- 1Key Laboratory of Aquatic Germplasm Resources of Zhejiang, College of Biological and Environmental Sciences, Zhejiang Wanli University, Ningbo, China
- 2College of Fisheries, Henan Normal University, Xinxiang, China
- 3Ninghai Institute of Mariculture Breeding and Seed Industry, Zhejiang Wanli University, Ningbo, China
Ammonia nitrogen is a common toxic substance in the aquatic system, which seriously threatens the survival and growth of clams. However, less is known about the ammonia metabolism and detoxification strategy in razor clam. In this study, the polymorphism of the Glutamine synthetase gene from Sinonovacula constricta (Sc-GS) was found to be related to ammonia tolerance. By comparing the coding sequence (CDS) region of Sc-GS from two geographical populations, a total of 14 and 12 single nucleotide polymorphisms (SNPs) were identified, respectively, of which 10 loci were shared between the two populations. Among them, the locus c.1133T > G exhibited an extremely significant and strong association with ammonia tolerance in both populations (P < 0.01), and it was missense mutation, which led to the amino acid change from leucine (Leu) to arginine (Arg). Furthermore, the results about H&E staining and immunohistochemistry of Sc-GS protein in gills and hepatopancreas revealed that it was specifically localized in the lateral cilia of gill filaments and the endothelial cells of hepatocytes. After inhibiting the Sc-GS expression by RNA interference (RNAi) technology, the transcript levels of Sc-GS were extremely significantly downregulated at 24, 48, 72, and 96 h (P < 0.01) in the hepatopancreas. Taken together, these results indicated that the Sc-GS gene may participate in ammonia metabolism. In addition, these results will help to demonstrate the role of Sc-GS in ammonia nitrogen metabolism and provide markers related to ammonia nitrogen tolerance for molecular marker-assisted selection (MAS) of the razor clam.
Introduction
As we all know, the bivalve aquaculture industry is one of the fastest-growing global food sectors, and 89% of bivalves produced all over the world are from aquaculture (FAO, 2020). At present, mollusk aquaculture has accounted for 70% (20 million tons) of the total production of aquatic animals (FAO, 2020). The razor clam Sinonovacula constricta, which is an economically and ecologically important marine bivalve, with an annual output of 860,265 tons in 2020 (The Ministry of Agriculture and Fishery of the People’s Republic of China, 2021), was mainly polycultured with shrimps, crabs, and fish in Zhejiang and Fujian provinces of China (Li et al., 2015). Under a high-intensive cultivation situation, clams were threatened by various complex contaminants, such as nitrite and ammonia, which were typically important toxicity effectors in aquaculture systems (Cheng et al., 2019). S. constricta, as a benthic bivalve, often lives in mudflats or under 30 ∼ 40 cm of mud in ponds (Frankic and Hershner, 2003), they tend to experience more severe ammonia stress than other aquatic organisms (Zhang et al., 2020). Although it has been demonstrated that mollusks have a high tolerance to ammonia nitrogen (Widman et al., 2008), there are few reports on their detoxification mechanism of ammonia.
Ammonia exists in seawater with two different forms: NH4+ (ionized form) and NH3 (unionized form) (Francis-Floyd et al., 2009), of which NH3 (ammonia nitrogen) is toxic to the health of aquatic organisms because it can easily diffuse across the cell membrane (Randall and Tsui, 2002). In addition, ammonia is a primary environmental factor that can rapidly increase the mortality rate in the aquaculture industry and often brings serious economic losses to the aquatic cultivation industry (Schock et al., 2013). The high-concentration ammonia can reach a lethal effect on most aquatic animals, especially for invertebrates such as Manila clam Ruditapes philippinarum (Cong et al., 2017), Pacific white shrimp Litopenaeus vannamei (Zhang L. et al., 2018), and giant river prawn Macrobrachium rosenbergii (Dong et al., 2020). In addition, the different tissues from the same species differently respond to ammonia stress. For instance, it has been reported that ammonia level was higher in the hepatopancreases and gills than hemolymph and muscle when black tiger prawn Penaeus monodon was exposed to 0.718 mM ammonia (Chen and Chen, 2000). Similarly, the ammonia contents in the liver of swamp eel (Monopterus albus) had the highest levels under the 0.75 mM ammonia exposure (Ip et al., 2004). Furthermore, accumulating evidence has shown that their gills suffered significant physiological and histological damage when mollusk was exposed to toxic substances (e.g., the concentration of pH or ammonia exceeds the normal level) (Saravana and Geraldine, 2000; Henry et al., 2012). Accordingly, the lesion degree of the liver and gill is often used to assess the toxicity degree of harmful substances of aquatic animals (Mishra and Mohanty, 2008). In recent years, it has been reported the protein localization of genes related to ammonia-nitrogen excretion (Bucking et al., 2013; Boo et al., 2018; Sunga et al., 2020). However, at present, less is known about the protein localization of genes related to ammonia metabolism in the gill and hepatopancreas of benthic mollusk exposed to high ammonia.
Previous studies found that the synthesis of glutamine (Gln) was one of the most critical strategies to defend against ammonia toxicity, which was adopted by many invertebrates and vertebrate organisms, such as yellow catfish Pelteobagrus fulvidraco (Li et al., 2016), sea cucumber Apostichopus japonicus (Wang et al., 2014), swimming crab Portunus trituberculatus (Si et al., 2018), and L. vannamei (Qiu et al., 2018). Non-toxic Gln can be stored in the body and is used for other anabolic processes (Zhang et al., 2020). Gln was composed of glutamate (Glu) and ammonium (NH4+), which was catalyzed by Glutamine synthetase (GS) (Tok et al., 2011). GS was a multifunctional enzyme in nitrogen metabolism, which was a gateway to the conversion of inorganic nitrogen into organic nitrogen (Ding et al., 2018). However, few studies focus on the role of the GS gene on ammonia detoxification metabolism in mollusks. Zhang et al. (2020) found that the mRNA and protein expression levels of the GS gene from S. constricta (Sc-GS) were significantly increased under ammonia challenge (P < 0.05), which were higher in gills and hepatopancreases than foot, mantle, adductor muscle and haemocytes. Therefore, the Sc-GS gene was considered as the main object of the study, and its correlation with ammonia tolerance was analyzed by probing its gene polymorphism in two typical geographical populations of S. constricta. In addition, we tried to explore the histocellular localization and function of Sc-GS protein in response to ammonia nitrogen stress. Then, we identified the mRNA expression levels of Sc-GS at different times after RNA interference (RNAi) technology. These results provide a basis for screening the candidate genes of ammonia-nitrogen tolerance and for studying the molecular mechanism of ammonia-nitrogen metabolism regulation.
Materials and Methods
Ethics Statement
The razor clams used in this study were from the Genetic Breeding Research Center of Zhejiang Wanli University, China, and all experimental procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of Zhejiang Wanli University, China.
Sample Collection and Challenge Experiments
Two geographical populations of the razor clam S. constricta (shell length = 51.49 ± 4.32 mm, wet weight = 7.58 ± 1.45 g), consisting of 3,000 individuals at the age of 1 year, were sampled from Zhejiang (ZJ) and Fujian (FJ) provinces in China, respectively. Then, these clams were used for single nucleotide polymorphism (SNP) analysis, H&E, and immunohistochemistry test of the Sc-GS gene. All the clams were maintained in a 500-L recirculating seawater tank with a temperature of 23.5°C and a salinity of 20 with aeration for 3 days before experimentation and were fed with golden-brown algae two times per day. Before the ammonia stress experiment, 6 healthy clams were selected from the ZJ population for histological morphology study. During the ammonia challenge experiment, 2,400 clams from each population were randomly divided into 6 tanks (500 L), 3 control groups (CGs), and 3 ammonia stress groups (AGs). The CGs were filled with natural seawater, and the stress concentrations of ammonia in AGs were 180 mg/L (actual concentration = 174.00 ± 5.81 mg/L, the corresponding NH3 concentrations was 7.23 ± 0.17 mg/L, according to the 96-h LC50 values of the study by Zhang et al., 2020). The ammonia concentration was adjusted by diluting 1,000 mg/L total ammonia stock solution that is disposed of NH4Cl (Sangon, Shanghai, China). All the clams were monitored at intervals of 2 h for the removal of dead individuals in time, and at the same time, the pH of AGs and CGs were detected to maintain at about 8 for ammonia exposure for 120 h.
