- 1Key Laboratory of South China Sea Fishery Resources Exploitation & Utilization, Ministry of Agriculture, South China Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Guangzhou, China
- 2National Demonstration Center for Experimental Fisheries Science Education, Shanghai Engineering Research Center of Aquaculture, Shanghai Ocean University, Shanghai, China
- 3Tianjin Key Laboratory of Aqua-ecology and Aquaculture, College of Fisheries, Tianjin Agricultural University, Tianjin, China
Leucine-responsive regulatory protein (Lrp) is an essential transcriptional regulator in prokaryotes. However, the regulatory role of lrp in Vibrio alginolyticus has still not been studied in detail. In this study, an lrp mutant strain was constructed to gain insight into the role of lrp in Vibrio alginolyticus. The absence of lrp significantly enhances swarming motility, biofilm formation, extracellular protease secretion activity, and tolerance to copper ions. The cumulative mortality of zebrafish (Danio rerio) challenged by intraperitoneal injection against the lrp mutant strain reached 68.89%, significantly higher than the 40.00% suffered by fish injected with the wild-type strain. The expression levels of lrp decreased gradually with increasing culture time under the influence of various physicochemical factors. The expression level of lrp was significantly increased after two hours of culture at pH 5, 22°C, 5% NaCl, the presence of 1 mM Cu2+, 1/4/7 mM ferric citrate, 0.1 mg/L NaNO3, and 0.1 mg/L KH2PO4. The mRNA level of lrp decreased significantly after six hours of culture at 37°C, 1% and 5% NaCl, and the presence of 1/7 mM ferric citrate, 0.1/5 mg/L NaNO3, and 0.1/0.5/2.0 mg/L KH2PO4. The expression of lrp increased after ten hours of culture at pH 5/9, 22°C, 1% NaCl, and the presence of 1 mM Cu2+, 7 mM ferric citrate, 5 mg/L NaNO3, and 0.1 mg/L KH2PO4. Overall, this study indicates that lrp negatively controls the virulence of V. alginolyticus, probably by reducing its swarming motility, biofilm formation, extracellular protease secretion activity, and tolerance to copper ions, and that the expression of lrp is affected by numerous physicochemical factors, and is especially up-regulated after 2 hours of culture in bacterial growth.
1. Introduction
Vibrio alginolyticus, a Gram-negative bacterium found throughout the ocean (Ren et al., 2013), has emerged as the primary aquatic pathogenic bacterium in recent years (Li et al., 2021). Diseases caused by V. alginolyticus can lead to epidermal bleeding, whitening of the kidneys, and congestion of the internal organs of animals. Its pathogenicity increases significantly in warmer temperatures in summer (Zuo et al., 2019) when it causes severe economic damage to the aquaculture industry (Mohamad et al., 2019; Fahmy and Hamed, 2021; Yang et al., 2021). In humans, V. alginolyticus infections can cause septicemia, eardrum infections, and intraocular inflammation (Li et al., 2009; Gaüzère et al., 2016; Baker-Austin et al., 2018). In total, V. alginolyticus can not only lead to large-scale mortality in aquatic animals with considerable negative economic impacts, but is also a potential source of human-aquatic animal co-morbidity, with a risk to human health. Current studies have shown that the pathogenic effects of V. alginolyticus are due to virulence factors such as extracellular products (Wang et al., 2007; Wang et al., 2008; Rui et al., 2009), bacterial adhesion (Wang and Leung, 2000), cell motility (Liu et al., 2020), and biofilm formation (Liu et al., 2011; Liu et al., 2011; Gu et al., 2019b).
Physicochemical factors have an impact on bacterial growth and reproduction. They can also directly influence the expression of bacterial virulence or virulence-regulating genes (DeAngelis et al., 2018), thereby affecting the risks to host organisms (Huang et al., 2015). For example, the abundance of V. alginolyticus is affected by factors such as nutrients, temperature, salinity, and phytoplankton (Oberbeckmann et al., 2011). Furthermore, the metabolism of Vibrio is significantly influenced by the environmental levels of nitrate, ammonia nitrogen, and phosphate (Unemoto et al., 1977; Oliver et al., 1983; Davis et al., 2017). The formation of V. alginolyticus blooms can be promoted by Saharan dust nutrients, which drive changes in its population dynamics (Westrich et al., 2016). Likewise, environmental conditions can substantially impact biofilm formation and adhesion of bacteria (Luo et al., 2016; Toyofuku et al., 2016). For example, the expression of the cytochrome C oxidase gene is effected by temperature, pH, and salinity, which alter the capacity of V. alginolyticus to adhere to the host (Huang et al., 2019). Vibrio alginolyticus is a conditionally pathogenic bacterium, and external environmental changes can also modulate the expression of pathogenesis-related proteins (Hoe and Goguen, 1993). Outer membrane proteins adapt and respond to environmental stress by altering the expression levels of specific proteins (Abdallah et al., 2012). Environmental factors change periodically both seasonally and geographically. Aquaculture systems are vulnerable to human activities, including overfeeding, drug usage, industrial pollution, and domestic sewage. The degree of these impacts have changed over time, posing additional challenges and threats, including those which affect the occurrence and development of pathogen virulence (Halpern et al., 2019). Therefore, understanding the influence of environmental factors on bacterial virulence factors or virulence regulators is of great significance in monitoring bacterial virulence and predicting disease occurrence, and can provide a theoretical basis for establishing virulence prevention and control measures based on environmental regulation.
Leucine-responsive regulatory protein (Lrp) belongs to the Leucine-responsive regulatory protein/asparagine synthase C gene product family (Lrp/AsnC family) of globally specific transcriptional regulators, also known as feast/famine regulatory proteins (Brinkman et al., 2003; Yokoyama et al., 2006). Lrp binds regulatory DNA via a helix-turn-helix motif, which strongly influences Lrp-DNA binding (Hart et al., 2011). Lrp primarily responds to exogenous amino acid effectors and globally impacts cellular metabolism (Thaw et al., 2006). Early studies found that Lrp can respond to leucine in Escherichia. coli (Chen et al., 2001). Further research found that Lrp is involved in the regulation of various metabolic functions, such as amino acid synthesis and catabolism (Brinkman et al., 2003), fimbriae biosynthesis, and pili phase-variation (Nou et al., 1993; Huisman et al., 1994; van der Woude and Low, 1994). As a global regulator, Lrp is associated with cytotoxicity, chemotaxis, and iron uptake in Vibrio vulnificus (Ho et al., 2017) and chromosome replication in Vibrio cholerae (Ciaccia et al., 2018). Furthermore, numerous studies have shown that Lrp plays a vital role in regulating the virulence of bacteria (Lin et al., 2007; Alice and Crosa, 2012; Hussa et al., 2015; Chen et al., 2019). Lin et al. (2007) found that Lrp could regulate the quorum-sensing regulator AphA, and then activate the expression of the virulence gene. However, in Vibrio alginolyticus the lrp gene has not yet been the subject of any detailed research, and its function is still unknown.
To understand the function of lrp in V. alginolyticus, we constructed a null mutant of the lrp gene, studied the biological properties and pathogenicity of the Δlrp strain, and then used qRT-PCR to analyze the transcription levels of the lrp gene when stressed with different physicochemical factors. These results make a significant contribution to the functional study of lrp in V. alginolyticus, especially regarding the role of lrp in regulating the virulence of V. alginolyticus and the effects of physicochemical factors on lrp expression. The study will aid the monitoring of V. alginolyticus virulence to predict outbreaks of diseases caused by V. alginolyticus, as well as provide a theoretical basis for the establishment of the environmental regulation system of V. alginolyticus related diseases.
2. Materials and methods
2.1. Bacterial strains, plasmids, and construction of the lrp mutant strain
ZJ-T is a translucent variant of V. alginolyticus ZJ-51 (Deng et al., 2016a), and was used as the wild-type strain in this study. ZJ-T cells were cultured in LBS (Luria-Bertani medium with 3% sodium chloride) at 30°C. E. coli cells were cultured in LB medium at 37°C. Table 1 lists the bacterial strains and plasmids used in this study.
The lrp mutant strain was constructed by homologous recombination (Deng et al., 2016b). The plasmid pSW7848 was linearized with the pair primer pSW7848-F/R, and the two flanking regions of the desired deletion fragments of lrp were amplified using the primer pairs dlrp-U-F/R and dlrp-D-F/R, respectively (Table 2). The linearized plasmid and flanking fragments were assembled using a ClonExpress MultiS one-step cloning kit (Vazyme Biotech Co., Ltd., Nanjing, China) with an isothermal assembly. The recombinant plasmid was then introduced into E.coli II3813, followed by E.coli GEB883. After transformation, the integrity of the flanking fragments was examined using a pair primer Del-check-pSW7848-F/R. After that, the recombinant plasmid was transferred into V. alginolyticus ZJ-T by conjugation (Figure 1). The pSW7848 plasmid carries chloramphenicol resistance. D-glucose reduces while L-arabinose induces the expression of the virulence gene ccdB in the plasmid (Val et al., 2012). Two conjugation experiments were performed by screening colonies on TCBS plates with either D-glucose or L-arabinose. After conjugation, the mutant strains were obtained by PCR amplification and sequencing verification using the primer pairs Del-lrp-check-F/R.
