- 1School of Environmental Science and Engineering, Nanjing Tech University, Nanjing, China
- 2State Key Laboratory of Estuarine and Coastal Research, East China Normal University, Shanghai, China
- 3Fujian Provincial Key Laboratory for Plant Eco-Physiology, School of Geographical Sciences, School of Carbon Neutrality Future Technology, Fujian Normal University, Fuzhou, China
Methane (CH4) is an important greenhouse gas with a global warming potential 28 – 34 times that of CO2 over the 100-year horizon. Denitrifying anaerobic methane oxidation (DAMO) is a recently discovered process that potentially represents an important CH4 sink globally. This process involves two possible pathways: the nitrite-dependent DAMO mediated by NC10 bacteria and the nitrate-dependent DAMO by ANME-2d archaea. Both are widely detected in freshwater and coastal habitats using molecular tools. However, the distributions of these two processes and the functional microorganisms and their interactions with other N cycling pathways are far from clear. In this review, we conducted a scientometric analysis on a co-citation network consisting of 835 references derived from 354 citing articles closely related to the distribution of DAMO in the environment. Through this analysis, we found that current studies focus more on freshwater systems than coastal systems, and ANME-2d archaea are generally under-studied compared to NC10 bacteria. The emerging research topics in this area include AMO processes coupled to alternative electron acceptors and their role as CH4 sinks. We further reviewed papers focusing on DAMO distribution in freshwater and coastal environments guided by the result of the scientometric analysis. Finally, we identified several areas that require further research and proposed future research including comparisons of DAMO with other N cycling pathways and environmental conditions in the context of the river-estuary-sea continuum.
1 Introduction
Methane (CH4) is an important greenhouse gas with a 100-year global warming potential 28-34 times that of CO2 (Dean et al., 2018). CH4 is estimated to be responsible for 0.5°C of increase in global average temperature during 2010-2019 compared to 1850-1900, contributing to 16% of global warming (IPCC, 2021). Therefore, controlling CH4 emissions is the key to mitigating the global greenhouse effect and global warming. In ecosystems, the microbe-mediated CH4 oxidizing processes are important sinks for global CH4 emission. Anaerobic methane oxidation (AMO) has long been discovered in marine sediments, which is typically coupled with sulfate reduction, removing 90% of marine CH4 emissions before they leave the sea (Knittel and Boetius, 2009). This process plays a critical role in controlling atmospheric CH4 concentrations globally. In recent years, a related process, denitrifying anaerobic methane oxidation (DAMO), has been observed, drawing great attention as a novel CH4 cycling pathway. DAMO is an anaerobic process that couples anaerobic CH4 oxidation with and reduction, making it one of the linkages between C and N cycles, and can potentially remove CH4 and reactive N. Therefore, a better understanding of the DAMO processes in natural and engineered ecosystems can help us reduce greenhouse gas emissions and mitigate eutrophication in aquatic environments.
DAMO was first observed in an enrichment culture after long-term incubation retrieved from eutrophic freshwater sediments in Netherland (Raghoebarsing et al., 2006). Since then, many studies have focused on DAMO in freshwater systems, including sediments of lakes (Deutzmann and Schink, 2011; Kojima et al., 2012), reservoirs (Han and Gu, 2013), paddy fields (Hu et al., 2014; Vaksmaa et al., 2016), and wetlands (Hu et al., 2014; Shen et al., 2015b). Their distributions in coastal marine environments, such as coastal rivers, estuaries, and adjacent seas, however, have not been examined as much. Scientometric reviews based on the statistical analysis of the bibliometric records of a research field can help understand the intellectual structures of the underlying field and identify emerging directions in the research. This process is increasingly facilitated by available science mapping tools such as CiteSpace (Chen, 2006), VosViewers (van Eck and Waltman, 2010), etc. They have been widely used in scientometric reviews in areas such as medicine (Chen et al., 2014), greenhouse emission (Wang et al., 2021), ecosystem health (Yang et al., 2019), etc. However, a scientometric analysis of the vast body of DAMO literature has not been reported to the best of our knowledge.
In light of this, we reviewed studies focusing on DAMO distributions and their affecting factors in freshwater and coastal marine ecosystems, compared the abundance and dominating microorganisms in these two types of systems, identified knowledge gaps, and provided an outlook for this area. This review intends to give a comprehensive overview of the current research on DAMO and to provide insights into factors that help shape the distribution of DAMO processes in freshwater and coastal marine ecosystems, and their potential in removing CH4 emission and reactive N in these systems.
The specific objectives of this review were to (1) provide a brief overview of the DAMO processes, (2) identify influential references and emerging hot topics, (3) review the DAMO processes in freshwater and coastal systems in depth, and (4) point out limitations and propose future research directions. These objectives were addressed in each of the following sections of this review.
2 The DAMO processes
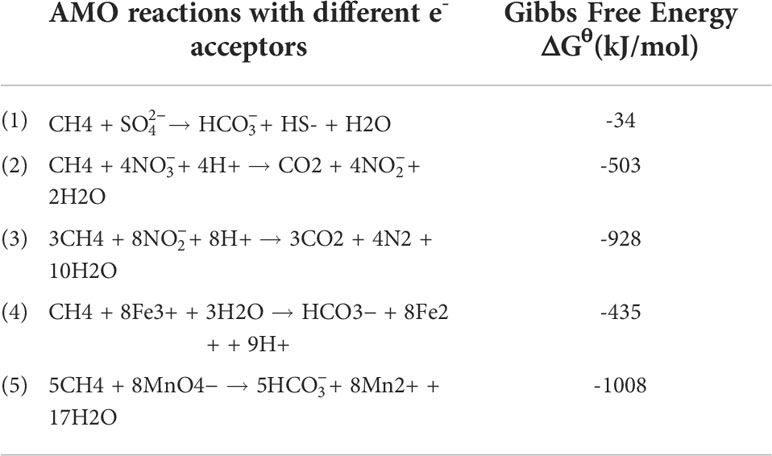
Scientists have long discovered that CH4 oxidation could be coupled to sulfate reduction under anoxic conditions (Barnes and Goldberg, 1976). This pathway has been recognized to remove more than 90% of CH4 emission from the ocean (Knittel and Boetius, 2009), which makes the ocean a minor contributor to global CH4 emission (6 – 2 Tg CH4 yr−1, accounting for 1 – 2% of the global emission), despite that it covers 70% of the Earth’s surface (Weber et al., 2019; Wallenius et al., 2021). The sulfate-dependent AMO uses as the electron (e-) acceptor and prevails in the open ocean, considering the mean sulfate concentration of 28 mmol/L in seawater (Jørgensen et al., 2019). However, the concentrations of other potential e- acceptors (e.g., and ) can be much higher in coastal and freshwater systems compared to the open ocean, and in theory, they can drive several different AMO pathways (Table 1). Because of anthropogenic activities, especially the use of synthetic fertilizers and fossil fuel combustion, large amounts of reactive N are delivered to freshwater and coastal environments. As a result, AMO coupled with or reduction, also known as the DAMO process, has drawn much attention. DAMO microorganisms were first enriched in the laboratory after 16 months’ incubation of highly eutrophic freshwater sediments (Raghoebarsing et al., 2006). The enriched culture contained bacteria and archaea, which performed AMO while using both and as e- acceptors. Ettwig et al. (2008); Ettwig et al. (2010) further enriched the DAMO bacteria Candidatus Methylomirabilis oxyfera (M. oxyfera), which are nitrite-dependent and belong to the NC10 clade. M. oxyfera can catalyze the nitrite-dependent DAMO process, which reduces to N2 while oxidizing CH4 to CO2 (Eqn. 3, Table 1). From the methane perspective, the nitrite-dependent DAMO by M.oxyfera follows the classical aerobic methane oxidation pathway, where methane is converted into methanol catalyzed by the particulate Methane monooxygenase (pMMO) enzyme (Wu et al., 2011). The produced methanol is further oxidized into formaldehyde, formate, and CO2 catalyzed by the methanol dehydrogenase, formaldehyde dehydrogenase, and formate dehydrogenase enzymes. The end-product CO2 is assimilated into the organism via the Calvin Benson Bassham cycle. On the nitrite side, the nitrite-dependent DAMO follows a novel denitrification pathway, where is metabolized into O2 and N2 via NO, catalyzed by enzymes including nitrite reductase and a postulated putative NO dismutase (Ettwig et al., 2010; Wu et al., 2012) (Figure 1A). This pathway is different from the canonical denitrification pathway, in that NO is not reduced to N2O as in the canonical pathway, catalyzed by the nitric oxide reductase enzyme (Ettwig et al., 2009). Rather, M.oxyfera lacks the genes responsible for N2O reduction to N2, but only a trace-amount of N2O was detected in nitrite-dependent DAMO by M.oxyfera (Ettwig et al., 2009; Ettwig et al., 2010; Reimann et al., 2015). To solve this apparent paradox, a unique dismutation reaction was proposed to convert NO into N2 and O2, catalyzed by the putative nitric oxide dismutase (NOD) enzyme (Ettwig et al., 2012). Through this pathway, 75% of the produced O2 is then used to oxidize CH4 through the classical aerobic methane oxidation pathway as described earlier (Ettwig et al., 2010; Wu et al., 2011). Because the source of the O2 is from the NC10 bacteria themselves, this aerobic methane oxidation process is obligate anaerobic and is also called an “intra-aerobic” (Wu et al., 2011). More research is required to thoroughly understand the proposed intra-aerobic pathway.
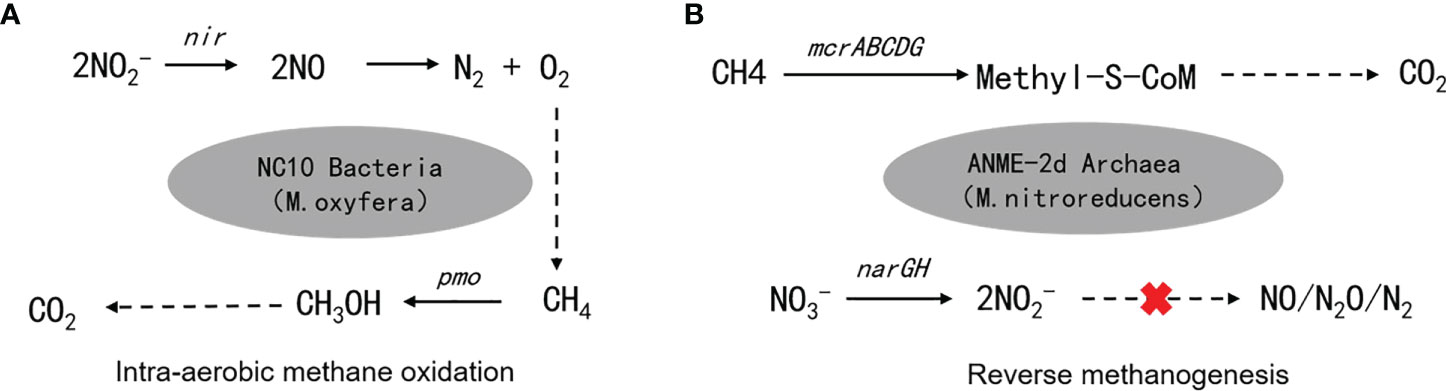
Figure 1 Metabolic pathways of (A) nitrite-dependent DAMO mediated by NC10 bacteria M. oxyfera and (B) nitrate-dependent DAMO mediated by ANME-2d archaea M.nitroreducens.
