- 1College of Marine Life Sciences, Institute of Evolution and Marine Biodiversity, Frontiers Science Center for Deep Ocean Multispheres and Earth System, Ocean University of China, Qingdao, China
- 2College of Environmental Science and Engineering, Ocean University of China, Qingdao, China
- 3Laboratory for Marine Ecology and Environmental Science, Pilot National Laboratory for Marine Science and Technology (Qingdao), Qingdao, China
- 4Key Laboratory of Marine Environment and Ecology, Ministry of Education, Ocean University of China, Qingdao, China
- 5OUC-UMT Joint Academic Centre for Marine Studies, Ocean University of China, Qingdao, China
- 6Department of Critical Care Medicine, the Affiliated Hospital of Qingdao University, Qingdao, China
Coastal eutrophication may be a vital inducement of green tide. Denitrification is an important nitrogen removal pathway that involves a series of enzymatic reactions. The rate-limiting step in the conversion of nitrite to nitric oxide is encoded by two functionally equivalent but structurally distinct genes, copper-containing nitrite reductase gene (nirK) and cytochrome cd1-containing nitrite reductase gene (nirS). Here, we used Illumina Miseq sequencing approach to examine the variations in denitrifying bacterial community characteristics and interactions during an Ulva prolifera green tide in coastal Qingdao areas. Our findings suggested that the variations in the denitrifying bacterial community structure during the green tide were closely related to the changes of chlorophyll a content, salinity and dissolved oxygen content. The nirK-type denitrifying bacteria were more sensitive to green tide than the nirS-type denitrifying bacteria. Additionally, the nirK-type denitrifying bacterial interactions were more stable and complex during the outbreak phase, while the nirS-type denitrifying bacterial interactions were more stable and complex during the decline phase. All of these characters demonstrated that the nirK-type and nirS-type denitrifying bacteria respond differently to the green tide, implying that they may occupy different niches during the green tide in coastal Qingdao areas.
Introduction
Green tide is a harmful algal bloom that occurs frequently in coastal areas. Since 2007, an annual green tide caused by Ulva prolifera has occurred in the Yellow Sea, which is thought to be the largest green tide ever (Liu et al., 2013; Zhang et al., 2019). In general, U. prolifera green tide appears along the Jiangsu coast from the middle of April to early May, migrates northward due to ocean currents and seasonal monsoons, reaches the coastal areas of the Shandong Peninsula in the middle of June, and then begins to decay in the middle of July (Zhang et al., 2019). During the migration period, U. prolifera multiplies rapidly and expands its coverage area, posing a major threat to coastal areas. It is reported that green tide alters not only the physicochemical factors of seawater, but also the microbial community in marine environments (van Alstyne et al., 2015; Lin et al., 2017; Qu et al., 2020). Qu et al. (2020), for example, stated that the outbreak of green tide reduced the abundance and diversity of bacterial community in Qingdao offshore areas. In turn, some microbial groups, such as Cytophaga-Flexibacter-Bacteroides, play a vital role in regulating the presence, development and decline of green tide (Marshall et al., 2006).
Green tide may be induced by coastal eutrophication caused by high nitrogen input (Li et al., 2017). Denitrification is regarded as the most effective nitrogen removal pathway, with the potential to minimize nitrogen pollution and eutrophication (Zumft, 1997; Falkowski et al., 2008; Guo et al., 2014). U. prolifera convert dissolved inorganic nitrogen (DIN) to dissolved organic nitrogen (DON) during the outbreak phase, and release quantities of DON and ammonium into seawater during the decline phase, as a result, U. prolifera green tide is regarded as a “nitrogen pump” and may have an impact on the denitrification process in coastal areas (Zhang et al., 2021a). Furthermore, U. prolifera competes for nitrate with denitrifying bacteria, which may affect the denitrification process and associated microbial activity (Christensen et al., 2000; Sun et al., 2020). Therefore, it is of importance to investigate variations in the denitrification process and associated microbial communities during the U. prolifera green tide.
As previously stated, denitrification is an important nitrogen removal pathway, accounting for 77% of total global nitrogen removal (Dalsgaard et al., 2012). According to reports, denitrification dominates the nitrogen removal in marine environments, rather than other processes such as anaerobic ammonium oxidation (Dalsgaard et al., 2005; Babbin et al., 2014). Denitrification is the process that contains a series of enzymatic reactions including nitrate reduction to nitrite, nitrite reduction to nitric oxide, nitric oxide reduction to nitrous oxide and nitrous oxide reduction to nitrogen, which are catalyzed sequentially by nitrate reductase (Nar), nitrite reductase (Nir), nitrogen oxide reductase (Nor) and nitrous oxide reductase (Nos) (Zumft, 1997). Due to the great phylogenetic diversity, studies on the denitrifying bacterial communities in natural environments should target the functional genes involved in the denitrification process (Lee and Francis, 2017).
The reduction of nitrite to nitric oxide is a rate-limiting step in the denitrification pathway, as well as the initial step in producing gaseous nitrogen products (Zumft, 1997). Copper-containing nitrite reductase gene (nirK) and cytochrome cd1-containing nitrite reductase gene (nirS) have been widely used as molecular markers to study the characteristics of denitrifying bacterial communities in a variety of habitats (Wolsing and Priemé, 2004; Santoro et al., 2006; Oakley et al., 2007; Francis et al., 2013; Liu et al., 2020). Though functionally equivalent, the nirK and nirS genes have distinct evolutionary histories and represent two ecologically separate groups of denitrifying bacteria (Jones and Hallin, 2010). Studies have shown that the community richness and abundance of the nirS-type denitrifying bacteria was greater than that of the nirK-type denitrifying bacteria in sediments of the San Francisco Bay (Mosier and Francis, 2010), whereas the nirK-type denitrifying bacterial community presented greater richness and more abundant in agricultural soils (Yoshida et al., 2009; Zhou et al., 2011; Yang et al., 2018). Bothe et al. (2000) pointed out that the nirS-type denitrifying bacteria has a more widely distribution than the nirK-type denitrifying bacteria. The nirS-type denitrifying bacteria is more susceptible to environmental changes than the nirK-type denitrifying bacteria (Yang et al., 2018). Overall, the nirK-type and nirS-type denitrifying bacteria may have different responses to environmental changes, implying that the two types of denitrifying bacteria may occupy different niches in the same environment (Yuan et al., 2012; Wei et al., 2015). However, it is unknown if the nirK-type and nirS-type denitrifying bacteria respond differently to the U. prolifera green tide.
In the present study, variations of the nirK-type and nirS-type denitrifying bacterial communities during an U. prolifera green tide in coastal Qingdao areas were studied using Illumina Miseq sequencing approach of the nirK and nirS genes. The prime goal of this study was to compare the responses of the nirK-type and nirS-type denitrifying bacteria to an U. prolifera green tide in coastal Qingdao areas. Furthermore, the possible critical physicochemical factors regulating variations of the nirK-type and nirS-type denitrifying bacterial communities during the green tide were also discussed in the present study.
Materials and methods
Sample collection and analysis of physicochemical factor
An U. prolifera green tide affected the coastal areas of the Yellow Sea from late April to early September 2019, according to Bulletin of China Marine Disaster, with a maximum distribution area and coverage area of 55699 km2 and 508 km2, respectively. The green tide arrived in the coastal areas of the Shandong Peninsula in late May, began to decline around the end of July, and vanished in early September. In comparison to the previous green tide events, the green tide in 2019 vanished at the latest and had the second largest distribution areas, only second to the distribution areas of the green tide in 2016.
Three sampling stations in coastal Qingdao areas were chosen, and named XG (36.07°E, 120.30°N), ZQ (36.06°E, 120.31°N) and MD (36.05°E, 120.42°N) (Figure 1). Among the sampling stations, ZQ and MD were significantly influenced by the green tide, whereas XG was less likely to be influenced. Samples were taken between June and September 2019 (ten sampling time points). Twenty liters of seawater were filtered through an 800-mesh plankton net and a 0.22-μm pore-size polycarbonate membrane (Millipore, 47 mm in diameter), then the filters were stored at -80°C until DNA extraction. We also collected seawater samples to determine physicochemical factors, including temperature, salinity, dissolved oxygen (DO) content, pH, chlorophyll a (chla) content, nitrate (NO3-) concentration, nitrite (NO2-) concentration, ammonium (NH4+) concentration and phosphate (PO43-) concentration. The measurement methods of physicochemical factors have been previously described (Zhao et al., 2022), and results were displayed in Supplementary Figure 1. All samples were separated into two phases based on the chla content: the outbreak phase (samples collected in 13 June, 27 June, 11 July, 18 July and 22 July) and the decline phase (samples collected in 6 August, 16 August, 23 August, 29 August and 5 September).
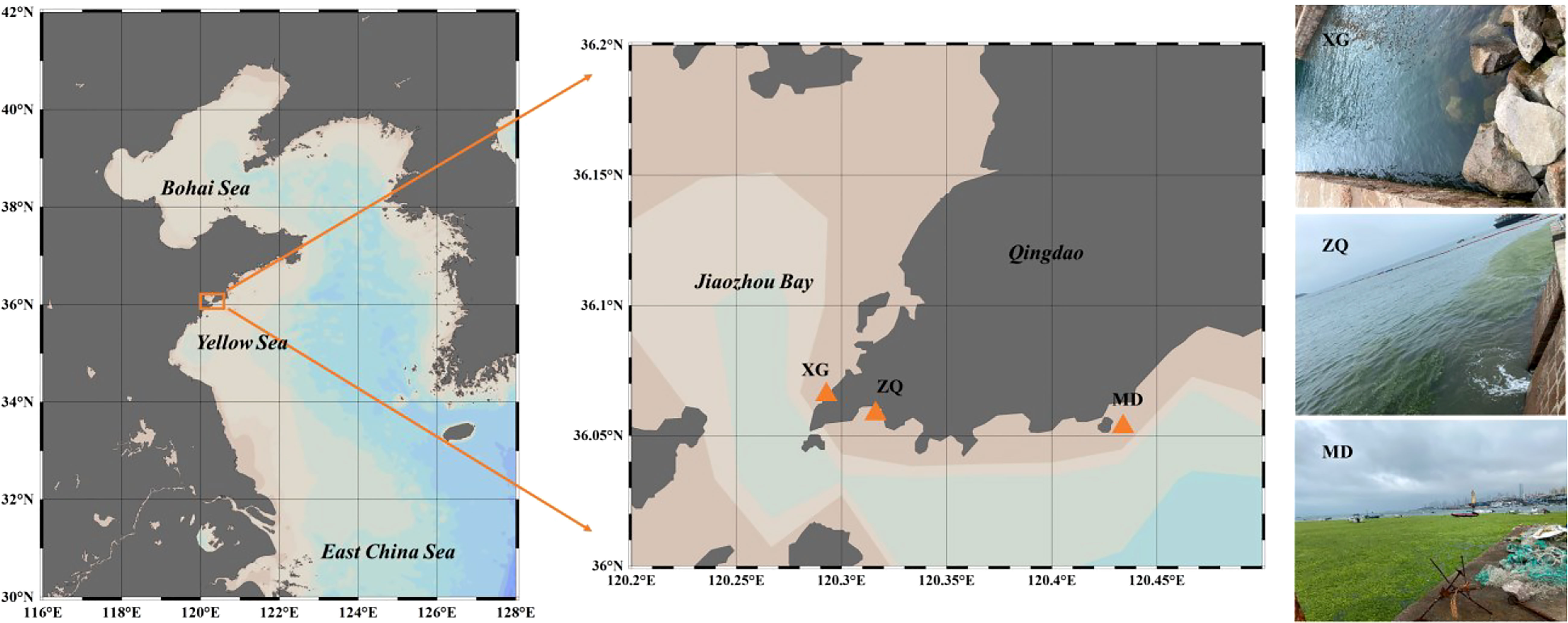
Figure 1 Locations of sampling stations chosen in coastal Qingdao areas during an U. prolifera green tide, and the right panel was the photos captured on 11 July, 2019 at three sampling stations.