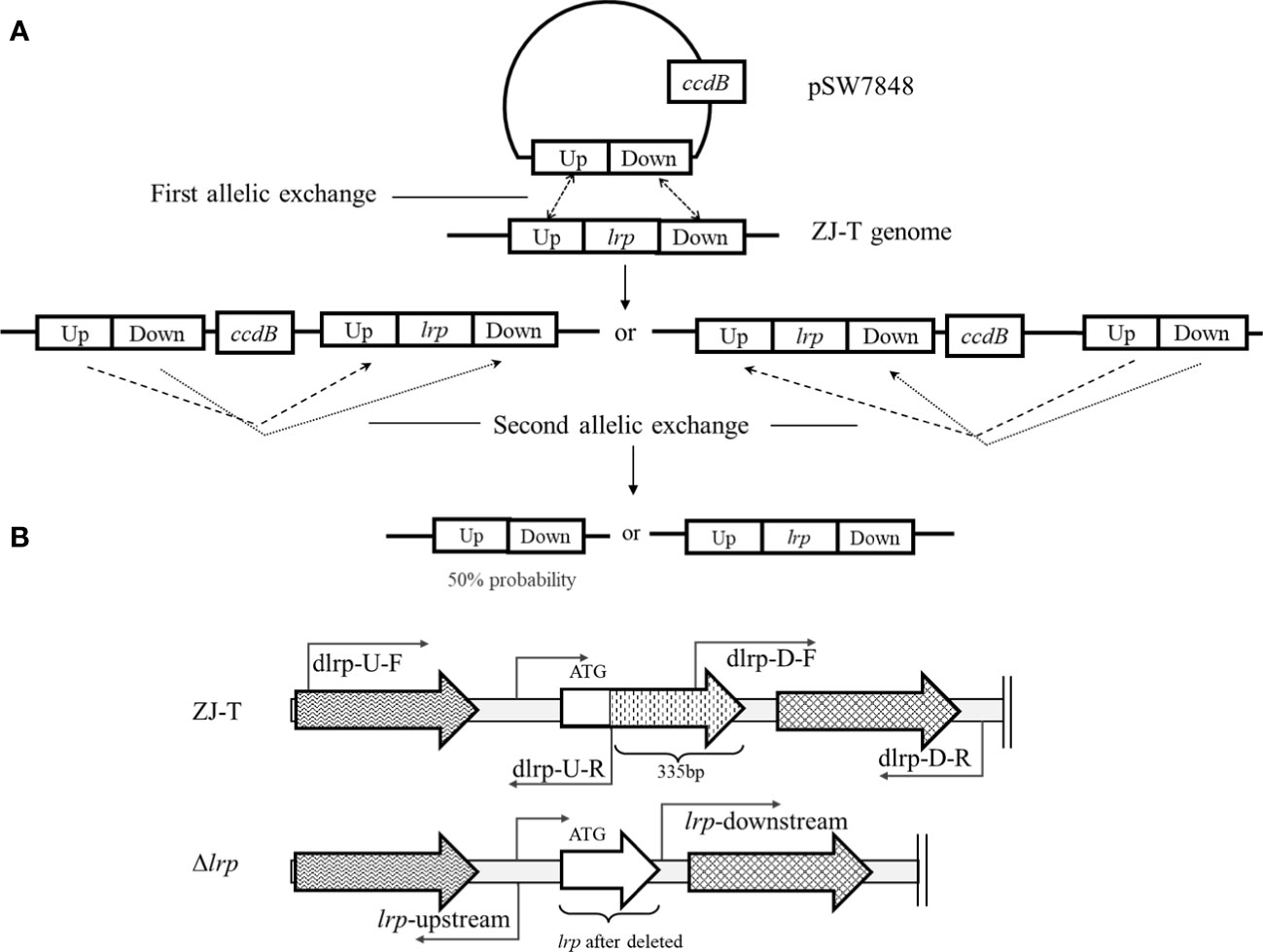
Figure 1 The conjugation between recombinant plasmid and V. alginolyticus genome (A), and the structure of the lrp locus (B). A: TCBS with 0.2% D-glucose and 20 μg/mL of Cm was used to select transconjugants in the first allelic exchange. TCBS with 0.2% L-arabinose was used to select the transconjugants that underwent plasmid excision in the second allelic exchange. The carried ccdB gene expressed was induced by L- arabinose, then pSW7848 was excised. B: 335 bp of the lrp ORF were deleted from the 33rd codon to the stop codon, giving rise to the Δlrp strain.
2.2. Bacterial growth measurement
The monoclonal clones of V. alginolyticus ZJ-T (WT) and the lrp mutant strain (Δlrp) were inoculated into LBS liquid medium and cultured overnight at 30°C with 200 rpm shaking. The overnight cultures were diluted to 0.01 at an optical density of OD600 nm (OD600 nm) in conical flasks and re-cultured at 30°C with 200 rpm continuous shaking. The change of absorbance during 36 h of bacterial growth was measured using a spectrophotometer (INESA Co., Ltd., Shanghai, China). The growth of bacteria in the LBS medium was observed. The experiments were conducted in triplicate, and the results were presented graphically. One-way ANOVA analysis of covariance was performed to analyze the effect of lrp deletion with time as a covariate against the OD600 nm values.
2.3. Iron absorption, copper ion and H2O2 resistance
The overnight cultures (described in 2.2) were diluted to OD600 nm = 5.0 and then gradient diluted in 10-fold steps (100 to 109) with fresh LBS medium. After that, 5 μL of each dilution were simultaneously spotted onto LBS plates alone or with either 0.003% H2O2, 4.5 mM CuSO4, or 150 μM 2,2’-dipyridyl (DIP). Colony growth was observed after incubation at 30°C for 24 h.
2.4. Hemolytic activity
The effect of lrp on the hemolytic activity of V. alginolyticus was determined using the Mueller-Hinton blood agar plate method (HuanKai Microbial Sci&Tech. Co., Ltd., Guangdong, China). If the bacteria had caused hemolytic activity, hemolytic circles would appear on the plate. The overnight strain cultures (described in 2.2) were streaked onto Mueller-Hinton blood agar plates and hemolysis was observed after incubation at 30°C for 24 h.
2.5. Analysis of swimming and swarming motility
Overnight cultures of the WT and Δlrp strains were prepared as described in 2.2. The cultures were diluted with fresh LBS medium to OD600 nm = 1.0. Three parallel 5 μL samples of the diluted WT and Δlrp strains were spotted onto 0.3% agar and 1.5% agar LBS plates and the plates were incubated at 30°C for 16 h and 24 h, respectively. The culture diameters were measured to compare the differences in motility between the WT and Δlrp strains.
2.6. Analysis of biofilm formation activity
Overnight cultures of the WT and Δlrp strains were prepared as described in 2.2, and the bacteria were diluted to OD600 nm = 1.0. Samples were pipetted into sterilized 24-well plates with 1 mL per well. The plates were then incubated at 30°C for 48 h and used for biofilm quantification (O’Toole, 2011). After incubation, the wells were aspirated and fixed by adding 10% methanol. Then the biofilms were stained with 1% crystal violet and dissolved in 95% ethanol, followed by measurement of absorbance values of the biofilms at 550 nm using a microplate reader. The experiment was performed in triplicate and repeated three times.
2.7. Extracellular protease secretion assay
In this experiment, the extracellular protease secretion activity of the WT and Δlrp strains were determined following Swift et al. (1999) with some modifications. Overnight cultures of the WT and Δlrp strains were prepared as described in 2.2. The cultures were diluted to OD600 nm = 5.0, and 5 μL of the diluted cultures were spotted three times in parallel onto LBS plates containing 1% skimmed milk and then incubated at 30°C. The relative enzyme activity was calculated following Usman et al. (2021). The relative secretion abilities were measured as the diameter of the protein decomposition zone/the diameter of the colony after 24 h.
2.8. Antibiotic susceptibility assays
The effect of lrp deficiency on the susceptibility of V. alginolyticus to 23 antibiotics was determined using the Kirby-Bauer disk diffusion method (Yao et al., 2021). Overnight cultures of the WT and Δlrp strains were prepared as described in 2.2. 200 μL overnight cultures were mixed with 10 mL fresh LBS liquid medium and poured onto LBS plates. After 10 min wetting, the remaining bacterial solution was poured off. The plates were air-dried, and antibiotic loaded paper pills were placed onto them and then incubated at 30°C. After 24 h, the diameter of the inhibition circle on the plate was recorded, and the sensitivity results were determined by referring to the drug sensitivity instructions.
2.9. Zebrafish challenge assay
Zebrafish (average body weight, 0.35 ± 0.05 g) were purchased from an aquarium store in Guangzhou, Guangdong Province and acclimated for two weeks in an indoor laboratory at 30°C with continuous aeration.
Zebrafish challenge experiments were performed to determine the effect of lrp on the virulence of V. alginolyticus. The WT and Δlrp monoclonal strains were inoculated into LBS agar slant culture medium, then incubated overnight at 30°C. The colonies were then eluted with 3 mL of 0.85% sterile saline, and the bacterial concentration was determined and diluted to OD600 nm = 0.3. The zebrafish were challenged by intraperitoneal injection with 40 μL/fish (2.55~2.71×108 CFU/fish). Each strain was injected into 60 fish equally divided into three tanks. The negative control was injected with 40 μL/fish of 0.85% sterile saline. Mortality was monitored every 24 h following the challenge, and the cumulative mortalities of four days (96 h) were calculated.
2.10. Effect of physicochemical factors on the expression of lrp during growth
The overnight cultures of the WT strain was diluted to OD600 nm = 0.01 in LBS medium with different pH (pH 5, 7, and 9), temperature (22°C, 30°C, and 37°C), salinity (1%, 3%, and 5% sodium chloride), copper ions (0.1 mg/L, 1.0 mg/L, and 5.0 mg/L copper sulfate), iron ions (1 mM, 4 mM, and 7 mM ferric citrate), nitrate (0.1 mg/L, 0.5 mg/L, and 2.0 mg/L sodium nitrate), and phosphate (1 mM, 4 mM, and 7 mM potassium dihydrogen phosphate) levels. The pH group, salinity group, and the groups with different concentration of added copper sulfate, ferric citrate, sodium nitrate, and potassium dihydrogen phosphate were cultured at 30°C with 200 rpm shaking. The temperature group were cultured at the different test temperature conditions with 200 rpm shaking. Bacteria cultured in LBS medium at pH 7, 30°C, and 3% NaCl were considered as the controls for the copper ions group, iron ions group, nitrate group, and phosphate group (the pH group use pH 7 as control, temperature group use 30°C as control, and salinity group use 3% NaCl as control). The lrp transcription levels of the bacteria cultured in the various conditions were analyzed after 2, 6, and 10 h of growth. RNAiso Plus reagent (Takara Bio Inc., Beijing, China) was used to extract total RNA from the cultures. Reverse transcription was performed using an Evo M-MLV Mix Kit with gDNA Clean (Accurate Biology Co., Ltd., Hunan, China). RT-PCR amplification was performed to assess the expression of lrp, using a SYBR® Green Premix Pro Taq HS qPCR kit (Accurate Biology Co., Ltd., Hunan, China). The 16S rRNA gene was used as a control in the qRT-PCR. The primers used for RT-PCR are listed in Table 2. Gene expression analysis was performed using the 2-ΔΔCT method (Livak and Schmittgen, 2001).
