
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 30 November 2021
Sec. Aquatic Physiology
Volume 8 - 2021 | https://doi.org/10.3389/fmars.2021.784845
This article is part of the Research Topic Omics Approaches in Aquatic Nutritional Physiology View all 7 articles
A correction has been applied to this article in:
Corrigendum: Liver Transcriptomic Analysis of the Effects of Dietary Fish Oil Revealed a Regulated Expression Pattern of Genes in Adult Female Spotted Scat (Scatophagus argus)
 Tuo Wang1
Tuo Wang1 Dongneng Jiang1
Dongneng Jiang1 Hongjuan Shi1
Hongjuan Shi1 Umar Farouk Mustapha1
Umar Farouk Mustapha1 Siping Deng1
Siping Deng1 Zhilong Liu1
Zhilong Liu1 Wanxin Li1
Wanxin Li1 Huapu Chen1
Huapu Chen1 Chunhua Zhu1,2
Chunhua Zhu1,2 Guangli Li1*
Guangli Li1*
Despite the significance of n-3 long-chain polyunsaturated fatty acid (n-3 LC-PUFA) in fish oil in promoting the maturation of female broodstocks, the detailed mechanism of the effect on the expression of hepatic reproduction-related genes is still unclear. In this study, transcriptome sequencing was used to analyze the effect of the higher dietary n-3 LC-PUFA level on gene expression in the liver of adult females spotted scat. Two-year-old female spotted scat (average weight, 242.83 ± 50.90 g) were fed with diets containing 8% fish oil (FO) or 8% soybean oil (SO) for 40 days. The fatty acid profile in the serum, liver, and ovary was analyzed, and high proportions of n-3 LC-PUFA were observed in the FO group. The final average fish body weight and gonadosomatic index were similar between the FO and SO groups. The serum vitellogenin (Vtg) and hepatosomatic index (HSI) of the FO group were significantly higher and lower than that of the SO group, respectively. Comparatively, the liver transcriptome analysis showed 497 upregulated and 267 downregulated genes in the FO group. Among them, the expression levels of three estrogen-regulated genes (i.e., Vtga, Vtgb, and Zp4) were significantly higher in the FO than in the SO group. This expression pattern could be related to the upregulation of Hsd17b7 (the key gene for the synthesis of liver steroid hormone) and the downregulation of the Hsp90 (the estrogen receptor chaperone). The expression levels of Foxo1a and Lep, which are involved in the lipid metabolism, decreased significantly in the FO group, which may be related to the lower level of HSI in the FO group. The genes related to liver LC-PUFA absorption and transport, Fabp2 and Mfsd2ab, were significantly upregulated in the FO group, indicating that fish actively adapt to high-fish-oil diets. In brief, high-fish-oil diets can influence the expression of genes related to liver n-3 LC-PUFA metabolism and reproduction, inhibit the accumulation of liver fat, and promote the liver health and gonad development. This study will contribute to clarifying the mechanism of dietary n-3 LC-PUFA on promoting reproductive development in teleost fish.
The effects of essential fatty acids in feed on the reproduction of broodstocks are among the main research areas of artificial fish breeding in recent years. Long-chain polyunsaturated fatty acids (LC-PUFA) supplied in fish oil, including docosahexaenoic acid (DHA, 22:6n-3), eicosapentaenoic acid (EPA, 20:5n-3), and arachidonic acid (ARA, 20:4n-6), are essential nutrients for the normal growth and development of fish and can play key roles in the lipid metabolism, synthesis of sex steroid hormones, and reproductive regulation (Zhou et al., 2011; Glencross et al., 2015; Xu et al., 2017). For example, in yellowfin sea bream (Acanthopagrus latus) and flame angelfish (Centropyge loriculus), the relative fecundity, hatching rate, and larval survival rate were increased by the supplementing higher level of n-3 LC-PUFA in diet (Zakeri et al., 2011; Callan et al., 2012). When Japanese flounder (Paralichthys olivaceus) were fed with diets containing three different levels of n-3 LC-PUFA (i.e., 2.1, 4.8, and 6.2%), egg production increased in 6.2% n-3 LC-PUFA group. At the same time, the hatching rate and percentage of normal larvae were higher in 2.1% n-3 LC-PUFA group, indicating that excessive n-3 LC-PUFA in the diet would reduce the egg quality (Furuita et al., 2002). Consistently, in Plectorhynchus cinctus broodstock fed with four different levels of n-3 LC-PUFA, the moderate level of n-3 LC-PUFA improved the reproductive performance. In comparison, too high or too low n-3 LC-PUFA levels caused a reduction in reproductive performance (Li et al., 2004).
The liver is a key metabolic organ that is widely involved in the bioconversion of fatty acid and the regulation of lipid homeostasis in fish (Lazzarotto et al., 2018; Esmaeili et al., 2021). The liver is also the most important active tissue for gonadal development in oviparous vertebrates. In teleost fish, most of the yolk is synthesized and secreted by liver cells in the form of a protein precursor: the vitellogenin (Vtg) (Bemanian et al., 2004). In addition, some zona pellucida (Zp) genes encoding Zp glycoproteins, which constitute the protective coat surrounding oocytes, were also expressed in the teleost fish liver (Wu et al., 2018). Hence, the liver is a vital protein synthesis “factory” for the normal ovary development in fish. Previous studies have shown that the addition of fish oil to increase the dietary n-3 LC-PUFA level can promote the expression of Vtg in the liver of female fish, including the spotted scat (Scatophagus argus) (Zhang et al., 2013; Peng et al., 2015). However, the regulatory mechanism of the reproductive-related gene caused by the dietary n-3 LC-PUFA level in the liver of broodstock is still unclear, even though very pertinent in breeding programs.
In artificial breeding, the primary step is to get broodstocks with healthy oocytes and sperm in females and males, respectively. Also, the usual way to obtain mature fish with a better reproductive ability is by nutrient supplementation in the fish diet. Nutrigenomics utilizes high-throughput genomics approaches to unravel how dietary nutrients regulate gene expression at the mRNA/protein level and ultimately modulate cellular and organism metabolism. The most widely applied technique of nutrigenomics is the transcriptome analysis via next-generation sequencing (gene expression patterns at the mRNA level), also known as RNA-seq (Afman and Müller, 2012). Studies of the transcriptional effects of LC-PUFA on liver gene expression in fish have been carried out in postsmolts Atlantic salmon (Salmo salar) (Glencross et al., 2015; Xue et al., 2020) and 2-month-old golden pompano (Trachinotus ovatus) (Lei et al., 2019). Nonetheless, evaluating the effects of n-3 LC-PUFA on livers of female broodstocks using the transcriptional method is lacking. This study focused on the hepatic transcriptome analysis to comprehensively understand the detailed molecular mechanisms of the effects of fish oil on reproductive-related genes in the liver of adult female spotted scat.
Spotted scat is an important food fish mainly produced in the South and South-East Asian countries. Due to its good nutrient quality, high protein content, and delicious taste, the fish has gained high demand and become an important aquaculture species. It is also a popular aquaria fish due to its colorful appearance, strong adaptability, and easiness to be cultured indoors (Gupta, 2016). For these reasons, studies focusing on its reproduction biology are on the rise, including reproduction endocrinology (Jiang et al., 2017; Zhang et al., 2018; Mandal et al., 2020), sex determination and differentiation (Mustapha et al., 2018, 2021; He et al., 2019), and artificial breeding (Cai et al., 2010; Gandhi et al., 2014; Mandal et al., 2021). Its artificial breeding is successful in several countries, including China (Cai et al., 2010) and India (Mandal et al., 2021). To obtain matured spotted scat broodstock, appropriate nutritional factors should be maintained to enhance gonad development and induce reproduction (Cai et al., 2010). Although fish oil provides adequate nutrition for broodstock, the regulatory mechanism is unclear. Also, there is a delay in female gonad maturation compared with that of male (Gandhi et al., 2014). Therefore, it is important to advance studies on the effects of fish oil on the expressions of liver genes to gain insight into gene regulatory mechanisms in female spotted scat. The findings will provide essential knowledge for further functional studies on fish oil in promoting teleost fish reproduction.
