
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 26 September 2022
Sec. Marine Biology
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.1014810
This article is part of the Research Topic Impact of Climate Change: Physiological responses to environmental changes seen through Biomineralization View all 4 articles
 Caixia Cai1†
Caixia Cai1† Qi He1†
Qi He1† Bingyi Xie1
Bingyi Xie1 Zhijie Xu1
Zhijie Xu1 Cheng Wang1
Cheng Wang1 Chuangye Yang1,2,3,4
Chuangye Yang1,2,3,4 Yongshan Liao2,3,4
Yongshan Liao2,3,4 Zhe Zheng1,2,3,4*
Zhe Zheng1,2,3,4*Marine environmental change directly affects bivalve growth and survival. Exoskeleton formation, the main energy dissipation in the physiological metabolism, typically reflects the body growth of the bivalve. However, how bivalves regulate the biomineralization of the exoskeleton under environmental stressors is not yet clear. Long non-coding RNA regulates various life processes through complex mechanisms in vertebrates and invertebrates. In this research, we cloned the complete sequence of a mantle-specific expressed long non-coding RNA (designated as LncMPEG1) from a pearl oyster, Pinctada fucata martensii. A quantitative real-time PCR analysis showed that LncMPEG1 expression was significantly high in early umbo larvae and juveniles, which would be the critical periods of shell development. LncMPEG1 was identified in the outer epithelium of the middle fold from the mantle edge, mantle pallial, and mantle center by using in situ hybridization. Additionally, the expression of LncMPEG1 was stimulated by shell damage, alien invasion, heat and cold temperature stress, and hypoxia stress. In the mantle, a decreased in LncMPEG1 expression was detected by RNA interference, which can cause the irregular growth of crystals on the inner surface of the prismatic layer and nacre in the shells. Therefore, we propose that LncMPEG1 could be a key regulator in biomineralization and responds to environmental stress in the mantle .
Marine benthic communities are subjected to changing environmental conditions (Couce et al., 2020). Mollusca is the second most bio-diverse extant molluscan phylum, in which shelled mollusks are distinguished by a vast array of biomineralization fabricated architectures (Williams, 2007). Since the Cambrian (543–490 mya), biomineralization has undergone significant diversification, resulting in the formation of mineralized skeletons in the majority of classes, such as Bivalvia and Gastropoda. These shelled mollusks successfully adapted to the oceanic environmental changes and continued to evolve after the Cambrian (Kouchinsky et al., 2012). Bivalves are primarily protected by their shells from predators, infections, and, to a lesser extent, adverse environmental factors such as drouth, wave action, and iceberg detriment. (Trussell, 2000; Wilke and Falniowski, 2001; Ji et al., 2018). Records in carbonate biominerals have shown the response of mollusks to climatic events through biological processes during active mineralization (Kennett and Voorhies, 1995). For example, the Mg/Ca ratio rises as the results of the increase of magnesium concentration to stabilize amorphous calcium carbonate, which is independent to the rise in sea surface temperature (Pérez-Huerta et al., 2013). Although marine biodiversity has undergone extensive evolution, anthropogenic-induced environmental change still has an impact on it, especially on mollusks and other creatures with calcified exoskeletons (Khan et al., 2020). This is made worse by the fact that these animals are having a harder time surviving in oceans due to changes in salinity, pH, and temperature (Zheng et al., 2020a). With rapid and frequently fluctuating environmental stresses, it may become more energy intensive to create marine skeletons, and the increasingly erosion conditions in seawater are anticipated to cause the corrosion of these exterior skeletons. Molluscan shell formation is regulated by gene expression and requires energy because of the ATP demand of physiological processes related to biomineralization, including mineral and ion transport and Extracellular matrix (ECM) protein biosynthesis, which are affected by changes in temperature, salinity, and pH (Ivanina et al., 2020). However, it remains unclear how shelled molluscs regulate gene expression patterns to a degree that alters biologic mineralization processes in response to biotic/abiotic environmental stress.
Long noncoding RNAs (LncRNAs) are defined as RNA genes that are greater than 200 bp, do not have protein-coding potential, and play an important role in regulating noncoding RNAs (Gong et al., 2012). LncRNAs are classified into sense, antisense, bidirectional, intergenic, and intronic LncRNAs based on their positions and transcription directions of LncRNAs and protein encoding genes on the genome (Rinn and Chang, 2012). According to the research, LncRNAs can regulate gene expression at the epigenetic, transcription, post-transcriptional levels and involved in many crucial regulatory processes, including transcriptional activation and suppression, chromatin modification, genomic imprinting, and nuclear transport (Qiu et al., 2013; Yoon et al., 2013). As signals, decoys, guides, and scaffolds, LncRNAs can not only regulate gene expression through interacting with RNA, DNA, and proteins as well as regulate post-transcriptional gene expression via regulating splicing, mRNA translation, and degradation as microRNA(miRNA) precursor molecules (Dykes and Emanueli, 2017). LncRNAs are predicted to be important functions in the response to environmental stressors and regulate body growth in mollusks including exoskeleton biomineralization. For example, differential LncRNA expression profiles reveal the regulatory roles of LncRNAs in the against viruses and bacteria immune response of abalones, oysters, and mussels via links to multiple immune-related pathways, for instance, the toll-like receptor, NF-kappaB signaling pathway, pattern recognition receptors, lectin immune-related pathways, fibrinogen, and O-glycan biosynthesis (Sun and Feng, 2018; Lu et al., 2021; Pereiro et al., 2021). Also, LncRNAs are reported to participate in controlling shell formation and body growth by affecting pigmentation, shell matrix gene expression, and glycogen accumulation (Sun and Feng, 2018). In addition, our previous study proposed that the biomineralization process and defense function of the mantle tissue are crosslinked and regulated by LncRNAs in the pearl oyster (Zheng et al., 2020b; Zheng et al., 2020c). As mentioned above, changes in environmental conditions could induce responses at multiple levels in mollusk species, including affecting the biomineralization process. Thus, crosslinked regulatory roles of LncRNAs between anti-environmental stress and biomineralization-related processes should be considered.
Pinctada fucata martensii is the main species that is cultured for production of marine pearls in China and Japan (Jiao et al., 2012). In this study, we cloned a novel LncRNA which was a mantle-specific expressed LncRNA, named LncMPEG1. We found that it not only participated in response to environmental stress, such as LPS stimulation but it also regulated the biomineralization of the shell. These findings can provide light on how anti-environmental stress and biomineralization in pearl oysters are regulated by LncRNAs in a cross-linked manner.