2.11. Statistical analysis
The effects of lrp deletion on the phenotypes and virulence of V. alginolyticus and the effects of the various physicochemical factors on the expression of lrp were analyzed using Graphpad Prism 9 (Graphpad Software, San Diego, CA, USA). Mean ± SD values was calculated for the growth, motility, biofilm formation, extracellular protease, zebrafish challenge assay, and lrp expression experiments. Independent two-sample t-tests were used to analyze changes in motility, biofilm formation, and extracellular protease secretion activity following lrp deletion. A one-way ANOVA was conducted to assess the growth curves and lrp expression level assays in the same conditions at the three different time periods. Differences in cumulative mortalities caused by the WT and Δlrp strains were analyzed using the Log-rank (Mantel-Cox) test. P-values < 0.05 were considered statistically significant.
3. Results
3.1. Construction of the V. alginolyticus ZJ-T lrp mutant strain
The lrp deletion strain was constructed using homologous recombination following Deng et al. (2016b) with some modifications. The linearized fragment of plasmid pSW7848 (3309 bp), and the upstream and downstream fragments of lrp (1025 bp and 904 bp) were successfully amplified using PCR (Figure 2, lane 1-3). The upstream and downstream fragments were then assembled with the linearized suicide vector pSW7848, and the fusion action was transformed into E. coli II3813. The fusion was identified using the primer pairs Del-check-pSW7848-F/R, resulting in a fragment of 2214 bp (Figure 2, lane 4). The resulting recombination plasmid pSW7848-lrpup-lrpdown was subsequently transformed into E. coli GEB883. The allelic exchange was performed after the recombinant plasmid was transferred from E. coli GEB883 to V. alginolyticus ZJ-T by conjugation. The lrp disruptant was finally confirmed by sequencing, and the strain was named Δlrp (Figure 2, lane 5-6).
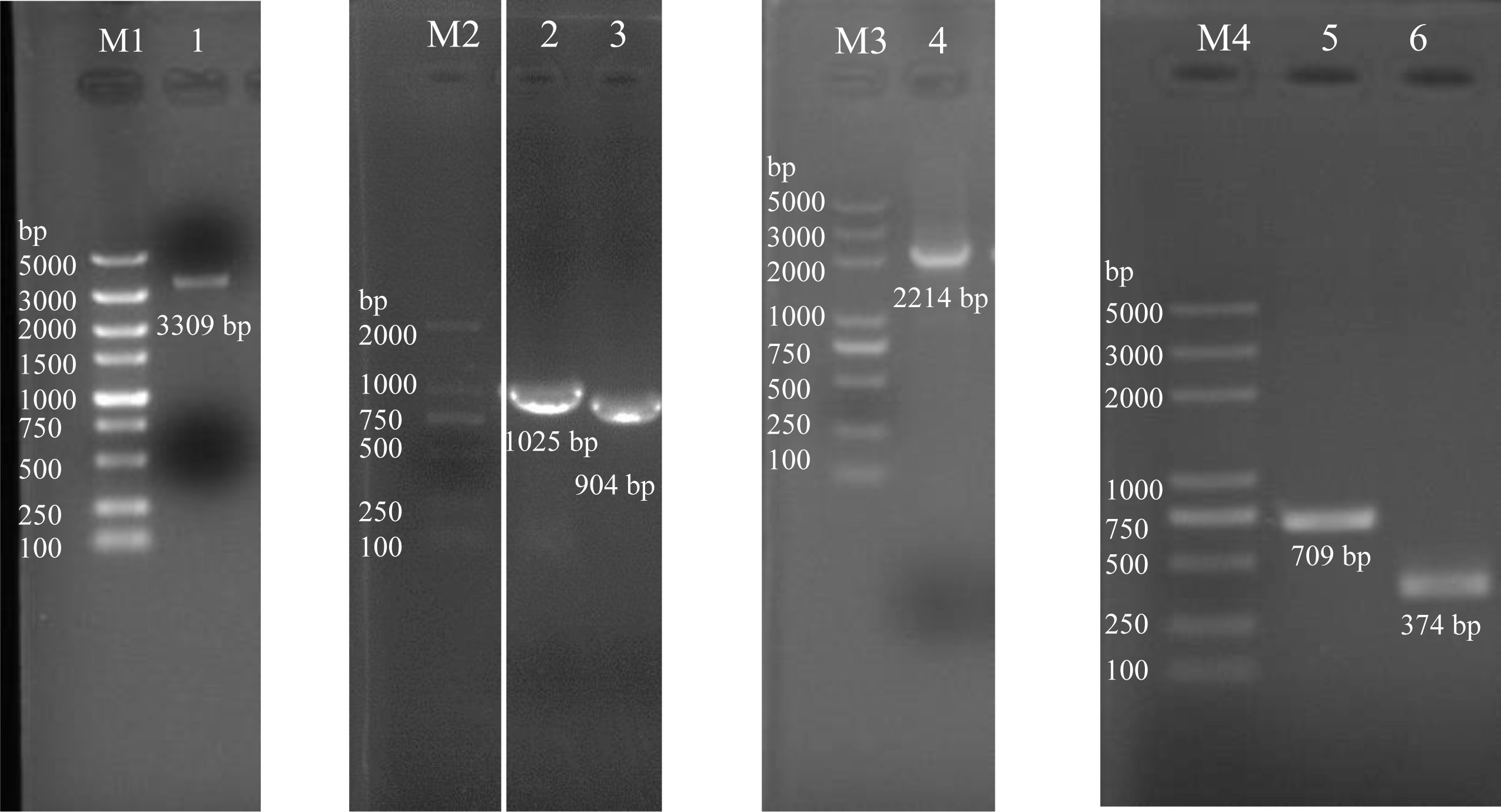
Figure 2 PCR identification of Δlrp construction in V. alginolyticus. M1: 5,000 DNA marker (Guangzhou Dongsheng Biotech Co., Ltd.); M2: 2,000 DNA marker (Guangzhou Dongsheng Biotech Co., Ltd.); M3 and M4: 5,000 DNA marker (Tsingke Biotechnology Co., Ltd.); lane 1: pSW7848 linearized plasmid; lane 2: upstream fragment of lrp; lane 3: downstream fragment of lrp; lane 4: detecting of the recombination plasmid of pSW7848-lrpup-lrpdown; lane 5: amplification fragment of wild strain with primers of Del-lrp-check-F/R; lane 6: amplification fragment of Δlrp with primers of Del-lrp-check-F/R. This figure is the combination result of four gel pictures, and they are M1/1, M2/2/3, M3/4, and M4/5/6, respectively. In the gel M1/1, we cut all the lanes on the right of lane 1; in the gel M2/2/3, we cut a lane between lane M2 and lane 2, and cut all the lanes on the right of lane 3, then we spliced the lane M2 and lane 2/3, and there is a space between M2 and lane 2 showing the splicing; in the gel M3/4, we cut all the lanes on the right of lane 4; in the gel M4/5/6, we cut all the lanes on the right of lane 6.
3.2. Effect of lrp on the growth of V. alginolyticus under different conditions
The WT and Δlrp strains were cultured in LBS medium. The growth of Δlrp did not differ significantly from WT during any growth period. During the 36 h of incubation, the first hour presented the latent period. The log phase began at 2 h and the stable phase was reached after 12 hours incubation (Figure 3A).
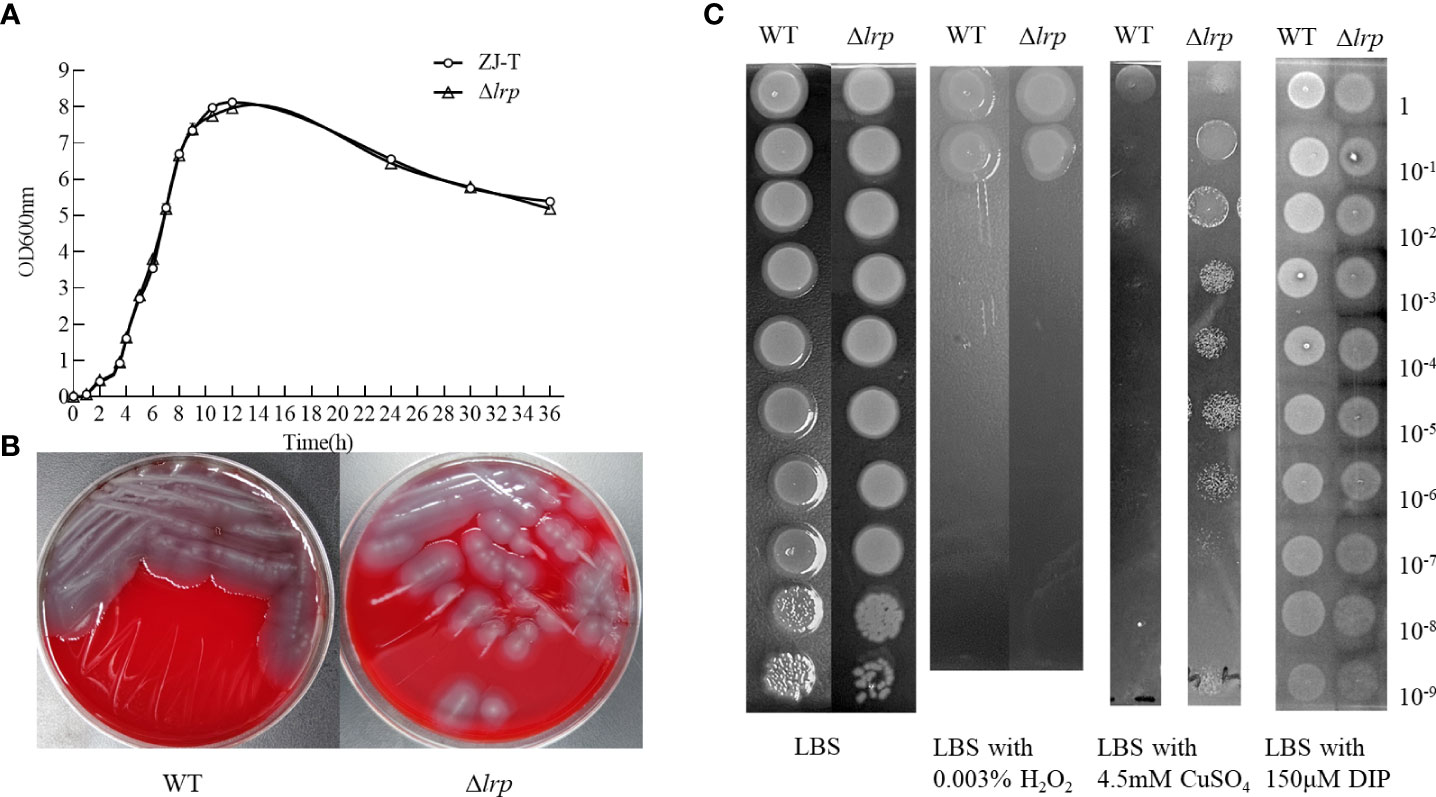
Figure 3 Growth, hemolysis, iron uptake capacity, copper ion and H2O2 tolerance of the WT and Δlrp strains. The growth curve of WT and (A), and the hemolytic activity on sheep blood agar plate (B), as well as their sensitivity of them to H2O2, CuSO4, and DIP on the LBS agar plate (C).
