- 1Key Laboratory of Marine Biotechnology of Fujian Province, Institute of Oceanology, College of Marine Sciences, Fujian Agriculture and Forestry University, Fuzhou, China
- 2Southern Marine Science and Engineering Guangdong Laboratory (Zhuhai), Zhuhai, China
The overuse of antibiotics leads to the emergence of bacterial resistance, which poses a serious threat to aquaculture. Antimicrobial peptides (AMPs) display excellent antimicrobial activity and are thought to be the most feasible replacements for antibiotics. The usage of AMPs as feed additives has great application prospects in aquaculture. In this study, large yellow croaker interferon-γ related gene (IFNG1R) was cloned, and a 17-amino acids (aa) short peptide named SKL17-2 was synthesized based on its protein sequence. The synthesized SKL17-2 peptide possessed a strong antimicrobial activity against Pseudomonas plecoglossicida, which could cause visceral white nodules disease (VWND) in cultured marine fish, with a minimum inhibitory concentration (MIC) of 2 μM. SKL17-2 peptide also showed weak antimicrobial activities against other tested bacteria, indicating its narrow-spectrum antimicrobial activity. This suggested that SKL17-2 peptide may not kill probiotics in intestinal flora when used as the feed additive. Furthermore, SKL17-2 had broad temperature and pH stability, low cytotoxicity, and negligible hemolysis, indicating its good biosafety and stability. Mechanistically, the synthesized SKL17-2 peptide can form α-helical structure in a membrane environment and destroy P. plecoglossicida through membrane disruption. Thus, our data showed that SKL17-2 peptide may represent a potential feed additive used for prevention and treatment of VWND.
Introduction
Aquaculture has made an important contribution to food security, but bacterial infections pose a serious threat to aquaculture, as they may result in high mortality and reduced productivity (Katzenback, 2015). Antibiotics can be used to prevent bacterial diseases. However, the overuse of antibiotics has hastened the evolution of multi-drug resistant bacteria (Roope et al., 2019). New antimicrobial drugs are urgently needed due to the high cost of conventional antibiotics used in aquaculture and the rise of drug-resistant bacteria.
Antimicrobial peptides (AMPs), also known as host defense peptides (HDPs) (Shafee et al., 2017), have the ability to kill tumor cells, parasites, viruses, fungi, bacteria, and other microorganisms (Zhong et al., 2017). Additionally, AMPs can mediate cell chemotaxis and apoptosis as well as increase cell immunity and promote wound repair (Zhong et al., 2017). AMPs are peptides that are amphiphilic (with more than 40% hydrophobic amino acids), mostly cationic (often between +2 and +10) even though some AMPs in fish shown to be anionic (Lai et al., 2002; Valero et al., 2020), and short (approximately 10 to 60 amino acids) (Rahman et al., 2018; Chaturvedi et al., 2020). Based on their structural characteristics, AMPs can be categorized into four groups, the α-helical, β-sheet, loop, and extended peptides. It is worth noting that most AMPs belong to α-helical and β-sheet peptides (Chaturvedi et al., 2020). The total net positive charge and hydrophobicity of AMPs are critical for their ability to kill bacteria. Although the compositions of gram-negative and gram-positive bacterial membranes differ, lipopolysaccharide (LPS) on the outer membrane of gram-negative bacteria and teichoic acid on the cell wall surface of gram-positive bacteria are anionic, and thus can interact with the net positive charge of AMPs (Strömstedt et al., 2009). It has been demonstrated that raising the overall net positive charge of AMPs improves their binding ability to the surface of bacterial membranes, hence boosting antimicrobial activity (Zhu et al., 2014). The hydrophobicity of AMPs can also alter their antimicrobial action. When AMPs attach to the surface of bacteria, the hydrophobic amino acids interact with the phospholipids in the bacterial cell membrane, thus damaging the cell membrane integrity (Schmidtchen et al., 2014). In general, high hydrophobicity will improve the antimicrobial ability of AMPs, but excessive hydrophobicity may damage the host cell membranes (Wood et al., 2014). AMPs can also kill bacteria via non-membrane targeting mechanisms, such as the suppression of nucleic acid and protein production and the reduction of enzymatic activity in bacteria (Brogden et al., 2005).
Based on the membrane and non-membrane targeting mechanisms, AMPs display broad-spectrum antimicrobial activity and low selection of resistance, and are considered as the most feasible replacements for antibiotics (Yasir et al., 2018; Mwangi et al., 2019). Nonetheless, endogenous AMPs have several drawbacks, such as high host toxicity and poor resistance to temperature, pH, and protease (Kim et al., 2014; Anunthawan et al., 2015). Furthermore, the expense of producing large amounts of amino acid residues restricts the therapeutic application of AMPs (Kim et al., 2014; Anunthawan et al., 2015; Huan et al., 2020). In order to overcome these drawbacks, researchers have recently concentrated on developing artificial AMPs with strong antimicrobial activity, low toxicity to host cells, high temperature and pH stability, and low production costs (Huan et al., 2020; Tan et al., 2021). The methods of AMP design include template-based design, site-directed mutation, de novo created peptides, computer design, and rational design (Huan et al., 2020). The template-based design technique uses natural protein sequence templates as a starting point, and then prefers and modifies peptide sequence according to the residue types, such as residue charge, polarity or hydrophobicity (Zelezetsky and Tossi, 2006; Pizzo et al., 2018; Yang et al., 2019). After alteration, peptide characteristics including cationicity, amphiphilicity, and hydrophobicity can be systematically adjusted to produce optimal AMPs (Huan et al., 2020). This method can lower the cost of design and synthesis while preserving natural peptide sequence information. For species whose genomes have been sequenced, for instance, the large yellow croaker (Larimichthys crocea) (Ao et al., 2015; Mu et al., 2018), novel AMPs may be generated based on the genomic sequences using the template-based design technique.
Pseudomonas plecoglossicida is a gram-negative bacterium that can cause visceral white nodules disease (VWND) in the large yellow croaker (L. crocea), orange-spotted grouper (Epinephelus coioides), and rainbow trout (Oncorhynchus mykiss) (Zhang et al., 2018). This illness, characterized by white nodules in the kidney, liver, and spleen of an infected fish, has resulted in massive economic losses in the cage-cultured large yellow croaker (Zhang et al., 2014). It has been demonstrated that an AMP named β-defensin has bactericidal effects against P. plecoglossicida (Li et al., 2021), suggesting that AMP could be used to prevent and cure VWND in fish. It is worth noting that most AMPs can indiscriminately kill pathogenic bacteria and probiotics, thereby disrupting gut flora and destroying the balance between healthy microbiota and the immune system (Eckert et al., 2012; Tan et al., 2021). As a result, AMPs with a narrow-spectrum antimicrobial activity against P. plecoglossicida would be ideal antimicrobial drugs against VWND.
In this study, large yellow croaker IFNG1R was cloned, and SKL17-2, a rationally designed peptide using the template-based design technique, was synthesized based on SKL17, a 17-aa peptide with total net positive charge and hydrophobicity existing in IFNG1R protein sequence. The synthesized SKL17-2 peptide possessed a strong antimicrobial activity against P. plecoglossicida with a MIC of 2 μM, but weak antimicrobial activity against other tested bacteria, indicating its narrow-spectrum antimicrobial activity. SKL17-2 was found to have broad temperature and pH stability, low cytotoxicity, and negligible hemolysis. Further researches showed that the α-helical structure of SKL17-2 was crucial for cell membrane disruption and antimicrobial activity against P. plecoglossicida.
Materials and methods
Complete cDNA cloning of large yellow croaker IFNG1R
RNA isolation and cDNA synthesis were performed as previously described (Zhang et al., 2022). Based on the gene sequence of IFNG1R identified in the large yellow croaker genomic sequence (GenBank accession number: NC_040020.1), primers (F: CGTTTGTATCGAAGCGGTCCATT, R: TGATTCATGATTTCTGTTTTTATTCG) were designed. And Eastep Super Total RNA Extraction Kit (Promega) was used to extrat total RNA, first-strand cDNA was synthesized by using Eastep RT Master Mix (Promega), and PCR was then performed to amplify the open reading frame (ORF) of large yellow croaker IFNG1R. PCR products were sequenced at Sangon Biotech Co., Ltd. (Shanghai, China). Subsequently, ClustalW software (Version 1.83) was used to perform the multiple sequence alignments. Protein identification was conducted using the Expert Protein Analysis System (http://www.expasy.org/tools/), and the signal peptide was predicted using the SignalP program (http://www.cbs.dtu.dk/services/SignalP/). Neighbor-joining (NJ) method of the MEGA program (version 11.0.11) was used to construct phylogenetic tree with 1000 bootstrap replicates.