The surviving clams from the 6 AGs (i.e., 3 AGs in each population) throughout the ammonia challenge experiment were divided into an ammonia tolerant group (TG). Then, the hepatopancreases of individuals in the TGs were dissected for RNA extraction. At the same time, gills and hepatopancreases of five clams were collected from AG and CG of the FJ population for the histocellular localization study.
RNA Interference Experiment
Live and healthy clams (1-year old, shell length = 52.15 ± 4.12 mm, and wet weight = 7.73 ± 1.38 g) were collected at the Ningbo Marine and Fishery Science and Technology Innovation Base (Zhejiang province, China) for RNAi assays. The control conditions of these clams acclimatized in seawater were the same as mentioned earlier.
For the Sc-GS RNAi assay, 300 healthy clams were selected and divided into 6 groups, in which three groups were treated as AGs and the remaining three groups as the CGs (the treatments of AGs and CGs were the same as mentioned earlier) for 120 h. The siRNA-negative control (NC, Table 1) and diethyl pyrocarbonate (DEPC)-treated water (DEPC-W) were used as a CG. Sc-GS small interfering RNA (siRNA-GS, Sangon, Shanghai, China, Table 1) was diluted to 5,000 ng/μl with DEPC-W and injected into the adductor muscle of clams in one group of AGs, and the remaining two groups were injected into NC and DEPC-W, respectively. The same treatment was used for the CGs. Then, the hepatopancreases of six clams from each group were collected at 0, 24, 48, 72, 96, and 120 h postinjection. These selected hepatopancreases were cut into pieces for RNA extraction.
RNA Extraction and cDNA Synthesis
The quality and quantity of RNA samples extracted using TRIzol Reagent (Omega, R6830-02, United States) were evaluated by 1% agarose gel electrophoresis and were quantified with UV spectrophotometers. For each sample, 2 μg DNase I-treated RNA was reverse-transcribed using the RT-PCR Kit (Takara, Dalian, China). The thermal cycles were conducted in a PCR machine (Bio-Rad, California, United States).
Primers and PCR Amplification
We designed the gene-specific primers using Primer 5 software based on the sequence of Sc-GS gene (GenBank accession No. MK451701). The PCR conditions and purification methods were same as those reported by Sun et al. (2021).
Association Analysis Between Polymorphisms and Ammonia Tolerance
The nucleotide sequences of the Sc-GS gene in different individuals were aligned using MEGA 7 and Mutation Surveyor software. The base locus with two peaks at the nearly same height was defined as a heterozygous locus. The genotype frequencies and their associations with ammonia tolerance were analyzed by SPSS version 22. The information of these SNPs between two populations was calculated by PopGen32.
Paraffin Section, H&E, and Immunofluorescence Staining
The gills and hepatopancreases of clams were first fixed in 4% paraformaldehyde at 4°C, then dehydrated, and finally embedded in paraffin sections. The paraffin sections (5 μm) were dewaxed with xylene and rehydrated with gradient ethanol, then stained with hematoxylin (10 min) and eosin (30 s), respectively. Additionally, immunofluorescence staining was carried out to analyze the expression changes of Sc-GS. The dewaxed paraffin sections were sealed with 5% skimmed milk powder solution at room temperature for 1.5 h after the antigen retrieval by ethylenediaminetetraacetic acid (EDTA), and then, the sections were added with primary antibody (Sc-GS, 1:250) overnight at 4°C, washing three times [0.01 M phosphate-buffered saline (PBS), 10 min per time]. Fluorescein isothiocyanate (FITC)-labeled murine anti-rabbit IgG as a secondary antibody (1:150, containing 4’,6-diamidino-2-phenylindole (DAPI)) was added in a dark room and protected from light for 1 h at room temperature and washed three times (0.01 M PBS, 10 min per time). Finally, a fluorescence microscope (Nikon 80i, Japan) was used to take photographs.
Quantitative Real-Time PCR
The expression profiles of Sc-GS were analyzed by using triplicate quantitative real-time PCR (qRT-PCR). Primers Sc-GS-F1/R1 (Table 1) were designed by Primer 5, and the Ribosomal protein S9 (RS9, Table 1) gene was selected as an internal reference gene (Zhao et al., 2018). A 7500 Fast Real-Time PCR Machine (ABI, United States) was used for PCR, and the relative value of 2–ΔΔCt was adopted for data processing (Montresor et al., 2013). SPSS version 22 was used to analyze the quantitative differences of fluorescence results. One-way ANOVA was used to compare the difference between groups.
Results
Single Nucleotide Polymorphism Identification and Association Analysis of Glutamine Synthetase Gene From Sinonovacula constricta Gene Exons
In the ammonia challenge experiment, the first dead clam was observed at 22 h in the FJ population. The clams in both populations who survived after 120 h ammonia stress were contemplated as the TG, overall, 130 and 116 surviving clams were collected from ZJ and FJ populations, respectively. During the entire experimental period, no dead clam was found in the CG.
In this study, 14 and 12 SNPs were genotyped successfully in FJ and ZJ populations, respectively. Then, the genotyping results of TG and CG individuals were used for the statistics of genotype frequencies. It was worth noting that only SNP c.1133T > G was strongly and significantly associated with the ammonia tolerance in both two populations (P < 0.01) (Table 2). Notably, 6 out of the 12 SNPs in the ZJ population were transitions, while there were 7 transitions out of the 14 SNPs in the FJ population (Table 2). It was noteworthy that c.1133T > G was a non-synonymous mutation, resulting in an amino acid exchange from leucine (Leu) to arginine (Arg).
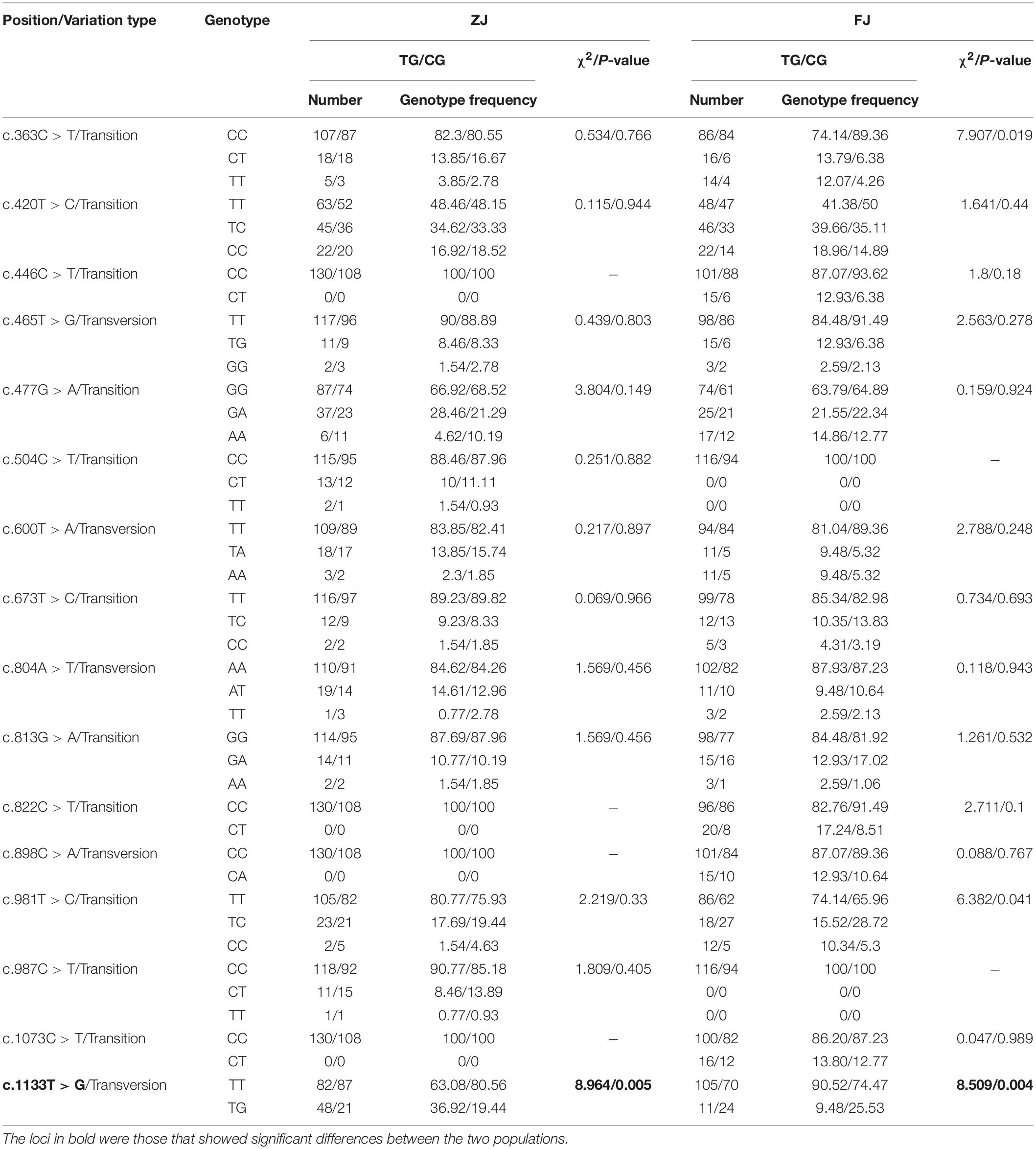
Table 2. Genotype, variation type, and gene frequency of single nucleotide polymorphisms (SNPs) in Glutamine synthetase gene from Sinonovacula constricta (Sc-GS).
