- 1Wuxi Fisheries College, Nanjing Agricultural University, Wuxi, China
- 2Key Laboratory of Integrated Rice-Fish Farming Ecology, Ministry of Agriculture and Rural Affairs, Freshwater Fisheries Research Center, Chinese Academy of Fishery Sciences, Wuxi, China
Endogenous water pollution and excessive nitrogen in ponds have always been prominent problems threatening aquaculture. To solve this problem, a 70-day indoor simulation experiment was conducted. The snail Bellamya purificata was stocked at four different densities: 0, 15, 30, and 60 individuals/tank, represented as CON (control), LD (low density), MD (medium density), and HD (high density), respectively, to investigate the effects of B. purificata on the nitrogen occurrence forms and bacterial communities in the sediment. At the end of the experiment, the nitrate concentration was significantly higher, while the total nitrogen content was significantly lower in the MD group than in the other three groups. Ammonia monooxygenase activity was significantly lower in the CON group than in the other three groups, and hydroxylamine oxidase activity was significantly higher in the HD group than in the other three groups. The CON and MD groups showed the highest and lowest values in nitrate reductase activity, respectively. The hydroxylamine reductase activity decreased significantly with increasing density. Through 16S ribosomal RNA (rRNA) high-throughput sequencing, significantly affected bacterial communities by B. purificata were found. Alpha diversity results showed that, a significantly lower Shannon index was observed in the MD group than in the other three groups. The LD and MD groups showed the highest and lowest Chao1 index values, respectively. Phyla Nitrospinae and family Nitrosomonadaceae were significantly enriched in the HD and MD groups, respectively. Redundancy analysis (RDA) indicated a significant correlation between differential bacterial taxa and TN content. Predicted functional analysis based on FAPROTAX (Functional annotation of prokaryotic taxa) database showed that functional groups aerobic ammonia oxidation and aerobic nitrite oxidation were significantly enriched in the MD group. Overall, B. purificata significantly altered the bacterial community composition, increased hydroxylamine oxidase and ammonia monooxygenase activities, enhanced the bacterial nitrification process, and promoted the transformation of total nitrogen to nitrate. Moreover, B. purificata stocked at 30 and 60 individuals/tank appeared to have a significant promotion effect on the bacterial community and nitrogen occurrence forms in the sediment. Hence, co-culturing B. purificata may be a feasible and effective ecological restoration method to alleviate excess nitrogen and reduce water eutrophication.
1 Introduction
Pond aquaculture is the most important method of freshwater aquaculture in China (Zhang et al., 2020; Liu et al., 2021). In 2020, the total production and area of pond aquaculture accounted for 59.12% and 37.31% of total freshwater aquaculture, respectively (MARA, 2021). Aquaculture pond ecosystems are important freshwater ecosystems that provide valuable ecological services and economic benefits to humans (Li et al., 2009; Herbeck et al., 2021; Guo et al., 2021). However, traditional pond aquaculture is a labour-intensive industry, and the improvement of production and economic benefits relies on excessive feed input and high stocking density. Excessive nitrogen input severely affects pond nitrogen cycling and causes serious environmental problems (Zhu et al., 2019; Li J et al., 2022; Cao et al., 2020). With the continuous accumulation of organic nitrogen in aquaculture ponds, the sediment, as a carrier for the migration and accumulation of biogenic elements, can release nutrients and toxic substances into the upper water, thus causing endogenous water pollution and eutrophication of the pond ecosystem (Decaestecker et al., 2007; Gold et al., 2017; Woodman et al., 2021).
To address this dilemma and reduce the risk of endogenous water pollution and eutrophication, researchers have attempted to alleviate organic waste accumulation and reduce nitrogen pollution in pond sediments by co-culturing economical macrobenthos according to their feeding habits and abilities to utilise organic debris (Yuan et al., 2008; Ge, 2014). Yuan et al. (2008) used the sea cucumber Apostichopus japonicus to remove organic particulate waste and promote the regeneration of nutrients in sediments of shellfish culture systems. Ge (2014) attempted to improve resource utilisation efficiency and purify the culture environment by co-culturing freshwater prawns, Macrobrachium nipponense, in aquaculture ponds.
Benthic fauna plays an important role in aquatic ecosystems, especially in the biogeochemical cycle at the sediment-water interface, as they can lead to variations in the physical structure and chemical properties of the sediment and affect nitrogen migration and transformation (Dong et al., 2009; Löfgren et al., 2014; Li et al., 2015; Zou et al., 2022). In addition to their impacts on biogeochemistry, benthic fauna also contributes to the construction of most ecological communities through the redistribution of available resources (Modenutti and Pérez, 2001; Parro et al., 2019).
Microorganisms are the main driving factors involved in nitrogen migration and transformation (Kuypers et al., 2018; Zhang et al., 2022). Bacterial nitrogen cycling processes related to nitrogen elimination play an important role in controlling nitrogen balance in pond ecosystems (Feng et al., 2017; Wang et al., 2018; Wei et al., 2019). Bacteria produce various enzymes through metabolic processes to control nitrogen transformation. For example, ammonia monooxygenase and hydroxylamine oxidoreductase are involved in the first stage of nitrification, which converts ammonia to nitrite (Xu et al., 2016; Xiang et al., 2020). Previous studies report that the microbial community composition and function can also be regulated using benthic animals by altering the nutrient transport and physicochemical properties of sediments (Tuikka et al., 2016; Wu et al., 2017; Saxena et al., 2018). It may be an effective way to understand the pathways and mechanisms of benthic fanua affecting nitrogen transformation and transportation in sediments through a comprehensive study of nitrogen occurrence forms, extracellular enzyme activity, and bacterial community.
Studies on the effects of macrobenthos on nitrogen and its driving mechanisms from the perspective of bacteria and extracellular enzyme activity, however, are still limited.
The snail, Bellamya purificata, is an important freshwater macrobenthic organism with wide distribution in China and high nutritional value (Meng et al., 2013; Huang et al., 2021). The B. purificata is not only a popular aquatic product with high economic value but also an important ecological species (Liang et al., 2013). B. purificata can effectively improve water quality and provide important ecosystem services (Chen et al., 2012; Zhao, 2014). Hou et al. (2021) demonstrated that B. purificata can significantly promote the degradation of organic waste in sediments. However, there are few reports on how B. puificata affects the occurrence forms, migration, and transformation of nitrogen and the bacterial community. Determining the effects of B. puificata bioturbation on the nitrogen occurrence forms, extracellular enzyme activity, and bacterial communities in sediment can provide new ideas and theories for mitigating nitrogen pollution from the perspectives of polyculture and bioremediation.
Hence, the aims of our study were to investigate the effects of B. puificata on the nitrogen occurrence forms, related extracellular enzyme activity, and benthic bacterial communities, and to explore the pathways and mechanisms of B. puificata affecting nitrogen transformation. In addition, we hope to provide theoretical support for mitigating nitrogen pollution and preventing endogenous water pollution in aquaculture ponds and surrounding aquatic ecosystems.
2 Materials and methods
2.1 Culture experiment
The experiment in this study was conducted at the Key Laboratory of Integrated Rice-Fish Farming Ecology, Ministry of Agriculture and Rural Affairs, Freshwater Fisheries Research Center, Chinese Academy of Fishery Sciences (Wuxi, China). Bellamya purificata and experimental sediments were collected from the Dapu and Qiting aquaculture facilities, respectively (119°55’56.06”E, 31°18’41.69”N; 119°49’17.47”E, 31°23’55.32”N). The B. purificata were healthy and energetic with an average wet weight of 2.05 ± 0.28 g, shell length of 2.03 ± 0.15 cm, and shell height of 1.43 ± 0.18 cm. Before the experiment started, all snails were kept in glass tanks for 14 days to acclimatise them to laboratory conditions. In order to maintain the consistency and homogeneity of the experimental sediments, we dried all the sediments in the sun, grinded and passed all the sediments through a 100 µm mesh sieve, and mixed all the sediments evenly before use. A 10 cm thick layer of sediment was spread on the bottom of each experimental tank (63 × 30 × 39 cm), after which they were filled with filtered freshwater. The sediment in each tank were allowed to settle and stabilise for 14 days before the start of the experiment. Three treatments and one control were set up, with three replicates for each treatment. Snails were cultured in the experimental tanks at four different densities: 0, 15, 30, and 60 individuals/tank, represented as CON (control), LD (low density), MD (medium density), and HD (high density), respectively. Apart from the differences in stocking density, other experimental conditions were kept strictly consistent. During the experiment, the snails were fed at 16:00 every day. The feeding amount (2% of the total weight of snails in MD group) was identical for all tanks. Commercial feed (Zhejiang Haida Feed Co., Ltd, China) was used as experimental diets after being ground and sieved through a 100-µm mesh, which contained total organic matter (119.57 ± 2.31 mg/g), total nitrogen (41.02 ± 0.98 mg/g), and total phosphorus (7.07 ± 0.87 mg/g). One-third of the water was changed every two days. The water temperature was maintained at 25.0 ± 0.5°C. The experiment was conducted under a natural light: dark cycle for 70 days.
2.2 Sample collection
Since the experimental sediments have been pre-processed for keeping consistency and homogeneity before the experiment started. We only collected the sediment samples at the end of the experiment to avoid the disturbances caused by sampling operation. Surface sediment samples (0-1 cm depth) were collected at nine random sampling points in each tank using a plastic centrifuge tube with a diameter of 2 cm. Then, all the surface sediment samples collected from the same tank were mixed well and stored at -80°C for further determination of bacterial community, nitrogen occurrence forms, and extracellular enzyme activities.
2.3 Laboratory analyses
2.3.1 Nitrogen form
Before determination, the sediment samples were dried in a lyophiliser (CHRIST LYO Alpha 1-4 LD plus, German) and ground to a powder in a mortar. Ammonia, nitrate, nitrite, total nitrogen (TN), fixed ammonium (FA) and exchangeable nitrogen (EN) concentrations were determined. Ammonia nitrogen content was determined using Nessler’s reagent (Kołacińska and Koncki., 2014). Nitrate nitrogen was determined using phenol disulfonic acid spectrophotometry (Kendüzler and Türker., 2005). Nitrite nitrogen was determined using naphthalenediamine spectrophotometry (Pozzobon et al., 2021). TN was determined using alkaline potassium persulfate (McCarthy et al., 1980). FA and EN were determined as described by Silva and Bremner (1966) and Brodrick et al. (1987), respectively.
2.3.2 Enzyme activity
Five enzymes related to nitrification and denitrification were identified: nitrate reductase, nitrite reductase, hydroxylamine reductase, ammonia monooxygenase, and hydroxylamine oxidase. Nitrate reductase, nitrite reductase, and hydroxylamine reductase activities were determined using an enzyme activity kit from Grace Biotech International Group Co. Ltd. (Suzhou, China). Ammonia monooxygenase and hydroxylamine oxidase levels were determined using an ELISA kit from Shanghai Enzyme-linked Biotechnology Co. Ltd. (Shanghai, China).
2.3.3 DNA extraction and PCR amplification
The selected sequencing region was 16S-V3-V4 in this experiment. All samples were tested as required. According to the manufacturer’s protocols, E.Z.N.A. Soil DNA Kit (Omega Bio-tek, Norcross, GA, USA) was used to extracted the bacterial DNA from the collected sediment samples. We evaluated the DNA quality and quantity using a NanoDrop 2000 Spectrophotometer (Bio-Rad Laboratories Inc., USA). The V3-V4 region of the bacterial 16S ribosomal RNA gene was amplified according to the polymerase chain reaction (PCR) method with the primers 341F 5’- CCTAYGGGRBGCASCAG-3’ and 806R 5’-GGACTACNNGGGTATCTAAT-3’ as reported in Hou et al. (2021). After purification and quantification using AxyPrep DNA Gel Extraction Kit (Axygen Biosciences,Union City, CA, USA) and QuantiFluor™ -ST (Promega, USA), the PCR products extracted from all the sediment samples were pooled averagely, and then used to Illumina Pair-End library based on Illumina’s genomic DNA library preparation procedure. The amplicon library was paired-end sequenced (2 × 250) on an Illumina MiSeq platform (Shanghai BIOZERON Co., Ltd.) according to standard protocols. Raw reads were deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) database (PRJNA863036).
