- 1Cooperative Institute for Marine and Atmospheric Studies, University of Miami, Miami, FL, United States
- 2Ocean Chemistry and Ecosystems Division, NOAA’s Atlantic Oceanographic and Meteorological Laboratory, Miami, FL, United States
- 3Department of Oceanography and Coastal Sciences, College of the Coast & Environment, Louisiana State University, Baton Rouge, LA, United States
Stony coral tissue loss disease (SCTLD) was first observed in 2014 near Virginia Key in Miami-Dade County, Florida. Field sampling, lab experiments, and modeling approaches have suggested that reef sediments may play a role in SCTLD transmission, though a positive link has not been tested experimentally. We conducted an ex situ transmission assay using a statistically-independent disease apparatus to test whether reef sediments can transmit SCTLD in the absence of direct contact between diseased and healthy coral tissue. We evaluated two methods of sediment inoculation: batch inoculation of sediments collected from southeast Florida using whole colonies of diseased Montastraea cavernosa, and individual inoculations of sediments following independent, secondary infections of ∼5 cm2 coral fragments. Healthy fragments of the coral species Orbicella faveolata and M. cavernosa were exposed to these diseased sediment treatments, as well as direct disease contact and healthy sediment controls. SCTLD transmission was observed for both batch and individual diseased sediment inoculation treatments, albeit with lower proportions of infected individuals as compared to disease contact controls. The time to onset of lesions was significantly different between species and among disease treatments, with the most striking infections occurring in the individual diseased sediment treatment in under 24 h. Following infection, tissue samples were confirmed for the presence of SCTLD signs via histological examination, and sediment subsamples were analyzed for microbial community variation between treatments, identifying 16 SCTLD indicator taxa in sediments associated with corals experiencing tissue loss. This study demonstrated that reef sediments can indeed transmit SCTLD through indirect exposure between diseased and healthy corals, and adds credence to the assertion that SCTLD transmission occurs via an infectious agent or agents. This study emphasizes the critical need to understand the roles that sediment microbial communities and coastal development activities may have on the persistence of SCTLD throughout the endemic zone, especially in the context of management and conservation strategies in Florida and the wider Caribbean.
Introduction
First observed in 2014 off Virginia Key in Miami-Dade County, Florida, stony coral tissue loss disease (SCTLD) has become perhaps the most damaging described coral disease to date (Walton et al., 2018; Gintert et al., 2019; Spadafore et al., 2021). Since then, it has spread throughout the entirety of Florida’s Coral Reef, notably reaching the Dry Tortugas in 2021 (NOAA, 2018; Dobbelaere et al., 2020b; Muller et al., 2020; Roth et al., 2020; Sharp et al., 2020), and has now been observed in a growing number of Caribbean territories (Alvarez-Filip et al., 2019; Meiling et al., 2020; Roth et al., 2020; Dahlgren et al., 2021; Estrada-Saldívar et al., 2021; Heres et al., 2021). The rapid spread of SCTLD among coral colonies and regions was likely exacerbated by its highly-infectious nature, including broad susceptibility across an estimated 24 scleractinian coral species (NOAA, 2018) and rapid progression of lesions leading to colony mortality within days to weeks (NOAA, 2018; Aeby et al., 2019; Landsberg et al., 2020). The disease has also remained persistent across much of the endemic zone since the initial outbreak, with little evidence to support seasonal patterns in disease prevalence (Meiling et al., 2020; Muller et al., 2020).
As a result, SCTLD remains a substantial threat to the persistence of corals throughout Florida’s Coral Reef and the wider Caribbean. Despite a coordinated effort among researchers, resource managers, and animal husbandry experts, no pathogens have been identified to date, nor have the modes of transmission that have allowed the disease to persist or spread across oceanographically-isolated regions been comprehensively characterized. Evidence from histological examination suggests that the disease primarily affects the coral’s symbiotic algae of the family Symbiodiniaceae, as lesions tend to originate in the basal body wall gastrodermis and involve dysbiosis (Landsberg et al., 2020; Meiling et al., 2021). The reported successes of antibiotic application in slowing or halting lesion progression in infected corals (Neely et al., 2020; Shilling et al., 2021; Walker et al., 2021) suggests a bacterial pathogen, or bacterial component to a pathogenic consortium. Likewise, examination of microbial communities in SCTLD-affected corals and their surrounding environments (e.g., water and sediments) have identified indicator bacterial taxa that distinguish healthy and infected individuals, including some previously-described coral pathogens (Meyer et al., 2019; Rosales et al., 2020; Ushijima et al., 2020; Becker et al., 2021). A recent microscopy-based study also posited that viruses may play a role in the onset of the disease through infection of Symbiodiniaceae (Work et al., 2021), although this hypothesis has not been tested experimentally.
Etiological models and transmission experiments indicate that SCTLD is transmitted through water (Dobbelaere et al., 2020b; Muller et al., 2020; Sharp et al., 2020; Meiling et al., 2021), but knowledge of potential disease vectors and reservoirs is limited. Several hypotheses exist regarding modes of transmission, including local spread through non-coral intermediary species (Noonan and Childress, 2020), transport via neutrally-buoyant particles (Dobbelaere et al., 2020a,b), pathogen residence in surrounding sediments (Rosales et al., 2020), and long-range transfer to SCTLD-naïve regions through ballast water transfers (Dahlgren et al., 2021; Rosenau et al., 2021). Given the observed persistence of the SCTLD outbreak throughout the endemic zone and continued new observations among reefs in the wider Caribbean, controlled experimental studies are necessary to characterize the specific modes of transmission for SCTLD.
Mitigation of future disease spread is contingent on a thorough understanding of SCTLD dynamics, including modes of transmission and vectors/reservoirs. Nearshore reef environments in Florida are subjected to disturbance associated with coastal development and water pollution, such as dredging operations for port expansion and beach renourishment projects, stormwater runoff, and wastewater discharges (Finkl and Charlier, 2003; Lirman and Fong, 2007; Sinigalliano et al., 2019). Dredging operations in southeast Florida, such as the Port of Miami expansion that occurred in 2013–2015, can cause the release of sediments onto local coral reef habitats (Walker et al., 2012; Miller et al., 2016; Cunning et al., 2019), which can lead to smothering of colonies in extreme cases, as well as sedimentation-associated stress and mortality (Vargas-Ángel et al., 2006, 2007; Erftemeijer et al., 2012; Shore-Maggio et al., 2018). While it is difficult to disentangle coral mortality due to sedimentation, SCTLD, or a prior thermal stress event, several studies suggest that sedimentation played a role in sublethal and lethal effects on local coral populations due to the dredging of Port of Miami (Miller et al., 2016; Cunning et al., 2019; Gintert et al., 2019; Spadafore et al., 2021). Additionally, the plume was observed through remote sensing platforms over an estimated 228 km2 in southeast Florida (Barnes et al., 2015), suggesting the potential for spread of the pathogen(s) through local currents and sediment transport to northward reefs, including potential interactive effects of sedimentation on coral health (Dobbelaere et al., 2020a,b). Ultimately, the ability of sediments to act as a SCTLD vector and/or reservoir throughout the endemic zone will likely impact local disease prevalence and coral mortality, potentially prolonging the outbreak, and representing a threat of reemergence following disturbance events, such as storms and future coastal development activities. We therefore conducted an ex situ laboratory experiment to determine whether reef sediments can serve as a vector for SCTLD, enabling downstream disease transmission without direct contact between diseased and healthy corals, or disease-inoculated water and healthy corals.
Materials and Methods
Experimental Apparatus
The transmission experiment was conducted in the Experimental Reef Laboratory at the University of Miami’s Cooperative Institute for Marine and Atmospheric Studies and NOAA’s Atlantic Oceanographic and Meteorological Laboratory. A laser-cut acrylic rack system was designed and constructed to support individual 0.5 L coral vessels, half-suspended in 250 L flow-through raceways (Figure 1). Incoming seawater was sourced from Biscayne Bay via the University of Miami’s Marine Technology & Life Sciences Seawater Complex and pre-filtered to 25 μm, with an in-lab manifold system to deliver independent water sources to coral vessels at a consistent flow rate of ∼200 mL/min. Temperature was maintained for raceways and coral vessels based on local ambient reef temperatures at 27°C using in-line chillers and 500 W submersible heaters. Light was provided by three Aqua Illumination Hydra 52 HD 135 W LED arrays per raceway, with 3-h dawn and dusk ramps and a 6-h static day, set at a peak photosynthetically active radiation (PAR) of 275 μmol m–2 s–1 based on local reef measurements.
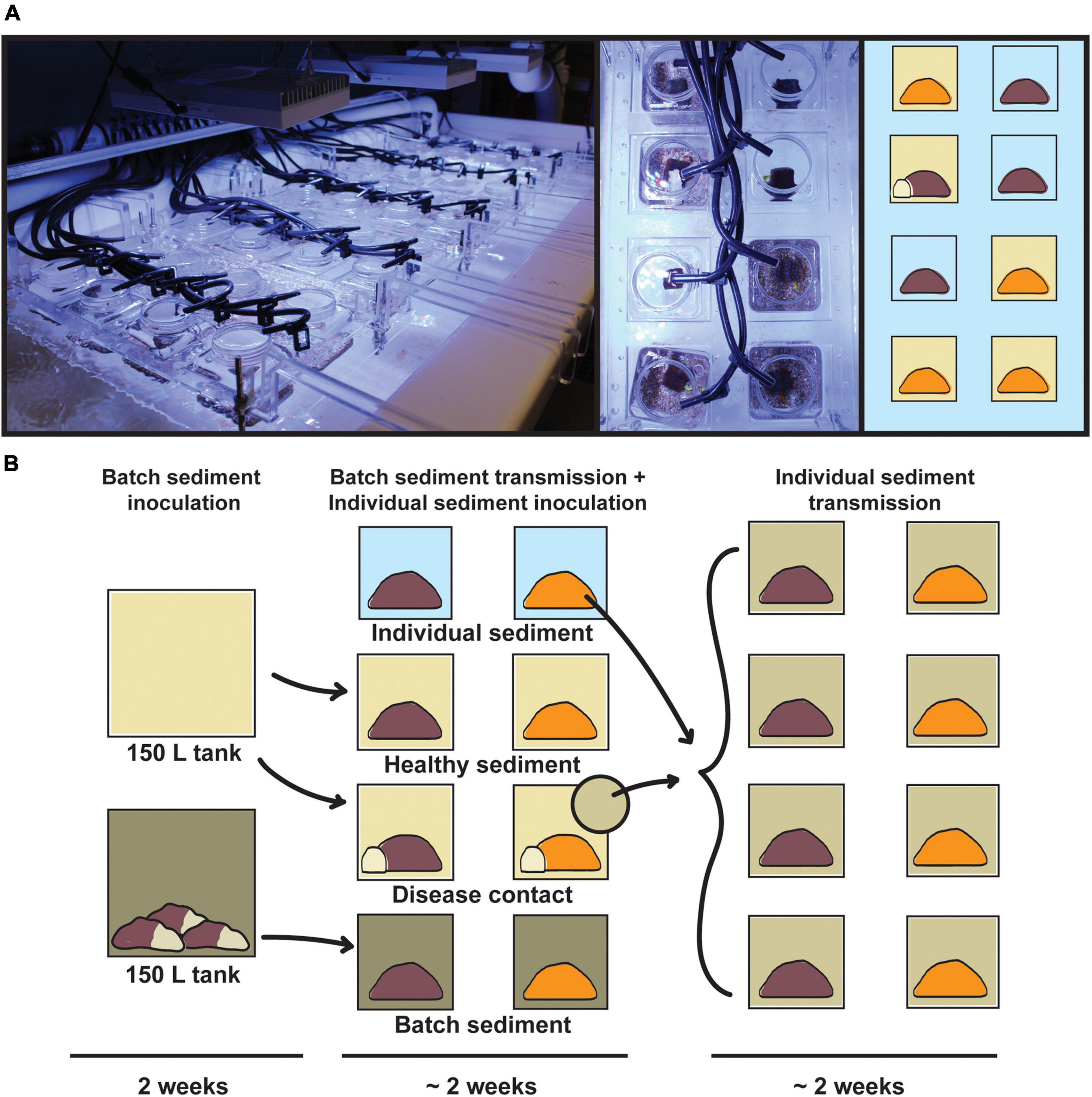
Figure 1. Infographic of the transmission experiment apparatus and design. (A, left) Acrylic rack system containing 40 0.5 L coral vessels in flow-through raceway, with individual seawater sources; (right) top-down view of randomized treatments for Orbicella faveolata (orange) and Montastraea cavernosa (brown). (B) Experimental design consisting of batch sediment inoculation (14 days), exposure of batch diseased sediments to experimental coral fragments (∼14 days), and exposure of individual diseased sediments to initial control fragments (∼14 days). Vessel background colors correspond to disease treatments: healthy sediment (beige), disease contact (beige with white coral fragment), batch diseased sediment (green), and individual diseased sediment (light green).
