- 1Yellow Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Qingdao, China
- 2Laboratory for Marine Biology and Biotechnology, Pilot National Laboratory for Marine Science and Technology (Qingdao), Qingdao, China
- 3College of Fisheries and Life Science, Dalian Ocean University, Dalian, China
- 4College of Fisheries and Life Science, Shanghai Ocean University, Ministry of Education, Shanghai, China
The tiger puffer (Takifugu rubripes) is an important economic fish species in northern China. However, it is a warm-temperature species, and low winter temperatures can result in high mortality in aquaculture. Understanding the mechanisms of cold resistance in tiger puffers will thus provide critical information to help cope with winter cold. In this study, we performed transcriptome analysis of livers from puffer fish kept at different temperatures (18°C, 13°C, and 8°C) to identify the key pathways and genes involved in the response to low-temperature stress. We also detected serum levels of proteases, arginine, and proline to obtain further information on the response to cold adaption. Totals of 51, 942, and 195 differentially expressed genes were identified in the 18°C vs 13°C, 18°C vs 8°C, and 13°C vs 8°C groups, respectively. Pathway analysis showed that significantly enriched pathways were mainly related to digestion, metabolism, and environmental adaptation. Most genes in the pathways related to digestion and metabolism were down-regulated, while most genes in the pathways related to environmental adaptation were up-regulated. Serum levels of proteases were significantly lower in the low-temperature groups (13°C and 8°C) compared with the control group (18°C), while arginine and proline levels were significantly higher in the 8°C group compared with the other two groups. These results suggest that low temperature caused digestive and metabolic disorders, as well as adaptive changes to low temperature in tiger puffers. On this premise, we found that some up-regulated genes in the pancreatic secretion pathway, arginine and proline metabolism pathway, and circadian rhythm pathway played important roles in the survival, growth, and development of tiger puffers under low-temperature stress. The accumulation of arginine and proline can maintain metabolism and circulation and resist cold stress. The circadian rhythm is closely related to digestion and metabolism, which is an adaptive change and plays a positive role in the resistance to low temperature. The results of this study provide new insights and a theoretical basis for the study of cold tolerance in tiger puffers.
Introduction
Water temperature plays an important role in the survival of fish by directly affecting their physiology, biochemical state, and behavior (Beitinger et al., 2000; Zhou L.Y et al., 2019). The tiger puffer (Takifugu rubripes) is a eurythermal, euryhaline carnivorous fish that is suitable for cage culture in northern China, with a growth-temperature range of 16–25°C. However, lower water temperatures in winter may result in reduced growth performance or even death. Tiger puffers cultivated in northern China inevitably experience the process of the land-sea relay breeding method and the overwintering process, due to the decrease of winter temperature, which requires heating to raise the water temperature, which will increase the cost and pressure of environmental-protection pressures. Economic losses caused by low temperatures can be reduced by breeding lines with cold-tolerant traits, and understanding the regulatory mechanisms at low temperature can provide a strong foundation for related research.
Recent advancements in sequencing technology mean that Illumina RNA-seq technology has played an increasingly important role in analyzing the regulation of economic traits, including the study of fish under environmental stress. For example, both low salinity and high temperature were shown to disrupt lipid metabolism in the liver of turbot (Scophthalmus maximus), and the corresponding key genes were screened using transcriptomics technology (Liu et al., 2021; Zhao et al., 2021). Low temperature also affected renal immune function-related pathways in Nile tilapia (Oreochromis niloticus) (Zhou T et al., 2019) and amino acid metabolism-related pathways in European perch (Dicentrarchus labrax) (Zhang et al., 2021). In addition, high temperature resulted in immune disorders and metabolic imbalance in crayfish (Procambarus clarkii) (Luo et al., 2021). These studies demonstrate the important effects of temperature on the metabolic responses of fish. The liver is an important metabolic organ in fish and plays an important role in environmental adaptation, coping with environmental disturbance, and homeostasis of the internal environment (Ibarz et al., 2010; Jiao et al., 2020). The liver is thus a suitable organ in which to analyze the metabolic adaptability of fish to changes in temperature.
Considering the importance of cold tolerance in tiger puffers in aquaculture, we studied the transcriptomic changes in livers from tiger puffers subjected to cold stress. We carried out RNA sequencing of livers from puffer fish kept at normal temperature (18°C) and low temperatures (13°C and 8°C) using Illumina sequencing technology, followed by Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses to determine the key pathways. We also examined the physiological responses in the blood to clarify the metabolic effects of temperature. This study aimed to investigate the adaptive molecular regulatory mechanisms in tiger puffers maintained in a long-term low-temperature environment, and identify key signaling pathways and genes. The results further our understanding of the molecular consequences of low-temperature exposure in puffer fish, and lay the foundation for selective breeding programs aimed at improving cold tolerance in puffer fish.
Materials and methods
Ethics statement
The study protocol was approved and supervised by the Institutional Animal Care and Use Committee of the Yellow Sea Fisheries Research Institute (Qingdao, China).
Experimental design and sample collection
Tiger puffers were maintained by our research group at an aquatic farm in Tangshan, China. The fish had no surface damage and showed active swimming. Fish used in the experiment was 8 months with a body weight of 106.6 ± 4.9 (M ± SD) g and body length of 15.0 ± 0.2 (M ± SD) cm. We set up three groups of fish in constant-temperature recirculating aquaculture systems, with three experimental barrels (1000 L) per group. Each experimental barrel included 30 randomly selected fish, and was kept at 18°C for 1 week before the experiment. Barrels in the three groups were then set at temperatures of 18°C (group A), 13°C (group B), and 8°C (group C), respectively, with 18°C used as the control group and 13°C and 8°C used as the low-temperature groups. Three parallel groups were set for each temperature. At the beginning of the experiment, the water temperatures in the two low-temperature groups were adjusted to 8°C and 13°C within 12 h, respectively, and the control group was kept at 18°C. The fish were then stocked at a constant water temperature under natural light for 4 weeks, with salinity 30 ppt, dissolved oxygen > 8 mg/L and ammonia nitrogen < 0.2 mg/L. Three fish were randomly selected from each barrel at the fourth week, anesthetized in 200 mg/L MS-222 (Sigma-Aldrich Corporation, St. Louis, MO, USA), and immediately dissected on ice. Blood samples were collected from the caudal vein, transferred into RNase-free tubes, and stored at 4°C for 6 h. Afterward, the supernatant was collected by centrifugation at 2000×g for 10 min at 4°C. The liver was removed from each fish and immediately placed in a microtube, flash frozen in liquid nitrogen, and then stored at −80°C prior to RNA extraction.
RNA extraction, cDNA library construction, and sequencing
RNA from 3 fish in the same barrel was combined into one sample. Each temperature group had three biological replicates. Total RNA was extracted from the liver using the TRIzol (Invitrogen, Carlsbad, CA, USA) method, and its concentration and purity were detected using a Nanodrop2000 (NanoDrop Technologies, LLC, Wilmington, DE, USA). RNA integrity was detected by 1% agarose gel electrophoresis. Each sample was prepared using 1 μg total RNA (Concentration ≥ 35ng/μL, OD260/280 ≥ 1.8, OD260/230 ≥ 1.0) for library construction using an Illumina Truseq™ RNA Sample Prep Kit. The mRNA was then enriched by the oligo(dT) method, followed by adding fragmentation buffer, and the RNA was randomly fractured into small fragments of about 300 bp. First and second strands of cDNA were synthesized using reverse transcriptase, random primers, and mRNA templates. The viscous ends of double-stranded cDNA were filled using End Repair Mix and an A base was added to the 3’ end. The product was purified and sorted. Polymerase chain reaction (PCR) amplification was performed using the sorted product and the library was obtained after purification. After QuantiFluor® dsDNA System quantification, sequencing was performed using the Illumina platform.
Analysis of differentially expressed genes and functional enrichment analysis
Expression levels of genes and transcripts were analyzed quantitatively using the RSEM method (Li and Dewey, 2011) (fragments per kilobase of exon per million mapped reads). Differences in expression levels between samples were analyzed using EdgeR software (Love et al., 2014). Genes with a false discovery rate < 0.05 and log2 fold-change ≥ 1 were considered as DEGs. DEGs were further examined for gene function enrichment using GO (http://www.geneo-ntology.org/) and KEGG (http://www.genome.jp/kegg/) analyses. DEGs were significantly enriched when the adjusted P value was ≤ 0.05.
Real-time PCR verification of Illumina sequencing data
We selected 15 DEGs with roles in pancreatic secretion, circadian rhythm, and arginine and proline metabolism pathways for verification by quantitative PCR (qPCR). Primers were designed using Primer 5 based on the obtained sequences (Table 1). β-actin was used as an internal reference gene. All RNA samples were reverse-transcribed to cDNA using a FastKing RT Kit and the obtained cDNA was subjected to qPCR using an Applied Biosystems StepOnePlus PCR instrument. The reaction was performed using the TOROGreen qPCR Master Mix kit, with a 20 μL reaction containing Master Mix (10.0 μL), RNase-free water (6.4 μL), forward and reverse primer (0.8 μL), and first-strand cDNA (2 μL). The reaction procedure included a pre-denaturation at 95°C for 60 s, followed by 40 cycles of denaturation at 95°C for 10 s and annealing at 60°C for 30 s in a 20 μL reaction mixture. Relative gene quantification was calculated by the 2-ΔΔCt method (Livak and Schmittgen, 2001). Two-way ANOVA was used to detect the statistical differences between the groups. Differences were considered statistically significant at P < 0.05. The consistency between the RNA-seq and qPCR results was analyzed by correlation scatterplots using GraphPad Prism 6.0.