Neither the WT nor Δlrp strains produced hemolytic circles on the Mueller-Hinton blood agar plate (Figure 3B). In the absence of lrp, there was no significant difference in the growth of V. alginolyticus on either the LBS plate or on the LBS plate with 150 μM DIP or with 0.003% H2O2 (Figure 3C). As shown in Figure 3C, there was a 105-fold difference in survival rate between the WT and the lrp mutant strains on the LBS agar plate containing 4.5 mM CuSO4. Resistance of V. alginolyticus to copper ions was strengthened when lrp was absent (Figure 3C).
3.3. Absence of lrp increased the motility, biofilm formation, and extracellular secretion activity of V. alginolyticus
Both the WT and Δlrp strains showed swimming motility on the 0.3% agar LBS plates, but no differences in extension diameter were observed. However, the swarming motility of the Δlrp strain on the 1.5% agar LBS plates was significantly greater than the WT (Figure 4A and Figure 4B).
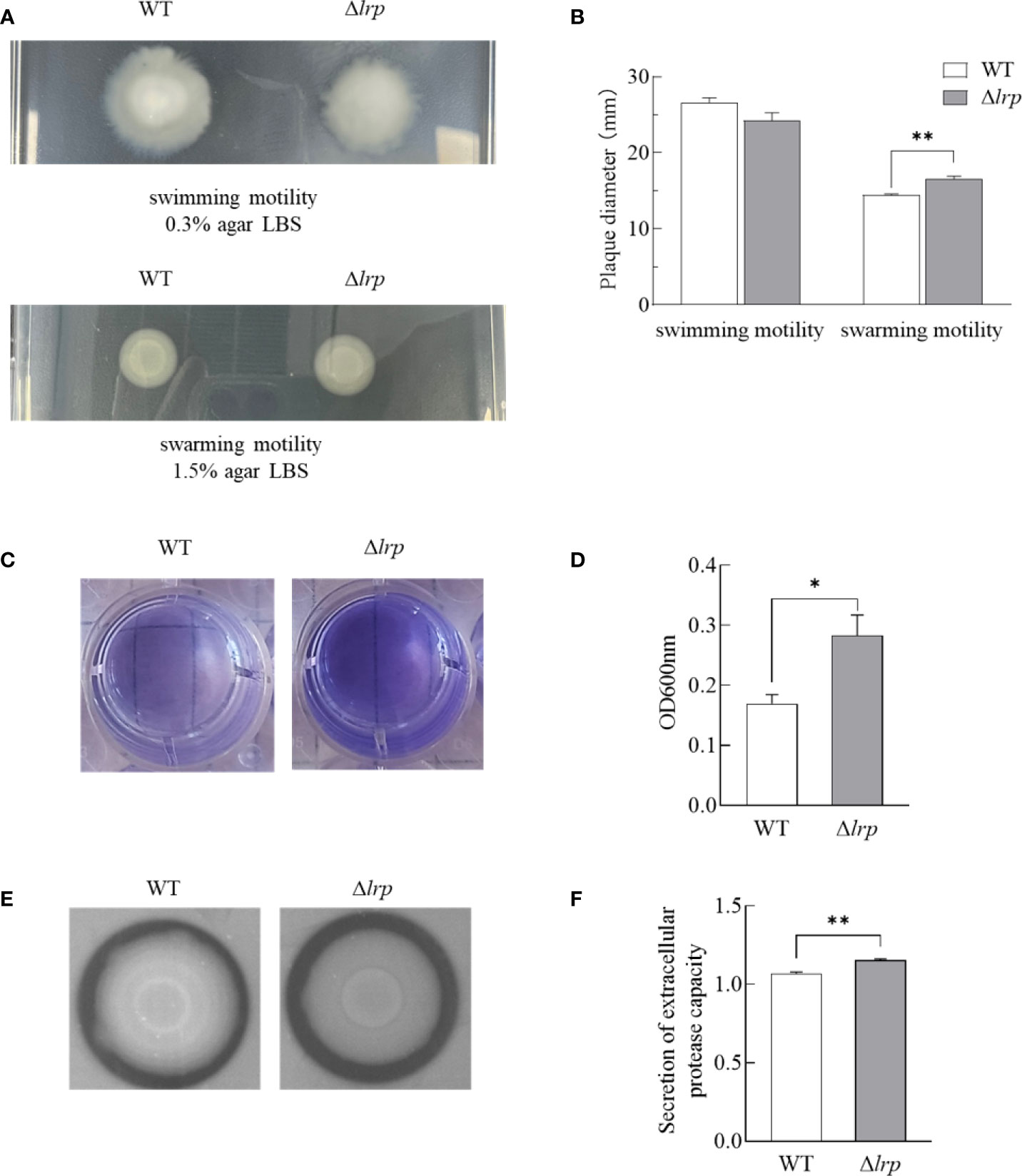
Figure 4 Motility, biofilm formation and extracellular protease secretion activity of WT and Δlrp strains. (A): the growth of the strains on the 0.3% and 1.5% agar LBS plate; (B): the colony diameter statistical analysis; (C): dissolved crystal violet-stained biofilm; (D): the absorbance value of biofilm staining; (E): strains growth on the skim milk plate; (F): graph of extracellular protease secretion capacity, vertical coordinate is the ratio of outer circle diameter to inner circle diameter. (**, P<0.01; *, P<0.05).
The biofilm circles were stained with crystal violet and eluted with acetic acid (Figure 4C). The purple biofilm precipitation could be seen at the bottom of the culture wells. The mean amount of Δlrp strain biofilm formed was significantly greater than the WT strain (Figure 4D).
The ratio of the transparent proteolytic circle diameter to the colony diameter indicates the capacity for extracellular protease secretion (Figure 4E). The secretion activity of Δlrp was significantly greater than that of WT (Figure 4F).
3.4. No significant effect of lrp on antibiotic resistance of V. alginolyticus
The bacterial inhibition circles produced around the 23 antibiotic paper tablets revealed that the absence of lrp did not significantly change the antibiotic resistance of V. alginolyticus. The results showed that the WT and Δlrp strains were resistant to 13 antibiotics, including rifampicin, amoxicillin, sulfafurazole, medimycin, cephalexin, enrofloxacin, furazolidone, compound sulfamethoxazol, streptomycin, cefazolin, tetracycline, ampicillin, and vancomycin. In addition, the absence of lrp did not alter the susceptibility of V. alginolyticus to 10 antibiotics, including claricid, doxycycline, florfenicol, tobramycin, ciprofloxacin, gentamicin, erythromycin, chloramphenicol, ofloxacin, and norfloxacin (Table S1).
3.5. Enhanced virulence of the lrp mutant strain to zebrafish
The WT and Δlrp strains were used to challenge experimental zebrafish. The cumulative zebrafish mortality rates in the WT group were 31.11%, 37.78%, 40.00%, and 40.00% for 24 h, 48 h, 72 h, and 96 h after intraperitoneal injection, respectively. The mortality of zebrafish after injection with Δlrp appeared to be greater than for the WT, with cumulative mortality rates of 64.44%, 66.67%, 68.89%, and 68.89% for 24 h, 48 h, 72 h, and 96 h after intraperitoneal injection, respectively (Figure 5). The mortality difference between the WT and Δlrp groups was statistically significant according to a Log Rank (Mantel-Cox) analysis. The results of the challenge experiment showed that the deletion of lrp enhanced the virulence of V. alginolyticus in zebrafish, and that lrp may negatively regulate V. alginolyticus virulence.
