L., Gao D., Li X., Lu K., et al. (2017). Nitrogen losses in sediments of the East China Sea: spatiotemporal variations, controlling factors, and environmental implications. J. Geophys. Res.: Biogeosci. 122 (10), 2699–2715. doi: 10.1002/2017jg004036
Lisa J. A., Song B., Tobias C. R., Duernberger K. A. (2014). Impacts of freshwater flushing on anammox community structure and activities in the new river estuary, USA. Aquat. Microbial. Ecol. 72 (1), 17–31. doi: 10.3354/ame01682
Liu S., Chen Y., Xiao L. (2021). Metagenomic insights into mixotrophic denitrification facilitated nitrogen removal in a full-scale A2/O wastewater treatment plant. PloS One 16 (4), e0250283. doi: 10.1371/journal.pone.0250283
Liu X., Hu H.-W., Liu Y.-R., Xiao K.-Q., Cheng F.-S., Li J., et al. (2015). Bacterial composition and spatiotemporal variation in sediments of jiaozhou bay, China. J. Soils Sediments 15 (3), 732–744. doi: 10.1007/s11368-014-1045-7
Liu Y., Liu J., Yao P., Ge T., Qiao Y., Zhao M., et al. (2018). Distribution patterns of ammonia-oxidizing archaea and bacteria in sediments of the eastern China marginal seas. Syst. Appl. Microbiol. 41 (6), 658–668. doi: 10.1016/j.syapm.2018.08.008
Li H., Zhang Y., Wang T., Feng S., Ren Q., Cui Z., et al. (2019). Responses of soil denitrifying bacterial communities carrying nirS, nirK, and nosZ genes to revegetation of moving sand dunes. Ecol. Indic. 107, 105541. doi: 10.1016/j.ecolind.2019.105541
Lovley D. R., Phillips E. J. P. (1987). Rapid assay for microbially reducible ferric iron in aquatic sediments. Appl. Environ. Microbiol. 53 (7), 1536–1540. doi: 10.1128/aem.53.7.1536-1540.1987
Medhi K., Singhal A., Chauhan D. K., Thakur I. S. (2017). Investigating the nitrification and denitrification kinetics under aerobic and anaerobic conditions by paracoccus denitrificans ISTOD1. Biores. Technol. 242, 334–343. doi: 10.1016/j.biortech.2017.03.084
Ming H., Fan J., Chen Q., Su J., Song J., Yuan J., et al. (2021). Diversity and abundance of denitrifying bacteria in the sediment of a eutrophic estuary. Geomicrobiol. J. 38 (3), 199–209. doi: 10.1080/01490451.2020.1822959
Mosier A. C., Francis C. A. (2010). Denitrifier abundance and activity across the San Francisco bay estuary. Environ. Microbiol. Rep. 2 (5), 667–676. doi: 10.1111/j.1758-2229.2010.00156.x
Mulder A., De Graaf V., Astfid A., Robertson L. A., Kuenen J. G. (1995). Anaerobic ammonium oxidation discovered in a denitrifying fluidized bed reactor. FEMS Microbiol. Ecol. 16 (3), 177–183. doi: 10.1111/j.1574-6941.1995.tb00281.x
Murray C. C., Agbayani S., Ban N. C. (2015). Cumulative effects of planned industrial development and climate change on marine ecosystems. Global Ecol. Conserv. 4, 110–116. doi: 10.1016/j.gecco.2015.06.003
Nicol G. W., Leininger S., Schleper C., Prosser J. I. (2008). The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ. Microbiol. 10 (11), 2966–2978. doi: 10.1111/j.1462-2920.2008.01701.x
Oshiki M., Mizuto K., Kimura Z.-I., Kindaichi T., Satoh H., Okabe S. (2017). Genetic diversity of marine anaerobic ammonium-oxidizing bacteria as revealed by genomic and proteomic analyses of ‘Candidatus scalindua japonica’. Environ. Microbiol. Rep. 9 (5), 550–561. doi: 10.1111/1758-2229.12586
Peralta A. L., Matthews J. W., Kent A. D. (2010). Microbial community structure and denitrification in a wetland mitigation bank. Appl. Environ. Microbiol. 76 (13), 4207–4215. doi: 10.1128/AEM.02977-09
Philippot L., Andert J., Jones C. M., Bru D., Hallin S. (2011). Importance of denitrifiers lacking the genes encoding the nitrous oxide reductase for N2O emissions from soil. Global Change Biol. 17 (3), 1497–1504. doi: 10.1111/j.1365-2486.2010.02334.x
Ravishankara A. R., Daniel J. S., Portmann R. W. (2009). Nitrous oxide (N2O): the dominant ozone-depleting substance emitted in the 21st century. Science 326 (5949), 123–125. doi: 10.1126/science.1176985
