
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 23 November 2022
Sec. Coral Reef Research
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.1009407
This article is part of the Research Topic Stony Coral Tissue Loss Disease in the Caribbean View all 50 articles
Since 2014, corals throughout Florida’s Coral Reef have been plagued by an epizootic of unknown etiology, colloquially termed stony coral tissue loss disease (SCTLD). Although in Florida the movement of this waterborne coral disease has been consistent with natural transport via water currents, outbreaks in the Caribbean have been more sporadic, with infections occurring in locations inconsistent with spread via natural means. Often Caribbean outbreaks have been clustered near ports, potentially implicating ships as mediators of SCTLD into new regions. Biofilms attached to ship hulls, ballast tank walls, or other surfaces could represent a possible vector for the disease. We investigated whether bacteria shed by healthy and SCTLD-diseased corals would form distinct biofilms, and whether a SCTLD signal would be detectable within biofilm bacterial communities. Stainless steel plates serving as proxies for ship hulls, ballast tank walls, and other colonizable surfaces were incubated for three days in filtered seawater mesocosms containing healthy or SCTLD-infected corals. Resulting biofilm bacterial communities were characterized through sequencing of the V4 region of the 16S rRNA gene. We determined that bacteria shed by healthy and diseased corals formed significantly different biofilms consisting of highly diverse taxa. Comparison with 16S data from previous SCTLD investigations spanning different coral species, collection locations, years, and source material revealed the presence of numerous genetically identical sequences within the biofilm bacterial communities formed during exposure to SCTLD-infected corals, including several previously identified as possible SCTLD bioindicators. These results suggest ship-associated biofilms may have the potential to be vectors for the transmission of SCTLD into new regions.
Since the first coral diseases were documented in the 1970s (Antonius, 1973; Garrett and Ducklow, 1975), disease outbreaks on coral reefs have grown increasingly common and severe (Weil and Rogers, 2011), such that today disease is a major contributor to coral reef decline (Bourne et al., 2009). Of all the coral diseases, arguably the most destructive is also the most recent: an ongoing tissue loss disease epidemic impacting scleractinian corals, colloquially referred to as stony coral tissue loss disease (SCTLD). This devastating disease is unprecedented in its geographic and temporal extent, the numbers of species impacted, and the rapidity with which it decimates entire reefs (Precht et al., 2016; Walton et al., 2018; Estrada-Saldívar et al., 2020; Meiling et al., 2020; Brandt et al., 2021; Estrada-Saldívar et al., 2021). SCTLD was first observed off the coast of Miami in 2014 (Precht et al., 2016), and has since spread to infect the entirety of Florida’s Coral Reef (Dobbelaere et al., 2022). In some of the most heavily impacted regions of Florida, some coral species have experienced mortality exceeding 97% of their pre-outbreak populations (Precht et al., 2016). More recently, SCTLD outbreaks have also been observed in the Caribbean (Alvarez-Filip et al., 2019; Weil et al., 2019; Brandt et al., 2021; Dahlgren et al., 2021; Heres et al., 2021).
Despite the severity of this epidemic and considerable research effort, little is known about the etiology of this destructive disease. It has been established that SCTLD has a waterborne causative agent; experiments in laboratory settings have demonstrated indirect exposure to an infected coral can transmit the disease to a healthy colony (Aeby et al., 2019). The specific identity of this causative agent, however, remains unknown and a research topic of intense effort. While the possibility of a viral component has been proposed (Work et al., 2021), to date the majority of the molecular investigations of microorganisms associated with SCTLD have focused on bacteria (Meyer et al., 2019; Rosales et al., 2020; Ushijima et al., 2020; Becker et al., 2021; Clark et al., 2021; Iwanowicz et al., 2021; Evans et al., 2022; Huntley et al., 2022; Studivan et al., 2022), as treatment with various classes of antibiotics have been demonstrated to successfully halt progression of the disease (Aeby et al., 2019; Neely et al., 2020; Neely et al., 2021; Shilling et al., 2021; Walker et al., 2021) and reduce the probability of reinfection (Neely et al., 2021). Collectively, these investigations of SCTLD-associated bacterial communities have identified specific bacterial taxa that are enriched in SCTLD lesions compared to healthy tissue, and more recently, specific amplicon sequence variants (ASVs) have been identified as consistently associated with the disease. Although it is currently unclear whether any of these recurring ASVs include the causative agent(s) of SCTLD, their pervasiveness across multiple SCTLD datasets spanning different geographic regions, collection years, source material (i.e., coral tissue/mucus, seawater, sediments), and host coral species (Meyer et al., 2019; Rosales et al., 2020; Becker et al., 2021; Clark et al., 2021; Evans et al., 2022; Huntley et al., 2022; Studivan et al., 2022) underscores their utility as SCTLD “bioindicators” (Becker et al., 2021).
Within Florida’s Coral Reef, the spread of SCTLD has been consistent with movement of the unknown pathogen via natural ocean currents (Dobbelaere et al., 2020; Muller et al., 2020). Within the wider Caribbean, however, SCTLD outbreaks have occurred far more sporadically, often suddenly appearing in disparate regions inconsistent with transport via natural means (Kramer et al., 2019). SCTLD’s arrival in the Bahamas, for example, initially occurred on two islands located more than 200 km apart, with no incidence of disease in the areas between them (Dahlgren et al., 2021). Dahlgren and colleagues further determined that disease incidence increased in closer proximity to large commercial shipping ports, suggesting those areas had been impacted first. Taken together, the authors concluded that SCTLD was likely introduced to the region by commercial shipping (Dahlgren et al., 2021). Within ships, contaminated ballast water, biofouling (including biofilms), and sediments have all been proposed as potentially contributing to the spread of SCTLD (Rosenau et al., 2021). Though ballast water is frequently cited as a potential vector for the introduction of nonnative macro- and microorganisms (Holeck et al., 2004; Klein et al., 2010), including pathogens (Ruiz et al., 2000), the potential role of biofilms in mediating disease spread is less well understood.
Biofilms are aggregates of microbes that form when free-living bacteria attach to a surface and begin excreting a sticky matrix of extracellular polymeric substances. As additional microbes encounter and join the biofilm, the biofilm microbial community grows in size and complexity, such that within the marine environment a biofilm can form in a few hours (Siboni et al., 2007). Microorganisms also exit from these biofilms, either as free-living microbes or within clumps of detached biofilm material containing viable cells (Stoodley et al., 2001). Within the context of ships, any surface in contact with seawater provides a suitable substrate for biofilm growth, with hulls and ballast tanks providing the largest surface area for biofilm formation. Biofilms provide bacteria with protection from external stressors (Yin et al., 2019) such as desiccation (Frösler et al., 2017), ultraviolet (UV) radiation (de Carvalho, 2017), and antibiotics (Hathroubi et al., 2017), and consequently it seems reasonable that some treatments imposed upon ballast tanks to disinfect ballast water (e.g., UV radiation) may prove ineffective in completely eradicating biofilms. Further, biofouled ship hulls have long been recognized as likely contributing to the spread of nonnative species into new regions (Carlton, 1987; Bailey et al., 2020). However, regulations on biofouling generally include the regular removal of visible organisms. For example, New Zealand’s biofouling regulations for international vessels allow for the presence of a “slime layer” (i.e., biofilm) of microorganisms to be considered as meeting “clean hull” standards (MPI, 2018). Thus, even in the face of considerable regulations, biofilms may still represent a possible vector for the introduction of nonnative microbes to new regions.
Given the apparent waterborne transmission route of SCTLD (Aeby et al., 2019; Dobbelaere et al., 2020; Muller et al., 2020), the unknown pathogen(s) must be shed by infected corals into the surrounding water. It has been established that biofilm community composition may be influenced by the close proximity of benthic organisms, including corals (Remple et al., 2021). It therefore seems reasonable that the causative agent of SCTLD could join a biofilm microbial assemblage. If this biofilm formed on a ship-associated surface, whether the hull, ballast tank, or some other surface, this SCTLD pathogen would then have the capability to be transported into new regions, effectively facilitating the spread of the disease. However, the extent to which coral-shed pathogens may infiltrate biofilms is unclear. Thus, our objectives in this study were: (1) to characterize the microbial communities of biofilms formed in mesocosms containing healthy or diseased corals; (2) to determine if the health state of the coral was a stronger influence on biofilm microbial community composition than coral species; and (3) to assess the biofilm microbial communities for the presence of previously identified SCTLD bioindicators. Our findings suggest the potential for biofilms to serve as vectors of SCTLD.
In March 2021, portions of nine colonies exhibiting visual signs of SCTLD (tissue loss) were collected from two reef sites south of Vaca Key, FL (24.6855 -81.0435 or 24.6846 -81.0475) under collection permit FKNMS-2019-160. Of these, eight were specimens of Colpophyllia natans, and one was a specimen of Pseudodiploria strigosa. One additional mixed C. natans sample (CnD-25) comprised of four smaller pieces of multiple colonies that had broken off during collection was also opportunistically collected, for a total of ten diseased coral samples. Using a VcpA RapidTest, as described by Ushijima et al. (2020), diseased donor corals were negative for the presence of Vibrio coralliilyticus, a species thought to enhance lesion progression (Ushijima et al., 2020). CnD-25 was not tested due to its opportunistic collection.
Healthy corals were also collected to serve as controls. However, recent studies have suggested that apparently healthy corals remaining on endemic reefs, with no visual symptoms consistent with SCTLD, may actually represent asymptomatic infections of the disease (Work et al., 2021). Therefore, this experiment instead used apparently healthy corals (n = 4) that had been collected before SCTLD reached their collection sites to reduce the likelihood that control corals had any prior exposure to SCTLD. All four colonies were collected from Florida reefs and included two colonies of C. natans collected from the Key West Nursery in January 2019 under permit FKNMS-2017-128-A2 and two colonies of P. strigosa collected from the Dry Tortugas in January 2020 under permit FKNMS-2019-160. Following collection, all healthy corals were maintained in large indoor quarantine tanks located at the Smithsonian Marine Station in Ft. Pierce, Florida that each contained ~570 L of recirculating and temperature-controlled (25.5°C ± 0.3°C) filtered seawater. Additional details on these facilities and coral husbandry are described by Ushijima et al. (2020). All healthy donors exhibited no visual signs of SCTLD at the time of our experiment or during their prior time in aquaria, and all were negative for the presence of Vibrio coralliilyticus based on a VcpA RapidTest.
