Abstract
Iron is essential for all living things, especially marine organisms, due to its low availability in the marine environment. Iron regulation is key in all vertebrates and is controlled by hepcidin–ferroportin. To improve the knowledge of iron homeostasis in fish, an iron overload was induced in gilthead seabream (Sparus aurata), which was chosen as a study species because of its high interest in Mediterranean aquaculture. The amount of iron in different tissues was measured to determine its biodistribution and/or bioaccumulation. Since the liver is directly involved in iron metabolism, the morphological changes induced in this organ as a consequence of the iron increase were studied. The bactericidal activity of fish skin mucus was also determined, observing that it decreased in fish with high iron levels compared to control fish. In addition, to better understand iron regulation, the gene expression of hepcidin, ferroportin, transferrin, and ferritin was evaluated in the head kidney (the main hematopoietic organ in this species) and in the liver. Special interest was taken in the study of the multiple copies of the hamp2 gene present in the gilthead seabream genome. Bioinformatic analysis of the protein sequences derived from these hepcidin genes allowed us to determine the presence of one type I hepcidin and 12 type II hepcidins, all of them with antimicrobial potential. This number of mature hepcidin sequences found in gilthead seabream is the highest within Eupercaria described to date. All the results obtained indicate that the modulation of iron in seabream seems to be much more complicated than in other vertebrates, probably due to the possible involvement of the different hepcidins as mediators between iron metabolism and host immune response.
Introduction
The transition metal iron is essential for life because it plays an important role in the organism, being crucial in vital processes such as oxygen transport and energy metabolism. The regulation of iron metabolism is a serious challenge for all living organisms due to its poor bioavailability. In fact, Fe+3 tends to be insoluble when bound to oxygen, making it completely necessary for it to be found in protein-binding complexes (Bury and Grosell, 2003). For this reason, proteins involved in iron regulation have been narrowly selected for well-defined functions (Ilbert and Bonnefoy, 2013; Wallace, 2016). In addition to this, iron uptake mechanisms are critical for regulating iron balance, as no active iron excretion system has been determined to control iron overload (Kohgo et al., 2008). Instead, it is possible to balance iron in the body through the hepcidin–ferroportin axis (Rodrigues et al., 2006; Neves et al., 2015; Wallace, 2016). These genes involved in iron homeostasis are highly expressed in the liver, as a modulatory organ of iron storage, and in hepatocytes, enterocytes, and macrophages at the cellular level (Vogt et al., 2021). Moreover, transferrin forms protein complexes to transport iron into the circulatory system, whereas the storage function in tissues is performed by ferritin (Ponka et al., 1998).
Hepcidin–ferroportin modulation is shared between mammals and teleosts; the mature 25-residue human hepcidin is a negative modulator of the only iron exporting channel, ferroportin, which is degraded upon binding to hepcidin (Rodrigues and Pereira, 2004; Nemeth et al., 2006; Hilton and Lambert, 2008; Link et al., 2021). In contrast to mammals (except mice), multiple hepcidin genes have been found in fish. The single gene hamp1 (human orthologue) is involved in iron homeostasis, while the duplicate hamp2 gene plays immune functions as an antimicrobial peptide (Rodrigues et al., 2006; Neves et al., 2017; Boumaiza and Abidi, 2019). This hepcidin–ferroportin modulation has been elucidated using a teleost fish (European sea bass, Dicentrarchus labrax) based on acute iron overload. These studies indicated that hamp1 performed a negative control on ferroportin and not hamp2. In addition to iron overload, the effects of iron on hepatocyte morphology and massive iron accumulation in the liver were characterised (2017; Rodrigues and Pereira, 2004; Neves et al., 2015). Moreover, hepcidin was shown to modulate a coordination between iron control and innate antimicrobial immunity in zebrafish, as reduced iron availability limited the proliferation of pathogenic microorganisms (Lemos and Osorio, 2007; Jiang et al., 2017). Thus, iron regulation by hepcidin provides a firm bridge between iron metabolism and the host immune response (Rodrigues et al., 2006; Michels et al., 2015; Jiang et al., 2017).
The hepcidin family has been investigated by our research group from an immunological/antimicrobial perspective in gilthead seabream (Sparus aurata) (Cuesta et al., 2008; Serna-Duque et al., 2022). However, no advances have been reported on the regulatory functions of hepcidins and iron in this farmed marine fish. Therefore, the aim of the present work is to gain insight into the involvement of multiple type I and II hepcidins in the iron metabolism of gilthead seabream by developing an in vivo iron overload study. In addition, the effects of iron overload on liver morphology, iron storage in different organs, and the antimicrobial activity present in skin mucus was evaluated. Finally, a complete bioinformatic study of hepcidin gene-coded proteins was carried out, observing that gilthead seabream is the one that has developed the highest number of antimicrobial hepcidins within Eupercaria.
Methodology
Animals and experimental design
Twelve specimens of the seawater teleost gilthead seabream (S. aurata, 25 g mean weight) were maintained at the Marine Fish Facilities of the University of Murcia. The fish were kept in 200-L aquaria of running seawater (28‰ salinity) at 24°C ± 2°C and an artificial photoperiod of 12 h light:12 h dark. The animals were fed 1% of their body weight daily with a commercial pelleted diet (Skretting, Burgos, Spain) and acclimatised for 30 days prior to the experiments. All experimental procedures were evaluated and approved by the Ethical-Scientific Committee for Animal Experimentation of the University of Murcia (protocol CEEA 701/2021) in accordance with the European Directive 2010/63/UE on the protection of animals used for scientific purposes. The animals were randomly placed into four aquaria (three animals per aquarium). Six specimens from two aquaria were then injected intraperitoneally (i.p.) with 50 µl of sterile phosphate-buffered saline (PBS, Invitrogen) (control group). The other six fish (in two different aquaria) were injected with 50 µl of 10 mg ml−1 iron dextran (Sigma-Aldrich) diluted in sterile PBS (iron overload group) (Rodrigues and Pereira, 2004; Neves et al., 2015). At 72 h post-injection, five fish were randomly selected from each of the experimental groups and were anaesthetised and sacrificed by a benzocaine overdose (0.21 mM). Skin mucus samples were collected by gently scraping the surface of the gilthead seabream specimens with a plastic cell scraper, avoiding contamination of the samples with blood, urinogenital, or intestinal excretions (Guardiola et al., 2014). Blood samples were collected from the caudal vein with heparin-treated insulin syringes; after dissection, liver, head kidney (HK), anterior and posterior intestine, gill, eye, skin, spleen, and muscle samples were collected in ice and stored at −80°C until used.
Liver histology
Macroscopic images of the livers were obtained and immediately, liver samples (from both the control and iron overload group) were fixed in 10% formalin (Panreac) at room temperature for 24 h. After serial dehydration steps with ethanol, the samples were embedded in Paraplast (Thermo Fisher Scientific). Liver blocks were sectioned at 5 μm (Microm), and the sections were routinely stained with hematoxylin-eosin (H-E) and mounted in DPX (Sigma). Images were acquired with a Leica DFC280 digital camera coupled to a light microscope (Leica 6000B).
