
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 18 October 2022
Sec. Aquatic Microbiology
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.1017136
This article is part of the Research TopicMicrobes from Marine Distinctive EnvironmentsView all 19 articles
 Josefien Van Landuyt1
Josefien Van Landuyt1 Kankana Kundu1
Kankana Kundu1 Sven Van Haelst2
Sven Van Haelst2 Marijke Neyts3
Marijke Neyts3 Koen Parmentier3
Koen Parmentier3 Maarten De Rijcke2
Maarten De Rijcke2 Nico Boon1,4*
Nico Boon1,4*Historic shipwrecks form an anthropogenic landmark in marine environment, yet their influence on the local geochemistry and microbiology remains largely unexplored. In this study, sediment and steel hull samples were taken around the V-1302 John Mahn, a World War II shipwreck, at increasing distance from the wreck, in different directions. Polycyclic aromatic hydrocarbons (PAH’s), explosives, and heavy metal levels were determined and related to the microbial composition. Benz(a)anthracene and fluoranthene remain present at the mg kg-1 level, probably originating from the coal bunker. These PAH’s indicate that the wreck is still influencing the surrounding sediments however the effects are very dependent on which side of the wreck is being studied. Known PAH degrading taxa like Rhodobacteraceae and Chromatiaceae were more abundant in samples with high aromatic pollutant content. Moreover, sulphate reducing bacteria (such as Desulfobulbia), proven to be involved in steel corrosion, were found present in the biofilm. This study shows that even after 80 years, a historic shipwreck can still significantly steer the surrounding sediment chemistry and microbial ecology.
On the 12th of February 1942 Vorpostenboot V-1302 of the German Kriegsmarine sank to the bottom of the Belgian part of the North Sea (BPNS) during Operation Cerberus (The Channel Dash). The 48 m long ship was built in 1927 by the Reiherstieg Schiffwerke (Hamburg) as the fishing boat “John Mahn”. During World War II (WWII), the steam trawler was requisitioned by the German Navy and adapted to warfare. At 15h53 that day in February, V-1302 was attacked by six British Royal AirForce Hawker Hurricanes in front of the Belgian coast. Despite successfully downing one of the aircrafts, she was struck by two aerial bombs. The first bomb hit the funnel amidships and detonated in the boiler room, the second hit the aft section and destroyed the propeller shaft tunnel. V-1302 sank rapidly, heeling over after only half a minute, taking 11 sailors and all remaining munitions and coal reserves with her. Today the wreck lies slightly askew at the bottom of the BPNS, with a large hole on the port side due to the first bomb hit.
Shipwrecks on the seafloor worldwide contain hazardous substances such as explosives and petroleum products that, if released, may harm the marine environment. In contrast to artificial reefs (i.e. intentionally sunk vessels and structures), wartime shipwrecks were sunk without being stripped of hazardous substances, often having reserves of crude oil or other petroleum derivatives and unexploded munitions still on board. Even today, it is estimated that World War I and II shipwrecks around the globe, collectively, contain 2.5 to 20.4 million tonnes of petroleum products (Landquist et al., 2017). Rogowska et al. (2015) showed that latent, historic (WWII shipwreck) contamination with fossil fuels from point sources can influence the chemical composition of the marine sediment to this day. The risk that petroleum products and its derivatives pose to marine wildlife has been studied extensively, showing they can affect feeding, growth, reproduction and causes irreversible tissue damage in many marine organisms (Martínez-Gómez et al., 2010). In addition, up to 1.6 million tonnes of ammunition of all types (both attached to and separate from ships) were sunk or dumped in the Northern seas of Europe after both World Wars (Sternheim et al., 2019). The few studies performed on the subject suggest explosive compounds such as trinitrotoluene (TNT) and its derivatives (Koske et al., 2020), as well as chemical warfare agents (Czub et al., 2020), can have toxic effects on aquatic wildlife. However, the influence of these compounds on an ecological level remains unclear. Historic shipwrecks could also significantly affect the surrounding fauna and flora when leaching heavy metals and metalloids due to (biological) corrosion (Kelly et al., 2012). While wrecks are often considered biodiversity hotspots, the overall environmental impact on the seafloor of historic shipwrecks from the World Wars has only recently sparked interest (Landquist et al., 2013; Thomas et al., 2021).
As solid heterogeneous substrates, shipwrecks are rapidly colonized by microorganisms which allow other organisms to attach and form an assemblage with dynamic community interactions (Price et al., 2020). A recent study on historic (sunk >50 years ago) shipwrecks in shallow water performed by (Price et al., 2020) has shown that the hull microbiome composition differs even on the phylum level compared to the close-surrounding sediments. Another recent study, performed by Hamdan et al. (2018), showed a similar result in deep water. Where sediments are high in Deltaproteobacteria, the shipwrecks and sediments closer to the ships will show higher abundances in Alphaproteobacteria and Gammaproteobacteria. Several bacteria have been collected or identified from the hull of a shipwreck and marine steel fragments and proven to be involved in microbial induced corrosion (MIC) (Smith et al., 2019; Price et al., 2020). These bacteria were shown to be either members of the iron-oxidising bacterial groups like Mariprofundus ferrooxydans or sulphate-reducing/sulphur-oxidising bacteria (SRB/SOB) like Desulfobulbus propionicus. Sulphur cyclers such as Desulfovibrionaceae and Desulfobulbaceae combined made up 30 to 75% of mooring chain surface corrosion products formed after 10 years at 2 km deep (Rajala et al., 2022).
Not only prokaryotes, but also microalgae play a role in the structure and function of marine sediment (Baustian et al., 2011; Lake and Brush, 2011). The microphytobenthos and its microalgal community structure can be shaped and changed by disturbances due to anthropogenic, polluting compounds ranging from polycyclic aromatic hydrocarbons (PAH’s) to pesticides (Chapman et al., 2010; Sundbäck et al., 2010; Magnusson et al., 2013; Pinckney et al., 2013; Cibic et al., 2019). Sundbäck et al. (2010) showed that the effect of pyrene (a PAH) on the benthic microalgae in a microcosm study was two-fold. On the one hand, grazers were affected which resulted in higher benthic microalgae in total, while on the other hand the pyrene caused a shift in biodiversity, with the main structural differences found in the Bacillariophyceae (diatoms). Little to no research has been done to investigate the microalgal contribution to shipwreck microbial ecology. Investigating the shipwreck’s microbiome, both bacterial as well as microalgal, can give us a deeper insight into the corrosion (Price et al., 2020), the potential presence, and biodegradation of contaminants, as well as the micro biodiversity and macro biodiversity potential through biofilm-mediated settlement clues (Whalan and Webster, 2014).
This study investigates the microbial community composition of the biofilm on steel fragments of the historic World War II shipwreck V-1302 John Mahn and in the surrounding seabed to identify species related to bio-corrosion and PAH biodegradation. The bacterial and microalgal chloroplast diversity was determined using 16S rRNA gene sequencing. At the same time, the top layer (up to 15 cm deep) of the sediment surrounding the shipwreck was analysed for PAH’s, explosives and heavy metals using gas chromatography combined with mass spectrometry for the organics and inductively coupled plasma atomic emission spectroscopy for the metals. With this integrative study of the sediment chemistry and the microbial community we provide insights into the microbial ecology of historic artificial features in coastal waters which contributes to a deeper understanding of the effect of potentially polluting shipwrecks on the (local) microbiology at the bottom of the sea.
Chemical analysis in samples taken in four axial directions from the shipwreck was performed (Figure 1) to see whether pollution increased upon approaching the John Mahn shipwreck from any direction. Figures 1A, B show the gradients based on the heavy metal content. From bow to stern, we observe higher concentrations when approaching the ship. The highest metal and metalloid concentrations were found in the sample closest to the coal bunker, with specifically a high nickel, copper, and arsenic content. There is less of a trend on the starboard-port side axis when approaching the shipwreck. Most notably, the freshly deposited sediment in the wake of the wreck has a high metal content, which could be due to the deposition of metal flakes peeling off the wreck along with sediment particles. We also observed that the heavy metal signature is different in the fresh sediment relative to the other samples on the port-starboard axis, as it has a considerably higher nickel content and considerably lower chromium content. This freshly deposited sediment on the starboard side is being deposited by a dead zone in the underwater current. PAH concentrations tended to be inversely related to the distance to the ship on both the port and bow side (Figure 1C). The stern side samples, taken closest to the damaged coal bunker, show the highest PAH concentrations. Samples from the starboard side, where fresh sediment is deposited in the wake of the ship due to scour along the predominant direction of the tidal currents (Figure 1D), have a much lower aromatic compound content. Low concentrations (µg/kg) of explosive compounds were detected in mainly the port and stern side sediment samples. Both TNT and 1,3-dinitrobenzene (1,3-DNB), a biodegradation product of TNT (Supplementary Figure S1), were detected, however, only TNT data are shown in Figures 1E, F as 1,3-DNB was found in some of the blanks as well. When plotting the diversity based on the chemical content in a PCA plot (Figure 2A), it shows that the chemical composition determines some clustering of the stern side samples whereas the fresh sediment sample presents a clear outlier away from all important polluting drivers.
