- 1Key Laboratory of Marine Drugs, Chinese Ministry of Education, School of Medicine and Pharmacy, Ocean University of China, Qingdao, China
- 2Laboratory for Marine Drugs and Bioproducts, Pilot National Laboratory for Marine Science and Technology, Qingdao, China
- 3Lab of Marine Medicinal Resources Discovery, Marine Biomedical Research Institute of Qingdao, Qingdao, China
Four new ACTG-toxins like meroterpenoids tricycloalternarenes O-R were discovered from a marine-derived fungus Alternaria alternata JJY-32 by the overexpression of a LaeA-like global transcriptional regulator and a MpaB-like biosynthetic tailoring enzyme. Among those structures, tricycloalternarene O possesses a non-canonical cyclohexanone-pyran fused furan ring structure and tricycloalternarene R contains a truncated isoprenoid terminal. The structures were elucidated based on comprehensive 1D and 2D NMR data and assisted by ECD calculations. Tricycloalternarenes O-R showed potent anti-inflammatory activity in TLR4-transfected microphage cells (RAW264.7).
Introduction
Marine Microorganisms are remarkable producers of therapeutic compounds that significantly improve human health and well-being. However, the speed of new bioactive compound discovery has been slowing down due to the consistent employment of conventional cultivation and isolation procedure. Recently, benefiting from the development of genome sequencing, genome mining, and gene transfer and expressing technologies, a number of new approaches have been developed to alleviate this challenge (Albarano et al., 2020; Chu et al., 2020). Among those methods, the overexpression of the secondary metabolism regulator (Bayram and Braus, 2012; Gacek and Strauss, 2012) or critical biosynthetic genes (Shin et al., 2012; Li et al., 2016) has been demonstrated as an effective tool for triggering the expression of the inert biosynthetic pathway and providing diversified secondary metabolites.
The genus Alternaria produces a broad diversity of secondary metabolites with a large range of bioactivities (Bräse et al., 2009; Lou et al., 2013; Lou et al., 2013; Abdessemed et al., 2021; Wang et al., 2022). ACTG toxins are a family of meroterpenoids mainly discovered from Alternaria alternata and Alternaria citri. As a group of phytotoxins, the skeleton of ACTG toxins particularly contains a terpenoid part derived from a farnesyl unit and a non-terpenoid part which is usually present in the form of six-membered cyclic α, β-unsaturated ketone (Kono et al., 1986; Liebermann et al., 1997; Liebermann et al., 2000; Logrieco et al., 2009; Tsuge et al., 2013; Wang et al., 2013; Shen et al., 2018). ACTG toxin derivatives usually possess the ability to bind membrane proteins thus affecting the permeability of the membrane structure and acting on signaling receptors to exert a variety of biological activities such as antibacterial, antiparasitic, antitumor, and anti-inflammatory effects (Matsumoto et al., 1992; Horiuchi et al., 2006; Gozari et al., 2021).
During our previous work in searching bioactive secondary metabolites from marine-derived fungi, the strain A. alternata JJY-32 was obtained from a sea sponge sample collected from the South China Sea. One Strain Many Compounds (OSMAC) strategy guided chemical studies on JJY-32 have provided a series of ACTG toxin derivatives that showed promising anti-inflammatory activity (Yang et al., 2013; Zhang et al., 2013; Wang et al., 2020). In a recent work, with the intent to obtain diversified ACTG toxin structures, we sequenced the genome and processed genome mining on JJY-32. As a result, one gene analog of LaeA, a common global regulation gene of secondary metabolism in filamentous fungi (Kosalková et al., 2009; Jiang et al., 2016; Yu et al., 2019; Wang et al., 2020), and one analog of MpaB (Zhang et al., 2019), a terpenoid-trimming enzyme in mycophenolic acid biosynthesis, are screened out for the potential to modulate ACTG toxin production based on phylogenetic analysis (Figure S1 and Figure S2). The overexpression of the above genes successfully altered the metabolic profile of A. alternata and further chemical investigation afforded four new ACTG toxin derivatives with anti-inflammatory activity. Herein, we will describe the details of metabolic profile modulation through the overexpression of aLaeA and aMpaB, the isolation and characterization of new ACTG toxins, as well as bioactivity evaluation of obtained structures.
Results and discussion
Metabolic profile modulation by overexpression of aLaeA and aMpaB
A group of transcriptional regulators such as VeA, HogA, CmrA, and LaeA has been discovered for their ability in modulating growth morphology, asexual development, and mycotoxin production in A. alternata strains (Fetzner et al., 2014; Estiarte et al., 2016; Igbalajobi et al., 2019; Gao et al., 2022). We sequenced JJY-32 followed by genome mining using bioinformatics tools of LocalBLAST and InterPro analysis. A homologue of LaeA (named aLaeA) with a length of 1583 bp encoding 301 amino acids and one homolog of MpaB (named aMpaB) with a length of 2133 bp encoding 710 amino acids were identified from the genome of JJY-32 using LaeA (BAP58881.1) and MpaB (AJG44380.1) as references. Further protein sequence alignment and phylogenetic analysis revealed the S-adenosyl-L-methionine-dependent methyltransferase feature for aLaeA and the oxygenase feature for MpaB, which indicated their potent modulation functions in secondary metabolites production (Figure S1 and Figure S2). The genes aLaeA and aMpaB were PCR amplified from genomic DNA and inserted into the pHyg vector to generate OE-alaeA and OE-aMpaB (Figures S3 and S4). The recombinant vectors were transformed into wild-type JJY-32 to give the engineered strains JJY32-OE-alaeA and JJY32- OE-aMpaB.
Strains JJY32-OE-alaeA and JJY32-OE-aMpaB were fermented separately in 10 L PDB medium and the High-Performance Liquid Chromatography (HPLC) analysis of the crude extracts showed altered metabolic profiles for both engineered strains with new peaks presenting at 23.6 min for JJY32-OE-alaeA and at 21.6, 26.8 and 27.6 min for JJY32-OE-aMpaB (Figure 1A), which indicated the capacity for new secondary metabolites production.