Iron determination
Frozen samples of blood, liver, HK, anterior and posterior intestine, gill, eye, skin, spleen, and muscle were lyophilised, and 100–200 mg of the resulting powder were placed in Teflon vessels with 3 ml of water, 2 ml of concentrated H2O2, and 5 ml of concentrated HNO3 acid solution (Thermo Fisher Scientific). Sample digestion was performed with a Milestone ETHOS Plus Microwave system operating on a standard program (85°C, 200°C, 210°C, and 0°C during 2, 8, 10, and 20 min, respectively). Finally, the solution was diluted to 50 ml and used to determine iron concentration by flame atomic absorption spectrometry (FAAS) (VARIAN, SpectrAA 220Z ZEEMAN). The accuracy of the present results was evaluated by analysing two reference materials (DOLT-2 dogfish liver and DORM-2 dogfish muscle). Data are presented as parts per million (ppm) of Fe (milligrammes of Fe per kilogramme of dry-weight tissue), and the proportion (%) of each tissue compared to total iron in the body was determined.
Bactericidal activity of skin mucus
The marine pathogenic bacterium Vibrio angillarum (strain R-82) was used to determine the bactericidal activity present in the skin mucus samples. Bacteria were cultured in liquid tryptic soy broth (TSB, Sigma) under agitation (24 h, 25°C, 200–250 rpm). Bacteria were adjusted to 108 colony-forming unit (CFU) ml−1. Bactericidal activity was determined following the method of Stevens et al. (1991) with some modifications using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (García Beltrán et al., 2020). Samples of 20 μl of skin mucus or PBS (positive control) were placed in a 96-well U-bottom plate. Aliquots of 100 μl of the previously adjusted bacteria were added per well, and the samples were incubated (5 h, 25°C). Next, 25 μl of MTT (1 mg ml−1; Sigma) was added to each well and incubated for another 10 min. The plates were centrifuged (10,000 rpm, 10 min), and the supernatant was removed. The pellet was then resuspended in 200 μl of dimethyl sulfoxide (DMSO; Sigma), which was added to each well. Finally, 100 μl of the DMSO was transferred to a 96-well flat-bottom plate, and the absorbance of dissolved formazan was measured at 570 nm. Bactericidal activity was expressed as percentage of nonviable bacteria, calculated as the difference between absorbance of bacteria surviving in test samples compared to the absorbance of bacteria from positive controls (100% growth or 0% bactericidal activity). Samples without bacteria were used as blanks (negative control). Samples without skin mucus were used as positive controls (100% growth or 0% bactericidal activity). All samples were assayed in triplicate.
Gene expression in the liver and head kidney
Total RNA was isolated from frozen liver and HK samples with TRIzol reagent following the manufacturer’s instructions. One microgram of total RNA was treated with DNAse I (Invitrogen) to remove genomic DNA, and first-strand cDNA was synthesised by reverse transcription using ThermoScriptTM RNAse H− Reverse Transcriptase (Invitrogen) with an oligo-dT12–18 primer (Invitrogen), followed by treatment with RNAse H (Invitrogen). Fast qPCR was performed with a QuantStudio™ 5 Real-Time PCR System (Applied Biosystems) using SYBR Green PCR Core Reagents (Applied Biosystems), cDNA samples, and designed primers (Tables 1, 2) (Cordero et al., 2016a). The reaction mixtures were incubated for 10 min at 95°C, followed by 40 cycles of 15 s at 95°C, 1 min at 60°C, and finally, 15 s at 95°C, 1 min at 60°C, and 15 s at 95°C. The PCR products were subjected to melting curve analysis. Ct values of hepcidin mRNAs were normalised to the 18S housekeeping gene, and iron overload samples were compared to control cDNA for expression ratio (Livak and Schmittgen, 2001).
Table 1
| Hepcidin name | Gene |
|---|---|
| hamp1 | 115566792 |
| hamp2.1 | 115567002 |
| hamp2.2 | 115567007 |
| hamp2.3 | BK059173 |
| hamp2.4 | BK059174 |
| hamp2.5 | 115567006 |
| hamp2.6 | 115567004 |
| hamp2.7 | BK059175 |
| hamp2.8 | 115566998 |
| hamp2.9 | BK059176 |
| hamp2.10 | 115567005 |
| hamp2.11 | 115567000 |
| hamp2.12 | 115567003 |
| hamp2.13 | 115566999 |
| hamp2.14 | 115567001 |
Accession numbers of hamp genes in gilthead seabream (NCBI).
Table 2
| Target | Forward primer | Reverse primer |
|---|---|---|
| hamp1 | AAGCGTCAGAGCCACATCTC | AGTCAATGCGTCGGAGAAGG |
| hamp2.1 | ATGTGGTGTCTGCTGCACATT | GCAGCATGACCAAATCCAGAGAT |
| hamp2.2 | CCTGACACGACTGGATGTAATGT | GCATGACCAAATCCAGGAACATCC |
| hamp2.3 | AGCAGCTTTCCAAATTTCCTTAGT | TTAGGATGATACATCAGTTAGCACA |
| hamp2.4 | CAGCCTGGGGTTCACACAAC | ACTGATCACACATGAAGGAGGATG |
| hamp2.5 | AGATGGGGTATGGCAACAGG | AATGCAATTTGGAGAACTGTTTATG |
| hamp2.6 | TGCTGTCCCATTCACTAAGGT | CAAAACTTACACCTCCTGCG |
| hamp2.7 | GGGATTCACACAACAACCACTG | GAAGATTCTTGAGGATGATACAGTCAC |
| hamp2.8 | AGCCTGGGATTCACACAAC | GATTTGACACTTTCAGTTAAAAAGGTT |
| hamp2.9 | CGGTTGCTGTCCTAACATGA | GACCAAATCCAGATATTACATCCTC |
| hamp2.10 | CCGCTGGCTGTAAGTTTTGC | TTGTGTGAATCCCAGGCTGC |
| hamp2.11 | CTGGGATTCACACAACAACCA | TGCGACTGTATCACCTACAC |
| hamp2.12 | CCGCTGGCTGTAAGTTTTGT | TTGTGTGAATTCCAGACTGC |
| hamp2.13 | TGGGATTCACACAACAACAACTT | GTAGCGTGTGTTGGTGATACAGTC |
| hamp2.14 | AGCCCTGCTGACTGTGAGTT | ACAGCCACAAAAGGAGTGCAA |
| slc40a1 | TAAAGTGGCCCAGACCTCGC | GGATGTAGCAGGTCGTCAGAAT |
| tf | CAGGACCAGCAGACCAAGTT | TGGTGGAGTCCTTGAAGAGG |
| fth1a | CCTCAGAATGGCATGGCAGA | AGCCGGTATCATGCAGATGG |
| fth1b | CAAACACACCATGGCCGAAG | TGCAGTACATGATGGGGAGC |
| rps18 | CGCTCAACCTCCTCATCAGT | AGGGTGTTGGCAGACGTTAC |
| ef1a | CTTCAACGCTCAGGTCATCAT | GCACAGCGAAACGACCAAGGGGA |
Primers designed for gene expression analysis.