Detection the proteases, arginine and proline of serum
Serum levels of proteases, arginine, and proline were detected at different temperatures using protease enzyme-linked immunosorbent assay (ELISA), FISH proline ELISA, and FISH Arg ELISA kits (Shanghai Enzyme-linked Biotechnology Co., Ltd.). The kit numbers are YJ722032, YJ712912, and YJ729390, respectively.
Results
Sequencing results and analysis
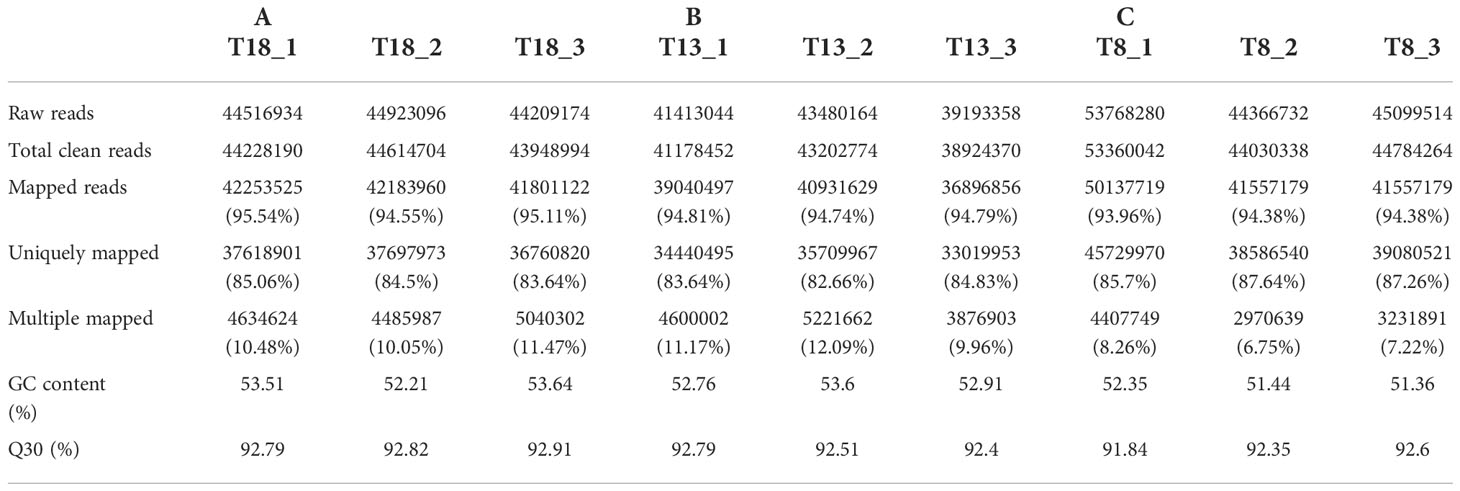
Nine libraries were sequenced using the Illumina sequencing platform, and 400,970,296 raw reads were obtained, including 133,649,204 in group A, 124,086,566 in group B, and 143,234,526 in group C. A total of 398,272,128 clean reads were obtained, including 132,791,888 in group A, 123,305,596 in group B, and 142,174,644 in group C. The Q30 base percentage was 91.84%– 92.91% and the GC content was 51.36%–53.64%. After mapping with the tiger puffer reference genome, a total of 376,359,666 mapped reads were obtained, including 338,645,140 multiple mapped reads (Table 2).
Analysis of DEGs
EdgeR identified DEGs between the control group (A: 18°C) and low-temperature groups (B: 13°C, C: 8°C). Comparative analysis identified 51 DEGs in the 13°C group compared with the control group (A vs B), including 33 up-regulated genes and 18 down-regulated genes; 942 DEGs were identified in the 8°C group compared with the control group (A vs C), including 384 up-regulated and 558 down-regulated genes; and 195 DEGs were identified in the 8°C group compared with the 13°C group (B vs C), including 54 up-regulated and 141 down-regulated genes (Figure 1).
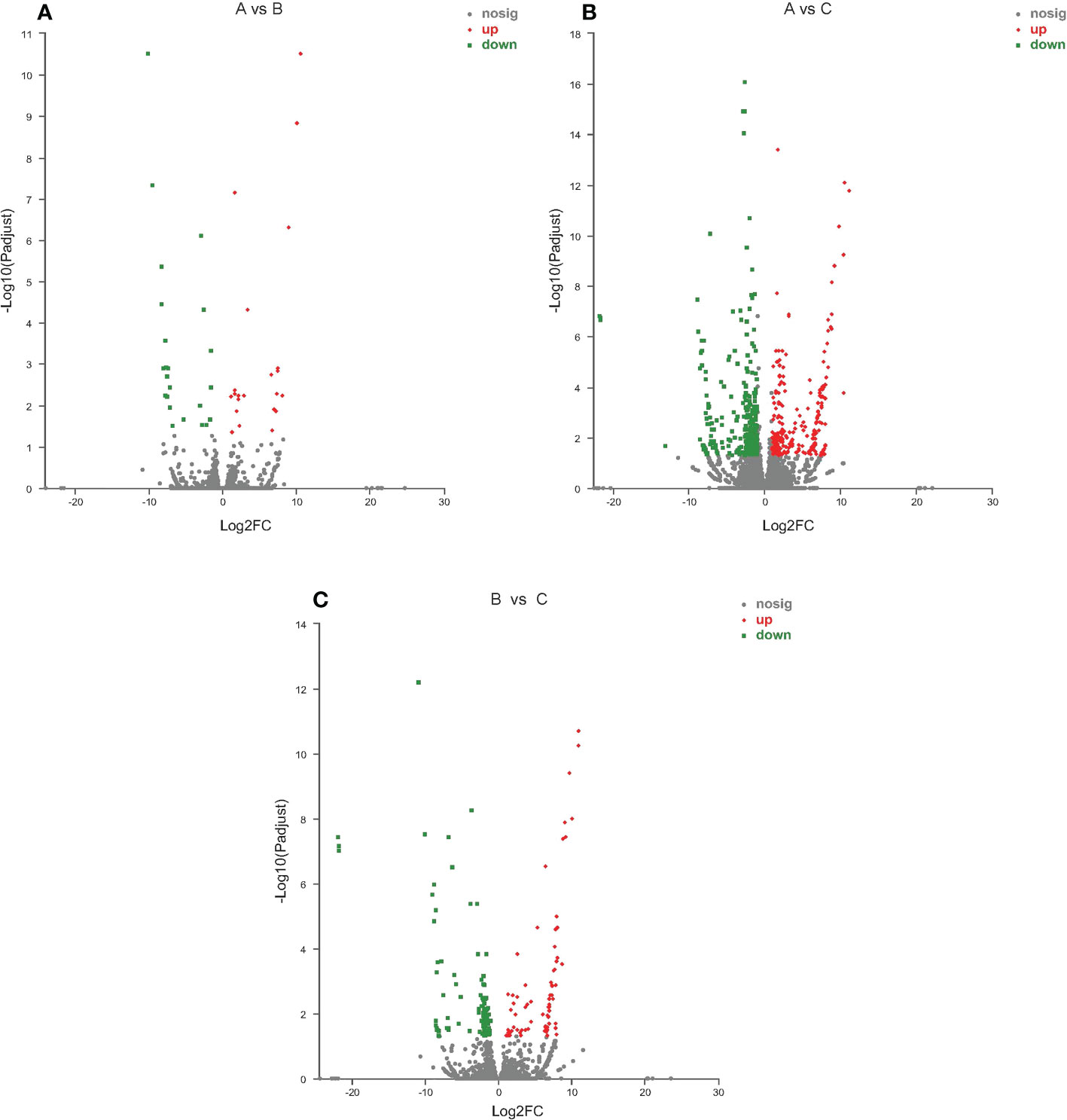
Figure 1 Volcano plot of differentially expressed genes identified from different temperature groups. (A) 13°C group compared with 18°C group; (B) 8°C group compared with 18°C group; (C) 8°C group compared with 13°C group. Each dot represents a gene, blue dots representing significantly down regulated genes, red dots representing significantly up regulated genes, and gray dots representing not significantly different genes.
