- 1State Key Laboratory of Marine Environmental Science, College of Ocean and Earth Scences, Xiamen University, Xiamen, China
- 2Fujian Key Laboratory of Genetics and Breeding of Marine Organisms, Xiamen University, Xiamen, China
- 3National Observation and Research Station for the Taiwan Strait Marine Ecosystem (Xiamen University), Zhangzhou, China
Trait separation which often appears in shellfish progeny, has been commercially used in shellfish breeding projects. A three-way cross hybrid abalone was produced with heterosis in growth performance and thermal tolerance, and with segregation in mantle marking. However, the inheritance mechanism of mantle marking is unclear. In this study, mantle marking was demonstrated to be a qualitative trait, following simple Mendelian inheritance, through inheritance pattern analysis of the presence of black markings on the mantle in five families of three-way cross of hybrid abalone. Thermal tolerance of abalone was assessed by the Arrhenius breakpoint temperature (ABT) of cardiac performance and a correlation between mantle marking and thermal tolerance of abalone was verified. A genome-wide association study (GWAS) for mantle marking was conducted using 603,067 high-quality single nucleotide polymorphisms (SNPs) derived from 57 individuals with mantle markings (Y group) and 58 individuals without mantle markings (N group). A total of 493 SNPs that were significantly (p < 2.32E–07) associated with mantle markings were mainly distributed on chromosome 15. In the potential significantly associated region, 80 genes, including pigmentation-related genes PTPRT, PTPRC, PNCA, and CALM4 were annotated. Transcriptome analysis of the two groups showed that the expression levels of these genes (PTPRT, DDT-b, ATOX1, SLC6A3, and GSTO1) were significantly different, and they may play important roles in the formation of mantle markings in the three-way cross hybrid abalone. Overall, our data provide valuable information for deciphering the phenotype differences of mantle marking in three-way cross hybrid abalone and help in the molecular marker-assisted breeding in abalone.
Introduction
Mendel’s law of segregation states that the two alleles for each trait segregate during the formation of gametes, and that the alleles will combine at random with other alleles during zygote formation (Harel et al., 2015). Allele differences often lead to phenotype changes. The phenomenon of trait separation in progenies often appears in organisms and has frequently been used by researchers (Spielman et al., 1990; Zhu et al., 2007; Zhou et al., 2017). For example, color polymorphism occurs in many aquatic species. It has been used to artificially select special individuals or populations since different colors are often associated with phenotypes. For example, the boring giant clam (Tridacna crocea) is polymorphic for mantle color, and the survival rate of the blue strain was significantly lower than the yellow-green strain (Zhou et al., 2021). In the black and white shell varieties of the Pacific oyster (Crassostrea gigas), the relationship between the color of the adductor muscle scars and the dried soft-body weight can be explained by the high hydroxyl free radical scavenging capacity of the muscle attached to the black adductor muscle scar (Yu et al., 2017). Genetic introgression caused by interspecific hybridization can lead to the segregation of pigment traits in progeny. Red tilapia has a higher market value than black blotched tilapia due to its brighter red flesh, faster growth rate, and lower feed coefficient (Fang et al., 2022).
Many studies have evaluated environmental and genetic factors that influence color formation and its relationship with other traits. High-throughput genome sequencing technologies provide tools useful for identifying genetic mechanisms underlying color formation. For example, a genome-wide association study (GWAS) in Salmo salar verified that its flesh color is strongly associated with two single nucleotide polymorphisms (SNPs) positioned on chromosome 26 (Helgeland et al., 2019). The same methods have also been used in mollusks. Two SNPs are significantly associated with the shell color of the Yesso scallop (Patinopecten yessoensis) (Zhao et al., 2017), and 469 SNPs are significantly associated with the shell color of the bay scallop (Argopecten irradians) (Zhao et al., 2017). A combination of genomic and transcriptomic approaches can be used to study key loci, genes and pathways. In the Yesso scallop, the differentially expressed gene between the white and orange muscles, PyBCO-like 1 encoding carotenoid oxygenase, is in the genomic region underlying carotenoid coloration in scallop muscle (Li et al., 2019b). The use of linkage mapping, fine mapping, and gene expression analyses indicated that the Pmel17 gene was responsible for golden and black skin colors in Mozambique tilapia (Liu et al., 2022).
Abalone is an economically important aquaculture species, and the Chinese abalone yield accounts for more than 90% of the global production (FAO, 2020). Based on the commercial demand for improved germplasm, abalone have become models for hybridization research and application practice. Due to the heterozygosity of genotypes and interactions between genes, hybrids often exhibit superior phenotypic characteristics, including enhanced growth rates and increased stress tolerance relative to their parents (Yu et al., 2021). This has been interpreted as heterosis or hybrid vigor. Heterosis research has been conducted in several hybrid abalone species (Hamilton et al., 2009; You et al., 2015), including the hybrid between the greenlip abalone (Haliotis. laevigata) and the blacklip abalone (H. rubra), the hybrid between Xishi abalone (H. gigantea) and the Pacific abalone (H. discus hannai), and the hybrid between Pacific abalone and green abalone (H. fulgens). In our observations of the morphology of pure abalones and hybrid abalones, we found an interesting phenomenon. There were mantle markings in H. fulgens and its hybrid with H. discus hannai, but there were no mantle markings in H. gigantea, H. discus hannai, and their hybrid. Thus, it is possible that the black mantle markings are inherited in green abalone. Previous studies have shown that three-way cross hybrid abalone exhibits heterosis in growth performance, thermal tolerance, and hypoxia tolerance (Xiao et al., 2022b). The offspring of the three-way cross abalone showed segregation of the mantle color trait, with some of the offspring having black markings on the mantle and some of the offspring lacking mantle markings. Segregation of mantle markings also occurs in the offspring of backcrossed abalone, in which individuals with mantle markings grow faster than those without markings (Xiao et al., 2022a). Although preliminary studies (Xiao et al., 2022a) have been made on the mantle markings of abalone, little is known about the genetic basis or the pattern of mantle markings. The juvenile abalone with black mantle markings can be sold at a higher price in China than those without black mantle markings. Color is not only a useful character for species identification but also can be related to traits such as growth and survival (Williams, 2017; Yan et al., 2019). Therefore, the mantle markings in three-way cross abalone may be related to economic traits such as growth and thermal tolerance. It is of theoretical significance and economic value to determine the mechanism behind the separation of mantle markings in abalone.
The objective of this study was to determine the molecular mechanisms of segregation of mantle markings in three-way cross abalone. First, five families of three-way cross abalone were designed to make a further exploration of the inheritance pattern of mantle markings, which will provide valuable information for the eficient selective breeding of mantle markings in abalone. Then thermal tolerance of three-way cross abalone families was assessed through Arrhenius break temperatures (ABT) of cardiac performance to validate the difference between individuals with mantle markings and individuals without mantle markings at physiological levels. Finally, GWAS and transcriptome analyses were performed to identify SNPs and candidate genes that were significantly associated with mantle markings in three-way cross abalone. These results could deepen our understanding of the molecular mechanisms of shellfish trait segregation and provide primary data for breeding.
Materials and methods
Proportion statistics and observation of mantle markings
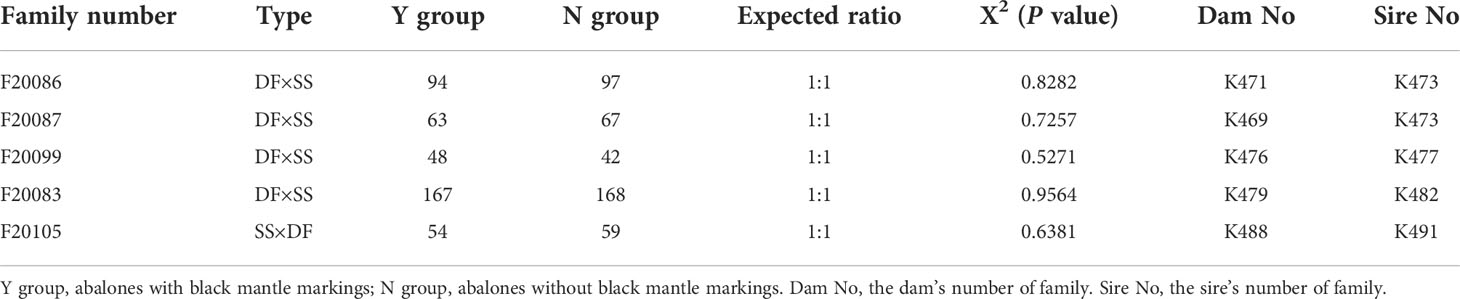
Five full-sib families, including four DF × SS [(H. discus hannai♀ × H. fulgens♂)♀ × H. gigantea♂] families (Family number: F20083, F20086, F20087 F20099) and one SS × DF (H. gigantea♀ × (H. discus hannai♀ × H. fulgens♂)♂) family (Family number: F20105) were established at Fuda Abalone Farm (Jinjiang, China) in November, 2020. FF (H. fulgens ♀ × H. fulgens♂) was introduced from the USA in 2007 and has been bred for five generations, SS (H. gigantea♀ × H. gigantea♂) was introduced from Japan in 2003 and has been bred for several generations. DD (H. discus hannai♀ × H. discus hannai♂) was a strain bred for seven generations in our laboratory, and DF was a hybrid between DD and FF. Selection of gonadally mature individuals as parents for family construction. Information about the sire and dam of each family was shown in the Table 1. After 8 months of culturing in the same environment, each family was divided into two groups (Y group: abalones with mantle markings; N group: abalones without mantle markings) and the number of abalones in each group was counted. The X2 test was used to determine whether the segregation proportion of mantle markings was in accordance with Mendel’s Law of segregation.