Schmid M. C., Risgaard-Petersen N., Van De Vossenberg J., Kuypers M. M. M., Lavik G., Petersen J., et al. (2007). Anaerobic ammonium-oxidizing bacteria in marine environments: widespread occurrence but low diversity. Environ. Microbiol. 9 (6), 1476–1484. doi: 10.1111/j.1462-2920.2007.01266.x
Seitzinger S., Harrison J. A., Böhlke J. K., Bouwman A. F., Lowrance R., Peterson B., et al. (2006). Denitrification across landscapes and waterscapes: a synthesis. Ecol. Appl. 16 (6), 2064–2090. doi: 10.1890/1051-0761(2006)016[2064:DALAWA]2.0.CO;2
Shen L.-D., Zheng P.-H., Ma S.-J. (2016). Nitrogen loss through anaerobic ammonium oxidation in agricultural drainage ditches. Biol. Fertil. Soils 52 (2), 127–136. doi: 10.1007/s00374-015-1058-4
Shi R., Huang H., Qi Z., Han T. (2019a). Distribution patterns of nirS-encoding and nirK-encoding denitrifiers in the surface sediment of the pearl river estuary. Russian J. Mar. Biol. 45 (6), 453–463. doi: 10.1134s1063074019060099
Shi R., Xu S., Qi Z., Huang H., Liang Q. (2019b). Seasonal patterns and environmental drivers of nirS-and nirK-encoding denitrifiers in sediments of daya bay, China. Oceanologia 61 (3), 308–320. doi: 10.1016/j.oceano.2019.01.002
Shrewsbury L. H., Smith J. L., Huggins D. R., Carpenter-Boggs L., Reardon C. L. (2016). Denitrifier abundance has a greater influence on denitrification rates at larger landscape scales but is a lesser driver than environmental variables. Soil Biol. Biochem. 103, 221–231. doi: 10.1016/j.soilbio2016.08.016
Smith J. M., Ogram A. (2008). Genetic and functional variation in denitrifier populations along a short-term restoration chronosequence. Appl. Environ. Microbiol. 74 (18), 5615–5620. doi: 10.1128/AEM.00349-08
Smith V. H., Schindler D. W. (2009). Eutrophication science: where do we go from here? Trends Ecol. Evol. 24 (4), 201–207.
Song G. D., Liu S. M., Kuypers M. M. M., Lavik G. (2016). Application of the isotope pairing technique in sediments where anammox, denitrification, and dissimilatory nitrate reduction to ammonium coexist. Limnol. Oceanogr.: Methods 14 (12), 801–815. doi: 10.1002/lom3.10127
Speth D. R., Lagkouvardos I., Wang Y., Qian P.-Y., Dutilh B. E., Jetten M. S. M. (2017). Draft genome of scalindua rubra, obtained from the interface above the discovery deep brine in the red Sea, sheds light on potential salt adaptation strategies in anammox bacteria. Microbial. Ecol. 74 (1), 1–5. doi: 10.1007/s00248-017-0929-7
Thamdrup B., Dalsgaard T. (2002). Production of N2 through anaerobic ammonium oxidation coupled to nitrate reduction in marine sediments. Appl. Environ. Microbiol. 68 (3), 1312–1318. doi: 10.1128/AEM.68.3.1312-1318.2002
Throbäck I. N., Enwall K., Jarvis Å, Hallin S. (2004). Reassessing PCR primers targeting nirS, nirK and nosZ genes for community surveys of denitrifying bacteria with DGGE. FEMS Microbiol. Ecol. 49 (3), 401–417. doi: 10.1016/j.femsec.2004.04.011
Trimmer M., Nicholls J. C. (2009). Production of nitrogen gas via anammox and denitrification in intact sediment cores along a continental shelf to slope transect in the north Atlantic. Limnol. Oceanogr. 54 (2), 577–589. doi: 10.4319/lo.2009.54.2.0577
Trimmer M., Nicholls J. C., Deflandre B. (2003). Anaerobic ammonium oxidation measured in sediments along the Thames estuary, united kingdom. Appl. Environ. Microbiol. 69 (11), 6447–6454. doi: 10.1128/AEM.69.11.6447-6454.2003
Wang J., Kan J., Qian G., Chen J., Xia Z., Zhang X., et al. (2019). Denitrification and anammox: Understanding nitrogen loss from Yangtze estuary to the east China sea (ECS). Environ. pollut. 252, 1659–1670. doi: 10.1016/j.envpol.2019.06.025
Wang Q., Rogers M. J., Ng S., He J. (2021). Fixed nitrogen removal mechanisms associated with sulfur cycling in tropical wetlands. Water Res. 189, 116619. doi: 10.1016/j.watres.2020.116619
