- 1Biodiversity Research Center, Academia Sinica, Taipei, Taiwan
- 2Penghu Marine Biology Research Center, Fisheries Research Institute, Penghu, Taiwan
- 3Faculty of Science and Marine Environment, Universiti Malaysia Terengganu, Kuala Nerus, Malaysia
- 4Tropical Marine Science Institute, National University of Singapore, Singapore, Singapore
- 5Department of Marine Biology and Environmental Sciences, Faculty of Agriculture, University of Miyazaki, Miyazaki, Japan
- 6Department of Life Science, National Taiwan Normal University, Taipei, Taiwan
- 7Department of Life Science, Tunghai University, Taichung, Taiwan
The east Taiwan Strait is largely fringed by sandy and muddy habitats. However, a massive algal reef made of crustose coralline algae has been found along the coast off Taoyuan city in northwestern Taiwan. The porous structure of Taoyuan Algal Reef harbors high abundance and diversity in marine organisms, including the ferocious reef crab, Eriphia ferox. Such a pivotal geographic location and unique ecological features make Taoyuan Algal Reef a potential stepping stone connecting biotic reefs in the east Taiwan Strait, South China Sea to the south, and even the high latitude of Japan to the north. In this study, we examined the population connectivity and historical demography of E. ferox by analyzing mitochondrial cytochrome oxidase I (COI) fragments of 317 individuals sampled from 21 localities in the northwestern Pacific. Our analyses of haplotype network and pairwise FST comparisons revealed a lack of phylogeographical structure among E. ferox populations, implying the existence of a migration corridor connecting the South and East China Seas through the east Taiwan Strait. Multiple lines of evidence, including significant values in neutrality tests, unimodally shaped mismatch distributions, and Bayesian skyline plots elucidated the rapid population growth of E. ferox following the sea-level rise after Last Glacial Maximum (ca. 2–10 Ka). Such demographic expansion in E. ferox coincided with the time when Taoyuan Algal Reef started to build up around 7,500 years ago. Coalescent migration analyses further indicated that the large and continuous E. ferox population exclusively found in Datan Algal Reef, the heart of Taoyuan Algal Reef, was a source population exporting migrants both northward and southward to the adjacent populations. The bidirectional gene flow should be attributed to larval dispersal by ocean currents and secondary contact due to historical population expansion. Instead of serving as a stepping stone, our results support that Taoyuan Algal Reef is an essential population source for biotic reef-associated species along the east Taiwan Strait, and highlight the importance of conserving such a unique ecosystem currently threatened by anthropogenic development.
Introduction
The Taiwan Strait, bounded between southeastern China and Taiwan, is a shallow passage bridging the South China Sea (SCS) and the East China Sea (ECS). Situated at the tectonic and geographic junction along the coast of eastern Eurasia, historical closure and opening of the Taiwan Strait were affected profoundly by sea-level fluctuations during the last glacial cycles. The interplay of paleoclimate and distinct topographical features of the Taiwan Strait has subsequently shaped the distribution patterns and genetic diversity of extant marine organisms in the northwestern Pacific (Ni et al., 2014 and references therein).
During glacial periods throughout the Pleistocene (2.6 Ma–11.7 ka), the recurrent desiccation and exposure of land mass in the Taiwan Strait culminated in transient disconnections between SCS and ECS basins (Wang, 1999; Kimura, 2000; Ludt and Rocha, 2015). Consequently, gene flow between the two marginal seas was blocked and divergence in marine organisms occurred. Distinct lineages were found to be concordant with the two sea basins, which should have served as independent refugia during glaciations (Ni et al., 2014; Wang et al., 2017; Qu et al., 2021 and references therein). As glaciers receded at the end of the Last Glacial Maximum (LGM, 17–19 Ka), sea level rose drastically and flooded the entire Taiwan Strait, establishing the coastlines and ocean current regimes that we know today (Li et al., 2014). The emerging waterway conceptually restores oceanic connectivity, and should facilitate genetic homogenization in marine organisms along the Taiwan Strait. Surprisingly, studies of moon turban snail Lunella granulata (Gmelin, 1791) and Portuguese oyster Magallana (formerly Crassostrea) angulata (Lamarck, 1819) revealed a significant subdivision in populations between the Chinese and Taiwanese coasts of the Taiwan Strait (Chiu et al., 2013; Hsiao et al., 2016). As our current understanding of marine phylogeography in the northwestern Pacific exclusively relies on studies conducted along the Chinese coast (reviewed in Ni et al., 2014), such cross-strait divergence highlights the necessity to explore the role of the uninvestigated west coast of Taiwan.
The west coast of Taiwan is mainly composed of sandy/muddy habitats (Chen and Shashank, 2009). Hard substrates constructed by reef-building corals appear only at the northern/southern tips and east coast of Taiwan (Dai and Cheng, 2020). The only biotic reef along the west coast of Taiwan is a massive algal reef built by crustose coralline algae (CCA) in the intertidal zone to a depth of around 10 m along a 27 km stretch of the coastline off Taoyuan city (Liou et al., 2017). The porous algal reef has an ancient origin, dated ca. 7,500 BP (Dai et al., 2009). Its complex structures provide shelter for diverse marine organisms, including crustaceans, polychaetes, sipunculans, and the largest population of the critically-endangered caryophylliid coral Polycyathus chaishanensis (Lin et al., 2012) (Liou et al., 2017; Kuo et al., 2020) in Taiwan. In addition, Taoyuan Algal Reef is reported to share specific marine species like Oulastrea crispata (Lamarck, 1816) and Epinephelus quoyanus (Valenciennes, 1830) with Penghu archipelago, which comprises as many as 90 islands and islets in the southeastern Taiwan Strait. These organisms are not found elsewhere in the Taiwan Strait (Chen, 2017). Its ancient origin and pivotal geographic location, supplemented with the relatively high diversity of marine benthos and species connectivity with the Penghu Islands, make Taoyuan Algal Reef a potential stepping stone connecting biotic reefs in the east Taiwan Strait (Chen, 2017; Kuo et al., 2020).
To examine the hypothesis of Taoyuan Algal Reef as a stepping stone, we investigated the genetic structure of the ferocious reef crab Eriphia ferox (Koh and Ng, 2008), a species recognized as Eriphia smithii (MacLeay, 1836) until 2008 (Koh and Ng, 2008). The relatively newly described E. ferox has a widespread distribution along the hard substrates of the northwestern Pacific, ranging from southern Japan in the north to the southern SCS in the south (Koh and Ng, 2008). In 2019, a significantly large and continuous population of E. ferox was found at Datan (Li et al., 2021), which is located within the Taoyuan Algal Reef. These biogeographical and ecological characteristics make E. ferox an ideal model organism to test the stepping stone hypothesis. We used the mitochondrial (mt) cytochrome oxidase I (COI) fragment to elucidate the genetic structure, direction of gene flow, and historical demography and connectivity of E. ferox. The findings of the present study should broaden our current understanding of marine phylogeography in the northwestern Pacific.