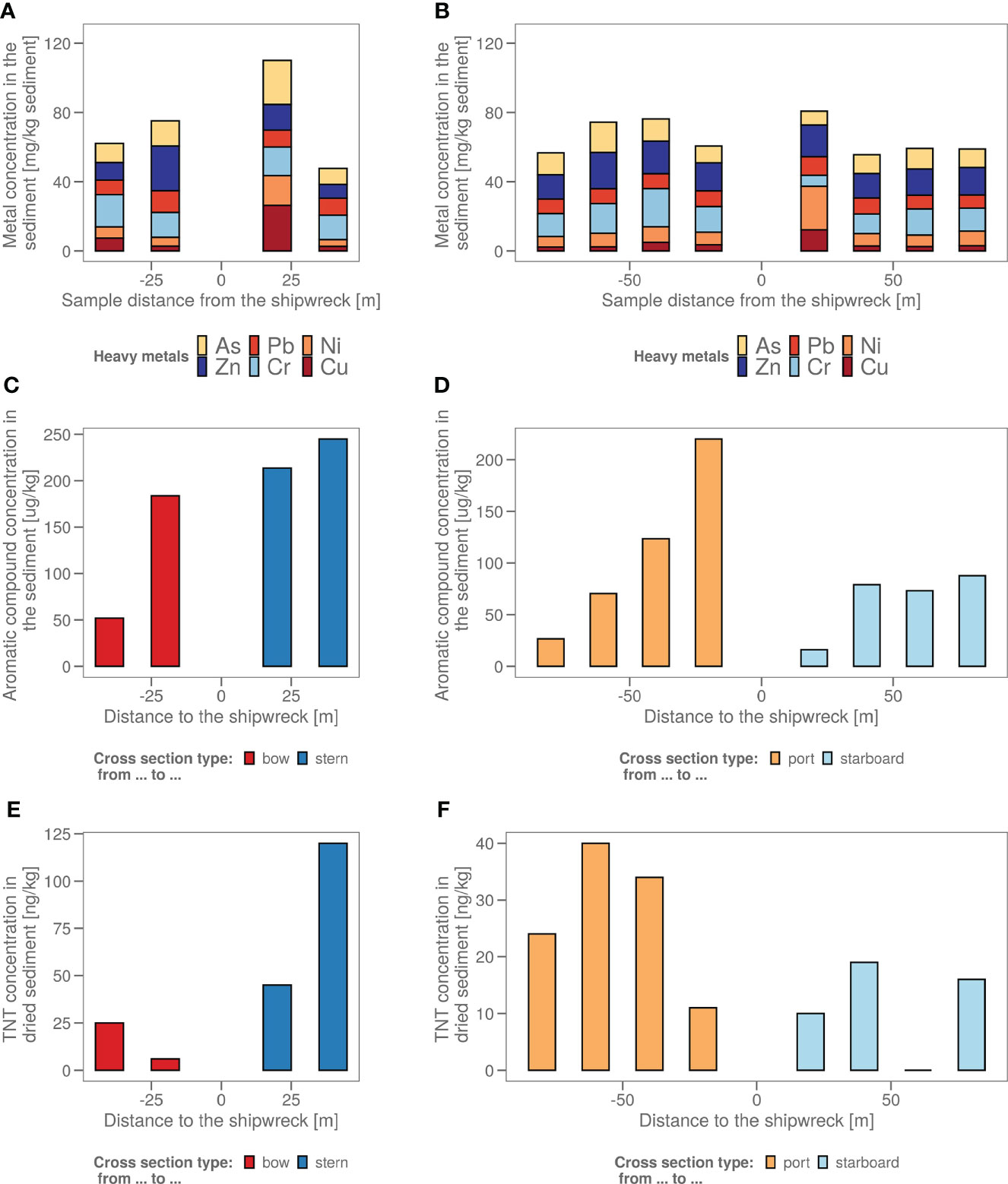
Figure 1 (A) The sediment samples from bow to stern and (B) from port side to starboard side with their total heavy metal content in mg heavy metals/kg wet sediment. The sample taken in the dead zone of the ship wreck, with freshly deposited sediment is indicated with a red arrow. (C) The sediment samples from bow to stern and (D) from port side to starboard side with their polycyclic aromatic hydrocarbon (PAH) content in µg aromatics/kg wet sediment. The sample taken in the dead zone of the ship wreck, with freshly deposited sediment is indicated with a red arrow. (E) The sediment samples from bow to stern and (F) from port side to starboard side with their total explosive content in ng TNT/kg dry sediment. (All values <17 are estimations as 17 ng/kg is the LOQ).
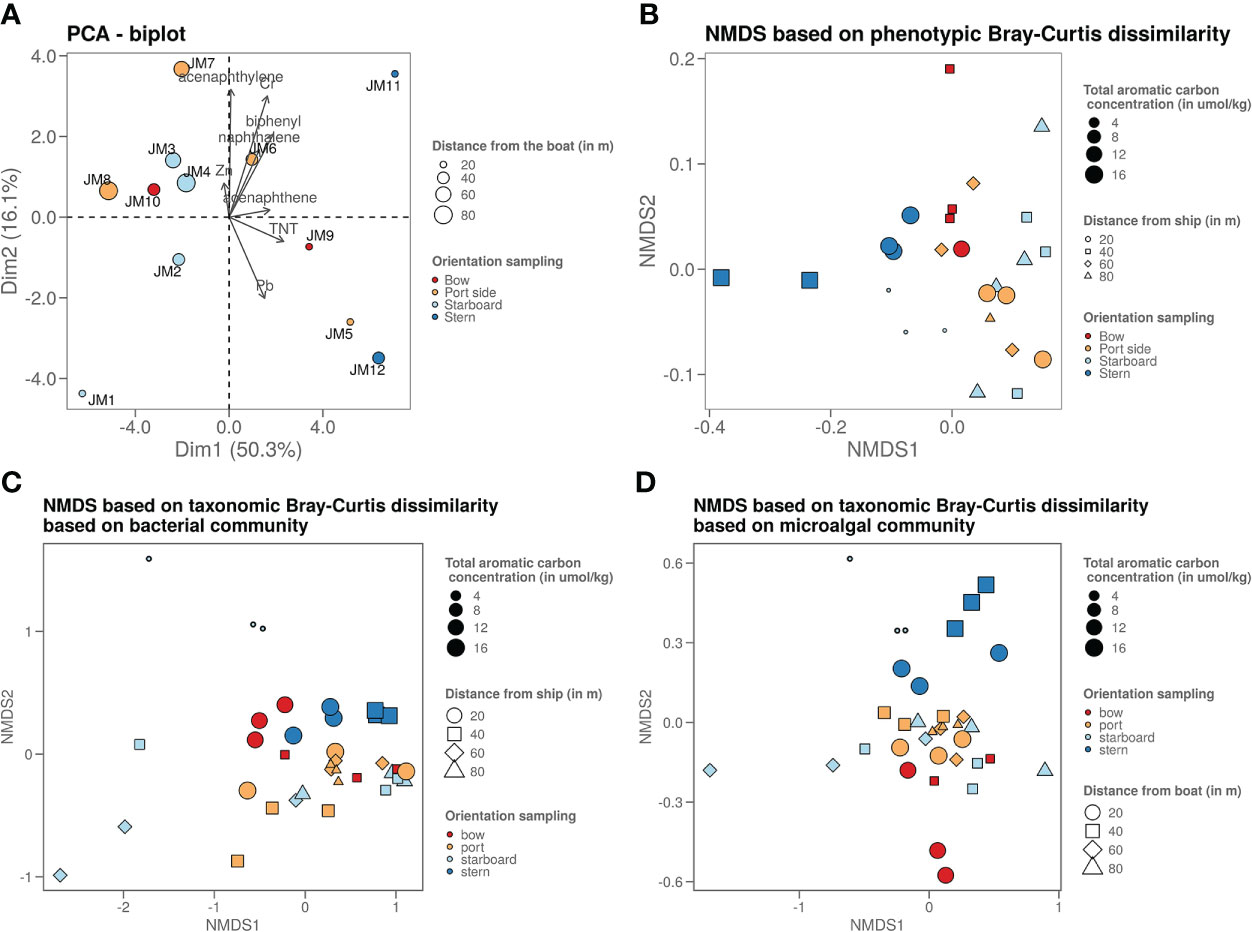
Figure 2 (A) Graph shows a PCA plot of the chemistry data where it shows that the samples taken on the stern side are the ones strongest polluted (highest chemical content). The sample closest to the ship on the starboard side is least polluted, as it is freshly deposited due to the dead zone. (B) Graph shows an NMDS visualisation of the microorganisms phenotypic betadiversity, (C) graph shows an NMDS visualisation of the bacterial taxonomic betadiversity and (D) graph shows an NMDS visualisation of the microalgal taxonomic betadiversity.
The number of microbial cells in the sediment were on average the same across all samples, namely around 106 cells per g wet sediment (Figures 3A, B). A biological analysis in four axial directions from the shipwreck of the study area was also made based on the bacterial and algal chloroplast composition in the sediment (Figures 3C–F) based on 16S rRNA gene analysis.
