
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 27 September 2022
Sec. Aquatic Physiology
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.1005401
The mitogen-activated protein kinase (MAPK) gene family performs crucial roles in cell division, migration, development, apoptosis, inflammatory response, and abiotic and biotic stress responses. However, very little information is available about the MAPKs in turbot (Scophthalmus maximus). In this study, 15 turbot MAPKs (SmMAPKs) were identified throughout the whole genome, and their basic chemical and physical properties and subcellular localization were illustrated. All SmMAPKs contained the serine/threonine protein kinases, catalytic domain (S_TKc, SMART00220). Phylogenetic analysis revealed that SmMAPKs were classified into three subfamilies, namely, c-Jun NH2-terminal kinase (JNK), extracellular signal-regulated kinase (ERK), and p38. Conserved motif and gene structure analysis revealed high levels of conservation within and between phylogenetic subfamilies. Expression patterns of MAPKs in distinct tissues and under diverse abiotic and biotic stresses were examined using the published available RNA-seq data sets. As a result, SmMAPKs showed obviously tissue-specific expression. Furthermore, 7 and 10 candidate stress-responsive MAPK genes were detected under abiotic and biotic stresses, respectively, among which five common MAPK genes, namely, SmMAPK4 (ERK4), SmMAPK6 (ERK3), SmMAPK11 (p38β), SmMAPK12b (p38γ), and SmMAPK15 (ERK7/8) showed extremely significant responses to both abiotic and biotic stresses, demonstrating their potential functions in comprehensive antistress. These results demonstrate that MAPKs might play vital roles in response to various abiotic and biotic stresses in turbot, which would contribute to making scientific preventive measures to environmental changes in the process of farming and promoting the development of selective breeding for comprehensive stress resistance in turbot.
Mitogen-activated protein kinase (MAPK) cascades are ubiquitous and highly evolutionarily conservative signal transduction pathways in all eukaryotes (Chang and Karin, 2001; Colcombet and Hirt, 2008; Kyriakis and Avruch, 2012), which respond to various extracellular stimuli to induce and regulate multiple cellular processes, such as proliferation, differentiation, development, stress response, survival, and apoptosis. (Keshet and Seger, 2010; Plotnikov et al., 2011). The canonical MAPK cascade is made up of three evolutionarily conserved sequentially acting kinases: MAPKKK (MAPK kinase kinase), MAPKK (MAPK kinase), and MAPK (Widmann et al., 1999). Among them, MAPKs, a cluster of serine/threonine protein kinases that play vital roles in signal transduction in response to variations in the cellular environment (Teramoto and Gutkind, 2013), are classified into three main subfamilies, namely, extracellular signal-regulated kinase (ERK), c-Jun NH2-terminal kinase (JNK), and p38 MAP kinases, which are based on the dual-phosphorylation site motif. In detail, ERKs, containing a TEY motif (Thr-Glu-Tyr), are coded by two genes, ERK1 (mapk3) and ERK2 (mapk1). Moreover, JNKs, having a TPY motif (Thr-Pro-Tyr), are coded by three genes, JNK1, 2, and 3, also called mapk8, 9, and 10, respectively, whereas, p38 MAPKs, comprising a TGY motif (Thr-Gly-Tyr), are encoded by mapk14 (p38α), 11 (p38β), 12 (p38γ), and 13 (p38δ) (L. and Razvan, 2002).
MAPKs can be activated by various stimuli (Zhang and Liu, 2002). In general, the ERK subfamily is preferentially activated by growth factors, cytokines, viruses, carcinogens, and pro-inflammatory stimuli (Kyriakis and Avruch, 2012). Activation of ERKs is involved in mediating cell division, migration, development, survival, innate immunity, and inflammation (Cowan and Storey, 2003; Raman et al., 2007). Furthermore, JNKs are more responsive to mitogens, pro-inflammatory cytokines, environmental stresses (ionizing radiation, heat shock, oxidants), genotoxins, pathogen-associated molecular patterns (PAMPs)/damage-associated molecular pattern (DAMPs), etc. (Kyriakis et al., 1994; Kyriakis and Avruch, 2012). Activation of JNK proteins participates in the inflammatory response, cytokine production, apoptosis, cell migration, and metabolism (Chen et al., 2001; Raman et al., 2007). Furthermore, p38 MAPKs can be activated by a variety of environmental stresses (such as UV radiation, hypoxia, and osmotic shock), pro-inflammatory cytokines, PAMPs, and DAMPs (Raman et al., 2007; Kyriakis and Avruch, 2012). Activation of p38 MAPKs mainly fulfills functions during the course of response to microbial infections, inflammatory responses, and environmental stresses (L. and Razvan, 2002).
In the last decades, genome-wide identification and the biological roles of MAPKs have been studied extensively in various plants and mammals (Widmann et al., 1999; Liu et al., 2000; Colcombet and Hirt, 2008; Kyriakis and Avruch, 2012). However, to date, genome-wide identification of MAPKs only has been reported in several teleost fishes, including zebrafish (Danio rerio) (Fujii et al., 2000; Krens et al., 2006a), spotted sea bass (Lateolabrax maculatus) (Tian et al., 2019), Japanese flounder (Paralichthys olivaceus) (Qiao et al., 2021), and seahorse (Hippocampus erectus) (Wang et al., 2021), whereas, an increasing number of functional studies about MAPKs of teleost have been performed in recent years, especially in the aspect of functional characterizations in response to distinct stresses. For instance, transcriptional expression analyses under cold and heat temperature stresses and Edwardsiella tarda infection indicated that eight MAPK genes were involved in the regulation of biotic and abiotic stress resistance in Japanese flounder (Paralichthys olivaceus) (Qiao et al., 2021). Several MAPKs in the JNK and p38 subfamilies showed differential expression after salinity and hypoxia challenge, illustrating their underlying roles in osmotic pressure and hypoxia responses in Lateolabrax maculatus (Tian et al., 2019). Moreover, it was observed that p38 MAPK is phosphorylated, activated, and involved in the induction of Hsp70 in the thermally stressed red blood cells of Sparus aurata (Feidantsis et al., 2012). In Dicentrarchus labrax, starvation stress was shown to affect the phosphorylation and activation of p38 MAPK (Antonopoulou et al., 2013). The expression patterns of MAPKs in seahorse (Hippocampus erectus) were analyzed postinfection with Vibrio fortis, uncovering their upregulation pattern in the brood pouch and other immune tissues (Wang et al., 2021). Following ammonia stress, lipopolysaccharide, and Aeromonas hydrophila challenge, the study results proved that the p38 MAPK gene had antistress properties and played key roles in protection against bacterial infection and inflammation in blunt snout bream (Megalobrama amblycephala) (Zhang et al., 2019). In addition, studies in euryhaline fish, killifish (Fundulus heteroclitus), indicated that ERK1, JNK, and p38 MAPK were important components of salinity adaptation and participated in osmotic regulation (Kultz and Avila, 2001; Marshall et al., 2005). All the above studies emphasize the important functions of MAPKs in biotic and abiotic stress responses in teleost.
Turbot (Scophthalmus maximus, FishBase ID: 1348), a commercially important cold-water marine flatfish species, is considered a cosmopolitan marine-culture flatfish species with delicious taste, high nutritional value, and the highest annual aquaculture production (Lei et al., 2012). However, due to continuous expansion of the intensive breeding scale coupled with increased frequency of extreme weather events in recent years, turbot is subject to various biotic and abiotic stresses, such as pathogen invasion, heat, oxygen, and salinity stress, which greatly impede the healthy and sustainable development of the turbot industry (Ronza et al., 2016; Cui et al., 2019; Cui et al., 2020; Huang et al., 2020; Nie et al., 2020; Gao et al., 2021; Ronza et al., 2021). Given the roles of MAPKs in coping with various stresses, therefore, systematically identifying MAPKs and investigating their functions underlying abiotic and biotic stresses have important practical significance in turbot. To date, no systematic analysis of MAPKs has been conducted in turbot. Therefore, in this study, we carried out the systematic identification of MAPKs based on a genome-wide search against turbot genome assembly. Then, basic physical and chemical properties, phylogenetic analysis, gene structure, and conserved motifs were characterized. Furthermore, the expression patterns of MAPKs in diverse tissues and under various abiotic and biotic stresses were investigated based on multiple published RNA-seq data sets from turbot. These results provide new insights into the biological roles of MAPKs in turbot under various biotic and abiotic stresses and provide valuable information for elucidating the environmental adaptation of turbot.
To do a genome-wide identification of all MAPKs in turbot, a high-quality, chromosome-level turbot genome assembly (GCA_022379125.1) (Xu et al., 2022) and MAPK protein sequences of Danio rerio, Oryzias latipes, Lepisosteus oculatus were downloaded from NCBI and UniProt. Then, D. rerio MAPK protein sequences were selected as a query database to search against the turbot genome using TBLASTN (Gertz et al., 2006) with an E-value of < 1e-10 and a minimum identity of 50% as the threshold. Then, all candidate sequences were conducted to analyze conserved domains with the HMMER software (Potter et al., 2018) with an E-value of 10-4 and Batch SMART tool in the Tbtools software (Chen et al., 2020) with default parameters. Some genes without MAPK-specific S_TKc domains were rejected. Next, the putative MAPKs functionally annotated as MAPKs in the UniProt database were manually selected as the final identified turbot MAPKs (SmMAPKs). Furthermore, the ExPASy ProtParam tool (https://web.expasy.org/protparam/) was performed to analyze the chemical and physical properties of MAPKs, including the numbers of amino acids, molecular weight (MW), and theoretical isoelectric point (pI). Finally, the subcellular localization was detected by the WoLF PSORT tool (https://www.genscript.com/wolf-psort.html).
To investigate the phylogenetic relationships of MAPK proteins among different teleost species, a phylogenetic tree was constructed based on the amino acid sequences of MAPKs from turbot, D. rerio, O. latipes, and L. oculatus. Multiple sequence alignments were first carried out through the ClustalW program with default parameters. Then, the phylogenetic tree was constructed using the MEGA 11.0 (Tamura et al., 2021) software by the neighbor-joining (NJ) method with the Jones-Taylor-Thornton (JTT) substitution model and 1,000 bootstrap replications. Finally, EvolView (2.0) (He et al., 2016) (https://www.evolgenius.info/evolview/) was used to visualize the phylogenetic tree.