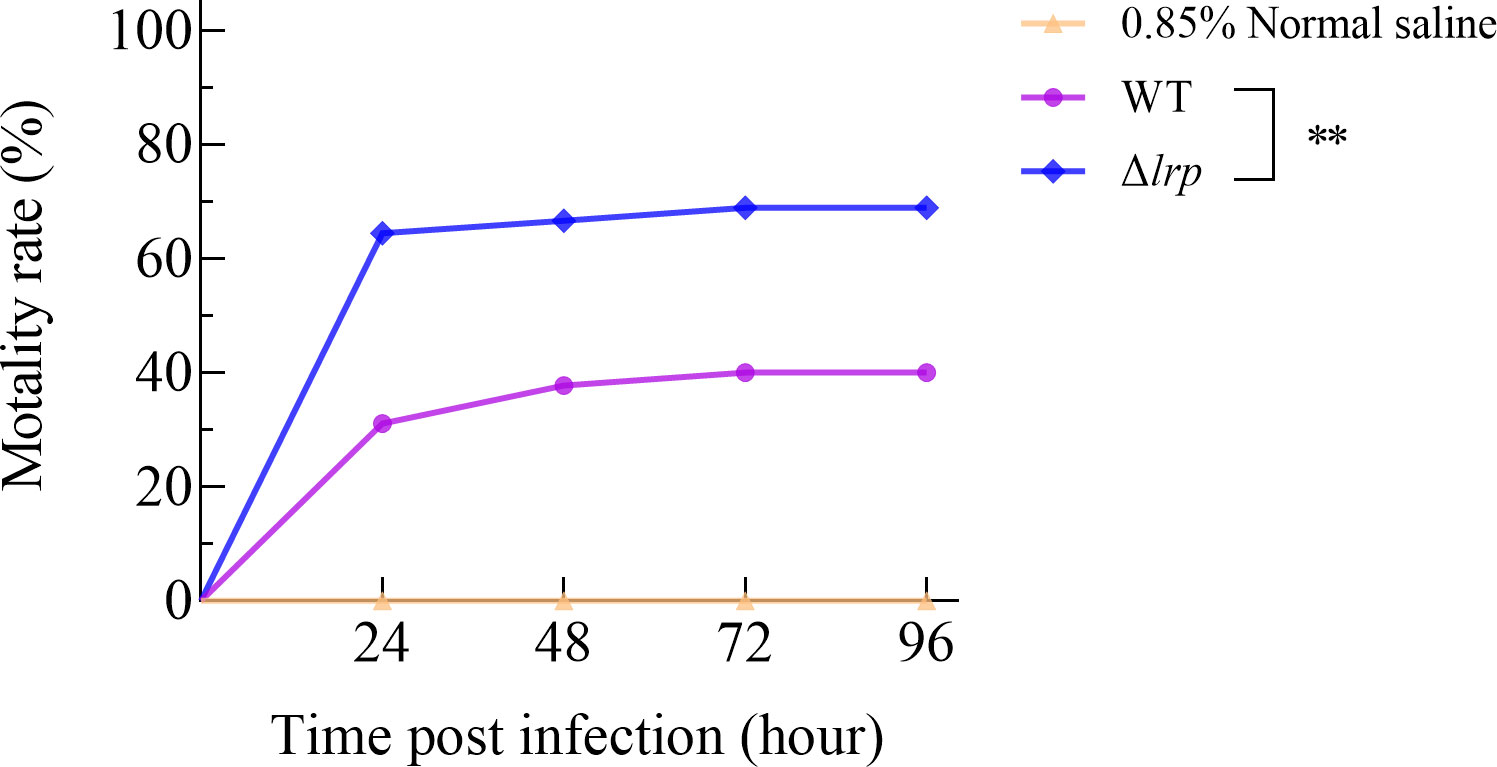
Figure 5 Cumulative mortality of zebrafish after injection of WT and Δlrp strains. Cumulative mortality was recorded every 24 h. Differences in cumulative mortalities were analyzed by Log-rank (Mantel-Cox) test (**, P<0.01).
3.6. The expression of lrp was impacted by various physicochemical factors and was gradually downregulated
In general, the expression of lrp showed a gradual decrease with increasing growth under the same cultural conditions. Compared with the control group (pH 7), the expression of lrp increased at pH 5 after 2 h (3.43-fold, P < 0.001) and 10 h (1.59-fold, P < 0.001) (Figure 6A). At the same incubation time, lrp was expressed most at pH 5, followed by pH 7 and pH 9. Meanwhile, the pH 9 group showed a higher lrp expression level (P < 0.01) at 10 h (Figure 6A). The mRNA of lrp increased at a temperature of 22°C at 2 h (3.47-fold, P < 0.01) and 10 h (1.52-fold, P < 0.05) but decreased at a temperature of 37°C at 2 h (1.75-fold, P < 0.05); at 6 h (3.84-fold, P < 0.001); and at 10 h (2.13-fold, P < 0.001). Culture conditions at a temperature of 37°C could inhibit the expression of lrp (Figure 6B). The expression of lrp was significantly increased in the 5% NaCl group at 2 h (1.78-fold, P < 0.01), but significantly decreased at 6 h (P < 0.05) and 10 h (P < 0.01). In the 1% NaCl group, its expression was significantly decreased at 6 h (2.63-fold, P < 0.001) but increased at 10 h (P < 0.05) (Figure 6C).
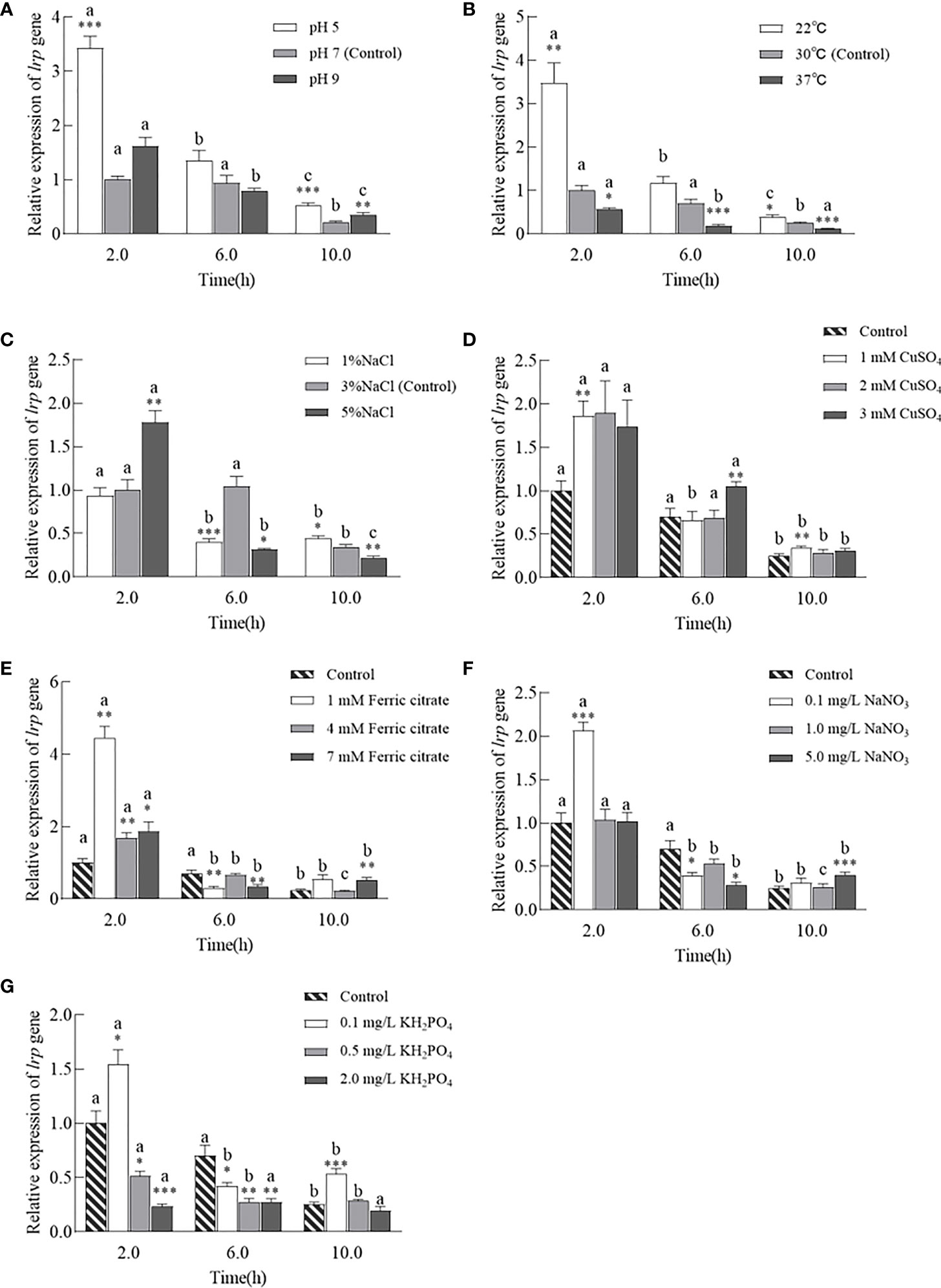
Figure 6 Expression profiles of WT strains under 7 incubate conditions. Expression levels of strains cultured in LBS with different pH (A), temperature (B), salinity (C), and addition of different concentrations of copper ion (D), iron ion (E), nitrate (F), phosphate (G). Data are expressed as means ± SE relative to control group. Significant differences between control group and treatment groups at the same growth period are indicated with asterisks (*, P<0.05; **, P<0.01; ***, P<0.001). Take strains with the same culture conditions as a group and use different letters to indicate significant differences in the growth period of the same group (P<0.05).
lrp mRNA was generally induced by CuSO4. The 1 mM CuSO4 group had a significantly higher lrp mRNA expression than the control when cultured for 2 h and 10 h (1.86-fold and 1.36-fold, P < 0.01, respectively). The expression of lrp increased at 6 h when the strain was grown in 3 mM CuSO4 (1.49-fold, P < 0.01) (Figure 6D). The bacterial cultures with added Fe3+ (1 mM, 4 mM, and 7mM) showed relatively higher expression at 2 h compared with the control group, with 4.46-fold (P < 0.01), 1.69-fold (P < 0.01), and 1.87-fold (P < 0.05) increases, respectively. Meanwhile, under the same conditions, lrp was usually most expressed at 2 h (Figure 6E). The expression of lrp with the addition of 1 mM Fe3+ and 7 mM Fe3+ were significantly lower than the expression of lrp at 6 h (P < 0.01). The 7 mM Fe3+ group increased lrp transcription at 10 h (2.03-fold, P < 0.01) (Figure 6E). With the same concentration of NaNO3, the expression of lrp was greatest at 2 h, and was significantly increased by the addition of 0.1 mg/L NaNO3 (2.07-fold, P < 0.001). Appropriate concentrations of NaNO3 decreased lrp expression at 6 h (0.1/5.0 mg/L NaNO3, P < 0.05), while the expression of lrp was increased at 10 h in 5.0 mg/L NaNO3 (1.59-fold, P < 0.001) (Figure 6F). The lrp expression of WT in LBS generally decreased with increasing concentrations of KH2PO4, except that the 0.1 mg/L KH2PO4 group significantly increased lrp expression at 2 h (1.55-fold, P < 0.05) and 10 h (2.10-fold, P < 0.001). The lrp expression of the 0.5 mg/L and 2.0 mg/L groups decreased at 2 h and 6 h, respectively (Figure 6G).
4. Discussion
In this study, a lrp mutant strain of V. alginolyticus was constructed to explore the role of Lrp and its biological properties and virulence. Biophenotype characterization revealed that the deletion of lrp did not significantly affect growth, iron absorption, radical oxygen resistance, hemolytic activity, antibiotic resistance, or swimming motility but increased swarming motility, biofilm formation, secretion of extracellular protease, and copper ion resistance. The virulence of V. alginolyticus to zebrafish was significantly enhanced after lrp deletion. lrp expression was affected by many physicochemical factors, including pH, temperature, salinity, and the presence of copper sulfate, ferric citrate, sodium nitrate, and potassium dihydrogen phosphate.