Peptide synthesis
Peptides were synthesized by Sangon Biotech Co., Ltd. (Shanghai, China). Crude peptide was purified by reverse phase-high performance liquid chromatography (RP-HPLC) to a final purity greater than 95%. The synthetic peptide was stored at −80°C in the form of dry lyophilized powder and resuspended in phosphate buffer saline (PBS) when used.
Bacterial strains
Escherichia coli (ATCC 25922), Pseudomonas aeruginosa (ATCC 9027), Salmonella typhimurium (ATCC 14028), Vibrio parahaemolyticus (ATCC 17802), Staphylococcus aureus (ATCC 25923), and Streptococcus agalactiae (ATCC 13813) were obtained from American Type Culture Collection (ATCC). P. plecoglossicida was isolated from diseased large yellow croaker and preserved in our laboratory (Li et al., 2020), and this strain was found to infect a variety of fish species, such as large yellow croaker, orange-spotted grouper, rainbow trout, ayu (Plecoglossus altivelis), pejerrey (Odontesthes bonariensis), spotted seabass (Lateolabrax maculatus), mandarinfish (Siniperca chuatsi), barramundi (Lates calcarifer), and zebrafish (Danio rerio) (Zhang et al., 2018; Sun et al., 2020). Bacteria were cultivated in tryptic soy broth (TSB), and the nutritional medium for V. paraheamolyticus was supplemented with 3% NaCl.
Antimicrobial activity assays
The antimicrobial activity of the peptide was determined using the microdilution method. Bacterial cells were grown to the mid-logarithmic phase before being diluted in TSB to a final concentration of 1×105 CFU/mL. Subsequently, 10 μL of synthetic peptide was combined with 90 μL of bacterial solution in a 96-well plate. After incubation for 18 h at 37°C (28°C for P. plecoglossicida), the minimum inhibitory concentrations (MICs) were determined at 600 nm using an Infinite M Nano spectrophotometer (Tecan, Switzerland). MICs were defined as the lowest peptide concentration that inhibited more than 95% of bacterial growth. Thereafter, 50 μL of each incubation mixture was transferred to tryptic soy agar (TSA) plates for overnight incubation to confirm the minimum bactericidal concentrations (MBCs). MBCs were defined as the lowest peptide concentration that killed more than 99.9% of the bacterial cells. Antimicrobial activity of SKL17-2 was also measured by a bacterial growth inhibition zone assay. In this assay, 60 μL of peptide with the concentration of 32 μM was spotted onto an TSA plate containing P. plecoglossicida (about 105 CFU/mL in TSA), and same volume of 10 mM PBS was set as a control. The plate was stored at 28°C for 16 h to observe the inhibition zone.
Kinetics of the peptide’s bactericidal activity
P. plecoglossicida was exposed to the peptide at a concentration of 1×MBC, and the cell mixture was collected at various time points, serially diluted, and transferred to TSA plates to determine the survival rate. The PBS-treated P. plecoglossicida served as a control. The survival rate of bacteria (% survival) was calculated by dividing the number of peptide-treated cells by the number of PBS-treated cells.
Cytotoxicity and hemolysis of peptide
A CCK-8 assay was conducted to investigate the effect of the peptide on the viability of large yellow croaker macrophages. In brief, 100 μL of LYC-FM cells (about 2×104 cells) were added to 96-well plates and cultivated at 28°C. Twelve hours later, the culture medium was replaced with 100 μL of fresh medium supplemented with a range of concentrations of the peptide. Cells were cultured for 48 h before adding CCK-8 solution (Biosharp, China) to the plates. After approximately 4 h of incubation, the absorbance was measured at 450 nm with the Infinite M Nano spectrophotometer (Tecan, Switzerland). Cells treated with 2% Triton X-100 served as the positive control (A100), whereas untreated cells served as the negative control (A0). The cell viability rate was calculated using the following formula: Cell viability rate (%) = (Apeptide−A100)/(A0−A100) ×100. The LYC-FM cells were cultured in the same manner as previously described (Zhang et al., 2022).
To assess peptide hemolysis, the quantity of hemoglobin in large yellow croaker red blood cells (RBCs) following peptide treatment was evaluated. In brief, fresh blood cells from large yellow croaker were centrifuged at 500 × g for 10 minutes at 4°C, washed three times with PBS, and resuspended in PBS to a final concentration of 1.5×108 cells/mL. Then, 120 μL of RBCs were mixed with 80 μL of a series of peptide solutions and incubated for 1 h at 28°C. After centrifuging the cell mixtures at 500 × g for 10 minutes, the absorbance of the supernatants was measured at 405 nm with the Infinite M Nano spectrophotometer (Tecan, Switzerland). RBCs treated with 2% Triton X-100 served as the positive control (A100), whereas untreated cells served as the negative control (A0). The peptide hemolysis percentage was calculated using the following formula: Hemolysis (%) = (Apeptide−A0)/(A100−A0) ×100.
Temperature and pH stability of the peptide
To determine the thermal stability, the peptide was incubated for 30 minutes at 20, 40, 60, 80, or 100°C. To evaluate the pH stability, the peptide was incubated for 4 h in 50 mM glycine-HCl buffer (pH 2.0), 50 mM sodium acetate buffer (pH 4.0), 50 mM 2-(N-morpholino) ethanesulfonic acid-NaOH buffer (pH 6.0), 50 mM Tris-HCl (pH 8.0), or 50 mM glycine-NaOH (pH 10.0). After treatment, 10 μL of the peptide solution was added to 90 μL of P. plecoglossicida solution in a 96-well plate, with a final concentration of peptide at 1×MBC. After 18 h of incubation at 28°C, bacterial cells were serially diluted and transferred to TSA plates to measure the survival rate. Bacterial cells treated with PBS served as a control. Bactericidal activity was calculated as 100% survival rate minus the survival rate of each treatment.
Structure measurement
A MOS-500 spectral polarizer (Bio-Logic, Grenoble, France) was used to measure the peptide’s circular dichroism (CD) spectra between 190 and 250 nm. The spectra of samples containing 0.2 mg/mL peptide in PBS or membrane environments were examined, with the membrane environment including 60 mM SDS micelles and 100% 2,2,2-trifluoroethanol (TFE). Three independent spectra were scanned, and an average was calculated to obtain three technical replicates. The scanned spectra were then transformed to mean molar ellipticity using the formula: θM= mdeg·M/(l·c·n), where θM is the molar ellipticity (deg·cm2·dmol−1); mdeg is the measured ellipticity corrected for the buffer at a given wavelength; M is the molar mass of the peptide; l is the path length (mm); c is the peptide concentration (mg/mL); and n is the number of amino acids. The HeliQuest analysis website (https://heliquest.ipmc.cnrs.fr/cgi-bin/ComputParams.py) was used to display the helical wheel projection of SKL17-2.
Membrane integrity testing
To determine bacterial membrane integrity following peptide treatment, 180 μL of P. plecoglossicida cells (1×105 CFU/mL) of mid-logarithmic phase was combined with 20 μL of peptide at the final concentrations of 4, 8,16, or 32 μM and incubated for 2 h at 28°C. After incubation, a final concentration of 6 μg/mL propidium iodide (PI, Sigma-Aldrich) was added. The influx of PI into bacterial cells was investigated using an Accuri C6 Plus flow cytometer (BD Biosciences) with 10,000 events. The cell-penetrating efficiency was analyzed using the FlowJo software package (Tree Star).
