- 1Department of Environmental Engineering, Korea Maritime and Ocean University, Busan, South Korea
- 2Interdisciplinary Major of Ocean Renewable Energy Engineering, Korea Maritime and Ocean University, Busan, South Korea
This study investigated the potential role of the concentration, shape, size, and polymer type of microplastics (MP), in addition to the microbial community in six coastal areas of Busan city, South Korea. The results showed that the average MP concentration was 94 ± 41 particles·m-3, and particles sized 20 μm –300 μm accounted for 67.5 ± 9.6% of the total MP abundance throughout the study sites. Further, polyethylene (PE), polypropylene (PP), and polystyrene (PS) polymers were abundant among the collected fragment- and fiber-shaped MPs, implying that highly populated and industrialized areas are major sources of MPs contamination. Moreover, the bacterial diversity and taxa-based on 16S rRNA gene sequencing was significantly different between MPs and seawater (SW) samples. Specifically, Psychrobacter, Pseudomonas, Flavobacterium, and Winogradskyella were significantly enriched in MPs compared with SW (p <0.05). Redundancy analysis revealed that nutrient concentrations, salinity, and temperature potentially contributed to shaping the microbial communities on MPs. These results indicate the dynamic and complicated interactions of MP contamination with the physiochemical and environmental factors of the surrounding area. Our findings would help in understanding of MP contamination levels in marine environments and their characteristics, as well as bacterial colonization on plastics in urban coastal areas of Busan, South Korea.
Introduction
The emergence and rapid spread of microplastics (MPs; diameter < 5 mm) has resulted in an increasing concern regarding environmental and public health risks (Barletta et al., 2019; Li et al., 2020). Most MPs are currently located in the global oceans as plastic pellets from terrestrial anthropogenic sources, and can enter directly into the marine environment (Horton et al., 2017; Mhiret Gela et al., 2022). One estimate has reported that MP pollution levels in marine environments can increase to 250 million metric tons by 2025 (Jambeck et al., 2015). A major concern is that suspended MPs in marine environments gradually disintegrate into smaller debris via ultraviolet (UV) radiation exposure, oxidation, and mechanical disruption (Shim et al., 2017). Plastic waste has accumulated in the vast space of the ocean, most notably in the Great Pacific Garbage Patch, with the lower trophic levels often consuming plastics as prey (Harrison et al., 2014; Jambeck et al., 2015). Accordingly, the ubiquitous dissemination of MPs has attracted an abundance of research to address this critical global environmental concern.
Interestingly, MPs provide habitats in marine environments for microorganisms, and have come to be termed the ‘plastisphere’ (Rummel et al., 2017; Amaral-Zettler et al., 2020). Previous studies have found that MPs are vectors for the selection and spread of marine microorganisms via colonization and biofilm formation on floating plastic debris (Jiang et al., 2018; Yang et al., 2019). Further, MPs can influence the prevalence of plastic pollution and marine microbial ecology due the small size of plastic debris, providing an environmental niche for colonization by biofilm forming bacteria and long-distance transportation (Kiessling et al., 2015; Amaral-Zettler et al., 2020). Accordingly, several studies have attempted to understand microbial communities on MP surfaces to characterize the relationships between MPs and surrounding seawater, revealing distinct patterns of microbial diversity and compositions on MPs compared to open seawater (Wright et al., 2020). Indeed, some current studies suggest that MP biofilms may serve as reservoirs for pathogens and antibiotic-resistant genes (Wu et al., 2019; Yang et al., 2019).
In Asia, MP pollution in marine environments have been extensively assessed in China (Li et al., 2015; Xu et al., 2019; Yang et al., 2019) and South Korea (Kang et al., 2015; Shim et al., 2018; Song et al., 2018; Jang et al., 2020), with a focus on the characterization of the physiochemical properties of the ocean environment. However, the colonization of the microbiome in MPs has not yet been characterized for South Korea. Owing to the ecological importance of the plastisphere, the microbial communities of coastal areas adjacent to urbanized developments could provide useful information on the potential plastic debris in marine ecosystems (Oberbeckmann et al., 2018; Li et al., 2019; Basili et al., 2020).
Previous studies have shown that MP contamination levels are significantly higher in coastal areas near urbanized or populated areas (Hurley et al., 2018; Jang et al., 2020); thus, MP contamination in marine environments is substantially influenced by land-based human activities, such as recreation, shipping, and fisheries. Busan is the second largest metropolitan area in South Korea, with 31.7% of Korean marine industries located in Busan’s global port. Further, Busan port has recently become a tourist attraction for leisure and travel. Busan also encompasses various popular recreational beaches and parks in South Korea, such as Gwanganli, Haeundae Beach located close to urbanized coastal areas. The island of Yeongdo in Busan is a growing representative area of marine urban tourism that simultaneously encompasses aspects of marine and urban leisure (Park et al., 2019; Zhan et al., 2021). Due to these geographical characteristics and marine industry activities, it was suspected that the coast of Busan city is highly contaminated by MPs (Kwon et al., 2020; Lee and Khim, 2022); however, to the best of the authors’ knowledge, no studies have characterized the microbial communities of MPs along the Korean coastal area.
To this end, the aim of the present study was to investigate the contamination levels of MPs and colonization of bacterial communities on MPs from six coastal locations near the urban environment of Busan, South Korea, and characterize the key bacterial groups using high-throughput sequencing. Furthermore, the potential relationships between the major bacterial taxa on the collected MPs and marine environmental parameters were explored.
Materials and methods
Study sites and sampling
A total of 24 MP and seawater samples were collected from six different coastal seawater samples off the southeast coast of Korea, between September and December 2020 (Figure 1). MP abundance and associated characteristics of the surrounding environment were investigated by selecting two types of coastal waters: populated and coastal industry (detailed sampling points such as latitude and longitude were described in Table 1). Sites HU (Haeundae beach) and GA (Gwanganli beach) were located in touristic areas with large populations; whereas GM (Gamman citizen park) had a similarly large population, but was adjacent to shipping and port industrial areas (Table 1). Comparatively, BS (Busan’s global port) maintains a small population, with large-scale shipping and port industrial activities, while YH (Yeongdo harbor) and KM (Korea Maritime and Ocean University Island) are also low populated areas centered around shipping routes and fishery activities. To collect MP samples, ~80 L of surface seawater (from depths of approximately 5−10 cm) from each of the six study sites were collected using sterile flasks and filtered through a stainless steel sieve (mesh size, 20 µm). Suspended plastic debris along the seawater surface were also captured using a 30 cm-wide plankton net (mesh size, 30 µm) as shown in Figure S1. After sampling, the plankton net was thoroughly rinsed with seawater to concentrate MPs in the cod-end bucket, and all samples were transferred into a 1 L bottle. Surface seawater samples were also collected at each of the six sampling sites (Figures 1; S1). All collected samples from the six sites were stored at 4°C in sterile containers placed in ice boxes, and transported immediately to the laboratory for further analysis. The natural substrate and other materials were visually classified and removed as all non-plastic particles following the previous study (Horton et al., 2017; Zhang et al., 2019; Wang et al., 2021a). Additional marine environmental parameters (Table S1), including temperature (T), salinity, pH, dissolved oxygen (DO), chemical oxygen demand (COD), total nitrogen (TN), total phosphorus (TP), suspended solids (SS), and chlorophyll-a (Chl-a) were obtained from the Korea Ministry of Oceans and Fisheries for each study area (https://www.meis.go.kr).
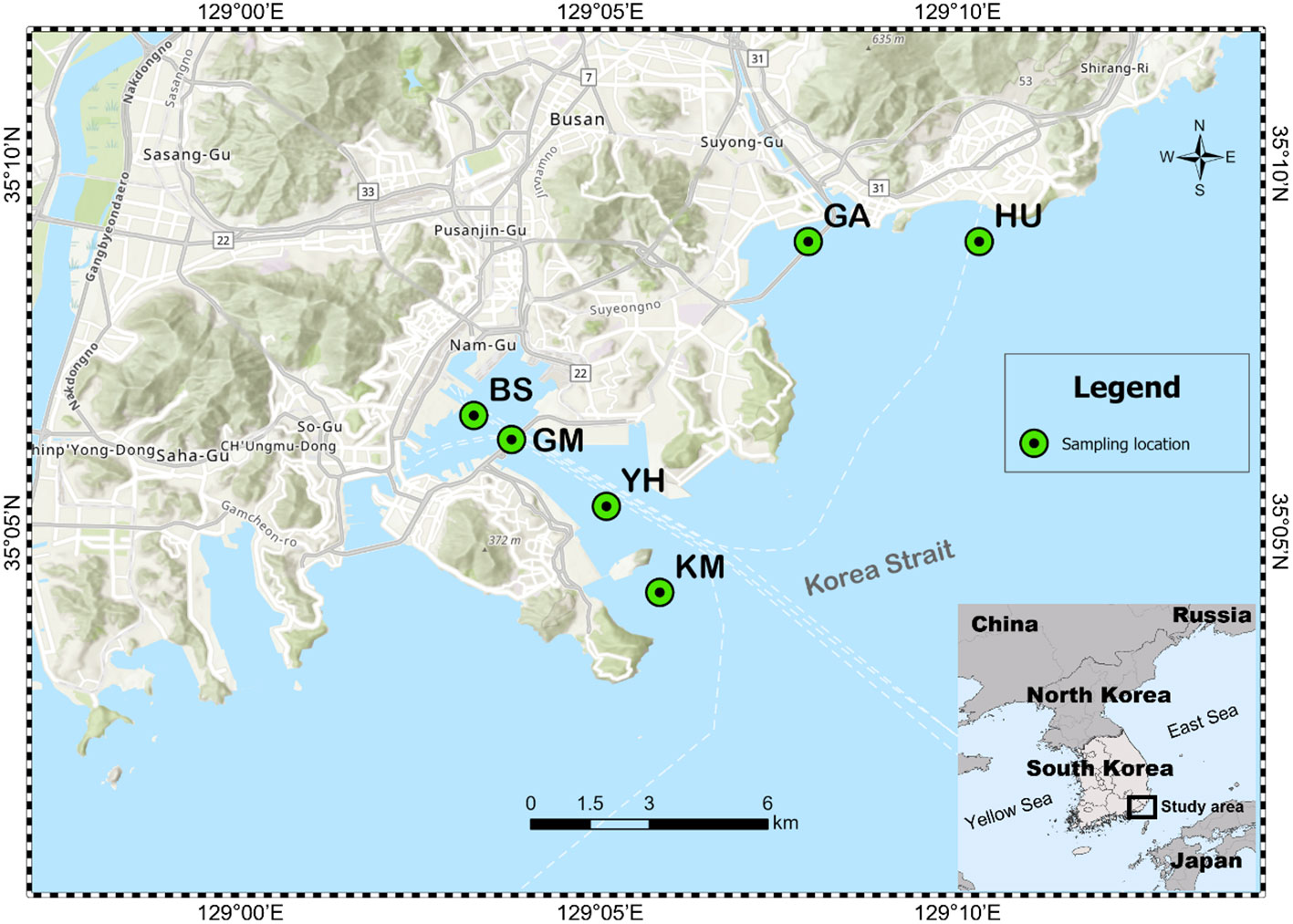
Figure 1 Six study sites selected for sampling microplastics in Busan City, South Korea. BS indicates Busan’s global port, YH indicates Yeongdo harbor; KM indicates Korea Maritime and Ocean University Island; HU indicates Haeundae beach; GM indicates Gamman citizen park; GA indicates Gwanganli beach.