Bioinformatic analysis of hepcidin mature peptides
Protein sequences were obtained from the UniProtKB database (https://www.uniprot.org/), using the entry Pfam PF06446 (hepcidin) as a query. Of the 811 obtained (September 2022), we focused on the 196 Eupercaria sequences. From these, incomplete sequences and duplicates were removed, resulting in the sequences used in the study (Supplementary Table S1). Mature hepcidin sequences were obtained after analysis of the signal peptide cleavage sites with SignalP 6.0 and mature peptides after furin action with ProP 1.0 (https://services.healthtech.dtu.dk/). Calculations and estimations on the physicochemical properties of the peptides were performed with PepCalc (https://pepcalc.com/), taking into account the formation of disulfide bonds between the cysteines present in the mature hepcidins. Sequences were aligned to default parameters using the MAFFT server (https://mafft.cbrc.jp/alignment/server/) and visualised with ESPript 4.0.10 (https://espript.ibcp.fr/ESPript) and Weblogo (https://weblogo.berkeley.edu/). Subsequently, the mature sequence tree was constructed using Mega 11 (https://www.megasoftware.net/) with the Neighbour-Joining (NJ) method, the JTT substitution model, heterogeneity between ignored sites (α = ∞) and a bootstrap of 1,000 replicates to increase the reliability of the obtained tree. The tree was visualised with iTOL (https://itol.embl.de/).
Statistical analysis
Results are presented in the figures as means ± standard error of the mean (SEM) or colour intensity in the heat maps. Statistical differences between control and iron overload groups were evaluated by a Student’s t-test in SPSS for Windows (version 15.0, SPSS Inc.). The significance level was 95% in all cases (p < 0.05).
Results
Effects of iron dextran injection in the liver of gilthead seabream
Since the liver is a critical organ in the regulation of iron metabolism in vertebrates, not only the iron concentration in this organ was determined but also its possible alterations at the macroscopic and histological levels. At the macroscopic level, the liver of gilthead seabream appeared as a reddish organ (control group), but in fish injected with iron dextran, the livers were always darker brown, with respect to the colour observed in the liver of fish in the control group (Figures 1A, B). Microscopically, the hepatocyte cords in the control livers showed the typical morphology of large, rounded polygonal cells. Also, the hepatocyte cords appeared interspersed with sinusoids filled with erythrocytes in their lumen. In addition, hepatocytes showed a central, clear, spherical nucleus (with a patent nucleolus) and eosinophilic cytoplasm (Figure 1C).
Figure 1

Representative macroscopic photographs of the liver of (A) control and (B) iron overload seabreams sampled 72 h postinjection. Bar = 2.5 cm. Representative histological sections of the liver of (C) control and (D) iron dextran-injected gilthead seabream sampled 72 h postinjection and stained with haematoxylin-eosin. Hemosiderin and iron-storage complex are indicated by arrows. Bar = 50 μm.
On the other hand, the liver of iron dextran-injected gilthead seabreams showed remarkable morphological alterations, with the appearance of the organ parenchyma being totally changed only 72 h postinjection. More specifically, the organisation of the liver parenchyma was lost, including the previously described corded distribution of hepatocytes (Figure 1D). These cells showed their nuclei displaced towards the cell periphery and a large vacuolation was evident in the cytoplasm of the cells. As for hepatic sinusoids, the presence of blood vessel congestion and the large vacuolation of the hepatocytes made it difficult to locate sinusoid-like structures in the liver of the iron dextran-injected specimens. However, the most notable difference between the cytoplasm of hepatocytes from the control and the iron overload group was the large amount of refractile brown spherical deposits formed by hemosiderin in the cytoplasm of hepatocytes from the iron overload group, giving rise to homochromatic livers (Figure 1D).
Loss of bactericidal activity in the skin mucus of iron overload gilthead seabream
Overall bactericidal activity was measured in the skin mucus of iron-overloaded fish because this constitutes the physical and chemical barrier against invading pathogens. A clear decrease in mucus protection against pathogenic Vibrio was observed with respect to the control, resulting in a 35% overgrowth of the pathogen, although a remarkable variability between samples was observed (Figure 2).
Figure 2

Bactericidal activity against pathogenic bacteria Vibrio anguillarum (strain R-82) of skin mucus of control and iron dextran-injected gilthead seabream. The bars represent the mean ± SEM. Different letters denote significant differences (Student’s t-test; p < 0.05) with control samples.
Distribution and accumulation of iron in gilthead seabream
The quantification of iron in several tissues and organs was performed by atomic absorption spectroscopy. The iron concentration of control fish was high in the anterior intestine (1,363.9 ppm, 54%), medium in the blood (571 ppm, 22.6%) and gills, an oxygen-retaining organ (445.1 ppm, 17.6%), low in the posterior intestine (3.6%), and very low in the liver and spleen (0.02%) (Figures 3A, B). In contrast, iron accumulated in the liver (20,481.5 ppm, 61%) and spleen (12,119 ppm, 36%), of the iron dextran-injected gilthead seabreams (Figure 3). Furthermore, it is noteworthy that the predominant iron storage organs in the control fish were completely relegated to the liver and spleen in the iron dextran-injected fish (Figure 3). In fact, no statistically significant changes in iron levels were found in either the skin or HK of control or iron dextran-injected fish.
Figure 3

(A) The distribution of iron in 10 gilthead seabream tissues, both in control and iron-overloaded fish (n = 3), was determined by atomic absorption spectroscopy and expressed in parts per million. (B) Iron distribution is shown as proportional concentration data (%) in the table. % Data from iron-storing tissues were highlighted in the control and treated samples.
Expression of genes involved in iron metabolism
The effect of iron overload on the expression of genes involved in iron metabolism, including the 15 hepcidin genes, was studied in both the liver (organ involved in iron accumulation and metabolism) and head kidney (the main haemopoietic organ in this fish species) of fish. While the expression of some hepcidin genes was significantly altered (p < 0.05) in the liver of fish injected i.p. with iron dextran with respect to the values recorded in control fish (Figure 4), no significant change in those genes could be detected in the HK (Figure 5). Thus, in the liver of iron dextran-injected fish, the expression of type I hepcidin (hamp1) was strongly upregulated (14-fold, Figure 4B), whereas type II hepcidins maintained their mRNA levels unchanged or were even downregulated (four-fold) as in the case of hamp2.4 and hamp2.5 (Figures 4C, D). In contrast, the expression of some genes related to iron storage did show remarkable changes in HK (Figure 5A), significantly decreasing the expression of ferroportin (slc40a1) (4-fold or four-fold., p < 0.05) (Figure 5B), transferrin (tf) (fivefold, p < 0.05) (Figure 5C), and the ferritin gene fth1b (more than sixfold, p < 0.05) (Figure 5D). These results show that the liver and HK exhibit differential expression patterns in hepcidin and iron storage modulator genes following iron overload.
Figure 4

Heatmap of the expression of hepcidin and modulators of iron metabolism in (A) the liver of gilthead seabream stimulated with iron overload at 72 h by real-time PCR. Data are shown as mean target gene expression relative to endogenous control 18s expression (n = 5). The color key indicates the intensity associated with the expression values. Significant differences (Student’s t-test; *p ≤ 0.05;) with the control are denoted. Significant results were expressed as the mean ± SEM (n = 5) of the relative mRNA expression of (B)hamp1, (C)hamp2.4 and (D)hamp2.5 in bar graphs. Significant differences (Student’s t-test; *p≤0.05) with control are denoted.
