- School of Marine Science, Ningbo University, Ningbo, China
Crustacean hyperglycemic hormone superfamily neuropeptides (CHHs) are typical crustacean eyestalk hormones that include the crustacean hyperglycemic hormone (CHH), moult-inhibiting hormone (MIH), vitellogenesis/gonad-inhibiting hormone (VIH/GIH) and mandibular organ-inhibiting hormone (MOIH), which are divided into two subfamilies: type I CHH (included CHH) and type II CHH (consisting of MIH, VIH/GIH, and MOIH). They are involved in various biological activities, such as metabolism, molting, reproduction, and osmotic regulation. Discovery of the ion transport peptide (ITP) in insects expanded the members of CHHs and revealed that CHHs are not restricted to crustaceans. In this study, we focused on three economically important crabs: the mud crab, Scylla paramamosain, the swimming crab, Portunus trituberculatus, and the Chinese mitten crab, Eriocheir sinensis. Their genomes, Pacbio full-length transcriptomic data as well as comparative RNA-seq data were obtained and used to analyze the genomic structures and expression patterns of CHHs and their putative receptors through bioinformatic methods. Two type I CHH members (CHH1 and CHH2) were identified, of which CHH1 had two splice variants, CHH1-v1 and CHH1-v2. One copy of type II CHH (MIH) was found in P. trituberculatus and E. sinensis. While most decapods, including S. paramamosain, have two copies of type II CHHs (MIH/VIH), these MIH/VIHs are adjacent to each other on the same chromosome. Besides type I and II CHH, ITP-like peptides have also been found in the three crabs, and they are mainly expressed in the eyestalk. Four, five, and three G protein-coupled receptors (GPCRs) were identified in S. paramamosain, P. trituberculatus, and E. sinensis, respectively, which might be putative CHH receptors. These GPCRs were divided into three groups. One group was composed of two contiguous genomic position GPCRs, and they were mainly expressed in the hepatopancreas. These findings provide a basis for further studies on CHHs receptor binding tests and on CHHs/GPCRs signaling pathways.
Introduction
The eyestalk ganglia, also referred to as the X-organ-sinus gland (XO-SG) complex is composed of medulla terminalis X-organs (XO), which is involved in the synthesis of neuropeptides, aggregation of XO terminals, sinus gland (SG), as well as in neuropeptide storage and release (reviews, Fingerman, 1997; Webster et al., 2012). Crustacean hyperglycemic hormone superfamily neuropeptides (CHHs) are classic XO-SG complex neuropeptides that are involved in reproduction (reviews, Chung et al., 2010; Jayasankar et al., 2020), molting (reviews, Chung et al., 2010; Webster, 2015a; Chen et al., 2020; Mykles and Chang, 2020; Mykles, 2021), metabolism (reviews, Chung et al., 2010; Chen et al., 2020), immunity (reviews, Chen et al., 2020; Tong et al., 2021), stress (reviews, Chen et al., 2020; Tong et al., 2021), and osmotic regulation (reviews, Chung et al., 2010; Webster, 2015b; Chen et al., 2020).
In 1989, a 72aa peptide was isolated from XO-SG complex of the shore crab, Carcinus maenas. This peptide regulated hemolymph glucose levels and was therefore named “crustacean hyperglycemic hormone (CHH)” which serves as a hyperglycemic factor (Kegel et al., 1989). Subsequently, the moult-inhibiting hormone (MIH) with 78aa was also isolated from the SG of C. maenas and it showed a conserved six cysteine motif with CHH. MIH, produced and secreted by the XO-SG complex, inhibits the synthesis of ecdysteroids during intermoult (Webster, 1991). In the same year, the vitellogenesis-inhibiting hormone (VIH) with 77aa was isolated from the SG of the American lobster, Homarus americanus (Soyez et al., 1991) and characterized. VIH, which is now also referred to as the gonad-inhibiting hormone (GIH), suppresses the onset of vitellogenesis. Later, two neuropeptides, mandibular organ-inhibiting hormone 1 (MOIH-1) and MOIH-2, consisting of 78 residues were isolated from SG extracts of the crab, Cancer pagurus. MOIHs inhibit methyl farnesoate synthesis by the mandibular organ (Wainwright et al., 1996). These are classic crustacean hyperglycemic hormone family peptides, derived from the decapod eyestalk and are 72-78aa in length with conserved six aligned cysteines that form three disulphide bridges (Ohira, 2021). Based on their amino acid sequences, CHHs are divided into two subgroups: type I CHHs and type II CHHs. Type I CHHs have a CHH precursor-related peptide (CHH-PRP) between the signal and mature peptides, while type II CHHs lack CHH-PRP and have an additional glycine in position 5 after the first cysteine residue (review, Chen et al., 2020; Ohira, 2021).
Not only are CHH-family peptides present in crustaceans, they have also been found in several insects. The first non-crustacean member of CHHs was isolated from the desert locust, Schistocerca gregaria, and since it is involved in ion transport, this neuropeptide was named ion transport peptide (ITP) (Audsley and Phillips, 1990; Audsley et al., 1992; Meredith et al., 1996). ITP contains a dibasic cleavage site and a CHH-PRP, which led to it being classified as type I CHH (Montagné et al., 2010). However, phylogenetic analyses revealed that all ITP orthologs are clustered into a clade, which formed a newly named group, type III CHH (Montagné et al., 2010; Toullec et al., 2017). Transcriptomic analyses identified new decapod CHH peptides, whose BLAST hit matched ITPs of insects, therefore, they were named ITP-like peptides (Toullec et al., 2017). These peptides have six conserved cysteine residues, but neither do they contain CHH-PRP nor a specific glycine. For this reason, they were also named CHH-MIH-like (Veenstra, 2016). Based on their phylogeny, ITP-like peptide orthologs constitute a new set, type IV CHH (review, Chen et al., 2020). Hence, as more CHHs sequences were discovered from RNA-seq, reclassification of the CHH family peptides became necessary.
To reveal the varied and complicated functions of CHH peptides, determination of their receptors is crucial (Chung et al., 2010). The G protein-coupled receptors (GPCRs) (BNGR-A24 and BNGR-A34) have been revealed to be silkworm, Bombyx mori ITP and ITP-L receptors (Nagai et al., 2014). Subsequently, several GPCRs belonging to the BNGR-A34 orthologs have been identified as candidate receptors for CHH or MIH in decapods, including the crayfish, Procambarus clarkii (Veenstra, 2015, Rump et al., 2021), the Eastern rock lobster, Sagmariasus verreauxi (Buckley et al., 2016), the blackback land crab, Gecarcinus lateralis (Tran et al., 2019), the rock lobster, Jasus edwardsii (Christie and Yu, 2019), the spiny lobster, Panulirus argus, the blue crab, Callinectes sapidus, H. americanus (Rump et al., 2021), and the swimming crab, Portunus trituberculatus (Tu et al., 2021). Compared to the vast array of CHHs, their candidate GPCRs should be evaluated more.
Recently, due to advances in sequencing technologies, several decapod genomes have been sequenced and assembled (Gutekunst et al., 2018; Zhang et al., 2019; Tan et al., 2020; Tang et al., 2020, 2021; Van Quyen et al., 2020; Cui et al., 2021; Uengwetwanit et al., 2021; Zhao et al., 2021), providing an important basis for comprehensive understanding and differentiation of decapod CHHs and their putative receptors, as well as for further exploration of CHHs genomic structures and characteristics. In this study, CHHs and their putative GPCRs were identified and characterized in three species of economically important crabs: the mud crab, Scylla paramamosain, P. trituberculatus, and the Chinese mitten crab, Eriocheir sinensis. Genomic mining, full-length transcriptome, reference transcriptomic assembly and phylogenetic analyses were performed to determine the genomic structures of CHHs and their putative receptors. Moreover, transcriptome data of the three crabs were analyzed to establish the expression levels of CHHs and their putative receptors in different tissues and under different treatments. Our findings provide a reference basis for the classification and functional studies of CHHs in decapods.
Materials and Methods
Genomic Data
Most of the decapod genome assemblies were downloaded from ncbi1 including those of: E. sinensis (GCA_003336515.1_ASM333651v1 and GCA_013436485.1_ASM1343648v1), P. trituberculatus (GCA_008373055.1_ASM837305v1), the Pacific white shrimp, Penaeus vannamei (GCA_003789085.1_ASM378908v1), the black tiger shrimp, Penaeus monodon (GCF_0152280 65.1_NSTDA_Pmon_1), the blue king crab, Paralithodes platypus (GCA_013283005.1_ASM1328300v1), the red claw crayfish, Cherax quadricarinatus (GCA_009761615.1_DU_Cquad_1.0), and the oriental river prawn, Macrobrachium nipponense (GCA_015104395.1_ASM1510439v1). The S. paramamosain genome was downloaded from the Genome WareHouse (2 GWH accession number: GWHALOH00000000). One of E. sinensis assembled version was downloaded from the3 website.
Transcriptomic Data
The full-length RNA-seq Pacbio data were downloaded from NCBI, including those of S. paramamosain (PRJNA565275 and PRJNA527462), P. trituberculatus (PRJNA656860), and E. sinensis (PRJNA663255). The Pacbio raw data were polished, and the Generic Feature Format (GFF) files of alternative spliced variants were generated using the SMRT Link software (v9.0.0). SRAs data were downloaded from NCBI4 or ENA5. Raw data for Illumina sequencing were quality trimmed using Trimmomatic-0.36 to generate clean reads, which were mapped and aligned to genome with Hisat2 (v2.1.0), The Binary Sequence Alignment/Map (BAM) files generated from Hisat2 and the GFF file were used to calculate transcripts per million (TPM) values of transcripts including alternative spliced variants in each biological replicate by StringTie (v2.1.5). The prepDE.py script was used to convert TPMs to raw read counts. EdgeR (R-3.2.0) was used to normalize raw read counts and estimate differences in transcript expressions. The thresholds for differentially expressed genes (DEGs) were logFC >1 (fold change >2 or fold change <–2) and p-value <0.01.
