- 1Department of Ecology, Jinan University, Southern Marine Science and Engineering Guangdong Laboratory (Zhuhai), Guangzhou, China
- 2Key Laboratory of Tropical Marine Bio-resources and Ecology and Guangdong Provincial Key Laboratory of Applied Marine Biology, South China Sea Institute of Oceanology, Chinese Academy of Sciences, Guangzhou, China
- 3Center for Evolution and Conservation Biology, Southern Marine Sciences and Engineering Guangdong Laboratory (Guangzhou), Guangzhou, China
- 4Sino-Danish Centre for Education and Research, University of Chinese Academy of Sciences, Beijing, China
- 5State Key Laboratory of Lake Science and Environment, Nanjing Institute of Geography and Limnology, Chinese Academy of Sciences, Nanjing, China
Increasing anthropogenic activities have caused serious environmental problems and undesirable ecological impacts on bay ecosystems. However, much remains to be learned regarding marine bacterial community assembly and its underlying mechanisms under intensive anthropogenic activities in subtropical bays. In this study, we used the community assembly theory to analyze bacterial community distributions in the subtropical Daya Bay, where the habitats are subject to serious thermal discharge and excessive nutrient load. We found the community assembly of bacterial in the Daya Bay was dominantly shaped by environmental factor of seawater phosphate, followed by temperature, and silicate. High phosphate concentration significantly increased the relative abundance of Actinobacteria, Cyanobacteria, Planctomycetes, and Gammaprotecteria, but reduced the relative abundance of Bacteroidetes and Alphaproteobacteria. Moreover, higher phosphate concentration was found significantly and positively correlated with higher bacterial alpha diversity. Compared with stochastic processes, we found higher phosphate concentration imposed stronger deterministic processes (primarily homogeneous selection) in structuring bacterial community assembly in the subtropical Daya bay, and bacterial communities tended to be higher phylogenetically clustered in higher phosphate-concentration habitats. In summary, we proposed that phosphate is a major environmental determinant in the subtropical Daya Bay and influenced the relative importance of deterministic and stochastic processes in bacterial community assembly.
1. Introduction
Bays are essentially important to human health and well-being for the services they provided, including shoreline protection and fish and wildlife habitat (Barbier et al., 2011; Temmink et al., 2020). However, due to the increasing and intensive anthropogenic activities around the bay ecosystems, currently, many bay ecosystems are facing serious environmental problems (Lefcheck et al., 2018; He and Silliman, 2019). Existing studies indicate that eutrophication, environmental pollution, and harmful algal bloom have occurred frequently in bay ecosystems and caused a decrease in biodiversity, a collapse of living organisms, and an ecological imbalance (McLusky and Elliott, 2004; Cloern and Jassby, 2010; Stauffer et al., 2020). The influence of environmental change on marine planktonic community has been a hot issue in the field of bay environmental protection (Matear et al., 2019). Bacteria, an integral component of marine planktonic community, play important roles in regenerating nutrients and decomposing organic matter of bays (Falkowski et al., 2008; Wang et al., 2020). Due to the short generation time and frequent ecological drift of bacteria, its community diversity may change synchronously with seawater environmental changes (Luo and Moran, 2015) and have influence on the resistance and resilience of other organisms to environmental change (Bissett et al., 2013). Despite the ecological functioning of bacterial community, much remains to be learned regarding the response of bacterial community assembly to environmental changes and its underlying mechanisms in the bay ecosystems.
It is a longstanding challenge to understand the mechanisms underlying bacterial community assembly in natural aquatic ecosystems (Quattrini et al., 2017; Shen et al., 2018; Aguilar and Sommaruga, 2020). One promising approach to understand the mechanisms underlying bacterial community assembly is the use of the phylogenetic framework (Webb et al., 2002; Bouwman et al., 2013; Ning et al., 2020). The changes in the relative influences of deterministic and stochastic processes across heterogeneous environmental conditions can be inferred in the phylogenetic framework, by comparing observed community phylogenetic structure with randomization procedures (Webb et al., 2002; Cavender-Bares et al., 2004; Quattrini et al., 2017; Ning et al., 2020). Based on the assumption that closely related phylogenetic taxa are more likely to have similar niches in phylogenetic framework, a nonrandom observed phylogenetic structure is expected to suggest the deterministic processes of community assembly (Webb, 2000; Wiens et al., 2010). Deterministic processes assume that bacterial community structure is controlled by deterministic factors such as environmental heterogeneity and species interactions. Meanwhile, stochastic processes suppose bacterial community structure is shaped by stochastic processes, such as probabilistic dispersal and random birth/death events (Hubbell, 2011). Because both deterministic and stochastic processes may play important roles in shaping bacterial community assembly, it comes up a question of that how their relative contributions of stochastic and deterministic processes change with environmental heterogeneity in natural bay ecosystems.
Daya Bay is located in the northern South China Sea (114.3°E to 114.5°E, 22.3°N to 22.5°N) between Shenzhen and Huizhou in Guangdong province near Hong Kong (Figure 1). It is one of the largest and most important semi-closed bays along the southern coast of China with an area around 600 km2, a depth between 6 and 15 m, and an average annual water temperature from 22.5- 23.5°C (Wang et al., 2008). As a core region of Pearl River Delta, the coastal area of this bay has been experiencing rapid economic development during the past decades. Since the operation of the first large-capacity commercial nuclear power plant, namely Daya Bay Nuclear Power Station in 1994, more and more agriculture, industries, and urbanization have been carried out around the Daya Bay (Tang et al., 2003; Wang et al., 2011). The increasingly anthropogenic activities around the bay ecosystems led to undesirable heterogeneous environmental problems and subsequent ecological impacts on the Daya bay ecosystems (Wu et al., 2017). It is showed that the Daya Bay are suffering long-term thermal pollution from the cooling systems of the two nuclear power stations, Daya Bay nuclear power station and Ling’ao nuclear power station (completed in 2002) (Tang et al., 2003; Wei et al., 2013). In addition, the bay is facing long-term eutrophication caused by the excessive nutrient input from the domestic sewage and aquaculture (Song et al., 2004; Sun et al., 2006). Long-term thermal pollution and nutrient (e.g., nitrogen and phosphorus) excess might be a “stress” event that acts as an environmental filter selecting bacterial taxa with suitable biological traits and filter out other members from the local species pool (Dai et al., 2017). Besides the deterministic processes of environmental factors, dispersal might simultaneously play important roles in shaping the patterns of the bacterial community assembly in the Daya Bay ecosystem, since exchanges of bacterial compositions across different patches through water flows and tides in the bay (Ren et al., 2019). The frequent dispersal of bacteria across different patches might result in the presence of bacterial taxa in places that are less suitable for them and weakening the deterministic effects of environmental factors (Juračka et al., 2016; Wisnoski and Lennon, 2021).

Figure 1 Map of sampling locations in the subtropical Daya Bay, Guangdong, China (A) and principal component analysis (PCA) depicting the relationships among all of the investigated environmental variables (B). T, temperature; DO, dissolved oxygen; : silicate, , phosphate; , nitrate, and , ammonium.
In this study, we used a phylogenetic framework to study bacterial community assembly and the underlying determining mechanisms based on 16S rRNA high-throughput sequencing collected from a set of habitats located in the Daya Bay sampled in a wet summer season. We hypothesized that environmental changes linked to increasing anthropogenic activities might regulate the interplay between deterministic and stochastic processes of bacterial community assembly in the subtropical Daya Bay, and significantly change bacterial community composition, diversity, and phylogenetic structure patterns. Our study might contribute to a broader understanding of the ecological effects of environmental changes to subtropical bay and benefit the management of the subtropical bays.
2. Materials and methods
2.1. Study area, sample collection and environment factor analyses
We set 11 sampling sites in the Daya Bay (Figure 1A), and sampled the seawater in August 2017, including the marine aquaculture area seriously affected by human activities (Fanhe harbour aquaculture area S1; Xiaoguiwan aquaculture area S5; Dapengao aquaculture area S10), nuclear power station thermal pollution area (S9), petrochemical pollution area (S6), the offshore site (S11), river inflow area (S2), and site S7 that was less affected by human activities.
At each sampling site, we set three replicates. 5 L surface seawater was collected at each replicate site and was filtered through 0.2-μm -pore-size Isopore filters (Millipore, Billerica, MA, USA). The filters with microbial biomass were stored at –70°C for further analysis. Water temperature, dissolved oxygen, salinity and pH were measured in situ at each replicate site by using multi-parameter water quality analyzer (YSI 6600, Yellow Springs, OH, USA). 500 mL surface seawater was further collected at each replicate site for the measurement of nutrient conditions. According to a standard procedure (GB 17378.4‐2007), seawater ammonium nitrogen, nitrate nitrogen, phosphate, and silicate concentrations were determined using a UV-visible spectrophotometer (UV2450, Shimadzu, Tokyo, Japan).
2.2. DNA extraction, PCR amplification, high-throughput sequencing and data processing
Microbial genomic DNA was extracted using PowerWater DNA Isolation Kit (MoBio Laboratories, Carlsbad, CA) and was purified by PowerClean DNA Clean up Kit (Mo Bio Laboratories, Carlsbad, CA, USA). With 60 ng DNA as template, PCR amplification was performed in the bacterial 16S rRNA V3 and V4 regions using the primer with 10 mM barcode and 25 μL 2 × PCR Premix Taq. The amplification primers were F341 (5 ‘-CCTACGGGAGGCAGCAG-3’) and R806 (3 ‘-GGACTACGGGTTCTAAT-5’). PCR reactions included an initial denaturation at 94°C for 5 min, followed by 30 cycles of 30 s at 94°C, 30 s at 52°C, and 30 s at 72°C and the final extension for 10 minutes (72°C). The positive amplicons were quantified using the PicoGreen dsDNA assay kit (Invitrogen Corporation, Carlsbad, CA, USA). Subsequently, the PCR products were equally combined and purified with Zymo’s Genomic DNA Clean & Concentrator kit (Zymo Research Corporation, Irvine, CA, USA). Finally, amplicons were sequenced using PE250 on Illumina Hiseq2500 platform.