2.3.4 Bioinformatics analysis
QIIME software was used to process sequencing data, and UCLOST was used to classify operational taxonomic units (OTUs) at the similarity level of 97%. The sequence with the highest abundance of each OTU was selected as the representative sequence of OTU. Then, based on the number of OTU sequences in each sample, a matrix file of OTU abundance in each sample was constructed. To ensure the reliability and accuracy of the analysis results, the barcode sequences information for each sample with the following criteria: (i) The 250bp reads were truncated at any site receiving an average quality score <20 over a 10 bp sliding window, discarding the truncated reads that were shorter than 50bp. (ii) Exact barcode matching, 2 nucleotide mismatch in primer matching, 0.2 in overlap, reads containing ambiguous characters were removed. (iii) The reference and denovo were combined to remove singletons, the database was Silval 138, 27466 reads were left for each sample after normalization. The Ribosomal Database Project (RDP) Classifier (version 2.2 http://rdp.cme.msu.edu/) was used to determine the taxonomic identities of the phylotypes.
2.4 Statistical analysis
All data were subjected to one-way analysis of variance (ANOVA), followed by Duncan’s test for multiple comparisons to determine the differences between groups using SPSS Statistics software (version 22.0, International Business Machines Corporation, USA). The level of significance (α) was 0.05 (where P < 0.05 was considered significant). Functional annotation of prokaryotic taxa (FAPROTAX) database was used to predict ecological and biological functions of bacteria based on the python. Principal component analysis (PCA) was using SIMCAP (version 11.5, Umetrics, MKS Instruments AB, USA) to analyze, and the distance was based on Bray-Curtis. Use Mothur (version 1.30.2, https://www.mothur.org/wiki/Downl, USA) to analyze Chao1, Shannon and Simpson index. Redundancy analysis (RDA, CANOCO 4.5 software) was applied to reveal the correlations between environmental factors and the differential bacterial taxa.
3 Results
3.1 Nitrogen occurrence forms in sediment
The different nitrogen occurrence forms in the sediment of the CON, LD, MD, and HD groups are shown in Figure 1. The nitrate nitrogen content in the MD group was significantly higher than that in the other three groups (P < 0.05). Ammonia nitrogen content was significantly higher in the CON and LD groups than in the MD and HD groups (P < 0.05). The total nitrogen content in the MD group was significantly lower than that in the other three groups (P < 0.05). There was no significant difference in the nitrite nitrogen concentration in the sediments among the four groups (P > 0.05). No significant differences in fixed ammonium and exchangeable nitrogen concentrations were observed among the four groups (P > 0.05).
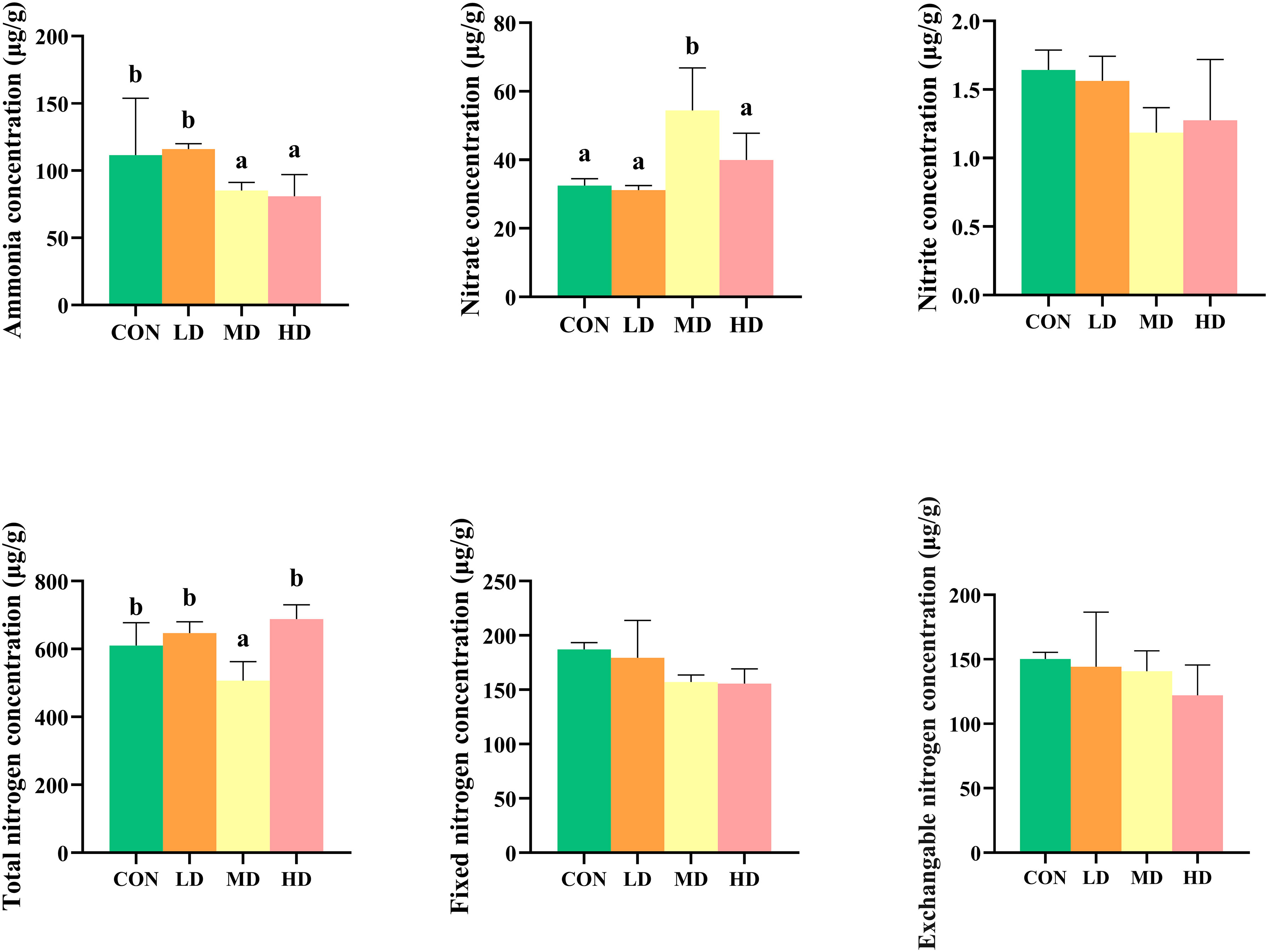
Figure 1 Differences in sedimentary nitrogen forms between the CON, LD, MD and HD groups during the experimental period, including ammonia, nitrate, and nitrite concentrations, total nitrogen, fixed nitrogen, and exchangeable nitrogen. Different lowercase letters represent significant differences between the CON, LD, MD, and HD groups (P < 0.05).
3.2 Extracellular enzyme activity in sediment
Extracellular enzyme activities, including nitrate reductase, nitrite reductase, hydroxylamine reductase, ammonia monooxygenase, and hydroxylamine oxidase in sediment of CON, LD, MD, and HD groups are shown in Figure 2. The CON and MD groups showed the highest and lowest nitrate reductase activities, respectively (P < 0.05). No significant differences in the nitrate reductase activity were observed between the LD and HD groups (P > 0.05). The CON and LD groups showed the lowest and highest nitrite reductase activity, respectively (P < 0.05). No significant differences in nitrite reductase activity were observed between the MD and HD groups (P > 0.05). The CON and HD groups showed the highest and lowest hydroxylamine reductase activity, respectively (P < 0.05). No significant differences in hydroxylamine reductase activity were observed between the MD and LD groups (P > 0.05). The HD and LD groups showed the highest and lowest hydroxylamine oxidase activity, respectively (P < 0.05). No significant differences in hydroxylamine oxidase activity were observed between the CON and MD groups (P > 0.05). Ammonia monooxygenase activity was significantly higher in the HD and MD groups than in the CON and LD groups (P < 0.05).
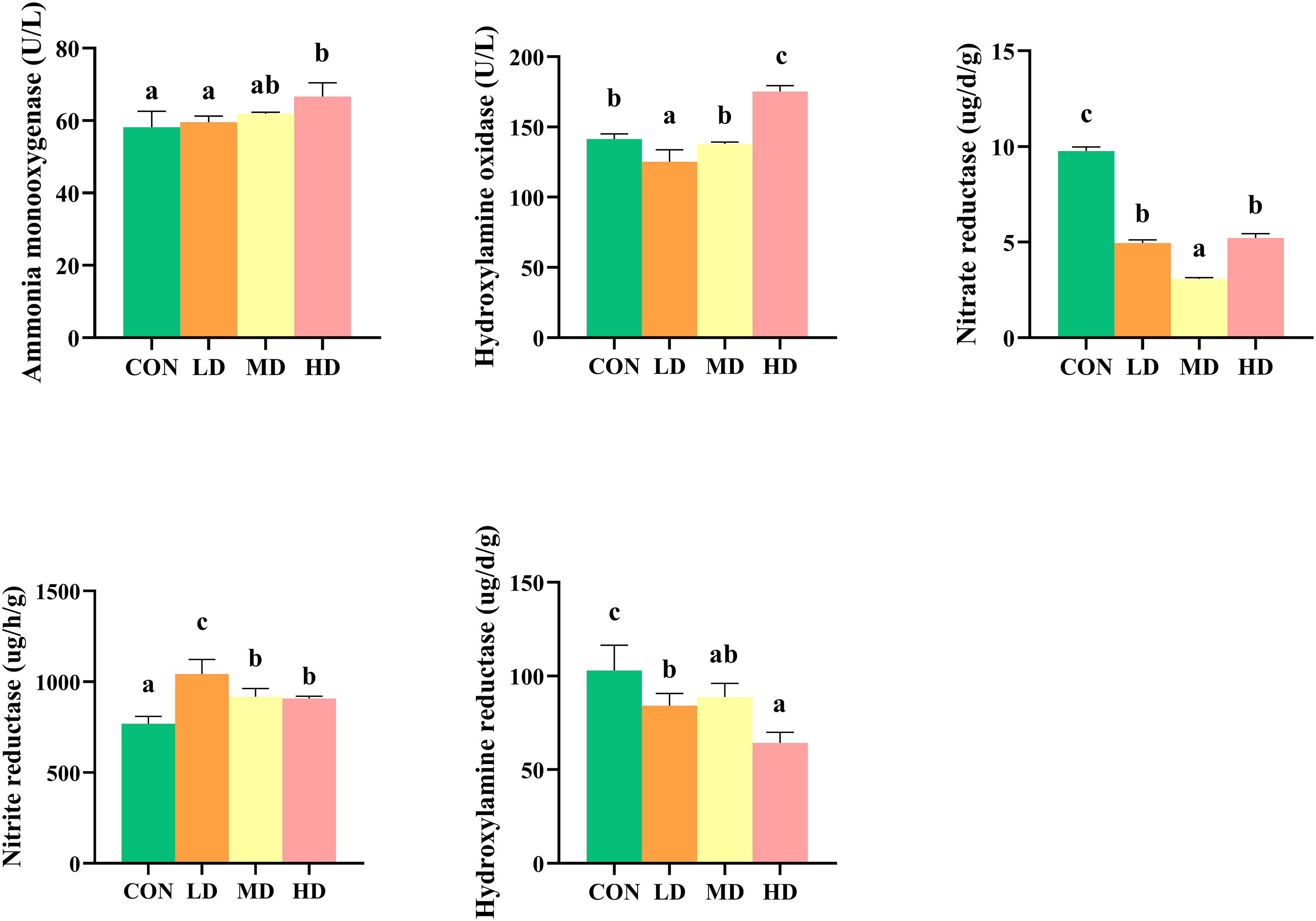
Figure 2 Differences in sedimentary extracellular enzyme activity between the CON, LD, MD and HD groups during the experimental period, including nitrate reductase, nitrite reductase, hydroxylamine oxidase, hydroxylamine reductase, and ammonia monooxygenase. Different lowercase letters represent significant differences between the CON, LD, MD, and HD groups (P < 0.05).