Materials and Methods
Sampling and DNA Sequencing
In the field, crabs were captured for tissue sampling and released afterward. From each crab, one pereiopod was removed and preserved in 95% ethanol for DNA extraction later. A total of 317 samples were collected from southern Japan, northern/northwestern Taiwan (mainland), the Penghu archipelago (Taiwan), Malaysia, and Singapore (Table 1). Genomic DNA was extracted from pereiopod muscle tissue using a Tissue and Cell Genomic DNA Purification Kit (GeneMark, Taiwan) according to the company’s protocol. A fragment containing the COI gene was amplified with the E. ferox specific primer set ErifeCOI_F (forward primer, 5′- GTA GGA ACT TCA CTA AGA CTA ATT ATT CG-3′) and ErifeCOI_R (reverse primer, 5′- GCA GGG TCA AAG AAG GAT G-3′), which was manually designed in this study. The COI sequence (HM638034.1) by Lai et al. (2014) was used as reference. The volume of the PCR mixture was 25 μL, containing 0.5–1.0 μL of genomic DNA template, 0.5 μL of each primer (10 μM), 12.5 μL of Taq DNA Polymerase Master Mix RED (Ampliqon, Denmark), and 10.5 μL of ddH2O. PCR was performed under the following conditions: initial denaturation at 95°C for 3 min, followed by 40 cycles at 94°C for 30 s, 50–55°C for 40 s, 72°C for 30 s, and a final extension at 72°C for 5 min. The resulting PCR products were sent to the company Genomics (Taipei, Taiwan) for Sanger sequencing.
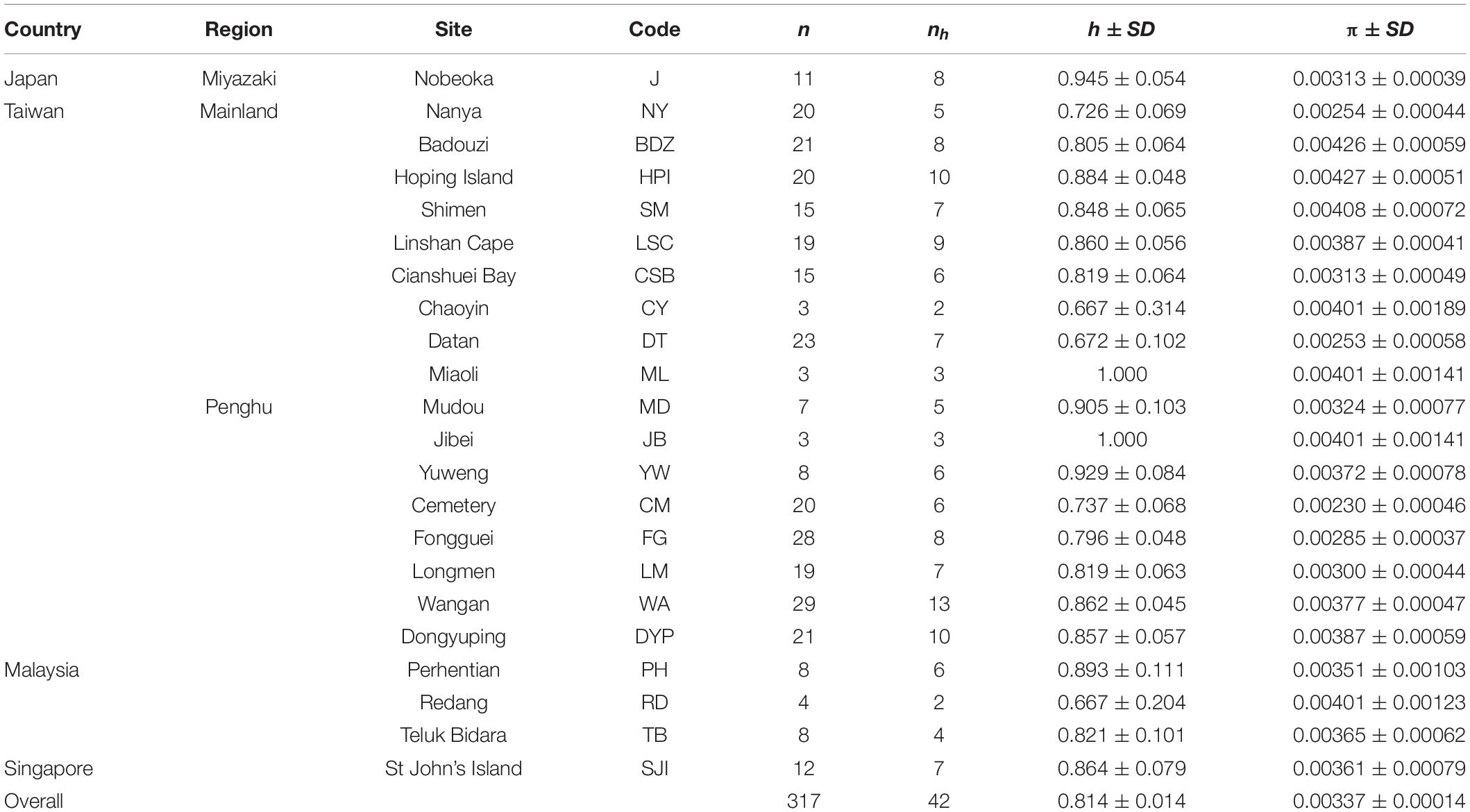
Table 1. Sampling sites, sample size (n), number of haplotypes (nh), haplotype diversity (h ± SD), and nucleotide diversity (π ± SD) from samples of Eriphia ferox collected along the west coast of the Pacific.
Data Analyses
Raw sequences were inspected, trimmed and assembled with the software Geneious version R10 (Biomatters, New Zealand). Cleaned consensus sequence of each sample was imported to MEGA X (Kumar et al., 2018) for sequence alignment using the built-in MUSCLE algorithm. Genetic diversity was evaluated for the number of haplotypes (nh), haplotype diversity (h), and nucleotide diversity (π) for each sampling site using DnaSP 6.12 (Rozas et al., 2017). The haplotype network was illustrated with the median-joining algorithm in POPART (Leigh and Bryant, 2015). Pairwise FST comparisons were calculated between sampling sites, with statistical significance assessed by performing 10,000 permutations using Arlequin 3.5.2 (Excoffier and Lischer, 2010). Statistical significance of FST estimates was subsequently adjusted by Benjamini and Hochberg’s FDR correction (Benjamini and Hochberg, 1995) in R version 3.6.1 (R Core Team, 2013). To investigate regional demographic history, a total of five groups were defined for the analyses: Japan, Taiwan (pooled northern/northwestern Taiwanese populations), Penghu (pooled Penghu populations), Malaysia (pooled Malaysian populations), and Singapore. Signatures of historical demography were inferred from neutrality tests, mismatch distributions, and Bayesian Skyline Plots. For neutrality tests, Tajima’s D and Fu’s Fs were calculated by Arlequin with 10,000 permutations (Tajima, 1989; Fu, 1997), while Ramos-Onsins and Roza’s R2 index (Ramos-Onsins and Rozas, 2002) were calculated using DnaSP. For every geographic population, mismatch distribution was established to test against a model of exponential population growth (Rogers and Harpending, 1992). The validity of the sudden demographic expansion model was examined in Arlequin by computing the Harpending’s raggedness index (rg, Harpending, 1994) and sum of square deviations (SSD) between the observed and expected mismatch distributions. The dynamics of effective population size (Ne) through time were visualized with Bayesian Skyline Plots (BSPs), which were performed using BEAST 2.6.5 (Bouckaert et al., 2019). The HKY model was selected as the best fit model by jModelTest 2.1.10 (Darriba et al., 2012), and a mutation rate of 4.87% per million years for Eriphia (Deli et al., 2019) was applied to a Markov chain Monte Carlo (MCMC) simulation with 100 million iterations and a 10% burn-in. The convergence of simulation and resulting BSPs were examined with Tracer 1.7.2 (Rambaut et al., 2018).