This experiment was undertaken in accordance with the guidance, care, and use of laboratory animals in China with approval from the Committee on the Ethics of Animal Experiments of Guangdong Ocean University.
Two-year-old female spotted scats were bought from the Zhuhai HengDa Cultivation Base (Guangdong, China). The sex ratio of spotted scats is approximately 1:1 (Mustapha et al., 2021). The female spotted scats were empirically selected from the mixed-sex fish by the sexual morphological difference in the head, anal fin, body color, body weight, and genital pore (Barry and Fast, 1992). The fish were transferred to the Donghai Island experimental base, Guangdong province, China. The fish were reared in a concrete pond (12 m × 5 m × 2 m) and adapted to the rearing condition for 2 weeks. Aeration was supplied to the tank through air stones, and the average water temperature was 31.1 ± 0.8°C. The salinity was 8‰, while pH and dissolved oxygen ranged from 7.5–8.5 and 6.0–7.0 mg/L, respectively. Fish were fed 2.5% of their body weight with commercial fish pellets consisting of 43% crude protein and 12% crude lipid. Fish were fed two times a day at 06:00 and 18:00 h throughout the adaptation period. The pond water was not recirculated. Approximately, 20% of the water in the pond was drained off and replaced with fresh water once in 3 days.
After the adaptation period, 100 females (average initial weights of 242.83 ± 50.90 g, and length of 19.48 ± 1.13 cm) were randomly distributed into two treatment groups (50 fish in each group) containing different levels of fish oil (0% and 8%; ingredients are listed in Table 1). The diet containing 8% fish oil was marked as the FO group, while the diet of the control group containing 8% soybean oil marked the SO group. Each treatment group was duplicated and contained 25 fish in each net cage (5 m × 3.5 m × 1.8 m). Two independent concrete ponds (12 m × 5 m × 2 m) were used; one concrete pond had two net cages for both FO and SO treatments. The fish were fed with the two diets for 40 days before sampling. The culturing condition was consistent with the adaptation stage.
At the termination of the experiment, the spotted scats were sampled after 24 h starvation. Five fish per cage were sacrificed for sampling. The final number of fish, body weight, body length, gut weight, liver weight, and gonad weight were taken individually. The survival rate (SR), condition factor (CF), viscerosomatic index (VSI), hepatosomatic index (HSI), and gonadosomatic index (GSI) were calculated using the following formulas:
SR (%) = final number of fish/initial number of fish × 100;
CF = final body weight (g)/final body length (cm) 3 × 100;
VSI (%) = final viscera weight (g)/final body weight (g) × 100;
HIS (%) = final liver weight (g)/final body weight (g) × 100;
GSI (%) = final gonad weight (g)/final body weight (g) × 100.
The liver and ovary samples of the fish were excised and quickly frozen in liquid nitrogen. Blood samples were collected from the caudal vein of the fish. The blood samples were stored at 4°C overnight and centrifuged (at 3,000 rpm for 10 min at 4°C), and the upper layer serums were collected. All samples were subsequently stored at −80°C for further analysis.
The total lipid from the ovary and liver (n = 6 per group) and feed were extracted using diethyl ether after acid hydrolysis, whereas lipid from the serum (n = 6 per group) was extracted with chloroform according to the procedure of the National Standards of the People’s Republic of China (5009.168—2016). Extracted lipid was saponified with NaOH-methanol. Their constitutive fatty acids were methylated through the action of boron trifluoride-methanol, separated using Agilent 7890B gas chromatograph (Agilent Technologies, Santa Clara, CA, United States) on a capillary column (SPTM-2560; 100 m × 0.25 mm × 0.25 mm), and quantified using glyceryl triundecanoate as an internal standard.
To evaluate the effects of fish oil on the genes expressed in the liver, the transcriptome analysis was carried out. First, total RNA (n = 6 per group) from the two groups was extracted from fish liver using RNeasy® Mini Kit reagents according to the instructions of the manufacturer (Qiagen GMBH, Hilden, Germany). The RNA quality was determined and quantified using an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, United States) and checked using the RNase-free agarose gel electrophoresis. The mRNA was enriched by oligo (dT) beads; the mRNA was interrupted. The first strand of cDNA was synthesized in M-MuLV reverse-transcriptase system, and the second strand was synthesized from dNTPs in DNA polymerase I system. Then, the cDNA fragments were ligated to Illumina sequencing adapters. The ligation products were size selected using agarose gel electrophoresis, PCR amplified, and sequenced using novaseq 6000 by Guangzhou Genedenovo Biotechnology Co. (Guangzhou, China). All clean libraries of sequencing data were submitted to the NCBI Sequence Read Archive (SRA) database (Accession No: PRJNA765892).
To ensure that high-quality data are achieved, the original data were filtered before analyzing. The raw reads were filtered using fastp (version 0.18.0). The adapters read with more than 10% unknown nucleotides and over 50% low-quality nucleotides (Q-value ≤ 20) were removed. The ribosome RNA (rRNA) reads were removed with the alignment tool Bowtie2 (version 2.2.8). Then, these clean reads were mapped to the spotted scat reference genome (Huang et al., 2021) with HISAT2.2.4. Genes with unknown transcripts or in intergenic regions were defined as novel genes. The fragments per kilobase of transcript per million mapped reads (FPKM) method was used to estimate the gene expression levels. Differentially expressed genes (DEGs) in two groups were identified using the EdgeR software package. In comparison, to avoid some important genes being filtered out, genes with | log2 (fold change)| > 1 and P < 0.05 were identified as significant DEGs. DEGs were annotated with the enrichment analysis of Gene Ontology (GO) functions and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. The KEGG pathway with P < 0.05 was regarded as a significant enrichment pathway.
To validate the gene expression profiles from the RNA-seq analysis, 24 differentially expressed genes related to fat metabolism and ovarian development were selected for the real-time PCR analysis. Real-time PCR was performed using the PerfectStartTM Green qPCR SuperMix (TRAN, China). The PCR reaction was carried out in a total volume of 20 μl with 10 μl of 2 × PerfectStartTM Green qPCR SuperMix, 0.4 μl of each primer, and 2 μl cDNA. The thermal cycling program included an initial denaturation at 94°C for 30 s, followed by 40 cycles of 94°C for 5 s, 60°C for 15 s, and 72°C for 10 s. The reference gene, β-actin, was used to normalize the expression values. The 2–ΔΔCt method was used to calculate the relative expression of the target genes. Each group contains 10 independent samples, and each sample was detected in triplicate. Primer sequences used for real-time PCR are listed in Supplementary Table 1.
The content of serum Vtg was detected using Grouper Vitellogenin ELISA Kit according to the instructions of the manufacturer (Cusabio, Wuhan, China). The serum samples from 10 fish in each group were analyzed.
All data are expressed as means ± standard error (SE), and an independent sample t-test was used with the significance level at P < 0.05. The statistical analysis of the data was performed using the SPSS 17.0 software (SPSS Inc., Chicago, IL, United States).
The DHA, EPA, and ARA contents in FO diet are higher than that of the SO diet. On the contrary, the oleic acid (C18:1n-9) and linoleic acid (C18:2n-6) contents in FO diet are lower than that of the SO diet (Table 2). The differences are mainly attributed to their different oil sources.
Table 3 shows fatty acid composition in different tissues of female spotted scat. In the ovary, fish oil treatment significantly increased (P < 0.05) the EPA concentration. Also, in the liver, fish oil treatment significantly decreased (P < 0.05) the C18:3n-6 concentration and increased the EPA concentration. In the serum, fish oil treatment significantly increased (P < 0.05) C14:0, C15:0, C16:1n-7, C17:0, C22:1n-9, EPA, C24:0, and DHA concentration, while fish oil treatment significantly decreased (P < 0.05) linoleic acid, C18:3n-6, C20:2-6, and C20:3n-6 concentration.