3.3 Bacterial community composition in sediment
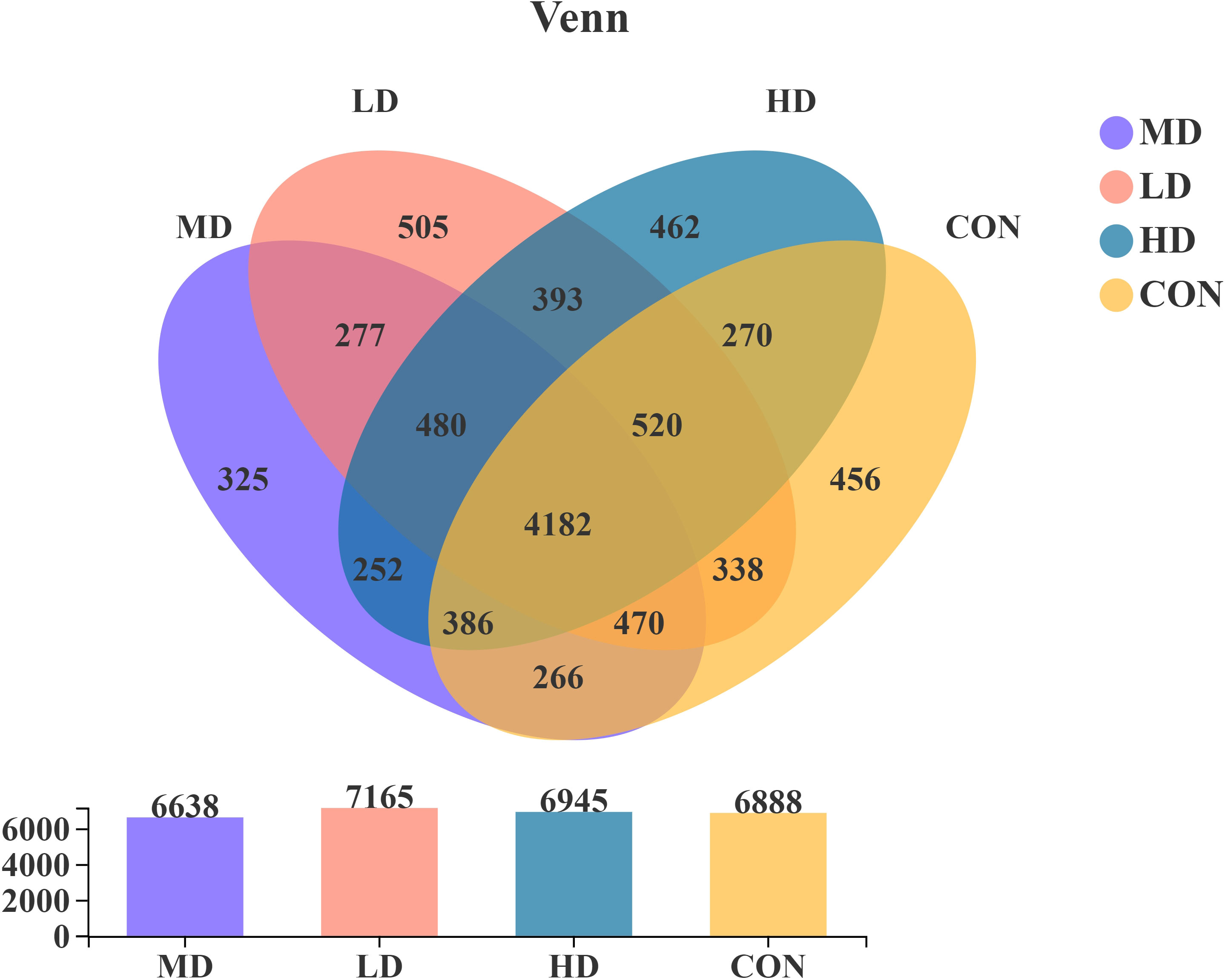
The present study obtained 9,582 OTUs from sediment samples using Illumina sequencing technology based on the bacterial 16S rRNA gene. The OTUs were assigned to 58 phyla, 606 families, and 1,066 genera. There were 456, 505, 325, and 462 OTUs unique to the CON, LD, MD, and HD groups, respectively (Figure 3). The CON, LD, MD, and HD groups contained 6,888, 7,165, 6,638, and 6,945 OTUs, respectively. The four groups shared 4,182 OTUs. Among these, there were 277 OTUs shared by the LD and MD groups, 393 OTUs shared between the LD and HD groups, and 252 OTUs shared between the MD and HD groups.
As shown in Figure 4, the ten most abundant phyla in the sediment at the end of the experiment were Proteobacteria, Acidobacteria, Bacteroidetes, Chlorofiexi, Nitrospirae, Verrucomicrobia, Actinobacteria, Nitrospinae, Cyanobacteria, and unclassified. The ten most abundant families in the sediment were Burkholderiaceae, Rhodocyclaceae, Thermodesulfovibrionia_norank, Syntrophaceae, Geobacteraceae, Nitrosomonadaceae, Steroidobacteraceae, Methylomonaceae, Pedosphaeraceae, and unclassified. The ten most abundant genera in the sediment were Burkholderiaceae_uncultured, Thermodesulfovibrionia_norank, Desulfatiglans, Steroidobacteraceae_uncultured, Subgroup 6, Anaerolineaceae_uncultured, Pedosphaeraceae_norank, P9X2b3D02_norank, Sva0485_norank, and unclassified.
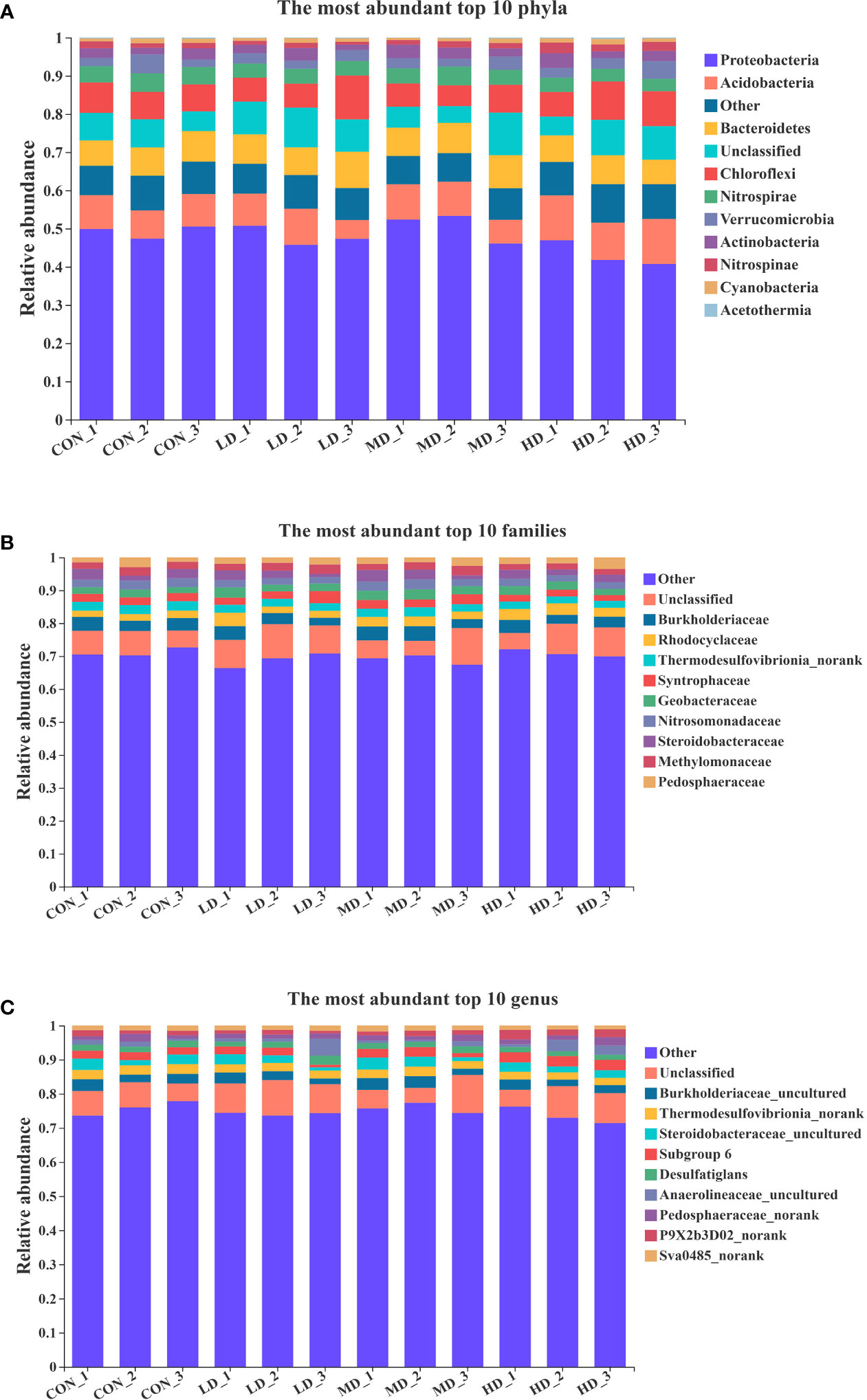
Figure 4 Ten most abundant phyla (A), families (B), and genera (C) in sediment of the CON, LD, MD, and HD groups at the end of the experiment.
Significant differences in the ten most abundant phyla, families, and genera among the CON, LD, MD, and HD groups were observed (Figure 5). At the phylum level, the relative abundance of Nitrospinae was significantly higher in the HD group than that in the other three groups (P < 0.05). The abundance of Proteobacteria was significantly lower in the HD group than in the other three groups (P < 0.05). At the family level, the relative abundance of Nitrosomonadaceae in the MD and CON groups was significantly higher than that in the LD and HD groups (P < 0.05). At the genus level, the HD and CON groups exhibited the significantly lowest and highest relative abundances in Thermodesulfovibrionia_norank, respectively (P < 0.05), while P9X2b3D02_norank was significantly more abundant in the HD group (P < 0.05).
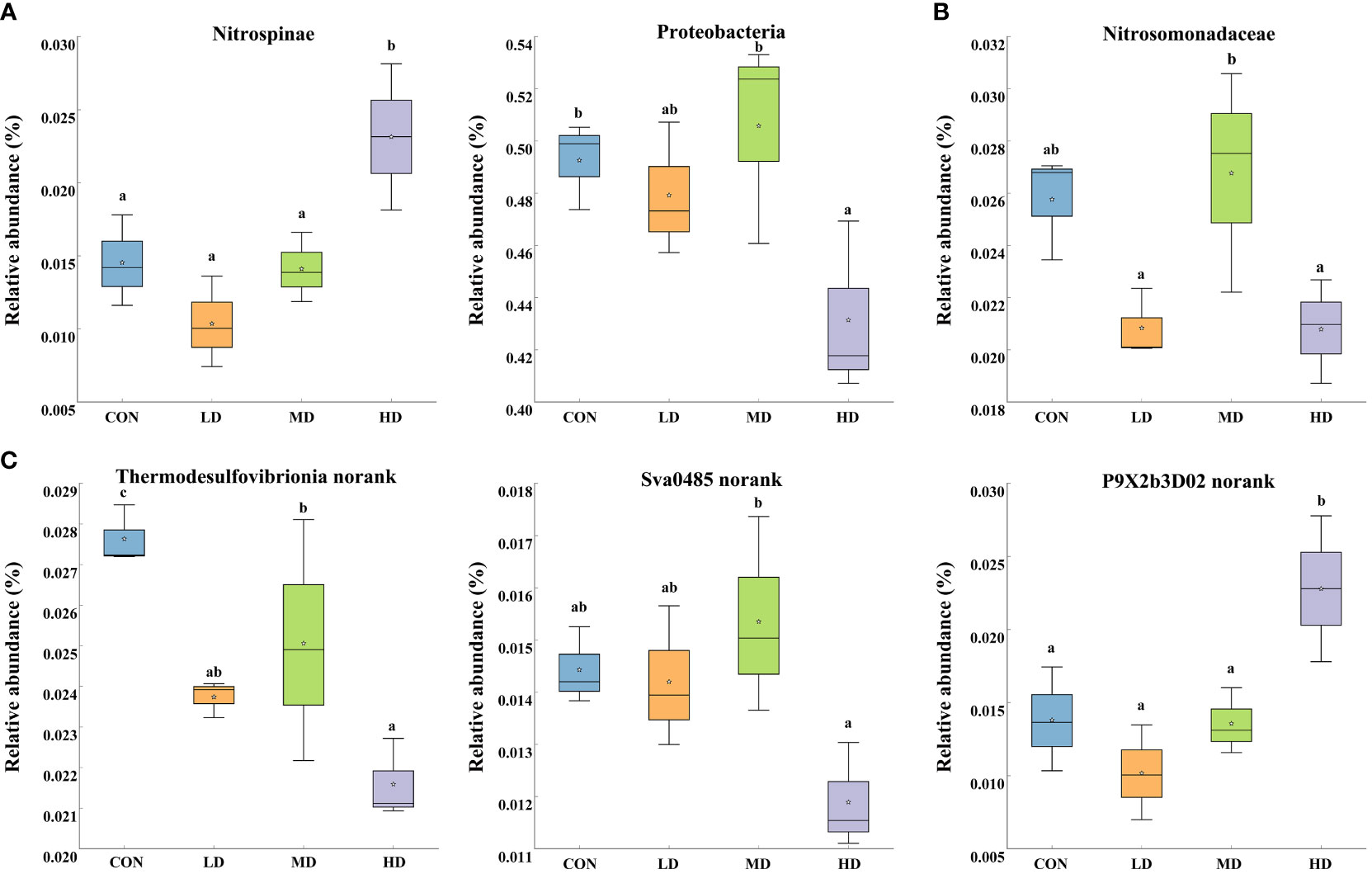
Figure 5 Relative abundances of different taxa in the ten most abundant phyla (A), families (B), and genera (C) among the CON, LD, MD and HD groups, respectively. Different lowercase letters represent significant differences between the CON, LD, MD, and HD groups (P < 0.05).