To better understand the chromosomal distribution of MAPKs in turbot, the sequences of SmMAPKs were aligned to the 22 turbot chromosomes based on the Generic Transfer Format (GTF) file. Then, the alignment results were visualized using the gene location visualization function in TBtools (Chen et al., 2020). Moreover, to analyze the conserved motifs of the SmMAPKs, MEME (5.4.1) (Bailey et al., 2015) (http://meme-suite.org/tools/meme) was used, with the number of motifs being 10 and the other parameters set to default. Meanwhile, multiple sequence alignment was also conducted to survey the conserved dual-phosphorylation site motif in SmMAPKs using the DNAMAN 10.3.3.126 software. Furthermore, the GTF file and phylogenetic tree of SmMAPKs constructed by MEGA 11.0 were submitted to the Gene Structure Display Server (GSDS, http://gsds.cbi.pku.edu.cn) to show the gene structure with the length and position of exons and introns. Finally, the combination of the phylogenetic tree, conserved motifs, and intron-exons structure was generated.
To illustrate the expression patterns of MAPKs in distinct tissues of turbot, multiple RNA-seq data sets of 12 different tissues from healthy turbot in the control group (under normal conditions), specifically, testis, ovary, gill, liver, intestine, blood, kidney, muscle, spleen, thymus, brain, and pyloric caeca, were collected from the NCBI Sequence Read Archive (SRA) database. Detailed information of these RNA-seq data sets are provided in Supplementary Table 1.
To investigate the expression patterns of MAPKs under abiotic stress in turbot, six published abiotic stress (heat, oxygen, salinity)-related RNA-seq data sets (Supplementary Table 2) were downloaded from the NCBI SRA database. In detail, for heat-stress expression analysis, two RNA-seq data sets were used. In the SRA study of SRP152627, kidney tissue in turbot exposed to heat stress (23°C, 25°C, and 28°C) for 24 h and kidney tissue in the control group maintained at 14°C was used for RNA-seq (Huang et al., 2020). In the SRA study of SRP273870, RNA-seq was conducted on liver tissue in the treatment group under heat stress (20°C, 24°C, and 28°C) for 24 h and in the control group maintained at 14°C (Zhao et al., 2021). The SRA study of SRP167318 was used for oxygen-stress expression analysis, in which gill tissues in turbot subjected to waterless treatment under different oxygen conditions (6.5 mg/L (water, control group), 300 mg/L (waterless, air group), and 1430 mg/L (waterless, oxygen)) were used for RNA-seq (Nie et al., 2020). For salinity-stress expression analysis, we downloaded three low- and high-salinity stress-related RNA-seq data sets. In the SRA study of SRP238143 and SRP153594, gill and kidney tissue samples under different salinity stresses [low salinity (5‰), natural seawater (30‰), and high salinity (50‰)] for 24 h were collected, respectively, for RNA-seq (Cui et al., 2019; Cui et al., 2020). In the SRA study of SRP277001, liver tissues subjected to low salinity and natural seawater were collected at 24 h and used for RNA-seq (Liu et al., 2021).
To elaborate the expression patterns of MAPKs under biotic stress in turbot, publicly available RNA-seq data sets correlated to pathogen stresses, encompassing parasite (Enteromyxum scophthalmi), bacteria (Vibrio anguillarum), and virus (Megalocytivirus) infection, were downloaded from the NCBI SRA database (Supplementary Table 2). For E. scophthalmi infection, four RNA-seq data sets focusing on five distinct tissues in healthy and parasitized turbot were acquired. In the SRA study of SRP065375 and SRP050607, head kidney, pyloric caeca, and spleen tissues at 24 and 42 days postinfection (dpi) with E. scophthalmi were obtained for RNA-seq (Robledo et al., 2014; Ronza et al., 2016). In the SRA study of SRP308109, RNA-seq was performed on blood in four levels of infected (incipient, slight, moderate, and severe) and not infected turbot (Ronza et al., 2021). In the SRA study of SRP255305, thymus tissues from control and infected turbot at 24 and 42 dpi were selected for RNA-seq (Ronza et al., 2020). In addition, five SRA studies were conducted under V. anguillarum challenge. For the SRA study of SRP191266, intestine tissues 0, 1, 4, and 12 h after injection with V. anguillarum were sampled for RNA-seq (Gao et al., 2021). In the SRA studies of SRP319434, SRP320422, SRP335896, and SRP336094, spleen, gill, head kidney, and liver tissues were collected, respectively, under V. anguillarum infection for RNA-seq (Song et al., 2022).
To perform expression analyses of SmMAPKs, raw reads of the above RNA-seq data sets were first aligned to the turbot genome using the STAR software with default parameters (Dobin et al., 2013). Following this, we used the featureCounts (Liao et al., 2014) software program in the Subread (Liao et al., 2013) package to construct reads count matrixes. Next, significant and differentially expressed (DE) (P < 0.05) SmMAPKs and extremely significant DE (false discovery rate (FDR) < 0.05 and |log2 fold change (FC)| > 1) SmMAPKs were identified using the edgeR software (Robinson et al., 2010). Then, expression patterns of SmMAPKs in diverse tissues and under biotic and abiotic stresses were analyzed based on the reads per kilobase per million mapped reads (RPKM) values. Finally, heat maps of expression patterns of MAPKs were visualized using the TBtools package (Chen et al., 2020) with normalized RPKM [log2 (RPKM + 1)].
In the present study, a total of 15 turbot MAPKs (named SmMAPKs) (Table 1 and Supplementary Table 3) were identified in the genome of S. maximus after detecting and validating the conserved domains of all candidate MAPKs. The detailed results of domain validation using the Batch SMART tool and HMMER software are provided in Supplementary Table 4 and Supplementary Table 5. All SmMAPKs were predicted to contain the serine/threonine protein kinases, catalytic domain (S_TKc, SMART00220) (Supplementary Figure 1). The characteristics of SmMAPKs are summarized in Table 1. In detail, the length of encoded proteins varied from 360 (SmMAPK12b) to 1,116 (SmMAPK7) amino acids with predicted MWs ranging from 41,577.52 Da (SmMAPK11) to 122,595.13 Da (SmMAPK7) and theoretical pI ranging from 4.92 (SmMAPK6) to 9.1 (SmMAPK15). Subcellular localization indicated SmMAPK proteins were universally located in the cytosol and nucleus except for SmMAPK3 in the cytoskeleton and SmMAPK10 in the mitochondrion.
To clarify the evolutionary relationships of MAPKs in a distinct teleost, an NJ phylogenetic tree was constructed utilizing the protein sequences of 15 SmMAPKs and 43 other teleost MAPKs. As shown in Figure 1, the tree shows that the 58 MAPKs were classified into three subfamilies, comprising ERK, JNK, and p38, and further divided into 13 distinct clades. The ERK subfamily contained 23 MAPKs, and the JNK subfamily included 15 MAPKs, whereas the p38 subfamily comprised 20 MAPKs. For turbot, the ERK subfamily consisted of six MAPKs (MAPK1, 3, 4, 6, 7, 15), and the JNK subfamily was composed of four MAPKs (MAPK 8a, 8b, 9, 10), whereas five MAPKs (11, 12a, 12b, 13, 14a, 15) were included in the p38 MAPK subfamily. In addition, not all teleost species had MAPKs from each clade, meanwhile, the numbers of MAPKs in each clade were varied. For example, L. oculatus did not include members in the MAPK11 and MAPK3 clades. D. rerio and O. latipes contained two members in the clade of MAPK14, whereas L. oculatus and S. maximus only had one member.
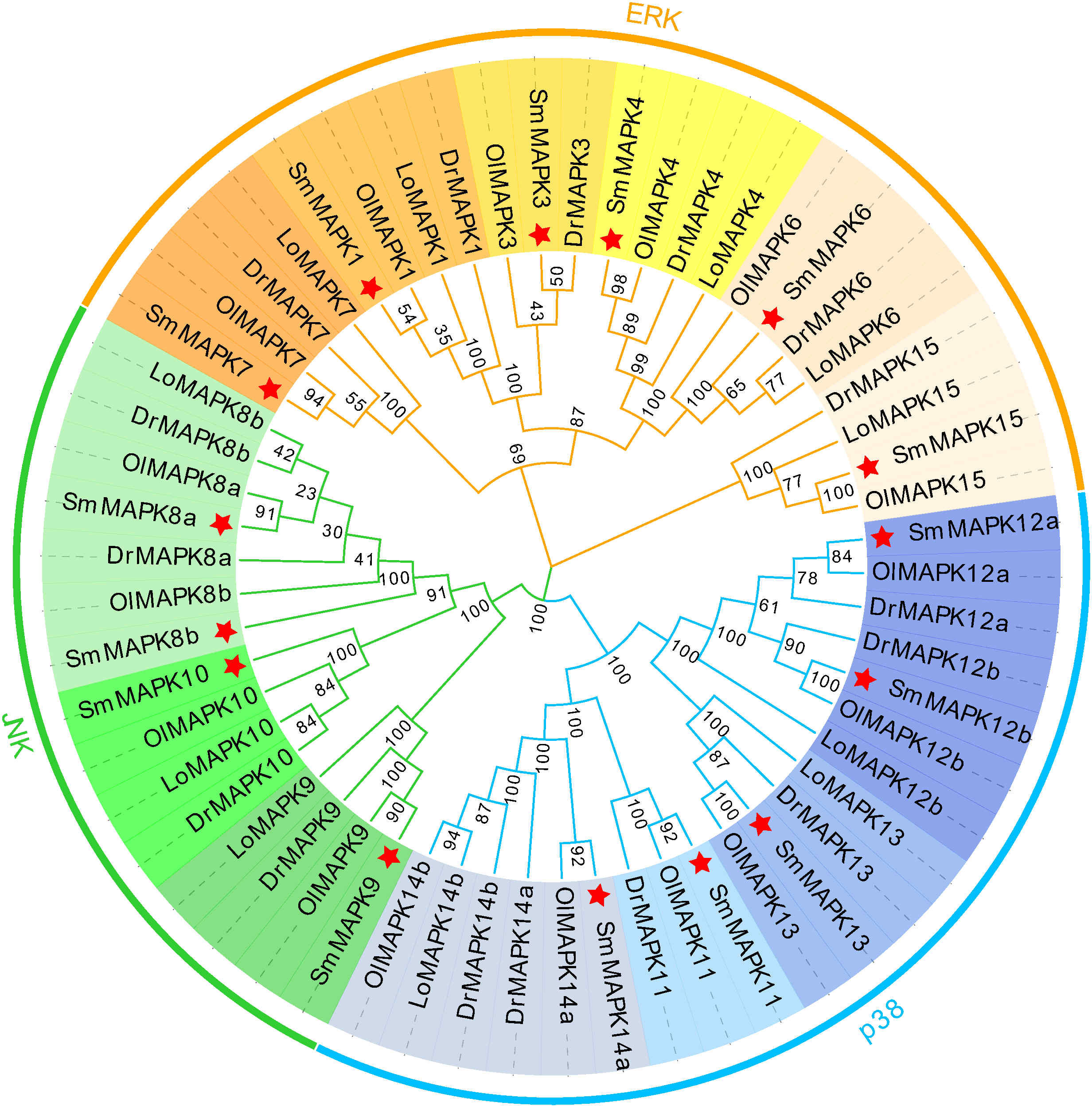
Figure 1 Phylogenetic analysis of mitogen-activated protein kinases (MAPKs) in diverse teleost species. A total of 58 MAPK protein sequences from four teleost species were used to create the neighbor-joining (NJ) tree using MEGA 11.0 with the JTT model and 1,000 bootstrap replications. Three subfamilies are labeled extracellular signal-regulated kinase (ERK), c-Jun NH2-terminal kinase (JNK), and p38. Thirteen clades are discriminated by different colors, and the MAPKs of Scophthalmus maximus are marked with a red asterisk. Dr, Danio rerio; Ol, Oryzias latipes; and Lo, Lepisosteus oculatus.