DNA extraction and quantitative PCR analysis
A FastDNA Spin kit for Soil (MP Biomedicals, OH, USA) was used to extract the genomic DNA. After being checked with agarose gel electrophoresis and spectrophotometer analysis, the eligible DNA was utilized to investigate the abundance of the nirK-type and nirS-type denitrifying bacterial communities during the green tide. The nirK and nirS genes were amplified separately using the primer pairs F1aCu (5’-ATC ATG GTS CTG CCG CG) and R3Cu (5’-GCC TCG ATC AGR TTG TGG TT), cd3aF (5’-GTS AAC GTS AAG GAR ACS GG) and R3cd (5’-GAS TTC GGR TGS GTC TTG A) (Hallin and Lindgren, 1999; Throbäck et al., 2004). The amplification started with a 10-minute activation at 95°C, followed by 40 cycles of 30 seconds at 95°C, 30 seconds at 55°C for the nirK gene (or 30 seconds at 53°C for the nirS gene) and 45 seconds at 72°C. The melting stage was added after amplification. The standard curve was generated by the quantification of the serially diluted standard plasmids containing the nirK or nirS gene. Each reaction was comprised of 10 μL FastStart Universal SYBR Green Master (ROX) (Roche, Germany), 0.3 μM forward primer, 0.3 μM reverse primer, 0.2 μg/μL bovine serum albumin (BSA) and 2.0 μL standard plasmid. All qPCR reactions were run in triplicate using SYBR Green I method on an ABI PRISM 7500 Sequence Detection System (Applied Biosystems, CA). The abundance of the nirK and nirS genes was examined under the above-mentioned procedures, and each reaction contained standard plasmids and negative controls to confirm an uncontaminated and stable amplification reaction.
Illumina Miseq sequencing analysis and sequence processing
The variations of the nirK-type and nirS-type denitrifying bacterial communities during the green tide were investigated by Illumina Miseq sequencing approach. The nirK and nirS genes were amplified with the specific primer pairs F1aCu/R3Cu and cd3aF/R3cd, respectively. Then, the amplicons were sequenced on an Illumina Miseq platform (Majorbio, Shanghai, China) after visualization, purification and quantification. Raw reads were submitted to the Sequence Read Archive (SRA) database of the National Center for Biotechnology Information (NCBI) under the accession numbers PRJNA781861 (nirK gene) and PRJNA781658 (nirS gene).
Raw reads were merged with Fast Length Adjustment of Short reads software (FLASH, version 1.2.11), and reads with overlap shorter than 10 bp and mismatch ratio less than 0.2 were removed (Magoč and Salzberg, 2011). Quantitative Insights into Microbial Ecology (QIIME, version 1.9.1) was used to screen the merged sequences (Caporaso et al., 2010). Operational taxonomic units (OTUs) were clustered using UPARSE software (version 7.0.1090) with a 97% sequence similarity cutoff, and chimeric sequences were examined and removed at the same time (Edgar, 2013). The most abundant sequence in each OTU was chosen as representative sequence, and then assigned by RDP classifier against the NCBI nr/nt database (ftp://ftp.ncbi.nih.gov/blast/db) to acquire the taxonomic information with a confidence level greater than 70%.
Statistical analysis
QIIME was used to calculate the Alpha-diversity of the nirK-type and nirS-type denitrifying bacterial communities, including Chao1 richness estimator, Shannon index and Good’s coverage. The differences in the denitrifying bacterial community structure during the green tide were investigated with principal components analysis (PCA). Correlations of abundance, richness, diversity and relative abundance of dominant taxa with physicochemical factors was estimated with SPSS software (version 26). After variance inflation factor (VIF) analysis, distance-based redundancy analysis (db-RDA) was utilized to assess the influence of physicochemical factors on the denitrifying bacterial communities during the green tide. Variations of the denitrifying bacterial communities during the green tide, including abundance, richness, diversity and relative abundance of dominant taxa, were evaluated with one-way analysis of variance (ANOVA).
Six categories of co-occurrence network were established in this study, including (i) the denitrifying bacterial networks during the green tide, the outbreak phase and the decline phase; (ii) the denitrifying bacterial networks at station XG, ZQ and MD; (iii) the nirK-type denitrifying bacterial networks during the green tide, the outbreak phase and the decline phase; (iv) the nirK-type denitrifying bacterial networks at station XG, ZQ and MD; (v) the nirS-type denitrifying bacterial networks during the green tide, the outbreak phase and the decline phase; (vi) the nirS-type denitrifying bacterial networks at station XG, ZQ and MD. Only OTUs that occur in more than 25% of all samples were displayed. The OTUs that were statistically significant (P < 0.01 and Q < 0.05) and had a spearman’s coefficient greater than |0.6| were subjected to further analysis using “fdrtool” and “igraph” packages of the R statistical software (version 4.0.4). The visualization of co-occurrence networks and the calculation of topological properties were generated with Gephi (version 0.9.2).
Results
Alpha-diversity of denitrifying bacterial communities
In total, we obtained 550946 and 641853 high-quality nirK and nirS gene sequences with average lengths of 425 bp and 371 bp, respectively. With a 97% sequence similarity cutoff, the high-quality sequences were clustered into 359 nirK-type denitrifying bacterial OTUs and 491 nirS-type denitrifying bacterial OTUs. Good’s coverage of the nirK-type and nirS-type denitrifying bacterial communities across all samples was greater than 99.29%, indicating that this study covered the majority of nirK-type and nirS-type denitrifying bacteria in our studied areas.
Alpha-diversity of the nirK-type and nirS-type denitrifying bacterial communities was shown in Table 1. During the green tide, the Chao1 richness estimator of the nirK-type and nirS-type denitrifying bacterial communities ranged from 72.00 to 258.00 and 71.81 to 351.63, while the corresponding Shannon index ranged from 1.25 to 3.97 and 0.28 to 3.91, respectively. Overall, the richness of the nirS-type denitrifying bacterial community was significantly greater than that of the nirK-type denitrifying bacterial community during the green tide (P < 0.01); for the Shannon index, the average value of the nirK-type denitrifying bacterial community was greater, but not significantly (P > 0.05).
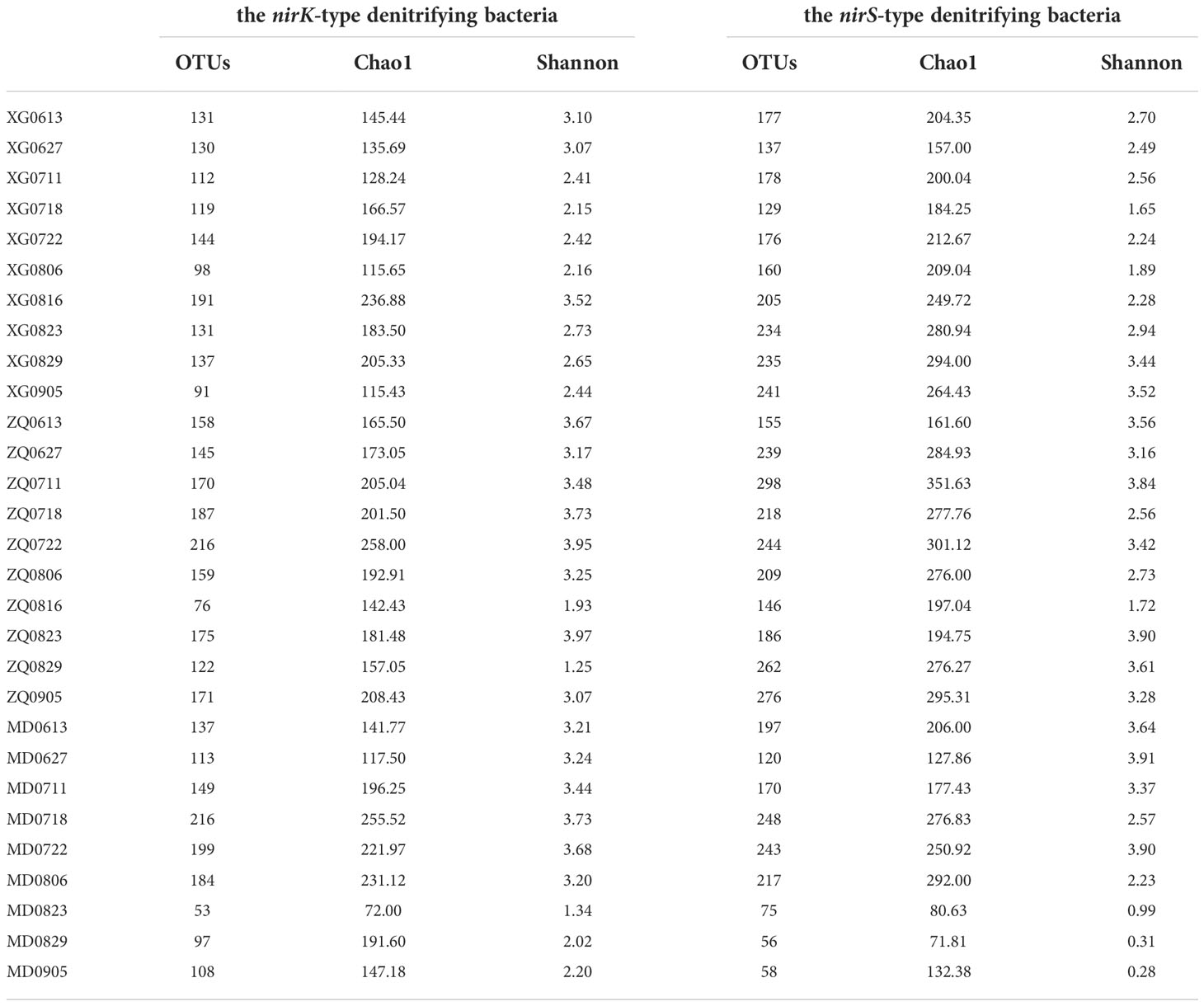
Table 1 Alpha-diversity of the nirK-type and nirS-type denitrifying bacterial communities during the green tide in Qingdao coastal areas.
The Chao1 richness of the nirK-type and nirS-type denitrifying bacterial communities varied from 117.50 to 258.00 and 127.86 to 351.63 during the outbreak phase, and from 72.00 to 236.88 and 71.81 to 295.31 during the decline phase, respectively. During the outbreak phase, the Shannon diversity of the nirK-type and nirS-type denitrifying bacterial communities separately varied from 2.15 to 3.95 and 1.65 to 3.91, while during the decline phase, it ranged from 1.25 to 3.97 and 0.28 to 3.90, respectively. The Chao1 richness of the nirK-type and nirS-type denitrifying bacteria exhibited comparable tendencies during the green tide, with a greater community richness during the outbreak phase. Similar trends have also been observed on variations in community diversity of the nirK-type and nirS-type denitrifying bacteria. This finding revealed that the outbreak phase may be beneficial to increase the community richness and diversity of denitrifying bacteria.