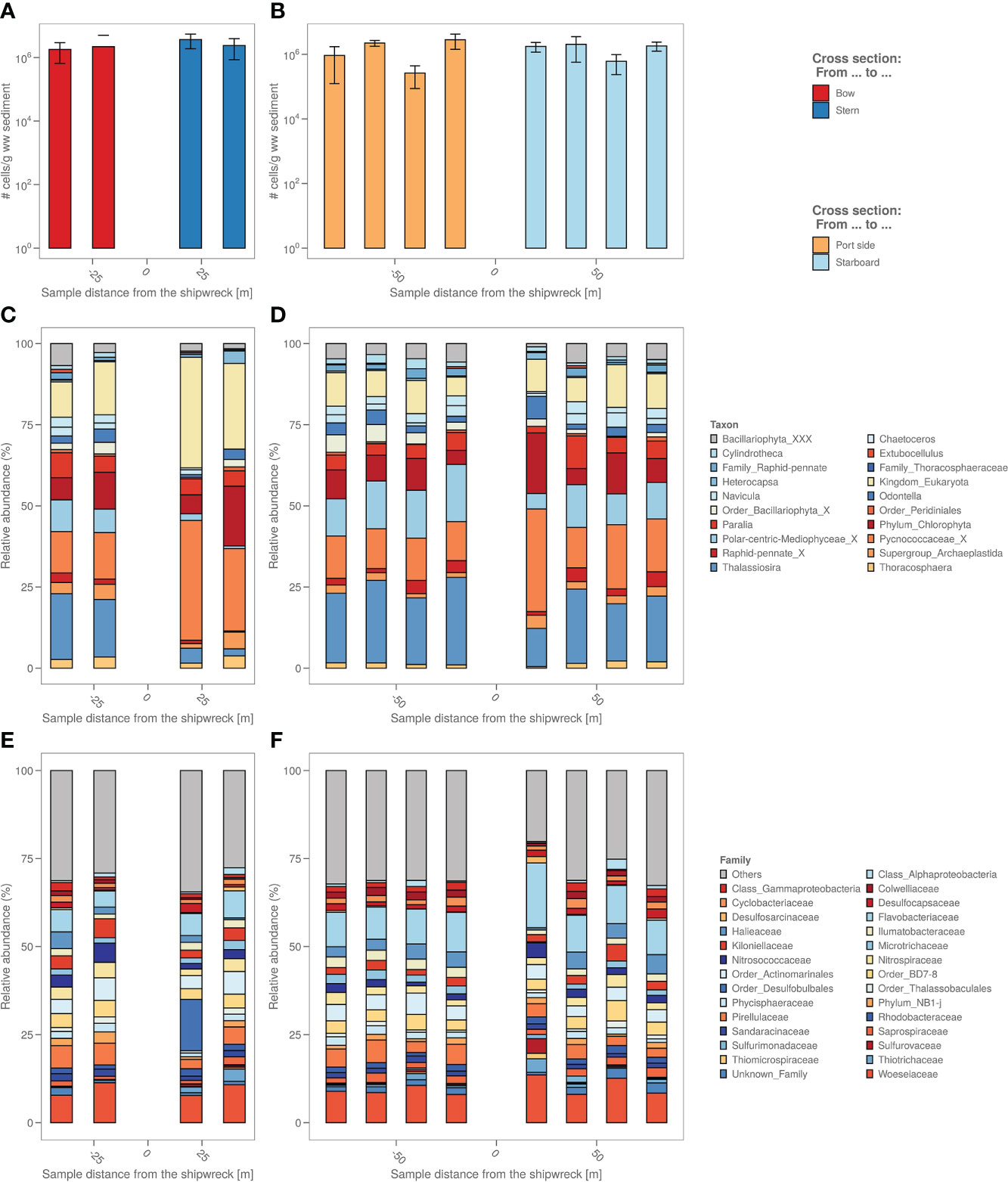
Figure 3 (A) The sediment samples from bow to stern and (B) from port side to starboard side with total number of cells per g wet weight sediment. (C) The sediment samples from bow to stern and (D) from port side to starboard side with their relative micro-algal abundances per sample site coloured by top 9 most abundant microalgae genus. “_X” and “_XXX” were added when classification could not be performed down to genus level, i.e. “unclassified”. (E) The sediment samples from bow to stern and (F) from port side to starboard side with their relative bacterial abundances per sample site coloured by top 19 most abundant bacterial family.
Algal chloroplast 16S rRNA gene sequences were used to identify the relative abundance of eukaryotic microalgae in the samples (Figures 3C, D). The analysis based on the algal chloroplast content shows a different composition of the samples taken closest to the coal bunker (stern side) with relatively more Pycnococcaceae (27.6 ± 7.4% vs. 19.5 ± 14.7%) as well as unidentified chloroplasts (Eukaryota kingdom) and relatively less Navicula (1.4 ± 1.9% vs. 4.9 ± 1.6%), Thallassiosira (4.3 ± 3.0% vs. 20.2 ± 5.0%) and chloroplasts from the Bacillariophyta order (2.5 ± 1.5% vs. 4.1 ± 1.9%) compared to the port and bow side. The freshly deposited sediment also appears to have a different microalgal composition, with relatively more Pycnococcaceae (33.0 ± 4.7%) and unidentified chloroplasts (Eukaryota kingdom) and relatively less Thallassiosira (11.7 ± 4.5%) compared to the port and bow side samples.
The relative abundances of the different bacterial taxonomic groups are different in the freshly deposited sediment (starboard sample at 20m) compared to the other sediment samples (Figures 3E, F); among others, there is relatively more Flavobacteriaceae (18.9 ± 1.8% vs. 9.1 ± 3.1%) and relatively less Halieaceae (0.5 ± 0.9% vs. 3.2 ± 1.6%). A representative of the Desulfobulbales (8.6 ± 14.2%) order is highly abundant in the sample closest to the coal bunker that is almost non-existent in the other samples (on average 0.2 ± 0.4%).
Flow cytometry data were used to plot a non-metric dimension scaling (NMDS) figure of the phenotypic microbial Bray-Curtis beta-diversity (Figure 2B). This plot revealed a slight grouping per sampling side. Mainly the stern side samples are most dissimilar from the other samples. When looking at the Bray-Curtis dissimilarity of the 16S rRNA gene sequencing data (Figures 2C, D), most samples are similarly based on the bacterial taxonomy. However, the freshly deposited sediment at the starboard side is an outlier. The samples with higher PAH content from the stern and bow side tend to cluster together. The micro algal taxonomy shows a similar pattern, with the higher PAH polluted (points with bigger size on the figure) stern and bow side samples clustering more clearly away from the other samples.
Comparing the lowest chemically contaminated samples (low: total PAH below 25 µg/kg, high: total PAH over 75 µg/kg) versus the highest contaminated samples, there are several bacterial amplicon sequence variants (ASV’s) significantly more abundant. Most of these ASV’s could be attributed to the following families: Rhodobacteraceae, Flavobacteriaceae and Chromatiaceae. From the top 100 most abundant bacterial ASV’s there are two ASV’s, one belonging to the Desulfobulbales order and one to the Marinilabiliaceae family (Figure 4A). With the 16S rRNA gene sequencing and the chemical data, a canonical component analysis (CCA) was performed to indicate what chemistry and which bacterial ASV’s contributed to the variance of the bacterial alpha and beta diversity amongst all locations (Figure 4B). The CCA visualisation shows that the PAH concentration drives the bacterial differences between the fresh sediment and exposed/aged sediments. For the bacterial ASV’s contributing most to the variance (based on the CCA), a Pearson’s correlation matrix was made (Supplementary Figure S2) to investigate whether the abundancy of these ASV’s was significantly correlated to the heavy metals, metalloids and aromatic compound concentrations measured in the samples. Bacterial ASV’s with the highest positive correlation to aromatic compound content and heavy metal concentrations were identified as representatives of the Woeseia, Thiotrichaceae, Ilumatobacter, Arenicella and Rhodobacteraceae (red dots on the figure). Bacterial ASV’s with negative correlations included members of Sulfurimonas and Woeseia (blue dots on the figure). A similar correlation matrix analysis (Supplementary Figure S3) was performed on the ASV’s that were identified as chloroplasts of algae. In red, we can see some algal chloroplast ASV’s correlated positively to the presence of PAH, similar to the chloroplasts of the families Pycnoccaceae and Thoracospharaceae. In blue, we marked a single negatively correlated algal chloroplast ASV that could not be identified beyond the Chlorophyta taxon.
