
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 01 November 2022
Sec. Marine Pollution
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.1030070
This article is part of the Research TopicNew Insights to Coastal Zone Environmental Pollution, Ecosystem Disturbance and RemediationView all 5 articles
The arborescent bryozoan Bugulina neritina is a cosmopolitan fouling species in sub-tropical to sub-temperate waters. The butenolide compound 5-octylfuran-2(5H)-one (hereafter named butenolide) reportedly inhibits larval settlement of B. neritina, but its effect on the larval behavioral and transcriptomic responses remained unclear. We report that 10 μg mL-1 or higher concentration of butenolide and/or prolonged treatment (10 h or longer) resulted in significant increase in larval mortality and prominent spiral larval swimming behavior. The transcriptomic analyses not only revealed up-regulation of typical stress-related protein genes in response to 10 mg mL-1 butenolide treatment, but also indicated up-regulation of mucin, synaptic genes and nitric oxide signaling genes. In addition, the expression of developmental genes and lipid biosynthesis genes were also affected. Overall, our larval behavior and transcriptome analyses reflected the impact of butenolide on the metabolism, neuronal and molecular signaling in B. neritina larvae.
Maritime industries face severe biofouling problem, referring to the undesirable and continuous growth of sessile marine invertebrates on submerged man-made surfaces. Biofouling communities form elaborated structures composed of fouling organisms which could either be soft-foulers, referring to those that have a lightly or non-calcified body, or hard-foulers, which contain a soft-body protected by a heavily calcified shell (Prevosto et al., 2012).
Marine bryozoans, which form dense colonies by asexual reproduction, are widely distributed worldwide, especially in the Southern Ocean (Marta and Costello, 2020) and are one of the most common foulers (Wood, 2001). Colonies of calcifying species such as those in the genus Watersipora grow horizontally and form heavily calcified mats that cover the substratum surface (Brock, 1985; Gordon and Mawatari, 1992). On the other hand, species which adult colonies lack a calcified shell seems to be less problematic, at least to the shipping industry, as these species are neither resistant to high current flow nor salinity changes and are heavily predated (Pratt, 2008). However, these soft bryozoans such as species in the genus Bugulina (previously known as Bugula) often exhibit strong seasonality. During the growing season, their colonies grow exponentially (Conradi et al., 2000), forming dense bushy colonies. Moreover, certain Bugulina species such as B. neritina reportedly releases purple color leachates that will chemically induce the larval settlement and subsequent accumulation of the tubeworm Hydroides elegans (Bryan et al., 1997; Bryan et al., 1998). As such, the arborescent colonies of soft non-calcified bryozoan species provide a highly elaborate 3D habitat structure and are therefore an important key component in the fouling community (Scheer, 1945; Dumont et al., 2011). For the stake-holders such as fishermen, the growth of soft bryozoans is still highly unwelcome as the bushy colonies often resulted in clogging of fish cages and accumulation of hard foulers (Wood et al., 2013).
Multiple bioactive compounds were found to be inhibitive to the settlement of B. neritina larvae, including di(1H-indol-3-yl)methane (DIM) compounds (Wang et al., 2015; Xu et al., 2020) and the butenolide compound 5-octylfuran-2(5H)-one (refers as butenolide hereafter), which was subsequently patented as an antifoulant (Zhang et al., 2011a; Zhang et al., 2012b). The mode of action and toxicology studies raised concern over the commercialization of DIM as an antifoulant, owing to the endocrine disrupting nature of the compound. Butenolide appears to be a promising, non-toxic and environmentally friendly antifoulant. In HeLa cells (Zhang et al., 2011b), marine medaka (Oryzias melastigma) (Chen et al., 2014b; Chen et al., 2015), the cypris larvae of the barnacle Amphibalanus amphitrite as well as the coronate larvae of B. neritina, butenolide mainly affected the gene expression of energy metabolic proteins, especially those associated with or directly involved in lipid or fatty acid metabolism.
For instance, Zhang et al. reported butenolide could bind to very long chain acyl-CoA dehydrogenase (ACADVL), actin, and glutathione S-transferases (GSTs) in B. neritina and acetyl-CoA acetyltransferase 1 (ACAT1) in A. amphitrite cypris larvae (Zhang et al., 2012b); Chen et al. reported that, in marine medaka, butenolide treatment resulted in increased reactive oxygen species (ROS) production and down-regulation of fatty acid-binding protein, but the effect was concentration dependent and appeared to be transient and reversible when the butenolide concentration was maintained at a low level (Chen et al., 2014a).
Although butenolide was suggested to affect larval energy metabolism, it remains unclear if the compound will trigger severe responses on behavioral and gene expression level. The molecular mode of action of butenolide on marine fouling organism larvae has been examined in multiple model or non-model organisms, including marine medaka (Oryzias melastigma) (Chen et al., 2014a; Chen et al., 2015), the cyprid larvae of acorn barnacle Amphibalanus amphitrite and bryozoan larvae (Zhang et al., 2011a; Zhang et al., 2012b).
Transcriptomic analyses were typically used to access gene expression changes relating to compound treatment (Heijne et al., 2005; Wu and Wang, 2018). In general, differentially expressed genes (DEGs) between treatment and control groups are identified by bioinformatics and the enriched gene functions among the DEGs are then linked to the compound’s effect (Xu et al., 2020). In the case of antifouling compound treatment, highlighted gene functions are generally linked to the inhibitive or inductive effect of the compound on larval settlement. Yet these research efforts focused on the anti-larval settlement effectiveness and the toxicity of the compound. None of these studies reported the immediate short-term effect of the treatment on larval behavior. As the antifouling compound coated on the antifouling paint will constantly release and being diluted by ambient seawater, the amount and duration of larval exposure to the antifouling compound could be highly sporadic.
Application of a chemical compound during the bioassay experiment may lead to acute response such as behavioral changes, including crawling, swimming and phototactic behavior in larvae (Pires and Woollacott, 1997; Shimizu et al., 2000; Zhang et al., 2011a). As gene expression is a highly dynamic process, observing the behavior of larvae in the bioassay experiment is as important as observing the larval settlement rate. We argue that larval behavioral data may provide novel perspectives to the interpretation of transcriptomic data.
In this study, we accessed the effect of the butenolide on B. neritina on both behavioral and transcriptomic levels. We firstly observed both larval swimming behavior and the larval settlement rate in B. neritina larvae treated with different concentrations of butenolide. Larval behavior was observed by video and the larval swimming track was reconstructed based on video data. Recovery assay was performed to determine the critical concentration and duration of butenolide treatment that caused non-recoverable effect on the larvae. Subsequently, larvae treated with butenolide at the critical concentration and duration, as well as larvae treated with a higher concentration were subjected to RNAseq. Finally, we sought for linkage between the transcriptomic results and behavioral changes.
Bugulina neritina adult colonies were collected from fishfarm rafts in the Daya Bay, Shenzhen, P. R. China (22°33′53.2″N, 114°32′09.0″E). The adult colonies were kept in dark, transported to the laboratory and maintained in seawater aquarium. To keep the activity of adult colonies, the container was constantly aerated and about one third of seawater was replaced daily. After applying bright light illumination for 30 min to the adult colonies, the released larvae were collected for the settlement bioassays. Butenolide powder was solubilized in 100% DMSO. Before the experiment, butenolide (BU) solutions were prepared in filtrated seawater (FSW) with the final concentrations of 2% DMSO and 2.5, 5, 10, 20, 40 μg mL-1 of BU. Five hundred microliters of seawater containing about 10 to 15 larvae were added to each of the well of a 24-well polystyrene plates, followed by the addition of an equal volume of BU solution. In total, seven treatment groups, namely FSW control, 1% DMSO FSW control, 1.25, 2.5, 5, 10 and 20 μg mL-1 BU were prepared in quadruplicate. The 24-well plates were kept in dark at room temperature (about 25°C) and the number of settled larvae was recorded after 3 hours using a dissecting microscope (Leica EZ4, Heerbrugg, Switzerland). Larval mortality was recorded at 3, 9, 24 hours. The full set of experiment was repeated three times using three different batches of B. neritina larvae released from different adult colonies.