The Chao1 richness of the nirK-type denitrifying bacterial community separately ranged from 115.43 to 236.88 at station XG, 142.43 to 258.00 at station ZQ, and 72.00 to 255.52 at station MD. The Shannon diversity of the nirK-type denitrifying bacterial community varied from 2.15 to 3.52 at station XG, 1.25 to 3.97 at station ZQ, and 1.34 to 3.73 at station MD, respectively. For the nirS-type denitrifying bacteria, the community richness separately ranged from 157.00 to 294.00 at station XG, 161.60 to 351.63 at station ZQ, and 71.81 to 292.00 at station MD; and the community diversity varied from 1.65 to 3.52 at station XG, 1.72 to 3.90 at station ZQ, and 0.28 to 3.91 at station MD, respectively. During the green tide, the nirK-type denitrifying bacteria had the lowest community Alpha-diversity at station XG and the highest at station ZQ, while the richness and diversity of the nirS-type denitrifying bacterial community exhibited the lowest average value at station MD and the highest average value at station ZQ. ANOVA analysis revealed that except for the community richness of the nirS-type denitrifying bacteria (P < 0.05), the community Alpha-diversity of the nirK-type denitrifying bacteria and the community diversity of the nirS-type denitrifying bacteria did not differ significantly among sampling stations (P > 0.05).
Composition and structure of denitrifying bacterial communities
Proteobacteria were the dominant phylum of the nirK-type and nirS-type denitrifying bacterial communities. Alphaproteobacteria and Gammaproteobacteria accounted for 59.28% and 34.98% of the total sequences in Proteobacteria for the nirK-type and nirS-type denitrifying bacteria, respectively. The compositions of the nirK-type and nirS-type denitrifying bacterial communities were studied at the family level in this study. The nirK-type denitrifying bacterial community was dominated by unclassified bacteria (9.28–90.55%), Rhodobacteraceae (0.99–46.69%), unclassified Proteobacteria (0.20–81.38%), unclassified Alphaproteobacteria (0.14–35.77%), Pseudomonadaceae (0.00–28.16%), Bradyrhizobiaceae (0.00–1.44%) and unclassified Rhizobiales (0.00–1.81%) during the green tide (Figure 2A). Rhodobacteraceae (P < 0.01) and unclassified Alphaproteobacteria (P > 0.05) exhibited greater relative abundance during the outbreak phase, whereas unclassified bacteria (P > 0.05), unclassified Proteobacteria (P < 0.05) and Pseudomonadaceae (P > 0.05) were more abundant during the decline phase. Sampling stations may also have an effect on the nirK-type denitrifying bacterial community, and markedly influenced the relative abundance of unclassified bacteria (P < 0.05) and unclassified Alphaproteobacteria (P < 0.01) during the green tide.
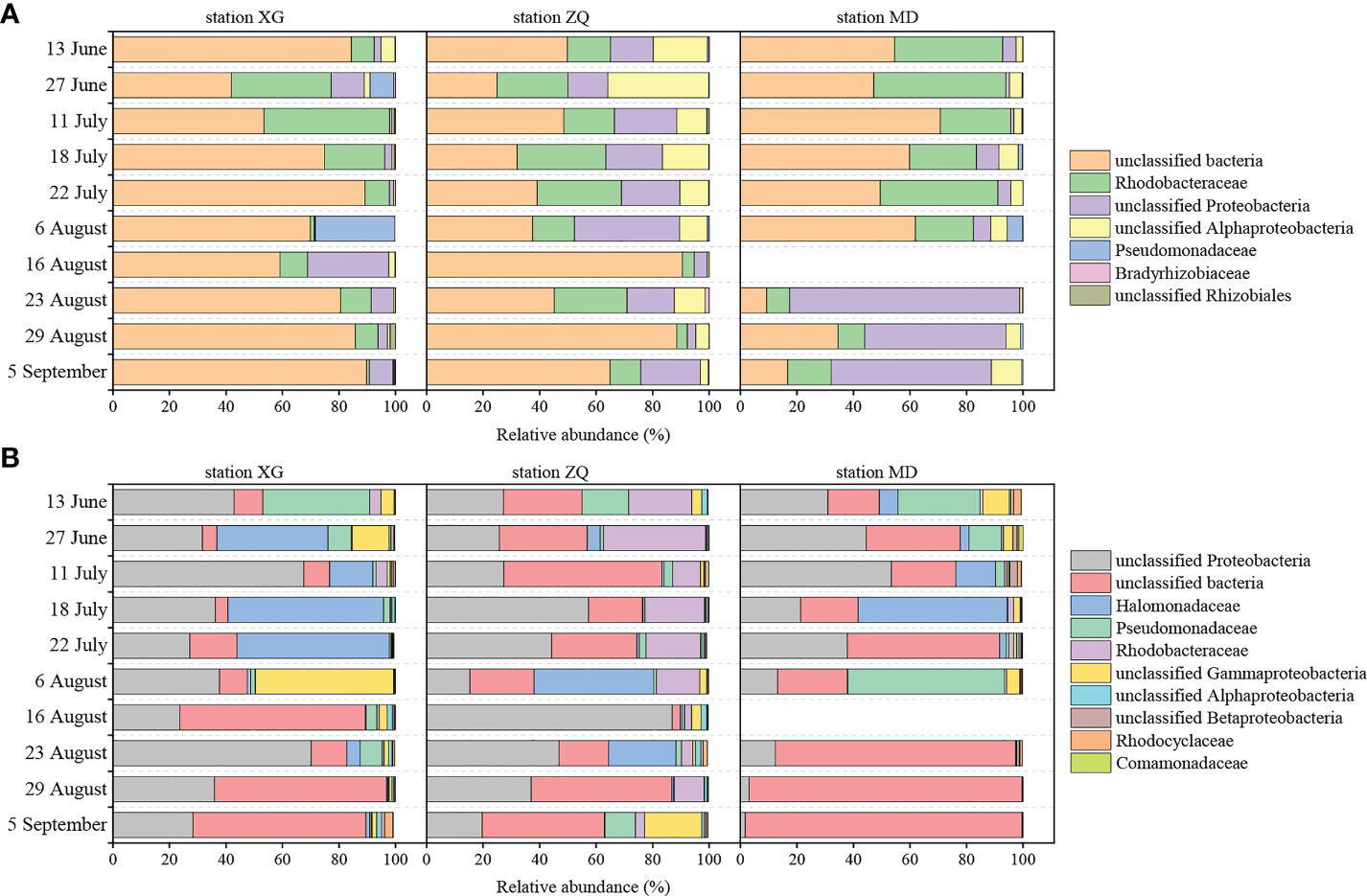
Figure 2 Relative abundance of the dominant nirK-type (A) and nirS-type (B) denitrifying bacteria (relative abundance greater than 1%) at the family level during the green tide in coastal Qingdao areas. Due to the failure of the nirK and nirS gene amplicon library construction on 16 August at station MD, the relevant community information was shown as blanks in the Figure.
The dominant nirS-type denitrifying bacterial community was comprised of unclassified Proteobacteria (1.68–86.92%), unclassified bacteria (2.89–97.86%), Halomonadaceae (0.00–55.00%), Pseudomonadaceae (0.02–55.41%), Rhodobacteraceae (0.08–36.06%) and unclassified Gammaproteobacteria (0.01–48.92%), and to a lesser extent, unclassified Alphaproteobacteria (0.00–2.04%), unclassified Betaproteobacteria (0.01–2.66%), Rhodocyclaceae (0.00–2.87%) and Comamonadaceae (0.00–1.46%, Figure 2B). Greater proportions of unclassified Proteobacteria, Halomonadaceae, Pseudomonadaceae, Rhodobacteraceae, unclassified Betaproteobacteria and Comamonadaceae were observed during the outbreak phase; in contrast, unclassified bacteria, unclassified Gammaproteobacteria and unclassified Alphaproteobacteria displayed greater average relative abundance during the decline phase. Only the relative abundance of unclassified bacteria differed significantly between the outbreak phase and the decline phase (P < 0.05). The sampling stations had a significant impact on the relative abundance of Rhodobacteraceae (P < 0.01) and unclassified Alphaproteobacteria (P < 0.05).
PCA was employed to examine the differences in denitrifying bacterial community structure during the green tide at the OTU level (Figure 3). Bloom phases had considerable impacts on both the nirK-type (P < 0.01, R = 0.32) and nirS-type (P < 0.01, R = 0.16) denitrifying bacterial communities, with the nirK-type denitrifying bacterial community having the most impact. Furthermore, sampling stations may influence both the nirK-type (P < 0.05, R = 0.28) and nirS-type (P < 0.05, R = 0.25) denitrifying bacterial communities, notably the nirK-type denitrifying bacterial community. Thus, it is possible to conclude that the nirK-type denitrifying bacteria may be more sensitive to the green tide.
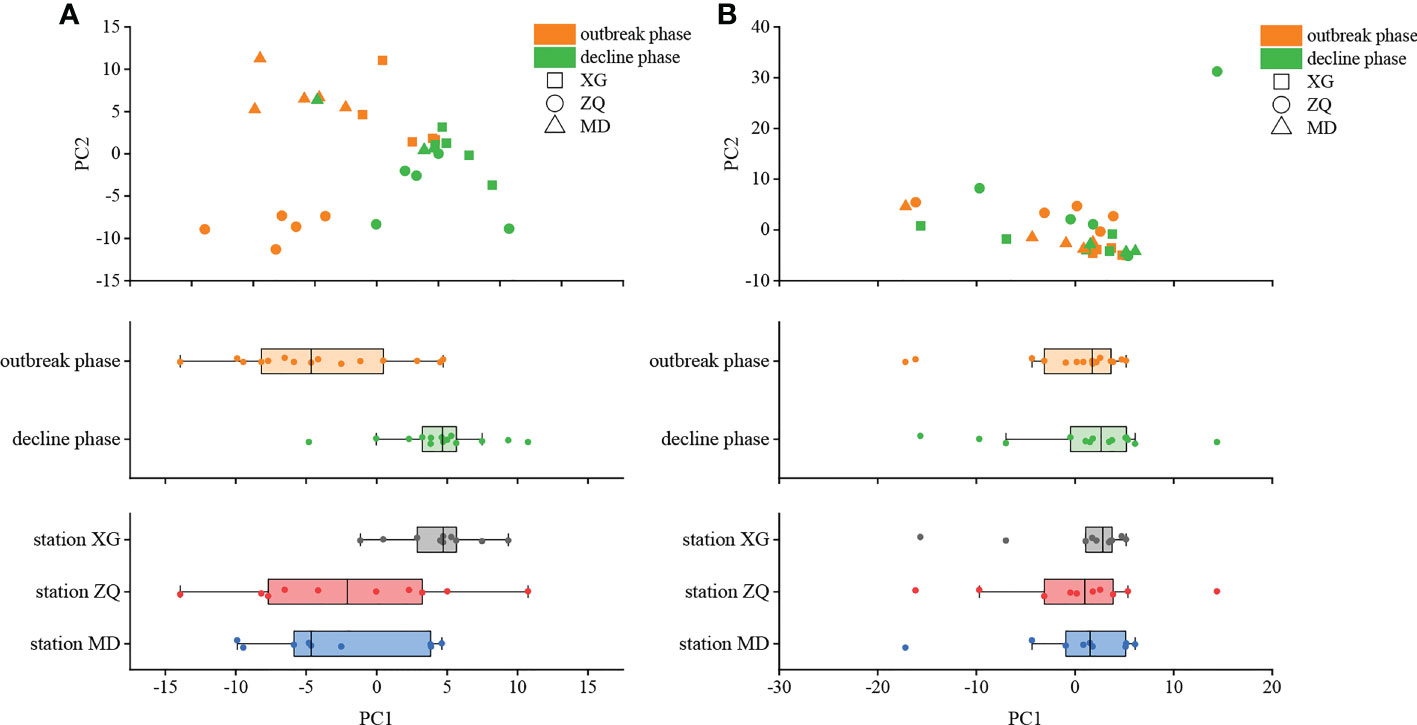
Figure 3 Principal component analysis (PCA) of the nirK-type (A) and nirS-type (B) denitrifying bacterial communities during the green tide in coastal Qingdao areas.