Figure 5
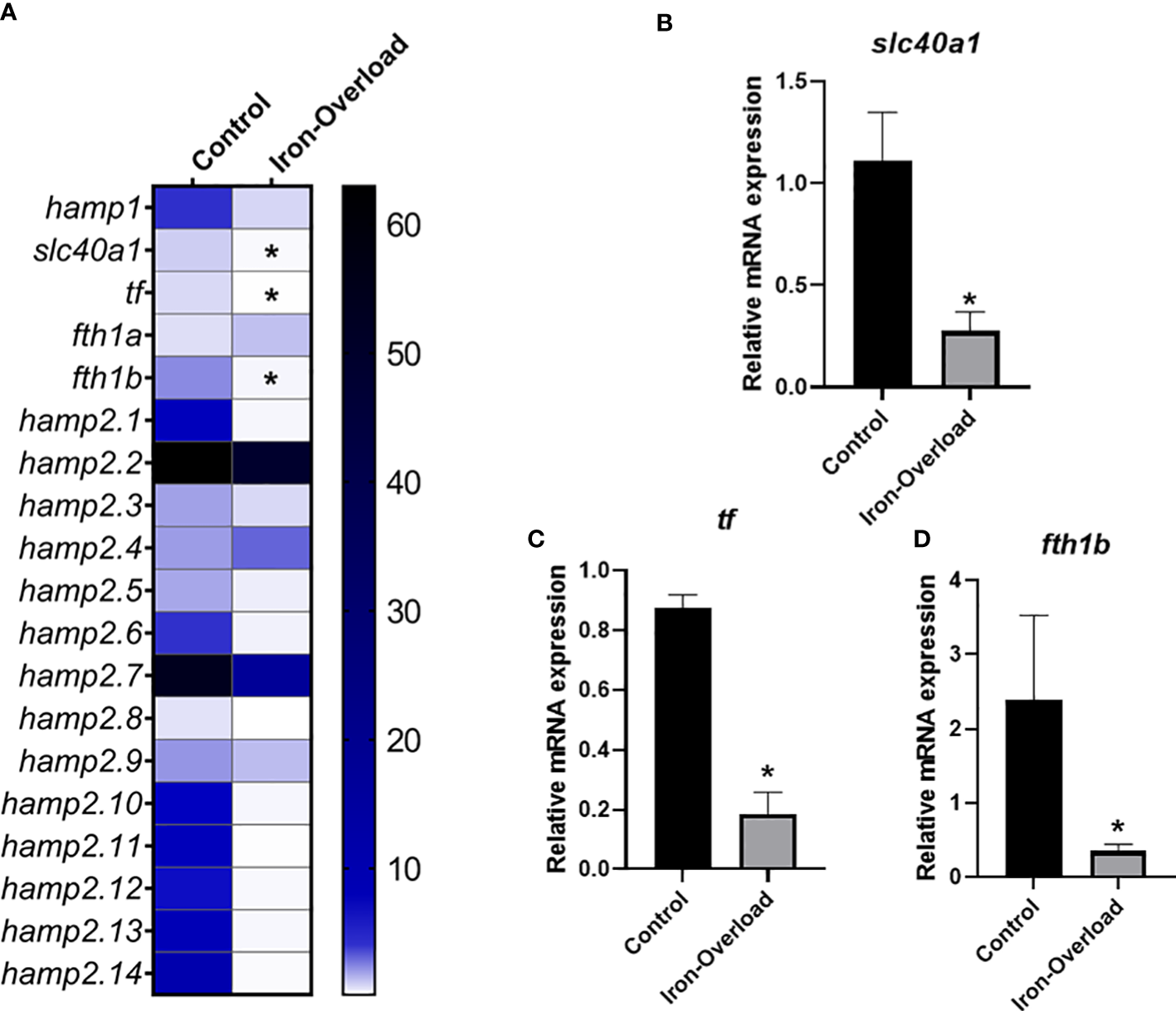
Heatmap of hepcidin and iron metabolism modulators expression (A) in the head-kidney of gilthead seabream stimulated with iron overload at 72 h by real-time PCR. Data are shown as mean target gene expression relative to the expression of control 18s expression (n = 5). The color key indicates the intensity associated with the expression values. Significant differences (Student’s t-test; *p ≤ 0.05;) with the control were denoted. Significant results were expressed as the mean ± SEM (n = 5) of the relative mRNA expression of (B)slc40a1, (C)tf and (D)fthb in bar graphs. Significant differences (Student’s t-test; *p≤0.05) with control are denoted.
Sparus aurata produces the largest variety of mature type II hepcidins within Eupercaria
Due to the large number of Sparus aurata hepcidin sequences found in UniProt (24) with respect to the previously described genes (Serna-Duque et al., 2022), we performed a detailed bioinformatics analysis not only of these sequences but also of the hepcidin sequences deposited in that database. The result showed the existence of 806 sequences in Gnathostomata (jawed vertebrates), of which more than half (468) are found in percomorph fishes (subdivision Percomorphaceae). Within the nine well-supported series (supraordinate groups) that group the diversity of percomorphs (Sanciangco et al., 2016), Ovalentaria (191) and Eupercaria (196) have the most sequences. By manually curating the sequences of the latter supraordinal group and removing duplicate or incomplete sequences, a total of 188 sequences were obtained (Supplementary Table S1). In these, the most representative orders were Perciformes (52.7%), Spariformes (21.3%), Tetraodontiformes (14.4%), and Centrarchiformes (5.9%) (Supplementary Figure S1). The Sparidae (21.3%), Tetraodontidae (12.8%), Sciaenidae (9.0%), and Nototheniidae (8.5%) were the most representative among the 29 families found in this study (Figure 6).
Figure 6
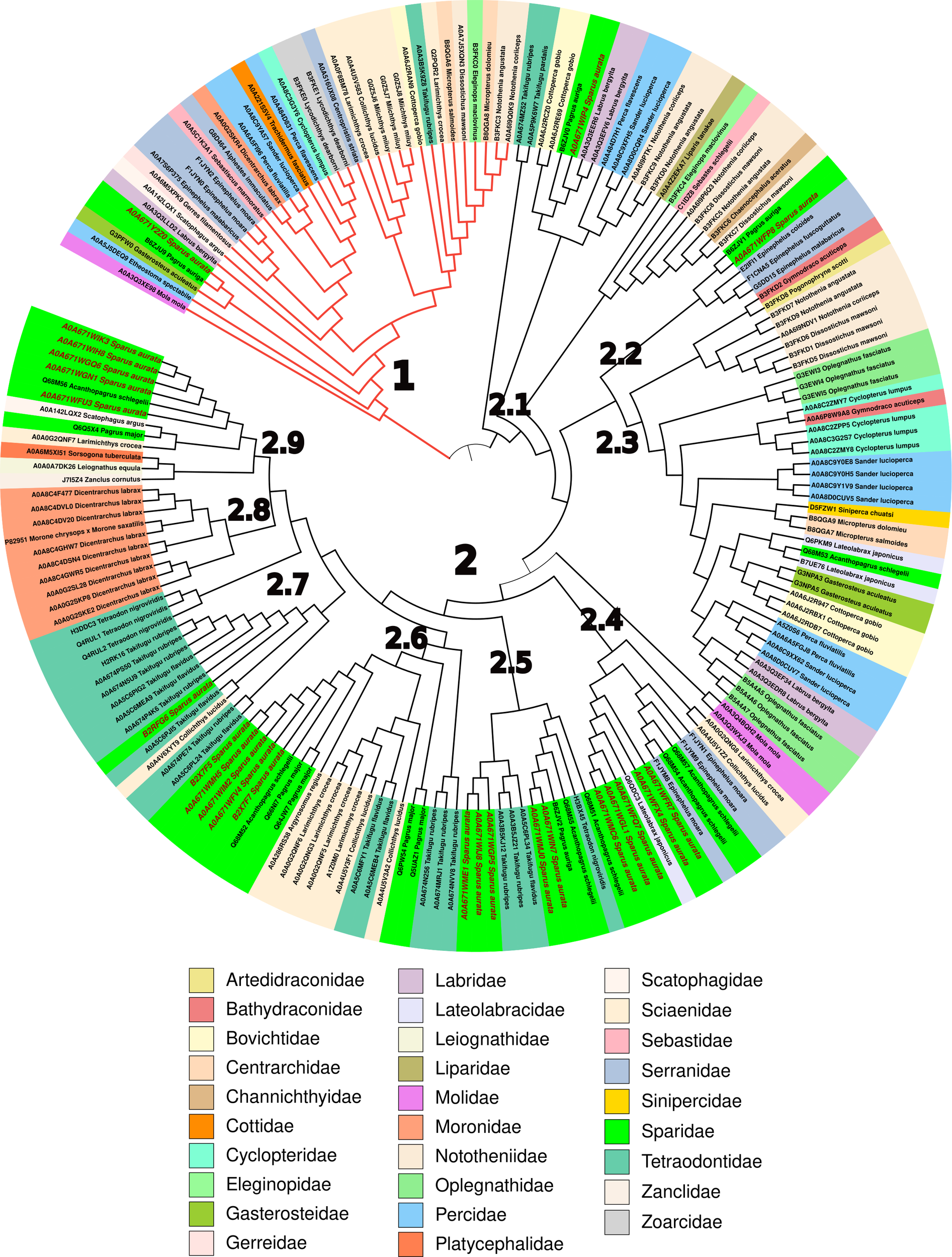
Phylogenetic analysis of mature hepcidins from different Eupercaria families. The neighbour-Joining (NJ) tree was obtained from 1,000 replicates as described in Materials and methods. The codes of the hepcidins are summarised in Supplementary Table S1.