After the enrichment of DAMO bacteria, DAMO archaea were also enriched in 2013, after incubating in an anaerobic bioreactor fed with , , and CH4 for 350 days (Haroon et al., 2013). The enriched archaea, which was named Methanoperedens nitroreducens (M. nitroreducens) belonging to ANME-2d, performed AMO through the reverse methanogenesis pathway using as the terminal e- acceptor (Figure 1B). The ANME-2d contains all required functional genes for a complete reverse methanogenesis pathway (Haroon et al., 2013; Leu et al., 2020). The first step of this pathway is catalyzed by the methyl-coenzyme M reductase (MCR) enzymes. This step requires energy and has drawn great research interest since it is considered the rate-limiting step of the reverse methanogenesis pathway; however, the kinetic properties of the MCR enzyme remain largely unknown (Cai et al., 2021). The rest of the reverse methanogenesis pathway involves another 6 steps, each catalyzed by a specific enzyme, which has been reviewed in Timmers et al., 2017. This reverse methanogenesis process is coupled to reduction to , catalyzed by the nitrate reductase subunit NarG (Haroon et al., 2013). ANME-2d cannot further reduce to N2; this process was finished by the syntrophic partner, anammox bacteria in the enriched co-culture (Haroon et al., 2013). Unlike M. oxyfera, ANME-2d reportedly can use multiple e- acceptors other than , including Fe (Cai et al., 2018) and Mn (Leu et al., 2020).
Recently, the development of molecular tools, such as polymerase chain reaction (PCR), denaturing gradient gel electrophoresis (DGGE), and fluorescence in situ hybridization (FISH), enables relatively rapid detection of both M. oxyfera-like bacteria and ANME-2d archaea in the environment. PCR is the most used tool. A large group of PCR primers targeting 16s rRNA has been developed for the detection of M. oxyfera-like bacteria in the environment. In addition, the M. oxyfera-like bacteria use particular methane monooxygenase (pMMO) for CH4 oxidation (Figure 1A). This allows scientists to use functional gene pmoA as biomarkers for the detection of M. oxyfera-like bacteria (Luesken et al., 2011). To date, a number of primers have been developed, and they have been summarized comprehensively in (Shen et al., 2015a), including the sequences and the PCR thermal profiles. Compared to M. oxyfera bacteria, the development of primers for DAMO archaea M. nitroreducens started later. In 2015, Ding and colleagues designed 16s rRNA primers for DAMO archaea in samples from several freshwater sediments and paddy soils (Ding et al., 2015), which marks the first PCR primers designed for DAMO archaea. The ANME-2d archaea possess the mcrA gene, which encodes the alpha-submit of the MCR enzyme, making it a popular functional gene for the environmental detection of ANME-2d (Vaksmaa et al., 2017a). Although primers targeting functional genes pmoA and mcrA have high specificity, Shen and colleagues (Shen et al., 2017; Shen et al., 2019a) also pointed out that clone library analysis based on 16s rRNA combined with high throughput sequencing can increase sequencing depths and result in more operational taxonomic units (OTUs), which helps with community diversity analysis. In addition to the PCR approach, early studies also employed the FISH methods to identify M. oxyfera in cultures, such as in (Raghoebarsing et al., 2006; Ettwig et al., 2008; Ettwig et al., 2009). The catalyzed reporter deposition-FISH method was used to detect NC10 bacteria in the sediments of Lake Biwa, which marks the first application of the FISH method in environmental samples. However, for the purpose of environmental detection, the FISH method is considered less sensitive compared with the PCR approach and could be limited if the abundance of the target microorganism is low in the environment (Chen et al., 2016).
Molecular evidence for DAMO bacteria and archaea is often complemented by potential rates measured using isotope tracing methods combined with slurry incubations. In general, 13C tracers are usually added with e- acceptors such as and to the incubated samples and are compared with the control treatments. The rates can be inferred from the production of 13CO2, measured by GC-IRMS. Measured this way, the original structure and gradients of the sediment is likely destroyed because of the homogenization of the slurry, and samples are fertilized with substrates such as CH4 and and during the incubation. Therefore, the rate should not be considered the true rate in the field, but rather, represents the potential rate of the measured process under the incubation conditions. Despite these limitations, 13C tracing combined with slurry incubation is widely used for measuring the potential DAMO rates in sediment samples (Hu et al., 2014; Shen et al., 2016; Zhang et al., 2018; Li et al., 2020). Techniques combining isotope tracing and sequencing, such as stable carbon isotope probing can result in a more nuanced understanding of the DAMO process and the role each specific microorganism plays in the process (Sharp et al., 2012; Rasigraf et al., 2014).
3 Scientometric analysis of the DAMO research
Scientometric is a branch of informatics focusing on quantitative analysis of research papers and journals to understand the development and mechanisms of science. Co-citation is a common measurement used in quantitative studies, as a group of scientific papers frequently being co-cited by citing documents that represent key knowledge, methods, or concepts in a research field. The co-citation patterns could be used to map out the relationships between key references, authors, or keywords, to reveal the knowledge structure behind a research field. In fact, science mapping represents an important research topic in the field of bibliometrics (Cobo et al., 2011).
In the field of DAMO research, since the prediction of the DAMO process and the successful enrichment of DAMO organisms in 2006 (Raghoebarsing et al., 2006), the number of publications has grown rapidly. To understand where we stand in the domain of DAMO, particularly the distributions of DAMO in the environment, we conducted a co-citation network analysis on a literature database collected from the Web of Science Core Collection. The goals of this section are to (1) reveal the structure of the co-citation network in this field, (2) identify influential references that shape the development of the field, and (3) detect the emerging hot topics and areas of DAMO research based on co-citation analysis on the references of the literature database.
The co-citation analysis and visualization were implemented using CiteSpace 6.1 R3. CiteSpace is a science mapping tool capable of exploring the literature co-citation network and visualizing the knowledge structure of a field. The foundation and application of CiteSpace in exploring and visualizing a research field have been demonstrated in a series of publications (Chen, 2004; Chen, 2006; Chen et al., 2012; Chen and Song, 2019; Chen, 2020), and it has been used to summarize research in ecology (Yang et al., 2019; Hu et al., 2019). For the current review work, we used CiteSpace to analyze the co-citation network with emphasis on three key aspects: (1) the structure and the timeline view of the co-citation network, (2) references with citation bursts (i.e., an abrupt change in citation counts), and (3) keywords with citation bursts. By focusing on these three aspects, we hope to achieve the three goals of this section as stated earlier. Several key concepts in CiteSpace that we will focus on include citation burst, clustering, and labeling. Briefly, a burst refers to a surge of frequencies of certain types of events, including the citation count, keyword appearance, or noun phrases in the titles, abstracts, or other parts of the publication. CiteSpace adopts the algorithm from Kleinberg 2002 to determine such burst events. CiteSpace also partitions the co-citation network into mutually-exclusive clusters such that references were tightly associated with each other but loosely connected with references outside the cluster using the spectral clustering algorithm formulated as an optimization problem (Chen et al., 2010). The technical details are described in (Shi and Malik, 2000; Ng et al., 2002; Chen et al., 2010). After the clusters are defined, they are automatically labeled by noun phrases extracted from the titles, abstracts, and indexed keywords of the citing publications and ranked using the log-likelihood ratio (LLR) test (Dunning, 1993).
For the current review, we built the citing article dataset focused on DAMO in the environment. We collect data from the Web of Science Core Collection using keywords “anaerobic oxidation of methane”, “anaerobic methane oxidation”, “nitrate-dependent anaerobic methane oxidation”, “nitrite-dependent anaerobic methane oxidation”, “denitrifying anaerobic methane oxidation”, “n-damo”, “damo”, “M. oxyfera”, “M. nitroreducens”, “NC10”, “ANME-2d”. The resulting literature was further refined using keywords including “river”, “lake”, “reservoir”, “estuary”, “sea”, “marine”, “sediment”, “soil”, and “wetland” to limit the search to studies on those environments. The resulting literature dataset contained 354 publications, including 316 research articles, 35 reviews, and 2 editorials, ranging from 2000 to 2022.
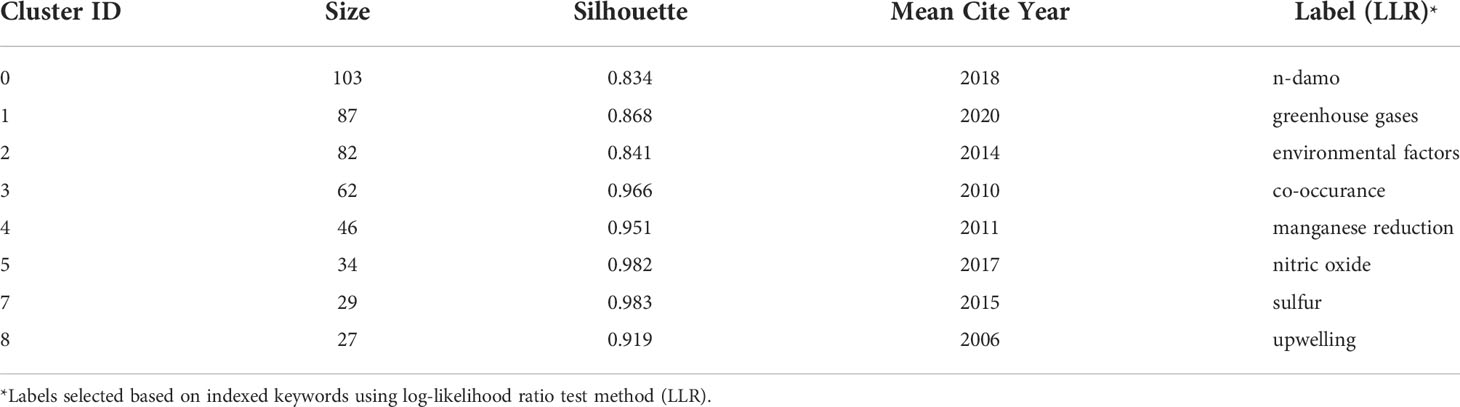
The literature database was then analyzed by CiteSpace for the 2000 – 2022 period, with configurations suggested by the developer (Chen et al., 2012). The CiteSpace project files and model configurations were available in Mendeley Data (10.17632/mwrkrmp3jn.2). A co-citation network of 835 nodes was derived from the citing article database. In terms of countries, China contributed the most, accounting for 34% of all references, followed by Germany and the USA, each contributing 13%, and Netherland ranked the 4th, which contributed 12% (Figure 2). The network was divided into 17 clusters such that the references were closely related to each other within a cluster and references were loosely connected between clusters (Figure 2). The top 8 largest clusters were listed in Table 2. They each have >20 member references, which can represent the sub-research area reasonably well. Their silhouette values were close to 1, suggesting a good homogeneity within each cluster (Table 2, Chen et al., 2012).