GO annotation analysis
GO terms can be categorized by biological process (BP), molecular function (MF), and cellular component (CC). The most obvious subclassification terms between different comparison groups are basically the same (Figure 2). DEGs between the 18°C and 13°C groups were mainly enriched in membrane part (12 genes), membrane (10 genes), and cell part (8 genes) in CC, cellular process (17 genes), biological regulation (15 genes), and metabolic process (13 genes) in BP, and binding (21 genes) and catalytic activity (17 genes) in MF. Comparing the 18°C and 8°C groups, DEGs were mainly enriched in membrane part (277 genes), membrane (251 genes), and cell part (213 genes) in CC, cellular process (326 genes), metabolic process (298 genes), and biological regulation (208 genes) in BP, and binding (450 genes), catalytic activity (362 genes), and transporter activity (48 genes) in MF. DEGs between the 13°C and 8°C groups were mainly enriched in membrane part (49 genes), membrane (42 genes), and cell part (36 genes) in CC, cellular process (76 genes), biological regulation (58 genes), and metabolic process (36 genes) in BP, and binding (84 genes), catalytic activity (83 genes), and transporter activity (16 genes) in MF.
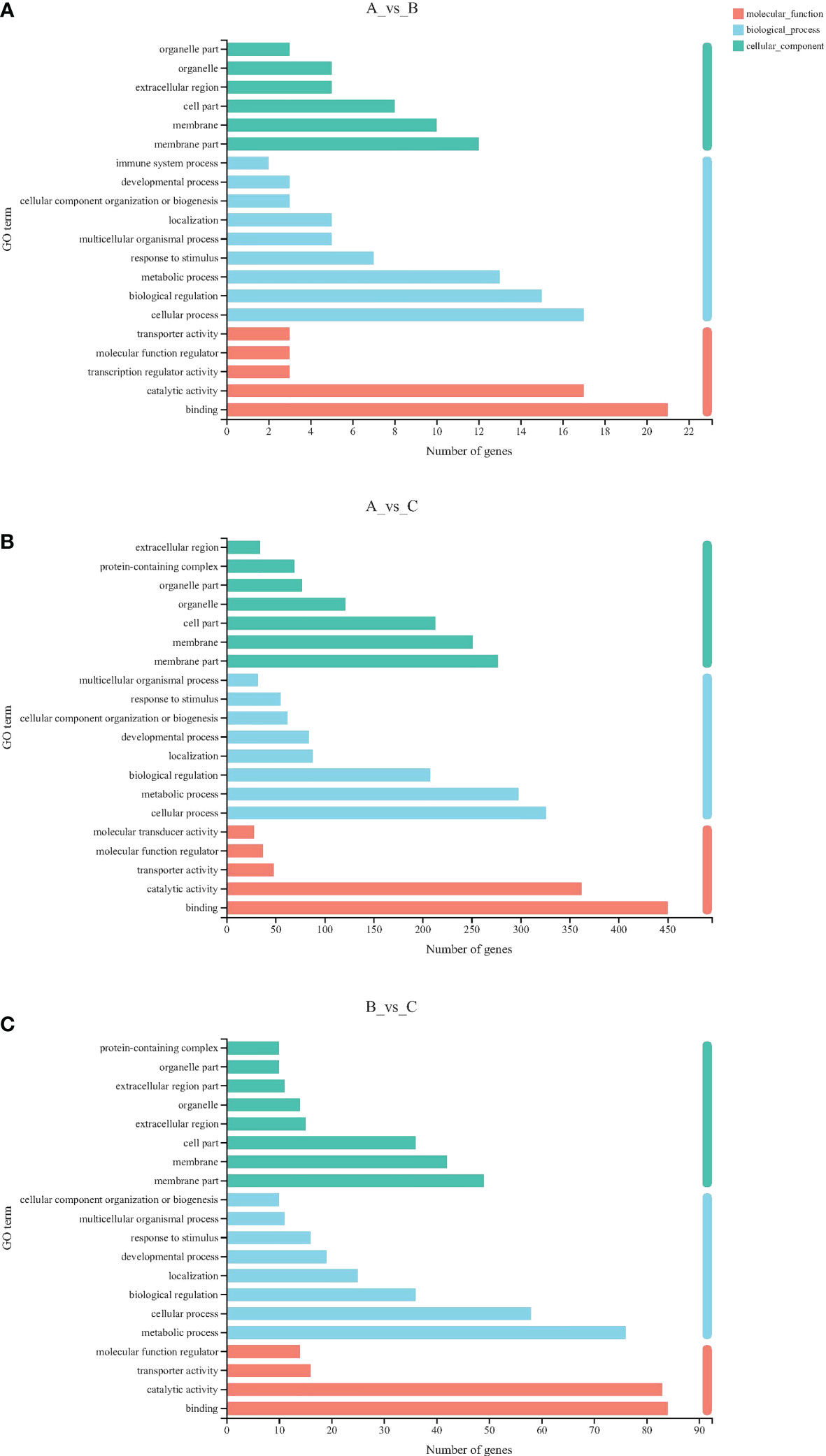
Figure 2 GO classification statistical histogram. (A) 13°C group compared with 18°C group; (B) 8°C group compared with 18°C group; (C) 8°C group compared with 13°C group. The Y-axis represents the secondary classification term for GO. The X-axis represents the number of genes. Three colors represent three categories.
KEGG enrichment analysis
We further elucidated the functions of the DEGs in signaling pathways by detecting significantly abundant pathways in the KEGG database (Figures 3, 4; Table 3). Six pathways were significantly enriched in the 13°C group compared with the 18°C group, including circadian rhythm-fly, circadian rhythm, peroxisome proliferator-activated receptor signaling pathway, fatty acid biosynthesis, adipocytokine signaling pathway, and transcriptional misregulation in cancer. There were 19 genes in these pathways, including 11 up-regulated genes and 8 down-regulated genes. The top three significantly enriched pathways were circadian rhythm-fly (3 up-regulated genes, 0 down-regulated genes), circadian rhythm (2 up-regulated genes, 1 down-regulated gene), and fatty acid biosynthesis (2 up-regulated genes, 0 down-regulated genes). Ten pathways were significantly enriched in the 8°C group compared with the 18°C group, including pancreatic secretion, protein digestion and absorption, influenza A, arginine and proline metabolism, complement and coagulation cascades, circadian rhythm-fly, circadian rhythm, glycine, serine and threonine metabolism, glycolysis/gluconeogenesis, and antigen processing and presentation. There were 176 genes in these pathways, including 33 up-regulated genes and 143 down-regulated genes. The top three significantly enriched pathways were circadian rhythm-fly (5 genes up-regulated, 1 down-regulated), pancreatic secretion (3 genes up-regulated, 36 down-regulated), and arginine and proline metabolism (3 genes up-regulated, 9 down-regulated). Ten pathways were significantly enriched in the 8°C group compared with the 13°C group, including pancreatic secretion, protein digestion and absorption, glycerolipid metabolism, influenza A, fat digestion and absorption, glycolysis/gluconeogenesis, circadian rhythm-fly, one-carbon pool by folate, steroid biosynthesis, and arginine and proline metabolism. There were 95 genes in these pathways, including 6 up-regulated genes and 89 down-regulated genes. The top three significantly enriched pathways were pancreatic secretion (0 up-regulated genes, 28 down-regulated genes), circadian rhythm-fly (3 up-regulated genes, 0 down-regulated genes), and protein digestion and absorption (1 up-regulated gene, 23 down-regulated genes). These pathways were mainly related to digestion, metabolism, the immune system, environmental adaptation, and the endocrine system, and the top three significantly enriched pathways were mainly related to digestion, metabolism, and environmental adaptation.