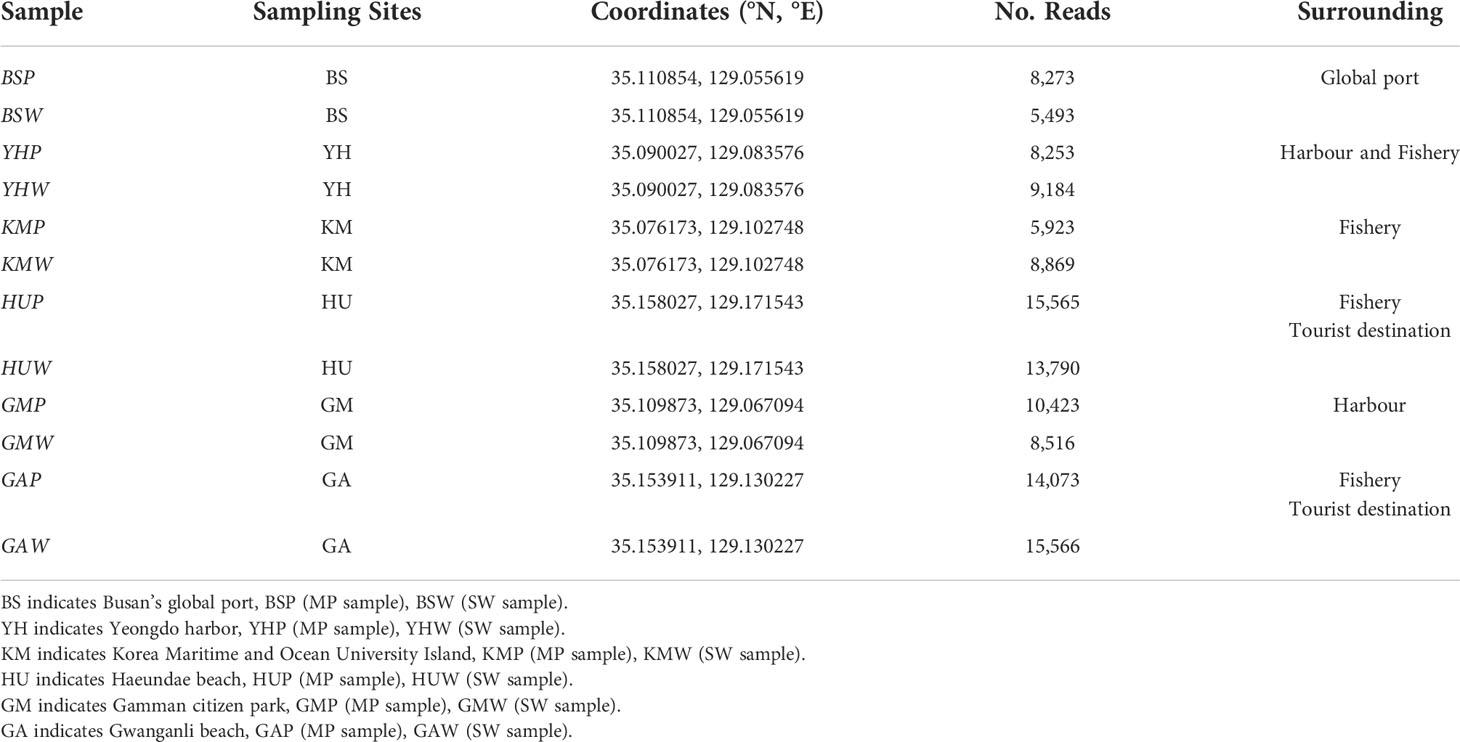
Table 1 Sampling location information, including surrounding environment characteristics and number of sequencing reads.
Microplastic identification
The protocol of the National Oceanic and Atmospheric Administration (Masura et al., 2015) was followed in using pretreatment methods for the identification of polymer types in collected MP samples. Suspected MP particles were dried at 70°C overnight in a laboratory drying oven. After drying, the organic matter was digested using 20 mL of a 35% H2O2 and Fe (II) solution on a hot plate (75°C, 180 rpm) for 30 min and filtered through stainless steel metal filters of 20 µm mesh size. Following pretreatment, the physical parameters of the MPs, such as size, shape, and morphology, were carefully observed and inspected using a stereoscopic microscope (SZX7, Olympus, Japan) (Qiu et al., 2016; Wang et al., 2021a). Identification was based on morphological features from the literature (Enders et al., 2015). Polymer characteristics of each collected MP sample were identified using Fourier transform infrared spectroscopy (FTIR) to determine their chemical composition. All FTIR measurements were performed in the attenuated total reflection (ATR) mode using a Nicolet FTIR iN10 spectrometer (Thermo Fisher Scientific, USA) with 256 scans. FTIR spectra were collected in the range of 400-4,000 cm−1, and the resulting sample spectra obtained through OMNIC™ Picta™ software (Thermo Fisher Scientific, USA) were compared with the reference OMNIC polymer spectra library for identifying the type of polymer. Only the measured similarities for spectral libraries of hit index ≥ 70% were deemed acceptable (Jang et al., 2020).
Cotton lab coats were worn at all times during analysis to prevent potential contamination. All glass vessels and materials used for the first time for one sample were covered after each step and cleaned using filtered Milli-Q water before reuse to avoid cross-contamination. The workplace for stereomicroscopic analysis was cleaned before opening and analyzing the Petri dishes in which MPs were stored.
DNA extraction and sequencing processing
The remaining particles or debris without used in pretreatment were used to extract microorganism DNA. The collected microplastics were rinsed with sterile distilled water and filtered. The biofilm attached to the microplastic surface was extracted from the collected particles on the filter using sterile tweezers. To minimize sample heterogeneity, triplicate samples from each site were pooled prior to DNA extraction. Seawater samples (2 L) were collected at each sampling site and run through nylon filters (0.2 µm pore size, Whatman, Marlborough, MA, USA). Genomic DNA was extracted from MP and seawater samples using a DNeasy PowerSoil DNA Kit (Qiagen, Valencia, CA, USA) according to the same protocols as recommend in manufacturer. The purity and concentration of the extracted DNA were determined via 1% agarose gel electrophoresis, and a Micro UV VIS spectrophotometer (Nano-300, Allsheng, China). The extracted DNA samples were stored at –80°C prior to sequencing.
The V3–V4 region of the 16S rRNA gene was amplified using primer sets 341F (CCTACGGGNGGCWGCAG) and 805R (GACTACHVGGGTATCTAATCC). Paired-end (2 × 150) 16S rRNA gene amplicon sequencing was performed using Illumina MiSeq platform (Macrogen Ltd., Seoul, South Korea). The obtained sequence datasets were deposited in the MG-RAST, under sample IDs mgm4926380 to mgm4967466.
Data and statistical analysis
The sequence alignment tool MOTHUR (version 1.44.3) (Schloss et al., 2009) was used to merge and segment the resulting sequences based on 97% sequence similarity (Kozich et al., 2013; Yoo et al., 2019). The adapter, barcode, and primer sequences were clipped. Further quality filtering of reads (permitted length = ~500 bp, max. number of ambiguous bases per sequence = 0, maximum number of homopolymers per sequence = 8, chimera filtering via vsearch (Edgar et al., 2011; Kozich et al., 2013; Yoo et al., 2018), taxonomy assignment (Wang classification, reference database SILVA SSU nr_v132 (Quast et al., 2012), required bootstrap value ≥80%, removal of chloroplasts, mitochondria, eukaryotes, unknown sequences) and picking of operational taxonomic units (OTUs, label = 0.03) were carried out (Edgar et al., 2011; Yoo et al., 2019). Sequences with the highest abundance in each operational taxonomic unit (OTU) were selected as the representative (Kozich et al., 2013; Kim et al., 2019). The OTU numerical statement and plotted output were implemented in R program (version 4.1.3).
Alpha diversity analyses were conducted to analyze the complexity of species assemblages for the two substrates (MPs and seawater—SW) using Shannon, Simpson, Chao, and ACE indices, and visualized in Rstudio and the phyloseq package (McMurdie and Holmes, 2013). Linear discriminant analysis (LDA) was used to identify similar genera between the MP and SW microbial communities using the LDA effective size analysis tool (Segata et al., 2011). Beta-diversity analyses, including principal coordinates analysis (PCoA) based on Bray-Curtis similarities, and permutational multivariate analysis of variance (PERMANOVA) using 10,000 permuted p-values were performed via the vegan package in R (v. 4.13) (Oksanen et al., 2019). Correlations between bacterial genera and marine environmental parameters were explored with a redundancy analysis (RDA) using vegan. Wilcoxon rank-sum tests were run in the ggpubr package of R for statistically evaluating the significant differences between MP and seawater samples (Kassambara, 2020).
Results
Microplastic characterization
To characterize MP contamination in the Busan coastal area, MP abundance, shape, size, and polymer type of six sampling locations were characterized (Figure 2). Particles classified by visual observation were analyzed in detail with a microscope and FT-IR, and particles that appeared to be MPs were counted. The average number of MPs across the six sites were 94 ± 41 particles·m-3 (Figure 2A). Among them, the highest concentrations were detected in GM (134 particles·m-3), HU (112 particles·m-3), and GA (106 particles·m-3), while KM (74 particles·m-3), BS (66 particles·m-3), and YH (53 particles·m-3) contained lower levels. All sites showed signs of MP pollution, with the highest levels occurring in more densely populated and urbanized areas located near coastal cities compared to others.
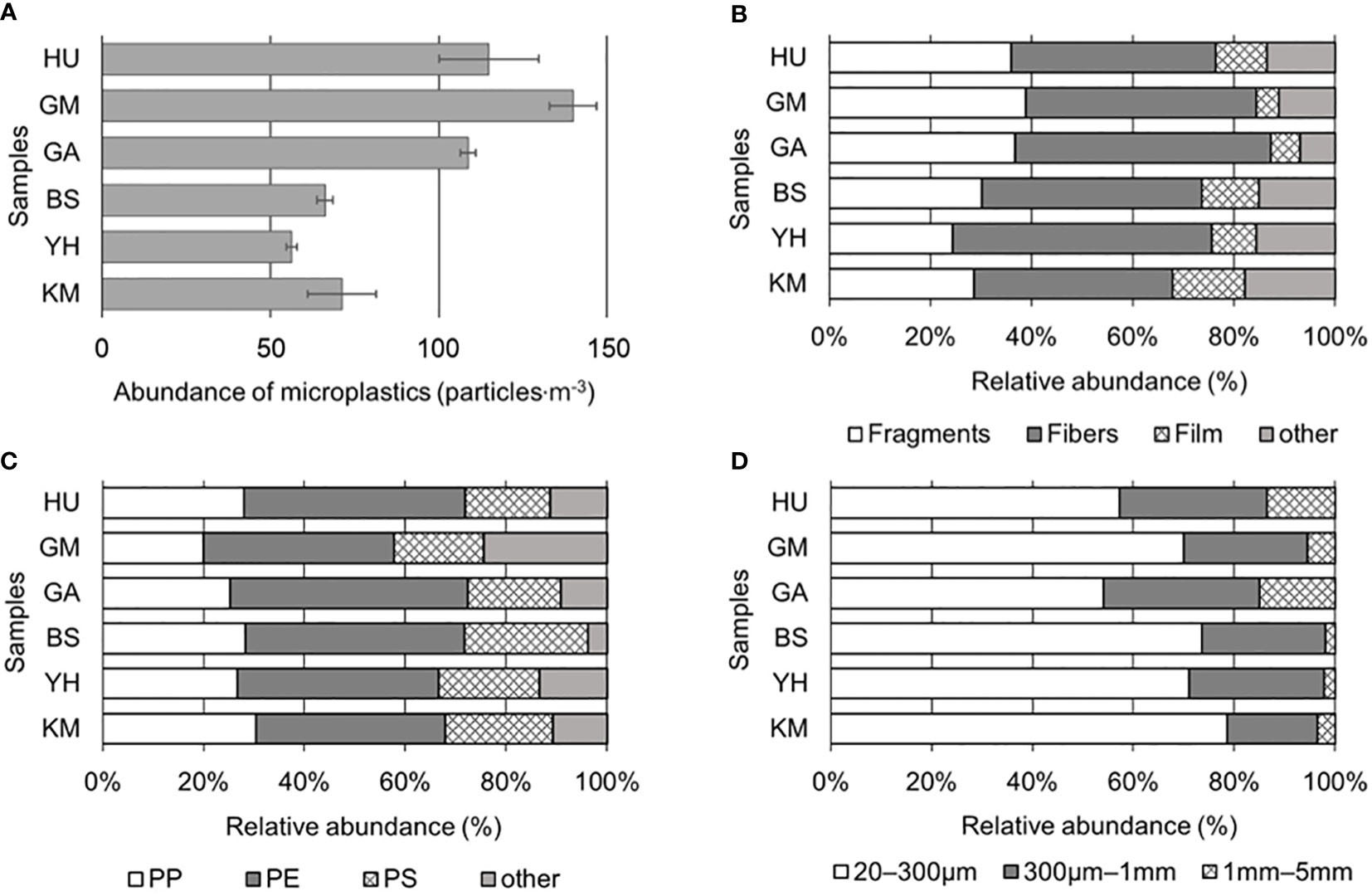
Figure 2 Microplastic (MP) compositions: (A) MP abundance (particles·L-1), (B) shapes, (C) polymer types (PP, polypropylene; PE, polyethylene; PS, polystyrene), and (D) sizes.