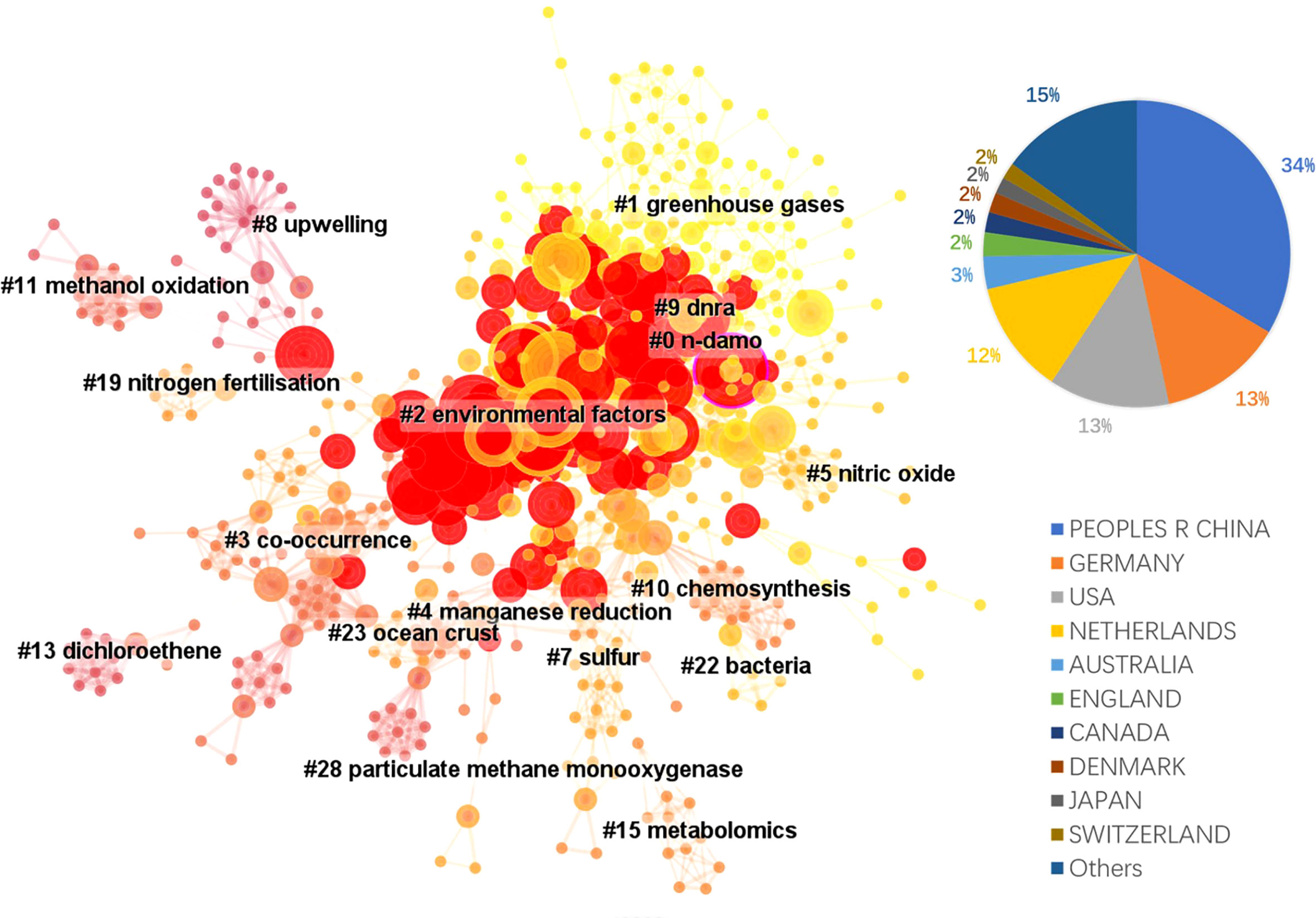
Figure 2 The landscape view of the reference co-citation network (total 835 nodes) derived from the 354 citing references from 2000 to 2022 and the country contributions of the citing references. Red circles represent references with citation bursts; the size of the circles reflects citation counts; links between nodes suggest that they were co-cited by citing references.
Clusters 0 and 2 were the two clusters with the most citation bursts. Cluster 0 was labeled n-damo based on the indexed keywords of the citing articles using the LLR test (Chen et al., 2012). This was the largest cluster containing 103 member references, and the representative references were focused on the distribution of DAMO and other AMO pathways in the environment. For example, in a series of works by Shen and colleagues, they demonstrated that nitrite-dependent DAMO and M.oxyfera were more active and abundant in freshwater marshes in East China (Shen et al., 2017). Later, in Shen et al., 2019a, AMO processes driven by different e- acceptors (including nitrate, nitrite, sulfate, and iron) across different riverbeds in England were compared, with nitrate- and nitrite-dependent AMO being the most active pathways. Moreover, the nitrite-dependent AMO activity was higher than the nitrate-dependent pathway, consistent with their findings for the freshwater marshes (Shen et al., 2017). These two papers were co-cited later by authors other than the Shen group focusing on similar topics, such as by Chen et al. (2021), who compared nitrite-/nitrate-dependent DAMO in estuaries and intertidal wetlands in the Yangtze Estuary.
The second largest cluster, cluster 2 was labeled as environmental factors based on the indexed keywords of the citing articles. Their co-cited reference network contained several fundamental references in this area, such as the series of research by Ettwig et al. (2008); Ettwig et al. (2009); Ettwig et al. (2010), where the bacteria community performing nitrite-dependent DAMO was enriched and the metabolic pathways were investigated. Cluster 2 also contained the milestone paper by Haroon et al. (2013), where the ANME-2d archaea N. nitroreducens capable of nitrate-dependent DAMO was enriched. We demonstrated some of the most representative articles (with citation bursts) of the two largest clusters in the area. The complete list of all cluster members and their citation data (including burstiness and citation counts) is available as the open data associated with this manuscript in Mendeley Data (10.17632/mwrkrmp3jn.2). This cluster also contained many representative papers reporting the DAMO community in various natural environments. For example, Shen and colleagues investigated the nitrite-dependent DAMO distributions in the sediments of the Jiaojiang Estuary (Shen et al., 2014b), Xiazhuhu wetland (Shen et al., 2015b) in China, and 7 permeable riverbeds in England (Shen et al., 2019a), and explored the effects of different environmental conditions on their distribution in these systems. A more detailed discussion on DAMO distributions in freshwater and coastal systems is presented in Section 4.
Taking a timeline view, cluster 8 first emerged around 2007 (Figure 3). The most cited paper (and with citation burst) in this cluster was the landmark reference Raghoebarsing et al., 2006, which reported the first enriched culture performing DAMO in the laboratory. Cluster 2 contained the most high-profile references (i.e., references with strong citation bursts, indicated by the large red rings), as discussed earlier for the landscape view of the co-citation network (Figure 2). The timeline view further revealed that these high-profile references emerged in two periods; first started around 2007 – 2010, which was featured by the series of research by Ettwig and colleagues, co-cited with the Raghoebarsing et al., 2006 reference (Figure 3). These references were focused on the metabolic pathways of the nitrite-dependent DAMO. The second cluster was around the famous Haroon et al., 2013 paper, where the enrichment of N. nitroreducens was reported. High-profile reference groups were also formed after 2013 in cluster 0, as highlighted in Figure 3. They include a highly cited review on nitrite- and nitrate-dependent DAMO by Welte et al. (2016), and studies on primer designs for DAMO archaea (Ding et al., 2015), DAMO bacteria and archaea distributions in paddy field (Vaksmaa et al., 2016; Ding et al., 2016), freshwater rivers (Long et al., 2017b; Shen et al., 2019b), reservoirs (Long et al., 2017a), marshes (Shen et al., 2017), and in coastal systems (Zhang et al., 2018). These references will be discussed in more depth in the later sections of the review. In addition to these two major clusters, cluster 1 was a notably new one, as the mean citing year was 2018 (Table 2). This recently formed cluster featured only a few high-profile references, which were focused on novel e- acceptors such as humic substances (Valenzuela et al., 2019; Bai et al., 2019) and metals (He et al., 2018). These novel processes are associated with ANME archaea. For example, recently, AMO processes coupled to Fe(III) and Mn(IV) were reported after long-term incubation. They were performed by novel species belonging to ANME-2d, which were named Candidatus Methanoperedens ferrireducens (Cai et al., 2018)and Candidatus Methanoperedens manganicus (Leu et al., 2020), respectively. The metal reduction was hypothesized to be conducted through the direct interspecies e- transfer (DIET) pathway catalyzed by the multi-heme c-type cytochrome (MHCs), similar to other common metal-reducing organisms (Shi et al., 2016; Cai et al., 2018; Leu et al., 2020). Other novel e- acceptors include As(V) (Shi et al., 2020) and humic acids (Bai et al., 2019) can also fuel AMO, supposedly through the MHC-dependent DIET process. This cluster represents one of the frontiers in the research of AMO processes, as the role of different e- acceptors and their interactions with DAMO can have important implications for the CH4 sink/source behaviors of a system.
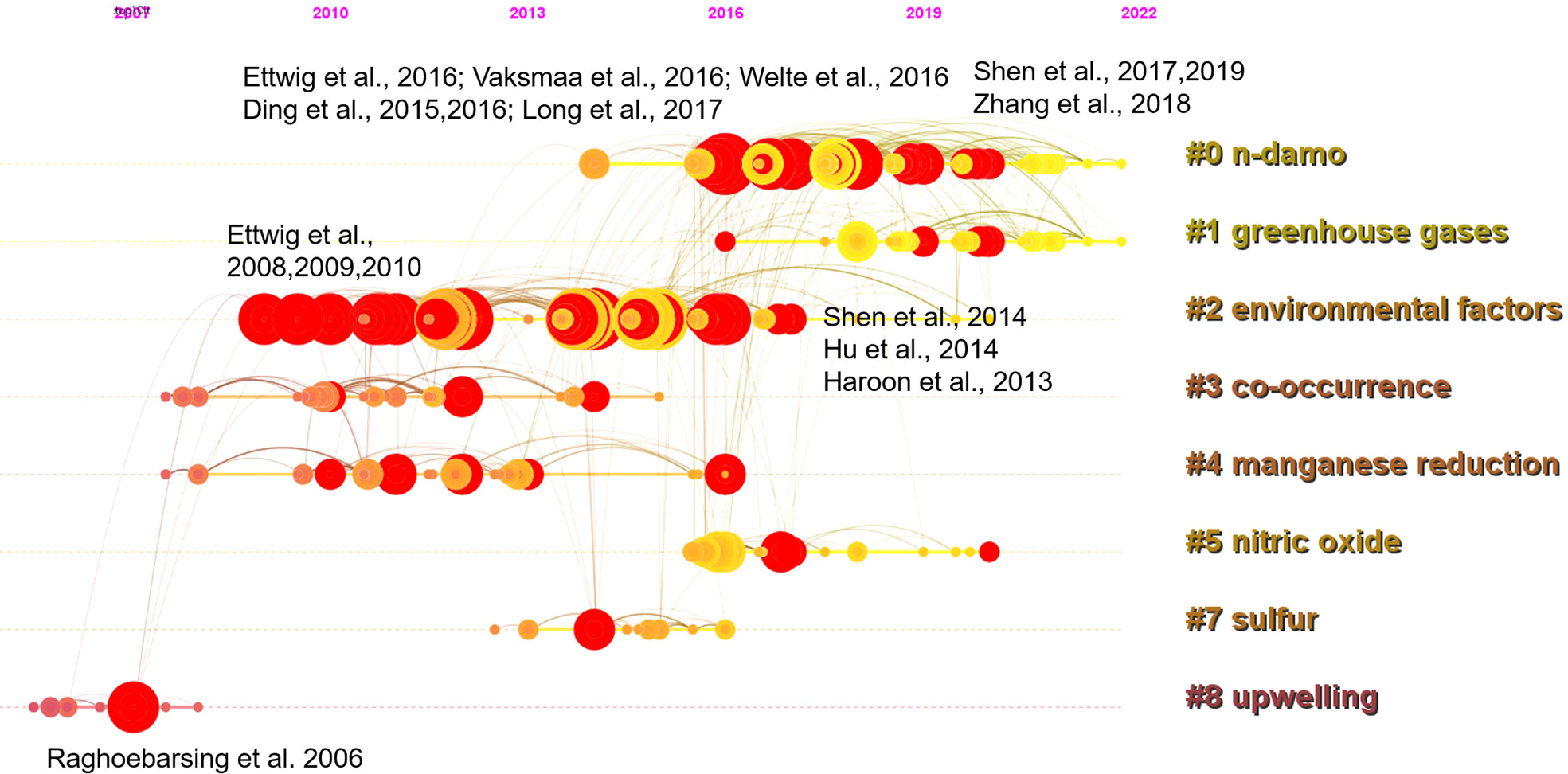
Figure 3 A timeline view of the co-citation network with the 8 largest clusters. Red circles represent references with citation bursts; the size of the circles reflects citation counts; links between nodes suggest that they were co-cited by citing references.