To determine the tolerability and efficacy of the concentration of BU solution during treatment, recovery assays were performed. After adding 20-30 larvae (500 μL in volume) to each well of the 24-well plates, equal volume of BU stock solutions was applied to the final concentration of 1.25, 2.5, 5, 10, 20 μg mL-1 with the seawater and DMSO (1%) as control. The plates were placed in dark for 3 hours and then the original solutions were replaced with same volume of fresh seawater twice. The numbers of settled larvae were recorded at 24 hours after incubation in dark.
To analyze the effect of BU treatment on larval behavior, three 10 seconds 12 frame per second (fps) video footages were taken using a digital camera (Olympus DP72, Tokyo, Japan) coupled with a stereo microscope (Leica MZ16, Heerbrugg, Switzerland) on each of the six replicates in the control (seawater) group and 5, 10 and 20 μg mL-1 BU-treated groups at different time points. For the control group, video footages were taken at 1.5 hours after the initiation of the settlement bioassay; for the 5, 10 and 20 μg mL-1 BU-treated groups, video footages were taken at 1.5, 6 hours and 1 hour after recovery from a 3 hours BU treatment. From each video footage, all frames were extracted into.jpeg format image using a python script (provided in Supplementary File 1). The extracted frames were then imported into Adobe Photoshop (version 2020). The initial frame image of a footage was imported firstly as background and all subsequent frame images were adjusted to dim-light mode and then overlaid on top of the initial frame image. Larval swimming behavior was then summarized based on the trajectory of each larva, as visible in the resulting overlaying image of each footage. Larvae with a circular swimming trajectory were concluded as exhibiting looping swimming behavior; larvae that exhibited spiral swimming trajectory were concluded as exhibiting forward looping behavior; larvae swimming trajectory pattern that did not match neither of the above swimming behavior were determined to be typical swimming behavior.
Five control larval biological replicate samples, four replicates of 5 μg mL-1 and 10 μg mL-1 butanolide treated samples were prepared for RNAseq experiment. To prepare RNAseq sample, approximately 300 larvae were treated with 5 or 10 μg mL-1 butenolide and the FSW was used as negative control. After incubation in dark for 3 hours, the unsettled larvae were collected for RNAseq analysis, and all samples were frozen with liquid nitrogen immediately and then kept in -80°C until RNA extraction. All treatments were repeated four times.
Total RNA of the larval samples was extracted with TRIzol (Bioer, Hangzhou, China) and the quality of the total RNA was tested using Bioanalyzer (Aglient, Santa Clara, CA, USA). After enrichment and fragmentation of mRNA, cDNA library construction and normalization were conducted using the Illumina Truseq v3 kit according to the standard protocol. The cDNA libraries generated from each sample were sequenced using Illumina HiSeq 2500.
All raw read data generated from the RNAseq of five control larval biological replicate samples, four replicates of 5 μg mL-1 and 10 μg mL-1 butanolide treated samples were deposited to the NCBI Short Read Archive (SRA) with the BioProject accession number PRJNA855486. Clean read data were firstly assembled using Trinity v2.6.2 (Haas et al., 2013) with the following specific parameters (–min_glue 3 –trimmomatic –max_memory 100G –group_pairs_distance 250 –path_reinforcement_distance 85 –min_kmer_cov 3). The obtained Trinity assembly was further clustered using Corset (Davidson and Oshlack, 2014) based on the result of read mapping and hierarchical clustering of the gene expression pattern of each Trinity transcript (unigene) across all the 12 samples. The clustered assembly was further collapsed at 95% sequence identity using CD-HIT-EST (Li and Godzik, 2006), generating the final B. neritina transcriptome assembly.
The final assembly was then annotated by BLASTx (NCBI-blast+ v.2.10.0) against NCBI non-redundant protein database (nr), NCBI nucleotide database (nt), EuKaryotic Orthologous Groups (KOG), Swiss-Prot, Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO). Protein domain assignment was also performed using HMMer against the Pfam HMM database (Finn et al., 2011). E-value cutoff was set as 1e-5 for homology search by BLASTx and for protein domain assignment by HMMer. In parallel, Illumina read mapping was performed using Bowtie2 (Langmead and Salzberg, 2012) and subsequent transcript abundant estimation was performed using Salmon (Patro et al., 2017). Transcript expression level was normalized to transcripts per million reads (TPM). Differential expression analysis was performed using DESeq2 (Love et al., 2014). Two pairwise comparisons, 5 μg mL-1 treatment vs control and 10 μg mL-1 treatment vs control, were performed. The threshold for differential expression was set at abs(log2(FoldChange))>1 (or at least 2 times differences) and adjusted p-value < 0.05. The differentially expressed unigenes were selected and further clustered by hierarchical clustering and k-mean clustering. Functional enrichment analysis of KEGG pathway was performed using KOBAS (2.0) (–fdr BH) (Xie et al., 2011), and the threshold for over-representation of a KEGG pathway was set at adjusted p-value < 0.05.
At 3 hours post-BU treatment, larval settlement rates at 10 μg mL-1 or higher concentrations were significantly lower (ANOVA p<0.001) than the FSW or 1% DMSO FSW control, indicating that BU have a strong and significant inhibitory effect on B. neritina larval settlement (Figure 1A), which is in accord with our previous studies (Zhang et al., 2012b). While more than 80% larvae have settled in the control, 1.25 and 2.5 μg mL-1 BU treatment groups in the first three hours of the bioassay experiment, persistent inhibitory effect on larval settlement was observed in 10 and 20 μg mL-1 BU treatment (Figure 1B). No mortality was observed in all treatment or control groups in the first 3 hours of treatment. However, after 9 hours of BU treatment, more than 74% (± 19%) mortality was observed in 20 μg mL-1 BU treatment group (Figure 1C). After 24 hours of BU treatment, all concentrations led to different degrees of larval mortality. Almost all larvae were dead in 10 and 20 μg mL-1 BU treatment groups. But in lower BU concentration treatments, only the 5 μg mL-1 BU treatment group resulted in significantly higher larval mortality rate than the seawater control group (ANOVA p<0.001) (Figure 1C). The mortality rates at 2.5 μg mL-1 and lower BU concentration treatments were not significantly different to the SW or DMSO control group.