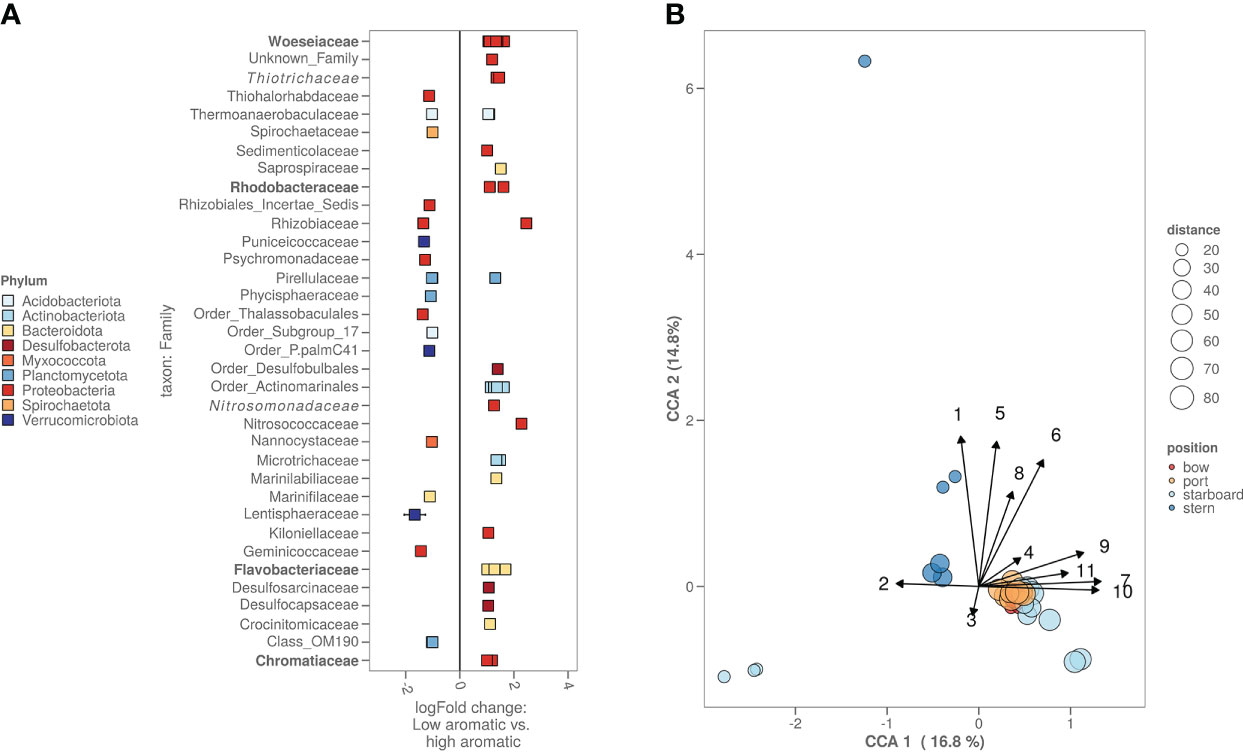
Figure 4 (A) High versus low polluted sediment based on the bacterial ASV’s. Cut-off for high polycyclic aromatic hydrocarbon (PAH) content was >75 PAH µg kg-1 sediment while cut-off for low was <25 µg kg-1 sediment. Families with known biodegradation pathways were highlighted in bold. Families with known enrichment in PAH polluted sediments were highlighted in italic. (B) CCA plot combining chemistry, bacterial and algal information. Starboard samples closest to the ship were freshly deposited sediment. Numbers on the arrows stand for the following chemicals: 1. methylphenanthrene, 2. Pb, 3. Zn, 4. acenaphthene, 5. fluorene, 6. dimethylnaphthalene, 7. organic carbon content in µg kg-1, 8. total PAH in umol kg-1, 9. biphenyl, 10. naphthalene and 11. acenaphthylene.
The biofilm on the shipwreck contained >40% of sulphur cycling bacteria from families including Desulfocapsaceae, Desulfosarcinaceae, Sulfurimonadaceae, Sulfurovaceae, Thiomicrospinaceae and Thiotrichaceae (Figure 5A). Total relative abundance of sulphur cyclers was on average almost 6 times more abundant in steel samples (>40%) compared to sediment samples (7%) (Figure 5A). Comparing the specific bacterial families found in sediment versus shipwreck biofilm samples using Analysis of Compositions of Microbiomes with Bias Correction (ancombc) confirmed a significantly higher abundance of families with members involved in sulphur cycling (Figure 5B). When looking at the taxonomic families composing the community structure of the samples, average relative abundance showed that about 2% of the families present in the shipwreck microbiome were specific to the shipwreck niche (i.e. not native to the sediment microbiome (<10-10%)). Alpha diversity analysis (Hill numbers, D2) showed a significantly lower bacterial biodiversity on the biofilm compared to the sediment samples (Wilcoxon Rank Sum test; p<0.05) (Figure 5C). Moreover, there did not seem to be a trend of higher diversity in the sediment closer to the shipwreck, and lower diversity when moving further away from the wreck, for some sampling directions an opposite trend may even be seen, as for bow and stern the diversity decreases closer to the wreck (Figure 5D).
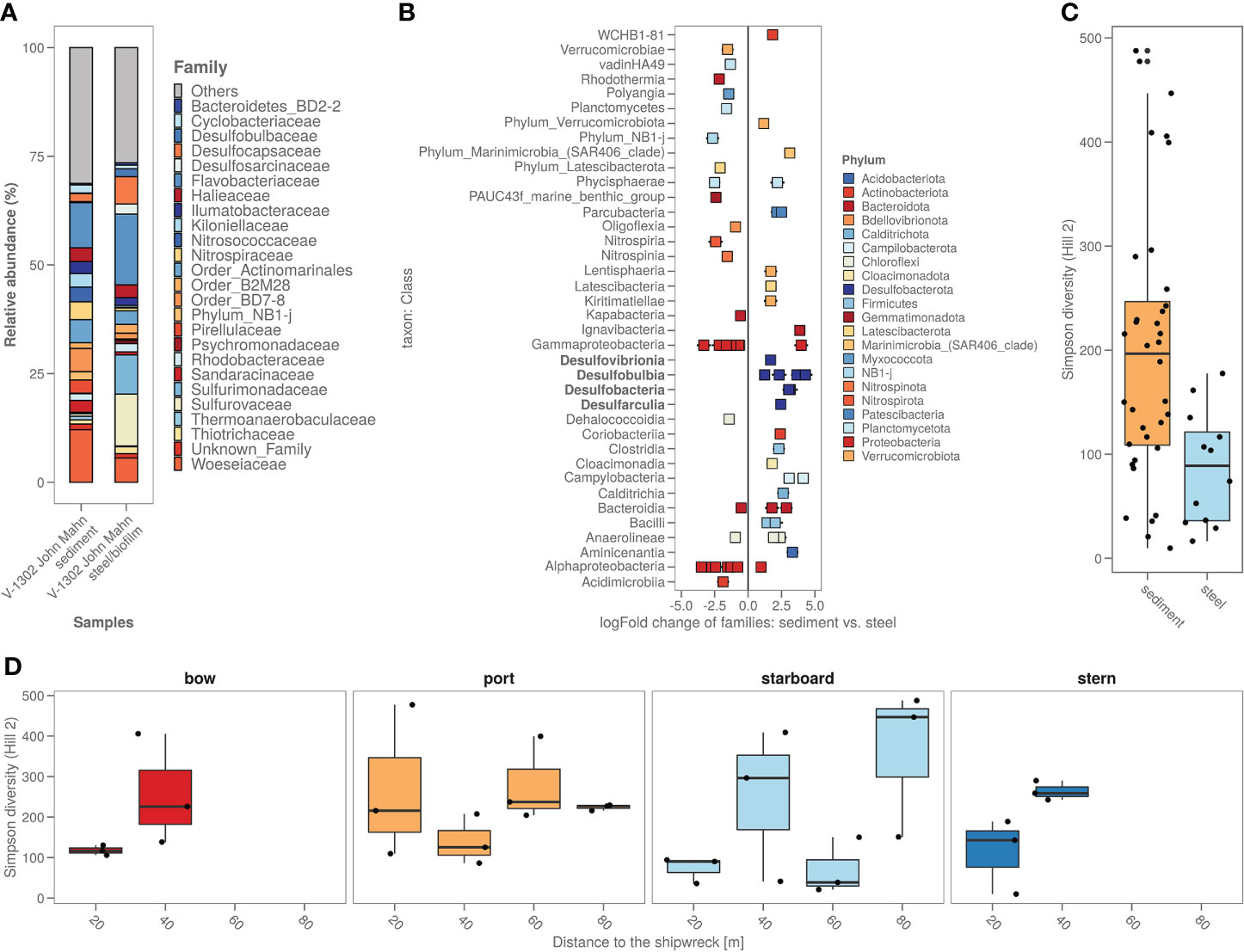
Figure 5 (A) Sediment around the shipwreck and the steel their bacterial composition (ASV’s) based on top 25 taxa based on family; (B) Steel versus sediment based on the bacterial families. Classes with known corrosion members were highlighted in bold; (C) Alpha diversity (as Simpson diversity calculated by Hill number q=2) of the sediment and steel samples; (D) Alpha diversity (as Simpson diversity calculated by Hill number q=2) per sampling direction and distance from the ship.
To the best of our knowledge, the present study is the first to combine a chemical and microbial characterization of a potentially polluting wreck in the Belgian part of the North Sea. The John Mahn shipwreck, even after almost 80 years on the seafloor, seems to still leach micropollutants into the sediment, both from the coal bunker (aromatic compound content) as well as munition (still) present on the wreck (TNT content) and the wreck itself (heavy metals). In comparison to the total PAH concentrations found around the s/s Stuttgart of the Polish coast in the Baltic Sea (Rogowska et al., 2010), i.e. from 16.05 mg/kg to 244.90 mg/kg sediment, the concentrations found around the V-1302 John Mahn are about 100 to 1000 times lower (µg/kg vs. mg/kg), which is in the same range as the concentrations found around the HMS Royal Oak in Scotland (Thomas et al., 2021) (closest samples taken 50 m from the HMS) and within background concentration levels for sediments in the OSPAR (Oslo/Paris convention for the Protection of the Marine Environment of the North-East Atlantic) marine area (Dataset Commission, O, 2017). Our sampling pattern revealed increasing concentrations when approaching the shipwreck (Figures 1C, D). The PAH composition showed a higher pyrogenic than petrogenic content, which is more indicative of exhaust pollution rather than un-burnt hydrocarbons leaching from a tank, contrasting with the concentration profiles that do suggest leaching from the wreck. However, leaching from ashes in the boiler, a flash fire from the bomb hit, and burned coal tar potentially used to preheat the boilers could also explain the higher pyrogenic rather than petrogenic PAH content. The heavy metal concentrations surrounding the V-1302 John Mahn, are in the same range as the ones found by Gwizdala et al. (2018) in the gulf of Gdańsk surrounding the small World War II shipwrecks, Munin and Abille, where samples were taken at 100 m from both wrecks. The higher arsenic combined with higher PAH content in the samples closest to the coal bunker suggests the arsenic might originate from the coal. Arsenic and copper could also be leaching from antifouling agents, as the paints used in that time period commonly contained arsenic, mercury or copper (Almeida et al., 2007). The TNT concentrations found in this study (up to 120 ng TNT/kg dry sediment) were 5 to 10 times lower than what was detected in sediments close to sunken artillery shells in Halifax, Canada, where they found concentrations ranging from 0.5 to >100 ug/kg (Rodacy et al., 2001), although, to compare the values of TNT in these different sediments in-depth, normalisation of the organic matter content should be performed. These results suggest either the remnants of a strong historic contamination or continuous slow leaching. The half-life in marine sediments of recalcitrant PAH components can range from several days up to decades (Marini and Frapiccini, 2013; Mandić and Vrančić, 2017) which means some of the components could be historic contamination however others, such as naphthalene (with a half-life up to 13 days according to Marini and Frapiccini (2013)) has probably been slowly leaching more recently.