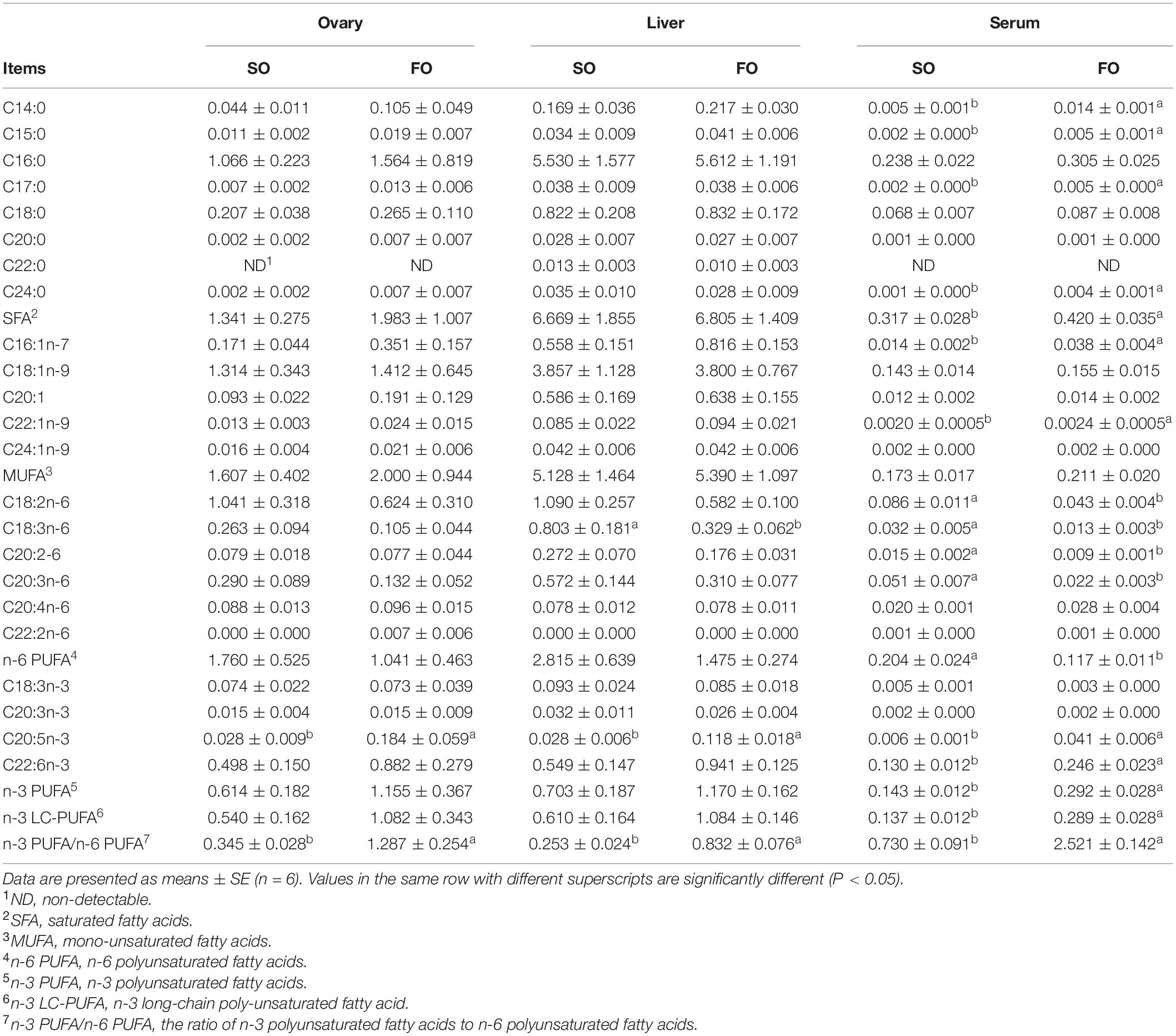
Table 3. Fatty acid composition in different tissues of the female spotted scat fed with different oil sources.
At the end of the experiment, no fish died in both FO and SO groups. The final body weight, body length, CF, GSI, and VSI of the SO and FO fish are similar (P > 0.05, Table 4). The HSI of fish in FO group was significantly lower than that of the SO group (P < 0.05).
Twelve cDNA libraries were constructed from livers from the SO and FO groups. In this experiment, a total of 534,586,540 (i.e., 289,243,632 in the SO group and 245,342,908 in the FO group) clean reads were obtained from the transcriptome sequencing. The percentages of Q30 bases were more than 93% for all the samples, indicating a high-quality sequence (Table 5). The results showed a total mapped reads of 497,792,649, and the percentage of mapped reads for each library ranged from 91.21 to 94.36% (Table 5). A total of 20,574 genes were detected, including 20,238 known genes and 336 novel genes, accounting for 83.44% of the reference genomes. Supplementary Table 2 shows the number of genes detected in each sample.
In this study, a total of 764 genes were annotated to be DEGs (P < 0.05, | log2(fold change)| > 1). Compared with the SO group, 497 genes were upregulated in the FO group, while 267 genes were downregulated in the FO group (Figure 1).
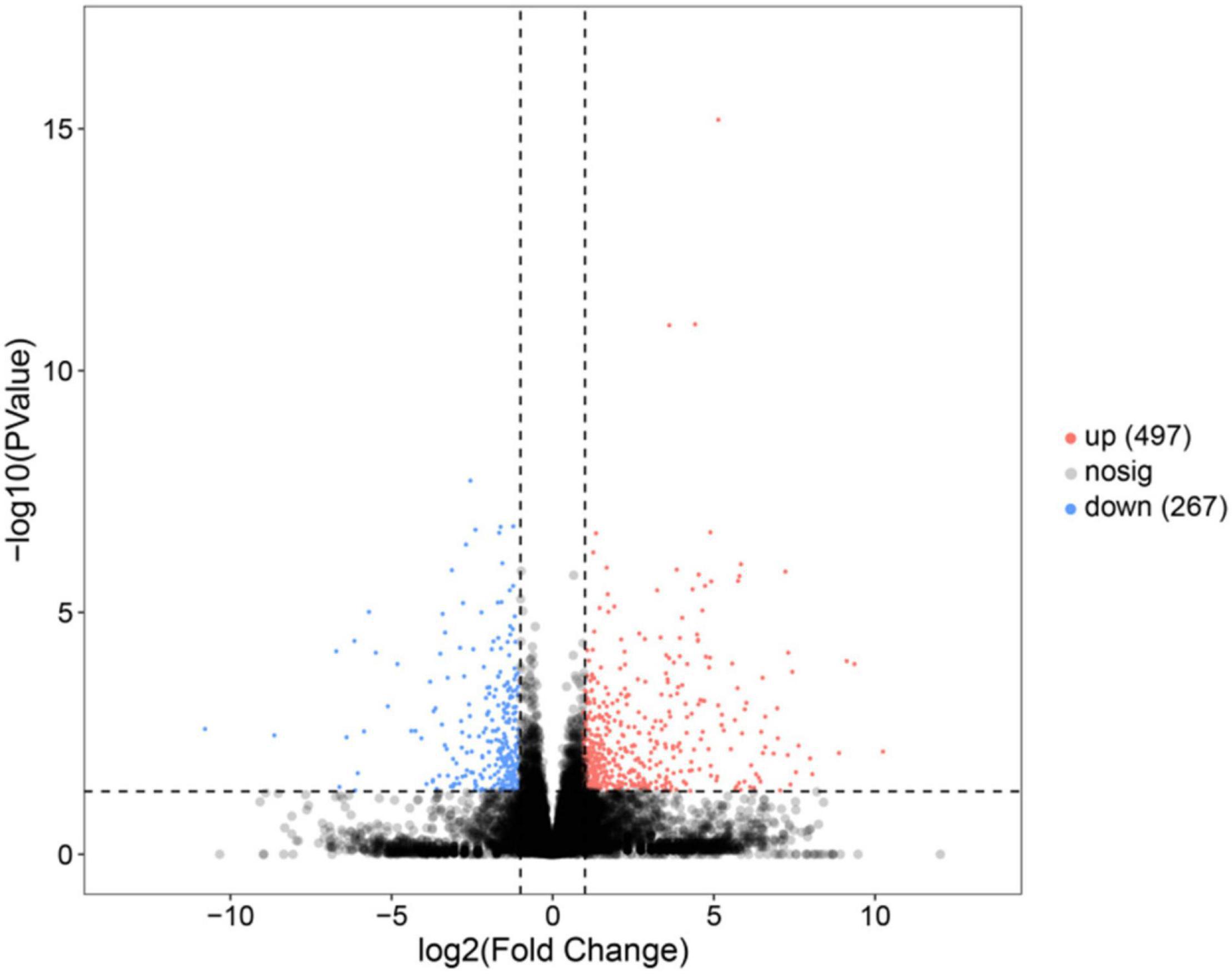
Figure 1. Volcano plot of the differentially expressed genes (DEGs) in the liver of female spotted scat fed with different oil sources.