Bioinformatics Analysis
Crustacean hyperglycemic hormone superfamily neuropeptides and their putative receptor sequences were searched and retrieved from their assemblies using BLAST (v2.6.0+) and Perl. Identification of CHHs and their putative receptors was done as previously described (Bao et al., 2015, 2018a, 2020). Signal peptides of CHHs precursors were predicted using SignalP 4.16. Prediction of prohormone cleavage sites and peptide structures was based on established propeptide processing schemes (Li et al., 2003; Dircksen et al., 2011; Derst et al., 2016). Visualization of genomic structures of CHHs and their putative GPCRs was done using the Exon-Intron Graphic Maker7 and Photoshop CS 6. Multiple sequence alignment was performed using ClustalX and the conserved sequence motifs highlighted by LaTEX TexShade (Beitz, 2000). The GPCRs amino acid sequences were used to build a phylogenetic tree. Phylogenetic analysis was conducted using PhyML (SeaView software) (Gouy et al., 2009) and the resultant phylogenetic tree visualized using Figtree v1.4.3 and Photoshop CS 6.
Results
Genome Mining
Scylla paramamosain
Four CHHs genes, including Sp-CHH2, Sp-MIH, Sp-VIH, and Sp-ITP-like peptide were identified from the S. paramamosain genome (Figures 1, 2). Sp-CHH2 is a recently identified peptide, has three exons and two introns, located on the LG17 chromosome (Figure 1). Sp-MIH and Sp-VIH as type II CHH peptides, located on the same chromosome (LG24), have three exons and two introns each, and they are adjacent to each other on the chromosome. This phenomenon is also present in type II CHH peptides of P. platypus, C. quadricarinatus, and M. nipponense (Figure 3). Moreover, tandem gene duplications of type II CHH peptides were found in P. vannamei and P. monodon, which have six and thirteen MIHs, respectively (Figure 3). Besides these CHHs, an Sp-ITP-like peptide was identified in S. paramamosain. This peptide has five exons and four introns and is located on the LG25 chromosome (Figure 1). The Sp-ITP-like peptide is composed of a 26aa signal peptide followed by a 102aa mature peptide with six conserved cysteine residues (Figure 2). These characteristics are consistent with those of decapod ITP-like peptides (Veenstra, 2016). Sp-CHH1 was not identified in the S. paramamosain genome, however, two splice variants of Sp-CHH1 (Sp-CHH1-v1 and Sp-CHH1-v2) were identified in the Pacbio full-length transcriptome. Full-length cDNA of both Sp-CHH1s have been cloned in S. paramamosain (Fu et al., 2016).
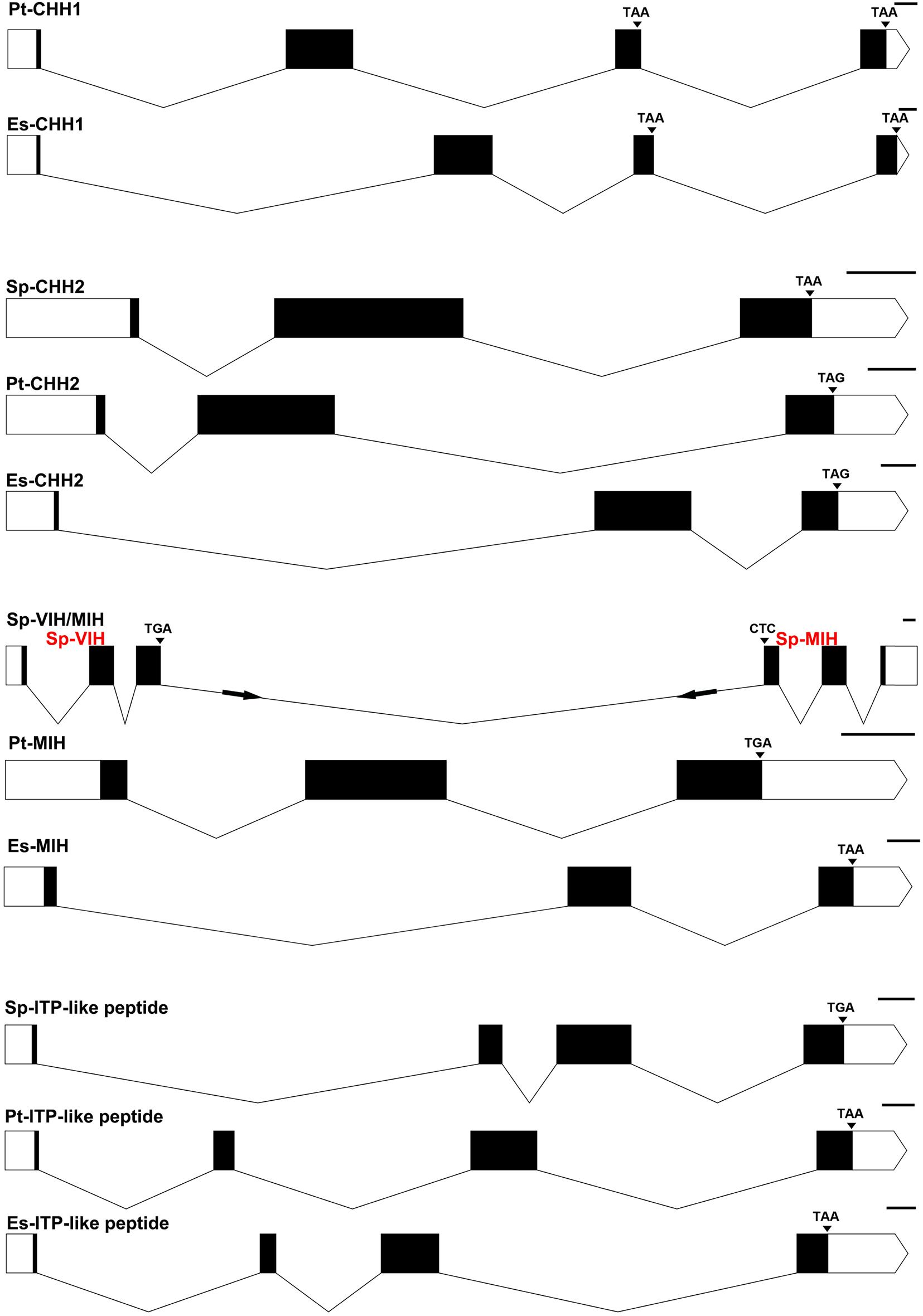
Figure 1. Gene structures of CHHs obtained from genomic assembles of S. paramamosain, P. trituberculatus, and E. sinensis. Schematic diagrams showing the genomic structures of CHH1, CHH2, MIH/VIH, and ITP-like peptide identified in S. paramamosain, P. trituberculatus, and E. sinensis. Blocks and lines represent exons and introns, respectively. Black areas of the blocks represent the coding sequence (CDs) of CHHs. Scale length is 100 base pair.
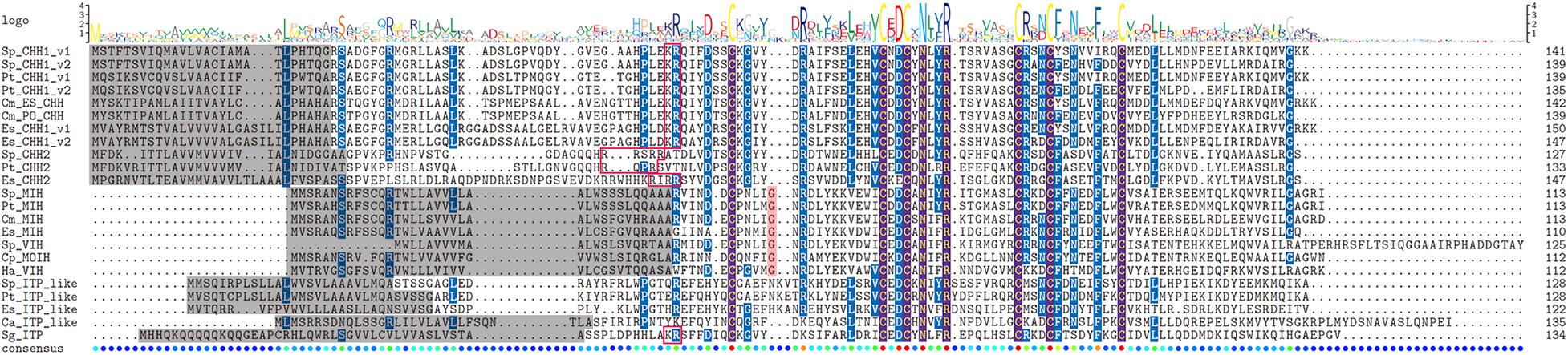
Figure 2. Sequence alignment of CHHs precursors. Comparison of sequence alignment of CHH1-v1, CHH1-v2, CHH2, MIH/VIH, and ITP-like peptide precursors in S. paramamosain, P. trituberculatus, and E. sinensis. Sequence logo of CHHs is shown above the alignment column. The gray highlighted represents signal peptide, pink highlighted represents the additional glycine of type II CHHs, red box represents the putative prohormone cleavage sites, the six conserved cysteines in the mature peptides were aligned and shown brown highlight.