Scanning electron microscopy imaging
The bacterial morphological alterations following peptide treatment were observed using a scanning electron microscopy. Ten microliters of peptide was added to 90 μL of P. plecoglossicida cells (1×108 CFU/mL in TSB) to a final peptide concentration of 4 μM. After incubation for 2 h at 28°C, the bacteria were fixed, dehydrated, vacuum dried, sputter coated with gold, and observed with a Sigma 300 field emission scanning electron microscopy (ZEISS) at an accelerating voltage of 5 kV. Bacteria treated with the same volume of PBS served as a control.
Statistical analysis
All experiments were carried out on three independent occasions unless otherwise stated. Data are presented as the mean ± standard error of mean (SEM). Statistical comparisons were carried out using one-way ANOVA or t-tests with SPSS software (version 19, IBM). Differences with a value of P < 0.05 were considered statistically significant.
Results
Complete cDNA sequence analysis of large yellow croaker IFNG1R
The complete cDNA sequence of large yellow croaker IFNG1R was cloned (GenBank accession number: ON997294) by PCR based on its genomic sequence (GenBank accession number: NC_040020.1). The ORF of IFNG1R consisted of 552 base pairs (bp), which encodes a protein of 183 amino acids (aa) (Figure 1A). According to the multiple sequence alignment results, the large yellow croaker IFNG1R shared the highest identity (about 54.2%) with mandarin fish IFNG1R and the lowest identity (about 5.4%) with common carp IFNG1R (Figure 1B). A phylogenetic tree was constructed to further confirm the identification of the large yellow croaker IFNG1R. Large yellow croaker IFNG1R was grouped together with other fish IFNG1R sequence (Figure 1C), indicating that the gene cloned here was large yellow croaker IFNG1R.
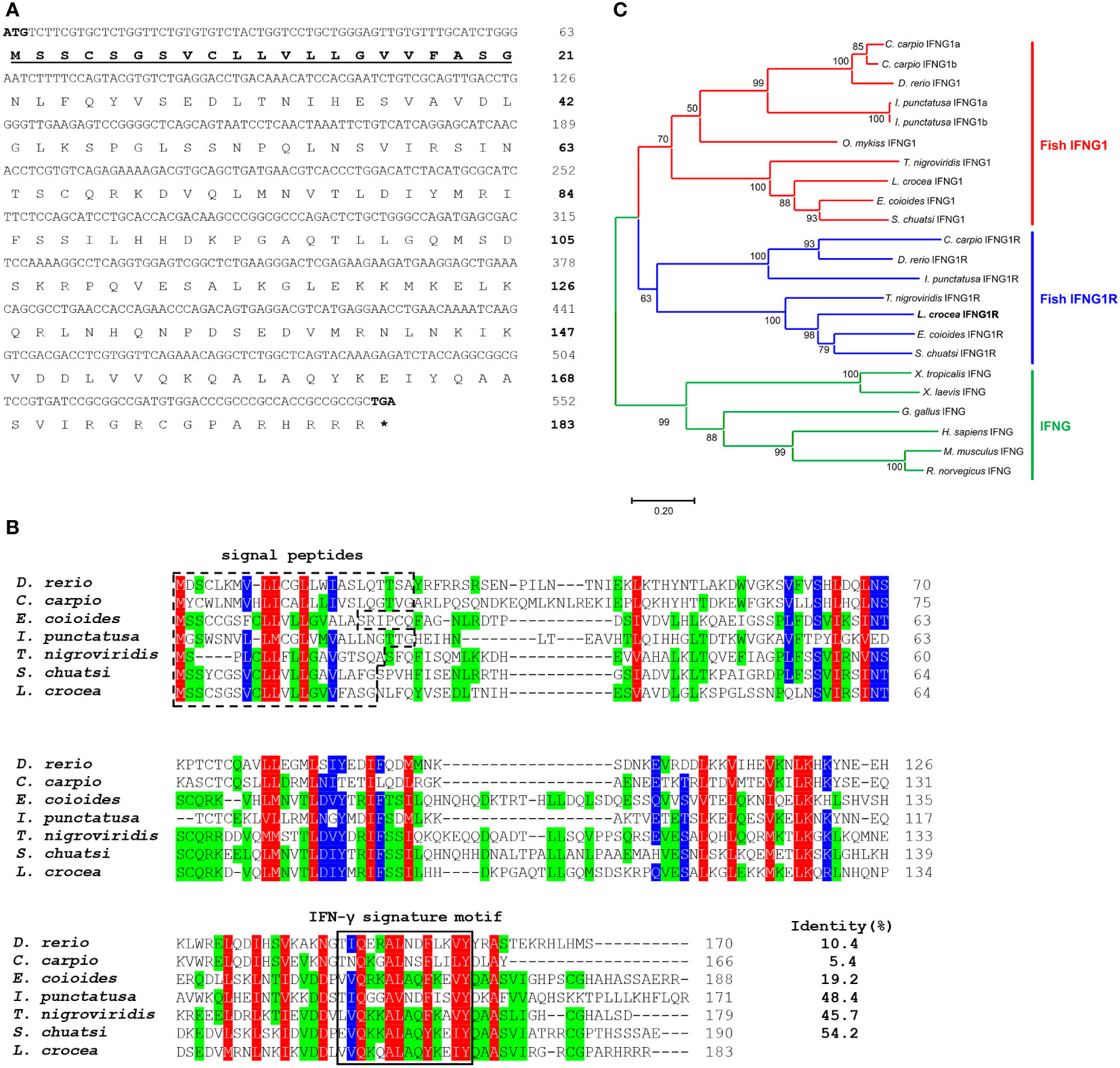
Figure 1 Sequence analysis of large yellow croaker IFNG1R molecule. (A) ORF and the deduced amino acid sequence of IFNG1R. The predicted signal peptides are in boldface and underlined. (B) Multiple sequence alignments of large yellow croaker IFNG1R with selected vertebrate IFNG1R. (C) An unrooted phylogenetic tree of vertebrate IFNG1R. Percentage values shown for each node represent 1000 bootstrap replications. In the multiple alignment and tree construction, GenBank accession numbers are as follows: human (Homo sapiens) IFNG, NP_000610.2; mouse (Mus musculus) IFNG, NP_032363.1; rat (Rattus norvegicus) IFNG, NP_620235.1; chicken (Gallus gallus) IFNG, NP_990480.1; African clawed frog (Xenopus laevis) IFNG, XP_018110090.1; tropical clawed frog (Xenopus tropicalis) IFNG, XP_002938555.1; zebrafish (Danio rerio) IFNG1, NP_998029.1; IFNG1R, BAD72865.1; common carp (Cyprinus carpio) IFNG1a, CAJ51088.1; IFNG1b, CAJ51089.1; IFNG1R, CAJ98867.1; orange-spotted grouper (Epinephelus coioides) IFNG1, AFM31242.1; IFNG1R, QEA72089.1; channel catfish (Ictalurus punctatus) IFNG1a, AAZ40505.1; IFNG1b, AAZ40506.1; IFNG1R, AAZ40504.1; spotted green pufferfish (Tetraodon nigroviridis) IFNG1, AHZ62714.1; IFNG1R, AHZ62713.1; mandarin fish (Siniperca chuatsi) IFNG1, QDO15115.1; IFNG1R, QDO15116.1; rainbow trout (Oncorhynchus mykiss) IFNG1, CAE82300.1; large yellow croaker (Larimichthys crocea) IFNG1, XP_010749999.2; IFNG1R, ON997294.
SKL17-2 possesses a strong antimicrobial activity against P. plecoglossicida
Using large yellow croaker IFNG1R protein sequence as a template, a 17-aa peptide SKL17 was synthesized. Unfortunately, the SKL17 peptide showed no antimicrobial activity against P. plecoglossicida (data not shown). Thereafter, a novel peptide known as SKL17-2 was rationally developed by replacing the seventh and twelfth glutamic acids in the SKL17 peptide with arginine (Figure 2A). Interestingly, the resultant SKL17-2 peptide possessed antimicrobial activity against P. plecoglossicida, with MIC and MBC values of 2 and 4 μM, respectively (Figure 2B). To more intuitively demonstrate the antimicrobial activity of SKL17-2, the inhibition zone method was utilized. As shown in Figure 2C, a distinct inhibition zone appeared around SKL17-2 compared with the control in the TSA plate. Subsequently, the kinetics of bactericidal activity of SKL17-2 against P. plecoglossicida were examined. The results showed that SKL17-2 could kill 90% of P. plecoglossicida within 8 h, and all bacteria were eradicated by SKL17-2 after 14 h (Figure 2D).