Although not the most prevalent (only 2 bacterial ASV’s from the top 100 most abundant ones), several bacterial ASV’s were more dominant in the more contaminated sediments (Figure 4A). These ASV’s belong to families which include members that are known to be able to degrade aromatic compounds, e.g. Rhodobacteraceae (Buchan et al., 2019), Flavobacteraceae (Trzesicka-Mlynarz and Ward, 1995), Chromataciaceae (Rochman et al., 2017) and Woeseiaceae (Bacosa et al., 2018), while others belonged to families previously shown to be enriched in PAH contaminated sediments, e.g. Thiotrichaceae (Dell’Anno et al., 2021) and Nitrosomonadaceae (Jiao et al., 2016). Specific (relatively low abundant) ASV’s belonging to the genus Woeseia and to the families Thiotrichaceae and Rhodobacteraceae showed a strong positive correlation with the PAH concentrations (Supplementary Figure S2). It is also suggested from both the algal and bacterial beta-diversity that the freshly deposited sediment is different from sediment surrounding the shipwreck (Figure 3). Overall, our results indicate that both the shipwreck itself and the PAH which are presumably leaching from the wreck to the sediment, despite the low concentrations, drive at least part of the bacterial community structure in the surrounding sediment.
The effects of the shipwreck on both chemical and microbial composition of the sediment are in contrast with the study performed by Thomas et al. (2021) on sediments surrounding the HMS Royal Oak. Although similar in time spent on/in the sediment (John Mahn: sank in 1942 vs. Royal Oak: sank in 1939), similar in depth (21-35 m vs. 12-33 m), similar in climate (both in between 50-60° N and between 3° W and 3° E, both in a sheltered Sea (BPNS vs. Scapa Flow), and similar in how far the samples were taken from the ship (40 m vs. 50 m), Thomas et al. (2021) found no leaching nor any clear bacterial enrichment due to PAH’s. The bacterial composition of sediment samples of the HMS Royal Oak showed similar higher taxonomic groups as reported here. Acidimicrobiia, Alphaproteobacteria, Bacteroidia, Gammaproteobacteria, Desulfobulbia, Nitrospira, Phycispaerae and Polyangia were the most significant contributors in both cases (Supplementary Figure S4). However, when looking into a lower taxonomic level, there was a clear difference based on relative abundances of families present, with Flavobacteriaceae, Desulfosarcinaceae, and Sulfurovaceae dominating the Royal Oak sediment while Woeseiaceae, Flavobacteriaceae, and BD-7-8 group dominated the John Mahn sediment. Although similar research aims were pursued, a big difference with the HMS Royal Oak study is that Thomas et al. (2021) focused on sediment further from the shipwreck (50 - 1000 m), whereas here the samples were taken close to and on the shipwreck (0 to 80 m). This could suggest that the effect of World War II shipwrecks (<50 m deep) in the North Sea sediment are limited to a small area around the wreck due to the dynamic nature of the BPNS, with strong currents from diverse directions. In addition, sandy sediment often lacks a high carrying capacity for contaminants. Furthermore, the Royal Oak contained fuel oil rather than coal, which was demonstrated to leak a lot during the 1960s and the 1990s and was partially cleaned during the 2000s, while to our knowledge the V-1302 John Mahn’s remaining coal reserves are mostly still in place, seemingly slow but continuously leaching to this date.
Phycisphaerae, Verrucomicrobia and Bacteroidetes were reported by Price et al. (2020) to be at a higher abundance on shipwreck samples compared to sediment. In this study, the abundance of Verrucomicrobia and Bacteroidetes are also generally higher on the shipwreck steel (Supplementary Figure S4). Upon closer look, several families have significantly higher abundances while other families from these phyla show a significantly lower abundance (Figure 4B).
A clear difference found in this study was the higher abundance of sulphur cyclers in the steel samples. The classes, highlighted in bold in Figure 5B, shown to be significantly more abundant on the steel fragments, are sulphate reducing bacteria (SRB) and sulphur-oxidising bacteria (SOB). When looking at solely the Zetaproteobacteria (main iron-oxidising bacterial group) only one ASV from the Mariprofundus genus is present, and only on the steel fragment samples (0.076% vs. 0%). Colonization of steel in marine sediments starts with an increase in abundance of iron-oxidizers, and as the biofilm matures, bacteria and other microbes, involved in speeding up the corrosion process, appear (McBeth and Emerson, 2016). Smith et al. (2019) performed a thorough analysis of marine steel fragments, obtained from a corroded steel surface in the UK, showing the involvement of both SRB and SOB in marine steel bio-corrosion. Iron sulphides are formed by reaction with hydrogen sulphide produced by the SRB causing primary corrosion. These iron sulphides are subsequently being oxidised through a series of sulphur oxidation states by the SOB, forming acid at all stages, causing even more corrosion. Enning et al. (2012) have suggested that some bacteria (such as SRB and SOB) are able to take up external electrons from abiotic sources like shipwreck steel, explaining the enrichment of both SRB and SOB and other heterotrophic bacteria on shipwreck steel. SRB and SOB would thus be expected in a mature steel hull bacterial biofilm.
Two genera from the algal class Bacillariophyta were more abundant in the steel samples compared to the sediment samples (Supplementary Figure S5), moreover, if we look at the total average % of diatoms in the samples, it shows that more diatoms were present on the steel fragments (57.2% ± 15.1% vs. 49.9% ± 18.6%) suggesting a role in settlement on/in the biofilm and ultimately a potential (in)direct effect on corrosion. Smith et al. (2019) showed diatomaceous algae cemented within the corrosion biofilm and iron oxide nodules as well, showing diatoms could be involved in the mechanism at some level. However, more research into what specific functional mechanism these microalgae contribute to corrosion is still needed.
In conclusion, this study demonstrated that the local microbiology and geochemistry on and around this World War II ship is still influenced by the wreck even after having been down there for almost 80 years. Both local bacteria as well as microalgae are influenced by the chemistry and structure this shipwreck provides.
Based on literature and incidental reports by recreational divers, an estimated 35 wrecks in the Belgian Part of the North Sea may still contain munitions. From this list, the V-1302 was selected for this study based on the known presence of pollutants (coal reserves, depth charges) and its favourable conditions for diving (depth, current speeds, average turbidity). When built, the ship was 48 m long and weighed 292 tonnes. As the ship was built in 1927, it was most likely built with Schiffbaustahl type I, which had a yield strength of 360-430 MPa and a 20% strain. It consisted of 0.25-0.3% carbon, 0.3-0.5% manganese and 0.3% silicon (Bauer et al., 1925). During adaptation from shipping boat to warfare, an 88mm naval gun, 20mm c/38 light anti-air artillery pieces, and multiple anti-submarine depth charges were added. Documents from the German Military Records in the Bundesarchiv in Freiburg revealed the fate of this vessel. The ship was hit twice after which it sank within 35 minutes. The ship wreck lies slightly askew with a large hole on the port side (due to the first bomb blast) at 51°28,937’ N and 2°41,339’ E between 21m and 35m of depth (Supplementary Figure S6). Visual inspections by scientific divers have shown that various munitions (88mm shells, 20mm cartridges, and at least six depth charges) are still present on the wreck, but the leakage of polyaromatic hydrocarbons (PAH's), munition compounds or heavy metals had not been investigated to date.
To investigate whether pollutants (organic aromatic compounds, heavy metals and explosives) are (still) present in the sediment surrounding the wreck, samples were taken in a cross-shaped sampling pattern around the V-1302 John Mahn. Eight sampling points were decided upon on the port/starboard axis, four on each side. Four sampling points were decided upon on the bow/stern axis, two on each side. Sampling points were chosen approximately 20 m apart, as shown in Figure 6. At each sampling point, a Van Veen grab with a sampling surface of 0.1 m², and a sampling depth of approximately 15 cm was lowered. Once aboard, the sediment of each point was homogenised by hand before the sample was split into four, providing ample sample for each of the different analyses. Two small pieces of loose wreckage were also recovered by divers during the sampling campaign. Before the microbial analysis, all the sub-samples for this analysis (sediment and wreckage pieces) were frozen at -20°C on board.