Bacteria move in highly viscous environments using their flagella, which help them to invade and colonize their hosts (McCarter and Silverman, 1990; Josenhans and Suerbaum, 2002). Many genes have been associated with bacterial motility (Yuan et al., 2022). Liu et al. (2021) showed that the RNA binding protein, CsrA, activates swarming motility by enhancing the expression of lateral flagellum-related genes. Deleting the quorum sensing-regulated extracellular protein gene pep leads to reduced motility and failure to generate lateral flagella (Cao et al., 2011). Biofilm formation is encouraged by bacterial spores and flagella (Yildiz and Visick, 2009) and helps bacteria to colonize of the host’s interior or the surface of aquatic plankton (Tamplin et al., 1990). Motility therefore plays a crucial role in biofilm formation, pathogenicity, and environmental adaptation in many bacteria (Gu et al., 2019a). The lateral flagella support bacterial swarming motility over surfaces or through viscous environments (McCarter, 2004). Gu et al. (2019a) reported that a GntR family transcription factor may increase swarming motility by enhancing the transcription of lateral flagella genes in Vibrio parahaemolyticus. The flagellin-homologous proteins (FHPs) in V. vulnificus could enhance the exopolysaccharide-enriched biofilm matrix through excretion to the extracellular milieu. Therefore, the presence of FHPs involved in biofilm enhancement could promote the pathogenicity of bacteria (Jung et al., 2019). In this study, bacterial swimming motility was not affected by lrp deletion, while swarming motility was significantly increased. The absence of lrp may induce the flagella to move when crossing surfaces or in sticky environments. Lrp may affect biofilm formation and swarming motility by regulating FHPs secreted into the extracellular environment. Changes in biofilm formation and cell motility may lead to enhanced virulence of V. alginolyticus in zebrafish. Therefore, because of the changes in motility and biofilm formation after lrp deletion found in this study, we hypothesize that lrp may affect the expression of the flagella gene in V. alginolyticus to regulate its swarming motility, and so influence biofilm formation and lead to changes in virulence.
Extracellular protease secretion activity also increased after deletion of the lrp gene. Extracellular proteases are pathogenic factors and play an essential role in the infection process of V. alginolyticus (Watnick et al., 2001). Extracellular production of the alkaline serine protease has been reported to be lethal to fish (Lee et al., 1997), and the alkaline serine protease gene asp is regulated by the quorum sensing system, resulting in changes in virulence (Rui et al., 2009). The response regulator protein LuxO has multiple roles in regulating extracellular products, and the extracellular protease activity was significantly enhanced after luxO deletion (Wang et al., 2007). The results of this study are similar to those described above. After lrp mutation, extracellular protease secretory activity was significantly increased, as were swarming motility and biofilm formation. Further research is needed to determine whether Lrp can regulate virulence by inducing changes in extracellular protease activity, and interact with biofilm formation or flagellar gene expression.
Copper, in its two oxidation states, is involved in essential redox reactions and promotes the formation of reactive oxygen species, leading to cellular damage (Karlin, 1993). Copper efflux is essential to bacterial pathogens for host colonization and virulence and is primary prerequisite for Vibrio intracellular phases and cytotoxicity to hemocytes (Kong et al., 2015; Vanhove et al., 2016). Kong et al. (2015) found that the adhesion capacity of V. alginolyticus was significantly reduced after treatment with copper ions (50 mg/L). In V. cholerae, copper resistance proteins were reduced in nqr (electrogenic, sodium ion-translocating NADH: quinone oxidoreductase gene) deletion strains, resulting in diminished copper resistance (Toulouse et al., 2018). Vanhove et al. (2016) found that the primary mechanism of copper ion resistance was to prevent the host phagocytes killing pathogens, and to induce the cytolysis of host immune cells and host colonization. In this study, the mutant strain Δlrp was less damaged by copper ion stress than the WT, indicating that its sensitivity to copper ions was reduced after the mutation ameliorated the toxicity of copper ions. It appears that Lrp may regulate the expression of copper resistance proteins in V. alginolyticus, decreasing its sensitivity to copper ions and enhancing its adhesion capacity.
Physicochemical factors can influence bacterial gene expression, thus regulating the biological properties relating to virulence. The attachment of V. alginolyticus was affected by temperature, which changed the expression of rstA and rstB. These two genes have a regulatory role in bacterial adhesion, biofilm production, motility, and hemolysis (Huang et al., 2018a). Appropriate salinity levels facilitated the adherence of V. alginolyticus to the skin mucus of Sparus aurata, and V. alginolyticus showed a higher adhesion rate in seawater (3.5% salinity) than in PBS and saline solution (Balebona et al., 1995). Studies have shown that the adhesion ability of V. alginolyticus was impaired under heavy metal (50 mg/L Cu2+, 100 mg/L Pb2+, 50 mg/L Hg2+) and acidic stresses (Kong et al., 2015). VspR is the primary regulator in V. alginolyticus, and enables the formation/maintenance of biofilms by directly sensing phosphate levels (Hsieh et al., 2022). In this study, the expression of lrp was generally up-regulated by different physicochemical factors, and lrp usually showed its highest expression after 2 h of culture time. Many combinations of adverse environmental stresses could increase the expression of lrp (pH 5, 22°C, 5% NaCl, 1/4/7 mM Fe3+, 0.1/5 mg/L NO3-, and 0.1 mg/L at 2 h; and pH 5, pH 9, 22°C, 1% NaCl, 1 mM Cu2+, 7 mM Fe3+, 5 mg/L , and 0.1 mg/L at 10 h). The virulence of V. alginolyticus was reduced in the presence of lrp, indicating that increased lrp expression of ZJ-T in different conditions might also decrease its virulence. Some conditions could decrease the transcription level of lrp at 6 h, including 37°C, 1% NaCl, 5% NaCl, 1/7 mM iron ions, 0.1/5 mg/L nitrate, 0.1/0.5/2 mg/L phosphate. The significant decrease in lrp expression indicated that the virulence of V. alginolyticus might increase over a similar time in these culture conditions. These culture conditions may lead to changes in biological characteristics similar to those found in Δlrp, such as motility, biofilm formation, and enhanced extracellular protease secretion. Therefore, various physicochemical factors that change the expression of lrp probably contribute to variations in adhesion, colonization, and toxin release of V. alginolyticus, thereby leading to changes in virulence. Especially after 2 h and 10 h of cultivation, the physicochemical factors tested tend to promote lrp expression, thereby probably inhibiting the virulence of V. alginolyticus. However, whether the increased expression of lrp in adverse environments is related to the maintenance of ZJ-T homeostasis or is involved in regulating the secretion of factors related to flagella, clearly requires further study.
5. Conclusions
After lrp deletion, V. alginolyticus displayed dramatically increased virulence, swarming motility, biofilm formation, extracellular protease activity, and resistance to copper ions. The expression of lrp showed a gradual decrease with time in any one cultural environment. After 2 h incubation time, cultivation with pH 5, 22°C, 5% NaCl, 1 mM Cu2+, 1/4/7 mM Fe3+, 0.1 mg/L , 0.1 mg/L increased lrp expression, and at 37°C, 0.5 mg/L and 2 mg/L lrp expression was inhibited. After 6 h of culture at 37°C, 1/5% NaCl, 1/7 mM Fe3+, 0.1/5 mg/L , 0.1/0.5/2 mg/L the expression of lrp was inhibited. lrp expression was promoted after 10 h of culture at pH 5/9, 22°C, 1% NaCl, 1 mM Cu2+, 7 mM Fe3+, 5 mg/L , and 0.1 mg/L , but was inhibited at 37°C and 5% NaCl. Therefore, Lrp was strongly affected by various physicochemical factors and may reduce the virulence of V. alginolyticus by inhibiting biofilm formation, swarming motility, and extracellular protease secretion. These results further the study of virulence regulation in V. alginolyticus. Future research should focus on: (1) how lrp affects motility, biofilm formation, and extracellular protease secretion, and ultimately the virulence of V. alginolyticus at the gene regulation level; and (2) the effects of various levels of virulence of V. alginolyticus on aquatic animals under different environmental conditions, including temperature, pH, salinity, and the presence of copper sulfate, ferric citrate, sodium nitrate, and potassium dihydrogen phosphate. The results of such work will help to establish precise virulence control strategies based on environmental regulation.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, CP016224.1.
Ethics statement
The animal study was reviewed and approved by the study protocol was approved by the Animal Care and Use Committee of the South China Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences and performed in accordance with institutional regulations and guidelines.