As shown in Figure 2, 15 SmMAPKs were unevenly distributed on 10 S. maximus chromosomes. In detail, three MAPK genes, namely, SmMAPK 12a, 13, and 14a, were located on chr7. There were two MAPK genes observed on chr 12, chr16, and chr 19, respectively. Moreover, only one SmMAPK was distributed on chr 4, chr 9, chr 10, chr 17, chr 20, and chr 22. In addition, gene structure analysis indicated that SmMAPKs are variable in length, ranging from 4,896 to 33,625 bp (Table 1 and Figure 3). Furthermore, the exon numbers of the SmMAPKs varied from 7 to 14 (Figure 3). On the whole, the numbers of exons in the ERK subfamily were fewer than the JNK and p38 MAPK subfamilies, whereas most members within the same subfamily shared similar numbers of exons. Among them, SmMAPK7 in the ERK subfamily had the fewest exons with only seven, whereas SmMAPK10 in the JNK subfamily, the longest MAPK (33,625 bp) in turbot, had the largest number of exons (14). The motif analysis results revealed a total of 10 motifs detected using the MEME tool (Figure 3), and the sequence logos of 10 motifs from the results of MEME are provided in Supplementary Figure 2. Therefore, motifs 1, 2, 4, 5, and 7 exist in all SmMAPKs, whereas motif 8 could only be observed in members of the JNK subfamily, and motif 10 could only be found in the p38 and ERK subfamilies. The motif numbers among MAPKs varied from 7 to 10, and most of SmMAPKs had nine motifs, demonstrating the similar protein structures and functions among SmMAPKs. Furthermore, multiple sequence alignment showed that all SmMAPKs contained the correct amino acid residue within the specific dual-phosphorylation site (Table 1 and Supplementary Figure 3). In detail, the ERK subfamily had a TEY (Thr-Glu-Tyr) activation motif, among which ERK3 (MAPK6) and ERK4 (MAPK4) contained the SEG (Ser-Glu-Gly) motif, and the p38 MAPK subfamily contained TGY (Thr-Gly-Tyr) conserved structures, and the JNKs have a TPY (Thr-Pro-Tyr) activation motif.
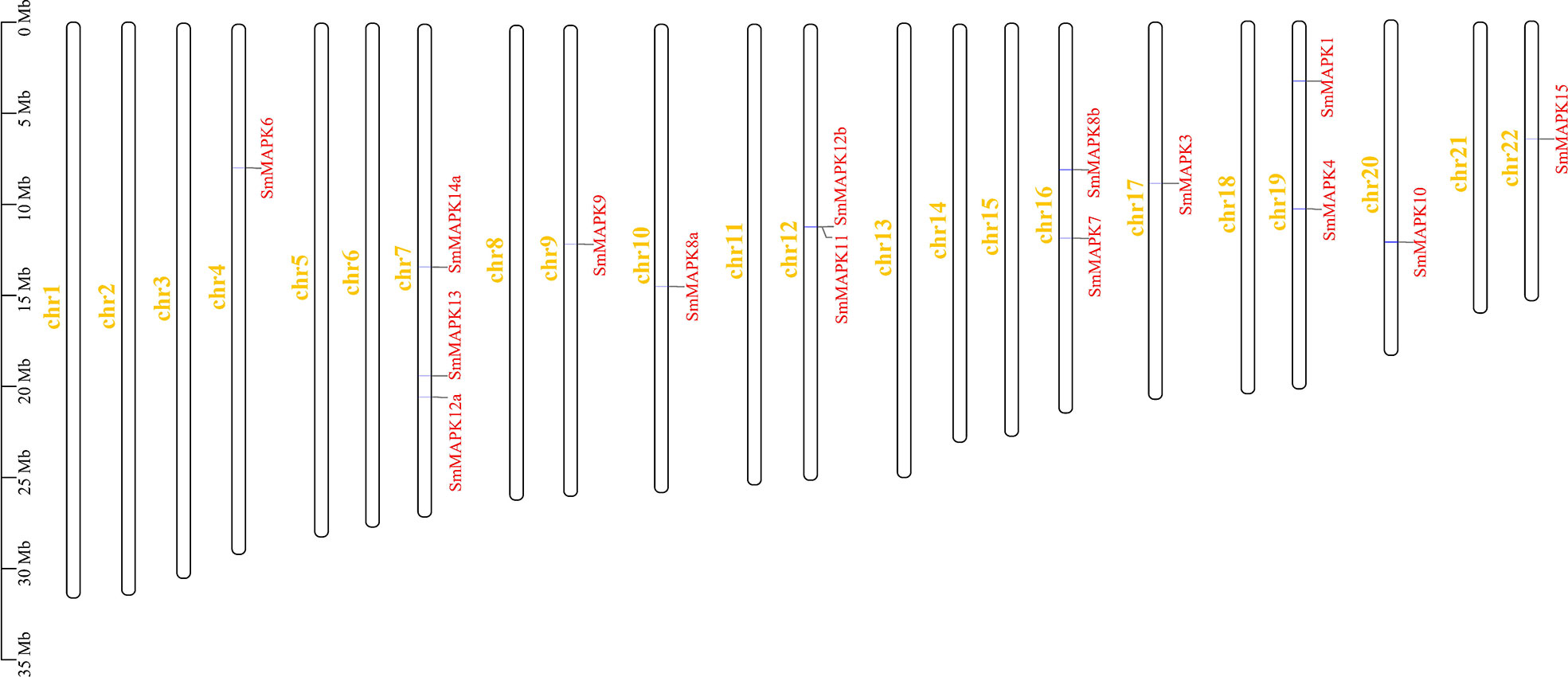
Figure 2 Chromosomal distribution of Scophthalmus maximus mitogen-activated protein kinase (MAPK) genes. The MAPK genes and chromosomes are highlighted in red and yellow, respectively.
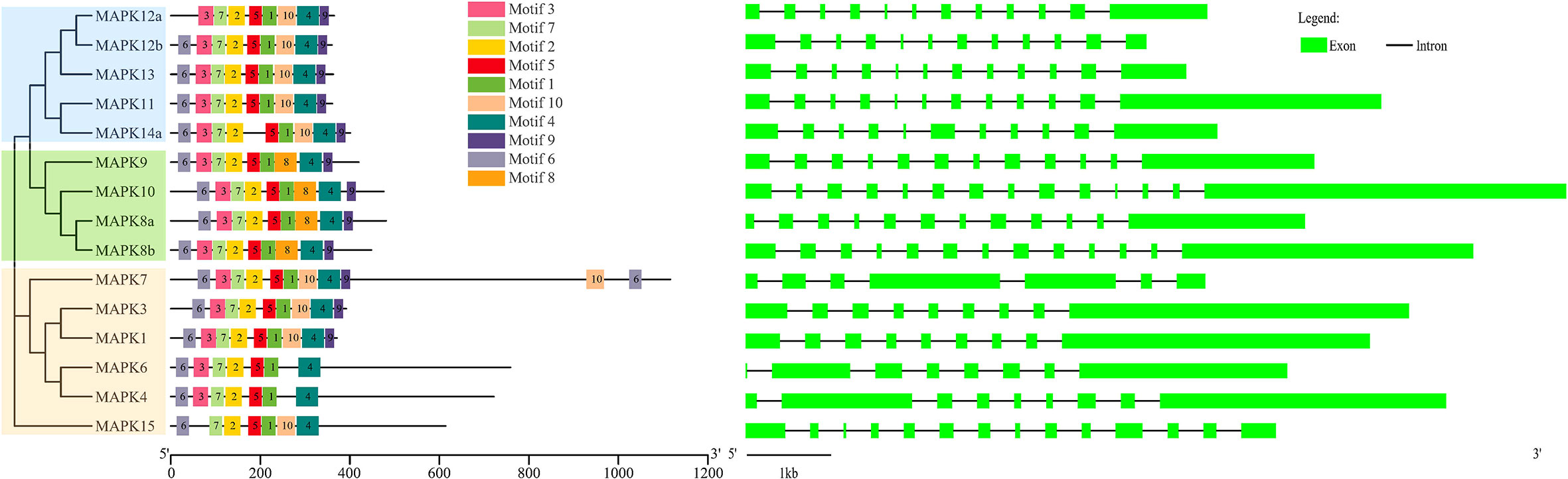
Figure 3 The combination of phylogenetic relationship, conserved motif, and exon–intron structure. The left of the graph shows the phylogenetic relationships. The middle of the graph shows the distribution of conserved motifs, and each colored rectangular box represents a motif. The right of the graph shows the exon–intron structure, and the lengths of introns in each SmMAPK are displayed in the same length.
The expression patterns of all SmMAPK members in 12 different tissues are shown in Figure 4. The expression data of SmMAPKs are provided in Supplementary Table 6. On the whole, MAPKs were widely expressed in all 12 tissues and, meanwhile, differentially expressed in some tissues. In detail, MAPKs showed the highest global expression levels in the brain, relatively higher global expression levels in the thymus and kidney, and the lowest global expression levels in the liver and muscle. Moreover, the global expression levels of SmMAPK3, 6, and 14a were higher than other members in almost all tissues. In addition, some genes presented obviously tissue-specific expression patterns; for instance, the expression of SmMAPK10, 4, and 8a was almost only detected in the brain. SmMAPK7, 15, 12a, and 12b showed higher expression levels than other members in the testis, ovary, thymus, and kidney, respectively.