The direction of gene flow was estimated using a Bayesian coalescent approach implemented in MIGRATE 4.4.4 (Beerli et al., 2019), which infers the mutation-scaled effective population size θ (= Neμ for mtDNA data, where Ne is effective population size and μ is mutation rate) and migration rate M (= m/μ, where m is the immigration rate per generation). Based on these parameters, the effective number of migrants per generation was then calculated as Nem = θM (Beerli et al., 2019). Because the distance between Malaysia/Singapore and the remaining populations is too far (>2,000 km Euclidean distance apart), samples of those two regions were excluded from gene flow estimation. Of the remaining populations, six were selected for analysis, which being two from Penghu (Dongyuping and Longmen), three from northwestern Taiwan (Datan, Shimen and Hoping Island), and one from Japan. To perform coalescent migration analyses, a HKY model was applied, and the other priors were set as default. The MCMC simulation consisted of one long chain with 500,000 recorded genealogies after a 10% burn-in, a sampling increment of 100, and a static heating scheme with four chains and temperatures {1.0, 1.5, 3.0, 106}. To test the stepping stone hypothesis, the analyses were conducted with (1) a stepping stone model with symmetric migrations and (2) a migration matrix model specified to allow full migrations between adjacent populations.
Results
Sequence Diversity and Population Differentiation
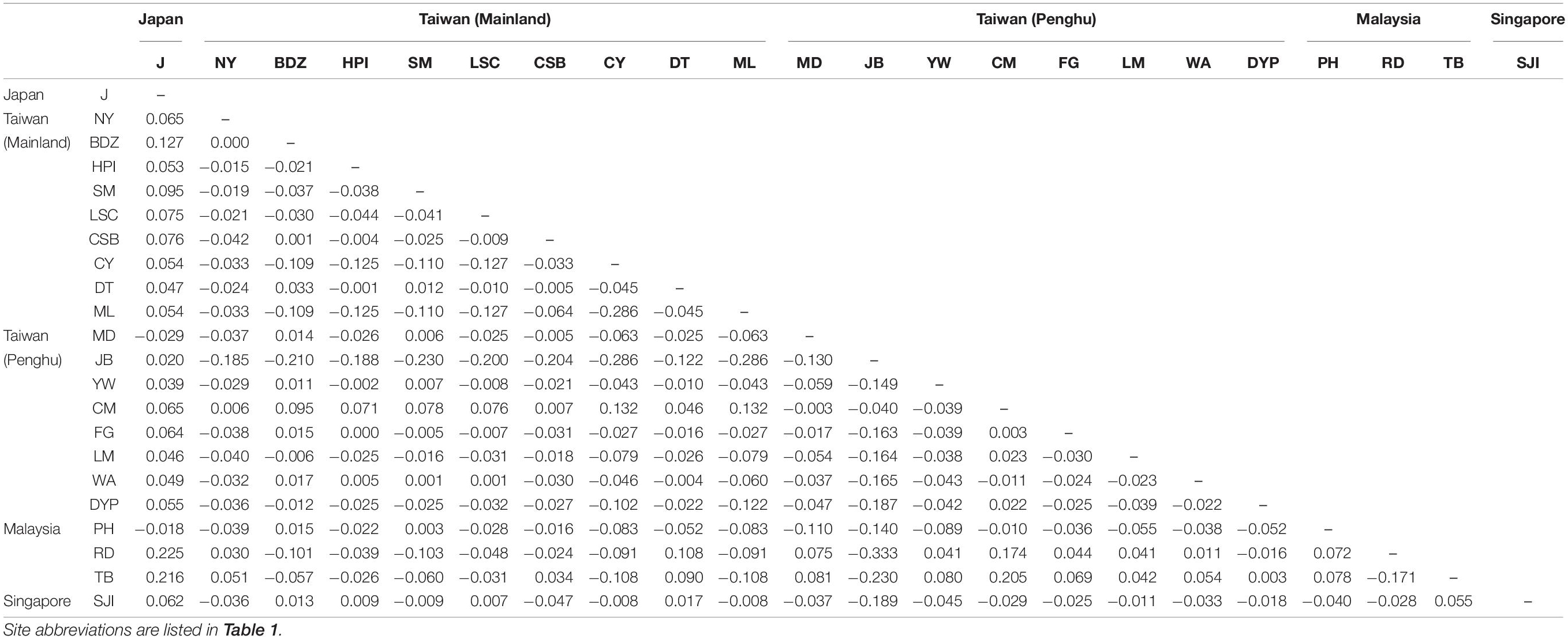
All 317 samples were amplified and sequenced successfully. The length of raw mtCOI sequences ranged from 499 bp to 536 bp. After trimming, 499 bp were retained for the following analyses. Nucleotide composition profiling demonstrated that sequences were A + T-rich (A, 33.43%; T, 27.38%). After alignment, 41 variable sites were detected, predominantly transition substitutions (Ti: Tv = 43.11), 16 of which were phylogenetically informative. A total number of 42 haplotypes were identified, with the highest found in Wangan (Table 1). The relative abundance of the haplotypes in each site is shown in Figure 1. High haplotype diversity (h) was detected in all populations, with a mean value of 0.814 ± 0.014. In contrast, nucleotide diversity (π) was relatively low, with a mean value of 0.00337 ± 0.00014 (Table 1). Median-joining network (MJN) analysis revealed a star-like network, consisting of one major dominant haplotype, from which two second dominant haplotypes and numerous singletons derived. The latter two also contained several derived singletons of their own. All haplotypes differed from each other by only one or two mutation steps (Figure 2). No distinct haplotype clustering with respect to geographical location was detected. Pairwise FST comparisons also suggested low genetic differentiation among E. ferox populations (Table 2), indicating little or no phylogeographical structure in E. ferox.