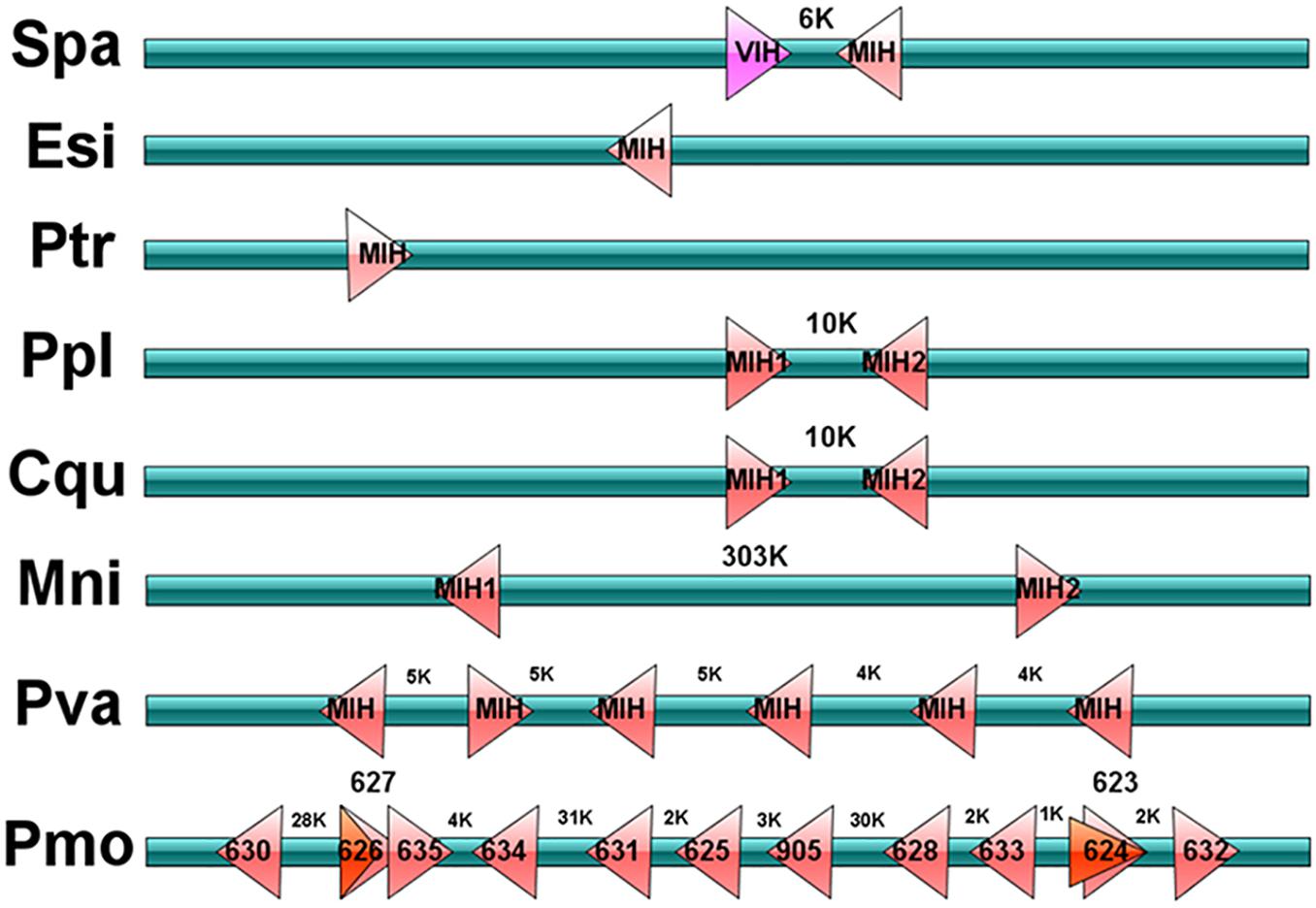
Figure 3. Schematic representation of type II CHH genes in decapods. Schematic diagram showing type II CHH (MIH and VIH) genes location in the scaffold of genomic assembles from S. paramamosain, P. trituberculatus, E. sinensis, P. platypus, C. quadricarinatus, M. nipponense, P. vannamei, and P. monodon. “K” represents Kilobase pair. In schematic of P. monodon, numbers in each block represents the designation of P. monodon type II CHHs, and these information are obtained from Supplementary Material.
In our previous study, through RNA-seq, we identified five transcripts encoding three complete coding sequences (CDs) (Sp-GPCR-A33, Sp-GPCR-A35, and Sp-GPCR-A36) and two partial CDs (Sp-GPCR-A34 and Sp-GPCR-A37) (Bao et al., 2018a). These GPCRs were grouped with ortholog of the B. mori ITP receptor (BNGR-A34) and the predicted CHH receptors from P. clarkii and S. verreauxi (Veenstra, 2015; Buckley et al., 2016; Bao et al., 2018a). In this study, we identified four GPCRs (Sp-GPCR-A34-A37) in the S. paramamosain genome (Figure 4). Among them, Sp-GPCR-A34 has eight exons and seven introns, Sp-GPCR-A35 has nine exons and eight introns, Sp-GPCR-A36 and Sp-GPCR-A37 are splice variants that share the last 6 exons, that is, Sp-GPCR-A36 has 11 exons (1–5, 7–12) while Sp-GPCR-A36 has 7 exons (6-12) (Figure 4).
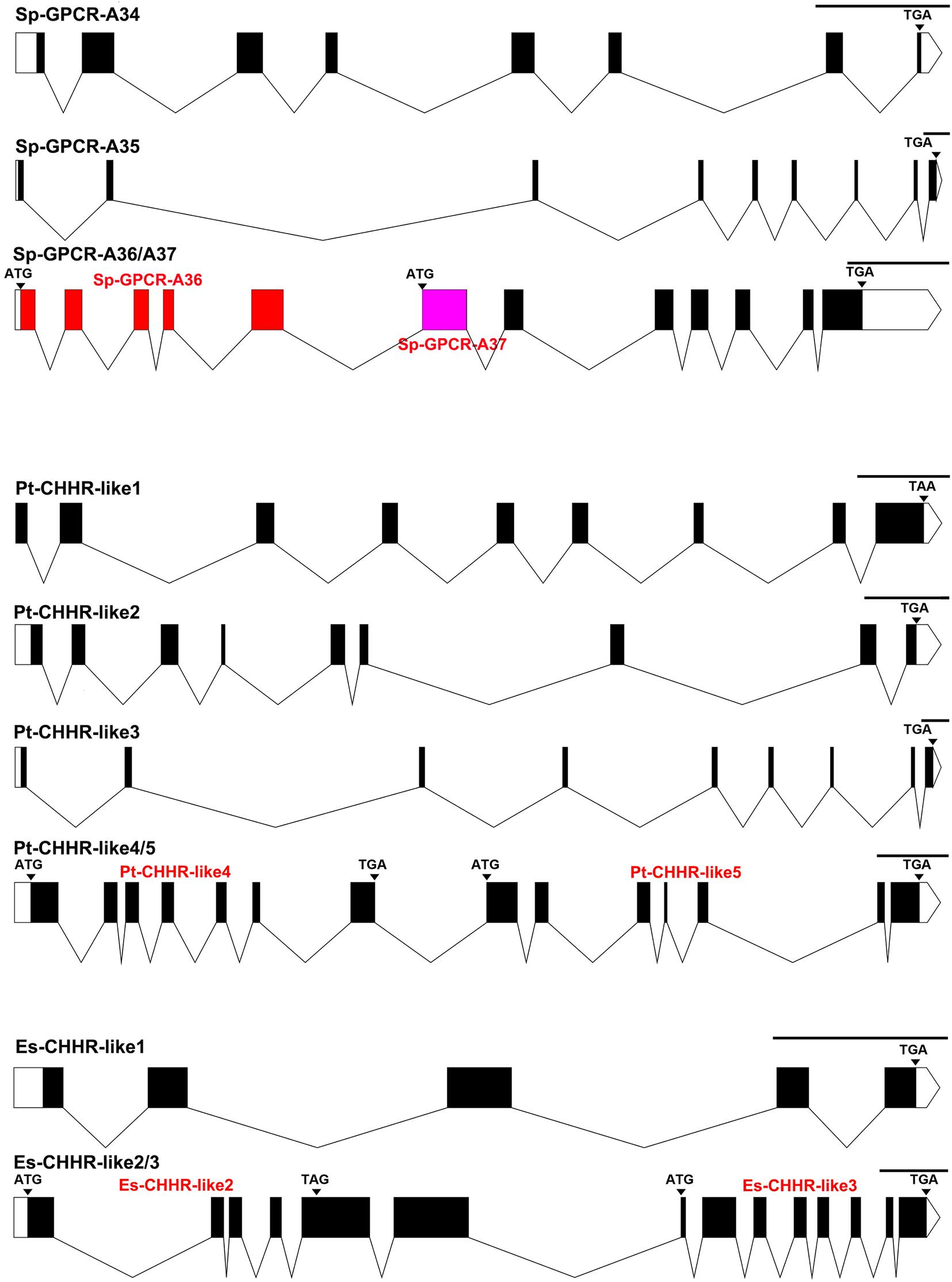
Figure 4. Gene structures of CHH GPCRs obtained from genomic assembles of S. paramamosain, P. trituberculatus, and E. sinensis. Schematic diagrams showing the putative CHH GPCRs genomic structures identified in S. paramamosain, P. trituberculatus, and E. sinensis. Blocks and lines represent exons and introns, respectively. Black areas of the blocks represent the CDs of CHHs. In the schematic diagram of Sp-GPCR-A36-A37, red blocks represent specific exons of Sp-GPCR-A36 gene, pink blocks represent specific exon of Sp-GPCR-A37 gene, and the black blocks represent their common areas. Scale length is 1000 base pair.
