- 1South China Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Guangzhou, China
- 2Guangdong Provincial Key Laboratory of Fishery Ecology and Environment, South China Sea Fisheries Research Institute, CAFS, Guangzhou, China
- 3College of Marine Science, Shanghai Ocean University, Shanghai, China
The remote coral reef in the deep sea is one of the most important nursery grounds for many marine fishes in the South China Sea. Diversity studies on larval fishes in this area are few, and the fish information on the coral reefs ecosystem is lacking. In this study, larval fishes were sampled during the summer of 2019 from the Zhongsha Atoll in the South China Sea and identified using DNA barcodes for the first time. Ninety-five larval fish species were recognized, belonging to 37 families and 12 orders based on morphological classification and DNA barcoding identification. The larval fish collected could be assigned to three categories as reef-associated, deep-sea, and pelagic. Most of the species were small fish with low commercial value but would play an essential role in the coral reef ecosystem. Some commercial fishes, including Auxis thazard, Euthynnus affinis, Sarda orientalis, Decapterus macarellus, Lutjanus viridis, and Centropyge vrolikii, were the dominant species higher than 2% total catch. The larval fish assemblage showed distinct spatial differences responding well with the geographical conditions. The most reef-associated fish occurred inside the Atoll, and the abyssal fish presented near the edge. In addition, larval fish spread over from the southwest to northeast may reflect the oceanography effect.
Introduction
Larval fish is the basis for the sustainable utilization of fishery resources, with survival directly influencing recruitment. Its investigations serve as an alternative approach for fish biodiversity, especially for some habitats, e.g., coral reefs and mangroves, where conventional fish sampling is challenging. The information on the distribution and abundance of larval fish provides a means to know the fish diversity, which is the base for fisheries management and conservation planning of the ecosystem (Almany et al., 2017).
Coral reefs are among the most biologically diverse and valuable ecosystems, providing habitat for over 4,000 species of fish at some point in their life cycle (Teh et al., 2013; Woodhead et al., 2019), especially in the early growth stages. Living coral reefs play as spawning areas, hatchery yards, nursery grounds, and recruitment sites (Jones et al., 2005; Cole et al., 2008; Almany et al., 2017). As under increasingly exposed to the adverse effects of global climate change and human activities (Comte and Pendleton, 2018), coral reefs distinct declined, and the population and biodiversity of fish are threatened (Jones et al., 2004; De'ath et al., 2012).
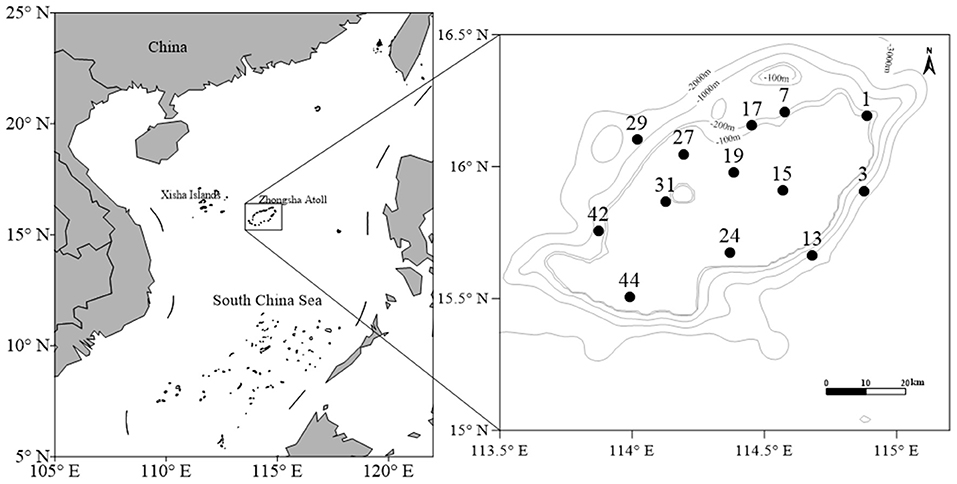
The South China Sea is the largest low-latitude marginal sea, hosting some of the most productive and prosperous coral reef ecosystems and the most diverse fish on the Earth (Wang et al., 2011; Shan et al., 2021). The coral reef in the South China Sea covers ~8,000 km2, spreading over along its entire coastline and the islands in the deep sea, with the largest concentration around the relatively remote Zhongsha Atoll (Yu, 2012). The Zhongsha Atoll (also known as Macclesfield Bank) is one of the largest drowned atolls in the world. It locates in the central South China Sea, about 140 km away from Xisha islands and 550 km apart from Hainan Islands, comprising many submerged reefs in the seamount around by deep sea over 1,000 m in depth (Figure 1). The entire Zhongsha Atoll covers an area of about 23,500 km2, with a collection of entirely submerged banks, seamounts, and shoals. Its discontinuous marginal reefs surrounded a lagoon with a water depth of up to 50–70 m. The platform slopes extend from the atoll margins to the deep-sea basin cut by submarine canyons, which promote the development of upwelling currents and may provide the hydrodynamic conditions required for reef-building corals and other organisms to flourish (Huang et al., 2020). The Zhongsha Atoll plays a vital role in the ecology and fish diversity of the South China Sea, supporting a diverse fish fauna in the sea around the Atoll (Sun et al., 2006; Chen et al., 2007).