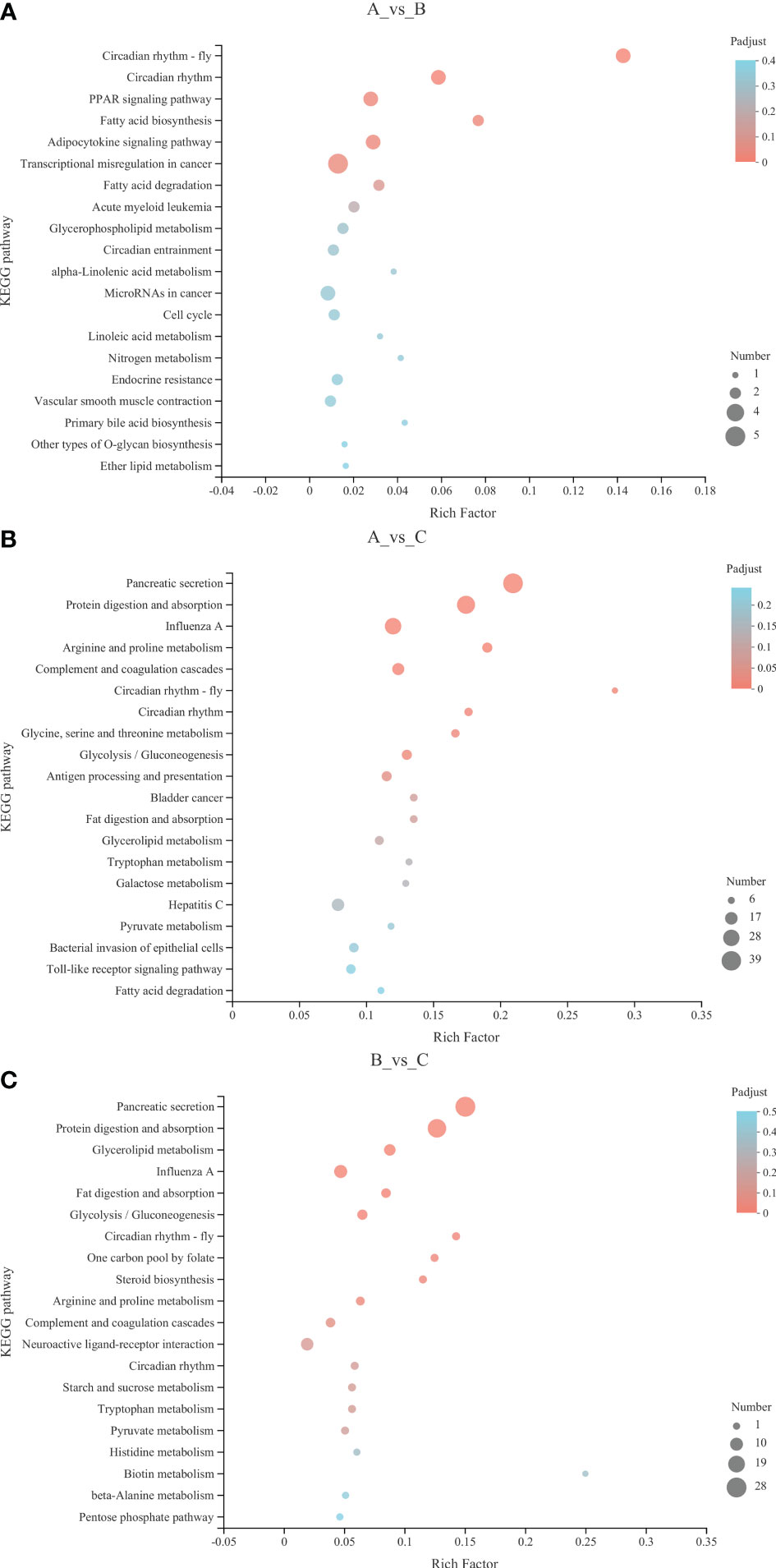
Figure 3 KEGG enrichment analysis bubble diagram. (A) 13°C group compared with 18°C group; (B) 8°C group compared with 18°C group; (C) 8°C group compared with 13°C group. The Y-axis represents the pathway name. The X-axis represents the number of genes. The size of the points represents the number of genes in the pathway. The colors of the points correspond to different Padjust ranges.
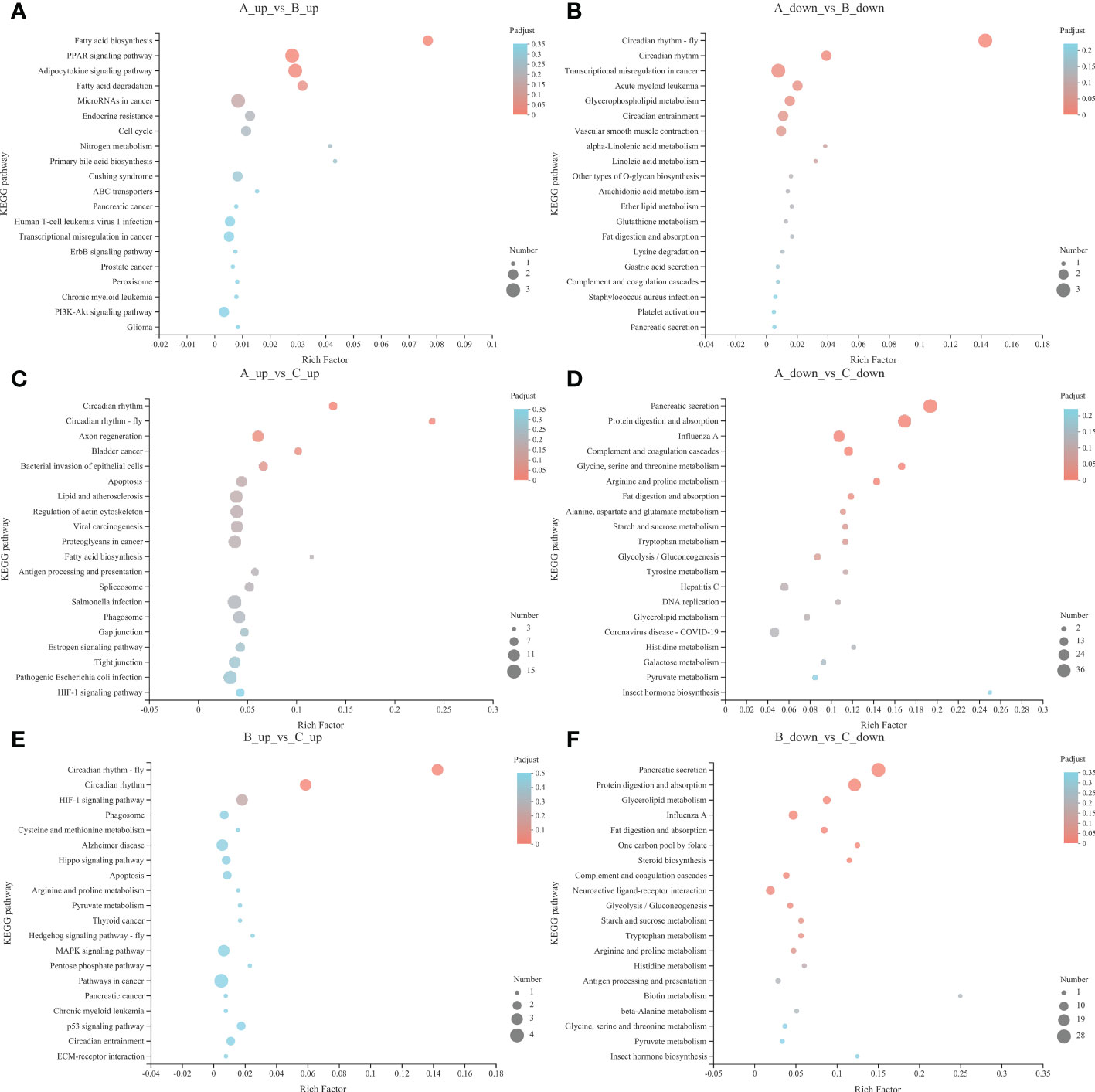
Figure 4 Up-regulated pathways KEGG enrichment analysis bubble diagram of 13°C group compared with 18°C group (A), 8°C group compared with 18°C group (C) and 8°C group compared with 13°C group (E). Down-regulated pathways KEGG enrichment analysis bubble diagram of 13°C group compared with 18°C group (B), 8°C group compared with 18°C group (D) and 8°C group compared with 13°C group (F). The Y-axis represents the pathway name. The X-axis represents the number of genes. The size of the points represents the number of genes in the pathway. The colors of the points correspond to different Padjust ranges.

Table 3 The number of up-regulated and down-regulated genes in the top three significantly enriched pathways.
qPCR analysis
The pathways that were significantly enriched in this study mainly involved digestion, metabolism, and environmental adaptation. We selected 15 genes with roles in pancreatic secretion, arginine and proline metabolism, and circadian rhythm for verification by qPCR. The RNA-seq results indicated that low temperature resulted in up-regulation of nitric oxide synthase 1 (nos1), ornithine decarboxylase 1 (odc1), proline dehydrogenase 1 (prodha), aryl hydrocarbon receptor nuclear translocator-like 1a (arntl1a), clock circadian regulator a (clocka), aryl hydrocarbon receptor nuclear translocator-like 2 (arntl2), neuronal PAS domain protein 2 (npas2), ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2 (atp2a2), ras homolog gene family, member Ab (rhoab), and period circadian protein homolog 2-like (per2), and down-regulation of guanidinoacetate N-methyltransferase (gamt), cryptochrome circadian regulator 5 (cry5), carboxypeptidase A5 (cpa5), elastase 2 (ela2), and carboxypeptidase B1 (cpb1). We created a histogram of expression levels based on the qPCR results and combined these with the RNA-seq results to produce a correlation scatter plot. The results showed that the expression levels determined by qPCR were consistent with the RNA-seq results, with a significant positive correlation (0.7705≤ r ≤0.9985, P<0.05). This suggested that the RNA-seq results were credible (Figures 5, 6).