Given that hepcidins are expressed as a precursor (preprohepcidin) (Barton and Acton, 2019), which must first be processed by removing its endoplasmic signal peptide from the N-ter (prohepcidin) and subsequently converted into the mature peptide by the action of furin (a subtilisin-like endopeptidase), it was necessary to determine bioinformatically which peptide is the mature peptide for each of the sequences (Supplementary Figure S2A). This allowed us to determine for the first time which were the consensus sequences for both signal peptide cleavage (SSA↓xP/S; Supplementary Figure S2B) and furin cleavage (RxKR↓xx; Supplementary Figure S2B). However, in the case of this protease, not only the basic amino acids of its cleavage site must be taken into account but also its clear preference for certain amino acids at positions −7 (Y), −6 (N), and −5 (N) with respect to the cleavage site (Supplementary Figure S2B).
The phylogenetic tree obtained from the mature hepcidin sequences clearly shows two main clades, one corresponding to type I hepcidins (29) (Figure 6, red branch) and the other corresponding to type II hepcidins (Figure 6). Clade 1 consists of hepcidins that bind to the ferroportin receptor, triggering its internalisation and degradation of the receptor–ligand complex, and whose consensus sequence is clearly conserved (Q-S-x-[IL]-S-[FLM]-C-x(2)-C-x(0,2)-C-x(0,1)-C-x(4)-G-x-G-x-C-C-C-C-[KR]-F) (Supplementary Figure S3), unlike the nine subclades (2.1–2.9) into which type II hepcidins are divided. Thus, subclade 2.1 has only the eight highly conserved cysteine characteristics of the hepcidin motif (C-x(2)-C-C-C-C-x-C-C-C-x(3,6)-C-x(2)-C-C) (Supplementary Figure S4), which stabilise the hairpin motif formed by the two antiparallel β-strands (Supplementary Figure S2). This minimal motif is reinforced in subclade 2.2 with another conserved area (GVCG) in the loop upstream of the second β-strand (C-x(2)-C-C-C-C-C-x-C-C-C-x(2)-G-V-C-G-x-C-C-C) (Supplementary Figure S5), which in turn is extended in clade 2.3 by the appearance of basic amino acids distributed throughout the mature peptide (G-[IFV]-K-C-C-[RK]-[FV]-C-C-x-C-C-C-x(2)-G-V-C-G-G-x-C-C-C-C-[RK]-[FK]-R-[RF]-G) (Supplementary Figure S6). Subclade 2.4 has a characteristic signature in its N-ter represented by SPAD (S-P-[AK]-[DK]-C-[EQR]-F-C-x-C-C-C-[PT]-[DEN]-x(2)-G-C-[GN]-x-C-x-x-[FHY]) (Supplementary Figure S7), whereas in subclade 2.5 this signature is in the C-ter with the sequence GCGTCCKF (C-R-F-C-C-C-C-x-C-C-C-x(0,1)-P-x-M-x-G-C-G-G-T-C-K-F) (Supplementary Figure S8). Subclade 2.6 is arginine-rich, with a highly conserved central core that includes both the β-strands and the loop connecting them (C-[HKR]-[FL]-C-C-R-C-C-C-x(1,2)-M-x-G-G-G-C-G-[ILV]-C-C), although, within this subclade, the QGSPAR sequence in the N-ter of members of the family Sciaenidae and Tetraodontidae particularly stands out (Supplementary Figure S9).
Finally, the next three subclades (2.7–2.9) are quite similar to each other (C-[KR]-F-C-C-C-x-C-C-C-P-[GDN]-M-x-G-C-G-G-G-[LV]-C-C-R-F), except for the N-ter, which ranges from the S-P-x-x of subclade 2.7 (Supplementary Figure S10) to the S-P-A-G of subclade 2.9 (Supplementary Figure S11), whereas subclade 2.8 (Supplementary Figure S12) is highly conserved with only three positions with small mutations, belonging basically to the nine type II hepcidins of the European sea bass Dicentrarchus labrax and the single one of the hybrid striped bass Morone chrysops x Morone saxatilis.
In general, the tree shows, with some exceptions as in the case of European sea bass, that there is no family pattern within the type II hepcidins, but rather, depending on the number of mature sequences that each fish has, there is a dispersion of sequences in several subclades, implying a great variability of sequences that use as a base the same tertiary structure (two-strand antiparallel β-sheet stabilised by disulfide bridges) that gives rise to peptides with a great diversity of properties. This is clearly seen in the diversity of the 12 mature S. aurata type II hepcidin peptides (Supplementary Table S2), ranging from poor to good water solubility and from a negative (−2 in A0A671WFX4) to a positive charge (+6 in B2X7F7), through the neutrality shown by A0A671WIJ8 (Supplementary Table S2).