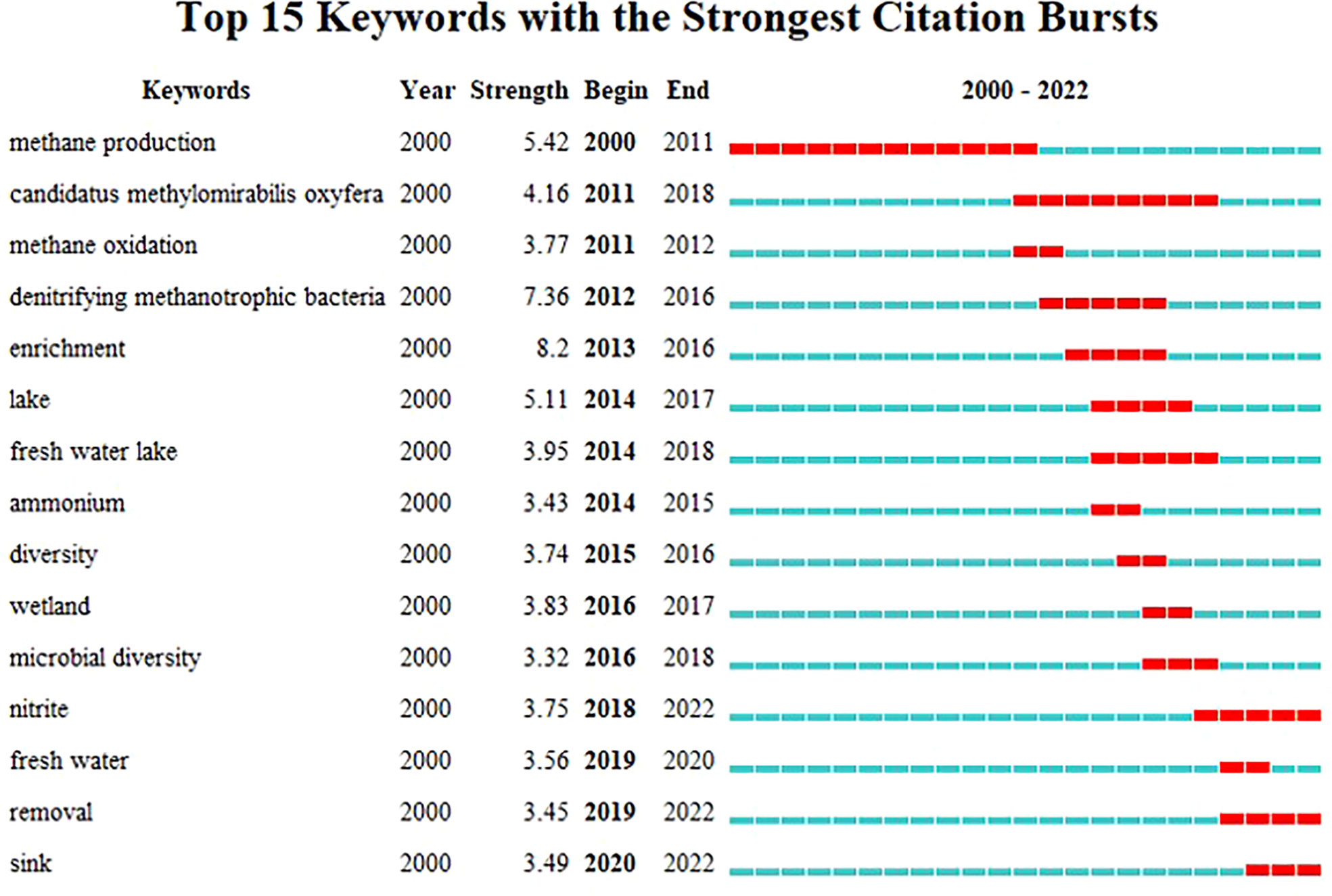
The emerging trends of the DAMO research were further investigated through the co-occurrence analysis of reference keywords. Before the successful enrichment and the naming of the DAMO bacteria M.oxyfera, “methane production” was the keyword with a long burst duration (2000 – 2011, Figure 4), as the scientific community had a strong interest in understanding CH4 emission in various types of ecosystems, such as rice paddies and peatlands (e.g., Chidthaisong and Conrad, 2000; Liesack et al., 2000 in our citing article database). “M.oxyfera” and “denitrifying methanotrophic bacteria” were the two keywords refer to the DAMO bacteria, with the latter term bursting for a shorter period from 2012 – 2016 (Figure 4). The term “enrichment” became popular in 2013-2016, reflecting the research effort to investigate the DAMO bacteria and archaea through enriched cultures (e.g., Ettwig et al., 2010; Ettwig et al., 2016). Later on, keywords about natural ecosystems started to experience citation bursts, including “lake”, “freshwater lake”, and “wetland”. The most recent keywords were “sink” and “removal”, which referred to DAMO as N and CH4 sinks in ecosystems including wetlands (Xie et al., 2020; Yang et al., 2022; Wang et al., 2022) and aquatic systems (Shen et al., 2020; Li et al., 2020). These new trends suggest that the distribution of DAMO in the environment and the potential contributions to CH4 and N sinks are becoming the current focus of research and like to continue in the future. Comparing freshwater and coastal systems, keywords contained “freshwater”, “lake”, “river”, “reservoir”, and “paddy” resulted in a total of 100 occurrences, whereas keywords contained “coastal”, “coast”, “estuary”, “bay”, “marine”, “ocean”, and “sea” resulted in 41 occurrences. Keywords related to M.oxyfera also occurred more frequently than the keywords related to ANME-2d archaea. The keywords and their occurrence frequencies were available together with other CiteSpace documents in Mendeley Data as stated earlier. This result suggests that freshwater systems are currently more studied than coastal systems, and DAMO bacteria is more studied than their archaeal counterparts.
In summary, the co-citation analysis of the literature database helped us identify the important areas in the field of DAMO in the environment. We identified a number of key references with abrupt increases in citation counts over the 2000 – 2022 period, which were focused on DAMO metabolic pathways, laboratory enrichment, and environmental distributions in environments including wetlands, rice paddies, and freshwater and coastal systems. The timeline view also revealed the evolution of research in this area, from studies on enriched cultures to their distributions in the environment. The latest trends include the role of DAMO as N and CH4 sinks and alternative e- acceptors of AMO, such as humic substances and metals. In the next sections, we will be focused on DAMO distribution in freshwater and coastal environments, and analyze the relevant literature in more depth guided by the results of the co-citation analysis.
4 DAMO in freshwater and coastal systems
4.1 Freshwater systems
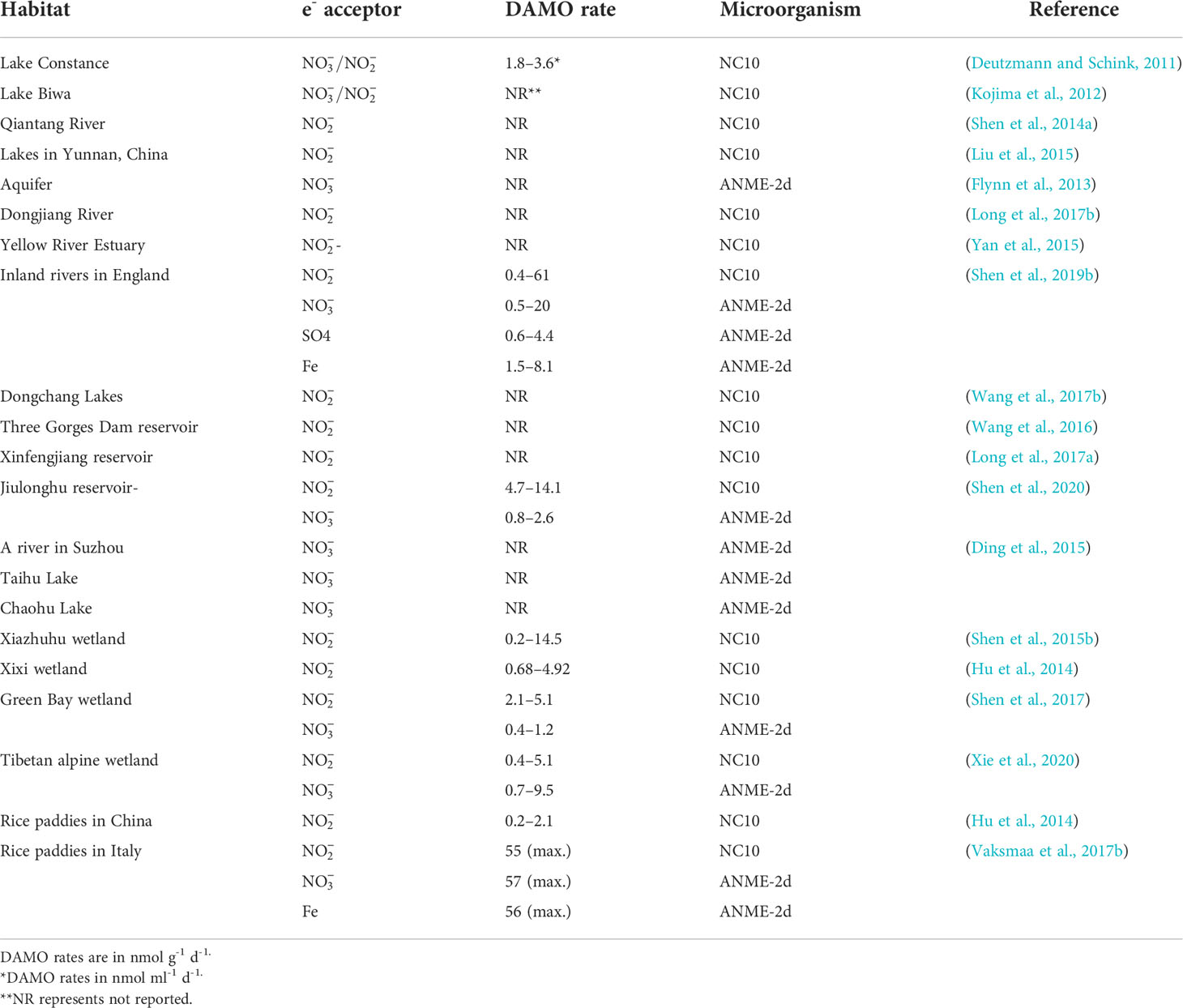
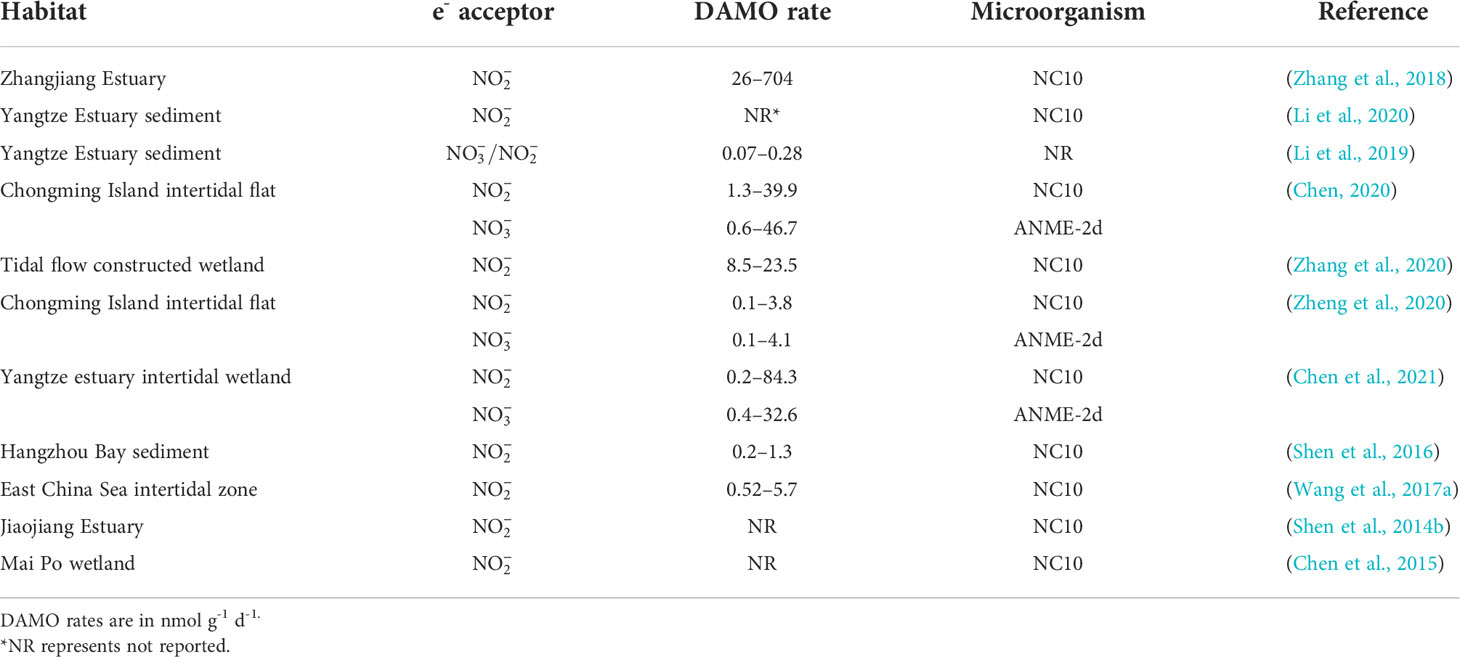
As revealed by the co-citation analysis, in early studies, DAMO was often investigated in the laboratory with enriched cultures. However, as molecular tools evolved and primers for DAMO bacteria and archaea were designed, their distribution and activities in the fields have been studied more. To date, DAMO microorganisms and activities have been reported in a wide range of natural and engineered habitats (Table 3). For freshwater systems, Hu et al. (2014) estimated that in freshwater wetlands, the potential rates of nitrite-dependent DAMO can reach 4.1 – 6.1 Tg CH4 m-2 yr-1, accounting for 2 – 6% of CH4 emission by global wetlands, suggesting that DAMO could be an important but overlooked CH4 sink. Zhu et al. (2015) compared 91 soil samples from 24 different wetlands in China and proved that M. oxyfera-like bacteria are widely distributed in freshwater wetlands. These results show that M. oxyfera-like bacteria are widely distributed in freshwater environments. Common environmental factors in freshwater systems affecting the distribution of DAMO bacteria include oxygen levels, CH4 concentrations, and nutrient and organic matter conditions. For example, Shen et al. (2015b) studied DAMO bacteria and the corresponding nitrite-dependent DAMO rates in soils of Xiazhuhu wetland, China, and found that DAMO bacteria abundance and activities were higher in deep soil layers, where the oxygen levels were low and CH4 concentrations were high. As described in Section 2, the M.oxyfera employs an intra-aerobic pathway, and thus, detrimental effects from the external supply of O2 can be expected. Evidence supporting this idea comes from laboratory experiments with M. oxyfera cultures, where DO concentrations of 1 or 4 mg/L suppressed the CH4 and metabolism (Luesken et al., 2012). However, exposed at lower oxygen levels, increases in both CH4 and consumption rates were observed (Kampman et al., 2018), suggesting that micro-oxygenic environments might stimulate the M. oxyfera metabolism. Dissolved CH4 concentrations represent another major factor that can regulate the metabolism of DAMO. The NC10 bacteria typically have higher CH4 affinity than ANME-2d archaea, as the CH4 affinity constant can be 1 – 2 orders of magnitudes lower for the NC10 bacteria than their archaeal counterparts (He et al., 2013; Lu et al., 2019). Long et al. (2017b) compared DAMO bacteria abundance and community structures in Dongjiang and its tributaries from May to August and identified that , , and C/N affected the community structure. In addition, Long et al. (2017a) also examined the vertical and horizontal distributions of DAMO bacteria in reservoir sediments and pointed to , total organic C, P, and N as key factors contributing to the variations in DAMO bacteria abundance and rates.