Six individuals with mantle markings and six individuals without mantle markings were selected and their mantle tissues were dissected. The morphology of the pigment cells on the mantle was observed under a stereomicroscope (Leica MDG41). Photomicrographs were obtained using a Leica DFC495 camera connected to the stereomicroscope, using the Leica ApplicatiOn Suite 4.5.0.418 software.
ABT of cardiac performance
Twenty-four abalones with mantle markings (Y group) and twenty-four abalones without mantle markings (N group) from each DF×SS family were selected and used to determine their ABT value. The non-invasive ABT method was used for heart-rate measurement (Chen et al., 2016). A total of 144 individuals (shell length: 65.18 ± 4.30 mm) from three DF×SS families (Family number: F20083, F20086, F20099) were acclimated in a thermos-controlled seawater recirculating system for seven days. After acclimation, abalones were placed in a transparent plastic box (30.0 × 20.0 × 15.0 cm) that was immersed in a thermo-controlled water bath. The seawater in the plastic box was aerated, and its temperature was increased at a rate of 0.1°C/min starting from 20°C. A thermometer (Fluke 54II, Fluke calibration, Everett, USA) was used for temperature monitoring. An infrared sensor (Krazy Glue, Westerville, OH, USA) was glued to the shell above the heart of the abalone. Then the fluctuations of heart beats were amplified, filtered, and recorded by an infrared signal amplifier (AMP03, Newshift, Leiria, Portugal) and Powerlab (8/35, ADInstruments, March-Hugstetten, Australia). Heart beats were monitored and analyzed by LabChart v8.0. The ABT value was defined as the temperature at which the heart rate decreased dramatically, determined by using regression analyses to generate the best fit line on both sides of a putative break point. To construct Arrhenius plots, heart rates were transformed to the natural logarithm of beats min−1. Temperatures are shown as 1000/K (Kelvin temperature). Student’s t-tests were conducted by SPSS v24.0 to compare the differences in ABT between the Y group and the N group. A P < 0.05 was considered to be significant.
GWAS analysis for mantle markings in abalone
In this study, considering the complexity and completeness of the genotypes of the individuals in three-way crosses abalone, we finally selected all individuals from one family for the GWAS analysis. A total of 115 14-month-old individuals were selected from F20087 family and divided into two groups according to the presence (Y group, 57 individuals) or absence (N group, 58 individuals) of mantle markings. Their foot muscle tissues were dissected, immediately frozen in liquid nitrogen, and stored at −80°C for DNA extraction.
Genomic DNA was extracted using the DNeasy 96 Blood & Tissue Kit (Qiagen, Shanghai, China). DNA quality and quantity were assessed using agarose gel electrophoresis and a Nanodrop2000 (Thermo Scientific, Wilmington, DE, USA), respectively. Paired-end libraries with an insert size of 350 bp were constructed for the individual samples and sequenced on an Illumina NovaSeq 6000 platform (Illumina, USA) using the 150 bp pair-end strategy at Novogene Corporation (Beijing, China).
The sequence reads for each sample were quality controlled using an in-house Perl script, removing the following reads: those aligned to the barcode adaptor; those with ≥ 10% unidentified nucleotides (N); and low-quality reads (those where > 50% of all bases had phred quality scores ≤ 20). The clean reads were aligned to the green abalone genome (unpublished) using the BWA mem tool (Li and Durbin, 2009) with default parameters. Variant detection and filtering of the generated BAM files were performed using the SAMtools (Li et al., 2009) pipeline to generate a set of raw SNPs. Raw SNPs were further quality controlled by Vcftools (Danecek et al., 2011). All non-biallelic SNPs were removed. Then, the following SNPs and samples were discarded: average sequencing depth < 8×(-minDP); SNPs with minor allele frequency (-maf) < 0.05; SNPs with deviation from Hardy-Weinberg equilibrium < 0.001 (-hwe); and individuals with a missing data rate (-mind) > 0.2.
To reduce the false-positive rate, population structure analysis was performed to provide principal components analysis (PCA) through PLINK software (Purcell et al., 2007) and a kinship matrix was estimated by Gemma software (Zhou and Stephens, 2012). Genome-wide association analysis between genotypes and mantle markings was conducted using Gemma software by employing a mixed linear model with these two covariates to detect the associated SNPs. The statistic p-value for each SNP was calculated and summarized in ascending order. The threshold p-value for genome-wide significance was calculated using the Bonferroni correction based on the number of qualified makers. When an SNP scored less than the significance cutoff (2.32E–07), the sequence of each maker was extracted and used as a query to exert BLASTN to search the draft genome of green abalone. In addition, the Manhattan plot was drawn by the R CMplot package, and these potential SNPs (p < 2.32E–07) were captured to conduct bioinformatics analysis.
Transcriptomic analysis for mantle markings in abalone
Three individuals with mantle markings (Y group) and three individuals without mantle markings (N group) in F20087 family were dissected for mantle tissues. In this study, the mantle tissues sampling locations of RNA-seq were shown in the Figure 1B. Mantle tissues were immediately frozen in liquid nitrogen and stored at −80°C for RNA extraction. Total RNA was extracted using the Trizol method. To check the purity and integrity of RNA, the NanophotometerR spectrophotometer (Implen, Westlake Village, CA, USA) and RNA Nano 6000 Assay Kit of the Agilent Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA) were used. Library preparation, and sequencing were performed by Novogene (Beijing, China). RNA-seq libraries were constructed according to the manufacturer’s protocol of the Vazyme mRNA-seq library preparation kit (Vazyme) and were sequenced to generate 150-nucleotide paired-end reads on an HiSeq platform (Illumina).
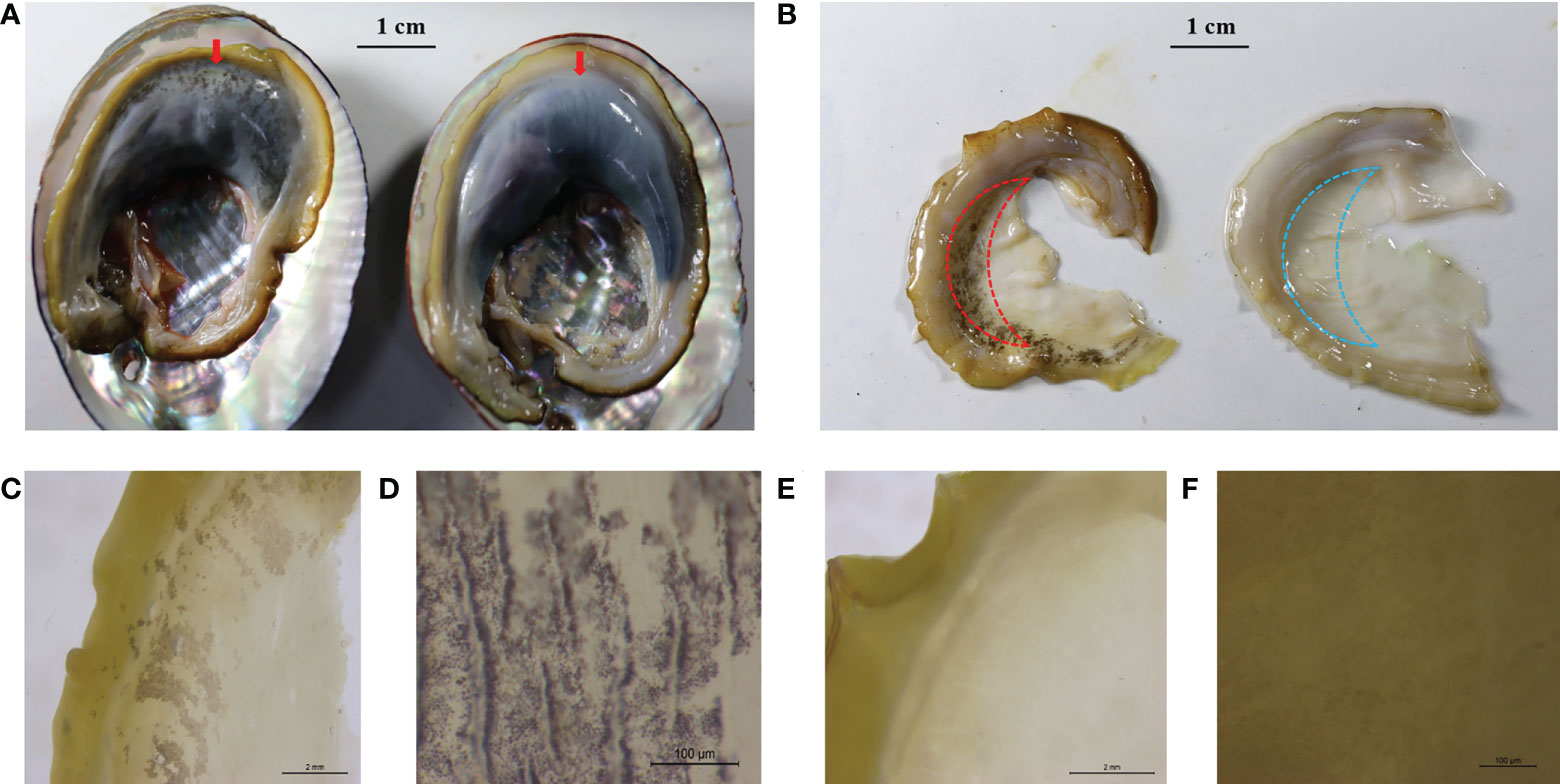
Figure 1 Representative mantle marking morphs. (A) An individual with black mantle markings (left, the red arrow) and an individual without black mantle markings (right, the red arrow) recorded with a digital camera, scale 1 cm. (B) Mantle with black markings (left) and mantle without black markings (right) recorded with a digital camera, scale 1 cm. Red and blue dashed lines indicate the sampling locations of RNA-seq. (C) Black mantle markings under the microscope, scale 2 mm. (D) Black mantle markings under the microscope, scale 100 μm. (E) Mantle without black markings under the microscope, scale 2 mm. (F) Mantle without black markings, scale 100 μm.