In this study, MP shapes were divided into four principal categories: fragments (mean 32.5 ± 5.6%), fibers (45.1 ± 5.0%), films (9.1 ± 3.6%), and other (13.3 ± 3.9%). Fragments and fibers were predominant in all samples (Figure 2B). Fragments were most abundant at GM (38.9%), followed by GA (36.8%), HU (35.9%); whereas fibers were most abundant at YH (51.1%), followed by GA (50.6%), GM (45.6%). Films were relatively higher at KM (14.3%) and BS (11.3%) compared HU (10.1%), GA (5.7%), and GM (4.4%) sites. Notably, there were no significant differences in MP shapes detected among the sites (p > 0.05). Diverse colors, such as blue, brown, black, green, yellow, and red, were detected across all MPs from the collected samples, accounting for 70% of all MPs, while the remaining 30% was comprised of white or transparent MPs. Specifically, fiber samples contained more varied colors compared to the fragment and film shapes; although, no significant differences in color were observed among shapes or sampling sites (p > 0.05).
To confirm whether MPs were plastics, FT-IR analysis was performed that identified eight polymer types among collected particles on the filter (Figures 2C; S2). It was observed that PE and PP were predominant across the six sites, where overall averages of MP polymers were PE (41.6 ± 3.8%), PP (26.5 ± 3.6%), polystyrene (PS; 19.8 ± 2.8%), and others (12.1 ± 6.8%), including polyester, polyvinyl chloride (PVC), polyethylene terephthalate (PET), polytetrafluoroethylene (PTFE), and polyamide (PA). PP was most abundant at KM (30.4%), while PE and PS were the most abundant at GA (47.1%) and BS (24.5%), respectively. At HU, GA, and GM, average abundance for PE types was the highest (42.9%), followed by PP (24.4%), and PS (17.7%). Although PE was most abundant in BS, YH, and KM, PP and PS concentrations were also significantly increased compared to HU, GA, and GM sites (p < 0.05). This study found that low-density MPs, such as PE and PP were predominant in seawater (density, 1.02 g·cm-3) compared to high-density polymers (nylon, PVC, PS, and PET) (Figure 2C).
The MPs collected from Busan coastal area were divided into three size classes (Figure 2D), where the abundance of the 20–300 μm size group (>20 μm, ≤300 μm) accounted for 67.5 ± 9.6% of the total MPs collected throughout the six study sites. MPs from 300 μm to 1 mm (>300 μm, ≤1 mm) accounted for 25.6 ± 4.6% of the total, while 1–5 mm MPs (>1 mm, ≤5 mm) maintained 6.9 ± 5.8% of the total. KM (78.6%) held the highest level of abundance in the 20–300 μm size group, followed by BS (73.6%), YH (71.1%), GM (70.0%), HU (57.3%), and GA (54.0%). In the 300 μm–1 mm size group, GA (31.0%), HU (29.2%), and GM (24.4%) sites showed relatively higher abundances compared to YH (26.7%), BS (24.5%), and KM (17.9%). Lastly, in the 1–5 mm size group, GA (14.9%), HU (13.5%), and GM (5.6%) were higher than KM (3.6%), YH (2.2%), and BS (1.9%).
Microbial community diversity and composition
The taxonomic compositions of the microbial communities in the MP and SW samples were identified using 16S rRNA gene sequencing. After quality filtering, 50,843 and 74,052 high-quality reads were obtained from the MP and SW samples, respectively, with corresponding OTU averages of 10,418 and 10,236. Based on the OTUs, bacterial α-diversities were calculated using Shannon, Simpson, Chao1, and ACE indices (Figure 3). All MP bacterial α-diversity indices were significantly higher than those of the SW samples (p < 0.05). The corresponding Shannon diversity index values for MP and SW were 4.80 and 3.56, Simpson index values were 0.978 and 0.916; whereas the species richness estimator Chao1 values were 1,239 and 1,121, and ACE values were 1,212 and 1,057, respectively, across the six sites.
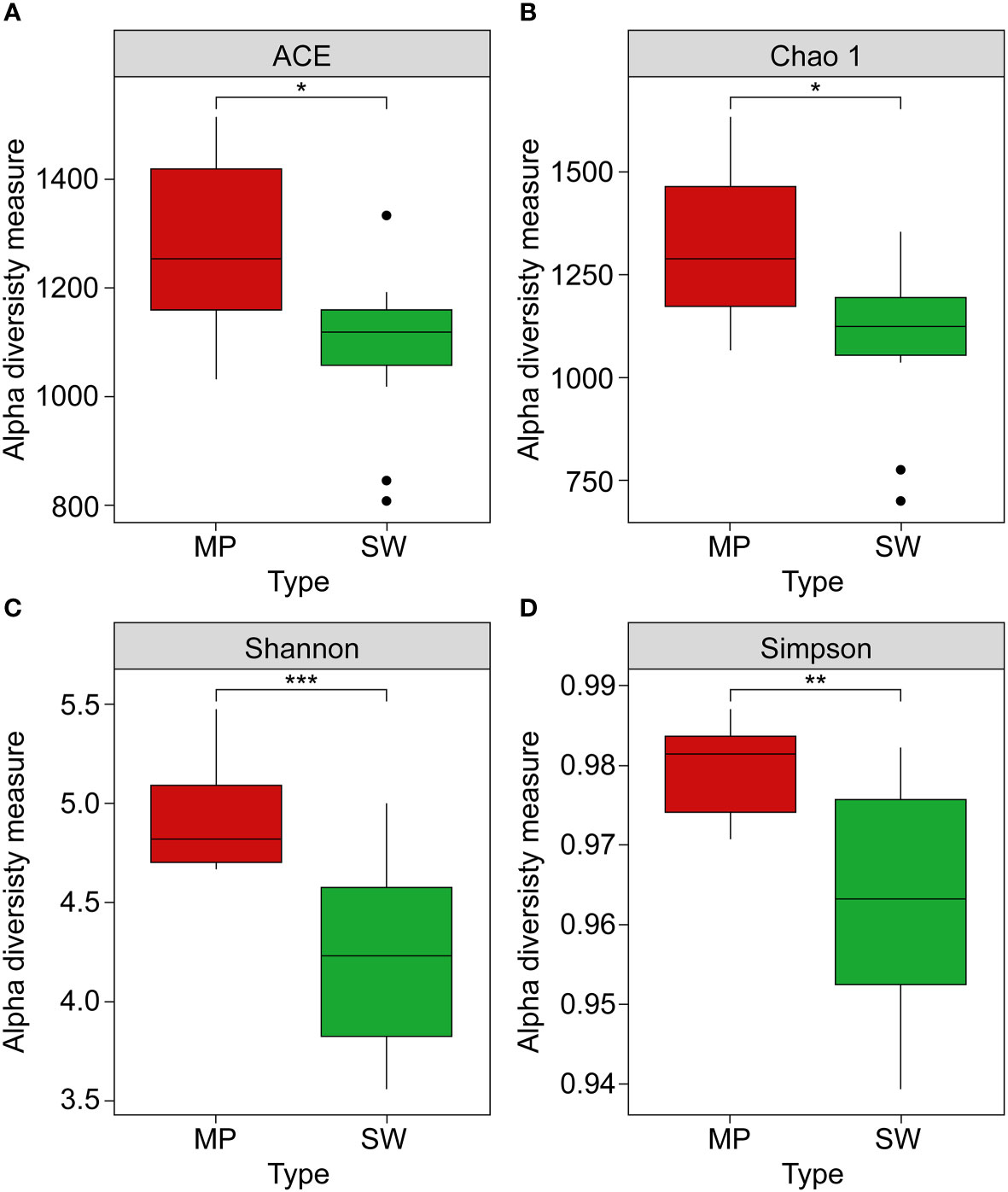
Figure 3 Alpha diversity measurements: (A) ACE, (B) Chao1, (C) Shannon, and (D) Simpson indices of microbial communities between marine plastics (MPs, red boxes) and seawater (SW green boxes) samples. Asterisks indicate a statistically significant difference between the two groups *, **, *** refer to p < 0.05, 0.01, and 0.001, respectively).
The taxonomic profiles of the MP and SW bacterial communities were characterized at the phylum, family, and genus levels (Figures 4; S3). Proteobacteria were the most abundant phylum (37.1–71.6%) in all collected samples, followed by Bacteroidetes (12.0–32.3%), Firmicutes (0.2–26.0%), and Actinobacteria (0.9–12.3%). Further, both MP and SW samples were dominated by Gammaproteobacteria (30.0% in MP and SW), followed by Alphaproteobacteria (16.2% in MP, 27.5% in SW), and Deltaproteobacteria (2.5% in MP and 1.2% in SW). Interestingly, Firmicutes accounted for 7.9% of the total OTUs from MPs at all six sites, but represented < 0.9% of the SW samples, suggesting different types of bacterial colonization in MPs. At the family level, Flavobacteriaceae and Rhodobacteraceae were relatively abundant in all sample types, with the former being the most dominant family (0.2–25.1% total reads), followed by Rhodobacteraceae (2.1–23.2%), Alteromonadaceae (0–7.2%), and Saprospiraceae (0.005–11.8%). There were no significant differences in the bacterial communities at the family level between the MP and SW samples (p > 0.05). At the genus-level, Arcobacter accounted for the highest proportion (0.1–6.5% in MP samples), followed by Pseudoalteromonas (0.02–7.9%), Zymophilus (0.03–8.9%), Acinetobacter (0.006–5.8%), Maribacter (0.002–4.1%), and Simplicispira (0–3.9%). In SW samples, Glaciecola was the most abundant genus (0.6–6.6%), followed by Arcobacter (0.5–8.1%), Aurantivirga (0.7–3.7%), Planktomarina (0.4–7.1%), Amylibacter (0.6–3.0%), and Persicirhabdus (0.01–6.0%).
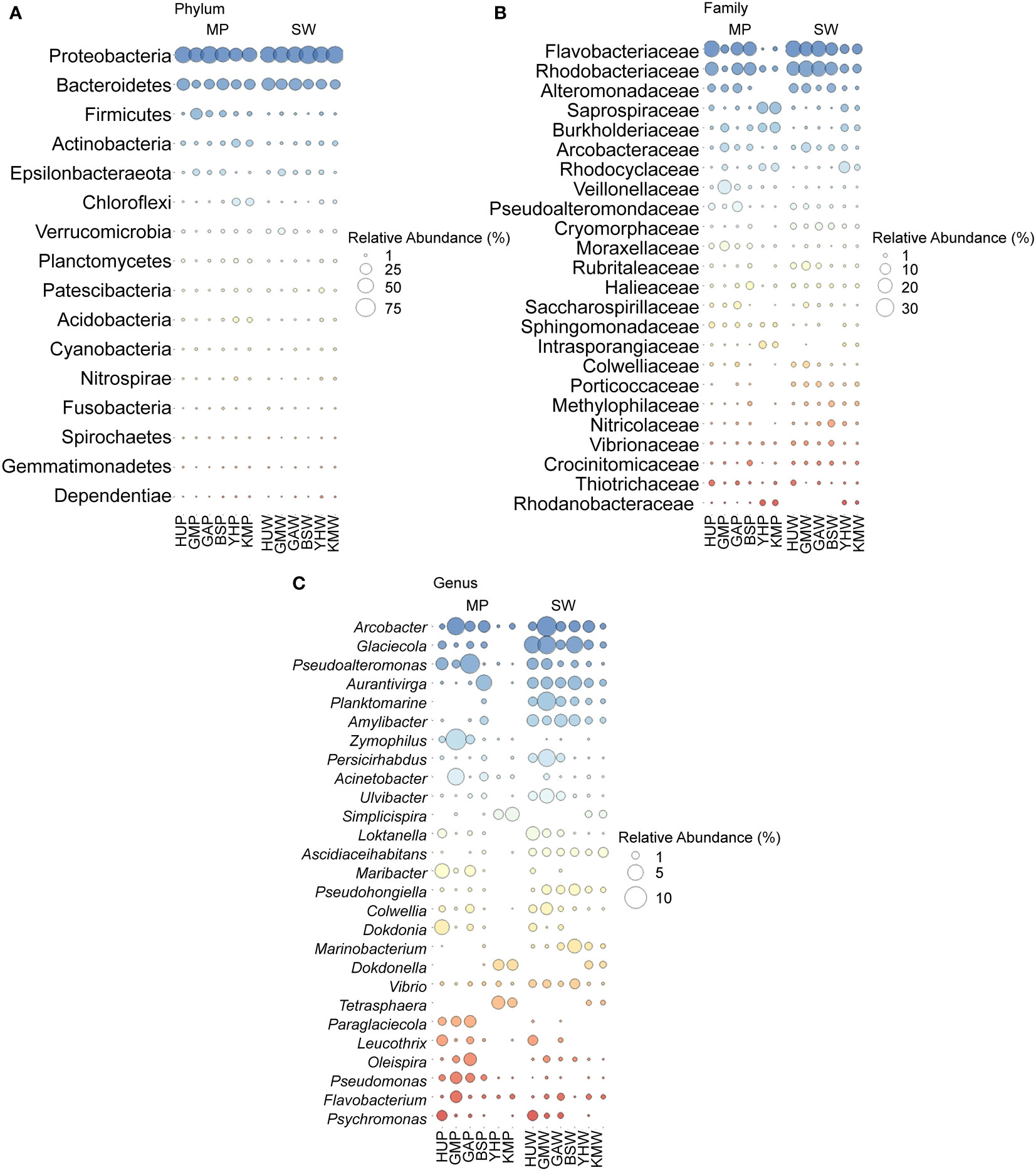
Figure 4 (A) Major phyla of bacterial communities (top 16) in MP and SW from six sites. (B) Major families of bacterial communities (top 24) in MP and SW from six sites. (C) Major genera of bacterial communities (top 27) in MP and SW from six sites. All bacterial taxa levels were expressed as mean value from September and December samples in this study. HUP, GMP, GAP, BSP, YHP, KMP indicate the MP particle samples; HUW, GMW, GAW, BSW, YHW, KMW indicate the SW samples.