Discussion
Iron overload in gilthead seabream (Sparus aurata) has been studied using multiple approaches to learn about the regulation and storage of iron because, despite the interest in iron metabolism and how it is regulated, it has yet to be studied in this farmed fish species. In this work, we have not only verified some observations found in other fish, but we have also provided new advances in the knowledge of iron metabolism and its relationship with the immune system in teleosts. Iron dextran was injected into the fish because it keeps iron solubilised and prevents its precipitation. In addition, it is rapidly absorbed after injection into the peritoneal cavity (Rodrigues and Pereira, 2004). The liver has a predominant role in iron metabolism, and this was the reason for studying the microscopic appearance of hepatocytes and the iron accumulation in this organ. In fact, our data agree with other studies on gilthead seabream, sea bass, and trout that reported iron deposits in the cytoplasm of hepatocytes after iron overload (Carriquiriborde et al., 2004; Rodrigues and Pereira, 2004; Rodrigues et al., 2006). However, we have described important microscopic changes in the hepatocytes of fish injected with iron dextran, such as displacement of the nucleus to the periphery of the hepatocytes and the widespread vacuolisation of their cytoplasm, together with congestion of the sinusoids. All these changes are consistent with those described in the liver histology of fish reared in water contaminated with heavy metals (Savassi et al., 2020). Likewise, the increase in the proportion of iron present in the liver and spleen of fish injected with iron dextran supports the fact that iron accumulates in these organs and that the spleen is an organ involved in the recycling of this micronutrient (Carpenè et al., 1999; Bury and Grosell, 2003; Rodrigues and Pereira, 2004). As for the results of iron in the intestine and gills of the fish in the control group, they coincided with previous findings in the literature. In other words, it is known that the gut and gills play an important role in iron uptake and that the blood contains different heme groups in red blood cells and transferrin (Carpenè et al., 1999; Bury and Grosell, 2003). However, in this study, it was clear that tissue iron ratios shifted from the gut, gills, and blood of control fish to the liver and spleen of iron dextran-injected fish. In addition, the skin and head kidney were ruled out as iron stores, in contrast to other classical works that described the role of the kidney in iron storage (Carpenè et al., 1999; Carriquiriborde et al., 2004). However, further studies are needed to know whether iron could accumulate in the central part of the kidney or the caudal zone (excretory part) (Leknes, 2001). Since excess iron is toxic, despite being a vital micronutrient for teleost fish, both its metabolism and possible storage should be very finely regulated by the liver and spleen to avoid this possible toxicity.
In addition to a different accumulation of iron in some organs after iron overload in gilthead seabream, the bactericidal activity of the skin mucus, which is the first line of host defence, was also altered; more concretely, it was considerably reduced against the pathogen V. anguillarum R-82. In fact, the bacteria grew better in this mucus than in that of the control group. This bacterial overgrowth might be associated with a higher release of secreted iron in the mucus, as this does not accumulate in the skin according to our FAAS results (Jiang et al., 2017; Martínez et al., 2017; Díaz et al., 2021). Thus, in the mucosa of iron-overloaded fish, competition for iron occurs between bacterial siderophores and transferrin (Raeder et al., 2007; Arezes et al., 2015; Cordero et al., 2016b), with iron remaining available despite overexpression of hamp1, downregulation of iron storage genes in HK, and accumulation in hepatocytes.
As previously described, hepcidin–ferroportin modulation is key to the control of iron metabolism. For this reason, we have studied the expression of hepcidin, ferroportin, transferrin, and ferritin genes in iron-overloaded gilthead seabream. While the hamp1 gene was found to be strongly upregulated in the liver, consistent with other studies, however, the lack of changes in the ferroportin gene was unexpected (2017; Neves et al., 2015). Nevertheless, the downregulation of ferroportin could be induced earlier in gilthead seabream liver compared to sea bass, and for that reason, we decide to use a common primer for two copies of slc40a1 gene (Chen et al., 2021). Likewise, this observation is in agreement with previous results obtained in turbot injected with synthetic hepcidin. All these results seem to suggest that ferroportin regulation may vary among fish species (Yang et al., 2013). In contrast to the studies performed in sea bass, in this work, we encountered a major challenge in the study of hepcidin regulation due to the massive duplications of hepcidin II in gilthead seabream. Although most hamp2 genes maintained their mRNA levels after iron injection, surprisingly, hamp2.4 and 2.5 genes were downregulated, and, contrary to expectations, hamp2 expression was not altered by iron overload (Shi and Camus, 2006; Neves et al., 2015; Neves et al., 2017). As suggested, hepcidin II expression could be decreased for “resource-sparing” reasons or might be involved in modulating iron metabolism (Hirono et al., 2005; Mu et al., 2018). However, these questions should be investigated in future studies. In contrast, hepcidin expression was not induced by iron overload in the HK, which reinforced the role of the liver as a major contributor to hepcidin synthesis (Neves et al., 2015).
In contrast, ferroportin, transferrin, and ferritin-1b genes appeared downregulated in the primary immune organ, such as the HK, but not in the iron-regulating organ, the liver, in fish studied 72 h after iron injection. These results are consistent with those obtained after in vivo synthetic hepcidin injection in turbot, which indicated the same downregulation of ferroportin and transferrin genes in the HK but not in the liver (Yang et al., 2013). Indeed, this decreased expression of genes involved in iron storage is likely related to the immunometabolic response to limit iron for pathogens (Vibrio and Photobacterium) (Lemos and Balado, 2020), which is consistent with FAAS findings in this organ. Thus, the HK not only shows local regulation of iron metabolism but may also be priming its leucocytes for potential microbial attack (Nairz et al., 2009; Grayfer et al., 2014; Neves et al., 2015; Díaz et al., 2021).
From the above discussion, there remains an intriguing and novel detail in this work, and that is the downregulation of the hamp2.4 and hamp2.5 genes. From the bioinformatic analysis carried out with all the hepcidin sequences in Eupercaria, it is clear that the type I ones related to the interaction with ferroportin have a great similarity, but not so for the type II ones, which are distributed in multiple clades as a specific response of each one to the possible pathogens faced by each fish. Gilthead seabream is the most promiscuous example with up to 12 different mature peptides to better adapt against different pathogens. These type II hepcidins of this fish species range from anionic and neutral peptides to highly cationic peptides, such as those corresponding to the hamp2.4 (+5, A0A671WMH5) and hamp2.5 (+6, B2X7F7) genes. At least in the case of the unique hamp2.5 sequence, its downregulation could be explained by the high Boman index (2.48 kcal/mol), which estimates the potential of a protein to bind other proteins (Boman, 2003). In other words, a high value of the Boman index indicates that an antimicrobial peptide will be multifunctional or play a variety of different roles within the cell due to its ability to interact with a wide range of proteins (Azad et al., 2011), and in this case, its downregulation would allow dysregulation of other mechanisms to counteract the iron overload to which the gilthead seabream is being subjected (Valenzuela-Muñoz et al., 2020). Iron overdose is known to alter the immune response of fish (Valenzuela-Muñoz et al., 2020). However, it should be noted that the present results may not reflect the usual metabolic and immune responses to Fe, since these responses were obtained under the experimental conditions already mentioned (very high iron concentration and iron exposure was by i.p. injection, which delivers a high concentration of Fe directly to the fish organs).
In conclusion, induced iron overload in gilthead seabream has been used to investigate the role of iron from multiple approaches. In particular, the molecular regulation of genes involved in iron control, especially multiple hepcidins, has been integrated with the effects of iron overload in fish, such as iron storage in hepatocytes, changes in tissue distribution of iron, and loss of skin mucosal bactericidal activity. This multipolar integration has supported the involvement of type I and II hepcidins in iron metabolism and suggests an immunometabolic role for iron control. Furthermore, extensive bioinformatic analysis shows that, within Eupercaria, gilthead seabream has a plethora of hepcidins, whose antimicrobial capacity should be studied not only for their implications in aquaculture but also for their possible interaction with other proteins involved in iron metabolism.