Abundance of denitrifying bacterial communities
The abundance of the nirK and nirS genes ranged from 1.72×103 to 1.18×106 copies/L and 2.31×103 to 2.19×106 copies/L during the green tide, respectively (Figure 4). The nirK gene was more abundant during the outbreak phase than the decline phase, while the nirS gene exhibited greater abundance during the decline phase, according to our data, but ANOVA analysis revealed no significant difference in the abundance of the nirK and nirS genes between the outbreak phase and the decline phase (P > 0.05). Meanwhile, we discovered that the abundance of the nirK and nirS genes followed a similar pattern across sampling stations, with lower abundance at station XG which was less likely to be influenced by the green tide, and greater abundance at station ZQ and MD which were significantly affected by the green tide. ANOVA analysis revealed that there were no obvious differences in the abundance of the nirK and nirS genes among sampling stations (P > 0.05).
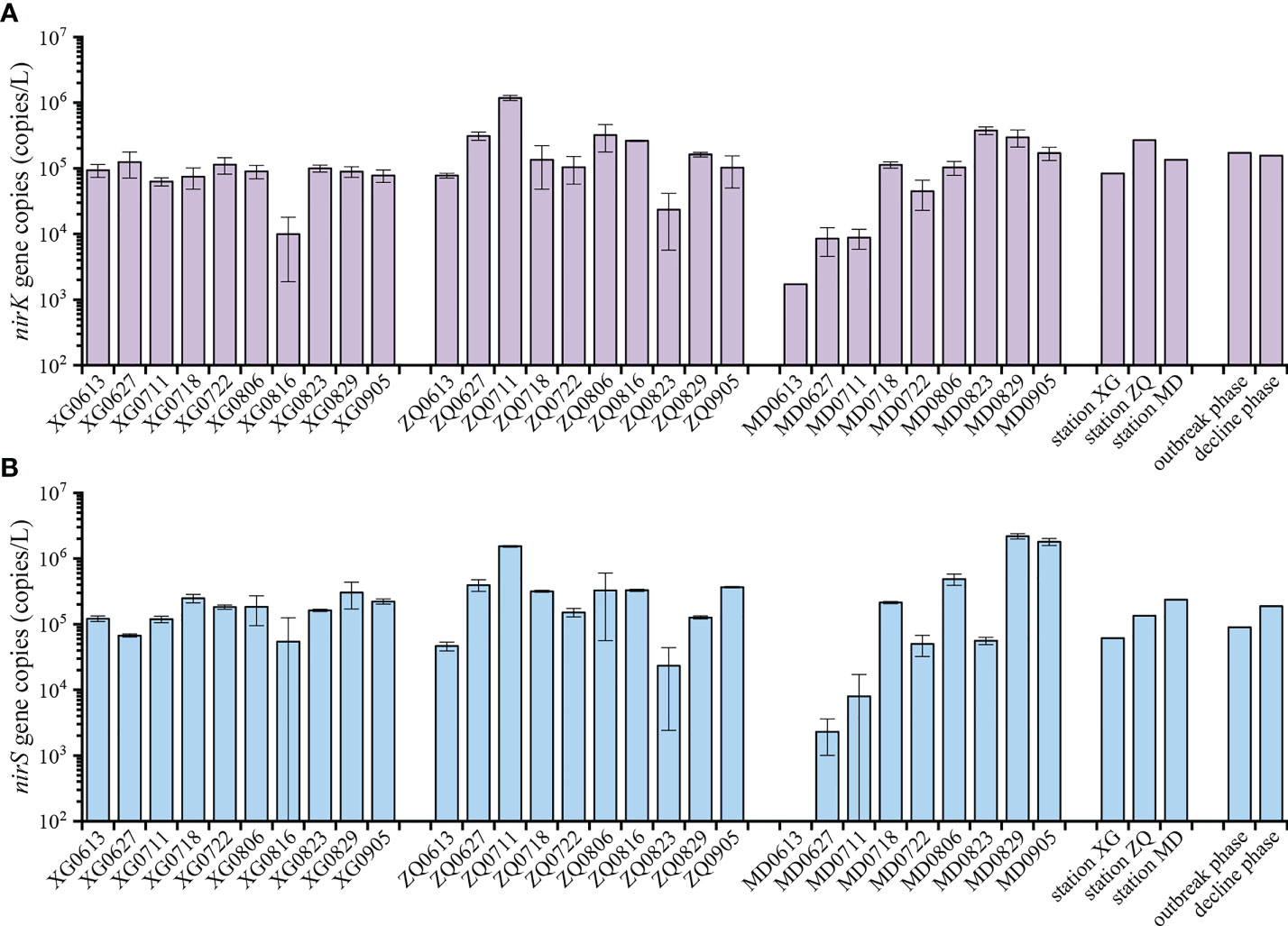
Figure 4 Quantitative analysis of the nirK (A) and nirS (B) genes during the green tide in coastal Qingdao areas.
Links between the denitrifying bacterial communities and physicochemical factors
Distance-based redundancy analysis (db-RDA) was used to illustrate the links between physicochemical factors and the denitrifying bacterial communities at the OTU level. The nirK-type denitrifying bacterial community structure was strongly related to chla content (R2 = 0.37, P < 0.01), salinity (R2 = 0.30, P < 0.05) and DO content (R2 = 0.25, P < 0.05), and the nirS-type denitrifying bacterial community structure was likewise clearly associated with salinity (R2 = 0.32, P < 0.05), DO content (R2 = 0.31, P < 0.05) and chla content (R2 = 0.28, P < 0.05) (Figure 5). Chla content is an important indicator of phytoplankton biomass and eutrophication level. According to the results of Spearman correlation analysis, chla content affected the denitrifying bacterial communities mostly through impacting the relative abundance of dominant taxa. For the nirK-type denitrifying bacterial community, chla content had a substantial effect on the relative abundance of unclassified Alphaproteobacteria (R = 0.512, P < 0.05) during the green tide (Figure 6; Supplementary Figure 2). In regard to the nirS-type denitrifying bacterial community, chla content correlated significantly with the relative abundance of Rhodobacteraceae (R = 0.532, P < 0.05) and unclassified Gammaproteobacteria (R = -0.556, P < 0.05) during the green tide, and was clearly related to the relative abundance of unclassified bacteria (R = 0.881, P < 0.01) and unclassified Gammaproteobacteria (R = -0.790, P < 0.05) during the outbreak phase (Figure 6; Supplementary Figure 3). Furthermore, chla content may influence the Alpha-diversity of the denitrifying bacterial communities. In the current study, however, only the Chao1 richness of the nirK-type (R = 0.905, P < 0.01) and nirS-type (R = 0.857, P < 0.01) denitrifying bacterial communities during the outbreak phase was clearly linked with chla content.
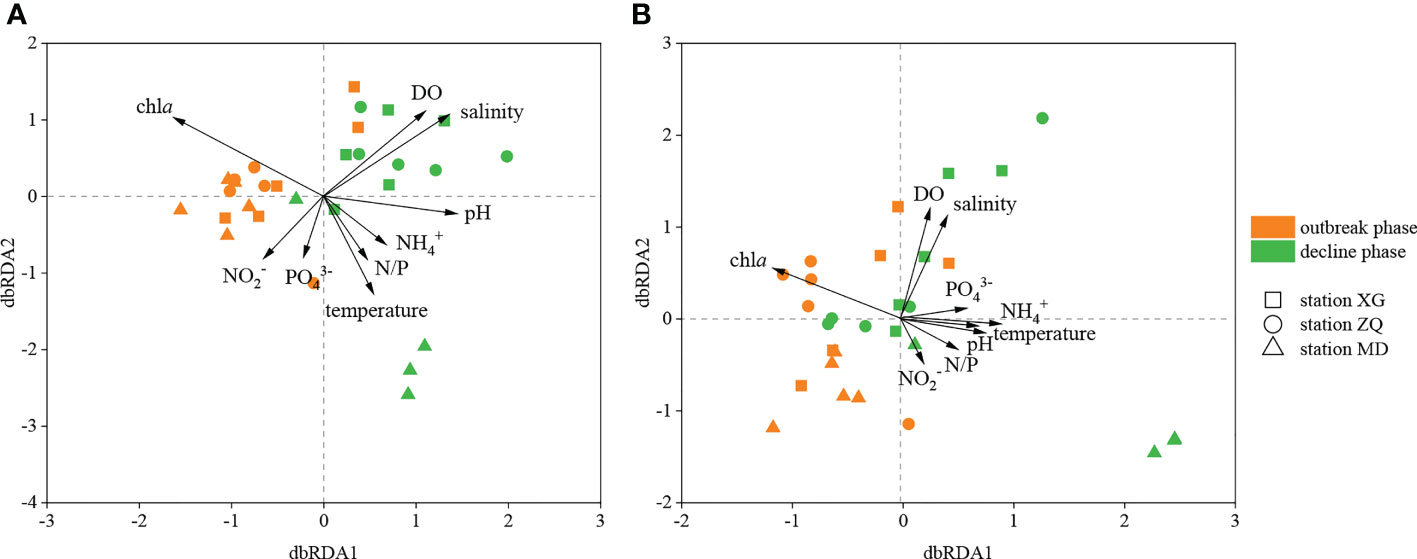
Figure 5 db-RDA analysis of the nirK-type (A) and nirS-type (B) denitrifying bacterial communities with physicochemical factors during the green tide in coastal Qingdao areas.