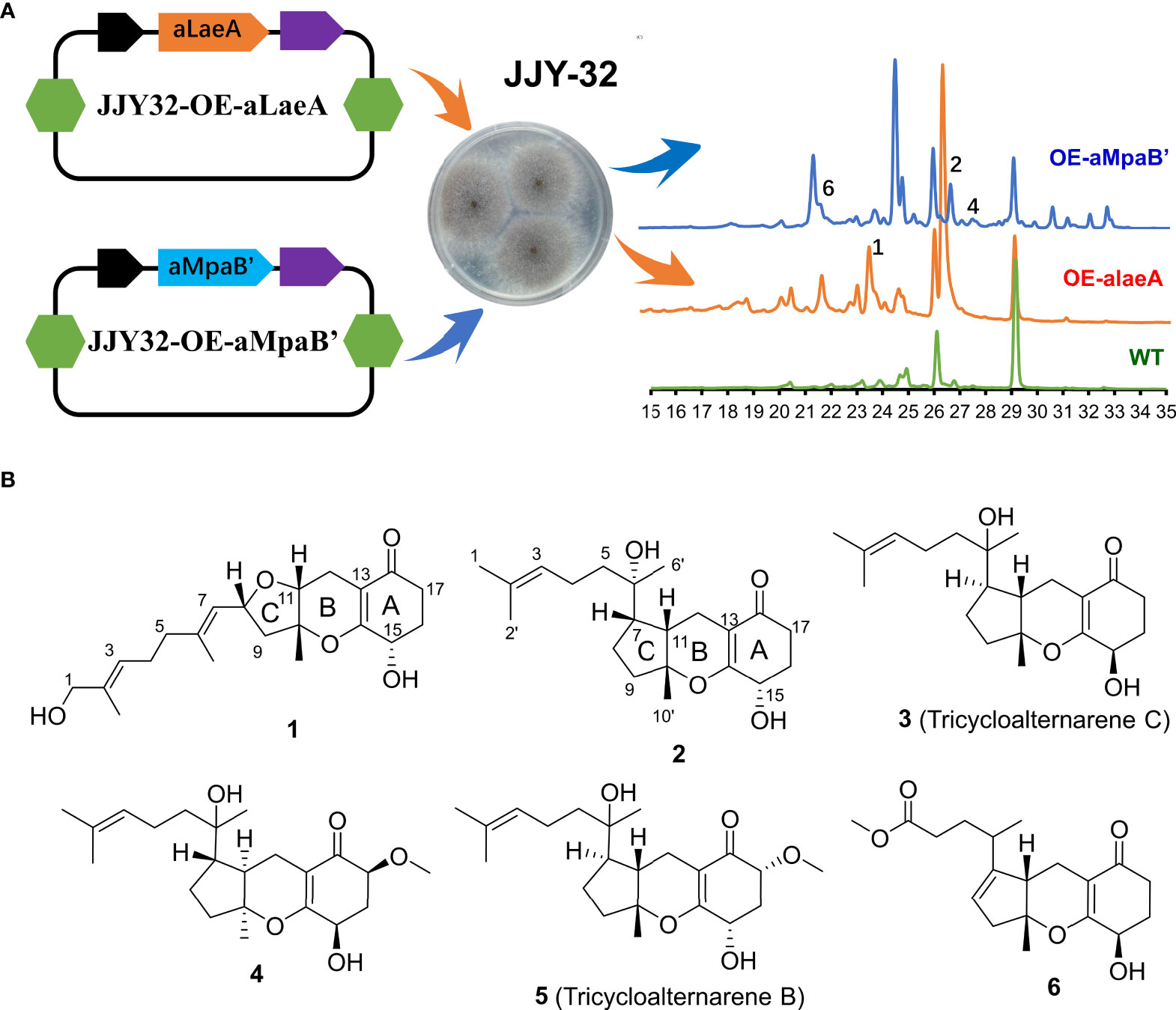
Figure 1 The overexpression of aLaeA and aMpaB in A alternata JJY-32. (A) Heterologous expression of aLaeA and aMpaB altered the metabolic profile of JJY-32 and led to the production of compounds 1-2, 4, and 6 by HPLC (mobile phase: 5% methanol, 5 min; 100% methanol, 35 min); (B) Structure of compounds obtained from cultures of JJY32-OE-alaeA and JJY32- OE-aMpaB.
Compound isolation and structure elucidation
The EtOAc extracts of strain JJY32-OE-alaeA (10.0 g) and JJY32-OE-aMpaB (15.0 g) were subjected to vacuum-liquid chromatography (VLC), Medium Pressure Liquid Chromatography (MPLC) column chromatography and HPLC semi-preparative column chromatography for purification. Compound 1 was obtained from the crude extract of JJY32-OE-alaeA, and compounds 2-6 were isolated from the extract of JJY32-OE-aMpaB.
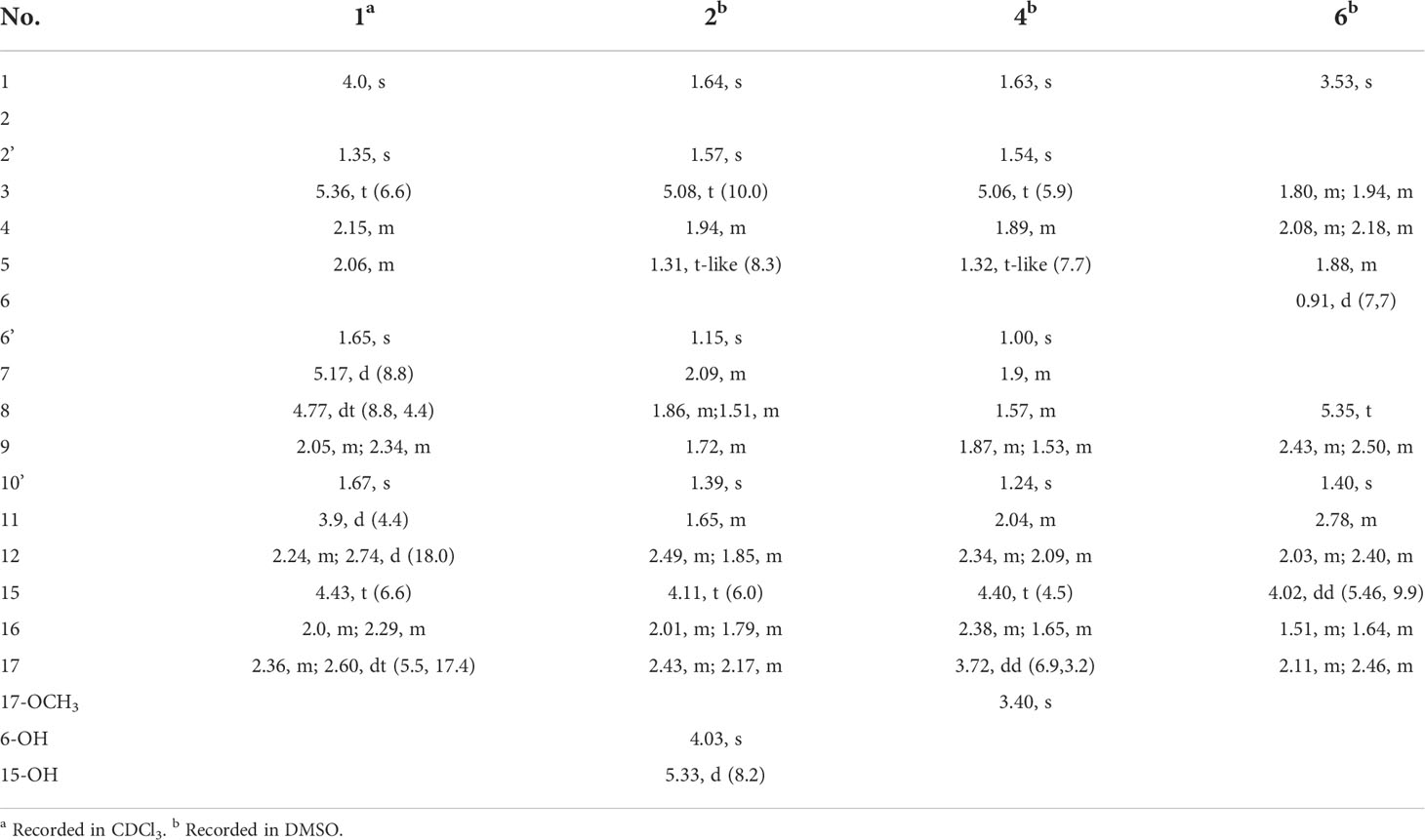
Compound 1 was obtained as a colorless oil with the [M + H]+ ion at m/z 363.2186 in HRESIMS indicative of a molecular formula of C21H30O5, requiring seven double bond equivalents (DBEs). The 1H NMR and 13C NMR spectroscopic data of compound 1 displayed three methyls, seven methylenes with one attached to oxygen, three oxygen-attached sp3 methines, two sp2 methines, and six quaternary carbons including one conjugated carbonyl carbon, four sp2 quaternary carbons, and one oxygenated sp3 quaternary carbon (Tables 1 and 2). Above information counted for four DBEs, which indicated a tricyclic structure for 1. The 1H-1H COSY correlations of H-3/H-4/H-5, H-7/H-8/H-9, H-11/H-12, and H-15/H-16/H-17, together with the HMBC correlations from H3-2′ to C-1, C-2 and C-3, from H3-6′ to C-5, C-6 and C-7, from H3-10′ to C-9, C-10 and C-11, from H2-12 to C-13, C-14 and C-18, from H2-15 to C-9, C-10, and from H2-17 to C-18 established the cyclohexanone-sesquiterpenoid skeleton as shown in Figure 2A. Based on the formula and the chemical shifts of C-10 (δC 84.3)/C-14 (δC 166.4), and C-8 (δC 70.3)/C-11 (δC 76.4), C-10 and C-8 were respectively linked to C-14 and C-11 via an ether bond to form the rest rings (Rings B and C), thus completing the planar structure of compound 1 (Figure 2A), which was the first example of the ACTG-toxin-like compounds with a cyclohexanone-pyran fused furan ring system. The NOESY correlations of H-1/H-3 and H-5/H-7 defined the E stereochemistry of the double bonds at C-2 and C-6. The NOESY correlations of H8/H10′/H11 assigned the 8R*, 10R*, 11R* relative configuration. To facilitate computational processing and ECD spectra fitting (Nugroho and Morita, 2014), compound 1 was truncated as 1a-1d, where the C-6 side chain was replaced by a methyl group. The ECD spectra of (8R, 10R, 11R, 15R)-1a, (8R, 10R, 11R, 15S)-1b, (8S, 10S, 11S, 15S)-1c, and (8S, 10S, 11S, 15R)-1d were produced using density functional theory (DFT) calculations performed at the B3LYP/6-31+G(d) level. The experimental spectra of compound 1 showed an identical trend with that of (8R, 10R, 11R, 15R)-1a (Figure S6 and Table S5), thus determining the absolute configuration at C-8, C-10, C-11, and C-15. Compound 1 was then named tricycloalternarene O.