Coral Sampling
Corals displaying no tissue loss, paling, or visible signs of stress were sourced from Fisher Island in the Port of Miami by the Florida Fish and Wildlife Conservation Commission and the Florida Department of Environmental Protection’s Coral Rescue Team. Three Orbicella faveolata and two Montastraea cavernosa colonies were collected and transported to the Experimental Reef Laboratory in August 2020 and cut into 5 cm2 fragments using a Gryphon AquaSaw following acclimation to the tank conditions. Coral fragments were quarantined in the flow-through raceways for approximately 3 months prior to the transmission experiment, with periodic observations to ensure no introduction of disease occurred following field collections. Corals were fed nightly with 0.3 g of phytoplankton food (Polyplab Reef-Roids) until the start of the experiment.
Disease donor M. cavernosa colonies were harvested on October 30, 2020 from Lauderdale-by-the-Sea reef site BC1 (26.1479, −80.0939) in Broward County, Florida. Three colonies with active SCTLD lesions were collected from a depth of ∼6.4 m and were transported back to the lab using bubble wrap and ambient seawater in insulated coolers. Additionally, ∼36 L of aragonite sand was collected from the same reef site and was transported back to the lab in sealed buckets. Reef sediments were autoclaved at 121°C for 20 min prior to use in the experiment.
Sediment Inoculation and Transmission Treatments
Sediments were divided into roughly equal aliquots (∼13–16 kg each) and dispensed into two 75 L acrylic tanks with independent recirculating 75 L sumps (150 L total per tank system). The three disease donor M. cavernosa colonies were added to one of the tanks (disease-inoculated sediment), while the other sediment tank was not exposed to SCTLD (healthy sediment). Both tanks were incubated for 14 days, with temperature and light regimes as described previously.
Following the SCTLD inoculation period, sediments in each of the two tanks were separately homogenized, then 150 g aliquots were dispensed into the respective 0.5 L coral vessels by treatment. The experiment consisted of four treatments with three replicates of each of the healthy O. faveolata and M. cavernosa colonies per treatment (nofav = 9 and nmcav = 6 per treatment, total n = 60; Table 1): (1) healthy sediment (HS) – corals were placed on sediments that were never in contact with diseased corals, (2) disease contact (DC) – corals were placed in direct contact with disease donor coral fragments on top of uninoculated sediments, (3) batch diseased sediment (BDS) – corals were placed on sediments that were inoculated with entire disease donor colonies, and (4) individual diseased sediment (IDS) – corals were placed on sediments that were inoculated with individual diseased coral fragments obtained from the disease contact treatment (Figure 1). Vessel placement within acrylic racks was randomized across treatments. Prior to transfer of the corals into the experimental apparatus, vessels were flushed for 1 h (a ∼24 × volume refresh) to minimize chances of disease transmission due to waterborne pathogens. Disease donor fragments for the disease contact treatment (∼1 cm × 3 cm strips containing lesions) were cut using the diamond band saw from one of the diseased M. cavernosa colonies used to generate batch diseased sediments. Contact was maintained between diseased fragments and experimental corals in this treatment for the duration of the observation period, and donor fragments were replaced as needed (following complete mortality/sloughing of the donor tissue). Corals were monitored daily for a period of 4 weeks with top-down photographs and observations to quantify time to infection (visible lesion formation) and signs (e.g., mesenterial filaments, liquefactive necrosis, tissue loss; definitions in Supplementary Dataset 1).
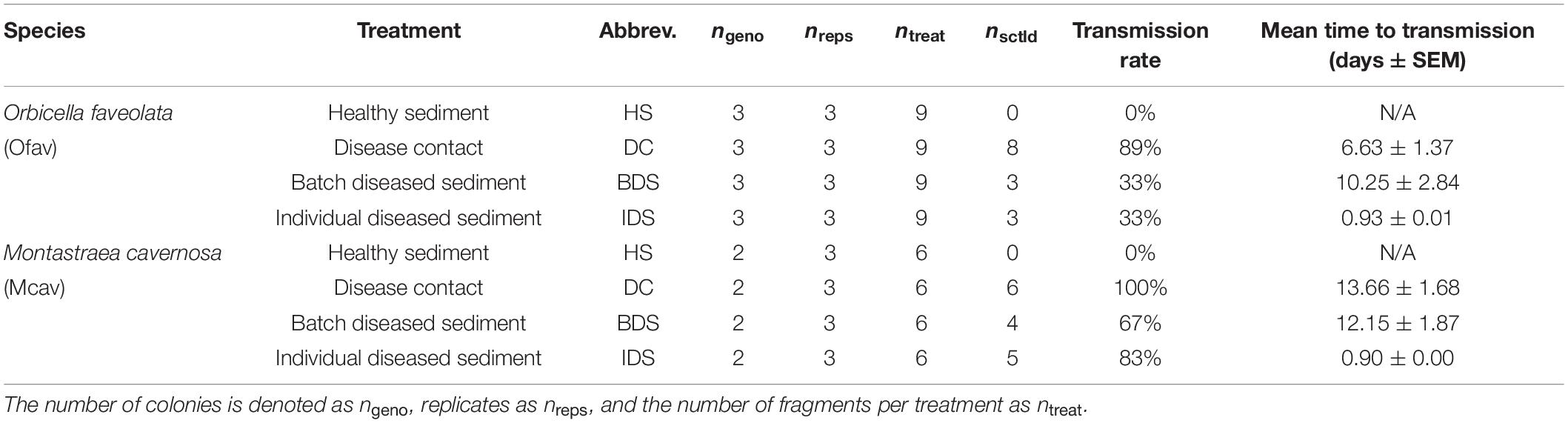
Table 1. Sample sizes of Orbicella faveolata and Montastraea cavernosa fragments used for the four experimental treatments, with corresponding transmission rates and the mean time to infection by disease treatment.
When SCTLD lesions were observed to have progressed approximately 50% across individual corals, the respective fragments were removed from the apparatus, photographed, and preserved in Anatech Z-fix for histology. Prior to shipment to Louisiana State University for processing and analysis, histology samples were rinsed with RO water for 24 h, then stored in 70% ethanol at room temperature. For corals in the batch diseased sediment treatment, the corresponding colony fragment from the healthy sediment treatment was removed from the apparatus and sampled at the same time. Aliquots (∼2 mL) of sediments were also collected from the sediment treatments at time of coral sampling, preserved in an equal volume of Zymo DNA and RNA Shield, and flash-frozen in a liquid nitrogen dry shipper prior to storage at −80°C. Any corals remaining in the apparatus at the end of the 4-week monitoring period [e.g., samples demonstrating no active infection (NAI) and corresponding healthy controls] were sampled as described previously.
Transmission Rate Analyses
Mean time to infection for each treatment was quantified as the number of days between initial contact to diseased sediments/corals and visible signs of lesion formation. All statistical analyses were done in the R statistical environment (R Core Team, 2019). Following assumption testing for data normality, time data were analyzed for differences among species, colonies, and disease treatments using a three-way ANOVA with pairwise Tukey tests among levels of significant factors. Transmission rates were calculated as the proportion of corals that exhibited disease signs within each treatment. To quantify the relative risk of infection among disease treatments, a survivorship analysis was conducted using time to infection and transmission rate data using the R package survival (Therneau, 2021), and survminer (Kassambara et al., 2021) for visualization. A fit proportional hazards regression model was applied to compare risk (hazard ratios) between the two diseased sediment treatments using the disease contact treatment as a reference comparison.
Histological Analyses
Tissue/skeletal samples preserved for histology were decalcified using 1% EDTA HCl solution, and tissue areas that excluded obvious lesions were processed using a Leica ASP6025 tissue processor, embedded in wax blocks on a Leica EG1150H embedding machine, sectioned on a Leica RM2125RTS microtome, and slides were stained with hematoxylin and eosin on a Leica ST5020. Slides were analyzed on an Olympus BX41 microscope with SC180 camera attachment. Five serial slides, separated by 500 μm, were analyzed per individual coral sample.
Ten photos were taken per sample across all five slides, ensuring various parts of the coral fragment were analyzed. Photographs were analyzed using ImageJ software (Abràmoff et al., 2004; Schneider et al., 2012) and a 12-cell grid was overlaid on each image; each grid-cell had an area of 5,000 μm2. A random number generator was used to select one grid-cell per pictograph for image analysis, and was repeated for every image used. Within the grid-cell of interest, all Symbiodiniaceae and their vacuole areas were measured until 25 symbiont and vacuole areas were captured for each sample. Additionally, within the grid-cell of interest, the number of symbionts exocytosed from coral gastrodermal cells was counted and the proportion calculated. Finally, when abnormal separation of the gastrodermis from the mesoglea was observed (which indicated tissue degradation), the maximum separation distance was measured. Where mesoglea was not in the image, an NA was noted. For proportion exocytosis and gastrodermal separation, all ten pictographs were used. Symbiont cell and vacuole areas required a variable number of pictographs for the 25 measurements.
Mean measurements (symbiont-to-vacuole area ratio, proportion of exocytosis, and gastrodermal separation) per sample were used for all analyses. For evaluation of symbiont-to-vacuole ratio as a predictor of sample health condition (healthy or diseased), a binomial generalized linear model (GLM) was conducted for each coral species. To test for overall species and treatment effects in symbiont-to-vacuole ratio and proportion of symbiont cells exocytosed, two-way beta regressions were applied using the R package betareg (Cribari-Neto and Zeileis, 2010), since these responses were bound between zero and one. Additionally, one-way beta regressions were applied to compare pairwise differences between disease treatments and the healthy sediment treatment within each species. A two-way ANOVA was used to test the effects of species and treatment on gastrodermal separation.