Raw sequences were processed with the mothur software package (version 1.30.0, 2013) (http://www.mothur.org) according to the MiSeq standard operating procedure (Kozich et al., 2013). Briefly, the raw sequences were combined, denoised, trimmed, quality-filtered, and aligned to the SILVA v132 databases using mothur (Kozich et al., 2013). The high-quality sequences were then classified using the SILVA v132 databases at the recommended bootstrap threshold of 80% (Wang et al., 2007). The lineages belonging to chloroplasts, mitochondria, archaea or eukaryotes were removed and the high-quality sequences were then clustered into operational taxonomic units (OTUs) at a 97% similarity level. All singletons of OTUs and OTUs occurring in less than two samples were removed in the further analyses for minimizing bias caused by sequencing depth. In total, we obtained 949 OTUs though the whole sample set. Moreover, the minimum number of sequences (24100 sequences) was randomly subsampled in the whole sample set to correct for differences in sequencing depth.
2.3. Phylogenetic community assembly framework
To evaluate the phylogenetic signal of bacterial community assembly, we carried out a mantel correlogram based on Pearson correlation coefficients between differences in phylogenetic distances and environmental traits using mantel.correlog in R package of “Vegan” (Stegen et al., 2013). In addition, we performed community assembly mechanisms by phylogenetic bin-based null model analysis (iCAMP) framework modified by Ning et al. (Ning et al., 2020) from a previous framework (Stegen et al., 2013). When the βNRI value > +1.96 or< -1.96, it means the predominance of deterministic processes (heterogeneous or homogeneous selection). In contrast, a |βNRI | ≤ 1.96 indicates stochastic processes with |RCbray| > 0.95 representing of homogenizing dispersal or dispersal limitation, and |RCbray| ≤ 0.95 meaning ecological drift and other processes, such as speciation, weak selection, and/or weak dispersal (Ning et al., 2020). The relative importance of heterogeneous selection, homogeneous selection, dispersal limitation, homogenizing dispersal, and drift and other processes was calculated over the whole sample set and in the sample groups of different phosphate concentration in R package of “iCAMP” (Ning et al., 2020).
In addition, the mean-nearest-taxon-distance (MNTD) and the standardized-effect size of MNTD (SES.MNTD) were calculated by using package “picante” in R. By comparing the difference between the observed community and the random community, the main ecological processes of bacterial community assembly was determined in the Daya Bay. SES.MNTD means the differences in MNTD between the observed communities and the mean value of the 999 null communities divided by the standard deviation of the MNTD in the 999 null communities (Webb, 2000). When SES.MNTD greater than 2, it indicates phylogenetic overdispersion (taxa are more distantly related than would be expected random). If SES.MNTD less than 2, it suggests phylogenetic clustering (taxa are more closely related than would be expected at random). When SES.MNTD between -2 and 2, it represents that stochastic processes dominate the bacterial community assembly (Webb, 2000; Webb et al., 2002).
2.4. Statistical analyses
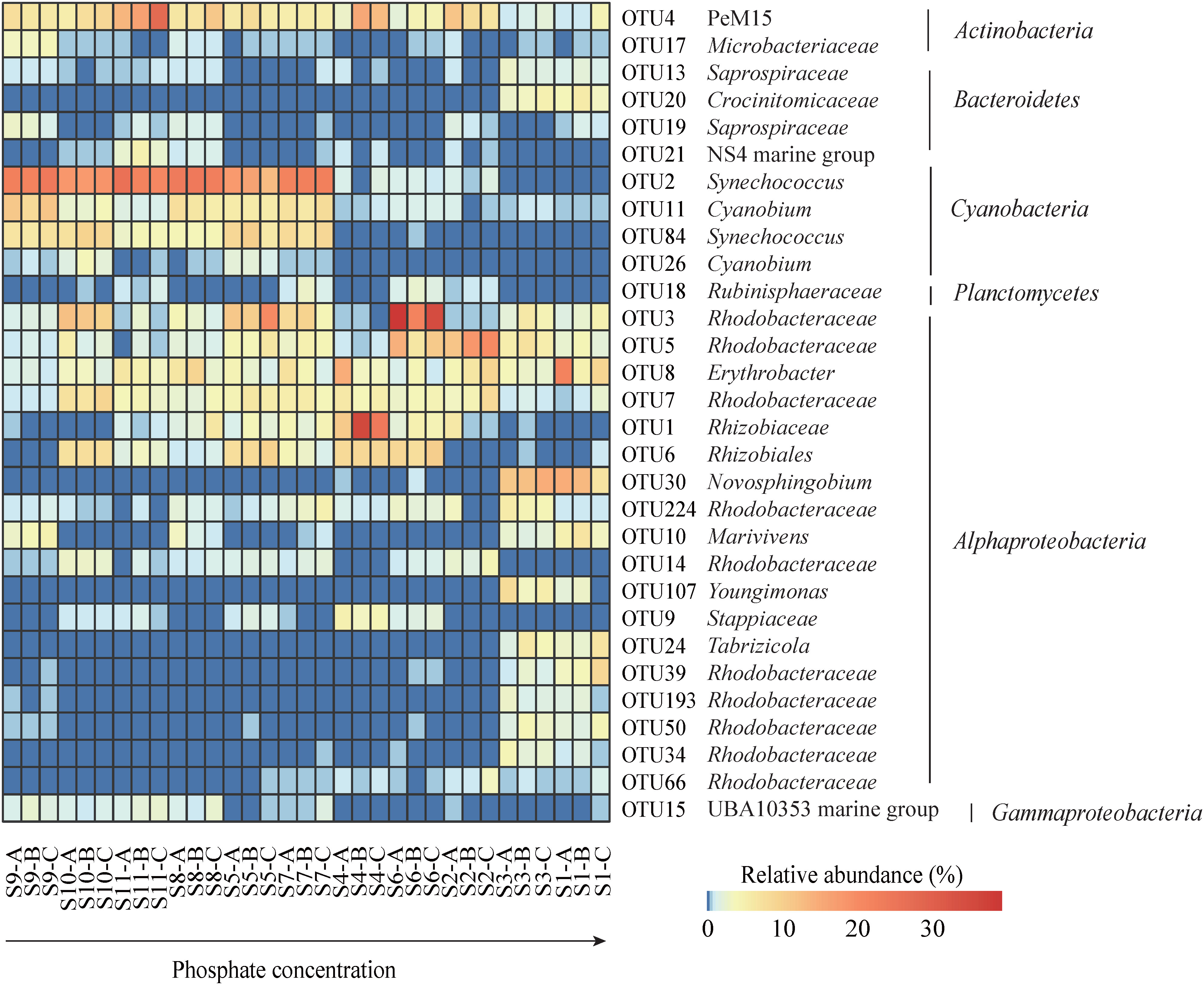
The spatial distributions of the studied environmental factors and bacterial relative abundance in the Daya Bay were showed in ArcGIS software. In addition, the relationships among all of the studied environmental factors were analyzed by performing principal component analysis (PCA) in the R package of vegan. Bacterial OTU richness and Pielou’s evenness were calculated using the “vegan” packages in R. The dissimilarities of the bacterioplanton community compositions (BCCs) between sites were performed based on both of the Bray-Curtis and the inter-community mean nearest taxon (MNTD) distance by using the “vegan” and “picante” packages, respectively, in R. The distributions of BCCs in the Daya Bay were further visualized by using nonmetric multidimensional scaling (NMDS) in the vegan package in R. Redundancy analysis (RDA) and multiple regression tree (MRT) were carried out to find the key environmental factors in determining the spatial pattern of BCCs by using the “vegan” and “mvpart” packages in R. Permutational multivariate analysis of variance (PERMANOVA) and pairwise PERMANOVA were performed to test significant differences of BCCs among different phosphate groups. Variance partitioning analysis (VPA) was carried out to find the effects of pure environments (E.), pure spatial (S.) factors and their mixed effect (Mix.) in explaining the bacterial community structure by using adjusted R-squared in redundancy analysis (RDA) in the “vegan” packages in R. The relative importance of the investigated environmental factors in determining bacterial community structure was further determined via Mantel test in the “vegan” packages in R. The relative abundances of the main bacterial lineages or clades of the top 30 OTUs were depicted along the phosphate gradient using the pheatmap command in the “pheatmap” package in R.
3. Results
3.1. Physicochemical characteristics in the Daya Bay
The average water temperature in the subtropical Daya Bay was 29.78 ± 1.14°C with the highest at site S9 (32.03°C), which was strongly affected by thermal pollution of the cooling system of the nuclear power stations (Figure S1). The average salinity of the Daya Bay was 28.84 ± 3.03‰, with the highest at site S11 (33.33‰) and the lowest at S1 (22.78‰) (Figure S1). The concentration of phosphate, nitrate, ammonium and silicate showed a decreased tendency from the top (site S1 and S3) to the offshore site (S11) of the Daya Bay (Figure S1). Principal component analysis (PCA) showed that the studied environmental factors could be reduced to two principal components (PC1 and PC2) describing 66.7% and 13.4% of the total variation of the environmental factors, respectively (Figure 1B). Moreover, seawater phosphate concentration was found positively correlated with silicate, nitrate, and ammonium concentration, but negatively linked to seawater dissolved oxygen, temperature, pH, and salinity (Figure 1B).
