
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 11 February 2022
Sec. Aquatic Microbiology
Volume 8 - 2021 | https://doi.org/10.3389/fmars.2021.787879
This article is part of the Research Topic Microbial Ecology of the Arabian/Persian Gulf View all 9 articles
 Thirumahal Muthukrishnan1
Thirumahal Muthukrishnan1 Christiane Hassenrück2,3
Christiane Hassenrück2,3 Dhikra Al Fahdi1
Dhikra Al Fahdi1 Liny Jose4
Liny Jose4 Fahad Al Senafi5
Fahad Al Senafi5 Huda Mahmoud4
Huda Mahmoud4 Raeid M. M. Abed1*
Raeid M. M. Abed1*
Marine biofouling is a complicated process involving changes within micro- and macro-fouling community, species co-occurrence, and inter-taxa association patterns. An investigation of all above-mentioned aspects has rarely been conducted so far. Our study aimed to compare the monthly succession of the biofouling community developed at two locations each in the north- (Kuwait) and south-west (Oman) of the Arabian Gulf (AG) over 6 months, and to explore the association patterns within microfouling and between micro- and macro-fouling communities on a temporal and spatial scale. Spatio-temporal effects on the abundance and composition of micro- and macro-fouling communities were detected based on total biomass, bacterial and phototroph abundances, macrofouling coverage and 16S rRNA gene sequencing. We documented the development of distinct ecological niches within the fouling community resulting in fundamentally different succession patterns depending on location. Network analysis revealed nine clusters of highly interconnected co-occurring fouling bacterial taxa (M1-M9), with strong association (both positive and negative) to microalgae and macrofoulers in both Kuwait and Oman. Early stages of Kuwait biofilm showed M7 (cyanobacterial OTUs) positively and negatively associated with the majority of diatoms and macroalgae (Cladophoraceae), respectively, unlike the later stages where M5 (composed of Vibrio spp.) was positively associated with polychaetes (Hydroides elegans). While the causal relationships behind the observed inter-taxa associations remain unknown, our study provided insights into the underlying dynamics of biofouling processes encountered in the north- and south-west of the AG. Comprehensive future investigations encompassing transcriptomic or metabolomic tools may be required to address the challenge of interpreting such complicated dynamics over time and space in a continuously changing environment.
Marine biofouling, defined as the undesirable accumulation of benthic organisms on submerged surfaces, is a multistage process that involves surface conditioning, followed by formation of microbial biofilms (e.g., bacteria, diatoms, and algae), and the settlement of sessile (e.g., barnacles, hydroids, mussels, sponges, and bryozoans) and mobile (e.g., crabs, shrimps, and snails) macrofouling organisms (Dang and Lovell, 2016). The structure and speed of development of micro- and macro-fouling communities are influenced by many environmental variables, such as current flow, light availability, nutrient load (Qian et al., 2007; Dang and Lovell, 2016; de Carvalho, 2018), as well as by biological factors, such as the availability, growth rate and metabolic state of fouling species and inter-species interactions (Dang and Lovell, 2016; Pollet et al., 2018; Zhang et al., 2019). Such combination of environmental and biological factors leads to temporal and spatial variations in the composition of fouling communities (Dang and Lovell, 2000, 2016; Oberbeckmann et al., 2016; Navarrete et al., 2020).
Owing to the complexity of the biofouling process, and the involvement of many exogenous and endogenous factors, most studies related to temporal succession of biofouling communities have focused on either micro- (Rampadarath et al., 2017; Pollet et al., 2018; Antunes et al., 2020) or macro-fouling (Naser, 2017; Briand et al., 2018; Navarrete et al., 2020) communities, rather than both together. Short-term, i.e., hourly, and daily succession of microfouling communities has been investigated to identify primary and secondary colonizers (Dang and Lovell, 2000; Pollet et al., 2018; Antunes et al., 2020). Long-term succession studies mostly involved analyzing microfouling communities that were developed for two (Satheesh and Wesley, 2012; Elifantz et al., 2013) or four weeks per season (Antunes et al., 2020), while analyses pertaining to monthly succession were quite limited (Pinto et al., 2019). Conversely, succession of macrofouling communities has been extensively investigated beginning from two until a maximum of 84 months (Macleod et al., 2016; Naser, 2017; Navarrete et al., 2020). The exploration of spatial effects on micro- (Briand et al., 2012; Muthukrishnan et al., 2017; Rampadarath et al., 2017) and macro-fouling (Macleod et al., 2016; Navarrete et al., 2020) community development extended to several ocean basins, such as the Arabian Gulf (AG), and the Indian and Atlantic oceans. Despite the extensive literature on temporal and spatial changes in biofouling communities, the concurrent spatio-temporal effects on both micro- and macro-fouling communities have been underexplored, except for two studies in the Atlantic Ocean (Dziubińska and Janas, 2007; De Tender et al., 2017).
The co-occurrence of either different micro- or macro-fouling species or both together at any stage of biofouling can be partially driven by non-specific (forced co-existence due to concurrent taxa availability) and/or specific associations involving the establishment of positive (e.g., symbiotic) and negative (e.g., parasitic) relationships (Flemming et al., 2016; Rampadarath et al., 2017; Price et al., 2021). Co-occurrent species allow for the development of ecological niches within the biofouling community (Rampadarath et al., 2017; Pollet et al., 2018). The identification of such co-occurrence networks has become possible using weighted correlation network analyses of high throughput sequence datasets (Milici et al., 2016; Weiss et al., 2016; Hirano and Takemoto, 2019). Previous studies using co-occurrence network analyses were mostly conducted on freshwater (Liu et al., 2014; Lin et al., 2019), and terrestrial (Barberán et al., 2012; Cobo-Díaz et al., 2019; Price et al., 2021) ecosystems, whereas marine ecosystem-based network analyses focused mainly on the planktonic bacterial communities of the Atlantic and Arctic oceans (Guidi et al., 2016; Milici et al., 2016; Fadeev et al., 2018). Co-occurrence network analyses on micro- and macro-fouling communities have been rarely reported.
Biofouling has serious impacts on the maritime and oil-refinery industries in the AG region (Stachowitsch et al., 2002; Naser, 2017; Al Senafi et al., 2020). The environment of AG is unique due to its low-lying slopes, high temperatures, high evaporation rates (0.2–2 m yr–1) and salinities (up to 70 ppt), strong monsoon and dry Shamal winds resulting in frequent dust storms (Piontkovski et al., 2012; Al Senafi et al., 2019; Vaughan et al., 2019). The geomorphologies of the north- and the south-west of AG are different resulting in region-specific hydrological regimes and clear differences in the physico-chemical seawater characteristics (Pous et al., 2004; Piontkovski and Queste, 2016; Al Senafi et al., 2020). The north-west of AG has a comparatively more flattened seabed, higher water temperatures and is prone to recurrent dust deposition compared to the south-west, which has a naturally more wedged seabed near the Strait of Hormuz and Sea of Oman resulting in differing seawater densities and current flow patterns (Al-Zaidan et al., 2003; Piontkovski and Queste, 2016; Ahd et al., 2019). Research on biofouling in AG is required for the establishment of more efficient strategies to reduce biofouling and related environmental impacts on marine infrastructure.
In this study, we hypothesize that given the environmental differences between the north- and south-west of the AG, the development and succession of micro- and macro-fouling communities and inter-taxa associations will exhibit spatio-temporal variation. The main objectives of the study were: (1) to compare the biofouling community development between the north- and south-west of AG monthly over a period of 6 months and, (2) to study the association patterns within microfouling communities and between micro- and macro-fouling communities over time and space.
The spatial and temporal succession of biofouling communities was investigated in four marinas: Fintas (29°09′52.9″N 48°07′35.6″E) and Salmiya (29°20′30.2″N 48°03′10.1″E) in Kuwait (north-west of the AG), and Al Mouj (23°37′50.4″N 58°16′11.1″E) and Bandar Rowdha (23°34′55.6″N 58°36′24.6″E) in Oman (south-west of the AG; Supplementary Figure 1). In each location, a total of 18 marine grade acrylic panels (15 × 20 cm) were randomly fastened onto 2 m long polyvinyl chloride (PVC) pipes and were deployed at 2 m depth using nylon ropes. Biofouling communities developed on three replicate panels were sampled at the end of each month between August 2017 and January 2018 (total number of panels = 72; 4 locations × 3 replicas × 6 time points). The chosen time period spanned a transition from the summer to winter seasons in all locations. The physico-chemical parameters of the seawater including temperature, pH, salinity, and turbidity were monitored every month in the four locations using thermometer, pH meter, refractometer, and turbidity meter (HI 98703, HANNA, United States), respectively.
The panels were carefully pulled out of the water and each panel was immediately photographed to calculate the percentage coverage of macrofoulers using the ImageJ v.1.00 software, relative to the total surface area of each panel (Schneider et al., 2012). Obtained photos were later used to identify the major macroalgal groups and macrofouling species based on the Global Invasive Species Database1 and previous taxonomic records of the region (Dobretsov, 2015; Naser, 2017). The total biomass developed on each panel was scraped off using a sterile scalpel and was collected in 50 ml sterile Falcon tubes (Falcon®, United States). To estimate the bacterial abundance, a homogenous suspension was prepared by mixing 50 mg of biofilm (n = 3) with 100 μl of sterile distilled water. A volume of 10 μl of this mixture was then spread evenly on a clean glass slide and stained with 5 μl of 4′,6-diamidino- 2-phenylindole (DAPI, Sigma, Munich, Germany) solution (Dobretsov and Thomason, 2011). Bacterial abundance was calculated by directly counting cells in 20 randomly selected fields of view using epifluorescence microscopy (Olympus BX51, United States, magnification 1000×; λEx = 359 nm, λEm = 441 nm). To estimate the abundance of phototrophs, the concentration of chlorophyll a was measured (Ahmad et al., 2015). Briefly, 100–300 mg of biofilm (n = 3) were mixed with 90% acetone and incubated in the dark for 24 h at 4°C. The obtained extracts were spectrophotometrically analyzed at 645, 662, and 750 nm (Ahmad et al., 2015). Data of the physico-chemical parameters of the seawater samples and the biofouling community abundance are available on Pangaea doi.org/10.1594/PANGAEA.925308, doi.org/10.1594/PANGAEA.925310.
Biofilm samples (wet weight = 0.1–0.2 g) from three replicates per time point at each location were subjected to DNA extraction using a previously described protocol (Lueders et al., 2004) with modifications in the cell lysis step (Muthukrishnan et al., 2018). Approximately 0.1 g of glass beads (diameter; 0.2–0.3 mm) was added to each of the replicate biofilm samples. A total volume of 750 μl of NaPO4 buffer (120 mM, pH 8.0) and 250 μl of TNS solution (500 mM Tris–HCl, 100 mM NaCl, 10% SDS (w/v), pH 8.0) were added to each sample-bead mixture. Each sample was vortexed using the horizontal vortex adaptor (Vortex- Genie 2, Mo Bio, United States) for 5 min and incubated at 65°C for 30 min. The heated sample mixture was vortexed briefly and centrifuged for 5 min (16000 × g, 4°C). A total volume of 900 μl of the supernatant was transferred to a 2 ml Eppendorf tube and an equal volume of phenol-chloroform-isoamyl alcohol [25:24:1 (vol/vol/vol), Sigma-Aldrich, United States] was added. The mixture was briefly vortexed and centrifuged for 5 min (16000 × g, 4°C), following which the upper clear phase was carefully pipetted (650 μl) into a new 2 ml Eppendorf tube and mixed thoroughly with 2 volumes (1300 μl) of polyethylene glycol (PEG; 40% w/v) to precipitate the DNA. The sample was centrifuged for 30 min (16000 × g, 4°C) and the liquid was decanted gently thereafter. A total volume of 500 μl of ice cold 70% ethanol was added to wash the pellet, followed by centrifugation for 4 min (16000 × g, 4°C). The pellet was air dried at room temperature and resuspended in 20–30 μl of elution buffer (10 mM Tris–HCl, pH 8.5). The concentration and purity of DNA extracts obtained were measured on a NanoDrop (Thermo Fisher Scientific, United States).