3.4 Bacterial community diversity in sediment
Bacterial community alpha diversity, including Chao1, Shannon, and Simpson indices, in the sediment of the CON, LD, MD, and HD groups are shown in Figure 6. A significantly lower Shannon index was observed in the MD group than in the other three groups (P < 0.05). The LD and MD groups had the highest and lowest Chao1 index, respectively (P < 0.05). However, no significant differences in the Simpson index were found among the four groups (P > 0.05).
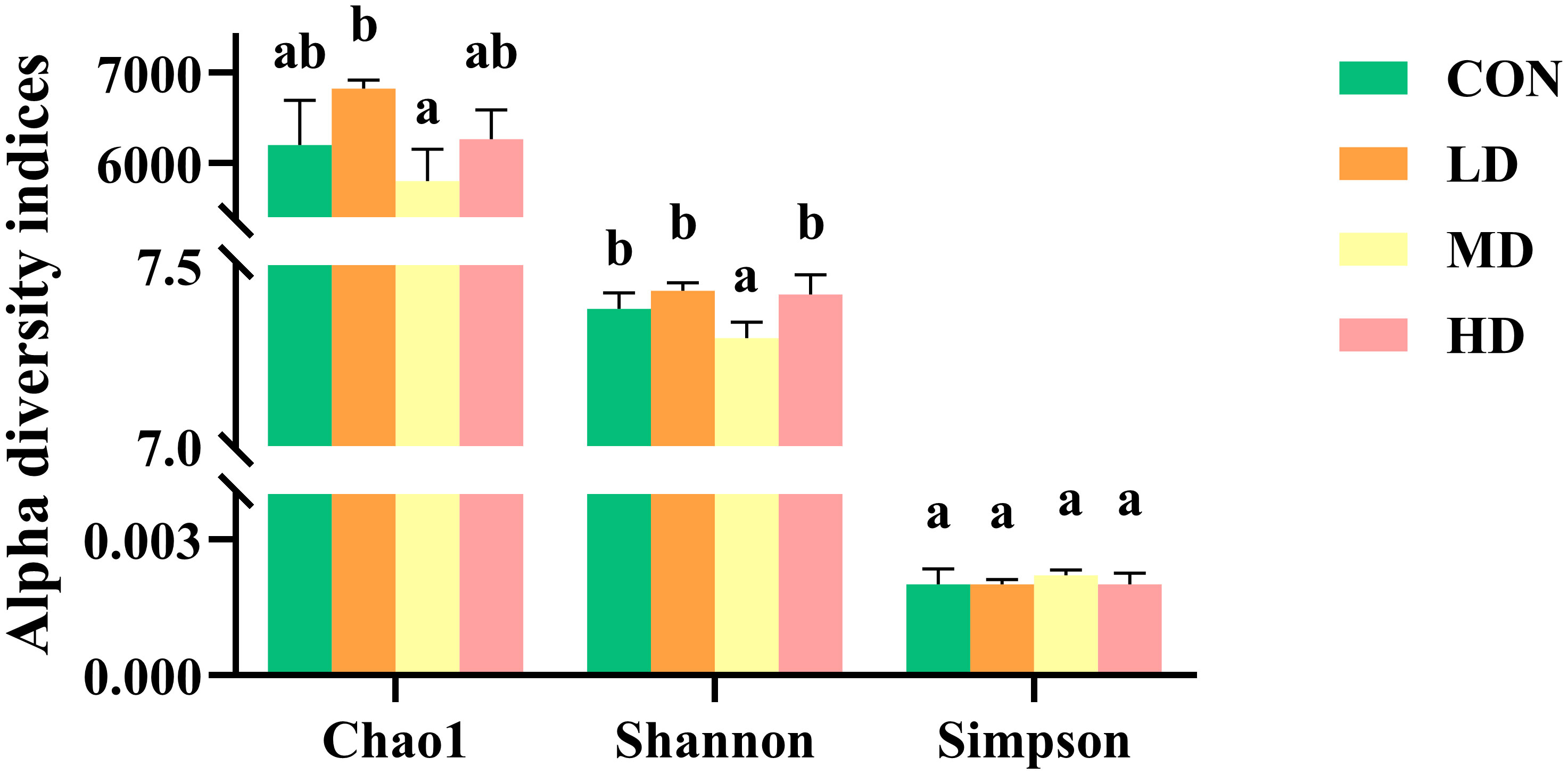
Figure 6 Bacterial community alpha diversity indices, including Chao1, Shannon, and Simpson in sediment of the CON, LD, MD, and HD groups. Different lowercase letters in the same column represent significant differences between the CON, LD, MD, and HD groups (P < 0.05).
Principal component analysis (PCA) was used to investigate the differences in benthic bacterial communities among the CON, LD, MD, and HD groups, based on the Bray-Curtis distance. As illustrated in Figure 7, the PC1 axis explained 39.154% of the difference in the bacterial community composition, and the PC2 axis explained 15.118%. The bacterial communities of all sediment samples were categorised into four groups, as revealed by the PCA results. Visible differences in bacterial communities among the CON, LD, MD, and HD groups were observed at the end of the experiment.
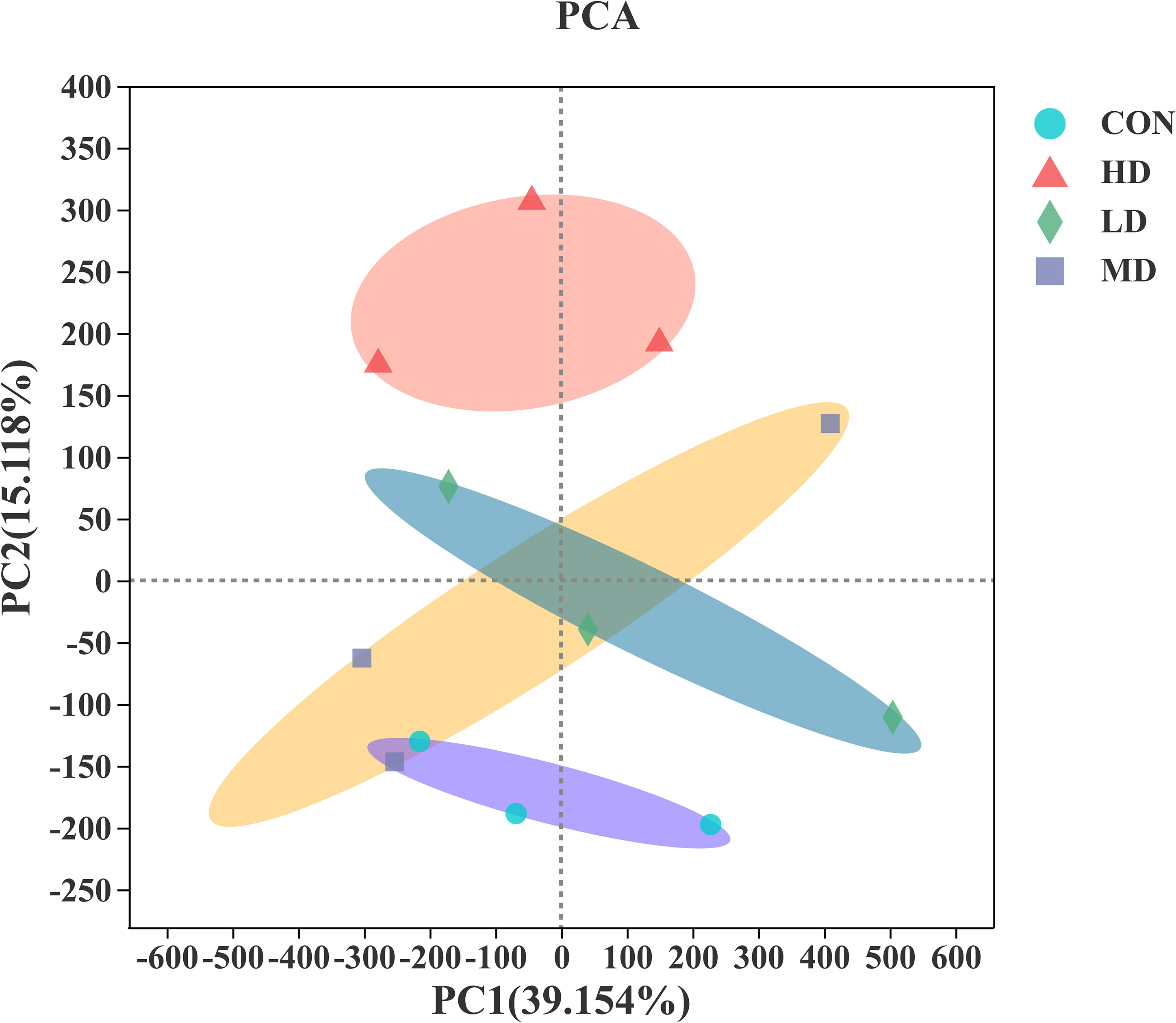
Figure 7 Principal component analysis (PCA) of the results based on the Bray-Curtis distance over all samples.
3.5 Correlation analysis between bacterial community and environmental factors
RDA analysis in the sediment of the CON,LD,MD and HD groups are shown in Figure 8. RDA1 and RDA2 accounted for 97.44% and 2.41% of the community changes, respectively. Based on the RDA, relationships between bacteria and each environmental factor were as follows: ammonia (r2=0.258, P=0.251), nitrate (r2=0.080, P=0.702), nitrite (r2=0.166, P=0.457), TN (r2=0.568, P=0.02), EN (r2=0.339, P=0.149), FA (r2=0.217, P=0.329). The above results showed that, TN had significant correlation with the bacterial community structure (P < 0.05), while other environmental factors had no significant correlation with the bacterial community structure (P > 0.05). Specifically, at the phylum level, TN was negatively correlated with Proteobacteria and Nitrospinae. At the family level, TN was negatively correlated with Nitrosomonadaceae. At the genus level, TN was negatively correlated with Thermodesulfovibrionia_norank and P9X2b3D02_norank.