Microbial Community Profiling
Total genomic DNA was extracted from preserved sediment samples using a Qiagen Power Soil DNA extraction kit following the manufacturer’s protocol, with a slight modification to the lysis step which was carried out in an MP FastPrep-24 with a beating step of 6.0 m/s for 1 min. DNA samples were sent to the Michigan State University (MSU) Genomic Core for 16S library preparation and sequencing, with positive (ZymoBIOMICS Microbial Community DNA Standard [Cat# D6305]) and negative (no template DNA) control reactions included during library preparation. The V4 hypervariable region of the 16S rRNA gene was amplified using dual-indexed Illumina primers 515F/806R following the protocol developed by Kozich et al. (2013). PCR products were batch normalized using Invitrogen SequalPrep DNA Normalization plates, pooled, then cleaned and concentrated using a QIAquick PCR Purification column followed by AMPureXP magnetic beads. The pool was QC’d and quantified using a combination of Qubit dsDNA HS, Agilent 4200 TapeStation HS DNA1000, and Invitrogen Collibri Library Quantification qPCR assays. The pool was sequenced on a MiSeq v2 standard flow cell in a 2 × 250 bp paired end format using a MiSeq v2 500 cycle reagent cartridge. Custom sequencing and index primers complementary to the 515F/806R (GTGCCAGCMGCCGCGGTAA and GGACTACHVGGGTWTCTAAT, respectively) oligomers were added to appropriate wells of the reagent cartridge. Base calling was done by Illumina Real Time Analysis (RTA) v1.18.54 and output of RTA was demultiplexed and converted to FastQ format with Illumina Bcl2fastq v2.20.0.
Raw fastq files were analyzed using the QIIME 2 platform (Bolyen et al., 2019) and alternative sequence variances (ASVs) were generated using the DADA2 plugin (Callahan et al., 2016). The mock community sample (positive control) was analyzed first to optimize all parameters in the pipeline. Based on the results of the mock community, we applied a low frequency filter by which any ASVs represented by less than 20 sequences were eliminated. The taxonomy assignment for each ASV was completed based on the SILVA v138 database (Quast et al., 2013; Yilmaz et al., 2014) and the resulting tree was used for phylogenetic diversity calculations. ASVs assigned to chloroplast or mitochondrial rRNA, as well as taxonomically unassigned ASVs, were removed from further analyses. Differentially abundant ASVs among sediment treatments were identified using beta binomial regression models with the package corncob (Martin et al., 2020), with visualization of Bray–Curtis dissimilarity using PCoA within the packages vegan and ape (Oksanen et al., 2015; Paradis and Schliep, 2019). To narrow these analyses to observed differences in disease transmission among corals, we conducted additional differential abundance tests of sediment samples between three groups corresponding to coral condition (C, control; NAI, no active infection; and TL, tissue loss). These analyses were then repeated using a dataset first collapsed by genus to identify differentially-abundant genera across treatments and coral conditions. Overall tests of significant differences in microbial communities between species and among treatments/coral conditions were conducted using separate two-factor PERMANOVAs in vegan, with pairwise tests of significant factors using pairwiseAdonis (Martinez Arbizu, 2020). To identify differentially-abundant taxa in disease-inoculated sediments while eliminating origin effects on sediment communities, the dataset was then filtered to remove BDS samples (i.e., comparing HS and IDS samples only), and the analyses were repeated as described above. Last, a cross-study comparison of significant ASVs was carried out using the MUSCLE alignment plugin (Edgar, 2004) with the software Geneious v.10.2.61.
Results
Stony Coral Tissue Loss Disease Transmission
No disease transmission was observed for either species in the healthy sediment treatment over the course of the experiment. Transmission occurred in 89% of O. faveolata and 100% of M. cavernosa within the disease contact (DC) treatment, and the onset of visible lesion formation occurred after 6.6 ± 1.4 and 13.7 ± 1.3 days, respectively (mean ± SEM; Table 1). SCTLD transmission was also observed in the batch diseased sediment (BDS) treatment, albeit with lower transmission rates compared to disease contact corals: 33% of O. faveolata and 67% of M. cavernosa fragments. Lesion formation occurred after similar exposure times between species, with 10.3 ± 2.8 and 12.2 ± 1.9 days, respectively (Table 1). Corals that were exposed to individual diseased sediments (IDS), however, demonstrated distinct transmission metrics compared to both other disease treatments. Transmission rates remained relatively low for O. faveolata (33%, same as in the batch diseased sediment treatment), but were higher than the batch diseased sediment treatment in M. cavernosa (83%). The mean time to onset of lesions was remarkably lower for both species in the individual diseased sediment treatment, with 0.93 days for O. faveolata and 0.90 days for M. cavernosa (Table 1). A three-way ANOVA indicated that the time to onset of disease was significantly different between species (p < 0.018), among coral colonies (p < 0.039), and among disease treatments (p < 0.001), with a significant species:treatment effect (p < 0.046; Table 2 and Figure 2). Pairwise post hoc tests determined that the species effect was attributed to differences within the disease contact treatment, while the treatment effect was due to differences between individual diseased sediment and both disease contact and batch diseased sediment treatments for M. cavernosa (Tukey’s: both p < 0.001; Table 2 and Figure 2). A fit proportional hazards regression model revealed that risk of infection was only significantly different among disease treatments for M. cavernosa (log-rank: z2,17 = 16.19, p < 0.001), though pairwise comparisons could not be completed due to lack of model convergence (Supplementary Figure 1). Risk of infection was statistically indistinguishable among disease treatments for O. faveolata.
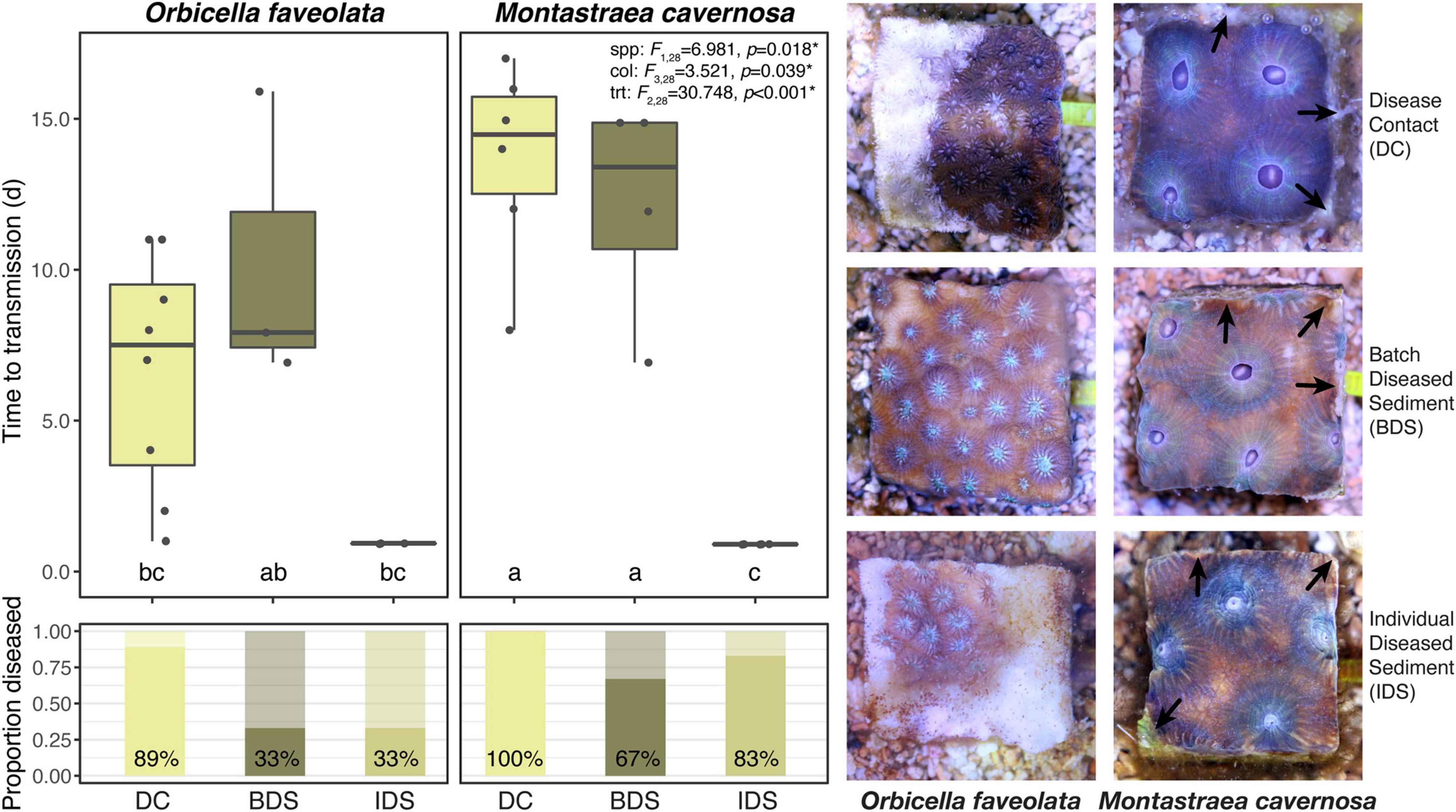
Figure 2. Mean time to transmission (initial observation of lesions) and transmission rates (proportion diseased) by species and disease treatments, with representative photographs. Error bars denote standard error of the mean, test statistics indicate the results of three-way ANOVA among species, colonies, and treatments, and different letters denote significant pairwise differences as generated by Tukey’s test. Healthy sediment treatment not shown or analyzed due to no observed cases of transmission in the treatment. Treatment abbreviations as in Table 1, and treatment colors as in Figure 1. Arrows in photo panel indicate excess mucus production (Top right), tissue necrosis (Center right), and tissue retraction (Bottom right) observed more commonly in M. cavernosa fragments.
Signs of SCTLD infection were similar among disease treatments but varied between species. Early signs including discharge of mesenterial filaments, tissue swelling, and tissue paling were common, ranging from ∼24 h to several days prior to visible lesion formation. Characteristic white lesions and tissue loss (Aeby et al., 2019; Landsberg et al., 2020) were more prevalent in O. faveolata, with rapid progression across entire fragments within a few days (Figure 2). Lesions were often less defined in M. cavernosa fragments, with smaller patches of necrosis and tissue loss that appeared to progress slowly over several days to a week. In both disease-inoculated sediment treatments, lesion formation was more likely to occur on the vertical sides of the fragment, where there was close contact between coral tissue and the sediments. In these cases, tissue loss was more likely to occur along the fragment margins, often accompanied by excess mucus production in M. cavernosa (Figure 2).