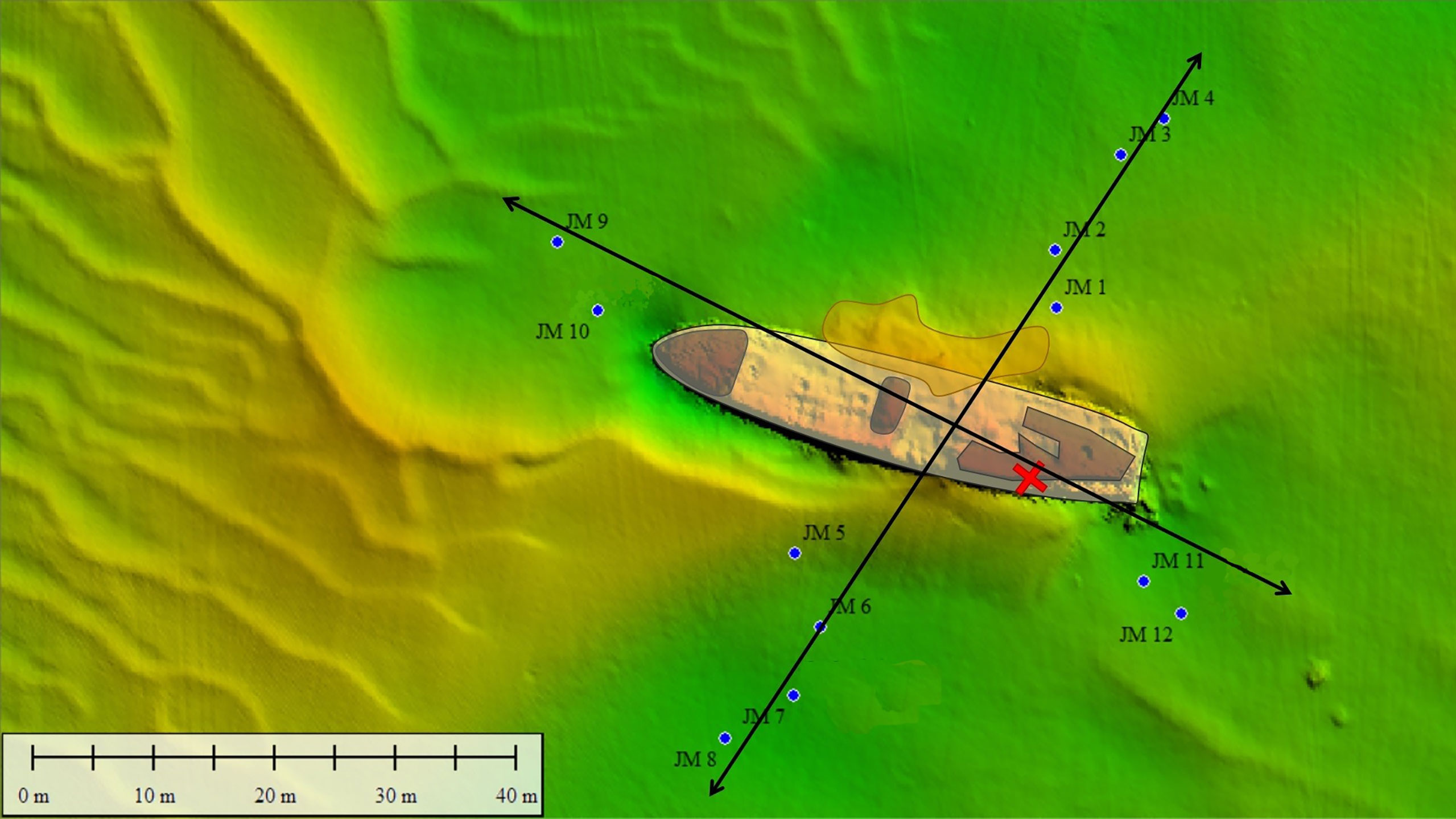
Figure 6 Sediment (JM xx) samples were taken around the John Mahn shipwreck on the four transects radiating Northeast (four sediment samples and one water sample), Southeast (two sediment samples and one water sample), Southwest (four sediment samples and one water sample) and Northwest (two sediment samples and one water sample) from the ship’s centre. The coal bunker was located in the back of the ship on the port/stern side indicated by the red cross. The bombs the ship was equipped with were predominantly found at the front and back of the vessel.
A broad-spectrum analysis of 23 PAHs was used to compare the chemical composition and pollution of various sampling stations. The analysis included naphthalene, 1-methylnaphthalene, 2,6-dimethylnaphthalene, 2,3,5-trimethylnaphthalene, biphenyl, acenaphthylene, acenaphthene, fluorene, fluoranthene, phenanthrene, 1-methylphenanthrene, anthracene, pyrene, chrysene, benz(a)anthracene, dibenz(a,h)anthracene, benzo(b)fluoranthene, benzo(k)fluoranthene, benzo(e)pyrene, benzo(a)pyrene, perylene, benzo(ghi)perylene, and indeno(1,2,3-cd)pyrene. First, PAHs were extracted from the sediment with dichloromethane in an Accelerated solvent extractor at a pressure of 13.8 MPa. Gel permeation chromatography (columns used: ENVIROGEL 19X300MM and ENVIROGEL 19X150MM) was then used for the clean-up. The analysis was done by gas chromatography with mass spectrometry (GC-MS; Thermo TSQ8000) with the conditions for the column (Rxi-5silMS 20 m, 0.18 mm diameter, 0.18 µm internal diameter) as follows: 35°C for 2 min, heat to 250°C at 25° min-1, hold for 0 min, heat to 300°C at 10°C min-1, and a final hold for 10 min; The GC-MS is equipped with a programmable temperature vaporising (PTV) injector, using helium as a carrier gas in the constant flow mode (1 mL min-1). The conditions of the PTV were 40°C for 0.01 min, heating to 300°C at 14.5°C s-1, holding for 2 min. A split flow of 20 ml min-1 was used. Explosives were extracted in the same way as the PAHs with an additional clean-up with aluminium oxide (10% deactivated) and a limited pressure (10.3 MPa). The analysis was done by GC-MS (Thermo TSQ8000). The conditions for the column (Rxi-5silMS 20 m, 0.18 mm diameter, 0.18 µm internal diameter) were as follows: 30°C for 4 min, heat to 120°C at 20°C min-1, heat to 150°C at 7°C min-1, heat to 300°C at 20°C min-1 heat to 300°C at 10°C min-1, and a final hold for 1 min; Large volume PTV injection carrier gas helium in constant flow mode (1 mL min-1). The conditions of the PTV were 40°C for 0.30 min with a pressure of 20 kPa at a flow of 70 ml mincript-1. The heating to 260°C was performed at 1.3°C sec-1 and at a pressure of 250 kPa. Final hold was for 1 min. A split flow of 20 ml min-1 was used.
During flow cytometry analysis, the samples were first diluted in 0.22 µm-filtered Instant Ocean® artificial seawater and stained with 1 vol% SYBR® Green I (SG, 100x concentrate in 0.22 µm-filtered DMSO, Invitrogen) for total cell analysis. Staining was performed as described previously, with an incubation period of 20 min at 37°C in the dark (Chatzigiannidou et al., 2018). Samples were analysed immediately after incubation on an Attune NXT (Thermo Fisher Scientific, Waltham, MA, USA) flow cytometer, equipped with a blue (488 nm) and red (637 nm) laser with Focusing Fluid (Thermo Fisher Scientific, Waltham, MA, USA) as sheath fluid. The instrument performance was verified daily using Attune™ Performance Tracking Beads (Thermo Fisher Scientific, Waltham, MA, USA). The gating strategy can be found in Supplementary Figure S7.
For 16S rRNA gene sequencing, triplicate subsamples of approximately 100 mg were taken from the -20°C frozen sediment samples. Two swabs per -20°C frozen iron piece of the wreckage were taken for 16S rRNA gene sequencing as well. Subsequent DNA extraction was performed by adding about 1 mL Tris/HCl (100 mM, pH 8) lysis buffer and 200 mg glass beads (0.11 mm Sartorius, Göttingen, Germany) to each swab after which cells were mechanically lysed by multidirectional beating for 5 mins at 4500 rpm in a PowerLyzer instrument (Mo Bio laboratories). Glass beads were removed by centrifugation for 5 min at 18 000 g, DNA was purified by a phenol‐chloroform extraction and precipitated by adding 1 volume ice‐cold isopropyl alcohol and 1:10 volume 3 M sodium acetate for at least 1 hr at -20°C, followed by centrifugation for 30 min at 18 000 g. The supernatant was discarded and the DNA pellet was dried and suspended in 100 µL 1× TE buffer (10 mM Tris, 1 mM EDTA). A DNA Clean and Concentrator kit (Zymo Research, BaseClear, Netherlands) was used according to the producer’s protocol to remove any contaminants able to interfere with PCR and sequencing.
For subsequent DNA extract quality control, the 16S rRNA gene V3-V4 hypervariable regions were amplified by PCR with Taq DNA Polymerase and the Fermentas PCR Master Mix Kit according to the manufacturers’ specifications (Thermo Fisher Scientific, Waltham, MA, USA) using primers 341F (5’-CCT ACG GGN GGC WGC AG -3’) and 785Rmod (5’-GAC TAC HVG GGT ATC TAA KCC-3’). The reverse primer was adapted from Klindworth et al. (2013), to increase coverage. The V3-V4 hypervariable region of the 16SrRNA gene was chosen as these regions have the best “environments” for the best pick-up rate during 2 x 250 base pair Illumina Miseq sequencing Klindworth et al. (2013). The obtained PCR product was run along with the DNA extract as well as a GeneRuler DNA Ladder Mix (Thermo Fisher Scientific, Waltham, MA, USA) on a 2% agarose gel for 30 minutes at 100 V as a control. The genomic DNA of the samples was then sent out to LGC genomics GmbH (Berlin, Germany) for library preparation and sequencing on an Illumina Miseq platform with V3 chemistry. The microbial data from the HMS Royal Oak sediments were downloaded from the European Nucleotide Archive database under accession number PRJEB37440, these data were obtained by Thomas et al. (2021).