Statements
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The animal study was reviewed and approved by Comité de Ética de la Universidad de Murcia.
Author contributions
JS-D: methodology, investigation, and writing original draft. CE: methodology, formal analysis, and writing—review and editing. SML: methodology and investigation. ÁS-F: conceptualisation, validation, data curation, visualisation, supervision, and writing—review and editing. ME: term, conceptualisation, resources, project administration, funding acquisition, and writing—review and editing. All authors contributed to the article and approved the submitted version.
Funding
This work has been funded by the proyecto PID2020-113637RB-C21 de investigación financiado por MCIN/AEI/10.13039/501100011033 and form part of the ThinkInAzul programme supported by MCIN with funding from European Union Next Generation EU (PRTR-C17.I1) and by the Comunidad Autónoma de la Región de Murcia-Fundación Séneca (Spain).
Acknowledgments
JASD has a predoctoral contract (FPU19/02192).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.1073060/full#supplementary-material
References
1
Arezes J. Jung G. Gabayan V. Valore E. Ruchala P. Gulig P. A. et al . (2015). Hepcidin-induced hypoferremia is a critical host defense mechanism against the siderophilic bacterium Vibrio vulnificus. Cell Host Microbe17, 47–57. doi: 10.1016/j.chom.2014.12.001
2
Azad M. A. Huttunen-Hennelly H. E. K. Ross Friedman C. (2011). Bioactivity and the first transmission electron microscopy immunogold studies of short de novo-designed antimicrobial peptides. Antimicrob. Agents Chemother.55, 2137–2145. doi: 10.1128/AAC.01148-10
3
Barton J. C. Acton R. T. (2019). Hepcidin, iron, and bacterial infection. Vitam. Horm.110, 223–242. doi: 10.1016/bs.vh.2019.01.011
4
Boman H. G. (2003). Antibacterial peptides: basic facts and emerging concepts. J. Intern. Med.254, 197–215. doi: 10.1046/j.1365-2796.2003.01228.x
5
Boumaiza M. Abidi S. (2019). “Chapter one - hepcidin cDNA evolution in vertebrates,” in Vitamins and hormones iron metabolism: Hepcidin. Ed. LitwackG. (Cambridge, USA: Academic Press), 1–16. doi: 10.1016/bs.vh.2019.01.001
6
Bury N. Grosell M. (2003). Iron acquisition by teleost fish. Comp. Biochem. Physiol. C Toxicol. Pharmacol.135, 97–105. doi: 10.1016/s1532-0456(03)00021-8
7
Carpenè E. Serra R. Manera M. Isani G. (1999). Seasonal changes of zinc, copper, and iron in gilthead sea bream (Sparus aurata) fed fortified diets. Biol. Trace Elem. Res.69, 121–139. doi: 10.1007/BF02783864
8
Carriquiriborde P. Handy R. D. Davies S. J. (2004). Physiological modulation of iron metabolism in rainbow trout (Oncorhynchus mykiss) fed low and high iron diets. J. Exp. Biol.207, 75–86. doi: 10.1242/jeb.00712
9
Chen J. Jiang W. Xu Y.-W. Chen R.-Y. Xu Q. (2021). Sequence analysis of hepcidin in barbel steed (Hemibarbus labeo): QSHLS motif confers hepcidin iron-regulatory activity but limits its antibacterial activity. Dev. Comp. Immunol.114, 103845. doi: 10.1016/j.dci.2020.103845
10
Cordero H. Mauro M. Cuesta A. Cammarata M. Esteban M. Á. (2016a). In vitro cytokine profile revealed differences from dorsal and ventral skin susceptibility to pathogen-probiotic interaction in gilthead seabream. Fish. Shellfish Immunol.56, 188–191. doi: 10.1016/j.fsi.2016.07.018
11
Cordero H. Morcillo P. Cuesta A. Brinchmann M. F. Esteban M. A. (2016b). Differential proteome profile of skin mucus of gilthead seabream (Sparus aurata) after probiotic intake and/or overcrowding stress. J. Proteomics132, 41–50. doi: 10.1016/j.jprot.2015.11.017
12
Cuesta A. Meseguer J. Esteban M. Á. (2008). The antimicrobial peptide hepcidin exerts an important role in the innate immunity against bacteria in the bony fish gilthead seabream. Mol. Immunol.45, 2333–2342. doi: 10.1016/j.molimm.2007.11.007
13
Díaz R. Troncoso J. Jakob E. Skugor S. (2021). Limiting access to iron decreases infection of Atlantic salmon SHK-1 cells with bacterium Piscirickettsia salmonis. BMC Vet. Res.17, 155. doi: 10.1186/s12917-021-02853-6
14
García Beltrán J. M. Silvera D. G. Ruiz C. E. Campo V. Chupani L. Faggio C. et al . (2020). Effects of dietary Origanum vulgare on gilthead seabream (Sparus aurata l.) immune and antioxidant status. Fish. Shellfish Immunol.99, 452–461. doi: 10.1016/j.fsi.2020.02.040
15
Grayfer L. Hodgkinson J. W. Belosevic M. (2014). Antimicrobial responses of teleost phagocytes and innate immune evasion strategies of intracellular bacteria. Dev. Comp. Immunol.43, 223–242. doi: 10.1016/j.dci.2013.08.003
16
Guardiola F. A. Cuesta A. Arizcun M. Meseguer J. Esteban M. A. (2014). Comparative skin mucus and serum humoral defence mechanisms in the teleost gilthead seabream (Sparus aurata). Fish. Shellfish Immunol.36, 545–551. doi: 10.1016/j.fsi.2014.01.001
17
Hilton K. B. Lambert L. A. (2008). Molecular evolution and characterization of hepcidin gene products in vertebrates. Gene415, 40–48. doi: 10.1016/j.gene.2008.02.016
18
Hirono I. Hwang J.-Y. Ono Y. Kurobe T. Ohira T. Nozaki R. et al . (2005). Two different types of hepcidins from the Japanese flounder Paralichthys olivaceus. FEBS J.272, 5257–5264. doi: 10.1111/j.1742-4658.2005.04922.x
19
Ilbert M. Bonnefoy V. (2013). Insight into the evolution of the iron oxidation pathways. Biochim. Biophys. Acta1827, 161–175. doi: 10.1016/j.bbabio.2012.10.001
20
Jiang X. Liu Z. Lin A. Xiang L. Shao J. (2017). Coordination of bactericidal and iron regulatory functions of hepcidin in innate antimicrobial immunity in a zebrafish model. Sci. Rep.7, 4265. doi: 10.1038/s41598-017-04069-x
21
Kohgo Y. Ikuta K. Ohtake T. Torimoto Y. Kato J. (2008). Body iron metabolism and pathophysiology of iron overload. Int. J. Hematol.88, 7–15. doi: 10.1007/s12185-008-0120-5
22
Leknes I. L. (2001). The uptake of foreign ferritin by macrophages in the spleen, trunk kidney and liver of platy. J. Fish. Biol.59, 1412–1415. doi: 10.1006/jfbi.2001.1728
23
Lemos M. L. Balado M. (2020). Iron uptake mechanisms as key virulence factors in bacterial fish pathogens. J. Appl. Microbiol.129, 104–115. doi: 10.1111/jam.14595
24
Lemos M. L. Osorio C. R. (2007). Heme, an iron supply for vibrios pathogenic for fish. Biometals20, 615–626. doi: 10.1007/s10534-006-9053-8
25
Link C. Knopf J. D. Marques O. Lemberg M. K. Muckenthaler M. U. (2021). The role of cellular iron deficiency in controlling iron export. Biochim. Biophys. Acta Gen. Subj.1865, 129829. doi: 10.1016/j.bbagen.2020.129829
26
Livak K. J. Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods25, 402–408. doi: 10.1006/meth.2001.1262
27
Martínez D. Oyarzún R. Pontigo J. P. Romero A. Yáñez A. J. Vargas-Chacoff L. (2017).Nutritional immunity triggers the modulation of iron metabolism genes in the Sub-Antarctic notothenioid Eleginops maclovinus in response to Piscirickettsia salmonis. (Accessed March 16, 2022).