Raw reads were filtered using fastp (Chen et al., 2018) with the following parameters: -w 16 -z 6 -q 20 -u 30 -n 10 -l 150. The clean reads were aligned to the reference genome of the green abalone using HISAT2 (Kim et al., 2015), generating BAM files. The BAM files were sorted using SAMtools and then used to estimate abundances of annotated genes in the reference genome using StringTie (Pertea et al., 2015) with the following parameters: -e –B. The gene read counts were extracted using the pre-pDE.py script included in StringTie. The gene expression profiles were derived from the TMM normalization of the read counts with rlog transformation using R DESeq2 package. The differentially expressed genes (DEGs) were identified with a false discovery rate (FDR) < 0.05 and |log2FoldChange| > 1. The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of genes was conducted using R clusterProfiler package (Yu et al., 2012), based on a modified Fisher’s exact test with p < 0.05 and FDR cutoff < 0.05.
Results
Descriptive statistics for mantle markings in three-way cross abalone
Based on the mantle markings (Figure 1), individuals from each family were reclassified into Y group and N group. Under the microscope, the mantle surface of the Y group was found to have black deposited pigment granules, whereas the mantle surface of the N group was pale yellow without black deposited granules (Figure 1). As shown in Table 1, the number of individuals with black mantle markings and without black mantle markings corresponded to a 1:1 ratio, both in DF×SS and SS×DF families.
Correlation between mantle markings and thermal tolerance
The ABT values differed between the two groups in three families (Figure 2). The ABT values of the Y group (F20086-Y: 32.66 ± 0.58°C; F20083-Y: 32.61 ± 0.73°C; F20099-Y: 32.42 ± 0.75°C) were significantly higher than the N group (F20086-N: 32.14 ± 0.66°C; F20083-N: 31.87 ± 0.72°C; F20099-N: 31.80 ± 0.64°C) in three DF×SS families (p < 0.05). This indicated that individuals with mantle markings are more heat-tolerant than those without mantle markings.
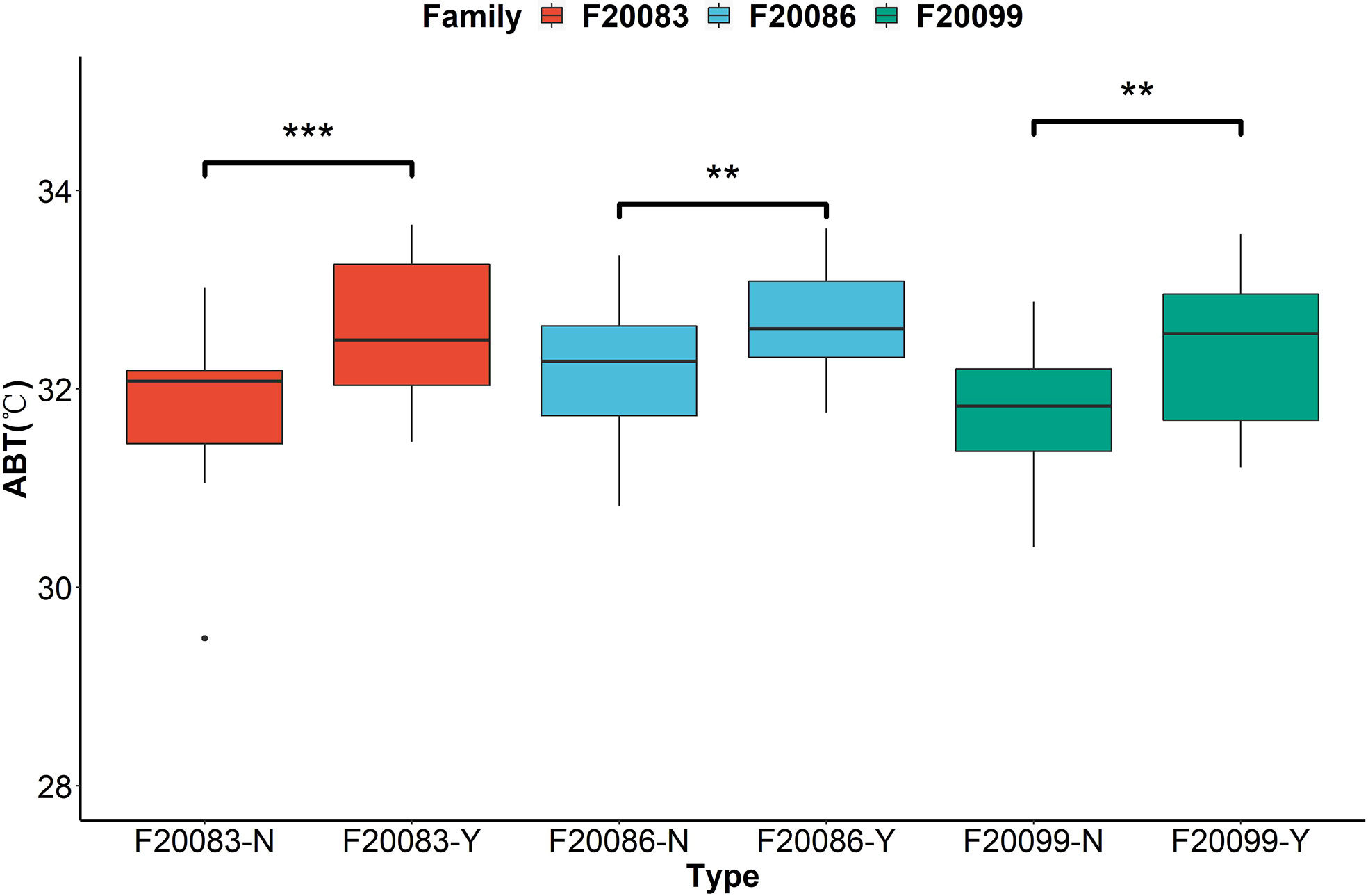
Figure 2 The comparisons of thermal tolerance between the individuals with black mantle markings (Y) and individuals without black mantle markings (N) in three families based on the assessment of Arrhenius break temperatures (ABT) of cardiac performance. **indicates P < 0.01. ***indicates P < 0.001.
Results of GWAS for mantle markings
Whole genome resequencing of 115 individuals generated a total of 1545.16 Gb of raw data. After the removal of low-quality reads, 1502.18 Gb of clean data remained. As the total length of the assembled green abalone reference genome is approximately 1.4 Gb, the average sequencing depth for each sample was 9.28X. After quality control, a total of 603,067 SNPs and 115 samples remained for further analysis. On average, the marker density was approximately 2,320 bp/SNP across the whole genome (Figure 3).