Pearl oysters used in this research were breeding at the coastal area of Liushawan, Zhanjiang, Guangdong Province, China (20°25’ N, 109°57’ E). The two-year-old pearl oysters with a size of 5-6cm were selected as the experimental material. Before performing experiments, pearl oysters breed in seawater for 2 days at 25–27°C.
In this study, we tested the expression pattern of LncMPEG1 in six tissues by qRT-PCR, including the mantle edge (ME), mantle pallial (MP), mantle center (MC), hepatopancreas (HE), adductor muscles (A), and gills (Gi). We also detected the expression trends of LncMPEG1 at different development stages, including unfertilized eggs (E), fertilized ovum at 30 min (Fe), blastocyst at 5 h (B), gastrula at 6 h (G), early trochelminth larva at 8 h (ET), trochelminth larva at 16 h (T), D-stage larvae at 19 h (D), D-stage larvae prior to feeding at 4 d (DF), early umbo larvae at 14 d (EU), eyed larvae at 28 d (EL), spat at 40 d (S), and juveniles at 90 d (J) after fertilization. All samples were drown in liquid nitrogen several minites and then kept in −80°C freezer for storage.
After being temporarily cultured at 22°C for 2 d, 96 normal P. f. martensii were randomly divided into three groups and cultured in three 300 L experimental buckets with water temperatures of 17°C (low temperature group), 22°C (control group), and 32°C (high temperature group) as stress treatments. The temperatures were monitored three times a day in all the aquaria and were adjusted if needed. During the culture process, the same amount of unicellular algae was fed and 50% of the water was changed every day. At 1 d, 3 d, 5 d, and 10 d after the beginning of the experimental culture, eight individuals were randomly selected from each experimental barrel, and the mantle tissues were used to detect gene expression.
Before the hypoxia treatment, 64 healthy and energetic pearl oysters were cultured in an aquaculture system with a controlled water temperature (32°C and 6.0 mg/L DO) for 7 d. The normoxic group (0 h) was set at 6.0 mg/L DO. In the hypoxia group, 2.0 mg/L DO was used for the hypoxia conditions. The oxygen concentration in the aquarium was continuously monitored during the experiment using an oxygen dissolving apparatus (Aqua TROLL@400 instrument, USA). The pearl oysters were suddenly exposed to the hypoxia treatment and the mantle tissues were collected at 12 h, 24 h, 2 d, 5 d, 10 d, 15 d, and 25 d after processing.
The shell powder was obtained by milling the pearl shell to a nano-scale, sterilized and added the disinfected filtered seawater to obtain a shell powder paste. The shellfish was placed face down to allow for the flow of the water to remove the impurities in the pearl shell, it was made sure that the mouth was open, and an appropriate amount of shell powder was injected into the side film at the junction of the pearl shell and the shell. The pearl shellfish that was injected with shell powder paste was placed upside down, kept aside, and placed in seawater for cultivation. The MP was harvested at 12 h, 1 d, 2 d, 5 d, and 10 d after stimulation. The mantle tissues from the pearl oyster withou injection treatments were used as blank control.
The shell damage assays were performed on 40 normal individuals with a V-shape notch breach cut in the shell down to the nacreous layer. The MPs of eight pearl oysters were collected at 6 h, 12 h, 24 h, 36h, and 48 h after shell damage respectively. The MPs from the untreated pearl oyster were considered as the negative control. Then, the expression profile of LncMPEG1 at the different notching times was detected by qRT-PCR.
The total RNA was isolated via the Trizol reagent (Thermo Fisher Scientific, USA) and managed to remove genomic DNA by DNase I (Promega, USA). Then, the integrities of the prepared RNA samples were tested by a 1.0% garose gel electrophoresis assay. RNA concentration was detected by NanoDrop ND 1000 Spectrophotometer (Thermo Fisher Scientific) and RNA purity was estimated via OD260/OD280. The cDNA templates for gene amplication and RACE were prepared by random primers using M-MLV reverse transcriptase (Promega) according to the manufacturer’s instruction. The cDNA template for rapid amplification of cDNA ends (RACE) was synthesized using the SMARTer RACE 5′/3′ kit (TaKaRa, China). All the cDNAs were stored at −20°C until use.
The reference sequence of LncMPEG1 for RACE, qRT-PCR and template preparation of dsRNA was obtained by Strand-specific transcriptome sequencing (unpublished). The primers used for gene amplification were shown in table gene-specific primers for amplification which are shown in Table S1. The coding ability of the sequence that was obtained with validation was predicted via the online software coding potential assessment tool that refers to the zebrafish (CPAT; http://lilab.research.bcm.edu/cpat/index.php). And the CPAT refers to the zebrafish model. Furthermore, the secondary structure prediction was performed by the online MFOLD (http://mfold.rna.albany.edu/?q=mfold/RNA-Folding-Form).
In this research, the expression level detection of LncMPEG1 by qRT-PCR was performed using the LnRcute LncRNA SYBR Green qPCR Kit (TIANGEN, China) according to its protocol. The cDNA template used for LncMPEG1 amplification was synthesed by LnRcute LncRNA First-Strand cDNA Synthesis Kit (TIANGEN, China). In addition, amplification and fluorescence detection was performed using the Applied Biosystems 7500/7500 Fast Real-Time System (Applied Biosystems, USA). The relative expression level of LncMPEG1 was calculated using the 2-△ct or 2-△△ct method, with GAPDH as the reference gene. The differential expression analysis was performed using SPSS 22.0 software.
To localize the expression of LncMPEG1 in the mantle of P. f. martensii, in situ hybridization (ISH) was performed. The RNA probes for ISH were synthesized in vitro using T7 High Efficiency Transcription Kit (Tansgene, Beijing) and digoxigenin RNA Labeling Mix (Roche, USA). The mantle tissue samples were fixed using paraformaldehyde containing 0.1% diethyl dicarbonate (Sigma-Aldrich, USA) for approximately 1.5 h and were embedded in paraffin. Then, the embedded compound was sliced into 7 μm sections. Finally, the obtained tissue sections were constructed for ISH after being dewaxed and rehydrated. The detailed experimental operation procedures are detailed in Zheng et al. (2020c).