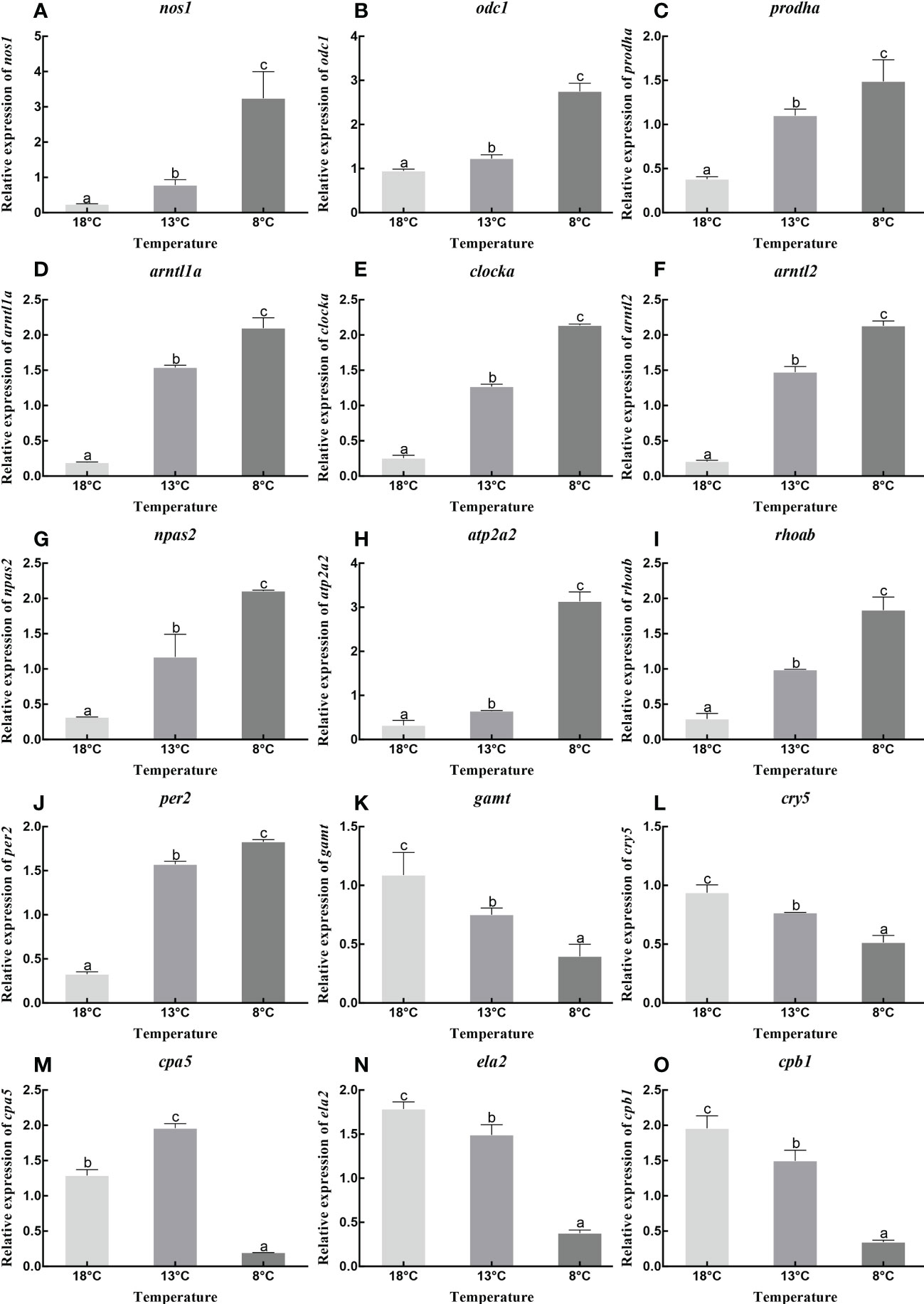
Figure 5 Relative expression of nos1 (A), odc1 (B), prodha (C), arntl1a (D), clocka (E), arntl2 (F), npas2 (G), atp2a2 (H), rhoab (I), per2 (J), gamt (K), cry5 (L), cpa5 (M), ela2 (N) and cpb1 (O) under different temperature. Data were presented as the mean ± SEM. Different letters indicate significant differences between groups (P<0.05).
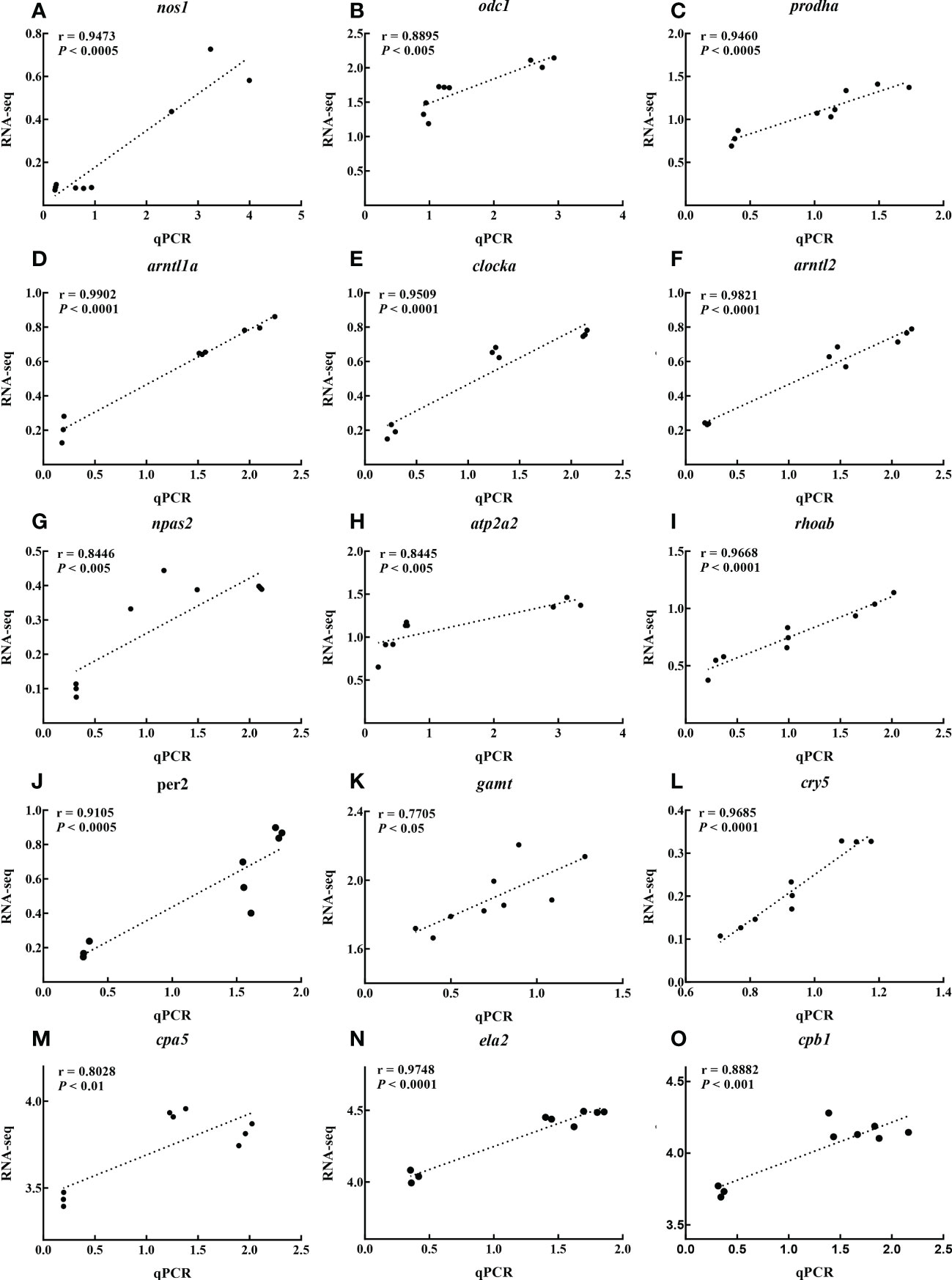
Figure 6 Scatter plot of correlation between RNA-seq and qPCR results of nos1 (A), odc1 (B), prodha (C), arntl1a (D), clocka (E), arntl2 (F), npas2 (G), atp2a2 (H), rhoab (I), per2 (J), gamt (K), cry5 (L), cpa5 (M), ela2 (N) and cpb1 (O) under different temperature.
Serum levels of proteases, arginine and proline
The protease content was significantly higher in the control group compared to the low-temperature groups (P<0.05), and increased with increasing temperature. Serum levels of arginine and proline were significantly higher in the 8°C group compared to the other two groups (P<0.05), and decreased with increasing temperature (Figure 7).
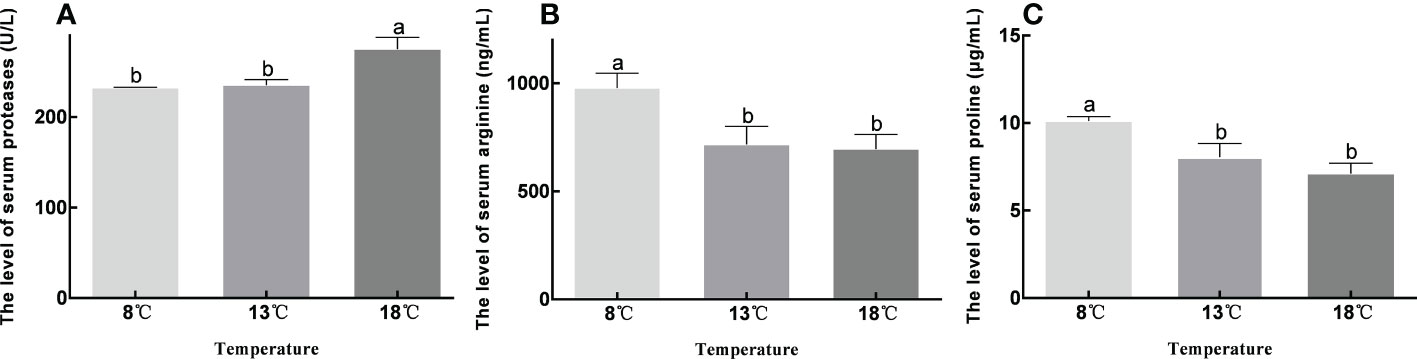
Figure 7 Contents of proteases, arginine and proline in serum at different temperature groups. Data were presented as the mean ± SEM. Different letters indicate significant differences between groups (P<0.05).