3.2. The distribution of bacterial community compositions in the Daya Bay
In the subtropical Daya Bay, the dominant bacterial phyla across all sampled habitats were Alphaproteobacteria, Cyanobacteria, Bacteroidetes, and Actinobacteria, representing approximately 51.0%, 22.8%, 9.8% and 9.7% of the total sequences, respectively (Figures S2, Figure 2). In addition, Gammaproteobacteria (3.3%), Planctomycetes (2.5%), Deltaproteobacteria (0.16%), Firmicutes (0.13%), and Acidobacteria (0.04%) were present in most sampled habitats but with low relative abundance (Figure S2). We observed that bacterial (sub)phylum compositions at S1 and S3 stations was similar, and the BCCs was mainly composed of Alphaproteobacteria (Figures S2, Figure 2E). The relative abundance of Bacteroides was much higher in S1, S3, and S11 than the other sites (Figures S2, Figure 2B). However, Cyanobacteria had the highest relative abundance at S7, S8, S9, and S10, where was strongly affected by the thermal pollution from the nuclear power plants (Figures S2, Figure 2C). In contrast with other phyla, Actinobacteria were mainly found in the estuary of the bay (Figures S2, Figure 2A).
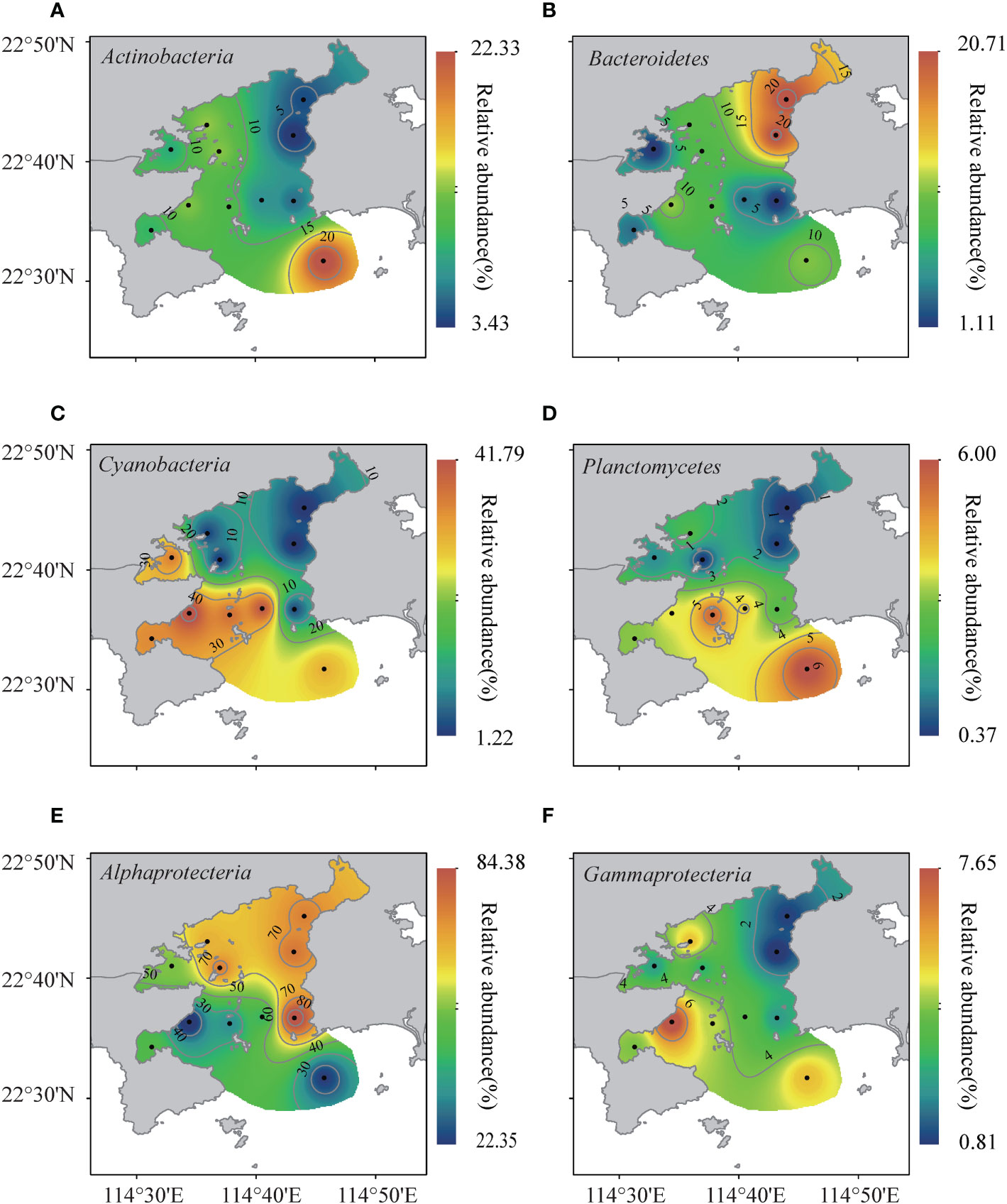
Figure 2 The distributions of dominant bacterial taxa in the Daya Bay, including Actinobacteria (A), Bacteroidetes (B), Cyanobacteria (C), Planctomycetes (D), Alphaproteobacteria (E), and Gammaproteobacteria (F).
3.3. Relating bacterial community compositions with environmental factors
Among the measured environmental factors, seawater phosphate had the highest correlation with BCCs in the Daya Bay, followed by temperature, and silicate as revealed by both redundancy analysis (RDA, FigureS3) and multiple regression tree (MRT, Figure S4). Phosphate concentration was significantly correlated with differences in BCCs across different bacterial phyla and subphyla (Figure 3A). Therefore, we further explored the relationship between the phosphate concentration and the relative abundance of the dominant phyla and subphyla. The relative abundances of Actinobacteria (Figure 3B), Cyanobacteria (Figure 3D), Planctomycetes (Figure 3E), and Gammaprotecteria (Figure 3G) decreased linearly with phosphate concentration, while the relative abundances of Bacteroidetes (Figure 3C) and Alphaproteobacteria (Figure 3F) were positively correlated with phosphate concentration. At the site with high phosphate concentration, the main lineage or clades were mostly composed of the bacteria Saprospiraceae and Croinitomicaceae of Bacteriodetes, and Rhodobacteraceae, Novosphingobium and Tabrizicola of Alphaproteobacteria (Figure 4), while NS4 marine group of Bacteriodetes, Synechococcus and Cyanobium of Cyanobacteria, and UBA10353 marine group of Gammaproteobacteria dominated in low phosphate concentration (Figure 4). The NMDS plot showed that bacterial spatial distributions did not show obvious geographical patterns, but was arranged according to the phosphate concentration as the low concentration (LPC, phosphate< 0.5 mg L-1), the medium concentration (MPC, 0.5 mg L-1< phosphate< 1.0 mg L-1), and the high concentration (HPC, phosphate > 1.0 mg L-1) groups (Figures 5A, B). Among these three phosphate groups, significant BCC differences were observed as revealed by permutational multivariate analysis of variance (PERMANOVA) and pairwise PERMANOVA (Table 1). Variance partitioning analysis revealed that pure environmental variables explained 33.2% of bacterial community structure, much higher than pure spatial effect (3.8%) (Figure S5). Moreover, seawater phosphate was found that had the highest Spearman’s correlations (R = 0.669, p< 0.001) with BCCs as determined via Mantel test (Table 2). We further observed that the BCCs of different spatial habitats but with the approximate phosphate concentration tend to be similar, suggesting the existence of strong linkages between phosphate concentration and community structure (Figure 5C, D). Between pairwise sampling habitats, the BCC dissimilarity both based on Bray-Curtis and inter-community mean nearest taxon (MNTD) distance significantly increased with the differences in seawater phosphate concentration (Figures 5C, D).

Figure 3 The relationships between environmental factors and their correlations with the community structure of the dominant bacterial (sub)phyla. Edge width means the R values of the Mantel’s statistic, and edge color represents the statistical significance based on 9,999 permutations (A). The relative abundances of Actinobacteria (B), Bacteroidetes (C), Cyanobacteria (D), Planctomycetes (E) Alphaproteobacteria (F), and Gammaproteobacteria (G) along the phosphate gradient.