Due to the limitation of geographical location and sampling conditions, the historical survey data of this sea area are concise, and the biology information of fish is limited. The precise nature and composition of the larval fishes across the entire Zhongsha Atoll are, as yet, poorly known (Chen, 1979). It is necessary to better understand the larval fish diversity and assemblages of the Atoll for guiding conservation policy for coral reefs of the South China Sea. The present study attempted to investigate the fish diversity in the Zhongsha Atoll using morphology and molecular tools for the first time and assess the ecological function of coral reef in the Atoll by assemblages and distribution patterns analysis to enhance our understanding of this ecosystem.
Data and Methods
Sample Collection
Larval fishes were sampled from the Zhongsha Atoll aboard the Yuezhanyuke 10 on May 9–29, 2019 (Figure 1). All sampling processes were conducted in the daytime. Samples were collected using a plankton net with a mouth diameter of 0.8 m, length of 2.8 m, and mesh size of 330 μm equipped with a centrally mounted flow meter (Hydro-Bios, Kiel-Holtenau, Germany). The net was trawling vertically from 200 m (or 5 m above the bottom at stations with depth <200 m) to surface with a speed of 1 m s−1. After the net was retrieved, the samples were stored in 95% ethanol solution and frozen at −20°C. Each larval fish was sorted and identified to the lowest possible taxonomic level based on their morphological characteristics in the laboratory following guilds of Leis and Rennis (1983), Okiyawa (1988), and Wan and Zhang (2016).
DNA Data Collection
Then molecular identification method was performed with sample categorized (Lin et al., 2016). Before DNA extraction, larvae were macerated and then washed in ultrapure water to remove attachments. Total genomic DNA was extracted from tissue using a TIANamp Marine Animals DNA Kit (Tiangen, China) following manufacturer protocols. Partial of the mitochondrial cytochrome oxidase I (COI) sequences was amplified from total genomic DNA using the PCRs with the primers FishBCL and FishBCH (Baldwin et al., 2009).
PCRs were run in a final volume of 30 μl, containing 3 μl of 10 × PCR buffer, 3 μl of dNTPs mix (10 mM), 0.3 μl of E × Taq polymerase (Takara Bio. Inc.), 0.9 μl of each primer (10 μM), 2–3 μl of genomic DNA, and distilled water. PCR was carried out in an Eppendorf thermal cycler with 5 min initial denaturation at 95°C, followed by 35 cycles of 30 s at 95°C, 30 s at 52°C, 45 s at 72°C, and a final step at 72°C for 5 min on a 2720 Thermal Cycler (Applied Biosystems, Waltham, MA, USA). PCR products were visualized in 1.5% agarose gel, and samples amplified successfully were selected for sequencing. Sequences were obtained using an automated DNA sequencing device 3500 (Life Technologies, USA).
Data Analysis
The consensus DNA sequences were checked, and ambiguous bases were removed using the DNASTAR (DNASTAR, Inc., Madison, WI, USA) and then aligned in MEGA ver. 7.0.26 software (Kumar et al., 2016). Sequences were uploaded to the ORF finder (https://www.ncbi.nlm.nih.gov/orffinder/) to check for the reading frame, as no indels were observed. Identification was performed firstly by comparing with databases of National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov/) with basic local-alignment search tool (BLAST) with a similarity threshold of 98% to assign specimens to species was followed (Ward et al., 2005). These sequences were then entered in FASTA format to the Barcode of life data systems (BOLD, http://www.boldsystems.org/) with the BOLD Identification Tool. Species-level identification would be returned when one is possible. All sequences have been deposited in GenBank (Accession Numbers OL512811—OL512944).
For those sequences that could not recognize into species, their best-matched sequences in the databases were downloaded for the following analysis. A Maximum likelihood (ML) tree of analyzed DNA barcode sequences based on the Kimura two-parameter (K2P) distance was performed using MEGA ver. 7.0.26 software, with 5,000 bootstrap replications (Kumar et al., 2016). Majority rule consensus trees were reconstructed after discarding a burn-in of 500 and displayed with TreeView v.1.6.6 (Page, 1996). The K2P genetic distances for defining the species, genus, and family levels were based on Ward et al. (2005).