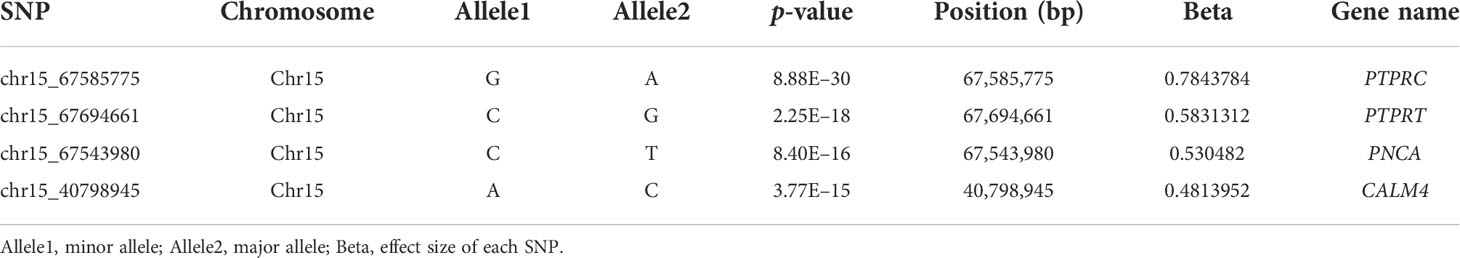
The GWAS was carried out using 603,067 SNPs and 115 samples from one full-sib family. After conducting Bonferroni correction and 0.05 cutoff (p < 2.32E−07), we detected 493 genome-wide SNPs that were significantly associated with mantle markings (Figure 4A). Because of a linkage with a major gene, the majority of association signals were located on chromosome 15 (Figure 4B). Based on the position information of the significant SNPs, the genes located upstream and downstream of the significant SNPs, and the nearest genes within the 50 Kb range were selected as candidate genes related to mantle markings of the abalone. Finally, a total of 80 genes were annotated. Further analysis revealed that the gene with the greatest number of highly significant SNPs was PTPRC (receptor-type tyrosine-protein phosphatase C), containing 95 SNPs, followed by PTPRT (receptor-type tyrosine-protein phosphatase T), containing 82 SNPs. Other genes, such as CALM4 (calmodulin-4) and PNCA (pyrazinamidase/nicotinamidase) (Table 2), may also be involved in the formation of mantle markings in three-way cross hybrid abalone.
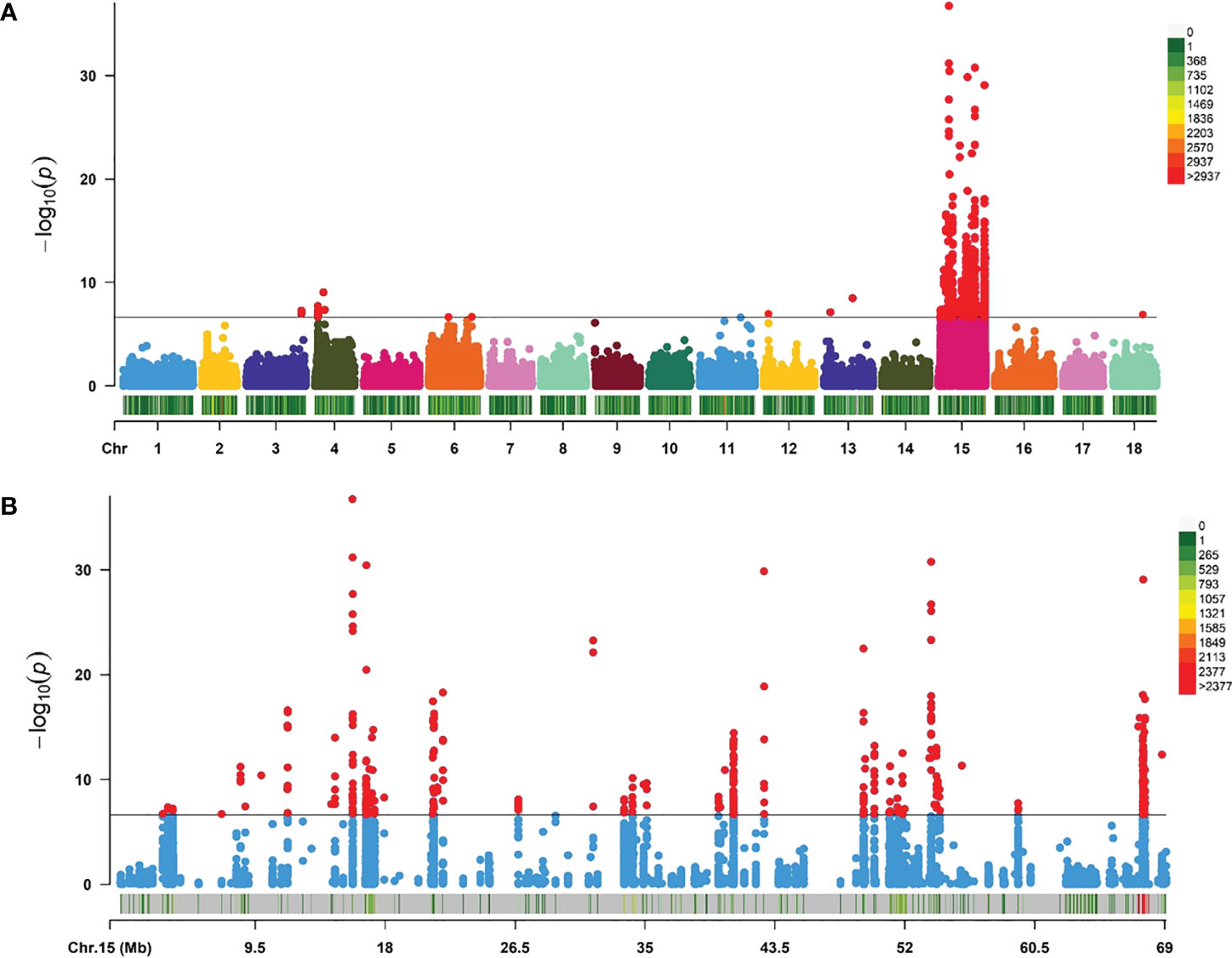
Figure 4 Manhattan plots of genome-wide association study (GWAS) analysis. (A) GWAS for mantle markings with 603,067 single-nucleotide polymorphisms (SNPs) in three-way cross abalone (p < 2.32E–07). (B) GWAS for mantle markings with 10,116 SNPs on Chromosomes 15 in three-way cross abalone (p < 2.32E–07).
Transcriptome sequencing and identification of DEGs
In total, an average of 32.66 million raw reads and 31.13 million clean reads per sample were obtained through RNA-sequencing. A PCA based on the whole-genome gene expression profiles showed that two groups were clearly separated in the PC1-PC2 score plot (Figure 5A), with PC1 explaining 36% and PC2 explaining 27% of the total variance. Between the Y and N groups, a total of 413 DEGs were identified. Of these, 252 genes were upregulated and 161 were downregulated in group Y relative to group N (Figure 5B). In addition, 276 of the DEGs could be annotated on the green abalone genome, including PTPRT (receptor-type tyrosine-protein phosphatase T), DDT-b (D-dopachrome decarboxylase-B), ATOX1 (copper transport protein ATOX1), SLC6A3(sodium-dependent dopamine transporter), and GSTO1(glutathione S-transferase omega-1) (Figure 5C). The results of the KEGG pathway enrichment analysis showed that those 413 DEGs were enriched in vitamin digestion and absorption, cysteine and methionine metabolism, and peroxisome and folate biosynthesis (Figure 5D).