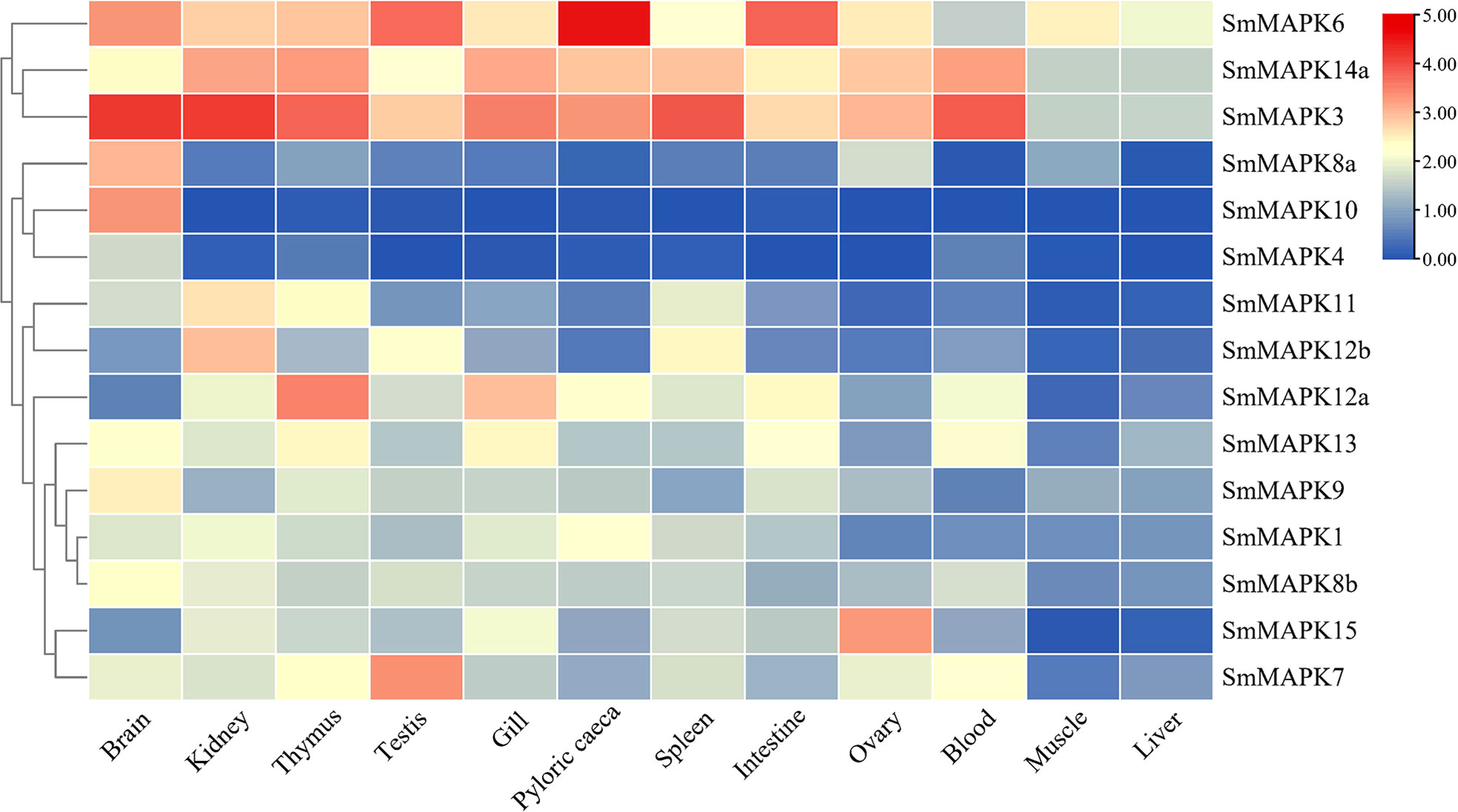
Figure 4 Heat map of mitogen-activated protein kinase (MAPK) expression in 12 different tissues in S. maximus. Each cell with a different color corresponds to a different expression level, which was normalized into log2 (RPKM + 1).
The expression patterns of SmMAPKs in the kidney and liver under heat stress were detected based on RNA-seq data sets related to heat stress (Figure 5A). The expression data are provided in Supplementary Table 7. On the whole, nearly all MAPKs exhibited obviously higher expression levels in the kidney than in the liver in both the control and heat treatment groups except for SmMAPK4, 10, and 8a. In the head kidney, five MAPK genes showed significant differential expression, among which SmMAPK6, 12b, and 15 showed extremely significant differential expression (Figure 5A). In detail, SmMAPK6 was first downregulated continuously at 23°C and 25°C and then extremely significantly upregulated at 28°C compared to 25°C. SmMAPK12b was extremely significantly downregulated at 28°C compared to 23°C. SmMAPK15 was significantly upregulated at 25°C and then extremely significantly 28°C compared to 25°C. In the liver, only SmMAPK13 showed extremely significant differential expression (Figure 5A). SmMAPK13 was significantly downregulated under all heat treatments, and extremely significantly downregulated in the groups with 20°C vs. 14°C and 28°C vs. 14°C. To sum up, SmMAPK6 and SmMAPK15 (both in the head kidney) in the ERK subfamily and SmMAPK12b (in the head kidney) and SmMAPK13 (in the liver) in the p38 subfamily were extremely sensitive in response to heat stress in turbot.
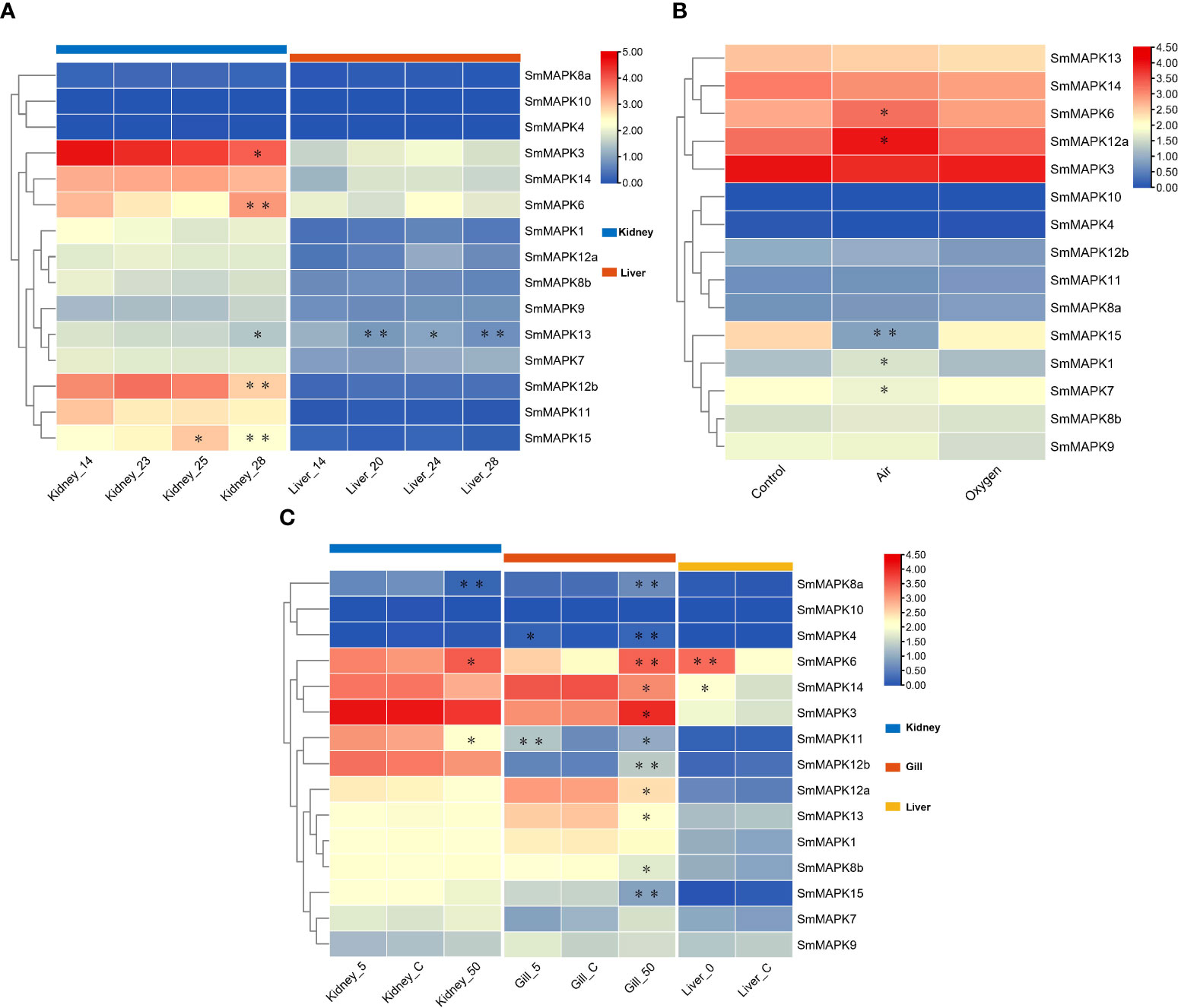
Figure 5 Heat map of mitogen-activated protein kinase (MAPK) expression in Scophthalmus maximus under three abiotic stresses. (A) Expression of MAPKs in kidney and liver tissues under heat stress: 14 °C: control group; 20 °C, 23 °C, 24 °C, 25 °C, and 28 °C: heat stress group. (B) Expression of MAPKs in gill tissues under oxygen stress: Control: 6.5 mg/L of O2, water; Air: 300 mg/L of O2, waterless; and oxygen: 1,430 mg/L of O2, waterless. (C) Expression of MAPKs in kidney, gill, and liver tissues under salinity stress: Control: 30‰ salinity (natural seawater); 50: 50‰ salinity; 5: 5‰ salinity; and 0: fresh water. Each cell with a different color corresponds to a different expression level, which was normalized into log2 (FPKM + 1). *Significantly differentially expressed MAPKs with P < 0.05. **Extremely significant differential expression of MAPKs with false discovery rate (FDR) < 0.01 and |log2 fold change (FC)| > 1.
The expression patterns of SmMAPKs in the kidney and liver under heat stress were detected based on RNA-seq data sets related to heat stress (Figure 5A). The expression data are provided in Supplementary Table 7. On the whole, nearly all MAPKs exhibited obviously higher expression levels in the kidney than in the liver in both the control and heat treatment groups except for SmMAPK4, 10, and 8a. In the head kidney, five MAPK genes showed significant differential expression, among which SmMAPK6, 12b, and 15 showed extremely significant differential expression (Figure 5A). In detail, SmMAPK6 was first downregulated continuously at 23°C and 25°C and then extremely significantly upregulated at 28°C compared to 25°C. SmMAPK12b was extremely significantly downregulated at 28°C compared to downregulation at 23°C. SmMAPK15 was significantly upregulated at 25°C and then extremely significantly 28°C compared to 25°C. In the liver, only SmMAPK13 showed extremely significant differential expression (Figure 5A). SmMAPK13 was significantly downregulated under all heat treatments, and extremely significantly downregulated in the groups with 20°C vs. 14°C and 28°C vs. 14°C. To sum up, SmMAPK6 and SmMAPK15 (both in the head kidney) in the ERK subfamily and SmMAPK12b (in the head kidney) and SmMAPK13 (in the liver) in the p38 subfamily were extremely sensitive in response to heat stress in turbot.