In this study, GO and KEGG enrichment analyses were carried out to explore the biological functions of the DEGs. The DEGs were annotated into GO categories of the cellular component, biological process, and molecular function (Figure 2). In the cellular component categories, organelle (61), cell (74), cell part (74), membrane part (45), and membrane (50) were the most enriched GO terms. In the biological process categories, cellular process (144), single-organism process (128), metabolic process (124), biological regulation (91), and response to stimulus (60) were the most enriched GO terms. In the molecular function categories, binding (140), catalytic activity (89), molecular transducer activity (22), and nucleic acid binding transcription factor activity (17) were the most enriched GO terms.
The top 20 biological pathways revealed in KEGG analysis are shown in Figure 3. The DEGs are mainly classified into cellular processes, environmental information processing, metabolism, organismal systems, and human diseases. Key genes for steroid biosynthesis (i.e., Lss, Hsd17b7, Tm7sf2, and Nsdhl, etc.), oocyte meiosis (i.e., Ccnb2, Ppp2ca, etc.), cell cycle (i.e., Cdc14ab, Myc, Cdkn1a, and Gadd45b, etc.), FoxO signaling pathway (i.e., Pck1, Foxo1a, Sgk3, Bnip3, and Bcl6, etc.), MAPK signaling pathway (i.e., Dusp8, Jund, Hsp70, Epha2, Jun, Fos, Relb, Mapk8ip1, and Fgfr1a, etc.), PPAR signaling pathway [i.e., Acsbg2, Stearoyl-CoA desaturase 1b (Scd1b), and Fabp2, etc.], phagosome (i.e., Calr, Mr1, etc.), as well as additional crucial genes that play roles in amino sugar and nucleotide sugar metabolism (i.e., Cmah, Chia, etc.) were identified. In addition, DEGs fell into the categories of the interaction of cytokine-cytokine receptor (i.e., tumor necrosis factor receptor superfamily member 6B-like isoform X2, Ackr3, etc.), Herpes simplex infection [i.e., Socs3, aryl hydrocarbon receptor nuclear translocator-like protein 2 (Arntl2), etc.], and progesterone-mediated oocyte maturation (i.e., Hsp90a.1, etc.).
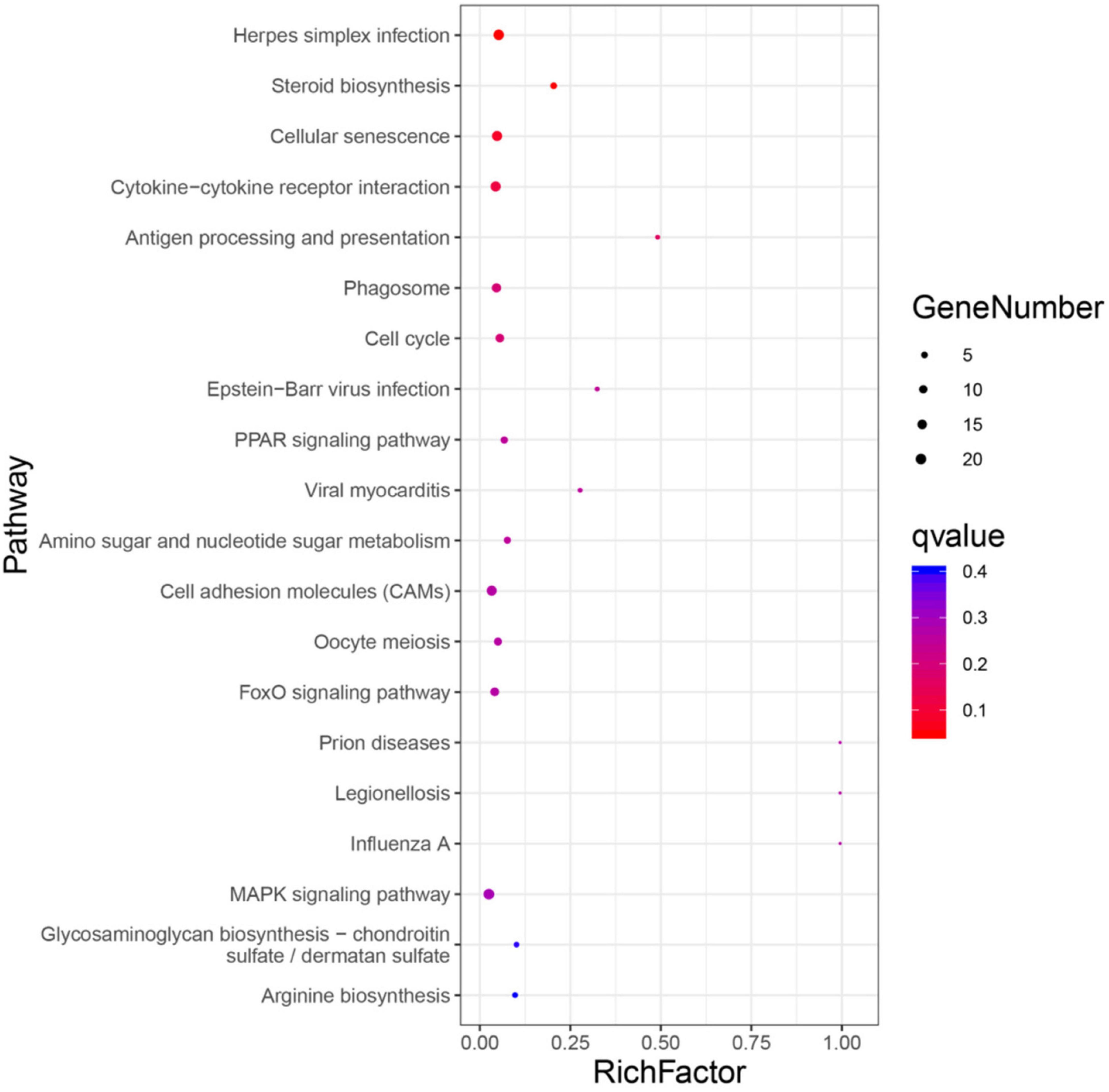
Figure 3. Top 20 pathway enrichments. The x-axis indicates the ratio of the number of genes in the pathway of the DEGs and all genes. The y-axis indicates the pathway.
Twenty-four genes related to ovarian development and fat metabolism from RNA-seq were verified using real-time PCR (Figure 4). The results showed that the expression levels of the selected genes were consistent with RNA-seq, suggesting that the RNA-seq results are reliable.
The mRNA expression levels of Vtga and Vtgb in livers of the FO group are higher than that of the SO group. The Vtg protein level in the serum was detected. The results showed that the serum Vtg level in the fish from the FO group was significantly higher than that of the SO group (P < 0.05; Figure 5). The results at the mRNA and protein levels are consistent.