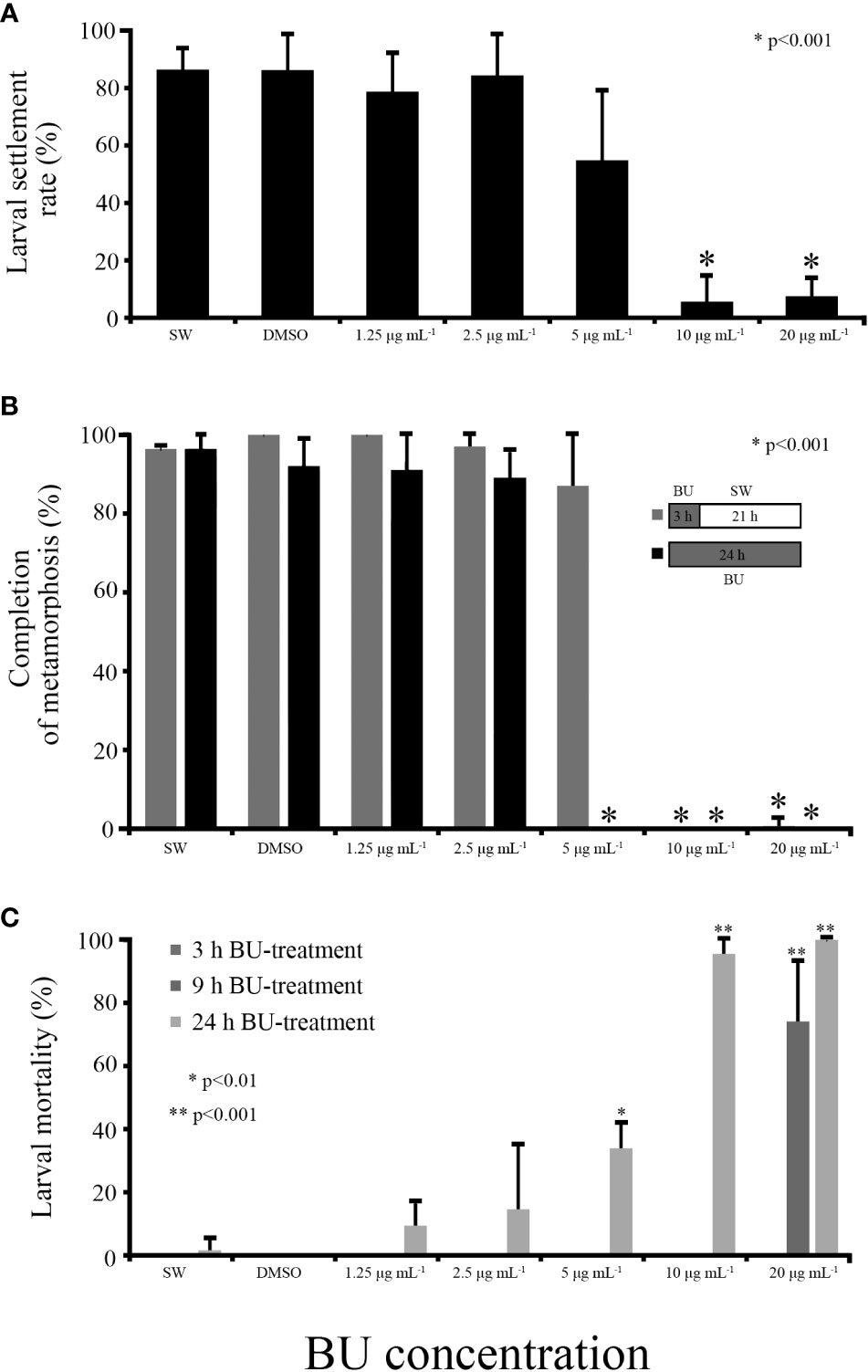
Figure 1 Bugulina neritina larval settlement bioassay for butenolide treatment. (A) Larval settlement rate after 3 hours of BU treatment. “*” indicates ANOVA p value < 0.001. (B) Rate of metamorphosis at 24 hours after the initiation of the bioassay experiment. Larvae in BU treatment groups were treated with BU for 3 hours, after which BU was replaced with autoclaved filtered seawater and incubated for 21 hours in dark until observation. “*” indicates ANOVA p value < 0.001. (C) Larval mortality at 3, 9 and 24 hours with different BU concentration treatment. “*” and “**” indicate ANOVA p value <0.01 and < 0.001.
In the recovery bioassay, BU solution was replaced by seawater after 3 hours BU treatment and the rate of larvae completed metamorphosis in the recovered wells was observed after 24 hours (3 hours BU+ 21 hours seawater) and compared to that in wells that experienced prolonged 24 hours BU treatment (no recovery). As expected, larvae in the control groups, 1.25 μg mL-1 and 2.5 μg mL-1 BU treatment groups completed metamorphosis within 24 hours in both the recovery and prolonged BU treatment experiments; larvae in 10 and 20 μg mL-1 BU treatment groups could not complete metamorphosis in both the recovery and prolonged BU treatment experiments, suggesting that larvae were unable to recovered from a 3 hours 10 μg mL-1 or higher BU concentration treatment; larvae in prolonged 5 μg mL-1 BU treatment group could not complete metamorphosis, but removal of BU at 3 hours allowed 87% (± 18%) BU-treated larvae to recover and complete metamorphosis (Figure 1B).
To analyze the effect of BU treatment on B. neritina larval swimming behavior, we took video footages of B. neritina larvae in seawater (SW control), in BU treatments and in recovery experiments (replacement of BU solution with SW). By analyzing the larval movement trajectory, we observed circular looping (see Figure 2A) and spiral swimming behavior (see Figure 2B) among larvae treated with 5 μg mL-1 or higher concentrations of BU, but seldom in 1.25 and 2.5 μg mL-1 BU-treated or control larvae, which typically exhibited movement with large displacement distances (see Figure 2C).
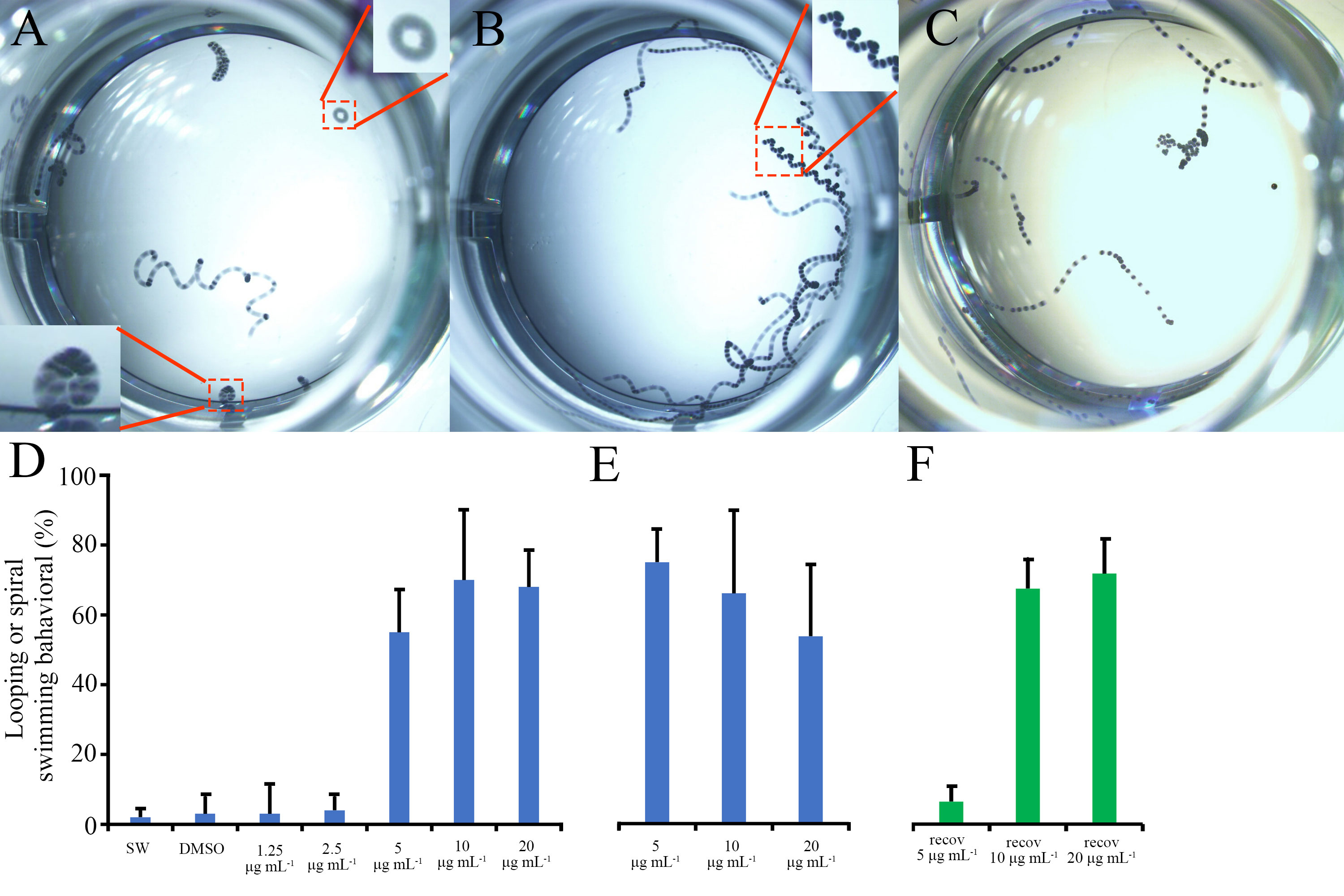
Figure 2 Bugulina neritina larval swimming behavior analysis. (A–C) Representative overlaid images of all frames of a 10 seconds video which recorded the larval swimming behavior after 1.5 hours of BU treatment. (A) Looping swimming behavior of B. neritina larvae in 10 μg mL-1 BU treatment. (B) Spiral swimming trajectory of B. neritina larvae in 5 μg mL-1 BU treatment. (C) The swimming trajectory of B. neritina larvae in the control group. (D, E) The percentage of larvae exhibiting looping or spiral swimming behavior at (D) 1.5 hours and (E) 6 hours of BU treatments. (F) The percentage of larvae exhibiting looping or spiral swimming behavior at 1 hour after recovery from 3 hours BU treatments in the recovery group.