Author contributions
WS: Completed experiment, Data curation, investigation, software, and writing. YD: Funding acquisition, conceptualization, resources, writing-review, and editing. SZ: Aided experiment conduct and validation, data collation. SG: Visualization, data curation, and validation. JF: Funding acquisition, project administration, supervision, writing-review and editing, validation. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Central Public-Interest Scientific Institution Basal Research Fund, CAFS (2022GH03), the Guangzhou Science and Technology Program (202201010162), the National Natural Science Foundation of China (NSFC) (31902415), Central Public-interest Scientific Institution Basal Research Fund, South China Sea Fisheries Research Institute, CAFS (2021SD15), the Natural Science Fund of Guangdong (2019A1515011833), and the China Agriculture Research System (CARS-48).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.1042899/full#supplementary-material
References
Abdallah F. B., Lagha R., Ellafi A., Namane A., Rousselle J.-C., Lenormand P., et al. (2012). Identification of outer membrane proteins altered in response to UVC-radiation in vibrio parahaemolyticus and vibrio alginolyticus. Indian J. Microbiol. 52, 660–665. doi: 10.1007/s12088-012-0299-2
Alice A. F., Crosa J. H. (2012). The TonB3 system in the human pathogen vibrio vulnificus is under the control of the global regulators lrp and cyclic AMP receptor protein. J. Bacteriol. 194, 1897–1911. doi: 10.1128/JB.06614-11
Baker-Austin C., Oliver J. D., Alam M., Ali A., Waldor M. K., Qadri F., et al. (2018). Vibrio spp. infections. Nat. Rev. Dis. Primer 4, 8. doi: 10.1038/s41572-018-0005-8
Balebona M. C., Moriñigo M. A., Faris A., Krovacek K., Månsson I., Bordas M. A., et al. (1995). Influence of salinity and pH on the adhesion of pathogenic vibrio strains to sparus aurata skin mucus. Aquaculture 132, 113–120. doi: 10.1016/0044-8486(94)00376-Y
Brinkman A. B., Ettema T. J. G., de Vos W. M., van der Oost J. (2003). The lrp family of transcriptional regulators. Mol. Microbiol. 48, 287–294. doi: 10.1046/j.1365-2958.2003.03442.x
Cao X., Wang Q., Liu Q., Rui H., Liu H., Zhang Y. (2011). Identification of a luxo-regulated extracellular protein pep and its roles in motility in vibrio alginolyticus. Microb. Pathog. 50, 123–131. doi: 10.1016/j.micpath.2010.12.003
Chen K.-Y., Rathod J., Chiu Y.-C., Chen J.-W., Tsai P.-J., Huang I.-H. (2019). The transcriptional regulator lrp contributes to toxin expression, sporulation, and swimming motility in clostridium difficile. Front. Cell. Infect. Microbiol. 9. doi: 10.3389/fcimb.2019.00356
Chen S., Rosner M. H., Calvo J. M. (2001). Leucine-regulated self-association of leucine-responsive regulatory protein (lrp) from escherichia coli. J. Mol. Biol. 312, 625–635. doi: 10.1006/jmbi.2001.4955
Ciaccia P. N., Ramachandran R., Chattoraj D. K. (2018). A requirement for global transcription factor lrp in licensing replication of vibrio cholerae chromosome 2. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.02103
Davis B. J. K., Jacobs J. M., Davis M. F., Schwab K. J., DePaola A., Curriero F. C. (2017). Environmental determinants of vibrio parahaemolyticus in the Chesapeake bay. Appl. Environ. Microbiol. 83, e01147–e01117. doi: 10.1128/AEM.01147-17
DeAngelis C. M., Saul-McBeth J., Matson J. S. (2018). Vibrio responses to extracytoplasmic stress. Environ. Microbiol. Rep. 10, 511–521. doi: 10.1111/1758-2229.12693
Deng Y., Chen C., Zhao Z., Huang X., Yang Y., Ding X. (2016a). Complete genome sequence of vibrio alginolyticus ZJ-T. Genome Announc. 4, e00912–e00916. doi: 10.1128/genomeA.00912-16
Deng Y., Chen C., Zhao Z., Zhao J., Jacq A., Huang X., et al. (2016b). The RNA chaperone hfq is involved in colony morphology, nutrient utilization and oxidative and envelope stress response in vibrio alginolyticus. PloS One 11, e0163689. doi: 10.1371/journal.pone.0163689
Fahmy N. M., Hamed E. S. A. E. (2021). Isolation and characterization of vibrio alginolyticus strain Hat3 causing skin ulceration disease in cultured Sea cucumber holothuria atra (jaeger 1833). Egypt. J. Aquat. Res. 48, 75–81. doi: 10.1016/j.ejar.2021.11.007
Gaüzère B.-A., Chanareille P., Vandroux D. (2016). Septicémie À vibrio alginolyticus au décours d’une presque noyade À la réunion (océan indien). Bull. Société Pathol. Exot. 109, 151–154. doi: 10.1007/s13149-016-0505-2
Gu D., Meng H., Li Y., Ge H., Jiao X. (2019a). A GntR family transcription factor (VPA1701) for swarming motility and colonization of vibrio parahaemolyticus. Pathogens 8, 235. doi: 10.3390/pathogens8040235
Gu D., Zhang J., Hao Y., Xu R., Zhang Y., Ma Y., et al. (2019b). Alternative sigma factor RpoX is a part of the RpoE regulon and plays distinct roles in stress responses, motility, biofilm formation, and hemolytic activities in the marine pathogen vibrio alginolyticus. Appl. Environ. Microbiol. 85, e00234–e00219. doi: 10.1128/AEM.00234-19
Halpern B. S., Frazier M., Afflerbach J., Lowndes J. S., Micheli F., O’Hara C., et al. (2019). Recent pace of change in human impact on the world’s ocean. Sci. Rep. 9, 11609. doi: 10.1038/s41598-019-47201-9
Hart B. R., Mishra P. K., Lintner R. E., Hinerman J. M., Herr A. B., Blumenthal R. M. (2011). Recognition of DNA by the helix-Turn-Helix global regulatory protein lrp is modulated by the amino terminus. J. Bacteriol. 193, 3794–3803. doi: 10.1128/JB.00191-11
Hoe N. P., Goguen J. D. (1993). Temperature sensing in yersinia pestis: Translation of the lcrf activator protein is thermally regulated. J. Bacteriol. 175, 7901–7909. doi: 10.1128/jb.175.24.7901-7909.1993
Ho Y.-C., Hung F.-R., Weng C.-H., Li W.-T., Chuang T.-H., Liu T.-L., et al. (2017). Lrp, a global regulator, regulates the virulence of vibrio vulnificus. J. Biomed. Sci. 24, 54. doi: 10.1186/s12929-017-0361-9
Hsieh M.-L., Kiel N., Jenkins L. M. M., Ng W.-L., Knipling L., Waters C. M., et al. (2022). The vibrio cholerae master regulator for the activation of biofilm biogenesis genes, vpsr, senses both cyclic di-gmp and phosphate. Nucleic Acids Res. 50, 4484–4499. doi: 10.1093/nar/gkac253
Huang X., Chen C., Ren C., Li Y., Deng Y., Yang Y., et al. (2018b). Identification and characterization of a locus putatively involved in colanic acid biosynthesis in vibrio alginolyticus zj-51. Biofouling 34, 1–14. doi: 10.1080/08927014.2017.1400020
Huang L., Huang L., Zhao L., Qin Y., Su Y., Yan Q. (2019). The regulation of oxidative phosphorylation pathway on vibrio alginolyticus adhesion under adversities. MicrobiologyOpen 8, e00805. doi: 10.1002/mbo3.805
Huang L., Hu J., Su Y., Qin Y., Kong W., Ma Y., et al. (2015). Identification and characterization of three vibrio alginolyticus non-coding rnas involved in adhesion, chemotaxis, and motility processes. Front. Cell. Infect. Microbiol. 5. doi: 10.3389/fcimb.2015.00056
Huang L., Xu W., Su Y., Zhao L., Yan Q. (2018a). Regulatory role of the rstb-rsta system in adhesion, biofilm production, motility, and hemolysis. MicrobiologyOpen 7, e00599. doi: 10.1002/mbo3.599
Huisman T. T., Bakker D., Klaasen P., de Graaf F. K. (1994). Leucine-responsive regulatory protein, Is1 insertions, and the negative regulator faea control the expression of the fae (k88) operon in escherichia coli. Mol. Microbiol. 11, 525–536. doi: 10.1111/j.1365-2958.1994.tb00333.x
Hussa E. A., Casanova-Torres A. M., Goodrich-Blair H. (2015). The global transcription factor lrp controls virulence modulation in xenorhabdus nematophila. J. Bacteriol. 197, 3015–3025. doi: 10.1128/JB.00272-15
Josenhans C., Suerbaum S. (2002). The role of motility as a virulence factor in bacteria. Int. J. Med. Microbiol. IJMM 291, 605–614. doi: 10.1078/1438-4221-00173
Jung Y.-C., Lee M.-A., Lee K.-H. (2019). Role of flagellin-homologous proteins in biofilm formation by pathogenic vibrio species. mBio 10, e01793–e01719. doi: 10.1128/mBio.01793-19
Karlin K. D. (1993). Metalloenzymes, structural motifs, and inorganic models. Science 261, 701–708. doi: 10.1126/science.7688141
Kong W., Huang L., Su Y., Qin Y., Ma Y., Xu X., et al. (2015). Investigation of possible molecular mechanisms underlying the regulation of adhesion in vibrio alginolyticus with comparative transcriptome analysis. Antonie Van Leeuwenhoek 107, 1197–1206. doi: 10.1007/s10482-015-0411-9
Lee K. K., Yu S. R., Liu P. C. (1997). Alkaline serine protease is an exotoxin of vibrio alginolyticus in kuruma prawn, penaeus japonicus. Curr. Microbiol. 34, 110–117. doi: 10.1007/s002849900153
Le Roux F., Binesse J., Saulnier D., Mazel D. (2007). Construction of a vibrio splendidus mutant lacking the metalloprotease gene vsm by use of a novel counterselectable suicide vector. Appl. Environ. Microbiol. 73, 777–784. doi: 10.1128/AEM.02147-06
Lin W., Kovacikova G., Skorupski K. (2007). The quorum sensing regulator hapr downregulates the expression of the virulence gene transcription factor apha in vibrio cholerae by antagonizing lrp- and vpsr-mediated activation. Mol. Microbiol. 64, 953–967. doi: 10.1111/j.1365-2958.2007.05693.x
Liu B., Gao Q., Zhang X., Chen H., Zhang Y., Sun Y., et al. (2021). Csra regulates swarming motility and carbohydrate and amino acid metabolism in vibrio alginolyticus. Microorganisms 9, 2383. doi: 10.3390/microorganisms9112383