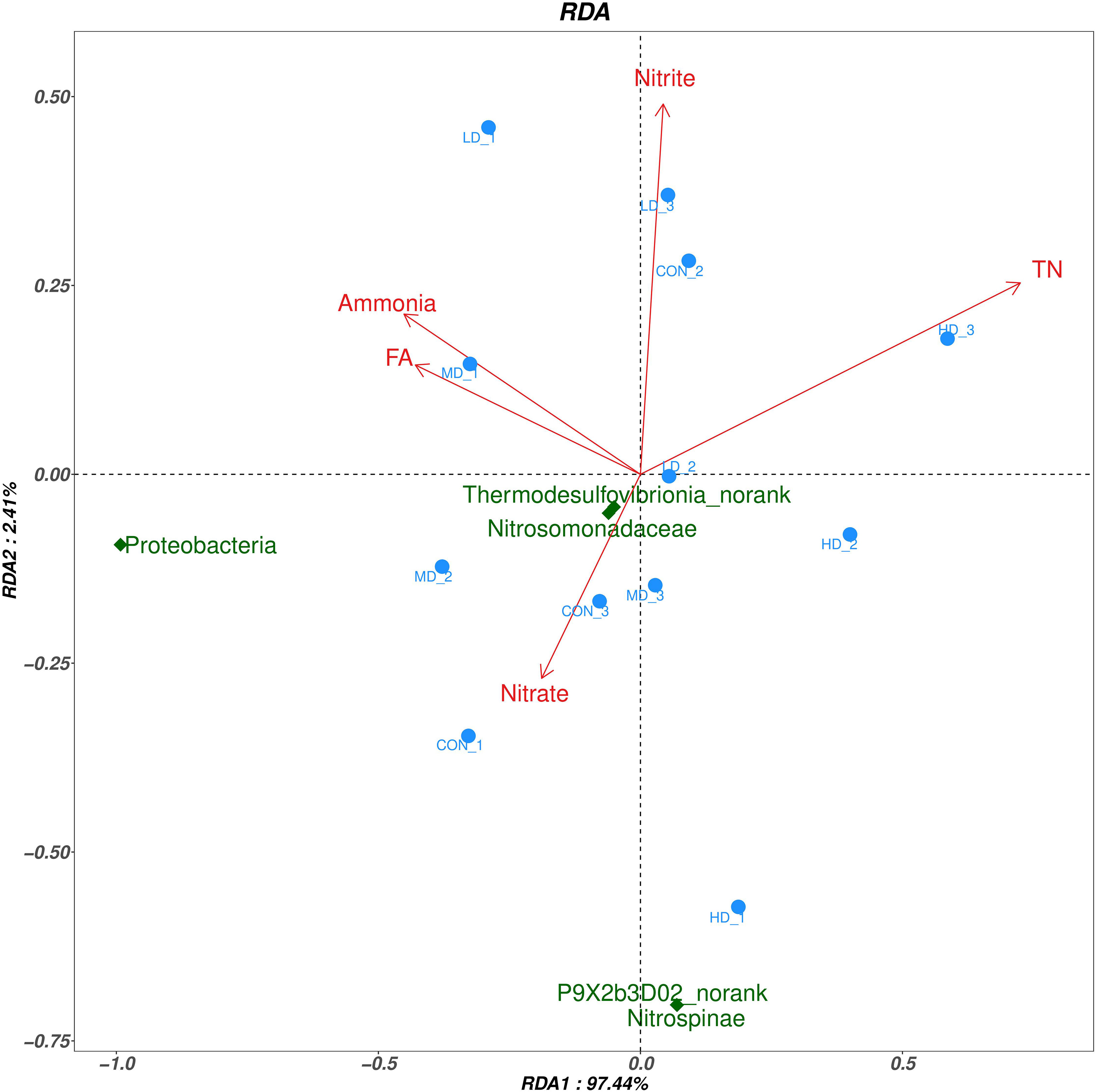
Figure 8 Redundancy analysis (RDA) to reveal the correlations between environmental factors and the differential bacterial taxa.
3.6 Bacterial community function prediction in sediment
Based on the functional annotation of prokaryotic taxa (FAPROTAX) database, we found 61 functional groups in total, twelve of which were related to the nitrogen cycle: aerobic ammonia oxidation, aerobic nitrite oxidation, nitrification, nitrate denitrification, nitrite denitrification, nitrous oxide denitrification, denitrification, nitrogen fixation, nitrite respiration, nitrate respiration, nitrate reduction, and nitrogen respiration (Figure 9). Among the nitrogen cycling pathways mentioned above, two significantly different functional groups were observed in the CON, LD, MD, and HD groups (Figure 10). The MD group showed the highest aerobic ammonia oxidation (P < 0.05). Aerobic nitrite oxidation was also significantly higher in the MD group than in the other three groups (P < 0.05).
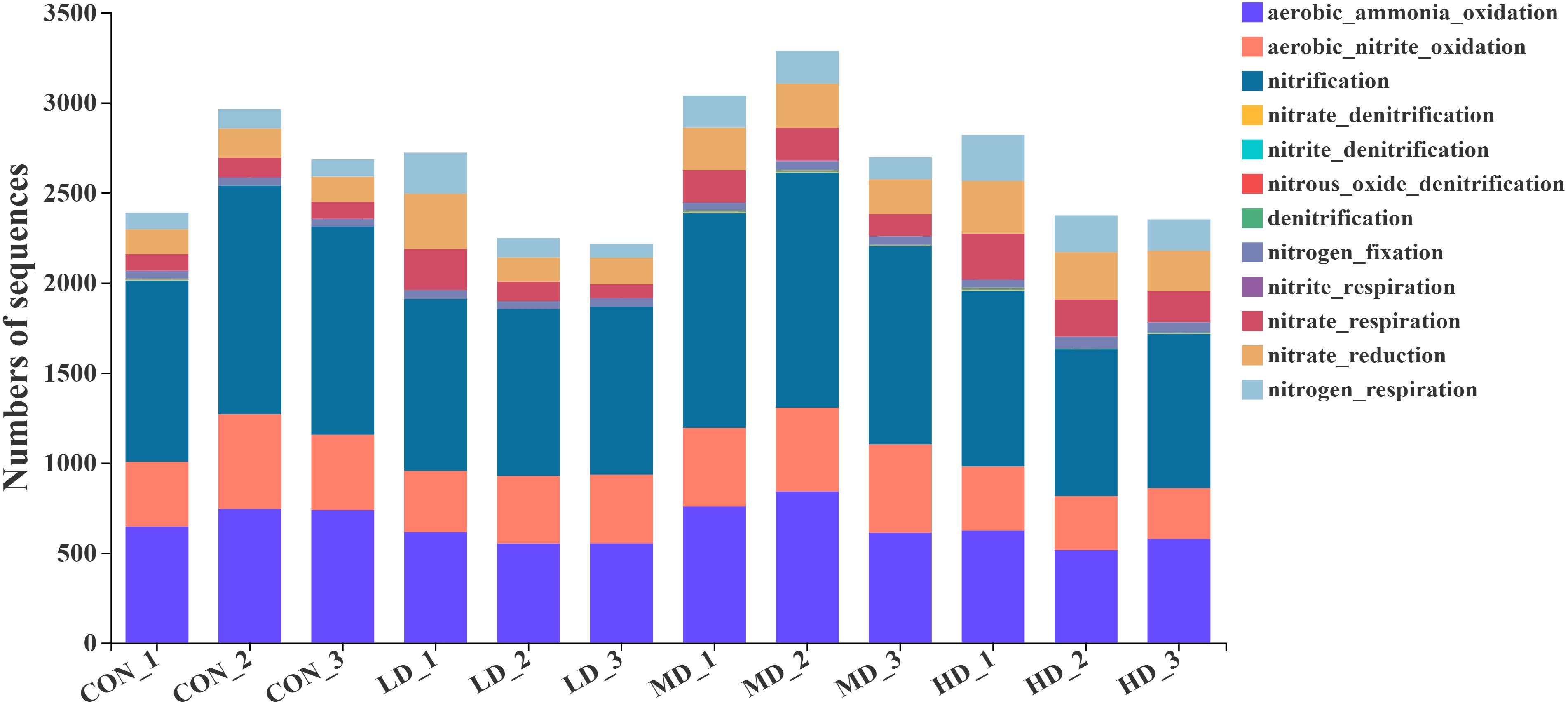
Figure 9 Functional groups related to nitrogen cycling in sediments of the CON, LD, MD and HD groups, respectively.
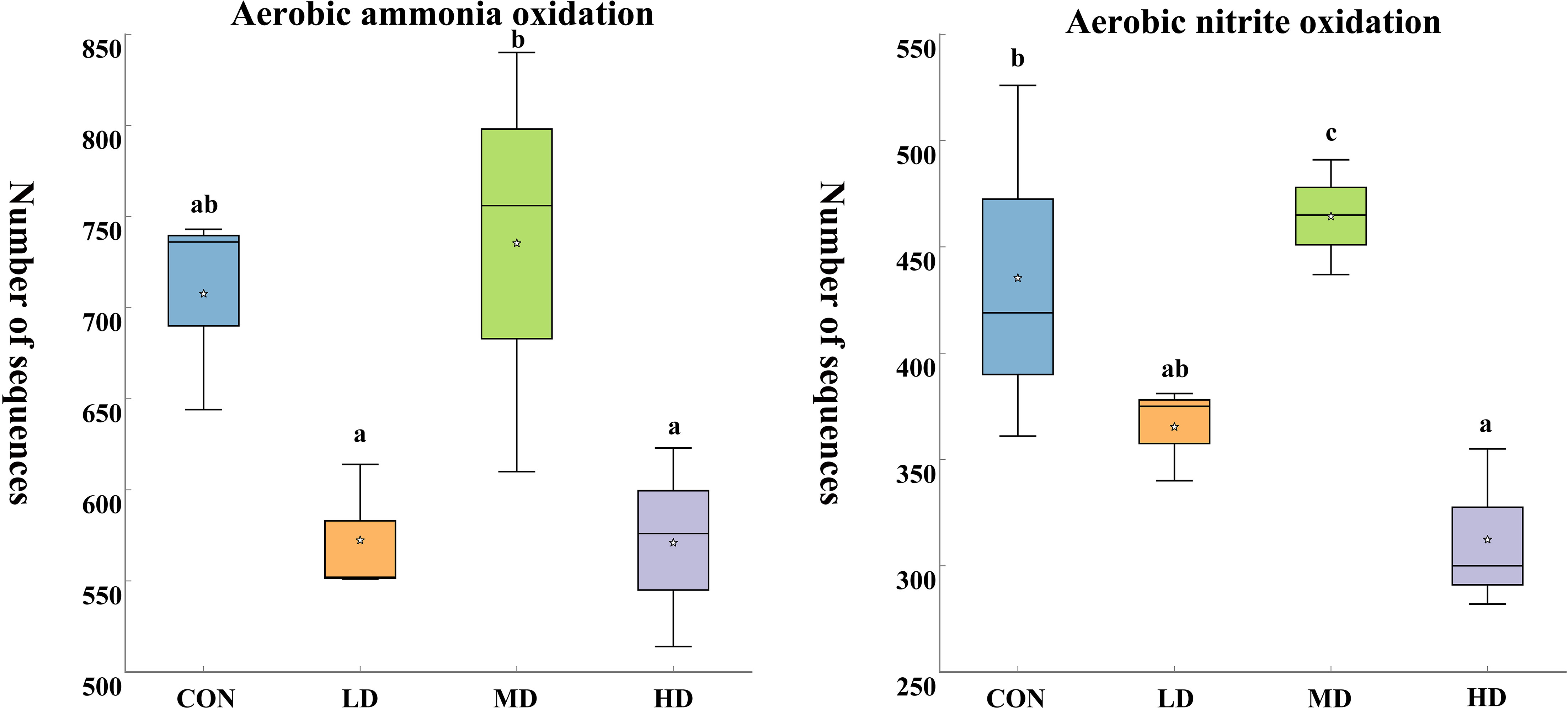
Figure 10 Significantly different functional groups related to nitrogen cycling in sediments of the CON, LD, MD, and HD groups at the end of the experiment. Different lowercase letters represent significant differences between the CON, LD, MD, and HD groups (P < 0.05).