Seawater for use in mesocosms was collected via intake pipe ~1,600 m offshore of South Hutchinson Island, Florida (latitude 27.347, longitude -80.235) and prefiltered through a 20 µm filter, UV-sterilized using a Coralife Turbo-Twist 12x UV Sterilizer with a 36 W UVC lamp and a radiation wavelength of 254 nm, and then sequentially filtered through 0.35 µm and 0.22 µm filters (Ushijima et al., 2020; Evans et al., 2022). The low microbial abundance seawater generated by this process is hereafter referred to as filtered seawater (FSW). We have previously visually confirmed by microscopy that this series of treatments results in a low microbial background (Evans et al., 2022). Some of this FSW was then stored in covered outdoor holding tanks and used to fill 15 5-gallon high-density polyethylene (HDPE) plastic buckets with ~18L of FSW per bucket. Fourteen of these bucket mesocosms received one of the diseased (n = 10) or healthy (n = 4) donor corals described above, and a weighted airline to circulate and oxygenate the water. These buckets were placed into insulated water tables containing circulating temperature-controlled freshwater to maintain the mesocosms at ~28°C. Water tables were located under a shade cloth canopy to provide some light attenuation. Separate water tables were used for healthy and diseased mesocosms to reduce the potential for cross-contamination. Additional details on these facilities are described by Ushijima et al. (2020).
Coral mesocosms (n = 14) were incubated 48-72 hours with either diseased or healthy corals to enrich the mesocosm water with coral-shed microbes. Note that based on prior microscopy assessments of similar mesocosms (Evans et al., 2022), the overall bacterial load is similar or slightly elevated compared to ordinary seawater. After this initial incubation, cleaned and autoclaved marine grade (316) stainless steel plates measuring ~7.5 cm x ~5.1 cm were added to the mesocosms and positioned in sterile petri dishes to allow for water contact on both sides. An example of the coral mesocosm setup is depicted in Figure 1. A final mesocosm was also created at this time containing only FSW and a steel plate in a petri dish to serve as a seawater control and assist in identifying biofilm constituents forming naturally from exposure to any microbes remaining within the UV-treated FSW or sourced from the local environment during the incubation period. Water quality was visually assessed and assigned a value from 1 (clear) to 3 (cloudy, visible color change). After addition of the steel plates, mesocosms (n = 15) were incubated three days (72 hours), during which time all coral mesocosms (n = 14) received one 100% water change with FSW. Following the conclusion of the experiment, all plates (n = 15) were placed in individual sterile whirl-pak bags, immersed in RNAlater solution (Invitrogen), stored at 4°C overnight, transported from Ft. Pierce, FL to St. Petersburg, FL on cold packs, and then stored at -20°C until processing.
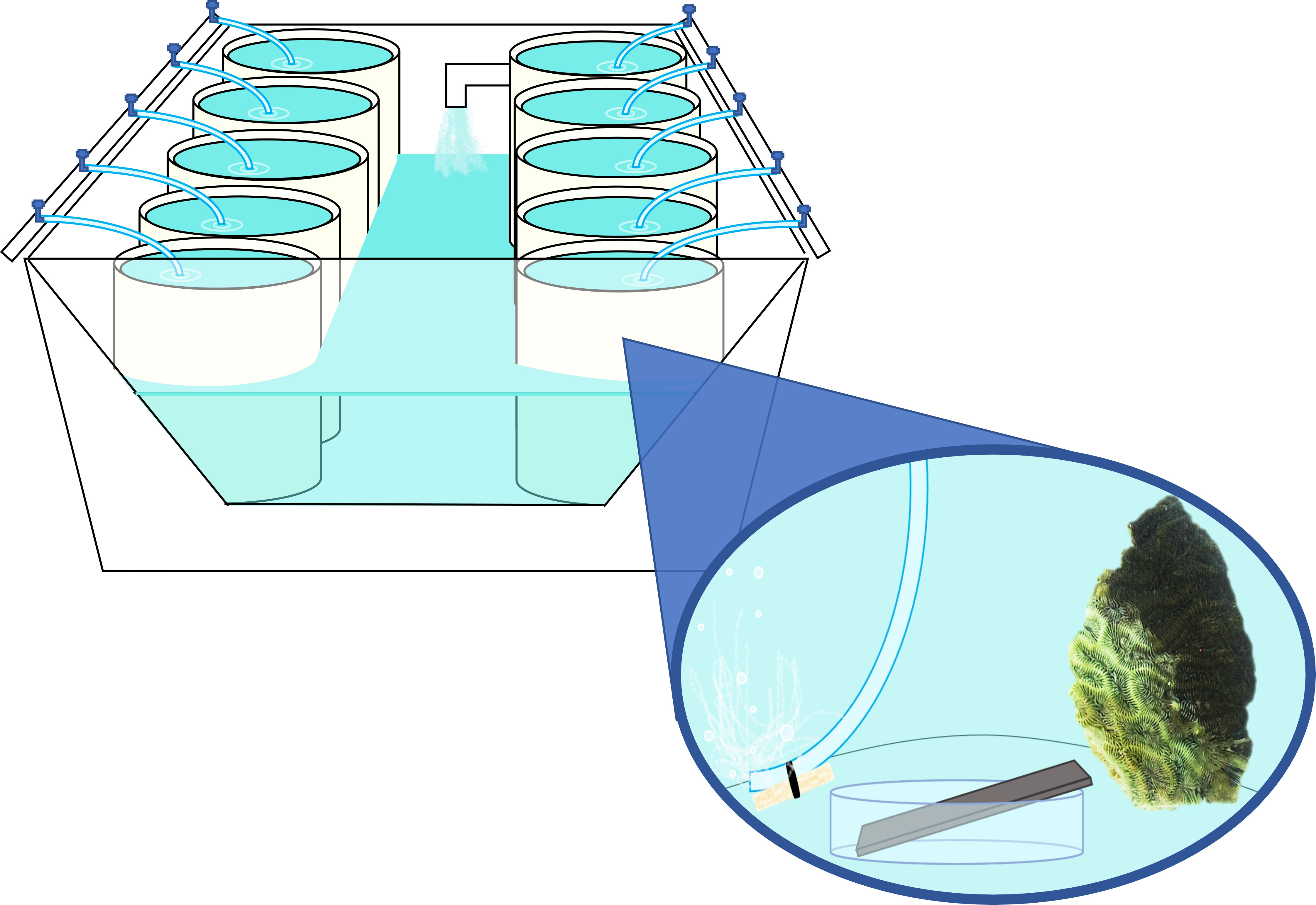
Figure 1 Mock-up of mesocosm water table, exemplifying arrangement of individual coral mesocosms. Each water table contained ~ 570 L of recirculating, temperature-controlled freshwater. Separate water tables were used to hold diseased and healthy coral mesocosms to reduce cross-contamination. Superimposed image depicts individual mesocosm setup, with each mesocosm containing ~18L of filtered and UV-treated seawater (FSW), a “donor” coral, a weighted airline for aeration and water circulation, and a stainless steel plate arranged on a sterile petri dish to allow for water contact on all sides.
Biofilms were removed from plates by first scraping both sides of the plate with a flame-sterilized razor blade, then rinsing both sides of the plate and razor blade with sterile 1x phosphate-buffered saline (PBS). DNA extraction was performed using a DNeasy PowerBiofilm kit (Qiagen), following the manufacturer’s QuickStart Protocol (November 2016 version), with the following modifications: 1.) Step 1: instead of weighing biofilm material for processing, all solution resulting from the rinse (i.e., PBS + biofilm) was transferred into 2 mL tubes, then centrifuged as directed. 2.) Step 2: for any samples resulting in multiple tubes from step 1, the 350 μL of lysis buffer (Qiagen’s MBL solution) was added to the first tube to resuspend the centrifuged material. This ~350 μL of resuspended material + buffer was then used to sequentially resuspend the material in any remaining tubes for that sample, before transferring the final combined solution to the bead tube. 3.) Step 5: each sample was bead beat at 2500 rpm for 30 s instead of 3200 rpm for 30 s. An extraction kit blank (nothing added) and a PBS blank (~4 mL, processed the same way as the biofilm samples) were processed through the Qiagen kit to serve as reagent contamination controls.
DNA extractions were sent to the Michigan State University RTSF Genomics Core for additional processing and sequencing, along with a mock community (MSA-3001 ABRF-MGRG 10 Strain Even Mix Genomic Material (ATCC)). Amplicon libraries were created based on a dual-index sequencing strategy (Kozich et al., 2013) using the 16S rRNA gene (V4 region) primers 515F (5’-GTG CCA GCM GCC GCG GTA A-3’) and 806R (5’-GGA CTA CHV GGG TWT CTA AT-3’) (Caporaso et al., 2011). Batch normalization was conducted using Invitrogen SequalPrep DNA Normalization plates, and resulting products were pooled together, concentrated via Amicon spin column, and subsequently purified using AMPure XP magnetic SPRI beads. Sequencing was performed on an Illumina MiSeq v2 Standard flow cell in a 2 x 250 base pair (bp) paired end format with a v2 500 cycle reagent cartridge. Illumina Real Time analysis (v 1.18.54) was used for base calling. Illumina Bcl2fastq (v 2.20.0) was used for demultiplexing. All raw sequencing files were deposited in the NCBI Sequence Read Archive (SRA) under BioProject PRJNA828575 and are available in a U.S. Geological Survey data release (Kellogg et al., 2022).
Demultiplexed raw sequences were imported into QIIME2 v2021.4 (Bolyen et al., 2019). Denoising was performed with DADA2 (Callahan et al., 2016) with truncation at position 200, and sequences were assigned to amplicon sequence variants (ASVs) based on 100% sequence identity matches. Sequences were aligned with MAFFT v7.0 (Katoh and Standley, 2013) and assigned to a phylogenetic tree with FastTree 2 (Price et al., 2010). Sequences were assigned taxonomy using the pre-trained naïve Bayes classifier SILVA-138-99-515-806 (Bokulich et al., 2018), and all sequences identified as chloroplasts or mitochondria were removed from the dataset.