PCoA based on Bray-Curtis dissimilarities were conducted to understand if the main genera were influenced by the bacterial colonization of MPs (Figure 5). As such, the bacterial compositions of the 24 MP and SW samples were clustered, indicating high levels of heterogeneity in bacterial structures and taxa. Additionally, analysis of similarities (ANOSIM) results showed significantly different bacterial compositions between the MPs and SW samples (R = 0.519, p < 0.01). The first two principal components explained >76% of the total variance in the data matrix. In particular, samples located in different MP habitats appeared dissimilar, although SW samples were clustered.
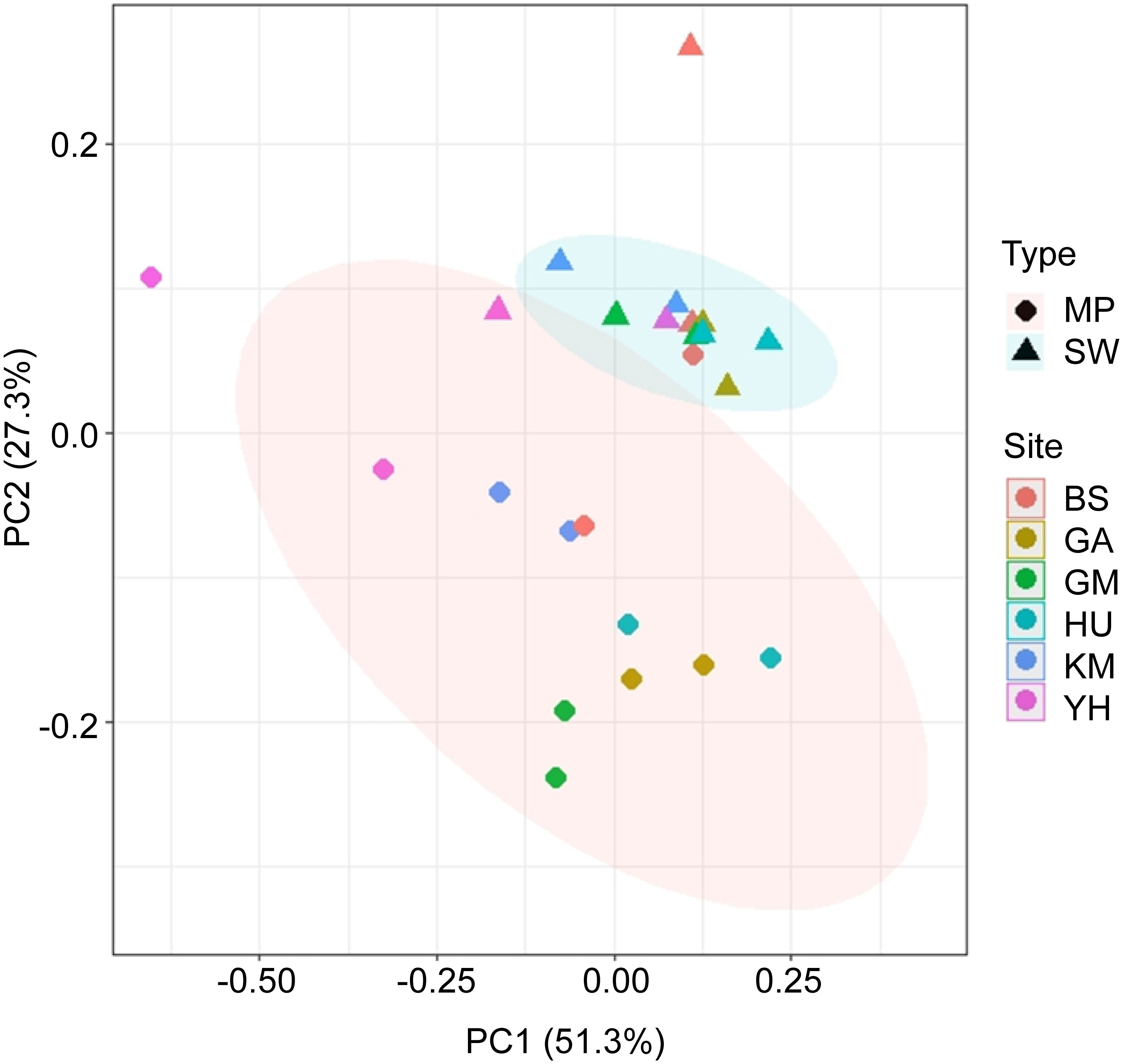
Figure 5 Principal coordinate analysis (PCoA) plot based on Bray–Curtis dissimilarity of genus-level relative abundances between MP and SW samples. The red and blue ellipses represent the clustering of the MP and SW samples, respectively.
To identify the genus-level taxonomic community differences between MP and SW samples, LefSe analyses revealed significant differences in Sneathiella, Flavobacterium, Lactobacillus, Desulfovibrio, Paraglaciccola, Winogradskyella, Pseudomonas, and Psychrobacter (Figure 6); however, the SW samples had fewer species with significant differences compared to MP samples, including Colwella, Arcobacter, and Glaceicola. The results reveal that MPs can enhance the bacterial community.
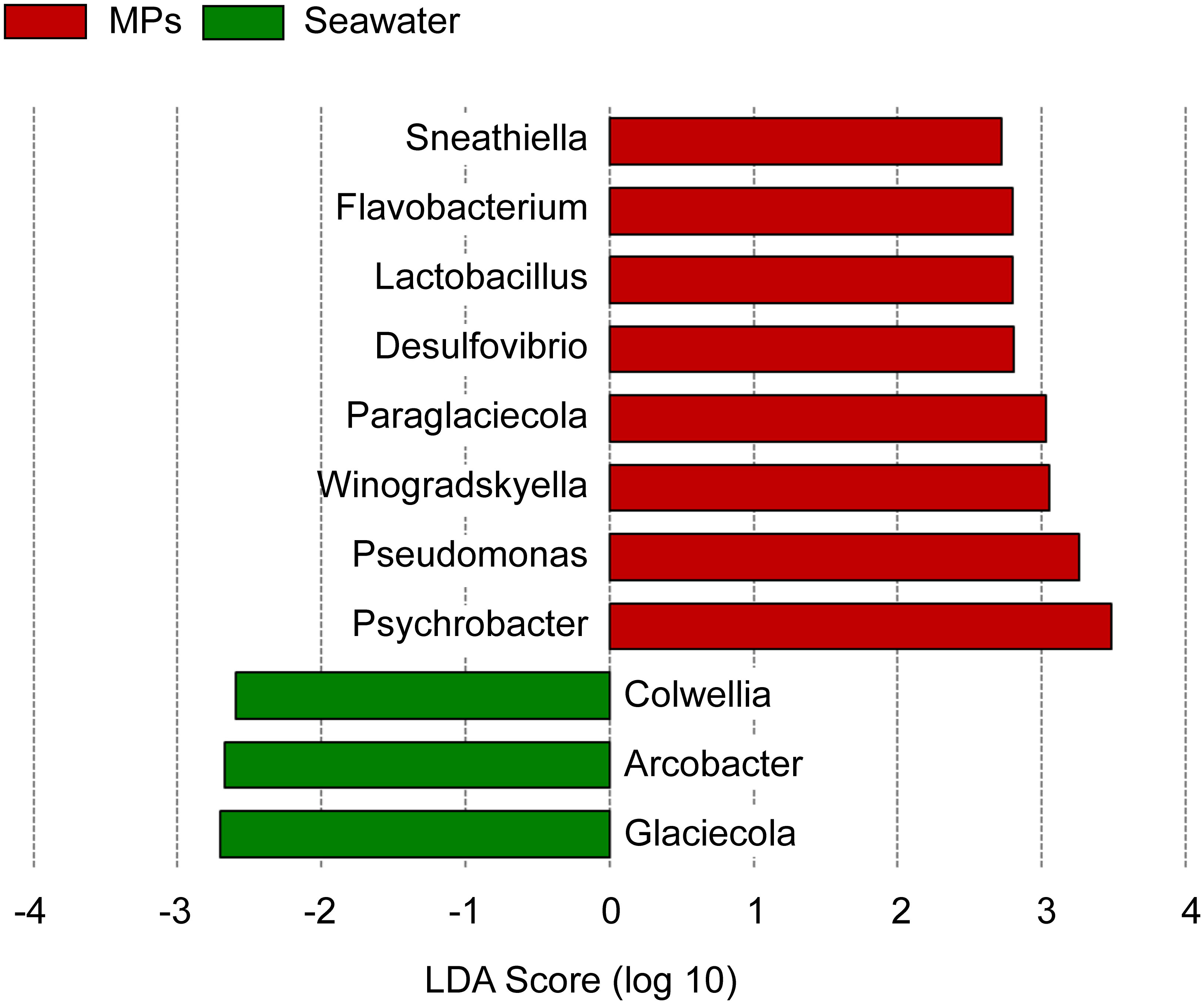
Figure 6 Linear discriminant analysis (LDA) effect size (LEfSe) for the bacterial genera showed the most divergent bacterial communities between the MPs and SW samples.
Potential relationships between MPs and marine environment
RDA was conducted to investigate potential relationships between the identified MP bacterial genera and marine environmental parameters (Figure 7). The analysis revealed that TN, TP, Chl-a, temperature, COD, DO, SS, and salinity were important factors affecting variations in MP bacterial community composition. Here, TN and TP were negatively correlated with most genera, including Paraglaciccola, Pseudomonas, Lactobacillus, and Psychrobacter. Polysaccharide-degrading bacteria, such as Dokdonia, Maribacter, Flavobacterium, and Leucothrix, were positively associated with temperature and Chl-a.; whereas COD, DO, SS, and salinity of the surface water were positively correlated with the distribution of Winogradskyella and Colwellia. These results imply that the environmental conditions in coastal areas influence the MP-attachment of bacterial communities in marine environments.