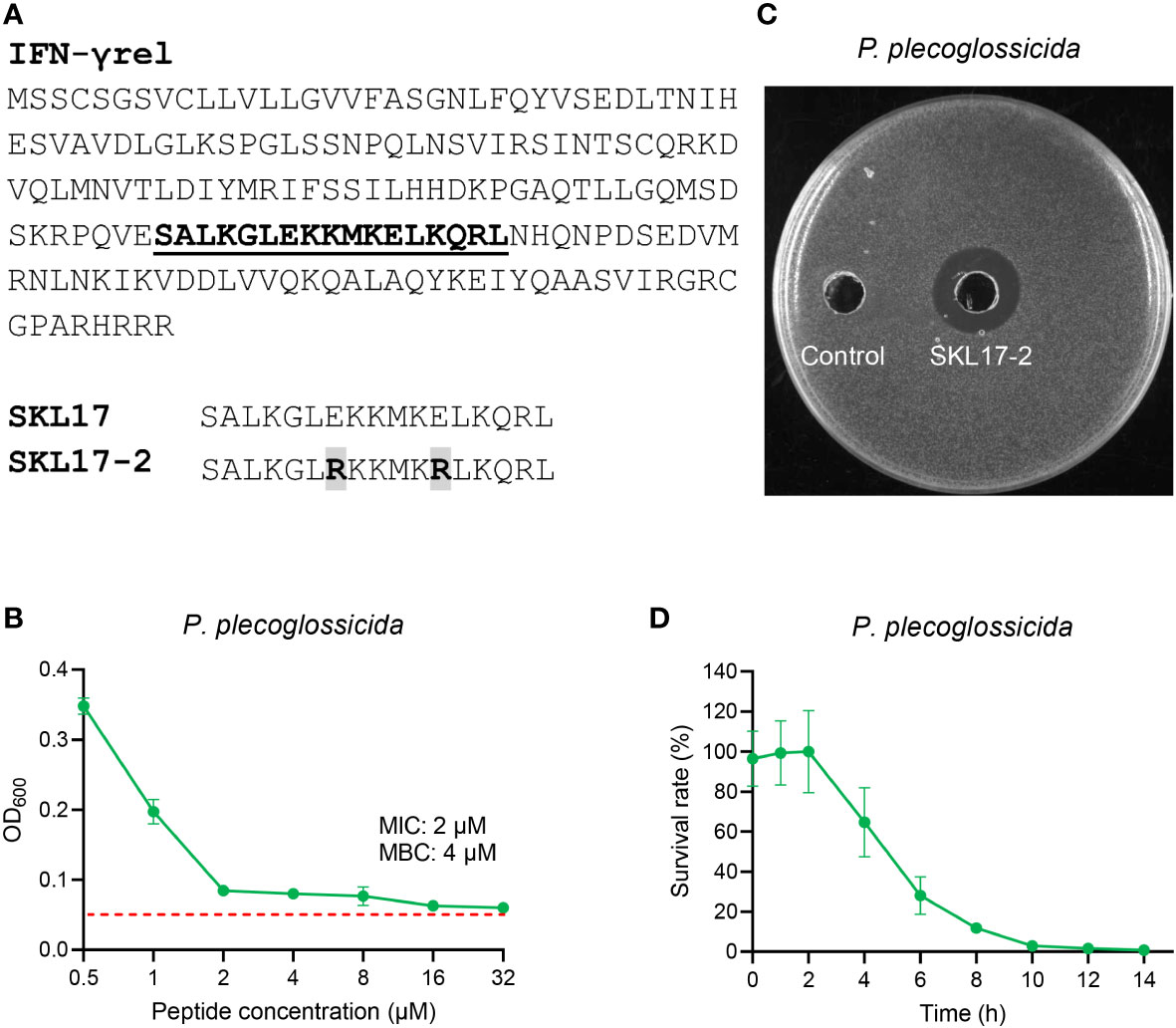
Figure 2 Antimicrobial activity of SKL17-2 against P. plecoglossicida. (A) Peptide sequence of SKL17-2. (B) Antimicrobial activity of SKL17-2 against P. plecoglossicida. The red dotted line represents the absorbance values of the medium. (C) SKL17-2 inhibition zone against P. plecoglossicida. (D) Time-kill kinetic curves of the SKL17-2 against P. plecoglossicida at 1×MBC. The kinetic graphs were the mean values of three independent experiments.
SKL17-2 possesses weak antimicrobial activity against other tested bacteria
The above results demonstrated that SKL17-2 had a strong antimicrobial activity against P. plecoglossicida, and then its activity against other bacteria was further investigated. When the peptide concentration was within 32 μM, SKL17-2 showed negligible antimicrobial activities against E. coli, P. aeruginosa, V. parahaemolyticus, and S. agalactiae, and weak antimicrobial activities against S. typhimurium and S. aureus (Figure 3).
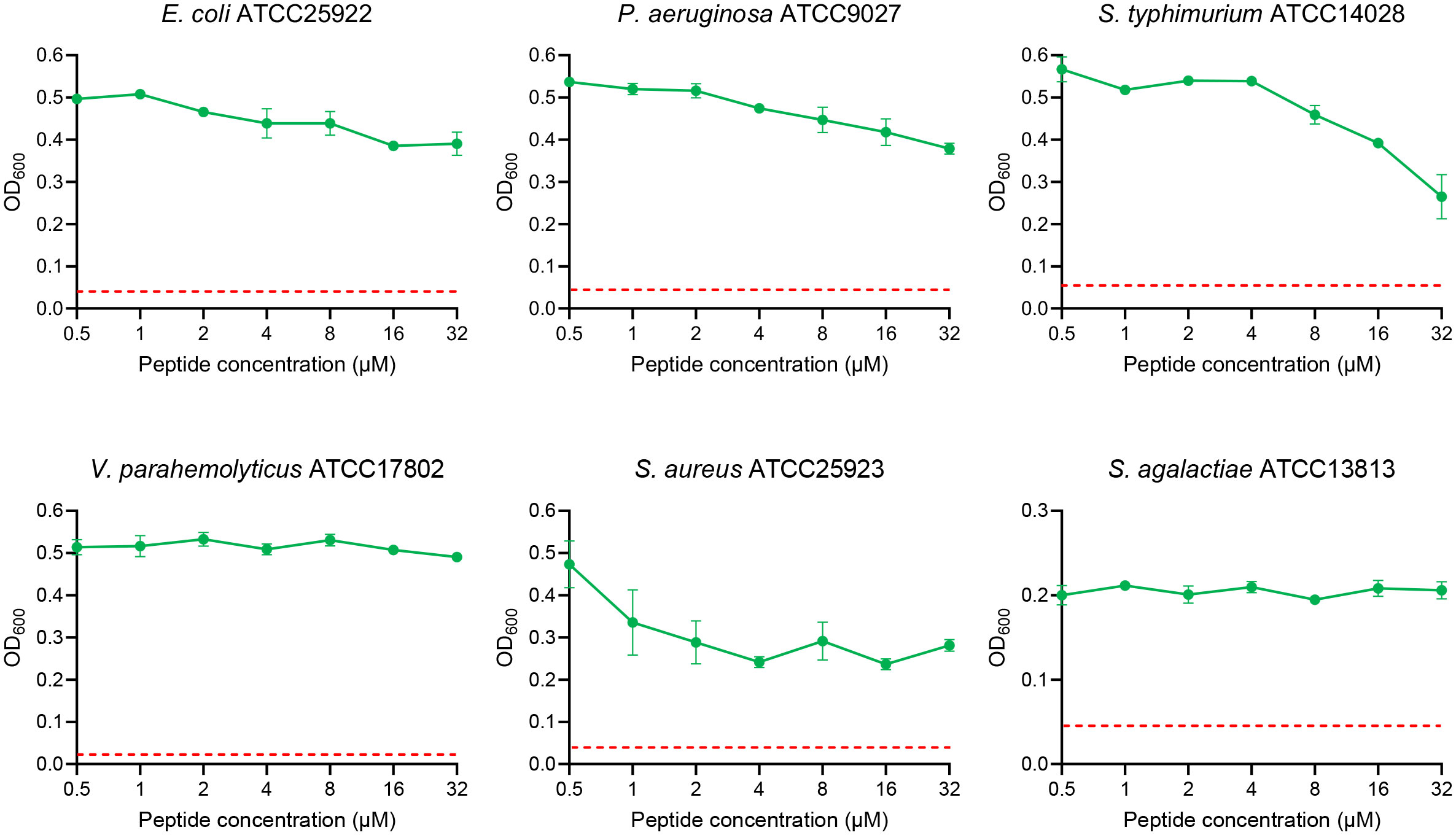
Figure 3 Antimicrobial activity of SKL17-2 against E. coli, P. aeruginosa, S. typhimurium, V. parahemolyticus, S. aureus, and S. agalactiae. The red dotted line represents the absorbance values of the medium.