Results
Species Identification and Fish Diversity
Totally 445 larval fish were collected, 146 specimens were used for molecular experiments after morphological classification, and 12 samples failed in DNA extraction and PCR process. The rest (91.78%) sequences of specimens were acquired, yielding a partial region of the COI mitochondrial gene sequences length of 648–722 bp without deletions, insertions, or stop codons. The detailed comparison was based on the aligned tool from the Genbank and BOLD systems, with the overall genetic similarity ranging from 81.87% to 100%. Comparing each of the COI sequences obtained from our samples with sequences deposited in databases online, 90 sequences (67.16%) were recovered as species of marine fish (sequence similarity > 98%). Altogether 44 sequences from 31 species showed low or no similarity matches with the databases, indicating that the reference sequences of these species were not barcoded ever. These unrecognized sequences belonged to small fishes, including Diogenichthys sp., Naso sp., and Pseudanthias sp. could be aligned into Diogenichthys atlanticus, Naso minor, and Rabaulichthys squirei, with private or early-released data in the BOLD systems (Table 1).
The ML tree based on K2P genetic divergence was constructed using the un-matched sequences and their most matched sequences in the databases (Figure 2). Most of the specimens were identified into genus without one affirmed into the family. Based on molecular identification, 95 species of fish larvae were identified from the 134 samples, of which 1, 30, and 64 were identified to family, genus, and species levels, respectively (Table 1). Perciformes was the most abundant order, with 69.66% of total specimens belonging to 55 taxa. Myctophiformes (10.11% of samples belonging to 17 taxa), Beryciformes (4.27% of specimens belonging to 5 taxa), and the other orders contributed 15.96% of specimens and 18 taxa (Table 1). Four orders contained only one species: Histrio histrio (Lophiiformes), Engyprosopon latifrons (Pleuronectiformes), Sebastapistes sp. (Scorpaeniformes), and Netuma thalassina (Siluriformes). The most diverse family was Myctophidae attributing to 17 species, followed by Gobiidae with 13 species, and Scombridae with 6 species. There were 11 and 24 predominant species with a higher than 2% and 1% of the total catch, respectively. The most dominant taxa was Auxis thazard, accounting for 8.99% of all specimens, followed by Paracheilinus carpenter, Euthynnus affinis, Sarda orientalis, and Pseudanthias sp., with 8.54%, 8.09%, 6.29%, and 4.72% in the catch. While about 38.95% (37 of 95) of all species were represented by a single specimen.
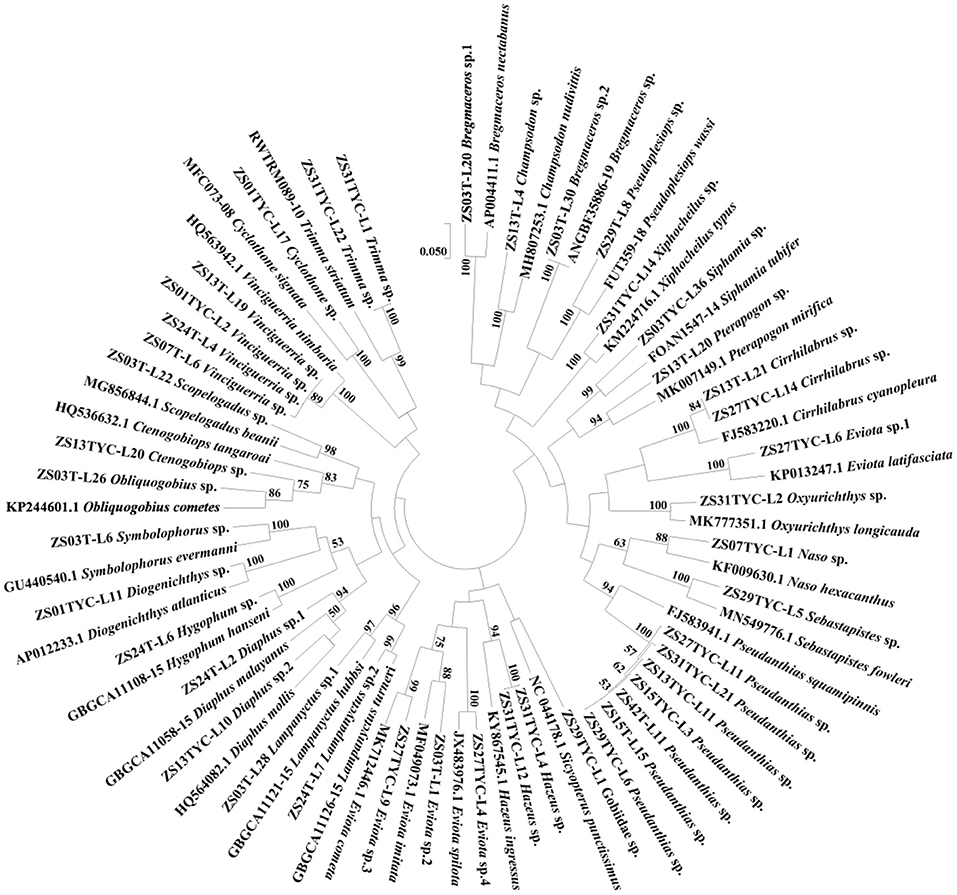
Figure 2. Kimura two-parameter (K2P) distance ML tree of un-matched sequences and their best-matched sequences from the NCBI and BOLD systems with the ID of the samples and species given. Bootstrap values >50 are shown. ML, Maximum likelihood; K2P, the Kimura two-parameter; NCBI, National Center for Biotechnology Information; BOLD systems, the Barcode of life data systems.