To illustrate the expression patterns of MAPKs in turbot under oxygen stress, one RNA-seq data set of gill tissues under different oxygen conditions was used. The expression data are provided in Supplementary Table 8. As shown in Figure 5B, all MAPKs are shown obviously upregulated or downregulated under different oxygen conditions except for SmMAPK4 and 10. In detail, five genes showed significant differential expression, of which SmMAPK15 showed extremely significant differential expression (Figure 5B). First, SmMAPK15 was extremely significantly downregulated in the air vs. control groups, and then significantly upregulated in the oxygen vs. air groups. In conclusion, MAPKs in the air group showed more significant differential expression than in the oxygen group when compared to the control group, among which SmMAPK15 was extremely susceptible to air exposure stress.
The expression patterns of SmMAPKs under salinity stress in the head kidney, gill, and liver of turbot were illuminated using low- and high-salinity-related RNA-seq data sets (Figure 5C). The expression data are provided in Supplementary Table 9. On the whole, all SmMAPKs exhibited obviously distinct expression levels in the kidney, gill, and liver except for SmMAPK10 that had the highest holistic expression levels in the kidney, relatively higher in the gill, and lowest in the liver. For low-salinity stress, no MAPK genes showed significant differential expression in the head kidney. In the gill, two genes were significantly upregulated, of which SmMAPK11 was extremely significantly upregulated (Figure 5C). In the liver, two MAPK genes were significantly upregulated, among which SmMAPK6 was extremely significantly upregulated (Figure 5C). Under high-salinity stress, 3 and 11 MAPK genes showed significant differential expression in the kidney and gill, respectively, of which one and five genes showed extremely significant differential expression (Figure 5C). In the head kidney, SmMAPK8a was extremely significantly downregulated and upregulated, respectively. In the gill, SmMAPK4, 6, 8a, 12b, and 15 were extremely significantly upregulated. SmMAPK11 and SmMAPK12b (both in the gill) in the p38 MAPK subfamily; SmMAPK6 (in the gill and liver) and SmMAPK15 and SmMAPK4 (both in the gill) in the ERK subfamily; and SmMAPK8a (in the head kidney and gill) in the JNK subfamily were extremely impressible in response to salinity stress.
Using pathogen (parasite, bacteria, and virus) challenged RNA-seq data sets, the involvement of SmMAPKs in different tissues under distinct pathogen infections is elucidated in Figure 6. The expression data are provided in Supplementary Table 10. Under E. scophthalmi infection, expression patterns of SmMAPKs in five different tissues were elaborated. In detail, in the head kidney, three MAPK genes were significantly influenced by E. scophthalmi infection, among which SmMAPK6 was extremely significantly upregulated at 42 dpi (Figure 6). In the pyloric caeca, two MAPK genes showed significant differential expression, of which SmMAPK14a was extremely significantly downregulated at 42 dpi (Figure 6). In the spleen, five MAPK genes in response to E. scophthalmi infection were detected, among which SmMAPK4 and 10 were extremely significantly upregulated and downregulated at 24 dpi, respectively (Figure 6). In the thymus, three significantly DE MAPK genes were determined (Figure 6). In the blood, eight significantly DE MAPK genes were only detected in the moderate and severe infection groups, of which four extremely significant DE MAPK genes were detected in the severe infection group (Figure 6). Specifically, SmMAPK12a, 12b, and 11 were extremely significantly upregulated in severe infection. SmMAPK15 was extremely significantly downregulated in severe infection. Collectively, SmMAPK11, 12a, 12b, and 14a in the p38 MAPK subfamily; SmMAPK4, 6, and 15 in the ERK subfamily; and SmMAPK10 in the JNK subfamily were extremely prone to E. scophthalmi infection.
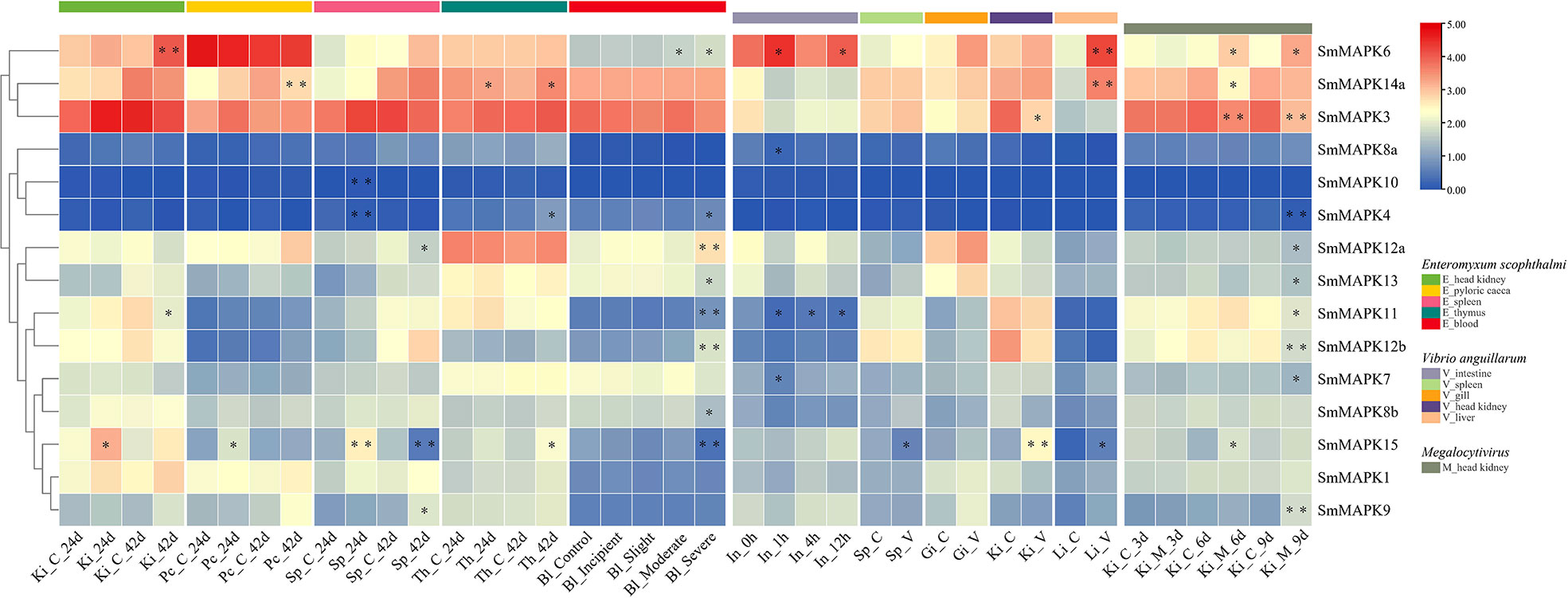
Figure 6 Heat map of mitogen-activated protein kinases (MAPK) expression in Scophthalmus maximus under pathogen (Enteromyxum scophthalmi, Vibrio anguillarum, and Megalocytivirus) infection. Ki, Pc, Sp, Th, Bl, In, Gi, and Li represent the head kidney, pyloric caeca, spleen, thymus, blood, intestine, gill, and liver, respectively. C represents the control group, V represents the V. anguillarum infection, and M represents the Megalocytivirus infection. Each cell with a different color corresponds to a different expression level, which was normalized into log2 (FPKM + 1). *Significantly differentially expressed of MAPK genes with P < 0.05. **Extremely significant differential expression of MAPK genes with false discovery rate (FDR) < 0.01 and |log2 fold change (FC)| > 1.
Using bacteria (V. anguillarum) infected RNA-seq data sets, the involvement of SmMAPKs in five distinct tissues was expounded. In the intestine, spleen, and gill, there were four, one, and zero significantly affected MAPK genes detected (Figure 6), respectively, whereas no extremely significant DE MAPK genes were found. In the head kidney, two genes showed significant differential expression, of which SmMAPK15 showed extremely significant differential expression (Figure 6). In the liver, three MAPK genes that were significantly upregulated were identified among which SmMAPK6 and 14a were extremely significantly upregulated (Figure 6). Given the above, SmMAPK14a in the p38 MAPK subfamily and SmMAPK6 and 15 in the ERK subfamily showed an extremely significant response to V. anguillarum infection.
In addition, to investigate the expression patterns of SmMAPKs under virus infection, an RNA-seq data set of the head kidney under Megalocytivirus infection was used. Based on the results, nine MAPK genes showed significant differential expression and four MAPK genes showed extremely significant differential expression (Figure 6). In detail, SmMAPK4 and 12b were extremely significantly downregulated at 9 dpi. SmMAPK3 was extremely significantly downregulated at 6 and 9 dpi. SmMAPK9 was extremely significantly upregulated at 9 dpi. SmMAPK9 in the JNK subfamily, SmMAPK3 and 4 in the ERK subfamily, and SmMAPK12b in the p38 MAPK subfamily were extremely significantly affected by Megalocytivirus challenge.
MAPKs, a series of mitogen-activated protein kinases lying within the MAPK cascade, are unique to eukaryotes, which play key roles in transducing upstream signals to the target proteins, and thus, induce various cellular responses (Teramoto and Gutkind, 2013). Recently, an increasing number of studies have shown that MAPKs participate in various biological processes in teleost, including embryogenesis and cell death as well as response to environmental stresses (toxicant, heat, hypoxia, oxidative, salinity, and pathogen infection) (Krens et al., 2006a; Urushibara et al., 2009; Shi and Zhou, 2010; Tian et al., 2019; Wang et al., 2021). However, systematic identification and functional investigation of MAPKs have not been reported in turbot up to now. In the present study, we first identified and characterized SmMAPKs at a genome-wide level systematically and then we performed phylogenetic analysis and gene structure and conserved motif analysis. Finally, we investigated their expression patterns in diverse tissues and divergent functions on abiotic and biotic stress responses based on multiple RNA-seq data sets.