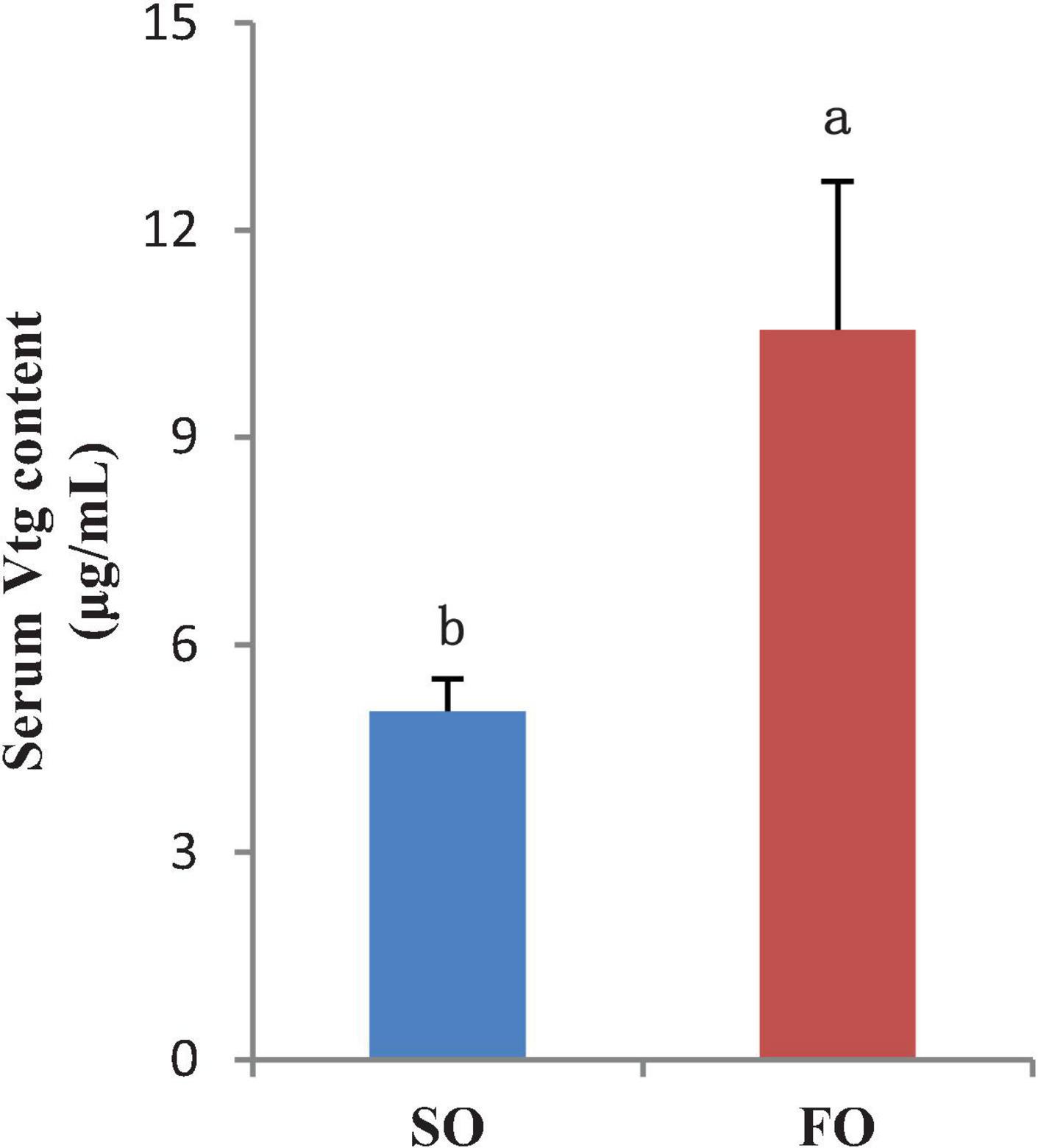
Figure 5. Serum vitellogenin (Vtg) content of the female spotted scat fed with different oil sources (μg/ml, means ± SE, n = 10). Different letters denote statistically significant difference (P < 0.05).
The growth of the fish was not affected by the oil source during the 40-day experimental stage. In sharpsnout seabream (Diplodus puntazzo), replacing fish oil with vegetable oil did not affect growth after 3 months of feeding (Piedecausa et al., 2007). However, in cobia (Rachycentron canadum), the proportional replacement of fish oil with soybean oil gradually increased (i.e., 0% SO, 33% SO, 67% SO, and 100% SO) and resulted in a gradual decrease in weight gain (Trushenski et al., 2011). The discrepancy could be related to the differences in fatty acid requirement among species (Xu et al., 2015). The current study suggests that spotted scat can be fed with a diet containing lower fish oil for normal growth. That is probably because spotted scat is omnivorous fish species. Of course, we could not conclude that the spotted scat does not need LC-PUPA for normal growth, as residual fish oil in fish meal could supply the LC-PUPA in the SO group. Decreased HSI in the FO group reveals that a high level of dietary n-3 PUFAs significantly reduced fat accumulation in the liver. It is well known that adult zebrafish suffer from hepatic adipose infiltration, which might influence reproduction and offspring (Virote et al., 2020). The liver with less fat tissue should be healthier, and the healthy liver might have a stronger ability to synthesize the proteins and lipids, such as the Vtg and Zp proteins required for gonadal development. In female spotted scat, the higher GSI values were observed in the reproduction season, indicating the maturing of the ovary (Gandhi et al., 2014). Consistently, the GSI in the FO group is relatively higher than that of SO group, although there was no statistical difference (P = 0.306). The possible reason for the large standard error between groups is the genetic background of the fish affected the regulation of lipid metabolism (Morais et al., 2011). These results suggest that a high level of dietary n-3 LC-PUFA might be beneficial to a healthy liver and promote ovarian development in female spotted scat.
The liver transcriptome analysis was carried out to confirm whether the high level of LC-PUPA would benefit the reproduction via influencing the liver genes. Vitellogenesis- and zonagenesis-related genes are induced by the estrogen in the liver to synthesize Vtg and zona radiata proteins (Zrp), respectively. Vtg and Zrp are secreted and transported from blood to the ovary and are taken up by the maturing oocytes (Arukwe and Goksøyr, 2003; Levi et al., 2009). Vtga, Vtgb, and Vtgc are all expressed increasingly during the ovary maturation from phase II to phase IV in the female liver of spotted scat (Cui et al., 2017). In this study, both transcriptome and real-time PCR data showed that Vtga and Vtgb are highly expressed in the FO group. Consistently, the serum Vtg protein is also higher in the FO group than that of the SO group. Zp4 is one of the genes controlling the making of Zp (in mammals) or vitelline envelope (in fish, amphibians, and birds), a glycoprotein layer surrounding the oocytes (Laine et al., 2019). In addition, Zp4 expression is significantly upregulated in the livers of FO fish. Taken together, the fish oil might promote ovary development by upregulating genes critical for vitellogenesis (i.e., Vtga and Vtgb) and zonagenesis (i.e., Zp4) in spotted scat.
The estrogen signaling pathway is mediated by the nuclear receptors. Three estrogen receptors, erα, erβ1, and erβ2, were characterized in many fish species, including spotted scat (Cui et al., 2017). Although, the three estrogen receptors are expressed in the female livers, their expressions were not affected by the high level of LC-PUPA in the presents study. In contrast, the liver estrogen level might be increased by the upregulation of the Hsd17b7. Hsd17b7 could transform estrone to estradiol in vitro (Ohnesorg et al., 2006). Similarly, fish oils upregulate 17β-hsd gene expression in the testicular tissues of rats (Mohammad et al., 2015). The regulatory mechanism of fish oil on Hsd17b7 expression remains to be elucidated.
Hsp90 is a ubiquitously expressed molecular chaperone of estrogen receptors and is involved in the signal transduction process of estrogen receptors (Chang et al., 2014). Herein, the Hsp90 expression was downregulated in the FO group. Whether less Hsp90 would either promote or inhibit the estrogen signaling pathway is still unclear. Considering the upregulation of Vtgs and Zp4 expressions in the FO group, the results suggested that less Hsp90 might enhance the estrogen signaling pathway. The possible mechanism is that decreasing Hsp90 would increase the number of free estrogen receptors. Those free receptors would combine with estrogen to be translocated into the nuclear to take action. The aryl hydrocarbon nuclear translocator (Arnt) has been found to be a coactivator of erα- and erβ-dependent transcription (Brunnberg et al., 2003). The Arntl2 gene has been considered a candidate gene in the molecular regulation of reproduction seasonality (Zhao W. et al., 2020). However, its expression was decreased in the FO group. So, the n-3 LC-PUPA might have a diverse influence on the expression of the genes involved in the network of the estrogen signaling pathway. The total output of the estrogen signaling pathway in the female liver should be enhanced in the group that contains higher level n-3 LC-PUPA.