Purified DNA extracts were submitted to Molecular Research (MR) DNA laboratory2 (Shallowater, TX, United States) for paired-end Illumina MiSeq sequencing of the V3-V4 hypervariable region of the bacterial 16S rRNA gene (forward primer: CCTACGGGNGGCWGCAG, reverse primer: GACTACNVGGGTATCTAATCC; Klindworth et al., 2013). Sequences were generated on two separate MiSeq runs for the biofilm samples collected in August/September and October to January, respectively, using a single indexing strategy. Samples were demultiplexed, and sequencing adapters, barcodes, and primers were removed using the fastqprocessor developed by MRDNA (Shallowater, TX, United States, date accessed: 23.02.2019), which further ensured the correct orientation of all reads as forward (R1) and reverse (R2). After demultiplexing, intact read pairs were extracted using pairfq_lite.3 These sequences were deposited to the European Nucleotide Archive (ENA) under the accession number PRJEB41883 in compliance with the Minimal Information about any (X) Sequence (MIxS) standard (Yilmaz et al., 2011) using the brokerage service GFBio (Diepenbroek et al., 2014). Further processing was conducted in R v3.5.2 (R Core Team, 2018) using the R package dada2 v1.10.1 (Callahan et al., 2016) with default parameters if not otherwise indicated. Quality filtering was conducted at a maximum expected error rate of 3 for both forward and reverse reads after reads were truncated to 230 bp. Error learning (based on at least 108 bases) and denoising (pooling all sequences per run) were executed separately for each MiSeq run. Forward and reverse reads of each sample were merged with a minimum overlap of 10 bp, and chimera detection was performed with the method ‘consensus’ on the whole dataset. Furthermore, only sequences between 399 and 430 bp as well as those occurring at least twice in the data set were retained. Taxonomic classification was conducted using the SILVA NGS web-service (date accessed 12.04.2019) with the SILVA ribosomal database (version 132; Quast et al., 2012). Only bacterial sequences classified on phylum level with a sequence similarity of at least 93% to the reference database and not affiliated with chloroplasts and mitochondria were used for the further analysis. Operational taxonomic units (OTUs) were defined as unique amplicon sequence variants.
Statistical analysis was conducted in R 3.5.1 using R studio 1.0.153 (R Studio Team, 2016). As the monthly environmental parameters in every marina were not conducted in replicates, only differences between location (Oman vs. Kuwait) and marinas (nested within location) were tested accounting for the variability over time using a sequential general linear mixed model with sampling month as random factor (lmerTest 3.0.1; Kuznetsova et al., 2017). Patterns in biological parameters (biomass, chlorophyll a, coverage, bacterial counts), where replicate measurements were available, were tested using a full-factorial ANOVA model, again with marina nested within location. Normality and homoscedasticity were assessed based on residual versus fitted plots, and parameters were reciprocal or log transformed if required to meet the assumptions of the applied tests.
Alpha diversity was assessed based on the number of OTUs (richness), and the inverse Simpson index (effective species richness; Chao et al., 2014). These indices were calculated after rarefying the OTU table repeatedly (100 times) to 25191 sequences per sample. Differences in alpha diversity between sampling month, location, and marina were tested using the same ANOVA model as described above. Non-metric multidimensional scaling (NMDS) based on Bray-Curtis dissimilarities of relative OTU proportions was performed to explore patterns in beta diversity. PERMANOVA based on square-root transformed Bray-Curtis dissimilarities was then used to test for differences in community composition between sampling month, location, and marina as described above. Furthermore, the contribution of sampling month and marina to explaining the variation in bacterial community composition at each sampling location was explored by separate PERMANOVA tests based on the R2 value.
Sparse Inverse Covariance for Ecological Statistical Inference (SPIECEASI 1.0.2, phyloseq 1.26.0; McMurdie and Holmes, 2013; Kurtz et al., 2015) was applied to construct co-occurrence networks. Prior to network analysis, rare and low coverage OTUs were removed from the analysis, i.e., OTUs had to be present with a sequence proportion of at least 0.01% in at least 10% of the samples and constitute at least 0.1% in three samples. This data set reduction did not alter beta diversity trends (Mantel test, r = 0.98, p = 0.001). Louvain clustering (igraph 1.2.2; Csardi and Nepusz, 2006) generated nine network modules (i.e., sub-communities of co-occurring OTUs). Module eigengenes were calculated to obtain a representative average OTU profile for each module (weighted correlation network analysis; WGCNA 1.66; Langfelder and Horvath, 2008). A PCA was conducted with the eigengenes to explore the associations between modules and sampling month and marinas. Characteristic OTUs for each module were selected based on a high sequence proportion and correlation to their respective eigengenes.
The microalgal community was characterized using the chloroplast sequences of the 16S data set (Yeh et al., 2021). Taxonomy was assigned based on the best BLAST (Camacho et al., 2009) hit to a custom chloroplast database with a mean of sequence identity and query coverage of at least 93. The reference database was generated from chloroplast sequences downloaded from NCBI with the following search query: (chloroplast[All Fields] AND 16S[All Fields] AND ribosomal[All Fields] AND complete[All Fields]) AND (protists[filter] OR plants[filter]) AND chloroplast[filter]) (date accessed 15.01.2019). After taxonomic classification, non-marine taxa were removed from the dataset, and only those taxa with an occurrence of at least 10 sequences per sample were considered for the presence/absence-based analysis to avoid the detection of microalgae that were present by chance and which were not tightly associated with the settlement panels. The association between the characteristic OTUs for each of the bacterial network modules (eigengenes) and the microalgal/macrofouling taxa was explored based on the biserial correlation coefficient. For this analysis, the data set was split by sampling location to account for unknown/unobserved factors, which may differ between Oman and Kuwait, and which may influence the association between bacteria and microalgae/macrofoulers. Only microalgae and macrofoulers present and absent in at least three samples were considered for the correlation analysis to reduce bias due to unbalanced observations.
The metabolic potential of the bacterial communities was estimated using pathway prediction by phylogenetic placement (PAPRICA 0.5.1; Bowman and Ducklow, 2015) with default parameters. Predicted metabolic pathways, which did not contribute 0.001% in at least 10 samples or 0.01% in at least 3 samples were removed from the dataset. The remaining pathways were grouped based on the MetaCyc ontology4 and the proportion of the following metabolic functions was compared across samples: Autoinducer Biosynthesis, Cell Structure Biosynthesis, CO2 Fixation, Nitrogen fixation, Nitrate Reduction, Sulfide Oxidation, Sulfite Oxidation, Phosphorus Compound Metabolism, Polysaccharide Biosynthesis, Antibiotic Biosynthesis, Aromatic Compound Degradation, Detoxification, Polysaccharide Degradation, and Sugar Degradation (Dang and Lovell, 2016; Rampadarath et al., 2017). Other pathways of potential relevance in the studied system, such as Ammonia Oxidation, Iron Metabolism, Dissimilatory Sulfate Reduction, and Siderophore Biosynthesis were not detected according to PAPRICA. The bash and R scripts for the data analysis are available as supplement to the data set on PANGAEA doi.org/10.1594/PANGAEA.925308
Although the temperature, salinity, turbidity, and pH of the seawater showed changes over time in both Kuwait and Oman, significant differences were detected between the locations (Supplementary Tables 1,2). The water temperature in both Kuwait and Oman decreased from August to January with comparable values recorded during summer (August and September; 30–34°C) in all marinas. However, a 16°C drop during winter resulted in colder waters in Kuwait (17–19°C), than in Oman (24–25°C). Seawater salinity showed lowest values in November in all marinas, except Al Mouj, with overall values ranging between 24–43 ppt in Kuwait waters and 19–36 ppt in Oman waters (Supplementary Table 1). Seawater turbidity exhibited temporal changes depending on the location/marina with a maximum turbidity of 77 NTU recorded in Salmiya in December (Supplementary Table 1). Significant differences were observed between Kuwait marinas (i.e., 1–5 and 5–77 NTU in Fintas and Salmiya, respectively) rather than between Oman marinas, which showed comparable average values of 1–2 NTU. There were no significant differences in pH values at either location or marina, although lowest values were recorded in November in all marinas except Bandar Rowdha.
Mature biofouling communities developed on all submerged panels in Kuwait and Oman (Figure 1). Time, location, and marina significantly affected the total biomass, abundance of bacteria and phototrophs, and the macrofouler coverage on the panels (Figure 2 and Supplementary Table 3). After 6 months, the average biomass developed on panels was significantly higher in Oman (0.40 ± 0.22 g cm–2 at Al Mouj) than in Kuwait (0.14 ± 0.01 g cm–2 at Salmiya; Figure 2A, Supplementary Table 3). Bacterial counts showed more prominent fluctuations over time in Kuwait than in Oman with the highest abundance detected after 6 months in the latter location (Figure 2B). The abundance of phototrophs estimated by chlorophyll a concentration gradually increased over time in both marinas in Oman and Kuwait (only until December in Fintas). The highest abundance of phototrophs in all marinas was detected after 6 months in Salmiya (Figure 2C). Percentage coverage of macrofoulers increased with time and was significantly higher on Oman panels (>86% of the panel’s total surface area) than on the Kuwait panels (<24%) after 6 months (Figure 2D and Supplementary Table 3).
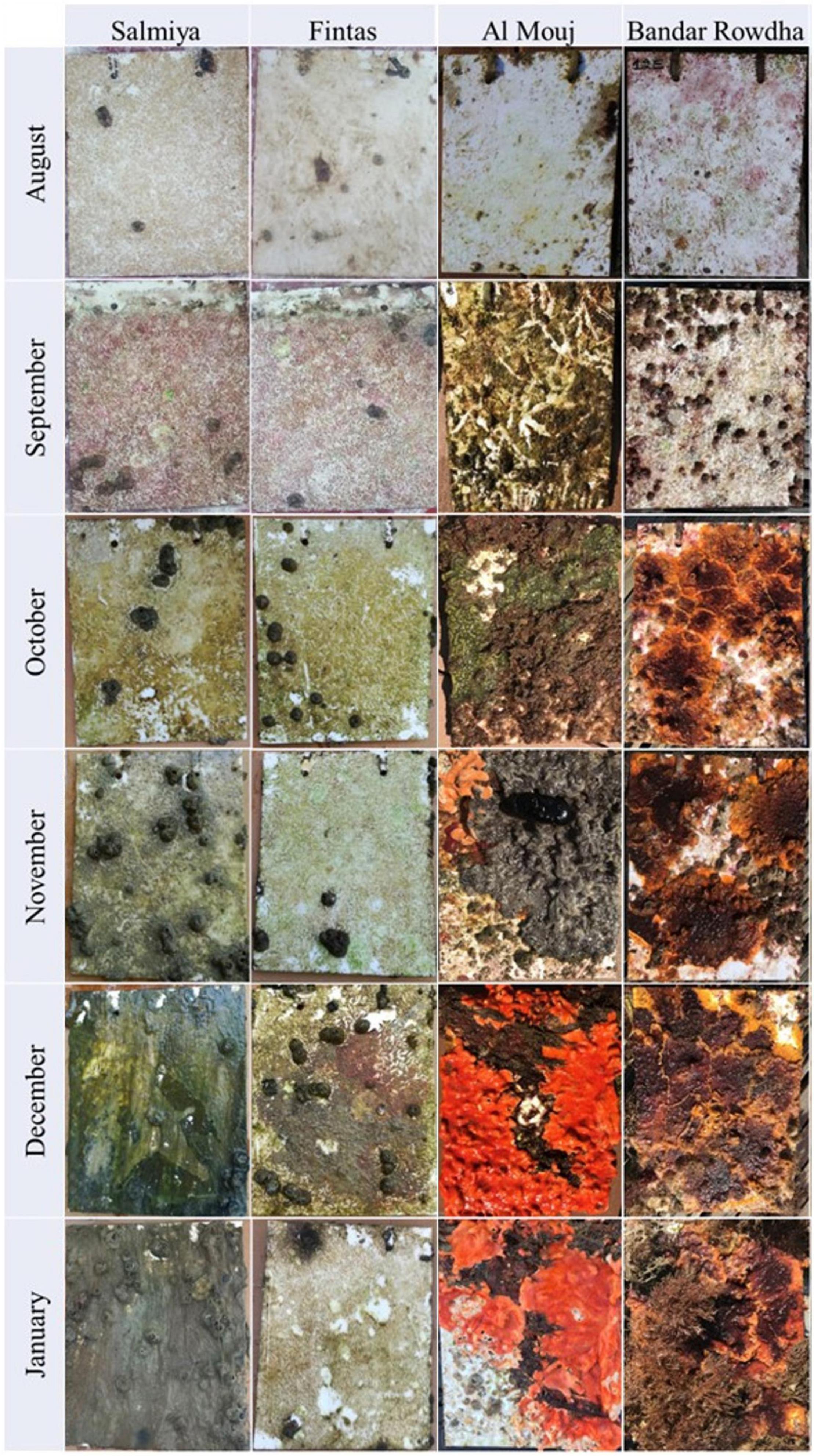
Figure 1. Development of mature biofouling community on acrylic panels at the sampling locations in Kuwait (Fintas and Salmiya) and Oman (Bandar Rowdha and Al Mouj) during the sampling period of 6 months from August 2017 to January 2018.