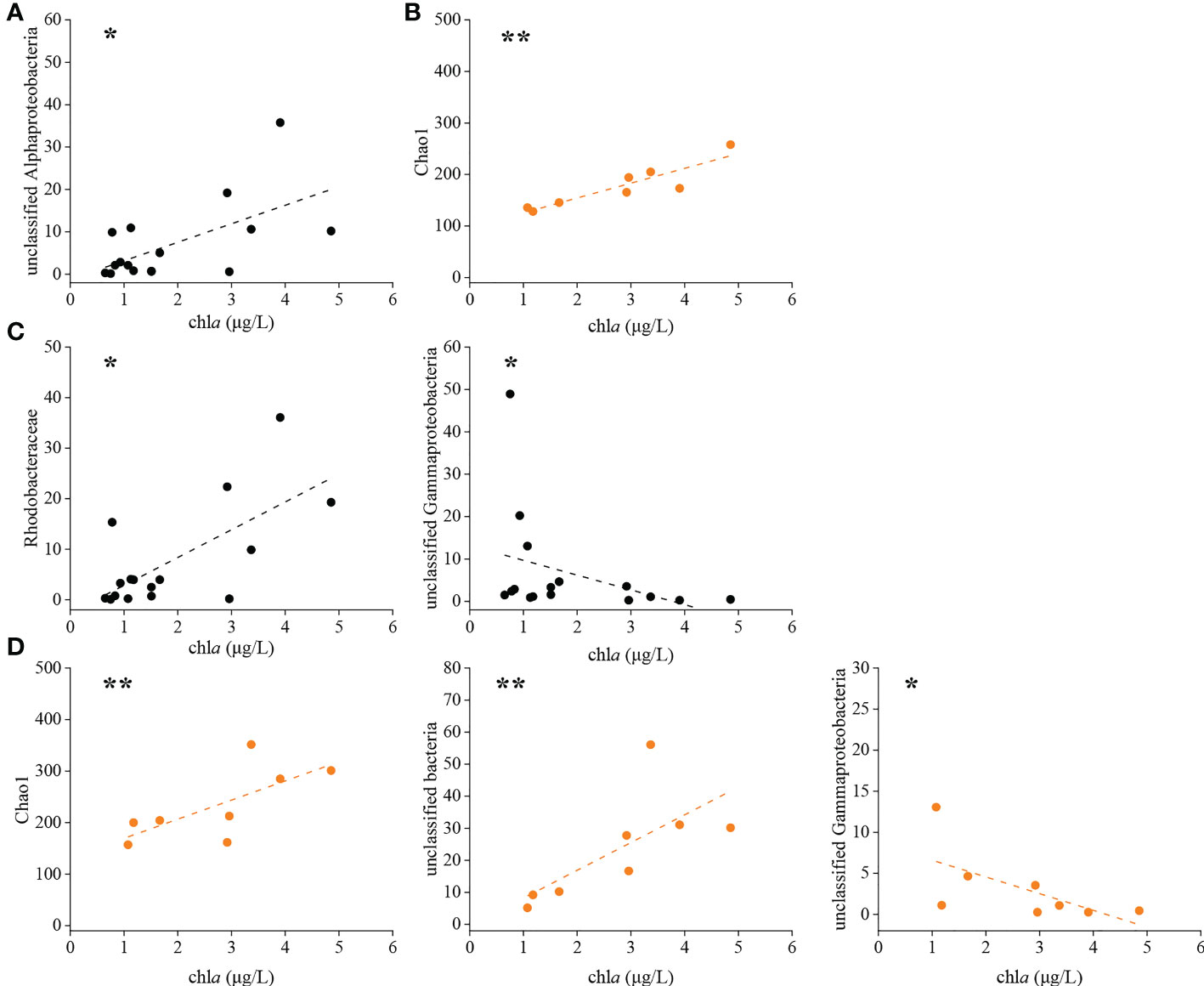
Figure 6 Significant relationships between chla content and the nirK-type denitrifying bacterial community during the green tide (A) and the outbreak phase (B), and the nirS-type denitrifying bacterial community during the green tide (C) and the outbreak phase (D). *P < 0.05, **P < 0.01.
In addition, Spearman correlation analysis was also conducted to explore the associations of abundance, Alpha-diversity and relative abundance of dominant taxa with other physicochemical factors (Supplementary Figure 4). For the nirK-type denitrifying bacterial community during the green tide, Rhodobacteraceae was clearly correlated with temperature (R = -0.57, P < 0.01), NO2- concentration (R = -0.56, P < 0.01), NO3- concentration (R = -0.52, P < 0.01) and NOx- concentration (R = -0.52, P < 0.01); unclassified Proteobacteria and unclassified Alphaproteobacteria were positively related to DIN concentration (R = 0.41, P < 0.05) and salinity (R = 0.43, P < 0.05), respectively; unclassified Rhizobiales was negatively correlated with temperature (R = -0.44, P < 0.05). During the outbreak phase, temperature (R = 0.76, P < 0.01) was clearly associated with the Chao1 richness of the nirK-type denitrifying bacterial community, and the abundance of the nirK gene was negatively related to DO content (R = -0.71, P < 0.01). During the decline phase, the abundance of the nirK gene was clearly associated with NO2- concentration (R = -0.66, P < 0.01), NO3- concentration (R = -0.55, P < 0.05) and NOx- concentration (R = -0.55, P < 0.05); Pseudomonadaceae, Bradyrhizobiaceae and unclassified Rhizobiales were the major nirK-type denitrifying bacteria, and were strongly related to pH (R = -0.56, P < 0.05), NO2- concentration (R = 0.56, P < 0.05) and salinity (R = -0.69, P < 0.01), respectively. In regard to the nirS-type denitrifying bacterial community during the green tide, Shannon diversity was related to pH (R = 0.53, P < 0.01); Pseudomonadaceae (R = -0.52, P < 0.01), unclassified Gammaproteobacteria (R = -0.37, P < 0.05) and unclassified Betaproteobacteria (R = -0.37, P < 0.05) were negatively related to temperature; and Rhodobacteraceae was strongly related to salinity (R = 0.51, P < 0.01), NO2- concentration (R = -0.43, P < 0.05), NO3- concentration (R = -0.43, P < 0.05) and NOx- concentration (R = -0.43, P < 0.05). Numerous physicochemical factors had discernible effects on the abundance, Alpha-diversity and dominant taxon of the nirS-type denitrifying bacterial community during the outbreak phase, for example, temperature (R = 0.59, P < 0.05) had a significant impact on the Chao1 richness, whereas pH (R = 0.64, P < 0.05), DO content (R = 0.58, P < 0.05) and NO2- concentration (R = -0.56, P < 0.05) were critical to the Shannon diversity; Pseudomonadaceae was significantly related to temperature (R = -0.73, P < 0.01), salinity (R = 0.78, P < 0.01) and pH (R = 0.60, P < 0.05); Rhodocyclaceae was markedly connected to NO2- concentration (R = -0.78, P < 0.01), NO3- concentration (R = -0.72, P < 0.01) and NOx- concentration (R = -0.73, P < 0.01). During the decline phase, unclassified Proteobacteria and unclassified Gammaproteobacteria were respectively shown to be negatively related to NH4+ concentration (R = -0.58, P < 0.05) and temperature (R = -0.54, P < 0.05), whereas unclassified bacteria was found to be significantly related to NH4+ concentration (R = 0.58, P < 0.05) and DIN concentration (R = 0.58, P < 0.05).
Co-occurrence network analysis of denitrifying bacteria
Networks were constructed to explore the co-occurrence patterns of denitrifying bacteria during the green tide, the outbreak phase and the decline phase. The majority of correlations in the networks were positive, indicating that mutualism may be the most important interaction among denitrifying bacteria during the green tide, as well as the outbreak phase and the decline phase (Figure 7). Meanwhile, we discovered that the nodes and edges in network of the outbreak phase (450 and 1702) were greater than those in network of the decline phase (436 and 1221), indicating a more complex denitrifying bacterial co-occurrence network during the outbreak phase (Figures 7B, C). The modularity of network was 0.517 and 0.710 during the outbreak phase and the decline phase, implying a more modular denitrifying bacterial network during the decline phase. Thus, a more complex and less modular denitrifying bacterial network was observed during the outbreak phase. Moreover, the nodes with greater relative abundance in each module were specific to a particular phase, for example, the majority of OTUs in module 6 had greater relative abundance during the outbreak phase, whereas most of OTUs in module 1 exhibited relatively greater abundance during the decline phase (Figure 7A).
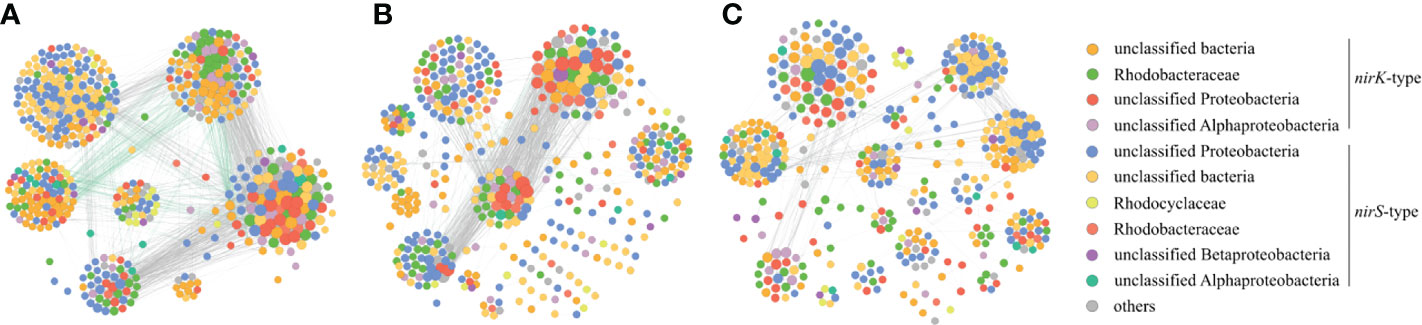
Figure 7 Co-occurrence networks of the denitrifying bacteria during the green tide (A), the outbreak phase (B) and the decline phase (C) in coastal Qingdao areas.
The nirK-type denitrifying bacterial networks during the outbreak phase and the decline phase were separately built to discern the differences in microbial interactions between bloom phases, and the same analysis was also performed on the nirS-denitrifying bacteria in the current study. For the nirK-type denitrifying bacteria, the nodes and edges in network of the outbreak phase (208 and 797) were greater than those in network of the decline phase (169 and 371), indicating that the nirK-type denitrifying bacterial network was more complex during the outbreak phase (Table 2; Figure 8). The modularity of the network was 0.524 and 0.682 during the outbreak phase and the decline phase, respectively, implying that a more modular nirK-type denitrifying bacterial network was observed during the decline phase. In comparison to the outbreak phase, the network of the decline phase had lower average degree (5.549 versus 3.643), lower graph density (0.037 versus 0.026), lower clustering coefficient (0.528 versus 0.440) and longer path length (4.771 versus 5.555), suggesting a more associated nirK-type denitrifying bacterial network during the outbreak phase. As a result, the nirK-type denitrifying bacterial community may be more complex, more associated and less modular during the outbreak phase than the decline phase.
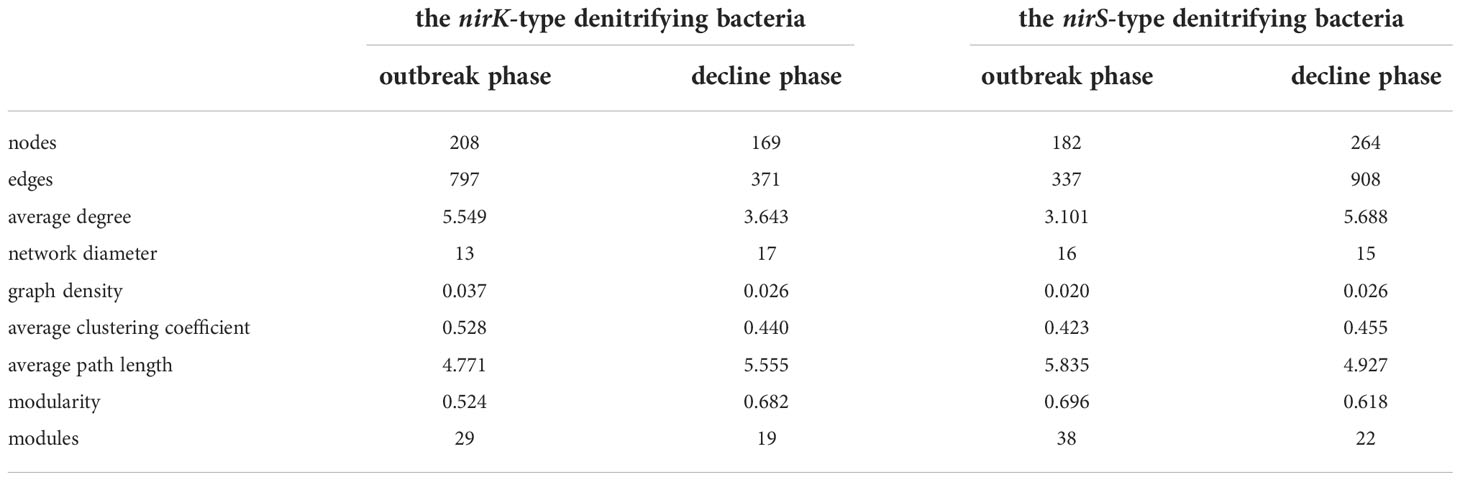
Table 2 Topological properties of the denitrifying bacterial co-occurrence networks during the green tide in coastal Qingdao areas.