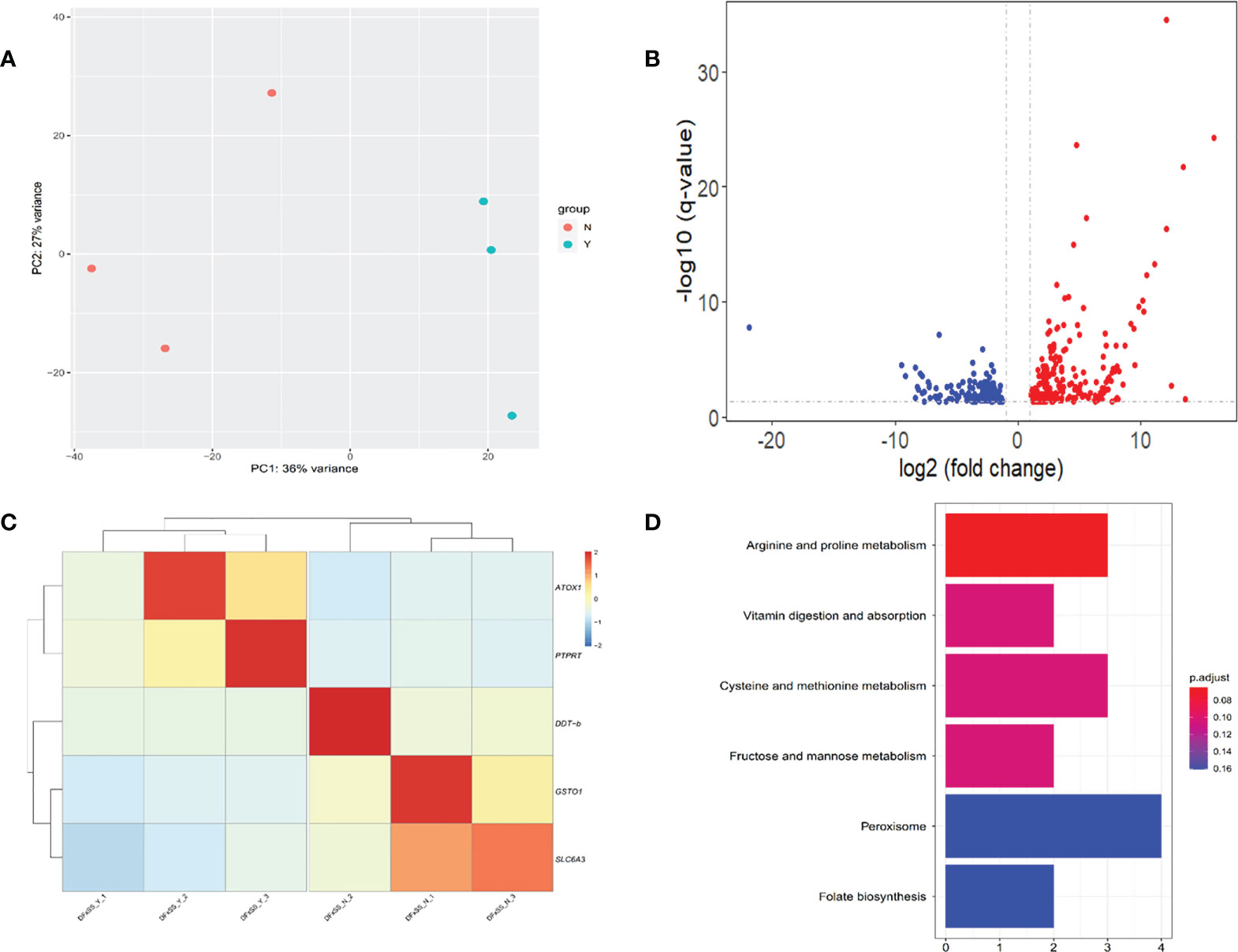
Figure 5 Transcriptomic analysis between individuals with black mantle markings (Y group) and individuals without black mantle markings (N group). (A) Principal component analysis plot based on the whole-genome gene expression profiles. (B) Volcano plot of gene expression in the mantle of Y group and N group. The up-regulated and down-regulated differentially expressed genes (DEGs) are shown in red and blue dots, respectively. (C) Heatmaps of candidate DEGs. (D) KEGG enrichment pathways of the DEGs.
Discussion
Color traits are heritable in most aquatic species. Examples include the flesh color in Atlantic salmon (Norris and Cunningham, 2004), red/black skin of tilapia (Thodesen et al., 2013), carapace/hepatopancreas color in crab (Li et al., 2019a), black/white shell color in oyster (Xu et al., 2017; Xu et al., 2019), and white/orange in scallop adductor muscle (Li et al., 2019b). Mantle pigmentation is inherited in C. gigas according to Mendelian inheritance with “lighter” and “darker” mantles in a 3:1 ratio (Wang et al., 2015). In scallops, analysis of F1 and F2 families showed that carotenoid content in muscle is controlled by a single Mendelian factor (Li et al., 2019b). The color pattern in the mantle is also an important trait for species identification in abalone, indicating it is likely an inherited trait. In the present study, the number of individuals with black mantle markings and individuals without black mantle markings were present in a 1:1 ratio in both DF×SS and SS×DF families (Table 1). There are mantle markings in H. fulgens and its hybrid with H. discus hannai (H. discus hannai♀×H. fulgens♂ and H. fulgens♀×H. discus hannai♂), but there are no mantle markings in H. gigantea, H. discus hannai, and their hybrid (H. gigantea♀×H. discus hannai♂ and H. discus hannai♀×H. gigantea♂). Thus, it is possible that the black mantle markings are inherited in green abalone and its offspring as a qualitative trait, possibly dominant by one genetic locus or a linkage between genomic regions. In rainbow trout, skin color characters are controlled by one locus or very few loci, in a dominant, recessive, or co-dominant inheritance mode (Blanc et al., 2006). Most of the significant SNPs associated with mantle color were primarily allocated to chromosome 15 of three-way cross abalone. Similarity to the bay scallop, most of the significant SNPs associated with shell color were primarily allocated to chromosome 7 (Zhu et al., 2021). In the Yesso scallop, the major SNPs associated with shell color are located on chromosome 11 (Zhao et al., 2017), and almost all the significant SNPs associated with carotenoid coloration in scallop adductor muscle are distributed on chromosome 8 (Li et al., 2019b). Since most qualitative traits are usually determined by one or several key genes, we speculated that the color variants in the mantle of abalone may be regulated by a limited number of candidate genes that are closely linked to genomic regions of the specific linkage groups.
In aquatic animals, color variation is of interest to both scientists and breeders. Color is an important commercial trait, given that it influences consumer acceptance, and affects the commercial value (Colihueque, 2010). In the Chinese abalone industry, juvenile abalone with black mantle markings sell at much higher prices than those without black mantle markings. However, it is unknown if selection for mantle markings has adverse effects on other commercial traits, such as growth, survival, and environmental adaptability. In Fujian Province, which produces 80% of the abalone in China, abalone faces serious threats from high summer temperatures. An increase in thermal resistance could play important role in increase abalone survival. It is therefore important to breed heat tolerant strains of abalone (Xiao et al., 2021). In this study, the correlation between mantle markings and thermal tolerance was assessed by the non-invasive Arrhenius breakpoint temperature method in three DF×SS families. There was a significant correlation in mantle markings and ABT values. Individuals with black mantle markings had better thermal tolerance than those without black mantle markings. At the same time, the results of the KEGG pathway enrichment analysis showed that those DEGs were enriched in vitamin digestion and absorption pathway and peroxisome and folate biosynthesis pathway. Studies have shown that the peroxisome plays a key role in the heat stress response. High temperature induce oxidative stress in organism by accelerating the generation of reactive oxygen species (ROS) (Sharma et al., 2012). The peroxisome is one of the key components of the ROS scavenging system. The peroxisome contains an efficient ROS scavenging system including glutathione peroxidase, peroxidase, superoxide dismutase and catalase that scavenges ROS to reduce damage caused by heat stress to the organism (Corpas et al., 2017). Many studies have demonstrated the importance of vitamins in heat stress responses. For example, the vitamin digestion and absorption pathway has an important function in insect adaptation to high temperatures. After heat stress, up-regulation of the expression of some genes involved in the vitamin digestion and absorption pathway can be detected during the signaling of ROS (Liu et al., 2017). This may be due to the similarity of processes in the biological and metabolic pathways involved in the control of mantle markings and thermal tolerance in abalone. Similar studies have shown strong positive genetic correlations (correlation coefficient range of 0.59–0.70) between growth related traits and body color in banana shrimp (Fenneropenaeus merguiensis) (Nguyen et al., 2014). In the Pacific oyster, the adductor muscle scar color was correlated with the soft-body dry weight, so the black adductor muscle scar may be a potential breeding character that could be used to obtain a larger soft-body dry weight (Yu et al., 2017). In some species of Drosophila, the melanin in the body is associated with heat resistance and has wound healing and immune functions (Rajpurohit et al., 2008). Additionally, melanin has bioactivity as an antioxidant and may be involved in the process of inflammatory reactions and immune responses in abalone. This suggests that black mantle markings may be a new breeding trait, which could be used to obtain abalone with increased heat tolerance.