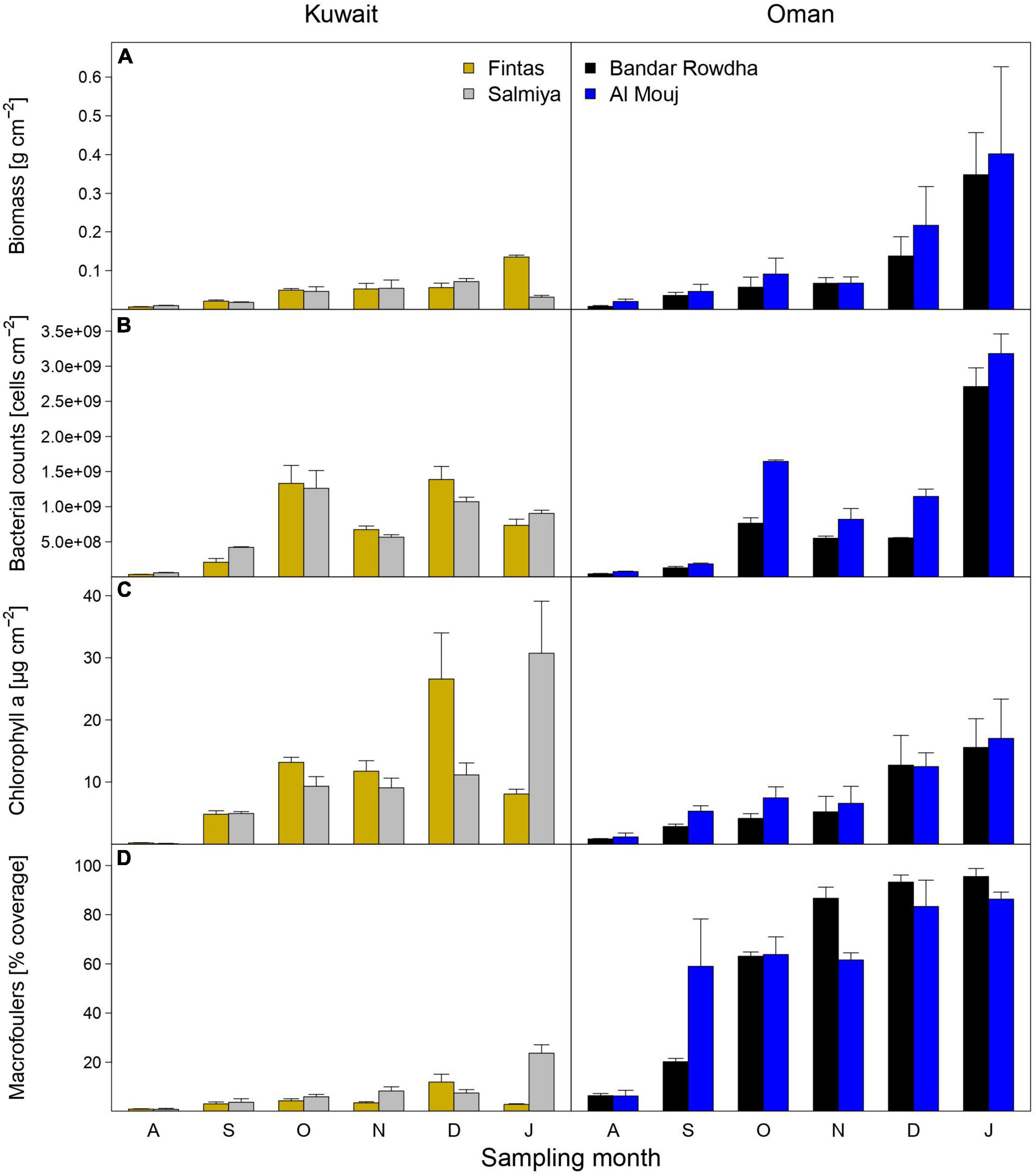
Figure 2. The abundance of biofouling communities as estimated using measurements of (A) total biomass, (B) bacterial counts, (C) chlorophyll a concentration, and (D) macrofouler coverage at the sampling locations in Kuwait (Fintas and Salmiya) and Oman (Bandar Rowdha and Al Mouj) during the sampling period of 6 months from August 2017 to January 2018 (abbreviated as A, S, O, N, D, J). Data presented as mean ± standard deviation (n = 3).
Richness (OTU number) and Inverse Simpson Index estimated for the fouling bacterial communities changed significantly over time in both Kuwait and Oman, with different patterns displayed in each location (Figures 3A,B and Supplementary Table 3). In Kuwait, the highest diversity was detected after one month in both marinas, followed by a steep decline after the second month and a constant increase thereafter until the end of the experiment, except for November. However, in Oman, the changes in bacterial diversity were complex since the variability within each month was higher than between months compared to Kuwait (Figures 3A,B). Diversity patterns differed in each marina wherein Bandar Rowdha did not show a clear trend while a diversity spike was recorded in November in Al Mouj (Figures 3A,B). NMDS based on Bray-Curtis dissimilarities of relative OTU proportions showed a clear separation of bacterial communities by location (Figure 3C). In general, bacterial communities were more heterogeneous in Kuwait and mainly clustered by sampling time rather than marina. Conversely, community composition in Oman was less heterogeneous and more strongly affected by differences between marinas. These patterns were confirmed to be significant by PERMANOVA, with sampling time exhibiting the highest explanatory power in Kuwait, while marina was attributed more importance in Oman (Figure 3C and Supplementary Table 4).
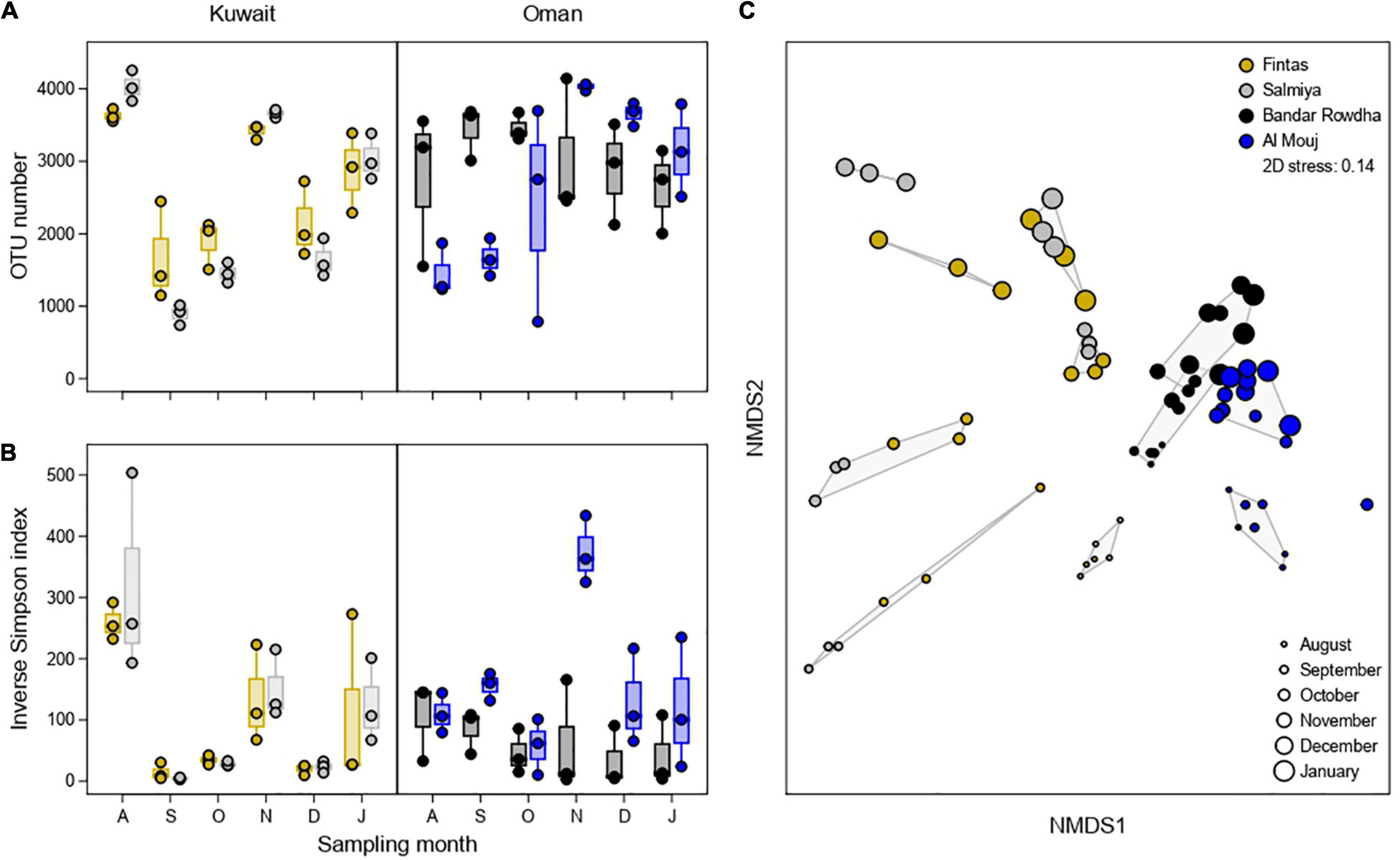
Figure 3. Alpha diversity of the fouling bacterial community based on (A) OTU number and (B) Inverse Simpson Index at the sampling locations in Kuwait (Fintas and Salmiya) and Oman (Bandar Rowdha and Al Mouj) during the sampling period of 6 months from August 2017 to January 2018 (abbreviated as A, S, O, N, D, J). (C) NMDS ordination plot showing the segregation of fouling bacterial communities based on location and sampling time. Shaded hulls indicate groups of samples with a Bray-Curtis dissimilarity of <0.8.