Portunus trituberculatus
Four CHHs genes, including Pt-CHH1, Pt-CHH2, Pt-MIH, and Pt-ITP-like peptide were identified in the P. trituberculatus genome (Figures 1,2). Among them, Pt-CHH1 has 4 exons and 3 introns and is located in scaffold_2, there are two splice variants (Pt-CHH1-v1 and Pt-CHH1-v2) generated from the Pt-CHH1 gene. The Pt-CHH1-v1 transcript has three exons (1, 2, and 4), while the Pt-CHH1-v2 transcript has four exons (1, 2, 3, and 4), therefore, Pt-CHH1-v2 is also referred to as CHH-L (CHH long isoform) (Figures 1,2). Pt-CHH1-v1 was blocked at the C-terminal with the conserved motif: XV-amide and was mainly expressed in the eyestalk. Pt-CHH1-v2 was not blocked by the C-terminal and was expressed in various non-eyestalk tissues (Figure 2). Pt-CHH2 has three exons and two introns, located in scaffold_50 (Figure 1). Compared to S. paramamosain, only one type II CHH peptide, Pt-MIH, was found in P. trituberculatus. Pt-MIH has three exons and two introns and is located in scaffold_24 (Figure 1). The protein sequence of Pt-MIH is 91% identity to that of Sp-MIH. The Pt-ITP-like peptide has four exons and three introns (Figure 1). The Pt-ITP-like peptide is composed of a 31aa signal peptide and a 97aa mature peptide (Figure 2), and just like the Sp-ITP-like peptide, it has a highly conserved sequence, that is, it is 77% identity in protein sequence to the Sp-ITP-like peptide.
Five orthologs of possible CHH receptors were identified in the P. trituberculatus genome (Figure 4). Among them, Pt-CHHR-like1 exhibited conserved regions with Sp-GPCR-A33, which has nine exons and lacks the N terminal in the first exon. Pt-CHHR-like2 is the ortholog of Sp-GPCR-A34 and has nine exons (Figure 4). Pt-CHHR-like3 exhibited a highly conserved sequence and genomic structure with Sp-GPCR-A35, which has nine exons. Both Pt-CHHR-like3 and Pt-CHHR-like1 are located on scaffold_36. Pt-CHHR-like4 and Pt-CHHR-like5 exhibited highly conserved regions with Sp-GPCR-A36 and Sp-GPCR-A37. However, compared to Sp-GPCR-A36 and Sp-GPCR-A37, Pt-CHHR-like4 and Pt-CHHR-like5 do not share any exons, and they are adjacent to each other in scaffold_17, with six exons, respectively (Figure 4).
Eriocheir sinensis
Four CHHs genes, including Es-CHH1, Es-CHH2, Es-MIH, and Es-ITP-like peptide were identified in the E. sinensis genome (Figures 1,2). Es-CHH1 has 3 introns and 4 exons and is located in scaffold_90439; Es-CHH2 has three exons and two introns, located on scaffold_4002; Es-MIH has three exons and two introns and is located in scaffold_4920; Es-ITP-like peptide has four exons and three introns while Es-ITP-like peptide is composed of a 28aa signal peptide and a 94aa mature peptide (Figure 2).
Three Es-CHHR-like GPCRs were identified in the E. sinensis genome (Figure 4). Es-CHHR-like1 has five exons and showed high sequence similarity with Sp-GPCR-A33. Es-CHHR-like2 and Es-CHHR-like3 are orthologs of Sp-GPCR-A36/A37 and Pt-CHHR-like 4/5. Es-CHHR-like2 and Es-CHHR-like3 were located in the same scaffold (scaffold_8205), with five and eight exons, respectively (Figure 4). Phylogenetic analysis showed that CHHR-likes from three crabs can be mainly divided into three clades, that is, Sp-GPCR-A36, Sp-GPCR-A37, Pt-CHHR-like4, Pt-CHHR-like5, Es-CHHR-like2, Es-CHHR-like3, and four putative decapod CHH receptors were clustered into one group. Sp-GPCR-A34, Pt-CHHR-like2 and one P. clarkii GPCR (Pcla11008355) formed a clade. The third clade consisted of three subgroups, which Sp-GPCR-A33, Pt-CHHR-like1 and three predicted crab CHH receptors formed a subgroup; Sp-GPCR-A35, Pt-CHHR-like3 and three predicted crab CHH receptors formed a subgroup, which was separated from the third subgroup was formed by five predicted lobster and crayfish CHH receptors. Moreover, two J. edwardsii CHHRs clustered with BNGR-A34, and they were separated from these clades (Figure 5). Since Sp-GPCR-A36 and its orthologs were majorly expressed in the hepatopancreas (Figure 6 and Supplementary Material), we referred to these GPCRs as hepatopancreatic CHHR-likes.
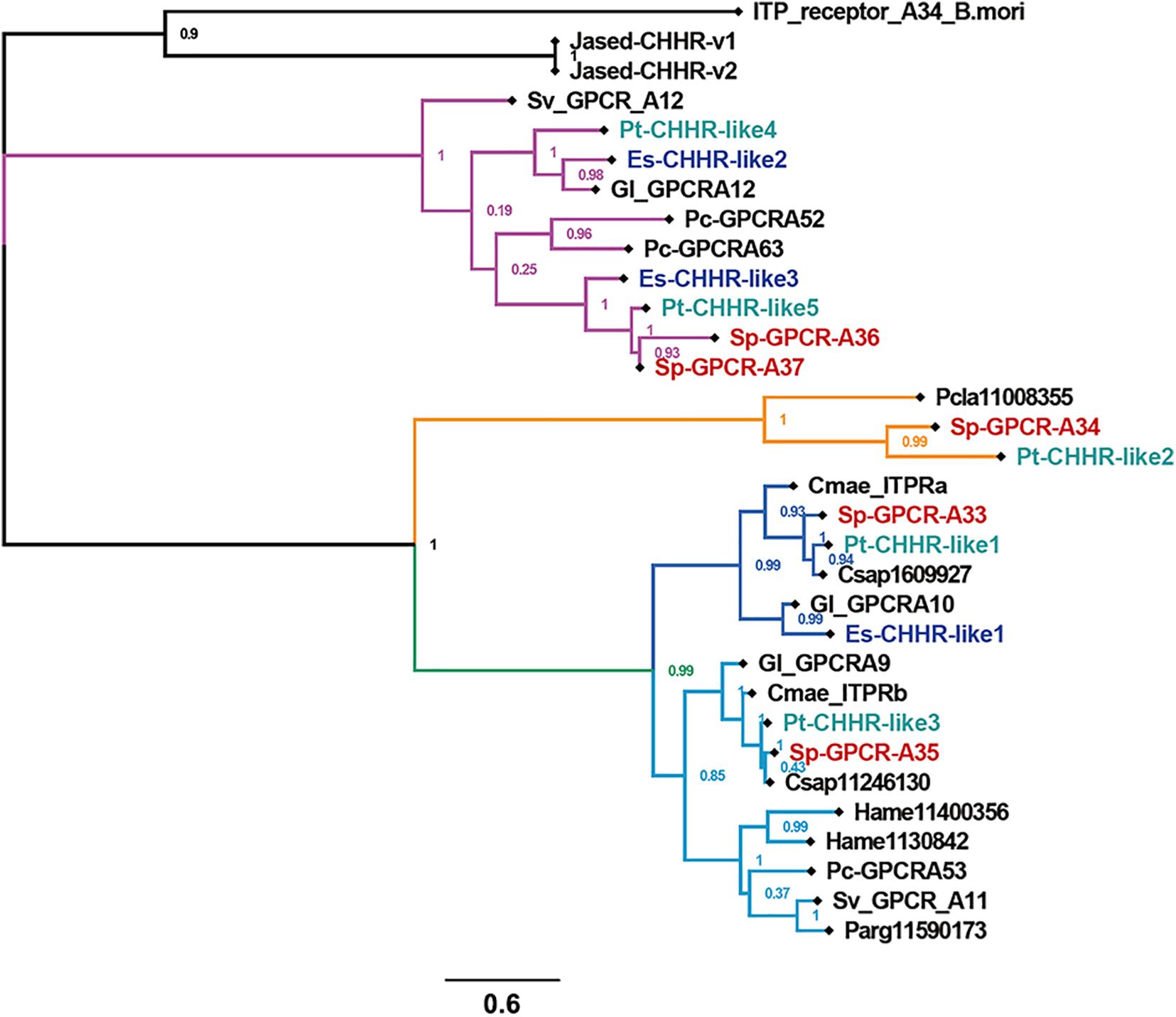
Figure 5. The phylogeny of CHH GPCRs. Cladogram of putative CHH GPCRs showing connections in the clustermap of putative CHH GPCRs. Sv_GPCR, Sagmariasus verreauxi GPCR; Pc-GPCR, Procambarus clarkii GPCR; ITP_receptor_A34_B.mori, Bombyx mori ITP receptor (BNGR-A34); Gl, Gecarcinus lateralis; Pcla, Procambarus clarkii. Cmae, Carcinus maenas; Hame, Homarus americanus; Csap, Callinectes sapidus; Parg, Panulirus argus; Jased, Jasus edwardsii.