Quality controls (mock community and reagent controls; n = 3) were removed from the dataset prior to statistical analyses. All remaining samples (n = 15) were rarefied to 24,793 sequences prior to diversity analyses, and a rarefaction curve was constructed to confirm that this sampling depth captured sufficient diversity across all samples, as evidenced by a flattening of the curve for all samples. Alpha and beta diversity analyses were performed in QIIME2 using the q2-diversity plugin and following the “core-metrics-phylogenetic” approach. Alpha diversity metrics for richness (observed features), evenness (Pielou’s evenness; Pielou, 1966), and diversity [Shannon’s diversity index (Shannon, 1948) and Faith’s phylogenetic diversity (Faith, 1992)] were calculated using the alpha-group-significance plugin and statistically analyzed with Kruskal-Wallis tests. For beta diversity, principal coordinate analyses (PCoA) were performed based on weighted and unweighted UniFrac distance matrices (Lozupone and Knight, 2005), and results were statistically analyzed within QIIME2 using the beta-group-significance plugin and permutational multivariate analyses of variance (PERMANOVA), with subsequent pairwise PERMANOVAs performed for all statistically significant PERMANOVA tests (Anderson and Walsh, 2013). The small sample size of seawater controls (n = 1) precluded its ability to be statistically compared to coral biofilm communities using the pairwise comparisons, so Emperor PCoA plots were generated instead to visualize the relationships between different samples, including seawater. PERMDISP tests in QIIME2 were used to determine whether any significant PERMANOVA tests were the result of unequal dispersion within groups.
In order to determine the presence or absence of a SCTLD signal within our biofilm microbial communities, sequences previously identified as SCTLD bioindicators (Becker et al., 2021) were searched against our unrarefied data set. ASVs identified as 100% identical base pair matches over 100% query coverage (126/126 identities, 0 gaps) were considered positive matches with SCTLD bioindicators. To further assess the robustness of a SCTLD signal in our data set, all ASVs identified as matches for SCTLD bioindicators were further compared against other SCTLD data sets (Meyer et al., 2019; Rosales et al., 2020; Clark et al., 2021) using the NCBI BLASTn sequence alignment service. A 100% identity match with 100% query coverage (252/252 or 253/253 identities, depending on sequence length, and 0 gaps) was considered to represent the same ASV across these other studies. Biofilm ASVs were further assessed for their potential to be “SCTLD-obligate” (present in at least 70% of the disease-associated biofilms) and “SCTLD-specific” (present only in biofilms from diseased mesocosms and not the healthy mesocosms, seawater control, reagent controls, or the mock community). The 70% cut-off is arbitrary and intended to balance the ability to detect rarer microbes undergoing a multi-step process; e.g., that a specific bacterium is shed by the coral, successfully encounters the steel plate, successfully forms or joins the biofilm, and is still detected after DNA extraction and sequencing.
A total of 727,783 raw sequences were acquired from our 18 samples. Of these, 692,543 were derived from biofilms (n = 15), while 31,741 corresponded to the mock community (n = 1), and 3,499 to the reagent controls (n = 2). Of the biofilm-derived sequences, 529,666 remained following bioinformatic processing, with an average of ~35,311 sequences obtained per mesocosm (max = 49,917, min = 24,793). Average sequence return was relatively comparable across the three mesocosm types (diseased, healthy, and seawater control), with biofilms containing on average ~36,860 and ~32,355 sequences per mesocosm for diseased and healthy samples respectively, and 31,643 sequences acquired from the seawater-control biofilm. A total of 3,224 ASVs were detected across all samples, with 3,146 of these present in biofilm microbial communities. Of the reagent control-derived sequences, 2,084 sequences spanning 96 ASVs remained following bioinformatic analysis. Of these, 72 ASVs were unique to the reagent controls, indicating the vast majority of biofilm ASVs were not reagent contaminants.
Biofilm microbial communities were highly diverse and included 37 bacterial and two archaeal (Crenarchaeota and Nanoarchaeota) phyla. Alpha diversity analyses revealed no significant differences in richness, evenness, or diversity between the biofilm microbial communities based on donor health status (conditions “healthy,” “diseased,” or “na” [seawater]; Kruskal-Wallis; p > 0.05) or donor type (conditions “P. strigosa,” “C. natans,” or “seawater”; Kruskal-Wallis; p > 0.05). The PCoA eigenvalues revealed that the weighted UniFrac distance matrix captured the most variation within the data (Figure 2).
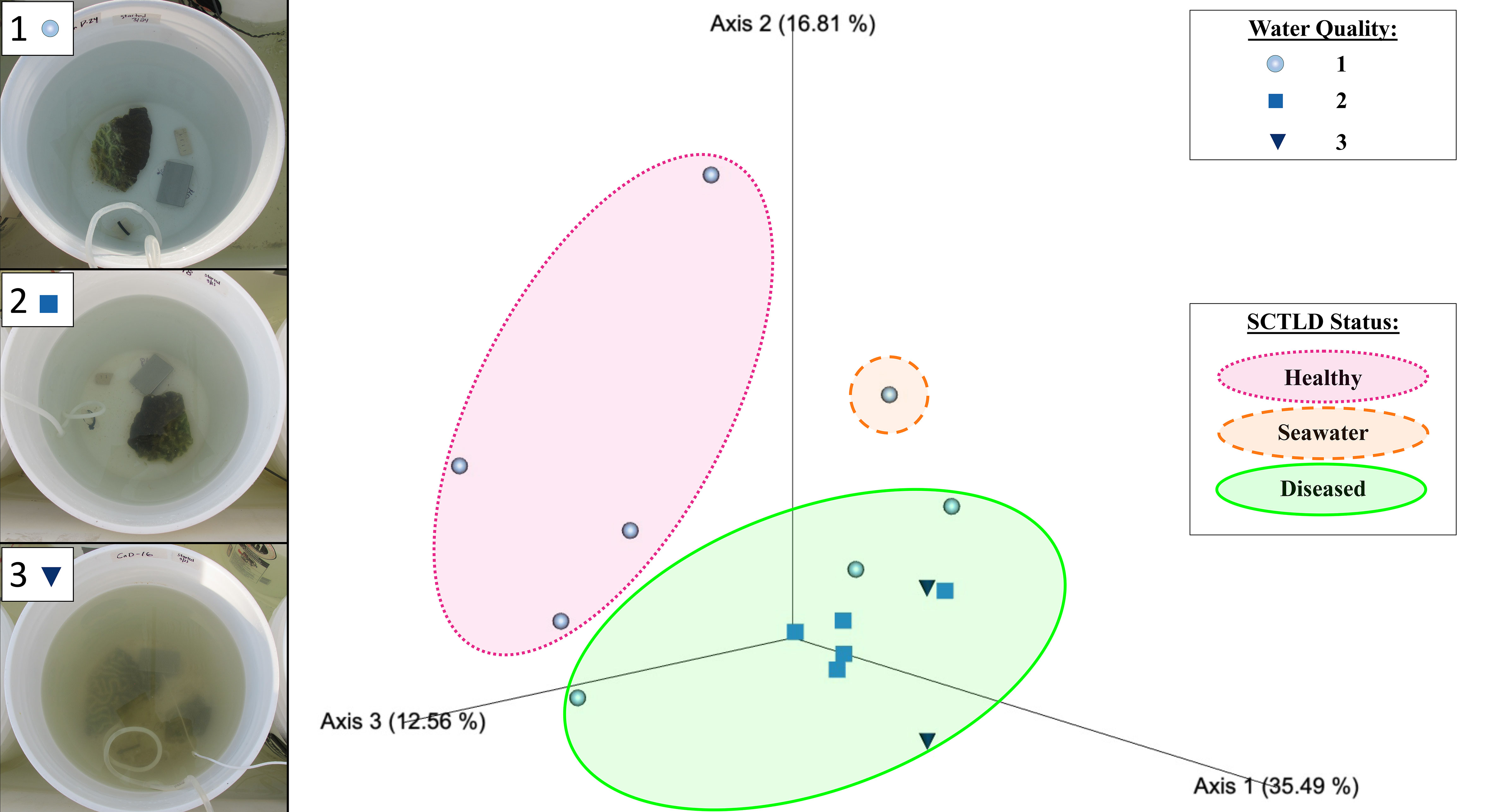
Figure 2 Emperor principal coordinates analysis plot based on a weighted UniFrac distance matrix, depicting the relationships between the biofilm prokaryotic microbial communities resulting from different mesocosms, with mesocosm water quality and donor coral health status indicated. Water quality values assigned included 1 ● (clear), 2 ■ (mild turbidity and color change), and 3 ▼ (cloudy, visible color change), as exemplified in the photos on the left. Overlays indicate biofilm samples sourced from healthy donor coral mesocosms (in pink), the filtered seawater (FSW) mesocosm (in orange), and diseased donor coral mesocosms (in green). Axis percentage values indicate the percent variation explained by that axis.
Beta diversity analyses based on the weighted UniFrac distance matrix revealed significant differences in biofilm microbial community structure based on donor health status (PERMANOVA, p = 0.003; Table 1). Pairwise comparisons revealed significant differences between biofilms sourced from diseased and healthy donor corals (PERMANOVA, p = 0.002; Table 1). The PCoA plot revealed that samples clustered in response to donor coral health status, with the filtered seawater biofilm community appearing to cluster away from the coral biofilms (Figure 2). However, it should be noted that because we used aquarium-maintained corals for our healthy individuals, the conditions “donor health status” (heathy vs. diseased) and “donor source” (aquarium vs. reef) represent identical comparisons in terms of coral groupings, and therefore it may be impossible to distinguish the individual influence of each condition. Donor type (conditions “P. strigosa,” “C. natans,” or “seawater”) was found to have no significant effect on biofilm microbial community structure. No significant differences in dispersion were detected for any of the comparisons (Table 1). Although all diseased mesocosm samples generally clustered together, visually an apparent effect of mesocosm water quality was also evident, with less turbid diseased mesocosm biofilm communities clustering closer to those from the healthy coral and seawater mesocosms and the cloudier, more “polluted” mesocosms (Figure 2) appearing to cluster further away. However, we were unable to statistically assess this variation due to the small number of samples within some of the water quality classifications. No significant differences in dispersion were detected for any of the comparisons (PERMDISP, p >0.05; Table 1).