Distinct temporal and spatial changes were detected in the proportion of the major groups of fouling bacteria, including Proteobacteria, Bacteroidia, Acidimicrobiia, Parcubacteria, Chlorobia, Planctomycetacia, and Verrucomicrobiae (Figure 4A). In Kuwait, the bacterial community was mainly represented by Bacteroidia, Alpha- and Gammaproteobacteria. These groups showed similar patterns over time in Fintas and Salmiya with only minor variation between the marinas. The contribution of Alpha- and Gammaproteobacteria to bacterial community composition strongly fluctuated over time, with highest values detected during November and September with approx. 50 and 95% of the total number of sequences, respectively (Figure 4A). Bacteroidia proportions ranged between 5 and 30% with lower proportions only encountered in September. Oxyphotobacteria (now renamed to Cyanobacteriia) and Campylobacteria only occurred sporadically in high proportions in both marinas, with the former reaching 25–30% in August, and the latter reaching 25–40% in December (Figure 4A).
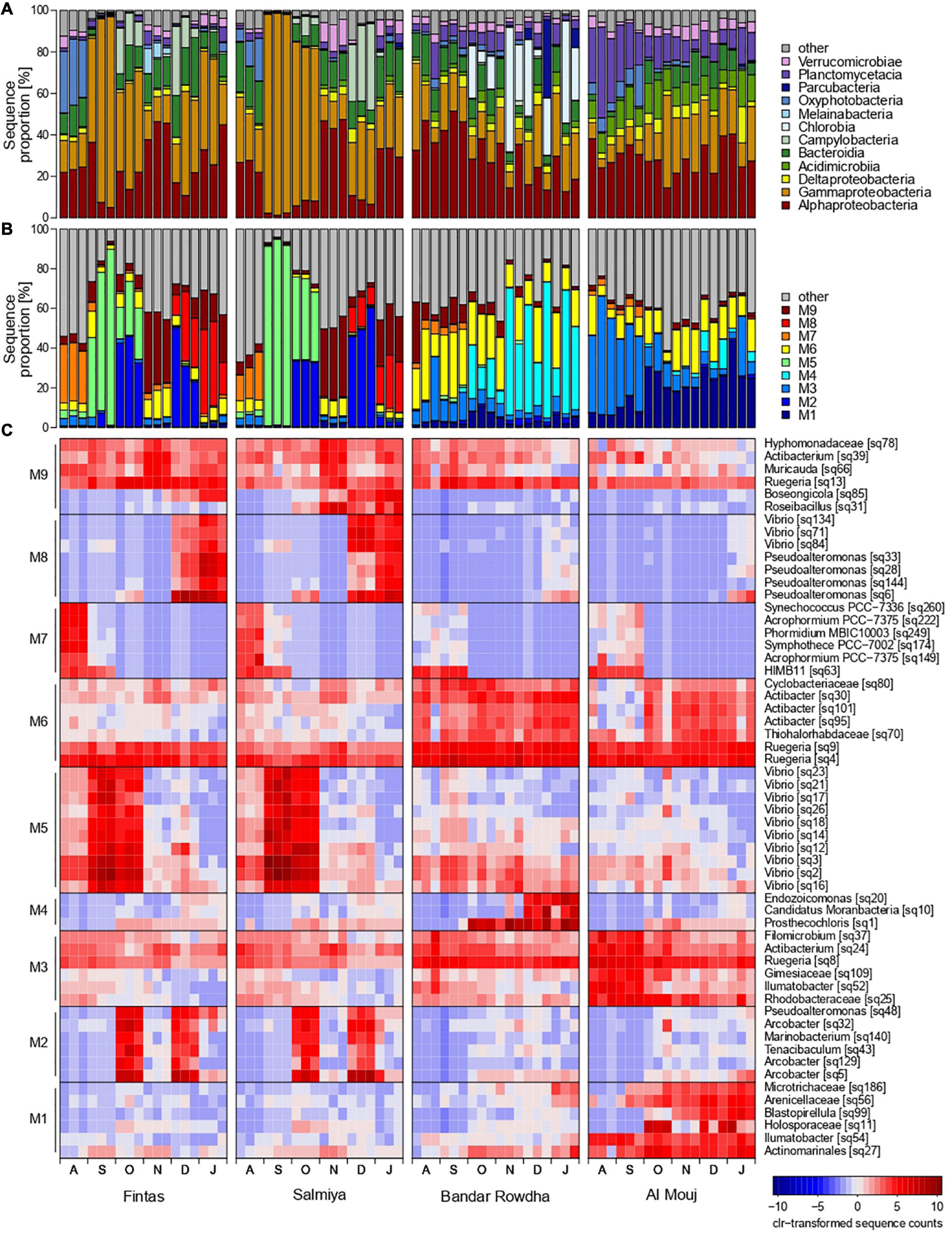
Figure 4. Changes in the proportion of bacterial sequences affiliated to (A) dominant classes, (B) network modules, and (C) OTU representatives of each module at the sampling locations in Kuwait (Fintas and Salmiya) and Oman (Bandar Rowdha and Al Mouj) during the sampling period of 6 months from August 2017 to January 2018 (abbreviated as A, S, O, N, D, J).
In Oman, Proteobacteria constituted more than 50% of the total sequences in both marinas throughout the sampling period, but each of the dominant proteobacterial classes, i.e., Alpha- and Gammaproteobacteria, exhibited distinct temporal shifts in their proportion depending on the marina (Figure 4A). In Bandar Rowdha, Alphaproteobacteria accounted for 40–50% of the total sequences in August and September but then gradually decreased to 10–30% in the later months. However, this was not the case in Al Mouj where comparable proportions (20–35%) were detected throughout the experiment. Conversely, Gammaproteobacteria showed relatively similar proportions in Bandar Rowdha (10–40%) while exhibiting an increasing trend over time in Al Mouj (Figure 4A). Other dominant bacterial classes, such as Acidimicrobia, Bacteroidia, Chlorobia, and Planctomycetacia also showed marina-dependent shifts in their proportions over time (Figure 4A). For example, Planctomycetacia did not display strong changes over time in Bandar Rowdha (5–10%) but decreased from 35–40% to less than 15% in Al Mouj. Chlorobia were only present in high proportions in Bandar Rowdha in October to January reaching 25–60% (Figure 4A).
Based on Louvain clustering, sub-communities of co-occurring bacterial OTUs were separated into nine network modules M1-9 (Figures 4B,C and Supplementary Figure 2). Each of these modules was represented by less than 10 characteristic bacterial OTUs that accounted for a high sequence proportion and showed a high correlation with their respective module eigengenes (Figures 4B,C). A PCA based on the module eigengenes revealed that the five modules M2, M5, M7-9 were mainly detected in Kuwait, while the other modules M1, M3, M4, and M6 were more prevalent in Oman (Supplementary Figure 2). These patterns were also evident when mapping the module membership onto the network graph (Supplementary Figure 2). The contributions of each module and its representative OTUs to community composition further depended on the sampling month and marina in each location (Figures 4B,C and Supplementary Figure 2). For example, Kuwait-specific modules M5 and M7 were mainly detected in the early months of biofouling, while M2, M8-9 were mostly detected in the latter months, in both marinas in Kuwait. In August, M7 was prevalent and constituted sequences affiliated with the cyanobacterial genera Symphothece, Synechococcus, Phormidium, and Acrophormium and the alphaproteobacterial clade HIMB11. M5 was dominant in September and October and exhibited a strong co-occurrence of 10 different Vibrio OTUs. The later months of biofouling in the Kuwait marinas demonstrated strong associations between bacterial OTUs belonging to diverse groups such as Proteobacteria, Bacteroidia, and Verrucomicrobiae. For example, in the December and January biofilms, co-occurring OTUs affiliated with Vibrio and Pseudoalteromonas accounted for the highest proportion of sequences in M8, while alphaproteobacterial OTUs were associated with Muricauda (Bacteroidia) and Roseibacillus (Verrucomicrobiae) in M9. Module M2, mainly represented by three different OTUs affiliated with the genus Arcobacter (Campylobacteria) contributed to a considerably high proportion of sequences in the October and December biofilms (Figure 4C).
In Oman, the bacterial network module M3 was mainly detected in August and September with comparatively higher proportions in Al Mouj than Bandar Rowdha (Figures 4B,C and Supplementary Figure 2). The majority of the characteristic OTUs detected in M3 belonged to the taxa Ruegeria, Ilumatobacter, and Rhodobacteraceae. The modules M1 and M4 were mainly encountered in the latter months of October–January in Al Mouj and Bandar Rowdha, respectively. M1 was mainly comprised of Blastopirellula (Planctomycetacia) and Alpha- and Gammaproteobacteria-related OTUs. M4 was more dominant in Bandar Rowdha and constituted OTUs belonging to the gammaproteobacterial genus Endozoicomonas and the genera Prosthecochloris (Chlorobia) and Candidatus Moranbacteria (Parcubacteria), respectively. Furthermore, regardless of the marina or month, M6 was detected throughout the biofouling period with Ruegeria-affiliated OTUs accounting for the highest sequence proportion (Figure 4C).
Among the predicted metabolic pathways of high relevance in the studied system (Supplementary Figure 3), sugar and polysaccharide degradation pathways were the most prevalent throughout the sampling period at all four marinas, although with a decreasing contribution to the metabolic potential of the bacterial community toward the end of the experiment in Al Mouj. Aromatic compound degradation was predicted in similar proportions as sugar degradation, with marina-dependent temporal patterns and slightly higher values in Oman. An increased prevalence of carbon and nitrogen fixation pathways was detected in August and September in Kuwait, presumably related to the higher proportion of cyanobacterial OTUs (module M7) at these sampling times. Interestingly, these two pathways showed an opposite temporal pattern in Bandar Rowdha in Oman. The prevalence of autoinducer biosynthesis pathways coincided with increased proportions of Vibrio (modules M5 and M8) in September and December in Kuwait (Figure 4C and Supplementary Figure 3).
A total of 44 types of microalgae were detected in the 16S dataset based on their chloroplast sequences. Kuwait-specific microalgae were comprised of seven taxa: Mantoniella, Dictyochophyceae, and Pinguiochrysidaceae, and diatom genera Odontella, Grammonema, Pleurosigma, and Nitzschia that were mainly detected in the August and September biofilms (Supplementary Figure 4). Oman-specific microalgal taxa comprising of Vitrella, Chloroparvula, and Prasinococcales were detected throughout the sampling period (Supplementary Figure 4).
A total of 18 macrofouling taxa belonging to three types of macroalgae (green, red, brown) and seven major groups of invertebrates (barnacles, encrusting bryozoans, sea squirts, hydrozoan, sponge, mollusk, and polychaete) were collectively identified based on visual analysis of all panels (Figure 1 and Supplementary Figure 5). The composition of macrofouling communities differed by location with Kuwait panels showing the lowest diversity compared to Oman (Supplementary Figure 5). Only three macrofouling taxa, including green algae (Cladophoraceae), barnacles (Balanus sp. 2), and polychaetes (Hydroides elegans), were detected in Kuwait with increasing coverage over time in both marinas (Supplementary Figure 5). In Oman, only Balanus spp., Ciona sp., Phallusia sp., and Pinctada sp. showed similar presence and dominance patterns over time in both marinas, while the other 13 detected taxa exhibited pronounced differences (Supplementary Figure 5). The encrusting bryozoan Schizoporella sp. was dominant from October onward in Bandar Rowdha, but only rarely found until December in Al Mouj. Unlike in Bandar Rowdha, all macroalgal groups (Cladophoraceae, Gelidium sp., and Phaeophyceae) were dominant during August–October in Al Mouj. Certain macrofouling taxa in Oman occurred exclusively only in one marina. For example, the sea squirt Eudistoma sp. and the hydrozoan Obelia sp. were only detected on Bandar Rowdha panels, while other types of sea squirts including Botrylloides spp., Polyclinum sp., and the sponge Pione vastifica were detected only on Al Mouj panels.