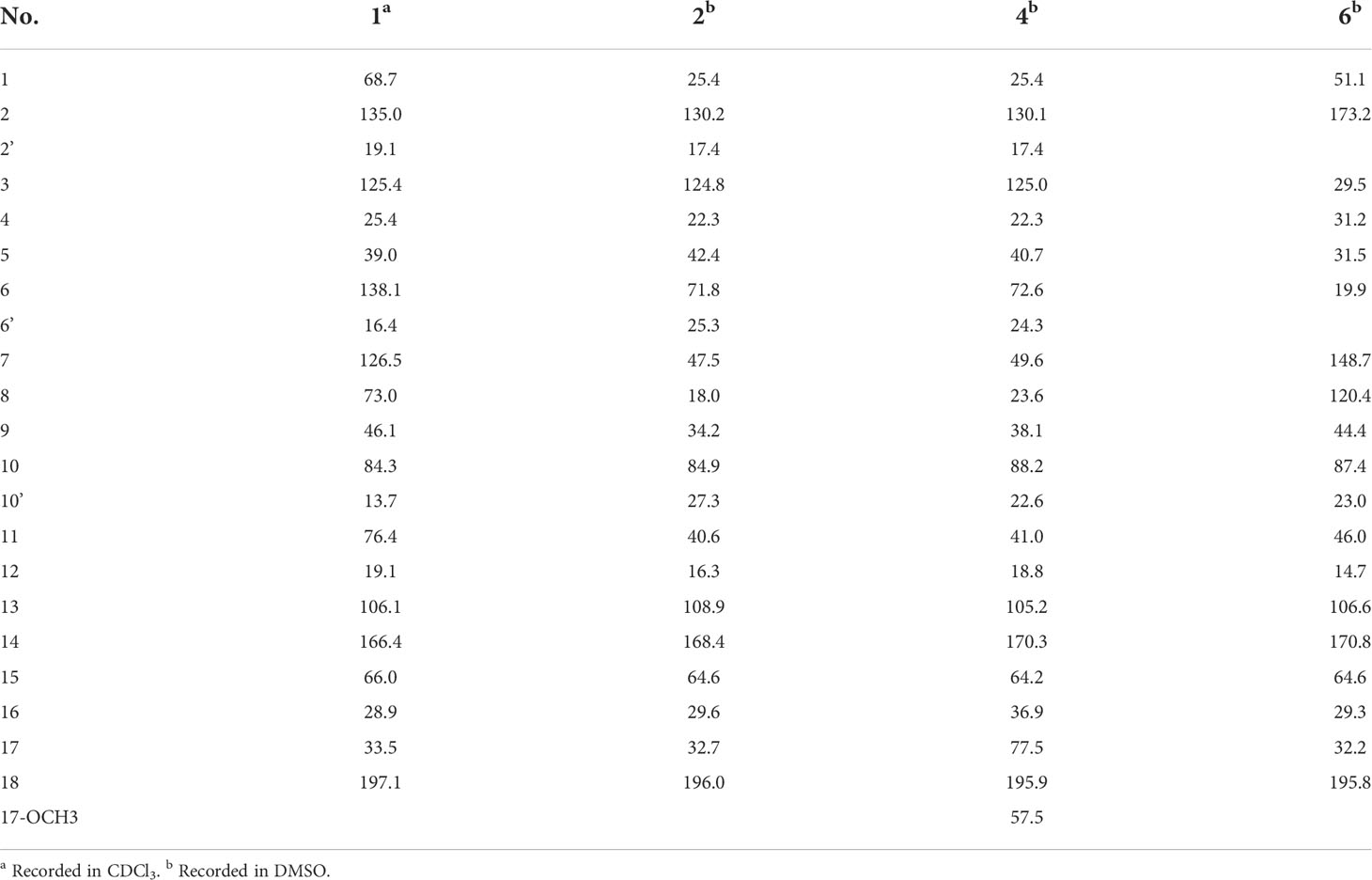
Compound 2 was obtained as a colorless oil. The HRESIMS [M + H]+ peak at m/z 349.2383 indicated a molecular formula of C21H32O4. The 1H and 13C NMR spectra indicated four methyls, seven sp3 methylenes, three sp3 methines, one olefinic methine, two oxygenated sp3 quaternary carbons, and four sp2 quaternary carbons including a conjugated carbonyl carbon (Tables 1 and 2). The above information showed considerable resemblance to that of tricycloalternarene C (3), an ACTG toxin derivative isolated from JJY-32 by our previous study (Zhang et al., 2013). The COSY correlations of OH-15/H-15/H-16/H-17, H-9/H-8/H-7/H-11/H-12, and H-3/H-4/H-5, together with the HMBC correlations from OH-15 to C-13 and C-14, from H2-17 to C-18, from H-1 and H-2′ to C-3, from H3-6′ to C-5, C-6, and C-7, from H3-10′ to C-9, C-10, and C-11, and from H2-12 to C-13, C-14, and C-18 confirmed that compound 2 possess the same planar structure as 3 (Figure 2B), while the different CD spectra (Zhang et al., 2013) indicated the different absolute configurations at the stereo-centers (Figure 3B).