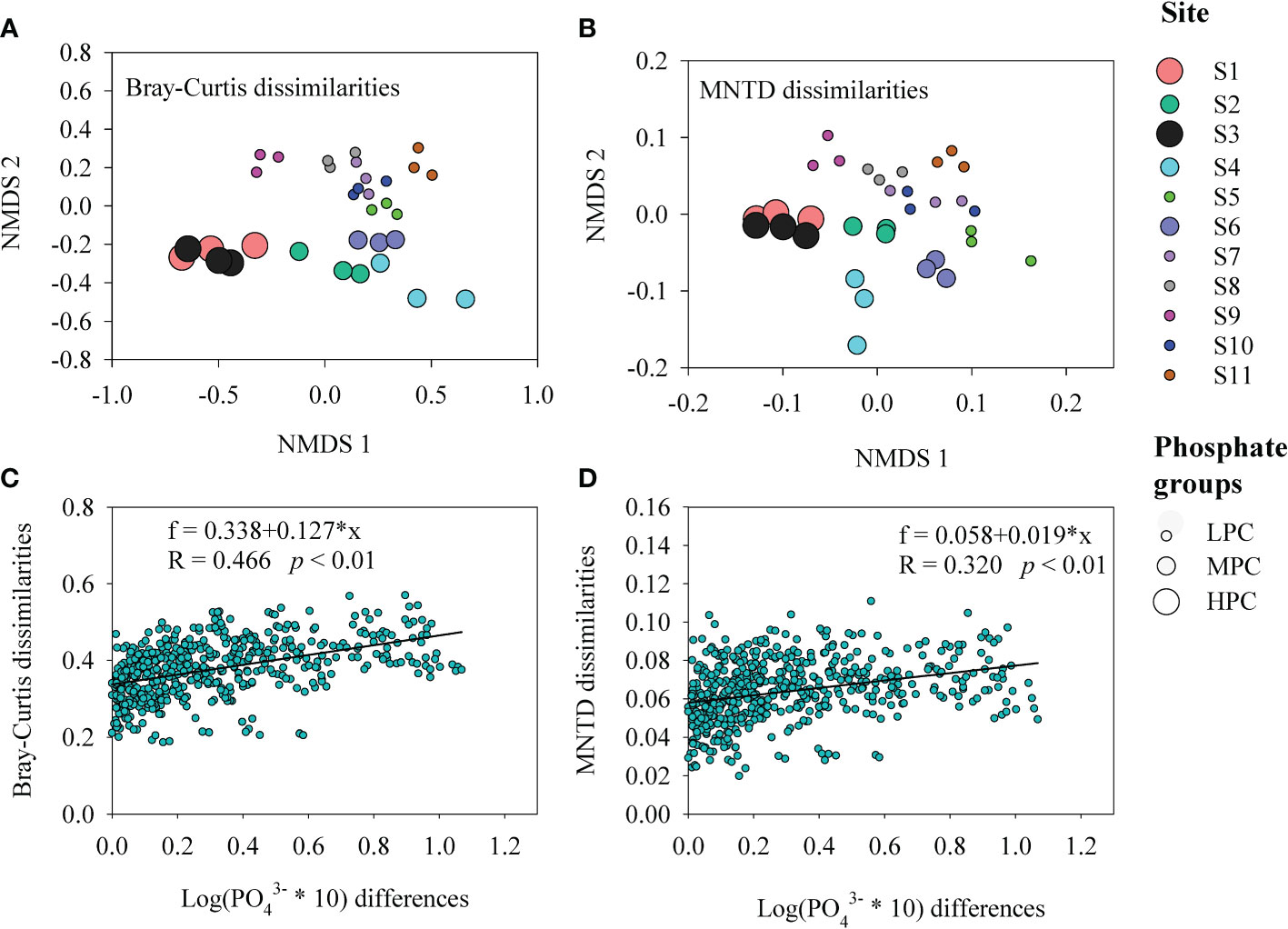
Figure 5 Non-metric multidimensional scaling (NMDS) ordinations showing the distributions of bacterial community compositions (BCCs) in the Daya Bay according to the phosphate gradient both based on BCC Bray-Curtis dissimilarity (A) and inter-community mean nearest taxon (MNTD) dissimilarity (B). Relationships between BCC Bray-Curtis (C) and MNTD (D) dissimilarities with the differences of phosphate concentration. The formulas, R values, and significances are given in each panel. LPC: the low phosphate concentration (phosphate< 0.5 mg L-1), MPC: the medium phosphate concentration (0.5 mg L-1< phosphate< 1.0 mg L-1), and HPC: the high phosphate concentration (phosphate > 1.0 mg L-1).
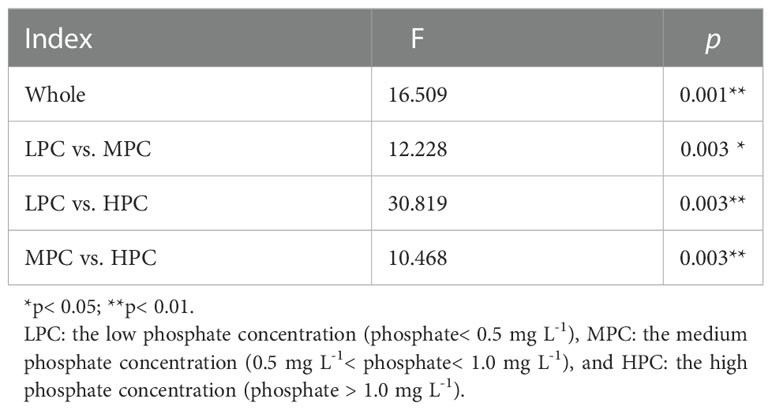
Table 1 Permutational multivariate analysis of variance (PERMANOVA) and pairwise PERMANOVA of seawater bacterial community structure according to different phosphate groups.
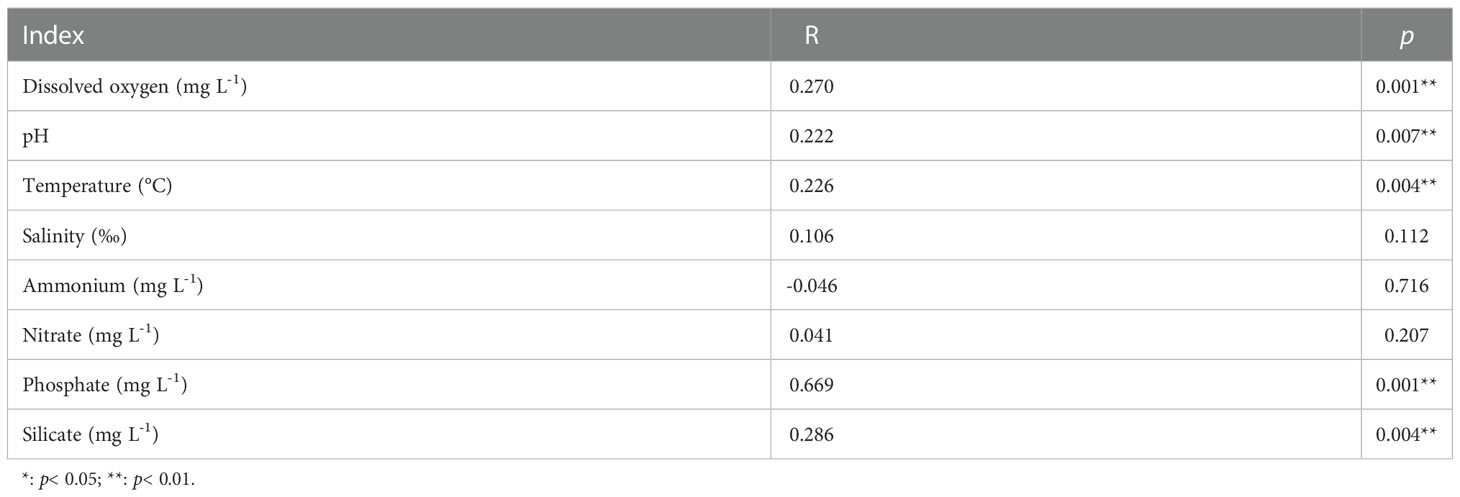
Table 2 Spearman’s correlations (R values) between transformed environmental variables and water bacterial community compositions as determined via Mantel test.
3.4. The distributions of bacterial alpha diversity and phylogenetic structure in the Daya Bay
We found seawater phosphate concentration had significantly positive correlations with bacterial alpha diversity, including OTU richness (R = 0.399, p< 0.05 Figure 6A) and Pielou’s evenness (R = 0.411, p< 0.05; Figure 6B). To understand the mechanisms underlying bacterial diversity pattern in the bay, the phylogenetic framework was performed, and we observed in contrast to alpha diversity pattern, phosphate concentration had significantly negative correlations with MNTD (R = 0.622, p< 0.05, Figure 6C), indicating that higher phosphate concentration linked to closer phylogenetic relationships between the bacterial taxa. Moreover, we observed that, the standardized-effect size of the MNTD (SES.MNTD) was significantly lower than zero (p< 0.05 in both cases) and decreased linearly with increasing phosphate concentration (R = 0.666; p< 0.05, Figure 6D). Bacterial phylogenetic signal was found across relatively short phylogenetic distance that was lower than 10% of the maximum phylogenetic distance (Figure 7A). In addition, among the deterministic and stochastic assembly processes (homogeneous and heterogeneous selection, dispersal limitations, homogenizing dispersal and the drift and other fractions), homogeneous selection were found contributing the largest fraction to the community assembly (40.5%), followed by dispersal limitation (34.3%) and drift and other processes (24.0%), which mostly consist of ecological drift, weak selection and diversification (Figure 7B). When calculating the relative importance of these processes by grouping samples according to phosphate concentration (LPC, MPC, and HPC), we found homogeneous selection played the highest relative importance in HPC, while drift and other processes dramatically decreased its relative importance in HPC (Figure 7C).