Low cytotoxicity and negligible hemolysis of SKL17-2
The CCK-8 assays revealed that the cell survival rate was greater than 70% following peptide administration, indicating that SKL17-2 was only weakly cytotoxic to LYC-FM cells (Figure 4A). The hemolysis of SKL17-2 towards large yellow croaker RBCs was less than 0.5% at the tested dose, indicating that SKL17-2 had negligible hemolytic activity (Figure 4B). Thus, SKL17-2 exhibited good biosafety, further assuring the security of its clinical application.
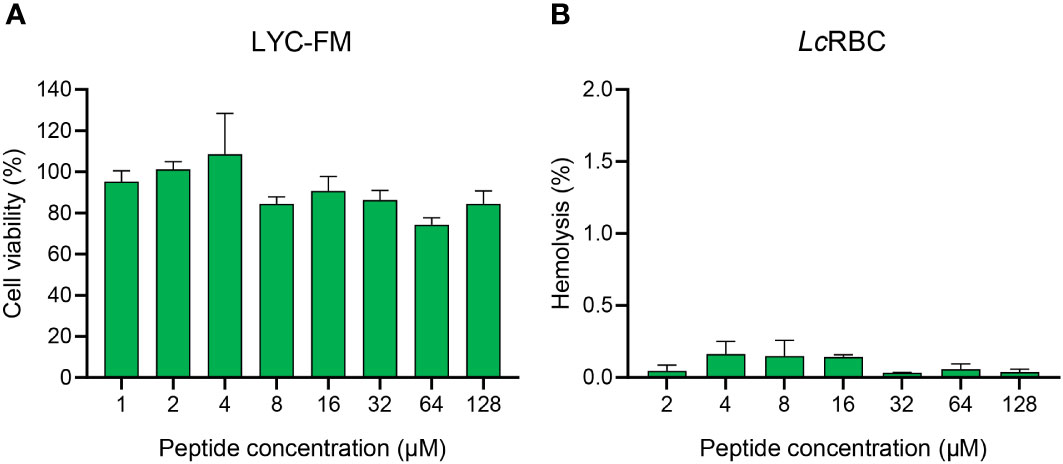
Figure 4 Cytotoxicity and hemolytic activity of SKL17-2. (A) Cytotoxicity of SKL17-2 against LYC-FM cells. (B) Hemolytic activity of SKL17-2 against large yellow croaker RBCs. The data are shown as the mean ± SEM of three independent experiments performed in triplicate.
High stability of SKL17-2 to temperature and pH
SKL17-2 displayed strong antimicrobial activity against P. plecoglossicida, a clinical pathogen that seriously affects fish aquaculture, thus suggesting its application potential in the treatment of VWND in fish. In practical applications, the stability of SKL17-2 in various conditions of temperature and pH must be considered. The antimicrobial activity of SKL17-2 was unaltered when exposed to 20, 40, 60, 80, or 100°C (Figure 5A). SKL17-2 exhibited robust antimicrobial activity at pH levels ranging from 2 to 10 (Figure 5B). These results indicated that SKL17-2 was well tolerant to different temperature and pH, even extreme conditions of temperature or pH.
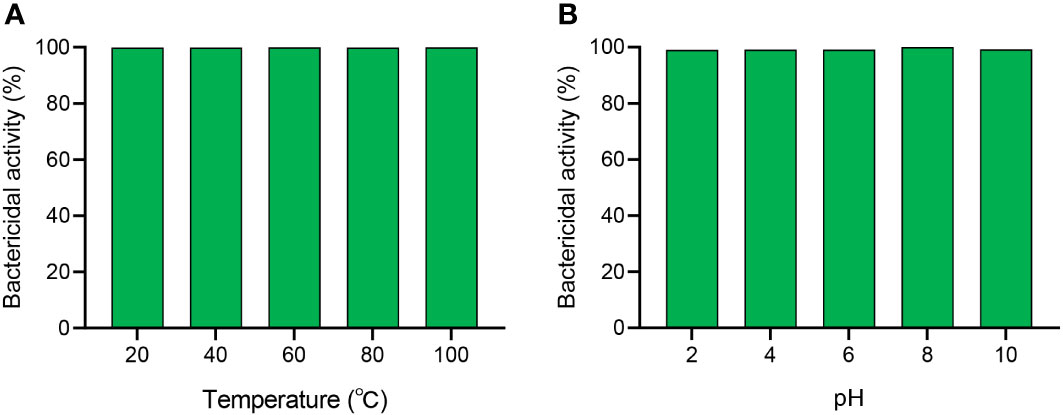
Figure 5 The effects of temperature (A) and pH (B) on the antimicrobial activity of SKL17-2. The final concentration of SKL17-2 was 4 μM (1×MBC). The data are presented as the mean ± SEM of three independent experiments performed in triplicate.
Structure analysis of SKL17-2
The secondary structure in the membrane environment is essential for peptide antimicrobial activity, and the structure of SKL17-2 in different solutions was determined using CD spectroscopy. As shown in Figure 6A, the secondary structure of SKL17-2 in PBS was characterized by a coil, with a negative minimum at 200 nm. Interestingly, the CD spectrum of SKL17-2 in SDS and TFE, which mimicked the bacterial cell membrane environment, showed two negative minima at 208 and 222 nm, implying that α-helix was the primary structure of SKL17-2. Moreover, the wheel diagram (Figure 6B) revealed that SKL17-2 exhibited a hydrophobic face and a cationic face.
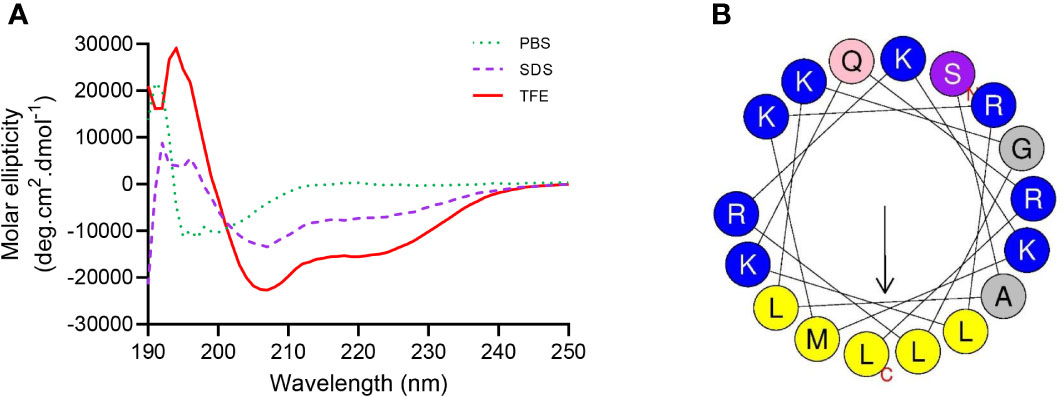
Figure 6 Structure analysis of SKL17-2. (A) Circular dichroism spectra of SKL17-2. The data from the three scans were averaged per sample, and the peptide concentration was set at 0.2 mg/mL. (B) Helical wheel projection of SKL17-2. The output of the helical wheel projection shows charged residues as blue, hydrophobicity residues as yellow, uncharged residues as light pink, alanine and glycine as gray, and serine as gray purple by default.