MAPKs are widespread in all eukaryotes, whereas the numbers of MAPKs vary greatly among species. In the plant genome, different numbers of MAPKs have been found, such as 10 in Ziziphus jujuba Mill. (Liu et al., 2017), 21 in Manihot esculenta Crantz (Yan et al., 2016), 32 in Glycine max (McNeece et al., 2019), and 54 in Triticum aestivum L. (Zhan et al., 2017), whereas only 13 MAPKs were detected in mammal species, such as mouse and human (Widmann et al., 1999). In the present study, a total of 15 MAPKs were identified in the S. maximus genome, which is similar to the numbers of identified MAPKs in other teleost species, such as P. olivaceus (14) (Qiao et al., 2021) and L. maculatus (14) (Tian et al., 2019). Previous studies also showed that the total numbers of MAPKs ranged from 13 to 16 in teleost, which might be a reflection of evolutionary conservation of MAPKs in teleost. In addition, the gene copy number of MAPKs also showed variation among teleost based on the phylogenetic analysis of four representative teleost species. For example, no MAPK members of L. oculatus were included in the MAPK11 and MAPK3 clades. Moreover, D. rerio and O. latipes included two members in the clade of MAPK14, whereas L. oculatus and S. maximus only had one member. A varied gene copy number might be the result of gene gain or loss in the process of evolution. Furthermore, except for MAPK8, 12, 14 containing two copies, most MAPKs in the species of this study had only a single copy. Although members in each subfamily varied in distinct teleost, they were more similar to each other than to other subfamilies, indicating that MAPKs were highly conserved during evolutionary history (Qiao et al., 2021).
Gene structure can also provide evidence for assessing evolutionary relationships within the gene family (Wang et al., 2015). In the present study, gene structures and conserved motifs of SmMAPKs were in keeping with the subfamily classification. In detail, the number of exons in the ERK subfamily, with an average of nine, was fewer than the JNK (with 13) and p38 subfamilies (with 12), and most of the members within the same subfamily shared similar numbers of exons and introns. Furthermore, motif analysis showed that all members in the JNK subfamily contained motif 8, whereas motif 10 could only be observed in almost all members in the p38 and ERK subfamilies. In addition, multiple sequence alignment indicated that the same subfamily contained the same TXY (Thr-X-Tyr) dual-phosphorylation site motif except for ERK3 and ERK4 that contained an alternative dual-phosphorylation motif SEG; even so, the SEG motif still contains an intermediate glutamic acid (E) residue, which is in accord with zebrafish and other vertebrates (Krens et al., 2006a; Krens et al., 2006b). SmMAPKs showed similar exon–intron distribution patterns, conserved motif constitution, and the same dual-phosphorylation site motif within members in the same subfamilies, demonstrating that they were highly evolutionarily conserved in gene structure and might represent highly similar features and functions. This might be the consequence of gene duplication and expansion and also provides dependable support for the identification and classification of MAPKs.
According to the expression patterns of SmMAPKs in different tissues, we concluded that SmMAPKs were expressed in a tissue-specific manner. For example, SmMAPK4, 8a, and 10 are almost only expressed in the brain, and global expression levels were lower in the liver and muscle than in other tissues. Studies in Japanese flounder also demonstrate that MAPKs showed obviously tissue-specific expression patterns; meanwhile, hardly any MAPKs were expressed in liver tissues, which is in accord with our results (Qiao et al., 2021). In addition, nearly all MAPKs in both control and treatment groups under all stresses exhibited the highest global expression levels in the head kidney. As is well known, the head kidney is an essential organ which comprises cortisol-producing inter-renal cells (Best and Gilmour, 2022), whereas cortisol is the primary glucocorticoid released when teleosts are exposed to stress and it plays key roles in the stress response of teleosts (Bordin and Freire, 2021). Hence, we hold the opinion that the head kidney should be selected as the main tissue for stress studies in teleosts.
In recent years, an increasing number of studies have demonstrated that MAPKs have enormous effects on the response of teleosts to cope with abiotic stresses (Marshall et al., 2005; Feidantsis et al., 2012; Tian et al., 2019; Wen et al., 2019; Qiao et al., 2021). In our study, SmMAPK6 (ERK3), 15 (ERK7/8), 12b (p38γ), and 13 (p38δ) were extremely and significantly affected by heat stress. A similar result was also found that the activation of p38 MAPK was indispensable for adaptation to thermal stress in the red blood cells of S. aurata through the induction of Hsp70 (Feidantsis et al., 2012). Moreover, heat stress increased the expression of p38 MAPK in Argyrosomus regius (Antonopoulou et al., 2020). Furthermore, it was reported that gene CGA1 was implicated in the regulation of resistance to heat or salinity stress via HSP70A and MAPK6 in Chlamydomonas reinhardtii (Lee et al., 2017). SmMAPK15 was also extremely and significantly influenced by air exposure in our study. A previous study in humans indicated that MAPK15 could protect from oxidative stress-dependent cellular senescence by inducing the mitophagic process (Franci et al., 2022). In addition, six DE SmMAPKs in response to salinity stress were identified, namely, SmMAPK4, 6, 8a, 11, 12b, and 15. Among them, MAPK8 (JNKl) and 11 (p38β) were demonstrated to be significantly sensitive to salinity challenge (Tian et al., 2019). Furthermore, transcriptome analysis showed that MAPK6 (ERK3) played a crucial role in salt acclimation in Arabidopsis (Shen et al., 2014). Moreover, MAPK4 (ERK4) overexpressed transgenic soybean and showed significantly increased tolerance to salt stress. In addition, phosphorylated JNK and p38 obviously increased under hypertonic shock and hypotonic stress in Fundulus heteroclitus (Marshall et al., 2005). The activity of the three subgroups of MAPK (ERK1, JNK1, and p38) significantly decreased and increased in response to high- and low-salinity stress in gill epithelium cells of F. heteroclitus, respectively, indicating that MAPKs were key components of salinity adaptation and involved in osmosensory signaling pathways in euryhaline fish (Kultz and Avila, 2001). Combining our results with other available evidence, we detected seven candidate MAPK genes (SmMAPK4, 6, 8a, 11, 12b, 13, and 15) responsive to abiotic stress.
Herein, functions of MAPKs in response to biotic stress were also investigated by examining the expression patterns of SmMAPKs under diverse pathogen infections. Based on the expression analysis, we concluded that SmMAPKs could show apparent differentially expressed patterns in distinct tissues under the same pathogen infection, and the expression would vary in a time-dependent manner. For instance, under E. scophthalmi infection, SmMAPK4 and 10 were detected differentially expressed at 24 dpi in the spleen, and DE SmMAPK15 were detected at 24 and 42 dpi in the spleen, whereas DE SmMAPK6 in the head kidney and SmMAPK14a in the pyloric caeca were only detected at 42 dpi, and no DE SmMAPKs were identified in the thymus. In addition, four DE SmMAPKs, namely, SmMAPK11, 12a, 12b, and 15, were identified in severely infected blood. Furthermore, following V. anguillarum challenge, DE SmMAPKs were only found in the liver and head kidney. Postinoculation with Megalocytivirus, 0, 1, and 4 DE SmMAPKs were identified at 3, 6, and 9 dpi, respectively. Moreover, among all of the above DE SmMAPKs, SmMAPK6, 4, 12b, 14a, and 15 were extremely significant differentially expressed under at least two pathogens’ infection, manifesting their important functions in preventing invasions of different pathogens.
Currently, many previous studies have demonstrated the functions of the above DE MAPK genes identified under diverse biotic stresses. For instance, a study in Takifugu rubripies showed that MAPK6 (ERK6) was located at the center of the regulatory networks conferred by interferon gamma, which was critical for innate and adaptive immunity in fish (Kong et al., 2018). MAPK6 (ERK6), 11 (p38β), and 14a (p38α) were reported as significantly influenced following Edwardsiella tarda challenge in Japanese flounder (Qiao et al., 2021). It was also proved that the mRNA levels of MAPK11 (p38β) and 14 (p38α) in the head kidney of Oplegnathus fasciatus were all upregulated under the PAMPs challenge (Umasuthan et al., 2015). Moreover, infection with Vibrio fortis led to significant upregulation of MAPK9 (JNK2) and 10 (JNK3) in different tissues of the lined seahorse (Wang et al., 2021). Furthermore, MAPK4-silenced plants of Glycine max were more resistant to Soybean mosaic virus and downy mildew in contrast to vector control plants, revealing that MAPK4 (ERK4) negatively regulated defense response (Liu et al., 2011). For the intestine transcriptomic profiling of the grass carp treated with Bacillus subtilis H2, MAPK12b (p38γ) was significantly upregulated, which showed the effect of MAPK12b in response to B. subtilis infection. Based on the results in our study coupled with findings of the abovementioned studies, we concluded that SmMAPK3, 4, 6, 9, 10, 11, 12a, 12b, 14a, and 15 might play crucial roles in responding to pathogen infection.
It is worth noting that SmMAPK11 and 12b in the p38 MAPK subfamily and SmMAPK4, 6, and 15 in the ERK subfamily showed extremely significant differential expression under both biotic and abiotic stresses. Among them, SmMAPK15 (ERK7/8) showed differential expression under all stresses (heat, oxygen, salinity, and parasitic and bacterial infections) except for viral infection. The above results demonstrate that these MAPK genes, especially SmMAPK15 (ERK7/8), might play crucial antistress roles in response to both biotic and abiotic challenges. MAPK15, originally also known as ERK7/8, was the atypical MAPK identified recently and the least studied to date (Lau and Xu, 2019). Recent studies show that MAPK15 participated in multifarious cellular activities, such as regulating cell proliferation and division, promoting cell transformation and apoptosis, maintaining genome stability, responding to heat and oxygen stresses, and so on (Colecchia et al., 2012; Yang et al., 2013; Kazatskaya et al., 2017; Lau and Xu, 2019). In addition, using Entamoeba invadens, a reptilian parasite as a model organism, a stress-responsive MAPK15 that was upregulated under different stress conditions was identified (Singh et al., 2018), whereas functions of MAPK15 in response to various stresses acting on teleosts deserve more experiments and validations.
In this study, we identified MAPKs in turbot at a genome-wide level and investigated their functions in response to abiotic and biotic stresses. A total of 15 MAPK gene family members were identified in turbot and were divided into three subfamilies. Furthermore, phylogenetic relationship, motif compositions, and gene structure analysis illustrated that the MAPKs were highly evolutionarily conservative across various teleost species. Based on the expression profiles, 7 and 10 candidate stress-responsive MAPK genes were identified under abiotic (heat, oxygen, and salinity) and biotic (parasitic (E. scophthalmi), bacterial (V. anguillarum), and viral (Megalocytivirus) infections) stresses. It is particularly necessary to point out that SmMAPK11 and 12b in the p38 MAPK subfamily and SmMAPK4, 6, and 15 in the ERK subfamily were common MAPK genes in response to both biotic and abiotic stresses, of which MAPK15 (ERK7/8) was in response to all stresses except for Megalocytivirus infection. This study provides the first systematic analysis of MAPKs in turbot, which not only improves our understanding of the functions of MAPKs in response to abiotic and biotic stresses in turbot, but also lays the foundation for research on the molecular adaptation mechanisms of turbot under environmental stress.