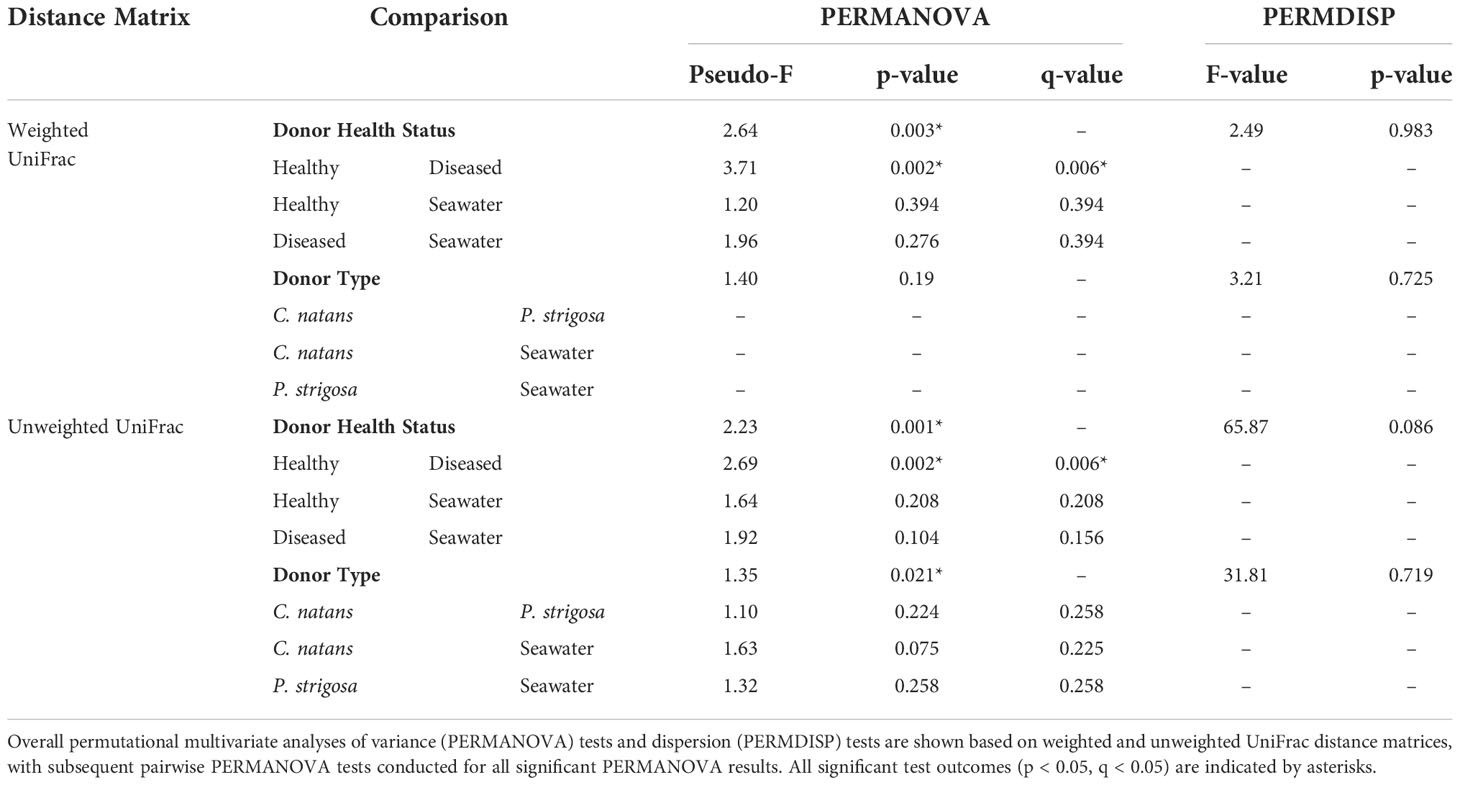
Table 1 Comparisons of microbial community structure and dispersion between samples based on Donor Health Status (conditions “diseased,” “healthy,” or “seawater”) and Donor Type (conditions “Colpophyllia natans,” “Pseudodiploria strigosa,” or “seawater”).
Beta diversity analyses based on the unweighted UniFrac distance matrix again revealed significant differences in biofilm microbial community structure based on donor health status and donor source (PERMANOVA, p = 0.001 for both comparisons; Table 1). Pairwise comparisons revealed significant differences between healthy and diseased mesocosms (PERMANOVA, p = 0.002; Table 1). A significant effect of donor type (coral species or seawater) was also detected (PERMANOVA, p = 0.021), but post-hoc pairwise tests revealed no significant difference between any of the donor types. The PCoA plot revealed that this observed difference was likely driven by apparent strong differences between coral-sourced biofilms and the filtered seawater biofilm, as samples did not appear to cluster in response to coral species, while the filtered seawater sample clustered away from coral samples (Figure 3).
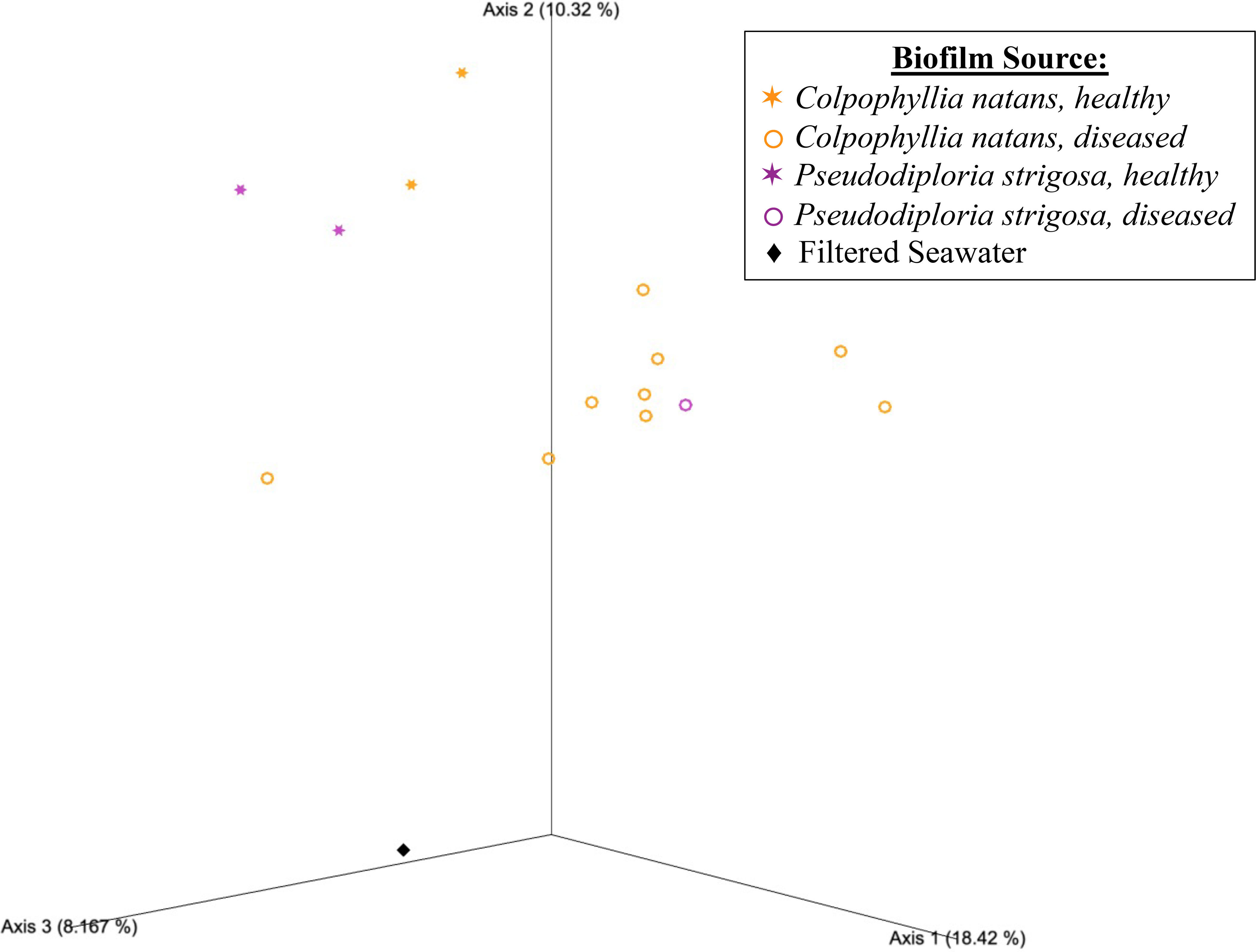
Figure 3 Emperor principal coordinates analysis plot based on an unweighted UniFrac distance matrix, illustrating the relationships between the biofilm bacterial/archaeal communities sourced from mesocosms containing different donor types, including Colpophyllia natans (in orange), Pseudodiploria strigosa (in purple), and filtered seawater (FSW; black diamond). Health status of the donor coral is also indicated, with healthy corals represented by stars (✶) and diseased corals by open circles (〇). Axis percentage values indicate the percent variation explained by that axis.
Comparisons to the 25 previously identified SCTLD bioindicators (Becker et al., 2021) revealed the presence of 19 of these ASVs within our biofilm dataset (Table 2). The vast majority of the corresponding biofilm ASVs were detected exclusively in diseased mesocosms (Table 2). In a few cases, bioindicator ASVs were enriched in biofilms from diseased mesocosms compared to biofilms from healthy mesocosms (Table 2). Most of these biofilm-associated ASVs were also previously detected in association with at least one SCTLD lesion by previous studies (Meyer et al., 2019; Rosales et al., 2020; Clark et al., 2021). Of these ASVs matching SCTLD bioindicators, two, ASV179 (genus Vibrio) and ASV261 (order Alteromonadales), were identified as simultaneously “SCTLD-obligate-and-specific” (present in at least 70% of diseased mesocosm biofilms, but no other mesocosms or controls). A BLASTn search with the Entrez query “coral” revealed that ASV179 exhibited 100% sequence similarity with the overlapping region (253 bp) of an ASV previously associated with another coral disease (Séré et al., 2015).