The sediment subsamples of each 3-4 grams sediment were dried for 48 h at 55°C and weighed again. Metal content analysis was performed according to the official 2010 VITO (Dataset VITO, 2010; Dataset VITO, 2020) destruction method for solid material (CMA/2/II/A.3). In short, 0.5 g of dried sediment was digested and metals were extracted with a mixture of HCl, HF and HNO3 in a 20 min during temperature cycle with a microwave, followed by the addition of a Hipt3BO3 microwaved for another 3 minutes. Samples were subsequently filtered and run on an inductively coupled plasma-optical emission spectroscopy (Thermo Fisher iCap Q). The accuracy of the analysis was verified using BCR-277R estuarine sediment (certified reference material, European Commission).
All data analysis was performed in R (version 4.0.3). The DADA2 R package was used to process the amplicon sequence data according to the pipeline tutorial (Callahan et al., 2016). In a first quality control step, the primer sequences were removed and reads were truncated at a quality score cut-off (truncQ = 2). Besides trimming, additional filtering was performed to eliminate reads containing any ambiguous base calls or reads with high expected errors (maxEE = 2,2). After dereplication, unique reads were further denoised using the Divisive Amplicon Denoising Algorithm (DADA) error estimation algorithm and the selfConsist sample inference algorithm (with the option pooling = TRUE). The obtained error rates were inspected and after approval, the denoised reads were merged. Subsequently, the amplicon sequence variant (ASV) table obtained after chimera removal was used for taxonomy assignment using the Naive Bayesian Classifier and the DADA2 formatted Silva v138 (Quast et al., 2013). ASV’s mapping back to anything other than ‘Bacteria’ were filtered out as they were considered technical noise. Singletons, reads occurring only once in all samples were considered noise and were removed (McMurdie and Holmes, 2014). In all analyses of bacteria, we removed chloroplast/mitochondria sequences. In analyses of phytoplankton, we only used chloroplast/mitochondria sequences. All the ‘chloroplast’ ASV’s were separately reassigned using the 16S rRNA sequence taxonomy from the plastid, apicoplast, chromatophore and mitochondrion part of the pr2 database (Decelle et al., 2015; Derelle et al., 2016; Gaonkar et al., 2018; Adl et al., 2019) to identify the chloroplasts present in the sample as a proxy of photosynthetic eukaryotes as performed in a similar workflow by both Fuentes et al. (2019) and Tamayo-Leiva et al. (2021).
Further data analysis was performed using statistical packages like Phenoflow (v1.1.2) (Props et al., 2016) for flow cytometric data analysis, Phyloseq (v1.22.3) (McMurdie and Holmes, 2013) for ASV’s data handling, vegan (v2.5.6) (Dixon, 2003) and betapart for diversity analysis of ASV’s, ancombc (v2) (Lin and Peddada, 2020) for significant higher/lower abundance of ASV’s. To assess significant changes due to the imposed environmental conditions, (PERM)ANOVA analysis was used, using a cut-off of (adjusted) p-value<0.05. Furthermore, a Pearson’s correlation was performed in R to correlate environmental conditions (heavy metal concentration, organic pollutant concentration) with the abundance of specific ASV’s.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: BioProject number PRJNA800643 in the NCBI database (https://www.ncbi.nlm.nih.gov/).
JV: Formal analysis, Investigation, Writing - original draft, Visualization. KK: Methodology, Writing - review and editing, Supervision. SV: Methodology, Investigation. MN: Methodology, Investigation. KP: Methodology, Writing - review and editing, Supervision. MD: Conceptualization, Methodology, Writing - review and editing, Supervision, Project administration, Funding acquisition. NB: Conceptualization, Methodology, Writing - review and editing, Supervision, Project administration, Funding acquisition. All authors contributed to the article and approved the submitted version.
The research leading to the results presented in this publication was carried out with infrastructure funded by the BOF.GOA.2015.0002.01 BOF15/GOA/006 project awarded by BOF and the “North Sea Wrecks” project awarded by the EU Interreg North Sea Region Programme.
Dr. Ruth Plets for her help in the acquisition and processing of multibeam imagery. Volunteers of the VLIZ Scientific diving team (Marjan Steppe, Jessica Vandevelde). The crew of the RV Simon Stevin. Uwe Wilchert for providing eye-witness reports on the sinking of V-1302 from the German Military Records in the Bundesarchiv in Freiburg, Germany. The EU Interreg North Sea Region programme for their financial support of the “North Sea Wrecks” project. Dr. Karel Folens for his help with the metal analysis. Dr. Frederiek-Maarten Kerckhof for his suggestions and his help on the microbial data analysis.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.1017136/full#supplementary-material
Adl S. M., Bass D., Lane C. E., Lukeš J., Schoch C. L., Smirnov A., et al. (2019). Revisions to the classification, nomenclature, and diversity of eukaryotes. J. Eukaryotic Microbiol. 66, 4–119. doi: 10.1111/jeu.12691
Almeida E., Diamantino T. C., de Sousa O. (2007). Marine paints: The particular case of antifouling paints. Prog. Organic Coatings 59, 2–20. doi: 10.1016/j.porgcoat.2007.01.017
Bacosa H. P., Erdner D. L., Rosenheim B. E., Shetty P., Seitz K. W., Baker B. J., et al. (2018). Hydrocarbon degradation and response of seafloor sediment bacterial community in the northern gulf of Mexico to light Louisiana sweet crude oil. ISME J. 12, 2532–2543. doi: 10.1038/s41396-018-0190-1
Bauer B., Buzek C., Daeves D., Cremer T, Daeves K, Dornhecker K, et al. (1925). “Handbuch der eisen- und stahlgießerei,” in Erster band grundlagen (Berlin, Heidelberg: Springer Berlin Heidelberg). zweite, erweiterte auflage edn.
Baustian M. M., Rabalais N. N., Morrison W. L., Turner R. E. (2011). Seasonal microphytobenthos on the hypoxic northern gulf of Mexico continental shelf. Mar. Ecol. Prog. Ser. 436, 51–66. doi: 10.3354/meps09262
Buchan A., González J. M., Chua M. J. (2019). “Aerobic hydrocarbon-degrading alphaproteobacteria: Rhodobacteraceae (Roseobacter),” in Taxonomy, genomics and ecophysiology of hydrocarbon-degrading microbes. Ed. McGenity T. J. (Cham: Springer International Publishing), 93–104. doi: 10.1007/978-3-030-14796-9_8
Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J. A., Holmes S. P. (2016). DADA2: High-resolution sample inference from illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Chapman M., Tolhurst T., Murphy R., Underwood A. (2010). Complex and inconsistent patterns of variation in benthos, micro-algae and sediment over multiple spatial scales. Mar. Ecol. Prog. Ser. 398, 33–47. doi: 10.3354/meps08328
Chatzigiannidou I., Props R., Boon N. (2018). Drinking water bacterial communities exhibit specific and selective necrotrophic growth. NPJ Clean Water 1, 1–4. doi: 10.1038/s41545-018-0023-9
Cibic T., Fazi S., Nasi F., Pin L., Alvisi F., Berto D., et al. (2019). Natural and anthropogenic disturbances shape benthic phototrophic and heterotrophic microbial communities in the po river delta system. Estuarine Coast. Shelf Sci. 222, 168–182. doi: 10.1016/j.ecss.2019.04.009
Czub M., Nawała J., Popiel S., Dziedzic D., Brzeziński T., Maszczyk P., et al. (2020). Acute aquatic toxicity of sulfur mustard and its degradation products to daphnia magna. Mar. Environ. Res. 161, 105077. doi: 10.1016/j.marenvres.2020.105077
Dataset Commission, O (2017). Status and trends in the concentrations of polycyclic aromatic hydrocarbons (PAHs) in sediment.
Dataset VITO (2010). Gesloten en semi-open microgolfoven destructiemethode met salpeterzuur, zoutzuur en waterstoffluoride.
Dataset VITO (2020). Compendium voor monsterneming en analyses van afvalstoffen en bodem (CMA). EMIS.