Histological Structure and Histocellular Localization of Glutamine Synthetase Gene From Sinonovacula constricta Protein
The histological structure, expression level and histocellular localization of Sc-GS in the gills and hepatopancreas were determined by paraffin section, HE staining and immunohistochemistry assay. The gills were made up of two gill flaps on the left and right sides. A pair of gill flaps in the middle of both sides of the mantle cavity was inner gill flaps, while a pair on both sides was outer gill flaps. Each gill flap was composed of ascending and descending gill lamellae, which was composed of numerous gill filaments in dense clusters perpendicular to the longitudinal axis of the body. These gill filaments could be subdivided into flat cells and columnar cells of frontal cilia and lateral cilia according to their different positions (Figure 1A). Then, the FITC-labeled antibody selected in this study could specifically bind to Sc-GS protein, which led to the protein expression region showing green fluorescence. In view of this, we found that the columnar cells of lateral cilia of gills in the AG showed stronger positive signals than those in CG (Figure 2).
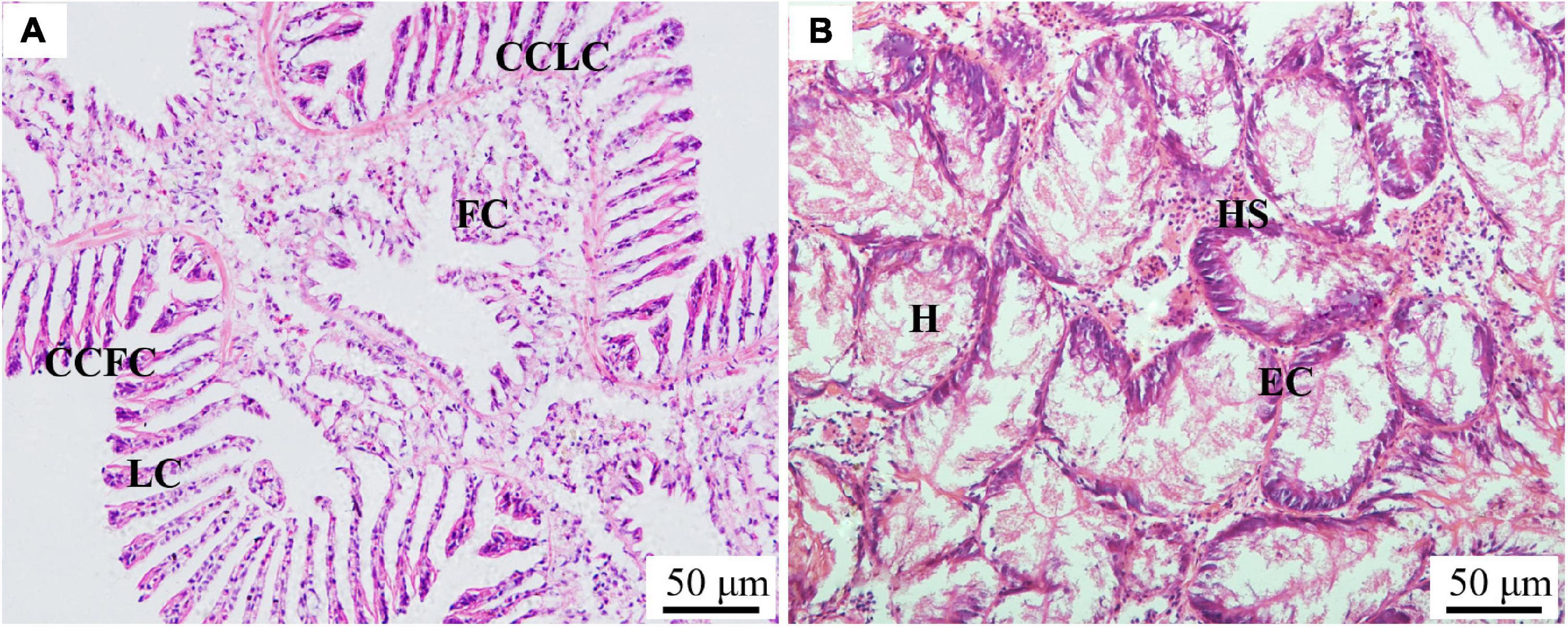
Figure 1. Paraffin section observation of gills (A) and the hepatopancreas (B) in the Sinonovacula constricta. CCFC, columnar cells of frontal cilia; CCLC, columnar cells of lateral cilia; FC, flat cells; HS, hepatic sinusoid; EC, endothelial cells; H, hepatocytes. Scale bars were 50 μm.