Compared with DAMO bacteria, DAMO archaea are in general less studied for their environmental distributions (Table 3). As the conditions for nitrate- and nitrite-dependent DAMO processes to occur are similar, and is more prevalent in natural environments, ANME-2d and M. oxyfera-like bacteria are likely to co-exist in natural habitats. According to the reaction kinetics, compared to , as the e- acceptor produces more energy (Table 1), and some studies do show that DAMO bacteria have high abundance as well as potential rates than archaea, such as in wetlands (Shen et al., 2017) and reservoirs (Shen et al., 2020). However, opposite results also exist, such as in a Tibetan alpine wetland (Xie et al., 2020). Oxygen levels may be one of the factors that cause niche differentiation between DAMO bacteria and archaea, as M. oxyfera bacteria are hypothesized to better tolerate oxygen (Wallenius et al., 2021). In their study, the interaction of M. oxyfera with other N cycling microorganisms was found to be affected by multiple factors, including temperature, N, and P contents, whereas the interaction of M. nitroreducens with other microorganisms was solely affected by N compounds (Xie et al., 2020). These results suggest that multiple mechanisms may play a role in shaping distributions of DAMO bacteria and archaea, requiring further studies. Overall, using molecular tools, researchers found that DAMO and the functional microorganisms are widely distributed in natural habitats, especially in freshwater systems, including paddy soils. However, M. oxyfera and nitrite-dependent DAMO are generally more reported than ANME-2d and nitrate-dependent DAMO, and factors and mechanisms controlling their distribution patterns, relative abundance, and niche separation are far from clear.
4.2 Coastal systems
Lying between the land-ocean interface, coastal systems, including river networks, estuaries, and adjacent seas are routinely submerged in water and due to the terrestrial input of nutrient and organic matter, often have high and availabilities, making them preferable habitats for DAMO processes to occur (Kraft et al., 2014; Lin and Lin, 2022). However, DAMO in these coastal systems is far under-studied than in freshwater systems, as suggested by the keywords frequencies from the scientometric analysis. Currently, studies on coastal systems are focused on tidal flats and mangroves. For example, Zhang et al. (2018) measured nitrite-dependent DAMO ranging from 26 – 704 nmol CH4 g-1 d-1 in the mangrove sediments of Zhangjiang Estuaries, even higher than some rice paddies (Table 3). The DAMO bacteria abundances and activities were higher in the surface layer not deeper, contrary to the reported freshwater wetland and lake systems (Kojima et al., 2012; Hu et al., 2014; Xie et al., 2020). The opposite observation was explained by the micro-oxygen environments in the surface layer of the mangrove system, as trace-level oxygen can be beneficial to M. oxyfera (Luesken et al., 2012). The detail of this mechanism and how it is different from the freshwater systems need further investigation. Wang et al. (2017a) compared the spatial and temporal distribution of nitrite-dependent DAMO process in low, median, high tidal flats, and compared DAMO to CH4 removal, and pointed out that bacterial DAMO can be important sinks for CH4 and reactive N in tidal flat systems. Compared with M. oxyfera, studies that also considered ANME-2d are fewer for coastal systems (Table 4). The limited studies considered both DAMO processes show that their relative abundances and activities can vary from different habitats even within the same system. For example, Chen et al. (2021) sampled multiple locations along the salinity gradient of the Yangtze Estuary and studied the distributions of both nitrate- and nitrite-dependent DAMO. Their results show that intertidal wetlands can be hotspots for DAMO, where both M. oxyfera-like bacteria and ANME-2d archaea exist, but bacteria dominated the microorganism community. As a result, nitrite-dependent DAMO contributed more to CH4 removal, and total DAMO rates decreased along the gradient, significantly correlated with pH, total organic C, Fe(II), and Fe(III). On the other hand, in a parallel study (Zheng et al., 2020) on the intertidal marshes on an island of the Yangtze Estuary, results show that NC10 bacteria and ANME-2d archaea had similar abundances with comparable potential rates, suggesting that both processes can be important in intertidal marshes of the Yangtze Estuary. Affected by oxygen levels, NC10 bacteria and nitrite-dependent DAMO were higher in the water-sediment interface, whereas ANME-2d and nitrate-dependent DAMO occupied deeper layers. An interesting observation was that in the layers where ANME-2d abundance was high, concentrations were actually low, which leaves open the possibility of ANME-2d exploiting alternative e- acceptors such as Fe(III) and , requiring further investigation (Zheng et al., 2020).
Compared to the organic-rich tidal flats and mangroves, the sediments of coastal rivers, estuaries, and adjacent seas may contain lower levels of nutrients and organic matter, and the DAMO microorganism abundance and rates may be lower. For example, Shen et al. (2016) studied nitrite-dependent DAMO and M. oxyfera bacteria in Hangzhou Bay and found DAMO rates ranged from 0.2 – 1.3 nmol CH4 g-1 dry soil d-1, accounting for 2 – 9.4% of CH4 oxidation. This rate is ~1 – 2 orders of magnitudes lower than in the coastal wetlands (Table 4), but considering the vast area of an estuary, the role of estuary sediments as potential CH4 sink should not be overlooked.
In summary, DAMO in coastal systems is generally understudied compared to freshwater systems. However, existing research shows that in some coastal wetlands, DAMO rates can be even higher than in some highly eutrophic freshwater systems, such as rice paddies. The highly dynamic environments and the existence of salinity gradients in coastal systems can result in different distribution patterns compared to freshwater systems. The mechanisms controlling niche differentiation and interactions between NC10 bacteria and AMNE-2d archaea still require further study.
5 Limitation and outlook
Studies on the DAMO process and microorganisms have been expanding over the past years. However, research focusing on DAMO in coastal environments is still very limited. In this review, we attempted to identify several areas in studies on coastal DAMO that require more research efforts. First, existing research mainly focuses on M. oxyfera and nitrite-dependent DAMO process, whereas the relative abundance and activities of ANME-2d archaea and nitrate-dependent DAMO in coastal systems are largely overlooked. More field- and laboratory-based studies are needed to understand the environmental factors and mechanisms controlling the distributions and interactions between DAMO bacteria and archaea (Wallenius et al., 2021). Second, ANME-2d archaea have the potential to use multiple e- acceptors, such as Fe, Mn, and sulfate. In certain coastal habitats, their contents can be higher than . AMO driven by different e- acceptors and their relative contributions to CH4 removal remains unclear for coastal systems. We can improve our understanding of ANME-2d in coastal environments by comparing their AMO activities driven by different e- acceptors.