The RNA interference (RNAi) experiment was set for the experimental and control groups. The test group was injected with dsRNA-LncMPEG1. The control group was injected with dsRNA red fluorescent protein. For muscle injection, pearl oystersper group were injected with a dose of 100 μL per shell, and the concentration of dsRNA in test group was 600 ng/μL. After 6 d, the detected tissues including ME, mantle pallial (MP), and MC were cut to test gene expression of LncMPEG1. The shells collected from each groups were washed and dried, and then cutted into small pieces. The inner surface of shells was observed by using scanning electron microscope (SEM) to analyze the ultrastructure of novel nacreous layer and prismatic layer.
The full length of LncMPEG1 was obtained using RACE, and was 2735 bp (Figure 1A). The encoding ability of LncMPEG1 was predicted using the CPAT. The total coding probability was about 0.0057 (Figure 1B), without the coding tag, indicating that LncMPEG1 does not have the potential to encode proteins. Increasing evidence suggests that RNA-protein interactions are related to the secondary structure of RNA. To explore whether LncMPEG1 has a specific secondary structure, we predicted the secondary structure of LncMPEG1 using MFOLD online and found that there were three main branches folded into a clover-leaf secondary structure (Figure 2).

Figure 1 The full-length sequence and prediction encoding capability of LncMPEG1 in the pearl oyster P. (f.) martensi. (A), the full-length sequence of LncMPEG1. (B), the prediction of the LncMPEG1 encoding capability using the coding potential assessment tool.
The LncMPEG1 expression level in the ME, MC, A, HE, and Gi of P. f. martensii was detected using qRT-PCR. The results showed that the expression level of LncMPEG1 in the MC was significantly higher than that in the other tissues (P < 0.05; Figure 3A). The expression of LncMPEG1 was also detected at different development stages. The results have shown that LncMPEG1 could be tested at all the development stages and exhibited three peaks in the gastrula stage at early development, eyed larvae stage before metamorphosis, and juvenile stage (Figure 3B).
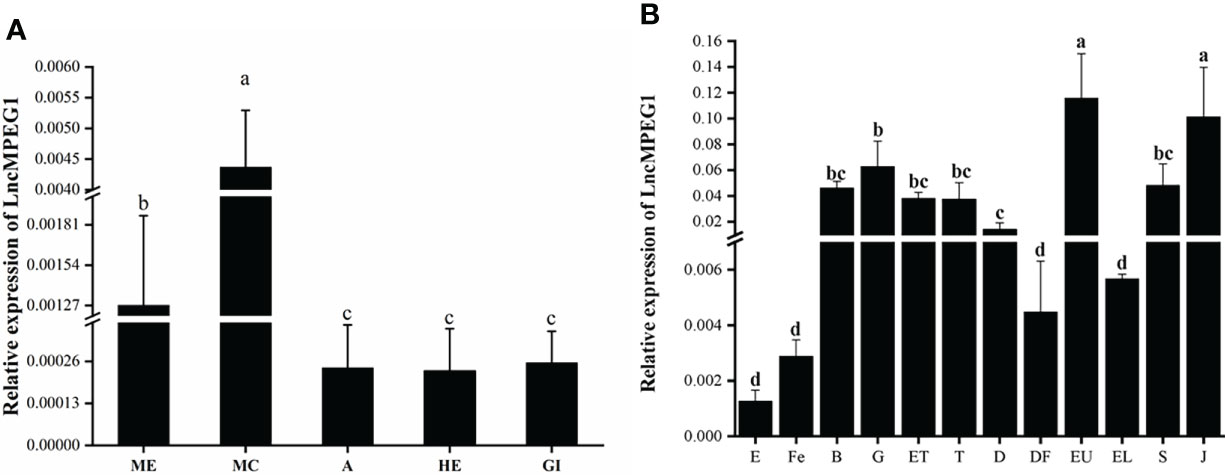
Figure 3 The normal expression profile of LncMPEG1 in the different tissues (A) and at different development stages (B) of P. (f.) martensii. a: ME, mantle edge; MC, mantle center; A, adductor muscle; HE, hepatopancreas; and Gi, gill. b: the expression pattern of LncMPEG1 during the larvae development of P. f. martensii. E, egg; Fe, fertilized ovum; B, blastocyst; G, gastrula; ET, early trochelminth larva; T, trochelminth larva, D, D-stage larvae; DF, D-stage larvae prior to feeding; EU, early umbo larvae; EL, eyed larvae; S, spat; and J, juvenile; different letters indicate significant differences (P < 0.05).
In this study, we used ISH technology to detect the localization of LncMPEG1 in the mantle tissue. In the experimental group (Figure 4C), the outer epithelium of the MP and MC displayed strong positive hybridization signals, while the outer epithelium of the middle fold displayed weak positive hybridization signals (Figures 4D, E). In the control group (Figure 4A), there was no red fluorescence hybridization signal and only a blue fluorescence nuclear staining signal (Figure 4B).