4 Discussion
4.1 Nitrogen form and enzyme activity in sediments
Extracellular enzyme activities, such as ammonia monooxygenase, nitrate reductase, and hydroxylamine reductase, play an important role in the migration and transformation of nitrogen, by participating in nitrification and denitrification (Sun et al., 2020; Song et al., 2020; Zhao et al., 2021; Hong et al., 2019). Ammonia monooxygenase, which oxidises ammonia to hydroxylamine, is involved in the first step of nitrification (Aakra et al., 2001; Shinozaki & Fukui, 2002; Lin et al., 2015). Nitrate reductase and hydroxylamine reductase contribute to denitrification, nitrate reductase reduces nitrate ions to nitrite ions, and hydroxylamine reductase reduces hydroxylamine to ammonia (Jansson et al., 2008; Boutrin et al., 2012). In this study, the ammonia monooxygenase activity in the sediment was significantly increased with B. purificata bioturbation, whereas the nitrate reductase and hydroxylamine reductase activities were significantly decreased. This may indicate that B. purificata bioturbation could enhance the nitrification process and weaken denitrification in the sediment by affecting the related extracellular enzyme activities.
In addition, the results of nitrogen occurrence forms in this experiment were consistent with the results of extracellular enzyme activities discussed above. Nitrification refers to a reaction process in which ammonia nitrogen is oxidised to nitrite, which is further oxidised to form nitrate. The significantly higher nitrate content in the sediment of the MD group indicates the promotion of nitrification by B. purificata. Moreover, previous studies have reported that macrobenthos, including snails, can effectively promote degradation of organic matter in sediments (He et al., 2019; Hou et al., 2021; Blackburne et al., 2007). Organic nitrogen is degraded by ammonification to produce ammonia, which is the initial substrate for nitrification (Cui et al., 2019; Hu et al., 2021; Hou et al., 2021). Hence, the significantly decreased TN content in the sediment of the MD group may indicate that B. purificata promotes the transformation of total nitrogen to nitrate by enhancing nitrification and decomposition.
4.2 Bacterial community structure and functional prediction
Microorganisms are highly sensitive to changes in the ecological environment, and bacterial diversity is closely related to environmental changes (Escalas et al., 2017; Crowther et al., 2019; Sokol et al., 2022). Nitrogen transformation is closely associated with soil microorganisms (Yang et al., 2015; Pu et al., 2020; Shi et al., 2020). In this study, differences in PCA, phylum, family, and genus among the four groups revealed large impacts of B. purificata on the bacterial communities. Nitrospinae and Nitrosomonadaceae were significantly enriched in the HD and MD groups, respectively (P < 0.05). Previous studies have shown that members of the phylum Nitrospinae are the most abundant and widely known nitrite oxidizing bacteria (NOB), and are distributed in oceans as well as in freshwater bodies (Black and Just, 2018; Duan et al., 2019; Mueller et al., 2021; Liu et al., 2021). NOB are key players in the global nitrogen and carbon cycle, and can cooperate with microorganisms to improve the nitrification capacity of the system and enhance the degradation of ammonia nitrogen (Spieck et al., 2014; Pachiadaki et al., 2017). Nitrosomonadaceae belong to the most abundant ammonia-oxidising bacteria (AOB) and ammonia-oxidising archaea (AOA) at the family level (Straka et al., 2019; Louca et al., 2016). Functionally, Nitrosomonadaceae results in nitrogen removal after aeration and contributes to ammonia removal (Clark et al., 2021; Wong et al., 2021; Li J et al., 2022; Zhang et al., 2022). Hence, significantly higher Nitrosomonadaceae in the MD group indicates a strong ability to oxidise ammonia to nitrite (Wang et al., 2021; Morse et al., 2018), and significantly higher Nitrospinae in the HD group indicates an enhanced nitrite oxidation ability (Sorokin et al., 2014). The bacterial communities in the sediment influenced by B. purificata demonstrated a stronger ability to remove ammonia and nitrogen. Significantly negative correlations between TN and Nitrospinae and Nitrosomonadaceae also supported this discussion.
In addition, the results of the FAPROTAX function prediction in the present study were consistent with the discussion above. Aerobic ammonia oxidation and aerobic nitrite oxidation were significantly enriched in the MD group. Aerobic ammonia oxidation and aerobic nitrite oxidation are key functions of nitrification (Koch et al., 2014; Beman et al., 2021; Wu et al., 2022). Studies have shown that the potential application of these two microbial processes in wastewater treatment is of great significance (Chen et al., 2005; Rabaey et al., 2008). Specifically, aerobic ammonia oxidation converts NH4+ to NO2- under aerobic conditions (Xia et al., 2013; Trenerry et al., 2021). Aerobic nitrite oxidation converts NO2- to NO3- under aerobic conditions (Xu et al., 2011; Ning et al., 2016). Their activities directly or indirectly affect sediment nutrient cycling, water eutrophication, greenhouse gases, and ecosystem functions (Oguz et al., 2007; Schlicht et al., 2019). This explains the higher nitrification capacity in the MD and HD groups than that in the CON group. The analysis of bacterial community composition and the FAPROTAX function prediction showed that the bioturbation caused by B. purificata was beneficial for the enrichment of aerobic nitrification bacteria and promoted the nitrification process.
Besides, oxygen is crucial for nitrification (Fan et al., 2021; Rodríguez-Gómez et al., 2021; Zhu et al., 2021), as it is carried out under aerobic conditions, with oxygen as the electron acceptor and nitrogen as the electron donor (Daims et al., 2015; van Kessel et al., 2015). Previous studies have shown that macrobenthos bioturbation can affect the distribution of oxygen in sediments through bioturbation (Satoh and Okabe, 2013). During macrobenthos bioturbation processes, upper water with high dissolved oxygen is pumped into the sediments, which can increase the dissolved oxygen content and improve the redox potential in the sediment and then affect the bacterial activities and promote the nitrogen, phosphorus, and sulfur cycles at the sediment-water interface (Shull et al., 2009; Nguyen et al., 2022; Zhang et al., 2022). Meanwhile, extracellular enzyme activities in sediments are closely related to bacterial communities in sediments (Rietl et al., 2016). Therefore, in this study, significantly enhanced nitrification was reflected by the changes of extracellular enzyme activity and bacterial community structure. This may be a bioturbation from B. purificata.
5 Conclusion
In conclusion, B. purificata could significantly alter the bacterial community composition and functional groups related to nitrification and increase the hydroxylamine oxidase and ammonia monooxygenase activities, thus enhancing the nitrification process in sediment and promoting the transformation of total nitrogen to nitrate. Moreover, B. purificata stocked at 30 and 60 individuals/tank appears to have a better promotion effect on the bacterial community and forms of nitrogen occurrence. Hence, co-culturing B. purificata at suitable stocking densities may be a feasible and effective ecological restoration method to alleviate excess nitrogen and reduce water eutrophication.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author contributions
YZ: Methodology, formal analysis, data curation, and original draft preparation. YH: Conceptualisation, methodology, formal analysis, and original draft preparation. RJ and BL: Visualisation and investigation. JZ and XG: Resources, writing, reviewing, editing, and supervision. All authors contributed to the article and approved the submitted version.
Funding
This work was financially supported by the China Agriculture Research System of MOF and MARA (grant No. CARS-45), the National Natural Science Foundation of China (grant No. 31802302), and the Central Public-interest Scientific Institution Basal Research Fund, CAFS (grant No. 2022XT0504).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aakra A., Utåker J. B., Nes I. F. (2001). Comparative phylogeny of the ammonia monooxygenase subunit a and 16S rRNA genes of ammonia-oxidizing bacteria. FEMS Microbiol. Lett. 205 (2), 237–242. doi: 10.1111/j.1574-6968.2001.tb10954.x
Beman J. M., Vargas S. M., Wilson J. M., Perez-Coronel E., Karolewski J. S., Vazquez S., et al. (2021). Substantial oxygen consumption by aerobic nitrite oxidation in oceanic oxygen minimum zones. Nat. Commun. 12 (1), 7043. doi: 10.1038/s41467-021-27381-7
Blackburne R., Vadivelu V. M., Yuan Z., Keller J. (2007). Kinetic characterisation of an enriched nitrospira culture with comparison to nitrobacter. Water Res. 41 (14), 3033–3042. doi: 10.1016/j.watres.2007.01.043
Black E. M., Just C. L. (2018). The genomic potentials of NOB and comammox nitrospira in river sediment are impacted by native freshwater mussels. Front. Microbiol. 9, 2061. doi: 10.3389/fmicb.2018.02061
Boutrin M. C., Wang C., Aruni W., Li X., Fletcher H. M. (2012). Nitric oxide stress resistance in Porphyromonas gingivalis is mediated by a putative hydroxylamine reductase. Bacteriol 194 (6), 1582–1592. doi: 10.1128/JB.06457-11
Brodrick S., Cullen P., Maher W. (1987). Determination of exchangeable inorganic nitrogen species in wetlands soils. Bull. Environ. Contam Toxicol. 38 (3), 377–380. doi: 10.1007/BF01606601
Cao J., Zhang T., Wu Y., Sun Y., Zhang Y., Huang B., et al. (2020). Correlations of nitrogen removal and core functional genera in full-scale wastewater treatment plants: Influences of different treatment processes and influent characteristics. Bioresour Technol. 297, 122455. doi: 10.1016/j.biortech.2019.122455
Chen Y. C., Lan H. X., Zhan H. Y., Fu S. Y. (2005). Simultaneous anaerobic-aerobic biodegradation of halogenated phenolic compound under oxygen-limited conditions. Environ. Sci. (China) 17 (5), 873–875.