GWAS is a powerful method for elucidating the genetic variations associated with complex economic traits, providing candidate genes and markers for selective breeding programs (Visscher et al., 2012). Functional genes can often be found around significant SNPs in GWAS (Pharoah et al., 2013). The Manhattan plot results showed an extremely significant association between some SNPs and mantle markings. The value of −log10 (P value) was over 15, which indicated that false positives were rare (Figure 4). Because of linkage with major gene, more association signals involve chromosome 15. In the potential region we identified the genes PTPRC, PTPRT, PNCA, and CALM4 whose functions involve regulation of migration and transport. In this study, we hypothesized that the black mantle markings might result from melanin precipitation. Previous studies have shown the presence of melanin in the muscles and mantles of the abalone (Hao, 2018). At the same time, we extracted the pigment of the mantle with black marking and scanned the UV spectrum in the range of 190-500nm (unpublished). The results showed that the extracted pigment in mantle of abalone and the melanin standard sample had the same spectral characteristics. They had obvious absorption characteristics at UV wavelengths with a maximum absorption peaks at 210-220 nm, and their absorbance values decreased with increasing wavelengths, which was the same as the absorption characteristics of melanin in the UV spectrum reported by Bell (Bell, 1986). Therefore, it can be tentatively concluded that the mantle contains melanin. The synthesis of melanin is a complex biochemical process involving a series of enzymatic reactions. First, tyrosine is converted into DOPA (dihydroxyphenylalanine), which is then used to produce DOPA melanin. Alternatively, DOPA can be further converted into dopamine by DOPA decarboxylase. Finally, phenol oxidases convert dopamine into its quinones, which are converted into dopamine melanin (Lemonds et al., 2016). The black pigments on the mantle and muscle of H. iris were identified as melanin (Hao, 2018). Melanin biosynthesis is likely to be regulated by tyrosinase that contributes to the black-brown shell pigmentation in most mollusks (Sun et al., 2017). PTPRT is a classical protein tyrosine phosphatase that has a core cysteine residue in the active site and is localized in the plasma membrane by its trans-membrane domain (Sallee et al., 2006). PTPRT has been linked to signal transduction, cell adhesion, and neurite extension, and regulates synapse formation by interacting with cell adhesion molecules and protein tyrosine kinase (Lim et al., 2009). In this study, expression of PTPRT in the mantle was higher in the Y group than in the N group (Figure 5C), suggesting that PTPRT may promote the production of melanin. CALM4 is present in stratified epithelia, vibrissae hair follicles, and perichondral osteoblasts (Hwang et al., 2005). CALM4 is a calcium-dependent regulatory protein that is widely present in all eukaryotic cells and has a conserved structure. It stimulates the activity of calmodulin-dependent protein kinase, which promotes melanin synthesis and involved in the melanogenesis pathway (Lessard et al., 2015). PNCA is a nicotinamidase encoded by the PNCA gene and functions in the NAD salvage pathway by converting nicotinamide into nicotinic acid (Shang et al., 2018). Nicotinamide can inhibit the formation of melanin particles and inhibit melanosome transfer from melanocytes to keratinocytes. It can promote skin lightening in vivo and is widely used in the cosmetics industry (Hakozaki et al., 2002).
The mantles of DF×SS were collected and the gene expression patterns were analyzed in different mantles. A series of genes that may be related to melanin synthesis were found to be differentially expressed. DDT-b encodes DDT-b that is a cytokine and a member of the macrophage migration inhibitory factor (MIF) protein superfamily. MIF activates the ERK1/2 MAPK pathway, induces cell proliferation and inhibits apoptosis. The MAPK pathway has also been shown to have an important role in melanogenesis (Jung et al., 2016). DDT-b converts d-dopachrome into 5,6-dihydroxyindole, and is involved in melanin biosynthesis. Dopaquinone can spontaneously convert into dopachrome, which can be decarboxylated by dopachrome conversion enzyme into form 5,6-dihyroxyindole. The resulting compound will polymerize to form melanin. Dopamine, derived from dopa via the action of dopa decarboxylase, can also serve as the catecholamine substrate for melanin production (Christensen et al., 2005). ANTOX1, the encoding production of ATOX1, is a small cytosolic protein and plays an essential role in copper homeostasis by acting as a copper carrier and facilitating the transfer of copper to the secretory pathway (Hatori and Lutsenko, 2016). Copper is essential in the pigmentation process as a cofactor of enzymes involved in electron transfer, oxygen transport, and redox reactions (Contreras-Moreno et al., 2020). The active center of tyrosinase contains two copper ions. The absence of copper ions will greatly reduce the activity of tyrosinase and hinder the conversion of tyrosine into melanin. Copper ions therefore play a vital role in the production of melanin catalyzed by tyrosinase (Ghani, 2019). In the present study, the expression levels of ATOX1 in the mantle were higher in the Y group than in the N group (Figure 5C), suggesting that ATOX1 may increase tyrosinase activity and promote melanin production. GSTO1 as a member of the glutathione S-transferase family genes (Wang et al., 2019). Glutathione S-transferase (GST) is the key enzyme in the glutathione binding reaction and catalyzes the initiation step of the glutathione binding reaction, which plays an active role in the development of drug resistance by numerous cancer cells, including melanoma cells (Ji et al., 2019). Glutathione scavenges free radicals, directly inhibits tyrosinase activity, and tilts the melanin synthesis pathway toward pheomelanin by hydrolysis to cysteine. In this study, the expression levels of GSTO1 in the mantle were lower in the Y group than in the N group (Figure 5C), suggesting that GSTO1 may inhibit tyrosinase activity. SLC6A3 encodes dopamine transporter. The dopamine transporter then mediates the reuptake of the neurotransmitter dopamine from the extracellular space into the presynaptic dopaminergic neuron, and serves an important function in terminating dopaminergic signaling (Borre et al., 2014). Dopamine is a precursor for melanin synthesis, inducing melanosome aggregation through alpha-adrenergic receptors and thus promoting melanogenesis (IGA, 1980). In this study, the expression level of SLC6A3 in the mantle was lower in the Y group than in the N group (Figure 5C), suggesting that SLC6A3 may terminate dopaminergic signaling and reduce melanin formation. The KEGG pathway enrichment analysis showed that those 413 DEGs were enriched in vitamin digestion and absorption, cysteine and methionine metabolism, and peroxisome and folate biosynthesis. Vitamins are involved in the regulation of the organs’ metabolism, so digestion and absorption of vitamin are important for the normal functioning of the organism. Vitamin D is an essential hormone synthesized in the skin and is responsible for skin pigmentation. Studies have shown that vitamin D promotes melanin proliferation, and can helps regulate the immune response to skin lesions. Vitamin D also has the function of promoting melanocyte proliferation and migration, restoring calcium homeostasis inside and outside the cells, and relieving the inhibition of melanin synthesis by oxidative stress (Alghamdi et al., 2013). Cysteine and methionine are sulfur-containing amino acids. When consumed together with vitamin C, cysteine inhibits melanin production and deposition. During the biosynthesis process of melanin, cysteine accelerates the pathway directed to formation of pheomelanin, which produces yellowish or reddish colors and blocks formation of eumelanin that produces dark colors (Solano, 2014). Peroxisomes participate in various metabolic pathways including biosynthesis of cholesterol, and detoxification of xenobiotic and reactive oxygen species. Studies have shown that cells can reduce the intensity of color by activating lysosomes and peroxisomes, which reduce the amount of melanin (Park et al., 2016). However, since the interspecific three-way cross abalones were used for the GWAS and transcriptome analysis, the identified genetic signals could be due to the differences in the three purebred genomes (i.e., different alleles in each species). Future studies will need to sequence and assemble the genome of three-way cross abalone to overcome the limitations of the reference genome used here.
Conclusion
In conclusion, the mantle marking of three-way cross abalone was demonstrated to be a qualitative trait controlled by a genetic locus or linkage genomic region. The mantle markings of three-way cross abalone were significantly correlated with thermal tolerance. This genetic relationship should be considered when simultaneously incorporating them into breeding objectives. We also identified, using GWAS, one genomic region on chromosome 15 that is associated with mantle markings in abalone. In this potential region, four genes (PTPRC, PTPRT, PNCA, and CALM4) may be important in the mantle markings of three-way cross hybrid abalone. Transcriptomic analysis showed that the expression levels of five genes (PTPRT, DDT-b, ATOX1, SLC6A3, and GSTO1) were significantly different in individuals with and without mantle markings. These data collectively provide useful information for deciphering the phenotype differences of mantle markings and will aid in molecular marker-assisted breeding of abalone.
Data availability statement
The sequencing reads for both whole genome-sequencing and transcriptome sequencing are archived in the China National GeneBank DataBase (CNGB) with BioProject accession CNP0003526.
Ethics statement
The animal study was reviewed and approved by Xiamen University Committee on the Use and Care of Animals.
Author contributions
CK, WP, XuL, WY, and QX conceived and designed the experiment. QX, XiL, HJ, and YG conducted the experiment. QX, SG, YL, ZeH, and ZhH involved in data analysis. QX, YS, WY, and CK wrote the in manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from National Natural Science Foundation of China (No. 31872564 and 32102775) and National Key R&D Program of China (2018YFD0901401); Key S & T Program of Fujian Province (No. 2020NZ08003); Earmarked fund for CARS (CARS-49); and the Special Fund for Ocean and Fisheries of Xiamen (No.21CZY018HJ01).