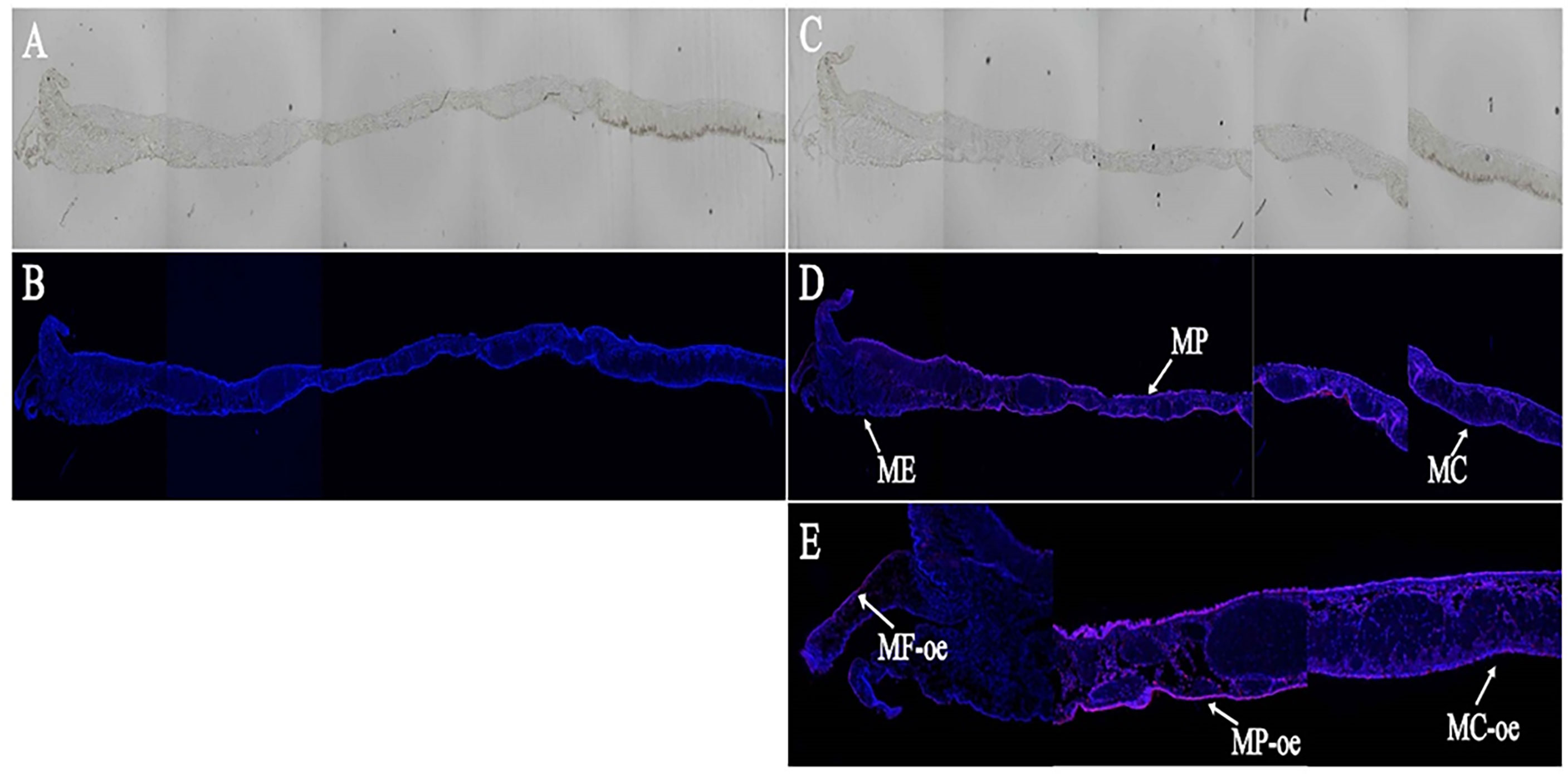
Figure 4 The in situ hybridization results of LncMPEG1 in the outer epidermal cells of the mantle. (A, B) control group; (C, D) experiment group; (E) the local enlarged mantle tissue of the experiment group; (A, C) general observation; (B, D, E) fluorescence observation; ME, mantle edge; MP, mantle pallial; MC, mantle center; MF, middle fold; and oe, outer epithelium. The red fluorescence and arrow indicate the in situ hybridization signal of LncMPEG1; the blue fluorescence indicates the nucleus.
The LncMPEG1 expression level in the ME, MP, and MC was investigated by using real-time PCR after RNAi.In the tested group, the expression level of LncMPEG1 in the ME, MP, and MC of was significantly down-regulated as compared to the control group (P < 0.05, Figures 5A–C). This indicates that LncMPEG1-dsRNA significantly inhibited the expression level of the LncMPEG1 gene in mantle of P. f. martensii. Moreover, the SEM observation of the ultrastructure of the shell after RNAi showed that the control group’s crystal surface was smooth and had a regular arrangement. However, in the tested group, the crystal growth was disordered the crystal plate became smaller, and the surface was rough and hollow (Figure 5D).
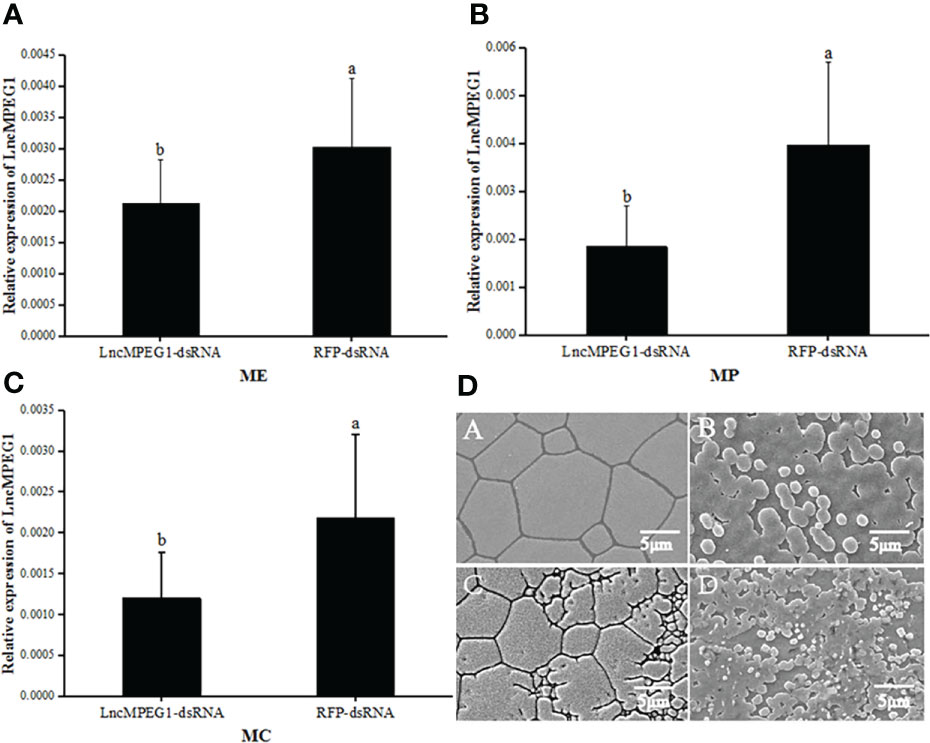
Figure 5 The expression level of LncMPEG1 in the mantle tissue (A–C) and the ultrastructure of the inner surface of the shell (D) after RNAi. The negative control group: red fluorescent protein (RFP); The experimental group: LncMPEG1-dsRNA,; ME, mantle edge; MP, mantle pallial; and MC, mantle center; the mean values with different letters indicate significant difference (P < 0.05); A–C the prismatic layer of the control and experiment group, respectively; B–D, the nacreous layer of the control and experimental group, respectively.