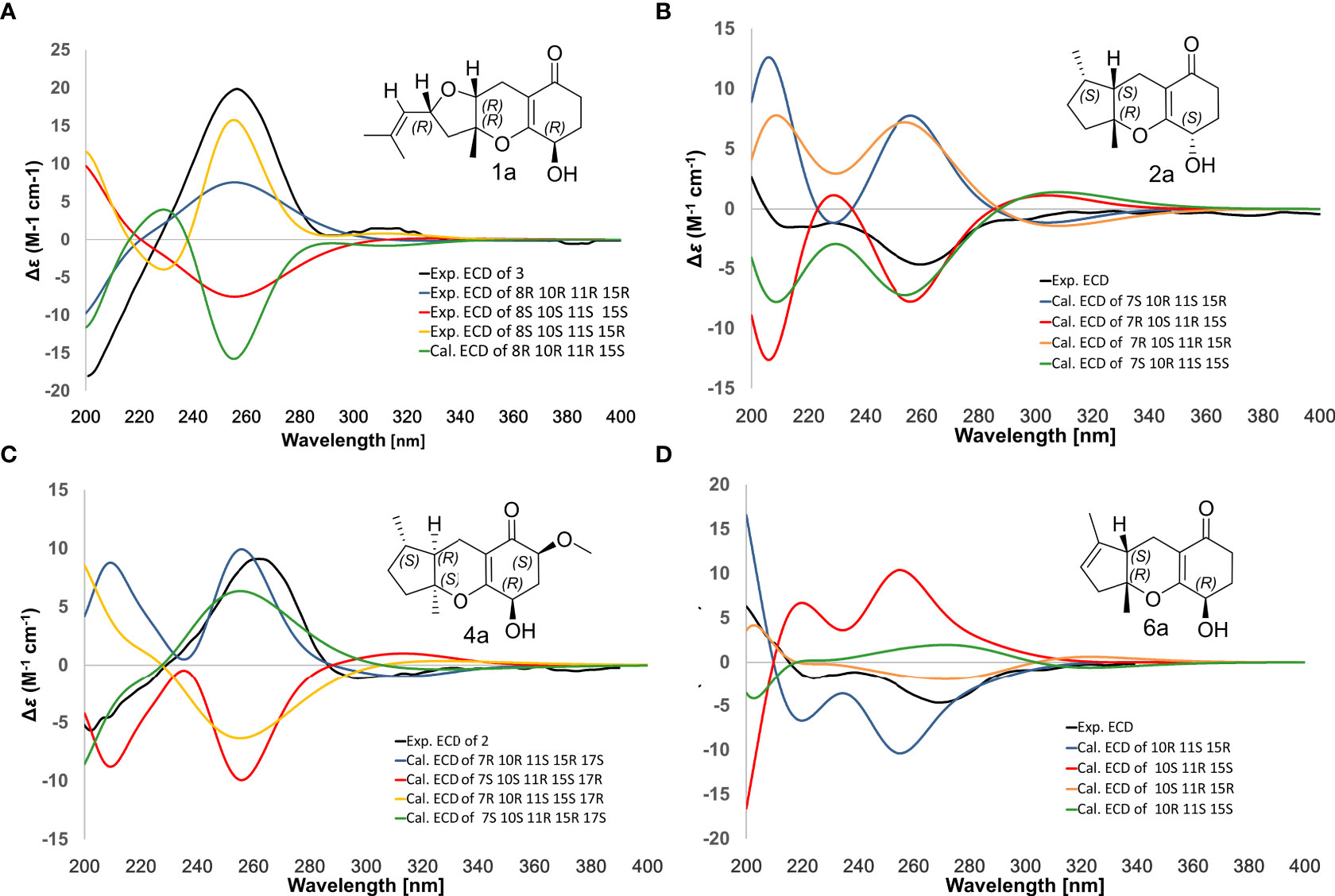
Figure 3 Comparison of experimental/calculated ECD spectra for compounds 1 (A), 2 (B), 4 (C) and 6 (D).
The NOESY correlation between H3-10′ and H-11 revealed the cis fusion between rings B and C, which was inconsistent with ACTG toxin structures from literature reports (Figure 3). NOESY correlation between H3-10′ and H-7 further suggested the 7S*, 10R*, 11S* relative configuration. The electronic circular dichroism (ECD) spectra calculation method was adopted to solve the absolute configuration. By comparing the experimental ECD spectrum with the four output curves (7S, 10R, 11S, 15S; 7R, 10S, 11R, 15S; 7S, 10R, 11S, 15R; 7R, 10S, 11R, 15R) from DFT computational process, the absolute configuration of 2 was determined as 7S, 10R, 11S, 15S.
The configuration at C-6 was tentatively proposed based on a detailed conformational analysis (Figure 2B). In the 2D NOESY spectrum, consistent correlations were observed from H3-6′ (δH 1.15) to H-11 (δH 1.65), H2-12 (δH 2.49 and 1.85) and H-7 (δH 2.09), and from H-7 (δH 2.09) to H2-5 (δH 1.31). However, there was no apparent NOESY correlation between H-7 and 6-OH (δH 4.03), and between H3-6′ and H2-8 (δH 1.86 and 1.51). The above data indicated the 6S configuration. Compound 2 was named tricycloalternarene P.
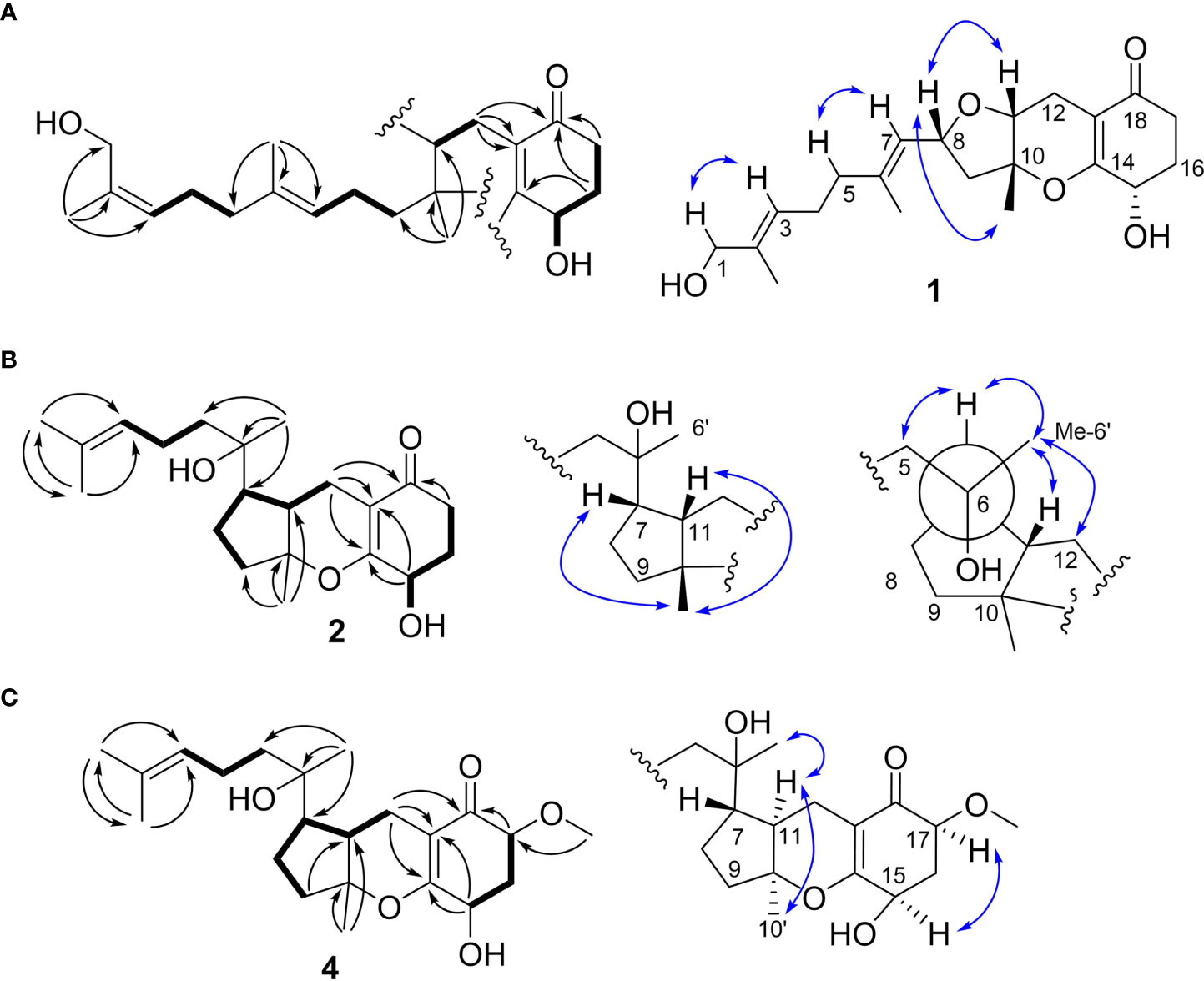
Compound 4, named tricycloalternarene Q, was also obtained as a colorless oil. The molecular composition was determined to be C22H34O5 by HRESIMS and 1D NMR data (Tables 1 and 2). Careful comparison of its NMR spectroscopic data with that of 2 revealed the absence of a methylene and the presence of an extra methine and a methoxyl group. The HMBC correlation from 17-OCH3 to C-17 attached the methoxyl groups to C-17. Further 2D NMR data of 4 completed the same planar structure as tricycloalternarene B (5), an ACTG toxin derivative previously isolated from JJY-32 (Zhang et al., 2013) (Figure 1B). The NOESY correlations between H3-10′ and H-11, H-11 and H3-6′ indicated the 7S*, 10S*, 11R* relative configuration; the NOESY correlations from H-15 to H-17 indicated the 15R*, 17S* relative configuration (Figure 2C). As evidenced from the ECD fitting experiments using the truncated structure 4a as a model structure, the absolute configuration of compound 4 was determined as 7S, 10S, 11R, 15R, 17S.