The data sets analyzed for this study can be found in NCBI: https://www.ncbi.nlm.nih.gov/. The accession numbers are SRP152627, SRP273870, SRP167318, SRP277001, SRP238143, SRP153594, SRP308109, SRP255305, SRP065375, SRP050607, SRP191266, SRP336094, SRP335896, SRP320422, SRP319434, SRP347383, SRP136753, SRP287484, SRX3568427, SRX3568428, SRX3568438, SRX5528155, SRX5528156, SRX5528159, SRX3568429, SRX3568430, SRX3568437, SRX1798798, SRX1798799, SRX1798800, SRX1798792, SRX1798793, and SRX1798794.
SC, Y-JL, and X-WX applied, designed, and supervised the project. WZ, X-wX, and ZE conducted the experiments and analyzed the data. WZ, X-wX, ZE, SC, and YL wrote and revised the manuscript. All authors read and approved the final manuscript.
This work was funded by the Key Research and Development Project of Shandong Province (2021LZGC028), the Central Public-interest Scientific Institute Basal Research Fund, CAFS (No. 2020TD19), and the Taishan Scholar Project of Shandong Province.
We thank the contributors for all the above published RNA-seq data sets.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.1005401/full#supplementary-material
Antonopoulou E., Chatzigiannidou I., Feidantsis K., Kounna C., Chatzifotis S. (2020). Effect of water temperature on cellular stress responses in meagre (Argyrosomus regius). Fish. Physiol. Biochem. 46 (3), 1075–1091. doi: 10.1007/s10695-020-00773-0
Antonopoulou E., Kentepozidou E., Feidantsis K., Roufidou C., Despoti S., Chatzifotis S. (2013). Starvation and re-feeding affect hsp expression, MAPK activation and antioxidant enzymes activity of European Sea bass (Dicentrarchus labrax). Comp. Biochem. Physiol. a-Molecular Integr. Physiol. 165 (1), 79–88. doi: 10.1016/j.cbpa.2013.02.019
Bailey T. L., Johnson J., Grant C. E., Noble W. S. (2015). The MEME suite. Nucleic Acids Res. 43 (W1), W39–W49. doi: 10.1093/nar/gkv416
Best C., Gilmour K. M. (2022). Regulation of cortisol production during chronic social stress in rainbow trout. Gen. Comp. Endocrinol. 325, 114056. doi: 10.1016/j.ygcen.2022.114056
Bordin D., Freire C. A. (2021). Remarkable variability in stress responses among subtropical coastal marine teleosts. Mar. Biol. 168 (8), 122. doi: 10.1007/s00227-021-03929-5
Chang L., Karin M. (2001). Mammalian MAP kinase signalling cascades. Nature 410 (6824), 37–40. doi: 10.1038/35065000
Chen C., Chen H., Zhang Y., Thomas H. R., Frank M. H., He Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13 (8), 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen Z., Gibson T. B., Robinson F., Silvestro L., Pearson G., Xu B.-e., et al. (2001). MAP kinases. Chem. Rev. 101 (8), 2449–2476. doi: 10.1021/cr000241p
Colcombet J., Hirt H. (2008). Arabidopsis MAPKs: A complex signalling network involved in multiple biological processes. Biochem. J. 413 (2), 217–226. doi: 10.1042/BJ20080625
Colecchia D., Strambi A., Sanzone S., Iavarone C., Rossi M., Dall'Armi C., et al. (2012). MAPK15/ERK8 stimulates autophagy by interacting with LC3 and GABARAP proteins. Autophagy 8 (12), 1724–1740. doi: 10.4161/auto.21857
Cowan K. J., Storey K. B. (2003). Mitogen-activated protein kinases: new signaling pathways functioning in cellular responses to environmental stress. J. Exp. Biol. 206 (7), 1107–1115. doi: 10.1242/jeb.00220
Cui W., Ma A., Huang Z., Wang X.a., Sun Z., Liu Z., et al. (2019). Transcriptomic analysis reveals putative osmoregulation mechanisms in the kidney of euryhaline turbot Scophthalmus maximus responded to hypo-saline seawater. J. Oceanology Limnology 38 (2), 467–479. doi: 10.1007/s00343-019-9056-2
Cui W., Ma A., Wang X. (2020). Response of the PI3K-AKT signalling pathway to low salinity and the effect of its inhibition mediated by wortmannin on ion channels in turbot Scophthalmus maximus. Aquacult. Res. 51 (7), 2676–2686. doi: 10.1111/are.14607
Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29 (1), 15–21. doi: 10.1093/bioinformatics/bts635
Feidantsis K., Poertner H. O., Markou T., Lazou A., Michaelidis B. (2012). Involvement of p38 MAPK in the induction of Hsp70 during acute thermal stress in red blood cells of the gilthead sea bream, Sparus aurata. J. Exp. Zoology Part a-Ecological Integr. Physiol. 317A (5), 303–310. doi: 10.1002/jez.1725
Franci L., Tubita A., Bertolino F. M., Palma A., Cannino G., Settembre C., et al. (2022). MAPK15 protects from oxidative stress-dependent cellular senescence by inducing the mitophagic process. Aging Cell. 21 (7), e13620 doi: 10.1111/acel.13620
Fujii R., Yamashita S., Hibi M., Hirano T. (2000). Asymmetric p38 activation in zebrafish: Its possible role in symmetric and synchronous cleavage. J. Cell Biol. 150 (6), 1335–1347. doi: 10.1083/jcb.150.6.1335
Gao C., Cai X., Cao M., Fu Q., Yang N., Liu X., et al. (2021). Comparative analysis of the miRNA-mRNA regulation networks in turbot (Scophthalmus maximus l.) following Vibrio anguillarum infection. Dev. Comp. Immunol. 124, 104164 doi: 10.1016/j.dci.2021.104164
Gertz E. M., Yu Y.-K., Agarwala R., Schaffer A. A., Altschul S. F. (2006). Composition-based statistics and translated nucleotide searches: Improving the TBLASTN module of BLAST. BMC Biol. 4, 41. doi: 10.1186/1741-7007-4-41
He Z., Zhang H., Gao S., Lercher M. J., Chen W.-H., Hu S. (2016). Evolview v2: An online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res.W236-W241 44, W236–W241. (W1). doi: 10.1093/nar/gkw370
Huang Z., Ma A., Yang S., Liu X., Zhao T., Zhang J., et al. (2020). Transcriptome analysis and weighted gene co-expression network reveals potential genes responses to heat stress in turbot Scophthalmus maximus. Comp. Biochem. Physiol. Part D Genomics Proteomics 33, 100632. doi: 10.1016/j.cbd.2019.100632
Kazatskaya A., Kuhns S., Lambacher N. J., Kennedy J. E., Brear A. G., McManus G. J., et al. (2017). Primary cilium formation and ciliary protein trafficking is regulated by the atypical MAP kinase MAPK15 in Caenorhabditis elegans and human cells. Genetics 207 (4), 1423–1440. doi: 10.1534/genetics.117.300383
Keshet Y., Seger R.J.M.M.B. (2010). The MAP kinase signaling cascades: A system of hundreds of components regulates a diverse array of physiological functions. Methods Mol. Biol. 661, 3–38. doi: 10.1007/978-1-60761-795-2_1
Kong D., Cui J., Wang Z., Zang L., Sun H., Hu Z., et al. (2018). The regulatory networks conferred by IFN-gamma in the kidney of Takifugu rubripes. Int. J. Agric. Biol. 20 (10), 2189–2195. doi: 10.17957/ijab/15.0758
Krens S. F. G., He S., Spaink H. P., Snaar-Jagalska B. E. (2006a). Characterization and expression patterns of the MAPK family in zebrafish. Gene Expression Patterns 6 (8), 1019–1026. doi: 10.1016/j.modgep.2006.04.008
Krens S. F. G., Spaink H. P., Snaar-Jagalska B. E. (2006b). Functions of the MAPK family in vertebrate-development. FEBS Lett. 580 (21), 4984–4990. doi: 10.1016/j.febslet.2006.08.025
Kultz D., Avila K. (2001). Mitogen-activated protein kinases are in vivo transducers of osmosensory signals in fish gill cells. Comp. Biochem. Physiol. B-Biochemi. Mol. Biol. 129 (4), 821–829. doi: 10.1016/s1096-4959(01)00395-5
Kyriakis J. M., Avruch J. (2012). Mammalian MAPK signal transduction pathways activated by stress and inflammation: A 10-year update. Physiol. Rev. 92 (2), 689–737. doi: 10.1152/physrev.00028.2011
Kyriakis J. M., Banerjee P., Nikolakaki E., Dai T., Rubie E. A., Ahmad M. F., et al. (1994). The stress-activated protein kinase subfamily of c-jun kinases. Nature 369 (6476), 156–160. doi: 10.1038/369156a0
Lau A. T. Y., Xu Y.-M. (2019). Regulation of human mitogen-activated protein kinase 15 (extracellular signal-regulated kinase 7/8) and its functions: A recent update. J. Cell. Physiol. 234 (1), 75–88. doi: 10.1002/jcp.27053
Lee C. S., Ahn J. W., Choi Y. E. (2017). The G-protein alpha-subunit gene CGA1 is involved in regulation of resistance to heat and osmotic stress in Chlamydomonas reinhardtii. Cell. Mol. Biol. 63 (2), 29–39. doi: 10.14715/cmb/2017.63.2.5
Lei J. L., Liu X. F., Guan C. T. (2012). Turbot culture in China for two decades: achievements and prospect. Prog. Fishery Sci. 33 (4), 123–130. doi: 10.1007/s11783-011-0280-z
Liao Y., Smyth G. K., Shi W. (2013). The subread aligner: Fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 41 (10):e108. doi: 10.1093/nar/gkt214
Liao Y., Smyth G. K., Shi W. (2014). FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30 (7), 923–930. doi: 10.1093/bioinformatics/btt656
Liu J.-Z., Horstman H. D., Braun E., Graham M. A., Zhang C., Navarre D., et al. (2011). Soybean homologs of MPK4 negatively regulate defense responses and positively regulate growth and development. Plant Physiol. 157 (3), 1363–1378. doi: 10.1104/pp.111.185686
Liu Z., Ma A., Yuan C., Zhao T., Chang H., Zhang J. (2021). Transcriptome analysis of liver lipid metabolism disorders of the turbot Scophthalmus maximus in response to low salinity stress. Aquaculture 534, 736273. doi: 10.1016/j.aquaculture.2020.736273