Bacterial membrane permeability of SKL17-2
PI staining of nucleic acid in cells can reflect a damaged cell membrane structure (Yang et al., 2019). This is a comprehensive method for determining the integrity of the cell membrane. According to the relative fluorescence intensity, the penetration efficiency of SKL17-2 to bacteria is shown in Figure 7. The results indicated that the control (no peptide) resulted in only 0.3% PI-positive cells, while SKL17-2 treatment resulted in positive nucleic acid staining of 14.1% (4 μM), 15.8% (8 μM), 41.1% (16 μM), and 50.9% (32 μM), suggesting that SKL17-2 damaged the bacterial membrane in a dose-dependent manner. These results revealed that SKL17-2 killed bacteria through membrane-permeabilizing action.
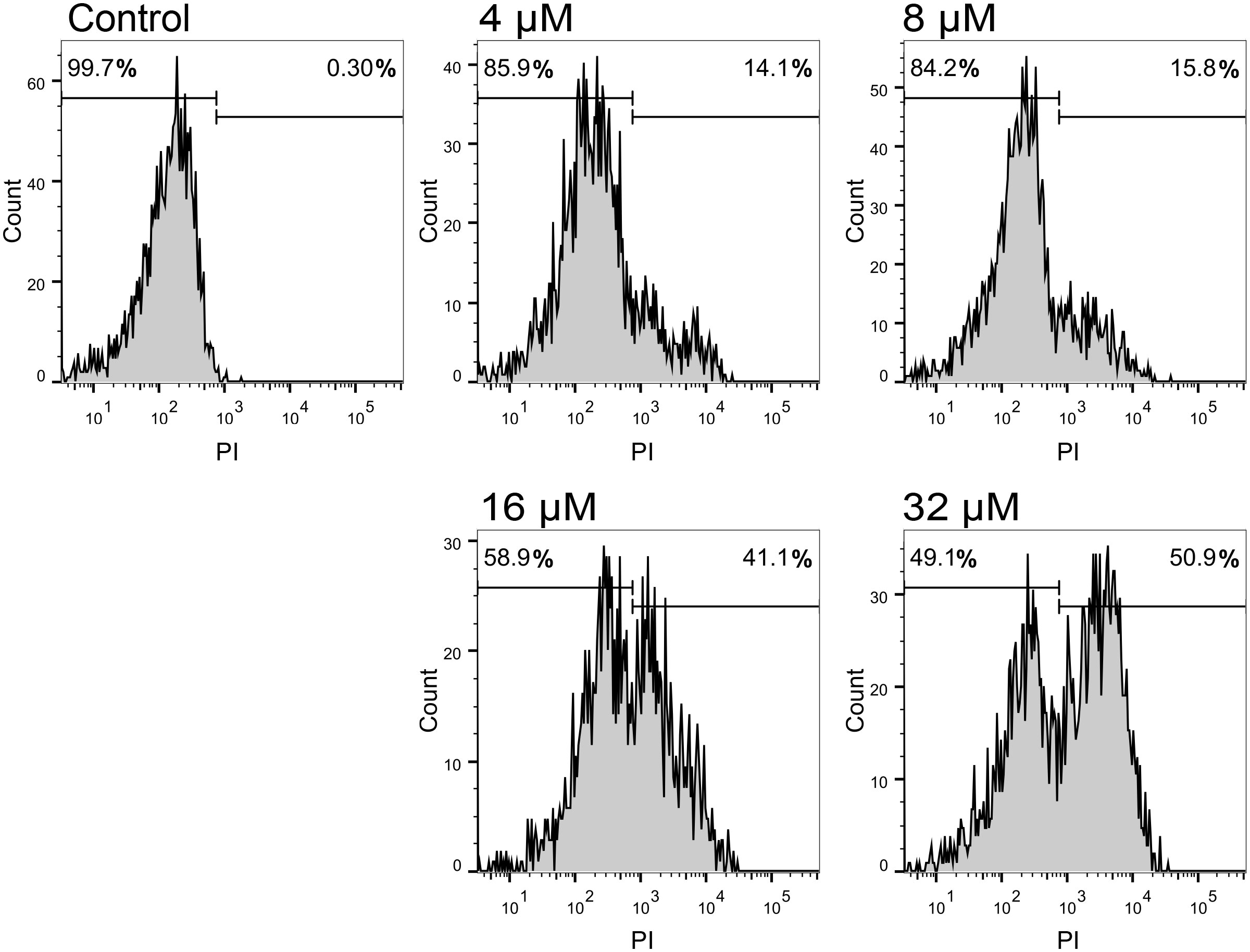
Figure 7 Flow cytometric analysis of P. plecoglossicida treated with SKL17-2 at different doses. P. plecoglossicida were incubated with SKL17-2 for 2 h at 28°C. The influx of PI into bacterial cells was then examined using flow cytometry. The cell-penetrating efficiency was shown as percentage. Data are representative of three independent experiments.
Observation of the bacteria morphology treated with SKL17-2
Scanning electron microscopy was used to observe the morphological changes in bacteria following treatment with SKL17-2. The membrane integrity of P. plecoglossicida was severely impaired by SKL17-2 treatment (Figure 8). The control bacterial cells exhibited smooth surface and normal morphologies, but P. plecoglossicida treated with SKL17-2 displayed clear morphological changes. The membrane surfaces of peptide-treated cells were extensively disrupted, becoming noticeably roughened and damaged.

Figure 8 Scanning electron microscopy images of P. plecoglossicida treated for 2 h at 28°C with 4 μM SKL17-2 or 10 mM PBS (control). Following that, the bacteria were fixed and collected on polycarbonate filters. After dehydration and vacuum drying, filters with bacteria were placed onto aluminum stubs, coated with gold, and examined under scanning electron microscope. Scale bars, 300 nm.
Discussion
P. plecoglossicida is a lethal pathogen that can cause VWND and consequent high mortality in fish (Zhang et al., 2018; Li et al., 2020). In China, fluoroquinolone antibiotics are frequently used in the treatment of fish diseases (He et al., 2012). However, the rise of drug-resistant bacteria as a result of antibiotic overuse poses a substantial threat to aquaculture and has created intractable clinical treatment bottlenecks (Katzenback, 2015). At present, developing novel antimicrobial agents is critical to combat antibiotic resistance in aquaculture. AMPs, which have unique antimicrobial activity that disrupts bacterial membranes, show the potential to replace traditional antibiotics as a novel alternative (Wang et al., 2018). However, the downsides of naturally occurring AMPs include weak efficacy, hemolytic or cytotoxic effects on host cells, and high synthetic cost. By comparison, de novo synthesized peptides may have the drawbacks of decreased bacteriostatic effectiveness and possible drug resistance (Campoccia et al., 2010). To circumvent the aforementioned drawbacks, short peptides generated artificially from natural peptide templates are thought to be an effective strategy (Luo et al., 2017).
Based on large yellow croaker IFNG1R protein sequence, a 17-aa short peptide named SKL17-2 that showed a strong antimicrobial activity was synthesized. Positive charges and overall net charges of AMPs have been shown to be crucial for electrostatic adsorption between peptides and anionic molecules on the bacterial membrane surface (Chou et al., 2016). It is worth noting that suitable positive charge of an AMP boosts its antimicrobial activity, whereas excessive positive charge may decrease its antimicrobial activity (Chou et al., 2016). Generally, a positive charge ranging from +4 to +6 is optimal for peptide’s antimicrobial activity, and positive charges comprise contributions from positively charged residues such as lysine (K), arginine (R), and histidine (H) (Yang et al., 2019). Through the amine groups on its side chain, lysine can interact with the negatively charged phosphatidylglycerol, lipopolysaccharides, and lipoteichoic acid of bacterial membranes, and the lengthy aliphatic side chain helps to localize the peptide in the lipid bilayer of the bacterial plasma membrane (Bhat et al., 2022). With its side chain guanidinium groups, arginine can form strong bidentate hydrogen bonds with the phosphor-rich membrane surface of bacteria, promoting deeper membrane insertion and making AMPs more capable of membrane disruption (Yang et al., 2019). SKL17-2 contains eight positive charges, including five lysine and three arginine residues, compared to four positive charges in SKL17, and displays strong antimicrobial activity against P. plecoglossicida, while SKL17 has no antimicrobial activity (data not shown). This may be because SKL17 lacks sufficient positive charge to adsorb to bacterial membranes. Surprisingly, SKL17-2 showed strong antimicrobial activity against P. Plecoglossicida, while exhibiting weak or negligible antimicrobial activity against other bacteria. The possible reason is that the structure of LPS varies among different gram-negative bacteria and cell walls composition in gram-positive bacteria is different (Li et al., 2018). These results indicated that total net positive charge affected AMPs’ antimicrobial activity.