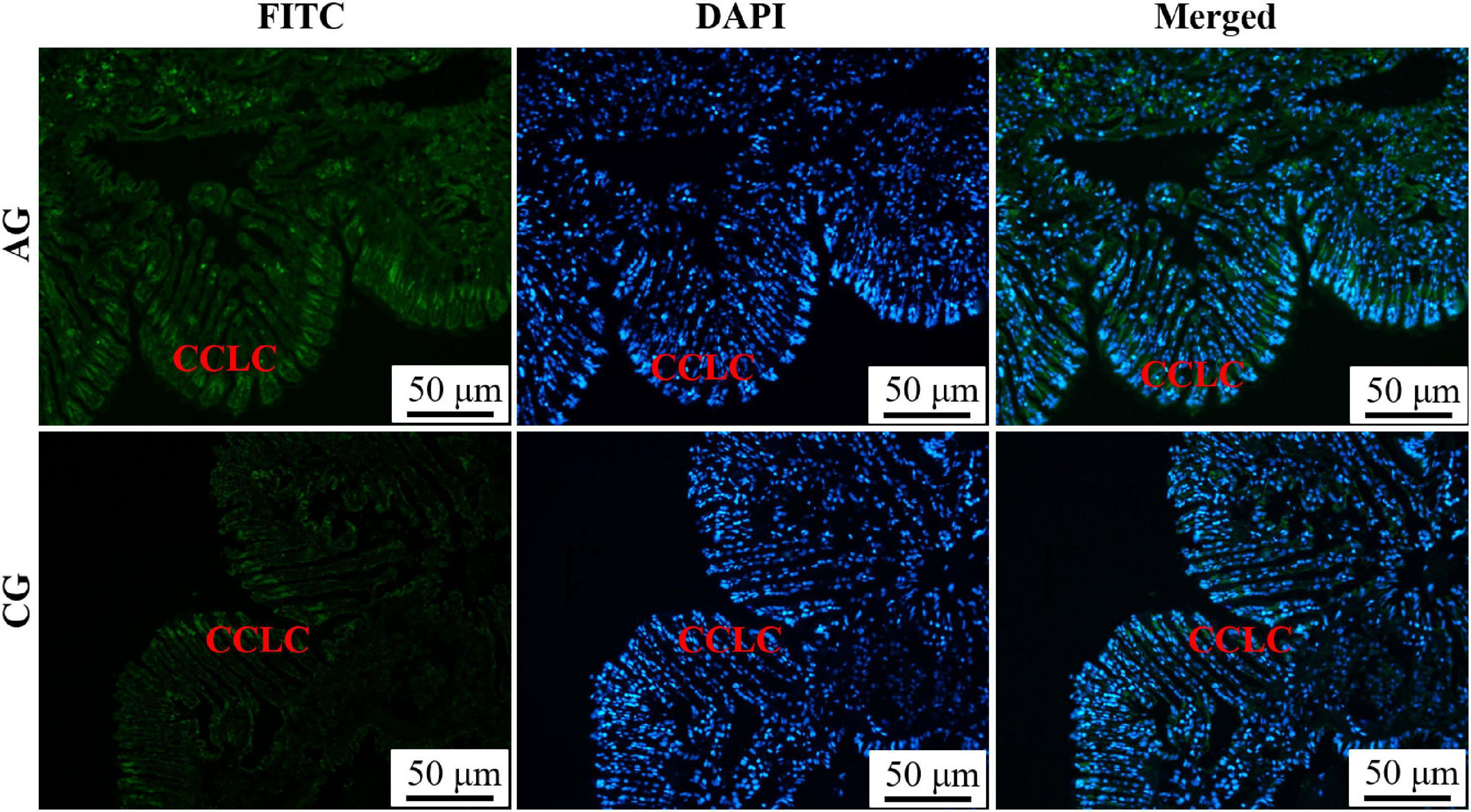
Figure 2. Immunofluorescence of Glutamine synthetase gene from Sinonovacula constricta (Sc-GS) in the gills. Sc-GS protein was stained with the anti-GS antibody in green, and nuclei were stained with DAPI in blue. The merged images were colocalized with Sc-GS protein (green) and DAPI (blue) in the columnar cells of lateral cilia of gills. Scale bars were 50 μm.
The hepatopancreas surrounded the sides of the stomach and was made up of individual hepatocyte cells. The hepatic sinusoid formed between hepatocytes cells was composed of endothelial cells (Figure 1B). On this basis, the endothelial cells of a hepatic sinusoid in the AG were showed a positive signal in relation to CG (Figure 3). Furthermore, the combination of green and blue signals showed that the two colors were not coincident, indicating that Sc-GS protein was not expressed in the nucleus (Figures 2, 3).
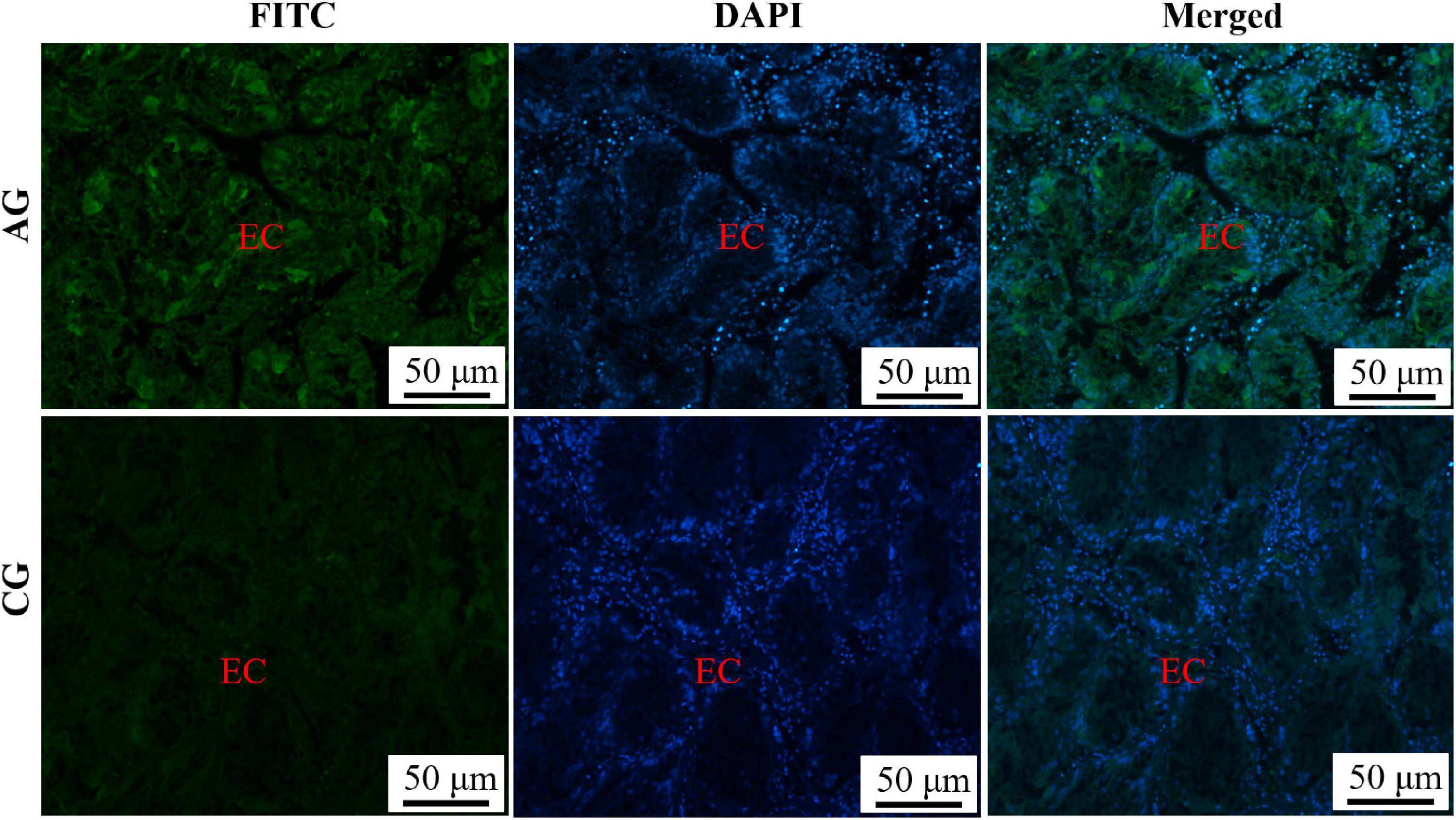
Figure 3. Immunofluorescence of Sc-GS in hepatopancreas. Sc-GS protein was stained with the anti-GS antibody in green, and nuclei were stained with DAPI in blue. The merged images were colocalized with Sc-GS protein (green) and DAPI (blue) in the endothelial cells of hepatic sinusoid. Scale bars were 50 μm.
Expression of Glutamine Synthetase Gene From Sinonovacula constricta After RNA Interference Silencing
The function of Sc-GS in hepatopancreas was further investigated by RNAi. In AGs, the mRNA expression level of Sc-GS in hepatopancreas was extremely significantly (P < 0.01) lower at 24, 48, 72, and 96 h in siRNA-GS than that in NC, which downregulated to approximately 86.44, 89.10, 91.17, and 84.45%, respectively (Figure 4A). Similarly, the mRNA expression level of Sc-GS in CGs was extremely significantly (P < 0.01) lower at 24, 48, 72, and 96 h in siRNA-GS compared to that in NC, which downregulated to approximately 71.69, 74.44, 80.44, and 80.03%, respectively (Figure 4B). In addition, the variation trend of the mRNA expression level of Sc-GS genes in DEPC-W and NC in each group (AG or CG) was roughly the same.