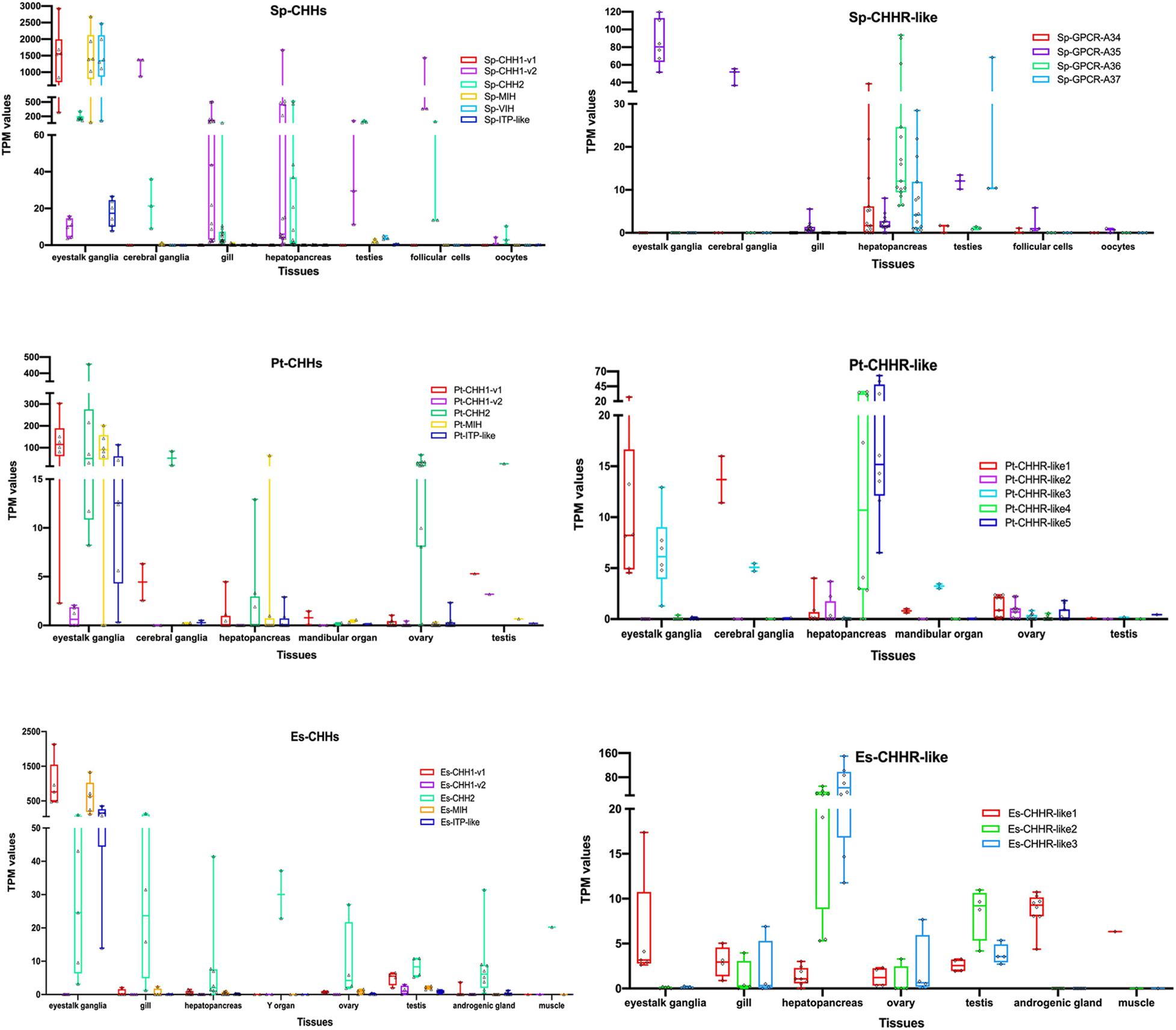
Figure 6. Expression patterns of CHHs and their putative GPCRs. CHHs and their putative GPCRs expressions from different RNA-seq in S. paramamosain, P. trituberculatus and E. sinensis. The boxplot consists of the minimum and maximum range values. The upper boundary of the box indicates the 75th percentile and the lower boundary of the box indicates the 25th percentile, the line in the box is the median. Whiskers above and below the box indicate maximun value and minimun value, respectively. Each triangle or diamond symbol in the box indicates one sample. More sample sources were deposited in Supplementary Material.
Expression of RNA-Seq
Scylla paramamosain
Sp-CHH1-v1 and Sp-CHH1-v2 exhibited different expression patterns (Figure 6 and Supplementary Material). Sp-CHH1-v1 was highly expressed in eyestalk and its TPM values were approximately 1500. Moreover, Sp-CHH1-v2 was expressed in the eyestalk, but it exhibited lower expression levels, and its TPM value was less than 5. Sp-CHH1-v2 was expressed in various tissues, including the cerebral ganglia, gill, hepatopancreas, testis and ovary. In the ovary, Sp-CHH1-v2 exhibited high expression levels in follicular cells (TPM values more than 170). In salinity stress tests, the hepatopancreatic Sp-CHH1-v2 transcript exhibited a significant increase when mud crabs were challenged in low salinity stress (25 to 4‰) (Supplementary Material). The Sp-CHH2 transcript exhibited a similar expression pattern to that of Sp-CHH1-v2, which was detected in all RNA-seq samples. Moreover, the Sp-CHH2 transcript was highly expressed in follicular cells under low salinity stress (Figure 6 and Supplementary Material). Just like Sp-CHH1-v1, Sp-MIH and Sp-VIH transcripts were majorly detected in eyestalk, and their TPM values were more than 1000. Sp-ITP-like was also mainly detected in the eyestalk. Sp-GPCR-A34 was detected in the hepatopancreas and gonads, and it exhibited higher expression levels in the hepatopancreas at post-molt stage and under low salinity treatment (Supplementary Material). Sp-GPCR-A35 was expressed in various tissues and showed higher expression levels in the eyestalk. Sp-GPCR-A36 and Sp-GPCR-A37 were mainly expressed in the hepatopancreas, with Sp-GPCR-A36 showing higher TPM values than Sp-GPCR-A37. The Sp-GPCR-A36 transcript was significantly expressed in low salinity stress conditions (25 to 4‰) (Supplementary Material).
Portunus trituberculatus
Two splice variants of Pt-CHH1 showed varying expression patterns (Figure 6 and Supplementary Material). Pt-CHH1-v1 was mainly expressed in eyestalk ganglia and had higher TPM values. Moreover, it was detected in the cerebral ganglia, hepatopancreas, mandibular organ, testis and ovary. Pt-CHH1-v2 was only detected in eyestalk ganglia, testis and in one ovary sample with relative lower TPM values (Figure 6 and Supplementary Material). Just like Sp-CHH2, Pt-CHH2 was expressed in various tissues, including eyestalk ganglia, cerebral ganglia, hepatopancreas, mandibular organ, testis and ovary (Figure 6 and Supplementary Material). Pt-MIH was highly expressed in the eyestalk ganglia, had higher TPM values and was detected in other tissues with low TPM values (Figure 6 and Supplementary Material). Compared to other tissues, Pt-ITP-like had higher TPM values in the eyestalk ganglia. Pt-CHHR-like1 was detected in various tissues and it exhibited relatively higher TPM values in the eyestalk ganglia and cerebral ganglia. Pt-CHHR-like2 was only expressed in the hepatopancreas and ovary. Pt-CHHR-like3 was detected in the eyestalk ganglia, cerebral ganglia, mandibular organ, testis, ovary and in one hepatopancreatic sample. Pt-CHHR-like4 and Pt-CHHR-like5 were mainly expressed in the hepatopancreas (Figure 6 and Supplementary Material).
Eriocheir sinensis
Es-CHH1-v1 was mainly expressed in the eyestalk and it exhibited higher TPM values (Figure 6 and Supplementary Material), which were comparable to the expression patterns of Sp-CHH1-v1 and Pt-CHH1-v1. However, Es-CHH1-v2 did not exhibit extensive tissue distributions and had lower TPM values (Figure 6 and Supplementary Material). Es-CHH2 was expressed in various tissues, including the eyestalk, gill, muscle, hepatopancreas, androgenic gland, Y organ, testis and in the ovary. The Es-MIH transcript was mainly expressed in the eyestalk, and their TPM values were more than 100. Es-ITP-like was highly expressed in the eyestalk (Figure 6 and Supplementary Material). The Es-CHHR-like1 transcript was detected in various tissues, while their TPM values were less than 10. Es-CHHR-like2 and Es-CHHR-like3 were mainly expressed in the hepatopancreas. In addition, they were expressed in the ovary, gill and even eyestalk, but their TPM values were low (Figure 6 and Supplementary Material).
Discussion
Several CHHs and their putative GPCRs from crabs are described here. We focused on their genomic structures and expression patterns. The RNA-seq is a powerful technique for identifying mud crab neuropeptides and their GPCRs (Bao et al., 2015, 2018a). We combined genomic assembly, full-length transcriptome sequencing and RNA-seq analyses to identify crabs CHHs and their putative GPCRs.
In typical classification, CHHs were divided into type I CHH and type II CHH. Alternative splicing of type I CHH gene has been shown in almost all reported decapod species (review, Webster et al., 2012). The type I CHH gene is composed of four exons, one splice variant lacks the 3rd exon and transcript end in the 4th exon. The other one includes all exons, and is hereby named CHH-L. However, CHH-L transcript ends in the 3rd exon, therefore, it exhibits similar amino acid lengths with the first splice variant (review, Webster et al., 2012). These features match the findings from the CHH1 genes of P. trituberculatus and E. sinensis in this study. However, we did not identify CHH1 genomic sequences in the S. paramamosain genome, which might be attributed to incomplete genomic assembly (Zhao et al., 2021). There are three indications suggesting that the Sp-CHH1 gene has four exons and generates two splice variants. First, consistency of the CHH1 gene structure in several decapods. Second, in a previous study, two CHH isoforms (“Sp-CHH1” and “Sp-CHH2”) exhibited identical nucleotide sequences, except the insert (equivalent to the 3rd exon) in the “Sp-CHH2” isoform (Fu et al., 2016). Finally, full-length transcriptome analyses revealed that two transcripts that are identical to the above reported CHH isoforms exist in mud crab (Fu et al., 2016). Given another type I CHH gene was identified from crabs in this study, we named this typical type I CHH as “CHH1,” and the new-found type I CHH was referred to as “CHH2.” CHH1-v1 and CHH1-v2 were used to describe the C-terminal amidated CHH peptide and long-isoform CHH peptide, respectively. Generally, CHH1-v1 transcripts were mainly detected in the neurons of the X-organ, which are located in the eyestalk, while CHH1-v2 transcripts were found in the pericardial organ, thoracic ganglia, antennal glands, gills and heart (review, Webster et al., 2012). Tissues expression profiles of the CHH1 gene in S. paramamosain has also been reported (Fu et al., 2016). However, common areas from two splice variants were selected to design qRT-PCR primers, resulting in expression patterns of two splice variants that could not be distinguished (Fu et al., 2016). We used the StringTie software to re-analyze the expression patterns of two splice variants that originate from the Sp-CHH1 gene. Our results were consistent with those of previously reported species (review, Webster et al., 2012). The Sp-CHH1-v1 transcript was majorly expressed in the eyestalk while the Sp-CHH1-v2 transcript was mainly detected in various tissues. Just like S. paramamosain, two splice variants from P. trituberculatus and E. sinensis CHH1 gene have also shown differential expression patterns. CHH1-v1 was expressed in the eyestalk throughout decapods, suggesting that it might be functionally conserved. Non-eyestalk splice variant was expressed in a variety of tissues, their expression patterns in three crabs were different. The Sp-CHH1-v2 transcript was highly expressed in the cerebral ganglia, hepatopancreas under salinity stress and post-molt, and in ovarian follicular cells, while Pt-CHH1-v2 and Es-CHH1-v2 exhibited low expression levels in our study. These findings suggest that CHH1-v2 might have species specific functions. The expression levels of Sp-CHH1-v2 significantly increased when mud crabs were subjected to salinity changes, which might speculatively implicate the hepatopancreas in, as yet undefined, roles in osmo/iono regulation. Elevated expression levels of Sp-CHH1-v2 in the follicular cells were comparable to those of Sp-BMP7 (Shu et al., 2016) and Sp-sNPF (Bao et al., 2018b) in the ovary, which are involved in vitellogeneses and ovarian maturation (Shu et al., 2016; Bao et al., 2018b).