At 1.5 hours, the percentage of larvae that exhibited circular looping or spiral swimming behavior in 5, 10 and 20 μg mL-1 BU treatment (55% ± 12%, 70% ± 20%, 68 ± 10%) groups were significantly higher than in 1.25 and 2.5 μg mL-1 BU-treated [3 ± 8% and 4 ± 4%)] larvae and the control groups (SW:2 ± 2%; DMSO 3 ± 5%) (Figure 2D).
At 6 hours, more than 95% larvae in the control groups (SW and 1% DMSO), 1.25 and 2.5 μg mL-1 BU treatment groups already settled and initiated metamorphosis and hence no video was taken for these experimental groups. Similar percentage of unsettled larvae in 5, 10 and 20 μg mL-1 BU treatment groups (67% ± 8%, 59% ± 21%, 48 ± 18%) exhibited looping or spiral swimming behavior as at the 1.5 hours time point (Figure 2E).
In the recovery assay, after replacement of BU (after 3 hours treatments) by SW for 1 hour, the percentage of larvae exhibiting looping or spiral swimming behavior in 10 and 20 μg mL-1 BU-treated groups (67% ± 8%, 63% ± 9%) remained at a similar level as at 1.5 and 6 hours. In contrast, percentage of larvae exhibited looping or spiral swimming behavior in the recovery assay was significantly lower in the 5 μg mL-1 BU treatment group (6 ± 4%) (Figure 2F).
Based on the above bioassay results, we define 5 μg mL-1 and 10 μg mL-1 as “low” and “high” BU concentration (low [BU], high [BU]) treatments. To understand the influence of these BU treatments on larvae, four low and four high [BU] treated larval samples, as well as five untreated “control” larval samples were submitted for cDNA library preparation and HiSeq sequencing. After quality trimming, these 13 samples generated 635,547,702 paired-end reads, composing of 95.33 Gbp. De novo assembling of the RNAseq data of all the 13 samples generated a transcriptome assembly containing 138,636 transcripts, which were further clustered into 48,001 unigenes based on the result of read mapping and transcript abundance using the Corset algorithm (Davidson and Oshlack, 2014). The transcript length N50 of the assembly is 1,893 bp and the average transcript length is 1,214 bp (Figure S1).
Among the 48,001 unigenes, 22,298 (or 46.45%) were annotated (by BLASTx, e-value cutoff at 1e-5 for homology search, OR, by HMMer for protein domain assignment) by at least one of the public databases, including NCBI nucleotide (nt) sequence database, the NCBI non-redundant (nr) database, SwissProt, Pfam, Gene Ontology (GO), EuKaryotic Orthologous Groups (KOG), KEGG Orthrology (KO) and the KEGG database (Figure S2). Consistent to our previous RNAseq study (Xu et al., 2020), close to 22% of the nr-annotated unigenes were best-matched (share the highest sequence homology) to the brachiopod Lingula anatina (Figure S3), once again indicating a potential close phylogenetic relationship between bryozoans and brachiopods, as suggested by an earlier study (Luo et al., 2015).
The functional annotation of the unigenes exhibited different tendency in different functional annotation methods. GO classification of the unigene highlighted the dominance of genes relating to “Binding” and/or “Catalytic activity”, in terms of molecular function; “Macromolecule complex”, in terms of Cellular Component; “Signaling” and “Regulation of biological process” in terms of Biological Process (Figure S4). On the other hand, KEGG classification indicated the dominance of genes relating to signal transduction and translation (Figure S5).
The transcriptome profiles of high [BU] and low [BU] treated larvae exhibited major differences comparing to the control untreated larvae, as revealed by the Pearson correlation coefficient analysis of the gene expression profile of each sample replicates. However, high [BU] and low [BU] treatments have led to a very similar larval transcriptome profile, as indicated by the outstanding Pearson correlation coefficient (r2 = 0.845 to 0.911) between high [BU] and low [BU] treated larval samples (Figure S6).
To further characterize the effect of BU on larval gene expression, the larval gene expression levels at high [BU] and low [BU] treatments were compared to that of the control. 8,040 genes were differentially expressed in both high [BU]-to-control and low [BU]-to-control comparisons while only 2,995 were specific to high [BU]-to-control (Figure S7) and 1,757 DEGs to low [BU]-to-control comparisons (Figure S8).
To further understand the larval response to BU treatment, the functional enrichment analysis was performed based on the over-representation of enriched KEGG and GO terms among the list of DEG in high [BU]-to-control and low [BU]-to-control comparisons. In the GO enrichment analysis, the GOBP term “DNA integration” and the GOMF term “ATP activity” were both over-represented in both high [BU]- and low [BU]-to-control DEGs (Figures 3A, B), and the fold of enrichment of both GO terms were more significant in the up-regulated DEGs than in the down-regulated DEGs. The GOBP term “xenobiotic metabolic process”, which is mainly related to the metabolism of foreign compounds, or in other words, detoxification, was over-represented in the list of up-regulated DEGs in the low [BU]-to-control comparison (Figure 3A).