Decelle J., Romac S., Stern R. F., Bendif E. M., Zingone A., Audic S., et al. (2015). PhytoREF: a reference database of the plastidial 16S rRNA gene of photosynthetic eukaryotes with curated taxonomy. Mol. Ecol. Resour. 15, 1435–1445. doi: 10.1111/1755-0998.12401
Dell’Anno F., Rastelli E., Tangherlini M., Corinaldesi C., Sansone C., Brunet C., et al. (2021). Highly contaminated marine sediments can host rare bacterial taxa potentially useful for bioremediation. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.584850
Derelle R., López-García P., Timpano H., Moreira D. (2016). A phylogenomic framework to study the diversity and evolution of stramenopiles (=Heterokonts). Mol. Biol. Evol. 33, 2890–2898. doi: 10.1093/molbev/msw168
Dixon P. (2003). VEGAN, a package of r functions for community ecology. J. Vegetation Sci. 14, 927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x
Enning D., Venzlaff H., Garrelfs J., Dinh H. T., Meyer V., Mayrhofer K., et al. (2012). Marine sulfate-reducing bacteria cause serious corrosion of iron under electroconductive biogenic mineral crust. Environ. Microbiol. 14, 1772–1787. doi: 10.1111/j.1462-2920.2012.02778.x
Fuentes S., Arroyo J. I., Rodríguez-Marconi S., Masotti I., Alarcón-Schumacher T., Polz M. F., et al. (2019). Summer phyto- and bacterioplankton communities during low and high productivity scenarios in the Western Antarctic peninsula. Polar Biol. 42, 159–169. doi: 10.1007/s00300-018-2411-5
Gaonkar C. C., Piredda R., Minucci C., Mann D. G., Montresor M., Sarno D., et al. (2018). Annotated 18S and 28S rDNA reference sequences of taxa in the planktonic diatom family chaetocerotaceae. PLoS One 13, e0208929. doi: 10.1371/journal.pone.0208929
Gwizdala M., Jelenska M., Leczynski L. (2018). “Surface sediments pollution around small shipwrecks (Munin and abille) in the gulf of gdańsk: Magnetic and heavy metals study,” in Magnetometry in environmental sciences: Studying environmental structure changes and environmental pollution. Eds. Jelenska M., Leczynski L., Ossowski T. (Cham: Springer International Publishing), 37–50. GeoPlanet: Earth and Planetary Sciences. doi: 10.1007/978-3-319-60213-4_3
Hamdan L. J., Salerno J. L., Reed A., Joye S. B., Damour M. (2018). The impact of the deepwater horizon blowout on historic shipwreck-associated sediment microbiomes in the northern gulf of Mexico. Sci. Rep. 8, 9057. doi: 10.1038/s41598-018-27350-z
Jiao S., Chen W., Wang E., Wang J., Liu Z., Li Y., et al. (2016). Microbial succession in response to pollutants in batch-enrichment culture. Sci. Rep. 6, 21791. doi: 10.1038/srep21791
Kelly L. W., Barott K. L., Dinsdale E., Friedlander A. M., Nosrat B., Obura D., et al. (2012). Black reefs: iron-induced phase shifts on coral reefs. ISME J. 6, 638–649. doi: 10.1038/ismej.2011.114
Klindworth A., Pruesse E., Schweer T., Peplies J., Quast C., Horn M., et al. (2013). Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41, e1. doi: 10.1093/nar/gks808
Koske D., Goldenstein N. I., Rosenberger T., Machulik U., Hanel R., Kammann U. (2020). Dumped munitions: New insights into the metabolization of 2,4,6-trinitrotoluene in Baltic flatfish. Mar. Environ. Res. 160, 104992. doi: 10.1016/j.marenvres.2020.104992
Lake S. J., Brush M. J. (2011). The contribution of microphytobenthos to total productivity in upper Narragansett bay, Rhode island. Estuarine Coast. Shelf Sci. 95, 289–297. doi: 10.1016/j.ecss.2011.09.005
Landquist H., Hassellöv I. M., Rosén L., Lindgren J. F., Dahllöf I. (2013). Evaluating the needs of risk assessment methods of potentially polluting shipwrecks. J. Environ. Manage. 119, 85–92. doi: 10.1016/j.jenvman.2012.12.036
Landquist H., Norrman J., Lindhe A., Norberg T., Hassellöv I. M., Lindgren J. F., et al. (2017). Expert elicitation for deriving input data for probabilistic risk assessment of shipwrecks. Mar. Pollut. Bull. 125, 399–415. doi: 10.1016/j.marpolbul.2017.09.043
Lin H., Peddada S. D. (2020). Analysis of compositions of microbiomes with bias correction. Nat. Commun. 11, 3514. doi: 10.1038/s41467-020-17041-7
Magnusson M., Heimann K., Ridd M., Negri A. P. (2013). Pesticide contamination and phytotoxicity of sediment interstitial water to tropical benthic microalgae. Water Res. 47, 5211–5221. doi: 10.1016/j.watres.2013.06.003
Mandić J., Vrančić M. P. (2017). Concentrations and origin of polycyclic aromatic hydrocarbons in sediments of the middle Adriatic Sea. Acta Adriatica 58, 3–24. doi: 10.32582/aa.58.1.1
Marini M., Frapiccini E. (2013). Persistence of polycyclic aromatic hydrocarbons in sediments in the deeper area of the northern Adriatic Sea (Mediterranean Sea). Chemosphere 90, 1839–1846. doi: 10.1016/j.chemosphere.2012.09.080
Martínez-Gómez C., Vethaak A. D., Hylland K., Burgeot T., Köhler A., Lyons B. P., et al. (2010). A guide to toxicity assessment and monitoring effects at lower levels of biological organization following marine oil spills in European waters. ICES J. Mar. Sci. 67, 1105–1118. doi: 10.1093/icesjms/fsq017
McBeth J. M., Emerson D. (2016). In situ microbial community succession on mild steel in estuarine and marine environments: Exploring the role of iron-oxidizing bacteria. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.00767
McMurdie P. J., Holmes S. (2013). Phyloseq: An r package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8, e61217. doi: 10.1371/journal.pone.0061217
McMurdie P. J., Holmes S. (2014). Waste not, want not: Why rarefying microbiome data is inadmissible. PLoS Comput. Biol. 10, e1003531. doi: 10.1371/journal.pcbi.1003531
Pinckney J. L., Hagenbuch I. M., Long R. A., Lovell C. R. (2013). Sublethal effects of the antibiotic tylosin on estuarine benthic microalgal communities. Mar. pollut. Bull. 68, 8–12. doi: 10.1016/j.marpolbul.2013.01.006
Price K. A., Garrison C. E., Richards N., Field E. K. (2020). A shallow water ferrous-hulled shipwreck reveals a distinct microbial community. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.01897
Props R., Monsieurs P., Mysara M., Clement L., Boon N. (2016). Measuring the biodiversity of microbial communities by flow cytometry. Methods Ecol. Evol. 7, 1376–1385. doi: 10.1111/2041-210X.12607
Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., et al. (2013). The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Rajala P., Cheng D.-Q., Rice S. A., Lauro F. M. (2022). Sulfate-dependant microbially induced corrosion of mild steel in the deep sea: A 10-year microbiome study. Microbiome 10, 4. doi: 10.1186/s40168-021-01196-6
Rochman F. F., Sheremet A., Tamas I., Saidi-Mehrabad A., Kim J.-J., Dong X., et al. (2017). Benzene and naphthalene degrading bacterial communities in an oil sands tailings pond. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.01845
Rodacy P., Reber S., Walker P., Andre J. (2001). Chemical sensing of explosive targets in the Bedford basin, Halifax, Nova Scotia. Tech. Rep. SAND2001-3569, 789594. doi: 10.2172/789594
Rogowska J., Kudłak B., Tsakovski S., Gałuszka A., Bajger-Nowak G., Simeonov V., et al. (2015). Surface sediments pollution due to shipwreck s/s “Stuttgart”: a multidisciplinary approach. Stoch Environ. Res. Risk Assess. 29, 1797–1807. doi: 10.1007/s00477-015-1054-0
Rogowska J., Wolska L., Namieśnik J. (2010). Impacts of pollution derived from ship wrecks on the marine environment on the basis of s/s “Stuttgart” (Polish coast, Europe). Sci. Total Environ. 408, 5775–5783. doi: 10.1016/j.scitotenv.2010.07.031
Smith M., Bardiau M., Brennan R., Burgess H., Caplin J., Ray S., et al. (2019). Accelerated low water corrosion: the microbial sulfur cycle in microcosm. NPJ Mater Degrad 3, 1–11. doi: 10.1038/s41529-019-0099-9
Sternheim J., Rytkönen J., Helavuori M.. (2019). SUBMERGED – HELCOM: Warfare materials in the Baltic Sea. Finland: techreport. pp. 141. Available at: https://portal.helcom.fi/meetings/RESPONSE%2028-2020-743/MeetingDocuments/9-2%20Att.2%20Comments%20on%20the%20draft%20Submerged%20Assessment.pdf
Sundbäck K., Alsterberg C., Larson F. (2010). Effects of multiple stressors on marine shallow-water sediments: Response of microalgae and meiofauna to nutrient–toxicant exposure. J. Exp. Mar. Biol. Ecol. 388, 39–50. doi: 10.1016/j.jembe.2010.03.007
Tamayo-Leiva J., Cifuentes-Anticevic J., Aparicio-Rizzo P., Arroyo J. I., Masotti I., Díez B. (2021). Influence of estuarine water on the microbial community structure of Patagonian fjords. Front. Mar. Sci., 8. doi: 10.3389/fmars.2021.611981
Thomas G. E., Bolam S. G., Brant J. L., Brash R., Goodsir F., Hynes C., et al. (2021). Evaluation of polycyclic aromatic hydrocarbon pollution from the HMS royal oak shipwreck and effects on sediment microbial community structure. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.650139
Trzesicka-Mlynarz D., Ward O. P. (1995). Degradation of polycyclic aromatic hydrocarbons (PAHs) by a mixed culture and its component pure cultures, obtained from PAH-contaminated soil. Can. J. Microbiol. 41, 470–476. doi: 10.1139/m95-063
Keywords: microbial ecology, shipwreck microbiome, heavy metal contamination, aromatic hydrocarbon, microalgal chloroplasts
Citation: Van Landuyt J, Kundu K, Van Haelst S, Neyts M, Parmentier K, De Rijcke M and Boon N (2022) 80 years later: Marine sediments still influenced by an old war ship. Front. Mar. Sci. 9:1017136. doi: 10.3389/fmars.2022.1017136
Received: 11 August 2022; Accepted: 15 September 2022;
Published: 18 October 2022.
Edited by:
Shan He, Ningbo University, ChinaReviewed by:
Erin Field, East Carolina University, United StatesCopyright © 2022 Van Landuyt, Kundu, Van Haelst, Neyts, Parmentier, De Rijcke and Boon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nico Boon, Tmljby5Cb29uQFVHZW50LmJl
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.