Chen J., Song G. T., Wang X., She L., Wu S., Wu M., et al. (2012). The efficiency of water purification by different densities of bellamya aeruginosa and Bellamya purificata. J. Anhui Agric. 40, 11708–11709. doi: 10.13989/j.cnki.0517-6611.2012.23.011
Clark I. M., Hughes D. J., Fu Q., Abadie M., Hirsch P. R. (2021). Metagenomic approaches reveal differences in genetic diversity and relative abundance of nitrifying bacteria and archaea in contrasting soils. Sci. Rep. 11 (1), 15905. doi: 10.1038/s41598-021-95100-9
Crowther T. W., van den Hoogen J., Wan J., Mayes M. A., Keiser A. D., Mo L., et al. (2019). The global soil community and its influence on biogeochemistry. Science 365 (6455), 1–10. doi: 10.1126/science.aav0550
Cui P., Liao H., Bai Y., Li X., Zhao Q., Chen Z., et al. (2019). Hyperthermophilic composting reduces nitrogen loss via inhibiting ammonifiers and enhancing nitrogenous humic substance formation. Sci. Total Environ. 692, 98–106. doi: 10.1016/j.scitotenv.2019.07.239
Daims H., Lebedeva E. V., Pjevac P., Han P., Herbold C., Albertsen M., et al. (2015). Complete nitrification by nitrospira bacteria. Nature 528 (7583), 504–509. doi: 10.1038/nature16461
Decaestecker E., Gaba S., Raeymaekers J. A., Stoks R., Van Kerckhoven L., Ebert D., et al. (2007). Host-parasite ‘red queen’ dynamics archived in pond sediment. Nature 450 (7171), 870–873. doi: 10.1038/nature06291
Dong L. F., Smith C. J., Papaspyrou S., Stott A., Osborn A.M., Nedwell D.B., et al. (2009). Changes in benthic denitrification, nitrate ammonification, and anammox process rates and nitrate and nitrite reductase gene abundances along an estuarine nutrient gradient (The colne estuary, united kingdom). Appl. Environ. Microbiol. 75 (10), 3171–3179. doi: 10.1128/AEM.02511-08
Duan H., Ye L., Wang Q., Zheng M., Lu X., Wang Z., et al. (2019). Nitrite oxidizing bacteria (NOB) contained in influent deteriorate mainstream NOB suppression by side stream inactivation. Water Res. 162, 331–338. doi: 10.1016/j.watres.2019.07.002
Escalas A., Troussellier M., Yuan T., Bouvier T., Bouvier C., Mouchet M. A., et al. (2017). Functional diversity and redundancy across fish gut, sediment and water bacterial communities. Environ. Microbiol. 19 (8), 3268–3282. doi: 10.1111/1462-2920.13822
Fan Y., Chen X., Yao Z., Li H., Wang D., Tian M., et al. (2021). A novel inhibition mechanism of aniline on nitrification: Aniline degradation competes dissolved oxygen with nitrification. Sci. Total Environ. 770, 145205. doi: 10.1016/j.scitotenv.2021.145205
Feng J., Zhu X., Wu H., Ning C., Lin G. (2017). Distribution and ecological risk assessment of heavy metals in surface sediments of a typical restored mangrove-aquaculture wetland in shenzhen, China. Mar. pollut. Bull. 124 (2), 1033–1039. doi: 10.1016/j.marpolbul.2017.01.004
Ge X. (2014). Progress in research and development of recirculating aquaculture technology for conventional freshwater fish. Sci. Fish Farming 08, 2–4. doi: 10.14184/j.cnki.issn1004-843x.2014.08.006
Gold A. C., Thompson S. P., Piehler M. F. (2017). Coastal stormwater wet pond sediment nitrogen dynamics. Sci. Total Environ. 609, 672–681. doi: 10.1016/j.scitotenv.2017.07.213
Guo C., Gao J., Zhou B., Yang J. (2021). Factors of the ecosystem service value in water conservation areas considering the natural environment and human activities: A case study of funiu mountain, China. Int. J. Environ. Res. Public Health 18 (21), 11074. doi: 10.3390/ijerph182111074
He Y., Men B., Yang X., Li Y., Xu H., Wang D. (2019). Relationship between heavy metals and dissolved organic matter released from sediment by bioturbation. J. Environ. Sci. (China) 75, 216–223. doi: 10.1016/j.jes.2018.03.031
Herbeck L. S., Krumme U., Nordhaus I., Jennerjahn T. C. (2021). Pond aquaculture effluents feed an anthropogenic nitrogen loop in a SE Asian estuary. Sci. Total Environ. 756, 144083. doi: 10.1016/j.scitotenv.2020.144083
Hong Y., Wu J., Guan F., Yue W., Long A. (2019). Nitrogen removal in the sediments of the pearl river estuary, China: Evidence from the distribution and forms of nitrogen in the sediment cores. Mar. pollut. Bull. 138, 115–124. doi: 10.1016/j.marpolbul.2018.11.040
Hou Y., Li B., Feng G., Zhang C., He J., Li H., et al. (2021). Responses of bacterial communities and organic matter degradation in surface sediment to Macrobrachium nipponense bioturbation. Sci. Total Environ. 759, 143534. doi: 10.1016/j.scitotenv.2020.143534
Hou Y., Li B., Luo J., Zhang C., He J., Zhu J., et al. (2021). Effect of Bellamya purificata on organic matter degradation in surface sediment as revealed by amino acids. Aquacult Environ. Interact. 13, 1–12. doi: 10.3354/aei00382
Huang S., Jiang H., Zhang L., Gu Q., Wang W., Wen Y., et al. (2021). Integrated proteomic and transcriptomic analysis reveals that polymorphic shell colors vary with melanin synthesis in Bellamya purificata snail. J. Proteomics. 230, 103950. doi: 10.1016/j.jprot.2020.103950
Hu J., Zhao Y., Yao X., Wang J., Zheng P., Xi C., et al. (2021). Dominance of comammox nitrospira in soil nitrification. Sci. Total Environ. 780, 146558. doi: 10.1016/j.scitotenv.2021.146558
Jansson E. A., Huang L., Malkey R., Govoni M., Nihlen C., Olsson A., et al. (2008). A mammalian functional nitrate reductase that regulates nitrite and nitric oxide homeostasis. Nat. Chem. Biol. 4 (7), 411–417. doi: 10.1038/nchembio.92
Kendüzler E., Türker A. R. (2005). Determination of trace cadmium in waters by flame atomic absorption spectrophotometry after preconcentration with 1-nitroso-2-naphthol-3,6-disulfonic acid on ambersorb 572. Ann. Chim. 95 (1-2), 77–85. doi: 10.1002/adic.200590009
Koch H., Galushko A., Albertsen M., Schintlmeister A., Gruber-Dorninger C., Lücker S., et al. (2014). Growth of nitrite-oxidizing bacteria by aerobic hydrogen oxidation. Science 345 (6200), 1052–1054. doi: 10.1126/science.1256985
Kołacińska K., Koncki R. (2014). A novel optoelectronic detector and improved flow analysis procedure for ammonia determination with nessler’s reagent. Anal. Sci. 30 (10), 1019–1022. doi: 10.2116/analsci.30.1019
Kuypers M. M. M., Marchant H. K., Kartal B. (2018). The microbial nitrogen-cycling network. Nat. Rev. Microbiol. 16 (5), 263–276. doi: 10.1038/nrmicro.2018.9
Liang L., Wang Y. Y., Yao J. J. (2013). Analysis and evaluation on nutritional composition of three freshwater snails in jinjiang river. Sci. Guizhou Agric. 41, 131–133.
Li S., Guo Z. X., Yang H. Y., Yang Z.Y. (2009). Evaluation of cultural service value of aquaculture pond ecosystem: a case study in a water conservation area of shanghai. Ying Yong Sheng Tai Xue Bao. 20 (12), 3075–3083.
Li J., Nedwell D. B., Beddow J., Dumbrell A. J., McKew B. A., Thorpe E. L., et al. (2015). amoA gene abundances and nitrification potential rates suggest that benthic ammonia-oxidizing bacteria and not archaea dominate n cycling in the colne estuary, united kingdom. Appl. Environ. Microbiol. 81 (1), 159–165. doi: 10.1128/AEM.02654-14
Lin J. J., Zhang S., Yang Z. Y., He L. P., Zhou N., Zhang Q. R., et al. (2015). Effect of drought and subsequent re-wetting cycles on transferable nitrogen and its form distribution in the sediment of water level fluctuating zone in the tributary of three gorge reservoir areas. Huan Jing Ke Xue. 36 (7), 2459–2464. doi: 10.13227/j.hjkx.2015.07.018
Liu S., Cai H., Wang J., Wang H., Zheng T., Chen Q., et al. (2021). In-situ expressions of comammox nitrospira along the Yangtze river. Water Res. 200, 117241. doi: 10.1016/j.watres.2021.117241
Liu X. G., Wang J., Wu Z. F., Cheng G. F., Gu Z. J. (2021). Anaerobic ammonium oxidation bacteria in a freshwater recirculating pond aquaculture system. Int. J. Environ. Res. Public Health 18 (9), 4941. doi: 10.3390/ijerph18094941
Li Y., Wang M., Teng K., Dong D., Liu Z., Zhang T., et al. (2022). Transcriptome profiling revealed candidate genes, pathways and transcription factors related to nitrogen utilization and excessive nitrogen stress in perennial ryegrass. Sci. Rep. 12 (1), 3353. doi: 10.1038/s41598-022-07329-7
Li J., Zeng W., Liu H., Zhan M., Miao H. (2022). Achieving deep autotrophic nitrogen removal in aerated biofilter driven by sponge iron: Performance and mechanism. Environ. Res. 213, 113653. doi: 10.1016/j.envres.2022.113653
Löfgren S., Grandin U., Stendera S. (2014). Long-term effects on nitrogen and benthic fauna of extreme weather events: Examples from two Swedish headwater streams. Ambio 43 Suppl 1, 58–76. doi: 10.1007/s13280-014-0562-3
Louca S., Parfrey L. W., Doebeli M. (2016). Decoupling function and taxonomy in the global ocean microbiome. Science 353 (6305), 1272–1277. doi: 10.1126/science.aaf4507
McCarthy I. D., Sharafi A Oxby C. B., Oxby C. B, Burkinshaw L. (1980). The accuracy of total body nitrogen determined by neutron activation analysis. Phys. Med. Biol. 25 (5), 849–863. doi: 10.1088/0031-9155/25/5/003
Meng F. G., Shi H. Z., Wang J., Liu H., Guo Q. S., Gong F. L., et al. (2013). Correlation analysis between nutritive components of Whitmania pigra and bellamya purificata. Zhongguo Zhong YaoZa Zhi 38 (12), 1915–1918.
Modenutti B. E., Pérez G. L. (2001). Planktonic ciliates from an oligotrophic south Andean lake, morenito lake (Patagonia, Argentina). Braz. J. Biol. 61 (3), 389–395. doi: 10.1590/S1519-69842001000300007
Morse N., Payne E., Henry R., Hatt B., Chandrasena G., Shapleigh J., et al. (2018). Plant-microbe interactions drive denitrification rates, dissolved nitrogen removal, and the abundance of denitrification genes in stormwater control measures. Environ. Sci. Technol. 52 (16), 9320–9329. doi: 10.1021/acs.est.8b02133
Mueller A. J., Jung M. Y., Strachan C. R., Herbold C. W., Kirkegaard R. H., Wagner M., et al. (2021). Genomic and kinetic analysis of novel nitrospinae enriched by cell sorting. ISME J. 15 (3), 732–745. doi: 10.1038/s41396-020-00809-6
Nguyen T. P., Koyama M., Nakasaki K. (2022). Effects of oxygen supply rate on organic matter decomposition and microbial communities during composting in a controlled lab-scale composting system. Waste Manage. 153, 275–282. doi: 10.1016/j.wasman.2022.09.004
Ning X. S., Wang M. M., Yao C. Z., Chen X. M., Kang Y. B. (2016). Tert-butyl nitrite: Organic redox cocatalyst for aerobic aldehyde-selective Wacker-tsuji oxidation. Org Lett. 18 (11), 2700–2703. doi: 10.1021/acs.orglett.6b01165
Oguz M. T., Robinson K. G., Layton A. C., Sayler G. S. (2007). Concurrent nitrite oxidation and aerobic denitrification in activated sludge exposed to volatile fatty acids. Biotechnol. Bioeng. 97 (6), 1562–1572. doi: 10.1002/bit.21379
Pachiadaki M. G., Sintes E., Bergauer K., Brown J. M., Record N. R., Swan B., et al. (2017). Major role of nitrite-oxidizing bacteria in dark ocean carbon fixation. Science 358 (6366), 1046–1051. doi: 10.1126/science.aan8260
Parro V., Puente-Sánchez F., Cabrol N. A., Gallardo-Carreño I., Moreno-Paz M., Blanco Y., et al. (2019). Microbiology and nitrogen cycle in the benthic sediments of a glacial oligotrophic deep Andean lake as analog of ancient martian lake-beds. Front. Microbiol. 10, 929. doi: 10.3389/fmicb.2019.00929