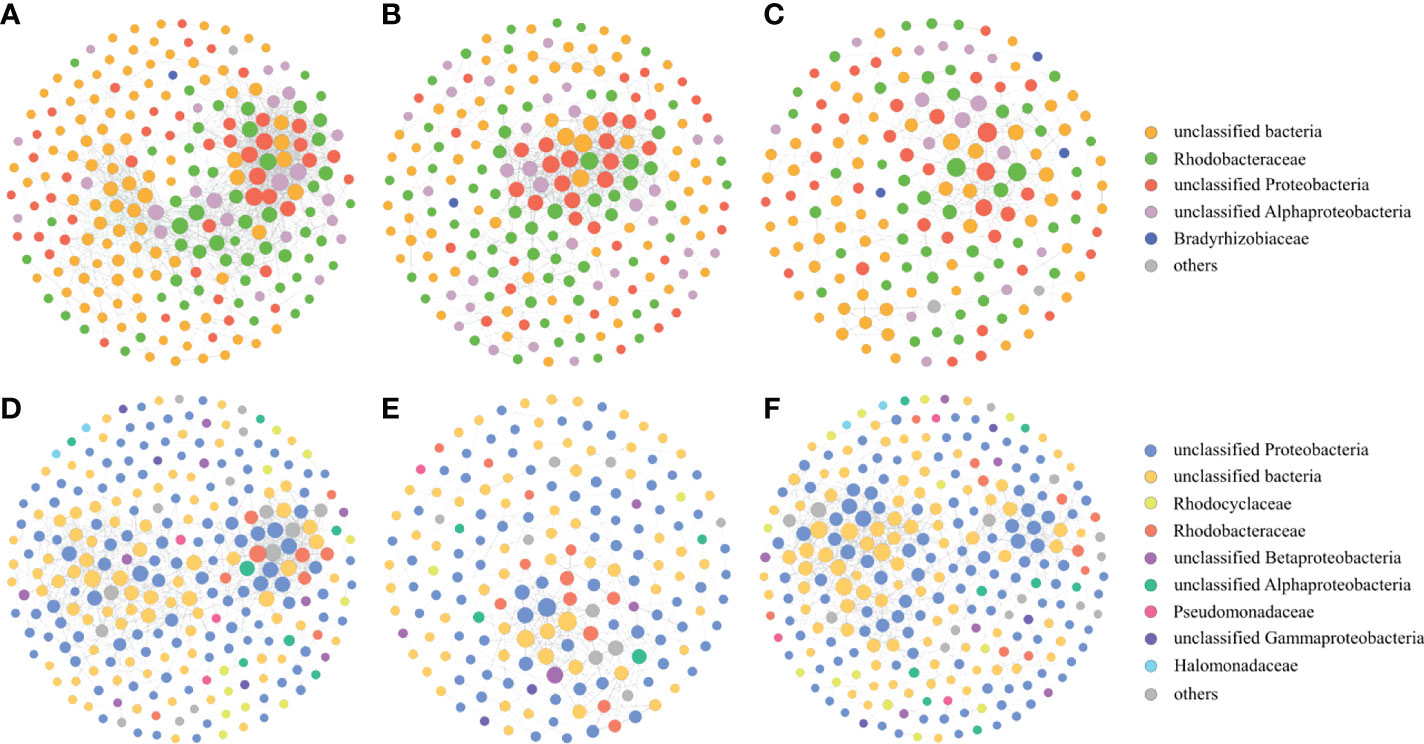
Figure 8 Co-occurrence networks of the nirK-type denitrifying bacteria during the green tide (A), the outbreak phase (B) and the decline phase (C), and co-occurrence networks of the nirS-type denitrifying bacteria during the green tide (D), the outbreak phase (E) and the decline phase (F).
The nirS-type denitrifying bacterial network contained 182 nodes and 337 edges during the outbreak phase, and 264 nodes and 908 edges during the decline phase (Table 2; Figure 8). In comparison to the decline phase, the network of the nirS-type denitrifying bacteria exhibited lower average degree (3.101 versus 5.688), longer path length (5.835 versus 4.929), lower graph density (0.020 versus 0.026), lower clustering coefficient (0.423 versus 0.455) and greater modularity (0.683 versus 0.618) during the outbreak phase. All of these characters manifested that the nirS-type denitrifying bacterial community was less complex, less associated and more modular during the outbreak phase than the decline phase. In comparison to the nirK-type denitrifying bacterial network, the nirS-type denitrifying bacterial network had more nodes (224 versus 261), more edges (1016 versus 1454), lower average degree (7.679 versus 5.049), lower clustering coefficient (0.514 versus 0.386) and longer path length (3.513 versus 3.687), indicating that nirS-type denitrifying bacterial network was more complex and associated than the nirK-type denitrifying bacterial network during the green tide. Taken together, it is hypothesized that the nirK-type and nirS-type denitrifying bacteria behave differently to the U. prolifera green tide, and may occupy different niches during the green tide in coastal Qingdao areas.
In addition, we also constructed the denitrifying bacterial networks, the nirK-type denitrifying bacterial networks and the nirS-type denitrifying bacterial networks at three sampling stations, respectively. The nodes and edges of the denitrifying bacterial network at station XG were less than those at two other stations, and the proportions of negative links at station XG were also less than those at two other stations, indicating that the denitrifying bacterial network was more complex and stable at station ZQ and MD which were greatly impacted by the green tide (Figures 9A–C). Both the nirK-type denitrifying bacterial networks and nirS-type denitrifying bacterial networks displayed a similar trend across sampling stations, with greater complexity and stability networks at station ZQ and MD (Figures 9D–I).
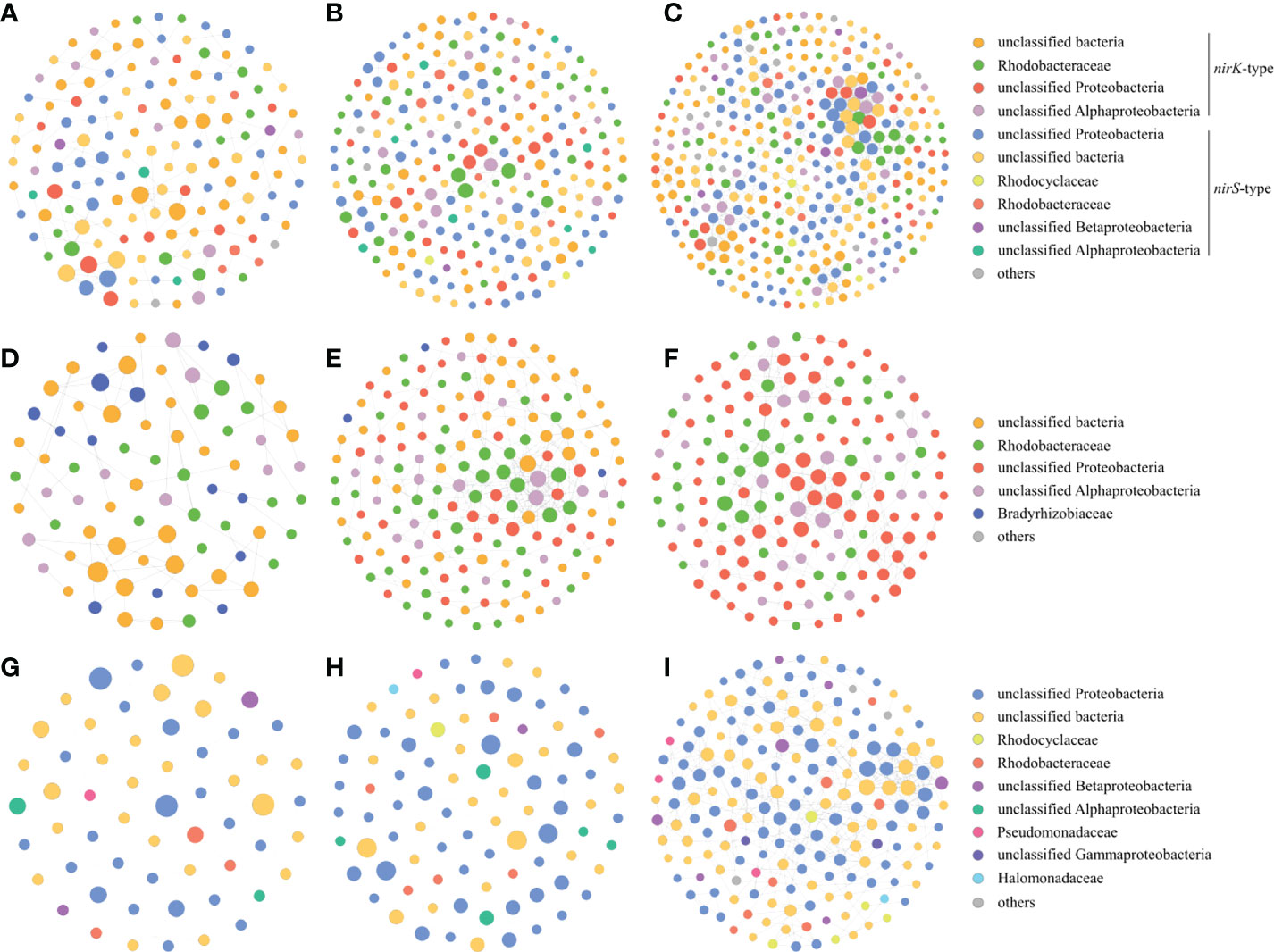
Figure 9 Co-occurrence networks of the denitrifying bacteria during the green tide (A), the outbreak phase (B) and the decline phase (C), co-occurrence networks of the nirK-type denitrifying bacteria during the green tide (D), the outbreak phase (E) and the decline phase (F), and co-occurrence networks of the nirS-type denitrifying bacteria during the green tide (G), the outbreak phase (H) and the decline phase (I).
Discussion
Through Illumina Miseq sequencing, we explored variations of the nirK-type and nirS-type denitrifying bacterial communities during an U. prolifera green tide in coastal Qingdao areas. Chen et al. (2020) pointed out that a higher sequence similarity cutoff level may be preferable for obtaining more accurate and reliable information on community compositions and structures, so a 97% clustering level was used in the current study to investigate the community characteristics of denitrifying bacteria. According to our results, the structure and diversity of denitrifying bacterial communities, as well as the organization and structure of microbial co-occurrence networks, varied during an U. prolifera green tide in coastal Qingdao areas. Furthermore, we noticed that the nirK-type and nirS-type denitrifying bacteria respond differently to the green tide, implying that they may occupy different niches during the green tide in coastal Qingdao areas.