Most of the fish (64 of 95) collected belonged to subsistence fisheries, and only 32 taxa accounting for 46.07% of total specimens were commercial in fisheries. Twelve taxa of collected larvae were commercially valuable fishes. Taxa belonging to Scombridae (Auxis thazard, Euthynnus affinis, and Sarda orientalis), Carangidae (Decapterus macarellus, D. macrosoma, and Selar crumenophthalmus), Lutjanidae (Lutjanus viridis), and Lethrinidae (Lethrinus rubrioperculatus) accounted for 31.34% of the total specimens (Table 1). Account for the biology groups, 53 taxa were reef-associated fishes; 30 of them were fishes of the deep sea (such as bathy-demersal fish and bathy-mesopelagic fish), and the rest of the species collected were pelagic fishes, such as coastal-pelagic fish and oceanic-pelagic fish. Most of the un-matched specimens belonged to the reef-associated and the deep-sea fishes, whose adults were shy and tiny. They were limited to inhabiting heterogeneous coral reefs, such as species of Gobiidae, and abysmal sea (such as Myctophidae). Specimens of these fishes were hard to collect or identify by morphology.
Distribution and Assemblage Patterns
The sampling stations were divided into internal, edge, and outer groups according to their location at the Atoll, with depth <100, 100–200, and greater than 200 m, respectively (Table 2). The overall mean taxa amount was 15.62 ± 8.77, and the abundance was 571.85 ± 556.10 ind/1,000 m−3. The lowest species number, abundance, diversity index (H'), and evenness index (J') with minimum variation ranges are presented in the edge area. The highest average abundance occurred in the outer area, while changing tremendously between stations. The species number and abundance in the internal area were relatively lower than the outer area, while higher H' and J' had a high fluctuation in these stations.
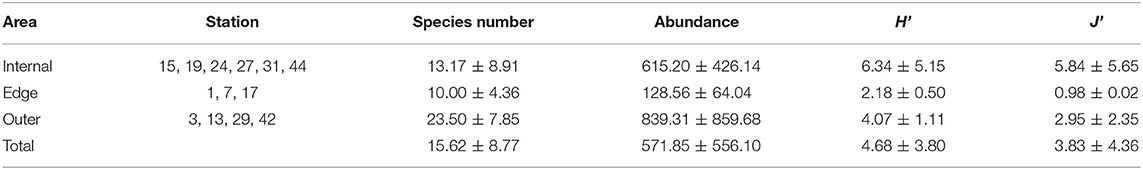
Table 2. Average species number, abundance (ind/1000 m3), Shannon-Weaver diversity index (H'), and Pielou's evenness index (J') of larval fishes from different areas.
The spatial distribution of species number and abundance of larval fish is also shown in Figure 3. Many more species are distributed in the western (stations 31 and 42) and central area, rather than the near edge in the southern and northeastern of the Zhongsha Atoll. At the same time, a high species number also appeared in the southeastern region (stations 3 and 13) outside the edge (Figure 3A). The distribution of abundance was in accordance with that of species number, except for stations 3 and 13 with related low density of individuals occurred (Figure 3B).