Pozzobon V., Cui N., Moreaud A., Michiels E., Levasseur W. (2021). Nitrate and nitrite as mixed source of nitrogen for chlorella vulgaris: Growth, nitrogen uptake and pigment contents. Bioresour Technol. 330, 124995. doi: 10.1016/j.biortech.2021.124995
Pu J. H., Jiang N., Juan Y. H., Chen L. J. (2020). Effects of freeze-thaw on dissolved nitrogen pool, nitrogen transformation processes and diversity of bacterial community in temperate soils. Ying Yong Sheng Tai Xue Bao. 31 (9), 2893–2902. doi: 10.13287/j.1001-9332.202009.033
Rabaey K., Read S. T., Clauwaert P., Freguia S., Bond P. L., Blackall L. L., et al. (2008). Cathodic oxygen reduction catalyzed by bacteria in microbial fuel cells. ISME J. 2 (5), 519–527. doi: 10.1038/ismej.2008.1
Rietl A. J., Overlander M. E., Nyman A. J., Jackson C. R. (2016). Microbial community composition and extracellular enzyme activities associated with juncus roemerianus and spartina alterniflora vegetated sediments in Louisiana saltmarshes. Microb. Ecol. 71 (2), 290–303. doi: 10.1007/s00248-015-0651-2
Rodríguez-Gómez L. E., Rodríguez-Sevilla J., Hernández A., Álvarez M. (2021). Factors affecting nitrification with nitrite accumulation in treated waste water by oxygen injection. Environ. Technol. 42 (5), 813–825. doi: 10.1080/09593330.2019.1645742
Satoh H., Okabe S. (2013). Spatial and temporal oxygen dynamics in macrofaunal burrows in sediments: a review of analytical tools and observational evidence. Microbes Environ. 28 (2), 166–179. doi: 10.1264/jsme2.ME12182
Saxena G., Mitra S., Marzinelli E. M., Xie C., Wei T. J., Steinberg P. D., et al. (2018). Metagenomics reveals the influence of land use and rain on the benthic microbial communities in a tropical urban waterway. Msystems 3 (3), e00136–e00117. doi: 10.1128/mSystems.00136-17
Schlicht K., Krattenmacher N., Lugert V., Schulz C., Thaller G., Tetens J. (2019). Estimation of genetic parameters for growth and carcass traits in turbot (Scophthalmus maximus). Arch. Anim. Breed. 62 (1), 265–273. doi: 10.5194/aab-62-265-2019
Shinozaki H., Fukui M. (2002). Comparison of 16S rRNA, ammonia monooxygenase subunit a and hydroxylamine oxidoreductase gene, in chemolithotrophic ammonia-oxidizing bacteria. J. Gen. Appl. Microbiol. 48 (3), 173–176. doi: 10.2323/jgam.48.173
Shi Y., Zhang X., Wang Z., Xu Z., He C., Sheng L., et al. (2020). Shift in nitrogen transformation in peatland soil by nitrogen inputs. Sci. Total Environ. 764, 142924. doi: 10.1016/j.scitotenv.2020.142924
Shull D., Benoit J., Wojcik C., Senning J. (2009). Infaunal burrow ventilation and pore-water transport in muddy sediments. Estuarine Coast. Shelf Sci. 83, 277–286. doi: 10.1016/j.ecss.2009.04.005
Silva J. A., Bremner J. M. (1966). Determination and isotope-ratio analysis of different forms of nitrogen in soil: 5. fixed ammonium. Soil Sci. Soc. Am. J. 30 (5), 587–594. doi: 10.2136/sssaj1966.03615995003000050017x
Sokol N. W., Slessarev E., Marschmann G. L., Nicolas A., Blazewicz S. J., Brodie E. L., et al. (2022). Life and death in the soil microbiome: how ecological processes influence biogeochemistry. Nat. Rev. Microbiol. 20 (7), 415–430. doi: 10.1038/s41579-022-00695-z
Song X., Razavi B. S., Ludwig B., Zamanian K., Zang H., Kuzyakov Y., et al. (2020). Combined biochar and nitrogen application stimulates enzyme activity and root plasticity. Sci. Total Environ. 735, 139393. doi: 10.1016/j.scitotenv.2020.139393
Sorokin D. Y., Vejmelkova D., Lücker S., Streshinskaya G. M., Rijpstra W. I. C., Sinninghe Damsté J. S., et al. (2014). Nitrolancea hollandica gen. nov., sp. nov., a chemolithoautotrophic nitrite-oxidizing bacterium isolated from a bioreactor belonging to the phylum Chloroflexi. Int. J. Syst. Evol. Microbiol. 64 (Pt 6), 1859–1865. doi: 10.1099/ijs.0.062232-0
Spieck E., Keuter S., Wenzel T., Bock E., Ludwig W. (2014). Characterization of a new marine nitrite oxidizing bacterium, Nitrospina watsonii sp. nov, a member of the newly proposed phylum “Nitrospinae. Syst. Appl. Microbiol. 37 (3), 170–176. doi: 10.1016/j.syapm.2013.12.005
Straka L. L., Meinhardt K. A., Bollmann A., Stahl D. A., Winkler M. H. (2019). Affinity informs environmental cooperation between ammonia-oxidizing archaea (AOA) and anaerobic ammonia-oxidizing (Anammox) bacteria. ISME J. 13 (8), 1997–2004. doi: 10.1038/s41396-019-0408-x
Sun W., Liu S. J., Feng J. C., Wang P. T., Ma D. Y., Xie Y., et al. (2020). Effects of water and nitrogen source types on soil enzyme activity and nitrogen utilization efficiency of wheat. Ying Yong Sheng Tai Xue Bao. 31 (8), 2583–2592. doi: 10.13287/j.1001-9332.202008.023
Trenerry M. J., Wallen C. M., Brown T. R., Park S. V, Berry J. F. (2021). Spontaneous N2 formation by a diruthenium complex enables electrocatalytic and aerobic oxidation of ammonia. Nat. Chem. 13 (12), 1221–1227. doi: 10.1038/s41557-021-00797-w
Tuikka A. I., Leppänen M. T., Akkanen J., Sormunen A. J., Leonards P. E., van Hattum B., et al. (2016). Predicting the bioaccumulation of polyaromatic hydrocarbons and polychlorinated biphenyls in benthic animals in sediments. Sci. Total Environ. 563-564, 396–404. doi: 10.1016/j.scitotenv.2016.04.110
van Kessel M. A., Speth D. R., Albertsen M., Nielsen P. H., Op den Camp H. J., Kartal B., et al. (2015). Complete nitrification by a single microorganism. Nature 528 (7583), 555–559. doi: 10.1038/nature16459
Wang J., Fu Z., Chen G., Zou G., Song X., Liu F. (2018). Runoff nitrogen (N) losses and related metabolism enzyme activities in paddy field under different nitrogen fertilizer levels. Environ. Sci. pollut. Res. Int. 25 (27), 27583–27593. doi: 10.1007/s11356-018-2823-3
Wang L., Qiu S., Guo J., Ge S. (2021). Light irradiation enables rapid start-up of nitritation through suppressing nxrB gene expression and stimulating ammonia-oxidizing bacteria. Environ. Sci. Technol. 55 (19), 13297–13305. doi: 10.1021/acs.est.1c04174
Wei H., Ting-Mei L., Lu-Lu C., Lu L., Lu-Ji Y., Zhao-Xu P. (2019). Effect of adding microorganism and carbon source to substrate on nitrogen removal treating the drainage of WWTP. Water Sci. Technol. 79 (10), 1947–1955. doi: 10.2166/wst.2019.192
Wong W. W., Greening C., Shelley G., Lappan R., Leung P. M., Kessler A., et al. (2021). Effects of drift algae accumulation and nitrate loading on nitrogen cycling in a eutrophic coastal sediment. Sci. Total Environ. 790, 147749. doi: 10.1016/j.scitotenv.2021.147749
Woodman S. G., Khoury S., Fournier R. E., Emilson E. J. S., Gunn J. M., Rusak J. A., et al. (2021). Forest defoliator outbreaks alter nutrient cycling in northern waters. Nat. Commun. 12 (1), 6355. doi: 10.1038/s41467-021-26666-1
Wu H., Cui M., Yang N., Liu Y., Wang X., Zhang L., et al. (2022). Aerobic biocathodes with potential regulation for ammonia oxidation with concomitant cathodic oxygen reduction and their microbial communities. Bioelectrochemistry 144, 107997. doi: 10.1016/j.bioelechem.2021.107997
Wu H., Liu J., Bi X., Lin G., Feng C. C., Li Z., et al. (2017). Trace metals in sediments and benthic animals from aquaculture ponds near a mangrove wetland in southern China. Mar. pollut. Bull. 117 (1-2), 486–491. doi: 10.1016/j.marpolbul.2017.01.026
Xiang Y., Shao Z., Chai H., Ji F., He Q. (2020). Functional microorganisms and enzymes related nitrogen cycle in the biofilm performing simultaneous nitrification and denitrification. Bioresour Technol. 314, 123697. doi: 10.1016/j.biortech.2020.123697
Xia X., Tokash J. C., Zhang F., Liang P., Huang X., et al (2013). Oxygen-reducing biocathodes operating with passive oxygen transfer in microbial fuel cells. Environ. Sci. Technol. 47 (4), 2085–2091. doi: 10.1021/es3027659
Xu D., Gu J., Li Y., Zhang Y., Howard A., Guan Y., et al. (2016). Purifying capability, enzyme activity, and nitrification potentials in December in integrated vertical flow constructed wetland with earthworms and different substrates. Environ. Sci. pollut. Res. Int. 23 (1), 273–281. doi: 10.1007/s11356-015-5734-6
Xu G., Xu X., Yang F., Liu S. (2011). Selective inhibition of nitrite oxidation by chlorate dosing in aerobic granules. J. Hazard Mater. 185 (1), 249–254. doi: 10.1016/j.jhazmat.2010.09.025
Yang S., Li X. B., Wang R. Z., Cai J. P., Xu Z. W., Zhang Y. G., et al. (2015). Effects of nitrogen and water addition on soil bacterial diversity and community structure in temperate grasslands in northern China. Ying Yong Sheng Tai Xue Bao. 26 (3), 739–746. doi: 10.13287/j.1001-9332.20150122.003
Yuan X. T., Yang H. S., Zhou Y., Mao Y. Z., Xu Q., Wang L. L., et al. (2008). Bioremediation potential of apostichopus japonicus (Selenka) in coastal bivalve suspension aquaculture system. Ying Yong Sheng Tai Xue Bao. 19 (4), 866–872. doi: 10.13287/j.1001-9332.2008.0222
Zhang D., Li M., Yang Y., Yu H., Xiao F., Mao C., et al. (2022). Nitrite and nitrate reduction drive sediment microbial nitrogen cycling in a eutrophic lake. Water Res. 220, 118637. doi: 10.1016/j.watres.2022.118637
Zhang W., Yu D., Zhang J., Miao Y., Zhao X., Ma G., et al. (2022). Start-up of mainstream anammox process through inoculating nitrification sludge and anammox biofilm: Shift in nitrogen transformation and microorganisms. Bioresour Technol. 347, 126728. doi: 10.1016/j.biortech.2022.126728
Zhang K., Zheng X., He Z., Yang T., Shu L., Xiao F., et al. (2020). Fish growth enhances microbial sulfur cycling in aquaculture pond sediments. Microb. Biotechnol. 13 (5), 1597–1610. doi: 10.1111/1751-7915.13622
Zhao F. (2014). Studies on the community structure and some ecological function of macrozoobenthos in pond (Wuhan: Huazhong Agricultural University). PhD dissertation. doi: 10.7666/d.Y2565768
Zhao Z., Shao S., Liu N., Liu Q., Jacquemyn H., Xing X. (2021). Extracellular enzyme activities and carbon/nitrogen utilization in mycorrhizal fungi isolated from epiphytic and terrestrial orchids. Front. Microbiol. 12, 787820. doi: 10.3389/fmicb.2021.787820
Zhu B., Chen S., Zhao C., Zhong W., Zeng R., Yang S. (2019). Effects of Marichromatium gracile YL28 on the nitrogen management in the aquaculture pond water. Bioresour Technol. 292, 121917. doi: 10.1016/j.biortech.2019.121917
Zhu L., Shi W., Zhou J., Yu J., Kong L., Qin B. (2021). Strong turbulence accelerates sediment nitrification-denitrification for nitrogen loss in shallow lakes. Sci. Total Environ. 761, 143210. doi: 10.1016/j.scitotenv.2020.143210
Keywords: Bellamya purificata, bioturbation, nitrogen occurrence forms, extracellular enzyme activity, bacterial community, sediment
Citation: Zhang Y, Hou Y, Jia R, Li B, Zhu J and Ge X (2022) Nitrogen occurrence forms and bacterial community in sediment influenced by Bellamya purificata bioturbation. Front. Mar. Sci. 9:1028716. doi: 10.3389/fmars.2022.1028716
Received: 26 August 2022; Accepted: 01 November 2022;
Published: 24 November 2022.
Edited by:
Bin Xia, Qingdao Agricultural University, ChinaReviewed by:
Ji Liu, Qingdao Agricultural University, ChinaYunfei Sun, Shanghai Ocean University, China
Copyright © 2022 Zhang, Hou, Jia, Li, Zhu and Ge. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xianping Ge, Z2V4cEBmZnJjLmNu; Jian Zhu, emh1akBmZnJjLmNu
†These authors contributed equally to this work and share first authorship
 Yiyun Zhang
Yiyun Zhang Yiran Hou
Yiran Hou Rui Jia
Rui Jia Bing Li
Bing Li Jian Zhu
Jian Zhu Xianping Ge
Xianping Ge