Histological Analysis
Histological analysis of experimental samples identified gross differences in coral tissues exposed to SCTLD versus those in the healthy sediment treatment. Signs associated with SCTLD (e.g., symbiont exocytosis, body wall breakage, liquifying necrosis; Landsberg et al., 2020; Meiling et al., 2021) were confirmed more commonly in diseased versus healthy treatments, with evidence of subtle variation between species (Supplementary Figure 2). Health condition (diseased or healthy) of corals was predictable using symbiont cell-to-vacuole area ratio in O. faveolata (binomial GLM; z1,23 = −2.638, p < 0.008; Supplementary Figure 3), but not in M. cavernosa (z1,35 = −1.293, p > 0.196). When the O. faveolata symbiont cell-to-vacuole ratio was 0.56, there was a 50% chance of the model predicting the coral was diseased. Above 0.56, the coral was likely healthy, and below, likely diseased. Vacuolization was significantly different between species (two-way beta regression; F1,59 = 4.340, p < 0.037) and among treatments (F3,59 = 5.778, p < 0.001), with significant pairwise differences between all diseased treatments and the healthy sediment treatment (all p < 0.003) in O. faveolata, but with no significant pairwise differences in M. cavernosa (all p > 0.06; Supplementary Figure 4). The proportion of symbiont cells exocytosed was not significantly different between species (F1,59 = 1.442, p > 0.230) or among treatments (F3,59 = 1.583, p > 0.191), although there was a significant difference between individual diseased sediment and healthy sediment treatments in O. faveolata only (p < 0.002; Supplementary Figure 4). There were no significant effects of species (ANOVA: F1,59 = 1.187, p > 0.281) or treatment (F3,59 = 1.244, p > 0.303) on gastrodermal separation distance (Supplementary Figure 4).
Microbial Community Profiling
A total of 5,385 ASVs were identified across all sediment samples belonging to 2 domains (Bacteria and Archaea), 46 phyla, 91 classes, 212 orders, 331 families, and 477 genera (Supplementary Dataset 3); with evidence of community differences among treatments (Figure 3). Healthy sediment and individual diseased sediment treatment samples clustered along coordinate 1 (33.4% variation explained), with batch diseased sediment samples representing a separate group. Additionally, there was variation along coordinate 2 (11.4%) that may represent variation in sediment microbial communities due to coral condition (C, control; NAI, no active infection; TL, tissue loss; Figure 3). Binomial regressions among sediment treatments revealed 221 ASVs that were significantly differentially abundant, which corresponded to 121 genera when data were first collapsed to genus-level taxonomy. Analysis of microbial communities using disease status of the corresponding coral fragment as a factor resulted in 20 ASVs that were significantly different, including 11 ASVs showing enrichment in sediments associated with corals exhibiting tissue loss (Table 3). When collapsed by genus prior to testing, there were 30 genera significantly differentially abundant among coral conditions, including 16 genera for which the abundance was higher in tissue loss-associated samples (Figure 4).
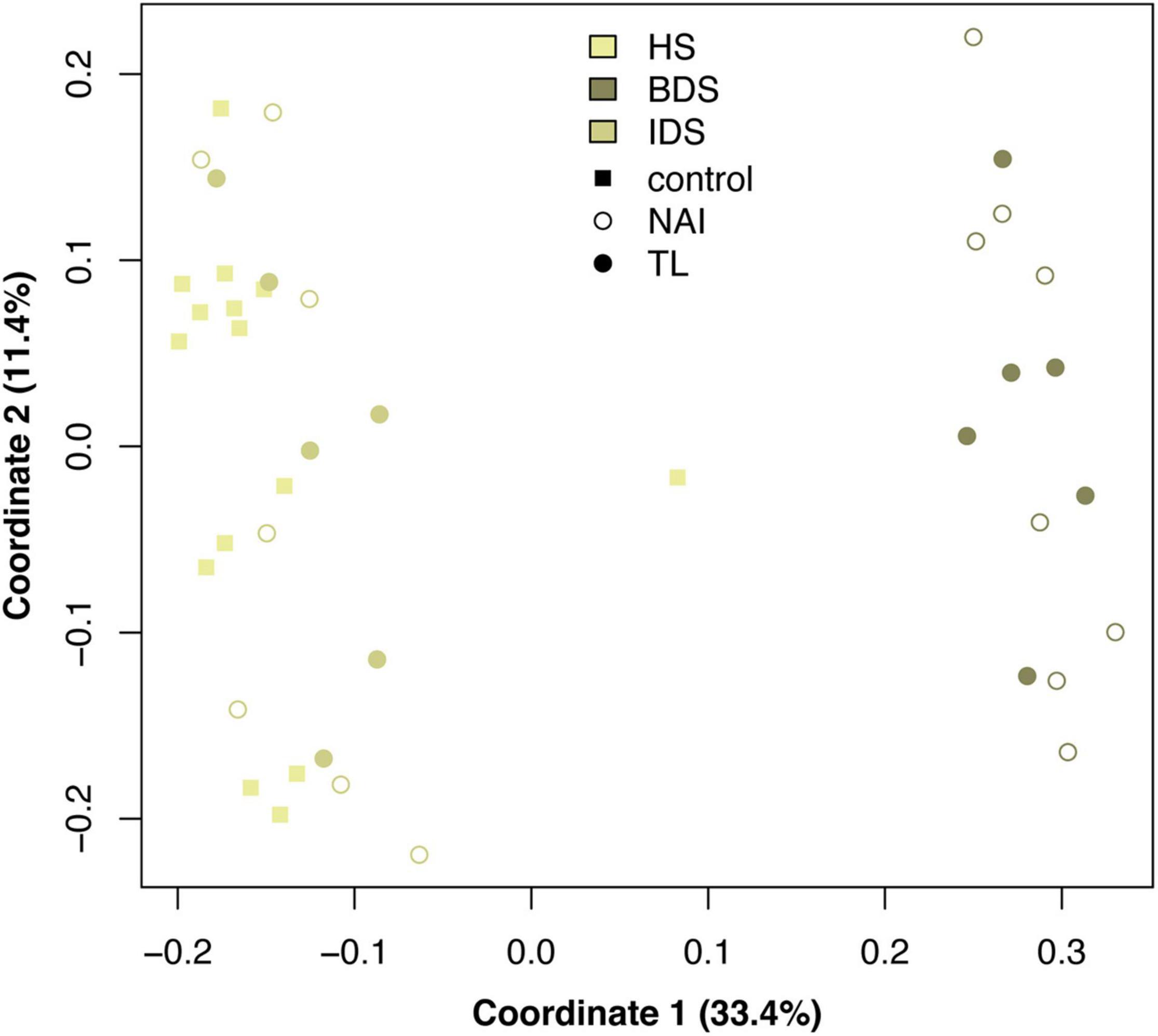
Figure 3. Principal coordinates analysis of microbial communities among sediment treatments and sample conditions, based on Bray–Curtis dissimilarity among samples. Treatment abbreviations as in Table 1 and treatment colors as in Figure 1.
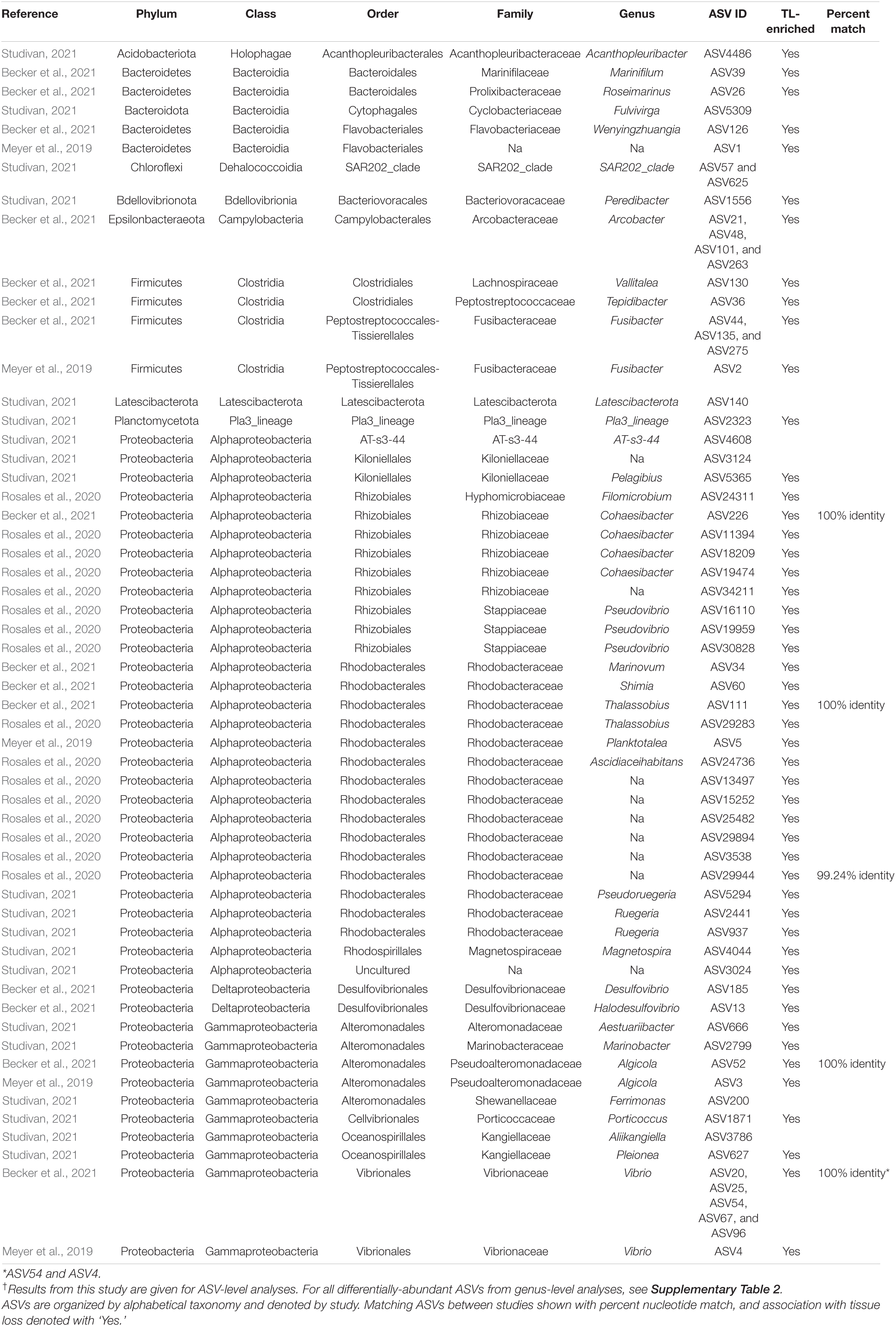
Table 3. Differentially abundant ASVs identified across sediment samples by coral condition in our experiment†, as well as those found in association with SCTLD from previous studies.