Compound 6, trivially named tricycloalternarene R, exhibited [M + H]+ ion at m/z 335.1801 accompanied by [M + Na]+ion at m/z 357.1681 in HRESIMS, indicating a molecular formula of C19H26O5. The 1H NMR spectrum (Table 1) showed one methyl doublet at δH 0.91 (J = 7.68 Hz, H-6′), one methyl singlet at δH 1.40 (H-10′), one methoxy singlet at δH 3.53 (CH3O-3), one broad double doublet at δH 4.02 (J = 9.91, 5.46 Hz, H-15) attributable to an oxymethine and one broad singlet at δH 5.35 (H-8) due to an olefinic proton. The 13C NMR spectrum (Table 2) along with DEPT and HSQC data demonstrated the presence of three methyls, six methylenes, four methines and six unprotonated carbons. The above data showed considerable similarity with that of nortricycloalternarate, except for the upfield shift of the chemical shifts for CH-15 (δH 4.02 vs δH 4.38; δC 64.6 vs δC 66.6), indicating the same planar structure as nortricycloalternarate (Shi et al., 2018). However, the CD spectrum of 6 showed the opposite trend at around 280-300 nm compared with nortricycloalternarate, which indicated that 6 was an isomer of nortricycloalternarate. Finally, on considering the conserved co-facial orientation of H-11 and H3-10′, and by comparing the experimental ECD spectrum with the calculated curves presenting the four possible configurations (Figure 3D), the absolute configuration at C-10, C-11, and C-15 was proposed to be 10R, 11S, 15R.
Anti-inflammatory activity evaluation
Toll-like receptors (TLR) associated inflammatory responses usually involve the over-activation of macrophages, which may cause damage to the host by excessive release of various inflammatory mediators such as NO and PGE2, and pro-inflammatory cytokines such as iNOS, COX-2, and IL-6 (Lee et al., 1992; Surh et al., 2001; Lin and Karin, 2007). During this process, NF-κB acts as a key transcription factor complex controlling the expression of cell survival genes as well as those pro-inflammatory cytokines and enzymes (Brown et al., 2007).
The new structures 1-2, 4, and 6 were subjected to cytotoxicity tests using K562 and RAW264 cancer cells, and no cytotoxicity was observed for all the above compounds. Then 1-2, 4, and 6 were evaluated for the anti-inflammatory effects using TLR4-transfected microphage cells (RAW264.7) and human embryonic kidney 293T cells. First, the inhibitive activity of nitric oxide (NO) and prostaglandin E2 (PGE2) production was tested in LPS-activated RAW264.7 cells. As shown in Table 3, the compounds showed moderate inhibition of NO production with IC50 values ranging from 17.5 to 30.6 μM and of PGE2 production with IC50 values ranging from 14.5 to 25.2 μM (curcumin as a positive control with an IC50 value of 5.2 μM), while compounds 1 and 6, whose isopentenyl terminal are oxidized, were revealed to be more potent than compounds 2 and 4 (Table 3).
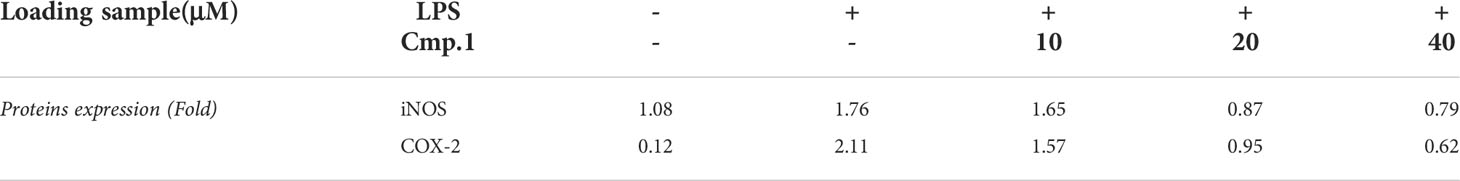
To determine whether the decrease in the production of NO and PGE2 mediated by ACTG-toxins was due to the suppressed activity of upstream signaling molecules, compound 1 was selected to examine the inhibitive effects on LPS-induced expression of the key regulator NF-κB and the pro-inflammatory cytokines of iNOS COX-2 and IL-6 in 293T cells. The NF-κB luciferase activity induced by the overexpression of TLR4 together with its driving adaptor molecule MyD88 (Pasare and Medzhitov, 2004) was suppressed under the presence of 1 (20 μM) (Figures 4A, B). The expression of iNOS and COX-2, two enzymes directly related to the production of NO and PGE2, were also found to be dose-dependent and inhibited by 1 with concentrations from 10 to 40 μM (Figure 4C and Table 4). In addition, the LPS-induced expression of IL-6 was also potently abated by compound 1 treatments (Figure 4D). The above results indicated the potent inhibitory effects of 1 on MyD88-dependent TLRs signaling pathways.
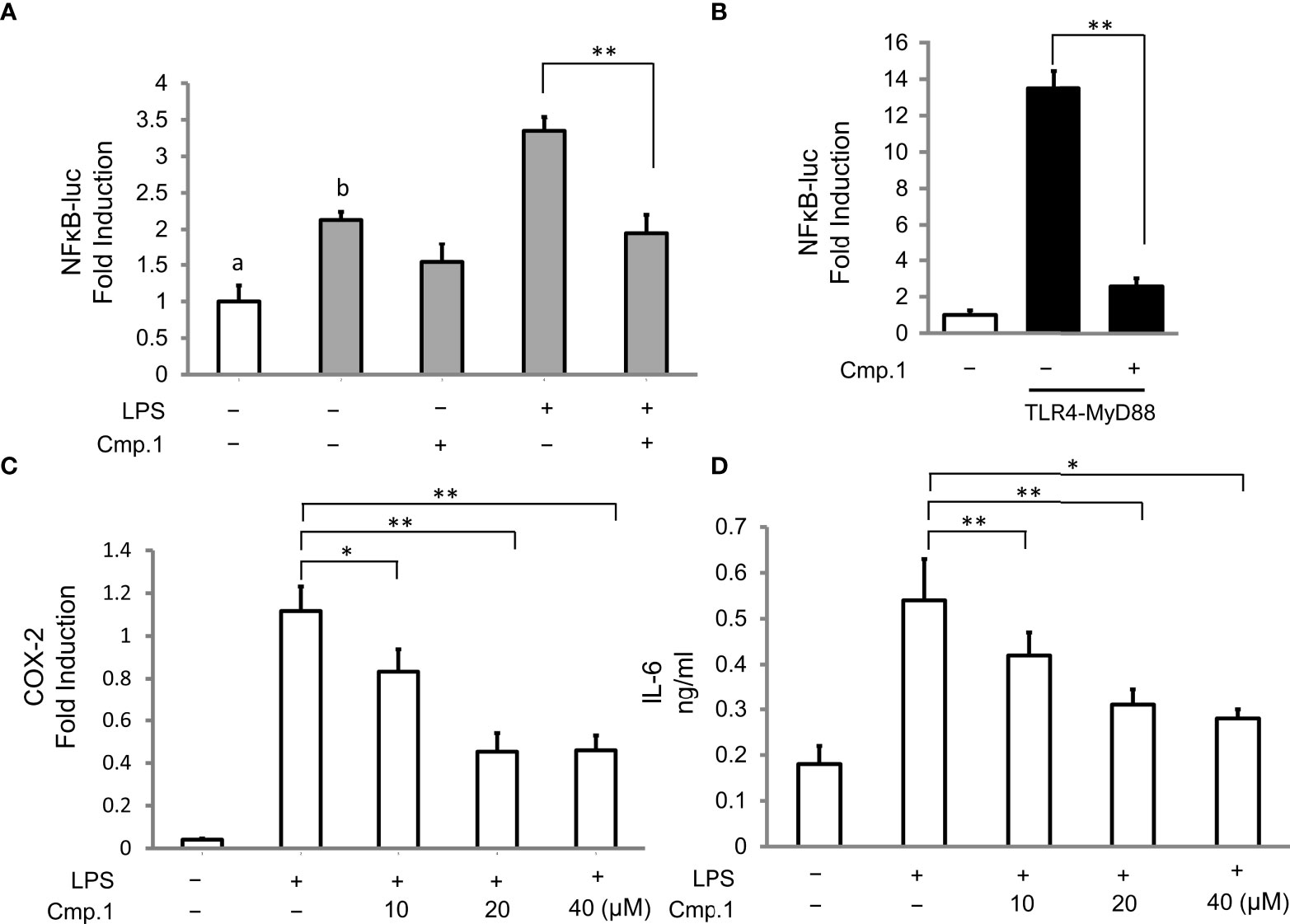
Figure 4 The inhibitive activity of compound 1 on NF-κB, COX-2, and IL-6 production. 293T cells were transfected with the expression of TLR4 (A) or TLR4 plus MyD88 (B) and then stimulated with or without LPS (0.1 μg/mL) in the presence of 1 (20 μM) for 16 (h) 293T cells without TLR4 transfection are marked with “a”, and 293T cells with TLR4 transfection are marked with “b”; RAW264.7 cells were pretreated with the indicated amount of 1 followed by stimulation with LPS (0.1 μg/mL) for 16 (h) COX-2 production was analyzed by Western blotting and presented as relative light units from immunofluorescence microscopy (C), and IL-6 production was determined by ELISA as the cytokine accumulation in the medium after treatment (D). Double asterisks denote a statistically significant difference from the control group at P < 0.01 and a single asterisk denotes P < 0.05 by Tukey’s test. Data are representative of at least three independent experiments.