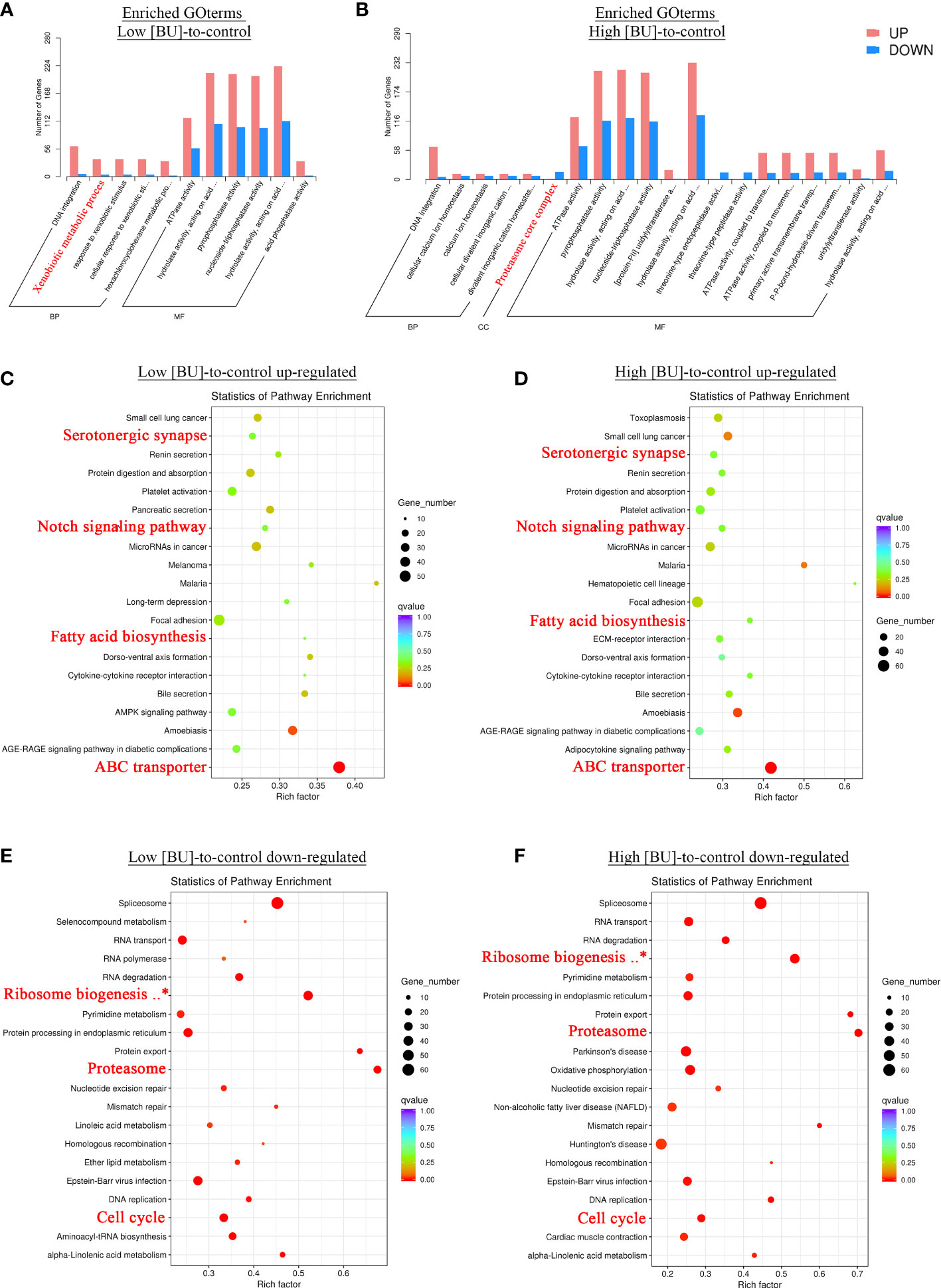
Figure 3 GO and KEGG enrichment analyses of differentially expressed genes in response to BU treatment. (A, B) Results of GO enrichment analysis. Enriched GO terms among the DEGs in (A) low concentration (5 μg mL-1) and (B) high concentration (10 μg mL-1) BU treatment were shown. (C–F) Results of KEGG enrichment analysis. The enrichment q-values of each enriched KEGG pathway among (C) up-regulated and (D) down-regulated DEGs in low concentration BU treatment, (E) up-regulated and (D) down-regulated DEGs in high concentration BU treatment were shown in the dot plots. Pathways that are not relating to human disease and broadly implicated across metazoan taxa were highlighted in red.
The list of enriched KEGG terms among the DEG in the low and high [BU]-treated larvae were highly similar. However, the enriched KEGG terms in the up- and down-regulated DEGs were remarkably different. For the up-regulated DEGs, the nervous system related term “Serotonergic synapse”, the detoxification related term “ABC transporter”, the lipid biosynthesis related term “fatty acid biosynthesis”, and the development pathway “Notch signaling pathway” were over-represented (Figures 3C, D); for the down-regulated DEGs, some of the essential functional terms such as “Cell cycle”, “Proteasome” and “Ribosome biogenesis in eukaryotes” were over-represented (Figures 3E, F). The results of GO and KEGG enrichment analyses indicated both low and high [BU] treatments suppressed de novo synthesis, cell proliferation and developmental processes, and at the same time triggered detoxification and increased energy production.
It is worth to note that B. neritina is a non-model organism and some of the differentially expressed genes that have important functions may not be included in the GO or KEGG functional categories. Therefore, we decided to investigate unigenes involved in nitric oxide signaling and other important functional categories that were implicated in larval settlement.
The enrichment of the KEGG term “serotonergic synapse” among up-regulated DEGs in both low and high [BU]-treated larvae suggested the effect of BU treatment on larval nervous system. The list of up-regulated DEGs relating to “serotonergic synapse” included the synaptic enzyme aromatic-L-amino-acid decarboxylase (AADC, or DOPA decarboxylase) which catalyzes the formation of dopamine from L-DOPA, neurotransmitter receptors Gamma-aminobutyric acid receptor (subunit beta) and 5-hydroxytryptamine receptor. The selected list also included a number of intracellular components directly relating to intracellular calcium signaling, including inositol 1,4,5-triphosphate receptor, protein kinase A, phosphatidylinositol phospholipase C and various types of voltage-dependent calcium channels (Tables 1, S1). The above unigenes were up-regulated by 1.2 to 4.3 folds in low and/or high [BU] treated larvae (Table S1).
Five and six nitric oxide synthase (NOS) unigenes were up-regulated in low and high [BU]-treated larvae, with a magnitude of up-regulation ranged from 1.5- to 2.9-fold. In addition to the up-regulation of NOS unigenes, the unigenes encoding downstream components cGMP-dependent protein kinase (PKG), soluble guanylate cyclase (sGC) and guanylate cyclase (GC) were also up-regulated in BU-treated larvae. The soluble guanylate cyclase unigene (Cluster-12452.419) was found to be up-regulated by 1-fold only in low [BU]-treated while the guanylate cyclase unigene (Cluster-13761.0) was up-regulated by 3.5- and 3.1-fold in low and high [BU]-treated larvae; the unigene of PKG was up-regulated by 1.4 and 1.5 fold in low and high [BU]-treated larvae. The expression of the unigene of arginine kinase, which uses the same substrate (L-arginine) as NOS and hence affects one of the upstream components the NO signaling pathway, was not found to exhibit significant variation (data not shown).
The developmental KEGG term “Notch signaling process” was over-represented among up-regulated DEGs, implying that developmental processes in the larvae were likely to be affected. We therefore investigated in detail the expression pattern of developmental pathway genes. Surprisingly, except Notch, which was up-regulated by almost two folds in both low and high [BU] treated larvae, other major developmental morphogens were all significantly down-regulated by BU treatment. Wnt ligands wnt10 (Cluster-12452.10908), wnt16-like (Cluster-9233.0), an WNT antagonist sfrp-5 (Cluster-12452.10592), and the Wnt pathway key player beta-catenin (beta-catenin-like protein 1, Cluster-12452.22208) were either not detected in low [BU] or significantly down-regulated in both high [BU] and low [BU] treated larvae. In addition, the BMP ligand BMP5 (Cluster-12452.1991), the Shh ligand ihh (Cluster-12452.18656), were also not detected in low [BU] or significantly down-regulated in the high [BU] treated larvae (Table 1).