At each location, strong positive and negative association patterns were detected between certain bacterial sub-populations (co-occurrence network modules) and microalgae/macrofoulers (Figure 5). Kuwait biofilms demonstrated a strong positive association between M7 (characterized by cyanobacterial OTUs) and the majority of microalgal taxa (mainly diatoms) with joint occurrences in August, while exhibiting a mutually exclusive relationship with green macroalgae (Cladophoraceae). Further associations were detected between bacterial module M5 (characterized by Vibrio OTUs) and the two microalgal taxa Aureoumbra and Pelagomonadaceae, which were negatively associated, and module M8 (Vibrio and Pseudoalteromonas OTUs) and the polychaete (H. elegans), which were positively associated.
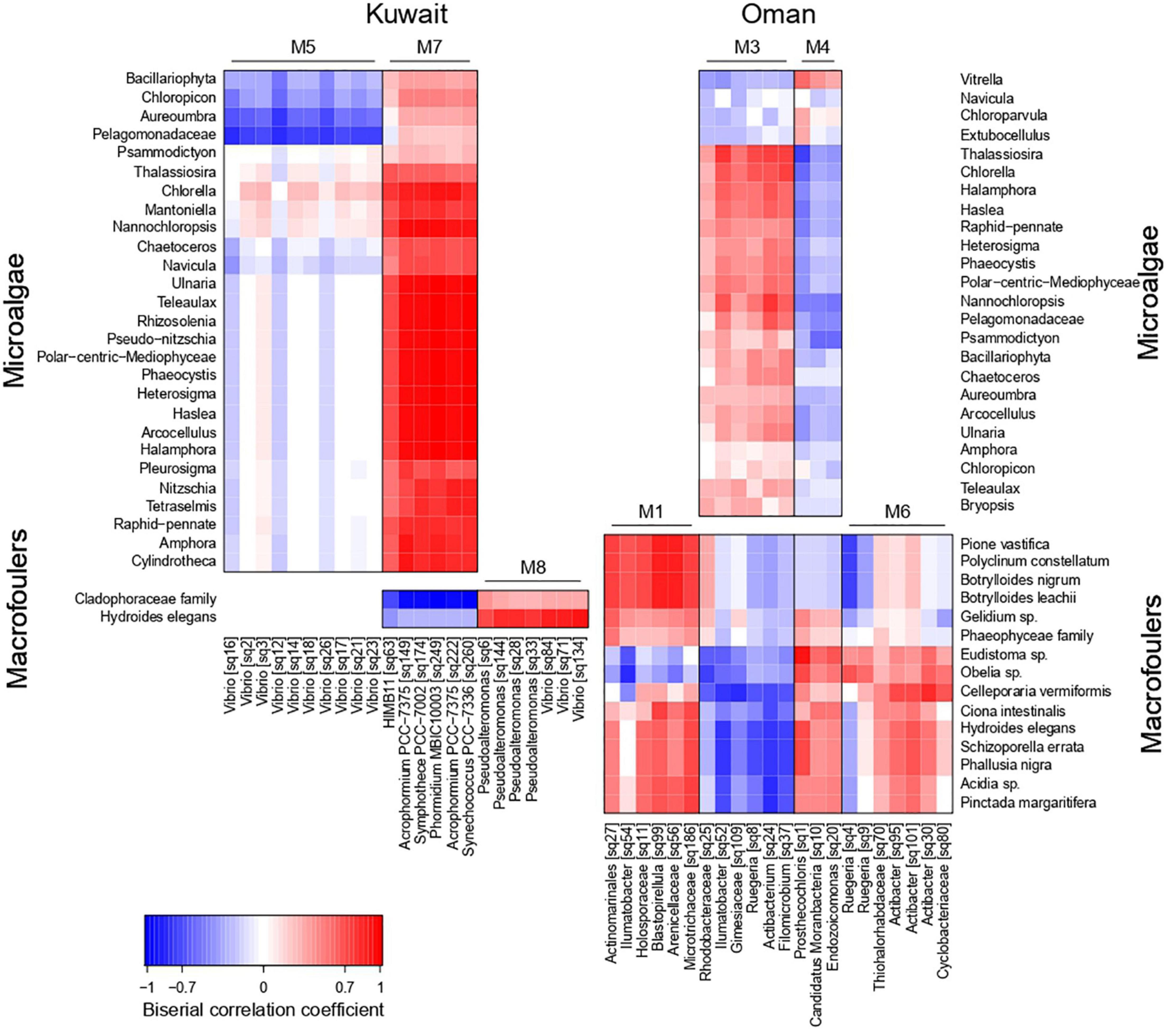
Figure 5. Heatmap showing association patterns between bacterial co-occurrence network modules (represented by their characteristic OTUs) and microalgae (upper heatmaps) and macrofoulers (lower heatmaps) in Kuwait (left hand side) and Oman (right hand side).
In Oman, the bacterial modules M3 and M4 exhibited opposite association patterns with microalgae (Figure 5), with M3 (Ruegeria, Filomicrobium, Actibacterium, and Ilumatobacter OTUs) being positively correlated to microalgal taxa, e.g., Thalassiosira, Nannochloropsis, and Chlorella, occurring during the early stages of biofouling. Association patterns with macrofoulers in Oman were strongly influenced by the marina-dependent presence of macrofoulers (Figure 5). M4 (Endozoicomonas, Candidatus Moranbacteria, and Prosthecochloris OTUs) showed a positive association with various invertebrate macrofoulers, e.g., Eudistoma sp. and Obelia sp., both occurring predominantly during the later stages of biofouling in Bandar Rowdha. Other invertebrates, e.g., Botrylloides spp., Polyclinum constellatum, and P. vastifica, found on the biofouling panels at Al Mouj in October to January, were positively associated with M1 (mainly Arenicellaceae, and Blastopirellula OTUs). Furthermore, the presence of the bryozoan Celleporaria vermiformis in Oman was associated with M6 (Ruegeria and Actibacter OTUs).
Although not previously reported for the AG, the overall effects of time and space on micro- and macro-fouling communities were consistent with reports from other regions (Dziubińska and Janas, 2007; De Tender et al., 2017). The detection of unique clusters of co-occurring fouling bacterial species, changes in the predicted metabolic potential of the microfouling communities, and location-dependent associations with macrofouling organisms indicated that the Kuwait- and Oman-specific biofouling communities followed fundamentally different succession patterns over time. To the best of our knowledge, co-occurrence patterns of fouling bacteria, taxa associations and predicted functionality on a spatio-temporal scale have been rarely reported elsewhere. The measured location-specific timely changes in temperature, salinity, and turbidity of seawater over the 6 months deployment period may contribute, among other environmental parameters, to the observed changes in the biofouling communities in accordance with previous research demonstrating the effects of these environmental variables on the structure of fouling communities (Chiu et al., 2008; Hödl et al., 2011; Bellou et al., 2012; Fuhrman et al., 2015).
Total biomass, bacterial counts and macrofouling coverage were strongly affected by time and space, with lower estimates in Kuwait than in Oman. Due to the natural sustenance of a larger abundance of macrofauna in the Sea of Oman (Coles, 1997; Ghazanfar, 1999), it is highly likely that macrofoulers in Oman waters are more available for colonizing artificial surfaces than in Kuwait waters. The anatomical structure of each macrofouler taxon plays a role in determining the overall weight of the developing biofilm. For example, only barnacles and polychaetes settled on Kuwait panels, whereas Oman panels exhibited settlement of large and calcareous/hydrated macrofouling organisms (bryozoans, sponges, mollusks, hydrozoans). The surface morphology of any macrofouler also determines the available surface area for settlement of bacteria. For instance, barnacles and polychaetes on the Kuwait panels provide a smaller surface area for attachment and retention of bacterial cells compared with sponges and bryozoans encountered on Oman panels. This can explain the significantly lower bacterial counts detected in Kuwait biofilms (Bengtsson et al., 2010; Dobretsov et al., 2013).
The higher amounts of phototrophs detected on Kuwait than on Oman panels may be explained by the availability of more propagules in Kuwait waters contributed from extensive soft mudflats covered with cyanobacterial/algal mats (Al-Zaidan et al., 2006; Mohanna et al., 2007; Al-Awadhi et al., 2012). Additionally, the recurrent deposition of desert-borne, nutrient-rich dust particles in the Kuwait marinas owing to the frequently occurring dust storms in December can also stimulate algal blooms (Zohdi and Abbaspour, 2019). Conversely, although attributable to the high abundance of macrofoulers, the significant temporal changes in the abundance of biofouling communities on Oman panels may also be enhanced by the unavoidable seasonal changes in the current flow patterns of the Sea of Oman. Previous oceanographic surveys in this region have pointed out the increased proliferation of microorganisms as a consequence of mixing of nutrient-rich and nutrient-deficit waters from Indian Ocean and AG (Pous et al., 2004; Burt et al., 2016; Piontkovski and Queste, 2016; Ahd et al., 2019; Piontkovski et al., 2019).
Since the attachment, detachment and re-attachment of bacteria is expected during the latter stages of biofouling per se (Dang and Lovell, 2000, 2016; Pollet et al., 2018; Abed et al., 2019; Pinto et al., 2019; Navarrete et al., 2020), we assume that the fouling bacterial community described here will most likely represent secondary or tertiary rather than primary colonizers. The detection of several sub-assemblages of co-occurring bacteria among these secondary and tertiary colonizers added another dimension to our investigation, enabling the analysis of compositional shifts in the fouling bacterial community structure by ecological co-occurrence patterns independent of taxonomic grouping. However, the interpretation of such patterns was quite challenging given the fact that co-occurrences may result from positive or negative inter-species interactions, environmental influences, or the coincidence of being present at the same time and space.
We detected high proportions of Vibrio OTUs in Kuwait in September/October and December/January, closely associated only with other Vibrio OTUs and Pseudoalteromonas-related OTUs, respectively. Concurrent changes were also detected in the predicted functional potential, mainly attributable to the ability of Vibrio spp. to secrete autoinducer or signaling compounds (Linthorne et al., 2015). The assignment of Vibrio spp. to separate co-occurrence clusters during September/October, and December/January additionally indicated the development of unique time-dependent niches within the bacterial community and may point to a different ecological role of the detected Vibrio OTUs. Such ecological niches within the fouling bacterial community can be attributed to specific or non-specific interactions or environmental factors, such as atmospheric deposition, temperature, or light intensity changes, nutrient availability owing to seasonal upwelling or eddy formation (Fuhrman et al., 2015; Dang and Lovell, 2016; Flemming et al., 2016; Rampadarath et al., 2017; Price et al., 2021). Previous studies have reported that when Vibrio spp. are exposed to physico-chemical changes and nutrient overload, rapid proliferation occurs through the regulation of central carbon metabolism and morphological changes (Vattakaven et al., 2006; Rutherford et al., 2011; Kaberdin et al., 2015; Pérez-Reytor et al., 2017; Montánchez et al., 2019; Gorrasi et al., 2020). Substantial temperature changes in the Kuwaiti sea due to the dominant dry northwesterly winds (Shamal) and the excessive evaporation rate over precipitation cause hyper-saline conditions with salinities up to 70 ppt (Rezai and Savari, 2004; Thoppil and Hogan, 2010; Vaughan et al., 2019; Al Senafi and Anis, 2020). A relatively higher turbidity in Kuwait waters can also result from the thick dust storms occurring at a rate of 10 storms per year especially in December and the turbid water/sewage discharges from the Shatt-Al-Arab River occurring at a rate of 880 m3/s (Al Senafi and Anis, 2015). Apart from large scale environmental conditions, potentially favoring the proliferation of Vibrio spp., the biofilm community itself is modifying its environment, creating micro-niches where conditions can be highly heterogeneous and markedly different from the surrounding water column (Dang and Lovell, 2016). For instance, the high proportions of Vibrio spp., exhibiting a heterotropic lifestyle, may have been triggered by an increased availability of organic compounds after the initial settlement by microalgae and cyanobacteria (Asplund et al., 2011).