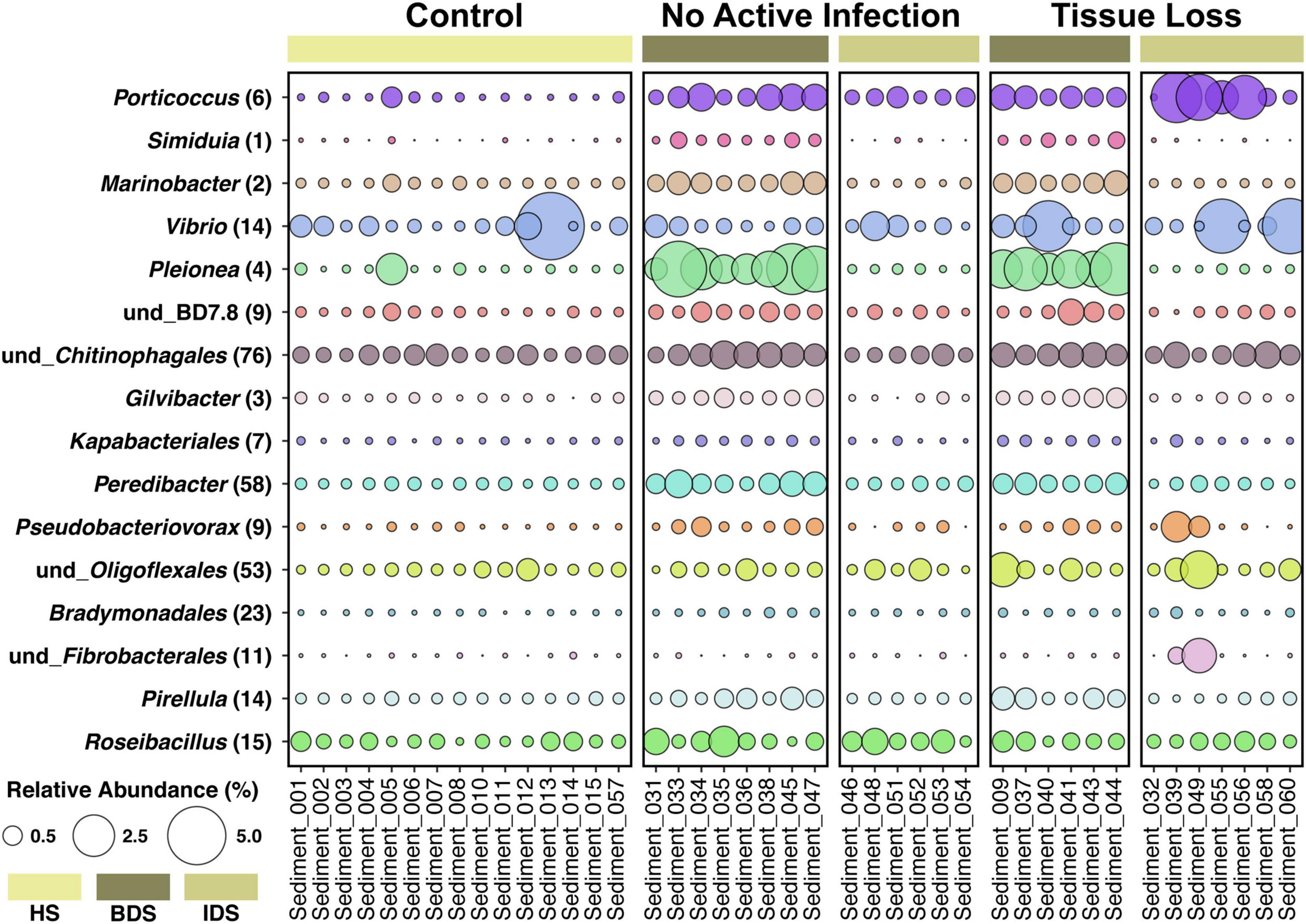
Figure 4. Relative abundance of 16 bacterial genera enriched in diseased sediment samples associated with coral fragments exhibiting no active infection (NAI) and tissue loss (TL), in comparison to healthy sediment associated with control coral fragments exhibiting non-symptomatic conditions. Each genus is followed by the number of differentially-abundant ASVs identified in that genus. Treatment abbreviations as in Table 1, colors as in Figure 1; and sample IDs as in Supplementary Dataset 1.
PERMANOVAs indicated that there was significant variation in overall microbial communities among treatments (p < 0.001) attributed to pairwise differences between HS/BDS and BDS/IDS treatments (p < 0.002 for both), and among coral conditions (p < 0.003) attributed to pairwise differences between control/NAI and control/TL conditions (p < 0.003 for both; Supplementary Table 1). Species did not have a significant effect on communities for either test, and treatment explained more of the variance in microbial communities (33.8%) compared to coral condition (13.0%). Based on these results, differential abundance tests were repeated using a filtered dataset that removed BDS samples to specifically identify significant ASVs following individual SCTLD inoculations, while removing origin effects on microbial communities (IDS versus HS). Only one ASV (ASV 625) was found to be differentially abundant, but was less abundant in individually-inoculated samples compared to healthy controls. When collapsed by genus prior to testing, the number of differentially-abundant genera rose to six, and of these, four were found in higher abundance in IDS samples. Likewise, there were no significant effects of treatment and coral condition on the filtered dataset, though there was a significant species effect for both tests (p < 0.036 and p < 0.041, respectively; Supplementary Table 1).
Discussion
Stony Coral Tissue Loss Disease Transmission
The metrics quantified in this study were consistent with previous transmission experiments (as reported in Dobbelaere et al., 2020b) and indicate that the diseased coral colonies used in the sediment inoculation treatments were indeed infectious. The proportion of diseased corals among treatments suggest that transmission of SCTLD was less likely to occur from sediments than direct contact between diseased and healthy coral tissue, and that M. cavernosa fragments were more likely to become infected than O. faveolata across all treatments. There is evidence of a species-specific response with regards to transmission rates and species susceptibility as has been demonstrated with previous experiments (NOAA, 2018; Aeby et al., 2019; Dobbelaere et al., 2020b; Meiling et al., 2021). The higher rates of transmission observed for M. cavernosa are likely related to the fact that the disease donors were also M. cavernosa, despite higher relative SCTLD susceptibility reported for O. faveolata (NOAA, 2018; Meiling et al., 2021). It is also important to note that the disease contact treatment is likely not representative of an ecologically relevant scenario since few populations exist with sufficient coral density to achieve close association of susceptible colonies. Therefore, transmission rates from waterborne or sediment vectors are more likely to accurately estimate risk of transmission in a reef environment (Dobbelaere et al., 2020b; Muller et al., 2020).
This transmission experiment demonstrated that reef sediments can serve as a SCTLD vector. We also observed significant variation in time to visible lesion formation between disease-inoculated sediment treatments. It does not appear that this variation was due to the ratio of diseased tissue area used for inoculation to sediment mass (i.e., disease “dose”), as the estimated ratio was higher for the batch diseased sediment treatment (∼1,050 cm2 coral tissue to ∼13,000 g sediment, or 0.08:1) versus individual inoculation (∼5 cm2 coral tissue to ∼150 g sediment, 0.03:1). The inoculation time was higher for the individual diseased sediment treatment: 14 days for batch diseased sediment and 19.6 ± 2.3 days for individual diseased sediment, the latter depending on the time to lesion formation for individual fragments in the preceding disease contact treatment. This suggests that there may be a temporal component to the infectiousness of inoculated sediments, but this hypothesis requires further testing in a controlled experiment specifically incorporating time treatments. Likewise, it is possible that necrotic tissue sloughing off disease donor corals or biofilms were more abundant in the semi-enclosed vessels in the individual diseased sediment treatment versus the larger batch diseased sediment tank, perhaps due to perturbation of the sediment boundary layer following homogenization in the latter treatment. The presence of necrotic tissue and/or biofilms in sediments was not examined for this study, but remains a potential contributing factor to the reservoir potential of sediments. Necrotic tissues in particular are a suspected vector of SCTLD among colonies and even reefs through transport due to oceanographic currents (Dobbelaere et al., 2020b; Muller et al., 2020).
Last, it is of note that while we could not eliminate the potential for waterborne transmission to confound the observed results in this experiment, efforts were made to reduce any confounding SCTLD vectors through extensive flushing of potentially disease-laden water prior to coral fragments’ exposure to diseased sediments. Prior experimental evidence suggests that the probability of waterborne transmission is far lower than with direct contact treatments (Dobbelaere et al., 2020b), and that time to lesion formation is greater than observed in this study (pers. obs.). It is of course possible that SCTLD pathogens are continuously shed into the water column from inoculated sediments and/or infected coral tissue, potentially exacerbating the risk of transport among corals (Dobbelaere et al., 2020b).
Histological Analysis
The presence of SCTLD signs in all disease treatments was confirmed through histological examination of coral tissues. Additionally, there was limited evidence of variation between diseased and healthy sediment treatments in O. faveolata for three metrics examined, mainly symbiont-to-vacuole ratio. Vacuolization of the symbionts has been shown to occur during SCTLD infection, presumably due to a breakdown of coral-algal symbiosis (Landsberg et al., 2020). However, the current results suggest there is a species- or symbiont-specific vacuolization response, potentially indicating holobiont-specific SCTLD-associated dysbiosis (Supplementary Figure 4), which could indicate immune or physiological mechanisms that determine disease trajectories. Symbiont vacuolization in M. cavernosa was not significantly different across treatments, and the range of vacuolization was narrower than that of O. faveolata. Diseased O. faveolata demonstrated significantly higher vacuolization (lower symbiont:vacuole area ratios) than healthy corals, which may indicate a more extreme disease response in this species, as has been reported in previous field and laboratory studies (Landsberg et al., 2020; Meiling et al., 2021). Exocytosis was also more extreme in diseased O. faveolata, but not for M. cavernosa. Symbiont communities have not been examined for the corals used in this experiment, but remain a potential factor in the observed differences between species. There was no difference in gastrodermal separation between disease and healthy treatment corals for both species. While gastrodermal separation has previously been recognized as a sign of SCTLD infection, it may also be an artifact of histological sampling and/or processing (Landsberg et al., 2020; Meiling et al., 2021), which may confound these results.
Microbial Community Profiling
Analysis of the sediment samples among treatments suggested that the dominant driver of microbial community structure was due to origin/bottleneck effects of initial sediment inoculation (Figure 3 and Supplementary Table 1). Despite subsequent exposure of sediments to SCTLD in the individual diseased sediment treatment, overall microbial communities were more similar to the healthy sediment treatment than to the batch diseased sediment treatment. Therefore, analysis of differentially abundant taxa among coral condition groups (control, no active infection, and tissue loss) rather than treatments provided a more targeted examination of microbes associated with SCTLD, and demonstrated subtle variation in community structure likely related to the disease. Six of the tissue loss-associated genera belonged to the Gammaproteobacteria including Porticoccus (6 ASVs), Marinobacter (2 ASVs), Vibrio (14 ASVs), Pleionea (4 ASVs), and an unidentified genus within phylogenetic cluster BD7-8 (9 ASVs). Based on genome-wide phylogeny, Porticoccus belongs to the newly-designated order of Cellvibrionales of marine gammaproteobacteria (Spring et al., 2015), and are hydrocarbon degrading bacteria reported to be associated with marine phytoplankton including Symbiodiniaceae (Lawson et al., 2018). One of the Porticoccus ASVs (ASV1871; Table 3) showed 100% identity to five clonal sequences previously found to be associated with white plague disease type II in O. faveolata (Sunagawa et al., 2009) which have been currently assigned to the genus Porticoccus by the Silva v138 database. Since the pathogenic agent of white plague II is Aurantimonas corallicida, lesion enrichment with other bacterial taxa in that study was explained by possible opportunistic succession of a variety of bacteria following initial infection (Sunagawa et al., 2009). There was only one ASV belonging to the genus Simiduia (Simiduia agarivorans), which is an agarolytic bacterium (Shieh et al., 2008), and its potential relationship in reef sediment communities is unknown.