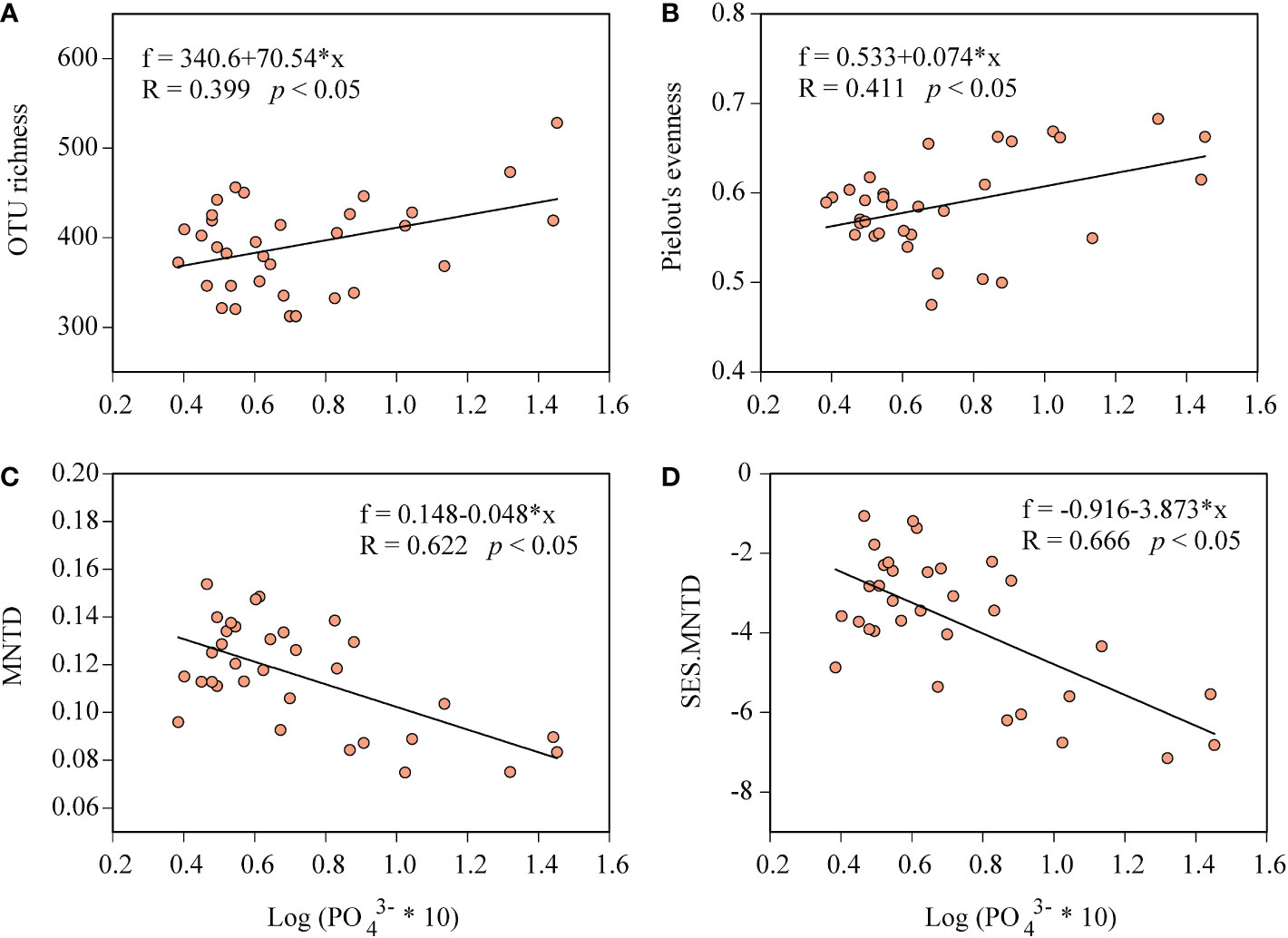
Figure 6 The distributions of the bacterial OTU richness (A), Pielou’s evenness (B), the mean nearest taxon distance (MNTD, C) and the standardized-effect size of MNTD (SES.MNTD, D) along the phosphate gradient in the Daya Bay.
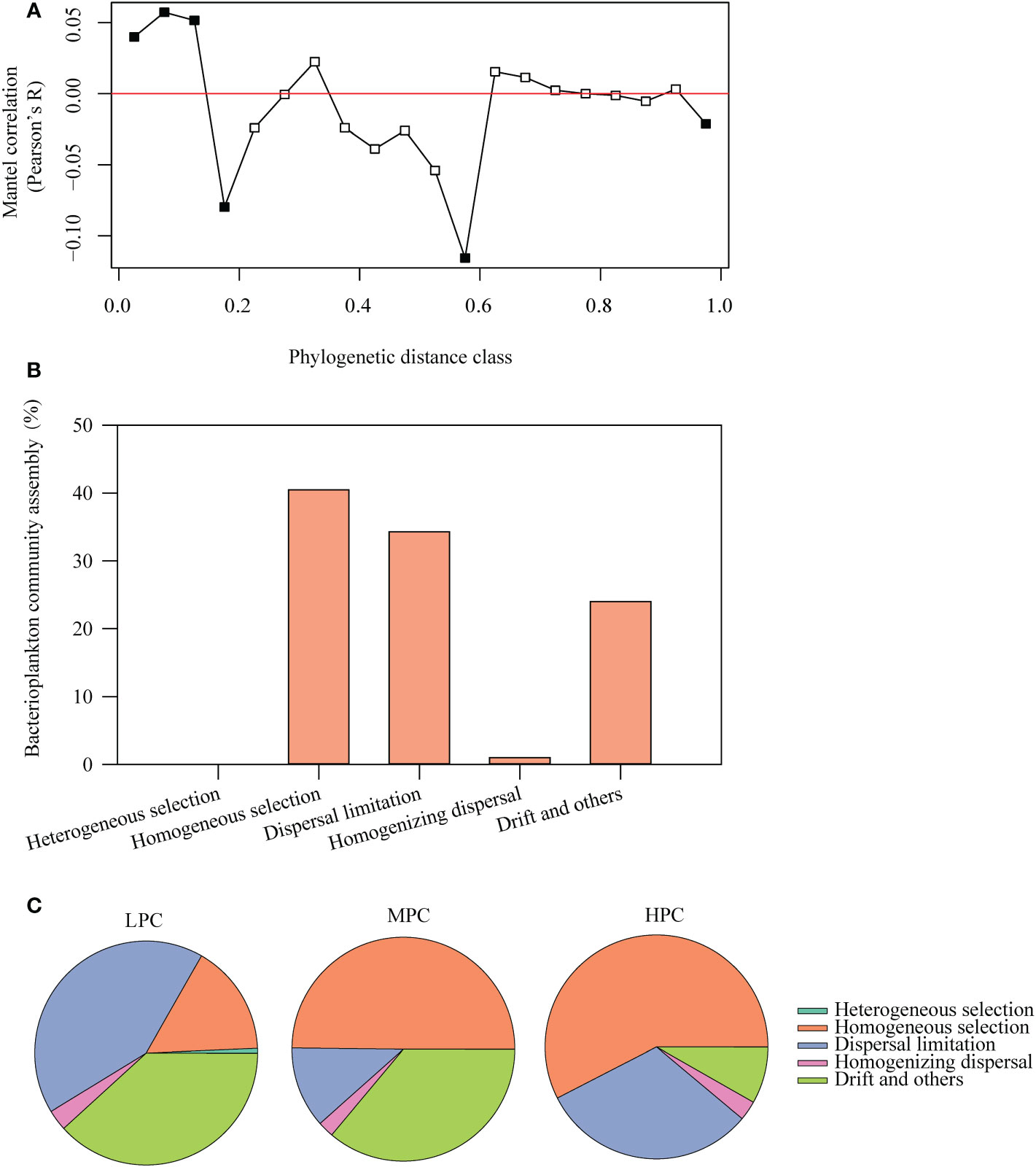
Figure 7 Pearson’s correlations from the Mantel correlogram (999 permutations) between the pairwise matrix of OTU niche distances and phylogenetic distances of bacterial community (A), and the relative importance of bacterial community assembly processes over the whole sample set (B) and in the sample groups of different phosphate concentration (C). The phylogenetic signal in niche associations was indicated by significant correlations (p< 0.05, solid circles). The community assembly processes included homogeneous and variable selection, dispersal limitations, homogenizing dispersal and the drift and other fractions. LPC: the low phosphate concentration (phosphate< 0.5 mg L-1), MPC: the medium phosphate concentration (0.5 mg L-1< phosphate< 1.0 mg L-1), and HPC: the high phosphate concentration (phosphate > 1.0 mg L-1).
4. Discussion
In this study, we examined seawater bacterial community assembly and the underlying mechanisms in the subtropical semi-enclosed Daya Bay ecosystem. We found that the spatial pattern of BCCs in the Daya Bay was dominantly shaped by environmental factor of seawater phosphate, followed by temperature, and silicate. Moreover, apparent phosphate concentration-related patterns of community compositions, bacterial diversity, and phylogenetic structure were observed in the Daya Bay.
4.1. Bacterial community assembly was dominantly shaped by environmental determinant of seawater phosphate in the Daya Bay
Dispersal plays an important role in determining the community assembly of bacteria among a set of interconnected habitats (Leibold et al., 2004). High dispersal rates may result in bacterial taxa surviving in habitats that less suitable for their growth, and in contrast, low dispersal rates of bacterial taxa may lead to dispersal limitation and thus lead to significant spatial variations (Juračka et al., 2016; Wisnoski and Lennon, 2021). In this study, we found that bacterial dispersal among the sampling habitats by water flows and tides in the Daya Bay did not result in BCC spatial homogeneity. In contrast, significant heterogeneity in BCCs was found across different sampling sites in the Daya Bay. Among the environmental factors, phosphate concentration was found to be the key environmental determinant of the assembly patterns of marine bacterial communities and apparent phosphate concentration-related patterns of BCCs were observed in the Daya Bay. This finding was in line with previous studies (Shen et al., 2018; Ren et al., 2019; Aguilar and Sommaruga, 2020), suggesting that bacterial community assembly in interconnected aquatic habitats was dominantly shaped by deterministic processes of some key environmental factors, followed by dispersal and other processes, such as ecological drift, weak selection and diversification. A number of studies have demonstrated that BCCs can rapidly track changes in the environment (Van Der Gucht et al., 2001; Muylaert et al., 2002). Thus, the persistence of newly arrived populations of bacteria migrating through flowing water was more likely to be determined by deterministic processes of heterogeneous habitats (Logue and Lindström, 2010).