Acknowledgments
Thanks for Germplasm resources sharing platform of aquatic species in Fujian Province and XMUMRB abalone research center support of the experiment.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alghamdi K., Kumar A., Moussa N. (2013). The role of vitamin d in melanogenesis with an emphasis on vitiligo. Indian J. Dermatol Venereol Leprol 79 (6), 750–758. doi: 10.4103/0378-6323.120720
Bell A. (1986). Biosynthesis and functions of fungal melanins. Annu. Rev. Phytopathol. 24 (1), 411–451. doi: 10.1146/annurev.phyto.24.1.411
Blanc J. M., Poisson H., Quillet E. (2006). A blue variant in the rainbow trout, Oncorhynchus mykiss walbaum. J. Hered 97 (1), 89–93. doi: 10.1093/jhered/esj010
Borre L., Andreassen T. F., Shi L., Weinstein H., Gether U. (2014). The second sodium site in the dopamine transporter controls cation permeation and is regulated by chloride. J. Biol. Chem. 289 (37), 25764–25773. doi: 10.1074/jbc.M114.574269
Chen N., Luo X., Gu Y., Han G., Dong Y., You W., et al. (2016). Assessment of the thermal tolerance of abalone based on cardiac performance in Haliotis discus hannai , H. gigantea and their interspecific hybrid. Aquaculture 465, 258–264. doi: 10.1016/j.aquaculture.2016.09.004
Chen S., Zhou Y., Chen Y., Gu J. (2018). Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34 (17), i884–i890. doi: 10.1093/bioinformatics/bty560
Christensen B. M., Li J., Chen C. C., Nappi A. J. (2005). Melanization immune responses in mosquito vectors. Trends Parasitol. 21 (4), 192–199. doi: 10.1016/j.pt.2005.02.007
Colihueque N. (2010). Genetics of salmonid skin pigmentation: clues and prospects for improving the external appearance of farmed salmonids. Rev. Fish Biol. Fisheries 20 (1), 71–86. doi: 10.1007/s11160-009-9121-6
Contreras-Moreno F. J., Munoz-Dorado J., Garcia-Tomsig N. I., Martinez-Navajas G., Perez J., Moraleda-Munoz A. (2020). Copper and melanin play a role in myxococcus xanthus predation on sinorhizobium meliloti. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.00094
Corpas F. J., Barroso J. B., Palma J. M., Rodriguez-Ruiz M. (2017). Plant peroxisomes: a nitro-oxidative cocktail. Redox Biol. 11, 535–542. doi: 10.1016/j.redox.2016.12.033
Danecek P., Auton A., Abecasis G., Albers C. A., Banks E., DePristo M. A., et al. (2011). The variant call format and VCFtools. Bioinformatics 27 (15), 2156–2158. doi: 10.1093/bioinformatics/btr330
Fang W., Huang J., Li S., Lu J. (2022). Identification of pigment genes (melanin, carotenoid and pteridine) associated with skin color variant in red tilapia using transcriptome analysis. Aquaculture 547. doi: 10.1016/j.aquaculture.2021.737429
FAO (2020). The state of world fisheries and aquaculture, (2020). Sustainability in action (Rome: FAO). doi: 10.4060/ca9229en
Ghani U. (2019). Carbazole and hydrazone derivatives as new competitive inhibitors of tyrosinase: Experimental clues to binuclear copper active site binding. Bioorg Chem. 83, 235–241. doi: 10.1016/j.bioorg.2018.10.026
Hakozaki T., Minwalla L., Zhuang J., Chhoa M., Matsubara A., Miyamoto K, et al. (2002) The effect of niacinamide on reducing cutaneous pigmentation and suppression of melanosome transfer. Br. J. Dermatol. 147 (1), 20–31. doi: 10.1046/j.1365-2133.2002.04834.x
Hamilton M. G., Kube P. D., Elliott N. G., McPherson L. J., Krsinich A. (2009). Development of a breeding strategy for hybrid abalone. Aquaculture 18, 350–353.
Hao S. (2018). Preliminary research of black mantle and muscle of the new Zealand haliotis iris. Shanghai: Shanghai Ocean University.
Harel T., Pehlivan D., Caskey C. T., Lupski J. R. (2015). “Mendelian, non-mendelian, multigenic inheritance, and epigenetics,” in Rosenberg's molecular and genetic basis of neurological and psychiatric disease (Fifth Edition) 2015, 3–27. doi: 10.1016/B978-0-12-410529-4.00001-2
Hatori Y., Lutsenko S. (2016). The role of copper chaperone Atox1 in coupling redox homeostasis to intracellular copper distribution. Antioxidants (Basel) 5 (3), 1–16. doi: 10.3390/antiox5030025
Helgeland H., Sodeland M., Zoric N., Torgersen J. S., Grammes F., von Lintig J., et al. (2019). Genomic and functional gene studies suggest a key role of beta-carotene oxygenase 1 like (bco1l) gene in salmon flesh color. Sci. Rep. 9 (1), 20061. doi: 10.1038/s41598-019-56438-3
Hwang M., Kalinin A., Morasso M. I. (2005). The temporal and spatial expression of the novel ca++-binding proteins, scarf and Scarf2, during development and epidermal differentiation. Gene Expr Patterns 5 (6), 801–808. doi: 10.1016/j.modgep.2005.03.010
IGA (1980). Melanosome aggregatimg action of dopamime on fish melanophores. Memoirs Faculty Sci. Shimane Univ. 14, 95–101.
Ji Y., Dai F., Yan S., Shi J. Y., Zhou B. (2019). Identification of catechol-type diphenylbutadiene as a tyrosinase-activated pro-oxidative chemosensitizer against melanoma A375 cells via glutathione s-transferase inhibition. J. Agric. Food Chem. 67 (32), 9060–9069. doi: 10.1021/acs.jafc.9b02875
Jung E., Kim J. H., Kim M. O., Jang S., Kang M., Oh S. W., et al. (2016). Afzelin positively regulates melanogenesis through the p38 MAPK pathway. Chem. Biol. Interact. 254, 167–172. doi: 10.1016/j.cbi.2016.06.010
Kim D., Langmead B., Salzberg S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12 (4), 357–360. doi: 10.1038/nmeth.3317
Lemonds T. R., Liu J., Popadic A. (2016). The contribution of the melanin pathway to overall body pigmentation during ontogenesis of periplaneta americana. Insect Sci. 23 (4), 513–519. doi: 10.1111/1744-7917.12356
Lessard J. C., Kalinin A., Bible P. W., Morasso M. I. (2015). Calmodulin 4 is dispensable for epidermal barrier formation and wound healing in mice. Exp. Dermatol. 24 (1), 55–57. doi: 10.1111/exd.12568
Li H., Durbin R. (2009). Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 25 (14), 1754–1760. doi: 10.1093/bioinformatics/btp324
Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., et al. (2009). The sequence Alignment/Map format and SAMtools. Bioinformatics 25 (16), 2078–2079. doi: 10.1093/bioinformatics/btp352
Li Q., Liu Q., Zhang C., Cheng Y., Wu X. (2019a). Can color-related traits in the Chinese mitten crab (Eriocheir sinensis) be improved through quantitative genetic inheritance? Aquaculture 512. doi: 10.1016/j.aquaculture.2019.734355
Lim S. H., Kwon S. K., Lee M. K., Moon J., Jeong D. G., Park E., et al. (2009). Synapse formation regulated by protein tyrosine phosphatase receptor T through interaction with cell adhesion molecules and fyn. EMBO J. 28 (22), 3564–3578. doi: 10.1038/emboj.2009.289
Liu Y., Su H., Li R., Li X., Xu Y., Dai X., et al. (2017). Comparative transcriptome analysis of glyphodes pyloalis walker (lepidoptera: pyralidae) reveals novel insights into heat stress tolerance in insects. BMC Genomics 18 (1), 974. doi: 10.1186/s12864-017-4355-5
Liu F., Sun F., Kuang G. Q., Wang L., Yue G. H. (2022). Identification of Pmel17 for golden skin color using linkage mapping in Mozambique tilapia. Aquaculture 548. doi: 10.1016/j.aquaculture.2021.737703
Li X., Wang S., Xun X., Zhang M., Wang S., Li H., et al. (2019b). A carotenoid oxygenase is responsible for muscle coloration in scallop. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1864 (7), 966–975. doi: 10.1016/j.bbalip.2019.03.003
Nguyen H. N., Quinn J., Powell D., Elizur A., Thoa N. P., Nocillado J., et al. (2014). Heritability for body colour and its genetic association with morphometric traits in banana shrimp (Fenneropenaeus merguiensis). BMC Genet. 15, 132. doi: 10.1186/s12863-014-0132-5
Norris A. T., Cunningham E. P. (2004). Estimates of phenotypic and genetic parameters for flesh colour traits in farmed Atlantic salmon based on multiple trait animal model. Livestock Production Sci. 89 (2-3), 209–222. doi: 10.1016/j.livprodsci.2004.02.010