Lastly, the coastal environments are not isolated sections, but rather hydraulically connected continua along which physical and biological factors evolve. Current research focuses more on DAMO distribution in a specific coastal habitat, such as a wetland, or an estuary, whereas comparisons among different types of habitats along the river-estuary-sea continuum are quite limited. Direct comparisons of DAMO community and activities among different habitats along the river-estuary-sea continuum are rarely reported. However, such comparisons are necessary to understand how DAMO microorganisms and processes vary along the continuum as a whole to make comprehensive management decisions. In fact, some studies have shown that factors including oxygen levels, nutrient conditions, organic matter contents, pH, Fe, and sulfate, as well as N cycling processes, can change dramatically along the river-estuary-sea continuum (Lin et al., 2016; Lin et al., 2017b; Lin et al. 2017a; Wei et al., 2020; Wei and Lin, 2021), most of which are also sensitive factors for DAMO as we reviewed.
In light of this, we call for more research comparing DAMO microorganisms, activities, and interactions with other N cycling processes, among different types of habitats along the river-estuary-sea continuum to better understand the environmental factors shaping their distributions in coastal systems, and their contributions to CH4 and reactive N removal (Figure 5). Specifically, we propose for direct comparisons of DAMO community composition and activity using consistent methods among hydraulically connected representative habitats (e.g., tidal freshwater rivers, estuaries, and adjacent seas). We also propose to compare the DAMO processes with other N cycling pathways, such as denitrification, anaerobic ammonium oxidation, and dissimilatory nitrate reduction to ammonium, and with AMO processes coupled to other e- acceptors, such as the metal- and humic acid-dependent AMO.
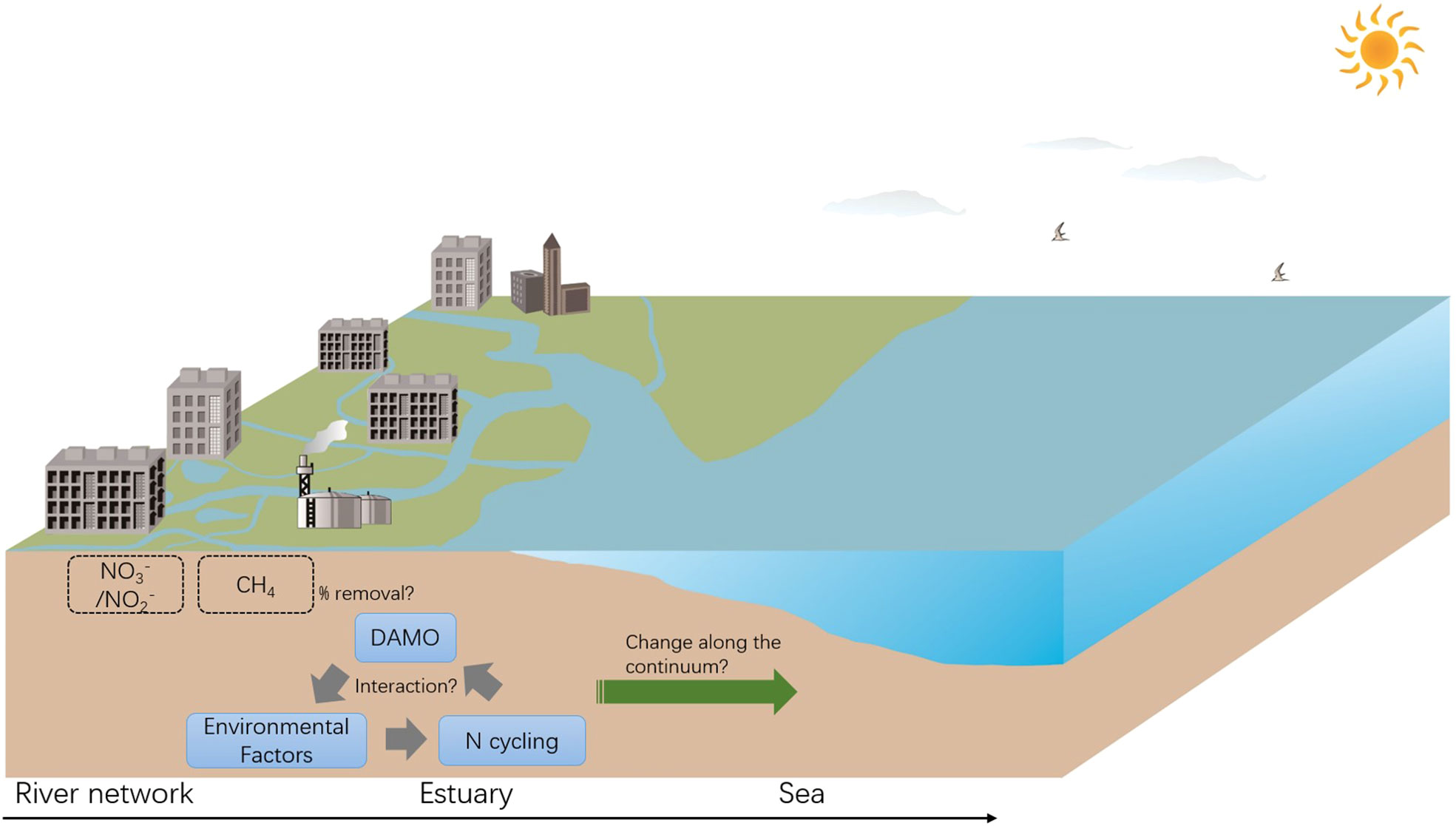
Figure 5 Proposed studies focusing on the interactions between DAMO, other N cycling processes, and environmental factors, and their changes along the river-estuary-sea continuum, with implications for CH4 and reactive N removal.
6 Conclusion
In this study, we reviewed the existing research on environmental distributions of DAMO microorganisms and activities. Through the co-citation analysis, we identified the active areas, key references, and emerging topics in the study of DAMO. Under the guidance of the co-citation analysis, we further reviewed the literature on DAMO in freshwater and coastal environments in more depth. Current studies show that DAMO bacteria and archaea can be widely distributed in freshwater and coastal systems, including rivers, lakes, reservoirs, wetlands, tidal flats, mangroves, and estuarine and sea sediments. However, compared to freshwater systems, fewer studies focus on coastal systems. NC10 bacteria and nitrite-dependent DAMO are more studied than ANME-2d and nitrate-dependent DAMO. In general, environmental factors and mechanisms that control the distributions of nitrite- and nitrate-DAMO in freshwater and coastal systems are far from clear. We identified several areas that require further research and called for more research focusing on direct comparisons of DAMO community and activity in the context of the river-estuary-sea continuum. These kinds of information can improve our understanding of DAMO distributions in the coastal zones and help us make more comprehensive management decisions.
Author contributions
HW conducted the scientometric analysis, reviewed the literature, and wrote the manuscript. MW and MY conducted the scientometric analysis. CX conducted the scientometric analysis and reviewed the literature. All authors contributed to the article and approved the submitted version.
Funding
This work is supported by the following funding sources, the National Science Foundation of China (41601008), the General Program of Natural Science Foundation of Fujian Province of China (2018J01737), the project of Fujian Forestry Bureau (2021FKJ29), Natural Science Foundation of Jiangsu Province (BK20190483), and the Natural Science Foundation of the Jiangsu Higher Education Institutions of China (22KJB170013).
Acknowledgments
We would like thank Zhichao Wang and Yalan Zhao for their help with CiteSpace and suggestions throughout this work.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Bai Y., Wang X., Wu J., Lu Y., Fu L., Zhang F., et al. (2019). Humic substances as electron acceptors for anaerobic oxidation of methane driven by ANME-2d. Water Res. 164, 114935. doi: 10.1016/j.watres.2019.114935
Barnes R. O., Goldberg E. D. (1976). Methane production and consumption in anoxic marine sediments. Geology 4, 297. doi: 10.1130/0091-7613(1976)4<297:MPACIA>2.0.CO;2
Cai C., Leu A. O., Xie G.-J., Guo J., Feng Y., Zhao J.-X., et al. (2018). A methanotrophic archaeon couples anaerobic oxidation of methane to Fe(III) reduction. ISME J. 12, 1929–1939. doi: 10.1038/s41396-018-0109-x
Cai C., Zhang X., Wu M., Liu T., Lai C. Y., Frank J., et al. (2021). Roles and opportunities for microbial anaerobic oxidation of methane in natural and engineered systems. Energy Environ. Sci. 14, 4803–4830. doi: 10.1039/d1ee00708d
Chen C. (2004). Searching for intellectual turning points: Progressive knowledge domain visualization. Proc. Natl. Acad. Sci. 101, 5303–5310. doi: 10.1073/pnas.0307513100
Chen C. (2006). CiteSpace II: Detecting and visualizing emerging trends and transient patterns in scientific literature. J. Am. Soc. Inf. Sci. Technol. 57, 359–377. doi: 10.1002/asi.20317
Chen J., Dick R., Lin J. G., Gu J. D. (2016). Current advances in molecular methods for detection of nitrite-dependent anaerobic methane oxidizing bacteria in natural environments. Appl. Microbiol. Biotechnol. 100, 9845–9860. doi: 10.1007/s00253-016-7853-5
Chen C., Dubin R., Kim M. C. (2014). Emerging trends and new developments in regenerative medicine: A scientometric updat -2014). Expert Opin. Biol. Ther. 14, 1295–1317. doi: 10.1517/14712598.2014.920813
Chen C., Ibekwe-SanJuan F., Hou J. (2010). The structure and dynamics of cocitation clusters: A multiple-perspective cocitation analysis. J. Am. Soc. Inf. Sci. Technol. 61, 1386–1409. doi: 10.1002/asi.21309
Chen C. (2020). A glimpse of the first eight months of the COVID-19 literature on Microsoft academic graph: Themes, citation contexts, and uncertainties. Front. Res. Metr. Anal. 5. doi: 10.3389/frma.2020.607286
Chen C., Hu Z., Liu S., Tseng H. (2012). Emerging trends in regenerative medicine: a scientometric analysis in CiteSpace. Expert Opin. Biol. Ther. 12, 593–608. doi: 10.1517/14712598.2012.674507
Chen C., Song M. (2019). Visualizing a field of research: A methodology of systematic scientometric reviews. PloS One 14, e0223994. doi: 10.1371/journal.pone.0223994
Chen F., Zheng Y., Hou L., et al. (2020). Denitrifying anaerobic methane oxidation in marsh sediments of chongming eastern intertidal flat. Mar. pollut. Bull. 150, 110681. doi: 10.1016/j.marpolbul.2019.110681
Chen F., Zheng Y., Hou L., Niu Y., Gao D., An Z., et al. (2021). Microbial abundance and activity of nitrite/nitrate-dependent anaerobic methane oxidizers in estuarine and intertidal wetlands: Heterogeneity and driving factors. Water Res. 190, 116737. doi: 10.1016/j.watres.2020.116737
Chen J., Zhou Z., Gu J. D. (2015). Complex community of nitrite-dependent anaerobic methane oxidation bacteria in coastal sediments of the mai po wetland by PCR amplification of both 16S rRNA and pmoA genes. Appl. Microbiol. Biotechnol. 99, 1463–1473. doi: 10.1007/s00253-014-6051-6
Chidthaisong A., Conrad R. (2000). Turnover of glucose and acetate coupled to reduction of nitrate, ferric iron and sulfate and to methanogenesis in anoxic rice field soil. FEMS Microbiol. Ecol. 31, 73–86. doi: 10.1111/j.1574-6941.2000.tb00673.x
Cobo M. J., López-Herrera A. G., Herrera-Viedma E., Herrera F. (2011). Science mapping software tools: Review, analysis, and cooperative study among tools. J. Am. Soc. Inf. Sci. Technol. 62, 1382–1402. doi: 10.1002/asi.21525
Dean J. F., Middelburg J. J., Röckmann T., Aerts R., Blauw L. G., Egger M., et al. (2018). Methane Feedbacks to the Global Climate System in a Warmer World. Rev. Geophysics 56, 207–250. doi: 10.1002/2017RG000559
Deutzmann J. S., Schink B. (2011). Anaerobic oxidation of methane in sediments of lake constance, an oligotrophic freshwater lake. Appl. Environ. Microbiol. 77, 4429–4436. doi: 10.1128/AEM.00340-11
Ding J., Ding Z. W., Fu L., Lu Y. Z., Cheng S. H., Zeng R. J., et al. (2015). New primers for detecting and quantifying denitrifying anaerobic methane oxidation archaea in different ecological niches. Appl. Microbiol. Biotechnol. 99, 9805–9812. doi: 10.1007/s00253-015-6893-6
Ding J., Fu L., Ding Z., Lu Y.-Z., Cheng S. H., Zeng R. J., et al. (2016). Environmental evaluation of coexistence of denitrifying anaerobic methane-oxidizing archaea and bacteria in a paddy field. Appl. Microbiol. Biotechnol. 100, 439–446. doi: 10.1007/s00253-015-6986-2
Dunning T. (1993). Accurate methods for the Statistics of Surprise and Coincidence. Comput. Linguistics 19.