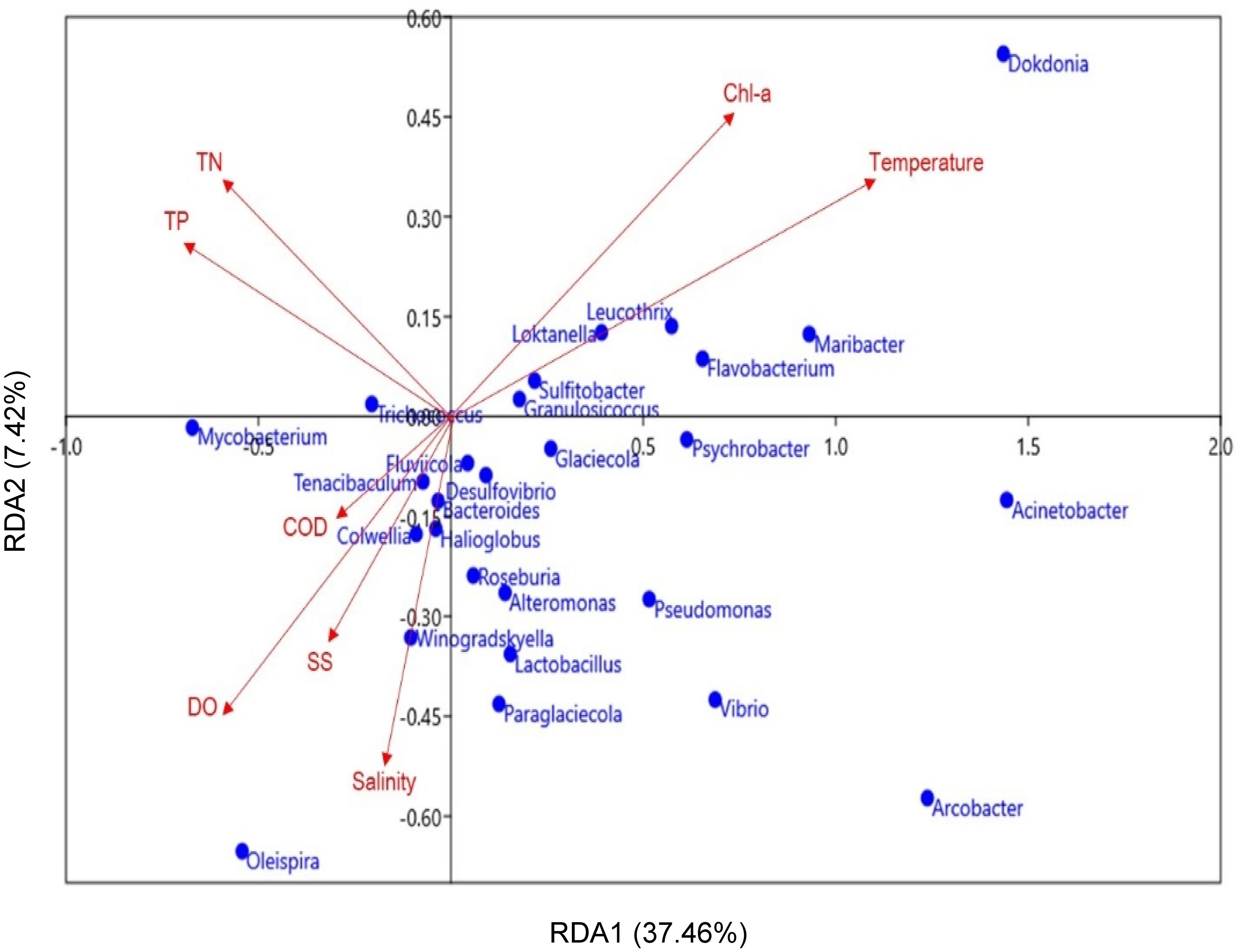
Figure 7 Redundancy analysis (RDA) plot of the correlations between the major MP bacteria (genus-level, blue dots) and marine environmental parameters (red arrows). Significant differences in environmental parameters (p < 0.05) were only included in the present study.
Discussion
Recently, various studies have been conducted to understand the marine ecology of plastispheres according to the spatiotemporal diversity and composition of MPs in sediments, bays, and coastal marine environments (Zhao et al., 2018; Zhang et al., 2019; Teng et al., 2020). However, to date, no studies have investigated the microbial communities on suspended plastic debris in the Busan coastal area. Although it offers some of the most attractive marine tourism spots in South Korea, such as beaches, islands, and recreational markets, limited information is available on MP contamination, particularly the specialized colonization of different MP types in the country. In this study, the distribution of MP shape, polymer type, and size were investigated, and the corresponding microbial community dynamics were compared across six representative coastal sites. Further, potential relationships between bacterial communities on MPs and marine environmental parameters were analyzed.
Microplastic contamination in coastal area
As expected, the results indicated that MP abundance was relatively higher in densely populated and more active areas, where PP, PE, and PS fragments and fiber shapes were predominant (Figure 2). Overall, the results showed a relatively high abundance of MPs (94 ± 41 particles·m-3) in the Busan coastal area. Previous studies have reported that approximately small-sized plastic (MPs < 1 mm) with abundance of 378 ± 284 particles·m-3 were detected in domestic coastal regions, fish farms, and marine sediment, whereas small-sized plastic concentrations of ~545 ± 282 particles·m-3 were observed in surface seawaters of the Yellow Sea (Kang et al., 2015; Song et al., 2018; Kwon et al., 2020). Moreover, previous Busan area studies showed MP concentrations ranging from 1.35 particles·m-3 to 1020 ± 279 particles·m-3 (Song et al., 2018; Kwon et al., 2020). The abundances of microplastics in the surface seawaters of Mediterranean regions were low compared to this study and had 0.29 ± 0.35 particles·m-3 in the Menorca Channel (Spain) (Ruiz-Orejón et al., 2019), 0.246 ± 0.076 particles·m-3 in the Italian Minor Islands (De Lucia et al., 2018), and 4.3 ± 2.2 particles·m-3 in the Lebanese coast (Eastern Mediterranean Basin) (Kazour et al., 2019). The abundance of MPs in Nanxun reef and Ma’an Islands (500–1,773 items/m3) is lower than that in the South China Sea, Maowei Sea, and Yangtze Estuary (2,569-4,500 items/m3) (Jiang et al., 2022). The estuaries and coastal areas are generally closer to residential and industrial areas than islands and reef areas. The results suggest that heavily populated areas with extensive anthropogenic activities enhance MP accumulation and concentration due to its close correlation with human activity levels on land and sea (Anderson et al., 2017). Even when measured in the same area, the degree of accumulation showed different results depending on the collection period. The significant changes in MPs’ circulation and dispersion due to the complex interactions between natural and anthropogenic impact might explain the differences in the MP concentration in the same area (Tsang et al., 2017). Other potential factors are the ocean current, waves, effluents from treated wastewater nearby the coast area, and weather conditions, such as rain and wind that might influence the MP’s abundance (Zhang et al., 2017; Gela and Aragaw, 2022). Presently, direct comparison of the MP abundance in the same and different regions is difficult due to the lack of standardization methods for assessing small-sized plastics, including collection, pretreatment processes, and unit display methods, in addition to difficulties in monitoring of any changes in the marine and coastal environments at the time of study (Shim et al., 2018; Song et al., 2018).
Fiber-based MP shapes were predominant (> 90%) in the marine environmental samples from coastal seawaters and sediments in China and Korea, as well as in the surface and subsurface Arctic waters from Norway (Lusher et al., 2015; Nel and Froneman, 2015; Xu et al., 2019; Jang et al., 2020). Because fibers have a relatively low densities compared to other MP shapes (e.g., fragments and films), they are often the most abundant MP shape in seawater samples (Li et al., 2015; Kwon et al., 2020). Generally, discharged laundry water from residential areas are suspected as a major point source of fibers (Hartline et al., 2016; Hernandez et al., 2017). Washing of clothes potentially contributes to the high abundance of fiber, as HU, GA, and GM regions have more people living and staying in accommodations near the coast than in BS, KM, and YH. In addition, the high abundance of fiber shapes observed in coastal waters could be partly explained by fishing materials, recreational boating, airborne contaminants, industrial wastewater, and wastewater treatment plant effluents (GESAMP 2015; Ramasamy et al., 2022). BS, KM, and YH locations also predominantly have fiber-shaped MPs. It is suspected that fishing materials, such as fishing gear and fish cages or nylon rope, and rope wear material are potential sources of MPs due to fishing activities, in addition to activities such as the movement and anchoring of trade ships. The primary sources of fragments are considered to be riverine discharge from land or ship spillages along coastal areas (GESAMP 2015; Eo et al., 2018). Furthermore, the breakdown of plastic containers, packaging materials, and cosmetics are also regarded as potential sources (Yan et al., 2019). Films have been reported as the secondary highest abundances of MP shapes due to their low density, leading to high detection levels in marine environments (Zhou et al., 2020). As films mainly arise from fragments of plastic bags composed of PE or PP (Zhou et al., 2020), the concentration of film-shaped microplastics is likely to be increased due to the many uses of plastic packaging in areas close to fisheries or industries. Since the BS area is an international shipping and logistics complex and the KM and YH are areas with many fishing activities, it is possible that plastic packaging or products can contribute to increasing films observed in BS samples compared to other samples (GESAMP 2015; Zhou et al., 2020). Thus, the characteristic MP shapes in Busan were mostly because of its proximity to local vessel activity, waste from nearby tourist attractions, and consequent land-based sources. MPs < 300 μm in size were detected in the surface of seawater and sediment area in South Korea according to previous studies (Eo et al., 2018; Song et al., 2018; Jang et al., 2020). It is suspected that smaller MPs (20–300 μm) are potentially derived from the movement of ships, ocean currents, waves, or UV-degradation (GESAMP 2015; Oberbeckmann et al., 2018).
According to results of MP polymer type composition analyses, PE and PP accounted for the highest proportions among MPs. Elsewhere, it has been observed that PE and PP are the most common polymers in seawater, marine sediments, and marine organisms worldwide (Zhang et al., 2017; Phuong et al., 2018). The high abundance of PE and PP polymer types and fragment and fiber shapes is due to the surrounding environment of the coastal area being urbanized and industrialized (Oberbeckmann et al., 2018; Ramasamy et al., 2022). Urban stormwater runoff can carry MP as dust-associated, construction activities, artificial turf, landfill leaching, degraded litter, etc., and the main types of polymers of non-point source MPs observed are PE, PP, and PS (Bailey et al., 2021; Tan et al., 2022). Also, the most prominent MP polymer in the final wastewater effluent is PE, mostly from plastic bags entering sewage treatment plants (Gela et al., 2022; Ramasamy et al., 2022). Urban rainwater runoff and wastewater discharge may have affected the MP distribution of HU and GA samples near urban areas. Recent study from Korea reported that PP, PE, and PET were identified as major MP polymers in wastewater treatment plants (WWTPs) from Seoul and Busan metropolitan city (Park et al., 2020). Jang et al. (2020) found that PP (33%) and PE (28%) were dominant in seawaters and sediments collected from the semi-enclosed Masan Bay, Korea, which is surrounded by a densely populated Changwon and Masan city. In the present study, the sampling area is a location of frequent trade and leisure activities; thus, there is the strong possibility that an abundance of plastic waste was generated, as PP and PP/PE copolymer types have been previously correlated with these activities in the Korean coastal area (Kang et al., 2015; Kwon et al., 2020).
Bacterial community relationships to MP characteristics
The present study reports the first efforts to explore the MP bacterial communities from the coastal area of Busan city in South Korea. The results showed that alpha diversity, such as the Shannon index and Chao1, was significantly higher in MPs than in the surrounding seawater (Figure 3), as has been observed in bay of Brest, France and Bohai sea, China (Frère et al., 2018; Zhao et al., 2018). The significantly higher Shannon index of MP bacterial communities were likely due to the high percentage of rare OTUs in the plastic matrix. These results suggest that certain marine microorganisms selectively grow on the surface of MPs because they could provide living habitat in the marine environment (Huang et al., 2019). These organisms can interact with biofilm-forming bacteria, selectively enriching plastispheres in the surrounding seawater (Roller et al., 2016; Oberbeckmann et al., 2018). Large concentrations of small- (20–300 μm) and medium-sized (300 μm–1 mm) plastics were found in the present study, suggesting that MPs could provide a potential habitat for microorganisms (Zettler et al., 2013; Shen et al., 2022). Relatively high abundances of alpha diversity on MP samples may explain that MP quickly adsorbs organic and inorganic nutrients from SW to proceed colonization via biofilm forming process (De Tender et al., 2017; Pollet et al., 2018). During this process, microorganisms quickly attach to use nutrients on the MP surface and accelerate biofilm formation, resulting in colonization by many microorganisms by the complex interactions between the species abundance and increased number of species (Oberbeckmann et al., 2015; Kirstein et al., 2016). These suggested that the alpha diversity was relatively higher in the MP samples than in the SW samples.