Another type I CHH gene (CHH2) from several decapods has also been found (Veenstra, 2016). In S. paramamosain, full-length Sp-CHH2 cDNA was cloned, and it was shown to regulate hyperglycemic activity (Liu et al., 2019). In this study, the expression levels of hepatopancreatic Sp-CHH2 were found to be significantly elevated as a result of salinity stress, adding to the evidence that Sp-CHH2 plays a key role in metabolism (Liu et al., 2019). This expression profile is comparable to that of Sp-CHH1-v2. Furthermore, Sp-CHH2 mRNA was highly expressed in follicular cells, indicating that Sp-CHH1-v2 and Sp-CHH2 play synergetic roles in S. paramamosain. The functions of Pt-CHH2 and Es-CHH2 have not been verified, however, their expressions in various tissues indicate that they have multiple functions.
Even though type II CHH has three classifications (MIH, VIH/GIH and MOIH), most decapods have two type II CHH genes (review, Webster et al., 2012). For instance, C. pagurus has two MOIHs (Wainwright et al., 1996). In the spider crab, Libinia emarginata, three peptides were purified to show MOIH activity (Liu and Laufer, 1996), however, one of them belonged to type I CHH (Liu et al., 1997). These designations were based on their biological activities when they were identified for the first time. Two type II CHH genes were also been found in S. paramamosain, and were named MIH and VIH, respectively as their first identifications (Huang et al., 2015; Liu et al., 2018). However, only one MIH was found in P. trituberculatus and E. sinensis, respectively. Despite we searched the type II CHH in all accessible genome, Pacbio full-length transcriptome and RNA-seq. We postulate that there is only one type II CHH gene in P. trituberculatus and E. sinensis. This could be because one copy of type II CHHs might have been lost from the genome during evolution. Pt-MIH expression has been negatively correlated with the ecdysteroid (20-hydroxyecdysone, 20E) titer during molt stages (Wang et al., 2013). MIH has also been identified in E. sinensis, and the recombinant Es-MIH (rEs-MIH) inhibits ecdysteroid secretion by Y-organs in this species (Zhang et al., 2011). It is now timely to determine whether the single copy type II CHHs have inhibitory roles in gonad maturation or methyl farnesoate (MF) synthesis by mandibular organs. Alternatively, we postulate that the lost roles of type II CHHs may have been replaced by type I CHHs. Conversely, six type II CHH genes were found in the P. vannamei genomic assembly (Zhang et al., 2019). Expansion of type II CHH seems to be universal phenomenon in the Penaeidae, as we also found thirteen type II CHH genes in P. monodon. In C. pagurus, MIH and MOIH were clustered in the same chromosome (Lu et al., 2000). Due to the lack of assembled genome, this feature had not been found in other decapods until six tandem MIH genes were determined in the P. vannamei genome (Zhang et al., 2019). In this study, all identified type II CHHs (more than one copy) from every species were located in adjacent positions or showed tandem structures in the same chromosome. This characteristic of type II CHH is ubiquitous in decapods.
The ITP-like peptide exhibited a higher sequence similarity with ITP isoforms of insects rather than CHHs or MIHs (Toullec et al., 2017). In addition, 4 exon/3 intron genomic structure of the ITP-like peptide is identical to that of ITP. The 4-exon structure of the ITP gene can generate splice variants, just like CHH1 (reviews, Webster et al., 2012; Chen et al., 2020). Nevertheless, in this study none were found in the PacBio generated full-length transcriptomes. Whilst deeper level transcriptome sequencing might show alternative splice variants, it is also possible that such transcripts are expressed at extremely low levels, which could have no functional relevance. Specific expressions in eyestalk indicate that the ITP-like peptide has a conserved function in decapods. However, it should be determined whether the newly identified ITP-like peptide functions as an ion transport factor.
Biochemical characterization of receptor binding characteristics using 125-I labeled CHH, MIH and membrane preparations in C. maenas (Webster, 1993) and C. sapidus (Chung et al., 2010) has been used to identify receptor binding sites in various tissues. In C. sapidus, CHH1-v1 (ES-CHH) and CHH1-v2 (PO-CHH or CHH-L) have multiple binding sites from various tissues, including hepatopancreas, gills, abdominal muscles, scaphognathites, hindgut, midgut, and heart (Katayama and Chung, 2009). The Y-organs of juveniles and hepatopancreas of females were confirmed to be binding sites for C. sapidus MIH (Zmora et al., 2009). In C. maenas, CHH1-v1 and MIH have highly specific binding sites of Y-organs (Webster, 1993). In this study, only the transcriptome data for E. sinensis Y-organs were available in public databases, and all Es-CHHR-likes were not detected in this tissue. Therefore, more YO RNA-seq data should be generated to investigate the expression profiles of CHHR-likes in other crabs. In addition, C. maenas CHH1-v1 has many binding sites in the hepatopancreas, gills and hindgut (Kummer and Keller, 1993; Webster, 1993; Chung and Webster, 2006). Hepatopancreas membranes from crayfish, Orconectes limosus and C. maenas were shown to have a CHH1-v1 binding specificity. Membranes from the hepatopancreas of C. maenas had lower affinities for O. limosus CHH1-v1, while C. maenas CHH1-v1 did not bind O. limosus CHH1-v1 (Kummer and Keller, 1993). This species-specificity of CHH1-v1 is consistent with its hyperglycaemic activity in different crabs, and this was attributed to co-evolution of CHH1-v1/receptors (review, Chen et al., 2020). In this study, although hepatopancreatic CHHR-like GPCRs were clustered in a clade, they exhibited differences in terms of their amino acid sequences. These differences might have led to protein structural differences, leading to CHH species-specificity in the hepatopancreas. Differences in genomic structures of hepatopancreatic CHHR-like GPCRs were that copies of Sp-GPCR-A36 and Sp-GPCR-A37 overlapped on the chromosome, while hepatopancreatic GPCRs from P. trituberculatus and E. sinensis showed an isolated copy located in adjacent positions in the same chromosome, which might be evolutionary traces. Based on previous findings, three decapods (C. sapidus, C. maenas, and O. limosus) CHH1-v1 have common binding tissues, hepatopancreas, the major metabolic site (Kummer and Keller, 1993; Webster, 1993; Chung and Webster, 2006). The newly identified hepatopancreatic GPCRs from S. paramamosain, P. trituberculatus, and E. sinensis might be receptors for CHH1-v1, which regulates hyperglycemic activity. This postulate was supported by the significant increase in expression levels of Sp-GPCR-A36 in salinity stress conditions. Hepatopancreas is a major vitellogenin source and a metabolic site, and the female hepatopancreas for C. sapidus is a target tissue for MIH (Zmora et al., 2009), suggesting that hepatopancreatic CHHR-like GPCRs bind MIH/VIHs in this species. However, in C. maenas specific binding of MIH to hepatopancreas membrane preparations was not observed (Webster, 1993). Two copies of hepatopancreatic CHHR-like GPCRs were differentially expressed, indicating that they play different functions in hepatopancreas. However, it has not been determined whether one of them is a CHH1-v1 receptor and another a MIH/VIH receptor. Furthermore, Sp-GPCR-A34 and its ortholog, Pt-CHHR-like2, were specifically expressed in the hepatopancreas and in the ovary. Sp-GPCR-A34, which showed higher expressions in the hepatopancreas during the post-molt stage, exhibited the MIH receptor feature (reviews, Chung et al., 2010; Chen et al., 2020). Expressions of Sp-GPCR-A33, Sp-GPCR-A35 and their orthologs in various tissues suggests pleiotropic functions, and it should be determined whether they were activated by the multi-tissue expressed CHH1-v2/CHH2 or by the pleiotropic CHH1-v1. Previous review revealed that decapod putative CHHRs were clustered into two clades (CHHR1 and CHHR2) (review, Mykles, 2021). In this study, more decapod putative CHHR sequences were collected to construct phylogenetic tree to clarify their clustering. Of which, our hepatopancreatic CHHR-like clade was consistent with the CHHR2 clade, and four putative CHHRs (Sv_GPCR_A12, Gl_GPCRA12, Pc-GPCRA52, and Pc-GPCRA63) in the CHHR2 clade also expressed in hepatopancreas (Veenstra, 2015; Buckley et al., 2016; Tran et al., 2019). In the review, only four CHHRs in the CHHR1 clade (review, Mykles, 2021), our analysis expanded this clade, which three subgroups were separated from each other. Moreover, three putative CHHRs constitute a new CHHR set.