Proteasome is one of the over-represented KEGG term in the list of down-regulated DEGs. In fact, there were 29 proteasome DEGs down-regulated in both low and high [BU]-treated larvae and one proteasome DEG down-regulated exclusively in high [BU] treated larvae. These proteasome DEGs were annotated as different subunits, regulatory subunits or non-ATPase regulatory subunit of 20S and 26S proteasomes, proteasomes inhibitor subunits, or proteasome assembly chaperone. The magnitude of down-regulation ranged from 1- to 2.18-fold.
Although the KEGG term “fatty acid biosynthesis” was enriched among up-regulated DEGs in both low and high [BU]-treatment groups, it was rather surprising to find that only two lipid metabolic proteins acetyl-CoA carboxylase and long-chain-fatty-acid–CoA ligase were up-regulated. While acetyl-CoA carboxylase catalyzes the production malonyl CoA, the building block for fatty acid biosynthesis, the unigene of fatty acid synthase (Cluster-12452.14482) was down-regulated by 1- and 2-fold in low and high [BU]-treated larvae; the unigene of elongation of very long chain fatty acids protein (Cluster-12452.16696, Cluster-12452.7219) was down-regulated by 1.2-fold in high [BU]-treated larvae; the unigene of fatty acid 2-hydroxylase-like, which is an important lipid metabolic protein in neural tissue, was drastically down-regulated by 5.5- and 5.6-fold in low and high [BU]-treated larvae.
More surprisingly, four vitellogenin-like unigenes were drastically down-regulated in both low and high [BU] treated larvae. The magnitude of down-regulation ranges from 5.6- to 7-fold. These four B. neritina vitellogenin-like unigenes were two short sequences (Cluster-12452.14875, Cluster-12452.15808) that were shorter than 500 bp and two long sequences (Cluster-12452.20127, Cluster-12452.15767) that were longer than 4,500 bp. Nevertheless, the deduced ORFs of all four unigenes contained the lipoprotein domain (PF01347) (or Vitellogenin N-terminal domain).
Bugulina neritina could settle and metamorphose in as short as 30 min after spawning (Wendt and Woollacott, 1995). The larval nervous system, which links the locomotory coronate ciliated cells, the pyriform organ, the sensory vibration tuft, the eyespots and the adult presumptive tissue internal sac, was suggested to be the key player in regulating larval settlement (Reed, 1988; Wendt and Woollacott, 1995). For instance, the larval nervous system was suggested to be the key to a coordinated phototactic behavioral response (Reed, 1988). The sensory input from the eye spots and sensory vibration tuft is transduced to the locomotory coronated ciliated cells and the pyriform organ to enable the larvae to perceive and swim toward the light source. Larval contact with the substratum was perceived by the vibration tuft as a mechanical signal and then transduced to the pyriform organ to trigger mucous secretions. Continuous secretion of mucus results in the formation of silk-like thread that stick to the bottom, anchoring the larva to the side or bottom of the well and thereby leading to the cyclic swimming behavior (Young, 2020).
Here, from the video record, we observed BU-treated larvae exhibited spiral or, in certain larvae, concentric swimming behavior. Bugulina neritina larvae are phototactic and would swim toward light with a rather direct trajectory. Spiral swimming behavior normally does not occur. Such swimming behavior might indicate failure of the nervous system in coordinating the locomotory organ. Shimizu and colleagues have reported that biogenic amine neurotransmitter, especially serotonin and dopamine, have a major effect on the larval settlement of B. neritina (Shimizu et al., 2000). Interestingly, in our gene expression analyses, the KEGG term “Serotonergic synapse” was over-represented and multiple receptors for different neurotransmitters were up-regulated in response to BU treatments. The result overall suggested that the larval nervous system of B. neritina might be directly targeted or indirectly affected by BU.
While the spiral swimming behavior might be relating to the nervous system control, the concentric swimming behavior may relate to the pyriform organ mucous secretion. Mucous secretions and anchored swimming larvae can normally be observed during the substratum exploration phase. In fact, the pyriform organ, which is responsible for these secretions, not only contains secretory tissue cells, but is also equipped with a series of specialized sensory cilia (Miyamoto et al., 2013; Wirth et al., 2019). The sticky secretions that form silk-like threads is likely to be released upon mechanical contact with foreign objects and thereby assisting larval attachment at the initial stage. In the laboratory condition, when the larval density was too high and the larvae were too crowded (over 200 larvae in a well of 24-well plate with 2 ml of seawater), the swimming larvae always tended to form a big clump and the larvae in the center of the clump would be trapped within a patch of mucous secretion (unpublished observations). These larval secretions were obviously triggered by mechanical contact between different swimming larvae. However, in our bioassay experiment, the larval density in each well was maintained at 10 larvae per ml of seawater each well. As our transcriptomic analysis indicated that the up-regulation of mucin genes was associated with BU treatment, we propose that BU treatment may not only result in abnormal swimming behavior, but also triggered improper mucus secretion even the larvae were not ready for settlement. As a result, some larvae were anchored to the substratum but at the same time continued to swim vigorously, resulting in the concentric swimming trajectory as observed in the BU bioassay experiment.
NO signal was previously detected in the equatorial nerve ring of B. neritina larvae and we have experimentally established that high endogenous or exogenous NO level inhibits larval settlement (Yang et al., 2018b). In addition, the gene expression level of NOS and sGC or GC were all up-regulated during the larval settlement process (Wang et al., 2016; Yang et al., 2018a). As NO in vertebrate systems could act as a neurotransmitter, it was suggested that the inhibitory effect of NO might be mediated through the nervous system (Yang et al., 2018a). Since NOS is the major enzyme producing NO and the enzyme was reportedly expressing in proximity to the equatorial nerve ring (Yang et al., 2018a), BU induced up-regulation of NOS gene further suggested the role of nervous system in mediating the inhibitory effect of BU.
Our transcriptomic data also reported the up-regulation of lipid metabolic genes and down-regulation of lipid storage genes. These results, especially the up-regulation of genes encoding acetyl-CoA carboxylase and long-chain-fatty-acid–CoA ligase, are consistent with the finding by Zhang and colleagues that BU targets the energy metabolism of B. neritina larvae (Zhang et al., 2012a). However, as we examine the transcriptomic signature of other lipid metabolic genes, it is obvious that there are more down-regulated lipid metabolic and storage genes than up-regulated genes. The result revealed the shortcoming of molecular target identification using chemical pull-down experiments, which lacks a comprehensive view and may lead to mis-conclusion on the effect of the antifouling compound on the larvae.
While BU induced certain behavioral and gene expression changes in B. neritina larvae that were previously not reported, the compound did induce consistent down-regulation of proteasome and developmental genes, which were also down-regulated by the treatment of another antifouling compound DIM (Xu et al., 2020). As the chemical structures of BU and DIM differ substantially, they may have very different molecular targets as well as mode of actions. Consistent down-regulation of proteasome and developmental genes may indicate a general larval response to chemical treatment by adjusting its physiology and halting their developmental program. While differential expression of proteasome might be a common stress response, down-regulation of developmental genes including Wnt10, Wnt16, Shh, sfrp, Hox4, goosecoid might be more species specific, as down-regulation of these development genes were not reported in BU-treated barnacle larvae (Zhang et al., 2011a).