Ettwig K. F., Butler M. K., le Paslier D., Pelletier E., Mangenot S., Kuypers M. M.M., et al. (2010). Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464, 543–548. doi: 10.1038/nature08883
Ettwig K. F., Shima S., van de Pas-Schoonen K. T., Kahnt J., Medema M. H., Op Den Camp H. J.M., et al. (2008). Denitrifying bacteria anaerobically oxidize methane in the absence of archaea. Environ. Microbiol. 10, 3164–3173. doi: 10.1111/j.1462-2920.2008.01724.x
Ettwig K. F., van Alen T., van de Pas-Schoonen K. T., Jetten M. S.M., Strous M. (2009). Enrichment and molecular detection of denitrifying methanotrophic bacteria of the NC10 phylum. Appl. Environ. Microbiol. 75, 3656–3662. doi: 10.1128/AEM.00067-09
Ettwig K. F., Speth D. R., Reimann J., Wu M. L., Jetten M. S. M., Keltjens J. T. (2012). Bacterial oxygen production in the dark. Front. Microbiol. 3. doi: 10.3389/fmicb.2012.00273
Ettwig K. F., Zhu B., Speth D., Keltjens J. T., Jetten M. S.M., Kartal B., et al. (2016). Archaea catalyze iron-dependent anaerobic oxidation of methane. Proc. Natl. Acad. Sci. 113, 12792–12796. doi: 10.1073/pnas.1609534113
Flynn T. M., Sanford R. A., Ryu H., Bethke C. M., Levine A. D., Ashbolt N. J., et al. (2013). Functional microbial diversity explains groundwater chemistry in a pristine aquifer. BMC Microbiol. 13, 146. doi: 10.1186/1471-2180-13-146
Han P., Gu J. (2013). A newly designed degenerate PCR primer based on pmoA gene for detection of nitrite-dependent anaerobic methane-oxidizing bacteria from different ecological niches. Appl. Microbiol. Biotechnol. 97, 10155–10162. doi: 10.1007/s00253-013-5260-8
Haroon M. F., Hu S., Shi Y., Imelfort M., Keller J., Hugenholtz P., et al. (2013). Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature 500, 567–570. doi: 10.1038/nature12375
He Z., Cai C., Geng S., Lou L., Xu X., Zheng P., et al. (2013). Modelling a nitrite-dependent anaerobic methane oxidation process: Parameters identification and model evaluation. Bioresour Technol. 147, 315–320. doi: 10.1016/j.biortech.2013.08.001
He Z., Zhang Q., Feng Y., Luo H., Pan X., Gadd G. M., et al. (2018). Microbiological and environmental significance of metal-dependent anaerobic oxidation of methane. Sci. Total Environ. 610–611, 759–768. doi: 10.1016/j.scitotenv.2017.08.140
Hu W., Li C., Ye C., Wang J., Wei W., Deng Y., et al. (2019). Research progress on ecological models in the field of water eutrophication: CiteSpace analysis based on data from the ISI web of science database. Ecol. Modell 410, 108779. doi: 10.1016/j.ecolmodel.2019.108779
Hu B., Shen L., Lian X., Zhu Q., Liu S., Huang Q., et al. (2014). Evidence for nitrite-dependent anaerobic methane oxidation as a previously overlooked microbial methane sink in wetlands. Proc. Natl. Acad. Sci. 111, 4495–4500. doi: 10.1073/pnas.1318393111
IPCC (2021). “Summary for Policymakers,” IPCC: Climate change 2021: The physical science basis. Contribution of Working Group I to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change, eds. Masson-Delmotte V., Zhai P., Pirani A. S., Connors L., Péan C., et al eds. (Cambridge, United Kingdom and New York, NY, USA: Cambridge University Press) 3−32. doi: 10.1017/9781009157896.001
Jørgensen B. B., Findlay A. J., Pellerin A. (2019). The biogeochemical sulfur cycle of marine sediments. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.00849
Kampman C., Piai L., Temmink H., Hendrickx T. L. G., Zeeman G., Buisman C. J. N. (2018). Effect of low concentrations of dissolved oxygen on the activity of denitrifying methanotrophic bacteria. Water Sci. Technol. 77, 2589–2597. doi: 10.2166/wst.2018.219
Knittel K., Boetius A. (2009). Anaerobic oxidation of methane: Progress with an unknown process. Annu. Rev. Microbiol. 63, 311–334. doi: 10.1146/annurev.micro.61.080706.093130
Kojima H., Tsutsumi M., Ishikawa K., Iwata T., Mußmann M., Fukui M., et al. (2012). Distribution of putative denitrifying methane oxidizing bacteria in sediment of a freshwater lake, lake biwa. Syst. Appl. Microbiol. 35, 233–238. doi: 10.1016/j.syapm.2012.03.005
Kraft B., Tegetmeyer H. E., Sharma R., Klotz M. G., Ferdelman T. G., Hettich R. L., et al. (2014). The environmental controls that govern the end product of bacterial nitrate respiration. Science 345, 676–679. doi: 10.1126/science.1254070
Leu A. O., Cai C., McIlroy S. J., Southam G., Orphan V. J., Yuan Z, et al. (2020). Anaerobic methane oxidation coupled to manganese reduction by members of the methanoperedenaceae. ISME J. 14, 1030–1041. doi: 10.1038/s41396-020-0590-x
Liesack W., Schnell S., Revsbech N. P. (2000). Microbiology of flooded rice paddies. FEMS Microbiol. Rev. 24, 625–645. doi: 10.1111/j.1574-6976.2000.tb00563.x
Li X., Gao D., Liu M. (2019). Composition, diversity and abundance of candidatus m. oxyfera-like bacteria in response to the estuary salinity gradient. Biogeochemistry 143, 1–14. doi: 10.1007/s10533-019-00545-w
Li X., Lai D., Gao D. (2020). Anaerobic oxidation of methane with denitrification in sediments of a subtropical estuary: Rates, controlling factors and environmental implications. J. Environ. Manage 273, 111151. doi: 10.1016/j.jenvman.2020.111151
Lin X., Li X., Gao D., Liu M., Cheng L. (2017a). Ammonium production and removal in the sediments of shanghai river networks: Spatiotemporal variations, controlling factors, and environmental implications. J. Geophys. Res. Biogeosci. 122, 2461–2478. doi: 10.1002/2017JG003769
Lin G., Lin X. (2022). Bait input altered microbial community structure and increased greenhouse gases production in coastal wetland sediment. Water Res. 218, 118520. doi: 10.1016/j.watres.2022.118520
Lin X., Liu M., Hou L., Gao D., Li X., Lu K., et al. (2017b). Nitrogen losses in sediments of the East China Sea: Spatiotemporal variations, controlling factors, and environmental implications. J. Geophys. Res. Biogeosci. 122, 2699–2715. doi: 10.1002/2017JG004036
Lin X., Liu M., Li X., Zheng Y., Yin G., Hou L., et al. (2016). Nitrogen mineralization and immobilization in sediments of the East China Sea: Spatiotemporal variations and environmental implications. J. Geophys. Res. Biogeosci. 121, 2842–2855. doi: 10.1002/2016JG003499
Liu Y., Zhang J., Zhao L., Li Y., Yang Y., Xie S., et al. (2015). Aerobic and nitrite-dependent methane-oxidizing microorganisms in sediments of freshwater lakes on the yunnan plateau. Appl. Microbiol. Biotechnol. 99, 2371–2381. doi: 10.1007/s00253-014-6141-5
Long Y., Guo Q., Li N., Li B., Tong T., Xie S., et al. (2017a). Spatial change of reservoir nitrite-dependent methane-oxidizing microorganisms. Ann. Microbiol. 67, 165–174. doi: 10.1007/s13213-016-1247-x
Long Y., Jiang X., Guo Q., Li B., Xie S. (2017b). Sediment nitrite-dependent methane-oxidizing microorganisms temporally and spatially shift in the dongjiang river. Appl. Microbiol. Biotechnol. 101, 401–410. doi: 10.1007/s00253-016-7888-7
Lu P., Liu T., Ni B.-J., Guo J., Yuan Z., Hu S. (2019). Growth kinetics of Candidatus ‘Methanoperedens nitroreducens’ enriched in a laboratory reactor. Sci. Total Environ. 659, 442–450. doi: 10.1016/j.scitotenv.2018.12.351
Luesken F. A., van Alen T. A., van der Biezen E., Frijters C., Toonen G., Kampman C., et al. (2011). Diversity and enrichment of nitrite-dependent anaerobic methane oxidizing bacteria from wastewater sludge. Appl. Microbiol. Biotechnol. 92, 845–854. doi: 10.1007/s00253-011-3361-9
Luesken F. A., Wu M. L., Op den Camp H. J. M., Keltjens J. T., Stunnenberg H., Francoijs K.-J., et al. (2012). Effect of oxygen on the anaerobic methanotroph ‘Candidatus methylomirabilis oxyfera’: kinetic and transcriptional analysis. Environ. Microbiol. 14, 1024–1034. doi: 10.1111/j.1462-2920.2011.02682.x
Ng A. Y., Jordan M. I., Weiss Y. (2002). On spectral clustering: Analysis and an algorithm. in. Adv. Neural Inf. Process. Systems.