Park D. J., Sekhon S. S., Yoon J., Kim Y. H., Min J. (2016). Color reduction of melanin by lysosomal and peroxisomal enzymes isolated from mammalian cells. Mol. Cell. Biochem. 413 (1-2), 119–125. doi: 10.1007/s11010-015-2645-2
Pertea M., Pertea G. M., Antonescu C. M., Chang T. C., Mendell J. T., Salzberg S. L. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33 (3), 290–295. doi: 10.1038/nbt.3122
Pharoah P. D., Tsai Y. Y., Ramus S. J., Phelan C. M., Goode E. L., Lawrenson K., et al. (2013). GWAS meta-analysis and replication identifies three new susceptibility loci for ovarian cancer. Nat. Genet. 45 (4), 362–370, 370e361-362. doi: 10.1038/ng.2564
Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A., Bender D., et al. (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81 (3), 559–575. doi: 10.1086/519795
Rajpurohit S., Parkash R., Ramniwas S. (2008). Body melanization and its adaptive role in thermoregulation and tolerance against desiccating conditions in drosophilids. Entomol Res. 38 (1), 49–60. doi: 10.1111/j.1748-5967.2008.00129.x
Sallee J. L., Wittchen E. S., Burridge K. (2006). Regulation of cell adhesion by protein-tyrosine phosphatases: II. cell-cell adhesion. J. Biol. Chem. 281 (24), 16189–16192. doi: 10.1074/jbc.R600003200
Shang F., Chen J., Wang L., Jin L., Zou L., Bu T., et al. (2018). Crystal structure of the nicotinamidase/pyrazinamidase PncA from bacillus subtilis. Biochem. Biophys. Res. Commun. 503 (4), 2906–2911. doi: 10.1016/j.bbrc.2018.08.067
Sharma P., Jha A. B., Dubey R. S., Pessarakli M. (2012). Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J. Bot. 2012, 1–27. doi: 10.1155/2012/217037
Solano F. (2014). Melanins: Skin pigments and much more–types, structural models, biological functions, and formation routes. New J. Sci. 2014, 1–28, 498276. doi: 10.1155/2014/498276
Spielman L. J., Sweigard J. A., Shattock R., Fry W. E. (1990). The genetics of phytophthora infestans segregation of allozyme markers in F2 and backcross progeny and the inheritance of virulence against potato resistance genes R2 and R4 in F1 progeny. Exp. Mycol 14 (1), 57–69. doi: 10.1016/0147-5975(90)90086-9
Sun X., Wu B., Zhou L., Liu Z., Dong Y., Yang A. (2017). Isolation and characterization of melanin pigment from yesso scallop patinopecten yessoensis. J. Ocean Univ. China 16 (2), 279–284. doi: 10.1007/s11802-017-3162-6
Thodesen J., Rye M., Wang Y.-X., Li S.-J., Bentsen H. B., Yazdi M. H., et al. (2013). Genetic improvement of tilapias in China: Genetic parameters and selection responses in growth, survival and external color traits of red tilapia (Oreochromis spp.) after four generations of multi-trait selection. Aquaculture 416-417, 354–366. doi: 10.1016/j.aquaculture.2013.09.047
Visscher P. M., Brown M. A., McCarthy M. I., Yang J. (2012). Five years of GWAS discovery. Am. J. Hum. Genet. 90 (1), 7–24. doi: 10.1016/j.ajhg.2011.11.029
Wang Q. Z., Li Q., Cong R. H., Kong N., Kong L. F., Yu H. (2015). Inheritance of mantle pigmentation in selected families of the pacific oyster Crassostrea gigas. Mar. Sci. 39 (11), 86–90. doi: 10.11759/hykx20150805002
Wang L. K., Yue H. L., Peng X. J., Zhang S. J. (2019). GSTO1 regards as a meritorious regulator in cutaneous malignant melanoma cells. Mol. Cell Probes 48, 101449. doi: 10.1016/j.mcp.2019.101449
Williams S. T. (2017). Molluscan shell colour. Biol. Rev. Camb Philos. Soc. 92 (2), 1039–1058. doi: 10.1111/brv.12268
Xiao Q., Gan Y., Yu F., Boamah G. A., Shen Y., Wang Y., et al. (2022a). Study of hybrid and backcross abalone populations uncovers trait separation and their thermal resistance capacity. Aquacult Res. 53 (7), 2619–2628. doi: 10.1111/are.15779
Xiao Q., Huang Z., Shen Y., Gan Y., Wang Y., Gong S., et al. (2021). Transcriptome analysis reveals the molecular mechanisms of heterosis on thermal resistance in hybrid abalone. BMC Genomics 22 (1), 650. doi: 10.1186/s12864-021-07954-y
Xiao Q., Shen Y., Gan Y., Wang Y., Zhang J., Huang Z., et al. (2022b). Three-way cross hybrid abalone exhibit heterosis in growth performance, thermal tolerance, and hypoxia tolerance. Aquaculture 555. doi: 10.1016/j.aquaculture.2022.738231
Xu L., Li Q., Yu H., Kong L. (2017). Estimates of heritability for growth and shell color traits and their genetic correlations in the black shell strain of pacific oyster Crassostrea gigas. Mar. Biotechnol. (NY) 19 (5), 421–429. doi: 10.1007/s10126-017-9772-6
Xu C., Li Q., Yu H., Liu S., Kong L., Chong J. (2019). Inheritance of shell pigmentation in pacific oyster Crassostrea gigas. Aquacult 512 doi: 10.1016/j.aquaculture.2019.734249
Yan X., Nie H., Huo Z., Ding J., Li Z., Yan L., et al. (2019). Clam genome sequence clarifies the molecular basis of its benthic adaptation and extraordinary shell color diversity. iScience 19, 1225–1237. doi: 10.1016/j.isci.2019.08.049
You W., Guo Q., Fan F., Ren P., Luo X., Ke C. (2015). Experimental hybridization and genetic identification of pacific abalone Haliotis discus hannai and green abalone H. fulgens. Aquaculture 448, 243–249. doi: 10.1016/j.aquaculture.2015.05.043
Yu D., Gu X., Zhang S., Dong S., Miao H., Gebretsadik K., et al. (2021). Molecular basis of heterosis and related breeding strategies reveal its importance in vegetable breeding. Hortic. Res. 8 (1), 120. doi: 10.1038/s41438-021-00552-9
Yu W., He C., Cai Z., Xu F., Wei L., Chen J., et al. (2017). A preliminary study on the pattern, the physiological bases and the molecular mechanism of the adductor muscle scar pigmentation in pacific oyster Crassostrea gigas. Front. Physiol. 8. doi: 10.3389/fphys.2017.00699
Yu G., Wang L. G., Han Y., He Q. Y. (2012). clusterProfiler: an r package for comparing biological themes among gene clusters. OMICS 16 (5), 284–287. doi: 10.1089/omi.2011.0118
Zhao L., Li Y., Li Y., Yu J., Liao H., Wang S., et al. (2017). A genome-wide association study identifies the genomic region associated with shell color in yesso scallop, Patinopecten yessoensis. Mar. Biotechnol. (NY) 19 (3), 301–309. doi: 10.1007/s10126-017-9751-y
Zhou Z., Li J., Ma H., Li Y., Qin Y., Wei J., et al. (2021). The evaluation of culture performance and mantle coloration of two boring giant clam (Tridacna crocea) strains. Aquacult Rep. 20. doi: 10.1016/j.aqrep.2021.100646
Zhou T., Liu S., Geng X., Jin Y., Jiang C., Bao L., et al. (2017). GWAS analysis of QTL for enteric septicemia of catfish and their involved genes suggest evolutionary conservation of a molecular mechanism of disease resistance. Mol. Genet. Genomics 292 (1), 231–242. doi: 10.1007/s00438-016-1269-x
Zhou X., Stephens M. (2012). Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 44 (7), 821–824. doi: 10.1038/ng.2310
Zhu C., Wang F., Wang J., Li G., Zhang H., Zhang Y. (2007). Reconstruction of linkage maps in the distorted segregation populations of backcross, doubled haploid and recombinant inbred lines. Chin. Sci. Bull. 52 (12), 1648–1653. doi: 10.1007/s11434-007-0244-7
Keywords: genome-wide association study, transcriptome, abalone, mantle marking, trait separation, three-way cross
Citation: Xiao Q, Gong S, Shen Y, Lu Y, Lai X, Peng W, Huang Z, Han Z, Ji H, Gan Y, Luo X, You W and Ke C (2022) Genome-wide association and transcriptome studies reveal the segregation mechanism of mantle markings in three-way cross hybrid abalone. Front. Mar. Sci. 9:1013447. doi: 10.3389/fmars.2022.1013447
Received: 07 August 2022; Accepted: 30 September 2022;
Published: 19 October 2022.
Edited by:
Yuehuan Zhang, South China Sea Institute of Oceanology (CAS), ChinaReviewed by:
Qingzhi Wang, Liaoning Ocean and Fisheries Research Institute, ChinaLinlin Zhang, Institute of Oceanology (CAS), China
Copyright © 2022 Xiao, Gong, Shen, Lu, Lai, Peng, Huang, Han, Ji, Gan, Luo, You and Ke. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weiwei You, d3d5b3VAeG11LmVkdS5jbg==; Caihuan Ke, Y2hrZUB4bXUuZWR1LmNu
 Qizhen Xiao1,2
Qizhen Xiao1,2 Xinlian Lai
Xinlian Lai Zekun Huang
Zekun Huang Xuan Luo
Xuan Luo Weiwei You
Weiwei You Caihuan Ke
Caihuan Ke