4.2. Distributions of bacterial community compositions along phosphate gradients in the Daya Bay
We found that the sites with higher phosphate concentration were dominated by Alphaproteobacteria. Alphaproteobacteria are an “opportunistic group” that respond quickly to nutritional pulses, and they are found that dominated in plenty eutrophic water (Pinhassi and Berman, 2003). At the lineage or clade level of the top 30 OTUs, we also observed a larger BCC variation at higher phosphate-concentration sites by compared to the other sites. For higher phosphate-concentration sites, the relative abundance of Rhodobacteraceae, Novosphingobium and Saprospiraceae was obviously increased. We found the OTUs with the highest relative abundance belonged to the Rhodobacteraceae family (Alphaproteobacteria), which was one of the nine most widely distributed bacterial lineages in marine habitats (Garrity et al., 2015). Many bacterial taxa of Rhodobacteraceae are aerobic heterotrophs and have been identified as key players in organic carbon cycling (González et al., 1999; Moran et al., 2003; Newton et al., 2010). It has been reported that Rhodobacteraceae often existed in an environment rich in nitrogen/phosphorus nutrients, and it can use the soluble organic matter released by phytoplankton to sustain its own growth (Jones et al., 2007; Beck et al., 2014). Furthermore, recent studies suggest that the Rhodobacteraceae showed the highest response to the phosphorus amendment in northwest coastal Mediterranean waters (Sánchez et al., 2017). Novosphingobium was another abundant taxon in high phosphorus habitats. Previous study revealed that the main polar lipids of Novosphingobium were bisphosphatidylglycerol, phosphatidylethanolamine, phosphatidylcholine (Chen et al., 2015). Bacterial taxa belonging to Novosphingobium can therefore utilize surrounding phosphorus efficiently and tend to have high environmental abundance. Many studies about Saprospiraceae indicated that they were associated with the degradation of organic matter (Schauer et al., 2005; Chen et al., 2014). Shifts in the relative abundance of these bacterial taxa under higher phosphorus habitats may alter the overall bacterial community structure (Lebaron et al., 2001).
4.3. Phosphate increased bacterial alpha diversity in the Daya Bay
The nutrient condition in the Daya Bay has changed from previous nitrogen (N) limited to current phosphorus (P) limited for the excessive input of nitrogen (He et al., 2005; Zhang et al., 2020). Previous studies have revealed the potential phosphorus limitation of phytoplankton growth in Daya Bay (Wang et al., 2008; Wu et al., 2017; Zhang et al., 2018). Phosphorus limitation exists not only for phytoplankton, but also for bacteria. Compared to phytoplankton, bacteria can assimilate phosphorus more effectively because of their larger specific surface area, and they have a lower half-saturation constant for phosphate than phytoplankton (Currie and Kalff, 1984). In addition, the N:P ratio of phytoplankton biomass was generally about 16:1, which is much higher than that of heterotrophic bacteria (Kirchman, 2000), resulting in a higher efficiency of phosphorus requirement for bacteria than phytoplankton. It is suggested that bacterial cells were difficult to divide when phosphate concentration were limit (Zweifel et al., 1993; Yuan et al., 2011). The addition of phosphate in seawater might therefore enhance bacterial cell division and support for higher abundance and diversity of bacterial community (Yuan et al., 2011).
In our study, we found bacterial alpha diversity (out richness and Pielou’s evenness) was significant increased with phosphate concentration, and the S1 site with the highest phosphate concentration also had the highest bacterioutOTU richness and Pielou’s evenness. The S1 site in our study was located near the Fanhe harbor aquaculture area. Due to the rapid development of marine aquaculture industry, aquaculture was carried out since the 1990s on mangrove beaches and nutrients were fed into the water body in the form of bait (Wu et al., 2014). It was reported that only about 33% of phosphate was assimilated by fish during cage fish-farming, with the rest being retained in various forms in the cultured environment (Bouwman et al., 2013). It is suggested that sufficient nutrient supply is generally associated with enhanced primary productivity (Whitney et al., 2005). This enhanced primary productivity may lead to increased bacterial alpha diversity for the reduced resource competitive exclusion as “the larger pie can be divided into more pieces” (Brown, 1981; Fuhrman et al., 2008).
4.4. Phosphate influenced the relative importance of deterministic and stochastic processes in bacterial community assembly in the Daya Bay
It is important to elucidate the ecological mechanisms underlying microbial community assembly in community ecology (Webb et al., 2002; Zhou and Ning, 2017). In the phylogenetic framework, the relative influences of deterministic and stochastic processes in determining bacterial community assembly can be inferred by comparing observed community phylogenetic structure with randomization procedures (Webb et al., 2002; Quattrini et al., 2017; Ning et al., 2020). In this study, we observed significant negative correlations of MNTD with the phosphate concentration, suggesting that bacterial communities tended to be higher phylogenetically clustered in higher phosphate habitats. The phylogenetic clustering of bacterial community has been interpreted as evidence of environmental filtering, where a group of closely related species share a trait, or suite of traits, that allow them to persist in a given habitat (Webb et al., 2002). In this study, bacterial phylogenetic signal was also observed across relatively short phylogenetic distances, suggesting that the closely related taxa (OTUs) were niche conservatism. This finding was further confirmed by the spatial distributions of top 30 OTUs, from which, we found the closely related 8 OTUs of Rhodobacteraceae, which might share similar trait in phosphate utilization; all increased their relative abundance in high phosphate habitats (Figure 4).
We further compared the phylogenetic structure with randomization procedures, and found the standardized-effect size of MNTD was significantly lower than zero and decreased linearly with increasing the phosphate concentration. This finding indicates that phosphate in the Daya Bay influences the relative importance of deterministic and stochastic processes in bacterial community assembly, and compared with stochastic processes, high phosphate concentration imposed stronger deterministic processes in structuring bacterial community assembly in this subtropical Daya bay. This finding was further confirmed by phylogenetic bin-based null model analysis. It revealed that deterministic processes (primarily homogeneous selection) contributing the largest fraction to the overall community assembly dramatically increased in high- phosphate habitats.
It is not surprising that the higher of phosphate concentration linked to the stronger of deterministic assembly processes. Long term phosphate excess in a subtropical bay impacted by thermal discharge might be considered”as a ““tress” that select species from the regional species pool with suitable biological traits and excluding (filtering out) other members (Fodelianakis et al., 2014; Zhang et al., 2015). In this study, we found higher phosphate-concentration sites had higher relative abundance of Rhodobacteraceae, Novosphingobium and Saprospiraceae, which have been confirmed that have special biological traits in utilizing organic matter (Moran et al., 2003; Schauer et al., 2005; Newton et al., 2010; Chen et al., 2014) or phosphate (Chen et al., 2015). In addition, phosphate can stimulate the growth of protozoa (Caron and Countway, 2009), which might cause an increase in predation pressure on bacterial taxa, and resulted in enhanced deterministic processes in bacterial community assembly in high phosphate habitats (Chen et al., 2016). Our finding is in line with previous studies, which also showed that with the increase of nutrient load, the deterministic processes in aquatic bacterial community assembly were enhanced (Horner-Devine and Bohannan, 2006; Mykrä et al., 2016; Dai et al., 2017).
5. Conclusion
Our results showed that the bacterial community assembly was dominantly shaped by deterministic processes, and seawater phosphate was a major environmental determinant in the subtropical Daya Bay. We observed that high phosphate concentration not only significantly increased the diversity of bacterial communities, but also largely shifted the community compositions. Compared stochastic processes, high phosphate concentration imposed stronger effects of deterministic processes in shaping marine bacterial community assembly in the subtropical Daya Bay, and bacterial communities tended to be higher phylogenetically clustered in high phosphate habitats. We therefore proposed that among the heterogeneous habitats in Daya Bay, phosphate was a major environmental determinant, influencing the relative importance of deterministic and stochastic processes and shaping the patterns of marine bacterial diversity. Our research might contribute to a broader understanding of the ecological effects of environmental disturbance to subtropical bay and benefit the management of the subtropical bay.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found on: https://www.ncbi.nlm.nih.gov/, PRJNA765469.
Ethics statement
Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author contributions
LR and XS conceived the study, conducted the fieldwork, analyzed the data, wrote the manuscript, and provided funding. YC, GM, and CW analyzed the data and wrote the manuscript. QW and B-PH contributed to the manuscript revision and editing. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (42293264, 32171517, 41890853), Project of Southern Marine Science and Engineering Guangdong Laboratory (Guanzhou, GML20220017), and State Key Laboratory of Lake Science and Environment (2018SKL007).