On the other hand, multiple ASVs of Vibrio have been shown to be enriched in tissue lesions (Meyer et al., 2019; Ushijima et al., 2020; Becker et al., 2021). A cross-study comparison of ASVs among our data to previous SCTLD studies (Meyer et al., 2019; Rosales et al., 2020; Becker et al., 2021) revealed that 15 out the 25 ASVs identified by Becker et al. (2021) were present in our sediment samples, including 4 ASVs assigned to the genus Vibrio and one to genus Algicola (ASV1497, ASV3388, ASV187, ASV343, ASV3483, Supplementary Dataset 3). While these ASVs were not differentially abundant between coral conditions most likely due to their relative rarity, the genus Vibrio (14 ASVs) did show significant enrichment in the TL-associated sediment samples (Figure 4). It should also be noted that the ASV3388 sequence showed 100% nucleotide identity to Vibrio coralliilyticus for which at least one pathogenic strain has been shown to contribute to SCTLD progression (Ushijima et al., 2020).
In the present study, Alphaproteobacteria was represented by two differentially abundant ASVs within the family Rhodobacteraceae (ASV5294 and ASV937), two ASVs belonging to genera Pelagibius and Magnetopsira (ASV5365 and ASV4044), and one undescribed bacterium (ASV3024; Table 3). The ASV5294 from the current study was most closely related to ASV29944 from Rosales et al. (2020), with the sequences showing 99.24% nucleotide identity within the order Rhodobacterales (Table 3). Rhodobacterales have been found to be enriched in multiple coral diseases as well as SCTLD, including multiple white syndrome-like diseases, white band disease, and black band disease (Sunagawa et al., 2009; Miller and Richardson, 2011; Meyer et al., 2019; Gignoux-Wolfsohn et al., 2020; Rosales et al., 2020; MacKnight et al., 2021), demonstrating an opportunistic type of colonization and infection. Despite repeated analyses on a filtered dataset to remove origin/bottleneck effects on microbial communities, only one additional ASV (ASV3332) was found to be enriched in sediment samples following SCTLD inoculation, besides those already identified in analysis of the full dataset (Supplementary Table 2). This ASV corresponded to Shimia sp., which is also within Rhodobacteraceae, and has been previously found in association with corals infected with Porites white patch syndrome (Séré et al., 2013) and Australian subtropical white syndrome (Godwin et al., 2012).
The final three genera showing enrichment in sediment samples associated with corals exhibiting tissue loss belonged to two taxonomically distinct genera and one undescribed group: Peredibacter (phylum Bdellovibrionota), Acanthopleuribacter (phylum Acidobacteriota), and uncultured Planctomycetess clade referred to as Pla3_lineage (ASV1556, ASV4486, and ASV2323; Table 3). Bacteria in the genus Peredibacter are closely related to Bdellovibrio-and-like organisms (BALOs) which are predators of other bacteria (Davidov and Jurkevitch, 2004; Welsh and Thurber, 2016; Welsh et al., 2016); hence its abundance would be expected to increase with enrichment of other bacteria. The type species of the genus Acanthopleuribacter was isolated from foot tissue of chiton (Fukunaga et al., 2008), and it is unknown whether this genus is also found in association with corals. Lastly, little is known about the recently established phylum Planctomycetes of diverse and mostly aquatic bacteria, with many representatives comprising less than 5% of bacterioplankton in coastal waters (Yilmaz et al., 2016). None of the previous SCTLD 16S rDNA profiling studies (Meyer et al., 2019; Rosales et al., 2020; Becker et al., 2021), however, have documented enrichment of the above three bacterial taxa in diseased coral tissue, suggesting that increased abundance in the SCTLD-inoculated sediment samples in the present study may represent opportunistic growth of bacteria associated with organic matter in the sediment/and or interstitial seawater from necrotic coral tissue.
Microbial data from the present and previous SCTLD studies has suggested that opportunistic infections of multiple taxa may be occurring, instead of individual pathogenic strains (Table 3 and Figure 4; Meyer et al., 2019; Rosales et al., 2020; Becker et al., 2021). There is evidence of strong variation among locations, host species, and/or microenvironments (i.e., coral tissue versus sediments), given that each of the studies mentioned above represent sample sets from diverse locations, times, and species. It has also been hypothesized that most coral diseases including SCTLD are the result of a dysbiosis of normal bacterial microbiomes, caused by either an environmental immunosuppression or other primary pathogenic agent (e.g., virus or microbial eukaryote; Meyer et al., 2019; Thurber et al., 2020). Similar to the results of the study by Rosales et al. (2020) that also examined sediments in association with SCTLD-infected corals, however, we identified members of Rhodobacteraceae and Rhizobiales in our dataset (136 and 81 ASVs, respectively; Supplementary Dataset 3), including two taxa within Rhodobacteraceae with increased abundance in tissue loss-associated samples (Table 3). In addition, 15 of the 25 SCTLD microbial indicators identified by Becker et al. (2021) within eight genera including Vibrio, Algicola, Shimia, Cohaesibacter, Halarcobacter, Tepidibacter, Fusibacter, and Marinifilum were detected in the sediment samples of the present study. Furthermore, broad sampling of water and corals on reefs in southeast Florida identified taxa within Flavobacteriaceae and Rhodobacteraceae at higher abundance within/near local ports, inlets, and sewage outfalls (Staley et al., 2017; Laas et al., 2021). Taken together, this reinforces the notion that sediments may serve as a reservoir for SCTLD pathogens, and that co-infecting bacteria of this disease exist in other environments beyond diseased coral tissue.
Conclusion
This experiment demonstrated that reef sediments can in fact transmit SCTLD, and therefore act as a disease vector. The results of this study also pose the hypothesis that sediments may serve as disease reservoirs or vectors at local scales under natural conditions (i.e., in the absence of human disturbance). Experimental and modeling approaches have identified that water may serve as a transmission vector, particularly over regional geographic scales (Aeby et al., 2019; Dobbelaere et al., 2020b; Muller et al., 2020; Sharp et al., 2020; Meiling et al., 2021). We observed similar transmission rates in the diseased sediment treatment compared to waterborne transmission rates reported in the aforementioned studies. The continued persistence of the SCTLD outbreak within the endemic zone, especially in the context of severely reduced host density in some areas, may also suggest that the pathogen(s) are persisting within local sediments. Disease-inoculated sediments may pose additional threats to corals, such as resuspension, transport via local currents, and tidal flushing that may “seed” new areas with SCTLD (Dobbelaere et al., 2020a,b; Rosales et al., 2020). Nearshore coral reef environments in Florida are known to be exposed to increased rates of sedimentation (Lirman and Fong, 2007), and sedimentation has been shown experimentally to cause stress responses in corals (Vargas-Ángel et al., 2006, 2007). Sediments may also cause physical injury to coral tissues via sand scouring as a result of strong tidal forces and storm events, potentially providing a wound conducive to SCTLD infection, but this remains to be tested experimentally. The additive or synergistic effects of sedimentation on SCTLD transmission and progression are currently unknown.
It does remain to be tested how long disease-inoculated sediments can remain infectious, or if the SCTLD pathogen(s) beyond the microbial indicators identified in this and other studies (Table 3; Rosales et al., 2020; Becker et al., 2021) naturally occur in such micro-habitats. Future ex situ transmission studies are recommended to quantify longevity of infectiousness, and address species-specific trends in transmission rates. The former has implications for ongoing and future port expansion and dredge projects, as further spread of SCTLD may be mitigated in coastal zones through sufficient handling of dredged sediments and resulting plumes. Dredging operations already have environmental impact assessment and disposal requirements that aim to minimize impacts to local habitats including coral reefs (described in Miller et al., 2016). In the future, however, additional considerations may be necessary for those operations that may cause significant sediment resuspension in the context of mitigating the spread of SCTLD.
Sediments may have played a critical role in the uncharacteristically rapid spread of SCTLD throughout Florida’s Coral Reef, as well as the persistence of the disease for the past 7 years. For instance, it is possible that sediments have facilitated transmission to oceanographically-isolated areas of the Caribbean through exchanges of ballast water and associated sediments from endemic to naïve regions (Rosenau et al., 2021). Therefore, continued disease prediction and mitigation approaches should consider sediments both as a SCTLD vector and potential reservoir. Treatment methods should also be evaluated to reduce disease spread due to natural sediment reservoirs, and following coastal development activities that transport sediments. Effective and comprehensive management strategies in the context of the ongoing SCTLD outbreak are contingent on a holistic understanding of modes of transmission and targeted actions to reduce further disease spread; here we recommend that consideration be given to the impacts that sediments and associated development activities may have on coral reefs in Florida and beyond.
Data Availability Statement
Datasets generated from this study can be found in the Supplementary Material, and analysis scripts are available as a GitHub repository release (Studivan, 2021). Raw sequence files from the microbial community analysis are archived in an NCBI SRA (Accession PRJNA779487).
Author Contributions
IE and MS secured funding for this research, with additional funding support to DH and AR. MS and IE designed the experiments. MS, NS, AR, and ER performed the research and data collection. MS, AR, and ER conducted the data analyses. All authors contributed to the article and approved the submitted version.
Funding
Funding was provided to IE and MS through the NOAA Coral Reef Conservation Program (#31252) and NOAA OAR ‘Omics Program.’ Funding was provided to DH and AR through the Louisiana Board of Regents Research Support Fund Research Competitiveness Subprogram [#LEQSF(2020-23)-RD-A-06].
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Graham Kolodziej for field assistance and Rob Ruzicka from the Florida Fish and Wildlife Conservation Commission and the Florida Department of Environmental Protection’s Coral Rescue Team for coordinating the collection of corals used in the experiment. Allyson DeMerlis created the infographic in Figure 1. Jocelyn Karazsia from the NOAA National Marine Fisheries Service and Victoria Barker from the NOAA Coral Reef Conservation Program provided critical feedback and support on the design of this study. Disease donor corals and reef sediments were collected under a permit from the Florida Fish and Wildlife Conservation Commission to IE (Special Activity License SAL-20-2116C-SCRP).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.815698/full#supplementary-material
Supplementary Figure 1 | Survivorship curves for disease treatments within species based on mean time to transmission (initial observation of lesions) and transmission rates (proportion diseased). Shaded areas denote 95% CI and test statistics indicate the results of fit proportional hazards regression models for each species. Treatment abbreviations as in Table 1 and treatment colors as in Figure 1.