Taxonomic profiles of the MP and SW bacterial communities at the phylum-, family-, and genus-levels were also compared in this study. Proteobacteria, Bacteroidetes, and Firmicutes were the dominant phyla in both MP and SW samples, consistent with previous microbial community studies on marine MPs (Oberbeckmann et al., 2016; Xu et al., 2019; Li et al., 2020). As Proteobacteria are a major bacterium in the marine environment (Figure 4), they were generally dominant in the initial phase of biofilm development (Elifantz et al., 2013; Pollet et al., 2018). Gammaproteobacteria and Alphaproteobacteria are considered early colonizers on polyurethane and acrylic-based plastics, and are known to be dominant in MP and seawater (Xu et al., 2019). Surface attachment and biofilm development are well-known strategies for bacteria to survive in marine environments because of their ability to form stable consortia, and its beneficial properties for horizontal gene exchange, nutrient accumulation, and protection against toxic substances (Rummel et al., 2017). Here, identified bacteria in MP samples significantly increased compared to SW samples (Figure 6), as has been found in other previous marine and coastal area MP studies (Gong et al., 2019; Hu et al., 2021; Szabó et al., 2021). The increasing influx of MPs into seawater could be affected by the potential contribution of land plastic waste-associated sources. Additionally, in MPs distributed along the seaside, the wide-reaching disturbance and vortexing effects of ship movements can facilitate the redistribution of MPs in surface waters and below, potentially altering plastisphere communities between MPs and SW samples (Urbanek et al., 2018; Anjana et al., 2020). Flavobacerium, Pseudomonas, Lactobacillus, and Psychrobacter genera have been reported to the various marine plastics, namely PP, PE, PS, and PET, respectively (Oberbeckmann et al., 2015; Anjana et al., 2020; Tu et al., 2020). These bacteria are known to be selectively enriched through anthropogenic land-based and maritime activities, while marine environmental conditions can also contribute to the high-level contamination of nearshore sediment and seawater in coastal areas. Such bacteria were predominantly observed in the present study. Notably, previous studies have found that suspended plastic packaging waste and plastic bags in seawater can potentially affect the bacterial community structure through physiochemical processes over time (Urbanek et al., 2018; Zhang et al., 2019). It is necessary to investigate the relationship between the surface properties of polymers and MP microbial communities, but unlike MP biofilm studies through culture methods, field-based studies still have limitations to identify specific correlations between collected polymers and microbial communities. Therefore, more studies are required to better understanding the shaping plastisphere on polymers.
In particular, some of the identified bacterial taxa (e.g., Flavobacterium and Pseudomonas spp.) were capable of degrading plastics (Figure 6). Previous studies have suggested that marine microorganisms can degrade MPs by colonizing the surface of suspended plastics (Debroas et al., 2017; Urbanek et al., 2018; Hansen et al., 2021). Known polymer and hydrocarbon degraders, such as Flavobacterium, Pseudomonas, and Paraglaciecola spp., have been found in plastic attached bacterial communities (Egelkamp et al., 2017; Hansen et al., 2021). Marine microorganisms can produce enzymes with the capacity to degrade MP as a potential energy source (Zettler et al., 2013; Yoshida et al., 2016). Chitinase enzymes are produced by certain bacterial genera, including Flavobacterium, Pseudomonas, and Vibrio spp., while chitin hydrolyzation breaks down the polymer structure (Souza et al., 2011). Therefore, the results suggest a potential role for plastispheres in the degradation of plastic polymers. Although the main metabolic pathway remains largely unknown, Pseudomonas spp. are well-known degraders of plastic particles (Rogers et al., 2020). Interestingly, previous studies have found that Vibrio spp., a ubiquitous genus of marine bacteria, were significantly more abundant in MPs than in seawater samples (Zettler et al., 2013; Oberbeckmann et al., 2015); whereas other studies, including that presented here, rarely identified Vibrio spp. in the MP samples (Bryant et al., 2016; Debroas et al., 2017). Generally, the high abundance of Vibrio spp. in seawater and nearshore areas may be related to background environmental conditions, such as relatively higher temperatures and organic matter concentrations (Oberbeckmann et al., 2018). There is a possibility to the low abundances of Vibrio spp. observed here was affected by low temperature during sampling time.
Environmental factors affecting microbial community in microplastics
Previous studies have suggested that MP microbial communities are strongly grouped according to geographical characteristics rather than polymer type (Oberbeckmann et al., 2016; Chen et al., 2019). Environmental factors of the geographic area would thus strongly affect the bacterial community developed on MPs. The results here showed that nutrients (TN and TP), commonly regarded as major aquatic parameters to develop bacterial growth on MP surfaces, negatively influenced the genera of Pseudomonas, Roseburia, Vibrio, and Alteromonas in this marine environment (Figure 7). MPs biofilms have been reported to play an essential part in biogeochemical cycles of nitrogen and phosphorous in marine environment (Shen et al., 2022). Previous studies reported that the biofilm attached to the MP surface can promote the oxidation and denitrification of ammonia nitrogen and nitrite but can also enrich nitrogen fixation compared to SW microbial community (Li et al., 2019; Xu et al., 2019). Additionally, MPs could promote the release and absorption of phosphorous by phosphorous-accumulating bacteria and inhibit the removal of total inorganic nitrogen by reducing the content of denitrifying bacteria (Shen et al., 2022). However, information on the direct impact of MPs on the nitrogen and phosphorous in the marine environment is very limited because the potential mechanism for releasing or adsorption N and P from MP is relatively complex (Li et al., 2019; Shen et al., 2022). As the open ocean environment is nutrient-poor, and it is believed that biofilms are favorably formed in these environmental conditions; however, coastal areas, such as the study sites here, typically receive higher inputs of nutrients and waste debris from the surrounding terrestrial environment (Oberbeckmann et al., 2015; Keswani et al., 2016), as organic matter input and high nutrient (N, P) concentrations are released from urban areas via anthropogenic activities (Oberbeckmann et al., 2015; Harrison et al., 2018). Actually, Vibrio and Pseudomonas genera is one of dominant marine bacteria, and they highly affected specific environmental factors, such as the high temperature, low salinity, nutrients, and Chl-a (Oberbeckmann et al., 2015). In addition, microplastics could aggregate more Vibrio, and Alteromonas genera can be aggregated in microplastics when a favorable environment is provided because amino acid metabolism and metabolism of cofactors and vitamins in surface colonizing capacity on microplastics would be increased compared with other substrates (Wang et al., 2021b). However, unfortunately, detailed information on the bacterial function and metabolic pathway was not obtained because this study focused on the microbial community diversity, compositions, and structures. Therefore, further investigation is quite necessary to interpret the environmental effects of microplastic bacterial colonization. Accordingly, the results of the present study were highly correlated with the MP sampling sites and season. Despite relatively low temperatures and biomass levels observed at the study sites due to the seasonal effect, the observed high abundance of bacteria may be explained by the urban-based nutrient inflow.
Temperature, one of the most common marine environmental monitoring parameters, and are often found to be key to shaping MP microbial communities (De Tender et al., 2015; Harrison et al., 2018; Oberbeckmann et al., 2018). According to the RDA results here (Figure 7), Chl-a and temperature parameters were relatively less correlated with MP bacterial communities. Relatively, lower temperatures, low nutrient contents, and shorter sunshine duration hours in the autumn and winter may also negatively affect bacterial proliferation development (Oberbeckmann et al., 2018; Basili et al., 2020). However, some bacteria, such as Sulfitobacter and Flavobacterium genera, had positive relationships (Figure 7). It is suggested that they may have interactions along eutrophication gradients because they are ubiquitous species probably able to have specific metabolic roles such as aerobic/anaerobic or anoxygenic photosynthesis (Elifantz et al., 2013; Basili et al., 2020; Shen et al., 2022). This seasonal environmental effect may have significantly influenced the plastispheres in the present study, and should be assessed in future analyses.
The salinity parameter was the most important marine environmental factor affecting MP bacterial community in previous study (Oberbeckmann et al., 2018; Li et al., 2020), while temperature and nutrients (TN and TP) were found to significantly correlate to marine bacterial diversity and structure in this study. One previous study has reported that the MP bacterial communities were positively associated with salinity, as well as other marine environmental parameters, such as pH and DO (Li et al., 2020); whereas other studies have shown that temperature and salinity are weakly correlated with bacterial abundance and diversity (Oberbeckmann et al., 2015; Harrison et al., 2018). Although several marine environmental parameters (salinity, temperature, and nutrients) potentially contributed to the bacterial community on PE and PS (Harrison et al., 2018; Jiang et al., 2018), the presence and variety of microbial communities, as well as their biofilm-framing characteristics in MPs remains largely unknown. Additionally, considerable uncertainty remains regarding the ecological role of the plastisphere based on the seasonal and marine environmental effects, such as during transport by currents, ship operation, UV exposure, waves, and winds. Environmental effects are often highly complex, and can interact with other environmental parameters, thereby making it difficult to decipher the precise relationships between microorganisms and the surrounding area. Therefore, additional studies are required to better characterize the potential relationships among microbial communities, marine environmental parameters, and the effects in plastispheres.
Conclusion
This field study in an urban coastal area revealed that high levels of MP concentrations (mean: 94 ± 41 particles·m-3), and small sized MPs (20–300 μm) were dominantly detected (67.5% ± 9.6%) at six sites in the Busan coastal area. In addition, the microbial community was significantly different between MP and SW, and MP-associated microbial diversity was higher than that in SW. The most abundant taxonomic profiles of the bacterial communities in MPs comprised phylum Proteobacteria, class Gammaproteobacteria, and families Flavobacteriaceae and Rhodobacteraceae. LEfSe results showed that Sneathiella, Flavobacterium, Lactobacillus, Desulfovibrio, Winogradskyella, Pseudomonas, and Psychrobacter genera were abundant in MP samples compared with SW samples. Moreover, temperature and nutrients, such as TN-TP, were found to be potential variables that could influence bacterial community in MPs in urban coastal area. Our results can help characterize the abundance and occurrence of MP contamination, and their influence on the plastispheres of marine environments.
The study here, however, had several notably limitations. Due to the limited sampling period, any seasonal variations and influences were not considered; thus, larger sample sizes across longer temporal scales should be monitored to further understand MP plastispheres. To date, we do not know much about the dynamics and successional changes in microplastic-associated assemblages that occur as the particles are subjected to different environments characterized by different local communities. Therefore, future research should address the relationship between MP contamination and biofilm-forming bacterial communities, as the diverse microbial composition of marine biofilms can lead to interspecific interactions affecting the fates of marine MPs. Although current research on microplastics in the marine environment is increasing significantly, there is a limit to obtaining systematic scientific information owing to the lack of standard methods for intrinsically considering and analyzing multi-layered factors. Therefore, it is necessary to develop a standard method to intensively investigate fibers and fragments’ microplastics because they are predominant MPs in marine environment. These approaches could contribute to an in-depth understanding of the current status and effects of microplastic pollution in the coastal waters of Busan. In addition, this information can be used as primary data for future research on tracking the source of this pollution, characterization of microplastic degradation along the coastal areas, and the impact on ecosystems.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.mg-rast.org/ , mgm4926380 to mgm4967466.