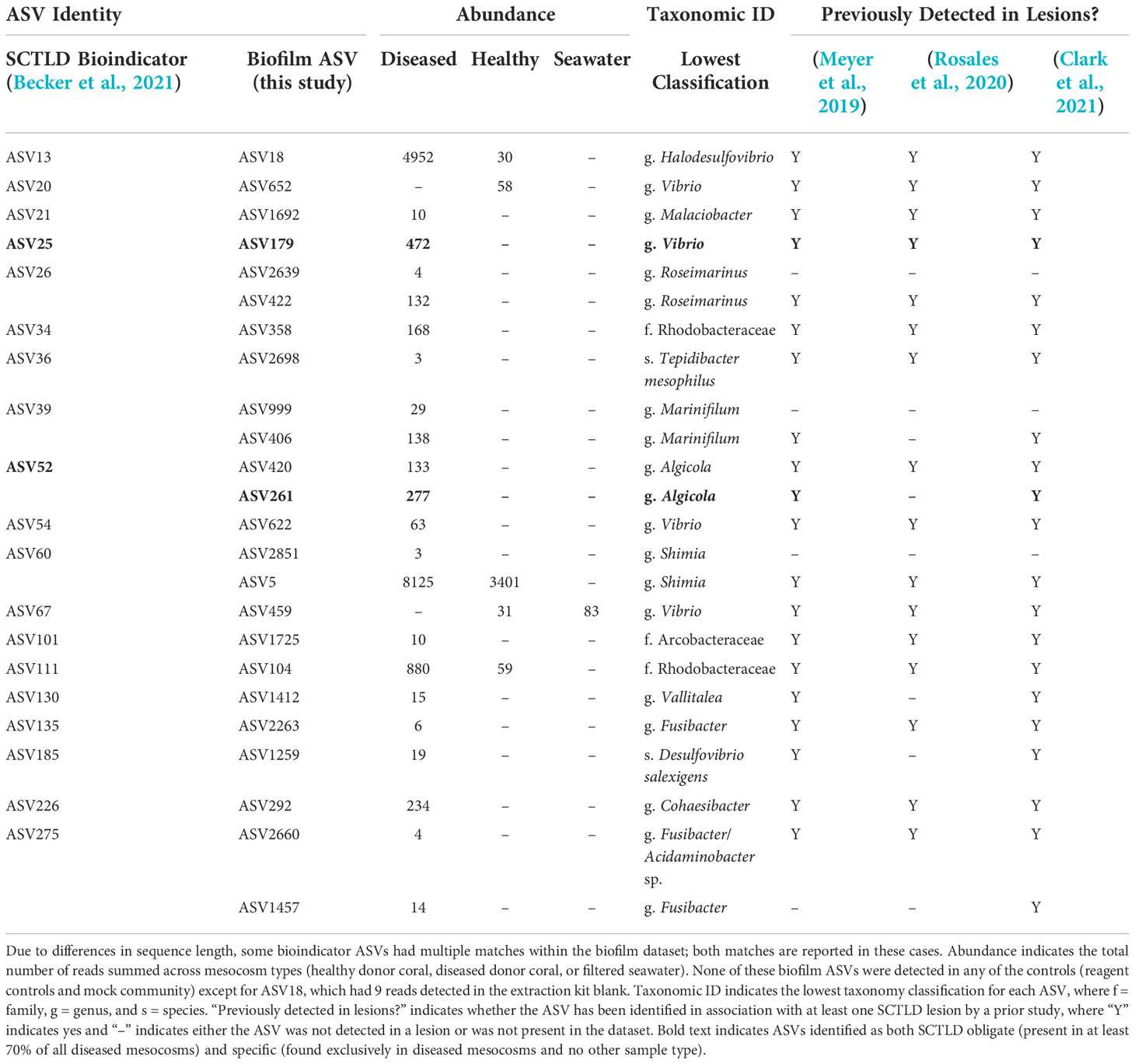
Table 2 Previously identified stony coral tissue loss disease (SCTLD) “bioindicator” amplicon sequence variants (ASVs) (Becker et al., 2021) that also represent 100% sequence identity matches for the overlapping region of ASVs present in biofilms (this study).
From the full dataset, 42 additional ASVs were SCTLD-obligate-and-specific, with three of these (ASVs 23, 26, and 47) detected in all ten diseased mesocosm biofilms (Table 3). BLASTn searches with the Entrez query “coral” revealed that one of these, ASV23, represented a 100% match for the overlapping region of three clone library sequences associated with white plague disease-infected coral (Roder et al., 2014). One additional ASV, ASV32 (family Rhodobacteraceae) was identified as also obligate to all ten diseased mesocosm biofilms, with ~302 reads per sample on average (maximum: n = 948, minimum: n = 30), but was also detected in the DNA extraction kit reagent blank (n = 43). A BLASTn search with the Entrez query “corals” identified this ASV as a 100% match for the overlapping region of a clone library sequence previously linked to Black Band Disease (Arotsker et al., 2015).
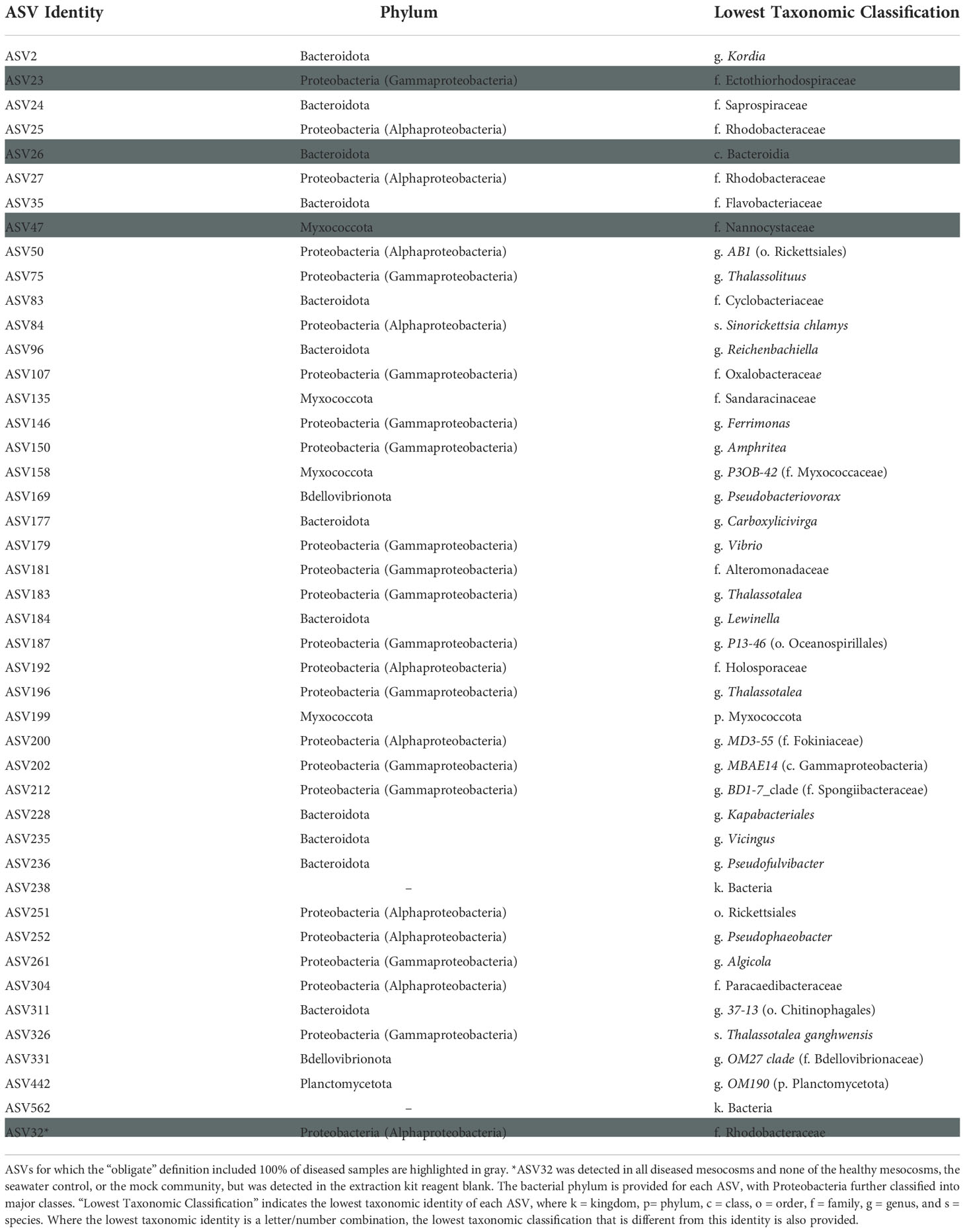
Table 3 Biofilm amplicon sequence variants (ASVs) that were both obligate (present in at least 70% of diseased samples) and specific (found only in diseased samples).
Our findings suggest that bacteria and archaea “shed” by corals may be capable of forming biofilms on stainless steel. Within 72 hours, our biofilm microbial communities were distinguishable from one another based on the health status of the source coral. We also identified numerous SCTLD bioindicator ASVs in our diseased coral mesocosm biofilms that have been previously detected within SCTLD lesions by prior studies. Taken together, these results indicate that SCTLD-associated bacteria, which could potentially include the disease’s unknown causative agent, may be capable of forming biofilms on ship-associated surfaces. While our experimental mesocosms represent simplified systems compared to the conditions faced by biofilms forming on ships (e.g., shear stress, fluctuating external conditions between sites, lengthy transport times), this work represents an important first step in our understanding of the potential role of biofilms in transporting SCTLD.
Given the presence of SCTLD throughout the majority of Florida’s Coral Reef at the time of this study, acquiring truly healthy corals from the reef may be impossible, as histological work has suggested that even apparently healthy colonies may represent asymptomatic infections (Work et al., 2021). Consequently, here we used apparently healthy corals that had been collected ahead of the disease front and held in aquaria for a minimum of one year without SCTLD symptoms. Given the quick progression of tissue loss and subsequent death associated with SCTLD (NOAA, 2018), diseased corals must be collected from the reef immediately prior to their experimental use. Some studies have determined that a transition from the field to aquaria settings may not influence coral microbiomes as strongly as other characteristics of the host coral (Damjanovic et al., 2020). Conversely, other research has found that corals moved from the reef to an artificial environment experience shifts in their associated microbial communities and subsequently develop distinctly different, apparently stable microbial associations (Kooperman et al., 2007; Pratte et al., 2015; Röthig et al., 2017). Given that diseased corals in our study were exclusively from the reef and healthy corals were sourced from aquaria, we acknowledge that multiple variables associated with the donor corals align, such as health status and source, whose individual influences upon biofilm microbial assemblages are impossible to entirely disentangle.