Supplementary Figure 2 | Micrographs of coral tissue stained with hematoxylin and eosin. Columns denote transmission treatments (from left to right): healthy sediment, disease contact, diseased sediment, and diseased sediment (acute). (A) Tissue from healthy sediment treatment: Symbiodiniaceae in vacuoles, some exocytosis of symbiont cells in the gastrodermis, no major gastrodermal separation or necrosis shown. (B) Tissue from disease contact transmission: body wall breakage apparent in epidermis as well as exocytosis of symbionts in the gastrodermis. Necrosis of basal gastrodermis is apparent behind broken epidermis. (C) Tissue from diseased sediment transmission: mucus and fungus/sponge cells/spicules surround epidermis with extensive liquifying necrosis. Symbiodiniaceae in larger vacuoles and some apparent exocytosis. (D) Tissue from diseased sediment (acute): body wall breakage of epidermis and necrosis are apparent. Symbiodiniaceae present in large vacuoles and some apparent exocytosis. (E) Tissue from healthy sediment: fungal cells outside of epidermis, Symbiodiniaceae in vacuoles with some exocytosis. Some potential necrotic tissues. (F) Tissue from disease contact transmission: epidermis mostly intact, paling of mesoglea, as well as gastrodermal separation from the mesoglea and extensive symbiont exocytosis. (G) Tissue from diseased sediment: mucus surrounds the epidermis, and gastrodermal separation from mesoglea, body wall breakage, and necrosis are apparent. Some Symbiodiniaceae are within vacuoles, but also extensive exocytosis is apparent. Necrosis of basal gastrodermis is apparent behind broken epidermis. (H) Tissue from disease sediment (acute): no clear epidermis (apparently entirely necrotic), extensive Symbiodiniaceae exocytosis within necrotic tissue. S, Symbiodiniaceae; m, mesoglea; g, gastrodermis; e, epidermis; xs, symbiont exocytosis; bwb, body wall breakage; N, necrosis; f, fungus or sponge cells or spicules; Mu, mucus; gs, gastrodermal separation (with lines showing separation distance). Micrographs taken at 20× magnification; scale bar is 40 μm.
Supplementary Figure 3 | Logistic regression of health condition on symbiont-to-vacuole ratio for Orbicella faveolata. Dashed lines denote where model has 50% accuracy in predicting health condition. Test statistics indicate results of one-way binomial regression analysis, and error bars denote standard error of the mean. Treatment abbreviations as in Table 1. Treatment colors as in Figure 1, except for healthy sediment (HS) shown as gray to distinguish from disease contact (DC).
Supplementary Figure 4 | Boxplots of symbiont-to-vacuole ratio, proportion of exocytosis, and gastrodermal separation in each treatment for each species. Test statistics indicate results of two-way beta regressions, and asterisks denote significant differences in treatments relative to healthy controls following a one-way beta regression for each species (p < 0.05). Error bars indicate standard error of the mean. Treatment abbreviations as in Table 1. Treatment colors as in Figure 1, except for healthy sediment (HS) shown as gray to distinguish from disease contact (DC).
Supplementary Table 1 | Test statistics for comparisons of variation in microbial communities among species, treatments, and coral conditions. Non-significant p-values denoted as ns. Treatment abbreviations for post hoc comparisons as in Table 1.
Supplementary Table 2 | Differentially abundant ASVs identified across sediment samples by coral condition and the individually-inoculated sediment (IDS) treatment in our experiment. ASVs are organized by alphabetical taxonomy, and association with tissue loss/IDS samples denoted with ‘yes.’
Supplementary Dataset 1 | Dataset of time to lesion and transmission rate data across species and disease treatments, including general transmission signs. Transmission symptom definitions: mesenterial filaments (MF), mucus production (Mucus), liquefactive necrosis (Necrosis), tissue paling/bleaching (Paling), tissue swelling (Swelling), and tissue loss (TL).
Supplementary Dataset 2 | Dataset of histological measurements across species and disease treatments, including symbiont-to-vacuole ratio (Zooxarea), proportion of symbiont cells exocytosed (Exopercent), and gastrodermal separation distance (Gastrosep).
Supplementary Dataset 3 | Dataset of ASVs identified in sediment samples, including taxonomic assignments and relative abundance across samples. Sample IDs as in Supplementary Dataset 1.
Footnotes
References
Abràmoff, M. D., Magalhães, P. J., and Ram, S. J. (2004). Image processing with imagej. Biophotonics Int. 11, 36–41. doi: 10.1117/1.3589100
Aeby, G. S., Ushijima, B., Campbell, J. E., Jones, S., Williams, G. J., Meyer, J. L., et al. (2019). Pathogenesis of a tissue loss disease affecting multiple species of corals along the florida reef tract. Front. Mar. Sci. 6:1–18. doi: 10.3389/fmars.2019.00678
Alvarez-Filip, L., Estrada-Saldívar, N., Pérez-Cervantes, E., Molina-Hernández, A., Gonzalez-Barrios, F. J., Alvarez-Filip, L., et al. (2019). A rapid spread of the stony coral tissue loss disease outbreak in the mexican caribbean. PeerJ 7:e8069. doi: 10.7717/peerj.8069
Barnes, B. B., Hu, C., Kovach, C., and Silverstein, R. N. (2015). Sediment plumes induced by the port of miami dredging: analysis and interpretation using landsat and MODIS data. Remote Sens. Environ. 170, 328–339. doi: 10.1016/j.rse.2015.09.023
Becker, C. C., Brandt, M., Miller, C. A., and Apprill, A. (2021). Microbial bioindicators of stony coral tissue loss disease identified in corals and overlying waters using a rapid field-based sequencing approach. Environ. Microbiol. doi: 10.1111/1462-2920.15718
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Core Team, R. (2019). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Cribari-Neto, F., and Zeileis, A. (2010). Beta regression in R. J. Stat. Softw. 34, 129–150. doi: 10.1201/9781315119403-7
Cunning, R., Silverstein, R. N., Barnes, B. B., and Baker, A. C. (2019). Extensive coral mortality and critical habitat loss following dredging and their association with remotely-sensed sediment plumes. Mar. Pollut. Bull. 145, 185–199. doi: 10.1016/j.marpolbul.2019.05.027
Dahlgren, C., Pizarro, V., Sherman, K., Greene, W., and Oliver, J. (2021). Spatial and temporal patterns of stony coral tissue loss disease outbreaks in the Bahamas. Front. Mar. Sci. 8:682114. doi: 10.3389/fmars.2021.682114
Davidov, Y., and Jurkevitch, E. (2004). Diversity and evolution of Bdellovibrio-and-like organisms (BALOs), reclassification of Bacteriovorax starrii as Peredibacter starrii gen. nov., comb. nov., and description of the Bacteriovorax-Peredibacter clade as Bacteriovor. Int. J. Syst. Evol. Microbiol. 54, 1439–1452. doi: 10.1099/ijs.0.02978-0
Dobbelaere, T., Muller, E. M., Gramer, L. J., Holstein, D. M., and Hanert, E. (2020b). Coupled epidemio-dydrodynamic modeling to understand the spread of a deadly coral disease in Florida. Front. Mar. Sci. 7:591881. doi: 10.3389/fmars.2020.591881
Dobbelaere, T., Muller, E. M., Gramer, L., and Holstein, D. (2020a). Report on the Potential Origin of the SCTLD in the Florida Reef Tract. Miami, FL: Florida DEP, 15.
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Erftemeijer, P. L. A., Riegl, B., Hoeksema, B. W., and Todd, P. A. (2012). Environmental impacts of dredging and other sediment disturbances on corals: a review. Mar. Pollut. Bull. 64, 1737–1765. doi: 10.1016/j.marpolbul.2012.05.008
Estrada-Saldívar, N., Quiroga-García, B. A., Pérez-Cervantes, E., Rivera-Garibay, O. O., and Alvarez-Filip, L. (2021). Effects of the stony coral tissue loss disease outbreak on coral communities and the benthic composition of cozumel reefs. Front. Mar. Sci. 8:632777. doi: 10.3389/fmars.2021.632777
Finkl, C. W., and Charlier, R. H. (2003). Sustainability of subtropical coastal zones in southeastern Florida: challenges for urbanized coastal environments threatened by development, pollution, water supply, and storm hazards. J. Coast. Res. 19, 934–943.
Fukunaga, Y., Kurahashi, M., Yanagi, K., Yokota, A., and Harayama, S. (2008). Acanthopleuribacter pedis gen. nov., sp. nov., a marine bacterium isolated from a chiton, and description of Acanthopleuribacteraceae fam. nov., Acanthopleuribacterales ord. nov., Holophagaceae fam. nov., Holophagales ord. nov. and Holophagae class. Int. J. Syst. Evol. Microbiol. 58, 2597–2601. doi: 10.1099/ijs.0.65589-0
Gignoux-Wolfsohn, S. A., Precht, W. F., Peters, E. C., Gintert, B. E., and Kaufman, L. S. (2020). Ecology, histopathology, and microbial ecology of a white-band disease outbreak in the threatened staghorn coral Acropora cervicornis. Dis. Aquat. Organ. 137, 217–237. doi: 10.3354/dao03441
Gintert, B. E., Precht, W. F., Fura, R., Rogers, K., Rice, M., Precht, L. L., et al. (2019). Regional coral disease outbreak overwhelms impacts from a local dredge project. Environ. Monit. Assess. 191:630. doi: 10.1007/s10661-019-7767-7
Godwin, S., Bent, E., Borneman, J., and Pereg, L. (2012). The role of coral-associated bacterial communities in Australian subtropical white syndrome of Turbinaria mesenterina. PLoS One 7:e44243. doi: 10.1371/journal.pone.0044243
Heres, M. M., Farmer, B. H., Elmer, F., and Hertler, H. (2021). Ecological consequences of stony coral tissue loss disease in the turks and caicos Islands. Coral Reefs 40, 609–624. doi: 10.1007/s00338-021-02071-4
Kassambara, A., Kosinski, M., and Biecek, P. (2021). survminer: Drawing Survival Curves Using “ggplot2”. R Package Version 0.4.9.
Kozich, J. J., Westcott, S. L., Baxter, N. T., Highlander, S. K., and Schloss, P. D. (2013). Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 79, 5112–5120. doi: 10.1128/AEM.01043-13
Laas, P., Ugarelli, K., Absten, M., Boyer, B., Briceño, H., and Stingl, U. (2021). Composition of prokaryotic and eukaryotic microbial communities in waters around the Florida reef tract. Microorganisms 9:1120. doi: 10.3390/microorganisms9061120
Landsberg, J. H., Kiryu, Y., Peters, E. C., Wilson, P. W., Perry, N., Waters, Y., et al. (2020). Stony coral tissue loss disease in Florida is associated with disruption of host–zooxanthellae physiology. Front. Mar. Sci. 7:1090. doi: 10.3389/fmars.2020.576013
Lawson, C. A., Raina, J. B., Kahlke, T., Seymour, J. R., and Suggett, D. J. (2018). Defining the core microbiome of the symbiotic dinoflagellate. Symbiodinium. Environ. Microbiol. Rep. 10, 7–11. doi: 10.1111/1758-2229.12599
Lirman, D., and Fong, P. (2007). Is proximity to land-based sources of coral stressors an appropriate measure of risk to coral reefs? An example from the Florida reef tract. Mar. Pollut. Bull. 54, 779–791. doi: 10.1016/j.marpolbul.2006.12.014
MacKnight, N. J., Cobleigh, K., Lasseigne, D., Chaves-Fonnegra, A., Gutting, A., Dimos, B., et al. (2021). Microbial dysbiosis reflects disease resistance in diverse coral species. Commun. Biol. 4:679. doi: 10.1038/s42003-021-02163-5
Martin, B. D., Witten, D., and Willis, A. D. (2020). Modeling microbial abundances and dysbiosis with beta-binomial regression. Ann. Appl. Stat. 14, 94–115. doi: 10.1214/19-AOAS1283
Martinez Arbizu, P. (2020). pairwiseAdonis: Pairwise Multilevel Comparison Using Adonis. R Package Version 0.4.