Liu Q., Zhang G. Y., Shinozaki K. (2000). The plant mitogen-activated protein (MAP) kinase. Acta Botanica Sin. 42 (7), 661–667. doi: 0577-7496(2000)07-0661-07
Liu Z., Zhang L., Xue C., Fang H., Zhao J., Liu M. (2017). Genome-wide identification and analysis of MAPK and MAPKK gene family in Chinese jujube (Ziziphus jujuba mill.). BMC Genomics 18, 855. doi: 10.1186/s12864-017-4259-4
L. J. G., Razvan L. (2002). Mitogen-activated protein kinase pathways medicated by ERK, JNK, and p38 protein kinases. Science 298, 1911–1912. doi: 10.1126/science.1072682
Marshall W. S., Ossum C. G., Hoffmann E. K. (2005). ). Hypotonic shock mediation by p38 MAPK, JNK, PKC, FAK, OSR1 and SPAK in osmosensing chloride secreting cells of killifish opercular epithelium. J. Exp. Biol. 208 (6), 1063–1077. doi: 10.1242/jeb.01491
McNeece B. T., Sharma K., Lawrence G. W., Lawrence K. S., Klink V. P. (2019). The mitogen activated protein kinase (MAPK) gene family functions as a cohort during the glycine max defense response to Heterodera glycines. Plant Physiol. Biochem. 137, 25–41. doi: 10.1016/j.plaphy.2019.01.018
Nie X., Zhang C., Jiang C., Li S., Hong W., Chen S., et al. (2020). Characterizing transcriptome changes in gill tissue of turbot (Scophthalmus maximus) for waterless preservation. Aquaculture 518, 734830. doi: 10.1016/j.aquaculture.2019.734830
Plotnikov A., Zehorai E., Procaccia S., Seger R. (2011). The MAPK cascades: Signaling components, nuclear roles and mechanisms of nuclear translocation. Biochim. Et Biophys. Acta-Molecular Cell Res. 1813 (9), 1619–1633. doi: 10.1016/j.bbamcr.2010.12.012
Potter S. C., Luciani A., Eddy S. R., Park Y., Lopez R., Finn R. D. (2018). HMMER web server: 2018 update. Nucleic Acids Res. 46 (W1), W200–W204. doi: 10.1093/nar/gky448
Qiao Y., Yan W., He J., Liu X., Zhang Q., Wang X. (2021). Identification, evolution and expression analyses of mapk gene family in Japanese flounder (Paralichthys olivaceus) provide insight into its divergent functions on biotic and abiotic stresses response. Aquat. Toxicol. 241, 106005. doi: 10.1016/j.aquatox.2021.106005
Raman M., Chen W., Cobb M. H. (2007). Differential regulation and properties of MAPKs. Oncogene 26 (22), 3100–3112. doi: 10.1038/sj.onc.1210392
Robinson M. D., McCarthy D. J., Smyth G. K. (2010). edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26 (1), 139–140. doi: 10.1093/bioinformatics/btp616
Robledo D., Ronza P., Harrison P. W., Losada A. P., Bermudez R., Pardo B. G., et al. (2014). RNA-Seq analysis reveals significant transcriptome changes in turbot (Scophthalmus maximus) suffering severe enteromyxosis. BMC Genomics 15, 1149. doi: 10.1186/1471-2164-15-1149
Ronza P., Alvarez-Dios J. A., Robledo D., Losada A. P., Romero R., Bermudez R., et al. (2021). Blood transcriptomics of turbot Scophthalmus maximus: A tool for health monitoring and disease studies. Anim. (Basel) 11 (5), 1296. doi: 10.3390/ani11051296
Ronza P., Robledo D., Bermudez R., Losada A. P., Pardo B. G., Sitja-Bobadilla A., et al. (2016). RNA-Seq analysis of early enteromyxosis in turbot (Scophthalmus maximus): New insights into parasite invasion and immune evasion strategies. Int. J. Parasitol. 46 (8), 507–517. doi: 10.1016/j.ijpara.2016.03.007
Ronza P., Robledo D., Losada A. P., Bermudez R., Pardo B. G., Martinez P., et al. (2020). The teleost thymus in health and disease: new insights from transcriptomic and histopathological analyses of turbot, Scophthalmus maximus. Biol. (Basel) 9 (8), 221. doi: 10.3390/biology9080221
Shen X., Wang Z., Song X., Xu J., Jiang C., Zhao Y., et al. (2014). Transcriptomic profiling revealed an important role of cell wall remodeling and ethylene signaling pathway during salt acclimation in Arabidopsis. Plant Mol. Biol. 86 (3), 303–317. doi: 10.1007/s11103-014-0230-9
Shi X., Zhou B. (2010). The role of Nrf2 and MAPK pathways in PFOS-induced oxidative stress in zebrafish embryos. Toxicological Sci. 115 (2), 391–400. doi: 10.1093/toxsci/kfq066
Singh T., Agarwal T., Ghosh S. K. (2018). Identification and functional analysis of a stress-responsive MAPK15 in Entamoeba invadens. Mol. Biochem. Parasitol. 222, 34–44. doi: 10.1016/j.molbiopara.2018.05.002
Song Y., Dong X., Hu G. (2022). Transcriptome analysis of turbot (Scophthalmus maximus) head kidney and liver reveals immune mechanism in response to Vibrio anguillarum infection. J. Fish. Dis. 45 (7), 1045–1057. doi: 10.1111/jfd.13628
Tamura K., Stecher G., Kumar S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38 (7), 3022–3027. doi: 10.1093/molbev/msab120
Teramoto H., Gutkind J. S. (2013). Mitogen-activated protein kinase family. Encyclopedia Biol. Chem. 30 (1), 176–180. doi: 10.1097/00003246-200201001-00010
Tian Y., Wen H., Qi X., Zhang X., Li Y. (2019). Identification of mapk gene family in Lateolabrax maculatus and their expression profiles in response to hypoxia and salinity challenges. Gene 684, 20–29. doi: 10.1016/j.gene.2018.10.033
Umasuthan N., Bathige S. D. N. K., Noh J. K., Lee J. (2015). Gene structure, molecular characterization and transcriptional expression of two p38 isoforms (MAPK11 and MAPK14) from rock bream (Oplegnathus fasciatus). Fish. Shellfish Immunol. 47 (1), 331–343. doi: 10.1016/j.fsi.2015.09.018
Urushibara N., Mitsuhashi S., Sasaki T., Kasai H., Yoshimizu M., Fujita H., et al. (2009). JNK and p38 MAPK are independently involved in tributyltin-mediated cell death in rainbow trout (Oncorhynchus mykiss) RTG-2 cells. Comp. Biochem. Physiol. C-Toxicol. Pharmacol. 149 (4), 468–475. doi: 10.1016/j.cbpc.2008.10.109
Wang J., Pan C., Wang Y., Ye L., Wu J., Chen L., et al. (2015). Genome-wide identification of MAPK, MAPKK, and MAPKKK gene families and transcriptional profiling analysis during development and stress response in cucumber. BMC Genomics 16 (1), 386. doi: 10.1186/s12864-015-1621-2
Wang K., Wang X., Zou Q., Jiang H., Zhang R., Tian Y., et al. (2021). Genome-wide evolution of MAPKs family and their expression in response to bacterial infection in seahorse Hippocampus erectus. J. Oceanology Limnology 39 (6), 2309–2321. doi: 10.1007/s00343-020-0332-y
Wen X., Zhang X., Hu Y., Xu J., Wang T., Yin S. (2019). iTRAQ-based quantitative proteomic analysis of takifugu fasciatus liver in response to low-temperature stress. J. Proteomics 201, 27–36. doi: 10.1016/j.jprot.2019.04.004
Widmann C., Gibson S., Jarpe M. B., Johnson G. L. (1999). Mitogen-activated protein kinase: Conservation of a three-kinase module from yeast to human. Physiol. Rev. 79 (1), 143–180. doi: 10.1152/physrev.1999.79.1.143
Xu X.-w., Zheng W., Meng Z., Xu W., Liu Y., Chen S. (2022). Identification of stress-related genes by co-expression network analysis based on the improved turbot genome. Sci. Data 9 (1), 374. doi: 10.1038/s41597-022-01458-4
Yang S.-W., Huang H., Gao C., Chen L., Qi S.-T., Lin F., et al. (2013). The distribution and possible role of ERK8 in mouse oocyte meiotic maturation and early embryo cleavage. Microscopy Microanalysis 19 (1), 190–200. doi: 10.1017/s1431927612013918
Yan Y., Wang L., Ding Z., Tie W., Ding X., Zeng C., et al. (2016). Genome-wide identification and expression analysis of the mitogen-activated protein kinase gene family in cassava. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01294
Zhang W., Liu H. T. (2002). MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 12, 9–18. doi: 10.1038/sj.cr.7290105
Zhang C.-N., Rahimnejad S., Lu K.-L., Zhou W.-H., Zhang J.-L. (2019). Molecular characterization of p38 MAPK from blunt snout bream (Megalobrama amblycephala) and its expression after ammonia stress, and lipopolysaccharide and bacterial challenge. Fish. Shellfish Immunol. 84, 848–856. doi: 10.1016/j.fsi.2018.10.074
Zhan H., Yue H., Zhao X., Wang M., Song W., Nie X. (2017). Genome-wide identification and analysis of MAPK and MAPKK gene families in bread wheat (Triticum aestivum l.). Genes 8 (10), 284. doi: 10.3390/genes8100284
Keywords: Scophthalmus maximus, MAPK, biotic stress, abiotic stress, stress resistance
Citation: Zheng W, Xu X-w, Zechen E, Liu Y and Chen S (2022) Genome-wide identification of the MAPK gene family in turbot and its involvement in abiotic and biotic stress responses. Front. Mar. Sci. 9:1005401. doi: 10.3389/fmars.2022.1005401
Received: 29 July 2022; Accepted: 25 August 2022;
Published: 27 September 2022.
Edited by:
Rui Jia, Freshwater Fisheries Research Center (CAFS), ChinaReviewed by:
Licao Cui, Jiangxi Agricultural University, ChinaCopyright © 2022 Zheng, Xu, Zechen, Liu and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yingjie Liu, bHlqQGNhZnMuYWMuY24=; Songlin Chen, Y2hlbnNsQHlzZnJpLmFjLmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.