The finding that BU and DIM induce the down-regulation of developmental genes has an important environmental implication. Developmental genes typically act as master regulator in the developmental program across metazoans. For example, the WNT ligands are key morphogens that regulate cellular polarity, axial patterning and cell fate determination. The molecular signal transduction pathway is conserved from sponge to human (Adamska et al., 2010). In B. neritina, WNT ligands were suggested to play an important role in the patterning and development of the ancestrula feeding apparatus lophophore (Fuchs et al., 2011; Wong et al., 2012; Wong et al., 2014). For the lecithotrophic larvae of fouling organisms like those of B. neritina, which has a very short free-swimming period (as short as 30 min) and which settlement and metamorphosis are simultaneous, the expression of developmental genes should either remains at a low level or be maintained at a constant level during the larval period. Differential expression of these genes may imply that the normal developmental program were likely being disrupted. It is noteworthy, however, that our transcriptomic results reflected the effect of at least 5 μg mL-1 BU treatment for at least 3 hours, while in the field, marine larvae may not experience such high concentration and prolonged exposure to the chemical. Nonetheless, we argue that chemicals that induce differential expression of developmental genes deserve special alert to its application. With more compound being tested in the future, we suggest that the gene expression level of developmental genes could be developed into the biomarkers for assessing the degree of impact of antifouling compound on lecithotrophic larvae of marine invertebrates.
In conclusion, we combined behavioral and transcriptomic analyses to acquire a clearer picture on the inhibitory mechanisms of BU on larval settlement. We suggest that BU might not only act on the larval energy metabolism, but could also affect the larval nervous system and mucus secretion in a direct or indirect manner, resulting in the specific looping swimming patterns in B. neritina larvae. Besides the specific effect of BU on larval behavior and gene expression, both BU and DIM appeared to trigger down-regulation of developmental genes and up-regulation of proteasome, suggesting important environmental implications on the application of antifouling compounds in the field. Our results once again suggest the commercialization of BU should proceed with cautions.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Conceptualization: J-YL, Y-FC, YZ, YX and YHW. Data curation: J-YL, Y-FC. Formal analysis: J-YL, Y-FC, YZ, YX and YHW. Funding acquisition: YX, YZ, and YHW. Investigation: J-YL, Y-FC, YX, YZ, and YHW. Methodology: J-YL, Y-FC, YZ, YX and YHW. Project administration: YX, YZ, and YHW. Supervision: YX, YZ, and YHW. Validation: YX, J-YL, YHW. Roles/Writing - original draft: J-YL, Y-FC, YZ, and YHW. Writing - review & editing: J-YL, YX, YZ, and YHW. All authors contributed to the article and approved the submitted version.
This study was supported grants from Scientific and Technical Innovation Council of Shenzhen and Guangdong Natural Science Foundation (JCYJ20210324093412035 and 2020A1515011117) awarded to YZ; the Innovation Team Project of Universities in Guangdong Province (2020KCXTD023), Natural Science Foundation of Shenzhen University award (2019019) and the Natural Science Foundation of China (41906091) awarded to YHW.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.1030070/full#supplementary-material
Adamska M., Larroux C., Adamski M., Green K., Lovas E., Koop D., et al. (2010). Structure and expression of conserved wnt pathway components in the demosponge Amphimedon queenslandica. Evol. Dev. 12 (5), 494–518. doi: 10.1111/j.1525-142X.2010.00435.x
Brock B. J. (1985). South Australian fouling bryozoans. Bryozoa: Ordovician to Recent. (Fredensborg, Denmark: Olsen & Olsen), 45–49.
Bryan P. J., Kreider J. L., Qian P. Y. (1998). Settlement of the serpulid polychaete Hydroides elegans (Haswell) on the arborescent bryozoan Bugula neritina (L.): Evidence of a chemically mediated relationship. J. Exp. Mar. Biol. Ecol. 220 (2), 171–190.
Bryan P. J., Qian P. Y., Kreider J. L., Chia F. S. (1997). Induction of larval settlement and metamorphosis by pharmacological and conspecific associated compounds in the serpulid polychaete Hydroides elegans. Mar. Ecol-Prog. Ser. 146, 81–90. doi: 10.3354/meps146081
Chen L., Ye R., Xu Y., Gao Z., Au D. W., Qian P. Y. (2014a). Comparative safety of the antifouling compound butenolide and 4,5-dichloro-2-n-octyl-4-isothiazolin-3-one (DCOIT) to the marine medaka (Oryzias melastigma). Aquat. Toxicol. 149, 116–125. doi: 10.1016/j.aquatox.2014.01.023
Chen L., Zhang H., Sun J., Wong Y. H., Han Z., Au D. W., et al. (2014b). Proteomic changes in brain tissues of marine medaka (Oryzias melastigma) after chronic exposure to two antifouling compounds: Butenolide and 4,5-dichloro-2-n-octyl-4-isothiazolin-3-one (DCOIT). Aquat. Toxicol. 157, 47–56. doi: 10.1016/j.aquatox.2014.09.010
Chen L., Sun J., Zhang H., Au D. W., Lam P. K., Zhang W., et al. (2015). Hepatic proteomic responses in marine medaka (Oryzias melastigma) chronically exposed to antifouling compound butenolide [5-octylfuran-2(5H)-one] or 4,5-dichloro-2-N-octyl-4-isothiazolin-3-one (DCOIT). Environ. Sci. Technol. 49 (3), 1851–1859. doi: 10.1021/es5046748
Conradi M., López-González P. J., Cervera J. L., García-Gómez J. C. (2000). Seasonality and spatial distribution of peracarids associated with the bryozoan Bugula neritina in algeciras bay, Spain. J. Crustacean Biol. 20 (2), 334–349. doi: 10.1163/20021975-99990045
Davidson N. M., Oshlack A. (2014). Corset: Enabling differential gene expression analysis for de novo assembled transcriptomes. Genome Biol. 15 (7), 410. doi: 10.1186/s13059-014-0410-6
Dumont C. P., Harris L. G., Gaymer C. F. (2011). Anthropogenic structures as a spatial refuge from predation for the invasive bryozoan Bugula neritina. Mar. Ecol-Prog. Ser. 427, 95–103. doi: 10.3354/meps09040
Finn R. D., Clements J., Eddy S. R. (2011). HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37. doi: 10.1093/nar/gkr367
Fuchs J., Martindale M. Q., Hejnol A. (2011). Gene expression in bryozoan larvae suggest a fundamental importance of pre-patterned blastemic cells in the bryozoan life-cycle. Evodevo 2 (1), 13. doi: 10.1186/2041-9139-2-13
Gordon D. P., Mawatari S. F. (1992). Atlas of marine-fouling bryozoa of new-Zealand ports and harbours. Miscellaneous Publ. New Z. Oceanographic Institute 107, 1–52.
Haas B. J., Papanicolaou A., Yassour M., Grabherr M., Blood P. D., Bowden J., et al. (2013). De novo transcript sequence reconstruction from RNA-seq using the trinity platform for reference generation and analysis. Nat. Protoc. 8 (8), 1494–1512. doi: 10.1038/nprot.2013.084
Heijne W. H., Kienhuis A. S., Van Ommen B., Stierum R. H., Groten J. P. (2005). Systems toxicology: Applications of toxicogenomics, transcriptomics, proteomics and metabolomics in toxicology. Expert Rev. Proteomics 2 (5), 767–780. doi: 10.1586/14789450.2.5.767
Langmead B., Salzberg S. L. (2012). Fast gapped-read alignment with bowtie 2. Nat. Methods 9 (4), 357–359. doi: 10.1038/nmeth.1923
Li W., Godzik A. (2006). Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22 (13), 1658–1659. doi: 10.1093/bioinformatics/btl158
Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15 (12), 550. doi: 10.1186/s13059-014-0550-8
Luo Y. J., Takeuchi T., Koyanagi R., Yamada L., Kanda M., Khalturina M., et al. (2015). The Lingula genome provides insights into brachiopod evolution and the origin of phosphate biomineralization. Nat. Commun. 6, 8301. doi: 10.1038/ncomms9301
Marta P.-E., Costello M. J. (2020). “The biology, ecology and societal importance of marine bryozoa,” in Encyclopedia of the world's biomes. Eds. Goldstein M. I., DellaSala D. A. (Oxford: Elsevier), 499–503.