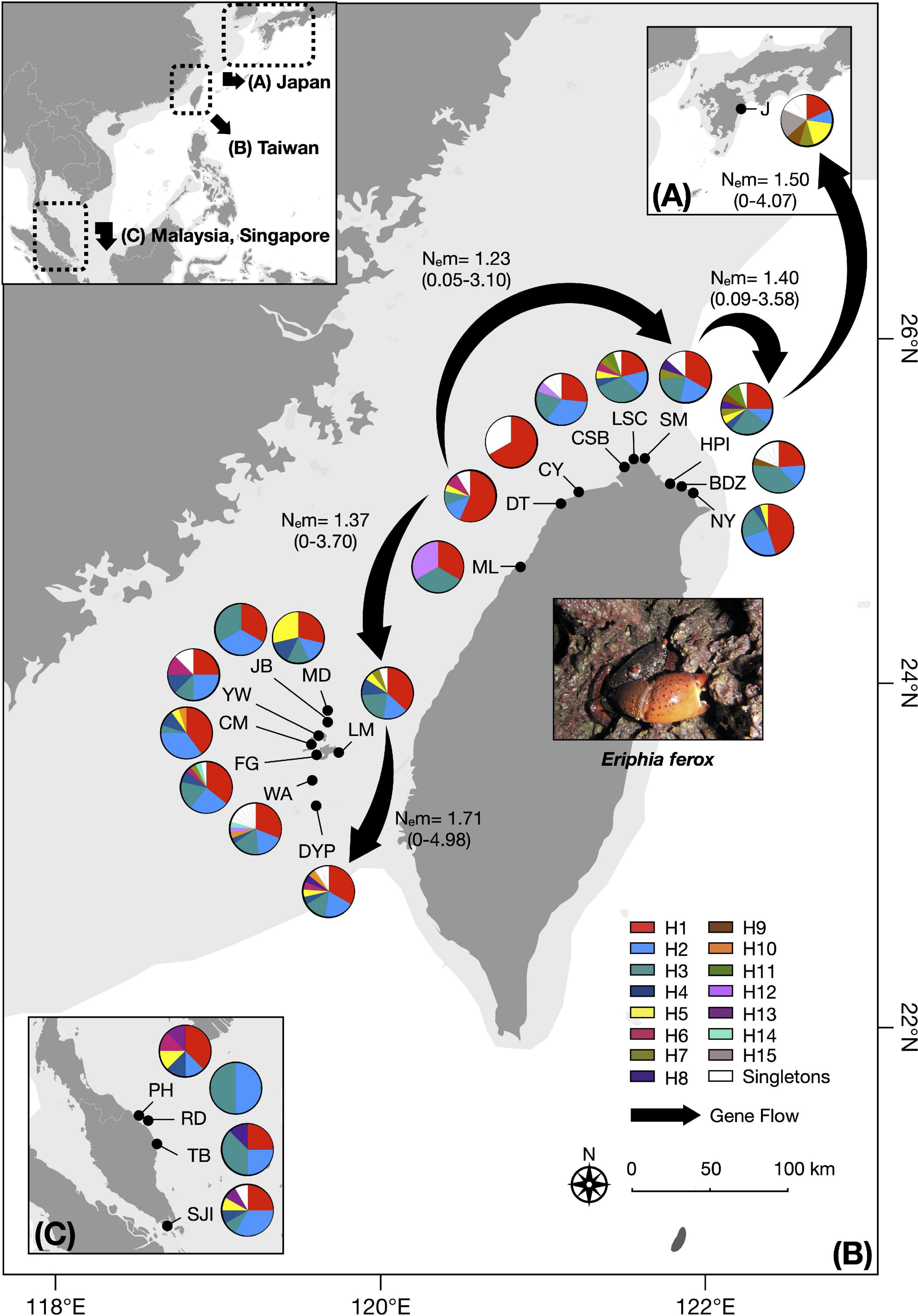
Figure 1. Haplotype distribution and frequency map for Eriphia ferox in (A) Japan, (B) Taiwan, and (C) Malaysia and Singapore. Site abbreviations are listed in Table 1. Different colors refer to different haplotypes. The landmass exposed during the Last Glacial Maximum (Ludt and Rocha, 2015) is shaded light gray. The directions of gene flow estimated between adjacent E. ferox populations are indicated with black arrows. Only six representative populations were examined (J: Japan; HPI: Hoping Island; SM: Shimen; DT: Datan; LM: Longmen; Dongyuping). Gene flows are presented by the effective number of migrants per generation (Nem), with 95% confidence intervals shown in parentheses.
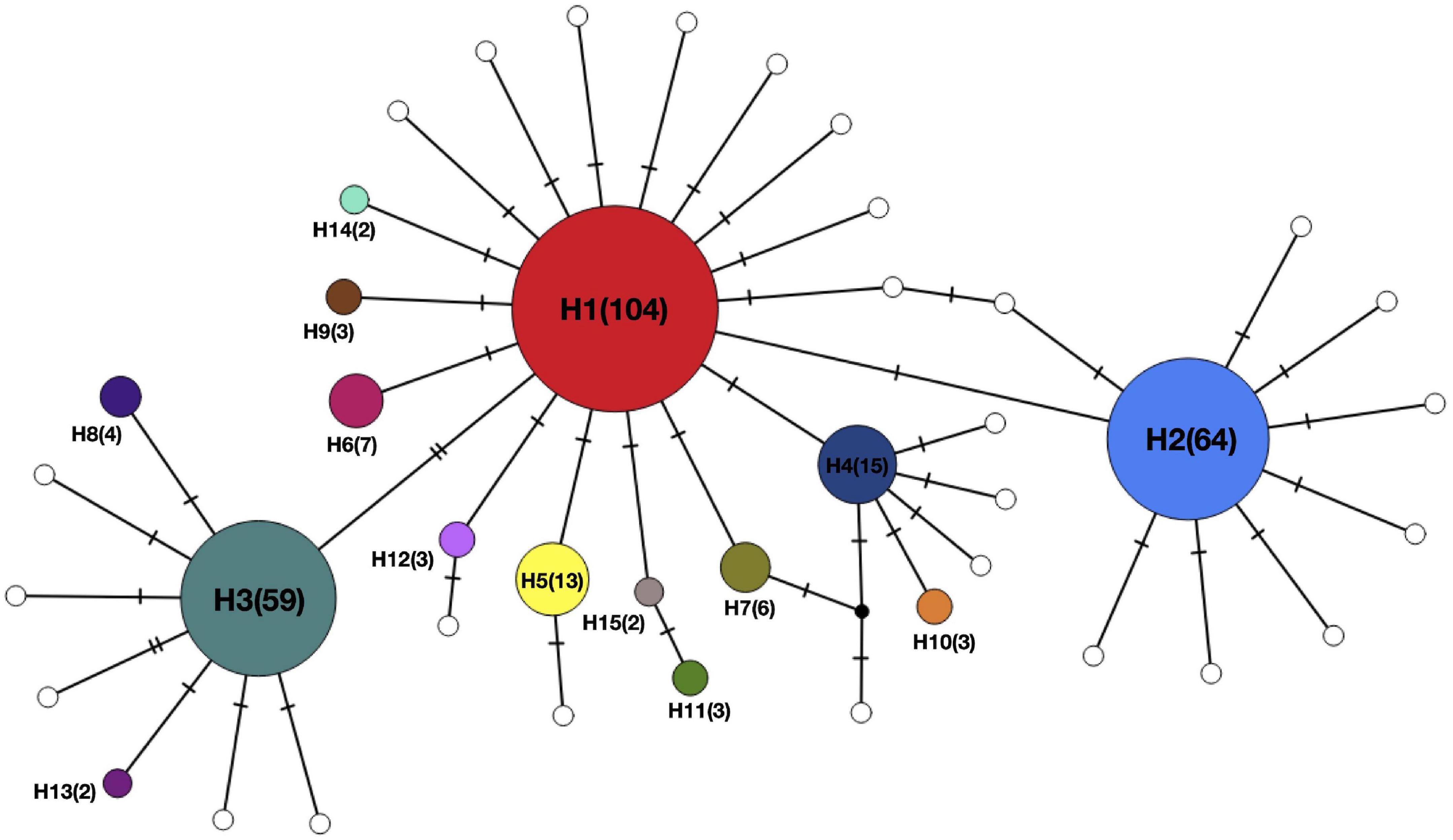
Figure 2. Median-joining network of Eriphia ferox. Each circle represents a unique haplotype. Circle sizes are proportional to haplotype frequencies shown in parentheses. Each slash across a line connecting haplotypes indicates a single mutation step.