The control group: red fluorescent protein (RFP); The tested group: LncMPEG1-dsRNA,; Abbreviations: ME, mantle edge; MP, mantle pallial; and MC, mantle center; the mean values with different letters indicate significant difference (P < 0.05); A, C: the prismatic layer of the control and tested group, respectively; B, D: the nacreous layer of the control and tested group, respectively.
We detected the pattern of LncMPEG1 expression in mantle under abiotic stressors including heat and cold temperature stress and hypoxia stress. The changes in LncMPEG1 in the mantle under low temperature (17°C) and high temperature (32°C) stress were investigated using qRT-PCR; LncMPEG1 significantly increased after 3 d and 5 d in the low temperature group (Figure 6A). Additionally, the expression of LncMPEG1 was significantly up-regulated after 1 d and 3 d (Figure 6B). This finding indicates that LncMPEG1 responded to heat and cold stress in the mantle tissue. In addition, we found that there was an upward trend in LncMPEG1 after 5 d and it was significantly up-regulated after 25 d under hypoxic stress (P < 0.05; Figure 6C).
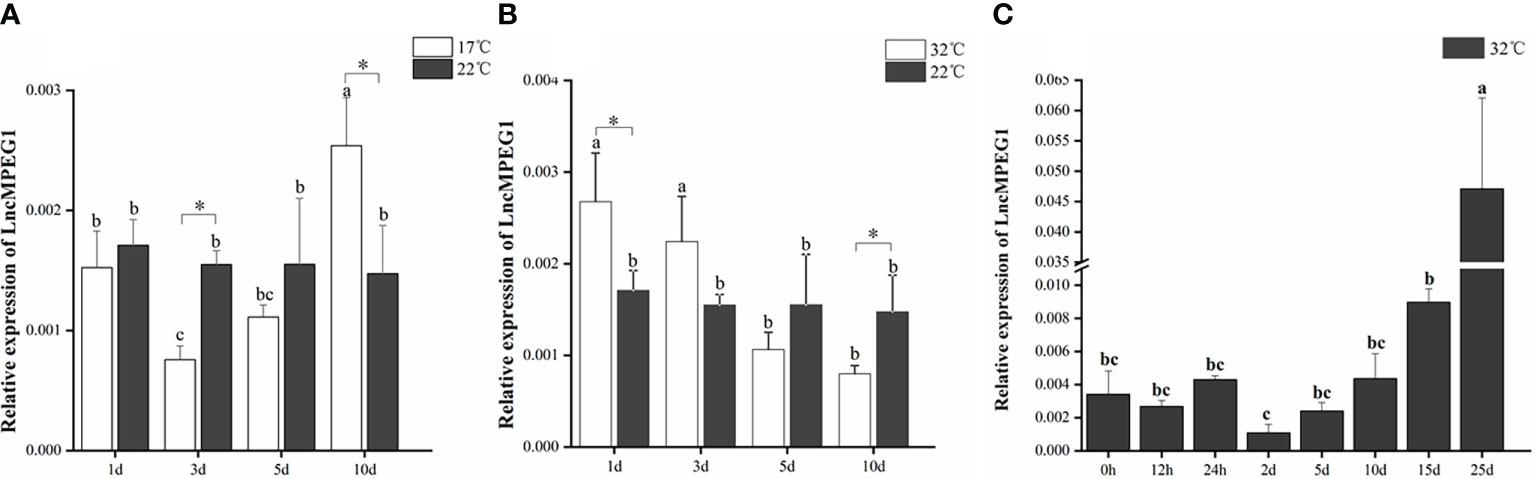
Figure 6 The relative expression of LncMPEG1 in the mantle tissue after the temperature treatment and hypoxic stress. The expression pattern of LncMPEG1 in the mantle tissue of P. f. martensii under 17°C stress (A), under 32°C stress (B), and under hypoxia treatment at a high temperature (C). The mean values with * indicate significantly different (P < 0.05). The mean values with different letters indicate significant differences (P < 0.05).
We performed shell damage and simulated the biotic stressors of body damage and alien invasion. Shell damage causes mantle exposure to the seawater environment and fluid loss. The expression of LncMPEG1 was significantly up-regulated at 12 h after shell damage (P < 0.05), and the expression level remained relatively stable from 12 h to 36 h, with the highest expression level at 48 h (Figure 7A). The expression of LncMPEG1 significantly increased at 1 d (P < 0.05), remained relatively stable until 5 d, and fell back to the initial level at 10 d (Figure 7B).
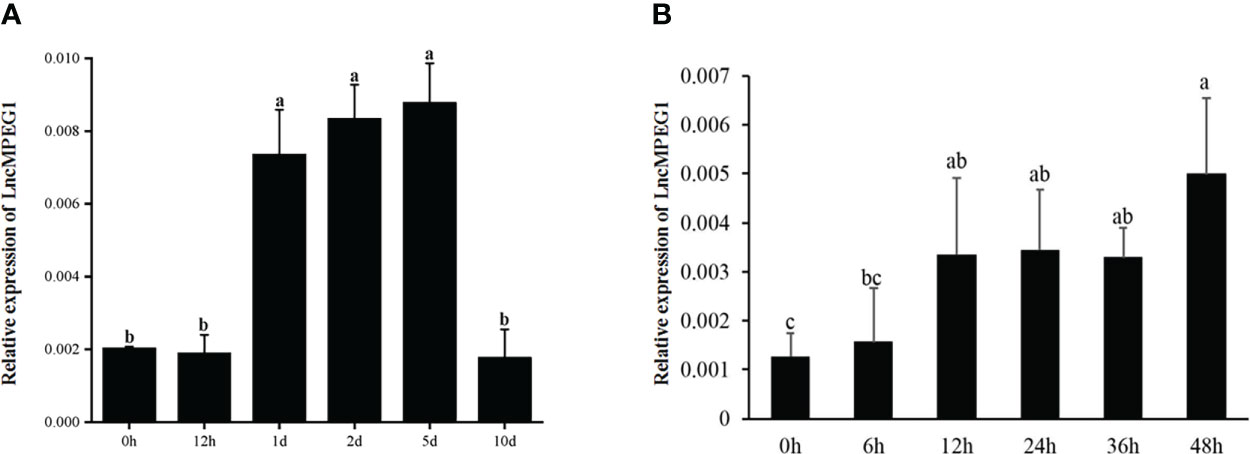
Figure 7 The relative expression of LncMPEG1 due to powder stimulation and shell damage. (A) the sequential expression of LncMPEG1 in the mantle with injected shell powder in EPS. (B) the sequential expression of LncMPEG1 in the mantle of Pinctada fucata martensii after shell damage. The mean values with different letters indicate significant differences (P < 0.05).