The forkhead transcription factor Foxo1 is a critical regulator of hepatic lipid and glucose metabolism, and it functions in hepatic lipid homeostasis (Shin et al., 2012). The DHA supplemented diets reduced the expression of Foxo1 in the liver of pigs after 30-day feeding experiment (Chen et al., 2012). In contrast, EPA was more effective in reducing Foxo1 expression than DHA in rats after an 8-week feeding experiment (Hong et al., 2019). DHA or EPA alone or together could reduce the expression of Foxo1 in 3T3-L1 adipocytes in vitro (Martins et al., 2020). In mice, gain and loss of function proved that Foxo1 could promote the insulin sensitivity and lipid synthesis (Matsumoto et al., 2006). Foxo1a was highly expressed in insulin-responsive tissues (e.g., adipose tissue, liver, and muscle) in grass carp (Ctenopharyngodon idella) (Sun et al., 2017). In contrast, the addition of bovine insulin in vitro stimulates Vtg expression in hepatopancreatic explants from the mud crab (Scylla paramamosain) (Huang et al., 2017). The decreasing of the HIS in the FO group could be explained by the DHA/EPA in the fish oil decreasing the Foxo1a expression and reducing the lipid synthesis. Whether the increase of Vtg in FO group is affected by Foxo1-mediated insulin needs further verification.
Leptin (Lep) is a protein hormone that regulates body weight and energy homeostasis (Paolucci et al., 2020). In fish, the liver seems to be the primary organ for Lep synthesis (Frøiland et al., 2010). In large yellow crookeder (Larimichthys crocea), Lep levels in blood were significantly lower in the FO group than in palm oil (Wang et al., 2019). Similarly, the liver Lep mRNA in the fish of FO group was significantly lower than that of SO group in the present study. The high level of LC-PUPA might decrease the Lep expression level in the liver in fish. However, there was a contrary relationship between liver lipid content and liver Lep mRNA expression in Arctic charr (Salvelinus alpinus) (Frøiland et al., 2010). The upregulation of the Lep mRNA expression in the SO group containing high HIS might be a negative feedback mechanism to regulate the liver lipid. The E2-induced Vtg mRNA transcript was influenced negatively by recombinant Lep in the rainbow trout (Oncorhynchus mykiss) in vitro (Paolucci et al., 2020). Thereafter, the induction of the Vtgs expression by higher n-3 LC-PUPA level might also depend on the decreasing of Lep.
The Ppar-related gene expression could be regulated by dietary fat that affects hepatic lipid storage and maintenance of metabolic health (Soni et al., 2015). The fatty acid-binding protein (Fabp) is the DEG enriched by the KEGG analysis in the PPAR signaling pathway (ko03320). The long-chain fatty acids are assisted by some fatty acid transporters to move through the cell membrane and are then transported by Fabps to the corresponding metabolic target sites (Lei et al., 2019). A Fabp2–/– zebrafish decreased intestinal LC-PUPA and increased hepatic lipid accumulation compared with wild-type zebrafish. At the same time, transgenic overexpression Fabp2 significantly increased intestinal LC-PUPA (Zhao Y. et al., 2020). These results suggested that Fabp2 also promoted LC-PUPA absorption in fish. Consistently, Fabp2 expression was upregulated in the FO group, which contains high LC-PUFA than the SO group in the present study. In golden pompano (Trachinotus ovatus), the liver expressions of the Fabp -1, -4, and -6 were higher in the 2.1% LC-PUFA than that of 1.0% LC-PUFA feed group, suggesting that Fabps may be involved in LC-PUFA metabolism in this species (Lei et al., 2019). Taken together, Fabp gene family members may be essential for the LC-PUFA uptake and deposition in fish, while different subtypes of Fabp may be used for gene duplication and functional diversification in different species. In addition, the major facilitator superfamily domain-containing 2a (Mfsd2a) is confirmed as a transporter of DHA as lysophosphatidylcholine-DHA and is highly expressed in the liver (Pauter et al., 2017). Consistently, Mfsd2ab is also upregulated in the FO group in spotted scat. Therefore, the n-3 LC-PUFA supplementation affected the genes involved in LC-PUFA metabolism in spotted scat; hence, the regulator mechanism of these genes should be studied in the future.
The dietary supplementation of fish oil will significantly increase the deposition of n-3 LC-PUFA in spotted scat. The fish growth rates were similar between the FO and SO groups, while the HIS of the FO group was significantly lower than that of the SO group. The dietary fish oil supplementation increased the serum Vtg level, which is required for the ovary development. The liver transcription analysis showed that 764 genes were differentially expressed between the FO and SO groups. These data indicate that, the physiological response in the liver is associated with dietary oil. The expression of liver genes analyzed by RNA-seq gives us some new implications on how the dietary n-3 LC-PUFA supplied from the fish oil could promote the reproductive development of marine teleost.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA765892.
The animal study was reviewed and approved by Committee on the Ethics of Animal Experiments of Guangdong Ocean University.
TW and DJ designed and took part in the whole process of the experiment and wrote the draft of this manuscript. HS, SD, HC, and CZ co-conceived the experiment. ZL participated in the experiments. UM and WL revised the draft. GL supervised the experiments. All authors read and approved the final manuscript.
This study was supported by grants from the Key Project of “Blue Granary Science and Technology Innovation” of the Ministry of Science and Technology (2018YFD0901203), the National Natural Science Foundation of China (Nos. 31972775 and 32002367), Natural Science Foundation of Guangdong Province (2018B030311050), Guangdong Basic and Applied Basic Research Foundation (2019A1515012042 and 2021A1515010430), Guangdong Provincial Special Fund for Modern Agriculture Industry Technology Innovation Teams (2019KJ149), and Department of Education of Guangdong Province (2018KTSCX090).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.784845/full#supplementary-material
Afman, L. A., and Müller, M. (2012). Human nutrigenomics of gene regulation by dietary fatty acids. Prog. Lipid Res. 51, 63–70. doi: 10.1016/j.plipres.2011.11.005
Arukwe, A., and Goksøyr, A. (2003). Eggshell and egg yolk proteins in fish: hepatic proteins for the next generation: oogenetic, population, and evolutionary implications of endocrine disruption. Comp. Hepatol. 2:4. doi: 10.1186/1476-5926-2-4
Barry, T. P., and Fast, A. W. (1992). Biology of the Spotted scat (Scatophagus argus) in the Philippines. Asian Fish. Sci. 5, 163–179.
Bemanian, V., Male, R., and Goksøyr, A. (2004). The aryl hydrocarbon receptor-mediated disruption of vitellogenin synthesis in the fish liver: cross-talk between AHR- and ERα-signalling pathways. Comp. Hepatol. 3:2. doi: 10.1186/1476-5926-3-2
Brunnberg, S., Pettersson, K., Rydin, E., Matthews, J., Hanberg, A., and Pongratz, I. (2003). The basic helix–loop–helix–PAS protein ARNT functions as a potent coactivator of estrogen receptor-dependent transcription. Proc. Natl. Acad. Sci. U.S.A. 100, 6517–6522. doi: 10.1073/pnas.1136688100
Cai, Z. P., Wang, Y., Hu, J. W., Zhang, J. B., and Lin, Y. G. (2010). Reproductive biology of scatophagus argus and artificial induction of spawning. J. Trop. Oceanogr. 29, 180–185. doi: 10.3969/j.issn.1009-5470.2010.05.029
Callan, C. K., Laidley, C. W., Kling, L. J., Breen, N. E., and Rhyne, A. L. (2012). The effects of dietary HUFA level on flame angelfish (Centropyge loriculus) spawning, egg quality and early larval characteristics. Aquac. Res. 45, 1176–1186. doi: 10.1111/are.12063
Chang, Z., Lu, M., Kim, S. S., and Park, J. S. (2014). Potential role of HSP90 in mediating the interactions between estrogen receptor (ER) and aryl hydrocarbon receptor (AhR) signaling pathways. Toxicol. Lett. 226, 6–13. doi: 10.1016/j.toxlet.2014.01.032
Chen, Y. J., Chen, C. C., Li, T. K., Wang, P. H., Liu, L. R., Chang, F. Y., et al. (2012). Docosahexaenoic acid suppresses the expression of FoxO and its target genes. J. Nutr. Biochem. 23, 1609–1616. doi: 10.1016/j.jnutbio.2011.11.003
Cui, X. F., Zhao, Y., Chen, H. P., Deng, S. P., Jiang, D. N., Wu, T. L., et al. (2017). Cloning, expression and functional characterization on vitellogenesis of estrogen receptors in Scatophagus argus. Gen. Comp. Endocr. 246, 37–45. doi: 10.1016/j.ygcen.2017.03.002
Esmaeili, M., Carter, C. G., Wilson, R., Walker, S. P., Miller, M. R., Bridle, A., et al. (2021). Proteomic investigation of liver and white muscle in efficient and inefficient Chinook salmon (Oncorhynchus tshawytscha): fatty acid metabolism and protein turnover drive feed efficiency. Aquaculture 542:736855. doi: 10.1016/j.aquaculture.2021.736855
Frøiland, E., Murashita, K., Jørgensen, E. H., and Kurokawa, T. (2010). Leptin and ghrelin in anadromous Arctic charr: cloning and change in expressions during a seasonal feeding cycle. Gen. Comp. Endocr. 165, 136–143. doi: 10.1016/j.ygcen.2009.06.010
Furuita, H., Tanaka, H., Yamamoto, T., Suzuki, N., and Takeuchi, T. (2002). Effects of high levels of n-3 HUFA in broodstock diet on egg quality and egg fatty acid composition of Japanese flounder, Paralichthys olivaceus. Aquaculture 210, 323–333. doi: 10.1016/S0044-8486(01)00855-9
Gandhi, V., Venkatesan, V., and Ramamoorthy, N. (2014). Reproductive biology of the spotted scat Scatophagus argus (Linnaeus, 1766) from Mandapam waters, south-east coast of India. Indian J. Fish. 61, 55–59.