Discussion
Water temperature is one of the most common abiotic stress factors (Long et al., 2013) and has been extensively investigated in aquatic organisms such as turbot (Ma et al., 2018), Nile tilapia (Qiang et al., 2013), yellow drum (Nibea albiflora) (Jiao et al., 2020), and pacific white shrimp (Litopenaeus vannamei) (Wen et al., 2017). Temperature is also an important factor limiting the survival, growth, and development of tiger puffers in culture (Kikuchi et al., 2006; Liu et al., 2022). However, most studies of the effects of low temperature on tiger puffers to date have focused on short-term low-temperature stress (Liu et al., 2022), and a lack of understanding of the molecular mechanisms involved in long-term stress has limited progress in breeding tiger puffers for cold tolerance. In the present study, we carried out liver transcriptome analysis at different temperatures to analyze the molecular mechanisms of adaptation under chronic low-temperature stress. GO functional annotation and KEGG enrichment were used to analyze the functions and pathways of DEGs. Fifty-one and 942 DEGs were found in the 13°C and 8°C groups compared with the control group, respectively, and 195 DEGs were found in the 8°C group compared with the 13°C group. These results indicated that a fall in temperature to 13°C had little effect on tiger puffers, while a temperature of 8°C had a greater impact. KEGG analysis produced similar results. Ten were significantly enriched in 8°C group compared with other two groups, while six pathways were significantly enriched in the 13°C group compared with the control group. The significantly enriched signaling pathways in the three comparison groups were related to digestion, metabolism, and environmental adaptation. Four pathways were significantly enriched in the 8°C group compared with the other two groups, including circadian rhythm, pancreatic secretion, protein digestion and absorption, and arginine and proline metabolism, while only circadian rhythm was significantly enriched in the 13°C group compared with the control group.
Previous studies found that extreme temperature conditions affected fish metabolism and digestion, while low temperatures reduced fish growth and metabolic rates (Chang et al., 2006; Teodósio et al., 2021). And the digestive rate of fish is also affected at non-adaptive temperature (Yúfera et al., 2019; Xing et al., 2022). Similarly, change in temperature also affected the circadian rhythm in fish (Liu et al., 2020; Bernal et al., 2022). In the present study, most genes in the enriched pathways related to digestion, metabolism, and environmental adaptation were down-regulated at low temperature and only a few genes were up-regulated, suggesting that low temperature inhibited normal digestion and metabolism in tiger puffers, and changed their circadian rhythm. These up-regulated genes may play an important role in response to low-temperature stress.
Genes in the pancreatic secretion pathway were mostly down-regulated, including cpa1, cpb1, and ela2, which are associated with proteolysis. cpa1 and cpb1 are biomarkers related to proteolysis, and their content in plasma were shown to be higher under normal proteolysis conditions compared with proteolysis inhibition (Dieden et al., 2021). Deletion of ela2 resulted in the destruction of proteolytic activity (Salipante et al., 2007). Genes enriched in the protein digestion and absorption pathway were mainly down-regulated with decreased temperature, suggesting that low temperature may inhibit the expression of genes related to proteolysis in pancreatic secretion, leading to disruption of protein digestion and absorption. In addition, serum protease levels were significantly lower in the low-temperature groups compared with the control group, thus confirming the reliability of the study conclusion. Furthermore, the expression levels of genes related to ATP hydrolysis and cell processes, including atp2a2 and rhoab, increased at low temperature. The atp2a2 gene encodes SERCA Ca2+-ATPases, which catalyze the hydrolysis of ATP and the transport of calcium ions from the cytoplasm to the sarcoplasmic reticulum lumen (Kabashima et al., 2020; Muslimova et al., 2020). The rhoab gene was shown to enable Rho GTPase activity in zebrafish (Salas-Vidal et al., 2007), which plays an important role in a series of cellular processes, including proliferation, apoptosis, cell migration, cell polarization, membrane trafficking, cytoskeleton rearrangements, and transcriptional regulation. All known Rho GTPases are involved in normal development in animals (Salas-Vidal et al., 2005). Low temperature disrupts pancreatic secretion, and normal metabolism will be affected by inhibition of protein digestion and absorption (Hou et al., 2015). However, the up-regulation of genes related to energy expenditure and cell function at low temperature may be important for improving the survival, growth, and development of tiger puffers.
Arginine and proline metabolism was significantly enriched in many metabolomics studies of temperature tolerance in fish (Jiao et al., 2020; Zhang et al., 2021). Arginine and proline have been shown to have a protective effect against temperature changes in some fish species (Zhao et al., 2015; Cheng et al., 2021). In addition, arginine and proline were shown to improve the cold resistance of crops and reduce damage caused by freezing (Zhang et al., 2010; Alvarez et al., 2022). These results suggest that arginine and proline metabolism may play an important role in the response of tiger puffers to low temperature. In the present study, genes involved in the arginine and proline metabolism, and glycine, serine, and threonine metabolism pathways were generally down-regulated at low temperatures, suggesting that low temperature can inhibit amino acid metabolism. However, prodha, odc1, and nos1 genes involved in arginine and proline metabolism were up-regulated at low temperature. Proline was previously shown to accumulate in plants in response to environmental stress, and was rapidly oxidized by proline dehydrogenase (ProDH) when the stress was relieved (Cabassa-Hourton et al., 2016). Not only the accumulation of proline, but also continuous proline metabolism or turnover was required to cope with drought in arabidopsis (Arabidopsis thaliana) (Sharma et al., 2011). This explains why prodha expression increases in response to cold stress: during cold tolerance, both the continuous accumulation of proline and the continuous oxidation of proline by prodha is required to maintain normal metabolism and turnover in tiger puffers during cold tolerance. Under the action of arginase, arginine was decomposed into ornithine, and then ornithine was catalyzed by ornithine decarboxylase to synthesize putrescine, which has been shown to alleviate cold damage (Kovács et al., 2010; Zuo et al., 2021). Nitric oxide synthase is a key enzyme in arginine catabolism, and decomposes nitric oxide (NO) from arginine (Martínez, 1995), while low temperature makes plants accumulate more NO to reduce cold damage (Plohovska et al., 2019). This also indicates that increased arginine accumulation is required to supply ornithine and NO production at low temperatures. Moreover, serum levels of arginine and proline were significantly higher at 8°C than at the higher temperatures, and increased with decreasing temperature, further confirming that prodha, odc1, and nos1 in arginine and proline metabolism were essential components of the response to cold stress in tiger puffers, even though low temperatures results in metabolic disorders.
Our current understanding of the molecular mechanism of the circadian system in fish is mainly based on zebrafish, in which the interaction of clock gene products is similar to that in mammals (Cahill, 2002). In the present study, several genes, including arntl1a, clocka, arntl2, per2, and cry5, were involved in the circadian rhythm pathway, and all except cry5 increased as the temperature decreased. Studies of circadian rhythms in mammals and fish found that these genes formed a transcription–translation feedback loop: heterodimeric interactions of clock transcription factors (CLOCK (orthologous to fish clocka) and BMAL1 (ARNTL)) drive the transcription and translation of PERIOD1-3 (PER1-3) and CRYPTO-CHROME 1-2 (CRY1-2), which are translated into proteins and form heterodimers themselves, and are recycled from the cytoplasm back to the nucleus to inhibit BMAL1 and CLOCK (Cahill, 2002; Zhdanova and Reebs, 2006; Anea et al., 2018). Global loss of the BMAL1 or PER genes in this conserved transcription–translation feedback loop impairs vascular endothelial cell layer function, and has acute and chronic adverse effects on vascular system adaptation (Viswambharan et al., 2007; Somanath et al., 2011). Overexpression of clock components can also prevent the generation of deleterious vascular phenotypes (Qin and Deng, 2015). Circadian rhythms also regulate metabolism and digestion primarily via through this feedback loop (Zhong et al., 2018; Segers and Depoortere, 2021), which controls and drives metabolism, mainly including neurotransmitters, hormones, amino acids, and lipids (Han et al., 2022). Meanwhile, the circadian rhythm and metabolism interact with each other to maintain normal physiological activities (Reinke and Asher, 2019; Takahashi, 2019; Sinturel et al., 2020). Previous studies showed that circadian clock genes caused diurnal fluctuations in the digestive system by altering the activities of various digestive enzymes (Maouyo et al., 1993; Espinosa-Chaurand et al., 2017). Therefore, the circadian rhythm has a complex relationship with digestion and metabolism, and the regulation of circadian rhythm can alleviate the damage caused by digestive and metabolic disorders, which is an adaptive change and plays a positive role in the resistance to low temperature.