Westrich J. T., Berner R. A. (1984). The role of sedimentary organic matter in bacterial sulfate reduction: The G model tested 1. Limnol. Oceanogr. 29 (2), 236–249. doi: 10.4319/lo.1984.29.2.0236
Worm B., Barbier E. B., Beaumont N., Duffy J.E., Folke C., Halpern B. S., et al. (2006). Impacts of biodiversity loss on ocean ecosystem services. Science 314 (5800), 787–790. doi: 10.1126/science.1132294
Wu J., Hong Y., Chang X., Jiao L., Li Y., Liu X., et al. (2019). Unexpectedly high diversity of anammox bacteria detected in deep-sea surface sediments of the south China Sea. FEMS Microbiol. Ecol. 95 (3), fiz013. doi: 10.1093/femsec/fiz013
Wu J., Hong Y., Liu X., Hu Y. (2021). Variations in nitrogen removal rates and microbial communities over sediment depth in daya bay, China. Environ. pollut. 286, 117267. doi: 10.1016/j.envpol.2021.117267
Wu J., Hong Y., Wen X., Li Y., Wang Y., Chang X. (2020). Activity, abundance, and community composition of anaerobic ammonia–oxidizing (anammox) bacteria in sediment cores of the pearl river estuary. Estuar. Coasts 43 (1), 73–85. doi: 10.1007/s12237-019-00668-1
Xie H., Hong Y., Liu H., Jiao L., Wu J., Wang L. (2020). Spatio-temporal shifts in community structure and activity of nirS-type denitrifiers in the sediment cores of pearl river estuary. PloS One 15 (4), e0231271. doi: 10.1371/journal.pone.0231271
Yang Y., Li M., Li H., Li X.-Y., Lin J.-G., Denecke M., et al. (2020). Specific and effective detection of anammox bacteria using PCR primers targeting the 16S rRNA gene and functional genes. Sci. Total Environ. 734, 139387. doi: 10.1016/j.scitotenv.2020.139387
Zhang Y., Ji G., Wang C., Zhang X., Xu M. (2019). Importance of denitrification driven by the relative abundances of microbial communities in coastal wetlands. Environ. pollut. 244, 47–54. doi: 10.1016/j.envpol.2018.10.016
Zhang X., Zhang Q., Yang A., Hou L., Zheng Y., Zhai W., et al. (2018). Incorporation of microbial functional traits in biogeochemistry models provides better estimations of benthic denitrification and anammox rates in coastal oceans. J. Geophys. Res.: Biogeosci. 123 (10), 3331–3352. doi: 10.1029/2018jg004682
Zhao Y., Chen W., Wen D. (2020b). The effects of crude oil on microbial nitrogen cycling in coastal sediments. Environ. Int. 139, 105724. doi: 10.1016/j.envint.2020.105724
Zhao R., Mogollón J. M., Abby S. S., Schleper C., Biddle J. F., Roerdink D. L., et al. (2020a). Geochemical transition zone powering microbial growth in subsurface sediments. Proc. Natl. Acad. Sci. 117 (51), 32617–32626. doi: 10.1073/pnas.2005917117
Zhao Y., Xia Y., Kana T. M., Wu Y., Li X., Yan X. (2013). Seasonal variation and controlling factors of anaerobic ammonium oxidation in freshwater river sediments in the taihu lake region of China. Chemosphere 93 (9), 2124–2131. doi: 10.1016/j.chemosphere.2013.07.063
Zheng Y., Hou L., Liu M., Gao J., Yin G., Li X., et al. (2015). Diversity, abundance, and distribution of nirS-harboring denitrifiers in intertidal sediments of the Yangtze estuary. Microbial. Ecol. 70 (1), 30–40. doi: 10.1007/s00248-015-0567-x
Zheng Y., Hou L., Liu M., Yin G. (2019). Dynamics and environmental importance of anaerobic ammonium oxidation (anammox) bacteria in urban river networks. Environ. pollut. 254, 112998. doi: 10.1016/j.envpol.2019.112998
Keywords: denitrification, anammox, nirS/nirK, AMX 16S rRNA, 15N isotope
Citation: Teng Z, Zhen Y, Yu Z, Mi T and Cai T (2022) Nitrogen-loss and associated microbial communities in sediments from the Yangtze Estuary and adjacent sea. Front. Mar. Sci. 9:1078653. doi: 10.3389/fmars.2022.1078653
Received: 24 October 2022; Accepted: 14 November 2022;
Published: 01 December 2022.
Edited by:
Jing Wei, Sun Yat-sen University, ChinaCopyright © 2022 Teng, Zhen, Yu, Mi and Cai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yu Zhen, emhlbnl1QG91Yy5lZHUuY24=
 Zhenzhen Teng1,2,3
Zhenzhen Teng1,2,3 Yu Zhen
Yu Zhen Zhigang Yu
Zhigang Yu