Historical Demography and Gene Flow Estimation
Neutrality tests indicated that E. ferox experienced a sudden population expansion, with support from significant negative values of Tajima’s D (−2.08071, p < 0.01) and Fu’s Fs indices (−27.53360, p < 0.001), together with a significantly positive value of Ramo-Onsins and Roza’s R2 index (0.01973, p < 0.05) in a combined analysis of all populations in the northwestern Pacific (Table 3). At the regional scale, results showed that all populations experienced sudden population growth (significant negative D and Fu’s Fs, significant positive R2) except for Malaysia (Table 3). Population expansion in E. ferox was further illustrated by unimodal mismatch distributions (Figure 3). Non-significant SSD statistics and rg indices were observed in populations from Taiwan, Malaysia, and Singapore, implying that the polymorphism pattern in these populations conformed to unimodal frequency distribution, except for Penghu (Table 3). An SSD estimate and rg index were not available for the E. ferox population in Japan due to highly differentiated sequences that Arlequin failed to process. Bayesian skyline plot (BSP) analysis revealed that the initiation of overall E. ferox population growth occurred approximately 10,000 years ago, and the effective population size reached a plateau around 2,000 years ago (Figure 4A). At the regional scale, the effective population sizes of E. ferox remained stable in Japan, Malaysia, and Singapore, probably because the sample sizes of those populations were too small (≤20 individuals each) to estimate fluctuation through time (Grant, 2015). In contrast, populations in Taiwan and Penghu both experienced recent population expansions over the past 20,000 years. The Taiwanese population started expanding roughly 16,000 years ago, and reached a stable effective population size about 6,000 years ago (Figure 4B). Population expansion in Penghu began approximately 10,000 years ago, and saturated ca. 2,000 years ago (Figure 4C). For coalescent migration analyses, the migration matrix model allowing full migrations between adjacent populations (model 2) was selected as the more probable model over the stepping stone model (model 1) using log Bayes factors (LBFs). The log marginal likelihood for model 1 and model 2 were −1134.916 and −1113.828, respectively. The value of LBF was −42.176. Hence, the stepping stone hypothesis was rejected. Our results demonstrated a highly asymmetric gene flow between adjacent E. ferox populations along the east Taiwan Strait (Figure 1). Centered at Datan, gene flow occurred in two directions: one migrated northward with approximately 1.23 effective migrants (Nem) per generation from Datan to Shimen, 1.4 Nem from Shimen to Hoping Island, and then 1.5 Nem from Hoping Island to Japan (Figure 1). The other moved southward with 1.37 Nem from Datan to Longmen, and then 1.71 Nem from Longmen to Dongyuping in Penghu (Figure 1).
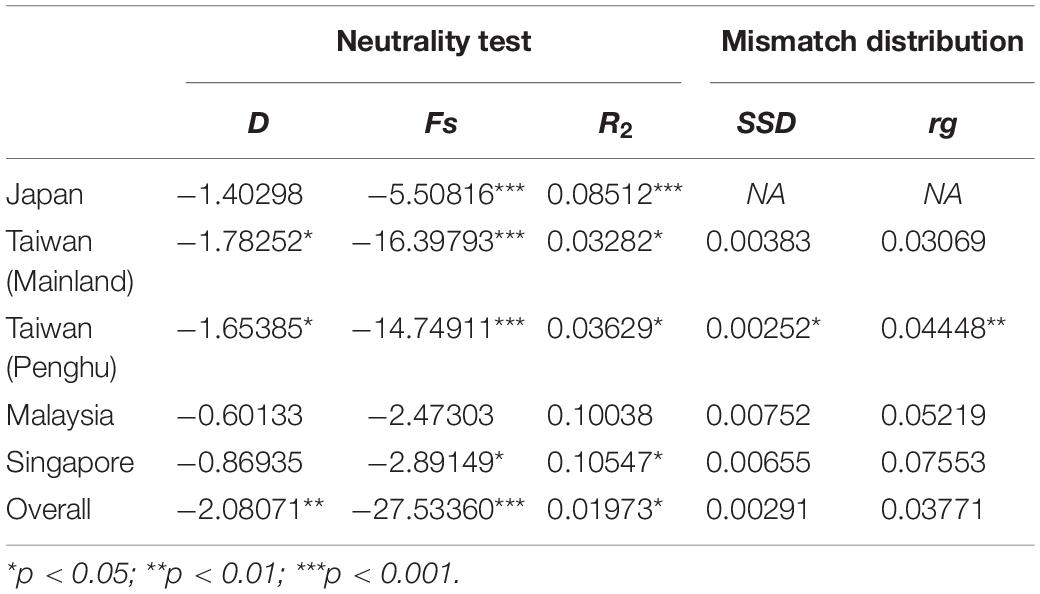
Table 3. Summary statistics of historical demography for Eriphia ferox populations under investigation.
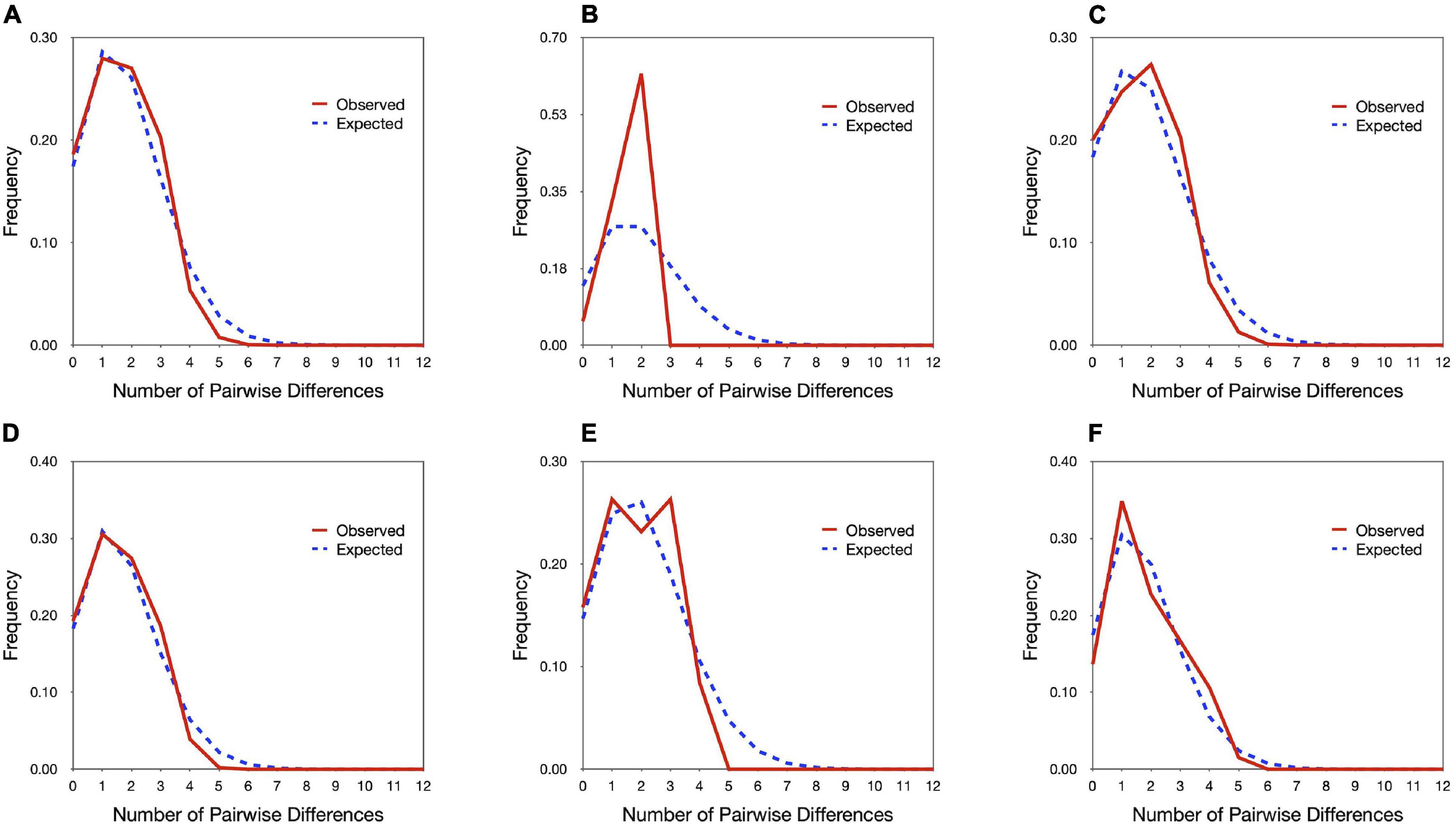
Figure 3. Mismatch distribution of (A) overall Eriphia ferox under investigation; Eriphia ferox populations in (B) Japan, (C) Taiwan (Mainland), (D) Taiwan (Penghu), (E) Malaysia, and (F) Singapore. The solid red line represents observed frequency and the dashed blue line specifies modeled frequency.