Nonetheless, suggesting that health status of the donor coral played a particularly strong role in shaping the biofilm microbial communities was the detection of numerous ASVs that were 100% identical matches for the overlapping region of ASVs previously linked to SCTLD lesions (Meyer et al., 2019; Rosales et al., 2020; Becker et al., 2021; Clark et al., 2021). The majority of these SCTLD “bioindicators” (Becker et al., 2021) were detected exclusively within the diseased mesocosms, highlighting the influence of donor coral health state in the resulting biofilm microbial assemblages. Further, these comparison studies span different collection locations, sampling years, coral species, and source material (tissue/mucus vs. biofilm). Considering that coral microbiomes can exhibit intraspecific variability (Damjanovic et al., 2020) and even variability across different compartments (e.g. mucus, tissue, skeleton) of the same colony (Sweet et al., 2011; Apprill et al., 2016), the extent of the cross-study overlap observed here seems especially noteworthy. In addition, although coral species may be a strong driver of microbial community composition in some cases (Rohwer et al., 2002), it did not appear to play a strong role in shaping the biofilm microbial communities (e.g., Figure 3), further suggesting some other factor(s) were more greatly influencing biofilm development.
Of the 25 previously identified SCTLD bioindicators, 19 were detected within our biofilm dataset, with matches to 24 biofilm ASVs. Four of these (ASVs 179, 459, 622, and 652) classified to the genus Vibrio, a genus that includes known coral pathogens. However, two of these, ASV459 and ASV652, were not detected in any of our diseased mesocosms. The corresponding bioindicator ASV for ASV459 (ASV67; Becker et al., 2021) was similarly found in greater abundance in healthy compared to SCTLD-diseased mesocosms in a previous study examining mesocosm water (Evans et al., 2022), while the corresponding bioindicator for ASV652 (ASV20; Becker et al., 2021) was found in association with both diseased and healthy mesocosms in the same study (Evans et al., 2022). Of the two Vibrio ASVs found in disease-associated biofilms here (ASVs 179 and 622), ASV622 was a 100% match for the overlapping region of seven clone library sequences previously associated with Black Band Disease mats (Klaus et al., 2011). ASV179 represented a 100% sequence match for the overlapping region of a Vibrio ASV previously demonstrated through inoculation experiments to elicit an apparent disease response and fulfilling several components of Koch’s postulates for the coral disease Porites white patch syndrome (Séré et al., 2015). Interestingly, although here ASV179 was SCTLD-obligate-and-specific (present in at least 70% of diseased mesocosms and no other mesocosms or controls), a prior study using a similar mesocosm approach to compare microbial communities found in seawater identified the corresponding SCTLD bioindicator ASV (ASV25; Becker et al., 2021) exclusively in one of the two healthy mesocosms and none of the diseased mesocosms (n = 4). Such conflicting results as these highlight the importance of investigating marine disease etiology through the use of multiple approaches in order to help narrow the scope of possible causative agents.
Analysis of SCTLD-obligate-and-specific ASVs identified 44 ASVs as potentially associated with SCTLD (Table 3). Two of these, ASV179 (genus Vibrio) and ASV261 (genus Algicola) were 100% matches for the overlapping region of SCTLD bioindicator ASVs (Becker et al., 2021). Meyer et al. (2019) identified five ASVs that were enriched in SCTLD lesions compared to apparently healthy tissue of three species: Montastraea cavernosa, Diploria labyrinthiformis, and Dichocoenia stokesii. Our ASV261 was a 100% sequence identity match for one of these, an Algicola ASV, which the authors note was previously linked to Black Band Disease (Arotsker et al., 2015; Meyer et al., 2019). Similarly, of the three SCTLD-obligate-and-specific ASVs detected in all ten diseased mesocosm biofilm microbial communities, one (ASV23, family Ectothiorhodospiraceae) matched 100% with the overlapping region of sequences previously linked to White Plague Disease (Roder et al., 2014). ASV32 (family Rhodobacteraceae) was also SCTLD-obligate-and-specific for all ten diseased mesocosm biofilms, and was a 100% match for the overlapping region of a clone library sequence previously associated with Black Band Disease (Arotsker et al., 2015). However, ASV32 also had reads (n = 43) within one of the DNA extraction kit reagent control blanks, suggesting it is possibly a kit contaminant, though this small number of reads could also be the result of sequencing artifacts. Nevertheless, because this ASV was otherwise detected exclusively within the diseased mesocosms, classified to a family previously suggested to possibly be linked to SCTLD (Rosales et al., 2020), and matched to an ASV previously associated with another coral disease (Arotsker et al., 2015), we have opted to still highlight it here as potentially associated with SCTLD. While the connection to other coral diseases identified for several biofilm ASVs may suggest these particular ASVs are opportunistic pathogens not specifically associated with SCTLD infection, this overlap further highlights the ability of biofilms formed in association with diseased corals to acquire a disease signal.
During the initial incubation period, some diseased mesocosms became visually more polluted, presumably due to sloughing tissue, expulsion of zooxanthellae, decomposition, and resulting microbial blooms. Hypothesizing that these more “polluted” conditions represent more nutrient-rich environments for microbial growth, blooms of opportunistic prokaryotes responding to localized nutrient-enrichment may at least partially explain some of the differences that we observed between microbial biofilms associated with apparently healthy and SCTLD-infected corals. Nutrient enrichment has previously been shown to significantly influence marine biofilm microbial communities, though not as much as other variables such as exposure to different macroorganisms (Remple et al., 2021). However, looking exclusively at biofilms formed in association with corals, Remple et al. (2021) found that nutrient enrichment resulted in an increase in species richness and a decrease in species evenness, as well as increases in Flavobacteriaceae, a family which was considerably more enriched in diseased compared to healthy mesocosms (average n = ~4,260 and 121 reads, respectively) and which we do see represented within our SCTLD-obligate-and-specific ASVs (ASVs 35 and 236). This suggests that the apparent water quality deterioration observed in some diseased mesocosms may indeed have resulted in a nutrient shift that impacted the resulting mesocosm and biofilm microbial communities.
Although no definitive link between SCTLD and nutrient enrichment has been made, an increased nutrient load has been shown to increase the severity of disease in other coral diseases (Bruno et al., 2003; Vega Thurber et al., 2014), suggesting that a positive feedback loop of nutrient enrichment and coral degradation may play some role in progression of the disease. Potentially supporting this hypothesis is the fact that three biofilm ASVs representing 100% matches for the overlapping region of SCTLD bioindicator ASVs (ASV420, genus Algicola; ASV1725, family Arcobacteraceae; and ASV2660, genus Fusibacter/Acidaminobacter sp.) were detected exclusively in the biofilms from one of the two most polluted mesocosms (water quality condition “3”), highlighting that some disease signals were detected only in those mesocosms with presumably enhanced nutrient levels.
Biofilm microbial communities within ballast tanks have previously been demonstrated to include pathogenic bacteria (Drake et al., 2005), and potential coral pathogens have previously been detected in the ballast water of ocean-going vessels discharging in close proximity to a coral reef system (Aguirre-Macedo et al., 2008). Though management strategies intended to limit the spread of micro- and macroorganisms retained within ballast water exist (Rosenau et al., 2021), it has been suggested that some treatment methods may be ineffective at eradicating pathogens (Petersen et al., 2019), and some approved ballast water treatment systems may actually enhance bacterial growth (Cohen and Dobbs, 2015). Further, commonly utilized approaches such as ballast water exchange may not be fully effective at eliminating microbial biofilms from ballast tank walls (Baier et al., 2014), and ballast tank-sourced biofilms have been demonstrated to be capable of spreading from the source biofilm to colonize new surfaces (Baier et al., 2014). Ship hull biofilms have similarly been demonstrated to include pathogenic bacteria (Shikuma and Hadfield, 2010), and while ship hull antifoulants may alter biofilm microbial community composition, they do not eliminate its formation altogether (Papadatou et al., 2021). Here we determined that stainless steel plates exposed to SCTLD-diseased and healthy corals form distinctly different biofilms. We further identified a clear SCTLD signal within almost exclusively the biofilm microbial communities that formed on plates in association with diseased corals, though additional investigation into the transmissibility of SCTLD from biofilms to healthy corals will be necessary to conclusively establish links between microbial biofilms and SCTLD infection. Nevertheless, our work suggests that biofilms could represent a reservoir for SCTLD pathogens that, when attached to ships, could allow for the spread of the disease into new regions.
All claims expressed in this article are solely those of the authors and the U.S. Geological Survey but do not necessarily represent those of the publisher, the editors and the reviewers. This article has been peer reviewed and approved for publication consistent with USGS Fundamental Science Practices (https://pubs.usgs.gov/circ/1367/).
The sequence data and associated metadata presented in the study are deposited in the NCBI Sequence Read Archive (SRA), PRJNA828575 and are also available through a U.S. Geological Survey data release (https://doi.org/10.5066/P9T6NW4V).
CK designed the study. JE, VP, and CK performed the experiment and collected the data. JE performed the laboratory work and data analysis. JE and CK drafted the initial manuscript. All authors contributed to the article and approved the submitted version.
This work was funded by the USGS Coastal and Marine Hazards Resources Program of the Natural Hazards Mission Area, the USGS Ecosystems Mission Area Biological Threats Program, and the Florida Department of Environmental Protection (grant number B7C0F5 to VP).