Whether they are antibiotics or AMPs, ideal antimicrobial agents should target specific pathogens. If the antimicrobial agents have broad-spectrum antimicrobial activity, they may kill probiotics, causing intestinal flora diseases and disrupting the balance between healthy microbiota and the immune system (Tan et al., 2021). Interestingly, SKL17-2 peptide designed here could kill P. plecoglossicida but had a weak or negligible antimicrobial activity against other bacteria, indicating its narrow-spectrum antimicrobial activity. In contrast, β-defensin, a natural AMP, showed broad-spectrum antimicrobial activities against P. plecoglossicida as well as other bacteria (Li et al., 2021). The narrow-spectrum antimicrobial activity of SKL17-2 makes it an ideal therapeutic agent for combating P. plecoglossicida infection in aquaculture.
Many investigations have demonstrated that the formation of a stable spatial secondary structure is a significant element in AMP antimicrobial activity at the molecular level (Yang et al., 2019). In SDS and TFE solutions, our CD findings indicated that SKL17-2 had a distinct and stable α-helical secondary structure. Furthermore, SKL17-2 had an amphipathic α-helical structure with hydrophobic residues on one side and cationic residues on the other side. It has been demonstrated that the amphipathic character of AMPs is critical for their interaction with bacterial cell membranes and for their antimicrobial activity (Zelezetsky and Tossi, 2006). This effect was consistent with the membrane permeabilization and scanning electron microscopy data obtained here. P. plecoglossicida treated with SKL17-2 displayed positive nucleic acid staining, and the proportion of positive staining increased with peptide concentration, suggesting the membrane permeabilization of P. plecoglossicida following SKL17-2 treatment. Furthermore, the scanning electron microscopy data explicitly indicated that SKL17-2 caused morphological alterations of bacterial membranes such as roughening and corruption, suggesting that SKL17-2 may exert its strong antimicrobial effect primarily by destroying bacterial membrane structure.
Before clinical application, the toxicity of AMPs to eukaryotic cells should be evaluated. In our study, SKL17-2 had a low cytotoxicity to LYC-FM cells as well as a poor hemolytic reaction with red blood cells with a hemolytic rate less than 0.5%, suggesting that SKL17-2 may maintain a cell selectivity against P. plecoglossicida and would be safe for application in large yellow croaker. These findings further support the potential of SKL17-2 administration in the treatment of VWND. Our long-term goal is to use SKL17-2 as a feed additive in aquaculture, and thus antimicrobial activity variation in different temperature and pH should be evaluated due to high temperatures and complicated condition changes during feed procesing. In our investigation, SKL17-2 demonstrated good temperature and pH stability, hinting that it has an excellent therapeutic potential in clinical applications.
In summary, a peptide targeting P. plecoglossicida was developed and produced based on a large yellow croaker IFNG1R protein sequence. The peptide has low cytotoxicity and negligible hemolytic activity as well as high temperature and pH stability, indicating promising therapeutic potential in the treatment of VWND caused by P. plecoglossicida. As SKL17-2 demonstrated strong antimicrobial activity against P. plecoglossicida in vitro, the therapeutic potential of SKL17-2 as a feed additive will require further investigations in the future.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Author contributions
YL performed most of the experiments, analyzed the data, and wrote the manuscript. SY and XW contributed to antibacterial assays. RX, JC, and TH help with experimental operations. X-YZ and XC designed the research and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the National Key R&D Program of China (2018YFD0900505), National Natural Science Foundation of China (32102837), China Agriculture Research System of MOF and MARA (Grant CARS-47), Natural Science Foundation of Fujian Province (2021J05021), Fujian Science and Technology Department (2021N5008), and Training Program of Innovation and Entrepreneurship for Undergraduates (X202210389122).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Anunthawan T., de la Fuente-Núñez C., Hancock R. E. W., Klaynongsruang S. (2015). Cationic amphipathic peptides KT2 and RT2 are taken up into bacterial cells and kill planktonic and biofilm bacteria. Biochim. Biophys. Acta 1848, 1352–1358. doi: 10.1016/j.bbamem.2015.02.021
Ao J., Mu Y., Xiang L.-X., Fan D., Feng M., Zhang S., et al. (2015). Genome sequencing of the perciform fish Larimichthys crocea provides insights into molecular and genetic mechanisms of stress adaptation. PloS Genet. 11, e1005118. doi: 10.1371/journal.pgen.1005118
Bhat R. A. H., Khangembam V. C., Thakuria D., Pant V., Tandel R. S., Tripathi G., et al. (2022). Antimicrobial activity of an artificially designed peptide against fish pathogens. Microbiol. Res. 260, 127039. doi: 10.1016/j.micres.2022.127039
Brogden K. A., Guthmiller J. M., Salzet M., Zasloff M. (2005). The nervous system and innate immunity: The neuropeptide connection. Nat. Immunol. 6, 558–564. doi: 10.1038/ni1209
Campoccia D., Montanaro L., Speziale P., Arciola C. R. (2010). Antibiotic-loaded biomaterials and the risks for the spread of antibiotic resistance following their prophylactic and therapeutic clinical use. Biomaterials 31, 6363–6377. doi: 10.1016/j.biomaterials.2010.05.005
Chaturvedi P., Bhat R. A. H., Pande A. (2020). Antimicrobial peptides of fish: Innocuous alternatives to antibiotics. Rev. Aquac 12, 85–106. doi: 10.1111/raq.12306
Chou S., Shao C., Wang J., Shan A., Xu L., Dong N., et al. (2016). Short, multiple-stranded β-hairpin peptides have antimicrobial potency with high selectivity and salt resistance. Acta Biomater 30, 78–93. doi: 10.1016/j.actbio.2015.11.002
Eckert R., Sullivan R., Shi W. (2012). Targeted antimicrobial treatment to re-establish a healthy microbial flora for long-term protection. Adv. Dent. Res. 24, 94–97. doi: 10.1177/0022034512453725
He X., Wang Z., Nie X., Yang Y., Pan D., Leung A. O. W., et al. (2012). Residues of fluoroquinolones in marine aquaculture environment of the pearl river delta, south China. Environ. Geochem Health 34, 323–335. doi: 10.1007/s10653-011-9420-4
Huan Y., Kong Q., Mou H., Yi H. (2020). Antimicrobial peptides: Classification, design, application and research progress in multiple fields. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.582779
Katzenback B. A. (2015). Antimicrobial peptides as mediators of innate immunity in teleosts. Biol. (Basel) 4, 607–639. doi: 10.3390/biology4040607
Kim H., Jang J. H., Kim S. C., Cho J. H. (2014). De novo generation of short antimicrobial peptides with enhanced stability and cell specificity. J. Antimicrob. Chemother. 69, 121–132. doi: 10.1093/jac/dkt322