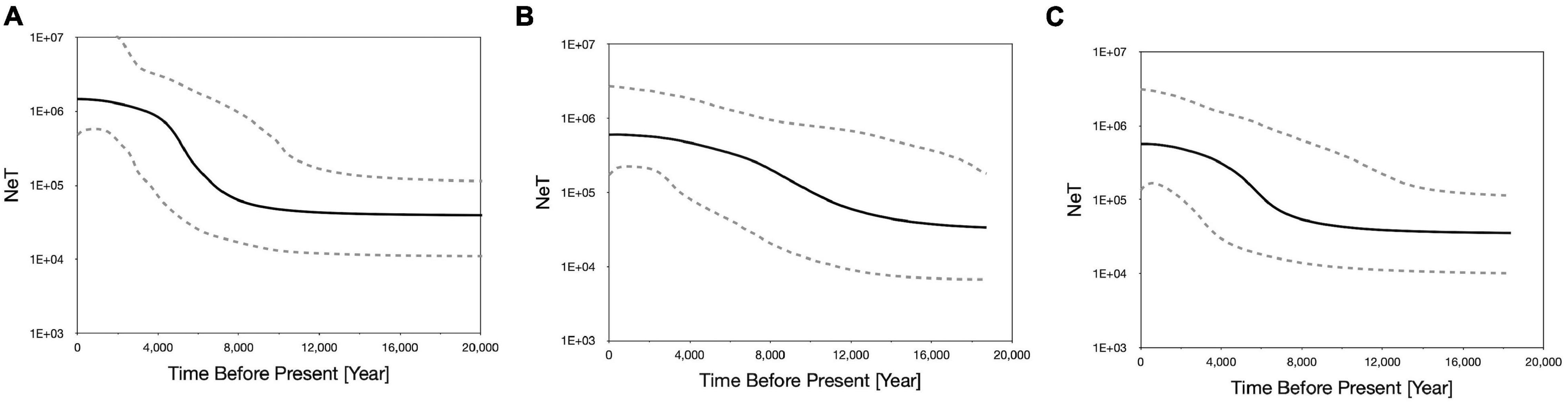
Figure 4. Bayesian skyline plotting of (A) overall Eriphia ferox under investigation; Eriphia ferox populations in (B) Taiwan (Mainland), (C) Taiwan (Penghu). The y-axis refers to the product of effective population size (Ne) and generation time (T). The black line represents the median estimate, and the two dashed lines bound the 95% highest posterior density (HPD) region.
Discussion
The Taiwan Strait plays an important role in shaping marine phylogeography across marginal seas in the northwestern Pacific. Yet, previous broad-scale research in the region preferentially emphasized the west Taiwan Strait (i.e., Chinese coast), leaving the east Taiwan Strait barely explored. In the present study, we analyzed 317 partial mtCOI sequences of the ferocious reef crab Eriphia ferox collected from 21 sites, spanning nearly 4,500 km (Euclidean distance) within the northwestern Pacific. With specific focus on the populations inhabiting the northwestern Taiwan and Penghu archipelago, we unveil the detailed historical connectivity and demography of E. ferox through the east Taiwan Strait. Despite mtCOI reveals only partial information, it is still the benchmark for non-model organisms lack of genetic information, e.g., E. ferox in this study.
Our analyses of the haplotype network and pairwise FST comparisons demonstrated a lack of phylogeographical structure in E. ferox populations, implying the existence of a migration corridor connecting the South and East China Seas (SCS and ECS) through the east Taiwan Strait. Previous studies along the Chinese coast of the west Taiwan Strait have reported significant genetic structures, e.g., mollusks and crustaceans (reviewed in Ni et al., 2014), suggesting that there is no such corridor along the west Taiwan Strait. Our discovery of the corridor along the east Taiwan Strait is probably the first ever reported in this region. Moreover, instead of serving as a stepping stone, our analysis of coalescent migration demonstrated that Taoyuan Algal Reef, the only biotic reef in the west coast of Taiwan, is not just essential, but also the only population source that exports migrants both northward and southward to adjacent populations. Our results indicated that northward gene flow propagated sequentially from Datan to Shimesn, Hoping Island, and then Japan. Southward gene flow extended from Datan to Longmen (northern Penghu) and then Donyuping (southern Penghu). Previous studies indicated that the migration rates approximating one effective migrant per generation that we estimated in this study were sufficient to maintain genetic diversity and population viability (Wang, 2004; Nathan et al., 2017). The migration of benthic invertebrates mostly depends on larval dispersal via ocean currents. The panmictic population structure of E. ferox across the northwestern Pacific observed in our study suggested its potential for long distance dispersal. Although so far there is no estimate of the pelagic larval duration for E. ferox, larvae of its congeneric species E. verrucosa (Forskål, 1775) were reported to last for 49 days, contributing to remarkably long-distance connectivity across the East Atlantic and Mediterranean Sea (Deli et al., 2019).
Based on the information above, we speculated that the lack of migration corridor along the west Taiwan Strait results from the difference in coastal hydrology along the two sides of the Taiwan Strait while the dispersal pattern of E. ferox centered at Datan is shaped by the complex current system in the east Taiwan Strait. In the east Taiwan Strait, the Kuroshio’s branch or loop current flows unidirectionally northward year-round along the west coast of Taiwan through the Penghu channel (Hu et al., 2010). Both seawater temperature and salinity are rather warm and stable in the east Taiwan Strait, providing a smooth pathway for the dispersal of tropical/subtropical marine faunas like E. ferox. The reproduction season of E. ferox is reported to occur from July to October (previously recognized as E. smithii in Tomikawa and Watanabe, 1992), thus the northward gene flow from Datan should be attributed to the conveyance of pelagic larvae by Kuroshio’s loop current along the east Taiwan Strait in summer. Conversely, the southward gene flow originating from Datan is thus more likely to arise from secondary contact due to post-LGM population expansion. The dispersal direction from Taiwan to Penghu is congruent with our BSP estimation that population expansion in Taiwan occurred before the population expansion in Penghu. Contrarily, surface currents in the west Taiwan Strait are influenced more substantially by regional monsoons, which reverse in direction on a semiannual basis. In summer, the extension of the South China Sea Warm Current (SCSWC), together with the Yuedong Coastal Current driven by the southwestern monsoon, flows northward along the Chinese coast in the west Taiwan Strait (Hu et al., 2010). In winter, the counter-wind SCSWC tends to be weaker or even disappears in the upper layer and current direction changes from northward to southwestward when the southwestward Zhemin Coastal Current (ZCC) propelled by the northeastern monsoon dominates in the region. Moreover, the ZCC brings in diluted cold water originating from the Changjiang estuary (Hu et al., 2010), creating a steep sea surface temperature (SST) gradient, which in turn forms a dispersal barrier in the west Taiwan Strait and contributes to the genetic differentiation of lineages between the SCS and ECS along the Chinese coast (Ni et al., 2012; Xu and Chu, 2012; Wang et al., 2017; Qu et al., 2021).