Despite more evident spatial effects on the composition of fouling bacterial communities in Oman rather than in Kuwait, the increasing homogeneity in both Oman marinas from October onward may be attributed to the thriving of similar groups of macrofoulers (Bengtsson et al., 2010; Dobretsov et al., 2013). The continuous attachment of similar types of macrofoulers such as macroalgae, barnacles, bryozoans and sponges indirectly suggests the dominance of the same macrofouler-associated bacterial taxa such as Ruegeria sp. and Arenicellaceae (Lee et al., 2009; Elifantz et al., 2013; Muthukrishnan et al., 2018; Abed et al., 2019). Similar composition of fouling bacteria was previously reported for 30-days old biofilms developed in the same marinas of Oman although at a different time of the year (Muthukrishnan et al., 2018). It is quite possible that man-made disturbances such as tourist boat traffic between the marinas during our study period may contribute to the dispersion of similar propagules for community development in both marinas, thereby indirectly contributing to the homogeneity of the biofilm communities. The chemotactic behavior and rapid surface-mechanosensing capability of Ruegeria spp. (Buchan et al., 2005; Miller and Belas, 2006) can favor the rapid settlement and dominance of this bacterial genus over other bacterial genera during biofilm development as previously reported by studies conducted in the coastal and offshore areas of Atlantic and Pacific oceans (Dang and Lovell, 2002; Jones et al., 2007; Elifantz et al., 2013).
In Kuwait, the strong positive associations between M7 (characterized by cyanobacterial OTUs) and diatoms in the absence of other major co-occurring bacterial clusters or macrofoulers in August indicated the establishment of a microphytobenthic biofilm. This was also evident from the higher prevalence of predicted carbon and nitrogen fixation pathways, which indirectly suggested higher primary productivity in the August biofilm than in the biofilms from latter months. On the contrary, the clear negative associations between diatoms and green macroalgae for August biofilms could hint to the secretion of algicidal metabolites by diatoms (Paul and Pohnert, 2011). In later stages of Kuwait biofilms’ development, a positive association was encountered between M8 (Vibrio and Pseudoalteromonas-related OTUs) and the polychaete H. elegans. This may be due to the ability of these bacteria to stimulate larval settlement, as previously shown (Maki et al., 1989; Holmström and Kjelleberg, 1999; Holmström et al., 2002; Lau et al., 2002; Huang and Hadfield, 2003; Vijayan et al., 2019). Conversely, the weaker positive association between M8 (Vibrio-related OTUs) and green macroalgae may suggest that these types of Vibrio spp. may not strongly favor algal colonization (Mok et al., 2003; Tait et al., 2009).
In Oman, the positive associations detected between Ruegeria spp. and microalgal communities in the early biofilms were probably due to the versatile ability of these bacterial species to feed on nutrient exudates from microalgae and establish mutualistic relationships (Wagner-Döbler and Biebl, 2006; Geng and Belas, 2010; Fuentes et al., 2016). The positive associations detected between Endozoicomonas and sea squirt-related genus Eudistoma in the later biofilms may indicate the prevalence of a facultative symbiotic relationship between these two taxa (Hyun et al., 2014; Schreiber et al., 2016). Previous studies reported that this bacterium was also able to co-exist as a commensal in the mucus lining of the pharynx of sea squirts (Schreiber et al., 2016). Although positive associations were also detected between Blastopirellula and other types of sea squirts Polyclinum and Botrylloides in the later biofilms, the extent of association, i.e., commensal, mutualistic, or coincidental, remains unknown. Blastopirellula has been known to thrive as a secondary colonizer in biofilms by being associated with macrofoulers based on previous studies (Lee et al., 2009; Elifantz et al., 2013; Muthukrishnan et al., 2018; Abed et al., 2019). Higher prevalence of potentially epibiotic bacterial taxa along with their associated macrofoulers concurrent with significantly higher bacterial counts in Oman biofilms compared to Kuwait warrants thorough investigations in the future. Of particular interest in this context is the hypothesis, that the macrofouler-associated microbiome may contribute via the abundance of the macrofouler to the increase in overall bacterial abundances within biofilms beyond a mere surface-area effect. Owing to the morphological differences between different macrofoulers, it is expected that there would be a competition for space and nutrients among the bacterial epibionts thereby contributing to the overall spatio-temporal dynamics of biofilm development.
Spatio-temporal influences on the succession of micro- and macro-fouling communities developed on artificial surfaces inevitably concurrent with the environmental changes is not an exception to previous research. Nevertheless, our findings related to the temporal changes in the co-occurring bacterial species and their association with macrofoulers provides us with a glimpse into the underlying dynamics of complex biofouling processes encountered in the north- and south-west of the AG. Strong co-occurrences indicative of unique ecological niches within a maturing biofouling community and subsequent changes to the functional potential of the bacterial community are quite difficult to interpret on a spatio-temporal scale. Whether these associations between different species occur conventionally or due to specific interactions or environmental influences is still unknown. The potential impact of such co-occurrence and functionality patterns within the biofouling community on nutrient cycling in the marine ecosystem remains also unknown. Comprehensive investigations spanning co-cultivation of identified fouling species in the laboratory may shed some light on possible species interactions. However, to gain clear information under natural conditions, future studies will need to utilize transcriptomic or metabolomic tools to examine all components of the biofouling community, i.e., microbes, macroalgae, and macrofoulers, at the same time and in a given space.
The sequencing data in this study has been archived on the European Nucleotide Archive (ENA) under the project accession number PRJEB41883. The environmental data and data analysis scripts have been deposited on PANGAEA (doi: 10.1594/PANGAEA.925308 and doi: 10.1594/PANGAEA.925310).
RA, TM, HM, and FA designed the project and experiments. TM and DA did the experiments in Oman. LJ and HM did the experiments in Kuwait. TM and CH analyzed the data and wrote the manuscript. All others read the manuscript and contributed to the final text.
This research was financially supported by the Gulf Cooperation Council (GCC) grant collaborating between Kuwait University, Kuwait (grant #2017003) and Sultan Qaboos University, Oman (grant #SQU-GCC/CL/17/02).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.787879/full#supplementary-material
Abed, R. M. M., Al Fahdi, D., and Muthukrishnan, T. (2019). Short-term succession of marine microbial fouling communities and the identification of primary and secondary colonizers. Biofouling 35, 526–540. doi: 10.1080/08927014.2019.1622004
Ahd, A. S., Mehrfar, H., and Pasand, M. M. (2019). The physical study of vertical structure of temperature, salinity and density layer structure between Persian Gulf and Oman Se. Mod. App. Ocean. Petrochem. Sci. 2, 157–161.
Ahmad, A. S., Siong, Y. J., Syamsumir, D. F., Zin, N. A., Radzi, S. A., Islamiah, M. N., et al. (2015). The potential of carotenoids from marine tropical microalgae in the healing process of gastritis. J. Sustain. Sci. Manag. 10, 92–106.
Al Senafi, F., Abed, R. M. M., Muthukrishnan, T., Anis, A., Al Rawahi, A. N., and Mahmoud, H. (2020). Development and diversity of bacterial biofilms in response to internal tides, a case study off the Coast of Kuwait. Front. Mar. Sci. 7:21. doi: 10.3389/fmars.2020.00021
Al Senafi, F., and Anis, A. (2015). Shamals and climate variability in the Northern Arabian/Persian Gulf from 1973 to 2012. Int. J. Climatol. 35, 4509–4528. doi: 10.1002/joc.4302
Al Senafi, F., and Anis, A. (2020). Wind-driven flow dynamics off the Northwestern Arabian Gulf Coast. Estuar. Coast. Shelf Sci. 233, 106511. doi: 10.1016/j.ecss.2019.106511
Al Senafi, F., Anis, A., and Menezes, V. (2019). Surface heat fluxes over the Northern Arabian Gulf and the Northern Red Sea: evaluation of ECMWF-ERA5 and NASA-MERRA2 reanalyses. Atmosphere 10:504. doi: 10.3390/atmos10090504
Al-Awadhi, H., Al-Mailem, D., Dashti, N., Khanafer, M., and Radwan, S. (2012). Indigenous hydrocarbon-utilizing bacterioflora in oil-polluted habitats in Kuwait, two decades after the greatest man-made oil spill. Arch. Microbiol. 194, 689–705. doi: 10.1007/s00203-012-0800-7
Al-Zaidan, A. S., Jones, D. A., Al-Mohanna, S. Y., and Meakins, R. (2003). Endemic macrofauna of the Sulaibikhat Bay salt marsh and mudflat habitats, Kuwait: status and need for conservation. J. Arid Environ. 54, 115–124. doi: 10.1006/jare.2001.0886
Al-Zaidan, A. S., Kennedy, H., Jones, D. A., and Al-Mohanna, S. Y. (2006). Role of microbial mats in Sulaibikhat Bay (Kuwait) mudflat food webs: evidence from δ13C analysis. Mar. Ecol. Prog. Ser. 308, 27–36. doi: 10.3354/meps308027
Antunes, J. T., Sousa, A. G., Azevedo, J., Rego, A., Leão, P. N., and Vasconcelos, V. (2020). Distinct temporal succession of bacterial communities in early marine biofilms in a Portuguese Atlantic Port. Front. Microbiol. 11:1938. doi: 10.3389/fmicb.2020.01938
Asplund, M. E., Rehnstam-Holm, A. S., Atnur, V., Raghunath, P., Saravanan, V., Härnström, K., et al. (2011). Water column dynamics of Vibrio in relation to phytoplankton community composition and environmental conditions in a tropical coastal area. Environ. Microbiol. 13, 2738–2751. doi: 10.1111/j.1462-2920.2011.02545.x
Barberán, A., Bates, S. T., Casamayor, E. O., and Fierer, N. (2012). Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 6, 343–351. doi: 10.1038/ismej.2011.119
Bellou, N., Papathanassiou, E., Dobretsov, S., Lykousis, V., and Colijn, F. (2012). The effect of substratum type, orientation and depth on the development of bacterial deep-sea biofilm communities grown on artificial substrata deployed in the Eastern Mediterranean. Biofouling 28, 199–213. doi: 10.1080/08927014.2012.662675
Bengtsson, M. M., Sjøtun, K., and Øvreås, L. (2010). Seasonal dynamics of bacterial biofilms on the kelp Laminaria hyperborea. Aquat. Microb. Ecol. 60, 71–83. doi: 10.3354/ame01409
Bowman, J. S., and Ducklow, H. W. (2015). Microbial communities can be described by metabolic structure: a general framework and application to a seasonally variable, depth-stratified microbial community from the coastal West Antarctic Peninsula. PLoS One 10:e0135868. doi: 10.1371/journal.pone.0135868
Briand, J. F., Djeridi, I., Jamet, D., Coupé, S., Bressy, C., Molmeret, M., et al. (2012). Pioneer marine biofilms on artificial surfaces including antifouling coatings immersed in two contrasting French Mediterranean coast sites. Biofouling 28, 453–463. doi: 10.1080/08927014.2012.688957
Briand, J. F., Pochon, X., Wood, S. A., Bressy, C., Garnier, C., Réhel, K., et al. (2018). Metabarcoding and metabolomics offer complementarity in deciphering marine eukaryotic biofouling community shifts. Biofouling 34, 657–672. doi: 10.1080/08927014.2018.1480757
Buchan, A., González, J. M., and Moran, M. A. (2005). Overview of the marine Roseobacter lineage. Appl. Environ. Microbiol. 71, 5665–5677. doi: 10.1128/AEM.71.10.5665-5677.2005
Burt, J. A., Coles, S., Van Lavieren, H., Taylor, O., Looker, E., and Samimi-Namin, K. (2016). Oman’s coral reefs: a unique ecosystem challenged by natural and man-related stresses and in need of conservation. Mar. Pollut. Bull. 105, 498–506. doi: 10.1016/j.marpolbul.2015.11.010
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421
Chao, A., Gotelli, N. J., Hsieh, T. C., Sander, E. L., Ma, K. H., Colwell, R. K., et al. (2014). Rarefaction and extrapolation with Hill numbers: a framework for sampling and estimation in species diversity studies. Ecol. Monogr. 84, 45–67. doi: 10.1890/13-0133.1
Chiu, J. M., Zhang, R., Wang, H., Thiyagarajan, V., and Qian, P. Y. (2008). Nutrient effects on intertidal community: from bacteria to invertebrates. Mar. Ecol. Prog. Ser. 358, 41–50.