Conclusion
Four new ACTG-toxin-like meroterpenoids were discovered from a marine-sponge-associated fungus Alternaria sp. JJY-32 using metabolism modulation methods involving overexpression of a metabolism regulator aLaeA and a biosynthetic tailoring enzyme aMpaB. Among those structures, compound 1 was the first example of ACTG-toxin-like compounds with a cyclohexanone-pyran fused furan ring structure, while compound 6 represented a non-canonical ACTG-toxin whose terpenoid part was truncated by losing three carbons, which responded to the modifying function of aMpaB. Meanwhile, compounds exhibited potent anti-inflammatory activity by interfering with MyD88-dependent TLRs signaling pathways in LPS-activated 293T cells. The above results showed overexpression of metabolism regulators and pathway-specific tailoring enzymes could be an efficient method to develop structural diversity of secondary metabolites produced in marine-derived Alternaria strains.
Experimental section
General experimental procedures
The specific rotations were measured on a JASCO P-1020 digital polarimeter. The UV spectra were recorded on a Beckman DU 640 spectrophotometer. The NMR spectra were obtained on an Agilent 500 MHz DD2 spectrometer with TMS as an internal standard. The ESIMS spectra were obtained on a Thermo Scientific LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific) or a Micromass Q-TOF ULTIMA GLOBAL GAA076 LC Mass spectrometer (Waters Corporation). Semi-preparative HPLC was performed on an ODS column (HPLC (YMC-Pack ODS-A, 10 × 250 mm, 5 µm, 3 mL/min)) (YMC Co., Ltd.). MPLC was performed on a Bona-Agela CHEETAHTM HP100 (Beijing Agela Technologies Co., Ltd.).
Fungal material
The wild-type fungus Alternaria alternata JJY-32 was isolated from a sponge-derived sample collected in Qionghai, Hainan (China). JJY-32 was deposited at the Key Laboratory of Marine Drugs, the Ministry of Education of China, School of Medicine and Pharmacy, Ocean University of China, Qingdao, People’s Republic of China.
Construction of the aLaeA and aMpaB expression vector
The integrative vector pHyg which mainly contains a constitutive promoter gpdA, ampicillin, and hygromycin-resistant genes was digested with restriction endonucleases XmaI and XbaI (New England Biolabs, NEB). The LaeA gene was amplified from the genomic DNA library of JJY-32 using specific primers containing KpnI and XbaI restriction sites and the MpaB gene was amplified from the genomic DNA library of JJY-32 using specific primers containing XmaI and XbaI restriction sites (Table S12) via PCR catalyzed by TransStart® Fastpfu DNA Polymerase (Transgen Biotech, Beijing, China). After digested with the corresponding endonucleases above mentioned, the LaeA gene and MpaB gene were placed downstream of the gpdA promoter within the pHyg vector to generate plasmids OE-alaeA and OE-aMpaB respectively (Figures S3 and S4). The constructed plasmids were transformed into E. coli DH10B competent cell for propagation and then transformed into wild-type JJY-32 to give the engineered strains JJY32-OE-alaeA and JJY32- OE-aMpaB.
Transformation and Overexpression of aLaeA and aMpaB
The strain Alternaria alternata JJY-32 was inoculated on PDA plates and cultured at 28 °C for 4 days to generate mycelium. Fresh mycelia were selected and added to 50 mL of MR medium (2% Sucrose, 1% KNO3, 0.25% MgSO4•6H2O, 0.5% KH2PO4, 0.1% Yeast Extract, 0.6 M Mannitol and 0.5 M NaCl add water to a constant volume 1 L). They were cultured at 200 rpm at 28 °C for 36 hours, then transferred from the culture medium to a 50 mL centrifuge tube and centrifuged at 4000 rpm for 15 min. The supernatant was discarded and mycelium was collected, weighed, and cleaned twice by washing buffer (1 M NaCl and 10 mM CaCl2). The resulting mycelium was then treated with osmotic buffer (10 mM Na2HPO4, pH 5.8, 20 mM CaCl2, 1.2 M NaCl) containing a mixture of cell wall-degrading enzymes (4 mg ml-1 driselase, 0.2 mg ml-1 lyticase, 4 mg ml-1 cellulase and 8 mg ml-1 glusulase). Every 1 g mycelium was treated with 10 mL osmotic buffer.
At 30 °C and 100 rpm, the enzymolysis status was checked at 2.5 h, and the mycelium could be collected when the enzymolysis rate reached 70% under the microscope. The enzymolysis solution was passed through three layers of mirror paper, the filtrate was collected and centrifuged at 4000 rpm for 15 min, the supernatant was discarded, and the protoplast was precipitated. It was then rinsed gently with a small amount of STC (1.2 M sorbitol, 10 mM CaCl2, 10 mM pH 7.5 Tris-HCl) twice and then added to the STC to re-hang.
The constructed plasmids were amplified by large extract, freeze-drying, and dissolved in 30 μl of STC and added into 100 mL protoplasts, mixed lightly, and left to rest on ice for 1 h. Next, 800 μL of PEG solution (60% PEG, 50 mM calcium chloride, and 50 mM pH 7.5 Tris-HCl) was added to the protoplasts, and let stand at room temperature for 30 min. The mixture was coated on a PSA (PDA medium with 1.2 M sorbitol and 100 μg/mL hygromycin) medium and cultured at 28 °C for 3 days until the single colony grew.
Fermentation and extraction
The engineered strains containing aLaeA and aMpaB genes were cultured under rotary shaker conditions at 180 rpm at 28°C in 500 mL Erlenmeyer flasks containing 200 mL of PDB. After 11 days of shaking, the supernatant and mycelium were separated by filtration through gauze. The supernatant part was extracted three times with ethyl acetate, and the mycelium part extracted with methanol three times, concentrated under reduced pressure to give an aqueous solution, and then extracted with ethyl acetate three times. All ethyl acetate solutions were combined and concentrated under reduced pressure to give an extraction.