Liu H., Wang Q., Liu Q., Cao X., Shi C., Zhang Y. (2011). Roles of hfq in the stress adaptation and virulence in fish pathogen vibrio alginolyticus and its potential application as a target for live attenuated vaccine. Appl. Microbiol. Biotechnol. 91, 353–364. doi: 10.1007/s00253-011-3286-3
Liu R., Zheng R., Liu G., Sun C. (2020). The cyclic lipopeptides suppress the motility of vibrio alginolyticus Via targeting the na+ -driven flagellar motor component motx. Environ. Microbiol. 22, 4424–4437. doi: 10.1111/1462-2920.15144
Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Li X. C., Xiang Z. Y., Xu X. M., Yan W. H., Ma J. M. (2009). Endophthalmitis caused by vibrio alginolyticus. J. Clin. Microbiol. 47, 3379–3381. doi: 10.1128/JCM.00722-09
Li X., Zhang C., Wei F., Yu F., Zhao Z. (2021). Bactericidal activity of a holin-endolysin system derived from vibrio alginolyticus phage Hh109. Microb. Pathog. 159, 105135. doi: 10.1016/j.micpath.2021.105135
Luo G., Huang L., Su Y., Qin Y., Xu X., Zhao L., et al. (2016). Flra, flrb and flrc regulate adhesion by controlling the expression of critical virulence genes in vibrio alginolyticus. Emerg. Microbes Infect. 5, 1–11. doi: 10.1038/emi.2016.82
McCarter L. L. (2004). Dual flagellar systems enable motility under different circumstances. Microb. Physiol. 7, 18–29. doi: 10.1159/000077866
McCarter L., Silverman M. (1990). Surface-induced swarmer cell differentiation of vibrio parahaemoiyticus. Mol. Microbiol. 4, 1057–1062. doi: 10.1111/j.1365-2958.1990.tb00678.x
Mohamad N., Mohd Roseli F. A., Azmai M. N. A., Saad M. Z., Md Yasin I. S., Zulkiply N. A., et al. (2019). Natural concurrent infection of vibrio harveyi and v. alginolyticus in cultured hybrid groupers in Malaysia. J. Aquat. Anim. Health 31, 88–96. doi: 10.1002/aah.10055
Nguyen A. N., Disconzi E., Charrière G. M., Destoumieux-Garzón D., Bouloc P., Le Roux F., et al. (2018). csrB gene duplication drives the evolution of redundant regulatory pathways controlling expression of the major toxic secreted metalloproteases in vibrio tasmaniensis LGP32. mSphere 3, e00582–e00518. doi: 10.1128/mSphere.00582-18
Nou X., Skinner B., Braaten B., Blyn L., Hirsch D., Low D. (1993). Regulation of pyelonephritis-associated pili phase-variation in escherichia coli: Binding of the papi and the lrp regulatory proteins is controlled by dna methylation. Mol. Microbiol. 7, 545–553. doi: 10.1111/j.1365-2958.1993.tb01145.x
Oberbeckmann S., Wichels A., Wiltshire K. H., Gerdts G. (2011). Occurrence of vibrio parahaemolyticus and vibrio alginolyticus in the German bight over a seasonal cycle. Antonie Van Leeuwenhoek 100, 291–307. doi: 10.1007/s10482-011-9586-x
Oliver J. D., Warner R. A., Cleland D. R. (1983). Distribution of vibrio vulnificus and other lactose-fermenting vibrios in the marine environment. Appl. Environ. Microbiol. 45, 985–998. doi: 10.1128/aem.45.3.985-998.1983
O’Toole G. A. (2011). Microtiter dish biofilm formation assay. J. Vis. Exp. JoVE 47, 2437. doi: 10.3791/2437
Ren C., Hu C., Jiang X., Sun H., Zhao Z., Chen C., et al. (2013). Distribution and pathogenic relationship of virulence associated genes among vibrio alginolyticus from the mariculture systems. Mol. Cell. Probes 27, 164–168. doi: 10.1016/j.mcp.2013.01.004
Rui H., Liu Q., Wang Q., Ma Y., Liu H., Shi C., et al. (2009). Role of alkaline serine protease, asp, in vibrio alginolyticus virulence and regulation of its expression by luxo-luxr regulatory system. J. Microbiol. Biotechnol. 19, 431–438. doi: 10.4014/jmb.0807.404
Swift S., Lynch M. J., Fish L., Kirke D. F., Tomás J. M., Stewart G. S. A. B., et al. (1999). Quorum sensing-dependent regulation and blockade of exoprotease production in aeromonas hydrophila. Infect. Immun. 67, 5192–5199. doi: 10.1128/IAI.67.10.5192-5199.1999
Tamplin M. L., Gauzens A. L., Huq A., Sack D. A., Colwell R. R. (1990). Attachment of vibrio cholerae serogroup O1 to zooplankton and phytoplankton of Bangladesh waters. Appl. Environ. Microbiol. 56, 1977–1980. doi: 10.1128/aem.56.6.1977-1980.1990
Thaw P., Sedelnikova S. E., Muranova T., Wiese S., Ayora S., Alonso J. C., et al. (2006). Structural insight into gene transcriptional regulation and effector binding by the Lrp/Asnc family. Nucleic Acids Res. 34, 1439–1449. doi: 10.1093/nar/gkl009
Toulouse C., Metesch K., Pfannstiel J., Steuber J. (2018). Metabolic reprogramming of vibrio cholerae impaired in respiratory NADH oxidation is accompanied by increased copper sensitivity. J. Bacteriol. 200, e00761–e00717. doi: 10.1128/JB.00761-17
Toyofuku M., Inaba T., Kiyokawa T., Obana N., Yawata Y., Nomura N. (2016). Environmental factors that shape biofilm formation. Biosci. Biotechnol. Biochem. 80, 7–12. doi: 10.1080/09168451.2015.1058701
Unemoto T., Hayashi M., Terao K. (1977). Lysis of halophilic vibrio alginolyticus and vibrio costicolus induced by chaotropic anions. Biochim. Biophys. Acta 500, 425–431. doi: 10.1016/0304-4165(77)90033-2
Usman A., Mohammed S., Mamo J. (2021). Production, optimization, and characterization of an acid protease from a filamentous fungus by solid-state fermentation. Int. J. Microbiol. 2021, 6685963. doi: 10.1155/2021/6685963
Val M.-E., Skovgaard O., Ducos-Galand M., Bland M. J., Mazel D. (2012). Genome engineering in vibrio cholerae: A feasible approach to address biological issues. PloS Genet. 8, e1002472. doi: 10.1371/journal.pgen.1002472
van der Woude M. W., Low D. A. (1994). Leucine-responsive regulatory protein and deoxyadenosine methylase control the phase variation and expression of the sfa and daa pili operons in escherichia coli. Mol. Microbiol. 11, 605–618. doi: 10.1111/j.1365-2958.1994.tb00340.x
Vanhove A. S., Rubio T. P., Nguyen A. N., Lemire A., Roche D., Nicod J., et al. (2016). Copper homeostasis at the host vibrio interface: Lessons from intracellular vibrio transcriptomics. Environ. Microbiol. 18, 875–888. doi: 10.1111/1462-2920.13083
Wang X. H., Leung K. Y. (2000). Biochemical characterization of different types of adherence of vibrio species to fish epithelial cells. Microbiol. Read. Engl. 146 (Pt 4), 989–998. doi: 10.1099/00221287-146-4-989
Wang Q., Liu Q., Cao X., Yang M., Zhang Y. (2008). Characterization of two tonb systems in marine fish pathogen vibrio alginolyticus: Their roles in iron utilization and virulence. Arch. Microbiol. 190, 595–603. doi: 10.1007/s00203-008-0407-1
Wang Q., Liu Q., Ma Y., Rui H., Zhang Y. (2007). LuxO controls extracellular protease, haemolytic activities and siderophore production in fish pathogen vibrio alginolyticus. J. Appl. Microbiol. 103, 1525–1534. doi: 10.1111/j.1365-2672.2007.03380.x
Watnick P. I., Lauriano C. M., Klose K. E., Croal L., Kolter R. (2001). The absence of a flagellum leads to altered colony morphology, biofilm development and virulence in vibrio cholerae O139. Mol. Microbiol. 39, 223–235. doi: 10.1046/j.1365-2958.2001.02195.x
Westrich J. R., Ebling A. M., Landing W. M., Joyner J. L., Kemp K. M., Griffin D. W., et al. (2016). Saharan Dust nutrients promote vibrio bloom formation in marine surface waters. Proc. Natl. Acad. Sci. U. S. A. 113, 5964–5969. doi: 10.1073/pnas.1518080113
Yang B., Zhai S., Li X., Tian J., Li Q., Shan H., et al. (2021). Identification of vibrio alginolyticus as a causative pathogen associated with mass summer mortality of the pacific oyster (crassostrea gigas) in China. Aquaculture 535, 736363. doi: 10.1016/j.aquaculture.2021.736363
Yao H., Liu J., Jiang X., Chen F., Lu X., Zhang J. (2021). Analysis of the clinical effect of combined drug susceptibility to guide medication for carbapenem-resistant klebsiella pneumoniae patients based on the Kirby-Bauer disk diffusion method. Infect. Drug Resist. 14, 79–87. doi: 10.2147/IDR.S282386
Yildiz F. H., Visick K. L. (2009). Vibrio biofilms: So much the same yet so different. Trends Microbiol. 17, 109–118. doi: 10.1016/j.tim.2008.12.004
Yokoyama K., Ishijima S. A., Clowney L., Koike H., Aramaki H., Tanaka C., et al. (2006). Feast/Famine regulatory proteins (ffrps): Escherichia coli lrp, asnc and related archaeal transcription factors. FEMS Microbiol. Rev. 30, 89–108. doi: 10.1111/j.1574-6976.2005.00005.x
Yuan B., Zhao L.-M., Zhuang Z.-X., Wang X.-R., Fu Q., Huang H.-B., et al. (2022). Transcriptomic and metabolomic insights into the role of the flgK gene in the pathogenicity of pseudomonas plecoglossicida to orange-spotted grouper (Epinephelus coioides). Zool. Res. 43, 952–965. doi: 10.24272/j.issn.2095-8137.2022.216
Keywords: Vibrio alginolyticus, lrp, biological characteristics, virulence regulation, physicochemical factors
Citation: Su W, Deng Y, Zang S, Gao S and Feng J (2022) The global role of Lrp in Vibrio alginolyticus and its response to diverse physicochemical factors. Front. Mar. Sci. 9:1042899. doi: 10.3389/fmars.2022.1042899
Received: 13 September 2022; Accepted: 06 December 2022;
Published: 19 December 2022.
Edited by:
Claudia R. Serra, University of Porto, PortugalReviewed by:
Min Yu, Ocean University of China, ChinaQingpi Yan, Jimei University, China
Dan Gu, Yangzhou University, China
Copyright © 2022 Su, Deng, Zang, Gao and Feng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yiqin Deng, eWlxaW5kZEAxMjYuY29t; Juan Feng, anVhbmZlbmdAc2NzZnJpLmFjLmNu
 Wenxiao Su
Wenxiao Su Yiqin Deng
Yiqin Deng Shujun Zang
Shujun Zang Si’ao Gao1,3
Si’ao Gao1,3 Juan Feng
Juan Feng