Biomineralization is one of the most important actions in shelled mollusks. Shell formation is controlled by the organic matrix via cellular-mediated complex regulation mechanisms. Then, LncRNA is a cluster of epigenetic regulators that have an important role in biomineralization, immunity, and stress resistance (Yu et al., 2016; Feng et al., 2018; Huang et al., 2018; Zheng et al., 2019). Previously, LncRNAs had been reported to play a role in biomineralization of pearl oysters, such as LncMSEN1, LncMSEN2, and LncRunt (Zheng et al., 2019; Zheng et al., 2020a; Zheng et al., 2020b). In this study, we cloned a novel LncRNA that is highly expressed in the mantle epidermis cells of P. f. martensii. Bioinformatics predicted the encoding ability of LncMPEG1 using CPAT, with a total encoding probability of about 0.0057, without a coding tag. This indicated that LncMPEG1 did not have the potential to encode proteins. Long noncoding RNA can perform biological functions by folding to form secondary structures, and it could produce specific binding, allosteric and catalytic effects (Novikova et al., 2012). Multiple stem-loop structures of LncMPEG1 indicated the potential interactions with proteins may depend on the specific RNA secondary structure.
The mantle is recognized as a biomineralization-related tissue in shelled mollusks (Suzuki and Nagasawa, 2013). Numerous genes or protiens involved in biomineralization directly (including temptin gene and chitinase) were proved to expressed in high levels in the mantle and demonstrated have a vitro mineralization function (Weiner and Addadi, 2011; Liao et al., 2021). Therefore, it is possible to assume that LncMPEG1 is connected to shell formation based on its high expression in the MC and ME. In bivalves, Shell formation begins early in the larval stage. The trochelminth larva, D-shaped, umbonal, eyespot, and spat stages, where the calcium carbonate crystal polymorphism and shell structure change significantly, are the five stages that contribute to early shell formation. And the shell gland starts secreting organic shell components in the late trochophore stage (Rose and Baker, 1994; Fujimura et al., 1995; Andres Aranda-Burgos et al., 2014; Zhao et al., 2018). The highest expression of LncMPEG1 in the early umbo larvae and juveniles indicates that LncMPEG1 participates in dissoconch and shell formation. The mantle is an important organ that is responsible for shell mineralization. Different regions of the mantle form different shell structures (Nakahara and Bevelander, 1971; Kinoshita et al., 2011). The ME is mainly responsible for the shell prismatic layer formation, while the MP and MC are mainly responsible for the shell nacre formation (Awaji and Machii, 2011). The ISH demonstrated that LncMPEG1 was located in the outer epithelium of the middle fold from the ME, MP, and MC. To further verify its functions of shell mineralization, we reduced the expression of LncMPEG1 using RNAi and found obviously disordered growth in both the prismatic and nacre layers of the shell. These findings suggested that LncMPEG1 is important for the regulation of biomineralization in P. f. martensii.
Shell formation in bivalves reflects their body growth and living conditions. Shell mineralization is extremely energetically expensive. Mantle cells need to transcribe and translate shell matrix proteins and synthesize other matrices such as glycosaminoglycan to induce and control the extracellular biomineralization of the shell. In the environment, marine bivalves can suffer from multiple environmental stressors, affecting the energy and protein metabolism of the mantle cells and depressing shell growth. High or low water temperatures deeply influence shell growth and its microstructure (MacDonald and Thompson, 1986; Wilson, 1987). Furthermore, temperature elevation also damage the microstructure of abalone via expression changes in the crystal induction genes in Haliotis discus hannai (Zheng et al., 2020a). In this study, we found the expression pattern of LncMPEG1 obviously induced under low temperature and inhibited under high temperature, which is an indicator of regulator under temperature stress. As mentioned above, LncMPEG1 could significantly regulated the formation of novel calficied layer of shell. So we proposed that the regulation roles of LncMPEG1 to temperature stress may be mediated by biocalfication related pathway. Temperature always coupled to dissolved oxygen to affeact aquatic animals’ survival (Joos et al., 2003). Oyster could adapt hypoxia stress via metabolism depression and anaerobic glycolysis metabolism (Meng et al., 2018). In sea cucumbers LncRNA such as MSTRG.34610 and MSTRG.10941 could regulated HIF-1α gene and responed to hypoxia stress (Huo et al., 2020). In this study, we also dected LncMPEG1 in hypoxia stress under hight temperature and found that LncMPEG1 significant increase after 15d. Therefore, we proposed that expression changes of LncMPEG1 under environment stresses results in the coupled regulation roles of biomineralization and the stress response in the mantle of pearl oyster.
Beyond fabricating shells to control body growth, the mantle has a screening function as the epidermis and mucosa. Shell damage, sandy or particulate matter (PM) insertion, and bacteria and parasite invasion can cause a serious rejection reaction and immune response by the mantle cells. It has been reported that LncMSEN1 and LncMSEN2 participate in biomineralization and the immune response in mantle of the pearl oyster (Zheng et al., 2020a; Zheng et al., 2020b). In this study, we injected shell powder to simulate sandy or PM insertion in EPS and found that LncMPEG1 responded to the shell powder with a high expression pattern. Shell damage also induced LncMPEG1 expression after 12 h. These two stressors are part of the immune defense against pathogenic microorganism invasion after the exoskeleton is damaged, and the subsequent shell reconstruction is mediated by biomineralization. Thus, LncMPEG1 may be a core regulator in response to environmental stressors by changing the gene expression of the mantle tissue during biomineralization or controlling the expression of stress-related proteins, therefore, affecting the dominant energy for biomineralization and the stress response.
In conclusion, we cloned the novel LncRNA, LncMPEG1, from the pearl oyster, P. f. martensii. It was highly expressed in mantle and widely observed in all the detected development stages. LncMPEG1 repression caused the disordered growth of the shell. LncMPEG1 also participated in multiple abiotic and biotic stressors by changing its expression pattern, which indicates a vital regulation role in mediating biomineralization and anti-environmental stress in the pearl oyster.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
CC: Conceptualization, data curation, writing original draft. QH: Data curation, writing original draft. BX: Methodology, data curation, writing original draft. ZX: Conceptualization. CW: Validation. CY: Resources, validation. YL: Resources. ZZ: Project administration, writing - review & editing, funding acquisition. All authors contributed to the article and approved the submitted version.