Cobo-Díaz, J. F., Baroncelli, R., Le Floch, G., and Picot, A. (2019). Combined metabarcoding and co-occurrence network analysis to profile the bacterial, fungal and Fusarium communities and their interactions in maize stalks. Front. Microbiol. 10:261. doi: 10.3389/fmicb.2019.00261
Coles, S. L. (1997). Reef corals occurring in a highly fluctuating temperature environment at Fahal Island, Gulf of Oman (Indian Ocean). Coral Reefs 16, 269–272.
Csardi, G., and Nepusz, T. (2006). The igraph software package for complex network research. Int. J. Complex Syst. 1695, 1–9. doi: 10.1186/1471-2105-12-455
Dang, H., and Lovell, C. R. (2000). Bacterial primary colonization and early succession on surfaces in marine waters as determined by amplified rRNA gene restriction analysis and sequence analysis of 16S rRNA genes. Appl. Environ. Microbiol. 66, 467–475. doi: 10.1128/AEM.66.2.467-475.2000
Dang, H., and Lovell, C. R. (2002). Seasonal dynamics of particle-associated and free-living marine Proteobacteria in a salt marsh tidal creek as determined using fluorescence in situ hybridization. Environ. Microbiol. 4, 287–295. doi: 10.1046/j.1462-2920.2002.00295.x
Dang, H., and Lovell, C. R. (2016). Microbial surface colonization and biofilm development in marine environments. Microbiol. Mol. Biol. Rev. 80, 91–138. doi: 10.1128/MMBR.00037-15
de Carvalho, C. C. (2018). Marine biofilms: a successful microbial strategy with economic implications. Front. Mar. Sci. 5:126. doi: 10.3389/fmars.2018.00126
De Tender, C., Devriese, L. I., Haegeman, A., Maes, S., Vangeyte, J., Cattrijsse, A., et al. (2017). Temporal dynamics of bacterial and fungal colonization on plastic debris in the North Sea. Environ. Sci. Technol. 51, 7350–7360. doi: 10.1021/acs.est.7b00697
Diepenbroek, M., Glöckner, F. O., Grobe, P., Güntsch, A., Huber, R., König-Ries, B., et al. (2014). “Towards an integrated biodiversity and ecological research data management and archiving platform: the German federation for the curation of biological data (GFBio),” in Informatik, eds E. Plödereder, L. Grunske, E. Schneider, and D. Ull (Bonn: Gesellschaft für Informatik), 1711–1721.
Dobretsov, S. (2015). Biofouling on artificial substrata in Muscat waters. J. Agri. Mar. Sci. 20, 24–29. doi: 10.24200/jams.vol20iss0pp24-29
Dobretsov, S., Abed, R. M. M., and Voolstra, C. R. (2013). The effect of surface colour on the formation of marine micro and macrofouling communities. Biofouling 29, 617–627. doi: 10.1080/08927014.2013.784279
Dobretsov, S., and Thomason, J. C. (2011). The development of marine biofilms on two commercial non-biocidal coatings: a comparison between silicone and fluoropolymer technologies. Biofouling 27, 869–880. doi: 10.1080/08927014.2011.607233
Dziubińska, A., and Janas, U. (2007). Submerged objects-a nice place to live and develop. Succession of fouling communities in the Gulf of Gdańsk, Southern Baltic. Oceanol. Hydrobiol. Stud. 36, 65–78.
Elifantz, H., Horn, G., Ayon, M., Cohen, Y., and Minz, D. (2013). Rhodobacteraceae are the key members of the microbial community of the initial biofilm formed in Eastern Mediterranean coastal seawater. FEMS Microbiol. Ecol. 85, 348–357. doi: 10.1111/1574-6941.12122
Fadeev, E., Salter, I., Schourup-Kristensen, V., Nöthig, E. M., Metfies, K., Engel, A., et al. (2018). Microbial communities in the east and west fram strait during sea ice melting season. Front. Mar. Sci. 5:429. doi: 10.3389/fmars.2018.00429
Flemming, H. C., Wingender, J., Szewzyk, U., Steinberg, P., Rice, S. A., and Kjelleberg, S. (2016). Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 14, 563–575. doi: 10.1038/nrmicro.2016.94
Fuentes, J. L., Garbayo, I., Cuaresma, M., Montero, Z., González-del-Valle, M., and Vílchez, C. (2016). Impact of microalgae-bacteria interactions on the production of algal biomass and associated compounds. Mar. Drugs 14:100. doi: 10.3390/md14050100
Fuhrman, J. A., Cram, J. A., and Needham, D. M. (2015). Marine microbial community dynamics and their ecological interpretation. Nat. Rev. Microbiol. 13, 133–146. doi: 10.1038/nrmicro3417
Geng, H., and Belas, R. (2010). Expression of tropodithietic acid biosynthesis is controlled by a novel autoinducer. J. Bacteriol. 192, 4377–4387. doi: 10.1128/JB.00410-10
Gorrasi, S., Pasqualetti, M., Franzetti, A., Pittino, F., and Fenice, M. (2020). Vibrio communities along a salinity gradient within a marine saltern hypersaline environment (Saline di Tarquinia, Italy). Environ. Microbiol. 22, 4356–4366. doi: 10.1111/1462-2920.15041
Guidi, L., Chaffron, S., Bittner, L., Eveillard, D., Larhlimi, A., Roux, S., et al. (2016). Plankton networks driving carbon export in the oligotrophic ocean. Nature 532, 465–470. doi: 10.1038/nature16942
Hirano, H., and Takemoto, K. (2019). Difficulty in inferring microbial community structure based on co-occurrence network approaches. BMC Bioinformatics 20:329. doi: 10.1186/s12859-019-2915-1
Hödl, I., Hödl, J., Wörman, A., Singer, G., Besemer, K., and Battin, T. J. (2011). Voronoi tessellation captures very early clustering of single primary cells as induced by interactions in nascent biofilms. PLoS One 6:e26368. doi: 10.1371/journal.pone.0026368
Holmström, C., and Kjelleberg, S. (1999). Marine Pseudoalteromonas species are associated with higher organisms and produce biologically active extracellular agents. FEMS Microbiol Ecol. 30, 285–293. doi: 10.1111/j.1574-6941.1999.tb00656.x
Holmström, C., Egan, S., Franks, A., McCloy, S., and Kjelleberg, S. (2002). Antifouling activities expressed by marine surface associated Pseudoalteromonas species. FEMS Microbiol Ecol. 41, 47–58.
Huang, S., and Hadfield, M. G. (2003). Composition and density of bacterial biofilms affect metamorphosis of the polychaete Hydroides elegans. Mar. Ecol. Prog. Ser. 260, 161–172. doi: 10.3354/meps260161
Hyun, D. W., Shin, N. R., Kim, M. S., Oh, S. J., Kim, P. S., Whon, T. W., et al. (2014). Endozoicomonas atrinae sp. nov., isolated from the intestine of a comb pen shell Atrina pectinata. Int. J. Syst. Evol. Microbiol. 64, 2312–2318. doi: 10.1099/ijs.0.060780-0
Jones, P. R., Cottrell, M. T., Kirchman, D. L., and Dexter, S. C. (2007). Bacterial community structure of biofilms on artificial surfaces in an estuary. Microb. Ecol. 53, 153–162. doi: 10.1007/s00248-006-9154-5
Kaberdin, V. R., Montánchez, I., Parada, C., Orruño, M., Arana, I., and Barcina, I. (2015). Unveiling the metabolic pathways associated with the adaptive reduction of cell size during Vibrio harveyi persistence in seawater microcosms. Microb. Ecol. 70, 689–700. doi: 10.1007/s00248-015-0614-7
Klindworth, A., Pruesse, E., Schweer, T., Peplies, J., Quast, C., Horn, M., et al. (2013). Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next generation sequencing-based diversity studies. Nucleic Acid Res. 41:e1. doi: 10.1093/nar/gks808
Kurtz, Z. D., Müller, C. L., Miraldi, E. R., Littman, D. R., Blaser, M. J., and Bonneau, R. A. (2015). Sparse and compositionally robust inference of microbial ecological networks. PLoS Comput. Biol. 11:e1004226. doi: 10.1371/journal.pcbi.1004226
Kuznetsova, A., Brockhoff, P. B., and Christensen, R. H. (2017). lmerTest package: tests in linear mixed effects models. J. Stat. Softw. 82, 1–26.
Langfelder, P., and Horvath, S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9:559. doi: 10.1186/1471-2105-9-559
Lau, S. C., Mak, K. K., Chen, F., and Qian, P. Y. (2002). Bioactivity of bacterial strains isolated from marine biofilms in Hong Kong waters for the induction of larval settlement in the marine polychaete Hydroides elegans. Mar. Ecol. Prog. Ser. 226, 301–310. doi: 10.3354/meps226301
Lee, J., Jung, J. Y., Kim, S., Chang, I. S., Mitra, S. S., and Kim, I. S. (2009). Selection of the most problematic biofoulant in fouled RO membrane and the seawater intake to develop biosensors for membrane biofouling. Desalination 247, 125–136.