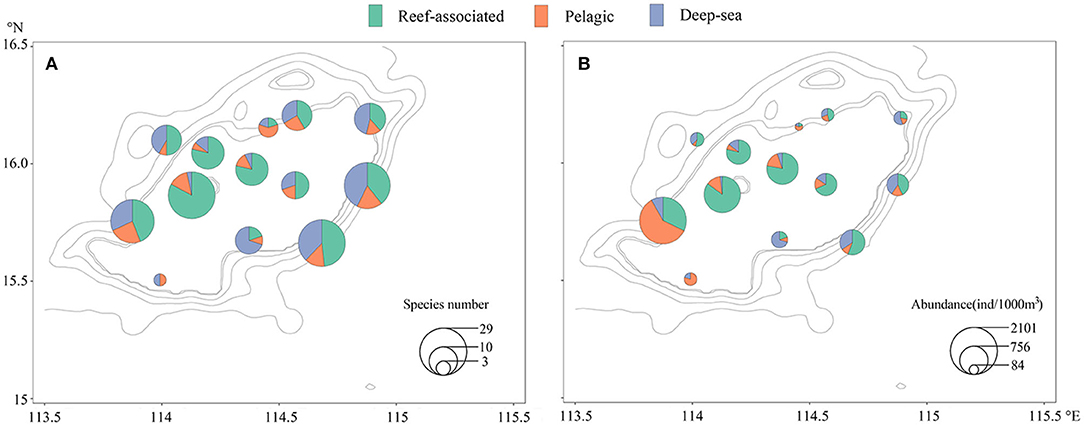
Figure 3. (A,B) Distribution of species number and abundance of larval fish account for taxa groups as reef-associated, deep-sea, and pelagic fishes.
The reef-associated fishes appeared in most sampling stations without station 44 on the southern edge. The species number and percentage increased from the southwestern to the inner Atoll and then decreased gradually to the northeastern (Figure 3A). In comparison, the abundance showed a roughly opposite trend of the species number, with a higher proportion of reef-associated fishes in the northeastern (Figure 3B). The pelagic fishes distributed in all the stations with the species number and their proportion were both low (Figure 3A), while the highest values were found in the southwestern station and lower to northeast account for the abundance and its ratio (Figure 3B). The deep-sea fishes occurred in all stations showing an apparent distribution pattern that most deep-sea fishes appeared at off or near the edge of the Atoll. More minor species and individuals dispersed over the Atoll internal (Figure 3).
Discussion
Identification and Diversity of Fish
It is most commonly accepted that fish biological diversity in the coral reef of the South China Sea is immensely rich (Arai, 2015). In face to the coral decline, understanding the present status of coral reef fishes in the South China Sea in terms of their biodiversity and abundance is essential for the sustainable protection and restoration of coral reef ecosystems (Jones et al., 2004; Brandl et al., 2019). While with the limitations of sampling and habitat heterogeneity, many fish species in the coral reef are undiscovered ever, especially for small species with their all life stages confined to a minimal area (Brandl et al., 2018; Coker et al., 2018). It is hard to comprehensively understand the fish diversity by the traditional sampling gear, such as gill nets, hand-lines, long lines, hydroacoustic techniques, or visual investigation, especially in the areas that are hard to reach. The diversity of coral reefs is supposed to underestimate (Hubert et al., 2012). The ichthyoplanktons are easy to collect. Their investigation has become an alternative way of fish diversity in the coral reef (Carassou et al., 2009).
Few studies investigate the fish in the early life stage in the coral reef in the South China Sea. There is hardly any information on the early life-history stages of fish from the Zhongsha Atoll, the biggest atoll in the South China Sea. The only reference we could find is about pelagic eggs and larvae of fish in the Zhongsha Islands by Chen (1979). We used DNA barcoding to identify the species in the Zhongsha Atoll for the first time and some unknown species (such as 12 taxa of Gobiidae) previously coral reef fish surveys (Table 1). We also found that the coral reefs support many deep-sea fish larvae, for their adult would not be collected in the shallow coral reef waters. The larval fish survey helped to improve our understanding of fish diversity on the coral reef in the Zhongsha Atoll. While 44 sequences from 31 species showed no similarity matches with the database, indicating that the reference sequences of these species were not barcoded ever. It is necessary to acquire genome information of coral reef fishes continuously. Even though we found 95 fish taxa in the Zhongsha Atoll in one cruise, the actual fish species number was likely to be far higher than this current study. More investigation is necessary to better known fish diversity in the coral reef of Zhongsha Atoll.
Distribution and Assemblage Patterns of Larval Fish
The coral reef serves as a critical ground to marine fishes for spawning, nursing, and feeding (Brandl et al., 2019; Woodhead et al., 2019; Wagner et al., 2020). Coral reefs are compound habitats, with complex water flow and water environments, with noticeable regional and temporal variations. The distribution of marine larvae could reflect the adaptation to habitats. Even though many studies focused on the distribution and community of larval fishes, the larval fish assemblage pattern in the coral reef waters is still unknown (Drew and Amatangelo, 2017; Edmunds et al., 2018).