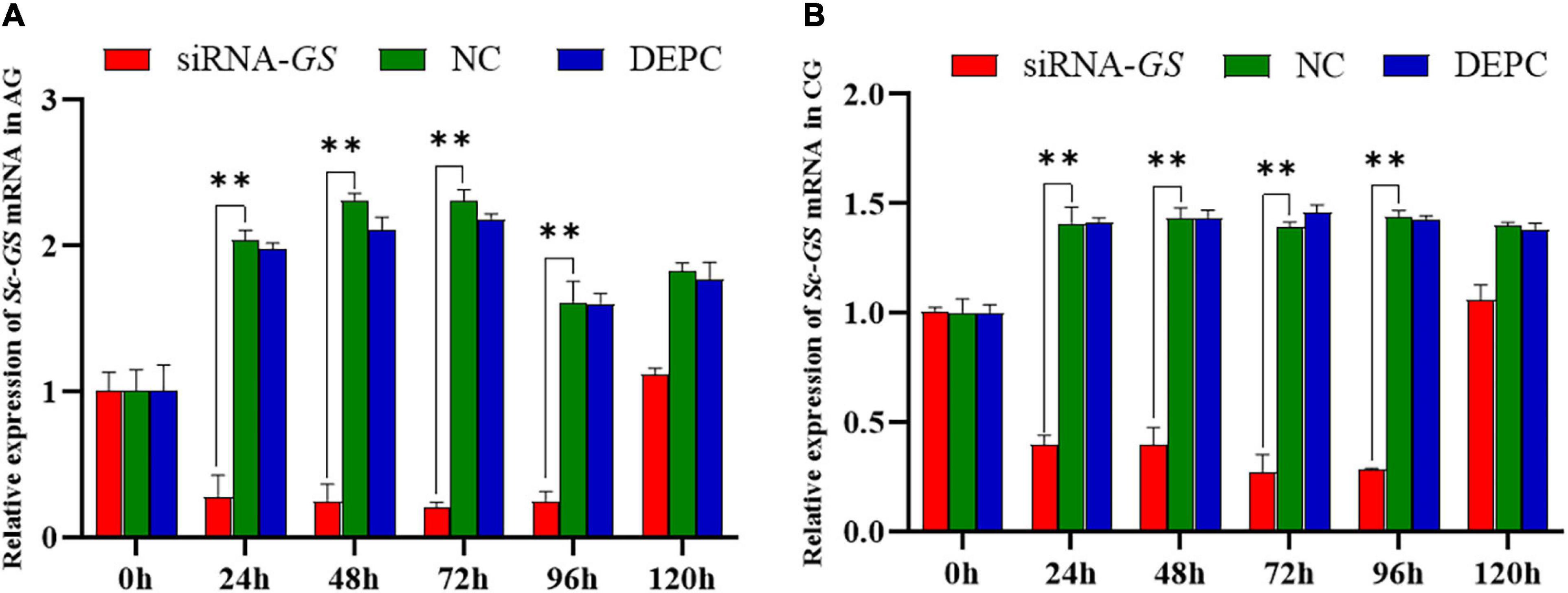
Figure 4. Relative expression of Sc-GS genes in the hepatopancreas after RNA interference (RNAi) Time-course expression of Sc-GS genes in ammonia stress group (AG) after RNAi (A). Time-course expression of Sc-GS genes in the control group (CG) after RNAi (B). Vertical bars represent the mean ± SD (n = 4). ∗∗ Denotes an extremely significant difference between Sc-GS small interfering RNA (siRNA-GS) and negative control (NC) (P < 0.01).
Discussion
Ammonia was a major toxicant in the aquatic system, which led to adverse effects such as growth reduction (Sinha et al., 2012), oxidative stress and damage (Zhang M. et al., 2018), immune suppression (Li et al., 2016), histological changes (Cheng et al., 2019), and mortality. However, it is still quite limited to the understanding of the molecular mechanism of ammonia tolerance in aquatic animals. It is well known that GS can catalyze the synthesis of Gln from NH4+ and Glu, which in turn provides a substrate for the synthesis of urea-related formula phosphate synthetase III (i.e., it plays a central role in the urea cycle) (Wang and Walsh, 2000). So far, the research on the GS genes in aquatic organisms has mainly focused on fish and crustaceans (Ip et al., 2005; Qiu et al., 2018; Dong et al., 2020). On the contrary, research on the ammonia nitrogen detoxification metabolism of Sc-GS is very scarce. In this study, our objective was to identify the polymorphisms in the Sc-GS gene and to investigate the association between its polymorphisms and tolerance to ammonia. In addition, we explored the histocellular localization and function of the Sc-GS gene in response to ammonia tolerance using H&E staining, immunohistochemistry, and RNAi technology.
Studies on L. vannamei (Qiu et al., 2018) and pharaoh cuttlefish Sepia pharaonis (Peng et al., 2017) have shown that the mRNA and protein expression level of GS in some tissues (i.e., liver, muscle, intestine, and gills) was increased significantly when exposed to ammonia. Our previous studies showed that Sc-GS could be involved in ammonia detoxification (Zhang et al., 2020). In this study, 12 SNPs were detected in the Sc-GS gene (1 per 87 bp) of the ZJ population, while 14 SNPs (1 per 74 bp) were found in the FJ population. The difference in the numbers of SNPs between the ZJ and FJ populations may be due to the population genetic divergence or the deviation of sequencing. To our knowledge, the highest levels of DNA polymorphism were reported in the animal kingdom such as nematode Caenorhabditis remanei (Cutter et al., 2006) and ascidian Ciona savignyi (Small et al., 2007) with 1 per 20 bp. In addition to that, the density of SNPs in insect species was often highly polymorphic, e.g., 1 per 50 bp in fruit fly Drosophila (Shapiro et al., 2007) and 1 per 125 bp in mosquitoes Aedes aegypti (Morlais and Severson, 2003). Similarly, accumulating evidence has shown that a high density of DNA polymorphism was detected in mollusks (Saavedra and Bachère, 2006). This study reached the same conclusion as mentioned earlier, indicating that bivalves have abundant genetic variation. Notably, we found two types of mutation, namely, transversion and transition in two populations. In this study, the frequency of the transition of C-T was much higher than that of A-G, which can be explained by the fact that C in the C&G sequence has been often methylated and can be spontaneously deaminated to form T (Yoon et al., 2001).
The discovery of SNPs with missense mutation provides a new possibility for molecular marker-assisted selection (MAS) of aquatic animal varieties with economic traits to relieve environmental stress (Syvänen, 2001; Wang et al., 2013). It has been shown that three SNPs about factor inhibiting HIF-1 (Fih-1) found in Wuchang bream Megalobrama amblycephala were significantly associated with hypoxia traits (Zhang et al., 2016). A study on the association between candidate gene and heat tolerance in bay scallop Argopecten irradians showed that SNP all-53308-760T/C may be related to heat (Du et al., 2014). Similarly, SNPs in other candidate genes for related heat tolerance, such as metallothionein 1 and heat shock protein 90, have been reported in bay scallops (Yang et al., 2013, 2015). Notably, high-concentration ammonia is a harmful environmental factor in the survival, development, and growth of aquatic animals. In this regard, it is significant to explore the molecular markers associated with ammonia tolerance. It has been reported that one SNP (C > T545) of hemocyanin may be served as a marker for selecting high growth performance in ammonia-tolerant L. vannamei (Janpoom et al., 2020). Another study on L. vannamei showed that 12 SNP loci related to ammonia resistance may be significantly associated with disease resistance, which can provide the critical potential for genetic selection to increase the production of shrimp (Lu et al., 2018). These findings highlight the usefulness of missense mutation SNP in environmental resistance-related MAS. In this study, one missense mutation (c.1133T > G) was identified in the Sc-GS gene, which mutated from Leu to Arg between two populations. Furthermore, the protein homology analysis showed that c.1133T > G had high homology in mollusk, suggesting that this locus plays an important role in the normal structure and function of Sc-GS (Zhang et al., 2020). As we all know, Leu was a non-polar hydrophobic amino acid (Madeira et al., 2013), and Arg was a polar hydrophilic amino acid with side chain charged (Harms et al., 2011), which changes the conformation of proteins. It has been shown that heterozygous variant c.35G > T (i.e., a substitution of Arg to Leu) identified in exon 1 of CRYAA gene was closely related to the human congenital cataract and microphthalmia (Song et al., 2018). Therefore, we speculate that the mutation c.1133T > G may affect the secondary structure of the mutant protein, leading to changes in its folded conformation and stability, eventually resulting in changes in the function of the GS gene. At present, the genetic selection program for S. constricta with high ammonia tolerance is still in its infancy. Our findings suggest that c.1133T > G, which was found associated with ammonia tolerance in both populations, can be used as a universal marker for further breeding in S. constricta.