Purification
The EtOAc extract (5.0 g) of JJY32-OE-alaeA was separated by MPLC using a gradient elution of MeOH-H2O (5%-100%) yielding five subfractions (fractions 1-5). Compound 1 (10.0 mg, tR 23.6 min) was isolated from Fraction 3 by Sephadex LH-20 Column Chromatography and followed by semi-preparative HPLC with a gradient elution of MeOH-H2O (60%-100%). The EtOAc extract (8.0 g) of JJY32-OE-aMpaB was separated by MPLC using a gradient elution of MeOH-H2O (5%-100%) yielding eight subfractions (fractions 1-8). Guided by the retention time and ion peaks of UPLC-MS data, fractions 6 and 7 were selected. Fraction 6 was separated by semi-preparative HPLC eluting with MeOH-H2O (50: 50) to furnish compounds 6 (9.0 mg, tR 21.6 min) and 4 (5.0 mg, tR 27.6 min). Fraction 7 was fractionated by semi-preparative HPLC with a gradient elution of MeOH-H2O (60%-100%) to give compounds 2 (13.0 mg, tR 26.8 min), 3 (5.0 mg, tR 24.5 min) and 5 (15.0 mg, tR 26.0 min), respectively.
Compound 1: Colorless oil; (c 1.0, MeOH); UV (MeOH) λmax (log ϵ) 250 (4.13); IR (KBr) νmax cm-1 1699, 1653, 1593, 1527; HRESIMS m/z 363.1958 [M+H]+, 1H and 13C NMR data see Table 1 and 2.
Compound 2: Colorless oil; (c 1.0, MeOH); UV (MeOH) λmax (log ϵ) 265 (4.10); IR (KBr) νmax 3850, 3746, 3671, 3647, 3564, 1699, 1649, 1539, 1508, 1457, 1034, 506 cm-1 ;1H and 13C NMR see Table 1 and 2; HRESIMS m/z 349.2379 [M+H]+ (calcd [M+H]+ 349.2383).
Compound 4: Colorless oil; ; c 1.0, MeOH), UV (MeOH) λmax (log ϵ) 264 (4.07); IR (KBr) νmax cm-1 3850, 3673, 3615, 1747, 1649, 1540, 1521, 1458, 504, 420 cm-1; 1H and 13C NMR see Table 1 and 2; HRESIMS m/z 401.2304 [M+Na]+ (calcd [M+ Na]+ 401.2304).
Compound 6: Colorless oil; (c 0.1, MeOH); IR(KBr)cm-1: 1739, 1613, 1381, 1242, 1156, 977; HRESIMS m/z 335.2 [M+H]+, 1H and 13C NMR data see Table 1 and 2.
Assay of Cytotoxicity Inhibitory Activity: As previously reported (Yang et al., 2013).
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Materials. Further inquiries can be directed to the corresponding author.
Author contributions
GL and DLiu drafted the molecular biology work and natural product chemistry experiments; ZL contributed to the plasmid construction and bioinformatic analysis; JJ and XH performed the compounds isolation and structure elucidation; XZ performed the biological evaluation; QC, TZ and DLi contributed to the critical reading of the manuscript; GZ designed the study, supervised the laboratory work, and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by the National Natural Science Foundation of China (81991522, 41976105), Marine S&T Fund of Shandong Province for Pilot National Laboratory for Marine Science and Technology(Qingdao)(No.2022QNLM030003-2), Taishan Scholar Youth Expert Program in Shandong Province (tsqn 202103153), Major Basic Research Programs of Natural Science Foundation of Shandong Province (ZR2019ZD18), the Fundamental Research Funds for the Central Universities (201941001), the NSFC-Shandong Joint Fund (U1906212), the Marine S&T Fund of Shandong Province for Pilot National Laboratory for Marine Science and Technology (Qingdao) (No. 2018SDKJ0401-2) and the Youth Innovation Plan of Shandong province (2019KJM004).
Conflict of interest
After completion of the research, JJ was employed by Sino Silliker Testing Services Qingdao Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.1015684/full#supplementary-material
References
Abdessemed N., Staropoli A., Zermane N., Vinale F. (2021). Metabolic profile and mycoherbicidal activity of three Alternaria alternate isolates for the control of Convolvulus arvensis, Sonchus oleraceus, and xanthium strumarium. Pathogens 10 (11), 1448. doi: 10.3390/pathogens10111448
Albarano L., Esposito R., Ruocco N., Costantini M. (2020). Genome mining as mew challenge in natural products discovery. Mar. Drugs 18 (4), 199. doi: 10.3390/md18040199
Bayram O., Braus G. H. (2012). Coordination of secondary metabolism and development in fungi: the velvet family of regulatory proteins. FEMS Microbiol. Rev. 36 (1), 1–24. doi: 10.1111/j.1574-6976.2011.00285
Bräse S., Encinas A., Keck J., Nising C. F. (2009). Chemistry and biology of mycotoxins and related fungal metabolites. Chem. Rev. 109 (9), 3903–3990. doi: 10.1021/cr050001f
Brown K. L., Cosseau C., Gardy J. L., Hancock R. E. (2007). Complexities of targeting innate immunity to treat infection. Trends Immunol. 28 (6), 260–266. doi: 10.1016/j.it.2007.04.005
Chu L., Huang J., Muhammad M., Deng Z., Gao J. (2020). Genome mining as a biotechnological tool for the discovery of novel marine natural products. Crit. Rev. Biotechnol. 40 (5), 571–589. doi: 10.1080/07388551.2020.1751056
Estiarte N., Lawrence C. B., Sanchis V., Ramos A. J., Crespo-Sempere A. (2016). LaeA and VeA are involved in growth morphology, asexual development, and mycotoxin production in Alternaria alternata. Int. J. Food Microbiol. 238, 153–164. doi: 10.1016/j.ijfoodmicro.2016.09.003
Fetzner R., Seither K., Wenderoth M., Herr A., Fischer R. (2014). Alternaria alternata transcription factor CmrA controls melanization and spore development. Microbiology 160 (Pt 9), 1845–1854. doi: 10.1099/mic.0.079046-0
Gacek A., Strauss J. (2012). The chromatin code of fungal secondary metabolite gene clusters. Appl. Microbiol. Biotechnol. 95 (6), 1389–1404. doi: 10.1007/s00253-012-4208-8
Gao J., Wenderoth M., Doppler M., Schuhmacher R., Marko D., Fischer R. (2022). Fungal melanin biosynthesis pathway as source for fungal toxins. mBio 13 (3), e0021922. doi: 10.1128/mbio.00219-22
Gozari M., Alborz M., El-Seedi H. R., Jassbi A. R. (2021). Chemistry, biosynthesis and biological activity of terpenoids and meroterpenoids in bacteria and fungi isolated from different marine habitats. Eur. J. Med. Chem. 210, 112957. doi: 10.1016/j.ejmech.2020.112957
Horiuchi M., Tokuda H., Ohnishi K., Yamashita M., Nishino H., Maoka T. (2006). Porritoxins, metabolites of Alternaria porri, as anti-tumor-promoting active compounds. Nat. Prod. Res. 20 (2), 161–166. doi: 10.1080/14786410500046646
Igbalajobi O., Yu Z., Fischer R. (2019). Red- and blue-light sensing in the plant pathogen Alternaria alternata depends on phytochrome and the white-collar protein LreA. mBio 10 (2), e00371–e00319. doi: 10.1128/mBio.00371-19
Jiang T., Wang M., Li L., Si J., Song B., Zhou C., et al. (2016). Overexpression of the global regulator LaeA in Chaetomium globosum leads to the bosynthesis of chaetoglobosin z. J. Nat. Prod. 79 (10), 2487–2494. doi: 10.1021/acs.jnatprod.6b00333