The distribution of larval fishes changed obviously in the Atoll. More species and individuals occurred in the western and central parts of the Zhongsha Atoll (Table 2 and Figure 3), respectively. Oceanographic features, such as eddies and fronts, promoted by monsoons or topography in the coral reef waters, are confirmed in enhancing the spread and survival of marine fish larvae (Shulzitski et al., 2016). The South China Sea is strongly affected by monsoon systems, influenced by the southwest monsoon causing southwest-northeast current in summer (Luo et al., 2016). The upwelling would generate at a sudden seafloor elevation in the western Zhongsha Atoll (Figure 1). The upwelling water might also elevate the diffusion efficiency and provide more food sources, supporting highly diverse fish larvae in the internal Atoll downstream. The habitat complexity would be afforded coral reefs as a shelter for larval fishes, maintaining a higher survival. It would make the internal Atoll a suitable habitat to live in for larval fishes, with the highest species diversity and evenness. However, taxa number, abundance, indexes H' and J' of larval fishes in the edge area were lower than the near parts (Table 2). It might be the integrated result of oceanographic and geomorphic over the shallow coral reef and the deep sea. The larval fishes of reef-associated, deep-sea, and pelagic co-occurrence in the Zhongsha Atoll, but their distribution patterns differed greatly (Figure 3). The reef-associated larval fishes seemed likely to distribute within or near the Atoll area, while most deep-sea larval fishes dispersed near the edge area. The pelagic fishes scattered in all the stations with the species number and its proportion were both low. The species number and abundance also showed a roughly consistent trend with the southwest monsoon. Therefore, the geographical variation shown from distribution and assemblage patterns of larval fish might result from topography and the monsoon effect. Faced with the deteriorating condition of deep-sea coral reefs habitats (Roberts and Hirshfield, 2004; Roder et al., 2013; Rogers, 2019), there is a pressing need to better understand reef ichthyoplankton populations, community dynamics, and the key ecological processes effects in the coral reefs.
Conclusion
The larval fish investigation provided an alternative way to investigate fish diversity in the coral reef of Zhongsha Atoll. Ninety-five larval fish species were identified based on morphological classification and DNA barcoding for the first time, reflecting the rich diversity of larval fish species in the Atoll. Most taxa were non-economic small fishes but would play an essential role in the coral reef ecosystem. Some commercial species, including Auxis thazard, Euthynnus affinis, Sarda orientalis, Decapterus macarellus, Lutjanus viridis, and Centropyge vrolikii, were the dominant species higher than 2% total catch. The larval fish assemblage in the Atoll was assigned categories to reef-associated, deep-sea, and pelagic, respectively. The distribution of larval fishes changed distinctly in the Atoll. The abundance of larval fish was high in the outer Atoll, indexes of species diversity and species evenness were high in the internal area, while the abundance, species number, and indexes of species diversity and species evenness were low in the edge stations. The most reef-associated fish occurred inside the Atoll, and the abyssal fish presented near the edge. The larval fish assemblage distribution might be the result of the oceanographic and geomorphologic effect.
Data Availability Statement
The datasets presented in this study can be found in online repositories. Sequence data for specimens were submitted to the NCBI with the accession numbers as OL512811—OL512944.
Ethics Statement
The animal study was reviewed and approved by Ethics Committee of the Laboratory of Animal Welfare and Ethics of South China Sea Fisheries Research Institute.
Author Contributions
DH designed the work, collected the samples, analyzed the data, and wrote the manuscript. JC performed the DNA barcoding analysis and drafted the manuscript. LX, JN, YL, LW, and SL were involved in revising critically for important intellectual content. ZL participated in the design of the work. FD participated in the work design and approved the final version. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Science and Technology Basic Resources Investigation Program of China (2018FY100105 and 2017FY201405), the Central Public-interest Scientific Institution Basal Research Fund (2018YB04), and the Fund of Guangdong Provincial Key Laboratory of Fishery Ecology and Environment (FEEL-2020-11).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We express our sincere thanks to the NCBI and BOLD systems for the easy access to fish barcoding data. We also thank the officers and the crew of the Yuezhanyuke 10 for their assistance in sampling.