We elucidated a rapid population growth following a population bottleneck throughout E. ferox demographic history, inferring from the combination of high mean haplotype diversity and low mean nucleotide diversity (Grant and Bowen, 1998; Avise, 2000). The recent or sudden population growth of E. ferox is supported by our demographic analyses of neutrality test and mismatch distribution. Bayesian skyline plots (BSPs) further indicated that the E. ferox population expanded during the post-LGM period, when a rapid rise in sea level submerged the Taiwan Strait and stabilized to the present level approximately 7,000 years ago (Kimura, 2000; Li et al., 2014). Such population expansion of E. ferox occurred first at Taiwan and followed at Penghu, possibly reflecting the direction of seawater intrusion into the east Taiwan Strait after the end of the LGM. The demographic expansion of E. ferox also coincided with the time that Taoyuan Algal Reef started to form around 7,500 years ago. Hence, E. ferox might have colonized Taoyuan Algal Reef since then and gradually grew to its present substantial population size.
In conclusion, our phylogeographical study of E. ferox based on mtCOI sequences identifies a potential migration corridor connecting SCS and ECS through the east Taiwan Strait. Instead of serving as a stepping stone, we showed that Taoyuan Algal Reef, represented by Datan Algal Reef, is confirmed as an essential and also the only population source along this migration pathway for reef-associated biotic species in the northwestern Pacific. To complete the picture of marine phylogeography in this region, it is necessary to involve more reef-associated marine taxa with different life histories in future studies. The gaps between Penghu and Malaysian/Singapore populations should be filled by increasing the number of sampling sites in between. The pattern of demographic history and genetic connectivity of E. ferox described in this study should also be compared by additional nuclear markers and multi-locus genetic approaches; e.g., microsatellites or single nucleotide polymorphisms (SNP). Still, our study, along with other pioneer research (Kuo et al., 2020; Heard et al., 2021), demonstrates the uniqueness and pivotal role of the Taoyuan Algal Reef along the east Taiwan Strait. Such a distinctive ecosystem with an ancient origin is now confronting catastrophic disturbance. The construction of an industrial port and terminals to receive and store liquefied natural gas has undermined the integrity of the algal reefs, leaving the survival of its marine inhabitants in jeopardy (Liou et al., 2017; Silver, 2018). As this ecosystem is severely understudied to date, further research is urgently needed to reduce the impacts of construction and enable conservation management strategies that ensure the sustainability of Taoyuan Algal Reef.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, OK509858-OK510174.
Author Contributions
CAC conceived the project. M-HC, Y-YH, and CAC coordinated the experiments and wrote the manuscript. M-HC, Y-YH, B-YH, HH, JL, MN, and HF assisted in wild crab collections. M-HC and Y-YH contributed to data analyses and interpretation of the results. All authors provided feedback and approved the submitted version.
Funding
This study was funded by the Forestry Bureau, Council of Agriculture, Executive Yuan of Taiwan, through grant 107RR-2.2.6-1.1-COA-001-01(Z).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the Forestry Bureau, Council of Agriculture, Executive Yuan of Taiwan, for financial support. We also thank Ching-Yu Liou for providing the photo of Eriphia ferox. Samples from Malaysia were permitted from Department of Marine Park Malaysia [JTLM 630-7Jld. 9(9)]. Samples from Singapore were permitted from the National Parks Board, Singapore (NP/RP17-062).
References
Avise, J. C. (2000). Phylogeography: The History And Formation Of Species. Cambridge: Harvard university press. doi: 10.2307/j.ctv1nzfgj7
Beerli, P., Mashayekhi, S., Sadeghi, M., Khodaei, M., and Shaw, K. (2019). Population genetic inference with MIGRATE. Curr. Protoc. Bioinform. 68:e87. doi: 10.1002/cpbi.87
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Bouckaert, R., Vaughan, T. G., Barido-Sottani, J., Duchêne, S., Fourment, M., Gavryushkina, A., et al. (2019). BEAST 2.5: an advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 15:e1006650. doi: 10.1371/journal.pcbi.1006650
Chen, C. A. (2017). The migration of global marine creatures The stepping-stone for the migration pathway in the Taiwan Strait. Sci. Mon. 573, 692–699.
Chen, C. A., and Shashank, K. (2009). Taiwan as a connective stepping-stone in the Kuroshio triangle and the conservation of coral ecosystems under the impacts of climate change. Kuroshio Sci. 3, 15–22.
Chiu, Y.-W., Bor, H., Tan, M.-S., Lin, H.-D., and Jean, C.-T. (2013). Phylogeography and genetic differentiation among populations of the moon turban snail Lunella granulata Gmelin, 1791 (Gastropoda: Turbinidae). Int. J. Mol. Sci. 14, 9062–9079. doi: 10.3390/ijms14059062
Dai, C. F., and Cheng, Y. R. (2020). Corals of Taiwan Vol.1: Scleractinia Fauna. Taipei: Owl Publishing House Co., Ltd.
Dai, C. F., Wang, S. W., and Chang, J. S. (2009). Taoyuan Guanyin Algal Reef Ecological Guide Manual (A). Taiwan: CPC Corporation, 98.
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9, 772–772. doi: 10.1038/nmeth.2109
Deli, T., Kiel, C., and Schubart, C. D. (2019). Phylogeographic and evolutionary history analyses of the warty crab Eriphia verrucosa (Decapoda, Brachyura, Eriphiidae) unveil genetic imprints of a late Pleistocene vicariant event across the Gibraltar Strait, erased by postglacial expansion and admixture among refugial lineages. BMC Evol. Biol. 19:105. doi: 10.1186/s12862-019-1423-2
Excoffier, L., and Lischer, H. E. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
Fu, Y.-X. (1997). Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147, 915–925. doi: 10.1093/genetics/147.2.915
Grant, W., and Bowen, B. W. (1998). Shallow population histories in deep evolutionary lineages of marine fishes: insights from sardines and anchovies and lessons for conservation. J. Heredity 89, 415–426. doi: 10.1093/jhered/89.5.415
Grant, W. S. (2015). Problems and cautions with sequence mismatch analysis and Bayesian skyline plots to infer historical demography. J. Heredity 106, 333–346. doi: 10.1093/jhered/esv020
Harpending, H. (1994). Signature of ancient population growth in a low-resolution mitochondrial DNA mismatch distribution. Hum. Biol. 66, 591–600.