Immunohistochemistry has become the most effective method for protein expression localization due to its advantages of visualization of target proteins (Lugos et al., 2020). Hepatopancreas was known to be the essential digestion and detoxification organ (Wu et al., 2013). At high levels of ammonia, the hepatopancreases of aquatic animals are injured in different degrees and even cause death (Mishra and Mohanty, 2008). Similarly, in this study, the distribution of Sc-GS protein was changed after the ammonia challenge, which was highly expressed in the cytoplasm of endothelial cells, and the same conclusions were found in the livers of vertebrates such as chicken (Smith and Campbell, 1988; Ueberham et al., 2004). Thus, it can be concluded that Sc-GS protein in hepatopancreas from razor clam may play a key role in the detoxification process of dealing with high ammonia concentration in the organism.
The RNA interference was first discovered in the nematode worm Caenorhabditis elegans as a response to double-stranded RNA (dsRNA), which leads to the silencing of homologous genes (Fire et al., 1998). Fabioux et al. (2009) successfully induced Pacific oyster Crassostrea gigas sterility by injecting dsRNA of the Oyvlg gene into an oyster, which was the first report that RNAi induced homologous gene silencing in mollusk. In addition, dsRNA of PfN23 gene was injected into the adult pearl oyster (Pinctada fucata), resulting in a disordered of nacre surface, suggesting that PfN23 gene may be a key accelerator in the control of crystal growth in nacre (Fang et al., 2012). Similarly, the expression of the BMP7 gene was successfully suppressed by injecting dsRNA into the razor clam, indicating that the BMP7 gene plays a vital role in repairing shell damage (Zhao et al., 2020). Moreover, in our previous study, the silencing of Glutamate dehydrogenase, a gene related to ammonia nitrogen detoxification, was induced by the injection of specific siRNA into the razor clam (Sun et al., 2021). Beyond that, a study in GS RNAi transgenic rice showed that GS contributed an important role in the amelioration of NH4+ toxicity (Le et al., 2018). In addition, the extended developmental duration and pupal weight losses in fruit fly could be associated with a disruption in ammonia metabolism after GS gene silence (Hazel et al., 2003; Zhang M. Y. et al., 2018). In this study, the mRNA expression of Sc-GS genes in the hepatopancreas was successfully inhibited by injecting specific siRNA into S. constricta, thus providing a practical method to characterize the function of Sc-GS in response to ammonia stress.
Conclusion
The genetic improvement for stress tolerance is extremely beneficial for the sustainable development of the clam aquaculture industry. In this study, a significant difference in the SNP with the missense mutation (c.1133T > G) in Sc-GS was detected between the two geographical populations. Further results of H&E staining, immunohistochemistry, and RNAi indicated that the Sc-GS gene plays a vital role in the detoxification of ammonia in razor clam. Collectively, these findings could provide candidate markers for the molecular selective breeding of ammonia-tolerant clams in the future and could provide a basis for the detoxification metabolism of bivalves under ammonia stress.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Ethics Statement
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of Zhejiang Wanli University.
Author Contributions
YD and JH conceived and designed the experiments. GS performed the experiments under the support of HY and CS. GS and YD analyzed the data and plot. GS, YD, WD, and ZL wrote and revised the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by the National Key Research and Development Program of China (2018YFD0901405), the Zhejiang Major Program of Science and Technology (2021C02069-7), the Ningbo Major Project of Science and Technology (2019B10005), and the National Marine Genetic Resource Center Program.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Boo, M. V., Hiong, K. C., Goh, E. J. K., Choo, C. Y. L., Wong, W. P., Chew, S. F., et al. (2018). The ctenidium of the giant clam, Tridacna squamosa, expresses an ammonium transporter 1 that displays light-suppressed gene and protein expression and may be involved in ammonia excretion. J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 185, 765–777. doi: 10.1007/s00360-018-1161-6
Bucking, C., Edwards, S. L., Tickle, P., Smith, C. P., McDonald, M. D., and Walsh, P. J. (2013). Immunohistochemical localization of urea and ammonia transporters in two confamilial fish species, the ureotelic gulf toadfish (Opsanus beta) and the ammoniotelic plainfin midshipman (Porichthys notatus). Cell Tissue Res. 352, 623–637. doi: 10.1007/s00441-013-1591-0
Chen, J. M., and Chen, J. C. (2000). Study on the free amino acid levels in the hemolymph, gill, hepatopancreas and muscle of Penaeus monodon exposed to elevated ambient ammonia. Aquat. Toxicol. 50, 27–37. doi: 10.1016/s0166-445x(99)00095-8
Cheng, C. H., Ma, H. L., Su, Y. L., Deng, Y. Q., Feng, J., Xie, J. W., et al. (2019). Ammonia toxicity in the mud crab (Scylla paramamosain): the mechanistic insight from physiology to transcriptome analysis. Ecotoxicol. Environ. Saf. 179, 9–16. doi: 10.1016/j.ecoenv.2019.04.033
Cong, M., Wu, H., Yang, H., Zhao, J., and Lv, J. (2017). Gill damage and neurotoxicity of ammonia nitrogen on the clam Ruditapes philippinarum. Ecotoxicology 26, 459–469. doi: 10.1007/s10646-017-1777-4
Cutter, A. D., Baird, S. E., and Charlesworth, D. (2006). High nucleotide polymorphism and rapid decay of linkage disequilibrium in wild populations of Caenorhabditis remanei. Genetics 174, 901–913. doi: 10.1534/genetics.106.061879
Ding, Y., Zang, X., Shi, J., Hou, L., He, B., Dong, M., et al. (2018). cDNA cloning of gs, gogat, and gdh from Haematococcus pluvialis and transcription and enzyme level analysis in different nitrogen concentration. J. Appl. Phycol. 31, 183–190. doi: 10.1007/s10811-018-1564-5
Dong, X., Liu, Q., Kan, D., Zhao, W., Guo, H., and Lv, L. (2020). Effects of ammonia-N exposure on the growth, metabolizing enzymes, and metabolome of Macrobrachium rosenbergii. Ecotoxicol. Environ. Saf. 189:110046. doi: 10.1016/j.ecoenv.2019.110046
Du, X., Li, L., Zhang, S., Meng, F., and Zhang, G. (2014). SNP identification by transcriptome sequencing and candidate gene-based association analysis for heat tolerance in the bay scallop Argopecten irradians. PLoS One 9:e104960. doi: 10.1371/journal.pone.0104960
Fabioux, C., Corporeau, C., Quillien, V., Favrel, P., and Huvet, A. (2009). In vivo RNA interference in oyster -vasa silencing inhibits germ cell development. FEBS J. 276, 2566–2573. doi: 10.1111/j.1742-4658.2009.06982.x
Fang, D., Pan, C., Lin, H., Lin, Y., Zhang, G., Wang, H., et al. (2012). Novel basic protein, PfN23, functions as key macromolecule during nacre formation. J. Biol. Chem. 287, 15776–15785. doi: 10.1074/jbc.M112.341594
FAO (2020). The State of World Fisheries and Aquaculture (2020). Sustainability in Action. Rome: FAO.
Fire, A., Xu, S., Montgomery, M. K., Kostas, S. A., Driver, S. E., and Mello, C. C. (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811. doi: 10.1038/35888
Francis-Floyd, R., Watson, C., Petty, D., and Pouder, D. (2009). Ammonia in aquatic systems. Univ. Fla. Inst. Food Agric. Sci. 16, 1–5.