Kono Y., Gardner J. M., Suzuki Y., Takeuchi S. (1986). Studies on host-selective toxins produced by a pathotype of alternaria citri causing brown spot disease of mandarins. Agri. Biol. Chem. 50, 1597–1606. doi: 10.1271/bbb1961.50.1597
Kosalková K., García-Estrada C., Ullán R. V., Godio R. P., Feltrer R., Teijeira F., et al. (2009). The global regulator LaeA controls penicillin biosynthesis, pigmentation and sporulation, but not roquefortine c synthesis in Penicillium chrysogenum. Biochimie 91 (2), 214–225. doi: 10.1016/j.biochi.2008.09.004
Lee S. H., Soyoola E., Chanmugam P., Hart S., Sun W., Zhong H., et al. (1992). Selective expression of mitogen-inducible cyclooxygenase in macrophages stimulated with lipopolysaccharide. J. Biol. Chem. 267 (36), 25934–25938. doi: 10.1016/S0021-9258(18)35698-9
Liebermann B., Ellinger R., Günther W. H., Ihn W., Gallander H. (1997). Tricycloalternarenes produced by Alternaria alternata related to ACTG-toxins. Phytochemistry 46, 297–303. doi: 10.1016/S0031-9422(97)00278-1
Liebermann B., Nussbaum R. P., Günther W. (2000). Bicycloalternarenes produced by the phytopathogenic fungus Alternaria alternata. Phytochemistry 55 (8), 987–992. doi: 10.1016/s0031-9422(00)00268-5
Li X. J., Guo X., Zhou Y. H., Shi K., Zhou J., Yu J. Q., et al. (2016). Overexpression of a brassinosteroid biosynthetic gene dwarf enhances photosynthetic capacity through activation of Calvin cycle enzymes in tomato. BMC Plant Biol. 16, 33. doi: 10.1186/s12870-016-0715-6
Lin W. W., Karin M. (2007). A cytokine-mediated link between innate immunity, inflammation, and cancer. J. Clin. Invest. 117 (5), 1175–1183. doi: 10.1172/JCI31537
Logrieco A. F., Moretti A., Solfrizzo M. (2009). Alternaria toxins and plant diseases: an overview of origin, occurrence and risks. World Mycotoxin. J. 2, 129–140. doi: 10.3920/WMJ2009.1145
Lou J., Fu L., Peng Y., Zhou L. (2013). Metabolites from Alternaria fungi and their bioactivities. Molecules 18 (5), 5891–5935. doi: 10.3390/molecules18055891
Matsumoto M., Matsutani S., Sugita K., Yoshida H., Hayashi F., Terui Y., et al. (1992). Depudecin: a novel compound inducing the flat phenotype of NIH3T3 cells doubly transformed by ras- and src-oncogene, produced by Alternaria brassicicola. J. Antibiot. (Tokyo). 45 (6), 879–885. doi: 10.7164/antibiotics.45.879
Nugroho A. E., Morita H. (2014). Circular dichroism calculation for natural products. J. Nat. Med. 68 (1), 1–10. doi: 10.1007/s11418-013-0768-x
Pasare C., Medzhitov R. (2004). Toll-like receptors: linking innate and adaptive immunity. Microbes Infect. 6 (15), 1382–1387. doi: 10.1016/j.micinf.2004.08.018
Shen L., Tian S. J., Song H. L., Chen X., Guo H., Wan D., et al. (2018). Cytotoxic tricycloalternarene compounds from endophyte alternaria sp. W-1 associated with Laminaria japonica. Mar. Drugs 16 (11), 402. doi: 10.3390/md16110402
Shi Z. Z., Fang S. T., Miao F. P., Ji N. Y. (2018). Two new tricycloalternarene esters from an alga-epiphytic isolate of Alternaria alternata. Nat. Prod. Res. 32 (21), 2523–2528. doi: 10.1080/14786419.2017.1423312
Shin G. H., Veen M., Stahl U., Lang C. (2012). Overexpression of genes of the fatty acid biosynthetic pathway leads to accumulation of sterols in Saccharomyces cerevisiae. Yeast 29 (9), 371–383. doi: 10.1002/yea.2916
Surh Y. J., Chun K. S., Cha H. H., Han S. S., Keum Y. S., Park K. K., et al. (2001). Molecular mechanisms underlying chemopreventive activities of anti-inflammatory phytochemicals: down-regulation of COX-2 and iNOS through suppression of NF-kappa b activation. Mutat. Res., 480, 243–268. doi: 10.1016/s0027-5107(01)00183-x
Tsuge T., Harimoto Y., Akimitsu K., Ohtani K., Kodama M., Akagi Y., et al. (2013). Host-selective toxins produced by the plant pathogenic fungus alternaria alternata. FEMS Microbiol. Rev. 37 (1), 44–66. doi: 10.1111/j.1574-6976.2012.00350.x
Wang Q. X., Bao L., Yang X. L., Guo H., Ren B., Guo L. D., et al. (2013). Tricycloalternarenes f-h: three new mixed terpenoids produced by an endolichenic fungus ulocladium sp. using OSMAC method. Fitoterapia 85, 8–13. doi: 10.1016/j.fitote.2012.12.029
Wang H., Guo Y., Luo Z., Gao L., Li R., Zhang Y., et al. (2022). Recent advances in Alternaria phytotoxins: A review of their occurrence, structure, bioactivity, and biosynthesis. J. Fungi. (Basel). 8 (2), 168. doi: 10.3390/jof8020168
Wang L., Jiao J., Liu D., Zhang X., Li J., Che Q., et al. (2020). Cytotoxic meroterpenoids from the fungus alternaria sp. JJY-32. Chem. Biodivers. 17 (7), e2000226. doi: 10.1002/cbdv.202000226
Wang L., Zhang X., Zhang K., Zhang X., Zhu T., Che Q., et al. (2020). Overexpression of global regulator pbrlaeA leads to the discovery of new polyketide in fungus Penicillium brocae HDN-12-143. Front. Chem. 8. doi: 10.3389/fchem.2020.00270
Yang X., Zhang G., Tang X., Jiao J., Kim S. Y., Lee J. Y., et al. (2013). Toll-like receptor 4/nuclear factor-κB signaling pathway is involved in ACTG-toxin h-mediated anti-inflammatory effect. Mol. Cell Biochem. 374 (1-2), 29–36. doi: 10.1007/s11010-012-1502-9
Yu J., Han H., Zhang X., Ma C., Sun C., Che Q., et al. (2019). Discovery of two new sorbicillinoids by overexpression of the global regulator LaeA in a marine-derived fungus Penicillium dipodomyis YJ-11. Mar. Drugs 17 (8), 446. doi: 10.3390/md17080446
Zhang W., Du L., Qu Z., Zhang X., Li F., Li Z., et al. (2019). Compartmentalized biosynthesis of mycophenolic acid. Proc. Natl. Acad. Sci. U. S. A. 116 (27), 13305–13310. doi: 10.1073/pnas.1821932116
Keywords: Alternaria alternata, meroterpenoid, transcriptional regulator, tailoring enzyme, anti-inflammatory
Citation: Liu G, Liu D, Li Z, Jiao J, Hou X, Zhang X, Che Q, Zhu T, Li D and Zhang G (2022) Overexpression of transcriptional regulator and tailoring enzyme leads to the discovery of anti-inflammatory meroterpenoids from marine-derived fungus Alternaria alternata JJY-32. Front. Mar. Sci. 9:1015684. doi: 10.3389/fmars.2022.1015684
Received: 10 August 2022; Accepted: 20 September 2022;
Published: 28 October 2022.
Edited by:
Usama Ramadan Abdelmohsen, Minia University, EgyptReviewed by:
Reinhard Fischer, Karlsruhe Institute of Technology (KIT), GermanyHui Cui, Guangzhou University of Chinese Medicine, China
Copyright © 2022 Liu, Liu, Li, Jiao, Hou, Zhang, Che, Zhu, Li and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guojian Zhang, emhhbmdndW9qaWFuQG91Yy5lZHUuY24=
†Present address: Jieying Jiao, Sino Silliker Testing Services (Qingdao) Co., Ltd, Qingdao, Shandong, China
‡These authors have contributed equally to this work
 Guowei Liu
Guowei Liu Dan Liu1‡
Dan Liu1‡ Tianjiao Zhu
Tianjiao Zhu Dehai Li
Dehai Li Guojian Zhang
Guojian Zhang