Miyamoto N., Shinozaki A., Fujiwara Y. (2013). Neuroanatomy of the vestimentiferan tubeworm Lamellibrachia satsuma provides insights into the evolution of the polychaete nervous system. PloS One 8 (1), e55151. doi: 10.1371/journal.pone.0055151
Patro R., Duggal G., Love M. I., Irizarry R. A., Kingsford C. (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14 (4), 417–419. doi: 10.1038/nmeth.4197
Pires A., Woollacott R. M. (1997). Serotonin and dopamine have opposite effects on phototaxis in larvae of the bryozoan Bugula neritina. Biol. Bull. 192 (3), 399–409. doi: 10.2307/1542749
Pratt M. C. (2008). Living where the flow is right: How flow affects feeding in bryozoans. Integr. Comp. Biol. 48 (6), 808–822. doi: 10.1093/icb/icn052
Prevosto M., Davies P., Compére C., Olagnon M. (2012). “Constraints of the marine environment,” in Marine renewable energy handbook (Hoboken, New Jersey: Wiley-ISTE) 23–42.
Reed C. G. (1988). Organization of the nervous system and sensory organs in the larva of the marine bryozoan Bowerbankia gracilis (Ctenostomata: Vesiculariidae): Functional significance of the apical disc and pyriform organ. Acta Zool 69 (3), 177–194. doi: 10.1111/j.1463-6395.1988.tb00915.x
Scheer B. T. (1945). The development of marine fouling communities. Biol. Bull. 89 (1), 103–121. doi: 10.2307/1538088
Shimizu K., Hunter E., Fusetani N. (2000). Localisation of biogenic amines in larvae of Bugula neritina (Bryozoa: Cheilostomatida) and their effects on settlement. Mar. Biol. 136 (1), 1–9. doi: 10.1007/s002270050001
Wang K. L., Xu Y., Lu L., Li Y., Han Z., Zhang J., et al. (2015). Low-toxicity diindol-3-ylmethanes as potent antifouling compounds. Mar. Biotechnol. (NY) 17 (5), 624–632. doi: 10.1007/s10126-015-9656-6
Wang K. L., Zhang G., Sun J., Xu Y., Han Z., Liu L. L., et al. (2016). Cochliomycin a inhibits the larval settlement of Amphibalanus amphitrite by activating the NO/cGMP pathway. Biofouling 32 (1), 35–44. doi: 10.1080/08927014.2015.1121245
Wendt D. E., Woollacott R. M. (1995). Induction of larval settlement by KCl in three species of Bugula (Bryozoa). Invertebr. Biol. 114 (4), 345–351. doi: 10.2307/3226843
Wirth M., Wolff J. O., Appel E., Gorb S. N. (2019). Ultrastructure of spider thread anchorages. J. Morphol. 280 (4), 534–543. doi: 10.1002/jmor.20962
Wong Y. H., Ryu T., Seridi L., Ghosheh Y., Bougouffa S., Qian P. Y., et al. (2014). Transcriptome analysis elucidates key developmental components of bryozoan lophophore development. Sci. Rep. 4, 6534. doi: 10.1038/srep06534
Wong Y. H., Wang H., Ravasi T., Qian P. Y. (2012). Involvement of wnt signaling pathways in the metamorphosis of the bryozoan Bugula neritina. PloS One 7 (3), e33323. doi: 10.1371/journal.pone.0033323
Wood T. S. (2001). “14 - bryozoans,” in Ecology and classification of north American freshwater invertebrates (Second edition). Eds. Thorp J. H., Covich A. P. (San Diego: Academic Press), 505–525.
Wood A. C., Rowden A. A., Compton T. J., Gordon D. P., Probert P. K. (2013). Habitat-forming bryozoans in new Zealand: Their known and predicted distribution in relation to broad-scale environmental variables and fishing effort. PloS One 8 (9), e75160. doi: 10.1371/journal.pone.0075160
Wu Y., Wang G. (2018). Machine learning based toxicity prediction: From chemical structural description to transcriptome analysis. Int. J. Mol. Sci. 19 (8), 2358. doi: 10.3390/ijms19082358
Xie C., Mao X., Huang J., Ding Y., Wu J., Dong S., et al. (2011). KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 39(Web Server issue), W316–W322. doi: 10.1093/nar/gkr483
Xu Y., Zhang L., Wang K. L., Zhang Y., Wong Y. H. (2020). Transcriptomic analysis of the mode of action of the candidate anti-fouling compound di(1H-indol-3-yl)methane (DIM) on a marine biofouling species, the bryozoan Bugula neritina. Mar. pollut. Bull. 152, 110904. doi: 10.1016/j.marpolbul.2020.110904
Yang X. X., Wong Y. H., Zhang Y., Zhang G., Qian P. Y. (2018a). Exploring the regulatory role of nitric oxide (NO) and the NO-p38MAPK/cGMP pathway in larval settlement of the bryozoan Bugula neritina. Biofouling 34 (5), 545–556. doi: 10.1080/08927014.2018.1470240
Yang X. X., Zhang Y., Wong Y. H., Qian P. Y. (2018b). HSP90 regulates larval settlement of the bryozoan Bugula neritina through the nitric oxide pathway. J. Exp. Biol. 221 (Pt 8), jeb167478. doi: 10.1242/jeb.167478
Young C. M. (2020). “Behavior and locomotion during the dispersal phase of larval life,” in Ecology of marine invertebrate larvae (Boca Raton: CRC Press), 249–277.
Zhang Y. F., Wang G. C., Ying X., Sougrat R., Qian P. Y. (2011a). The effect of butenolide on behavioral and morphological changes in two marine fouling species, the barnacle Balanus amphitrite and the bryozoan Bugula neritina. Biofouling 27 (5), 467–475. doi: 10.1080/08927014.2011.583985
Zhang Y. F., Xiao K., Chandramouli K. H., Xu Y., Pan K., Wang W. X., et al. (2011b). Acute toxicity of the antifouling compound butenolide in non-target organisms. PloS One 6 (8), e23803. doi: 10.1371/journal.pone.0023803
Zhang Y. F., Kitano Y., Nogata Y., Zhang Y., Qian P. Y. (2012a). The mode of action of isocyanide in three aquatic organisms, Balanus amphitrite, Bugula neritina and Danio rerio. PloS One 7 (9), e45442. doi: 10.1371/journal.pone.0045442
Keywords: butenolide, transcriptome, antifouling (AF), larval swimming behavior, bryozoan Bugulina neritina
Citation: Liang J-Y, Cheng Y-F, Huang X-L, Xu Y, Wong YH and Zhang Y (2022) Behavioral and transcriptomic changes in butenolide treated larvae of the cosmopolitan fouling bryozoan Bugulina (Bugula) neritina. Front. Mar. Sci. 9:1030070. doi: 10.3389/fmars.2022.1030070
Received: 28 August 2022; Accepted: 21 September 2022;
Published: 01 November 2022.
Edited by:
André Ricardo Araújo Lima, Center for Marine and Environmental Sciences (MARE), PortugalReviewed by:
Gen Zhang, Hong Kong University of Science and Technology, Hong Kong SAR, ChinaCopyright © 2022 Liang, Cheng, Huang, Xu, Wong and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yue Him Wong, dGltd29uZ0BzenUuZWR1LmNu; Yu Zhang, YmlvenlAc3p1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.