Glencross, B. D., Santis, C. D., Bicskei, B., Taggart, J. B., Bron, J. E., Betancor, M. B., et al. (2015). A comparative analysis of the response of the hepatic transcriptome to dietary docosahexaenoic acid in Atlantic salmon (Salmo salar) post-smolts. BMC Genomics 16:684. doi: 10.1186/s12864-015-1810-z
Gupta, S. (2016). An overview on morphology, biology, and culture of spotted scat Scatophagus argus (Linnaeus 1766). Rev. Fish Sci. Aquac. 24, 203–212. doi: 10.1080/23308249.2015.1119800
He, F. X., Jiang, D. N., Huang, Y. Q., Mustapha, U. F., Yang, W., Cui, X. F., et al. (2019). Comparative transcriptome analysis of male and female gonads reveals sex-biased genes in spotted scat (Scatophagus argus). Fish Physiol. Biochem. 45, 1963–1980. doi: 10.1007/s10695-019-00693-8
Hong, L., Zahradka, P., Cordero-Monroy, L., Wright, B., and Taylor, C. G. (2019). Dietary Docosahexaenoic Acid (DHA) and Eicosapentaenoic Acid (EPA) operate by different mechanisms to modulate hepatic steatosis and hyperinsulemia in fa/fa zucker rats. Nutrients 11:917. doi: 10.3390/nu11040917
Huang, X., Feng, B., Huang, H., and Ye, H. (2017). In vitro stimulation of vitellogenin expression by insulin in the mud crab, Scylla paramamosain, mediated through PI3K/Akt/TOR pathway. Gen. Comp. Endocrinol. 250, 175–180. doi: 10.1016/j.ygcen.2017.06.013
Huang, Y. Q., Mustapha, U. F., Huang, Y., Tian, C. X., Yang, W., Chen, H. P., et al. (2021). A chromosome-level genome assembly of the spotted scat (Scatophagus argus). Genome Biol Evol. 13:evab092. doi: 10.1093/gbe/evab092
Jiang, D. N., Li, J. T., Tao, Y. X., Chen, H. P., Deng, S. P., Zhu, C. H., et al. (2017). Effects of melanocortin-4 receptor agonists and antagonists on expression of genes related to reproduction in spotted scat, Scatophagus argus. J. Comp. Physiol. B 187, 603–612. doi: 10.1007/s00360-017-1062-0
Laine, V. N., Verhagen, I., Mateman, A. C., Pijl, A., Williams, T. D., Gienapp, P., et al. (2019). Exploration of tissue-specific gene expression patterns underlying timing of breeding in contrasting temperature environments in a song bird. BMC Genomics 20:693. doi: 10.1186/s12864-019-6043-0
Lazzarotto, V., Me’dale, F., Larroquet, L., and Corraze, G. (2018). Long-term dietary replacement of fishmeal and fish oil in diets for rainbow trout (Oncorhynchus mykiss): effects on growth, whole body fatty acids and intestinal and hepatic gene expression. PLoS One 13:e0190730. doi: 10.1371/journal.pone.0190730
Lei, C. X., Li, M. M., Tian, J. J., Li, Y. Y., and Wen, J. K. (2019). Transcriptome analysis of golden pompano (Trachinotus ovatus) liver indicates a potential regulatory target involved in HUFA uptake and deposition. Comp. Biochem. Physiol. D. 33:100633. doi: 10.1016/j.cbd.2019.100633
Levi, L., Pekarski, I., Gutman, E., Fortina, P., Hyslop, T., Biran, J., et al. (2009). Revealing genes associated with vitellogenesis in the liver of the zebrafish (Danio rerio) by transcriptome profiling. BMC Genomics 10:141. doi: 10.1186/1471-2164-10-141
Li, Y. Y., Chen, W. Z., Sun, Z. W., Chen, J. H., and Wu, K. G. (2004). Effects of n-3HUFA content in broodstock diets on reproductive performance and seasonal changes of plasma sex steroids levels in Plectorhynchus cinctus. Zool. Res. 25, 249–255. doi: 10.3321/j.issn:0254-5853.2004.03.011
Mandal, B., Kailasam, M., Bera, A., Sukumaran, K., Hussain, T., Biswas, G., et al. (2021). Standardization of oocyte size during artificial fertilization and optimization of stocking density during indoor larval and outdoor nursery rearing of captive spotted scat (Scatophagus argus) for a viable juvenile production system. Aquaculture 534:736262. doi: 10.1016/j.aquaculture.2020.736262
Mandal, B., Kailasam, M., Bera, A., Sukumaran, K., Hussain, T., Makesh, M., et al. (2020). Gonadal recrudescence and annual reproductive hormone pattern of captive female Spotted Scats (Scatophagus argus). Anim. Reprod. Sci. 213:106273. doi: 10.1016/j.anireprosci.2020.106273
Martins, F. F., Aguila, M. B., and Mandarim-de-Lacerda, C. A. (2020). Eicosapentaenoic and docosapentaenoic acids lessen the expression of PPARγ/Cidec affecting adipogenesis in cultured 3T3-L1 adipocytes. Acta Histochem. 122:151504. doi: 10.1016/j.acthis.2020.151504
Matsumoto, M., Han, S., Kitamura, T., and Accili, D. (2006). Dual role of transcription factor FoxO1 in controlling hepatic insulin sensitivity and lipid metabolism. J. Clin. Invest. 116, 2464–2472. doi: 10.1172/JCI27047
Mohammad, N. S., Arafa, M. H., and Atteia, H. H. (2015). Coenzyme Q10 and fish oil synergistically alleviate aluminum chloride-induced suppression of testicular steroidogenesis and antioxidant defense. Free Radic. Res. 49, 1319–1334. doi: 10.3109/10715762.2015.1069290
Morais, S., Pratoomyot, J., Taggart, J. B., Bron, J. E., Guy, D. R., Bell, J. G., et al. (2011). Genotype-specific responses in Atlantic salmon (Salmo salar) subject to dietary fish oil replacement by vegetable oil:a liver transcriptomic analysis. BMC Genomics 12:255. doi: 10.1186/1471-2164-12-255
Mustapha, U. F., Huang, Y., Huang, Y. Q., Assan, D., Shi, H. J., Jiang, M. Y., et al. (2021). Gonadal development and molecular analysis revealed the critical window for sex differentiation, and E2 reversibility of XY-male spotted scat, Scatophagus argus. Aquaculture 544:737147. doi: 10.1016/j.aquaculture.2021.737147
Mustapha, U. F., Jiang, D. N., Liang, Z. H., Gu, H. T., Yang, W., Chen, H. P., et al. (2018). Male-specific Dmrt1 is a candidate sex determination gene in spotted scat (Scatophagus argus). Aquaculture 495, 351–358. doi: 10.1016/j.aquaculture.2018.06.009
Ohnesorg, T., Keller, B., Angelis, M. H. D., and Adamski, J. (2006). Transcriptional regulation of human and murine 17β-hydroxysteroid dehydrogenase type-7 confers its participation in cholesterol biosynthesis. J. Mol. Endocrinol. 37, 185–197. doi: 10.1677/jme.1.02043