This research was funded by National Natural Science Foundation of China (32002369), Guangdong Basic and Applied Basic Research Foundation (2019A1515011096, 2022A1515010030), Innovation Team Project (2021KCXTD026) from the Department of Education of Guangdong Province, Supported by the earmarked fund for CARS-49, and the Graduate Education Innovation Program of Guangdong Ocean University (Grant No. 202158).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.1014810/full#supplementary-material
Andres Aranda-Burgos J., Da Costa F., Novoa S., Ojea J., Martinez-Patino D. (2014). Embryonic and larval development of Ruditapes decussatus (Bivalvia: Veneridae): A study of the shell differentiation process. J. Molluscan Stud. 80, 8–16. doi: 10.1093/mollus/eyt044
Awaji M., Machii A. (2011). Fundamental studies on in vivo and in vitro pearl formation–contribution of outer epithelial cells of pearl oyster mantle and pearl sacs. Aqua-BioSci Monogr. 4, 1–39. doi: 10.5047/absm.2011.00401.0001
Couce E., Engelhard G. H., Schratzberger M. (2020). Capturing threshold responses of marine benthos along gradients of natural and anthropogenic change. J. Appl. Ecol. 57 (6), 1137–1148. doi: 10.1111/1365-2664.13604
Dykes I. M., Emanueli C. (2017). Transcriptional and post-transcriptional gene regulation by long non-coding RNA. Genomics Proteomics Bioinf. 15 (3), 177–186. doi: 10.1016/j.gpb.2016.12.005
Feng D., Li Q., Yu H., Kong L., Du S. (2018). Transcriptional profiling of long non-coding RNAs in mantle of Crassostrea gigas and their association with shell pigmentation. Sci. Rep. 8. doi: 10.1038/s41598-018-19950-6
Fujimura T., Wada K., Iwaki T. (1995). Development and morphology of the pearl oyster larvae, Pinctada fucata. J. Malacol Venus. 54 (1), 25–48.
Gong C., Popp M. W.-L., Maquat L. E. (2012). Biochemical analysis of long non-coding RNA-containing ribonucleoprotein complexes. Methods. 58 (2), 88–93. doi: 10.1016/j.ymeth.2012.06.020
Huang X.-D., Dai J.-g., Lin K.-t., Liu M., Ruan H.-t., Zhang H., et al. (2018). Regulation of IL-17 by lncRNA of IRF-2 in the pearl oyster. Fish Shellfish Immunol. 81, 108–112. doi: 10.1016/j.fsi.2018.07.020
Huo D., Sun L., Storey K. B., Zhang L., Liu S., Sun J., et al. (2020). The regulation mechanism of lncRNAs and mRNAs in sea cucumbers under global climate changes: Defense against thermal and hypoxic stresses. Sci. Total Environ. 709, 136045. doi: 10.1016/j.scitotenv.2019.136045
Ivanina A. V., Jarrett A., Bell T., Rimkevicius T., Beniash E., Sokolova I. M. (2020). Effects of seawater salinity and pH on cellular metabolism and enzyme activities in biomineralizing tissues of marine bivalves. Comp. Biochem. Physiol. a-Molecular Integr. Physiol. 248. doi: 10.1016/j.cbpa.2020.110748
Jiao Y., Wang H., Du X., Zhao X., Wang Q., Huang R., et al. (2012). Dermatopontin, a shell matrix protein gene from pearl oyster Pinctada martensii, participates in nacre formation. Biochem. Biophys. Res. Commun. 425 (3), 679–683. doi: 10.1016/j.bbrc.2012.07.099
Ji H. M., Liang S. M., Li X. W., Chen D. L. (2018). A self-assembled smart architecture against drilling predation in a Pinctada maxima shell: Protective mechanisms. J. Mat Sci. 53 (5), 3417–3426. doi: 10.1007/s10853-017-1782-2
Joos F., Plattner G.-K., Stocker T. F., Körtzinger A., Wallace D. W. R. (2003). Trends in marine dissolved oxygen: Implications for ocean circulation changes and the carbon budget. Eos Trans. Am. Geophys Union 84 (21), 197–201. doi: 10.1029/2003EO210001
Kennett D., Voorhies B. (1995). Middle Holocene periodicities in rainfall inferred from oxygen and carbon isotopic fluctuations in prehistoric tropical estuarine mollusc shells. Archaeometry. 37 (1), 157–170. doi: 10.1111/j.1475-4754.1995.tb00734.x
Khan F. U., Hu M., Kong H., Shang Y., Wang T., Wang X., et al. (2020). Ocean acidification, hypoxia and warming impair digestive parameters of marine mussels. Chemosphere. 256. doi: 10.1016/j.chemosphere.2020.127096
Kinoshita S., Wang N., Inoue H., Maeyama K., Okamoto K., Nagai K., et al. (2011). Deep sequencing of ESTs from nacreous and prismatic layer producing tissues and a screen for novel shell formation-related genes in the pearl oyster. PloS One. 6 (6). doi: 10.1371/journal.pone.0021238
Kouchinsky A., Bengtson S., Runnegar B., Skovsted C., Steiner M., Vendrasco M. (2012). Chronology of early Cambrian biomineralization. Geol Magazine. 149 (2), 221–251. doi: 10.1017/s0016756811000720
Liao Q., Qin Y., Zhou Y., Shi G., Li X., Li J., et al. (2021). Characterization and functional analysis of a chitinase gene: Evidence of ch-chit participates in the regulation of biomineralization in Crassostrea hongkongensis. Aquac Rep. 21, 100852. doi: 10.1016/j.aqrep.2021.100852
Lu G., Yao C., Zhang X., Sun Y., Wang Y., Zhang Z. (2021). Differentially expressed lncRNAs involved in immune responses of Haliotis diversicolor and H. discus hannai challenged with vibrio parahaemolyticus. Comp. Biochem. Physiol. D-Genomics Proteomics. 40. doi: 10.1016/j.cbd.2021.100873
MacDonald B. A., Thompson R. J. (1986). Influence of temperature and food availability on the ecological energetics of the giant scallop Placopecten magellanicus. Mar. Biol. 93 (1), 37–48. doi: 10.1007/BF00428653