In summary, we show the genomic structures and expressions of CHHs and their putative receptors in S. paramamosain, P. trituberculatus, and E. sinensis. Our findings provide an important basis for subsequent studies on CHHs binding sites and on CHHs/GPCRs signaling pathways.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
YY: conceptualization, editing, and sample collection. YX: software support. PZ: editing. ZC: supervision. CB: conceptualization, sample collection, bioinformatics analysis, writing–original draft, and supervision. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (grant number: 31902350) and the K. C. Wong Magna Fund in Ningbo University.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Simon G. Webster and Donald Mykles for their valuable comments and suggestions that helped improve the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.787007/full#supplementary-material
Footnotes
- ^ www.ncbi.nlm.nih.gov/genome/?term=decapoda
- ^ https://ngdc.cncb.ac.cn/gwh/
- ^ http://www.genedatabase.cn/esi_genome.html
- ^ https://www.ncbi.nlm.nih.gov/
- ^ https://www.ebi.ac.uk/ena/browser/home
- ^ http://www.cbs.dtu.dk/services/SignalP/
- ^ http://wormweb.org/exonintron
References
Audsley, N., and Phillips, J. E. (1990). Stimulants of ileal salt transport in neuroendocrine system of the desert locust. Gen. Comp. Endocrinol. 80, 127–137. doi: 10.1016/0016-6480(90)90156-g
Audsley, N., McIntosh, C., and Phillips, J. E. (1992). Isolation of a neuropeptide from locust corpus cardiacum which influences ileal transport. J. Exp. Biol. 173, 261–274. doi: 10.1242/jeb.173.1.261
Bao, C., Liu, F., Yang, Y., Lin, Q., and Ye, H. (2020). Identification of peptides and their GPCRs in the peppermint shrimp Lysmata vittata, a protandric simultaneous hermaphrodite species. Front. Endocrinol. 11:226. doi: 10.3389/fendo.2020.00226
Bao, C., Yang, Y., Huang, H., and Ye, H. (2015). Neuropeptides in the cerebral ganglia of the mud crab, Scylla paramamosain: transcriptomic analysis and expression profiles during vitellogenesis. Sci. Rep. 5, 1–15. doi: 10.1038/srep17055
Bao, C., Yang, Y., Huang, H., and Ye, H. (2018a). Inhibitory role of the mud crab short neuropeptide F in vitellogenesis and oocyte maturation via autocrine/paracrine signaling. Front. Endocrinol. 9:390. doi: 10.3389/fendo.2018.00390
Bao, C., Yang, Y., Zeng, C., Huang, H., and Ye, H. (2018b). Identifying neuropeptide GPCRs in the mud crab, Scylla paramamosain, by combinatorial bioinformatics analysis. Gen. Comp. Endocrinol. 269, 122–130. doi: 10.1016/j.ygcen.2018.09.002
Beitz, E. (2000). TeXshade: shading and labeling of multiple sequence alignments using LaTeX2e. Bioinformatics 16, 135–139. doi: 10.1093/bioinformatics/16.2.135
Buckley, S. J., Fitzgibbon, Q. P., Smith, G. G., and Ventura, T. (2016). In silico prediction of the G-protein coupled receptors expressed during the metamorphic molt of Sagmariasus verreauxi (Crustacea: Decapoda) by mining transcriptomic data: RNA-seq to repertoire. Gen. Comp. Endocrinol. 228, 111–127. doi: 10.1016/j.ygcen.2016.02.001
Chen, H. Y., Toullec, J. Y., and Lee, C. Y. (2020). The crustacean hyperglycemic hormone superfamily: progress made in the past decade. Front. Endocrinol. 11:769. doi: 10.3389/fendo.2020.578958
Christie, A. E., and Yu, A. (2019). Identification of peptide hormones and their cognate receptors in Jasus edwardsii–a potential resource for the development of new aquaculture management strategies for rock/spiny lobsters. Aquaculture 503, 636–662. doi: 10.1016/j.aquaculture.2018.11.059
Chung, J. S., and Webster, S. G. (2006). Binding sites of crustacean hyperglycemic hormone and its second messengers on gills and hindgut of the green shore crab, Carcinus maenas: a possible osmoregulatory role. Gen. Comp. Endocrinol. 147, 206–213. doi: 10.1016/j.ygcen.2006.01.002
Chung, J. S., Zmora, N., Katayama, H., and Tsutsui, N. (2010). Crustacean hyperglycemic hormone (CHH) neuropeptides family: functions, titer, and binding to target tissues. Gen. Comp. Endocrinol. 166, 447–454. doi: 10.1016/j.ygcen.2009.12.011
Cui, Z., Liu, Y., Yuan, J., Zhang, X., Ventura, T., Ma, K. Y., et al. (2021). The Chinese mitten crab genome provides insights into adaptive plasticity and developmental regulation. Nat. Commun. 12, 1–13. doi: 10.1038/s41467-021-22604-3
Derst, C., Dircksen, H., Meusemann, K., Zhou, X., Liu, S., and Predel, R. (2016). Evolution of neuropeptides in non-pterygote hexapods. BMC Evol. Biol. 16:51. doi: 10.1186/s12862-016-0621-4
Dircksen, H., Neupert, S., Predel, R., Verleyen, P., Huybrechts, J., Strauss, J., et al. (2011). Genomics, transcriptomics, and peptidomics of Daphnia pulex neuropeptides and protein hormones. J. Proteome Res. 10, 4478–4504. doi: 10.1021/pr200284e
Fingerman, M. (1997). Crustacean endocrinology: a retrospective, prospective, and introspective analysis. Physiol. Zool. 70, 257–269. doi: 10.1086/639593
Fu, C., Huang, X., Gong, J., Chen, X., Huang, H., and Ye, H. (2016). Crustacean hyperglycaemic hormone gene from the mud crab, Scylla paramamosain: cloning, distribution and expression profiles during the moulting cycle and ovarian development. Aquac. Res. 47, 2183–2194. doi: 10.1111/are.12671
Gouy, M., Guindon, S., and Gascuel, O. (2009). SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27, 221–224. doi: 10.1093/molbev/msp259
Gutekunst, J., Andriantsoa, R., Falckenhayn, C., Hanna, K., Stein, W., Rasamy, J., et al. (2018). Clonal genome evolution and rapid invasive spread of the marbled crayfish. Nat. Ecol. Evol. 2, 567–573. doi: 10.1038/s41559-018-0467-9
Huang, H., Fu, C., Chen, X., Gong, J., Huang, X., and Ye, H. (2015). Molt-inhibiting hormone (MIH) gene from the green mud crab Scylla paramamosain and its expression during the molting and ovarian cycle. Aquac. Res. 46, 2665–2675. doi: 10.1111/are.12421
Jayasankar, V., Tomy, S., and Wilder, M. N. (2020). Insights on molecular mechanisms of ovarian development in decapod Crustacea: focus on vitellogenesis-stimulating factors and pathways. Front. Endocrinol. 11:577925. doi: 10.3389/fendo.2020.577925
Katayama, H., and Chung, J. S. (2009). The specific binding sites of eyestalk-and pericardial organ-crustacean hyperglycaemic hormones (CHHs) in multiple tissues of the blue crab, Callinectes sapidus. J. Exp. Biol. 212, 542–549. doi: 10.1242/jeb.022889
Kegel, G., Reichwein, B., Weese, S., Gaus, G., Peter-Kataliníc, J., and Keller, R. (1989). Amino acid sequence of the crustacean hyperglycemic hormone (CHH) from the shore crab, Carcinus maenas. FEBS Lett. 255, 10–14. doi: 10.1016/0014-5793(89)81051-8
Kummer, G., and Keller, R. (1993). High-affinity binding of crustacean hyperglycemic hormone (CHH) to hepatopancreatic plasma membranes of the crab Carcinus maenas and the crayfish Orconectes limosus. Peptides 14, 103–108. doi: 10.1016/0196-9781(93)90016-a
Li, L., Kelley, W. P., Billimoria, C. P., Christie, A. E., Pulver, S. R., Sweedler, J. V., et al. (2003). Mass spectrometric investigation of the neuropeptide complement and release in the pericardial organs of the crab, Cancer borealis. J. Neurochem. 87, 642–656. doi: 10.1046/j.1471-4159.2003.02031.x
Liu, A., Liu, J., Chen, X., Lu, B., Zeng, C., and Ye, H. (2019). A novel crustacean hyperglycemic hormone (CHH) from the mud crab Scylla paramamosain regulating carbohydrate metabolism. Comp. Biochem. Physiol. A 231, 49–55. doi: 10.1016/j.cbpa.2019.01.015
Liu, C., Jia, X., Zou, Z., Wang, X., Wang, Y., and Zhang, Z. (2018). VIH from the mud crab is specifically expressed in the eyestalk and potentially regulated by transactivator of Sox9/Oct4/Oct1. Gen. Comp. Endocrinol. 255, 1–11. doi: 10.1016/j.ygcen.2017.09.018
Liu, L., and Laufer, H. (1996). Isolation and characterization of sinus gland neuropeptides with both mandibular organ inhibiting and hyperglycemic effects from the spider crab Libinia emarginata. Arch. Insect Biochem. Physiol. 32, 375–385.