Paolucci, M., Coccia, E., Imperatore, R., and Varricchio, E. (2020). A cross-talk between leptin and 17β-estradiol in vitellogenin synthesis in rainbow trout Oncorhynchus mykiss liver. Fish Physiol. Biochem. 46, 331–344. doi: 10.1007/s10695-019-00720-8
Pauter, A. M., Trattner, S., Gonzalez-Bengtsson, A., Talamonti, E., Asadi, A., Dethlefsen, O., et al. (2017). Both maternal and offspring Elovl2 genotypes determine systemic DHA levels in perinatal mice. J Lipid Res. 58, 111–123. doi: 10.1194/jlr.M070862
Peng, S., Gao, Q., Shi, Z., Zhang, C., Wang, J., Yin, F., et al. (2015). Effect of dietary n-3 LC-PUFAs on plasma vitellogenin, sex steroids, and ovarian steroidogenesis during vitellogenesis in female silver pomfret (Pampus argenteus) broodstock. Aquaculture 444, 93–98. doi: 10.1016/j.aquaculture.2015.03.031
Piedecausa, M. A., Mazón, M. J., García, B. G., and Hernández, M. D. (2007). Effects of total replacement of fish oil by vegetable oils in the diets of sharpsnout seabream (diplodus puntazzo). Aquaculture 263, 211–219. doi: 10.1016/j.aquaculture.2006.09.039
Shin, D. J., Joshi, P., Hong, S. H., Mosure, K., Shin, D. G., and Osborne, T. F. (2012). Genome-wide analysis of FoxO1 binding in hepatic chromatin: potential involvement of FoxO1 in linking retinoid signaling to hepatic gluconeogenesis. Nucleic Acids Res. 40, 11499–11509. doi: 10.1093/nar/gks932
Soni, N. K., Nookaew, I., Sandberg, A. S., and Gabrielsson, B. G. (2015). Eicosapentaenoic and docosahexaenoic acid-enriched high fat diet delays the development of fatty liver in mice. Lipids Health Dis. 14:74. doi: 10.1186/s12944-015-0072-8
Sun, J., Xiao, P. Z., Chang, Z. G., Ji, H., Du, Z. Y., and Chen, L. Q. (2017). Forkhead box O1 in grass carp Ctenopharyngodon idella: molecular characterization, gene structure, tissue distribution and mRNA expression in insulin-inhibited adipocyte lipolysis. Comp. Biochem. Physiol. A. 204, 76–84. doi: 10.1016/j.cbpa.2016.11.011
Trushenski, J., Schwarz, M., Lewis, H., Laporte, J., Delbos, B., Takeuchi, R., et al. (2011). Effect of replacing dietary fish oil with soybean oil on production performance and fillet lipid and fatty acid composition of juvenile cobia Rachycentron canadum. Aquacult Nutr. 17, e437–e447. doi: 10.1111/j.1365-2095.2010.00779.x
Virote, B. D. C. R., Moreira, A. M. S., Souza, J. G. D. S., Castro, T. F. D., Melo, N., Carneiro, W. F., et al. (2020). Obesity induction in adult zebrafish leads to negative reproduction and offspring effects. Reproduction 160, 833–842.
Wang, Q., Mu, H., Shen, H., Gu, Z., Liu, D., Yang, M., et al. (2019). Comparative analysis of glucose metabolism responses of large yellow croaker Larimichthys crocea fed diet with fish oil and palm oil. Fish Physiol. Biochem. 45, 1603–1614. doi: 10.1007/s10695-019-00646-1
Wu, T., Cheng, Y., Liu, Z., Tao, W., Zheng, S., and Wang, D. (2018). Bioinformatic analyses of zona pellucida genes in vertebrates and their expression in Nile tilapia. Fish Physiol. Biochem. 44, 435–449. doi: 10.1007/s10695-017-0434-4
Xu, H., Cao, L., Wei, Y., Zhang, Y., and Liang, M. (2017). Effects of different dietary DHA:EPA ratios on gonadal steroidogenesis in the marine teleost, tongue sole (Cynoglossus semilaevis). Br. J. Nutr. 118, 179–188. doi: 10.1017/S0007114517001891
Xu, H., Zhang, Y., Wang, J., Zuo, R., Mai, K., and Ai, Q. (2015). Replacement of fish oil with linseed oil or soybean oil in feeds for Japanese Seabass, Lateolabrax japonicus: effects on growth performance, immune response, and tissue fatty acid composition. J. World Aquacult. Soc. 46, 349–362. doi: 10.1111/jwas.12205
Xue, X., Hall, J. R., Caballero-Solares, A., Eslamloo, K., Taylor, R. G., Parrish, C. C., et al. (2020). Liver transcriptome profiling reveals that dietary DHA and EPA levels influence suites of genes involved in metabolism, redox homeostasis, and immune function in Atlantic salmon (Salmo salar). Mar. Biotechnol. 22, 263–284. doi: 10.1007/s10126-020-09950-x
Zakeri, M., Kochanian, P., Marammazi, J. G., Yavari, V., Savari, A., and Haghi, M. (2011). Effects of dietary n-3 HUFA concentrations on spawning performance and fatty acids composition of broodstock, eggs and larvae in yellowfin sea bream, Acanthopagrus latus. Aquaculture 310, 388–394. doi: 10.1016/j.aquaculture.2010.11.009
Zhang, G., Wang, W., Su, M., and Zhang, J. (2018). Effects of recombinant gonadotropin hormones on the gonadal maturation in the spotted scat, Scatophagus argus. Aquaculture 483, 263–272. doi: 10.1016/j.aquaculture.2017.10.017
Zhang, M. Z., Li, G. L., Zhu, C. H., and Deng, S. P. (2013). Effects of fish oil on ovarian development in spotted scat (Scatophagus argus). Anim. Reprod. Sci. 141, 90–97. doi: 10.1016/j.anireprosci.2013.06.020
Zhao, W., Yuan, T., Fu, Y., Niu, D., Chen, W., Chen, L., et al. (2020). Seasonal differences in the transcriptome profile of the Zhedong white goose (Anser cygnoides) pituitary gland. Poultry Sci. 100, 1154–1166. doi: 10.1016/j.psj.2020.10.049
Zhao, Y., Cao, X., Fu, L., and Gao, J. (2020). n-3 PUFA reduction caused by fabp2 deletion interferes with triacylglycerol metabolism and cholesterol homeostasis in fish. Appl. Microbiol. Biotechnol. 104, 2149–2161. doi: 10.1007/s00253-020-10366-9
Keywords: spotted scat, female broodstock, fish oil, liver, transcriptome, reproduction, lipid metabolism, vitellogenin
Citation: Wang T, Jiang D, Shi H, Mustapha UF, Deng S, Liu Z, Li W, Chen H, Zhu C and Li G (2021) Liver Transcriptomic Analysis of the Effects of Dietary Fish Oil Revealed a Regulated Expression Pattern of Genes in Adult Female Spotted Scat (Scatophagus argus). Front. Mar. Sci. 8:784845. doi: 10.3389/fmars.2021.784845
Received: 28 September 2021; Accepted: 03 November 2021;
Published: 30 November 2021.
Edited by:
Erchao Li, Hainan University, ChinaReviewed by:
Zhenhua Ma, South China Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences (CAFS), ChinaCopyright © 2021 Wang, Jiang, Shi, Mustapha, Deng, Liu, Li, Chen, Zhu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guangli Li, bGlnbEBnZG91LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.