Meng J., Wang T., Li L., Zhang G. (2018). Inducible variation in anaerobic energy metabolism reflects hypoxia tolerance across the intertidal and subtidal distribution of the pacific oyster (Crassostrea gigas). Mar. Environ. Res. 138, 135–143. doi: 10.1016/j.aqrep.2021.100852
Nakahara H., Bevelander G. (1971). The formation and growth of the prismatic layer of Pinctada radiata. Calcified Tissue Res. 7 (1), 31–45. doi: 10.1007/BF02062591
Novikova I. V., Hennelly S. P., Sanbonmatsu K. Y. (2012). Sizing up long non-coding RNAs: do lncRNAs have secondary and tertiary structure? Bioarchitecture. 2 (6), 189–199. doi: 10.4161/bioa.22592
Pereiro P., Moreira R., Novoa B., Figueras A. (2021). Differential expression of long non-coding RNA (lncRNA) in Mediterranean mussel (Mytilus galloprovincialis) hemocytes under immune stimuli. Genes. 12 (9). doi: 10.3390/genes12091393
Pérez-Huerta A., Etayo-Cadavid M. F., Andrus C. F. T., Jeffries T. E., Watkins C., Street S. C., et al. (2013). El Nino impact on mollusk biomineralization-implications for trace element proxy reconstructions and the paleo-archeological record. PloS One. 8 (2). doi: 10.1371/journal.pone.0054274
Qiu M.-T., Hu J.-W., Yin R., Xu L. (2013). Long noncoding RNA: An emerging paradigm of cancer research. Tumor Biol. 34 (2), 613–620. doi: 10.1007/s13277-013-0658-6
Rinn J. L., Chang H. Y. (2012). Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 81, 145–166. doi: 10.1146/annurev-biochem-051410-092902
Rose R. A., Baker S. B. (1994). Larval and spat culture of the Western Australian silver- or goldlip pearl oyster, Pinctada maxima jameson (Mollusca: Pteriidae). Aquaculture. 126 (1), 35–50. doi: 10.1016/0044-8486(94)90246-1
Sun W., Feng J. (2018). Differential lncRNA expression profiles reveal the potential roles of lncRNAs in antiviral immune response of Crassostrea gigas. Fish Shellfish Immunol. 81, 233–241. doi: 10.1016/j.fsi.2018.07.032
Suzuki M., Nagasawa H. (2013). Mollusk shell structures and their formation mechanism. Can. J. Zool-Revue Can. Zool 91 (6), 349–366. doi: 10.1139/cjz-2012-0333
Trussell G. C. (2000). Phenotypic clines, plasticity, and morphological trade-offs in an intertidal snail. Evolution; Int. J. organic Evol. 54 (1), 151–166. doi: 10.1111/j.0014-3820.2000.tb00016.x
Weiner S., Addadi L. (2011). Crystallization pathways in biomineralization. Annu. Rev. Mat Res. 41 (1), 21–40. doi: 10.1146/annurev-matsci-062910-095803
Wilke T., Falniowski A. (2001). The genus adriohydrobia (Hydrobiidae: Gastropoda): polytypic species or polymorphic populations? J. Zoological Syst Evol Res. 39 (4), 227–234. doi: 10.1046/j.1439-0469.2001.00171.x
Williams S. T. (2007). Origins and diversification of indo-West pacific marine fauna: evolutionary history and biogeography of turban shells (Gastropoda, turbinidae). Biol. J. Linn. Soc. 92 (3), 573–592. doi: 10.1111/j.1095-8312.2007.00854.x
Wilson J. H. (1987). Environmental parameters controlling growth of ostrea edulis l. and Pecten maximus l. in suspended culture. Aquaculture. 64 (2), 119–131. doi: 10.1016/0044-8486(87)90348-6
Yoon J.-H., Abdelmohsen K., Gorospe M. (2013). Posttranscriptional gene regulation by long noncoding RNA. J. Mol. Biol. 425 (19), 3723–3730. doi: 10.1016/j.jmb.2012.11.024
Yu H., Zhao X., Li Q. (2016). Genome-wide identification and characterization of long intergenic noncoding RNAs and their potential association with larval development in the pacific oyster. Sci. Rep. 6. doi: 10.1038/srep20796
Zhao R., Takeuchi T., Luo Y.-J., Ishikawa A., Kobayashi T., Koyanagi R., et al. (2018). Dual gene repertoires for larval and adult shells reveal molecules essential for molluscan shell formation. Mol. Biol. Evol. 35 (11), 2751–2761. doi: 10.1093/molbev/msy172
Zheng X., Lei S., Zhao S., Ye G., Ma R., Liu L., et al. (2020a). Temperature elevation and acidification damage microstructure of abalone via expression change of crystal induction genes. Mar. Environ. Res. 162. doi: 10.1016/j.marenvres.2020.105114
Zheng Z., Li W., Xu J., Xie B., Yang M., Huang H., et al. (2020b). LncMSEN1, a mantle-specific LncRNA participating in nacre formation and response to polyI:C stimulation in pearl oyster Pinctada fucata martensii. Fish Shellfish Immunol. 96, 330–335. doi: 10.1016/j.fsi.2019.12.015
Zheng Z., Xie B., Cai W., Yang C., Du X. (2020c). Identification of a long non-coding RNA (LncMSEN2) from pearl oyster and its potential roles in exoskeleton formation and LPS stimulation. Fish Shellfish Immunol. 103, 403–408. doi: 10.1016/j.fsi.2020.05.040
Keywords: Pinctada fucata martensii, LncMPEG1, biomineralization, environmental stressor, LncRNA
Citation: Cai C, He Q, Xie B, Xu Z, Wang C, Yang C, Liao Y and Zheng Z (2022) Long non-coding RNA LncMPEG1 responds to multiple environmental stressors by affecting biomineralization in pearl oyster Pinctada fucata martensii. Front. Mar. Sci. 9:1014810. doi: 10.3389/fmars.2022.1014810
Received: 09 August 2022; Accepted: 05 September 2022;
Published: 26 September 2022.
Edited by:
Menghong Hu, Shanghai Ocean University, ChinaReviewed by:
Xiaohui Cai, Beibu Gulf University, ChinaCopyright © 2022 Cai, He, Xie, Xu, Wang, Yang, Liao and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhe Zheng, emhlbmd6aGVAZ2RvdS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.