Liu, L., Laufer, H., Gogarten, P. J., and Wang, M. (1997). cDNA cloning of a mandibular organ inhibiting hormone from the spider crab Libinia emarginata. Invert. Neurosci. 3, 199–204. doi: 10.1007/BF02480375
Lu, W., Wainwright, G., Webster, S. G., Rees, H. H., and Turner, P. C. (2000). Clustering of mandibular organ-inhibiting hormone and moult-inhibiting hormone genes in the crab, Cancer pagurus, and implications for regulation of expression. Gene 253, 197–207. doi: 10.1016/S0378-1119(00)00282-1
Meredith, J., Ring, M., Macins, A., Marschall, J., Cheng, N. N., Theilmann, D., et al. (1996). Locust ion transport peptide (ITP): primary structure, cDNA and expression in a baculovirus system. J. Exp. Biol. 199, 1053–1061. doi: 10.1242/jeb.199.5.105
Montagné, N., Desdevises, Y., Soyez, D., and Toullec, J. Y. (2010). Molecular evolution of the crustacean hyperglycemic hormone family in ecdysozoans. BMC Evol. Biol. 10:62. doi: 10.1186/1471-2148-10-62
Mykles, D. L. (2021). Signaling pathways that regulate the crustacean molting gland. Front. Endocrinol. 12:674711. doi: 10.3389/fendo.2021.674711
Mykles, D. L., and Chang, E. S. (2020). Hormonal control of the crustacean molting gland: insights from transcriptomics and proteomics. Gen. Comp. Endocrinol. 294:113493. doi: 10.1016/j.ygcen.2020.113493
Nagai, C., Mabashi-Asazuma, H., Nagasawa, H., and Nagata, S. (2014). Identification and characterization of receptors for ion transport peptide (ITP) and ITP-like (ITPL) in the silkworm Bombyx mori. J. Biol. Chem. 289, 32166–32177. doi: 10.1074/jbc.M114.590646
Ohira, T. (2021). “Chapter 76 – crustacean hyperglycemic hormone,” in Handbook of Hormones, eds H. Ando, K. Ukena, and S. Nagata (Cambridge, MA: Academic Press), 731–733. doi: 10.1016/B978-0-12-801028-0.00053-2
Rump, M. T., Kozma, M. T., Pawar, S. D., and Derby, C. D. (2021). G protein-coupled receptors as candidates for modulation and activation of the chemical senses in decapod crustaceans. PLoS One 16:e0252066. doi: 10.1371/journal.pone.0252066
Shu, L., Yang, Y., Huang, H., and Ye, H. (2016). A bone morphogenetic protein ligand and receptors in mud crab: a potential role in the ovarian development. Mol. Cell. Endocrinol. 434, 99–107. doi: 10.1016/j.mce.2016.06.023
Soyez, D., Le Caer, J. P., Noel, P. Y., and Rossier, J. (1991). Primary structure of two isoforms of the vitellogenesis inhibiting hormone from the Iobster Homarus americanus. Neuropeptides 20, 25–32. doi: 10.1016/0143-4179(91)90036-i
Tan, M. H., Gan, H. M., Lee, Y. P., Grandjean, F., Croft, L. J., and Austin, C. M. (2020). A giant genome for a giant crayfish (Cherax quadricarinatus) with insights into cox1 pseudogenes in decapod genomes. Front. Genet. 11:201. doi: 10.3389/fgene.2020.00201
Tang, B., Wang, Z., Liu, Q., Wang, Z., Ren, Y., Guo, H., et al. (2021). Chromosome-level genome assembly of Paralithodes platypus provides insights into evolution and adaptation of king crabs. Mol. Ecol. Resour. 21, 511–525. doi: 10.1111/1755-0998.13266
Tang, B., Zhang, D., Li, H., Jiang, S., Zhang, H., Xuan, F., et al. (2020). Chromosome-level genome assembly reveals the unique genome evolution of the swimming crab (Portunus trituberculatus). GigaScience 9:giz161. doi: 10.1093/gigascience/giz161
Tong, R. X., Pan, L. Q., Zhang, X., and Li, Y. F. (2021). Neuroendocrine-immune regulation mechanism in crustaceans: a review. Rev. Aquac. 1–21. doi: 10.1111/raq.12603
Toullec, J. Y., Corre, E., Mandon, P., Gonzalez-Aravena, M., Ollivaux, C., and Lee, C. Y. (2017). Characterization of the neuropeptidome of a Southern Ocean decapod, the Antarctic shrimp Chorismus antarcticus: focusing on a new decapod ITP-like peptide belonging to the CHH peptide family. Gen. Comp. Endocrinol. 252, 60–78. doi: 10.1016/j.ygcen.2017.07.015
Tran, N. M., Mykles, D. L., Elizur, A., and Ventura, T. (2019). Characterization of G-protein coupled receptors from the blackback land crab Gecarcinus lateralis Y organ transcriptome over the molt cycle. BMC Genomics 20:74. doi: 10.1186/s12864-018-5363-9
Tu, S., Xu, R., Wang, M., Xie, X., Bao, C., and Zhu, D. (2021). Identification and characterization of expression profiles of neuropeptides and their GPCRs in the swimming crab, Portunus trituberculatus. PeerJ 9:e12179. doi: 10.7717/peerj.12179
Uengwetwanit, T., Pootakham, W., Nookaew, I., Sonthirod, C., Angthong, P., Sittikankaew, K., et al. (2021). A chromosome-level assembly of the black tiger shrimp (Penaeus monodon) genome facilitates the identification of growth-associated genes. Mol. Ecol. Resour. 21, 1620–1640. doi: 10.1111/1755-0998.13357
Van Quyen, D., Gan, H. M., Lee, Y. P., Nguyen, D. D., Nguyen, T. H., Tran, X. T., et al. (2020). Improved genomic resources for the black tiger prawn (Penaeus monodon). Mar. Genomics. 52:100751. doi: 10.1016/j.margen.2020.100751
Veenstra, J. A. (2015). The power of next-generation sequencing as illustrated by the neuropeptidome of the crayfish Procambarus clarkii. Gen. Comp. Endocrinol. 224, 84–95. doi: 10.1016/j.ygcen.2015.06.013
Veenstra, J. A. (2016). Similarities between decapod and insect neuropeptidomes. PeerJ 4:e2043. doi: 10.7717/peerj.2043
Wainwright, G., Webster, S. G., Wilkinson, M. C., Chung, J. S., and Rees, H. H. (1996). Structure and significance of mandibular organ-inhibiting hormone in the crab, Cancer pagurus: involvement in multihormonal regulation of growth and reproduction. J. Biol. Chem. 271, 12749–12754. doi: 10.1074/jbc.271.22.12749
Wang, C., Zhu, D., Qi, Y., Hu, Z., Xie, X., and Shen, J. (2013). Molt-inhibiting hormone levels and ecdysteroid titer during a molt cycle of Portunus trituberculatus. Acta Hydrobiol. Sin. 37, 22–28. doi: 10.7541/2013.22
Webster, S. G. (1991). Amino acid sequence of putative moult-inhibiting hormone from the crab Carcinus maenas. Proc. R. Soc. Lond. Ser. B. 244, 247–252. doi: 10.1098/rspb.1991.0078
Webster, S. G. (1993). High-affinity binding of putative moult-inhibiting hormone (MIH) and crustacean hyperglycaemic hormone (CHH) to membrane-bound receptors on the Y-organ of the shore crab Carcinus maenus. Proc. R. Soc. London, Ser. B. 251, 53–59. doi: 10.1098/rspb.1993.0008
Webster, S. G. (2015a). “Endocrinology of metabolism and water balance: crustacean hyperglycemic hormone,” in Physiology, eds E. S. Chang and M. Thiel (Oxford: Oxford Press), 36–67.
Webster, S. G. (2015b). “Endocrinology of molting,” in Physiology, eds E. S. Chang and M. Thiel (Oxford: Oxford Press), 1–35.
Webster, S. G., Keller, R., and Dircksen, H. (2012). The CHH-superfamily of multifunctional peptide hormones controlling crustacean metabolism, osmoregulation, moulting, and reproduction. Gen. Comp. Endocrinol. 175, 217–233. doi: 10.1016/j.ygcen.2011.11.035
Zhang, X., Yuan, J., Sun, Y., Li, S., Gao, Y., Yu, Y., et al. (2019). Penaeid shrimp genome provides insights into benthic adaptation and frequent molting. Nat. Commun. 10, 1–14. doi: 10.1038/s41467-018-08197-4
Zhang, Y., Sun, Y., Liu, Y., Geng, X., Wang, X., Wang, Y., et al. (2011). Molt-inhibiting hormone from Chinese mitten crab (Eriocheir sinensis): cloning, tissue expression and effects of recombinant peptide on ecdysteroid secretion of YOs. Gen. Comp. Endocrinol. 173, 467–474. doi: 10.1016/j.ygcen.2011.07.010
Zhao, M., Wang, W., Zhang, F., Ma, C., Liu, Z., Yang, M. H., et al. (2021). A chromosome-level genome of the mud crab (Scylla paramamosain estampador) provides insights into the evolution of chemical and light perception in this crustacean. Mol. Ecol. Resour. 21, 1299–1317. doi: 10.1111/1755-0998.13332
Zmora, N., Sagi, A., Zohar, Y., and Chung, J. S. (2009). Molt-inhibiting hormone stimulates vitellogenesis at advanced ovarian developmental stages in the female blue crab, Callinectes sapidus 2: novel specific binding sites in hepatopancreas and cAMP as a second messenger. Saline Syst. 5, 1–11. doi: 10.1186/1746-1448-5-7
Keywords: CHHs, GPCRs, genome, transcriptome, bioinformatics, decapod
Citation: Yang Y, Xu Y, Zhang P, Cui Z and Bao C (2021) Comparative Genomic and Transcriptomic Analyses of CHHs and Their Putative Receptors in Scylla paramamosain, Portunus trituberculatus, and Eriocheir sinensis. Front. Mar. Sci. 8:787007. doi: 10.3389/fmars.2021.787007
Received: 30 September 2021; Accepted: 01 November 2021;
Published: 22 November 2021.
Edited by:
Haihui Ye, Jimei University, ChinaReviewed by:
Simon G. Webster, Bangor University, United KingdomDonald Mykles, Colorado State University, United States
Copyright © 2021 Yang, Xu, Zhang, Cui and Bao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chenchang Bao, YmFvY2hlbmNoYW5nQG5idS5lZHUuY24=
 Yanan Yang
Yanan Yang Yuanfeng Xu
Yuanfeng Xu Chenchang Bao
Chenchang Bao