Heard, J., Tung, W. C., Pei, Y. D., Lin, T. H., Lin, C. H., Akamatsu, T., et al. (2021). Coastal development threatens Datan area supporting greatest fish diversity at Taoyuan Algal Reef, northwestern Taiwan. Aquat. Conserv. 31, 590–604. doi: 10.1002/aqc.3477
Hsiao, S.-T., Chuang, S.-C., Chen, K.-S., Ho, P.-H., Wu, C.-L., and Chen, C. A. (2016). DNA barcoding reveals that the common cupped oyster in Taiwan is the Portuguese oyster Crassostrea angulata (Ostreoida; Ostreidae), not C. gigas. Sci. Rep. 6:34057. doi: 10.1038/srep34057
Hu, J., Kawamura, H., Li, C., Hong, H., and Jiang, Y. (2010). Review on current and seawater volume transport through the Taiwan Strait. J. Oceanogr. 66, 591–610. doi: 10.1007/s10872-010-0049-1
Kimura, M. (2000). Paleogeography of the Ryukyu Islands. Tropics 10, 5–24. doi: 10.1016/j.ympev.2018.02.011
Koh, S., and Ng, P. K. (2008). A revision of the shore crabs of the genus Eriphia (Crustacea: Brachyura: Eriphiidae). Raffles Bull. Zool. 56, 327–355.
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kuo, C.-Y., Keshavmurthy, S., Chung, A., Huang, Y.-Y., Yang, S.-Y., Chen, Y.-C., et al. (2020). Demographic census confirms a stable population of the critically-endangered caryophyllid coral Polycyathus chaishanensis (Scleractinia; Caryophyllidae) in the Datan Algal Reef, Taiwan. Sci. Rep. 10:10585. doi: 10.1038/s41598-020-67653-8
Lai, J. C., Thoma, B. P., Clark, P. F., Felder, D. L., and Ng, P. K. (2014). Phylogeny of eriphioid crabs (Brachyura, Eriphioidea) inferred from molecular and morphological studies. Zool. Scr. 43, 52–64. doi: 10.1111/zsc.12030
Leigh, J. W., and Bryant, D. (2015). popart: full-feature software for haplotype network construction. Methods Ecol. Evol. 6, 1110–1116. doi: 10.1111/2041-210x.12410
Li, G., Li, P., Liu, Y., Qiao, L., Ma, Y., Xu, J., et al. (2014). Sedimentary system response to the global sea level change in the East China Seas since the last glacial maximum. Earth Sci. Rev. 139, 390–405. doi: 10.1016/j.earscirev.2014.09.007
Li, K.-C., Liu, H.-C., and Lin, H.-C. (2021). Multiple environmental factors increase the niche complexity and species diversity of brachyuran crabs in an intertidal algal reef ecosystem in northwestern Taiwan. Zool. Stud. 60:73.
Liou, C.-Y., Yang, S.-Y., and Chen, C. A. (2017). Unprecedented calcareous algal reefs in northern Taiwan merit high conservation priority. Coral Reefs 36, 1253–1253. doi: 10.1007/s00338-017-1619-0
Ludt, W. B., and Rocha, L. A. (2015). Shifting seas: the impacts of Pleistocene sea-level fluctuations on the evolution of tropical marine taxa. J. Biogeogr. 42, 25–38. doi: 10.1111/jbi.12416
Nathan, L. R., Kanno, Y., and Vokoun, J. C. (2017). Population demographics influence genetic responses to fragmentation: a demogenetic assessment of the ‘one migrant per generation’ rule of thumb. Biol. Conserv. 210, 261–272.
Ni, G., Li, Q., Kong, L., and Yu, H. (2014). Comparative phylogeography in marginal seas of the northwestern Pacific. Mol. Ecol. 23, 534–548. doi: 10.1111/mec.12620
Ni, G., Li, Q., Kong, L., and Zheng, X. (2012). Phylogeography of bivalve Cyclina sinensis: testing the historical glaciations and Changjiang River outflow hypotheses in northwestern Pacific. PLoS One 7:e49487. doi: 10.1371/journal.pone.0049487
Qu, J., Shi, B., Xu, Y., Li, R., Sun, X., Guo, X., et al. (2021). Phylogeography of Patelloida pygmaea along the China coast. Mar. Biodivers. 51:38. doi: 10.1007/s12526-021-01164-1
R Core Team (2013). R: A Language And Environment For Statistical Computing. Vienna: R Foundation for Statistical Computing.
Rambaut, A., Drummond, A. J., Xie, D., Baele, G., and Suchard, M. A. (2018). Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 67, 901–904. doi: 10.1093/sysbio/syy032
Ramos-Onsins, S. E., and Rozas, J. (2002). Statistical properties of new neutrality tests against population growth. Mol. Biol. Evol. 19, 2092–2100. doi: 10.1093/oxfordjournals.molbev.a004034
Rogers, A. R., and Harpending, H. (1992). Population growth makes waves in the distribution of pairwise genetic differences. Mol. Biol. Evol. 9, 552–569. doi: 10.1093/oxfordjournals.molbev.a040727
Rozas, J., Ferrer-Mata, A., ánchez-DelBarrio, J. C. S., Guirao-Rico, S., Librado, P., Ramos-Onsins, S. E., et al. (2017). DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 34, 3299–3302. doi: 10.1093/molbev/msx248
Silver, A. (2018). Taiwanese Scientists Fight Construction Of A New Port They Say Would Damage A Unique Reef. Available Online at: https://www.sciencemag.org/news/2018/11/taiwanese-scientists-fight-construction-new-port-they-say-would-damage-unique-reef [accessed Sep 1, 2021].
Tajima, F. (1989). Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595. doi: 10.1093/genetics/123.3.585
Tomikawa, N., and Watanabe, S. (1992). Reproductive ecology of the xanthid crab Eriphia smithii McLeay. J. Crustacean Biol. 12, 57–67. doi: 10.2307/1548719
Wang, J. (2004). Application of the one-migrant-per-generation rule to conservation and management. Conserv. Biol. 18, 332–343.
Wang, P. (1999). Response of Western Pacific marginal seas to glacial cycles: paleoceanographic and sedimentological features. Mar. Geol. 156, 5–39.
Wang, X., Kong, L., Chen, J., Matsukuma, A., and Li, Q. (2017). Phylogeography of bivalve Meretrix petechialis in the Northwestern Pacific indicated by mitochondrial and nuclear DNA data. PLoS One 12:e0183221. doi: 10.1371/journal.pone.0183221
Keywords: crustacean, Eriphia ferox, mitochondrial cytochrome oxidase I (COI), Taiwan Strait, northwestern Pacific, Taoyuan Algal Reef, population genetics, historical demography
Citation: Chen M-H, Huang Y-Y, Huang B-Y, Hsieh HJ, Lee JN, Neo ML, Fukami H and Chen CA (2022) Historical Connectivity and Demography of the Ferocious Reef Crab, Eriphia ferox (Crustacea; Eriphiidae), Demonstrate That Taoyuan Algal Reef Is an Essential Population Source Along the East Taiwan Strait. Front. Mar. Sci. 8:799989. doi: 10.3389/fmars.2021.799989
Received: 22 October 2021; Accepted: 21 December 2021;
Published: 12 January 2022.
Edited by:
Rachel Collin, Smithsonian Tropical Research Institute (SI), United StatesReviewed by:
Maria Pia Miglietta, Texas A&M University at Galveston, United StatesPedro A. Peres, Florida International University, United States
Copyright © 2022 Chen, Huang, Huang, Hsieh, Lee, Neo, Fukami and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ya-Yi Huang, allantoma@gate.sinica.edu.tw; Chaolun Allen Chen, cac@gate.sinica.edu.tw
 Mu-Han Chen
Mu-Han Chen Ya-Yi Huang
Ya-Yi Huang Bi-Ying Huang1
Bi-Ying Huang1 Jen Nie Lee
Jen Nie Lee Mei Lin Neo
Mei Lin Neo Hironobu Fukami
Hironobu Fukami Chaolun Allen Chen
Chaolun Allen Chen