Conclusions
In the present study, we analyzed the effects of long-term cold stress on the liver transcriptome in tiger puffers. Totals of 942 and 195 DEGs were identified in the 8°C group compared with the 18°C and 13°C groups, respectively. Further characterization showed that most DEGs were enriched in pathways related to digestion, metabolism, and environmental adaptation, in which arginine and proline metabolism, pancreatic secretion, and circadian rhythm were considered to be the primary signaling pathways. Regarding the effects of low temperature on metabolic and digestive disorders, genes involved in arginine and proline metabolism and pancreatic secretion were up-regulated at low temperatures, which may help to improve cold resistance in tiger puffers. Serum tests confirmed protease inhibition and the accumulation of arginine and proline at low temperatures. In addition, low temperature induced adaptive changes in the circadian rhythm in tiger puffers, which may also improve their low-temperature tolerance. To the best of our knowledge, this was the first study to expose tiger puffers to long-term cold stress, and the results provide new insights into the mechanism of cold tolerance in this species. The selected key genes may provide potential biomarkers for cold response in tiger puffers, to allow the screening of populations with cold tolerance.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The animal study was reviewed and approved by Institutional Animal Care and Use Committee of the Yellow Sea Fisheries Research Institute.
Author contributions
All authors certify that they have participated sufficiently in the work to take public responsibility for the content, including participation in the concept, design, analysis, writing, or revision of the manuscript. Furthermore, each author certifies that this material or similar material will not be submitted to or published in any other publication before its appearance in the Frontiers in Marine Science. ZL and LZ participated in the experimental design, data processing and manuscript writing of this research. AM participated in the discussion of experimental results. XW, SL, HC, ZS, FX and HZ helped to perform the low temperature stress experiment and carried out sample preparation. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Key Research and Development Plan of Nation (Grant No. 2018YFD0900301-12); National Nature Science Foundation of China (Grant No. 32002362); Key Research and Development Plan of Nation (Grant No. 2018YFD0900102); Shandong Provincial Natural Science Foundation, China (Grant No. ZR2019BC089); Central Public-interest Scientific Institution Basal Research Fund, CAFS (Grant No. 2020TD25).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alvarez M. E., Savouré A., Szabados L. (2022). Proline metabolism as regulatory hub. Trends Plant Sci. 27, 39–55. doi: 10.1016/j.tplants.2021.07.009
Anea C. B., Merloiu A. M., Fulton D. J. R., Patel V., Rudic R. D. (2018). Immunohistochemistry of the circadian clock in mouse and human vascular tissues. Vessel Plus 2, 16. doi: 10.20517/2574-1209.2018.46
Beitinger T. L., Bennett W. A., McCauley R. W. (2000). Temperature tolerances of north American freshwater fishes exposed to dynamic changes in temperature. Environ. Biol. Fishes 58, 237–275. doi: 10.1023/A:1007676325825
Bernal M. A., Schmidt E., Donelson J. M., Munday P. L., Ravasi T., Ravasi T. (2022). Molecular response of the brain to cross-generational warming in a coral reef fish. Front. Mar. Sci. 9 doi: 10.3389/fmars.2022.784418
Cabassa-Hourton C., Schertl P., Bordenave-Jacquemin M., Saadallah K., Guivarc'h A., Lebreton S., et al. (2016). Proteomic and functional analysis of proline dehydrogenase 1 link proline catabolism to mitochondrial electron transport in Arabidopsis thaliana. Biochem. J. 473:2623–2634. doi: 10.1042/BCJ20160314
Cahill G. M. (2002). Clock mechanisms in zebrafish. Cell Tissue Res. 309, 27–34. doi: 10.1007/s00441-002-0570-7
Chang Y. M., Kuang Y. Y., Cao D. C., Liang L., Sun X. W., Lei Q. Q. (2006). Effects of cooling temperature stress on hematology and serum chemistry values of. J. Fisheries China 30, 701–706. doi: 10.1360/aps050066
Cheng Y., Li X., Wang L., Lu K., Song K., Ai Q., et al. (2021). Effects of dietary arginine levels on growth, immune function of physical barriers and serum parameters of spotted seabass (Lateolabrax maculatus) reared at different water temperatures. Aquaculture 541, 736812. doi: 10.1016/j.aquaculture.2021.736812
Dieden A., Malan L., Mels C., Lammertyn L., Wentzel A., Nilsson P., et al. (2021). Exploring biomarkers associated with deteriorating vascular health using a targeted proteomics chip: The SABPA study. Medicine 100:e25936. doi: 10.1097/MD.0000000000025936
Espinosa-Chaurand D., Vega-Villasante F., Carrillo-Farnés O., Nolasco-Soria H. (2017). Effect of circadian rhythm, photoperiod, and molt cycle on digestive enzymatic activity of macrobrachium tenellum juveniles. Aquaculture 479, 225–232. doi: 10.1016/j.aquaculture.2017.05.029
Han H., Dou J. M., Hou Q. Q., Wang H. J. (2022). Role of circadian rhythm and impact of circadian rhythm disturbance on the metabolism and disease. J. Cardiovasc. Pharmacol. 79, 254–263. doi: 10.1097/FJC.0000000000001178
Hou Y., Yin Y., Wu G. (2015). Dietary essentiality of “nutritionally non-essential amino acids” for animals and humans. Exp. Biol. Med. (Maywood N.J.) 240, 997–1007. doi: 10.1177/1535370215587913
Ibarz A., Martín-Pérez M., Blasco J., Bellido D., de Oliveira E., Fernández-Borràs J. (2010). Gilthead sea bream liver proteome altered at low temperatures by oxidative stress. PROTEOMICS 10, 963–975. doi: 10.1002/pmic.200900528
Jiao S., Nie M., Song H., Xu D., You F. (2020). Physiological responses to cold and starvation stresses in the liver of yellow drum (Nibea albiflora) revealed by LC-MS metabolomics. Sci. Total Environ. 715, 136940. doi: 10.1016/j.scitotenv.2020.136940
Kabashima Y., Ogawa H., Nakajima R., Toyoshima C. (2020). What ATP binding does to the Ca 2+ pump and how nonproductive phosphoryl transfer is prevented in the absence of Ca 2+. Proc. Natl. Acad. Sci. 117, 18448–18458. doi: 10.1073/pnas.2006027117
Kikuchi K., Iwata N., Kawabata T., Yanagawa T. (2006). Effect of feeding frequency, water temperature, and stocking density on the growth of tiger puffer, takifugu rubripes. J. World Aquaculture Soc. 37, 12–20. doi: 10.1111/j.1749-7345.2006.00002.x
Kovács Z., Simon-Sarkadi L., Szűcs A., Kocsy G. (2010). Differential effects of cold, osmotic stress and abscisic acid on polyamine accumulation in wheat. Amino Acids 38, 623–631. doi: 10.1007/s00726-009-0423-8
Li B., Dewey C. N. (2011). RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinf. 12, 323. doi: 10.1186/1471-2105-12-323
Liu Z. F., Ma A. J., Yuan C. H., Zhao T. T., Chang H. W., Zhang J. S. (2021). Transcriptome analysis of liver lipid metabolism disorders of the turbot scophthalmus maximus in response to low salinity stress. Aquaculture 534, 736273. doi: 10.1016/j.aquaculture.2020.736273
Liu Z. F., Ma A., Yuan C. H., Zhu L., Chang H. W., Yu L. L., et al. (2022). Kidney transcriptome analysis of tiger puffer (Takifugu rubripes) uncovers key pathways and genes in response to low-temperature stress. Aquaculture Rep. 26, 101287. doi: 10.1016/j.aqrep.2022.101287
Liu L., Zhang R., Wang X., Zhu H., Tian Z. (2020). Transcriptome analysis reveals molecular mechanisms responsive to acute cold stress in the tropical stenothermal fish tiger barb (Puntius tetrazona). BMC Genomics 21, 737. doi: 10.1186/s12864-020-07139-z
Livak K. J., Schmittgen T. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-DDCt method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Long Y., Song G., Yan J., He X., Li Q., Cui Z. (2013). Transcriptomic characterization of cold acclimation in larval zebrafish. BMC Genomics 14, 612. doi: 10.1186/1471-2164-14-612
Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi: 10.1186/s13059-014-0550-8