Frankic, A., and Hershner, C. (2003). Sustainable aquaculture: developing the promise of aquaculture. Aquac. Int. 11, 517–530. doi: 10.1023/B:AQUI.0000013264.38692.91
Harms, M. J., Schlessman, J. L., Sue, G. R., and García-Moreno, B. (2011). Arginine residues at internal positions in a protein are always charged. Proc. Natl. Acad. Sci. U.S.A. 108, 18954–18959. doi: 10.1073/pnas.1104808108
Hazel, M. H., Christensen, R. J., and O’Donnell, M. J. (2003). Inhibition of the SAPK/JNK pathway blocks the stimulatory effects of glutamine on fluid secretion by the Malpighian tubules of Rhodnius prolixus. J. Insect Physiol. 49, 897–906. doi: 10.1016/s0022-1910(03)00138-0
Henry, R. P., Lucu, C., Onken, H., and Weihrauch, D. (2012). Multiple functions of the crustacean gill: osmotic/ionic regulation, acid-base balance, ammonia excretion, and bioaccumulation of toxic metals. Front. Physiol. 3:431. doi: 10.3389/fphys.2012.00431
Ip, Y. K., Leong, M. W. F., Sim, M. Y., Goh, G. S., Wong, W. P., and Chew, S. F. (2005). Chronic and acute ammonia toxicity in mudskippers, Periophthalmodon schlosseri and Boleophthalmus boddaerti: brain ammonia and glutamine contents, and effects of methionine sulfoximine and MK801. J. Exp. Biol. 208, 1993–2004. doi: 10.1242/jeb.01586
Ip, Y. K., Tay, A. S. L., Lee, K. H., and Chew, S. F. (2004). Strategies for surviving high concentrations of environmental ammonia in the swamp eel Monopterus albus. Physiol. Biochem. Zool. 77, 390–405. doi: 10.1086/383510
Janpoom, S., Kaewduang, B., Prasertlux, S., Rongmung, P., Ratdee, O., Lirdwitayaprasit, T., et al. (2020). A SNP of the hemocyanin gene (LvHc) is a marker for high growth and ammonia-tolerance in Pacific white shrimp Litopenaeus vannamei. Fish Shellfish Immunol. 106, 491–501. doi: 10.1016/j.fsi.2020.07.058
Le, T. N., Lee, B. B., Kyoungwhan, K., Young, S., and Cheong, H. (2018). Coordinated expression of cytosolic and chloroplastic glutamine synthetase during reproductive stage and its impact in GS1 RNAi transgenic rice. Rice Sci. 25, 250–260. doi: 10.1016/j.rsci.2018.08.001
Li, L., Yan, B., Li, S., Xu, J., and An, X. (2015). A comparison of bacterial community structure in seawater pond with shrimp, crab, and shellfish cultures and in non-cultured pond in Ganyu, Eastern China. Ann. Microbiol. 66, 317–328. doi: 10.1007/s13213-015-1111-4
Li, M., Gong, S., Li, Q., Yuan, L., Meng, F., and Wang, R. (2016). Ammonia toxicity induces glutamine accumulation, oxidative stress and immunosuppression in juvenile yellow catfish Pelteobagrus fulvidraco. Comp. Biochem. Physiol. C 183–184, 1–6. doi: 10.1016/j.cbpc.2016.01.005
Lu, X., Kong, J., Meng, X., Cao, B., Luo, K., Dai, P., et al. (2018). Identification of SNP markers associated with tolerance to ammonia toxicity by selective genotyping from de novo assembled transcriptome in Litopenaeus vannamei. Fish Shellfish Immunol. 73, 158–166. doi: 10.1016/j.fsi.2017.12.005
Lugos, M. D., Davou, G. I., Choji, T. P. P., Jugu, K. P., Ashi, R. R., Oyero, S. K., et al. (2020). Using immunohistochemistry without linkers to determine the optimum concentrations of primary antibodies for immunofluorescence staining of formalin-fixed paraffin-embedded tissue sections. Appl. Immunohistochem. Mol. Morphol. 28, 249–257. doi: 10.1097/PAI.0000000000000718
Madeira, P. P., Bessa, A., Álvares-Ribeiro, L., Aires-Barros, M. R., Rodrigues, A. E., and Zaslavsky, B. Y. (2013). Analysis of amino acid-water interactions by partitioning in aqueous two-phase systems. I–amino acids with non-polar side-chains. J. Chromatogr. A 1274, 82–86. doi: 10.1016/j.chroma.2012.11.080
Mishra, A. K., and Mohanty, B. (2008). Acute toxicity impacts of hexavalent chromium on behavior and histopathology of gill, kidney and liver of the freshwater fish, Channa punctatus (Bloch). Environ. Toxicol. Pharmacol. 26, 136–141. doi: 10.1016/j.etap.2008.02.010
Montresor, L. C., Miranda-Filho, K. C., Paglia, A., Luz, D. M. R., Araújo, J. M., Silva, M. J. D. S., et al. (2013). Short-term toxicity of ammonia, sodium hydroxide and a commercial biocide to golden mussel Limnoperna fortune (Dunker, 1857). Ecotoxicol. Environ. Saf. 92, 150–154. doi: 10.1016/j.ecoenv.2013.03.016
Morlais, I., and Severson, D. W. (2003). Intraspecific DNA variation in nuclear genes of the mosquito Aedes aegypti. Insect Mol. Biol. 12, 631–639. doi: 10.1046/j.1365-2583.2003.00449.x
Peng, R. B., Le, K. X., Wang, P. S., Wang, Y., Han, Q. X., and Jiang, X. M. (2017). Detoxification pathways in response to environmental ammonia exposure of the cuttlefish, Sepia pharaonis: glutamine and urea formation. J. World Aquac. Soc. 48, 342–352. doi: 10.1111/jwas.12341
Qiu, L., Shi, X., Yu, S., Han, Q., Diao, X., and Zhou, H. (2018). Changes of ammonia-metabolizing enzyme activity and gene expression of two strains in shrimp Litopenaeus vannamei under ammonia stress. Front. Physiol. 9:211. doi: 10.3389/fphys.2018.00211
Randall, D. J., and Tsui, T. K. N. (2002). Ammonia toxicity in fish. Mar. Pollut. Bull. 45, 17–23. doi: 10.1016/s0025-326x(02)00227-8
Saavedra, C., and Bachère, E. (2006). Bivalve genomics. Aquaculture 256, 1–14. doi: 10.1016/j.aquaculture.2006.02.023
Saravana, B. P., and Geraldine, P. (2000). Histopathology of the hepatopancreas and gills of the prawn Macrobrachium malcolmsonii exposed to endosulfan. Aquat. Toxicol. 50, 331–339. doi: 10.1016/s0166-445x(00)00096-5
Schock, T. B., Duke, J., Goodson, A., Weldon, D., Brunson, J., Leffler, J. W., et al. (2013). Evaluation of Pacific white shrimp (Litopenaeus vannamei) health during a super intensive aquaculture growout using NMR-based metabolomics. PLoS One 8:e59521. doi: 10.1371/journal.pone.0059521
Shapiro, J. A., Huang, W., Zhang, C., Hubisz, M. J., Lu, J., Turissini, D. A., et al. (2007). Adaptive genic evolution in the Drosophila genomes. Proc. Natl. Acad. Sci. U.S.A. 104, 2271–2276. doi: 10.1073/pnas.0610385104
Si, L., Pan, L., Wang, H., and Zhang, X. (2018). Identification of the role of Rh protein in ammonia excretion of swimming crab Portunus trituberculatus. J. Exp. Biol. 221:184655. doi: 10.1242/jeb.184655
Sinha, A. K., Diricx, M., Chan, L. P., Liew, H. J., Kumar, V., Blust, R., et al. (2012). Expression pattern of potential biomarker genes related to growth, ion regulation and stress in response to ammonia exposure, food deprivation and exercise in common carp (Cyprinus carpio). Aquat. Toxicol. 122–123, 93–105. doi: 10.1016/j.aquatox.2012.05.013
Small, K. S., Brudno, M., Hill, M. M., and Sidow, A. (2007). Extreme genomic variation in a natural population. Proc. Natl. Acad. Sci. U.S.A. 104, 5698–5703. doi: 10.1073/pnas.0700890104
Smith, D. D. Jr., and Campbell, J. W. (1988). Distribution of glutamine synthetase and carbamoyl-phosphate synthetase I in vertebrate liver. Proc. Natl. Acad. Sci. U.S.A. 85, 160–164. doi: 10.1073/pnas.85.1.160
Song, Z., Si, N., and Xiao, W. (2018). A novel mutation in the CRYAA gene associated with congenital cataract and microphthalmia in a Chinese family. BMC Med. Genet. 19:190. doi: 10.1186/s12881-018-0695-5
Sun, G., Dong, Y., Sun, C., Yao, H., and Lin, Z. (2021). Vital role of Glutamate dehydrogenase gene in ammonia detoxification and the association between its SNPs and ammonia tolerance in Sinonovacula constricta. Front. Physiol. 12:664804. doi: 10.3389/fphys.2021.664804
Sunga, J., Wilson, J. M., and Wilkie, M. P. (2020). Functional re-organization of the gills of metamorphosing sea lamprey (Petromyzon marinus): preparation for a blood diet and the freshwater to seawater transition. J. Comp. Physiol. B 190, 701–715. doi: 10.1007/s00360-020-01305-1
Syvänen, A. C. (2001). Accessing genetic variation: genotyping single nucleotide polymorphisms. Nat. Rev. Genet. 2, 930–942. doi: 10.1038/35103535
The Ministry of Agriculture and Fishery of the People’s Republic of China. (2021). Chinese Fishery Statistical Yearbook 2021. Beijing: China Agriculture Press.