Lin, Y., Zhao, D., Zeng, J., Cao, X., and Jiao, C. (2019). Network analysis reveals seasonal patterns of bacterial community networks in Lake Taihu under aquaculture conditions. Water 11:1868. doi: 10.3390/w11091868
Linthorne, J. S., Chang, B. J., Flematti, G. R., Ghisalberti, E. L., and Sutton, D. C. (2015). A direct pre-screen for marine bacteria producing compounds inhibiting quorum sensing reveals diverse planktonic bacteria that are bioactive. Mar. Biotechnol. 17, 33–42. doi: 10.1007/s10126-014-9592-x
Liu, L., Yang, J., Lv, H., and Yu, Z. (2014). Synchronous dynamics and correlations between bacteria and phytoplankton in a subtropical drinking water reservoir. FEMS Microbiol. Ecol. 90, 126–138. doi: 10.1111/1574-6941.12378
Lueders, T., Manefield, M., and Friedrich, M. W. (2004). Enhanced sensitivity of DNA-and rRNAbased stable isotope probing by fractionation and quantitative analysis of isopycnic centrifugation gradients. Environ. Microbiol. 6, 73–78. doi: 10.1046/j.1462-2920.2003.00536.x
Macleod, A. K., Stanley, M. S., Day, J. G., and Cook, E. J. (2016). Biofouling community composition across a range of environmental conditions and geographical locations suitable for floating marine renewable energy generation. Biofouling 32, 261–276. doi: 10.1080/08927014.2015.1136822
Maki, J. S., Rittschof, D., Schmidt, A. R., Snyder, A. G., and Mitchell, R. (1989). Factors controlling attachment of bryozoan larvae: a comparison of bacterial films and unfilmed surfaces. Biol. Bull. 177, 295–302. doi: 10.2307/1541944
McMurdie, P. J., and Holmes, S. (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217
Milici, M., Deng, Z. L., Tomasch, J., Decelle, J., Wos-Oxley, M. L., Wang, H., et al. (2016). Co-occurrence analysis of microbial taxa in the Atlantic Ocean reveals high connectivity in the free-living bacterioplankton. Front. Microbiol. 7:649. doi: 10.3389/fmicb.2016.00649
Miller, T. R., and Belas, R. (2006). Motility is involved in Silicibacter sp. TM1040 interaction with dinoflagellates. Environ. Microbiol. 8, 1648–1659. doi: 10.1111/j.1462-2920.2006.01071.x
Mohanna, S. Y., George, P., and Subrahmanyam, M. N. (2007). Benthic microalgae on a sheltered intertidal mudflat in Kuwait Bay of the Northern Arabian Gulf. J. Mar. Biol. Ass. India. 49, 27–34.
Mok, K. C., Wingreen, N. S., and Bassler, B. L. (2003). Vibrio harveyi quorum sensing: a coincidence detector for two autoinducers controls gene expression. EMBO J. 22, 870–881. doi: 10.1093/emboj/cdg085
Montánchez, I., Ogayar, E., Plágaro, A. H., Esteve-Codina, A., Gómez-Garrido, J., Orruño, M., et al. (2019). Analysis of Vibrio harveyi adaptation in sea water microcosms at elevated temperature provides insights into the putative mechanisms of its persistence and spread in the time of global warming. Sci. Rep. 9:289. doi: 10.1038/s41598-018-36483-0
Muthukrishnan, T., Al Khaburi, M., and Abed, R. M. M. (2018). Fouling microbial communities on plastics compared with wood and steel: are they substrate-or location-specific? Microb. Ecol. 78, 361–374. doi: 10.1007/s00248-018-1303-0
Muthukrishnan, T., Dobretsov, S., De Stefano, M., Abed, R. M. M., Kidd, B., and Finnie, A. A. (2017). Diatom communities on commercial biocidal fouling control coatings after one year of immersion in the marine environment. Mar. Environ. Res. 129, 102–112. doi: 10.1016/j.marenvres.2017.05.001
Naser, H. A. (2017). Variability of marine macrofouling assemblages in a marina and a mariculture centre in Bahrain, Arabian Gulf. Reg. Stud. Mar. Sci. 16, 162–170. doi: 10.1016/j.rsma.2017.09.005
Navarrete, S. A., Parragué, M., Osiadacz, N., Rojas, F., Bonicelli, J., Fernández, M., et al. (2020). Susceptibility of different materials and antifouling coating to macrofouling organisms in a high wave-energy environment. J. Ocean Technol. 15, 72–91.
Oberbeckmann, S., Osborn, A. M., and Duhaime, M. B. (2016). Microbes on a bottle: substrate, season and geography influence community composition of microbes colonizing marine plastic debris. PLoS One 11:e0159289. doi: 10.1371/journal.pone.0159289
Paul, C., and Pohnert, G. (2011). Interactions of the algicidal bacterium Kordia algicida with diatoms: regulated protease excretion for specific algal lysis. PLoS One 6:e21032. doi: 10.1371/journal.pone.0021032
Pérez-Reytor, D., Plaza, N., Espejo, R. T., Navarrete, P., Bastías, R., and Garcia, K. (2017). Role of non-coding regulatory RNA in the virulence of human pathogenic Vibrios. Front. Microbiol. 7:2160. doi: 10.3389/fmicb.2016.02160
Pinto, M., Langer, T. M., Hüffer, T., Hofmann, T., and Herndl, G. J. (2019). The composition of bacterial communities associated with plastic biofilms differs between different polymers and stages of biofilm succession. PLoS One 14:e0217165. doi: 10.1371/journal.pone.0217165
Piontkovski, S. A., Al-Gheilani, H. M. H., Jupp, B. P., Al-Azri, A., and Al-Hashmi, K. A. (2012). Interannual changes in the Sea of Oman ecosystem. Open Mar. Biol. J. 6, 38–52. doi: 10.2174/1874450801206010038
Piontkovski, S. A., and Queste, B. Y. (2016). Decadal changes of the Western Arabian sea ecosystem. Int. Aquat. Res. 8, 49–64.
Piontkovski, S. A., Hamza, W. M., Al-Abri, N. M., Al-Busaidi, S. S., and Al-Hashmi, K. A. (2019). A comparison of seasonal variability of Arabian Gulf and the Sea of Oman pelagic ecosystems. Aquat. Ecosyst. Health Manag. 22, 108–130.
Pollet, T., Berdjeb, L., Garnier, C., Durrieu, G., Le Poupon, C., Misson, B., et al. (2018). Prokaryotic community successions and interactions in marine biofilms: the key role of Flavobacteriia. FEMS Microbiol. Ecol. 94:fiy083. doi: 10.1093/femsec/fiy083
Pous, S. P., Carton, X., and Lazure, P. (2004). Hydrology and circulation in the Strait of Hormuz and the Gulf of Oman—Results from the GOGP99 Experiment: 2. Gulf of Oman. J. Geophys. Res. Oceans 109, 57–78. doi: 10.1029/2003JC002146
Price, G. W., Langille, M. G., and Yurgel, S. N. (2021). Microbial co-occurrence network analysis of soils receiving short-and long-term applications of alkaline treated biosolids. Sci. Total Environ. 751:141687. doi: 10.1016/j.scitotenv.2020.141687
Qian, P. Y., Lau, S. C., Dahms, H. U., Dobretsov, S., and Harder, T. (2007). Marine biofilms as mediators of colonization by marine macroorganisms: implications for antifouling and aquaculture. Mar. Biotechnol. 9, 399–410. doi: 10.1007/s10126-007-9001-9
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, 590–596. doi: 10.1093/nar/gks1219
R Core Team (2018). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
R Studio Team (2016). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Rampadarath, S., Bandhoa, K., Puchooa, D., Jeewon, R., and Bal, S. (2017). Early bacterial biofilm colonizers in the coastal waters of Mauritius. Electron. J. Biotechnol. 29, 13–21. doi: 10.1016/j.ejbt.2017.06.006
Rezai, H., and Savari, A. (2004). Observation on reef fishes in the coastal waters off some Iranian Islands in the Persian Gulf. Zool. Middle East. 31, 67–76.
Rutherford, S. T., Van Kessel, J. C., Shao, Y., and Bassler, B. L. (2011). AphA and LuxR/HapR reciprocally control quorum sensing in vibrios. Genes Dev. 25, 397–408. doi: 10.1101/gad.2015011
Satheesh, S., and Wesley, S. G. (2012). Temporal changes of diatoms in marine biofilm developed on acrylic panels submerged in a tropical coast. Ocean Sci. J. 47, 509–517. doi: 10.1007/s12601-012-0046-y
Schneider, C. A., Rasband, W. S., and Eliceiri, K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. doi: 10.1038/nmeth.2089
Schreiber, L., Kjeldsen, K. U., Funch, P., Jensen, J., Obst, M., López-Legentil, S., et al. (2016). Endozoicomonas are specific, facultative symbionts of sea squirts. Front. Microbiol. 7:1042. doi: 10.3389/fmicb.2016.01042
Stachowitsch, M., Kikinger, R., Herler, J., Zolda, P., and Geutebrück, E. (2002). Offshore oil platforms and fouling communities in the southern Arabian Gulf (Abu Dhabi). Mar. Pollut. Bull. 44, 853–860. doi: 10.1016/s0025-326x(02)00085-1
Tait, K., Williamson, H., Atkinson, S., Williams, P., Cámara, M., and Joint, I. (2009). Turnover of quorum sensing signal molecules modulates cross-kingdom signalling. Environ. Microbiol. 11, 1792–1802. doi: 10.1111/j.1462-2920.2009.01904.x
Thoppil, P. G., and Hogan, P. J. (2010). Persian Gulf response to a wintertime shamal wind event. Deep Sea Research Part I Oceanogr. Res. Pap. 57, 946–955. doi: 10.1016/j.dsr.2010.03.002
Vattakaven, T., Bond, P., Bradley, G., and Munn, C. B. (2006). Differential effects of temperature and starvation on induction of the viable-but-nonculturable state in the coral pathogens Vibrio shiloi and Vibrio tasmaniensis. Appl. Environ. Microbiol. 72, 6508–6513. doi: 10.1128/AEM.00798-06
Vaughan, G. O., Al-Mansoori, N., and Burt, J. A. (2019). “The Arabian Gulf,” in World seas: An Environmental Evaluation, ed. S. Charles (London: Academic Press), 1–23.
Vijayan, N., Lema, K. A., Nedved, B. T., and Hadfield, M. G. (2019). Microbiomes of the polychaete Hydroides elegans (Polychaeta: Serpulidae) across its life-history stages. Mar. Biol. 166:19. doi: 10.1007/s00227-019-3465-9
Wagner-Döbler, I., and Biebl, H. (2006). Environmental biology of the marine Roseobacter lineage. Annu. Rev. Microbiol. 60, 255–280. doi: 10.1146/annurev.micro.60.080805.142115
Weiss, S., Van Treuren, W., Lozupone, C., Faust, K., Friedman, J., Deng, Y., et al. (2016). Correlation detection strategies in microbial data sets vary widely in sensitivity and precision. ISME J. 10, 1669–1681. doi: 10.1038/ismej.2015.235
Yeh, Y.-C., McNichol, J., Needham, D. M., Fichot, E. B., Berdjeb, L., and Fuhrman, J. A. (2021). Comprehensive single-PCR 16S and 18S rRNA community analysis validated with mock communities, and estimation of sequencing bias against 18S. Environ. Microbiol. 23, 3240–3250. doi: 10.1111/1462-2920.15553
Yilmaz, P., Kottmann, R., Field, D., Knight, R., Cole, J. R., Amaral-Zettler, L. et al. (2011). Minimum information about a marker gene sequence (MIMARKS) and minimum information about any (x) sequence (MIxS) specifications. Nat. Biotechnol. 29, 415–420. doi: 10.1038/nbt.1823
Zhang, W., Ding, W., Li, Y. X., Tam, C., Bougouffa, S., Wang, R., et al. (2019). Marine biofilms constitute a bank of hidden microbial diversity and functional potential. Nat. Commun. 10:517. doi: 10.1038/s41467-019-08463-z
Keywords: biofouling, bacteria, macrofoulers, co-occurrence, association, Arabian Gulf
Citation: Muthukrishnan T, Hassenrück C, Al Fahdi D, Jose L, Al Senafi F, Mahmoud H and Abed RMM (2022) Monthly Succession of Biofouling Communities and Corresponding Inter-Taxa Associations in the North- and South-West of the Arabian Gulf. Front. Mar. Sci. 8:787879. doi: 10.3389/fmars.2021.787879
Received: 01 October 2021; Accepted: 27 December 2021;
Published: 11 February 2022.
Edited by:
Rachel Ann Foster, Stockholm University, SwedenReviewed by:
Veronica Molina, Universidad de Playa Ancha, ChileCopyright © 2022 Muthukrishnan, Hassenrück, Al Fahdi, Jose, Al Senafi, Mahmoud and Abed. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Raeid M. M. Abed, cmFiZWRAbXBpLWJyZW1lbi5kZQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.