The authors wish to thank the following for their contributions to this work: Keys Marine Laboratory staff performed field collections of the diseased coral colonies. Mote Marine Laboratory staff and Smithsonian Marine Station staff performed field collections of healthy corals. Smithsonian Marine Station staff members J. Houk, K. Pitts, and T. Vekich further assisted with experimental setup and maintenance. J. Voelschow assisted with preparing the data release and NCBI submission. A. Aunins and B. Ushijima provided constructive feedback on the manuscript. Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aeby G. S., Ushijima B., Campbell J. E., Jones S., Williams G. J., Meyer J. L., et al. (2019). Pathogenesis of a tissue loss disease affecting multiple species of corals along the Florida reef tract. Front. Mar. Sci. 6. doi: 10.3389/fmars.2019.00678
Aguirre-Macedo M. L., Vidal-Martinez V. M., Herrera-Silveira J. A., Valdés-Lozano D. S., Herrera-Rodríguez M., Olvera-Novoa M. A. (2008). Ballast water as a vector of coral pathogens in the gulf of Mexico: the case of the cayo arcas coral reef. Mar. pollut. Bull. 56 (9), 1570–1577. doi: 10.1016/j.marpolbul.2008.05.022
Alvarez-Filip L., Estrada-Saldívar N., Pérez-Cervantes E., Molina-Hernández A., González-Barrios F. J. (2019). A rapid spread of the stony coral tissue loss disease outbreak in the Mexican Caribbean. PeerJ 7, e8069. doi: 10.7717/peerj.8069
Anderson M. J., Walsh D. C. I. (2013). PERMANOVA, ANOSIM, and the mantel test in the face of heterogeneous dispersions: What null hypothesis are you testing? Ecol. Monogr. 83 (4), 557–574. doi: 10.1890/12-2010.1
Antonius A. (1973). “New observations on coral destruction in reefs,” in Tenth meeting of the association of island marine laboratories of the Caribbean (Mayaguez, Puerto Rico: University of Puerto Rico Mayaguez) 10, 3.
Apprill A., Weber L. G., Santoro A. E. (2016). Distinguishing between microbial habitats unravels ecological complexity in coral microbiomes. mSystems 1 (5), e00143–e00116. doi: 10.1128/mSystems.00143-16
Arotsker L., Kramarsky-Winter E., Ben-Dov E., Siboni N., Kushmaro A. (2015). Changes in the bacterial community associated with black band disease in a red Sea coral, Favia sp., in relation to disease phases. Dis. Aquat. Organisms 116, 47–58. doi: 10.3354/dao02911
Baier R. E., Forsberg R. L., Meyer A. E., Lundquist D. C. (2014). Ballast tank biofilms resist water exchange but distribute dominant species. Manage. Biol. Invasions 5 (3), 241–244. doi: 10.3391/mbi.2014.5.3.07
Bailey S. A., Brown L., Campbell M. L., Canning-Clode J., Carlton J. T., Castro N., et al. (2020). Trends in the detection of aquatic non-indigenous species across global marine, estuarine and freshwater ecosystems: A 50-year perspective. Diversity Distributions 26 (12), 1780–1797. doi: 10.1111/ddi.13167
Becker C. C., Brandt M., Miller C. A., Apprill A. (2021). Microbial bioindicators of stony coral tissue loss disease identified in corals and overlying waters using a rapid field-based sequencing approach. Environ. Microbiol 24, 1166–1182. doi: 10.1111/1462-2920.15718
Bokulich N. A., Kaehler B. D., Rideout J. R., Dillon M., Bolyen E., Knight R., et al. (2018). Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2's q2-feature-classifier plugin. Microbiome 6, 90. doi: 10.1186/s40168-018-0470-z
Bolyen E., Rideout J. R., Dillon M. R., Bokulich N. A., Abnet C. C., Al-Ghalith G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37 (8), 852–857. doi: 10.1038/s41587-019-0209-9
Bourne D. G., Garren M., Work T. M., Rosenberg E., Smith G. W., Harvell C. D. (2009). Microbial disease and the coral holobiont. Trends Microbiol. 17 (12), 554–562. doi: 10.1016/j.tim.2009.09.004
Brandt M. E., Ennis R. S., Meiling S. S., Townsend J., Cobleigh K., Glahn A., et al. (2021). The emergence and initial impact of stony coral tissue loss disease (SCTLD) in the united states virgin islands. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.715329
Bruno J. F., Petes L. E., Harvell C. D., Hettinger A. (2003). Nutrient enrichment can increase the severity of coral diseases. Ecol. Lett. 6 (12), 1056–1061. doi: 10.1046/j.1461-0248.2003.00544.x
Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J. A., Holmes S. P. (2016). DADA2: High-resolution sample inference from illumina amplicon data. Nat. Methods 13 (7), 581–583. doi: 10.1038/nmeth.3869
Caporaso J. G., Lauber C. L., Walters W. A., Berg-Lyons D., Lozupone C. A., Turnbaugh P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U S A 108, 4516–4522. doi: 10.1073/pnas.1000080107
Carlton J. T. (1987). Patterns of transoceanic marine biological invasions in the pacific ocean. Bull. Mar. Sci. 41 (2), 452–465.
Clark A. S., Williams S. D., Maxwell K., Rosales S. M., Huebner L. K., Landsberg J. H., et al. (2021). Characterization of the microbiome of corals with stony coral tissue loss disease along florida’s coral reef. Microorganisms 9 (11), 2181. doi: 10.3390/microorganisms9112181
Cohen A. N., Dobbs F. C. (2015). Failure of the public health testing program for ballast water treatment systems. Mar. pollut. Bull. 91 (1), 29–34. doi: 10.1016/j.marpolbul.2014.12.031
Dahlgren C., Pizarro V., Sherman K., Greene W., Oliver J. (2021). Spatial and temporal patterns of stony coral tissue loss disease outbreaks in the Bahamas. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.682114
Damjanovic K., Blackall L. L., Peplow L. M., van Oppen M. J. H. (2020). Assessment of bacterial community composition within and among Acropora loripes colonies in the wild and in captivity. Coral Reefs 39 (5), 1245–1255. doi: 10.1007/s00338-020-01958-y
de Carvalho C. C. C. R. (2017). “Biofilms: Microbial strategies for surviving UV exposure,” in Ultraviolet light in human health, diseases and environment. Ed. Ahmad S. I. (Cham: Springer International Publishing), 233–239.
Dobbelaere T., Holstein D. M., Muller E. M., Gramer L. L., McEachron L., Williams S. D., et al. (2022). Connecting the dots: Transmission of stony coral tissue loss disease from the marquesas to the dry tortugas. Front. Mar. Sci. 9. doi: 10.3389/fmars.2022.778938
Dobbelaere T., Muller E. M., Gramer L. J., Holstein D. M., Hanert E. (2020). Coupled epidemio-hydrodynamic modeling to understand the spread of a deadly coral disease in Florida. Front. Mar. Sci. 7. doi: 10.3389/fmars.2020.591881
Drake L. A., Meyer A. E., Forsberg R. L., Baier R. E., Doblin M. A., Heinemann S., et al. (2005). Potential invasion of microorganisms and pathogens via ‘Interior Hull fouling’: Biofilms inside ballast water tanks. Biol. Invasions 7, 969–982. doi: 10.1007/s10530-004-3001-8
Estrada-Saldívar N., Molina-Hernández A., Pérez-Cervantes E., Medellín-Maldonado F., González-Barrios F. J., Alvarez-Filip L. (2020). Reef-scale impacts of the stony coral tissue loss disease outbreak. Coral Reefs 39 (4), 861–866. doi: 10.1007/s00338-020-01949-z
Estrada-Saldívar N., Quiroga-García B. A., Pérez-Cervantes E., Rivera-Garibay O. O., Alvarez-Filip L. (2021). Effects of the stony coral tissue loss disease outbreak on coral communities and the benthic composition of cozumel reefs. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.632777
Evans J. S., Paul V. J., Ushijima B., Kellogg C. A. (2022). Combining tangential flow filtration and size fractionation of mesocosm water as a method for the investigation of waterborne coral diseases. Biol. Methods Protoc. 7 (1), 1–8. doi: 10.1093/biomethods/bpac007
Faith D. P. (1992). Conservation evaluation and phylogenetic diversity. Biol. Conserv. 61 (1), 1–10. doi: 10.1016/0006-3207(92)91201-3
Frösler J., Panitz C., Wingender J., Flemming H.-C., Rettberg P. (2017). Survival of deinococcus geothermalis in biofilms under desiccation and simulated space and Martian conditions. Astrobiology 17 (5), 431–447. doi: 10.1089/ast.2015.1431
Hathroubi S., Mekni M. A., Domenico P., Nguyen D., Jacques M. (2017). Biofilms: Microbial shelters against antibiotics. Microb. Drug Resist. 23 (2), 147–156. doi: 10.1089/mdr.2016.0087
Heres M. M., Farmer B. H., Elmer F., Hertler H. (2021). Ecological consequences of stony coral tissue loss disease in the Turks and Caicos islands. Coral Reefs 40 (2), 609–624. doi: 10.1007/s00338-021-02071-4
Holeck K. T., Mills E. L., MacIsaac H. J., Dochoda M. R., Colautti R. I., Ricciardi A. (2004). Bridging troubled waters: Biological invasions, transoceanic shipping, and the laurentian great lakes. BioScience 54 (10), 919–929. doi: 10.1641/0006-3568(2004)054[0919:Btwbit]2.0.Co;2
Huntley N., Brandt M. E., Becker C. C., Miller C. A., Meiling S. S., Correa A. M. S., et al. (2022). Experimental transmission of stony coral tissue loss disease results in differential microbial responses within coral mucus and tissue. ISME Commun. 2 (1), 46. doi: 10.1038/s43705-022-00126-3
Iwanowicz D. D., Schill W. B., Woodley C. M., Bruckner A., Neely K., Briggs K. M. (2021). Exploring the stony coral tissue loss disease bacterial pathobiome (Preprint). bioRxiv. doi: 10.1101/2020.05.27.120469
Katoh K., Standley D. M. (2013). MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30 (4), 772–780. doi: 10.1093/molbev/mst010
Kellogg C. A., Evans J. S., Voelschow J. J. (2022). Prokaryotic communities from marine biofilms formed on stainless steel plates in coral mesocosms – raw and processed data. U.S. Geological Survey Data Release. doi: 10.5066/P9T6NW4V
Klaus J. S., Janse I., Fouke B. W. (2011). Coral black band disease microbial communities and genotypic variability of the dominant cyanobacteria (CD1C11). Bull. Mar. Sci. 87 (4), 795–821. doi: 10.5343/bms.2010.1050
Klein G., MacIntosh K., Kaczmarska I., Ehrman J. M. (2010). Diatom survivorship in ballast water during trans-pacific crossings. Biol. Invasions 12 (5), 1031–1044. doi: 10.1007/s10530-009-9520-6
Kooperman N., Ben-Dov E., Kramarsky-Winter E., Barak Z., Kushmaro A. (2007). Coral mucus-associated bacterial communities from natural and aquarium environments. FEMS Microbiol. Lett. 276 (1), 106–113. doi: 10.1111/j.1574-6968.2007.00921.x
Kozich J. J., Westcott S. L., Baxter N. T., Highlander S. K., Schloss P. D. (2013). Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq illumina sequencing platform. Appl. Environ. Microbiol. 79 (17), 5112–5120. doi: 10.1128/Aem.01043-13
Kramer P. R., Roth L., Lang J. (2019) Map of stony coral tissue loss disease outbreak in the Caribbean. Available at: www.agrra.org (Accessed August 30 2021).