Tok, C. Y., Chew, S. F., and Ip, Y. K. (2011). Gene cloning and mRNA expression of glutamate dehydrogenase in the liver, brain, and intestine of the swamp eel, Monopterus albus (Zuiew), exposed to freshwater, terrestrial conditions, environmental ammonia, or salinity stress. Front. Physiol. 2:100. doi: 10.3389/fphys.2011.00100
Ueberham, E., Arendt, E., Starke, M., Bittner, R., and Gebhardt, R. (2004). Reduction and expansion of the glutamine synthetase expressing zone in livers from tetracycline controlled TGF-beta1 transgenic mice and multiple starved mice. J. Hepatol. 41, 75–81. doi: 10.1016/j.jhep.2004.03.024
Wang, G., Li, X., and Li, J. (2013). Association between SNPs in interferon regulatory factor 2 (IRF-2) gene and resistance to Aeromonas hydrophila in freshwater mussel Hyriopsis cumingii. Fish Shellfish Immunol. 34, 1366–1371. doi: 10.1016/j.fsi.2013.02.006
Wang, G., Pan, L., and Ding, Y. (2014). Defensive strategies in response to environmental ammonia exposure of the sea cucumber Apostichopus japonicus: glutamine and urea formation. Aquaculture 432, 278–285. doi: 10.1016/j.aquaculture
Wang, Y., and Walsh, P. J. (2000). High ammonia tolerance in fishes of the family Batrachoididae (Toadfish and Midshipmen). Aquat. Toxicol. 50, 205–219. doi: 10.1016/s0166-445x(99)00101-0
Widman, J. C., Meseck, S. L., Sennefelder, G., and Veilleux, D. J. (2008). Toxicity of un-ionized ammonia, nitrite, and nitrate to juvenile bay scallops, Argopecten irradians irradians. Arch. Environ. Contam. Toxicol. 54, 460–465. doi: 10.1007/s00244-007-9051-z
Wu, H., Ji, C., Wei, L., Zhao, J., and Lu, H. (2013). Proteomic and metabolomic responses in hepatopancreas of Mytilus galloprovincialis challenged by Micrococcus luteus and Vibrio anguillarum. J. Proteomics 94, 54–67. doi: 10.1016/j.jprot.2013.09.001
Yang, C., Wang, L., Jiang, Q., Wang, J., Yue, F., Zhang, H., et al. (2013). The polymorphism in the promoter region of metallothionein 1 is associated with heat tolerance of scallop Argopecten irradians. Gene 526, 429–436. doi: 10.1016/j.gene.2013.05.033
Yang, C., Wang, L., Liu, C., Zhou, Z., Zhao, X., and Song, L. (2015). The polymorphisms in the promoter of HSP90 gene and their association with heat tolerance of bay scallop. Cell Stress Chaperones 20, 297–308. doi: 10.1007/s12192-014-0546-z
Yoon, J. H., Smith, L. E., Feng, Z. H., Tang, M. S., Lee, C. S., and Pfeifer, G. P. (2001). Methylated CpG dinucleotides are the preferential targets for G-to-T transversion mutations induced by benzo[a]pyrene diol epoxide in mammalian cells: similarities with the p53 mutation spectrum in smoking-associated lung cancers. Cancer Res. 61, 7110–7117. doi: 10.1097/00002820-200110000-00014
Zhang, B., Chen, N., Huang, C., Huang, C., Chen, B., Liu, H., et al. (2016). Molecular response and association analysis of Megalobrama amblycephala fih-1 with hypoxia. Mol. Genet. Genomics 291, 1615–1624. doi: 10.1007/s00438-016-1208-x
Zhang, H., Sun, G., Lin, Z., Yao, H., and Dong, Y. (2020). The razor clam Sinonovacula constricta uses the strategy of conversion of toxic ammonia to glutamine in response to high environmental ammonia exposure. Mol. Biol. Rep. 47, 9579–9593. doi: 10.1007/s11033-020-06018-w
Zhang, L., Pan, Q., Xu, L., and Si, L. (2018). Effects of ammonia-N exposure on the concentrations of neurotransmitters, hemocyte intracellular signaling pathways and immune responses in white shrimp Litopenaeus vannamei. Fish Shellfish Immunol. 75, 48–57. doi: 10.1016/j.fsi.2018.01.046
Zhang, M., Li, M., Wang, R., and Qian, Y. (2018). Effects of acute ammonia toxicity on oxidative stress, immune response and apoptosis of juvenile yellow catfish Pelteobagrus fulvidraco and the mitigation of exogenous taurine. Fish Shellfish Immunol. 79, 313–320. doi: 10.1016/j.fsi.2018.05.036
Zhang, M. Y., Wei, D., Li, R., Jia, H. T., Liu, Y. W., Taning, C. N. T., et al. (2018). Cytoplasmic glutamine synthetase gene expression regulates larval development in Bactrocera dorsalis (Hendel). Arch. Insect Biochem. Physiol. 97:e21447. doi: 10.1002/arch.21447
Zhao, J., Cui, B., Yao, H., Lin, Z., and Dong, Y. (2020). A potential role of bone morphogenetic protein 7 in shell formation and growth in the razor clam Sinonovacula constricta. Front. Physiol. 11:1059. doi: 10.3389/fphys.2020.01059
Zhao, X. L., Fu, J. P., Jiang, L. T., Zhang, W. W., Shao, Y. N., Jin, C. H., et al. (2018). Transcriptome-based identification of the optimal reference genes as internal controls for quantitative RT-PCR in razor clam (Sinonovacula constricta). Genes Genomics 40, 603–613. doi: 10.1007/s13258-018-0661-9
Keywords: Sinonovacula constricta, ammonia tolerance, Glutamine synthetase, SNP, immunohistochemistry, RNAi
Citation: Sun G, Sun C, He J, Yao H, Dai W, Lin Z and Dong Y (2021) Characterizing the Role of Glutamine synthetase Gene on Ammonia Nitrogen Detoxification Metabolism of the Razor Clam Sinonovacula constricta. Front. Mar. Sci. 8:793118. doi: 10.3389/fmars.2021.793118
Received: 11 October 2021; Accepted: 28 October 2021;
Published: 06 December 2021.
Edited by:
Yuehuan Zhang, South China Sea Institute of Oceanology, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Shoubao Yang, Shaoxing University, ChinaXiujun Sun, Chinese Academy of Fishery Sciences (CAFS), China
Copyright © 2021 Sun, Sun, He, Yao, Dai, Lin and Dong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jing He, aGVqaW5nMjI2QDEyNi5jb20=; Yinghui Dong, ZG9uZ3lpbmdodWkxMThAMTI2LmNvbQ==
 Gaigai Sun1,2
Gaigai Sun1,2 Changsen Sun
Changsen Sun Hanhan Yao
Hanhan Yao Yinghui Dong
Yinghui Dong