28
Michels K. Nemeth E. Ganz T. Mehrad B. (2015). Hepcidin and host defense against infectious diseases. PloS Pathog.11, e1004998. doi: 10.1371/journal.ppat.1004998
29
Mu Y. Huo J. Guan Y. Fan D. Xiao X. Wei J. et al . (2018). An improved genome assembly for Larimichthys crocea reveals hepcidin gene expansion with diversified regulation and function. Commun. Biol.1, 1–12. doi: 10.1038/s42003-018-0207-3
30
Nairz M. Fritsche G. Crouch M.-L. V. Barton H. C. Fang F. C. Weiss G. (2009). Slc11a1 limits intracellular growth of Salmonella enterica sv. Typhimurium by promoting macrophage immune effector functions and impairing bacterial iron acquisition. Cell Microbiol.11, 1365–1381. doi: 10.1111/j.1462-5822.2009.01337.x
31
Nemeth E. Preza G. C. Jung C.-L. Kaplan J. Waring A. J. Ganz T. (2006). The n-terminus of hepcidin is essential for its interaction with ferroportin: structure-function study. Blood107, 328–333. doi: 10.1182/blood-2005-05-2049
32
Neves J. V. Caldas C. Vieira I. Ramos M. F. Rodrigues P. N. S. (2015). Multiple hepcidins in a teleost fish, Dicentrarchus labrax : Different hepcidins for different roles. J.Immunol195, 2696–2709. doi: 10.4049/jimmunol.1501153
33
Neves J. V. Ramos M. F. Moreira A. C. Silva T. Gomes M. S. Rodrigues P. N. S. (2017). Hamp1 but not Hamp2 regulates ferroportin in fish with two functionally distinct hepcidin types. Sci. Rep.7, 14793. doi: 10.1038/s41598-017-14933-5
34
Ponka P. Beaumont C. Richardson D. R. (1998). Function and regulation of transferrin and ferritin. Semin. Hematol.35, 35–54.
35
Raeder I. L. U. Paulsen S. M. Smalås A. O. Willassen N. P. (2007). Effect of fish skin mucus on the soluble proteome of Vibrio salmonicida analysed by 2-d gel electrophoresis and tandem mass spectrometry. Microb. Pathog.42, 36–45. doi: 10.1016/j.micpath.2006.10.003
36
Rodrigues P. N. S. Pereira F. A. (2004). A model for acute iron overload in sea bass (Dicentrarchus labrax l.). Lab. Anim.38, 418–424. doi: 10.1258/0023677041958909
37
Rodrigues P. N. S. Vázquez-Dorado S. Neves J. V. Wilson J. M. (2006). Dual function of fish hepcidin: Response to experimental iron overload and bacterial infection in sea bass (Dicentrarchus labrax). Dev. Comp. Immunol.30, 1156–1167. doi: 10.1016/j.dci.2006.02.005
38
Sanciangco M. D. Carpenter KE Betancur R.-R. . (2016). Phylogenetic placement of enigmatic percomorph families (Teleostei: Percomorphaceae). Mol Phylogenet Evol. 94(Pt B):565–576. doi: 10.1016/j.ympev.2015.10.006.
39
Savassi L. A. Paschoalini A. L. Arantes F. P. Rizzo E. Bazzoli N. (2020). Heavy metal contamination in a highly consumed Brazilian fish: immunohistochemical and histopathological assessments. Environ. Monit. Assess.192, 542. doi: 10.1007/s10661-020-08515-8
40
Serna-Duque J. A. Cuesta A. Esteban M. Á. (2022). Massive gene expansion of hepcidin, a host defense peptide, in gilthead seabream (Sparus aurata). Fish. Shellfish Immunol.124, 563–571. doi: 10.1016/j.fsi.2022.04.032
41
Shi J. Camus A. C. (2006). Hepcidins in amphibians and fishes: Antimicrobial peptides or iron-regulatory hormones? Dev. Comp. Immunol.30, 746–755. doi: 10.1016/j.dci.2005.10.009
42
Stevens M. G. Kehrli M. E. Canning P. C. (1991). A colorimetric assay for quantitating bovine neutrophil bactericidal activity. Vet. Immunol. Immunopathol.28, 45–56. doi: 10.1016/0165-2427(91)90042-B
43
Valenzuela-Muñoz V. Valenzuela-Miranda D. Gonçalves A. T. Novoa B. Figueras Huerta A. (2020). Induced−iron overdose modulate the immune response in Atlantic salmon increasing the susceptibility to Piscirickettsia salmonis infection. Aquaculture521, 735058. doi: 10.1016/j.aquaculture.2020.735058
44
Vogt A.-C. S. Arsiwala T. Mohsen M. Vogel M. Manolova V. Bachmann M. F. (2021). On iron metabolism and its regulation. Int. J. Mol. Sci.22, 4591. doi: 10.3390/ijms22094591
45
Wallace D. F. (2016). The regulation of iron absorption and homeostasis. Clin. Biochem. Rev.37, 51–62.
46
Yang C.-G. Liu S.-S. Sun B. Wang X.-L. Wang N. Chen S.-L. (2013). Iron-metabolic function and potential antibacterial role of hepcidin and its correlated genes (Ferroportin 1 and transferrin receptor) in turbot (Scophthalmus maximus). Fish. Shellfish Immunol.34, 744–755. doi: 10.1016/j.fsi.2012.11.049
Summary
Keywords
hepcidin, host-defence peptide, iron, immunometabolism, Gilthead seabream (Sparus aurata L.), evolution, teleosts
Citation
Serna-Duque JA, Espinosa Ruiz C, Martínez Lopez S, Sánchez-Ferrer Á and Esteban MÁ (2022) Immunometabolic involvement of hepcidin genes in iron homeostasis, storage, and regulation in gilthead seabream (Sparus aurata). Front. Mar. Sci. 9:1073060. doi: 10.3389/fmars.2022.1073060
Received
18 October 2022
Accepted
22 November 2022
Published
08 December 2022
Volume
9 - 2022
Edited by
Rodrigo Vidal, University of Santiago, Chile
Reviewed by
Xianyong Bu, Ocean University of China, China; Itziar Estensoro, Spanish National Research Council (CSIC), Spain
Updates
Copyright
© 2022 Serna-Duque, Espinosa Ruiz, Martínez Lopez, Sánchez-Ferrer and Esteban.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Álvaro Sánchez-Ferrer, alvaro@um.es; Maria Ángeles Esteban, aesteban@um.es
This article was submitted to Marine Fisheries, Aquaculture and Living Resources, a section of the journal Frontiers in Marine Science
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.