Author contributions
KY: Conceptualization, Validation, Writing – review and editing, Funding acquisition. SB: Investigation, Data curation, Visualization, Writing – original draft. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (Ministry of Science, ICT & Future Planning) (No.2020R1C1C1008951).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.1030476/full#supplementary-material
References
Amaral-Zettler L. A., Zettler E. R., Mincer T. J. (2020). Ecology of the plastisphere. Nat. Rev. Microbiol. 18 (3), 139–151. doi: 10.1038/s41579-019-0308-0
Anderson P. J., Warrack S., Langen V., Challis J. K., Hanson M. L., Rennie M. D. (2017). Microplastic contamination in lake Winnipeg, Canada. Environ. pollut. 225, 223–231. doi: 10.1016/j.envpol.2017.02.072
Anjana K., Hinduja M., Sujitha K., Dharani G. (2020). Review on plastic wastes in marine environment–biodegradation and biotechnological solutions. Mar. pollut. Bull. 150, 110733. doi: 10.1016/j.marpolbul.2019.110733
Bailey K., Sipps K., Saba G. K., Arbuckle-Keil G., Chant R. J., Fahrenfeld N. (2021). Quantification and composition of microplastics in the raritan Hudson estuary: comparison to pathways of entry and implications for fate. Chemosphere 272, 129886. doi: 10.1016/j.chemosphere.2021.129886
Barletta M., Lima A. R., Costa M. F. (2019). Distribution, sources and consequences of nutrients, persistent organic pollutants, metals and microplastics in south American estuaries. Sci. Total Environ. 651, 1199–1218. doi: 10.1016/j.scitotenv.2018.09.276
Basili M., Quero G. M., Giovannelli D., Manini E., Vignaroli C., Avio C. G., et al. (2020). Major role of surrounding environment in shaping biofilm community composition on marine plastic debris. Front. Mar. Sci. 262. doi: 10.3389/fmars.2020.00262
Bryant J. A., Clemente T. M., Viviani D. A., Fong A. A., Thomas K. A., Kemp P., et al. (2016). Diversity and activity of communities inhabiting plastic debris in the north pacific gyre. MSystems 1 (3), e00024–e00016. doi: 10.1128/mSystems.00024-16
Chen X., Xiong X., Jiang X., Shi H., Wu C. (2019). Sinking of floating plastic debris caused by biofilm development in a freshwater lake. Chemosphere 222, 856–864. doi: 10.1016/j.chemosphere.2019.02.015
Debroas D., Mone A., Ter Halle A. (2017). Plastics in the north Atlantic garbage patch: a boat-microbe for hitchhikers and plastic degraders. Sci. total Environ. 599, 1222–1232. doi: 10.1016/j.scitotenv.2017.05.059
De Lucia G. A., Vianello A., Camedda A., Vani D., Tomassetti P., Coppa S., et al. (2018). Sea Water contamination in the vicinity of the Italian minor islands caused by microplastic pollution. Water 10 (8), 1108. doi: 10.3390/w10081108
De Tender C. A., Devriese L. I., Haegeman A., Maes S., Ruttink T., Dawyndt P. (2015). Bacterial community profiling of plastic litter in the Belgian part of the north Sea. Environ. Sci. Technol. 49 (16), 9629–9638. doi: 10.1021/acs.est.5b01093
De Tender C., Schlundt C., Devriese L., Mincer T., Zettler E., Amaral-Zetter L. (2017). A review of microscopy and comparative molecular-based methods to characterize “Plastisphere” communities. Analytical Methods 9 (14), 2132–2143. doi: 10.1021/acs.est.5b01093
Edgar R. C., Haas B. J., Clemente J. C., Quince C., Knight R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27 (16), 2194–2200. doi: 10.1093/bioinformatics/btr381
Egelkamp R., Schneider D., Hertel R., Daniel R. (2017). Nitrile-degrading bacteria isolated from compost. Front. Environ. Sci. 5, 56. doi: 10.3389/fenvs.2017.00056
Elifantz H., Horn G., Ayon M., Cohen Y., Minz D. (2013). Rhodobacteraceae are the key members of the microbial community of the initial biofilm formed in Eastern Mediterranean coastal seawater. FEMS Microbiol. Ecol. 85 (2), 348–357. doi: 10.1111/1574-6941.12122
Enders K., Lenz R., Stedmon C. A., Nielsen T. G. (2015). Abundance, size and polymer composition of marine microplastics≥ 10 μm in the Atlantic ocean and their modelled vertical distribution. Mar. pollut. Bull. 100 (1), 70–81. doi: 10.1016/j.marpolbul.2015.09.027
Eo S., Hong S. H., Song Y. K., Lee J., Lee J., Shim W. J. (2018). Abundance, composition, and distribution of microplastics larger than 20 μm in sand beaches of south Korea. Environ. pollut. 238, 894–902. doi: 10.1016/j.envpol.2018.03.096
Frère L., Maignien L., Chalopin M., Huvet A., Rinnert E., Morrison H., et al. (2018). Microplastic bacterial communities in the bay of Brest: Influence of polymer type and size. Environ. pollut. 242, 614–625. doi: 10.1016/j.envpol.2018.07.023
Gela M. S., Aragaw T. A. (2022). Abundance and characterization of microplastics in main urban ditches across the bahir dar city, Ethiopia. Front. Environ. Sci. 35. doi: 10.3389/fenvs.2022.831417
GESAMP. (2015). Sources, fate and effects of microplastics in the marine environment: A global assessment. In Kershaw P. J. Ed Rep Stud GESAMP90, 96.
Gong M., Yang G., Zhuang L., Zeng E. Y. (2019). Microbial biofilm formation and community structure on low-density polyethylene microparticles in lake water microcosms. Environ. pollut. 252, 94–102. doi: 10.1016/j.envpol.2019.05.090
Hansen J., Melchiorsen J., Ciacotich N., Gram L., Sonnenschein E. C. (2021). Effect of polymer type on the colonization of plastic pellets by marine bacteria. FEMS Microbiol. Lett. 368 (5), fnab026. doi: 10.1093/femsle/fnab026
Harrison J. P., Hoellein T. J., Sapp M., Tagg A. S., Ju-Nam Y., Ojeda J. J. (2018). “Microplastic-associated biofilms: a comparison of freshwater and marine environments,” in Freshwater microplastics (Cham: Springer), 181–201.
Harrison J. P., Schratzberger M., Sapp M., Osborn A. M. (2014). Rapid bacterial colonization of low-density polyethylene microplastics in coastal sediment microcosms. BMC Microbiol. 14 (1), 1–15. doi: 10.1186/s12866-014-0232-4
Hartline N. L., Bruce N. J., Karba S. N., Ruff E. O., Sonar S. U., Holden P. A. (2016). Microfiber masses recovered from conventional machine washing of new or aged garments. Environ. Sci. Technol. 50 (21), 11532–11538. doi: 10.1021/acs.est.6b03045
Hernandez E., Nowack B., Mitrano D. M. (2017). Polyester textiles as a source of microplastics from households: A mechanistic study to understand microfiber release during washing. Environ. Sci. Technol. 51 (12), 7036–7046. doi: 10.1021/acs.est.7b01750
Horton A. A., Walton A., Spurgeon D. J., Lahive E., Svendsen C. (2017). Microplastics in freshwater and terrestrial environments: evaluating the current understanding to identify the knowledge gaps and future research priorities. Sci. total Environ. 586, 127–141. doi: 10.1016/j.scitotenv.2017.01.190
Huang Y., Zhao Y., Wang J., Zhang M., Jia W., Qin X. (2019). LDPE microplastic films alter microbial community composition and enzymatic activities in soil. Environ. pollut. 254, 112983. doi: 10.1016/j.envpol.2019.112983
Hurley R., Woodward J., Rothwell J. J. (2018). Microplastic contamination of river beds significantly reduced by catchment-wide flooding. Nat. Geosci. 11 (4), 251–257. doi: 10.1038/s41561-018-0080-1
Hu K., Tian W., Yang Y., Nie G., Zhou P., Wang Y., et al. (2021). Microplastics remediation in aqueous systems: Strategies and technologies. Water Res. 198, 117144. doi: 10.1016/j.watres.2021.117144
Jambeck J. R., Geyer R., Wilcox C., Siegler T. R., Perryman M., Andrady A., et al. (2015). Plastic waste inputs from land into the ocean. Science 347 (6223), 768–771. doi: 10.1126/science.126035
Jang M., Shim W. J., Cho Y., Han G. M., Song Y. K., Hong S. H. (2020). A close relationship between microplastic contamination and coastal area use pattern. Water Res. 171, 115400. doi: 10.1016/j.watres.2019.115400
Jiang Y., Yang F., Kazmi S. S. U. H., Zhao Y., Chen M., Wang J. (2022). A review of microplastic pollution in seawater, sediments and organisms of the Chinese coastal and marginal seas. Chemosphere 286, 131677. doi: 10.1016/j.chemosphere.2021.131677
Jiang P., Zhao S., Zhu L., Li D. (2018). Microplastic-associated bacterial assemblages in the intertidal zone of the Yangtze estuary. Sci. total Environ. 624, 48–54. doi: 10.1016/j.scitotenv.2017.12.105
Kang J.-H., Kwon O. Y., Lee K.-W., Song Y. K., Shim W. J. (2015). Marine neustonic microplastics around the southeastern coast of Korea. Mar. pollut. Bull. 96 (1-2), 304–312. doi: 10.1016/j.marpolbul.2015.04.054
Kassambara A. (2020) ggpubr:'ggplot2'Based publication ready plots. r package version 0.4. 0. computer software. Available at: https://cran-r-project.org/web/packages/ggpubr/indes.html.
Kazour M., Jemaa S., Issa C., Khalaf G., Amara R. (2019). Microplastics pollution along the Lebanese coast (Eastern Mediterranean basin): Occurrence in surface water, sediments and biota samples. Sci. total Environ. 696, 133933. doi: 10.1016/j.scitotenv.2019.133933
Keswani A., Oliver D. M., Gutierrez T., Quilliam R. S. (2016). Microbial hitchhikers on marine plastic debris: human exposure risks at bathing waters and beach environments. Mar. Environ. Res. 118, 10–19. doi: 10.1016/j.marenvres.2016.04.006
Kiessling T., Gutow L., Tiel M. (2015). Marine litter as habitat and dispersal vector.In Marine Anthropogenic Litter Eds Bergmann M., Gutow L., Klages M. (Cham:Springer Open; Elsevier), 141–181. doi: 10.1007/978-3-319-16510-3_6
Kim Y. K., Yoo K., Kim M. S., Han I., Lee M., Kang B. R., et al. (2019). The capacity of wastewater treatment plants drives bacterial community structure and its assembly. Sci. Rep. 9 (1), 1–9. doi: 10.1038/s41598-019-50952-0
Kirstein I. V., Kirmizi S., Wichels A., Garin-Fernandez A., Erler R., Löder M., et al. (2016). Dangerous hitchhikers? evidence for potentially pathogenic vibrio spp. on microplastic particles. Mar. Environ. Res. 120, 1–8. doi: 10.1016/j.marenvres.2016.07.004
Korea Ministry of Oceans and Fisheries (2021) Marine environmental monitoring network information. Available at: https://www.meis.go.kr.
Kozich J. J., Westcott S. L., Baxter N. T., Highlander S. K., Schloss P. D. (2013). Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq illumina sequencing platform. Appl. Environ. Microbiol. 79 (17), 5112–5120. doi: 10.1128/AEM.01043-13
Kwon O. Y., Kang J.-H., Hong S. H., Shim W. J. (2020). Spatial distribution of microplastic in the surface waters along the coast of Korea. Mar. pollut. Bull. 155, 110729. doi: 10.1016/j.marpolbul.2019.110729
Lee J., Khim J. S. (2022). Revisited a sediment quality triad approach in the Korean coastal waters: Past research, current status, and future directions. Environ. pollut. 292, 118262. doi: 10.1016/j.envpol.2021.118262
Li J., Huang W., Jiang R., Han X., Zhang D., Zhang C. (2020). Are bacterial communities associated with microplastics influenced by marine habitats? Sci. Total Environ. 733, 139400. doi: 10.1016/j.scitotenv.2020.139400
Li J., Yang D., Li L., Jabeen K., Shi H. (2015). Microplastics in commercial bivalves from China. Environ. pollut. 207, 190–195. doi: 10.1016/j.envpol.2015.09.018
Li W., Zhang Y., Wu N., Zhao Z., Xu W., Ma Y., et al. (2019). Colonization characteristics of bacterial communities on plastic debris influenced by environmental factors and polymer types in the haihe estuary of bohai bay, China. Environ. Sci. Technol. 53 (18), 10763–10773. doi: 10.1021/acs.est.9b03659
Lusher A. L., Tirelli V., O’Connor I., Officer R. (2015). Microplastics in Arctic polar waters: the first reported values of particles in surface and sub-surface samples. Sci. Rep. 5 (1), 1–9. doi: 10.1038/srep14947
Masura J., Baker J., Foster G., Arthur C. (2015). Laboratory methods for the analysis of microplastics in the marine environment: Recommendations for quantifying synthetic particles in waters and sediments, (NOAA Technical Memorandum NOS-OR&R-48), doi: 10.25607/OBP-604
McMurdie P. J., Holmes S. (2013). Phyloseq: an r package for reproducible interactive analysis and graphics of microbiome census data. PloS One 8 (4), e61217. doi: 10.1371/journal.pone.0061217
Mhiret Gela S., Aragaw T. A. (2022). Abundance and characterization of microplastics in main urban ditches across the bahir dar city, ethiopia. Front. Environ. Sci. 35
Nel H. A., Froneman P. W. (2015). A quantitative analysis of microplastic pollution along the south-eastern coastline of south Africa. Mar. pollut. Bull. 101 (1), 274–279. doi: 10.1016/j.marpolbul.2015.09.043
Oberbeckmann S., Kreikemeyer B., Labrenz M. (2018). Environmental factors support the formation of specific bacterial assemblages on microplastics. Front. Microbiol. 8, 2709. doi: 10.3389/fmicb.2017.02709
Oberbeckmann S., Löder M. G., Labrenz M. (2015). Marine microplastic-associated biofilms–a review. Environ. Chem. 12 (5), 551–562. doi: 10.1071/EN15069
Oberbeckmann S., Osborn A. M., Duhaime M. B. (2016). Microbes on a bottle: substrate, season and geography influence community composition of microbes colonizing marine plastic debris. PloS One 11 (8), e0159289. doi: 10.1371/journal.pone.0159289
Oksanen J., Blanchet F. G., Friendly M., Kindt R., Legendre P., McGlinn D., et al. (2019). “Vegan: Community ecology package (version 2.5-6),” in The comprehensive r archive network, http://CRAN.R-project.org/package=vegan.