References
Almany, G. R., Planes, S., Thorrold, S. R., Berumen, M. L., Bode, M., Saenz-Agudelo, P., et al. (2017). Larval fish dispersal in a coral-reef seascape. Nat. Ecol. Evol. 1, 1–7. doi: 10.1038/s41559-017-0148
Arai, T. (2015). Diversity and conservation of coral reef fishes in the Malaysian South China Sea. Rev. Fish Biol. Fish. 25, 85–101. doi: 10.1007/s11160-014-9371-9
Baldwin, C. C., Mounts, J. H., Smith, D. G., and Weigt, L. A. (2009). Genetic identification and color descriptions of early life-history stages of Belizean Phaeoptyx and Astrapogon (Teleostei: Apogonidae) with Comments on identification of adult Phaeoptyx. Zootaxa (2008) 26, 1–22. doi: 10.5281/zenodo.185742
Brandl, S. J., Goatley, C. H., Bellwood, D. R., and Tornabene, L. (2018). The hidden half: ecology and evolution of cryptobenthic fishes on coral reefs. Biol. Rev. 93, 1846–1873. doi: 10.1111/brv.12423
Brandl, S. J., Rasher, D. B., Cote, I. M., Casey, J. M., Darling, E. S., Lefchecku, J. S., et al. (2019). Coral reef ecosystem functioning: eight core processes and the role of biodiversity. Front. Ecol. Environ. 17, 445–453. doi: 10.1002/fee.2088
Carassou, L., Mellin, C., and Ponton, D. (2009). Assessing the diversity and abundances of larvae and juveniles of coral reef fish: a synthesis of six sampling techniques. Biodiv. Conservat. 18, 355–371. doi: 10.1007/s10531-008-9492-3
Chen, G., Li, Y., and Chen, X. (2007). Species diversity of fishes in the coral reefs of South China Sea. Biodiv. Sci. 15, 373–381. doi: 10.1360/biodiv.060268
Chen, Z. (1979). Floating fish eggs and larvae of Xisha and Zhongsha Islands. Aquat. Sci. Technol. Inform. 4, 11–13.
Coker, D. J., DiBattista, J. D., Sinclair-Taylor, T. H., and Berumen, M. L. (2018). Spatial patterns of cryptobenthic coral-reef fishes in the Red Sea. Coral Reefs 37, 193–199. doi: 10.1007/s00338-017-1647-9
Cole, A. J., Pratchett, M. S., and Jones, G. P. (2008). Diversity and functional importance of coral-feeding fishes on tropical coral reefs. Fish Fisheries 9, 286–307. doi: 10.1111/j.1467-2979.2008.00290.x
Comte, A., and Pendleton, L. H. (2018). Management strategies for coral reefs and people under global environmental change: 25 years of scientific research. J. Environ. Manag. 209, 462–474. doi: 10.1016/j.jenvman.2017.12.051
De'ath, G., Fabricius, K. E., Sweatman, H., and Puotinen, M. (2012). The 27–year decline of coral cover on the Great Barrier Reef and its causes. Proc. Natl. Acad. Sci. U. S. A. 109, 17995–17999. doi: 10.1073/pnas.1208909109
Drew, J. A., and Amatangelo, K. L. (2017). Community assembly of coral reef fishes along the Melanesian biodiversity gradient. PLoS ONE 12:186123. doi: 10.1371/journal.pone.0186123
Edmunds, P. J., Mcilroy, S. E., Adjeroud, M., Ang, P. O., Bergman, J. L., Carpenter, R. C., et al. (2018). Critical information gaps impeding understanding of the role of larval connectivity among coral reef islands in an era of Global Change. Front. Mar. Sci. 5:290. doi: 10.3389/fmars.2018.00290
Huang, X., Betzler, C., Wu, S., Bernhardt, A., Eagles, G., Han, X., et al. (2020). First documentation of seismic stratigraphy and depositional signatures of Zhongsha Atoll (Macclesfield Bank), South China Sea. Mar. Petrol. Geol. 117:104349. doi: 10.1016/j.marpetgeo.2020.104349
Hubert, N., Meyer, C. P., Bruggemann, H. J., Guerin, F., Komeno, R. J., Espiau, B., et al. (2012). Cryptic diversity in Indo-Pacific coral-reef fishes revealed by DNA-barcoding provides new support to the centre-of-overlap hypothesis. PLoS ONE 7:e28987. doi: 10.1371/journal.pone.0028987
Jones, G. P., McCormick, M. I., Srinivasan, M., and Eagle, J. V. (2004). Coral decline threatens fish biodiversity in marine reserves. Proc. Natl. Acad. Sci. U. S. A. 101, 8251–8253. doi: 10.1073/pnas.0401277101
Jones, G. P., Planes, S., and Thorrold, S. R. (2005). Coral reef fish larvae settle close to home. Curr. Biol. 15, 1314–1318. doi: 10.1016/j.cub.2005.06.061
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Leis, J. M., and Rennis, D. S. (1983). The Larvae of Indo-Pacific Coral Reef Fishes. Hawaii: University Hawaii Press. doi: 10.2307/1444356