Raghoebarsing A. A., Pol A., van de Pas-Schoonen K. T., Smolders A. J.P., Ettwig K. F., Rijpstra W. I. C., et al. (2006). A microbial consortium couples anaerobic methane oxidation to denitrification. Nature 440, 918–921. doi: 10.1038/nature04617
Rasigraf O., Kool D. M., Jetten M. S. M., Sinninghe Damsté J. S., Ettwig K. F. (2014). Autotrophic carbon dioxide fixation via the Calvin-Benson-Bassham cycle by the denitrifying methanotroph “Candidatus methylomirabilis oxyfera.” Appl. Environ. Microbiol. 80, 2451–2460. doi: 10.1128/AEM.04199-13
Reimann J., Jetten M. S. M., Keltjens J. T. (2015). “Metal Enzymes in ‘Impossible’ Microorganisms Catalyzing the Anaerobic Oxidation of Ammonium and Methane,” 257–313. doi: 10.1007/978-3-319-12415-5_7
Sharp C. E., Stott M. B., Dunfield P. F. (2012). Detection of autotrophic verrucomicrobial methanotrophs in a geothermal environment using stable isotope probing. Front. Microbiol. 3. doi: 10.3389/fmicb.2012.00303
Shen L., He Z., Wu H., Gao Z. (2015a). Nitrite-dependent anaerobic methane-oxidising bacteria: unique microorganisms with special properties. Curr. Microbiol. 70, 562–570. doi: 10.1007/s00284-014-0762-x
Shen L., Huang Q., He Z., Lian X., Liu S., He Y., et al. (2015b). Vertical distribution of nitrite-dependent anaerobic methane-oxidising bacteria in natural freshwater wetland soils. Appl. Microbiol. Biotechnol. 99, 349–357. doi: 10.1007/s00253-014-6031-x
Shen L., Hu B., Liu S., Chai X., He Z., Ren H., et al. (2016). Anaerobic methane oxidation coupled to nitrite reduction can be a potential methane sink in coastal environments. Appl. Microbiol. Biotechnol. 100, 7171–7180. doi: 10.1007/s00253-016-7627-0
Shen L., Liu S., Zhu Q., Li X., Cai C., Cheng D., et al. (2014a). Distribution and diversity of nitrite-dependent anaerobic methane-oxidising bacteria in the sediments of the qiantang river. Microb. Ecol. 67, 341–349. doi: 10.1007/s00248-013-0330-0
Shen L., Ouyang L., Zhu Y., Trimmer M. (2019a). Active pathways of anaerobic methane oxidation across contrasting riverbeds. ISME J. 13, 752–766. doi: 10.1038/s41396-018-0302-y
Shen L., Ouyang L., Zhu Y., Trimmer M. (2019b). Spatial separation of anaerobic ammonium oxidation and nitrite-dependent anaerobic methane oxidation in permeable riverbeds. Environ. Microbiol. 21, 1185–1195. doi: 10.1111/1462-2920.14554
Shen L., Tian M., Cheng H., Liu X., Yang Y., Liu J., et al. (2020). Different responses of nitrite- and nitrate-dependent anaerobic methanotrophs to increasing nitrogen loading in a freshwater reservoir. Environ. pollut. 263, 114623. doi: 10.1016/j.envpol.2020.114623
Shen L., Wu H., Liu X., Li J. (2017). Cooccurrence and potential role of nitrite- and nitrate-dependent methanotrophs in freshwater marsh sediments. Water Res. 123, 162–172. doi: 10.1016/j.watres.2017.06.075
Shen L., Zhu Q., Liu S., Du P., Zeng J., Cheng D., et al. (2014b). Molecular evidence for nitrite-dependent anaerobic methane-oxidising bacteria in the jiaojiang estuary of the East Sea (China). Appl. Microbiol. Biotechnol. 98, 5029–5038. doi: 10.1007/s00253-014-5556-3
Shi L., Dong H., Reguera G., Beyenal H., Lu A., Liu J., et al. (2016). Extracellular electron transfer mechanisms between microorganisms and minerals. Nat. Rev. Microbiol. 14, 651–662. doi: 10.1038/nrmicro.2016.93
Shi L.-D., Guo T., Lv P.-L., Niu Z.-F., Zhou Y.-J., Tang X.-J., et al. (2020). Coupled anaerobic methane oxidation and reductive arsenic mobilization in wetland soils. Nat. Geosci 13, 799–805. doi: 10.1038/s41561-020-00659-z
Shi J., Malik J. (2000). Normalized cuts and image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 22, 888–905. doi: 10.1109/34.868688
Timmers P. H. A., Welte C. U., Koehorst J. J., Plugge C. M., Jetten M. S. M., Stams A. J. M. (2017). Reverse Methanogenesis and Respiration in Methanotrophic Archaea. Archaea 2017, 1–22. doi: 10.1155/2017/1654237
Vaksmaa A., Jetten M. S. M., Ettwig K. F., Lüke C. (2017a). McrA primers for the detection and quantification of the anaerobic archaeal methanotroph ‘Candidatus methanoperedens nitroreducens. Appl. Microbiol. Biotechnol. 101, 1631–1641. doi: 10.1007/s00253-016-8065-8
Vaksmaa A., Lüke C., van Alen T., Valè G., Lupotto E., Jetten M. S.M., et al. (2016). Distribution and activity of the anaerobic methanotrophic community in a nitrogen-fertilized Italian paddy soil. FEMS Microbiol. Ecol. 92, 1–11. doi: 10.1093/femsec/fiw181
Vaksmaa A., van Alen T. A., Ettwig K. F., Lupotto E., Valè G., Jetten M. S.M., et al. (2017b). Stratification of diversity and activity of methanogenic and methanotrophic microorganisms in a nitrogen-fertilized Italian paddy soil. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.02127
Valenzuela E. I., Avendaño K. A., Balagurusamy N., Arriaga S., Nieto-Delgado C., Thalasso F., et al. (2019). Electron shuttling mediated by humic substances fuels anaerobic methane oxidation and carbon burial in wetland sediments. Sci. Total Environ. 650, 2674–2684. doi: 10.1016/j.scitotenv.2018.09.388
van Eck N. J., Waltman L. (2010). Software survey: VOSviewer, a computer program for bibliometric mapping. Scientometrics 84, 523–538. doi: 10.1007/s11192-009-0146-3
Wallenius A. J., Dalcin Martins P., Slomp C. P., Jetten M. S. M. (2021). Anthropogenic and environmental constraints on the microbial methane cycle in coastal sediments. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.631621
Wang X., Bai J., Xie T., Wang W., Zhang G., Yin S., et al. (2021). Effects of biological nitrification inhibitors on nitrogen use efficiency and greenhouse gas emissions in agricultural soils: A review. Ecotoxicol Environ. Saf. 220, 112338. doi: 10.1016/j.ecoenv.2021.112338
Wang Y., Huang P., Ye F., Jiang Y., Song L., Op den Camp H. J. M., et al. (2016). Nitrite-dependent anaerobic methane oxidizing bacteria along the water level fluctuation zone of the three gorges reservoir. Appl. Microbiol. Biotechnol. 100, 1977–1986. doi: 10.1007/s00253-015-7083-2
Wang S., Liu Y., Liu G., Huang Y., Zhou Y. (2017b). A new primer to amplify pmoa gene from NC10 bacteria in the sediments of dongchang lake and dongping lake. Curr. Microbiol. 74, 908–914. doi: 10.1007/s00284-017-1260-8
Wang J., Shen L., He Z., Hu J., Cai Z., Zheng P., et al. (2017a). Spatial and temporal distribution of nitrite-dependent anaerobic methane-oxidizing bacteria in an intertidal zone of the East China Sea. Appl. Microbiol. Biotechnol. 101, 8007–8014. doi: 10.1007/s00253-017-8521-0
Wang J., Yao X., Jia Z., Zhu L., Zheng P., Kartal B., et al. (2022). Nitrogen input promotes denitrifying methanotrophs’ abundance and contribution to methane emission reduction in coastal wetland and paddy soil. Environ. pollut. 302, 119090. doi: 10.1016/j.envpol.2022.119090
Weber T., Wiseman N. A., Kock A. (2019). Global ocean methane emissions dominated by shallow coastal waters. Nat. Commun. 10, 4584. doi: 10.1038/s41467-019-12541-7
Wei H., Gao D., Liu Y., Lin X. (2020). Sediment nitrate reduction processes in response to environmental gradients along an urban river-estuary-sea continuum. Sci. Total Environ. 718, 137185. doi: 10.1016/j.scitotenv.2020.137185
Wei H., Lin X. (2021). Shifts in the relative abundance and potential rates of sediment ammonia-oxidizing archaea and bacteria along environmental gradients of an urban river–estuary–adjacent sea continuum. Sci. Total Environ. 771, 144824. doi: 10.1016/j.scitotenv.2020.144824
Welte C. U., Rasigraf O., Vaksmaa A., Versantvoort W., Arshad A., Op den Camp H. J. M., et al. (2016). Nitrate- and nitrite-dependent anaerobic oxidation of methane. Environ. Microbiol. Rep. 8, 941–955. doi: 10.1111/1758-2229.12487
Wu M. L., Ettwig K. F., Jetten M. S. M., Strous M., Keltjens J. T., van Niftrik L., et al. (2011). “A new intra-aerobic metabolism in the nitrite-dependent anaerobic methane-oxidizing bacterium Candidatus ‘Methylomirabilis oxyfera’, in Biochem. Soc. Trans. 39, 243–248. doi: 10.1042/BST0390243
Wu M. L., van Teeseling M. C. F., Willems M. J. R., van Donselaar E. G., Klingl A., Rachel R., et al. (2012). Ultrastructure of the denitrifying methanotroph “Candidatus methylomirabilis oxyfera,” a novel polygon-shaped bacterium. J. Bacteriol. 194, 284–291. doi: 10.1128/JB.05816-11
Xie F., Ma A., Zhou H., Liang Y., Yin J., Ma K., et al. (2020). Niche differentiation of denitrifying anaerobic methane oxidizing bacteria and archaea leads to effective methane filtration in a Tibetan alpine wetland. Environ. Int. 140, 105764. doi: 10.1016/j.envint.2020.105764
Yang H., Chen X., Zhang C., Zhao M., Zhao X., Perry D. C., et al. (2022). Nitrogen removal by eutrophic coastal wetlands accomplished with CH4 emission reduction. J. Clean Prod. 332, 130082. doi: 10.1016/j.jclepro.2021.130082
Yang H., Shao X., Wu M. (2019). A review on ecosystem health research: A visualization based on CiteSpace. Sustainability 11, 4908. doi: 10.3390/su11184908
Yan P., Li M., Wei G., Li H., Gao Z. (2015). Molecular fingerprint and dominant environmental factors of nitrite-dependent anaerobic methane-oxidizing bacteria in sediments from the yellow river estuary, China. PloS One 10, e0137996. doi: 10.1371/journal.pone.0137996
Zhang M., Huang J.-C., Sun S., Rehman M. M.U., He S. (2020). Depth-specific distribution and significance of nitrite-dependent anaerobic methane oxidation process in tidal flow constructed wetlands used for treating river water. Sci. Total Environ. 716, 137054. doi: 10.1016/j.scitotenv.2020.137054
Zhang M., Luo Y., Lin L., Lin X., Hetharua B., Zhao W., et al. (2018). Molecular and stable isotopic evidence for the occurrence of nitrite-dependent anaerobic methane-oxidizing bacteria in the mangrove sediment of zhangjiang estuary, China. Appl. Microbiol. Biotechnol. 102, 2441–2454. doi: 10.1007/s00253-017-8718-2
Zheng Y., Hou L., Chen F., Zhou J., Liu M., Yin G., et al. (2020). Denitrifying anaerobic methane oxidation in intertidal marsh soils: Occurrence and environmental significance. Geoderma 357, 113943. doi: 10.1016/j.geoderma.2019.113943
Keywords: DAMO, scientometric analysis, M. oxyfera, M. nitroreducens, review, CiteSpace
Citation: Wei H, Wang M, Ya M and Xu C (2022) The denitrifying anaerobic methane oxidation process and microorganisms in the environments: A review. Front. Mar. Sci. 9:1038400. doi: 10.3389/fmars.2022.1038400
Received: 07 September 2022; Accepted: 29 September 2022;
Published: 13 October 2022.
Edited by:
Jing Wei, Sun Yat-sen University, ChinaCopyright © 2022 Wei, Wang, Ya and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chaobin Xu, eHVjaGFvYmluQGZqbnUuZWR1LmNu
 Hengchen Wei
Hengchen Wei Mengxin Wang1
Mengxin Wang1 Chaobin Xu
Chaobin Xu