Luo L., Huang J. H., Liu D. L., Jiang S. G., Zhou F. L., Jiang S., et al. (2021). Transcriptome reveals the important role of metabolic imbalances, immune disorders and apoptosis in the treatment of procambarus clarkii at super high temperature. Comp. Biochem. Physiol. Part D: Genomics Proteomics 37, 100781. doi: 10.1016/j.cbd.2020.100781
Maouyo D., Sarfati P., Guan D., Morisset J., Adelson J. W. (1993). Circadian rhythm of exocrine pancreatic secretion in rats: major and minor cycles. Am. J. Physiol. 264, G792–G800. doi: 10.1152/ajpgi.1993.264.4.g792
Martínez A. (1995). Nitric oxide synthase in invertebrates. Histochemical J. 27, 770–776. doi: 10.1007/BF02388302
Ma A. J., Wang X. A., Huang Z. H., Liu Z. F., Cui W. X., Qu J. B. (2018). Estimation of genetic parameters for upper thermal tolerance and growth-related traits in turbot scophthalmus maximus using the Bayesian method based on Gibbs sampling. Acta Oceanologica Sin. 37, 40–46. doi: 10.1007/s13131-018-1185-5
Muslimova E. F., Rebrova T. Y., Kondratieva D. S., Akhmedov S. D., Afanasiev S. A. (2020). Expression of the Ca2+-ATPase SERCA2a (ATP2A2) gene and the ryanodine receptor (RYR2) gene in patients with chronic heart failure. Russian J. Genet. 56, 843–848. doi: 10.1134/S1022795420070108
Plohovska S. H., Krasylenko Y. A., Yemets A. I. (2019). Nitric oxide modulates actin filament organization in arabidopsis thaliana primary root cells at low temperatures. Cell Biol. Int. 43, 1020–1030. doi: 10.1002/cbin.10931
Qiang J., Yang H., Wang H., Kpundeh M. D., Xu P. (2013). Interacting effects of water temperature and dietary protein level on hematological parameters in Nile tilapia juveniles, oreochromis niloticus (L.) and mortality under streptococcus iniae infection. Fish Shellfish Immunol. 34, 8–16. doi: 10.1016/j.fsi.2012.09.003
Qin B., Deng Y. (2015). Overexpression of circadian clock protein cryptochrome (CRY) 1 alleviates sleep deprivation-induced vascular inflammation in a mouse model. Immunol. Lett. 163, 76–83. doi: 10.1016/j.imlet.2014.11.014
Reinke H., Asher G. (2019). Crosstalk between metabolism and circadian clocks. Nat. Rev. Mol. Cell Biol. 20, 227–241. doi: 10.1038/s41580-018-0096-9
Salas-Vidal E., Cheng X., Cui C., Li X., Schnabel D., Meijer A. H., et al. (2007). Functional genomics of zebrafish rhoab, cdc42c and rac1a rho small GTPases. Dev. Biol. 306, 380. doi: 10.1016/j.ydbio.2007.03.353
Salas-Vidal E., Meijer A. H., Cheng X., Spaink H. P. (2005). Genomic annotation and expression analysis of the zebrafish rho small GTPase family during development and bacterial infection. Genomics 86, 25–37. doi: 10.1016/j.ygeno.2005.03.010
Salipante S. J., Benson K. F., Luty J., Hadavi V., Kariminejad R., Kariminejad M. H., et al. (2007). Double de novo mutations of ELA2 in cyclic and severe congenital neutropenia. Hum. Mutat. 28, 874–881. doi: 10.1002/humu.20529
Segers A., Depoortere I. (2021). Circadian clocks in the digestive system. Nat. Rev. Gastroenterol. Hepatol. 18, 239–251. doi: 10.1038/s41575-020-00401-5
Sharma S., Villamor J., Verslues P. (2011). Essential role of tissue-specific proline synthesis and catabolism in growth and redox balance at low water potential. Plant Physiol. 157, 292–304. doi: 10.1104/pp.111.183210
Sinturel F., Petrenko V., Dibner C. (2020). Circadian clocks make metabolism run. J. Mol. Biol. 432, 3680–3699. doi: 10.1016/j.jmb.2020.01.018
Somanath P. R., Podrez E. A., Chen J., Ma Y., Marchant K., Antoch M., et al. (2011). Deficiency in core circadian protein Bmal1 is associated with a prothrombotic and vascular phenotype. J. Cell. Physiol. 226, 132–140. doi: 10.1002/jcp.22314
Takahashi J. S. (2019). Circadian clock genes and the transcriptional architecture of the clock mechanism. IBRO Rep. 6, S2–S3. doi: 10.1016/j.ibror.2019.07.171
Teodósio R., Aragão C., Colen R., Carrilho R., Dias J., Engrola S. (2021). A nutritional strategy to promote gilthead seabream performance under low temperatures. Aquaculture 537, 736494. doi: 10.1016/j.aquaculture.2021.736494
Viswambharan H., Carvas J. M., Antic V., Marecic A., Jud C., Zaugg C. E., et al. (2007). Mutation of the circadian clock gene Per2 alters vascular endothelial function. Circulation 115, 2188–2195. doi: 10.1161/CIRCULATIONAHA.106.653303
Wen H., Ren C., Li H., Da H., Wang Y., Jiang X., et al. (2017). Transcriptomic analyses on muscle tissues of litopenaeus vannamei provide the first profile insight into the response to low temperature stress. PLoS One 12, 1–21. doi: 10.1371/journal.pone.0178604
Xing Q., Tu H., Yang M., Chen G., Tang Q., Yi S., et al. (2022). Evaluation of cold tolerance and gene expression patterns associated with low-temperature stress in giant freshwater prawn macrobrachium rosenbergii. Aquaculture Rep. 24, 101172. doi: 10.1016/j.aqrep.2022.101172
Yúfera M., Nguyen M. V., Navarro-Guillén C., Moyano F. J., Jordal A. E. O., Espe M., et al. (2019). Effect of increased rearing temperature on digestive function in cobia early juvenile. Comp. Biochem. Physiol. Part A: Mol. Integr. Physiol. 230, 71–80. doi: 10.1016/j.cbpa.2019.01.007
Zhang X., Shen L., Li F., Zhang Y., Meng D., Sheng J. (2010). Up-regulating arginase contributes to amelioration of chilling stress and the antioxidant system in cherry tomato fruits. J. Sci. Food Agric. 90, 2195–2202. doi: 10.1002/jsfa.4070
Zhang Z. Q., Zhou C., Fan K. P., Zhang L., Liu Y., Liu P. F. (2021). Metabolomics analysis of the effects of temperature on the growth and development of juvenile European seabass (Dicentrarchus labrax). Sci. Total Environ. 769, 145155. doi: 10.1016/j.scitotenv.2021.145155
Zhao X.-l., Han Y., Ren S.-T., Ma Y.-M., Li H., Peng X.-X. (2015). L-proline increases survival of tilapias infected by streptococcus agalactiae in higher water temperature. Fish Shellfish Immunol. 44, 33–42. doi: 10.1016/j.fsi.2015.01.025
Zhao T. T., Ma A. J., Huang Z. H., Liu Z. F., Sun Z. B., Zhu C. Y., et al. (2021). Transcriptome analysis reveals that high temperatures alter modes of lipid metabolism in juvenile turbot (Scophthalmus maximus) liver. Comp. Biochem. Physiol. Part D: Genomics Proteomics 40, 100887. doi: 10.1016/j.cbd.2021.100887
Zhdanova I., Reebs S. (2006). Circadian rhythms in fish. Fish Physiol. 24, 197–238. doi: 10.1016/S1546-5098(05)24006-2
Zhong X., Yu J., Frazier K., Weng X., Li Y., Cham C. M., et al. (2018). Circadian clock regulation of hepatic lipid metabolism by modulation of m6A mRNA methylation. Cell Rep. 25, 1816–1828. doi: 10.1016/j.celrep.2018.10.068
Zhou L. Y., Fu S. J., Fu C., Ling H., Li X. M. (2019). Effects of acclimation temperature on the thermal tolerance, hypoxia tolerance and swimming performance of two endangered fish species in China. J. Comp. Physiol. B 189, 237–247. doi: 10.1007/s00360-018-01201-9
Zhou T., Gui L., Liu M. L., Li W. H., Hu P., Duarte D. F. C., et al. (2019). Transcriptomic responses to low temperature stress in the Nile tilapia, oreochromis niloticus. Fish Shellfish Immunol. 84, 1145–1156. doi: 10.1016/j.fsi.2018.10.023
Keywords: Takifugu rubripes, low temperature, liver, transcriptome, qPCR
Citation: Liu Z, Zhu L, Wang X, Liu S, Ma A, Chang H, Sun Z, Xu F and Zhao H (2022) Application of transcriptome analysis to investigate the effects of long-term low temperature stress on liver function in the tiger puffer (Takifugu rubripes). Front. Mar. Sci. 9:1069711. doi: 10.3389/fmars.2022.1069711
Received: 14 October 2022; Accepted: 09 November 2022;
Published: 29 November 2022.
Edited by:
Zhao Yunpeng, Dalian University of Technology, ChinaReviewed by:
Rui Jia, Freshwater Fisheries Research Center (CAFS), ChinaHuanhuan Huo, Jiangxi Agricultural University, China
Copyright © 2022 Liu, Zhu, Wang, Liu, Ma, Chang, Sun, Xu and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aijun Ma, bWFhakB5c2ZyaS5hYy5jbg==
†These authors have contributed equally to this work
 Zhifeng Liu
Zhifeng Liu Liguang Zhu1,2,3†
Liguang Zhu1,2,3† Xinan Wang
Xinan Wang Aijun Ma
Aijun Ma