Lozupone C., Knight R. (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71 (12), 8228–8235. doi: 10.1128/aem.71.12.8228-8235.2005
Meiling S., Muller E. M., Smith T. B., Brandt M. E. (2020). 3D photogrammetry reveals dynamics of stony coral tissue loss disease (SCTLD) lesion progression across a thermal stress event. Front. Mar. Sci. 7. doi: 10.3389/fmars.2020.597643
Meyer J. L., Castellanos-Gell J., Aeby G. S., Hase C. C., Ushijima B., Paul V. J. (2019). Microbial community shifts associated with the ongoing stony coral tissue loss disease outbreak on the Florida reef tract. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.02244
MPI (2018). Craft risk management standard: Biofouling on vessels arriving to new Zealand (Wellington, New Zealand: CRMS-BIOFOUL).
Muller E. M., Sartor C., Alcaraz N. I., van Woesik R. (2020). Spatial epidemiology of the stony-Coral-Tissue-Loss disease in Florida. Front. Mar. Sci. 7. doi: 10.3389/fmars.2020.00163
Neely K. L., Macaulay K. A., Hower E. K., Dobler M. A. (2020). Effectiveness of topical antibiotics in treating corals affected by stony coral tissue loss disease. Peerj 8, e9289. doi: 10.7717/peerj.9289
Neely K. L., Shea C. P., Macaulay K. A., Hower E. K., Dobler M. A. (2021). Short- and long-term effectiveness of coral disease treatments. Front. Mar. Sci. 8 (1031). doi: 10.3389/fmars.2021.675349
NOAA (2018) Case definition: Stony coral tissue loss disease (SCTLD). Available at: https://floridadep.gov/rcp/coral/documents/stony-coral-tissue-loss-disease-sctld-case-definition (Accessed 11/17/2020).
Papadatou M., Robson S. C., Dobretsov S., Watts J. E. M., Longyear J., Salta M. (2021). Marine biofilms on different fouling control coating types reveal differences in microbial community composition and abundance. MicrobiologyOpen 10 (4), e1231. doi: 10.1002/mbo3.1231
Petersen N. B., Madsen T., Glaring M. A., Dobbs F. C., Jørgensen N. O. G. (2019). Ballast water treatment and bacteria: Analysis of bacterial activity and diversity after treatment of simulated ballast water by electrochlorination and UV exposure. Sci. Total Environ. 648, 408–421. doi: 10.1016/j.scitotenv.2018.08.080
Pielou E. C. (1966). The measurement of diversity in different types of biological collections. J. Theor. Biol. 13, 131–144. doi: 10.1016/0022-5193(66)90013-0
Pratte Z. A., Richardson L. L., Mills D. K. (2015). Microbiota shifts in the surface mucopolysaccharide layer of corals transferred from natural to aquaria settings. J. Invertebrate Pathol. 125, 42–44. doi: 10.1016/j.jip.2014.12.009
Precht W. F., Gintert B. E., Robbart M. L., Fura R., van Woesik R. (2016). Unprecedented disease-related coral mortality in southeastern Florida. Sci. Rep. 6, 31374. doi: 10.1038/srep31374
Price M. N., Dehal P. S., Arkin A. P. (2010). FastTree 2 - approximately maximum-likelihood trees for Large alignments. PLoS One 5 (3). doi: 10.1371/journal.pone.0009490
Remple K. L., Silbiger N. J., Quinlan Z. A., Fox M. D., Kelly L. W., Donahue M. J., et al. (2021). Coral reef biofilm bacterial diversity and successional trajectories are structured by reef benthic organisms and shift under chronic nutrient enrichment. NPJ Biofilms Microbiomes 7, 84. doi: 10.1038/s41522-021-00252-1
Roder C., Arif C., Bayer T., Aranda M., Daniels C., Shibl A., et al. (2014). Bacterial profiling of white plague disease in a comparative coral species framework. ISME J. 8 (1), 31–39. doi: 10.1038/ismej.2013.127
Rohwer F., Seguritan V., Azam F., Knowlton N. (2002). Diversity and distribution of coral-associated bacteria. Mar. Ecol. Prog. Ser. 243, 1–10. doi: 10.3354/meps243001
Rosales S. M., Clark A. S., Huebner L. K., Ruzicka R. R., Muller E. M. (2020). Rhodobacterales and Rhizobiales are associated with stony coral tissue loss disease and its suspected sources of transmission. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.00681
Rosenau N. A., Gignoux-Wolfsohn S., Everett R. A., Miller A. W., Minton M. S., Ruiz G. M. (2021). Considering commercial vessels as potential vectors of stony coral tissue loss disease. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.709764
Röthig T., Roik A., Yum L. K., Voolstra C. R. (2017). Distinct bacterial microbiomes associate with the deep-Sea coral Eguchipsammia fistula from the red Sea and from aquaria settings. Front. Mar. Sci. 4. doi: 10.3389/fmars.2017.00259
Ruiz G. M., Rawlings T. K., Dobbs F. C., Drake L. A., Mullady T., Huq A., et al. (2000). Global spread of microorganisms by ships. Nature 408 (6808), 49–50. doi: 10.1038/35040695
Séré M. G., Tortosa P., Chabanet P., Quod J.-P., Sweet M. J., Schleyer M. H. (2015). Identification of a bacterial pathogen associated with porites white patch syndrome in the Western Indian ocean. Mol. Ecol. 24 (17), 4570–4581. doi: 10.1111/mec.13326
Shannon C. E. (1948). The mathematical theory of communication. Bell System Tech. J. 27, 379–423. doi: 10.1002/j.1538-7305.1948.tb01338.x
Shikuma N. J., Hadfield M. G. (2010). Marine biofilms on submerged surfaces are a reservoir for Escherichia coli and Vibrio cholerae. Biofouling 26 (1), 39–46. doi: 10.1080/08927010903282814
Shilling E. N., Combs I. R., Voss J. D. (2021). Assessing the effectiveness of two intervention methods for stony coral tissue loss disease on Montastraea cavernosa. Sci. Rep. 11, 8566. doi: 10.1038/s41598-021-86926-4
Siboni N., Lidor M., Kramarsky-Winter E., Kushmaro A. (2007). Conditioning film and initial biofilm formation on ceramics tiles in the marine environment. FEMS Microbiol. Lett. 274 (1), 24–29. doi: 10.1111/j.1574-6968.2007.00809.x
Stoodley P., Wilson S., Hall-Stoodley L., Boyle J. D., Lappin-Scott H. M., Costerton J. W. (2001). Growth and detachment of cell clusters from mature mixed-species biofilms. Appl. Environ. Microbiol. 67 (12), 5608–5613. doi: 10.1128/AEM.67.12.5608-5613.2001
Studivan M. S., Rossin A. M., Rubin E., Soderberg N., Holstein D. M., Enochs I. C. (2022). Reef sediments can act as a stony coral tissue loss disease vector. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.815698
Sweet M. J., Croquer A., Bythell J. C. (2011). Bacterial assemblages differ between compartments within the coral holobiont. Coral Reefs 30 (1), 39–52. doi: 10.1007/s00338-010-0695-1
Ushijima B., Meyer J. L., Thompson S., Pitts K., Marusich M. F., Tittl J., et al. (2020). Disease diagnostics and potential coinfections by Vibrio coralliilyticus during an ongoing coral disease outbreak in Florida. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.569354
Vega Thurber R. L., Burkepile D. E., Fuchs C., Shantz A. A., McMinds R., Zaneveld J. R. (2014). Chronic nutrient enrichment increases prevalence and severity of coral disease and bleaching. Global Change Biol. 20 (2), 544–554. doi: 10.1111/gcb.12450
Walker B. K., Turner N. R., Noren H. K. G., Buckley S. F., Pitts K. A. (2021). Optimizing stony coral tissue loss disease (SCTLD) intervention treatments on Montastraea cavernosa in an endemic zone. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.666224
Walton C. J., Hayes N. K., Gilliam D. S. (2018). Impacts of a regional, multi-year, multi-species coral disease outbreak in southeast Florida. Front. Mar. Sci. 5. doi: 10.3389/fmars.2018.00323
Weil E., Hernández-Delgado E. A., Gonzalez M., Williams S., Suleimán-Ramos S., Figuerola M., et al. (2019). Spread of the new coral disease “SCTLD“ into the Caribbean: implications for Puerto Rico. Reef Encounter 34, 38–43.
Weil E., Rogers C. S. (2011). “Coral reef diseases in the Atlantic-Caribbean,” in Coral reefs: An ecosystem in transition. Eds. Dubinsky Z., Stambler N. (Dordrecht: Springer Netherlands), 465–491.
Work T. M., Weatherby T. M., Landsberg J. H., Kiryu Y., Cook S. M., Peters E. C. (2021). Viral-like particles are associated with endosymbiont pathology in Florida corals affected by stony coral tissue loss disease. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.750658
Keywords: SCTLD, biofilm, bacteria, coral disease, transmission, biofouling, 16S, stony coral tissue loss disease
Citation: Evans JS, Paul VJ and Kellogg CA (2022) Biofilms as potential reservoirs of stony coral tissue loss disease. Front. Mar. Sci. 9:1009407. doi: 10.3389/fmars.2022.1009407
Received: 01 August 2022; Accepted: 17 October 2022;
Published: 23 November 2022.
Edited by:
Aldo Cróquer, The Nature Conservancy, Dominican RepublicReviewed by:
Emmanuel Hanert, Université Catholique de Louvain, BelgiumCopyright © 2022 Evans, Paul and Kellogg. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christina A. Kellogg, Y2tlbGxvZ2dAdXNncy5nb3Y=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.