Acknowledgments
We acknowledge Meiting Tan, Fuwu Xie, Yadong Huang, Jiaxing Liu, Chenhui Xiang, Weiwei Liu, and Kaizhi Li for assistance with experimental sampling, data analyses and manuscript revision. Zweifel, U. L., Norrman, B., Hagstrom, A. (1993). Consumption of dissolved organic carbon by marine bacteria and demand for inorganic nutrients. Mar. Ecol-Prog. Ser. 101, 23–23. doi: 10.3354/meps101023
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.1065973/full#supplementary-material
References
Aguilar P., Sommaruga R. (2020). The balance between deterministic and stochastic processes in structuring lake bacterioplankton community over time. Mol. Ecol. 29, 3117–3130. doi: 10.1111/mec.15538
Barbier E. B., Hacker S. D., Kennedy C., Koch E. W., Stier A. C., Silliman B. R. (2011). The value of estuarine and coastal ecosystem services. Ecol. Monogr. 81 (2), 169–193. doi: 10.1890/10-1510.1
Beck D. A., McTaggart T. L., Setboonsarng U., Vorobev A., Kalyuzhnaya M. G., Ivanova N., et al. (2014). The expanded diversity of Methylophilaceae from lake Washington through cultivation and genomic sequencing of novel ecotypes. PloS One 9, e102458. doi: 10.1371/journal.pone.0102458
Bissett A., Brown M. V., Siciliano S. D., Thrall P. H. (2013). Microbial community responses to anthropogenically induced environmental change: towards a systems approach. Ecol. Lett. 16, 128–139. doi: 10.1111/ele.12109
Bouwman L., Goldewijk K. K., van der Hoek K. W., Beusen A. H., Van Vuuren D. P., Willems J., et al. (2013). Exploring global changes in nitrogen and phosphorus cycles in agriculture induced by livestock production over the 1900–2050 period. Proc. Natl. Acad. Sci. U.S.A. 110, 20882–20887. doi: 10.1073/pnas.1012878108
Brown J. H. (1981). Two decades of homage to Santa rosalia: toward a general theory of diversity. Am. Zool. 21, 877–888. doi: 10.1093/icb/21.4.877
Caron D. A., Countway P. D. (2009). Hypotheses on the role of the protistan rare biosphere in a changing world. Aquat. Microb. Ecol. 57, 227–238. doi: 10.3354/ame01352
Cavender-Bares J., Ackerly D. D., Baum D., Bazzaz F. (2004). Phylogenetic overdispersion in Floridian oak communities. Am. Nat. 163, 823–843. doi: 10.1890/00129658(2006)87[109:PSOFPC]2.0.CO;2
Chen Z. R., Lei X. Q., Lai Q. L., Li Y., Zhang B. Z., Zhang J. Y., et al. (2014). Phaeodactylibacter xiamenensis gen. nov., sp. nov., a member of the family Saprospiraceae isolated from the marine alga Phaeodactylum tricornutum. Int. J. Syst. Evol. Microbiol. 64, 3496–3502. doi: 10.1099/ijs.0.063909-0
Chen X. X., Wang K., Guo A. N., Dong Z. Y., Zhao Q. F., Qian J., et al. (2016). Excess phosphate loading shifts bacterioplankton community composition in oligotrophic coastal water microcosms over time. J. Exp. Mar. Biol. Ecol. 483, 139–146. doi: 10.1016/j.jembe.2016.07.009
Chen N., Yu X. J., Yang J. S., Wang E. T., Li B. Z., Yuan H. L. (2015). Novosphingobium tardum sp. nov., isolated from sediment of a freshwater lake. Antonie. Van. Leeuwenhoek. 108, 51–57. doi: 10.1007/s10482-015-0463-x
Cloern J. E., Jassby A. D. (2010). Patterns and scales of phytoplankton variability in estuarine–coastal ecosystems. Estuaries Coast. 33, 230–241. doi: 10.1007/s12237-009-9195-3
Currie D. J., Kalff J. (1984). The relative importance of bacterioplankton and phytoplankton in phosphorus uptake in freshwater. Limnol. Oceanogr. 29, 311–321. doi: 10.4319/lo.1984.29.2.0311
Dai W. F., Zhang J. J., Tu Q. C., Deng Y., Qiu Q. F., Xiong J. B. (2017). Bacterioplankton assembly and interspecies interaction indicating increasing coastal eutrophication. Chemosphere 177, 317–325. doi: 10.1016/j.chemosphere.2017.03.034
Falkowski P. G., Fenchel T., Delong E. F. (2008). The microbial engines that drive earth’s biogeochemical cycles. Science 320, 1034–1039. doi: 10.1126/science.1153213
Fodelianakis S., Papageorgiou N., Pitta P., Kasapidis P., Karakassis I., Ladoukakis E. D. (2014). The pattern of change in the abundances of specific bacterioplankton groups is consistent across different nutrient-enriched habitats in Crete. Appl. Environ. Microbiol. 80, 3784–3792. doi: 10.1128/AEM.00088-14
Fuhrman J. A., Steele J. A., Hewson I., Schwalbach M. S., Brown M. V., Green J. L., et al. (2008). A latitudinal diversity gradient in planktonic marine bacteria. Proc. Natl. Acad. Sci. U.S.A. 105, 7774–7778. doi: 10.1073/pnas.0803070105
Garrity G., Belland J. A., Lilburn T., Family I (2015). “Rhodobacteraceae fam. nov,” in Bergey’s manual of systematic bacteriology, vol. 2, part c: The alpha-, beta-, delta-, and epsilonproteobacteria, second ed., vol. 2. Eds. Brenner D. J., Krieg N. R., Staley J. T. (New York: Springer). Garrity.
González J. M., Kiene R. P., Moran M. A. (1999). Transformation of sulfur compounds by an abundant lineage of marine bacteria in the α-subclass of the class Proteobacteria. appl. Environ. Microbiol. 65, 3810–3819. doi: 10.1128/AEM.65.9.3810-3819.1999
He Y. X., Huang X. P., Huang L. M., Xu Z. Z., Yue W. Z., Zhang J. L. (2005). Annual variation and analysis of nutrients in aquaculture area of daya bay. Mar. Environ. Sci. 24, 20–23. doi: 10.1007/11428831_101
He Q., Silliman B. R. (2019). Climate change, human impacts, and coastal ecosystems in the anthropocene. Curr. Biol. 29, R1021–R1035. doi: 10.1016/j.cub.2019.08.042
Horner-Devine M. C., Bohannan B. J. (2006). Phylogenetic clustering and overdispersion in bacterial communities. Ecology 87, S100–S108. doi: 10.1890/0012-9658(2006)87[100:PCAOIB]2.0.CO;2
Hubbell S. P. (2011). The unified neutral theory of biodiversity and biogeography (MPB-32) (Princeton: Princeton University Press).
Jones P. R., Cottrell M. T., Kirchman D. L., Dexter S. C. (2007). Bacterial community structure of biofilms on artificial surfaces in an estuary. Microb. Ecol. 53, 153–162. doi: 10.1007/s00248-006-9154-5
Juračka P. J., Declerck S. A., Vondrák D., Beran L., Černý M., Petrusek A. (2016). A naturally heterogeneous landscape can effectively slow down the dispersal of aquatic microcrustaceans. Oecologia 180, 785–796. doi: 10.1007/s00442-015-3501-5
Kirchman D. L. (2000). “Uptake and regeneration of inorganic nutrients by marine heterotrophic bacteria,” in Microbial ecology of the oceans. Ed. Kirchman D. L. (USA: Wiley-Liss), 261–288.
Kozich J. J., Westcott S. L., Baxter N. T., Highlander S. K., Schloss P. D. (2013). Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq illumina sequencing platform. Appl. Environ. Microbiol. 79, 5112–5120. doi: 10.1128/AEM.01043-13
Lebaron P., Servais P., Troussellier M., Courties C., Muyzer G., Bernard L., et al. (2001). Microbial community dynamics in Mediterranean nutrient-enriched seawater mesocosms: changes in abundances, activity and composition. FEMS. Microbiol. Ecol. 34, 255–266. doi: 10.1111/j.1574-6941.2001.tb00776.x
Lefcheck J. S., Orth R. J., Dennison W. C., Wilcox D. J., Murphy R. R., Keisman J., et al. (2018). Long-term nutrient reductions lead to the unprecedented recovery of a temperate coastal region. Proc. Natl. Acad. Sci. U.S.A. 115, 3658–3662. doi: 10.1073/pnas.1715798115
Leibold M. A., Holyoak M., Mouquet N., Amarasekare P., Chase J. M., Hoopes M. F., et al. (2004). The metacommunity concept: a framework for multi-scale community ecology. Ecol. Lett. 7, 601–613. doi: 10.1111/j.1461-0248.2004.00608.x
Logue J. B., Lindström E. S. (2010). Species sorting affects bacterioplankton community composition as determined by 16S rDNA and 16S rRNA fingerprints. ISME. J. 4, 729–738. doi: 10.1038/ismej.2009.156
Luo H. W., Moran M. A. (2015). How do divergent ecological strategies emerge among marine bacterioplankton lineages? Trends. Microbiol. 23, 577–584. doi: 10.1016/j.tim.2015.05.004
Matear L., Robbins J. R., Hale M., Potts J. (2019). Cetacean biodiversity in the bay of Biscay: suggestions for environmental protection derived from citizen science data. Mar. Policy. 109, 103672. doi: 10.1016/j.marpol.2019.103672
McLusky D. S., Elliott M. (2004). The estuarine ecosystem: ecology, threats and management (Oxford: Oxford University Press).