Park E., Choi B.-K., Lee T. J. (2019). The role and dimensions of authenticity in heritage tourism. Tourism Manage. 74, 99–109. doi: 10.1016/j.tourman.2019.03.001
Park H.-J., Oh M.-J., Kim P.-G., Kim G., Jeong D.-H., Ju B.-K., et al. (2020). National reconnaissance survey of microplastics in municipal wastewater treatment plants in Korea. Environ. Sci. Technol. 54 (3), 1503–1512. doi: 10.1021/acs.est.9b04929
Phuong N. N., Poirier L., Pham Q. T., Lagarde F., Zalouk-Vergnoux A. (2018). Factors influencing the microplastic contamination of bivalves from the French Atlantic coast: location, season and/or mode of life? Mar. pollut. Bull. 129 (2), 664–674. doi: 10.1016/j.marpolbul.2017.10.054
Pollet T., Berdjeb L., Garnier C., Durrieu G., Le Poupon C., Misson B., et al. (2018). Prokaryotic community successions and interactions in marine biofilms: the key role of flavobacteriia. FEMS Microbiol. Ecol. 94 (6), fiy083. doi: 10.1093/femsec/fiy083
Qiu Q., Tan Z., Wang J., Peng J., Li M., Zhan Z. (2016). Extraction, enumeration and identification methods for monitoring microplastics in the environment. Estuarine Coast. Shelf Sci. 176, 102–109. doi: 10.1016/j.ecss.2016.04.012
Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., et al. (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41 (D1), D590–D596. doi: 10.1093/nar/gks1219
Ramasamy R., Aragaw T. A., Balasaraswathi Subramanian R. (2022). Wastewater treatment plant effluent and microfiber pollution: Focus on industry-specific wastewater. Environ. Sci. pollut. Res. 29(34), 51211–51233 doi: 10.1007/s11356-022-20930-7
Rogers K. L., Carreres-Calabuig J. A., Gorokhova E., Posth N. R. (2020). Micro-by-micro interactions: How microorganisms influence the fate of marine microplastics. Limnology Oceanography Lett. 5 (1), 18–36. doi: 10.1002/lol2.10136
Roller B. R., Stoddard S. F., Schmidt T. M. (2016). Exploiting rRNA operon copy number to investigate bacterial reproductive strategies. Nat. Microbiol. 1 (11), 1–7. doi: 10.1038/nmicrobiol.2016.160
Ruiz-Orejón L. F., Mourre B., Sardá R., Tintoré J., Ramis-Pujol J. (2019). Quarterly variability of floating plastic debris in the marine protected area of the menorca channel (Spain). Environ. pollut. 252, 1742–1754. doi: 10.1016/j.envpol.2019.06.063
Rummel C. D., Jahnke A., Gorokhova E., Kühnel D., Schmitt-Jansen M. (2017). Impacts of biofilm formation on the fate and potential effects of microplastic in the aquatic environment. Environ. Sci. Technol. Lett. 4 (7), 258–267. doi: 10.1021/acs.estlett.7b00164
Schloss P. D., Westcott S. L., Ryabin T., Hall J. R., Hartmann M., Hollister E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75 (23), 7537–7541. doi: 10.1128/AEM.01541-09
Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12 (6), 1–18. doi: 10.1186/gb-2011-12-6-r60
Shen M., Song B., Zhou C., Almatrafi E., Hu T., Zeng G., et al. (2022). Recent advances in impacts of microplastics on nitrogen cycling in the environment: a review. Sci. Total Environ. 815, 152740. doi: 10.1016/j.scitotenv.2021.152740
Shim W. J., Hong S. H., Eo S. E. (2017). Identification methods in microplastic analysis: A review. Analytical Methods 9 (9), 1384–1391. doi: 10.1039/C6AY02558G
Shim W. J., Hong S. H., Eo S. (2018). “Marine microplastics: abundance, distribution, and composition,” in Microplastic contamination in aquatic environments (Elsevier), 1–26. doi: 10.1016/B978-0-12-813747-5.00001-1
Song Y. K., Hong S. H., Eo S., Jang M., Han G. M., Isobe A., et al. (2018). Horizontal and vertical distribution of microplastics in Korean coastal waters. Environ. Sci. Technol. 52 (21), 12188–12197. doi: 10.1021/acs.est.8b04032
Souza C. P., Almeida B. C., Colwell R. R., Rivera I. N. (2011). The importance of chitin in the marine environment. Mar. Biotechnol. 13 (5), 823–830. doi: 10.1007/s10126-011-9388-1
Szabó I., Al-Omari J., Szerdahelyi G. S., Farkas M., Al-Omari Y., Szabó P. M., et al. (2021). In situ investigation of plastic-associated bacterial communities in a freshwater lake of Hungary. Water Air Soil pollut. 232 (12), 1–17. doi: 10.1007/s11270-021-05445-0
Tan Y., Dai J., Wu X., Wu S., Zhang J. (2022). Characteristics, occurrence and fate of non-point source microplastic pollution in aquatic environments. J. Cleaner Production 341, 130766. doi: 10.1016/j.jclepro.2022.130766
Teng J., Zhao J., Zhang C., Cheng B., Koelmans A. A., Wu D., et al. (2020). A systems analysis of microplastic pollution in laizhou bay, China. Sci. Total Environ. 745, 140815. doi: 10.1016/j.scitotenv.2020.140815
Tu C., Chen T., Zhou Q., Liu Y., Wei J., Waniek J. J., et al. (2020). Biofilm formation and its influences on the properties of microplastics as affected by exposure time and depth in the seawater. Sci. Total Environ. 734, 139237. doi10.1016/j.scitotenv.2020.139237
Tsang Y.Y, Mak C.W., Liebich C., Lam S.W, Sze E.T.P, Chan K.M.. (2017). Microplastic pollution in the marine waters and sediments of Hong Kong. Mar. Pollut. Bull. 115(1), 2028. doi: 10.1016/j.marpolbul.2016.11.003
Urbanek A. K., Rymowicz W., Mirończuk A. M. (2018). Degradation of plastics and plastic-degrading bacteria in cold marine habitats. Appl. Microbiol. Biotechnol. 102 (18), 7669–7678. doi: 10.1007/s00253-018-9195-y
Wang X., Bolan N., Tsang D. C., Sarkar B., Bradney L., Li Y. (2021a). A review of microplastics aggregation in aquatic environment: Influence factors, analytical methods, and environmental implications. J. Hazardous Materials 402, 123496. doi: 10.1016/j.jhazmat.2020.123496
Wang J., Guo X., Xue J. (2021b). Biofilm-developed microplastics as vectors of pollutants in aquatic environments. Environ. Sci. Technol. 55 (19), 12780–12790. doi: 10.1021/acs.est.1c04466
Wright R. J., Erni-Cassola G., Zadjelovic V., Latva M., Christie-Oleza J. A. (2020). Marine plastic debris: A new surface for microbial colonization. Environ. Sci. Technol. 54 (19), 11657–11672. doi: 10.1021/acs.est.0c02305
Wu X., Pan J., Li M., Li Y., Bartlam M., Wang Y. (2019). Selective enrichment of bacterial pathogens by microplastic biofilm. Water Res. 165, 114979. doi: 10.1016/j.watres.2019.114979
Xu X., Wang S., Gao F., Li J., Zheng L., Sun C., et al. (2019). Marine microplastic-associated bacterial community succession in response to geography, exposure time, and plastic type in china's coastal seawaters. Mar. pollut. Bull. 145, 278–286. doi: 10.1016/j.marpolbul.2019.05.036
Yang Y., Liu G., Song W., Ye C., Lin H., Li Z., et al. (2019). Plastics in the marine environment are reservoirs for antibiotic and metal resistance genes. Environ. Int. 123, 79–86. doi: 10.1016/j.envint.2018.11.061
Yan M., Nie H., Xu K., He Y., Hu Y., Huang Y., et al. (2019). Microplastic abundance, distribution and composition in the pearl river along guangzhou city and pearl river estuary, China. Chemosphere 217, 879–886. doi: 10.1016/j.chemosphere.2018.11.093
Yoo K., Yoo H., Lee J. M., Shukla S. K., Park J. (2018). Classification and regression tree approach for prediction of potential hazards of urban airborne bacteria during Asian dust events. Sci. Rep. 8 (1), 1–11. doi: 10.1038/s41598-018-29796-7
Yoo K., Han I., Ko K., Lee S., K T., Yoo H., et al (2019). Bacillus-dominant airborne bacterial communities identified during asian dust events. Microb. Ecol. 78 (3), 677–687. doi: 10.1007/s00248-019-01348-0
Yoshida S., Hiraga K., Takehana T., Taniguchi I., Yamaji H., Maeda Y., et al. (2016). A bacterium that degrades and assimilates poly (ethylene terephthalate). Science 351 (6278), 1196–1199. doi: 10.1126/science.aad635
Zettler E. R., Mincer T. J., Amaral-Zettler L. A. (2013). Life in the “plastisphere”: microbial communities on plastic marine debris. Environ. Sci. Technol. 47 (13), 7137–7146. doi: 10.1021/es401288x
Zhan Z., Ali L., Sarwat S., Godil D. I., Dinca G., Anser M. K. (2021). A step towards environmental mitigation: Do tourism, renewable energy and institutions really matter? a QARDL approach. Sci. Total Environ. 778, 146209. doi: 10.1016/j.scitotenv.2021.146209
Zhang B., Wu D., Yang X., Teng J., Liu Y., Zhang C., et al. (2019). Microplastic pollution in the surface sediments collected from sishili bay, north yellow Sea, China. Mar. pollut. Bull. 141, 9–15. doi: 10.1016/j.marpolbul.2019.02.021
Zhang W., Zhang S., Wang J., Wang Y., Mu J., Wang P., et al. (2017). Microplastic pollution in the surface waters of the bohai Sea, China. Environ. pollut. 231, 541–548. doi: 10.1016/j.envpol.2017.08.058
Zhao J., Ran W., Teng J., Liu Y., Liu H., Yin X., et al. (2018). Microplastic pollution in sediments from the bohai Sea and the yellow Sea, China. Sci. Total Environ. 640, 637–645. doi: 10.1016/j.scitotenv.2018.05.346
Keywords: microplastics, plastisphere, bacterial community, coastal area, surface colonization
Citation: Bae S and Yoo K (2022) Microplastic contamination and microbial colonization in coastal area of Busan City, Korea. Front. Mar. Sci. 9:1030476. doi: 10.3389/fmars.2022.1030476
Received: 30 August 2022; Accepted: 27 October 2022;
Published: 16 November 2022.
Edited by:
Hideyuki Kanematsu, Suzuka College, JapanReviewed by:
Tadele Assefa Aragaw, Bahir Dar University, EthiopiaZhihua Feng, Jiangsu Ocean Universiity, China
Akiko Ogawa, Suzuka College, Japan
Copyright © 2022 Bae and Yoo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Keunje Yoo, a2p5b29Aa21vdS5hYy5rcg==
 Suyeon Bae
Suyeon Bae Keunje Yoo
Keunje Yoo