Salinity, DO content and chla content were the most important factors influencing the structure of the nirK-type and nirS-type denitrifying bacterial communities during the green tide (Figure 5). Salinity strongly contributed to the nirK-type and nirS-type denitrifying bacterial communities during the green tide, which was consistent with previous reports on the dominant position of salinity in shaping the denitrifying bacterial community structure (Abell et al., 2010; Francis et al., 2013; Zheng et al., 2015). DO content has also been proven to be an important element in determining the structure of the denitrifying bacterial communities (Liu et al., 2003; Hanning et al., 2006). In the processes of green tide, rapid photosynthesis of U. prolifera during daytime could increase the DO content in seawater, whereas respiration of U. prolifera at night, as well as accumulation and decomposition of U. prolifera, could consume the DO content in seawater, resulting in the changes of DO content in seawater and further leading to the variations of the denitrifying bacterial community structure and abundance during the green tide. Variations of the denitrifying bacterial communities could also be explained by chla content. Significant positive correlations between chla content and denitrifying bacterial community structure were observed in the current study during the outbreak phase, probably because the high density of U. prolifera at this phase provides superior conditions for the growth and metabolism of denitrifying bacteria, which was consistent with the previous result conducted by Chen et al. (2016). In addition, other physicochemical factors, such as temperature, NO2- concentration and NO3- concentration, can influence the denitrifying bacterial communities. Temperature sensitivity differed among denitrifying bacterial taxa, indicating that temperature has a significant impact on the community structure of denitrifying bacteria, mostly through impacting their growth rate and metabolic activity. Temperature fluctuations may be one of the factors influencing the structure and diversity of the denitrifying bacterial communities in the current study (Supplementary Figure 4). As the substrate supply and electron acceptor of denitrification, NO2- and NO3- concentrations were regarded as critical factors influencing the denitrifying bacterial communities. In this study, we discovered that NO2- and NO3- concentrations have observable effects on the denitrifying bacterial communities, and comparable results have been seen in the pelagial of the central Baltic Sea and the San Francisco Bay (Hanning et al., 2006; Lee and Francis, 2017).
Three sampling stations were chosen in order to analyze how the impact degree of green tides influenced the denitrifying bacterial networks and community characteristics in different region of coastal Qingdao areas. The denitrifying bacteria’s community richness, diversity and abundance, as well as their network complexity and stability, were greater at station ZQ and MD, suggesting that higher impact degree of green tide might result in higher community richness, community diversity, community abundance, and network complexity and stability of the denitrifying bacteria. Based on the relevant analysis of the nirK-type and nirS-type denitrifying bacteria, similar results were obtained in this study.
All of the denitrifying bacteria identified in the current study were Proteobacteria. Alphaproteobacteria and Gammaproteobacteria accounted for 59.28% and 34.98% of the total sequences in Proteobacteria for the nirK-type and nirS-type denitrifying bacteria, respectively. At the family level, unclassified bacteria, Rhodobacteraceae, unclassified Proteobacteria and unclassified Alphaproteobacteria dominated the nirK-type denitrifying bacterial community, and the dominant nirS-type denitrifying bacterial community was comprised of unclassified Proteobacteria, unclassified bacteria, Halomonadaceae, Pseudomonadaceae and Rhodobacteraceae (Figure 2). According to our findings, the outbreak phase may benefit the richness and diversity of denitrifying bacterial communities (Table 1). During the outbreak phase, the community richness of the nirK-type and nirS-type denitrifying bacteria was strongly positively related to chla content which is an essential indicator of phytoplankton biomass (P < 0.01), according to the results of Spearman correlation analysis (Figure 6). The accumulation of U. prolifera may lead to a high nitrogen and low oxygen environment suitable for the growth of denitrifying bacteria, which may explain their greater community richness and diversity during the outbreak phase (Lin et al., 2017; Qu et al., 2020). The abundance of the nirK and nirS genes also displayed different patterns between the outbreak phase and the decline phase, with a higher abundance of the nirK gene during the outbreak phase but the nirS gene being more abundant during the decline phase (Figure 4). During the green tide, both bloom phases and sampling stations influenced the nirK-type and nirS-type denitrifying bacterial communities, but the effects varied. Compared with the nirS-type denitrifying bacterial community, the nirK-type denitrifying bacterial community was well separated, both in terms of bloom phases and sampling stations, as validated by PCA analysis (Figure 3). These findings revealed that the nirK-type and nirS-type denitrifying bacteria behave differently to the green tide, and the nirK-type denitrifying bacteria may be more sensitive to the U. prolifera green tide in coastal Qingdao areas, which was consistent with previous reports (Jones et al., 2008; Yoshida et al., 2010).
For the denitrifying bacterial communities, the nodes and edges in network of the outbreak phase were greater than those in network of the decline phase (Figures 7B, C), which was consistent with the above result that a greater community richness and diversity was observed during the outbreak phase, indicating that the outbreak of green tide may increase not only the interactions among denitrifying bacteria, but also the complexity and stability of denitrifying bacterial networks. Moreover, greater proportions of negative links during the outbreak phase may enhance the stability of denitrifying bacterial communities under disturbances of the green tide, which was consistent with past observations that more negative interactions may mean more stable microbial networks and may help the microbial communities return to stability more quickly under disturbances (Coyte et al., 2015; de Vries et al., 2018).
Studies have shown that the topological properties of network can influence how microbial communities respond to environmental changes (de Vries et al., 2012; de Vries et al., 2018). The topological properties of the nirK-type denitrifying bacterial networks differed significantly between the outbreak phase and the decline phase, as did the nirS-type denitrifying bacterial networks (Table 2). Some topological properties of the nirK-type denitrifying bacterial networks, such as nodes, edges, average degree, average clustering coefficient and centrality, decreased from the outbreak phase to the decline phase, while others, such as average path length and modularity, increased. Topological properties of the nirS-type denitrifying bacterial networks, on the other hand, displayed the opposite trend from the outbreak phase to the decline phase when compared to the nirK-type denitrifying bacterial networks. As a result, the nirK-type denitrifying bacterial network possessed topological properties which indicated higher complexity, higher association and higher stability during the outbreak phase, whereas the nirS-type denitrifying bacterial network was more complex, more associated and more stable during the decline phase. In addition to the topological properties, the composition and structure of network also showed clear distinctions in both the nirK-type and nirS-type denitrifying bacteria during the green tide. Modules are clusters of closely connected microorganisms within communities, and more modules may be associated with stronger niche differentiation and greater community stability (Williams et al., 2014; Hou et al., 2018; Zhang et al., 2021b). The modules in the nirK-type denitrifying bacterial network were greater during the outbreak phase, whereas the modules of the nirS-type denitrifying bacterial network were greater during the decline phase (Table 2). Therefore, the nirK-type and nirS-type denitrifying bacteria displayed stronger niche differentiation and greater community stability during the outbreak phase and the decline phase, respectively. These results suggested that the nirK-type and nirS-type denitrifying bacteria respond differently to the green tide, and may display different environmental adaptation mechanisms during the green tide in coastal Qingdao areas. Some microorganisms with nirK and nirS genes, such as Bradyrhizobium oligotrophicum S58 (Sánchez and Minamisawa, 2018), have recently been discovered, and how these non-traditional denitrification pathways respond to the green tide warrants investigation. Meanwhile, N2O is one of the important products in denitrification pathways, the impacts of green tide on N2O emissions also need to be investigated further.
Conclusion
We studied the variations of the nirK-type and nirS-type denitrifying bacterial interactions and community characteristics during the green tide in coastal Qingdao areas in this study. According to our findings, the abundance, diversity and structure of the nirK-type and nirS-type denitrifying bacterial communities shifted during the green tide. The topological properties and organization of the nirK-type and nirS-type denitrifying bacterial networks have also changed during the green tide. During the outbreak phase, the nirK-type denitrifying bacteria displayed greater abundance, network complexity and network stability; whereas the nirS-type denitrifying bacteria exhibited greater abundance, network complexity and network stability during the decline phase. The nirK-type and nirS-type denitrifying bacteria respond differently to the green tide and may occupy different niches during the green tide in coastal Qingdao areas.
Data availability statement
The data presented in the study are deposited in the SRA repository, accession number PRJNA781861 and PRJNA781658.
Author contributions
GZ: investigation, methodology, data curation, formal analysis, and writing-original draft. HH: conceptualization, writing-review and editing, supervision, and funding acquisition. MY: methodology, and data curation. HW: conceptualization, writing-review and editing, and supervision. HS: conceptualization, and supervision. MW: conceptualization, writing-review and editing, supervision, and funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by the National Natural Science Foundation of China (Grant Nos. 41806131, 41976117, and 42120104006), and Fundamental Research Funds for the Central Universities (Grant No. 202172002).
Acknowledgments
We thanked Ding Zhang for the determination of physicochemical factors. We also appreciated the computing resources provided by IEMB-1, a high-performance computation cluster operated by the Institute of Evolution and Marine Biodiversity.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.1063585/full#supplementary-material
References
Abell G. C., Revill A. T., Smith C., Bissett A. P., Volkman J. K., Robert S. S. (2010). Archaeal ammonia oxidizers and nirS-type denitrifiers dominate sediment nitrifying and denitrifying populations in a subtropical macrotidal estuary. ISME J. 4, 286–300. doi: 10.1038/ismej.2009.105
Babbin A. R., Keil R. G., Devol A. H., Ward B. B. (2014). Organic matter stoichiometry, flux, and oxygen control nitrogen loss in the ocean. Science 344, 406–408. doi: 10.1126/science.1248364
Bothe H., Jost G., Schloter M., Ward B. B., Witzel K. P. (2020). Molecular analysis of ammonia oxidation and denitrification in natural environments. FEMS Microbiol. Rev. 24 (5), 673–690. doi: 10.1111/j.1574-6976.2000.tb00566.x
Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., et al. (2010). QIIME allows analysis of high-throughput community sequence data. Nat. Methods 7 (5), 335–336. doi: 10.1038/nmeth.f.303
Chen Q., Fan J., Ming H., Su J., Wang Y., Wang B. (2020). Effects of environmental factors on denitrifying bacteria and functional genes in sediments of bohai Sea, China. Mar. Pollut. Bull. 160, 111621. doi: 10.1016/j.marpolbul.2020.111621
Chen X., Jiang H., Sun X., Zhu Y., Yang L. (2016). Nitrification and denitrification by algae-attached and free-living microorganisms during a cyanobacterial bloom in lake taihu, a shallow eutrophic lake in China. Biogeochemistry 131, 135–146. doi: 10.1007/s10533-016-0271-z
Christensen P. B., Rysgaard S., Sloth N. P., Dalsgaard T., Schwærter S. (2000). Sediment mineralization, nutrient fluxes, denitrification and dissimilatory nitrate reduction to ammonium in an estuarine fjord with sea cage trout farms. Aquat. Microb. Ecol. 21, 73–84. doi: 10.3354/ame021073
Coyte K. Z., Schluter J., Foster K. R. (2015). The ecology of the microbiome: networks, competition, and stability. Science 350, 663–666. doi: 10.1126/science.aad2602
Dalsgaard T., Thamdrup B., Canfield D. E. (2005). Anaerobic ammonium oxidation (anammox) in the marine environment. Res. Microbiol. 156, 457–464. doi: 10.1016/j.resmic.2005.01.011
Dalsgaard T., Thamdrup B., Farias L., Revsbech N. P. (2012). Anammox and denitrification in the oxygen minimum zone of the eastern south pacific. Limnol. Oceanogr. 57 (5), 1331–1346. doi: 10.4319/lo.2012.57.5.1331
de Vries F. T., Griffiths R. I., Bailey M., Craig H., Girlanda M., Gweon H. S., et al. (2018). Soil bacterial networks are less stable under drought than fungal networks. Nat. Commun. 9, 3033. doi: 10.1038/s41467-018-05516-7
de Vries F. T., Liiri M. E., Bjørnlund L., Bowker M. A., Christensen S., Setälä H. M., et al. (2012). Land use alters the resistance and resilience of soil food webs to drought. Nat. Climate Change 2, 276–280. doi: 10.1038/nclimate1368
Edgar R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Falkowski P. G., Fenchel T., Delong E. F. (2008). The microbial engines that drive earth’s biogeochemical cycles. Science 320, 1034–1039. doi: 10.1126/science.1153213
Francis C. A., O'mullan G. D., Cornwell J. C., Ward B. B. (2013). Transitions in nirS-type denitrifier diversity, community composition, and biogeochemical activity along the Chesapeake bay estuary. Front. Microbiol. 4, 237. doi: 10.3389/fmicb.2013.00237
Guo L., Hu Z., Fang F., Liu T., Chuai X., Yang L. (2014). Trophic status determines the nirS-denitrifier community in shallow freshwater lakes. Ann. Microbiol. 64, 999–1006. doi: 10.1007/s13213-013-0737-3
Hallin S., Lindgren P. E. (1999). PCR detection of genes encoding nitrile reductase in denitrifying bacteria. Appl. Environ. Microbiol. 65 (4), 1652–1657. doi: 10.1128/AEM.65.4.1652-1657.1999
Hanning M., Braker G., Dippner J., Jürgens K. (2006). Linking denitrifier community structure and prevalent biogeochemical parameters in the pelagial of the central Baltic proper (Baltic Sea). FEMS Microbiol. Ecol. 57 (2), 260–271. doi: 10.1111/j.1574-6941.2006.00116.x
Hou L., Xie X., Wan X., Kao S. J., Jiao N., Zhang Y. (2018). Niche differentiation of ammonia and nitrite oxidizers along a salinity gradient from the pearl river estuary to the south China Sea. Biogeosciences 15, 5169–5187. doi: 10.5194/bg-15-5169-2018
Jones C. M., Hallin S. (2010). Ecological and evolutionary factors underlying global and local assembly of denitrifier communities. ISME J. 4, 633–641. doi: 10.1038/ismej.2009.152
Jones C. M., Stres B., Rosenquist M., Hallin S. (2008). Phylogenetic analysis of nitrite, nitric oxide, and nitrous oxide respiratory enzymes reveal a complex evolutionary history for denitrification. Mol. Biol. Evol. 25 (9), 1955–1966. doi: 10.1093/molbev/msn146
Lee J. A., Francis C. A. (2017). Spatiotemporal characterization of San Francisco bay denitrifying communities: a comparison of nirK and nirS diversity and abundance. Microbial Ecol. 73, 271–284. doi: 10.1007/s00248-016-0865-y
Lin G., Sun F., Wang C., Zhang L., Zhang X. (2017). Assessment of the effect of enteromorpha prolifera on bacterial community structures in aquaculture environment. PloS One 12 (7), e0179792. doi: 10.1371/journal.pone.0179792
Liu D., Keesing J. K., He P. M., Wang Z. L., Shi Y. J., Wang Y. J. (2013). The world’s largest macroalgal bloom in the yellow Sea, China: formation and implications. Estuar. Coast. Shelf Sci. 129, 2–10. doi: 10.1016/j.ecss.2013.05.021
Liu N., Liao P., Zhang J., Zhou Y., Luo L., Huang H., et al. (2020). Characteristics of denitrification genes and relevant enzyme activities in heavy-metal polluted soils remediated by biochar and compost. Sci. Total Environ. 739, 139987. doi: 10.1016/j.scitotenv.2020.139987
Liu X., Tiquia S. M., Holguin G., Wu L., Nold S. C., Devol A. H., et al. (2003). Molecular diversity of denitrifying genes in continental margin sediments within the oxygen-deficient zone off the pacific coast of Mexico. Appl. Environ. Microbiol. 69 (6), 3549–3560. doi: 10.1128/AEM.69.6.3549-3560.2003
Li H. M., Zhang Y. Y., Tang H. J., Shi X. Y., Rivkin R. B., Legendre L. (2017). Spatiotemporal variations of inorganic nutrients along the jiangsu coast, China, and the occurrence of macroalgal blooms (green tides) in the southern yellow Sea. Harmful Algae 63, 164–172. doi: 10.1016/j.hal.2017.02.006
Magoč T., Salzberg S. L. (2011). FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Marshall K., Joint I., Callow M. E., Callow J. A. (2006). Effect of marine bacterial isolates on the growth and morphology of axenic plantlets of the green alga ulva linza. Microbial Ecol. 52, 302–310. doi: 10.1007/s00248-006-9060-x
Mosier A. C., Francis C. A. (2010). Denitrifier abundance and activity across the San Francisco bay estuary. Environ. Microbiol. Rep. 2 (5), 667–676. doi: 10.1111/j.1758-2229.2010.00156.x
Oakley B. B., Francis C. A., Roberts K. J., Fuchsman C. A., Srinivasan S., Staley J. T. (2007). Analysis of nitrite reductase (nirK and nirS) genes and cultivation reveal depauperate community of denitrifying bacteria in the black Sea suboxic zone. Environ. Microbiol. 9 (1), 118–130. doi: 10.1111/j.1462-2920.2006.01121.x
Qu T. F., Zhao X. Y., Hao Y., Zhong Y., Guan C., Hou C. Z., et al. (2020). Ecological effects of ulva prolifera green tide on bacterial community structure in qingdao offshore environment. Chemosphere 244, 125477. doi: 10.1016/j.chemosphere.2019.125477
Sánchez C., Minamisawa K. (2018). Redundant roles of bradyrhizobium oligotrophicum Cu-type (nirK) and cd1-type (nirS) nitrite reductase genes under denitrifying conditions. FEMS Microbiol. Lett. 365, fny015. doi: 10.1093/femsle/fny015
Santoro A. E., Boehm A. B., Francis C. A. (2006). Denitrifier community composition along a nitrate and salinity gradient in a coastal aquifer. Appl. Environ. Microbiol. 72 (3), 2102–2109. doi: 10.1128/AEM.72.3.2102-2109.2006
Sun F., Wang C., Chen H., Zheng Z. (2020). Metagenomic analysis of the effect of enteromorpha prolifera bloom on microbial community and function in aquaculture environment. Curr. Microbiol. 77, 816–825. doi: 10.1007/s00284-019-01862-x
Throbäck I. N., Enwall K., Jarvis A., Hallin S. (2004). Reassessing PCR primers targeting nirS, nirK and nosZ genes for community surveys of denitrifying bacteria with DGGE. FEMS Microbiol. Ecol. 49 (3), 401–417. doi: 10.1016/j.femsec.2004.04.011
van Alstyne K. L., Nelson T. A., Ridgway R. L. (2015). Environmental chemistry and chemical ecology of “green tide” seaweed blooms. Integr. Comp. Biol. 55 (3), 518–532. doi: 10.1093/icb/icv035
Wei W., Isobe K., Nishizawa T., Zhu L., Shiratori Y., Ohte N., et al. (2015). Higher diversity and abundance of denitrifying microorganisms in environments than considered previously. ISME J. 9, 1954–1956. doi: 10.1038/ismej.2015.9
Williams R. J., Howe A., Hofmockel K. S. (2014). Demonstrating microbial co-occurrence pattern analyses within and between ecosystems. Front. Microbiol. 5, 358. doi: 10.3389/fmicb.2014.00358
Wolsing M., Priemé A. (2004). Observation of high seasonal variation in community structure of denitrifying bacteria in arable soil receiving artificial fertilizer and cattle manure by determining T-RFLP of nir gene fragments. FEMS Microbiol. Ecol. 48 (2), 261–271. doi: 10.1016/j.femsec.2004.02.002
Yang Y. D., Hu Y. G., Wang Z. M., Zeng Z. H. (2018). Variations of the nirS-, nirK-, and nosZ-denitrifying bacterial communities in a northern Chinese soil as affected by different long-term irrigation regimes. Environ. Sci. pollut. Res. 25, 14057–14067. doi: 10.1007/s11356-018-1548-7
Yoshida M., Ishiii S., Otsuka S., Senoo K. (2010). nirK-harboring denitrifiers are more responsive to denitrification-inducing conditions in rice paddy soil than nirS-harboring bacteria. Microbes Environments 25 (1), 45–48. doi: 10.1264/jsme2.ME09160
Yoshida M., Ishii S., Otsuka S., Senoo K. (2009). Temporal shifts in diversity and quantity of nirS and nirK in a rice paddy field soil. Soil Biol. Biochem. 41 (10), 2044–2051. doi: 10.1016/j.soilbio.2009.07.012
Yuan Q., Liu P., Lu Y. (2012). Differential responses of nirK- and nirS-carrying bacteria to denitrifying conditions in the anoxic rice field soil. Environ. Microbiol. Rep. 4 (1), 113–122. doi: 10.1111/j.1758-2229.2011.00311.x
Zhang Y. Y., He P. M., Li H. M., Li G., Liu J. H., Jiao F. L., et al. (2019). Ulva prolifera green-tide outbreaks and their environmental impact in the yellow Sea, China. Natl. Sci. Rev. 6, 825–838. doi: 10.1093/nsr/nwz026
Zhang C., Liu Q., Li X., Wang M., Liu X., Yang J., et al. (2021b). Spatial patterns and co-occurrence networks of microbial communities related to environmental heterogeneity in deep-sea surface sediments around yap trench, Western pacific ocean. Sci. Total Environ. 759, 143799. doi: 10.1016/j.scitotenv.2020.143799
Zhang P. Y., Xin Y., Zhong X. S., Yan Z. W., Jin Y. M., Yan M. J., et al. (2021a). Integrated effects of ulva prolifera bloom and decay on nutrients inventory and cycling in marginal sea of China. Chemosphere 264, 128389. doi: 10.1016/j.chemosphere.2020.128389
Zhao G., He H., Wang H., Liang Y., Guo C., Shao H., et al. (2022). Variations in marine bacterial and archaeal communities during an ulva prolifera green tide in coastal qingdao areas. Microorganisms 10, 1204. doi: 10.3390/microorganisms10061204
Zheng Y., Hou L., Liu M., Gao J., Yin G., Li X., et al. (2015). Diversity, abundance, and distribution of nirS-harboring denitrifiers in intertidal sediments of the Yangtze estuary. Microbial Ecol. 70, 30–40. doi: 10.1007/s00248-015-0567-x
Zhou Z. F., Zheng Y. M., Shen J. P., Zhang L. M., He J. Z. (2011). Response of denitrification genes nirS, nirK, and nosZ to irrigation water quality in a Chinese agricultural soil. Environ. Sci. pollut. Res. Int. 18 (9), 1644–1652. doi: 10.1007/s11356-011-0482-8
Keywords: Ulva profilera green tide, denitrifying bacteria, community structure, cooccurrence network, Illumina Miseq sequencing approach
Citation: Zhao G, He H, Yue M, Wang H, Shao H and Wang M (2022) Differential responding patterns of the nirK-type and nirS-type denitrifying bacterial communities to an Ulva prolifera green tide in coastal Qingdao areas. Front. Mar. Sci. 9:1063585. doi: 10.3389/fmars.2022.1063585
Received: 07 October 2022; Accepted: 11 November 2022;
Published: 02 December 2022.
Edited by:
Xiyang Dong, Third Institute of Oceanography of the Ministry of Natural Resources, ChinaReviewed by:
Haikun Zhang, Yantai Institute of Coastal Zone Research (CAS), ChinaLing-Dong Shi, University of California, Berkeley, United States
Copyright © 2022 Zhao, He, Yue, Wang, Shao and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hui He, aGVodWlAb3VjLmVkdS5jbg==; Min Wang, bWluZ3dhbmdAb3VjLmVkdS5jbg==
 Guihua Zhao1
Guihua Zhao1 Hui He
Hui He Hualong Wang
Hualong Wang Hongbing Shao
Hongbing Shao Min Wang
Min Wang