Moran M. A., González J. M., Kiene R. P. (2003). Linking a bacterial taxon to sulfur cycling in the sea: studies of the marine Roseobacter group. Geomicrobiol. J. 20, 375–388. doi: 10.1080/01490450303901
Muylaert K., van der Gucht K., Vloemans N., Meester L. D., Gillis M., Vyverman W. (2002). Relationship between bacterial community composition and bottom-up versus top-down variables in four eutrophic shallow lakes. Appl. Environ. Microbiol. 68, 4740–4750. doi: 10.1128/aem.68.10.4740-4750.2002
Mykrä H., Tolkkinen M., Markkola A. M., Pirttilä A. M., Muotka T. (2016). Phylogenetic clustering of fungal communities in human-disturbed streams. Ecosphere 7, e01316. doi: 10.1002/ecs2.1316
Newton R. J., Griffin L. E., Bowles K. M., Meile C., Gifford S., Givens C. E., et al. (2010). Genome characteristics of a generalist marine bacterial lineage. ISME. J. 4, 784–798. doi: 10.1038/ismej.2009.150
Ning D., Yuan M., Wu L., Zhang Y., Guo X., Zhou X., et al. (2020). A quantitative framework reveals ecological drivers of grassland microbial community assembly in response to warming. Nat. Commun. 11, 4717. doi: 10.1038/s41467-020-18560-z
Pinhassi J., Berman T. (2003). Differential growth response of colony-forming α- and γ-proteobacteria in dilution culture and nutrient addition experiments from lake kinneret (Israel), the eastern Mediterranean Sea, and the gulf of eilat. Appl. Environ. Microbiol. 69, 199–211. doi: 10.1128/AEM.69.1.199-211.2003
Quattrini A. M., Gómez C. E., Cordes E. E. (2017). Environmental filtering and neutral processes shape octocoral community assembly in the deep sea. Oecologia 183, 221–236. doi: 10.1007/s00442-016-3765-4
Ren L. J., Song X. Y., He D., Wang J. J., Tan M. T., Xia X. M., et al. (2019). Bacterioplankton metacommunity processes across thermal gradients: weaker species sorting but stronger niche segregation in summer than in winter in a subtropical bay. Appl. Environ. Microbiol. 85, e02088–e02018. doi: 10.1128/AEM.02088-18
Sánchez O., Gasol J. M., Ferrera I. (2017). Effects of grazing, phosphorus and light on the growth rates of major bacterioplankton taxa in the coastal NW Mediterranean. Environ. Microbiol. Rep. 9, 300–309. doi: 10.1111/1758-2229.12535
Schauer M., Kamenik C., Hahn M. W. (2005). Ecological differentiation within a cosmopolitan group of planktonic freshwater bacteria (SOL cluster, Saprospiraceae, Bacteroidetes). Appl. Environ. Microbiol. 71, 5900–5907. doi: 10.1128/AEM.71.10.5900-5907.2005
Shen D. D., Jürgens K., Beier S. (2018). Experimental insights into the importance of ecologically dissimilar bacteria to community assembly along a salinity gradient. Environ. Microbiol. 20, 1170–1184. doi: 10.1111/1462-2920.14059
Song X. Y., Huang L. M., Zhang J. L., Huang X. P., Zhang J. B., Yin J. Q., et al. (2004). Variation of phytoplankton biomass and primary production in daya bay during spring and summer. Mar. pollut. Bull. 49, 1036–1044. doi: 10.1016/j.marpolbul.2004.07.008
Stauffer B. A., Sukhatme G. S., Caron D. A. (2020). Physical and biogeochemical factors driving spatially heterogeneous phytoplankton blooms in nearshore waters of Santa Monica bay, USA. Estuaries. Coasts 43, 909–926. doi: 10.1007/s12237-020-00704-5
Stegen J. C., Lin X., Fredrickson J. K., Chen X., Kennedy D. W., Murray C. J., et al. (2013). Quantifying community assembly processes and identifying featuresthat impose them. ISME. J. 7, 2069–2079. doi: 10.1038/ismej.2013.93
Sun C. C., Wang Y. S., Sun S., Zhang F. Q. (2006). Analysis dynamics of phytoplankton community characteristics in daya bay. Acta Ecol. Sin. 26, 3948–3958. doi: 10.3321/j.issn:1000-0933.2006.12.005
Tang D. L., Kester D. R., Wang Z. D., Lian J. S., Kawamura H. (2003). AVHRR satellite remote sensing and shipboard measurements of the thermal plume from the daya bay, nuclear power station, China. Remote. Sens. Environ. 84, 506–515. doi: 10.1016/S0034-4257(02)00149-9
Temmink R. J., Christianen M. J., Fivash G. S., Angelini C., Boström C., Didderen K., et al. (2020). Mimicry of emergent traits amplifies coastal restoration success. Nat. Commun. 11, 3668. doi: 10.1038/s41467-020-17438-4
Van Der Gucht K., Sabbe K., De Meester L., Vloemans N., Zwart G., Gillis M., et al. (2001). Contrasting bacterioplankton community composition and seasonal dynamics in two neighbouring hypertrophic freshwater lakes. Environ. Microbiol. 3, 680–690. doi: 10.1046/j.1462-2920.2001.00242.x
Wang Q., Garrity G. M., Tiedje J. M., Cole J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Wang Y. S., Lou Z. P., Sun C. C., Sun S. (2008). Ecological environment changes in daya bay, China, from 1982 to 2004. Mar. pollut. Bull. 56, 1871–1879. doi: 10.1016/j.marpolbul.2008.07.017
Wang Z. H., Mu D. H., Li Y. F., Cao Y., Zhang Y. J. (2011). Recent eutrophication and human disturbance in daya bay, the south China Sea: dinoflagellate cyst and geochemical evidence. Estuar. Coast. Shelf. Sci. 92, 403–414. doi: 10.1016/j.ecss.2011.01.015
Wang Y. M., Pan J., Yang J., Zhou Z. C., Pan Y. P., Li M. (2020). Patterns and processes of free-living and particle-associated bacterioplankton and archaeaplankton communities in a subtropical river-bay system in south China. Limnol. Oceanogr. 65, S161–S179. doi: 10.1002/lno.11314
Webb C. O. (2000). Exploring the phylogenetic structure of ecological communities: an example for rain forest trees. Am. Nat. 156, 145–155. doi: 10.1086/303378
Webb C. O., Ackerly D. D., McPeek M. A., Donoghue M. J. (2002). Phylogenies and community ecology. Annu. Rev. Ecol. Syst. 33, 475–505. doi: 10.1146/annurev.ecolsys.33.010802.150448
Wei X., Ni P. T., Zhan H. G. (2013). Monitoring cooling water discharge using Lagrangian coherent structures: A case study in daya bay, China. Mar. pollut. Bull. 75, 105–113. doi: 10.1016/j.marpolbul.2013.07.056
Whitney F., Crawford W., Harrison P. (2005). Physical processes that enhance nutrient transport and primary productivity in the coastal and open ocean of the subarctic NE pacific. Deep. Sea. Res. Part. II. Top. Stud. Oceanogr. 52, 681–706. doi: 10.1016/j.dsr2.2004.12.023
Wiens J. J., Ackerly D. D., Allen A. P., Anacker B. L., Buckley L. B., Cornell H. V., et al. (2010). Niche conservatism as an emerging principle in ecology and conservation biology. Ecol. Lett. 13, 1310–1324. doi: 10.1111/j.1461-0248.2010.01515.x
Wisnoski N. I., Lennon J. T. (2021). Microbial community assembly in a multi-layer dendritic metacommunity. Oecologia 195, 13–24. doi: 10.1007/s00442-020-04767-w
Wu H., Peng R. H., Yang Y., He L., Wang W. Q., Zheng T. L., et al. (2014). Mariculture pond influence on mangrove areas in south China: Significantly larger nitrogen and phosphorus loadings from sediment wash-out than from tidal water exchange. Aquaculture 426, 204–212. doi: 10.1016/j.aquaculture.2014.02.009
Wu M. L., Wang Y. S., Wang Y. T., Yin J. P., Dong J. D., Jiang Z. Y., et al. (2017). Scenarios of nutrient alterations and responses of phytoplankton in a changing daya bay, south China Sea. J. Mar. Syst. 165, 1–12. doi: 10.1016/j.jmarsys.2016.09.004
Yuan X. C., He L., Yin K. D., Pan G., Harrison P. J. (2011). Bacterial distribution and nutrient limitation in relation to different water masses in the coastal and northwestern south China Sea in late summer. Cont. Shelf. Res. 31, 1214–1223. doi: 10.1016/j.csr.2011.04.012
Zhang Y., Li H. L., Guo H. M., Zheng C. M., Wang X. J., Zhang M., et al. (2020). Improvement of evaluation of water age and submarine groundwater discharge: A case study in daya bay, china. J. Hydrol 586, 124775. doi: 10.1016/j.jhydrol.2020.124775
Zhang Q. F., Tang F. Y., Zhou Y. J., Xu J. R., Chen H. P., Wang M. K., et al. (2015). Shifts in the pelagic ammonia-oxidizing microbial communities along the eutrophic estuary of yong river in ningbo city, China. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.01180
Zhang X., Zhang J. P., Shen Y., Zhou C. H., Huang X. P. (2018). Dynamics of alkaline phosphatase activity in relation to phytoplankton and bacteria in a coastal embayment daya bay, south China. Mar. pollut. Bull. 131, 736–744. doi: 10.1016/j.marpolbul.2018.05.008
Zhou J. Z., Ning D. L. (2017). Stochastic community assembly: does it matter in microbial ecology? Microbiol. Mol. Biol. Rev. 81, e00002–e00017. doi: 10.1128/MMBR.00002-17
Keywords: seawater phosphate, bacterial community, phylogenetic assembly, environmental filter, subtropical bay
Citation: Chen Y, Ma G, Wu C, Peng Y, Song X, Wu QL, Han B-P and Ren L (2023) Bacterial communities exhibit apparent phosphate concentration-related patterns of community composition, alpha diversity, and phylogenetic structure in the subtropical Daya Bay. Front. Mar. Sci. 9:1065973. doi: 10.3389/fmars.2022.1065973
Received: 10 October 2022; Accepted: 28 December 2022;
Published: 19 January 2023.
Edited by:
Jinbo Xiong, Ningbo University, ChinaReviewed by:
Huajun Zhang, Ningbo University, ChinaHuang Honghui, South China Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences (CAFS), China
Copyright © 2023 Chen, Ma, Wu, Peng, Song, Wu, Han and Ren. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lijuan Ren, bGlqdWFucmVuQGpudS5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
 Ye Chen1†
Ye Chen1† Xingyu Song
Xingyu Song Qinglong L. Wu
Qinglong L. Wu Lijuan Ren
Lijuan Ren