Meiling, S. S., Muller, E. M., Lasseigne, D., Rossin, A., Veglia, A. J., MacKnight, N., et al. (2021). Variable species responses to experimental stony coral tissue loss disease (SCTLD) exposure. Front. Mar. Sci. 8:670829. doi: 10.3389/fmars.2021.670829
Meiling, S., Muller, E. M., Smith, T. B., and Brandt, M. E. (2020). 3D photogrammetry reveals dynamics of stony coral tissue loss disease (SCTLD) lesion progression across a thermal stress event. Front. Mar. Sci. 7:1128. doi: 10.3389/fmars.2020.597643
Meyer, J. L., Castellanos-Gell, J., Aeby, G. S., Häse, C. C., Ushijima, B., and Paul, V. J. (2019). Microbial community shifts associated with the ongoing stony coral tissue loss disease outbreak on the Florida reef tract. Front. Microbiol. 10:2244. doi: 10.3389/fmicb.2019.02244
Miller, A. W., and Richardson, L. L. (2011). A meta-analysis of 16S rRNA gene clone libraries from the polymicrobial black band disease of corals. FEMS Microbiol. Ecol. 75, 231–241. doi: 10.1111/j.1574-6941.2010.00991.x
Miller, M. W., Karazsia, J., Groves, C. E., Griffin, S., Moore, T., Wilber, P., et al. (2016). Detecting sedimentation impacts to coral reefs resulting from dredging the port of Miami, Florida USA. PeerJ 4:e2711. doi: 10.7717/peerj.2711
Muller, E. M., Sartor, C., Alcaraz, N. I., and van Woesik, R. (2020). Spatial epidemiology of the stony-coral-tissue-loss disease in Florida. Front. Mar. Sci. 7:163. doi: 10.3389/fmars.2020.00163
Neely, K. L., Macaulay, K. A., Hower, E. K., and Dobler, M. A. (2020). Effectiveness of topical antibiotics in treating corals affected by stony coral tissue loss disease. PeerJ 8:9289. doi: 10.7717/peerj.9289
NOAA (2018). Stony Coral Tissue Loss Disease Case Definition. Washington, DC: NOAA, Silver Spring, 1–10.
Noonan, K. R., and Childress, M. J. (2020). Association of butterflyfishes and stony coral tissue loss disease in the Florida keys. Coral Reefs 39, 1581–1590. doi: 10.1007/s00338-020-01986-8
Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P. R., O’Hara, R., et al. (2015). vegan: Community Ecology Package. R Package Version 2.0-10.
Paradis, E., and Schliep, K. (2019). ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35, 526–528. doi: 10.1093/bioinformatics/bty633
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Rosales, S. M., Clark, A. S., Huebner, L. K., Ruzicka, R. R., and Muller, E. M. (2020). Rhodobacterales and Rhizobiales are associated with stony coral tissue loss disease and its suspected sources of transmission. Front. Microbiol. 11:681. doi: 10.3389/fmicb.2020.00681
Rosenau, N. A., Gignoux-Wolfsohn, S., Everett, R. A., Miller, A. W., Minton, M. S., and Ruiz, G. M. (2021). Considering commercial vessels as potential vectors of stony coral tissue loss disease. Front. Mar. Sci. 8:709764. doi: 10.3389/fmars.2021.709764
Roth, L., Kramer, P., Doyle, E., and O’Sullivan, C. (2020). Caribbean SCTLD Dashboard. Available online at: www.agrra.org. (accessed March 6, 2021)
Schneider, C. A., Rasband, W. S., and Eliceiri, K. W. (2012). NIH image to imagej: 25 years of image analysis. Nat. Methods 9, 671–675. doi: 10.1038/nmeth.2089
Séré, M. G., Tortosa, P., Chabanet, P., Turquet, J., Quod, J. P., and Schleyer, M. H. (2013). Bacterial communities associated with porites white patch syndrome (PWPS) on three western Indian ocean (WIO) coral reefs. PLoS One 8:e83746. doi: 10.1371/journal.pone.0083746
Sharp, W. C., Shea, C. P., Maxwell, K. E., Muller, E. M., and Hunt, J. H. (2020). Evaluating the small-scale epidemiology of the stony-coral-tissue-loss-disease in the middle Florida Keys. PLoS One 15:e0241871. doi: 10.1371/journal.pone.0241871
Shieh, W. Y., Liu, T. Y., Lin, S. Y., Jean, W. D., and Chen, J. S. (2008). Simiduia agarivorans gen. nov., sp. nov., a marine, agarolytic bacterium isolated from shallow coastal water from Keelung, Taiwan. Int. J. Syst. Evol. Microbiol. 58, 895–900. doi: 10.1099/ijs.0.65371-0
Shilling, E. N., Combs, I. R., and Voss, J. D. (2021). Assessing the effectiveness of two intervention methods for stony coral tissue loss disease on Montastraea cavernosa. Sci. Rep. 11:8566. doi: 10.1038/s41598-021-86926-4
Shore-Maggio, A., Aeby, G. S., and Callahan, S. M. (2018). Influence of salinity and sedimentation on vibrio infection of the hawaiian coral Montipora capitata. Dis. Aquat. Organ. 128, 63–71. doi: 10.3354/dao03213
Sinigalliano, C. D., Enochs, I. C., Stamates, S. J., Jones, P. R., Featherstone, C. M., Gidley, M. L., et al. (2019). Water Quality and Coral Reef Monitoring Along the Southeast Florida Coast. NOAA Tech. Report, OAR-AOML-47. 164.
Spadafore, R., Fura, R., Precht, W. F., and Vollmer, S. V. (2021). Multi-variate analyses of coral mortality from the 2014–2015 stony coral tissue loss disease outbreak off miami-dade county, Florida. Front. Mar. Sci. 8:1–39. doi: 10.3389/fmars.2021.723998
Spring, S., Scheuner, C., Göker, M., and Klenk, H.-P. (2015). A taxonomic framework for emerging groups of ecologically important marine gammaproteobacteria based on the reconstruction of evolutionary relationships using genome-scale data. Front. Microbiol. 6:281. doi: 10.3389/fmicb.2015.00281
Staley, C., Kaiser, T., Gidley, M. L., Enochs, I. C., Jones, P. R., Goodwin, K. D., et al. (2017). Differential impacts of land-based sources of pollution on the microbiota of southeast Florida coral reefs. Appl. Environ. Microbiol. 83, e03378-16. doi: 10.1128/AEM.03378-16
Studivan, M. S. (2021). mstudiva/SCTLD-Sediment-Transmission: Stony Coral Tissue Loss Disease (SCTLD) Sediment Transmission Version 1.0.0. doi: 10.5281/zenodo.5703024
Sunagawa, S., Desantis, T. Z., Piceno, Y. M., Brodie, E. L., Desalvo, M. K., Voolstra, C. R., et al. (2009). Bacterial diversity and white plague disease-associated community changes in the caribbean coral Montastraea faveolata. ISME J 3, 512–521. doi: 10.1038/ismej.2008.131
Thurber, R. V., Mydlarz, L. D., Brandt, M., Harvell, D., Weil, E., Raymundo, L., et al. (2020). Deciphering coral disease dynamics: integrating host, microbiome, and the changing environment. Front. Ecol. Evol. 8:575927. doi: 10.3389/fevo.2020.575927
Ushijima, B., Meyer, J. L., Thompson, S., Pitts, K., Marusich, M. F., Tittl, J., et al. (2020). Disease diagnostics and potential coinfections by Vibrio coralliilyticus during an ongoing coral disease outbreak in Florida. Front. Microbiol. 11:2682. doi: 10.3389/fmicb.2020.569354
Vargas-Ángel, B., Peters, E. C., Kramarsky-Winter, E., Gilliam, D. S., and Dodge, R. E. (2007). Cellular reactions to sedimentation and temperature stress in the caribbean coral Montastraea cavernosa. J. Invertebr. Pathol. 95, 140–145. doi: 10.1016/j.jip.2007.01.003
Vargas-Ángel, B., Riegl, B., Gilliam, D. S., and Dodge, R. E. (2006). An experimental histopathological rating scale of sedimentation stress in the caribbean coral Montastraea cavernosa. 1168–1173. Available online at: https://nsuworks.nova.edu/occ_facpresentations/49 (accessed May 12, 2020).
Walker, B. K., Turner, N. R., Noren, H. K. G., Buckley, S. F., and Pitts, K. A. (2021). Optimizing stony coral tissue loss disease (SCTLD) intervention treatments on Montastraea cavernosa in an endemic zone. Front. Mar. Sci. 8:666224. doi: 10.3389/fmars.2021.666224
Walker, B., Gilliam, D., Dodge, R., and Walczak, J. (2012). Dredging and shipping impacts on southeast florida coral reefs. 9–13. Available online at: https://nsuworks.nova.edu/occ_facpresentations (accessed September 29, 2021).
Walton, C. J., Hayes, N. K., and Gilliam, D. S. (2018). Impacts of a regional, multi-year, multi-species coral disease outbreak in Southeast Florida. Front. Mar. Sci. 5:323. doi: 10.3389/fmars.2018.00323
Welsh, R. M., and Thurber, R. V. (2016). Bacterial predators in host microbiomes: halobacteriovorax bacteria are marine predators that drive the dynamics of their host microbiomes, in turn modulating the health of their animal hosts. Microbe 11, 61–67. doi: 10.1128/microbe.11.61.1
Welsh, R. M., Zaneveld, J. R., Rosales, S. M., Payet, J. P., Burkepile, D. E., and Thurber, R. V. (2016). Bacterial predation in a marine host-associated microbiome. ISME J 10, 1540–1544. doi: 10.1038/ismej.2015.219
Work, T. M., Weatherby, T. M., Landsberg, J. H., Kiryu, Y., Cook, S. M., and Peters, E. C. (2021). Viral-like particles are associated with endosymbiont pathology in Florida corals affected by stony coral tissue loss disease. Front. Mar. Sci. 8:750658. doi: 10.3389/fmars.2021.750658
Yilmaz, P., Parfrey, L. W., Yarza, P., Gerken, J., Pruesse, E., Quast, C., et al. (2014). The SILVA and “all-species living tree project (LTP)” taxonomic frameworks. Nucleic Acids Res. 42, D643–D648. doi: 10.1093/nar/gkt1209
Keywords: disease transmission, disease vector, ballast water, histology, 16S, microbial communities, disease reservoir, sedimentation
Citation: Studivan MS, Rossin AM, Rubin E, Soderberg N, Holstein DM and Enochs IC (2022) Reef Sediments Can Act As a Stony Coral Tissue Loss Disease Vector. Front. Mar. Sci. 8:815698. doi: 10.3389/fmars.2021.815698
Received: 15 November 2021; Accepted: 17 December 2021;
Published: 13 January 2022.
Edited by:
Christina A. Kellogg, United States Geological Survey (USGS), United StatesReviewed by:
William F. Precht, Dial Cordy and Associates, Inc., United StatesRyan McMinds, University of South Florida, United States
Copyright © 2022 Studivan, Rossin, Rubin, Soderberg, Holstein and Enochs. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael S. Studivan, studivanms@gmail.com
 Michael S. Studivan
Michael S. Studivan