Lin, H.-Y., Chiu, M.-Y., Shih, Y.-M., Chen, I.-S., Lee, M.-A., and Shao, K.-T. (2016). Species composition and assemblages of ichthyoplankton during summer in the East China Sea. Continent. Shelf Res. 126, 64–78. doi: 10.1016/j.csr.2016.07.016
Luo, M., Leung, Y., Graf, H. F., Herzog, M., and Zhang, W. (2016). Interannual variability of the onset of the South China Sea summer monsoon. Int. J. Climatol. 36, 550–562. doi: 10.1002/joc.4364
Page, R. D. (1996). Tree view: an application to display phylogenetic trees on personal computers. Bioinformatics 12, 357–358. doi: 10.1093/bioinformatics/12.4.357
Roberts, S., and Hirshfield, M. (2004). Deep-sea corals: out of sight, but no longer out of mind. Front. Ecol. Environ. 2, 123–130. doi: 10.1890/1540-9295(2004)002[0123:DCOOSB]2.0.CO;2
Roder, C., Berumen, M. L., Bouwmeester, J., Papathanassiou, E., Al-Suwailem, A., and Voolstra, C. R. (2013). First biological measurements of deep-sea corals from the Red Sea. Sci. Rep. 3:srep02802. doi: 10.1038/srep02802
Rogers, A. D. (2019). “Chapter 23 - threats to seamount ecosystems and their management,” in World Seas: an Environmental Evaluation, 2nd Edn, ed C. Sheppard (Cambridge, MA: Academic Press), 427–451. doi: 10.1016/B978-0-12-805052-1.00018-8
Shan, B., Liu, Y., Yang, C., Zhao, Y., Zhang, G., Wu, Q., et al. (2021). DNA barcoding of fish in mischief reef—fish diversity of a reef fish community from Nansha Islands. Front. Mar. Sci. 7:618954. doi: 10.3389/fmars.2020.618954
Shulzitski, K., Sponaugle, S., Hauff, M., Walter, K. D., and Cowen, R. K. (2016). Encounter with mesoscale eddies enhances survival to settlement in larval coral reef fishes. Proc. Natl. Acad. Sci. U. S. A. 113, 6928–6933. doi: 10.1073/pnas.1601606113
Sun, D., Qiu, Y., Lin, Z., and Wang, X. (2006). Preliminary studies on the composition of coral reef fish resources in the waters of Zhongsha Island in spring. Trans. Oceanol. Limnol. 3, 85–92. doi: 10.1016/j.ejps.2006.05.004
Teh, L. S., Teh, L. C., and Sumaila, U. R. (2013). A global estimate of the number of coral reef fishers. PLoS ONE 8:e65397. doi: 10.1371/journal.pone.0065397
Wagner, D., Friedlander, A. M., Pyle, R. L., Brooks, C. M., Gjerde, K. M., and Wilhelm, T. A. (2020). Coral reefs of the high seas: hidden biodiversity hotspots in need of protection. Front. Mar. Sci. 7:567428. doi: 10.3389/fmars.2020.567428
Wan, R., and Zhang, R. (2016). Fish Eggs, Larvae and Juveniles in the Offshore Waters of China and Their Adjacent Waters. Shanghai: Shanghai Scientific & Technical Publishers.
Wang, X., Du, F., Lin, Z., Sun, D., Qiu, Y., and Huang, S. (2011). Fish species diversity and community pattern in coral reefs of the Xisha Islands, South China Sea. Biodiv. Sci. 19, 463–469. doi: 10.3724/SP.J.1003.2011.07267
Ward, R. D., Zemlak, T. S., Innes, B. H., Last, P. R., and Hebert, P. D. (2005). DNA barcoding Australia's fish species. Philos. Trans. Royal Soc. B Biol. Sci. 360, 1847–1857. doi: 10.1098/rstb.2005.1716
Woodhead, A. J., Hicks, C. C., Norström, A. V., Williams, G. J., and Graham, N. A. J. (2019). Coral reef ecosystem services in the Anthropocene. Funct. Ecol. 33, 1023–1034. doi: 10.1111/1365-2435.13331
Keywords: larval fish assemblages, Zhongsha Atoll, South China Sea, coral reef, distribution patterns
Citation: Huang D, Chen J, Xu L, Wang X, Ning J, Li Y, Wang L, Liu S, Lin Z and Du F (2022) Larval Fish Assemblages and Distribution Patterns in the Zhongsha Atoll (Macclesfield Bank, South China Sea). Front. Mar. Sci. 8:787765. doi: 10.3389/fmars.2021.787765
Received: 01 October 2021; Accepted: 22 November 2021;
Published: 06 January 2022.
Edited by:
Yuan Li, Ministry of Natural Resources, ChinaReviewed by:
Mohammad Reza Shokri, Shahid Beheshti University, IranKaizhi Li, South China Sea Institute of Oceanology (CAS), China
Copyright © 2022 Huang, Chen, Xu, Wang, Ning, Li, Wang, Liu, Lin and Du. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Feiyan Du, ZmVpeWFuZWdnQDE2My5jb20=
 Delian Huang
Delian Huang Jing Chen1,3
Jing Chen1,3 Xuehui Wang
Xuehui Wang Yafang Li
Yafang Li Feiyan Du
Feiyan Du