Lai R., Liu H., Hui Lee W., Zhang Y. (2002). An anionic antimicrobial peptide from toad Bombina maxima. Biochem. Biophys. Res. Commun. 295, 796–799. doi: 10.1016/s0006-291x(02)00762-3
Li K., Li W., Chen X., Luo T., Mu Y., Chen X. (2021). Molecular and functional identification of a β-defensin homolog in large yellow croaker (Larimichthys crocea). J. Fish Dis. 44, 391–400. doi: 10.1111/jfd.13324
Li A., Schertzer J. W., Yong X. (2018). Molecular dynamics modeling of pseudomonas aeruginosa outer membranes. Phys. Chem. Chem. Phys. 20, 23635–23648. doi: 10.1039/c8cp04278k
Li C., Wang S., Ren Q., He T., Chen X. (2020). An outbreak of visceral white nodules disease caused by pseudomonas plecoglossicida at a water temperature of 12°C in cultured large yellow croaker (Larimichthys crocea) in China. J. Fish Dis. 43, 1353–1361. doi: 10.1111/jfd.13206
Luo Y., McLean D. T. F., Linden G. J., McAuley D. F., McMullan R., Lundy F. T. (2017). The naturally occurring host defense peptide, LL-37, and its truncated mimetics KE-18 and KR-12 have selected biocidal and antibiofilm activities against Candida albicans, Staphylococcus aureus, and Escherichia coli in vitro. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.00544
Mu Y., Huo J., Guan Y., Fan D., Xiao X., Wei J., et al. (2018). An improved genome assembly for Larimichthys crocea reveals hepcidin gene expansion with diversified regulation and function. Commun. Biol. 1, 195. doi: 10.1038/s42003-018-0207-3
Mwangi J., Hao X., Lai R., Zhang Z.-Y. (2019). Antimicrobial peptides: new hope in the war against multidrug resistance. Zool Res. 40, 488–505. doi: 10.24272/j.issn.2095-8137.2019.062
Pizzo E., Pane K., Bosso A., Landi N., Ragucci S., Russo R., et al. (2018). Novel bioactive peptides from PD-L1/2, a type 1 ribosome inactivating protein from phytolacca dioica l. evaluation of their antimicrobial properties and anti-biofilm activities. Biochim. Biophys. Acta Biomembr 1860, 1425–1435. doi: 10.1016/j.bbamem.2018.04.010
Rahman M. A., Bam M., Luat E., Jui M. S., Ganewatta M. S., Shokfai T., et al. (2018). Macromolecular-clustered facial amphiphilic antimicrobials. Nat. Commun. 9, 1–10. doi: 10.1038/s41467-018-07651-7
Roope L. S., Smith R. D., Pouwels K. B., Buchanan J., Abel L., Eibich P., et al. (2019). The challenge of antimicrobial resistance: What economics can contribute. Science 364, eaau4679. doi: 10.1126/science.aau4679
Schmidtchen A., Pasupuleti M., Malmsten M. (2014). Effect of hydrophobic modifications in antimicrobial peptides. Adv. colloid Interface Sci. 205, 265–274. doi: 10.1016/j.cis.2013.06.009
Shafee T., Lay F. T., Phan T. K., Anderson M. A., Hulett M. D. (2017). Convergent evolution of defensin sequence, structure and function. Cell. Mol. Life Sci. 74, 663–682. doi: 10.1007/s00018-016-2344-5
Strömstedt A. A., Pasupuleti M., Schmidtchen A., Malmsten M. (2009). Evaluation of strategies for improving proteolytic resistance of antimicrobial peptides by using variants of EFK17, an internal segment of LL-37. Antimicrobial Agents chemother 53, 593–602. doi: 10.1128/AAC.00477-08
Sun Y., Zhu Z., Weng S., He J., Dong C. (2020). Characterization of a highly lethal barramundi (Lates calcarifer) model of Pseudomonas plecoglossicida infection. Microb. pathogenesis 149, 104516. doi: 10.1016/j.micpath.2020.104516
Tan P., Fu H., Ma X. (2021). Design, optimization, and nanotechnology of antimicrobial peptides: From exploration to applications. Nano Today 39, 101229. doi: 10.1016/j.nantod.2021.101229
Valero Y., Saraiva-Fraga M., Costas B., Guardiola F. A. (2020). Antimicrobial peptides from fish: Beyond the fight against pathogens. Rev. Aquacult 12, 224–253. doi: 10.1111/raq.12314
Wang J., Chou S., Yang Z., Yang Y., Wang Z., Song J., et al. (2018). Combating drug-resistant fungi with novel imperfectly amphipathic palindromic peptides. J. Med. Chem. 61, 3889–3907. doi: 10.1021/acs.jmedchem.7b01729
Wood S. J., Park Y. A., Kanneganti N. P., Mukkisa H. R., Crisman L. L., Davis S. E., et al. (2014). Modified cysteine-deleted tachyplesin (CDT) analogs as linear antimicrobial peptides: influence of chain length, positive charge, and hydrophobicity on antimicrobial and hemolytic activity. Int. J. Pept. Res. Ther. 20, 519–530. doi: 10.1007/s10989-014-9419-7
Yang Z., He S., Wang J., Yang Y., Zhang L., Li Y., et al. (2019). Rational design of short peptide variants by using kunitzin-RE, an amphibian-derived bioactivity peptide, for acquired potent broad-spectrum antimicrobial and improved therapeutic potential of commensalism coinfection of pathogens. J. Med. Chem. 62, 4586–4605. doi: 10.1021/acs.jmedchem.9b00149
Yasir M., Willcox M. D. P., Dutta D. (2018). Action of antimicrobial peptides against bacterial biofilms. Materials (Basel) 11, E2468. doi: 10.3390/ma11122468
Zelezetsky I., Tossi A. (2006). Alpha-helical antimicrobial peptides-using a sequence template to guide structure-activity relationship studies. Biochim. Biophys. Acta 1758, 1436–1449. doi: 10.1016/j.bbamem.2006.03.021
Zhang B., Luo G., Zhao L., Huang L., Qin Y., Su Y., et al. (2018). Integration of RNAi and RNA-seq uncovers the immune responses of Epinephelus coioides to L321_RS19110 gene of Pseudomonas plecoglossicida. Fish Shellfish Immunol. 81, 121–129. doi: 10.1016/j.fsi.2018.06.051
Zhang J. T., Zhou S. M., An S. W., Chen L., Wang G. L. (2014). Visceral granulomas in farmed large yellow croaker, Larimichthys crocea (Richardson), caused by a bacterial pathogen, pseudomonas plecoglossicida. J. Fish Dis. 37, 113–121. doi: 10.1111/jfd.12075
Zhang X.-Y., Zhuo X., Cheng J., Wang X., Liang K., Chen X. (2022). PU.1 regulates cathepsin s expression in Large yellow croaker (Larimichthys crocea) macrophages. Front. Immunol. 12. doi: 10.3389/fimmu.2021.819029
Zhong G., Cheng J., Liang Z. C., Xu L., Lou W., Bao C., et al. (2017). Short synthetic β-sheet antimicrobial peptides for the treatment of multidrug-resistant Pseudomonas aeruginosa burn wound infections. Adv. Healthc Mater 6, 1601134. doi: 10.1002/adhm.201601134
Keywords: antimicrobial peptides, IFNG1R, visceral white nodules disease, large yellow croaker (Larimichthys crocea), aquaculture
Citation: Lin Y, Yang S, Wang X, Xie R, Cheng J, He T, Chen X and Zhang X-Y (2022) A synthetic peptide based on large yellow croaker (Larimichthys crocea) IFNG1R protein sequence has potential antimicrobial activity against Pseudomonas plecoglossicida. Front. Mar. Sci. 9:1038013. doi: 10.3389/fmars.2022.1038013
Received: 06 September 2022; Accepted: 05 December 2022;
Published: 23 December 2022.
Edited by:
Lei Wang, Institute of Oceanology (CAS), ChinaReviewed by:
Yulema Valero, University of Murcia, SpainYibin Yang, Yangtze River Fisheries Research Institute (CAFS), China
Su-Ming Zhou, Ningbo University, China
Copyright © 2022 Lin, Yang, Wang, Xie, Cheng, He, Chen and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiang-Yang Zhang, enh5QG53YWZ1LmVkdS5jbg==; Xinhua Chen, Y2hlbnhpbmh1YUB0aW8ub3JnLmNu
 Yijun Lin
Yijun Lin Shunzhe Yang1
Shunzhe Yang1 Tianliang He
Tianliang He Xinhua Chen
Xinhua Chen Xiang-Yang Zhang
Xiang-Yang Zhang